
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 11 February 2020
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01783
This article is part of the Research Topic Linking Stomatal Development and Physiology: From Stomatal Models to Non-Model Species and Crops View all 15 articles
Rice (Oryza sativa L.) contributes to the diets of around 3.5 billion people every day and is consumed more than any other plant. Alarmingly, climate predictions suggest that the frequency of severe drought and high-temperature events will increase, and this is set to threaten the global rice supply. In this review, we consider whether water or heat stresses in crops — especially rice — could be mitigated through alterations to stomata; minute pores on the plant epidermis that permit carbon acquisition and regulate water loss. In the short-term, water loss is controlled via alterations to the degree of stomatal “openness”, or, in the longer-term, by altering the number (or density) of stomata that form. A range of molecular components contribute to the regulation of stomatal density, including transcription factors, plasma membrane-associated proteins and intercellular and extracellular signaling molecules. Much of our existing knowledge relating to stomatal development comes from research conducted on the model plant, Arabidopsis thaliana. However, due to the importance of cereal crops to global food supply, studies on grass stomata have expanded in recent years, with molecular-level discoveries underscoring several divergent developmental and morphological features. Cultivation of rice is particularly water-intensive, and there is interest in developing varieties that require less water yet still maintain grain yields. This could be achieved by manipulating stomatal development; a crop with fewer stomata might be more conservative in its water use and therefore more capable of surviving periods of water stress. However, decreasing stomatal density might restrict the rate of CO2 uptake and reduce the extent of evaporative cooling, potentially leading to detrimental effects on yields. Thus, the extent to which crop yields in the future climate will be affected by increasing or decreasing stomatal density should be determined. Here, our current understanding of the regulation of stomatal development is summarised, focusing particularly on the genetic mechanisms that have recently been described for rice and other grasses. Application of this knowledge toward the creation of “climate-ready” rice is discussed, with attention drawn to the lesser-studied molecular elements whose contributions to the complexity of grass stomatal development must be understood to advance efforts.
Feeding a global population expected to reach 9.8 billion by 2050 is one of the greatest challenges of our time (McGuire, 2015; United Nations, 2017). Together with rising temperatures and dwindling reserves of natural resources such as freshwater, the problem is exasperated by growing climate instabilities (Stott, 2016). The frequency and intensity of extreme weather events are projected to increase under future climate scenarios, which will likely lead to substantial losses of crop yields (Seneviratne et al., 2012; Challinor et al., 2014). Thus, to alleviate the risk of crop failures, the development of “climate-ready” crops that can withstand future climatic stresses should be prioritized.
Rice (Oryza sativa L.) is a major source of food and income for billions worldwide. In Asia, where over 90% of rice is grown and consumed (Brar and Khush, 2013; Elert, 2014), the primary climatic factor for the majority of the growing region is the monsoon, which contributes >80% of annual rainfall within a few months (Haefele et al., 2016). Rainfed cultivation accounts for ~20% of global rice supply yet, as a consequence of this irregular rainfall, productivity in rainfed regions is already constrained by drought (Wassmann et al., 2009; Maclean et al., 2013; Swain et al., 2017). Rice is also susceptible to heat stress, particularly during the reproductive and grain filling stages (Matsui et al., 2000; Lin et al., 2010), and there are concerns that both high-temperature events and droughts are expected to become more prevalent across the world’s rice cultivation regions (Krishnan et al., 2011). Furthermore, there is an increasing requirement to limit water use for crop irrigation, which already accounts for around 70% of global freshwater usage (Foley et al., 2011). It is therefore important that water use efficiency (WUE; defined here as the amount of carbon assimilated per unit of water lost) and tolerances to heat-stress and drought are improved to ready rice for future climates.
Plants use microscopic epidermal openings called stomata to regulate the uptake of CO2 and the release of water between internal tissue surfaces and the environment (Buckley, 2005). Typically consisting of a pair of specialised turgor-driven guard cells (GCs) surrounding a central pore, stomata also function to regulate plant temperature and to move solutes and water internally via the transpiration stream. Under water-limiting conditions stomata close, thereby restricting water loss via reduced stomatal conductance (gs), with CO2 uptake for photosynthesis (A), evaporative cooling, and nutrient transfer often diminished as a result (Arve et al., 2011; Urban et al., 2017). Because less cooling occurs, stomatal closure exacerbates the risk of plant organs reaching lethally high temperatures which, particularly for rice in lower latitudes, may lead to substantial crop losses (Long and Ort, 2010; Sánchez et al., 2014).
Adjustments to the size of the stomatal pore permit a near-immediate plant response to a change in the surrounding environment (Brownlee, 2001). However, when sustained environmental stimuli arise over longer durations, alterations to stomatal density (SD; the number of stomata that form per unit area) and size may also occur (Casson and Gray, 2008; Qi and Torii, 2018). By increasing SD, gs might be increased, which in turn may lead to greater evaporative cooling and increases in A (Tanaka et al., 2013). Conversely, by reducing SD, gs can be lowered, which may curb water loss and result in improved WUE and drought tolerance (Doheny-Adams et al., 2012; Caine et al., 2019). If reductions in SD do not have a significant detrimental impact on A or evaporative cooling, the overall benefits of increased water conservation could represent a viable strategy to protect certain crops, including rice, against future extreme weather events (Hepworth et al., 2018).
A deep understanding of the stomatal development program in rice is required for genetic manipulation of SD. The majority of our existing knowledge stems from the model plant Arabidopsis (Arabidopsis thaliana), and this is beginning to be translated into grass species (to name a few: Raissig et al., 2016; Raissig et al., 2017; Yin et al., 2017; Abrash et al., 2018; Lu et al., 2019; Mohammed et al., 2019; Wu et al., 2019). In this review, our current understanding of the molecular-level control of stomatal development in rice will be summarized, drawing comparisons and distinctions between Arabidopsis, the model grass Brachypodium (Brachypodium distachyon), and other grass species of agricultural importance such as maize (Zea mays) and barley (Hordeum vulgare). Then, translating this knowledge into action, recent advances towards “climate-ready” rice with altered SD will be discussed.
Rice and other cereal crops such as maize, barley, and wheat (Triticum aestivum) are monocot grasses and, relative to eudicot plants such as Arabidopsis, are defined by a number of distinctive features relating to where and how stomata develop (Raissig et al., 2016; Conklin et al., 2018; Hepworth et al., 2018). In grass leaves, stomata are organized into evenly distributed parallel files which are interspersed either side of leaf veins, with the origin of stomatal development primarily occurring at the leaf base (Conklin et al., 2018). Whereas stomatal development is primarily initiated at the leaf base in Arabidopsis, the location of development is not as constrained as in grasses and the spatial distribution of stomata appears more random as a result (Conklin et al., 2018). The culmination of stomatal development for grasses is a cellular complex composed of a pair of dumbbell-shaped GCs adjoined on either side by subsidiary cells (SCs) (Figure 1). These complexes are typically separated by at least one epidermal cell and thus development follows the one-cell spacing rule (Sachs, 1991; Chen et al., 2017; Hepworth et al., 2018). In Arabidopsis, stomatal GCs are kidney-shaped (Figure 1) and are without flanking SCs but, like grasses, the one-cell spacing rule is maintained (Hara et al., 2007).
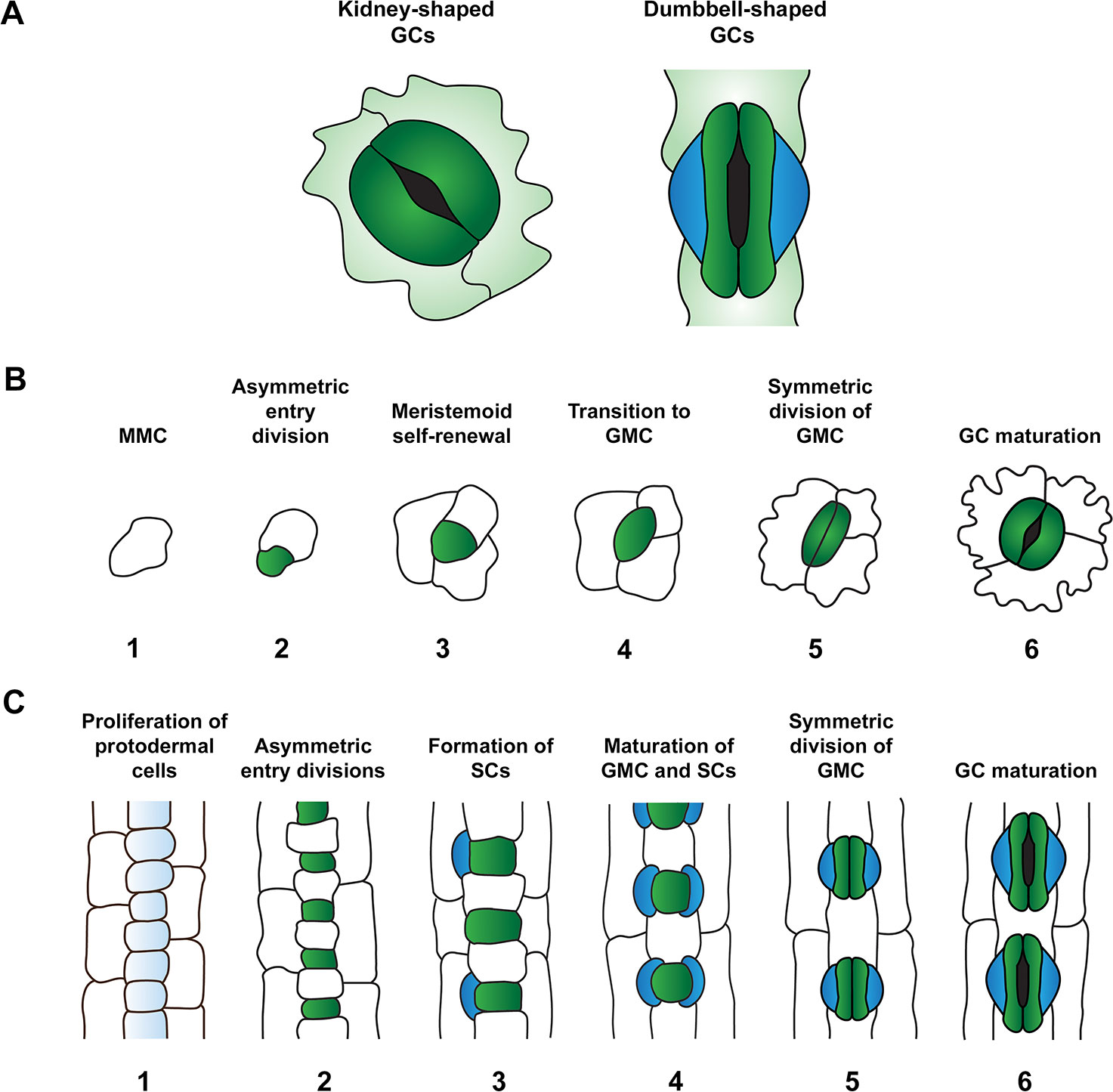
Figure 1 Comparison of stomatal morphology and development in the leaves of the eudicot Arabidopsis thaliana and monocot grasses. (A) Comparison of guard cell (GC) morphology: kidney-shaped GCs of A. thaliana (left) and dumbbell-shaped GCs typical of monocot grasses (right). GCs and subsidiary cells (SCs) are depicted in dark green and blue respectively. (B) Stomatal development in A. thaliana. Meristemoid mother cells (MMCs) (1) divide asymmetrically to form meristemoids (green) (2). This process of asymmetric division is repeated in the self-renewing meristemoid phase (3). Note, asymmetric spacing divisions of MMCs occur around this developmental stage, with newly formed satellite meristemoids forming at least one-cell away from pre-existing meristemoids and stomata. Asymmetric divisions are then terminated and meristemoids transition to guard mother cell (GMC) state (4). GMCs undergo a single symmetric division to produce a pair of GCs (5) which mature to define the stomatal aperture amid the tessellated pavement cells of the leaf epidermis (6). (C) Stomatal development in grasses (modelled from barley, Hordeum vulgare). At the beginning of stomatal development, protodermal cells start to proliferate at a faster rate than surrounding cell files (1). Protodermal cells undergo just one round of asymmetric division, generating smaller GMCs (green) and larger epidermal cells (2). GMCs become enlarged and provide cues to induce asymmetric divisions in flanking subsidiary mother cells (SMCs) (3) which result in the production of SCs (blue) (4). Analogous to eudicot stomatal development, GMCs undergo a single symmetric division (5) to produce a pair of daughter cells that differentiate into mature GCs surrounded by columnar pavement cells (6).
