
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 March 2020
Sec. Plant Proteomics and Protein Structural Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00232
AtNHR2A (Arabidopsis thaliana nonhost resistance 2A) and AtNHR2B (Arabidopsis thaliana nonhost resistance 2B) are two proteins that participate in nonhost resistance, a broad-spectrum mechanism of plant immunity that protects plants against the majority of potential pathogens. AtNHR2A and AtNHR2B are localized to the cytoplasm, chloroplasts, and other subcellular compartments of unknown identity. The multiple localizations of AtNHR2A and AtNHR2B suggest that these two proteins are highly dynamic and versatile, likely participating in multiple biological processes. In spite of their importance, the specific functions of AtNHR2A and AtNHR2B have not been elucidated. Thus, to aid in the functional characterization of these two proteins and identify the biological processes in which these proteins operate, we used immunoprecipitation coupled with mass spectrometry (IP-MS) to identify proteins interacting with AtNHR2A and AtNHR2B and to generate their interactome network. Further validation of three of the identified proteins provided new insights into specific pathways and processes related to plant immunity where AtNHR2A and AtNHR2B participate. Moreover, the comprehensive analysis of the AtNHR2A- and AtNHR2B-interacting proteins using published empirical information revealed that the functions of AtNHR2A and AtNHR2B are not limited to plant immunity but encompass other biological processes.
Plant-pathogen interactions are determined by the genetic characteristics of the biological partners that participate in the interaction: a specific plant species, and a specific pathogen species or its biological variants. Depending on the combination of plant and pathogen, plants are classified as hosts and nonhosts. A host plant is the one that due to its genetic characteristics provides a suitable environment for pathogen growth, and as a result, succumbs to infections by such pathogen. In contrast, a nonhost plant is one that does not provide a suitable environment for pathogen growth and, consequently, is resistant to pathogen infections. This species-specific and broad-spectrum mechanism of resistance against pathogens is called nonhost resistance (Heath, 2000). Nonhost resistance is a complex process that comprises preformed and inducible defenses, as well as the participation of hundreds of genes, many of which remain to be identified and characterized (Senthil-Kumar and Mysore, 2013). Identifying all the components of nonhost resistance and understanding how it operates is of paramount importance toward generating crops with broad resistance to microbial diseases (Lee et al., 2016).
We previously identified AtNHR2A (Arabidopsis thaliana nonhost resistant protein 2A) and AtNHR2B (Arabidopsis thaliana nonhost resistance 2B) as two proteins that participate in nonhost resistance (Singh et al., 2018). We showed that these proteins are synthesized in response to infection by bacterial pathogens and contribute to the strengthening of the plant cell wall through the deposition of the ß- 1,3-glucan polymer, callose (Singh et al., 2018). AtNHR2A and AtNHR2B fusions to the green fluorescent protein (GFP) revealed that both proteins localized predominantly to cytoplasm, chloroplasts, and other subcellular components of unknown identity (Singh et al., 2018). Because the detailed functions of AtNHR2A and AtNHR2B are still unknown, the main objective of this work is to gain insight into their function by investigating their respective protein–protein interaction network (interactome). Investigating protein–protein interactions are powerful tools to gain insight into the function of proteins of interest given that most biological processes occur by the formation of protein complexes (Struk et al., 2018).
Since AtNHR2A and AtNHR2B are not expressed during development but induced upon exposure to pathogens or pathogen-derived elicitors (Singh et al., 2018), unraveling their respective interactomes necessitates the use of in vivo approaches that capture the appropriate biological context of where and when these proteins are actually present. Those approaches have mostly relied on mass spectrometry (MS), in particular affinity purification (AP) coupled with mass spectrometry (AP-MS) (Gingras et al., 2007). In an AP-MS approach, the AP step involves the purification of the protein of interest (bait) by enrichment with a ligand coupled to a solid support. The most frequently used ligands are antibodies that either directly recognize the bait or recognize an epitope tag fused to such bait. Enrichment of the bait by antibodies (immunoprecipitation), co-immunoprecipitates the interacting partners (preys) (Dunham et al., 2012). The co-immunoprecipitated samples are then proteolytically digested, and the resulting peptides are further identified by MS, hence IP-MS (Wendrich et al., 2017; Junkova et al., 2018; Zhang et al., 2019).
Here, we used IP-MS as a tool to screen for proteins that specifically interact with AtNHR2A and AtNHR2B in the biological context when AtNHR2A and AtNHR2B are optimally expressed. IP-MS of AtNHR2A-GFP and AtNHR2B-GFP revealed that AtNHR2A- and AtNHR2B-interacting proteins form complex networks spanning distinct biological processes and occurring in distinct cellular compartments. Intriguingly, in spite of the 65% amino acid sequence identity between AtNHR2A and AtNHR2B, most of the interacting proteins interacted with either AtNHR2A or AtNHR2B and only a few interacted with both proteins. The distinct interactomes of AtNHR2A and AtNHR2B highlight unique functions for each protein, and also supports our previous hypothesis that both proteins act together during plant immunity (Singh et al., 2018). Interestingly, the data revealed that in addition to interacting with proteins that participate in plant immunity, AtNHR2A and AtNHR2B also interact with proteins not implicated in plant immunity, suggesting that AtNHR2A and AtNHR2B integrate plant immunity with other biological processes.
Pseudomonas syringae pathovar tabaci was cultured in King’s B (KB) medium at 30°C supplemented with rifampicin (25 μg/mL). Escherichia coli (E. coli) strains DH5α and Rosetta were cultured in Luria-Bertani (LB) medium at 37°C supplemented with appropriate antibiotics. Agrobacterium tumefaciens was cultured at 28°C in LB medium supplemented with rifampicin (25 μg/mL) and kanamycin (50 μg/mL).
Arabidopsis complementation lines expressing AtNHR2A-GFP and AtNHR2B-GFP in their respective mutant backgrounds (Singh et al., 2018) and a line expressing 35Spro:GFP were used for co-immunoprecipitation experiments. Seeds were surface-sterilized, plated on Murashige and Skoog (MS) media, and grown for 4 weeks at 21°C in a growth chamber with 8/16 h light/dark cycle.
N. benthamiana plants used for transient expression were grown in a growth chamber under 8/16 h light/dark cycle at 25°C.
Four-week-old complementation lines expressing AtNHR2A-GFP and AtNHR2B-GFP in the Atnhr2a and Atnhr2b mutant backgrounds, respectively (Singh et al., 2018), and one line expressing GFP were flood-inoculated (Ishiga et al., 2011) with P. syringae pv. tabaci at 1 × 106CFU/mL to induce expression of AtNHR2A-GFP and AtNHR2B-GFP. Inoculated leaves were collected at 6 hpi and flash frozen in liquid nitrogen. Approximately 1 g of tissue (from 20 plants/genotype/treatment) was manually ground and homogenized in 6 mL of Co-IP extraction buffer [100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 10% Glycerol, 0.2% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM dithiothreitol (DTT), and 1X Proteinase inhibitor cocktail (Sigma Aldrich, St. Louis, MO, United States)]. Extracts were incubated in ice for 30 min and centrifuged twice at 13,000 rpm for 30 min at 4°C. Clear supernatants were transferred to new pre-chilled 50-mL falcon tubes and total protein concentrations were measured using Bradford Assay (BioRad, Hercules, CA, United States). Protein expression was confirmed by Western blot using anti-GFP-HRP (1:1000 dilution; Miltenyi Biotec, Auburn, CA, United States) and detected by luminol solution (ImmunoCruz, SantaCruz Biotechnology Inc., Dallas, TX, United States) (Supplementary Figure S1A).
Five milligrams of total protein extracts (in 5 ml) were mixed with 200 μl of GFP-Trap A beads (ChromoTek, Germany) and incubated overnight at 4°C with end to end rocking. After incubation, beads were washed once with TEN buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA) followed by two washes with higher stringency TEN buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.5 mM EDTA). Proteins bound to the beads were resuspended in 2 ml of TEN buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA). We conducted two independent co-immunoprecipitation experiments for AtNHR2A (AtNHR2A interactors 1st screen, AtNHR2A interactors 2nd screen) (Supplementary Table S1), and two independent co-immunoprecipitation experiments for AtNHR2B (AtNHR2B interactors 1st screen, AtNHR2B interactors 2nd screen) (Supplementary Table S2). For each co-immunoprecipitation experiment that included AtNHR2A-GFP, a parallel co-immunoprecipitation was done for GFP. Similarly, for each co-immunoprecipitation experiment that included AtNHR2B-GFP, a parallel co-immunoprecipitation was done for GFP. Co-immunoprecipitated samples were processed for on-bead trypsin digestion or, in-gel trypsin digestion (Figure 1A).
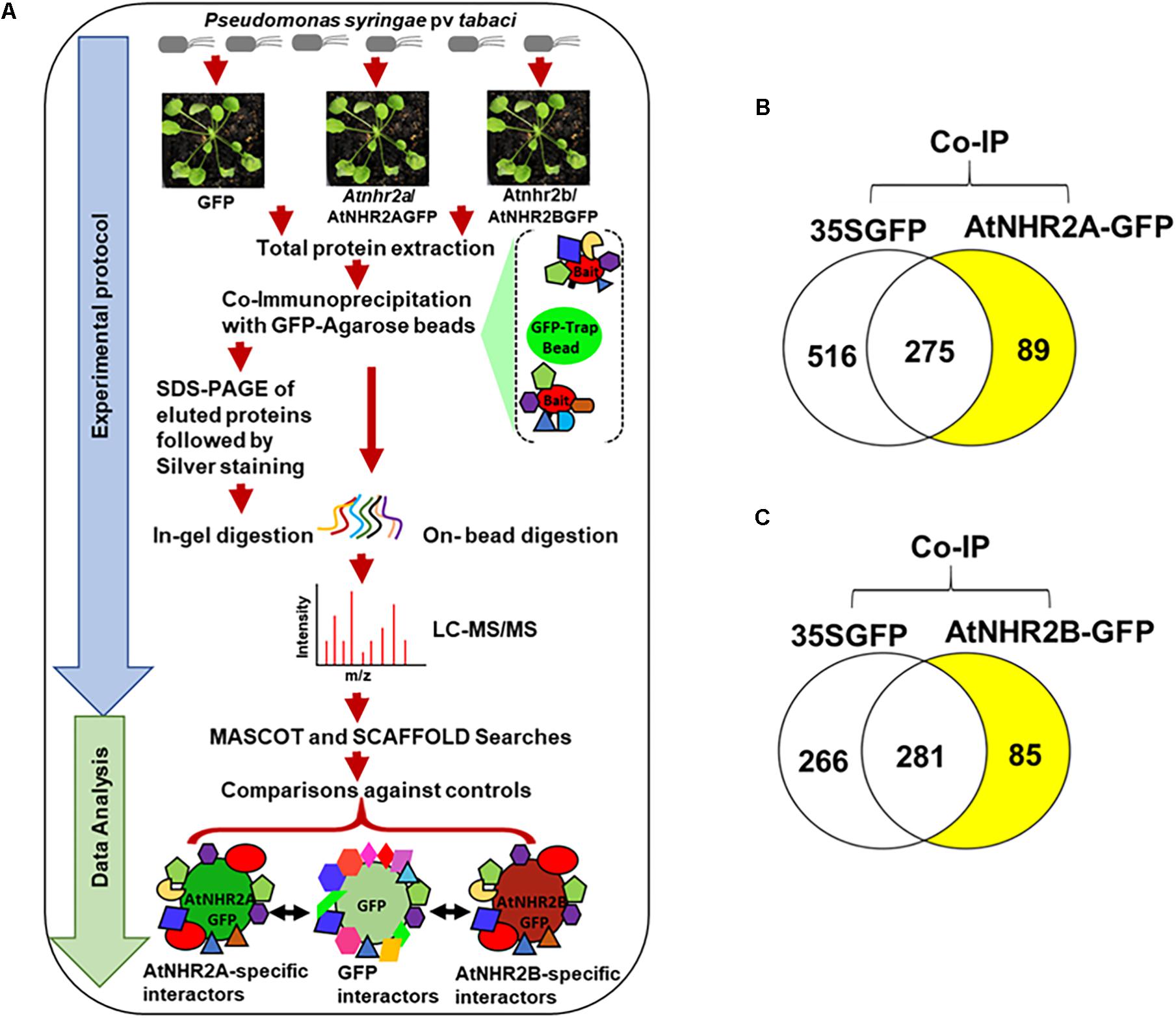
Figure 1. Co-immunoprecipitation/mass spectrometry analysis to unravel the AtNHR2A and AtNHR2B Interactomes. Co-IP/MS workflow summarizing experimental protocol and data analysis. For the experimental protocol, four-week-old transgenic Arabidopsis thaliana plants expressing AtNHR2A-GFP and AtNHR2B-GFP GFP were inoculated with Pseudomonas syringae pv tabaci at 1 × 106 CFU/mL. At 6 hpi, plants were harvested for total protein extraction. Extracted proteins from each sample were subjected to co-immunoprecipitation using GFP Trap A beads. GFP-bound protein complexes were eluted with 2X SDS sample buffer and resolved by SDS-PAGE for silver staining and in-gel trypsin digestion, or directly processed for on-bead trypsin digestion. Both in-gel and on-bead tryptic digested products were analyzed by LC-MS/MS. For data analysis, identified peptide spectra were then analyzed by Mascot using the Arabidopsis thaliana protein database as reference, and identified peptides were further validated and compared using Scaffold to identify peptides representing proteins that specifically interact with AtNHR2A-GFP or AtNHR2B-GFP but not with GFP. Peptide identification were done by setting peptide hits ≥ 2 with the false discovery rate (FDR) below 1% based on decoy database (A). Venn diagrams show specific proteins interacting with AtNHR2A-GFP (B) and AtNHR2B-GFP (C) (shaded in yellow).
