
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 October 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01348
 Yangyang Diao1†
Yangyang Diao1† Jingjing Zhan1†
Jingjing Zhan1† Yanyan Zhao1
Yanyan Zhao1 Lisen Liu1
Lisen Liu1 Peipei Liu2
Peipei Liu2 Xi Wei1
Xi Wei1 Yanpeng Ding2
Yanpeng Ding2 Muhammad Sajjad1
Muhammad Sajjad1 Wei Hu1
Wei Hu1 Peng Wang1
Peng Wang1 Xiaoyang Ge1*
Xiaoyang Ge1*Transcription factors (TFs) and transcriptional regulators are important switches in transcriptional networks. In recent years, the transcriptional regulator TIE1 (TCP interactor containing EAR motif protein 1) was identified as a nuclear transcriptional repressor which regulates leaf development and controls branch development. However, the function and regulatory network of GhTIE1 has not been studied in cotton. Here, we demonstrated that GhTIE1 is functionally conserved in controlling shoot branching in cotton and Arabidopsis. Overexpression of GhTIE1 in Arabidopsis leads to higher bud vigor and more branches, while silencing GhTIE1 in cotton reduced bud activity and increased branching inhibition. Yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays showed that GhTIE1 directly interacted with subclass II TCPs (GhBRC1, GhBRC2, and GhTCP13) in vivo and in vitro. Overexpression of GhBRC1, GhBRC2, and GhTCP13 in mutant brc1-2 partially rescued the mutant phenotype and decreased the number of branches, showing that these TCPs are functionally redundant in controlling branching. A transient dual-luciferase reporter assay indicated that GhTIE1 repressed the protein activity of GhBRC1 and GhTCP13, and thereby decreased the expression of their target gene GhHB21. Gene expression level analysis in GhTIE1-overexpressed and silenced plants also proved that GhTIE1 regulated shoot branching via repressing the activity of BRC1, HB21, HB40, and HB53. Our data reveals that shoot branching can be controlled via modulation of the activity of the TIE1 and TCP proteins and provides a theoretical basis for cultivating cotton varieties with ideal plant types.
Branch development plays a decisive role in controlling aboveground plants, and the aboveground parts of crops are closely related to growth and yield (Kebrom et al., 2012). The development of shoots begins with axillary meristem and then develops into small shoots (McSteen and Leyser, 2005; Pasternak et al., 2019; Wang et al., 2019). The regulatory network consists of plant hormones and transcription factors (TFs) that can break the dormancy of small shoots and eventually develop into lateral branches (Rameau et al., 2015; Wang and Jiao, 2018). Cotton is an infinitely growing crop with a complex branching pattern consisting of fruit branches, vegetative shoots, and axillary buds. Developing and cultivating the ideal plant type not only affects cotton production but also saves a lot of labor. Therefore, the study of cotton branching control provides an important theoretical basis for improvement of cotton plants.
At present, with increased research on plant branch development, TCP (TEOSINTE BRANCHED1 (TB1) from maize, BRC1 from Arabidopsis, and PROLIFERATING CELL FACTOR from rice) family TFs regulating branch development have been successively discovered. TB1 was first found to be a key gene controlling maize branching (Dong et al., 2017); subsequently, the homologous genes of TB1 in other species, such as rice FINE CULM 1/OsTB1 (Takeda et al., 2010), ArabidopsisBRANCHED1 (BRC1) (Aguilar-Martínez et al., 2007), pea PsBRC1 (Nils et al., 2012), potato BRANCHED1 (Nicolas et al., 2015), and tomato BRC1-like genes (Martı´n-Trillo et al., 2011) were identified. Similar to corn TB1, overexpression of these genes inhibits branching, while loss-of-function mutations lead to increased branching (Gaudin et al., 2014; Dong et al., 2017). Transgenic rice plants overexpressing OsTB1 showed significantly reduced branching (Takeda et al., 2010). The Arabidopsis thalianaBRC1 gene was mainly expressed in axillary buds (axillary meristem, bud primordium, and nourishing vascular tissue), and the expression level decreased with the growth of buds (Finlayson, 2007). The loss-of-function BRC1 mutant brc1-2 is characterized by an accelerated onset of axillary meristems with faster shoot development and more branches. Related studies have also demonstrated that BRC1 is an integrator that controls the internal and external factors of bud activity (Aguilar-Martínez et al., 2007). Hormone strigolactone positively regulates BRC1 at the transcriptional level and inhibits plant branching (Chevalier et al., 2014; Minakuchi et al., 2010). Cytokinin negatively regulates BRC1 expression, thereby promoting branch growth in rice and peas (Arite et al., 2010; Takeda et al., 2010; Dun et al., 2012; Nils et al., 2012).
In recent years, the network consisting of upstream genes regulating BRC1 and downstream targets of BRC1 has been revealed. Rice TF IPA1 acts as an upstream gene, which promotes the expression of OsTB1 by directly binding to the promoter region (Lu et al., 2013). BRC1 downstream target genes were initially discovered, including three HD-ZIP TFs—HOMEOBOX protein (HB) 21, HB40, and HB53—which are directly regulated by BRC1 in Arabidopsis (González-Grandío et al., 2017). Recently, a new transcriptional regulator, TIE1 (TCP interactor containing EAR motif protein 1), was identified to control Arabidopsis branching by controlling the activity of BRC1 protein and then repressing the expression of BRC1 target genes HB21, HB40, and HB53. In the development of branching, overexpression of TIE1 results in higher shoot vigor and more branches, while disruption of TIE1 reduces shoot activity and increases branch inhibition (Tao et al., 2013; Yang et al., 2018).
Studies have shown that the developmental control of branches in different species is both conserved and specific (Wang et al., 2018). There are, however, no studies on the function of GhTIE1 in regulating cotton branching. Our research indicates that GhTIE1 is a transcriptional repressor that is expressed in both the nucleus and cytoplasm. Arabidopsis overexpressing GhTIE1 produces more branches, and gene silencing of GhTIE1 leads to decreased branching in cotton. GhBRC1, GhBRC2, and GhTCP13 interact with GhTIE1in vitro and in vivo. Overexpression of GhBRC1, GhBRC2, and GhTCP13 in the mutant brc1-2 partially rescued the branching phenotype, suggesting their redundant function in controlling branching. Expression level analysis supported that GhTIE1 indirectly inhibits the expression of BRC1 target genes HB21, HB40, and HB53 by inhibiting the activity of BRC1. The results of our study show that GhTIE1 is functionally conserved in controlling branching in cotton and Arabidopsis, and shoot branching was controlled by modulating the activity of the TIE1 and TCP proteins.
The cotton materials used in this study were Gossypium hirsutum cultivar ZM24. Cotton seeds were immersed in sterile H2O for 24 h, then planted in mixed soils including trophic soils and vermiculite (1:1, v/v), and grown with regular watering under adjusted conditions of 27/20°C, 14/10 h, and humidity of 75%.
Arabidopsisthaliana ecotype Columbia-0 (Col-0) was used in this study. The mutant brc1-2 was obtained from the laboratory of Dr Genji Qin. The Arabidopsis seeds were sterilized by commercially diluted (1:1, v/v) NaOCl, followed by several rinses with sterile water. Germination was carried out on sterile Murashige and Skoog (MS) medium. After 7 days, seedlings were transferred to soil and grown at 22°C under long-day conditions (16-h light and 8-h dark, 70% relative humidity).
We obtained the apical new tissue of cotton, including stem, leaves, and axillary buds. For A. thaliana, we used the rosette disc with a bit of the stem and then extracted RNA using the RNAprep Pure Plant Kit (Tiangen). We then used the PrimeScript RT reagent Kit with gDNA Eraser (Takara) to produce cDNAs.
For RT-PCR, the following parameters were used: 94°C for 5 min, 27 cycles at 98°C for 15 s, 60°C for 15 s, and 68°C for 30 s. Diluted cDNA was used for quantitative RT-PCR (qRT-PCR) with SYBR Premix Ex Taq (Tli RNaseH Plus, TAKARA) on an ABI 7900 qRT-PCR System (Applied Biosystems). A three-step method was used with the following PCR conditions: 40 cycles at 95°C for 30 s, 95°C for 5 s, and 60°C for 30 s. We checked the dissociation curves of each reaction and used the cycle threshold (CT) 2–∆∆Ct method to calculate the expression level of each target gene (Livak and Schmittqen, 2001). Each reaction was performed with at least three biological replicates. All the primers used in this study are listed in Supplementary Table 1. All the qPCR data are showed in Supplementary Table 2.
In our study, we constructed all vectors using a one-step cloning method. This method uses a primer with a homologous arm and a double restriction site to amplify the gene fragment, which is then ligated to the vector with the same homology arm and restriction site. All vectors were converted to Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method.