For most plant species, including Arabidopsis and grasses, the stomatal lineage progresses in much the same way; originating from a set of undifferentiated epidermal cells at the base of the leaf known as protodermal cells (Stebbins and Shah, 1960; Yang et al., 1995; Luo et al., 2012; Pillitteri and Dong, 2013; Raissig et al., 2016; Hepworth et al., 2018; Endo and Torii, 2019). The stomatal lineage is initiated when individual protodermal cells (known in Arabidopsis as meristemoid mother cells, MMCs) gain the capacity to undergo asymmetric cell divisions (Figure 1). In both Arabidopsis and grasses an asymmetric entry division leads to the formation of one smaller daughter cell and one larger daughter cell (Figure 1). The larger cell in Arabidopsis is termed a stomatal lineage ground cell (SLGC), while in grasses this cell is typically referred to simply as a sister cell or long cell. The smaller cell in Arabidopsis is a meristemoid and retains the ability to undergo self-renewing amplifying divisions, often generating several new SLGCs as a result (Figure 1). Although Wu et al. (2019) refer to the smaller cells in grasses as meristemoids, there is no evidence in the literature to suggest that these cells possess the ability to undergo amplifying divisions; instead the smaller daughter cell expands slightly and transitions directly to a guard mother cell (GMC) (Raissig et al., 2016; Hepworth et al., 2018). For clarity, in this review we will refer to the smaller cells derived from grass asymmetric divisions as GMCs. While Arabidopsis SLGCs have the ability to undergo a spacing division to form satellite meristemoids (Pillitteri and Dong, 2013), similar functionality has not been reported for the larger sister cells of grasses. However, since the proximal-distal development of grass stomata is not completely linear, and multiple large epidermal cells can develop adjacently without the presence of an intervening GMC (Hepworth et al., 2018), the possibility that larger sister cells can undergo infrequent satellite divisions to produce new GMCs cannot be ruled out.
The divergent developmental processes described above during the beginning of the stomatal lineage are briefly realigned when meristemoids in Arabidopsis transition to GMCs and take on a form similar to that of grass GMCs (Figure 1). However, while Arabidopsis GMCs are typically programmed to divide symmetrically to form a pair of GCs, grass GMC cell state is temporarily maintained (Luo et al., 2012; Pillitteri and Dong, 2013; Hepworth et al., 2018). This allows asymmetric divisions to occur in both of the adjacent subsidiary mother cells (SMCs), resulting in the production of a pair of SCs either side of the GMC. Once SCs have formed, grass GMCs — like Arabidopsis GMCs — undergo a single symmetric division to form a pair of GCs. The termination of stomatal development occurs when nascent Arabidopsis and grass GCs fully differentiate to become kidney-shaped and dumbbell-shaped cells respectively (Conklin et al., 2018) (Figure 1).
In Arabidopsis, five basic helix-loop-helix (bHLH) transcription factors are primarily responsible for regulating stomatal lineage entry and subsequent advancement to mature stomata. These bHLHs are known as SPEECHLESS (SPCH), its close paralogues MUTE and FAMA, and SCREAM (SCRM, also known as ICE1) and SCRM2 (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007; Kanaoka et al., 2008) (Figure 2). The formation of MMCs, asymmetric entry divisions and subsequent meristemoid self-renewal requires the first of these, SPCH (Pillitteri and Dong, 2013; Endo and Torii, 2019). MUTE activity is then required to terminate asymmetric divisions and promote the differentiation of meristemoids into GMCs (Pillitteri et al., 2007). Recent work by Han et al. (2018) has shown that MUTE also promotes the GMC-to-GC division by directly inducing the expression of a series of cell-cycle genes that trigger the symmetric divisions of GMCs. MUTE also promotes the transcriptional repressor of these genes, FAMA, which together with the closely related MYB proteins FOUR LIPS (FLP) and MYB88, acts to inhibit extraneous symmetric divisions of GMCs and promote GC maturation (Ohashi-Ito and Bergmann, 2006; Han et al., 2018). At each specific stage of cell division and transition, SPCH, MUTE, and FAMA form obligate heterodimer complexes with SCRM and SCRM2 (Kanaoka et al., 2008; Conklin et al., 2018). These complexes are crucial to the activity of SPCH, MUTE and FAMA; double Arabidopsis scrm scrm2 knockout plants fail to produce cells that enter the stomatal lineage (Kanaoka et al., 2008).
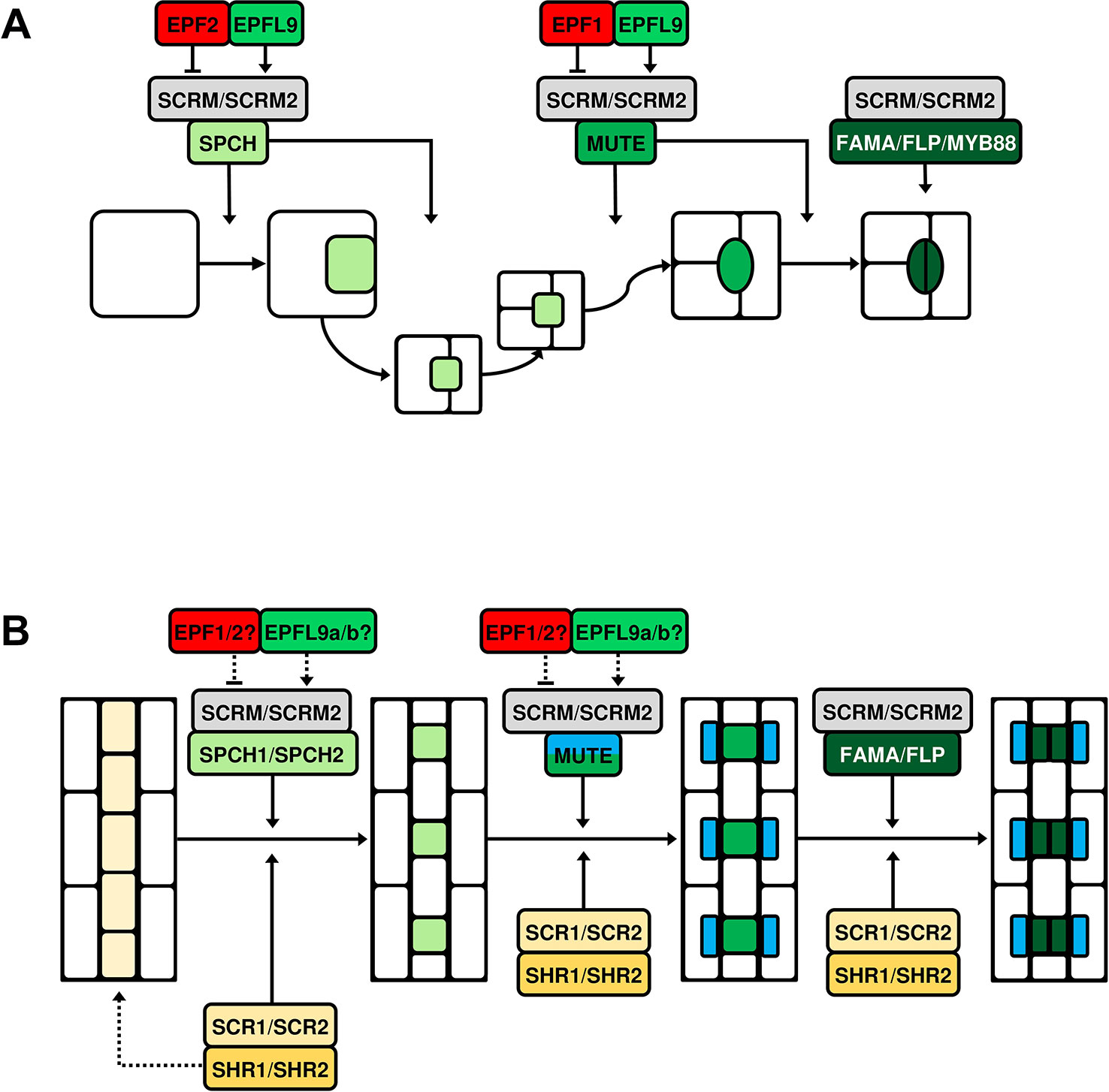
Figure 2 Simplified models of bHLH-mediated stomatal cell lineage transitions in Arabidopsis thaliana and grasses. Five basic helix-loop-helix (bHLH) transcription factors, SPEECHLESS (SPCH), MUTE, FAMA, and their heterodimeric partners SCREAM (SCRM) and SCRM2 exhibit control over stomatal cell type transitions and are conserved across land plants. (A) Stomatal development in A. thaliana. SPCH directs asymmetric divisions of meristemoid mother cells (MMCs) and promotes the meristemoid self-renewing divisions. Epidermal patterning factor 2 (EPF2) and EPF-like protein 9 (EPFL9) are antagonistic regulators of SPCH; competing to suppress and promote its activity respectively. MUTE terminates the meristemoid self-renewing state and promotes guard mother cell (GMC) differentiation and GMC symmetric division. EPF1 primarily targets MUTE; by restraining its activity extra symmetric divisions of GMCs are prevented. EPF1, like EPF2, is assumed to compete with EPFL9 for the regulation of MUTE. FAMA also prevents ectopic symmetric divisions of GMCs and ultimately promotes guard cell (GC) maturation together with the closely related MYB proteins FOUR LIPS (FLP) and MYB88. At each stage of transition, SPCH, MUTE and FAMA form obligate heterodimer complexes with SCRM or SCRM2. (B) The molecular control of stomatal development is ‘alternatively wired’ in monocotyledonous grasses such as rice (Oryza sativa) and Brachypodium (Brachypodium distachyon). It has been suggested that the transcription factors SHORTROOOT1 and SHORTROOT2 (SHR1/2) are involved in the establishment of stomatal lineage cell files (dashed arrow). SPCH has been duplicated and both homologues act to induce asymmetric divisions in these stomatal lineage cells, with SPCH2 the more predominant actor. MUTE promotes GMC differentiation and is involved in the formation of subsidiary cells (SCs) in neighboring subsidiary mother cells (SMCs). As in Arabidopsis, FAMA inhibits supernumerary symmetric divisions of GMCs, probably in combination with a single FLP protein. SCRM and SCRM2 also appear to form heterodimer complexes with the aforementioned bHLHs throughout the lineage, as is observed in Arabidopsis. SHR1/2 and their partner transcription factors SCARECROW1/2 (SCR1/2) exert regulatory influence at each stage of transition. EPFs/EPFLs are also present during grass stomatal development and, while they can demonstrably constrain or promote development, exactly where and how they regulate the grass bHLHs has not been determined (dashed arrows).
