
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 February 2020
Sec. Plant Abiotic Stress
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00046
This article is part of the Research Topic New Insights into Salinity Sensing, Signaling and Adaptation in Plants View all 26 articles
Understanding the regulation of proline metabolism necessitates the suppression of two Δ1-pyrroline-5-carboxylate synthetase enzyme (P5CS) genes performed in switchgrass (Panicum virgatum L.). The results reveal that overexpressing PvP5CS1 and PvP5CS2 increased salt tolerance. Additionally, transcript levels of spermidine (Spd) and spermine (Spm) synthesis and metabolism related genes were upregulated in PvP5CS OE-transgenic plants and downregulated in the PvP5CS RNAi transformants. According to salt stress assay and the measurement of transcript levels of Polyamines (PAs) metabolism-related genes, P5CS enzyme may not only be the key regulator of proline biosynthesis in switchgrass, but it may also indirectly affect the entire subset of pathway for ornithine to proline or to putrescine (Put). Furthermore, application of proline prompted expression levels of Spd and Spm synthesis and metabolism-related genes in both PvP5CS-RNAi and WT plants, but transcript levels were even lower in PvP5CS-RNAi compared to WT plants under salt stress condition. These results suggested that exogenous proline could accelerate polyamines metabolisms under salt stress. Nevertheless, the enzymes involved in this process and the potential functions remain poorly understood. Thus, the aim of this study is to reveal how proline functions with PAs metabolism under salt stress in switchgrass.
Plants frequently encounter various environmental biotic and abiotic stresses (i.e., pathogen infection, herbivore attack, drought, salt, cold, heat and toxic metals) (Zhu, 2016). Salt stress seriously limits plant development and productivity leading to both osmotic and ionic toxicity effects on cells and, eventually, disorganized membranes, non-functional proteins, and accumulation of reactive oxygen species (ROS) (Krasensky and Jonak, 2012; Zhu, 2016). ROS cause serious damage through peroxidation of membrane lipid components. These effects on cells can be mitigated by an active oxygen scavenger (e.g., proline, polyamines, glutathione, etc.) and enzymatic protectors (i.e., superoxide dismutase [SOD], catalase [CAT], ascorbate peroxidase [APX] and glutathione peroxidase [GPX]). SODs act as the first line of defense against ROS while CAT subsequently detoxifies H2O2 (Apel and Hirt, 2004; Krasensky and Jonak, 2012).
In plants, Polyamines (PAs) mainly exist in their free form playing essential roles in cellular function. The putrescine (Put), spermidine (Spd) and spermine (Spm) are the three major forms of polyamines. Putrescine is the central product of the common PAs biosynthetic pathway. It contains two amino groups and is a synthetic precursor of Spd and Spm. Spd and Spm are produced by the sequential addition of aminopropyl moieties to the putrescine skeleton through enzymatic reactions catalyzed by Spd and Spm synthases (SPDS and SPMS). The donor of the aminopropyl groups is decarboxylated S-adenosyl-methionine synthesized from S-adenosyl-methionine by S-adenosyl-methionine decarboxylase (SAMDC). PAs are catabolized by diamine oxidases (DAOs) and polyamine oxidases (PAOs) (Hanfrey et al., 2001; Shao et al., 2012). PAs play important roles in protecting plants against abiotic stress, such as osmotic adjustment, stabilizing membranes to prevent electrolyte leakage and maintaining normal concentrations of reactive oxygen species (ROS) (Bouchereau et al., 1999; Alcázar et al., 2010; Pál et al., 2015; Chen et al., 2018). On the one hand, ectopic expression of SPDS in Arabidopsis thaliana resulted a high concentration of Spd and plants showed resistance to salt, drought and cold stress (Kasukabe et al., 2004). On the other hand, overexpression of carnation SAMDC genes produced more amounts of PAs which increased abiotic stresses in Nicotiana tabacum (Wi et al., 2006). PA accumulation is coupled with an increase in the activity of PA metabolism genes. For instance, under normal condition, transgenic plants overexpressed with Ethylene responsive factor (MfERF1) induced proline enrichment associated with P5CS transcripts which was upregulated compared to those of wild type (Zhuo et al., 2018). Additionally, exogenous PAs treatments could increase proline content under osmotic stress (Pál et al., 2018).
Proline, acting as excellent osmolyte, antioxidative defense and signaling molecule, plays a highly beneficial role under environmental stresses (Hayat et al., 2012).When plants are exposed to stressful conditions, accumulated proline functions in maintaining cell osmotic balance, stabilizes membranes to prevent electrolyte leakage and serves as an antioxidant to regulate the level reactive oxygen species (ROS) (Delauney and Verma, 1993). Additionally, proline plays an important role in plant growth and development (Szabados and Savoure, 2010), particularly in the function of cell cycle transition in maize (Wang et al., 2014). Proline contents are determined by its biosynthesis, catabolism and transport. In plants, proline is generally synthesized through the glutamate pathway during osmotic stress. However, ornithine pathway serves as the other pathway to proline synthesis (Hu et al., 1992). In glutamate pathway, proline is produced from glutamate by Δ1-pyrroline-5-carboxylate synthetase (P5CS) and Δ1-pyrroline-5-carboxylate reductase (P5CR) enzymes (Hu et al., 1992). In the ornithine pathway, ornithine is transmitted by ornithine-delta-aminotransferase (OAT) producing GSA and P5C which is, thereafter, converted to proline (Roosens et al., 1998). Proline catabolism occurs in mitochondria when proline dehydrogenase (PDH) and P5C dehydrogenase (P5CDH) convert proline to glutamate (Verslues and Sharma, 2010). Intercellular transport of proline occurs in different cellular organelles as exhibited by the compartmentalization of proline metabolism. Proline biosynthesis enzymes (i.e., P5CS1, P5CS2, and P5CR) are predicted to be localized in the cytoplasmic whereas the proline catabolism enzymes (i.e., ProDH1, ProDH2, P5CDH, and OAT) are predicted to be localized in the mitochondria (Szabados and Savoure, 2010). In Arabidopsis, a p5cs1 mutant showed hypersensitivity to salt stress. Additionally, p5cs2 mutations cause embryo abortion during the latter stages of seed development. Thus, P5CS enzymes perform non-redundant functions (Székely et al., 2008). However, the transcriptional pattern of P5CS family genes varies among different species. For example, in Brassica napus, Phaseolus vulgaris and Oryza sativa, two P5CS family genes were induced by different stresses (Hur et al., 2004; Xue et al., 2009; Chen et al., 2013; Sripinyowanich et al., 2013). Furthermore, MtP5CS3, the third gene in Medicago truncatula, similar to the other two MtP5CS genes, also plays a crucial role in regulating proline accumulation under salinity stress (Kim and Nam, 2013). However, in switchgrass, the extent and functional status of P5CS family genes is not clear. Switchgrass (Panicum virgatum L.) is a perennial, warm season C4 model grass native to North America recognized as a dedicated bioenergy crop with significant tolerance to abiotic stresses (e.g., heat, cold, and draught) (Casler et al., 2007; Keshwani and Cheng, 2009). This research determined that the functional identification of two P5CS genes from Panicum virgatum, designated as PvP5CS1 and PvP5CS2, ectopically expressed lines in switchgrass and described their phenotypic and physiological characterization.
Switchgrass (Alamo, lowland-type, 2n = 4× = 36) was used as wild type for expression profiling and transformation of PvP5CS1 (Pavir.J02344.1) and PvP5CS2 (Pavir.J06546.1). Plants were grown in a greenhouse under either a 16h light or 8h dark photoperiod at 25 ± 2°C. According to the description of Hardin, developmental stages of switchgrass are divided into six elongation stages (i.e., E0, E1, E2, E3, E4, and E5) and three reproductive stages (i.e., R1, R2, and R3) (Hardin et al., 2013). At the first stage of elongation, there in none internode, so it is defined as E0. When switchgrass grows the internode, E1 to E5 are defined by the number of internodes present. At the beginning of the reproductive stage, flag leaf comes out, and it is defined as R1. When switchgrass has the fully emerged spikelets without peduncle, it arrived at R2 stage. The R3 stage is defined by fully emerged spikelets and visible peduncle. Plant height, tiller number, leaf length and internode diameter were measured at R1 stage. Each line had three biological replicates, and three tillers were measured in each biological replicate. The harvested above-ground tissues were dried in an oven at 80°C for 72h.
Based on the amino acid sequence of AtP5CS1 (NM_001202786.1) and AtP5CS2 (NM_115419.5), the researchers BLAST-searched the corresponding genes in the Switchgrass Genome Annotation Program (https://phytozome.jgi.doe.gov/pz/portal.html) and found the full-length cDNA sequences of switchgrass PvP5CS1 and PvP5CS2. For the phylogenetic analysis, full-length sequences of P5CS genes from different monocotyledon and dicotyledon plants were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/), the amino acid sequences of two P5CS genes from switchgrass and all those other species, were used to input in the software MEGA 5.0 to do the phylogenetic analysis. DNAMAN software was used to do a homology analysis of two PvP5CS genes. Their full-length cDNA sequences were used to do multiple sequence alignment, and the result was outputted in a form of EMF file. To further analyze the conserved domains of two PvP5CS genes, the researcher inputted their amino acid sequences into the website of NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?) and gained the results of conserved domains.
For overexpression transformants, cDNA sequences of PvP5CS1 and PvP5CS2 were amplified, were fused to binary vector Ubi1301 under ZmUbi1 promoter. To knockdown both PvP5CS1 and PvP5CS2, an RNAi fragment was amplified by PCR from the conserved regions of PvP5CS1 and PvP5CS2 and cloned into pVT1629 vector (Xu et al., 2011). The recombinant binary vector were introduced into the embryogenic callus (Liu et al., 2015). Wild type (WT) is the control plants. All primers are provided under Table S1.
P5CS activity was measured following the protocol of the P5CS ELISA Kit (Suzhou Comin Biotechnology Co. Ltd, China). For calibration purpose standard gradient was measured at 450 nm using a spectrophotometer to make a standard curve. Stop solution was used to change the color from blue to yellow and the intensity of the color was measured. ProDH activity was measured following the protocol prescribed by the kit (Suzhou Comin Biotechnology Co. Ltd, China). ProDH catalyzed dehydrogenation was detected by methyl isothiocyanate and measured at 600nm using a spectrophotometer.
Proline was measured following established protocol (Bates et al., 1973) with minor modifications. Briefly, the fresh sample was weighed and cut into pieces. After adding 5ml 3% sulfur-salicylic acid in a 10ml centrifuge tube, it was boiled for 10min, and the supernatant was used for the proline extract. Two milliliter supernatant was reacted with 2ml glacial acid and 3ml acid ninhydrin, boiling for 45min. when the temperature of reaction mixture reached to room temperature, extracting with 5ml toluene. The supernatant was measured at 520nm, and proline content was calculated from a standard curve.