We used two digestion methods to increase the recovery of proteins as each method provides specific advantages. The advantage of the on-bead digestion method is that it maximizes protein solubilization making them more susceptible to enzymatic cleavage (Chen et al., 2007). The advantage of in-gel digestion method is that it increases the number of protein identifications because it reduces the complexity of the sample by adding the size separation on the gel prior to MS (Feist and Hummon, 2015). The digestion strategy also determines the instrument used for mass spectrometry.
Proteins bound to the beads were pre-digested with 0.2% of RapiGest SF (Waters Corporation, Milford, MA, United States) in 50 mM ammonium bicarbonate (Schechter et al., 2014). After pre-digestion, samples were reduced with 10 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl) and alkylated with 25 mM iodoacetamide, followed by overnight digestion at 37°C with sequencing grade trypsin (Promega, Madison, WI, United States) in 1:50 trypsin/protein ratio. Digested samples were dissolved in 1% acetonitrile/0.1% formic acid.
Proteins bound to the beads were eluted with 100 μl of 2X SDS-buffer and boiled at 90°C for 8 min. Proteins co- immunoprecipitated with AtNHR2A-GFP, AtNHR2B-GFP, and GFP were separated by mass spectrometry compatible 4–20% Tris-Glycine precast gel (Thermo Fisher Scientific Inc., Carlsbad, CA, United States) then stained with PierceTM silver stain kit for Mass Spectrometry (Thermo Fisher Scientific Inc., Carlsbad, CA, United States). The stained gel lanes were then divided into four sections based on 4 ranges of molecular weight (Supplementary Figure S1B), and each section was sliced further into about 4–5 pieces of ca 1 mm2 size gel pieces to increase the trypsin contact surface area for efficient trypsin digestion. Gel pieces were first destained with 15 mM potassium ferricyanide and 50 mM sodium thiosulfate. Destained gel pieces were dehydrated using 100% HPLC grade acetonitrile and completely dried using a SpeedVac. Dried gel pieces were treated with 10 mM DTT in 25 mM ammonium bicarbonate (pH 7.8) at 60°C for 1 h to reduce proteins. Excess DTT was removed by pipetting, and gel pieces were treated with 20 mM iodoacetamide in 25 mM ammonium bicarbonate at room temperature for 1 h in the dark to alkylate the reduced proteins. After the iodoacetamide treatment, gel pieces were washed thoroughly with 25 mM ammonium bicarbonate, dehydrated with 100% acetonitrile, and completely dried using a SpeedVac. One hundred microliters of MS grade Trypsin (Thermo Fisher Scientific Inc., Carlsbad, CA, United States) in 25 mM ammonium bicarbonate were added to the dried gel pieces to a final concentration of 50 ng/μl. Samples were kept at 4°C for 30 min to ensure trypsin was efficiently absorbed to the dried gel pieces. One hundred microliters of additional 25 mM ammonium bicarbonate were added to each sample and incubated at 37°C for 24 h. Trypsin-digested samples were acidified with 5% formic acid (FA) in 60% acetonitrile (ACN) to quench trypsin enzyme activity and break down ammonium bicarbonate. The samples were further desalted using Pierce C18- spin columns (Thermo Fisher Scientific Inc., Carlsbad, CA, United States) following manufacturer’s instructions.
Five microliters of the on-bead protein digested samples were used for LC-MS/MS analysis using an LTQ-Orbitrap Velos Pro (Thermo Fisher Scientific, San Jose, CA, United States) coupled with a U3000 RSLCnano HPLC (Thermo Fisher Scientific, San Jose, CA, United States) using previously described conditions (Alvarez et al., 2013, 2014; Huang et al., 2016). The LTQ-Orbitrap mass analyzer was operated in the data-dependent acquisition mode in which each MS1 scan was followed by collision-induced dissociation (CID) of the 10 most intense precursor ions. The mass range for the MS1 done using the FTMS was 365 to 1800 m/z with resolving power set to 60,000 @ 400 m/z and the automatic gain control (AGC) target set to 1,000,000 ions with a maximum fill time of 100 ms. The selected precursors were fragmented in the ion trap using an isolation window of 1.5 m/z, an AGC target value of 10,000 ions, a maximum fill time of 100 ms, a normalized collision energy of 35, and activation time of 30 ms. Dynamic exclusion was performed with a repeat count of 1, exclusion duration of 45 s, and a minimum MS ion count for triggering MS/MS set to 5000 counts.
Five microliters of the in-gel desalted trypsin-digested peptides were analyzed by LC-MS/MS using an Agilent 1200 series microflow high-performance liquid chromatography (HPLC) coupled to a Bruker amaZon SL quadrupole ion trap mass spectrometer with a captive spray ionization source. Peptides were separated by reverse-phase high-performance liquid chromatography (RP-HPLC) using a Zorbax SB C18 column (150 × 0.3 mm, 3.5 μm particle size, 300Å pore size, Agilent Technologies) with a solvent flow rate of 4 μL/minute and a gradient of 5–38% consisting of 0.1% FA (solvent A) and ACN (solvent B) over a period of 320 min. MS analyses were performed in a positive ion mode using Bruker captive electrospray source with a dry nitrogen gas temperature of 200°C and a nitrogen flow rate of 3 L/minute. LC-MS/MS data were carried out in the Auto MS(n) mode. Optimized trapping condition for the ions at m/z 1000 was set. Regular MS level scans were performed using the enhanced scanning mode (8100 m/z/second), while MS/MS fragmentation (collision induced fragmentation) scans were performed automatically for top ten precursor ions with a set threshold for 1 min using UltraScan mode (32,500 m/z/second) (Karash et al., 2017; Rath et al., 2019). Immunoprecipitated samples (AtNHR2A 2nd screen and AtNHR2B 2nd screen) were injected twice into the latter instrument to generate a technical replicate (AtNHR2A 3rd screen and AtNHR2B 3rd screen) (Supplementary Tables S1, S2). Consolidated data and common interactors between AtNHR2A and AtNHR2B is presented in Supplementary Table S3.
The MS/MS spectra obtained from both methods were searched against the TAIR10_20101214 non-redundant Arabidopsis protein available in the Arabidopsis Information Resource (TAIR) database, containing 35,639 protein entries. MASCOT v 2.5 (Matrix Science, London, United Kingdom; version 2.5.1) database search software was used to search TAIR database using trypsin as the enzyme with one miss cleavage, carbamidomethyl fixed modification, and methionine oxidation as the variable modification. Protein identifications were accepted if they were represented by at least 2 unique peptides, each peptide showing probability higher than 80% (Mascot Ion Score higher than 20) and with a false discovery rate of < 1.0% as determined by the Scaffold Local FDR algorithm. Protein probabilities were assigned by the Protein Prophet algorithm (Keller et al., 2002). For each of the AtNHR2A screens, AtNHR2A interactors were those proteins identified only from AtNHR2A-GFP samples but not from GFP samples. Similarly, for each of the AtNHR2B screens, AtNHR2B interactors were those proteins identified only from the AtNHR2B-GFP samples and not from GFP samples. Proteins identified in all samples (AtNHR2A-GFP, AtNHR2B-GFP, and GFP) were considered non-AtNHR2A- or non-AtNHR2B- specific proteins and therefore, were removed from the list.
AtNHR2A and AtNHR2B interacting proteins were broadly classified into biological process, molecular function, and cellular components, according to the Gene Ontology (GO) annotation tool of the Arabidopsis thaliana reference genome using GO Slim available in the Arabidopsis Information Resource (TAIR) database1. A more detailed annotation was obtained by identifying convincing empirical data regarding protein subcellular localization and function in the published literature.
AtENGD-1 (At1g30580) was cloned into pDONR207 (Thermo Fisher Scientific Inc., Carlsbad, CA, United States) using primers 5′GGGGACAAGTTTGTACAAAAAAGCAGGCTYYA TGCCTCCGAAAGCCAAA3′ and 5′GGGGACCACTTTGTA CAAGAAAGCTGGGTYTCATTTCTTCCCACCACCG3′, and subsequently transferred to pET59-DESTTM (EMD Millipore, Burlington, VA, United States) to generate 6X-His-AtENGD-1.
A plasmid harboring AtRPN1A (At2g20580) epitope tagged with Myc (Myc-AtRPN1A) was obtained from the Arabidopsis Biological Resource Center (ABRC) stock collection2.
AtCCoAOMT1 (At4g34050) was cloned into pDONR201 (Thermo Fisher Scientific Inc., Carlsbad, CA, United States) using primers 5′GGGGACAAGTTTGTACAAAAAAGCAGGC TYYATGGCGACGACAACAACA3′ and 5′GGGGACAAGTTT GTACAAAAAAGCAGGCTYYATGCCTCCGAAAGCCAAA3′ and subsequently transferred to pEarleyGate201 (Earley et al., 2006) to generate HA-AtCCoAOMT1.