There are many hormonal stimuli that directly and/or indirectly regulate stomatal development. For example, the abiotic stress signal abscisic acid (ABA), is known to be an important regulator of both stomatal development and stomatal closure (Chater et al., 2014). However, since our understanding of hormonal regulation in grass stomatal development is limited with respect to our knowledge from Arabidopsis, here we will focus primarily on the core transcription factor-driven stomatal development module. For further information on the hormonal control of stomatal development, see Qi and Torii (2018) or Zoulias et al. (2018).
Several phylogenetic studies have concluded that SPCH, MUTE, FAMA, SCRM, and SCRM2 are highly conserved across land plants, with SPCH undergoing a duplication event in grass species (MacAlister and Bergmann, 2011; Ran et al., 2013; Chater et al., 2017; Qu et al., 2017) (Figure 3). Despite the divergence of stomatal morphology and development in grasses (Figure 1), bHLH orthologues have now been characterised in both rice (Liu et al., 2009; Wu et al., 2019) and Brachypodium (Raissig et al., 2016), with some discreet differences in functionality detected (Figure 2).
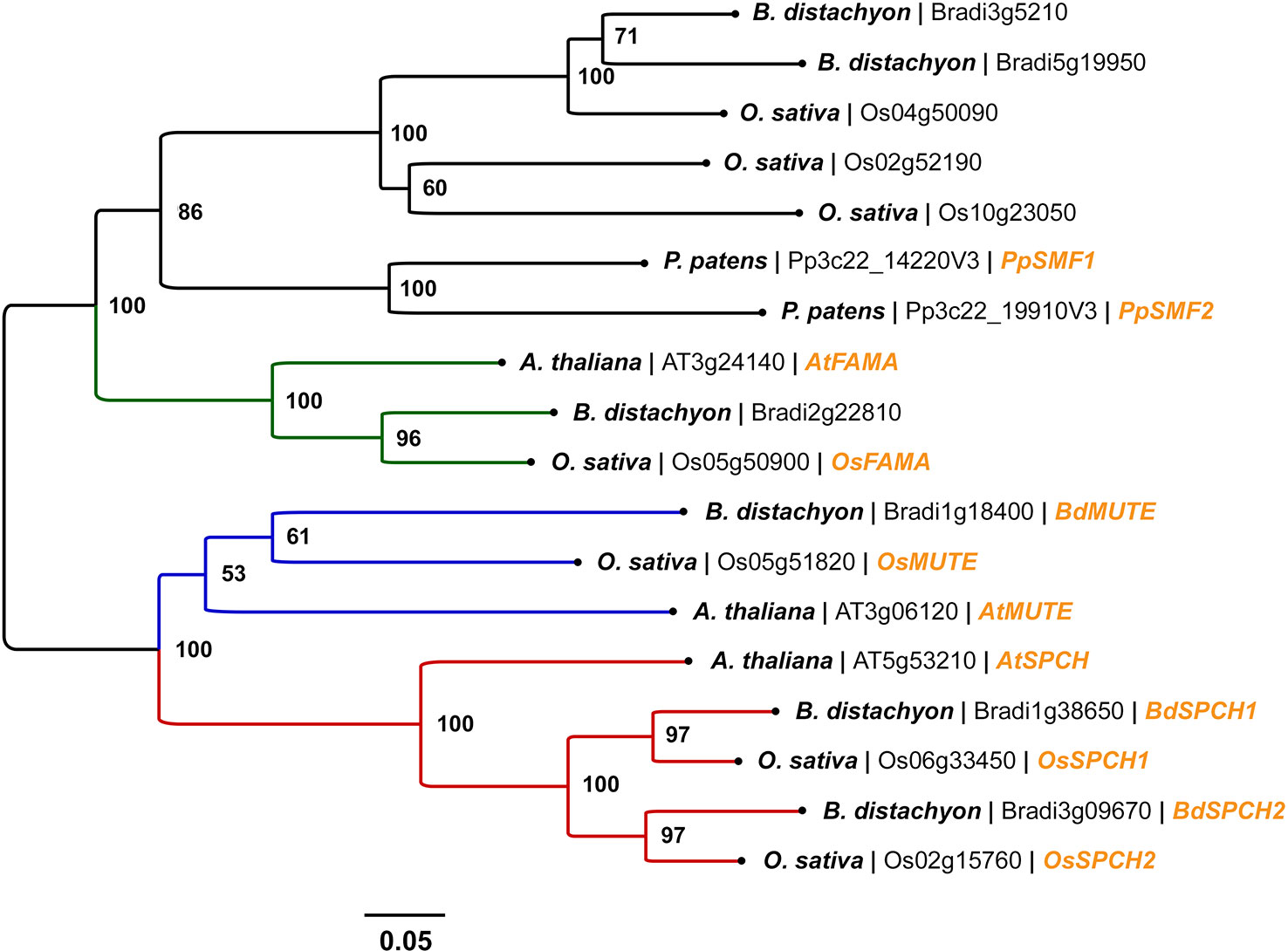
Figure 3 Phylogenetic analysis of SPEECHLESS (SPCH), MUTE, and FAMA peptide sequences in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and Brachypodium (Brachypodium distachyon). Peptide sequences were obtained via BLAST searches of the rice and Brachypodium genomes using Phytozome v12. All peptide sequences with expect values (E-values) < 1 x 10-30 against AtSPCH, AtMUTE, and AtFAMA were used in the analysis. Stomatal bHLH peptide sequences PpSMF1 and PpSMF2 from the non-vascular moss Physcomitrella patens were included for evolutionary context; obtained from Chater et al. (2017). A total of 18 peptide sequences were involved in the phylogenetic analysis. Peptide sequences were aligned using the MUSCLE algorithm (Edgar, 2004). The phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 4.05347046 is shown. A bootstrap test was performed (1,000 replicates) and the percentage of replicate trees that clustered taxa into the conformation displayed is shown adjacent to the nodes. Evolutionary distances between peptide sequences (in units of amino acid substitutions per site) were calculated using the Poisson correction method (Zuckerkandl and Pauling, 1965). The tree is drawn to scale, with branch lengths proportionate to the evolutionary distances used to infer the tree. All evolutionary analyses were performed in MEGA6. From left to right, species name, gene accession number and gene name (if characterised) are labeled at the branch tips. The clades comprising SPCH, MUTE, and FAMA orthologues are coloured red, blue and green respectively.
Recent research has shown that the two bHLH SPCH paralogues — OsSPCH1 and OsSPCH2 — both contribute to stomatal development in rice. Despite this, a reduction in GMC number and SD is only observed in single osspch2 knockout plants (Liu et al., 2009; Wu et al., 2019). Analysis of downstream gene expression in osspch1 and osspch2 single mutants support this finding as OsMUTE and OsFAMA are only downregulated in osspch2 plants (Wu et al., 2019). However, a more severe effect on development is observed in osspch1 osspch2 double knockouts where no GMCs or stomata form, suggesting that OsSPCH1 does contribute to early stomatal development in the absence of OsSPCH2 (Wu et al., 2019). Genetic reporter analyses in Brachypodium further emphasize the importance of the grass SPCH2 paralogue over SPCH1 (Raissig et al., 2016). A BdSPCH2 signal can be strongly detected from the initiation of stomatal development until SC formation, whereas BdSPCH1 displays a lower overall activity, with the strongest signal occurring after the initial asymmetric division and declining before SC formation begins. Relative to the mild stomatal phenotype of bdspch1 knockouts, bdspch2 plants have a drastically reduced SD, consolidating SPCH2 as the main player in initiating stomatal development in grasses (Raissig et al., 2016). Nevertheless, as with rice double knockouts, bdspch1 bdspch2 plants also fail to form GMCs or stomata, suggesting that in both grass species studied thus far, at least one SPCH gene must be present for cells to enter the stomatal lineage (Figure 2).
Functioning downstream of SPCH in Arabidopsis, MUTE stops meristemoid amplifying divisions, promotes GMC formation and subsequently promotes the symmetric GMC division which leads to the formation of GC pairs (Pillitteri et al., 2007; Han et al., 2018). Grasses are not thought to possess self-renewing meristemoids, yet by generating osmute gene knockout lines, Wu et al. (2019) reported that OsMUTE functions primarily to prevent ectopic divisions of GMCs (Figure 2), as GMCs appear to undergo further division(s) before arresting in osmute plants. Interestingly, like in rice, ZmMUTE has recently been shown to be responsible for preventing ectopic divisions in maize, with multiple aborted GMCs often found to be stacked within a stomatal file in maize zmmute (also termed bizui2) mutants (Wang et al., 2019). Ectopic divisions of GMCs often occur in irregular orientations in osmute plants and it is unclear whether the stacking phenotype that is observed in maize zmmute mutants also occurs in the rice mutants. In both osmute and zmmute, neither GCs or SCs form, which for zmmute plants results in seedling lethality around 14 days post-germination. Taken together, these observations show that if MUTE functionality is compromised it is possible to invoke meristemoid-like activity in grasses. However, as mentioned previously, to date no grass species surveyed has naturally exhibited such a cell type.
