
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 19 February 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00085
This article is part of the Research Topic Chromatin Stability and Dynamics: Targeting and Recruitment of Chromatin Modifiers View all 10 articles
FACT is a heterodimeric histone chaperone consisting of the SSRP1 and SPT16 proteins and is conserved among eukaryotes. It interacts with the histones H2A-H2B and H3-H4 as well as with DNA. Based on in vitro and in vivo studies mainly in yeast and mammalian cells, FACT can mediate nucleosome disassembly and reassembly and thus facilitates in the chromatin context DNA-dependent processes including transcription, replication and repair. In plants, primarily the role of FACT related to RNA polymerase II transcription has been examined. FACT was found to associate with elongating Arabidopsis RNA polymerase II (RNAPII) as part of the transcript elongation complex and it was identified as repressor of aberrant intragenic transcriptional initiation. Arabidopsis mutants depleted in FACT subunits exhibit various defects in vegetative and reproductive development. Strikingly, FACT modulates important developmental transitions by promoting expression of key repressors of these processes. Thus, FACT facilitates expression of DOG1 and FLC adjusting the switch from seed dormancy to germination and from vegetative to reproductive development, respectively. In the central cell of the female gametophyte, FACT can facilitate DNA demethylation especially within heterochromatin, and thereby contributes to gene imprinting during Arabidopsis reproduction. This review discusses results particularly from the plant perspective about the contribution of FACT to processes that involve reorganisation of nucleosomes with a main focus on RNAPII transcription and its implications for diverse areas of plant biology.
In eukaryotes, the nuclear DNA is packaged into nucleosomes, which represent the basic repeat unit of chromatin. The nucleosome contains 147 bp of DNA wrapped around an octamer composed of two copies each of the four core histones H2A, H2B, H3, and H4. Adjacent nucleosomes are connected by linker DNA of variable length (10–80 bp depending on cell type and species) that typically associates with linker histones such as H1 (McGhee and Felsenfeld, 1980; Luger and Richmond, 1998; Kornberg and Lorch, 1999). The general stability of nucleosome particles imposes major obstacles to transcription and other DNA-dependent processes (Li et al., 2007). Therefore, different mechanisms have evolved that facilitate chromatin transcription by destabilising/disassembly of nucleomes (Henikoff, 2008; Zhou et al., 2019). In the regulation of nucleosome dynamics, in addition to other factors so-called histone chaperones are critically involved. Histone chaperones are a heterogeneous class of proteins that functionally interact with core histones to assemble/disassemble nucleosome particles without consuming energy in form of ATP and they are not necessarily part of the final product (De Koning et al., 2007). There are various types of histone chaperones that contribute to different chromatin-related processes including transcription, replication, and DNA repair (Das et al., 2010; Avvakumov et al., 2011; Gurard-Levin et al., 2014; Hammond et al., 2017). Often histone chaperones are classified as either H2A-H2B or H3-H4 chaperones, reflecting their preferential interaction with different core histones. Some histone chaperones even display selectivity towards specific H3 or H2A isoforms such as replicative or replacement variants (De Koning et al., 2007; Hammond et al., 2017). Beyond that histone chaperones have been functionally linked with the occurrence of certain post-translational modifications of core histones and thus with the establishment, maintenance and propagation of specific chromatin states (Avvakumov et al., 2011).
Due to the extensive evolutionary conservation of the structure of the nucleosome particle, many of the histone chaperones that have been studied in detail in yeast and metazoa also occur in plants. Thus, a variety of H2A-H2B and H3-H4 chaperones have been identified throughout the plant kingdom (Tripathi et al., 2015; Zhou et al., 2015; Kumar and Vasudevan, 2020). By modulating local chromatin structure histone chaperones were found to contribute to the regulation of plant growth and development (Ramirez-Parra and Gutierrez, 2007; Otero et al., 2014; Takatsuka and Umeda, 2015; Zhou et al., 2015). Moreover, tuning of chromatin states by histone chaperones to mediate altered gene expression programs can assist plants to cope more efficiently with environmental stress conditions (Zhu et al., 2013; Liu et al., 2015; Probst and Mittelsten Scheid, 2015).
In this article, the current knowledge about the histone chaperone FACT will be summarised, particularly its role in Arabidopsis, as most studies in plants were performed using this model. At first, though the discovery of FACT and its mode of action in yeast and metazoa is introduced.
Originally, FACT (FAcilitates Chromatin Transcription) was identified in yeast and mammalian cells (Brewster et al., 1998; Orphanides et al., 1998; Orphanides et al., 1999; Wittmeyer et al., 1999). Its name originates from the finding that FACT promoted in vitro transcription from reconstituted chromatin templates by destabilising nucleosomes, facilitating RNA polymerase II passage during elongation (Orphanides et al., 1998; Orphanides et al., 1999; Belotserkovskaya et al., 2003). Over the years it became clear that besides chromatin transcription, FACT is also involved in other chromatin-dependent processes such as replication, recombination, and repair (Belotserkovskaya et al., 2004; Singer and Johnston, 2004; Winkler and Luger, 2011; Formosa, 2012; Gurova et al., 2018), and hence, the established name may well stand more broadly for facilitates chromatin transactions. FACT is a heterodimer consisting of the SSRP1 (Structure-Specific Recognition Protein 1; termed Pob3 in yeast) and SPT16 (SuPpressor of Ty 16). The main feature of FACT is its ability to disassemble and reassemble nucleosomes, and thus its involvement in overcoming and maintaining the nucleosomal barrier to DNA-dependent processes occurring in the chromatin context. Accordingly, FACT can interact with various nucleosomal targets including H2A-H2B dimers, H3-H4 tetramers and DNA (Jamai et al., 2009; Winkler et al., 2011; Hondele et al., 2013; Kemble et al., 2015). The nature of FACT-histone interactions has been further elucidated in a recent cryo-EM study of human FACT in complex with partially assembled sub-nucleosomes (Liu et al., 2020). This work illustrates that structure of FACT resembles a unicycle, consisting of a saddle and fork that is engaged in extensive interactions of SSRP1 and SPT16 with nucleosomal DNA and all histones. Competition between FACT and DNA for histone binding seems to be critical for reversible nucleosome reorganisation and uncoiling of the nucleosomal DNA from the histone core that generally leads to increased DNA accessibility (Xin et al., 2009; Hondele et al., 2013; Kemble et al., 2015; Tsunaka et al., 2016; Valieva et al., 2016; Wang et al., 2018). Following transient nucleosome destabilisation, for instance, during passage of transcribing RNA polymerase II, FACT promotes nucleosome reassembly that is important to maintain proper chromatin signature and to prevent aberrant transcriptional initiation from cryptic promoters (Kaplan et al., 2003; Mason and Struhl, 2003; Cheung et al., 2008; Jamai et al., 2009; Wang et al., 2018). Further intriguing molecular and structural details of numerous studies on yeast and metazoan FACT are summarised in various excellent review articles (Belotserkovskaya et al., 2004; Winkler and Luger, 2011; Formosa, 2012; Gurova et al., 2018).
