- Department of Applied Genetics and Cell Biology, BOKU—University of Natural Resources and Life Sciences, Vienna, Austria
Plants are exposed to a variety of abiotic and biotic stresses that may result in DNA damage. Endogenous processes - such as DNA replication, DNA recombination, respiration, or photosynthesis - are also a threat to DNA integrity. It is therefore essential to understand the strategies plants have developed for DNA damage detection, signaling, and repair. Alternative splicing (AS) is a key post-transcriptional process with a role in regulation of gene expression. Recent studies demonstrate that the majority of intron-containing genes in plants are alternatively spliced, highlighting the importance of AS in plant development and stress response. Not only does AS ensure a versatile proteome and influence the abundance and availability of proteins greatly, it has also emerged as an important player in the DNA damage response (DDR) in animals. Despite extensive studies of DDR carried out in plants, its regulation at the level of AS has not been comprehensively addressed. Here, we provide some insights into the interplay between AS and DDR in plants.
DNA Damage Response in Plants
The genomic integrity of living cells is perpetually challenged by a variety of environmental and internal cellular factors. Environmental stresses, such as drought, salinity, ultraviolet (UV), ionizing radiation, xenobiotic toxicity, heavy metals, and mutagenic chemicals damage DNA and affect its stability (Hu et al., 2016; Nisa et al., 2019). Cellular replication, recombination errors, and reactive oxygen species resulting as a byproduct of metabolism also cause DNA damage. A cell's reaction to genotoxic stress, referred to as DNA damage response (DDR), starts with cell cycle arrest and, in the case of plants, endoreplication (De Veylder et al., 2011). To ensure the repair of a variety of different types of DNA lesions, several DNA repair mechanisms are active and constitute the DNA repair phase of DDR. Should the repair of DNA damage not be sufficient, programmed cell death eliminates the damaged cell and ensures homeostasis (Manova and Gruszka, 2015; Kim et al., 2019). Due to their sessile nature, plants find themselves at increased risk to detrimental environmental factors. It has also been shown that light and temperature conditions affect DNA repair mechanisms such as homologous recombination and photoreactivation (Li et al., 2002; Boyko et al., 2005).
The repair of UV-induced lesions by photoreactivation appears to be an ancient conserved DNA damage repair mechanism. It relies on the activity of photolyase, utilizing the energy of UV-A or blue light to reverse UV damage in the DNA (Manova and Gruszka, 2015; Kavakli et al., 2017; Zhang et al., 2017a). Another mechanism of UV damage repair is nucleotide excision repair (NER), which identifies, removes, and repairs the damaged base(s) using the other DNA strands as a template. In addition to UV lesions, NER repairs bulky adducts that change the DNA conformation. Global genomic repair (GGR) and transcription-coupled repair (TCR), although differing in their mode of damage recognition, share similarities in their mechanisms of action (Hanawalt, 2002). The DNA glycosylases, which initiate base excision repair (BER) at damaged sites, facilitate the repair of a variety of DNA lesions (Wallace, 2014). There is evidence for BER being active in chloroplasts to counter the effects of reactive oxygen species production during photosynthesis (Gutman and Niyogi, 2009). The mismatch repair (MMR) pathway is responsible for the repair of replication errors, such as mismatches and indels, UV, and oxidative damage (Li et al., 2016; Liu et al., 2017; Belfield et al., 2018). Double-strand breaks (DSBs) are repaired via non-homologous end joining (NHEJ) and homologous recombination (HR). While HR requires homologous sequences to ensure efficient repair, NHEJ joins DSBs without considering sequence context and is, thus, an error prone mechanism, which can result in mutations and DNA changes (Manova and Gruszka, 2015).
Two protein kinases, ATM (ATAXIA-TELANGIECTASIA MUTATED) and ATR (ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED), initiate eukaryotic DDR. Once activated, they signal via checkpoint kinases 1 and 2 (CHK1 and CHK2), respectively. Human homologs of CHK1 and CHK2 activate p53, which in turn controls cell cycle arrest, DNA damage repair, and programmed cell death. While the downstream processes of ATM, ATR, and p53 have been studied extensively, data on their upstream activation and regulation remains scarce. Neither orthologs of CHK1 and CHK2, nor of p53, have been identified in plants so far. However, a functional homolog of p53, SUPPRESSOR OF GAMMA RESPONSE 1 (SOG1), transcriptionally regulating DDR downstream of ATM and ATR was found (Preuss and Britt, 2003; Yoshiyama et al., 2009; Yoshiyama, 2016). Indeed, SOG1 was identified as a master regulator transcription factor of the plant DDR, influencing expression of genes related to the cell cycle and DNA repair (Ogita et al., 2018). About 300 direct targets of SOG1 were identified, including transcription factors, DNA repair genes, and regulators of the cell cycle (Bourbousse et al., 2018).
A recent research update highlights the growing interest in DDR in plants but also serves to show that a role for alternative splicing (AS) remains to be established (Gimenez and Manzano-Agugliaro, 2017).
Overview of Alternative Splicing
Most messenger RNAs in higher eukaryotes are synthesized as precursors, which contain intervening sequences, known as introns. To provide a template for protein synthesis, messenger RNA (mRNA) introns have to be removed and exons joined in a process termed pre-mRNA splicing. However, exons and introns or their parts can be differentially included in mRNA by AS. AS produces transcript and protein variants from a single gene with different fates and functions, and is a fundamental aspect of RNA biology that has a key role in our understanding of gene expression regulation. Up to 95% of human and 70% of plant multi-exonic genes are alternatively spliced (Pan et al., 2008; Wang et al., 2008; Marquez et al., 2012; Chamala et al., 2015; Zhang et al., 2017b). Further studies report that about 50% of the genes in soybeans, 46% in rice, 40% in maize, and over 60% in tomatoes and barley undergo AS (Thatcher et al., 2014; Chamala et al., 2015; Clark et al., 2019; Rapazote-Flores et al., 2019), emphasizing its importance in crop plant development and environmental response. AS has a broad role in many aspects of plant biology, but its role in responding to DNA damage is mostly unknown and requires further investigation.
Pre-mRNA splicing requires the core splicing signals, which consist of the 5' and 3' splice sites and a branch site (Wang and Burge, 2008). However, multiple additional features, such as intronic and exonic splicing regulatory cis-elements (splicing enhancers and silencers), length of introns and exons, and differential guanine-cytosine content between exons and introns, affect the recognition and selection of the core splicing signals (Braunschweig et al., 2013). The secondary structure of the pre-mRNA can alter access to splicing signals and binding sites for splicing factors (SFs) or change the distance between these elements (Shepard and Hertel, 2008). Differential DNA methylation, histone modifications, and nucleosome positioning modulate RNA polymerase II elongation speed and recruitment of SFs, thus also resulting in alternative splice site selection [for a recent review see (Jabre et al., 2019)].
