
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 30 October 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01362
This article is part of the Research TopicAdvanced Microbial Biotechnologies For Sustainable AgricultureView all 39 articles
Fusarium head blight (FHB) caused by Fusarium pathogens are devastating diseases worldwide. Host-induced gene silencing (HIGS) which involves host expression of double-stranded RNA (dsRNA)-generating constructs directed against genes in the pathogen has been a potential strategy for the ecological sound control of FHB. In this study, we constructed transgenic Brachypodium distachyon lines carrying RNA interference (RNAi) cassettes to target two essential protein kinase genes Fg00677 and Fg08731, and cytochrome P450 lanosterol C14-α-demethylase (CYP51) encoding genes (CYP51A, CYP51B, and CYP51C) of Fusarium graminearum, respectively. Northern blotting confirmed the presence of short interfering RNAs (siRNA) derived from Fg00677, Fg08731, and CYP51 in transgenic B. distachyon plants, and the transcript levels of the corresponding genes were down-regulated in the F. graminearum colonizing B. distachyon spikes. All the corresponding independent, Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi transgenic T2 lines exhibited strong resistance to F. graminearum, suggesting that silencing molecules produced by transgenic plants inhibited the corresponding gene function by down-regulating its expression, thereby reducing pathogenicity. Our results indicate that Fg00677 and Fg08731 are effective targets for HIGS and can be applied to construct transgenic HIGS materials to enhance FHB resistance in wheat and other cereal crops.
Fusarium head blight (FHB), which is caused by the fungal pathogen Fusarium graminearum, is a devastating disease in wheat production around the world (Osborne and Stein, 2007). Wheat can be infected by F. graminearum from the seedling to the heading stage, causing seedling blight, head blight, basal stem rot, and stalk rot (Stenglein, 2009). F. graminearum produces mycotoxins, such as deoxynivalenol, that accumulate in wheat grains, detrimentally affect the food quality and thus pose a serious threat to human and animal health (Zhang et al., 2013). Current control methods are mainly dependent on chemical control, but fungicide resistance has become increasingly prominent due to long-term use of a single chemical fungicide (Chen et al., 2007). Breeding new wheat cultivars for resistance against FHB is the most effective measure to control the disease. Due to the narrow genetic base of wheat, breeding new cultivars against FHB by traditional breeding methods has been difficult (Liu et al., 2009; Loffler et al., 2009). Therefore, a more effective and stable method for enhancing the resistance against FHB must be developed.
RNA interference (RNAi) is extensively used in functional genomics studies (Carthew, 2001; Ketting, 2011). The RNAi process includes three steps. First, a double-stranded RNA (dsRNA) or a single-stranded RNA that can form a hairpin RNA (hpRNA) is recognized by the ribonuclease Dicer and cleaved into small interfering RNAs (siRNAs) (Fagard et al., 2000). Then, siRNAs are processed into the RNA-induced silencing complex (RISC) containing an Agonaute protein (Fagard et al., 2000). Finally, the RISC binds to the target mRNA through homologous pairing, down-regulating gene expression at the transcriptional or post-transcriptional level (Ghildiyal and Zamore, 2009; Liu and Paroo, 2010). The expression of hpRNA, dsRNA, or siRNA molecules directed against parasite transcripts has been used to decrease these parasite transcripts and then to control harmful parasitic organism in plants (Cai et al., 2018); this is referred to as host-induced gene silencing (HIGS) (Nunes and Dean, 2012). Later studies have confirmed that HIGS can be a new, pesticide-free, and potentially sustainable option to enhance resistance of crops against bacteria, viruses, fungi, insects, nematodes, and parasitic weeds (Patil et al., 2011; Saurabh et al., 2014; Zhu et al., 2017; Cai et al., 2018; Panwar et al., 2018; Qi et al., 2018). HIGS has been an effective strategy to confer resistance against Fusarium pathogens F. graminearum, F. oxysporum and F. culmorum (Koch et al., 2013; Ghag et al., 2014; Cheng et al., 2015; Hu et al., 2015; Chen et al., 2016). However, there are only a few studies on the application of HIGS to enhance the resistance against FHB in wheat, and it is urgent to identify new effective HIGS target genes in F. graminearum.
Due to the special challenges of wheat genetic transformation, including a long process, cumbersome procedures, and high cost, it will be beneficial to use effective HIGS targets for genetic transformation. Quickly determining the most effective HIGS target is the primary problem for developing wheat disease-resistant materials. As a monocotyledon that is widely grown in the temperate zone, Brachypodium distachyon is rich in germplasm resources. B. distachyon has become an ideal model plant for functional genomics study of monocotyledons because of its interesting characteristics, including a small size, a short life-cycle, a small genome, diploid inheritance, self-fertility, a routine genetic transformation procedure, and simple growth requirements (Mur et al., 2011). B. distachyon is closely related to the large-genome cereal grasses, such as rice and wheat, and it is more closely related evolutionarily to wheat than to rice (Draper et al., 2001). B. distachyon is the ideal model plant to study the mechanism of plant-pathogen interactions because it is a host for many cereal pathogens (Peraldi et al., 2011; Sandoya and de Oliveira Buanafina, 2014). For example, B. distachyon-F. graminearum interactions closely model the head blight in wheat caused by F. graminearum (Peraldi et al., 2011). Therefore, we tested whether B. distachyon can be used for rapidly identifying effective HIGS targets that will be applied in developing FHB resistance in wheat.
In our current study, we selected two genes (Fg00677 and Fg08731) as targets for HIGS analysis. Fg00677 and Fg08731, which encode alpha catalytic subunit of casein kinase 2 (CK2) and casein kinase 1 (CK1), respectively, are essential in F. graminearum and are up-regulated when F. graminearum infects wheat spikes (Wang et al., 2011). As the resistance to F. graminearum was enhanced through HIGS methods targeting CYP51 genes in barley and Arabidopsis (Koch et al., 2013), CYP51 was selected as a HIGS target. We show that the transgenic B. distachyon expressing Fg00677-RNAi, Fg08731-RNAi, or CYP51-RNAi construct confers resistance to F. graminearum, indicating that Fg00677 and Fg08731 can be used as ideal targets to enhance the resistance to FHB in wheat by HIGS methods, and HIGS applied in the B. distachyon-F. graminearum interaction is a valuable model to rapidly identify effective HIGS targets.
B. distachyon Bd21-3 was cultivated in a growth chamber at 22°C with a 16 h light/8 h darkness photoperiod. F. graminearum strain PH-1 was cultured on Potato Dextrose Agar Medium (PDA). Conidia were produced in Carboxymethyl Cellulose (CMC) broth as described previously (Duvick et al., 1992).
The sequence information of genes studied in this paper was acquired from Ensembl Fungi (http://fungi.ensembl.org/Fusarium_graminearum_gca_000240135/Info/Index). Gene numbers of F. graminearum genes were abbreviated by replacing FGSG_ with Fg. The sequences of Fg00677 and Fg08731 were analyzed using BLAST search, then the conserved domain of Fg00677 and Fg08731 was detected with InterProScan and NCBI Conserved Domain Search. Multiple sequence alignment was implemented with DNAMAN and CLUSTALX2.0 software. The phylogenetic tree was constructed with the MEGA 5.0 software (Tamura et al., 2011).
Specific partial sequences of CYP51A (Fg04092), CYP51B (Fg01000), and CYP51C (Fg11024) were used to prepare the CYP51-RNAi constructs as described by Koch et al. (2013). The 294 bp of CYP51A, 220 bp of CYP51B, and 238 bp of CYP51C were stacked into CYP51BAC by overlap PCR using the predesigned primer (Figure 2A; Table S1). The sequence-specific fragments of Fg00677 and Fg08731 used for the preparing the RNAi constructs were amplified from F. graminearum complementary DNA (cDNA) by PCR using the primers shown in Table S1. The length of Fg00677, Fg08731, and CYP51BAC fragments were 452, 481, and 752 bp, respectively (Figure 2B; Figure S2). These sequence-specific fragments were cloned into the gateway™ pDONR221 vector to generate the entry constructs (pDONR221-Fg00677, pDONR221-Fg08731, and pDONR221-CYP51BAC) by gateway BP reactions. The recombinant entry module can be used in gateway LR reaction to transfer the sequence-specific fragment to the pCAMBIA1300-based vector to generate Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi construct, respectively Figure 2B.
The RNAi vectors were transformed into Agrobacterium tumefaciens strain AGL1 through the electroporation method (Lazo et al., 1991). The A. tumefaciens was cultured in mannitol glutamate/luria-bertani (MG/L) medium (5 g/L tryptone, 2.5 g/L yeast extract, 5 g/L NaCl, 5 g/L mannitol, 1.2 g/L glutamic acid, 0.25 g/L K2HPO4, 0.1 g/L MgSO4, pH to 7.2 with 1N NaOH) supplemented with 50 mg/L rifampicin and 50 mg/L kanamycin to an OD600 of 0.6. The constructs contained a hygromycin phosphotransferase gene to confer hygromycin resistance to transformed plants. The Bd21-3 wild-type line was genetically transformed using the procedure of Vogel and Hill (2008).
