
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 05 February 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00036
This article is part of the Research Topic Organelle Autophagy in Plant Development View all 9 articles
Autophagy is a mechanism to recycle intracellular constituents such as amino acids and other carbon- and nitrogen (N)-containing compounds. Although autophagy-related (ATG) genes required for autophagy are encoded by many algal genomes, their functional importance in microalgae in nutrient-deficiency has not been appraised using ATG-defective mutants. Recently, by characterization of an insertional mutant of the ATG8 encoding a ubiquitin-like protein indispensable for autophagosome formation in a green alga Chlamydomonas reinhardtii, we have provided evidence that supports the following notions. ATG8 protein is required for the degradation of lipid droplets and triacylglycerol (TAG) triggered by resupply of N to cell culture in N-deficient conditions. ATG8 protein is also necessary for starch accumulation under phosphorus-deficient conditions. Algal autophagy is not necessary for inheritance of chloroplast and mitochondrial genomes. In this review, we discuss the physiological roles of algal autophagy associated with nutrient deficiency revealed by the genetic and biochemical analyses using disruption mutants and reagents that inhibit the fatty acid biosynthesis and vacuolar H+-ATPase.
Macroautophagy (hereafter autophagy) is a recycling system for degradation of cytoplasmic constituents in the vacuole or lysosomes (Mizushima et al., 2011; Liu and Bassham, 2012; Yang and Bassham, 2015). This pathway is conserved among eukaryotes (Liu and Bassham, 2012; Yang and Bassham, 2015). In order to understand the physiological functions of plant autophagy, the following strategies has been conducted, because the processes of autophagy, from autophagosome formation to degradation of target cellular components in the vacuole are very rapid (Yoshimoto, 2012): 1) analyzing the phenotype of autophagy-defective core ATG gene mutants (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005); 2) visualizing subcellular localization of ATG8 protein with a fluorescent protein (Yoshimoto et al., 2004; Contento et al., 2005; Merkulova et al., 2014); and 3) artificially halting the autophagic flux in the vacuole by treatment with concanamycin A, which inhibits H+-ATPase and thus inactivating the vacuolar acid hydrolases, leading to the autophagic body accumulating inside the vacuole (Yoshimoto et al., 2004).
The following findings have been reported regarding the regulation of the accumulation of photosynthetic assimilation products, such as lipids and starch, by autophagy. Firstly, starch synthesized in leaves by photosynthesis during the day is degraded at night, and phenotypic analysis of atg mutants in Arabidopsis thaliana suggested that autophagy facilitates this process (Wang et al., 2013). In addition, autophagy was shown to promote triacylglycerol (TAG) degradation under C-deficient conditions using the seedlings of A. thaliana atg mutants (Avin-Wittenberg et al., 2015). Recently, Fan et al. (2019) reported that basal autophagy is required for TAG biosynthesis by lipid turnover providing fatty acids from organellar membrane lipids in A. thaliana. Similarly, studies on atg mutants in rice which showed male sterility concluded that autophagy is necessary for TAG and starch accumulation and lipid droplet formation as well as normal reproductive post-meiotic anther development during pollen maturation (Kurusu et al., 2014). However, studies of plant autophagy have been limited to a few model species for which atg mutants are available, and the role of autophagy in the metabolism of carbon (C)-assimilation products throughout the plant kingdom needs to be further investigated. Because in algae the role of autophagy in the accumulation and metabolism of photosynthetic assimilation products remained unclear, studies using autophagy-deficient mutant strains have been considered necessary in addition to those using wild type cells treated with autophagy-inhibiting chemicals. In this review, the physiological functions of algal autophagy in response to nutrient deficiency will be discussed, based on recent reports of autophagy-defective mutants in Chlamydomonas reinhardtii (hereafter Chlamydomonas) and the effects of treatment of the wild-type Chlamydomonas cells with cerulenin and concanamycin A for inhibition of fatty acid synthesis and vacuolar lysosomal function, respectively.
