
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 18 September 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01136
Woody perennials enter seasonal dormancy to avoid unfavorable environmental conditions. Plant hormones are the critical mediators regulating this complex process, which is subject to the influence of many internal and external factors. Over the last two decades, our knowledge of hormone-mediated dormancy has increased considerably, primarily due to advancements in molecular biology, omics, and bioinformatics. These advancements have enabled the elucidation of several aspects of hormonal regulation associated with bud dormancy in various deciduous tree species. Plant hormones interact with each other extensively in a context-dependent manner. The dormancy-associated MADS (DAM) transcription factors appear to enable hormones and other internal signals associated with the transition between different phases of bud dormancy. These proteins likely hold a great potential in deciphering the underlying mechanisms of dormancy initiation, maintenance, and release. In this review, a recent understanding of the roles of plant hormones, their cross talks, and their potential interactions with DAM proteins during dormancy is discussed.
Bud dormancy is an essential adaptation, which allows temperate woody perennials to survive adverse environmental conditions during winter. During dormancy, plants experience arrested growth and reduced metabolic activities (Arora et al., 2003). According to the source of the signals that induce dormancy, dormancy can be categorized into three classes: paradormancy (PD), endodormancy (ED), and ecodormancy (ECD) (Lang, 1987). In PD, lateral bud growth is suppressed by the terminal bud, a phenomenon known as apical dominance. In ED, short days (SDs) and/or low temperatures trigger internal bud signals, which enable buds to become tolerant to temperatures well below freezing. Endodormant buds track chilling units and will not resume growth until the fulfillment of the chilling requirement. ECD marks the last stage of dormancy where buds resume the ability to grow but are inhibited by unfavorable weather conditions. While this classification allows convenient references to the different stages of the dormancy–growth cycle of deciduous perennials, the nomenclature of PD and ECD has raised some concerns and confusions as they lack many genetic and biological hallmarks that are characteristic of a true dormancy. At the core of a true dormancy is the “inability to resume growth from meristems under favorable conditions” (Rohde and Bhalerao, 2007), which apparently does not apply to PD or ECD. In view of this, several authors have advocated the prudent use of dormancy, especially in the discussion of dormancy mechanisms at the molecular or cellular levels (Rohde and Bhalerao, 2007; Cooke et al., 2012; Considine and Considine, 2016). In this review, we refer to ED as dormancy unless specified otherwise and PD or ECD per se. Dormancy is a highly regulated and complex process and is subjected to the influences of many internal and external factors. Plant hormones have been shown to be the most significant internal mediators in the control of dormancy cycle in deciduous trees.
Plant hormones, or phytohormones, are naturally occurring small signaling molecules that affect plant physiological metabolism at low concentrations (Davies, 2010). Plant hormones regulate developmental and growth processes throughout the plant’s life cycle and also trigger adaptive responses induced by external stimuli such as environmental changes and biotic or abiotic stresses. Conventional plant hormones include five major groups: abscisic acid (ABA), gibberellin (GA), ethylene (ET), auxin (indole-3-acetic acid, IAA), and cytokinin (CK). Other major plant-produced substances known to exert hormone-like functions in plants include jasmonates (JAs), brassinosteroids (BRs), strigolactones (SL), and salicylic acid (SA). Many of these plant hormones have been found to participate in the highly complex orchestration of bud dormancy. Thus far, our knowledge in deciphering the mechanisms by which dormancy is regulated by plant hormones still remains limited. However, several breakthroughs associated with mechanistic aspects of hormone signaling at the molecular and subcellular levels have elucidated hormone perception, signal transduction, and signal interplay in several major hormones (Chow and Mccourt, 2006; Santner and Estelle, 2009; Santner et al., 2009). Genetic mutagenesis in model plant species have also allowed us to examine the function of a different element of hormone biosynthesis and signaling pathways under various conditions, including dormancy. This, along with the recent advances in omics and bioinformatics that provide global views of all relevant genetic events at a specific time, has considerably advanced our knowledge of hormone regulation as it applies to bud dormancy. Such quick gains in research, knowledge, and technologies necessitate a summarization of recent findings and suggestions for future research. A comprehensive overview of the underlying molecular and biochemical mechanisms involved in bud dormancy will also help researchers design practical approaches to address critical issues in agriculture, horticulture, and forestry, such as global warming and spring frost. In this review, we focus on topics related to recent findings associated with bud dormancy, with an emphasis on the functions and interactions of plant hormones during bud dormancy, particularly in deciduous fruit trees.
Plant hormone ABA regulates a great number of aspects in plant growth and development and is also an important messenger of stress responses (Finkelstein, 2013). The primary role of ABA in plants is to repress growth and to promote organ senescence and abscission (Finkelstein, 2013; Zheng et al., 2015). In light of this, ABA is of particular importance in regulating dormancy, since dormancy in essence is the suspension of meristematic growth and successful dormancy establishment entails cessation of the overall plant growth (Cooke et al., 2012). The central role of ABA in regulating bud dormancy has been extensively documented in many physiological, genetic, and molecular studies.
It has been widely observed that endogenous ABA levels increase at dormancy establishment and decrease towards dormancy release (transition from ED to ECD). For example, ABA content in grapevine (Vitis vinifera) buds increases up to threefold at the onset of dormancy and then decreases gradually towards the release of dormancy, indicated by the increasing bud break rate from node cuttings in forcing conditions (Zheng et al., 2015). Similar results were also observed in some other woody species including peach (Prunus persica) (Wang et al., 2015), pear (Pyrus pyrifolia) (Tuan et al., 2017; Li et al., 2018), and sweet cherry (Prunus avium) (Chmielewski et al., 2018). It has been proposed that the increase of ABA content is triggered by SD photoperiod prior to the establishment of dormancy (Ruttink et al., 2007) and its decline is concomitant with chilling accumulation (Li et al., 2018). Such parallel relationship between ABA levels and the depth of dormancy suggests ABA’s role in the initiation and progression of the dormancy cycle and that ABA may also be involved in mediating environmental signals.
The importance of ABA in dormancy regulation is also evidenced by the precocious release of dormancy when ABA content in dormant buds is artificially reduced. Early study in rose (Rosa hybrida) indicated that application of ABA synthesis inhibitor fluridone to dormant buds initiated the growth of new leaf primordia (Le Bris et al., 1999), which led the authors to suggest that continued in situ ABA synthesis is required for the maintenance of bud dormancy. Furthermore, ABA catabolism in grapevine was effectively activated, and its content was reduced after treatment of buds with hydrogen cyanamide (HC), an efficient bud-breaking chemical (Ophir et al., 2009; Zheng et al., 2015). These findings are also supported by a transgenic study in pear (P. pyrifolia), in which dormancy release was accelerated when an ABA catabolism enzyme 8′-hydroxylase (ABA8ʹOH) was overexpressed (Li et al., 2018). These studies support the notion that ABA is an effective suppressor of primordia growth during dormancy and even though ABA is on the decline after dormancy has been established, basal levels of ABA and de novo ABA production may still be required to keep the buds at dormancy, and the continuous reduction of ABAs contributes to the release of dormancy.
Studies of exogenous ABA application have provided more insight into how ABA affects dormancy. In several woody species, exogenous ABA application was found to promote dormancy initiation and to delay bud break (Dutcher and Powell, 1972; Mielke and Dennis, 1978; Lionakis and Schwabe, 1984; Li et al., 2018). However, closer examination shows that the inhibitory effect of exogenous ABA on bud cuttings diminishes as dormancy intensifies and disappears after the dormancy is released. ABA inhibitory effect on bud break is also affected by chilling accumulation, as injection of ABA only inhibited bud break of Japanese pear (Pyrus fauriei) shoots that were exposed to 200–600 chilling hours, but not those exposed to 800–1,200 chilling hours (Tamura et al., 2002). Together, this proposes that a non-ABA-mediated regulatory mechanism, which controls the dormancy transition at certain phases or when given requirements have been met, may exist. This can be supported by the observation that the variation of ABA levels is not always in exact synchronization with dormancy progression. For instance, while the decrease in ABA levels was found to commence somewhat prior to the release of dormancy (Or et al., 2000; Zhang et al., 2018), Chmielewski et al. (2018) reported that ABA levels in sweet cherry did not decline until buds have entered ECD. Thus, it appears that even though ABA is a key regulator of dormancy, some separate regulatory networks, which respond more closely to environmental stimuli, may still be at play and override ABA’s regulatory action at certain stages.
