Corrigendum: Qualitative and Quantitative NAD+ Metabolomics Lead to Discovery of Multiple Functional Nicotinate N-Glycosyltransferase In Arabidopsis
- 1State Key Laboratory of Plant Genomics and National Center for Plant Gene Research (Beijing), Institute of Genetics and Developmental Biology, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing, China
- 2College of Advanced Agricultural Sciences, University of the Chinese Academy of Sciences, Beijing, China
The Preiss-Handler pathway, which salvages nicotinate (NA) for NAD synthesis, is a conserved biochemical pathway in land plants. We previously demonstrated that various NA conjugations (mainly methylation and glycosylation) shared the NA detoxification function in all tested plants. It remains unclear whether other NA conjugates with low abundance exist in plants. In this study, we discovered at least two additional NA N-glycosides in Arabidopsis, which was tentatively elucidated as nicotinate N-pentoside (NaNP) and NA N-rhamoside (NaNRha), using liquid chromatography triple-quadrupole mass spectrometry (LC-QQQ-MS) with precursor ion-scanning (PreIS). We further quantitatively profile the NAD+-related metabolites in 24 tissues of Arabidopsis. Biochemical assays of UGT76C family revealed that UGT76C5 (encoded by At5g05890, previously identified as NaNGT) was a multiple functional nicotinate N-glycosyltransferase, with high preference to UDP-xylose and UDP-arabinose. The deficiency of NaNP and NaNRha in ugt76c5 mutant suggested that UGT76C5 is responsible for biosynthesis of NaNP and NaNRha in planta. We also identify one amino acid difference in PSPG (plant secondary product glycosyltransferase) motif is responsible for the divergence of NaNGT (UGT76C4) and UGT76C5. Taken together, our study not only identifies a novel nicotinate N-glycosyltransferase but also paves the way for investigations of the in planta physiological functions of various NA conjugations.
Introduction
It has long been known that NAD is a coenzyme that serves as an electron carrier in hundreds of redox reactions. In land plants, NAD can be biosynthesized via both a de novo pathway starting from aspartate (Asp) or by the Preiss-Handler salvage pathway (Katoh et al., 2006; Noctor et al., 2006; Wang and Pichersky, 2007). The Preiss-Handler pathway, shared by land plants and most bacteria, starts with nicotinate (NA), which generated by nicotinamidase (NIC1, EC 3.5.1.19, encoded by At2g22570) from nicotinamide (NAM). In Arabidopsis, NA was catalyzed sequentially by nicotinate phosphoribosyltransferase (NaPRT, EC 6.3.4.21, encoded by At2g23420 and At4g36940), nicotinate mononucleotide adenylyltransferase (NaMNAT, EC 2.7.7.18, encoded by At5g55810), and NAD synthase (NADS, EC 6.3.5.1, encoded by At1g55090) (Figure 1) (Preiss and Handler, 1958a, Preiss and Handler, 1958b; Wang and Pichersky, 2007). Feeding experiments, typically using carbonyl [14C]-nicotinamide ([14C]-NAM) as a tracer, showed that NA in plants, in addition to being a precursor for NAD replenishment, can also be converted into a variety of NA conjugations (Figure 1). The over-accumulation of NA has been demonstrated to cause toxicity in plant cells (Zheng et al., 2005; Wang and Pichersky, 2007; Li et al., 2015; Li et al., 2017). We previously proposed that plant developed various strategies to deal with the NA toxicity based on the fact that various conjugates of NA (glycosylation or methylation at the N-position or carboxyl group of NA) had been detected in plants. We had demonstrated that NA O-glucosylation catalyzed by nicotinate O-glucosyltransferase (NaOGT, encoded by At2g43820), a process likely restricted to the Brassicaceae, functions to detoxify NA and involve into seed germination under stress conditions (Li et al., 2015). Two genes encoding NA N-glucosyltransferase (NaNGTs, EC 2.4.1.196, encoded by At5g05880 and At5g05890) were also cloned and functional characterized (Li et al., 2015). However, N-methylnicotinate, also called trigonelline, is a ubiquitous NA conjugate in land plants. NA detoxification conferred by nicotinate N-methyltransferase (NaNMT, EC 2.1.1.7, encoded by At3g53140) might have facilitated the retention of the Preiss-Handler pathway in land plants (Li et al., 2017). More recently, the reversible methylation at carboxyl group of NA had been characterized as long-distance transport of NAD precursors in Arabidopsis thaliana (Wu et al., 2018).
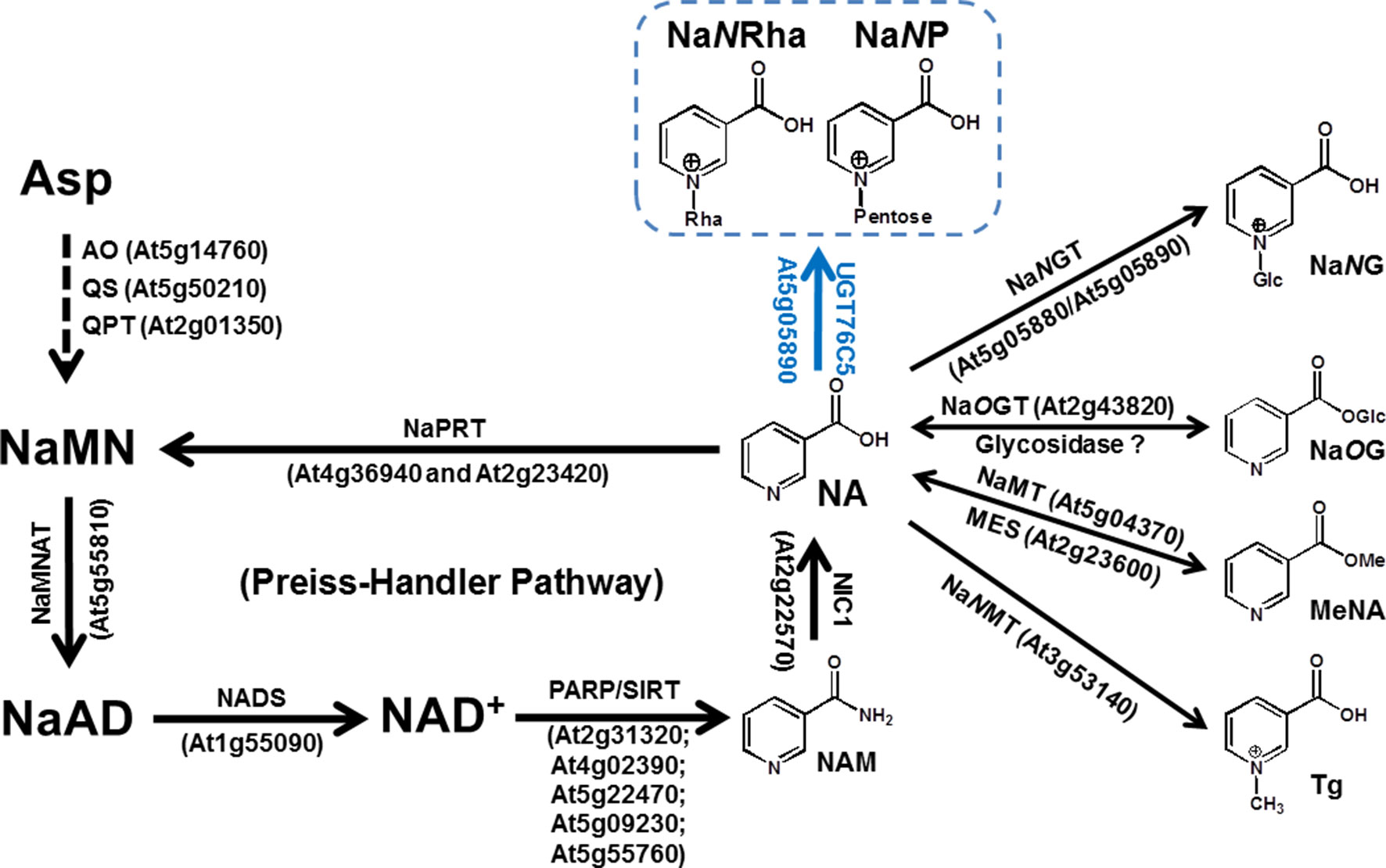
