- 1Biotechnology Research Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 2State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Temasek Life Sciences Laboratory, and the Department of Biological Sciences, National University of Singapore, Singapore, Singapore
- 4Marine and Agricultural Biotechnology Laboratory, Institute of Oceanography, Minjiang University, Fuzhou, China
The subcellular localization of proteins is a fundamental aspect of protein functions. Determining the subcellular localization is important for understanding the biological functions of proteins. Here, we developed a set of rice organelle marker lines, in which the expressing fluorescent organelle markers could be used as comparative standards in determining the subcellular localization of the protein of interest. We constructed green fluorescent protein (GFP)- and/or Discosoma sp. red fluorescent protein (DsRed)-tagged organelle markers targeted to the endoplasmic reticulum (ER), mitochondria, Golgi apparatus, peroxisome, actin cytoskeleton, plastid, tonoplast, plasma membrane, and nucleus, respectively. The utility of the rice marker lines for protein subcellular localization studies was demonstrated by detecting a nucleus-localized OsWRKY45 and a mitochondria-associated NbHxk1 in protoplasts of the GFP-OsH2B and the ScCOX4-DsRed lines, respectively. Using a sheath-inoculation method, followed by a live-cell imaging, we detected co-localization of a Magnaporthe oryzae PWL2:mCherry : NLS fusion with the nucleus marker in the GFP-OsH2B rice epidermal cells, confirming the translocation of the M. oryzae effector PWL2 into host cells, and further demonstrating the feasibility of using the organelle marker lines for studying dynamics of proteins in rice cells in the interactions between rice and pathogens. The set of organelle marker lines developed in the present study, provides a valuable resource for protein subcellular localization studies in rice.
Introduction
Eukaryotic cells are highly compartmentalized and various biological processes are implemented within specialized subcellular compartments (Nelson et al., 2007). In general, the subcellular localization of proteins is closely associated with functions related to the biological processes carried out in the corresponding organelles (Lunn, 2007; Mathur, 2007). Thus, information on the subcellular localization is helpful in understanding the biological functions of proteins (Martin et al., 2009; Tanz et al., 2013).
Protein subcellular localization analysis has been routinely employed as an experimental methodology for gene functional studies. Over the past decades, fluorescent protein (FP) tagging has become the dominant approach for determining the subcellular localization of proteins in living cells in different organisms, including plants (Chudakov et al., 2010; Tanz et al., 2013). Based on fusion of FPs with the well-established targeting proteins or motifs (Mathur, 2007), numerous sets of fluorescent organelle markers have been developed for Arabidopsis (Nelson et al., 2007; Geldner et al., 2009; Kim et al., 2013), Nicotiana benthamiana (Martin et al., 2009), Medicago truncatula (Luo and Nakata, 2012), maize (Wu et al., 2013; Krishnakumar et al., 2015), and rice (Wu et al., 2016; Dangol et al., 2017). Currently, the common strategy utilized to detect localization of FP-tagged proteins has been based on transient expression of fusion constructs in protoplasts through polyethylene glycol (PEG)-mediated transfection (Chen et al., 2006; Zhang et al., 2011), or in epidermal cells through biolistic bombardment (Dangol et al., 2017) or agro-infiltration (Lunn, 2007; Nelson et al., 2007). Although transient expression systems provide convenient and rapid assays for subcellular localization, the approaches have limitations in monitoring processes in a developmental context or under particular environmental conditions, such as in response to abiotic stresses, or pathogen attack (Geldner et al., 2009). Recently, transgenic lines stably expressing FP-tagged organelle markers have been generated in Arabidopsis, M. truncatula, maize, and rice (Geldner et al., 2009; Luo and Nakata, 2012; Kim et al., 2013; Wu et al., 2013; Krishnakumar et al., 2015; Wu et al., 2016). These transgenic marker lines provided valuable resources for protein subcellular localization studies, especially for studying the dynamics of proteins or organelles in developmental or environmental contexts (Geldner et al., 2009; Kim et al., 2013; Wu et al., 2016).
Rice is one of the most important food crops in the world, as well as a monocot model plant. More recently, sets of fluorescent organelle markers have been developed for protein subcellular localization studies in rice (Wu et al., 2016; Dangol et al., 2017). However, only green fluorescent protein (GFP)-based organelle marker lines have been generated in rice (Wu et al., 2016). In the present study, to provide more flexible combinations of organelle marker lines, we developed transgenic rice plants stably expressing GFP- and/or Discosoma sp. red fluorescent protein (DsRed)-tagged organelle markers. We demonstrated the utility of the marker lines for co-localization studies by detecting a nucleus-localized rice OsWRKY45 and a mitochondria-associated N. benthamiana hexokinase NbHxk1 in protoplasts of the GFP-OsH2B and the ScCOX4-DsRed rice lines, respectively. In addition, we demonstrated the feasibility of using the organelle marker lines for studying dynamics of proteins in rice cells in the interactions between rice and pathogens. Using a sheath-inoculation method, followed by a live-cell imaging (Khang et al., 2010; Park et al., 2012), we detected co-localization of a Magnaporthe oryzae PWL2:mCherry : NLS fusion with the nucleus marker in the GFP-OsH2B rice epidermal cells, confirming the translocation of the M. oryzae effector PWL2 into host cells. The organelle markers and marker lines developed in this study, along with previously developed organelle markers and marker lines (Wu et al., 2016; Dangol et al., 2017), therefore, provide valuable resources for protein localization studies in rice.
Materials and Methods
Generation of Organelle Marker Constructs
The sGFP(S65T) gene (Chiu et al., 1996) and the DsRed gene (Goodin et al., 2002) were used for the construction of fluorescent organelle marker constructs. GFP- or DsRed-fusion fragments were generated by overlapping PCR and were cloned into a plant binary vector, pCXSN (Chen et al., 2009) in which the GFP- or DsRed-fusions were driven by the CaMV 35S promoter. All constructs were confirmed by sequencing. Primers used for PCR or overlapping PCR were listed in Supplementary Table S1.
Agro-Infiltration Assay in Nicotiana benthamiana
The organelle marker constructs were introduced into the Agrobacterium tumefaciens strain GV3101. Transformed GV3101 bacteria were cultured in liquid yeast extract peptone media supplemented with kanamycin (50 ug/ml) and rifampicin (50 ug/ml). Suspensions of transformed GV3101 bacteria were adjusted to an OD600 of 0.7 in the infiltration buffer (10 mM MES, 10 mM MgCl2, and 150 µM acetosyringone), and were maintained at room temperature for 2 h. The suspension cultures were infiltrated into leaves of 4-week-old N. benthamiana plants grown in a growth chamber at 25°C under 16/8 h light/dark cycle.
Rice Transformation
The organelle marker constructs were introduced into A. tumefaciens LBA4404 by electroporation. Rice calli induced from the embryos of mature seeds of cv. Nipponbare were used for transformation via the Agrobacterium-mediated method as described previously (Hiei et al., 1994). Regenerated transgenic plants and their self-pollinated progeny were grown in a greenhouse.
