
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci., 07 February 2020
Sec. Plant Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01795
This article is part of the Research TopicChromatin Stability and Dynamics: Targeting and Recruitment of Chromatin ModifiersView all 10 articles
In June 2019, more than a hundred plant researchers met in Cologne, Germany, for the 6th European Workshop on Plant Chromatin (EWPC). This conference brought together a highly dynamic community of researchers with the common aim to understand how chromatin organization controls gene expression, development, and plant responses to the environment. New evidence showing how epigenetic states are set, perpetuated, and inherited were presented, and novel data related to the three-dimensional organization of chromatin within the nucleus were discussed. At the level of the nucleosome, its composition by different histone variants and their specialized histone deposition complexes were addressed as well as the mechanisms involved in histone post-translational modifications and their role in gene expression. The keynote lecture on plant DNA methylation by Julie Law (SALK Institute) and the tribute session to Lars Hennig, honoring the memory of one of the founders of the EWPC who contributed to promote the plant chromatin and epigenetic field in Europe, added a very special note to this gathering. In this perspective article we summarize some of the most outstanding data and advances on plant chromatin research presented at this workshop.
Last year, the Max Planck Institute for Plant Breeding Research in Cologne hosted the 6th European Workshop on Plant Chromatin (EWPC). A total of 110 researchers met to present the most recent focuses, advances, and challenges in the plant chromatin and epigenetics field during this 2-day workshop that comprised more than 25 standard talks and a similar number of short PechaKucha-style talks. Many other topics were talked over during the poster sessions in which the participants had the opportunity to discuss new discoveries and concepts in plant chromatin science in a thriving atmosphere.
Several talks emphasized the complexity of chromatin organization within the three-dimensional space of the nucleus and presented cutting-edge techniques developed to provide a deeper and higher-resolution view of chromatin structure (Figure 1). As in previous EWPCs, histone variants and histone marks were an important theme for many research laboratories. Considerable progress has been made in recent years to understand their links to transcriptional regulation. Also, of note have been the advances in our understanding of the proteins and complexes that are involved in the deposition of histone variants and marks, which, additionally, may act as readers of these chromatin features.
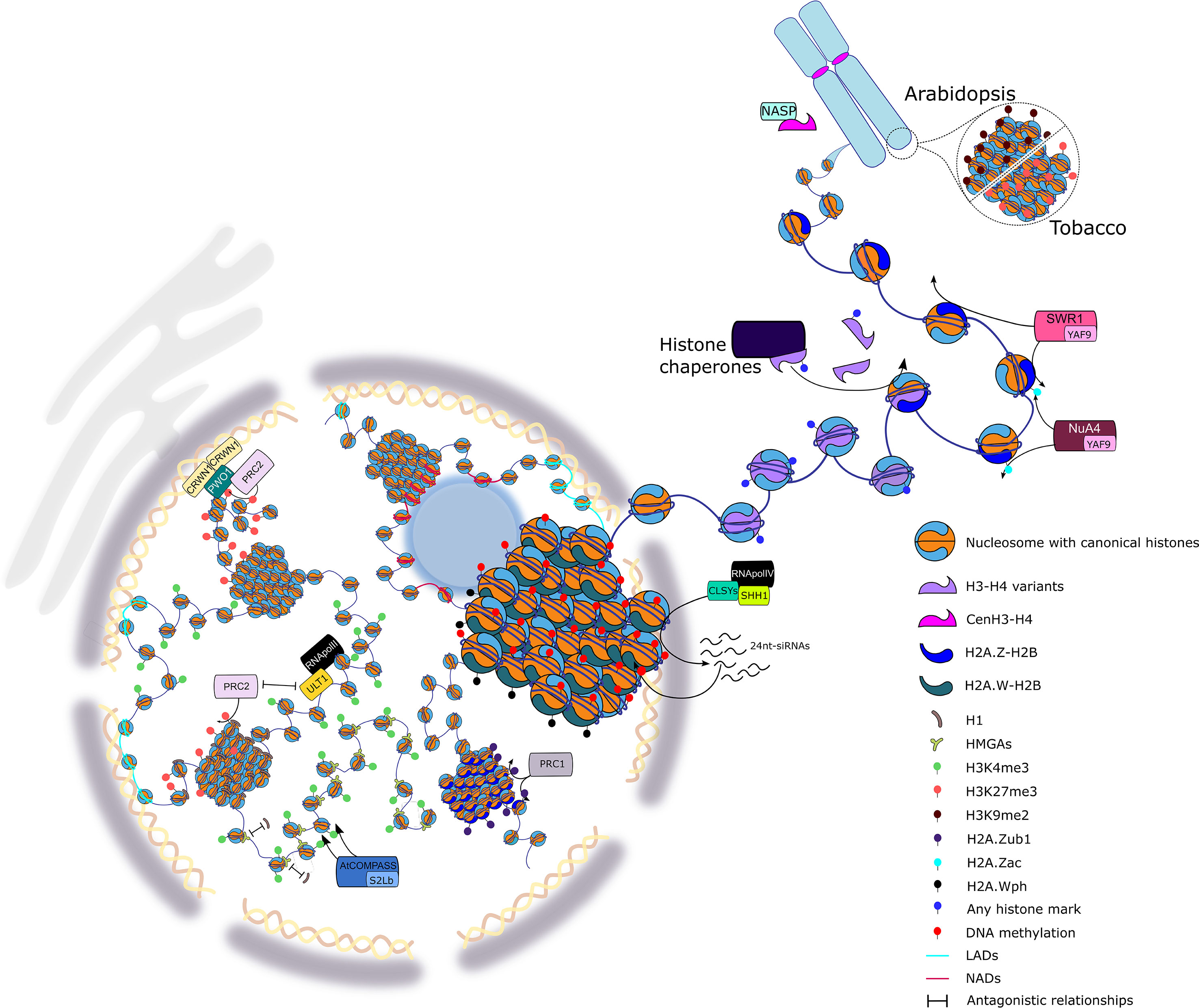
Figure 1 Highlights of the European Plant Chromatin Workshop 2019. Chromatin organization is a central player in controlling gene expression and concomitantly regulating plant development and plant responses to the environment. The scheme illustrates some of the aspects of plant chromatin organization presented at the EWPC ranging from local chromatin changes touching the bricks of the nucleosome to higher-order chromatin organization. At the level of the nucleosome, modifications of the DNA molecule and histone proteins were discussed, such as the regulation of DNA methylation involving the CLASSY (CLSY) proteins, the incorporation of specific variants of histones H1, H3, and H2A through dedicated histone chaperone complexes, and the dynamics of non-histone DNA-binding proteins, such as HIGH MOBILITY GROUP A (HMGA). Post-translational modifications of histones are set by specific complexes exemplified here by COMPLEX PROTEINS ASSOCIATED WITH SET1 (COMPASS), involved in H3K4me3, Nucleosome Acetyltransferase of Histone H4 (NuA4), in H2A.Z acetylation, and Polycomb Repressive Complex 1 (PRC1), in H2A.Z monoubiquitination. The role of chromatin remodelers in the deposition of histones is also depicted through H2A.Z-mediated deposition by the SWI/SNF-Related protein 1 (SWR1) complex. Modification of H2A.W by phosphorylation and its association to the heterochromatin was observed. The interplay between repressive modifications set by PRC2 and the antagonizing activity of ULTRAPETALA1 (ULT1) allows for a dynamic transcriptional regulation. Finally, the formation of specific chromatin domains in the nucleus, such as telomeres, nucleolus/lamina associated domains (NADs/LADs), or the association of chromatin domains via PWWP INTERACTOR OF POLYCOMB (PWO1) to CRWN1, a plant lamina component, were presented.
Current challenges that have arisen from issues such as food security and climate change have added a new dimension to the study of epigenetic regulation of plant traits and epigenetic inheritance of transcriptional stages. For that reason, the link between chromatin dynamics, gene expression, and plant developmental adaptation to the environment was also substantially addressed in the meeting. To advance in this field, analyses of chromatin architecture changes at different developmental stages and the tissue- or cell-specific level that have been technically challenging were presented.
Julie Law from the Salk Institute (La Jolla, USA) was invited to present the keynote lecture, which highlighted some of the most important past and present contributions to the DNA methylation field from her laboratory. Julie gave an overview of the crucial roles played by DNA methylation in gene regulation and transposon silencing. In addition, she reported that a family of four putative chromatin remodeling factors, CLASSY (CLSY) 1–4, associate with the RNA-directed DNA methylation (RdDM) pathway components Pol-IV and SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) (Law et al., 2011). Further recent studies showed that CLSY proteins function individually as locus-specific regulators of RdDM and in global regulation of DNA methylation patterns in Arabidopsis (Zhou et al., 2018). The next phase of the Law’s laboratory work aims to identify the roles of CLSY proteins in controlling DNA methylation patterns in a tissue-specific manner.