To construct the overexpression vector, the primers 35S-TIE1-F/R and 35S-BRC1-F/R (Supplementary Table 1) were used to ligate the amplified gene to a pCAMBIA2300 vector. A. thaliana inflorescences were immersed in a suspension of Agrobacterium to obtain transgenic A. thaliana (Clough and Bent, 1998).
For the VIGS experiments, the primers VTIE1-F/R and VBRC1-F/R (Supplementary Table 1) were used. A conserved 251-bp fragment of GhTIE1a and GhTIE1d was cloned into a pCLCrVA vector to generate pCLCrV::GhTIE1, and pCLCrV-CHL1 was used as a positive control. The constructs were then transformed to GV3101. The cotyledons of 7-day-old ZM24 cotton seedlings were injected with equal amounts of Agrobacterium expressing the CLCrV vectors. Two-week-old cotton leaves were sampled for real-time PCR to check the interference efficiency.
Analysis of the amino acid sequence of GhTIE1 identified the nuclear signal peptide. In order to verify whether it is expressed in the nucleus, we first cloned the gene with the primer QBV3-GhTIE1-F/R, then ligated it to the vector QBV3, and finally constructed the vector into the YFP fluorescent tag-containing vector pEG101 by an LR reaction to obtain the fused expression vector GhTIE1-YFP (Qian et al., 2013).
To confirm the cellular localization of GhTIE1, we injected tobacco leaves with Agrobacterium containing the GhTIE1-YFP vector to detect transient expression. After 48 h of inoculation, the infiltrated Nicotiana benthamiana leaves were observed by a laser scanning confocal microscope (OLYMPUS FV1200 confocal microscope). Localization of GhBRC1, GhBRC2, and GhTCP13 was obtained using the same method. Sequences of the genes in this study were retrieved from the CottonGen database (https://www.cottongen.org/) (Zhang et al., 2015).
To examine the interaction of TIE1 with BRC1, BRC2, and GhTCP13, we amplified the coding sequences for TIE1 and BRC1 and for BRC2 and GhTCP13 using BDTIE1 F/R, ADBRC1-F/R, ADBRC2-F/R, and ADTCP13-F/R, respectively (Supplementary Table 1). The bait structure BD-TIE1 was cloned into the BD-pGBKT7 vector, and the prey structures AD-BRC1, AD-BRC2, and AD-GhTCP13 were ligated to the AD-pGADT7 vector. The bait structure and each prey were co-transformed into yeast strain AH109. Glucose-free medium (SD/-2) without Leu and Trp and glucose-free medium (SD/-4) without Leu, Trp, His, and Ade were used for culturing all transformed yeast cells.
The vectors pXY104 and pXY106 were used to produce constructs for the bimolecular fluorescence complementation (BiFC) assays, and they carried fragments encoding the C- and N-terminal halves of YFP (cYFP and nYFP), respectively (Yu et al., 2008). The cDNA fragments encoding GhBRC1 were fused to the fragment encoding the C-terminus of YFP, and the fragments encoding GhTIE1 were fused to the fragment encoding the N-terminus of YFP, then transformed into Agrobacterium strain GV3101. GV3101 cultures harboring constructs expressing nYFP fusion proteins and cYFP fusion proteins were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves through infiltration. YFP signals were detected by confocal microscopy as described above. For the luciferase (LUC) assay, a 1,500-bp promoter region of GhHB21 was constructed into a vector pGWB435 containing a Luc reporter gene to construct the reporter GhHB21pro-LUC. GhHB21pro-LUC and 35S-pCAMBIA2300-GFP were co-infiltrated into N. benthamiana leaves as a positive control; GhHB21pro-LUC, 35S-pCAMBIA2300-GhBRC1, and 35S-pCAMBIA2300-GhTIE1 were co-infiltrated into N. benthamiana leaves to confirm whether GhTIE1 represses the GhBRC1 activity; and 35S-pCAMBIA2300-GhTCP13, GhHB21pro-LUC, and 35S-pCAMBIA2300-GhTIE1 were co-infiltrated into N. benthamiana leaves to detect the interaction between TIE1 and BRC1 (Chen et al., 2008). The plants were placed in the dark for 12 h, followed by 48 h in a growth chamber under normal conditions. The infiltrated N. benthamiana leaves were sprayed with luciferin (100 mM) and kept in the dark for 10 min. The leaves were observed under a low-light cooled charge-coupled device (CCD) imaging apparatus Lumazone_1300B (Roper Bioscience) and then photos were taken. Quantitative analysis was done using a Tanon 5200 Multi chemiluminescent imaging system.
Using the AtTIE1 amino acid sequence as a query sequence, we searched the G. hirsutum protein database using the blastp program (https://www.cottongen.org/) to obtain two homologous genes located in At (GH_A05G0254) and Dt (GH_D05G0259) subgenomes, respectively, named GhTIE1a and GhTIE1d (Figure 1A). We chose GhTIE1a, GhTIE1d, AtTIE1(At4G28840.1), AtTIE2(At2G20080), AtTIE3(At1G29010), and AtTIE4(At2G34010) to make a phylogenetic tree. It showed that GhTIE1 are closer to AtTIE1 than the other AtTIE genes (Supplementary Figure 1). We cloned GhTIE1a and GhTIE1d from upland cotton; GhTIE1a and GhTIE1d encode proteins containing 199 and 202 amino acids, respectively. The sequence alignment showed that the similarities of GhTIE1a and GhTIE1d protein sequences to AtTIE1 were 45.95% and 45.74%, respectively. The similarity between GhTIE1a and GhTIE1d protein sequences reached 96.48%. Despite the low similarity of TIE1 between Arabidopsis and cotton, some of the conserved domains of the two GhTIE1s are highly similar to AtTIE1: the N-terminal region contains a helical region (between residues 46 and 55), and the C-terminal region contains a typical EAR motif sequence (DLELRL), showing that GhTIE1a and GhTIE1d may possess a similar function to AtTIE1 as a transcriptional repressor (Figure 1A) (Yang et al., 2018). The putative nuclear localization signal (KRGK) was located at the N-terminus of the GhTIE1a and GhTIE1d proteins, suggesting that GhTIE1a and GhTIE1d might be localized in the nucleus (Dingwall et al., 1988; Ohta et al., 2001). GhTIE1-YFP was infiltrated into tobacco leaves to detect transient expression, and the results showed that GhTIE1a and GhTIE1d proteins were localized in the nucleus and the cytoplasm, but this was more pronounced in the nucleus (Figures 1B, C).
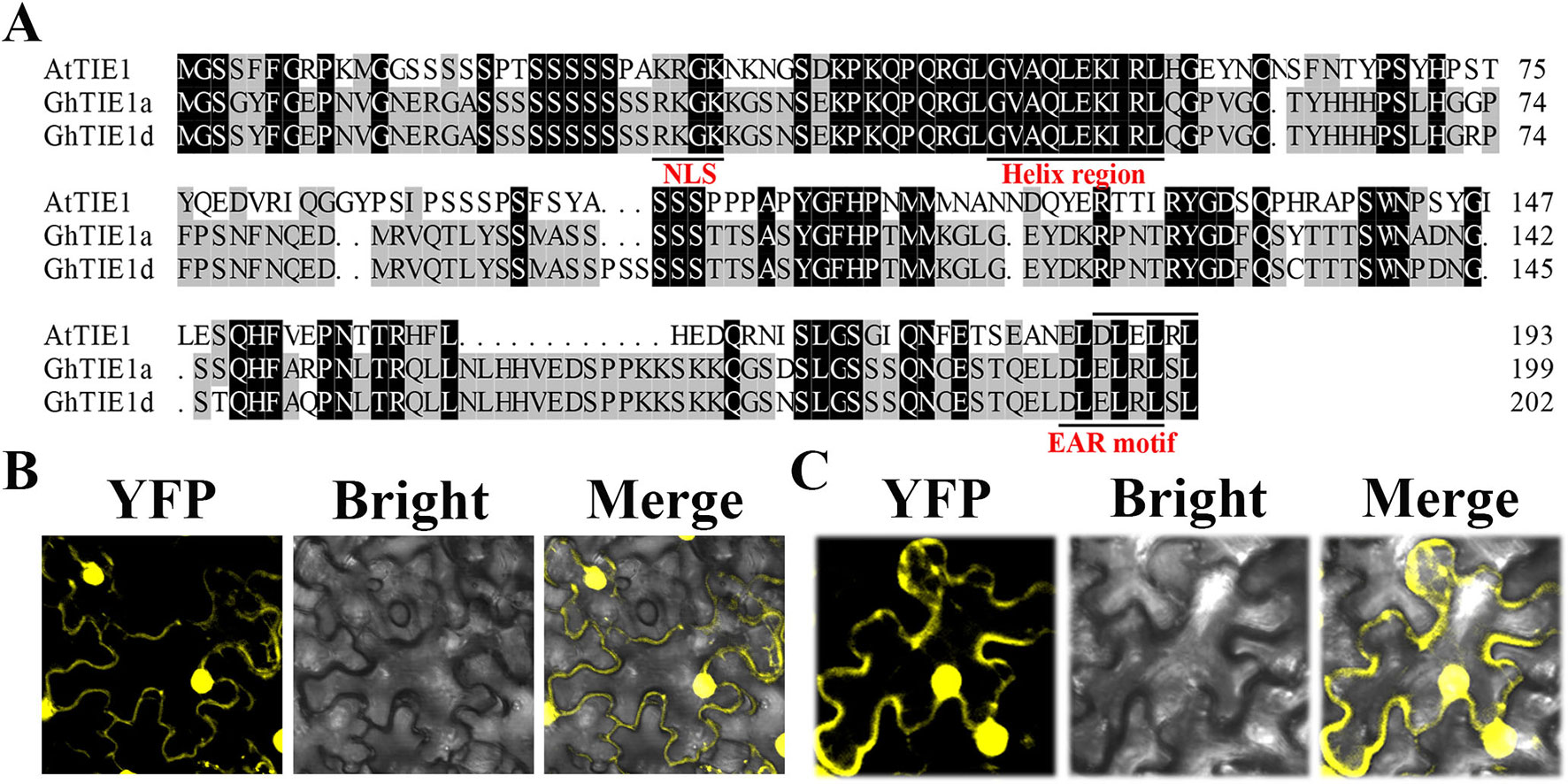
Figure 1 Sequence analysis and subcellular localization of GhTIE1. (A) Sequence alignment and domain analysis of TIE1 in Arabidopsis and upland cotton. Amino acid sequence alignment of AtTIE1 (At4g28840), GhTIE1d (GH_D05G0259), and GhTIE1a (GH_A05G0254). They all contain a nuclear localization signal, a helix region, and an EAR motif. (B)GhTIE1d and (C)GhTIE1a are localized in the nucleus and the cytoplasm, mostly in the nucleus.