Unicellular algae known as microalgae are photosynthetic eukaryotes, classified as one of protists. For the purpose of biofuel production, many microalgae have been nominated by screening, which accumulate high levels of C-storage compounds such as lipid and starch. Especially understanding the physiological culture conditions and the molecular mechanisms for accumulation of their lipid and starch is necessary to achieve realistic biofuel production. So far, it is reported that many algae accumulate these C-storage compounds in cells when exposed to nutrient-deficient stress conditions such as nitrogen (N)-deficiency after stopping their growth in these stress conditions but they do not die immediately and that instead, they maintain cell viability for a period of time. This cell survival under nutrient-deplete conditions suggests that autophagy is involved in the system for keeping the cell viability. Although autophagy is involved in the regulation of C metabolism in yeast, animals, and terrestrial plants, contribution of autophagy to lipid and starch metabolism in algae has not been fully understood. In several species of algae from Chlorophyta, Rhodophyta, and Chromalveolata, orthologs for known ATG genes have been found in genome databases, except for red algae whose genomes lack the ATG gene (Díaz-Troya et al., 2008a; Avin-Wittenberg et al., 2012; Jiang et al., 2012; Shemi et al., 2015). However, no autophagy-defective mutant has been reported in any species of algae so far.
In a model photosynthetic single-cell eukaryote, Chlamydomonas, a series of studies have been reported to provide molecular evidence of autophagy and autophagic activities responding to rapamycin, an inhibitor of the target of rapamycin (TOR) protein kinase (Pérez-Pérez et al., 2010), abiotic stresses such as endoplasmic reticulum (ER) stress, oxidative stress (Pérez-Pérez et al., 2010; Pérez-Martín et al., 2014), and metal toxicity (Pérez-Martín et al., 2015), and the following nutrient deficiency; N-deficiency (Pérez-Pérez et al., 2010; Goodenough et al., 2014), C-deficiency (Pérez-Pérez et al., 2010), and carotenoid deficiency (Pérez-Pérez et al., 2012). In the all mentioned stress conditions, the abundance of ATG8 protein and its PE-conjugated/lipidated form increases. Accumulation of ATG8 and ATG3 proteins increased in a conditional repression line of a chloroplast protease ClpP1, implying that chloroplast proteolysis systems and autophagy in Chlamydomonas partially complement each other (Ramundo et al., 2014). Furthermore, in vitro assays using recombinant ATG8 and ATG4 proteins and complementation tests with the corresponding yeast mutants revealed that correspond to the orthologs for ATG8 and ATG4 genes in Chlamydomonas, respectively, encode proteins with functions similar to those in other organisms (Pérez-Pérez et al., 2010; Pérez-Pérez et al., 2016).
Recently, we reported the functional importance of ATG8 and ATG3 proteins by using insertion mutants defect in corresponding genes in Chlamydomonas (Kajikawa et al., 2019). Using these autophagy-defective strains, contribution of autophagy on cell processes including photosynthesis, metabolism, and reproduction under various nutrient-deficient conditions was clarified.
The TOR kinase complex 1 (TORC1) is reported to be an important regulatory factor making balance between growth and autophagy in the eukaryotes (Noda and Ohsumi, 1998; Pattingre et al., 2008; Liu and Bassham, 2010). When nutrients are abundantly supplied to the cell, TOR is activated and promotes protein synthesis and cell growth, while negatively regulating autophagy by inhibiting the formation of the ATG1 complex (Mizushima et al., 2011). Land plants have TOR-dependent and TOR-independent pathways for regulation of autophagy (Pu et al., 2017). Activation of autophagy in nutrient deficiency, salt, and osmotic stress requires suppression of TOR. On the other hand, induction of autophagy under oxidative stress and ER stress conditions is suggested to be controlled independently by TOR (Pu et al., 2017; Soto-Burgos et al., 2018). Although there are still many unknown regarding algal TOR systems, it is reported that TOR, LST8 and Raptor genes, which are components of TORC1 protein complex, are widely conserved in algal genomes (Shemi et al., 2015).
