- 1Department of Plant Sciences, Crop Development Centre, University of Saskatchewan, Saskatoon, SK, Canada
- 2International Maize and Wheat Improvement Center (CIMMYT), Mexico City, Mexico
Genetic resistance in the host plant is the most economical and environmentally friendly strategy for controlling wheat leaf rust, caused by Puccinia triticina Eriks. The durum wheat lines Gaza (Middle East), Arnacoris (France) and Saragolla (Italy) express high levels of resistance to the Mexican races of P. triticina. Three recombinant inbred line (RIL) populations, derived from crosses of each of these resistance sources to the susceptible line ATRED #2, were evaluated for leaf rust reactions at CIMMYT’s leaf rust nurseries in Mexico. Genetic analyses of host reactions suggested oligogenic control of resistance in all populations. The F8 RILs from each cross were genotyped using the Illumina iSelect 90K array, and high-density genetic maps were constructed for each population. Using composite interval mapping, a total of seven quantitative trait loci (QTL) that provide resistance to leaf rust were identified. Two QTL designated as QLr.usw-6BS and QLr.usw-6BL were identified on chromosome 6B in Gaza, which explained up to 78.5% and 21.3% of the observed leaf rust severity variance, respectively. A major QTL designated as QLr.usw-7BL was detected on the long arm of chromosome 7B in Arnacoris, which accounted for up to 65.9% of the disease severity variance. Arnacoris also carried a minor QTL on chromosome 1BL, designated as QLr.usw-1BL.1 that explained up to 17.7% of the phenotypic variance. Three QTL conferred leaf rust resistance in Saragolla, namely QLr.usw-2BS, QLr.usw-3B, and QLr.usw-1BL.2, which accounted for up to 42.3, 9.4, and 7.1% of the phenotypic variance, respectively. Markers flanking each QTL were physically mapped against the durum wheat reference sequence and candidate genes involved in disease resistance were identified within the QTL intervals. The QTL identified in this study and their closely linked markers are useful resources for gene pyramiding and breeding for durable leaf rust resistance in durum wheat.
Introduction
The importance of leaf rust as a threat to global durum wheat (Triticum turgidum L. ssp durum) production has increased dramatically during the last decade, due to the occurrence of highly virulent races of Puccinia triticina and the breakdown of the resistance genes that were widely deployed (Singh et al., 2004; Huerta-Espino et al., 2011; Herrera-Foessel et al., 2014a; Kolmer, 2015b; Soleiman et al., 2016; Kolmer and Hughes, 2017). Genetic control offers a cost-effective and environmentally friendly alternative to avoid yield losses due to this pathogen. Resistance to wheat leaf rust is commonly categorized into two classes based on their genetic control and phenotypic effect: race-specific, all-stage resistance, which is usually expressed as a hypersensitive response leading to host cell death, and adult plant resistance (APR), which is usually expressed as a slow-rusting phenotype (Knott, 1989; Lagudah, 2011; Kolmer, 2013; Singh et al., 2016). To date, over 77 leaf rust resistance (Lr) genes have been characterized and catalogued in wheat (McIntosh et al., 2017). Most Lr genes have major effects and confer race-specific, all-stage near-immunity. However, this class of resistance is prone to rapid breakdown as the pathogen population evolves, and new virulent races emerge (Suenaga et al., 2003; Huerta-Espino et al., 2011; Lowe et al., 2011; Ellis et al., 2014; Herrera-Foessel et al., 2014b). The over-reliance on a single race-specific resistance gene has led to leaf rust epidemics and considerable losses in the state of Sonora in Mexico, when the new fungal race BBG/BN overcame Lr72 in 2001 (Singh et al., 2004; Herrera-Foessel et al., 2014a). Subsequently, loss of the resistance conferred by the complementary genes Lr27+Lr31 occurred in 2008, with the emergence of race BBG/BP (Singh et al., 2004; Huerta-Espino et al., 2011). In contrast to race-specific resistance genes, APR genes express at the adult stage and only provide partial resistance which results in longer latent periods and smaller and fewer fungal spores or uredinia (Knott, 1989; Herrera-Foessel et al., 2008c; Lagudah, 2011; Lowe et al., 2011). Currently, at least eight genes that confer APR to leaf rust have been characterized in hexaploid wheat, including Lr34 (Suenaga et al., 2003), Lr46 (Singh et al., 1998), Lr67 (Hiebert et al., 2010), Lr68 (Herrera-Foessel et al., 2012), Lr74 (McIntosh et al., 2015), Lr75 (Singla et al., 2017), Lr77 (Kolmer et al., 2018c), and Lr78 (Kolmer et al., 2018a). Only Lr46 has been reported in durum wheat (T. turgidum L. ssp. durum) (Herrera-Foessel et al., 2011).
Mapping quantitative trait loci (QTL) enables the detection of genes with both major and minor effects and the identification of linked molecular markers that could be used for gene stacking and breeding for durable rust resistance (Soriano and Royo, 2015). The advent of next generation sequencing technologies and high-throughput SNP genotyping platforms facilitated the development of high density genetic maps in wheat (Wang et al., 2014; Maccaferri et al., 2015; Winfield et al., 2016), enhancing our ability to dissect economically important traits such as disease resistance. Several studies have used QTL mapping to identify and tag genomic regions involved in leaf rust resistance in hexaploid wheat (Schnurbusch et al., 2004; Rosewarne et al., 2008; Rosewarne et al., 2012; Buerstmayr et al., 2014; Kolmer, 2015a; Soriano and Royo, 2015), however, only a few studies have been conducted to map QTL for leaf rust resistance in durum wheat (Marone et al., 2009; Singh et al., 2013a; Lan et al., 2017). The objective of this study was to use the iSelect 90K SNP array to characterize and map genetic loci conferring leaf rust resistance in three globally resistant durum wheat genotypes and to identify linked SNP markers useful for gene pyramiding and marker-assisted breeding.
Materials and Methods
Plant Materials and Field Phenotyping
Three sources of resistance to leaf rust (P. triticina), including the Middle Eastern landrace Gaza (unknown pedigree, CIMMYT genotype ID 233), the French cultivar Arnacoris (unpublished pedigree, CIMMYT genotype ID 6048080), and the Italian cultivar Saragolla (pedigree: Iride/Linea PSB 0114, CIMMYT genotype ID 255301), were identified by CIMMYT’s durum wheat breeding program, through extensive multi-race, multisite testing (data not shown). Each source was crossed to the susceptible line ATRED #2 (pedigree: Atil*2/LocalRed, CIMMYT genotype ID 5460557), and RIL populations of over 200 RILs each were developed by advancing generations through single-seed-descent, at the two locations used by CIMMYT’s breeding program: CENEB experimental station near Ciudad Obregon in Sonora (latitude 27.33, longitude -109.93, altitude 35 meters above sea level (masl)), with a wheat crop season from mid-November to late April, and El Batán experimental station, northeast of Mexico City (latitude 19.53, longitude -98.84, altitude 2250 masl), with a wheat crop season from mid-May to mid-October. Reactions to the widely virulent race of P. triticina BBG/BP (Huerta-Espino et al., 2011; Loladze et al., 2014) were scored on the F2 progenies during the spring of 2011 (CENEB). During summer 2011, the F2-derived F3 families (F2:3) were space planted in double 1.2-meter-long rows at El Batán. In 2013, F6 families were grown in replicated 1.2 m rows at the CENEB station, while paired 1.2 m rows of the F8 RILs were phenotyped at El Batán, in summer 2014. In all experiments, parental lines and progenies were inoculated at the tillering stage of plant development, using a mineral oil suspension of urediniospores of race BBG/BP of P. triticina, at a concentration of 5 to 10 mg of urediniospores per 5 ml of oil (Soltrol 170). Susceptible spreader rows surrounding plots and consisting of a mixture of the cultivars Banamichi C2004 and Jupare C2001 (susceptible only to race BBG/BP in Mexico) were also inoculated. The race BBG/BP of P. triticina is the predominant durum-specific race in Mexico, with the following avirulence/virulence formula: Lr1, 2a, 2b, 2c, 3, 3ka, 3bg, 9, 13, 14a, 15, 16, 17, 18, 19, 21, 22a, 24, 25, 26, 28, 29, 30, 32, 35, 37/Lr10, 11, 12, 14b, 20, 23, 27 + 31, 33, 72 (Huerta-Espino et al., 2008; Huerta-Espino et al., 2011; Loladze et al., 2014).
For the F2:3 families and the F6 RIL populations from the crosses Gaza/ATRED #2, Arnacoris/ATRED #2, and Saragolla/ATRED #2, as well as the F8 RILs from the Gaza/ATRED #2 cross, the modified Cobb scale was used to visually estimate the percentage of pustule-infected leaf area or leaf rust severity (LRS) on the parental lines and their progenies (Peterson et al., 1948). Three LRS scores were recorded at weekly intervals, starting at 14 days post inoculation, and the area under the disease progress curve (AUDPC) was calculated as before (Maccaferri et al., 2010). Host reactions were also recorded using four categories: “R” to indicate resistance or uredinia traces, “MR” to indicate moderate resistance with small uredinia surrounded by necrosis, “MS” to indicate moderate susceptibility expressed as chlorosis surrounding moderate sized uredinia, and “S” to indicate full susceptibility with large uredinia lacking necrosis or chlorosis (Roelfs et al., 1992). Based on the host reactions of plants within each family, the F2:3 families were categorized as homozygous resistant (R), homozygous susceptible (S), and heterozygous (Het). The F8 RILs from the crosses Arnacoris/ATRED #2 and Saragolla/ATRED #2 were scored as R or S. The chi-square (χ2) test was used to estimate the number of genes involved in the inheritance of leaf rust resistance in these populations. For analyses with a single degree of freedom, the chi-square values were adjusted with the Yates’s correction for continuity (Yates, 1934).
Seedling Stage Evaluations of the F2:3 Families
Seedlings of F2:3 families from the crosses Gaza/ATRED #2, Arnacoris/ATRED #2, and Saragolla/ATRED #2 were evaluated for resistance to race BBG/BP of P. triticina, under controlled conditions in CIMMYT’s greenhouse, at El Batán experimental station. Approximately 25 to 35 seedlings from each F2:3 family were grown in 7-by-7-by-10 cm pots and inoculated with a suspension of urediniospores of race BBG/BP in light mineral oil (Loladze et al., 2014). Infection types (IT) were recorded using the 0–4 scale where “0” = no visible leaf rust symptoms; “;” = hypersensitive flecks without any uredinia; “1” = small uredinia surrounded by necrosis; “2” = small to medium uredinia surrounded by chlorosis or necrosis; “3” = medium-sized uredinia with or without chlorosis; “4” = large uredinia without chlorosis or necrosis; “X” = random distribution of variable-sized uredinia (mesothetic reaction), and “+” and “-” were used when uredinia were somewhat larger or smaller than the average for the IT class (McIntosh et al., 1995). ITs of 3, 3+, and 4 indicate susceptible host reactions, whereas all of the other ITs were considered resistant. Based on their ITs at 10 to 12 days after inoculation, the F2:3 families were classified as homozygous resistant “R,” homozygous susceptible “S,” or heterozygous “Het.” The χ2 test was used to estimate the number of genes involved in the inheritance of leaf rust resistance at the seedling stage.
Allelism Tests Between Gaza and Carriers of Known Lr Genes
Allelism between the resistance genes in Gaza and the known Lr genes Lr61 and LrCamayo was investigated using 177 F2 plants from the cross Gaza/Sooty_9/Rascon_37//Guayacan INIA, and 273 F2 plants from the cross Gaza/Cirno C2008, respectively. Allelism to the Lr genes in the durum lines Geromtel_3 and Tunsyr_2 was also studied in populations of 326 and 181 F2 plants, respectively. The F2 progenies were tested for their field reaction to race BBG/BP of P. triticina, at El Batán. The resulting resistant/susceptible ratios were tested for goodness of fit to various gene models, using chi-square analysis. When no susceptible recombinants were detected in the F2 progenies, it was assumed that the two resistant parents carried allelic or closely linked leaf rust resistance genes.