Figure 1 NAD Metabolism and Nicotinate Conjugations in Arabidopsis thaliana. The reaction catalyzed by UGT76C5 and its products, NaNP (nicotinate N-pentoside) and NaNRha (nicotinate N-rhamnose), are highlighted. The genes, which encode characterized enzymes in Arabidopsis, are also indicated. Abbreviations: AO, aspartate oxidase; MeNA, methyl nicotinate; MES, methylesterase; NA, nicotinate; NaAD, nicotinate adenine dinucleotide; NAD, nicotinamide adenine dinucleotide; NADS, NAD synthase; NAM, nicotinamide; NaMN, nicotinate mononucleotide; NaMNAT, NaMN adenylyltransferase; NaMT, nicotinate methyltransferase; NaNMT, nicotinate N-methyltransferase; NaOG, nicotinate O-glucoside; NaOGT, nicotinate O-glucosyltransferase; NaNG, nicotinate N-glucoside; NaNGT, nicotinate N-glucosyltransferase; NaPRT, nicotinate phosphoribosyltransferase; NIC1, nictinamidase 1; PARP, poly(ADP-ribose) polymerase; QPT, quinolinate phosphoribosyltransferase; QS, quinolinate synthase; SIRT, sirtuins (NAD dependent protein deacetylases); Tg, trigonelline.
Besides the above-mentioned NA conjugations, it is an open question whether additional NA conjugates exist in Arabidopsis thaliana. Given the low resolution of TLC analysis used in previous studies on NA metabolism, the discovery of unknown NA conjugates, especially for those which are in low abundance, is really limited. Recently, the mass spectrometry with precursor ion-scanning (PreIS) becomes an alternative choice to discover a series of derivatives with a common substructure (such as the NA moiety in NA conjugates) (Du et al., 2018). The QQQ-MS with PreIS provides much higher resolution and selectivity than the classical TLC analysis.
In this study, we first discovered two types of NA N-glycosides in Arabidopsis thaliana by using LC-QQQ-MS with PreIS; one peak was identified as NA N-pentoside, and another one was tentatively identified as NA N-rhamoside. Quantitative NAD+ metabolomics of 24 Arabidopsis tissues were determined using LC-QQQ-MS with the optimized MRM (multiple reaction monitoring) conditions. We further determine UGT76C5 (encoded by At5g05890) as a multiple functional NA N-glycosyltransferase with sugar-donor promiscuity. One amino acid in PSPG (plant secondary product glycosyltransferase) motif is responsible for the biochemical difference of UGT76C4 (NaNGT) and UGT76C5.
Materials and Methods
Plant Materials and Chemicals
The wild-type and transgenic Arabidopsis thaliana (Col-0 ecotype) lines were grown on soil at 22°C under a 16-h light (120 µmol m−2 s−1)/8-h dark cycle, 60% relative humidity. To harvest young seedlings, the sterilized seeds were sown on ½ MS plates (0.8% agar). The plates were incubated at 4°C for 3 days and then transferred into growth chamber with long-day condition (16-h light/8-h dark cycle, 60 µmol m−2 s−1, 60% relative humidity). All chemicals used in this study were purchased from Sigma-Aldrich (USA) except the radiolabeled (carboxyl-14C)-NA (55 mCi/mmol) were purchased from American Radiolabeled Chemicals (USA). UDP-xylose and UDP-arabinose were purchased from Sugars Tech company (Qingdao, China).
LC-QQQ-MS Analysis With Precursor Ion Scan and MRM
The extraction and measurement of NA, its conjugates, and other NAD-related chemicals in Arabidopsis are carried out as described previously (Li et al., 2015; Wu et al., 2018). An Agilent 1290 ultra-high performance liquid chromatography coupled with an Agilent 6495 Triple Quadrupole Mass Spectrometer was applied for metabolites analysis. Samples were separated on an ACQUITY UPLC BEH Amide column (1.7 μm, 2.1 mm i.d. × 100 mm) (Waters Corporation). The mobile phase consisted of 50% acetonitrile (A) and 95% acetonitrile (B). The mobile phases A and B contained 0.15% ammonium hydroxide and 5mM ammonium acetate. A linear gradient elution program was used: 0–0.5 min, 98% B; 0.5–6 min, 98–75% B; and 6–7 min, 75–25% B. The column temperature was 35°C. The flow rate was 0.35 ml/min. Precursor ion scan mode was used for identifying the unknown NA N-glycosides, and MRM mode was used for the quantification of the interested metabolites. The MRM parameters were listed in Supplemental Table 1. The MS acquisition and precursor ion scan parameters were as follows: high-purity nitrogen (N2) was used as the nebulizer (12 L/min, 200°C, 30 psig) and sheath gas (12 L/min, 350°C). The parent ion ranges from m/z 50 to 800. The precursor ion was m/z 124, fragment was 380 v, and collision voltage was 20 v. Data were analyzed by MassHunter Software B07.01 (Agilent).
