
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 December 2019
Sec. Plant Breeding
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01493
This article is part of the Research Topic Translational Research for Cucurbit Molecular Breeding: Traits, Markers, and Genes View all 22 articles
Fruit bitterness is a serious problem threatening the bottle gourd (Lagenaria siceraria [Mol.] Standl.) industry worldwide. Previous genetic studies indicated that fruit bitterness in the bottle gourd was controlled by a pair of complementary genes. In this study, based on two non-bitter landraces “Hangzhou Gourd” and “Puxian Gourd,” each of which carries a single bitterness gene, and their derived segregation populations, we mapped the complementary genes causing fruit bitterness. Quantitative trait locus (QTL) scanning based on an F2 population detected two QTLs, which was QBt.1 locating in a 17.62-cM interval on linkage group (LG)2 corresponding to a 1.6-Mb region on chromosome 6, and QBt.2 mapped to a 8.44-cM interval on LG9 corresponding to a 1.9-Mb region on chromosome 7. An advanced bulked segregant analysis (A-BSA) well validated the QTL mapping results. Sequence-based comparative analysis showed no syntenic relationship between QBt.1/QBt.2 and the known bitterness genes in cucumber, melon, and watermelon, suggesting that causal genes underlying QBt.1 and QBt.2 were not direct orthologs of the reported cucurbit bitterness genes. Our results shed light on the molecular genetic mechanisms underlying fruit bitterness in the bottle gourd and is useful to guide breeders to properly select parental lines to avoid the occurrence of bitter fruits in breeding programs.
Bottle gourd or calabash (Lagenaria siceraria [Mol.] Standl.) (2n = 2x = 22), a member of the genus Lagenaria of the Cucurbitaceae family (Beevy and Kuriachan, 1996), is recognized as indigenous to Africa and domesticated independently in Asia (Erickson et al., 2005). Having a cultivation history of over 8,000 years by man (Crawford, 1992), today, the bottle gourd is grown all over the tropics and subtropics for its immature fruits used as a vegetable or hard-shelled mature fruits used as containers, musical instruments, or handicrafts. The bottle gourd seedlings are widely used as a rootstock for grafting with watermelon to defend soil-borne diseases and to increase low-temperature tolerance (Davis et al., 2008; King et al., 2008).
While young fruits of bottle gourd are traditionally consumed as a delightful culinary vegetable in many areas of Asia including China, the undesirable occurrence of fruit bitterness in this crop as documented in the ancient medical books Ben Cao Jing Ji Zhu and Ben Cao Gang Mu has long been threatening the bottle gourd industry. Fruit bitterness not only affects the economic value of the bottle gourd but causes severe food poisoning symptoms such as nausea, vomiting, diarrhea, and abdominal cramps in humans and livestock (Zhang, 1981). Fruit bitterness appears to be a common trait to the Cucurbitaceae family, in which the compounds cucurbitacins having the function of defending against insects and herbivores cause the bitterness phenotype (Balkema-Boomstra et al., 2003). In cucumber, the effective cucurbitacin component is cucurbitacin C (CuC). It has been clear that nine genes are involved in the CuC biosynthetic pathway, and two genes, Bl (Bitter leaf) and Bt (Bitter fruit), regulate the bitterness/non-bitterness phenotype in leaves and fruits, respectively. The selection of natural mutations on Bi and Bt has played an important role in the domestication of ancient wild cucumber to form the present-day non-bitter cultivars (Shang et al., 2014). The major gene clusters for cucurbitacin biosynthesis were found to be highly conserved in cucumber, melon, and watermelon, while the regulatory genes seemed divergent among the three crops (Zhou et al., 2016).
Previous studies through traditional genetic analysis indicated that fruit bitterness in the bottle gourd was controlled by a pair of complementary genes (Zhang, 1981); however, the genome locations of the genes remain unknown. Genome-wide scan for quantitative trait loci (QTLs) is an effective approach for mapping traits governed by multi-genes including digenic genes (Zhang et al., 2013). Alternatively, Wu and Huang (2006) developed an advanced bulked segregant analysis (A-BSA hereafter) method to specifically identify DNA markers linked to two interactive genes. This method relies on the construction of a DNA pool mixed with homozygous genotypes (homo-pool) and another pool mixed from individuals with heterozygous genotypes showing contrasting phenotypes (heter-pool). By comparing marker genotypes among the parental lines and the two pools, DNA markers linked to the trait-determining genes as well as their parental allele origins can be inferred.