In OsMUTE-overexpressing plants, Wu et al. (2019) report that all epidermal cells aside from silica and cork cells take on GC-like identity although, upon inspection of the images provided, this suggestion is difficult to interpret as the cells that form appear to be undifferentiated. This is especially apparent when the given examples are contrasted with overexpression studies of MUTE in Arabidopsis. In Arabidopsis, ectopic GCs form as part of a stoma and are more clearly GC-like (Pillitteri et al., 2007). To truly determine the identity of the cells generated via OsMUTE overexpression — and thus understand more about OsMUTE functionality and governance of cell state — it would be useful to repeat the genetic manipulation of OsMUTE in reporter plants that convey GMC, SC, and GC identity.
Whereas neither GCs or SCs form in osmute and zmmute knockout mutant plants, GC pairs do form in bdmute knockout mutant plants, but these GCs are devoid of SCs (Raissig et al., 2017). By generating YFP-BdMUTE reporter lines, it has been demonstrated that the BdMUTE protein is mobile and can promote asymmetric SC entry divisions by entering SMCs from neighboring GMCs (Raissig et al., 2017). Despite the phenotypic discrepancy between bdmute mutants and zmmute and osmute mutants, Wang et al. (2019) showed that ZmMUTE and OsMUTE are also capable of travelling from GMCs to SMCs. Similarly to what was initially described in Brachypodium YFP-BdMUTE reporter lines, YFP-ZmMUTE and YFP-OsMUTE signals are detectable in both GMCs and neighboring SMCs immediately prior to SC-forming divisions in maize and rice, respectively (Raissig et al., 2017; Wang et al., 2019). Yet, unlike osmute and zmmute, the course of GMC and GC development is less perturbed in bdmute knockout plants, and thus MUTE’s involvement in the regulation of GMC cell fate and divisions appears to show some variation amongst different grass species.
OsFAMA function has been analyzed via the production of both T-DNA and CRISPR osfama mutants, with both methods revealing that OsFAMA acts to promote GC maturation in rice (Liu et al., 2009; Wu et al., 2019) (Figure 2). Intriguingly, ectopic GMC divisions (as in osmute) and, in some cases, undivided SMC cells are detectable in osfama knockout plants (Wu et al., 2019). This implies that FAMA, perhaps in combination with MUTE, contributes to the regulation of SMC asymmetric divisions and GMC symmetric divisions in rice. There are no reports yet that describe how FAMA functions during Brachypodium or maize stomatal development, and so at present it is not possible to compare the function of other grass FAMA orthologues with that of OsFAMA. It will be interesting to learn whether BdFAMA and ZmFAMA also partake in events upstream of GC formation as has been shown in OsFAMA.
Genes with high sequence similarity to Arabidopsis SCRM and SCRM2 have been identified in rice (Os11g32100 and Os01g71310 respectively), suggesting that the heterodimerisation of bHLH transcription factors observed in Arabidopsis also occurs in rice (Liu et al., 2009) (Figure 2). The proposed protein-protein interactions of these gene products — OsSCRM and OsSCRM2 — with the aforementioned stomatal development genes (OsSPCH1/2, OsMUTE, and OsFAMA) have now been confirmed in vitro via bimolecular fluorescent complementation (BiFC) and yeast two-hybrid (Y2H) assays (Wu et al., 2019). Like Arabidopsis SCRM and SCRM2, OsSCRM and OsSCRM2 act redundantly during stomatal development, with double scrm scrm2 knockouts leading to the absence of all stomatal lineage cell types and stomata in both species (Kanaoka et al., 2008; Wu et al., 2019). Whereas osscrm CRISPR knockout mutants fail to produce stomata (and only a few meristemoids form in early development), Arabidopsis T-DNA knockdown plants with little-to-no SCRM expression produce a range of stomatal lineage cells (at various aborted stages) as well as stomata (Kanaoka et al., 2008). However, when equivalent osscrm T-DNA knockouts are examined in rice (which display a weaker phenotype than CRISPR osscrm plants), the function of OsSCRM appears to more closely mirror that of AtSCRM. This is because aborted stomatal lineage cells can be observed throughout the development stages, implying that, together with OsSPCH1/2 in early development, OsSCRM probably associates with OsMUTE and OsFAMA in vivo (Wu et al., 2019). In contrast to scrm knockouts, stomatal development is unaffected in both atscrm2 and osscrm2 knockout mutants, suggesting that these paralogous proteins play a minor role in the development of stomata in their respective species (Kanaoka et al., 2008; Wu et al., 2019).
In Brachypodium, the roles of BdSCRM and BdSCRM2 are more distinct. Both bdscrm and bdscrm2 single mutants produce a seedling lethal phenotype, owing to an inability to produce mature stomata (Raissig et al., 2016). While bdscrm plants fail to produce any stomatal lineage cells, entry to the stomatal lineage is not blocked in bdscrm2 mutants; instead aborted four-cell complexes form which fail to progress to mature stomata (Raissig et al., 2016). This indicates that BdSCRM2 might fulfil a novel function later in development which is independent of BdSCRM (Raissig et al., 2016). It is conceivable that this function is to specifically interact with FAMA and drive the formation of mature stomata. Thus, it will be interesting to learn whether the stomatal phenotype of bdfama matches that of bdscrm2. Considered across the species discussed, there are clear similarities and some subtle differences in the function and coordination of the bHLH transcription factors; both between the grasses and Arabidopsis and within the grasses themselves.
For plants to adjust their leaf SD, succinct titration of bHLH activity is essential. This is particularly true of SPCH, which has a diverse range of transcriptional targets in Arabidopsis, with chromatin immunoprecipitation-sequencing (ChIP-Seq) experiments identifying over 8,000 genomic binding sites (Lau et al., 2014). To coordinate the activity of SPCH (and other bHLHs) during stomatal development, intra- and extracellular signalling are particularly important and, in Arabidopsis, much research has been conducted into these signalling modules.
Arabidopsis uses apoplastic cysteine-rich signalling peptides called epidermal patterning factors (EPFs) and EPF-like (EPFL) peptides to convey extracellular signals between stomatal lineage cells, mesophyll cells, and cells destined to become pavement cells (Simmons and Bergmann, 2016) (Figure 2). EPF2 and EPF1 negatively regulate SD; EPF2 primarily by preventing stomatal lineage entry and meristemoid amplifying divisions (Hara et al., 2009; Hunt and Gray, 2009); EPF1 by restricting meristemoid identity and facilitating the correct orientation of SLGC spacing divisions (Hara et al., 2007; Qi et al., 2017). On the other hand, EPFL9 promotes stomatal development in opposition to EPF2 and EPF1 (Hunt et al., 2010; Sugano et al., 2010). At the plasma membrane, EPF/EPFL signals are perceived by members of the ERECTA family (ERECTA, ER; ERECTA-LIKE1, ERL1; and ERECTA-LIKE2, ERL2) of receptor kinases along with the receptor protein TOO MANY MOUTHS (TMM) (Nadeau and Sack, 2002; Shpak et al., 2005; Lin et al., 2017).The binding of EPF2 to an ER/TMM complex in the presence of somatic embryogenesis receptor kinases (SERKs) promotes signal transduction across the plasma membrane into the cytoplasm; thereby activating the intracellular mitogen-activated protein kinase (MAPK) pathway (Meng et al., 2015). When activated, this pathway culminates in the phosphorylation and subsequent degradation of SPCH (Lampard et al., 2008). EPFL9 acts antagonistically to EPF2 by competing to bind to the ER/TMM complex (Lee et al., 2015). If successful in doing so, EPFL9 can prevent SPCH degradation by blocking activation of the MAPK pathway (Bergmann et al., 2004; Lampard et al., 2008). Rather than an ER/TMM complex, EPF1 preferentially binds to ERL1, which also associates with TMM and, like EPF2, probably competes with EPFL9 for binding (Lee et al., 2015).
Like with the bHLH transcription factors, functional orthologues of EPF/EPFLs are also present in grasses (Tanaka et al., 2013; Caine et al., 2016; Hughes et al., 2017; Yin et al., 2017; Lu et al., 2019) (Figure 2). Moreover, phylogenetic analysis also suggests that ERECTA and TMM genes are present, with OsERECTA already shown to be a governor of heat tolerance in rice (Shen et al., 2015). As in Arabidopsis, there are probably two EPF genes involved in negatively regulating stomatal development in grasses (Hepworth et al., 2018; Lu et al., 2019). However, rather than one EPFL9 gene, grasses normally have two; EPFL9a and EPFL9b (Hepworth et al., 2018; Lu et al., 2019). Using CRISPR-Cas9 and CRISPR-Cpf1 genome editing, Yin et al. (2017) have shown that OsEPFL9a plays a major role in regulating rice stomatal development, with osepfl9a knockout plants having a ~90% reduction in SD. Conversely, overexpression of OsEPFL9a has been shown to moderately increase SD and reduce stomatal size (Mohammed et al, 2019). Interestingly, little-to-no clustering of stomata occurs upon OsEPFL9a overexpression in rice, whereas the large increase in density in EPFL9-overexpressing Arabidopsis plants is driven by stomatal clustering (Hunt et al., 2010). Despite this, exactly how, where and when OsEPFL9a contributes to stomatal lineage progression is still not well understood and requires further study. While no equivalent OsEPFL9b knockout or overexpressing rice plants have been generated thus far, recent work in Arabidopsis has shown that OsEPFL9b can moderately increase SD when overexpressed (Lu et al., 2019). In contrast to OsEPFL9a overexpression, OsEPFL9b-overexpressing plants have a tissue-specific clustering phenotype, with stomatal clusters forming in hypocotyls but not rosette leaves (Lu et al., 2019). Nevertheless, based on evidence that is currently available, it seems that OsEPFL9a is the dominant player in stomatal development, with OsEPFL9b perhaps playing a minor role.