Full length AtNHR2B and truncated AtNHR2B1–140 in pDONR201 were transferred to pSITEnEYFPC1 (Martin et al., 2009). AtCCoAOMT1 was cloned into pSITEcEYF PC1(Martin et al., 2009).
Plasmids harboring Myc-AtRPN1A, HA-AtCCoAOMT1, AtCCoAOMT1-cEYFP, AtNHR2B-nEYFP, and AtNHR2B(1–140)-nEYFP were transformed into Agrobacterium tumefaciens strain GV2260 by electroporation for transient gene expression in N. benthamiana. pET59-DEST:AtENGD-1 was transformed into E. coli strain Rosetta for protein expression and purification.
Escherichia coli harboring plasmid pET59-DEST:AtENGD-1 was grown in 5 mL of LB broth supplemented with ampicillin (50 μg/mL). The overnight culture was then subcultured into 250 mL of LB supplemented with ampicillin (50 μg/mL). When the culture reached an OD600 of 0.5, protein expression was induced by adding 0.5 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) followed by incubation for additional 6 h at 28°C. Bacterial cells were harvested by centrifugation at 12,000 rpm at 4°C, and proteins were purified on nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Thermo Fisher Scientific Inc., Carlsbad, CA, United States), following manufacturer’s instructions. Proteins were then separated in a 12% SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue to confirm IPTG induction. Protein expression was confirmed by Western blotting using anti-His antibodies (1:500 dilution; Cell Signaling Technology, Danvers, MA, United States).
The predicted functional association network of Arabidopsis thaliana was downloaded from the STRING database available at: https://stringdb-static.org/download/protein.links.v11.0/3702.protein.links.v11.0.txt.gz. All evidence channels (genomic context, coexpression, text mining, experimental evidence, curated protein databases) (Szklarczyk et al., 2019) were used for the analysis. The downloaded network had ∼11 million total edges with scores ranging from 150 to 900. The edges were sorted based on the increasing order of their edge score with the top 10% edges (combined minimum score of 626) selected as high confidence edges and the rest filtered. This top 10% network was searched with the 154 proteins that we found to be interacting with either AtNHR2A or AtNHR2B or with both proteins. The predicted links in STRING network and the interactions identified by IP-MS were assembled using Cytoscape v3.3.0 and including subcellular localization and functional category based on literature searches (Supplementary Tables S4–S6).
Agrobacterium tumefaciens strains harboring HA-AtCCoAOMT1, Myc-AtRPN1A, and AtNHR2B-GFP were grown overnight at 28°C with constant shaking. Overnight cultures were harvested by centrifugation at 6,000 rpm for 10 min, resuspended in induction buffer [20mM 2-N-morpholino ethanesulfonic acid (MES) pH 5.5; 3% sucrose, 200μM acetosyringone], and incubated at room temperature with constant shaking for 4 h. Induced cultures were adjusted to an OD600 = 0.3. A. tumefaciens harboring AtNHR2B-GFP was co-infiltrated with A. tumefaciens harboring HA-AtCCoAOMT1 or Myc-AtRPN1A into fully expanded leaves of 3-week-old N. benthamiana plants using a needless syringe. A. tumefaciens harboring a construct encoding GFP was co-infiltrated with either HA-AtCCoAOMT1 or Myc-AtRPN1A and used as negative control. After 3 days, infiltrated leaves were harvested for protein extraction. Proteins isolated from leaves co-expressing AtNHR2B-GFP/HA-AtCcoAOMT and GFP/HA-AtCcoAOMT were immunoprecipitated with the PierceTM HA Epitope Tag Antibody conjugated to agarose beads (Thermo Fisher Scientific Inc., Carlsbad, CA, United States). Proteins isolated from leaves co-expressing AtNHR2B-GFP/Myc-AtRPN1A and GFP/Myc-AtRPN1A were precleared with protein A-sepharose beads (GE Healthcare, Chicago, IL, United States) at 4°C for 2 h with gentle shaking. Precleared samples were immunoprecipitated with Myc antibodies (Miltenyi Biotec, Bergisch Gladbach, Germany) coupled with protein A-sepharose beads. Co-immunoprecipitated samples were washed and eluted in 2× SDS protein loading buffer, ran in a SDS-PAGE gel, and transferred to nitrocellulose membranes for Western blot and probed with anti-GFP-HRP (1:1000 dilution; Miltenyi Biotec, Bergisch Gladbach, Germany), or anti-Myc-HRP (1:1000 dilution; Miltenyi Biotec, Bergisch Gladbach, Germany), or anti-HA-HRP (1:1000 dilution; Thermo Fisher Scientific Inc., Carlsbad, CA, United States), depending on the protein combination. The proteins were detected by chemiluminescence (SantaCruz Biotechnology, Dallas, TX, United States).
To validate the interaction between AtNHR2A-GFP and His-AtENGD-1, AtNHR2A-GFP and GFP were transiently expressed in N. benthamiana, and proteins extracted as described above. His-AtENGD-1 was expressed and purified in E. coli. Thereafter, 5 μg of the purified His-AtENGD-1 were mixed with 100 μg of total protein extracted from N. benthamiana plants transiently expressing AtNHR2A-GFP or GFP, and subjected to co-immunoprecipitation with GFP Trap-A beads (Chromotek, Germany), as described above. The co-immunoprecipitated samples were washed, eluted in 2 × SDS protein loading buffer, resolved by SDS-PAGE, and transferred to a nitrocellulose membrane for Western blotting using anti-His (1:500 dilution; Cell Signaling Technology) or anti-GFP-HRP (1:1000 dilution; Miltenyi Biotec, Bergisch Gladbach, Germany) antibodies.
AtNHR2B fused to the N-terminal-encoding half of the enhanced yellow fluorescent protein (EYFP), and AtCCoAOMT1 fused to the C-terminal-encoding half of EYFP were co-infiltrated in N. benthamiana for transient expression. Leaf samples were collected at 3 days after infiltration for live-cell image analysis using a Leica TCS SP5 II confocal microscope (Leica Microsystems, Buffalo Grove, IL, United States) at an excitation wavelength of 514 nm (argon laser) and an emission wavelength of 500 to 530 nm.
To identify the AtNHR2A- and AtNHR2B- interactomes, this work relied on IP-MS. The experiments used transgenic Arabidopsis lines expressing AtNHR2A-GFP and AtNHR2B-GFP in the Atnhr2a and Atnhr2b mutant backgrounds, respectively (Singh et al., 2018). To provide the appropriate biological context, we inoculated plants with P. syringae pv. tabaci to induce expression of AtNHR2A and AtNHR2B (Figure 1A). Because the maximum induction of both AtNHR2A and AtNHR2B occurs at 6 hours post inoculation (hpi) (Singh et al., 2018), we chose this time point to harvest inoculated plants for protein extraction. Thus, we are confident that these conditions ensured endogenous levels of AtNHR2A-GFP and AtNHR2B-GFP and reduced the occurrence of spurious results. The combination of the two approaches increased protein recovery, while the stringent data analyses allowed us to confidently identify true interactors for AtNHR2A and AtNHR2B.
The experiments using AtNHR2A-GFP as bait yielded a total of 881 nonredundant proteins from both methods. From those 881 proteins, 516 proteins interacted with the GFP tag, 275 proteins interacted with both the GFP tag and AtNHR2A-GFP, and 89 interacted only with AtNHR2A-GFP (Figure 1B and Supplementary Table S1). Similarly, co-immunoprecipitation experiments using AtNHR2B-GFP yielded 632 nonredundant proteins from both methods. Out of those 632 proteins, 266 interacted with the GFP tag, 281 interacted with both the GFP tag and AtNHR2B-GFP, and 85 interacted only with AtNHR2B-GFP (Figure 1C and Supplementary Table S2). Out of the 89 proteins interacting with AtNHR2A and the 85 proteins interacting with AtNHR2B, we found 20 proteins that interacted with both AtNHR2A and AtNHR2B (Supplementary Table S3). The AtNHR2A- and AtNHR2B- specific interactors were considered high confidence interactors because: (i) they were not identified in the negative control, (ii) were represented by two or more unique peptides, and (iii) they had an FDR less than 1.0%.
We initially classified the proteins interacting with AtNHR2A and AtNHR2B using the GO categories biological process, molecular function, and cellular component, and found that AtNHR2A- and AtNHR2B- interacting proteins were placed in similar functional categories (Supplementary Figures S2A,B), yet visual inspection indicated striking differences between the two datasets. Therefore, to extract more meaningful information from the data, we manually annotated the proteins using published literature that convincingly reported subcellular localization and/or specific function. That specific function was then assigned to a broader functional category. Only proteins with known subcellular localization and function were included (Tables 1–3).

Table 1. AtNHR2A specific interactors with known subcellular localization and function, ordered by gene ID.

Table 2. AtNHR2B specific interactors with known subcellular localization and function, ordered by gene ID.
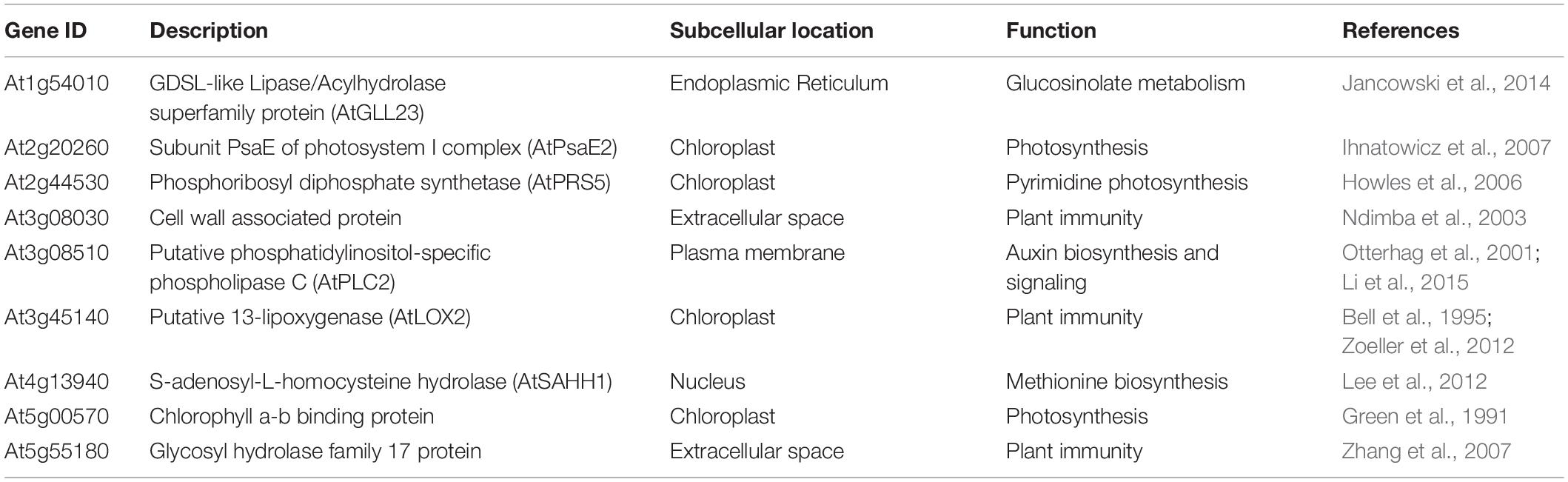
Table 3. List of proteins interacting with AtNHR2A and AtNHR2B with known subcellular localization and function, ordered by gene ID.