In contrast to applications of exogenous ABA to cuttings, effects of ABA applied on whole plants produced inconsistent results, with application timing appearing to be critical. Foliar application of ABA to dormant peach trees prior to budburst slightly accelerated bloom progression (Parker et al., 2012). Similarly, spring application of ABA on field-grown grapevine produced little or no inhibitory effect on budburst (Hellman et al., 2006). On the other hand, fall application of ABA on nursery apple trees (Malus domestica) promoted the occurrence of the physiological events preceding dormancy commencement such as N mobilization from leaves to stems, cold acclimation in stems, and shoot growth cessation (Guak and Fuchigami, 2015). ABA application on grapevines was more effective in inducing deeper dormancy during early autumn between the veraison and post veraison stages compared to mid-autumn applications (Li and Dami, 2016). Furthermore, leaf age was also found to influence the effectiveness of exogenous ABA, as grapevines with older leaves were more responsive to exogenous ABA in inducing dormancy compared to those with younger leaves (Zhang et al., 2011). Plant responses to exogenous ABA depend on the successful penetration of ABA into tissue (Addicott and Lyon, 1969). This may explain why cuttings are more consistently responsive to ABA applications compared to whole-plant applications, as cuttings are usually incubated with ABA solution for an extended period of time to facilitate cuticular penetration of ABA. In contrast, uptake of ABA by whole-plant application is more dependent on successful cuticular penetration, frequency of applications, and several environmental variables, which may all contribute to the inconsistency of ABA effect on whole-plant application.
In higher plants, ABA metabolism is finely coordinated to ensure proper growth and effective stress responses. The ABA biosynthesis pathway has been reviewed extensively (Marin et al., 1996; Milborrow, 2001; Finkelstein, 2013; Liao et al., 2018). The first step of ABA production is the epoxidation of zeaxanthin to antheraxanthin by zeaxanthin epoxidase (ZEP). Next, antheraxanthin is converted to neoxanthin or violaxanthin, which are both cleaved to form xanthoxin by 9-cis-epoxycarotenoid dioxygenase (NCED) (Liotenberg et al., 1999; Taylor et al., 2000). In the following two steps, xanthoxin is first dehydrated by alcohol dehydrogenase and then oxidized to ABA by aldehyde oxidase (Bittner et al., 2001; Cheng et al., 2002). Though ZEP has been shown to affect ABA biosynthesis in some species (Nambara and Marion-Poll, 2005), severe zep mutation could not completely eliminate the production of ABA, suggesting the existence of a ZEP-independent minor pathway for ABA biosynthesis (Barrero et al., 2005). In contrast, the NCED-mediated violaxanthin cleavage is a rate-limiting and committed step, constituting a regulation pivot in controlling ABA biosynthesis, and has received substantial attention in the ABA-related studies (Nambara and Marion-Poll, 2005).
Accumulating evidence has indicated that ABA biosynthesis is involved in controlling dormancy. Upregulation of ABA synthetic enzyme NCED at the onset of dormancy and its downregulation during dormancy release were observed in many species such as peach (Wang et al., 2015), pear (P. pyrifolia) (Li et al., 2018), and grapevine (Zheng et al., 2015), to name a few. However, it was noted that various NCED homologs follow distinct expression patterns in these plant species during dormancy. For example, NCED1 in peach is expressed more in vegetative buds than in floral buds, while NCED2 has higher expression in floral buds (Wang et al., 2015). In pear, NCED2 and NCED3 are highly expressed at the initiation of dormancy, but NCED1 peaks only towards the dormancy release (Li et al., 2018). Among the three homologs of NCED genes in grapevine, only NCED1 was detectable during dormancy (Zheng et al., 2015). These findings indicate the existence of a complex regulatory network of ABA biosynthesis, in which NCED genes are probably regulated by relatively independent mechanisms and are expressed in an organ-specific manner.
In catabolism, ABA is primarily degraded through the 8′ position hydroxylation by enzyme ABA8′OH, a cytochrome P450 monooxygenase, encoded by the CYP707A gene family (Kushiro et al., 2004; Umezawa et al., 2006). ABA8′OH is a key regulator of ABA catabolism, and mutation of CYP707A can lead to accumulation of high levels of ABA (Finkelstein, 2013). ABA hydroxylation produces two major catabolites: phaseic acid (PA) and dihydrophaseic acid (DPA), both of which are commonly used to indicate the level of ABA catabolism (Destefano-Beltran et al., 2006). Recent studies indicate that ABA8′OH is involved in the control of dormancy release, and its synthesis is regulated at the transcription level. In grapevine, transcript levels of VvCYP707A4, an ABA8′OH gene, drastically increased about 20 days before the onset of dormancy release and remained steady throughout the dormancy release period (Zheng et al., 2015). The upregulation of VvCYP707A4 was concomitant with the accumulation of ABA catabolites, such as neoPA, PA, and DPA. Similarly, PpCYP707A2 and PpCYP707A3 in peach and CYP707A in Japanese pear were all found to have heightened expression during the release of dormancy (Wang et al., 2015; Tuan et al., 2017; Li et al., 2018). Li et al. (2018) noted that an increase of PpCYP707A3 in flower buds of white pear (P. pyrifolia) during dormancy was related to chilling accumulation, further confirming the link between ABA and the fulfillment of chilling requirements.
Homeostasis of ABA in plants is essential for normal growth and development, in which buds are both the target site for ABA to act upon and the principal location of ABA metabolism and catabolism. Transcription studies indicate that the alteration of one process is often accompanied by an opposite change in the other process, suggesting that these two processes are closely co-regulated. For example, VvNCED expression in grape peaks during early dormancy while expression of VvCYC707A4 remains very low. In contrast, during late dormancy, VvNCED expression starts to decline as VvCYC707A4 rapidly increases (Zheng et al., 2015). Furthermore, Li et al. (2018) found that PpCYP707A1-5 expression was drastically upregulated in ABA-treated pear (P. pyrifolia) trees while PpNCED expression remained unaltered. This observation suggests that ABA may stimulate its own degradation via a negative feedback pathway. In contrast, positive feedback of ABA regulation has been observed in Arabidopsis where ABA synthesis genes, including NCED, were induced by exogenous ABA (Cheng et al., 2002; Xiong and Zhu, 2003). This mechanism seems to function only under certain genetic backgrounds (such as aba mutants) and stress conditions (such as drought) (Xiong et al., 2002). As a plant growth regulator, ABA can be used in agricultural practices to manipulate dormancy release and bloom date, thus saving early-bloom varieties and species from potential spring freezes. However, the systems by which endogenous ABA and its signal pathway are affected by exogenous ABA during dormancy need further elucidation.
ABA deactivation can also be achieved through conjugation. In this process, an ABA molecule typically conjugates with glucosyl ester to form ABA glucosyl ester (ABA-GE) by the enzyme glucosyltransferase (Piotrowska and Bajguz, 2011). ABA-GE lacks a direct biological function and is generally believed to serve as the storage form of ABA, which can be relocated and disrupted to release ABA in response to stresses, such as dehydration (Sauter et al., 2002; Lee et al., 2006; Xu et al., 2012). A recent study has indicated that the glucosyl ester may also act as an ABA antagonist by regulating ABA supply during bud dormancy. In sweet cherry buds, ABA-GE levels were lower than ABA levels during dormancy but increased as buds transitioned from dormancy to ECD (Chmielewski et al., 2018). Similarly, a homolog of glucosyltransferase was found to be highly expressed in Prunus mume during later stages of dormancy (Zhang et al., 2018). While these reports confirm the role of ABA glucosylation in ABA deactivation during dormancy release, it still remains unclear to what extent this mechanism supplements the oxidative catabolism of ABA and how it is spatially and temporally regulated.
In addition to ABA metabolism, ABA responses are also regulated at the signal transduction level. The ABA signaling pathway consists of three key components: ABA receptors, protein phosphate 2Cs (PP2Cs), and SNF1-related protein kinase 2s (SnRK2s). The three types of high-affinity ABA-binding proteins that have been recently identified are regulatory pyrabactin resistance (PYR), pyrabactin like (PYL), and components of ABA receptors 1 (RCAR) (Raghavendra et al., 2010). ABA forms a complex with PP2Cs after binding to the RCAR/PYR/PYL receptor and thus deactivates PP2Cs, which suppress the activity of SnRK2s through dephosphorylation. Phosphorylated SnRK2s activate the ABA-responsive element (ABRE)-binding protein/ABRE-binding factors (AREB/ABF) transcription factors, which subsequently trigger the downstream ABA responses by binding to the ABREs (Soon et al., 2012).
Genetic studies have indicated that transcript levels of these central components of ABA signaling pathway are modulated by environmental signals and levels of ABA as well. In grapevine, some RCAR and PP2C genes are highly induced by cold, drought, salt, and exogenous ABA, and their induction differs greatly across organs (Boneh et al., 2012). In a transcriptomic study with pear, Li et al. (2018) showed that the expression of ABA receptor PYL and the positive regulator SnRK2s was upregulated at the entrance of dormancy; and in contrast, the expression of PP2Cs, the suppressor of ABA signaling, remained low during dormancy and increased as ABA content decreased towards the end of the dormancy. Furthermore, exogenous ABA promoted the expression of SnRK2s but suppressed that of PP2Cs. A similar result was also obtained in Japanese pear, in which PP2C genes were upregulated and SnRK2s downregulated after buds exited from dormancy (Bai et al., 2013). Ruttink et al. (2007) indicated that changes of ABA signaling component expression were induced in response to the SD photoperiod. Taken together, these results suggest that the ABA signaling pathway is subject to the influence of both seasonal variation and ABA contents during dormancy.
