- 1School of Biological Sciences, Nanyang Technological University, Singapore, Singapore
- 2National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China
- 3Department of Plant and Soil Sciences, Kentucky Tobacco Research and Development Center, University of Kentucky, Lexington, KY, United States
Most plant species generate and store triacylglycerol (TAG) in their seeds, serving as a core supply of carbon and energy to support seedling development. Plant seed oils have a wide variety of applications, from being essential for human diets to serving as industrial renewable feedstock. WRINKLED1 (WRI1) transcription factor plays a central role in the transcriptional regulation of plant fatty acid biosynthesis. Since the discovery of Arabidopsis WRI1 gene (AtWRI1) in 2004, the function of WRI1 in plant oil biosynthesis has been studied intensively. In recent years, the identification of WRI1 co-regulators and deeper investigations of the structural features and molecular functions of WRI1 have advanced our understanding of the mechanism of the transcriptional regulation of plant oil biosynthesis. These advances also help pave the way for novel approaches that will better utilize WRI1 for bioengineering oil production in crops.
Introduction
Plants biosynthesize and store fatty acids mostly as triacylglycerol (TAG) in their seeds to support seedling development as carbon and energy resource. TAG (often familiar to many people as vegetable oils) is a highly energy-rich natural resource, as it has higher energy compared to carbohydrates and proteins. Vegetable oils are not only vital for the human diet, but also have other important applications such as the production of detergents and lubricants. Vegetable oils are also used to produce biodiesel. The global demand for plant oils is rapidly increasing, and is estimated to double by 2030 (Chapman and Ohlrogge, 2012). The growing demand for vegetable oil increases the need for higher plant oil production.
TAG biosynthesis consists of two major steps, fatty acid biosynthesis and TAG assembly, requiring the collaboration between cellular compartment plastids and the endoplasmic reticulum (ER) (Ohlrogge and Chapman, 2011). Fatty acids are first synthesized in the plastids and then exported to ER to complete the TAG assembly. Fatty acid biosynthesis initiates with acetyl-CoA carboxylase (ACC) that catalyzes the carboxylation to convert acetyl-CoA and bicarbonate into malonyl-CoA. Fatty acids are next elongated by the fatty acid synthase (FAS) complex, with two carbon increments. Upon completion of fatty acids assembly, mediated by FAS, the synthesized fatty acids are exported from plastids to ER, in forms of acyl-CoA. The subsequent TAG synthesis in ER mainly occurs through the eukaryotic phospholipid biosynthetic pathway (Bates et al., 2009). In the last step of TAG synthesis, diacylglycerol (DAG) is converted to TAG (either using acyl-CoA or phospholipids), which is catalyzed by diacylglycerol acyltransferase (DGAT) or phosphatidylcholine:diacylglycerol acyltransferase (PDAT) (Zou et al., 1999; Dahlqvist et al., 2000; Zhang et al., 2009). TAG biosynthesis is sophisticated and not fully understood; therefore, investigation of regulatory mechanisms of TAG biosynthesis is essential, not only for advancing the basic research of plant lipids, but also for bioengineering novel oil crops with increased oil content.
Crucial Role of WRI1 in Transcriptional Control of Plant Oil Biosynthesis
Numerous studies have shown that WRINKLED1 (WRI1) is vital for transcriptional control of plant oil biosynthesis. The discovery of wri1-1 mutant (the Arabidopsis loss-of-function mutant of AtWRI1) was reported in 1998, leading to subsequent characterization of AtWRI1. Compared to the wild-type (WT), seed oil content in wri1-1 is reduced by 80% (Focks and Benning, 1998). The WRI1 gene encodes a transcription factor which belongs to the APETALA2 (AP2) transcription factor family (Figure 1A) (Cernac and Benning, 2004; Masaki et al., 2005). Transcriptomic (microarray) analysis of WT and wri1-1 using developing seeds found that the majority of genes with reduced expression level in the wri1-1 mutant encode enzymes in late glycolysis and fatty acid biosynthesis (Ruuska et al., 2002). Subsequent studies confirmed that a number of genes in late glycolysis and fatty acid biosynthesis are indeed AtWRI1 targets (Baud et al., 2007; Maeo et al., 2009). The AW-box ([CnTnG](n)7[CG]) was characterized as an AtWRI1 binding element (Maeo et al., 2009). WRI1 was hence considered a “master regulator” in transcriptional control of TAG biosynthesis (Chapman and Ohlrogge, 2012).