Site-Directed Mutagenesis and Glycosyltransferase Assays
Mutated UGT76C4 and UGT76C5 were generated by using a previously described PCR-mediated method (Ho et al., 1989). The primers used are detailed in Supplemental Table 2.
NaNGT proteins, including UGT76C4, UGT76C5, and their mutants, expression in E. coli, purification of recombinant NaNGTs’ and GT assays are carried out as described previously (Li et al., 2015).
Results and Discussion
Novel NA Conjugates Detected in Arabidopsis thaliana
Based on mass spectrometry cleavage rules of known NA conjugates (such as NaNGT and NaOGT), the m/z 124.0 fragment (NA moiety, C6H6NO2+, the calculated m/z 124.0398) was thus chosen as signature ion for PreIS to screen unknown NA conjugates (Li et al., 2015). We firstly applied LC-QQQ-MS with PreIS of m/z 124 to detect unknown NA conjugates from the root of 15-day-old Arabidopsis seedlings. As shown in Figure 2A, five compounds probably with NA moiety were detected on the chromatogram. Among these five putative NA conjugates, peaks 1, 2, and 5 were elucidated as NaOG, NaR (nicotinate riboside), and NaNG by comparison with authentic compounds. The chemical structures of peaks 3 and 4 were further elucidated by product ion mode. The [M+H]+ ions for peaks 3 and 4 display monoisotopic m/z at 270.1 (270.0975 on qTOF-MS) and 256.1 (256.0817 on qTOF-MS), respectively. The lost neutral fragment for peak 3 has a predicted molecular weight of 146.0 (C6H10O4) consistent with a rhamnose moiety (Figure 2B–C). However, the lost neutral fragment for peak 4 has a predicted molecular weight of 132.0 (C5H8O4), consistent with a pentose moiety (Figure 2D–E). Therefore, peaks 3 and 4 were tentatively elucidated as nicotinate N-rhamnoside and nicotinate N-pentoside, respectively, based on the polarity of their chemical structures and their behavior in our HPLC analytical system (Figure 2A). The pentose moiety attached to nicotinate could be either xylose or arabinose, both of which shared the same chemical formula (C5H10O5) and commonly detected in plants (Reiter, 2008; Caputi et al., 2012). It is noteworthy that NA N-arabinoside and NA N-xyloside could not be well separated on our LC-QQQ-MS system (Supplemental Figure 1); we thus use NA N-pentoside (NaNP) to express the sum of NA N-arabinoside and NA N-xyloside for further analysis.
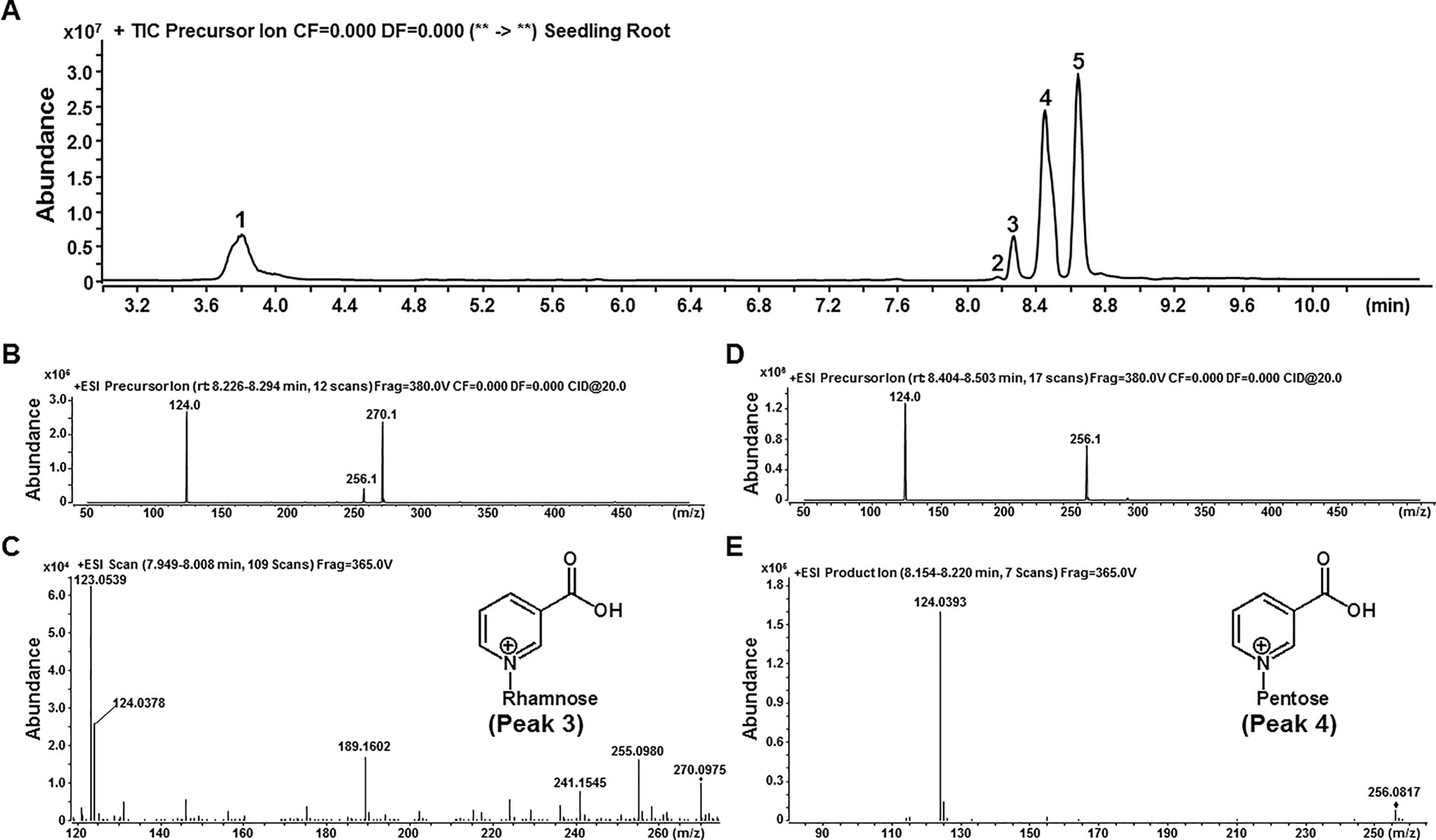
Figure 2 Two novel NA glycosides detected by using LC-QQQ-MS with PreIS of m/z 124. (A) Liquid chromatograph of total five different NA glycosides, which detected in the root tissue of 15-day-old Arabidopsis seedlings. Peaks 1, 2, and 5 were elucidated as NaOG, NaR (nicotinate riboside), and NaNG by comparison with authentic compounds. (B and C). Mass spectra of Peak 3, which generated with QQQ-MS (B) and qTOF-MS (C). (D and E). Mass spectra of Peak 4, which generated with QQQ-MS (D) and qTOF-MS (E).