Rice Protoplast Isolation and Transfection
Protoplasts were isolated from 2-week-old rice seedlings of transgenic rice organelle marker lines or wild type rice cv. Nipponbare. PEG-mediated protoplast transfection was performed as described previously (Chen et al., 2006). Protoplasts were incubated under dark at room temperature for 16–24 h.
Rice Leaf Sheath Inoculation With Magnaporthe oryzae
A transgenic M. oryzae isolate carrying a PWL2:mCherry : NLS expression cassette (Khang et al., 2010) was used for rice sheath inoculation. Rice leaf sheath inoculation with M. oryzae was performed as described previously (Khang et al., 2010; Park et al., 2012). In brief, leaf sheaths were detached from 4-week-old rice seedlings, and were cut into 5–6 cm segments. Inoculation was performed by incubating fungal spores (1 × 105 conidial/ml) in inner interior of leaf sheaths. The inoculated sheaths were placed in a petri dish and were incubated under high humidity (95%) and at 26°C for about 2 days before microscopic inspection.
Fluorescence Visualization
Fluorescence imaging was performed on a Leica DMi8 Laser Scanning Confocal microscope (Leica, Wetzlar, Germany). Excitation/emission wavelengths were 488/535 nm for GFP, and 552/610 nm for RFP.
Results
Plant Expression Cassettes for Green Fluorescent Protein –/Discosoma sp. Red Fluorescent Protein-Fusion Organelle Markers
To develop rice marker lines with flexibility for co-localization assays, two commonly used FP genes, GFP and DsRed, were used to generate plant expression cassettes for organelle markers. The fusion markers were designed to target the endoplasmic reticulum (ER), mitochondria, Golgi apparatus, peroxisome, actin cytoskeleton, plastid, tonoplast, plasma membrane, and nucleus, respectively (Figure 1). The targeting signal sequences for the ER [signal peptide of AtWAK2 (He et al., 1999; Nelson et al., 2007) at the N-terminus and a His-Asp-Glu-Leu (HDEL) motif at the C-terminus of FPs], mitochondria (first 29-AA transit peptide of ScCOX4) (Köhler et al., 1997; Nelson et al., 2007), Golgi apparatus (C-terminal 125-AA residues of AtCASP) (Renna et al., 2005), peroxisomes [peroxisomes targeting signal 1 (PTS1), Ser-Lys-Leu motif] (Reumann, 2004; Nelson et al., 2007), and actin cytoskeleton [C-terminal residues (AA 2345-2541) of mouse mTalin] (McCann and Craig, 1997; Kost et al., 1998) were achieved according to previous reports. As for developing markers targeted to the plastid, tonoplast, plasma membrane, and nucleus, the coding sequences of rice orthologs of the first 48-AA transit peptide of rubisco activase small subunit (OsCTP1), the vacuolar membrane aquaporin (Osγ-TIP), the plasma membrane aquaporin (OsPIP2), and the histone 2B (OsH2B) were fused with GFP or DsRed, respectively (Figure 1). The GFP- or DsRed-fusions were placed under the control of the CaMV 35S promoter, allowing for high-level expression in both dicot and monocot plants (Chen et al., 2009).
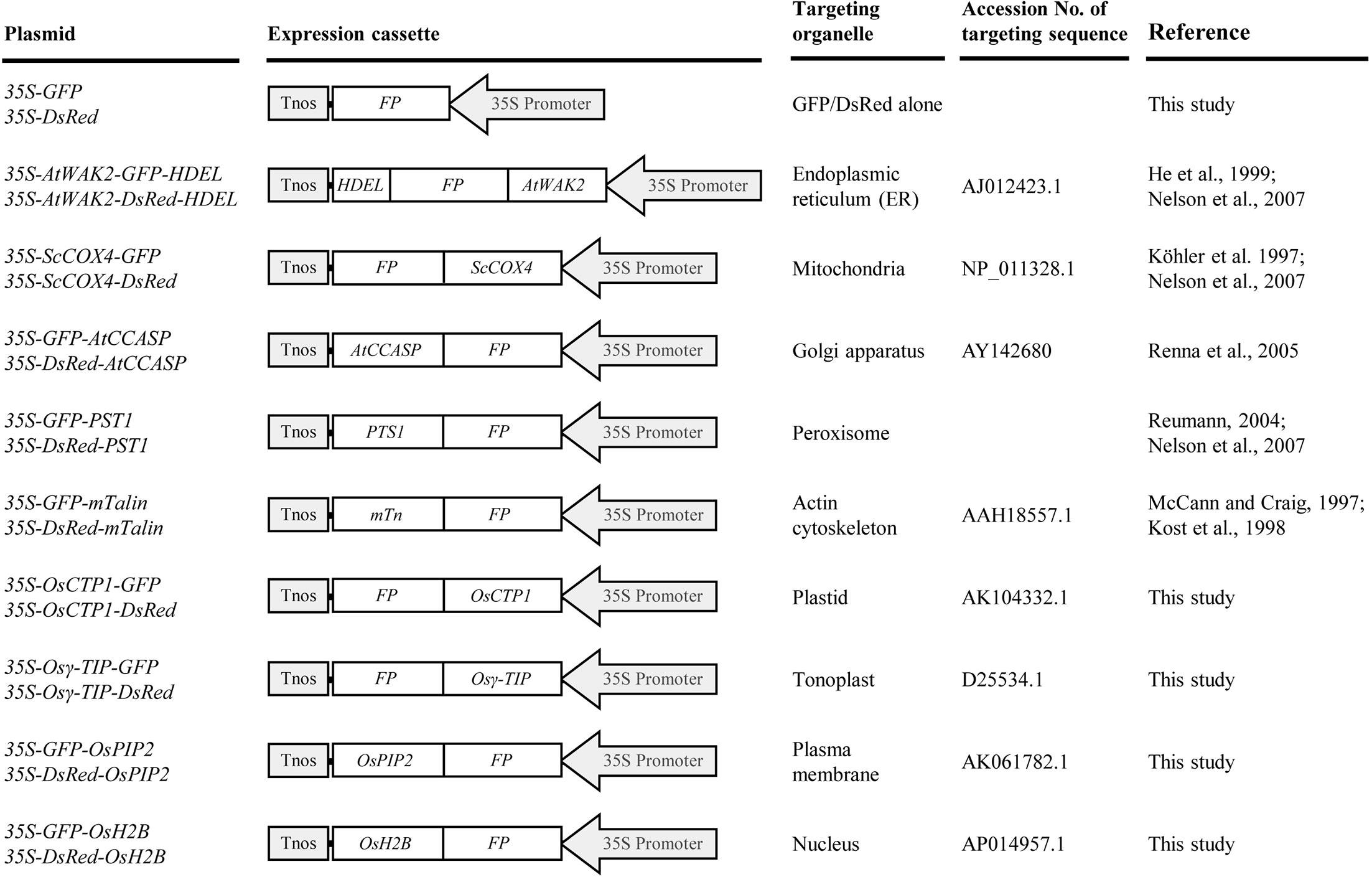
Figure 1 List of green fluorescent protein (GFP)- and Discosoma sp. red fluorescent protein (DsRed)-tagged organelle markers developed in this study. All chimeric GFP or DsRed fusion genes were placed under the control of a CaMV 35S promoter. 35S Promoter, CaMV 35S promoter; FP, fluorescent protein gene GFP or DsRed; AtWAK2, signal peptide sequence of AtWAK2 gene; HDEL, ER retention signal sequence; ScCOX4, transit sequence of ScCOX4; AtCCASP, coding sequence of C-terminal region of AtCASP; PTS1, peroxisomes targeting signal 1 sequence; mTn, coding sequence of C-terminal residues of mouse mTalin; OsCTP1, transit peptide sequence of rice rubisco activase small subunit; Osγ-TIP, coding sequence of rice vacuolar membrane aquaporin; OsPIP2, coding sequence of rice plasma membrane aquaporin; OsH2B, coding sequence of rice histone 2B; Tnos, nopaline synthase terminator.
