
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 18 November 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01453
 Xiang-Zhan Zhang1,2†
Xiang-Zhan Zhang1,2† Wei-Jun Zheng1†
Wei-Jun Zheng1† Xin-You Cao3
Xin-You Cao3 Xi-Yan Cui4
Xi-Yan Cui4 Shu-Ping Zhao1
Shu-Ping Zhao1 Tai-Fei Yu2
Tai-Fei Yu2 Jun Chen2
Jun Chen2 Yong-Bin Zhou1
Yong-Bin Zhou1 Ming Chen2
Ming Chen2 Shou-Cheng Chai1*
Shou-Cheng Chai1* Zhao-Shi Xu2*
Zhao-Shi Xu2* You-Zhi Ma2
You-Zhi Ma2Stress associated proteins (SAPs) containing A20/AN1 zinc finger domains have emerged as novel regulators of stress responses. In this study, 27 SAP genes were identified in soybean. The phylogenetic relationships, exon–intron structure, domain structure, chromosomal localization, putative cis-acting elements, and expression patterns of SAPs in various tissues under abiotic stresses were analyzed. Among the soybean SAP genes, GmSAP16 was significantly induced by water deficit stress, salt, and abscisic acid (ABA) and selected for further analysis. GmSAP16 was located in the nucleus and cytoplasm. The overexpression of GmSAP16 in Arabidopsis improved drought and salt tolerance at different developmental stages and increased ABA sensitivity, as indicated by delayed seed germination and stomatal closure. The GmSAP16 transgenic Arabidopsis plants had a higher proline content and a lower water loss rate and malondialdehyde (MDA) content than wild type (WT) plants in response to stresses. The overexpression of GmSAP16 in soybean hairy roots enhanced drought and salt tolerance of soybean seedlings, with higher proline and chlorophyll contents and a lower MDA content than WT. RNA inference (RNAi) of GmSAP16 increased stress sensitivity. Stress-related genes, including GmDREB1B;1, GmNCED3, GmRD22, GmDREB2, GmNHX1, and GmSOS1, showed significant expression alterations in GmSAP16-overexpressing and RNAi plants under stress treatments. These results indicate that soybean SAP genes play important roles in abiotic stress responses.
Adverse environmental stresses, such as drought, salt, and extreme temperatures, greatly affect plant growth, reduce agricultural productivity, and threaten food security (Guo et al., 2018; Qi et al., 2018). A biotechnology-mediated approach by editing or overexpressing stress-responsive genes has been proven to be a prospective tool for the improvement of abiotic stress-tolerant crop cultivars and has provided new insights into the mechanisms of stress tolerance (Umezawa et al., 2006; Hirayama and Shinozaki, 2010; Scheben et al., 2017).
Plants have developed a range of complex and efficient signaling networks to respond to various abiotic stresses (Shinozaki and Yamaguchi-Shinozaki, 2007; Kidokoro et al., 2015; Zhu, 2016; Yang and Guo, 2018). A number of genes involved in abiotic stress-related defenses have been identified, including regulatory proteins for conducting signals and transcriptional regulating stress response genes, such as mitogen-activated protein kinases (MAPKs), protein phosphatases, NAC, bZIP, AP2/ERF, MYB, MYC transcription factors, and stress associated proteins (SAPs) (Mukhopadhyay et al., 2004; Umezawa et al., 2009; Cristina et al., 2010; Xu et al., 2011; Joshi et al., 2016). Recently, the SAP family has been identified as novel regulatory proteins that participate in multiple abiotic stresses in plants (Giri et al., 2013; Dixit et al., 2018).
The SAPs belong to the zinc finger protein family, which are characterized by containing N-terminal A20 zinc finger domains, often in combination with the AN1 zinc finger domain and/or the Cys2–His2 zinc finger domain at the C-terminus (Giri et al., 2013). Increasing evidence has shown that the SAP family is involved in the regulation of environmental stress responses. Rice OsISAP1 was the first SAP gene isolated in plants and was induced by various abiotic stresses, such as cold, drought, salt, heavy metal, wounding, and abscisic acid (ABA). The overexpression of OsISAP1 in transgenic tobacco conferred stress tolerance at different developmental stages (Mukhopadhyay et al., 2004). Recently, Arabidopsis AtSAP13 was reported to provide drought, salt, and toxic metal tolerance (Dixit et al., 2018). The overexpression of AtSAP5 in cotton improved drought and heat tolerance by regulating stress-responsive genes (Hozain et al., 2012). The overexpression of the halophyte grass AlSAP gene in rice showed broad tolerance to abiotic stresses and increased rice grain yield under drought stress (Ben Saad et al., 2012).
In addition, SAP genes are involved in immune regulation, phytohormone response, and plant development. For example, transgenic tobacco expressing the OsSAP1 gene improved basal resistance by upregulating defense-related genes (Tyagi et al., 2014). Pha13, the homolog of AtSAP5, acts as an important regulator in plant antiviral immunity by coordinating with A20 and/or AN1 domains and modulating the activity of the E3 ligase (Chang et al., 2018). The overexpression of the rice A20/AN1 zinc-finger protein-coding genes OsZFP185 and OsDOG exhibited a semi-dwarf phenotype with reduced cell elongation and decreased gibberellic acid content (Liu et al., 2011; Zhang et al., 2015).
Accumulating studies have revealed that SAP members usually interact with other proteins to regulate specific responses. It has been reported that rice OsSAP1 can interact with itself and other proteins, including OsSAP11, receptor-like cytoplasmic kinase (OsRLCK253), aminotransferase (OsAMTR1), and pathogenesis-related protein (OsSCP), to form homo- or heterodimers through the A20 domain to regulate downstream stress-responsive genes (Giri et al., 2011; Kothari et al., 2016). Arabidopsis AtMBP interacted with the E3 ubiquitin ligase AtSAP5 to modulate the degradation of AtMBP and thus regulate ABA responses (Kang et al., 2013). It has been reported that AtSAP9 interacts with Rad23d to mediate biotic and abiotic stresses via the proteasome pathway (Kang et al., 2017). Prunus PpSAP1 strongly interacted with polyubiquitin proteins in a yeast two-hybrid system, and the overexpression of PpSAP1 increased water retention under drought stress (Lloret et al., 2017). Wheat TaSAP5 was reported to alter the drought stress response by promoting the degradation of DRIP proteins (Zhang et al., 2017).
Soybean (Glycine max), one of the most important crops worldwide, provides vegetable oil and protein meal to humans. With abiotic stress constraints in soybean production and an increasing global demand for soybean products, it is necessary to develop abiotic stress-tolerant soybean cultivars. Considering the potential role of SAPs in mediating stress resistance and the lack of information on soybean SAPs, we conducted a comprehensive genome-wide analysis of SAP genes in the soybean genome. A total of 27 SAP members were identified, and expression patterns under various stress treatments were analyzed. The GmSAP16 gene was functionally identified to be responsive to multiple stresses. This study contributes to expanding our understanding of the molecular mechanism underlying stress responses and tolerance.
The sequences of SAPs from Arabidopsis and rice were obtained from the TAIR database (https://www.arabidopsis.org/index.jsp) and the rice genome database (http://rice.plantbiology.msu.edu), respectively. The BLASTP program was used to search the SAP sequences against the soybean genome database Phytozome V10.3 (http://phytozome.jgi.doe.gov). After the removal of redundant sequences, candidate SAPs were confirmed by using the Pfam database (http://pfam.xfam.org/). The theoretical isoelectric points (pI) and molecular weights (Mw) of soybean SAP members were predicted on the ExPASy website (https://web.expasy.org/compute_pi/).
The sequences of SAPs in Arabidopsis, rice, and soybean were aligned by Clustal X 2.0 and then imported into MEGA6.0 to construct the phylogenetic tree using the neighbor-joining method with a bootstrap value of 1,000 repeats, as previously described (Zhao et al., 2017). The location information of soybean SAP genes was obtained from the Phytozome database. All soybean SAP genes were mapped on soybean chromosomes by using MapInspect software (http://mapinspect.software.informer.com) (Song et al., 2016).
The conserved domain and exon–intron structure of soybean SAPs were analyzed using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/). The promoter regions (2 kb upstream of the translation initiation codon) of the soybean SAP genes were obtained from the soybean genome database Phytozome, and the cis-acting elements were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Total RNA was isolated using RNAiso Plus reagent (Takara, Japan) according to the manufacturer’s manual. First-stand complementary DNA (cDNA) synthesis was performed using the PrimeScript II 1st Strand cDNA Synthesis Kit (Takara). Real-time quantitative PCR (RT-qPCR) was conducted with an ABI7500 real-time PCR system (ABI, USA) using TransStart Top Green qPCR SuperMix (Transgen, China). The data were analyzed as previously described (Liu et al., 2013). Arabidopsis actin2 (At3g18780) and soybean CYP2 (Glyma12g02790) were used as internal controls. Three biological replicates were applied in the experiments. The specific primers are listed in Supplemental Table S1.
The coding sequence of GmSAP16 (without a termination codon) was inserted into the N-terminus of green fluorescent protein (GFP), which was driven by the cauliflower mosaic virus (CaMV) 35S promoter. For subcellular localization, the GmSAP16-GFP recombinant vector was transformed into Arabidopsis protoplasts as described previously (Cui et al., 2019). After incubation in the dark at 23′C for 18–24 h, the protoplasts were observed using a confocal laser scanning microscope (Carl Zeiss LSM 700, Germany). GFP fluorescence was excited by argon laser at 488 nm and fluorescence emission was captured between 495 and 540 nm. RFP fluorescence was excited at 561 nm by HeNe laser and fluorescence emission was captured between 580 and 620 nm. The primer sequences used in the research are listed in Supplementary Table S1.
To obtain GmSAP16 transgenic Arabidopsis, the open reading frame (without a termination codon) of GmSAP16 was cloned into pCAMBIA1302 under the control of the CaMV35S promoter. The recombinant plasmid was validated by sequencing and then introduced into Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis Columbia-0 (Col-0) by the floral dip method as described previously (Clough and Bent, 1998).
Wild-type (WT) and transgenic Arabidopsis were grown on 1/2 Murashige and Skoog (MS) medium with a photoperiod of 16 h light/8 h dark at 22′C. For germination assays, seeds of WT and GmSAP16-overexpressing lines were surface sterilized and sown on 1/2 MS medium containing 6% PEG6000 (m/v), 80 mM NaCl, and ABA (0.1 and 0.15 µM). Germination was recorded at the indicated time points based on radicle protrusion after stratification at 4′C for 3 days. To determine the germination rates, at least 80 seeds of WT and each transgenic lines were measured, three independent biological repeats were performed for the seed germination assays.
For root length assays, the 5-day-old uniformly germinated seeds were transferred onto 1/2 MS medium with different concentrations of PEG6000 (0, 6, and 9%) and NaCl (0, 100, and 125 mM). Root length and fresh weight were evaluated after 7 days of treatment. Three biological repeats were performed, and at least 20 seedlings of each genotype were measured. To evaluate the drought and salt tolerance in soil, 3-week-old seedlings were subjected to drought stress by withholding water for 2 weeks and then rewatered for 3 days. For salt stress, seedlings growing under normal conditions were then treated with 250 mM NaCl for 1 week. The survival rate was recorded, and three independent biological repeats were performed.
Soybean (Zhonghuang 39) seedlings were grown in pots containing vermiculite at 25′C in a greenhouse. Two-week-old seedlings were treated with different stresses, including water deficit, salinity, and exogenous ABA. For drought stress, soybean seedlings were transferred onto filter paper for water deficit stress. For salinity stress and ABA treatment, soybean seedlings were transferred to Hoagland’s solution containing 250 mM NaCl or 200 µM ABA, respectively. The seedlings were harvested at the indicated time points 0, 1, 2, 4, 8, 12, and 24 h. Sampled seedlings were frozen in liquid nitrogen and stored at −80′C before RNA extraction.
For the measurement of the proline and malondialdehyde (MDA) contents, 3-week-old seedlings of WT and transgenic Arabidopsis were subjected to drought for 2 weeks or treated with salt stress for 5 days. Soybean seedlings were subjected to drought stress for 1 week or treated with 250 mM NaCl for 3 days. A fresh weight of 0.1 g seedlings from each genotype was sampled, and the proline, chlorophyll, and MDA contents were assayed as proposed by the manufacturer’s instructions (Comin, China). Absorbance values were measured using a Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific, USA). Each experiment included three biological replicates.
To obtain transgenic plants in soybean, the coding sequence of GmSAP16 was inserted into the plant transformation vector pCAMBIA3301 under the CaMV35S promoter to generate the GmSAP16-overexpressing (GmSAP16-OE) vector. For the GmSAP16 RNA interference (GmSAP16-RNAi) construct, a 554 bp specific fragment, including a 208 bp GmSAP16 fragment and its antisense sequence and a 146 bp maize alcohol dehydrogenase gene as spacer between the repeats, was synthesized (Augct, China) and inserted into pCAMBIA3301. The recombinant vectors and empty pCAMBIA3301 vector (CK) were introduced into A. rhizogenes strain K599 and then transformed into soybean hairy roots by A. rhizogenes-mediated transformation as reported previously (Kereszt et al., 2007). The soybean cultivar Williams 82 was used for the transformation. The primers used for cloning are listed in Supplementary Table S1.