Tissue Specificity of NAD+ Metabolomics
As it is hard to separate these two novel NA glycosides from NaNG using radio-TLC as we performed previously, we thus develop a targeted LC-QQQ-MS method with optimized MRM mode (Supplemental Table 1) to quantity these two chemicals and other NAD-related chemicals in 24 different tissues of Arabidopsis thaliana (detailed sample information see Supplemental Table 3). The analytical results showed that both two novel NA N-glycosides were highly accumulated in root tissues, from either young seedlings or adult plants (Figure 3A). This pattern is similar to NaNG, which suggested that the genes responsible for the production of novel NA N-glycosides is highly expressed in Arabidopsis roots.
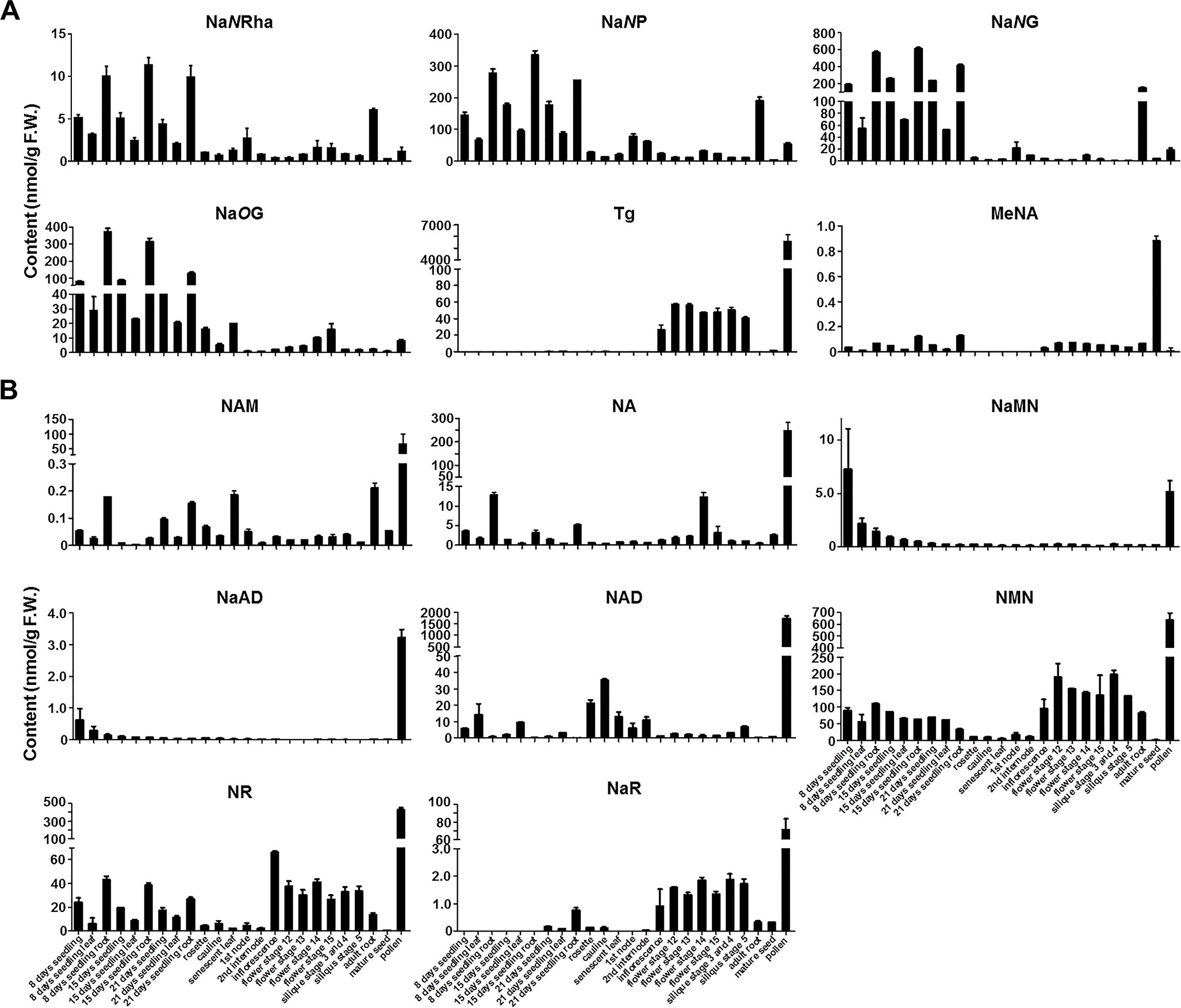
Figure 3 Measurement of endogenous NA conjugates (A) and NAD-related compounds (B) in 24 different tissues of Arabidopsis. It is noteworthy that MeNA was measured using different methods (detailed see Wu et al., 2018). F.W., fresh weight. The abbreviations for chemical name see Figure 1 legend. Bars show means ± SDs (n = 3). F.W., fresh weight. Other abbreviations: NaNRha, nicotinate N-rhamnoside; NaNP, nicotinate N-pentoside; NaR, nicotinate riboside; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside.