Chlamydomonas has all the components of the TORCl complex (Crespo et al., 2005; Díaz-Troya et al., 2008b). The arrest of cell cycle, bleaching, and vacuolization have been shown to be proceeded by the addition of rapamycin to the Chlamydomonas cells, indicating that Chlamydomonas has a rapamycin-sensitive TOR signaling network (Crespo et al., 2005). Addition of rapamycin also increases the expression of ATG8, led to detection of lipidated ATG8, and altered the subcellular localization of ATG8, suggesting that the rapamycin-sensitive TOR signaling network regulates autophagy in Chlamydomonas (Pérez-Pérez et al., 2010; Pérez-Pérez et al., 2017). Recently, Mubeen et al. (2018) demonstrated that rapamycin-induced TOR inhibition increases de novo amino acid synthesis due to upregulated ammonium assimilation in Chlamydomonas. It has been also reported that TOR inhibition by addition of TOR inhibitors, such as rapamycin, AZD8055, or Torin1, activates expression of genes encoding TAG biosynthesis-related enzymes and increased TAG accumulation in Chlamydomonas and a red alga Cyanidioschyzon merolae (Imamura et al., 2015; Imamura et al., 2016). Furthermore, it has been suggested that TOR in Chlamydomonas regulates the biosynthesis of the signaling molecules, inositol polyphosphates (InsPs) (Couso et al., 2016). A mutant, vip1-1, of the inositol polyphosphate kinase (VIP1) gene involved in the biosynthesis of the InsPs, showed hypersensitivity to rapamycin and increased TAG accumulation even under nutrient replete conditions (Couso et al., 2016). In addition, these reports indicate an interaction between TOR signaling network and lipid metabolism in algae (Pérez-Pérez et al., 2017). Most recently, through the characterization of a hypomorphic mutant of the LST8 gene, lst8-1 under P deficiency, Couso et al. (2020) demonstrated that P deficiency reduces the abundance of LST8 proteins and downregulates activity of TORC1 to activate autophagy and TAG synthesis in Chlamydomonas (Figure 1). In the N-deficient conditions, any significant phenotype was not observed in the lst8-1 mutant.
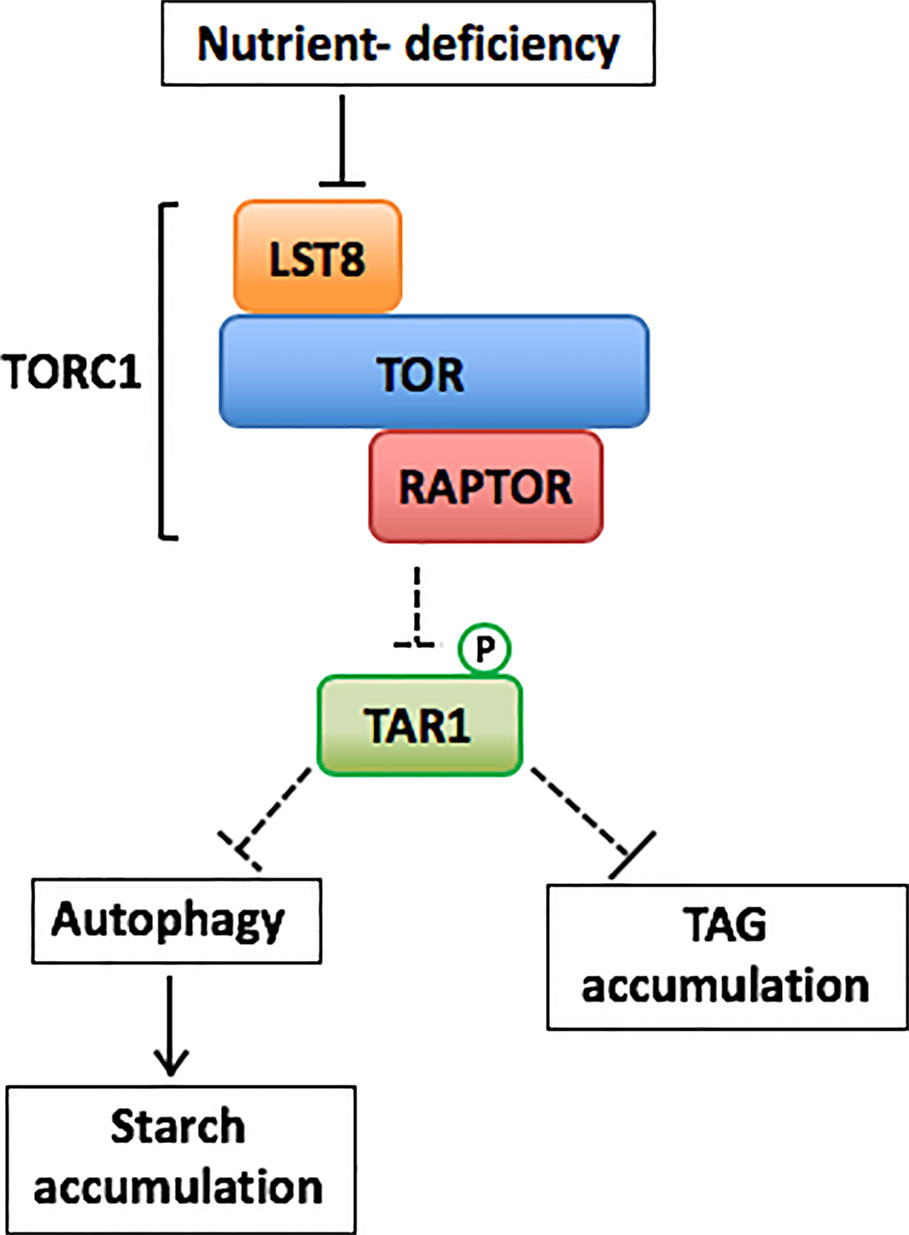
Figure 1 One of the possible models for TORC1-mediated regulation of autophagy and carbon metabolism in Chlamydomonas in response to nutrient deficiency based on models by Forzani et al. (2019); Couso et al. (2020) and Shinkawa et al. (2019). Unidentified but possible regulatory steps are shown by dashed arrows.