Total RNA was isolated from the leaves at different stages (i.e., elongation and reproductive stages) by the TRIzol reagent method (Invitrogen, Carlsbad, CA, USA) then subjected to reverse transcription with the PrimeScript RT reagent Kit (Takara, Shiga, Japan). SYBR Green (Takara, Shiga, Japan) was used as the reporter dye. Switchgrass ubiquitin1 (PvUBQ1) gene was used as the internal control. The relative expression levels of genes were calculated using the 2−△△CT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are provided under Table S1.
Salt treatment was performed according to (Jha et al., 2013). Briefly, the I3 leaves at R1 stage were cut into 4cm long pieces and soaked in either 0mM or 350mM NaCl solution for 30 days. Phenotypic changes of the leaf pieces were recorded. Furthermore, whole transgenic and WT plants with salinity stress in sand culture were also treated. Uniform tillers of both transgenic and control plants were treated with 1/2 × Hoagland nutrient solution supplemented with 0mM or 350mM NaCl, respectively, refreshed every two days. After 15 days, the researchers measured the physiological indexes (i.e., relative water content [RWC], electrolyte leakage [EL], chlorophyll, proline, Na+ and K+ contents, and root vigor). RWC and EL were measured following the methods (Li et al., 2010). Na+, K+ contents, and chlorophyll were measured following the methods as previously described (Li et al., 2010). Root vigor was evaluated following established protocol using the method of TTC (2, 3, 5-Tripheyl Tetrazolium Chloride) (Kausar et al., 2009). The roots were immersed and placed in the dark for 1 ~ 3h at 37°C, then they were ground with 3 to 4ml ethyl acetate and a small amount of quartz sand. Finally, the liquid was measured with spectrophotometer at wavelength of 485nm. Additionally, we also applied exogenous proline (10mM) to the RNAi transgenic plants to determine whether external proline could rescue their salt-sensitive phenotypes. SAMDC, SPDS and SPMS are the enzymes for PAs synthesis, and PAs are catabolized by PAO (Hanfrey et al., 2001; Shao et al., 2010). Based on the amino acid sequence of AtSAMDC (AT3G02470), AtSPDS (AT1G70310), AtSPMS (AT5G53120) and AtPAO (NM_105256.4) (Urano et al., 2003), the researchers BLAST-searched them in the Switchgrass Genome Annotation Program and found the full-length cDNA sequences of switchgrass PvSAMDC, PvSPDS, PvSPMS and PvPAO. Then we designed the primers used for qRT-PCR, and primers are provided under Table S1. Finally, we measured the relative expression level of PvSAMDC (Pavir.Ab02264.1), PvSPDS (Pavir.Ba02363.1), PvSPMS (Pavir.Ab01042.1) and PvPAO (Pavir.J39070.1) in the transgenic and WT plants.
H2O2 accumulation was evaluated by 3, 3ʹ-Diaminobenzidine (DAB) assay as described previously (Fryer et al., 2002). Additionally, H2O2 quantification was measured according to the method (Zhang et al., 2014). PvCAT and PvSOD play crucial role in ROS scavenging when plants experienced salt stress. Based on the amino acid sequence of AtCAT (AT1G20630) and AtSOD (AT1G08830) (Du et al., 2017), the researchers BLAST-searched them in the Switchgrass Genome Annotation Program and found the full-length cDNA sequences of switchgrass PvCAT (Pavir.J03636.1) and PvSOD (Pavir.Ib01670.1). Then we designed the primers used for qRT-PCR, and primers are provided under Table S1. Finally, we measured the relative expression level of PvCAT and PvSOD in the transgenic and WT plants.
Data from each trait were subjected to analysis of variance (ANOVA). The significance of treatments was tested at the level of P < 0.05. Standard errors were provided in all tables and figures when needed. All statistical analyses were performed using the SPSS statistical software package (SPSS 20.0, IBM Company, USA).
To understand how PvP5CSs function in switchgrass, we BLAST-searched the switchgrass genome referring to Arabidopsis sequences in the Phytozome version 12.1 and identified two putative PvP5CS candidate genes. Amino acid sequences of P5CS from NCBI were used to construct the phylogenetic tree. The cladogram was divided into three major groups (Figure S1). P5CS from monocots are clustered into two distinct groups (i.e., P5CS1 and P5CS2 groups), the result showed that PvP5CS1 is clustered into P5CS1 and PvP5CS2 is clustered into P5CS2. Next, the result of multiple sequence alignment showed that the sequence homology is 80.66% between AtP5CS and PvP5CS genes. Additionally, the conserved domains analysis showed that PvP5CS2 has a specific hit that not existed in PvP5CS1, it is AAK_P5CS_ProBA (Figure S1), similar in AtP5CS genes. For further analyze the difference between the two PvP5CS genes, we described PvP5CS1 and PvP5CS2 using temporal and spatial expression patterns by reverse transcriptase quantitative PCR (RT-qPCR) (Figure 1). The results showed that PvP5CS1 highly expressed in the stem and panicle while PvP5CS2 highly expressed in the roots and panicle. Additionally, PvP5CS1 was expressed higher at E0, E1, and E3 stages; while PvP5CS2 showed higher expression at E4, R1 and R3 stages (Figures 1A, B). These results showed that the two PvP5CS genes have different expression patterns in the growth and development of switchgrass. To evaluate if the P5CS are related to salt tolerance, expression levels of two PvP5CS genes during different salt concentrations treatment were examined. The results showed that the expression level of both genes peaked at 350mM NaCl concentration. Besides, expression levels of PvP5CS1 were higher than that of PvP5CS2 at four different salt stress concentrations (Figure 1C). To further clarify the additional influence of salt stress on transcripts of PvP5CSs, the expression patterns of two PvP5CS genes at various time points under 350mM NaCl treatment were investigated and the results demonstrated that the expression levels of PvP5CS1 and PvP5CS2 were significantly induced by salt treatment (Figure 1D) thereby suggesting PvP5CSs response and function in salt treatment.
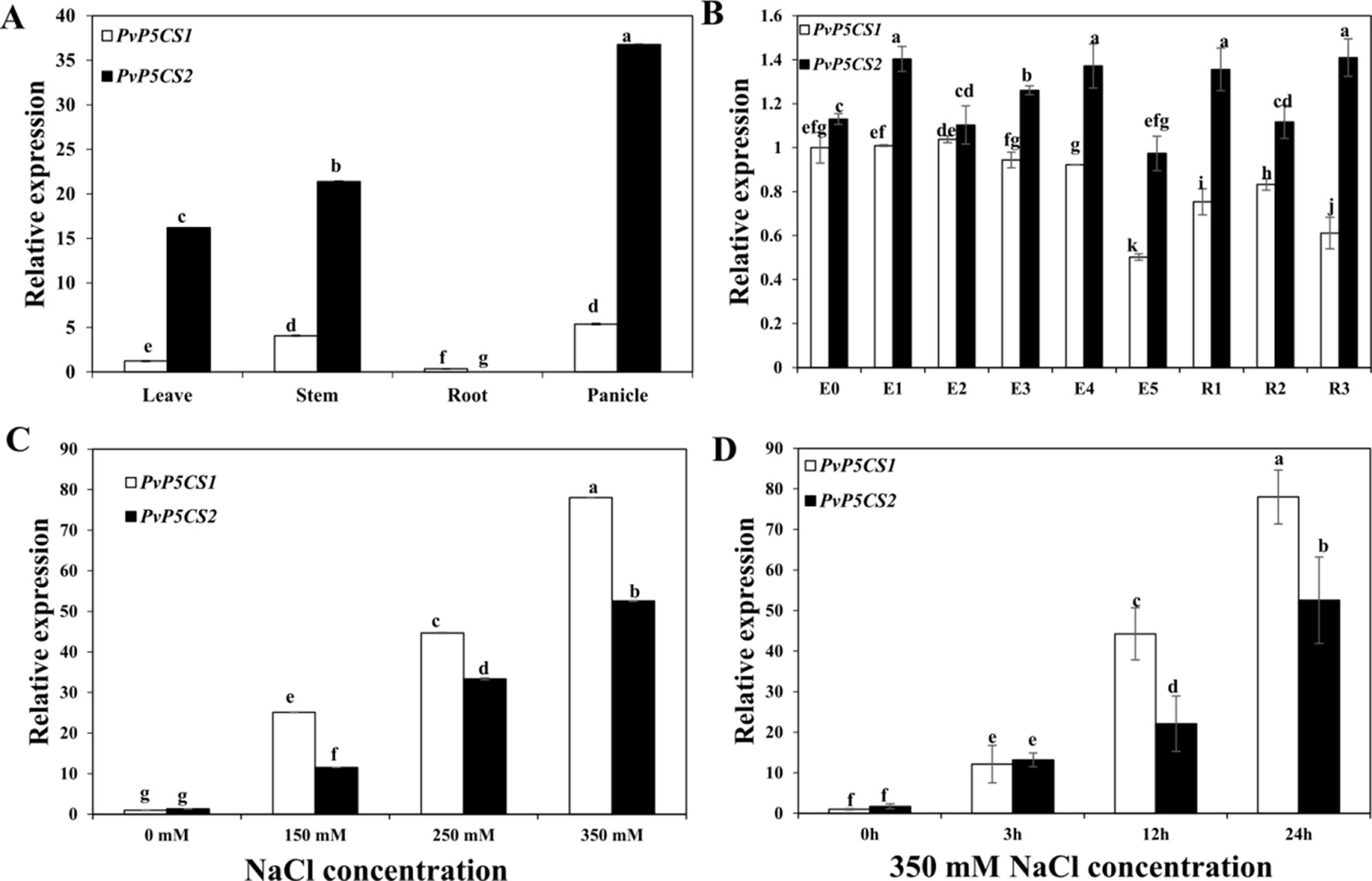
Figure 1 Expression pattern of two PvP5CS genes (PvP5CS1 and PvP5CS2) in switchgrass. Relative expression level of two PvP5CS genes in different tissues (A). Relative expression level of two PvP5CS genes in different development stages (B). Analysis of PvP5CS genes expression under different NaCl concentrations (0 mM, 150mM, 250mM, and 350mM) after 24h salt treatment (C). Transcripts of PvP5CS genes under 350 mM NaCl at the indicated time points (D). Switchgrass UBQ1 was used as the reference for normalization. At the first stage of elongation, there in none internode, so it is defined as E0. When switchgrass grows the internode, E1 to E5 are defined by the number of internodes present. At the beginning of the reproductive stage, flag leaf comes out, and it is defined as R1. When switchgrass has the fully emerged spikelets without peduncle, it arrived at R2 stage. The R3 stage is defined by fully emerged spikelets and visible peduncle. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test). WT: wild type control.