Stomatal aperture assays were performed as described previously with modifications (Ryu et al., 2010). Rosette leaves of 3-week-old WT and GmSAP16-overexpressing Arabidopsis were treated with stomata opening buffer (100 mM CaCl2, 10 mM KCl, and 10 mM MES, pH 6.1) for 2 h and then subjected to buffers containing different concentrations of ABA (0, 5, and 10 µM) for 2 h. Images were taken using a confocal laser scanning microscope (Zeiss LSM 700, Germany) and analyzed by ImageJ software (Schneider et al., 2012). The stomatal aperture was calculated by the ratio of width/length, and at least 30 stomata for each genotype were measured. For the water content measurements, 1.0 g rosette leaves of 3 -week-old WT and GmSAP16-overexpressing plants were placed on filter paper at room temperature, and the fresh weight was measured at the indicated time points. Three independent biological replicates were performed for the measurements.
All data were obtained from three independent biological replicates for statistical analysis, and the values are shown as the mean ± standard deviation (SD). ANOVA analysis was conducted by statistical software SPSS 17.0, and the significance (P < 0.05) was labeled with different letters. Student’s t test was conducted using Excel 2010 to determine significance, which was labeled *, P < 0.05; and **, P < 0.01.
To identify SAP genes in soybean, the full-length amino acid sequences of 14 AtSAPs from Arabidopsis and 18 OsSAPs from rice were used as queries to search the Phytozome database using the BLASTP program. After the elimination of repeated sequences and confirmation of the A20/AN1 domain by the Pfam database, a total of 27 candidate SAP genes were identified in soybean (Table 1). The soybean SAP genes were named according to the A20/AN1 domains and their accession numbers. The protein length of soybean SAPs ranged from 123 amino acids (13.25 kDa) to 292 amino acids (32.47 kDa), and the predicted isoelectric points ranged from 6.50 (GmSAP11) to 8.98 (GmSAP6).
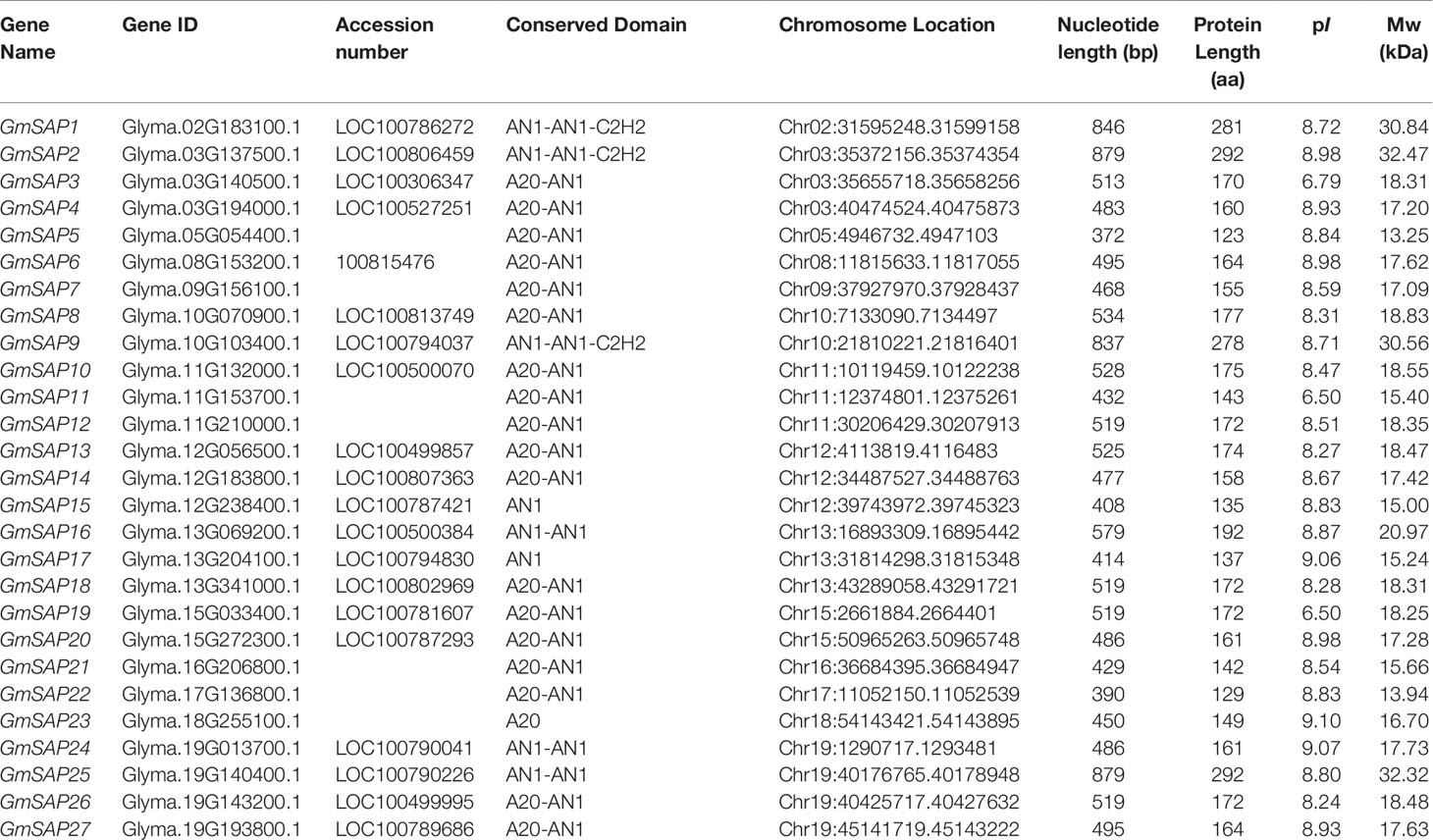
Table 1 Detailed information of the stress associated proteins (SAP) genes identified in the soybean genome.
To determine the phylogenetic and evolutionary relationships of soybean SAPs, a phylogenetic analysis was conducted with full-length sequences of soybean SAPs, Arabidopsis SAPs, and rice SAPs (Figure 1). Based on the intron presence, position, and splicing phase, soybean SAPs were mainly grouped into two clades (Figure 2A). There was an abundance of soybean SAPs in clade I, most SAPs in clade I contained typical A20 and AN1 domains and had no introns (Figures 2B, C). GmSAP23 contained a single A20 domain, GmSAP15 and GmSAP17 had only one AN1 domain. Soybean SAPs in clade II had two conserved AN1 domains, only one intron and a large splicing phase. GmSAP1, GmSAP2, and GmSAP9 contained additional C2H2 zinc finger domains (Figures 2B, C) (Supplementary Figure S1). Phylogenetic analysis showed that GmSAP6 and GmSAP20 were closely related to AtSAP5, which is homology to OsSAP1 and exhibited E3 ligase activity (Kang et al., 2013; Tyagi et al., 2014). GmSAP6 and GmSAP20 contains the conserved Cys- and His-residues that may be related to ubiquitin ligase activity, indicating that GmSAP6 and GmSAP20 may also function as ubiquitin ligases (Supplementary Figure S1).
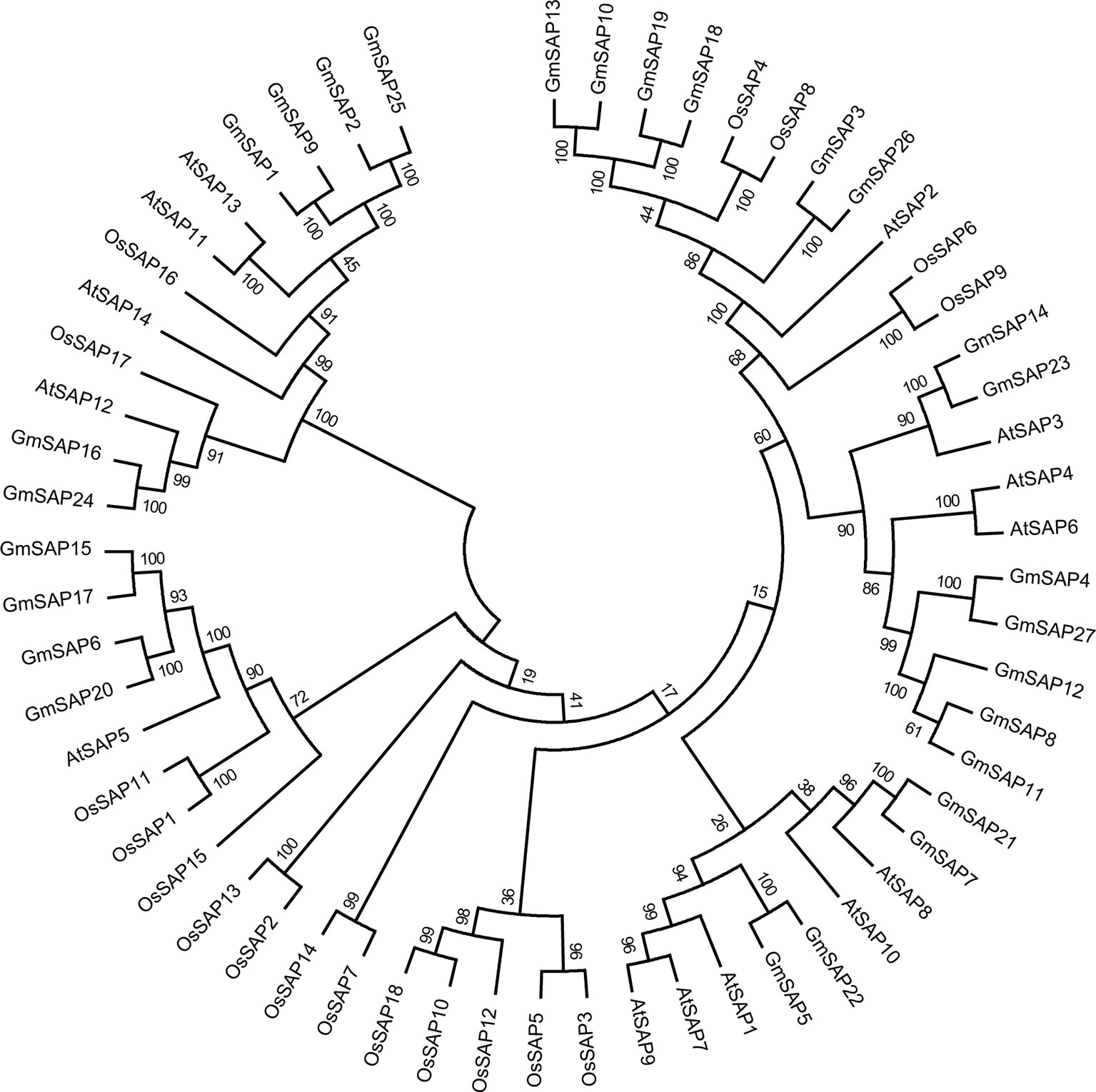
Figure 1 Phylogenetic analyses of stress associated proteins (SAPs) from Arabidopsis, rice, and soybean. The full-length amino acid sequences of SAPs were aligned by Clustal X 2.0, and a phylogenetic tree was constructed using MEGA 6.0 by the neighbor-joining method with 1,000 bootstrap replicates.
