- 1Temasek Life Sciences Laboratory, National University of Singapore, Singapore, Singapore
- 2Department of Biological Sciences, Faculty of Science, National University of Singapore, Singapore, Singapore
The orchid family (Orchidaceae) represents the second largest angiosperm family, having over 900 genera and 27,000 species in almost all over the world. Orchids have evolved a myriad of intriguing ways in order to survive extreme weather conditions, acquire nutrients, and attract pollinators for reproduction. The family of MADS-box transcriptional factors have been shown to be involved in the control of many developmental processes and responses to environmental stresses in eukaryotes. Several findings in different orchid species have elucidated that MADS-box genes play critical roles in the orchid growth and development. An in-depth understanding of their ecological adaptation will help to generate more interest among breeders and produce novel varieties for the floriculture industry. In this review, we summarize recent findings of MADS-box transcription factors in regulating various growth and developmental processes in orchids, in particular, the floral transition and floral patterning. We further discuss the prospects for the future directions in light of new genome resources and gene editing technologies that could be applied in orchid research and breeding.
Introduction
The orchid family (Orchidaceae) is currently the second largest angiosperm family, having over 900 genera and 27,000 species in almost all parts of the world except Antarctica. New genera in the orchid family are being discovered at a rate of around 13 per year for over the past decade (Schuiteman, 2004; Chase et al., 2015). Nevertheless, many wild species are at the brink of extinction because of illegal trading activities (Wijnstekers, 2001; Hossain et al., 2013). Orchids have their own ecological niches through their relationships with mycorrhizal fungi, specialized pollinators and host trees (Fay and Chase, 2009). Of all the orchid species, 70% are epiphytic (growing on trees), 25% are terrestrial (growing on ground), and the remaining 5% are found on various supports such as rocks (Atwood, 1986). Thus, it is important to conserve orchid species through generating awareness and increasing our understanding on the species physiology and diversity (Cribb et al., 2003).
Being a class of valuable ornamental plants with distinct and attractive flowers, orchid is viewed as a high value commodity in the global flower cultivation and landscaping industries. They are also highly sought for as food and traditional medicine (Arditti, 1992; Bulpitt et al., 2007). Besides their great economic values, orchids are also exclusive genetic resources for studying plant developmental processes, including floral transition, floral development, flower pigmentation, and senescence, because of the specialized reproductive structures and the unique strategies for reproduction (Yu and Goh, 2001; Gutiérrez, 2010; Da Silva et al., 2014). However, the orchid research as well as orchid breeding have been challenging due to the long vegetative developmental period before switching to flower development, and technical limitations in transformation and obtaining transgenic lines in various orchid species. Currently, to investigate the function of orchid genes, several methods are being used including the heterologous expression of gene of interest under the strong constitutive CaMV 35S promoter in Arabidopsis (Arabidopsis thaliana) or tobacco and virus-induced gene silencing (VIGS). Sometimes, transgenic orchids are also generated to study gene function. So far, the genetic transformation of orchids using the Agrobacterium-mediated approach on protocorm-like bodies or rhizomes has been reported in Cymbidium, Oncidium, Dendrobium, Phalaenopsis, and other orchids (Belarmino and Mii, 2000; Yu and Goh, 2000; Chai et al., 2002; Chen, 2002; Sjahril and Mii, 2006; Shrestha et al., 2007; Zhang et al., 2010; Ding et al., 2013).
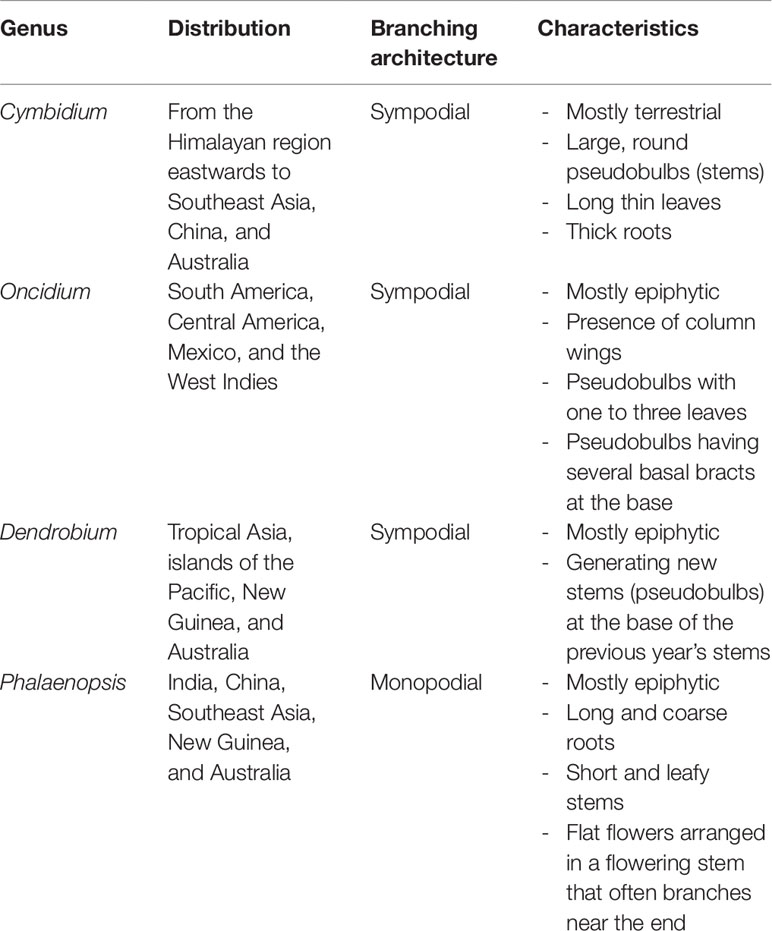
The orchids including Phalaenopsis, Dendrobium, Cymbidium, and Oncidium from the Epidendroideae subfamily are used as orchid plant models for research and biotechnology. Most of these orchids are predominantly found to be growing in tropical Asia to Australia with the exception of Oncidium in the West (Table 1). Orchids are either monopodial or sympodial in their growing habits. Monopodial orchids, such as Phalaenopsis, grow as a single erect “stem” with alternating leaves on opposing parts of the center. They store water in their thick leaves and roots but have no pseudobulbs. Sympodial orchids, such as Dendrobium, Cymbidium, and Oncidium, grow from a horizontal stem called rhizome and have pseudobulbs to store water and grow new leaves. After blooming, the plant will resume growth at axillary buds at the base of the previous pseudobulbs.
Recent findings from different orchid species have elucidated that a group of MADS-box transcription factors sharing a greatly conserved N-terminal DNA binding domain (MADS-box) exert important functions in controlling orchid growth and development, in particular, the floral transition and floral patterning. In this review, we discuss the biological roles of these MADS-box proteins and the mechanisms how they contribute to flowering and floral organ formation in orchids. We further elaborate about the prospects for research and development in light of new genome resources and gene editing technologies that could be applied in orchid research and breeding.
The Mads-Box Protein Family
In orchids and other angiosperms, there is a family of MADS-box transcription factors that have been identified to control many plant developmental processes, including floral transition, floral patterning, as well as male and female gametophyte development (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Yu and Goh, 2000; Acri-Nunes-Miranda and Mondragón-Palomino, 2014; Valoroso et al., 2019). The MADS-box family proteins are conserved in nearly all eukaryotes. The MADS-box acronym is derived from the yeast MINICHROMOSOME MAINTENANCE 1 (MCM1) (Passmore et al., 1988), the Arabidopsis AGAMOUS (AG) (Yanofsky et al., 1990), the Antirrhinum DEFICIENS (DEFA) (Schwarz-Sommer et al., 1990), and the mammalian SERUM RESPONSE FACTOR (SRF) (Norman et al., 1988; Gramzow et al., 2010). All identified MADS-box proteins each contain a MADS-box domain of ∼58 amino acid at the N-terminus that binds to a consensus CC[A/T]6GG sequence, termed as the “CarG-box” motif (Hayes et al., 1988; Riechmann et al., 1996). Interestingly, flowering plants (angiosperms) have more of these genes (e.g. 107 in Arabidopsis; 51 in Phalaenopsis equestris) compared to yeast (e.g. 4 in Saccharomyces cerevisiae) and mammals (e.g. 5 in Homo sapiens) (Becker and Theißen, 2003; Messenguy and Dubois, 2003; Par˘enicová et al., 2003; Cai et al., 2015). The MADS-box family of genes form two major lineages, namely type I of SRF-like genes and type II of MEF2-like genes, which is resulted from an ancient event of gene duplication prior to the divergence of the kingdoms of plants and animals (Alvarez-Buylla et al., 2000). In plants, type II genes of MADS-box, also called as MIKC-type genes, feature four distinct protein domains arranged from the N-terminal end to C-terminal end. They are the highly conserved DNA binding MADS-box (M) domain, the less-conserved intervening domain (I) for conferring interaction specificity between different MADS-box transcription factors and/or other proteins, the keratin-like coiled-coil (K) domain for conferring protein–protein interactions, and a highly variable C-terminal (C) domain for regulating gene transcription or multimeric protein complexes formations (Shore and Sharrocks, 1995; Theißen et al., 1996; Riechmann and Meyerowitz, 1997; Honma and Goto, 2001; Becker and Theißen, 2003; Hill et al., 2008).
The MIKC-type genes are specifically present in plants and many of these genes have been shown to control key processes of plant development including vegetative growth and reproductive organ development with complex cascades of events and networks (Theißen, 2001; Kaufmann et al., 2005; Adamczyk and Fernandez, 2009). Particularly, their functions in determining plant reproductive development are more remarkable as they regulate the development of consecutive reproductive processes, namely the floral transition, floral meristem specification, floral patterning, pollen growth, and development of ovules and seeds (Figure 1). The MADS-box genes that act in the regulation of flowering time and floral patterning in plants will be elaborated more in the latter sections (Table 2). The floral homeotic genes that play crucial roles in specifying reproductive floral organ identities are among the best characterized MADS-box genes. The extensive study of mutants with floral homeotic defects has resulted in the birth of the “ABCE model,” which explains how the genes of A, B, C, D and E classes act jointly to determine floral organs identities (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Theissen and Saedler, 2001). All floral homeotic genes in Arabidopsis belong to the MADS-box family except for the A-class gene, APETALA2 (AP2). In addition to determining the identities of floral organs, MADS-box proteins also regulate floral meristem specification, the process of which involves four meristem identity genes, namely LEAFY (LFY) and three MADS-box genes closely related to each other, APETALA1 (AP1), CAULIFLOWER (CAL), and FRUITFULL (FUL) (Ferrándiz et al., 2000; Ng and Yanofsky, 2001). Furthermore, studies of MADS-box proteins have also demonstrated their function in seed and silique growth. For example, three MADS-box genes, SHATTERPROOF 1 (SHP1), and its close homologs SHP2 and SEEDSTICK (STK) contribute to normal growth and development of carpels and fruits (Liljegren et al., 2000; Pinyopich et al., 2003). In addition, several MADS-box genes belonging to type I, such as PHERES 1 (PHE1), AGAMOUS-LIKE 80 (AGL80), DIANA (AGL61), etc., are involved in embryo and seed growth (Kohler et al., 2003; Portereiko et al., 2006; Bemer et al., 2008; Colombo et al., 2008; Kang et al., 2008).

Table 2 A summary of MADS-box regulators involved in reproductive development in the model plant Arabidopsis and orchids.