The FACT heterodimer consisting of SSRP1 (71.6 kDa) and SPT16 (120.6 kDa) was demonstrated by reciprocal coimmunoprecipitation from Arabidopsis cells (Duroux et al., 2004). SPT16 comprises an N-terminal domain, a dimerisation domain, a middle domain, and an acidic C-terminal domain (Figure 1), and the overall domain organisation of plant SPT16 closely resembles the counterparts of other eukaryotes (Supplementary Figure S1). SSRP1 contains an N-terminal domain that mediates dimerisation with SPT16, a middle domain, an acidic domain, and a C-terminal HMG-box domain (Figure 1). Metazoan SSRP1 differs from the plant orthologues by a more pronounced C-terminal extension, while the fungal orthologues lack the HMG-box domain (Supplementary Figure S2) that in yeast is provided by separate small HMGB-box proteins termed Nhp6a/b (Formosa, 2012; Gurova et al., 2018). Proteins closely related to Arabidopsis SSRP1 and SPT16 are encoded by monocot and dicot plants, as well as by Selaginella and Physcomitrella (Figure 2).
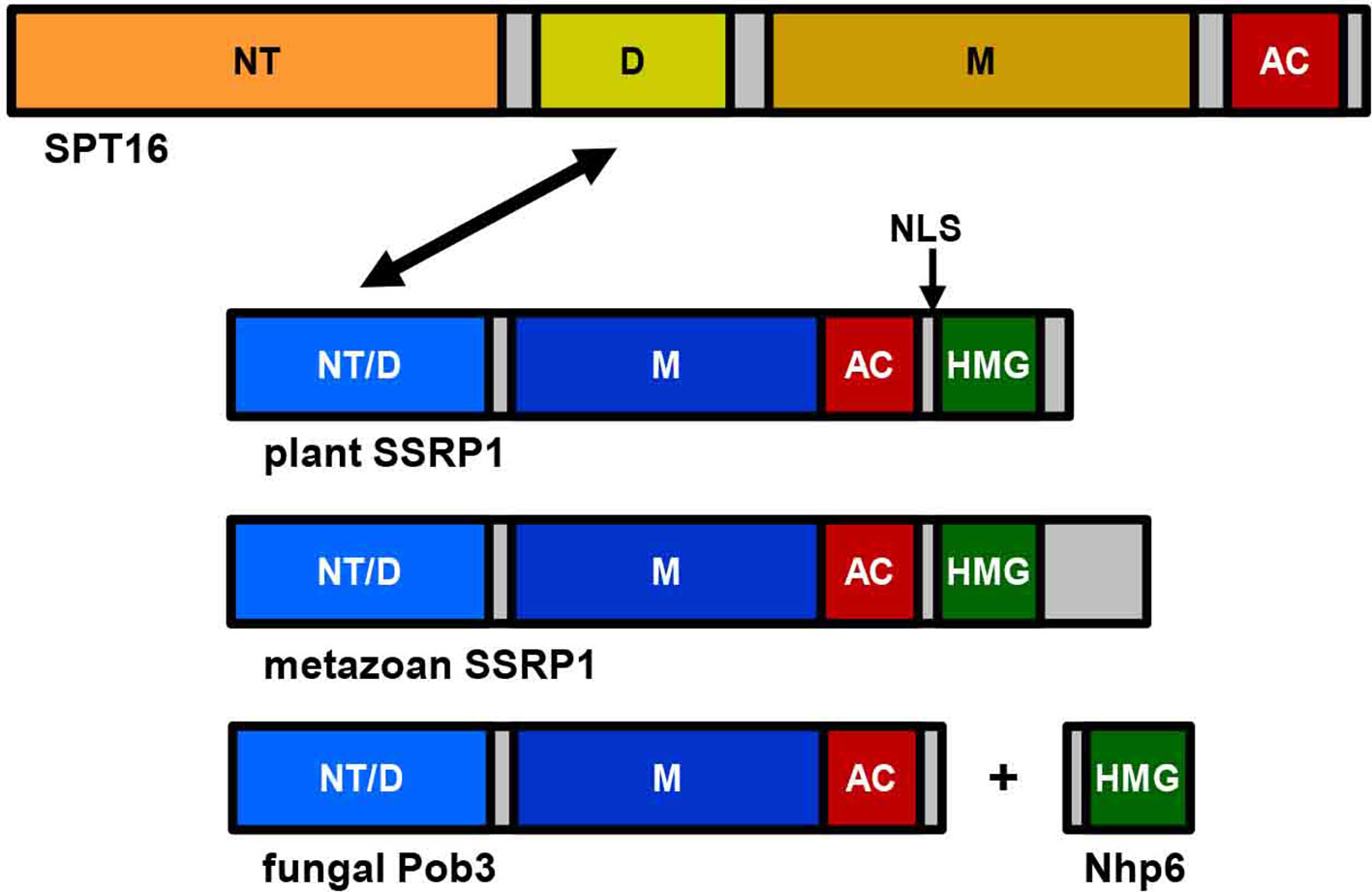
Figure 1 Schematic representation of the FAcilitates Chromatin Transcription (FACT) subunits SPT16 and SSRP1. While the overall structure of SPT16 is essentially conserved throughout eukaryotes, there are differences in the C-terminal region of SSRP1 (Pob3 in fungi). SPT16 consists of N-terminal domain (NT), dimerisation domain (D), middle domain (M), and acidic C-terminal domain (AC), while SSRP1/Pob3 proteins of different eukaryotes are composed of N-terminal domain (that is required for heterodimerisation (NT/D) with SPT16, indicated by an arrow), middle domain (M), acidic region (AC), and HMG-box domain (HMG), which in case of yeast Pob3 is provided by the separate protein(s) Nhp6a/b. Plant SSRP1 contains a nuclear localisation signal (NLS, indicated by an arrow) within a short basic region linking the acidic domain and the HMG-box (Röttgers et al., 2000).