DNA Extraction and Illumina iSekect 90K SNP Array Genotyping
Genomic DNA was extracted from the parental lines and the F8 RILs from the three crosses Gaza/ATRED #2, Arnacoris/ATRED #2, and Saragolla/ATRED #2, using a BIOMEK FXp liquid handling station and the Sbeadex mini plant kit (LGC, Teddington, Middlesex, UK) (Dreisigacker et al., 2016). The Illumina iSelect 90K SNP array (Illumina, San Diego, CA, USA) was used for genotyping of the RILs and the parental lines (Wang et al., 2014). Genotype calling was performed using GenomeStudio software (Illumina, San Diego, CA, USA). The genotyping data for all three biparental populations are provided as Supplementary Materials. Prior to mapping, data filtering was carried out and markers that showed significant segregation distortion or more than 10% missing values were excluded.
Construction of Linkage Maps and QTL Mapping
Curated SNP data were used to build linkage maps for each of the three populations. Initial linkage groups (LG) were obtained using the MSTMap software (Wu et al., 2008) with a stringent cutoff p-value of 1E-10 and a maximum distance between markers of 15.0 centimorgan (cM). The LGs were assigned to individual wheat chromosomes based on existing high-density SNP maps (Cavanagh et al., 2013; Wang et al., 2014; Maccaferri et al., 2015) and the tetraploid wheat reference sequences of wild emmer wheat (WEW) (Avni et al., 2017) and the modern durum wheat cultivar Svevo (Maccaferri et al., 2019). Once LGs were attributed to chromosomes, their genotypic data were pooled on a chromosome-by-chromosome basis and final genetic maps were constructed using MapDisto 1.7.7 software (Lorieux, 2012) at threshold values of recombination frequency = 0.3 and logarithm of the odds (LOD) = 3. Markers sharing the same segregation pattern (co-segregating) were identified in each LG, and the marker with the lowest percentage of missing data was chosen to represent each cluster or bin. Double recombinants were corrected using the functions “Show double recombinants,” “Show error candidates,” and “Replace error candidates by flanking genotype” as implemented in the MapDisto software (Lorieux, 2012). The order of the markers was determined using the Order, Ripple, and Check inversions commands. The Kosambi function was used to convert the recombination fractions to cM (Kosambi, 1943).
QTL detection was performed using the composite interval mapping (CIM) method implemented in QGene 4.4.0 software (Joehanes and Nelson, 2008). QTL were identified at a scan interval of 1 cM. The stepwise regression method was used to select cofactors and the LOD thresholds for determining statistically significant QTL were calculated by 1000 permutations with P < 0.05. The additive effect and percentage of phenotypic variance explained by each QTL were obtained from the final CIM results. Phenotypic traits analyzed included LRS, AUDPC and the host reaction (HR) recorded for the F8 RILs from the crosses Arnacoris/ATRED #2 and Saragolla/ATRED #2.
QTL Interaction Tests in Each Population
A single marker with the highest LOD score was selected from each QTL to estimate its phenotypic effect. The mean phenotypic data for all the RILs from each population were grouped based on their genotype combinations at these selected loci and mean separation was performed using Fisher’s protected least significant difference (LSD) test in SAS 9.4 software (SAS Institute Inc., Cary, NC, USA).
Genotyping of Molecular Markers Linked to Known Lr Genes
To test for presence of the APR gene Lr46 (http://maswheat.ucdavis.edu/protocols/Lr46), the two simple sequence repeat (SSR) markers Xwmc44 and Xgwm259 (William et al., 2006) and a kompetitive allele specific polymerase chain reaction (KASP) marker were used to genotype the parental lines Arnacoris, Saragolla, and ATRED #2, as well as subsets of resistant and susceptible RILs from each population. The durum cultivars Kofa, CDC Verona, and Strongfield were used as checks. Three SSR markers (i.e. Xwmc764, Xgwm210, and Xwmc661) and two KASP markers (i.e. kwm677 and kwm744) linked to the resistance gene Lr16 (McCartney et al., 2005; Kassa et al., 2017) were used to genotype both parents and selected lines from the Saragolla/ATRED #2 population, as well as the check cultivar AC Domain. The parental lines Gaza and ATRED #2 and check cultivars including the Lr61-carrying Guyacan INIA were genotyped using the SSR marker Xwmc487 linked to Lr61 (Herrera-Foessel et al., 2008a). The four parental lines Gaza, Arnacoris, Saragolla, and ATRED #2 were also screened using the nucleotide-binding site leucine-rich repeat (NBS-LRR)-specific primers 4406F/4840R, previously determined to be linked to Lr14a (Kthiri et al., 2018). The durum cultivars Sachem and Somayoa were used as positive controls that carry Lr14a. The amplification reactions were performed according to published protocols (McCartney et al., 2005; William et al., 2006; Herrera-Foessel et al., 2008a; Kthiri et al., 2018). Polymorphisms for the SSR markers and the NBS-LRR-specific primers were scored on 2% agarose gels.
Physical Mapping to the Durum Wheat Reference Genome
The program GMAP (Wu and Watanabe, 2005) was used to align the sequences of the SNP markers that localized within each QTL LOD plot area to the genome sequence of the durum wheat cultivar “Svevo” (Maccaferri et al., 2019). Putative physical intervals for each QTL were identified using a cut-off value of 98% for sequence identity and sequence coverage. Genes that mapped within these physical intervals were identified using the available annotations for the durum wheat reference genome (Maccaferri et al., 2019).
Results
The Inheritance of Leaf Rust Resistance in Gaza, Arnacoris, and Saragolla
Leaf rust symptoms developed adequately for all the field evaluation trials at the CENEB and El Batán experimental stations. The RILs from the three mapping populations expressed a wide range in disease severity, with the resistant parents Gaza, Arnacoris, and Saragolla showing the lowest scores for leaf rust severity (0–5%), and the highest scores (90–100%) being observed on the susceptible parent ATRED #2. No transgressive segregation was observed among the RILs from the three crosses, which confirmed that the susceptible parent ATRED #2 does not contribute any genes for leaf rust resistance that could be detected under the present phenotyping conditions.
The F2:3 seedlings from the cross Gaza/ATRED #2 segregated at 63R:94Het:50S, which is a good fit to the 1:2:1 ratio expected for a single dominant seedling resistance gene, based on p-value (P(< 0.05) = 0.18) of the χ2 test at a 95% level of confidence (Table 1). However, field-based segregation ratio of 7R:8Het:1S (P(< 0.05) = 0.12) of the F3 adult plants suggested the presence of two resistance genes in Gaza (Table 1). This discrepancy between seedlings and adult plants evaluation results led to the conclusion that Gaza could carry one APR and one seedling resistance gene. The involvement of an APR gene was further investigated by selecting 10 F2:3 families that were uniformly resistant in the field at the adult stage and uniformly susceptible at the seedling stage and testing them again at the adult plant stage, under greenhouse conditions. The results from these tests confirmed that the 10 selected families were indeed resistant at the adult stage. The F8 RILs from the cross Gaza/ATRED #2 showed a segregation of 1R:1S consistent with the segregation pattern of a single resistance gene (P (< 0.05) = 0.65) (Table 1). This may be explained by the fact that APR genes are less effective in El Batán compared to the CENEB station (K. Ammar and A. Loladze, unpublished).
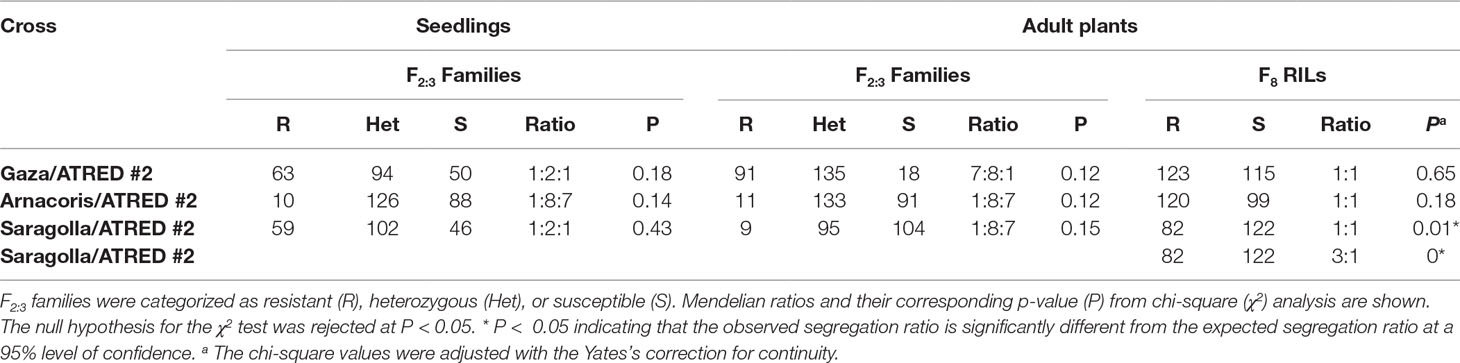
Table 1 Segregation ratios of the F2:3 and F8 progenies from three crosses of durum wheat lines evaluated for leaf rust resistance to race BBG/BP of P. triticina, at the seedling and adult plant stages.
Similar results were observed for the Saragolla/ATRED #2 population since segregation of the F2:3 seedlings fit a 1R:2Het:1S ratio (P(< 0.05) = 0.43) expected for a single resistance gene, while the F2:3 adult plants segregation ratio fit a 1R:8Het:7S (P(< 0.05) = 0.15) ratio, expected for the segregation of two resistance genes (Table 1). The screening of the F8 RILs, however, resulted in a distorted segregation of 82R:122S, which did not fit the expected ratios for Mendelian inheritance of one (P(< 0.05) = 0.01) or two (P(< 0.05) = 0) resistance genes (Table 1).
The 1R:8Het:7S segregation ratio observed for the F2:3 progenies from the cross Arnacoris/ATRED #2 at both the seedling (P(< 0.05) = 0.14) and adult plant (P(< 0.05) = 0.12) stages suggested the presence of two resistance genes in Arnacoris. However, the segregation ratio of 1R:1S (P(< 0.05) = 0.18) observed in the F8 RILs was more consistent with that expected for a single resistance gene (Table 1).
The frequency distributions of the LRS and the AUDPC were determined for the three RIL populations (Figure 1). Although both disease severity and AUDPC data of the three populations showed continuous distributions, there was an obvious tendency of skewness towards resistance, which suggested that, while leaf rust resistance in Gaza, Arnacoris, and Saragolla was not monogenically inherited, there may be major gene effects in these populations.
