- State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China
Theanine, a unique and abundant non-proteinogenic amino acid in tea, confers to the tea infusion its umami taste and multiple health benefits. Its content in new tea shoots is dynamic in winter and spring. However, its seasonal accumulation pattern and the underlying regulation mechanism of tea plants remain largely unknown. In this study, we measured the theanine contents in the roots and leaf buds of 13 tea cultivars at four time points from winter to spring (Dec. 12, Mar. 1, Mar. 23, and Apr. 13). We found theanine accumulated significantly in the roots to as high as ∼6% dry weight. We found theanine content in the roots was constant or slightly decreased on Mar. 1 compared with Dec.12 but increased consistently on Mar. 23 and then decreased on Apr. 13 in all 13 cultivars. In the leaf buds, theanine content kept increasing from Mar. 1 to Mar. 23 and decreasing from Apr. 13 in most of the 13 cultivars, meaning it was probably both season- and developmental stage-dependent. The expression of theanine biosynthesis and amino acid transporter genes in the roots and buds at the four time points was then examined. The correlation analyses between the gene expression and theanine content suggested the expression of theanine-biosynthesis genes was generally and negatively correlated with theanine content; however, the expression of amino acid transporter genes including CsLHT was generally and positively correlated with theanine contents. Finally, we showed that CsLHT has theanine transport activity. Taken together, this study provided insight into the seasonal regulation of theanine biosynthesis and transport in tea plants during winter and spring.
Introduction
Tea plants are unique; an abundance of health-promoting and pleasant flavor-conferring components, including catechins, caffeine, aroma, and theanine, accumulate in a single tea leaf. This makes tea one of the most popular beverages in the world. Theanine, a non-proteinogenic amino acid, is the most abundant free amino acid in tea, and it confers the tea infusion its umami taste (Ashihara, 2015). Recently, it was shown that theanine derivatives are also critical components of the aroma of oolong tea and large-leaf yellow tea (Guo et al., 2018; Guo et al., 2019). Impressively, many health-promoting effects of theanine, such as its anti-anxiety, anti-tumor, neuron-protecting, and memory improving properties alongside its antagonistic effect on the negative action of caffeine among other things, have been studied intensively (Sharma et al., 2018). Therefore, theanine content in tea leaves is highly related to the quality and price of teas, especially green teas (Yamaguchi and Ninomiya, 2000).
The substrates for theanine biosynthesis are glutamate (Glu) and ethylamine (EA). The enzyme catalyzing theanine synthesis is theanine synthetase (TS) (Sasaoka et al., 1965). Recently Wei and his colleagues identified the TS gene (CsTSI) in the tea plant (Camellia sinensis L.) (Wei et al., 2018). However, Cheng et al. showed that the glutamine synthetases (GS) of tea plants and Arabidopsis can also catalyze theanine biosynthesis from Glu and EA (Cheng et al., 2017). In bacteria, theanine can be synthesized from glutamine (Gln) and EA by gamma-glutamyltranspeptidase (GGT) (Suzuki and Kumagai, 2002; Suzuki et al., 2002; Yao et al., 2006; Shuai et al., 2010).
EA is derived from the decarboxylation of alanine catalyzed by alanine decarboxylase (AIDA) (Takeo, 1978). Arginine decarboxylase (ADC) was suggested to act as AIDA in tea plants (Shi et al., 2011). Glu is synthesized from Gln and ɑ-ketoglutarate under the catalysis of glutamate synthase (GOGAT). Glu can also be synthesized from ammonium and ɑ-ketoglutarate by Glu dehydrogenase (GDH) in tea plants (Takeo, 1979). The biosynthesis pathway and the enzymes involved in the process are illustrated in Figure 1A. Genes coding for enzymes in theanine biosynthesis have been widely identified in tea plants along with the completion of the genome sequencing (Xia et al., 2017; Wei et al., 2018).
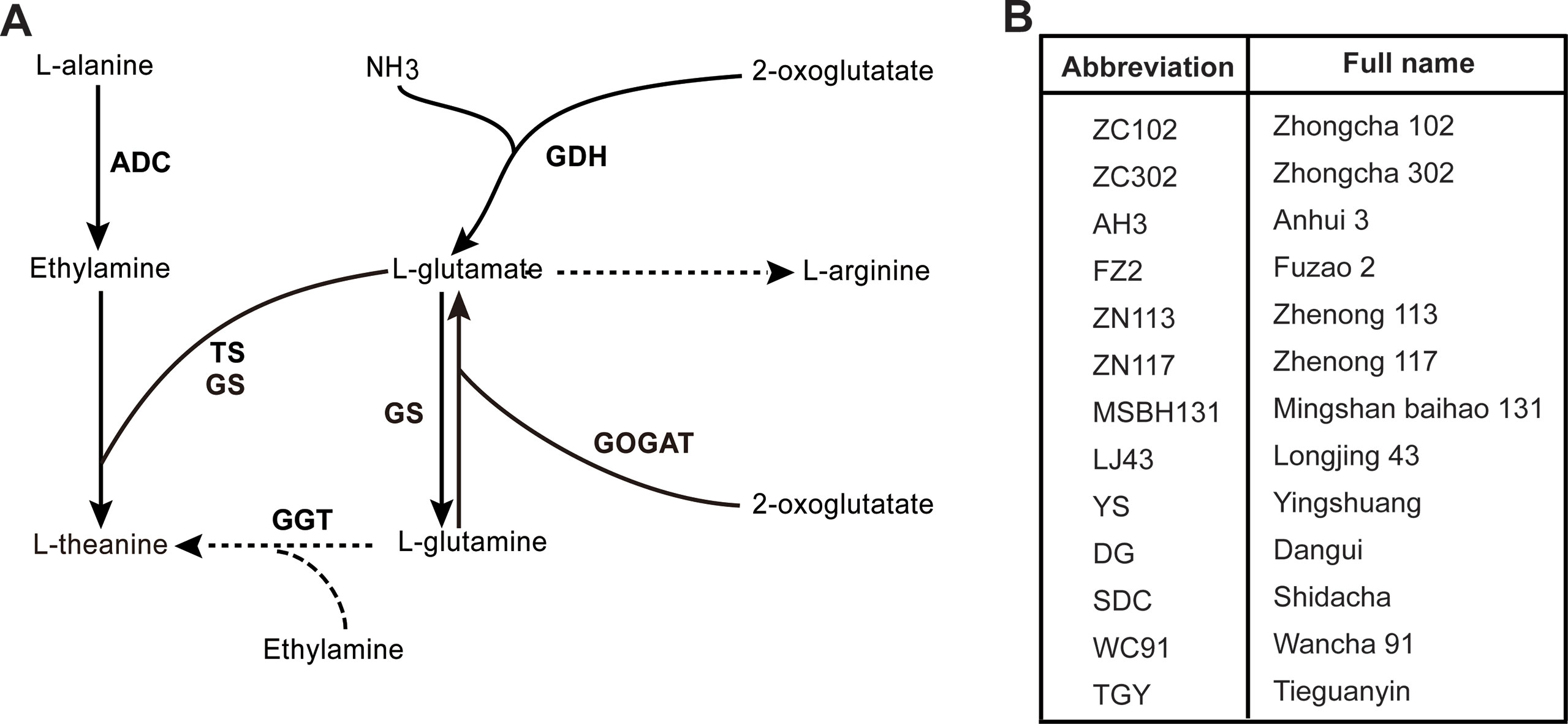
Figure 1 Putative theanine biosynthesis pathway in tea plant and cultivars used in this study. (A) Putative theanine biosynthesis pathway and the enzymes in tea plant. GDH, glutamate dehydrogenase; GOGAT, glutamate synthase; TS, theanine synthetase; GS, glutamine synthetase; GGT, glutamyltranspeptidase; ADC, alanine decarboxylase. (B) The 13 tea cultivars used in this study. All cultivars were 8-year-olds and were grown in the same tea plantation under the same management.
Theanine catabolism is catalyzed by theanine hydrolase in tea plants (Tsushida and Takeo, 1985). This process occurs mainly in leaves and is regulated by light (Kito et al., 1968). However, the gene encoding theanine hydrolase has not been identified yet.
In general, plants assimilate nitrogen (N) into Glu, Gln, Asp, and Asn as the main transport forms of N (Fisher et al., 1998). Tea plants are unique for the assimilation and storing of most N in the form of theanine in their roots (Oh et al., 2008). Theanine can account for 60–70% of free amino acid content in the roots and new shoots of tea plants (Takeo, 1981). Tea plants mainly transport Gln, Theanine, and Glu from root to shoot (Oh et al., 2008). Thus, theanine is also a transport in the form of N in tea plants. However, transporters that mediate theanine transport have not been identified in tea plants. As theanine has a similar structure to Gln, amino acid transporters transporting Gln likely have theanine transport activity.
In plants, there are two superfamilies of amino acid transporters: the amino acid/auxin permease (AAAP) and the Amino acid-Polyamine-Choline transporter (APC) family (Tegeder et al., 2007). The AAAP superfamily consists of amino acid permeases (AAPs), lysine, and histidine transporters (LHTs), proline transporters (ProTs), ɣ-aminobutyric acid transporters (GATs), auxin transporters (AUXs), and aromatic and neutral amino acid transporters (ANTs). Vesicular aminergic-associated transporters (VAATs) were recently identified and were shown to modulate amine neurotransmission in animals (Larhammar et al., 2014). The homologs of animal VAATs were identified in the potato genome (Ma et al., 2016). Amino acid transporter-like (ATLs) were also identified in potato and soybean (Cheng et al., 2016; Ma et al., 2016). Both VAATAs and ATLs were classified into the AAAP superfamily. The APC superfamily includes cationic amino acid transporters (CATs), amino acid/choline transporters (ACTs), and polyamine H+-symporters (PHSs) (Tegeder et al., 2007). Recently, it is reported that members of the Usually Multiple Acids Move In and out Transporters family (UMAMIT) can also transport amino acids (Muller et al., 2015; Besnard et al., 2016, 2018). Members of AAPs, LHTs, ANTs, CATs, and UMAMITs are reported to have varied affinities for Gln. The conserved homologs of these amino acid transporters are the candidate transporters mediating theanine transport from root to new shoot as theanine has a similar structure to Gln.
Previous studies have suggested that theanine is primarily synthesized in the root in the winter (Konishi and Kasai, 1968; Konishi and Takahashi, 1969). Ruan et al. (2012) also observed that theanine concentration in xylem sap increased in the spring. However, how the biosynthesis and transport are regulated during winter and spring is largely unknown. In this study, we investigated the dynamic changes of theanine content and expression of related genes were in the roots and buds of 13 cultivars at four time points of tea leaf bud development. The results demonstrated theanine accumulation patterns in the roots and buds during winter and spring and also provided insight into the regulation of theanine biosynthesis and transport.