In the current study, we mapped the fruits bitterness genes in the bottle gourd genome and revealed their relationships with known bitterness genes in related food cucurbits. Our study also demonstrates the usefulness of combining QTL scanning and A-BSA as a fast and efficient approach to map and validate complementary genes controlling a trait.
Two bottle gourd landraces “Hangzhou Gourd” (HZ hereafter) and “Puxian Gourd” (PX hereafter), their F1 progenies, and F2 populations derived from selfing independent F1s in the years 2013, 2014, and 2016 were used in this study. The population size for the three F2 populations (2013F2, 2014F2, and 2016F2) were 102, 169, and 101, respectively. For A-BSA, 24 individuals showing a bitter-fruit phenotype from the 2013F2 population were screened for their F3 progenies phenotypes, and those showing no fruit bitterness segregation in F3 generation were selected to construct the homozygous bitter pool (homo-pool). Six F2 individuals with non-bitter fruits were randomly selected to construct a heter-pool (Figure 1). All plants were grown in tunnel houses in 30-m rows spaced 0.5 m apart. Ambient temperature and light as well as normal management were applied.
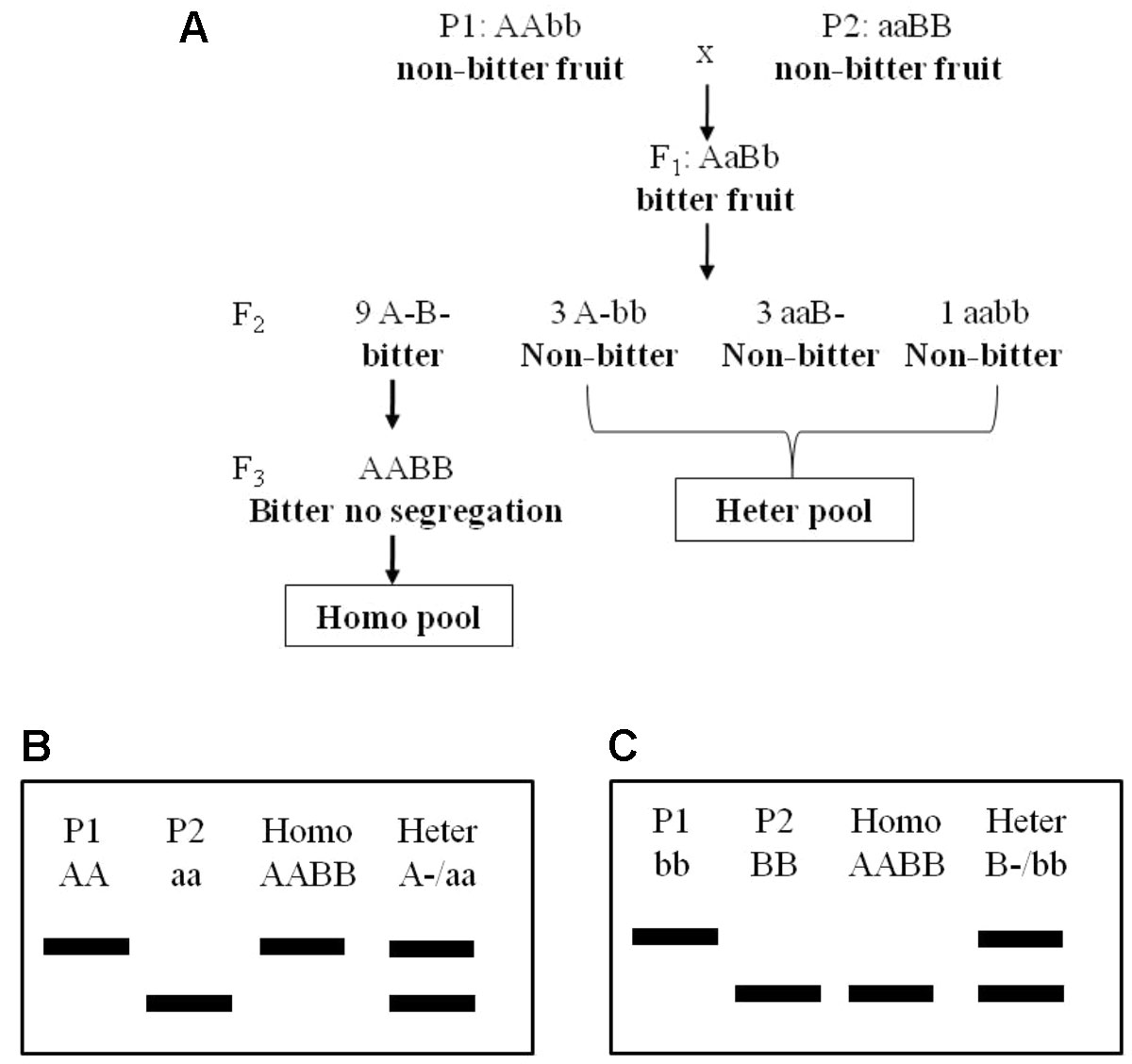
Figure 1 The genetic model of fruit bitterness in the F2 population (A) and advanced BSA method using in this study (B, C). BSA, bulked segregant analysis.
