- Key Laboratory of Urban Agriculture (South) Ministry of Agriculture, Plant Biotechnology Research Center, Fudan-SJTU-Nottingham Plant Biotechnology R&D Center, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
Artemisinin-based therapies are the only effective treatment for malaria, which reached to 219 million cases and killed 435,000 people in 2017. To meet the growing demand for artemisinin and make it accessible to the poorest, genetic engineering of Artemisia annua becomes one of the most promising approaches to improve artemisinin yield. In this work, AabZIP9 transcription factor has been identified and characterized. The expression profile of AabZIP9 revealed that it was clustered with the artemisinin specific biosynthetic pathway genes ADS, CYP71AV1, DBR2, and ALDH1. Furthermore, the transiently dual-LUC analysis showed that the activation of ADS promoter was enhanced by AabZIP9. Meanwhile, yeast one-hybrid assay showed that AabZIP9 was able to bind to the “ACGT” cis-element present in both ADS and CYP71AV1 promoters. AabZIP9 gene was driven by the constitutive CaMV35S promoter and the glandular trichome specific CYP71AV1 promoter and stably transformed into A. annua plants. The transcript level of AabZIP9 was increased in both of the 35S and CYP71AV1 driven transgenic plants compared with the wild type or GUS control plants. All the transgenic A. annua plants overexpressing AabZIP9 showed elevated transcript level of ADS, but the transcription levels of CYP71AV1, DBR2, and ALDH1 have no significant change in both types of transgenic plants. The significantly upregulated ADS promoted the accumulation of artemisinin, dihydroartemisinic acid, and artemisinic acid biosynthesis in the transgenic A. annua plants. These results suggest that AabZIP9 can positively regulate the biosynthesis of artemisinin.
Introduction
For decades, malaria has been a pandemic disease, threatening people in tropical and subtropical regions of the world, compromising millions of lives. The WHO (World Health Organization) reported 219 million cases of malaria in 2017 and 435,000 deaths (WHO, 2018). Artemisinin, a secondary metabolite produced in the wild plant Artemisia annua, along with its derivatives is the main ingredient of artemisinin combination therapies (ACTs), which is currently the only effective cure for malaria (Okell et al., 2014). The discovery of artemisinin by Prof. Youyou Tu, who was further awarded the Nobel Prize in Physiology and Medicine in 2015, could significantly elevate the current antimalaria therapies and save millions of lives. In addition to antimalarial activity and due to its diverse promising functions, including anticancer (Tin et al., 2012), viral (Obeid et al., 2013), inflammatory (Chadwick et al., 2013), diabetic (Li et al., 2017), and tuberculosis therapies (Zheng et al., 2017), the demand for artemisinin has risen in the recent years.
Many studies related to artemisinin production have been carried out in various areas in order to elucidate the biosynthetic production of artemisinin through the regulation of its multi enzymatic biosynthetic pathway. The enzymatic conversion of farnesyl pyrophosphate (FPP) into amorpha-4, 11-diene by amorpha diene synthase (ADS) is the first committed step of artemisinin specific biosynthetic pathway (Bouwmeester et al., 1999), which is followed by oxidization of amorpha-4,11-diene to artemisinic alcohol and subsequently to the artemisinic aldehyde by the cytochrome P450 monooxygenase CYP71AV1 (Teoh et al., 2006). The artemisinic aldehyde is then further oxidized to artemisinic acid by CYP71AV1 or alternatively, is reduced to dihydroartemisinic aldehyde by the double bond reductase 2 (DBR2) (Zhang et al., 2008). The later stage of the pathway is then mediated by aldehyde dehydrogenase 1 (ALDH1) that will oxidize dihydroartemisinic aldehyde into dihydroartemisinic acid (DHAA), which is the direct precursor of artemisinin (Teoh et al., 2009). The final stage of artemisinin production is considered to be a photosensitized related, nonenzymatic oxidation reaction (Brown and Sy, 2007; Czechowski et al., 2016).
Despite the fact that artemisinin biosynthetic pathway is relatively well elucidated, few researches and efforts have been carried out regarding the genetic regulation of the pathway. Recently several transcription factors including AaWRKY1 (Ma et al., 2009; Han et al., 2014), AabHLH1 (Ji et al., 2014), AabZIP1 (Zhang et al., 2015), AaMYC2 (Shen et al., 2016), AaNAC1 (Lv et al., 2016), AaMYB1 (Matías-Hernández et al., 2017), AaGSW1 (Chen et al., 2017) as well as various members of AP2/ERF transcription factors (TFs) including, AaERF1, AaERF2, AaORA, and AaTAR1 (Yu et al., 2012; Lu et al., 2013; Tan et al., 2015) have been reported which significantly participate in and regulate various stages of artemisinin biosynthetic pathway. Judd et al., reported that artemisinin is also produced in the nonglandular cells of self-pollinated inbred A. annua plants, which means that the regulation of artemisinin biosynthesis may not be limited to only glandular trichomes (Judd et al., 2019). The bZIP transcription factor family is one of the biggest families in plants, which plays important roles in plant growth and development. A variety of important bZIP genes were identified in model plants such as Arabidopsis and rice. To date, only two bZIP transcription factors (TF) have been identified in A. annua which regulate the biosynthesis of artemisinin. (Zhang et al., 2015; Zhong et al., 2018). The function of this important TF family in artemisinin biosynthesis remains obscure and needs to be addressed and characterized to enable a full understanding of bZIP functions, especially in regulating plant secondary metabolism.
In this study, we aimed to identify bZIP TFs involved in artemisinin biosynthesis in A. annua. A total of 86 putative AabZIP TFs genes were retrieved for coexpression analysis. One candidate bZIP TF, designated as AabZIP9, was highly expressed in trichome-enriched tissues (young-leaf, bud, and flower) and grouped with the artemisinin specific biosynthetic pathway genes.
Materials and Methods
Plant Materials
The seeds of A. annua (cultivar “Huhao 1”) were bred by our lab for several years in Shanghai, China. The seeds were sown in pots and grown in a growth chamber under standard conditions of light, temperature, and humidity (16/8 light/dark period, 25 ± 2°C, 50%–70% relative humidity). The Nicotiana benthamiana (a close relative of tobacco) seeds were also sown in pots and grown under the same conditions as for A. annua.
Bioinformatics Analysis and Gene Isolation
The transcriptome sequences of seven different tissues of A. annua were generated by our laboratory (Shen et al., 2018) and translated into protein sequences in six frames. The AabZIP homologs were identified by the HMM search against the A. annua protein sequence database with E value < 1×10-5 using conserved bZIP domain (PF00170) which was obtained from the Pfam database (Punta et al., 2012). The reads of each bZIP TFs in seven different tissues (young leaf, old leaf, flower, bud, root, stem, and seed) were obtained by BLASTN search against each database with E value < 1 × 10-6, and read counts were normalized by calculating the value of reads per kilobase per million (RPKM). To predict potential AabZIPs transcription factors that could be involved in the biosynthesis of artemisinin, coexpression analysis was performed based on RNA-seq data, using Multi-Experiment Viewer (MeV4.9.0) software (Saeed et al., 2003). Sample clustering was carried out using the HCL method and the evolutionary distances were computed with the Poisson correction and single linkage clustering (Eisen et al., 1998). The bZIP protein sequences of the Arabidopsis (retrieved from TAIR, The Arabidopsis Information Resource) and A. annua were aligned with ClustalW (Larkin et al., 2007). The unrooted phylogenetic tree was generated with MEGA6.1 software (Tamura et al., 2013), according to the neighbor-joining method and bootstrapping with 10,000 replicates to evaluate the accuracy of phylogenetic construction. The evolutionary distances were computed with the Poisson correction and partial deletion option method. Nuclear localization signal of AabZIP9 was predicted via NucPred online analysis tool (http://nucpred.bioinfo.se/nucpred/) (Brameier et al., 2007).
For gene isolation, the gene-specific primers, AabZIP9-F, and AabZIP9-R, were designated according to the assembled RNA-seq data. PCR was performed according to the manufacturer’s instructions for KOD DNA polymerase (Toyobo, Japan), using A. annua young leaf cDNA as template. The PCR products were cloned into the pLB vector (Tiangen, China) and sequenced (Biosune, China). The primers used in this study are listed in Supplemental Table 2.