Materials and Methods
Plant Samples
Leaf buds and unlignified lateral roots were collected from 13 tea plant (Camellia sinensis L.) cultivars grown in the Guohe Tea Plantation (Lujiang, Anhui, China) on December 12, 2017 (Dec. 12), March 1, 2018 (Mar. 1), March 23, 2018 (Mar. 23), and April 13, 2018 (Apr. 13). Leaf buds of the representative shoots within the top crown area were sampled. These representative shoots were at a similar developmental stage as the same cultivar. These cultivars included Zhongcha 102 (ZC102), Zhongcha 302 (ZC302), Anhui 3 (AH3), Fuzao 2 (FZ2), Zhenong 113 (ZN113), Zhenong 117 (ZN117), Mingshanbaihao 131 (MSBH131), Longjing 43 (LJ43), Yingshuang (YS), Dangui (DG), Shidacha (SDC), Wancha 91 (WC91), and Tieguanyin (TGY). These tea plant cultivars were all 8-year-old stem cutting clones and the growth status was similar. These tea plants were routinely managed: they were pruned for ∼50 cm and given 450 kg of inorganic compound fertilizer (N: P2O5: K2O, 15:15:15) and 150 kg urea per hectare in May, 2017; given 2,250 kg soybean cake fertilizer and 450 kg urea per hectare in October, 2017. The collected samples were immediately frozen in liquid nitrogen and stored at -80°C.
Determination of L-Theanine Content
Theanine was exacted and detected as previously described (Tai et al., 2015) with slight modification. The samples were grounded into a fine powder in liquid nitrogen before being freeze-dried. Then, 50 mg root or 200 mg bud was used to extract theanine, using 5 ml double distilled water, by boiling at 100°C for 20 min and cooling to room temperature. After centrifuging at 6,000 rpm for 10 min, the supernatant was filtered through a 0.22-µm membrane and analyzed by a Waters e2695 HPLC system equipped with 2489 UV/Vis detector. A reverse-phase C18 column (5 µm, 250 mm × 4.6 mm) was used and the column temperature was set at 28°C. The wavelength was set at 210 nm. The mobile phase A was water and phase B was 100% acetonitrile. The gradient elution was as follows: 0 min, 100% A, 0% B; 7 min, 100% A, 0% B; 9 min, 40% A, 60% B; 15 min, 100% A, 0% B; 20 min, 100% A, 0% B. The HPLC system was injected with 10 µl of the extraction for measurement. The amount of theanine was determined according to a calibration curve of authentic theanine standard, purchased from Sigma-Aldrich Chemical Company (St. Louis, MO, USA).
Total RNA Extraction and Real-Time Quantitative RT-PCR Analysis
Total RNA was extracted using a RNAprep Pure Plant Kit (Polysaccharides&Polyphenolics-rich) (TIANGEN, China) according to manufacturer instructions. First-strand cDNA was synthesized using the HiScript® II One Step RT-PCR Kit with gDNA Eraser (Vazyme, China). The 20-µl quantitative real time polymerase chain reaction (qRT-PCR) analysis contained 0.4 µl of forward and reverse primers (10 µM), 1 µl of eight-fold diluted first strand cDNA as template, and 10 µl of SYBR Green Supermix (Vazyme, China). Reactions were performed in a 96-well optical plate at 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s on a Bio-Rad CFX96. A melting curve was generated for each sample after each run to ensure the purity of the amplified products. No-template controls were included for each primer pair in each run. Relative transcript levels were calculated based on the 2-ΔCt method (Schmittgen et al., 2000), using CsGAPDH as an internal control. All primers used for qRT-PCR are listed in Table S4.
Identification and Selection of Theanine Biosynthesis and Transport-Related Genes for qRT-PCR Analysis
Theanine biosynthesis pathway genes, including CsGDHs, CsGOGAOTs, CsADCs, CsTSI, CsGGT, and CsGSs were identified and named by Wei et al. (2018). Theanine transport-related genes were identified and named by sequence homology with genes encoding amino acid transporters with glutamine transport activity in Arabidopsis. The nucleotide sequences of all these genes were downloaded from a tea plant genome database (Xia et al., 2019, http://teaplant.org/).
Statistical Analysis
The data were expressed as the mean ± standard deviation (SD) of three independent biological replicates. The correlation coefficient between theanine content and gene expression was analyzed via Pearson correlation.
Yeast Expression, Transformation, and Theanine Transport Measurement
The ORF of CsLHT was amplified by PCR from first-strand cDNA of RNA extracted from roots of tea plant (Camellia sinenesis, cv, Shuchazao) and cloned with HindIII/EcoRI in the yeast expression vector pYES2. The yeast strain 22Δ10α mutant (23344c background) was transformed with pYES2-CsLHT6 or the empty vector pYES2 using the yeast transformation kit (ZYMO RESEARCH, USA). Transformants were selected on yeast base media (DifcoTM Yeast Nitrogen Base without Amino Acids and Ammonium Sulfate, BD, USA) lacking uracil and supplemented with 2 mM ammonia sulfate.
For theanine transport activity determination, yeast cells were grown on a YNB medium to OD600 = 0.8. Yeast cells were collected and washed three times. Before theanine feeding, cells were cultured in YNB liquid medium with no nitrogen source for 1 h, at 30°C. This yeast solution was aliquoted into 4 ml tubes and added to 2 mM or 5 mM theanine for 5 min. The collected yeast cells were washed four times with deionized water. Then, 1 ml deionized water was used to resuspend cells and heated at 98°C for 1 h to extract theanine. After centrifugation at 13,000 rpm for 5 min, the supernatant was collected and filtered through a 0.22 µm membrane for a HPLC-based measurement of theanine.
Results
Theanine Accumulation in the Roots of 13 Tea Cultivars During Winter and Spring
To investigate the dynamic theanine accumulation in roots during winter and spring, we measured theanine contents in the roots of 13 tea cultivars on Dec. 12, Mar. 1, Mar. 23, and Apr. 13. The cultivars and the abbreviations of their names are listed in Figure 1B. These four time points are representative dates of winter time (Dec.12), leaf bud sprout (Mar. 1), leaf bud harvested for high-quality green tea production (Mar. 23), and leaf quality decreasing for green tea production (Apr. 13), respectively, in the major zone of green tea production in China.
Interestingly, we observed a consistently changing pattern of theanine content in the roots of these 13 cultivars (Figures 2A, B): slightly decreased on Mar. 1 compared with Dec. 12; obviously increased and reached the highest on Mar. 23 in all these cultivars; deceased on Apr. 13. These results suggested theanine accumulation in the roots of the 13 cultivars was dynamically and consistently regulated.
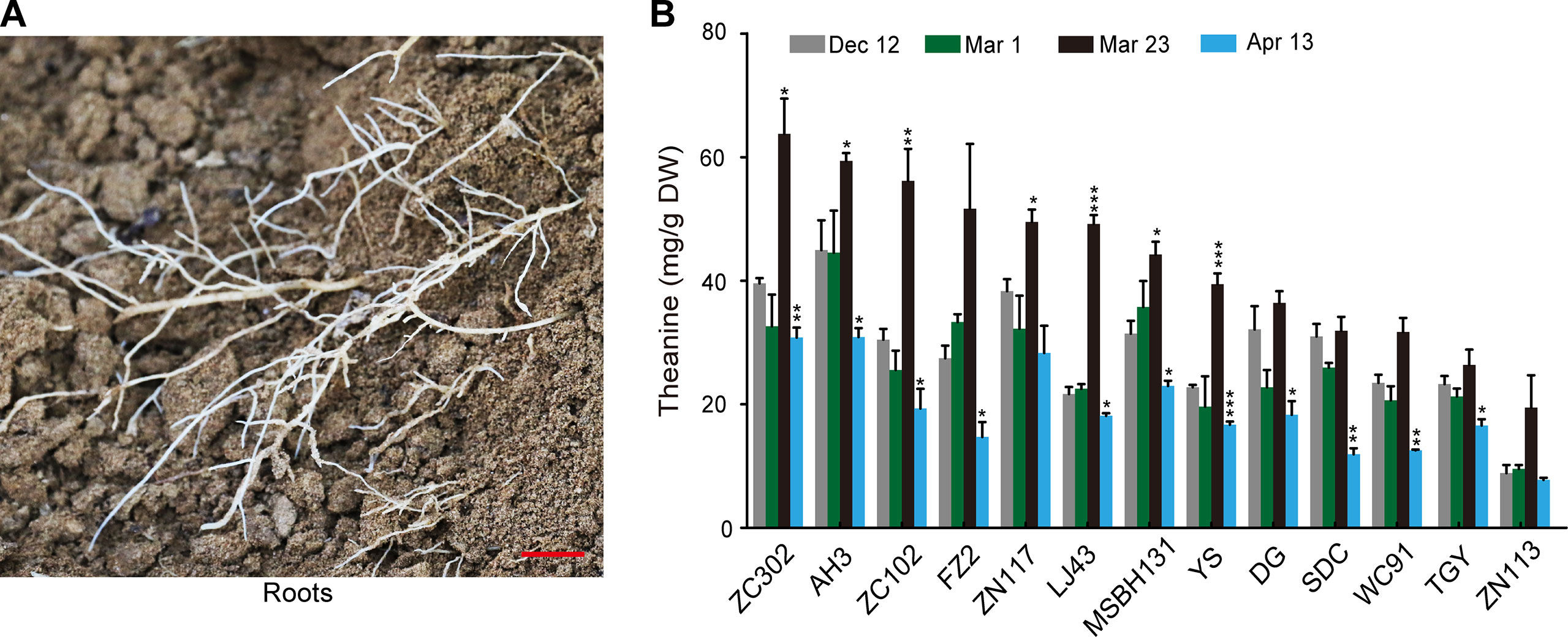
Figure 2 Theanine content in the roots of 13 cultivars at four time points. (A) Representative root sample collected. Scale bar: 1 cm (B) Theanine content in the roots. Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01, ***p 0.01).
Expression of Theanine Biosynthesis Pathway Genes in Roots During Winter and Spring
To explore the regulation of the dynamic changes of theanine accumulation in roots, we examined the expression of theanine biosynthesis pathway genes in five cultivars with high (AH3), medium (DG, TGY and Shuchazao [SCZ]), and low (ZN113) levels of theanine in the roots. The contents of theanine in the roots of SCZ at the four time points were reported by Dong et al. (2019). These theanine-biosynthesis genes included three CsGDHs, three CsGOGAOTs, three CsADCs, CsTSI, CsGGT, and four CsGSs, which were named by Wei et al. (2018).
The expression of genes encoding CsGDHs and CsGOGATs, which catalyze Glu synthesis, was examined first. We found CsGDHs were generally increased in spring (Mar. 1, Mar. 23, and Apr. 13) compared with Dec. 12, especially that of CsGDH2 and CsGDH3 in these cultivars (Figure 3A). Interestingly, CsGDH4 expression increased on Mar. 1 or Mar. 23 and then reduced on Apr. 13 in most of these cultivars. This changing pattern of CsGDH4 expression was similar to that of theanine content (Figure 2B). By contrast, CsGOGAT1 expression was highest on Mar. 1 and then reduced on Mar. 23 in four out of five cultivars; next, it increased on Apr. 13 in three out of five cultivars. Thus, the changing pattern of CsGOGAT1 expression was opposite to that of theanine content during these four time points. In addition, CsGOGAT2 and CsGOGAT3 expression was relatively stable during winter and spring. These results indicated that different homologs of CsGDHs and CsGOGATs respond differently to seasonal factors, suggested that there exist varied roles in glutamate and theanine synthesis during winter and spring in tea plants.