Early studies in Arabidopsis have demonstrated that transcriptional factors from the ABA-insensitive (ABI) group can individually or collaboratively mediate the expression of ABA-inducible genes (Nakamura et al., 2001). Among them, ABI3 is believed to play an important role in seed embryo maturation and dormancy by positively regulating the ABA signaling pathway (Nambara et al., 1995; Finkelstein et al., 2008). In poplar (Populus trichocarpa), ABI3 was found to be expressed in the embryonic leaves inside the bud during bud set, and the overexpression or silencing of ABI3 caused alterations in bud development, indicating its crucial role in bud formation (Rohde et al., 2002). Broader functions of ABI3 were also revealed in vegetative tissue, in which ABI3 regulates plastid differentiation (Rohde et al., 2000) and induces ABA responses through targeting the ABREs (Nakashima et al., 2006).
Several recent studies have indicated that ABA may regulate dormancy through modulating the intercellular communication. In plants, the cell-to-cell transport in the symplastic continuum relies on the connectivity of specialized channels between adjacent cells called plasmodesmata. The passage of mobile molecules across the plasmodesmata is primarily controlled by the deposition and degradation of callose, a β-1,3-glucan polymer, which are catalyzed by callose synthase (CALS1) and glucanase, respectively (Wu et al., 2018b). Symplastic closure caused by callose deposition is a major mechanism in plants to defend pathogen invasion. In fact, it is also a critical step in the establishment of dormancy and is triggered by SD photoperiod events (Rinne and Schoot, 2003). This indicates that plasmodesmata constriction is a common mechanism shared by defense and dormancy and plasmodesmata is “at the crossroads” of these two events (Rinne and Schoot, 2003). In a recent study, ABA-mediated plasmodesmata constriction in hybrid aspen was shown to prevent dormancy release by limiting the passage of growth factors such as flowering locus T (FT) into the dormant buds (Tylewicz et al., 2018). In this study, an ABA-insensitive mutant (abi1-1) failed to produce plasmodesmatal callose and exhibited compromised dormancy in SD treatment, whereas the plasmodesmata closure and dormancy were restored through downregulation of a chromodomain remodeling factor PICKLE (PKL) or ectopic expression of plasmodesmata located protein 1 (PDLP1); PKL is a chromatin remodeler that facilitates epigenetic marks (e.g. histone H3 lysine 27) and represses expression of tissue-specific genes associated with developmental transitions (Zhang et al., 2012a), and PDLPs are important regulators of plasmodesmata permeability and symplastic transport (Lim et al., 2016). It has been shown that ABA can also induce the expression of callose synthase 1 (CALS1) by suppressing the expression of PKL (Singh et al., 2019). Taken together, these results suggest that plasmodesmata blockage is an integral mechanism in the establishment of dormancy and ABA is central to the regulation to this process (Figure 1).
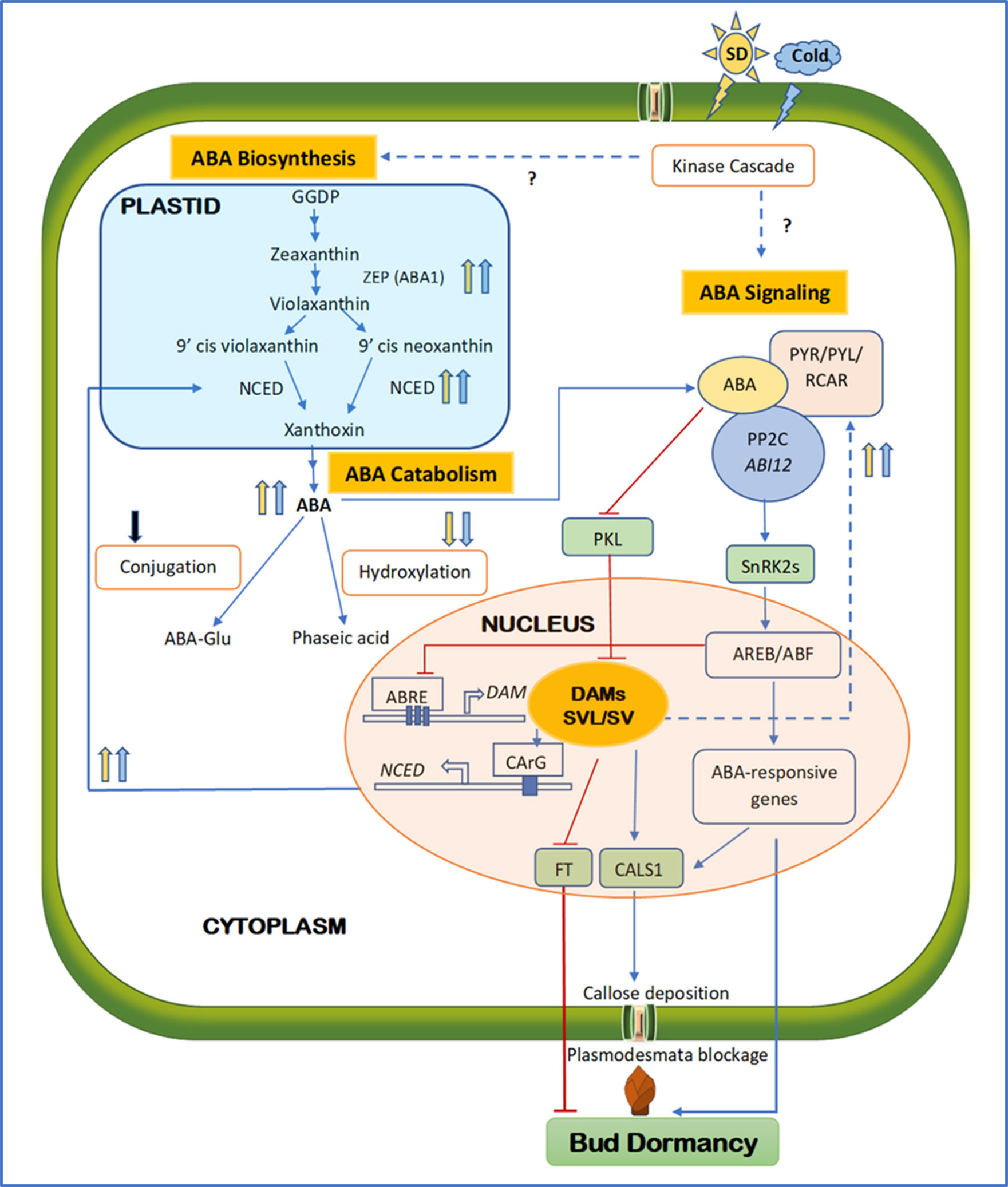
Figure 1 Schematic diagram integrating major components of ABA biosynthesis, signaling, and catabolism during the establishment of bud endodormancy. Solid and dashed lines indicate direct and indirect regulation; respectively. Arrowed blue and barred red lines indicate activation and inhibition, respectively. Upward and downward arrows indicate upregulation and downregulation, respectively. Blue and yellow arrows indicate cold- and SD-mediated activation, respectively, whereas the black arrow indicates an undetermined signal source. Question marks indicate the unconfirmed mechanisms. Towards the end of the growing season, SD photoperiod and short-term low temperature activate both ABA biosynthesis and signaling, possibly via kinase cascades. As a result, the key components in ABA biosynthesis and signaling such as ZEP, NCED, PYR/PYL/RCAR, and PP2C are upregulated, whereas ABA catabolism through hydroxylation or conjugation is downregulated. High ABA levels activate DAM and SVP/SVL transcription factors through repressing PKL. DAM promotes ABA synthesis by binding to the CArG motif of the NCED promoter, whereas SVL enhances ABA signaling by upregulating ABA receptor RCAR/PLY. The positive feedback loop between ABA and DAM/SVL can be balanced by the ABA signaling component ABRE, which represses the activity of DAM by binding to the three ABRE motifs in its promoter region. SVL can induce the expression of CALS1 and FT by binding to their promoters. The ABA signaling pathway can also upregulate CALS1, which in turn produces callose at the plasmodesmata to block the intercellular communication, contributing to the establishment of endodormancy. Activated ABA responses, repressed FT, and callose deposition all lead to the establishment of endodormancy. ABA, abscisic acid; FT, flowering locus T; SD, short day; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; PYR, regulatory pyrabactin resistance; PYL, pyrabactin like; RCAR, components of ABA receptors 1; PP2C, protein phosphate 2C; DAM, dormancy-associated MADS; SVP, short vegetative phase; SVL, SVP-like
Cell cycle is closely related to bud dormancy. In cell cycle, G1 is the interphase at which cells accomplish most of the growth and preparation for DNA synthesis, and it is also the phase where the cells may exit from cell cycle and enter stasis of G0. Previous studies indicate that cell cycle arrests at the G1 stage in the dormant buds (Devitt and Stafstrom, 1995; Gutierrez et al., 2002). Swiatek et al. (2002) found that ABA prevents DNA replication, keeping the cells in the G1 stage, and this effect of ABA on cell cycle seemed to be limited to the transition from the G1 to S phases. Further, SD photoperiod has been shown to keep more cells in the G1 stage than the G2 stage and is associated with an increase in ABA levels (Rohde et al., 1997; Welling et al., 1997). Thus, this ABA-regulated G1/S transition appears to be a primary checkpoint for the control of ED. Previous literature also indicates that cell cycle progression is primarily controlled by cyclins (especially types A, B, and D) and cyclin-dependent kinases (CDKs), whose activity can be repressed by inhibitor of CDK/kip-related protein (ICK) and thus induces arrested or delayed cell cycle (Yamaguchi et al., 2000; Verkest et al., 2005; Lipavska et al., 2011; Torres Acosta et al., 2011) (Figure 1). Recently, Vergara et al. (2017) showed that ABA suppresses the expression of CDK (e.g. VvCDKB1 and VvCDKB2) and cyclins (e.g. VvCYCA1, VvCYCA2, and VvCYCA3) and upregulates the expression of ICKs (e.g. VvICK5) in grapevine (V. vinifera) buds. Furthermore, treatments buds by the bud-breaking chemical HC reduced the content of ABA and upregulated the expression of CCG, and this effect was restored by the application of exogenous ABA. Together, these findings indicate that ABA-modulated cell cycle arrest may be central to the overall development of ED in woody buds.