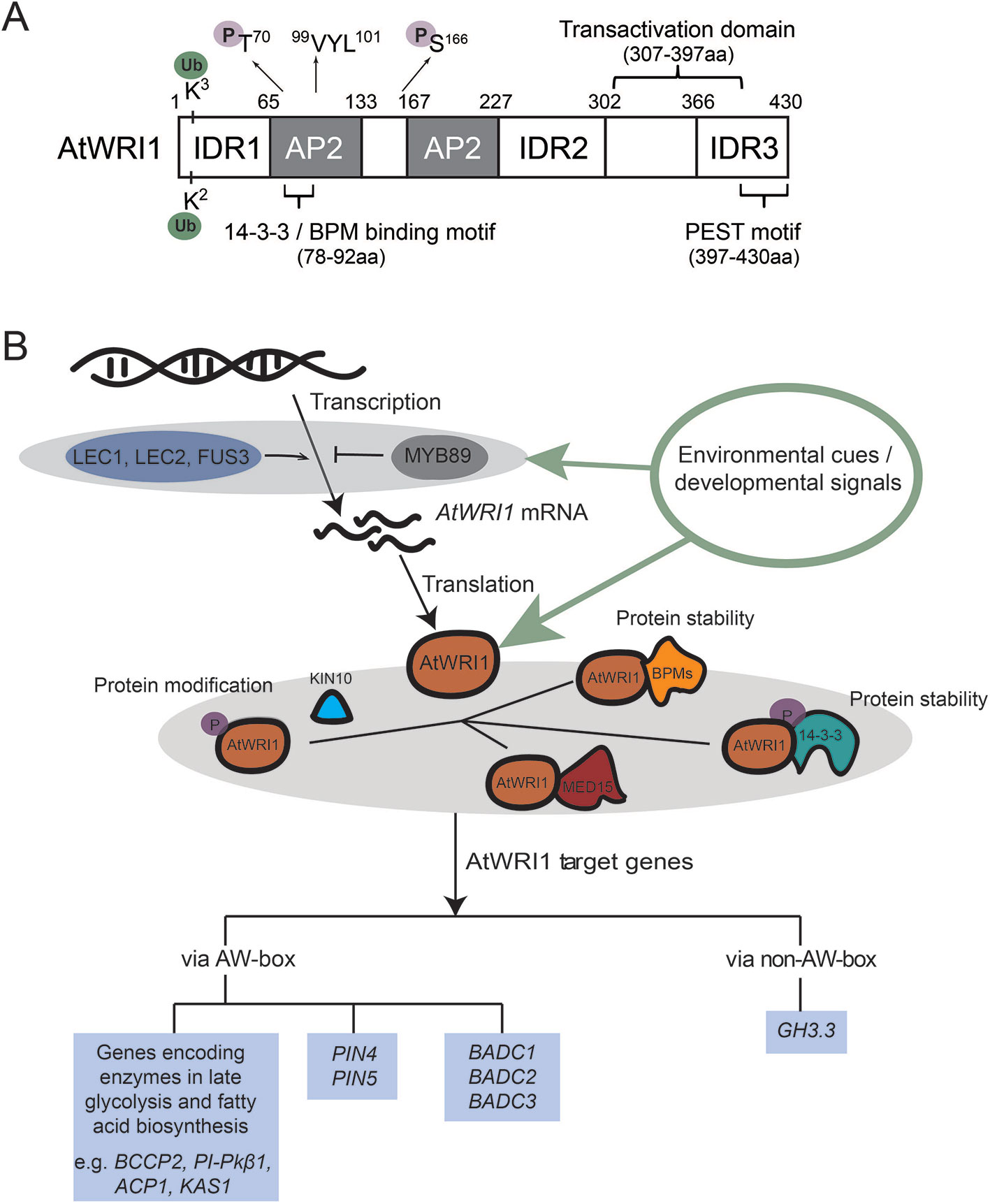
Figure 1 AtWRI1 structural characteristics and molecular mechanism of AtWRI1-regulated gene expression. (A) Schematic diagram of AtWRI1. AtWRI1 protein comprises of two AP2 domains, three intrinsically disordered regions (IDRs), a functional motif of “VYL”, the transactivation domain (TAD), the 14-3-3 and E3 ligase adaptor (BPM) binding motifs, the PEST motif, the ubiquitination sites, and the KIN10 phosphorylation sites. (B) Regulatory mechanism of AtWRI1 target genes. LEC1, LEC2, and FUS3 are positive regulators which mediate AtWRI1 expression. MYB89 acts as a negative regulator of AtWRI1 expression. At the protein level, AtWRI1 is regulated by post-translational modifications (such as phosphorylation and ubiquitination) and AtWRI1-interacting regulators, such as 14-3-3s, mediator subunit 15 (MED15), BPMs, and kinase KIN10. In the perception of environmental or developmental signals, the regulators fine-tune the protein stability and transcriptional activity of AtWRI1, through protein complex assembly and modifications, leading to mediation of the expression of AtWRI1 target genes.
Regulators Controlling the Expression of WRI1
Two central seed developmental regulators, LEAFY COTYLEDON1 (LEC1) and LEAFY COTYLEDON2 (LEC2), were hypothesized as essential transcriptional regulators which regulate AtWRI1 expression (Figure 1B). Expression of AtWRI1 is increased in the LEC1 gain-of-function mutant, tnp (Casson and Lindsey, 2006). In addition, expression of AtWRI1 is elevated in transgenic plants overexpressing LEC1 (Mu et al., 2008), suggesting LEC1 is possibly an upstream transcriptional regulator which activates the expression of AtWRI1 (Santos-Mendoza et al., 2008; Marchive et al., 2014). Induction of LEC2 in inducible LEC2-overexpression lines leads to the activation of AtWRI1, whereas AtWRI1 expression is decreased in lec2 mutant compared to WT plants (Baud et al., 2007). Therefore, the genetic and molecular evidence indicates that LEC2 functions as another transcriptional regulator controlling AtWRI1. In addition, transgenic maize overexpressing maize LEC1 (ZmLEC1) results in increased expression of maize WRI1 (ZmWRI1). ZmLEC1 is also shown to increase the GUS expression of promoterZmWRI1:GUS in maize cell culture (Shen et al., 2010). Transgenic soybean plants overexpressing soybean LEC2 (GmLEC2) results in elevated expression of soybean WRI1 (GmWRI1) (Manan et al., 2017). The evidence suggested a conserved mechanism controlling WRI1 expression via LEC1 and LEC2 in different plant species. How LEC1 or LEC2 binds the AtWRI1 promoter has not been determined (Santos-Mendoza et al., 2008; Marchive et al., 2014). Importantly, recent ChIP followed by DNA microarray (ChIP-chip) experiments demonstrated that AtWRI1 is a direct target gene of LEC1 (Pelletier et al., 2017).
FUSCA3 (FUS3) is another transcriptional regulator which mediates the expression of AtWRI1 (Figure 1B). Comparative transcriptome analysis using microarray showed that expression of AtWRI1 is reduced in a fus3 mutant (Yamamoto et al., 2010). FUS3 has been speculated to regulate the expression of AtWRI1 similar to LEC2, creating the functional redundancy between FUS3 and LEC2 (Yamamoto et al., 2010). Recent ChIP-Chip evidence confirmed that AtWRI1 is a direct target of FUS3 (Wang and Perry, 2013). Sucrose treatment induced AtWRI1 expression (Masaki et al., 2005). Expression of some sugar-responsive genes is stimulated by AtWRI1, suggesting the potential role of WRI1 in sugar signaling from carbon flow to oil production (Masaki et al., 2005). FUS3 does not affect LEC1/LEC2 expression, suggesting that activation of AtWRI1 by sucrose is via FUS3 instead of LEC1/LEC2 (Zhang et al., 2016a).