Fruit bitterness evaluation was conducted at the stage of 8–12 days after pollination. Bitterness phenotype of each individual was determined by manually tasting the sliced fruit sarcocarp by three trained tasters according to a classic tasting method described by Andeweg and DeBruyn (1959). The bitterness phenotype was scored as 1 for bitter and 0 for non-bitter. Only those results that were consistent among all three tasters were considered trustable and used in further analysis.
Genomic DNA was extracted from young leaves of 2-week-old seedlings using a DNA extraction kit (TIANGEN Co. Ltd, Beijing) following the manufacturer’s instructions. For single-nucleotide polymorphism (SNP) genotyping of the mapping populations, our previous RAD-Seq data from a set of bottle gourd accessions including HZ and PX were revisited (Wu et al., 2017a), from which 192 out of the 684 SNPs between the two parents that were evenly distributed in the genome were selected. Kompetitive allele-specific PCR (KASP) assays were used to genotype the mapping populations. KASP primers were designed using the Kraken™ software system (https://www.biosearchtech.com/support/tools/genotyping-software/kraken). Each KASP reaction was carried out in a final volume of 10 µl containing 20–40 ng of genomic DNA, 5 µl of 2× premade KASP master mix (LGC, Middlesex, UK), and 0.14 µl of primer mix. PCR amplification was performed in a Hydrocycler2 water bath thermal cycler following LGC parameters: 95°C, 15 min for hot-start Taq DNA polymerase activation, followed by a touchdown profile of 10 cycles at 94°C for 20 s and 61°C for 1 min with a 0.6°C reduction per cycle, and followed by 26 cycles at 94°C for 20 s and 55°C for 1 min. End-point fluorescent images were visualized using the BMG FLUOstar Omega (https://www.biosearchtech.com/products/instruments-and-consumables/genotyping-instruments/snpline-genotyping-automation/plate-reading), and allele calls for each genotype were obtained using the KlusterCaller™ software (LGC, UK).
A genetic linkage map for SNPs was constructed based on the 2014F2 population using the software QTL IciMapping (http://www.isbreeding.net). A likelihood of odds (LOD) threshold of 3.0 was used to determine the linkage groups (LGs), and an nnTwoOpt algorithm was used to determine the maker orders in each LG. The software MapQTL V5 (Van Ooijen, 2004) was used to detect bitterness QTL. Firstly, the interval mapping (IM) model was applied to detect QTLs for fruit bitterness, and then a multiple-QTL model (MQM) was used to scan for new QTLs with the markers closest to the original QTLs being implemented as cofactors. The mapping parameters were as follows: step size = 1.0, maximum number of neighboring markers = 5, maximum number of iterations = 200, and function tolerance = 1.0e−08. A genome-wide permutation test with 1,000 random permutations was conducted to obtain an empirical LOD score threshold for significance (P < 0.05).
The coding DNA sequence (CDS) of the bitterness genes Bt, Bi, and Bl from cucumber, melon, and watermelon (Shang et al., 2014; Zhou et al., 2016) were Blastn searched against the latest Hangzhou Gourd reference genome assembly V2.0 (Wang et al., 2018) to locate their syntenic region and orthologous genes in the bottle gourd genome with an e-value cutoff of 1e−10. The annotation of genes in the bottle gourd QTL intervals was retrieved from GourdBase (Wang et al., 2018).