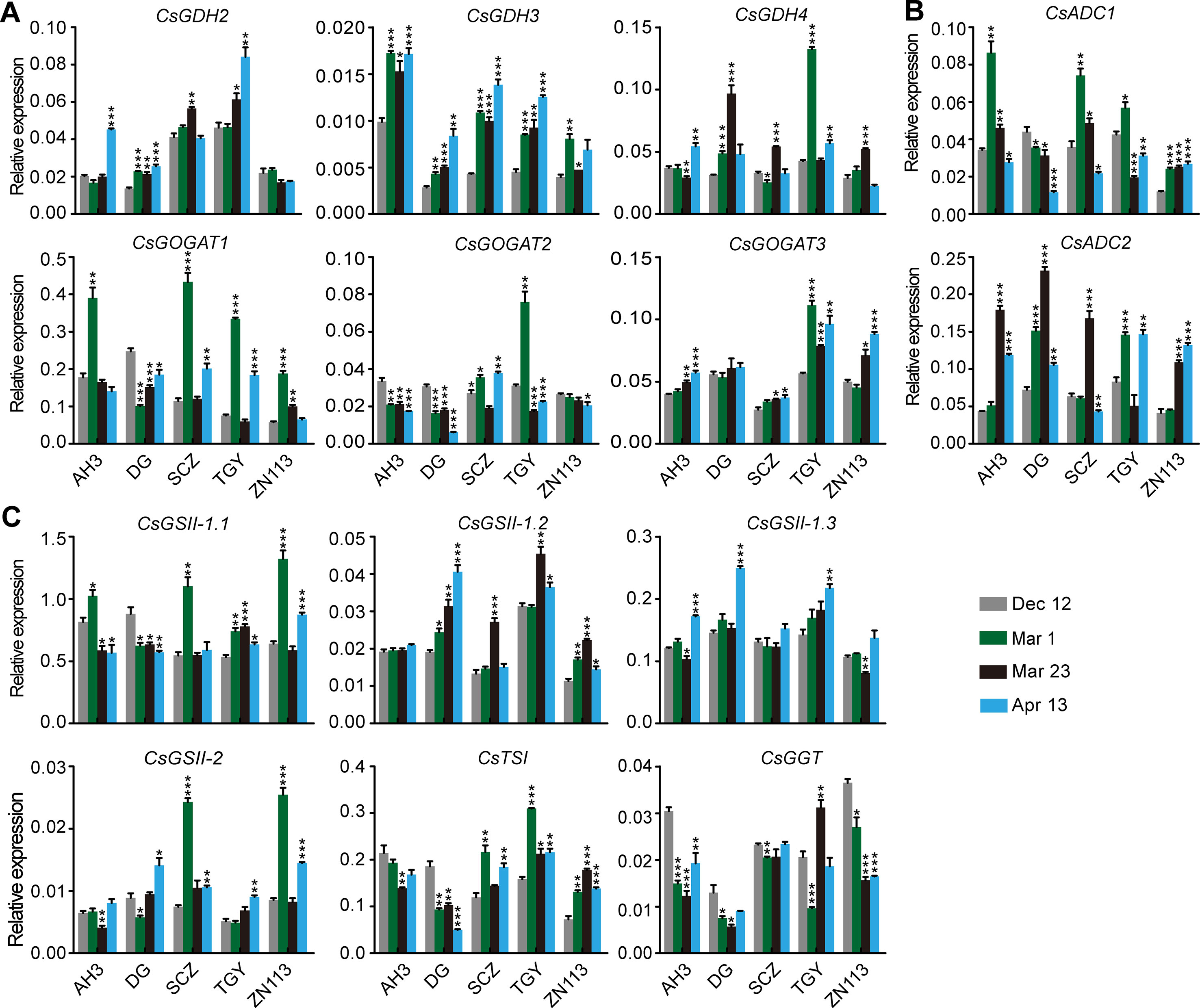
Figure 3 Relative expression of genes in theanine biosynthesis pathway in the roots of five representative cultivars at the four time points. (A) Relative expression of CsGHDs and CsGOGATs. (B) Relative expression of genes CsADCs. (C) Relative expression of CsGSs, CsTSI, and CsGGT. CsGAPDH was used as an internal control.Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01, ***p 0.01).
CsADCs were proposed to catalyze EA synthesis (Shi et al., 2011). CsADC1 and CsADC2 expression was then examined. Interestingly, CsADC1 expression was generally induced on Mar. 1 and was then gradually repressed on Mar. 23 and Apr. 13, whereas CsADC2 expression was continuously induced on Mar. 1 and Mar. 23 and then repressed on Apr. 13 in most of these cultivars (Figure 3B). Thus, the expression of CsADC2 generally followed the pattern of theanine content in the roots.
CsTSI and CsGSs are the characterized enzymes catalyzing theanine biosynthesis from Glu and EA (Cheng et al., 2017; Wei et al., 2018). CsGGT is a putative enzyme catalyzing Gln and EA into theanine. Compared with other theanine biosynthesis pathway genes, the expression of CsGSs, CsTS1, and CsGGT was relatively stable in the roots of these cultivars at these four time points (Figure 3C), suggesting these genes do not obviously respond to seasonal factors during winter and spring.
Correlation Between the Expression of Theanine Biosynthesis Pathway Genes and the Theanine Contents in Roots at Each Time Point
To get further insight into the regulation of theanine biosynthesis in the roots, we next analyzed the correlation between the gene expression and the theanine contents in the roots of AH3, DG, SCZ, TGY, and ZN113 at each time point. The correlation coefficient and p values are listed in Table S1. Here, we found the expression of these theanine biosynthesis pathway genes was generally and positively correlated with the theanine contents, with 10 out of 14 genes showing positive correlation on Dec. 12 (Figure 4A). In great contrast, the expression of most of these genes showed negative correlation with theanine contents on Mar. 1 and Mar. 23. Interestingly, on Apr. 13, most of these genes returned to a positive correlation with theanine contents. Given that the theanine level was relatively lower on Dec. 12, increased in Mar., and decreased on Apr. 13, the pattern of correlation between gene expression and theanine contents suggested theanine biosynthesis is generally feedback regulated in the roots.
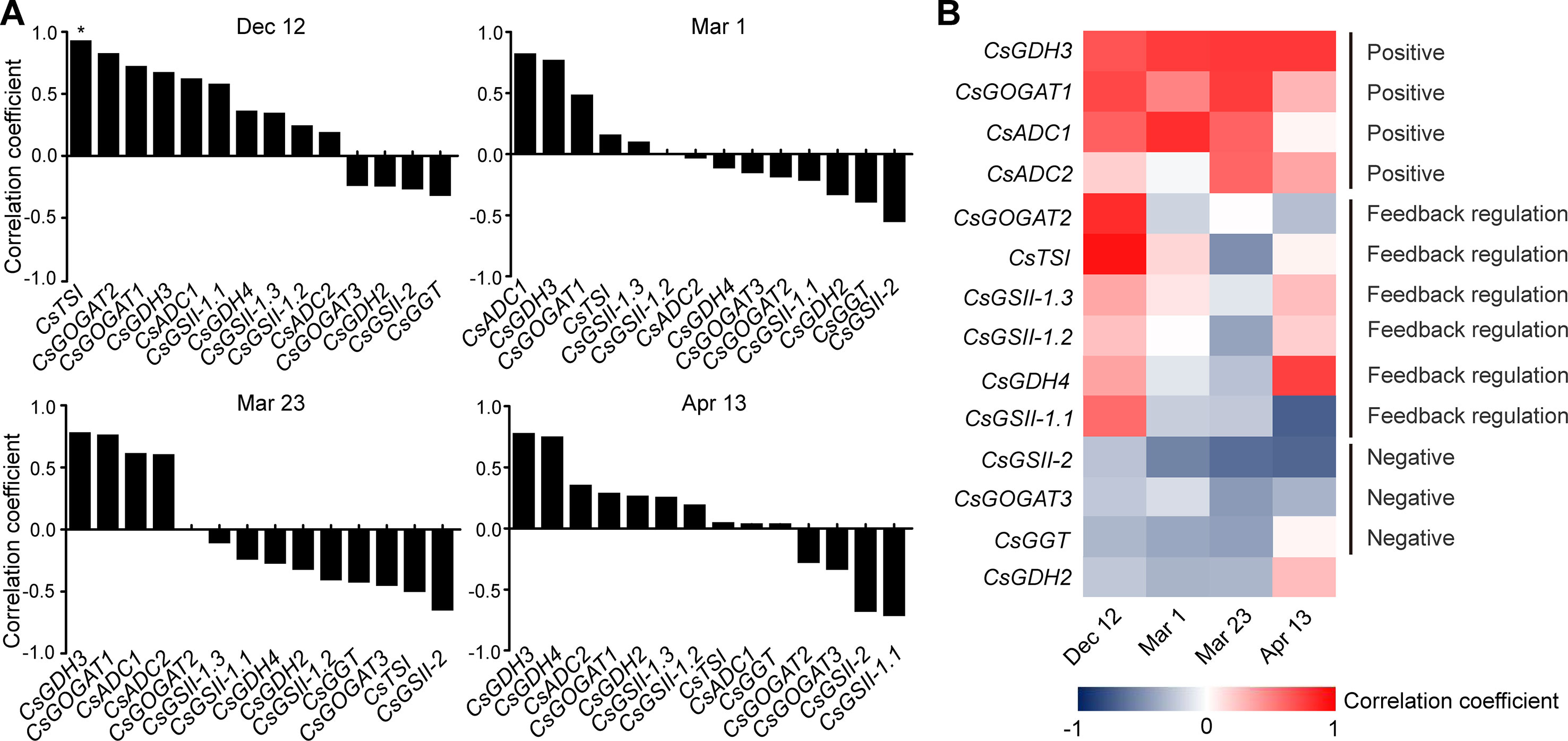
Figure 4 Correlation coefficient between the expression of theanine-biosynthesis pathway genes and the theanine content in the roots of five representative cultivars at each time point. (A) Correlation coefficients at each time point. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05). (B) Heat map of the Correlation coefficients.
Although the expression of the genes examined was generally feedback related with theanine contents in the roots, we found CsGDH3 and CsGOGAT1 always positively, and CsGSII-2 and CsGOGAT3 always negatively, correlated with theanine content in the roots at all four time points (Figure 4B). These results suggested the positive role of CsGDH3 and CsGOGAT1 and negative role of CsGSII-2 and CsGOGAT3 in theanine synthesis in the roots.
Further, we observed that the expression of CsGOGAT2 and CsTSI showed positive, reduced positive or negative, and higher negative correlation with the theanine contents in the roots on Dec. 12, Mar. 1, and Mar. 23, respectively. Subsequently, CsTSI showed a positive correlation with theanine contents on Apr. 13 (Figure 4B) when theanine contents decreased in the roots (Figure 2B). These results suggested CsGOGAT2 and CsTSI was involved in the feedback regulation of theanine biosynthesis.
Dynamic Accumulation Pattern of Theanine in the Buds of the 13 Cultivars During Winter and Spring
Leaf buds contain the highest concentration of theanine within the tissues in shoots (Feldheim et al., 1986) and are the main tissue harvested for high-quality green tea production. To explore how theanine accumulation in leaf buds changes during winter and spring, we next measured the theanine content in the buds of the 13 cultivars at the four time points (Dec. 12, Mar. 1, Mar. 23 and Apr. 13) (Figure 5A). We observed that theanine content in the buds was low (∼5 mg/g dry weight) on Dec. 12 in all these cultivars (Figure 5B). The content increased on Mar. 1, reached the highest levels on Mar. 23, and decreased on Apr. 13 in most of these cultivars. These results indicated theanine content in the buds generally started to increase in early Mar. and increase to the highest levels in late Mar. or early Apr. before decreasing. This is probably why the quality of green tea produced in Mar. and early Apr. is the highest, only for it to decrease from the middle of Apr.