GhTIE1 and AtTIE1 share conserved domains including an EAR motif and a helix region and are both mainly located in the nucleus, indicating that GhTIE1 may have a similar function as AtTIE1 in controlling shoot branching. To confirm the function of GhTIE1, overexpressed vectors 35S-GhTIE1a and 35S-GhTIE1d were constructed to transform Arabidopsis. These overexpression lines with 35S-GhTIE1a and 35S-GhTIE1d both showed increased rosette-leaf branching and cauline-leaf branching (Figure 2 and Supplementary Figure 2), suggesting that GhTIE1a and GhTIE1d are functionally redundant in regulating shoot branching.
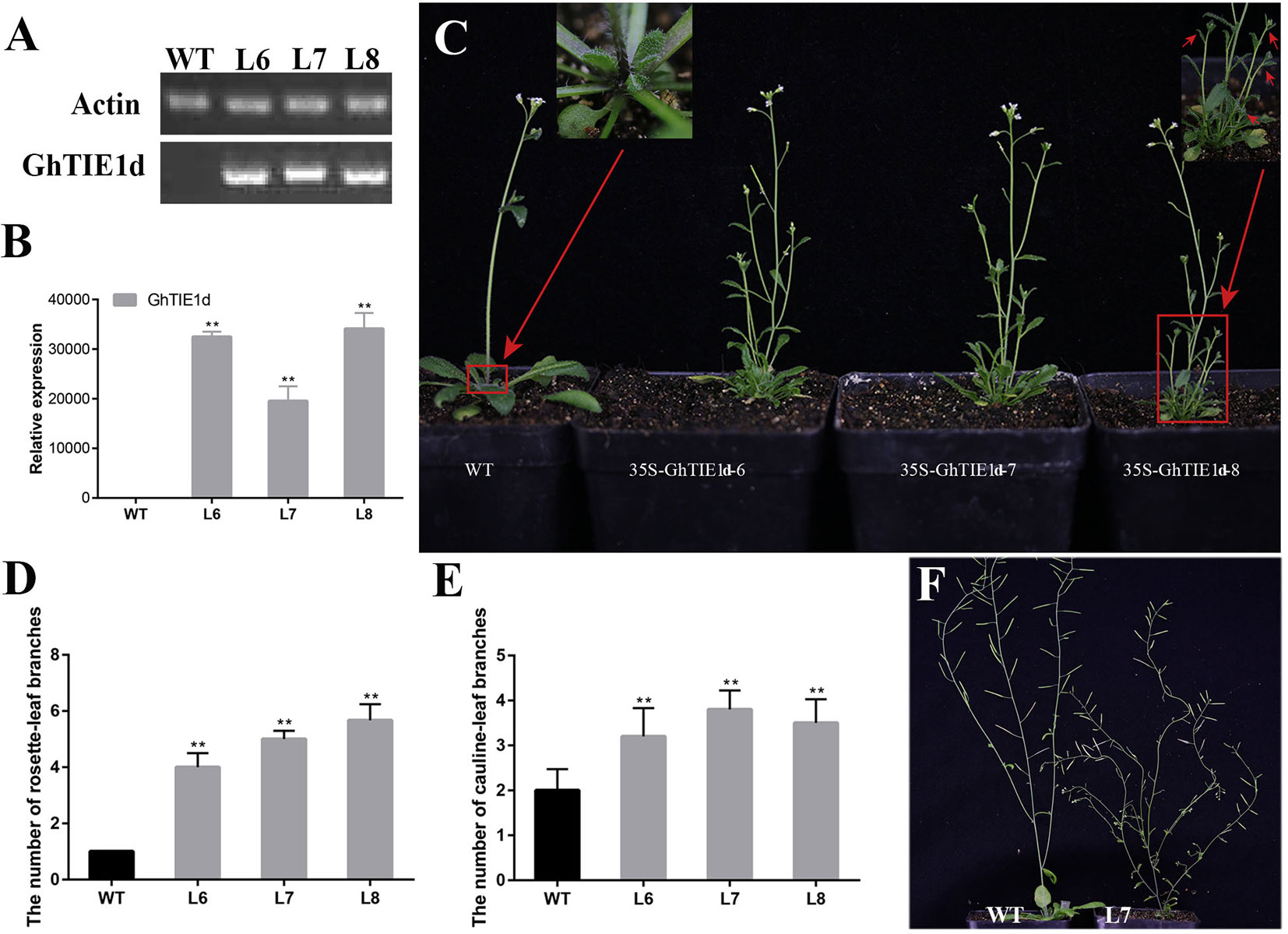
Figure 2 Overexpression of GhTIE1d causes excessive branching. (A)GhTIE1d was not expressed in WT, but was highly expressed in three overexpressed lines based on semi-quantitative PCR and (B) qRT-PCR. (C) Branching phenotypes of 35-day-old WT plants and three overexpressed 35S-GhTIE1d lines. Overexpressing GhTIE1d in Arabidopsis produced more branches than in wild-type plants. (D) Quantitative analysis of rosette-leaf branches in 35-day-old wild-type and three overexpressed lines. (E) Quantitative analysis of cauline-leaf branches in 80-day-old wild-type and three overexpressed lines. (F) Cauline-leaf branching phenotypes of 80-day-old WT plants and overexpressed 35S-GhTIE1d-7 lines. Overexpressing GhTIE1d in Arabidopsis produced more cauline-leaf branches than in wild-type plants. The rosette-leaf branches have been removed for better visualization of the main stem. The rosette-leaf branches and cauline-leaf branches of the overexpressed lines are significantly increased. L6, L7, and L8 are the three different overexpressed lines, 35S-GhTIE1d-6, 35S-GhTIE1d-7, and 35S-GhTIE1d-8, respectively. WT wild type (**p < 0.01). Data represent the mean ± SD from three biological replicates.
Considering the functional redundancy of GhTIE1a and GhTIE1d, only GhTIE1d-overexpressed Arabidopsis plants were further analyzed. Three homozygous lines—35S-GhTIE1d-6, 35S-GhTIE1d-7, and 35S-GhTIE1d-8, were selected to analyze the expression level and shoot branch phenotype. GhTIE1d was highly expressed, as confirmed by semi-quantitative PCR and qRT-PCR in these homozygous lines (Figures 2A, B). The shoot branch including a cauline-leaf branch and a rosette-leaf branch was obviously increased in GhTIE1d-overexpressed Arabidopsis compared to that of wild type (WT) in 35-day-old seedlings, where there was no observed rosette-leaf branch in WT. Most rosette-leaf buds remained small or dormant. Almost all the rosette-leaf buds in overexpressed (OE) lines grew out to form rosette-leaf branches, and more than four rosette-leaf branches were observed in OE lines (Figures 2C, D). Similarly, the cauline-leaf branch was obviously increased in OE lines compared to WT, where only one cauline-leaf branch was initially grown in WT but more than two cauline-leaf branches were observed in OE lines in 35-day-old seedlings (Figure 2C), suggesting that overexpression of GhTIE1d promoted axillary bud activity and the initiation and growth of cauline-leaf branch. In later reproductive stages, the cauline-leaf branch number was also increased in OE lines compared to WT in 80-day-old seedlings, where there were only about two cauline-leaf branches in WT but more than three cauline-leaf branches in OE lines (Figures 2E, F). These results showed that GhTIE1 positively regulates shoot branching growth vigor and shoot branch number.