Point inoculation and evaluation of symptoms followed the procedure of Pasquet et al. (2016) and Peraldi et al. (2011). Point inoculation was performed by pipetting 300 conidia in 0.01% Tween-20 (3 µL of a 105 conidia per mL suspension) into a central floral cavity of the second spikelet, numbered from the top of the spike of different lines in mid-anthesis stages (approximately 40–45 days after sowing). Inoculated plants were covered with clear plastic bags. During the first 24 h, inoculated heads were kept in the dark, then incubated with a photoperiod of 16 h light/8 h darkness at 25°C with the same light intensities as those used for plant development. Application of 0.01% Tween-20 was performed as the control condition for each inoculation experiment. At 9 days after inoculation, symptoms were evaluated with a scoring scale for each inoculated spike from 0 to 4 as follows: 0, no symptoms; 1, only the inoculated floret was symptomatic (most frequently, only browning); 2, extension of symptoms to additional florets of the inoculated spikelet (browning or bleaching); 3, symptoms on the entire inoculated spikelet; and 4, extension of the symptoms to the entire inoculated spikelet and at least one adjacent spikelet. Three independent biological replicates were performed.
Plant DNA was isolated by the cetyltrimethylammonium bromide (CTAB) method (Hormaza, 2002). The transgenic plants were identified by PCR using gene-specific primers (00677-RNAi-F/00677-RNAi-R, 08731-RNAi-F/08731-RNAi-R, and CYPBAC-RNAi-F/CYPBAC-RNAi-R) and universal primer (Bar-F/Bar-R) (Table S1). Total RNA was extracted from spikelets using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and transcribed into cDNA for quantitative real-time PCR (qRT-PCR). qRT-PCR analysis was carried out with a CFX96TM Real-Time PCR machine (Bio-Rad). Transcript levels of Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C were measured using cDNA prepared from total RNA isolated from five spikes of every corresponding transgenic line or non-transformed control plant inoculated with F. graminearum by qRT-PCR as described previously (Pasquet et al., 2016). Specific primers for qRT-PCR analysis of each gene were designed using the Primer 5.0 program (Table S1). Quantification results were analyzed using the comparative 2-ΔΔCT method. To assess fungal transcript levels, the F. graminearum beta-tubulin gene was used as the normalizing reference gene. Three independent biological replicates were performed. To quantify fungal biomass, the ratio of single copy beta-tubulin and the BdUBC18 was assessed in genomic DNA isolated from infected spikes using gene-specific primers (Table S1) (Cheng et al., 2015; Wang et al., 2017). The relative amounts of PCR product of beta-tubulin and BdUBC18 in F. graminearum-infected samples were calculated using generated gene-specific standard curves to quantify the F. graminearum and B. distachyon gDNA, respectively. Three independent biological replicates were performed. Northern blotting was used to detect accumulation of siRNAs as described previously (Zhu et al., 2017). The fragments derived from Fg00677, Fg08731, and CYP51BAC used in RNAi vector construction were produced by PCR, and then used as probe labeled by the random priming method to detect the siRNA derived from the transgenic RNAi plants (Supplementary Table S1). The isolated total RNAs were separated in 19% polyacrylamide/7M urea gels and transferred to Hybond N+ membranes (Amersham) using a mini trans-blot (Bio-Rad, Hercules, CA). The membranes were UV cross-linked in a UV crosslinker (CX-2000, UVP, Upland, CA, USA). The membranes were prehybridized with PerfectHyb TM (Sigma-Aldrich, St. Louis, MO, USA), and hybridized with the P32-labeled DNA probes overnight in PerfectHyb buffer.
Fg00677 and Fg08731 encode the alpha catalytic subunit of CK2 and CK1, respectively (Wang et al., 2011). Sequence analysis indicated that Fg00677 has an open reading frame (ORF) of 1,023 bp, encoding a putative protein composed of 340 amino acids with a molecular weight of 39.72 kDa and an isoeletric point (pI) of 7.55. Fg08731 has an ORF of 1,194 bp, encoding a putative protein composed of 397 amino acids with a molecular weight of 45.05 kDa and a pI of 9.58. The multi-sequence alignment of CK2 proteins from different organisms in NCBI database revealed that Fg00677 is 85%, 73%, 50%, 55%, and 64% identical to Aspergillus nidulans CK2, Ustilago maydis CK2, S. cerevisiae CKA1, S. cerevisiae CKA2, and Schizosaccharomyces pombe Cka1, respectively, and contain the STKc_CK2_alpha conserved domain (Figure S1A). The multi-sequence alignment of CK1 proteins from different organisms in NCBI database revealed that Fg08731 is 82%, 67%, 64%, 52%, and 43% identical to A. nidulans CK1, U. maydis CK1, S. pombe Hhp1, S. pombe Hhp2, and S. cerevisiae HRR25, respectively, and contain the STKc_CK1_delta_epsilon conserved domain (Figure S1B). Phylogenetic analysis confirmed that Fg00677 is orthologous to A. nidulans CK2, U. maydis CK2, S. cerevisiae CKA1, S. cerevisiae CKA2, and S. pombe Cka1 (Figure 1A), whereas Fg008731 is orthologous to A. nidulans CK1, U. maydis CK1, S. pombe Hhp1, S. pombe Hhp2, and S. cerevisiae HRR25 (Figure 1B). These results indicate that Fg00677 and Fg08731 are highly conserved in filamentous fungi.
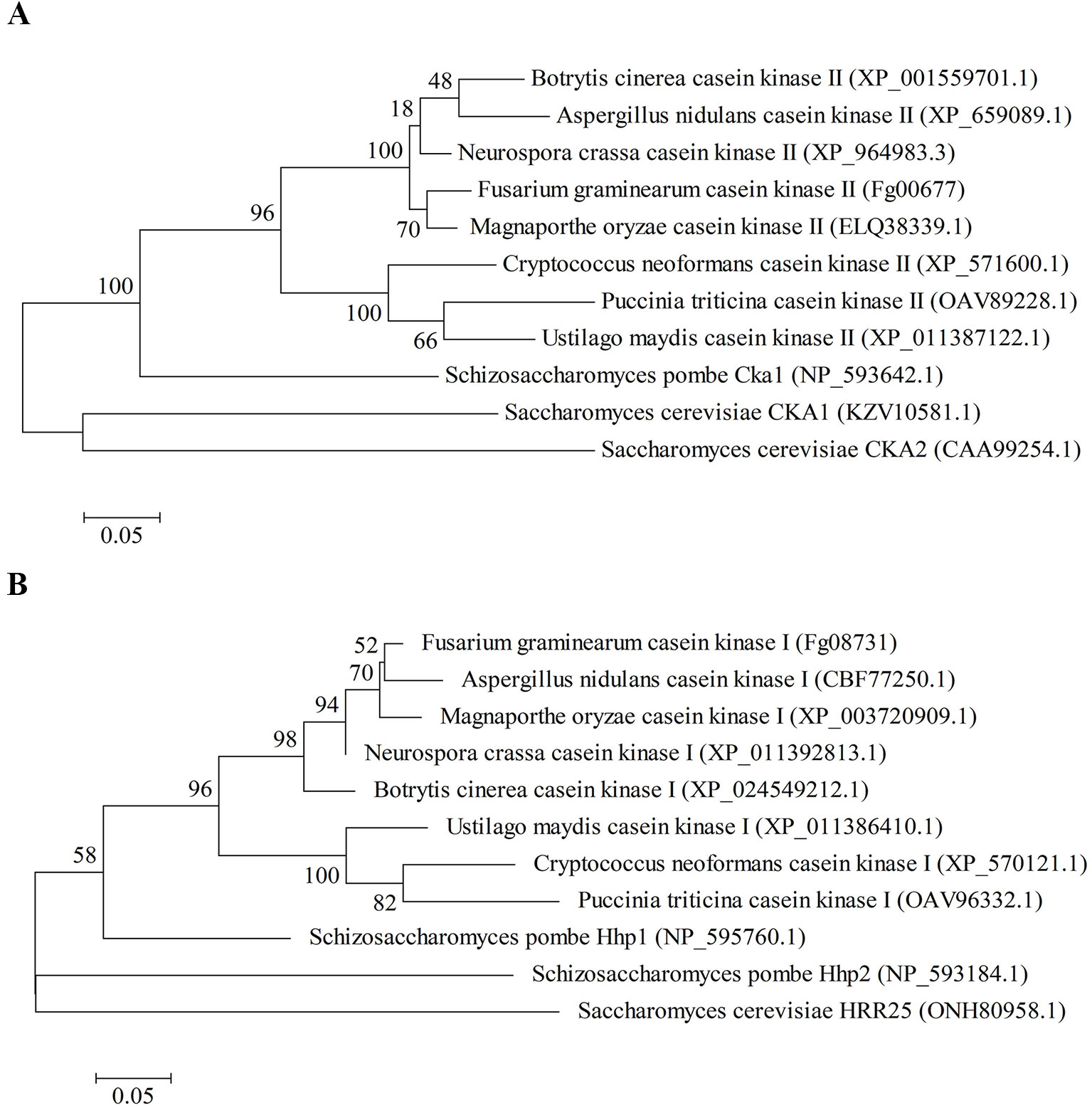
Figure 1 Phylogenetic analysis of Fg00677 (A) and Fg08731 (B) with their homologs in other fungal species. Phylogenetic analysis was carried out with the MEGA5 software by the maximum likelihood tree-building methods.