Generation of Plasmid Vectors
In order to obtain plant overexpression vector and perform dual Luciferase reporter gene assay (dual-LUC), AabZIP9 ORF was amplified with primers P3+P4, and cloned into pHB vector (driven by CaM35S), and this vector was also used for A. annua stable transformation. The promoters of ADS, CYP71AV1, DBR2, and ALDH1, previously generated by our lab (Hao et al., 2017), were also cloned into pGreenII 0800-LUC reporter plasmid (Hellens et al., 2005). For subcellular localization analysis, AabZIP9 was amplified with primers P5+P6 and cloned into Gateway vector pENTR-TOPO (Invitrogen, USA) and subsequently recombined into pEarleyGate104 (Earley et al., 2006) by LR reaction (Invitrogen, USA) to generate pEarleyGate104-YFP-AabZIP9. For gene overexpression in A. annua plant, the AabZIP9 was amplified with primers P7+P8, and cloned into a pHB-proCYP vector (BamHI/XbaI, the CaM35S promoter was replaced by the trichome specific CYP71AV1 promoter) via ClonExpress® II One Step Cloning (Vazyme, China). For yeast one-hybrid assay, the open reading frames (ORFs) of AabZIP9 was amplified with primers P9+P10 and then cloned into pB42AD (EcoRI/XhoI) by using ClonExpress II (Vazyme, China). The artificial synthesized triplicate cis-element promoter segments contain possible bZIP binding element from ADS, CYP71AV1, DBR2, and ALDH1 were inserted into pLacZ (EcoRI/XhoI) to generate reporter vector. The sequences of artificially synthesized triplicate cis-elements were listed in Supplemental Table 2.
Subcellular Localization Analysis and Dual-LUC Assay via N. benthamiana Leaf Infiltrations
The leaf infiltration assay was carried out as described by Moses et al. with minor modifications (Moses et al., 2015). Briefly, the plant expression constructs for expression in N. benthamiana were transformed into Agrobacterium tumefaciens strain GV3101. For dual-LUC analysis, the four pGreenII 0800-LUC reporter plasmids were each cotransformed with the helper plasmid pSoup19 into GV3101. All strains were first incubated overnight (180 rpm) at 28°C in 5 ml LB broth medium, supplemented with antibiotics (50 mg/L rifampicin, 50 mg/L kanamycin, and 20 mg/L gentamycin) and containing 10-mM MES (pH 5.7) and 40-µM acetosyringone. All the Agrobacterium cultures were collected by centrifugation (at 4,000 g), and then resuspended in 5 ml of infiltration buffer (200-µM acetosyringone, 10-mM MgCl2, and 10-mM MES, pH 5.7) to reach the OD600 = 0.6 and then incubated at room temperature for 3 h. For subcellular localization analysis, the strains were infiltrated into fully expanded 4-week-old N. benthamiana leaves. After 3 days, a piece of infiltrated leaf was cut and the images were obtained under LAS AF Lite laser scanning confocal microscopy (Leica, Germany), with argon laser excitation at 488 nm and a 505–550-nm emission filter set. Subcellular localization was done in three biological replicates. For dual-LUC analysis, the reporter strains and effector strain were mixed at the ratio 1:3, then the bacteria mixtures were infiltrated into fully expanded 4-week-old N. benthamiana leaves. The pHB-GFP was used as negative control. After 48 h, the infiltrated leaf discs (1 cm in diameter) were collected for dual-LUC assay using dual-LUC reaction reagents according to the manufacturer (Promega, USA). Four biological replicates were measured for each sample.
Yeast One-Hybrid Assay
The yeast one-hybrid assay was performed as described previously (Yan et al., 2017). The possible cis-elements in the promoters of ADS, CYP71AV1, DBR2, and ALDH1 were predicted via PlantCARE online tools (bioinformatics.psb.ugent.be/webtools/plantcare/html/). In brief, the pB42AD-AabZIP9 plasmid was cotransformed into yeast strain EGY48a with each pLacZ-Boxes, respectively, via LiAc mediated method. The cotransformed of pB42AD-AabZIP9 and pLacZ-control was set as negative control. The transformants were cultivated on synthetic minimal double dropout medium deficient in Trp and Ura (SD/-Leu/-Ura) at 30°C for 3 days. Yeast cells grown on SD/-Leu/-Ura medium plates were transferred and suspended with sterile water, then dropped on a selected medium SD-GAL/RAF with X-Gal plate at 30°C for 24–36 h. Six biological replicates were measured for each yeast transformant.
Plant Transformation of A. annua
The plant overexpression construct (pHB-AabZIP9, pHB-pCYP-AabZIP9, and pHB-GUS) were introduced into Agrobacterium tumefaciens strain EHA105, and the resulting strains were used in the transformation of A. annua. The transgenic plants of A. annua were generated as described previously (Shen et al., 2012). The rooted transformed A. annua plantlets were transferred into soil pots in the growth chamber for one month and then transplanted into the greenhouse. The T0 transgenic plants of A. annua were confirmed by genomic DNA-based PCR on both AabZIP9 and Hyg (Hygromycin) resistant genes by using primers P11+P12, P13+P14, and Hyg-F+ Hyg-R, respectively. All the primers used in genomic DNA-based PCR were listed in Supplemental Table 2.
Relative Expression Analysis via Quantitative Real-Time PCR
Leaf samples were collected as described previously (Yan et al., 2017). Leaf1 to leaf5 (Figure 5A) were collected for gene expression analysis. Samples were picked and immediately frozen in liquid nitrogen and stored at –80°C. The total RNA was extracted by RNAprep pure Plant Kit according to the manufacturer (Tiangen, China). 1.0 µg of RNA was used to synthesize the cDNA using PrimeScript RT Master Mix Kit (TaKaRa, China). qRT-PCR assays were performed by using SuperReal PreMix SYBR Green Kit (Tiangen, China) on the LightCycle96 machine (Roche, Switzerland) as reported before (Shen et al., 2016). The relative expression levels of genes were compared with the expression of A. annua β-actin and calculated by the 2-ΔCt. Three biological×four technical replicates were measured for each sample. All the primers used in qRT-PCR were listed in Supplemental Table 2.
HPLC Analysis of Metabolites in A. annua
Four months old plant leaves were collected for HPLC analysis, samples of the transgenic plants and control plants were prepared as described previously (Lu et al., 2013). Leaf samples were dried in a drying oven at 45°C and then ground to powder and extracted with methanol in an ultrasonic processor under the conditions of 25°C and 50W for 30 min. The samples were centrifuged for 10 min at 10,000 g and then the supernatants were passed through a 0.25-µm membrane.
The filtrates were then used for metabolites analysis by Waters Alliance 2695 HPLC system coupled with a Waters 2420 ELSD detector. The conditions for HPLC were set as described previously (Lu et al., 2013). Standard of artemisinin was purchased from Sigma and standard of dihydroartemisinic acid and artemisinic acid were bought from Guangzhou Honsea Sunshine Bio Science and Technology Co. Ltd (Honsea Sunshine Bio, China). Three biological replicates were measured for each sample.
Results
Global Expression Analysis of bZIP Transcription Factors in A. annua
The transcriptomic sequencing data of seven different tissues of A. annua was generated via RNA-seq in our lab (Shen et al., 2018). The genes encoding putative bZIP proteins in A. annua were identified using conserved bZIP domain (PF00170) through the HMM (hidden Markov model) search program providing the 86 unique genes encoding proteins containing the bZIP domain (Supplementary Table 1). Since the biosynthesis of plant secondary metabolites is usually species or tissue specific (Goossens, 2014), to identify transcription factors involved in the regulation of artemisinin biosynthesis we looked at their expression across tissues. We identified several TFs of the bZIP family which showed expression profiles similar to that of genes of the artemisinin biosynthetic pathway such as ADS, CYP71AV1, DBR2, and ALDH1 (Figure 1A and Supplementary Figure 1). AabZIP9 is one of the candidates that clustered with those pathway genes bearing a higher expression level in buds and young leaves, which could be associated with its involvement in artemisinin biosynthesis.

Figure 1 Global expression profile, sequence alignment, and phylogenetic analysis of AabZIP9 transcription factor in A. annua. (A) Hierarchical cluster analysis of bZIP TFs (partial) in A. annua. The color scale at the top represents the value of RPKM (Reads per Kilobase per Million mapped reads); (B) Alignment of AabZIP9 with related bZIP proteins from Helianthus annuus, Nicotiana tabacum, Medicago truncatula, Arabidopsis thaliana, Solanum lycopersicum, etc. The amino acid sequence of AabZIP9 was aligned with C.cardunculus bZIP9 (KVH91346.1), H.annuus bZIP9 (XP_022029119.1), N.tabacum bZIP9 (XP_016449896.1), M.truncatula bZIP9 (XP_003588595.1), G.max bZIP9 (XP_006601038.1), S.lycopersicum bZIP9 (XP_004244469.1), E.grandis bZIP9 (XP_010049789.1), G.hirsutum bZIP9 (XP_016720593.1), and A. thaliana bZIP9 (NP_568457.1) using the Vector NTI 10.3 software. The amino acid residues shaded with yellow, blue, and green, respectively, the high, intermediate and low sequence similarity of AabZIP9 with respect to the other species. The conserved bZIP domain region was marked with red box, and the nuclear localization signal of AabZIP9 was marked with red inverted triangle. (C) Phylogenetic analysis of AabZIP9 and all the bZIP TFs from Arabidopsis. The amino acid sequences of Arabidopsis bZIP TFs were obtained from the Arabidopsis Information Resource database (http://www.arabidopsis.org). The phylogenetic tree presented here was analyzed by MEGA 6.1 software, according to the neighbor-joining method. The type-c bZIP subfamily was highlighted by blue background.