GhTIE1a and GhTIE1d are highly functionally redundant in regulating Arabidopsis shoot branching. Considering the functional redundancy caused by two copies of GhTIE1, the highly conserved CDS fragment between GhTIE1a and GhTIE1d was cloned into a CLCrV vector to simultaneously silence these two copies. We generated a CLCrV : GhTIE1 construct to transform the cotyledon of cotton. Under the constant conditions of a 16-h light/8-h dark cycle at 25°C, most of the control plants produced about 13 branches, while almost all cotton plants with the GhTIE1 silenced showed approximately nine branches (Supplementary Table 3). Three GhTIE1-silenced plants with a relatively higher gene silencing efficiency, where the GhTIE1 expression level was less than 50%, exhibited less than nine shoot branches (Figures 3A–C). Based on the GhTIE1 inferring efficiency, the GhTIE1-silenced plants were grouped into two classes, where the shoot branch number was less than 9 when the silencing efficiency was more than 50%. In contrast, the shoot branch number was between 10 and 11 when the silencing efficiency was less than 50% (Figure 3D). These results indicate that, like AtTIE1, GhTIE1 positively regulates shoot branching number in cotton.
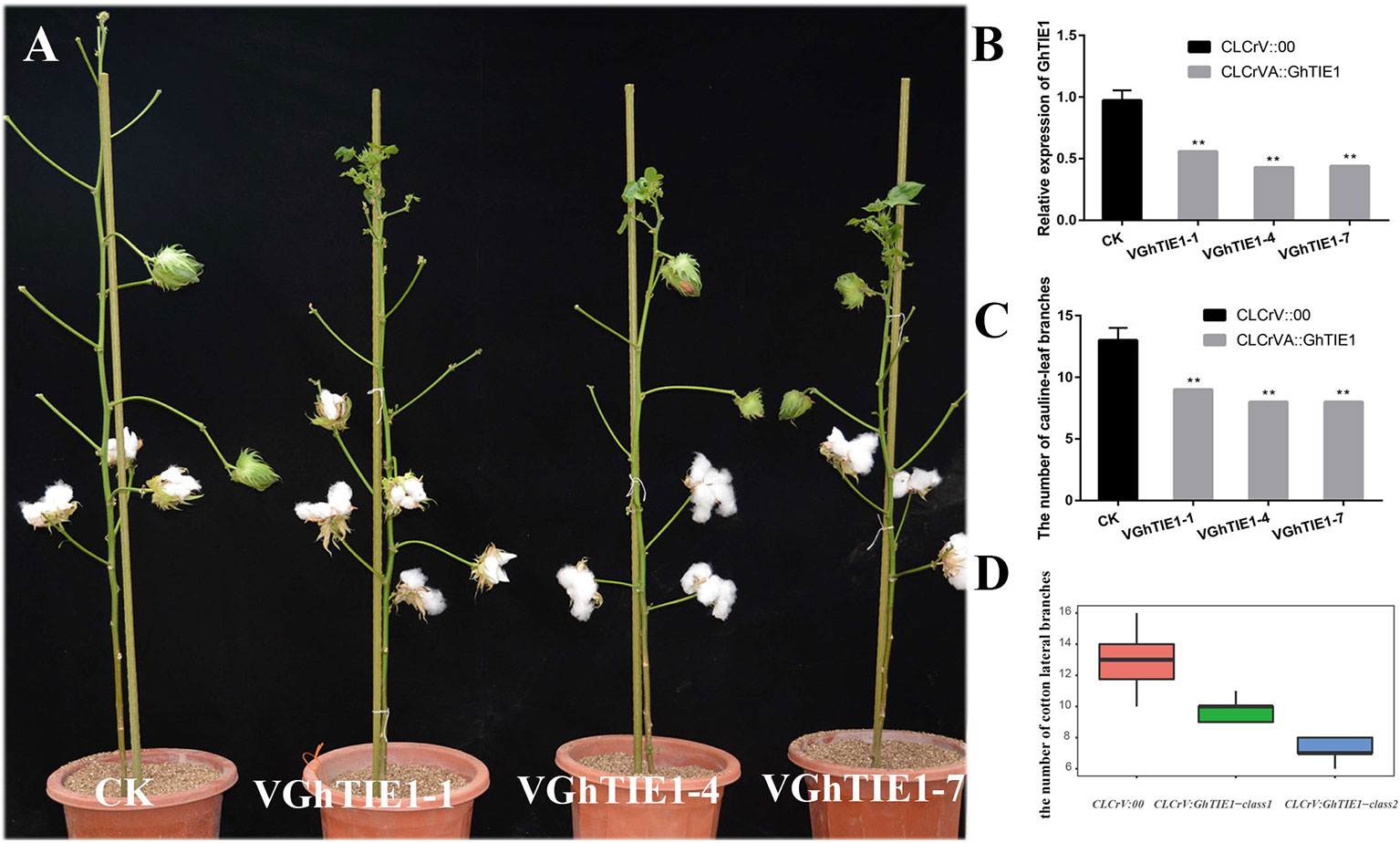
Figure 3 Silencing of GhTIE1 in cotton seriously decreases shoot branching. (A) Branching phenotypes of cotton grown for 4 months of control (CLCrV:00) and GhTIE1-silenced (VGhTIE1-1, VGhTIE1-4, and VGhTIE1-7) plants. (B) Analysis of the GhTIE1 gene expression in CK and VIGS plants by qRT-PCR in three different lines: VGhTIE1-1, VGhTIE1-4, and VGhTIE1-7. (C) Quantitative analysis of branches of CLCrV:00 and GhTIE1-silenced plants (**p < 0.01). VGhTIE1-1, VGhTIE1-4, and VGhTIE1-7 are the three different silenced cotton plants. CK, the empty vector CLCrV:00. (D) Analysis of the number of branches in 20 different CLCrV:00 and GhTIE1-silenced cotton plants. CLCrV : GhTIE1-class1, the cotton plants with a silencing efficiency less than 50%; CLCrV : GhTIE1-class2, the cotton plants with a silencing efficiency more than 50%. Data represent the mean ± SD from three biological replicates.
In Arabidopsis, AtTIE1 interacted with AtBRC1 and inhibited AtBRC1 activity to activate bud activity (Yang et al., 2018). To reveal the molecular mechanism of GhTIE1 regulating shoot branching in cotton, cotton axillary buds, shoot branches, and leaves were collected to construct a library. The GhTIE1 interacting proteins were screened via the yeast two-hybrid system. Several class II TCP genes, including GhBRC1 (TCP18), GhTCP13, and GhBRC2 (TCP12), potentially interacted with GhTIE1. The protein blast results and phylogenetic analysis showed that four GhBRC1 proteins were homologous to AtBRC1 and two GhBRC2 proteins were homologous to AtBRC2 (Supplementary Figures 3 and 4). To further verify the interaction between GhTIE1d and GhTCPs, the full-length CDS sequence of candidate GhTCPs were cloned into AD vectors to perform yeast two-hybrid assays. The results confirmed that two copies of GhBRC1 (GH_D11G0067, GH_A11G0062, GH_D12G2879, and GH_A12G2854) and GhBRC2 (GH_A12G1898 and GH_D12G1898) and one copy of GhTCP13 (GH_A05G3929) interacted with GhTIE1d (Figure 4A and Supplementary Figure 5A). To study the interaction of the above proteins in vivo, BiFC assays were performed. Two copies of GhBRC1, two copies of GhBRC2, and one copy of GhTCP13 were interacted with GhTIE1 in the nucleus, but GhBRC2 was shown to interact with GhTIE1d in the cell cytoplasm (Figure 4B and Supplementary Figure 5B). Consistent with the BiFC results, GhBRC1, GhBRC2, and GhTCP13 were all localized in the nucleus and cytoplasm (Supplementary Figure 6).