The EWPC was also the perfect venue for honoring the memory of Professor Lars Hennig who has recently passed away (Mozgová et al., 2018a). Together with Claudia Köhler and Valérie Gaudin, he established the EWPC in 2009 as one of the main gathering platforms for the plant epigenetic research community in Europe, bringing his enthusiasm and passion on plant science to these workshop series. During this tribute session, Lars’ colleagues and alumni shared with the audience his impact and vision on the chromatin and epigenetics field.
This perspective article summarizes the main topics discussed during the EWPC 2019 and provides insight into the future paths that the plant epigenetic community will follow in the next years. We thank all the laboratories, which have contributed to the EWPC with recently published or unpublished data, and we apologize to the researchers whose work could not be cited due to space limitations.
The key bearer of genetic information in eukaryotic cells is chromatin, which is non-randomly distributed inside the nucleus and shows an extraordinary degree of compaction and spatial organization. Nuclear organization is achieved by many factors, including histone proteins, modifiers and readers, as well as structural components of the nuclear periphery and nuclear bodies, which together dynamically control the nuclear architecture and may form nuclear domains (Sexton and Cavalli, 2015). The first session of the EWPC meeting dealt with the role of these factors in chromatin and nuclear organization.
The core histones have been structurally conserved through evolution and have evolved to accomplish two conflicting and yet vital tasks: on one hand, the long DNA molecules have to be packaged within the limits of the eukaryotic nucleus, preventing knots and tangles and protecting the genome from physical damage; on the other hand, the information that is encoded in the DNA needs to be accessed at appropriate times (Rosa and Shaw, 2013). The linker DNA between nucleosomes is bound by linker histones H1 (Rutowicz et al., 2015; Kotliński et al., 2017) whose role is much less understood than core histones. A recent study presented by Célia Baroux (Zurich, Switzerland) provided a multi-scale functional analysis of Arabidopsis linker histones. The work, done in collaboration with the laboratories from Andrzej Jerzmanowski (Warsaw, Poland) and Fredy Barneche (Paris, France), showed that H1-deficient plants are viable but exhibit phenotypes in seed dormancy, flowering time, as well as lateral root and stomata formation. In addition to a role in heterochromatin compaction, H1 seems to regulate nucleosome distribution over gene bodies. Yet, the authors showed that H1-mediated chromatin organization may act downstream of transcriptional control for a large number of loci in Arabidopsis. In addition, a new connection was found between H1 and H3K27me3. The findings suggest that H1 may act as a chromatin organizer favoring the maintenance of this epigenetic mark as well as others (Rutowicz et al., 2019).
Frédéric Pontvianne (Perpignan, France) focused on the nucleolus, the largest nuclear body, which is well known as the site of ribosomal RNA (rRNA) gene transcription, rRNA processing, and ribosome biogenesis (Boisvert et al., 2007). In a previous study, Frédéric and co-workers identified chromatin regions associated with the nucleolus, termed Nucleolus Associated Domains (NADs). NADs are primarily genomic regions with heterochromatic signatures and include transposable elements (TEs), sub-telomeric regions, and mostly inactive protein-coding genes (Pontvianne et al., 2016). Recent data now suggest that the rRNA gene copy number impacts the organization of NADs, and this suggests a role of nucleolus organizer regions (NORs) in establishing domains of inactive chromatin associated with the nucleolus (Picart-Picolo et al., 2019).
Similar to the nucleolus, the nuclear periphery is another compartment within the nucleus that plays a crucial role in chromatin organization and nuclear architecture. Kalyanikrishna (Berlin, Germany) presented data showing a putative link between Polycomb Group (PcG)-mediated repression and the nuclear periphery in Arabidopsis. PWWP INTERACTOR OF POLYCOMB (PWO1) is a PWWP-domain containing protein able to interact with any of the three possible POLYCOMB REPRESSIVE COMPLEX 2 (PRC2) histone methyltransferases in Y2H, and PWO1-CURLY LEAF (CLF) interaction was confirmed in planta (Hohenstatt et al., 2018; Mikulski et al, 2019). Among the putative interactors of PWO1, CROWDED NUCLEI1 (CRWN1) has been identified (Mikulski et al, 2019). CRWN1 is a coiled coil analog of lamin proteins, whose absence alters nuclear morphology (Wang et al., 2013), and a set of H3K27me3 targets were upregulated in crwn1 crwn2 double mutants in Arabidopsis. The interaction between PWO1 and CRWN1 suggests a role of the nuclear periphery in PRC2-mediated gene regulation in Arabidopsis (Mikulski et al., 2019). The Schubert laboratory continues to work on characterizing putative interactors involved in this pathway.
The post-translational modifications of telomere histones in plants have been investigated by Katerina Adamusova (Brno, Czech Republic). Among the canonical and non-canonical telomeres in plants, the authors found two kinds of epigenetic patterns regardless of the differences in telomere length and telomeric sequences used. One of them corresponds to the Arabidopsis-like pattern, where telomere histones are marked predominantly with H3K9me2. The other one is the tobacco-like pattern marked predominantly with H3K27me3 (Adamusová et al., 2019).
Hua Jiang (Gatersleben, Germany) discussed the role of AT-hook proteins in the regulation of gene expression by mediating the H3K9me2 heterochromatic mark at the nuclear matrix-associated regions (MARs). They identified AT-Hook Like 10 (AHL10), a member of the AT-hook family in Arabidopsis, and the SET domain containing SU(VAR)3-9 homolog (SUVH9) as interacting partners of ADMETOS (ADM), which functions in establishing the postzygotic hybridization barrier in Arabidopsis. Significantly increased expression of ADM and AHL10 in Arabidopsis triploid seeds results in H3K9me2 hypermethylation in MARs. Furthermore, AHL10-mediated H3K9me2 hypermethylation at MARs is independent of DNA methylation (Jiang et al., 2017). Apart from AHL10, the authors found that the overexpression line of another AHL also has increased H3K9me2 levels at TEs in sporophytic tissues, indicating a similar role for other members of this family.
Recent advances in our understanding of inter-generational inheritance of epigenetic and chromatin marks have revealed a variety of plant peculiarities, rendering this topic an exciting field of study with impact on our fundamental understanding of inheritance, phenotypic plasticity, population dynamics, and evolution (Köhler and Springer, 2017; Miryeganeh and Saze, 2019). Nevertheless, many open questions remain concerning what epigenetic information is inherited, the mechanisms of inheritance, and the processes involved in eventual reprogramming to prevent inheritance. To shed light on these questions, an enhanced understanding of gene regulation in gametophytes is vital. Sara Simonini (Zurich, Switzerland) focused her presentation on gene regulation in the female gametophyte and during early seed development by analyzing interaction partners and direct targets of the PRC2 methyltransferase MEDEA (MEA). Previous works have implicated MEA in the repression of seed development before fertilization and in endosperm cellularization (Chaudhury et al., 1997; Grossniklaus et al., 1998; Köhler et al., 2003a). The new unpublished data indicate that MEA interacts with histone deacetylases (HDACs), and that plants depleted in HDACs display similar abnormal phenotypes as mea mutants, suggesting an interplay between histone methylation and acetylation during early seed development.
The double fertilization process of plants generates an additional complication in the understanding of trans-generational inheritance and maternal and paternal contributions to the next generation. Thus, being able to distinguish events taking place in the endosperm from other plant tissues will be crucial to understand the peculiarities of this triploid tissue. An exciting technical advance in this direction was presented by Vikash Kumar Yadav (Uppsala, Sweden). He performed modified high-throughput chromatin conformation (mHi-C) on purified endosperm nuclei isolated by the INTACT method (Moreno-Romero et al., 2017), thus enabling Hi-C analysis on a limited number of nuclei. With this technique, he was able to observe elevated chromatin interaction levels in endosperm tissue compared to leaf tissue and discover that self-looping genes are on average expressed at a higher level compared to non-self-looping genes.
Heinrich Bente (Vienna, Austria) focused his presentation on yet another aspect of epigenetic inheritance, the phenomenon of paramutation, characterized by interallelic communication between epialleles at a single locus that results in stable and heritable silencing. Employing an epigenetically regulated resistance marker for hygromycin in Arabidopsis, Heinrich and his co-workers found that paramutation becomes apparent in F2 progeny of tetraploid hybrids but not in diploid ones. Small RNA profiles differ between the two epialleles, as do DNA methylation and chromatin marks. The fact that the paramutation is not observed at low temperatures, where also small RNA production is reduced (Baev et al., 2014), supports the assumption that small RNAs may be involved in paramutations.