Despite these energetic studies in this field, any direct molecular evidence that algal autophagy is involved in interaction between TOR signaling network and lipid metabolism has not been provided and needs to be examined experimentally in future. By quantitative phosphoproteomic approach, total 258 of phosphosites from 219 unique phosphopeptides were detected in Chlamydomonas wild-type cells as significantly modulated by inhibition of TOR (Werth et al., 2019). The key target factors downstream of TOR that are involved in autophagy induction and regulation of lipid turnover and link each other might be discovered from proteins composed of such phosphopeptides.
Chlamydomonas accumulates high levels of neutral lipids and starch in the absence of nutrients such as N, P, and sulfur (S). In addition, the Chlamydomonas genome contains ATG genes related to autophagy. Therefore, we attempted to isolate mutants that lack the ATG8 and ATG3 genes, which are required for the formation of autophagosomal membranes—an early process of autophagy (Kajikawa et al., 2019). For both genes, only one copy was present in the Chlamydomonas genome, and we predicted that their deletion would lead to an autophagy defect. DNA tag-insertion mutants of each gene were selected, one by one, from approximately 4,600 mutant strain libraries, and antibodies were used to show that these were functionally deficient mutants. In the atg8 and atg3 mutants, the cell viability under N-deficient conditions was significantly reduced. After two weeks, the survival rate of the wild-type cells was 80%, whereas the survival rate in both mutant strains decreased to less than 5%, indicating that autophagy is necessary for algal cell survival.
The role of autophagy in the control of C metabolic processes was determined by examining the accumulation of neutral lipids (TAG) and starch under nutrient-deficient conditions, using atg8 mutants as a representative strain. Under N- and S-deficient conditions, the atg8 mutant accumulated starch in the same way as did the wild-type strain during the early stress phase. However, since the accumulated starch in the atg8 mutant was broken down after the second day of stress, this suggests that autophagy was required to maintain the accumulated starch content. In addition, when the medium was replaced with N-deficient conditions before being returned to the N-replete condition, TAG and starch degraded rapidly in the wild-type strain, while degradation of TAG and oil droplets in the atg8 cells occurred approximately 6 h later than in the wild strain. However, the rate of starch degradation in the atg8 mutant was unaffected, suggesting that autophagy is required for rapid degradation of TAG.
The green algae accumulated starch and TAG under P-deficient conditions, as well as under N- and S-deficient conditions. However, although the atg8 mutants accumulated TAG at rates similar to that of the wild-type strain, starch accumulation did not occur from the beginning of the deficiency period, suggesting that autophagy was required for starch accumulation in response to P deficiency (Figure 1).