To evaluate the physiological functions and mechanisms of two PvP5CS genes, PvP5CS1 and PvP5CS2 overexpression lines and PvP5CS-RNAi lines were generated. A total of 26 transgenic lines (T0 generation) overexpressing PvP5CS1, 20 transgenic lines (T0 generation) overexpressing PvP5CS2 and 39 PvP5CS-RNAi lines (T0 generation) were generated and tested as positive by genomic PCR analysis (Figure S2). In relation to WT plants, the expression levels of PvP5CS1 and PvP5CS2 were significantly higher in the most of PvP5CS1 and PvP5CS2 overexpression lines except OE-PvP5CS1 line 6, and that were lower in the all PvP5CS-RNAi lines (Figure S3). According to their expression levels, the researchers selected three independent PvP5CS1 overexpression lines (designated as OE-2, OE-14, and OE-19, respectively), three PvP5CS2 overexpression lines (designated as OE-5, OE-6, and OE-10, respectively), and six independent RNAi lines (designated as RNAi-2, RNAi-7, RNAi-9, RNAi-11, RNAi-18, and RNAi-32, respectively) for further analysis. The selected OE and RNAi transgenic lines used for the experiment were labeled with the stars in the Figure S3. The enzyme activity of P5CS and proline content were higher in the PvP5CS overexpression lines but lower in PvP5CS-RNAi lines when compared to the WT plants (Figures 2A, C). The results showed that PvP5CS was highly expressed in the PvP5CS overexpression lines and downregulated in the PvP5CS-RNAi lines. Consequently, to further evaluate the effect of P5CS enzyme on proline metabolism, ProDH activity was tested in PvP5CSs overexpressing and in RNAi-PvP5CS lines. The result showed that the enzyme activity of ProDH was higher in the PvP5CS-RNAi lines and lower in the PvP5CS1 and PvP5CS2 overexpression lines. Thus, the proline metabolism is changed both in the overexpression lines and RNAi transgenic plants (Figure 2B).
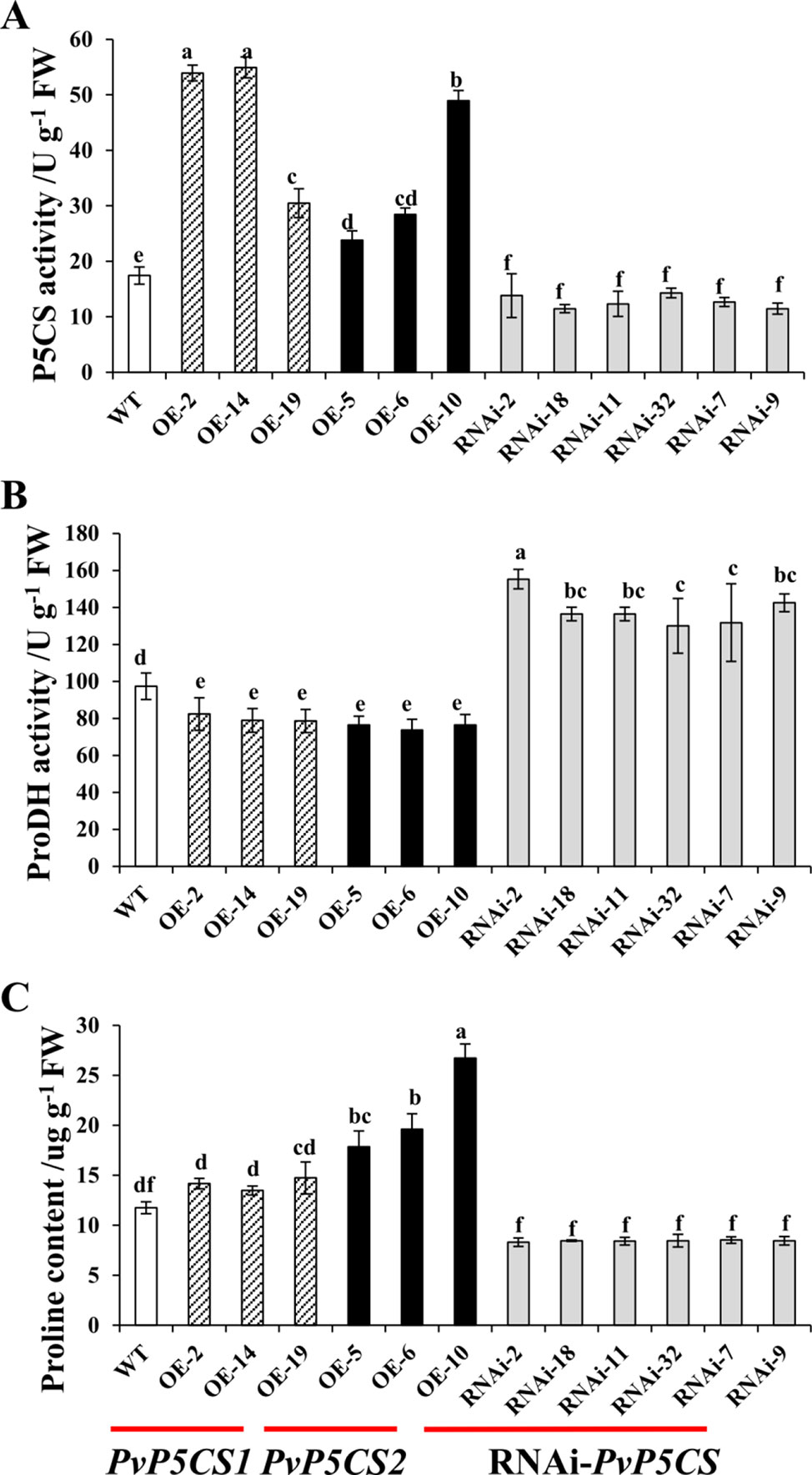
Figure 2 P5CS, ProDH activity and proline content in PvP5CS OE and RNAi transgenic plants. P5CS (A) and ProDH (B) activity and Proline content (C) in PvP5CS OE and RNAi transgenic plants. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
Interestingly, PvP5CS overexpression lines grew faster in greenhouses than WT plants. Phenotypic traits (i.e., plant height, tiller number, leaf length, internode length, and diameter) were measured in transgenic and WT plants (Figures 3A, B; Table S2). When compared to the WT, PvP5CS1 and PvP5CS2 overexpression lines showed on average 38.2% and 37.3% higher plant height, 30.3% and 21.5% more number of tillers, 51.1% and 34.9% longer leaf length, 58.1% and 47.4% wider internode diameter and 36.3% and 23.8% increase in internode length, respectively. The dry weight biomass of plants at five months old was measured and the result showed the biomass of PvP5CS1 overexpression lines were 140–180% higher than WT plants while PvP5CS2 overexpression lines were a 68–98% higher than WT (Figure 3C). Thus, both overexpressing PvP5CS1 and PvP5CS2 genes improve the biomass in transgenic switchgrass. In addition, it was found that the flowering time is different between transgenic and WT plants. The data showed that flowering time of PvP5CS2 overexpression lines are shorter by an average of 30 days than WT plants while no obvious difference was manifested for the PvP5CS1 overexpression lines (Figure 3D). To further evaluate the flowering related genes, we identified two flowering-related genes which are PvFT and PvFLC and the expression level of PvFT was higher in OE-PvP5CS2 transgenic plants. However, the expression level of PvFLC was lower in both PvP5CS1 and PvP5CS2 overexpression lines (Figure S4). These results suggested that PvP5CS2 gene may play an important role in regulating the flowering time in Switchgrass.
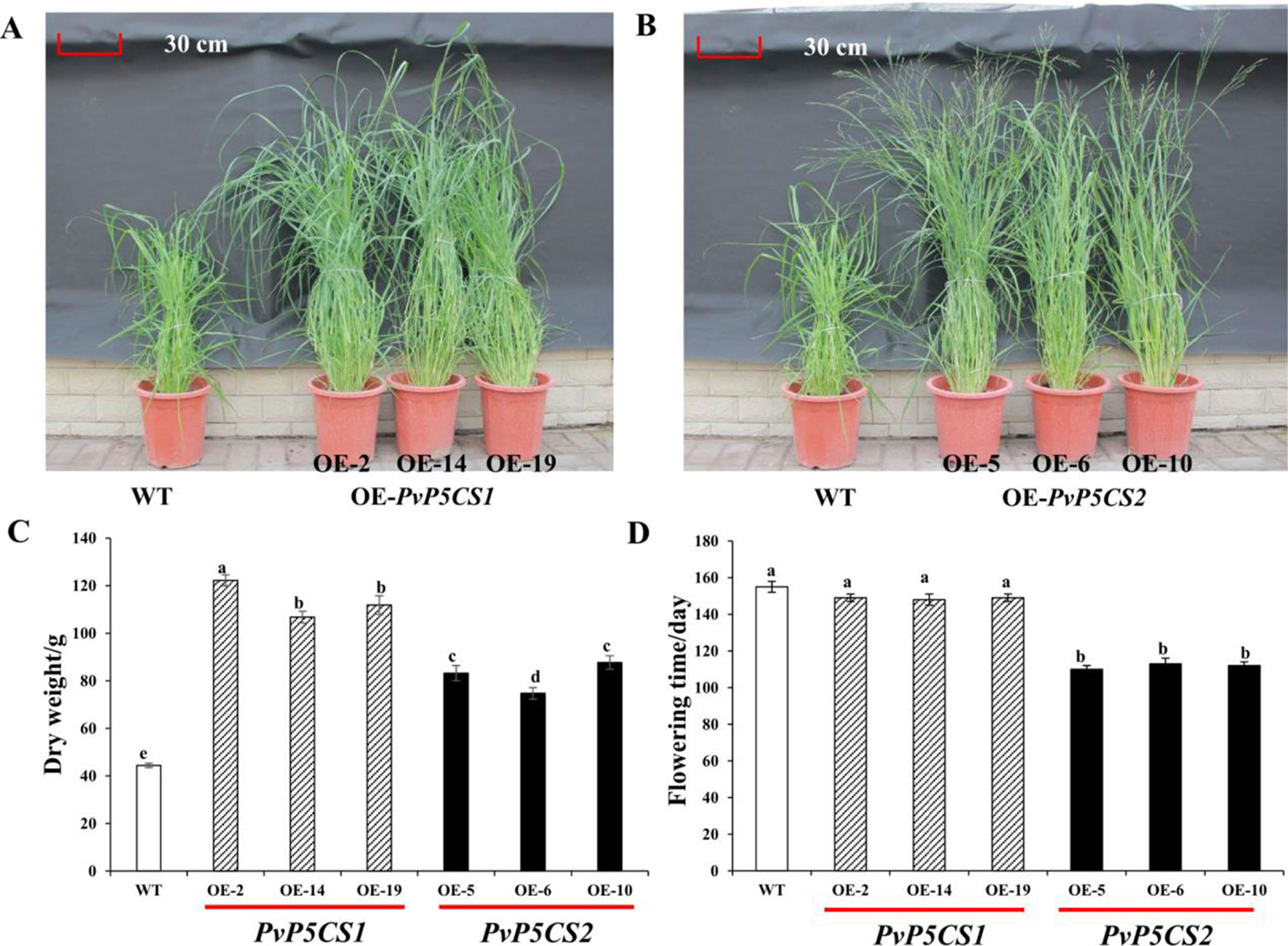
Figure 3 Phenotypic comparation and biomass analysis among PvP5CS OE transgenic plants. Phenotypes of three independent transgenic lines overexpressing PvP5CS1 (A) and PvP5CS2 (B). Total above ground dry weight of transgenic lines overexpressing PvP5CS genes (C). Flowering time in OE-PvP5CS1 and OE-PvP5CS2 transgenic switchgrass lines (D). Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
Based on the previous results that the expression levels of PvP5CSs were upregulated under salt stress, it was theorized that the two PvP5CS genes may play the significant role in salt stress response. To verify this hypothesis, salt tolerance experiments were performed on PvP5CS overexpression and RNAi transgenic switchgrass. After 30 days, leaves of the WT plants almost turned yellow with the overexpression lines losing their green color under 350mM NaCl salt stress (Figure S5). Taken together, PvP5CS1 and PvP5CS2 overexpression lines are involved in salt tolerance. Additionally, this result is supported by less chlorophyll bleaching in the cut-leaf float assay while large areas of leaves of RNAi transgenic lines lose their green color. However, there are no obvious phenotypic difference between the WT, PvP5CS overexpression and RNAi transgenic plants under normal growth conditions (Figure 4A). Only when treated with 350mM NaCl, the WT and PvP5CS-RNAi transgenic plants showed serious leaf damage after 15 days whereas PvP5CS OE-transgenic lines retained its green color and showed better performance (Figure 4B). This suggested that the PvP5CS1 and PvP5CS2 genes are involved in salt tolerance.