Before epigenetic marks such as DNA methylation can be inherited between generations, they need to be maintained during cell divisions in the parents. Especially for asymmetric CHH methylation, maintenance is coupled to RdDM (Law and Jacobsen, 2010). Gergely Molnar (Tulin, Austria) reported the characterization of freak show (fks), a novel missense mutant of the RNA Polymerase V-specific subunit NRPE5A (Ream et al., 2009). The mutation displayed loss of transposon silencing due to generally reduced CG DNA methylation as well as hypermethylation at other loci, together leading to abnormal phenotypes, including flowering time defects and homeotic transformations. The findings seem to contrast canonical RNA Pol-V function in RdDM only, which mainly affects CHG and CHH methylation, and suggest a connection between an RdDM component and CG methylation maintenance.
To modulate nucleosome properties, including DNA accessibility and interactions between nucleosomes or even chromatin fibers, different histone variants can be incorporated. Recent years have seen accumulating evidence for the functional importance of these different histone variants for processes ranging from gene expression control and reprogramming to DNA repair processes in mammals and plants (Jiang and Berger, 2016; Buschbeck and Hake, 2017; Dabin and Polo, 2017).
Intriguing examples for these roles, reported by Anna Schmücke (Vienna, Austria), are the plant-specific histone variants H2A.W.6, H2A.W.7, and H2A.W.12, highly enriched in heterochromatin and involved in chromatin fiber–fiber interactions (Yelagandula et al., 2014). These histone variants are distinguished by a highly conserved KSPKK motif in their C-terminal tail. In response to DNA damage in heterochromatin, one of the three H2A.W variants, namely H2A.W.7, is phosphorylated at its SQE motif, and this phosphorylation is required for an appropriate DNA damage response (DDR) (Lorković et al., 2017). New evidence now indicates that only H2A.W.6, and not H2A.W.7, is phosphorylated in the conserved KSPKK motif in a cell cycle-dependent manner in Arabidopsis. Through a synthetic approach in fission yeast, she demonstrated that the phosphorylation of the KSPKK motif in addition to the phosphorylated SQE motif impairs a proper DNA damage response. This exemplifies a highly complex relationship between histone variants, their post-translational modification status, and their biological function. Another interesting H2A variant is H2A.Z, which has been associated both with transcriptional activation and repression depending on its position within a gene (Coleman-Derr and Zilberman, 2012; Sura et al., 2017). Wiam Merini (Seville, Spain) presented recent data resolving part of the mystery of this dual role of H2A.Z in transcription. She showed that, similar to canonical H2A, H2A.Z can be mono-ubiquitinated by PRC1 and that this post-translational modification plays an important role in transcriptional repression independent of PRC2 activity (Gómez-Zambrano et al., 2019). Indeed, complementation with a ubiquitination-resistant H2A.Z protein failed to rescue expression of upregulated genes in h2a.z mutant plants revealing the importance of H2A.Z ubiquitination. In contrast, H2A.Z ubiquitination seems to be dispensable to the rescue expression of the genes downregulated in h2a.z mutant plants; these genes may simply require H2A.Z incorporation. Alternatively, other post-translational modifications may play a role; in yeast, H2A.Z is acetylated by the NuA4 complex (Lu et al., 2009). Indeed, the confirmation that H2A.Z acetylation occurs in plants was provided by José A. Jarillo (Madrid, Spain). He studied the plant homologues of YEAST ALL1-FUSED GENE FROM CHROMOSOME 9 (YAF9) proteins, which are common components of the SWR1 complex involved in H2A.Z deposition and the NuA4 complex. In the absence of YAF9 proteins, H2A.Z acetylation is reduced at the FLC chromatin, and FLC expression is repressed, while H2A.Z incorporation as such is unaffected at this locus (Crevillén et al., 2019).
Given the emerging roles of the different histone variants in gene expression control and DNA repair reported at this conference, it becomes clear that histone deposition needs to be tightly controlled in time and space, and histone chaperones play an important role in this process. As an example, loss of H3 histone chaperones, such as HISTONE REGULATOR A (HIRA) (Nie et al., 2014; Duc et al., 2015) and the Arabidopsis ALPHA THALASSEMIA-MENTAL REDARDATION X-LINKED (ATRX) homologue (Duc et al., 2017), which function in complementary pathways of histone H3.3 deposition, results in altered gene expression. Aline V. Probst (GReD, France) discussed work from her laboratory, showing that ATRX loss-of-function affects H3.3 deposition at genes characterized both by elevated H3.3 occupancy and high expression levels, whereas hira mutants show reduced nucleosomal occupancy both at genes and in heterochromatin translating into reactivation of transposable elements. While some H3 histone chaperones are highly conserved, species-specific chaperones deposit the centromeric histone CenH3 (Müller and Almouzni, 2014). So far, the factor responsible for escort and deposition of plant CenH3 has remained enigmatic. Inna Lermontova (Gatersleben, Germany) reported on the collaborative effort to search for histone CenH3 interactors and the identification of the plant homologue of NUCLEAR AUTOANTIGENIC SPERM PROTEIN (NASP) as a CenH3 binding protein. Previously shown to bind histone H3 monomers or H3-H4 dimers (Maksimov et al., 2016), the nuclear NASP protein interacts both with the N-terminal tail as well as with the histone fold domain of CenH3 and reduced NASP expression negatively affects CenH3 levels, suggesting that NASP functions as a CenH3 escort protein (Le Goff et al., 2019).
Professor Lars Hennig passed away last year, leaving a gap in the fields of chromatin biology and plant development (Mozgová et al., 2018a). Session 4 of the meeting gave a homage remembering him, not only as a valuable colleague, friend, and mentor, but also by highlighting his scientific contributions and how his work will impact future research.
Among many other topics, one of Lars’ main interests were histone variants and chaperones. He contributed to the identification of MULTICOPY SUPRESSOR OF IRA 1 (MSI1) as one subunit of the CHROMATIN ASSEMBLY FACTOR 1 (CAF-1) chaperone complex (Hennig et al., 2003). Lars further showed that transgenerational aggravation of the CAF-1 mutant phenotype was related to a global change in DNA methylation (Mozgová et al., 2018b). Following this curiosity on DNA methylation levels during development, Minerva Trejo-Arellano (Uppsala, Sweden), a former PhD student of Lars, reported on changes of DNA methylation during dark-induced leaf senescence (Figure 2A). She showed that senescent leaves had expanded chromocenters, which is indicative of heterochromatin de-condensation. These chromatin changes were accompanied by a concerted downregulation of genes involved in epigenetically mediated silencing pathways and a deregulation of transposable elements. Surprisingly, no genome-wide changes in DNA methylation were detected, only localized differentially methylated regions (DMRs), especially in the CHH context (Trejo-Arellano et al., 2019). Among the epigenetic changes that occur during developmental transitions, Lars soon focused his attention on Polycomb activity. He contributed to the identification of MSI1 as part of the PRC2 (Köhler et al., 2003b) and explored its role in embryo-to-seedling transitions, a work developed by Iva Mozgová (České Budějovice, Czech Republic) during her postdoc in Lars’ group. She found that the characteristic embryonic phenotype of the double mutant of clf and swinger (clf swn) (Chanvivattana et al., 2004; Mozgová et al., 2017), which is affected in two of the three possible methyltransferases of PRC2, depends on the presence of sucrose. This finding fits with the idea that, during this developmental transition, plant nutrition shifts from heterotrophic to autotrophic growth. Following this research line, Iva presented a progressive degradation of chloroplasts and an increase in Reactive Oxygen Species (ROS) in clf swn and, accordingly, the mitigation of the phenotype under reduced light intensities. Therefore, these data suggest an unexplored role of PRC2 in mediating the establishment and/or maintenance of photoautotrophic growth in Arabidopsis.