There are three other intriguing results from the chemical analysis. First, NMN is highly accumulated in all tested tissues except the mature seeds, although the physiological function is unclear thus far. NMN could be re-utilized for NAD biosynthesis catalyzed by NaMNAT (encoded by At5g55810), which adenylates both NMN and NaMN in vitro (Hashida et al., 2007). Whether NR (nicotinamide riboside) and NaR could be utilized for NAD production is also an open question to be further clarified. Second, trigonelline content was extremely high in pollen when compared with other tissues (Figure 3A). However, the gene encoding trigonelline synthase (also called nicotinate N-methyltransferase, At3g53140) was barely expressed in pollen (Li et al., 2017), which suggested that a transporter protein might be involved in trigonelline accumulation in pollen. Previously, a NA/trigonelline transporter protein (encoded by At3g13050) had been characterized by using a comparative genomics strategy (Jeanguenin et al., 2012). Interestingly, At3g13050 was predominantly expressed in mature pollen based on the data from TAIR website (https://www.arabidopsis.org/; Supplemental Figure 2; (Winter et al., 2007)). Third, the chemicals involved in NAD salvage pathway, including NAM, NA, NaMN, NaAD, and NAD, are predominantly accumulated in mature pollens (Figure 3B). This phenomenon is consistent with a previous report (Hashida et al., 2013). High-NAD accumulation in dry pollen and fast decrease in germinated pollen suggested that NAD biosynthetic pathway (de novo and salvage pathway) plays an important role in pollen germination and pollen tube elongation. Further experiments are needed to reveal the mechanism of this phenomenon.
UGT76C5 Is a Multiple Functional Nicotinate N-Glycosyltransferase in Arabidopsis
Previously, we had identified UGT76C4 and UGT76C5 as NA N-glucosyltransferase; both UGT76C4 and UGT76C5 also showed root-predominant expression pattern. The promiscuous property of plant small molecule glycosyltransferase led us to test whether UGT76C4 and UGT76C5 are responsible for NA pentosylation and/or rhamnosylation in Arabidopsis. UGT76C3 (encoded by At5g05900), which is homologous to UGT76C4 and UGT76C5, without a known function thus far, was also included in the NA glycosyltransferase assays using [14C]-NA and UDP-arabinose or UDP-xylose as co-substrate. The assays showed that UGT76C5, rather than UGT76C3 and UGT76C4, catalyzed the NA pentosylation with a high efficiency. Consistent with previous results, both UGT76C4 and UGT76C5 could utilize UDP-glucose and NA as substrate (Figure 4A). We did not test rhamnosyltransferase activity since UDP-rhamnose is not commercially available. To the best of our knowledge, UGT76C5 is the first multiple functional N-glycosyltransferase identified from plant species. Other UDP-sugars, e.g., UDP-Gal (UDP-galactose), UDP-GlcA (UDP-glucuronic acid), and UDP-GlcNA (UDP-N-acetylglucosamine), are not included in this study, as both UGT76C4 and UGT76C5 showed little activity toward these sugar donors (Li et al., 2015).
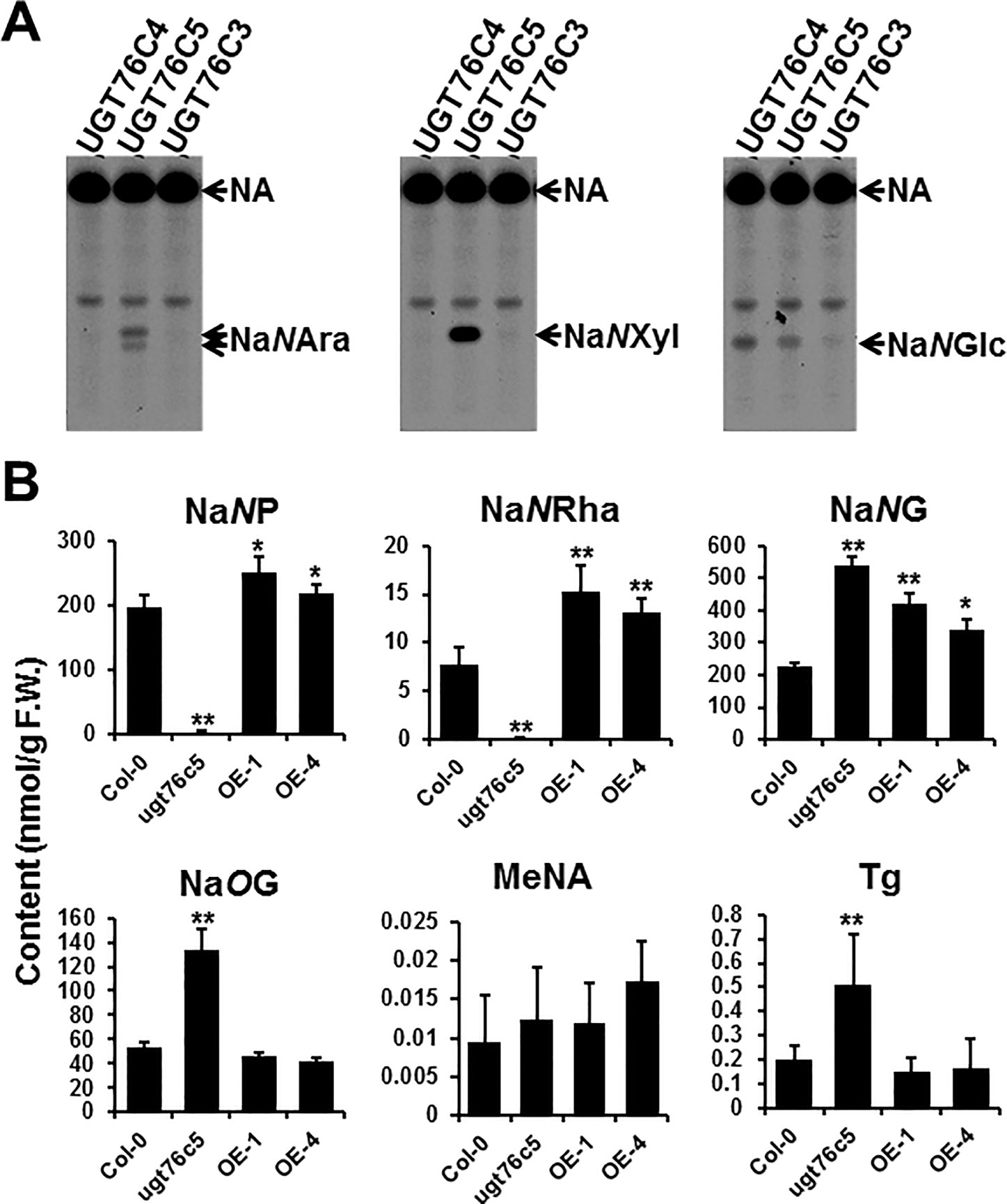
Figure 4 Characterization of UGT76C5 in Arabidopsis thaliana. (A) Glycosyltransferase assays using purified recombinant UGT76C proteins incubated with [14C]-NA and UDP-arabinose, UDP-xylose, or UDP-glucose as substrates. (B) Chemical analysis of the 10-day-old whole seedlings of UGT76C5 transgenic plants. It is noteworthy that MeNA was measured using different method (detailed see Wu et al., 2018). Bars show means ± SDs (n = 3). Asterisks indicate significant differences from the Col-0 plants; *P < 0.05, **P < 0.01 (two-tailed Student’s t test). F.W., fresh weight.