Cellular organelles, such as chloroplasts and mitochondria, have their own genomes, and only the genomes from one parent are inherited during mating—a process known as maternal inheritance. The Chlamydomonas cell undergoes a shift from vegetative growth to reproductive growth, and gametogenesis occurs meiotically under N-deficient conditions. Because autophagy is involved in the maternal inheritance of mitochondria in mice (Al Rawi et al., 2011) and nematode (Sato and Sato, 2011), experiments were conducted using the atg8 mutant to determine whether autophagy was involved in the maternal inheritance of algae. The results showed that maternal inheritance continued as normal during mating of both mitochondrial genomes, and that algal autophagy is not involved in maternal inheritance.
Heredia-Martínez et al. (2018) reported that chloroplast damage in Chlamydomonas cells induced by the addition of the fatty acid biosynthesis inhibitor, cerulenin, triggers autophagy. In addition, Couso et al. (2018) reported that changes in cells were suppressed when the protein degradation system was suppressed by treating the cells with concanamycin A to stop vacuole flux. The advantage of using such inhibitors is to be able to avoid downstream effects and to examine the effects on cells when a particular in vivo pathway is inhibited by time-limited conditions. In both cases, the addition of the inhibitor increased the expression of the ATG8 protein, which suggested autophagy was activated. When cerulenin was added to the cells, chloroplast shrinkage and thylakoid membrane aggregation was observed along with a decrease in photosynthetic proteins and ribosomal proteins, and a decrease in maximum quantum yield. Furthermore, the amount of ROS increased, while the thylakoid membrane lipid, MGDG, decreased. Both qRT-PCR and RNAseq analyses showed that the expression of genes involved in protein degradation, stress response, and signal transduction were altered.
Heredia-Martínez et al. (2018) have proposed a model demonstrating that when fatty acid synthesis is inhibited by cerulenin, the amount of MGDG is reduced, thereby impairing chloroplast function and causing thylakoid membrane aggregation and generation of ROS. This chloroplast damage propagates up to the nucleus as a retrograde signal and causes activation of autophagy with changes in gene expression, as well as the activation of proteolytic systems. Future research will be necessary to further verify the time-based vertical relationship and causality of these multiple-response reactions. In addition, by comparing the effect of treatment with cerulenin between the wild-type strain and the autophagy-deficient strain, it is possible to verify to what extent autophagy affects these chloroplast injuries.
Couso et al. (2018) reported that development of lipid droplets and TAG accumulation under N- or P-deficient conditions were repressed when the lytic digestion in the vacuolar or lysosomal lumen was inhibited in Chlamydomonas WT cells after treatment with concanamycin A, an inhibitor of vacuolar H+-ATPase activity. Moreover, treatment with concanamycin A during these nutrient-deficient periods suppressed the degradation of ribosomal proteins such as RPS6 and RPL37. This suggested that concanamycin A inhibits autophagic flux (Couso et al., 2018). By the live-cell imaging using the Chlamydomonas transgenic line expressing red fluorescent protein (mCherry)-tagged ATG8, the interactions and fusion between mCherry-labeled structures and lipid droplets were observed in the N deficiency (Tran et al., 2019). Based on this observation, Tran et al. (2019) suggests that autophagy-related pathway might be involved in lipid droplet turnover in the alga. On the other hand, genetic inhibition of autophagy by knockout of the core ATG genes compromises mobilization of various cytoplasmic components into the vacuolar or lysosomal lumen (Kajikawa et al., 2019). The atg mutants formed lipid droplets and accumulated TAG under both N- and P-deficient conditions as the WT cells did. In the red alga C. merolae which do not have any core ATG gene, mRNA abundance of genes encoding glycerol-3-phosphate acyltransferase (GPAT) and acyl-CoA:diacylglycerol acyltransferase (DGAT)in TAG biosynthesis and the level of TAG content increase under TOR-inhibition condition (Imamura et al., 2015). These reports suggest that TOR signaling regulates development of lipid droplets and TAG accumulation without though activation of autophagic pathway in the algae (Figure 1).