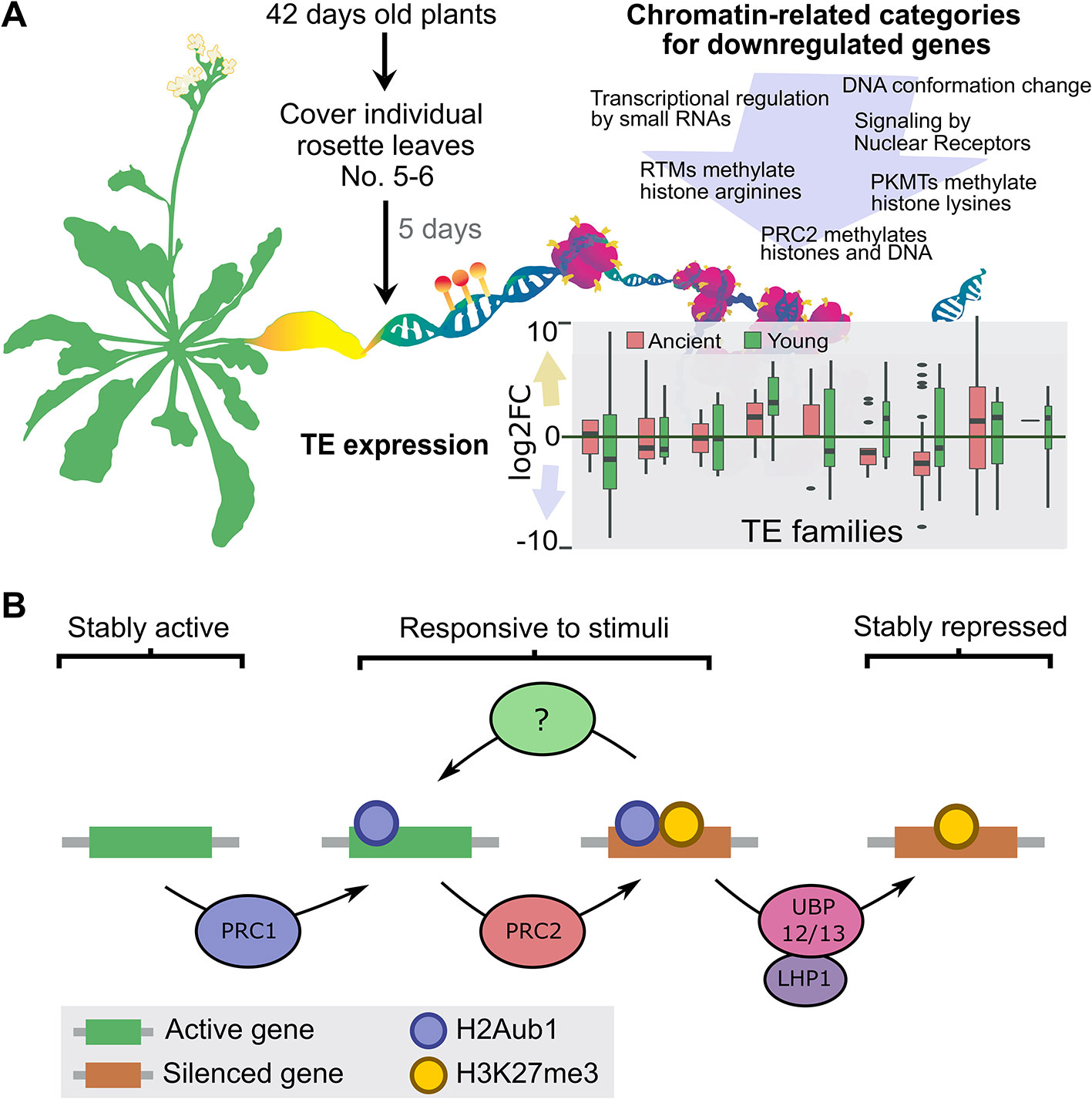
Figure 2 Overview of recent contributions from former Lars’ PhD students. (A) Dark-induced senescence causes localized changes in DNA methylation in Arabidopsis. Senescence was induced by covering individual Arabidopsis leaves. The yellowing of the covered-senescent leaves was accompanied by changes in the expression of transposable elements that depending on the TE family can be unaltered, up- or downregulated. Moreover, GO and pathway categories related with the maintenance of chromatin structure were enriched among the downregulated genes (for the complete analysis see Trejo-Arellano et al., 2019). Overall, the global DNA methylation landscape of the senescent leaves remained remarkably stable with only few localized DNA methylation changes detected, particularly in the CHH context (B) Working model for the UBP12/13-mediated gene repression. PRC2 causes silencing via deposition of H3K27me3, which in the majority of the cases is dependent on PRC1. However, by a mechanisms that remain to be resolved the product of PRC1 activity, H2Aub1, also creates an unstable state in which genes can be rapidly reactivated in response to a stimulus. Stable repression requires removal of H2Aub1 by LHP1-interacting UBP12/13. Figure courtesy of (A) M. Trejo, designed of Paulina Velasco, and (B) L. Kralemann.
To further understand the multiple functions of PcG proteins, Lars’ laboratory found a direct interaction between MSI1 and LIKE HETEROCHROMATIN PROTEIN1 (LHP1) (Derkacheva et al., 2013). Studying LHP1 protein interactors, UBIQUITIN SPECIFIC PROTEASES (UBP) 12 and 13 were found, and it was demonstrated that UBP12 mediates the deubiquitination of H2A (Derkacheva et al., 2016). However, it has been shown that H2A ubiquitination (H2Aub) by PRC1 is largely independent of PRC2 activity (Zhou et al., 2017). To further understand the link between H2Aub and H3K27me3, Lars’ former PhD student Lejon Kralemann (Uppsala, Sweden) presented a genome-wide analysis of these marks in mutants deficient for UBP12 and 13. The data suggest that H2Aub removal is required for preventing the loss of H3K27me3. In that model, LHP1 recruits UBP12/13 to deubiquitinate H2Aub and to stabilize H3K27me3-mediated repression (Figure 2B). Miyuki Nakamura (Uppsala, Sweden), a postdoc in Lars’ former group, reported another role of LHP1 through its interaction with DEK proteins (Derkacheva et al., 2013), which are linked to chromatin and associated with DNA topoisomerase 1α (TOP1α) (Waidmann et al., 2014). Miyuki presented that DEKs genetically interact with LHP1 by enhancing the early flowering of the lhp1 mutant, which is similar to what occurs in the top1α lhp1 mutant (Liu et al., 2014). She proposed that LHP1 interaction with DEKs and TOP1α is important for PcG target gene regulation. These works exemplify the direction where Lars’ research has lead the PcG field: finding new players of PcG activity and identifying mechanisms for target-specific PcG recruitment. In that direction, Justin Goodrich (Edinburgh, United Kingdom), through a second invited lecture, presented new data about ANTAGONIST OF LHP1 (ALP1), which was identified in a suppressor screening of the clf mutant (Liang et al., 2015). The interaction of ALP1 with PRC2 depends on ALP2, which interacts directly both with ALP1 and with MSI1, a core subunit of PRC2. To explain PcG antagonist function, Justin proposed that ALP1/ALP2 could compete for the core PRC2 complexes with other PcG “accessory proteins”. Interestingly, ALP proteins are likely inactive Harbinger-type transposases that are already demonstrated for ALP1 (Liang et al., 2015). As Harbinger transposases are encoded as part of the sequence of the ‘cut-and-paste’ Harbinger transposon superfamily (Kapitonov and Jurka, 2004), this is an example of how transposon domestication could provide novel genes for the hosts, in particular as components of PRC2.
Session 5 focused on different mechanisms that are involved in inducing a more relaxed and open chromatin structure, which usually correlates with an active transcription. For instance, Julia Engelhorn (Cologne, Germany) presented a very elegant approach in which Fluorescence Activated Cell Sorting (FACS) was combined with the Assay for Transposase-Accessible Chromatin with high-throughput sequencing (ATAC-seq), which allowed for the creation of maps with higher resolution than with DNase-seq from low cell numbers. Lines expressing the pDORNRÖSCHEN-LIKE::GFP in the apetala1 cauliflower mutant background were used (Wellmer et al., 2006), which allows for cell sorting of identical and highly synchronized Lateral Organ Founder Cells (LOFCs) (Frerichs et al., 2016). LOFCs-associated changes in chromatin accessibility were positively associated with transcriptional changes. In addition, highly accessible chromatin to the transposase corresponded well with previously described enhancer and conserved transcription factor (TF)-binding elements in promoters. These results also demonstrated that this approach can be further applied for genome-wide identification of novel transcriptional enhancers in plant specific cells (Frerichs et al., 2019).
