- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2China Agricultural University, College of Plant Protection, Beijing, China
Basic leucine zipper (bZIP) membrane-bound transcription factors (MTFs) play important roles in regulating plant growth and development, abiotic stress responses, and disease resistance. Most bZIP MTFs are key components of signaling pathways in endoplasmic reticulum (ER) stress responses. In this study, a full-length cDNA sequence encoding bZIP MTF, designated TabZIP74, was isolated from a cDNA library of wheat near-isogenic lines of Taichung29*6/Yr10 inoculated with an incompatible race CYR32 of Puccinia striiformis f. sp. tritici (Pst). Phylogenic analysis showed that TabZIP74 is highly homologous to ZmbZIP60 in maize and OsbZIP74 in rice. The mRNA of TabZIP74 was predicted to form a secondary structure with two kissing hairpin loops that could be spliced, causing an open reading frame shift immediately before the hydrophobic region to produce a new TabZIP74 protein without the transmembrane domain. Pst infection and the abiotic polyethylene glycol (PEG) and abscisic acid (ABA) treatments lead to TabZIP74 mRNA splicing in wheat seedling leaves, while both spliced and unspliced forms in roots were detected. In the confocal microscopic examination, TabZIP74 is mobilized in the nucleus from the membrane of tobacco epidermal cells in response to wounding. Knocking down TabZIP74 with barley stripe mosaic virus-induced gene silencing (BSMV-VIGS) enhanced wheat seedling susceptibility to stripe rust and decreased drought tolerance and lateral roots of silenced plants. These findings demonstrate that TabZIP74 mRNA is induced to splice when stressed by biotic and abiotic factors, acts as a critically positive regulator for wheat stripe rust resistance and drought tolerance, and is necessary for lateral root development.
Introduction
Plant pathogens, including fungi, viruses, bacteria, oomycetes, and nematodes, cause severe yield losses in crop production. To defend themselves against disease, plants have evolved different defense mechanisms against attackers. The first layer of defense is activated by athogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs). This kind of defense is referred to as PAMP triggered immunity (PTI) or MAMP triggered immunity (MTI) and defend the plant against non-specialized pathogens. This type of defense is perceived by plasma membrane receptors, which rely on protein maturation and endoplasmic reticulum (ER) protein folding quality control (Li et al., 2009; Lu et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009) and the secretion of anti-microbial proteins to the apoplast (van Loon et al., 2006).
Secretory proteins are synthesized and folded in ERs, and proper folding is needed to transport the proteins to their final destinations. Perturbations of this folding process result in ER stress, which is critical for rapid and effective basal immune responses of plant hosts (Wang et al., 2005). Regulation of ER capacity is important for immune signaling (Korner et al., 2015). The ER stress triggers cytoprotective signaling pathways, titled the unfold protein response (UPR), this signaling pathway restores and maintains ER homeostasis. There is growing recognition that ER stress responses are involved in normal plant development (Deng et al., 2016; Kim et al., 2018; Bao et al., 2019); for example, ER stress is involved in the cells synthesizing and secreting materials comprising the pollen coat (Deng et al., 2016).
There are two arms of the ER stress signaling pathway in plants, one of which encompass membrane-associated transcription factors (MTFs) through proteolytic cleavage, such as the MTFs of AtbZIP17 (Liu et al., 2007b), AtbZIP28 (Liu et al., 2007a; Tajima et al., 2008), ZmbZIP17 (Yang et al., 2013), and OsbZIP39 (Takahashi et al., 2012), which are mobilized from ERs to golgi apparatus in plant cell where they are released by site 1 and site 2 proteases (S1P and S2P) (Liu et al., 2007a; Sun et al., 2015). The other ER stress signaling pathway functions through inositol requiring enzyme 1 (IRE1) and the splicing of target RNA, such as bZIP60, which encodes an MTF of basic leucine zipper (bZIP) (Liu and Howell, 2010; Howell, 2013). Splicing of target mRNA lead a frame shift of ORF and produces a new protein without a transmembrane domain (TMD) in its C-terminal. Many MTF genes, such as XBP1 in yeast (Yoshida et al., 2001), AtbZIP60 in Arabidopsis (Deng et al., 2011), OsbZIP50/OsbZIP74 in rice (Hayashi et al., 2012), and ZmbZIP60 in maize (Li et al., 2012) are activated by the IRE1 depended mRNA splicing pathway.
The bZIP TF family is one of the largest families in plants, with the most diverse biological functions (Jakoby et al., 2002); several of its members, including AtbZIP60, NtbZIP60, and OsbZIP50, are membrane-associated bZIP factors, which fundamentally contribute to ER stress in plant basal immune responses. For example, Arabidopsis thaliana gene bZIP60 encoding an MTF is strongly induced to express by tunicamycin, an ER stress-inducing chemicals (Iwata and Koizumi, 2005), before translocating its protein without TMD into the cell nucleus and upregulating mRNA expression levels of several ER-resident chaperones, sucn as binding protein (BiP) and protein-disulfide isomerases (Iwata and Koizumi, 2005; Lu and Christopher, 2008). In tobacco, expression of NtbZIP60 was significantly upregulated upon infection with a non-host pathogen Pseudomonas cichorii, but not induced by a compatible pathogen Pseudomonas syringae pv. Tabaci (Tateda et al., 2008). Defense-related plant hormones salicylic acid, induced mRNA splicing of AtbZIP60 and OsbZIP50/OsbZIP74 (Hayashi et al., 2012; Lu et al., 2012; Moreno et al., 2012; Parra-Rojas et al., 2015), being the hallmark of IRE1-linking activation of an ER stress regulation defense responses in both Arabidopsis and rice (Oryza sativa L.). Abiotic stresses of heat and drought may also lead to splicing of ZmbZIP60 and TabZIP60, (Geng et al., 2018; Li et al., 2018) and BhbZIP60 mRNAs (Wang et al., 2017), respectively.
In this study, we characterized a gene designated TabZIP74 from common wheat (Triticum aestivum L.), encoding a homologous TF protein of AtbZIP60 or OsbZIP74. The results indicated that the mRNA sequence of TabZIP74 was spliced in the progress of Pst infection and drought stress, which encoded a nucleus-localized factor by frame shift. The spliced mRNA was also detected in stem nodes, roots, and stigmas during normal wheat development. Knocking down the spliced form of TaZIP74 increased the susceptibility level to stripe rust and decreased drought tolerance. Thus, TaZIP74 functions as a positive regulator for stripe rust infection and drought tolerance.
Materials and Methods
Plant Growth, Biotic Stress, and Chemical Treatments
Wheat near-isogenic lines (NILs) containing the resistant gene Yr10 (Taichung 29*6/Yr10) are resistant to some races of Pst in China, while its backcross parent Taichung 29 is highly susceptible (Wan et al., 2004; Chen et al., 2014). We constructed a full-length cDNA library of NIL Taichung 29*6/Yr10 infected with Pst races CYR34 (compatible race) and CYR17 (incompatible race). Wheat seedlings were grown in 8-cm pots and cultivated at 20°C under a 14 h/10 h day/night photoperiod cycle. Seven-day-old seedlings were inoculated with Pst races CYR34 and CYR17, or two-week-old wheat seedlings were treated with 5 µg ml–1 tunicamycin (TM, ER stress agent) for 4 h. Samples were collected at 0, 6, 12, 24, 36 and 48 h post-inoculation (hpi). To analyze the expression patterns of TabZIP74 under exogenous plant hormone application and drought stress, 10-day-old seedlings of Taichung 29*6/Yr10, cultured in fresh quarter-strength Hoagland solution, were treated with 0.1 mM abscisic acid (ABA) or 20% polyethylene glycol (PEG). Both the leaves and roots of treated plants were sampled at 0, 3, and 6 h post-treatment (hpt). Flag leaves, anthers, stigmas, stem internodes, stem nodes, and roots were sampled at the flowering stage (Feeks 10.5.1). All samples were immediately frozen in liquid nitrogen and stored at –80°C for RNA isolation.
Gene Expression Analysis
Total RNA of each wheat sample was extracted using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen, USA). The RNA was used to synthesize first-strand cDNA using a TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech). Reverse transcription (RT)-PCR was performed using TransTaq HiFi PCR SuperMix (TransGen Biotech) and detected by 1.5% agarose gel for gene-spliced assays (Wang et al., 2015). Two pairs of specific primers flanking the splice site were designed to distinguish unspliced (bzipSPassayf1/bzipSPassayr1) and spliced (bzipSPassayf1/bzipSPassayr2) forms of TabZIP74. Ta54227 transcripts of the AAA-superfamily of ATPases (Paolacci et al., 2009) and NbEF1 were used as controls in the semi-quantitative RT-PCR analyses of unspliced and spliced mRNA forms for expression in wheat or tobacco. Primer sequences are listed in Table 1.