In addition to the transcriptional regulators LEC1, LEC2, and FUS3, other plants species may have alternative upstream regulators which contribute to mediating the expression of WRI1. In oil palm mesocarp, where most palm oil is derived from and EgWRI1 displays high expression, the inability to detect expression of key regulator genes, such as EgLEC1, EgLEC2, and EgFUS3, suggests that expression of EgWRI1 involves additional fruit-specific upstream regulators (Bourgis et al., 2011). EgNF-YA3, EgNF-YC2, and EgABI5 have been recently found to be capable of binding to the promoter region and activate the expression of EgWRI1 (Yeap et al., 2017).
Arabidopsis transcription factor MYB89 is a newly identified transcriptional repressor of AtWRI1 (Figure 1B). Transgenic plants overexpressing MYB89 shows decreased AtWRI1 expression, and a myb89 loss-of-function mutant displays elevated AtWRI1 expression compared to WT (Li et al., 2017a). ChIP experiments also showed that MYB89 binds to the AtWRI1 promoter directly, suggesting that AtWRI1 is a direct target gene of MYB89 (Li et al., 2017a).
Regulatory Mechanisms Mediating WRI1 Protein Stability and Activity
WRI1 activity is vital for oil biosynthesis as well as some developmental processes, such as seed germination and seedling establishment (Focks and Benning, 1998; Cernac and Benning, 2004; Cernac et al., 2006). Hence, it is not surprising that recent work uncovered several regulatory mechanisms that modulate WRI1 activity at different levels.
To prevent hyper-activation of key biological processes, the activities of transcription factors controlling metabolic pathways are often fine-tuned through protein-interacting regulators (Mouchiroud et al., 2014; Pireyre and Burow, 2015; Hafner et al., 2019). The interaction of AtWRI1 with CULLIN3-based E3 ligase adaptor BTB/POZMATH 1 (BPM1) in yeast-two-hybrid (Y2H) screening identified BPM1 as an AtWRI1-interacting partner (Chen et al., 2013). It is also demonstrated that AtWRI1 physically interacts with other BPM proteins. AtWRI1 assembles with E3 ligase adaptor BPMs, which mediates the degradation of AtWRI1 via 26S proteasome (Chen et al., 2013), connecting E3 ligase with plant oil production through the mediation of WRI1 protein stability.
Many proteins lack fixed three-dimensional structures, and intrinsic disordered regions (IDRs) have been found in eukaryotic proteins (Dyson and Wright, 2005; Kragelund et al., 2012; Valsecchi et al., 2013). IDRs tend to facilitate different confirmation requirements for varied specificities that are often regulated by post-translational modifications. Recent in silico analysis predicted three IDRs in the AtWRI1 protein (Ma et al., 2015). Functional analysis led to the characterization of an IDR3-located PEST motif (related to proteolysis), and a transactivation domain (TAD). Both TAD and IDR3-PEST motif are in the C-terminal region of AtWRI1, although not overlapping (Figure 1A). AtWRI1 variants, including IDR3-PEST deletion and mutations in putative phosphorylation residues of the IDR3-PEST motif, display enhanced protein stability and the expression of these variants elevated oil biosynthesis compared to that of AtWRI1 WT (Ma et al., 2015); therefore, phosphorylation is speculated to regulate AtWRI1 stability and activity (Ma et al., 2015).
In recent years, new AtWRI1-interacting regulators have been identified, expanding our understanding of the AtWRI1 regulatory network (summarized in Figure 1B). AtWRI1 physically interacts with 14-3-3 proteins in yeast and plant cells (Ma et al., 2016). 14-3-3 proteins are phosphopeptide-binding factors that are involved in various biological and physiological processes, such as metabolism, membrane transport, signal transduction, and protein modification (Jaspert et al., 2011; Paul et al., 2012; de Boer et al., 2013). Overexpression of 14-3-3 elevated oil production by increasing transcriptional activity and protein stability of AtWRI1 (Ma et al., 2016). Subsequent functional analysis identified overlapping 14-3-3- and BPM- binding motifs in AtWRI1. A hypothetical model suggests the interaction between AtWRI1 and 14-3-3 hinders either BPMs binding to AtWRI1 or the detachment of BPMs from AtWRI1. The AtWRI1–14-3-3 complex possesses elevated stability and transcriptional activity (Ma et al., 2016; Kong and Ma, 2018a).
It is often the case that plant gene regulation does not require de novo synthesis of transcription factor proteins; rather, it is mediated by post-transcriptional modification pathways. Among such pathways, protein phosphorylation mediated by kinases is perhaps the best studied process. KIN10 is an SNF1-related protein kinase which serves as a central regulator in transcriptional control of plant energy signal transduction pathways (Baena-Gonzalez et al., 2007). Recently, KIN10 has been shown to physically interact with and phosphorylate AtWRI1, leading to AtWRI1 degradation (Zhai et al., 2017). Phosphorylation deficient mutations at two residues in AtWRI1 (T70 and S166) impair KIN10-mediated phosphorylation and increase the AtWRI1 stability. 14-3-3 binding motif and KIN10 phosphorylation sites in AtWRI1 are neighboring, suggesting the possibility that the two may have overlapping proteasome-mediated protein degradation pathways (Zhai et al., 2017). In addition, trehalose 6-phosphate (T6P) was found to contribute to WRI1 stabilization and increase the generation of fatty acids via suppression of KIN10 activity (Zhai et al., 2018).
Recruitment of mediator complexes by transcription factors to accomplish their functions is a conserved mechanism of transcriptional regulation in eukaryotes (Taatjes, 2010). Physical interaction of Arabidopsis mediator complex MED15 subunit with AtWRI1 was demonstrated in vivo and in vitro (Kim et al., 2016). Transgenic Arabidopsis plants overexpressing MED15 displayed increased expression levels of oil biosynthetic genes targeted by AtWRI1. ChIP assays revealed that MED15 is capable of binding to promoters of AtWRI1 targets (Kim et al., 2016). However, the elevated expression of these AtWRI1 target genes in transgenic wri1 plants overexpressing MED15 suggested that MED15 interacts with other transcriptional regulators to regulate the gene expression of AtWRI1 targets (Kim et al., 2016).