Fruit bitterness assessment of parental lines, their F1 progenies, and F2 populations showed consistent phenotypes in the three biological replicates in 2013, 2014, and 2016. HZ and PX always showed a non-bitter phenotype, whereas their F1 progenies were always bitter. For each of the three F2 sub-populations, the bitter- and non-bitter-fruited individuals fit a 9:7 segregating ratio (Table 1), which suggested two complementary genes controlling this trait as previously reported in other cultivars of this species (Zhang, 1981).
Of the 192 intended KASP assays, 174 successfully detected signals and 153 called polymorphic genotypes in the F2 population in 2014. Based on these 153 SNPs data, a genetic linkage map containing 147 SNP loci distributed on 11 LGs were constructed. In line with the digenic mode of fruit bitterness inheritance in this population, two QTLs, designated as QBt.1 and QBt.2, were detected through IM followed by MQM (Table 2, Figure 2 and Supplementary Table 1). QBt.1 was located in a 17.62-cM interval between the SNP markers BGReSe_09031 and BGReSe_09068 on LG2, which explained 18% of the phenotype variance; QBt.2 was mapped to an 8.44-cM interval defined by the SNP markers BGReSe_11107 and BGReSe_11032 on LG9, which accounted for 27.7% of the phenotype variance. QTL epistatic interaction was detected between QBt. 1and QBt.2, and the phenotypic variation explained reached 99.32%, indicating that QBt.1and QBt.2 were the pair of complementary genes controlling fruit bitterness in this population.
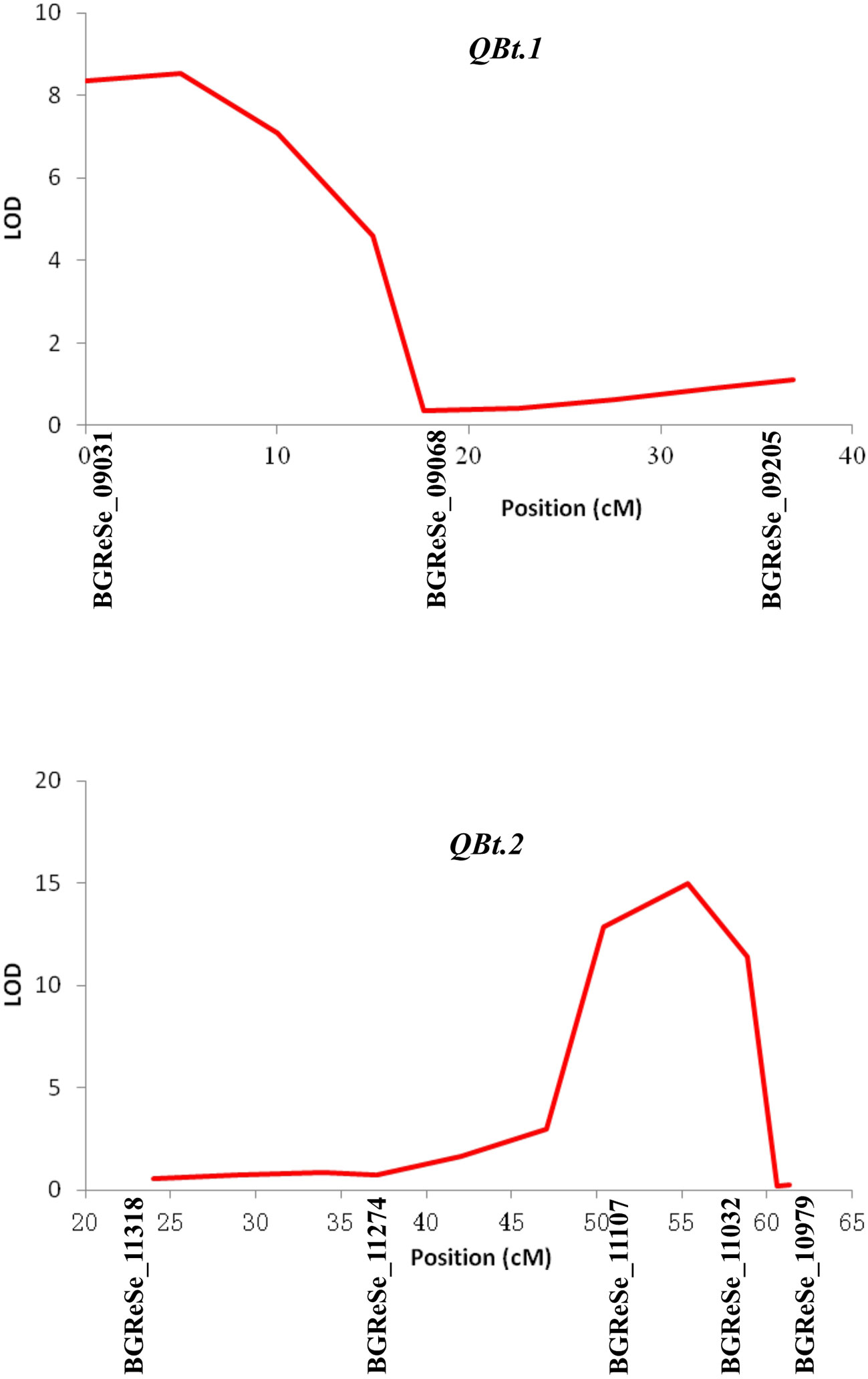
Figure 2 QTLs detected in the F2 population under a multiple-QTL model using MAPQTL V5. QTL, quantitative trait locus.