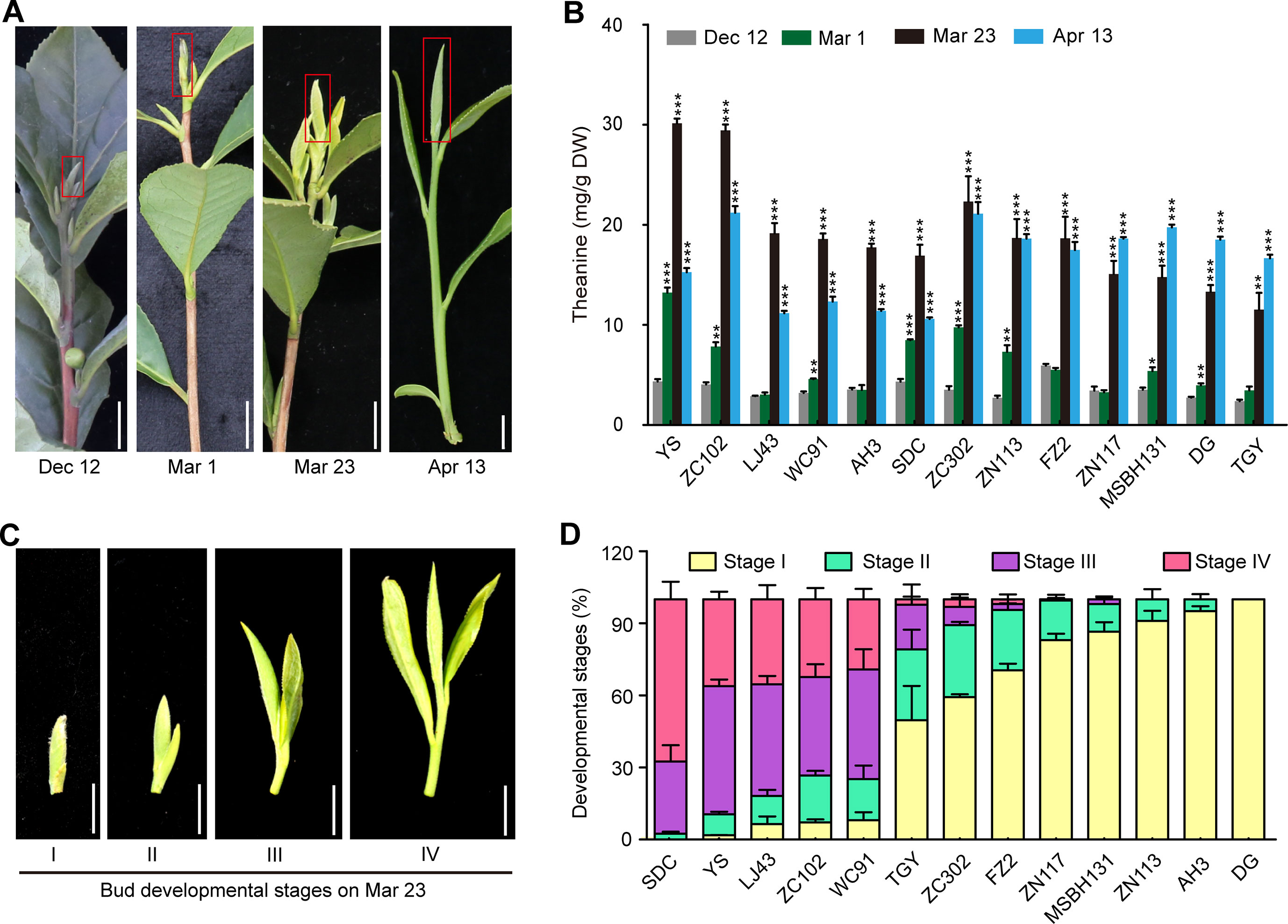
Figure 5 Theanine contents in the buds of 13 cultivars at four time points and the developmental stages of the buds on Mar. 23. (A) Representative bud samples collected at four time points. Red boxes highlighted the buds collected. (B) Theanine content in the buds at the four time points. Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01, ***p 0.01). (C) Buds of 13 cultivars on Mar. 23 were classified into four developmental stages, as shown. (D) Statistic results of the buds of the 13 cultivars at four developmental stages on Mar. 23. Data represent means ± SD of three biological replicates. Scale bars in (A) and (C), 1 cm.
However, we found that, in ZN117, MSBH131, DG, and TGY, theanine contents in the buds continued to increase on Apr. 13 (Figure 5B). In addition, in ZC302, ZN113, and FZ2, theanine content in buds just slightly decreased on Apr. 13 compared with that of Mar. 23. In contrast, in YS, ZN102, LJ43, WC91, AH3, and SDC, theanine content greatly decreased on Apr. 13. We hypothesized that this inconsistency was caused by the different developmental stages of the buds of these cultivars. To test this hypothesis, we evaluated the development stages of buds on Mar. 23 in these cultivars. We defined bud developmental stages I, II, III, and IV, as illustrated in Figure 5C. Interestingly, we found most of the buds of YS, ZN102, LJ43, and WC91 were in Stage III and IV, suggesting an early sprout time for the buds of these cultivars. However, most of the buds of ZN117, MSBH131, DG, TGY, ZC302, ZN113, and FZ2 were in Stage I and II (Figure 5D), suggesting a late sprout time for the buds of these cultivars. Therefore, the dynamic change of theanine content in these buds is highly associated with both seasons and bud developmental stages.
Expression of Theanine Biosynthesis Pathway Genes in Buds During Winter and Spring
Although theanine is mainly synthesized in the root, Deng and colleagues showed that tea plant shoots also have a weak theanine synthesis capacity (Deng et al., 2009). To explore the molecular mechanism of the dynamic changes of theanine in the buds, we next investigated the expression of theanine biosynthesis pathway genes in the buds of AH3, ZN113, TGY, DG, and SCZ. Theanine content in the buds of SCZ at the four time points was reported by Dong et al. (2019).
We observed that the expression of CsGDH4, CsGOGAT1, CsGOGAT3, CsTSI, and CsGGT was suppressed on Mar. 1 and Mar. 23 and was increased on Apr. 13 (Figures 6A–C). These results—indicated in the changed pattern of the expression of these genes—were generally opposite to those of the theanine content in the buds (Figure 5B) and implied that the expression of these genes was probably feedback regulated by theanine content in the buds.
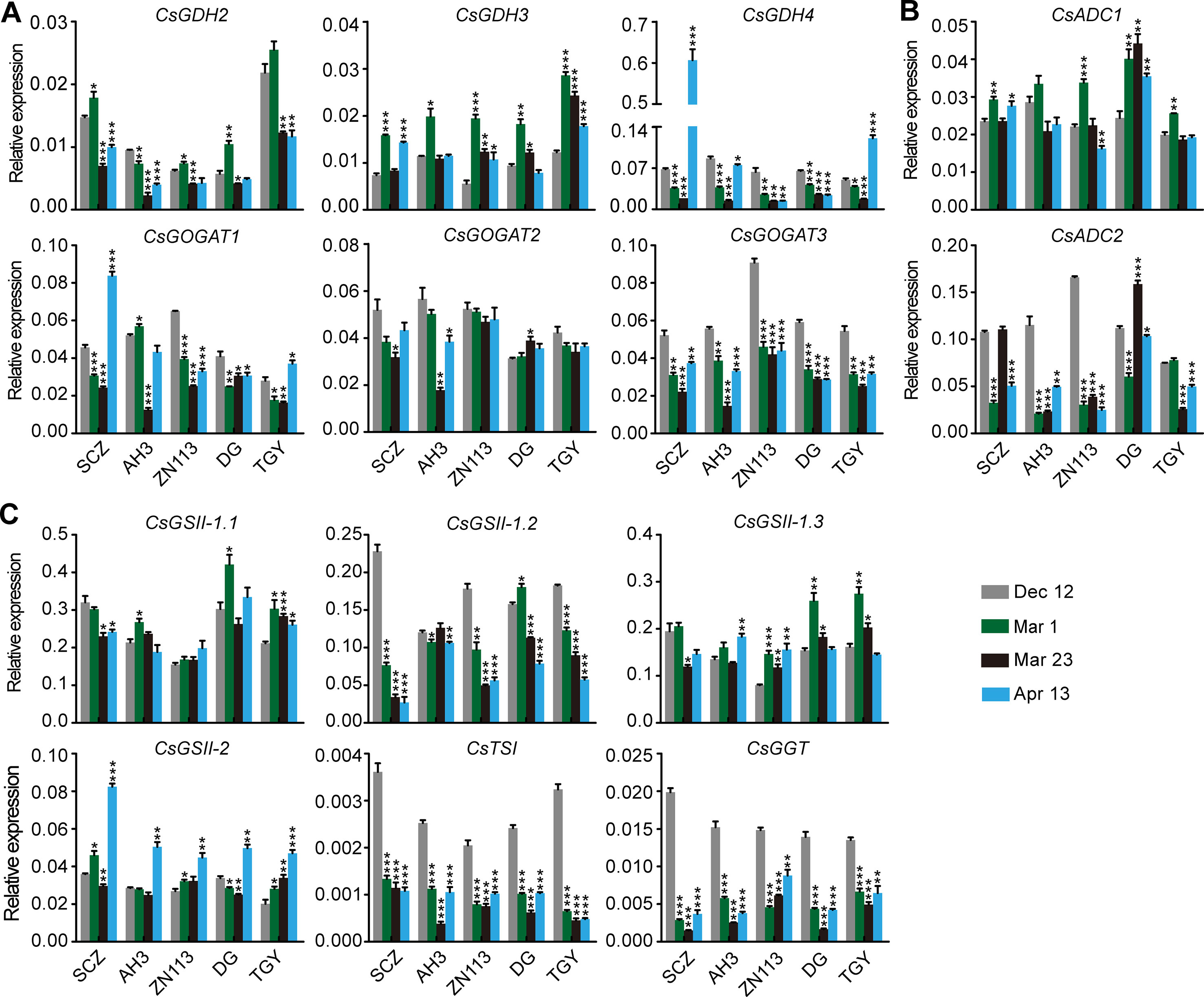
Figure 6 Relative expression of genes in theanine-biosynthesis pathway in the buds of five representative cultivars, at the fourtime points. (A) Relative expression of CsGHDs and CsGOGATs. (B) Relative expression of genes CsADCs. (C) Relative expression of CsGSs, CsTSI, and CsGGT. CsGAPDH was used as an internal control. Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01, ***p 0.01).
In contrast to the feedback-regulated genes, CsGSII-1.2 was gradually repressed in the buds from Dec. 12 to Apr. 13 (Figure 6C). CsGSII-2 expression, however, was stable on Dec. 12, Mar. 1, and Mar. 23 and was induced on Apr. 13 in all five cultivars. Therefore, CsGSII-1.2 and CsGSII-2 expression in the buds was likely regulated by seasonal factors, including temperature and light, but was not regulated by the theanine content in buds. In addition, CsGOGAT2 and CsGSII-1.1 expression in the buds was generally stable at all these four time points (Figures 6A, C). This result suggested CsGOGAT2 and CsGSII-1.1 was likely not regulated by theanine content in the bud.
