- Department of Microbe-Plant Interactions, Faculty of Biology and Chemistry, CBIB (Center for Biomolecular Interactions Bremen), University of Bremen, Bremen, Germany
Research on the interaction between the non-nodule-forming bacterial endophytes and their host plants is still in its infancy. Especially the understanding of plant control mechanisms which govern endophytic colonization is very limited. The current study sets out to determine which hormonal signaling pathway controls endophytic colonization in rice, and whether the mechanisms deviate for a pathogen. The endophyte Azoarcus olearius BH72—rice model was used to investigate root responses to endophytes in comparison to the recently established pathosystem of rice blight Xanthomonas oryzae pv. oryzae PXO99 (Xoo) in flooded roots. In the rice root transcriptome, 523 or 664 genes were found to be differentially expressed in response to Azoarcus or Xoo colonization, respectively; however, the response was drastically different, with only 6% of the differentially expressed genes (DEGs) overlapping. Overall, Xoo infection induced a much stronger defense reaction than Azoarcus colonization, with the latter leading to down-regulation of many defense related DEGs. Endophyte-induced DEGs encoded several enzymes involved in phytoalexin biosynthesis, ROS (reactive oxygen species) production, or pathogenesis-related (PR) proteins. Among putative plant markers related to signal transduction pathways modulated exclusively during Azoarcus colonization, none overlapped with previously published DEGs identified for another rice endophyte, Azospirillum sp. B510. This suggests a large variation in responses of individual genotypic combinations. Interestingly, the DEGs related to jasmonate (JA) signaling pathway were found to be consistently activated by both beneficial endophytes. In contrast, the salicylate (SA) pathway was activated only in roots infected by the pathogen. To determine the impact of SA and JA production on root colonization by the endophyte and the pathogen, rice mutants with altered hormonal responses were employed: mutant cpm2 deficient in jasmonate synthesis, and RNA interference (RNAi) knockdown lines of NPR1 decreased in salicylic acid-mediated defense responses (NPR1-kd). Only in cpm2, endophytic colonization of Azoarcus was significantly increased, while Xoo colonization was not affected. Surprisingly, NPR1-kd lines showed slightly decreased colonization by Xoo, contrary to published results for leaves. These outcomes suggest that JA but not SA signaling is involved in controlling the Azoarcus endophyte density in roots and can restrict internal root colonization, thereby shaping the beneficial root microbiome.
Introduction
For land plants the primary site of interactions with microbes are roots; here the tissues commonly harbor the largest numbers of microbes (Reinhold-Hurek et al., 2015). The tight association with microbes often improves plants’ nutrient uptake, protects them against pathogens or even promotes their growth by the release of phytohormone-like substances (Berendsen et al., 2012). In order to profit from distinct microbial functions, plants actively establish a beneficial microbial community inside and, on the root, as well as in the rhizosphere soil e.g., by releasing metabolites and energy sources (Peiffer et al., 2013). However, the current understanding of the complex plant-microbe interactions in the rhizosphere is still in its infancy.
Among the root and rhizosphere microbes, endophytic bacteria are expected to have a particularly tight interaction with their host plant. They reside within the living tissue of a plant without substantively harming it, in a symptomless association which remained for a long time undetected. Their endophytic lifestyle is remarkable. High numbers of culturable bacterial cells in roots have been reported (up to 108/g root dry weight), particularly in rice and flooded plants (Reinhold et al., 1986; Barraquio et al., 1997). In the gradient from bulk soil to the rhizosphere and endorhizosphere, the microbial community tends to have lower diversity and a higher degree of specialization toward the root interior (Reinhold-Hurek et al., 2015). Thus, endophytic bacteria are of high interest to study fundamental questions of molecular interactions but are also a mostly untapped reservoir for agro-biotechnological applications, e.g., for improvement of plant growth and health (Berendsen et al., 2012; Khare et al., 2018), phytoremediation (Barac et al., 2004), or as biofertilizer.
As plant cells can commonly detect and react to bacterial molecular components (MAMPs) through plant’s innate immunity-regulated defense responses (Macho and Zipfel, 2014), it is puzzling how endophytes can overcome these responses and colonize the root interior (Reinhold-Hurek et al., 2015). Mechanisms which enable plants to select endophytic cooperative partners over pathogens are still enigmatic. Phytohormones are highly relevant for the control of plant defense responses. It has been suggested that there are three key defense-related hormones: salicylic acid (SA), jasmonate (JA), and ethylene (ET) (Grosskinsky et al., 2012). SA mainly triggers plant defense against biotrophic or hemibiotrophic, JA against necrotrophic pathogens, though there are a few exceptions (Pieterse et al., 2012; Yang et al., 2015). SA acts as one of the systemic acquired resistance inducers in leaf (Gao et al., 2015), while mostly JA and ET regulate induced systemic resistance triggered by beneficial PRGR (plant growth-promoting rhizobacteria) (Pieterse et al., 2014). Some progress has been made in elucidating how plants shape their microbiome in the model plant Arabidopsis thaliana, where plant factors such as salicylic acid (Lebeis et al., 2015), coumarins (Voges et al., 2019), or the plant’s phosphate status (Hiruma et al., 2016) were shown to impact the microbial communities. However, not much is known about plant factors that shape the microbiome of important crop plants, including cereals such as rice. Moreover, gene functions related to plant immune response and secondary metabolism partly differ between rice and Arabidopsis (De Vleesschauwer et al., 2013; Tamaoki et al., 2013; Miyamoto et al., 2016), making studies on this cereal worthwhile. Bacterial endophytes can be assessed in this model system very well, as endophytic colonization and activity are documented for rice roots beyond doubt (Hurek et al., 1994; Reinhold-Hurek and Hurek, 1998; Egener et al., 1999).
Therefore, as a model for endophyte-rice interactions, Azoarcus olearius BH72 was chosen, an abundant nitrogen-fixing endophyte of Kallar grass roots (Reinhold et al., 1986; Reinhold-Hurek et al., 1993b; Hurek et al., 2002), which also colonizes rice densely and fixes nitrogen in the root cortex (Hurek et al., 1994; Hurek et al., 1997; Egener et al., 1999). Azoarcus’ root ingress is an active process to which many bacterial factors contribute, including cellulases (Reinhold-Hurek et al., 2006), type IV pili (Dörr et al., 1998) and their twitching motility (Böhm et al., 2007), type VI protein secretion (Sarkar et al., 2017), and cyclic-di-GMP-synthesizing proteins (Shidore et al., 2012). Interestingly, flagella of A. olearius BH72 are promoting endophytic rice root colonization, rather than acting as plant defense-inducing MAMPs (Shidore et al., 2012). With respect to the plant side, the common signaling pathway shared by nitrogen-fixing root nodule symbioses and arbuscular mycorrhizal symbioses is apparently not recruited for the establishment of the Azoarcus in rice (Chen et al., 2015). How rice is governing endophytic interactions, and which rice signaling cascades may facilitate or restrict endophytic colonization is still unclear. As plants can induce different panels of gene transcription during colonization by beneficial or detrimental microbes (Plett and Martin, 2018), it was hypothesized that rice root reactions to endophytic and pathogenic bacteria deviate and thus allow to filter out endophyte-specific plant responses.
Thus, the main objective of this study was to disclose differences in rice root responses to beneficial endophytic and pathogenic bacteria, in order to reveal putative endophyte-specific pathways which control colonization. For this, root transcriptomic responses of rice to A. olearius colonization were analyzed. This required an experimental strategy to allow a direct comparison under identical experimental settings under flooded conditions typical for paddy rice. Therefore, a reference model for pathogenic plant-bacterial interactions using the highly virulent leaf blight pathogen Xanthomonas oryzae pv. oryzae PXO99 (Xoo) was developed in a previous study (Chen et al., 2015). Although for X. oryzae mostly leaf responses were studied up to now, some cell death occurs also in roots upon incubation with the pathogen (Jalmi and Sinha, 2016). It was established by us that without external artificial wounding Xoo is able to infect rice roots, forms colonies inside root tissues, and causes no visible damage within 14 days of infection (Chen et al., 2015). Based on the transcriptome results, two rice mutants deficient in jasmonate synthesis (cpm2) or exhibiting reduced salicylic acid-mediated defense responses (NPR1-kd) were used to study if these hormonal pathways govern colonization levels of the endophyte or the pathogen.
Materials and Methods
Plant and Bacterial Material
For global transcriptome experiments, rice cultivar Oryza sativa cv. Nipponbare (japonica type) was used (accession IRGC 136196, IRRI International Rice Research Institute, Philippines). The cpm2 mutant was isolated from γ-ray-mutagenized M2 line of japonica type rice O. sativa cv. Nihonmasari, and the cpm2 homozygotes with longer coleoptile under continuous light (Riemann et al., 2013) were applied for bacterial colonization experiments. NPR1-knockdown (NPR1-kd) lines #1 and #7 are RNA interference mutation lines (Sugano et al., 2010).
For microarray analysis and root colonization tests, A. olearius BH72 (Reinhold et al., 1986) and X. oryzae pv. oryzae PXO99 originating from Philippines (Adhikari et al., 1995) were applied. Reporter strain A. olearius BHGN3.1 carried a transcriptional nifH::gusA fusion in the chromosome (Egener et al., 1999) and was used for visualizing physiologically successful rice colonization, under which the cells can derepress nitrogenase genes and actively fix nitrogen.
Plant Cultivation and Inoculation
Dehusked rice grains were surface sterilized, washed, and germinated on agar plates as described previously (Hurek et al., 1994) with the following modifications. Washing steps were extended to three times 1 h each. For germinating O. sativa japonica, rice grains were incubated in germination agar in Magenta boxes GA7 (Sigma-Aldrich, USA; 1% agar, Difco, Becton and Dickinson Company, USA) for 3 days at 30°C in the dark at ambient humidity without humidity control, followed by 2 days in light in the phytotron (see conditions below, end of paragraph). Azoarcus inoculation for transcriptome analysis and visualization was done as previously described in plant medium-flooded quartz sand (Egener et al., 1999), with bacterial inoculum of 2 x 108 cells per plant; medium was supplemented (per liter) with 20 mg of neutralized DL-malic acid as starter carbon source, as well as potassium phosphate buffer adjusted to pH 6.8 (0.88 g KH2PO4/1.12 g K2HPO4 at pH 6.8). For timeline experiments, seedlings were instead placed on top of plastic adaptors in hydroponic jars containing 300 ml (1 h; 4 h incubation) or 450 ml (24 h, 72 h incubation) of plant medium described above. X. oryzae pv. oryzae PXO99 was grown at 28°C on agar plates containing modified Wakimoto’s medium (Karnagilla and Natural, 1973). For infection of O. sativa with the pathogen Xoo PXO99, roots of seedlings were dipped for 5 min into a bacterial suspension of 5 x 109 cells/ml. Afterwards infected seedlings as well as non-infected seedlings (control) were grown gnotobiotically as described above. Plants were incubated in the phytotron at 30°C, 60% humidity, and 14/10-h light-dark cycle (approximately 170 µmol photons/m−2 x s−1).