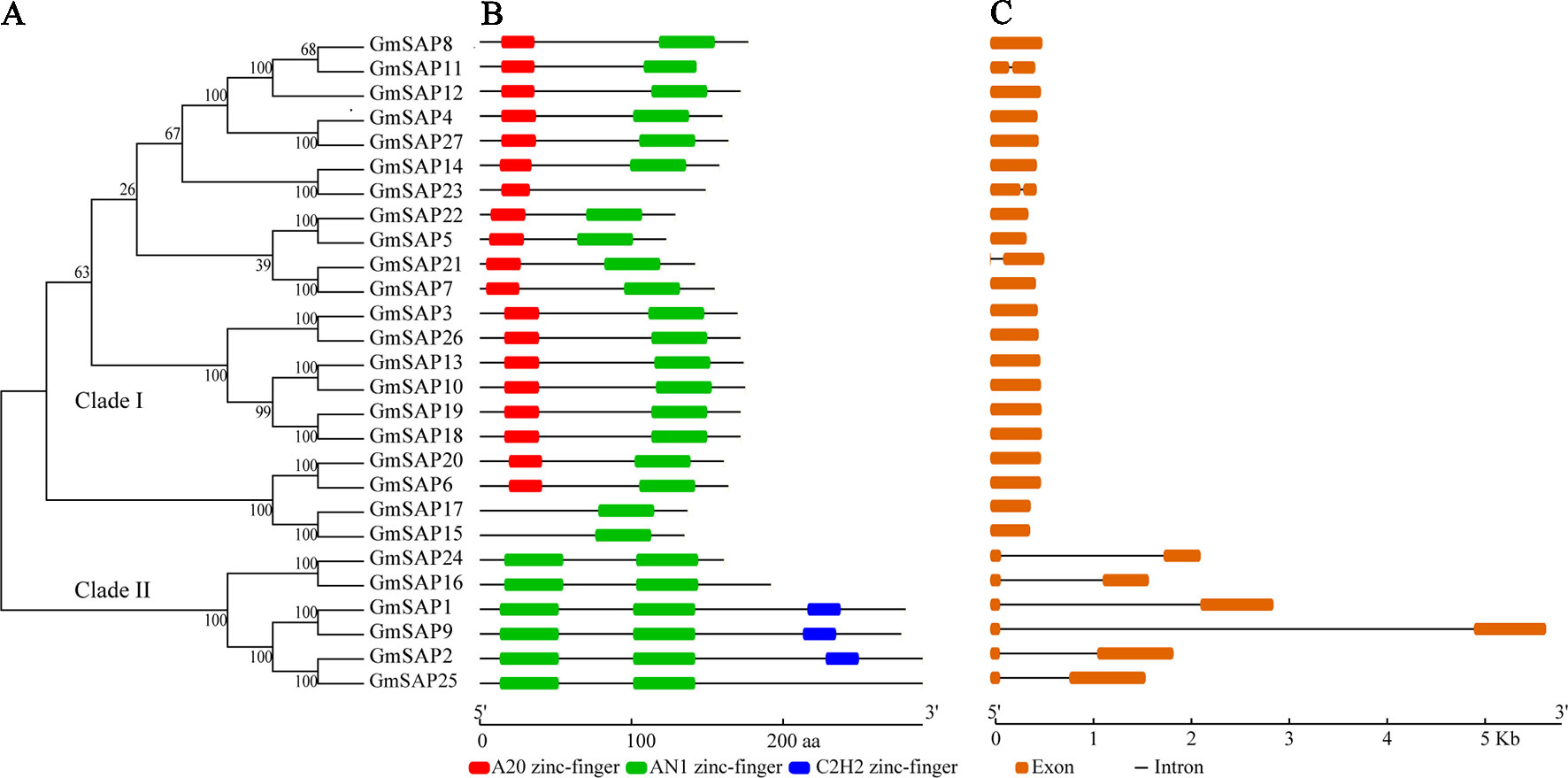
Figure 2 (A) Phylogenetic relationships; (B) conserved domain analysis; and (C) exon–intron structure analysis of soybean SAPs. The phylogenetic tree was conducted using full-length amino acid sequences by MEGA 6.0 with neighbor-joining for 1,000 bootstrap replicates. Exon–intron structure organization was performed using the Gene Structure Display Server online tool. The A20, AN1, and C2H2 zinc finger domains are indicated by red, green, and blue boxes, respectively. The orange boxes and black lines represent exons and introns, respectively. The sizes of exons and conserved domains are indicated at the bottom of the figures.
Cis-element analysis revealed that the soybean SAP genes contained cis-acting elements in their promoter regions, including the abiotic stress-related cis-elements MBS, LTR, MYC, MYB, STRE, and ABRE; the pathogen-related cis-elements TC-rich repeats, ARE, W-box, TCA, and CGTCA-motifs; and the phytohormone-responsive cis-elements ERE, GARE-motifs, and TGA elements (Supplementary Figure S2).
Based on the annotations of the soybean genomic locations, 27 soybean SAP genes were distributed among 14 chromosomes, excluding chromosomes 1, 4, 6, 7, 14, and 20 (Supplementary Figure S3). Four soybean SAP genes, including GmSAP24, GmSAP25, GmSAP26, and GmSAP27, were clustered on chromosome 19, which possesses the highest number of soybean SAP genes among all the soybean chromosomes.
The expression patterns of the soybean SAP genes in various tissues and organs were analyzed. The expression data were obtained from the soybean database, and eight soybean tissues, including flower, leaf, nodule, pod, root, root hair, seed, and stem tissues, were comprehensively analyzed (Figure 3). The results showed that most of the soybean SAP genes, including GmSAP2, GmSAP11, GmSAP21, GmSAP22, GmSAP23, GmSAP24, and GmSAP25, showed low transcript levels in various tissues. Several genes, including GmSAP10 and GmSAP13, showed high transcript levels in various tissues. Most soybean SAP genes showed different transcript levels in various tissues, such as GmSAP4, GmSAP10, GmSAP13, GmSAP14, GmSAP20, and GmSAP27, which showed high transcript levels in flowers. GmSAP21 was not expressed in flowers. The transcript profiles suggested that the soybean SAP genes may be involved in diverse tissue-dependent regulatory networks.

Figure 3 Patterns of soybean stress associated protein (SAPs) transcript accumulation in various soybean tissues (flower, leaf, nodule, pod, root, root hair, seed, and stem). The transcript level data of the soybean SAP genes were obtained from Phytozome. A heatmap was generated using MeV_4_9_0 software. Transcript levels are indicated by different colors on the scale bar.
According to our previous research, RNA-seq was performed using soybean seedlings exposed to various abiotic stresses, including water deficit, NaCl, and ABA (Shi et al., 2018). To study the potential functions of the soybean SAP genes in response to abiotic stresses, we analyzed the transcript profiles of the soybean SAP genes using RNA-seq data. The results showed that most of the soybean SAP genes were induced by abiotic stresses, especially water deficit stress (Figure 4).
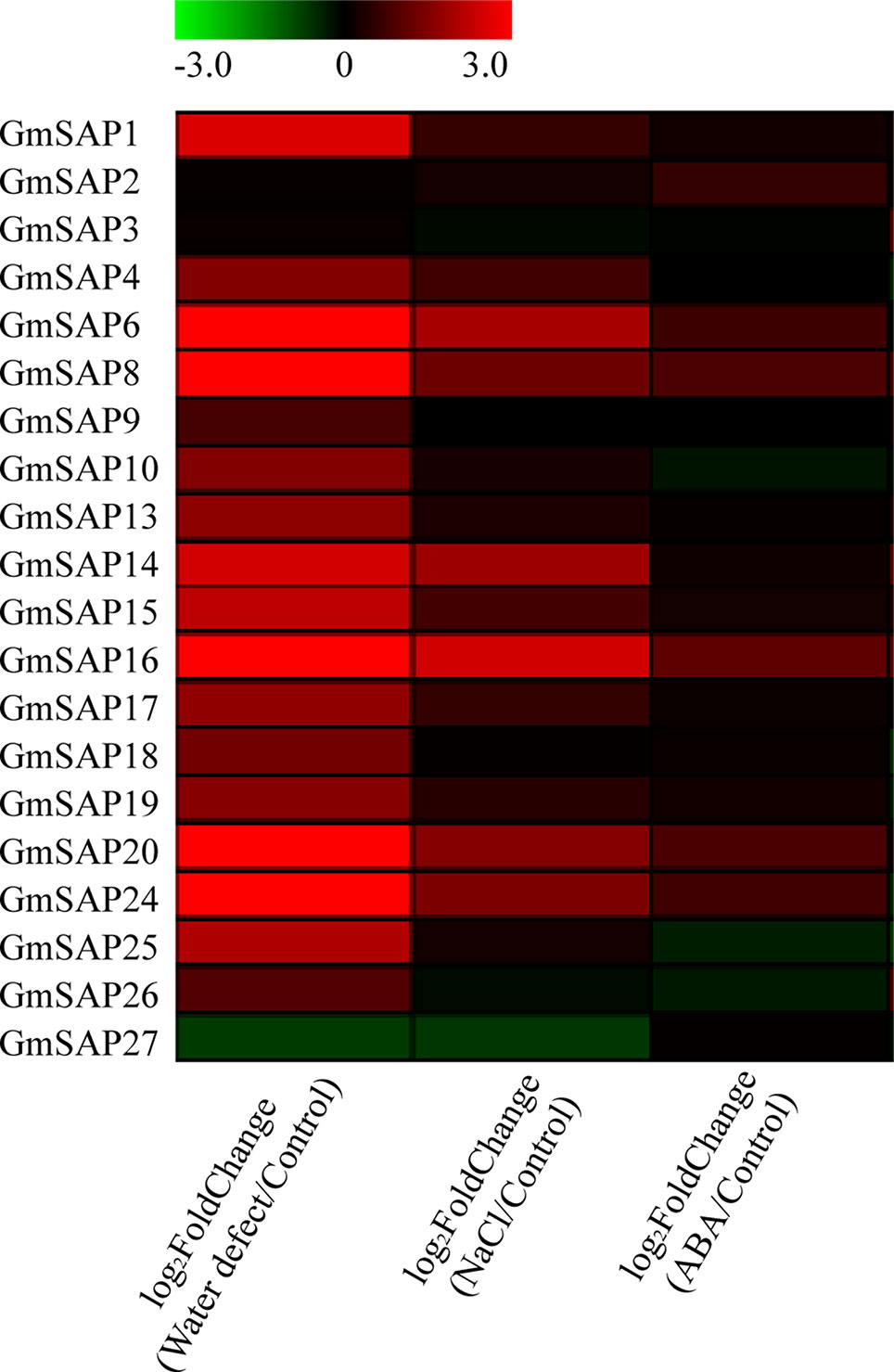
Figure 4 RNA-seq analysis of the soybean stress associated protein (SAP) genes under water deficit, salt, and abscisic acid (ABA) treatments. RNA-seq data were extracted from our previous research (Shi et al., 2018). The heatmap was produced by MeV_4_9_0 software, and different colors on the scale bar represent transcript levels.
To further confirm the transcript levels of the soybean SAP genes in response to abiotic stresses, we examined their expression patterns in response to water deficit, NaCl, and ABA by RT-qPCR analysis. Most of the soybean SAP genes were induced by water deficit stress and showed elevated transcript levels, except for GmSAP4, GmSAP7, GmSAP17, and GmSAP27. Among them, the transcript peak of GmSAP16 reached more than 300-fold (Figure 5). Similarly, most of the soybean SAP genes were induced by salt stress (Figure 6). For ABA treatment, most of the soybean SAP genes showed increased transcript levels, except for GmSAP7, GmSAP10, GmSAP20, and GmSAP22 (Figure 7). Combined with the results of RNA-seq and RT-qPCR, GmSAP16 was significantly induced by water deficit stress, and it also showed increased transcript level under salt stress and ABA treatment. Therefore, GmSAP16 was selected for further analysis.
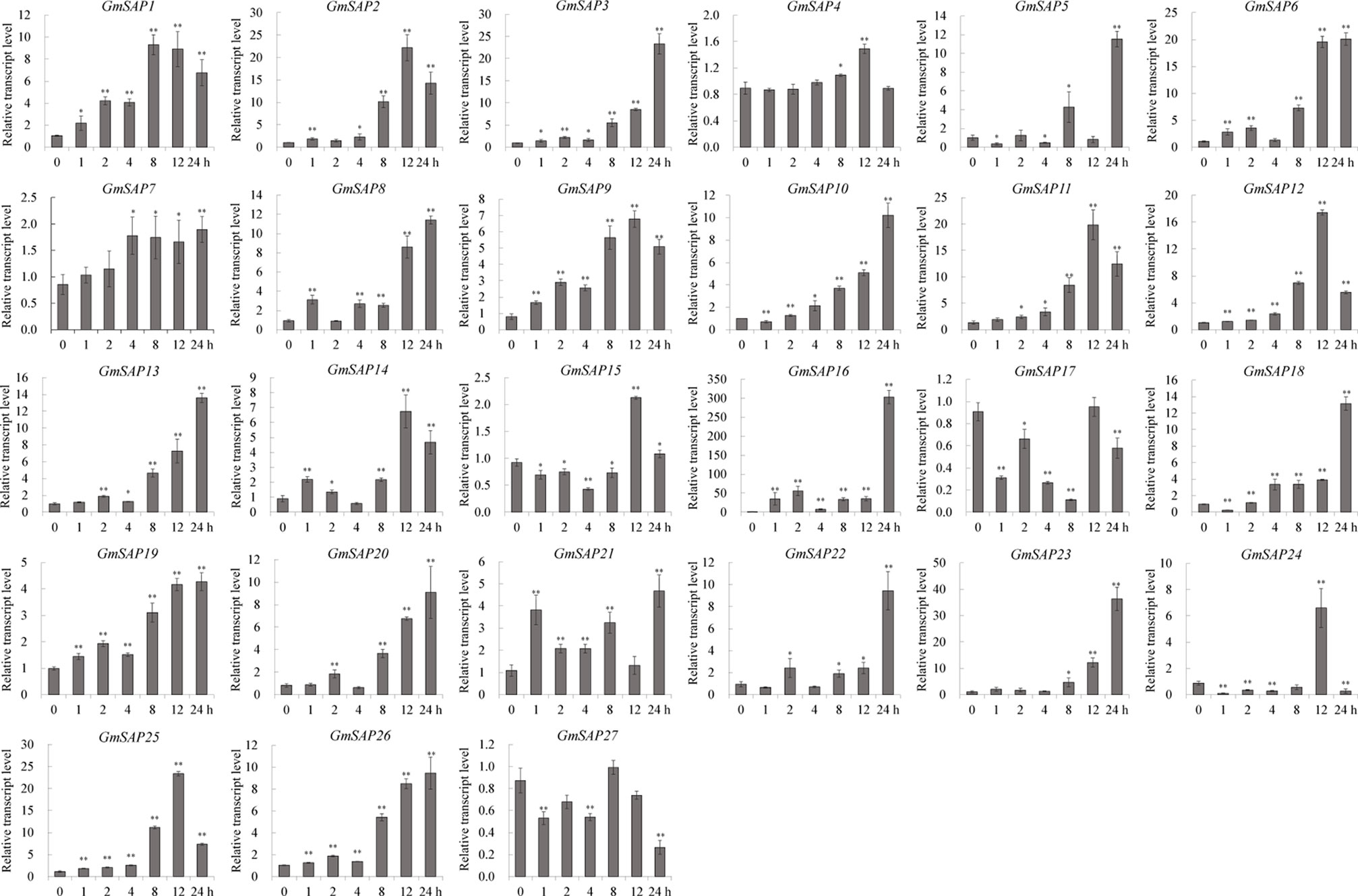
Figure 5 Patterns of soybean stress associated proteins (SAPs) transcript accumulation under water deficit stress. The data were normalized to the soybean internal control gene GmCYP2. Three biological replicates were performed, and the values are presented as the means ± SD. Asterisks indicate statistical significance (*, P 0.05; and **, P 0.01; Student’s t test) compared with the corresponding controls.