Interestingly, a dual-specificity tyrosine phosphorylation-regulated kinase, TAG accumulation regulator 1 (TAR1) has been reported to be necessary for the degradation of chlorophyll and photosynthesis-related proteins and acetate-dependent TAG accumulation in photomixotrophic N-deficient conditions (Kajikawa et al., 2015). In addition, in photoautotrophic N-deficient conditions, the tar1-defective mutant maintained higher levels of cell viability, TAG, and starch with lower accumulation of hydrogen peroxide compared with those of wild type (WT) cells with bubbling of air containing 5% carbon dioxide (Shinkawa et al., 2019). Recently, Forzani et al. (2019) and Barrada et al. (2019) reported that TOR represses an A. thaliana TAR1 ortholog, YAK1 to promote meristem activity and plant growth. YAK1 interacts with RAPTOR and is directly phosphorylated by TOR complex in A. thaliana (Forzani et al., 2019). These findings open a possibility that the Chlamydomonas TAR1 is also directly regulated by TOR-mediated phosphorylation (Figure 1). Relationship of the Chlamydomonas TAR1 kinase and regulation of algal autophagy and TOR signaling network could further deepen the understanding of the regulatory mechanism of cellular responses to nutrient-deplete stress conditions. Especially analyses of protein-protein interactions among regulatory kinases and autophagy components could contribute to better understanding of autophagy-related processes in photosynthetic organisms.
The research into autophagy-defective mutants and the effects of inhibitor addition have clarified the role of algal autophagy in the accumulation and degradation of neutral lipids and starch under various nutritional conditions. In the future, by combining these approaches—i.e. by adding rapamycin, cerulenin, concanamycin A, and other inhibitors of a specific pathway to the autophagy-defective mutants, and examining the resultant time-dependent phenotypic changes—we can verify the mechanism that works in parallel with autophagy, and which plays a role in complementing autophagy in algae.
MK and HF wrote this manuscript.
This work was supported by JSPS KAKENHI grant number 16H04805 (to HF), 17K07753 (to MK), JST ALCA program JPMJAL1105 (to HF), and NEDO project number P10010 (to HF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Al Rawi, S., Louvet-Vallée, S., Djeddi, A., Sachse, M., Culetto, E., Hajjar, C., et al. (2011). Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. doi: 10.1126/science.1211878
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Brock, R., et al (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell. 27, 306–322. doi: 10.1105/tpc.114.134205
Avin-Wittenberg, T., Honig, A., Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and animals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Barrada, A., Djendli, M., Desnos, T., Mercier, R., Robaglia, C., Montané, M. H., et al. (2019). A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development 146, dev171298. doi: 10.1242/dev.171298
Contento, A. L., Xiong, Y., Bassham, D. C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFPAtATG8e fusion protein. Plant J. 42, 598–608. doi: 10.1111/j.1365-313X.2005.02396.x
Couso, I., Evans, B., Li, J., Liu, Y., Ma, F., Diamond, S., et al. (2016). Synergism between inositol polyphosphates and TOR kinase signaling in nutrient sensing, growth control and lipid metabolism in Chlamydomonas. Plant Cell. 28, 2026–2042. doi: 10.1105/tpc.16.00351
Couso, I., Pérez-Pérez, M. E., Martínez-Force, E., Kim, H.-S., He, Y., Umen, J. G., et al. (2018). Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J. Exp. Bot. 69, 1355–1367. doi: 10.1093/jxb/erx372
Couso, I., Perez-Perez, M. E., Ford, M. M., Martinez Force, E., Hicks, L. M., Umen, J. G., et al. (2020). Phosphorus availability regulates TORC1 signaling via LST8 in Chlamydomonas. Plant Cell. 32, 69–80. doi: 10.1105/tpc.19.00179