Subcellular Localizations of Rice OsCTP1, Osγ-TIP, OsPIP2, and OsH2B in Nicotiana benthamiana
To validate the subcellular localization patterns of the four rice ortholog organelle markers, GFP-fusions of rice OsCTP1, Osγ-TIP, OsPIP2, and OsH2B were transiently expressed and investigated in N. benthamiana leaves, respectively. Microscopy of leaf epidermal cells showed that, while N. benthamiana leaves infiltrated with the 35S-GFP expression cassette displayed green fluorescence distributed uniformly throughout the whole cells (Figures 2B, C), the expression of OsCTP1-GFP, Osγ-TIP-GFP, GFP-OsPIP2, or GFP-OsH2B showed specific fluorescence patterns as expected: green fluorescence emitted from OsCTP1-GFP accumulated at the granular plastids (Figures 2E, F); Osγ-TIP-GFP signal appeared in the shape of small rings surrounds the vacuole (Figures 2H, I); fluorescent signal of GFP-OsPIP2 located at the periphery of the cells (Figures 2K, L); and GFP-OsH2B produced green fluorescence targeted to the nucleus of the epidermal cells (Figures 2N, O). These results also suggested that the four rice orthologs, OsCTP1, Osγ-TIP, OsPIP2, and OsH2B had conserved subcellular localization patterns across dicot and monocot species.
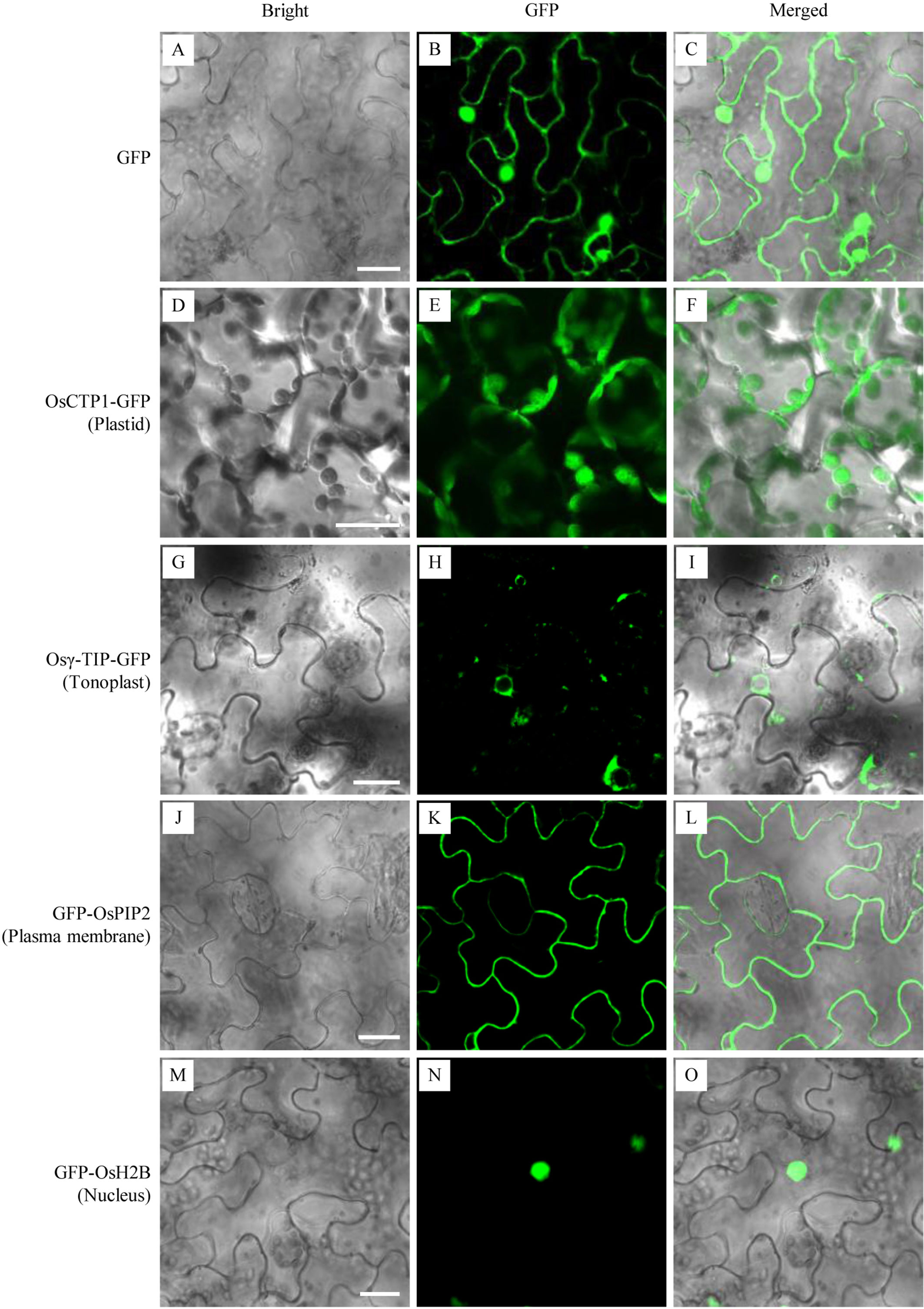
Figure 2 Transient validation of green fluorescent protein (GFP) fusions of four rice orthologs. The binary vectors harboring expression cassettes of GFP, OsCTP1-GFP, Osγ-TIP-GFP, GFP-OsPIP2, and GFP-OsH2B were transiently expressed in N. benthamiana epidermal cells, respectively. The subcellular localizations of GFP alone throughout the whole cells (A–C), OsCTP1-GFP at the granular plastids (D–F), Osγ-TIP-GFP in the shape of small rings surrounds the vacuole (G–I), GFP-OsPIP2 at the periphery of the cells (J–L), and GFP-OsH2B targeted to the nucleus (M–O) were shown. Scale bars, 10 μm.