GAs are a large group of tetracyclic diterpenoid compounds that exert significant effects on a broad spectrum of biological processes in plants. Of all the numerically coded GAs, only a few have been found to have bioactivity. These include GA1, GA3, GA4, and GA7, with GA3 being referred to as the gibberellic acid (Sun, 2008). GAs promote both vegetative and reproductive growth in plants by modulating leaf morphology, stem elongation, sex expression, seed germination, floral development, and dormancy (Takatsuka and Umeda, 2014; Hedden and Sponsel, 2015).
GAs are a leading phytohormone that modulates bud dormancy as significant changes of bioactive GA levels before and after dormancy have been widely noted (Cooke et al., 2012). In general, GA levels are downregulated at the induction of dormancy and upregulated during dormancy release or bud burst, and such dynamics of GA contents have been reported in many woody species such as sweet cherry (P. avium), hybrid aspen (Populus tremula × Populus tremuloides), grapevine, and Japanese apricot (P. mume) (Duan et al., 2004; Rinne et al., 2011; Zhuang et al., 2013; Zhuang et al., 2015; Zheng et al., 2018). At the early stage of dormancy initiation, decline of GA levels induces growth cessation and bud set, and growth cessation can be aborted by application of exogenous GA even with SD treatment (Mølmann et al., 2005). Interestingly, chemical inhibition of GA biosynthesis combined with low night temperature was unable to induce dormancy under long-day (LD) photoperiod in hybrid aspen (Mølmann et al., 2005), suggesting that GAs may function downstream the SD photoperiodic perception-mediated pathways for dormancy initiation. The role of GAs (especially GA3 and GA4) in promoting bud dormancy release is also evident and has been demonstrated in several woody species (Zhuang et al., 2013). In Populus, exogenous GA4 was found to substitute for the effect of chilling by upregulating several chilling-responsive genes (e.g. FT) and induce bud burst (Rinne et al., 2011). In a recent study, however, GAs’ effect in enhancing primordial growth and bud break was found to be restricted after meristem activation, and premature increase of GA rendered inhibitory effects on bud break, indicating the distinct functions of GA at the dormancy–growth transition (Zheng et al., 2018).
Studies examining exogenous GAs’ effects on dormancy release have proposed several mechanisms that GAs activate. First, GAs control dormancy cycle through modulating the intercellular communication. In plants, dormancy cycle progression is highly dependent on the mobility of molecules such as FT, auxin, sugars, and possibly ABA (Cooke et al., 2012), and thus, the permeability of plasmodesmata may play a role in controlling dormancy. A recent study indicated that SD photoperiod can trigger an ABA-mediated callose accumulation and GA catabolism (Singh et al., 2019). When active GA content is extremely low during the deep dormancy period, the rate of substance exchange in buds with adjacent organs declines remarkably (Hao et al., 2017). The connectivity of the plasmodesmata channel can be restored by GA4, which can induce the expression of β-1,3-glucanase to hydrolyze callose (Rinne et al., 2011). In a recent RNAi study, Singh et al. (2019) proposed a model that links GA to the ABA-mediated plasmodesmata closure. In this model, the authors proposed that in the absence of ABA, PKL suppresses the expression of SVL, a transcription factor orthologous to Arabidopsis floral repressor (SVP), which activates the expression of CALS1 and GA2 oxidase (a GA catabolic gene). Upon exposure to SD photoperiod, elevated ABA level suppresses the expression of PKL, which in turn leads to the activation of SVL and subsequently GA catabolism and callose accumulation (Singh et al., 2019). Secondly, GA was found to enhance the production of reactive oxygen species (ROS), which are of particular importance in dormancy breaking (Zhuang et al., 2013). In grapevine (V. vinifera), Sudawan et al. (2016) showed that bud break rates are highly correlated to the rapid accumulation of ROS in the spring. Production of ROS at dormancy release has been documented in several plant systems and considered to play a central role in bud break (Sudawan et al., 2016; Liu et al., 2017; Beauvieux et al., 2018). Finally, GA may activate the metabolic pathways leading to dormancy release. For instance, in Japanese apricot (P. mume), GA4 treatment led to the enhancement of many energy metabolism pathways, including those associated with sugar metabolism (Zhuang et al., 2015). Soluble sugars are considered to be an important energy source to sustain bud growth during dormancy release. Additionally, sucrose is a potential signaling element that can indirectly enhance the expression of the genes that are related to cell division and cell cycle (Ruan et al., 2010).
The pathways of GA biosynthesis and catabolism have been extensively investigated by a combination of biochemical and molecular techniques. In GA biosynthesis, three classes of enzymes have been identified that correspond to the three stages of conversion. In the first stage, GA precursor geranyl geranyl diphosphate (GGDP) is converted to ent-kaurene via copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS) in plastids. Next, ent-kaurene is oxidized to GA12 through stepwise oxidation via two cytochrome P450 monooxygenases: ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO). In the final step, GA12 is converted to bioactive GAs including GA1, GA3, and GA4 by dioxygenases 20-oxidase (GA20ox) and 3-oxidase (GA3ox) (Sun, 2008). The GA biosynthesis occurs primarily within the vicinity of its action (Kaneko et al., 2003; Yamaguchi, 2008), so higher transcript levels of the majority of GA biosynthetic genes can be found in the actively growing tissues including the bursting buds. Unlike the GA biosynthetic pathway that involves various enzymes and multiple stages of conversion, GA catabolism occurs mainly through 2-β hydroxylation via enzyme GA 2-oxidase (GA2ox) (Fleet and Sun, 2005). Two novel mechanisms that were found to reduce the bioactivity of GAs are the epoxidation by a P450 monooxygenase (Zhu et al., 2006) and methylation (Varbanova et al., 2007).
The metabolic enzymes of GA are crucial players in the maintenance of GA homeostasis and the regulation bud dormancy. Specifically, the synthetic genes GA20ox and GA3ox and the catabolism gene GA2ox are of particular importance in regulating GA levels, and they are all encoded by multigene families. Transcription studies have shown the close correlation between the expression of GA20ox, GA3ox, and GA2ox and the GA level variation during bud dormancy in woody species such as rose (Rosa sp.) (Choubane et al., 2012), Japanese apricot (P. mume) (Wen et al., 2016), and grapevine (Zheng et al., 2018). In particular, the expression of GA2ox was found to be significantly upregulated by SD photoperiod, and its overexpression resulted in accelerated bud set and delayed bud flush (Zawaski et al., 2011). This indicates that the GA2ox-mediated catabolism is the key mechanism that reduces GA levels for the establishment and maintenance of bud dormancy. In a comprehensive transcription study in the tea plant Camellia sinensis, the expression of the GA synthetic enzyme genes KAO and KO was in line with the progression of bud dormancy (Yue et al., 2018). When multiple genes from the GA20oxs, GA3oxs, and GA2oxs families were examined, some genes were expressed in correlation with the bud dormancy progression, and others showed differential expression (Yue et al., 2018). Similarly, these genes were differentially expressed when tea plants were treated with exogenous GA3. Such redundant roles and possibly specialized functions of GA metabolism gene families contribute to the complexity of the underlying mechanism of GA metabolism.