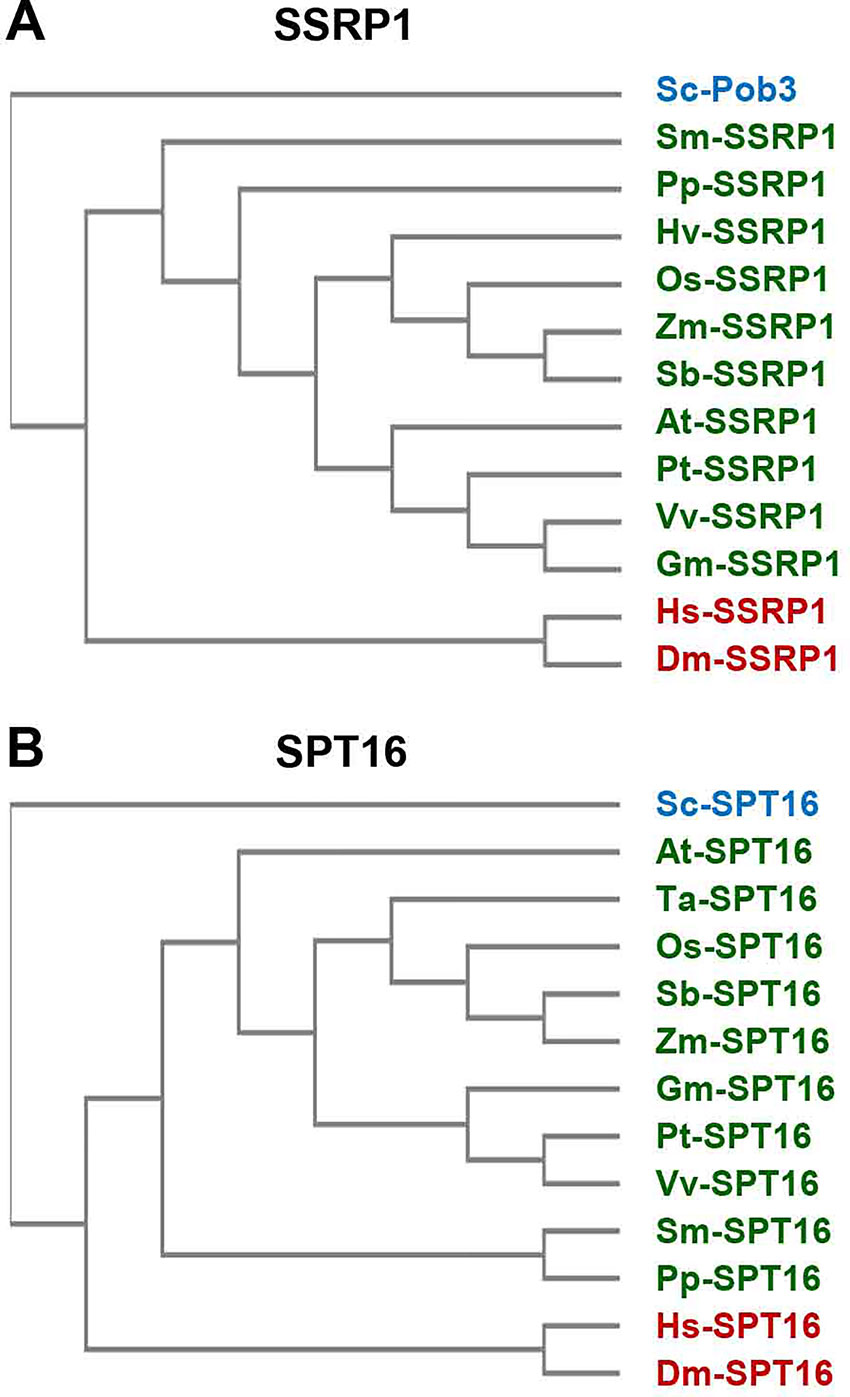
Figure 2 Sequence similarity of FAcilitates Chromatin Transcription (FACT) subunits. The amino acid sequences of SSRP1 (A) and SPT16 (B) proteins from various organisms (Arabidopsis thaliana (At), Drosophila melanogaster (Dm), Glycine max (Gm), Homo sapiens (Hs), Hordeum vulgare (Hv), Oryza sativa (Os), Physcomitrella patens (Pp), Populus trichocarpa (Pt), Saccharomyces cerevisiae (Sc), Selaginella moellendorfii (Sm), Sorghum bicolor (Sb), Triticum aestivum (Ta), Vitis vinifera (Vv), Zea mays (Zm)) were aligned by multiple sequence alignment (cf. Supplementary Figures S1, S2) using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) that served to cluster the sequences (unweighted pair group method with arithmetic mean). The sequences of plants are indicated in green and those of metazoa in red, while the yeast sequence is labelled blue.
