
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 April 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00423
Autophagy is an efficient degradation system for maintaining cellular homeostasis when plants are under environmental stress. ATG9 is the only integral membrane protein within the core ATG machinery that provides a membrane source for autophagosome formation. In this study, we isolated an ATG9 homologs gene in apple, MdATG9, from Malus domestica. The analysis of its sequence, subcellular localization, promoter cis-elements, and expression patterns revealed the potential function of MdATG9 in response to abiotic stressors. Overexpression of MdATG9 in apple callus conferred enhanced tolerance to nitrogen depletion stress. During the treatment, other important MdATGs were expressed at higher levels in transgenic callus than in the wild type. Furthermore, more free amino acids and increased sucrose levels were found in MdATG9-overexpression apple callus compared with the wild type in response to nitrogen starvation, and the expression levels of MdNRT1.1, MdNRT2.5, MdNIA1, and MdNIA2 were all increased higher in transgenic lines. These data suggest that, as an important autophagy gene, MdATG9 plays an important role in the maintenance of amino acids and sugars in response to nutrient starvation in apple.
Plants frequently encounter various types of harsh environmental factors, such as water shortages, ion toxicity, carbon deficiency, or nitrogen depletion in the soil. To survive under these adverse conditions, plants have evolved intricate regulatory mechanisms to protect themselves against potential damage. Autophagy is one of the key biological processes for maintaining cellular homeostasis when plants are subjected to stress, which entails the degradation of superfluous or damaged organelles and aberrant proteins for recycling in cells (Li and Vierstra, 2012). Three different types of autophagy, such as micro-autophagy, macro-autophagy, and selective autophagy, have been identified in plants (Bassham et al., 2006; Marshall and Vierstra, 2018). In all three molecular routes, cytoplasmic constituents are eventually transported to the vacuole for degradation (van Doorn and Papini, 2013). Here, we focused on macro-autophagy (hereafter termed autophagy), which has been widely studied in many organisms.
Autophagy is initiated with the formation of cup-shaped phagophores elongating into a closed double-membrane vesicle called an autophagosome, which then sequesters cargo delivered to the vacuole for degradation and recycling (Bassham, 2009; Noda and Inagaki, 2015). The autophagy process relies on a dedicated set of autophagy-related (ATG) proteins which were originally identified in yeast and are largely conserved in all eukaryotes (Matsuura et al., 1997; Kang et al., 2018). These ATGs can be subdivided into several major functional clusters, including the ATG1 kinase complex, the autophagy-specific phosphatidylinositol 3-kinase complex, the Atg9 recycling system, and the Atg12 and Atg8 ubiquitin-like conjugation systems (Mizushima et al., 2011; Kim et al., 2012). ATG9, which represents the third class of ATG proteins, is the only integral membrane protein within the core ATG machinery. It has six central membrane-spanning segments with two cytosolically oriented termini involved in interactions with other ATG components (Young et al., 2006). ATG9-mediated delivery between the endomembrane and the phagophore assembly site provides an essential membrane source for vesicle nucleation (Lai et al., 2019; Zhuang et al., 2017).
Similar to autophagy studies in other organisms, isolating and identifying ATG genes in plants helps us to understand their molecular mechanisms. Genes for most core ATG proteins have been analyzed in plants, and various Arabidopsis atg-mutants, such as atg2, atg4a/4b, atg5, atg7, atg9, atg10, and atg18a, have been isolated to research their phenotypes (Phillips et al., 2008; Han et al., 2011). One of the major physiological roles of autophagy in plants is protecting plant cells from nitrogen depletion or carbon starvation. Several Arabidopsis atg-mutants are hypersensitive to nutrient-limiting (low nitrate) conditions by displaying earlier leaf senescence and lower rosette biomass (Guiboileau et al., 2013). In contrast, emerging evidence indicates that autophagy can be highly induced by abiotic or biotic stressors, and act as a central biological process involved in the plant defense system. For example, atg5 and atg7 mutants have been used to confirm the importance role of autophagy in heat and drought tolerance (Zhou et al., 2013).
In addition to Arabidopsis, the ATG genes in other plant species have been isolated and analyzed. OsATG10b has been demonstrated to play an important role in the survival of rice cells under oxidative stress (Shin et al., 2009). ATG5- or ATG7-silenced tomato mutants display compromised heat tolerance compared with the wild type (Zhou et al., 2013). SiATG8a from foxtail millet plays a vital role conferring tolerance to both nitrogen starvation and drought stress (Li et al., 2015). The maize autophagy mutant atg12 has been used to reveal the specific aspects of maize metabolism that are controlled by autophagy (McLoughlin et al., 2018). In our previous study, we characterized MdATG18a in apple, and demonstrated that it enhances plant resistance to drought stress (Sun et al., 2018a), nitrogen depletion (Sun et al., 2018b), and heat stress (Huo et al., 2020a).
The autophagic recycling process contributes to remobilize nutrients under a nutrient deficient condition, providing amino acids, fatty acids, and sugars for plant survival (Signorelli et al., 2019). In response to a nutrient limitation, autophagy-mediated amino acid metabolism seems to play an important role in plant resistance. For example, in cases of limited carbohydrates, amino acids produced from protein degradation are an important source of alternative substrates for energy supply, and autophagy has a potential role in energetic maintenance under this condition (Avin-Wittenberg et al., 2015; Barros et al., 2017). Moreover, decreased levels of free amino acids in Arabidopsis seedlings, particularly in atg mutants, have been reported following carbon limitation (Avin-Wittenberg et al., 2015). These results exemplify the essential role of autophagy for maintaining free amino acid levels during starvation conditions. In addition to amino acid metabolism, the modulation of carbon metabolism is also important for plant cells to survive under starvation conditions. In our previous study, leaves of MdATG18a-overexpressing apple plants accumulated more soluble sugars compared with the wild type under a nitrogen-depletion treatment (Sun et al., 2018b).
In both mammals and yeast, ATG9 deficiency inhibits the formation of autophagosomes, particularly atg9-knockout mice present an embryonic lethal condition (Saitoh et al., 2009; Yamamoto et al., 2012). Consistent with the essential role of ATG9 in mammals and yeast, depleting ATG9 in plants leads to an extensive accumulation of abnormal autophagosome-related tubular structures when autophagy is induced, indicating the essential and specific role of ATG9 in the autophagic degradation pathway (Zhuang et al., 2017). Here, we isolated an ATG9 homologs gene, MdATG9, in apple. After analyzing its sequence, subcellular localization, and promoter cis-elements as well as its expression patterns in apple plants under abiotic stress, we revealed that, as an important autophagy gene, MdATG9 might possess the conserved function in response to abiotic stressors. Overexpression of MdATG9 in apple callus enhanced their tolerance to a nitrogen starvation treatment. Furthermore, the levels of free amino acids were less reduced and sucrose content increased more in MdATG9-overexpressing apple callus compared with the wild type under low-nitrogen stress.
Mature leaves were collected from three 2 years old apple (Malus domestica Borkh. “Golden Delicious”) plants growing at the Horticultural Experimental Station of Northwest A&F University (Yangling, Shaanxi, China) for cloning MdATG9 and its promoter.
Seeds of M. hupenensis were stratified at 4°C for 60 days, and the seedlings were grown in pots for 3 months in a greenhouse (25°C, 14 h photoperiod). The response of seedlings to H2O2, methyl viologen (MV), or exogenous abscisic acid (ABA) were examined by spraying the leaves with 10 mM H2O2, 50 μM MV, or 100 μM ABA, respectively. The response of seedlings to salinity was examined by irrigating the plants to saturation with 200 mM NaCl. Seedlings were exposed to 4 or 42°C to test their responses to low or high temperature, respectively. For each assay mentioned above, leave samples were collected at 0, 2, 4, 8, 12, and 24 h. Drought stress was induced by withholding irrigation from plants that had previously been fully watered. Seedlings for the nitrogen-starvation assay were transferred to a chamber for hydroponic culture. After a 15 days preincubation, Ca(NO3)2 and KNO3 in Hoagland’s nutrient solution were replaced with CaCl2 and KCl, respectively. In both cases, leaves were sampled on days 0, 2, 4, 6, 8, and 10. In all treatment types mentioned above, three biological replicates were prepared with five seedlings combined as one replicate. All collected samples were frozen rapidly in liquid nitrogen and stored at −80°C.
Seedlings of wild-type Arabidopsis thaliana L. (Columbia) and two homozygous T3 transgenic lines were used. They were sterilized and germinated on a standard Murashige and Skoog (MS) medium and cultured under a 14 h photoperiod at 25°C. Five days old uniformly sized seedlings grown on MS agar plates were transferred vertically into unmodified media or nitrogen-limited media (nitrogen concentration reduced from 60 to 10 mM) for the nitrogen-starvation and osmotic stress assays. Fresh weights and root lengths were measured from 5 plates on day 15 after transfer.