Gene Cloning and Identification
The full length of AabZIP9 (GenBank accession: MG584701) was found to be a 762 bp open reading frame, coding 254 amino acids, isolated from the cDNA of young leaves from A. annua “Huhao 1” cultivar. The BLAST results of bZIP9 proteins from other plants species are shown in Figure1B. The results indicate that AabZIP9 has 83% of protein sequence identity with Cynara cardunculus CabZIP9, 80% with Helianthus annuus HabZIP9, 62% with Nicotiana tabacum NtZIP9, 55% with Solanum lycopersicum SlbZIP9, and 52% with Arabidopsis thaliana AtbZIP9. The analysis of the AabZIP9 protein sequence performed with the NucPred online program (Brameier et al., 2007) revealed the presence of a putative nuclear localization signal (NLS; prediction score = 0.92.) indicated with an inverted red triangle in Figure 1B. Arabidopsis bZIP TFs (AtbZIP) are subdivided into ten groups accordingly to the similarity of their core domain and additional conserved motifs (Jakoby et al., 2002). Based upon sequence similarity, AabZIP9 groups together with type-C AtbZIPs which includes AtbZIP9 (At5g24800), AtbZIP10 (At4g02640), AtbZIP25 (At3g54620), and AtbZIP63 (At5g28770) genes (Figure 1C).
AabZIP9 Localizes to the Nucleus Where it Activates the Expression of the ADS Gene
The subcellular localization of AabZIP9 was examined via transient expression in epidermal cell of N. benthamiana of the AabZIP9 gene fused with the YFP reporter (35S:AabZIP9-YFP). The fusion protein (35S:AabZIP9-YFP) showed strong YFP fluorescence in the nucleus, whereas the control YFP (35S:YFP) signal was distributed in both the cytoplasm and the nucleus (Figure 2). This confirms that AabZIP9 localizes to the nucleus.
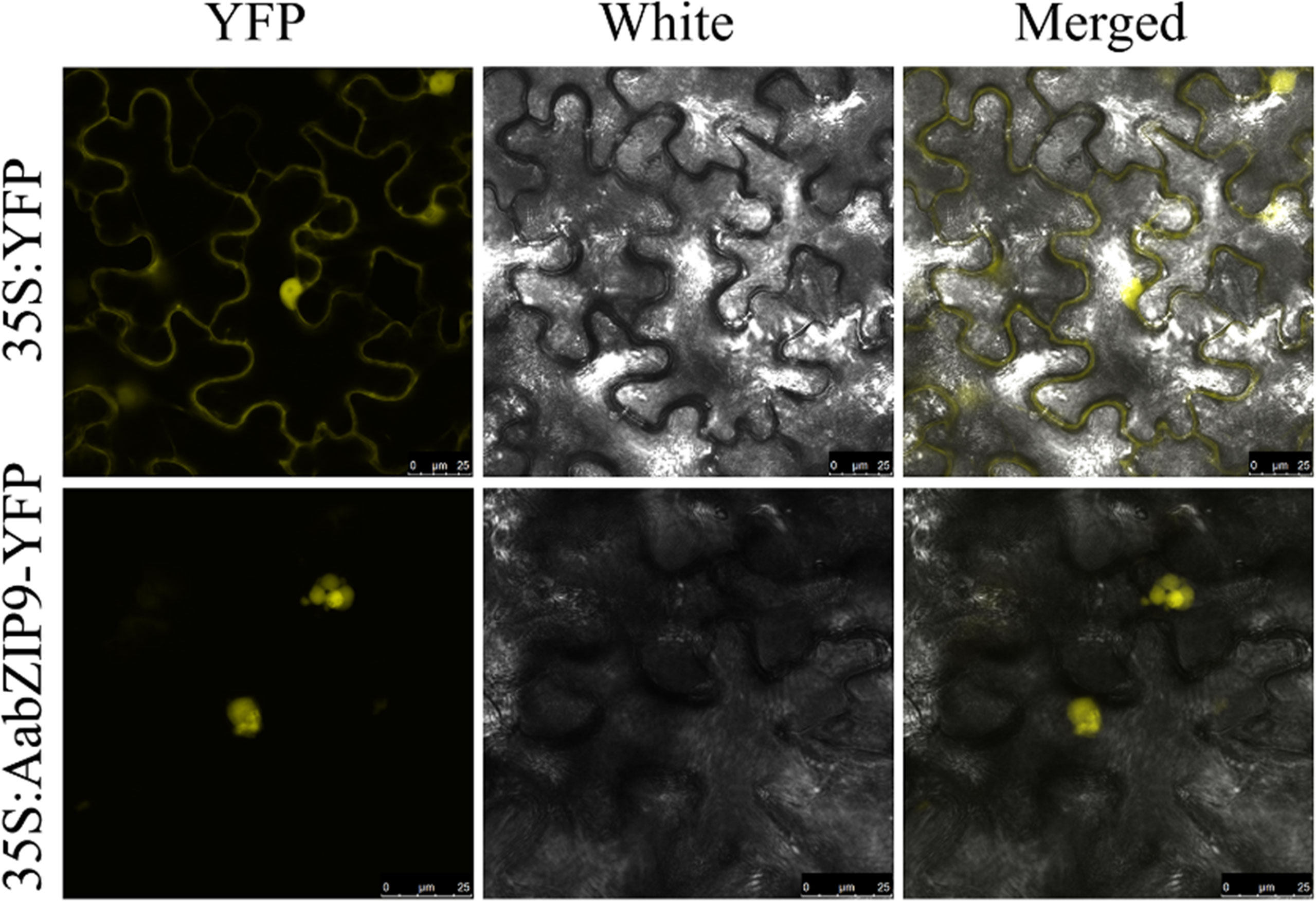
Figure 2 Nuclear localization of AabZIP9 proteins in N. benthamian. The coding sequence of AabZIP9 was in-frame fused with YFP, and under the control of the 35S promoter, then transferred into Agrobacterium GV3101 and infiltrated into leaves of N. benthamiana. The Agrobacterium strain harboring empty vector was used as control. Fluorescence (left), bright field (middle), and the corresponding merged (right) images of cells were observed under LAS AF Lite laser scanning confocal microscopy (Leica, Germany).
To determine the activation specificity of AabZIP9, a dual-luciferase (dual-LUC) assay was performed in N. benthamiana leaves. The dual-LUC assay system provides an efficient means of performing two reporter assays. In this assay, the firefly and renilla luciferases are simultaneously expressed and measured sequentially from a single sample to improve experimental accuracy. The full-length cDNA of AabZIP9 was inserted into the plant overexpression vector pHB as an effector construct (pHB-AabZIP9) (Figure 3A). The four constructs containing promoters of genes ADS, CYP71AV1, DBR2, and ALDH1 in the pGreenII 0800-LUC vector were previously generated by our lab and used as reporter constructs. The effector and reporter constructs were transiently expressed in N. benthamiana leaves using Agrobacterium tumefaciens-mediated coinfiltration. The pHB-GFP construct containing the green fluorescent protein (GFP) reporter was also coinfiltrated with each constructs as a negative control. The dual-LUC assay showed that only the ADS promoter’s activity was significantly enhanced by AabZIP9 transcription factor when compared with the GFP control (5.1-fold, **P <0.01), whereas the activity of CYP71AV1, DBR2, and ALDH1 promoters exhibited nonsignificantly differences (Figure 3B).