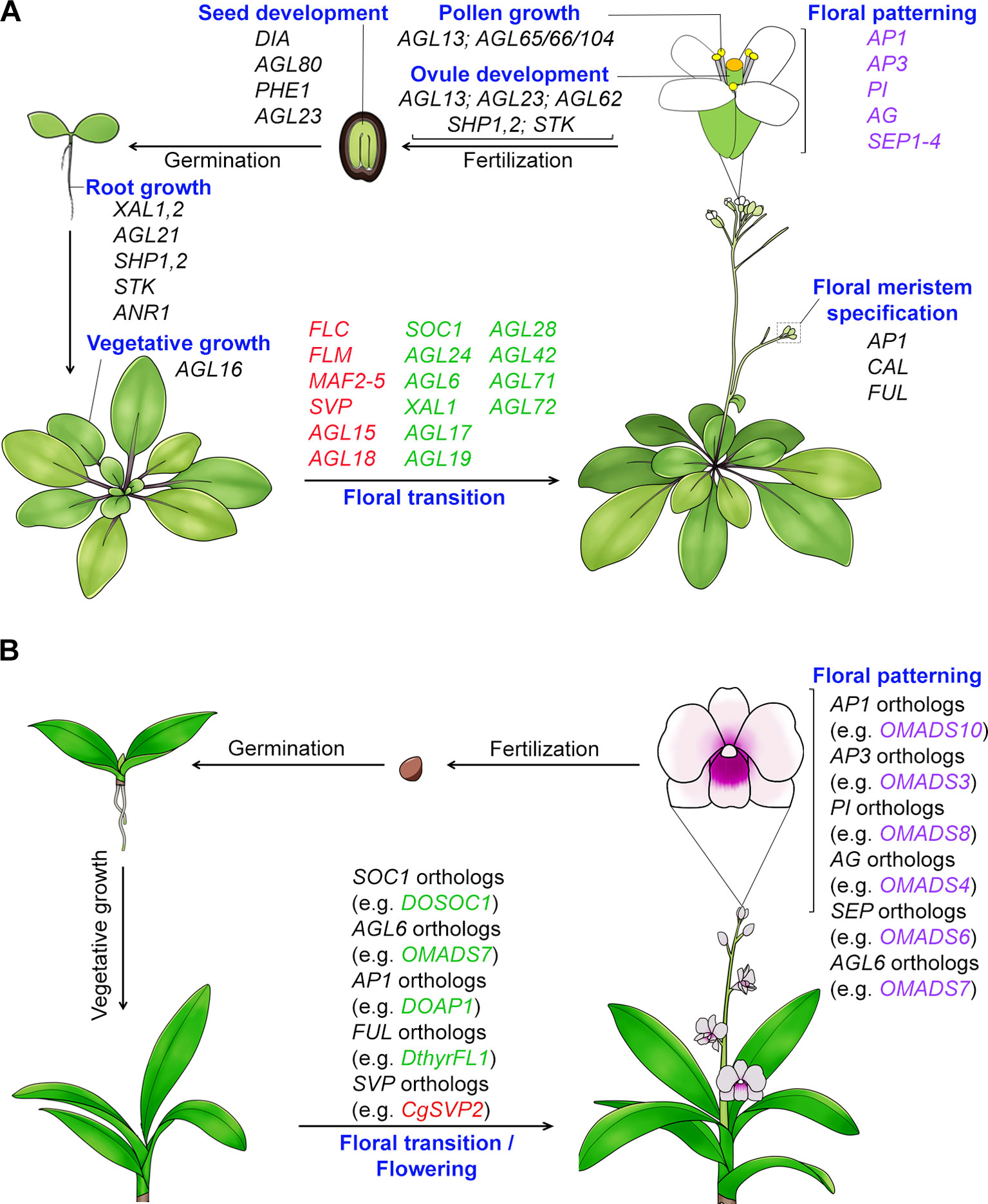
Figure 1 Function of MADS-box proteins in the whole plant life cycle. (A) MADS-box genes regulate Arabidopsis development throughout its life cycle. Many MADS-box genes mediate the transition to flowering. The flowering time repressor genes, including FLC (Michaels and Amasino, 1999), FLM (Ratcliffe et al., 2001), MAF2-5 (Ratcliffe et al., 2003; Gu et al., 2013), SVP (Hartmann et al., 2000; Li et al., 2008), and AGL15/18 (Adamczyk et al., 2007), are shown in red color, whereas the flowering time promoter genes, including SOC1 (Lee et al., 2000), AGL24 (Yu et al., 2002), AGL6 (Yoo et al., 2011), XAL1/AGL12 (Tapia-Lopez et al., 2008), AGL17 (Han et al., 2008), AGL19 (Schonrock et al., 2006), AGL28 (Yoo et al., 2006), and AGL42/71/72 (Dorca-Fornell et al., 2011), are shown in green color. All the identified floral organ identity genes except AP2 encode MADS-box transcription factors. MADS-box genes are also involved in root growth (e.g. XAL1, XAL2, AGL21, ANR1, SHP1,2, and STK) (Zhang and Forde, 1998; Tapia-Lopez et al., 2008; Moreno-Risueno et al., 2010; Garay-Arroyo et al., 2013; Yu et al., 2014), vegetative growth (e.g. AGL16’s function in stomata development) (Kutter et al., 2007), pollen maturation and tube growth (AGL65/66/104) (Adamczyk and Fernandez, 2009), ovule development (e.g. AGL13, AGL23, AGL62, SHP1,2 and STK) (Liljegren et al., 2000; Pinyopich et al., 2003; Colombo et al., 2008; Kang et al., 2008; Hsu et al., 2014), and embryo and seed development (e.g. DIA, AGL80, AGL23, and PHE1) (Kohler et al., 2003; Portereiko et al., 2006; Bemer et al., 2008; Colombo et al., 2008). (B) Functions of MADS-box genes in orchid development. Orchid MADS-box proteins have been shown to regulate flowering and floral organ formation. AG, AGAMOUS; AGL6, AGAMOUS-LIKE 6; AGL15, AGAMOUS-LIKE 15; AGL16, AGAMOUS-LIKE 16; AGL17, AGAMOUS-LIKE 17; AGL18, AGAMOUS-LIKE 18; AGL19, AGAMOUS-LIKE 19; AGL21, AGAMOUS-LIKE 21; AGL23, AGAMOUS-LIKE 23; AGL24, AGAMOUS-LIKE 24; AGL28, AGAMOUS-LIKE 28; AGL42, AGAMOUS-LIKE 42; AGL65, AGAMOUS-LIKE 65; AGL66, AGAMOUS-LIKE 66; AGL71, AGAMOUS-LIKE 71; AGL72, AGAMOUS-LIKE 72; AGL80, AGAMOUS-LIKE 80; AGL104, AGAMOUS-LIKE 104; ANR1, ARABIDOPSIS NITRATE REGULATED 1; AP1, APETALA1; AP3, APETALA3; CAL, CAULIFLOWER; CO, CONSTANS; DIA, DIANA; FLC, FLOWERING LOCUS C; FLM, FLOWERING LOCUS M; FT, FLOWERING LOCUS T; FUL, FRUITFULL; MAF2-5, MADS AFFECTING FLOWERING 2-5; PHE1, PHERES1; PI, PISTILLATA; SEP1-4, SEPALLATA1-4; SHP1,2, SHATTERPROOF1,2; SOC1, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1; STK, SEEDSTICK; SVP, SHORT VEGETATIVE PHASE; XAL1, XAANTAL 1; XAL2, XAANTAL 2.
Mads-Box Proteins in Orchid Flowering
The Floral Transition of Orchid
The floral transition, a developmental transition from vegetative to reproductive phase, is one of the key developmental transitions in the plant life cycle. The timing of floral transition greatly affects the success of plant reproduction. In the model plant Arabidopsis, the vegetative shoot apex, from which leaves are generated, is converted into the inflorescence meristem, from which flowers are generated, during the floral transition. In orchids, the length of the vegetative phase can vary from one to thirteen years between different species, but the average time for most species is between two to three years (Hew and Yong, 2004). The process of flowering is induced in the meristem of dormant axillary buds. In sympodial orchids such as Dendrobium and Oncidium, the formation of bud primordia occurs at the axils of the leaves. In the orchid Cymbidium, the inflorescence will be developed from the dormant axillary buds at the base of the pseudobulb. For monopodial orchids such as Phalaenopsis and Vandas, they typically have at least two dormant bud primordia at each leaf axil which can grow into inflorescences or keikis (new orchid plantlets).
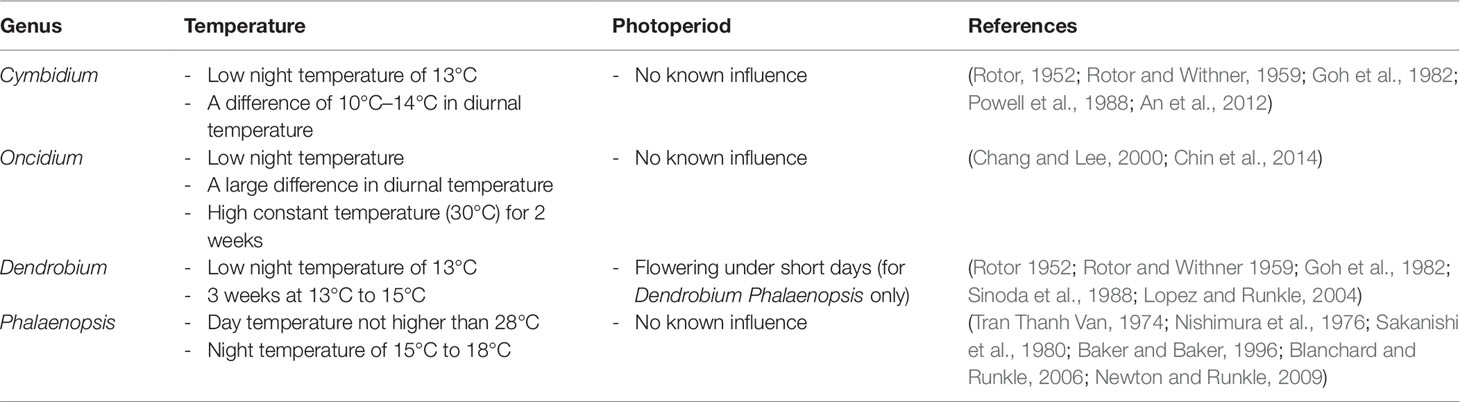
With the increasing demand for whole orchids and cut flowers, modern horticulturists and breeders are learning about the flowering behavior of different species to control the time of blooming so as to maximize their economic value. Each orchid species has a time of the year when it will bloom naturally. Most orchids will grow in the abundance of sunlight and moisture during summer and bloom in the fall, winter, or spring. Significant progress has been made to determine the effects of environmental conditions, such as temperature and day length, in inducing flowering in different species (Table 3). Studies on Cymbidium, Oncidium, Dendrobium, and Phalaenopsis have shown that a low night temperature of 13°C and a large fluctuation of 10°C to 14°C in daily diurnal temperature are sufficient to induce flowering (Rotor, 1952; Rotor and Withner, 1959). A high day temperature of more than 28°C for 8 h or longer promotes vegetative growth and inhibit the process of flowering in Phalaenopsis (Blanchard and Runkle, 2006; Newton and Runkle, 2009). However, a prolonged exposure to a constant elevated temperature of 30°C induces flowering by activating the thermal stress response (Chin et al., 2014). Since orchids are likely to be shaded by leaves on trees, the length of daylight is not known to influence flowering with the exception of Dendrobium phalaenopsis flowering under short days (Rotor, 1952; Rotor and Withner, 1959; Lopez and Runkle, 2004). It is noteworthy to mention that flowering is an intricate process and many environmental conditions including light intensity and humidity can affect the initiation of floral spikes.
Floral Pathway Integrators
For the model plant Arabidopsis, the timing of floral transition is regulated by a complex system consisting of several flowering pathways—photoperiod, vernalization, thermosensory, gibberellins (GA), autonomous, and age—that perceive both environmental and endogenous flowering signals (Mouradov et al., 2002; Simpson and Dean, 2002; Blázquez et al., 2003; Boss et al., 2004; Wang et al., 2009). For environmental flowering signals, the photoperiod pathway perceives the daylength in seasonal changes; the vernalization pathway measures the period of the plant’s exposure to cold; the thermosensory pathway mediates the ambient temperature effect. For endogenous flowering signals, the GA pathway promotes flowering under non-inductive photoperiod, while the autonomous pathway is a photoperiod-independent pathway that induces flowering through perceiving the internal signals at various stages of development. These genetic pathways regulate the transcription of two main integrators of floral pathways, FLOWERING LOCUS T (FT) and the MADS-box gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which then activate the expression of AP1 and LFY, two floral meristem identity genes, to start the process of floral meristem formation (Figure 2) (Kardailsky et al., 1999; Kobayashi et al., 1999; Blázquez and Weigel, 2000; Lee et al., 2000; Samach et al., 2000; Liu et al., 2009). Several MADS-box proteins including SOC1, AGAMOUS-LIKE 24 (AGL24), AGL6, and AGL17, promote flowering (Lee et al., 2000; Yu et al., 2002; Han et al., 2008; Yoo et al., 2011), whereas MADS-box regulators including FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), MADS AFFECTING FLOWERING 1/FLOWERING LOCUS M (MAF1/FLM), and MAF2/3/4/5 form various complexes to repress flowering (Figure 2) (Helliwell et al., 2006; Searle et al., 2006; Li et al., 2008; Gu et al., 2013; Lee et al., 2013; Pose et al., 2013; Mateos et al., 2015). Another three MADS-box transcription factors that are closely related to each other, AP1, CAL, and FUL are also involved in the activation of LFY in promoting flowering and floral meristem specification as the triple mutant of these genes generates leafy shoots in place of floral organs (Ferrándiz et al., 2000).