Generation of Stable Rice Lines Expressing Green Fluorescent Protein/Discosoma sp. Red Fluorescent Protein-Fusion Organelle Markers
The binary vectors harboring expression cassettes for GFP/DsRed-fusion organelle markers were introduced into the rice cultivar Nipponbare by Agrobacterium-mediated transformation, and more than 20 independent T0 plants were generated for each construct. T0 plants were screened by detecting fluorescent signals from the sliced-sheath tissues, and T1 lines derived from T0 plants with good fluorescent signal were used for detailed analyses.
Microscopic analysis of sheath cells showed that most of the GFP/DsRed-fusions could be stably expressed in rice (Figure 3). Fluorescent signals in transgenic rice expressing AtWAK2-GFP-HDEL or AtWAK2-DsRed-HDEL exhibited typical ER localization patterns, forming reticulate networks throughout the cytoplasm (Figures 3C, D). The mitochondria targeting ScCOX4-GFP or ScCOX4-DsRed appeared as small spots, randomly distributed in the cytoplasm (Figures 3E, F). GFP-AtCCASP showed punctate fluorescence corresponding to the typical Golgi localization pattern (Figure 3G). GFP or DsRed fused with the C-terminal peroxisome-tagging PTS1 motif, Ser-Lys-Leu, both appeared as spherical dots dispersed in the cytosol (Figures 3I, J). Red fluorescence labeling of the actin cytoskeleton was observed in DsRed-mTn-transgenic rice lines, showing as a dense filamentous network (Figure 3L). The fluorescence patterns of OsCTP1-GFP (Figure 3M), Osγ-TIP-GFP (Figure 3O), GFP-OsPIP2 (Figure 3Q), and GFP-OsH2B (Figure 3S) in transgenic rice were consistent, respectively, with the plastid-, tonoplast-, plasma membrane-, and nucleus-localization patterns observed in N. benthamiana. The OsCTP1-DsRed, Osγ-TIP-DsRed, and DsRed-OsPIP2 markers displayed similar localization patterns (Figures 3N, P, R) as OsCTP1-GFP, Osγ-TIP-GFP, and GFP-OsPIP2, respectively. Unexpectedly, we were unable to obtain DsRed-AtCCASP-, GFP-mTn-, or DsRed-OsH2B-transgenic rice plants expressing obvious fluorescent signals (data not shown). Protoplasts isolated from the above transgenic rice lines were subjected to microscopic analysis, and the results were consistent with the observations in sheath cells (Figure 4).
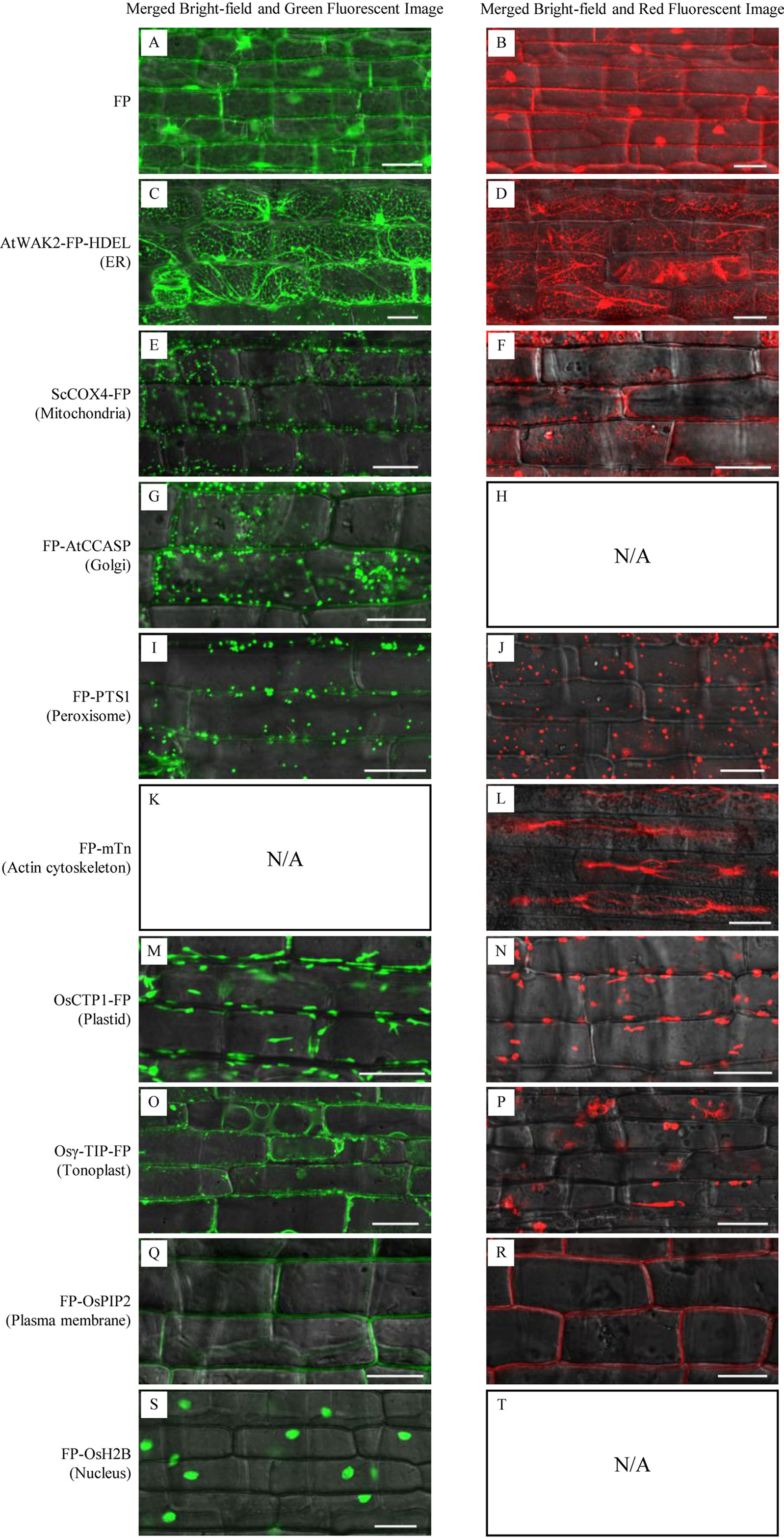
Figure 3 Subcellular localization of green fluorescent protein (GFP)/Discosoma sp. red fluorescent protein (DsRed)-fusion organelle markers in epidermal cells of stable transgenic rice plants. FP, fluorescent protein GFP or DsRed. Sheath epidermal cells of GFP-fusion (left panel) and DsRed-fusion (right panel) transgenic lines were examined by confocal microscopy. (A, B) GFP or DsRed alone; (C, D) ER marker AtWAK2-GFP-HDEL or AtWAK2-DsRed-HDEL; (E, F) Mitochondrial marker ScCOX4-GFP or ScCOX4-DsRed; (G) Golgi marker GFP-AtCCASP; (I, J) Peroxisome marker GFP-PTS1 or DsRed-PTS1; (L) cytoskeleton marker DsRed-mTn; (M, N) Plasmid marker CTP1-GFP or CTP1-DsRed; (O, P) Tonoplast marker Osγ-TIP-GFP or Osγ-TIP-DsRed; (Q, R) Plasma membrane marker GFP-OsPIP2 or DsRed-OsPIP2; (S) Nucleus marker GFP-OsH2B. N/A, No DsRed-AtCCASP- (H), GFP-mTn- (K), or DsRed-OsH2B-transgenic rice plants expressing obvious fluorescent signals (T) were obtained. Scale bars, 25 μm.