Using the above-mentioned LC-QQQ-MS method, we further analyzed the knock-out mutants and overexpression lines of UGT76C5 (all overexpression lines are in Col-0 genetic background), which had been characterized in our previous study (Li et al., 2015). The results showed that the content of NaNP and NaNRha are significantly decreased in 76c5 mutant (Figure 4B). Detection of low abundance of NaNP and NaNRha also suggested that other glycosyltransferases might also catalyze the NA N-pentosylation or NA N-rhamnosylation. It is noteworthy that NaNP and NaNRha did not increase too much in UGT76C5 overexpressor plants when compared with wild type. In contrast, overexpression of UGT76C4 leads to around 200-fold higher NaNG content than wild-type plants (Li et al., 2015). One possible explanation is that the limited availability of UDP-xylose and UDP-arabinose, rather than UDP-glucose, in Arabidopsis prevents the accumulation of NaNP and NaNRha. Moreover, NaNG content was increased by two-fold in the ugt76c5 mutant compared to that of WT. In a biochemist’s view, NaNP or/and NaNRha may repress UGT76C4’s (NaNGT) activity in Arabidopsis and remove of NaNP and NaNRha in ugt76c5 mutant increase the UGT76C4’s activity to some extent.
We also tested whether NaNP or/and NaNRha could be secreted from plants, mainly from root tissue. The results clearly showed that all nicotinate conjugates, except MeNA, can be detected in the liquid medium (Supplemental Figure 3). Consistent with the chemical profiling in planta (Figure 4B), little NaNP and NaNRha were found in the liquid medium cultured with ugt76c5 mutant (Supplemental Figure 3).
One Amino Acid Determines the Sugar Donor Specificities of Arabidopsis Nicotinate N-Glycosyltransferases
Both UGT76C4 and UGT76C5 shared 79.6% identity on protein level and recognized NA as sugar acceptor. The difference in sugar donor specificity between UGT76C4 (UDP-glucose) and UGT76C5 (UDP-glucose, UDP-xylose, and UDP-arabinose, and probably also UDP-rhamnose) prompted us to further study the mechanism underlying this phenomenon. PSPG motif (total 44 amino acids in almost all identified family 1 GTs) is highly conserved in all glycosyltransferases involved in a plant specialized metabolism, as this motif is thought to correspond to a UDP-binding site (Caputi et al., 2012). Variations in PSPG motif had been demonstrated to play an important role in the UDP-sugar donor specificity of plant family 1 GTs (Noguchi et al., 2009; Ono et al., 2010; Zong et al., 2019). There are three amino acids difference in the PSPG motif of UGT76C4 and UGT76C5: position 6 (D in UGT76C5 vs. E in UGT76C4), 23 (S in UGT76C5 vs. N in UGT76C4), and 32 (A in UGT76C5 and G in UGT76C4) in a total 44 amino acids in the common (Figure 5A). In order to further study the sugar donor specificity of UGT76C4 and UGT76C5, the different residues in PSPG motif were interconverted between these two nicotinate N-glycosyltransferases (Figure 5A). The UGT76C4 mutant N23S (the amino acids are re-numbered in PSPG motif) completely lost the NA N-glucosyltransferases activity (Figure 5B). Substitution of E6 (E6D) and G32 (G32A) showed a slight change in the NaNGT activity: E6D mutant had a slight increase (25%; n = 3), while G32A had a slight decrease (6%; n = 3) compared to wild-type UGT76C4 activity. It is noteworthy that S23N mutation in UGT76C5 led to around five-fold increase NaNGT activity, and D6E and A32G mutants had little effect on the NaNGT activity of UGT76C5 (Figures 4B, C and Table 1). These results reveal that N23 in PSPG motif plays a critical role for NaNGT activity. Comprehensive sequence analysis of Family 1 GTs showed that N23 is highly conserved in all tested GT proteins with the exception of group-A GTs, in which N23 was replaced with G (glycine) or S (serine). Altogether, N23 is important for the maximal catalytic efficiency of glucosyltransfer activity, whereas G or S at the same position is probably required for recognition of other sugar donors (Caputi et al., 2012), like the NaNPT in this study. Meanwhile, all mutants of UGT76C5 did not significantly change in NA N-pentosyltransferase activity, using either UDP-arabinose or UDP-xylose as sugar donor (Figure 5C).