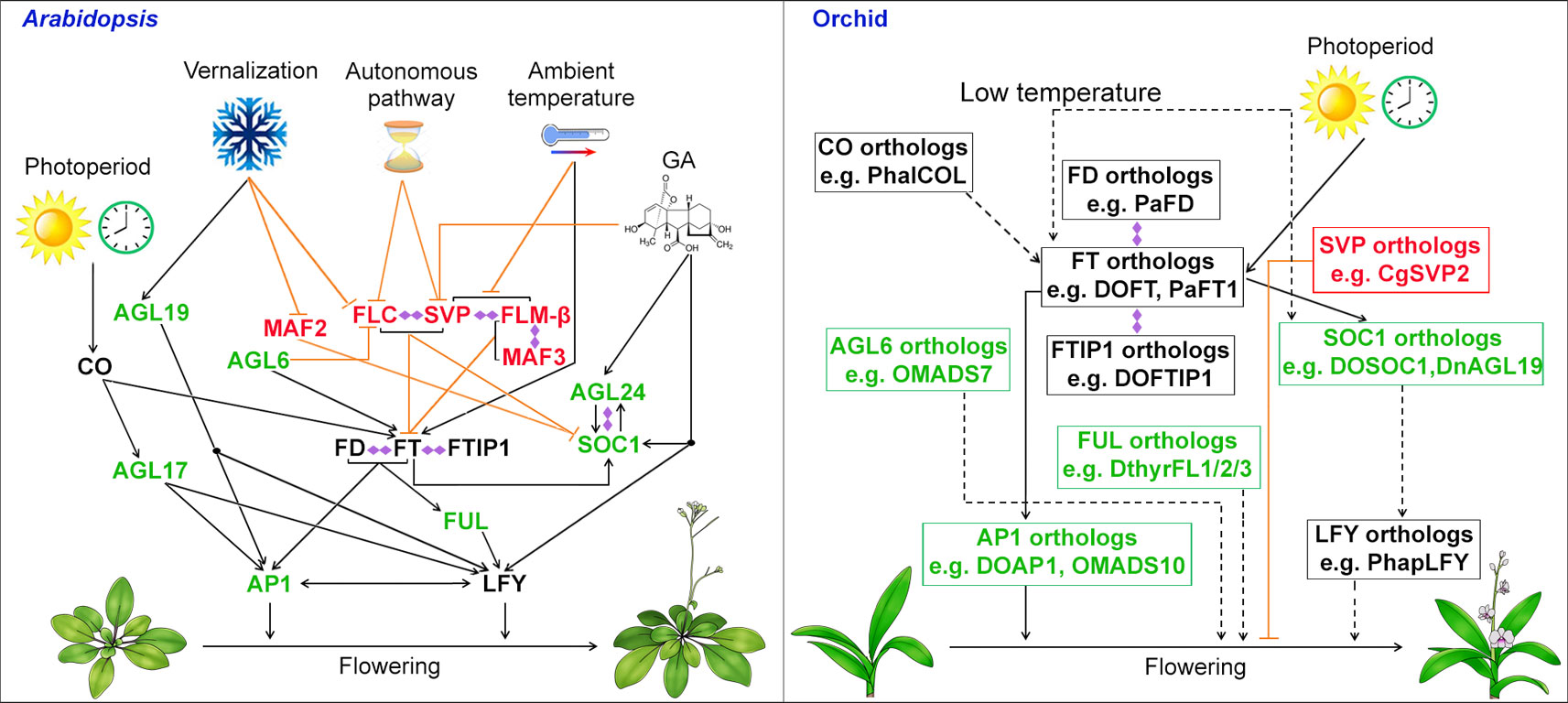
Figure 2 Biological roles of MADS-box genes in controlling flowering in the model plant Arabidopsis and orchid. In Arabidopsis, the MADS-box genes including SOC1, FLC, SVP and AGL24 integrates signals for flowering from environmental and endogenous cues. In orchid, orthologous genes of SOC1, AGL6, SVP, and AP1 have been isolated and functionally characterized either in heterologous system (e.g. Arabidopsis) or orchid and shown to be involved in promoting flowering. MADS-box transcription factors that function as flowering activators and suppressors are shown in green and red, respectively, whereas other flowering regulators are shown in black. Promoting and repressive effects are indicated by black arrows and orange T bars, respectively. The dashed lines with arrows indicate possible positive regulation based on the studies using heterologous systems. Double-ended diamond arrows indicate protein–protein interactions. AGL6, AGAMOUS-LIKE 6; AGL17, AGAMOUS-LIKE 17; AGL19, AGAMOUS-LIKE 19; AGL24, AGAMOUS-LIKE 24; AP1, APETALA1; CO, CONSTANS; FLC, FLOWERING LOCUS C; FLM, FLOWERING LOCUS M; FT, FLOWERING LOCUS T; FTIP1, FT-INTERACTING PROTEIN 1; FUL, FRUITFULL; LFY, LEAFY; MAF2, MADS AFFECTING FLOWERING 2; SOC1, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1; SVP, SHORT VEGETATIVE PHASE.
In orchids, the process of the floral transition occurs in the axillary buds where the bud primordia will develop into a more convex shape upon entering reproductive phase. Recent works have identified and examined many orthologs of flowering integrators and other MADS-box genes in orchids (Figure 2). As one of the major floral pathway integrator genes, FT, encoding a small globular protein, is transcriptionally activated by CONSTANS (CO) in companion cells in the leaf veins and the FT protein moves to the shoot apical meristem (Kardailsky et al., 1999; Kobayashi et al., 1999; An et al., 2004; Corbesier et al., 2007; Liu et al., 2012; Nakamura et al., 2014; Zhu et al., 2016). In orchids for instance, Oncidium, Dendrobium, and Cymbidium, the expression of FT orthologs was predominantly expressed in the leaves and axillary buds. In addition, the expression of FT orthologs has been found to be influenced by daylength in Oncidium and Cymbidium, showing a similar photoperiodic pattern like FT in Arabidopsis (Hou and Yang 2009; Huang et al., 2012). Ectopic expression of FT orthologs, OnFT, DnFT, DOFT, CeFT, CgFT, CsFT, and PaFT1 from the orchids Oncidium, Dendrobium, Cymbidium, and Phalaenopsis, respectively, results in a precocious flowering phenotype in transgenic plants of Arabidopsis or tobacco (Hou and Yang, 2009; Huang et al., 2012; Li et al., 2012; Xiang et al., 2012; Jang, 2015; Wang et al., 2017). More importantly, downregulation of DOFT delays flowering in Dendrobium orchids, whereas overexpression of DOFT accelerates flowering in orchids (Wang et al., 2017). Interestingly, low temperature treatment specifically induces the expression of FT in leaves in both Dendrobium and Phalaenopsis, suggesting FT is the main floral inducer under floral inductive low temperature regime (Li et al., 2012; Jang et al., 2015).
MADS-Box Genes and Orchid Flowering
SOC1 encodes a MADS-box transcription factor that is a member of the Tomato MADS-box gene 3 (TM3)-like genes subfamily from angiosperms and gymnosperms (Lee et al., 2000; Becker and Theißen, 2003; Cseke et al., 2003; Nakamura et al., 2005). SOC1 expression is detected in both leaves and shoot apices and is regulated by several floral pathways (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Moon et al., 2003). In Dendrobium nobile, the expression of a close SOC1 ortholog, DnAGL19, has been found to be increased after vernalization (Liang et al., 2012). In the orchid Dendrobium Chao Praya Smile, the expression of the SOC1 ortholog DOSOC1 is highly detected in reproductive organs, such as inflorescence apex, pedicel, floral buds and open flowers (Ding et al., 2013). DOSOC1 expression is upregulated in the whole seedlings upon the floral transition (Ding et al., 2013). Overexpression of DOSOC1 shows early flowering in both Arabidopsis and Dendrobium orchids, implying the evolutionary conserved functions of SOC1-like genes as activators of flowering (Figure 2) (Ding et al., 2013). Moreover, DOSOC1 expression is downregulated in DOFT knockdown Dendrobium orchid, whereas its expression is upregulated in DOFT overexpression orchid (Wang et al., 2017), indicating a conserved regulatory mechanism of SOC1-like genes expression. Intriguingly, DOSOC1 overexpression in Dendrobium results in abnormal floral organ development with formation of immature perianth organs only, indicating the role of DOSOC1 in maintaining the identity of floral meristem and formation floral organs (Ding et al., 2013). This is in line with the function of SOC1-like genes in flower development in some plant species (Liu et al., 2013; Teo et al., 2014). Additionally, another MADS-box gene FUL has been shown to act redundantly with SOC1 in regulation of flowering time in Arabidopsis (Melzer et al., 2008). Mutations in ful only slightly delay flowering, while in combination with soc1 mutants, the flowering is further delayed as compared with both single mutants. Three FUL-like genes have been isolated in the orchid Dendrobium thyrsiflorum, namely DthyrFL1/2/3 (Skipper et al., 2005). These three genes are upregulated during orchid reproductive development, yet their involvement in orchid flowering remains unknown.
SOC1 expression is repressed by a floral repressor protein complex formed by FLC and SVP, which are also MADS-box proteins (Li et al., 2008). The orchid SVP orthologs have been reported in Cymbidium orchids, whereas no FLC orthologs have been isolated so far in monocots. The expression levels of CgSVP1/2/3, SVP orthologs, are greatly reduced upon cold treatment in Cymbidium goeringii (Yang et al., 2019). Moreover, transient overexpression of CgSVP2 results in retarded flower bud growth, indicating its role as a repressor of flower bud formation (Yang et al., 2019). However, the involvement of SVP orthologs in orchid floral transition and its potential regulation of SOC1 need further investigation.
AGL6 encodes another MADS-box transcription factor which regulates the transition to flowering in Arabidopsis (Yoo et al., 2011). Knockdown of AGL6 by artificial microRNA leads to late flowering, in contrast, agl6-1D wherein AGL6 is activated by the 35S enhancer shows early flowering. Two AGL6-like genes, OMADS1 (OAGL6-2) and OMADS7 (OAGL6-1), have been found in the Oncidium Gower Ramsey orchid, and overexpression of either gene leads to early flowering in Arabidopsis (Hsu et al., 2003; Chang et al., 2009), implying a conserved function of AGL6-like genes in mediating flowering.
As mentioned above, AP1, a MADS-box protein, specifies the identity of floral meristem as well as sepal and petal identity (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993). In ap1 mutants, flowers exhibit a homeotic transformation that sepals develop into bracts and petals fail to develop. By comparison, AP1 overexpression causes early flowering and conversion of the inflorescence meristem into a determinate floral meristem (Mandel and Yanofsky, 1995). Orthologs of AP1 have been identified and characterized in Oncidium, Dendrobium, and Cymbidium. The expression of AP1 orthologs can be detected in both vegetative tissues and reproductive structures such as floral buds and pedicel (Yu and Goh, 2000; Chen et al., 2007; Chang et al., 2009; Tian et al., 2013; Sawettalake et al., 2017). Overexpression of AP1 orthologs, such as OMADS10 from Oncidium Gower Ramsey and DOAP1 from Dendrobium Chao Praya Smile, causes early flowering as well as conversion of inflorescence meristems to determinate floral meristems in Arabidopsis (Chang et al., 2009; Sawettalake et al., 2017). Transgenic Dendrobium orchids overexpressing DOAP1 also show accelerated flowering as compared to wild-type orchid and conversion of inflorescence meristems to determinate floral meristems (Sawettalake et al., 2017). In addition, DOAP1 expression is promoted by DOFT in Dendrobium orchids, which is a conserved regulation as that in Arabidopsis (Wang et al., 2017). These studies suggest that orchid AP1 orthologs have conserved functions in promoting the floral transition and determination of floral meristems.