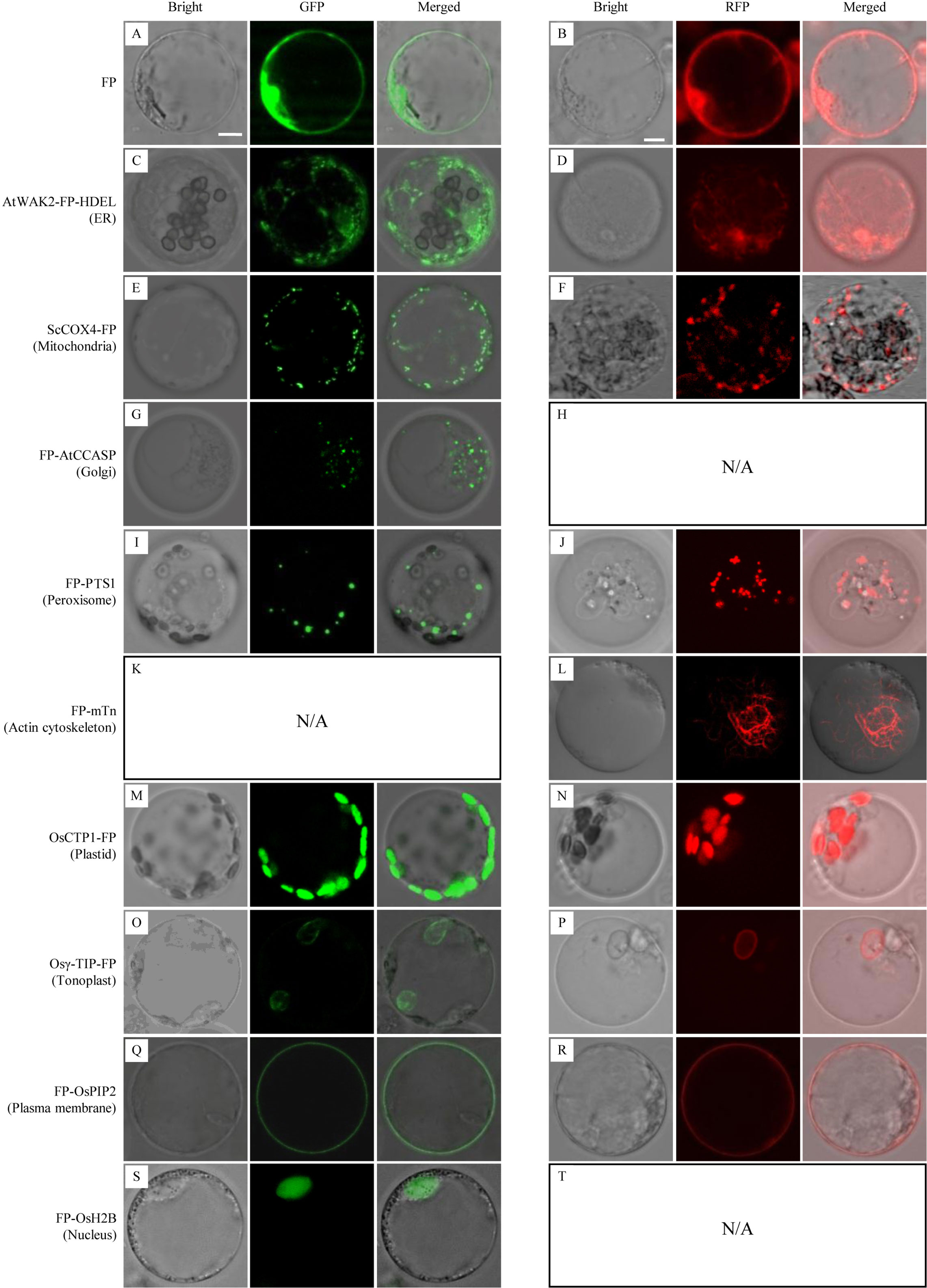
Figure 4 Subcellular localization of green fluorescent protein (GFP)/Discosoma sp. red fluorescent protein (DsRed)-fusion organelle markers in protoplasts of stable transgenic rice plants. FP, fluorescent protein GFP or DsRed. Protoplasts of GFP-fusion (left panel) and DsRed-fusion (right panel) transgenic lines were examined by confocal microscopy. (A, B) GFP or DsRed alone; (C, D) ER marker AtWAK2-GFP-HDEL or AtWAK2-DsRed-HDEL; (E, F) Mitochondrial marker ScCOX4-GFP or ScCOX4-DsRed; (G) Golgi marker GFP-AtCCASP; (I, J) Peroxisome marker GFP-PTS1 or DsRed-PTS1; (L) cytoskeleton marker DsRed-mTn; (M, N) Plasmid marker CTP1-GFP or CTP1-DsRed; (O, P) Tonoplast marker Osγ-TIP-GFP or Osγ-TIP-DsRed; (Q, R) Plasma membrane marker GFP-OsPIP2 or DsRed-OsPIP2; (S) Nucleus marker GFP-OsH2B. N/A, No DsRed-AtCCASP- (H), GFP-mTn- (K), or DsRed-OsH2B-transgenic rice plants expressing obvious fluorescent signals (T) were obtained. Scale bars, 10 μm.
To rule out the possibility that the DsRed-AtCCASP, GFP-mTn, and DsRed-OsH2B cassettes were not working for expression, we further transiently evaluated the three expression cassettes in N. benthamiana leaf cells and in rice protoplasts. Transient expression assays showed that the DsRed-AtCCASP, GFP-mTn, and DsRed-OsH2B fusions could be expressed in N. benthamiana leaves and rice protoplasts, producing fluorescent signals targeted to the Golgi apparatus, actin cytoskeleton, and nucleus, respectively (Supplementary Figure S1). These results indicated that the DsRed-AtCCASP, GFP-mTn, and DsRed-OsH2B cassettes were working for expression. Thus, it remains to be determined whether the three fusion proteins had harmful effects or there are other reasons that resulted in the failure to obtain transgenic rice lines stably expressing DsRed-AtCCASP, GFP-mTn, or DsRed-OsH2B.