Correlation Between the Expression of Theanine Biosynthesis Pathway Genes and Theanine Content in the Buds at Each Time Point
To investigate the regulation of theanine-biosynthesis gene expression in the buds, we also analyzed the correlation between gene expression and theanine content at each time point. The correlation coefficient and p values are listed in Table S2. Similarly to the situation in the roots, most of the genes had a positive correlation with theanine content in the buds (Figure 7A), when theanine content in the buds was low; next, 10 and 12 out of 14 genes had a negative correlation on Mar. 1 and Mar. 23, when theanine content in the buds increased, respectively. Furthermore, the number of genes that had a negative correlation reduced to six on Apr. 13 when theanine content decreased. These results suggested the expression of theanine biosynthesis pathway genes was also generally feedback regulated by the theanine content in buds.
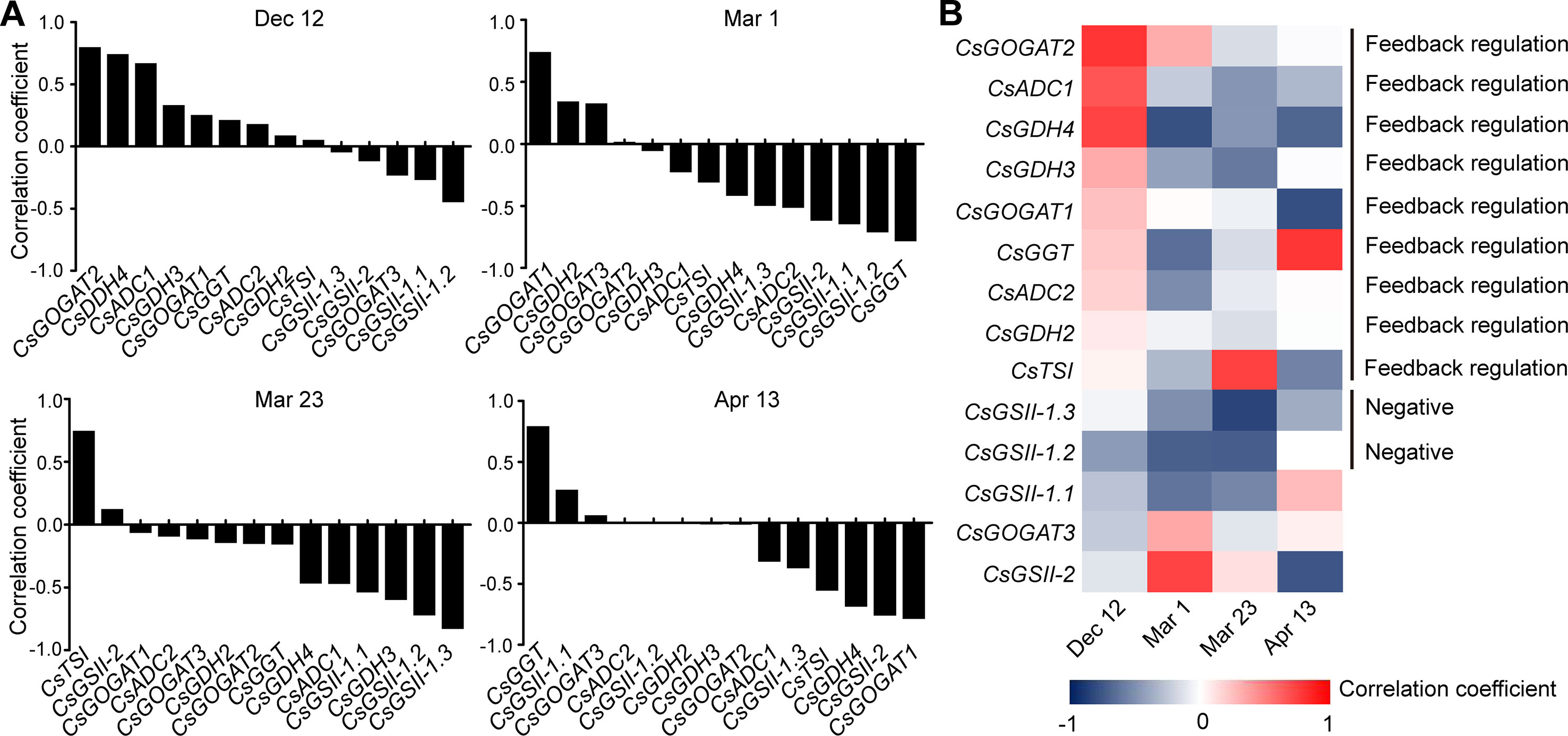
Figure 7 Correlation coefficient between the expression of theanine-biosynthesis pathway genes and the theanine content in the buds of five representative cultivars at each time point. (A) Correlation coefficients at each time point. Statistical analysis was performed and there was no statistical significance, but the correlation pattern is consistent and valuable. (B) Heat map of the correlation coefficients.
Next, we analyzed the correlation of individual genes. We observed that CsGDH2/3/4, CsGOGAT1/2, CsADC1/2, and CsGGT showed positive correlation when theanine content was low, negative correlation when theanine content increased, and positive correlation when theanine content decreased in the buds (Figure 7B). These results suggested that the expression of these eight genes was typically feedback regulated by theanine levels in the buds.
Interestingly, CsGSII-1.1/2/3 was almost always negatively correlated with theanine contents in the buds (Figure 7B), suggesting a negative role of these three genes in theanine biosynthesis in the buds. Finally, the fact that no gene showed continuously positive correlation with theanine contents in the buds supported the notion that buds have a minor role in theanine biosynthesis in tea plants.
Expression of Amino Acid Transporter Genes in the Roots During Winter and Spring
Amino acid long-distance transport is mediated by amino acid transporters (Tegeder, 2014). However, the molecular mechanism of theanine root-to-shoot transport is still largely unknown. To explore this mechanism, we next examined the expression of amino acid transporter genes in the roots of the five cultivars. These examined amino acid transporter genes included members of AAP, LHT, ProT, PUT, AUX, UMAMIT, VAAT, and ATL families.
We found that the expression of CsAAP9, CsVAAT, CsAUX, and CsATL was gradually induced in the roots of most of these cultivars through the four time points (Figure 8). By contrast, the expression of CsProT and CsPUT2 was gradually repressed. Although CsLHT was expressed at different levels in these cultivars, its expression levels were relatively stable through the four time points. Similarly, CsAAP2 expressed differentially but stably in these cultivars on Dec. 12, Mar. 1, and Mar. 23, but was great repressed on Apr. 13.
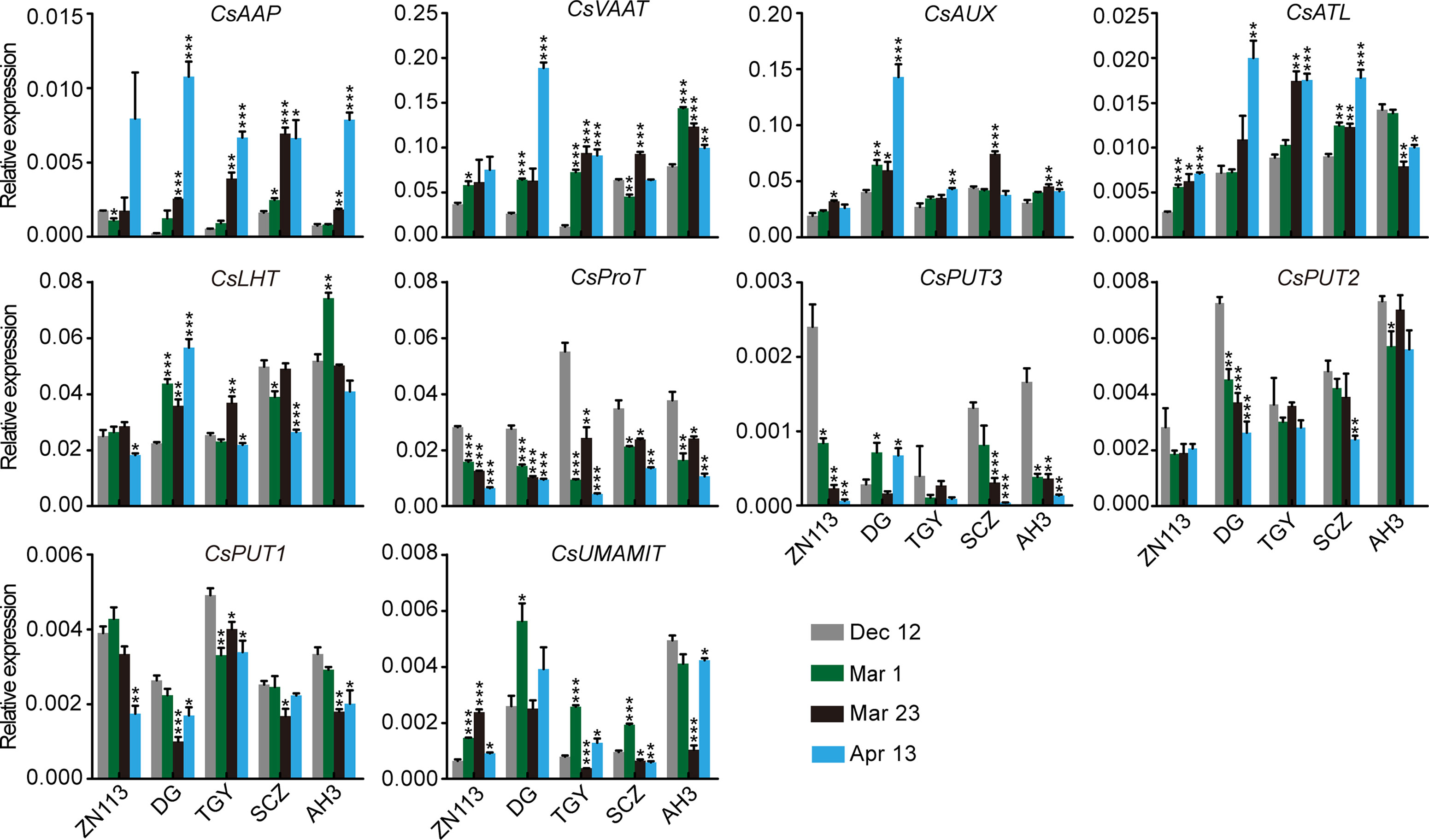
Figure 8 Relative expression of amino acid transporter genes in the roots of five representative cultivars, at the four time points. CsGAPDH was used as an internal control. Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01, ***p 0.01).
It is worth noting that theanine content in roots as slightly decreased on Mar. 1, increased on Mar. 23, and then decreased on Apr. 13 (Figure 2B). Thus, the change in the expression of these genes was generally different from that of theanine content in the roots. One exception was CsUMAMIT; this was generally induced when theanine content decreased and was repressed when theanine content increased in the roots (Figure 8), suggesting feedback regulation of its expression by theanine content in the root.
Correlation of Amino Acid Transporter Gene Expression With Theanine Content in the Roots During Winter and Spring
To further investigate the regulation of theanine transport, we next analyzed the correlation of CsAATs expression with theanine content at each time point in the roots of the five representative cultivars. The correlation coefficient and p values are listed in Table S3. Different from theanine biosynthesis pathway genes, most of the CsAATs showed positive correlation with theanine content in the roots at all these four time points (Figures 9A, B).