Since SSRP1 contains an HMG-box domain that typically mediates DNA-interactions (Antosch et al., 2012; Malarkey and Churchill, 2012), the DNA-binding properties of maize SSRP1 were examined. These experiments revealed that SSRP1 does not interact with DNA sequence-specifically, but according to a binding-site selection assay, it binds preferentially to sequences containing deformable dinucleotide steps (Röttgers et al., 2000). In line with this finding, mediated by its HMG-box domain SSRP1 can bend linear DNA and bind selectively to certain DNA structures (Röttgers et al., 2000; Pfab et al., 2018b). Furthermore, SSRP1 is phosphorylated by protein kinase CK2 and phosphorylation of two residues C-terminal of the HMG-box domain modulates the structure-specific interaction with DNA (Krohn et al., 2003). The HMG-box domain of SSRP1 is not only important for DNA-binding, but contributes also to nucleosome interactions (Lichota and Grasser, 2001; Pfab et al., 2018b). In view of the relevance of the HMG-box domain for in vitro DNA/nucleosome interactions, it was surprising that based on fluorescence recovery after photobleaching experiments intact SSRP1 and SSRP1 lacking its HMG-box domain (termed SSRP1∆HMG) displayed the same mobility within nuclei of Arabidopsis cells. Beyond that, expression of SSRP1∆HMG was almost as efficient as that of intact SSRP1 in supporting normal growth and development of the otherwise nonviable ssrp1-1 mutant (Pfab et al., 2018b). This suggested that the HMG-box domain, which is conserved among SSRP1 proteins of plants and metazoa, is not critical in Arabidopsis under standard growth conditions. Possibly, FACT containing SSRP1∆HMG (or intact SSRP1) functionally interacts with small abundant HMGB proteins similar to the mechanism reported for yeast FACT. Yeast Pob3 lacks the C-terminal HMG-box domain (thus structurally resembling Arabidopsis SSRP1∆HMG; cf. Figure 1) and the HMG-box function is provided by small Nhp6a/b HMG-box proteins (Brewster et al., 2001; Formosa et al., 2001). However, fusing a C-terminal HMG-box domain to Pob3 is insufficient for full, Nhp6-independent activity. Both yeast FACT containing the Pob3-HMG fusion and human FACT were dependent on the presence of Nhp6 for efficient nucleosome reorganisation (McCullough et al., 2018). Collectively, these findings suggest that SSRP1-SPT16 of plants/metazoa may need assistance of small HMGB proteins in a way analogous to the cooperation of Pob3/SPT16 with Nhp6 in yeast. However, this issue requires further investigations.
Both SSRP1 and SPT16 are nuclear proteins and are ubiquitously expressed in all/most Arabidopsis tissues, but expression is not detectable in certain terminally differentiated cells such as mature trichoblasts or cells of the root cap (Duroux et al., 2004; Ikeda et al., 2011; Pfab et al., 2018a). Consistent with the enrichment of SSRP1 in the highly micrococcal nuclease-sensitive fraction of chromatin (Lichota and Grasser, 2001), both SSRP1 and SPT16 are detected by indirect immunofluorescence microscopy in the euchromatin of interphase nuclei, but not in heterochromatic chromocenters (Duroux et al., 2004). Using chromatin immunoprecipitation SSRP1-SPT16 was detected along the transcribed region of genes transcribed by RNAPII, but not at loci transcribed by RNA polymerases I and III or intergenic regions. Moreover, association with the chromatin of active protein-coding genes occurred in a transcription-dependent manner (Duroux et al., 2004; Perales and Más, 2007; Lolas et al., 2010; Antosz et al., 2017). In accordance with that an affinity-purification approach combined with mass spectrometry demonstrated that FACT efficiently copurified with elongating RNAPII (CTD-phosphorylated at residues S2P, S5P) from Arabidopsis cells as well as with known transcript elongation factors including TFIIS, SPT4/SPT5 and PAF1C (Antosz et al., 2017). Moreover, SSRP1 and SPT16 genetically interact with TFIIS encoding a modulator of RNAPII activity and with HUB1/2, encoding factors catalysing elongation-related mono-ubiquitination of histone H2B (Lolas et al., 2010; Antosz et al., 2017). Taken together these findings indicate a role of Arabidopsis FACT in RNAPII transcriptional elongation (Van Lijsebettens and Grasser, 2014; Zhou et al., 2015; Grasser and Grasser, 2018), in line with the function of FACT as regulator of mRNA synthesis in other organisms (Reinberg and Sims, 2006; Formosa, 2012; Gurova et al., 2018), although the exact mechanism in vivo is still obscure.
Intriguing insight provided a study analysing genome-wide intragenic transcriptional initiation from cryptic promoters in Arabidopsis. Thousands of discrete, mostly exonic transcriptional start site positions were mapped in ssrp1 and spt16 mutants and the majority of these sites were detected in both mutants (Nielsen et al., 2019). This suggested that FACT is required for repression of aberrant intragenic transcript initiation, whereas no evidence was found for an involvement in repression of cryptic transcription by other elongation factors such as PAF1C, Elongator and the SDG8 H3K36-methyltransferase. At FACT-repressed intragenic start sites an enrichment of the RNAPII elongation signature H3K4me1 was detected that may contribute to suppress intragenic transcriptional initiation (Nielsen et al., 2019). Since FACT has been implicated in repressing cryptic transcription also in other organisms (Kaplan et al., 2003; Mason and Struhl, 2003; Cheung et al., 2008; Jamai et al., 2009), ensuring transcriptional fidelity by restricting transcript initiation to promoters may be a key function of FACT.
In various organisms including Arabidopsis, FACT is essential for viability (Cao et al., 2003; Lolas et al., 2010; Formosa, 2012; Frost et al., 2018). Arabidopsis mutant plants expressing reduced amounts of SSRP1 or SPT16 similarly show various defects in vegetative and reproductive development. Thus, the mutant plants display an increased number of leaves and inflorescences as well as altered leaf architecture (Lolas et al., 2010). In addition, these plants are early bolting, have abnormal flower morphology and a severely reduced seed set. The early bolting phenotype is associated with reduced expression of the floral repressor FLC in ssrp1 and spt16 plants relative to the wild type controls (Lolas et al., 2010). Germination assays with freshly harvested seeds demonstrated that in contrast to the wild type control, ssrp1 mutant seeds germinated efficiently without stratification (Figure 3), indicating reduced seed dormancy (Michl-Holzinger et al., 2019). In line with this phenotype, ssrp1 seeds exhibit decreased transcript levels of the DOG1 gene, which is a known quantitative trait locus of seed dormancy. Introduction of an additional copy of DOG1 into ssrp1 resulted in increased DOG1 transcript levels and consistently in more robust seed dormancy (Michl-Holzinger et al., 2019). Therefore, SSRP1 is required for efficient expression of DOG1 and FACT is a modulator of seed dormancy in Arabidopsis. These findings reveal that FACT is involved in two of the most important plant developmental switches, namely, the transition from seed dormancy to germination and from vegetative to reproductive development. Interestingly, both processes in addition to FACT are regulated by other modulators of transcriptional elongation and chromatin structure. Thus, factors including FACT, PAF1C, SWR1, SDG8 and HUB1/2 contribute to the expression of FLC in the induction to flowering (He et al., 2004; Oh et al., 2004; Zhao et al., 2005; Choi et al., 2007; Cao et al., 2008; Lázaro et al., 2008; Lolas et al., 2010), while factors such as FACT, TFIIS, H2B-monoubiquitinases, and H3-methylases influence the expression of DOG1 in the switch from seed dormancy to germination (Liu et al., 2007; Zheng et al., 2012; Molitor et al., 2014; Michl-Holzinger et al., 2019). Furthermore, FACT was identified as cofactor of the transcriptional regulation of circadian rhythms in Arabidopsis. Initially, it was observed that FACT rhythmically associates with the circadian oscillator gene TOC1 (Perales and Más, 2007). Subsequently, protein interactions were detected between FACT, elongating RNAPII and clock-related components termed LNKs. By interaction between LNKs and the MYB factor RVE8 the transcription machinery is recruited to target promoters, leading to rhythmic occupancy of clock gene promoters (Ma et al., 2018). FACT could be involved in this scenario facilitating the transition from RNAPII transcript initiation to productive elongation.