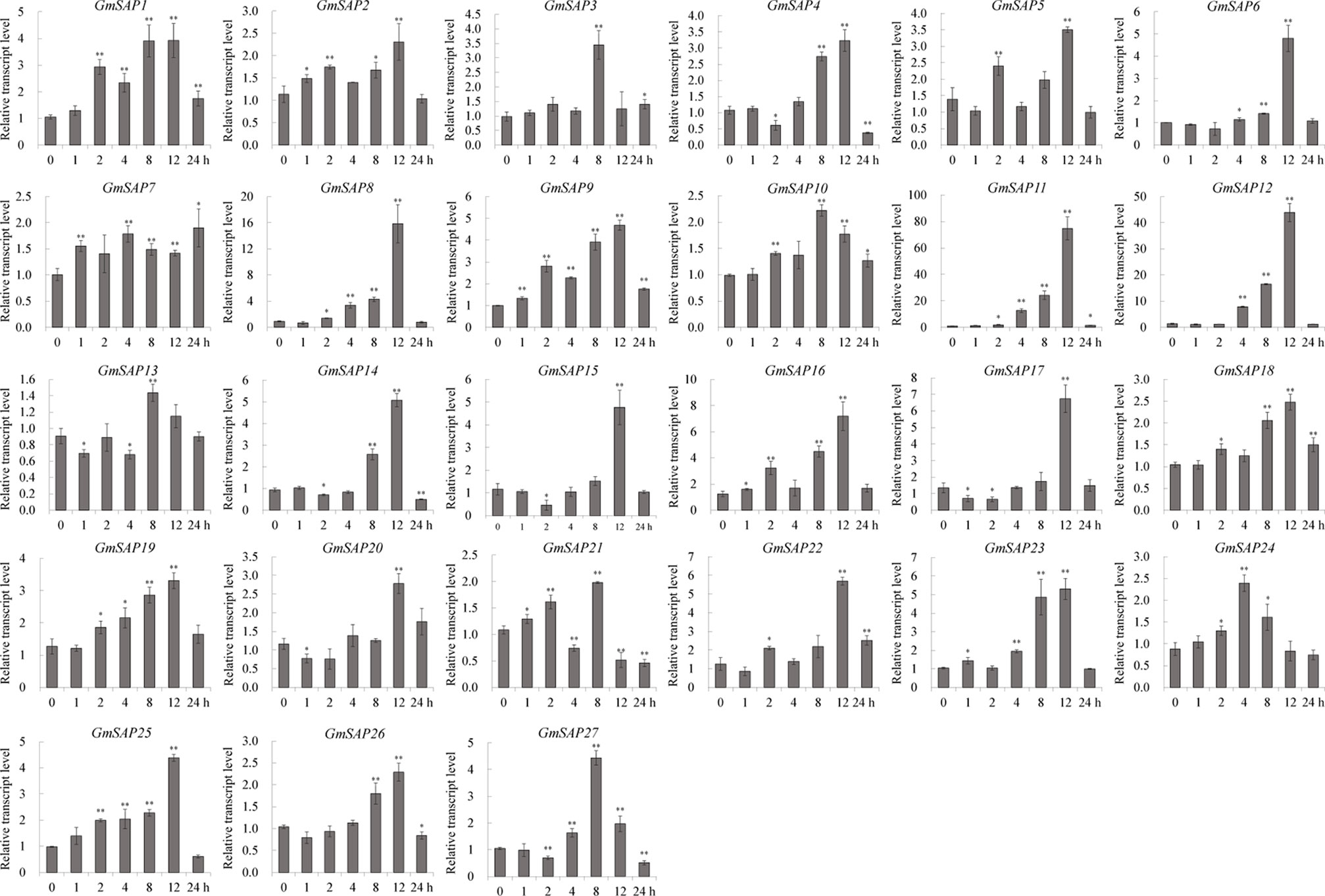
Figure 6 Patterns of soybean stress associated proteins (SAPs) transcript accumulation under salt stress. The data were normalized to the soybean internal control gene GmCYP2. Three biological replicates were performed, and the values are presented as the means ± SD. Asterisks indicate statistical significance (*, P 0.05; and **, P 0.01; Student’s t test) compared with the corresponding controls.
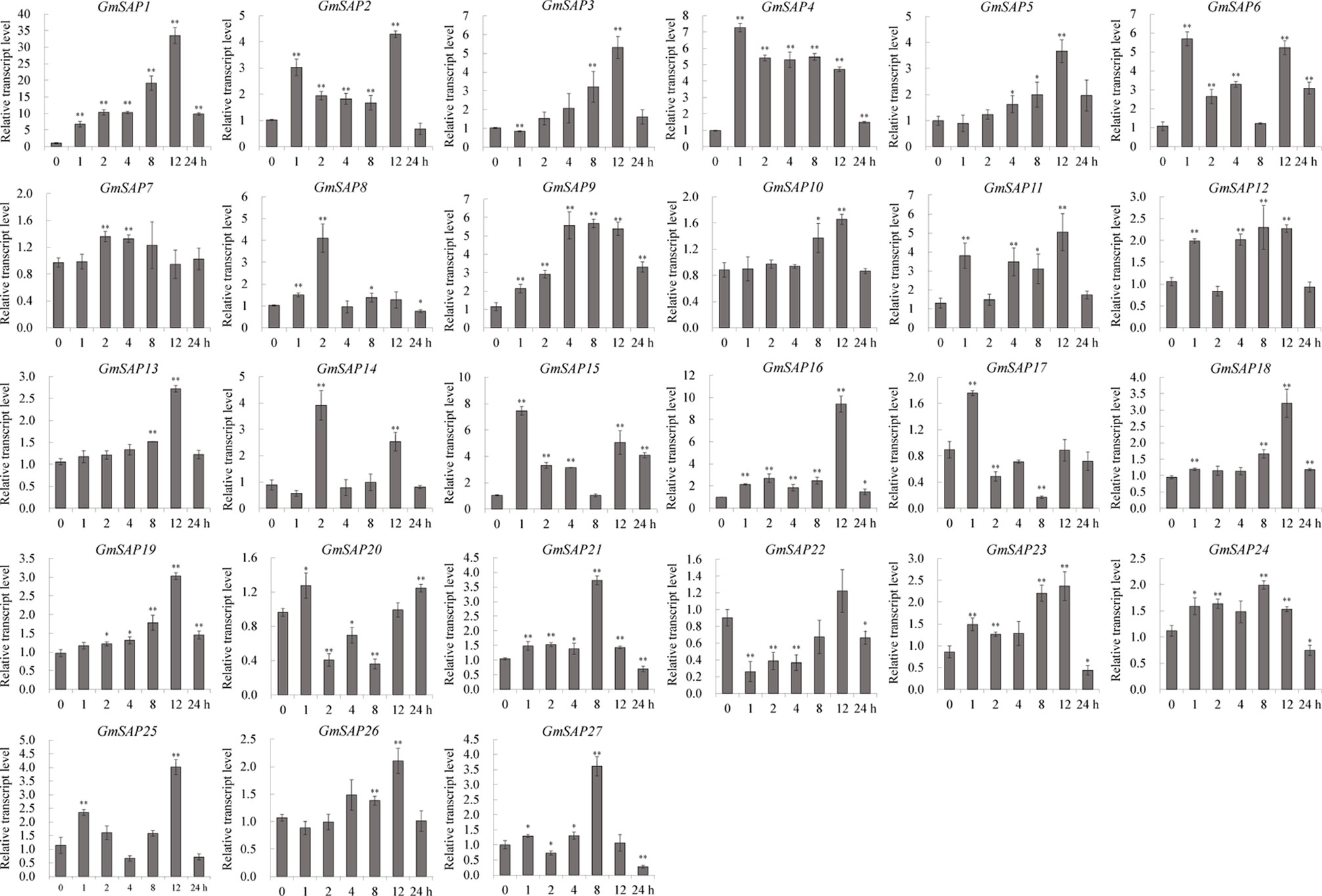
Figure 7 Patterns of soybean stress associated proteins (SAPs) transcript accumulation under abscisic acid (ABA) treatment. The data were normalized to the soybean internal control gene GmCYP2. Three biological replicates were performed, and the values are presented as the means ± SD. Asterisks indicate statistical significance (*, P 0.05; and **, P 0.01; Student’s t test) compared with the corresponding controls.
To examine the subcellular localization of GmSAP16, the cDNA sequence of GmSAP16 was cloned into the 16318hGFP vector under the control of the CaMV35S promoter. The empty vector 16318hGFP was used as a control, and nuclear localization signal–red fluorescent protein was used as the nucleus marker as reported previously (Ivanov and Harrison, 2014). GFP fluorescence was observed throughout cells. GmSAP16 was colocated with nuclear localization signal–red fluorescent protein in the nucleus, and the GmSAP16-GFP fusion protein also showed fluorescence in the cytoplasm, indicating that GmSAP16 was located in the nucleus and cytoplasm (Figure 8).
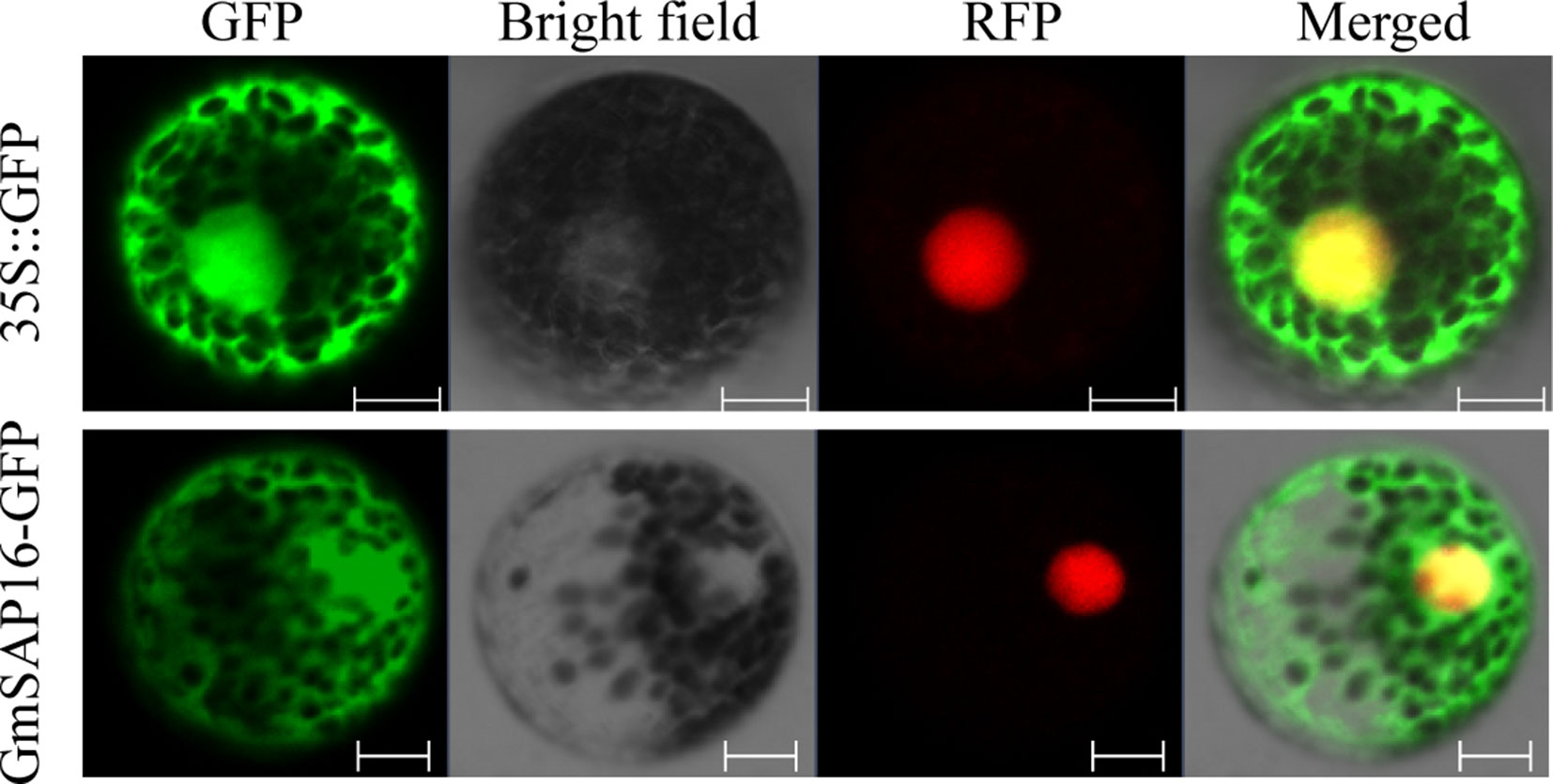
Figure 8 Subcellular localization of GmSAP16. 35S::GFP was used as a control, and nuclear localization signal–red fluorescent protein (NLS-RFP) was used as a nuclear marker. Green and red fluorescence signals were detected by confocal laser scanning. Scale bars = 10 µm.