Koch et al. (2013) confirmed that expression of CYP3RNA, a double-stranded (ds) RNA complementary to CYP51A, CYP51B, and CYP51C, caused co-silencing of CYP51A, CYP51B, and CYP51C in F. graminearum, thereby effectively inhibiting pathogen development and enhancing resistance in Arabidopsis and barley. The CYP3RNA was selected as the positive control to evaluate the efficiency of disease resistance in B. distachyon. The stacking CYP51BAC fragment, including 220 bp of CYP51B, 294 bp of CYP51A, and 238 bp of CYP51C was constructed in the pCAMBIA1300-based vector to produce CYP51-RNAi, which was transformed into B. distachyon Bd21-3 to express the same CYP3RNA as reported by Koch et al. (2013) (Figure 2A). To identify whether Fg00677 and Fg08731 are HIGS-effective targets, the selected 452 and 481 bp fragments from Fg00677 and Fg08731 were constructed into the pCAMBIA1300-based vector to produce Fg00677-RNAi and Fg08731-RNAi construct, respectively (Figure 2B; Figure S2). No off-targets were detected in F. graminearum, B. distachyon, wheat, and Homo sapiens for the selected fragments of Fg00677 and Fg08731 (Table S2). Each of these constructs contained an inverted repeat that, after transcription, is expected to result in a dsRNA sequence with a hairpin structure. Transgenic lines containing the RNAi construct were generated in the B. distachyon Bd21-3 by Agrobacterium-mediated transformation (Figure 3). In total, 21 putative T0 transgenic plants were produced from several transformation experiments. Molecular analysis of putative transgenic plants from T2 generations was performed by PCR analysis, which confirmed the presence of 452, 481, and 752 bp amplicons from the Fg00677, Fg08731, and CYP51BAC in the corresponding T2 transgenic line. No amplification bands were observed in the non-transgenic control plants (Figure 4). In addition, integration of the Bar gene segment in T2 transgenic plants was verified by PCR (Figure 4). The result indicated that the Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi constructs were integrated into the B. distachyon plant genome successfully by Agrobacterium-mediated transformation.
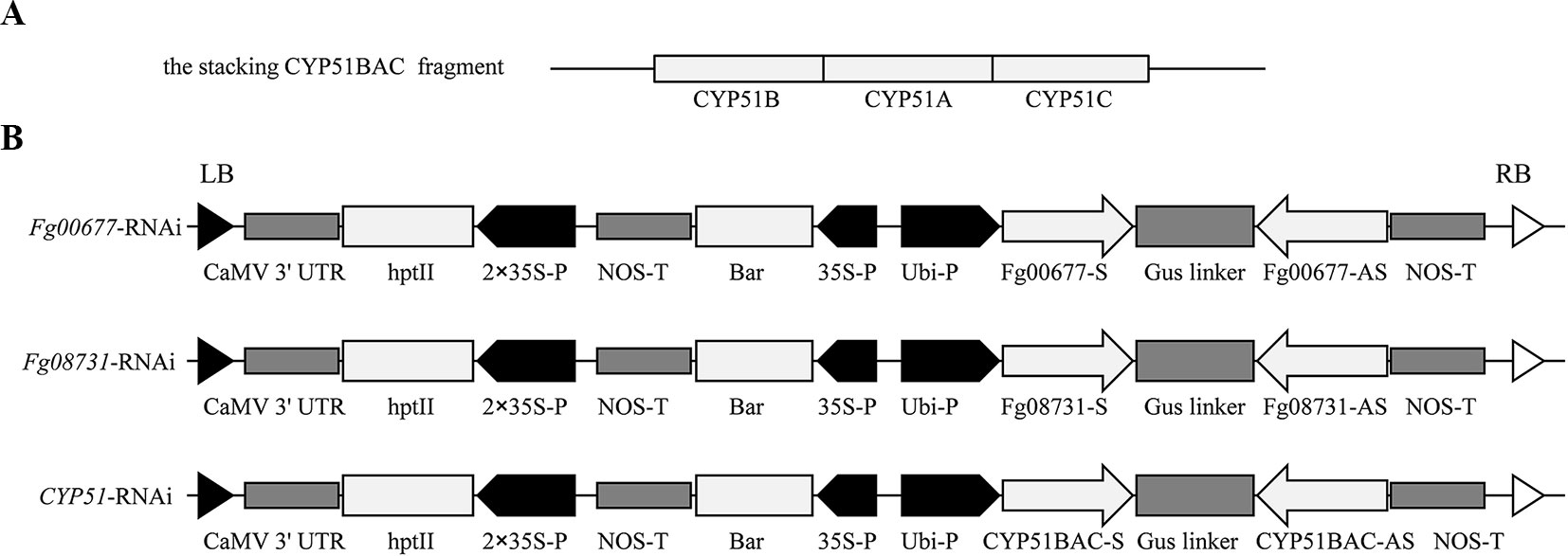
Figure 2 Structure of the RNAi construct. (A) The CYP51BAC fragment was generated by fragment stacking strategy from CYP51A, -B, and -C partial sequences for CYP51-RNAi cassette construction; (B) structure of the Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi cassette for stable B. distachyon transformation. Each fragment derived from Fg00677, Fg08731, and CYP51 in the sense (S) and antisense (AS) orientations was constructed such that the gus linker sequence was inserted between the S and AS sequences. Ubi-P, maize ubiquitin promoter; hpt II, hygromycin resistance gene; 2 × 35S-P, 2× CaMV 35S Promoter; Nos-T, Nos terminator; Bar, Biolaphos resistance gene.
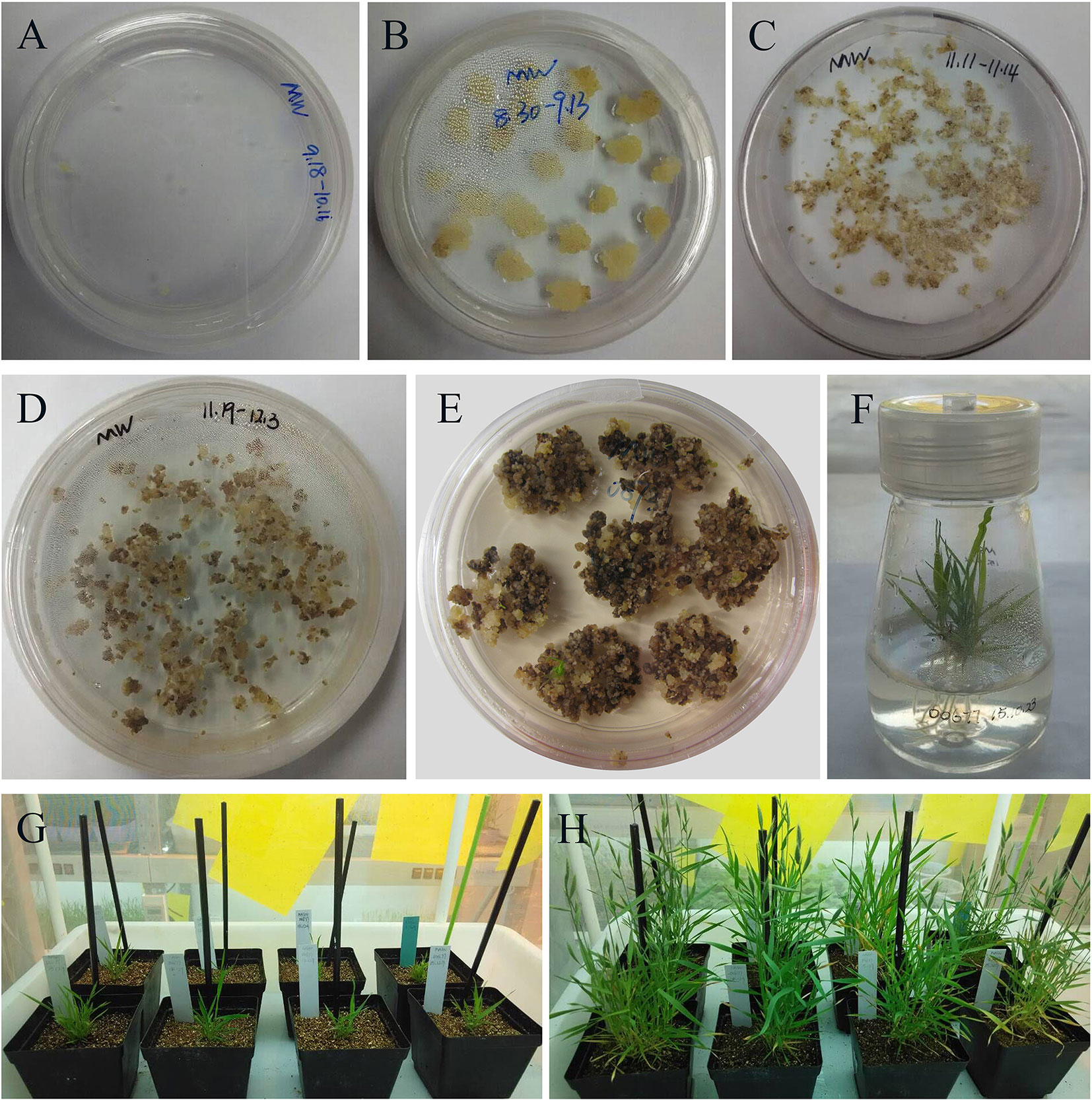
Figure 3 Stable genetic transformation of B. distachyon (genotype Bd21-3). (A) Immature embryos grown on callus induction medium; (B) embryo after 3 weeks on callus induction medium; (C) co-cultivation compact embryogenic callus with Agrobacterium on filter paper; (D) co-cultured compact embryogenic callus on selection medium; (E) regenerating callus on regeneration medium; (F) rooted putative transformants on rooting medium; putative transformant grown in soil for 2 weeks (G) and 8 weeks (H), respectively.