Crespo, J. L., Díaz-Troya, S., Florencio, F. J. (2005). Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 139, 1736–1749. doi: 10.1104/pp.105.070847
Díaz-Troya, S., Pérez-Pérez, M. E., Florencio, F. J., Crespo, J. L. (2008a). The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4, 851–865. doi: 10.4161/auto.6555
Díaz-Troya, S., Florencio, F. J., Crespo, J. D. (2008b). Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryot. Cell. 7, 212–222. doi: 10.1128/EC.00361-07
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Fan, J., Yu, L., Xu, C. (2019). Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell. 31, 1598–1613. doi: 10.1105/tpc.19.00170
Forzani, C., Duarte, G. T., Van Leene, J., Clément, G., Huguet, S., Paysant-Le-Roux, C., et al. (2019). Mutations of the AtYAK1 kinase suppress TOR deficiency in Arabidopsis. Cell Rep. 27, 3696–3370. doi: 10.1016/j.celrep.2019.05.074
Goodenough, U., Blady, I., Casero, D., Gallaher, S. D., Goodson, C., Johnson, S., et al. (2014). The path to triacylglycerol obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot. Cell. 13, 591–613. doi: 10.1128/EC.00013-14
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Heredia-Martínez, L. G., Andrés-Garrido, A., Martínez-Force, E., Pérez-Pérez, M. E., Crespo, J. L. (2018). Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Physiol. 178, 1112–1129. doi: 10.1104/pp.18.00630
Imamura, S., Kawase, Y., Kobayashi, I., Sone, T., Era, A., Miyagishima, S. Y., et al. (2015). Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol. Biol. 89, 309–318. doi: 10.1007/s11103-015-0370-6
Imamura, S., Kawase, Y., Kobayashi, I., Shimojima, M., Ohta, H., Tanaka, K. (2016). TOR (target of rapamycin) is a key regulator of triacylglycerol accumulation in microalgae. Plant Signal. Behav. 11, e1149285. doi: 10.1080/15592324.2016.1149285
Jiang, Q., Zhao, L., Dai, J., Wu, Q. (2012). Analysis of autophagy genes in microalgae: chlorella as a potential model to study mechanism of autophagy. PloS One 7, e41826. doi: 10.1371/journal.pone.0041826
Kajikawa, M., Sawaragi, Y., Shinkawa, H., Yamano, T., Ando, A., Kato, M., et al. (2015). Algal dual-specificity tyrosine-phosphorylation-regulated kinase TAR1 regulates accumulation of triacylglycerol in nitrogen- or sulfur-deficiency. Plant Physiol. 168, 752–764. doi: 10.1104/pp.15.00319
Kajikawa, M., Yamauchi, M., Shinkawa, H., Tanaka, M., Hatano, K., Nishimura, Y., et al. (2019). Isolation and characterization of Chlamydomonas autophagy-related mutants in nutrient-deficient conditions. Plant Cell Physiol. 60, 126–138. doi: 10.1093/pcp/pcy193
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice post-meiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Liu, Y., Bassham, D. C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PloS One 5, e11883. doi: 10.1371/journal.pone.0011883
Liu, Y., Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Merkulova, E. A., Guiboileau, A., Naya, L., Masclaux-Daubresse, C., Yoshimoto, K. (2014). Assessment and optimization of autophagy monitoring methods in Arabidopsis roots indicate direct fusion of autophagosomes with vacuoles. Plant Cell Physiol. 55, 715–726. doi: 10.1093/pcp/pcu041
Mizushima, N., Yoshimori, T., Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Mubeen, U., Jüppner, J., Alpers, J., Hincha, D. K., Giavalisco, P. (2018). Target of rapamycin inhibition in Chlamydomonas reinhardtii triggers de novo amino acid synthesis by enhancing nitrogen assimilation. Plant Cell. 30, 2240–2254. doi: 10.1105/tpc.18.00159
Noda, T., Ohsumi, Y. (1998). Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966. doi: 10.1074/jbc.273.7.3963
Pérez-Martín, M., Pérez-Pérez, M. E., Lemaire, S. D., Crespo, J. L. (2014). Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in Chlamydomonas reinhardtii. Plant Physiol. 166, 997–1008. doi: 10.1104/pp.114.243659