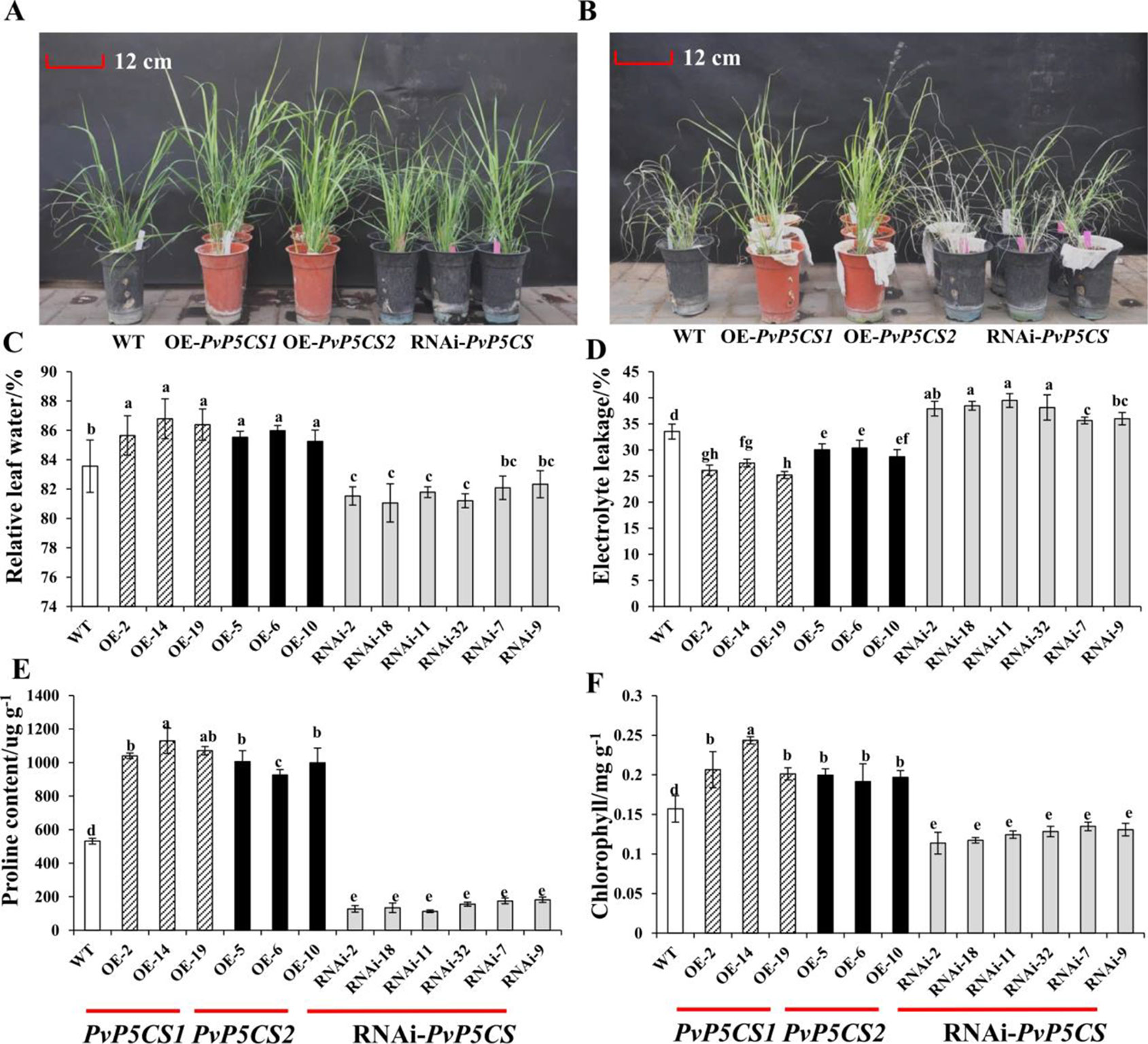
Figure 4 The salt tolerant evaluation of PvP5CS OE and RNAi transgenic plants under salt treatment. Phenotypes of PvP5CS OE and RNAi transgenic plants under 0 mM NaCl (A) and 350 mM NaCl (B) treatment for 15 days. Relative leaf water content (C), Electrolyte leakage (D), proline (E) and Chlorophyll (F) of transgenic lines OE-PvP5CS1, OE-PvP5CS2 and RNAi-PvP5CS in (B) after 350 mM NaCl treatment for 15 days. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
To see the effect of salt stress on plasma membrane integrity and membrane permeability, we measured the relative water content and electrolyte leakage. The result showed, there was no difference in RWC and EL in all lines under normal condition (Figures S6A, B). However, under salt-stressed condition, PvP5CS overexpressing lines were shown to contain less RWC and EL than the WT and PvP5CS-RNAi transgenic plants (Figures 4C, D). Meanwhile, Chlorophyll content was indistinguishable between WT and PvP5CS1 overexpression transgenic plants under normal condition while RNAi transgenic plants had relatively lower chlorophyll content (Figure S6C). When treated by salt stress, chlorophyll content decreased sharply in RNAi transgenic plants compared to WT plants (Figure 4F). Additionally, it was found that ion imbalance, especially intracellular K+ and Na+, directly affect carbon fixation and biomass production in plants under salt stress. Shoots Na+ and K+ contents in transgenic and WT plants were measured. Na+ and K+ contents in shoots were similar between transgenic and WT plants under normal conditions. When treated with 350mM salt stress, RNAi transgenic lines accumulated more Na+ but lower K+ content. Na+ content was lower and K+ content was higher in the PvP5CS overexpression transgenic lines relative to WT plants (Figure S7). Enrichment of proline significantly altered salt stress response which also happened in PvP5CS overexpression lines. Proline content in PvP5CS overexpression lines showed approximately a 2-fold increase comparing to WT, but around 4-fold decrease in RNAi transformants (Figure 4E). These results suggest that upregulated expression of PvP5CS1 and PvP5CS2 is critical for salt stress tolerance.
Salt stress leads to serious damage to roots thereby affecting the growth and arrangement of roots (Julkowska and Testerink, 2015). To evaluate whether the PvP5CS genes have relationship with root development during salt stress, the growth situation of transgenic plants was tested. The roots of PvP5CS1 and PvP5CS2 overexpression transgenic lines survived under salt stress. In contrast, the growth of roots in RNAi transgenic lines were suppressed and a different arrangement was formed (Figure 5A). To further analyze the extent of root damage under salt stress, the root vigor was measured. Root vigor of overexpression transgenic lines was higher than the ones in WT plants. An approximate increase of 21% in PvP5CS1 overexpression transgenic lines and 17% in PvP5CS2 overexpression transgenic lines were observed, while the PvP5CS-RNAi transgenic plants showed an approximate 43% decrease compared to WT plants (Figure 5B). Moreover, to see the relationship between salt stress with Na+ and K+, we measured the Na+ and K+ contents in roots and the result revealed no difference in Na+ and K+ contents in roots in normal condition. However, in those under the salt-stressed conditions, Na+ accumulation in roots increased significantly in all transgenic plants and WT while the K+ levels in roots started to decline (Figures 5C, D). In relation to the wild type, the increase in Na+ content was lower by more than 32% and 23% in the roots of PvP5CS1 and PvP5CS2 overexpression transgenic plants, respectively. Conversely, the increase in Na+ content was higher by more than 28% in the roots of RNAi transgenic lines (Figure 5C). Similarly, the decrease in the K+ content was faster by more than 7% in the roots of RNAi transgenic lines while the decrease in the K+ content was slower by 13% and 14% in the roots of PvP5CS1 and PvP5CS2 transgenic plants, respectively, compared to WT plants (Figure 5D). These results indicated that there is a significant change of Na+ and K+ contents between transgenic and WT plants. These further suggest that PvP5CS1 and PvP5CS2 overexpression transgenic lines reduced root damage by regulating Na+ and K+ contents under salt stress.