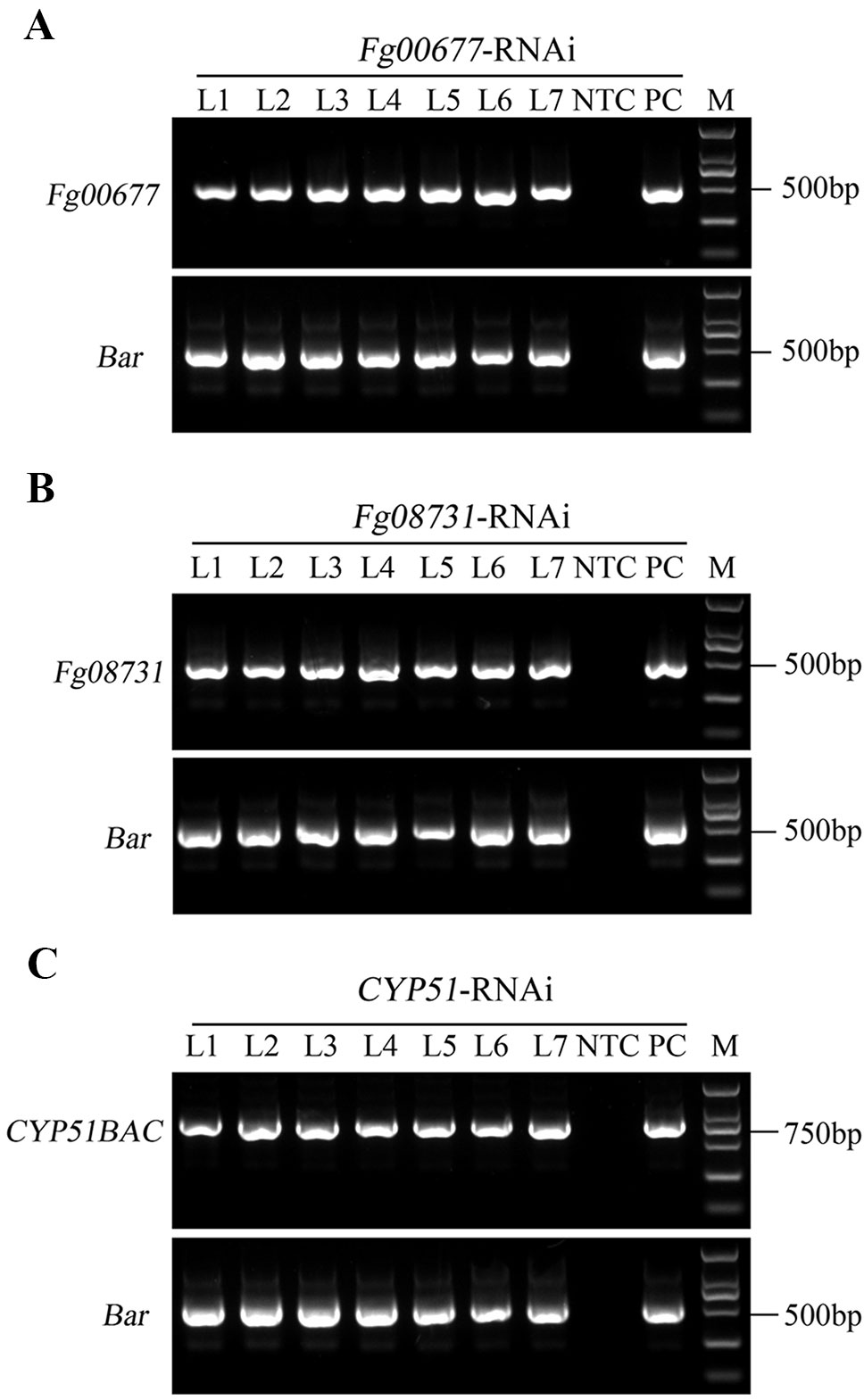
Figure 4 Molecular analysis of transgenic B. distachyon. (A) Integration of Fg00677-RNAi construct in transgenic plants analyzed by PCR amplification using Fg00677 and Bar gene-specific primers. (B) Integration of Fg08731-RNAi construct in transgenic plants analyzed by PCR amplification using Fg08731 and Bar gene-specific primers. (C) Integration of CYP51-RNAi construct in transgenic plants analyzed by PCR amplification using CYP51BAC and Bar gene-specific primers. The genomic DNA from non-transformed control (NTC) plants was used as negative control. The plasmid DNA, including FG00677-RNAi construct, FG08731-RNAi construct, or CYP51-RNAi construct was used as positive control (PC); M indicates DL2000 DNA marker.
To determine the presence of homologous siRNA in transgenic Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi lines, northern blot analysis was performed using the sequence-specific probes. The result showed that the siRNA molecules derived from Fg00677-RNAi, Fg08731-RNAi, and CYP51BAC-RNAi constructs were present in the transgenic T2 lines (Fg00677-RNAi-L1, -L2, Fg08731-RNAi -L3, -L4, -L6, CYP51-RNAi -L3, -L4) prior to F. graminearum inoculation (Figure 5; Figure S3). No hybridization signal was detected in the other transgenic T2 lines (Fg00677-RNAi -L3, -L4, -L5, -L6, -L7, Fg08731-RNAi –L1, -L2, -L5, -L7 and CYP51-RNAi -L1, -L2, -L5, -L6, -L7) or non-transformed control plants (Figure S3). The results showed that some RNAi transgenic plants express enough siRNA, and the other RNAi transgenic plants could not express enough siRNAs which can be detected by northern blot. In addition, the results indicated that the dsRNA derived from respective Fg00677-RNAi, Fg08731-RNAi, or CYP51-RNAi constructs were produced in the transgenic plants and processed by the host silencing machinery into siRNA molecules. Thus, the transgenic B. distachyon lines (Fg00677-RNAi-L1, -L2, Fg08731-RNAi -L4, -L6, CYP51-RNAi -L3, -L4) which produced more siRNAs were selected for further analysis.