In barley and wheat, Hughes et al. (2017) and Dunn et al. (2019) have shown that overexpression of HvEPF1 and TaEPF1B leads to inhibition of multiple stages of the stomatal lineage, including asymmetric entry divisions, GMC development and the formation of SCs. Overexpression of HvEPF1 in Arabidopsis produces a similar phenotype to that of Arabidopsis EPF1 overexpression, whereby meristemoid differentiation is perturbed, resulting in a clustering of small cells around a central meristemoid (Hughes et al., 2017). Given that cells derived from asymmetric entry divisions in grasses are not believed to take on meristemoid-like identity, it is intriguing that HvEPF1 is capable of regulating meristemoid fate in Arabidopsis. This finding, together with findings from wheat, suggests that EPF1 orthologues probably act relatively early in the stomatal lineage in grasses, but further investigation is required to ascertain exactly where and how these gene products are functioning. Subsequent work by Caine et al. (2019) and Lu et al. (2019), where OsEPF1 was overexpressed in both rice and Arabidopsis, produced similar results to those in barley and in wheat, with strong overexpression of OsEPF1 in rice resulting in the majority of protodermal cells failing to enter the stomatal lineage (Caine et al., 2019). As with OsEPF1, increasing the expression of OsEPF2 effectively reduces the SD of both rice and Arabidopsis, with phenotypic similarities to OsEPF1 overexpression in each instance (Lu et al., 2019). Clearly, assessment of how EPF/EPFLs regulate SD in rice has provided insight to their function, but more research (including generation of osepf1, osepf2 single knockout and osepf1 osepf2 double knockout plants) is required to enable a more succinct understanding of how rice stomatal development unfolds.
Little is known about how EPF signals are transduced to nuclear-residing bHLH transcription factors during grass stomatal development. However, a recent report has confirmed that like in Arabidopsis, a member of the MAPK pathway, YODA, is involved in regulating stomatal development in Brachypodium (Abrash et al., 2018). Abrash et al. (2018) show that bdyda1 knockout mutants have a large increase in SD and a severe stomatal clustering phenotype, whereby approximately 86% of stomata are involved in cell clusters. Interestingly, rather than arising as a result of faulty physical asymmetry in cell divisions (as in Arabidopsis yda mutants), cell clusters in bdyda mutants result from a failure of differentiation and fate reinforcement after asymmetric entry divisions have already occurred (Bergmann et al., 2004; Abrash et al., 2018). Thus, although YDA proteins influence entry to the stomatal lineage in both Brachypodium and Arabidopsis, their roles appear to be distinct. In Arabidopsis, YDA is a pre-division regulator, whereas BdYDA acts post-division to reinforce cell fate in Brachypodium (Abrash et al., 2018).
Until recently, the positional signals that prompt the specification of grass stomatal cell files remained unclear. Schuler et al.’s (2018) study of rice and maize SHR orthologues has shed new light onto this area. In Arabidopsis, SHR interacts with another transcription factor, SCARECROW (SCR), in the shoots to position bundle sheath cells around the developing vasculature (Cui et al., 2014). Given that (i) vascular patterning and stomatal patterning are coordinated, (ii) stomatal cell files are specified (laterally) adjacent to procambial centres in grasses, and (iii) OsSHR1 and OsSCR1 are expressed in stomatal lineage cells, it was proposed that SHR-SCR interactions might contribute to the specification and spacing of stomatal cell files in grasses (Kamiya et al., 2003; Schuler et al., 2018). Indeed, ectopic expression of ZmSHR1 in the rice epidermis (chosen to avoid potentially silencing another SHR gene, OsSHR2) does lead to the formation of extra stomatal cell files positioned further from the leaf veins (Schuler et al., 2018). In line with these findings, double knockout osshr1 osshr2 rice plants have been shown to have reduced SD, with phenotypes suggesting that OsSHR1/2 act redundantly at multiple points during the stomatal lineage (Wu et al., 2019) (Figure 2). Despite these findings suggesting that OsSHR1/2 promote stomatal formation, Wu et al.’s (2019) study does not indicate that overexpression of OsSHR1 or OsSHR2 alters the number of stomatal cell files, nor does SD increase when either gene is overexpressed. It is unclear whether this is because of gene silencing (as alluded to above) or whether OsSHR1 or OsSHR2 are genuinely incapable of promoting new stomatal files in rice.
Wu et al. (2019) also investigated the function of OsSCR1 and OsSCR2 during rice stomatal development and found that like OsSHR1/2, OsSCR1 and OsSCR2 positively regulate SD at multiple stages (Figure 2). While OsSCR1 has a prominent role, OsSCR2 function is only noticeable in osscr1 osscr2 double knockouts and not in single knockout osscr2 mutants. Binding assays have confirmed the existence of OsSHR-OsSCR interactions in vitro, suggesting that rice replicates the functional link between SHRs and SCRs that is observed in Arabidopsis. Further analysis of the genomic targets of SHRs and SCRs in grasses might help to clarify exactly how and when they regulate stomatal development and thus may also have implications for the optimisation of SD in rice.
Whereas the rice stomatal bHLHs are thought to invoke and regulate specific cell identities and divisions in the rice stomatal lineage, the precise mitotic control of these divisions has not been well-studied. Across life, specific cyclin-CDK (cyclin-dependent kinase) complexes are known to govern cell cycle transitions (Harashima et al., 2013; Han and Torii, 2019). In the Arabidopsis stomatal lineage, complexes of A2-type cyclins (CYCA2s) and a B-type CDK, CDKB1;1, promote symmetric divisions of GMCs (Boudolf et al., 2009; Vanneste et al., 2011). Undivided GCs are common in triple knockouts of CYCA2s (cyca2;134 and cyca2;234) and in cdkb1;1 mutants, and the nuclear DNA content of cells from the former is double that of normal GCs, suggesting that they are arrested at the G2-to-M phase (Boudolf et al., 2004; Vanneste et al., 2011).
While four CYCA2s are found in Arabidopsis, only one CYCA2 gene, OsCYCA2;1, is present in the rice genome (La et al., 2006; Qu et al., 2018). BiFC assays have shown that OsCYCA2;1 and OsCDKB1;1 (orthologue of Arabidopsis CDKB1;1) form a functional complex in rice (Qu et al., 2018). Yet, in contrast to Arabidopsis, when OsCYAC2;1 or OsCDKB1;1 are knocked down, GMC divisions are uninterrupted (Qu et al., 2018). Intriguingly, fewer asymmetric entry divisions are observed in stomatal cell files in OsCYCA2;1-RNAi plants, leading to an overall reduction in SD (Qu et al., 2018). Nevertheless, both OsCYCA2;1 and OsCDKB1;1 can rescue the defective phenotypes of Arabidopsis cyca2 and cdkb1 mutants respectively (Qu et al., 2018). Thus, the functions of CYCA2s and CDKB1;1 must be somewhat conserved between rice and Arabidopsis despite the divergence in the timing of their activity (Qu et al., 2018).
Connections between the core bHLH module and cell-cycle regulators in the Arabidopsis stomatal lineage are beginning to be identified (Xie et al., 2010; Lau et al., 2014; Adrian et al., 2015; Han et al., 2018). In the early lineage, CYCD3s are upregulated in response to SPCH expression (Adrian et al., 2015), with CYCD3;1 targeted directly by SPCH (Lau et al., 2014). Later in development, inducers of symmetric GMC divisions — CYCD5;1, CYCA2s, and CDKB1;1 — are directly upregulated by MUTE; a recent finding that has implicated MUTE as a governor of the GMC symmetric division (Han et al., 2018). FAMA and FLP/MYB88 are responsible for negatively regulating this symmetric division by ensuring that GCs do not undergo further divisions (Lai et al., 2005; Lee et al., 2014). Whereas MUTE enhances the expression of CDKB1;1, FLP represses its expression by binding to a cis-regulatory region in its promoter sequence (Xie et al., 2010). A single orthologue of FLP/MYB88, OsFLP, is present in the rice genome (Wu et al., 2019). Akin to its role in Arabidopsis, OsFLP appears to be involved in the regulation of symmetric divisions in rice, as knocking out its expression leads to abnormal division of GMCs (Wu et al., 2019) (Figure 2). However, as described earlier, CDKB1;1 does not influence the GMC-GC transition in rice, so the genomic targets of OsMUTE and OsFLP must have diverged to some extent. Thus, exactly how the functions of other cyclins and CDKs are programmed in the rice stomatal lineage must now be determined. Furthermore, by investigating the cyclin-CDK-mediated regulation of asymmetric SC entry divisions, insight will be gained as to how the regulatory machinery of the plant cell-cycle has been adjusted in grasses to accommodate these unique cell divisions.
Our extensive knowledge of stomatal development in Arabidopsis is gradually being translated into rice and other grasses. Now, by harnessing this knowledge, efforts are being made to alter the stomatal development of rice to create crops that might be better suited to future climate conditions. Decreasing the number of stomata on the leaves of rice could maintain its productivity in the future hotter, drier climate (Caine et al., 2019). Reducing SD in Arabidopsis, maize, and barley demonstrably improves WUE and/or drought tolerance (Hepworth et al., 2015; Liu et al., 2015; Hughes et al., 2017). However, until Caine et al.’s (2019) study, it was unclear whether this finding could be replicated in rice given the potentially negative impact of constraining the transpiration rate of such a water-intensive crop (Hoekstra and Chapagain, 2006). To test this hypothesis, Caine et al. (2019) engineered a rice cultivar (IR64) to overexpress OsEPF1, creating plants with up to an 88% reduction in stomata density (relative to wildtype SD). Direct examinations of whole-plant water use have shown that OsEPF1-overexpressing (OsEPF1oe) plants consume around 40% less water than control plants over equivalent periods, due primarily to reductions in gs (Caine et al., 2019).
In future predictions of the climate, plants will be faced with a dilemma: close stomata to save water or keep them open to stay cool (Chaves et al., 2016). Whereas both elevated atmospheric CO2 or reduced water availability trigger stomatal closure in plants, exposure to increased temperatures could lead to lethal overheating or photoinhibition if transpirational cooling is not maintained (Bertolino et al., 2019). By this logic, plants engineered to have fewer stomata should be more susceptible to heat-stress (Urban et al., 2017). However, thermal imaging of OsEPF1oe and comparable control plants subjected to droughted conditions reveals a heightened capacity for evaporative cooling in OsEPF1oe plants during late-stage drought. Essentially, OsEPF1oe plants are able to restrict transpiration during the early stages of drought, meaning more water is available for maintenance of evaporative cooling later in the drought (Caine et al., 2019). Moreover, under well-watered conditions at elevated temperature, OsEPF1oe plants can increase the aperture of their stomata, thereby mitigating the potentially detrimental effect of having fewer stomata when conditions are more favorable (Caine et al., 2019).
Although results have not been replicated in the field, during laboratory-simulated drought treatments OsEPF1oe grain yields are at least equivalent to controls (Caine et al., 2019). Interestingly, when drought is introduced during flowering (after 88 days of growth), lines with moderate stomatal reductions (OsEPF1oeW) outperform both control plants and those with more severe reductions (OsEPF1oeS). In these plants, both grain yield and above-ground biomass are increased, suggesting that subtle adjustments to SD might be beneficial during drought at the flowering stage (Caine et al., 2019). After the same treatment, the 1,000 grain weight of all OsEPF1oe lines is significantly higher than that of controls (Caine et al., 2019). Exposure to high temperatures during grain filling is known to reduce the content of starch molecules and storage proteins in rice grains (Lin et al., 2010). Therefore, the observed maintenance of grain weight in OsEPF1oe lines in droughted conditions might be due to prolonged transpirational cooling of the heat-sensitive flowers (Morita et al., 2016).