Wild type and transgenic “Orin” apple calluses were used, based on treatments described previously (Wang et al., 2017a). Briefly, 0.1 g portions of 15 days old similar and well-grown calluses of wild type and transgenic lines were transferred to MS control media or low-nitrogen media with the nitrogen concentration lowered from 60to 5 mM. Fresh weights were measured from 10 plates on day 15 after transfer. Frozen samples were collected from 15 plates and stored at −80°C.
Homologous sequences and putative conserved domains were predicted from the NCBI BLASTp programs. The sequences were aligned with homologs sequences from other species using DNAMAN. Phylogenetic trees were constructed with MEGA 5.0 software (Tamura et al., 2011), using the neighbor-joining method with 1,000 bootstrap replicates. Putative cis-regulatory elements in the MdATG9 promoter region were examined with the PlantCARE program.
To construct the vectors for subcellular localization and for the transgenic lines, the MdATG9 coding region was introduced into the pGWB405-GFP and pCambia2300 vectors, both driven by the CaMV 35S promoter and carried by the kanamycin (Kan) selectable marker in plants. The sequencing-confirmed plasmids were transformed into Agrobacterium tumefaciens strain EHA105 by electroporation. The primers used for constructing the vector are listed in Supplementary Table S2.
For subcellular localization, leaves of 5 weeks old tobacco (Nicotiana tabacum) were transiently transformed as described previously (Yang et al., 2000). Three days later, green fluorescent protein (GFP) signals in transformed tobacco leaves were observed using confocal microscopy, and the images were processed with FV10-ASW software.
The “Columbia” Arabidopsis ecotype was transformed using the floral dip method with Agrobacterium contained MdATG9-pCambia2300 vector (Zhang et al., 2006). Transgenic seeds (T1) were selected on MS medium supplemented with 50 mg L–1 Kan. After confirmation by polymerase chain reaction (PCR), T2 homozygous lines were obtained after selecting transgenic lines at a 3:1 segregation ratio, and the T3 seeds were collected.
Calluses of “Orin” apple were used for apple callus transformation. The calluses were cultured on solid MS medium containing 1.0 mg L–1 2,4-D, 1.0 mg L–1 6-BA, and 8 g L–1 agar in the dark at 25°C. The apple calluses were transformed referring to a method described previously (Wang et al., 2016, 2018). Briefly, 7 days old suspension callus grown in a liquid medium were mixed with Agrobacterium contained MdATG9-pCambia2300 vector and rotated gently for 10 min at 25°C for transformation. After 2 days co-cultivation on solid medium, the callus was washed three times with sterile water containing 400 mg L–1 cefotaxime, and then transferred to a selection medium supplemented with 400 mg L–1 cefotaxime and 30 mg L–1 kanamycin for transgene selection. After 3–5 serial subcultures, resistant calluses showing stable growth were subjected to PCR and qRT-PCR for analysis of the transgene.
Total RNA was extracted using the Wolact plant RNA isolation kit (Wolact, Hong Kong, China), and first-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, United States) with 1 μg total RNA. Genomic DNA was isolated with the Wolact Plant Genomic DNA purification kit. qRT-PCR was performed according to the SYBR Premix Ex Taq II Kit (Takara, Dalian, China) on a LightCycler 96 (Roche, Basel, Switzerland). Three biological replicates were tested in each assay, and representative data from one repetition were shown. Values were calculated using malate dehydrogenase (MDH) as the endogenous control (Perini et al., 2014). The relative expression level of each gene was calculated according to the 2–ΔΔCT method (Livak and Schmittgen, 2001), and a dissociation curve analysis was performed to determine the specificity of each gene. The gene-specific primers are shown in Supplementary Table S2.
Soluble sugars were extracted and derivatized sequentially with methoxamine hydrochloride and N-methyl-N-trimethylsilyl-trifluoroacetamide, as described previously (Hu et al., 2018). The metabolites were analyzed with a Shimadzu GCMS-2010 SE (Shimadzu Corp., Kyoto, Japan). Values were calculated based on their corresponding standard curves and internal standards.
Amino acids were extracted and measured as described previously (Huo et al., 2020b). Briefly, 200 mg of frozen leaf samples were extracted in 2 ml 50% ethanol (including 0.1 M HCl) and centrifuged at 13,000 g for 10 min. The supernatant was added to methanol at a final volume of 10 ml. The samples were filtered through a 0.22 μm filter to analyze the metabolites with a liquid chromatography-mass spectrometry system (QTRAP5500; SCIEX, Concord, ONT, Canada) equipped with an Inertsil ODS-4 C18 column (4.6 × 250 mm, 5 μm) at a flow rate of 0.3 ml/min. The solvent system consisted of water containing 0.1% (v/v) formic acid (A) and acetonitrile (B). Data were quantified by comparing the peak surface areas with those obtained using standard amino acids (Sigma-Aldrich, St. Louis, MO, United States).
SPSS 16.0 software (SPSS Inc., Chicago, IL, United States) was used for the statistical analysis. Three independent replicates were used for each determination. Experimental data are presented as mean ± standard error. The statistical analysis was performed by one-way analysis of variance followed by Tukey’s multiple range test. A p < 0.05 was considered significant.
We identified a homologous sequence of AtATG9 from M. domestica through homologous cloning and named it MdATG9. The gene contained a 2,622 bp open reading frame (ORF) and encoded an 874 amino acid-deduced protein. Protein alignment revealed high homology between the MdATG9 protein and other ATG9 proteins from A. thaliana (64%), Glycine max (70%), Nicotiana tabacum (67%), Populus euphratica (72%), and Solanum lycopersicum (66%) (Figure 1A). All of these aligned sequences had the conserved autophagy protein APG9 superfamily domain. The phylogenetic tree analysis indicated that the MdATG9 protein formed a close cluster with PeATG9 and GmATG9 (Figure 1B).
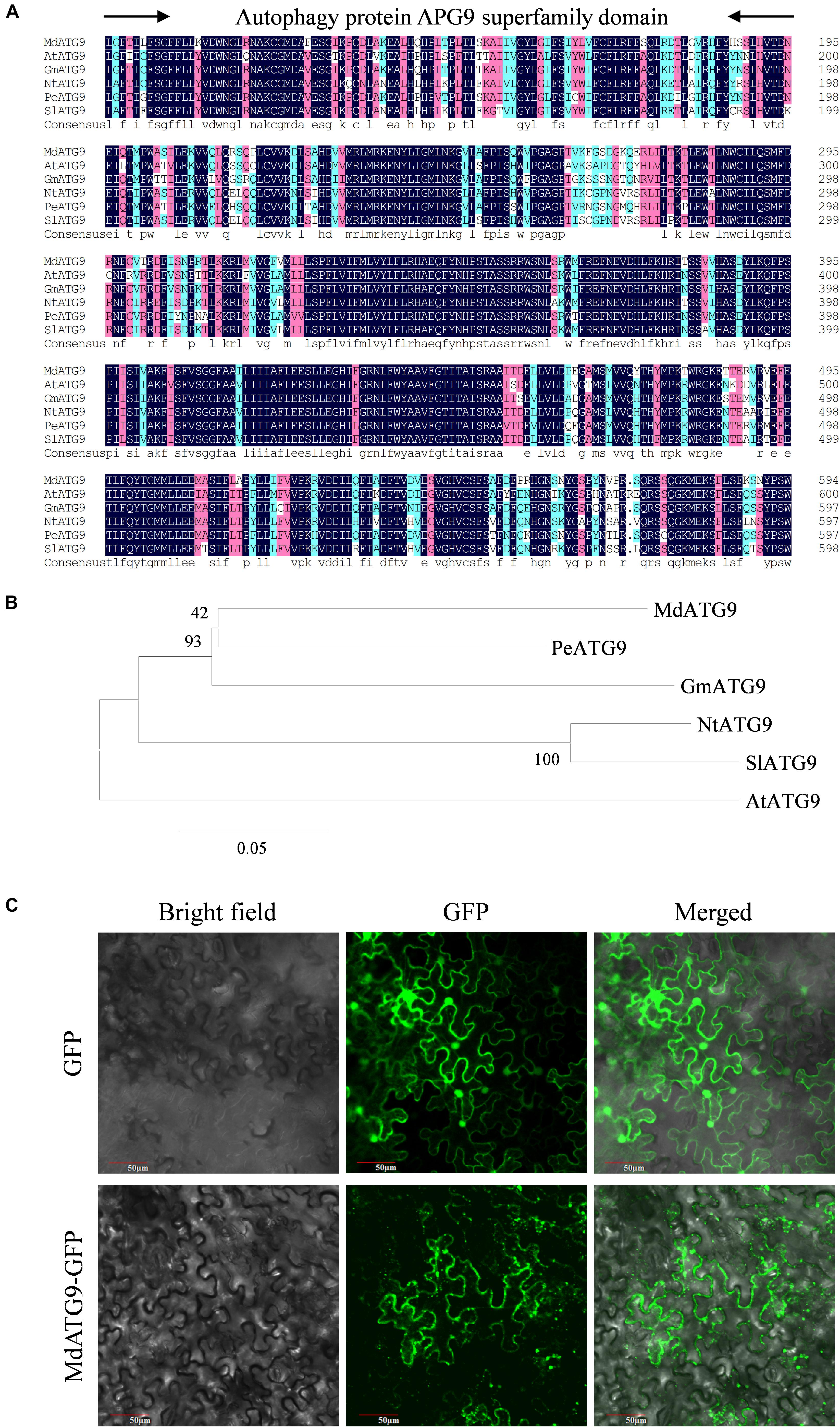
Figure 1. Sequence analysis and cellular localization of MdATG9. (A) Alignment of deduced ATG9 amino acid sequence from Malus domestica (Md), Arabidopsis thaliana (At), Populus euphratica (Pe), Glycine max (Gm), Nicotiana tabacum (Nt), and Solanum lycopersicum (Sl). (B) Phylogenetic analysis of MdATG9 protein with ATG9 proteins from other species. (C) Subcellular localization analysis of MdATG9-GFP fusion protein in tobacco epidermal cells.