Genome-wide approaches were also used to identify light-induced chromatin dynamics that occur at a very specific developmental switch, such as photomorphogenesis, which corresponds to the first perception of light after germination (Casal, 2013; Wu, 2014; Seluzicki et al., 2017). DE-ETIOLATED1 (DET1) is an atypical and conserved DAMAGED DNA BINDING PROTEIN 1 (DDB1)-CULLIN4 Associated Factor (DCAF) involved in the transcriptional reprogramming that occurs during photomorphogenesis (Chory et al., 1989; Pepper et al., 1994; Schroeder et al., 2002; Ma et al., 2003). Sandra Fonseca (Madrid, Spain) showed that DET1 and light control genome-wide levels and distribution of H2B ubiquitination (H2Bub) indirectly through degradation of a deubiquitination trimeric module (DUBm). One of the components of DUBm is UBP22, which acts as a major H2B deubiquitinase in the plant. Thus, DET1-mediated proteolytic degradation of DUBm is essential for chromatin reprogramming during photomorphogenesis (Nassrallah et al., 2018).
High Mobility Group A (HMGA) proteins have also been proposed to create a more permissive chromatin structure competing with linker histone H1 (Catez and Hock, 2010; Ozturk et al., 2014). Simon Amiard’s (Clermont Ferrand, France) presentation focused on GH1-HMGA1 and GH1-HMGA2 proteins, which comprise a conserved central globular domain (GH1) as well as AT-hook domains (Kotliński et al., 2017). Both GH1-HMGA1 and GH1-HMGA2-GFP fusion proteins are present in interphase and mitotic nuclei but are excluded from chromocenters and centromeres, and protein–protein interaction studies indicate possible GH1-HMGA1 homodimerization and heterodimerization with GH1-HMGA2. Mutants affected in the GH1-HMGA1 gene were impaired in development, with an overall size reduction due to smaller roots and leaves and a decrease in stem length, while gh1-hmga2 mutants were phenotypically normal. gh1-hmga1 mutants also showed shorter telomeres as a result of telomere instability (Charbonnel et al., 2018), and a transcriptome analysis of gh1-hmga1 mutants suggested a contribution of GH1-HMGA1 proteins to gene expression control.
Another epigenetic hallmark of active chromatin is H3K4me3. In yeast, SET DOMAIN GROUP 1 (SET1) adds this mark as part of COMPlex of proteins ASsociated with SET1 (COMPASS). Another subunit of this complex, Swd2, is needed for the recruitment of COMPASS to specific chromatin domains enriched in H2Bub (Sun and Allis, 2002; Kim et al., 2009). In Arabidopsis, a more complex scenario may exist since H3K4me3 can be placed by different histone methyltransferases (Baumbusch et al., 2001; Thorstensen et al., 2011; Zhang and Ma, 2012), and the function of At-COMPASS-like complexes have not been fully characterized yet (Fromm and Avramova, 2014; Xiao et al., 2016). Clara Bourbosse (Paris, France) reported recent results that showed that SET DOMAIN GROUP 2 (SDG2)/ARABIDOPSIS TRITHORAX 3 (ATX3), which has a main role in the deposition of H3K4me3 in Arabidopsis (Berr et al., 2010; Guo et al., 2010), binds to SWD2-like b (S2Lb), which interacts with core subunits of AtCOMPASS in a high-molecular weight complex. In addition, S2Lb, together with SDG2, is required for deposition of H3K4me3 and directly targets highly expressed genes. However, mutations in S2Lb affect the steady state levels of only a few of its target genes. Therefore, as part of AtCOMPASS, S2Lb may be required for appropriate transcriptional dynamics but is not essential for gene expression. Interestingly, S2Lb recruitment and H3K4me3 deposition at target genes are independent of H2Bub, indicating that AtCOMPASS-S2Lb activity does not require H2Bub in contrast to yeast. Whether there is a crosstalk between this histone mark and other methyltransferases is still an open question (Fiorucci et al., 2019).
Whether it is by changing large-scale chromatin conformation, nucleosome composition, and occupancy or histone post-translational modifications, chromatin regulation can impact plant development in a variety of ways, as highlighted in the previous sessions. This complexity becomes more evident when, despite being highly conserved among the plant species, the function of chromatin regulators, as well as their target genes, also depends on the context in which they act (Hennig et al., 2005; Merini et al., 2017). The last session of the meeting focused on the interplay between different chromatin regulators and accessory factors as well as their role in transcription regulation and impact on plant development.
Chromatin-based regulation allows us to quickly and reversibly switch genes on and off through the concerted action of antagonistic regulators. One example was presented by Cristel Carles (Grenoble, France) with her latest work on ULTRAPETALA1 (ULT1). It was known that ULT1 antagonizes the activity of PRC2 and regulates levels of H3K27me3 at genes involved in flowering and meristem determination (Carles and Fletcher, 2009). The new work showed that ULT1 genome-wide targets strongly overlap with those of the H3K27me3 methyltransferase CLF but not with the genes targeted by the demethylase RELATIVE OF EARLY FLOWERING 6 (REF6). ULT1 interacts with RNA Pol II (RNAPII) and several chromatin remodelers, suggesting that it might be involved in their recruitment, preventing binding of PcG proteins at specific loci.
TFs have also been shown to play a role in recruiting chromatin-associated regulators to control different aspects of plant development (Vachon et al., 2018). Pawel Mikulski (Norwich, United Kingdom) presented his work on VP1/ABI3-LIKE 1 (VAL1), a transcriptional repressor that promotes histone deacetylation at the FLC locus and is required for PRC2 nucleation in cold-induced vernalization (Questa et al., 2016). VAL1 was found to interact with subunits of the PRC1 (Yang et al., 2013; Questa et al., 2016), PRC2 (Chen et al., 2018), and LHP1 (Yuan et al., 2016), but no differences were observed in H2Aub in the mutants. Interestingly, the authors found that VAL1 influences nucleosome mobility around the region of the PRC2 nucleation site, suggesting it may act through the recruitment of chromatin remodelers. Other ways to achieve specificity include the formation of alternative chromatin-associated complexes or the interaction of core components with specific accessory proteins (Förderer et al., 2016). One example of the former was presented by Hernan Lopez-Marin (Cologne, Germany) with the identification of SUPER DETERMINANT 1 (SDE1), a new regulator of axillary meristem initiation in tomato. The sde1 mutation was mapped to a gene closely related to the PRC1 core components BMI1 and RING1 but which lacks the RING-finger domain required for depositing H2Aub (Buchwald et al., 2006). SDE1 interacts with LHP1, another component of the PRC1, suggesting it may be part of a new PcG complex involved in regulating axillary meristem initiation in tomato. Additionally, Sara Farrona (NUI, Galway) presented work from her laboratory on the identification of UBP5 as a new interactor of PWO1 and PRC2 subunits. As discussed in the first session of the meeting, PWO1 is itself an interactor of PRC2 methyltransferases and is involved in recruiting CLF to foci associated with the nuclear lamina (Hohenstatt et al., 2018; Mikulski et al., 2019). ubp5 mutants show a pleiotropic phenotype and de-repression of several meristem identity genes, known targets of PRC2, suggesting that UBP5 acts together with PcG proteins to regulate plant development.
Chromatin environments are also crucial for correct gene expression since they can modulate RNA polymerase II (RNAPII) and TF accessibility to target DNA. An interesting example was presented by Sebastian Marquardt (Copenhagen, Denmark), who showed that the histone chaperone complex FACT is required for the repression of cryptic intragenic Transcriptional Start Sites (TSSs) during RNAPII-mediated transcription. In their repressed state, these TSSs are enriched in H3K4me1, a hallmark for RNAPII elongation, while, in the fact mutants, they show increased levels of H3K4me3, similar to promoter TSSs, indicating a role for FACT in the regulation of transcript isoform diversity (Nielsen et al., 2019). Moreover, a computational approach presented by Dmitry Lapin (Cologne, Germany) helped to define chromatin features predicting dependency of gene expression on the immunity regulator Enhanced Disease Susceptibility 1 (EDS1) in Arabidopsis. Machine-learning methods were used to test whether this dependency can be inferred from binding of TFs and occupancy of histone modifications from public ChIP-seq data. A neural network model provided the highest accuracy (up to 85%). Under non-stress conditions, EDS1-dependent loci have low H3K36me3 and RNAPII levels. Authors proposed that initial chromatin status contributes to the specificity of gene expression regulation in immunity. On the other hand, taking advantage of epigenetic hybrids (epiHybrids) from crosses with decrease in dna methylation1 (ddm1)-derived epigenetic recombinant inbred lines (epiRILs), Ioanna Kakoulidou (Munich, Germany) showed that chromatin states can also impact subsequent generations. Previous work from the laboratory had shown that epiHybrids exhibit strong heterosis in several developmental traits, which correlates to DMRs in the parental lines (Lauss et al., 2018). Recently, the authors have used a high throughput phenotyping system to analyze 382 epiHybrids, and they were able to confirm that epigenetic divergence in the parents is sufficient to cause heterosis in the progeny. Future methylome, transcriptome, and small RNA-seq analyses of these epiHybrids are expected to contribute to a better understanding of how the parental epigenetic states affect the progeny.