Functional Conservation of WRI1 in Different Plant Species
A high degree of cross-species conservation is a hallmark of important master transcriptional regulators. A number of WRI1 orthologs have been found in various monocot and dicot plant species, including Brassica napus (Liu et al., 2010), Brachypodium distachyon (Yang et al., 2015), Camelina sativa (An et al., 2017), Zea mays (Shen et al., 2010; Pouvreau et al., 2011), Elaeis guineensis (Bourgis et al., 2011; Ma et al., 2013), Glycine max (Manan et al., 2017; Zhang et al., 2017; Chen et al., 2020), Ricinus communis (Tajima et al., 2013; Ji et al., 2018), Avena sativa (Grimberg et al., 2015), Cocos nucifera (Sun et al., 2017), Cyperus esculentus (Grimberg et al., 2015), Gossypium spp (Qu et al., 2012), Jatropha curcas (Ye et al., 2018), Persea americana (Kilaru et al., 2015), Solanum tuberosum (Grimberg et al., 2015), and Oryza sativa (Mano et al., 2019). Expressing AtWRI1 or WRI1 orthologs in wri1 loss-of-function mutants complemented the decreased seed oil phenotype of wri1 (Cernac and Benning, 2004; Pouvreau et al., 2011; Ma et al., 2013; An et al., 2017; Ye et al., 2018; Chen et al., 2020). Numerous WRI1 orthologs display high expression levels in developing seeds, correlating closely to the AtWRI1 expression patterns. Nonetheless, some WRI1s have been found to show high expression in non-seed tissues. For example, the expression of oil palm WRI1 (EgWRI1) is high in mesocarp and substantially increases in the ripening processes of fruits (Bourgis et al., 2011). In addition, PaWRI1 and CeWRI1 exhibit high expression levels in avocado mesocarp and nutsedge stem tubers, respectively (Grimberg et al., 2015; Kilaru et al., 2015). Structural features and functional domains/motifs, which have been identified in the AtWRI1 protein, including “VYL” (Ma et al., 2013), IDR (Ma et al., 2015), and the PEST motif (Ma et al., 2015), are conserved in other WRI1 proteins (Ma et al., 2013; Ma et al., 2015; Yang et al., 2015; An et al., 2017; Kong and Ma, 2018b; Ye et al., 2018; Chen et al., 2020; Kong et al., 2019; Snell et al., 2019; Tang et al., 2019). WRI1 orthologs identified in Ricinus connunis and Oryza sativa encode WRI1 proteins lacking a “VYL” motif but still trigger oil biosynthesis in planta by activating the expression of WRI1 target genes (Ji et al., 2018; Mano et al., 2019). Recent work on RcWRI1-B and OsWRI1-1 suggested that “VYL” might not be important for some WRI1s (Ji et al., 2018; Mano et al., 2019), despite its conservation in numerous WRI1s and its functional importance in both AtWRI1 (Ma et al., 2013) and Arabidopsis AP2 transcription factor, ANT (Krizek, 2003).
WRI1 Target Genes Which are Not Involved in Oil Biosynthesis
In addition to its role as a transcriptional regulator controlling expression of genes in late glycolysis and oil biosynthesis, WRI1 has been found to target genes in pathways other than oil biosynthesis. AtWRI1 is not only able to bind to the promoters of PINs (PIN-FORMEDs) through the AW-box, but also binds to the promoter of GH3.3 (a gene involved in auxin degradation) via a non-AW-box element, suggesting a role in modifying auxin homeostasis (Kong et al., 2017) (also see Figure 1B). The altered expression of auxin-related genes was also found in roots of soybean GmWRI1s-overexpressing plants or GmWRI1s-RNAi plants compared to WT control (Chen et al., 2020). Furthermore, AtWRI1 is a homolog of CitAP2.10 (encoding a Citrus sinensis AP2 transcription factor which is in charge of (+)-valencene biosynthesis), and AtWRI1 is capable of transactivating the C. sinensis terpene synthase 1 (CsTPS1) promoter in a dual-luciferase assay (Shen et al., 2016). How regulation of the AtWRI1 targets is linked to plant growth and development, such as root growth (Kong et al., 2017), is currently under investigation. It is important to consider the alternative targets when utilizing WRI1 to bioengineer crops for plant oil production. However, convincing evidence exists demonstrating WRI1 as one of the most effective genes for plant oil bioengineering.
Importance of WRI1 for Plant Oil Production
Transgenic plants overexpressing AtWRI1 or WRI1 orthologs show significantly increased seed oil content in a number of studies (Cernac and Benning, 2004; Liu et al., 2010; Shen et al., 2010; An and Suh, 2015; Yang et al., 2015; Sun et al., 2017; Ye et al., 2018). Transgenic Arabidopsis and Brachypodium seedlings, overexpressing AtWRI1 and BdWRI1 respectively, have increased oil content in vegetative tissues (Sanjaya et al., 2011; Yang et al., 2015). Selection of proper promoters to drive WRI1 expression is also critical for oil bioengineering. For instance, significantly increased oil content in seeds was observed in transgenic maize with embryo-preferred OLEOSIN (OLE) promoter driven ZmWRI1, but there was no distinguishable oil increase in transgenic maize with the starch endosperm-specific 19 KD ZEIN promoter driven ZmWRI1 (Shen et al., 2010). Another example is transgenic Arabidopsis transformed by a vector that confers FUS3 promoter driven AtWRI1 expression, which turns out to be effective in increasing seed oil production due to an extension of oil biosynthesis during the mid-phase seed developmental process (Kanai et al., 2016).