To examine the subcellular localization of MdATG9, we fused its ORF with the GFP at the N-terminus under control of the CaMV35S promoter. For transient expression in tobacco epidermal cells, the MdATG9–GFP fusion protein was detected in the cytoplasm, while GFP alone was distributed throughout the cell (Figure 1C).
In addition, we analyzed the promoter region of MdATG9 using the PlantCARE database and revealed the presence of several recognized stress-responsive cis-elements (Supplementary Figure S1 and Supplementary Table S1).
To investigate the possible functions of MdATG9, we examined it expression in the plants of M. hupenensis in response to several abiotic stressors. In response to H2O2 or MV, the expression of MdATG9 increased by approximately twofold at 8 h (Figures 2A,B). ABA or NaCl treatment also induced it expression, with both transcripts were upregulated more than 2.7-fold after 8 h of exposure (Figures 2C,D). Under the low temperature treatment (4±C), expression increased by approximately twofold 8 h after treatment and then decreased below the initial level (Figure 2E). High temperature (45±C) upregulated the transcripts continually, with expression being almost two and threefold higher at 2 and 12 h, respectively (Figure 2F). In response to nitrogen starvation, MdATG9 expression was induced approximately fourfold on day 6. In addition, the transcripts were significantly upregulated in response to drought treatment, by more than 2.5-fold on day 8 (Figure 2G). These data suggest that MdATG9 was induced by most stress conditions, particularly by nitrogen starvation.
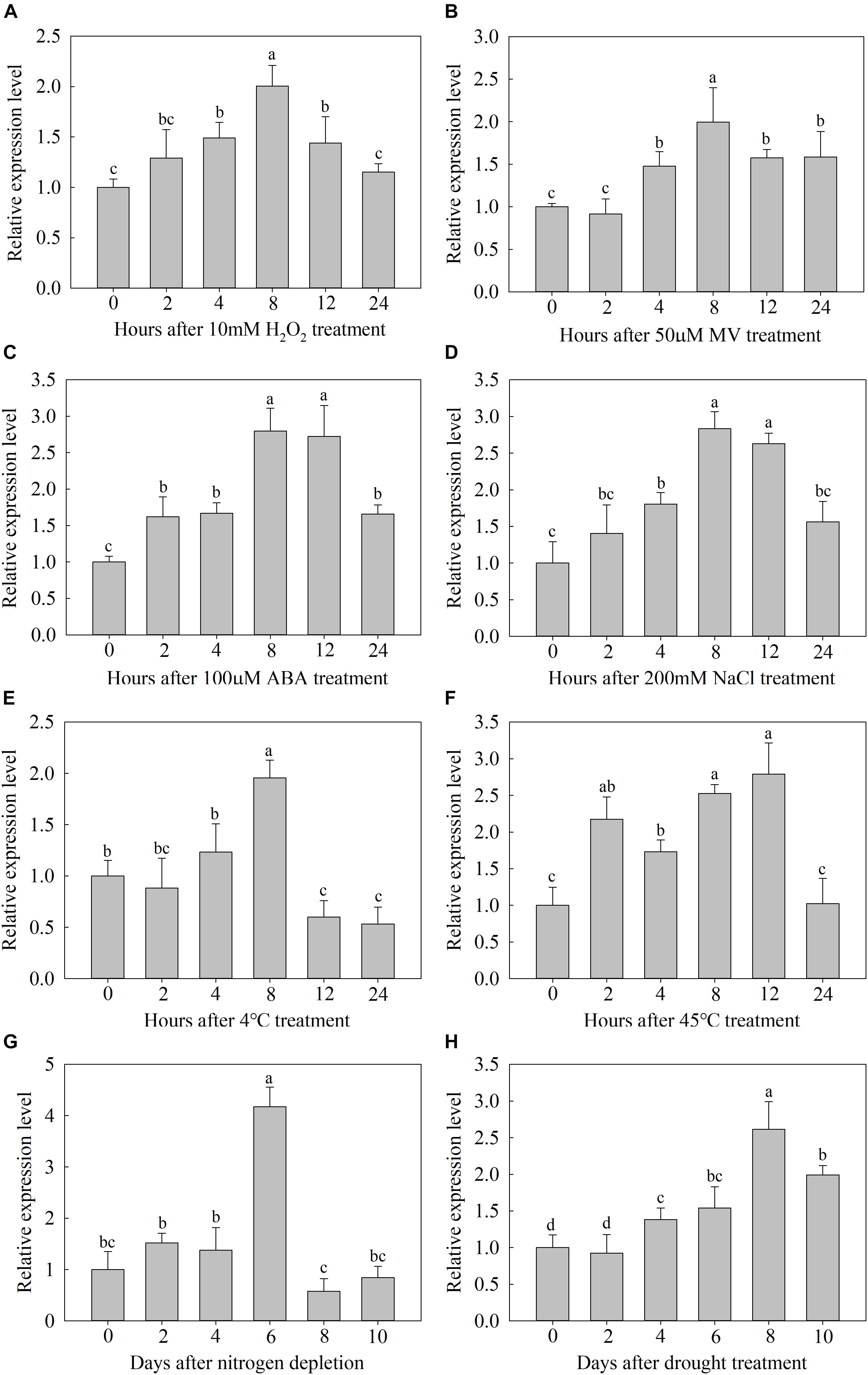
Figure 2. Expression analysis of MdATG9. Relative expression (A) under H2O2 treatment, (B) MV treatment, (C) ABA treatment, (D) NaCl stress, (E) low temperature, (F) high temperature, (G) nitrogen depletion treatment, and (H) drought stress. Total RNA was extracted from leaf samples. Data are the means of three replicates with SEs. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
As several stress-responsive cis-acting elements were found in the MdATG9 promoter region, and its transcription was induced by several abiotic stressors, we generated overexpressing (OE) “Orin” lines that over-expressed MdATG9, as confirmed by PCR with gDNA and qRT-PCR (Figures 3A,B). The mRNA transcripts increased by 7.7- and 6.3-fold in OE-3 and OE-10, respectively (Figure 3B).
