
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 November 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01400
This article is part of the Research TopicNovel Plant Molecules Regulating the Interaction with Pathogenic and Beneficial FungiView all 12 articles
 Yi Zhang1,2†
Yi Zhang1,2† Kedong Xu1,2†
Kedong Xu1,2† Dongli Pei3†
Dongli Pei3† Deshui Yu1,2
Deshui Yu1,2 Ju Zhang1,2
Ju Zhang1,2 Xiaoli Li1,2
Xiaoli Li1,2 Guo Chen1,2
Guo Chen1,2 Hui Yang1,2
Hui Yang1,2 Wenjie Zhou1,2
Wenjie Zhou1,2 Chengwei Li4*
Chengwei Li4*A previous complementary cDNA-amplified fragment length polymorphism (cDNA-AFLP) analysis examined responses to the powdery mildew pathogen Oidium neolycopersici (On) of the resistant cultivar Solanum habrochiates G1.1560, carrying the Ol-1 resistance gene, and susceptible cultivar S. lycopersicum Moneymaker (MM). Among other findings, a differentially expressed transcript-derived fragment (DE-TDF) (M14E72-213) was upregulated in near isogenic line (NIL)-Ol-1, but absent in MM. This DE-TDF showed high homology to a gene of unknown function, which we named ShORR-1 (Solanum habrochaites Oidium Resistance Required-1). However, MM homolog of ShORR-1 (named ShORR-1-M) was still found with 95.26% nucleic acid sequence similarity to ShORR-1 from G1.1560 (named ShORR-1-G); this was because the cut sites of restriction enzymes in the previous complementary cDNA-AFLP analysis was absent in ShORR-1-M and differs at 13 amino acids from ShORR-1-G. Transient expression in onion epidermal cells showed that ShORR-1 is a membrane-localized protein. Virus-induced gene silencing (VIGS) of ShORR-1-G in G1.1560 plants increased susceptibility to On. Furthermore, overexpressing of ShORR-1-G conferred MM with resistance to On, involving extensive hydrogen peroxide accumulation and formation of abnormal haustoria. Knockdown of ShORR-1-M in MM did not affect its susceptibility to On, while overexpressing of ShORR-1-M enhanced MM’s susceptibility to On. We also found that changes in transcript levels of six well-known hormone signaling and defense-related genes are involved in ShORR-1-G-mediated resistance to On. The results indicate that ShORR-1-M and ShORR-1-G have antagonistic effects in tomato responses to On, and that ShORR-1 is essential for Ol-1-mediated resistance in tomato.
Plants have evolved a multilayered immune system that prevents or hinders colonization by most potential pathogens. To date, two types of innate immune response have been recognized in plants. One is pathogen-associated molecular pattern-triggered immunity, which is activated by a number of pathogen-associated molecular patterns such as flagellin, EF-Tu, and chitin, and perceived by pattern recognition receptors (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Peng et al., 2018). The other is effector-triggered immunity, which is modulated by recognition of pathogen-derived avirulence effectors by plant R proteins (Jones and Dangl, 2006; Dodds and Rathjen, 2010).
Powdery mildew caused by Oidium neolycopersici (On) is one of the most severe diseases of tomato. Six resistance genes (termed Ol-X) and three quantitative resistance loci from wild tomato species have been identified, all of which mediate various resistance responses to On (Bai et al., 2003; Bai et al., 2004; Bai et al., 2005; Li, 2005). Ol-1, one of the resistance genes, derived from Solanum habrochaites G1.1560 (Lindhout et al., 1994), mediates partial resistance to On, including a slow hypersensitive response (HR) (Li et al., 2007). Ol-4, introgressed from wild tomato species S. peruvianum LA2172, confers complete resistance to On and is associated with a rapid HR (Bai et al., 2004). In previous studies, we found that near-isogenic lines (NILs) carrying ol-2, Ol-1 and Ol-4 genes in the genetic background of the susceptible cultivar MM had varying degrees of resistance to On (Li et al., 2006; Li et al., 2007). We also examined gene expression patterns in these lines by cDNA-amplified fragment length polymorphism (cDNA-AFLP) analysis. Transcript-derived fragments (TDFs) showing differential presence or intensity between resistant tomato NILs and susceptible MM after mock-inoculation and inoculation with On were identified (Li et al., 2006; Li et al., 2007). UniGene sequences in the Solanaceae Genomics Network (SGN) database showing high homology to each differentially expressed TDF were then identified. Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) constructs targeting these UniGenes were subsequently generated, and used to determine whether silencing the targeted genes altered the On resistance of relevant genotypes. These efforts revealed that acetolactate synthase (ALS) (Gao et al., 2014), a glutathione S-transferase (GST) gene (Pei et al., 2011a), and an NADP-malic enzyme gene (Pei et al., 2011b) are required for Ol-1-mediated resistance to On.
In the study presented here, we focused on another differentially expressed TDF (M14E72-213) and analyzed its involvement in On resistance. M14E72-213 is present in NIL-Ol-1, but not MM or NIL-Ol-4 (Li et al., 2006; Li et al., 2007). We found that silencing it resulted in loss of resistance, to varying degrees, in S. habrochaites G1.1560 (which carries the Ol-1 gene). Microscopic observation showed that the pathogen could complete its life cycle on leaves of the plants with silenced Ol-1. In addition, the HR was slow in epidermal cells of the leaves from this line, while rapid HR in attacked epidermal cells of control plants prevented completion of On’s life cycle. Thus, the gene is apparently required for Ol-1 mediated resistance to On, and was named ShORR-1 (S. habrochaites Oidium Resistance Required gene). According to open reading frame (ORF) finder, it encodes a putative 268 amino acid protein, which has 93% identity with an uncharacterized gene (XM_004242006), suggesting that it may be a novel gene. The results indicated that differences in ShORR-1 variants of susceptible and resistant tomato account for at least some of the differences in their resistance. Further analyses indicate that increases in H2O2 accumulation and formation of abnormal haustoria are involved in Ol-1-mediated resistance to powdery mildew in tomato, which requires an appropriate variant of ShORR-1, such as ShORR-1-G.
The On-susceptible tomato S. lycopersicum Mill (MM) and resistant cultivar S. habrochaites G1.1560, which carries the Ol-1 gene, were grown in a greenhouse providing 16 h and 23°C day/8 h and 20°C night cycles, with constant 80% relative humidity (RH). The tomato powdery mildew used in this research was identified as On isolate China (Li et al., 2008) on the basis of its morphological, histological, and molecular characteristics. The fungus was maintained on the susceptible tomato cultivar MM. For inoculation of On, whole plants were sprayed with a suspension of spores (5 × 103 conidia/ml) collected from infected tomato plants in a climate chamber in the conditions mentioned above, except that the RH was 85 and 95% during the day and night periods, respectively. Control plants were sprayed with sterile water.
The target DE-TDF was amplified by reverse transcription PCR (RT-PCR) with primers listed in Table 1. The recombinant vector TRV-LIC-ShORR-1 carrying the target sequence was constructed as described elsewhere (Dong et al., 2007). The vectors TRV1 and TRV2-LIC-ShORR-1 were introduced into Agrobacterium tumefaciens strain GV3101 by heat shock and cocultured overnight. Overnight cultures (5 ml) were grown at 28°C in appropriate antibiotic selection medium in 15 ml glass tubes for 1 day, then briefly centrifuged, and the collected cells were resuspended in infiltration medium (10 mM MES, 10 mM MgCl2, 200 µM acetosyringone) to an OD600 of 1. After incubation at room temperature for 3 h, the cultures were used for agro-infiltration, as previously reported (Liu et al., 2010). Briefly, plants at the four-leaf stage were infiltrated with a 1:1 mixture of TRV1 and TRV2-LIC-ShORR-1 fragments, and plants treated with cultures with empty vector provided negative controls. Ten days after agro-infiltration, plants were inoculated with On. Four plants per trial were inoculated and at least three trials were conducted.
An endogenous peroxidase-dependent 3,3-diaminobenzidine (DAB) assay was used to investigate H2O2 production in plants (Thordal-Christensen et al., 2010). To detect H2O2 accumulation, leaflets taken 65 h after On infection were immersed in DAB solution (1 mg/ml, pH 3.8) for 8–12 h until DAB staining was visible at the vein of the top leaflet. The DAB-stained leaflets were fixed and stained with Coomassie Brilliant Blue R250 in methanol (0.6%, w/v), following Li (2005) with minor modifications. Samples were observed under a differential-interference contrast microscope (Carl Zeiss, Germany), and images were acquired with a Color Video Camera equipped with image analysis software (Image-Pro Plus 4.1, Media Cybernetics, L.P.). In each microscopically examined sample, we observed more than 200 primary haustoria and secondary haustoria, and recorded percentages of host cells showing HR. Three biological replicates of microscopic samples of both ShORR-1 silenced and control plants were used in these examinations.
RNA was extracted from G1.1560 and MM tomato leaves with TRIzol reagent (Life Technologies, Grand Island, NY) following the manufacturer’s recommendations. After extraction, the RNA samples were treated with DNase I (TaKaRa) to eliminate trace contaminants of genomic DNA. cDNA was synthesized using a PrimeScript RT Perfect Real Time reagent kit (TaKaRa), and the resulting cDNAs were used as templates in the following PCR reactions. The TDF fragment M14E72-213 was aligned to the tomato genome (Consortium, 2012) and ShORR-1-F1/R1 primers (Table 1) were designed to clone the complete ORF based on the UniGene with highest identity. To detect genes homologous to ShORR-1, sequences from G1.1560 and MM, respectively designated ShORR-1-G and ShORR-1-M, were amplified with the ShORR-1-F2/R2 primers (Table 1). The PCR reactions involved denaturation at 94°C for 5 min, followed by 30 amplification cycles of 30 s at 94°C, 45 s at 58°C, 1 min at 72°C, and a final extension step of 10 min at 72°C. The resulting amplicons were inserted into the pMD18-T vector (TaKaRa) and recombinant plasmids were sequenced using the universal T7 primer.
ORFs of ShORR-1-G and ShORR-1-M were used to construct transient expression vectors by insertion into the pSAT6-GFP-N1 vector (Xu et al., 2014), which contains a gene encoding a modified green fluorescent protein (GFP) at Nco I–Xba I sites, with ShORR-1-F3/R3 primers (Table 1). pSAT6-GFP-N1 was digested with Kpn I and BamH I to construct pSAT6-GFP-N1-ShORR-1-G and pSAT6-GFP-N1-ShORR-1-M vectors, then the recombinant plasmids were transformed into onion epidermal cells by an Agrobacterium-mediated in planta transient transformation protocol (Xu et al., 2014). Microscopic observations and image acquisition were performed on a fluorescence microscope (Olympus BX61, Japan).