Quantitative RT-PCR (qRT-PCR) was performed on the basis of reported by Wang et al., 2015, GoTaq® qPCR Master Mix (Promega) and ABI7500 Real-Time PCR System (Applied BioSystems) were used. Dissociation curves were generated to ensure specific amplification for each reaction. Each PCR reaction was performed three times. The threshold values (CT) were used to quantify relative gene expression using the comparative threshold (2-ΔΔCT) method (Schmittgen and Livak, 2008). Ta54227 transcripts of the AAA-superfamily of ATPases (Paolacci et al., 2009) were used as a control for the qRT-PCR analyses of the expression level of TabZIP74 in VIGS plants. Each experiment were performed three replicates. The statistic software SPSS 16.0 (SPSS Inc., USA, http://spss.en.softonic.com/) was used to assy significant differences using one-way ANOVA, taking P < 0.05 as significant according to Duncan’s multiple range test.
Sequence Analysis of TabZIP74
Gene sequences were analyzed on the basis of reported by Wang et al., 2015, mainly including DNAMAN software (Lynnon Biosoft, USA) and on line analysis by BLAST and ORF Finder on the NCBI website (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignments were deduced, and a phylogenetic tree generated using the neighbor-joining method with Clustal X version 2.0 (Larkin et al., 2007). The phylogenetic comparison of isolated full-length TabZIP74, reported bZIP proteins (Sornaraj et al., 2015) and those derived from GenBank were constructed from the neighbor-joining algorithm using MEGA7 program (Kumar et al., 2016), the bootstrap re-sampling analysis was performed with bootstrap trials = 1000. The sequence of TabZIP74 was blasted on the Ensembl Plants website (http://plants.ensembl.org/index.html) to get chromosome location of TabZIP74.
Subcellular Localization
To confirm subcellular localization of TabZIP74, fusion-plasmid expression vectors of TabZIP74-eGFP, TabZIP74-eGFPΔC, and eGFP-TabZIP74, containing either a complete or C-terminal truncated sequence of TabZIP74 cDNA, were constructed (Figure 3A). For TabZIP74-eGFP expression vector construction, the encoding region delete the stop codon was amplified by PCR with forward primer sequence TabZIP74subf (5’-TAGCATCCATGGACACCGACCTCGACCT-3’, BamH | site in italics) and reverse primer sequence TabZIP74subr (5’-TATCTAGACTAGCAAGCGGCAGCTGCA-3’, Xba | site in italics). For the C-terminal deleted sequence expression vector (TabZIP74-eGFPΔC) construction, the forward primer TabZIP74subf and reverse primer sequence TabZIP74ΔCsubr (5’-TATTCTAGACGAAAGTACGGCAGACTCCT-3’, Xba | site in italics) were used. To construct the eGFP-TabZIP74 expression vector, the encoding region delete the stop codon was amplified by high-fidelity DNA polymerase HIFI Taq (TransGen Biotech) using forward primer eG-bZIP74f (5’-TAGGATCCATGGACACCGACCTCGACCT-3’, BamH | site in italics) and reverse primer eG-bZIP74r (5’-ATTCTAGACTAGCAGCGGCAGCTGCA-3’, Xba | site in italics). The PCR product was cloned into the binary vectors with eGFP in front or in the back of inserted sequences to produce different fusion vectors. These fusion vector plasmids were introduced into A. tumefaciens strain GV3101. Tobacco epidermal cells were infiltrated with A. tumefaciens strain GV3101 containing a binary vector encoding GFP-fusion construct for transient expression. 24 h post incubation at 25°C, fluorescence of the GFP images of the transformed tobacco epidermal cells was observed with a confocal microscope (Zeiss LSM 880 Confocal Microscope).
Transcriptional Activation Analysis in Yeast
To investigate the transcriptional activity of TabZIP74, the complete open reading frame (ORF) and C-terminal deleted cDNA fragment of TabZIP74 were amplified using the primer combinations (for complete ORF primers: TF1 5’- ATAGTCGACATGGACACCGACCTCGAC -3’ and TR1 5’- TACTGCAGCTAGCAAGCGGCAGCTGCA -3’; for C-terminal deleted sequence primers: TF1 5’- ATAGTCGACATGGACACCGACCTCGAC -3’ and primer TR2 5’- TACTGCAGGTTCTGGCGCAGTGCCATGTT -3’, Pst I and Sal I sites in italics) and fused in the encoding region of GAL4 DNA-binding domain (GAL4-BD) in yeast expression vector pGBKT7, and vectors of pGBKT7-TabZIP74 and pGBKT7-TabZIP74ΔC were constructed for analyzing transcriptional activity of TabZIP74. The empty vector pGBKT7 was used as a negative control. All the vectors were transformed into yeast strain AH109. The different transformants were streaked on medium plates containing SD/Trp- (yeast synthetic drop-out medium supplement without tryptophan) (Clontech, USA) or SD/Trp-/His-/Ade- (yeast synthetic drop-out medium supplement without tryptophan, histidine, or adenine) (Clontech, USA). Incubation at 28°C for 3 d, then evaluated the growth status of the transformants.
Functional Analysis in Response to Pst Infection
Wheat Barley Stripe Mosaic Virus (BSMV)-induced gene silencing assay was conducted as described by Yuan et al. (2011). Specific sequences of wheat TaPDS (primer pairs: vTaPDSf, 5’-AAGGAAGTTTAACTGCATAAACGCTTAAAAG-3,’ and vTaPDSr, 5’-AACCACCACCACCGTTCTCCAGTTATTTGAG-3’, LIC adapters in italics), TabZIP74 (primer pairs: vTabZIP74f 5’-AAGGAAGTTTAACCAACCGAAGTCTGGTGGCT-3’ and vTabZIP74r, 5’-AACCACCACCACCGTCTAGCAAGCGGCAGCTGCA-3’, ligation-independent cloning (LIC) adapters in italics) were amplified and inserted into vector pCa-γbLIC. For gene function analysis with VIGS, two-week-old common wheat cultivar Fengchan 3 were planted in a growth chamber under a 16 h/8 h day/night photoperiod cycle at 16 ± 2°C. The second leaf surface was inoculated with Nicotiana benthamiana leaf sap containing BSMV particles (Mock, BSMV, BSMV-TabZIP74, BSMV-TabZIP74sp) by gently sliding pinched fingers from the leaf base to the tip. Treated with sterile water as Mock control. The inoculated seedlings were placed in a growth chamber in the dark at 60% humidity, 22 ± 2°C and kept under a 16 h/8 h day/night photoperiod. Nine d after virus inoculation, the phenotypes of wheat seedlings were observed and photographed. Fengchan 3 is highly susceptible to the virulent Pst race of CYR32, and its seedlings pre-inoculated with BSMV were successfully infected. Fresh urediniospores of stripe rust race CYR34 were inoculated onto the surface of the third leaves with a paintbrush 14 d after pre-inoculating with the virus. Three independent biological replications were performed for each treatment. The Pst infection phenotypes were recorded and photographed from 8 to 16 d post-inoculation (dpi). The latend period and number of uredinia on 6-cm long inoculated leaf fragments was recorded at 16 dpi.
Functional Analysis in Response to Drought
To examine whether TabZIP74 is involved in wheat responses to drought stress, the leaf relative water content (RWC) of Mock, BSMV, and BSMV-TabZIP74 plants was determined (Mao et al., 2012). Different virus-pretreated plants were subjected to drought stress by withholding water supply for 10 d, after 10 d rewatering. Development of the treated plants was routinely monitored by record the symptoms of leaf rolling and leaf RWC.
Measurement of Primary and Lateral Root Lengths
To evaluate the role of TabZIP74 in the wheat rooting pattern, five-day-old seedlings of Mock, BSMV, and BMSV-TabZIP74 were transferred to fresh quarter-strength Hoagland solution and grown vertically for 20 d at 22°C, under a 16 h/8 h day/night photoperiod. Photographs were recorded with a digital camera, lateral root number in the distal end of primary roots were determined by ImageJ software (http://rsbweb.nih.gov/ij/download.html). The number of lateral roots on 10 plants was calculated, and the mean of lateral roots was used to measure lateral root growth. Three biological replications were performed.
Results
Sequence Analysis of Putative TabZIP74
According to the EST sequence of a differentially expressed bZIP gene in responsing to stripe rust infection, the sequence was cloned from a cDNA library of NIL Taichung 29*6/Yr10. The 1,096- base-pair (bp) cDNA clone contains a 909-bp ORF, encoding a putative bZIP74 TF of 302 amino acid residues sharing high identity with MTFs such as OsbZIP74, ZmbZIP60, NtbZIP60, HvbZIP74, and SibZIP50 (Figure 1A). Furthermore, genomic sequence blast results indicated TabZIP74 is located on wheat chromosome 7D.