Ectopic expression of AtWRI1 or WRI1 orthologs in a transient tobacco expression system is also able to productively trigger TAG accumulation in tobacco leaves (Vanhercke et al., 2013; Grimberg et al., 2015; Ma et al., 2015). Transient co-expression of AtWRI1 and DGAT1 in tobacco leaves has led to considerably augmented oil content compared to expression of WRI1 alone, which suggests a synergistic effect between these two genes (Vanhercke et al., 2013). Transient overexpression of engineered AtWRI1 variants in tobacco leaves, either deletion of IDR3-PEST motif or phosphorylation deficient mutations in the IDR3-PEST motif, resulted in production of stabilized WRI1 proteins and enhanced oil production (Ma et al., 2015). Transient co-expression of AtWRI1 and 14-3-3 in tobacco leaves resulted in increased stability of AtWRI1 and enhanced oil production (Ma et al., 2016). The AtWRI1K2RK3R variant, with mutations in two ubiquitination residues, resulted in increased protein stability and oil biosynthesis using a tobacco transient expression system (Zhai et al., 2017). In metabolic engineering to enhance oil production in vegetative tissues, the strategy of “push, pull, package, and protect” is to upregulate (push) the de novo fatty acid pathway using a key transcription factor, combined with pulling the precursors toward the end products using rate-limiting enzymes, packaging TAG in oil bodies, and protecting TAG from degradation. WRI1 overexpression has been shown to be pivotal in such designed strategy (Vanhercke et al., 2019).
Transgenic plants, overexpressing transcription factors which can upregulate WRI1 expression, also display elevated seed oil content. For instance, transgenic Arabidopsis plants that overexpress GmZF351 or GmDREBL have increased oil content in seeds. Both soybean transcription factors are capable of binding to the AtWRI1 promoter and activate AtWRI1 expression (Zhang et al., 2016b; Li et al., 2017b). Overexpression of ZmLEC1 (an activator of WRI1) also effectively increases seed oil in various plants species including Arabidopsis, Camelina, and maize (Shen et al., 2010; Zhu et al., 2018).
Unusual fatty acids, such hydroxy fatty acids (HFAs), have high value for industrial applications because of their distinctive physical and chemical properties. Nonetheless, transgenic Arabidopsis plants which overexpress a hydroxylase gene only yield low amount of HFA with reduction of total seed oil (Bates and Browse, 2011; Bates et al., 2014). A feedback inhibition of fatty acid biosynthesis was believed to be the main reason for the decreased seed oil content (Bates et al., 2014). In order to overcome this bottleneck, Adhikari et al. attempted to generate Arabidopsis transgenic plants which co-express AtWRI1 with RcFAH12 (encoding a castor fatty acid hydroxylase). The resulting transgenic Arabidopsis plants have significantly elevated proportions of HFA and entire seed oil content, demonstrating WRI1’s role for effective circumventing feedback inhibition of fatty acid synthesis when ectopic expressing a hydroxylase gene (Adhikari et al., 2016).
Aberrant growth or cell death was observed in some transgenic plants which overexpress WRI1 (Cernac and Benning, 2004; Marchive et al., 2014; Yang et al., 2015). Choosing suitable promoters for certain plant species hence becomes a necessary consideration for the development of oil crops.
Perspectives
Since the discovery of the wri1-1 mutant, multiple studies have established the central role of WRI1 in transcriptional regulation of plant oil biosynthesis. However, key questions remain, including what are the upstream regulators of WRI1 and how do the master regulators of seed development, such as LEC1, LEC2, and FUS3, mediate the expression of WRI1? Alternative transcriptional regulators could play important roles in controlling WRI1 expression in response to various developmental or environmental cues. Several transcriptional regulators of EgWRI1 have been recently identified in oil palm (Yeap et al., 2017). Whether the newly expanded regulatory network in oil palm is conserved in other plant species is unclear. More detailed analyses of the WRI1 promoters from various plant species may allow us to generalize the conserved nature of combinatorial WRI1 transcriptional regulation.
AtWRI3 and AtWRI4 are WRI1-like proteins identified in Arabidopsis. Seed-specific overexpression of AtWRI3 or AtWRI4 in the wri1 mutant complemented the low seed-oil phenotype (To et al., 2012). AtWRI3 and AtWRI4 seem to share overlapping function with AtWRI1 in providing acyl precursors for cutin biosynthesis in floral tissues; however, their roles in seed oil biosynthesis are less certain as they are predominantly expressed in non-seed tissues such as stems and flowers (To et al., 2012). It should be noted that the wri1 wri3 wri4 triple mutant displayed no apparent changes in vegetative growth, raising the speculation of the involvement of other unidentified transcriptional regulators that control fatty acid biosynthesis in vegetative tissues (To et al., 2012; Marchive et al., 2014). Investigation of the unique and overlapping functions of WRI1-like factors will shed light on the complex transcriptional control of fatty acid biosynthesis, especially in vegetative tissues.
Structural analyses of WRI1 variants and orthologs have revealed that the IDR3-PEST motif, binding motifs of E3 ligase adaptor BPMs and 14-3-3s, and phosphorylation residues (T70/S166) are relevant for AtWRI1 stability and transcriptional activity, consequently affecting plant oil production (Ma et al., 2013; Ma et al., 2015; Ma et al., 2016; Zhai et al., 2017). Recent studies on the missing “VYL” motif of RcWRI1-B and OsWRI1-1 also open the discussion about the importance of “VYL” in WRI1 proteins (Ji et al., 2018; Mano et al., 2019). Generation of the three-dimensional structure, in combination of computer modeling, will further advance our understanding of WRI1 function.
The emerging picture shows crosstalk between the WRI1-interacting regulators which are also post-translationally regulated, highlighting the cooperative activity of the WRI1 transcriptional machinery (Figure 1). Phosphorylation has been proposed to play dual roles in mediating AtWRI1 protein stability during embryo development (Ma et al., 2016; Kong and Ma, 2018a). Therefore, one possible mechanism to fine-tune WRI1 activity is through phosphorylation and dephosphorylation by various kinases and phosphatases in response to different developmental and environmental cues. Recent progress in WRI1 interactome facilitates the identification of novel WRI-interacting factors; however, the molecular mechanisms mediating the interplay among these regulators remain to be elucidated.