It has been shown that changes to stomatal size and density can be correlated with gs, and so it follows that genetic manipulation of SD has the potential to increase photosynthetic gas exchange (Franks et al., 2012). However, as pointed out by Harrison et al. (2019), such a coupling between SD and gas exchange does not always exist in practice. Nevertheless, Arabidopsis EPFL9-overexpressing plants (with ~600% increased SD) show enhanced A at both ambient and elevated CO2, probably due to a much increased gs (Tanaka et al., 2013). This is despite the occurrence of stomatal clustering, a trait that has been shown to negatively impact photosynthetic performance (Dow et al., 2014). In rice plants that overexpress OsEPFL9a, gas exchange (both A and gs) is unchanged, although this may be a signature of the smaller increase in SD (~20%–30%) in these plants relative to Arabidopsis EPFL9-overexpressing plants (Mohammed et al., 2019). A similarly modest increase in SD in rice has been generated via the overexpression of a maize SHORTROOT gene, ZmSHR1 (Schuler et al., 2018). Like with OsEPFL9a overexpression, higher SD in ZmSHR1-overexpressing plants does not lead to an enhancement of A or gs (Schuler et al., 2018). For OsEPFL9a overexpression, the lack of increase in either parameter may be explained by a concurrent reduction in stomatal size (Mohammed et al., 2019). Perhaps if changes to the stomatal number were more significant — as in Tanaka et al. (2013) — the compensatory effect of reduced stomatal size in OsEPFL9a-overexpressing plants might have less of an impact in preventing gs from increasing.
As discussed previously, manipulating SD will create a trade-off between the uptake of CO2 and the control of transpirational water flow. Indeed, in Arabidopsis, enhancement of A in Tanaka et al.’s (2013) AtEPFL9-overexpressing plants is not translated to biomass gains. This is likely due to the plants being more prone to water loss, as transgenic plants with greater SD have significantly greater transpiration rates (Tanaka et al., 2013). Relative to Arabidopsis and other plants, rice might be more able to afford an increased rate of water loss, as the majority (~75%) of production is sourced from irrigated lowland systems (Zhao et al., 2011). Moreover, amongst lowland rice varieties, A is correlated with SD when plants are grown in flooded soils (Tsunoda and Fukoshima, 1986). In fact, the SD and gs of 3 high-yielding rice cultivars (IR72, Takanari and LYPJ) are significantly higher than the corresponding average values from 69 lower-yielding accessions (Ohsumi et al., 2007).
Genetic manipulation of SD leads to a fixed change in the number of stomata that form per unit area in the epidermis. The stomata of Arabidopsis SD mutants (of multiple genotypic backgrounds) retain the capacity to open and close and changes in SD are often accompanied by inverse changes to stomatal size; i.e., reductions in SD are linked to increases in stomatal size and vice versa (Doheny-Adams et al., 2012). However, this size-density response is not consistently seen in grasses. For both barley and the IR64 rice cultivar (subspecies indica) the opposite response is observed, whereby reductions in SD result in co-reductions in stomata size (Hughes et al., 2017; Caine et al., 2019). In the Nipponbare rice cultivar (subspecies japonica), reduced SD leads to increased stomatal size and increased SD leads to reduced size; responses similar to those previously observed in Arabidopsis (Mohammed et al., 2019). Thus, within different subspecies of rice, the stomatal size responses to genetic manipulation of SD are different. Why this occurs is unclear, although there is evidence in grasses that smaller stomata can improve WUE by opening and closing quicker in response to environmental fluctuations (McAusland et al., 2016; Lawson and Vialet-Chabrand, 2019). For both subspecies, it would be interesting to assess how the stomata of low-SD OsEPF1oe plants perform under fluctuating conditions to ascertain the impact that size and density alterations have on the “speed” of stomatal movements and WUE.
By targeting the mechanisms that govern guard cell ion transportation across the plasma membrane and tonoplast, it is possible to effectively alter stomatal opening and closing in response to both abiotic and biotic stress (Huang et al., 2009; Daszkowska-Golec and Szarejko, 2013; Li et al., 2017; Lawson and Vialet-Chabrand, 2019) and to enhance plant biomass accumulation (Papanatsiou et al., 2019). Because the SCs of grasses are thought to both enable a swifter transport of ions and osmolytes to GCs and to lend a mechanical advantage to their stomatal movements, grass stomatal complexes are viewed as being more responsive than other stomatal morphologies (Franks and Farquhar, 2006; Cai et al., 2017; Nunes et al., 2019). Indeed, the stomata of bdmute plants (which are devoid of SCs) respond more slowly to changes in light intensity (Raissig et al., 2017). However, to date no genetic manipulations that enhance the speed of grass stomata have been reported, and it is therefore not possible to compare the potential water savings that could be achieved by low-SD rice with those of rice manipulated to have enhanced stomatal closure. To address this unknown, future efforts to manipulate rice stomata should aim to target short-term stomatal responses, perhaps individually and in combination with SD, so that a more complete picture of the control of WUE in grass plants can be developed.
Rice is the world’s most important human food crop and yields must be protected against future climate instabilities. As the “gatekeepers” of transpiration and carbon uptake, stomata represent an obvious target to improve A or water retention in rice (Lawson and Blatt, 2014). Although more detailed examinations of rice stomatal development will be required to match our understanding of stomatal development in Arabidopsis, the advances to our understanding of grass stomatal development discussed here have enabled high- and low-SD rice plants to be developed (Schuler et al., 2018; Caine et al., 2019; Mohammed et al., 2019). Contrary to expectation, rice plants with increased SD do not exhibit corresponding increases in A (Schuler et al., 2018; Mohammed et al., 2019), although this could be due to altered stomatal size and or aperture size. Thus, rice plants with more substantial increases in SD will be required to test the efficacy (or lack thereof) of targeting stomatal development for the enhancement of either A or evaporative cooling. On the other hand, given that freshwater insecurities and exposure to drought will likely become increasingly prevalent in areas of rice cultivation (Kang et al., 2009; Quentin Grafton, 2017), rice crops that use water more efficiently might be of greater importance in the future climate. This will be especially important in Africa where, as a result of water limitations, rainfed upland rice production is steadily increasing, despite this method being particularly susceptible to drought (Saito et al., 2018). Promisingly, OsEPF1oe lines with decreased SD exhibit improved water conservation and tolerance to drought without yield penalties (Caine et al., 2019). While this development is encouraging, these findings have not yet been reproduced in the field, and so it remains to be seen whether real-world fluctuations in environmental variables will influence the performance of these plants that have thus far only been tested under laboratory conditions.
CB, RC, and JG wrote the article and CB prepared the figures. All authors checked and approved the article before publication.
RC and JG acknowledge the BBSRC and Newton fund for the financial support. RC also acknowledges the University of Sheffield QR GCRF fellowship (Research England institutional allocation) for the fellows.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abrash, E., Anleu Gil, M. X., Matos, J. L., Bergmann, D. C. (2018). Conservation and divergence of YODA MAPKKK function in regulation of grass epidermal patterning. Dev. (Cambridge England) 145 (14), dev.165860. doi: 10.1242/dev.165860
Adrian, J., Chang, J., Ballenger, C. E., Bargmann, B. O. R., Alassimone, J., Davies, K. A., et al. (2015). Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev. Cell 33 (1), 107–118. doi: 10.1016/j.devcel.2015.01.025
Arve, L., Torre, S., Olsen, J., Tanino, K. (2011). “Stomatal Responses to Drought Stress and Air Humidity,” in Abiotic Stress in Plants - Mechanisms and Adaptations (Rijeka: IntechOpen), 268–280.
Bergmann, D. C., Lukowitz, W., Somerville, C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304 (5676), 1494–1497. doi: 10.1126/science.1096014
Bertolino, L. T., Caine, R. S., Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10, 225. doi: 10.3389/fpls.2019.00225
Boudolf, V., Vlieghe, K., Beemster, G. T. S., Magyar, Z., Torres Acosta, J. A., Maes, S., et al. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 (10), 2683–2692. doi: 10.1105/tpc.104.024398
Boudolf, V., Lammens, T., Boruc, J., Van Leene, J., Van Den Daele, H., Maes, S., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150 (3), 1482–1493. doi: 10.1104/pp.109.140269
Brar, D. S., Khush, G. S. (2013). Biotechnological approaches for increasing productivity and sustainability of rice production. In Progress and Prospects in Crop Research, (Cambridge, MA: Academic Press), 151–175.