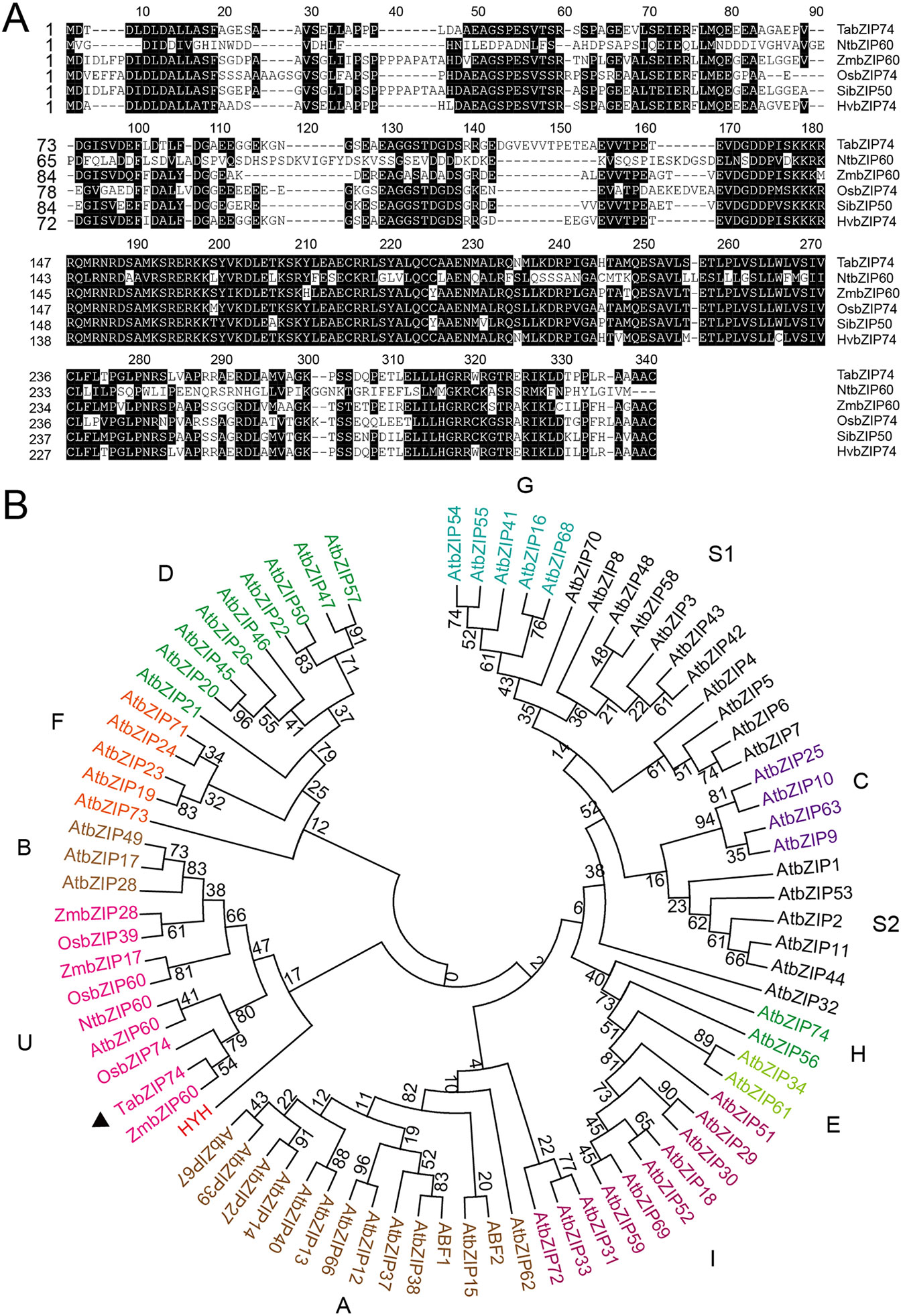
Figure 1 Phylogenetic tree of bZIP transcription factors and alignment of amino acid sequences of TabZIP74. (A) Amino acid alignment of TabZIP74 with other bZIP family members of NtbZIP60, OsbZIP74, ZmbZIP60, HvbZIP74, and SibZIP50 from tobacco (Nicotiana tabacum, NP_001311663.1), rice (Oryza sativa, XP_015641141.1), maize (Zea mays, NP_001147256.1), barley (Hordeum vulgare, BAJ96708.1), and millet (Setaria italica, XP_004965858.1), respectively. The numbers on the left indicate amino acid positions. Identical amino acid residues are shaded black, representing >50% similarity of conserved amino acids. (B) Phylogenetic tree of TabZIP74 with selected bZIP TFs from rice (OsbZIPs), Arabidopsis (AtbZIPs), tobacco (NtbZIPs), and maize (ZmbZIPs). Genbank accession numbers of the factors are listed in Supplementary Table 1. The subgroups were designated as A, B, C, D, E, F, G, H, S1, S2, and U, according to the analysis results (Jakoby et al., 2002; Zhou et al., 2017).
The phylogenetic tree constructed with TabZIP74 protein and other reported bZIP protein in Arabidopsis and rice indicated that different groups of bZIP factors were distinguished and named based on their phylogenetic relationships and functional divergence. The maximum likelihood analysis of bZIP proteins, including TabZIP74, identified 10 bZIP factors from rice, three from maize, one from tobacco and 73 from Arabidopsis in 11 distinct clades (A–I, S, and U), all of which had high bootstrap value support (Figure 1B).
The phylogenetic analysis revealed that TabZIP74 was most homologous to OsbZIP74 (LOC_Os06g41770) in the rice genome (Correa et al., 2008) and ZmbZIP60 in maize (Zea mays L.) (Figures 1A, B). OsbZIP74 also named as bZIP50 (Os06g0622700) in the Rice Annotation Project Database (Wakasa et al., 2011) and is an important ER stress regulator in rice (Lu et al., 2012).
TabZIP74 mRNA Splicing in Response to Biotic and Abiotic Factors
TabZIP74 has a high level of identity to OsbZIP74 with regard to amino acid sequences. Interestingly, the mRNA sequence of TabZIP74 can form two kissing hairpin loops through an RNA secondary structure prediction program Centroidfold (http://rtools.cbrc.jp/centroidfold/) (Hamada et al., 2009), and potentially produce its unspliced (TabZIP74-USP) and spliced (TabZIP74-SP) forms. The spacer between two hairpin loops in TabZIP74 comprises only two nucleotides, the same as OsbZIP74 (Figures 2A, B).
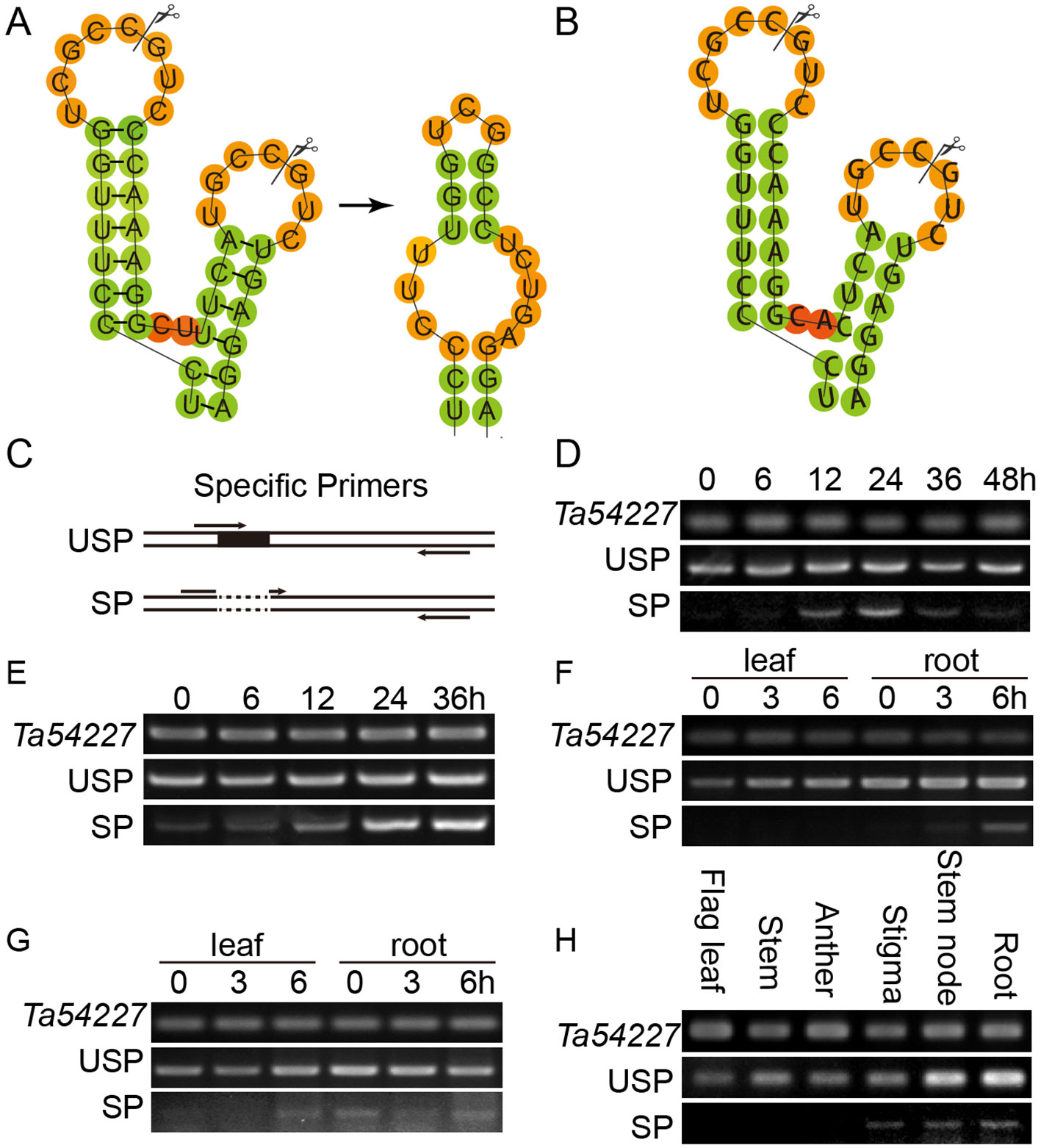
Figure 2 Splicing prediction and molecular assay of TabZIP74 in response to Pst infection, abiotic treatments, and development. (A, B) Predicted twin hairpin loop structures of unspliced (USP, left) and spliced (SP, right) forms of TabZIP74 (A) and its homolog of OsbZIP74 mRNA (B). Each structure contains two stems and two loops. The spliced and predicted cleavage sites are highlighted with scissors. Schematic representation of the USP and SP primers (C), Time-course experiments of the TabZIP74 splicing in response to infection by Pst with an avirulent race CYR17 (D) and a virulent race CYR32 (E). Splicing test in wheat seedling leaves and roots under PEG-induced drought stress (F) and ABA (G). (H) Splicing test in flag leaves, stems, anthers, stigmas, stem nodes, and roots of adult wheat plants.