Assessment of Colonization
Endophytic bacterial colonization (inside the roots) was quantified as described previously (Böhm et al., 2007); briefly, 14 days after inoculation, roots were treated by ultrasonication to remove surface bacteria, homogenized, and the number of colony forming units (cfu) per milligram of root fresh weight was estimated for both bacteria, Azoarcus (according to Böhm et al., 2007) and PXO99. Pathogen PXO99 was counted on Wakimoto’s medium agar plates (Karnagilla and Natural, 1973; Chen et al., 2015).
For histochemical detection of ß-glucuronidase (GUS) activity, roots were harvested 13 days post-inoculation and stained for up to 6 h as previously described (Egener et al., 1999). Roots from three independent experiments were inspected.
Ribonucleic Acid Extraction and Transcript Analysis by Real-Time Polymerase Chain Reaction
Plants were harvested 14 days post-inoculation, and roots were frozen in liquid nitrogen prior to RNA extraction from pools of plants. RNA applied for microarray experiments was extracted by using a hexadecyl trimethyl-ammonium bromide (CTAB)-based method: 0.5 g rice roots homogenized in liquid nitrogen were suspended in 18 ml of extraction buffer (2% hexadecyl trimethyl-ammonium bromide (CTAB), 2% polyvinylpyrrolidone, 100 mM Tris-HCl pH 8.0, 25 mM EDTA, 2 M NaCl, 0.5 g/L spermidine, and 2% β-mercaptoethanol, incubated at 65°C for 5 minutes; 18 ml of chloroform was added and mixed with the suspension; after centrifugation at 10,000 x g for 5 min, the supernatant was treated with chloroform again; lithium chloride (LiCl) was added to the final supernatant to a final concentration of 2 M and kept overnight at 4°C for RNA precipitation; RNA was pelleted at 10,000 x g for 30 min at 4°C and dissolved in RNase-free water.
RNA applied for timeline experiments was extracted with the RNAeasy Plus Mini Kit after homogenizing the root samples in liquid nitrogen. Samples contaminated with genomic DNA were subjected to DNase I treatment (Sigma-Aldrich, St. Louis, Missouri, USA).
For quantitative real time (RT)-PCR analysis, accession numbers of the respective rice genes and primer sequences are given in Table S1. The reverse transcription step was performed using Thermo Scientific RevertAid Premium Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions with and 1 μg RNA applied for 20 μl reaction volume. Real-time PCR was carried out either with Bio-Rad SsoAdvanced SYBR Green Supermix, with 300 nM of each primer and 2 μl of complementary DNA (cDNA) added. Real-time PCR reactions were performed either using a CFX96 Touch Real-Time Detection System (Bio-Rad, Munich, Germany) at 30 s of initial denaturation at 95°C, followed by 40 cycles of denaturation for 10 s at 95°C, annealing for 30 s, and extension for 30 s at 72°C. At the end of the amplification, a melting curve was recorded between 55 to 95°C in steps of 1°C, to ensure that the signal corresponded to a single PCR product. As the efficiency of each real-time PCR amplification was close to 100%, relative gene expression was calculated with 2-ΔΔCT method (Livak and Schmittgen, 2001).
Microarray Hybridization and Data Collection
For microarray hybridization, rice RNA samples were subjected to quality control using an Agilent Bioanalyzer 2100. Only those showing no degradation and clear 28S ribosomal RNA (rRNA) and 18S rRNA peaks were used. A two-color microarray-based analysis with Low Input Quick Amp Labeling kit and 4×44 k 60-mer microarrays (Agilent; Böblingen, Germany) was carried out according to the company’s instructions. The data extractions were performed with Feature Extraction Software version 9.5 (Agilent; Böblingen, Germany) and GeneSpring software (Agilent; Böblingen, Germany). For each experiment three biological replicates were performed, and for hybridization one dye-swap and a technical replicate were included. Genes showing equal or larger than 1.5-fold up- or down-regulation in all three experiments were regarded as differently regulated. Data were deposited at GEO (Gene Expression Omnibus) (GSE136706 and GSE136707).
Results and Discussion
Highly Divergent Global Transcriptomic Response of Rice Roots Toward Bacterial Endophyte or Pathogen
The current study aimed to compare plant responses to beneficial and pathogenic bacteria with similar surface characteristics or microbe-associated molecular patterns, such as an outer membrane with lipopolysaccharides which is typical for Gram-negative bacteria. As counterpart for A. olearius, a Gram-negative endophyte of rice roots, the Gram-negative leaf pathogen X. oryzae pv. oryzae strain PXO99 (Xoo) was chosen. It was possible to utilize this strain for root responses, because it was previously demonstrated (Chen et al., 2015) that it can also colonize rice roots in high numbers. Root transcriptomic responses were analyzed by two-color 4×44 k rice microarrays (Agilent; Böblingen, Germany). They were examined 2 weeks post-inoculation with bacteria under gnotobiotic conditions when nitrogen-fixing endophytic colonization is well detectable (Egener et al., 1999; Chen et al., 2015). This allowed simultaneous analysis of both, local early and late responses of roots to new local infections and fully established endophytes. As differentially expressed genes (DEGs), genes which showed at least 1.5 fold difference in all three replicates were considered.
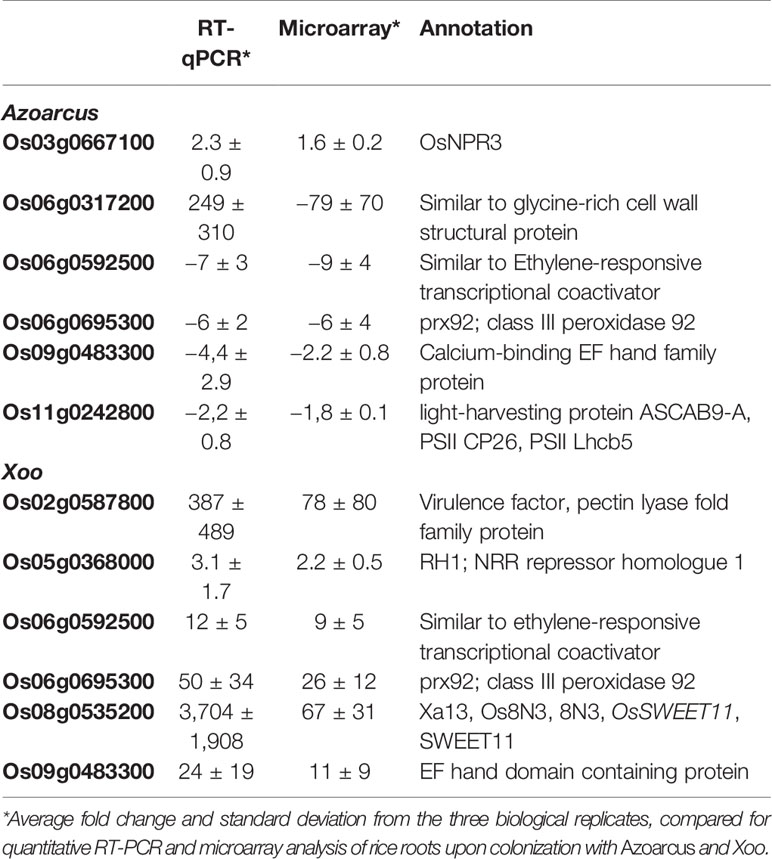
In total 523 genes were found to be differently regulated in response to the endophyte A. olearius BH72 compared to non-infected, sterile seedlings, with 260 up-regulated and 263 down-regulated genes. For the pathogen X. oryzae strain PXO99 (Xoo), the number of modulated genes (664) was almost equal to Azoarcus, albeit five times more genes were up- than down-regulated (549 versus 116) (Figure 1A). Six DEGs each, for pathogen and endophyte, were chosen for real-time polymerase chain reaction (PCR) to validate transcriptional changes (Table 1).
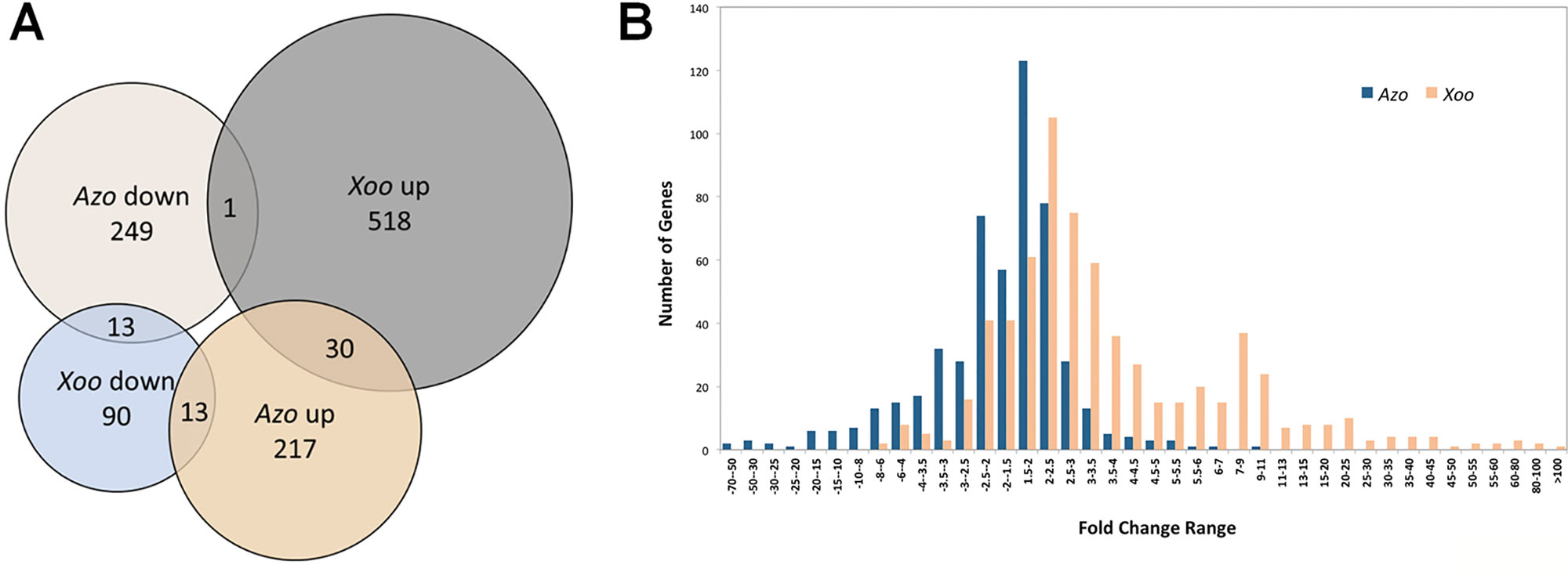
Figure 1 Overview of rice genes differentially regulated in roots in response to colonization by Azoarcus olearius BH72 (Azo) and Xanthomonas oryza pv. oryzae PXO99 (Xoo). (A) Venn gram of genes differently regulated in response to Azoarcus and Xoo. In total 1,227 genes were differentially regulated. Differentially expressed genes (DEGs) that were up-regulated are referred to as “up,” down-regulated as “down.” (B) Gene number distribution at different fold change ranges. Orange color represents number of differently regulated genes in response to Azoarcus, blue color in response to Xoo.