Common types of AS events include exon skipping, usage of alternative 5' and 3' splice sites, mutually exclusive exons, and intron retention. Exon skipping is the predominant event in animals, whereas it is infrequent in plants (Marquez et al., 2012; Braunschweig et al., 2013). Intron retention is widespread both in plants and animals (Marquez et al., 2012; Braunschweig et al., 2014). Interestingly, intron retention transcripts are often not substrates for nonsense-mediated mRNA decay due to their nuclear localization (James et al., 2012; Kalyna et al., 2012; Leviatan et al., 2013; Gohring et al., 2014). Retention of introns may regulate protein abundance during developmental transitions and in response to stress (including DNA damage). When transcripts with retained introns are recognized as incompletely processed they remain in the nucleus until a change in the cellular environment results in post-transcriptional splicing (Yap et al., 2012; Boothby et al., 2013; Boutz et al., 2015; Brown et al., 2015). Microexons (ultra-short exons of 3-30 nucleotides) found in hundreds of animal genes, and recently identified exitrons (alternatively spliced internal regions of protein-coding exons), which occur in ~7% of Arabidopsis and 4% of human protein-coding genes, complement the repertoire of AS events (Marquez et al., 2012; Irimia et al., 2014; Marquez et al., 2015; Staiger and Simpson, 2015; Sibley et al., 2016; Ustianenko et al., 2017; Zhang et al., 2017b).
Hundreds of proteins participate in the splicing process (Chen and Moore, 2015). However, the modulation of splice site recognition is mainly governed by two families of SFs - serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) - through binding to regulatory cis-elements in the pre-mRNA (Barta et al., 2010; Manley and Krainer, 2010; Yeap et al., 2014; Howard and Sanford, 2015). SR proteins and hnRNPs act as activators and repressors of splice site selection, respectively, however, the effect often depends on their binding position. Expression levels, localization, and post-translational modifications (PTMs) (phosphorylation, acetylation, ubiquitination, and sumoylation) of SFs in a particular cell are one of the components of the splicing code, which governs the AS outcomes (Barash et al., 2010; Baralle and Baralle, 2018). Interestingly, SR proteins and hnRNPs participate in multiple cellular processes, such as mRNA export, RNA stability and quality control, and translation.
Alternative Splicing and DNA Damage Response, Insights From Studies in Animals
It is becoming clear that RNA-binding proteins and AS are important in DDR. One of the first pieces of evidence that SFs may have a role in DDR came from a study which demonstrated that the depletion of a canonical human SR protein, SRSF1 (SF2/ASF), resulted in increased DSB formation and genome instability (Li and Manley, 2005). Several studies in animals have unexpectedly identified SFs and other RNA processing proteins associated with response to irradiation and DNA damaging chemicals. For example, genome-wide siRNA knockdown of multiple genes have shown that splicing and RNA processing factors are the most enriched functional category within factors whose depletion mediates DNA damage (Paulsen et al., 2009; Lackner et al., 2011). Studies of individual SFs, including SR proteins, have demonstrated changes in their expression levels, AS profiles, phosphorylation state, and subcellular distribution in response to DNA damage (Matsuoka et al., 2007; Busa et al., 2010; Sakashita and Endo, 2010; Ip et al., 2011; Adamson et al., 2012; Leva et al., 2012). The importance of AS and splicing factors in DDR in animals has been reviewed extensively (Naro et al., 2015; Shkreta and Chabot, 2015; Giono et al., 2016; Kai, 2016; Mikolaskova et al., 2018).
The interplay between DDR and AS occurs at multiple levels (Figure 1). One of the most rapid responses to stress and DNA damage is the change in activity of already translated proteins by PTMs. Multiple SFs have been identified in DDR-regulated phosphoproteomes (Bennetzen et al., 2010; Bensimon et al., 2010; Beli et al., 2012). The kinases ATM and ATR are directly activated by DNA lesions and phosphorylate hundreds of proteins in response to ionizing radiation, including several hnRNPs and SR proteins (Matsuoka et al., 2007). Studies using the treatment of mammalian cells with several genotoxic agents revealed reduced SR protein phosphorylation levels affecting their accumulation in nuclear granules. These studies also found differential AS of genes involved in DNA repair, cell cycle control, and apoptosis (Bennetzen et al., 2010; Leva et al., 2012; Shkreta et al., 2016). Remarkably, detained introns, a recently identified subgroup of retained introns, are enriched in genes involved in DDR. Moreover, DNA damage and the activity of certain Clk kinases, which maintain the hyperphosphorylated status of SR proteins, can modulate splicing of detained introns (Boutz et al., 2015). Changes in the activity of SR proteins also have been associated with their acetylation state in response to cisplatin-induced DNA damage (Edmond et al., 2011; Nakka et al., 2015). Interestingly, acetyltransferases can indirectly impact the translocation of SR proteins via the modification of SR protein kinases (Edmond et al., 2011). Recent studies also demonstrated the acetylation of hnRNPs in response to DNA damage (Magni et al., 2019; Siam et al., 2019). Ubiquitination, besides its regulatory activity during spliceosome assembly, affects SFs upon DNA damage (Lu and Legerski, 2007). Genotoxic agents cause deubiquitylation and sumoylation of hnRNPs (Vassileva and Matunis, 2004).