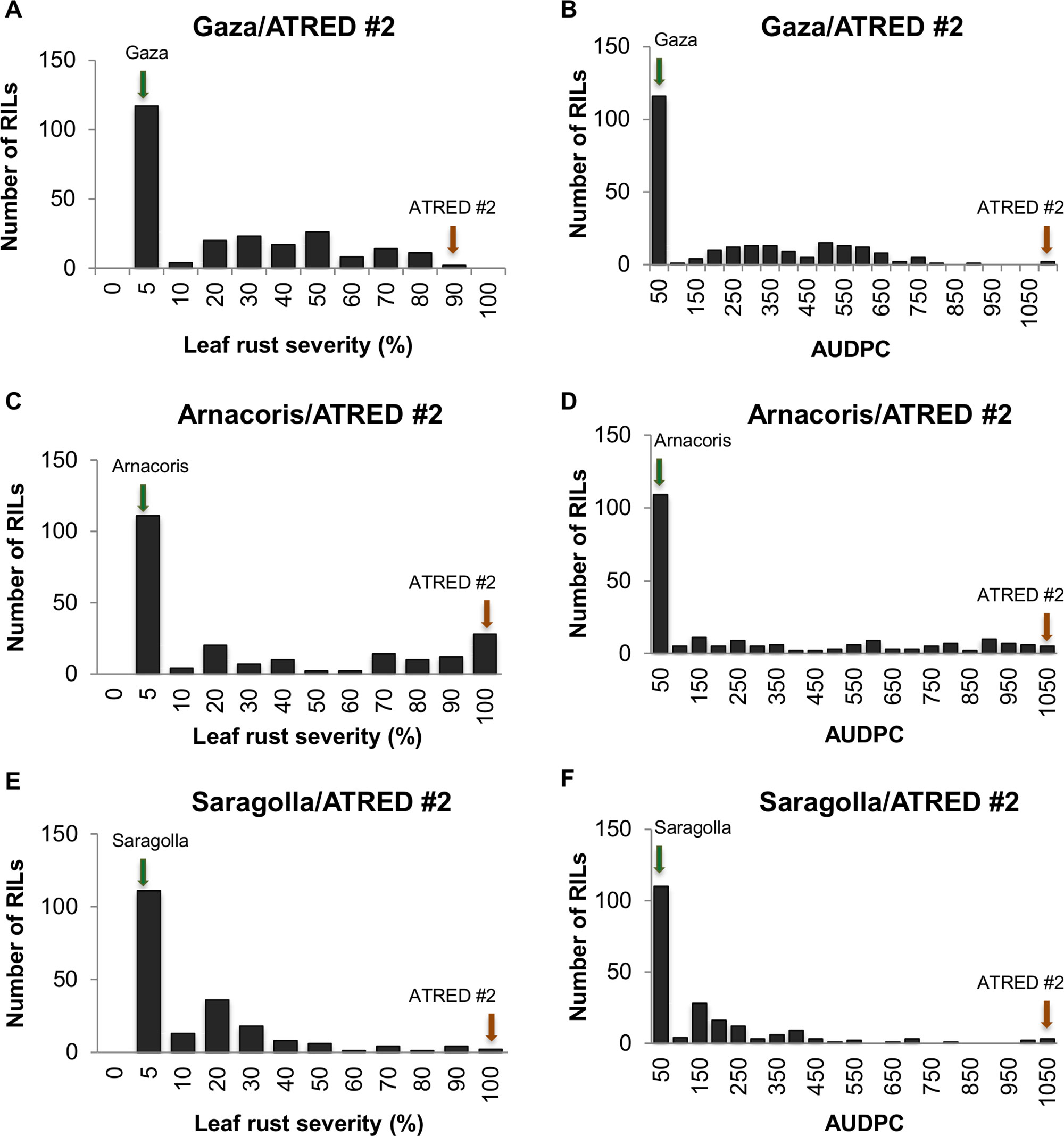
Figure 1 Frequency distributions of leaf rust severity and AUDPC in adult plants of the three RIL populations, evaluated for leaf rust reaction to the Mexican race BBG/BP in field plot tests. Panels represent (A) LRS for Gaza/ATRED #2, (B) AUDPC for Gaza/ATRED #2, (C) LRS for Arnacoris/ATRED #2, (D) AUDPC for Arnacoris/ATRED #2, (E) LRS for Saragolla/ATRED #2, (F) AUDPC for Saragolla/ATRED #2. Green arrows indicate positions of the resistant parents, and red arrows indicate positions of the susceptible parent ATRED #2.
QTL Analyses Detect Several Leaf Rust Resistance Loci in Gaza, Arnacoris, and Saragolla
Polymorphic SNP markers with call frequencies ≥ 90% that fit the expected segregation ratio of 1:1 were considered reliable for mapping and were subsequently used to construct linkage maps for each of the three F8 RIL populations from the following crosses: Gaza/ATRED #2 (6248 SNP), Arnacoris/ATRED #2 (7315 SNP), and Saragolla/ATRED #2 (5345 SNP) (Table 2). The number of LGs ranged from 29 to 35, with all 14 durum chromosomes represented by at least one LG (Table 2). Comparison across genomes indicated that the maximum number of markers mapped to the B genome, except for Gaza where 51.2% of the SNPs mapped to the A genome. The high proportion of co-segregating SNP markers significantly reduced the final number of bins (unique marker loci) in the three maps. Marker density was greatest for the Arnacoris/ATRED #2 population, with an average inter-bin interval of 2.4 cM. However, the Gaza/ATRED #2 population produced the longest map with a total length of 4476.2 cM and the lowest marker density. The Saragolla/ATRED #2 population produced the shortest map with a total length of 3647.2 cM (Table 2). Final genetic maps for the three RIL populations are available as supplementary material (Supplementary File S1).
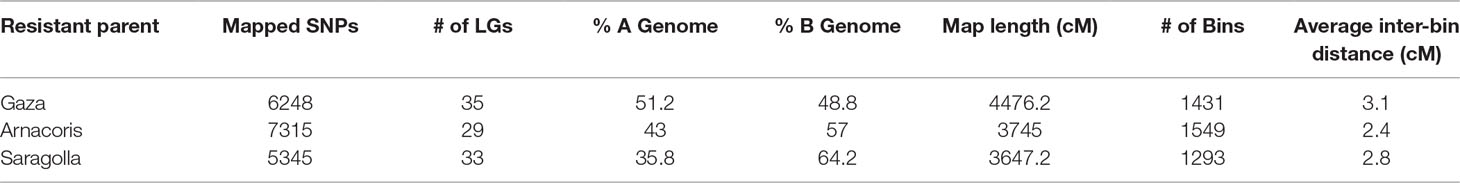
Table 2 Statistics of the three linkage maps from the three RIL populations Gaza/ATRED #2, Arnacoris/ATRED #2 and Saragolla/ATRED #2.
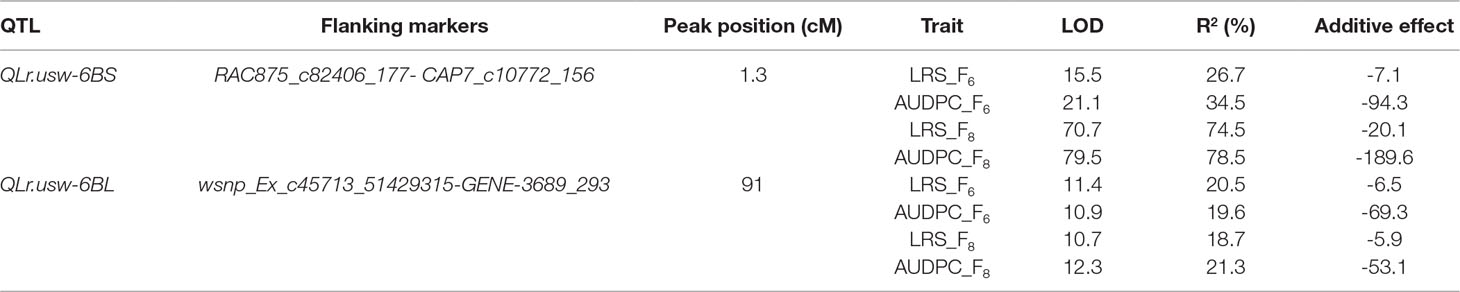
QTL in the Gaza/ATRED #2 Population
Two QTL on chromosome 6B were associated with leaf rust resistance in the Gaza/ATRED #2 population. Both were derived from the resistant parent Gaza. The QTL were detected at a LOD significance threshold of 3.5, based on 1,000 permutation tests at a type I error rate of α < 0.05 (Table 3). The first QTL, QLr.usw-6BS, peaked at the locus CAP7_c10772_156 and was detected in both F6 and F8 RILs, which were evaluated for leaf rust at CENEB in 2013 and at El Batán in 2014, respectively. This QTL was detected for both traits analyzed (LRS and AUDPC) at both locations, and explained up to 34.5% of the total phenotypic variance for AUDPC at CENEB (LOD 21.1) and up to 78.5% at El Batán (LOD 79.5) (Table 3). The second QTL detected in the Gaza/ATRED #2 progeny was designated as QLr.usw-6BL and peaked at the SNP marker GENE-3689_293. QLr.usw-6BL was also detected at both locations for all the traits analyzed, and accounted for 20.5% (LOD 11.4) and 18.7% (LOD 10.7) of the final LRS variance at CENEB and at El Batán, respectively (Table 3).
The RILs from the cross Gaza/ATRED #2 were grouped into four categories based on the allelic state of SNP markers CAP7_c10772_156 and GENE-3689_293 and mean separation was performed using Fisher’s LSD test. As expected, RILs that carried no resistance alleles scored the highest LRS and AUDPC of the allelic combinations (Figures 2A, B). QLr.usw-6BS had the strongest effect, since the mean LRS and AUDPC expressed by carriers of this QTL did not exceed 6% and 50.2, respectively (Figures 2A, B). QLr.usw-6BL was also able to reduce the disease symptoms, and though the reduction was not as strong as with QLr.usw-6BS, it was statistically significant. In general, the presence of QLr.usw-6BS reduced leaf rust symptoms to its lowest significant level, thereby masking the potential expression of QLr.usw-6BL.
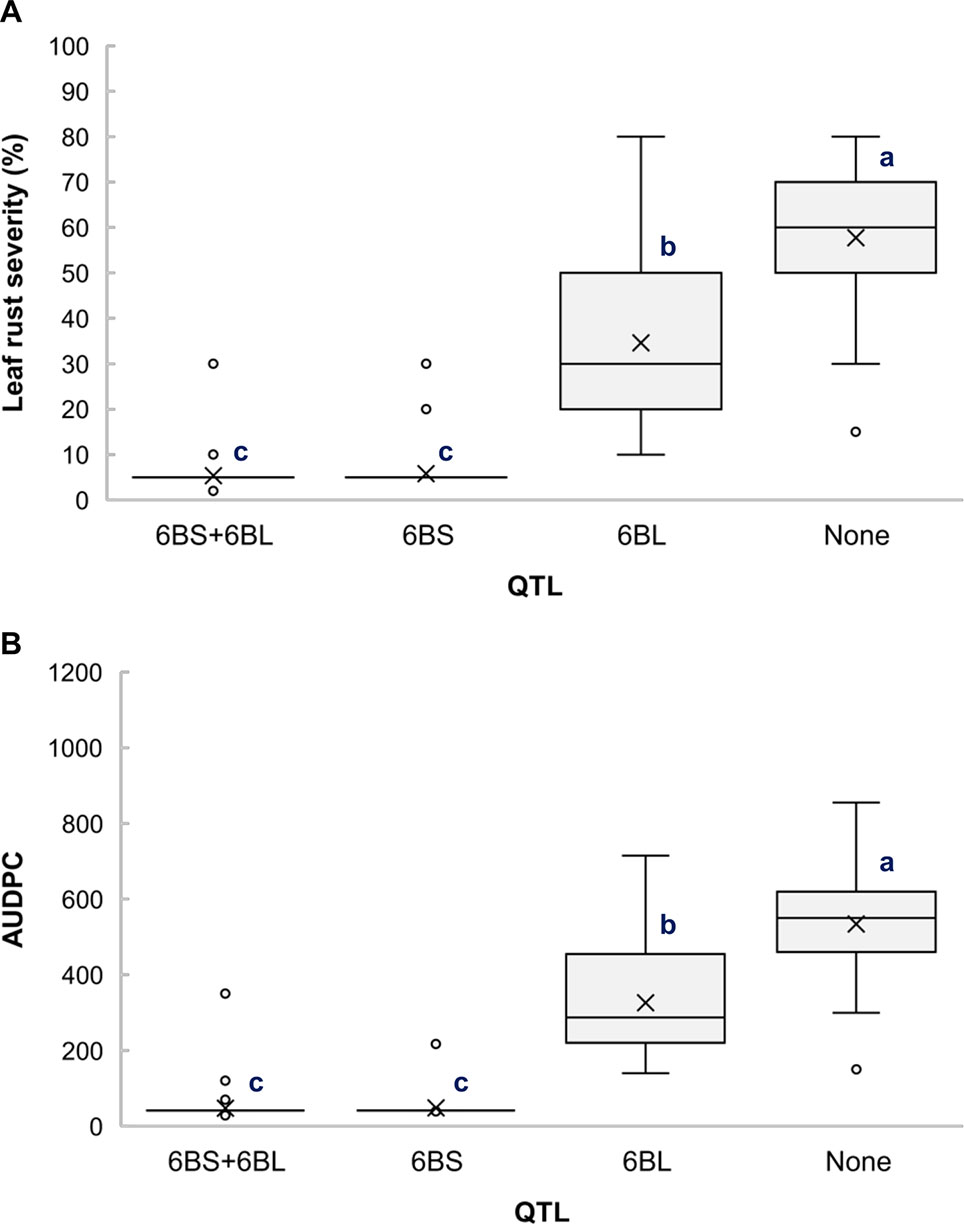
Figure 2 Boxplots illustrating the effects of QLr.usw-6BS and QLr.usw-6BL on (A) LRS and (B) AUDPC in the Gaza/ATRED #2 population. The QTL effects are shown in isolation and in combination. Means with the same letter are not significantly different from one another at P < 0.05.