Differentially regulated genes were highly divergent between endophyte and pathogen. With 6.3% of the pathogen-modulated genes, only very few DEGs overlapped in both interactions: 30 genes (2.3 %) were up-regulated and 13 genes (1%) were down-regulated. Several genes (14) were affected in the opposite way (Figure 1A, Table S2). Generally, the up-regulated genes in response to Xoo infection showed a much higher induction ratio than in response to Azoarcus. In contrast, down-regulated genes were generally more strongly repressed by the endophyte (Figure 1B). The root responses were also distinct with respect to functional categories of differentially regulated genes deduced by Kyoto Encyclopedia of Genes and Genomes (KEGG) (Figure S1). In the case of Xoo infection, a high cumulative fold change was found for gene induction in almost all functional categories. Contrastingly, the majority of categories showed a high cumulative fold change for genes repressed by Azoarcus. This included genes in functional categories related to the cell wall, stress, or major CHO metabolism, which were largely down-regulated in the presence of endophytic bacteria. This demonstrates that rice responds with different patterns of gene regulation to colonization by beneficial or detrimental bacteria.
Moderate Plant Defense Signaling Toward the Endophyte in Comparison to the Pathogen
Plant defense reactions were induced both by the endophyte and the pathogen colonization. However, the defense responses were weaker toward the endophyte, with respect to the number and type of DEGs as well as in the degree or trend of modulation (Table S3, Figure 2).
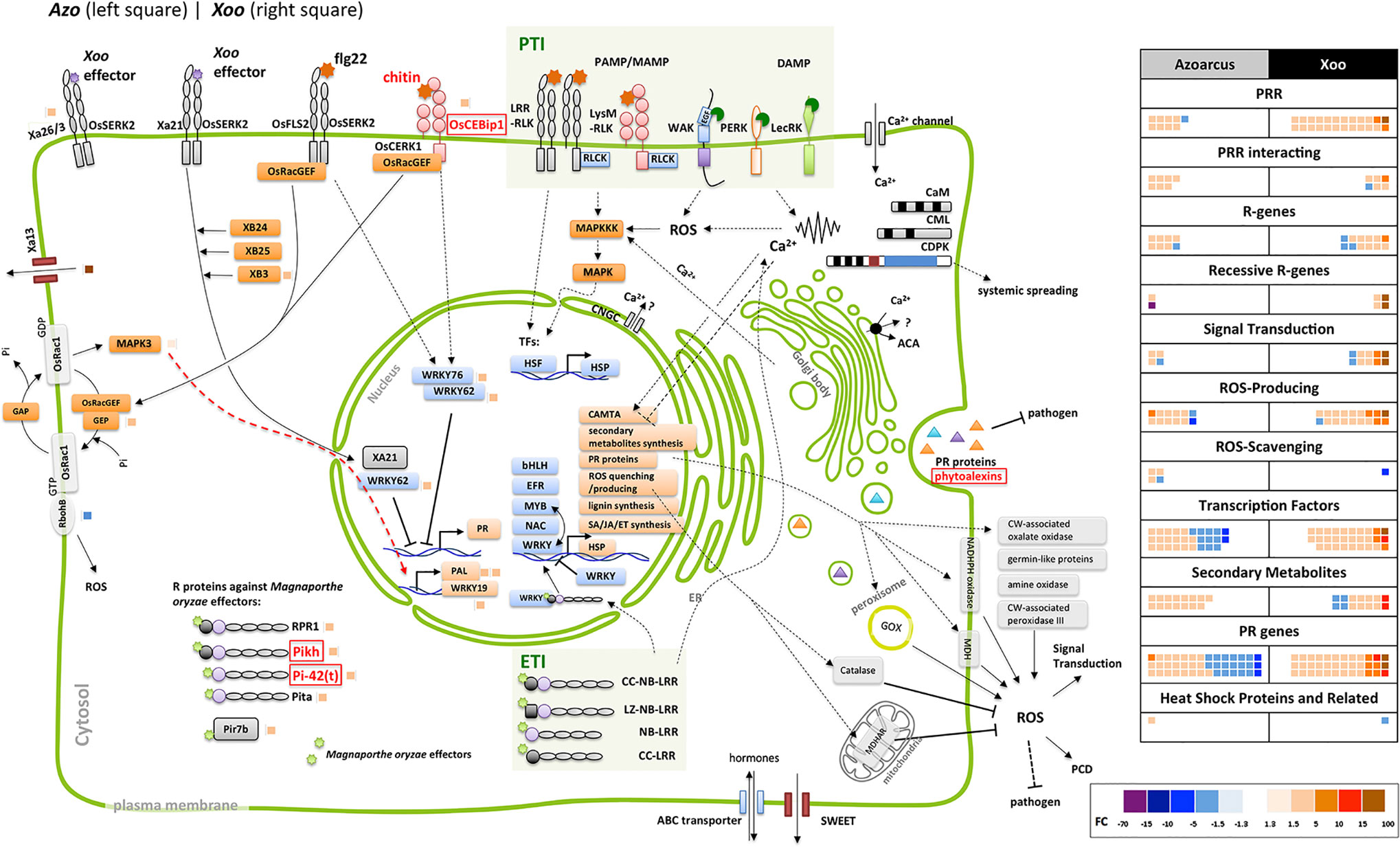
Figure 2 Elements of plant defense reactions modulated in Azoarcus- and Xoo-rice interactions in roots. Sketch of putative rice cellular defense network (left side). Dashed line: direct interaction not verified or indirect interaction; continuous line: direct interaction; arrow: induction, blunt end: inhibition. Small blocks beside rice protein names: Fold change of the differentially expressed gene (DEG) (right side of the vertical line: modulated by Xoo, left side: by Azoarcus) indicated by its color according to the color scale below. Genes or pathways up-regulated by Azoarcus olearius labeled in red. Right side, summary of defense-related DEGs modulated by Azo or Xoo, respectively; each colored block represents a modulated gene, its color indicating the fold change to the color scale below. ABA, abscisic acid; ABC, ATP-binding cassette; ACA, auto-inhibited calcium ATPase; bHLH, basic helix-loop-helix protein; BR, brassinosteroid; CaM, calmodulin; CAMTA, Ca2+/CaM-binding transcription factors; CC, coiled coil; CDPK, Ca2+-dependent protein kinases; CEBip, chitin elicitor-binding protein; CERK, chitin elicitor receptor kinase; CK, cytokinin; CML, calmodulin-like; CNGC, cyclic nucleotide-gated channels; CW, cell wall; DAMP, damage-associated molecular pattern; ER, endoplasmic reticulum; ERF, ethylene response transcription factor; ET, ethylene; flg, flagellin; FLS, flagellin-sensing; GA, gibberellic acid; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GEP, GDP/GTP exchange protein; GOX, glycolate oxidases; HSF, heat stress transcription factor; HSP, heat shock protein; IAA, indole-3-acetic acid; JA, jasmonate; LecRK, lectin receptor kinase; LRR, leucine rich repeat; LysM, lysine motif domain; LZ, leucine zipper nucleotide-binding site; MAPK, mitogen-activated protein kinase; MDHAR, monodehydroascorbate reductase; M.o., Magnaporthe oryzae MYB, myeloblastosis transcription factor family; NAC, no apical meristem (NAM); ATAF, Arabidopsis transcription activation factor; CUC and cup-shaped cotyledon transcription factor family; NBS, nucleotide-binding site; Os, Oryza sativa; PAL, phenylalanine ammonia lyase; PAMP/MAMP, pathogen/microbe-associated molecular pattern; PCD, programmed cell death; PERK, proline extension-like receptor kinase1; Pi, Pyricularia oryzae resistance; PR genes, pathogenesis-related genes; PRR, pattern recognition receptors; PTI, PRR-triggered immunity; R, resistance; Rac, Ras-related C3 botulinum toxin substrate; RbohB, respiratory burst oxidase homolog B; RLCK, receptor-like cytoplasmic kinases; RLK, receptor like kinases; ROS, reactive oxygen species; RPR, rice probenazol responsible; SA, salicylic acid; SERK, somatic embryogenesis receptor kinase; SWEET, sugars will eventually be exported transporters; syn, synthesis; TF, transcription factors; WAK, cell wall-associated kinase; Xa, Xanthomonas campestris pv. oryzae resistance; XB, XA21 binding proteins.
The first set of analyses was aimed at comparing the expression patters of genes encoding pattern recognition receptors (PRRs). These receptors mediate the first line of plant defense response. PRRs recognize microbe- or pathogen-associated molecular patterns from invading microbe (MAMPs/DAMPs) or damage-associated molecular patterns (DAMPs) released from plants upon damage by invading microbes (Macho and Zipfel, 2014), which induces PAMP-triggered immunity (PTI). The largest group of detected PRR-related DEGs encoded leucine-rich-repeat receptor-like kinases (LRR-RLKs), typically involved in the perception of classical MAMPs/PAMPs like e.g., bacterial flagellin, elongation factor Tu (EF-Tu), or endogenous Pep peptides (Ma et al., 2012; Macho and Zipfel, 2014). Interestingly, endophytic colonization led to moderate modulation of expression of only five DEGs encoding LRR-RLKs, while pathogen infection stipulated strong upregulation of 11 DEGs encoding LRR-RLKs (Table S3). Expression of only one gene (Os04g0227000) was upregulated by both, endophyte and pathogen.
Another group of detected DEGs encoding PRRs included lectin receptor kinases (LecRKs) known for their role in binding various carbohydrates, WAK proteins found to bind glycine-rich proteins (GRPs), pectin or oligogalacturonides (OGs) released from cell walls (Brutus et al., 2010; Delteil et al., 2016), and proline extension-like receptor kinase 1 (PERK1) for MAMP/PAMP and/or DAMP detection (Silva and Goring, 2002). Similarly, genes encoding lysine motif domain (LysM) domain-containing receptor kinases, out of which some detect peptidoglycan (PGN) or chitin like OsCERK1, OsLYP4, and OsLYP6 (Ao et al., 2014), were differently modulated. DEGs encoding all abovementioned groups of PRRs were strongly induced by pathogen infection, while only one member of each group was weakly modulated by Azoarcus colonization (Table S3). Therefore, it can be speculated that endophytic colonization may be less damaging to rice roots in comparison to pathogen infection (less DAMPs), or DAMP or PAMP signaling in mutualistic interactions might be masked or blocked as suggested previously (Plett and Martin, 2018).
Azoarcus perception might involve chitin perception receptor OsCEBiP1 (Akamatsu et al., 2013) and downstream Mitogen-Activated Protein Kinase 3 (MAPK3), as genes encoding both of these receptors were upregulated during Azoarcus colonization (Figure 2). Interestingly, in rice suspension culture chitin is able to induce production of jasmonic acid and phytoalexin (Kaku and Shibuya, 2016), which aligns with the transcriptional activation of genes related to JA and phytoalexin biosynthesis observed during rice colonization by Azoarcus. The LYP4 and LYP6 participating in peptidoglycan perception (Liu et al., 2012) were, however, not induced.
Pathogen infection induced more DEGs encoding receptor-like cytoplasmic kinases (RLCK, 5 DEGs) than endophytes (only one DEG) (Table S3). The signal transduction during PTI typically requires PRRs to phosphorylate RLCKs which, in turn, leads to mitogen-activated protein kinase (MAPK)-dependent or -independent ROS burst and defense gene expression (Figure 2) (Macho and Zipfel, 2014).