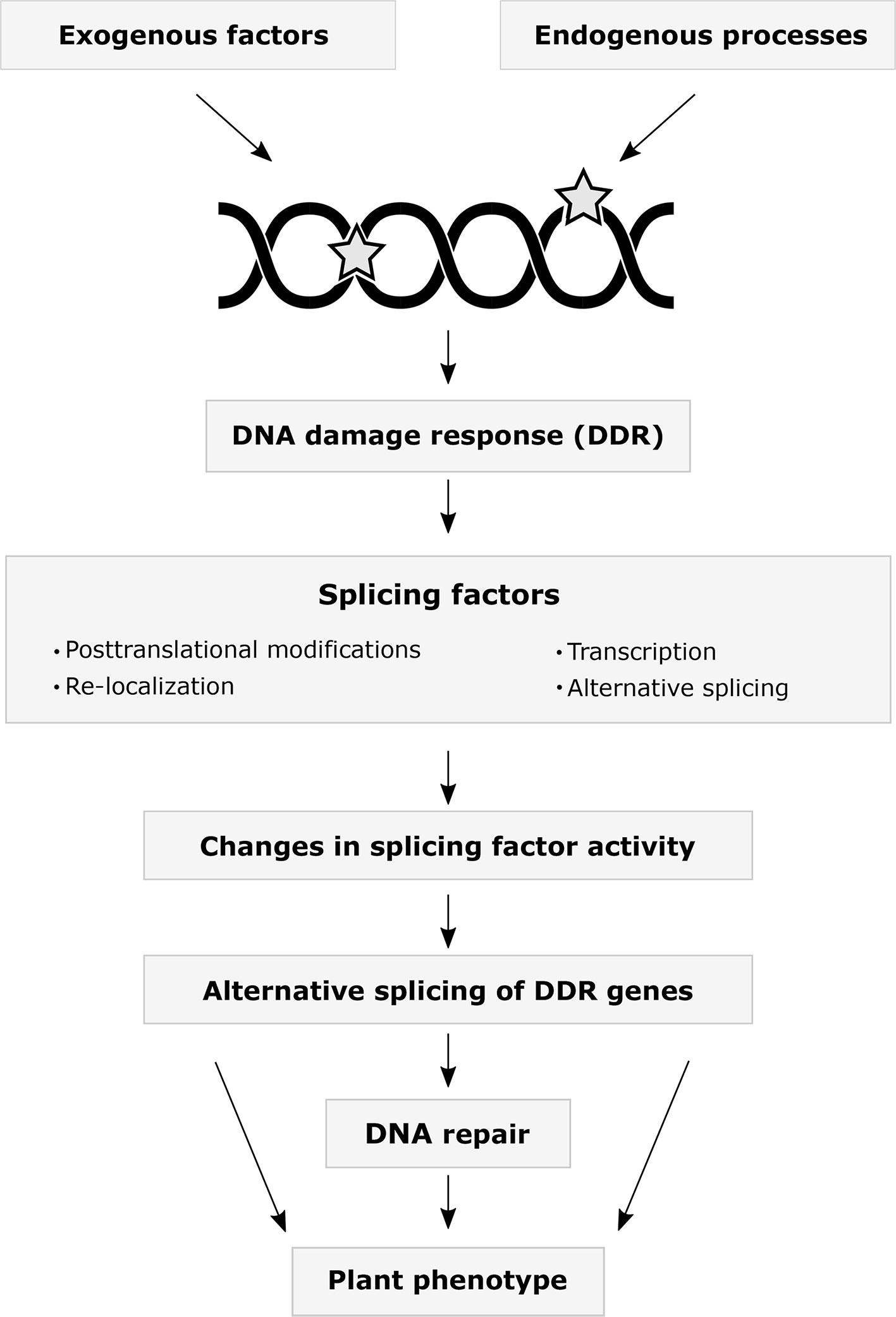
Figure 1 The interplay between the DNA damage response and alternative splicing. A variety of exogenous environmental stress factors and endogenous cellular processes may result in DNA damage. Numerous studies on animals have demonstrated that splicing factors change their expression levels, alternative splicing patterns, post-translational modification states, and subcellular localization in response to DNA damage. Altered expression and activities of splicing factors may regulate DNA repair by modulating alternative splicing of DDR genes. Current data indicates that many plant DDR genes undergo alternative splicing. Which plant splicing factors are involved in the DDR, how they are regulated, what are their target genes, and how the splicing changes are translated into the plant phenotype remains to be addressed in the future.
As localization and shuttling of SFs is highly dependent on their phosphorylation state, it is not surprising that DNA damage-induced nuclear translocation of SR protein kinases results in the hyperphosphorylation and subsequent nuclear accumulation of certain SR proteins (Edmond et al., 2011). UV irradiation also affects the redistribution of SFs into the cytoplasm, therefore impacting AS (van der Houven van Oordt et al., 2000; Llorian et al., 2005; Guil et al., 2006). The DNA damage-induced re-localization of SFs appears to be dependent on cell type and genotoxic treatment (Tissier et al., 2010; Wong et al., 2013).
In plants, members of different Arabidopsis SR protein sub-families localize into distinct populations of nuclear speckles (Lorkovic et al., 2008), with their localization dependent on their phosphorylation status (Ali et al., 2003; Tillemans et al., 2005). Different classes of kinases (such as SR protein kinases, PRP4 kinases, Cdc2-like or LAMMER-type kinases, and mitogen-activated protein kinases) phosphorylate plant SFs, including SR proteins and hnRNPs (Golovkin and Reddy, 1999; Savaldi-Goldstein et al., 2000; Feilner et al., 2005; de la Fuente van Bentem et al., 2006; de la Fuente van Bentem et al., 2008; Kanno et al., 2018), suggesting that DNA damage in plants could lead to altered SF activities and changes in AS. However, to which extent this occurs, which SFs are affected and the roles of different PTMs remain the subject of further studies.
In addition to the post-translational regulation of SFs during DDR, their activity can be altered by changes in their AS. Studies in animal cells have illustrated the impact DNA damage has on the AS of SF genes (Solier et al., 2010; Ip et al., 2011; Leva et al., 2012). Munoz and colleagues describe a mechanism by which AS is regulated during DDR (Munoz et al., 2009; Munoz et al., 2017). The hyperphosphorylation of the C-terminal domain of RNA polymerase II (RNAPII) is associated with a decrease in RNAPII elongation speed. This slowing down of RNAPII favors the selection of weaker splice sites as the time window for their recognition by the splicing machinery is extended before stronger downstream sites are synthesized. The hyperphosphorylation and slowdown of RNAPII in response to UV exposure leads to differential exon skipping events in multiple genes associated with apoptosis, cell cycle, and cancer (Munoz et al., 2009; Munoz et al., 2017). These findings raise questions regarding the mechanisms and PTMs affecting RNAPII elongation speed and the subsequent changes in splicing outcomes during DDR in plants. Which plant SFs are alternatively spliced during DDR, how their transcript isoforms differ in their function, and how their AS influences DDR itself also remains to be addressed in the future.
Alternative Splicing, a New Player in the Plant DNA Damage Response?