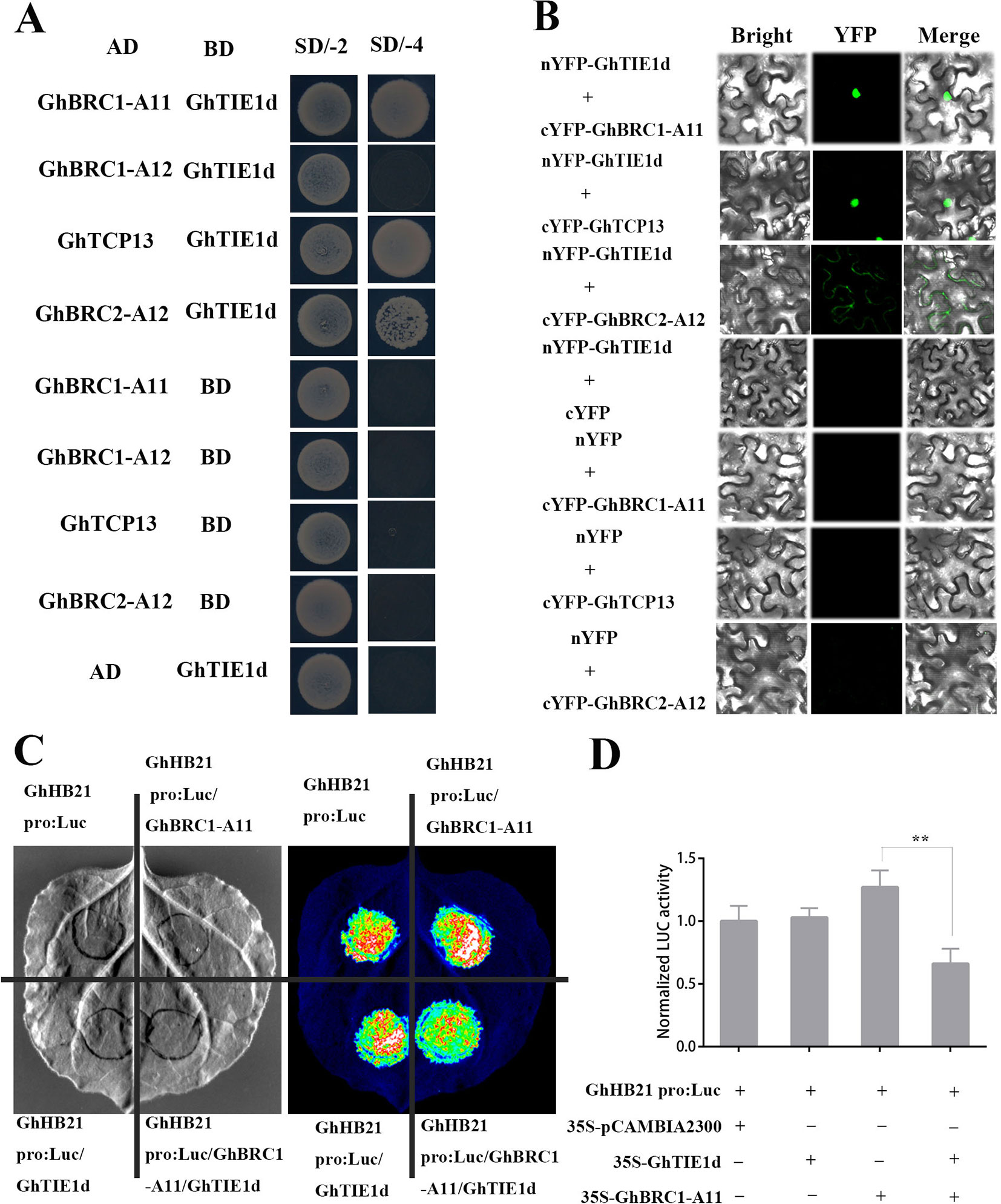
Figure 4 GhTIE1 interacts with GhBRC1, GhBRC2, and GhTCP13 and represses their activity. (A) Yeast two-hybrid assays of GhTIE1d (GH_D05G0259) with GhBRC1-A11 (GH_A11G0062), GhBRC1-A12 (GH_A12G2854), GhBRC2-A12 (GH_A12G1898), and GhTCP13 (GH_A05G3929). (B) BiFC assay proved the interaction of GhTIE1 with GhBRC1-A11, GhBRC2-A12, and GhTCP13. (C) The promoter of the GhHB21 gene fused to LUC was used as a reporter for the transactivation assay. GhTIE1 inhibited GhHB21 expression via interaction with GhBRC1 (GH_A11G0062). (D) LUC activity was decreased when GhTIE1d, GhBRC1-A11, and GhHB21 pro:Luc were co-infiltrated into Nicotiana benthamiana leaves (**p < 0.01). SD/-2 means SD-Leu-Trp; SD/-4 means SD-Leu-Trp-His-Ade. Data represent the mean ± SD from three biological replicates.
AtHB21, AtHB40, and AtHB53 were the direct targets of AtBRC1, and AtTIE1 interacted with AtBRC1 to control the shoot branching via repressing AtHB21, AtHB40, and AtHB53 expression (Yang et al., 2018). To confirm whether GhTIE1 represses GhBRC1 transcriptional activity, the AtHB21 homologous gene GhHB21 was selected, and the 1,500-bp promoter region upstream of GhHB21 was fused to LUCIFERASE (LUC) to construct the reporter construct GhHB21pro-LUC. Then, 35S-GFP as control, 35S-GhBRC1, and 35S-GhTIE1 were co-infiltrated into N. benthamiana leaves with GhHB21pro-LUC. LUC activity analysis indicated that GhBRC1 directly activated the GhHB21pro-LUC. In contrast, when GhHB21pro-LUC and 35S-GhBRC1 were co-infiltrated with 35S-GhTIE1, the activity of GhHB21pro-LUC was inhibited, showing that GhTIE1 represses GhBRC1 transcriptional activity (Figures 4C, D). Similarly, in transient expression of tobacco leaves, GhTIE1 also inhibited GhTCP13 transcriptional activity, thereby repressing GhHB21 expression (Supplementary Figure 7). These results showed that GhTIE1 controlled shoot branching via repressing GhBRC1, GhBRC2, and GhTCP13 activity, and the GhTIE1 and GhBRC1 interaction model was conserved as described in Arabidopsis, where AtTIE1 interacted with AtBRC1 and repressed BRC1 protein activity. Moreover, the interactions of GhTIE1 and GhBRC2 and of GhTIE1 and GhTCP13 were specific in cotton and not reported in Arabidopsis, suggesting that the GhTIE1 regulatory network was more complex than AtTIE1 in regulating branching.
AtBRC1 and AtBRC2 both arrest axillary bud development and prevent axillary bud outgrowth, but the brc2 mutant phenotype was weaker than brc1 (Aguilar-Martínez et al., 2007). GhBRC1-A11 and GhBRC2-A12, the homologues of AtBRC1 and AtBRC2, were selected to rescue the brc1 mutant phenotype. As expected, GhBRC1-A11 overexpression in the brc1 mutant seriously decreased the rosette-leaf branch number compared with the brc1 mutant: there was an average of eight rosette-leaf branches in the brc1 mutant, but only about four rosette-leaf branches in the 35S-GhBRC1-A11-brc1-2 lines. Similarly, GhBRC2-A12 also restored the brc1 mutant phenotype, and there were about five rosette-leaf branches in 35S-GhBRC2-A12-brc1-2 plants (Figure 5). Ectopically expressing GhTCP13 also rescued the brc1 mutant phenotype (Figure 5A). Like GhBRC1 and GhBRC2, GhTCP13 also belongs to the TCP class II subfamily, shares the conversed TCP domain, and interacts with GhTIE1 to repress bud activity, supporting the likely essential role of GhTCP13 in regulating bud activity and shoot branching. The possible explanation was that overexpression of GhTCP13 in the brc1-2 mutant significantly promoted the protein accumulation of GhTCP13. AtTIE1 may mainly interact with GhTCP13, weakening the interaction of AtTIE1 and AtBRC1 and increasing the activity of AtBRC1 and expression of AtHB21, AtHB40, and AtHB53 to inhibit bud activity. Alternatively, an increased GhTCP13 transcript level in 35S-GhTCP13-brc1-2 plants may directly promote GhHB21 transcription, as shown in Supplementary Figure 5; where GhHB21pro-LUC and 35S-GhTCP13 were co-infiltrated, the activity of GhHB21pro-LUC was increased.