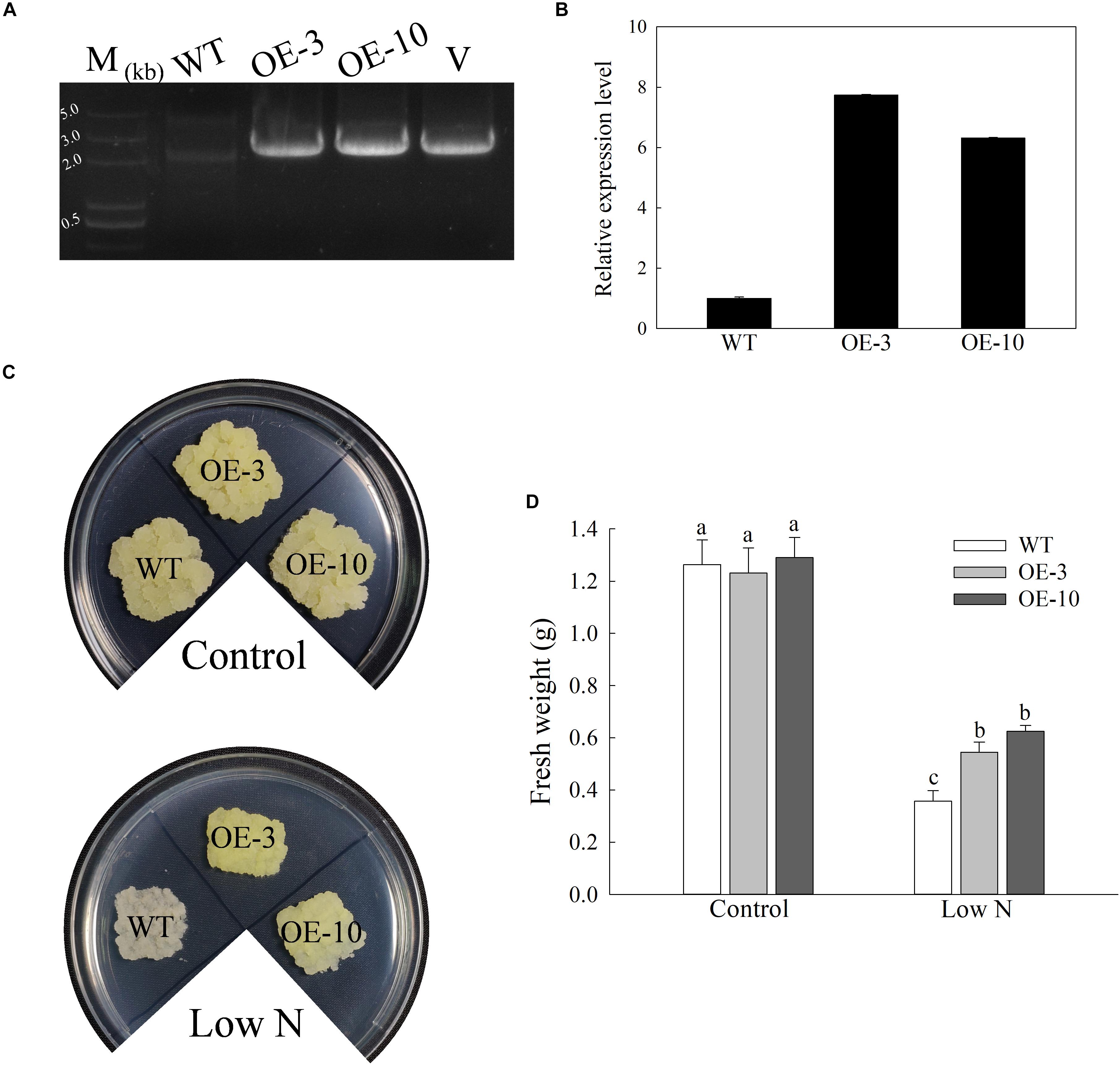
Figure 3. Overexpression of MdATG9 in apple callus confers enhanced tolerance to nitrogen starvation. (A) PCR with gDNA. Lanes M, molecular marker DL5000; WT, non-transformed wild-type; OE-3 and OE-10, MdATG9-transgenic callus lines; V, positive plasmid control. (B) qPCR analysis of MdATG9 transcripts in apple callus lines OE-3 and OE-10. (C) Phenotypes of WT and transgenic apple callus grew on normal MS media or nitrogen-limitation media for 15 days. (D) Fresh weights of callus after 15 days treatment. Data are the means of ten replicates with SE. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
After 15 days of growth on MS medium, the similar and well-grown calluses of the wild type and transgenic lines were transferred to MS control or low-nitrogen media (Figure 3C). The initial weight of all lines was controlled at 0.1 g, then we observed and measured their growth after 15 days of stress treatment. No significant difference was observed between the wild type (WT) and OE lines in terms of growth characteristics or fresh weights when grown on control MS medium. After 15 days of nitrogen deprivation, the WT callus had stopped growing and turned white, while the two transgenic lines grew faster and were slightly yellow (Figures 3C,D). These results show that overexpression of MdATG9 improved the tolerance of nitrogen starvation in apple callus.
Furthermore, we obtained two transgenic Arabidopsis lines for the further analyses on its potential functions under low-nitrogen condition. We selected the independent T3 homozygous lines and vertically transferred the 5 days old Arabidopsis seedlings of wild type and transgenic lines into unmodified media or nitrogen-limited media (Supplementary Figures S2A–C). After 15 days of nitrogen deprivation, both the fresh weights and root lengths of the OE lines were significantly higher than those of the wild type, and the transgenic lines grew more lateral roots than the wild type under the low-nitrogen treatment (Supplementary Figures S2C–E). These results suggest that ectopic expression of MdATG9 in Arabidopsis alleviated the negative effects of nitrogen starvation stress.
To investigate the occurrence of autophagy among genotypes under stress conditions, we used qRT-PCR to examine the expression patterns of 8 other important MdATGs. The expression of MdATGs was induced significantly by nitrogen starvation (Figure 4). Under the control condition, the expression of MdATG3a, MdATG5, MdATG7b, MdATG8c, MdATG8i, and MdATG10 did not differ among genotypes, except MdATG3b and MdATG7a, were expressed at higher levels in the OE lines than in the WT. Under the low-nitrogen treatment, the MdATGs were all expressed at higher levels in the OE lines, except for the expression of MdATG7b, which was lower in OE-3 than in the WT. These results suggest that the transcripts of other important ATG genes were more responsive to the nitrogen-starvation condition in the OE lines than in the WT.
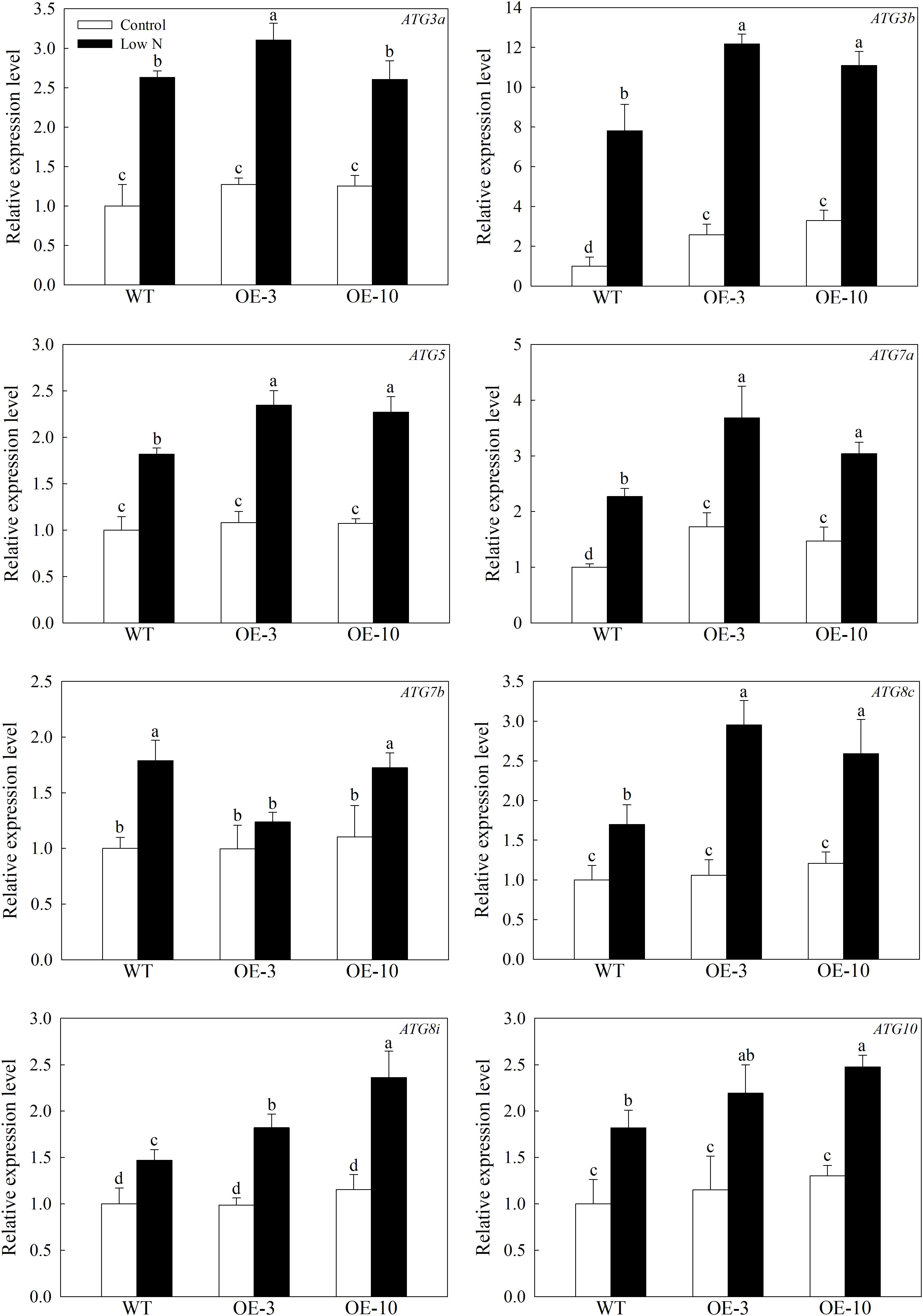
Figure 4. Changes in the expression of other MdATGs in WT and MdATG9-OE apple callus following nitrogen starvation. Data are the means of three replicates with SEs. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
The maintenance of free amino-acid levels is essential for plants under nutrient-starvation conditions (Onodera and Ohsumi, 2005). We measured the concentrations of 15 amino acids to further investigate whether MdATG9 was involved in amino acid metabolism in response to nitrogen starvation in the apple callus (Figure 5). The low-nitrogen treatment led to a significant decrease in the levels of most amino acids in the apple callus. Several amino acids with higher content in the apple callus under control conditions, such as Ala, Arg, Glu, and Ser, were reduced 10–50 times by the nitrogen-starvation treatment. However, at the end of the treatment, the concentration of each amino acid, except Asp, was significantly higher in the OE lines than those in the WT. For example, the levels of Ala and Tyr in the transgenic lines were almost 7- and 5-fold that of the WT, respectively after treatment. In addition, we observed that Pro content was reduced by the nitrogen-starvation treatment only in the WT callus, and it increased slightly in the OE lines. These results demonstrate that overexpression of MdATG9 alleviated the limitation of nitrogen-starvation to the size of the free amino-acid pool in the apple callus.