In summary, the EWPC2019 encouraged discussion about the most recent advances in epigenetics, chromatin-related mechanisms, and nuclear architecture in relation to the regulation of transcription and its impact on plants traits. Particularly, how the nuclear space is organized and how specific histones and structures within the nucleus, such as the nucleolus and the nuclear periphery, relate to specific chromatin domains was thoroughly discussed in various talks. However, we are still far from understanding the complexity that is shrouded by the nuclear envelope. The extent of interplay between DNA methylation, various histone modifications, histone variants, and regulatory RNAs taking place during epigenetic inheritance processes remains to be elucidated. Likewise, much remains to be understood on the importance of core histone variants and their chaperones in chromatin structure through the control of nucleosome assembly and occupancy or the role of linker histones and other dynamic DNA-binding proteins. While histone modifications have so far rarely been considered in a variant-specific manner, combinations of histone variants with their particular marks constitute an additional layer of complexity to fine-tune chromatin regulation that is now just emerging and that will most certainly require further studies. While different presentations exposed the complexity of chromatin-based regulatory mechanisms in plants, it became clear that we need to investigate how chromatin-associated proteins are regulated in different tissues, developmental stages, and under specific environmental conditions, in order to fully understand their role in transcriptional regulation. Novel technical advances making use of CRISPR/Cas9 based strategies or new developments in 3C techniques together with a deeper characterization of multi-subunit complexes and their functions will help to better our understanding of the organization of plant genomes and nuclear protein networks in the near future. Simultaneously, studying the interplay between different regulators with the help of emerging technologies, such as the development of imaging and image-processing solutions that take into account the challenges of plant systems (Dumur et al., 2019) and other cell-specific techniques, will certainly yield important new findings. Finally, while most work presented at the meeting used Arabidopsis as a model system, the fundamental mechanisms identified might in the future be applied to crop species by, for example, exploiting natural epigenetic diversity in plant breeding or induced epigenetic variation involved in stress priming and memory (Springer and Schmitz, 2017; Mozgová et al, 2019; Forestan et al., 2019). We expect to see some of these questions addressed in the future and exciting new data on chromatin regulation in model and crop plants to be presented in forthcoming EWPCs.
The memory of Lars Hennig imbued specifically one of the meeting sessions but was also present in many other talks, demonstrating that the contributions of this excellent scientist and mentor will last over time. His work had a tremendous impact on the understanding of chromatin regulation and plant development, particularly concerning our knowledge of the PcG pathway, and will perpetuate through the ongoing contributions of many of his alumni who are still actively investigating these questions.
SF and AP created Figure 1. JM-R edited Figure 2. Each author (JM-R, K, JE, IT, AP, and SF) wrote one session. JM-R, AP, and SF wrote introduction and conclusions. All authors contributed to edit text and figures. IT edited references. SF coordinated the manuscript.
JM-R is supported by funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 797473, financial support from the Spanish Ministry of Economy and Competitiveness through the “Severo Ochoa Program for Centres of Excellence in R&D” 2016-2019 (SEV‐2015‐0533), and by the CERCA Programme/Generalitat de Catalunya. AP, K, and SF acknowledge networking support from the COST Action CA16212 “Impact of nuclear domains on gene expression and plant traits (INDEPTH).” The EWPC was supported by grants from The German Research Foundation (TU126/14-1) and The Company of Biologists (EA1772). The organizers also acknowledge the generous contributions from industrial sponsors Zeiss Microscopy GmbH (Germany), Diagenode SA (Belgium), and Dispendix GmbH (Germany).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank all the authors that share with us their contribution. Finally, special thanks to Franziska Turck who organized the EWPC2019 and also contributed to this report.
Adamusová, K., Khosravi, S., Fujimoto, S., Houben, A., Matsunaga, S., Fajkus, J., et al. (2019). Two combinatorial patterns of telomere histone marks in plants with canonical and non-canonical telomere repeats. Plant J. doi: 10.1111/tpj.14653
Baev, V., Milev, I., Naydenov, M., Vachev, T., Apostolova, E., Mehterov, N., et al. (2014). Insight into small RNA abundance and expression in high- and low-temperature stress response using deep sequencing in Arabidopsis. Plant Physiol. Bioch. 84, 105–114. doi: 10.1016/j.plaphy.2014.09.007
Baumbusch, L. O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., et al. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29 (21), 4319–4333. doi: 10.1093/nar/29.21.4319
Berr, A., McCallum, E. J., Ménard, R., Meyer, D., Fuchs, J., Dong, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22 (10), 3232–3248. doi: 10.1105/tpc.110.079962
Boisvert, F.-M., van Koningsbruggen, S., Navascués, J., Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Bio. 8 (7), 574–585. doi: 10.1038/nrm2184
Buchwald, G., van der Stoop, P., Weichenrieder, O., Perrakis, A., van Lohuizen, M., Sixma, T. K. (2006). Structure and E3-ligase activity of the Ring-Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J. 25 (11), 2465–2474. doi: 10.1038/sj.emboj.7601144
Buschbeck, M., Hake, S. B. (2017). Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18 (5), 299–314. doi: 10.1038/nrm.2016.166
Carles, C. C., Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Gene Dev. 23, 2723–2728. doi: 10.1101/gad.1812609
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64 (1), 403–427. doi: 10.1146/annurev-arplant-050312-120221
Catez, F., Hock, R. (2010). Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta 1799 (1–2), 15–27. doi: 10.1016/j.bbagrm.2009.11.001
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y. H., Sung, Z. R., et al. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131 (21), 5263–5276. doi: 10.1242/dev.01400
Charbonnel, C., Rymarenko, O., Da Ines, O., Benyahya, F., White, C. I., Butter, F., et al. (2018). The linker histone GH1-HMGA1 is involved in telomere stability and DNA damage repair. Plant Physiol. 177 (1), 311–327. doi: 10.1104/pp.17.01789
Chaudhury, A. M., Ming, L., Miller, C., Craig, S., Dennis, E. S., Peacock, W. J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 94 (8), 4223–4228. doi: 10.1073/pnas.94.8.4223
Chen, N., Veerappan, V., Abdelmageed, H., Kang, M., Allen, R. D. (2018). HSI2/VAL1 Silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in arabidopsis. Plant Cell 30, 600–619. doi: 10.1105/tpc.17.00655
Chory, J., Peto, C., Feinbaum, R., Pratt, L., Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58 (5), 991–999. doi: 10.1016/0092-8674(89)90950-1000
Coleman-Derr, D., Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PloS Genet. 8 (10), e1002988. doi: 10.1371/journal.pgen.1002988
Crevillén, P., Gómez-Zambrano, A., López, J. A., Vázquez, J., Piñeiro, M., Jarillo, J. A. (2019). Arabidopsis YAF9 histone readers modulate flowering time through NuA4-complex-dependent H4 and H2A.Z histone acetylation at FLC chromatin. New Phytol. 222 (4), 1893–1908. doi: 10.1111/nph.15737
Dabin, J., Polo, S. E. (2017). Choreography of parental histones in damaged chromatin. Nucleus 8 (3), 255–260. doi: 10.1080/19491034.2017.1292192