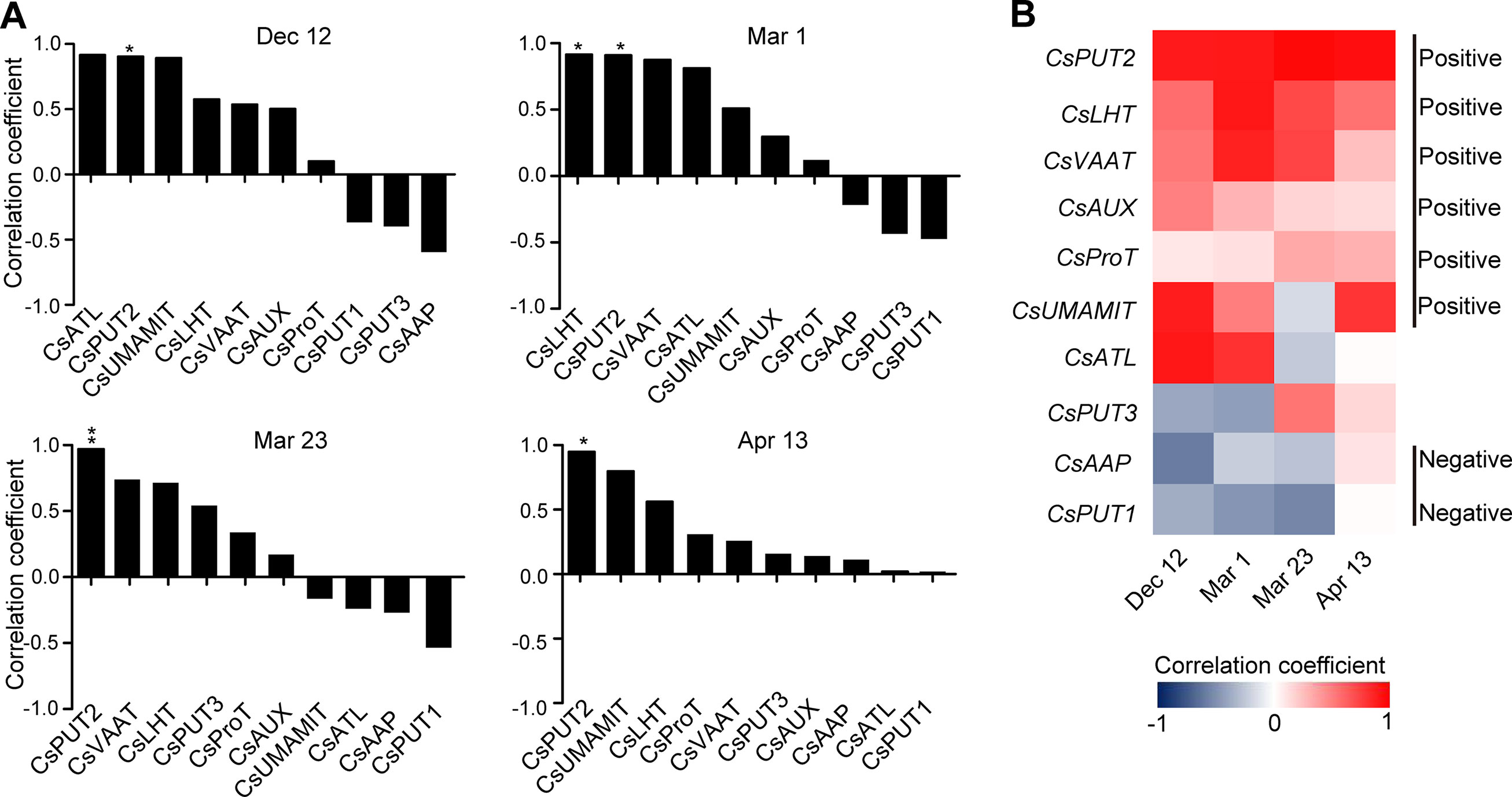
Figure 9 Correlation coefficient between the expression of amino acid transporter genes and the theanine contents in the roots of five representative cultivars at each time point. (A) Correlation coefficients at each time points. Asterisks represent statistical significance determined by Student’s t-test (*p 0.05, **p 0.01). (B) Heat map of the correlation coefficients.
When analyzed individually, we found CsPUT2 was always highly and positively correlated with theanine content in the roots at these four time points (Figures 9A, B), suggesting a continuous positive role of CsPUT2 in theanine transport. Although CsLHT and CsVAAT had positive correlation at all the four time points, they had higher positive correlation in Mar. and lower positive correlation on Apr. 13. This result suggested the positive role of CsLHT and CsVAAT in theanine transport was feedback promoted. Although most of these genes positively correlated with theanine contents in the roots, CsPUT1and CsAAP9 had negative correlation on Dec. 12, Mar. 1, and Mar. 23, suggesting CsPUT1 and CsAAP had a negative role in theanine transport.
CsLHT Can Transport Theanine
Theanine content in xylem sap increases in spring (Ruan et al., 2012), suggesting there is root-derived theanine transport to the shoot via the xylem. To explore the molecular mechanism of theanine root-to-shoot transport, it is necessary to identify what theanine transporter mediated this process. Given that CsPUT, CsLHT, and CsVAAT expression was positively correlated with theanine content in the roots (Figure 9B), CsPUT, CsLHT, and CsVAAT were candidates of theanine transporters. As LHT-family amino acid transporters have a high affinity for neutral and acidic amino acids and have been well studied in plants (Tegeder and Ward, 2012), we then examined whether CsLHT can transport theanine.
The yeast 22∆10∆α strain is an amino acid uptake mutant lacking an amino acid uptaking capacity due to the mutations of 10 amino acid transporter genes (Besnard et al., 2016). To test the theanine transport activity of CsLHT, we expressed CsLHT in 22∆10∆α with an empty vector pYES2 as a negative control and wild-type yeast strain 223344c as a positive control. Under the feeding of 2 or 5 mM theanine, we measured contents of theanine accumulated in these yeast strains. We found wild-type yeast accumulated the highest levels of theanine under both 2 and 5 mM theanine feeding (Figures 10A, B); CsLHT-expressing yeast cells only accumulated theanine under 5 mM theanine feeding. No theanine accumulation, however, was detected in the negative control strain. These results indicated that CsLHT can transport theanine under a higher theanine concentration.
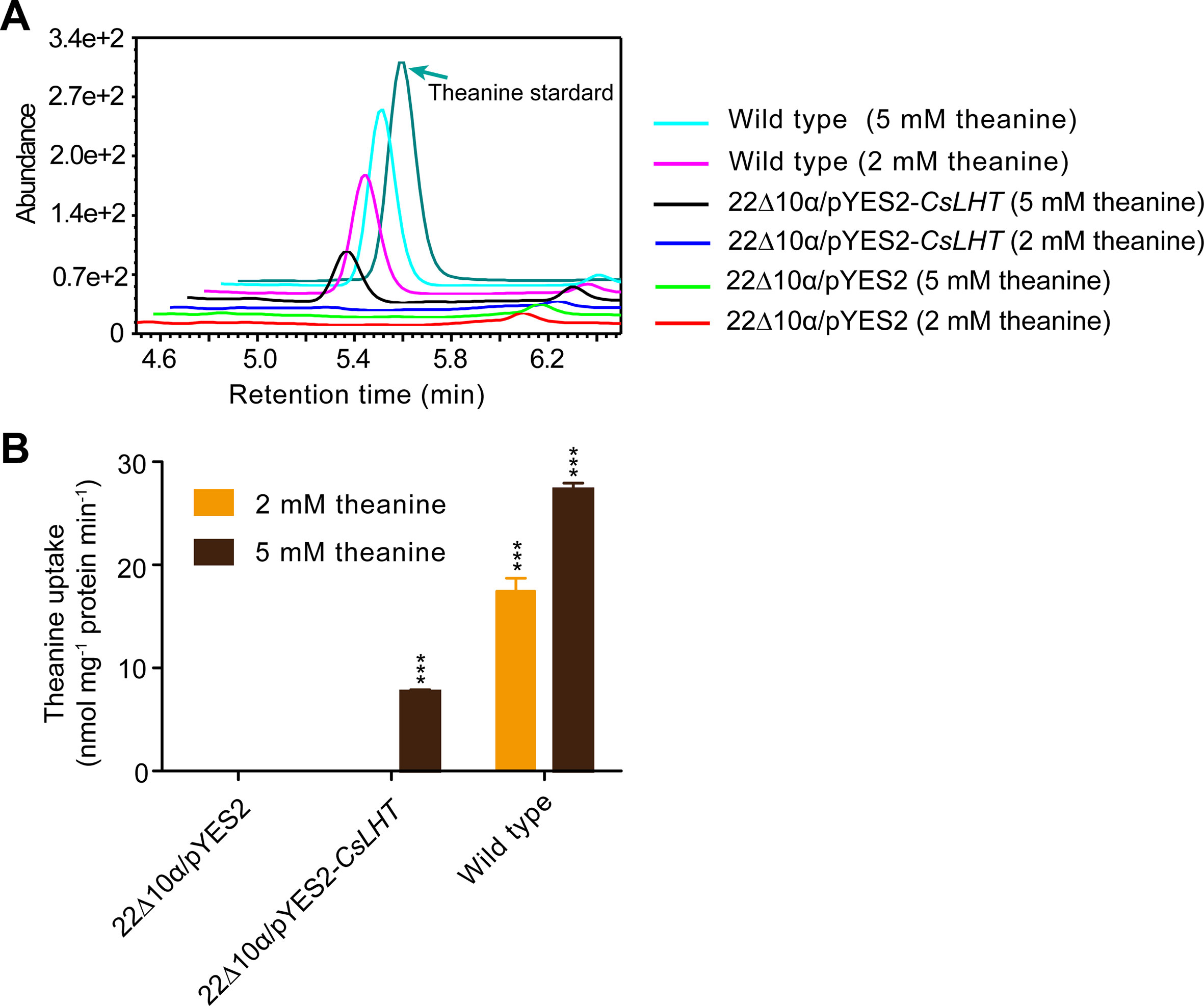
Figure 10 CsLHT transported theanine into yeast cells. (A) HPLC identification theanine in the yeast mutant strain 22Δ10α mediated by CsLHT. Yeast transformed with empty vector was used as a negative control and wild type yeast strain (23344c) as a positive control. (B) Determination of the theanine contents in 22Δ10α/pYES2, 22Δ10α/pYES2-CsLHT and wild-type yeast cells. Data represent means ± SD of three independent biological replicates. Asterisks represent statistical significance determined by Student’s t-test (***p 0.01).
Discussion
Theanine Accumulation Pattern in Root and Bud of Tea Plant in Winter and Spring
As one of the most important components of quality in green tea, theanine and its biosynthesis has been studied intently in tea plants since it was discovered by Yajiro Sakato in 1949 (Ashihara, 2015; Mu et al., 2015). These studies indicated that theanine accumulation was dynamically regulated by seasons (Konishi and Takahashi, 1969; Takeo, 1981; Janet et al., 2015), light (Yang et al., 2012; Deng et al., 2013; Chen et al., 2017; Ji et al., 2018; Sano et al., 2018), nutrient levels (Takeo, 1981; Ruan et al., 2007; Ruan et al., 2010; Ruan et al., 2012; Huang et al., 2018; Kc et al., 2018; Zhu et al., 2019), hormones (Li et al., 2016a), developmental stages (Feldheim et al., 1986; Li et al., 2016b; Liu et al., 2017), and stress (Deng et al., 2012; Wang et al., 2016). Theanine accumulation is also highly tissue-and cultivar-dependent (Fang et al., 2017; Liu et al., 2017; Chen et al., 2018a; Chen et al., 2018b). Theanine accumulation, therefore, is dynamic and highly regulated.