The F2 progenies from the crosses Gaza/Sooty_9/Rascon_37//Guayacan INIA and Gaza/Cirno C2008 segregated at 55R:9S (P(< 0.05) = 0.93) and 61R:3S (P(< 0.05) = 0.44), respectively (Supplementary Table S1), indicating that the leaf rust resistance genes in Gaza are neither allelic nor linked to Lr61 or LrCamayo. The absence of susceptible recombinants in the F2 progeny from the cross Gaza/Geromtel_3 suggests that these two cultivars carry either allelic or closely linked Lr genes. However, the 61R:3S (P(< 0.05) = 0.16) segregation ratio observed in the F2 population from the cross Gaza/Tunsyr_2 indicates that the Lr genes in these two cultivars are unrelated (Supplementary Table S1). The SSR marker Xwmc487 linked to Lr61 showed a clear difference in PCR product size between Gaza and the Lr61-carrying Guayacan INIA (Supplementary Figure S1).
QTL in the Arnacoris/ATRED #2 Population
Composite interval mapping revealed two QTL associated with resistance to leaf rust in the Arnacoris/ATRED #2 population (Table 4). A major QTL on the distal region of chromosome 7BL, designated as QLr.usw-7BL, with a peak LOD value at marker BS00010355_51, explained 65.9% of LRS variance (LOD 49.3) for the 2013 field trials at CENEB, and was highly significant for the AUDPC (LOD 46.1). QLr.usw-7BL was the only QTL detected for the host reactions of the F8 RILs evaluated at El Batán, and accounted for 99.6% of the phenotypic variance. A less significant QTL, designated as QLr.usw-1BL.1, was detected on the long arm of chromosome 1B for the 2013 field trials at CENEB (Table 4), explaining up to 17.7% of the phenotypic variance for the AUDPC (LOD 8.9) and BS00060686_51 was the most significant marker within the interval.
Based on the allelic state of the SNP markers BS00010355_51 and BS00060686_51, the RILs from the cross Arnacoris/ATRED #2 were classified into carriers and non-carriers of QLr.usw-7BL and QLr.usw-1BL.1, and mean separation for LRS and AUDPC was performed using Fisher’s LSD test. Clearly, QLr.usw-7BL had the strongest effect on all the traits and its presence alone conferred the highest level of resistance (Figures 3A, B). The results also showed that QLr.usw-1BL.1 was significant in reducing leaf rust symptoms only in the absence of QLr.usw-7BL (Figures 3A, B).
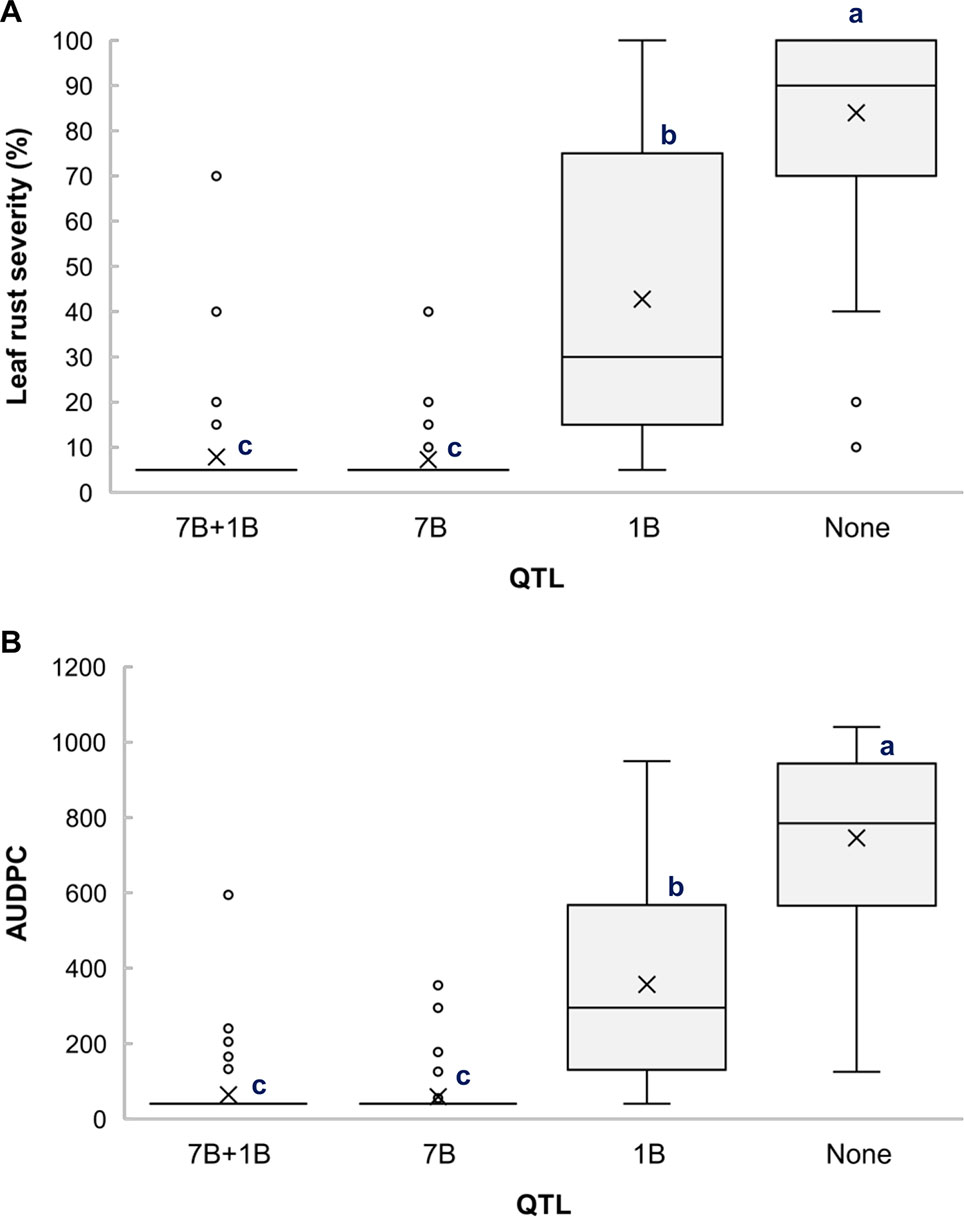
Figure 3 Boxplots illustrating the effects of QLr.usw-7BL and QLr.usw-1BL.1 on (A) LRS and (B) AUDPC in the Arnacoris/ATRED #2 population. The QTL effects are shown in isolation and in combination. Means with the same letter are not significantly different from one another at P < 0.05.
Molecular marker testing using the SSR markers Xwmc44 and Xgwm259 linked to the APR gene Lr46 on chromosome 1BL showed inconclusive results, since both positive (Kofa) and negative (CDC Verona) controls had PCR products of similar size for Xwmc44 (Supplementary Figure S2A), whereas marker Xwmg259 had no PCR product in Arnacoris (Supplementary Figure S2B). However, genotyping with the KASP marker linked to Lr46 showed that Arnacoris, as well as the RILs with and without QLr.usw-1BL.1, clustered with the susceptible parent ATRED #2 and the negative controls CDC Verona and Strongfield, suggesting that Arnacoris may not be carrying Lr46 (Supplementary Figure S3). PCR amplification using the NBS-LRR primers 4406F/4840R, which are linked to Lr14a, showed that the positive controls Somayoa and Sachem carry this marker, while Arnacoris had the null allele, which indicates that QLr.usw-7BL is different from Lr14a (Supplementary Figure S4).
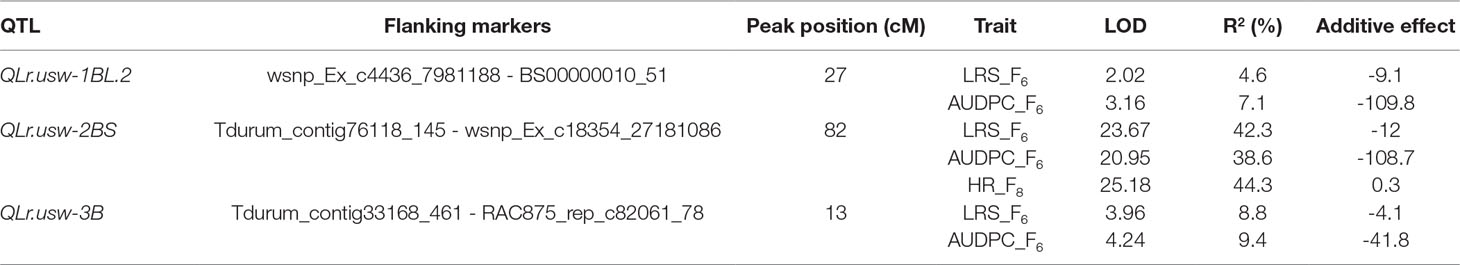
QTL in the Saragolla/ATRED #2 Population
Three QTL controlled leaf rust resistance in the Saragolla/ATRED #2 population, which were detected by CIM on chromosomes 1BL, 2BS, and 3B (Table 5). QLr.usw-2BS peaked at 82 cM on chromosome 2B with wsnp_Ex_c18354_27181086 being the most significant marker linked to this major QTL. QLr.usw-2BS explained up to 42.3% of the final leaf rust severity variance, and was the only QTL detected for the host reactions of the F8 RILs with a LOD score of 25.18. QLr.usw-3B peaked at marker RAC875_rep_c82061_78 on chromosome 3B and accounted for 9.4% of the AUDPC variance. The minor QTL on the long arm of chromosome 1B, designated as QLr.usw-1BL.2, was flanked by markers wsnp_Ex_c4436_7981188 and BS00000010_51, and explained up to 7.1% of the observed variance for AUDPC (Table 5).
Analysis of different combinations of QTL based on the allelic state of SNP markers BS00000010_51, wsnp_Ex_c18354_27181086, and RAC875_rep_c82061_78 showed that the RILs carrying all three QTL had the lowest averages for LRS (5.8%) (Figure 4A) and AUDPC (59) (Figure 4B), which indicates the additive effects of these QTL. It is also noticeable that QLr.usw-2BS had the major effect on leaf rust symptoms compared to both QLr.usw-3B and QLr.usw-1BL.2. However, both QLr.usw-3B and QLr.usw-1BL.2 significantly reduced the leaf rust symptoms in the absence of QLr.usw-2BS (Figure 4).