Mads-Box Proteins in Orchid Floral Patterning
The Orchid Flower
In angiosperms, the flowers are usually composed of four types of structures, which form two parts, namely the vegetative part and the reproductive part. While the morphology and elaboration can differ greatly among different species, the diversification of floral patterning has taken place in a relatively conserved manner in having similar general organization of four types of structures arranged in four concentric whorls. In Arabidopsis, the flower consists of four concentric whorls of floral organs from the outer to inner whorls: sepals (four), petals (four), stamens (six), and two fused carpels. In orchids, the flowers are usually bilaterally symmetrical (zygomorphic) with three outer sepals, two inner petals, and a highly specialized inner median petal named lip or labellum which acts as the main pollinator attractant (Figure 3A). In a number of orchid species, the outer sepals and inner petals are named as tepals as they cannot be distinguished from each other morphologically. The reproductive structure gynostemium or column is composed of fused male (stamen/anther) and female (carpel/pistil) organs.
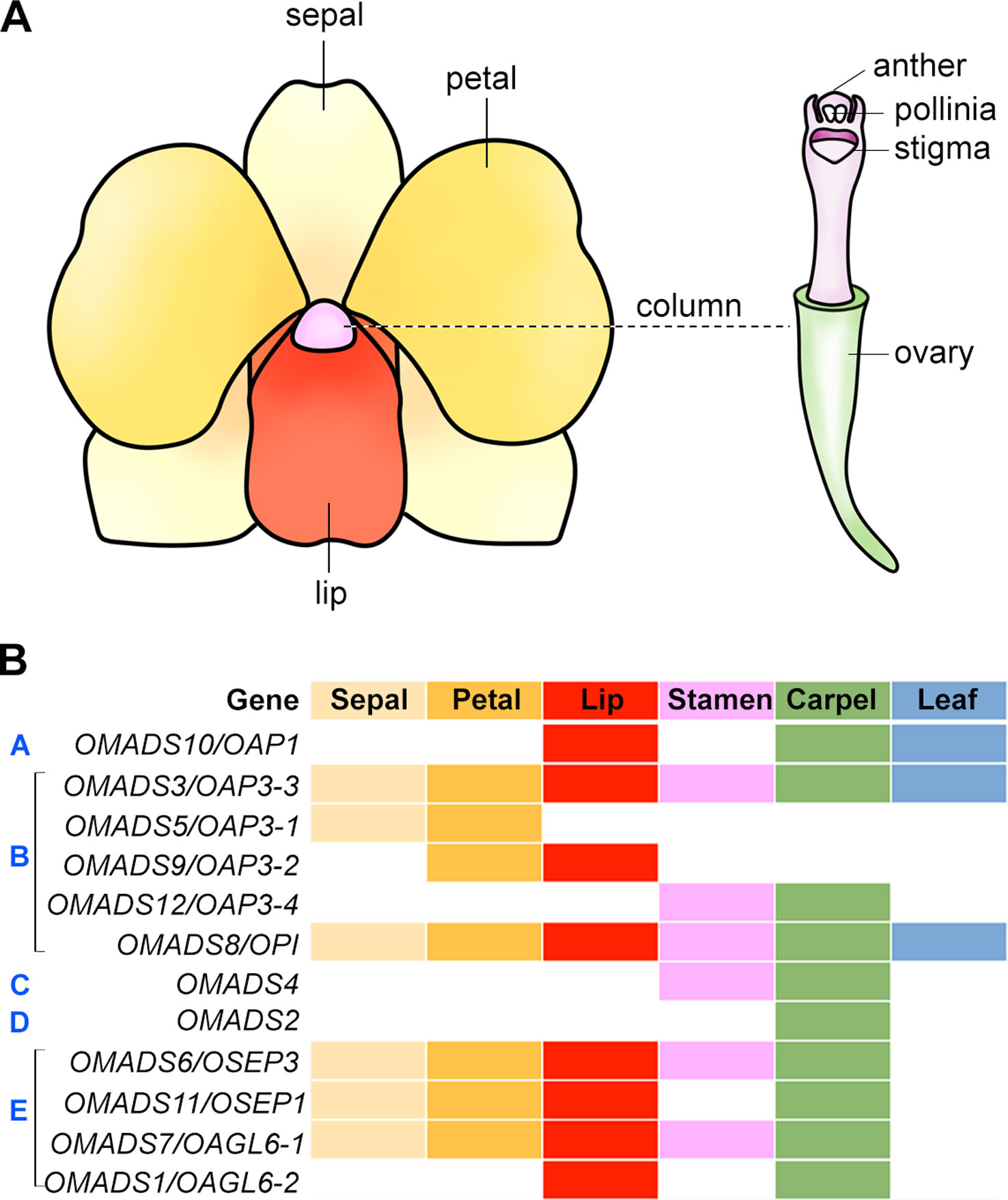
Figure 3 Floral organ identity genes in orchid. (A) An illustration showing a typical orchid flower structure. (B) Expression patterns of orthologs of floral organ identity genes in orchid. The floral organs, sepal, petal, lip, stamen, carpel, and leaf are color-coded, and presence of these colors indicates detected expression in these organs. The white color indicates no expression detected. The gene expression patterns are shown based on the studies in the Oncidium orchid (Chang et al., 2009; Chang et al., 2010; Hsu et al., 2010; Hsu et al., 2015).
ABCE and Floral Quartet Model
Understanding the specification of the distinct floral organs through genetic study in Arabidopsis and Antirrhinum majus (snapdragon) has resulted in the birth of “ABCE model” and “floral quartet model” (Figure 4A) (Meyerowitz et al., 1989; Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Irish, 2010; Theißen et al., 2016). In the classical “ABC model,” a combination of three gene classes specifies the four types of floral organs: sepal, petal, stamen, and carpel (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). In Arabidopsis, the A-class genes (AP1 and AP2) determine the sepal identify in the outermost whorl, A-class and B-class (AP3 and PI) genes together specify petals in the second whorl, B-class and C-class (AG) genes determine the identity of the male reproductive organ stamen in the third whorl, and the C-class gene specifies the female reproductive organ carpel in the innermost whorl. The expression domains of A-class and C-class genes are mutually exclusive. After discovering that the E-class genes (SEP1–4) are essential for the determination of all of the four whorls of floral organs, the classical “ABC model” was then extended to the “ABCE model.” Additionally, D-class genes (STK and SHP1/2) are needed for determining the identity of ovule (Pinyopich et al., 2003). Interestingly, except for AP2, all these genes belong to MADS-box gene family, and their proteins form tetrameric complexes. This was coined as the “floral quartet model” as the tetrameric complexes functions as a whole to direct the development of specific floral organs (Figure 4A) (Theissen and Saedler, 2001). In orchids, the study of MADS-box transcription factors related to floral patterning has been challenging as recent functional analyses have shown that these genes have functionally diversified in their own lineages, making the prediction of function on the basis of orthology difficult (Irish and Litt, 2005).
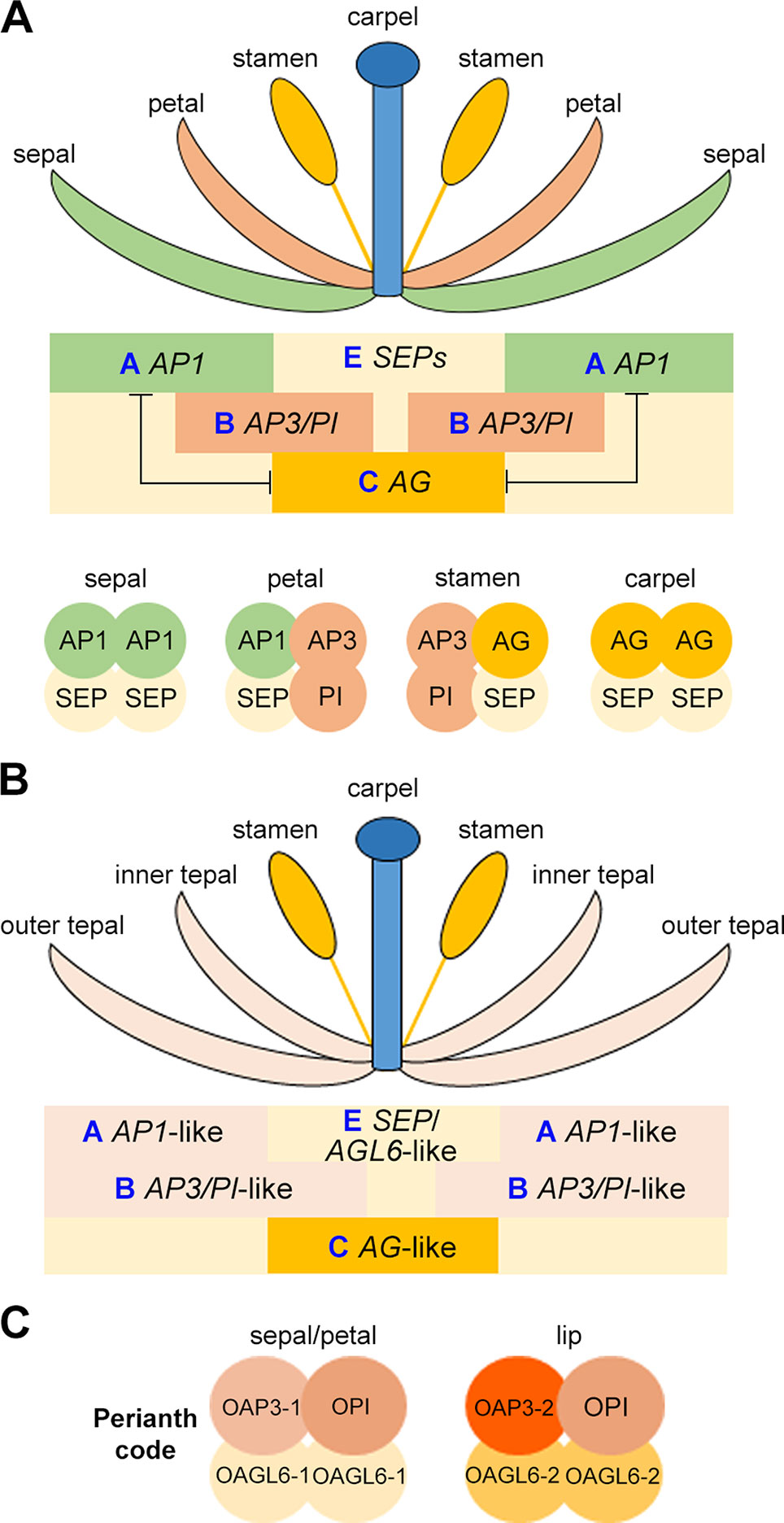
Figure 4 Schematic drawing showing the ABCE model (A), the modified ABC model (B), and the orchid Perianth code (C).