To investigate the biological function of GmSAP16, three homozygous T3 transgenic Arabidopsis lines (OE-2, OE-3, and OE-5) were confirmed by RT-qPCR for further analysis (Supplementary Figure S4). Seed germination assays were performed with 1/2 MS medium containing 6% PEG6000 or 80 mM NaCl. No obvious difference in germination rate was observed between WT and the GmSAP16 transgenic lines on 1/2 MS medium (Figures 9A, D). When subjected to 1/2 MS medium containing 6% PEG6000, the germination of WT and transgenic lines was inhibited, and GmSAP16-overexpressing lines showed higher germination rates than WT (Figures 9B, E). The seed germination of WT and transgenic Arabidopsis lines on 1/2 MS medium containing 80 mM NaCl was significantly inhibited compared with that on 1/2 MS medium, and WT seeds showed delayed germination and lower germination rates than GmSAP16 transgenic lines (Figures 9C, F).
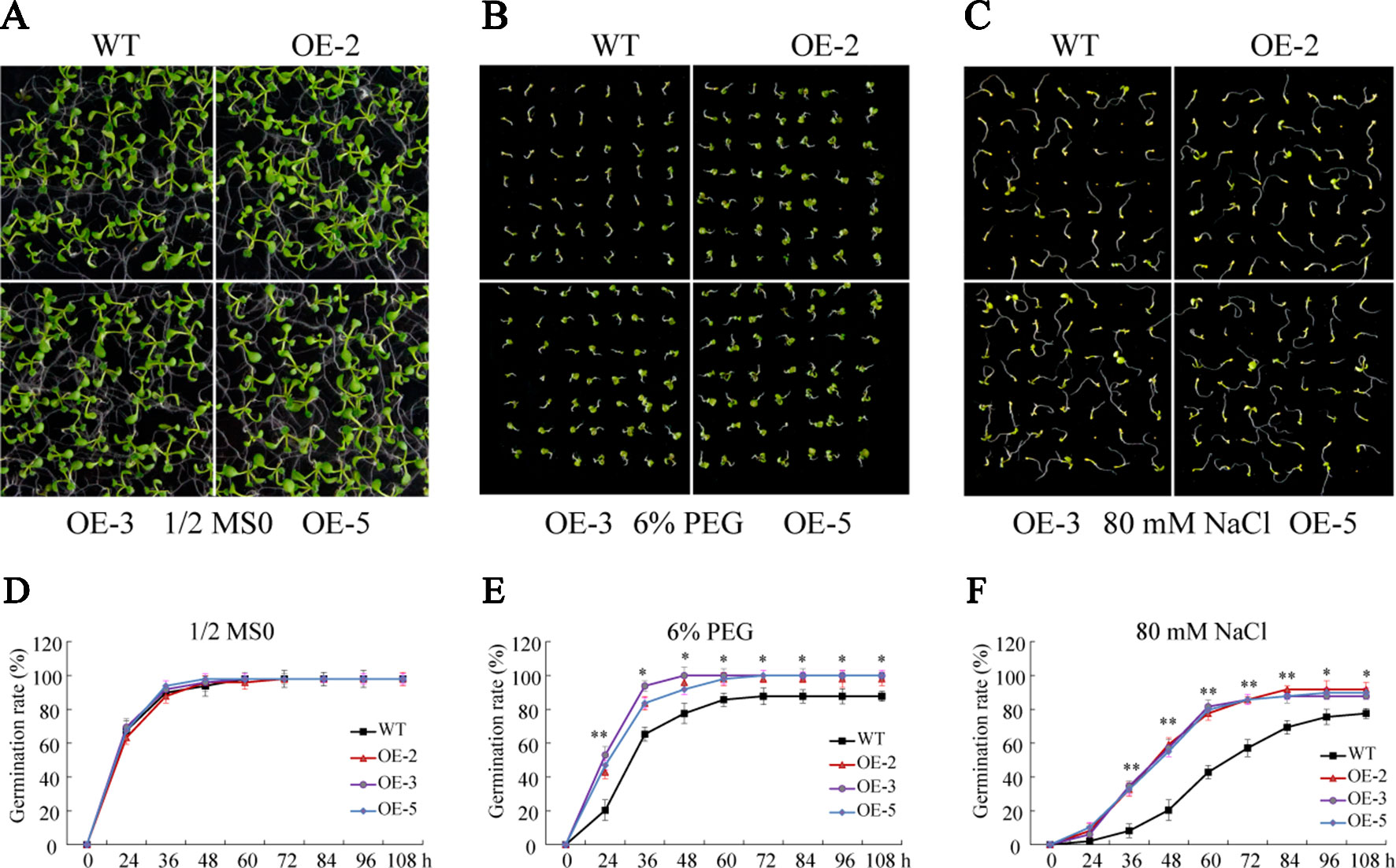
Figure 9 Germination assays of wild-type (WT) and GmSAP16 transgenic Arabidopsis seeds under polyethylene glycol (PEG) and NaCl treatments. Phenotypes of WT and GmSAP16 transgenic Arabidopsis seeds on 1/2 Murashige and Skoog (MS) medium (A), 1/2 MS medium containing 6% PEG (B), and 1/2 MS medium 80 mM NaCl (C). Germination rates of WT and GmSAP16 seeds on 1/2 MS medium (D), 1/2 MS medium containing 6% PEG (E), and 1/2 MS medium 80 mM NaCl (F). At least 80 seeds of each genotype were applied in the germination assays. Three biological replicates were performed, and error bars indicate ± SE of three replicates. Asterisks indicate statistical significance (*, P 0.05; and **, P 0.01; Student’s t test) compared with the corresponding controls.
To investigate the function of GmSAP16 under drought stress, 7-day-old uniformly germinated seeds of WT and GmSAP16-overexpressing lines were transferred to 1/2 MS medium containing 6% PEG6000 or 9% PEG6000 to simulate drought stress. After treatment for 1 week, the GmSAP16-overexpressing lines exhibited longer root lengths and greater fresh weights than WT (Figures 10A–C). All plants, including WT and transgenic Arabidopsis, showed no significant difference under normal conditions (Figures 10A–C).
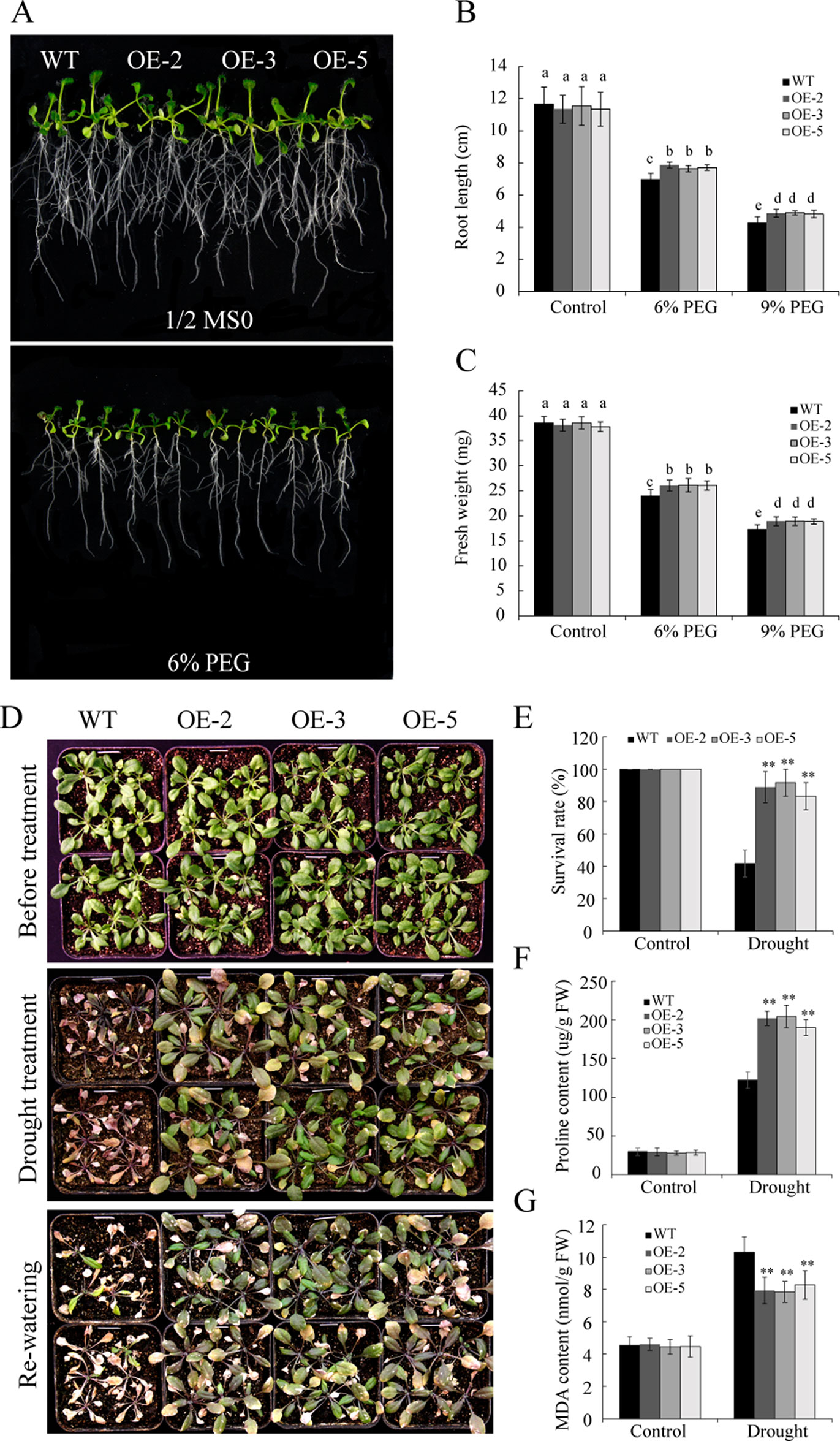
Figure 10 Overexpression of GmSAP16 enhances drought tolerance in Arabidopsis. (A) Root length assays of wild-type (WT) and GmSAP16-overexpressing plants under normal conditions and polyethylene glycol (PEG) treatment. Five-day-old seedlings of WT and GmSAP16-overexpressing lines were transferred to 1/2 Murashige and Skoog (MS) medium with different concentrations of PEG (0, 6%, and 9%) for root length assays. At least 20 seedlings were measured for (B) main root length and (C) fresh weight. (D) Drought tolerance phenotypes of WT and GmSAP16 transgenic Arabidopsis in soil. Three-week-old seedlings of WT and GmSAP16-overexpressing lines were dehydrated for 2 weeks and then rehydrated for 3 days. Statistical analysis of the survival rates (E), proline content (F), and MDA content (G). Data are presented as the mean ± SD of three independent replicates. The asterisks indicate significant differences between WT and GmSAP16-overexpressing lines (**, P 0.01; Student’s t test). Different letters indicate significant differences within treatments by ANOVA (P 0.05).
To assess the drought tolerance of GmSAP16 transgenic lines, 3-week-old seedlings of WT and GmSAP16 transgenic Arabidopsis were deprived of water or well-watered for 2 weeks. No significant differences were observed between WT and transgenic plants under normal conditions (Figure 10D). The GmSAP16-overexpressing lines exhibited less wilting and a higher survival rate than WT under drought stress (Figure 10E). To investigate potential physiological alterations for the enhanced drought tolerance of GmSAP16-overexpressing lines, the proline and MDA contents of WT plants and GmSAP16-overexpressing lines were measured. Result showed that all plants showed increased proline and MDA contents under drought stress conditions, and proline accumulation in the GmSAP16-overexpressing lines was significantly higher than that in WT plants under drought conditions (Figure 10F). In addition, WT plants accumulated higher MDA content than GmSAP16-overexpressing lines under drought stress (Figure 10G), indicating that the WT plants suffered more damages than GmSAP16-overexpressing plants under drought stress. WT plants and GmSAP16-overexpressing lines showed similar proline and MDA contents under normal conditions (Figures 10F, G).