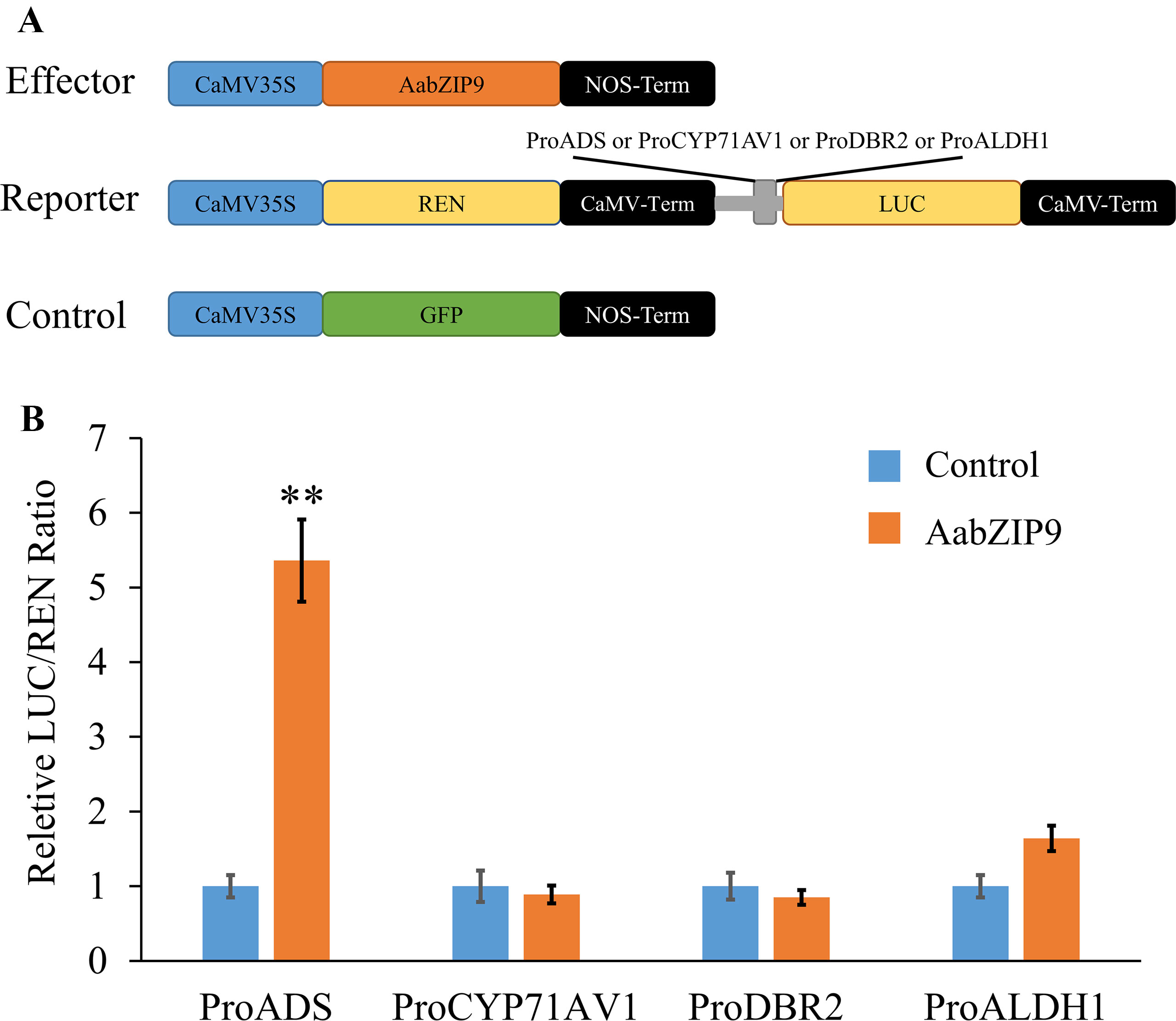
Figure 3 Transient dual-LUC analysis of AabZIP9 with four artemisinin biosynthesis specific pathway gene promoters. (A) Schematic representation of effector and reporter constructs used in the dual-LUC assays. Effector construct contains the AabZIP9 coding sequence driven by the CaMV35S promoter. ProADS : LUC, ProCYP71AV1:LUC, ProDBR2:LUC, and ProALDH1:LUC were used as reporter constructs. The REN (Renilla reniformis) was driven by the CaMV35S promoter and used as internal control. The effector construct contains green fluorescent protein (GFP) driven by CaMV35S promoter was used as negative control. (B) Dual-LUC analysis of AabZIP9 activating of amorpha diene synthase (ADS) promoter in N. benthamiana leaves. The values were reached by calculating the relative ratio of LUC activities to REN activities (LUC/REN), and the negative control was set to one. Error bars represent ± SD (n = 4), t-test, **P < 0.01).
AabZIP9 Bind to “ACGT” Cis-Elements Within the Promoters of ADS and CYP71AV1
Transcription factors have DNA binding domain for binding to the promoter of corresponding genes, thereby inducing or repressing transcription of downstream target genes. Several studies reported that plant bZIP proteins preferentially bind to DNA sequences with an “ACGT” core in the A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) (Jakoby et al., 2002). Therefore, we performed promoter cis-elements analysis in PlantCARE (Lescot et al., 2002), to identify the putative binding site of bZIP proteins in the promoter region of genes of artemisinin biosynthetic pathway. Sequence analysis revealed that the promoters of ADS, CYP71AV1, DBR2, and ALDH1 contained two, three, two, and two putative bZIP binding cis-elements, respectively (Figure 4A). To determine whether AabZIP9 binds to the cis-elements of the ADS, CYP71AV1, DBR2, and ALDH1 promoters in vivo, a yeast one-hybrid assay was performed. Each of these putative cis-elements along with the nucleotides in the flanking region was artificially synthesized into 3 × element triple fragment and inserted into the yeast one-hybrid pLacZ reporter vector (Supplementary Figure 2). The ORF of AabZIP9 was inserted into the pB42AD vector to generate effector construct (Supplementary Figure 2). The effector along with each respective reporter construct were cotransferred into the yeast EGY48 strain via LiAc mediated method as described before (Yan et al., 2017). As shown in Figure 4B, only the yeast cells harboring pB42AD-AabZIP9, proADS-box1, and proCYP71AV1-box1 were able to grow on the SD-Gal-Raf selective medium containing 20-mM X-gal as displayed by the development of the blue color. This indicates that AabZIP9 binds to the “ACGT” cis-elements within the promoters of genes ADS and CYP71AV1, but it fails to bind to the cis-elements of the other genes we tested. We think that this is due the flanking sequences of the cis-element which may affect the binding ability of AabZIP9 as it has previously been reported (Dombrecht et al., 2007).
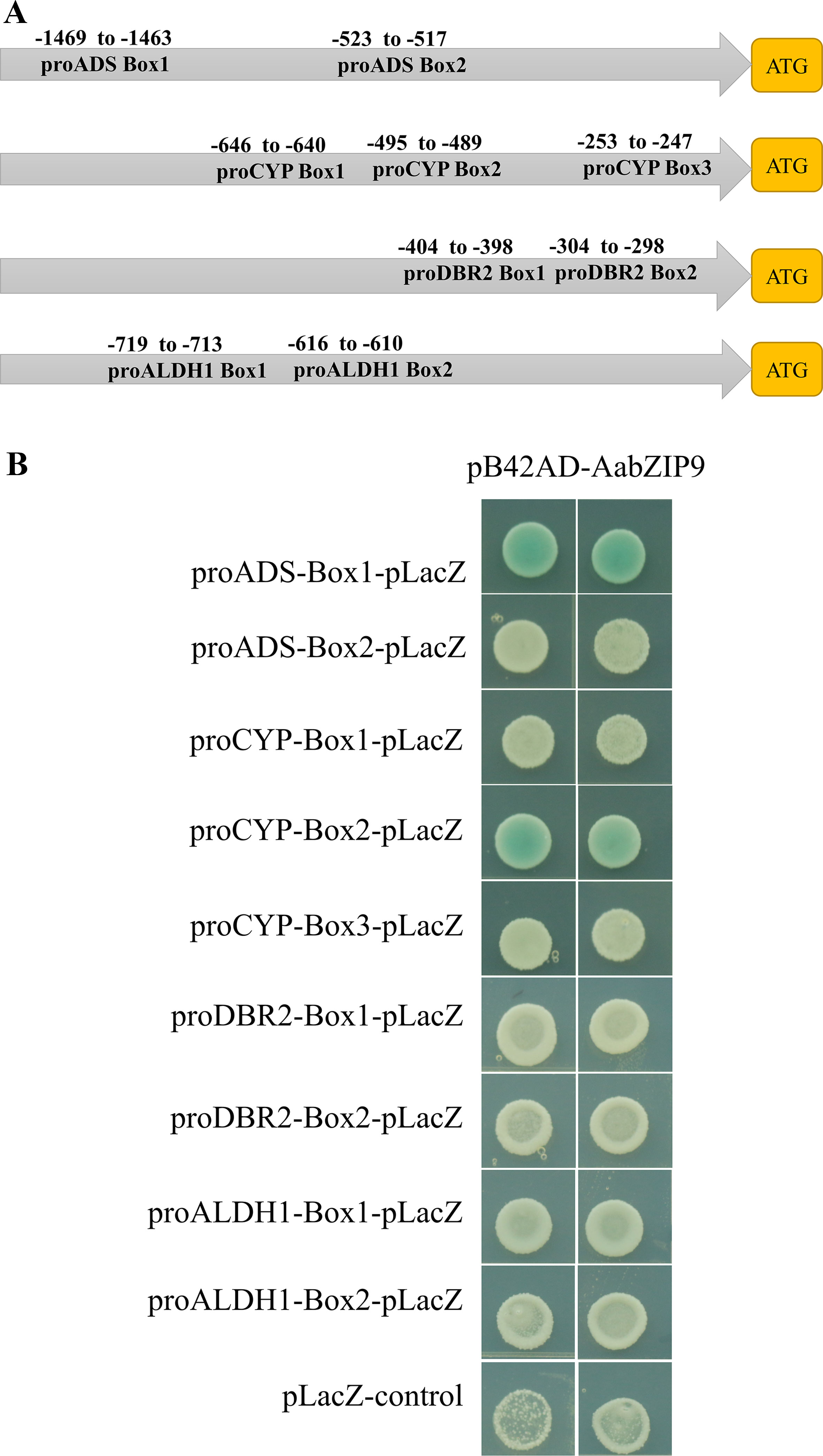
Figure 4 Binding assay of AabZIP9 to “ACGT” cis-elements in the promoter. (A) Schematic representation of all “ACGT” cis-elements boxes position in the four promoters. (B) The result of yeast one-hybrid assay for the interaction between AabZIP9 with DNA motifs. Triple of each “ACGT” cis-elements with flanking sequences were used as bait, respectively. The transformants were cultivated on selected medium SD-GAL/RAF with X-Gal at 30°C for 24–36 h.