Speicific primers flanking the predicted TabZIP74 splice sites was designed and used for RT-PCR analysis to assay the splicing of TabZIP74 (Figure 2C). When wheat seedling leaves were treated with 5 µg ml–1 tunicamycin (TM) for 4 h, one normal cDNA band and another smaller one were detected with RT-PCR in agarose gel electrophoresis. The sequencing result showed that a 20 bp fragment was spliced from the original TabZIP74 mRNA molecule in response to TM treatment (Figure 2A). The specific primers were also used to discriminate the unspliced (USP) and spliced (SP) forms of TabZIP74 after infection with Pst. When wheat seedlings were inoculated with two Pst races (CYR17, avirulent; CYR34, virulent), both induced TabZIP74 mRNA molecules for splicing (Figures 2D, E). In contrast, the abiotic factor PEG triggered TabZIP74 to be spliced in the roots of treated wheat seedlings (Figure 2F), while ABA induced splicing in both leaves and roots of stressed seedlings (Figure 2G). TabZIP74 mRNA splicing also occurred in stigmas, stem nodes, and roots of adult wheat plants (Figure 2H), but not in unstressed seedling leaves (Figures 2 D–F, H).
Subcellular Localization of TabZIP74
To investigate the subcellular localization of TabZIP74 in plant cells, different types of fusion protein with eGFP were constructed and these fusion vectors were transiently expressed in Nicotiana benthamiana leaf epidermal cells infiltrated with Agrobacterium tumefaciens cells of strain GV3101, containing a vector of TabZIP74-eGFP, TabZIP74-eGFPΔC or eGFP-TabZIP74 (Figure 3A). Confocal microscopic examination showed that the TabZIP74-GFP protein bound only to the plasma membrane and the TabZIP74-ΔC-GFP protein bound to the nuclei of tobacco epidermal cells, whereas eGFP-TabZIP74 was localized in both the plasma membrane and nucleus (Figure 3B). These results suggest that TabZIP74 is a membrane-bound and nucleus-localized protein.
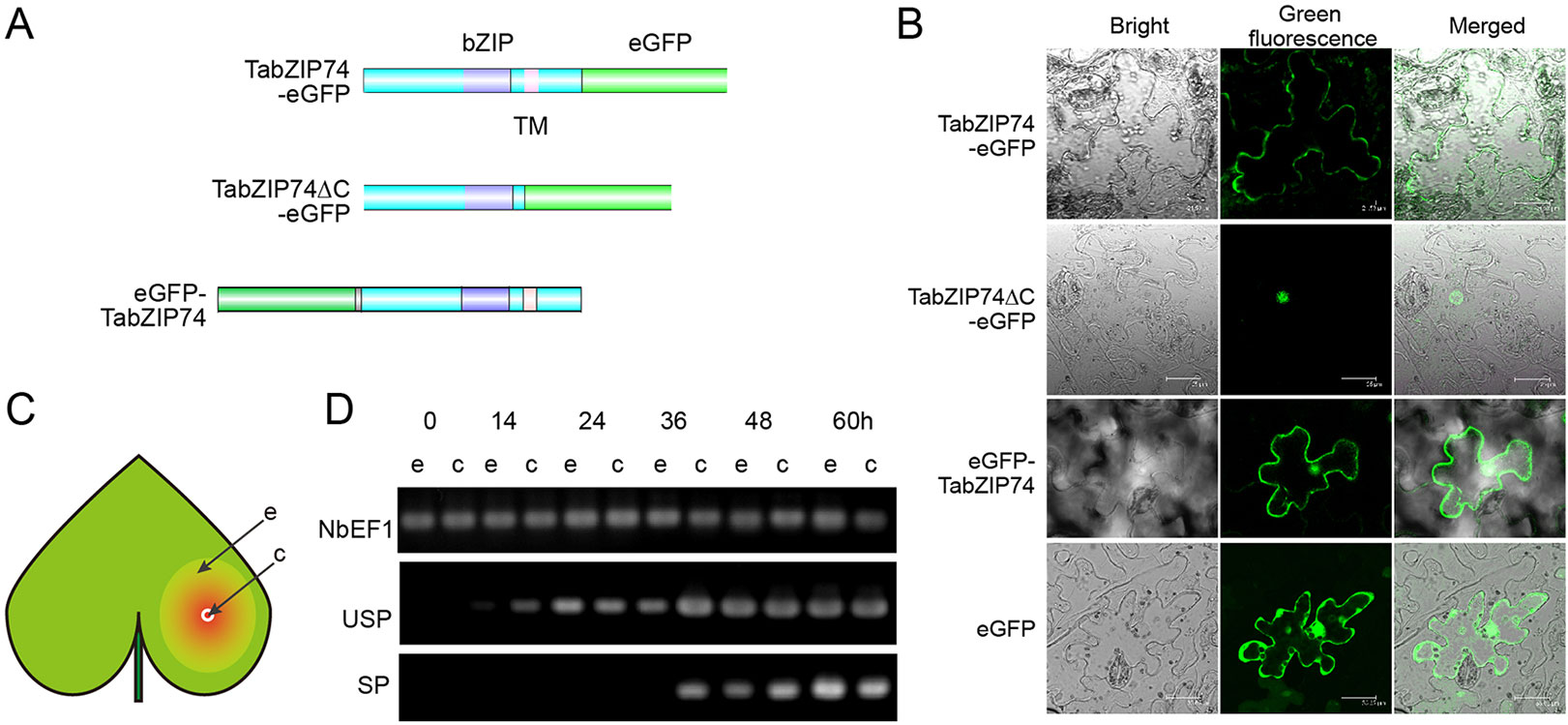
Figure 3 Subcellular localization of TabZIP74 and its mRNA splicing in tobacco leaf epidermal cells. (A) Plasmids encoding fusion proteins of eGFP with TabZIP74 were constructed, including two structures of full-length TabZIP74 fused to the N- or C-terminal of eGFP (top and bottom) and truncated TabZIP74(ΔC) (middle) fused to the N-terminal of eGFP protein. (B) Subcellular localization of TabZIP74 protein. (C) Schematic diagram of tobacco leaf infiltration. ‘c’ and ‘e’ represent the ‘center’ and the ‘edge’ of infiltrating area inoculated with A. tumefaciens cells containing GFP-fusion vectors, respectively. (D) mRNA splicing assay of TabZIP74 induced by wounding in tobacco leaf cells. USP and SP, unspliced, and spliced mRNA forms of TabZIP74. NbEF1 was used as control genes in the tobacco leaf RT-PCR analyses.
The mRNA levels of USP and SP forms of TabZIP74 in tobacco leaf epidermal cells infiltrated with the strain GV3101 with a fusion vector of eGFP-TabZIP74 was assayed by semi-RT-PCR. Fourteen hours after infiltrating, USP-type mRNA molecules of TabZIP74 were detected at the inoculation site center with a high expression level and at the edge of infiltrated leaves with a relatively lower level. The SP-type molecules of TabZIP74 were not detected at the inoculation site center until 36 h after infiltration, and no SP forms were found at the edge of the infiltrated area; both types of TabZIP74 mRNA were not detected at the inoculation site center or the edge of infiltrated leaves until 14 h after infiltration (Figures 3C, D). Therefore, wounding by infiltration possibly induces TabZIP74 mRNA splicing in tobacco cells, and the proteins encoded by the confusion gene eGFP-TabZIP74 were bound to the tobacco epidermal cell membrane and accumulated in the nuclei, while those encoded by TabZIP74-eGFP were only bound to the plasma membrane (Figure 3B).