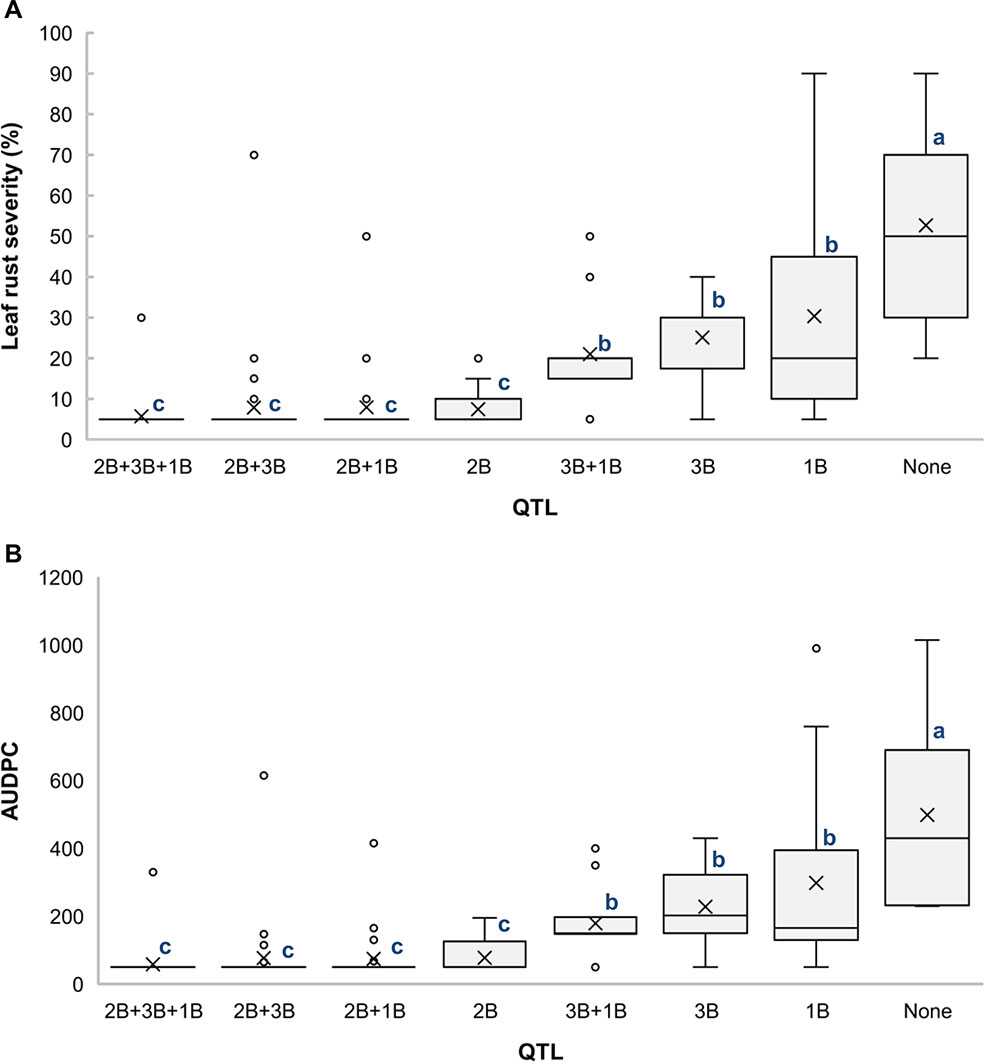
Figure 4 Boxplots illustrating the effects of QLr.usw-2BS, QLr.usw-3B, and QLr.usw-1BL.2 on (A) LRS and (B) AUDPC in the Saragolla/ATRED #2 population. The QTL effects are shown in isolation and in combination. Means with the same letter are not significantly different from one another at P < 0.05.
Both parents and selected lines from the Saragolla/ATRED #2 population were genotyped with three SSR markers (Xwmc764, Xgwm210, and Xwmc661) linked to Lr16 on chromosome 2BS (Supplementary Figure S5). The results showed polymorphism between the amplicons from Saragolla and AC Domain for the three markers, which suggests that QLr.usw-2BS is different from Lr16. The KASP markers kwm677 (Supplementary Figure S6A) and kwm744 (Supplementary Figure S6B) also showed polymorphism between Saragolla and the Lr16-carrying AC Domain.
Physical Mapping to the Durum Wheat Reference Genome
The DNA sequences associated with the SNP markers mapping within each QTL LOD plot area in Gaza, Arnacoris, and Saragolla were physically mapped to the durum wheat reference genome of Svevo (Maccaferri et al., 2019) to identify candidate genes for leaf rust resistance (Supplementary File S2). Several genes identified within these QTL intervals encode proteins with motifs known to be associated with disease resistance such as NBS-LRR receptor proteins, calcium-dependant lipid-binding (CaLB domain) proteins, ATP-binding cassette (ABC) transporter proteins, as well as several receptor-like protein kinases (RLKs) (Supplementary File S2).
Discussion
Diversification of the genetic basis of leaf rust resistance and breeding for durable resistance are both priorities in many durum wheat breeding programs worldwide, especially after the emergence of new virulent races of P. triticina. The goal of the present study was to characterize and map putatively uncharacterized genes for leaf rust resistance in the three durum genotypes: Gaza, Arnacoris, and Saragolla. Inheritance studies indicated the involvement of several loci that controlled leaf rust resistance in these lines, including at least one APR gene in each of Gaza and Saragolla. The segregation ratios of the F2:3 progenies from the Gaza/ATRED #2 population suggested one resistance gene in the seedlings evaluation and two in the adult plants evaluation. However, segregation of the F8 RILs adult plants suggested the presence of a single resistance gene. This discrepancy may be caused by the ineffectiveness of the Gaza APR gene in El Batán, since certain APR genes are known to be environment or temperature sensitive (Kaul and Shaner, 1989; McIntosh et al., 1995; Risk et al., 2012). For the Arnacoris/ATRED #2 population, the seedling and adult plant stage evaluations of the F2:3 progenies suggested the presence of two resistance genes, while the segregation ratio of the F8 RILs supported a single gene theory. Nonetheless, the F2:3 results were indeed supported by the QTL mapping results, since two QTL were detected in this population. The major phenotypic effect of QLr.usw-7BL suggests that this is an all-stage resistance gene, while QLr.usw-1BL.1 is likely an APR gene, with a minor phenotypic effect. The discrepancy between the segregation ratios could be due to the differences between the climatic conditions during the years of evaluations. The F2:3 progenies were evaluated in El Batán in 2011, while the F8 RILs evaluation was conducted in 2014, and it is possible that the APR gene was not effective in 2014. The environmental effects on this gene warrant for follow-up field studies under various conditions. Although the material was inoculated with the race BBG/BP of P. triticna in both years, it is also possible that the racial constitution of the natural field population of the pathogen has been different between 2011 and 2014, thus affecting the results. The Saragolla/ATRED #2 population showed similar results for the F2:3 progenies, with the seedling data suggesting the presence of a single resistance gene, while the adult plant data suggested the presence of at least one additional APR gene. However, the segregation ratio from the F8 generation could not fit neither one nor two-gene models and we could not find a reasonable explanation for this aberration, which also calls for further evaluation of this cultivar under various environments.
Composite interval mapping identified genomic regions on chromosomes 1BL, 2BS, 3B, 6BS, 6BL, and 7BL, which were associated with leaf rust resistance in these three durum cultivars. These may be particularly valuable in breeding since they can be strategically combined to produce more durable resistance. The SNP markers linked to all the leaf rust resistance QTL identified in the present study were converted into KASP markers (Supplementary Table S2) and are currently being validated at CIMMYT.
QTL on Chromosome 1B
The distal region of chromosome 1BL is known to carry the APR gene Lr46, identified in the wheat cultivar Pavon76 (Singh et al., 1998). The flanking SNP markers for QLr.usw-1BL.1 in Arnacoris, BS00060686_51, and Kukri_c46030_412, mapped at 152.5 cM and 162.3 cM, respectively, on chromosome 1BL in the SNP-based consensus map of tetraploid wheat (Maccaferri et al., 2015). Likewise, the SNP marker wsnp_Ex_c4436_7981188 that flanks QLr.usw-1BL.2 in Saragolla, mapped at 145.4 cM on 1BL. The two SSR markers linked to Lr46, Xwmc44 and Xgwm259 (William et al., 2006), mapped at 140.6 and 150.6 cM, respectively, in the same durum consensus map (Maccaferri et al., 2015). Despite the proximity between these marker intervals, further molecular marker analyses showed that Arnacoris may not be carrying Lr46 (Supplementary Figure S3). This is supported by the observation that QLr.usw-1BL.1 alone has a much stronger phenotypic effect on disease symptoms reduction (Figure 3) compared to Lr46, when present alone in durum wheat (K. Ammar, unpublished). However, Qlr.usw-1BL.2 present in Saragolla is likely Lr46, which is consistent with the observation that QLr.usw-1BL.2 has a very moderate effect on disease symptoms reduction (Figure 4).
Other leaf rust resistance genes that map to chromosome 1B include Lr33 (Dyck et al., 1987), Lr44 (Dyck and Sykes, 1994), Lr71 (Singh et al., 2013b), and Lr26 (Mago et al., 2002; Mago et al., 2005). However, virulence in durum specific races is common for Lr33, such as the Mexican race BBG/BP that is used in the present study (Herrera-Foessel et al., 2008b; Huerta-Espino et al., 2011; Lan et al., 2017). Genes Lr44 and Lr71 were both identified in spelt wheat (T. spelta) and no reports indicate their presence in durum wheat. Furthermore, the SSR markers Xgwm18 and Xbarc187, which are closely linked to Lr71 (Singh et al., 2013b), mapped at 35.6 and 35.7 cM in the tetraploid consensus map, respectively (Maccaferri et al., 2015). Based on the position of the SNP markers flanking QLr.usw-1BL.1 in the same consensus map at 152.5 and 162.3 cM, it is possible to conclude that Arnacoris does not carry Lr71. The short arm of chromosome 1R of rye (Secale cereale) carries the leaf rust resistance gene Lr26 and has been widely used in wheat breeding programs through the 1BL.1RS (wheat-rye) translocation (Mago et al., 2005). However, there is no indication of the presence of the 1BL.1RS translocation in Arnacoris. While it is highly unlikely that QLr.usw-1BL.1 harbor any of the three designated genes on chromosome 1B, definitive proof could not be obtained in the present study.
QTL on Chromosome 2B
QLr.usw-2BS mapped to a chromosome region known to carry at least six designated leaf rust resistance genes: Lr13 (Seyfarth et al., 2000), Lr16 (McCartney et al., 2005), Lr23 (McIntosh and Dyck, 1975), Lr35 (Seyfarth et al., 1999), Lr48 (Saini et al., 2002), and Lr73 (Park et al., 2014). Lr13, Lr35 and Lr48 are reportedly APR genes, whereas Saragolla likely carries a major seedling resistance gene, based on the seedling evaluations conducted at CIMMYT. In addition, the molecular markers Xbarc55 and IWB35283, previously reported to be linked to Lr13 (Zhang et al., 2016), mapped at 72.1 cM and 74.2 cM, respectively, in the tetraploid wheat consensus map (Maccaferri et al., 2015), while the SNP markers Tdurum_contig76118_145 and wsnp_Ex_c18354_27181086 flanking QLr.usw-2BS, mapped at 8.4 cM and 12.3 cM, respectively, in the same consensus map. Also, Lr35 is unlikely to be present in Saragolla since it was introgressed into hexaploid wheat from Aegilops speltoides and we did not detect molecular evidence that Saragolla is carrying this introgression. The SNP markers IWB31002, IWB39832, IWB34324, IWB72894, IWB36920, and IWB70147 are reported to be co-segregating with the leaf rust resistance gene Lr48 (Nsabiyera et al., 2016). However, none of these SNP markers are located within the genetic interval of QLr.usw-2BS in both the Saragolla/ATRED #2 genetic map (Supplementary File S1) and the tetraploid consensus map (Maccaferri et al., 2015). Therefore, it can be assumed that QLr.usw-2BS is different from Lr48. Lr23 is also an unlikely candidate for QLr.usw-2BS, since the race BBG/BP used in this study is virulent to Lr23 (Huerta-Espino et al., 2008; Huerta-Espino et al., 2011; Loladze et al., 2014). Lr73 was identified in Australia in the common wheat genotype Morocco, which is widely susceptible to isolates of P. triticina (Park et al., 2014), including race BBBQJ with a virulence phenotype and SSR genotype similar to BBG/BP (Aoun et al., 2017). Hence, it is unlikely that Lr73 is present in Sragolla. The all-stage resistance gene Lr16 maps to the distal end of chromosome 2BS, and is closely linked to the SSR markers Xwmc764, Xgwm210, and Xwmc661 (McCartney et al., 2005) and to KASP markers kwm677 and Kwm744 (Kassa et al., 2017). However, genotyping with these markers showed polymorphism between Saragolla and the Lr16-carrying AC Domain (Supplementary Figures S5, S6), which suggests that QLr.usw-2BS is different from Lr16. Aoun et al. (2017) identified LrPI244061 that confers resistance to race BBBQJ of P. triticina, on chromosome 2BS of the durum landrace PI 244061. Based on the tetraploid wheat consensus map of chromosome 2B (Maccaferri et al., 2015), markers IWB6117 and IWB72183 linked to LrPI244061 (Aoun et al., 2017) mapped at 42.8 cM and 45.8 cM, respectively, whereas markers Tdurum_contig76118_145 and wsnp_Ex_c18354_27181086, flanking QLr.usw-2BS, mapped at 8.4 cM and 12.3 cM, respectively. In addition, marker IWB72183 mapped at 133.8 cM on chromosome 2B_1, in the Saragolla/ATRED #2 genetic map (Supplementary File S1), while QLr.usw-2BS peaked at 82 cM. In summary, QLr.usw-2BS does not seem to include any of the six previously designated Lr genes and therefore is likely to harbor a previously uncharacterized gene.