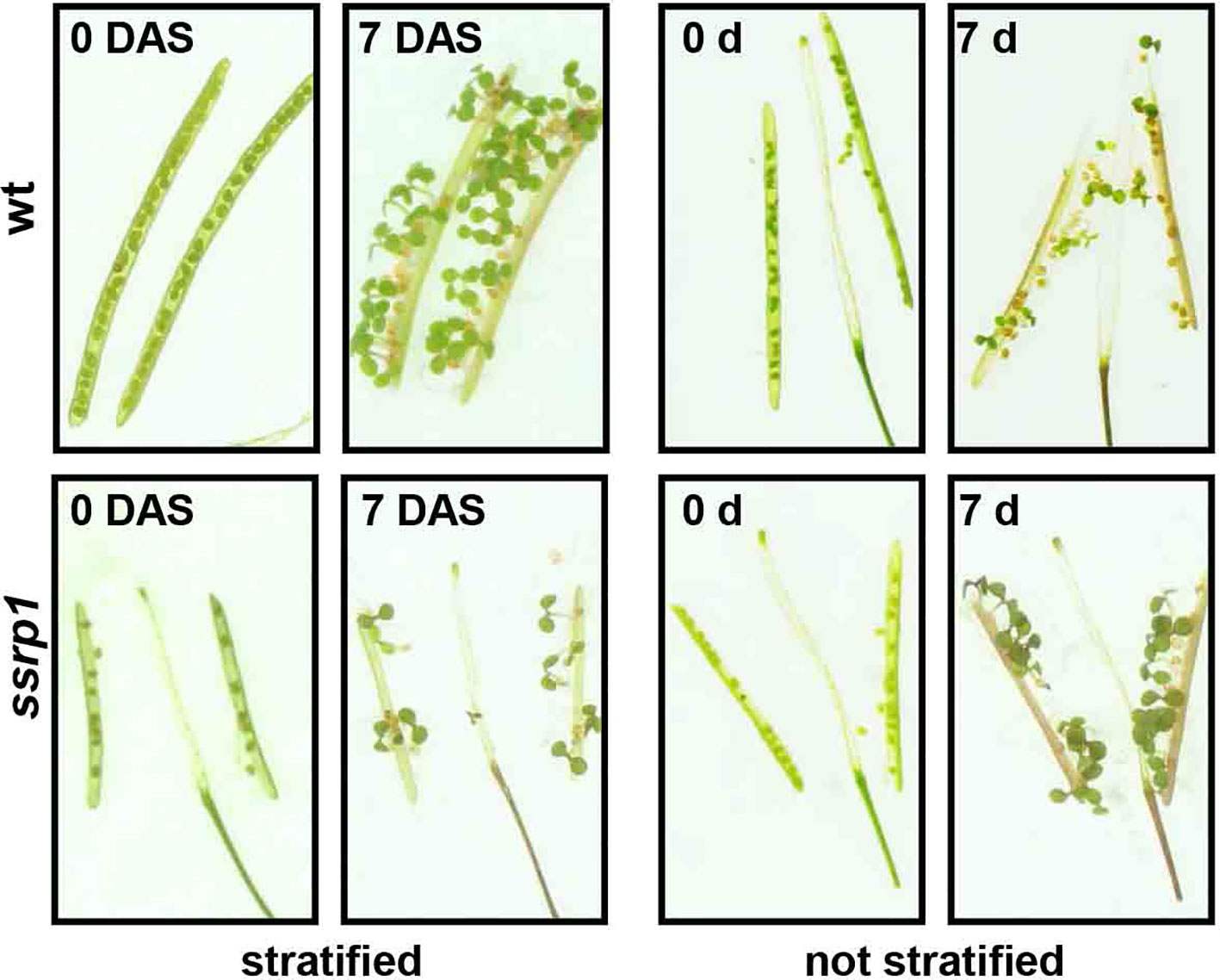
Figure 3 Reduced dormancy of ssrp1 seeds. Germination assays with opened siliques harvested 14 days after flowering. They are shown at day 0 and 7 days after incubation, either with or without prior stratification. Note the smaller siliques of ssrp1 containing a severely reduced number of seeds compared to wt. After stratification almost all seeds germinate, whereas without stratification wt seeds germinate inefficiently (< 50%), whereas ssrp1 seeds due to reduced seed dormancy germinate efficiently (~90%).
Recent transcript profiling of 10-day-old Arabidopsis ssrp1 and spt16 seedlings in comparison to the wild type demonstrated that a relatively small subset of genes is differentially expressed in the mutants (Pfab et al., 2018a). The alterations in the transcript profile of both mutants relative to wild type were very similar, consistent with the function of SSRP1 and SPT16 as a heterodimer. Among the downregulated genes, those encoding enzymes of anthocyanin biosynthesis were remarkably overrepresented. Upon exposure to moderate high-light stress several of the anthocyanin biosynthetic genes were induced in the ssrp1/spt16 plants to a lesser extent than in the wild type, and accordingly the mutant plants depleted in FACT accumulated lower amounts of anthocyanin pigments. Expression of SSRP1 and SPT16 was increased under these conditions (Pfab et al., 2018a). Therefore, FACT is required for transcriptional induction leading to anthocyanin accumulation in response to light stress.