To further gain insight into the role of GmSAP16 in salt stress, seedlings of 7-day-old WT and GmSAP16 transgenic Arabidopsis were transferred to 1/2 MS containing different concentrations of NaCl (0, 100, and 125 mM). No significant differences were observed between WT and the transgenic plants under normal conditions (Figure 11A). The GmSAP16 transgenic plants showed longer root lengths and greater fresh weights than WT after 1 week of salt treatment (Figures 11A–C).
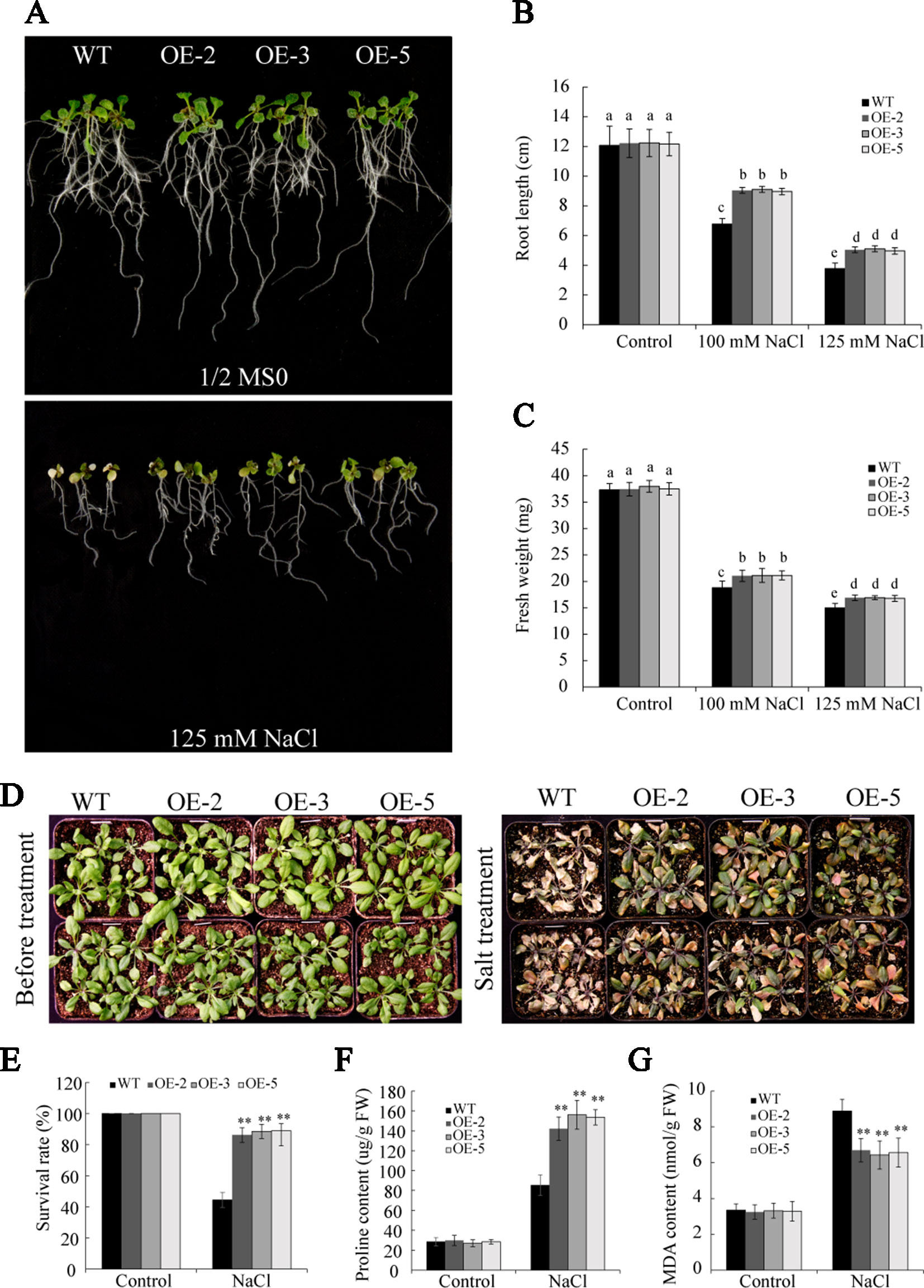
Figure 11 Overexpression of GmSAP16 improves salt tolerance in Arabidopsis. (A) Root length assays of wild-type (WT) and GmSAP16 transgenic Arabidopsis under salt treatment. Five-day-old seedlings of WT and GmSAP16-overexpressing lines were transferred to 1/2 Murashige and Skoog (MS) medium containing the indicated concentrations of NaCl (0, 100, and 125 mM). Measurements of main root length (B) and fresh weight (C) for at least 20 seedlings. (D) Salt tolerance phenotypes of WT and GmSAP16 transgenic lines in soil. Three-week-old seedlings of WT and GmSAP16 transgenic Arabidopsis were treated with 250 mM NaCl for 1 week. Statistical analysis of the survival rates (E), proline content (F), and MDA content (G). Error bars indicate the SD of three independent replicates. Asterisks represent significant differences between WT and GmSAP16-overexpressing lines (**, P 0.01; Student’s t test). Different letters indicate significant differences within treatments by ANOVA (P 0.05).
To investigate the performance of GmSAP16 in response to salt stress in soil, 3-week-old seedlings of WT and GmSAP16 transgenic lines were treated with 250 mM NaCl. After salt treatment for 1 week, the WT plants were severely wilted and showed albinism, whereas the GmSAP16-overexpressing plants were slightly wilted and remained green (Figure 11D). The survival rate of the GmSAP16-overexpressing Arabidopsis plants was significantly higher compared with that of the WT plants under salt treatment (Figure 11E). Additionally, we examined alterations in physiology that contributed to salt stress survival. The analysis of the proline and MDA contents showed that GmSAP16-overexpressing lines had a higher proline content and lower MDA content than WT (Figures 11F, G).
The analysis of RNA-seq and RT-qPCR demonstrated that ABA significantly induced the transcription of GmSAP16, indicating that GmSAP16 may be involved in ABA-mediated regulatory networks. Seed germination assays were performed with different concentrations of ABA (0, 0.1, and 0.15 µM). In the absence of ABA, the plants of WT and GmSAP16-overexpressing lines exhibited similar germination (Figures 12A, D). When treated with 1/2 MS medium containing ABA, germination was delayed in WT and overexpressing lines, and GmSAP16-overexpressing lines showed zWT (Figures 12B, E, F). The GmSAP16-overexpressing lines had a significantly lower cotyledon greening rate than the WT (Figure 12G). These results indicated that GmSAP16-overexpressing seeds were more sensitive to ABA than WT seeds at the germination stage.
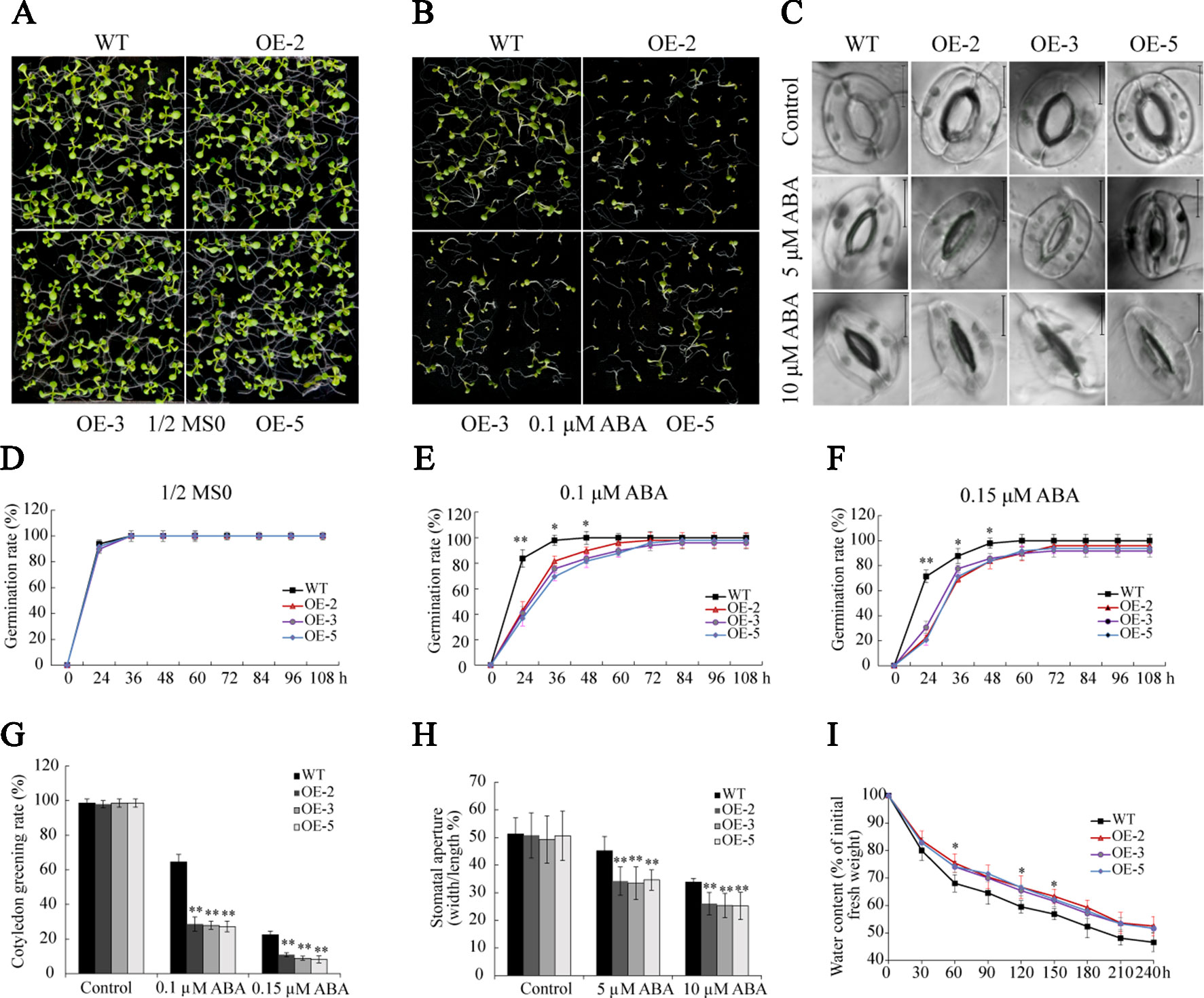
Figure 12 Overexpression of GmSAP16 increases abscisic acid (ABA) sensitivity in Arabidopsis. Germination assays of wild-type (WT) and GmSAP16 transgenic Arabidopsis seeds under normal conditions (A) and ABA treatment (B). Stomatal aperture assays were conducted with different concentrations of ABA (0, 5, 10 µM) (C). Germination rates of WT and GmSAP16-overexpressing seeds on 1/2 Murashige and Skoog (MS) medium (D), 1/2 MS medium containing 0.1 µM ABA (E), and 1/2 MS medium containing 0.15 µM ABA (F). At least 80 seeds of each genotype were used in the germination assays. (G) Measurement of cotyledon greening rate. (H) Measurement of stomatal aperture. At least 30 stomata were measured, and the aperture was calculated by width/length. (I) Relative water content of WT and GmSAP16-overexpressing Arabidopsis. A total of 1.0 g of fresh weight Arabidopsis seedlings was sampled for measurement at the indicated time points. Three biological replicates were performed, and error bars indicate ± SE. Asterisks represent significant differences compared with the corresponding controls. (*, P 0.05; and **, P 0.01; Student’s t test).
Given that GmSAP16-overexpressing lines were sensitive to ABA treatment in seed germination assays, we examined whether ABA induces stomatal closure in GmSAP16-overexpressing plants. Rosette leaves of 3-week-old WT and GmSAP16-overexpressing lines were treated with different concentrations of ABA when the stomata were fully opened. No obvious difference in the stomatal apertures was observed between WT and GmSAP16-overexpressing plants under normal conditions (Figures 12C, H). When treated with increasing ABA concentrations, the stomatal apertures of GmSAP16-overexpressing plants were significantly reduced compared with those of WT (Figures 12C, H). The relative water content was also assessed, as shown in Figure 12I, and the GmSAP16-overexpressing lines exhibited a lower water loss rate than WT. These results suggest that GmSAP16 is involved in ABA-dependent stomatal closure to reduce water loss.