To validate functions of ShORR-1-G and ShORR-1-M, the full length target genes were introduced into the pCAMBIA2300 vector with Kpn I and Sal I restriction enzymes. Then a 381 bp partial sequence of ShORR-1-M was cloned into RNAi vector pCAMBIA2300 using the sense and antisense strands. PCR products were amplified with KAPA HiFi PCR kits (Kapa Biosystems, USA), then transformed into Escherichia coli Trans T1 competent cells to generate recombinant plasmids, which were introduced into Agrobacterium strain GV3101. All applied oligonucleotide primers are listed in Table 1. Different homozygotes transgenic plants lines were produced by the Plant Genetic Transformation Center of the Henan Key Laboratory of Crop Molecular Breeding & Bioreactor. To investigate ShORR-1’s function in resistance to On, the positive transgenic plants were inoculated and microscopically analyzed, as previously described (Shen et al., 2007). Fresh leaves from wild-type and transgenic tomato plants were collected and placed on 1% agar plates containing 85 µM benzimidazole. The leaves were incubated in a climate chamber providing constant light at 20°C for at least 4 h, then inoculated with a suspension of On spores (5 × 103 conidia/ml). After allowing the On fungus to develop on the leaves for 65 h under the same conditions (Bai et al., 2005), the leaves were fixed in ethanol/acetic acid (1:1, v/v), stained with Coomassie Brilliant Blue R250 in methanol (0.6%, w/v) for 10 s, then rinsed in deionized water. Samples were subsequently observed with a BX61 microscope (Olympus, Tokyo, Japan), and microcolonies were counted. More than 1,000 germinated spores on each leaf segment from every plant were observed, and three biological replicates of control and transgenic plants were used.
To quantify levels of ShORR-1 transcript produced in response to On, susceptible and resistant tomato leaves were sampled at 0, 8, 24, 36, 72, and 120 h postinoculation (hpi), according to previous microscopic observations of tomato-On interaction (Gao et al., 2014). Estimates of fungal biomass in the samples were obtained by extracting On DNA and quantifying levels of the EF-1a gene following Trond and Cathrine (2009). To explore the resistance mechanisms involving ShORR-1, the expression of six marker genes associated with different disease resistance and hormone pathways was quantified in wild-type and ShORR-1-G-overexpressing plants in the presence and absence of On. For this, total RNA was extracted from samples of transgenic and wild-type plants with TRIzol reagent following recommendations of the manufacturer, and 1 µg total RNA was used for cDNA synthesis. It was then subjected to quantitative reverse transcription PCR (qRT-PCR) using a CFX96™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) with SYBR® Premix Ex Taq™ (Takara Bio Inc., Shiga, Japan). The amplification conditions consisted of 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, and 60°C for 30 s. Fold changes in levels of target transcripts were calculated using the 2−ΔΔCt and 2−ΔCt method (Livak and Schmittgen, 2001), with normalization against the tomato Actin (SlActin) transcript levels. Three biological and technical replicates were run for each cDNA sample. All the applied qRT-PCR primers are listed in Table 1.
Relative expression levels of genes were statistically analyzed using one-way analysis of variance (ANOVA) and Dunnett’s post hoc or Tukey’s HSD tests (P < 0.01 and P < 0.05). All analyses were performed using SPSS Statistics 17.0 following instructions in the SPSS Survival Manual.
In our previous cDNA-AFLP study, we found that TDF fragment M14E72-213—designated No. 25 by Li et al. (2007), Appendix 1)—was present in On-resistant NIL-Ol-1, but not in the On-susceptible cultivar MM. BLAST analysis indicated that the 134 bp sequence had been annotated in SGN before the tomato genome sequence became publicly available, and was described as having 97% identity with a UniGene of unknown function (SGN-U319851). Using the sequence of this UniGene, in the study presented here we cloned the 807 bp ORF designated ShORR-1-G from the Ol-1 resistant line G1.1560 (accession no. MK205292). We found it encodes a putative protein of 268 amino acids with a molecular weight of 30.55 kDa, isoelectric point (pI) of 9.53, and 95% identity to a protein of unknown function encoded by Solyc06g059860.2 according to a BLASTP search against the SGN database. In contrast, the ORF sequence of ShORR-1-M is 819 bp long (accession no. MK205293), and encodes a putative protein of 272 amino acids with a molecular weight of 30.96 kDa, isoelectric point (pI) of 9.48, and 99% identity with Solyc06g059860.2 (differing in only one nucleotide base). ShORR-1-G and ShORR-1-M have no conserved domain, according to a search of the NCBI database. The DNAMAN software package was applied to align the nucleotide and protein homologs of ShORR-1 cloned from the two tomato varieties (Figure 1). The ORF and protein sequence homologies of the susceptible and resistant species’ ShORR-1 variants were 95.26 and 94.87%, respectively. The alignment results showed that ShORR-1-G has one more amino acid residue (a lysine) than ShORR-1-M at the 5′ end, while there are five more amino acids in ShORR-1-M at the 3′ end, and seven other amino acids differ between them. Additionally, the amino acid sequences of ShORR-1-G and ShORR-1-M shared 95.22 and 97.42% identity with a hybrid signal transduction histidine kinase of S. pennellii with a BLASTP search in NCBI, respectively. The ShORR-1-G and ShORR-1-M sequences respectively contain 19 and 20 potential serine phosphorylation sites (http://www.dabi.temple.edu/disphos), and in both cases putative MAPK phosphorylation sites (Mao et al., 2011, Nobuaki et al., 2011) in the N terminus (Figure S1). Moreover, ShORR-1-M and ShORR-1-G proteins both have potential ubiquitination (Figure S2) and SUMOylation sites (Figure S3) by UbPred (http://www.ubpred.org/) and GPS-SUMO (http://sumosp.biocuckoo.org/online.php) (Radivojac et al., 2010; Zhao et al., 2014). Neither ShORR-1-M and ShORR-1-G proteins have any transmembrane helices according to TMHMM 2.0 predictions (http://www.cbs.dtu.dk/services/TMHMM/).

Figure 1 Nucleotide (A) and amino acid (B) sequences alignment of ShORR-1-M and ShORR-1-G share high homology using DNAMAN software.
To determine the subcellular localization of ShORR-1, the full-length sequences isolated from G1.1560 and MM were fused to the 5′ terminus of the GFP gene in the pSAT6-GFP vector, under control of the constitutive CaMV: 35S promoter. The ShORR-1 protein was transiently transferred into onion epidermal cells by A. tumefaciens-mediated transformation (Xu et al., 2014). Microscopic observation of the fluorescent signal emitted by the ShORR-1-GFP fusion protein revealed that ShORR-1-M and ShORR-1-G were both localized on the plasma membrane, while fluorescence signals from the control pSAT6-GFP vector were ubiquitous in examined cells (Figure 2). These results show that ShORR-1 is a membrane-localized protein.
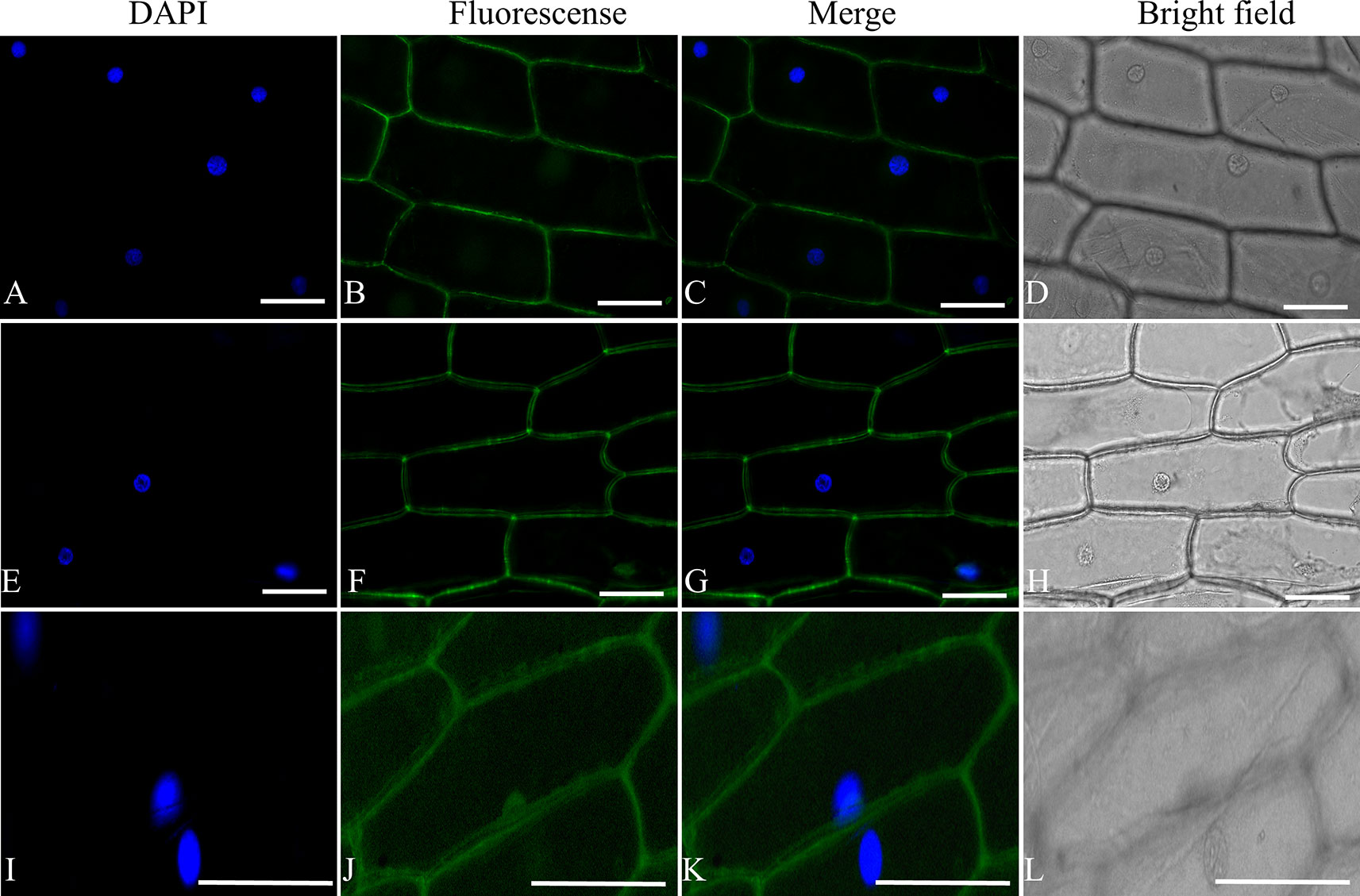
Figure 2 ShORR-1-M and ShORR-1-G both localized to the plasma membrane. Subcellular localization GFP fusions with ShORR-1 variants isolated from G1.1560 and MM in onion epidermal cells, as shown by 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (A, E, and I), green fluorescent protein (GFP) fluorescence (B, F, and J), merger of DAPI staining and fluorescence (C, G, and K), bright field microscopy (D, H, and L). The proteins displayed in panels (A–D), (E–H), and (I–L) were transiently expressed using pSAT6-GFP-N1-ShORR-1-G, pSAT6-GFP-N1-ShORR-1-M, and a construct designed to express GFP alone (as a control), respectively. Scale bar = 25 µm.