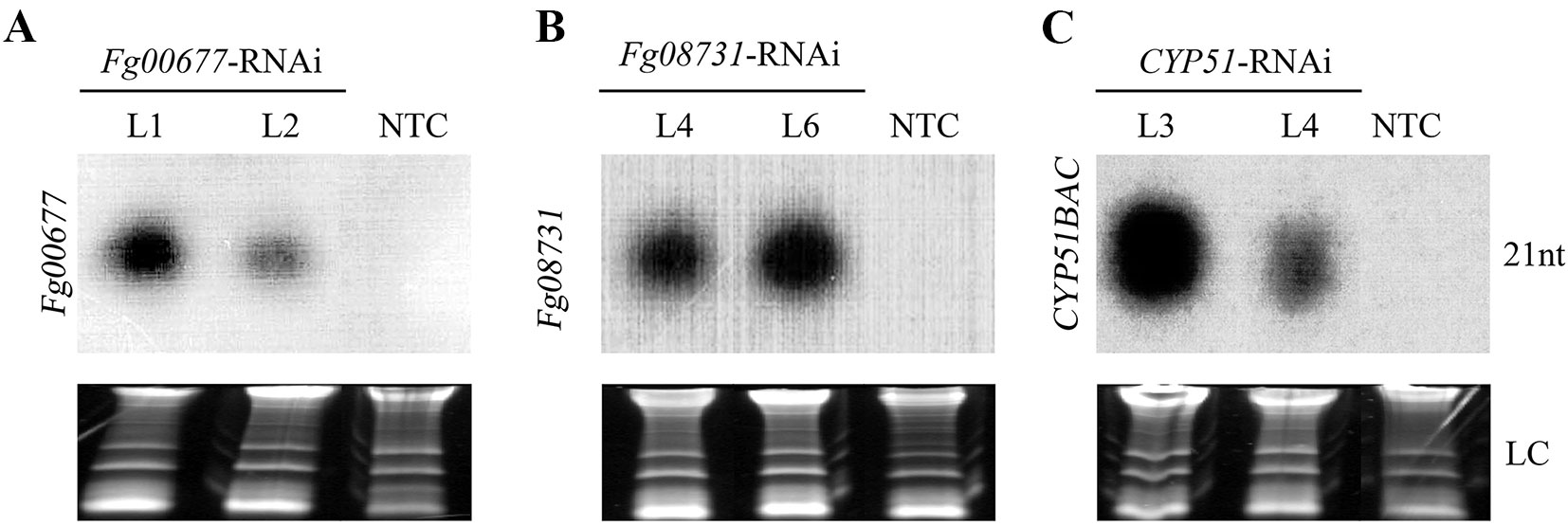
Figure 5 siRNA detection in transgenic B. distachyon lines. Northern blot analysis of sequence-specific siRNA molecules derived from Fg00677 (A), Fg08731 (B), and CYP51BAC (C) RNAi fragments in the T2 transgenic B. distachyon lines with Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi constructs. Ethidium bromide-stained rRNA served as loading control (LC). NTC, non-transformed control.
The transgenic B. distachyon plants that contained RNAi constructs displayed normal morphology (Figure S4), indicating no unintended effects in the RNAi plants. Two independent transgenic lines of the T2 generations were assayed for their resistance to F. graminearum strain PH-1 wild type of Bd21-3 was also inoculated and served as the control. At the flowering stage, the spikes of T2 transgenic plants of the same lines were inoculated by single-floret injection. After inoculations, obvious differences in disease symptoms on spikes were clearly discernible between the transgenic plants and the non-transgenic plants (Figure 6A). In the transgenic plants with Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi constructs, the FHB symptom occurred only in the partial florets of the inoculated spikelet. However, symptoms appeared on the entire inoculated spikelet and at least one adjacent spikelet in the non-transformed line Bd21-3 (Figure 6A). To evaluate the extent of spike colonization, the scoring method described by Pasquet et al. (2016) was used for scoring the symptoms at 9 days post-inoculation (dpi). The transgenic B. distachyon lines (Fg00677-RNAi -L1, -L2, Fg08731-RNAi -L4, -L6, CYP51-RNAi -L3, -L4) showed average disease symptom grades of 2.20, 2.54, 2.03, 2.32, 2.42, and 2.57, respectively, significantly lower than that of Bd21-3 which exhibited an average disease symptom grade of 3.72 (Figure 6B). The biomass of F. graminearum showed a significant decrease in lines Fg00677-RNAi -L1, -L2, Fg08731-RNAi -L4, -L6, CYP51-RNAi -L3, -L4 compared with Bd21-3 (Figure 6C; Figure S5). These results indicated significant increased resistance to F. graminearum in transgenic lines compared with that in the control plants.
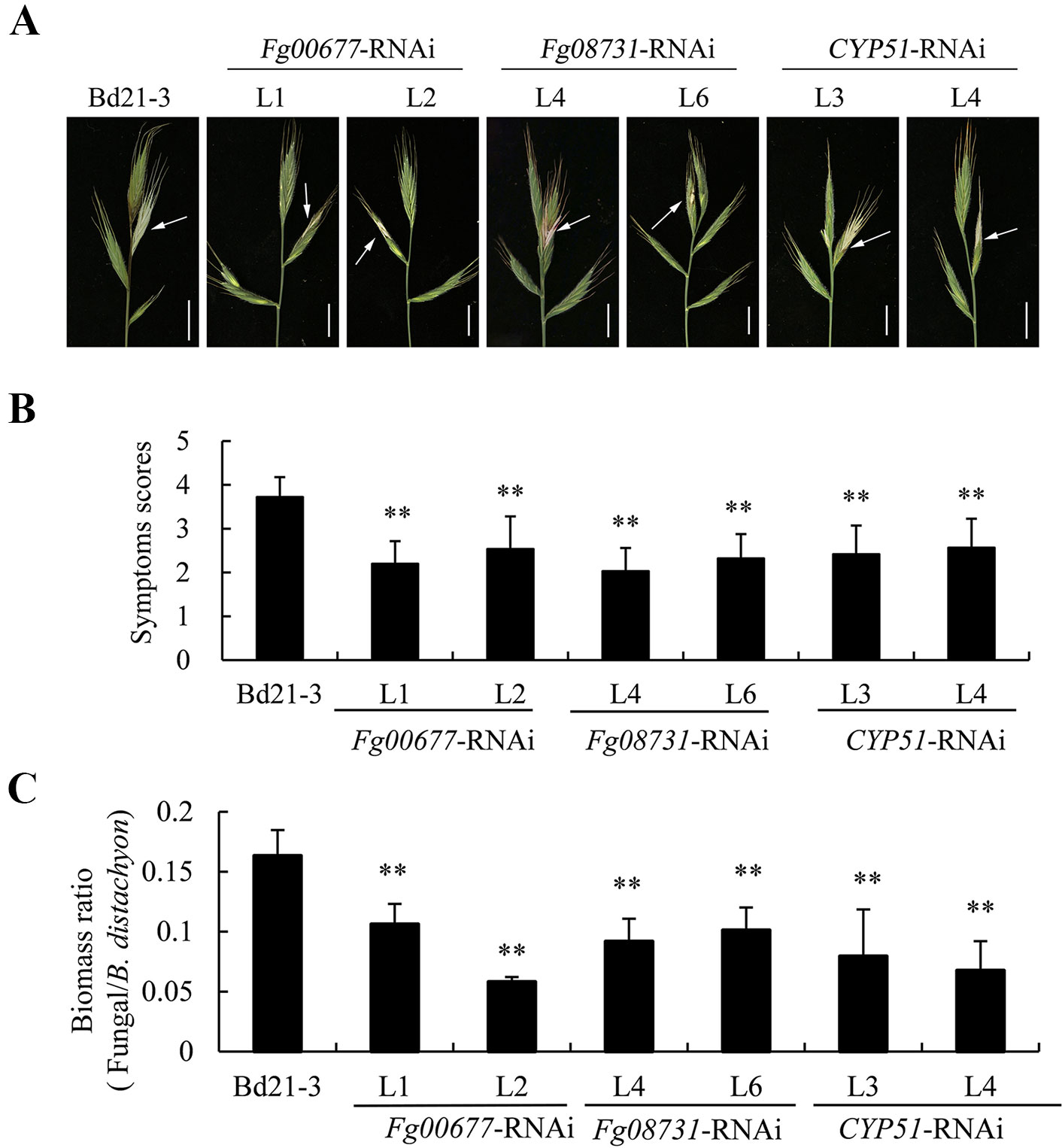
Figure 6 Evaluation of disease resistance of T2 transgenic B. distachyon lines with Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi constructs against F. graminearum. (A) The second spikelet from the top of the spike of transgenic B. distachyon lines and non-transgenic controls was point inoculated with F. graminearum PH-1 strain and photographed at 9 dpi. Bar, 1 cm. Arrows indicate the inoculation site. (B) Quantification of the disease symptoms using a scoring scale at 9 dpi. Values represent the means ± standard deviation of at least 30 plants. Similar results were obtained from three biological replicates. (C) F. graminearum and B. distachyon biomass ratio measured via total DNA content at 5 dpi by absolute quantification using the internal reference genes beta-tubulin and BdUBC18, respectively. Values represent the means ± standard deviation of three biological replicates. Differences were assessed using Student’s t tests. Double asterisks indicate P < 0.01.