Interestingly, it has been demonstrated that MAPK signaling can be negatively regulated by protein phosphatase 2C (PP2C), like in case of kinase-associated protein phosphatase (KAPP) interacting with FLS2 and reducing the flg22-induced immune responses (Gómez-Gómez et al., 2001; Park et al., 2008). A weak but stable upregulation of expression of four genes encoding PP2C homologues was detected upon Azoarcus colonization, and downregulation of another PP2C homologue in response to Xoo infection (Table S3). This specific modulation may be linked to the observed differences in strength of PTI response to a pathogen and an endophyte.
The second layer of the plant immune system is effector-triggered immunity (ETI). As a result of coevolution, plant pathogens produce virulence factors called effectors to modulate the PTI. Correspondingly, plants also evolved a family of the polymorphic intracellular nucleotide-binding site and leucine-rich repeat domain-containing proteins (NBS-LRRs or NLRs), known as resistance proteins (R proteins), to perceive pathogen effectors and induce ETI (Cui et al., 2015). Accordingly, the pathogen induced more and different R genes (eight DEGs) in comparison to the endophyte (three DEGs) (Table S3).
Divergent Signal Transduction in Endophyte- and Pathogen Induced Responses
As Ca2+ concentration change, activation of MAPKs and transcription factors are among earliest components of signaling pathways during plant defense responses (Meng and Zhang, 2013); the expression patterns of genes related to these processes between rice roots colonized by Xoo and Azoarcus were compared.
It was observed that Xoo infection led to a strong induction of 10 DEGs associated with calcium signaling, including two genes encoding EF-Hand type domain-containing proteins (Os09g0483500, Os09g0483100), exhibiting high FC of 20.1 and 55.8, respectively (Table S3). They are also strongly upregulated in rice overexpressing transcription factor OsERF71 which is linked to drought resistance, but not to biotic stress (Lee et al., 2016). Only four genes which belong to this category were moderately modulated by Azoarcus (Table S3). No overlapping DEGs were found for this category, which indicates differences in calcium signaling utilization in transcriptomic response to the endophyte and the pathogen.
Also, the expression patterns of plant transcription factor expression (TF) were hardly overlapping between rice roots colonized by the endophyte and plants colonized by the pathogen. Overall, more DEGs encoding for to AP2/ERF, bHLH, bZIP/TGA, MYB, and WRKY family were up-regulated by Xoo infection in each family, generally also with a higher FC. In contrast, genes belonging to these families were mostly down-regulated during Azoarcus colonization, some exhibiting high FC value (Table S3). One notable exception includes the DEGs encoding the NAC-TF-family, which were almost exclusively upregulated by Azoarcus. NAC TFs are a large group of genes, comprising 151 homologues in rice, playing various roles in rice biotic and abiotic responses. There is, however, very limited data from previous studies regarding Azoarcus-responsive NACs genes, with only two characterized DEGs including cold-induced OsONAC059 and salinity-stress induced OsONAC103 (Fang et al., 2008).
Genes encoding MAPKs which met the criteria for DEGs were not detected, however a gene encoding OsMAPK3 was exhibiting stable expression induction of 1.3-fold only in Azoarcus-colonized plants among all technical and biological replicates (left side of Figure 2 and Table S4). It remains to be investigated whether OsMAPK3 gene product, involved in resistance to abiotic stress like chilling (Zhang et al., 2017), is also involved in signal transduction in roots subjected to endophyte colonization.
Taken together, the analysis of signaling-related DEGs shows a strong difference in perception of pathogen and endophyte by the rice plant, with the latter inducing weaker responses, in many cases leading to transcriptional repression of signaling genes.
Most Downstream Defense Reactions Repressed in the Endophytic Interaction
PTI signaling leads to various cellular responses and physiological changes in plants including ROS production, induced cell wall fortification, biosynthesis of antimicrobial secondary metabolites, and upregulation of specific pathogen-related genes (Grosskinsky et al., 2012). Pronounced differences in expression patterns of genes governing these processes were observed between roots colonized by Xoo and Azoarcus. Many ROS-related DEGs encoding type III peroxidases, oxalate oxidases, germin-like proteins, and amine oxidases which were induced by the endophyte or the pathogen were detected (Table S3). Interestingly, endophyte induced less (12) DEGs with moderate FC (1.5–5.6x) than pathogen, which has induced more DEGs (18) with higher FC (2.1–14x). For the type III peroxidase gene OsPrx92 a very pronounced difference in expression was detected, as this gene was downregulated (−5.6x) by the endophyte and upregulated (+14.0x) by the pathogen. With respect to ROS scavenging, only one gene was upregulated by the endophyte (OsGRX9), while Os08g0470700 was strongly downregulated by the pathogen. ROS play a vital role in plant immunity as they prime plants against pathogens not only via localized oxidative bursts but also as a sustained ROS signaling system (Camejo et al., 2016). It can be speculated that weak ROS-related transcriptomic response in case of Azoarcus-colonized roots could further decrease defense-related systemic signaling, leading to lack of symptoms of pathogenicity in these roots.
To build up direct barrier against bacterial penetration, plants induce processes such as callose deposition and lignin synthesis. Azoarcus and Xoo both induced a group of genes related to lignin biosynthesis, though in Xoo infection to a much higher expression level (Figure 2, Tables S3 and S10). Several genes related to glycosyl hydrolases were down-regulated by Azoarcus only (Table S10), suggesting that by suppression of genes related to degradation, plant cell walls are strengthened against the endophyte.
Pathogenesis-related (PR) proteins are divergent set of proteins that are induced as a result of signaling upon pathogen infection. At least 17 groups of PR proteins are recognized in plants, and 13 groups of them were found differently regulated in Azoarcus and Xoo-rice interactions. Similarly, expression of this group of genes was more strongly upregulated by Xoo than by Azoarcus (Figure 2, Table S3). During Azoarcus colonization, genes coding for PR 1, 2, 3, 9, 10, 15, and 16 were mainly up-regulated. By Xoo infection, also genes coding for PR alpha, 1, 2, 3, 5, 8, 9, 10, 12, 13, 14, 15, and 16 were up-regulated, some of them strongly, like Os06g0695300 (encoding PR9, 14-fold), Os03g0700100 (encoding PR13, 44.1-fold), and Os07g0215500 (encoding PR14, 58.3-fold). Genes encoding PR 5 and 8 were only induced in case of Xoo infection. In contrast, genes encoding PR6, 13, and PR14 were mainly down-regulated by the endophyte (Table S3). Among 77 PR-encoding DEGs detected in Azoarcus-treated and Xoo-treated roots, only 5 DEGs exhibited similar expression pattern upon colonization by both bacteria, highlighting the strong difference between transcriptomic response to a pathogen and to an endophyte.
Defensin-like peptides called nodule-specific cysteine-rich peptides (NCR) can possess antimicrobial functions but also control rhizobial differentiation to increase efficiency of nitrogen fixation in root nodules of legumes (Maroti et al., 2015). Typical motifs for NCRs were found for Os04g0381500 (Figure S2), which was 1.5 fold upregulated by the endophyte but not modulated by Xoo (Table S3) and could have a potential role in Azoarcus-rice mutualism.
Another group of DEGs exhibiting strong difference in expression between the roots colonized by Xoo and Azoarcus, was a group of heat-shock protein-encoding genes. They are molecular chaperones, typically involved in heat resistance by disaggregating or degrading non-functional proteins and degrading irreversibly damaged polypeptides. They are also playing a role in resistance during HR (for example: OsHSP70, OsHSP40), or by functioning in HR, or interact with cytosolic R proteins (HSP90) (Guo et al., 2016). Interestingly, Azoarcus colonization led to down-regulation of two HSP-encoding genes (OsHSP70, OsHSP40), while Xoo strongly induced expression of OsHSP100 and OsHSP90, three small OsHSPs, and reduced expression of another OsHSP90 (Figure 2, Table S3).
Surprisingly, a group of defense-related genes encoding enzymes involved in phytoalexin biosynthesis was detected that was exclusively induced by the endophyte colonization. Phytoalexins are antimicrobial secondary metabolites which accumulate at sites of pathogen infection in plants (Yamane, 2013; Miyamoto et al., 2016). Azoarcus induced expression of genes involved in biosynthesis of phytoalexins such as momilactones, oryzalexins S, and phytocassanes (five DEGs). Contrastingly, Xoo colonization weakly modulated expression of two DEGs encoding ent-isokaurene C2-hydroxylase-like protein involved in phytoanticipin oryzalide A biosynthesis (Figure 3, Table S3). As previous studies reported enhanced expression of phytoalexin-biosynthesis enzymes upon Xoo infection in rice leaves (data retrieved from RiceXPro database), lack of strong induction of genes encoding these enzymes might be linked to differences in tissue-specific expression patterns.