In the GA signaling pathway, the GA-GID1-DELLA module is considered to be universal and highly conserved in angiosperms (Sun, 2011). In this model, DELLA proteins belong to the GRAS transcriptional factor family and negatively regulate GA responses in the absence of GA. GID1 (GA-insensitive dwarf 1) is the GA receptor and can form the GA-GID1 complex via binding with a GA-specific F-box protein SLEEPY1 (SLY1) to trigger rapid degradation of DELLA proteins via the ubiquitin-proteasome pathway (Itoh et al., 2003; Ariizumi et al., 2013; Hauvermale et al., 2014). Recently, DELLAs were also found to be involved in the GA feedback mechanism by upregulating the GA biosynthesis gene GA20ox (Yoshida and Ueguchi-Tanaka, 2014). In addition to GA metabolism, recent genetic studies also indicated the involvement of the GA signaling pathway in dormancy. For example, the expression of GID1 in Chinese cherry (Prunus pseudocerasus) is significantly downregulated at the early stage of dormancy development and rapidly increases when buds enter the ECD stage (Zhu et al., 2015). Similar increase of GID1 expression prior to bud break was also found in tea plant (Yue et al., 2018). The parallel relationship between GID1 expression and GA levels confirms the role of GID1 as a positive mediator of GA responses. In contrast to GID1, DELLA proteins mainly act as a negative mediator of GA responses. In poplar (P. tremula × Populus alba), DELLA genes (GAI and RGL) were found to be upregulated in response to SD photoperiod (Zawaski and Busov, 2014). Similarly, high DELLA expression in tea plant was observed during the induction of dormancy (Yue et al., 2018), and overexpression of C-repeat binding factors (CBFs), a regulatory gene responsible for cold acclimation, led to upregulation of DELLA and reduced growth in apple (Wisniewski et al., 2015). Thus far, it is still unclear if these GA signaling genes respond directly to the environmental signals or if their expression merely reflects the developmental events during dormancy.
The antagonism between ABA and GA marks the main feature of their interactions in modulating biological processes, in which the metabolism and signaling of these two phytohormones respond oppositely to environmental cues. In dormancy, the levels of GA and ABA are inversely correlated, with the ABA/GA ratio varying in parallel with the depth of dormancy (Duan et al., 2004). For example, in Japanese apricot, the decrease of ABA level is accompanied by a gradual increase in GA level from dormancy through dormancy release (Wen et al., 2016). Transgenic studies showed that the alternation of one hormone may affect the metabolism of the other. Mutations in the ABA pathway lead to higher GA content by promoting the GA synthesis genes GA3ox, and inhibition of GA upregulates the ABA synthetic genes (ABA1 and NCEDs) while downregulating the ABA catabolic gene CYP707A2 (Seo et al., 2006; Oh et al., 2007). These results suggest that ABA and GA are involved in the metabolic regulation of each other. Further evidence was obtained in a recent transcriptome study with tea plant (Yue et al., 2018), in which GA treatment was found to repress the expression of both ABA biosynthetic genes (ZEP, NCED1, and NCED1) and catabolic gene (CYP707A2). In the same study, ABA treatment was found to upregulate the GA synthetic genes (CsKS, CsKAO, and CsKO and GA3ox1, GA20ox1, and GA20ox2), while the catabolic gene GA2oxs showed different expression patterns upon exposure to ABA in a concentration-dependent manner.
The interaction between ABA and GA also occurs at the signal transduction level. In addition to playing a central role in the GA signaling pathway, DELLA proteins also act as a cross talk node that integrates the signaling pathways of several other hormones including ABA (Santner and Estelle, 2009; Davière and Achard, 2016). In mutants with impaired GA signaling, accumulated DELLAs upregulate the expression of XERICO, a RING zinc finger protein known to induce ABA synthesis (Zentella et al., 2007; Ariizumi et al., 2013) (Figure 2). In tea plant, GA treatment induces the expression of a negative mediator PP2C in ABA signaling, while represses the ABA receptor PYL8 (Yue et al., 2018). The fact that DELLA proteins can be stabilized by stress-induced ABA (Ariizumi et al., 2013) indicates that GA responses may be inhibited by high ABA levels at the dormancy initiation. Recently, Yue et al. (2018) showed that exogenous ABA represses the expression of GA receptor GID1 and induces the expression of DELLA genes. However, long hours of ABA treatment were found to suppress DELLA expression.
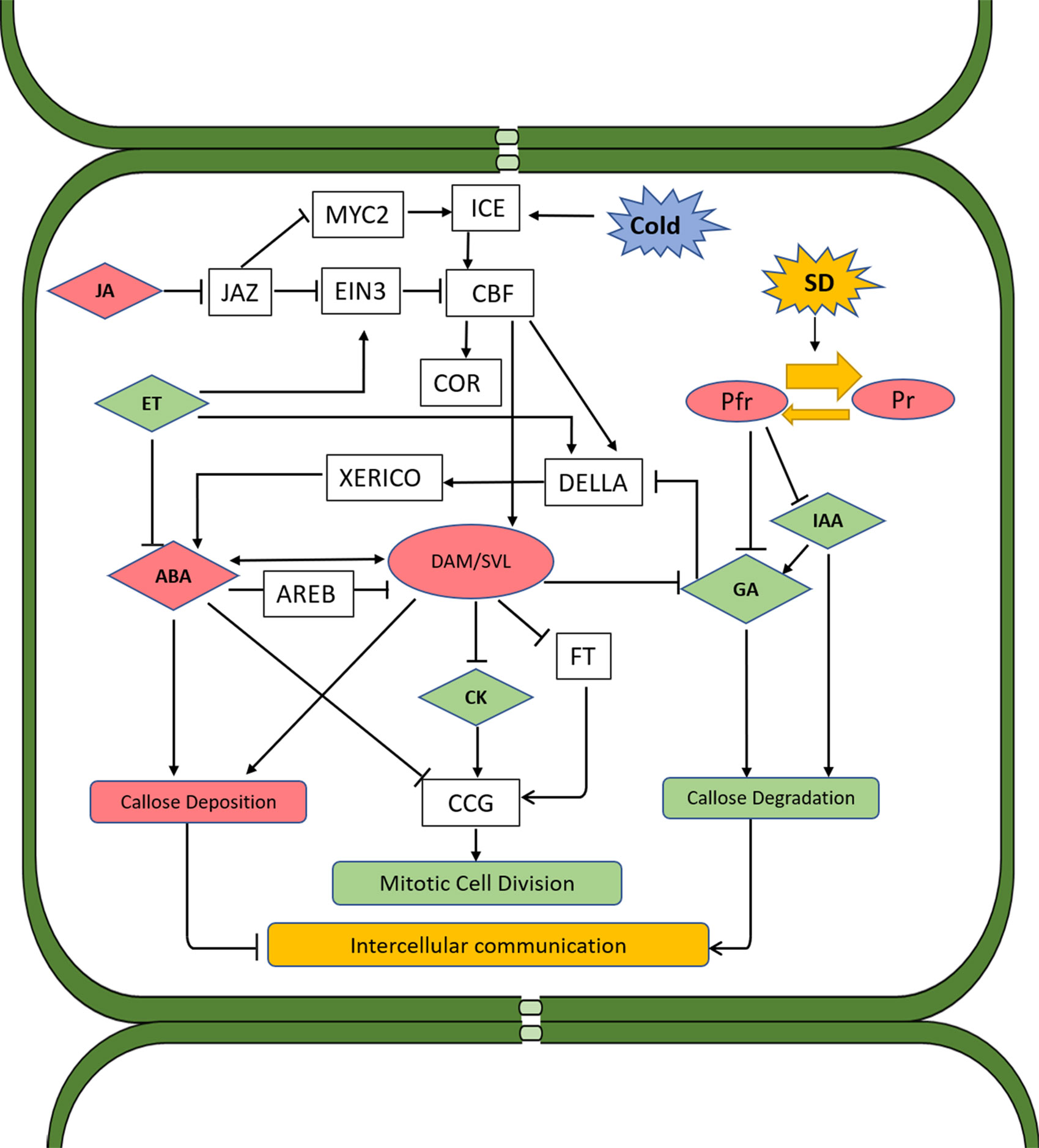
Figure 2 Proposed schematic model of hormone interactions during bud dormancy induction and release in woody perennials. Solid arrows and lines indicate actions or interactions among hormones, pathways, and environmental cues that have been documented in the literature. Red color indicates the substance or process that induces dormancy, and green color indicates those that promote dormancy release. Both SD photoperiod and low temperature induce CBF, which directly promotes the expression of DAM genes. DAM proteins regulate dormancy through ABA-dependent and ABA-independent pathways. In the former pathway, accumulation of DAMs reduces GA level, thus activating DELLA and subsequently XERICO proteins, which in turn promotes ABA synthesis. DAM genes can also upregulate ABA levels through upregulating the gene encoding NCED, a key enzyme in ABA synthesis. In addition to the control of DAMs over ABA biosynthesis and signaling, ABA can also negatively regulate the expression of DAM genes through SnRK2 and the ABA signaling component AREB. This pathway probably serves as a negative feedback regulation mechanism. In the ABA-independent regulation pathway, DAM proteins induce dormancy through negative regulation of FT, which in turn prevents CCG-mediated dormancy release. ABA can also repress CCG and inhibit the mitotic cell division. ABA was also found to suppress the intercellular communication during dormancy by enhancing the expression of callose synthase, leading to callose deposition and blockage of plasmodesmata. This alteration of plasmodesmata during dormancy is reversed by GA as buds transition to bud break. GAs induce the expression of glucanases which degrade callose, allowing for the passage of sugars and other growth-promoting factors. Under SD conditions, GA biosynthesis is inhibited through phytochrome and phytochrome-interacting factors. ET is induced by SD and has a negative impact on GA biosynthesis and signaling, which nominates it as a dormancy inducer. However, ET also inhibits ABA synthesis and signaling and negatively regulates CBF through activation of EIN3, which is also subject to the regulation of JA through JAZ proteins. JA induces bud dormancy by targeting JAZ proteins for degradation via the ubiquitination/26S proteasome, which in turn releases MYC2 and ICE1 from repression. Both MYC2 and ICE1 activate the expression of CBF. CK can repress ABA and promote dormancy release through inducing the expression of CCGs. IAA facilitates dormancy release through promoting GA biosynthesis and callose degradation. ABA, abscisic acid; CBF, C-repeat binding factor; CCG, cell cycle gene; CK, cytokinin; ET, ethylene; EIN3, ETHYLENE INSENSITIVE 3; FT, flowering locus T; GA, gibberellins; JA, jasmonates; IAA, indole-3-acetic acid; SD, short day; NCED, 9-cis-epoxycarotenoid dioxygenase; DAM, dormancy-associated MADS.