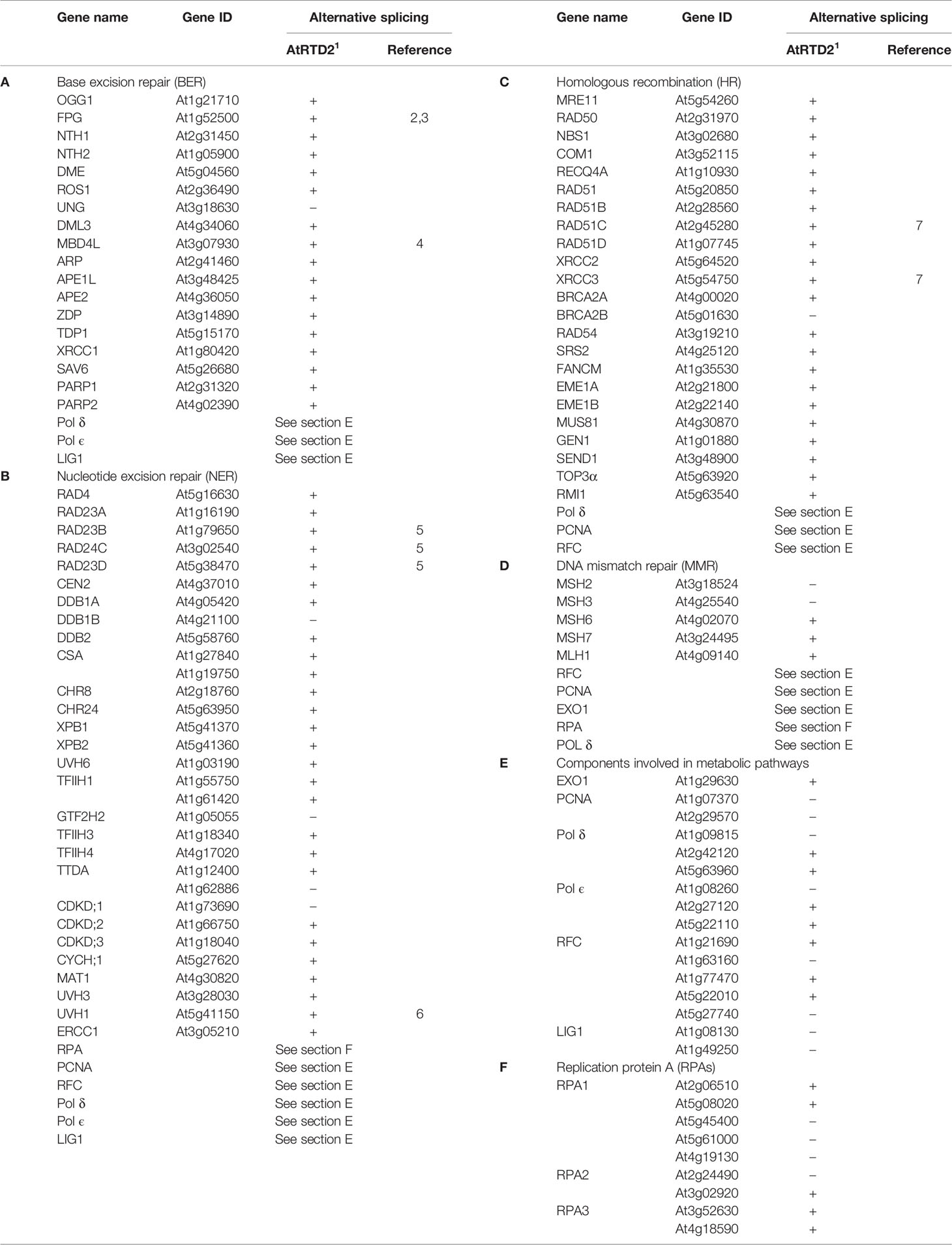
Despite extensive studies of DDR and AS in animals, comparatively little is known about this relationship in plants. The PubMed search with the terms “Splicing” and “DNA damage” or “DNA repair” returns a handful of papers in the plant field, which is in stark contrast to about 700 non-plant papers. The first papers describing AS of the Arabidopsis DNA damage/repair gene At-FPG/At-MMH DNA glycosylase were published about 20 years ago (Ohtsubo et al., 1998; Murphy and Gao, 2001). Since then, several key DNA repair genes have been reported to undergo AS, supporting the importance of AS in DDR in plants (Table 1). For example, genes encoding At-RAD1/UHV1 (homologous to yeast RAD1 and human XPF DNA repair endonuclease) and AtPOLK polymerase generate AS isoforms in a tissue-specific pattern (Vonarx et al., 2002; Garcia-Ortiz et al., 2004; Garcia-Ortiz et al., 2007). Two Arabidopsis translesion synthesis DNA polymerases, AtREV and AtPOLH, are regulated by AS, and complementation analysis of AtPOLH AS isoforms in Rad30-deficient yeast showed that the AtPOLH C-terminus is required for functional activity (Santiago et al., 2009). Several studies also reveal differential AS in DNA repair genes in crop plants, such as rice class II DNA photolyase (Hirouchi et al., 2003), endonuclease OsMUS81 (Mimida et al., 2007), and checkpoint protein OsRad9 (Li et al., 2017).
To estimate the extent of AS in DNA repair genes at the genome-wide level, we queried the Arabidopsis reference transcript dataset (AtRTD2), which contains 82,190 transcripts from 34,212 genes (Zhang et al., 2017b), with a list of 102 Arabidopsis DNA repair genes (Spampinato, 2017). Only nine genes from this list have previously been reported to be alternatively spliced. Remarkably, this survey revealed that more than 80% of these genes show evidence of AS in the AtRTD2 (Table 1). Further, key regulators of DDR in plants, SOG1, ATM, and ATR (not in the Spampinato, 2017 list), also undergo AS. Although this brief survey deals with a subset of DDR genes, it clearly illustrates a hidden potential for AS and regulation of DDR in plants. Plant mechanisms and SFs involved in DDR regulation remain to be investigated.
Conclusions
The cellular response to DNA damage must be tightly regulated. Numerous studies on animals reveal interactions between DDR and AS at multiple levels and demonstrate that AS has an important role in DDR. In plants, initial studies show that AS has a function in plant DDR, but many questions remain to be addressed. How is the expression and activity of plant SFs regulated in DDR, what are their target genes, and do RNAPII processivity or changes in chromatin structure convey DDR into differential splicing outcomes in plants? Comprehensive transcriptome analyses will identify genes that show differences in AS patterns in response to genotoxic stress. Moreover, SFs, RNA processing factors, and DNA repair genes that undergo changes in AS may be detected and help determine the complex interplay between DDR and AS in plants. Finally, the major stress factors restrict plant growth and decrease yield in crop plants. Recent studies report extensive AS in crop species, emphasizing the need for further investigations to establish AS involvement in the response mechanisms to stress exposure and DNA damage.