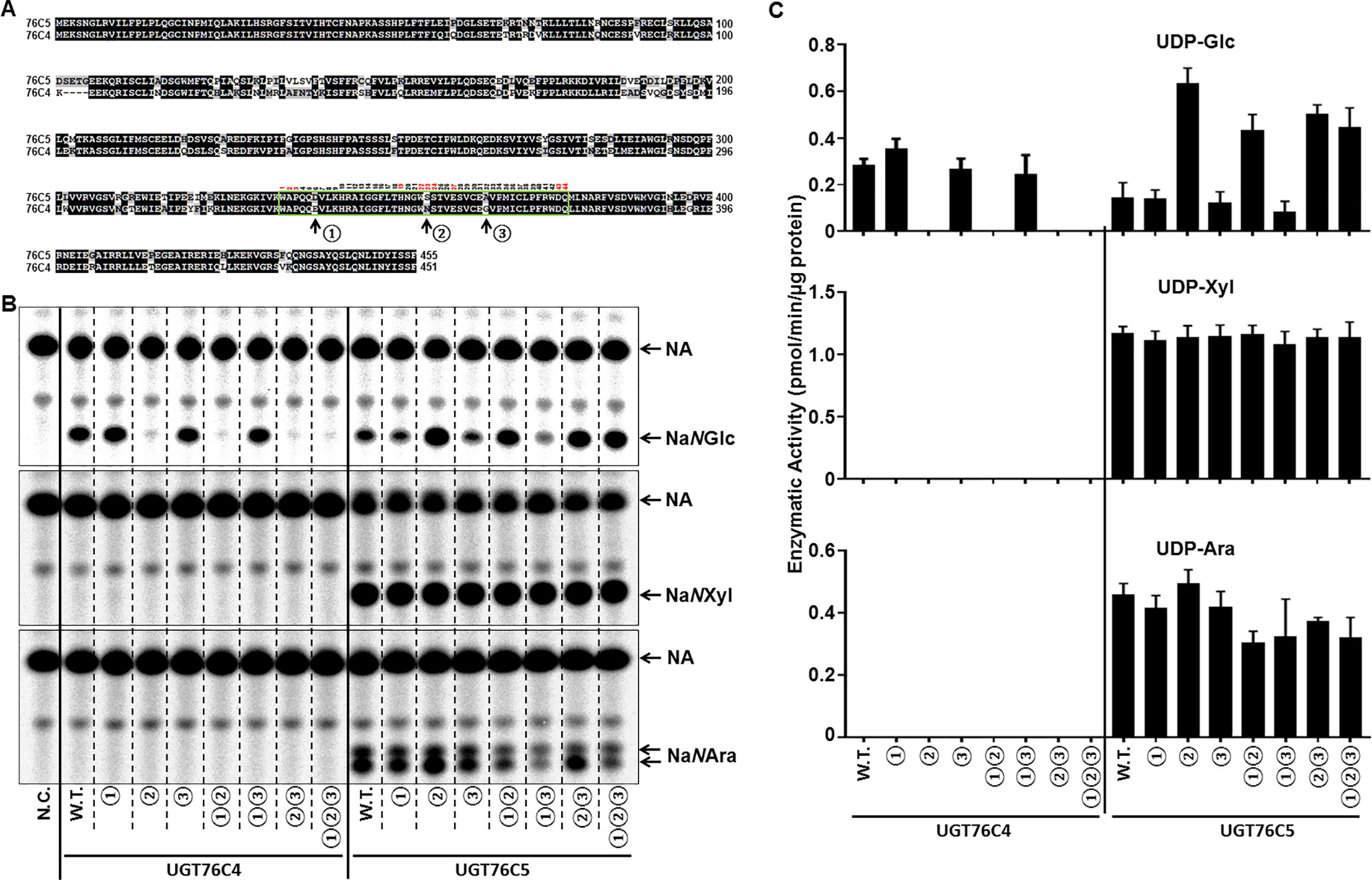
Figure 5 Glycosyltransferase assay of UGT76C4, UGT76C5, and their mutants. (A) Protein sequence alignment of UGT76C4 and UGT76C5. The PSPG motif was boxed in green, and the different amino acids in PSPG motif between UGT76C4 and UGT76C5 were indicated with black arrows and numbers ① (D340 in UGT76C5 or E336 in UGT76C4), ② (S357 in UGT76C5 or N353 in UGT76C4), and ③ (A366 in UGT76C5 or G362 in UGT76C4). Among 44 amino acids in PSPG motif, the amino acids marked with red number are putatively bound to UDP-sugars. (B) Biochemical assay of UGT76C4, UGT76C5, and their mutants with different sugar donors and [14C]-NA. N.C., negative control; only purified MBP tag protein was added in this reaction. W.T., wild-type. ①, amino acid interconversion between D and E; ②, amino acid interconversion between S and N; and ③, amino acid interconversion between A and G. (C) Comparisons of glycosyltransferase activity of UGT76C4 and UGT76C5 wild-type and mutant enzymes with various UDP-sugars. W.T., wild type. Bars show means ± SDs (n = 3).
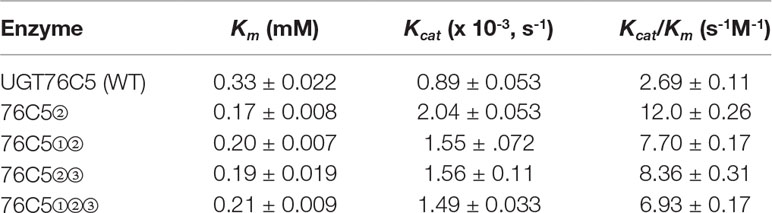
Table 1 Catalytic efficiency of wild-type UGT76C5 and mutants. Kinetic parameters for UDP-glucose were determined with 1.0 mM NA.
Moreover, other residues located outside the PSPG motif also played important role the recognition of UDP-sugars (Osmani et al., 2008; Ono et al., 2010; Louveau et al., 2018). Further experiments, e.g., crystal structural comparison of UGT76C4 and UGT76C5, are needed to elucidate the mechanism difference between UGT76C4 and UGT76C5.
In summary, we first performed the comprehensive NAD metabolomics across 24 tissues of Arabidopsis thaliana, which provided fundamental knowledge for studying the physiological functions of NA conjugates and the NAD salvage pathway. UGT76C5 is the first multiple functional N-glycosyltransferase with sugar-donor promiscuity characterized from plant species. One amino acid in PSPG motif is identified for sugar-donor specificity of UGT76C4 and UGT76C5. These results increase our understanding of the relationship between NAD biosynthesis and NA conjugations and should also allow for additional experiments to provide insights into NAD homeostasis when plants response and adapt to (a)biotic stresses. Two recent studies demonstrated that the plant specialized metabolites (both focus on terpenoids) played a key role to modulate the root microbiota (Chen et al., 2019; Huang et al., 2019). The data and materials generated in this study pave the way for further understanding of the physiological functions of NA conjugates in plants, e.g., whether these root-specific NA glycosides involved into root microbiota modulation in Arabidopsis thaliana.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
LL and GL performed the molecular and biochemical experiments and analyzed the data. FZ carried out LC-QQQ-MS analysis and analyzed the data. GW conceived the project, supervised the experiments, and completed the writing.