Although it is recognized as the ripening hormone, ET has wide-ranging effects on a number of other biological processes including, but are not limited to, seed germination, flowering, abscission, senescence, and stress responses (Bleecker and Kende, 2000). The function of ET in dormancy is closely related to its biosynthesis and signaling transduction. ET biosynthesis starts with the conversion of methionine to S-adenoysl-methionine (SAM) by SAM synthetase (Iqbal et al., 2013). In the subsequent rate-limiting and committed step, SAM is converted to 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase (ACS). In the final step, oxidization of ACC by ACC oxidase (ACO) produces ET, hydrogen cyanide (HCN) and CO2 (Wang et al., 2002a). In this biosynthetic pathway, enzymes ACS and ACO are the important regulators of endogenous content of ET and are both encoded by multi-gene families (Van de Poel and Van Der Straeten, 2014); (Johnson and Ecker, 1998). During high rates of ET biosynthesis, accumulated cyanide can become toxic to plants, and this toxicity can be alleviated by converting cyanide to β-cyanoalanine by enzyme β-cyanoalanine synthase (β-CAS) (Wang et al., 2002a). In the ET signaling pathway, five ET receptors ETR1, ETR2, EIN4, ERS1, and ERS2 have been identified in Arabidopsis (Wang et al., 2002a). In the absence of ET, these receptors recruit Constitutive Triple Response1 (CTR1) and inhibit a membrane spanning protein EIN2, a positive regulator of ET signaling (Gao et al., 2003; Qiu et al., 2012). EIN2 stabilizes the downstream transcription factors ETHYLENE INSENSITIVE 3 (EIN3)/EIN3-Line1 (EIL1) by inducing proteasomal degradation of F-box proteins (EBF1 and EBF2), which mediate proteolysis of EIN3/EIL1 in the absence of ET (An et al., 2010). Among the EIN3 direct targets are the ET RESPONSE FACTOR (ERF) genes, which belong to the AP2/ERF superfamily, members of which play pivotal roles in adaptation to various biotic and abiotic stresses (Mizoi et al., 2012).
The results of several studies have converged to indicate ET and its response pathway is involved in dormancy regulation. Early evidence showed that ET levels are elevated at both dormancy initiation and bud break stages and application of ET antagonist 2,5-norbornadiene (NBD) causes premature dormancy break in potato microtuber (Suttle, 2004). Later on, Ruonala et al. (2006) reported that ET-insensitive mutation (etr1-1) causes abolished formation of terminal buds and biosynthesis of ABA and delayed dormancy in European white birch (Betula pendula) in SD photoperiod condition. Similar result was also found in chrysanthemum, in which mutants with impaired ET receptor gene (DG-ERS1) fail to enter dormancy at dormancy inducing temperature (Sumitomo et al., 2008). The requirement of ET in the induction of dormancy was further confirmed by microarray and transcriptomic studies, in which ET biosynthesis gene set and signaling component genes (e.g. ETR2, EIN3 EIN4 and ERF) in poplar are upregulated in dormancy inducing conditions (Ruttink et al., 2007; Howe et al., 2015). Investigations into the interaction between ET and GA indicated that ET may modulate GA or GA signaling pathway to induce dormancy. It has been noted that in responses to environmental stresses, activated ET signaling induces GA deactivation and accumulation of DELLAs through enhancing the expression of ET Response Factors (ERF6) (Dubois et al., 2013; Dubois et al., 2015). Moreover, Achard et al. (2007) reported that ET can act on and stabilize DELLA via CTR1, independent of GA mediation. More evidence of the interaction between ET and GA was obtained when the interaction between ET action and phytochrome signaling was examined. In tobacco, low red to far-red light ratios (R:FR) was found to trigger ET biosynthesis and ET insensitive transgenic lines exhibit no shade avoidance, which can be rescued by application of GA3 (Pierik et al., 2004). This suggests that the early signal transduction of phytochrome-mediated light responses might trigger ET accumulation and GA reduction in response to SD, leading to growth cessation and dormancy inductions.
Interestingly, ET was also indicated to participate in the dormancy release. Application of ET signaling inhibitor NBD leads to increase of ABA levels in breaking buds, suggesting ET is required for the degradation of ABA and modulating of ABA signaling (Ophir et al., 2009; Zheng et al., 2015). In Arabidopsis, ET negatively regulates the cold tolerance gene CBF, through activating EIN3, which in turn represses CBF genes by binding to their promoters (Shi et al., 2012) (Figure 2). Further evidence was supported by the following observations in grape buds: 1) ET biosynthesis can be temporarily activated by dormancy break stimuli such as HC, heat shock and sodium azid; 2) exogenous ET application enhances bud break; and 3) dormancy release is severely delayed when of ET signaling is blocked by NBD (Ophir et al., 2009). These findings suggest that ET has complex actions during bud dormancy, in which ET interacts with ABA synergistically during dormancy initiation, but antagonistically during dormancy release. These studies also suggest that ET and its signaling pathway is essential for the development of dormancy, and both ABA and GA respond downstream of ET mediated dormancy regulation. How these opposite processes are integrated in a stage-dependent way warrants further investigation.
Increasing evidence has suggested that bud break is triggered by elevated levels of ROS, and ET is actively involved in this process. Differential expression of genes induced by HC revealed connection between dormancy release and oxidative stress, hypoxia, mitochondrial activity, ET biosynthesis and signaling pathways (Ophir et al., 2009; Sudawan et al., 2016; Ionescu et al., 2017). In response to HC, plants transiently elicit ROS, such as H2O2, and subsequently activating many pathways that are related to dormancy release, including antioxidant systems. ET biosynthesis has been suggested to increase oxidative stress in plants due to production of hydrogen cyanide (Ionescu et al., 2017), and hydrogen cyanide was found to be more effective than ET in breaking buds (Ionescu et al., 2017). On the other hand, ET can also act as a messenger molecule to activate the antioxidant system (such as CAT) to scavenge excessive ROS (Vergara et al., 2017). Indeed, the accumulation of ROS and their elimination has been proposed to be pivotal steps in releasing dormancy (Sudawan et al., 2016; Beauvieux et al., 2018).
CKs are a group of adenine-derived small compounds that play important roles in a variety of plant processes including cell division, cell differentiation, apical dominance, leaf senescence, and stress tolerance (Sakakibara, 2006; Zürcher and Müller, 2016). The effects of CKs are highly dependent on cell and tissue types, developmental stage and environmental conditions, thus CKs are particularly important in modulating meristem activity and morphogenesis. At the cellular level, CKs can activate cell cycle regulator, CDK by dephosphorylating its tyrosine (Tyr), and this CK effect is considered to be primary and required for the proper progression of cell cycle, which would otherwise arrest at the G2 phase (Zhang et al., 2005). Natural CKs differ greatly in the side chains, which are attached to the parental compound adenine, and this structural diversity provides high specificity of the interaction between CKs and the receptors (Kieber and Schaller, 2014). In CKs metabolism, the rate-limiting step in biosynthesis pathway is mediated by the adenosine phosphate-isopentenyltrasferase (IPT), and the major catabolic step is the oxidation of CKs that removes the side chains by the CK oxidase/dehydrogenase1 (CKX). Though CKs are highly mobile in plants and can be transported in the xylem sap over long distances, locally synthesized CKs were suggested to be critical in regulating dormancy (Tanaka et al., 2006).