Pérez-Martín, M., Blaby-Haas, C. E., Pérez-Pérez, M. E., Andrés-Garrido, A., Blaby, I. K., Merchant, S. S., et al. (2015). Activation of autophagy by metals in Chlamydomonas reinhardtii. Eukaryot. Cell. 14, 964–973. doi: 10.1128/EC.00081-15
Pérez-Pérez, M. E., Florencio, F. J., Crespo, J. L. (2010). Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 152, 1874–1888. doi: 10.1104/pp.109.152520
Pérez-Pérez, M. E., Couso, I., Crespo, J. L. (2012). Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8, 376–388. doi: 10.4161/auto.18864
Pérez-Pérez, M. E., Lemaire, S. D., Crespo, J. L. (2016). Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172, 2219–2234. doi: 10.1104/pp.16.01582
Pérez-Pérez, M. E., Couso, I., Crespo, J. L. (2017). The TOR signaling network in the model unicellular green alga Chlamydomonas reinhardtii. Biomolecules 7, 54. doi: 10.3390/biom7030054
Pattingre, S., Espert, L., Biard-Piechaczyk, M., Codogno, P. (2008). Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90, 313–323. doi: 10.1016/j.biochi.2007.08.014
Pu, Y., Luo, X., Bassham, D. C. (2017). TOR-dependent and -independent pathway regulate autophagy in Arabidopsis thaliana. Front. Plant Sci. 8, 1204. doi: 10.3389/fpls.2017.01204
Ramundo, S., Casero, D., Mühlhaus, T., Hemme, D., Sommer, F., Crèvecoeur, M., et al. (2014). Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell. 26, 2201–2222. doi: 10.1105/tpc.114.124842
Sato, M., Sato, K. (2011). Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144. doi: 10.1126/science.1210333
Shemi, A., Ben-Dor, S., Vardi, A. (2015). Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 11, 701–715. doi: 10.1080/15548627.2015.1034407
Shinkawa, H., Kajikawa, M., Nomura, Y., Ogura, M., Sawaragi, Y., Yamano, T., et al. (2019). Algal protein kinase, Triacylglycerol accumulation regulator 1, modulates cell viability and gametogenesis in carbon/nitrogen imbalanced conditions. Plant Cell Physiol. 60, 916–930. doi: 10.1093/pcp/pcz010
Soto-Burgos, J., Zhuang, X., Jiang, L., Bassham, D. C. (2018). Dynamics of autophagosome formation. Plant Physiol. 176, 219–229. doi: 10.1104/pp.17.01236
Thompson, A. R., Doelling, J. H., Suttangkakul, A., Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Tran, Q. G., Yoon, H. R., Cho, K., Lee, S. J., Crespo, J. L., Ramanan, R., et al. (2019). Dynamic interactions between autophagosomes and lipid droplets in Chlamydomonas reinhardtii. Cells 8, E992. doi: 10.3390/cells8090992
Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S., et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell. 25, 1383–1399. doi: 10.1105/tpc.112.108993
Werth, E. G., McConnell, E. W., Couso Lianez, I., Perrine, Z., Crespo, J. L., Umen, J. G., et al. (2019). Investigating the effect of target of rapamycin kinase inhibition on the Chlamydomonas reinhardtii phosphoproteome: from known homologs to new targets. New Phytol. 221, 247–260. doi: 10.1111/nph.15339
Yang, X., Bassham, D. C. (2015). “New insight into the mechanism and function of autophagy in plant cells,” in International Review of Cell and Molecular Biology. Ed. Jeon Kwang, W. (Burlington, MA: Academic Press), 1–40. doi: 10.1016/bs.ircmb.2015.07.005
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 16, 2967–2983. doi: 10.1105/tpc.104.025395
Keywords: autophagy, Chlamydomonas, lipid, nutrient deficiency, starch
Citation: Kajikawa M and Fukuzawa H (2020) Algal Autophagy Is Necessary for the Regulation of Carbon Metabolism Under Nutrient Deficiency. Front. Plant Sci. 11:36. doi: 10.3389/fpls.2020.00036
Received: 31 October 2019; Accepted: 13 January 2020;
Published: 05 February 2020.
Edited by:
Masanori Izumi, Riken, JapanReviewed by:
Xiaohong Zhuang, The Chinese University of Hong Kong, ChinaCopyright © 2020 Kajikawa and Fukuzawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hideya Fukuzawa, ZnVrdXphd2FAbGlmLmt5b3RvLXUuYWMuanA=
†Present address: Masataka Kajikawa, Faculty of Biology-Oriented Science and Technology, Kindai University, Kinokawa, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.