To further investigate whether GmSAP16 was responsible for multiple stress tolerances in soybean, A. rhizogenes-mediated soybean hairy root assays were conducted as previously described (Kereszt et al., 2007; Hao et al., 2011). One-week-old soybean seedlings were infected with GmSAP16-OE, GmSAP16-RNAi and empty pCAMBIA3301 (CK) expression constructs. To examine the transcript level of GmSAP16, the transgenic hairy roots of different genotypes were sampled for RT-qPCR analysis. The results showed that the expression of GmSAP16 was greatly enhanced in GmSAP16-OE plants and significantly reduced in GmSAP16-RNAi plants (Supplementary Figure S5). After removing the original main roots, the infected seedlings were subjected to drought stress by withholding water or to salt stress with 250 mM NaCl. The soybean seedlings harboring different constructs showed similar phenotypes under normal conditions (Figure 13A). Under drought or salt stress, the plants showed significant symptoms, including wilting, leaf chlorosis, or abscission, and the GmSAP16-RNAi plants in particular were more sensitive to stresses and displayed more yellow and shed leaves than the CK and GmSAP16-OE plants (Figure 13A). The chlorophyll content of these plants was also examined. All plants showed decreased chlorophyll under stress conditions compared with plants under normal conditions, and GmSAP16-OE plants showed higher chlorophyll content than CK plants, GmSAP16-RNAi plants had lower chlorophyll content than CK plants under stress conditions (Figure 13B). No obvious difference in the chlorophyll content among the plants was observed under normal conditions (Figure 13B). The results indicated that overexpression of GmSAP16 conferred drought and salt tolerance.
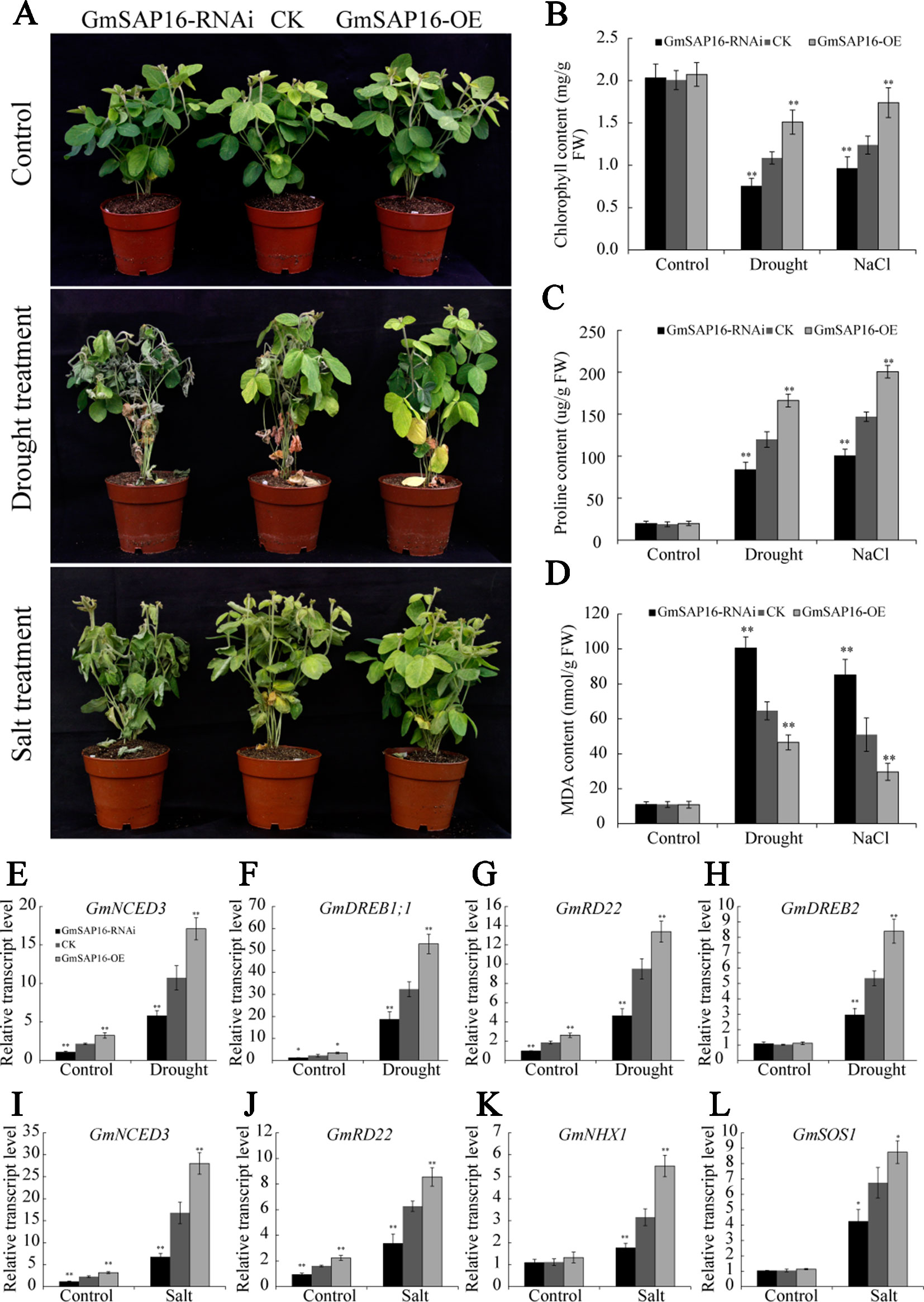
Figure 13 Overexpression of GmSAP16 in soybean root hairs improves drought and salt tolerance. (A) Drought and salt-tolerance phenotypes of GmSAP16 transgenic soybean. The plants of different genotypes with 2–5 cm infected hairy roots were cultured under normal conditions for 5 days and then dehydrated for 2 weeks or treated with 250 mM NaCl for 1 week. Statistical analysis of the chlorophyll (B), proline (C), and MDA (D) contents of CK and GmSAP16 transgenic plants under normal and stressed conditions. The leaves of different genotypes were detached and measured after treatments. (E–L) Expression profiles of several stress-related genes in empty control and transgenic soybean plants under drought or salt stress. Error bars indicate the SD of three biological replicates Asterisks indicate statistical significance (*, P 0.05; and **, P 0.01; Student’s t test) compared with the corresponding controls.
To investigate the physiological changes in GmSAP16-OE plants meditating drought and salt tolerance, soybean seedlings subjected to drought stress for 1 week or treated with 250 mM NaCl for three days were sampled to determine the MDA and proline contents. Drought and salt stress usually induce the accumulation of proline and MDA, proline acts as a protectant against osmotic stress and MDA reflects the oxidative damages. As shown in Figures 13C–D, all plants showed increased proline and MDA contents under drought or salt treatment compared with plants under normal conditions. GmSAP16-OE plants accumulated more proline than CK plants under stress condition, while GmSAP16-RNAi plants showed reduced proline content compared with CK plants (Figure 13C). When subjected to drought or salt stress, GmSAP16-RNAi plants showed higher content of MDA than CK plants under drought or salt treatment, GmSAP16-OE plants exhibited reduction of MDA contents, suggesting that GmSAP16-RNAi plants suffered more damages than CK and GmSAP16-OE plants under stress conditions (Figure 13D). All these plants showed no significant difference in the MDA and proline contents under normal conditions. These results suggest that overexpression of GmSAP16 conferred drought and salt tolerance.
To investigate the possible molecular mechanisms of GmSAP16 in multiple stress responses, we examined the expression changes in several reported soybean stress-responsive genes (Chen et al., 2007; Wang et al., 2012; Kidokoro et al., 2015; Wei et al., 2016; Cao et al., 2018; Li et al., 2019). Transgenic hairy roots treated with or without stresses were sampled for RT-qPCR analysis. The results showed that the transcript level of GmNCED3, GmDREB1;1, GmRD22, and GmDREB2 were significantly induced by drought stress. GmNCED3, GmDREB1;1, and GmRD22 showed increased transcript level in GmSAP16-OE plants and decreased transcript level in GmSAP16-RNAi plants under normal or drought conditions (Figures 13E–H). When subjected to salt stress, GmNCED3, GmRD22, GmHNX1, and GmSOS1 showed enhanced transcript level in GmSAP16-OE plants and reduced transcript level in GmSAP16-RNAi plants (Figures 13I–L). GmHNX1 and GmSOS1 showed no significant difference among GmSAP16-OE, CK, and GmSAP16-RNAi plants under normal conditions (Figures 13K, L). These results indicated that overexpression of GmSAP16 may activate the expression of drought or salt responsive genes to meditate stress responses.
SAPs, as a novel class of zinc-finger proteins, have been shown to be involved in immune responses in humans and stress responses in plants (Beyaert et al., 2000; Ströher et al., 2009). Soybean SAP genes have not been investigated, although SAP genes have been identified in many plant species (Vij and Tyagi, 2006; Solanke et al., 2009; Gao et al., 2016). The identification of stress-tolerant genes, including soybean SAPs, appears to be important for the breeding of stress-tolerant soybean cultivars. In the present study, we characterized and identified 27 SAP genes in soybean, and GmSAP16 improved stress tolerance in Arabidopsis and soybean.
It has been reported that exon–intron structural diversity often provides valuable information to understand the evolutionary mechanisms underlying gene families and significantly affects gene expression in various ways (Rogozin et al., 2005; Rose, 2008). SAP family members are typically characterized by intronless structures and lacking introns in different plant species. It was reported that approximately 61% of OsSAP genes and 82% of MdSAP genes have no introns, and 33% of OsSAP genes and 18% of MdSAP genes have only a single intron (Vij and Tyagi, 2006; Dong et al., 2018). Our study demonstrated that 67% of soybean SAP genes had no introns and that the other soybean SAP genes grouped into clade II had only one intron. There are indications that intronless gene families can reduce posttranscriptional processing and rapidly adjust transcript expression in response to environmental stimuli (Grzybowska, 2012).
Cis-acting elements play important roles in the regulation of many processes, including plant growth, development, and stress responses (Yamaguchi-Shinozaki and Shinozaki, 2005). Analysis of the promoters of the soybean SAP family revealed that many cis-acting elements related to abiotic stress responses, including ABRE, DREB, LTRE, MYB, MYC, HSE WUN-motifs, and TC-rich repeats, were present in the promoter regions, indicating that soybean SAP genes may be involved in different abiotic stresses. The cis-elements could associate with different transcription factor proteins via protein-DNA interactions to repress or activate stress-associated genes in response to environmental challenges (Narusaka et al., 2003). It has been reported that Arabidopsis AtSAP13 promoter fragments interact with several stress-related transcription factors, including ERF, bZIP, NAC, and DREB, by yeast one-hybrid assay (Dixit et al., 2018). These results imply that soybean SAP genes may be regulated by stress-related transcription factors via the interaction of specific cis-elements and proteins. Further studies are necessary to verify this hypothesis and to reveal the mechanism underlying the regulation of the expression of soybean SAP genes under abiotic stresses.
Pathogen-related cis-elements, such as CGTCA-motif, TCA-element, and W-box, were present in the promoters of many soybean SAP genes, suggesting the involvement of the soybean SAP genes in defense responses. A recent study revealed that the overexpression of OsSAP1 enhanced basal resistance against a bacterial pathogen in transgenic tobacco (Tyagi et al., 2014). Plants overexpressing Arabidopsis AtSAP9 had increased susceptibility to the nonhost pathogen Pseudomonas syringae pv. phaseolicola, indicating that AtSAP9 regulates disease response (Kang et al., 2017). The tomato SlSAP3 gene positively regulates immunity against Pseudomonas syringae pv. tomato (Pst) DC3000 (Liu et al., 2019). The results indicate that soybean SAP genes may be involved in biotic stress responses.
ABA is an important hormone that plays important roles in the regulation of seed germination, stomatal closure, plant growth, and abiotic stress responses (Finkelstein, 2013; Yoshida et al., 2014). When plants were challenged with stress treatments, the accumulating ABA content triggered stomatal closure and reduced water loss to adapt to stresses (Munemasa et al., 2015). Previous studies have reported that the SAP gene OsISAP1 was induced by ABA treatment and conferred drought and salt tolerance in tobacco (Mukhopadhyay et al., 2004). Arabidopsis AtSAP13 was significantly induced in response to ABA treatment, and the AtSAP13-overexpressing plants were resistant to drought and salt stresses (Dixit et al., 2018). Our study revealed that GmSAP16 was significantly induced by exogenous ABA. Phenotypic analyses demonstrated that GmSAP16 transgenic Arabidopsis lines were hypersensitive to ABA treatment with delayed seed germination and greater stomatal closure during ABA treatment. The GmSAP16-overexpressing lines also exhibited a lower water loss rate than WT plants, indicating that GmSAP16 improved drought tolerance by regulating stomatal closure. Additionally, GmSAP16 also affected the expression of ABA-related genes, including GmDREB1;1, GmNCED3, and GmRD22, implying that GmSAP16 may be involved in stress responses in an ABA-dependent manner.
All datasets for this study are included in the article/Supplementary Material.