AabZIP9 Regulates the Transcription of Genes in the Artemisinin Biosynthetic Pathway
Since AabZIP9 was able to bind to and activate ADS promoter, it was interesting to know whether overexpression of AabZIP9 could active ADS gene and therefore promote artemisinin biosynthesis. Meanwhile, in order to compare the efficiency of constitutive and trichome specific promoters, the promoter of CaMV35S and the promoter of gene CYP71AV1 were used for overexpressing AabZIP9 in A. annua plants respectively. We constructed the pHB-35S-AabZIP9 and pHB-proCYP-AabZIP9 plant overexpression vector and used pHB-35S-GUSplus vector as control one (Supplementary Figure 3). All these vectors were transformed into A. annua via the Agrobacterium-mediated method as described before (Zhang et al., 2009). In total, eight (driven by 35S promoter) and 10 (driven by the promoter of gene CYP71AV1) independent transgenic plants of A. annua were confirmed by genomic DNA-based PCR on both AabZIP9 and Hyg (Hygromycin) resistant genes.
Quantitative real-time PCR (qRT-PCR) was performed to measure the expression levels of AabZIP9 in the transgenic plants. Previous research on expression profile of CYP71AV1 shows that this promoter is highly expressed in young leaves and its expression gradually declines with age (Wang et al., 2012; Yu et al., 2012). Therefore, the expression levels of AabZIP9 in transgenic and wild-type plants were compared among five different development stages of leaves (from young to old) (Figure 5A). As shown in Figure 5B, the expression of AabZIP9 in 35S:: AabZIP9 lines is three to four times higher than in wild type and control plants. However, when the expression of AabZIP9 was driven by the trichome specific CYP71AV1 promoter, it showed a pattern of expression similar to that of CYP71AV1 with higher expression in young than in old leaves (Wang et al., 2012; Yu et al., 2012). Nevertheless, the expression levels of AabZIP9 (in leaf1 and leaf2) in the CYP71AV1 promoter driven transgenic lines were increased about 1.5- to 2-folds when compared with wild-type or GUS control plants (in leaf1 and leaf2), while the expression levels of AabZIP1 showed no significant difference between transgenic lines and wild-type or GUS control in the leaves from leaf3 to leaf5, Figure 5B.
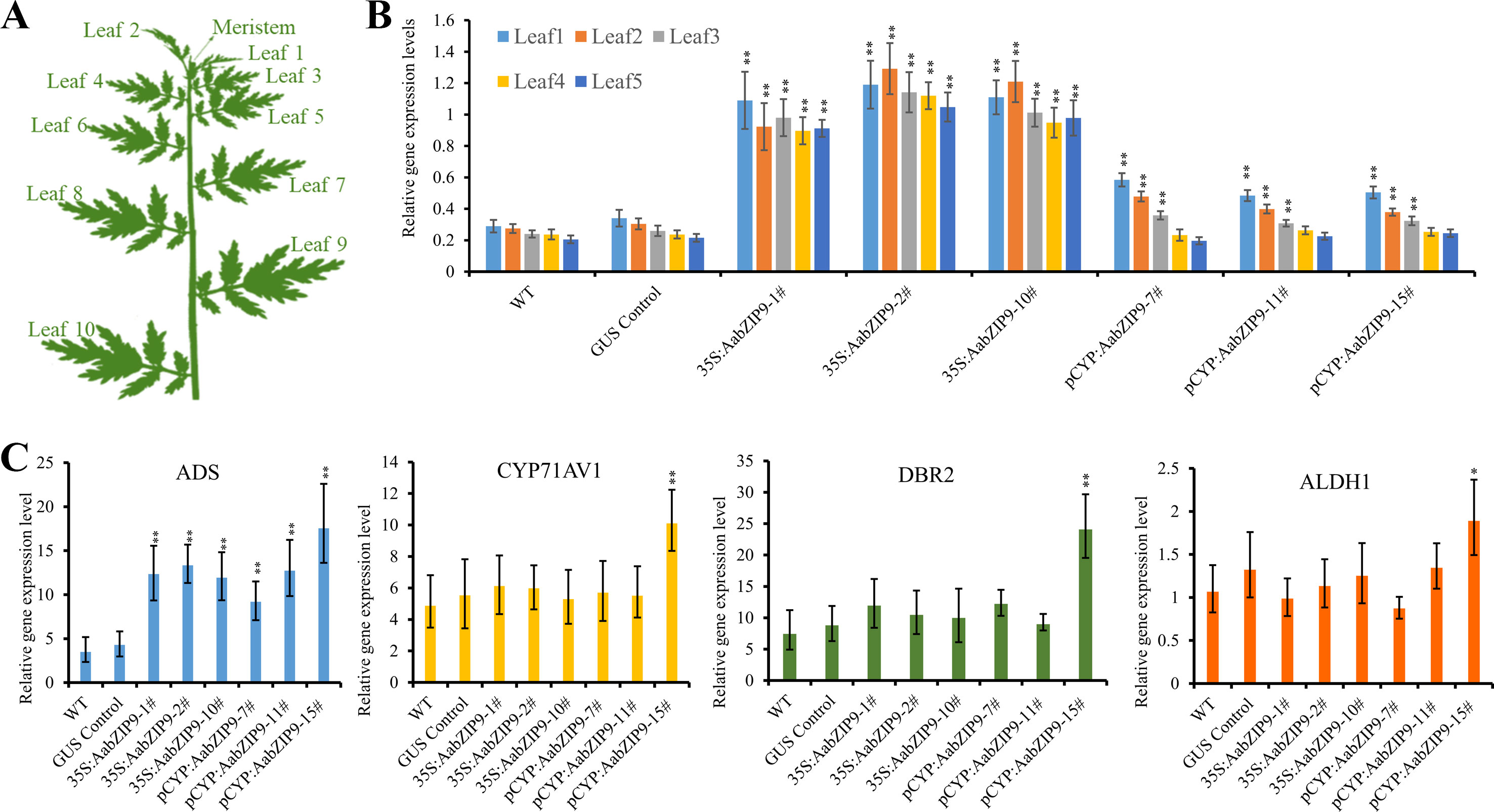
Figure 5 Overexpression of AabZIP9 upregulated expression levels of amorpha diene synthase (ADS) genes. (A) Schematic diagram of A. annua plant shows different positions of leaves (Yan et al., 2017). (B) The expression levels of AabZIP9 in five different position of leaves in the wild type, GUS control and 35S promoter or CYP71AV1 promoter driven overexpressed plants. β-actin was used as the internal standard. Error bars indicate ± SD of four technical replicates. Statistical significance was determined by Student’s t-test with paired and two-tailed distribution methods. Asterisks indicate the difference between overexpressing transgenic plants and wild type plants at the same position of each leaf (**, P < 0.01). (C) Relative expression of ADS, CYP71AV1, DBR2, and ALDH1 in leal2 of each wild type, GUS control and 35S promoter or CYP71AV1 promoter driven overexpressed plants. β-actin was used as the internal standard. Error bars indicate ± SD of three biological×four technical replicates. Statistical significance was determined by Student’s t-test with paired and two-tailed distribution methods. Asterisks indicate the difference between overexpressing transgenic plants and wild type plants (**, P < 0.01; *, P < 0.05)
Taking into account the expression pattern of AabZIP9 in different leaves, we used the second leaf from each transgenic and wild-type plants to measure the steady state level of genes in the artemisinin biosynthetic pathway. As expected, the expression level of ADS gene was upregulated in both 35S and CYP71AV1 promoters driven transgenic plants. Intriguingly, the expression levels of ADS in both 35S and CYP71AV1 promoters driven transgenic plants were similar, although the AabZIP9 gene showed higher expression levels in 35S promoter driven transgenic plants than in the CYP71AV1 promoter driven plants (Figure 5C). In most of the transgenic plants, the expression levels of CYP71AV1, DBR2, and ALDH1 showed no significant difference between the transgenic and wild-type plants, except in the pCYP : AabZIP9-15 plant (Figure 5C). ADS, CYP71AV1, DBR2, and ALDH1 were all upregulated in plant pCYP : AabZIP9-15 compared to wild-type and control (Figure 5C).