Funding
This work was financially supported by the National Key Research and Development Projects (2018YFA0900600), the “Priority Research Program” of the Chinese Academy of Science (ZDRW-ZS-2019-2-0202), and the State Key Laboratory of Plant Genomics of China (SKLPG2016A-13 and SKLPG2016B-13) to G.W.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01164/full#supplementary-material
References
Caputi, L., Malnoy, M., Goremykin, V., Nikiforova, S., Martens, S. (2012). A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 69, 1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x
Chen, Q., Jiang, T., Liu, Y. X., Liu, H., Zhao, T., Liu, Z., et al. (2019). Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci. 62, 947–958. doi: 10.1007/s11427-019-9521-2
Du, L. L., Xue, Y., Xie, J. (2018). Precursor ion-scan mode-based screening strategy for new amorfrutin derivatives from Amorpha fruticosa by UPLC-QQQ-MS and UPLC-Q-TOF-MS. Chromatographia 81, 769–776. doi: 10.1007/s10337-018-3501-9
Hashida, S. N., Takahashi, H., Kawai-Yamada, M., Uchimiya, H. (2007). Arabidopsis thaliana nicotinate/nicotinamide mononucleotide adenyltransferase (AtNMNAT) is required for pollen tube growth. Plant J. 49, 694–703. doi: 10.1111/j.1365-313X.2006.02989.x
Hashida, S. N., Takahashi, H., Takahara, K., Kawai-Yamada, M., Kitazaki, K., Shoji, K., et al. (2013). NAD(+) accumulation during pollen maturation in Arabidopsis regulating onset of germination. Mol. Plant 6, 216–225. doi: 10.1093/mp/sss071
Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., Pease, L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. doi: 10.1016/0378-1119(89)90358-2
Huang, A. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364, eaau6389. doi: 10.1126/science.aau6389
Jeanguenin, L., Lara-Nunez, A., Rodionov, D. A., Osterman, A. L., Komarova, N. Y., Rentsch, D., et al. (2012). Comparative genomics and functional analysis of the NiaP family uncover nicotinate transporters from bacteria, plants, and mammals. Funct. Integr. Genomic 12, 25–34. doi: 10.1007/s10142-011-0255-y
Katoh, A., Uenohara, K., Akita, M., Hashimoto, T. (2006). Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 141, 851–857. doi: 10.1104/pp.106.081091
Li, W., Zhang, F. X., Chang, Y. W., Zhao, T., Schranz, M. E., Wang, G. D. (2015). Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27, 1907–1924. doi: 10.1105/tpc.15.00223
Li, W., Zhang, F., Wu, R., Jia, L., Li, G., Guo, Y., et al. (2017). A novel N-methyltransferase in Arabidopsis appears to feed a conserved pathway for nicotinate detoxification among land plants and is associated with lignin biosynthesis. Plant Physiol. 174, 1492–1504. doi: 10.1104/pp.17.00259
Louveau, T., Orme, A., Pfalzgraf, H., Stephenson, M. J., Melton, R., Saalbach, G., et al. (2018). Analysis of two new arabinosyltransferases belonging to the carbohydrate-active enzyme (CAZY) glycosyl transferase family1 provides insights into disease resistance and sugar donor specificity. Plant Cell 30, 3038–3057. doi: 10.1105/tpc.18.00641
Noctor, G., Queval, G., Gakiere, B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57, 1603–1620. doi: 10.1093/jxb/erj202
Noguchi, A., Horikawa, M., Fukui, Y., Fukuchi-Mizutani, M., Iuchi-Okada, A., Ishiguro, M., et al. (2009). Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21, 1556–1572. doi: 10.1105/tpc.108.063826
Ono, E., Homma, Y., Horikawa, M., Kunikane-Doi, S., Imai, H., Takahashi, S., et al. (2010). Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera). Plant Cell 22, 2856–2871. doi: 10.1105/tpc.110.074625
Osmani, S. A., Bak, S., Imberty, A., Olsen, C. E., Moller, B. L. (2008). Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 148, 1295–1308. doi: 10.1104/pp.108.128256
Preiss, J., Handler, P. (1958a). Biosynthesis of diphosphopyridine nucleotide I. Identification of intermediates. J. Biol. Chem. 233, 488–492.
Preiss, J., Handler, P. (1958b). Biosynthesis of diphosphopyridine nucleotide II. Enzymatic aspects. J. Biol. Chem. 233, 493–500.
Reiter, W. D. (2008). Biochemical genetics of nucleotide sugar interconversion reactions. Curr. Opin. Plant Biol. 11, 236–243. doi: 10.1016/j.pbi.2008.03.009
Wang, G., Pichersky, E. (2007). Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 49, 1020–1029. doi: 10.1111/j.1365-313X.2006.03013.x
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. doi: 10.1371/journal.pone.0000718
Wu, R., Zhang, F., Liu, L. Y., Li, W., Pichersky, E., Wang, G. (2018). MeNA, controlled by reversible methylation of nicotinate, is an NAD precursor that undergoes long-distance transport in Arabidopsis. Mol. Plant 11, 1264–1277. doi: 10.1016/j.molp.2018.07.003
Zheng, X. Q., Hayashibe, E., Ashihara, H. (2005). Changes in trigonelline (N-methylnicotinic acid) content and nicotinic acid metabolism during germination of mungbean (Phaseolus aureus) seeds. J. Exp. Bot. 56, 1615–1623. doi: 10.1093/jxb/eri156
Zong, G., Fei, S., Liu, X., Li, J., Gao, Y., Yang, X., et al. (2019). Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-β-L-rhamnose and rhamnosylation mechanism. Plant J. 99, 257–269. doi: 10.1111/tpj.14321
Keywords: NAD, nicotinate, N-glycosyltransferase, Preiss-Handler pathway, Arabidopsis
Citation: Liu L, Zhang F, Li G and Wang G (2019) Qualitative and Quantitative NAD+ Metabolomics Lead to Discovery of Multiple Functional Nicotinate N-Glycosyltransferase in Arabidopsis. Front. Plant Sci. 10:1164. doi: 10.3389/fpls.2019.01164
Received: 28 May 2019; Accepted: 26 August 2019;
Published: 27 September 2019.
Edited by:
Shin-nosuke Hashida, Central Research Institute of Electric Power Industry (CRIEPI), JapanReviewed by:
Yuuma Ishikawa, Saitama University, JapanJeffrey Charles Waller, Mount Allison University, Canada
Copyright © 2019 Liu, Zhang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, gdwang@genetics.ac.cn
 Lingyun Liu
Lingyun Liu Fengxia Zhang1
Fengxia Zhang1 Guodong Wang
Guodong Wang