To investigate ShORR-1’s role in resistance to infection by the powdery mildew fungus On, we measured its transcript levels in tissues of leaves infected by On at selected time-points by qRT-PCR. Previous research had revealed that On haustoria form at 24–41 hpi (Huang et al., 1998; Bai et al., 2005). Expression of ShORR-1 was detected in both On-susceptible and -resistant species, but the transcription patterns and expression levels (relative to levels in controls) differed. Generally, relative expression levels of ShORR-1 were much higher in G1.1560 than in MM, although they were similar at 36 hpi (Figures 3A, B). The relative expression level of ShORR-1 in MM increased as early as 8 hpi, and peaked (at 7.8-fold) at 120 hpi, compared to levels in control leaves (Figure 3A), while the relative expression level of ShORR-1 in G1.1560 also increased at 8 dpi and peaked at 72 hpi (Figure 3B). These results indicate that ShORR-1 could be upregulated earlier and generally more strongly in resistant tomato plants than in susceptible tomato plants in response to On. Quantitative analysis of ShORR-1 expression patterns in MM tissues by qRT-PCR showed that it is expressed mainly in leaves and stems, and to a lesser extent in mature red fruits, roots, flowers, and green fruits (Figure 3C).
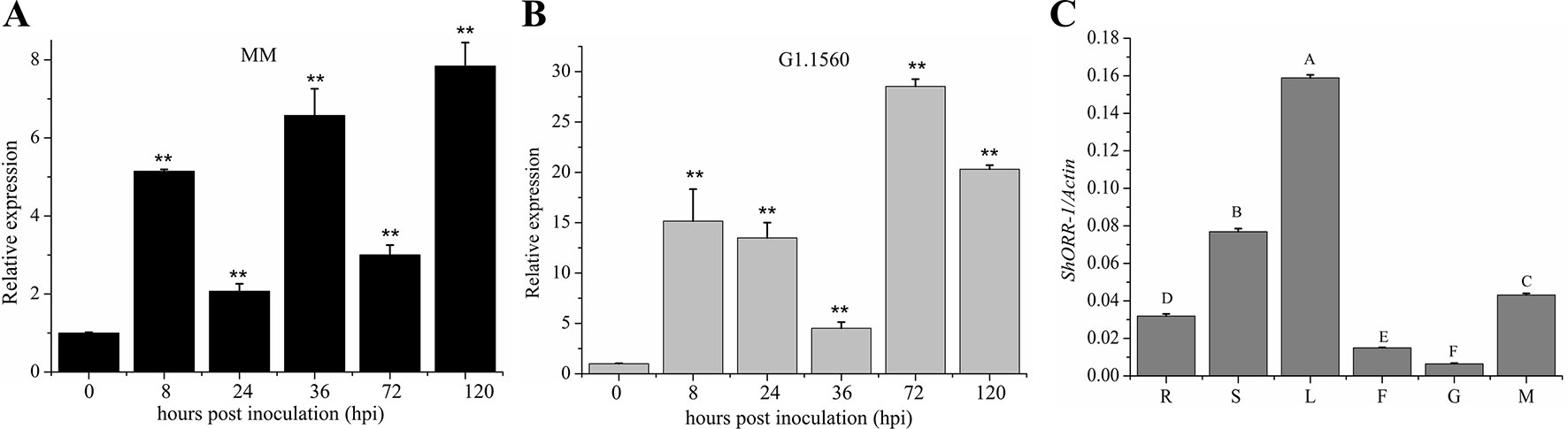
Figure 3 Expression profiles of ShORR-1 at indicated On infection stages and tissues in MM (A) and G1.1560 (B). Samples were harvested at 0, 8, 24, 36, 72, and 120 hpi (A–B). The relative expression levels of ShORR-1 at each time-point were normalized relative to SlActin. Asterisks indicate significant difference from the control determined by one-way ANOVA followed by an independent-samples Dunnett’s post hoc test (**: P < 0.01). (C) The transcript accumulation of ShORR-1 was examined by quantitative real-time PCR from MM tissues, including roots (R), stems (S), leaves (L), flowers (F), green fruits (G), and red fruits (M). Actin was used as an internal control. Mean and standard error were calculated using data from three independent biological replicates. Letters indicate significant differences between different tissues determined by one-way ANOVA followed by Tukey’s HSD test (P < 0.01).
For the TDF fragment M14E72-213, which had 97% identity with SGN-U319851, primers were designed based on U319851 to construct a TRV2-VIGS vector to silence ShORR-1 in S. habrochaites G1.1560. Twenty silenced plants of this previously resistant line were inoculated with On 10 days after VIGS infiltration. Seven days postinoculation (dpi) with the fungus, On colonies were clearly visible on leaves of all 20 silenced plants, with 5–30% of their foliar area affected, while no fungal colonies were observed on plants infiltrated with the empty vector (TRV2-EV) (Figure 4A). These results clearly indicate that silencing of ShORR-1 abolished Ol-1-mediated resistance to On at the macroscopic level. qRT-PCR analyses showed that ShORR-1 transcript levels were significantly lower in the silenced plants than in controls (Figure 4B). Moreover, microscopic analysis showed that numerous conidiophores with conidia were present on ShORR-1-silenced resistant plants, and the histological morphology of the fungus on ShORR-1-silenced resistant plants was similar to that on susceptible plants. Thus, the fungus successfully completed its life cycle on them, and silencing of ShORR-1 did not lead to morphological alteration of On. In contrast, fungal growth was clearly prevented, and no conidiospores formed, on TRV2-EV G1.1560 plants (Figure 4C). In conclusion, silencing ShORR-1 allowed fungal growth and sporulation, resulting in visible disease symptoms, indicating that ShORR-1 plays an important role in resistance to powdery mildew caused by On.
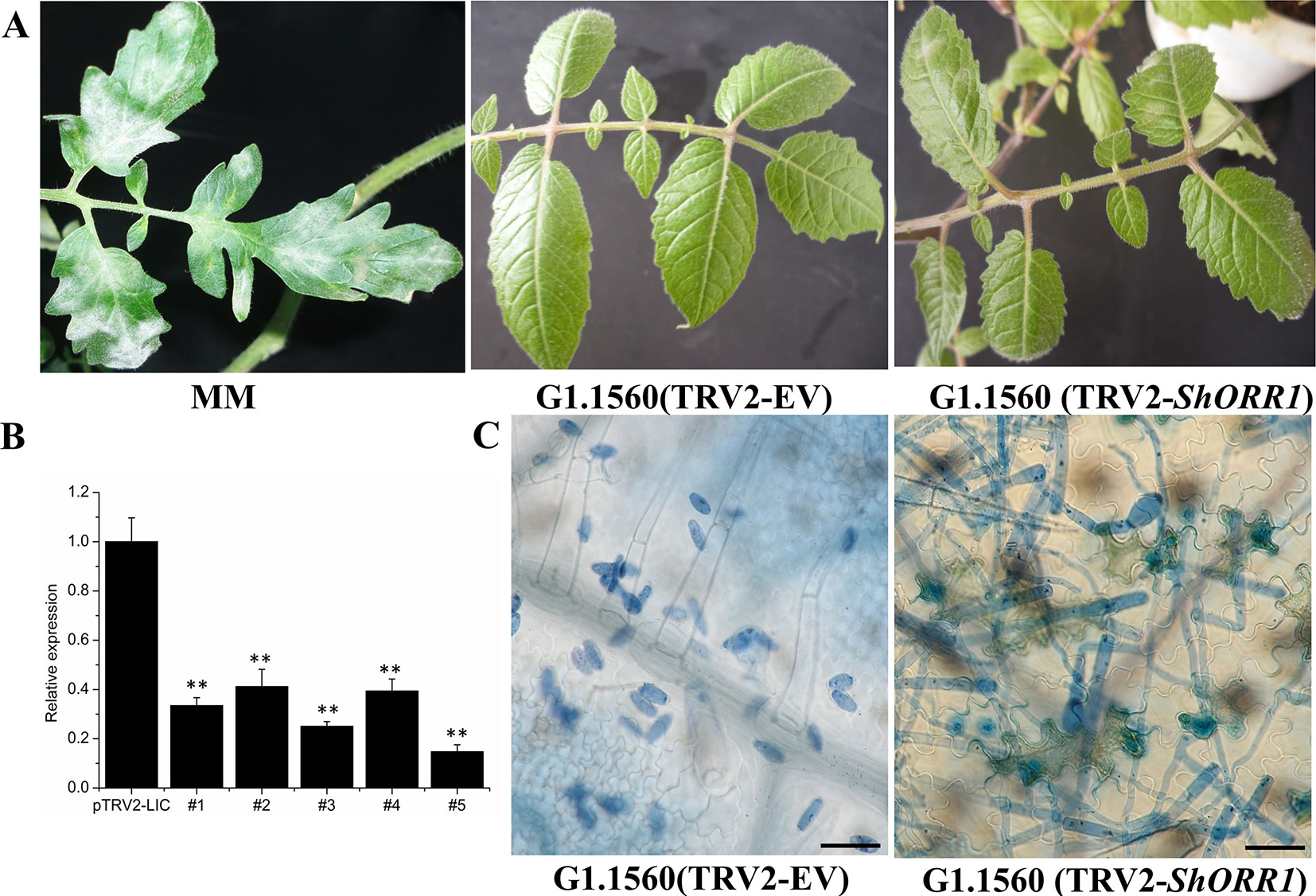
Figure 4 Downregulation of ShORR-1 by VIGS compromises Ol-1-mediated resistance. (A)On susceptible phenotypes of MM and ShORR-1-silenced (TRV2-ShORR-1) G1.1560 plants, and resistant phenotype of G1.1560 controls transformed with an empty vector (TRV2-EV). (B) Levels of ShORR-1 transcripts in control and TRV2-ShORR-1 leaves. Error bars represent standard deviations, obtained from analysis of three biological replicates. Asterisks indicate significant difference from the control by one-way ANOVA followed by an independent-samples Dunnett’s post hoc test (**: P < 0.01). (C) Microscopic morphologies of On in control and TRV2-ShORR-1 plants. Bar = 25 µm.