Z-SX conceived the project and revised the manuscript. X-ZZ performed the experiments and drafted the manuscript. W-JZ collected the dataset and conducted the bioinformatics analysis. X-YCa and X-YCu contributed to the RT-qPCR analysis, and S-PZ and T-FY helped with the physiology measurements and data analysis. MC and Y-BZ provided reagents. JC provided technical assistance. S-CC and Y-ZM contributed to valuable discussions. All authors read and approved the final manuscript.
This research was financially supported by the National Natural Science Foundation of China (31871611 and 31871624) and the National Transgenic Key Project of the Ministry of Agriculture of China (2018ZX0800909B and 2016ZX08002-002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Drs. Li-Juan Qiu and Shi Sun (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for kindly providing soybean seeds, and Dr. Hui Zhang (Institute of Crop Science, Chinese Academy of Agricultural Sciences) for providing the pCAMBIA3301 vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01453/full#supplementary-material
Figure S1 | Multiple alignments of soybean SAPs. DNAMAN 8.0 software was used for the alignments. A20, AN1 and C2H2 zinc finger domains are indicated in red, green, and blue boxes, respectively. Amino acids in different colors indicate sequence similarity. (Similarity: red = 100%; coral > 75%; pink > 50%).
Figure S2 | Main cis-acting elements in the promoter regions of soybean SAP genes. ABRE: ABA-responsive element; ARE: anaerobic-responsive element; CGTCA: MeJA responsiveness; ERE: ethylene-responsive element; GARE-motif: gibberellin-responsive element; LTR: low-temperature responsive element; MBS: MYB-binding site involved in drought response; MYB: response to drought and ABA signals; MYC: response to drought, ABA and cold signals; STRE: stress-response element; TC-rich repeat: defense and stress responsiveness; TCA: salicylic acid-responsive element; TGA element: auxin-responsive element; W-box: elicitation, wounding, and pathogen responsiveness/binding site of WRKY-type transcription factors; WUN-motif: wooden response.
Figure S3 | Chromosomal locations of the soybean SAP genes. The chromosomal locations of the soybean SAP genes refer to the genomic information provided on Phytozome v10.3. The scale bar represents megabases (Mb). Abbreviation: Chr, chromosome.
Figure S4 | Detection of GmSAP16 in WT and transgenic Arabidopsis lines. (A) Relative transcript level of GmSAP16 in WT and transgenic Arabidopsis lines determined by RT-qPCR. Error bars indicate the SD of three biological replicates. (B) The transcript level of GmSAP16 was detected in WT and transgenic plants by semi-quantitative PCR.
Figure S5 | Relative transcript level of GmSAP16 in CK and transgenic soybean hairy roots determined by RT-qPCR. The data were normalized to the soybean internal control (GmCYP2). Error bars indicate the SD of three biological replicates.
Table S1 | The primers and sequences used for the experiments.
Ben Saad, R., Fabre, D., Mieulet, D., Meynard, D., Dingkuhn, M., Al-Doss, A., et al. (2012). Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ. 35, 626–643. doi: 10.1111/j.1365-3040.2011.02441.x
Beyaert, R., Heyninck, K., Van Huffel, S. (2000). A20 and A20-binding proteins as cellular inhibitors of nuclear factor-κB-dependent gene expression and apoptosis. Biochem. Pharmacol. 60, 1143–1151. doi: 10.1016/S0006-2952(00)00404-4
Cao, D., Li, Y., Liu, B., Kong, F., Tran, L. S. P. (2018). Adaptive mechanisms of soybean grown on salt-affected soils. Land Degrad. Dev. 29, 1054–1064. doi: 10.1002/ldr.2754
Chang, L., Chang, H. H., Chang, J. C., Lu, H. C., Wang, T. T., Hsu, D. W., et al. (2018). Plant A20/AN1 protein serves as the important hub to mediate antiviral immunity. PloS Pathog. 14, e1007288. doi: 10.1371/journal.ppat.1007288
Chen, M., Wang, Q. Y., Cheng, X. G., Xu, Z. S., Li, L. C., Ye, X. G., et al. (2007). GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353, 299–305. doi: 10.1016/j.bbrc.2006.12.027
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cristina, M. S., Petersen, M., Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252
Cui, X. Y., Gao, Y., Guo, J., Yu, T. F., Zheng, W. J., Liu, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Dixit, A., Tomar, P., Vaine, E., Abdullah, H., Hazen, S., Dhankher, O. P. (2018). A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 41, 1171–1185. doi: 10.1111/pce.13103
Dong, Q., Duan, D., Zhao, S., Xu, B., Luo, J., Wang, Q., et al. (2018). Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 19, 2478. doi: 10.3390/ijms19092478
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis book/American Soc. Plant Biologists 11, e0166. doi: 10.1199/tab.0166
Gao, W., Long, L., Tian, X., Jin, J., Liu, H., Zhang, H., et al. (2016). Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genomics 291, 2199–2213. doi: 10.1007/s00438-016-1252-6
Giri, J., Dansana, P. K., Kothari, K. S., Sharma, G., Vij, S., Tyagi, A. K. (2013). SAPs as novel regulators of abiotic stress response in plants. Bioessays 35, 639–648. doi: 10.1002/bies.201200181
Giri, J., Vij, S., Dansana, P. K., Tyagi, A. K. (2011). Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 191, 721–732. doi: 10.1111/j.1469-8137.2011.03740.x
Grzybowska, E. A. (2012). Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 424, 1–6. doi: 10.1016/j.bbrc.2012.06.092
Guo, X., Liu, D., Chong, K. (2018). Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 60, 745–756. doi: 10.1111/jipb.12706
Hao, Y. J., Wei, W., Song, Q. X., Chen, H. W., Zhang, Y. Q., Wang, F., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. doi: 10.1111/j.1365-313X.2011.04687.x
Hirayama, T., Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Hozain, M. D., Abdelmageed, H., Lee, J., Kang, M., Fokar, M., Allen, R. D., et al. (2012). Expression of AtSAP5 in cotton up-regulates putative stress-responsive genes and improves the tolerance to rapidly developing water deficit and moderate heat stress. J. Plant Physiol. 169, 1261–1270. doi: 10.1016/j.jplph.2012.04.007
Ivanov, S., Harrison, M. J. (2014). A set of fluorescent protein-based markers expressed from constitutive and arbuscular mycorrhiza-inducible promoters to label organelles, membranes and cytoskeletal elements in Medicago truncatula. Plant J. 80, 1151–1163. doi: 10.1111/tpj.12706
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7, 1029. doi: 10.3389/fpls.2016.01029
Kang, M., Abdelmageed, H., Lee, S., Reichert, A., Mysore, K. S., Allen, R. D. (2013). AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 76, 481–493. doi: 10.1111/tpj.12312
Kang, M., Lee, S., Abdelmageed, H., Reichert, A., Lee, H. K., Fokar, M., et al. (2017). Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 40, 702–716. doi: 10.1111/pce.12892
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kidokoro, S., Watanabe, K., Ohori, T., Moriwaki, T., Maruyama, K., Mizoi, J., et al. (2015). Soybean DREB 1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 81, 505–518. doi: 10.1111/tpj.12746
Kothari, K. S., Dansana, P. K., Giri, J., Tyagi, A. K. (2016). Rice stress associated protein 1 (OsSAP1) interacts with aminotransferase (OsAMTR1) and pathogenesis-related 1a protein (OsSCP) and regulates abiotic stress responses. Front. Plant Sci. 7, 1057. doi: 10.3389/fpls.2016.01057
Li, S., Wang, N., Ji, D., Zhang, W., Wang, Y., Yu, Y., et al. (2019). A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell. 31, 2107–2130. doi: 10.1105/tpc.18.00662
Liu, P., Xu, Z. S., Lu, P. P., Hu, D., Chen, M., Li, L. C., et al. (2013). A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J. Exp. Bot. 64, 2915–2927. doi: 10.1093/jxb/ert133
Liu, S., Wang, J., Jiang, S., Wang, H., Gao, Y., Zhang, H., et al. (2019). Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 20, 815–830. doi: 10.1111/mpp.12793
Liu, Y., Xu, Y., Xiao, J., Ma, Q., Li, D., Xue, Z., et al. (2011). OsDOG, a gibberellin-induced A20/AN1 zinc-finger protein, negatively regulates gibberellin-mediated cell elongation in rice. J. Plant Physiol. 168, 1098–1105. doi: 10.1016/j.jplph.2010.12.013
Lloret, A., Conejero, A., Leida, C., Petri, C., Gil-Muñoz, F., Burgos, L., et al. (2017). Dual regulation of water retention and cell growth by a stress-associated protein (SAP) gene in Prunus. Sci. Rep. 7, 332. doi: 10.1038/s41598-017-00471-7
Mukhopadhyay, A., Vij, S., Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U. S. A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Munemasa, S., Hauser, F., Park, J., Waadt, R., Brandt, B., Schroeder, J. I. (2015). Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 28, 154–162. doi: 10.1016/j.pbi.2015.10.010
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Rogozin, I. B., Sverdlov, A. V., Babenko, V. N., Koonin, E. V. (2005). Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinform. 6, 118–134. doi: 10.1093/bib/6.2.118
Rose, A. (2008). Intron-mediated regulation of gene expression. Curr. Top. Microbiol. Immunol. 326, 277–290. doi: 10.1007/978-3-540-76776-3_15
Ryu, M. Y., Cho, S. K., Kim, W. T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154, 1983–1997. doi: 10.1104/pp.110.164749
Scheben, A., Wolter, F., Batley, J., Puchta, H., Edwards, D. (2017). Towards CRISPR/Cas crops-bringing together genomics and genome editing. New Phytol. 216, 682–698. doi: 10.1111/nph.14702
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Shi, W. Y., Du, Y. T., Ma, J., Min, D. H., Jin, L. G., Chen, J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19, 4087. doi: 10.3390/ijms19124087
Shinozaki, K., Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Solanke, A. U., Sharma, M. K., Tyagi, A. K., Sharma, A. K. (2009). Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genomics 282, 153–164. doi: 10.1007/s00438-009-0455-5
Song, H., Wang, P., Hou, L., Zhao, S., Zhao, C., Xia, H., et al. (2016). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7, 9. doi: 10.3389/fpls.2016.00009
Ströher, E., Wang, X. J., Roloff, N., Klein, P., Husemann, A., Dietz, K. J. (2009). Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant 2, 357–367. doi: 10.1093/mp/ssn084
Tyagi, H., Jha, S., Sharma, M., Giri, J., Tyagi, A. K. (2014). Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 225, 68–76. doi: 10.1016/j.plantsci.2014.05.016
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Vij, S., Tyagi, A. K. (2006). Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger (s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genomics 276, 565–575. doi: 10.1007/s00438-006-0165-1
Wang, H., Zhou, L., Fu, Y., Cheung, M. Y., Wong, F. L., Phang, T. H., et al. (2012). Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant Cell Environ. 35, 1932–1947. doi: 10.1111/j.1365-3040.2012.02526.x
Wei, P., Wang, L., Liu, A., Yu, B., Lam, H.-M. (2016). GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 7, 1082. doi: 10.3389/fpls.2016.01082
Xu, Z. S., Chen, M., Li, L. C., Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yamaguchi-Shinozaki, K., Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yang, Y., Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Yoshida, T., Mogami, J., Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, N., Yin, Y., Liu, X., Tong, S., Xing, J., Zhang, Y., et al. (2017). The E3 ligase TaSAP5 alters drought stress responses by promoting the degradation of DRIP proteins. Plant Physiol. 175, 1878–1892. doi: 10.1104/pp.17.01319
Zhang, Y., Lan, H., Shao, Q., Wang, R., Chen, H., Tang, H., et al. (2015). An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 67, 315–326. doi: 10.1093/jxb/erv464
Zhao, S. P., Xu, Z. S., Zheng, W. J., Zhao, W., Wang, Y. X., Yu, T. F., et al. (2017). Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front. Plant Sci. 8, 905. doi: 10.3389/fpls.2017.00905
Keywords: soybean, stress associated proteins, expression analysis, abiotic stresses, abscisic acid sensitivity
Citation: Zhang X-Z, Zheng W-J, Cao X-Y, Cui X-Y, Zhao S-P, Yu T-F, Chen J, Zhou Y-B, Chen M, Chai S-C, Xu Z-S and Ma Y-Z (2019) Genomic Analysis of Stress Associated Proteins in Soybean and the Role of GmSAP16 in Abiotic Stress Responses in Arabidopsis and Soybean. Front. Plant Sci. 10:1453. doi: 10.3389/fpls.2019.01453
Received: 31 July 2019; Accepted: 18 October 2019;
Published: 18 November 2019.
Edited by:
Randy D. Allen, Oklahoma State University, United StatesReviewed by:
Jitender Giri, National Institute of Plant Genome Research (NIPGR), IndiaCopyright © 2019 Zhang, Zheng, Cao, Cui, Zhao, Yu, Chen, Zhou, Chen, Chai, Xu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shou-Cheng Chai Y2hhaXNob3VjaGVuZ0AxMjYuY29t; Zhao-Shi Xu, eHV6aGFvc2hpQGNhYXMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.