Organelle Marker Lines for Co-Localization Studies
To test the utility of the GFP/DsRed-fusion organelle marker lines for protein co-localization studies, we selected a rice transcriptional factor OsWRKY45 (Shimono et al., 2007) and a N. benthamiana mitochondria-associated hexokinase NbHxk1 (Kim et al., 2006) for proof-of-concept testing. OsWRKY45 fused with a mCherry and NbHxk1 fused with a GFP, were expressed in protoplasts of the GFP-OsH2B and ScCOX4-DsRed rice lines, respectively. The results showed that the red fluorescence of OsWRKY45-mCherry was merged with the nucleus GFP-OsH2B marker, and the fluorescent signal of NbHxk1-GFP was partly merged with the mitochondria ScCOX4-DsRed marker (Figure 5). Overall, these experiments demonstrated that using protoplasts of the organelle marker lines can facilitate protein co-localization studies in rice cells.
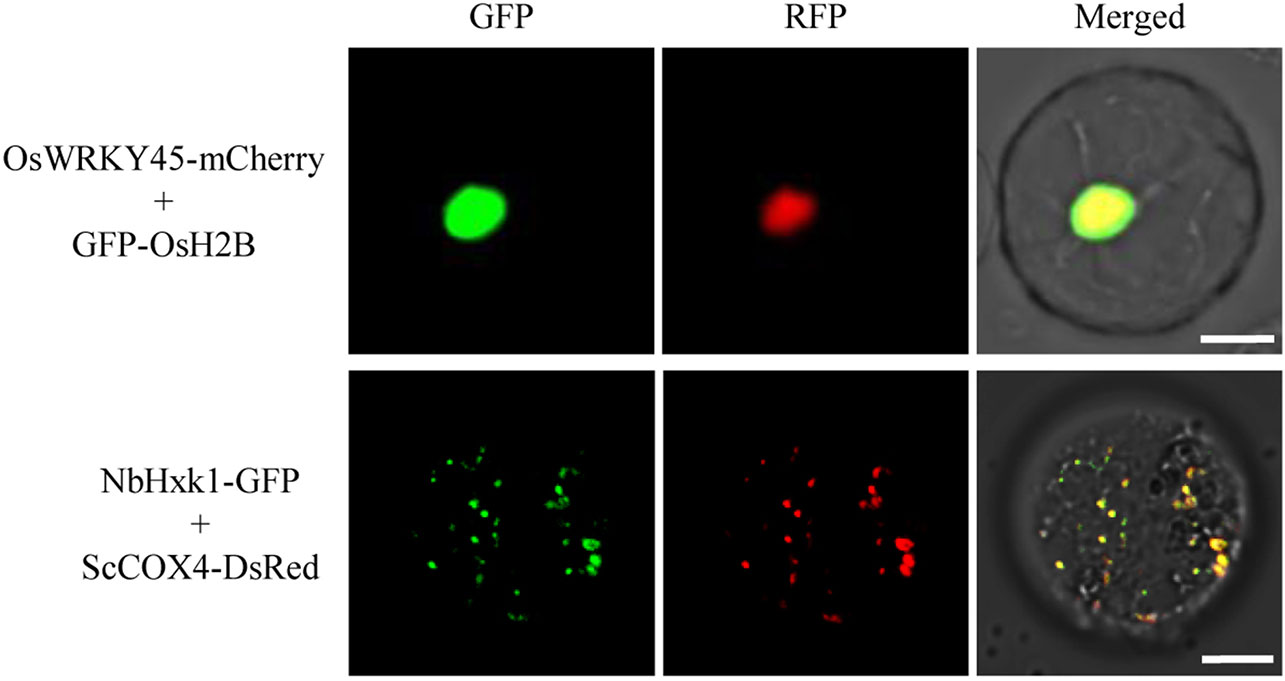
Figure 5 Co-localization analysis of OsWRKY45-mCherry and NbHxk1-green fluorescent protein (GFP) in protoplasts of the nuclear and mitochondrial marker lines. The OsWRKY45-mCherry and NbHxk1-GFP cassettes were transfected into protoplasts of the GFP-OsH2B (upper panel), and ScCOX4-DsRed (lower panel) rice lines, respectively. Scale bars, 10 μm.
Green Fluorescent Protein-OsH2B Marker Line for Detecting Translocation of Magnaporthe oryzae Effector Into Host Rice Cells
FP-tagged organelle marker plants provided powerful tool to study dynamics of proteins or organelles in plant cells in response to various stimuli, including pathogen attack. For example, Arabidopsis plants expressing various GFP-tagged organelles have been successfully used for investigating the subcellular reorganization of host cells in response to pathogen attack (Takemoto et al., 2003; Koh et al., 2005; Lu et al., 2012). In this study, we performed sheath-inoculation of the GFP-OsH2B rice plants with a transgenic M. oryzae isolate expressing a PWL2 effector fused with mCherry and a nuclear localization signal (PWL2:mCherry : NLS) (Khang et al., 2010). Our result confirmed that PWL2:mCherry : NLS was translocated into rice cells, as indicated by co-localization with the nucleus GFP-OsH2B marker (Figure 6). This experiment also demonstrated that the organelle marker lines will be useful resources for studying dynamics of proteins or organelles in rice cells in the interactions between rice and pathogens.