Author Contributions
MK designed the project. BN performed the survey of alternative splicing of Arabidopsis DNA repair genes and prepared the table and figure. The manuscript was written by BN, SR, and MK.
Funding
This work is supported by the Austrian Science Fund (FWF) (P26333 to MK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank Peter Venhuizen and Craig Simpson for their comments on the manuscript.
References
Adamson, B., Smogorzewska, A., Sigoillot, F. D., King, R. W., Elledge, S. J. (2012). A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14 (3), 318–328. doi: 10.1038/ncb2426
Ali, G. S., Golovkin, M., Reddy, A. S. (2003). Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 36 (6), 883–893. doi: 10.1046/j.1365-313X.2003.01932.x
Baralle, M., Baralle, F. E. (2018). The splicing code. Biosystems 164, 39–48. doi: 10.1016/j.biosystems.2017.11.002
Barash, Y., Calarco, J. A., Gao, W., Pan, Q., Wang, X., Shai, O., et al. (2010). Deciphering the splicing code. Nature 465 (7294), 53–59. doi: 10.1038/nature09000
Barta, A., Kalyna, M., Reddy, A. S. (2010). Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 22 (9), 2926–2929. doi: 10.1105/tpc.110.078352
Belfield, E. J., Ding, Z. J., Jamieson, F. J. C., Visscher, A. M., Zheng, S. J., Mithani, A., et al. (2018). DNA mismatch repair preferentially protects genes from mutation. Genome Res. 28 (1), 66–74. doi: 10.1101/gr.219303.116
Beli, P., Lukashchuk, N., Wagner, S. A., Weinert, B. T., Olsen, J. V., Baskcomb, L., et al. (2012). Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46 (2), 212–225. doi: 10.1016/j.molcel.2012.01.026
Bennetzen, M. V., Larsen, D. H., Bunkenborg, J., Bartek, J., Lukas, J., Andersen, J. S. (2010). Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell Proteomics 9 (6), 1314–1323. doi: 10.1074/mcp.M900616-MCP200
Bensimon, A., Schmidt, A., Ziv, Y., Elkon, R., Wang, S. Y., Chen, D. J., et al. (2010). ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci. Signal 3 (151), rs3. doi: 10.1126/scisignal.2001034
Boothby, T. C., Zipper, R. S., van der Weele, C. M., Wolniak, S. M. (2013). Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev. Cell 24 (5), 517–529. doi: 10.1016/j.devcel.2013.01.015
Bourbousse, C., Vegesna, N., Law, J. A. (2018). SOG1 activator and MYB3R repressors regulate a complex DNA damage network in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115 (52), E12453–e12462. doi: 10.1073/pnas.1810582115
Boutz, P. L., Bhutkar, A., Sharp, P. A. (2015). Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 29 (1), 63–80. doi: 10.1101/gad.247361.114
Boyko, A., Filkowski, J., Kovalchuk, I. (2005). Homologous recombination in plants is temperature and day-length dependent. Mutat. Res. 572 (1-2), 73–83. doi: 10.1016/j.mrfmmm.2004.12.011
Braunschweig, U., Gueroussov, S., Plocik, A. M., Graveley, B. R., Blencowe, B. J. (2013). Dynamic integration of splicing within gene regulatory pathways. Cell 152 (6), 1252–1269. doi: 10.1016/j.cell.2013.02.034
Braunschweig, U., Barbosa-Morais, N. L., Pan, Q., Nachman, E. N., Alipanahi, B., Gonatopoulos-Pournatzis, T., et al. (2014). Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 24 (11), 1774–1786. doi: 10.1101/gr.177790.114
Brown, J. W., Simpson, C. G., Marquez, Y., Gadd, G. M., Barta, A., Kalyna, M. (2015). Lost in translation: pitfalls in deciphering plant alternative splicing transcripts. Plant Cell 27 (8), 2083–2087. doi: 10.1105/tpc.15.00572
Busa, R., Geremia, R., Sette, C. (2010). Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic Acids Res. 38 (9), 3005–3018. doi: 10.1093/nar/gkq004
Chamala, S., Feng, G., Chavarro, C., Barbazuk, W. B. (2015). Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 3, 33. doi: 10.3389/fbioe.2015.00033
Chen, W., Moore, M. J. (2015). Spliceosomes. Curr. Biol. 25 (5), R181–R183. doi: 10.1016/j.cub.2014.11.059
Clark, S., Yu, F., Gu, L., Min, X. J. (2019). Expanding alternative splicing identification by integrating multiple sources of transcription data in tomato. Front. Plant Sci. 10, 689. doi: 10.3389/fpls.2019.00689
de la Fuente van Bentem, S., Anrather, D., Roitinger, E., Djamei, A., Hufnagl, T., Barta, A., et al. (2006). Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res. 34 (11), 3267–3278. doi: 10.1093/nar/gkl429
de la Fuente van Bentem, S., Anrather, D., Dohnal, I., Roitinger, E., Csaszar, E., Joore, J., et al. (2008). Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 7 (6), 2458–2470. doi: 10.1021/pr8000173
De Veylder, L., Larkin, J. C., Schnittger, A. (2011). Molecular control and function of endoreplication in development and physiology. Trends Plant Sci. 16 (11), 624–634. doi: 10.1016/j.tplants.2011.07.001
Edmond, V., Moysan, E., Khochbin, S., Matthias, P., Brambilla, C., Brambilla, E., et al. (2011). Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 30 (3), 510–523. doi: 10.1038/emboj.2010.333
Feilner, T., Hultschig, C., Lee, J., Meyer, S., Immink, R. G., Koenig, A., et al. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell Proteomics 4 (10), 1558–1568. doi: 10.1074/mcp.M500007-MCP200
Garcia-Ortiz, M. V., Ariza, R. R., Hoffman, P. D., Hays, J. B., Roldan-Arjona, T. (2004). Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 39 (1), 84–97. doi: 10.1111/j.1365-313X.2004.02112.x