A special role of FACT that was discovered in Arabidopsis is its involvement in parent-of-origin specific gene expression (genomic imprinting). Initially, SSRP1 was found to be required for DNA demethylation and activation/repression of parentally imprinted genes in the central cell of the female gametophyte (Ikeda et al., 2011). The authors proposed that SSRP1 might contribute to altering the chromatin state, facilitating demethylation by the DNA demethylase DEMETER (DME). Subsequently, bimolecular complementation assays indicated that SSRP1 and SPT16 colocalised with DME in the nucleus. Genome-wide analyses demonstrated that SSRP1 and SPT16 are required for demethylation at over half the DME-mediated demethylation sites in the central cell (Frost et al., 2018). DME demethylation sites that are particularly dependent on FACT occur in heterochromatic domains with high nucleosome occupancy and are enriched in H3K9me2 and H3K27me1 marks, whereas euchromatic DME targets apparently can be demethylated by the enzyme without assistance of FACT (Frost et al., 2018). Therefore, FACT may be required for DME targeting by facilitating its access to heterochromatic sites, but the exact molecular role of FACT in this process is unknown. Moreover, the authors suggest that the mode of FACT action in conjunction with DME during reproduction differs from that during transcriptional elongation.
There is substantial evidence that FACT in yeast and metazoa is involved in addition to transcription in various other DNA-dependent processes including replication, recombination and repair. To date essentially the role of plant FACT in transcription has been addressed, and therefore, broader approaches are required to gain insight to which extent it contributes to additional biologically crucial processes in plants. Open questions regarding FACT include how it is recruited to its target sites in chromatin. Analyses in yeast, for instance, indicate that FACT associates with the transcribed regions of all active RNAPII-transcribed genes (Mayer et al., 2010). However, various studies suggest that the absence of FACT causes rather moderate changes in gene expression of relatively small subsets of genes (Gurova et al., 2018). This raises the question, of why the transcription of certain genes is more dependent on FACT than the majority of other genes. Which gene characteristics determine the requirement for FACT action? There exist many potentially influencing parameters including DNA sequence, chromatin structural features, inducibility and expression level of the gene, RNAPII density and elongation rate, as well as cotranscriptional mRNA processing. Perhaps a combination of these and additional parameters defines the requirement of FACT for efficient transcription. Finally, because of the various functions of FACT in nucleosome reorganisation in different biological contexts it appears likely that FACT activity is regulated, but currently this is largely obscure. Although many facts about FACT have been elucidated in recent years, there remain important open questions.
KG wrote the manuscript.
German Research Foundation (DFG) through grants SFB960/A6 and Gr1159/14-2.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author thanks Philipp Michl-Holzinger for providing the images of Figure 3 and critical reading of the manuscript and Simon Obermeyer for bioinformatics analyses. Our research is funded by the German Research Foundation (DFG) through grants SFB960/A6 and Gr1159/14-2.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00085/full#supplementary-material
Antosch, M., Mortensen, S. A., Grasser, K. D. (2012). Plant proteins containing high mobility group box DNA-binding domains modulate different nuclear processes. Plant Physiol. 159, 875–883. doi: 10.1104/pp.112.198283
Antosz, W., Pfab, A., Ehrnsberger, H. F., Holzinger, P., Köllen, K., Mortensen, S. A., et al. (2017). Composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29, 854–870. doi: 10.1105/tpc.16.00735
Avvakumov, N., Nourani, A., Côté, J. (2011). Histone chaperones: modulators of chromatin marks. Mol. Cell 41, 502–514. doi: 10.1016/j.molcel.2011.02.013
Belotserkovskaya, R., Oh, S., Bondarenko, V. A., Orphanides, G., Studitsky, V. M., Reinberg, D. (2003). FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093. doi: 10.1126/science.1085703
Belotserkovskaya, R., Saunders, A., Lis, J. T., Reinberg, D. (2004). Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta 1677, 87–99. doi: 10.1016/j.bbaexp.2003.09.017
Brewster, N. K., Johnston, G. C., Singer, R. A. (1998). Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273, 21972–21979. doi: 10.1074/jbc.273.34.21972
Brewster, N. K., Johnston, G. C., Singer, R. A. (2001). A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21, 3491–3502. doi: 10.1128/MCB.21.10.3491-3502.2001
Cao, S., Bendall, H., Hicks, G. G., Nashabi, A., Sakano, H., Shinkai, Y., et al. (2003). The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol. Cell. Biol. 23, 5301–5307. doi: 10.1128/MCB.23.15.5301-5307.2003
Cao, Y., Dai, Y., Cui, S., Ma, L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20, 2586–2602. doi: 10.1105/tpc.108.062760
Cheung, V., Chua, G., Batada, N. N., Landry, C. R., Michnick, S. W., Hughes, T. R., et al. (2008). Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PloS Biol. 6, e277. doi: 10.1371/journal.pbio.0060277
Choi, K., Park, C., Lee, J., Oh, M., Noh, B., Lee, I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941. doi: 10.1242/dev.001891
Das, C., Tyler, J. K., Churchill, M. E. (2010). The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 35, 476–489. doi: 10.1016/j.tibs.2010.04.001