Derkacheva, M., Steinbach, Y., Wildhaber, T., Mozgová, I., Mahrez, W., Nanni, P., et al. (2013). Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 32 (14), 2073–2085. doi: 10.1038/emboj.2013.145
Derkacheva, M., Liu, S., Figueiredo, D. D., Gentry, M., Mozgova, I., Nanni, P., et al. (2016). H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat. Plants 2, 16126. doi: 10.1038/nplants.2016.126
Duc, C., Benoit, M., Le Goff, S., Simon, L., Poulet, A., Cotterell, S., et al. (2015). The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 81 (5), 707–722. doi: 10.1111/tpj.12758
Duc, C., Benoit, M., Détourné, G., Simon, L., Poulet, A., Jung, M., et al. (2017). Arabidopsis ATRX modulates H3.3 occupancy and fine-tunes gene expression. Plant Cell 29, 1773–1793. doi: 10.1105/tpc.16.00877
Dumur, T., Duncan, S., Graumann, K., Desset, S., Randall, R. S., Scheid, O. M., et al. (2019). Probing the 3D architecture of the plant nucleus with microscopy approaches: challenges and solutions. Nucleus 10 (1), 181–212. doi: 10.1080/19491034.2019.1644592
Förderer, A., Zhou, Y., Turck, F. (2016). The age of multiplexity: recruitment and interactions of Polycomb complexes in plants. Curr. Opin. Plant Biol. 29, 169–178. doi: 10.1016/j.pbi.2015.11.010
Fiorucci, A.-S., Bourbousse, C., Concia, L., Rougée, M., Deton-Cabanillas, A.-F., Zabulon, G., et al. (2019). Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination. Genome Biol. 20 (1), 100. doi: 10.1186/s13059-019-1705-4
Forestan, C., Farinati, S., Zambelli, F., Pavesi, G., Rossi, V., Varotto, S. (2019). Epigenetic signatures of stress adaptation and flowering regulation in response to extended drought and recovery in Zea mays. Plant Cell Environ. 43 (1), 55–75. doi: 10.1111/pce.13660
Frerichs, A., Thoma, R., Abdallah, A. T., Frommolt, P., Werr, W., Chandler, J. W. (2016). The founder-cell transcriptome in the Arabidopsis apetala1 cauliflower inflorescence meristem. BMC Genomics 17 (1), 855. doi: 10.1186/s12864-016-3189-x
Frerichs, A., Engelhorn, J., Altmüller, J., Gutierrez-Marcos, J., Werr, W. (2019). Specific chromatin changes mark lateral organ founder cells in the Arabidopsis inflorescence meristem. J. Exp. Bot. 70 (15), 3867–3879. doi: 10.1093/jxb/erz181
Fromm, M., Avramova, Z. (2014). ATX1/AtCOMPASS and the H3K4me3 marks: how do they activate Arabidopsis genes? Curr. Opin. Plant Biol. 21, 75–82. doi: 10.1016/j.pbi.2014.07.004
Gómez-Zambrano, Á., Merini, W., Calonje, M. (2019). The repressive role of Arabidopsis H2A.Z in transcriptional regulation depends on AtBMI1 activity. Nat. Commun. 10 (1), 2828. doi: 10.1038/s41467-019-10773-1
Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M. A., Gagliano, W. B. (1998). Maternal Control of Embryogenesis by MEDEA, a Polycomb Group Gene in Arabidopsis. Science 280 (5362), 446–450. doi: 10.1126/science.280.5362.446
Guo, L., Yu, Y., Law, J. A., Zhang, X. (2010). SET DOMAIN GROUP2 is the major histone H3 lysine 4 trimethyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 107 (43), 18557–18562. doi: 10.1073/pnas.1010478107
Hennig, L., Taranto, P., Walser, M., Schönrock, N., Gruissem, W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130 (12), 2555–2565. doi: 10.1242/dev.00470
Hennig, L., Bouveret, R., Gruissem, W. (2005). MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 15 (6), 295–302. doi: 10.1016/j.tcb.2005.04.004
Hohenstatt, M. L., Mikulski, P., Komarynets, O., Klose, C., Kycia, I., Jeltsch, A., et al. (2018). PWWP-DOMAIN INTERACTOR OF POLYCOMBS1 interacts with polycomb-group proteins and histones and regulates arabidopsis flowering and development. Plant Cell 30 (1), 117–133. doi: 10.1105/tpc.17.00117
Jiang, D., Berger, F. (2016). Histone variants in plant transcriptional regulation. BBA-Gene. Regul. Mech. 1860 (1), 123–130. doi: 10.1016/j.bbagrm.2016.07.002
Jiang, H., Moreno-Romero, J., Santos-González, J., De Jaeger, G., Gevaert., K., Van De Slijke, E., et al. (2017). Ectopic application of the repressive histone modification H3K9me2 establishes post-zygotic reproductive isolation in Arabidopsis thaliana. Gene Dev. 31 (12), 1272–1287. doi: 10.1101/gad.299347.117
Köhler, C., Springer, N. (2017). Plant epigenomics-deciphering the mechanisms of epigenetic inheritance and plasticity in plants. Genome Biol. 18, 132. doi: 10.1186/s13059-017-1260-9
Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., Grossniklaus, U. (2003a). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Gene Dev. 17, 1540–1553. doi: 10.1101/gad.257403
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., Gruissem, W. (2003b). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22 (18), 4804–4814. doi: 10.1093/emboj/cdg444
Kapitonov, V. V., Jurka, J. (2004). Harbinger transposons and ancient HARBI1 Gene Derived from a Transposase. DNA Cell Biol. 23 (5), 311–324. doi: 10.1089/104454904323090949
Kim, J., Guermah, M., McGinty, R. K., Lee, J. S., Tang, Z., Milne, T. A., et al. (2009). RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137 (3), 459–471. doi: 10.1016/j.cell.2009.02.027
Kotliński, M., Knizewski, L., Muszewska, A., Rutowicz, K., Lirski, M., Schmidt, A., et al. (2017). Phylogeny-based systematization of arabidopsis proteins with histone H1 globular domain. Plant Physiol. 174 (1), 27–34. doi: 10.1104/pp.16.00214
Lauss, K., Wardenaar, R., Oka, R., van Hulten, M. H. A., Guryev, V., Keurentjes, K. J. B., et al. (2018). Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 176, 1627–1645. doi: 10.1104/pp.17.01054
Law, J. A., Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Law, J. A., Vashisht, A. A., Wohlschlegel, J. A., Jacobsen, S. E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PloS Genet. 7 (7), e1002195. doi: 10.1371/journal.pgen.1002195
Le Goff, S., Keçeli, B. N., Jeřábková, H., Heckmann, S., Rutten, T., Cotterell, S., et al. (2019). The H3 histone chaperone NASPSIM3 escorts CenH3 in Arabidopsis. Plant J. tpj, 14518. doi: 10.1111/tpj.14518
Liang, S. C., Hartwig, B., Perera, P., Mora-García, S., de Leau, E., Thornton, H., et al. (2015). Kicking against the PRCs – a domesticated transposase antagonises silencing mediated by polycomb group proteins and is an accessory component of polycomb repressive complex 2. PloS Genet- 11 (12), 1–26. doi: 10.1371/journal.pgen.1005660
Liu, X., Gao, L., Dinh, T. T., Shi, T., Li, D., Wang, R., et al. (2014). DNA topoisomerase I affects polycomb group protein-mediated epigenetic regulation and plant development by altering nucleosome distribution in Arabidopsis. Plant Cell 26 (7), 2803–2817. doi: 10.1105/tpc.114.124941
Lorković, Z. J., Park, C., Goiser, M., Jiang, D., Kurzbauer, M. T., Schlögelhofer, P., et al. (2017). Compartmentalization of DNA damage response between heterochromatin and euchromatin is mediated by distinct H2A histone variants. Curr. Biol. 27 (8), 1192–1199. doi: 10.1016/j.cub.2017.03.002
Lu, P. Y. T., Lévesque, N., Kobor, M. S. (2009). NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem. Cell Biol. 87 (5), 799–815. doi: 10.1139/O09-062
Müller, S., Almouzni, G. (2014). A network of players in H3 histone variant deposition and maintenance at centromeres. BBA - Gene Regul. Mech. 1839 (3), 241–250. doi: 10.1016/j.bbagrm.2013.11.008
Ma, L., Zhao, H., Deng, X. W. (2003). Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130 (5), 969–981. doi: 10.1242/dev.00281
Maksimov, V., Nakamura, M., Wildhaber, T., Nanni, P., Ramström, M., Bergquist, J., et al. (2016). The H3 chaperone function of NASP is conserved in Arabidopsis. Plant J. 88 (3), 425–436. doi: 10.1111/tpj.13263
Merini, W., Romero-Campero, F. J., Gomez-Zambrano, A., Zhou, Y., Turck, F., Calonje, M. (2017). The arabidopsis polycomb repressive complex 1 (PRC1) components AtBMI1A, B, and C impact gene networks throughout all stages of plant development. Plant Physiol. 173 (1), 627–641. doi: 10.1104/pp.16.01259