Garcia-Ortiz, M. V., Roldan-Arjona, T., Ariza, R. R. (2007). The noncatalytic C-terminus of AtPOLK Y-family DNA polymerase affects synthesis fidelity, mismatch extension and translesion replication. FEBS J. 274 (13), 3340–3350. doi: 10.1111/j.1742-4658.2007.05868.x
Gimenez, E., Manzano-Agugliaro, F. (2017). DNA damage repair system in plants: a worldwide research update. Genes (Basel) 8 (11), e299. doi: 10.3390/genes8110299
Giono, L. E., Nieto Moreno, N., Cambindo Botto, A. E., Dujardin, G., Munoz, M. J., Kornblihtt, A. R. (2016). The RNA response to DNA damage. J. Mol. Biol. 428 (12), 2636–2651. doi: 10.1016/j.jmb.2016.03.004
Gohring, J., Jacak, J., Barta, A. (2014). Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell 26 (2), 754–764. doi: 10.1105/tpc.113.118075
Golovkin, M., Reddy, A. S. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem. 274 (51), 36428–36438. doi: 10.1074/jbc.274.51.36428
Guil, S., Long, J. C., Caceres, J. F. (2006). hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell Biol. 26 (15), 5744–5758. doi: 10.1128/MCB.00224-06
Gutman, B. L., Niyogi, K. K. (2009). Evidence for base excision repair of oxidative DNA damage in chloroplasts of Arabidopsis thaliana. J. Biol. Chem. 284 (25), 17006–17012. doi: 10.1074/jbc.M109.008342
Hanawalt, P. C. (2002). Subpathways of nucleotide excision repair and their regulation. Oncogene 21 (58), 8949–8956. doi: 10.1038/sj.onc.1206096
Hirouchi, T., Nakajima, S., Najrana, T., Tanaka, M., Matsunaga, T., Hidema, J., et al. (2003). A gene for a Class II DNA photolyase from Oryza sativa: cloning of the cDNA by dilution-amplification. Mol. Genet. Genomics 269 (4), 508–516. doi: 10.1007/s00438-003-0856-9
Howard, J. M., Sanford, J. R. (2015). The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6 (1), 93–110. doi: 10.1002/wrna.1260
Hu, Z., Cools, T., De Veylder, L. (2016). Mechanisms used by plants to cope with DNA damage. Annu. Rev. Plant Biol. 67, 439–462. doi: 10.1146/annurev-arplant-043015-111902
Ip, J. Y., Schmidt, D., Pan, Q., Ramani, A. K., Fraser, A. G., Odom, D. T., et al. (2011). Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 21 (3), 390–401. doi: 10.1101/gr.111070.110
Irimia, M., Weatheritt, R. J., Ellis, J. D., Parikshak, N. N., Gonatopoulos-Pournatzis, T., Babor, M., et al. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159 (7), 1511–1523. doi: 10.1016/j.cell.2014.11.035
Ishikawa, Y., Endo, M., Abe, K., Osakabe, K., Nakajima, N., Saji, H., et al. (2004). Isolation of four RAD23 Genes from Arabidopsis thaliana and detection of alternative splicing variants. Plant Biotechnol. 21 (1), 65–71. doi: 10.5511/plantbiotechnology.21.65
Jabre, I., Reddy, A. S. N., Kalyna, M., Chaudhary, S., Khokhar, W., Byrne, L. J., et al. (2019). Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 47 (6), 2716–2726. doi: 10.1093/nar/gkz121
James, A. B., Syed, N. H., Bordage, S., Marshall, J., Nimmo, G. A., Jenkins, G. I., et al. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24 (3), 961–981. doi: 10.1105/tpc.111.093948
Kai, M. (2016). Roles of RNA-binding proteins in DNA damage response. Int. J. Mol. Sci. 17 (3), 310. doi: 10.3390/ijms17030310
Kalyna, M., Simpson, C. G., Syed, N. H., Lewandowska, D., Marquez, Y., Kusenda, B., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40 (6), 2454–2469. doi: 10.1093/nar/gkr932
Kanno, T., Venhuizen, P., Wen, T. N., Lin, W. D., Chiou, P., Kalyna, M., et al. (2018). PRP4KA, a Putative Spliceosomal Protein Kinase, is important for alternative splicing and development in Arabidopsis thaliana. Genetics 210 (4), 1267–1285. doi: 10.1534/genetics.118.301515
Kavakli, I. H., Baris, I., Tardu, M., Gul, S., Oner, H., Cal, S., et al. (2017). The photolyase/cryptochrome family of proteins as DNA repair enzymes and transcriptional repressors. Photochem. Photobiol. 93 (1), 93–103. doi: 10.1111/php.12669
Kim, J. H., Ryu, T. H., Lee, S. S., Lee, S., Chung, B. Y. (2019). Ionizing radiation manifesting DNA damage response in plants: an overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 278, 44–53. doi: 10.1016/j.plantsci.2018.10.013
Lackner, D. H., Durocher, D., Karlseder, J. (2011). A siRNA-based screen for genes involved in chromosome end protection. PLoS One 6 (6), e21407. doi: 10.1371/journal.pone.0021407
Leva, V., Giuliano, S., Bardoni, A., Camerini, S., Crescenzi, M., Lisa, A., et al. (2012). Phosphorylation of SRSF1 is modulated by replicational stress. Nucleic Acids Res. 40 (3), 1106–1117. doi: 10.1093/nar/gkr837
Leviatan, N., Alkan, N., Leshkowitz, D., Fluhr, R. (2013). Genome-wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLoS One 8 (6), e66511. doi: 10.1371/journal.pone.0066511
Li, X., Manley, J. L. (2005). Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122 (3), 365–378. doi: 10.1016/j.cell.2005.06.008
Li, S., Paulsson, M., Bjorn, L. O. (2002). Temperature-dependent formation and photorepair of DNA damage induced by UV-B radiation in suspension-cultured tobacco cells. J. Photochem. Photobiol. B. 66 (1), 67–72. doi: 10.1016/S1011-1344(01)00277-9
Li, Z., Pearlman, A. H., Hsieh, P. (2016). DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38, 94–101. doi: 10.1016/j.dnarep.2015.11.019