Li (2005) noted that the TDF fragment M14E72-213 accumulated more rapidly, and to higher levels, in resistant tomato than in susceptible plants after inoculation with On. Microscopic analysis presented here showed that S. habrochaites G1.1560 control plants displayed a rapid HR following exposure to On. In this response, most cells invaded by primary haustoria of On rapidly accumulated H2O2 and died, thereby preventing further growth of On (Figure 5A). Contrary to expectations, On-induced cell death and H2O2 accumulation were observed in ShORR-1-silenced resistant plants (Figure 5A), although fungal growth and conidiophore formation were similar to those on susceptible plants. However, the proportion of dead cells among cells attacked by fungal haustoria was lower in ShORR-1-silenced resistant plants than in control resistant plants. We investigated more than 200 cells attacked by primary haustoria in each microscopic sample, and found that average percentages of cells showing HR in control and ShORR-1-silenced plants were about 68 and 30%, respectively, at 64 hpi. We then observed more than 200 cells attacked by secondary haustoria in each microscopic sample, and found that percentages of cells showing HR in control and ShORR-1-silenced plants were about 32 and 22%, respectively, at 147 hpi (Figure 5B). In contrast to those in resistant control plants, most cells invaded by fungal haustoria remained alive in ShORR-1-silenced resistant plants, and the haustoria showed normal morphology, resulting in further fungal growth and conidiophore formation in ShORR-1-silenced resistant plants. The results also indicated that the On-induced HR in the ShORR-1-silenced resistant plants was slower, and possibly weaker, than in the control resistant plants.
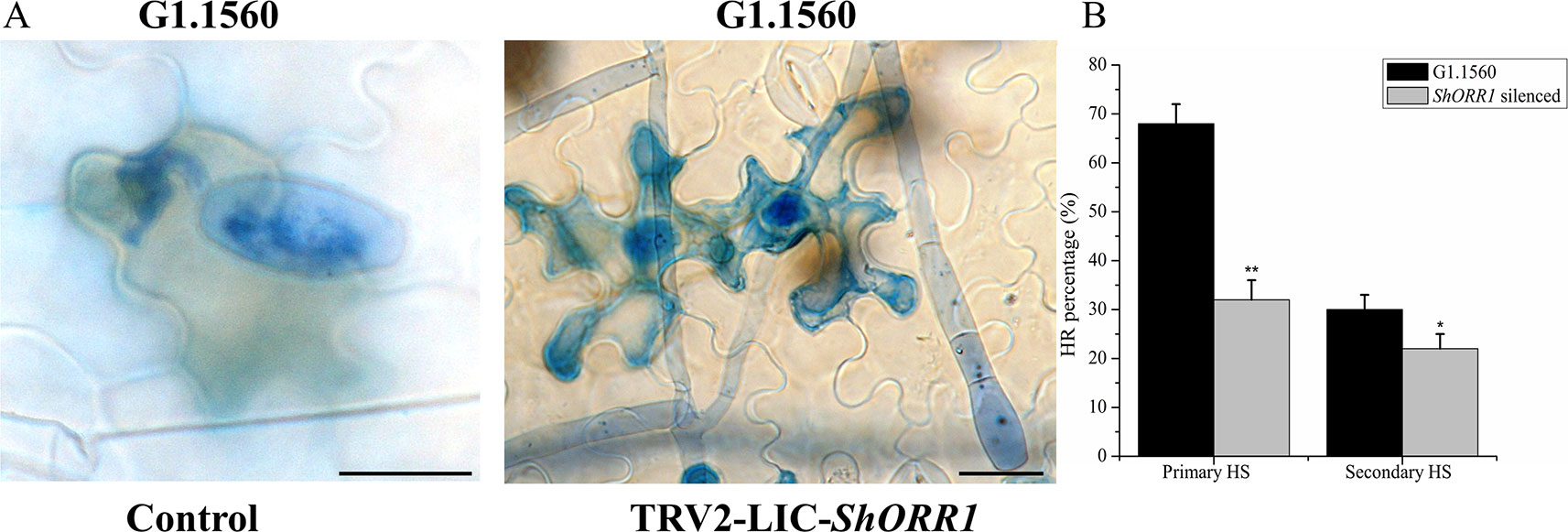
Figure 5 ShORR-1-silenced plants show decreased On-induced HR formation and H2O2 accumulation. (A) Micrographs of resistant S. habrochaites G1.1560 without ShORR-1 silencing (left) and ShORR-1-silenced S. habrochaites G1.1560 (right). Bar = 15 µm. (B) Percentages of HR-associated primary and secondary haustoria (HS) in ShORR-1-silenced and G1.1560 control plants. Asterisks indicate significant difference from the control by one-way ANOVA followed by an independent-samples Dunnett’s post hoc test (**: P < 0.01, *: P < 0.05). Three biological replicates of microscopic samples of both ShORR-1 silenced and control plants were observed.
To further investigate the role of ShORR-1-G in resistance to On, stable RNAi and overexpressing MM transformants of ShORR-1-G and ShORR-1-M were generated. Samples collected at 65 hpi were microscopically examined, and percentages of germinated On conidiospores that developed into microcolonies on them were recorded. There were significantly fewer On microcolonies on leaves of transgenic plants overexpressing ShORR-1-G than on leaves of wild-type plants (Figures 6A, C). In addition, numerous clear mycelial colonies were macroscopically observed on leaves of control plants, while few were found on leaves of ShORR-1-G overexpressing plants (Figure 6B). The efficiency of gene overexpressing was confirmed by qRT-PCR analyses, which showed that ShORR-1-G transcript levels were 6.5-fold higher in the overexpressing lines than in control plants (Figure 6D). Moreover, fungal biomass was 4- to 5-fold lower on the ShORR-1-G overexpressing plants, according to genetically based estimates (Figure 6E).
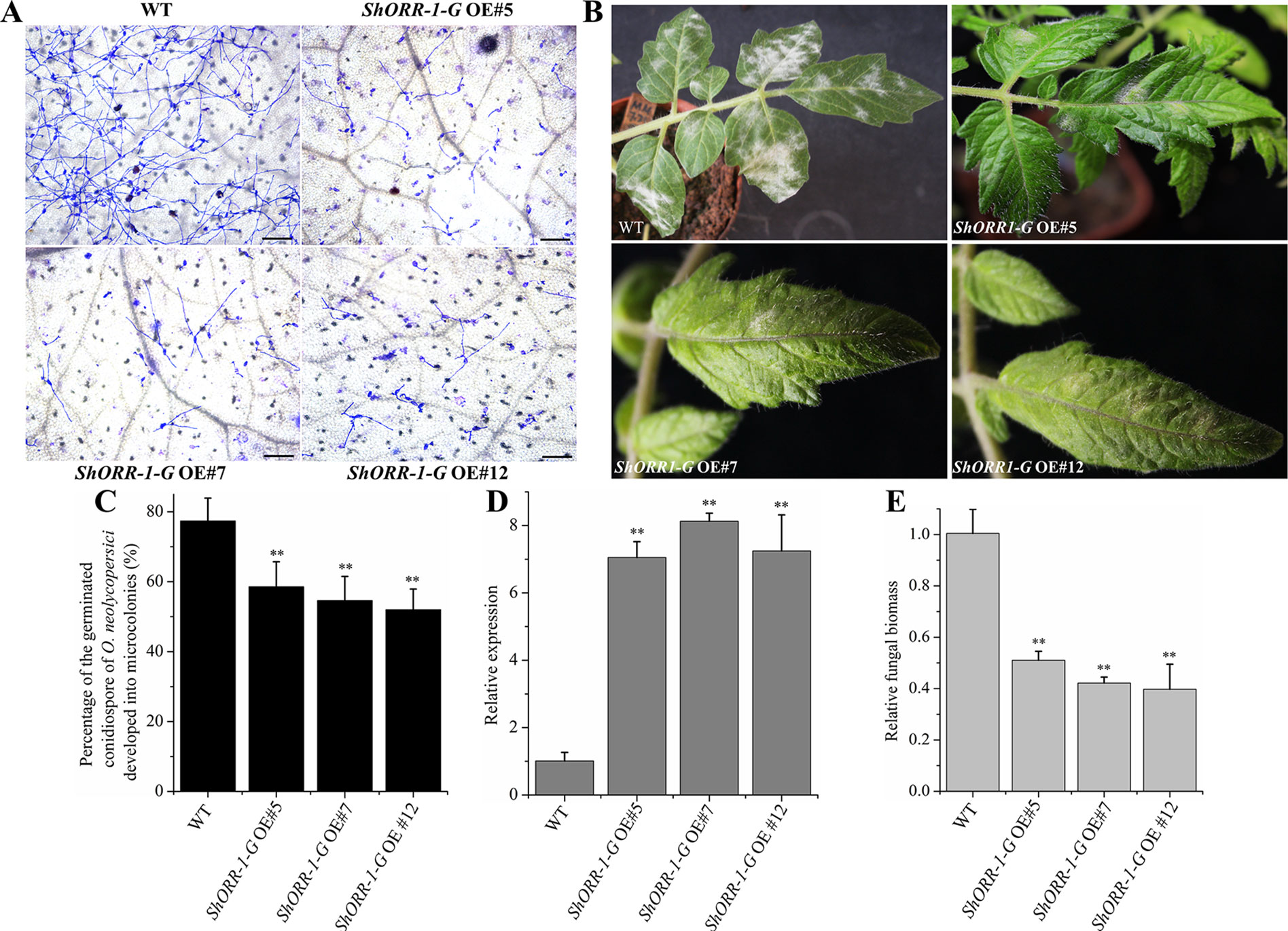
Figure 6 Overexpressing of ShORR-1-G enhanced tomato plants’ resistance to On. (A) Micrographs of powdery mildew on leaves of T2 ShORR-1-G overexpressing plants and untransformed MM plants, 65 h after inoculation. Scale bar = 25 µm. (B) Macroscopic phenotypes of On infected leaves of untransformed MM plants and T2 ShORR-1-G overexpressing plants. (C) Percentages of germinated On conidiospores on untransformed MM plants and T2 ShORR-1-G overexpressing plants at 65 h after infection. (D) Levels of ShORR-1 transcripts in three transformed plants and controls. Double asterisks indicate significant differences from the control by one-way ANOVA followed by an independent-samples Dunnett’s post hoc test (P < 0.01). (E) Estimated On fungal biomass on control plants and three transformed lines. All the above results are based on analyses of three biological replicates of control and transgenic plants.
However, microscopic observation indicated that overexpressing ShORR-1-M transgenic plants were more susceptible to On than control plants, while the plants with silenced ShORR-1-M had similar susceptibility to the controls (Figures 7A, C). Macroscopic observation confirmed these findings (Figure 7B). The gene silencing and overexpressing levels were confirmed by qRT-PCR analyses, which showed that ShORR-1-M transcript levels were dramatically lower in the gene-silenced plants and higher in the overexpressing transgenic plants than in the controls (Figure 7D). Quantification of fungal biomass confirmed that ShORR-1-M overexpressing increased the plants’ susceptibility, but silencing of ShORR-1-M did not increase resistance to On (Figure 7E). These results clearly indicate that ShORR-1-G is required for Ol-1 mediated resistance to On in tomato plants, while its homolog ShORR-1-M promotes susceptibility to On, presumably due to the differences in their sequences.