Overexpression of AabZIP9 Increases Artemisinin, Artemisinic Acid and Dihydroartemisinic Acid Biosynthesis in A. annua Transgenic Plants
Overexpression of AabZIP9 did not cause any visible morphological modification and all transgenic lines appear similar to the wild-type (Supplementary Figure 4). The artemisinin, dihydroartemisinic acid, and artemisinic acid contents in leaves of the 4-month-old A. annua plants were analyzed by high-performance liquid chromatography (HPLC). The results of the HPLC analysis revealed that in the 35S::AabZIP9 and CYP::AabZIP9 lines the content of artemisinin, dihydroartemisinic acid, and artemisinic acid level are 23.2%–67.1%, 34.5%–92.8%, and 40.4%–121.2% higher than in wild-type plants (Figures 6A, B). These results indicated that AabZIP9 is a positive regulator of artemisinin biosynthesis.
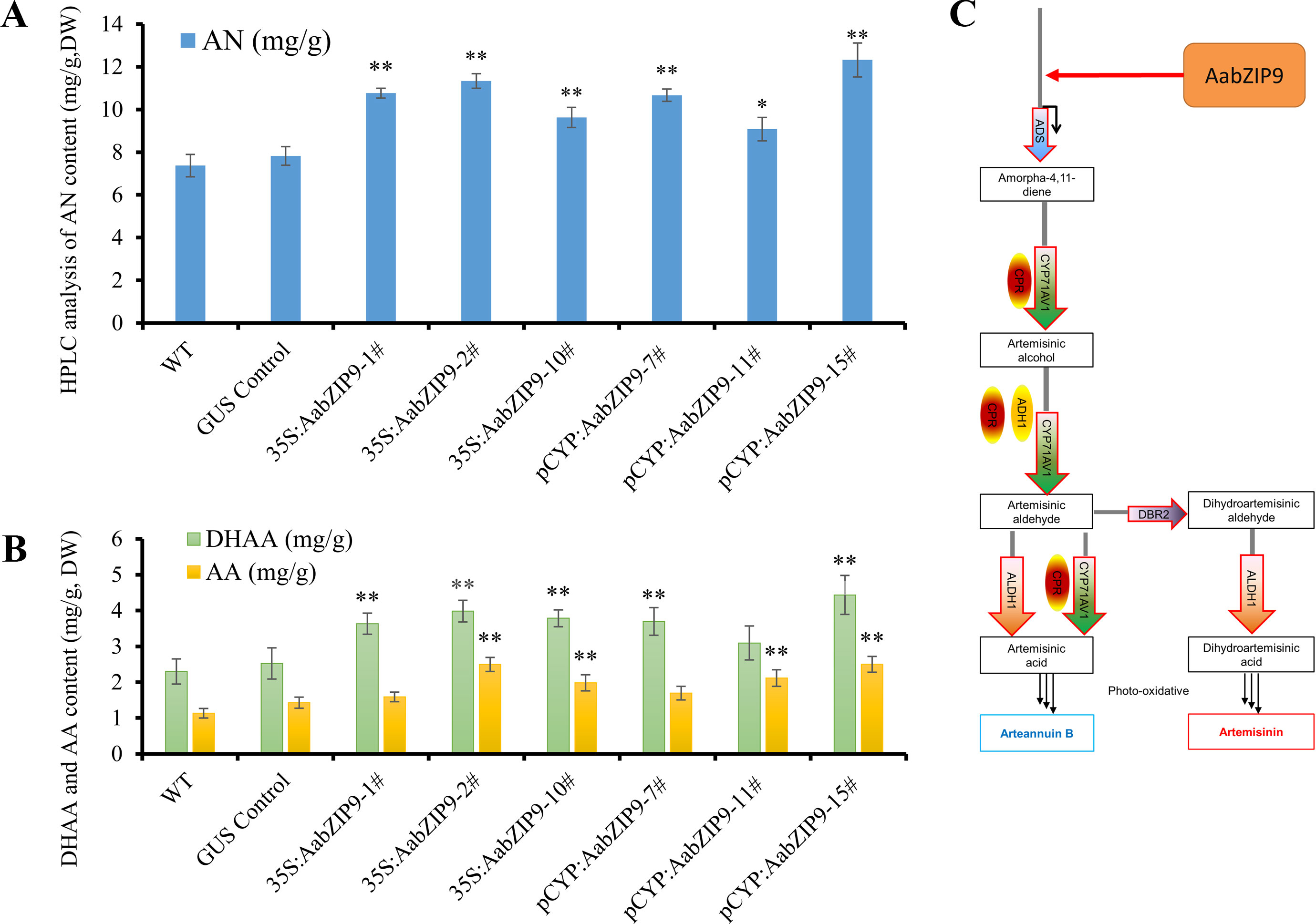
Figure 6 High-performance liquid chromatography (HPLC) analysis of metabolites in transgenic plants overexpressing AabZIP9. (A) Contents of artemisinin (AN) in the 4-month-old leaves of wild type, GUS control, and 35S promoter or CYP71AV1 promoter driven overexpressed plants, determined by HPLC. The contents of AN in transgenic plant leaves were compared to the wild type plants. Statistical significance was determined by t-test (**, P < 0.01; *, P < 0.05). Asterisks indicate the difference between overexpressing transgenic plants and wild type plants. (B) Contents of dihydroartemisinic acid (DHAA) and artemisinic acid (AA) in the 4-month-old leaves of wild type, GUS control, and 35S promoter or CYP71AV1 promoter driven overexpressed plants, determined by HPLC. The contents of DHAA and AA in transgenic plant leaves were compared to the wild type plants. Statistical significance was determined by t-test (**, P < 0.01; *, P < 0.05). Asterisks indicate the difference between overexpressing transgenic plants and wild type plants. Three biological replicates were measured for each sample. (C) A simplified model of action of AabZIP9 in regulating artemisinin biosynthesis in Artemisia annua.
Discussion
Due to the important medicinal properties of artemisinin, such as antimalarial, anticancer (Tin et al., 2012), antituberculosis (Zheng et al., 2017), understanding how the artemisinin biosynthetic pathway is regulated, is of extreme relevance. Despite the considerable progresses which have been achieved in producing artemisinin in microbes via semisynthetic synthesis (Ro et al., 2006; Paddon et al., 2013), A. annua is still the main resource for artemisinin (Peplow, 2016). However, the transcriptional regulation of this important pathway in A. annua is not yet well established. The bZIP transcription factors are one of the largest transcriptional regulators playing crucial roles in plant physiological processes, plant development, plant metabolism, and in numerous biotic/abiotic stress responses (Ang et al., 1998; Chen et al., 2016) (Osterlund et al., 2000; Tang et al., 2012). To date, the abscisic acid (ABA) response AabZIP1 transcription factor and AaABF3 are the only two bZIP TFs, which have been reported to play a significant role in the regulation of the artemisinin biosynthesis in A. annua (Zhang et al., 2015; Zhong et al., 2018). In this study, we isolated and characterized a new bZIP transcription factor, named AabZIP9, which regulates the biosynthesis of artemisinin.
Coexpression analysis has proved to be an efficient tool for successful gene discovery in plant secondary metabolism. Based on the RNA-seq data from different tissues of A. annua, 86 putative bZIP TFs in A. annua were retrieved and subject to coexpression analysis with the artemisinin specific biosynthetic pathway genes (ADS, CYP71AV1, DBR2, and ALDH1) (Figure 1A). Sequence alignment and phylogenetic analysis based on the Arabidopsis bZIP TFs classification, revealed that AabZIP9 belongs to the type-C subgroup of bZIP TFs (Figure 1C). There are only a few genes of this bZIP C-type subgroup in plants which have been cloned and characterized, including maize OPAQUE2 and parsley CPFR2. The bio-functions of this subgroup in plants are limited, and the information available on OPAQUE2 indicates that it regulates seed storage protein production (Vicente-Carbajosa et al., 1997), whereas CPFR2 might be involved in responses to light (Weisshaar et al., 1991). However, our AabZIP9 was found to be involved in plant secondary metabolism and it was of interest to elucidate if AabZIP9 regulates storage protein genes expression in the A. annua and responses to light signalling.
The plant bZIP proteins preferentially bind to DNA sequences with an “ACGT” core cis-element. The yeast one-hybrid result indicated that AabZIP9 interacts with the “ACGT” elements in the promoter region of the ADS and CYP71AV1 (Figure 4B). Furthermore, the in vivo transactivation of AabZIP9 to the ADS promoter was confirmed by transiently Agrobacterium infiltration in tobacco leaf and stably transformed in A. annua plants (Figures 3B and 5C). Yeast one-hybrid result showed that AabZIP9 binds to the “ACGT” element in the CYP71AV1 promoter (Figure 4B). However, the results of the dual-LUC assay indicate that AabZIP9 did not affect the activity of CYP71AV1 promoter (Figure 3B), and no significant upregulated gene expression of CYP71AV1 was found in almost all of the AabZIP9 overexpressed A. annua plants (both 35S and CYP71AV1 promoter driven transgenic plants) (Figure 5C). These results suggest that ADS is a target gene of AabZIP9 transcription factor (Figure 6C). AabZIP9 was founded only able to promote the expression level of ADS gene, which limited its functions in promoting the whole artemisinin biosynthetic pathway and yielded lower artemisinin content compared with our previous ones. An abnormal gene expression pattern was found in the pCYP : AabZIP9-15 transgenic plant as showed in Figure 5C. ADS, CYP71AV1, DBR2, and ALDH1 were all upregulated in plant pCYP : AabZIP9-15, and as a consequence, the highest content of artemisinin was detected in this plant (Figure 6A). According to our finding, we speculated that there is a possibility of silencing or even destruction of an artemisinin biosynthesis negative regulator gene during transformation. The identification of possible silenced negative regulator and its characterization may introduce and open a new insight in artemisinin metabolic engineering and its higher production which will remain for further investigation.