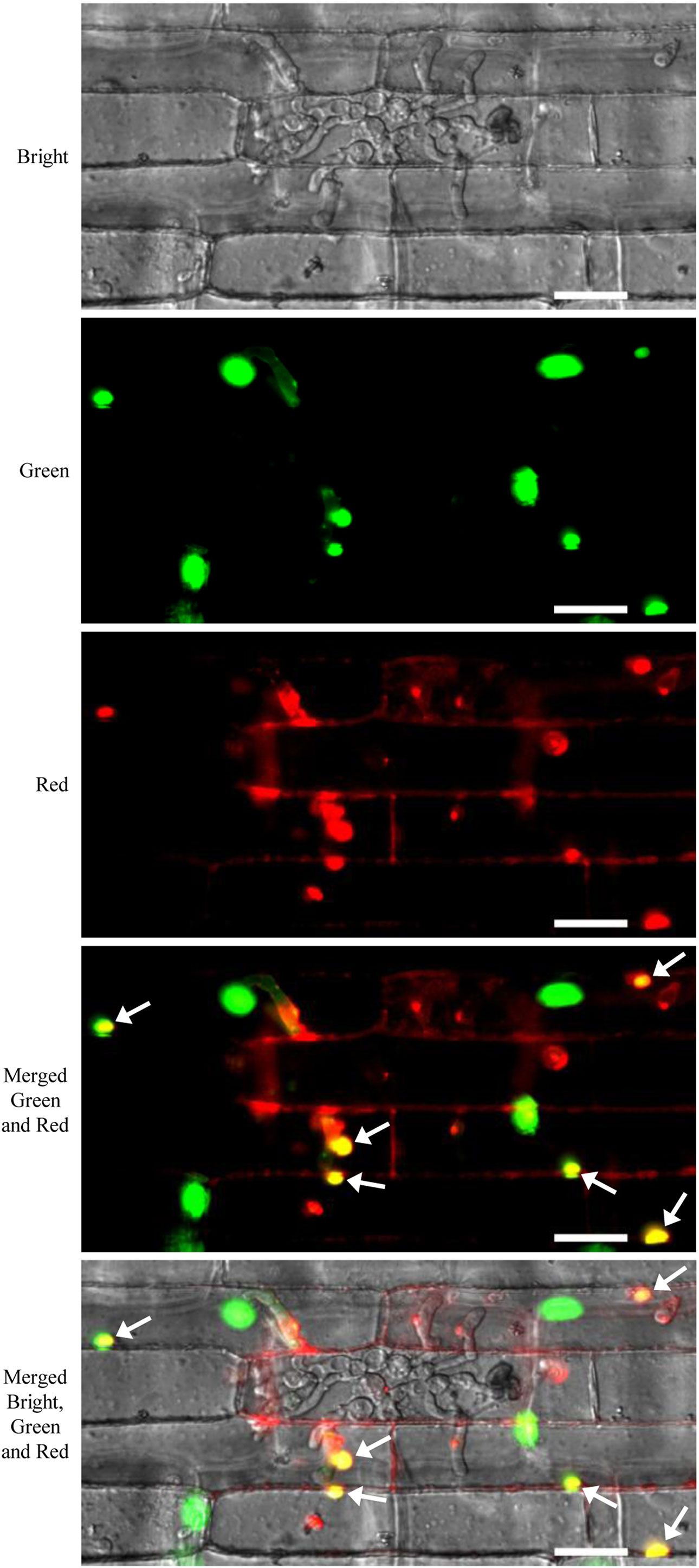
Figure 6 Live-cell imaging of Magnaporthe oryzae PWL2-mChery-NSL in rice cells of the green fluorescent protein (GFP)-OsH2B marker line. Rice leaf sheaths were inoculated with a transgenic M. oryzae isolate carrying a PWL2:mCherry : NLS expression cassette. Arrows indicate co-localization of PWL2-mCherry-NSL with the nuclear GFP-OsH2B marker. Scale bars, 25 μm.
Discussion
In this study, we developed a set of plant expression cassettes for GFP- and DsRed-fusion organelle markers. The markers designed to target ER, mitochondria, Golgi apparatus, peroxisome, or actin cytoskeleton were based on fusion of GFP or DsRed with the well-established targeting proteins or motifs (Mathur, 2007), and the plastid, tonoplast, plasma membrane, and nucleus markers were constructed by fusing GFP or DsRed to rice orthologs of the OsCTP1 transit peptide, Osγ-TIP, OsPIP2, and OsH2B, respectively. The binary constructs of the organelle markers were used to generate stable transgenic rice plants. Microscopic analysis of sheath cells and protoplasts of transgenic rice plants showed that most of the GFP- or DsRed-fusions could be stably expressed in rice cells. In addition, we observed no significant difference in morphological traits between wild-type and the organelle marker lines (data not shown). However, we were unable to obtain transgenic plants expressing fluorescent signals of DsRed-AtCCASP, GFP-mTn, or DsRed-OsH2B, although the DsRed-AtCCASP, GFP-mTn, and DsRed-OsH2B fusions could be transiently expressed in N. benthamiana leaf epidermal cells and in rice protoplasts (Supplementary Figure S1). Indeed, we also generated another set of expression cassettes of DsRed-AtCCASP, GFP-mTn, and DsRed-OsH2B, in which the fusion organelle marker genes were driven by a strong maize ubiquitin-1 promoter (Christensen et al., 1992; Chen et al., 2009). Similarly, no transgenic plants expressing obvious fluorescent signals of DsRed-AtCCASP, GFP-mTn, or DsRed-OsH2B were obtained with multiple tries (data not shown). While the reason for failure to obtain the DsRed-AtCCASP, GFP-mTn, or DsRed-OsH2B marker lines remains to be investigated, in the future, we will try to develop rice marker lines expressing green fluorescence targeted to the actin cytoskeleton, and lines expressing red fluorescence targeted to the Golgi apparatus and nucleus, respectively, using mCherry (Shaner et al., 2004) or other FP variants as reporters.
The availability of organelle marker lines will facilitate protein subcellular localization studies. Both our results and previous report (Wu et al., 2016) demonstrated that protoplasts from the rice organelle marker lines were suitable for transfection with fusion constructs of the protein of interest with FPs, and the markers could be used as comparative standards in determining the subcellular localization of the protein of interest. The use of the transgenic organelle marker lines also has an advantage over transient expression assays in studying the dynamics of proteins or organelles in developmental or environmental contexts (Geldner et al., 2009), because the transgenic lines are able to provide consistent, intact tissues for investigation under complicated conditions. For example, the Arabidopsis organelle marker lines have been used to study the interactions of Arabidopsis with oomycete (Takemoto et al., 2003; Lu et al.,2012), and powdery mildew fungus (Koh et al., 2005). In the present study, we presented an example application of using the leaf sheaths of the GFP-OsH2B marker line for detecting translocation of a M. oryzae PWL2 effector into rice cells, demonstrating the feasibility of the rice organelle marker lines as useful tools for dissection of protein dynamics or protein reorganization involved in rice-pathogen interactions. In addition, we generated both GFP- and/or DsRed-tagged organelle markers lines. These lines will allow more flexible design of FP combinations for co-localization studies in rice.
In summary, we developed a set of rice lines stably expressing GFP- and/or DsRed-tagged organelle markers, and demonstrated the utility of the marker lines for protein subcellular localization studies, especially for studying dynamics of proteins in rice cells in the interaction between rice with blast fungus. These marker lines, along with previously established organelle markers and marker lines (Wu et al, 2016; Dangol et al., 2017), provide important resources for the rice research community.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
ZW and SC conceived and designed the experiments. ZQC, WZ, LC, CL, TL, ZJC, HX, YH, LK, and XZ performed the experiments. ZQC, WZ, LC, CL, FW, and SC analyzed the data. ZQC, ZW, and SC wrote and revised the manuscript. All authors commented on the manuscript.