Regarding the subcellular localization of the interactors, we found that the proteins interacting with AtNHR2A and AtNHR2B localized to multiple subcellular compartments. However, the number and specific proteins in each compartment varies when comparing between AtNHR2A- and AtNHR2B-interactors. Among the AtNHR2A interactors, the subcellular compartment containing the largest number of proteins was the cytoplasm followed by the extracellular space (Table 1), while the subcellular compartments containing the largest number of AtNHR2B-interacting proteins were chloroplasts followed by cytoplasm (Table 2). Among the proteins that interacted with both AtNHR2A and AtNHR2B, the most common localization was to chloroplasts followed by extracellular space (Table 3). It was interesting to note that several of the AtNHR2A and AtNHR2B interactors have more than one localization, highlighting that several biological processes require interorganellar movement of proteins.
With regard to functional categories, the most common of them for AtNHR2A interactors was plant immunity and included 15 proteins with demonstrated function in pathogen recognition, regulation of immune responses, signaling, vesicle trafficking, secretion of defense secondary metabolites, and cell wall remodeling. The second most common category for AtNHR2A interactors included 10 proteins participating in enzymatic reactions during primary metabolism, such as synthesis of essential carbohydrates, lipids, or proteins, and occurring in cytoplasm, mitochondria, or chloroplasts. Other categories with significant number of proteins among the AtNHR2A interactors included those associated with RNA-mediated processes, such as transcription, splicing, and RNA stability, as well as those related to protein synthesis, the latter being largely represented by ribosomal proteins. Other categories with fewer proteins included protein folding and turnover, secondary metabolism, hormone-related processes, and abiotic stress (Table 1).
The most common functional category for AtNHR2B interactors was primary metabolism and included 19 proteins with enzymatic activities, as well as structural function, found to be essential in processes occurring in mitochondria and chloroplasts. The second most common category for AtNHR2B interactors was protein synthesis represented by 8 ribosomal proteins (Table 2). In contrast to the abundance of AtNHR2A interactors representing plant immunity, there are only 5 AtNHR2B interactors for the same category which included regulatory proteins, in addition to proteins functioning in pathogen recognition and, significantly, AtNHR2A (Table 2). Other categories represented among the AtNHR2B interactors, but with fewer proteins included vesicle trafficking, cell wall biosynthesis, protein folding and turnover, secondary metabolism, hormone-related processes, and abiotic stress. Two additional categories present among the AtNHR2B interactors but absent among the AtNHR2A interactors included one protein associated with the cytoskeleton and another protein associated with cell division (Table 2).
We found that the 20 proteins that interacted with both AtNHR2A and AtNHR2B can be grouped into the same functional categories as those assigned for proteins interacting with either AtNHR2A or AtNHR2B, those categories being primary metabolism and plant immunity. Other functional categories with fewer proteins were secondary metabolism and hormone-related processes (Table 3).
Although we were able to reliably assign subcellular localization and function to 62% of the proteins interacting with AtNHR2A, 66% of the proteins interacting with AtNHR2B, and 45% of the proteins interacting with both AtNHR2A and AtNHR2B, there were still many interacting proteins that were classified as unknown, either because they are truly unknown, or because published research on those proteins did not provide compelling evidence for their subcellular localization and/or function.
Altogether, the differences between the AtNHR2A and AtNHR2B interactomes demonstrates that AtNHR2A and AtNHR2B have distinct functions. However, the finding that they interact with common proteins, and that the non-common proteins participate in common functional processes, indicate that AtNHR2A and AtNHR2B act together as previously proposed (Singh et al., 2018).
To further confirm the accuracy of the IP-MS method, and to gain insight into the function of AtNHR2A and AtNHR2B in plant immunity, we chose to validate the interaction between AtNHR2A or AtNHR2B with proteins known to function in plant innate immunity such as AtENGD-1, AtRPN1A, and AtCCoAMT1 (Yao et al., 2012; Abdeeva et al., 2018; Xie et al., 2018). AtENGD-1 was fused to the 6× histidine (6× His) tag and expressed and purified from E. coli. Purified His- AtENGD-1 was added to protein extracts obtained from N. benthamiana plants expressing AtNHR2A-GFP. Immunoprecipitation of AtNHR2A-GFP with GFP antibodies, co-immunoprecipitated His- AtENGD-1, as shown by Western blot analysis using anti-His antibodies. Anti-GFP antibodies also immunoprecipitated GFP but did not co-immunoprecipitated His- AtENGD-1 (Figure 2A and Supplementary Figure S3).
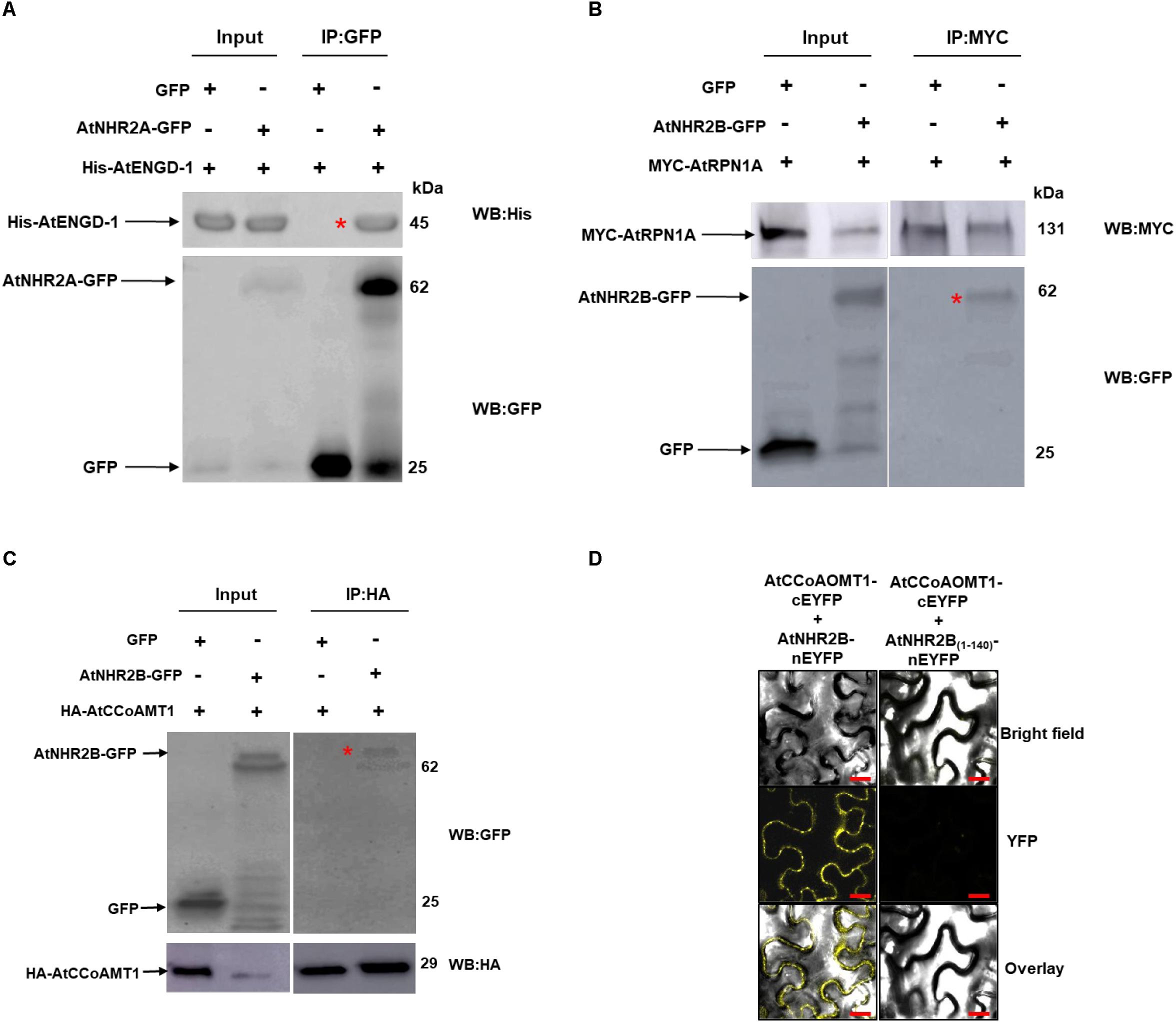
Figure 2. Validation of selected AtNHR2A and AtNHR2B interactions. Protein extracts of N. benthamiana transiently expressing AtNHR2A-GFP were mixed with His-AtENGD-1 purified from E. coli (Rosetta) and subjected to immunoprecipitation with anti-GFP antibodies. Co-immunoprecipitation of His-AtENGD-1 (red asterisk) was detected by Western blot using anti-His antibodies (A). AtNHR2B-GFP and Myc-AtRPN1A pair and AtNHR2B-GFP and HA-AtCCoAMT1 pair were transiently co-expressed in N. benthamiana and immunoprecipitated with anti-Myc and anti-HA antibodies, respectively. Co-immunoprecipitation of AtNHR2B-GFP in each experiment (red asterisks) were detected by Western blot using anti-GFP antibodies (B,C). Expected protein sizes are shown by arrows. AtNHR2B, or its non-functional version AtNHR2B(1–140) fused to the N-terminal fragment of EYFP were co-expressed in N. benthamiana with AtCCoAOMT1 fused to the C- terminal fragment of EYFP. The reconstitution of the EYFP signal was evaluated via laser scanning confocal microscopy at 3 days after infiltration (D). Images were taken using excitation wavelength of 514 nm and an emission wavelength of 500 to 530 nm. Bar = 10 μm.
The interaction between AtNHR2B-GFP and AtRPN1A, and that between AtNHR2B-GFP with AtCCoAOMT1 was validated by reciprocal co-IP, wherein AtRPN1A and AtCCoAOMT1 were cloned as fusions to the MYC and HA tags, respectively, and used to pull down AtNHR2B-GFP. Immunoprecipitation of MYC-AtRPN1A with anti-MYC antibodies co-immunoprecipitated AtNHR2B-GFP but not GFP alone (Figure 2B and Supplementary Figure S4). Immunoprecipitation of HA-AtCCoAOMT1 with anti-HA antibodies also co-immunoprecipitated AtNHR2B-GFP but not GFP alone (Figure 2C and Supplementary Figure S5).