Key questions also remain regarding WRI1 protein stability. Why do plants degrade WRI1? Understanding of the control mechanism of WRI1 degradation, particularly under different developmental stages and environmental conditions, will help address the biological questions of “when” and “how” the endogenous degradation system activates to degrade WRI1. The answers to these questions may ultimately be used to guide our efforts to increase plant oil production.
AtWRI1 activates the expression of several BIOTIN ATTACHMENT DOMAIN-CONTAINING (BADC) genes that inhibit fatty acid biosynthesis (Liu et al., 2019), suggesting an additional layer of regulation of WRI1-mediated fatty acid biosynthesis. In summary, while the recent insights on WRI1 significantly advance our understanding of the molecular mechanisms governing plant oil biosynthesis, our knowledge of the complex regulatory system is far from complete.
Author Contributions
QK, YY, and WM conceived the ideas, prepared the figures and wrote the first draft. LG, LY, and WM reviewed and edited the figures and manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by a Nanyang Technological University Startup grant and a Ministry of Education (MOE) of Singapore Tier 1 to WM (2018-T1-002-019).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to all authors whose important work is not cited due to space limitations.
References
Adhikari, N. D., Bates, P. D., Browse, J. (2016). WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol. 171 (1), 179–191. doi: 10.1104/pp.15.01906
An, D., Suh, M. C. (2015). Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 9, 137–148. doi: 10.1007/s11816-015-0351-x
An, D., Kim, H., Ju, S., Go, Y. S., Kim, H. U., Suh, M. C. (2017). Expression of Camelina WRINKLED1 Isoforms rescue the seed phenotype of the arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 8, 34. doi: 10.3389/fpls.2017.00034
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 (7156), 938–942. doi: 10.1038/nature06069
Bates, P. D., Browse, J. (2011). The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 68 (3), 387–399. doi: 10.1111/j.1365-313X.2011.04693.x
Bates, P. D., Durrett, T. P., Ohlrogge, J. B., Pollard, M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150 (1), 55–72. doi: 10.1104/pp.109.137737
Bates, P. D., Johnson, S. R., Cao, X., Li, J., Nam, J. W., Jaworski, J. G., et al. (2014). Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc. Natl. Acad. Sci. U. S. A 111 (3), 1204–1209. doi: 10.1073/pnas.1318511111
Baud, S., Mendoza, M. S., To, A., Harscoet, E., Lepiniec, L., Dubreucq, B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50 (5), 825–838. doi: 10.1111/j.1365-313X.2007.03092.x
Bourgis, F., Kilaru, A., Cao, X., Ngando-Ebongue, G. F., Drira, N., Ohlrogge, J. B., et al. (2011). Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. U. S. A 108 (30), 12527–12532. doi: 10.1073/pnas.1106502108
Casson, S. A., Lindsey, K. (2006). The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol. 142 (2), 526–541. doi: 10.1104/pp.106.080895
Cernac, A., Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40 (4), 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Cernac, A., Andre, C., Hoffmann-Benning, S., Benning, C. (2006). WRI1 is required for seed germination and seedling establishment. Plant Physiol. 141 (2), 745–757. doi: 10.1104/pp.106.079574
Chapman, K. D., Ohlrogge, J. B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287 (4), 2288–2294. doi: 10.1074/jbc.R111.290072
Chen, L., Lee, J. H., Weber, H., Tohge, T., Witt, S., Roje, S., et al. (2013). Arabidopsis BPM proteins function as substrate adaptors to a CULLIN3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25 (6), 2253–2264. doi: 10.1105/tpc.112.107292
Chen, B., Zhang, G., Li, P., Yang, J., Guo, L., Benning, C., et al. (2020). Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 18 (1), 155 –171. doi: 10.1111/pbi.13183
Dahlqvist, A., Stahl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., et al. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U. S. A 97 (12), 6487–6492. doi: 10.1073/pnas.120067297
de Boer, A. H., van Kleeff, P. J., Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250 (2), 425–440. doi: 10.1007/s00709-012-0437-z
Dyson, H. J., Wright, P. E. (2005). Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6 (3), 197–208. doi: 10.1038/nrm1589
Focks, N., Benning, C. (1998). wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118 (1), 91–101. doi: 10.1104/pp.118.1.91
Grimberg, A., Carlsson, A. S., Marttila, S., Bhalerao, R., Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15, 192. doi: 10.1186/s12870-015-0579-1
Hafner, A., Bulyk, M. L., Jambhekar, A., Lahav, G. (2019). The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20 (4), 199–210. doi: 10.1038/s41580-019-0110-x
Jaspert, N., Throm, C., Oecking, C. (2011). Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front. Plant Sci. 2, 96. doi: 10.3389/fpls.2011.00096
Ji, X. J., Mao, X., Hao, Q. T., Liu, B. L., Xue, J. A., Li, R. Z. (2018). Splice variants of the castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int. J. Mol. Sci. 19 (1), 146. doi: 10.3390/ijms19010146
Kanai, M., Mano, S., Kondo, M., Hayashi, M., Nishimura, M. (2016). Extension of oil biosynthesis during the mid-phase of seed development enhances oil content in Arabidopsis seeds. Plant Biotechnol. J. 14 (5), 1241–1250. doi: 10.1111/pbi.12489