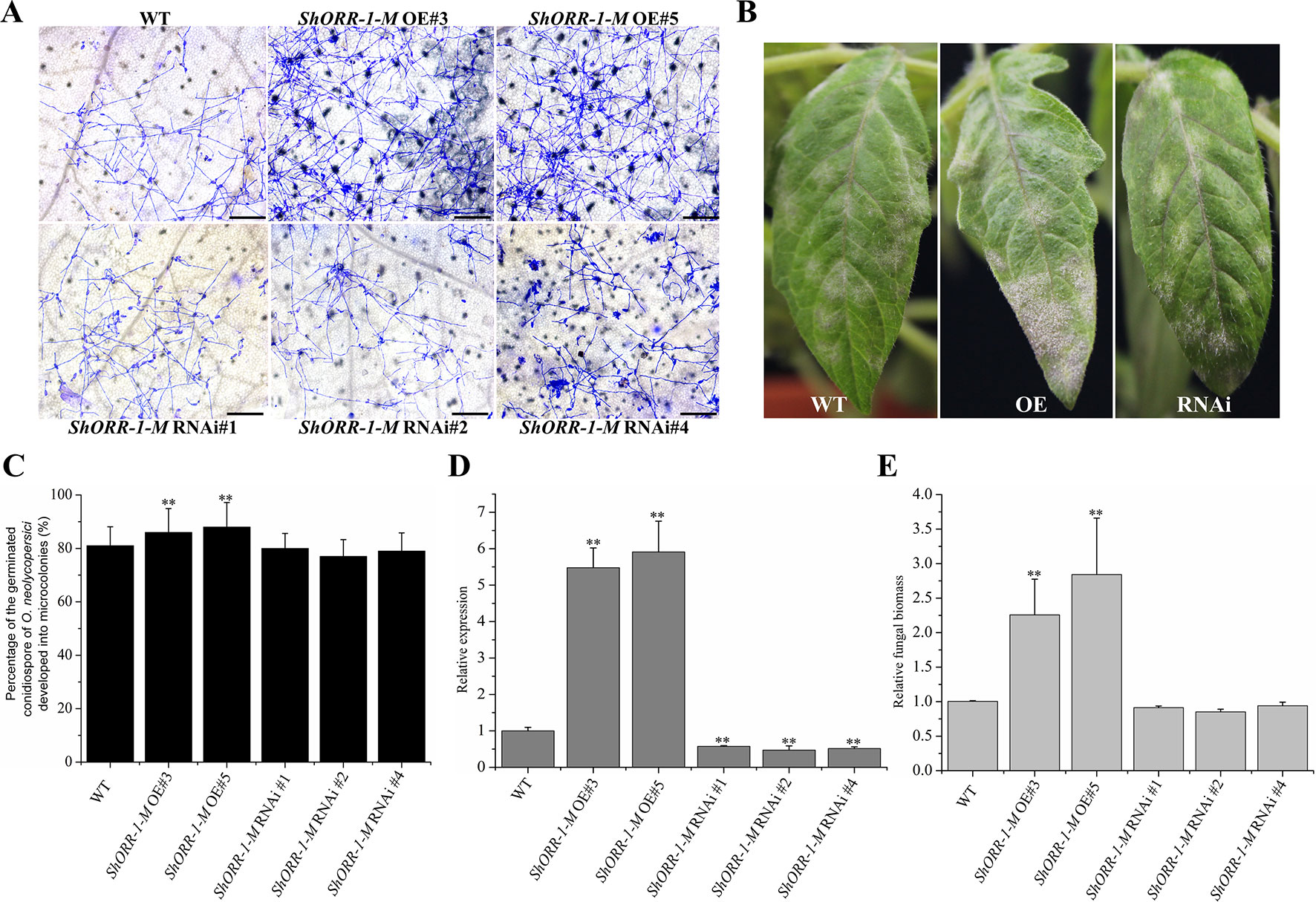
Figure 7 Overexpressing of ShORR-1-M increased susceptibility to On, but silencing it did not increase resistance to On. (A) Micrographs of powdery mildew on leaves of T2 ShORR-1-M overexpressing and silenced plants, compared with untransformed MM plants, 65 h after inoculation. Scale bar = 25 µm. (B) Macroscopic phenotypes of On infected leaves of T2 ShORR-1-M overexpressing and silenced plants. (C) Percentages of germinated On conidiospores on ShORR-1-M overexpressing and silenced plants, 65 h after infection. (D) Levels of ShORR-1 transcripts in transgenic plants and controls. Double asterisks indicate significant differences from the control by one-way ANOVA followed by an independent-samples Dunnett’s post hoc test (P < 0.01). (E) Estimated On fungal biomass on transgenic and control plants. All the above results are based on analyses of three biological replicates of control and transgenic plants.
In addition, DAB staining revealed that H2O2 levels were higher in epidermal cells of ShORR-1-G overexpressing plants than in those of wild-type plants at 65 hpi with On (Figure 8). Thus, ShORR-1-G positively regulates resistance to On in tomato, and the resistance is associated with H2O2 accumulation. Moreover, upregulation of ShORR-1-G resulted in more abnormal haustoria (Figure 9, red arrows) in attacked cells than in untransformed plants. These haustoria were irregular and oval, had high H2O2 contents, and some seemed to be plasmolyzed (Figure 9B).
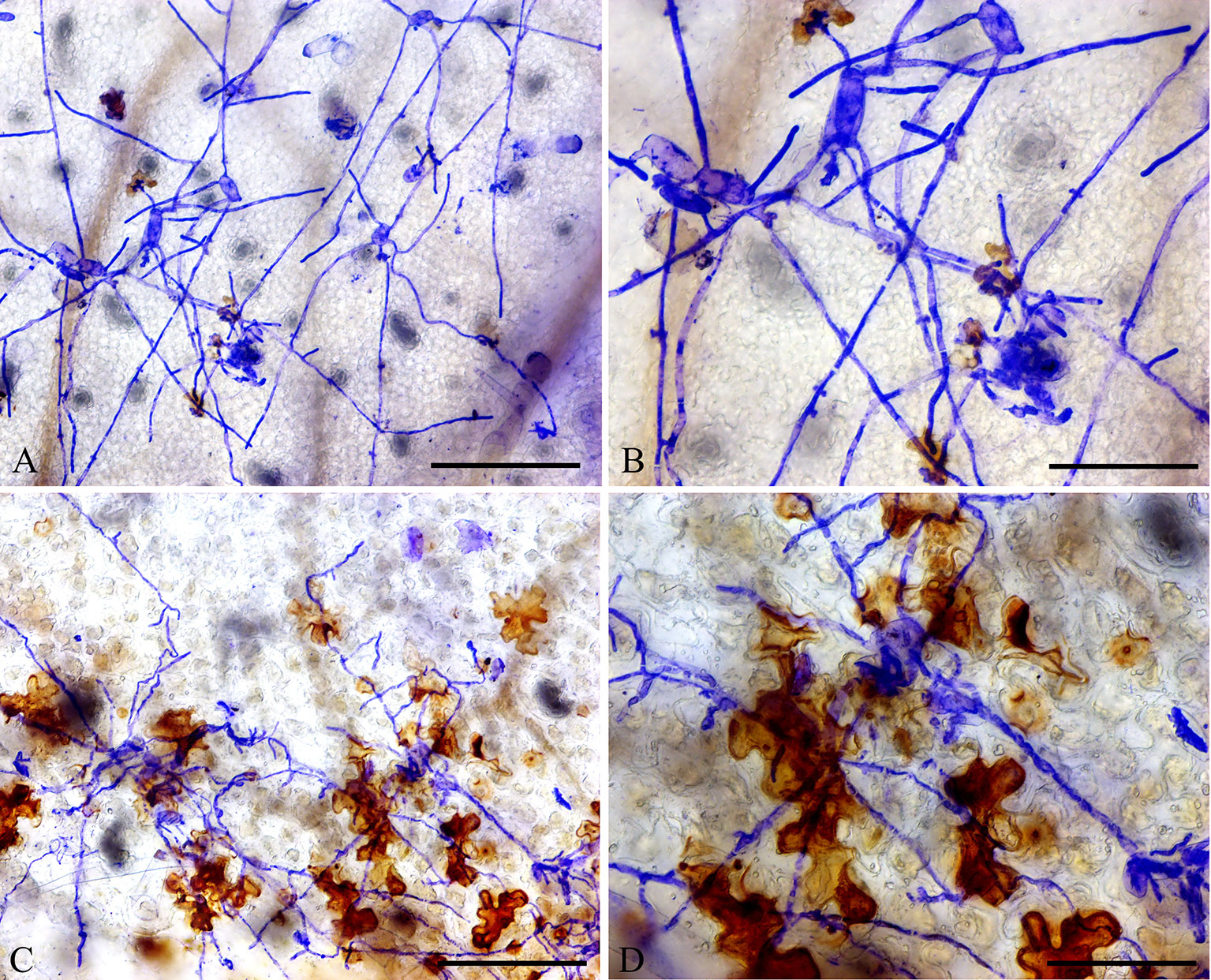
Figure 8 Overexpressing of ShORR-1-G increased H2O2 accumulation in transgenic tomatoes. (A) Wild-type tomato leaves were inoculated with On and sampled 65 hours after infection. (B) Enlarged view of A. (C)ShORR-1-G overexpressing tomato leaves challenged with On and sampled 65 h after infection. (D) Enlarged view of (C), showing H2O2 accumulation in epidermal cells, as manifested by 3,3-diaminobenzidine (DAB) staining. Scale bars = 25 µm in (A and C), 12.5 µm in (B and D).