We also used bimolecular fluorescence complementation (BiFC) in N. benthamiana to confirm the interaction between AtNHR2B-GFP and AtCCoAOMT1 in planta. For that purpose, full length AtNHR2B was fused to the N-terminal half of the enhanced yellow fluorescent protein (nEYFP), while AtCCoAOMT1 was fused to the C-terminal half of EYFP (cEYFP). Following the published guidelines for BIFC (Bock and Kudla, 2016), a truncated version of AtNHR2B was used as control containing only the first 140 amino acids (AtNHR2B1–140) as this truncation abolishes normal localization of AtNHR2B-GFP (Singh and Rojas, 2018). AtNHR2B1–140 was fused to nEYFP, and AtNHR2B-nEYFP or AtNHR2B(1–140)-nEYFP were transiently co-expressed with AtCCoAOMT1-cEYFP in N. benthamiana to examine the reconstitution of the EYFP fluorescence upon interaction between the two proteins. Co-expression of full length AtNHR2B-nEYFP with AtCCoAOMT1-cEYFP generated a yellow fluorescence signal indicating the reconstitution of the EYFP by the interaction between full length AtNHR2B and AtCCoAOMT1 in planta. Moreover, this BiFC data clearly demonstrated that the interaction between full length AtNHR2B and AtCCoAOMT1 occurs in the cytoplasm and punctae bodies where AtNHR2B has been consistently found (Singh et al., 2018). In contrast, co-expression of AtNHR2B(1–140) –nEYFP with AtCCoAOMT1-cEYFP did not show yellow fluorescence (Figure 2D) indicating that the truncation of AtNHR2B interferes with the interaction.
Our finding that AtNHR2A and AtNHR2B share a common set of interacting proteins, together with our accumulated genetic evidence (Singh et al., 2018), implies that AtNHR2A and AtNHR2B belong to the same protein network. Thus, we investigated if the observed links in our protein interaction data were also preserved in other protein networks. We reasoned that since proteins in a subnetwork are more likely to be functionally related, the interactions we observed in our IP-MS dataset should also be predictable from other data sources. To test this, we obtained high-confidence Arabidopsis protein network data from the STRING database (Szklarczyk et al., 2019). The protein links represented in the STRING database were computationally predicted and scored by combining probabilities depicting functional-relatedness from seven different evidence channels in Arabidopsis. Overlapping this predicted network with our data revealed that several of the AtNHR2A- and AtNHR2B- interactors that were experimentally identified in this study were also preserved in the STRING network data (Figure 3). Specifically, out of 155 proteins in our interactome, ∼40% are also connected to each other in the STRING network, indicating that these are genuine interactions. Remarkably, a large fraction of the proteins that overlapped with the predicted network are the proteins that are common for AtNHR2A and AtNHR2B with a large fraction of these proteins being involved in protein synthesis. The network also shows a preferred subcellular localization to chloroplasts among the AtNHR2B interactors.
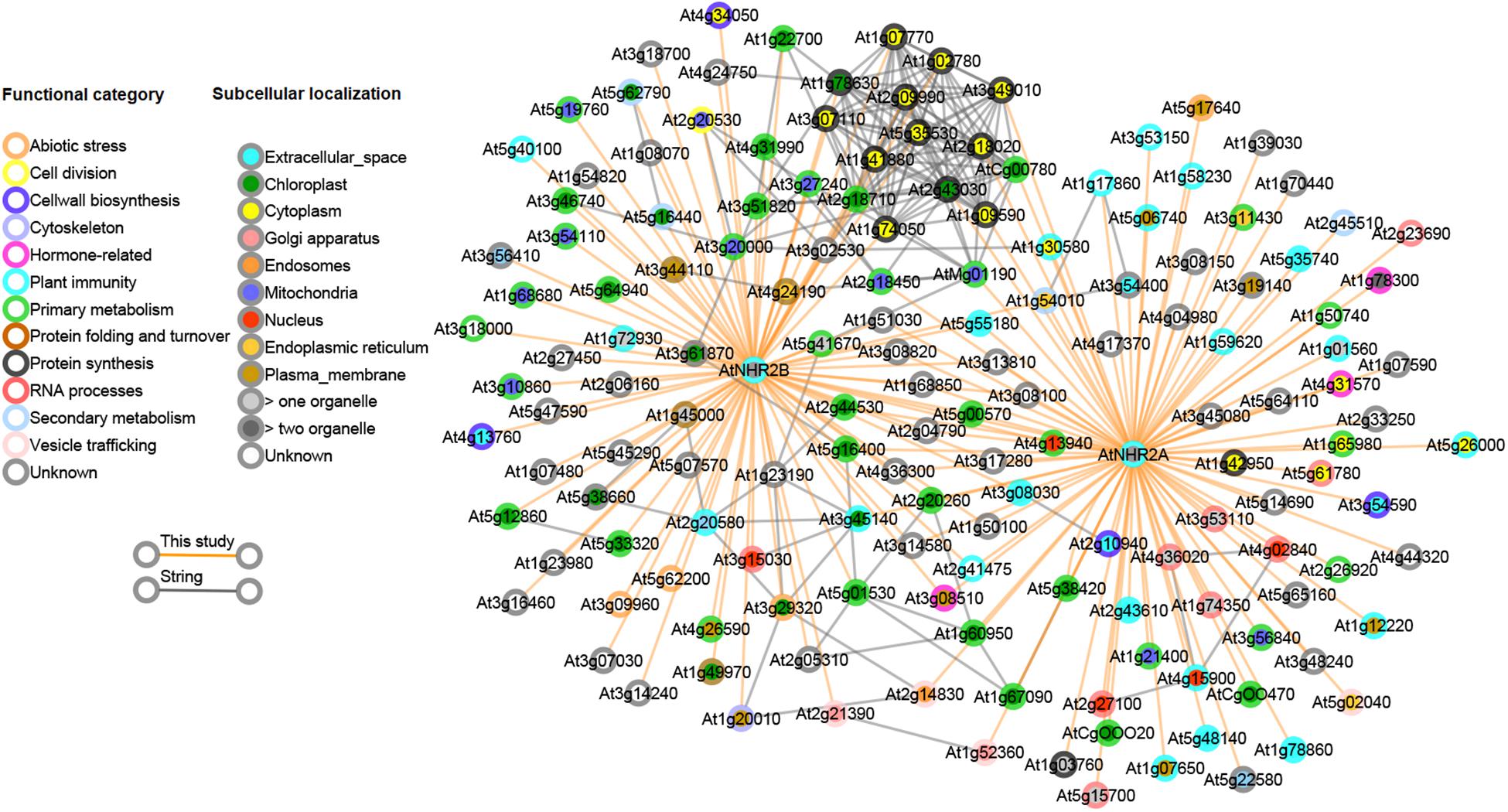
Figure 3. Combined interactome network for AtNHR2A and AtNHR2B. The AtNHR2A-AtNHR2B interactome was combined with predicted gene functional associations in the STRING database and visualized using Cytoscape v 3.3.0. Circles represent genes color-coded for functional category and subcellular localization based on literature searches. Orange lines show the interactions between each gene and AtNHR2A and/or AtNHR2B using IP-MS. Gray lines show predicted functional interactions using STRING network.
In this study, we unraveled the interactomes of AtNHR2A and AtNHR2B to gain further insight into their detailed function so as to integrate this knowledge with our previous live-cell imaging data and with the biological phenotypes for AtNHR2A and AtNHR2B. Our in vivo IP-MS used conditions that directly reflect AtNHR2A and AtNHR2B expression and their corresponding endogenous protein abundance while eliminating false positives due to non-specific interactions with the epitope tag (GFP). We found that proteins interacting with AtNHR2A and AtNHR2B are localized to diverse subcellular compartments with a preferred localization to cytoplasm and chloroplasts. These findings are not surprising considering our previous live-cell imaging data indicated that AtNHR2A and AtNHR2B localize to these compartments (Singh et al., 2018). The localization of AtNHR2A and AtNHR2B to chloroplasts is important in the context of plant immunity as this organelle provides key defense signaling molecules, such as hormones and reactive oxygen species (Serrano et al., 2016). However, none of the chloroplastic-localized proteins that interacted with AtNHR2A or AtNHR2B are known to play a role in chloroplast-mediated immune responses. Rather, all the AtNHR2A and AtNHR2B interactors that localized to chloroplasts are implicated in primary metabolism, functioning either as enzymes or as structural components. Thus, the localization of AtNHR2A and AtNHR2B to chloroplasts is still mysterious. Nevertheless, because AtNHR2B interacted with more chloroplastic proteins than AtNHR2A, it is likely that AtNHR2B plays a more prominent function than AtNHR2A in chloroplasts. Interestingly, chloroplasts were also the preferred localization for proteins that interacted with both AtNHR2A and AtNHR2B, and those common interactors can be seen as bridges integrating both independent interactomes.
One curious finding was the abundance of AtNHR2A- and AtNHR2B- interacting proteins localized to the extracellular space, particularly among the AtNHR2A interactors. Several of those extracellularly localized proteins included proteins functioning in plant immunity either as antimicrobials, such as chitinase (Kumar et al., 2018), or participating in cell wall biosynthesis or remodeling, such as pectin lyase and endo-1,3-beta-glucosidase (Cao, 2012; Houston et al., 2016). AtNHR2B also interacted with a putative polygalacturonase (Duan et al., 2016). Other AtNHR2A interactors localized to the extracellular space and functioning in cell wall biosynthesis included the extensin AtEXT2 (Liepman et al., 2010), and a proline-rich glycoprotein (Durufle et al., 2017). However, we did not find experimental evidence that these two proteins participate in plant immunity.
In addition to cell wall-related proteins localized to the extracellular space, AtNHR2A and AtNHR2B interacted with cytoplasmic proteins that also contribute to cell wall remodeling. AtNHR2A interacted with the thioglucoside glucohydrolase (AtTTG1), a myrosinase that functions in the hydrolysis of glucosinolates (Grubb and Abel, 2006) and the biosynthesis of callose (Clay et al., 2009). AtNHR2B interacted with Caffeoyl coenzyme A O-methyltransferase 1 (AtCCoAOMT1), an enzyme that participates in the biosynthesis of lignin, a component of the secondary plant cell wall (Liu et al., 2018). AtCCoAOMT1 has recently being implicated in plant immunity since this gene is induced by pathogens, and Atccoamt1 mutants are more susceptible to infections by P. syringae pv. tomato and Hyaloperonospora arabidopsidis (Yang Q. et al., 2017). The commonalities between AtNHR2B and AtCCoAOMT1 prompted us to validate this interaction in vivo by two independent methods. One method included transient co-expression of AtNHR2B-GFP and HA-AtCCoAOMT1 in N. benthamiana followed by co-immunoprecipitation, but using HA-AtCCoAOMT1 as bait to co-immunoprecipitate AtNHR2B-GFP in contrast to the original screen that used AtNHR2B-GFP as bait. We also validated this interaction in vivo by BiFC and demonstrated that AtNHR2B and AtCCoAOMT1 interacted in the cytoplasm and punctate bodies, which agrees with the expected localization of AtNHR2B. The validation of this interaction will pave the way to continue exploring how AtNHR2B and AtCCoAOMT1 work together during plant immunity.
Cell wall biosynthetic enzymes and antimicrobials are localized to the extracellular space because they are secreted proteins that transition through a secretory pathway involving the endomembrane system. Our findings that AtNHR2A and AtNHR2B interacted with extracellular proteins suggest either that AtNHR2A and/or AtNHR2B are also secreted proteins or that AtNHR2A and AtNHR2B are components of the secretory pathway. Although we do not have evidence yet for the secretion of AtNHR2A or AtNHR2B, our interactome data indicates that both AtNHR2A and AtNHR2B interacted with proteins associated with secretory processes. AtNHR2A interacted with the Prenylated Rab acceptor 1 (AtPRA1.A1) and with AtISTL10, two proteins that participate in membrane trafficking events through distinct mechanisms (Alvim Kamei et al., 2008; Buono et al., 2016). In addition, both AtNHR2A and AtNHR2B interacted with the coat protein complex I (COPI), a complex of 7 proteins (α, β.β’, γ,δ,ε,ζ) that transports cargo from the Golgi apparatus to the ER (Brandizzi and Barlowe, 2013). Interestingly, AtNHR2A interacted with the β subunit while AtNHR2B interacted with the α subunit, and the interaction between the α and β subunits was also uncovered with the interactome network. These results combined with our previous live-imaging data showing localization of AtNHR2A and AtNHR2B to small punctae resembling bodies of the endomembrane system (Singh et al., 2018) strongly suggests that AtNHR2A and AtNHR2B are involved in endomembrane trafficking related events and supports our previous hypothesis, based on genetic data, that AtNHR2A and AtNHR2B work together (Singh et al., 2018).