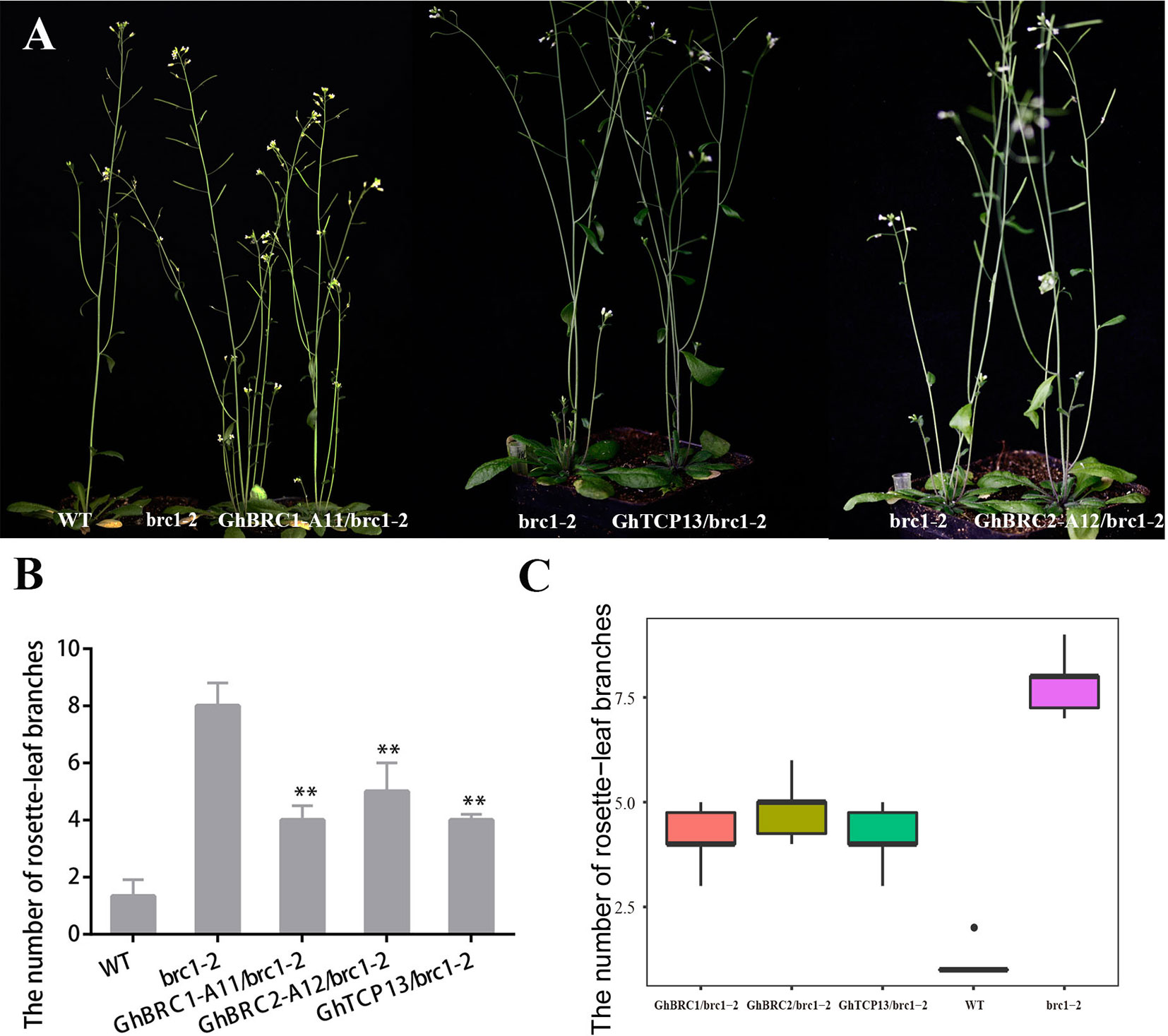
Figure 5 Overexpression of GhBRC1, GhBRC2, or GhTCP13 partially restores the phenotype of branching in Arabidopsis mutant brc1-2. (A) Phenotypes of a 45-day-old transgenic plant expressing 35S-GhBRC1-A11 (GH_A11G0062) to a mutant brc1-2, of a 40-day-old transgenic plant expressing 35S-GhTCP13 (GH_A05G3929) to a mutant brc1-2, and of a 40-day-old transgenic plant expressing 35S-GhBRC2-A12 (GH_A12G1898) to a mutant brc1-2. (B) Quantitative analysis of rosette-leaf branches in 45-day-old brc1-2 and transgenic plant. (C) Number of branches of 10 individual 45-day-old WT, brc1-2 plants, and transgenic Arabidopsis plants (**p < 0.01). Data represent the mean ± SD from three biological replicates.
To study the function of GhBRC1 in regulating cotton branch number, two copies of the GhBRC1 gene including GH_A11G0062 and GH_D11G0067 were simultaneously silenced using VIGS. This did not result in differences in the branch number between the silenced GhBRC1 plants and CLCrV:00 plants (the control line with the empty vector). The possible explanation was that there were four copies of GhBRC1 (Supplementary Figure 3) that showed low similarity excluding the TCP domain with AtBRC1. Only two copies including GH_A11G0062 and GH_D11G0067 were silenced in our study. The other two copies of GhBRC1 (GH_A12G2854 and GH_D12G2879) and the two copies of GhBRC2 and GhTCP13 were not simultaneously silenced, which may play a redundant function in regulating branch number.
HB21, HB40, and HB53 in Arabidopsis are directly regulated by AtBRC1, and AtTIE1 also indirectly repressed the transcription of these genes (Yang et al., 2018). Our hypothesis was that the transcriptional levels of these genes are also closely related to shoot branching number in cotton. Overexpression of GhTIE1 in Arabidopsis significantly increased the shoot branching number, including cauline-leaf branch and rosette-leaf branch, where AtBRC1, AtHB21, AtHB40, and AtHB53 transcriptions were more seriously inhibited than in WT (Figures 6A–D), suggesting that GhTIE1 may also directly or indirectly regulate AtBRC1 and three AtHB genes at the transcriptional level. GhBRC1 partially rescued the brc1-2 mutant phenotype and the shoot branching number was decreased compared to the brc1-2 mutant, but higher than that of the WT. Coinciding with the shoot branching phenotype, AtHB21, AtHB40, and AtHB53 expression levels were slightly higher in 35S-GhBRC1-brc1 plants than in the brc1 mutant (Figures 6B–D), showing that the expression of these three genes was negatively related to shoot branch number.
GhTIE1 was functionally conserved in regulating shoot branching in Arabidopsis and cotton. Silencing GhTIE1 obviously decreased the branch number in cotton. Considering that HB21, HB40, and HB53 played important roles in regulating shoot branching, homologous genes in cotton were analyzed. Based on the phylogenetic analysis, only GhHB21 and GhHB40 were identified. As expected, GhHB21 and GhHB40 were both unregulated in GhTIE1-silenced cotton plants, but slightly decreased in GhBRC1-silenced lines (Figures 6E, F), showing that the expression levels of these HB gene were positively regulated by GhBRC1 but negatively regulated by GhTIE1. Additionally, the expression levels of GhBRC1 and GhBRC2 were also regulated by GhTIE1. GhBRC1 showed an upregulated expression in GhTIE1-silenced plants compared to the control, while GhBRC2 expression level is similar to the control (Supplementary Figure 8), in link with the results of TIE1 function in controlling BRC1 expression in Arabidopsis (Yang et al., 2018).