Similar to its effect in releasing latent buds from PD, CKs are also implicated in the regulation of dormancy release (Faust, 1997). Early research showed that CK concentration in xylem sap increases rapidly in response to bud breaking chemicals and reaches a maximum level at the budburst in apple (Cutting et al., 1991). Genetic analysis demonstrated that forced condition either through LD photoperiod or treatment with HC can increase CK levels in the grapevine cuttings by activating the expression of CK biosynthesis genes (IPT and lonely Guy LOG1) while downregulating that of CK catabolism gene CKX (Noriega and Pérez, 2017). Furthermore, temporal expression analysis showed that the increase of CK levels induced by forced conditions can upregulate the expression of cell cycle genes and subsequently raises the rate of cell division and cellular respiration, which are the major events preceding the activation of dormant buds (Noriega and Pérez, 2017). In support of these findings, Hartmann et al. (2011) showed that high levels of CK in IPT-expressing potato tubers promoted early dormancy release, whereas reduced CK contents through CKX overexpression inhibited cellular metabolism and cell proliferation, rendering no response to GA3 and leading to prolonged dormancy period. Another evidence of CK’s effect in dormancy release lies in its mediation of light signal. Previous studies have identified several His kinases as CK receptors such as AHK2, AHK3 and AHK4, which positively regulate CK signals by phosphorylating His-containing phosphotransfer proteins (Higuchi et al., 2004; Mahonen et al., 2006; Riefler et al., 2006). Using loss-of-function approach, Tran et al. (2010) showed that these CK receptors negatively regulate the ABA responses in the presence of CK. Dobisova et al. (2017) demonstrated that light can regulate the transcription of these CK receptors to control the CK signaling. This result was supported by the finding in rose (Rosa hybrida), in which CK was found to participate in the initial responses of the light signaling pathway that promotes bud outgrowth (Roman et al., 2016). These results suggest the light-mediated increase of CKs during the dormancy release may contribute to reduce ABA levels. Taken together, these results suggest that CK is an essential regulator in the dormancy release and CK acts upstream of GA and ABA response pathways in stimulating meristematic activity.
Auxin has long been known to promote stem elongation and to suppress the growth of lateral buds, in a phenomenon of the apical dominance. Recent findings indicate auxin is also involved in plant senescence, blooming and stress responses (Di et al., 2016; Fendrych et al., 2016). Of all the four types of naturally occurring auxins, IAA is the most abundant and relatively well studied (Simon and Petrášek, 2011). IAA biosynthesis involves a two-step conversion: in the first step, IAA precursor tryptophan is converted to intole-3-pyrunvae (IPA) by an amino transferase; in the second step, IPA is oxidized to IAA by flavin monooxygenase (YUC) and this reaction is a rate-limiting step in IAA synthesis pathway (Zhao, 2012). Auxin biosynthesis occurs primarily in shoot apex and young leaves. Auxin is transported basipetally (from tip to base) through specialized membrane carriers, called PIN-FORMED proteins (PIN), which also play a role in maintaining auxin homeostasis (Friml and Palme, 2002; Muller and Leyser, 2011). In the auxin signaling transduction pathway, Aux/IAAs are transcriptional repressors that bind to and inhibit the activity of AUXIN RESPONSE FACTORs (ARFs), in the absence of auxin. ARF can bind to the Auxin Response Elements (AREs) in the promoter region of the auxin responsive genes, and both activaor AFRs and repressor ARFs have been identified (Leyser, 2018). In the presence of auxin, Aux/IAAs are subjected to the auxin triggered Ubiquitin degradation.
Being a major growth promoter, IAA has been implicated in the dormancy release in many species. Early study showed that exogenous auxin can promote the degradation of dormancy callose in the phloem of magnolia (Magnolia kobus) and lead to the restoration of the symplastic paths (Aloni and Peterson, 1997), which is the preparatory step of bud break. In tea plant (C. sinensis), Nagar and Sood (2006) found that IAA levels remain low during the entire stage of dormancy and increase steadily after dormancy release until bud break in the spring. This study also revealed that free IAA content changes in opposite to its conjugated form, suggesting conjugation may serve as a major mechanism in maintaining the homeostasis of endogenous IAA during dormancy. Increases of IAA levels during dormancy release were also reported in Chinese fir (Cunninghamia lanceolata) (Qiu et al., 2013) and Chinese plum (P. mume) (Zhang et al., 2018). Considering the fact that polarly transported IAA induces GA biosynthesis required for growth (Wolbang and Ross, 2001; Wolbang et al., 2004; Frigerio et al., 2006), IAA may likely facilitate dormancy release in collaboration with GA. Genetic study suggests that the IAA levels is mainly controlled at the metabolic level, as the of IAA synthetic gene YUC3 is upregulated during natural dormancy release or by HC; high levels of IAA, along with CK switches on the cell cycle machinery and release the buds from dormancy (Noriega and Pérez, 2017). Transcriptomic data revealed differential expression of the main components in the IAA signaling pathway during the transition from dormancy to active growth, and their potential roles remain yet to be further elucidated (Qiu et al., 2013; Zhang et al., 2018).
On the other hand, IAA also appears to be an integrator of environmental signals during the dormancy establishment. When exposed to low temperature or in combination with SD photoperiod, IAA levels in strawberry and the transcript levels of polar auxin transport (PAT)-related gene (e.g. PIN) decline significantly, accompanied by an increase of ABA levels and global genomic DNA methylation, indictive of dormancy initiation (Zhang et al., 2012b). In exposure to SD photoperiod, the auxin signaling repressor AUX/IAAs and PAT-related genes of hybrid aspen lose their responsiveness to auxin, accompanied by the downregulation of activator AFRs and induction of repressor ARFs (Baba et al., 2011). Transcriptome data implied that the majority of auxin-associated genes are downregulated during dormancy in Japanese apricot and poplar (Zhong et al., 2013; Howe et al., 2015). These results suggest that modulation of cellular auxin content, auxin responsiveness, auxin transport capacity, and conjugation could all be integrated in the regulation network of dormancy.
JAs are a class of lipid-based plant hormones that regulate diverse processes in plants development and defense (Browse, 2009). Bioactive forms of JA include jasmonic acids, its biosynthetic precursor 12-oxophytodienoic acid (OPDA) and the conjugate form JA-Ile, which all have been shown to be effective signaling compounds. Similar to ABA, JAs prevent plant growth by repressing meristem activity, and some stress related genes can be activated by both JAs and ABA, indicating their synergism in certain processes (Swiatek et al., 2002; Delker et al., 2006; Zhang and Turner, 2008; Browse, 2009). Jasmonic acids can arrest cell cycle of tobacco (Nicotiana tabacum) BY-2 cells at the G1 or G2 phases by repressing the activity of CDK (Swiatek et al., 2002). This may explain the reason that jasmonic acid and methyl JA (MeJA) inhibit seed germination in many species (Linkies and Leubner-Metzger, 2012). Indeed, JA can induce leaf senescence and control the expression of senescence-related genes in many species (Ueda and Kato, 1980; Shan et al., 2011; Yan et al., 2012; Lee et al., 2015). In Arabidopsis, JA biosynthesis is subject to the control of TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) transcription factors, which can directly activate JA biosynthetic gene LIPOXYGENASE2, and when TCP4 is targeted by microRNA (miR319), JA synthesis is repressed and leaf senescence is delayed and the expression of senescence associated genes decreased (Schommer et al., 2008). However, JAs seem to have opposite effects during bud dormancy. In beech trees (Fagus sylvatica), JA levels were found to increase remarkably during bud burst (Juvany et al., 2015). In agreement with this, contents of JA-Ile in potato tubers increase gradually as the buds transition from dormancy to active sprouting (Suttle et al., 2011; Juvany et al., 2015). Transcriptomic analysis showed that JA pathway is repressed during dormancy but activated during ECD stage and bud break (Hao et al., 2017). These results suggest that JAs may play a role other than inhibiting growth during dormancy release.
Recent research indicates that JA signaling pathway is actively involved in cold acclimation process, which is closely related to dormancy. In JA signaling pathway, JA ZIM-DOMAIN (JAZ) proteins are key repressors of JA signaling, and can form a JA coreceptor complex with F-box protein COI1 (CORONATINE INSENSITIVE1) (Sherif et al., 2015; Hu et al., 2017). Upon binding of JA to the complex, JAZ proteins undergo ubiquitin-mediated degradation and release several groups of transcription factors, such as the helix–loop–helix (bHLH) factors MYC2, MYC3 and MYC4 (Goossens et al., 2016). It has been shown that MYC2, as a key transcriptional activator of JA responses, can activate gene SAG29 (SENESCENCE-ASSOCIATED GENE29) from Arabidopsis by binding to its promotor and promote JA-induced leaf senescence, whereas another group of bHLH transcription factors (bHLH03, 13, 14 and 17) can bind to the promotor of SAG29 to counteract the enhancing effect of MYC2/MYC3/MYC4, thus these signaling components form an elegant feedback regulation mechanism to modulate JA-induced senescence (Qi et al., 2015). The pathway that is composed of the Inducer of CBF Expression (ICE), CBF transcription factors, and various Cold Regulated (COR) genes is the best-studied cold response pathway (Wang et al., 2017; Jin et al., 2018). Recently, more evidence was obtained for the direct interaction between JA signaling and the ICE-CBF-COR signaling cascade. First, the physical interactions between JA signaling repressors (JAZ proteins) and ICE proteins was demonstrated by yeast two-hybrid assays, and JA induced degradation of JAZ proteins led to the upregulation of the COR genes (Hu et al., 2013). Second, the JA-induced transcriptional activator, MYC2, was found to interact with ICE1 and activate the cold response pathway, reinforcing the notion that JA acts as an essential upstream signal of the ICE-CBF-COR pathway to positively regulate cold tolerance Zhao et al. (2013). Third, In Arabidopsis, it was noticed that exogenous JA application enhanced plant freezing tolerance and JA biosynthesis was triggered upon cold exposure (Hu et al., 2013). However, whether such cross talks between JA and cold-responsive pathways would function during dormancy, especially in woody species, still needs further validation. The interactions between JAZ proteins and other hormonal signaling components (e.g. DELLA and EIN3/EIL1) (Melcher et al., 2010; Zhu et al., 2011) indicate that JA could be widely and closely related to the regulation network of cold acclimation and dormancy, and its function will be better understood when examined in the context of its interaction with other hormones.