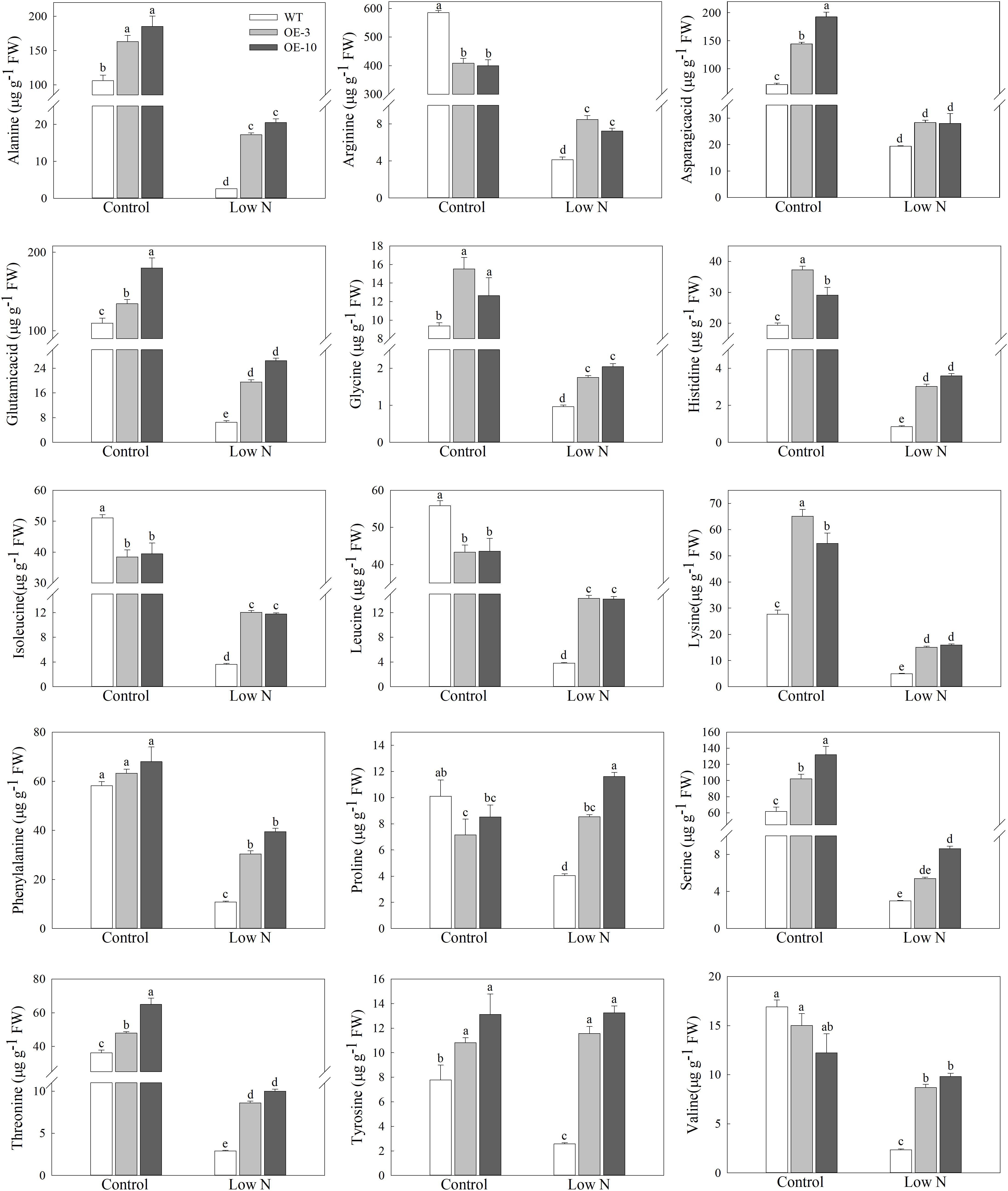
Figure 5. Amino acids levels in WT and MdATG9-OE apple callus after nitrogen starvation treatment, as measured by LC-MS. Data are the means of three replicates with SE. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
To examine whether MdATG9 influences N-signaling in apple, we investigated the transcript levels of several genes involved in nitrate uptake. As shown in Figure 6, the expression levels of MdNRT1.1, MdNRT2.5, and MdNIA1 increased under the low-nitrogen treatment, and they were all expressed higher in the OE lines than in the WT. For example, the MdNRT2.5 transcript level in the transgenic lines was almost three times that of the WT after treatment (Figure 6B). In addition, the MdNIA2 transcripts decreased threefold in the WT callus under treatment, whereas they were upregulated in the OE lines (Figure 6D). These data demonstrate that overexpression of MdATG9 upregulated some genes responsible for nitrate assimilation therefore increasing nitrate utilization in the apple callus.
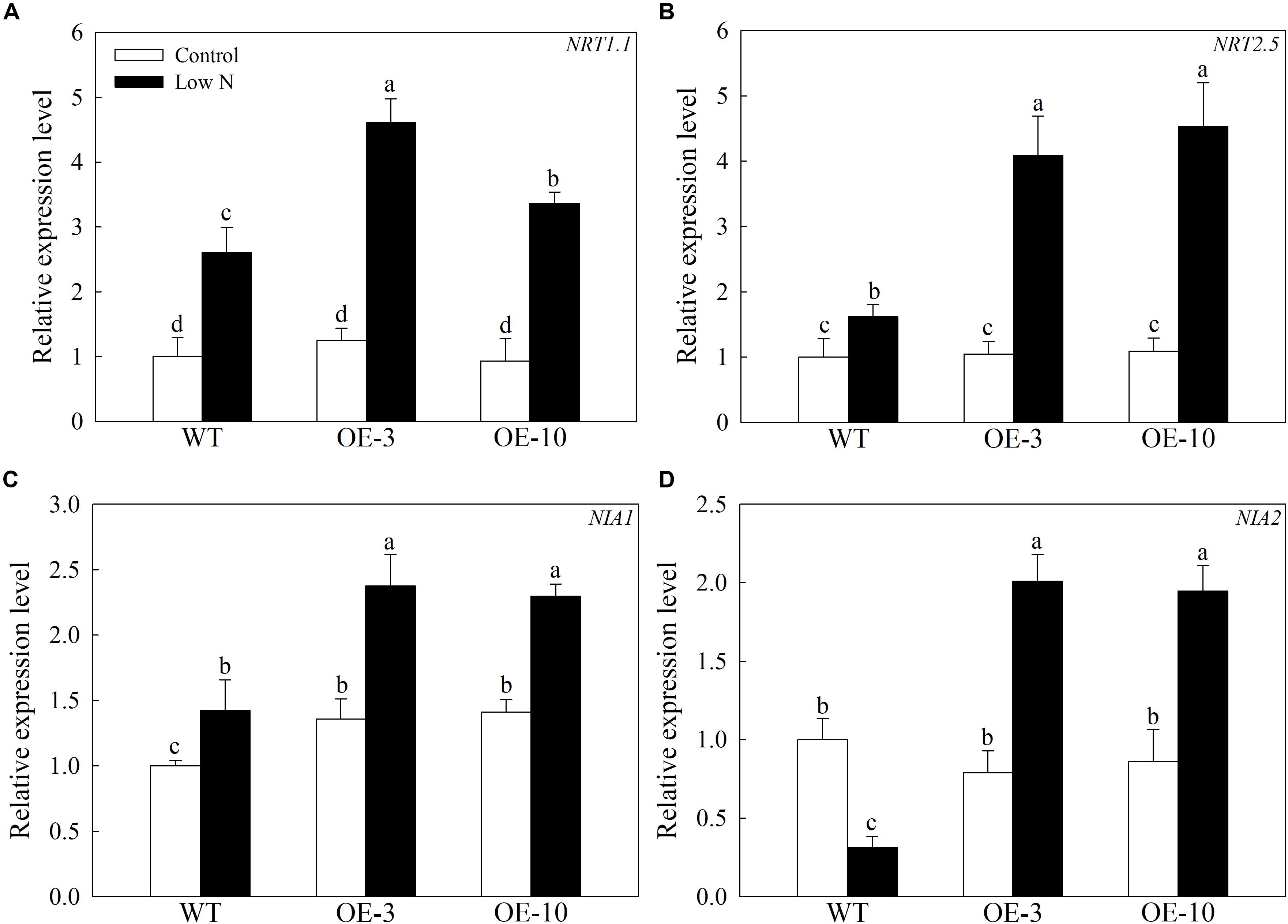
Figure 6. Changes in transcript levels of four genes involved in nitrate uptake following nitrogen starvation. Changes in expression of (A) MdNRT1.1, (B) MdNRT2.5, (C) MdNIA1, and (D) MdNIA2 under nitrogen-starvation stress. Data are the means of three replicates with SEs. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
In response to nitrogen starvation, the carbon metabolism of plant cells is remodeled to a certain extent, so we measured the concentrations of soluble sugars in apple calluses under a low-nitrogen treatment (Sun et al., 2018b). The nitrogen starvation treatment greatly increased the contents of fructose, glucose, and sucrose in the apple callus, but the changes in sorbitol concentration are inconspicuous (Figure 7). Although the levels of fructose rose more in the OE lines than in the WT, and the concentrations of glucose were lower in the OE lines than the WT after treatment (Figures 7A,C). In addition, the levels of sucrose were slightly higher in the OE lines even under the control conditions, but were approximately 1.8-fold higher when compared with WT calluses under the deficit treatment (Figure 7D). Furthermore, the expression levels of MdSUSY2 and MdSUSY3, which convert sucrose into fructose in apple, were induced by the low-nitrogen treatment and increased more in the transgenic lines than in the WT (Figures 7E,F). These data suggest that overexpression of MdATG9 improves the accumulation of sucrose in apple calluses in response to a low-nitrogen condition, with more conversion to fructose than glucose.