Transcription Activity of TabZIP74
The full-length ORF and its truncated cDNA fragment without TMD (TabZIP74ΔC) were fused into the GAL4-DB in the vector pGBKT7, and the constructs were transformed into yeast strain AH109 cells to assay TabZIP74 transcriptional activity. The yeast transformant cells containing the fusion plasmids of full-length cDNA of TabZIP74, TabZIP74ΔC, and the vector control pGBKT7 grew well on selection SD medium plate without tryptophan (Trp−) (Figure 4). However, only the yeast cells encoding the fusion protein of TabZIP74ΔC grew well on the selection medium without tryptophan, histidine, and adenine (Trp−/His−/Ade−) (Figure 4), indicating that TabZIP74ΔC had transcriptional activity in yeast cells.
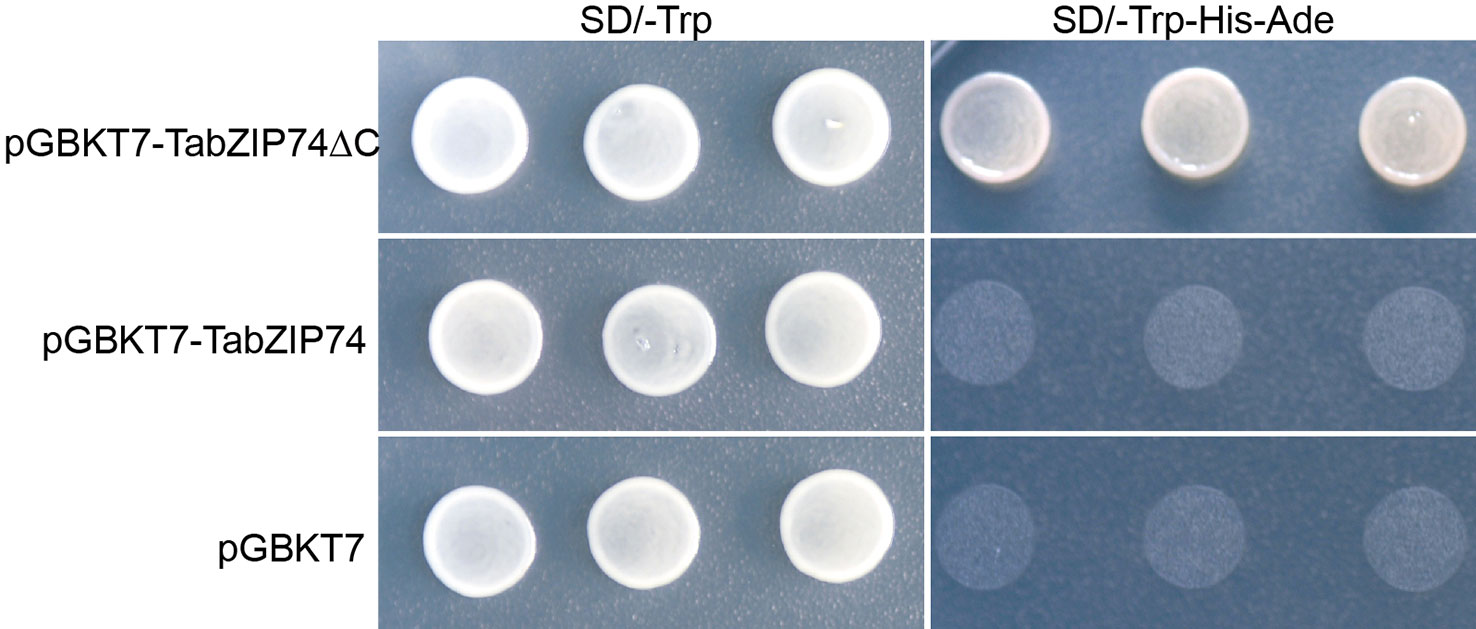
Figure 4 Transcription activity assay of TaZIP74 protein. The full-length encoding region and truncated cDNA fragment of TabZIP74 were fused with the GAL4 DNA-binding domain in vector pGBKT7. The plasmid containing the fusion gene and the empty vector control pGBKT7 were introduced into yeast cells of strain AH109. The yeast transformants were plated and incubated for 2 d at 28°C on SD/Trp− plate and SD/Trp−/His−/Ade− plate.
TabZIP74 Knockdown Plants Increased Susceptibility to Pst
To identify the regulatory roles of TabZIP74 in the wheat response to Pst infection, the specific C-terminal fragment of TabZIP74 was used to construct the BSMV-TabZIP74 fusion vector to silence TabZIP74 expression in wheat seedlings. Mild chlorotic mosaic symptoms were observed in the BSMV-inoculated plants 10 d post virus inoculated, and there was no obvious defects in further seedling leaf growth. Typical photobleaching was observed on the leaves of wheat seedlings pre-inoculated with BSMV-TaPDS (a specific fragment of wheat phytoene desaturase gene PDS) 14 d after virus inoculation, indicating the feasibility of the gene knockdown system applied in this study (Figure 5A). Interestingly, 11 dpi with the virulent Pst race of CYR 32, uredinium pustules erupted on the TabZIP74-knocked down seedling leaves of variety Fengchan 3, while no uredinia were visible on the Mock or vector control plants (Figure 5B). As a result, the latent period of Pst infection in silenced plants was significantly shorter than in the BSMV vector control and Mock seedlings (Figure 5C). Sixteen d post-inoculation, most Pst uredinia had matured and erupted from the seedling leaf surface (Figure 4B). There were no significant differences in the number of Pst uredinia on Mock, BSMV control and TabZIP74-knocked down seedling leaves. However, the TabZIP74-knocked down seedling leaves had longer uredinia than the Mock and BSMV control seedlings (Figure 5D). The expression level of TabZIP74 before Pst inoculation was ranked BSMV > Mock > TabZIP74-knocked down seedlings. At 48 hpi, no differences in expression level were detected between BSMV and Mock treatments, but the TabZIP74 expression level remained relatively low in its knockdown seedlings (Figure 5E).
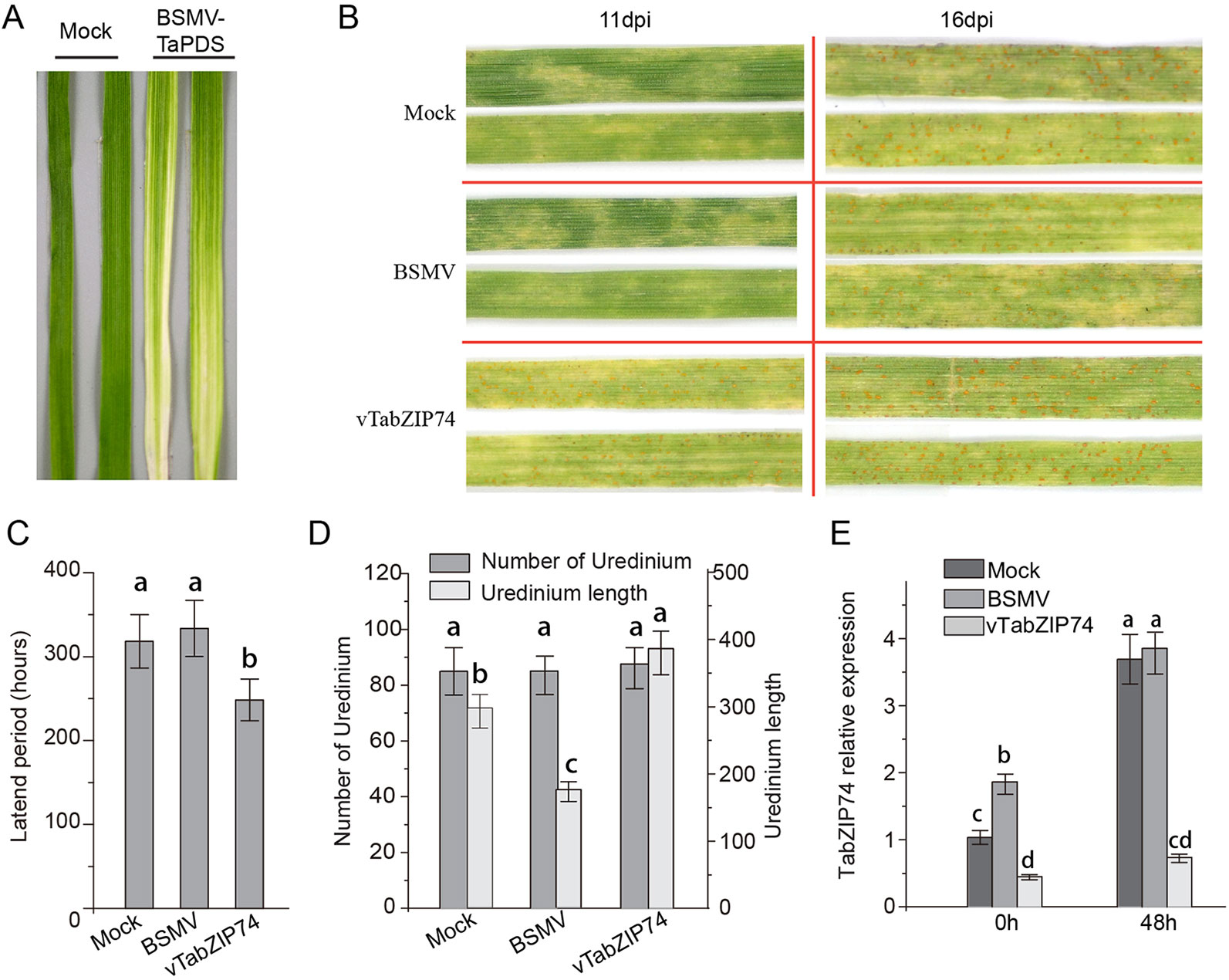
Figure 5 Functional characterization of TabZIP74 in response to Pst infection using the BSMV-VIGS system. (A) Third leaves of wheat seedlings of cultivar Fengchan 3 pre-inoculated with positive control vector (BSMV-TaPDS) at 14 d post-virus treatment (dpi). (B) Phenotypes for the third leaves inoculated with Pst race CYR32 were observed at 11 dpi and 16 dpi, respectively; the second leaves of these seedlings were pre-inoculated with water (Mock), empty BSMV vector and silencing vector of BSMV-vTabZIP74. (C) Pst latent infection period of different treatments. (D) Statistics of uredinium number and length on third leaves inoculated with Pst. Representative experiments were replicated three times (n = 3), at least 60 uredinia on 6-cm leaf segments from Mock, BSMV vector and BSMV-vTabZIP74 treated seedlings were counted in each replicate. Error bars indicate SD, letters indicate significant differences between Mock, BSMV, and BSMV-TabZIP74 samples determined by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range tests. (E) Silencing efficiency of TabZIP74 expression in knockdown leaves was determined by qRT-PCR; third leaves were sampled at 0 and 48 h after inoculation with BSMV virus. All experiments of TabZIP74 functions for Pst infection were replicated five times, and the results of three representatives were selected.