De Koning, L., Corpet, A., Haber, J. E., Almouzni, G. (2007). Histone chaperones: an escort network regulating histone traffic. Nat. Struct. Mol. Biol. 14, 997–1007. doi: 10.1038/nsmb1318
Duroux, M., Houben, A., Ruzicka, K., Friml, J., Grasser, K. D. (2004). The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 40, 660–671. doi: 10.1111/j.1365-313X.2004.02242.x
Formosa, T., Eriksson, P., Wittmeyer, J., Ginn, J., Yu, Y., Stillman, D. J. (2001). Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20, 3506–3517. doi: 10.1093/emboj/20.13.3506
Formosa, T. (2012). The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255. doi: 10.1016/j.bbagrm.2011.07.009
Frost, J. M., Kim, M. Y., Park, G. T., Hsieh, P. H., Nakamura, M., Lin, S. J. H., et al. (2018). FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E4720–E4729. doi: 10.1073/pnas.1713333115
Grasser, M., Grasser, K. D. (2018). The plant RNA polymerase II elongation complex: A hub coordinating transcript elongation and mRNA processing. Transcription 9, 117–122. doi: 10.1080/21541264.2017.1356902
Gurard-Levin, Z. A., Quivy, J. P., Almouzni, G. (2014). Histone chaperones: assisting histone traffic and nucleosome dynamics. Ann. Rev. Biochem. 83, 487–517. doi: 10.1146/annurev-biochem-060713-035536
Gurova, K., Chang, H. W., Valieva, M. E., Sandlesh, P., Studitsky, V. M. (2018). Structure and function of the histone chaperone FACT - Resolving FACTual issues. Biochim. Biophys. Acta 1861, 892–904. doi: 10.1016/j.bbagrm.2018.07.008
Hammond, C. M., Strømme, C. B., Huang, H., Patel, D. J., Groth, A. (2017). Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158. doi: 10.1038/nrm.2016.159
He, Y., Doyle, M. R., Amasino, R. M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18, 2774–2784. doi: 10.1101/gad.1244504
Henikoff, S. (2008). Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26. doi: 10.1038/nrg2206
Hondele, M., Stuwe, T., Hassler, M., Halbach, F., Bowman, A., Zhang, E. T., et al. (2013). Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114. doi: 10.1038/nature12242
Ikeda, Y., Kinoshita, Y., Susaki, D., Ikeda, Y., Iwano, M., Takayama, S., et al. (2011). HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev. Cell 21, 589–596. doi: 10.1016/j.devcel.2011.08.013
Jamai, A., Puglisi, A., Strubin, M. (2009). Histone chaperone Spt16 promotes redeposition of the original H3-H4 histones evicted by elongating RNA polymerase. Mol. Cell 35, 377–383. doi: 10.1016/j.molcel.2009.07.001
Kaplan, C. D., Laprade, L., Winston, F. (2003). Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099. doi: 10.1126/science.1087374
Kemble, D. J., McCullough, L. L., Whitby, F. G., Formosa, T., Hill, C. P. (2015). FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol. Cell 60, 294–306. doi: 10.1016/j.molcel.2015.09.008
Kornberg, R. D., Lorch, Y. (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryotic chromosome. Cell 98, 285–294. doi: 10.1016/S0092-8674(00)81958-3
Krohn, N. M., Stemmer, C., Fojan, P., Grimm, R., Grasser, K. D. (2003). Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J. Biol. Chem. 278, 12710–12715. doi: 10.1074/jbc.M300250200
Kumar, A., Vasudevan, D. (2020). Structure-function relationship of H2A-H2B specific plant histone chaperones. Cell Stress Chaperones In Press 25, 1–17. doi: 10.1007/s12192-019-01050-7
Lázaro, A., Gómez-Zambrano, A., López-González, L., Piñeiro, M., Jarillo, J. A. (2008). Mutations in the Arabidopsis SWC6 gene, encoding a component of the SWR1 chromatin remodelling complex, accelerate flowering time and alter leaf and flower development. J. Exp. Bot. 59, 653–666. doi: 10.1093/jxb/erm332
Li, B., Carey, M., Workman, J. L. (2007). The role of chromatin during transcription. Cell 128, 707–719. doi: 10.1016/j.cell.2007.01.015
Lichota, J., Grasser, K. D. (2001). Differential chromatin association and nucleosome binding of the maize HMGA, HMGB, and SSRP1 proteins. Biochemistry 40, 7860–7867. doi: 10.1021/bi010548y
Liu, Y., Koornneef, M., Soppe, W. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19, 433–444. doi: 10.1105/tpc.106.049221
Liu, J., Feng, L., Li, J., He, Z. (2015). Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6, 267. doi: 10.3389/fpls.2015.00267
Liu, Y., Zhou, K., Zhang, N., Wei, H., Tan, Y. Z., Zhang, Z., et al. (2020). FACT caught in the act of manipulating the nucleosome. Nature 577, 426–431. doi: 10.1038/s41586-019-1820-0
Lolas, I. B., Himanen, K., Grønlund, J. T., Lynggaard, C., Houben, A., Melzer, M., et al. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61, 686–697. doi: 10.1111/j.1365-313X.2009.04096.x
Luger, K., Richmond, T. J. (1998). DNA binding within the nucleosome core. Curr. Opin. Struct. Biol. 8, 33–40. doi: 10.1016/S0959-440X(98)80007-9
Ma, Y., Gil, S., Grasser, K. D., Mas, P. (2018). Targeted recruitment of the basal transcriptional machinery by LNK clock components controls the circadian rhythms of nascent RNAs in Arabidopsis. Plant Cell 30, 907–924. doi: 10.1105/tpc.18.00052
Malarkey, C. S., Churchill, M. E. (2012). The high mobility group box: the ultimate utility player of a cell. Trends Biochem. Sci. 37, 553–562. doi: 10.1016/j.tibs.2012.09.003
Mason, P. B., Struhl, K. (2003). The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23, 8323–8333. doi: 10.1128/MCB.23.22.8323-8333.2003
Mayer, A., Lidschreiber, M., Siebert, M., Leike, K., Söding, J., Cramer, P. (2010). Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278. doi: 10.1038/nsmb.1903
McCullough, L. L., Connell, Z., Xin, H., Studitsky, V. M., Feofanov, A. V., Valieva, M. E., et al. (2018). Functional roles of the DNA-binding HMGB domain in the histone chaperone FACT in nucleosome reorganization. J. Biol. Chem. 293, 6121–6133. doi: 10.1074/jbc.RA117.000199
McGhee, J. D., Felsenfeld, G. (1980). Nucleosome structure. Ann. Rev. Biochem. 49, 1115–1156. doi: 10.1146/annurev.bi.49.070180.005343