Another mapping approach, A-BSA, was also applied to screen for DNA markers linked to fruit bitterness. We screed 192 genome-wide distributed SNPs among the two parental lines, the homo-pool and the heter-pool, by using the KASP technology. It turned out that the marker BGReSe_09068 showed identical homozygous genotypes between HZ and the homo-pool, while it exhibited an alternative homozygous genotype in PX and, as expected, heterozygous genotype in the heter-pool (Table 3). Likewise, the marker BGReSe_11107 showed identical homozygous genotype in PX and the homo-pool but alternative homozygous genotype in HZ and heterozygous genotype in the heter-pool (Table 3). These results suggested that BGReSe_09068 and BGReSe_11107 each were linked to one of the complementary genes in HZ and PX, respectively. Coincidently, these two markers were found to fall into the QTLs intervals of QBt.1 and QBt.2, respectively. These results thus provide strong validation for the QTL mapping result and indicates that QBt.1 is one of the bitterness gene alleles from HZ and QBt.2 is the other from PX.
According to the physical locations of the QTL-flanking markers on the HZ reference genome V2.0, QBt.1 resides in a 1.62-Mb region (5,072,845–6,698,414 bp) on chromosome 6 with 142 predicted genes, and QBt.2 in a 1.89-Mb region (13,676,351–15,569,919 bp) containing 89 predicted genes on chromosome 7. To elucidate the relationship between the bottle gourd fruit bitterness QTLs and the known bitterness genes in related cucurbits, a cross-mapping analysis was performed. It showed that the cucumber bitterness genes CsBt and CsBi and the Bi regulator gene CsBl were syntenic to the genomic region on chromosome 6 in the bottle gourd (Figure 3, Supplementary Table 2). The four tissue-specific cucurbitacin regulator genes (CmBt and CmBr, ClBt and ClBr) in melon and watermelon were also found to be syntenic to the same region on chromosome 6 in bottle gourd except for CmBt. CmBi in melon and ClBi in watermelon found their orthologous genes in the distal region on chromosome 6 in bottle gourd (Figure 3, Supplementary Table 2). However, neither QBt.1 nor QBt.2 was located in or close to the syntenic region of these known bitterness genes. In addition, all the cucurbitacin biosynthetic genes in cucumber, melon, and watermelon found orthologs in the bottle gourd genome, but none of them were located in the QTL regions (Supplementary Table 2). These results suggest that the causal genes underlying QBt.1 and QBt.2 are unlikely to be direct orthologs of the bitterness genes in these related cucurbit crops.