TabZIP74 Knockdown Plants Decreased Drought Tolerance and Lateral Roots
Leaf RWC is an important indicator of plant drought response, reflecting the water balance in leaf tissues. Wheat seedlings withholding water for 10 d to drought stress, and then rewatering for 3 d, and the RWC of seedling leaves was assayed at different drought stages. There was no notable phenotypical difference in leaf rolling or wilting between treatments, but the drought-stressed plants pre-silenced by BSMV-vTabZIP74 had lower RWC than Mock and BSMV control plants; 3 d after rewatering, RWC did not differ between treatments (Figure 6A). There were clear differences in primary root length of treated plants, namely, TabZIP74 > Mock > BSMV (Figure 6B). Plants knocked down with TabZIP74 developed longer primary roots but had less drought tolerance than Mock- and BSMV-treated plants. Furthermore, 9 d post-virus inoculation, TabZIP74-knockdown plants had significantly fewer lateral roots than BSMV-infected and Mock plants (Figures 6C, D).
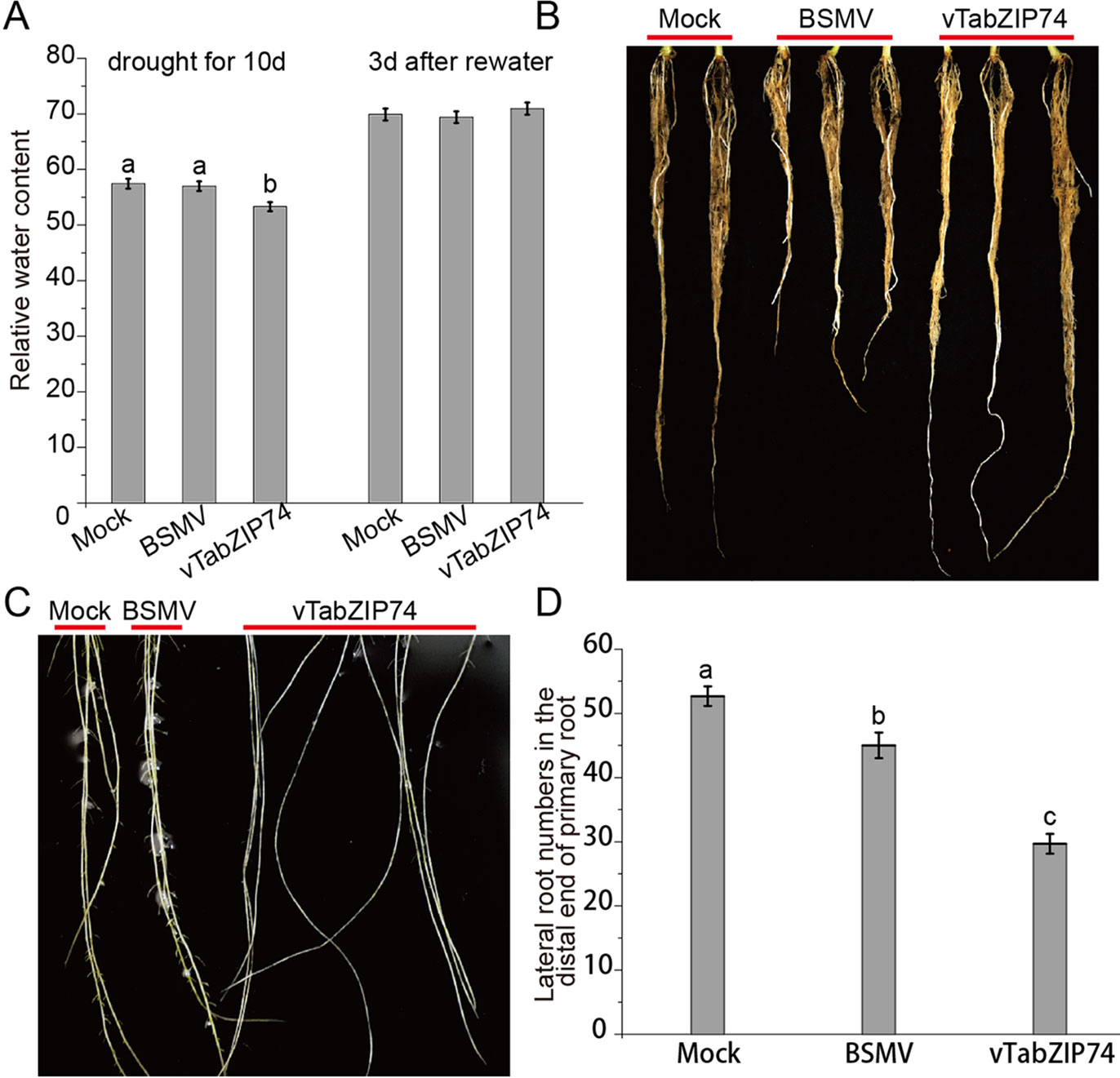
Figure 6 The effects of TabZIP74 silencing on wheat seedling drought tolerance and root development. (A) RWC of plants exposed to drought for 10 d and rewatered 3 d later. The experiments were performed in triplicate. Each value is the mean ± standard deviation of three independent measurements (SD, n = 3). Different letters indicate significant differences by one-way ANOVA, taking P < 0.05 level according to Duncan’s multiple range test for comparisons between treatment times. (B) Morphological differences in primary root development between Mock, BSMV, and BSMV-vTabZIP74 plants. (C) Morphological differences in lateral root development between Mock, BSMV, and BSMV-vTabZIP74 plants cultured in quarter-strength Hoagland solution for 20 d after sowing. (D) Lateral root numbers in the distal end of the primary root of Mock, BSMV, and BSMV-vTabZIP74 plants cultured in 10-cm pots for 20 d after sowing.
Discussion
Upon pathogen recognition, plants responsed rapid and complex immune responses. One type of plant defense response is the programmed burst in transcription and translation of pathogenesis-related proteins, most of which rely on ER processing (Korner et al., 2015). Two plant ER stress sensors, bZIP28 and IRE1, are involved in ER stress-induced signaling (Yoshida et al., 2001; Deng et al., 2011), but only IRE1 has been shown to operate in plant immune responses (Moreno et al., 2012). Plant bZIP TFs, such as AtbZIP60 and OsbZIP50/OsbZIP74, are involved in Regulated IRE1-Dependent Splicing (RIDS) in response to ER stress (Deng et al., 2011; Hayashi et al., 2012).
mRNA Splicing of TabZIP74 Was Initiated by Pst Infection and Wounding
TabZIP74 encodes an ORF consisting of 302 amino acids, beside bZIP DNA-binding domain there was a TMD in C terminal. The ER stress lead TabZIP74 mRNA splicing, and the spliced gene encode noval protein without TMD. The phylogenetic tree indicated that TabZIP74 belong to the subgroup U, most of its members have been characterized to regulate UPR. For example, ZmbZIP60 mRNA is spliced in maize in response to ER stress (Li et al., 2012), OsbZIP74 is an important ER stress regulator in rice (Lu et al., 2012), OsbZIP39 regulates the ER stress response in rice (Takahashi et al., 2012), and NtbZIP60 is an ER-localized transcription factor in tobacco (Tateda et al., 2008; Xu et al., 2013). Therefore, TabZIP74 may be involved in ER stress responses.