MADS-Box Genes and Orchid Floral Patterning
A-Class Genes
In orchids, there are several AP1 orthologs isolated in Cymbidium, Oncidium, Dendrobium, and Phalaenopsis (Table 2). In Dendrobium Madame Thong-In, the AP1 ortholog DOMADS2 is expressed during the transitional phase and floral development (Yu and Goh, 2000). In mature flowers, DOMADS2 is detected in the column and ovary but not in the pedicel, sepal, or petal. The differential expression pattern compared with AP1 expression in the sepal and petal in Arabidopsis suggests the functional divergence of AP1-like genes during floral patterning. In the Dendrobium Chao Praya Smile orchid, the AP1 ortholog DOAP1 is detected at high levels in the inflorescence meristem as well as flowers. In addition, the overexpression of DOAP1 can partially complement the Arabidopsis ap1 mutant in restoring petal formation, suggesting DOAP1 functions as a homeotic gene (Sawettalake et al., 2017). In Oncidium Gower Ramsey, the AP1 orthologs OMADS10 is expressed in the leaves, lip and carpel (Figure 3B). Overexpression of OMADS10 induces early flowering without any floral organs defects in Arabidopsis (Chang et al., 2009). Two AP1 orthologs, PaAP1-1 and PaAP1-2 has been isolated in the moth orchid Phalaenopsis aphrodite (Su et al., 2013b). PaAP1-1 is mainly expressed in the pollinia and pedicel, whereas PaAP1-2 is specifically expressed in the pedicel. The expression patterns of orchid AP1 orthologs are unlike the A-class genes in Arabidopsis which are only present in the sepals and petals, but is somehow similar to the monocot lily AP1 orthologs, LMADS5/6, which are expressed in the vegetative leaves and the innermost whorl carpel (Chen et al., 2008). This indicates the divergent function of orchid AP1-like genes in floral organ development.
B-Class Genes
The B-class genes are necessary for determining the identity of petals and stamens. There are two B-class genes in Arabidopsis, AP3 and PI, analogous to the A. majus DEFICIENS and GLOBOSA, respectively. Mutations in either AP3 or PI lead to similar phenotypes wherein petals and stamens are transformed into sepals and carpels, respectively (Bowman et al., 1989; Hill and Lord, 1989). Many studies have identified various numbers of B-class genes and studied their expression patterns in several orchid species (Table 2) (Hsu and Yang, 2002; Tsai et al., 2004; Tsai et al., 2005; Xu et al., 2006; Mondragón-Palomino and Theißen, 2008; Chang et al., 2009; Chang et al., 2010; Su et al., 2013b; Tsai et al., 2014; Hsu et al., 2015; Mao et al., 2015; Yang and Zhu, 2015). In Dendrobium crumenatum, the expression of DcOAP3A and DcOPI is detected in all parts of the mature flowers, but the expression of DcOAP3B is present in petals, lips, anthers, and column only (Xu et al., 2006). DcOAP3A/B can form heterodimers with DcOPI. In Oncidium Gower Ramsey, OMADS3 (OAP3-3), OMADS5 (OAP3-1), OMADS9 (OAP3-2), and OMADS12 (OAP3-4) belong to the AP3 lineage, while OMADS8 (OPI) belongs to the PI lineage. OMADS3 and OMADS8 are expressed in both vegetative tissues and all floral organs of mature flowers (Figure 3B) (Chang et al., 2010). OMADS5 is detected in both sepals and petals but not in the lip, whereas OMADS9 is detected in the petals and lips (Chang et al., 2010). OMADS12 is detected in the orchid reproductive floral organs including stamens and carpels, but not in the sepals, petals, and lips (Hsu et al., 2015). Overexpression of truncated OMADS3 in Arabidopsis results in ap2-like flowers with homeotic conversion from sepals and petals to carpel-like and stamen-like organs (Hsu and Yang, 2002), while overexpression of OMADS8, but not OMADS5/9, causes the transformation of sepals to expanded petal-like structures (Chang et al., 2010). While OMADS3, OMADS5, and OMADS9 can assemble into both homodimers and heterodimers within the same group, OMADS8 can only form heterodimer with OMADS3 (Chang et al., 2010). It has been proposed that OMADS3/5/8/9 is probably needed for the specification of sepals and petals and OMADS3/8/9 but the absence of OMADS5 leads to the formation of lips (Chang et al., 2010). In P. equestris, the MADS-box genes PeMADS2, PeMADS3, PeMADS4, and PeMADS5 belong to the AP3 lineage and PeMADS6 belongs to the PI lineage. They are all expressed in lips and columns with PeMADS2 also found in sepals and petals and PeMADS3 in petals (Tsai et al., 2004; Tsai et al., 2005). Similarly to those B-class genes in Dendrobium and Oncidium, PeMADS2-5 interacts with PeMADS6 to form heterodimers and binds to CarG boxes on DNA (Tsai et al., 2005).
C- and D-Class Genes
In Arabidopsis, the C-class gene AG is required for the normal development of the stamens and carpels found in the third and fourth whorls, respectively. Mutations in AG cause the homeotic transformation of stamens and carpels into petals and sepals (Yanofsky et al., 1990). In addition, since AG is also necessary for floral meristem determinacy, the flowers of ag mutants are indeterminate and show the “flower within a flower” phenotype of sepal-petal-petal reiteration. D-class genes are required for regulating ovule identify. C and D-class genes are members of the AG-like family and are resulted from an ancient gene duplication event (Becker and Theißen, 2003). Both C and D-class genes have been identfied from several orchid speices (Table 2).
In P. equestris, Phalaenopsis sp. “Hatsuyuki,” Cymbidium ensifolium, and Oncidium Gower Ramsey, the C-class genes PeMADS1, PhalAG1, CeMADS1, and OMADS4, respectively, are highly expressed in the floral buds and column in mature flowers (Figure 3B) (Song et al., 2006; Hsu et al., 2010; Wang et al., 2011; Chen et al., 2012). OMADS4 in the Oncidium Gower Ramsey orchid is specifically detected in the stamens and carpels, similar to the expression pattern of LMADS10 from Lilium longiflorum (Hsu et al., 2010). Both OMADS4 and LMADS10, when overexpressed in Arabidopsis, result in early flowering, whereas LMADS10 overexpression also leads to curly leaves and floral organ conversions, indicating the probable functional diversification of the monocot C-class genes. Several C-class genes from other orchid species have broader expression pattern. For examples, the D. crumenatum and D. thyrsiflorum AG orthologs, DcOAG1 and DthyrAG1, respectively, are expressed in all kinds of floral organs and are not confined to the reproductive organs (Skipper et al., 2006; Xu et al., 2006). This expression pattern is similar to the AG homolog from Illicium floridanum that is also expressed in the tepals and reproductive organs (Kim et al., 2005), suggesting the regulatory mechanisms involved in the regulation of the expression of these C-class genes have evolved independently. The ectopic expression of DcOAG1 in Arabidopsis accelerates flowering with abnormal floral organs in the first and second whorls (Xu et al., 2006).
OMADS2 in the orchid Oncidium Gower Ramsey, a D-class gene, is specifically detected in stigmatic cavity and ovary (Hsu et al., 2010). This expression pattern is close to that of LMADS2 from L. longiflorum, which is exclusively present in the carpel (Tzeng et al., 2002). OMADS2 forms homodimers and heterodimers with OMADS4. Overexpression of OMADS2 in Arabidopsis leads to early flowering without any floral organ converstion (Hsu et al., 2010).
E-Class Genes
The members of the SEP MADS-box subfamily belong to the E-class genes that are necessary for the formation of all floral organs and floral meristem determinacy in Arabidopsis. The triple mutant in SEP1/2/3/4 genes produce flowers with all floral organs converted to leaf-like organs (Pelaz et al., 2000; Ditta et al., 2004). SEP genes are present in angiosperms, but not gymnosperms, indicating that SEP genes may have been important for the existence of flowers (Nam et al., 2003). In D. crumenatum, the SEP ortholog DcOSEP1 is detected in all floral organs, similarly to Arabidopsis SEPs. DcOSEP1 is able to interact with the DcOAP3A-DcOPI and DcOAP3B-DcOPI heterodimers, but not with DcOAP3A and DcOPI individually, indicating that DcOSEP1 is able to form a higher order protein complex with DcOAP3A-DcOPI or DcOAP3B-DcOPI, similar to their counterparts in Arabidopsis (Honma and Goto, 2001; Theissen and Saedler, 2001; Xu et al., 2006). In the Dendrobium Madame Thong-In orchid, DOMADS1 and DOMADS3 encode MADS-box proteins closely related with SEP1 and SEP3, respectively. DOMADS1 is present in all the floral organs similarly to DcOSEP1, while DOMADS3 is only present in the pedicel (Yu and Goh, 2000). In the Oncidium Gower Ramsey orchid, OMADS6 (OSEP3) and OMADS11 (OSEP1) encode MADS-box proteins homologous to SEP3 and SEP1/2, respectively. Both genes are highly present in the sepal, petal, lip, and carpel, with weaker and undetectable expression in stamens for OMADS6 and OMADS11, respectively (Figure 3B) (Chang et al., 2009). Overexpression of OMADS6 in Arabidopsis leads to homeotic transformation of sepals into carpelliod structures and petals into stamen-like organs (Chang et al., 2009). In P. equestris, the four SEP-like PeSEP genes are expressed in flower buds with the expression of PeSEP2 higher in floral stalk and column and PeSEP3 in petals (Pan et al., 2014). Like the SEP proteins in D. crumenatum, PeSEP2, PeSEP3, and PeSEP4 proteins cannot interact with the B-class proteins PeMADS2, PeMADS4, PeMADS6, or the D-class protein PeMADS7 individually, but can form multimeric complexes with PeMADS2/6, PeMADS4/6 and PeMADS6/7. Only PeSEP1 is able to interact with PeMADS2, PeMADS4, PeMADS6, and PeMADS7 individually, and with PeMADS2/6, PeMADS4/6, and PeMADS6/7 (Pan et al., 2014). Silencing of PeSEP3 by VIGS results in the conversion of tepal to leaf-like organ in Phalaenopsis, whereas silencing of PeSEP2 does not greatly affect flower development (Pan et al., 2014), suggesting that these PeSEPs have divergent functions in orchid flower development.
The AGL6-like genes are similar to SEP-like genes and the AGL6-like gene in petunia functions like SEP genes in floral patterning (Rijpkema et al., 2009). It has been proposed to add AGL6-like genes to class-E genes. As described above, the Arabidopsis AGL6 functions as a flowering promoter (Yoo et al., 2011). Arabidopsis has another AGL6-like gene called AGL13, which acts similarly to E-class SEP genes in specifying male and female gametophytes (Hsu et al., 2014). The orchid Oncidium Gower Ramsey also has two AGL6-like genes, OMADS1 and OMADS7. OMADS7 is expressed in all the floral organs, similar to that of E-class gene including OMADS6 (Figure 3B) (Chang et al., 2009). OMADS1 shows a different expression pattern which is in the lip and carpel, but not in other floral organs (Figure 3B) (Chang et al., 2009). Besides being early flowering, the flowers of OMADS1 or OMADS7 overexpression show homeotic transformation of sepals into carpel-like structures (Chang et al., 2009), indicating their dual roles in promoting floral transition and regulating floral organ formation. In P. aphrodite, the AGL6-like gene PaAGL6-1 is expressed specifically in the lip, suggesting that PaAGL6 may play an important role in lip formation (Su et al., 2013b). Subsequent studies have further revealed that the orchid AGL6-like genes play important roles in determining sepal/petal/lip formation (discussed in the following section). Together, AGL6-like genes may have diverse function in all four whorls of floral organs.
The Orchid Perianth Code
Flowers in orchids and several other monocots such as lily, the sepals and petals are morphologically similar and are also collectively called tepals. This is different from Arabidopsis and other dicots flowers, in which sepals and petals have distinguished morphologies. To explain this difference in perianth organs specification, the modified ABC model has been proposed, in which the expression domain of B-class genes are extended to the outermost whorl of floral organs in many orchid species (Figure 4B) (Van Tunen et al., 1993; Bowman, 1997; Kramer et al., 2003; Mondragón-Palomino and Theißen, 2008). In the Oncidium Gower Ramsey orchid, the AP3-like gene OMADS3 and the PI-like gene OMADS8 are detected in all perianth organs (Figure 3B) (Chang et al., 2010).