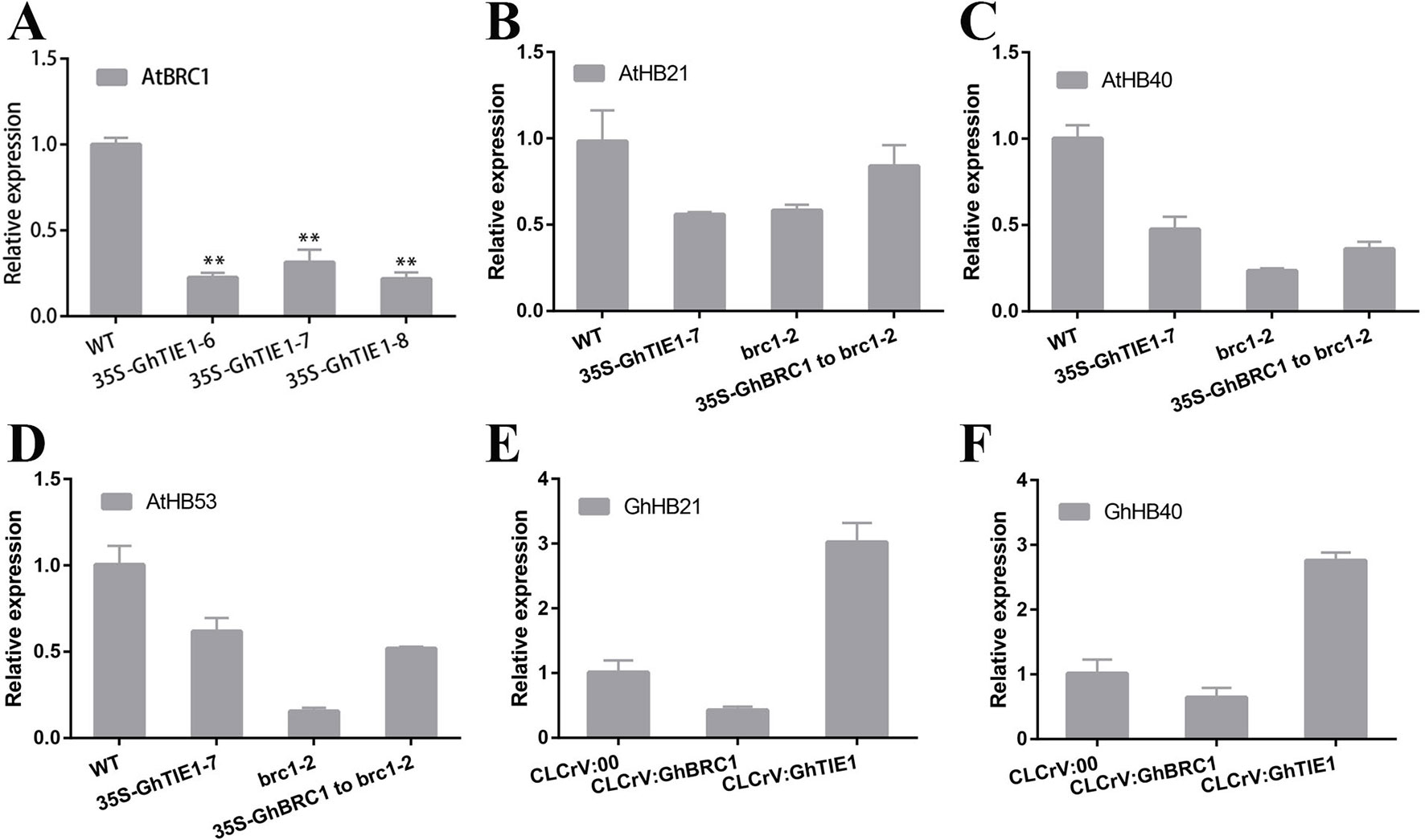
Figure 6 HB21, HB40, and HB53 expression levels are inhibited by TIE1 and BRC1. (A)AtBRC1 expression level in wild type (WT) and GhTIE1-overexpressing lines. (B)AtHB21 expression levels in WT, GhTIE1-overexpressing plant, and brc1-2 mutant and GhBRC1 overexpression in brc1-2 mutant plants. (C)AtHB40 expression levels in WT, GhTIE1-overexpressing plant, and brc1-2 mutant and GhBRC1 overexpression in brc1-2 mutant plants. (D)AtHB53 expression levels in WT, GhTIE1-overexpressing plant, and brc1-2 mutant and GhBRC1-A11 overexpression in brc1-2 mutant plants. (E)GhHB21 (GH_A02G1904) expression levels in CLCrV:00, CLCrV : GhBRC1, and CLCrV : GhTIE1 plants. (F)GhHB40 (GH_D11G1980) expression levels in CLCrV:00, CLCrV : GhBRC1, and CLCrV : GhTIE1 plants. Each reaction was performed with three biological replicates. Data represent the mean ± SD from three biological replicates.
Although the plant architecture between Arabidopsis and cotton is different, the shoot branch growth was controlled by bud activity in both species. AtTIE1 positively regulated shoot branching in Arabidopsis. Gain-of-function mutant tie1-D and overexpressing AtTIE1 in Arabidopsis produced significantly more branches (Tao et al., 2013). Two homologs of AtTIE1 in cotton, GhTIE1a and GhTIE1d, were also essential in regulating bud activity. Overexpression of GhTIE1a or GhTIE1d in Arabidopsis obviously increased the shoot branch number, including rosette-leaf branch and cauline-leaf branch, which presented a similar phenotype to tie1-D. Silencing the conversed sequence between GhTIE1a and GhTIE1d inhibited the bud activity, and shoot branch number was significantly decreased in CLCrV : GhTIE1 compared to CLCrV:00 plants, showing that GhTIE1 is functionally conserved in regulating shoot branching in plants. Silencing GhTIE1 generated fewer branches than the control. The VGhTIE1 plants appear to be younger than the wild type. The possible explanation is that there is a balance between vegetative and reproductive growth. Similar results were reported in Arabidopsis that disruption of TIE1 causes defects of shoot branching and axillary bud development; that is to say, TIE1-mEAR-VP16 plants produce fewer branches and appear to be younger than the wild type (Yang et al., 2018). Different from Arabidopsis architecture, including rosette-leaf branch and cauline-leaf branch, cotton is an infinitely growing crop with a complex branching pattern consisting of fruit branches and nutritional branches, which were developed from the cotton main stem (Chen et al., 2015). Decreased fruit branch number in silenced GhTIE1 plants partially inhibited the infinite growth, which facilitated the fiber maturation of later cotton bolls and improvement of cotton fiber length uniformity.
TCP activity is regulated at the protein level and transcription level and is essential in controlling plant development (Tao et al., 2013). Based on the TCP domain features, TCPs were divided into two classes, class I and class II (three subclass: CIN, CYC, and TB1) (Gaudin et al., 2014; Yin et al., 2018). Yeast two-hybrid screening revealed that GhTIE1 physically interacted with multiple GhTCP proteins, including GhTCP18 (GhBRC1), GhTCP12 (GhBRC2), and GhTCP13, and these GhTCPs were both grouped into class II. GhTCP18 and GhTCP12 were categorized into TB1 and CYC-like TCPs, respectively. GhTCP13 was categorized into CIN-like TCPs (Li et al., 2017). In Arabidopsis, TIE1 not only regulated leaf development via suppressing CIN-like TCP activity (Tao et al., 2013) but also controlled shoot branching by repressing BRC1 (TB1-like TCP) activity (Yang et al., 2018). Although there was no interaction between AtTIE1 and AtBRC2 in vitro, the brc2 mutant showed a similar branching phenotype to the brc1 mutant in Arabidopsis, suggesting that AtBRC2 activity may also be regulated directly or indirectly by AtTIE1. Different from Arabidopsis, GhTIE1 and subclass II TCPs (GhBCR1, GhBCR2, and GhTCP13) have strong interactions in vivo and in vitro, and these TCPs both partially rescued the phenotype of brc1 mutant in Arabidopsis. Previous researches found that CIN-type genes limit cell proliferation at the margins of the developing leaf primordia and regulate leave morphology in Antirrhinum, Arabidopsis, and tomato (Martín-Trillo and Cubas, 2009; Koyama et al., 2010). Different from the reported function of CIN-type genes, our research found that GhTCP13 was preferentially expressed in the leaves and axillary buds (Supplementary Figure 9), and GhTCP13 interacted with GhTIE1 and partially restored the brc1 mutant phenotype. These results supported that GhTCP13 may play a key role in regulating leaf and axillary bud development. Considering the reduced lateral branching phenotype caused by overexpression of GhTCP13 in the brc1 mutant, a possible explanation is that ectopically expressed GhTCP13 derived by the 35S promoter was ubiquitously expressed in the whole plant, which may cause the decreased branch number in the brc1 mutant, suggesting that GhTCP13 may partially function redundantly in regulating branching with AtBRC1, not AtTCP13. Alternatively, GhTCP13 may exhibit a dual function in controlling leaf and axillary bud development in cotton and function differently with AtTCP13.
These results show that these TCPs may be functionally redundant in controlling cotton branching, and the regulatory network involved in branching mediated by TIE1 and TCPs varied in different species.
Bud dormancy genes HB21, HB40, and HB53 were directly regulated by BRC1 and also modulated by TIE1 in Arabidopsis (Yang et al., 2018). These genes increased the ABA level to inhibit bud activity (Yao and Finlayson, 2015). Our results showed that GhTIE1 overexpression in Arabidopsis promoted more branches, along with the downregulated AtHB21, AtHB40, and AtHB53 compared to WT. GhBRC1 overexpression in the brc1-2 mutant decreased the shoot branch number, along with the increased AtHB21, AtHB40, and AtHB53 expression levels than in the brc1-2 mutant, showing that shoot branching controlled by GhTIE1 and GhBRC1 in Arabidopsis partially depended on the expression levels of AtHB21, AtHB40, and AtHB53. As expected, silenced GhTIE1 increased GhHB21 and GhHB53 expression, leading to a decreased branch number. Notably, AtBRC1 was downregulated in GhTIE1-overexpressed lines in A. thaliana, suggesting that GhTIE1 may also modulate directly or indirectly AtBRC1 at the transcriptional level. Meanwhile, AtBRC1 activity may be repressed by GhTIE1 via a direct interaction. The transcriptional and post-transcriptional regulation of AtBRC1 by GhTIE1 controlled the expressions of AtHB21, AtHB40, and AtHB53. Contrary to GhTIE1, silenced GhBRC1 repressed GhHB21 and GhHB40 transcription, but there were no obvious changes in branch number compared to the control. The possible reason was that GhBRC1, GhBRC2, and GhTCP13 were functionally redundant in regulating cotton branching, and GhBRC2 and GhTCP13 may compensate for the effect mediated by silenced GhBRC1. Genetic compensation was mediated by homologous genes in plants and animals. AtCLV1 paralogs compensate for the loss of CLV1 through transcriptional upregulation (Nimchuk et al., 2015). SlCLE9, the closest paralog of SlCLV3, contributes to stem cell homeostasis by compensating for the loss of SlCLV3 through transcriptional upregulation in tomato (Rodriguez-Leal et al., 2019). These results proved that the TIE1-BRC1-HB21/HB40/HB53 pathway was conserved in modulating bud activity and shoot branch growth in plants. More efforts should be made to reveal the molecular machinery that controls shoot branching via modulation of the activity of the TIE1 and TCP proteins, which will help us to further determine plant shoot architecture in response to environmental cues.