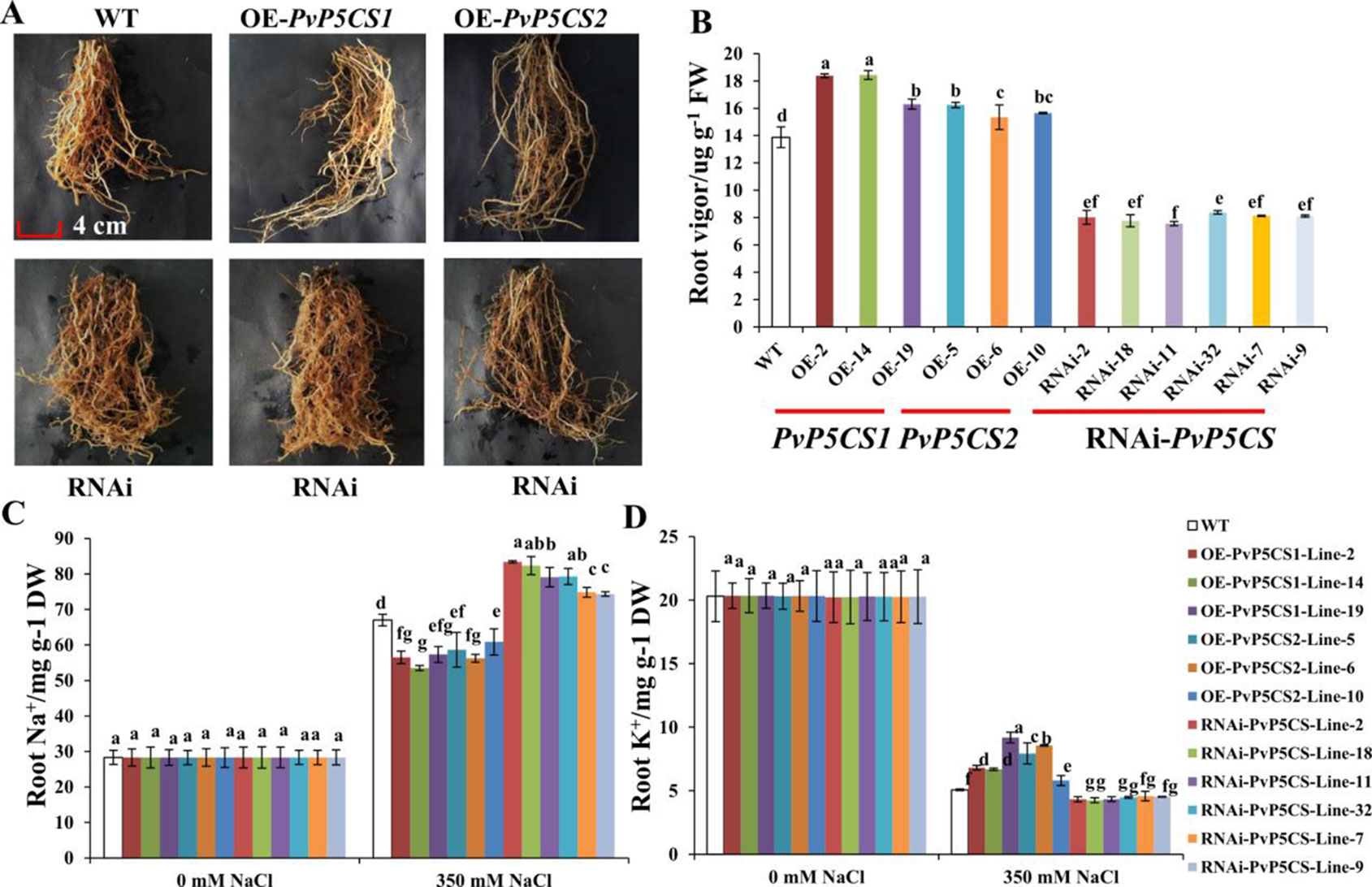
Figure 5 Phenotype and ion (Na+ and K+) contents of roots in PvP5CS OE and RNAi transgenic plants under salt stress. Phenotype of roots in PvP5CS OE and RNAi transgenic plants under 350 mM NaCl salt stress (A). Root vigor in PvP5CS OE and RNAi transgenic plants under 350 mM NaCl salt stress (B). Na+ (C) and K+ (D) contents of roots in PvP5CS OE and RNAi transgenic plants under 350 mM NaCl salt stress. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
ROS is one of the most important indicators of salt response. It can be analyzed by histochemical staining with DAB. Histochemical results did not show significant brown precipitations in all transgenic and WT plants under normal conditions (Figure 6A). Conversely, under salt-stressed conditions, the intensity of brown precipitations in WT leaves was stronger than those in PvP5CS OE-transgenic plants, but weaker than in RNAi transgenic plants (Figure 6B). To verify the histochemical staining, the levels of H2O2 were quantified using a detection kit. In consonance with the histochemical staining results, quantitative measurement showed that H2O2 levels were similar between the transgenic and WT plants under normal conditions (Figure 6C). While the levels of H2O2 in OE-transgenic lines were significantly lower than those in WT plants during the salt treatment, the H2O2 content in RNAi transgenic plants were significantly higher than in WT plants (Figure 6D). These results demonstrate the function of PvP5CSs in salt tolerance through ROS pathway. To confirm whether accumulation of ROS causes plant self-defensive ability, the transcript levels of two important enzyme genes (i.e., PvCAT and PvSOD) which play crucial role in ROS scavenging when plants experienced salt stress were examined. The transcription levels of PvCAT and PvSOD in PvP5CS OE-transgenic lines had no significant difference with WT under normal conditions. Meanwhile, the expression levels of PvCAT and PvSOD were significantly decreased in PvP5CS-RNAi transgenic plants compared to those in WT. Under salt stress, expression levels of PvCAT and PvSOD were significantly upregulated in PvP5CS OE-transgenic lines, but downregulated in PvP5CS-RNAi transgenic plants compared to WT plants (Figures 6E, F).
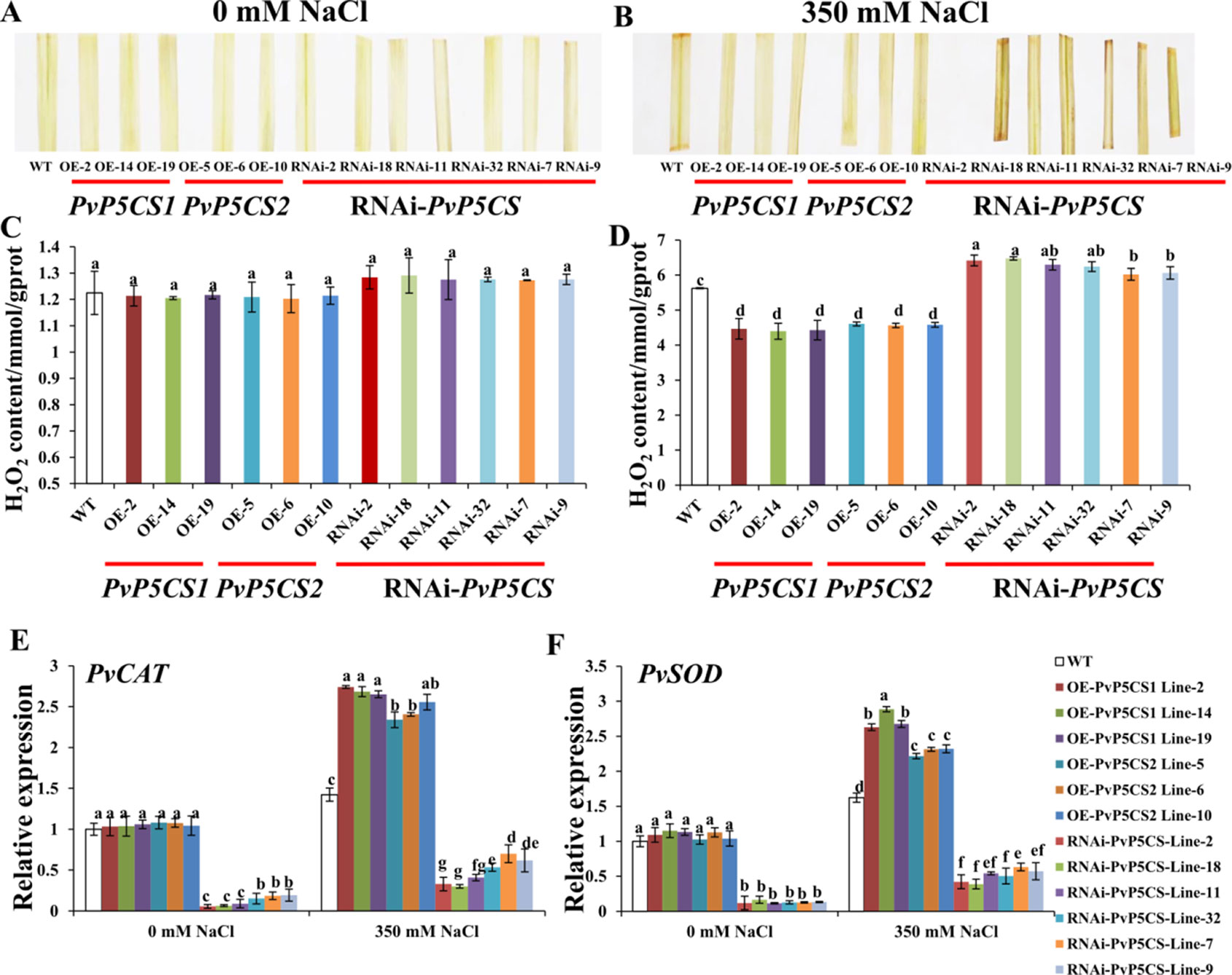
Figure 6 Overexpression of PvP5CS genes in switchgrass positively regulates the induction of ROS scavengers and membrane transporters, and reduces H2O2 accumulation under 350 mM NaCl treatment. Color of detached leaves after staining with diaminobenzidine (DAB) in wild type, PvP5CS OE and RNAi transgenic plants under 0 mM (A) and 350 mM (B) NaCl treatment. H2O2 content in the leaves of wild type, PvP5CS OE and RNAi transgenic plants under 0 mM NaCl (C) and 350 mM NaCl (D) treatment. Relative expression levels of PvCAT (E) and PvSOD (F) in wild type, PvP5CS OE and RNAi transgenic plants under 0 and 350 mM NaCl treatment. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
The researchers then further elucidated the underlying physiological mechanism relative to enhanced salt tolerance in the PvP5CS OE-transgenic plants. Polyamine is involved in adaptation to environmental signals (Alcázar et al., 2010). SAMDC, SPDS and SPMS are the enzymes for PAs synthesis, and PAs are catabolized by PAO (Hanfrey et al., 2001; Shao et al., 2010).The expression levels of these genes were examined by qRT-PCR. PvSAMDC, PvSPDS, PvSPMS, and PvPAO were all upregulated in PvP5CS1 overexpression transgenic lines under normal conditions, but only PvSPMS and PvPAO genes were elevated in the PvP5CS2 overexpression transgenic lines with all other genes decreasing in PvP5CS-RNAi transgenic plants compared to WT plants (Figure 7). Furthermore, Spd and Spm synthesis and metabolism related genes were induced by salt treatment in all plants, but the elevation in PvP5CS OE transgenic lines was much higher than that in WT plants while the opposite happened in PvP5CS-RNAi transgenic plants (Figure 7).