Brownlee, C. (2001). The long and the short of stomatal density signals. Trends Plant Sci. 6 (10), 441–442. doi: 10.1016/S1360-1385(01)02095-7
Buckley, T. N. (2005). The control of stomata by water balance. New Phytol. 168 (2), 275–292. doi: 10.1111/j.1469-8137.2005.01543.x
Cai, S., Papanatsiou, M., Blatt, M. R., Chen, Z.-H. (2017). Speedy grass stomata: emerging molecular and evolutionary features. Mol. Plant 10 (7), 912–914. doi: 10.1016/j.molp.2017.06.002
Caine, R. S., Chater, C. C., Kamisugi, Y., Cuming, A. C., Beerling, D. J., Gray, J. E., et al. (2016). An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143 (18), 3306–3314. doi: 10.1242/dev.135038
Caine, R. S., Yin, X., Sloan, J., Harrison, E. L., Mohammed, U., Fulton, T., et al. (2019). Rice with reduced stomatal density conserves water and has improved drought tolerance under future climate conditions. New Phytol. 221 (1), 371–384. doi: 10.1111/nph.15344
Casson, S., Gray, J. E. (2008). Influence of environmental factors on stomatal development. New Phytol. 178 (1), 9–23. doi: 10.1111/j.1469-8137.2007.02351.x
Challinor, A. J., Watson, J., Lobell, D. B., Howden, S. M., Smith, D. R., Chhetri, N. (2014). A meta-analysis of crop yield under climate change and adaptation. Nat. Climate Change 4 (4), 287–291. doi: 10.1038/nclimate2153
Chater, C. C., Oliver, J., Casson, S., Gray, J. E. (2014). Putting the brakes on: abscisic acid as a central environmental regulator of stomatal development. New Phytol. 202 (2), 376–391. doi: 10.1111/nph.12713
Chater, C. C. C., Caine, R. S., Fleming, A. J., Gray, J. E. (2017). Origins and evolution of stomatal development. Plant Physiol. 174 (2), 624–638. doi: 10.1104/pp.17.00183
Chaves, M. M., Costa, J. M., Zarrouk, O., Pinheiro, C., Lopes, C. M., Pereira, J. S. (2016). Controlling stomatal aperture in semi-arid regions—the dilemma of saving water or being cool? Plant Sci. 251, 54–64. doi: 10.1016/j.plantsci.2016.06.015
Chen, Z.-H., Chen, G., Dai, F., Wang, Y., Hills, A., Ruan, Y.-L., et al. (2017). Molecular evolution of grass stomata. Trends Plant Sci. 22 (2), 124–139. doi: 10.1016/j.tplants.2016.09.005
Conklin, P. A., Strable, J., Li, S., Scanlon, M. J. (2018). On the mechanisms of development in monocot and eudicot leaves. New Phytol. 221 (2), 706–724. doi: 10.1111/nph.15371
Cui, H., Kong, D., Liu, X., Hao, Y. (2014). SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J. 78 (2), 319–327. doi: 10.1111/tpj.12470
Daszkowska-Golec, A., Szarejko, I. (2013). Open or close the gate - Stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 4, 138. doi: 10.3389/fpls.2013.00138
Doheny-Adams, T., Hunt, L., Franks, P. J., Beerling, D. J., Gray, J. E. (2012). Genetic manipulation of stomatal density influences stomatal size, plant growth and tolerance to restricted water supply across a growth carbon dioxide gradient. Philosophic Transact. Royal Soc. B.: Biol. Sci. 367 (1588), 547–555. doi: 10.1098/rstb.2011.0272
Dow, G. J., Berry, J. A., Bergmann, D. C. (2014). The physiological importance of developmental mechanisms that enforce proper stomatal spacing in Arabidopsis thaliana. New Phytol. 201 (4), 1205–1217. doi: 10.1111/nph.12586
Dunn, J., Hunt, L., Afsharinafar, M., Meselmani, M. A., Mitchell, A., Howells, R., et al. (2019). Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 70(18), 4737–4748. doi: 10.1093/jxb/erz248
Edgar, R. C. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. doi: 10.1093/nar/gkh340
Elert, E. (2014). Rice by the numbers: a good grain. Nature 514 (7524), S50–S51. doi: 10.1038/514S50a
Endo, H., Torii, K. U. (2019). Stomatal development and perspectives toward agricultural improvement. Cold Spring Harbor Perspect. Biol. 11 (5), a034660. doi: 10.1101/cshperspect.a034660
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478 (7369), 337–342. doi: 10.1038/nature10452
Franks, P. J., Farquhar, G. D. (2006). The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol. 143 (1), 78–87. doi: 10.1104/pp.106.089367
Franks, P. J., Leitch, I. J., Ruszala, E. M., Hetherington, A. M., Beerling, D. J. (2012). Physiological framework for adaptation of stomata to CO2 from glacial to future concentrations. Philosophic Transact. Royal Soc. B.: Biol. Sci. 367 (1588), 537–546. doi: 10.1098/rstb.2011.0270
Haefele, S. M., Kato, Y., Singh, S. (2016). Climate ready rice: augmenting drought tolerance with best management practices. Field Crops Res. 190, 60–69. doi: 10.1016/J.FCR.2016.02.001
Han, S.-K., Torii, K. U. (2019). Linking cell cycle to stomatal differentiation. Curr. Opin. Plant Biol. 51, 66–73. doi: 10.1016/J.PBI.2019.03.010
Han, S.-K., Qi, X., Sugihara, K., Dang, J. H., Endo, T. A., Miller, K. L., et al. (2018). MUTE directly orchestrates cell-state switch and the single symmetric division to create stomata. Dev. Cell 45 (3), 303–315.e5. doi: 10.1016/j.devcel.2018.04.010
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C., Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21 (14), 1720–1725. doi: 10.1101/gad.1550707
Hara, K., Yokoo, T., Kajita, R., Onishi, T., Yahata, S., Peterson, K. M., et al. (2009). Epidermal cell density is Autoregulated via a secretory peptide, epidermal patterning factor 2 in Arabidopsis leaves. Plant Cell Physiol. 50 (6), 1019–1031. doi: 10.1093/pcp/pcp068
Harashima, H., Dissmeyer, N., Schnittger, A. (2013). Cell cycle control across the eukaryotic kingdom. Trends Cell Biol. 23 (7), 345–356. doi: 10.1016/J.TCB.2013.03.002
Harrison, E. L., Arce Cubas, L., Gray, J. E., Hepworth, C. (2019). The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J. tpj.14560. doi: 10.1111/tpj.14560
Hepworth, C., Doheny-Adams, T., Hunt, L., Cameron, D. D., Gray, J. E. (2015). Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol. 208 (2), 336–341. doi: 10.1111/nph.13598
Hepworth, C., Caine, R. S., Harrison, E. L., Sloan, J., Gray, J. E. (2018). Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 41, 1–7. doi: 10.1016/J.PBI.2017.07.009
Hoekstra, A. Y., Chapagain, A. K. (2006). “Water footprints of nations: Water use by people as a function of their consumption pattern,” in Integrated Assessment of Water Resources and Global Change (Dordrecht: Springer Netherlands), 35–48.
Huang, X. Y., Chao, D. Y., Gao, J. P., Zhu, M. Z., Shi, M., Lin, H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23 (15), 1805–1817. doi: 10.1101/gad.1812409
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174 (2), 776–787. doi: 10.1104/pp.16.01844
Hunt, L., Gray, J. E. (2009). The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr. Biol. 19 (10), 864–869. doi: 10.1016/j.cub.2009.03.069
Hunt, L., Bailey, K. J., Gray, J. E. (2010). The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 186 (3), 609–614. doi: 10.1111/j.1469-8137.2010.03200.x
Kamiya, N., Itoh, J.-I., Morikami, A., Nagato, Y., Matsuoka, M. (2003). The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36 (1), 45–54. doi: 10.1046/j.1365-313X.2003.01856.x
Kanaoka, M. M., Pillitteri, L. J., Fujii, H., Yoshida, Y., Bogenschutz, N. L., Takabayashi, J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20 (7), 1775–1785. doi: 10.1105/tpc.108.060848
Kang, Y., Khan, S., Ma, X. (2009). Climate change impacts on crop yield, crop water productivity and food security – a review. Prog. Nat. Sci. 19 (12), 1665–1674. doi: 10.1016/J.PNSC.2009.08.001
Krishnan, P., Ramakrishnan, B., Reddy, K. R., Reddy, V. R. (2011). High-temperature effects on rice growth, yield, and grain quality. Adv. Agron. 111, 87–206. doi: 10.1016/B978-0-12-387689-8.00004-7
La, H., Li, J., Ji, Z., Cheng, Y., Li, X., Jiang, S., et al. (2006). Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol. Genet. Genomics 275 (4), 374–386. doi: 10.1007/s00438-005-0093-5
Lai, L. B., Nadeau, J. A., Lucas, J., Lee, E.-K., Nakagawa, T., Zhao, L., et al. (2005). The Arabidopsis R2R3 MYB Proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17 (10), 2754–2767. doi: 10.1105/tpc.105.034116
Lampard, G. R., MacAlister, C. A., Bergmann, D. C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322 (5904), 1113–1116. doi: 10.1126/science.1162263
Lau, O. S., Davies, K. A., Chang, J., Adrian, J., Rowe, M. H., Ballenger, C. E., et al. (2014). Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells. Science 345 (6204), 1605–1609. doi: 10.1126/science.1256888
Lawson, T., Blatt, M. R. (2014). Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 164 (4), 1556–1570. doi: 10.1104/PP.114.237107
Lawson, T., Vialet-Chabrand, S. (2019). Speedy stomata, photosynthesis and plant water use efficiency. New Phytol. 221 (1), 93–98. doi: 10.1111/nph.15330
Lee, E., Lucas, J. R., Sack, F. D. (2014). Deep functional redundancy between FAMA and FOUR LIPS in stomatal development. Plant J. 78 (4), 555–565. doi: 10.1111/tpj.12489
Lee, J. S., Hnilova, M., Maes, M., Lin, Y.-C. L., Putarjunan, A., Han, S.-K., et al. (2015). Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522 (7557), 439–443. doi: 10.1038/nature14561
Li, J., Li, Y., Yin, Z., Jiang, J., Zhang, M., Guo, X., et al. (2017). OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 15 (2), 183–196. doi: 10.1111/pbi.12601
Lin, C.-J., Li, C.-Y., Lin, S.-K., Yang, F.-H., Huang, J.-J., Liu, Y.-H., et al. (2010). Influence of high temperature during grain filling on the accumulation of storage proteins and grain quality in rice (Oryza sativa L.). J. Agric. Food Chem. 58 (19), 10545–10552. doi: 10.1021/jf101575j
Lin, G., Zhang, L., Han, Z., Yang, X., Liu, W., Li, E., et al. (2017). A receptor-like protein acts as a specificity switch for the regulation of stomatal development. Genes Dev. 31 (9), 927–938. doi: 10.1101/gad.297580.117
Liu, T., Ohashi-Ito, K., Bergmann, D. C. (2009). Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136 (13), 2265–2276. doi: 10.1242/dev.032938
Liu, Y., Qin, L., Han, L., Xiang, Y., Zhao, D. (2015). Overexpression of maize SDD1 (ZmSDD1) improves drought resistance in Zea mays L. by reducing stomatal density. Plant Cell Tissue Organ Cult. (PCTOC) 122 (1), 147–159. doi: 10.1007/s11240-015-0757-8
Long, S. P., Ort, D. R. (2010). More than taking the heat: crops and global change. Curr. Opin. Plant Biol. 13 (3), 240–247. doi: 10.1016/J.PBI.2010.04.008
Lu, J., He, J., Zhou, X., Zhong, J., Li, J., Liang, Y.-K. (2019). Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 234–235, 18–27. doi: 10.1016/J.JPLPH.2019.01.010
Luo, L., Zhou, W.-Q., Liu, P., Li, C.-X., Hou, S.-W. (2012). The development of stomata and other epidermal cells on the rice leaves. Biol. Plant. 56 (3), 521–527. doi: 10.1007/s10535-012-0045-y
MacAlister, C. A., Bergmann, D. C. (2011). Sequence and function of basic helix-loop-helix proteins required for stomatal development in Arabidopsis are deeply conserved in land plants. Evol. Dev. 13 (2), 182–192. doi: 10.1111/j.1525-142X.2011.00468.x
MacAlister, C. A., Ohashi-Ito, K., Bergmann, D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445 (7127), 537–540. doi: 10.1038/nature05491
Maclean, J., Hardy, B., Hettel, G. (2013). Rice almanac: Source book for one of the most important economic activities on Earth (Los Baños: International Rice Research Institiute).