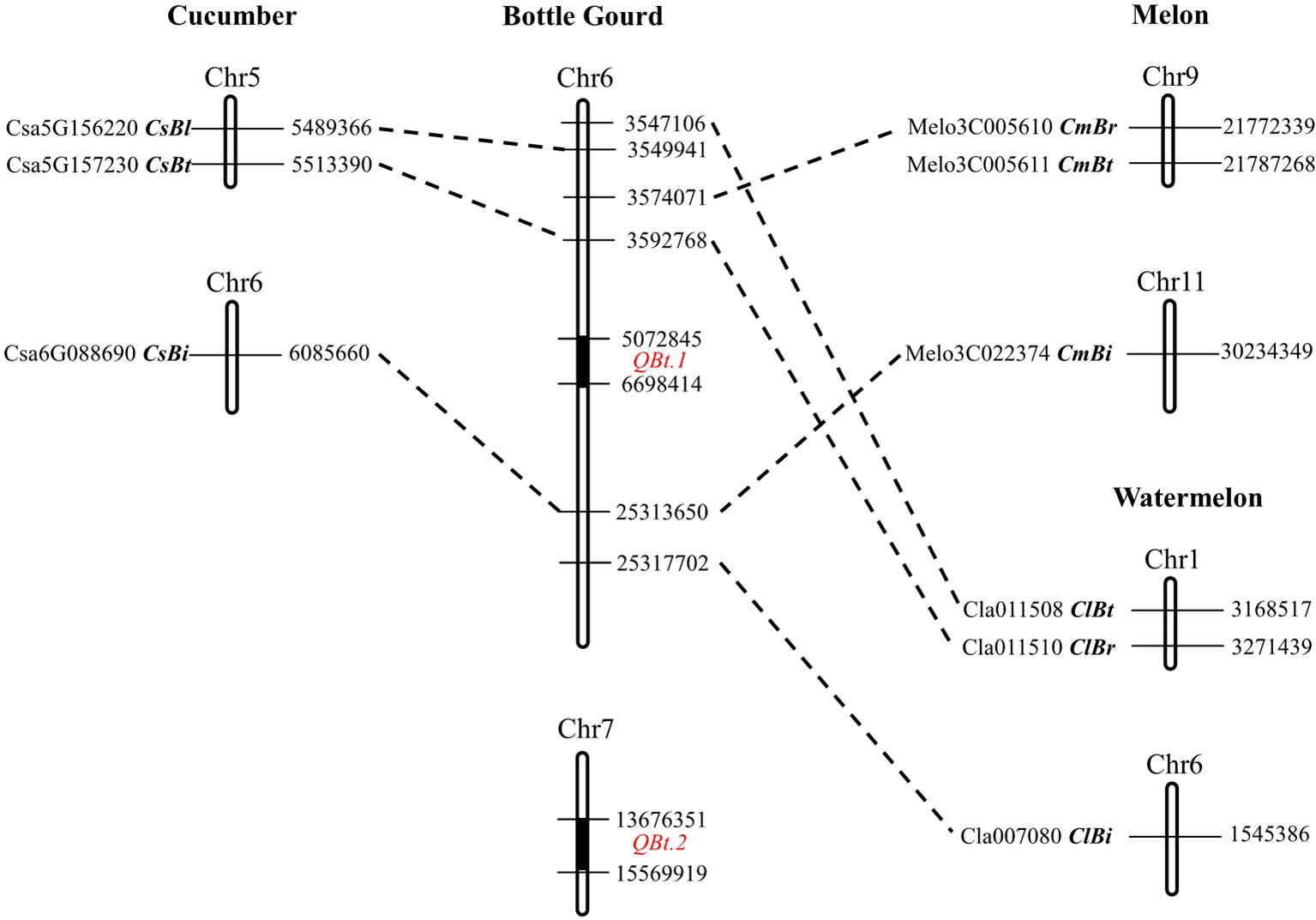
Figure 3 Genomic synteny of the bitterness genes between bottle gourd, cucumber, melon. and watermelon.
It has long been known that fruit bitterness in the bottle gourd is controlled by two interacting genes with complementary effect (Zhang, 1981). However, due to the lack of genomic resources, genetic mapping and elucidation of the bitterness genes in this species lag behind. Recently, plenty of SSR, Indel, and SNP markers information for the bottle gourd were released (Xu et al., 2011; Wu et al., 2017b; Wang et al., 2018), and reference genomes were available to the public (Xu et al., 2014; Wu et al., 2017c; Wang et al., 2018), allowing for more in-depth characterization of bitterness genes in this species. In this study, we initially mapped two QTLs, QBt.1 and QBt.2, each to a less than 2-Mb segment on chromosome 6 and chromosome 7, respectively. Then, by applying an advanced BSA method, we validated the QTL mapping results and elucidated that the functional alleles of QBt.1 and QBt.2 were from HZ and PX, respectively.