To determine the effect of HIGS strategy on the targeted F. graminearum genes (Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C), we measured the relative transcript levels of these genes in the pathogens infecting transgenic lines and compared them with the levels in pathogens infecting non-transformed plants. Total RNA was isolated from infected plant tissue 72 h after inoculation with F. graminearum PH-1 and cDNA was synthesized by reverse transcription. Gene-specific primers were designed for Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C outside the fragments used in the RNAi constructs to avoid possible artifacts. The transcript levels of F. graminearum beta-tubulin were also measured and used for normalization. Analysis of two independent transgenic lines showed a clear down-regulation of Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C in F. graminearum infecting transgenic B. distachyon lines as compared to F. graminearum infecting non-transgenic controls (Figure 7). In the F. graminearum-infected seedlings, the Fg00677 transcripts were reduced by 58% and 54%, respectively, in the two Fg00677-RNAi transgenic lines L1 and L2 relative to control seedlings (Figure 7A). Similarly, the Fg08731 transcripts were reduced by 60% and 55%, respectively, in the two F. graminearum-infected Fg08731-RNAi transgenic lines L4 and L6 relative to control seedlings (Figure 7B). The CYP51A, CYP51B, and CYP51C transcripts were reduced by 78%, 77%, and 70% in the F. graminearum-infected CYP51-RNAi transgenic line L3, and reduced by 75%, 78%, and 67% in the F. graminearum-infected transgenic line L4, respectively, relative to F. graminearum-infected control seedlings (Figure 7C). The results showed that the transcript levels of Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C were reduced significantly in fungi infecting corresponding transgenic lines. The results further indicated that the significant increased resistance to F. graminearum was due to the reduced transcript levels of corresponding genes in the corresponding transgenic plants with RNAi cassette.
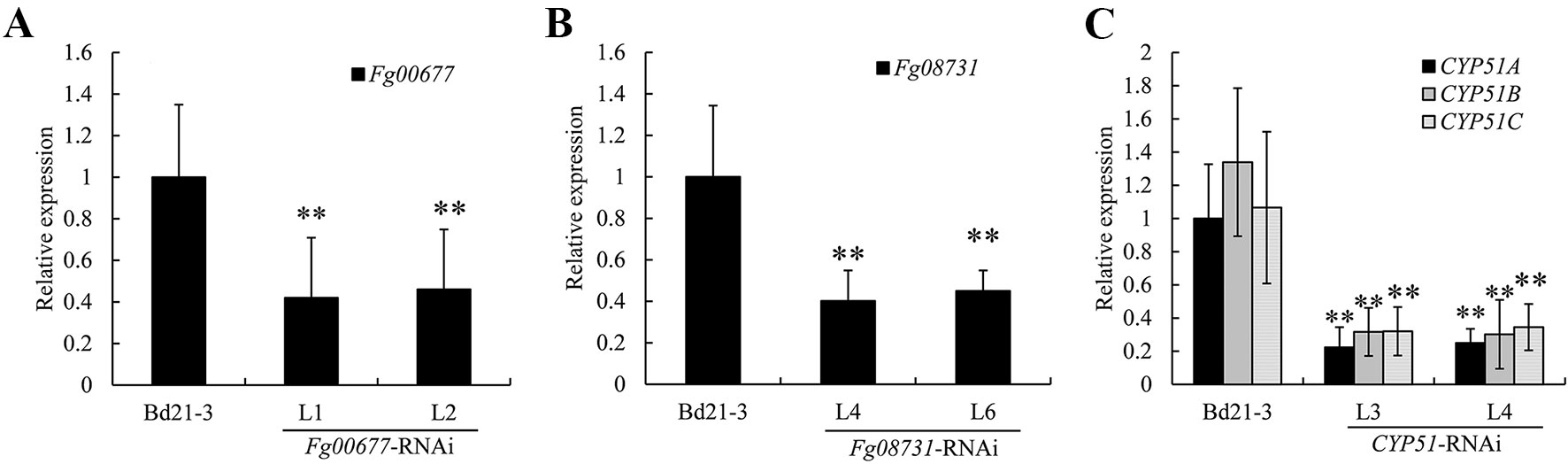
Figure 7 Relative transcript levels of Fg00677 (A), Fg08731 (B), CYP51A, CYP51B, and CYP51C (C) in F. graminearum-inoculated spikelets. The expression values were normalized to the level of the beta-tubulin of F. graminearum, and the expression level of target genes in F. graminearum-inoculated non-transformed plants set at 1. Bars represent the means ± standard deviation of three biological replicates. Differences were assessed using Student’s t test. Double asterisks indicate P < 0.01.
Genetic breeding for FHB disease resistance represents the most cost-effective control strategy. However, resistance breeding for FHB and reduced deoxynivalenol (DON) accumulation is complex and difficult, due to the limited disease-resistant germplasm sources and the rapid breakdown of resistance by new virulence races (Wegulo et al., 2015; Machado et al., 2018). Recently, HIGS has been applied in more plant pathogens, including FHB fungus F. graminearum, and provided a new strategy for the safe, pesticide-free and potentially sustainable plant protection against plant pathogen (Koch et al., 2013; Cheng et al., 2015; Chen et al., 2016). The objective of this study was to identify new effective HIGS target genes, and determine whether B. distachyon could be used as a model plant to identify the effective HIGS target of F. graminearum for using HIGS technology.
Increasing resistance against pathogens through HIGS technology in host depends on the effective HIGS targets which play a vital role in growth, development, and pathogenicity in fungi. Protein kinases play an important regulatory role in the pathogenesis and growth of plant pathogens. Earlier, it was reported that the two protein kinases encoding genes, CK2 orthologous gene Fg00677 and the CK1 orthologous gene Fg08731, are essential in F. graminearum, because deletion of them is lethal (Wang et al., 2011). CK2 and CK1 orthologous genes also play important roles in other organisms, especially in fungi. CK2 is involved in numerous cellular processes, including cellular morphology, signal transduction, ion homeostasis, cell viability, cellular polarity, and cell cycle progression (Tripodi et al., 2007; Lei et al., 2014; Wang et al., 2015). CK2 is essential for regulating cell cycle progression and proliferation in S. cerevisiae and S. pombe (Snell and Nurse, 1994; Rethinaswamy et al., 1998), and is required for growth and development in two filamentous fungi: N. crassa and A. nidulans with mutants resulting in microcolony and aerial mycelium-defective phenotype (Yang et al., 2002; He et al., 2006; Mehra et al., 2009; de Souza et al., 2013). CK1 enzymes have important roles in regulating several cellular processes, such as cell cycle, cell division, and cell differentiation, and the circadian clock in mammals, Drosophila, and S. cerevisiae (Brockman et al., 1992; Eide et al., 2005; Knippschild et al., 2005). CK1 orthologs characterized in S. cerevisiae, A. nidulans, and Neurospora crassa are proven to be essential for growth and development (Hoekstra et al., 1991; Hoekstra et al., 1994; Zhou and Elledge, 2000; Park et al., 2011; Apostolaki et al., 2012). Therefore, we selected these two genes for further analysis. We obtained the full-length ORFs of Fg00677 and Fg08731 from F. graminearum by PCR. Sequence analysis indicated that Fg00677 and Fg08731 contain the STKc_CK2_alpha conserved domain and STKc_CK1_delta_epsilon conserved domain, respectively. Multiple-alignment and phylogenetic analysis indicated that Fg00677 and Fg08731 are highly conserved in filamentous fungi. It is well known that the evolutionarily conserved proteins will play the critical functions usually. Therefore, Fg00677 and Fg08731 may be the effective HIGS target, which were identified using HIGS technology subsequently.
RNAi refers to a biological phenomenon that is capable of causing sequence-specific inhibition of gene expression at the transcriptional, post-transcriptional, or translational levels (Baulcombe, 2015). RNAi exists in plants, and the RNAi mechanism is conserved and widely involved in the growth and development of organisms (Plasterk, 2002). RNAi also exists in fungi, and the Dicer-dependent RNAi machinery was discovered in F. graminearum (Chen et al., 2015; Son et al., 2017). HIGS relies on the plant’s RNAi system to produce silencing molecules (dsRNA and siRNAs) which can be translocated into fungus through unclear mechanism, then relies on the interacting RNAi system of the fungus to down-regulate the expression of fungal genes (Cai et al., 2018). In this study, the siRNAs complementary to the respective fungal gene were detected by northern blot in the transgenic B. distachyon carrying the respective RNAi construct, indicating that dsRNAs were produced through transcription in the transgenic B. distachyon, and the dsRNAs were processed into siRNAs by plant’s RNAi system. The endogenous transcripts of the HIGS target genes (Fg00677, Fg08731, CYP51A, CYP51B, and CYP51C) were reduced in F. graminearum colonizing transgenic B. distachyon spikes carrying the respective RNAi construct, suggesting that the dsRNAs or siRNAs produced by transgenic B. distachyon may be translocated into the fungal cells during the infection process and serve as the silencing molecules that initiated in vivo post-transcriptional gene silencing in the colonizing fungus.