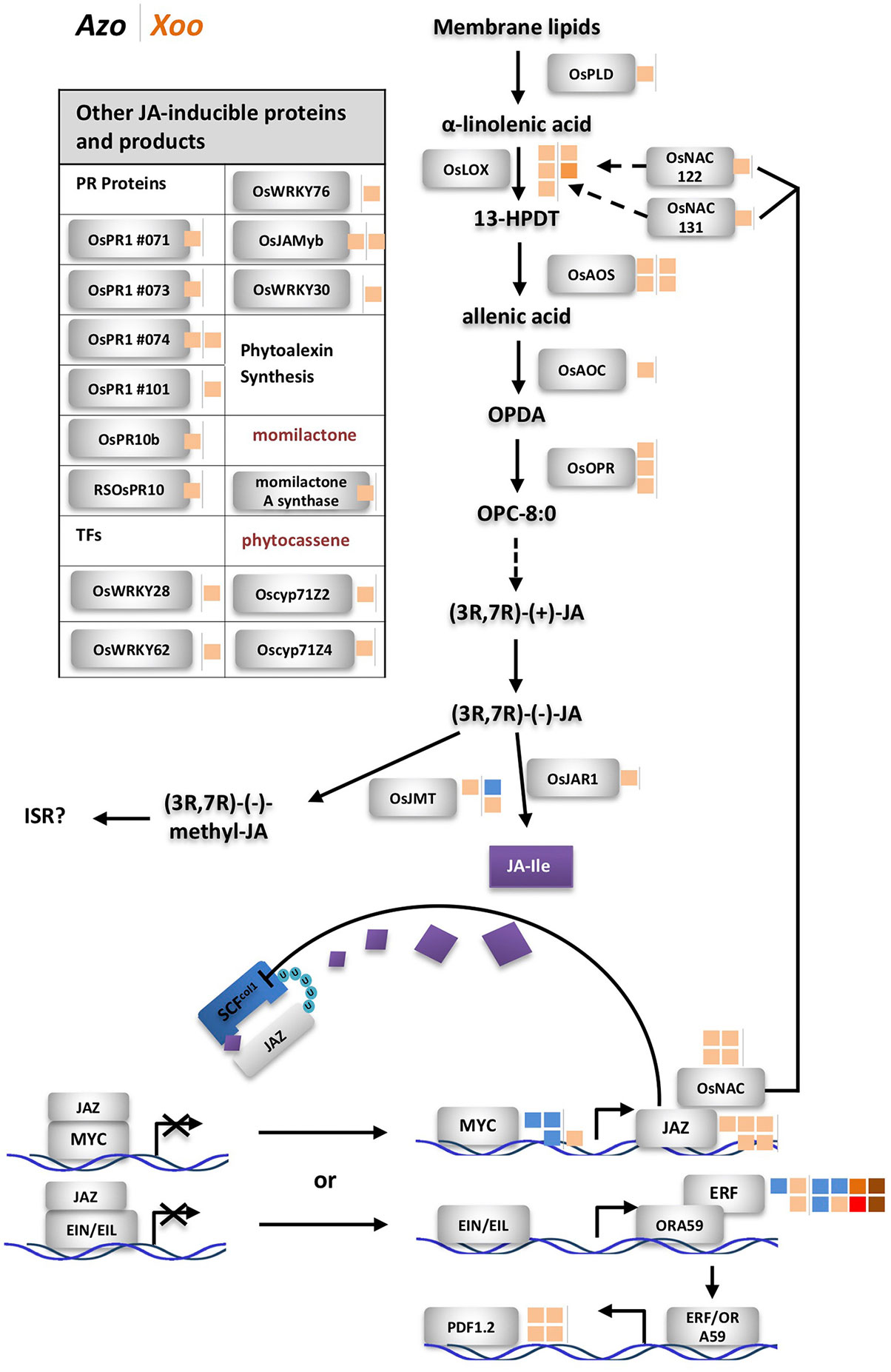
Figure 3 Differentially expressed genes related to jasmonate (JA) biosynthesis and downstream reactions. Next to rice protein names small blocks representing fold change of the differentially expressed gene (DEG) (right side of the vertical line, modulated by Xoo; left side, by Azoarcus) according to the color scale below. Dashed line: steps omitted, continuous line: direct reaction, arrow: reaction or induction, blunt end: inhibition. Left side, modulated DEGs not shown in the sketch; colored blocks indicating the fold change according to the color scale below. FC and annotation of DEGs from Table S5. AOC, allene oxide cyclase; AOS, allene oxide synthase; cyp, cytochrome P450; EIL, ethylene insensitive-3 (EIN3)-like; EIN, ethylene insensitive; ERF, ethylene response factor; FC, fold change; 13-HPDT, 13S-hydroperoxy-(9Z;11E;15)-octadecatrienoic acid; ISR, induced systemic resistance; JA, jasmonate; JA-Ile, jasmonoyl-isoleucine; JAMyb, JA-regulated myb transcription factor; JAR, jasmonate resistance; JAZ, jasmonate ZIM domain-containing; JMT, jasmonic acid methyl transferase; LOX, lipoxygenase; NAC, no apical meristem (NAM); OPC-8,0, 3-oxo-2-(20(Z)-pentenyl)-cyclopentane-1-octanoic acid; OPDA, oxophytodienoic acid; OPR, OPDA reductase; ORA, octadecanoid-responsive APETALA2 (AP2)/ERF; Os, Oryza sativa; PLD, phospholipase D; PR, pathogenesis related; RSOsPR, root-specific Oryza sativa PR; SCF, Skp1, Cullin, and F-box-containing complex; U, ubiquitinylated protein.
Also, alkaloids and anthocyanidins were identified by the Plant Metabolic Network tool as potential antimicrobial molecules (Chae et al., 2012). We have detected DEGs encoding strictosidine- and anthodyanidin-related enzymes, which were exclusively induced by Azoarcus colonization, and antioxidant-related DEGs which were only modulated by Xoo colonization (Figure 2, Table S3). Thus, also downstream defense reactions to the endophyte were weak, except for genes related to synthesis of secondary metabolites.
Dominating Role of Jasmonate-Related Defense Reactions Toward Endophytic Colonization
Jasmonate-related genes comprised the major DEGs affected by the endophyte, as summarized in Figure 3, from data in Table S5. Multiple DEGs encoding JA-biosynthesis genes were detected: phospholipase D (OsPLD), lipoxygenase (OsLOX), allene oxide synthase (OsAOS), allene oxide cyclase (OsAOC), oxophytodienoic acid (OPDA) reductase (OsOPR), and jasmonate resistance 1 (OsJAR1). Additionally, two genes OsNAC122 and 131 which positively regulate the expression of OsLOX, were up-regulated in roots colonized by Azoarcus. Also, upregulation of expression of genes encoding several JA-inducible proteins including PR-genes and genes encoding transcription factors was observed. Expression of some of them is uniquely governed by JA, such as OsPR1#71, #73, and #74 and OsJAmyb genes (Lee et al., 2001; Mitsuhara et al., 2008). Other hormonal pathways appeared unaffected or weakly affected by the endophyte (ethylene, ET; gibberellic acid, GA; cytokinins, CK; brassinosteroids, BR), or slightly down-regulated (abscisic acid ABA, auxin indole-3-acetic acid (IAA) known for competitive or antagonistic action to JA in plants (Wang and Irving; 2011) (Figure S3, Table S5). Therefore, it can be concluded that in Azoarcus-rice interactions, jasmonate appeared to play a dominating role in governing the defense response.
Interestingly, only few JA-related genes were induced by Xoo including two DEGs encoding allene oxide synthase (OsAOS) and two DEGs encoding lipoxygenase (OsLOX). Instead, Xoo infection had a strong impact on expression of salicylic acid-related genes, of which only few were moderately modulated by Azoarcus colonization (Figure S4, Table S5). Moreover, genes involved in BR and CK biosynthesis and signaling and GA pathways were up-regulated during Xoo colonization. This further underlines the hypothesis that pathogen and endophyte rewire hormonal responses differently.
The current observations on hormonal responses in response to the endophyte were validated by quantifying messenger RNA (mRNA) levels of genes participating in JA, SA, ET, and ABA hormone biosynthesis and corresponding downstream reactions by quantitative RT-PCR in a timeline of colonization (1 h, 24 h, 72 h, and 7 days post-inoculation). For each hormone, one gene encoding a protein involved in biosynthesis and one located downstream were chosen for the test: isochorismate synthase 1 (OsICS1) (Nahar et al., 2012; Choi et al., 2015) and OsWRKY45 ((Nahar et al., 2012) for SA, OsJAR1 (Svyatyna and Riemann, 2012; Lyons et al., 2013) and OsJAmyb (Lee et al., 2001) for JA, acyl-CoA synthetase 2 (OsACS2) (Helliwell et al., 2013) and SHR5 (Ma et al., 2013) for ET, and 9-cis-epoxycarotenoid dioxygenase 3 (OsNCED3) (Nahar et al., 2012) and OsMAPK5 (De Vleesschauwer et al., 2014) for ABA. Especially OsICS1 and OsACS2 were induced by pathogen infection as reported by previous studies (Nahar et al., 2012; Choi et al., 2015). During the infection process, only OsJAR1, representative for the JA-pathway [turning JA into active form jasmonoyl-isoleucine (JA-Ile)], responded significantly in three independent experiments: it was immediately up-regulated 1 h post-infection; though at 72 h, induction had seized, it rose again at later stage, 7 days post-infection (Figure S5). Expression of marker genes for other hormonal pathways did not respond consistently, except for OsNCED3 which was induced 1 h post-inoculation only (Figure S5).
Induction of JA-related defense responses appears to be a more general feature of bacterial endophytes. The JA pathway is induced in interactions between rice and many endophytic PGPR, though not all of them (Nadarajah, 2016). In our previous study on the same Japonica rice cultivar, RT-PCR analysis showed induction of marker genes OsJAR1 and OsJAmyb by A. olearius (Chen et al., 2015). In Indica varieties IR36 and IR42, JA-inducible proteins were overexpressed in proteome studies (Miché et al., 2006). Also, diazotrophic endophytes Azospirillum B510 (Drogue et al., 2014) and Gluconacetobacter diazotrophicus (Alquéres et al., 2013) induced JA-marker genes. In contrast, in Arabidopsis roots, JA-signaling was downregulated by Azospirillum brasilense 245 (Spaepen et al., 2014). How mutualistic microbes modulate defense responses—through effector proteins, small interfering RNAs (siRNAs) or other molecules, is still not clear (Plett and Martin, 2018).
Jasmonate-Related but not Salicylate-Related Pathways Control Endophytic Root Colonization of Azoarcus in Contrast to Xoo
In order to test whether JA- or SA-related pathways contribute to controlling endophytic colonization of roots, we employed well-characterized rice mutants with altered hormone levels or signaling cascades. First, rice mutant cpm2 (coleoptile photomorphogenesis) was tested, where the gene encoding allene oxide cyclase (AOC) in the JA synthesis pathway is disrupted, which results in a lack of JA production (Riemann et al., 2013). Colonization experiments were carried out in gnotobiotic culture systems with A. olearius BH72 or Xoo, respectively, and evaluated 14 days post-inoculation. The endophytic root colonization estimated by life cell counts was significantly increased (six-fold) in the jasmonate-deficient rice mutant in comparison to corresponding wild type cv. Nihonmasari (Figure 4C). Physiologically successful colonization was assessed by a reporter strain of A. olearius carrying a transcriptional fusion between the nitrogenase gene nifH and the ß-glucuronidase gene (Egener et al., 1999). Patterns of expression of nitrogen fixation genes were similar, with root tips and emergence points of lateral roots as main colonization and activity sites (Figure 4C.1), as well as intracellular (Figure 4C.2) and intercellular (Figure 4C.3) colonization. However, as expected from colonization quantification above, nif-gene expressing bacteria were more frequent and denser in cpm2-roots. In contrast, no significant effect of the cpm2 mutation on root colonization of the pathogen was detected (Figure 4A), as expected from expression profiling. This suggests that the JA pathway controls to some extent the density of internal colonization of roots by the endophyte, while the pathogen appeared to overcome this control.