QTL on Chromosome 3B
Chromosome 3B is known to carry the race-specific resistance gene Lr27 that requires the presence of the complementary gene Lr31 on chromosome 4B, to confer leaf rust resistance in wheat (Mago et al., 2011). However, race BBG/BP emerged in 2008 in the state of Sonora, in northwestern Mexico, after acquiring virulence to the adult plant race-specific resistance gene Lr12 and the seedling complementary resistance genes Lr27+Lr31 (Huerta-Espino et al., 2008; Huerta-Espino et al., 2011). Therefore, Lr27 is unlikely to be involved in the resistance to BBG/BP in Saragolla. The APR gene Lr74 was identified in the hexaploid wheat population Ning7840 × Clark, and mapped to the short arm of chromosome 3B, closely linked to the SNP markers IWA6651, IWA3724, IWA4654, IWA1702, IWA5203, IWA5202, and IWA5201 (Li et al., 2017). Kolmer et al. (2018b) identified a QTL for adult plant leaf rust resistance in the soft red winter wheat cultivar Caldwell, and mapped it very close to the Lr74 locus. Based on the tetraploid wheat consensus map (Maccaferri et al., 2015), all of the SNP markers linked to Lr74 mapped between positions 6 and 7.1 cM on chromosome 3BS, while markers Tdurum_contig33168_461 and RAC875_rep_c82061_78, which flank QLr.usw-3B in Saragolla, mapped at 87 and 88 cM, respectively, in the same consensus map. In addition, marker IWA3724 mapped at 15.9 cM on LG 3B_1 in the Saragolla/ATRED #2 map (Supplementary File S1), while QLr.usw-3B spanned the interval 11.5 to 13.5 cM on LG 3B_2. Therefore, we can assume that QLr.usw-3B is distinct from the APR gene Lr74. Recently, the new APR gene Lr77 was mapped to chromosome 3BL in the hard red winter wheat cultivar “Santa Fe” (Kolmer et al., 2018c). The SNP markers IWB32805 and IWB10344 co-segregating with Lr77, mapped respectively at 148.4 cM and 151.6 cM in the tetraploid wheat consensus map, approximately 62 cM distal to the markers flanking QLr.usw-3B (Maccaferri et al., 2015). Furthermore, marker IWB32805 mapped at 150.8 cM on LG 3B_2 in the Saragolla/ATRED #2 map, while QLr.usw-3B peaked at 13 cM on the same LG (Supplementary File S1). These large distances between the two intervals harboring Lr77 and QLr.usw-3B indicate that they are two distinct Lr genes. Lr79 is another newly mapped all-stage leaf rust resistance gene on chromosome 3BL, from the durum wheat landrace Aus26582 (Qureshi et al., 2018). Based on comparative analysis using the consensus 90K SNP genetic map (Wang et al., 2014) and the physical map of Chinese Spring (RefSeq v1.0), the authors estimated the distance between Lr77 and Lr79 at approximately 12 cM or 9.2 Mbp (Qureshi et al., 2018). Hence, it is possible to argue that Lr79 does also map at a large genetic distance from QLr.usw-3B, and that the latter may be an uncharacterized leaf rust resistance gene.
QTL on Chromosome 6B
Previous studies have reported three designated leaf rust resistance genes that map to the short arm of chromosome 6B: Lr36 (Dvorak and Knott, 1990), Lr53 (Marais et al., 2005), and Lr61 (Herrera-Foessel et al., 2008a). Genes Lr36 and Lr53 were introgressed into chromosome 6BS of hexaploid wheat from T. speltoides and T. dicoccoides, respectively (Dvorak and Knott, 1990; Marais et al., 2005), but no reports of the transfer of either of these genes into durum wheat are available. Hence, these two genes are unlikely candidates for the major QTL QLr.usw-6BS, identified on the short arm of chromosome 6B in Gaza. However, since T. dicoccoides (genome AABB) is the wild progenitor of durum wheat, further genetic analysis and allelism tests would be required to fully rule out Lr53 as a candidate. Lr61 was identified in the CIMMYT-derived Chilean durum wheat cultivar Guayacan INIA, with linkage to marker Xwmc487, and is effective against the P. triticina race BBG/BP, predominant in northwestern Mexico (Herrera-Foessel et al., 2008a; Loladze et al., 2014). The presence of susceptible plants in the F2 progeny from the cross between Gaza and the Lr61-carrying Sooty_9/Rascon_37//Guayacan INIA indicated that QLr.usw-6BS was neither allelic nor linked to Lr61 (Supplementary Table S1). Furthermore, Gaza showed polymorphism compared to the Lr61 carrier Guayacan INIA, for the SSR marker Xwmc487 (Supplementary Figure S1). Kthiri et al. (2018) identified two leaf rust resistance genes in the two durum cultivars Geromtel_3 and Tunsyr_2, conferring resistance to race BBG/BP of P. triticina, and mapping to the short arm of chromosome 6B. The authors showed that Lr_Geromtel_3 and Lr_Tunsyr_2 were either allelic or closely linked to each other and to Lr61 (Kthiri et al., 2018; Kthiri, 2017). Allelism tests showed that the resistance gene in Gaza is either allelic or closely linked to the gene in Geromtel_3 but not to the gene in Tunsyr_2 (Supplementary Table S1). The Australian breeding line PI 209274 carries resistance to races BBBQJ and BBB/BN_Lr61vir of P. triticina, on chromosome 6BS (Aoun et al., 2017). Molecular markers IWB39456 and IWB416, both linked to LrPI209274 (Aoun et al., 2017), mapped at 5.6 cM in the Gaza/ATRED #2 linkage map (Supplementary File S1), close to the peak position of QLr.usw-6BS at 1.3 cM. Additional fine mapping studies and allelism tests are required to determine the relationship between QLr.usw-6BS and LrPI209274.
Several Lr genes have also been previously identified on the long arm of chromosome 6B. These include Lr9, which was translocated into hexaploid wheat from Ae. umbellulata (Schachermayr et al., 1994). However, there are no reports of this gene being transferred into durum wheat, thus, Lr9 is unlikely a candidate gene for QLr.usw-6BL. The all-stage resistance gene Lr3 is a known locus on the long arm of chromosome 6B with four reported alleles: Lr3a, Lr3bg, Lr3ka, and Lr3d (Kolmer, 2015c). Herrera-Foessel et al. (2007) identified and mapped Lr3 and the closely linked gene LrCamayo in the two durum wheat lines Storlom and Camayo, respectively. Although these two closely linked genes on chromosome 6BL confer resistance telo P. triticina races prevalent on durum wheat in Northwestern Mexico, allelism tests between Gaza and Cirno C2008, a carrier of LrCamayo, suggested that the resistance to leaf rust in Gaza is unrelated to LrCamayo (Supplementary Table S1). Furthermore, analysis of seedling and adult plant evaluation results confirmed the involvement of an APR gene for leaf rust resistance in Gaza, while both Lr3 and LrCamayo are all-stage resistance genes. Recently, Lan et al. (2017) identified QLr.cim-6BL that confers adult plant resistance to race BBG/BP of P. triticina, in the CIMMYT durum wheat line Bairds. Likewise, Aoun et al. (2017) mapped LrPI387263 to the long arm of chromosome 6B, in the Ethiopian durum landrace PI 387263. Based on comparative mapping analysis, molecular markers linked to these genes mapped either very close to or overlapping the QLr.usw-6BL interval in the tetraploid wheat consensus map (Maccaferri et al., 2015) or the Gaza/ATRED #2 map (Supplementary File S1). Therefore, QLr.usw-6BL, QLr.cim-6BL, and LrPI387263 could possibly harbor the same gene.
QTL on Chromosome 7B
QLr.usw-7BL maps in a gene-dense region with several genes/QTL for resistance to rusts and other fungal diseases, including Lr14a, one of the most widely exploited Lr genes in wheat (Herrera-Foessel et al., 2008b; Terracciano et al., 2013), and the closely linked gene Lr14b (Dyck and Samborski, 1970), the slow-rusting APR gene Lr68 (Herrera-Foessel et al., 2012), LrBi16 which is allelic to Lr14a (Zhang et al., 2015), and LrFun, which is closely linked to Lr14a (Xing et al., 2014). The prevalent Mexican races of P. triticina, including race BBG/BP that is used in the present study, are virulent for Lr14b (Ordoñez and Kolmer, 2007; Herrera-Foessel et al., 2008b); therefore, it is unlikely that this gene is conferring leaf rust resistance in Arnacoris. Lr68 is an adult-plant resistance gene that confers a slow-rusting phenotype, however, QLr.usw-7BL had a major effect on leaf rust resistance in Arnacoris, at both the seedling and adult plant stages (Figure 3), making Lr68 an unlikely candidate for QLr.usw-7BL. When tested with the NBS-LRR primers 4406F/4840R, both parental lines Arnacoris and ATRED #2 had the null allele (Supplementary Figure S4), indicating that Lr14a is not segregating in this population. Further investigation will be required to identify the specific gene responsible for the leaf rust resistance conferred by QLr.usw-7BL.
Physical Mapping and Candidate Gene Identification
Anchoring of the SNP markers associated with the various QTL detected in the present study to the durum wheat reference genome identified several genes that encode for proteins known to be involved in plant pathogen interactions and disease resistance. So far, three race specific leaf rust resistance genes (Lr1, Lr10, and Lr21) have been cloned in wheat, and all three proteins contained NBS-LRR motifs (Feuillet et al., 2003; Huang et al., 2003; Cloutier et al., 2007). The APR gene Lr34 protein is a full-size ABC transporter (Krattinger et al., 2009) while Lr67 was shown to encode a recently evolved hexose transporter (Moore et al., 2015). Map-based cloning of Yr36, a gene that confers non-race-specific adult plant resistance to stripe rust in wild emmer wheat (T. turgidum ssp. dicoccoides), showed that it encodes a protein with a predicted kinase domain (Fu et al., 2009). The genes identified within the leaf rust resistance QTL intervals in Gaza, Arnacoris, and Saragolla have structures typical of disease resistance proteins and are excellent gene candidates for further studies.