Mikulski, P., Hohenstatt, M. L., Farrona, S., Smaczniak, C., Stahl, Y., Kalyanikrishna, et al. (2019). The chromatin-associated protein PWO1 interacts with plant nuclear lamin-like components to regulate nuclear size. Plant Cell 31 (5), 1141–1154. doi: 10.1105/tpc.18.00663
Miryeganeh, M., Saze, H. (2019). Epigenetic inheritance and plant evolution. Popul. Ecol. 62 (1), 17–27. doi: 10.1002/1438-390X.12018
Moreno-Romero, J., Santos-González, J., Hennig, L., Köhler, C. (2017). Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat. Protoc. 12 (2), 238–254. doi: 10.1038/nprot.2016.167
Mozgová, I., Muñoz-Viana, R., Hennig, L. (2017). PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PloS Genet. 13 (1), e1006562. doi: 10.1371/journal.pgen.1006562
Mozgová, I., Alexandre, C., Steinbach, Y., Derkacheva, M., Schäfer, E., Gruissem, W. (2018a). A tribute to lars hennig (1970–2018). J. Exp. Bot. 69 (21), 4989–4990. doi: 10.1093/jxb/ery337
Mozgová, I., Wildhaber, T., Trejo-Arellano, M. S., Fajkus, J., Roszak, P., Köhler, C., et al. (2018b). Transgenerational phenotype aggravation in CAF-1 mutants reveals parent-of-origin specific epigenetic inheritance. New Phytol. 220 (3), 908–921. doi: 10.1111/nph.15082
Mozgová, I., Mikulski, P., Pecinka, A., Farrona, S. (2019). “Epigenetic mechanisms of abiotic stress response and memory in plants,” in Epigenetics in plants of agronomic importance: fundamentals and applications. Eds. Alvarez-Venegas, De-la-Peña, Casas-Mollano (Cham: Springer). doi: 10.1007/978-3-030-14760-0_1
Nassrallah, A., Rougée, M., Bourbousse, C., Drevensek, S., Fonseca, S., Iniesto, E., et al. (2018). DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. eLife 7, e37892. doi: 10.7554/eLife.37892
Nie, X., Wang, H., Li, J., Holec, S., Berger, F. (2014). The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol. Open 3 (9), 794–802. doi: 10.1242/bio.20148680
Nielsen, M., Ard, R., Leng, X., Ivanov, M., Kindgren, P., Pelechano, V., et al. (2019). Transcription-driven chromatin repression of Intragenic transcription start sites. PloS Genet. 15 (2), 1–33. doi: 10.1371/journal.pgen.1007969
Ozturk, N., Singh, I., Mehta, A., Braun, T., Barreto, G. (2014). HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev.l Biol. 2, 5. doi: 10.3389/fcell.2014.00005
Pepper, A., Delaney, T., Washburn, T., Poole, D., Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78 (1), 109–116. doi: 10.1016/0092-8674(94)90577-0
Picart-Picolo, A., Picault, N., Pontvianne, F. (2019). Ribosomal RNA genes shape chromatin domains associating with the nucleolus. Nucleus 10 (1), 67–72. doi: 10.1080/19491034.2019.1591106
Pontvianne, F., Carpentier, M. C., Durut, N., Pavlištová, V., Jaške, K., Schořová, Š., et al. (2016). Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana Genome. Cell Rep. 16 (6), 1574–1587. doi: 10.1016/J.CELREP.2016.07.016
Questa, J. I., Song, J., Geraldo, N., An, H., Dean, C. (2016). Transcriptional repressor VAL1 triggers silencing of FLC during vernalization. Science 1 (6298), 1–5. doi: 10.1126/science.aaf7354
Ream, T. S., Haag, J. R., Wierzbicki, A. T., Nicora, C. D., Norbeck, A. D., Zhu, J. K., et al. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA Polymerase II. Mol. Cell 33 (2), 192–203. doi: 10.1016/j.molcel.2008.12.015
Rosa, S., Shaw, P. (2013). Insights into chromatin structure and dynamics in plants. Biology 2 (4), 1378–1410. doi: 10.3390/biology2041378
Rutowicz, K., Puzio, M., Halibart-Puzio, J., Lirski, M., Kotliński, M., Kroteń, M. A., et al. (2015). A specialized histone H1 variant is required for adaptive responses to complex abiotic stress and related DNA methylation in Arabidopsis. Plant Physiol. 169, 2080–2101. doi: 10.1104/pp.15.00493
Rutowicz, K., Lirski, M., Mermaz, B., Teano, G., Schubert, J., Mestiri, I., et al. (2019). Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biol. 20 (1), 157. doi: 10.1186/s13059-019-1767-3
Schroeder, D. F., Gahrtz, M., Maxwell, B. B., Cook, R. K., Kan, J. M., Alonso, J. M., et al. (2002). De-Etiolated 1 and damaged DNA binding protein 1 interact to regulate arabidopsis photomorphogenesis. Curr. Biol. 12 (17), 1462–1472. doi: 10.1016/S0960-9822(02)01106-5
Seluzicki, A., Burko, Y., Chory, J. (2017). Dancing in the dark: darkness as a signal in plants. Plant Cell Environ. 40 (11), 2487–2501. doi: 10.1111/pce.12900
Sexton, T., Cavalli, G. (2015). The Role of Chromosome Domains in Shaping the Functional Genome. Cell 160 (6), 1049–1059. doi: 10.1016/j.cell.2015.02.040
Springer, N. M., Schmitz, R. J. (2017). Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 18, 563–575. doi: 10.1038/nrg.2017.45
Sun, Z.-W., Allis, C. D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 (6893), 104–108. doi: 10.1038/nature00883
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Pawełoszek, L., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29 (4), 791–807. doi: 10.1105/tpc.16.00573
Thorstensen, T., Grini, P. E., Aalen, R. B. (2011). SET domain proteins in plant development. BBA- Gene Regu. Mech. Elsevier 1809 (8), 407–420. doi: 10.1016/J.BBAGRM.2011.05.008
Trejo-Arellano, M. S., Mehdi, S., de Jonge, J., Tomastíková, E. D., Köhler, C., Hennig, L. (2019). Dark-induced senescence causes localized changes in DNA methylation. Plant Physiol. doi: 10.1104/pp.19.01154
Vachon, G., Engelhorn, J., Carles, C. C. (2018). Interactions between transcription factors and chromatin regulators in the control of flower development. J. Exp. Bot. 69 (10), 2461–2471. doi: 10.1093/jxb/ery079
Waidmann, S., Kusenda, B., Mayerhofer, J., Mechtler, K., Jonak, C. (2014). A DEK domain-containing protein modulates chromatin structure and function in Arabidopsis. Plant Cell 26 (11), 4328–4344. doi: 10.1105/tpc.114.129254
Wang, H., Dittmer, T. A., Richards, E. J. (2013). Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13, 200. doi: 10.1186/1471-2229-13-200
Wellmer, F., Alves-Ferreira, M., Dubois, A., Riechmann, J. L., Meyerowitz, E. M. (2006). Genome-wide analysis of gene expression during early arabidopsis flower development. PloS Genet. 2 (7), e117. doi: 10.1371/journal.pgen.0020117
Wu, S.-H. (2014). Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu. Rev. Plant Biol. 65 (1), 311–333. doi: 10.1146/annurev-arplant-050213-040337
Xiao, J., Lee, U.-S., Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curre. Opin. Plant Biol. 34, 41–53. doi: 10.1016/J.PBI.2016.08.002
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., Calonje, M. (2013). VAL- and AtBMI1-Mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23 (24), 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yelagandula, R., Stroud, H., Holec, S., Zhou, K., Feng, S., Zhong, X., et al. (2014). The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158 (1), 98–109. doi: 10.1016/j.cell.2014.06.006
Yuan, W., Luo, X., Li, Z., Yang, W., Wang, Y., Liu, R., et al. (2016). A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nat. Genet. 48, 1527–1534. doi: 10.1038/ng.3712
Zhang, L., Ma, H. (2012). Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 195 (1), 248–263. doi: 10.1111/j.1469-8137.2012.04143.x
Zhou, Y., Romero-Campero, F. J., Gómez-Zambrano, A., Turck, F., Calonje, M. (2017). H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol. 18 (1), 69. doi: 10.1186/s13059-017-1197-z
Keywords: EWPC2019, chromatin, epigenetics, transcription, nucleus
Citation: Moreno-Romero J, Probst AV, Trindade I, Kalyanikrishna, Engelhorn J and Farrona S (2020) Looking At the Past and Heading to the Future: Meeting Summary of the 6th European Workshop on Plant Chromatin 2019 in Cologne, Germany. Front. Plant Sci. 10:1795. doi: 10.3389/fpls.2019.01795
Received: 24 October 2019; Accepted: 23 December 2019;
Published: 07 February 2020.
Edited by:
Bo Liu, University of California, Davis, United StatesReviewed by:
Li Pu, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2020 Moreno-Romero, Probst, Trindade, Kalyanikrishna, Engelhorn and Farrona. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara Farrona, c2FyYS5mYXJyb25hQG51aWdhbHdheS5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.