Most of the studies on theanine accumulation under various conditions were performed in one cultivar. In addition, when examining the effects of seasons, comparisons were made between spring, summer, and autumn. Furthermore, because new shoots or tender leaves are the harvested targets for tea, most of the studies focused on theanine accumulation in leaves. However, how theanine accumulation in roots and new shoots changes, during winter and spring, in various cultivars remains to be observed.
In this study, we clearly observed that theanine content in the roots of 13 cultivars slightly deceased or kept stable from Dec. 12 to Mar. 1 and then increased from Mar. 1 to Mar. 23, followed by an obvious decrease on Apr. 13 (Figure 2). This is slightly different from the Takeo’s report in 1979. He observed that the theanine content in roots slightly increased from Jan. to Feb. and then continuously decreased from Feb. to May; he subsequently concluded that theanine was mainly accumulated in winter. Our results, however, suggested that theanine content in roots is highest in late of Mar. or early Apr. and then decreases from the middle of Apr. This inconsistency may be caused by geographical factors or the origin area of the cultivars (Lee et al., 2010; Chen et al., 2018a). In addition, our results also indicated that the theanine levels in roots are likely controlled by genetic background; however, the changing pattern is mainly regulated by seasonal factors (Figure 2).
Different from that in roots, theanine content in buds was low on Dec. 12 and started to increase on Mar. 1 (Figure 5). Theanine content reached the highest in most of the cultivars on Mar. 23 and decreased on Apr. 13. But, in some cultivars, theanine content kept increasing on Apr. 13. Further analyses indicated the different changing patterns of theanine content on Mar. 23 and Apr. 13 were associated with bud sprout time and developmental stages. Therefore, theanine content in buds is regulated by genetic background, seasonal factors, and developmental stages. Here, it is also noteworthy that theanine content decreased quickly in April, and this is highly associated with the decline in green tea quality made from the new buds.
Regulation of the Expression of Theanine Biosynthesis and Amino Acid Transporter Genes
Many previous studies showed that the expression of theanine biosynthesis pathway genes was not always positively correlated with theanine content (Ruan et al., 2010; Fang et al., 2017; Kc et al., 2018; Zhu et al., 2019). Kc et al. found the expression of most theanine biosynthesis pathway genes was negatively correlated with theanine content in leaves (Kc et al., 2018). These results led us to explore the regulation of the expression of theanine biosynthesis and transport pathway genes in response to dynamic theanine accumulation in roots and buds.
In this study, we analyzed the correlation between the expression of theanine biosynthesis pathway genes and theanine content in roots and buds at four time points. These results demonstrated that the expression of theanine biosynthesis pathway genes is generally feedback correlated with theanine content in both roots and buds of tea plants (Figures 4 and 7). These results suggested that the expression of theanine biosynthesis genes is generally feedback regulated at transcription level by theanine levels. This is consistent with the regulation of amino acid biosynthesis genes in model plants (Pratelli and Pilot, 2014).
Although the expression of genes in theanine-synthesis pathways is generally feedback regulated in both roots and buds, the feedback regulation in buds is more profound than in roots. In the 14 genes examined, eight genes were feedback regulated in buds; in contrast, the number was six in the roots (Figures 4 and 7). More importantly, four genes were always positively correlated with theanine contents in the roots; however, no gene that was always positively correlated was identified in buds.
The molecular mechanisms of theanine transport are largely unknown in tea plants. In this study, we found the expression of most amino acid transporter genes examined was positively correlated with theanine content in the roots. These results implied that amino acid transporter genes are induced by theanine accumulation. This result also provided candidate genes for identifying theanine transporters.
Putative Key Genes Involved in the Regulation of the Dynamic Biosynthesis
In this study, we observed that the expression of genes in the same family correlated differently with theanine contents (Figures 4, 7, 9). For example, CsGDH3 showed continuously positive correlation with theanine content in the roots; CsGDH4, however, was always negatively correlated with theanine content in the roots. Similar correlation results were also observed by other reports (Li et al., 2016b; Liu et al., 2017; Zhu et al., 2019). These results indicated that genes in the same family were differently regulated and also suggested varied roles for these genes in theanine biosynthesis and transport.
In addition to this, the correlation degrees were also varied for different genes in theanine biosynthesis pathways and amino acid transporters (Figures 4, 7 and 9). These results suggested CsGDH3, CsGOGAT1, CsADC1, and CsADC2 have important roles in the positive regulation of theanine biosynthesis in roots by providing glutamate and ethylamine, two direct substrates of theanine biosynthesis.
CsLHT May Mediate Theanine Transport in Tea Plants
The high correlation of CsPUT2, CsLHT, and CsVAAT with theanine content in roots suggested these amino acid transporters have important roles in theanine transport. We further showed that CsLHT can transport theanine in yeast under 5 mM theanine feeding. This result supports the argument that CsLHT is a low affinity transporter for theanine. Considering that theanine content in roots is very high (∼60 mg/g dry weight, ∼150 mM/kg fresh weight), it is reasonable to have low affinity transport to mediate theanine transport. We will further biochemically characterize CsLHT in terms of its affinity for theanine and its function in mediating theanine root-to-shoot transport in the future. It is also interesting to test the theanine transport activity of CsPUT and CsVAAT.
Taken together study first uncovered the dynamic accumulation pattern of theanine in the roots and buds of tea plants during winter and spring. Next, the expression of theanine biosynthesis and transport genes in response to dynamic theanine accumulation was investigated in the roots and buds of 13 tea plant cultivars at four time points in winter and spring. Finally, a theanine transporter was identified in tea plants. These findings provided fundamental bases for further revealing the regulation mechanisms of theanine biosynthesis and transport in tea plants.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
ZZ, XW, and CW conceived the study and designed the experiments. FL, CD, TY, JM, and SZ carried out the experiments. FL, CD, TY, and ZZ analyzed the data. ZZ, XW, and FL wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFD1000601) and grants from the Department of Science and Technology of Anhui Province (17030701049 to ZZ) and the National Natural Science Foundation of China (31770731 to ZZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01397/full#supplementary-material
References
Ashihara, H. (2015). Occurrence, biosynthesis and metabolism of theanine (γ-glutamyl-L-ethylamide) in plants: a comprehensive review. Nat. Prod. Commun. 10, 803–810. doi: 10.1177/1934578X1501000525
Besnard, J., Pratelli, R., Zhao, C. S., Sonawala, U., Collakova, E., Pilot, G., et al. (2016). UMAMIT14 is an amino acid exporter involved in phloem unloading in arabidopsis roots. J. Exp. Bot. 67, 6385–6397. doi: 10.1093/jxb/erw412
Besnard, J., Zhao, C. S., Avice, J. C., Vitha, S., Hyodo, A., Pilot, G., et al. (2018). Arabidopsis UMAMIT24 and 25 are amino acid exporters involved in seed loading. J. Exp. Bot. 69, 5221–5232. doi: 10.1093/jxb/ery302
Chen, S., Li, M., Zheng, G., Wang, T., Lin, J., Wang, S., et al. (2018a). Metabolite profiling of 14 Wuyi rock tea cultivars using UPLC-QTOF MS and UPLC-QqQ MS combined with chemometrics. Molecules 23, 104. doi: 10.3390/molecules23020104
Chen, S., Lin, J., Liu, H., Gong, Z., Wang, X., Li, M., et al. (2018b). Insights into tissue-specific specialized metabolism in Tieguanyin tea cultivar by untargeted metabolomics. Molecules 23, 1817. doi: 10.3390/molecules23071817
Chen, Y., Fu, X., Mei, X., Zhou, Y., Cheng, S., Zeng, L., et al. (2017). Proteolysis of chloroplast proteins is responsible for accumulation of free amino acids in dark-treated tea (Camellia sinensis) leaves. J. Proteomics 157, 10–17. doi: 10.1016/jjprot2017.01.017
Cheng, L., Yuan, H. Y., Ren, R., Zhao, S. Q., Han, Y. P., Zhou, Q. Y., et al. (2016). Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine Max. Front. Plant Sci. 7, 515. doi: 10.3389/fpls.2016.00515
Cheng, S., Fu, X., Wang, X., Liao, Y., Zeng, L., Dong, F., et al. (2017). Studies on the biochemical formation pathway of the amino acid l-theanine in tea (Camellia sinensis) and other plants. J. Agric. Food Chem. 65, 7210–7216. doi: 10.1021/acs.jafc.7b02437
Deng, W. W., Fei, Y., Wang, S., Wan, X. C., Zhang, Z. Z., Hu, X. Y. (2013). Effect of shade treatment on theanine biosynthesis in Camellia sinensis seedlings. Plant Growth Regul. 71, 295–299. doi: 10.1007/s10725-013-9828-1
Deng, W. W., Ogita, S., Ashihara, H. (2009). Ethylamine content and theanine biosynthesis in different organs of Camellia sinensis seedlings. Z. Naturforsch C. 64, 387–390. doi: 10.1515/znc-2009-5-614
Deng, W. W., Wang, S., Chen, Q., Zhang, Z. Z., Hu, X. Y. (2012). Effect of salt treatment on theanine biosynthesis in Camellia sinensis seedlings. Plant Physiol. Biochem. 56, 35–40. doi: 10.1016/j.plaphy.2012.04.003
Dong, C., Li, F., Yang, T., Feng, L., Zhang, S., Li, F., et al. (2019). Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J. doi: 10.1111/tpj.14517
Fang, R., Redfern, S. P., Kirkup, D., Porter, E. A., Kite, G. C., Terry, L. A., et al. (2017). Variation of theanine, phenolic, and methylxanthine compounds in 21 cultivars of Camellia sinensis harvested in different seasons. Food Chem. 220, 517–526. doi: 10.1016/j.foodchem.2016.09.047
Feldheim, W., Yongvanit, P., Cummings, P. H. (1986). Investigation of the presence and significance of theanine in the tea plant. J. Sci. Food Agric. 37, 527–534. doi: 10.1002/jsfa.2740370604
Fisher, M., Zamir, A., Pick, U. (1998). Iron uptake by the halotolerant alga dunaliella is mediated by a plasma membrane transferrin. J. Biol. Chem. 273, 17553–17558. doi: 10.1074/jbc.273.28.17553
Guo, X., Ho, C. T., Schwab, W., Song, C., Wan, X. (2019). Aroma compositions of large-leaf yellow tea and potential effect of theanine on volatile formation in tea. Food Chem. 280, 73–82. doi: 10.1016/j.foodchem.2018.12.066
Guo, X., Song, C., Ho, C. T., Wan, X. (2018). Contribution of l-theanine to the formation of 2, 5-dimethylpyrazine, a key roasted peanutty flavor in Oolong tea during manufacturing processes. Food Chem. 263, 18–28. doi: 10.1016/j.foodchem.2018.04.117
Huang, H., Yao, Q., Xia, E., Gao, L. (2018). Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J. Agric. Food Chem. 66, 9828–9838. doi: 10.1021/acs.jafc.8b01995
Janet, T. C., John, W. K., Thomas, K., Kelvin, M. O., Francis, W. N. (2015). Effect of seasons on theanine levels in different kenyan commercially released tea cultivars and its variation in different parts of the tea shoot. Food Nutr. Sci. 6, 1450. doi: 10.4236/fns.2015.615149
Ji, H. G., Lee, Y. R., Lee, M. S., Hwang, K. H., Park, C. Y., Kim, E. H., et al. (2018). Diverse metabolite variations in tea (Camellia sinensis L.) leaves grown under various shade conditions revisited: a metabolomics study. J. Agric. Food Chem. 66, 1889–1897. doi: 10.1021/acs.jafc.7b04768
Kc, S., Liu, M., Zhang, Q., Fan, K., Shi, Y., Ruan, J. (2018). Metabolic changes of amino acids and flavonoids in tea plants in response to inorganic phosphate limitation. Int. J. Mol. Sci. 19, 3683. doi: 10.3390/ijms19113683
Kito, M., Kokura, H., Izaki, J., Sadaoka, K. (1968). Theanine, a precursor of the phloroglucinol nucleus of catechins in tea plants. Phytochemistry. 7, 599–603. doi: 10.1016/S0031-9422(00)88234-5
Konishi, S., Kasai, Z. (1968). Synthesis of theanine from 14CO2 in tea plants and sites of the synthesis. Jpn. J. Soil Sci. Plant Nutr. 39, 439–443.