Li, R., Wang, W., Li, F., Wang, Q., Wang, S., Xu, Y., et al. (2017). Response of alternative splice isoforms of OsRad9 gene from Oryza sativa to environmental stress. Z. Naturforsch. C. 72 (7-8), 325–334. doi: 10.1515/znc-2016-0257
Liu, D., Keijzers, G., Rasmussen, L. J. (2017). DNA mismatch repair and its many roles in eukaryotic cells. Mutat. Res. 773, 174–187. doi: 10.1016/j.mrrev.2017.07.001
Llorian, M., Beullens, M., Lesage, B., Nicolaescu, E., Beke, L., Landuyt, W., et al. (2005). Nucleocytoplasmic shuttling of the splicing factor SIPP1. J. Biol. Chem. 280 (46), 38862–38869. doi: 10.1074/jbc.M509185200
Lorkovic, Z. J., Hilscher, J., Barta, A. (2008). Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp. Cell Res. 314 (17), 3175–3186. doi: 10.1016/j.yexcr.2008.06.020
Lu, X., Legerski, R. J. (2007). The Prp19/Pso4 core complex undergoes ubiquitylation and structural alterations in response to DNA damage. Biochem. Biophys. Res. Commun. 354 (4), 968–974. doi: 10.1016/j.bbrc.2007.01.097
Magni, M., Buscemi, G., Maita, L., Peng, L., Chan, S. Y., Montecucco, A., et al. (2019). TSPYL2 is a novel regulator of SIRT1 and p300 activity in response to DNA damage. Cell Death Differ. 26 (5), 918–931. doi: 10.1038/s41418-018-0168-6
Manley, J. L., Krainer, A. R. (2010). A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 24 (11), 1073–1074. doi: 10.1101/gad.1934910
Manova, V., Gruszka, D. (2015). DNA damage and repair in plants - from models to crops. Front. Plant Sci. 6,885. doi: 10.3389/fpls.2015.00885
Marquez, Y., Brown, J. W., Simpson, C., Barta, A., Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22 (6), 1184–1195. doi: 10.1101/gr.134106.111
Marquez, Y., Hopfler, M., Ayatollahi, Z., Barta, A., Kalyna, M. (2015). Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 25 (7), 995–1007. doi: 10.1101/gr.186585.114
Matsuoka, S., Ballif, B. A., Smogorzewska, A., McDonald, ,. E. R., 3rd, Hurov, K. E., Luo, J., et al. (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316 (5828), 1160–1166. doi: 10.1126/science.1140321
Mikolaskova, B., Jurcik, M., Cipakova, I., Kretova, M., Chovanec, M., Cipak, L. (2018). Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways. Curr. Genet. 64 (5), 971–983. doi: 10.1007/s00294-018-0819-7
Mimida, N., Kitamoto, H., Osakabe, K., Nakashima, M., Ito, Y., Heyer, W. D., et al. (2007). Two alternatively spliced transcripts generated from OsMUS81, a rice homolog of yeast MUS81, are up-regulated by DNA-damaging treatments. Plant Cell Physiol. 48 (4), 648–654. doi: 10.1093/pcp/pcm029
Munoz, M. J., Perez Santangelo, M. S., Paronetto, M. P., de la Mata, M., Pelisch, F., Boireau, S., et al. (2009). DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137 (4), 708–720. doi: 10.1016/j.cell.2009.03.010
Munoz, M. J., Nieto Moreno, N., Giono, L. E., Cambindo Botto, A. E., Dujardin, G., Bastianello, G., et al. (2017). Major roles for pyrimidine dimers, nucleotide excision repair, and ATR in the alternative splicing response to UV irradiation. Cell Rep. 18 (12), 2868–2879. doi: 10.1016/j.celrep.2017.02.066
Murphy, T. M., Gao, M. J. (2001). Multiple forms of formamidopyrimidine-DNA glycosylase produced by alternative splicing in Arabidopsis thaliana. J. Photochem. Photobiol. B. 61 (3), 87–93. doi: 10.1016/S1011-1344(01)00172-5
Nakka, K. K., Chaudhary, N., Joshi, S., Bhat, J., Singh, K., Chatterjee, S., et al. (2015). Nuclear matrix-associated protein SMAR1 regulates alternative splicing via HDAC6-mediated deacetylation of Sam68. Proc. Natl. Acad. Sci. U. S. A. 112 (26), E3374–E3383. doi: 10.1073/pnas.1418603112
Naro, C., Bielli, P., Pagliarini, V., Sette, C. (2015). The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability. Front. Genet. 6, 142. doi: 10.3389/fgene.2015.00142
Nisa, M. U., Huang, Y., Benhamed, M., Raynaud, C. (2019). The plant DNA damage response: signaling pathways leading to growth inhibition and putative role in response to stress conditions. Front. Plant Sci. 10, 653. doi: 10.3389/fpls.2019.00653
Nota, F., Cambiagno, D. A., Ribone, P., Alvarez, M. E. (2015). Expression and function of AtMBD4L, the single gene encoding the nuclear DNA glycosylase MBD4L in Arabidopsis. Plant Sci. 235, 122–129. doi: 10.1016/j.plantsci.2015.03.011
Ogita, N., Okushima, Y., Tokizawa, M., Yamamoto, Y. Y., Tanaka, M., Seki, M., et al. (2018). Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 94 (3), 439–453. doi: 10.1111/tpj.13866
Ohtsubo, T., Matsuda, O., Iba, K., Terashima, I., Sekiguchi, M., Nakabeppu, Y. (1998). Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene, and analysis of functional domains of its product. Mol. Gen. Genet. 259 (6), 577–590. doi: 10.1007/s004380050851
Osakabe, K., Yoshioka, T., Ichikawa, H., Toki, S. (2002). Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana. Plant Mol. Biol. 50 (1), 71–81. doi: 10.1023/A:1016047231597
Pan, Q., Shai, O., Lee, L. J., Frey, B. J., Blencowe, B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40 (12), 1413–1415. doi: 10.1038/ng.259
Paulsen, R. D., Soni, D. V., Wollman, R., Hahn, A. T., Yee, M. C., Guan, A., et al. (2009). A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 35 (2), 228–239. doi: 10.1016/j.molcel.2009.06.021
Preuss, S. B., Britt, A. B. (2003). A DNA-damage-induced cell cycle checkpoint in Arabidopsis. Genetics 164 (1), 323–334.