Kilaru, A., Cao, X., Dabbs, P. B., Sung, H. J., Rahman, M. M., Thrower, N., et al. (2015). Oil biosynthesis in a basal angiosperm: transcriptome analysis of Persea Americana mesocarp. BMC Plant Biol. 15, 203. doi: 10.1186/s12870-015-0586-2
Kim, M. J., Jang, I. C., Chua, N. H. (2016). The mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol. 171 (3), 1951–1964. doi: 10.1104/pp.16.00664
Kong, Q., Ma, W. (2018a). WRINKLED1 as a novel 14-3-3 client: function of 14-3-3 proteins in plant lipid metabolism. Plant Signal Behav. 13, e1482176. doi: 10.1080/15592324.2018.1482176
Kong, Q., Ma, W. (2018b). WRINKLED1 transcription factor: how much do we know about its regulatory mechanism? Plant Sci. 272, 153–156. doi: 10.1016/j.plantsci.2018.04.013
Kong, Q., Ma, W., Yang, H., Ma, G., Mantyla, J. J., Benning, C. (2017). The Arabidopsis WRINKLED1 transcription factor affects auxin homeostasis in roots. J. Exp. Bot. 68, 4627–4634. doi: 10.1093/jxb/erx275
Kong, Q., Yuan, L., Ma, W. (2019). WRINKLED1, a “Master Regulator“ in transcriptional control of plant oil biosynthesis. Plants (Basel) 8 (7), 238. doi: 10.3390/plants8070238
Kragelund, B. B., Jensen, M. K., Skriver, K. (2012). Order by disorder in plant signaling. Trends Plant Sci. 17 (11), 625–632. doi: 10.1016/j.tplants.2012.06.010
Krizek, B. A. (2003). AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res. 31 (7), 1859–1868. doi: 10.1093/nar/gkg292
Li, D., Jin, C., Duan, S., Zhu, Y., Qi, S., Liu, K., et al. (2017a). MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 173 (2), 1211–1225. doi: 10.1104/pp.16.01634
Li, Q. T., Lu, X., Song, Q. X., Chen, H. W., Wei, W., Tao, J. J., et al. (2017b). Selection for a Zinc-finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 173 (4), 2208–2224. doi: 10.1104/pp.16.01610
Liu, J., Hua, W., Zhan, G., Wei, F., Wang, X., Liu, G., et al. (2010). Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 48 (1), 9–15. doi: 10.1016/j.plaphy.2009.09.007
Liu, H., Zhai, Z., Kuczynski, K., Keereetaweep, J., Schwender, J., Shanklin, J. (2019). WRINKLED1 regulates biotin attachment domain-containing proteins that inhibit fatty acid synthesis. Plant Physiol. 181 (1), 55–62. doi: 10.1104/pp.19.00587
Ma, W., Kong, Q., Arondel, V., Kilaru, A., Bates, P. D., Thrower, N. A., et al. (2013). Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PloS One 8 (7), e68887. doi: 10.1371/journal.pone.0068887
Ma, W., Kong, Q., Grix, M., Mantyla, J. J., Yang, Y., Benning, C., et al. (2015). Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 83 (5), 864–874. doi: 10.1111/tpj.12933
Ma, W., Kong, Q., Mantyla, J. J., Yang, Y., Ohlrogge, J. B., Benning, C. (2016). 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 88 (2), 228–235. doi: 10.1111/tpj.13244
Maeo, K., Tokuda, T., Ayame, A., Mitsui, N., Kawai, T., Tsukagoshi, H., et al. (2009). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60 (3), 476–487. doi: 10.1111/j.1365-313X.2009.03967.x
Manan, S., Ahmad, M. Z., Zhang, G., Chen, B., Haq, B. U., Yang, J., et al. (2017). Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 8, 1604. doi: 10.3389/fpls.2017.01604
Mano, F., Aoyanagi, T., Kozaki, A. (2019). Atypical splicing accompanied by skipping conserved micro-exons produces unique WRINKLED1, an AP2 domain transcription factor in rice plants. Plants (Basel) 8 (7), 207. doi: 10.3390/plants8070207
Marchive, C., Nikovics, K., To, A., Lepiniec, L., Baud, S. (2014). Transcriptional regulation of fatty acid production in higher plants: molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 116, 1332–1343. doi: 10.1002/ejlt.201400027
Masaki, T., Mitsui, N., Tsukagoshi, H., Nishii, T., Morikami, A., Nakamura, K. (2005). ACTIVATOR of Spomin::LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 46 (4), 547–556. doi: 10.1093/pcp/pci072
Mouchiroud, L., Eichner, L. J., Shaw, R. J., Auwerx, J. (2014). Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 20 (1), 26–40. doi: 10.1016/j.cmet.2014.03.027
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148 (2), 1042–1054. doi: 10.1104/pp.108.126342
Ohlrogge, J. B., Chapman, K. D. (2011). The seeds of green energy: expanding the contribution of plant oils as biofuels. Biochemist. (Lond.) 33, 34–38. doi: 10.1042/BIO03302034
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., Ferl, R. J. (2012). 14-3-3 phosphoprotein interaction networks - does isoform diversity present functional interaction specification? Front. Plant Sci. 3, 190. doi: 10.3389/fpls.2012.00190
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U. S. A 114 (32), E6710–E6719. doi: 10.1073/pnas.1707957114
Pireyre, M., Burow, M. (2015). Regulation of MYB and bHLH transcription factors: a glance at the protein level. Mol. Plant 8 (3), 378–388. doi: 10.1016/j.molp.2014.11.022
Pouvreau, B., Baud, S., Vernoud, V., Morin, V., Py, C., Gendrot, G., et al. (2011). Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156 (2), 674–686. doi: 10.1104/pp.111.173641
Qu, J., Ye, J., Geng, Y. F., Sun, Y. W., Gao, S. Q., Zhang, B. P., et al. (2012). Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160 (2), 738–748. doi: 10.1104/pp.112.198564
Ruuska, S. A., Girke, T., Benning, C., Ohlrogge, J. B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 (6), 1191–1206. doi: 10.1105/tpc.000877
Sanjaya, Durrett, T. P., Weise, S. E., Benning, C. (2011). Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9 (8), 874–883. doi: 10.1111/j.1467-7652.2011.00599.x