Funding
This work was supported by grants from National Natural Science Foundation of China (U1405212, 31171808, 31601596) and grant from Fujian Provincial Science and Technology Program (2017R1019-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01421/full#supplementary-material
References
Chen, S., Songkumarn, P., Liu, J., Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M. E., Umemura, K., Wang, G. L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7, 417–427. doi: 10.1111/j.1364-3703.2006.00346.x
Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. doi: 10.1016/s0960-9822(02)00483-9
Christensen, A. H., Sharrock, R. A., Quai, P. H. (1992). Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18, 675–689. doi: 10.1007/bf00020010
Chudakov, D. M., Matz, M. V., Lukyanov, S., Lukyanov, K. A. (2010). Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 90, 1103–1163. doi: 10.1152/physrev.00038.2009
Dangol, S., Singh, R., Chen, Y., Jwa, N. S. (2017). Visualization of multicolored in vivo organelle markers for co-localization studies in Oryza sativa. Mol. Cells 40, 828–836. doi: 10.14348/molcells.2017.0045
Geldner, N., Dénervaud-Tendon, V., Hyman, D. L., Mayer, U., Stierhof, Y. D., Chory, J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178. doi: 10.1111/j.1365-313X.2009.03851.x
Goodin, M. M., Dietzgen, R. G., Schichnes, D., Ruzin, S., Jackson, A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383. doi: 10.1046/j.1365-313X.2002.01360.x
He, Z. H., Cheeseman, I., He, D., Kohorn, B. D. (1999). A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol. Biol. 39, 1189–1196. doi: 10.1023/A:1006197318246
Hiei, Y., Ohta, S., Komari, T., Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Khang, C. H., Berruyer, R., Giraldo, M. C., Kankanala, P., Park, S. Y., Czymmek, K., et al. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22, 1388–1403. doi: 10.1105/tpc.109.069666
Kim, D. H., Xu, Z. Y., Hwang, I. (2013). Generation of transgenic Arabidopsis plants expressing mcherry-fused organelle marker proteins. J. Plant Bio 56, 399–406. doi: 10.1007/s12374-013-0348-3
Kim, M., Lim, J. H., Ahn, C. S., Park, K., Kim, G. T., Kim, W. T., et al. (2006). Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18, 2341–2355. doi: 10.1105/tpc.106.041509
Koh, S., André, A., Edwards, H., Ehrhardt, D., Somerville, S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44, 516–529. doi: 10.1111/j.1365-313X.2005.02545.x
Köhler, R. H., Zipfel, W. R., Webb, W. W., Hanson, M. R. (1997). The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 11, 613–621. doi: 10.1046/j.1365-313X.1997.11030613.x
Kost, B., Spielhofer, P., Chua, N. H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. doi: 10.1046/j.1365-313x.1998.00304.x
Krishnakumar, V., Choi, Y., Beck, E., Wu, Q., Luo, A., Sylvester, A., et al. (2015). A maize database resource that captures tissue-specific and subcellular-localized gene expression, via fluorescent tags and confocal imaging (Maize Cell Genomics Database). Plant Cell Physiol. 56, e12. doi: 10.1093/pcp/pcu178
Lu, Y. J., Schornack, S., Spallek, T., Geldner, N., Chory, J., Schellmann, S., et al. (2012). Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell Microbiol. 14, 682–697. doi: 10.1111/j.1462-5822.2012.01751.x
Lunn, J. E. (2007). Compartmentation in plant metabolism. J. Exp. Bot. 58, 35–47. doi: 10.1093/jxb/erl134
Luo, B., Nakata, P. A. (2012). A set of GFP organelle marker lines for intracellular localization studies in Medicago truncatula. Plant Sci. 188-189, 19–24. doi: 10.1016/j.plantsci.2012.02.006
Martin, K., Kopperud, K., Chakrabarty, R., Banerjee, R., Brooks, R., Goodin, M. M. (2009). Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 59, 150–162. doi: 10.1111/j.1365-313X.2009.03850.x
Mathur, J. (2007). The illuminated plant cell. Trends Plant Sci. 12, 506–513. doi: 10.1016/j.tplants.2007.08.017
McCann, R. O., Craig, S. W. (1997). The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. 94, 5679–5684. doi: 10.1073/pnas.94.11.5679
Nelson, B. K., Cai, X., Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x
Park., C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Renna, L., Hanton, S. L., Stefano, G., Bortolotti, L., Misra, V., Brandizzi, F. (2005). Identification and characterization of AtCASP, a plant transmembrane Golgi matrix protein. Plant Mol. Biol. 58, 109–122. doi: 10.1007/s11103-005-4618-4
Reumann, S. (2004). Specification of the peroxisome targeting signals type 1 and type 2 of plant peroxisomes by bioinformatics analyses. Plant Physiol. 135, 783–800. doi: 10.1104/pp.103.035584
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. doi: 10.1038/nbt1037
Shimono, M., Sugano, S., Nakayama, A., Jiang, C. J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Takemoto, D., Jones, D. A., Hardham, A. R. (2003). GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 33, 775–792. doi: 10.1007/s001220050734
Tanz, S. K., Castleden, I., Small, I. D., Millar, A. H. (2013). Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 4, 214. doi: 10.3389/fpls.2013.00214
Wu, Q., Luo, A., Zadrozny, T., Sylvester, A., Jackson, D. (2013). Fluorescent protein marker lines in maize: generation and applications. Int. J. Dev. Biol. 57, 535–543. doi: 10.1387/ijdb.130240qw
Wu, T. M., Lin, K. C., Liau, W. S., Chao, Y. Y., Yang, L. H., Chen, S. Y., et al. (2016). A set of GFP-based organelle marker lines combined with DsRed-based gateway vectors for subcellular localization study in rice (Oryza sativa L.). Plant Mol. Biol. 90, 107–115. doi: 10.1007/s11103-015-0397-8
Keywords: fluorescent protein, organelle marker lines, protein subcellular localization, rice, Magnaporthe oryzae
Citation: Chen Z, Zheng W, Chen L, Li C, Liang T, Chen Z, Xu H, Han Y, Kong L, Zhao X, Wang F, Wang Z and Chen S (2019) Green Fluorescent Protein- and Discosoma sp. Red Fluorescent Protein-Tagged Organelle Marker Lines for Protein Subcellular Localization in Rice. Front. Plant Sci. 10:1421. doi: 10.3389/fpls.2019.01421
Received: 17 July 2019; Accepted: 14 October 2019;
Published: 05 November 2019.
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Yuese Ning, Chinese Academy of Agricultural Sciences, ChinaWen-Ming Wang, Sichuan Agricultural University, China
Copyright © 2019 Chen, Zheng, Chen, Li, Liang, Chen, Xu, Han, Kong, Zhao, Wang, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Wang, d2ZAZmphZ2Uub3Jn; Zonghua Wang, d2FuZ3poQGZhZnUuZWR1LmNu; Songbiao Chen, c29uZ2JpYW9fY2hlbkBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Ziqiang Chen1†
Ziqiang Chen1† Wenhui Zheng
Wenhui Zheng Zaijie Chen
Zaijie Chen Zonghua Wang
Zonghua Wang Songbiao Chen
Songbiao Chen