The ancestor wild cucumber, melon, watermelon, and bottle gourd plants all exhibited an extreme bitter phenotype in fruits (Zhou et al., 2016). Common bitterness compounds and their biosynthetic pathways were found in these related cucurbit crops (Lester, 1997; Matsuo et al., 1999; Chen et al., 2005). Likely due to common human demands for fruit quality, the domestication roadmap from extremely bitter wild ancestor to non-bitter modern cultivars seems to be also similar among cucumber, melon, and watermelon (Zhou et al., 2016). It has been argued that the causative mutations at orthologous genes may underlie convergent changes in fruit bitterness in cucurbits. For example, the loss of fruit bitterness during domestication in cucumber, melon, and watermelon all were caused by mutations on Bt genes (CsBt, ClBt, and CmBt) that are syntenic between genomes (Zhou et al., 2016). The cucumber CsBt gene encodes a basic helix–loop–helix (bHLH) transcription factor that activates the foliar bitterness gene Bi and regulates CuC biosynthesis in the fruit (Shang et al., 2014). According to the HZ reference genome V2.0, a gene HG_GLEAN_10009202 was found to be the putative orthologous gene of Bt; however, this gene does not fall into the QTL intervals of QBt.1 nor QBt.2. In addition, the bottle gourd orthologs of the cucumber foliar bitterness genes CsBi and CsBl, and the melon and watermelon root bitterness genes CmBr and ClBr, were all found to be not coincident with QBt.1 or QBt.2. Therefore, the causal genes underlying the bitterness QTLs in the bottle gourd are unlikely to be direct orthologs of the cucumber CsBt, CmBr, or ClBr gene. However, this result does not exclude the possibility that similar genes or same gene family members are the bitterness gene in the bottle gourd. Of the 89 predicted genes in the QBt.2 region, there is a transcription factor bHLH35-like isoform X1 gene and three cytochrome P450 CYP73A100-like genes, which might be functionally related to known cucurbit bitterness genes. More future work is required to draw a clearer picture on the causal genes underlying bitterness QTLs in the bottle gourd.
Our results shed new light on the molecular genetic mechanisms underlying fruit bitterness in the bottle gourd. From the perspective of breeding, it will be useful to guide the selection of parental lines to avoid the occurrence of bitter fruits in breeding programs. Breeders could use the flanking markers to QBt.1 and QBt.2 to screen their germplasm and breeding lines to predict if the hybrids would show a bitterness phenotype without the need of making real crosses.
All datasets generated for this study are included in the article/Supplementary Material.
XYW, PX and GL conceived and designed the research. XYW performed the experiments and wrote the manuscript. XHW constructed the population and collected bitterness phenotypes. YW, BW and ZL carried out the field work. All authors analyzed the data and read and approved the final manuscript. PX revised the manuscript.
This study was partially supported by the National Natural Science Foundation of China (31401880), the Major Science and Technology Project of Plant Breeding in Zhejiang Province (2016C02051), and the project from State Key Laboratory for Quality and Safety of Agroproducts (2010DS700124-ZZ1808).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01493/full#supplementary-material
SUPPLEMENTARY TABLE 1 | Genotype data and phenotype data for QTL mapping in the F2 population.
SUPPLEMENTARY TABLE 2 | Colinearity analysis of the cucurbitacin biosynthetic and regulatory genes in four cucurbits.
Andeweg, J. M., DeBruyn, J. W. (1959). Breeding of non-bitter cucumbers. Euphytica 8, 13–20. doi: 10.1007/BF00022084
Balkema-Boomstra, A. G., Zijlstra, S., Verstappen, F. W. (2003). Role of cucurbitacin C in resistance to spider mite (Tetranychus urticae) in cucumber (Cucumis sativus L.). J. Chem. Ecol. 29, 225–235. doi: 10.1023/A:1021945101308
Beevy, S. S., Kuriachan, P. (1996). Chromosome numbers of south Indian Cucurbitaceae and a note on the cytological evolution in the family. J. Cytol. Genet. 31, 65–71.
Chen, J. C., Chiu, M. H., Nie, R. L., Cordell, G. A., Qiu, S. X. (2005). Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat. Prod. Rep. 22, 386–399. doi: 10.1039/b418841c
Crawford, G. W. (1992).Origins of Agriculture: An International Perspective Vol. pp. Cowan, CW, Watson, PJ editors. Washington, DC: Smithsonian Institution Press, 7–38.