Michl-Holzinger, P., Mortensen, S. A., Grasser, K. D. (2019). The SSRP1 subunit of the histone chaperone FACT is required for seed dormancy in Arabidopsis. J. Plant Physiol. 236, 108. doi: 10.1016/j.jplph.2019.03.006
Molitor, A. M., Bu, Z., Yu, Y., Shen, W. H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PloS Genet. 10, e1004091. doi: 10.1371/journal.pgen.1004091
Nielsen, M., Ard, R., Leng, X., Ivanov, M., Kindgren, P., Pelechano, V., et al. (2019). Transcription-driven chromatin repression of intragenic transcription start sites. PloS Genet. 15, e1007969. doi: 10.1371/journal.pgen.1007969
Oh, S., Zhang, H., Ludwig, P., van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16, 2940–2953. doi: 10.1105/tpc.104.026062
Orphanides, G., LeRoy, G., Chang, C.-H., Luse, D. S., Reinberg, D. (1998). FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116. doi: 10.1016/S0092-8674(00)80903-4
Orphanides, G., Wu, W.-H., Lane, W. S., Hampsey, M., Reinberg, D. (1999). The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288. doi: 10.1038/22350
Otero, S., Desvoyes, B., Gutierrez, C. (2014). Histone H3 dynamics in plant cell cycle and development. Cytogenet. Genome Res. 143, 114–124. doi: 10.1159/000365264
Perales, M., Más, P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19, 2111–2123. doi: 10.1105/tpc.107.050807
Pfab, A., Breindl, M., Grasser, K. D. (2018a). The Arabidopsis histone chaperone FACT is required for stress-induced expression of anthocyanin biosynthetic genes. Plant Mol. Biol. 96, 367–374. doi: 10.1007/s11103-018-0701-5
Pfab, A., Grønlund, J. T., Holzinger, P., Längst, G., Grasser, K. D. (2018b). The Arabidopsis histone chaperone FACT: role of the HMG-box domain of SSRP1. J. Mol. Biol. 430, 2747–2759. doi: 10.1016/j.jmb.2018.06.046
Probst, A. V., Mittelsten Scheid, O. (2015). Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27, 8–16. doi: 10.1016/j.pbi.2015.05.011
Röttgers, K., Krohn, N. M., Lichota, J., Stemmer, C., Merkle, T., Grasser, K. D. (2000). DNA-interactions and nuclear localisation of the chromosomal HMG domain protein SSRP1 from maize. Plant J. 23, 395–405. doi: 10.1046/j.1365-313x.2000.00801.x
Ramirez-Parra, E., Gutierrez, C. (2007). The many faces of chromatin assembly factor 1. Trends Plant Sci. 12, 570–576. doi: 10.1016/j.tplants.2007.10.002
Reinberg, D., Sims, R. J. (2006). de FACTo nucleosome dynamics. J. Biol. Chem. 281, 23297–23301. doi: 10.1074/jbc.R600007200
Singer, R. A., Johnston, G. C. (2004). The FACT chromatin modulator: genetic and structure/function relationships. Biochem. Cell Biol. 82, 419–427. doi: 10.1139/o04-050
Takatsuka, H., Umeda, M. (2015). Epigenetic control of cell division and cell differentiation in the root apex. Front. Plant Sci. 6, 1178. doi: 10.3389/fpls.2015.01178
Tripathi, A. K., Singh, K., Pareek, A., Singla-Pareek, S. L. (2015). Histone chaperones in Arabidopsis and rice: genome-wide identification, phylogeny, architecture and transcriptional regulation. BMC Plant Biol. 15, 42. doi: 10.1186/s12870-015-0414-8
Tsunaka, Y., Fujiwara, Y., Oyama, T., Hirose, S., Morikawa, K. (2016). Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686. doi: 10.1101/gad.274183.115
Valieva, M. E., Armeev, G. A., Kudryashova, K. S., Gerasimova, N. S., Shaytan, A. K., Kulaeva, O. I., et al. (2016). Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 23, 1111–1116. doi: 10.1038/nsmb.3321
Van Lijsebettens, M., Grasser, K. D. (2014). Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 19, 717–726. doi: 10.1016/j.tplants.2014.07.002
Wang, T., Liu, Y., Edwards, G., Krzizke, D., Scherman, H., Luger, K. (2018). The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci. Alliance 1, e201800107. doi: 10.26508/lsa.201800107
Winkler, D. D., Luger, K. (2011). The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286, 18369–18374. doi: 10.1074/jbc.R110.180778
Winkler, D. D., Muthurajan, U. M., Hieb, A. R., Luger, K. (2011). Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J. Biol. Chem. 286, 41883–41892. doi: 10.1074/jbc.M111.301465
Wittmeyer, J., Joss, L., Formosa, T. (1999). Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase a. Biochemistry 38, 8961–8971. doi: 10.1021/bi982851d
Xin, H., Takahata, S., Blanksma, M., McCullough, L., Stillman, D. J., Formosa, T. (2009). yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol. Cell 35, 365–376. doi: 10.1016/j.molcel.2009.06.024
Zhao, Z., Yu, Y., Meyer, D., Wu, C., Shen, W.-H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3K36. Nat. Cell Biol. 7, 1256–1260. doi: 10.1038/ncb1329
Zheng, J., Chen, F., Wang, Z., Cao, H., Li, X., Deng, X., et al. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193, 605–616. doi: 10.1111/j.1469-8137.2011.03969.x
Zhou, W., Zhu, Y., Dong, A., Shen, W.-H. (2015). Histone H2A/H2B chaperones: from molecules to chromatin-based functions in plant growth and development. Plant J. 83, 78–95. doi: 10.1111/tpj.12830
Zhou, K., Gaullier, G., Luger, K. (2019). Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol. 26, 3–13. doi: 10.1038/s41594-018-0166-x
Keywords: SSRP1, Pob3, SPT16, histone chaperone, Arabidopsis, chromatin
Citation: Grasser KD (2020) The FACT Histone Chaperone: Tuning Gene Transcription in the Chromatin Context to Modulate Plant Growth and Development. Front. Plant Sci. 11:85. doi: 10.3389/fpls.2020.00085
Received: 22 November 2019; Accepted: 21 January 2020;
Published: 19 February 2020.
Edited by:
Sara Farrona, National University of Ireland Galway, IrelandReviewed by:
Aline Valeska Probst, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2020 Grasser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Klaus D. Grasser, S2xhdXMuR3Jhc3NlckB1ci5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.