We also identified as AtNHR2A and AtNHR2B interactors, other proteins known to participate in plant immunity at multiple levels including pathogen recognition and regulation of immune processes. Regarding pathogen recognition, we found that AtNHR2A interacted with the lectins AtLecRK-S.5 and AtGAL2 (Hwang and Hwang, 2011; Lannoo and Van Damme, 2014) and three resistance genes: AtRPS5 (Qi et al., 2012), an NB-ARC type disease resistant protein (van Ooijen et al., 2008), and BRS650, a LRR-type receptor protein kinase functioning in brassinosteroid signaling during plant immunity (Xu et al., 2014). AtNHR2B also interacted with AtTN10, a type of nucleotide binding-leucine rich repeat (NLR) recognizing pathogen effectors (Nandety et al., 2013).
With respect to the interaction of AtNHR2A and AtNHR2B with proteins known to be important in regulation of plant immunity, we found that AtNHR2A interacted with AtPRL1, a protein that interacts with the plant defense transcription factor AtCDC5 (Palma et al., 2007), with the MAP kinase AtMPK11 (Carrasco et al., 2014), and with AtENGD-1 (Cheung et al., 2010). AtENGD-1 is a GTP-binding protein that participates in plant immunity regulating the production of ROS and interacting with the immunophilin AtCYP19, which is also important for plant defense responses against P. syringae (Cheung et al., 2010; Pogorelko et al., 2014; Abdeeva et al., 2018). We successfully validated the interaction between AtNHR2A and AtENGD-1 by semi- in vivo co-immunoprecipitation wherein His-AtENGD-1 purified from bacteria was successfully co-immunoprecipitated by AtNHR2A-GFP expressed and purified from N. benthamiana. AtNHR2B also interacted with proteins known to play regulatory roles in plant immunity, specifically the ATL-RING-H2-E3 ubiquitin ligase (Liu et al., 2008) and AtRPN1, a component of the 26S proteasome subunit (Brukhin et al., 2005). AtRPN1 is required for resistance against fungal and bacterial pathogens (Yao et al., 2012). We also validated the interaction between AtNHR2B and AtRPN1 by using Myc-AtRPN1 as a bait to co-immunoprecipitate AtNHR2B-GFP. The validations of two of these interactions between AtNHR2A or AtNHR2B and proteins known to participate in plant immunity not only supports the quality of the data obtained by IP-MS, but also provided further insight into the multiple functions of AtNHR2A and AtNHR2B in plant immunity.
Because AtNHR2A and AtNHR2B were initially identified as important components of plant immunity, the main focus of this research was to unravel immune-related processes in which these two proteins participate. Indeed, this study found that AtNHR2A and AtNHR2B interact with a significant number of proteins known to participate in plant immunity, and, remarkably, cover a wide range of processes encompassing pathogen recognition, regulation of immune responses, and deployment of defense molecules with antimicrobial properties or functioning in the strengthening of the plant cell wall. Although careful validation of other interactions is needed, we are confident that this study provided a solid ground to generate testable hypotheses that will enable us to further dissect the complexity of plant immunity. Intriguingly, this study also found several other proteins that do not appear to function in plant immunity, highlighting the dynamic and versatile nature of AtNHR2A and AtNHR2B, possibly working as hubs integrating plant immunity with other biological processes (Vandereyken et al., 2018).
The datasets generated for this study can be found in the PRIDE archive using accession number PXD015894.
RS, JL, and CR designed the study. RS and CR wrote the manuscript. RS and RL performed experiments, collected and analyzed data. CG and AP analyzed data. All authors read and approved the manuscript.
The LTQ-Velos Pro Orbitrap LC-MS/MS used in this manuscript was obtained by funding of the National Science Foundation (Grant No. DBI-0922879) to the Donald Danforth Plant Science Center. This publication was supported by the National Science Foundation CAREER award number 1842970 to CR and Grant Number P30 GM103450 from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) to JL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Elison Blancaflor for the 35S-GFP seeds, Drs. Shin-Cheng Tzeng and Bradley Evans at the Proteomics and Mass Spectrometry Facility at the Donald Danforth Plant Science Center for conducting mass spectrometry analysis, and Perla Rocha-Loyola for proofreading the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00232/full#supplementary-material
Abdeeva, I. A., Pogorelko, G. V., Maloshenok, L. G., Mokrykova, M. V., Fursova, O. V., and Bruskin, S. A. (2018). Search for partner proteins of A. thaliana immunophilins involved in the control of plant immunity. Molecules 23:953. doi: 10.3390/molecules23040953
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., and Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Alvarez, S., Roy Choudhury, S., Hicks, L. M., and Pandey, S. (2013). Quantitative proteomics-based analysis supports a significant role of GTG proteins in regulation of ABA response in Arabidopsis roots. J. Proteome Res. 12, 1487–1501. doi: 10.1021/pr301159u
Alvarez, S., Roy Choudhury, S., and Pandey, S. (2014). Comparative quantitative proteomics analysis of the ABA response of roots of drought-sensitive and drought-tolerant wheat varieties identifies proteomic signatures of drought adaptability. J. Proteome Res. 13, 1688–1701. doi: 10.1021/pr401165b
Alvim Kamei, C. L., Boruc, J., Vandepoele, K., Van den Daele, H., Maes, S., Russinova, E., et al. (2008). The PRA1 gene family in Arabidopsis. Plant Physiol. 147, 1735–1749. doi: 10.1104/pp.108.122226
Baldwin, A., Wardle, A., Patel, R., Dudley, P., Park, S. K., Twell, D., et al. (2005). A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol. 138, 715–733. doi: 10.1104/pp.105.063289
Batelli, G., Massarelli, I., Van Oosten, M., Nurcato, R., Vannini, C., Raimondi, G., et al. (2012). Asg1 is a stress-inducible gene which increases stomatal resistance in salt stressed potato. J. Plant Physiol. 169, 1849–1857. doi: 10.1016/j.jplph.2012.07.004
Bell, E., Creelman, R. A., and Mullet, J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92, 8675–8679. doi: 10.1073/pnas.92.19.8675
Bock, R., and Kudla, J. (2016). Lighting the way to protein-protein interactions: recommendations on best practices for bimolecular fluorescence complementation (BiFC) analyses. Plant Cell 28, 1002–1008. doi: 10.1105/tpc.16.00043
Borderies, G., Jamet, E., Lafitte, C., Rossignol, M., Jauneau, A., Boudart, G., et al. (2003). Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis 24, 3421–3432. doi: 10.1002/elps.200305608
Brandizzi, F., and Barlowe, C. (2013). Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell. Biol. 14, 382–392. doi: 10.1038/nrm3588
Brukhin, V., Gheyselinck, J., Gagliardini, V., Genschik, P., and Grossniklaus, U. (2005). The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17, 2723–2737. doi: 10.1105/tpc.105.034975
Buono, R. A., Paez-Valencia, J., Miller, N. D., Goodman, K., Spitzer, C., Spalding, E. P., et al. (2016). Role of SKD1 regulators LIP5 and IST1-LIKE1 in endosomal sorting and plant development. Plant Physiol. 171, 251–264. doi: 10.1104/pp.16.00240
Cain, P., Hall, M., Schroder, W. P., Kieselbach, T., and Robinson, C. (2009). A novel extended family of stromal thioredoxins. Plant Mol. Biol. 70, 273–281. doi: 10.1007/s11103-009-9471-4
Cao, J. (2012). The pectin lyases in Arabidopsis thaliana: evolution, selection and expression profiles. PLoS One 7:e46944. doi: 10.1371/journal.pone.0046944
Carrasco, J. L., Castello, M. J., Naumann, K., Lassowskat, I., Navarrete-Gomez, M., Scheel, D., et al. (2014). Arabidopsis protein phosphatase DBP1 nucleates a protein network with a role in regulating plant defense. PLoS One 9:e90734. doi: 10.1371/journal.pone.0090734
Carretero-Paulet, L., Ahumada, I., Cunillera, N., Rodriguez-Concepcion, M., Ferrer, A., Boronat, A., et al. (2002). Expression and molecular analysis of the Arabidopsis DXR gene encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase, the first committed enzyme of the 2-C-methyl-D-erythritol 4-phosphate pathway. Plant Physiol. 129, 1581–1591. doi: 10.1104/pp.003798
Carroll, A. J. (2013). The Arabidopsis cytosolic ribosomal proteome: from form to function. Front. Plant Sci. 4:32. doi: 10.3389/fpls.2013.00032
Chen, E. I., Cociorva, D., Norris, J. L., and Yates, J. R. III (2007). Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res. 6, 2529–2538. doi: 10.1021/pr060682a
Chen, X., Snyder, C. L., Truksa, M., Shah, S., and Weselake, R. J. (2011). sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signal. Behav. 6, 1695–1699. doi: 10.4161/psb.6.11.17777
Cheung, M. Y., Xue, Y., Zhou, L., Li, M. W., Sun, S. S., and Lam, H. M. (2010). An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein. J. Biol. Chem. 285, 37359–37369. doi: 10.1074/jbc.M110.172080
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Cohen, S., Zmudjak, M., Colas des Francs-Small, C., Malik, S., Shaya, F., Keren, I., et al. (2014). nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78, 253–268. doi: 10.1111/tpj.12466
de Bianchi, S., Betterle, N., Kouril, R., Cazzaniga, S., Boekema, E., Bassi, R., et al. (2011). Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23, 2659–2679. doi: 10.1105/tpc.111.087320
Duan, W., Huang, Z., Song, X., Liu, T., Liu, H., Hou, X., et al. (2016). Comprehensive analysis of the polygalacturonase and pectin methylesterase genes in Brassica rapa shed light on their different evolutionary patterns. Sci. Rep. 6:25107. doi: 10.1038/srep25107
Dunham, W. H., Mullin, M., and Gingras, A. C. (2012). Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics 12, 1576–1590. doi: 10.1002/pmic.201100523
Durufle, H., Herve, V., Balliau, T., Zivy, M., Dunand, C., and Jamet, E. (2017). Proline hydroxylation in cell wall proteins: is it yet possible to define rules? Front. Plant Sci. 8:1802. doi: 10.3389/fpls.2017.01802
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313x.2005.02617.x
Elvira-Matelot, E., Bardou, F., Ariel, F., Jauvion, V., Bouteiller, N., Le Masson, I., et al. (2016). The nuclear ribonucleoprotein SmD1 interplays with splicing, RNA quality control, and posttranscriptional gene silencing in Arabidopsis. Plant Cell 28, 426–438. doi: 10.1105/tpc.15.01045
Eschen-Lippold, L., Jiang, X., Elmore, J. M., Mackey, D., Shan, L., Coaker, G., et al. (2016). Bacterial AvrRpt2-Like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 171, 2223–2238. doi: 10.1104/pp.16.00336
Feist, P., and Hummon, A. B. (2015). Proteomic challenges: sample preparation techniques for microgram-quantity protein analysis from biological samples. Int. J. Mol. Sci. 16, 3537–3563. doi: 10.3390/ijms16023537
Figueroa, P., Leon, G., Elorza, A., Holuigue, L., Araya, A., and Jordana, X. (2002). The four subunits of mitochondrial respiratory complex II are encoded by multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol. Biol. 50, 725–734.