Conclusion
The present study identified six genomic regions involved in leaf rust resistance in durum wheat. The resistance to race BBG/BP of P. triticina in the Middle Eastern landrace Gaza was controlled by two QTL on chromosome 6B. Qlr.usw-6BS accounted for most of the phenotypic variance and is neither allelic nor linked to Lr61. The second QTL, QLr.usw-6BL, may be a new APR gene for leaf rust resistance in wheat. Likewise, the French cultivar Arnacoris carried two QTL for leaf rust resistance. QLr.usw-1BL.1 mapped to the Lr46 region; however, Arnacoris did not carry any of the Lr46 molecular markers. The major QTL on chromosome 7B in Arnacoris, QLr.usw-7BL, explained most of the leaf rust phenotypic variance and was shown to be different from the widely deployed gene Lr14a. The Italian durum variety Saragolla carried a major QTL on chromosome 2B, designated as QLr.usw-2BS, which accounted for most of the phenotypic variance, as well as two minor QTL on chromosomes 3B and 1BL. Molecular marker analysis suggested that QLr.usw-2BS is distinct from Lr16 while QLr.usw-1BL.2 is likely the APR leaf rust resistance gene Lr46, and QLr.usw-3B is a potentially uncharacterized leaf rust resistance gene. Physical mapping of the SNP markers associated with these QTL to the durum wheat reference sequence enabled the identification of candidate genes for leaf rust resistance in these cultivars. With the availability of SNP markers tightly linked to all these QTL, some with major and other with minor effects, the durum wheat lines used in the present study can be used as donors to strategically combine genes with different modes of action to produce a more durable leaf rust resistance in durum wheat.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
CP and KA conceived and designed the study. KA developed the populations. AL produced the purified inoculum and conducted phenotyping and data collection. AL and DK analyzed the phenotypic data. SD performed DNA extraction. DK performed the genotyping and QTL analysis and drafted the manuscript. AN’D built the genetic maps. KN conducted the physical mapping. SW revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
We are grateful for the financial support of Genome Canada, Genome Prairie, Western Grains Research Foundation, Saskatchewan Wheat Development Commission, Alberta Wheat Development Commission, the Saskatchewan Ministry of Agriculture, Viterra, the Saskatchewan Innovation and Opportunity Scholarship, and the Rene Vandeveld Postgraduate Scholarship. CIMMYT’s contribution to the present work has been made possible thanks to funding from “Patronato para la Investigación y Experimentación Agrícola del Estado de Sonora (PIEAES),” “Fundacíón Produce Sonora, México,” and the Durable Rust Resistance Wheat (DRRW) project as part of the Borlaug Global Rust Initiative (BGRI).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript is part of DK’s Ph.D. thesis at the University of Saskatchewan. This research was conducted as part of the Canadian Triticum Applied Genomics (CTAG2) project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01247/full#supplementary-material
References
Aoun, M., Kolmer, J. A., Rouse, M. N., Chao, S., Denbel Bulbula, W., Elias, E. M., et al. (2017). Inheritance and bulked segregant analysis of leaf rust and stem rust resistance in durum wheat genotypes. Phytopathology 107, 1496–1506. doi: 10.1094/PHYTO-12-16-0444-R
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Buerstmayr, M., Matiasch, L., Mascher, F., Vida, G., Ittu, M., Robert, O., et al. (2014). Mapping of quantitative adult plant field resistance to leaf rust and stripe rust in two European winter wheat populations reveals co-location of three QTL conferring resistance to both rust pathogens. Theor. Appl. Genet. 127, 2011–2028. doi: 10.1007/s00122-014-2357-0
Cavanagh, C. R., Chao, S., Wang, S., Huang, B. E., Stephen, S., Kiani, S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. 110, 8057–8062. doi: 10.1073/pnas.1217133110
Cloutier, S., McCallum, B. D., Loutre, C., Banks, T. W., Wicker, T., Feuillet, C., et al. (2007). Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol. Biol. 65, 93–106. doi: 10.1007/s11103-007-9201-8
Dreisigacker, S., Sehgal, D., Reyes-Jaimez, A. E., Luna-Garrido, B., Muñoz-Zavala, S., Núñez-Ríos, C., et al. (2016). CIMMYT wheat molecular genetics: laboratory protocols and applications to wheat breeding. Mexico, D.F: CIMMYT.
Dvorak, J., Knott, D. R. (1990). Location of a Triticum speltoides chromosome segment conferring resistance to leaf rust in Triticum aestivum. Genome 33, 892–897. doi: 10.1139/g90-134
Dyck, P. L., Kerber, E. R., Lukow, O. M. (1987). Chromosome location and linkage of a new gene (Lr33) for reaction to Puccinia recondita in common wheat. Genome 29, 463–466. doi: 10.1139/g87-080
Dyck, P. L., Samborski, D. J. (1970). The genetics of two alleles for leaf rust resistance at the Lr14 locus in wheat. Can. J. Genet. Cytol. 12, 689–694. doi: 10.1139/g70-091
Dyck, P. L., Sykes, E. E. (1994). Genetics of leaf-rust resistance in three spelt wheats. Can. J. Plant Sci. 74, 231–233. doi: 10.4141/cjps94-047
Ellis, J. G., Lagudah, E. S., Spielmeyer, W., Dodds, P. N. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5, 641. doi: 10.3389/fpls.2014.00641
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A. L., Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U. S. A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323, 1357–1360. doi: 10.1126/science.1166289
Herrera-Foessel, S., Huerta-Espino, J., Calvo-Salazar, V., Lan, C. X., Singh, R. P. (2014a). Lr72 confers resistance to leaf rust in durum wheat cultivar Atil C2000. Plant Dis. 98, 631–635. doi: 10.1094/PDIS-07-13-0741-RE
Herrera-Foessel, S., Lagudah, E. S., McIntosh, R., Salazar, V. C., Huerta-Espino, J., Singh, R. P., (2011). First report of slow rusting gene Lr46 in durum wheat. Proceedings of the Borlaug global rust initiative technical workshop, St. Paul, MN.
Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., Rosewarne, G. M., Periyannan, S. K., Viccars, L., et al. (2012). Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor. Appl. Genet. 124, 1475–1486. doi: 10.1007/s00122-012-1802-1
Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., William, H. M., Djurle, A., Yuen, J. (2008a). Molecular mapping of a leaf rust resistance gene on the short arm of chromosome 6B of durum wheat. Plant Dis. 92, 1650–1654. doi: 10.1094/PDIS-92-12-1650
Herrera-Foessel, S., Singh, R. P., Huerta-Espino, J., William, H. M., Garcia, V., Djurle, A., et al. (2008b). Identification and molecular characterization of leaf rust resistance gene Lr14a in durum wheat. Plant Dis. 92, 469–473. doi: 10.1094/PDIS-92-3-0469
Herrera-Foessel, S., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014b). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789. doi: 10.1007/s00122-013-2256-9
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Crossa, J., Djurle, A., Yuen, J. (2008c). Genetic analysis of slow-rusting resistance to leaf rust in durum wheat. Crop Sci. 48, 2132–2140. doi: 10.2135/cropsci2007.11.0606
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., William, M., Rosewarne, G., Djurle, A., et al. (2007). Identification and mapping of Lr3 and a linked leaf rust resistance gene in durum wheat. Crop Sci. 47, 1459–1466. doi: 10.2135/cropsci2006.10.0663
Hiebert, C. W., Thomas, J. B., McCallum, B. D., Humphreys, D. G., DePauw, R. M., Hayden, M. J., et al. (2010). An introgression on wheat chromosome 4DL in RL6077 (Thatcher*6/PI 250413) confers adult plant resistance to stripe rust and leaf rust (Lr67). Theor. Appl. Genet. 121, 1083–1091. doi: 10.1007/s00122-010-1373-y
Huang, L., Brooks, S. A., Li, W., Fellers, J. P., Trick, H. N., Gill, B. S. (2003). Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics 164, 655.
Huerta-Espino, J., Singh, R. P., German, S., McCallum, B. D., Park, R. F., Chen, W. Q., et al. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179, 143–160. doi: 10.1007/s10681-011-0361-x
Huerta-Espino, J., Singh, R. P., Herrera-Foessel, S. A., Pérez-López, J. B., Figueroa-López, P. (2008). First detection of virulence in Puccinia triticina to resistance genes Lr27 + Lr31 present in durum wheat in Mexico. Plant Dis. 93, 110–110. doi: 10.1094/PDIS-93-1-0110C
Joehanes, R., Nelson, J. C. (2008). QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 24, 2788–2789. doi: 10.1093/bioinformatics/btn523
Kassa, M. T., You, F. M., Hiebert, C. W., Pozniak, C. J., Fobert, P. R., Sharpe, A. G., et al. (2017). Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16. BMC Plant Biol. 17, 45. doi: 10.1186/s12870-017-0993-7
Kaul, K., Shaner, G. (1989). Effect of temperature on adult-plant resistance to leaf rust in wheat. Phytopathology 79, 391–394. doi: 10.1094/Phyto-79-391
Knott, D. R. (1989). The wheat rusts - Breeding for resistance. Berlin, Heidelberg: Springer-Verlag. doi: 10.1007/978-3-642-83641-1
Kolmer, J. A. (2013). Leaf rust of wheat: pathogen biology, variation and host resistance. Forests 4 (1), 70–84. doi: 10.3390/f4010070
Kolmer, J. A. (2015a). A QTL on chromosome 5BL in wheat enhances leaf rust resistance of Lr46. Mol. Breed. 35, 74. doi: 10.1007/s11032-015-0274-9
Kolmer, J. A. (2015b). First report of a wheat leaf rust (Puccinia triticina) phenotype with high virulence to durum wheat in the Great Plains region of the United States. Plant Dis. 99, 156–156. doi: 10.1094/PDIS-06-14-0667-PDN
Kolmer, J. A. (2015c). Leaf rust resistance in wheat line RL6062 is an allele at the Lr3 locus. Crop Sci. 55, 2186–2190. doi: 10.2135/cropsci2015.01.0031
Kolmer, J. A., Bernardo, A., Bai, G., Hayden, M. J., Chao, S. (2018a). Adult plant leaf rust resistance derived from toropi wheat is conditioned by Lr78 and three minor QTL. Phytopathology 108, 246–253. doi: 10.1094/PHYTO-07-17-0254-R
Kolmer, J. A., Chao, S., Brown-Guedira, G., Bansal, U., Bariana, H. (2018b). Adult plant leaf rust resistance derived from the soft red winter wheat cultivar ‘caldwell’ maps to chromosome 3BS. Crop Sci. 58, 152–158. doi: 10.2135/cropsci2017.05.0272
Kolmer, J. A., Hughes, M. E. (2017). Physiologic specialization of Puccinia triticina on wheat in the United States in 2015. Plant Dis. 101, 1968–1973. doi: 10.1094/PDIS-02-17-0200-SR
Kolmer, J. A., Su, Z., Bernardo, A., Bai, G., Chao, S. (2018c). Mapping and characterization of the new adult plant leaf rust resistance gene Lr77 derived from Santa Fe winter wheat. Theor. Appl. Genet. 131, 1553–1560. doi: 10.1007/s00122-018-3097-3
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Kthiri, D. (2017). Genetic and molecular characterization of leaf rust resistance from uncharacterized sources of durum wheat (Triticum turgidum L. ssp. durum). [dissertation/PhD. thesis]. Saskatoon (SK): University of Saskatchewan. http://hdl.handle.net/10388/7928.