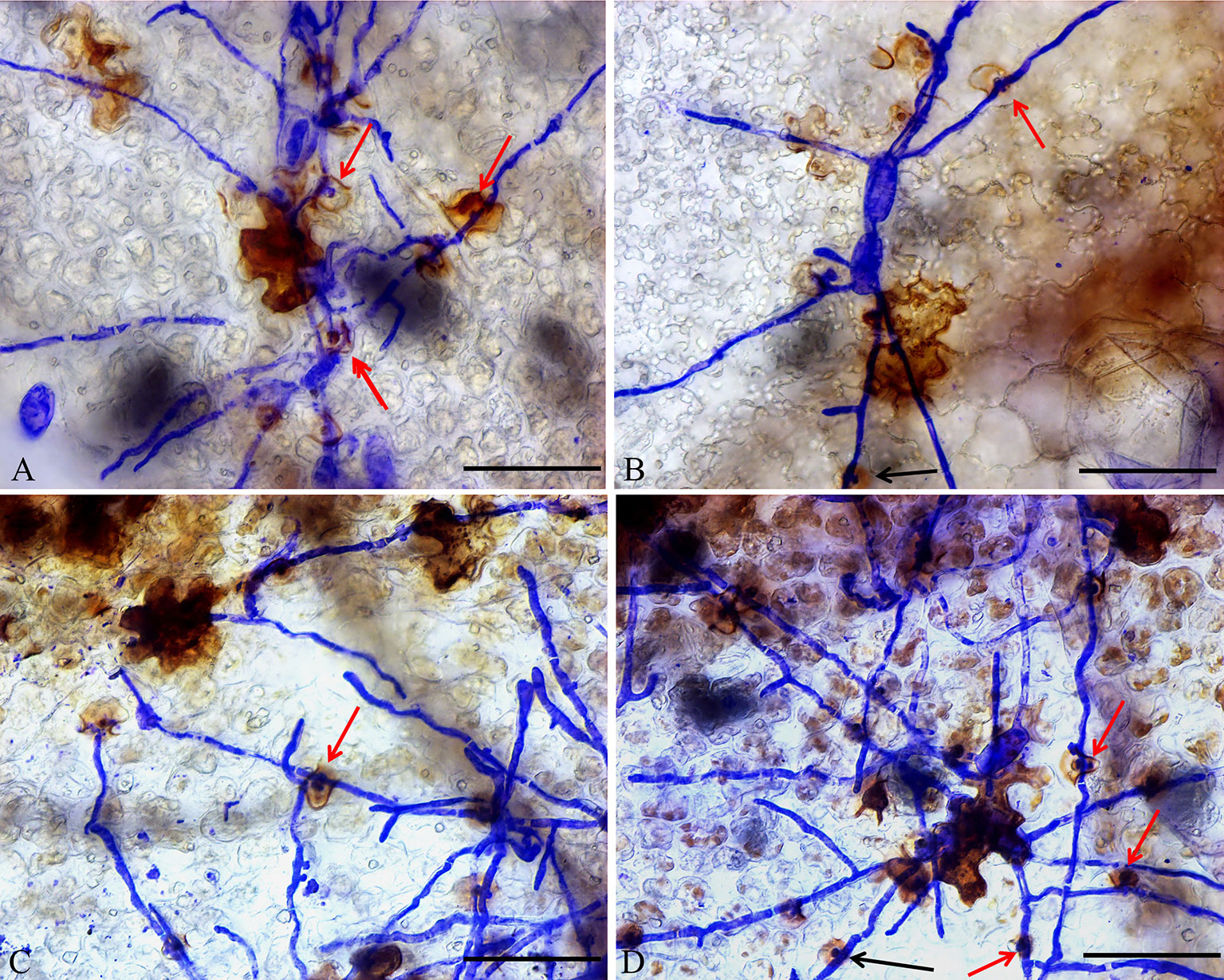
Figure 9 Micrographs of haustorium phenotypes in ShORR-1-G overexpressing plants. Red and black arrows show morphologically abnormal and normal haustoria with high H2O2 contents, respectively. Scale bar = 12.5 µm. Panels A–D show the different haustorium phenotypes in ShORR-1-G overexpressing lines 5, 7, 12 and 13.
Induced plant defenses are regulated by a highly interconnected signaling network in which the plant hormones jasmonic acid (JA) and salicylic acid (SA) play central roles (Pieterse et al., 2009, Pieterse et al., 2012). To investigate pathways that may be recruited in defenses against powdery mildew in ShORR-1-G overexpressing tomato plants, we used qRT-PCR to analyze the expression of six defense genes involved in JA (COI1) and SA (PR1, PR2) pathways, HR (HSR203J, BI1) and other important aspects of disease resistance (ROR2). Three ShORR-1-G overexpressing transgenic lines and MM were chosen to probe expression levels of these six marker genes in the presence and absence of On infection. The qRT-PCR results revealed that SA, JA, and HR might be involved in tomato resistance to On mediated by ShORR-1 (Figure 10).
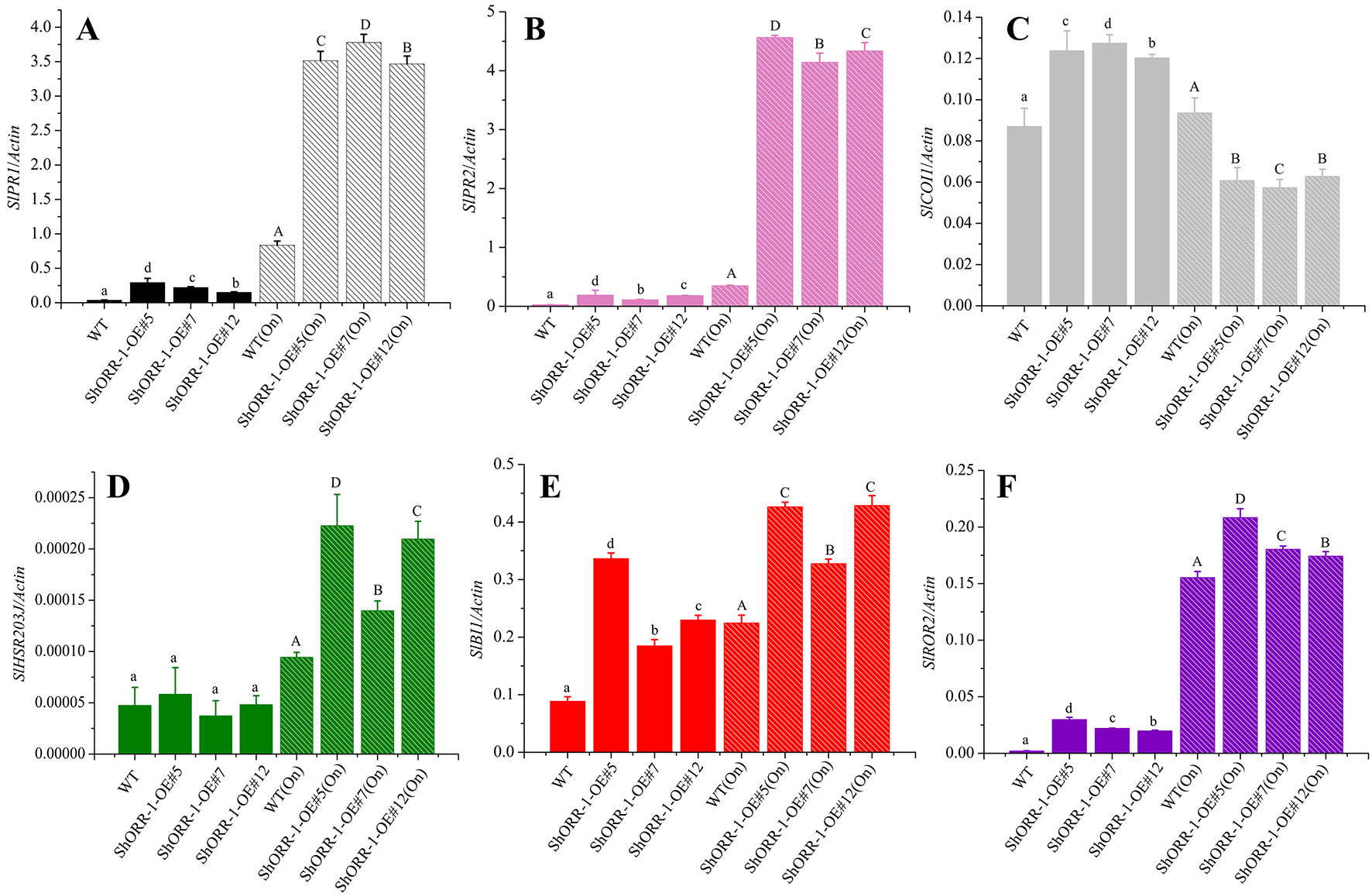
Figure 10 Quantitative real-time PCR analysis of expression levels of the following selected defense marker genes in wild-type and ShORR-1-G overexpressing (OE) transgenic tomatoes with and without On infection: (A)SlPR1, (B)SlPR2, (C)SlCOI1, (D)SlHSR203J, (E)SlROR2, (F)SlBI1. Actin was used as an internal control. Value for each sample is mean of three biological replicates. Mean and standard error were calculated using data from three independent biological replicates. Lowercase letters were used to indicate significant differences between lines in untreated (without On infection) conditions, and capital ones indicate significant differences between lines after On infection. Upper and lowercase letters indicate significant differences relative to control (WT) plants determined by one-way ANOVA followed by Tukey’s HSD test (P < 0.05).
PR1 and PR2 (encoding β-1,3-glucanase; also called BGL2) have been widely used as molecular marker genes for the SA hormone pathway in plants (Glazebrook, 2005; López-Cruz et al., 2017; Yang et al., 2018; Miao et al., 2019; Wang et al., 2019). Our results showed that expression levels of SlPR1 and SlPR2 were higher in On-infected wild-type plants and ShORR-1-G overexpressing plants than in wild-type plants without On infection. Furthermore, SlPR1 and SlPR2 expression levels were dramatically higher (350- and 400-fold, respectively) in On-infected transgenic plants. Thus, powdery mildew infection dramatically induced expression of SlPR1 and SlPR2, and overexpressing of ShORR-1 enhanced this induction, in accordance with previous findings that the powdery mildew Erysiphe orontii can elicit accumulation of PR1 and PR2 in Arabidopsis (Lynnereuber et al., 2010). However, On inoculation did not significantly affect transcript levels of SlCOI1, encoding coronatine insensitive 1, a key component of JA-mediated defense pathways (Li et al., 2004), in wild-type plants, while it substantially suppressed SlCOI1 expression in ShORR-1-G overexpressing plants. Moreover, we did not detect significant difference in expression levels of SlHSR203J and SlBI1—two important HR marker genes that are highly and rapidly induced in plant defense responses to various bacterial and fungal pathogens (Sanchez et al., 2000, Pontier et al., 2001)—between wild-type and ShORR-1-G overexpressing transgenic plants. However, SlBI1 and (especially) SlHSR203J were induced more strongly by On in ShORR-1-G transgenic plants than in wild-type plants, and their expression levels correlated with the extent of HR. Required for mlo-specified resistance (ROR2), an essential component of mlo-mediated basal penetration resistance to barley powdery mildew, has a specialized resistance function that is conserved between monocotyledons and dicotyledons (Collins et al., 2003). We found that SlROR2 was significantly induced in the ShORR-1-G overexpressing plants, with and without On inoculation, suggesting ShORR-1-G overexpression triggered this basal resistance.
Several DE-TDFs between powdery mildew-resistant NILs and the susceptible cultivar MM have been previously identified, including M14E72-213 (Li et al., 2006, Li et al., 2007). In the study presented here we found that M14E72-213 has high homology to a previously uncharacterized gene, which we named ShORR-1. By VIGS and overexpressing approaches, we demonstrated that a resistant variant of ShORR-1 is essential for Ol-1-based resistance (Figures 4 and 6). Moreover, its overexpressing is associated with H2O2 accumulation (Figure 8) and abnormal haustoria (Figure 9), and hence enhances the resistance.