Rapazote-Flores, P., Bayer, M., Milne, L., Mayer, C.-D., Fuller, J., Guo, W., et al. (2019). BaRTv1.0: an improved barley reference transcript dataset to determine accurate changes in the barley transcriptome using RNA-seq. BMC Genomics. 20 (1), 968. doi: 10.1186/s12864-019-6243-7
Sakashita, E., Endo, H. (2010). SR and SR-related proteins redistribute to segregated fibrillar components of nucleoli in a response to DNA damage. Nucleus 1 (4), 367–380. doi: 10.4161/nucl.1.4.12683
Santiago, M. J., Alejandre-Durán, E., Ruiz-Rubio, M. (2009). Alternative splicing of two translesion synthesis DNA polymerases from Arabidopsis thaliana. Plant Sci. 176 (4), 591–596. doi: 10.1016/j.plantsci.2009.01.018
Savaldi-Goldstein, S., Sessa, G., Fluhr, R. (2000). The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J. 21 (1), 91–96. doi: 10.1046/j.1365-313x.2000.00657.x
Shepard, P. J., Hertel, K. J. (2008). Conserved RNA secondary structures promote alternative splicing. RNA 14 (8), 1463–1469. doi: 10.1261/rna.1069408
Shkreta, L., Chabot, B. (2015). The RNA splicing response to DNA damage. Biomolecules 5 (4), 2935–2977. doi: 10.3390/biom5042935
Shkreta, L., Toutant, J., Durand, M., Manley, J. L., Chabot, B. (2016). SRSF10 connects DNA damage to the alternative splicing of transcripts encoding apoptosis, cell-cycle control, and DNA repair factors. Cell Rep. 17 (8), 1990–2003. doi: 10.1016/j.celrep.2016.10.071
Siam, A., Baker, M., Amit, L., Regev, G., Rabner, A., Najar, R. A., et al. (2019). Regulation of alternative splicing by p300-mediated acetylation of splicing factors. RNA 25 (7), 813–824. doi: 10.1261/rna.069856.118
Sibley, C. R., Blazquez, L., Ule, J. (2016). Lessons from non-canonical splicing. Nat. Rev. Genet. 17 (7), 407–421. doi: 10.1038/nrg.2016.46
Solier, S., Barb, J., Zeeberg, B. R., Varma, S., Ryan, M. C., Kohn, K. W., et al. (2010). Genome-wide analysis of novel splice variants induced by topoisomerase I poisoning shows preferential occurrence in genes encoding splicing factors. Cancer Res. 70 (20), 8055–8065. doi: 10.1158/0008-5472.CAN-10-2491
Spampinato, C. P. (2017). Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell Mol. Life Sci. 74 (9), 1693–1709. doi: 10.1007/s00018-016-2436-2
Staiger, D., Simpson, G. G. (2015). Enter exitrons. Genome Biol. 16 (1), 136. doi: 10.1186/s13059-015-0704-3
Thatcher, S. R., Zhou, W., Leonard, A., Wang, B. B., Beatty, M., Zastrow-Hayes, G., et al. (2014). Genome-wide analysis of alternative splicing in Zea mays: landscape and genetic regulation. Plant Cell 26 (9), 3472–3487. doi: 10.1105/tpc.114.130773
Tillemans, V., Dispa, L., Remacle, C., Collinge, M., Motte, P. (2005). Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells. Plant J. 41 (4), 567–582. doi: 10.1111/j.1365-313X.2004.02321.x
Tissier, A., Janel-Bintz, R., Coulon, S., Klaile, E., Kannouche, P., Fuchs, R. P., et al. (2010). Crosstalk between replicative and translesional DNA polymerases: PDIP38 interacts directly with Poleta. DNA Repair (Amst) 9 (8), 922–928. doi: 10.1016/j.dnarep.2010.04.010
Ustianenko, D., Weyn-Vanhentenryck, S. M., Zhang, C. (2017). Microexons: discovery, regulation, and function. Wiley Interdiscip. Rev. RNA 8 (4), e1418. doi: 10.1002/wrna.1418
van der Houven van Oordt, W., Diaz-Meco, M. T., Lozano, J., Krainer, A. R., Moscat, J., Caceres, J. F. (2000). The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149 (2), 307–316. doi: 10.1083/jcb.149.2.307
Vassileva, M. T., Matunis, M. J. (2004). SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell Biol. 24 (9), 3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004
Vonarx, E. J., Howlett, N. G., Schiestl, R. H., Kunz, B. A. (2002). Detection of Arabidopsis thaliana AtRAD1 cDNA variants and assessment of function by expression in a yeast rad1 mutant. Gene 296 (1-2), 1–9. doi: 10.1016/S0378-1119(02)00869-7
Wallace, S. S. (2014). Base excision repair: a critical player in many games. DNA Repair (Amst) 19, 14–26. doi: 10.1016/j.dnarep.2014.03.030
Wang, Z., Burge, C. B. (2008). Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14 (5), 802–813. doi: 10.1261/rna.876308
Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456 (7221), 470–476. doi: 10.1038/nature07509
Wong, A., Zhang, S., Mordue, D., Wu, J. M., Zhang, Z., Darzynkiewicz, Z., et al. (2013). PDIP38 is translocated to the spliceosomes/nuclear speckles in response to UV-induced DNA damage and is required for UV-induced alternative splicing of MDM2. Cell Cycle 12 (19), 3184–3193. doi: 10.4161/cc.26221
Yap, K., Lim, Z. Q., Khandelia, P., Friedman, B., Makeyev, E. V. (2012). Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 26 (11), 1209–1223. doi: 10.1101/gad.188037.112
Yeap, W. C., Namasivayam, P., Ho, C. L. (2014). HnRNP-like proteins as post-transcriptional regulators. Plant Sci. 227, 90–100. doi: 10.1016/j.plantsci.2014.07.005
Yoshiyama, K., Conklin, P. A., Huefner, N. D., Britt, A. B. (2009). Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 106 (31), 12843–12848. doi: 10.1073/pnas.0810304106
Yoshiyama, K. O. (2016). SOG1: a master regulator of the DNA damage response in plants. Genes Genet. Syst. 90 (4), 209–216. doi: 10.1266/ggs.15-00011
Zhang, M., Wang, L., Zhong, D. (2017a). Photolyase: dynamics and mechanisms of repair of sun-induced DNA damage. Photochem. Photobiol. 93 (1), 78–92. doi: 10.1111/php.12695
Keywords: alternative splicing, DNA repair, DNA damage response, Arabidopsis, plant, stress, splicing factor
Citation: Nimeth BA, Riegler S and Kalyna M (2020) Alternative Splicing and DNA Damage Response in Plants. Front. Plant Sci. 11:91. doi: 10.3389/fpls.2020.00091
Received: 29 July 2019; Accepted: 21 January 2020;
Published: 19 February 2020.
Edited by:
Paula Casati, CONICET Center for Photosynthetic and Biochemical Studies (CEFOBI), ArgentinaCopyright © 2020 Nimeth, Riegler and Kalyna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Kalyna, bWFyaXlhLmthbHluYUBib2t1LmFjLmF0
†ORCID ID: Barbara Anna Nimeth, orcid.org/0000-0001-8692-8413
Stefan Riegler, orcid.org/0000-0003-3413-1343
Maria Kalyna, orcid.org/0000-0003-4702-7625
‡Present Address: Stefan Riegler, Institute of Science and Technology Austria, Klosterneuburg, Austria
 Barbara Anna Nimeth
Barbara Anna Nimeth Stefan Riegler
Stefan Riegler Maria Kalyna
Maria Kalyna