IRE1 has been proved as a dual protein kinase/RNase. The predicted structure of the IRE1 splicing site was based on two ‘kissing’ hairpin loops with conserved bases, and its predicted cleavage sites located close to the ribonuclease catalytic sites in its cytosolic domain (Lee et al., 2008). TabZIP74 shares high similarity with OsbZIP74 in the nucleotide sequence, neither of which had a higher sequence identity than the bZIP TF of yeast HAC1 or mammalian XBP1 at nucleotide or protein levels in response to ER stress (Kawahara et al., 1998; Yoshida et al., 2001). TabZIP74 and OsbZIP60 mRNAs, being like HAC1 and XBP1 mRNAs, can fold and form an IRE1 recognition site with two stem loops, each containing the bases remarkably conserved from yeast to mammals at three positions.
The splicing of TabZIP74 mRNA in wheat leaves or roots was evident from RT-PCR and sequencing after ER stress, Pst infection, or drought and ABA treatments. The mRNA splicing of TabZIP74 was also detected in tobacco leaves infiltrated with A. tumefaciens cells containing the GFP-TabZIP74 fusion vector. Consequently, the TabZIP74 mRNA sequence in infiltrated tobacco leaves may have been spliced by IRE1 under ER stress triggered by wounding.
TabZIP74 mRNA Splicing Produces the Active Form of Transcription Factor bZIP74
The full length of TabZIP74 was located on the membrane, and the truncated form of TaZIP74 can enter the nucleus (Figure 3A); OsbZIP74, AtbZIP60, ZmbZIP60, and NtbZIP60 showed similar results. In this research, we fused eGFP in the N terminus and found the splicing of TabZIP74 lead to new proteins entering the nucleus. The IRE1 splicing in mammals produces the active form of the transcription factor by adding the transcriptional activation domain in the newly formed longer C-terminus (Lu et al., 2012). In this research, the truncated but not full length of TabZIP74 acts as a transcriptional activator in yeast; similar results were observed in NtbZIP60. However, the TMD in OsbZIP74 and AtbZIP74 did not affect activation activity in yeast. So, the splicing of TabZIP74 produces the active form of transcription factor bZIP74.
BSMV Infection Enhanced the Expression Level of TabZIP74
Plant-infecting viruses can activate the ER stress signaling mechanism. Upon infection, viruses hijack cellular machinery to replicate their genomes and translate viral proteins (Korner et al., 2015). In Arabidopsis, when leaves were infected with potyvirus Turnip mosaic virus (Gaguancela et al., 2016) the spliced bZIP60 mRNA were accumulated. ER stress marker genes were also induced in N. benthamiana infected with potexvirus Potato virus X (PVX) and fijivirus Rice black-streaked dwarf virus (Ye et al., 2011; Sun et al., 2013). And it has proved that ER stress activation acts as a positive regulator of virus replication (Korner et al., 2015).
Our results showed that the infection of BSMV induced a higher expression level of TabZIP74 than Mock or BSMV-TabZIP74 (Figure 4F). Moreover, the stripe rust disease severity of BSMV-treated seedlings was lower than Mock and BSMV-TabZIP74 seedlings (Figure 4E). As such, the wheat seedlings pre-infected with BSMV may reduce the disease severity of stripe rust.
TabZIP74 Involved in Wheat Drought Tolerance and Development
In plants, UPR is provoked by a heavy demand in anther tapetal cells to synthesize and secrete pollen coat materials (Deng et al., 2016). Our study is also showed that UPR contributes to normal plant development. Under normal development conditions, low levels of the spliced form of TabZIP74 were detected in wheat stem nodes, roots, and stigmas. It is interesting to note that TabZIP74 knockdown seedlings developed fewer lateral roots, while other studies have found that IRE1a and IRE1b support root growth in ways other than the splicing of bZIP60 mRNA (Deng et al., 2016). The growth of single or double bZIP60 mutants in combination with either bZIP17 or bZIP28 appear to be normal in unstressed conditions (Kim et al., 2018; Bao et al., 2019). Biotic stress, ER stress-inducing agent, ABA treatment, and drought could induce TabZIP74 mRNA splicing. However, in rice, the ER stress-inducing agent and SA treatment induced the splicing of OsbZIP74, the homolog of TabZIP74, whereas stress hormone ABA and drought did not. So differences in biological function may exist between TabZIP74 and OsbZIP74.
The wheat root system includes the primary root, crown, and lateral roots, and root hairs. When TabZIP74 expression was silenced with VIGS, the wheat seedlings developed longer primary roots, but with significantly fewer lateral roots. This may be the reason why the TabZIP74 knockdown plants showed less drought tolerance (Figure 5). Besides, TabZIP74 knockdown plants also increased susceptibility to Pst infection. Water deficiency, pathogen infection, or stress-induced agent ABA usually cause the disorder of protein synthesis, degradation, and folding. In the endoplasmic reticulum, it is termed ER stress. Thus, it is particularly important to maintain protein stability in plant cells. ABA plays a major role in abiotic stress signaling, in particular in drought and salinity stress responses. ABA also has a pivotal role in the regulation of the plant immune signaling network (Pieterse et al., 2012). In Arabidopsis, ABA signaling antagonizes plant immunity by suppressing SA-dependent defenses. Boea hygrometrica bZIP transcription factor, BhbZIP60, is a splicing-activated ER stress regulator involved in drought tolerance. So, we propose that the splicing form of TabZIP74 is involved in the ABA pathway to respond to abiotic and biotic stresses.
In brief, TabZIP74 encodes a membrane-associated bZIP-type transcription factor. Based on the results presented in this study, we conclude that TabZIP74 might positively regulate wheat defenses against Pst and drought stress tolerance and is necessary for lateral root development.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
RL and WC designed the experiment. RL and FW wrote the manuscript. FW, YL, PW, JF, and SX performed the experiments and analyzed the data.
Funding
The study was financially supported by the National Key Research and Development Program of China (projects 2018YFD0200501, 2018YFD0200401) and National Natural Science Foundation of China (31871949).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Zejian Guo and Professor Dawei Li of the Chinese Agricultural University for providing the eGFP expression vector and vectors of BSMV-VIGS system.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01551/full#supplementary-material
Supplementary Material includes one table listing Locus ID of bZIP genes.