However, the orchid flower has a median petal called lip, which has a highly diversified morphology and acts as the main attractor of pollinators. The specification of the lip cannot be simply explained by the modified “ABC model”. Several years of molecular studies of MADS-box proteins and orchid floral patterning have led to the discovery of the model of formation of perianth organs: the Perianth (P) code (Figure 4C) (Hsu et al., 2015). Based on this model, the two tetrameric MADS-box protein complexes, SP (sepal/petal) complex (OAP3-1/OAGL6-1/OAGL6-1/OPI) and L (lip) complex (OAP3-2/OAGL6-2/OAGL6-2/OPI), compete to promote the development of sepal/petal and lip, respectively. Different copies of B-class AP3-like genes (OAP3-1 and OAP3-2) and AGL6-like genes (OAGL6-1 and OAGL6-2) have different whorl-specific or whorl-biased expression patterns, providing the basis for the formation of SP and L complexes. Moreover, the relative levels of the two complexes may also determine the formation of various forms of intermediate lips or distinct lips in orchid.
Concluding Remarks and Future Perspectives
The orchid family is the second largest family of angiosperms and has delighted cultivators for their unsurpassed beauty and complexity. The study on orchids has come a long way since people began gathering and propagating them under controlled environment. Recent findings in orchids, mainly Cymbidium, Oncidium, Dendrobium, and Phalaenopsis, have revealed that MADS-box proteins play critical roles in orchid flowering and floral patterning. The unraveled molecular mechanisms underlying orchid flowering and floral development can be applied to both classical orchid breeding and targeted manipulation of orchids for desired flowering traits and floral patterns. Orchids have many MADS-box genes, for example, 51 in P. equestris and 63 in Dendrobium catenatum (Cai et al., 2015; Zhang et al., 2016), however, only several of the MADS-box genes have been characterized, and most of them are shown be involved in orchid flowering or floral development. A recent study shows that MADS-box regulators might be relevant with the development of seeds without endosperm and epiphytism in orchids (Zhang et al., 2017), but revealing the functions of MADS-box genes in other develomental processes needs more future endeavor. In addition, in this review, the comparisons of orchid MADS-box genes to Arabidopsis have been included as many orchid genes are named and share similar functions to their closest orthologs in Arabidopsis. With the increasing number of MADS-box genes being studied in model monocots such as rice, wheat, barley, maize, and lily (Callens et al., 2018), more detailed comparisons of MADS-box gene functions between orchid and other monocots can be performed and discussed in the near future.
Nowadays, the function of orchid MADS-box genes, in many cases, is studied in heterologous plant systems (e.g. Arabidopsis and tobacco). In only several studies, the function of MADS-box genes has been examined by using transient overexpression/knockdown in orchids by VIGS or generating transgenic orchid overexpressing or knocking down of target genes (Table 2). For examples, silencing of OAGL6-2 by VIGS has been done in Oncidium Gower Ramsey and Phalaenopsis amabilis hybrid to test the Perianth code, and DOAP1 when overexpressed in Dendrobium Chao Praya Smile, leads to early flowering. To better understand the function of orchid genes, there is a need for more reliable and faster genetic transformation systems of different orchid species in orchid study and targeted orchid breeding with desired traits. Moreover, with modern genomic editing tools such as CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats–Caspase 9), it is now possible to generate orchid mutants for in vivo functional characterizations (Kui et al., 2017). Indeed, CRISPR-Cas9 has been successfully used to create multiple mutants of MADS genes in the orchid P. equestris very recently (Tong et al., 2019).
With the advent of sequencing technologies, five orchid genomes have been released, including Apostasia shenzhenica, D. catenatum, Dendrobium officinale, P. equestris, and Vanilla planifolia (Cai et al., 2015; Yan et al., 2015; Zhang et al., 2016; Zhang et al., 2017; Hu et al., 2019). Moreover, the transcriptomes of several orchid species from different subfamilies are freely available in online databases, such as Orchidstra 2.0 (http://orchidstra2.abrc.sinica.edu.tw), OrchidBase 3.0 (http://orchidbase.itps.ncku.edu.tw) and OOGB (http://predictor.nchu.edu.tw/oogb) (Chang et al., 2011; Fu et al., 2011; Su et al., 2013a; Tsai et al., 2013; Chao et al., 2017; Tsai et al., 2017). Recently, the transcriptome of a Mediterranean orchid Orchis italica inflorescence have also been analyzed (Valoroso et al., 2019). Genome sequences and transcriptomic data have provided valuable information in aiding basic research and genomics-assisted horticultural breeding. The advent of molecular tools has allowed genomic analysis to determine the underlying mechanisms behind many morphological characteristics and developmental processes. With modern genomic editing tools available, it is now feasible to generate mutants or novel varieties in orchids. This would greatly help not only in the molecular genetic research of orchid biology, but also in generating novel orchid varieties with various desirable traits through targeted gene editing.
Author Contributions
ZT and LS wrote the manuscript. WZ made the drawings of Arabidopsis and orchid plants. All authors read and approved of the manuscript.
Funding
Preparation of the review has been supported by the intramural funding from Temasek Life Sciences Laboratory and National University of Singapore.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aceto, S., Gaudio, L. (2011). The MADS and the beauty: genes involved in the development of orchid flowers. Curr. Genomics 12, 342–356. doi: 10.2174/138920211796429754
Acri-Nunes-Miranda, R., Mondragón Palomino, M. (2014). Expression of paralogous SEP-, FUL-, AG-and STK-like MADS-box genes in wild-type and peloric Phalaenopsis flowers. Front. Plant Sci. 5, 76. doi: 10.3389/fpls.2014.00076
Adamczyk, B. J., Fernandez, D. E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149, 1713–1723. doi: 10.1104/pp.109.135806
Adamczyk, B. J., Lehti-Shiu, M. D., Fernandez, D. E. (2007). The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50, 1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x
Alvarez-Buylla, E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U. S. A. 97, 5328–5333. doi: 10.1073/pnas.97.10.5328
An, H., Roussot, C., Suarez-Lopez, P., Corbesier, L., Vincent, C., Pineiro, M., et al. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. doi: 10.1242/dev.01231
An, H. R., Kim, Y. J., Kim, K. S. (2012). Flower initiation and development in Cymbidium by night interruption with potassium and nitrogen. Hortic. Environ. Biotechnol. 53, 204–211. doi: 10.1007/s13580-012-0023-5
Atwood, J. T. (1986). The size of the Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana, 171–186. doi: 10.2307/41888801
Baker, M. L., Baker, C. O. (1996).Orchid species culture: Dendrobium. (Portland, Oregan. USA: Timber Press)
Becker, A., Theißen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Belarmino, M., Mii, M. (2000). Agrobacterium-mediated genetic transformation of a Phalaenopsis orchid. Plant Cell Rep. 19, 435–442. doi: 10.1007/s002990050752
Bemer, M., Wolters-Arts, M., Grossniklaus, U., Angenent, G. C. (2008). The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20, 2088–2101. doi: 10.1105/tpc.108.058958
Blázquez, M. A., Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. doi: 10.1038/35009125
Blázquez, M. A., Ahn, J. H., Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168. doi: 10.1038/ng1085
Blanchard, M. G., Runkle, E. S. (2006). Temperature during the day, but not during the night, controls flowering of Phalaenopsis orchids. J. Exp. Bot. 57, 4043–4049. doi: 10.1093/jxb/erl176
Borner, R., Kampmann, G., Chandler, J., Gleißner, R., Wisman, E., Apel, K., et al. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. doi: 10.1046/j.1365-313x.2000.00906.x
Boss, P. K., Bastow, R. M., Mylne, J. S., Dean, C. (2004). Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16, S18–S31. doi: 10.1105/tpc.015958
Bowman, J. L., Smyth, D. R., Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M., Smyth, D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743.
Bowman, J. L. (1997). Evolutionary conservation of angiospermflower development at the molecular and genetic levels. J. Biosci. 22, 13. doi: 10.1007/BF02703197
Bulpitt, C. J., Li, Y., Bulpitt, P. F., Wang, J. (2007). The use of orchids in Chinese medicine. J. R. Soc. Med. 100, 558–563. doi: 10.1177/0141076807100012014
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Callens, C., Tucker, M. R., Zhang, D., Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459. doi: 10.1093/jxb/ery086
Chai, M., Xu, C., Senthil, K., Kim, J., Kim, D. (2002). Stable transformation of protocorm-like bodies in Phalaenopsis orchid mediated by Agrobacterium tumefaciens. Sci. Hortic. 96, 213–224. doi: 10.1016/S0304-4238(02)00084-5
Chang, Y., Lee, N. (2000). Effect of temperature on growth of pseudobulb and inflorescences development of Oncidium‘Gower Ramsey’. J. Chin. Soc. Hortic. Sci. 46, 221–230.
Chang, Y.-Y., Chiu, Y.-F., Wu, J.-W., Yang, C.-H. (2009). Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 50, 1425–1438. doi: 10.1093/pcp/pcp087
Chang, Y.-Y., Kao, N.-H., Li, J.-Y., Hsu, W.-H., Liang, Y.-L., Wu, J.-W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152, 837–853. doi: 10.1104/pp.109.147116
Chang, Y.-Y., Chu, Y.-W., Chen, C.-W., Leu, W.-M., Hsu, H.-F., Yang, C.-H. (2011). Characterization of Oncidium‘Gower Ramsey’transcriptomes using 454 GS-FLX pyrosequencing and their application to the identification of genes associated with flowering time. Plant Cell Physiol. 52 (9), 1532–1545. doi: 10.1093/pcp/pcr101
Chao, Y.-T., Yen, S.-H., Yeh, J.-H., Chen, W.-C., Shih, M.-C. (2017). Orchidstra 2.0—A transcriptomics resource for the orchid family. Plant Cell Physiol. 58 (1), e9–e9. doi: 10.1093/pcp/pcw220
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., Van Den Berg, C., et al. (2015). An updated classification of Orchidaceae. Bot. J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Chen, D., Guo, B., Hexige, S., Zhang, T., Shen, D., Ming, F. (2007). SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 226, 369–380. doi: 10.1007/s00425-007-0488-0
Chen, M. K., Lin, I. C., Yang, C. H. (2008). Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiol. 49, 704–717. doi: 10.1093/pcp/pcn046
Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J., Lee, Y. I., et al. (2012). C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiol. 53, 1053–1067. doi: 10.1093/pcp/pcs048
Chen, L. (2002). High efficiency of Agrobacterium mediated transformation by using rhizome of Cymbidium (Orchidaceae: Maxillarieae). Lindleyana 17, 16–20.
Chin, D.-C., Shen, C.-H., Senthilkumar, R., Yeh, K.-W. (2014). Prolonged exposure to elevated temperature induces floral transition via up-regulation of cytosolic ascorbate peroxidase 1 and subsequent reduction of the ascorbate redox ratio in Oncidium hybrid orchid. Plant Cell Physiol. 55, 2164–2176. doi: 10.1093/pcp/pcu146
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31. doi: 10.1038/353031a0
Colombo, M., Masiero, S., Vanzulli, S., Lardelli, P., Kater, M. M., Colombo, L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54, 1037–1048. doi: 10.1111/j.1365-313X.2008.03485.x
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cribb, P. J., Kell, S. P., Dixon, K. W., Barrett, R. L. (2003). "Orchid conservation: a global perspective," in Orchid conservation. (Kota Kinabalu: Natural History Publications), 1–24.
Cseke, L. J., Zheng, J., Podila, G. K. (2003). Characterization of PTM5 in aspen trees: a MADS-box gene expressed during woody vascular development. Gene 318, 55–67. doi: 10.1016/S0378-1119(03)00765-0
Da Silva, J.a.T., Aceto, S., Liu, W., Yu, H., Kanno, A. (2014). Genetic control of flower development, color and senescence of Dendrobium orchids. Sci. Hortic. 175, 74–86. doi: 10.1016/j.scienta.2014.05.008
Ding, L., Wang, Y., Yu, H. (2013). Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 54, 595–608. doi: 10.1093/pcp/pct026
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Dorca-Fornell, C., Gregis, V., Grandi, V., Coupland, G., Colombo, L., Kater, M. M. (2011). The Arabidopsis SOC1-like genes AGL42, AGL71 and AGL72 promote flowering in the shoot apical and axillary meristems. Plant J. 67, 1006–1017. doi: 10.1111/j.1365-313X.2011.04653.x
Fay, M. F., Chase, M. W. (2009). Orchid biology: from Linnaeus via Darwin to the 21st century. Ann. Bot. 104, 359–364. doi: 10.1093/aob/mcp190
Ferrándiz, C., Gu, Q., Martienssen, R., Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734.