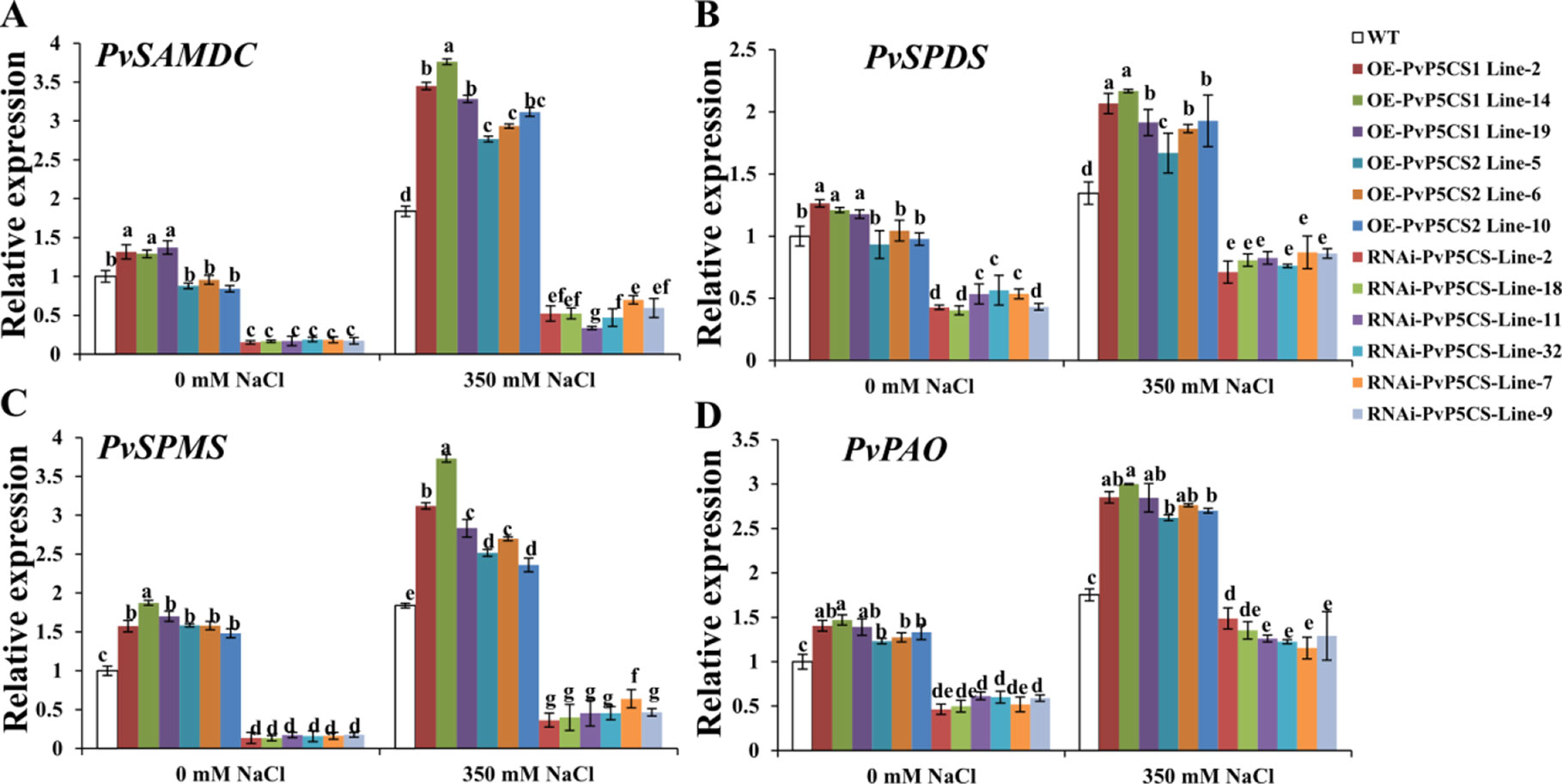
Figure 7 Transcript levels of the genes involved in polyamine synthesis (A–C) and catabolism (D) in PvP5CS OE and RNAi transgenic plants under control and salt stress conditions. Switchgrass UBQ1 was used as the reference for normalization. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
The researchers then attempted to analyze whether expression of polyamine synthesis genes was altered in PvP5CS-RNAi transgenic plants under 350mM salt stress with exogenous application of proline. Compared with the control (i.e., no exogenous proline under salt stress), transcript levels of Spd and Spm synthesis and metabolism genes in both PvP5CS-RNAi transgenic and WT plants were significantly increased under salt stress when applied with 10mM proline, but the extent of increase in PvP5CS-RNAi transgenic plants were much lower than that in WT plants (Figure 8). These results showed that exogenous proline could accelerate the expression levels of polyamines metabolism-related genes under salt stress. In summary, these results highlight the function of PvP5CS in Spd and Spm synthesis and metabolism during salt response.
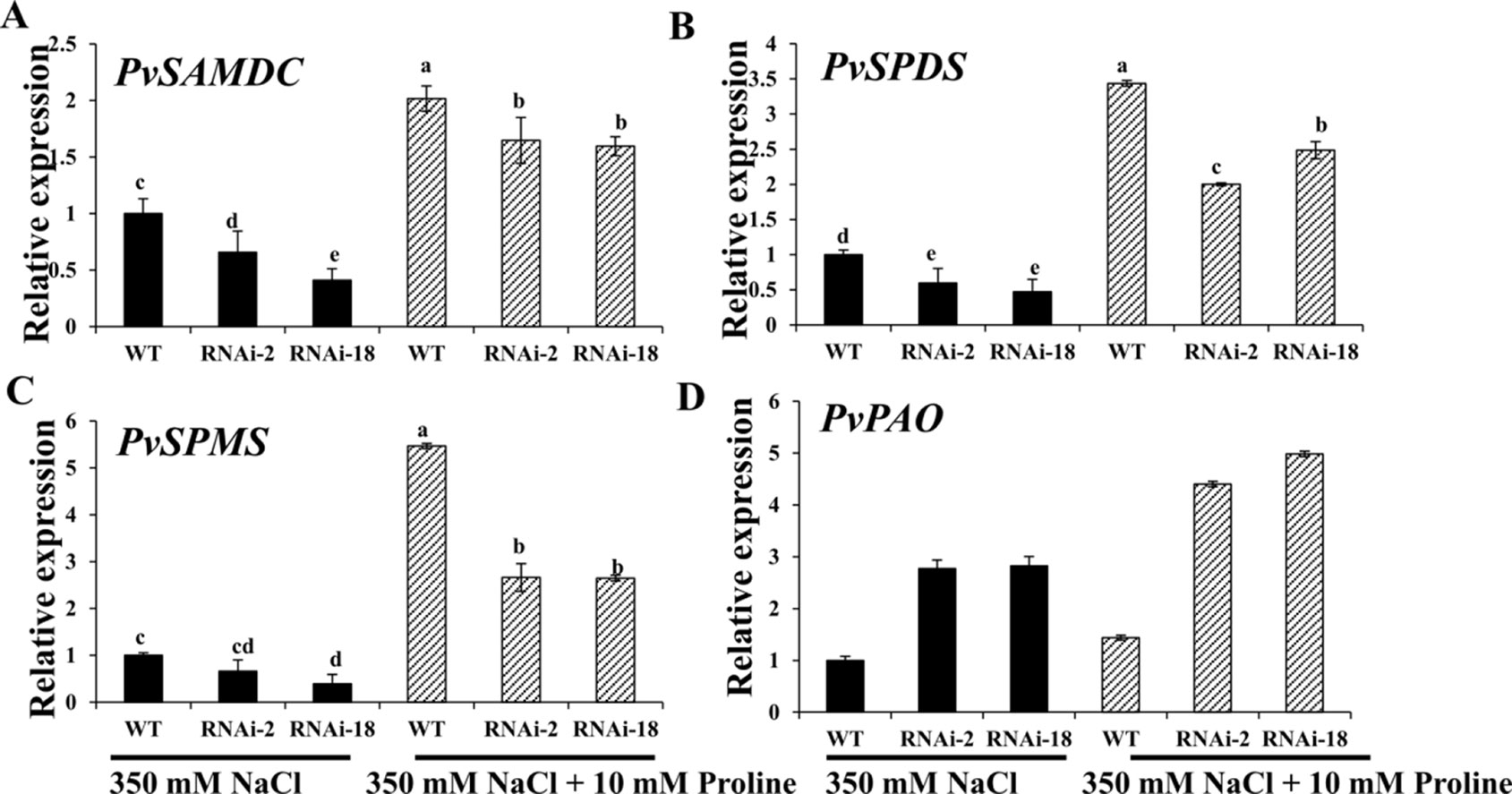
Figure 8 Exogenous proline can largely improve transcript levels of the genes involved in polyamine synthesis (A–C) and catabolism (D) in PvP5CS RNAi transgenic plants under 350 mM NaCl salt stress. Switchgrass UBQ1 was used as the reference for normalization. Data are mean values of three biological repeat, and significance of treatments was tested at the P < 0.05 level (one way ANOVA, Dunnett's test).
P5CS is a rate limiting enzyme in proline biosynthesis (Székely et al., 2008). Generally, P5CS enzyme is encoded by two homologous P5CS genes. P5CS genes have been isolated from various plants species including one housekeeping and one stress-specific P5CS isoform. Although the duplicated P5CS genes share a high level sequence homology in coding regions, their transcriptional regulations are different (Szabados and Savoure, 2010). In Arabidopsis, P5CS1-GFP is normally localized in cytosol of leaf mesophyll cells and P5CS2-GFP is predominantly localized in the cytosol. When cells are exposed to salt or osmotic stress, proline biosynthesis is regulated by the P5CS1 enzyme and its transcript is induced by different abiotic stress in the chloroplasts (Székely et al., 2008). Moreover, in rice although both OsP5CSs genes are upregulated by exogenous ABA, their kinetics differ with a faster and stronger upregulation of OsP5CS1 transcripts compared to that of OsP5CS2 in seedlings and different tissues of rice plants (Hur et al., 2004; Sripinyowanich et al., 2013). In switchgrass, PvP5CS1 and PvP5CS2 were induced by salt stress, whereas the transcript levels of PvP5CS1 showed a faster and stronger upregulation compared to the transcript levels of PvP5CS2. Furthermore, PvP5CS1 and PvP5CS2, were found to differ in tissue tropism and the kinetics and levels of transcript expression, where the PvP5CS1 showed higher expression at E0, E1, and E3 stages; while PvP5CS2 showed higher expression at R1 and R3 (Figures 1A, B). The expression patterns indicate that two switchgrass P5CS paralogs might have different functions.