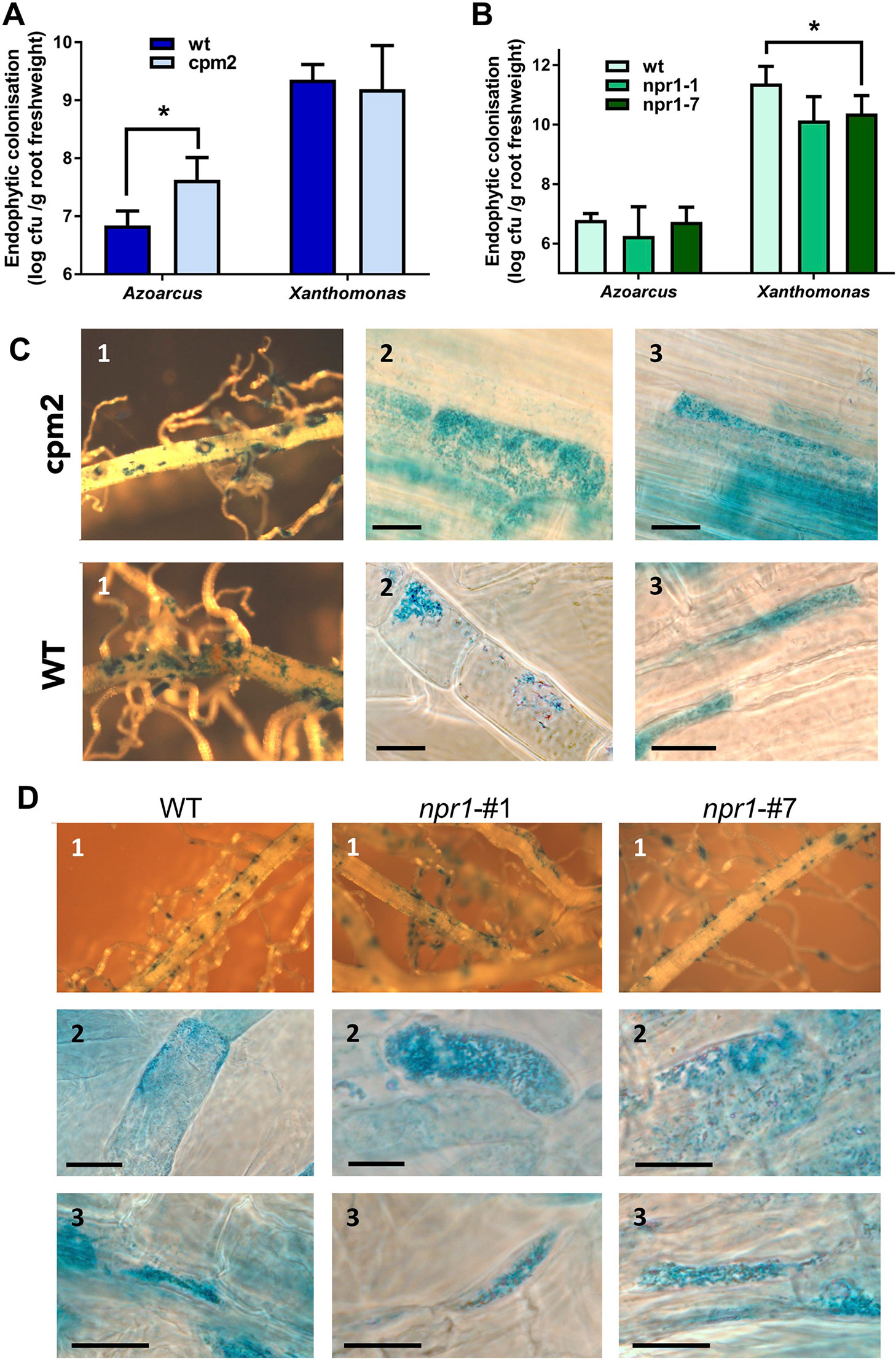
Figure 4 Root colonization of Azoarcus and Xoo in wild type and mutant rice altered in jasomonate (JA) and salicylate (SA) pathways. Jasmonate-deficient mutant cpm2 and parent Oryza sativa cv. Nihonmasare (A–C), or OsNPR1 knockdown mutant lines npr1#1 and npr1#7 and wild type cv. Nipponbare (A, B, D), respectively, were inoculated and harvested 14 days post-inoculation. (A) Quantitative assessment of endophytic root colonization of mutant and wild-type plants by Azoarcus olearius BH72, or by (B) Xanthomonas oryzae pv. PXO99. Bacteria colonizing the root interior were re-isolated after surface sterilization, and colony-forming units per gram root fresh weight were counted. Data from three independent biological experiments with 7–10 plants each (mean + SD). Significance according to two-tailed paired t-test (P < 0.05) is indicated by star*. Differences of cell counts in (A) were also significant within each of the three independent experiments. (C, D) Histochemical ß-glucuronidase (GUS) staining of roots inoculated with the nifH::gusA reporter strain A. olearius BHGN3.1. Examples from inspection of roots from three independent experiments. (C) Wild type rice (WT) (Nihonmasari) and cpm2 mutant; (D), wild type rice WT (Nipponbare), and mutant lines npr1#1 and npr1#7; 1, overview; 2, intracellular colonization; 3, intercellular colonization. Bars: 15 μm.
SA is playing a main role in plant defense against biotrophic or hemibiotrophic pathogens in Arabidopsis, with NPR1 (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1) acting as a central regulator of salicylic-acid (SA)-mediated defense signaling. OsNPR1 is the rice ortholog of AtNPR1. It has been shown in previous studies that over-expression of OsNPR1 conferred disease resistance to bacterial blight, but also enhanced herbivore susceptibility in transgenic plants (Yuan et al., 2007; Sugano et al., 2010). To test the impact of SA signaling on root colonization, two RNA interference (RNAi) knockdown mutants, OsNPR1-kd transgenic lines #1 and #7 (Sugano et al., 2010), were used. To verify the down-regulation of OsNPR1-expression in roots, transcript levels were quantified by RT-qPCR in our experimental system with and without inoculation of endophyte. In both lines, transcript levels were reduced as expected, even upon bacterial colonization (Figure S6). Endophytic colonization levels were not affected in the transgenic lines (Figure 4A), nor were nifH gene expression patterns altered (Figure 4D). Surprisingly, root colonization by the pathogen Xoo was decreased in knockdown lines, albeit only in line #7 at statistically significant levels (Figure 4B). In contrast, in rice leaves, RNAi lines showed enhanced disease susceptibility to X. oryzae (Yuan et al., 2007).
This highlights differences in defense responses in roots and shoots of rice. Also, in Arabidopsis thaliana and Brassica spp., the antagonistic interactions of the hormones JA and SA as well as their regulatory effects on defense genes was reported to differ between aerial and below-ground organs (Chuberre et al., 2018). For example, in rice, marker genes for defense responses PR-1 and PR-10 are transiently expressed during the early stages of root infection, while in leaves they continue to be transcribed during later stages of infection (Marcel et al., 2010). Concordantly, there were considerable differences in SA- and JA-related DEGs induced by Xoo according to our root data and published leaf data (RiceXpro, http://ricexpro.dna.affrc.go.jp/) (Table S6).
Putative Metabolic Responses Affected by Endophyte and Pathogen
X. oryzae injects transcription activator-like (TAL) effector proteins into plant host cells to modulate gene expression and thereby the plant response. Among the induced targets are sugars transporter (SWEET) genes, for example OsSWEET11 and OsSWEET14 in leaves (Strom et al., 2001; Baker et al., 2003; Wilkins et al., 2015), which may lead to increase of the sugar levels in the apoplast serving as carbon source for the pathogen. Partially these mechanisms appear to occur also in roots: OsSWEET11 and potential phosphate transporter encoding Os06g29790, which are only moderately induced in leaves (~10/2 fold) (Cernadas et al., 2014), were strongly induced in roots (70 fold, Table S3). Also, OsSWEET6a/6b which has not been reported to be affected in expression by Xoo in leaves, was upregulated by Xoo infection in roots. Interestingly, Azoarcus colonization resulted in down-regulation of two other OsSWEET genes, OsSWEET15 (16.6-fold) and OsSWEET1b (1.9-fold). OsSWEET15 was recently found to be able to support the Xoo virulence (Streubel et al., 2013). As Azoarcus does not grow on any carbohydrates (Reinhold-Hurek et al., 1993b) and would thus not profit from apoplastic sugars, we speculate that the endophyte could counteract carbohydrate supply to the pathogen.
Several other DEGs were also involved in the carbon metabolism (Figure S7A, Table S8). Xoo led to a strong induction of fermentative metabolism, indicated by a strong up-regulation of genes coding for PEP carboxykinase, lactate dehydrogenase ADH, and two pyruvate decarboxlyases. Also, the endophyte colonization induced alcohol dehydrogenase genes and decreased aldehyde dehydrogenase expression. This correlates well with the carbon sources preferences of A. olearius BH72: while malate is the preferred carbon source, ethanol is also readily metabolized, (Reinhold-Hurek et al., 1993a; Reinhold-Hurek et al., 1993b; Krause et al., 2011), especially during rice root colonization (Krause et al., 2011).
Among DEGs related to nitrogen metabolism, ammonium assimilation (glutamine synthetase, OsGS2), and aminotransferases expression was slightly decreased by the nitrogen-fixing endophyte, suggesting that at this stage, ammonium from nitrogen fixation might not be transferred. Interestingly, we have detected up-regulation of four genes encoding members of family of low affinity nitrate transporters/large peptide transporters (NTR1/PTR). While this family of transporters has 53 homologues in rice which exhibit various functions, it has been suggested (based on similarity to well-characterized members of this family in Arabidopsis) that Azoarcus-responsive genes OsNRT1.1C (5.2-fold up-regulated) and OsNRT1.2 (1.6-fold up-regulated) could encode actual nitrate transporters (Plett et al., 2010). Moreover, the expression of seven uncharacterized amino-acid transporters was also induced by Azoarcus (Table S7, Figure S7).
Comparison of Root Transcriptomic Responses to Other Microbes
In order to identify DEGs which might be specifically related to signal transduction in Azoarcus-rice interaction in contrast to pathogenic interactions, data for Xoo-rice leaf infection and Magnaporthe oryzae-rice root and leaf infection were included in the comparison (RiceXPro, ricexpro.dna.affrc.go.jp/). Only five DEGs were detected exclusively in the Azoarcus-rice root interaction: up-regulated DEGs coding for a LRR_RLK protein (Os11g0208900), OsWAK103 (Os10g0151100), OsERF86 (Os07g0410700), OsCDPK25 (Os11g0136600), and a down-regulated DEG coding for an EF hand domain-containing protein (Os09g0483300). Leucine-rich-repeat receptor-like kinases like Os11g0208900 are typically involved in perception of MAMPs/PAMPs. Whether any of these candidate proteins is involved in specific Azoarcus or endophyte perception and signal transduction will have to be tested in further experiments. As a first step the data presented here were compared with rice transcriptome results published for Azospirillum spp. (Drogue et al., 2014). They are root-associated diazotrophs that are well-known as phytostimulators (Okon and Labandera-Gonzalez, 1994; Cerezini et al., 2016), and plant growth promotion effects are mainly attributed to production of the phytohormone IAA and the modulation of the plant phytohormonal balance rather than nitrogen fixation (Steenhoudt and Vanderleyden, 2000; Somers et al., 2005). Azospirillum lipoferum 4B is an efficient rhizoplane colonizer (Drogue et al., 2014), while Azospirillum sp. B510 originates from surface-sterilized rice roots and is an endophyte of rice (Yasuda et al., 2009; Kaneko et al., 2010). Interestingly, there were no overlaps in genes related to signal perception and transduction modulated by both these strains and Azoarcus. However, comparison of DEGs in response to only endophytic strains BH72 and B510 revealed commonalities (Table S9). Both endophytes induced jasmonate-dependent responses, repressed DEGs for cell wall degradation, and downregulated genes related to photosynthesis. The latter is likely to be related to the jasmonate pathway, as both, nuclear and plastid photosynthetic genes, are repressed under the control of JA (Reinbothe et al., 2009). Although in contrast to Azospirillum, A. olearius is not known to produce IAA (Krause et al., 2006), both strains repressed OsIAA9, Os02g0805100 encoding an auxin responsive protein. The otherwise strongly strain-specific and cultivar-specific rice responses (Drogue et al., 2014) indicate consequences of different epiphytic and endophytic lifestyles, but also that individual genotypic variations of the host plants may be important driving forces in the cooperation with beneficial bacteria.