The reduction of the respective gene’s transcripts in colonizing F. graminearum by transgenic B. distachyon T2 lines expressing HIGS cassettes correlates with significant protection of B. distachyon against the disease. The resistance of Fg00677-RNAi and Fg08731-RNAi T2 lines reached the level equivalent to CYP51-RNAi lines. The engineered resistance trait existed in the T2 generation, indicating that the disease resistance traits can be transmitted to next generations. The HIGS cassettes of Fg00677 and Fg08731 do not contain an effective target of host genes in the B. distachyon genomic sequence according to the off-target prediction tool; thus, the probability of off-target silencing of the host gene is low. In this study, we did not observe a difference in growth status between the transgenic B. distachyon plants and the wild type Bd21-3. Although the expression levels of Fg08731 and Fg00677 were reduced partially in colonizing F. graminearum, the FHB resistance was enhanced significantly, indicating that the degree of silencing by HIGS is sufficient to attenuate pathogenicity of F. graminearum. Although Fg00677 and Fg08731 are essential in F. graminearum, we did not observe complete resistance to F. graminearum as the smaller browning or bleaching lesion occurred on the inoculated spikelet. This may be due to some degree of pathogenicity conferred by the residual transcripts, indicating that exploring HIGS technology for effectively improving disease resistance requires sufficient silencing of the target genes of phytopathogens. To increase the degree of resistance to pathogens, multiple genes can be silenced. Koch et al. (2013) and our study found that simultaneously silencing all three CYP51 genes in F. graminearum by HIGS completely eliminates fungal pathogenicity. Therefore, simultaneously silencing Fg00677 and Fg08731 may confer the enhanced resistance to F. graminearum. In conclusion, Fg00677 and Fg08731 are effective HIGS targets which can be used for improving resistance against FHB disease in wheat and other cereal crops.
The key genes of plant pathogen identified by knock-out technology were considered as candidate HIGS targets. However, whether these genes can be silenced effectively using HIGS technology is uncertain as the current mechanism of HIGS is not yet clear. Therefore, the candidate HIGS target genes need be validated in a plant-pathogen interaction system before being applied in disease-resistant breeding in wheat. However, it is not feasible to use transgenic wheat in screening of effective HIGS targets, because wheat genetic transformation is time consuming and costly, and wheat has a long growth cycle. B. distachyon-F. graminearum interactions closely model the head blight in wheat and barley caused by F. graminearum (Peraldi et al., 2011). B. distachyon as a grass crop, may be used as alternative plant for batch screening of HIGS targets, due to the shorter growth cycle and easier genetic transformation compared with wheat and other cereal crops. Koch et al. (2013) showed that HIGS targeting of the CYP51 genes rendered susceptible plants highly resistant to F. graminearum infection in Arabidopsis and barley. In this study, HIGS of CYP51 genes also confers efficient resistance against F. graminearum infection in B. distachyon. In addition, HIGS of two essential protein kinase encoding genes (Fg00677 and Fg08731) enhanced the resistance against F. graminearum infection in B. distachyon, significantly. Our data indicated that HIGS can be used in B. distachyon-F. graminearum interactions, and B. distachyon can be used as a model plant for identifying the effective HIGS targets of F. graminearum, with providing time and money saving benefits compared with wheat.
All datasets for this study are included in the article/Supplementary Material.
JG and ZK designed the experiment. FH, RZ, and JZ performed the experiments and analyzed the data. FH, TQ, and JG wrote the manuscript.
This study was supported by the National Key R&D Program of China (2018YFD0200400), Natural Science Basic Research Plan in Shaanxi Province of China (2017JM3007), and National Natural Science Foundation of China (31620103913).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01362/full#supplementary-material
Figure S1 | Amino acid sequence alignments of Fg00677 (A) and Fg08731 (B) with other fungal homologs. Amino acid identity (black boxes) and similarity (gray boxes) are shown within the protein kinase domain. The active sites are highlighted with a filled triangle. The ATP binding site is indicated with an arrow. Asterisks indicate the activation loop. Thick black horizontal line indicates the conserved domain of the corresponding protein kinase.
Figure S2 | The nucleotide sequences of Fg00677 and Fg08731 used for RNAi constructs. The full-length sequences represent the ORFs of Fg00677 (A) and Fg08731 (B). The nucleotides highlighted with bulue color of Fg00677 (A) and Fg08731 (B) were used for RNAi constructs.
Figure S3 | siRNA detection in transgenic B. distachyon lines. Northern blot analysis of sequence-specific siRNA molecules derived from Fg00677, Fg08731, and CYP51BAC RNAi fragments in the T2 transgenic B. distachyon lines with Fg00677-RNAi, Fg08731-RNAi, CYP51-RNAi constructs. Ethidium bromide-stained rRNA served as loading controls (LC). NTC, non-transformed control.
Figure S4 | Phenotypes of the Fg00677-RNAi, Fg08731-RNAi, and CYP51-RNAi lines. Transgenic lines compared with the wild-type Bd21-3 at 28 days (A) and at 40 days after sowing (B). Bars, 5 cm.
Figure S5 | The standard curve generated for the absolute quantification of F. graminearum and B. distachyon. The fragments of beta-tubulin and BdUBC18 were fused to the pMD18-T vector to generate the templates. Threshold cycles were plotted against the quantity of the templates. (A) Standard curve for beta-tubulin generated by using the quantity of the templates (101, 102, 103, 104, 105, 106, 107, 108). (B) Standard curve for BdUBC18 generated by using the quantity of the templates (104, 105, 106, 107, 108).
Table S1 | List of primers used in this study.
Table S2 | Prediction of off-target transcripts for Fg00677 and Fg08731.
Apostolaki, A., Harispe, L., Calcagno-Pizarelli, A. M., Vangelatos, I., Sophianopoulou, V., Arst, H. N., et al. (2012). Aspergillus nidulans CkiA is an essential casein kinase I required for delivery of amino acid transporters to the plasma membrane. Mol. Microbiol. 84, 530–549. doi: 10.1111/j.1365-2958.2012.08042.x
Baulcombe, D. C. (2015). VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 26, 141–146. doi: 10.1016/j.pbi.2015.06.007
Brockman, J. L., Gross, S. D., Sussman, M. R., Anderson, R. A. (1992). Cell cycle-dependent localization of casein kinase I to mitotic spindles. Proc. Natl. Acad. Sci. U. S. A. 89, 9454–9458. doi: 10.1073/pnas.89.20.9454
Cai, Q., He, B., Kogel, K.-H., Jin, H. (2018). Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 46, 58–64. doi: 10.1016/j.mib.2018.02.003
Carthew, R. W. (2001). Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 13, 244–248. doi: 10.1016/S0955-0674(00)00204-0
Chen, C., Wang, J., Luo, Q., Yuan, S., Zhou, M. (2007). Characterization and fitness of carbendazim-resistant strains of Fusarium graminearum (wheat scab). Pest Manag. Sci. 63, 1201–1207. doi: 10.1002/ps.1449
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
Chen, Y., Gao, Q., Huang, M., Liu, Y., Liu, Z., Liu, X., et al. (2015). Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 5, 12500. doi: 10.1038/srep12500
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., Qiu, X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. doi: 10.1111/pbi.12352
de Souza, C. P., Hashmi, S. B., Osmani, A. H., Andrews, P., Ringelberg, C. S., Dunlap, J. C., et al. (2013). Functional analysis of the Aspergillus nidulans kinome. PLoS One 8, e58008. doi: 10.1371/journal.pone.0058008
Draper, J., Mur, L. A., Jenkins, G., Ghosh-Biswas, G. C., Bablak, P., Hasterok, R., et al. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127, 1539–1555. doi: 10.1104/pp.010196
Duvick, J., Rood, T., Rao, A. G., Marshak, D. R. (1992). Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. J. Biol. Chem. 267, 18814–18820.