Silencing of ShORR-1 in the resistant tomato cultivar S. habrochaites G1.1560 resulted in a susceptible phenotype, as shown by whole plant disease assays and microscopic analysis of On-infected plants (Figure 4). Downregulation of ALS and ShGST, two other DE-TDFs induced in NIL-Ol-1, both compromised tomato’s resistance to powdery mildew caused by On, indicating that multiple resistance genes are involved in Ol-1-mediated resistance to On (Pei et al., 2011a; Gao et al., 2014).
We also found that silencing ShORR-1-G attenuated defense responses, i.e. rapid HR and H2O2 accumulation, which are normally induced strongly by infection with On in G1.1560 plants (Figure 5). The weakened responses (lacked by susceptible plants) were not sufficient to prevent growth and development of On. Similarly, knock-down of ShGST in G1.1560 tomato plants reduced resistance to On, and resulted in slow rather than rapid HR (Pei et al., 2011a). This response differed from the slow HR of the NIL-Ol-1, developed by backcrossing S. habrochaites G1.1560 with MM, in which cells invaded by primary fungal haustoria remained alive, and only cells invaded by secondary haustoria died (Li et al., 2006).
Furthermore, overexpressing of ShORR-1-G increased the On resistance of susceptible MM plants (Figure 6), and resulted in higher levels of H2O2 in their epidermal cells upon On attack than in wild-type plants (Figure 8). HR, which is frequently observed around the infection site when microbial pathogens attack, arrests development of the pathogen at the epidermal cell attacked (Perfect and Green, 2001; Salguero-Linares and Coll, 2019). H2O2 production closely accompanies HR, and is believed to be an important diffusive signal in programmed cell death. Our results suggest that ShORR-1-G (or another resistant variant of ShORR-1) is vital for resistance to powdery mildew caused by On in tomato, since its overexpressing enhanced HR, H2O2 levels and resistance to On infection, whereas there was less H2O2 accumulation in the highly susceptible MM. Moreover, in ShORR-1-G overexpressing plants we examined, and NIL-Ol-1 plants examined by Li (2005), more abnormal haustoria were observed than in wild-type counterparts (Figure 9). Haustoria are generally fungal pathogens’ main structures for nutrient uptake (Roman-Reyna and Rathjen, 2017) and signal exchange (especially delivery of virulence effectors) with host plants (Hacquard et al., 2013). Our results suggest that the haustorial abnormalities in ShORR-1-G overexpressing plants at least partially prevented the fungus from absorbing nutrients and water from the host, thus reducing the extent of its growth and infection. However, in the NIL-Ol-1 plants the abnormal haustoria were reportedly filled with small vesicles, which were not observed in the plants we examined, although abnormal plasmolyzed haustoria were found in both NIL-Ol-1 (Li, 2005) and ShORR-1 overexpressing plants. These findings suggest that the Ol-1 and ShORR-1 resistance mechanisms may have both similarities and differences.
M14E72-213 DE-TDF was found in NIL-Ol-1, but not either NIL-Ol-4 or MM plants (Li et al., 2006, Li et al., 2007). However, ORFs of ShORR-1 were isolated from both On-resistant NIL-Ol-1 and On-susceptible MM plants (Figure 1), with mutations at 13 amino acid sequence sites. It indicated the above contrary was caused by the following, sequence analysis of ShORR-1-M and ShORR-1-G suggested that the mutation between two homologs at the 234th nucleotide base of ShORR-1-M was just at the annealing sequence region of selective primer E72 designed based on the recognition site of restriction enzyme (EcoR I) and selective nucleotide, sequence analysis also indicated that E72 can anneal at digested fragments of ShORR-1-G but not at those of ShORR-1-M (Li, 2005), which resulted in the DE-TDF of M14E72-213 was only identified in ShORR-1-G but not in ShORR-1-M in the cDNA-AFLP analysis using M14 (designed based on the recognition site of restriction enzyme (Mse I) and selective nucleotide) and E72 selective primer combination. Functional analysis showed that overexpressing of ShORR-1-M increased susceptibility to On, while ShORR-1-M-silenced plants phenotypically resembled untransformed controls (Figure 7). All the results indicate that ShORR-1-G plays a vital role in resistance to On, but not ShORR-1-M. Similarly, the R gene PigmR confers broad-spectrum resistance (inter alia to the blast fungus Magnaporthe oryzae in rice), while PigmS (which differs in four amino acids) competitively attenuates PigmR homodimerization to suppress resistance (Deng et al., 2017). Whether the mutation sites in resistant lines play important roles in ShORR-1-mediated On resistance needs to be verified by site-specific mutation in future studies. In addition, sequences of ShORR-1-G and ShORR-1-M proteins respectively contain 20 and 19 serine phosphorylation sites (Figure S1), including in both cases a cluster of four potential MAPK phosphorylation sites (Ser-57, Ser-64, Ser-68, Ser-76) in the N terminus (Figure S1). Thus, ShORR-1 could be phosphorylated by MPKs, but this also requires verification.
Plants are not passive victims when attacked by microbial pathogens. SA-dependent signaling plays significant roles in plant resistance to biotrophic pathogens, especially powdery mildew, while the JA signaling pathway is important in resistance to necrotrophic pathogens, but there is complex crosstalk between the JA and SA signaling pathways (Glazebrook, 2005). When a plant is attacked by a biotrophic pathogen, this crosstalk leads to activation of the SA defense pathway and inhibition of the JA signaling pathway (Fu and Dong, 2013). We observed the same patterns in tomato plants infected by the powdery mildew On, especially ShORR-1-G overexpressing plants, in which the SA-related defense genes SlPR1 and SlPR2 were strikingly activated, but the JA-related defense gene SlCOI1 was repressed. Various biotrophic pathogens have evolved intricate mechanisms that enable them to evade plant defenses by hijacking the JA pathway (Yan and Xie, 2016). Moreover, increasing evidence indicates that they inject effectors and toxins into plant cells that prevent the triggering of host defenses by targeting JA pathway components (Cole et al., 2014, Gimenez-Ibanez et al., 2014). Whether ShORR-1 is associated with the SA- and JA-dependent pathways requires validation in further studies, for example by overexpressing ShORR-1-G in tomato lines with perturbances in the pathways, and/or screening for proteins that interact with ShORR-1 to elucidate the pathway(s) that ShORR-1 influences.
When attacked by fungal pathogens, HR is one of the most effective resistance responses (Salguero-Linares and Coll, 2019). The HR marker gene SlHSR203J is reportedly activated in tomato plants by the leaf mold pathogen Cladosporium fulvum (Pontier et al., 1998), and (in both susceptible and resistant tomato taxa) following exposure to On (Li, 2005). Our results confirmed that On significantly induced expression of SlHSR203J, accompanied by increases in numbers of epidermal cells displaying hypersensitive responses (Figure 9).
Another important gene in powdery mildew resistance is Mildew Locus O (MLO). Recessive loss-of-function (mlo) mutations in the gene confer resistance to powdery mildew in barley (Miklis et al., 2007), tomato (Bai et al., 2008), pepper (Zheng et al., 2013), pea (Pavan et al., 2011), wheat (Várallyay et al., 2012), and Arabidopsis (Consonni et al., 2006). The resistance mechanism is based on early abortion of fungal pathogenesis, with formation of papillae at attempted penetration sites (Bai et al., 2005). ROR2 plays a major role in mlo-mediated disease resistance, and the barley ror2 mutant reportedly has less penetration resistance but stronger HR than wild-type plants when attacked by barley powdery mildew (Collins et al., 2003). We found that both On infection and ShORR-1-G overexpressing induced expression of SlROR2, accompanied by higher frequencies of cells showing HR, especially in ShORR-1-G overexpressing tomatoes challenged by On. These findings suggest that upregulation of SlROR2 might lead to formation of papillae, resulting in enhancement of resistance to On, but this hypothesis requires further validation.
In summary, ShORR-1 (originally detected in the form of a DE-TDF in our previous cDNA-AFLP analysis) plays an essential role in Ol-1-mediated resistance. ShORR-1-G and ShORR-1-M, respectively cloned from G1.1560 and MM, have antagonistic roles in responses to On, presumably due to differences (13) in their amino acid sequences. Future research will decipher roles of mutation sites and analysis of the relationship of ShORR-1 and Ol-1 might provide new insights into the mechanisms of Ol-1-mediated tomato resistance to powdery mildew.
The datasets generated for this study can be found in the Genbank.
CL conceived and designed the research. YZ, KX, DP, GC, HY, and WZ performed the experiments. DY, JZ, and XL provided fungal materials, reagents, and analytical tools. YZ and DP analyzed the data, prepared figures, and wrote the paper. All authors read and reviewed the final manuscript.
This research was supported by the National Natural Science Foundation of China (grant nos. 31902030, 31571997, 31872129, 31071807, and 31272168), Natural Science Foundation of Henan province (grant no. 182300410058), Foundation of Henan Science and Technology Committee (grant nos. 192102110001 and 192102110124), and Training Program of Youth Backbone Teacher of Henan Province (grant no. 2017GGJS146).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge Professor David Baulcombe and Professor Liu Yule for providing the VIGS vectors and Dr. Bai Yuling for the seeds of wild-type tomato. We also thank the Plant Genetic Transformation Center of the Henan Key Laboratory of Crop Molecular Breeding & Bioreactor for providing the stable T2 transgenic tomatoes.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01400/full#supplementary-material
Figure S1 | Potential serine phosphorylation sites in ShORR-1-G (A) and ShORR-1-M (B) were predicted by DISPHOS (Version 1.3). The blue circles indicate putative MAPK phosphorylation sites in ShORR-1.
Figure S2 | Potential lysine ubiquitination sites of ShORR-1-G (A) and ShORR-1-M (B) were predicted by UbPred and graphed by IBS (Version 1.0). The threshold of confidence was set to be more than 0.69, and U indicates ubiquitination sites.
Figure S3 | Potential lysine SUMOylation sites of ShORR-1-G (A) and ShORR-1-M (B) were predicted by GPS-SUMO 2.0 Online Service and graphed by IBS (Version 1.0). Medium stringency (Ac> 89.12%, Sn> 67.93%, Sp> 90%, MCC> 0.3446, Pr> 21.96%) was chosen as the threshold for GPS-SUMO prediction, and S indicates SUMOylation sites.