References
Bao, Y., Bassham, D. C., Howell, S. H. (2019). A functional unfolded protein response is required for normal vegetative development. Plant Physiol. 179 (4), 1834–1843. doi: 10.1104/pp.18.01261
Chen, W., Wellings, C., Chen, X., Kang, Z., Liu, T. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. Tritici. Mol. Plant Pathol. 15 (5), 433–446. doi: 10.1111/mpp.12116
Correa, L. G., Riano-Pachon, D. M., Schrago, C. G., dos Santos, R. V., Mueller-Roeber, B., Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PloS One 3 (8), e2944. doi: 10.1371/journal.pone.0002944
Deng, Y., Humbert, S., Liu, J. X., Srivastava, R., Rothstein, S. J., Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 (17), 7247–7252. doi: 10.1073/pnas.1102117108
Deng, Y., Srivastava, R., Quilichini, T. D., Dong, H., Bao, Y., Horner, H. T., et al. (2016). IRE1, a component of the unfolded protein response signaling pathway, protects pollen development in Arabidopsis from heat stress. Plant J. 88 (2), 193–204. doi: 10.1111/tpj.13239
Gaguancela, O. A., Zuniga, L. P., Arias, A. V., Halterman, D., Flores, F. J., Johansen, I. E., et al. (2016). The IRE1/bZIP60 pathway and Bax Inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol. Plant Microbe Interact. 29 (10), 750–766. doi: 10.1094/MPMI-07-16-0147-R
Geng, X., Zang, X., Li, H., Liu, Z., Zhao, A., Liu, J., et al. (2018). Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 274, 252–260. doi: 10.1016/j.plantsci.2018.05.029
Hamada, M., Kiryu, H., Sato, K., Mituyama, T., Asai, K. (2009). Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics 25 (4), 465–473. doi: 10.1093/bioinformatics/btn601
Hayashi, S., Wakasa, Y., Takahashi, H., Kawakatsu, T., Takaiwa, F. (2012). Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69 (6), 946–956. doi: 10.1111/j.1365-313X.2011.04844.x
Howell, S. H. (2013). Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 64, 477–499. doi: 10.1146/annurev-arplant-050312-120053
Iwata, Y., Koizumi, N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. U.S.A. 102 (14), 5280–5285. doi: 10.1073/pnas.0408941102
Jakoby, M., Weisshaar, B., Droge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 (3), 106–111. doi: 10.1016/s1360-1385(01)02223-3
Kawahara, T., Yanagi, H., Yura, T., Mori, K. (1998). Unconventional splicing of HAC1/ERN4 mRNA required for the unfolded protein response. Sequence-specific and non-sequential cleavage of the splice sites. J. Biol. Chem. 273 (3), 1802–1807. doi: 10.1074/jbc.273.3.1802
Kim, J. S., Yamaguchi-Shinozaki, K., Shinozaki, K. (2018). ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176 (3), 2221–2230. doi: 10.1104/pp.17.01414
Korner, C. J., Du, X., Vollmer, M. E., Pajerowska-Mukhtar, K. M. (2015). Endoplasmic reticulum stress signaling in plant immunity–At the crossroad of life and death. Int. J. Mol. Sci. 16 (11), 26582–26598. doi: 10.3390/ijms161125964
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0 . Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, K. P., Dey, M., Neculai, D., Cao, C., Dever, T. E., Sicheri, F. (2008). Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132 (1), 89–100. doi: 10.1016/j.cell.2007.10.057
Li, J., Chu, Z. H., Batoux, M., Nekrasov, V., Roux, M., Roux, M., et al. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106 (37), 15973–15978. doi: 10.1073/pnas.0905532106
Li, Y., Humbert, S., Howell, S. H. (2012). ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes 5, 144. doi: 10.1186/1756-0500-5-144
Li, Z., Srivastava, R., Tang, J., Zheng, Z., Howell, S. H. (2018). Cis-effects condition the induction of a major unfolded protein response factor, ZmbZIP60, in response to heat stress in maize. Front. Plant Sci. 9, 833. doi: 10.3389/fpls.2018.00833
Liu, J. X., Howell, S. H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22 (9), 2930–2942. doi: 10.1105/tpc.110.078154
Liu, J. X., Srivastava, R., Che, P., Howell, S. H. (2007a). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28 . Plant Cell 19 (12), 4111–4119. doi: 10.1105/tpc.106.050021
Liu, J. X., Srivastava, R., Che, P., Howell, S. H. (2007b). Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51 (5), 897–909. doi: 10.1111/j.1365-313X.2007.03195.x
Lu, D. P., Christopher, D. A. (2008). Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genomics 280 (3), 199–210. doi: 10.1007/s00438-008-0356-z
Lu, X., Tintor, N., Mentzel, T., Kombrink, E., Boller, T., Robatzek, S., et al. (2009). Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl. Acad. Sci. U. S. A. 106 (52), 22522–22527. doi: 10.1073/pnas.0907711106
Lu, S. J., Yang, Z. T., Sun, L., Sun, L., Song, Z. T., Liu, J. X. (2012). Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol. Plant 5 (2), 504–514. doi: 10.1093/mp/ssr115
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63 (8), 2933–2946. doi: 10.1093/jxb/err462
Moreno, A. A., Mukhtar, M. S., Blanco, F., Boatwright, J. L., Moreno, I., Jordan, M. R., et al. (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PloS One 7 (2), e31944. doi: 10.1371/journal.pone.0031944
Nekrasov, V., Li, J., Batoux, M., Roux, M., Chu, Z. H., Lacombe, S., et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28 (21), 3428–3438. doi: 10.1038/emboj.2009.262
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10, 11. doi: 10.1186/1471-2199-10-11
Parra-Rojas, J., Moreno, A. A., Mitina, I., Orellana, A. (2015). The dynamic of the splicing of bZIP60 and the proteins encoded by the spliced and unspliced mRNAs reveals some unique features during the activation of UPR in Arabidopsis thaliana. PloS One 10 (4), e0122936. doi: 10.1371/journal.pone.0122936
Pieterse, C. M., Van Der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Saijo, Y., Tintor, N., Lu, X., Rauf, P., Pajerowska-Mukhtar, K., Haweker, H., et al. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28 (21), 3439–3449. doi: 10.1038/emboj.2009.263
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3 (6), 1101–1108. doi: 10.1038/nprot.2008.73
Sornaraj, P., Luang, S., Lopato, S., Hrmova, M. (2015). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta. 180 (1), 46–56. doi: 10.1016/j.bbagen.2015.10.014
Sun, Z., Yang, D., Xie, L., Sun, L., Zhang, S., Zhu, Q., et al. (2013). Rice black-streaked dwarf virus P10 induces membranous structures at the ER and elicits the unfolded protein response in Nicotiana benthamiana. Virology 447 (1), 131–139. doi: 10.1016/j.virol.2013.09.001
Sun, L., Zhang, S. S., Lu, S. J., Liu, J. X. (2015). Site-1 protease cleavage site is important for the ER stress-induced activation of membrane-associated transcription factor bZIP28 in Arabidopsis. Sci. China Life Sci. 58 (3), 270–275. doi: 10.1007/s11427-015-4807-6
Tajima, H., Iwata, Y., Iwano, M., Takayama, S., Koizumi, N. (2008). Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem. Biophys. Res. Commun. 374 (2), 242–247. doi: 10.1016/j.bbrc.2008.07.021
Takahashi, H., Kawakatsu, T., Wakasa, Y., Hayashi, S., Takaiwa, F. (2012). A rice transmembrane bZIP transcription factor, OsbZIP39, regulates the endoplasmic reticulum stress response. Plant Cell Physiol. 53 (1), 144–153. doi: 10.1093/pcp/pcr157
Tateda, C., Ozaki, R., Onodera, Y., Takahashi, Y., Yamaguchi, K., Berberich, T., et al. (2008). NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J. Plant Res. 121 (6), 603–611. doi: 10.1007/s10265-008-0185-5
van Loon, L. C., Rep, M., Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Wakasa, Y., Yasuda, H., Oono, Y., Kawakatsu, T., Hirose, S., Takahashi, H., et al. (2011). Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 65 (5), 675–689. doi: 10.1111/j.1365-313X.2010.04453.x
Wan, A., Zhao, Z., Chen, X., He, Z., Jin, S., Jia, Q., et al. (2004). Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002 . Plant Dis. 88 (8), 896–904. doi: 10.1094/PDIS.2004.88.8.896
Wang, D., Weaver, N. D., Kesarwani, M., Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308 (5724), 1036–1040. doi: 10.1126/science.1108791
Wang, F., Lin, R., Feng, J., Chen, W., Qiu, D., Xu, S. (2015). TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front. Plant Sci. 6, 108. doi: 10.3389/fpls.2015.00108
Wang, B., Du, H., Zhang, Z., Xu, W., Deng, X. (2017). BhbZIP60 from resurrection plant Boea hygrometrica is an mRNA splicing-activated endoplasmic reticulum stress regulator involved in drought tolerance. Front. Plant Sci. 8, 245. doi: 10.3389/fpls.2017.00245
Xu, H., Xu, W., Xi, H., Ma, W., He, Z., Ma, M. (2013). The ER luminal binding protein (BiP) alleviates Cd(2+)-induced programmed cell death through endoplasmic reticulum stress-cell death signaling pathway in tobacco cells. J. Plant Physiol. 170 (16), 1434–1441. doi: 10.1016/j.jplph.2013.05.017
Yang, Y. G., Lv, W. T., Li, M. J., Wang, B., Sun, D. M., Deng, X. (2013). Maize membrane-bound transcription factor Zmbzip17 is a key regulator in the cross-talk of ER quality control and ABA signaling. Plant Cell Physiol. 54 (12), 2020–2033. doi: 10.1093/pcp/pct142
Ye, C., Dickman, M. B., Whitham, S. A., Payton, M., Verchot, J. (2011). The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156 (2), 741–755. doi: 10.1104/pp.111.174110
Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107 (7), 881–891. doi: 10.1016/s0092-8674(01)00611-0
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PloS One 6 (10), e26468. doi: 10.1371/journal.pone.0026468
Keywords: common wheat, Puccinia striiformis f. sp. tritici, bZIP transcription factor, endoplasmic reticulum stress, mRNA splicing, disease resistance
Citation: Wang F, Lin R, Li Y, Wang P, Feng J, Chen W and Xu S (2019) TabZIP74 Acts as a Positive Regulator in Wheat Stripe Rust Resistance and Involves Root Development by mRNA Splicing. Front. Plant Sci. 10:1551. doi: 10.3389/fpls.2019.01551
Received: 20 July 2019; Accepted: 06 November 2019;
Published: 27 November 2019.
Edited by:
Laura Bertini, Università degli Studi della Tuscia, ItalyReviewed by:
Lei Huang, Purdue University, United StatesUrmil Bansal, University of Sydney, Australia
Lili Huang, Northwest A&F University, China
Copyright © 2019 Wang, Lin, Li, Wang, Feng, Chen and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiming Lin bGlucnVpbWluZ0BjYWFzLmNu
 Fengtao Wang
Fengtao Wang Ruiming Lin1*
Ruiming Lin1* Wanquan Chen
Wanquan Chen