Flugge, U. I., Hausler, R. E., Ludewig, F., and Gierth, M. (2011). The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 62, 2381–2392. doi: 10.1093/jxb/erq361
Frei dit Frey, N., Muller, P., Jammes, F., Kizis, D., Leung, J., Perrot-Rechenmann, C., et al. (2010). The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22, 1575–1591. doi: 10.1105/tpc.109.070680
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Fu, H., Doelling, J. H., Rubin, D. M., and Vierstra, R. D. (1999). Structural and functional analysis of the six regulatory particle triple-A ATPase subunits from the Arabidopsis 26S proteasome. Plant J. 18, 529–539. doi: 10.1046/j.1365-313x.1999.00479.x
Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2001). Leucine and its keto acid enhance the coordinated expression of genes for branched-chain amino acid catabolism in Arabidopsis under sugar starvation. FEBS Lett. 499, 161–165. doi: 10.1016/s0014-5793(01)02536-4
Ghahremani, M., Tran, H., Biglou, S. G., O’Gallagher, B., She, Y. M., and Plaxton, W. C. (2018). A glycoform of the secreted purple acid phosphatase AtPAP26 co-purifies with a mannose-binding lectin (AtGAL1) upregulated by phosphate-starved Arabidopsis. Plant Cell Environ. 42, 1139–1157. doi: 10.1111/pce.13432
Gingras, A. C., Gstaiger, M., Raught, B., and Aebersold, R. (2007). Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell. Biol. 8, 645–654. doi: 10.1038/nrm2208
Gong, Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B., and Zhu, J. K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. U.S.A. 99, 11507–11512. doi: 10.1073/pnas.172399299
Green, B. R., Pichersky, E., and Kloppstech, K. (1991). Chlorophyll a/b-binding proteins: an extended family. Trends Biochem. Sci. 16, 181–186. doi: 10.1016/0968-0004(91)90072-4
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Guirimand, G., Guihur, A., Phillips, M. A., Oudin, A., Glevarec, G., Mahroug, S., et al. (2012). Triple subcellular targeting of isopentenyl diphosphate isomerases encoded by a single gene. Plant Signal. Behav. 7, 1495–1497. doi: 10.4161/psb.21892
Hanke, G., and Mulo, P. (2013). Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 36, 1071–1084. doi: 10.1111/pce.12046
Heath, M. C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. doi: 10.1016/s1369-5266(00)00087-x
Hedtke, B., Borner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. doi: 10.1093/embo-reports/kvd086
Holscher, C., Lutterbey, M. C., Lansing, H., Meyer, T., Fischer, K., and von Schaewen, A. (2016). Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in Arabidopsis thaliana. Plant Physiol. 171, 192–205. doi: 10.1104/pp.15.01301
Horling, F., Koning, J., and Dietz, K. J. (2002). Type II peroxiredoxin C, a member of the peroxiredoxin family of Arabidopsis thaliana: its expression and activity in comparison with other peroxiredoxins. Plant Physiol. Biochem. 40, 491–499. doi: 10.1016/s0981-9428(02)01396-7
Houston, K., Tucker, M. R., Chowdhury, J., Shirley, N., and Little, A. (2016). The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7:984. doi: 10.3389/fpls.2016.00984
Howles, P. A., Birch, R. J., Collings, D. A., Gebbie, L. K., Hurley, U. A., Hocart, C. H., et al. (2006). A mutation in an Arabidopsis ribose 5-phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. 48, 606–618. doi: 10.1111/j.1365-313X.2006.02902.x
Huang, H., Alvarez, S., Bindbeutel, R., Shen, Z., Naldrett, M. J., Evans, B. S., et al. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15, 201–217. doi: 10.1074/mcp.M115.054064
Hudig, M., Maier, A., Scherrers, I., Seidel, L., Jansen, E. E., Mettler-Altmann, T., et al. (2015). Plants possess a cyclic mitochondrial metabolic pathway similar to the mammalian metabolic repair mechanism involving malate dehydrogenase and l-2-hydroxyglutarate dehydrogenase. Plant Cell Physiol. 56, 1820–1830. doi: 10.1093/pcp/pcv108
Hwang, I. S., and Hwang, B. K. (2011). The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 155, 447–463. doi: 10.1104/pp.110.164848
Ihnatowicz, A., Pesaresi, P., and Leister, D. (2007). The E subunit of photosystem I is not essential for linear electron flow and photoautotrophic growth in Arabidopsis thaliana. Planta 226, 889–895. doi: 10.1007/s00425-007-0534-y
Ishiga, Y., Ishiga, T., Uppalapati, S. R., and Mysore, K. S. (2011). Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7:32. doi: 10.1186/1746-4811-7-32
Ishiguro, S., Watanabe, Y., Ito, N., Nonaka, H., Takeda, N., Sakai, T., et al. (2002). SHEPHERD is the Arabidopsis GRP94 responsible for the formation of functional CLAVATA proteins. EMBO J. 21, 898–908. doi: 10.1093/emboj/21.5.898
Jancowski, S., Catching, A., Pighin, J., Kudo, T., Foissner, I., and Wasteneys, G. O. (2014). Trafficking of the myrosinase-associated protein GLL23 requires NUC/MVP1/GOLD36/ERMO3 and the p24 protein CYB. Plant J. 77, 497–510. doi: 10.1111/tpj.12408
Junkova, P., Danek, M., Kocourkova, D., Brouzdova, J., Kroumanova, K., Zelazny, E., et al. (2018). Mapping of plasma membrane proteins interacting with Arabidopsis thaliana flotillin 2. Front. Plant Sci. 9:991. doi: 10.3389/fpls.2018.00991
Karash, S., Liyanage, R., Qassab, A., Lay, J. O. Jr., and Kwon, Y. M. (2017). A comprehensive assessment of the genetic determinants in Salmonella typhimurium for resistance to hydrogen peroxide using proteogenomics. Sci. Rep. 7:17073. doi: 10.1038/s41598-017-17149-9
Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392. doi: 10.1021/ac025747h
Keren, I., Bezawork-Geleta, A., Kolton, M., Maayan, I., Belausov, E., Levy, M., et al. (2009). AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. doi: 10.1261/rna.1776409
Kim, J. S., Park, S. J., Kwak, K. J., Kim, Y. O., Kim, J. Y., Song, J., et al. (2007). Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35, 506–516. doi: 10.1093/nar/gkl1076
Kim, S., Schlicke, H., Van Ree, K., Karvonen, K., Subramaniam, A., Richter, A., et al. (2013). Arabidopsis chlorophyll biosynthesis: an essential balance between the methylerythritol phosphate and tetrapyrrole pathways. Plant Cell 25, 4984–4993. doi: 10.1105/tpc.113.119172
Kinoshita, H., Nagasaki, J., Yoshikawa, N., Yamamoto, A., Takito, S., Kawasaki, M., et al. (2011). The chloroplastic 2-oxoglutarate/malate transporter has dual function as the malate valve and in carbon/nitrogen metabolism. Plant J. 65, 15–26. doi: 10.1111/j.1365-313X.2010.04397.x
Kleffmann, T., Russenberger, D., von Zychlinski, A., Christopher, W., Sjolander, K., Gruissem, W., et al. (2004). The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362. doi: 10.1016/j.cub.2004.02.039
Koussevitzky, S., Stanne, T. M., Peto, C. A., Giap, T., Sjogren, L. L., Zhao, Y., et al. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63, 85–96. doi: 10.1007/s11103-006-9074-2
Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H. P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. doi: 10.1104/pp.010474
Kumar, M., Brar, A., Yadav, M., Chawade, A., Vivekanand, V., and Pareek, N. (2018). Chitinases_ Potential candidates for enhanced plant resistance towards fungal pathogens. Agriculture 8, 1–12.