All datasets generated for this study are included in the article/Supplementary Files.
XG conceived the original screening and research plan. YangD, JZ, PL, and WH performed the experiments. YZ, LL, XW, YanpD, and PW provided technical assistance to YangD and analyzed the data. XG, YangD and MS wrote the manuscript with all the authors.
This research was financially supported by the National Science Foundation of China (31621005, 31701476).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Professor GenJi Qin for providing the Arabidopsisbrc1-2 mutant.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01348/full#supplementary-material
Aguilar-Martínez, J. A., Poza-Carrión, C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 (2), 458–472. doi: 10.1105/tpc.106.048934
Arite, T., Iwata, H., Ohshima, K., Maekawa, M., Nakajima, M, M., Sakakibara, H., et al. (2010). DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 51 (6), 1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Chen, W., Yao, J., Chu, L., Yuan, Z., Li, Y., Zhang, Y. (2015). Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing. Theor. Appl. Genet. 128 (3), 539–547. doi: 10.1007/s00122-014-2452-2
Chen, H., Zhou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146 (2), 368–376. doi: 10.1104/pp.107.111740
Chevalier, F., Nieminen, K., Sánchez-Ferrero, J. C., Rodríguez, M. L., Chagoyen, M., Hardtke, C. S., et al. (2014). Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26 (3), 1134–1150. doi: 10.1105/tpc.114.122903
Koyama, T., Sato, F., Ohme-Takagi, M. (2010). A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci. Biotechnol. Biochem. 74 (10), 2145–2147. doi: 10.1271/bbb.100442
Dingwall, C., Robbins, J., Dilworth, S. M., Roberts, B., Richardson, W. D. (1988). The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107 (3), 841–849. doi: 10.1083/jcb.107.3.841
Dong, Z., Li, W., Unger-Wallace, E., Yang, J., Vollbrecht, E., Chuck, G. (2017). Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. U. S. A. 114 (41), E8656. doi: 10.1073/pnas.1714960114
Dun, E. A., de Saint Germain, A., Rameau, C., Beveridge, C. A. (2012). antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158 (1), 487–498. doi: 10.1104/pp.111.186783
Finlayson, S. A. (2007). Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48 (5), 667–677. doi: 10.1093/pcp/pcm044
Gaudin, A. C., Mcclymont, S. A., Soliman, S. S., Raizada, M. N. (2014). The effect of altered dosage of a mutant allele of Teosinte branched 1 (tb1-ref) on the root system of modern maize. BMC Genet. 15 (1), 23–23. doi: 10.1186/1471-2156-15-23
González-Grandío, E., Pajoro, A., Franco-Zorrilla, J. M., Tarancón, C., Immink, R. G. H., Cubas, P. (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proceed. Natl. Acad. Sci. 114 (2), E245–E254. doi: 10.1073/pnas.1613199114
Kebrom, T. H., Chandler, P. M., Swain, S. M., King, R. W., Richards, R. A., Spielmeyer, W. (2012). Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 160 (1), 308–318. doi: 10.1104/pp.112.197954
Livak, K. J., Schmittqen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif) 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li, W., Li, D. D., Han, L. H., Tao, M., Hu, Q. Q., Wu, W. Y., et al. (2017). Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 7 (1), 10118. doi: 10.1038/s41598-017-10609-2
Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., et al. (2013). Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 25 (10), 3743–3759. doi: 10.1105/tpc.113.113639
Martín-Trillo, M., Cubas, P. (2009). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 (1), 31–39. doi: 10.1016/j.tplants.2009.11.003
Martı´n-Trillo, M., Grandı´o, E. G., Serra, F., Marcel, F., Rodrı´guez-Buey, M. L., Schmitz, G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67, 701–714. doi: 10.1111/j.1365-313X.2011.04629.x
McSteen, P., Leyser, O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56 (1), 353–374. doi: 10.1146/annurev.arplant.56.032604.144122
Minakuchi, K., Kameoka, H., Yasuno, N., Umehara, M., Luo, L., Kobayashi, K., et al. (2010). FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 51 (7), 1127–1135. doi: 10.1093/pcp/pcq083
Nicolas, M., Rodríguez-Buey, M. L., Franco-Zorrilla, J. M., Cubas, P. (2015). A recently evolved alternative splice site in the BRANCHED1a gene controls potato plantarch itecture. Curr. Biol. 25 (14), 1799–1809. doi: 10.1016/j.cub.2015.05.053
Nils, B., Alexandre, D. S. G., Jean-Paul, P., Stéphanie, B. M., Marion, D., Ioanna, A., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158 (1), 225–238. doi: 10.1104/pp.111.182725
Nimchuk, Z. L., Zhou, Y., Tarr, P. T., Peterson, B. A., Meyerowitz, E. M. J. D. (2015). Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142 (6), 1043–1049. doi: 10.1242/dev.119677
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., Ohme-Takaqi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 (8), 1959–1968. doi: 10.1105/TPC.010127
Pasternak, T., Groot, E. P., Kazantsev, F., Teale, W., Omelyanchuk, N., Kovrizhnykh, V., et al. (2019). Salicylic acid affects root meristem patterning via auxin distribution in a concentration-dependent manner. Plant Physiol. 180 (3), 1725–1739. doi: 10.1104/pp.19.00130
Qian, Y., Lin, L., Xiong, Q., Flores, C., Wong, J., Shi, J., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45 (3), 330–333. doi: 10.1038/ng.2525
Rameau, C., Bertheloot, J., Leduc, N., Andrieu, B., Foucher, F., Sakr, S. (2015). Multiple pathways regulate shoot branching. Front. Plant Sci. 5, 741. doi: 10.3389/fpls.2014.00741
Rodriguez-Leal, D., Xu, C., Kwon, C.-T., Soyars, C., Demesa-Arevalo, E., Man, J., et al. (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51 (5), 786–792. doi: 10.1038/s41588-019-0389-8
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2010). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33 (3), 513–520. doi: 10.1046/j.1365-313X.2003.01648.x
Tao, Q., Guo, D., Wei, B., Zhang, F., Pang, C., Jiang, H., et al. (2013). The TIE1 Transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25 (2), 421–437. doi: 10.1105/tpc.113.109223
Wang, B., Smith, S. M., Li, J. (2018). Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 69, 1. doi: 10.1146/annurev-arplant-042817-040422
Wang, M., Le Moigne, M.-A., Bertheloot, J., Crespel, L., Perez-Garcia, M.-D., Ogé, L., et al. (2019). BRANCHED1: a key hub of shoot branching. Front. Plant Sci. 10, 76. doi: 10.3389/fpls.2019.00076
Wang, Y., Jiao, Y. (2018). Axillary meristem initiation - a way to branch out. Curr. Opin. Plant Biol. 41, 61–66. doi: 10.1016/j.pbi.2017.09.001
Yu, X., Li, L., Li, L., Guo, M., Chory, J., Yin, Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 105 (21), 7618–7623. doi: 10.1073/pnas.0802254105
Yang, Y., Nicolas, M., Zhang, J., Hao, Y. U., Qin, G. (2018). The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. Plos Genet. 14 (3), e1007296. doi: 10.1371/journal.pgen.1007296
Yao, C., Finlayson, S. A. (2015). Abscisic acid is a general negative regulator of Arabidopsis axillary bud growth. Plant Physiol. 169 (1), 611–626. doi: 10.1104/pp.15.00682
Yin, Z., Li, Y., Zhu, W., Fu, X., Han, X., Wang, J., et al. (2018). Identification, characterization, and expression patterns of tcp genes and microrna319 in cotton. Int. J. Mol. Sci. 19 (11), 1–13. doi: 10.3390/ijms19113655
Keywords: TIE1, shoot branching, TCP, plant type, cotton, Arabidopsis
Citation: Diao Y, Zhan J, Zhao Y, Liu L, Liu P, Wei X, Ding Y, Sajjad M, Hu W, Wang P and Ge X (2019) GhTIE1 Regulates Branching Through Modulating the Transcriptional Activity of TCPs in Cotton and Arabidopsis. Front. Plant Sci. 10:1348. doi: 10.3389/fpls.2019.01348
Received: 13 June 2019; Accepted: 27 September 2019;
Published: 28 October 2019.
Edited by:
Pilar Cubas, National Center of Biotechnology (CSIC), SpainReviewed by:
Genji Qin, Peking University, ChinaCopyright © 2019 Diao, Zhan, Zhao, Liu, Liu, Wei, Ding, Sajjad, Hu, Wang and Ge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyang Ge, Z2V4aWFveWFuZ0BjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.