Under normal conditions, change in free proline content was accompanied by plant growth and development especially in reproductive organs (Lehmann et al., 2010). To explore the extent of functional diversification of duplicated PvP5CSs genes in switchgrass, we evaluated the genetic and physiological characterizations of PvP5CS1 and PvP5CS2 overexpression transgenic lines for different developmental stages of the plant. PvP5CS2 OE lines bloomed earlier, and this result is coupled with a significant difference in relative expression of flowering-related genes PvFLC and PvFT between PvP5CS2 OE and WT plants. On the other hand, the promoter of Arabidopsis P5CS2 was an early target of CONSTANS (CO), which is involved in floral transition (Samach et al., 2000). And later, concurrent silencing of two AtP5CS genes resulted in stunted growth and resulted late flowering (Nanjo et al., 1999). Consequently, atp5cs2 mutant plants displayed a stronger late-flowering phenotype compared to that of atp5cs1 mutant plants, and only the embryos in atp5cs2 mutant plants showed alterations of the cellular division planes and inactive growth (Mattioli et al., 2009). Furthermore, under long-day conditions, atp5cs2 mutant plants were retarded in vegetative development whereas the growth and development of atp5cs1 mutant plants was normal (Funck et al., 2012). Together with these findings, our results suggested that PvP5CS2 also functions in reproductive growth stage in switchgrass. Nonetheless, it was also found that both PvP5CS1 and PvP5CS2 OE plants improved salt stress under high salt treatment. These results indicate that proline generates the suppression of NaCl-induced physiological damage as shown by the lower electrolyte leakage and ROS levels which further explain that proline significantly improves the physiological status of stressed plants (Figures 4 and 6). Similarly, our previous results also showed that overexpressing heterologous P5CS genes from Lolium perenne and Puccinellia chinampoensis in switchgrass improve the salt tolerance by reducing the electrolyte leakage and ROS levels (Guan et al., 2018; Guan et al., 2019). Therefore, no matter overexpressing heterologous or endogenous P5CS genes in switchgrass, which can increase the proline content and improve the salt stress. Additionally, increased proline content also improved the plant growth and development, especially in flowering time and biomass.
Reactive oxygen species is highly accumulated under stress conditions and endogenous plant cell sensors by recognizing the composite signaling system and transfers to kinases by secondary messengers (Waszczak et al., 2018). And also, the upregulation of proline biosynthesis results metabolic redistribution (Majumdar et al., 2016). Additionally, Polyamines also play essential roles in plant stress tolerance which partially reversed the NaCl-induced phenotypic and reprogramed the oxidative status (Tanou et al., 2014). Ornithine is a common precursor of PAs and proline and in plants, and putrescine can be synthesized from ornithine by ornithine decarboxylase (ODC). Despite lower concentrations of ornithine compared to PAs, Pro, and Glu, ornithine remains a gatekeeper in controlling PAs, Pro, and GABA Biosynthesis (Majumdar et al., 2016). Moreover, alteration of ornithine into PA biosynthesis affects the proline biosynthesis. Thus, it is meaningful to consider the relationship between PAs and proline metabolisms. Overexpressing MfERF1 induced proline and polyamine enrichment under cold tolerance, suggesting that proline and PAs jointly respond and improve cold tolerance (Zhuo et al., 2018). The results showed that overexpression of PvP5CS1 and PvP5CS2 elevate the transcript levels of Spd and Spm synthesis and metabolism genes under normal and salt stress conditions (Figure 7). Additionally, atpao5-2 and atpao5-3 mutants (mutants of Spd and Spm metabolism gene PAO) were found to exhibit higher proline biosynthesis and salt tolerance, and the proline content is higher in mutants under salt stress (Zarza et al., 2017). Thus, increased proline biosynthesis is directly sourced from glutamate by P5CS and the pathway from ornithine to proline is suppressed which divert more ornithine into PAs biosynthesis. On the other hand, to illuminate the mechanical connection between external proline application and PA metabolism under salt stress, we measured the expression levels of PAs biosynthesis and catabolism genes in leaves. By comparing proline plus NaCl treatment with NaCl-only treatment, application of proline leads to enhanced transcript levels of PAs metabolism related genes both in PvP5CS RNAi and in WT plants compared to NaCl-alone treatment, and the improved degree in PvP5CS RNAi plants is higher than in WT plants (Figure 8) indicating that PA metabolism is regulated by external proline application.
Based on the results, the researchers were able to summarize the model of the connections between PAs and proline metabolisms under normal and salt stress (Figure 9). Ornithine is a key metabolite involving in proline and PAs metabolism. In the ornithine pathway, ornithine is transmitted by ornithine-delta-aminotransferase (OAT) producing GSA and P5C which is later converted to proline (Roosens et al., 1998). PAs biosynthesis is controlled by enzymes activity and substrate. Ornithine is the substrate of Put biosynthesis and transmitted by ornithine decarboxylase (ODC) producing Put, then Put is converted to Spd and Spm. In our results, under both normal and stressed conditions, ornithine as a precursor may enter pathway of proline synthesis, and proline metabolism plays the main part compared with PAs metabolism in PvP5CS RNAi and WT plants. Overexpression of PvP5CS1 and PvP5CS2 improved the proline biosynthesis under normal and salt stress conditions, and an excess of proline produces toxic action to plants which result in ornithine, as a precursor, entering the synthesis pathway of PAs. In summary, proline metabolism is in cooperation with polyamines metabolism in switchgrass. If switchgrass has enough proline to confer salt stress, ornithine could be as substrate involved in PAs biosynthesis. However, under less proline accumulation to improve the salt tolerance of switchgrass, ornithine will be used as a precursor entering the pathway of proline synthesis. Altogether, proline and polyamine metabolism cooperated to improve salt tolerance in switchgrass.
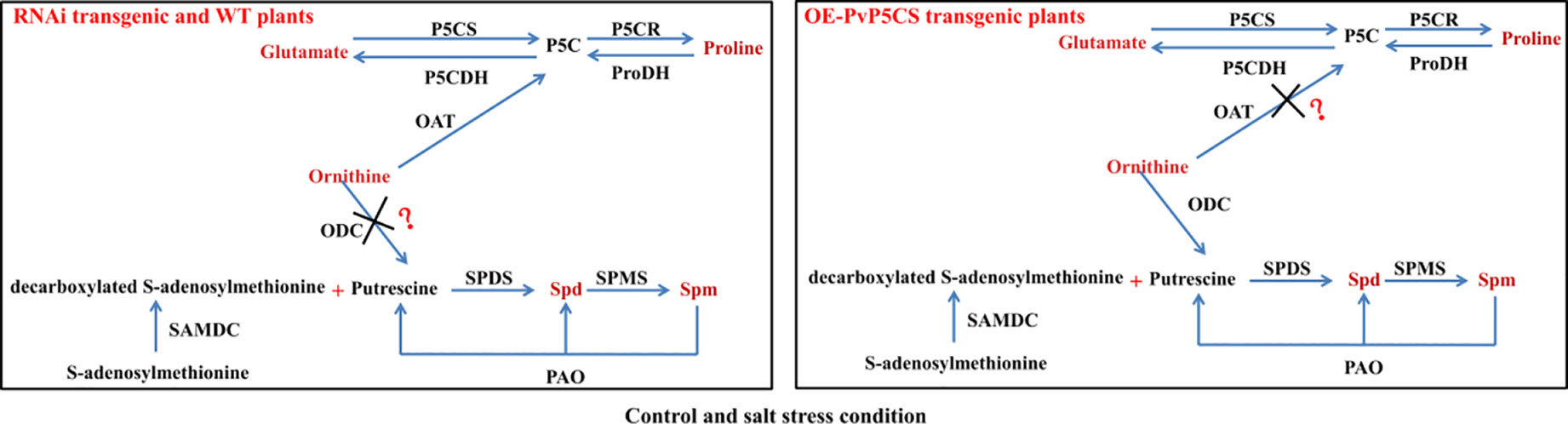
Figure 9 Schematic illustration of a working model depicting the relationship between proline and polyamine in wild type, PvP5CS OE and RNAi transgenic plants under control and salt stress conditions. In summary, proline metabolism is in cooperation with polyamines metabolism in switchgrass. If switchgrass has enough proline to confer salt stress, ornithine could be as substrate involved in PAs biosynthesis. However, under less proline accumulation to improve the salt tolerance of switchgrass, ornithine will be used as a precursor entering the pathway of proline synthesis. Altogether, proline and polyamine metabolism cooperated to improve salt tolerance in switchgrass.
Publicly available datasets were analyzed in this study. The Gene sequences are downloaded from Switchgrass Genome Annotation Program.