Langlois-Meurinne, M., Gachon, C. M., and Saindrenan, P. (2005). Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 139, 1890–1901. doi: 10.1104/pp.105.067223
Lannoo, N., and Van Damme, E. J. (2014). Lectin domains at the frontiers of plant defense. Front. Plant Sci. 5:397. doi: 10.3389/fpls.2014.00397
Lee, S., Doxey, A. C., McConkey, B. J., and Moffatt, B. A. (2012). Nuclear targeting of methyl-recycling enzymes in Arabidopsis thaliana is mediated by specific protein interactions. Mol. Plant 5, 231–248. doi: 10.1093/mp/ssr083
Lee, S., Whitaker, V. M., and Hutton, S. F. (2016). Mini review: potential applications of non-host resistance for crop improvement. Front. Plant Sci. 7:997. doi: 10.3389/fpls.2016.00997
Li, L., He, Y., Wang, Y., Zhao, S., Chen, X., Ye, T., et al. (2015). Arabidopsis PLC2 is involved in auxin-modulated reproductive development. Plant J. 84, 504–515. doi: 10.1111/tpj.13016
Liepman, A. H., Wightman, R., Geshi, N., Turner, S. R., and Scheller, H. V. (2010). Arabidopsis - a powerful model system for plant cell wall research. Plant J. 61, 1107–1121. doi: 10.1111/j.1365-313X.2010.04161.x
Lin, J. F., and Wu, S. H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628. doi: 10.1111/j.1365-313X.2004.02160.x
Lister, R., Chew, O., Lee, M. N., Heazlewood, J. L., Clifton, R., Parker, K. L., et al. (2004). A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol. 134, 777–789. doi: 10.1104/pp.103.033910
Liu, H., Zhang, H., Yang, Y., Li, G., Yang, Y., Wang, X., et al. (2008). Functional analysis reveals pleiotropic effects of rice RING-H2 finger protein gene OsBIRF1 on regulation of growth and defense responses against abiotic and biotic stresses. Plant Mol. Biol. 68, 17–30. doi: 10.1007/s11103-008-9349-x
Liu, Q., Luo, L., and Zheng, L. (2018). Lignins: biosynthesis and biological functions in plants. Int. J. Mol. Sci. 19:E335. doi: 10.3390/ijms19020335
Lubkowitz, M. (2011). The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol. Plant 4, 407–415. doi: 10.1093/mp/ssr004
Manara, A., DalCorso, G., Guzzo, F., and Furini, A. (2015). Loss of the atypical kinases ABC1K7 and ABC1K8 changes the lipid composition of the chloroplast membrane. Plant Cell Physiol. 56, 1193–1204. doi: 10.1093/pcp/pcv046
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., and Goodin, M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
Meyer, E. H., Taylor, N. L., and Millar, A. H. (2008). Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J. Proteome Res. 7, 786–794. doi: 10.1021/pr700595p
Millan-Zambrano, G., and Chavez, S. (2014). Nuclear functions of prefoldin. Open Biol. 4:140085. doi: 10.1098/rsob.140085
Millar, A. H., and Heazlewood, J. L. (2003). Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131, 443–453. doi: 10.1104/pp.009985
Minami, A., Fujiwara, M., Furuto, A., Fukao, Y., Yamashita, T., Kamo, M., et al. (2009). Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 50, 341–359. doi: 10.1093/pcp/pcn202
Mizutani, M. (2012). Impacts of diversification of cytochrome P450 on plant metabolism. Biol. Pharm. Bull. 35, 824–832. doi: 10.1248/bpb.35.824
Molesini, B., Pandolfini, T., Pii, Y., Korte, A., and Spena, A. (2012). Arabidopsis thaliana AUCSIA-1 regulates auxin biology and physically interacts with a kinesin-related protein. PLoS One 7:e41327. doi: 10.1371/journal.pone.0041327
Monne, M., Daddabbo, L., Gagneul, D., Obata, T., Hielscher, B., Palmieri, L., et al. (2018). Uncoupling proteins 1 and 2 (UCP1 and UCP2) from Arabidopsis thaliana are mitochondrial transporters of aspartate, glutamate, and dicarboxylates. J. Biol. Chem. 293, 4213–4227. doi: 10.1074/jbc.RA117.000771
Morris, K., Thornber, S., Codrai, L., Richardson, C., Craig, A., Sadanandom, A., et al. (2010). DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 22, 1118–1128. doi: 10.1105/tpc.109.066605
Murcha, M. W., Kubiszewski-Jakubiak, S., Wang, Y., and Whelan, J. (2014). Evidence for interactions between the mitochondrial import apparatus and respiratory chain complexes via Tim21-like proteins in Arabidopsis. Front. Plant Sci. 5:82. doi: 10.3389/fpls.2014.00082
Nandety, R. S., Caplan, J. L., Cavanaugh, K., Perroud, B., Wroblewski, T., Michelmore, R. W., et al. (2013). The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 162, 1459–1472. doi: 10.1104/pp.113.219162
Ndimba, B. K., Chivasa, S., Hamilton, J. M., Simon, W. J., and Slabas, A. R. (2003). Proteomic analysis of changes in the extracellular matrix of Arabidopsis cell suspension cultures induced by fungal elicitors. Proteomics 3, 1047–1059. doi: 10.1002/pmic.200300413
Otterhag, L., Sommarin, M., and Pical, C. (2001). N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 497, 165–170. doi: 10.1016/s0014-5793(01)02453-x
Palma, K., Zhao, Q., Cheng, Y. T., Bi, D., Monaghan, J., Cheng, W., et al. (2007). Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21, 1484–1493. doi: 10.1101/gad.1559607
Peltier, J. B., Emanuelsson, O., Kalume, D. E., Ytterberg, J., Friso, G., Rudella, A., et al. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236. doi: 10.1105/tpc.010304
Pogorelko, G. V., Mokryakova, M., Fursova, O. V., Abdeeva, I., Piruzian, E. S., and Bruskin, S. A. (2014). Characterization of three Arabidopsis thaliana immunophilin genes involved in the plant defense response against Pseudomonas syringae. Gene 538, 12–22. doi: 10.1016/j.gene.2014.01.029
Qi, D., DeYoung, B. J., and Innes, R. W. (2012). Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158, 1819–1832. doi: 10.1104/pp.112.194035
Raczynska, K. D., Stepien, A., Kierzkowski, D., Kalak, M., Bajczyk, M., McNicol, J., et al. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42, 1224–1244. doi: 10.1093/nar/gkt894
Rath, N. C., Gupta, A., Liyanage, R., and Lay, J. O. Jr. (2019). Phorbol 12-myristate 13-acetate-induced changes in chicken enterocytes. Proteomics Insights 10:1178641819840369. doi: 10.1177/1178641819840369
Schechter, M. A., Hsieh, M. K., Njoroge, L. W., Thompson, J. W., Soderblom, E. J., Feger, B. J., et al. (2014). Phosphoproteomic profiling of human myocardial tissues distinguishes ischemic from non-ischemic end stage heart failure. PLoS One 9:e104157. doi: 10.1371/journal.pone.0104157
Senthil-Kumar, M., and Mysore, K. S. (2013). Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu. Rev. Phytopathol. 51, 407–427. doi: 10.1146/annurev-phyto-082712-102319
Serrano, I., Audran, C., and Rivas, S. (2016). Chloroplasts at work during plant innate immunity. J. Exp. Bot. 67, 3845–3854. doi: 10.1093/jxb/erw088
Singh, R., Lee, S., Ortega, L., Ramu, V. S., Senthil-Kumar, M., Blancaflor, E. B., et al. (2018). Two chloroplast-localized proteins: AtNHR2A and AtNHR2B, contribute to Callose deposition during Nonhost disease resistance in Arabidopsis. Mol. Plant Microbe Interact. 31, 1280–1290. doi: 10.1094/MPMI-04-18-0094-R
Singh, R., and Rojas, C. M. (2018). Dissecting the functional domains of the Arabidopsis thaliana nonhost resistance 2B (AtNHR2B) protein. Plant Signal. Behav. 13:e1530024. doi: 10.1080/15592324.2018.1530024
Skalitzky, C. A., Martin, J. R., Harwood, J. H., Beirne, J. J., Adamczyk, B. J., Heck, G. R., et al. (2011). Plastids contain a second sec translocase system with essential functions. Plant Physiol. 155, 354–369. doi: 10.1104/pp.110.166546
Song, H., Dong, X., Yi, H., Ahn, J. Y., Yun, K., Song, M., et al. (2018). Genome-wide identification and characterization of warming-related genes in Brassica rapa ssp. pekinensis. Int. J. Mol. Sci. 19:E1727. doi: 10.3390/ijms19061727
Struk, S., Jacobs, A., Sanchez Martin-Fontecha, E., Gevaert, K., Cubas, P., and Goormachtig, S. (2018). Exploring the protein-protein interaction landscape in plants. Plant Cell Environ. 42, 387–409. doi: 10.1111/pce.13433
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Teh, O. K., Hatsugai, N., Tamura, K., Fuji, K., Tabata, R., Yamaguchi, K., et al. (2015). BEACH-domain proteins act together in a cascade to mediate vacuolar protein trafficking and disease resistance in Arabidopsis. Mol. Plant 8, 389–398. doi: 10.1016/j.molp.2014.11.015
van Ooijen, G., Mayr, G., Kasiem, M. M., Albrecht, M., Cornelissen, B. J., and Takken, F. L. (2008). Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 59, 1383–1397. doi: 10.1093/jxb/ern045
Vandereyken, K., Van Leene, J., De Coninck, B., and Cammue, B. P. A. (2018). Hub protein controversy: taking a closer look at plant stress response hubs. Front. Plant Sci. 9:694. doi: 10.3389/fpls.2018.00694
Vitlin Gruber, A., and Feiz, L. (2018). Rubisco assembly in the chloroplast. Front. Mol. Biosci. 5:24. doi: 10.3389/fmolb.2018.00024
Wendrich, J. R., Boeren, S., Moller, B. K., Weijers, D., and De Rybel, B. (2017). In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol. Biol. 1497, 147–158. doi: 10.1007/978-1-4939-6469-7_14
Wilkie, S. E., Lambert, R., and Warren, M. J. (1996). Chloroplastic aspartate aminotransferase from Arabidopsis thaliana: an examination of the relationship between the structure of the gene and the spatial structure of the protein. Biochem. J. 319(Pt. 3), 969–976. doi: 10.1042/bj3190969
Xie, M., Zhang, J., Tschaplinski, T. J., Tuskan, G. A., Chen, J. G., and Muchero, W. (2018). Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 9:1427. doi: 10.3389/fpls.2018.01427
Xu, P., Xu, S. L., Li, Z. J., Tang, W., Burlingame, A. L., and Wang, Z. Y. (2014). A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol. Plant 7, 441–444. doi: 10.1093/mp/sst105
Yang, H., Li, P., Zhang, A., Wen, X., Zhang, L., and Lu, C. (2017). Tetratricopeptide repeat protein Pyg7 is essential for photosystem I assembly by interacting with PsaC in Arabidopsis. Plant J. 91, 950–961. doi: 10.1111/tpj.13618
Yang, Q., He, Y., Kabahuma, M., Chaya, T., Kelly, A., Borrego, E., et al. (2017). A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–1372. doi: 10.1038/ng.3919
Yang, Y., and Karlson, D. (2013). AtCSP1 regulates germination timing promoted by low temperature. FEBS Lett. 587, 2186–2192. doi: 10.1016/j.febslet.2013.05.039
Yang, Y., Qin, Y., Xie, C., Zhao, F., Zhao, J., Liu, D., et al. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22, 1313–1332. doi: 10.1105/tpc.109.069609
Yao, C., Wu, Y., Nie, H., and Tang, D. (2012). RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J. 71, 1015–1028. doi: 10.1111/j.1365-313X.2012.05048.x
Yoon, G. M., and Kieber, J. J. (2013). 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell 25, 1016–1028. doi: 10.1105/tpc.113.110106
Zeeman, S. C., Thorneycroft, D., Schupp, N., Chapple, A., Weck, M., Dunstan, H., et al. (2004). Plastidial alpha-glucan phosphorylase is not required for starch degradation in Arabidopsis leaves but has a role in the tolerance of abiotic stress. Plant Physiol. 135, 849–858. doi: 10.1104/pp.103.032631
Zhang, T., Schneider, J. D., Lin, C., Geng, S., Ma, T., Lawrence, S. R., et al. (2019). MPK4 phosphorylation dynamics and interacting proteins in plant immunity. J. Proteome Res. 18, 826–840. doi: 10.1021/acs.jproteome.8b00345
Zhang, Z., Li, Q., Li, Z., Staswick, P. E., Wang, M., Zhu, Y., et al. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145, 450–464. doi: 10.1104/pp.107.106021
Zhao, Y., Wang, J., Liu, Y., Miao, H., Cai, C., Shao, Z., et al. (2015). Classic myrosinase-dependent degradation of indole glucosinolate attenuates fumonisin B1-induced programmed cell death in Arabidopsis. Plant J. 81, 920–933. doi: 10.1111/tpj.12778
Zoeller, M., Stingl, N., Krischke, M., Fekete, A., Waller, F., Berger, S., et al. (2012). Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378. doi: 10.1104/pp.112.202846
Keywords: nonhost resistance, co-immunoprecipitation, mass spectrometry, protein–protein interactions, interactome network
Citation: Singh R, Liyanage R, Gupta C, Lay JO Jr, Pereira A and Rojas CM (2020) The Arabidopsis Proteins AtNHR2A and AtNHR2B Are Multi-Functional Proteins Integrating Plant Immunity With Other Biological Processes. Front. Plant Sci. 11:232. doi: 10.3389/fpls.2020.00232
Received: 18 October 2019; Accepted: 14 February 2020;
Published: 04 March 2020.
Edited by:
Norbert Rolland, UMR 5168 Laboratoire de Physiologie Cellulaire Vegetale (LPCV), FranceReviewed by:
Tong Zhang, Pacific Northwest National Laboratory (DOE), United StatesCopyright © 2020 Singh, Liyanage, Gupta, Lay, Pereira and Rojas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clemencia M. Rojas, Y3IwMjJAdWFyay5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.