Fu, C.-H., Chen, Y.-W., Hsiao, Y.-Y., Pan, Z.-J., Liu, Z.-J., Huang, Y.-M. (2011). OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 52 (2), 238–243. doi: 10.1093/pcp/pcq201
Garay‐Arroyo, A., Ortiz‐Moreno, E., de la Paz Sánchez, M., Murphy, A.S., García‐Ponce, B., Marsch‐Martínez, N., et al. (2013). The MADS transcription factor XAL2/AGL14 modulates auxin transport during Arabidopsis root development by regulating PIN expression. EMBO J. 32 (21), 2884–2895.
Goh, C., Strauss, M., Arditti, J. (1982). "Flower induction and physiology in orchids," in Orchid biology: reviews and perspectives. (USA).
Gramzow, L., Ritz, M. S., Theißen, G. (2010). On the origin of MADS-domain transcription factors. Trends Genet. 26, 149–153. doi: 10.1016/j.tig.2010.01.004
Gu, X., Le, C., Wang, Y., Li, Z., Jiang, D., Wang, Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4, 1947. doi: 10.1038/ncomms2947
Gutiérrez, R. M. P. (2010). Orchids: a review of uses in traditional medicine, its phytochemistry and pharmacology. J. Med. Plants Res. 4, 592–638. doi: 10.5897/JMPR10.012
Han, P., Garcia-Ponce, B., Fonseca-Salazar, G., Alvarez-Buylla, E. R., Yu, H. (2008). AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 55, 253–265. doi: 10.1111/j.1365-313X.2008.03499.x
Hartmann, U., Hohmann, S., Nettesheim, K., Wisman, E., Saedler, H., Huijser, P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Hayes, T. E., Sengupta, P., Cochran, B. H. (1988). The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 2, 1713–1722. doi: 10.1101/gad.2.12b.1713
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W., Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Hew, C. S., Yong, J. W. (2004). The physiology of tropical orchids in relation to the industry (Singapore: World Scientific Publishing Company). doi: 10.1142/5505
Hill, J. P., Lord, E. M. (1989). Floral development in Arabidopsis thaliana: a comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 67, 2922–2936. doi: 10.1139/b89-375
Hill, K., Wang, H., Perry, S. E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53, 172–185. doi: 10.1111/j.1365-313X.2007.03336.x
Honma, T., Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409, 525. doi: 10.1038/35054083
Hossain, M. M., Kant, R., Van, P. T., Winarto, B., Zeng, S., Teixeira Da Silva, J. A. (2013). The application of biotechnology to orchids. Crit. Rev.Plant Sci. 32, 69–139. doi: 10.1080/07352689.2012.715984
Hou, C.-J., Yang, C.-H. (2009). Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol. 50, 1544–1557. doi: 10.1093/pcp/pcp099
Hsieh, M.-H., Lu, H.-C., Pan, Z.-J., Yeh, H.-H., Wang, S.-S., Chen, W.-H. (2013a). Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci. 201, 25–41. doi: 10.1016/j.plantsci.2012.11.003
Hsieh, M.-H., Pan, Z.-J., Lai, P.-H., Lu, H.-C., Yeh, H.-H., Hsu, C.-C. (2013b). Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J. Exp. Bot. 64 (12), 3869–3884. doi: 10.1093/jxb/ert218
Hsu, H.-F., Yang, C.-H. (2002). An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol. 43, 1198–1209. doi: 10.1093/pcp/pcf143
Hsu, H. F., Huang, C. H., Chou, L. T., Yang, C. H. (2003). Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 44, 783–794. doi: 10.1093/pcp/pcg099
Hsu, H.-F., Hsieh, W.-P., Chen, M.-K., Chang, Y.-Y., Yang, C.-H. (2010). C/D class MADS box genes from two monocots, orchid (Oncidium Gower Ramsey) and lily (Lilium longiflorum), exhibit different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 51, 1029–1045. doi: 10.1093/pcp/pcq052
Hsu, W. H., Yeh, T. J., Huang, K. Y., Li, J. Y., Chen, H. Y., Yang, C. H. (2014). AGAMOUS-LIKE13, a putative ancestor for the E functional genes, specifies male and female gametophyte morphogenesis. Plant J. 77, 1–15. doi: 10.1111/tpj.12363
Hsu, H.-F., Hsu, W.-H., Lee, Y.-I., Mao, W.-T., Yang, J.-Y., Li, J.-Y., et al. (2015). Model for perianth formation in orchids. Nat. Plants 1, 15046. doi: 10.1038/nplants.2015.46
Hu, Y., Resende, M. F. R., Bombarely, A., Brym, M., Bassil, E., Chambers, A. H. (2019). Genomics-based diversity analysis of Vanilla species using a Vanilla planifolia draft genome and genotyping-by-sequencing. Sci. Rep. 9, 3416. doi: 10.1038/s41598-019-40144-1
Huang, W., Fang, Z., Zeng, S., Zhang, J., Wu, K., Chen, Z., et al. (2012). Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int. J. Mol. Sci. 13, 11385–11398. doi: 10.3390/ijms130911385
Irish, V. F., Litt, A. (2005). Flower development and evolution: gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15, 454–460. doi: 10.1016/j.gde.2005.06.001
Irish, V. F., Sussex, I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. doi: 10.1105/tpc.2.8.741
Irish, V. F. (2010). The flowering of Arabidopsis flower development. Plant J. 61, 1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x
Jang, S., Choi, S.-C., Li, H.-Y., An, G., Schmelzer, E. (2015). Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PloS One 10, e0134987. doi: 10.1371/journal.pone.0134987
Jang, S. (2015). Functional characterization of PhapLEAFY, a FLORICAULA/LEAFY ortholog in Phalaenopsis aphrodite. Plant Cell Physiol. 56, 2234–2247. doi: 10.1093/pcp/pcv130
Kang, I. H., Steffen, J. G., Portereiko, M. F., Lloyd, A., Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Kaufmann, K., Melzer, R., Theißen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kim, S., Koh, J., Yoo, M. J., Kong, H., Hu, Y., Ma, H., et al. (2005). Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. 43, 724–744. doi: 10.1111/j.1365-313X.2005.02487.x
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. doi: 10.1101/gad.257403
Kramer, E. M., Di Stilio, V. S., Schluter, P. M. (2003). Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164, 11. doi: 10.1086/344694
Kui, L., Chen, H., Zhang, W., He, S., Xiong, Z., Zhang, Y., et al. (2017). Building a genetic manipulation tool box for orchid biology: identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 7, 2036. doi: 10.3389/fpls.2016.02036
Kutter, C., Schob, H., Stadler, M., Meins, F., Jr., Si-Ammour, A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19, 2417–2429. doi: 10.1105/tpc.107.050377
Lee, H., Suh, S.-S., Park, E., Cho, E., Ahn, J. H., Kim, S.-G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Lee, J. H., Ryu, H. S., Chung, K. S., Pose, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Li, D., Liu, C., Shen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Li, R., Wang, A., Sun, S., Liang, S., Wang, X., Ye, Q., et al. (2012). Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tissue Organ Cult. (PCTOC) 111, 143–151. doi: 10.1007/s11240-012-0178-x
Liang, S., Ye, Q.-S., Li, R.-H., Leng, J.-Y., Li, M.-R., Wang, X.-J., et al. (2012). Transcriptional regulations on the low-temperature-induced floral transition in an Orchidaceae species, Dendrobium nobile: an expressed sequence tags analysis. Comp. Funct. Genom. 2012. doi: 10.1155/2012/757801
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liu, C., Xi, W., Shen, L., Tan, C., Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Liu, L., Liu, C., Hou, X., Xi, W., Shen, L., Tao, Z., et al. (2012). FTIP1 is an essential regulator required for florigen transport. PloS Biol. 10, e1001313. doi: 10.1371/journal.pbio.1001313
Liu, C., Teo, Z. W. N., Bi, Y., Song, S., Xi, W., Yang, X., et al. (2013). A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 24, 612–622. doi: 10.1016/j.devcel.2013.02.013
Lu, H.-C., Chen, H.-H., Tsai, W.-C., Chen, W.-H., Su, H.-J., Chang, D. C.-N. (2007). Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 143 (2), 558–569. doi: 10.1104/pp.106.092742
Mandel, M. A., Gustafson-Brown, C., Savidge, B., Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273. doi: 10.1038/360273a0
Mao, W. T., Hsu, H. F., Hsu, W. H., Li, J. Y., Lee, Y. I., Yang, C. H. (2015). The C-terminal sequence and PI motif of the orchid (Oncidium Gower Ramsey) PISTILLATA (PI) ortholog determine its ability to bind AP3 orthologs and enter the nucleus to regulate downstream genes controlling petal and stamen formation. Plant Cell Physiol. 56, 2079–2099. doi: 10.1093/pcp/pcv129
Mateos, J. L., Madrigal, P., Tsuda, K., Rawat, V., Richter, R., Romera-Branchat, M., et al. (2015). Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 16, 31. doi: 10.1186/s13059-015-0597-1
Melzer, S., Lens, F., Gennen, J., Vanneste, S., Rohde, A., Beeckman, T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40, 1489–1492. doi: 10.1038/ng.253
Messenguy, F., Dubois, E. (2003). Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316, 1–21. doi: 10.1016/S0378-1119(03)00747-9
Meyerowitz, E. M., Smyth, D. R., Bowman, J. L. (1989). Abnormal flowers and pattern formation in floral development. Development 106, 209–217.
Michaels, S. D., Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. doi: 10.1105/tpc.11.5.949
Mondragón-Palomino, M., Theißen, G. (2008). MADS about the evolution of orchid flowers. Trends Plant Sci. 13 (2), 51–59. doi: 10.1016/j.tplants.2007.11.007
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Moreno-Risueno, M. A., Van Norman, J. M., Moreno, A., Zhang, J., Ahnert, S. E., Benfey, P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311. doi: 10.1126/science.1191937
Mouradov, A., Cremer, F., Coupland, G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14, S111–S130. doi: 10.1105/tpc.001362
Nakamura, T., Song, I.-J., Fukuda, T., Yokoyama, J., Maki, M., Ochiai, T., et al. (2005). Characterization of TrcMADS1 gene of Trillium camtschatcense (Trilliaceae) reveals functional evolution of the SOC1/TM3-like gene family. J. Plant Res. 118, 229–234. doi: 10.1007/s10265-005-0215-5
Nakamura, Y., Andrés, F., Kanehara, K., Liu, Y.-C., Dörmann, P., Coupland, G. (2014). Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 5, 3553. doi: 10.1038/ncomms4553
Nam, J., Depamphilis, C. W., Ma, H., Nei, M. (2003). Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 20, 1435–1447. doi: 10.1093/molbev/msg152
Newton, L. A., Runkle, E. S. (2009). High-temperature inhibition of flowering of Phalaenopsis and Doritaenopsis orchids. HortScience 44, 1271–1276. doi: 10.21273/HORTSCI.44.5.1271
Ng, M., Yanofsky, M. F. (2001). Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2, 186–195. doi: 10.1038/35056041
Nishimura, G., Kosugi, K., Furukawa, J. (1976). Flower bud formation in Phalaenopsis. Orchid Rev. 84, 175–179.
Norman, C., Runswick, M., Pollock, R., Treisman, R. (1988). Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003. doi: 10.1016/0092-8674(88)90244-9
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202 (3), 1024–1042. doi: 10.1111/nph.12723
Par˘enicová, L., De Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Passmore, S., Maine, G. T., Elble, R., Christ, C., Tye, B.-K. (1988). Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 204, 593–606. doi: 10.1016/0022-2836(88)90358-0
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200. doi: 10.1038/35012103
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Portereiko, M. F., Lloyd, A., Steffen, J. G., Punwani, J. A., Otsuga, D., Drews, G. N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18, 1862–1872. doi: 10.1105/tpc.106.040824
Pose, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Powell, C. L., Caldwell, K., Littler, R., Warrington, I. (1988). Effect of temperature regime and nitrogen fertilizer level on vegetative and reproductive bud development in Cymbidium orchids. J. Am. Soc. Hortic. Sci. 133, 552–556.