Konishi, S., Takahashi, E. (1969). Metabolism of 14C labeled theanine. [N-ethyl-14C] theanine and its metabolic redistribution in the tea plant. Jpn. J. Soil Sci. Plant Nutr. 40, 479–484.
Larhammar, M., Patra, K., Blunder, M., Emilsson, L., Peuckert, C., Arvidsson, E., et al. (2014). SLC10A4 is a vesicular amine-associated transporter modulating dopamine homeostasis. Biol. Psychiatry. 77, 526–536. doi: 10.1016/j.biopsych.2014.07.017
Lee, J. E., Lee, B. J., Chung, J. O., Hwang, J. A., Lee, S. J., Lee, C. H., et al. (2010). Geographical and climatic dependencies of green tea (Camellia sinensis) metabolites: a H-1 NMR-Based metabolomics study. J. Agric. Food Chem. 58, 10582–10589. doi: 10.1021/jf102415m
Li, C. F., Xu, Y. X., Ma, J. Q., Jin, J. Q., Huang, D. J., Yao, M. Z., et al. (2016). Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Bio. 16, 195. doi: 10.1186/s12870-016-0885-2
Li, X., Ahammed, G. J., Li, Z. X., Zhang, L., Wei, J. P., Shen, C., et al. (2016a). Brassinosteroids improve quality of summer tea (Camellia sinensis L.) by balancing biosynthesis of polyphenols and amino acids. Front. Plant Sci. 7, 1304. doi: 10.3389/fpls.2015.01304
Liu, Z. W., Wu, Z. J., Li, H., Wang, Y. X., Zhuang, J. (2017). L-Theanine content and related gene expression: novel insights into theanine biosynthesis and hydrolysis among different tea plant (Camellia sinensis L.) tissues and cultivars. Front. Plant Sci. 8, 498. doi: 10.3389/fpls.2017.00498
Ma, H., Cao, X., Shi, S., Li, S., Gao, J., Ma, Y., et al. (2016). Genome-wide survey and expression analysis of the amino acid transporter superfamily in potato (Solanum tuberosum L.). Plant Physiol. Biochem. 107, 164–177. doi: 10.1016/j.plaphy.2016.06.007
Mu, W., Zhang, T., Jiang, B. (2015). An overview of biological production of L-theanine. Biotechnol. Adv. 33, 335–342. doi: 10.1016/j.biotechadv.2015.04.004
Muller, B., Fastner, A., Karmann, J., Mansch, V., Hoffmann, T., Schwab, W., et al. (2015). Amino acid export in developing Arabidopsis seeds depends on umamit facilitators. Curr. Biol. 25, 3126–3131. doi: 10.1016/j.cub.2015.10.038
Oh, K., Kato, T., Xu, H. L. (2008). Transport of nitrogen assimilation in xylem vessels of green tea plants fed with NH4-N and NO3-N. Pedosphere. 18, 222–226. doi: 10.1016/s1002-0160(08)60010-7
Pratelli, R., Pilot, G. (2014). Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 65, 5535–5556. doi: 10.1093/jxb/eru320
Ruan, J., Gerendas, J., Haerdter, R., Sattelmacher, B. (2007). Effect of alternative anions (Cl- vs. SO42-) on concentrations of free amino acids in young tea plants. J. Plant Nutr. Soil Sci. 170, 49–58. doi: 10.1002/jpln.200625142
Ruan, J., Ma, L., Yang, Y. (2012). Magnesium nutrition on accumulation and transport of amino acids in tea plants. J. Sci. Food. Agric. 92, 1375–1383. doi: 10.1002/jsfa.4709
Ruan, J., Haerdter, R., Gerendás, J. (2010). Impact of nitrogen supply on carbon/nitrogen allocation: a case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants. Plant Biol. 12, 724–734. doi: 10.1111/j.1438-8677.2009.00288.x
Sano, T., Horie, H., Matsunaga, A., Hirono, Y. (2018). Effect of shading intensity on morphological and color traits and on chemical components of new tea (Camellia sinensis L.) shoots under direct covering cultivation. J. Sci. Food Agric. 98, 5666–5676. doi: 10.1002/jsfa.9112
Sasaoka, K., Kito, M., Onishi, Y. (1965). Some properties of the theanine synthesizing enzyme in tea seedlings. Agric. Biol. Chem. 29, 984–988. doi: 10.1271/bbb1961.29.984
Schmittgen, T. D., Zakrajsek, B. A., Mills, A. G., Gorn, V., Singer, M. J., Reed, M. W. (2000). Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285, 194–204. doi: 10.1006/abio.2000.4753
Sharma, E., Joshi, R., Gulati, A. (2018). L-Theanine: an astounding sui generis integrant in tea. Food Chem. 242, 601–610. doi: 10.1016/j.foodchem.2017.09.046
Shi, C. Y., Yang, H., Wei, C. L., Yu, O., Zhang, Z. Z., Jiang, C. J., et al. (2011). Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC Genomics. 12, 131. doi: 10.1186/1471-2164-12-131
Shuai, Y., Zhang, T., Jiang, B., Mu, W. (2010). Development of efficient enzymatic production of theanine by γ-glutamyltranspeptidase froma newly isolated strain of Bacillus subtilis, SK11.004. J. Sci. Food Agric. 90, 2563–2567. doi: 10.1002/jsfa.4120
Suzuki, H., Kumagai, H. (2002). Autocatalytic processing of γ-glutamyltranspeptidase. J. Biol. Chem. 277, 43536–43543. doi: 10.1074/jbc.M207680200
Suzuki, H., Miyakawa, N., Kumagai, H. (2002). Enzymatic production of γ-L-glutamyltaurine through the transpeptidation reaction of γ-glutamyltranspeptidase from Escherichia coli K-12. Enzyme Microb. Technol. 30, 883–888. doi: 10.1016/S0141-0229(02)00038-8
Tai, Y., Wei, C., Yang, H., Zhang, L., Chen, Q., Deng, W., et al. (2015). Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera). BMC Plant Biol. 15, 190. doi: 10.1186/s12870-015-0574-6
Takeo, T. (1978). L-alanine decarboxylase in camellia sinensis. Phytochem 17, 313–314. doi: 10.1016/s0031-9422(00)94173-6
Takeo, T. (1979). Glutamate dehydrogenase from tea rootlet. Agric. Biol. Chem. 43, 2257–2263. doi: 10.1271/bbb1961.43.2257
Takeo, T. (1981). Nitrogen metabolism pertaining to biosynthesis of theanine in tea plants. Jpn. Agric. Res. Q. 15, 110–116.
Tegeder, M., Ward, J. M. (2012). Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 3, 21. doi: 10.3389/fpls.2012.00021
Tegeder, M. (2014). Transporters involved in source to sink partitioning of amino acids and ureides: opportunities for crop improvement. J. Exp. Bot. 65, 1865–1878. doi: 10.1093/jxb/eru012
Tegeder, M., Tan, Q., Grennan, A. K., Patrick, J. W. (2007). Amino acid transporter expression and localisation studies in pea (Pisum sativum). Funct. Plant Biol. 34, 1019–10128. doi: 10.1071/FP07107
Tsushida, T., Takeo, T. (1985). An enzyme hydrolyzing L-theanine in tea leaves. Agric. Biol. Chem. 10, 2913–2917. doi: 10.1080/00021369.1985.10867191
Wang, W., Xin, H., Wang, M., Ma, Q., Wang, L., Kaleri, N. A., et al. (2016). Transcriptomic analysis reveals the molecular mechanisms of drought-stress-induced decreases in Camellia sinensis leaf quality. Front. Plant Sci. 7, 385. doi: 10.3389/fpls.2016.00385
Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C., Gao, L., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U. S. A. 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Xia, E. H., Li, F. D., Tong, W., Li, P. H., Wu, Q., Zhao, H. J., et al. (2019). Tea Plant Information Archive (TPIA): a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 17, 1938-1953. doi: 10.1111/pbi.13111
Xia, E. H., Zhang, H. B., Sheng, J., Li, K., Zhang, Q. J., Kim, C., et al. (2017). The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 10, 866–877. doi: 10.1016/j.molp.2017.04.002
Yamaguchi, S., Ninomiya, K. (2000). Umami and food palatability. J. Nut. 130, 921s–926s. doi: 10.1007/978-1-4615-4693-1_36
Yang, Z., Kobayashi, E., Katsuno, T., Asanuma, T., Fujimori, T., Ishikawa, T., et al. (2012). Characterisation of volatile and non-volatilemetabolites in etiolated leaves of tea (Camellia sinensis) plants in the dark. Food Chem. 135, 2268–2276. doi: 10.1016/j.foodchem.2012.07.066
Yao, Y. F., Weng, Y. M., Hu, H. Y., Ku, K. L., Lin, L. L. (2006). Expression optimization and biochemical characterization of a recombinant γ-glutamyltranspeptidase from escherichia coli novablue. Protein J. 25, 431–441. doi: 10.1007/s10930-006-9037-0
Keywords: Camellia sinensis, theanine, biosynthesis and transport, seasonal regulation, theanine transporter, CsLHT
Citation: Li F, Dong C, Yang T, Ma J, Zhang S, Wei C, Wan X and Zhang Z (2019) Seasonal Theanine Accumulation and Related Gene Expression in the Roots and Leaf Buds of Tea Plants (Camellia Sinensis L.). Front. Plant Sci. 10:1397. doi: 10.3389/fpls.2019.01397
Received: 18 April 2019; Accepted: 10 October 2019;
Published: 30 October 2019.
Edited by:
Xiaoya Chen, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2019 Li, Dong, Yang, Ma, Zhang, Wei, Wan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoliang Zhang, emhsemhhbmdAYWhhdS5lZHUuY24=; Xiaochun Wan, eGN3YW5AYWhhdS5lZHUuY24=
†These authors have contributed equally to this work
 Fang Li†
Fang Li† Chaoling Wei
Chaoling Wei Xiaochun Wan
Xiaochun Wan Zhaoliang Zhang
Zhaoliang Zhang