Early this year a report had indicated that the nonglandular trichomes cells can also produce trace amounts of artemisinin (Judd et al., 2019), however, the glandular trichomes were still cells that synthesized and stored of artemisinin. Moreover, the artemisinin specific synthetic pathway genes are all glandular trichome specific (Olsson et al., 2009). Therefore, overexpressing exogenous gene in A. annua by glandular trichome specific promoter seems more promising than using a constitutive promoter. In this study, we overexpressed the AabZIP9 gene by using 35S constitutive promoter and the glandular trichome specific CYP71AV1 promoter, respectively. However, based on gene expression analysis, we found that the 35S promoter driven transgenic plants showed even higher expression levels of AabZIP9 than the CYP71AV1 promoter driven transgenic ones, this results were consistent with Han’s study (Han et al., 2014). The downstream target gene (ADS) expression levels showed nearly the same abundance in two types of transgenic plants (Figure 5C). Then, we checked the GUS control plants which were also driven by the 35S promoter, the GUS staining assays showed that 35S promoter has very strong activities in most of the tissues such as the nonglandular trichomes, the leaf vein, the stomatal cells, and the mesophyll tissue (Supplementary Figure 5), but only few glandular trichomes were stained (Supplementary Figure 5). Due to the low expression of the 35S promoter in glandular trichome, the 35S promoter driven overexpressed AabZIP9 demonstrated limited regulation of target genes in A.annua (Figure 5C). We speculate that the reason for overexpressing exogenous genes via 35S promoter could increase the artemisinin content may partially depend on the nonglandular trichome pathway, which was proved by the previous report (Judd et al., 2019). The CYP71AV1 promoter region (GenBank: FJ870128.1) is glandular trichome specific, and the results obtained by Han et al. show that CYP71AV1 promoter is more efficient than 35S promoter in application for artemisinin metabolic engineering (Han et al., 2014). These results led us to use CYP71AV1 promoter for overexpressing AabZIP9 in A. annua plants. Indeed, the AabZIP9 gene expression levels were upregulated and the artemisinin contents were promoted in the transgenic plants while compared with the wild-type plants (Figures 5B and 6A). However, based on our results, it is too early to state that this glandular trichome-specific CYP71AV1 promoter was more effective than the constitutive 35S promoter. It has been reported that the CYP71AV1 gene was dramatically decreased during A. annua leaves development (Wang et al., 2012; Yu et al., 2012). Here, we analyzed the AabZIP9 transcript levels in five different developmental stages of leaves in the AabZIP9 overexpressed transgenic plants. The expression pattern of AabZIP9 that was driven by CYP71AV1 promoter revealed the same pattern as the CYP71AV1 gene, which was highly expressed in young leaves (leaf1 and leaf2) while lowly expressed in old leaves (leaf3 to leaf5) (Figure 5B). The promoters of genes ADS, DBR2, and ALDH1 have similar expression pattern to the promoter of gene CYP71AV1, so these promoters may not be ideal promoters for the regulation of the metabolism in the glandular trichomes. In spite of this, we believe that glandular trichomes specific promoters still have the great potential and will be powerful tools in plant metabolism regulation. The truncated ADS promoter, even in 400-bp length, is still displaying specific expression in A. thaliana trichomes and with a higher activity than the original one (Zhu et al., 2014). Moreover, by laser capture microdissection, we generated a RNA-seq database of trichome cells (Shen et al., 2018), it is hence worthwhile to screen, modify and achieve a series of compact trichome specific core promoters that both trichome specific and constitutively high expressing in all stages of leaves.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
QS, YLW, and KT conceived and coordinated the study and wrote the manuscript. QS, HH, and YZ performed the gene cloning, vector constructive, and plant transformation work. LX, QH, and YJZ did the gene expression, subcellular localization, luciferase analysis, and Y1H experiments. YTW and YLW helped with the metabolites analysis by HPLC. All authors reviewed the results and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation for Young Scientists of China (Grant No. 31600231), Major National Science and Technology Program of China for Innovative Drug (Grant No. 2017ZX09101002-003-002), Natural Science Foundation of Shanghai (Grant No. 18ZR1420600), and Natural Science Foundation and China Postdoctoral Science Foundation (Grant No. 2016M590356).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Dr. Danial Hassani (Shanghai Jiao Tong University, Shanghai, China) for kind suggestions and Ms.Pin Liu for her helping in HPLC analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01294/full#supplementary-material
Supplementary Table 1 | Amino acid sequences of putative bZIP TFs retrieved from A. annua transcriptome database
Supplementary Table 2 | Primers used in this investigation
Supplementary Figure 1 | Gene co-expression analysis of artemisinin biosynthetic pathway genes with bZIP transcription factor family by MeV 4.9 software. AabZIP9 (marked with red box) gene was clustered with ADS, CYP71AV1 and ALDH1 genes and had a high expression level in the bud, flower, and young leaf, suggesting it may involve in the regulation of artemisinin biosynthesis.
Supplementary Figure 2 | Schematic representation of effector and reporter constructs used in the yeast one-hybrid assays. Effector construct contains the AabZIP9 coding sequence driven by the GAL1 promoter while the reporter constructs contain the artificial synthesized triplicate cis-element promoter segments. The empty pLacZ vector was used as control.
Supplementary Figure 3 | Schematic of plant overexpression vector. The AabZIP9 was driven by CaM35S or CYP71AV1 promoter, respectively; Hyg (hygromycin phosphotransferase) was used as resistant selection gene for plant transformation.
Supplementary Figure 4 | The morphologies of 2-month-old wild-type, GUS control, 35S promoter and CYP71AV1 promoter driven overexpressed transgenic plants, and no morphology changed via transformation process; Bar, 5 cm.
Supplementary Figure 5 | Histochemical GUS staining of transgenic A. annua transformed using the 35S: GUS control plasmid. Glandular, nonglandular and stomata structures were pointed out by arrows. (A) 35S promoter has strong activities in most of the leaf tissues (B, C) only a few glandular trichomes get stained by using 35S promoter.