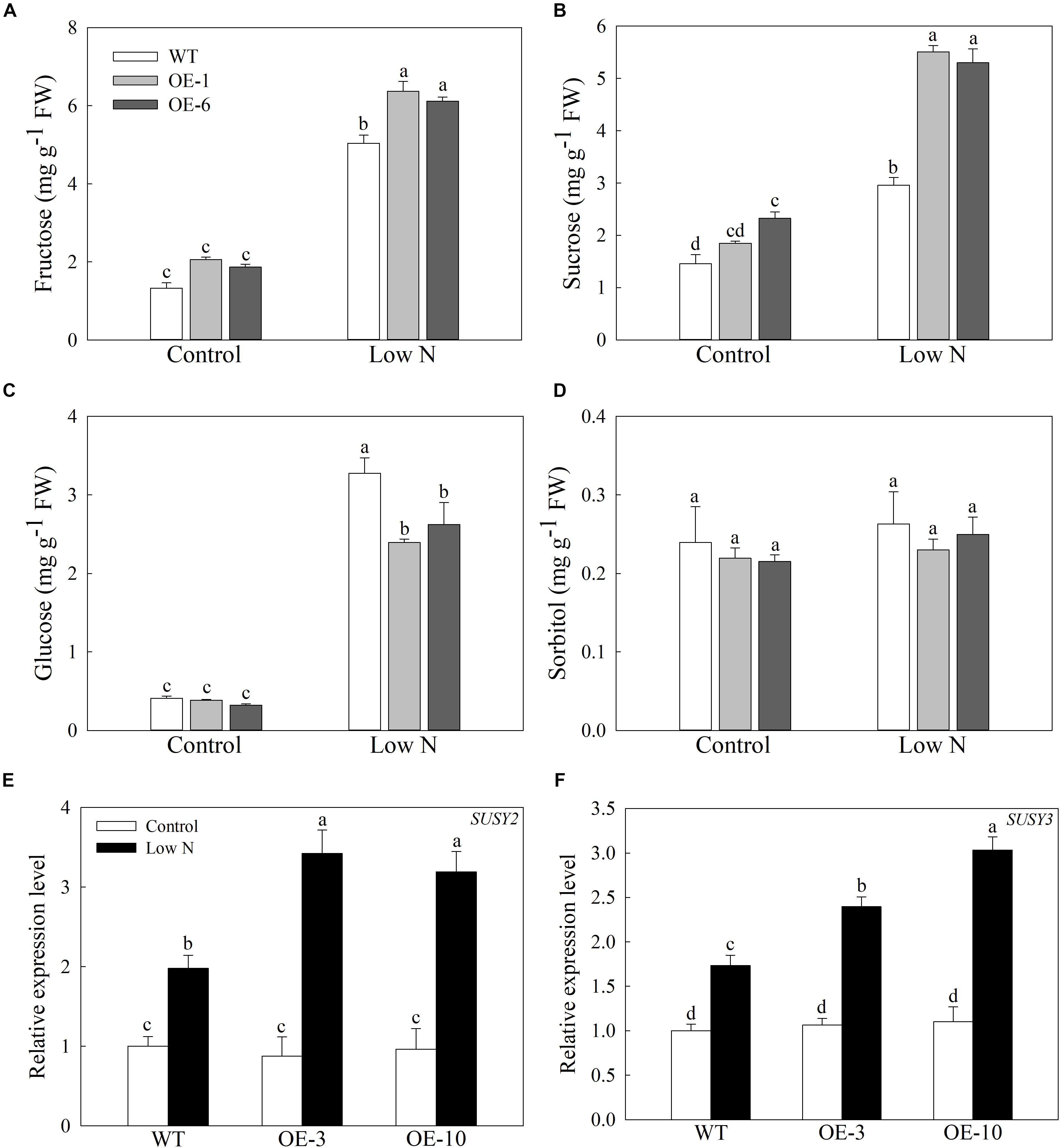
Figure 7. Accumulation of soluble sugars in apple callus under low-nitrogen treatment. (A) Fructose, (B) sucrose, (C) glucose, and (D) sorbitol contents in apple callus after nitrogen starvation treatment. Changes in expression of (E) MdSUSY2 and (F) MdSUSY3 under nitrogen-starvation stress. Data are the means of three replicates with SEs. Different letters indicate significant differences between treatments, according to one-way ANOVA and Tukey’s multiple range test (P < 0.05).
Autophagy is an intracellular catabolic process in which cellular components are enclosed by double membrane-bounded vesicular structures called autophagosomes and transported into vacuoles (Bassham, 2009; Noda and Inagaki, 2015). As the only transmembrane protein within the core ATG machinery, ATG9 has long been suggested to provide a membrane source for autophagosomes and to interact with several other functional ATG proteins at the site of autophagosome formation (Zhuang et al., 2017; Lai et al., 2019). Here, we isolated MdATG9 from apple, and characterized it function in response to nitrogen starvation stress in Arabidopsis and apple callus. The MdATG9 protein had a conserved autophagy protein APG9 superfamily domain and presented high homology with other ATG9s from various plant species (Figures 1A,B). Through a subcellular localization analysis, we found that the MdATG9–GFP fusion protein occurred in the cytoplasm of tobacco epidermal cells, with a multiple puncta appearance consistent with a previous report in yeast (Figure 1C; Noda et al., 2000). Then, we isolated the MdATG9 promoter region and bioinformatically analyzed several stress- or hormone-responsive cis-acting elements in the promoter (Supplementary Figure S1). Consistent with this, the expression analysis under abiotic stress conditions revealed that MdATG9 was induced by most stress conditions, particularly by nitrogen starvation (Figure 2).
For further evaluation of the function of MdATG9 in coping with stress, we generated transgenic Arabidopsis plants and apple calluses that overexpressed MdATG9. After treatment with the nitrogen starvation stress, overexpression of MdATG9 led to enhanced stress tolerance in both Arabidopsis plants and apple callus (Figure 3 and Supplementary Figure S2). Previous studies in Arabidopsis have reported that the atg9 mutant shows a relatively later and less severe phenotype of senescence compared with atg5-1 and atg7-2 mutants during an extended dark treatment (Barros et al., 2017). Although ATG9 is essential for the formation of autophagosomes, the atg9 mutant displayed a milder reduction in the autophagic process, which explains the relatively minor phenotype observed in this mutant line (Shin et al., 2014; Zhuang et al., 2017). In this study, we found that most detected genes were expressed at higher levels in the transgenic lines than in the WT after examining the expression patterns of other important MdATGs in response to low-nitrogen treatment in all apple callus lines (Figure 4). These results indicate that overexpression of MdATG9 in apple callus might promote the activity of other autophagy gene under nitrogen starvation stress.