Matsui, T., Omasa, K., Horie, T. (2000). High temperature at flowering inhibits swelling of pollen grains, a driving force for thecae dehiscence in rice (Oryza sativa L.). Plant Prod. Sci. 3 (4), 430–434. doi: 10.1626/pps.3.430
McAusland, L., Vialet-Chabrand, S., Davey, P., Baker, N. R., Brendel, O., Lawson, T. (2016). Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol. 211 (4), 1209–1220. doi: 10.1111/nph.14000
McGuire, S. (2015). FAO, IFAD, and WFP. The State of Food Insecurity in the World 2015: Meeting the 2015 International Hunger Targets: Taking Stock of Uneven Progress. Adv. Nutrition 6 (5), 623–624. doi: 10.3945/an.115.009936
Meng, X., Chen, X., Mang, H., Liu, C., Yu, X., Gao, X., et al. (2015). Differential function of Arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25 (18), 2361–2372. doi: 10.1016/j.cub.2015.07.068
Mohammed, U., Caine, R. S., Atkinson, J. A., Harrison, E. L., Wells, D., Chater, C. C., et al. (2019). Rice plants overexpressing OsEPF1 show reduced stomatal density and increased root cortical aerenchyma formation. Sci. Rep. 9 (1), 5584. doi: 10.1038/s41598-019-41922-7
Morita, S., Wada, H., Matsue, Y. (2016). Countermeasures for heat damage in rice grain quality under climate change. Plant Prod. Sci. 19 (1), 1–11. doi: 10.1080/1343943X.2015.1128114
Nadeau, J. A., Sack, F. D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 (5573), 1697–1700. doi: 10.1126/science.1069596
Nunes, T. D. G., Zhang, D., Raissig, M. T. (2019). Form, development and function of grass stomata. Plant J. tpj.14552. doi: 10.1111/tpj.14552
Ohashi-Ito, K., Bergmann, D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18 (10), 2493–2505. doi: 10.1105/tpc.106.046136
Ohsumi, A., Kanemura, T., Homma, K., Horie, T., Shiraiwa, T. (2007). Genotypic variation of stomatal conductance in relation to stomatal density and length in rice (Oryza sativa L.). Plant Prod. Sci. 10 (3), 322–328. doi: 10.1626/pps.10.322
Papanatsiou, M., Petersen, J., Henderson, L., Wang, Y., Christie, J. M., Blatt, M. R. (2019). Optogenetic manipulation of stomatal kinetics improves carbon assimilation, water use, and growth. Science 363 (6434), 1456–1459. doi: 10.1126/science.aaw0046
Pillitteri, L. J., Dong, J. (2013). Stomatal development in Arabidopsis. Arabidopsis Book 11, e0162. doi: 10.1199/tab.0162
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., Torii, K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445 (7127), 501–505. doi: 10.1038/nature05467
Qi, X., Torii, K. U. (2018). Hormonal and environmental signals guiding stomatal development. BMC Biol. 16 (1), 21. doi: 10.1186/s12915-018-0488-5
Qi, X., Han, S.-K., Dang, J. H., Garrick, J. M., Ito, M., Hofstetter, A. K., et al. (2017). Autocrine regulation of stomatal differentiation potential by EPF1 and ERECTA-LIKE1 ligand-receptor signaling. ELife 6, e24102. doi: 10.7554/eLife.24102
Qu, X., Peterson, K. M., Torii, K. U. (2017). Stomatal development in time: the past and the future. Curr. Opin. Genet. Dev. 45, 1–9. doi: 10.1016/J.GDE.2017.02.001
Qu, X., Yan, M., Zou, J., Jiang, M., Yang, K., Le, J. (2018). A2-type cyclin is required for the asymmetric entry division in rice stomatal development. J. Exp. Bot. 69 (15), 3587–3599. doi: 10.1093/jxb/ery158
Quentin Grafton, R. (2017). Responding to the ‘Wicked Problem’ of water insecurity. Water Resour. Manage. 31 (10), 3023–3041. doi: 10.1007/s11269-017-1606-9
Raissig, M. T., Abrash, E., Bettadapur, A., Vogel, J. P., Bergmann, D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U.S.A. 113 (29), 8326–8331. doi: 10.1073/pnas.1606728113
Raissig, M. T., Matos, J. L., Anleu Gil, M. X., Kornfeld, A., Bettadapur, A., Abrash, E., et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355 (6330), 1215–1218. doi: 10.1126/science.aal3254
Ran, J.-H., Shen, T.-T., Liu, W.-J., Wang, X.-Q. (2013). Evolution of the bHLH genes involved in stomatal development: implications for the expansion of developmental complexity of stomata in land plants. PloS One 8 (11), e78997. doi: 10.1371/journal.pone.0078997
Sánchez, B., Rasmussen, A., Porter, J. R. (2014). Temperatures and the growth and development of maize and rice: a review. Global Change Biol. 20 (2), 408–417. doi: 10.1111/gcb.12389
Saito, K., Asai, H., Zhao, D., Laborte, A. G., Grenier, C. (2018). Progress in varietal improvement for increasing upland rice productivity in the tropics. Plant Prod. Sci. 21 (3), 145–158. doi: 10.1080/1343943X.2018.1459751
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 (4), 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schuler, M. L., Sedelnikova, O. V., Walker, B. J., Westhoff, P., Langdale, J. A. (2018). SHORTROOT-mediated increase in stomatal density has no impact on photosynthetic efficiency. Plant Physiol. 176 (1), 757–772. doi: 10.1104/PP.17.01005
Seneviratne, S. I., Nicholls, N., Easterling, D., Goodess, C. M., Kanae, S., Kossin, J., et al. (2012). “Changes in Climate Extremes and their Impacts on the Natural Physical Environment,” in Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation. A Special Report of Working Groups I and II of the Intergovernmental Panel on Climate Change (IPCC) (Cambridge, UK: Cambridge University Press), 109–230.
Shen, H., Zhong, X., Zhao, F., Wang, Y., Yan, B., Li, Q., et al. (2015). Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33 (9), 996–1003. doi: 10.1038/nbt.3321
Shpak, E. D., McAbee, J. M., Pillitteri, L. J., Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 (5732), 290–293. doi: 10.1126/science.1109710
Simmons, A. R., Bergmann, D. C. (2016). Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant Biol. 29, 1–8. doi: 10.1016/j.pbi.2015.09.008
Stebbins, G. L., Shah, S. S. (1960). Developmental studies of cell differentiation in the epidermis of monocotyledons: II. Cytological features of stomatal development in the Gramineae. Dev. Biol. 2 (6), 477–500. doi: 10.1016/0012-1606(60)90050-6
Stott, P. (2016). How climate change affects extreme weather events. Science 352 (6293), 1517–1518. doi: 10.1126/science.aaf7271
Sugano, S. S., Shimada, T., Imai, Y., Okawa, K., Tamai, A., Mori, M., et al. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463 (7278), 241–244. doi: 10.1038/nature08682
Swain, P., Raman, A., Singh, S. P., Kumar, A. (2017). Breeding drought tolerant rice for shallow rainfed ecosystem of eastern India. Field Crops Res. 209, 168–178. doi: 10.1016/J.FCR.2017.05.007
Tanaka, Y., Sugano, S. S., Shimada, T., Hara-Nishimura, I. (2013). Enhancement of leaf photosynthetic capacity through increased stomatal density in Arabidopsis. New Phytol. 198 (3), 757–764. doi: 10.1111/nph.12186
Tsunoda, S., Fukoshima, M. T. (1986). Leaf properties related to the photosynthetic response to drought in upland and lowland rice varieties. Annals Bot. 58 (4), 531–539. doi: 10.1093/annbot/58.4.531
Urban, J., Ingwers, M. W., McGuire, M. A., Teskey, R. O. (2017). Increase in leaf temperature opens stomata and decouples net photosynthesis from stomatal conductance in Pinus taeda and Populus deltoides x nigra. J. Exp. Bot. 68 (7), 1757–1767. doi: 10.1093/jxb/erx052
Vanneste, S., Coppens, F., Lee, E., Donner, T. J., Xie, Z., Van Isterdael, G., et al. (2011). Developmental regulation of CYCA2s contributes to tissue-specific proliferation in Arabidopsis. EMBO J. 30 (16), 3430–3441. doi: 10.1038/emboj.2011.240
Wang, H., Guo, S., Qiao, X., Guo, J., Li, Z., Zhou, Y., et al. (2019). BZU2/ZmMUTE controls symmetrical division of guard mother cell and specifies neighbor cell fate in maize. PLOS Genet. 15 (8), e1008377. doi: 10.1371/journal.pgen.1008377
Wassmann, R., Jagadish, S. V. K., Sumfleth, K., Pathak, H., Howell, G., Ismail, A., et al. (2009). Regional vulnerability of climate change impacts on Asian rice production and scope for adaptation. Adv. Agron. 102, 91–133. doi: 10.1016/S0065-2113(09)01003-7
Wu, Z., Chen, L., Yu, Q., Zhou, W., Gou, X., Li, J., et al. (2019). Multiple transcriptional factors control stomata development in rice. New Phytol. 223 (1), 220–232. doi: 10.1111/nph.15766
Xie, Z., Lee, E., Lucas, J. R., Morohashi, K., Li, D., Murray, J. A. H., et al. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22 (7), 2306–2321. doi: 10.1105/tpc.110.074609
Yang, M., Sack, F. D., Lucas, J., Lee, E.-K., Nakagawa, T., Zhao, L., et al. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 (12), 2227–2239. doi: 10.1105/tpc.7.12.2227
Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36 (5), 745–757. doi: 10.1007/s00299-017-2118-z
Zhao, L., Wu, L., Wu, M., Li, Y. (2011). Nutrient uptake and water use efficiency as affected by modified rice cultivation methods with reduced irrigation. Paddy Water Environ. 9 (1), 25–32. doi: 10.1007/s10333-011-0257-3
Zoulias, N., Harrison, E. L., Casson, S. A., Gray, J. E. (2018). Molecular control of stomatal development. Biochem. J. 475 (2), 441–454. doi: 10.1042/BCJ20170413
Keywords: stomatal development, rice, crop yields, stomatal density, grasses, crop physiology
Citation: Buckley CR, Caine RS and Gray JE (2020) Pores for Thought: Can Genetic Manipulation of Stomatal Density Protect Future Rice Yields? Front. Plant Sci. 10:1783. doi: 10.3389/fpls.2019.01783
Received: 23 September 2019; Accepted: 20 December 2019;
Published: 11 February 2020.
Edited by:
Graham Dow, ETH Zürich, SwitzerlandReviewed by:
Yun-Kuan Liang, Wuhan University, ChinaCopyright © 2020 Buckley, Caine and Gray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie E. Gray, ai5lLmdyYXlAc2hlZmZpZWxkLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.