References
Ang, L. H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., et al. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. doi: 10.1016/s1097-2765(00)80022-2
Bouwmeester, H. J., Wallaart, T. E., Janssen, M. H., van Loo, B., Jansen, B. J., Posthumus, M. A., et al. (1999). Amorpha-4, 11-diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 52, 843–854. doi: 10.1016/S0031-9422(99)00206-X
Brameier, M., Krings, A., MacCallum, R. M. (2007). NucPred-Predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160. doi: 10.1093/bioinformatics/btm066
Brown, G. D., Sy, L.-K. (2007). In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron 63, 9548–9566. doi: 10.1016/j.tet.2007.06.062
Chadwick, M., Trewin, H., Gawthrop, F., Wagstaff, C. (2013). Sesquiterpenoids lactones: benefits to plants and people. Int. J. Mol. Sci. 14, 12780–12805. doi: 10.3390/ijms140612780
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Chen, X. B., Yao, Q. F., Gao, X. H., Jiang, C. F., Harberd, N. P., Fu, X. D. (2016). Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 26, 640–646. doi: 10.1016/j.cub.2015.12.066
Czechowski, T., Larson, T. R., Catania, T. M., Harvey, D., Brown, G. D., Graham, I. A. (2016). Artemisia annua mutant impaired in artemisinin synthesis demonstrates importance of nonenzymatic conversion in terpenoid metabolism. P. Natl. Acad. Sci. U. S. A. 113, 15150–15155. doi: 10.1073/pnas.1611567113
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Eisen, M. B., Spellman, P. T., Brown, P. O., Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. P. Natl. Acad. Sci. U. S. A. 95, 14863–14868. doi: 10.1073/pnas.95.25.14863
Goossens, A. (2014). It is easy to get huge candidate gene lists for plant metabolism now, but how to get beyond? Mol. Plant 8, 2–5. doi: 10.1093/mp/ssu099
Han, J., Wang, H., Lundgren, A., Brodelius, P. E. (2014). Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102, 89–96. doi: 10.1016/j.phytochem.2014.02.011
Hao, X., Zhong, Y., Fu, X., Lv, Z., Shen, Q., Yan, T., et al. (2017). Transcriptome analysis of genes associated with the artemisinin biosynthesis by jasmonic acid treatment under the light in Artemisia annua. Front. Plant Sci. 8, 971. doi: 10.3389/fpls.2017.00971
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. doi: 10.1186/1746-4811-1-13
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Ji, Y., Xiao, J., Shen, Y., Ma, D., Li, Z., Pu, G., et al. (2014). Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 55, 1592–1604. doi: 10.1093/pcp/pcu090
Judd, R., Bagley, M. C., Li, M., Zhu, Y., Lei, C., Yuzuak, S., et al. (2019). Artemisinin biosynthesis in non-glandular trichome cells of Artemisia annua. Mol. Plant 12, 704–714. doi: 10.1016/j.molp.2019.02.011
Larkin, M. A., Blackshields, G., Brown, N., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Casteels, T., Frogne, T., Ingvorsen, C., Honore, C., Courtney, M., et al. (2017). Artemisinins Target GABAA receptor signaling and impair a cell identity. Cell 168, 1–15. doi: 10.1016/j.cell.2016.11.010
Lu, X., Zhang, L., Zhang, F., Jiang, W., Shen, Q., Zhang, L., et al. (2013). AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198, 1191–1202. doi: 10.1111/nph.12207
Lv, Z., Wang, S., Zhang, F., Chen, L., Hao, X., Pan, Q., et al. (2016). Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 57, 1961–1971. doi: 10.1093/pcp/pcw118
Ma, D., Pu, G., Lei, C., Ma, L., Wang, H., Guo, Y., et al. (2009). Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. doi: 10.1093/pcp/pcp149
Matías-Hernández, L., Jiang, W., Yang, K., Tang, K., Brodelius, P. E., Pelaz, S. (2017). AaMYB1, and its orthologue AtMYB61, affect terpene metabolism and trichome development in Artemisia annua and Arabidopsis thaliana. Plant J. 90, 520–534. doi: 10.1111/tpj.13509
Moses, T., Pollier, J., Shen, Q., Soetaert, S., Reed, J., Erffelinck, M.-L., et al. (2015). OSC2 and CYP716A14v2 catalyze the biosynthesis of triterpenoids for the cuticle of aerial organs of Artemisia annua. Plant Cell 27, 286–301. doi: 10.1105/tpc.114.134486
Obeid, S., Alen, J., Pham, V. C., Meuleman, P., Pannecouque, C., Le, T. N., et al. (2013). Artemisinin analogues as potent inhibitors of in vitro hepatitis C virus replication. PloS One 8, e81783. doi: 10.1371/journal.pone.0081783
Okell, L. C., Cairns, M., Griffin, J. T., Ferguson, N. M., Tarning, J., Jagoe, G., et al. (2014). Contrasting benefits of different artemisinin combination therapies as first-line malaria treatments using model-based cost-effectiveness analysis. Nat. Commun. 5, 5606. doi: 10.1038/ncomms6606
Olsson, M. E., Olofsson, L. M., Lindahl, A.-L., Lundgren, A., Brodelius, M., Brodelius, P. E. (2009). Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70, 1123–1128. doi: 10.1016/j.phytochem.2009.07.009
Osterlund, M. T., Hardtke, C. S., Wei, N., Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Peplow, M. (2016). Synthetic biology’s first malaria drug meets market resistance. Nature 530, 389–390. doi: 10.1038/530390a
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2012). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301. doi: 10.1093/nar/gkr1065
Ro, D.-K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi: 10.1038/nature04640
Saeed, A., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. doi: 10.2144/03342mt01
Shen, Q., Chen, Y., Wang, T., Wu, S., Lu, X., Zhang, L., et al. (2012). Overexpression of the cytochrome P450 monooxygenase (cyp71av1) and cytochrome P450 reductase (cpr) genes increased artemisinin content in Artemisia annua (Asteraceae). Genet. Mol. Res. 11, 3298–3309. doi: 10.4238/2012.September.12.13
Shen, Q., Lu, X., Yan, T., Fu, X., Lv, Z., Zhang, F., et al. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210, 1269–1281. doi: 10.1111/nph.13874
Shen, Q., Zhang, L., Liao, Z., Wang, S., Yan, T., Shi, P., et al. (2018). The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant 11, 776–788. doi: 10.1016/j.molp.2018.03.015
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, H., Xiao, L., Gao, S., Li, Q., Chen, J., Xiao, Y., et al. (2015). TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development andartemisinin biosynthesis in Artemisia annua L. Mol. Plant 8, 1396–1411. doi: 10.1016/j.molp.2015.04.002
Tang, N., Zhang, H., Li, X. H., Xiao, J. H., Xiong, L. Z. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. doi: 10.1104/pp.111.190389
Teoh, K. H., Polichuk, D. R., Reed, D. W., Covello, P. S. (2009). Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87, 635–642. doi: 10.1139/B09-032
Teoh, K. H., Polichuk, D. R., Reed, D. W., Nowak, G., Covello, P. S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416. doi: 10.1016/j.febslet.2006.01.065
Tin, A. S., Sundar, S. N., Tran, K. Q., Park, A. H., Poindexter, K. M., Firestone, G. L. (2012). Antiproliferative effects of artemisinin on human breast cancer cells requires the downregulated expression of the E2F1 transcription factor and loss of E2F1-target cell cycle genes. Anti-Cancer Drug 23, 370–379. doi: 10.1097/CAD.0b013e32834f6ea8
Vicente-Carbajosa, J., Moose, S. P., Parsons, R. L., Schmidt, R. J. (1997). A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. P. Natl. Acad. Sci. U. S. A. 94, 7685–7690. doi: 10.1073/pnas.94.14.7685
Wang, H., Han, J., Kanagarajan, S., Lundgren, A., Brodelius, P. E. (2012). Trichome-specific expression of the amorpha-4, 11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion. Plant Mol. Biol. 81119, –138. doi: 10.1007/s11103-012-9986-y
Weisshaar, B., Armstrong, G. A., Block, A., da Costa e Silva, O., Hahlbrock, K. (1991). Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 10, 1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x
WHO, (2018). World Malaria Report 2018, World Health Organization [WHO], https://reliefweb.int/report/world/world-malaria-report-2018.
Yan, T., Chen, M., Shen, Q., Li, L., Fu, X., Pan, Q., et al. (2017). HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 213, 1145–1155. doi: 10.1111/nph.14205
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zhang, F., Fu, X., Lv, Z., Lu, X., Shen, Q., Zhang, L., et al. (2015). A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 8, 163–175. doi: 10.1016/j.molp.2014.12.004
Zhang, L., Jing, F., Li, F., Li, M., Wang, Y., Wang, G., et al. (2009). Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol. Appl. Bioc. 52, 199–207. doi: 10.1042/Ba20080068
Zhang, Y., Teoh, K. H., Reed, D. W., Maes, L., Goossens, A., Olson, D. J. H., et al. (2008). The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J. Biol. Chem. 283, 21501–21508. doi: 10.1074/jbc.M803090200
Zheng, H., Colvin, C. J., Johnson, B. K., Kirchhoff, P. D., Wilson, M., Jorgensen-Muga, K., et al. (2017). Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 13, 218–225. doi: 10.1038/nchembio.2259
Zhong, Y., Li, L., Hao, X., Fu, X., Ma, Y., Xie, L., et al. (2018). AaABF3, an abscisic acid–responsive transcription factor, positively regulates artemisinin biosynthesis in Artemisia annua. Front. Plant Sci. 9, 1777. doi: 10.3389/fpls.2018.01777
Zhu, M., Zhang, F., Lv, Z., Shen, Q., Zhang, L., Lu, X., et al. (2014). Characterization of the promoter of Artemisia annua amorpha-4, 11-diene synthase (ADS) gene using homologous and heterologous expression as well as deletion analysis. Plant Mol. Biol. Rep. 32, 406–418. doi: 10.1007/s11105-013-0656-2
Keywords: Artemisia annua, artemisinin, bZIP transcription factors, gene expression, metabolic regulation
Citation: Shen Q, Huang H, Zhao Y, Xie L, He Q, Zhong Y, Wang Y, Wang Y and Tang K (2019) The Transcription Factor Aabzip9 Positively Regulates the Biosynthesis of Artemisinin in Artemisia annua. Front. Plant Sci. 10:1294. doi: 10.3389/fpls.2019.01294
Received: 14 May 2019; Accepted: 18 September 2019;
Published: 07 November 2019.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Mather A. Khan, University of Missouri, United StatesMonica Borghi, Max Planck Institute of Molecular Plant Physiology, Germany
Copyright © 2019 Shen, Huang, Zhao, Xie, He, Zhong, Wang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuliang, Wang d2FuZ3l1bGlhbmdAc2p0dS5lZHUuY24=; Kexuan Tang, a3h0YW5nQHNqdHUuZWR1LmNu
 Qian Shen
Qian Shen Huayi Huang
Huayi Huang Yu Zhao
Yu Zhao Lihui Xie
Lihui Xie Yijun Zhong
Yijun Zhong Yuliang Wang
Yuliang Wang Kexuan Tang
Kexuan Tang