In our earlier investigation, we isolated some autophagy-related genes in apple, and found that the tolerance to limited supplies of nutrition increases after these genes were over-expressed, regardless of whether Arabidopsis or an apple callus was used in the tests (Wang et al., 2016, 2017a,b). In this study, we not only demonstrated that overexpression of MdATG9 increased the tolerance to a limited supply of nitrogen, but also detected higher transcript levels of other important MdATGs in MdATG9-OE apple calluses. We measured the concentrations of 15 amino acids, considering that the overexpression of MdATG9 might participate in amino acid metabolism in response to nitrogen starvation in apple callus. Although the low-nitrogen treatment led to a significant decrease in amino acid levels, the concentrations of most amino acids were significantly higher in the transgenic lines than in the WT. Protein synthesis is dramatically limited by intracellular amino acid levels under nitrogen starvation, and autophagy appears to be essential for the size of the free amino acid pool (Onodera and Ohsumi, 2005). Here, we discovered that overexpression of MdATG9 in apple calluses alleviated the limitation of nitrogen starvation to the free amino acid level (Figure 5); therefore, alleviating the limitation of protein synthesis, which contributed to a milder restriction on growth.
In addition, we found that although the levels of most of the detected amino acids were severely down-regulated by nitrogen starvation among genotypes, the contents of Pro and Try increased slightly in transgenic apple callus (Figure 5). Tyr is a substrate for dopamine, which was reported to promote the tolerance of apple to nutrient deficiency-induced stress (Liang et al., 2017). As a compatible solute, Pro has been reported to have antioxidant properties. Recent study reported that the role of plant autophagy in response to salt stress was associated with proline accumulation (Huo et al., 2020b). Combined with the results that the Tyr and Pro content was non-decreasing but increasing in transgenic apple callus in response to nitrogen starvation, the accumulation of both Tyr and Pro appears to contribute, at least in part to MdATG9-mediated tolerance to nitrogen starvation. However, the definite relationship between MdATG9 and Tyr and Pro accumulation requires further exploration.
The high-affinity nitrate transporter NRT2 in Arabidopsis accounts for most of the high-affinity nitrate influx activity under a nitrogen-limited condition (Leran et al., 2014). Here, we determined that the MdNRT2.5 transcript level was induced at higher levels by nitrogen starvation in the transgenic lines than in the WT (Figure 6). Nitrate reductase (NIA) is a key enzyme involved in nitrate assimilation after its uptake in plants (Nawaz et al., 2017). In the present study, MdNIA2 transcripts decreased in WT calluses under treatment, whereas they were upregulated in the OE lines. These results suggest that overexpression of MdATG9 in the callus might promote the absorption of a small amount of nitrate in low-nitrogen medium, and further promote nitrate assimilation; therefore, increasing nitrate utilization in apple callus.
In addition, as the sufficient sucrose was present in the medium, the limited amino acids in the plant cell might be re-utilized directly for protein synthesis rather than used as an energy source. As the energy source of callus growth, the concentrations of soluble sugars in apple calluses also changed in response to the low-nitrogen treatment. In our previous study, we demonstrated that MdATG18a-OE apple plants accumulate more soluble sugars compared with the WT under a nitrogen-depletion treatment (Sun et al., 2018b). Here, we found that fructose, glucose, and sucrose contents in apple calluses increased significantly under the low-nitrogen treatment (Figure 7). However, unlike these three sugars all accumulated higher in MdATG18a-OE apple plants after the deficit treatment, we found that the levels of glucose were lower in MdATG9-OE apple callus while the levels of sucrose in the OE lines were almost two times that of the WT in response to low nitrogen (Figures 7B,C). It might because that the sugars needed for growth of the callus are derived from absorbing the sucrose from the medium. We speculated that overexpression of MdATG9 might improve the sucrose accumulation in apple callus by enhancing absorption of sucrose from the low-nitrogen medium, with more conversion to fructose than glucose. Moreover, considering that both free amino acids and soluble sugars are important compatible osmolytes in the cytoplasm of plant cells (Barros et al., 2017), the significant increases of soluble sugars in apple callus might be able to alleviate the osmotic pressure caused by the large decrease in free amino acids in response to nitrogen starvation.
In summary, we isolated and characterized MdATG9 in apple and examined its function by overexpression in Arabidopsis and apple calluses. The results showed that overexpression of MdATG9 could enhance plant tolerance to nitrogen starvation stress (Figure 8). The free amino acid analysis revealed that MdATG9 overexpression lessened the reduction of most amino acid concentrations under the low-nitrogen treatment, and promoted nitrate assimilation and utilization. In addition, MdATG9 overexpression improved the accumulation of soluble sucrose in apple callus by enhancing the absorption of sucrose from low-nitrogen medium; therefore, resulting in better growth of the transgenic callus. Therefore, the current study characterized the function of MdATG9 under a nitrogen starvation condition, and explored the role of MdATG9 in response to nitrogen starvation from the perspectives of amino acid and sugar metabolism. These findings provide insight into the metabolic importance of MdATG9 acted as an important autophagy gene in response to nutrient starvation in apple.
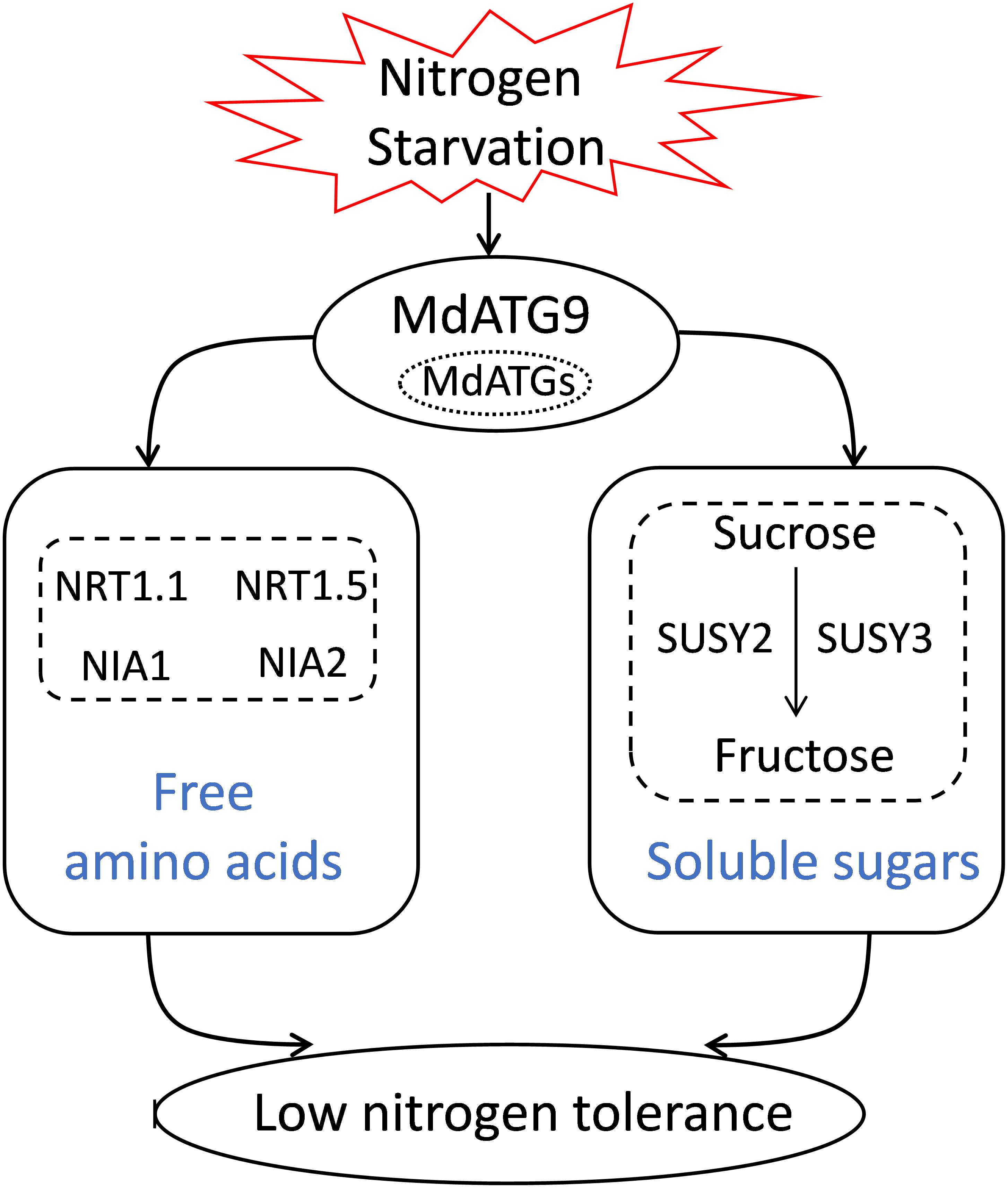
Figure 8. A proposed model for explaining the function of apple MdATG9 in response to nitrogen starvation.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
FM, XG, and LH designed the experiments. LH, ZG, and ZZ performed the experiments and analyzed the data, assisted by XJ, YS, XS, and PW. LH, XG, and FM wrote the manuscript with contributions from all authors.