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L., Riechmann, J. L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. doi: 10.1104/pp.126.1.122
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J., Riechmann, J. L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. doi: 10.1105/tpc.009506
Riechmann, J. L., Meyerowitz, E. M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101.
Riechmann, J. L., Wang, M., Meyerowitz, E. M. (1996). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24, 3134–3141. doi: 10.1093/nar/24.16.3134
Rijpkema, A. S., Zethof, J., Gerats, T., Vandenbussche, M. (2009). The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 60, 1–9. doi: 10.1111/j.1365-313X.2009.03917.x
Rotor, G. B., Withner, C. L. (1959). "The photoperiodic and temperature response of orchids," in The orchids: a scientific survey. Ed. Withner, C. L. (NY, USA: Ronald Press) 397–416.
Rotor, G. B. (1952). Daylength and temperature in relation to growth and flowering of orchids. Cornell Expt. Sta. Bul. 885, 3–47.
Sakanishi, Y., Imanishi, H., Ishida, G. (1980). Effect of temperature on growth and flowering of Phalaenopsis amabilis. Bull. Univ. Osaka Prefecture. Ser. B Agric. Biol. 32, 1–9.
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Sawettalake, N., Bunnag, S., Wang, Y., Shen, L., Yu, H. (2017). DOAP1 promotes flowering in the orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 8, 400. doi: 10.3389/fpls.2017.00400
Schonrock, N., Bouveret, R., Leroy, O., Borghi, L., Kohler, C., Gruissem, W., et al. (2006). Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 20, 1667–1678. doi: 10.1101/gad.377206
Schuiteman, A. (2004). Devogelia (Orchidaceae), a new genus from the Moluccas and New Guinea. Blumea - Biodivers. Evol. Biogeogr. Plants 49, 361–366. doi: 10.3767/000651904X484324
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Shore, P., Sharrocks, A. D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x
Shrestha, B. R., Chin, D. P., Tokuhara, K., Mii, M. (2007). Efficient production of transgenic plantls of Vanda through sonication-assisted Agrobacterium-mediated transformation of protocorm-like bodies. Plant Biotechnol. 24, 429–434. doi: 10.5511/plantbiotechnology.24.429
Simpson, G. G., Dean, C. (2002). Arabidopsis. Science 296, 285–289. doi: 10.1126/science.296.5566.285
Sinoda, K., Suto, K., Hara, M., Aoki, M. (1988). Effect of day and night temperature on the flowering of Dendrobium nobile-type cultivars. Bull. Natl. Res. Inst. Vegetables Ornam. Plants Tea. Ser. A. 2, 279–290.
Sjahril, R., Mii, M. (2006). High-efficiency Agrobacterium-mediated transformation of Phalaenopsis using meropenem, a novel antibiotic to eliminate Agrobacterium. J. Hortic. Sci. Biotechnol. 81, 458–464. doi: 10.1080/14620316.2006.11512088
Skipper, M., Pedersen, K. B., Johansen, L. B., Frederiksen, S., Irish, V. F., Johansen, B. B. (2005). Identification and quantification of expression levels of three FRUITFULL-like MADS-box genes from the orchid Dendrobium thyrsiflorum (Reichb. f.). Plant Sci. 169, 579–586. doi: 10.1016/j.plantsci.2005.04.011
Skipper, M., Johansen, L. B., Pedersen, K. B., Frederiksen, S., Johansen, B. B. (2006). Cloning and transcription analysis of an AGAMOUS-and SEEDSTICK ortholog in the orchid Dendrobium thyrsiflorum (Reichb. f.). Gene 366, 266–274. doi: 10.1016/j.gene.2005.08.014
Song, I.-J., Nakamura, T., Fukuda, T., Yokoyama, J., Ito, T., Ichikawa, H., et al. (2006). Spatiotemporal expression of duplicate AGAMOUS orthologues during floral development in Phalaenopsis. Dev. Genes Evol. 216, 301–313. doi: 10.1007/s00427-005-0057-0
Su, C.-l., Chao, Y.-T., Yen, S.-H., Chen, C.-Y., Chen, W.-C., Chang, Y.-C. A. (2013a). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54 (2), e11–e11. doi: 10.1093/pcp/pct004
Su, C.-l., Chen, W.-C., Lee, A.-Y., Chen, C.-Y., Chang, Y.-C. A., Chao, Y.-T. (2013b). A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PloS One 8 (11), e80462. doi: 10.1371/journal.pone.0080462
Tapia-Lopez, R., Garcia-Ponce, B., Dubrovsky, J. G., Garay-Arroyo, A., Perez-Ruiz, R. V., Kim, S. H., et al. (2008). An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 146, 1182–1192. doi: 10.1104/pp.107.108647
Teo, Z. W. N., Song, S., Wang, Y.-Q., Liu, J., Yu, H. (2014). New insights into the regulation of inflorescence architecture. Trends Plant Sci. 19, 158–165. doi: 10.1016/j.tplants.2013.11.001
Theißen, G., Kim, J. T., Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/BF02337521
Theißen, G., Melzer, R., Rümpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Theißen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Theissen, G., Saedler, H. (2001). Plant biology: floral quartets. Nature 409, 469. doi: 10.1038/35054172
Tian, Y., Yuan, X., Jiang, S., Cui, B., Su, J. (2013). Molecular cloning and spatiotemporal expression of an APETALA1/FRUITFULL-like MADS-box gene from the orchid (Cymbidium faberi). Sheng Wu Gong Cheng Xue Bao 29, 203–213.
Tong, C. G., Wu, F. H., Yuan, Y. H., Chen, Y. R., Lin, C. S. (2019). High-efficiency CRISPR/Cas-based editing of Phalaenopsis orchid MADS genes. Plant Biotechnol. J. doi: 10.1111/pbi.13264
Tran Thanh Van, M. (1974). Methods of acceleration of growth and flowering in a few species of orchids. Amer. Orchid Soc. Bul. 43, 699–707.
Tsai, W.-C., Kuoh, C.-S., Chuang, M.-H., Chen, W.-H., Chen, H.-H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 45, 831–844. doi: 10.1093/pcp/pch095
Tsai, W.-C., Lee, P.-F., Chen, H.-I., Hsiao, Y.-Y., Wei, W.-J., Pan, Z.-J., et al. (2005). PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 46, 1125–1139. doi: 10.1093/pcp/pci125
Tsai, W.-C., Fu, C.-H., Hsiao, Y.-Y., Huang, Y.-M., Chen, L.-J., Wang, M. (2013). OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54 (2), e7–e7. doi: 10.1093/pcp/pcs187
Tsai, W.-C., Pan, Z. J., Hsiao, Y. Y., Chen, L.-J., Liu, Z. J. (2014). Evolution and function of MADS-box genes involved in orchid floral development. J. Syst. Evol. 52, 397–410. doi: 10.1111/jse.12010
Tsai, W.-C., Dievart, A., Hsu, C.-C., Hsiao, Y.-Y., Chiou, S.-Y., Huang, H. (2017). Post genomics era for orchid research. Bot. Stud. 58 (1), 61. doi: 10.1186/s40529-017-0213-7
Tzeng, T. Y., Chen, H. Y., Yang, C. H. (2002). Ectopic expression of carpel-specific MADS box genes from lily and lisianthus causes similar homeotic conversion of sepal and petal in Arabidopsis. Plant Physiol. 130, 1827–1836. doi: 10.1104/pp.007948
Valoroso, M. C., Censullo, M. C., Aceto, S. (2019). The MADS-box genes expressed in the inflorescence of Orchis italica (Orchidaceae). PloS One 14, e0213185. doi: 10.1371/journal.pone.0213185
Van Tunen, A. J., Eikelboom, W., Angenent, G. C. (1993). Floral organogenesis in tulipa. Flow. Newsl. 16, 6.
Wang, J.-W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, S.-Y., Lee, P.-F., Lee, Y.-I., Hsiao, Y.-Y., Chen, Y.-Y., Pan, Z.-J., et al. (2011). Duplicated C-class MADS-box genes reveal distinct roles in gynostemium development in Cymbidium ensifolium (Orchidaceae). Plant Cell Physiol. 52, 563–577. doi: 10.1093/pcp/pcr015
Wang, Y., Liu, L., Song, S., Li, Y., Shen, L., Yu, H. (2017). DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya Smile. J. Exp. Bot. 68, 5759–5772. doi: 10.1093/jxb/erx400
Weigel, D., Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Wijnstekers, W. (2001). Evolution of Cites: a Reference to the Convention on International Trade in Endangered Species of Wild Fauna and Flora. (London, United Kingdom: Stationery Office Books)
Xiang, L., Li, X., Qin, D., Guo, F., Wu, C., Miao, L., et al. (2012). Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol. Biochem. 58, 98–105. doi: 10.1016/j.plaphy.2012.06.011
Xu, Y., Teo, L. L., Zhou, J., Kumar, P. P., Yu, H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46, 54–68. doi: 10.1111/j.1365-313X.2006.02669.x
Yan, L., Wang, X., Liu, H., Tian, Y., Lian, J., Yang, R., et al. (2015). The genome of Dendrobium officinale illuminates the biology of the important traditional chinese orchid herb. Mol. Plant 8, 922–934. doi: 10.1016/j.molp.2014.12.011
Yang, F., Zhu, G. (2015). Digital gene expression analysis based on de novo transcriptome assembly reveals new genes associated with floral organ differentiation of the orchid plant Cymbidium ensifolium. PloS One 10 (11), e0142434. doi: 10.1371/journal.pone.0142434
Yang, F., Zhu, G., Wei, Y., Gao, J., Liang, G., Peng, L., et al. (2019). Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genomics 20, 53. doi: 10.1186/s12864-019-5425-7
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35. doi: 10.1038/346035a0
Yoo, S. K., Lee, J. S., Ahn, J. H. (2006). Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem. Biophys. Res. Commun. 348, 929–936. doi: 10.1016/j.bbrc.2006.07.121
Yoo, S. K., Wu, X., Lee, J. S., Ahn, J. H. (2011). AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 65, 62–76. doi: 10.1111/j.1365-313X.2010.04402.x
Yu, H., Goh, C. J. (2000). Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123, 1325–1336. doi: 10.1104/pp.123.4.1325
Yu, H., Goh, C. J. (2001). Molecular genetics of reproductive biology in orchids. Plant Physiol. 127, 1390–1393. doi: 10.1104/pp.010676
Yu, H., Xu, Y., Tan, E. L., Kumar, P. P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. U. S. A. 99, 16336–16341. doi: 10.1073/pnas.212624599
Yu, L. H., Miao, Z. Q., Qi, G. F., Wu, J., Cai, X. T., Mao, J. L., et al. (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Zhang, H., Forde, B. G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279 (5349), 407–409. doi: 10.1126/science.279.5349.407
Zhang, L., Chin, D. P., Mii, M. (2010). Agrobacterium-mediated transformation of protocorm-like bodies in Cattleya. Plant Cell Tissue Organ Cult. (PCTOC) 103, 41–47. doi: 10.1007/s11240-010-9751-3
Zhang, G.-Q., Xu, Q., Bian, C., Tsai, W.-C., Yeh, C.-M., Liu, K.-W., et al. (2016). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 19029. doi: 10.1038/srep19029
Zhang, G.-Q., Liu, K.-W., Li, Z., Lohaus, R., Hsiao, Y.-Y., Niu, S.-C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379. doi: 10.1038/nature23897
Keywords: orchid, MADS-box transcription factors, floral transition, floral patterning, development
Citation: Teo ZWN, Zhou W and Shen L (2019) Dissecting the Function of MADS-Box Transcription Factors in Orchid Reproductive Development. Front. Plant Sci. 10:1474. doi: 10.3389/fpls.2019.01474
Received: 02 August 2019; Accepted: 23 October 2019;
Published: 15 November 2019.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Adriana Garay, National Autonomous, University of Mexico, MexicoPhilip Ruelens, Wageningen University & Research, Netherlands
Copyright © 2019 Teo, Zhou and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisha Shen, bGlzaGFAdGxsLm9yZy5zZw==
 Zhi Wei Norman Teo
Zhi Wei Norman Teo Wei Zhou
Wei Zhou Lisha Shen
Lisha Shen