Concluding Remarks
Plants are encountering a vast diversity of microorganisms in roots in comparison to the foliar region, including beneficial bacteria and fungi as well as both prokaryotic and eukaryotic pathogens (Mendes et al., 2013). The high density of bacterial colonization calls for a reduced sensitivity of roots toward microbial molecules and of defense responses, which may account for deviating hormonal responses in below- and above-ground tissues. Furthermore, different panels of host gene transcription are induced during root colonization by beneficial or detrimental microbes (Plett and Martin, 2018). For fungi having a pathogenic (Magnaporthe grisea and Fusarium moniliforme) or a symbiotic lifestyle (arbuscular mycorrhiza fungus Rhizophagus irregularis), an overlap of only 13% of DEGs was found in rice roots (Guimil et al., 2005). In case of the bacterial endophyte A. olearius BH72 compared to another Proteobacterium, X. oryzae, the overlap was even smaller (8% of the endophyte-, 6% of the pathogen-modulated DEGs), which demonstrates strong deviation of the lifestyle in a more “loose” beneficial interaction.
As observed for symbiotic interactions (Duplessis et al., 2005), defense reactions are provoked during the early phases of contact. A strong time-dependent modulation of expression of OsJAR1 gene was detected in experiments presented here, which raises the question: at which stages and how do endophytes attenuate defense? To elucidate pathways which could perceive and transduce signals specific for endophytic colonization, putative candidate genes should to be verified in transcriptomes at different time points and genotype combinations. How mutualistic microbes modulate defense responses—through effector proteins, siRNAs, or other molecules, is still not clear.
One of the key findings by using rice mutants with altered hormonal responses is that JA signaling is involved in controlling the Azoarcus endophyte density in roots and thus contributes to shaping the root microbiome. Colonization assays using rice mutants deficient in jasmonate synthesis (cpm2) or exhibiting reduced salicylic acid-mediated defense responses (NPR1-kd) suggested that endophytic colonization is controlled through mechanisms which involve JA-production and signaling and are SA-independent. Xoo colonization did not appear to be subject to these control mechanisms. Previous studies using plant mutants of Arabidopsis demonstrated that salicylic acid is the major hormonal pathway that modulates the community composition at roots (Lebeis et al., 2015). However, external addition of methyl jasmonate also affected the community structure of Arabidopsis rhizosphere soil (Carvalhais et al., 2013), and wheat roots (Liu et al., 2017). Unfortunately, these studies did not address quantitation of endophytic colonization. Furthermore, the endophytic compartment was not well differentiated because ultrasonication was used to remove surface bacteria, which is not very efficient in soil-based settings (Reinhold-Hurek et al., 2015). According to the presented data for Azoarcus, the JA pathway appears to restrict the internal root colonization, probably below a limit which may become harmful to the plant. Deeper knowledge of the molecular mechanisms, especially time-resolved responses, identification of endophyte-specific perception proteins, and bacterial signals involved, may help to modulate the endophyte microbiome for improved biotechnological applications.
Data Availability Statement
The datasets generated for this study can be found in the GEO (GSE136706 and GSE136707).
Author Contributions
XC and BR-H designed the experiments. XC carried out most experiments. MM performed the timeline experiments. XC, MM, and BR-H wrote the manuscript.
Funding
The work was supported by a grant of the Deutsche Forschungsgemeinschaft DFG to BR-H (Re756/18-1), and the University of Bremen (02/124/10).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Michael Riemann (KIT Karlsruhe Institute of Technology, Karlsruhe, Germany) for providing the rice cpm2 and advice how to select homozygotes, and also Hiroshi Takatsuji (National Institute of Agrobiological Sciences, Japan) for supplying OsNPR1-kd, lines #1, #7. We are also grateful to Florian Schnurrer (University of Bremen) for assisting in Xoo infection experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01758/full#supplementary-material
References
Adhikari, T. B., Cruz, C., Zhang, Q., Nelson, R. J., Skinner, D. Z., Mew, T. W., et al. (1995). Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl. Environ. Microbiol. 61, 966–971.
Akamatsu, A., Wong, H. L., Fujiwara, M., Okuda, J., Nishide, K., Uno, K., et al. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476. doi: 10.1016/j.chom.2013.03.007
Alquéres, S., Meneses, C., Rouws, L., Rothballer, M., Baldani, I., Schmid, M., et al. (2013). The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant-Microbe Interact. 26, 937–945. doi: 10.1094/MPMI-12-12-0286-R
Ao, Y., Li, Z. Q., Feng, D. R., Xiong, F., Liu, J., Li, J. F., et al. (2014). OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. doi: 10.1111/tpj.12710
Baker, G. C., Smith, J. J., Cowan, D. A. (2003). Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55, 541–555. doi: 10.1016/j.mimet.2003.08.009
Barac, T., Taghavi, S., Borremans, B., Provoost, A., Oeyen, L., Colpaert, J. V., et al. (2004). Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat. Biotechnol. 22, 583–588. doi: 10.1038/nbt960
Barraquio, W. L., Revilla, L., Ladha, J. K. (1997). Isolation of endophytic diazotrophic bacteria from wetland rice. Plant Soil 194, 15–24. doi: 10.1023/A:1004246904803
Berendsen, R. L., Pieterse, C. M., Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Böhm, M., Hurek, T., Reinhold-Hurek, B. (2007). Twitching motility is essential for endophytic rice colonization by the N2- fixing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 20, 526–533. doi: 10.1094/MPMI-20-5-0526
Brutus, A., Sicilia, F., Macone, A., Cervone, F., De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457. doi: 10.1073/pnas.1000675107
Camejo, D., Guzman-Cedeno, A., Moreno, A. (2016). Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Bioch. 103, 10–23. doi: 10.1016/j.plaphy.2016.02.035S0981-9428(16)30060-2
Carvalhais, L. C., Dennis, P. G., Badri, D. V., Tyson, G. W., Vivanco, J. M., Schenk, P. M. (2013). Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PloS One 8, e56457. doi: 10.1371/journal.pone.0056457
Cerezini, P., Kuwano, B. H., dos Santos, M. B., Terassi, F., Hungria, M., Nogueira, M. A. (2016). Strategies to promote early nodulation in soybean under drought. Field Crops Res. 196, 160–167. doi: 10.1016/j.fcr.2016.06.017
Cernadas, R. A., Doyle, E. L., Nino-Liu, D. O., Wilkins, K. E., Bancroft, T., Wang, L., et al. (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PloS Pathog. 10, e1003972. doi: 10.1371/journal.ppat.1003972PPATHOGENS-D-13-02542
Chae, L., Lee, I., Shin, J., Rhee, S. Y. (2012). Towards understanding how molecular networks evolve in plants. Curr. Opin. Plant Biol. 15, 177–184. doi: 10.1016/j.pbi.2012.01.006
Chen, X., Miché, L., Sachs, S., Wang, Q., Buschart, A., Yang, H., et al. (2015). Rice responds to endophytic colonization which is independent of the common symbiotic signaling pathway. New Phytol. 208, 531–543. doi: 10.1111/nph.13458
Choi, C., Hwang, S. H., Fang, I. R., Il Kwon, S., Park, S. R., Ahn, I., et al. (2015). Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis-related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 208, 846–859. doi: 10.1111/nph.13516
Chuberre, C., Plancot, B., Driouich, A., Moore, J. P., Bardor, M., Gügi, B., et al. (2018). Plant immunity is compartmentalized and specialized in roots. Front. Plant Sci. 9, 1692. doi: 10.3389/fpls.2018.01692
Cui, H. T., Tsuda, K., Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
De Vleesschauwer, D., Gheysen, G., Hofte, M. (2013). Hormone defense networking in rice: tales from a different world. Trends Plant Sci. 18, 555–565. doi: 10.1016/j.tplants.2013.07.002S1360-1385(13)00129-5
De Vleesschauwer, D., Xu, J., Hofte, M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5, 611. doi: 10.3389/fpls.2014.00611
Delteil, A., Gobbato, E., Cayrol, B., Estevan, J., Michel-Romiti, C., Dievart, A., et al. (2016). Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 16, 17. doi: 10.1186/s12870-016-0711-x
Dörr, J., Hurek, T., Reinhold-Hurek, B. (1998). Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30, 7–17. doi:10.1046/j.1365-2958.1998.01010.x
Drogue, B., Sanguin, H., Chamam, A., Mozar, M., Llauro, C., Panaud, O., et al. (2014). Plant root transcriptome profiling reveals a strain-dependent response during Azospirillum-rice cooperation. Front. Plant Sci. 5, 1–14. doi: 10.3389/Fpls.2014.00607
Duplessis, S., Courty, P. E., Tagu, D., Martin, F. (2005). Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol. 165, 599–611. doi: 10.1111/j.1469-8137.2004.01248.x
Egener, T., Hurek, T., Reinhold-Hurek, B. (1999). Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol. Plant-Microbe Interact. 12, 813–819. doi: 10.1094/MPMI.1999.12.9.813
Fang, Y., You, J., Xie, K., Xie, W., Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 280, 547–563. doi: 10.1007/s00438-008-0386-6
Gao, Q. M., Zhu, S. F., Kachroo, P., Kachroo, A. (2015). Signal regulators of systemic acquired resistance. Front. Plant Sci. 6, 228. doi: 10.3389/Fpls.2015.00228
Gómez-Gómez, L., Bauer, Z., Boller, T. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in arabidopsis. Plant Cell 13, 1155–1163. doi: 10.1105/tpc.13.5.1155
Grosskinsky, D. K., van der Graaff, E., Roitsch, T. (2012). Phytoalexin transgenics in crop protection-Fairy tale with a happy end? Plant Sci. 195, 54–70. doi: 10.1016/j.plantsci.2012.06.008
Guimil, S., Chang, H. S., Zhu, T., Sesma, A., Osbourn, A., Roux, C., et al. (2005). Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. U.S.A. 102, 8066–8070. doi: 10.1073/pnas.0502999102
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7, 114. doi: 10.3389/Fpls.2016.00114
Helliwell, E. E., Wang, Q., Yang, Y. N. (2013). Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 11, 33–42. doi: 10.1111/pbi.12004
Hiruma, K., Gerlach, N., Sacristan, S., Nakano, R. T., Hacquard, S., Kracher, B., et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028S0092-8674(16)30130-1
Hurek, T., Reinhold-Hurek, B., Van Montagu, M., Kellenberger, E. (1994). Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J. Bacteriol. 176, 1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994
Hurek, T., Egener, T., Reinhold-Hurek, B. (1997). Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the ß-subclass. J. Bacteriol. 179, 4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997
Hurek, T., Handley, L., Reinhold-Hurek, B., Piché, Y. (2002). Azoarcus grass endophytes contribute fixed nitrogen to the plant in an unculturable state. Mol. Plant-Microbe Interact. 15, 233–242. doi: 10.1094/MPMI.2002.15.3.233
Jalmi, S. K., Sinha, A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep-Uk 6, 37974. doi: 10.1038/srep37974
Kaku, H., Shibuya, N. (2016). Molecular mechanisms of chitin recognition and immune signaling by LysM-receptors. Physiol. Mol. Plant Pathol. 95, 60–65. doi: 10.1016/j.pmpp.2016.02.003
Kaneko, T., Minamisawa, K., Isawa, T., Nakatsukasa, H., Mitsui, H., Kawaharada, Y., et al. (2010). Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res. 17, 37–50. doi: 10.1093/dnares/dsp026
Karnagilla, A. D., Natural, M. P. (1973). A comparative study of culture media for Xanthomonas oryzae. Philipp. Agric. 57, 141–152.