Bai, Y., Huang, C. C., van der Hulst, R., Meijer-Dekens, F., Bonnema, G., Lindhout, P. (2003). QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorumG1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol. Plant-Microbe Interact. 16, 169–176. doi: 10.1094/MPMI.2003.16.2.169
Bai, Y., Pavan, S., Zheng, Z., Zappel, N. F., Reinstadler, A., Lotti, C., et al. (2008). Naturally occurringbroad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol. Plant-Microbe Interact. 21, 30–39. doi: 10.1094/MPMI-21-1-0030
Bai, Y., van der Hulst, R., Bonnema, G., Marcel, T. C., Meijer-Dekens, F., Niks, R. E., et al. (2005). Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2 . Mol. Plant-Microbe Interact. 18, 354–362. doi: 10.1094/MPMI-18-0354
Bai, Y., d., H. R., Huang, C. C., Wei, L., Stam, P., Lindhout, P. (2004). Mapping Ol-4, a gene conferring resistance to Oidium neolycopersici and originating from Lycopersicon peruvianum LA2172, requires multi-allelic, single-locus markers. Theor. Appl. Genet. 109, 1215–1223. doi: 10.1007/s00122-004-1698-5
Cole, S. J., Yoon, A. J., Faull, K. F., Diener, A. C. (2014). Host perception of jasmonates promotesinfection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugatedjasmonates. Mol. Plant Pathol. 15, 589–600. doi: 10.1111/mpp.12117
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L., et al. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nat. 425, 973–977. doi: 10.1038/nature02076
Consonni, C., Humphry, M. E., Hartmann, H. A., Livaja, M., Durner, J., Westphal, L., et al. (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 38, 716–720. doi: 10.1038/ng1806
Consortium, T. G. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nat. 485, 635–641. doi: 10.1038/nature11119
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Sci. 355, 962–965. doi: 10.1126/science.aai8898
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1126/science.aai8898
Dong, Y., Burchsmith, T. M., Liu, Y., Mamillapalli, P., Dineshkumar, S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 145, 1161–1170. doi: 10.2307/40065758
Fu, Z. Q., Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gao, D., Huibers, R. P., Loonen, A. E., Visser, R. G., Wolters, A. M., Bai, Y. (2014). Down-regulation of acetolactate synthase compromises Ol-1-mediated resistance to powdery mildew in tomato. BMC Plant Biol. 14, 32. doi: 10.1186/1471-2229-14-32
Gimenez-Ibanez, S., Boter, M., Fernandez-Barbero, G., Chini, A., Rathjen, J. P., Solano, R. (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792. doi: 10.1371/journal.pbio.1001792
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Hacquard, S., Kracher, B., Maekawa, T., Vernaldi, S., Schulzelefert, P., Themaat, E. V. L. V. (2013). Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc. Natl. Acad. Sci. USA 110, E2219–E2228. doi: 10.1073/pnas.1306807110
Huang, C. C., Groot, T., Meijer-Dekens, F., Niks, R. E., Lindhout, P. (1998). The resistance to powdery mildew (Oidium lycopersicum) in Lycopersicon species is mainly associated with hypersensitive response. Eur. J. Plant Pathol. 104, 399–407. doi: 10.1023/a:1008092701883
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nat. 444, 323–329. doi: 10.1038/nature05286
Li, C. (2005). “Transcriptional, microscopic and macroscopic investigations into monogenic and polygenic interactions of tomato and powdery mildew,” in Doctoral dissertation (Netherlands: Wageningen University).
Li, C., Bai, Y., Jacobsen, E., Visser, R., Lindhout, P., Bonnema, G. (2006). Tomato defense to the powdery mildew fungus: differences in expression of genes in susceptible, monogenic- and polygenic resistance responses are mainly in timing. Plant Mol. Biol. 62, 127–140. doi: 10.1007/s11103-006-9008-z
Li, C., Bonnema, G., Che, D., Dong, L., Lindhout, P., Visser, R., et al. (2007). Biochemical and molecular mechanisms involved in monogenic resistance responses to tomato powdery mildew. Mol. Plant-Microbe Interact. 20, 1161–1172. doi: 10.1094/mpmi-20-9-1161
Li, C., Pei, D., Wang, W., Ma, Y., Wang, L., Wang, F., et al. (2008). First report of powdery mildew caused by Oidium neolycopersici on tomato in China. Plant Dis. 92, 1370–1370. doi: 10.1094/PDIS-92-9-1370C
Li, L., Zhao, Y., Mccaig, B. C., Wingerd, B. A., Wang, J., Whalon, M. E., et al. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143. doi: 10.1105/tpc.017954
Lindhout, P., Beek, V. D. H., Pet, G. (1994). Wild Lycopersicon species as sources for resistance to powdery mildew (Oidium lycopersicum): mapping of the resistance gene Ol-1 on chromosome 6 of L hirsutum. Acta Hortic. 376, 387–394. doi: 10.17660/ActaHortic.1994.376.53
Liu, Y., Schiff, M., Marathe, R., Dinesh-Kumar, S. P. (2010). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001
López-Cruz, J., Óscar, C., Emma, F., Pilar, G., Carmen, G. (2017). Absence of Cud Carmen, G. (2dismutase BCSOD1 reduces Botrytis cinerea virulence in Arabidopsis and tomato plants, revealing interplay among reactive oxygen species, callose and signalling pathways. Mol. Plant Pathol. 18, 16–31. doi: 10.1111/mpp.12370
Lynnereuber, T., Plotnikova, J. M., Dewdney, J., Rogers, E. E., Wood, W., Ausubel, F. M. (2010). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. doi: 10.1046/j.1365-313x.1998.00319.x
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Miao, Y., Xu, L., He, X., Zhang, L., Shaban, M., Zhang, X., et al. (2019). Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. Plant J. 98, 329–345. doi: 10.1111/tpj.14222
Miklis, M., Consonni, C., Bhat, R. A., Lipka, V., Schulze-Lefert, P., Panstruga, R. (2007). Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144, 1132–1143. doi: 10.1104/pp.107.098897
Nobuaki, I., Reiko, Y., Miki, Y., Shinpei, K., Hirofumi, Y. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23, 1153–1170. doi: 10.1105/tpc.110.081794
Pavan, S., Schiavulli, A., Appiano, M., Marcotrigiano, A. R., Cillo, F., Visser, R. G. F., et al. (2011). Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 123, 1425–1431. doi: 10.1007/s00122-011-1677-6
Pei, D., Ma, H., Zhang, Y., Ma, Y., Wang, W., Geng, H., et al. (2011a). Virus-induced gene silencing of a putative glutathione S-transferase gene compromised Ol-1-mediated resistance against powdery mildew in tomato. Plant Mol. Biol. Rep. 29, 972–978. doi: 10.1007/s11105-011-0331-4
Pei, D. L., Ma, H. Z., Zhang, Y., Ma, Y. S., Wang, W. J., Geng, H. X., et al. (2011b). Silencing a putative cytosolic NADP-malic enzyme gene comprised tomato resistance to Oidium neolycopersici. Life Sci. J. 8, 652–657.
Peng, Y., Wersch, R. V., Zhang, Y. (2018). Convergent and divergent signaling in pamp-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 31, 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Pieterse, C. M., Leon-Reyes, A., Van der, E. S., Van Wees, S. C. (2009). Networking bysmall-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Pieterse, C. M., Van, d., D. D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Perfect, S. E., Green, J. R. (2001). Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2, 101–108. doi: 10.1046/j.1364-3703.2001.00055x
Pontier, D., Balagu, C., Bezombes-Marion, I., Tronchet, M., Deslandes, L., Roby, D. (2001). Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 26, 495–507. doi: 10.1046/j.1365-313x.2001.01049.x
Pontier, D., Tronchet, M., Rogowsky, P., Lam, E., Roby, D. (1998). Activation of hsr203, a plant gene expressed during incompatible plant-pathogen interactions, is correlated with programmed cell death. Mol. Plant-Microbe Interact. 11, 544–554. doi: 10.1094/MPMI.1998.11.6.544
Radivojac, P., Vacic, V., Haynes, C., Cocklin, R. R., Mohan, A., Heyen, J. W., et al. (2010). Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 78, 365–380. doi: 10.1002/prot.22555
Roman-Reyna, V., Rathjen, J. P. (2017). Apoplastic sugar extraction and quantification from wheat leaves infected with biotrophic fungi. Methods Mol Biol. 1659, 125–134. doi: 10.1007/978-1-4939-7249-4_11
Salguero-Linares, J., Coll, N. S. (2019). Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 70, 2087–2095. doi: 10.1093/jxb/erz030
Sanchez, P., De Torres Zabala, M., Grant, M. (2000). AtBI-1, a plant homologue of Bax Inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J. 21, 393–399. doi: 10.1046/j.1365-313x.2000.00690.x
Shen, Q., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Sci. 315, 1098–1103. doi: 10.1126/science.1136372
Thordal-Christensen, H., Zhang, Z., Wei, Y., Collinge, D. B. (2010). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Trond, L., Cathrine, L. (2009). Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 387, 238–242. doi: 10.1016/j.ab.2009.01.024
Várallyay, E., Giczey, G., Burgyán, J. (2012). Virus-induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch. Virol. 157, 1345–1350. doi: 10.1007/s00705-012-1286-y
Wang, L., Wen, R., Wang, J., Xiang, D., Wang, Q., Zang, Y., et al. (2019). Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol. 221, 919–934. doi: 10.1111/nph.15435
Xu, K., Huang, X., Wu, M., Wang, Y., Chang, Y., Liu, K., et al. (2014). A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS One 9, e83556. doi: 10.1371/journal.pone.0083556
Yan, C., Xie, D. (2016). Jasmonate in plant defence: sentinel or double agent? Plant Biotechnol. J. 13, 1233–1240. doi: 10.1111/pbi.12417
Yang, Y., Chen, T., Ling, X., Ma, Z. (2018). Gbvdr6, a Gene Encoding a Receptor-Like Protein of Cotton (Gossypium barbadense), Confers Resistance to Verticillium Wilt in Arabidopsis and Upland Cotton. Front. Plant Sci. 8, 2272. doi: 10.3389/fpls.2017.02272
Zhao, Q., Xie, Y., Zheng, Y., Jiang, S., Liu, W., Mu, W., et al. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42, W325–W330. doi: 10.1093/nar/gku383
Keywords: Oidium neolycopersici, Ol-1-mediated resistance, susceptible tomato, resistant tomato, H2O2 accumulation, abnormal haustoria
Citation: Zhang Y, Xu K, Pei D, Yu D, Zhang J, Li X, Chen G, Yang H, Zhou W and Li C (2019) ShORR-1, a Novel Tomato Gene, Confers Enhanced Host Resistance to Oidium neolycopersici. Front. Plant Sci. 10:1400. doi: 10.3389/fpls.2019.01400
Received: 14 June 2019; Accepted: 10 October 2019;
Published: 07 November 2019.
Edited by:
Richard Hickman, Utrecht University, NetherlandsReviewed by:
Rosa Rao, University of Naples Federico II, ItalyCopyright © 2019 Zhang, Xu, Pei, Yu, Zhang, Li, Chen, Yang, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengwei Li, bGljaGVuZ3dlaWhpc3RAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.