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54 (4), 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Shen, B., Allen, W. B., Zheng, P., Li, C., Glassman, K., Ranch, J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153 (3), 980–987. doi: 10.1104/pp.110.157537
Shen, S. L., Yin, X. R., Zhang, B., Xie, X. L., Jiang, Q., Grierson, D., et al. (2016). CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 67 (14), 4105–4115. doi: 10.1093/jxb/erw189
Snell, P., Grimberg, A., Carlsson, A. S., Hofvander, P. (2019). WRINKLED1 is subject to evolutionary conserved negative autoregulation. Front. Plant Sci. 10, 387. doi: 10.3389/fpls.2019.00387
Sun, R., Ye, R., Gao, L., Zhang, L., Wang, R., Mao, T., et al. (2017). Characterization and ectopic expression of CoWRI1, an AP2/EREBP domain-containing transcription factor from coconut (Cocos nucifera L.) Endosperm, changes the seeds oil content in transgenic arabidopsis thaliana and rice (Oryza sativa L.). Front. Plant Sci. 8, 63. doi: 10.3389/fpls.2017.00063
Taatjes, D. J. (2010). The human mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 35 (6), 315–322. doi: 10.1016/j.tibs.2010.02.004
Tajima, D., Kaneko, A., Sakamoto, M., Ito, Y., Hue, N. T., Miyazaki, M., et al. (2013). Wrinkled 1 (WRI1) Homologs, AP2-type transcription factors involving master regulation of seed storage oil Synthesis in Castor Bean (Ricinus communis L.). Am. J. Plant Sci. 4, 333–339. doi: 10.4236/ajps.2013.42044
Tang, T., Du, C., Song, H., Aziz, U., Wang, L., Zhao, C., et al. (2019). Genome-wide analysis reveals the evolution and structural features of WRINKLED1 in plants. Mol. Genet. Genomics 294 (2), 329–341. doi: 10.1007/s00438-018-1512-8
To, A., Joubes, J., Barthole, G., Lecureuil, A., Scagnelli, A., Jasinski, S., et al. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24 (12), 5007–5023. doi: 10.1105/tpc.112.106120
Valsecchi, I., Guittard-Crilat, E., Maldiney, R., Habricot, Y., Lignon, S., Lebrun, R., et al. (2013). The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol. Biosyst. 9 (9), 2282–2295. doi: 10.1039/c3mb70128j
Vanhercke, T., El Tahchy, A., Shrestha, P., Zhou, X. R., Singh, S. P., Petrie, J. R. (2013). Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 587 (4), 364–369. doi: 10.1016/j.febslet.2012.12.018
Vanhercke, T., Dyer, J. M., Mullen, R. T., Kilaru, A., Rahman, M. M., Petrie, J. R., et al. (2019). Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res. 74, 103–129. doi: 10.1016/j.plipres.2019.02.002
Wang, F., Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161 (3), 1251–1264. doi: 10.1104/pp.112.212282
Yamamoto, A., Kagaya, Y., Usui, H., Hobo, T., Takeda, S., Hattori, T. (2010). Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 51 (12), 2031–2046. doi: 10.1093/pcp/pcq162
Yang, Y., Munz, J., Cass, C., Zienkiewicz, A., Kong, Q., Ma, W., et al. (2015). Ectopic Expression of WRINKLED1 affects fatty acid homeostasis in brachypodium distachyon vegetative tissues. Plant Physiol. 169 (3), 1836–1847. doi: 10.1104/pp.15.01236
Ye, J., Wang, C., Sun, Y., Qu, J., Mao, H., Chua, N. H. (2018). Overexpression of a transcription factor increases lipid content in a woody Perennial Jatropha curcas. Front. Plant Sci. 9, 1479. doi: 10.3389/fpls.2018.01479
Yeap, W. C., Lee, F. C., Shabari Shan, D. K., Musa, H., Appleton, D. R., Kulaveerasingam, H. (2017). WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 91 (1), 97–113. doi: 10.1111/tpj.13549
Zhai, Z., Liu, H., Shanklin, J. (2017). Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29 (4), 871–889. doi: 10.1105/tpc.17.00019
Zhai, Z., Keereetaweep, J., Liu, H., Feil, R., Lunn, J. E., Shanklin, J. (2018). Trehalose 6-Phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30 (10), 2616–2627. doi: 10.1105/tpc.18.00521
Zhang, M., Fan, J., Taylor, D. C., Ohlrogge, J. B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21 (12), 3885–3901. doi: 10.1105/tpc.109.071795
Zhang, M., Cao, X., Jia, Q., Ohlrogge, J. (2016a). FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 88 (1), 95–107. doi: 10.1111/tpj.13233
Zhang, Y. Q., Lu, X., Zhao, F. Y., Li, Q. T., Niu, S. L., Wei, W., et al. (2016b). Soybean GmDREBL increases lipid content in seeds of transgenic Arabidopsis. Sci. Rep. 6, 34307. doi: 10.1038/srep34307
Zhang, D., Zhao, M., Li, S., Sun, L., Wang, W., Cai, C., et al. (2017). Plasticity and innovation of regulatory mechanisms underlying seed oil content mediated by duplicated genes in the palaeopolyploid soybean. Plant J. 90 (6), 1120–1133. doi: 10.1111/tpj.13533
Zhu, Y., Xie, L., Chen, G. Q., Lee, M. Y., Loque, D., Scheller, H. V. (2018). A transgene design for enhancing oil content in Arabidopsis and Camelina seeds. Biotechnol. Biofuels 11, 46. doi: 10.1186/s13068-018-1049-4
Keywords: WRI1, transcription factor, plant oil biosynthesis, intrinsically disordered region, post-translational modifications, protein stability, protein–protein interaction
Citation: Kong Q, Yang Y, Guo L, Yuan L and Ma W (2020) Molecular Basis of Plant Oil Biosynthesis: Insights Gained From Studying the WRINKLED1 Transcription Factor. Front. Plant Sci. 11:24. doi: 10.3389/fpls.2020.00024
Received: 03 November 2019; Accepted: 10 January 2020;
Published: 04 February 2020.
Edited by:
Xue-Rong Zhou, Commonwealth Scientific and Industrial Research Organisation, AustraliaReviewed by:
Enrique Martinez Force, Instituto de la Grasa (IG), SpainAruna Kilaru, East Tennessee State University, United States
Copyright © 2020 Kong, Yang, Guo, Yuan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ma, weima@ntu.edu.sg
 Que Kong1
Que Kong1