Kthiri, D., Loladze, A., MacLachlan, P. R., N’Diaye, A., Walkowiak, S., Nilsen, K., et al. (2018). Characterization and mapping of leaf rust resistance in four durum wheat cultivars. PLoS ONE 13, e0197317. doi: 10.1371/journal.pone.0197317
Lagudah, E. S. (2011). Molecular genetics of race non-specific rust resistance in wheat. Euphytica 179, 81–91. doi: 10.1007/s10681-010-0336-3
Lan, C., Basnet, B. R., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S., Ren, Y., et al. (2017). Genetic analysis and mapping of adult plant resistance loci to leaf rust in durum wheat cultivar Bairds. Theor. Appl. Genet. 130, 609–619. doi: 10.1007/s00122-016-2839-3
Li, C., Wang, Z., Li, C., Bowden, R., Bai, G., Su, Z., et al. (2017). Mapping of quantitative trait loci for leaf rust resistance in the wheat population Ning7840 × Clark. Plant Dis. 101, 1974–1979. doi: 10.1094/PDIS-12-16-1743-RE
Loladze, A., Kthiri, D., Pozniak, C., Ammar, K. (2014). Genetic analysis of leaf rust resistance in six durum wheat genotypes. Phytopathology 104, 1322–1328. doi: 10.1094/PHYTO-03-14-0065-R
Lorieux, M. (2012). MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30, 1231–1235. doi: 10.1007/s11032-012-9706-y
Lowe, I., Cantu, D., Dubcovsky, J. (2011). Durable resistance to the wheat rusts: integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica 179, 69–79. doi: 10.1007/s10681-010-0311-z
Maccaferri, M., Harris, N. S., Twardziok, S. O., Pasam, R. K., Gundlach, H., Spannagl, M., et al. (2019). Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 51, 885–895. doi: 10.1038/s41588-019-0381-3
Maccaferri, M., Ricci, A., Salvi, S., Milner, S. G., Noli, E., Martelli, P. L., et al. (2015). A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 13, 648–663. doi: 10.1111/pbi.12288
Maccaferri, M., Sanguineti, M. C., Mantovani, P., Demontis, A., Massi, A., Ammar, K., et al. (2010). Association mapping of leaf rust response in durum wheat. Mol. Breed. 26, 189–228. doi: 10.1007/s11032-009-9353-0
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112, 41–50. doi: 10.1007/s00122-005-0098-9
Mago, R., Spielmeyer, W., Lawrence, G., Lagudah, E., Ellis, J., Pryor, A. (2002). Identification and mapping of molecular markers linked to rust resistance genes located on chromosome 1RS of rye using wheat-rye translocation lines. Theor. Appl. Genet. 104, 1317–1324. doi: 10.1007/s00122-002-0879-3
Mago, R., Tabe, L., McIntosh, R. A., Pretorius, Z., Kota, R., Paux, E., et al. (2011). A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theor. Appl. Genet. 123, 615–623. doi: 10.1007/s00122-011-1611-y
Marais, G. F., Pretorius, Z. A., Wellings, C. R., McCallum, B., Marais, A. S. (2005). Leaf rust and stripe rust resistance genes transferred to common wheat from Triticum dicoccoides. Euphytica 143, 115–123. doi: 10.1007/s10681-005-2911-6
Marone, D., Del Olmo, A. I., Laidò, G., Sillero, J. C., Emeran, A. A., Russo, M. A., et al. (2009). Genetic analysis of durable resistance against leaf rust in durum wheat. Mol. Breed. 24, 25–39. doi: 10.1007/s11032-009-9268-9
McCartney, C. A., Somers, D. J., McCallum, B. D., Thomas, J., Humphreys, D. G., Menzies, J. G., et al. (2005). Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Mol. Breed. 15, 329–337. doi: 10.1007/s11032-004-5948-7
McIntosh, R., Dyck, P. L. (1975). Cytogenetical studies in wheat. VII Gene Lr23 for reaction to Puccinia recondita in gabo and related cultivars. Aust. J. Biol. Sci. 28, 201–211. doi: 10.1071/BI9750201
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., Appels, R., Xia, X. C.Catalogue of gene symbols for wheat: 2015-2016 supplement, 2015, Available at: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf.
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C., Xia, X. C.Catalogue of gene symbols for wheat: 2017 supplement, 2017, Available at: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2017.pdf.
McIntosh, R. A., Wellings, C. R., Park, R. F., (1995). Wheat rusts: an atlas of resistance genes. Netherlands: Springer. doi: 10.1071/9780643101463
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Nsabiyera, V., Qureshi, N., Bariana, H. S., Wong, D., Forrest, K. L., Hayden, M. J., et al. (2016). Molecular markers for adult plant leaf rust resistance gene Lr48 in wheat. Mol. Breed. 36, 65. doi: 10.1007/s11032-016-0488-5
Ordoñez, M. E., Kolmer, J. A. (2007). Virulence phenotypes of a worldwide collection of Puccinia triticina from durum wheat. Phytopathology 97, 344–351. doi: 10.1094/PHYTO-97-3-0344
Park, R. F., Mohler, V., Nazari, K., Singh, D. (2014). Characterisation and mapping of gene Lr73 conferring seedling resistance to Puccinia triticina in common wheat. Theor. Appl. Genet. 127, 2041–2049. doi: 10.1007/s00122-014-2359-y
Peterson, R. F., Campbell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26c, 496–500. doi: 10.1139/cjr48c-033
Qureshi, N., Bariana, H., Kumran, V. V., Muruga, S., Forrest, K. L., Hayden, M. J., et al. (2018). A new leaf rust resistance gene Lr79 mapped in chromosome 3BL from the durum wheat landrace Aus26582. Theor. Appl. Genet. 131, 1091–1098. doi: 10.1007/s00122-018-3060-3
Risk, J. M., Selter, L. L., Krattinger, S. G., Viccars, L. A., Richardson, T. M., Buesing, G., et al. (2012). Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnol. J. 10, 477–487. doi: 10.1111/j.1467-7652.2012.00683.x
Roelfs, A. P., Singh, R. P., Saari, E. E., (1992). Rust diseases of wheat: concepts and methods of disease management. Mexico: CIMMYT.
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S., Forrest, K. L., Hayden, M. J., et al. (2012). Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet x Pastor wheat population. Theor. Appl. Genet. 124, 1283–1294. doi: 10.1007/s00122-012-1786-x
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., Rebetzke, G. J. (2008). Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor. Appl. Genet. 116, 1027–1034. doi: 10.1007/s00122-008-0736-0
Saini, R. G., Kaur, M., Singh, B., Sharma, S., Nanda, G. S., Nayar, S. K., et al. (2002). Genes Lr48 and Lr49 for hypersensitive adult plant leaf rust resistance in wheat (Triticum aestivum L.). Euphytica 124, 365–370. doi: 10.1023/A:1015762812907
Schachermayr, G., Siedler, H., Gale, M. D., Winzeler, H., Winzeler, M., Keller, B. (1994). Identification and localization of molecular markers linked to the Lr9 leaf rust resistance gene of wheat. Theor. Appl. Genet. 88, 110–115. doi: 10.1007/BF00222402
Schnurbusch, T., Paillard, S., Schori, A., Messmer, M., Schachermayr, G., Winzeler, M., et al. (2004). Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor. Appl. Genet. 108, 477–484. doi: 10.1007/s00122-003-1444-4
Seyfarth, R., Feuillet, C., Schachermayr, G., Messmer, M., Winzeler, M., Keller, B. (2000). Molecular mapping of the adult-plant leaf rust resistance gene Lr13 in wheat (Triticum aestivum L.). J. Genet. Breed. 54, 193–198.
Seyfarth, R., Feuillet, C., Schachermayr, G., Winzeler, M., Keller, B. (1999). Development of a molecular marker for the adult plant leaf rust resistance gene Lr35 in wheat. Theor. Appl. Genet. 99, 554–560. doi: 10.1007/s001220051268
Singh, A., Pandey, M. P., Singh, A. K., Knox, R. E., Ammar, K., Clarke, J. M., et al. (2013a). Identification and mapping of leaf, stem and stripe rust resistance quantitative trait loci and their interactions in durum wheat. Mol. Breed. 31, 405–418. doi: 10.1007/s11032-012-9798-4
Singh, D., Mohler, V., Park, R. F. (2013b). Discovery, characterisation and mapping of wheat leaf rust resistance gene Lr71. Euphytica 190, 131–136. doi: 10.1007/s10681-012-0786-x
Singh, R. P., Huerta-Espino, J., Pfeiffer, W., Figueroa-Lopez, P. (2004). Occurrence and impact of a new leaf rust race on durum wheat in northwestern Mexico from 2001 to 2003. Plant Dis. 88, 703–708. doi: 10.1094/PDIS.2004.88.7.703
Singh, R. P., Mujeeb-Kazi, A., Huerta-Espino, J. (1998). Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88, 890–894. doi: 10.1094/PHYTO.1998.88.9.890
Singh, R. P., Singh, P. K., Rutkoski, J., Hodson, D. P., He, X., Jørgensen, L. N., et al. (2016). Disease impact on wheat yield potential and prospects of genetic control. Annu. Rev. Phytopathol. 54, 303–322. doi: 10.1146/annurev-phyto-080615-095835
Singla, J., Lüthi, L., Wicker, T., Bansal, U., Krattinger, S. G., Keller, B. (2017). Characterization of Lr75: a partial, broad-spectrum leaf rust resistance gene in wheat. Theor. Appl. Genet. 130, 1–12. doi: 10.1007/s00122-016-2784-1
Soleiman, N. H., Solis, I., Soliman, M. H., Sillero, J. C., Villegas, D., Alvaro, F., et al. (2016). Short communication: emergence of a new race of leaf rust with combined virulence to Lr14a and Lr72 genes on durum wheat. Span. J. Agric. Res. 14, e10SC02. doi: 10.5424/sjar/2016143-9184
Soriano, J. M., Royo, C. (2015). Dissecting the genetic architecture of leaf rust resistance in wheat by QTL meta-analysis. Phytopathology 105, 1585–1593. doi: 10.1094/PHYTO-05-15-0130-R
Suenaga, K., Singh, R. P., Huerta-Espino, J., William, H. M. (2003). Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 93, 881–890. doi: 10.1094/PHYTO.2003.93.7.881
Terracciano, I., Maccaferri, M., Bassi, F., Mantovani, P., Sanguineti, M. C., Salvi, S., et al. (2013). Development of COS-SNP and HRM markers for high-throughput and reliable haplotype-based detection of Lr14a in durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 126, 1077–1101. doi: 10.1007/s00122-012-2038-9
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
William, H. M., Singh, R. P., Huerta-Espino, J., Palacios, G., Suenaga, K. (2006). Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49, 977–990. doi: 10.1139/g06-052
Winfield, M. O., Allen, A. M., Burridge, A. J., Barker, G. L. A., Benbow, H. R., Wilkinson, P. A., et al. (2016). High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 14, 1195–1206. doi: 10.1111/pbi.12485
Wu, T. D., Watanabe, C. K. (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. doi: 10.1093/bioinformatics/bti310
Wu, Y., Bhat, P. R., Close, T. J., Lonardi, S. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet. 4, e1000212. doi: 10.1371/journal.pgen.1000212
Xing, L., Wang, C., Xia, X., He, Z., Chen, W., Liu, T., et al. (2014). Molecular mapping of leaf rust resistance gene LrFun in Romanian wheat line Fundulea 900. Mol. Breed. 33, 931–937. doi: 10.1007/s11032-013-0007-x
Yates, F. (1934). Contingency tables involving small numbers and the χ2 test. Supplement J. R. Stat. Soc. 1, 217–235. doi: 10.2307/2983604
Zhang, P., Hiebert, C. W., McIntosh, R. A., McCallum, B. D., Thomas, J. B., Hoxha, S., et al. (2016). The relationship of leaf rust resistance gene Lr13 and hybrid necrosis gene Ne2m on wheat chromosome 2BS. Theor. Appl. Genet. 129, 485–493. doi: 10.1007/s00122-015-2642-6
Keywords: durum wheat, leaf rust, Puccinia triticina, resistance, quantitative trait loci, single nucleotide polymorphism (SNP)
Citation: Kthiri D, Loladze A, N’Diaye A, Nilsen KT, Walkowiak S, Dreisigacker S, Ammar K and Pozniak CJ (2019) Mapping of Genetic Loci Conferring Resistance to Leaf Rust From Three Globally Resistant Durum Wheat Sources. Front. Plant Sci. 10:1247. doi: 10.3389/fpls.2019.01247
Received: 14 May 2019; Accepted: 06 September 2019;
Published: 08 October 2019.
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaReviewed by:
James Kolmer, Agricultural Research Service, United States Department of Agriculture, United StatesFilippo Maria Bassi, International Center for Agricultural Research in the Dry Areas (ICARDA), Morocco
Copyright © 2019 Kthiri, Loladze, N’Diaye, Nilsen, Walkowiak, Dreisigacker, Ammar and Pozniak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Curtis J. Pozniak, Y3VydGlzLnBvem5pYWtAdXNhc2suY2E=
 Dhouha Kthiri
Dhouha Kthiri Alexander Loladze
Alexander Loladze Amidou N’Diaye
Amidou N’Diaye Kirby T. Nilsen
Kirby T. Nilsen Sean Walkowiak
Sean Walkowiak Susanne Dreisigacker
Susanne Dreisigacker Karim Ammar
Karim Ammar Curtis J. Pozniak
Curtis J. Pozniak