Eide, E. J., Kang, H., Crapo, S., Gallego, M., Virshup, D. M., (2005). “Casein kinase I in the mammalian circadian clock,” in Methods in enzymology (Elsevier), 408–418. doi: 10.1016/S0076-6879(05)93019-X
Fagard, M., Boutet, S., Morel, J. B., Bellini, C., Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. U. S. A. 97, 11650–11654. doi: 10.1073/pnas.200217597
Ghag, S. B., Shekhawat, U. K., Ganapathi, T. R. (2014). Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. doi: 10.1111/pbi.12158
Ghildiyal, M., Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
He, Q., Cha, J., He, Q., Lee, H. C., Yang, Y., Liu, Y. (2006). CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20, 2552–2565. doi: 10.1101/gad.1463506
Hoekstra, M., Dhillon, N., Carmel, G., DeMaggio, A., Lindberg, R., Hunter, T., et al. (1994). Budding and fission yeast casein kinase I isoforms have dual-specificity protein kinase activity. Mol. Biol. Cell 5, 877–886. doi: 10.1091/mbc.5.8.877
Hoekstra, M. F., Liskay, R. M., Ou, A. C., DeMaggio, A. J., Burbee, D. G., Heffron, F. (1991). HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science 253, 1031–1034. doi: 10.1126/science.1887218
Hormaza, J. (2002). Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor. Appl. Genet. 104, 321–328. doi: 10.1007/s001220100684
Hu, Z., Parekh, U., Maruta, N., Trusov, Y., Botella, J. R. (2015). Down-regulation of Fusarium oxysporum endogenous genes by host-delivered RNA interference enhances disease resistance. Front. Chem. 3, 1. doi: 10.3389/fchem.2015.00001
Ketting, R. F. (2011). The many faces of RNAi. Dev. Cell 20, 148–161. doi: 10.1016/j.devcel.2011.01.012
Knippschild, U., Gocht, A., Wolff, S., Huber, N., Löhler, J., Stöter, M. (2005). The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17, 675–689. doi: 10.1016/j.cellsig.2004.12.011
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U. S. A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Lazo, G. R., Stein, P. A., Ludwig, R. A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Nat. Biotechnol. 9, 963–967. doi: 10.1038/nbt1091-963
Lei, Y., Liu, G., Li, Z., Gao, L., Qin, Y., Qu, Y. (2014). Functional characterization of protein kinase CK2 regulatory subunits regulating Penicillium oxalicum asexual development and hydrolytic enzyme production. Fungal Genet. Biol. 66, 44–53. doi: 10.1016/j.fgb.2014.02.007
Liu, Q., Paroo, Z. (2010). Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 79, 295–319. doi: 10.1146/annurev.biochem.052208.151733
Liu, S., Hall, M. D., Griffey, C. A., Mckendry, A. L. (2009). Meta-analysis of QTL associated with Fusarium head blight resistance in wheat. Crop Sci. 49, 1955–1968. doi: 10.2135/cropsci2009.03.0115
Loffler, M., Schon, C., Miedaner, T. (2009). Revealing the genetic architecture of FHB resistance in hexaploid wheat (Triticum aestivum L.) by QTL meta-analysis. Mol. Breed. 23, 473–488. doi: 10.1007/s11032-008-9250-y
Machado, A. K., Brown, N. A., Urban, M., Kanyuka, K., Hammond-Kosack, K. E. (2018). RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 74, 790–799. doi: 10.1002/ps.4748
Mehra, A., Shi, M., Baker, C. L., Colot, H. V., Loros, J. J., Dunlap, J. C. (2009). A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137, 749–760. doi: 10.1016/j.cell.2009.03.019
Mur, L. A. J., Allainguillaume, J., Catalan, P., Hasterok, R., Jenkins, G., Lesniewska, K., et al. (2011). Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytol. 191, 334–347. doi: 10.1111/j.1469-8137.2011.03748.x
Nunes, C. C., Dean, R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. doi: 10.1111/j.1364-3703.2011.00766.x
Osborne, L. E., Stein, J. M. (2007). Epidemiology of Fusarium head blight on small-grain cereals. Int. J. Food Microbiol. 119, 103–108. doi: 10.1016/j.ijfoodmicro.2007.07.032
Panwar, V., Jordan, M., McCallum, B., Bakkeren, G. (2018). Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 16, 1013–1023. doi: 10.1111/pbi.12845
Park, G., Servin, J. A., Turner, G. E., Altamirano, L., Colot, H. V., Collopy, P., et al. (2011). Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot. Cell 10, 1553–1564. doi: 10.1128/EC.05140-11
Pasquet, J. C., Changenet, V., Macadré, C., Boex-Fontvieille, E., Soulhat, C., Bouchabké-Coussa, O., et al. (2016). A Brachypodium UDP-glycosytransferase confers root tolerance to deoxynivalenol and resistance to Fusarium infection. Plant Physiol. 172, 559–574. 00371.02016. doi: 10.1104/pp.16.00371
Patil, B. L., Ogwok, E., Wagaba, H., Mohammed, I. U., Yadav, J. S., Bagewadi, B., et al. (2011). RNAi-mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. Plant Pathol. 12, 31–41. doi: 10.1111/J.1364-3703.2010.00650.X
Peraldi, A., Beccari, G., Steed, A., Nicholson, P. (2011). Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol. 11, 100. doi: 10.1186/1471-2229-11-100
Plasterk, R. H. (2002). RNA silencing: the genome’s immune system. Science 296, 1263–1265. doi: 10.1104/pp.010196
Qi, T., Zhu, X., Tan, C., Liu, P., Guo, J., Kang, Z., et al. (2018). Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 16, 797–807. doi: 10.1111/pbi.12829
Rethinaswamy, A., Birnbaum, M. J., Glover, C. V. (1998). Temperature-sensitive mutations of the CKA1 gene reveal a role for Casein Kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J. Biol. Chem. 273, 5869–5877. doi: 10.1074/jbc.273.10.5869
Sandoya, G. V., de Oliveira Buanafina, M. M. (2014). Differential responses of Brachypodium distachyon genotypes to insect and fungal pathogens. Physiol. Mol. Plant Pathol. 85, 53–64. doi: 10.1016/j.pmpp.2014.01.001
Saurabh, S., Vidyarthi, A. S., Prasad, D. (2014). RNA interference: concept to reality in crop improvement. Planta 239, 543–564. doi: 10.1007/s00425-013-2019-5
Snell, V., Nurse, P. (1994). Genetic analysis of cell morphogenesis in fission yeast–a role for casein kinase II in the establishment of polarized growth. EMBO J. 13, 2066–2074. doi: 10.1002/j.1460-2075.1994.tb06481.x
Son, H., Park, A. R., Lim, J. Y., Shin, C., Lee, Y. W. (2017). Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet. 13, e1006595. doi: 10.1371/journal.pgen.1006595
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tripodi, F., Zinzalla, V., Vanoni, M., Alberghina, L., Coccetti, P. (2007). In CK2 inactivated cells the cyclin dependent kinase inhibitor Sic1 is involved in cell-cycle arrest before the onset of S phase. Biochem. Biophys. Res. Commun. 359, 921–927. doi: 10.1016/j.bbrc.2007.05.195
Vogel, J., Hill, T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 27, 471–478. doi: 10.1007/s00299-007-0472-y
Wang, B., Wang, N., Song, N., Wang, W., Wang, J., Wang, X., et al. (2017). Overexpression of AtPAD4 in transgenic Brachypodium distachyon enhances resistance to Puccinia brachypodii. Plant Biol. 19, 868–874. doi: 10.1111/plb.12616
Wang, C., Zhang, S., Hou, R., Zhao, Z., Zheng, Q., Xu, Q., et al. (2011). Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 7, e1002460. doi: 10.1371/journal.ppat.1002460
Wang, M., Yang, H., Zhang, M., Liu, K., Wang, H., Luo, Y., et al. (2015). Functional analysis of Trichoderma reesei CKIIα2, a catalytic subunit of casein kinase II. Appl. Microbiol. Biotechnol. 99, 5929–5938. doi: 10.1007/s00253-015-6544-y
Wegulo, S. N., Baenziger, P. S., Nopsa, J. H., Bockus, W. W., Hallen-Adams, H. (2015). Management of Fusarium head blight of wheat and barley. Crop Prot. 73, 100–107. doi: 10.1016/j.cropro.2015.02.025
Yang, Y., Cheng, P., Liu, Y. (2002). Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev. 16, 994–1006. doi: 10.1101/gad.965102
Zhang, J., Wang, J., Gong, A., Chen, F., Song, B., Li, X., et al. (2013). Natural occurrence of Fusarium head blight, mycotoxins and mycotoxin-producing isolates of Fusarium in commercial fields of wheat in Hubei. Plant Pathol. 62, 92–102. doi: 10.1111/j.1365-3059.2012.02639.x
Zhou, B. B. S., Elledge, S. J. (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. doi: 10.1038/35044005
Keywords: Brachypodium distachyon, Fusarium graminearums, CYP51, essential protein kinase, host-induced gene silencing (HIGS)
Citation: He F, Zhang R, Zhao J, Qi T, Kang Z and Guo J (2019) Host-Induced Silencing of Fusarium graminearum Genes Enhances the Resistance of Brachypodium distachyon to Fusarium Head Blight. Front. Plant Sci. 10:1362. doi: 10.3389/fpls.2019.01362
Received: 30 April 2019; Accepted: 03 October 2019;
Published: 30 October 2019.
Edited by:
Christopher Rensing, Fujian Agriculture and Forestry University, ChinaReviewed by:
Guus Bakkeren, Agriculture and Agri-Food Canada, CanadaCopyright © 2019 He, Zhang, Zhao, Qi, Kang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Guo, Z3VvanVud2dxQG53c3VhZi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.