Khare, E., Mishra, J., Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9, 2732. doi: 10.3389/fmicb.2018.02732
Krause, A., Ramakumar, A., Bartels, D., Battistoni, F., Bekel, T., Boch, J., et al. (2006). Complete genome of the mutualistic, N2-fixing grass endophyte Azoarcus sp. strain BH72. Nat. Biotechnol. 24, 1385–1391. doi: 10.1038/nbt1243
Krause, A., Leyser, B., Miché, L., Battistoni, F., Reinhold-Hurek, B. (2011). Exploring the function of alcohol dehydrogeanses during the endophytic life of Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 24, 1325–1332. doi: 10.1094/MPMI-05-11-0139
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lee, M. W., Qi, M., Yang, Y. (2001). A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant-Microbe Interact. 14, 527–535. doi: 10.1094/MPMI.2001.14.4.527
Lee, D. K., Jung, H., Jang, G., Jeong, J. S., Kim, Y. S., Ha, S. H., et al. (2016). Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 172, 575–588. doi: 10.1104/pp.16.00379
Liu, B., Li, J. F., Ao, Y., Qu, J. W., Li, Z. Q., Su, J. B., et al. (2012). Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24, 3406–3419. doi: 10.1105/tpc.112.102475
Liu, H. W., Carvalhais, L. C., Schenk, P. M., Dennis, P. G. (2017). Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci. Rep-Uk 7, 1–8. doi: 10.1038/Srep41766
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 -DDCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lyons, R., Manners, J. M., Kazan, K. (2013). Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 32, 815–827. doi: 10.1007/s00299-013-1400-y
Ma, Y., Walker, R. K., Zhao, Y., Berkowitz, G. A. (2012). Linking ligand perception by PEPR pattern recognition receptors to cytosolic Ca2+ elevation and downstream immune signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 19852–19857. doi: 10.1073/pnas.1205448109
Ma, B., He, S. J., Duan, K. X., Yin, C. C., Chen, H., Yang, C., et al. (2013). Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 6, 1830–1848. doi: 10.1093/mp/sst087
Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Marcel, S., Sawers, R., Oakeley, E., Angliker, H., Paszkowski, U. (2010). Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 3177–3187. doi: 10.1105/tpc.110.078048
Maroti, G., Downie, J. A., Kondorosi, E. (2015). Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr. Opin. Plant Biol. 26, 57–63. doi: 10.1016/j.pbi.2015.05.031S1369-5266(15)00083-7
Mendes, R., Garbeva, P., Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Meng, X. Z., Zhang, S. Q. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314 doi: 10.1094/MPMI-19-0502
Miché, L., Battistoni, F., Gemmer, S., Belghazi, M., Reinhold-Hurek, B. (2006). Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant-Microbe Interact. 19, 502–511.
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genom. 279, 415–427. doi: 10.1007/s00438-008-0322-9
Miyamoto, K., Fujita, M., Shenton, M. R., Akashi, S., Sugawara, C., Sakai, A., et al. (2016). Evolutionary trajectory of phytoalexin biosynthetic gene clusters in rice. Plant J. 87, 293–304. doi: 10.1111/tpj.13200
Nadarajah, K. K. (2016). Induced systemic resistance in rice (Singapore: Springer Science+Business Media Singapore).
Nahar, K., Kyndt, T., Nzogela, Y. B., Gheysen, G. (2012). Abscisic acid interacts antagonistically with classical defense pathways in rice-migratory nematode interaction. New Phytol. 196, 901–913. doi: 10.1111/j.1469-8137.2012.04310.x
Okon, Y., Labandera-Gonzalez, C. A. (1994). Agronomic applications of Azospirillum - an evaluation of 20 years worldwide field inoculation. Soil Biol. Biochem. 26, 1591–1601. doi: 10.1016/0038-0717(94)90311-5
Park, C. J., Peng, Y., Chen, X., Dardick, C., Ruan, D., Bart, R., et al. (2008). Rice XB15, a protein phosphatase 2C, negatively regulates cell death and XA21-mediated innate immunity. PloS Biol. 6, e231. doi: 10.1371/journal.pbio.006023108-PLBI-RA-0879
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Pieterse, C. M., van der Does, D., Zamioudis, C., Leon-Reyes, A., van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Plett, J. M., Martin, F. M. (2018). Know your enemy, embrace your friend: using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 93, 729–746. doi: 10.1111/tpj.13802
Plett, D., Toubia, J., Garnett, T., Tester, M., Kaiser, B. N., Baumann, U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PloS One 5, e15289. doi: 10.1371/journal.pone.0015289
Reinbothe, C., Springer, A., Samol, I., Reinbothe, S. (2009). Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276, 4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x
Reinhold, B., Hurek, T., Niemann, E.-G., Fendrik, I. (1986). Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appl. Environ. Microbiol. 52, 520–526.
Reinhold-Hurek, B., Hurek, T. (1998). Life in grasses: diazotrophic endophytes. Trends Microbiol. 6, 139–144. doi: 10.1016/s0966-842x(98)01229-3
Reinhold-Hurek, B., Hurek, T., Claeyssens, M., Van Montagu, M. (1993a). Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J. Bacteriol. 175, 7056–7065. doi: 10.1128/jb.175.21.7056-7065.1993
Reinhold-Hurek, B., Hurek, T., Gillis, M., Hoste, B., Vancanneyt, M., Kersters, K., et al. (1993b). Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth) and description of two species Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int. J. Syst. Bacteriol. 43, 574–584. doi: 10.1099/00207713-43-3-574
Reinhold-Hurek, B., Maes, T., Gemmer, S., Van Montagu, M., Hurek, T. (2006). An endoglucanase is involved in infection of rice roots by the not cellulose-metabolizing endophyte Azoarcus sp. BH72. Mol. Plant-Microbe Interact. 19, 181–188. doi: 10.1094/MPMI-19-0181
Reinhold-Hurek, B., Bünger, W., Burbano, C. S., Sabale, M., Hurek, T. (2015). Roots shaping their microbiome: global hot spots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. doi: 10.1146/annurev-phyto-082712-102342
Riemann, M., Haga, K., Shimizu, T., Okada, K., Ando, S., Mochizuki, S., et al. (2013). Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238. doi: 10.1111/tpj.12115
Sarkar, A., Marszalkowska, M., Schäfer, M., Pees, T., Klingenberg, H., Macht, F., et al. (2017). Global expression analysis of the response to microaerobiosis reveals an important cue for endophytic establishment of Azoarcus sp. BH72. Environ. Microbiol. 19, 198–217. doi: 10.1111/1462-2920.13569
Shidore, T., Dinse, T., Öhrlein, J., Becker, A., Reinhold-Hurek, B. (2012). Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ. Microbiol. 14, 2775–2787. doi: 10.1111/j.1462-2920.2012.02777.x
Silva, N. F., Goring, D. R. (2002). The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol. 50, 667–685. . doi: 10.1023/A:1019951120788
Somers, E., Ptacek, D., Gysegom, P., Srinivasan, M., Vanderleyden, J. (2005). Azospirillum brasilense produces the auxin-like phenylacetic acid by using the key enzymes for indole-3-acetic acid biosynthesis. Appl. Environ. Microbiol. 71, 1803–1810. doi: 10.1128/AEM.71.4.1803-1810.2005
Spaepen, S., Bossuyt, S., Engelen, K., Marchal, K., Vanderleyden, J. (2014). Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 201, 850–861. doi: 10.1111/nph.12590
Steenhoudt, O., Vanderleyden, J. (2000). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 24, 487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv oryzae. New Phytol. 200, 808–819. doi: 10.1111/nph.12411
Strom, E. V., Dinarieva, T. Y., Netrusov, A. I. (2001). Methylobacillus flagellatus KT contains a novel cbo-type cytochrome oxidase. FEBS Lett. 505, 109–112. doi: 10.1016/s0014-5793(01)02795-8
Sugano, S., Jiang, C. J., Miyazawa, S., Masumoto, C., Yazawa, K., Hayashi, N., et al. (2010). Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. Rep. 74, 549–562. doi: 10.1007/s11103-010-9695-3
Svyatyna, K., Riemann, M. (2012). Light-dependent regulation of the jasmonate pathway. Protoplasma 249, 137–145. doi: 10.1007/s00709-012-0409-3
Tamaoki, D., Seo, S., Yamada, S., Kano, A., Miyamoto, A., Shishido, H., et al. (2013). Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal Behav. 8 (6), e24260. doi: 10.4161/psb.24260
Voges, M., Bai, Y., Schulze-Lefert, P., Sattely, E. S. (2019). Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U.S.A. 116, 12558–12565. doi: 10.1073/pnas.1820691116
Wilkins, K. E., Booher, N. J., Wang, L., Bogdanove, A. J. (2015). TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Front. Plant Sci. 6, 536. doi: 10.3389/fpls.2015.00536
Yamane, H. (2013). Biosynthesis of Phytoalexins and Regulatory Mechanisms of It in Rice. Biosci. Biotech. Biochem. 77, 1141–1148. doi: 10.1271/bbb.130109
Yang, Y. X., Ahammed, G. J., Wu, C. J., Fan, S. Y., Zhou, Y. H. (2015). Crosstalk among jasmonate, salicylate and ethylene signaling pathways in plant disease and immune responses. Curr. Protein Pept. Sci. 16, 450–461. doi: 10.2174/1389203716666150330141638
Yasuda, M., Isawa, T., Shinozaki, S., Minamisawa, K., Nakashita, H. (2009). Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in rice. Biosci. Biotech. Biochem. 73, 2595–2599. doi: 10.1271/bbb.90402
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Keywords: Oryza sativa, root endophytes, Azoarcus olearius, transcriptome, colonization, phytohormones, jasmonate, Xanthomonas oryzae pv. oryzae
Citation: Chen X, Marszałkowska M and Reinhold-Hurek B (2020) Jasmonic Acid, Not Salicyclic Acid Restricts Endophytic Root Colonization of Rice. Front. Plant Sci. 10:1758. doi: 10.3389/fpls.2019.01758
Received: 23 September 2019; Accepted: 16 December 2019;
Published: 29 January 2020.
Edited by:
Essaid Ait Barka, Université de Reims Champagne-Ardenne, FranceReviewed by:
Kiwamu Minamisawa, Tohoku University, JapanAvinash Chandra Pandey, Metahelix Life Sciences Limited, India
Copyright © 2020 Chen, Marszałkowska and Reinhold-Hurek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Reinhold-Hurek, YnJlaW5ob2xkQHVuaS1icmVtZW4uZGU=
 Xi Chen
Xi Chen Marta Marszałkowska
Marta Marszałkowska Barbara Reinhold-Hurek
Barbara Reinhold-Hurek