Davis, A. R., Perkins-Veazie, P., Sakata, Y., López-Galarza, S., Maroto, J. V., Lee, S., et al. (2008) Cucurbit grafting. CRC. Crit. Rev. Plant Sci. 27, 50–74. doi: 10.1080/07352680802053940
Erickson, L. D., Smith, D. B., Clarke, C. A., Sandweiss, H. D., Tuross, N. (2005). An Asian origin for a 10, 000-year-old domesticated plant in the Americas. Proc. Natl. Acad. Sci. U. S. A. 102, 18315–18320. doi: 10.1073/pnas.0509279102
King, S. R., Davis, A. R., Liu, W., Levi, A., King, S. R., Davis, A. R., et al. (2008). Grafting for disease resistance. HortScience 43, 1673–1676. doi: 10.21273/HORTSCI.43.6.1673
Lester, G. (1997). Melon (Cucumis melo L.) fruit nutritional quality and health functionality. HortTechnology 7, 222–227. doi: 10.21273/HORTTECH.7.3.222
Matsuo, K., DeMilo, A. B., Schroder, R. F. W., Martin, P. A. W. (1999). Rapid high-performance liquid chromatography method to quantitate elaterinide in juice and reconstituted residues from a bitter mutant of Hawkesbury watermelon. J. Agric. Food Chem. 47, 2755–2759. doi: 10.1021/jf9811572
Shang, Y., Ma, Y., Zhou, Y., Zhang, H., Duan, L., Chen, H. (2014). Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088. doi: 10.1126/science.1259215
Van Ooijen. (2004). MapQTL 5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen, Netherlands: Kyazma B.V.
Wang, Y., Xu, P., Wu, X., Wu, X., Wang, B., Huang, Y. (2018). GourdBase: a genome-centered multi-omics database for the bottle gourd (Lagenaria siceraria), an economically important cucurbit crop. Sci. Rep. 8, 3604. doi: 10.1038/s41598-018-22007-3
Wu, W., Huang, B. (2006). The strategy for mapping interaction genes using the bulked segregant analysis and Mapmaker/Exp software. Chin. Sci. Bull. 51 (18), 2134–2138. doi: 10.1007/s11434-006-2174-1
Wu, X., Xu, P., Wu, X., Wang, B., Lu, Z., Li, G. (2017a). Genome-wide association analysis of free glutamate content, a key factor conferring umami taste in the bottle gourd [Lagenaria siceraria (Mol.) Standl.]. Sci. Hortic. 225, 795–801. doi: 10.1016/j.scienta.2017.08.015
Wu, X., Xu, P., Wu, X., Wang, B., Lu, Z., Li, G. (2017b). Development of insertion and deletion markers for bottle gourd based on restriction site-associated DNA sequencing data. Hortic. Plant J. 3 (1), 13–16. doi: 10.1016/j.hpj.2017.01.011
Wu, S., Shamimuzzaman, M., Sun, H., Salse, J., Sui, X., Wilder, A. (2017c). The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 92, 963–975. doi: 10.1111/tpj.13722
Xu, P., Wu, X., Luo, J., Wang, B., Liu, Y., Ehlers, J. D., et al. (2011). Partial sequencing of the bottle gourd genome reveals markers useful for phylogenetic analysis and breeding. BMC Genomics 12, 467. doi: 10.1186/1471-2164-12-467
Xu, P., Xu, S., Wu, X., Tao, Y., Wang, B., Wang, S., et al. (2014). Population genomic analyses from low-coverage RAD-Seq data: a case study on the non-model cucurbit bottle gourd. Plant J. 77, 430–442. doi: 10.1111/tpj.12370
Zhang, S., Miao, H., Sun, R., Wang, X., Huang, S., Wehner, T. C., et al. (2013). Localization of a new gene for bitterness in cucumber. J. Hered. 104 (1), 134–139. doi: 10.1093/jhered/ess075
Zhang, G. (1981). Interaction of genes and the expression of bitterness in Lagenaria Sicereria. Acta Hortic. Sin. 8, 43–48.
Keywords: bottle gourd, fruit bitterness, complementary genes, genetic mapping, comparative analysis
Citation: Wu X, Wu X, Wang Y, Wang B, Lu Z, Xu P and Li G (2019) Molecular Genetic Mapping of Two Complementary Genes Underpinning Fruit Bitterness in the Bottle Gourd (Lagenaria siceraria [Mol.] Standl.). Front. Plant Sci. 10:1493. doi: 10.3389/fpls.2019.01493
Received: 15 June 2019; Accepted: 28 October 2019;
Published: 18 December 2019.
Edited by:
Feishi Luan, Northeast Agricultural University, ChinaReviewed by:
Jinfeng Chen, Nanjing Agricultural University, ChinaCopyright © 2019 Wu, Wu, Wang, Wang, Lu, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guojing Li, ligj@zaas.ac.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.