This work was supported by National Key Research and Development Program of China (2018YFD1000303) and by the Earmarked Fund for China Agriculture Research System (CARS-27).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00423/full#supplementary-material
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Bock, R., et al. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27, 306–322. doi: 10.1105/tpc.114.134205
Barros, J. A. S., Cavalcanti, J. H. F., Medeiros, D. B., Nunes-Nesi, A., Avin-Wittenberg, T., Fernie, A. R., et al. (2017). Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol. 175, 62–76. doi: 10.1104/pp.16.01576
Bassham, D. C. (2009). Function and regulation of macroautophagy in plants. Biochim. Biophys. Acta Mol. Cell Res. 1793, 1397–1403. doi: 10.1016/j.bbamcr.2009.01.001
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Guiboileau, A., Avila-Ospina, L., Yoshimoto, K., Soulay, F., Azzopardi, M., Marmagne, A., et al. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. doi: 10.1111/nph.12307
Han, S. J., Yu, B. J., Wang, Y., and Liu, Y. L. (2011). Role of plant autophagy in stress response. Protein Cell 2, 784–791. doi: 10.1007/s13238-011-1104-4
Hu, L. Y., Zhou, K., Li, Y. T. S., Chen, X. F., Liu, B. B., Li, C. Y., et al. (2018). Exogenous myo-inositol alleviates salinity-induced stress in Malus hupehensis Rehd. Plant Physiol. Biochem. 133, 116–126. doi: 10.1016/j.plaphy.2018.10.037
Huo, L. Q., Guo, Z. J., Wang, P., Zhang, Z. J., Jia, X., Sun, Y. M., et al. (2020a). MdATG8i functions positively in apple salt tolerance by maintaining photosynthetic ability and increasing the accumulation of arginine and polyamines. Environ. Exp. Bot. 172:103989. doi: 10.1016/j.envexpbot.2020.103989
Huo, L. Q., Sun, X., Guo, Z. J., Jia, X., Che, R. M., Sun, Y. M., et al. (2020b). MdATG18a overexpression improves basal thermotolerance in transgenic apple by decreasing damage to chloroplasts. Horticult. Res. 7:21. doi: 10.1038/s41438-020-0243-2
Kang, S., Shin, K. D., Kim, J. H., and Chung, T. (2018). Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 37, 653–664. doi: 10.1007/s00299-018-2258-9
Kim, S. H., Kwon, C., Lee, J. H., and Chung, T. (2012). Genes for plant autophagy: functions and interactions. Mol. Cells 34, 413–423. doi: 10.1007/s10059-012-0098-y
Lai, L. T. F., Yu, C. Y., Wong, J. S. K., Lo, H. S., Benlekbir, S., Jiang, L. W., et al. (2019). Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy 9, 575–583. doi: 10.1080/15548627.2019.1639300
Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Li, F. Q., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Li, W. W., Chen, M., Zhong, L., Liu, J. M., Xu, Z. S., Li, L. C., et al. (2015). Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 468, 800–806. doi: 10.1016/j.bbrc.2015.11.035
Liang, B. W., Li, C. Y., Ma, C. Q., Wei, Z. W., Wang, Q., Huang, D., et al. (2017). Dopamine alleviates nutrient deficiency-induced stress in Malus hupehensis. Plant Physiol. Biochem. 119, 346–359. doi: 10.1016/j.plaphy.2017.09.012
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marshall, R. S., and Vierstra, R. D. (2018). Autophagy: The master of bulk and selective recycling. Ann. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Matsuura, A., Tsukada, M., Wada, Y., and Ohsumi, Y. (1997). Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250. doi: 10.1016/s0378-1119(97)00084-x
McLoughlin, F., Augustine, R. C., Marshall, R. S., Li, F. Q., Kirkpatrick, L. D., Otegui, M. S., et al. (2018). Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. Plants 4, 1056–1070. doi: 10.1038/s41477-018-0299-2
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Ann. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Nawaz, M. A., Wang, L. M., Jiao, Y. Y., Chen, C., Zhao, L., Mei, M. J., et al. (2017). Pumpkin rootstock improves nitrogen use efficiency of watermelon scion by enhancing nutrient uptake, cytokinin content, and expression of nitrate reductase genes. Plant Growth Regul. 82, 233–246. doi: 10.1007/s10725-017-0254-7
Noda, T., Kim, J., Huang, W. P., Baba, M., Tokunaga, C., Ohsumi, Y., et al. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–479.
Onodera, J., and Ohsumi, Y. (2005). Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280, 31582–31586. doi: 10.1074/jbc.m506736200
Perini, P., Pasquali, G., Margis-Pinheiro, M., De Oliviera, P. R., and Revers, L. (2014). Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus x domestica Borkh.). Mol. Breed. 34, 829–842. doi: 10.1007/s11032-014-0078-3
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Saitoh, T., Fujita, N., Hayashi, T., Takahara, K., Satoh, T., Lee, H., et al. (2009). Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl. Acad. Sci. U.S.A. 106, 20842–20846. doi: 10.1073/pnas.0911267106
Shin, J. H., Yoshimoto, K., Ohsumi, Y., Jeon, J. S., and An, G. (2009). OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 27, 67–74. doi: 10.1007/s10059-009-0006-2
Shin, K. D., Lee, H. N., and Chung, T. (2014). A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol. Cells 37, 399–405. doi: 10.14348/molcells.2014.0042
Signorelli, S., Tarkowski, L. P., Van Den Ende, W., and Bassham, D. C. (2019). Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24, 413–430. doi: 10.1016/j.tplants.2019.02.001
Sun, X., Huo, L. Q., Jia, X., Che, R. M., Gong, X. Q., Wang, P., et al. (2018a). Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Horticult. Res. 5:10.
Sun, X., Jia, X., Huo, L. Q., Che, R. M., Gong, X. Q., Wang, P., et al. (2018b). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
van Doorn, W. G., and Papini, A. (2013). Ultrastructure of autophagy in plant cells: a review. Autophagy 9, 1922–1936. doi: 10.4161/auto.26275
Wang, H. B., Dong, Q. L., Duan, D. Y., Zhao, S., Li, M. J., Van Nocker, S., et al. (2018). Comprehensive Genomic Analysis Of The Tyrosine Aminotransferase (Tat) genes in apple (Malus domestica) allows the identification of MdTAT2 conferring tolerance to drought and osmotic stresses in plants. Plant Physiol. Biochem. 133, 81–91. doi: 10.1016/j.plaphy.2018.10.033
Wang, P., Sun, X., Jia, X., and Ma, F. W. (2017a). Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 256, 53–64. doi: 10.1016/j.plantsci.2016.12.003
Wang, P., Sun, X., Wang, N., Jia, X., and Ma, F. W. (2017b). Ectopic expression of an autophagy-associated MdATG7b gene from apple alters growth and tolerance to nutrient stress in Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 128, 9–23. doi: 10.1007/s11240-016-1070-x
Wang, P., Sun, X., Jia, X., Wang, N., Gong, X. Q., and Ma, F. W. (2016). Characterization of an Autophagy-Related gene MdATG8i from Apple. Front. Plant Sci. 7:720. doi: 10.3389/fpls.2016.00720
Yamamoto, H., Kakuta, S., Watanabe, T. M., Kitamura, A., Sekito, T., Kondo-Kakuta, C., et al. (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198, 219–233. doi: 10.1083/jcb.201202061
Yang, Y. N., Li, R. G., and Qi, M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22, 543–551. doi: 10.1046/j.1365-313x.2000.00760.x
Young, A. R. J., Chan, E. Y. W., Hu, X. W., Koch, R., Crawshaw, S. G., High, S., et al. (2006). Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900. doi: 10.1242/jcs.03172
Zhang, X. R., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B. F., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9: e1003196. doi: 10.1371/journal.pgen.1003196
Keywords: autophagy, MdATG9, nitrogen starvation, apple callus, amino acid, sugar
Citation: Huo L, Guo Z, Zhang Z, Jia X, Sun Y, Sun X, Wang P, Gong X and Ma F (2020) The Apple Autophagy-Related Gene MdATG9 Confers Tolerance to Low Nitrogen in Transgenic Apple Callus. Front. Plant Sci. 11:423. doi: 10.3389/fpls.2020.00423
Received: 08 November 2019; Accepted: 24 March 2020;
Published: 15 April 2020.
Edited by:
Luisa M. Sandalio, Spanish National Research Council, SpainReviewed by:
Tamar Avin-Wittenberg, Hebrew University of Jerusalem, IsraelCopyright © 2020 Huo, Guo, Zhang, Jia, Sun, Sun, Wang, Gong and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqing Gong, Z29uZ3hxMDEwM0Bud3N1YWYuZWR1LmNu; Fengwang Ma, ZndtNjRAc2luYS5jb20=; ZndtNjRAbndzdWFmLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.