Conceived and designed experiment: Y-WZ. Performed the experiments: CG, XC, H-YL, XL, and M-QL. Analyzed the data: CG, XC, H-YL, XL, M-QL, and Y-WZ. Wrote the manuscript: CG and YW-Z. All authors reviewed and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding for this work was provided by National Natural Science Foundation of China (31672478); the Ministry of Science And Technology, China (2014BAD23B03), (2012CB215301), (2012AA101801); Natural Science Foundation of Beijing (6162016). We thank Hui Wang for reading and reviewing the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00046/full#supplementary-material
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231 (6), 1237–1249. doi: 10.1007/s00425-010-1130-0
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bates, L. S., Waldren, R. P., Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bouchereau, A., Aziz, A., Larher, F., Martin-Tanguy, J. (1999). Polyamines and environmental challenges: recent development. Plant Sci. 140 (2), 103–125. doi: 10.1016/S0168-9452(98)00218-0
Casler, M. D., Vogel, K. P., Taliaferro, C. M., Ehlke, N. J., Berdahl, J. D., Brummer, E. C., et al. (2007). Latitudinal and longitudinal adaptation of switchgrass populations. Crop Sci. 47 (6), 2249–2260. doi: 10.2135/cropsci2006.12.0780
Chen, J. B., Yang, J. W., Zhang, Z. Y., Feng, X. F., Wang, S. M. (2013). Two P5CS genes from common bean exhibiting different tolerance to salt stress in transgenic Arabidopsis. J. Genet. 92, 461–469. doi: 10.1007/s12041-013-0292-5
Chen, D., Shao, Q., Yin, L., Younis, A., Zheng, B. (2018). Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01945
Delauney, A. J., Verma, D. P. S. (1993). Proline biosynthesis and osmoregulation in plants. Plant J. 4 (2), 215–223. doi: 10.1046/j.1365-313X.1993.04020215.x
Du, C., Zhao, P. P., Zhang, H. R., Li, N. N., Zheng, L. L., Wang, Y. C. (2017). The Reaumuria trigyna transcription factor RtWRKY1 confers tolerance to salt stress in transgenic Arabidopsis. J. Plant Physiol. 215, 48–58. doi: 10.1016/j.jplph.2017.05.002
Fryer, M. J., Oxborough, K., Mullineaux, P. M., Baker, N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. doi: 10.1093/jxb/53.372.1249
Funck, D., Winter, G., Baumgarten, L., Forlani, G. (2012). Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 12 (1), 191. doi: 10.1186/1471-2229-12-191
Guan, C., Huang, Y. H., Cui, X., Liu, S. J., Zhou, Y. Z., Zhang, Y. W. (2018). Overexpression of gene encoding the key enzyme involved in proline-biosynthesis (PuP5CS) to improve salt tolerance in switchgrass (Panicum virgatum L.). Plant Cell Rep. 37, 1187–1199. doi: 10.1007/s00299-018-2304-7
Guan, C., Huang, Y. H., Cen, H. F., Cui, X., Tian, D. Y., Zhang, Y. W. (2019). Overexpression of the Lolium perenne L. delta1-pyrroline 5-carboxylate synthase (LpP5CS) gene results in morphological alterations and salinity tolerance in switchgrass (Panicum virgatum L.). PLoS One. 14 (7), e0219669. doi: 10.1371/journal.pone.0219669
Hanfrey, C., Sommer, S., Mayer, M. J., Burtin, D., Michael, A. J. (2001). Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J. 27 (6), 551–560. doi: 10.1046/j.1365-313X.2001.01100.x
Hardin, C. F., Fu, C. X., Hisano, H., Xiao, X. R., Shen, H., Stewart, C. N., et al. (2013). Standardization of switchgrass sample collection for cell wall and biomass trait analysis. Bioenergy Res. 6, 755–762. doi: 10.1007/s12155-012-9292-1
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signaling Behav. 7 (11), 1456–1466. doi: 10.4161/psb.21949
Hu, C. A., Delauney, A. J., Verma, D. P. S. (1992). A bifunctional enzyme (delta 1-pyrroline-5-carboxylate synthetase) catalyses the first two steps in proline biosynthesis in plants. Proc. Natl. Acad. Sci. U. S. A. 89, 9354–9358. doi: 10.1073/pnas.89.19.9354
Hur, J., Jung, K. H., Lee, C. H., An, G. (2004). Stress-inducible OsP5CS2 gene is essential for salt and cold tolerance in rice. Plant Sci. 167, 417–426. doi: 10.1016/j.plantsci.2004.04.009
Jha, B., Mishra, A., Jha, A., Joshi, M. (2013). Developing Transgenic Jatropha using the SbNHX1 Gene from an Extreme Halophyte for Cultivation in Saline wasteland. PLoS One 8, e71136. 10.1371/journal.pone.0071136
Julkowska, M. M., Testerink, C. (2015). Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20 (9), 586–594. doi: 10.1016/j.tplants.2015.06.008
Kasukabe, Y., He, L., Nada, K., Misawa, S., Ihara, I., Tachibana, S. (2004). Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 45, 712–722. doi: 10.1093/pcp/pch083
Kausar, M., Mahmood, T., Basra, S. M. A., Arshad, M. (2009). Invigoration of low vigor sunflower hybrids by seed priming. Int. J. Agric. Biol. 11 (5), 521–528. doi: 10.1684/agr.2009.0328
Keshwani, D. R., Cheng, J. J. (2009). Switchgrass for bioethanol and other value-added applications: a review. Bioresour. Technol. 100 (4), 1515–1523. doi: 10.1016/j.biortech.2008.09.035
Kim, G. B., Nam, Y. W. (2013). A novel Δ1-pyrroline-5-carboxylatesynthetase gene of Medicago truncatula plays a predominant role in stress-induced proline accumulation during symbiotic nitrogen fixation. J. Plant Physiol. 170, 291–302. doi: 10.1016/j.jplph.2012.10.004
Krasensky, J., Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63 (4), 1593–1608. doi: 10.1093/jxb/err460
Lehmann, S., Funck, D., Szabados, L., Rentsch, D. (2010). Proline metabolism and transport in plant development. Amino Acids 39 (4), 949–962. doi: 10.1007/s00726-010-0525-3
Li, Z., Baldwin, C., Hu, Q., Liu, H., Luo, H. (2010). Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ. 33, 272–289. doi: 10.1111/j.1365-3040.2009.02080.x
Liu, Y. R., Cen, H. F., Yan, J. P., Zhang, Y. W., Zhang, W. J. (2015). Inside out: high-efficiency plant regeneration and Agrobacterium-mediated transformation of upland and lowland switchgrass cultivars. Plant Cell Rep. 34, 1099–1108. doi: 10.1007/s00299-015-1769-x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Majumdar, R., Barchi, B., Turlapati, S. A., Gagne, M., Minocha, R., Long, S., et al. (2016). Glutamate, ornithine, arginine, proline, and polyamine metabolic interactions: the pathway is regulated at the post-transcriptional level. Front. Plant Sci. 7, 78. doi: 10.3389/fpls.2016.00078
Mattioli, R., Falasca, G., Sabatini, S., Altamura, M. M., Costantino, P., Trovato, M. (2009). The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiol. Plant. 137 (1), 72–85. doi: 10.1111/j.1399-3054.2009.01261.x
Nanjo, T., Kobayashi, M., Yoshiba, Y., Sanada, Y., Wada, K., Tsukaya, H., et al. (1999). Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J. 18 (2), 185–193. doi: 10.1046/j.1365-313X.1999.00438.x
Pál, M., Szalaik, G., Janda, T. (2015). Speculation: polyamines are important in abiotic stress signaling. Plant Sci. 237, 16–23. doi: 10.1016/j.plantsci.2015.05.003
Pál, M., Tajti, J., Szalai, G., Peeva, V., Végh, B., Janda, T. (2018). Interaction of polyamines, abscisic acid and proline under osmotic stress in the leaves of wheat plants. Sci. Rep. 8 (1), 12839. doi: 10.1038/s41598-018-31297-6
Roosens, N. H., Thu, T. T., Iskandar, H. M., Jacobs, M. (1998). Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 117 (1), 263–271. doi: 10.1104/pp.117.1.263
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 (5471), 1613–1616. doi: 10.1126/science.288.5471.1613
Shao, L., Majumdar, R., Minocha, S. C. (2012). Profiling the amino-propyltransferases in plants: their structure, expression and manipulation. Amino Acids 42 (2-3), 813–830. doi: 10.1007/s00726-011-0998-8
Sripinyowanich, S., Klomsakul, P., Boonburapong, B., Bangyeekhun, T., Asami, T., Gu, H., et al. (2013). Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ. Exp. Bot. 86, 94–105. doi: 10.1016/j.envexpbot.2010.01.009
Székely, G., Ábrahám, E., Cséplő, Á., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53 (1), 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Szabados, L., Savoure, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15 (2), 89–97. doi: 10.1016/j.tplants.2009.11.009
Tanou, G., Ziogas, V., Belghazi, M., Christou, A., Filippou, P., Job, D., et al. (2014). Polyamines reprogram oxidative and nitrosative status and the proteome of citrus plants exposed to salinity stress. Plant Cell Environ. 37, 864–885. doi: 10.1111/pce.12204
Urano, K., Yoshiba, Y., Nanjo, T., Igarashi, Y., Seki, M., Sekiguchi, F., et al. (2003). Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ. 26, 1917–1926. doi: 10.1046/j.1365-3040.2003.01108.x
Verslues, P. E., Sharma, S. (2010). Proline metabolism and its implications for plant-environment interaction. Arabidopsis Book 8, e0140. doi: 10.1199/tab.0140
Wang, G., Zhang, J., Wang, G., Fan, X., Sun, X., Qin, H. (2014). Proline responding1 plays a critical role in regulating general protein synthesis and the cell cycle in Maize. Plant Cell 26 (6), 2582–2600. doi: 10.1105/tpc.114.125559
Waszczak, C., Carmody, M., Kangasjarvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 69, 209–236. doi: 10.1146/annurev-arplant-042817-040322
Wi, S. J., Kim, W. T., Park, K. Y. (2006). Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. 25 (10), 1111–1121. doi: 10.1007/s00299-006-0160-3
Xu, B., Escamillatreviño, L. L., Sathitsuksanoh, N., Shen, Z., Shen, H., Zhang, Y. H., et al. (2011). Silencing of 4-coumarate:coenzyme a ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 192 (3), 611–625. doi: 10.1111/j.1469-8137.2011.03830.x
Xue, X., Liu, A., Hua, X. (2009). Proline accumulation and transcriptional regulation of proline biothesynthesis and degradation in Brassica napus. BMB Rep. 42, 28–34. doi: 10.5483/BMBRep.2009.42.1.028
Zarza, X., Atanasov, K. E., Marco, F., Arbona, V., Carrasco, P., Kopka, J., et al. (2017). Polyamine oxidase 5 loss-of-function mutations in Arabidopsis thaliana trigger metabolic and transcriptional reprogramming and promote salt stress tolerance. Plant Cell Environ. 40, 527–542. doi: 10.1111/pce.12714
Zhang, H., Liu, Y., Wen, F., Yao, D., Wang, L., Guo, J., et al. (2014). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809. doi: 10.1093/jxb/eru313
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: switchgrass, PvP5CS1 and PvP5CS2, proline, polyamines, salt stress
Citation: Guan C, Cui X, Liu H-y, Li X, Li M-q and Zhang Y-w (2020) Proline Biosynthesis Enzyme Genes Confer Salt Tolerance to Switchgrass (Panicum virgatum L.) in Cooperation With Polyamines Metabolism. Front. Plant Sci. 11:46. doi: 10.3389/fpls.2020.00046
Received: 12 November 2019; Accepted: 14 January 2020;
Published: 14 February 2020.
Edited by:
Honghong Wu, Huazhong Agricultural University, ChinaReviewed by:
Jiangman He, University of California, Riverside, United StatesCopyright © 2020 Guan, Cui, Liu, Li, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun-wei Zhang, enl3ZWlAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.