The Dormancy-Associated MADS-box (DAM) has become increasingly prominent in recent studies on the bud dormancy. The first DAM genes were identified in a peach mutant Evergrowing (EVG), in which deletion of EVG locus rendered complete loss of growth cessation and bud formation in dormancy inductive conditions (Bielenberg et al., 2004). Later, six tandemly arranged MADS-box genes in the EVG locus of peach were identified, and they were believed to be the candidates that regulate dormancy, thus named Dormancy-Associated MADS-box (DAM) genes (Bielenberg et al., 2008). Phylogenetic analysis indicates these DAM genes are homologous to two transcription factor genes of Arabidopsis: SHORT VEGETATIVE PHASE (SVP) and AGAMOUS-LIKE 24 (AGL24) (Jimenez et al., 2009). Genes of SVP and AGL24 have antagonistic functions, in which SVP prevents flowing through inhibition of floral promoting gene FT, whereas AGL24 activates flowering by enhancing LEAFY (LFY) gene, a major regulator of floral development of land plants (Hartmann et al., 2000; Michaels et al., 2003; Lee et al., 2008). Up to date, numerous DAM-like or SVP-like (SVL) genes that potentially mediate in the control of dormancy have been identified in diverse species (Table 1). Based on the phylogenetic data, no consensus has been reached weather SVL genes represent a distinct group apart from DAMs or they belong to one group. Nevertheless, the fact that DAM and SVL both exhibit growth inhibitory effect across different species demonstrated by transgenic studies (Sasaki et al., 2011; Wu et al., 2017a; Wu et al., 2017b; Singh et al., 2018; Singh et al., 2019; Yamane et al., 2019) suggests that their potential roles may be similar or overlapping in mediating dormancy. Specifically, functional commonality in DAM and SVL was manifested in that they both promote ABA biosynthesis by upregulating NCED (Tuan et al., 2017; Singh et al., 2018) and delay bud break (Wu et al., 2017a; Singh et al., 2018; Yamane et al., 2019).

Table 1 The chronology of select major research findings related to identification of function of DAM/SVL genes.
Though DAM genes are highly conserved and closely arranged, their expression patterns are generally distinct in response to seasonal changes and developmental signals. In peach (P. persica), DAM1, DAM2 and DAM4 are upregulated during growth cessation and bud set, whereas DAM5 and DAM6 expression is strongly induced by SD photoperiod and their transcripts increase only during dormancy, then decline upon natural or chemical induction of dormancy release (Li et al., 2009; Jimenez et al., 2010; Yamane et al., 2011). It is thus suggested that DAM5 and DAM6 in peach operate downstream of the circadian perception of photoperiodic stimuli, which occurs prior to the onset of chilling. As the dormancy progresses, the expression of these genes decreases concomitant with chilling accumulation and reaches the minimum at bud break. In light of these findings, DAM5 and DAM6 are believed to be the primary internal regulators of dormancy induction and maintenance in peach and probably other stone fruit species.
Following the identification of peach DAM genes, identification and characterization of more DAM and SVP homologs have been reported in other perennial species including but are not limited to raspberry (Rubus idaeus) (Mazzitelli et al., 2007), Japanese pear (Ubi et al., 2010), Japanese apricot (P. mume) (Sasaki et al., 2011), kiwifruit (Actinidia spp.) (Wu et al., 2012), and pear (P. pyrifolia) (Niu et al., 2016). In a recent review, Falavigna et al. (2018) divided the documented DAM and SVP like genes into three groups according to their expression patterns. The majority of these DAM genes fall in the group that are highly expressed during the intensified stage of dormancy, characteristic of peach DAM 5 and DAM 6; whereas other DAM genes are asynchronous with the progression of dormancy. More definitive role of DAM genes in bud dormancy regulation were demonstrated in a number of recent transgenic studies. In poplar, Sasaki et al. (2011) showed that constitutive overexpression of Japanese apricot (P. mume) DAM6 gene induced premature growth arrest and bud set. Similarly, ectopic expression of DAM-like gene MdDAMb or MdSVP in apple causes delayed bud break (Wu et al., 2017a). Overexpression of DAM6 in Japanese apricot resulted in inhibited shoot growth, early bud set, repressed bud break competency and delayed bud break (Yamane et al., 2019). The high similarity of the DAM genes characteristics across species indicate that bud dormancy may be regulated by a highly conserved mechanism that is shared by most perennial species.
Though the role of DAM genes in dormancy regulation has been extensively verified, the underlying molecular processes are still largely unknown. Recent studies suggested that flowering regulating gene FT and cold response transcription factors CBFs may act as major mediators in the DAM regulation pathway. FT encodes a small globular protein that has been implicated in flowering, prevention of growth cessation and proper induction of dormancy (Bohlenius et al., 2006; Wigge, 2011). High expression of DAM genes was found to correlate with the downregulation of FT in leafy spurge (Euphorbia esula) and pear (P. pyrifolia) (Hao et al., 2015; Niu et al., 2016), in a manner similar to the repression of FT by the flowering inhibiting gene, SVP, in Arabidopsis (Lee et al., 2007). Similarly, expression of FT1 in aspen was reduced by the overexpression of SVL, and enhanced by silencing SVL (Singh et al., 2018). The CBF genes can be quickly activated by cold stress, and in turn trigger many downstream genes involving growth cessation, cold acclimation and dormancy (Kendall et al., 2011). Overexpression of CBF was found to upregulate two DAM genes (MdDAM1 and MdDAM3) in bud tissues of apple (Wisniewski et al., 2015). In a transient transformation and luciferase assay, Saito et al. (2015) demonstrated that CBF2 protein from Japanese pear (PpCBF2) regulates DAM expression through binding to the C-Repeat/Drought Response Element (CRT/DRE) motifs on the DAM genes. Confirmation of the direct regulation of DAM genes by CBF in Japanese pear (P. pyrifolia) (Niu et al., 2016; Li et al., 2019) and Japanese apricot (P. mume) (Zhang et al., 2018) prompted the hypothesis of a cold dependent CBF-DAM-FT model, in which cold temperature activates CBF, and CBF directly activates DAM genes, which repress the FT to arrest plant growth and result in the establishment of dormancy (Niu et al., 2016) (Figure 2).
Increasing evidence indicates the close interaction between DAM/SVL genes and plant hormones, especially ABA, suggesting that these hormones may be integral components in the DAM/SVL regulation cascade. For example, overexpression of DAM6 (35S:PmDAM6) in Japanese apricot induced high accumulation of ABA and decreased CK contents in the terminal buds (Yamane et al., 2019). Transient assay and electrophoretic mobility shift assay (EMSA) indicate the DAM-induced ABA accumulation may be due to the binding of DAM to the CArG motif in the promoter of the ABA biosynthesis enzyme NCED (Tuan et al., 2017). This notion was supported by Singh et al. (2018)’s finding, in which NCED3 was highly expressed in the transgenic aspen line with overexpression of SVL. On the other hand, DAM/SVL also mediates ABA signaling pathway. Using chromatin immunoprecipitation and ChIP-seq technique, Wu et al. (2018a) demonstrated that the SVP2 from kiwifruit, which suppresses meristem activity and suppress bud break, can bind to the intron regions of ABA insensitive ring protein 2, and modulate the downstream genes. In aspen, overexpression of SVL positively regulated ABA receptor genes (RCAR/PYL1 and RCAR/PYL2) genes (Singh et al., 2018). While DAM modulates ABA biosynthesis and signaling, ABA is also capable of regulating the expression of DAM/SVL. Early study in Arabidopsis reported that ABA can induce the expression of CBF, a positive regulator of DAM genes, via the CRT promoter element (Knight et al., 2004). Recent investigation in aspen showed that ABA application induces SVL expression and a mutation with reduced ABA response remarkably reduces SVL expression (Singh et al., 2018). Thus, ABA and DAM/SVL form a positive feedback loop in mediating dormancy. However, this notion was challenged by Tuan et al. (2017)’s finding in which an ABA singling gene AREB1 represses the expression of PpDAM1 by binding to the ABRE motifs in its regulatory region, indicating existence of a negative feedback regulation of ABA on DAM. To reconcile this contradiction, it is hypothesized that the ABA and DAMs interaction is dependent on the stage of dormancy: during dormancy induction, ABA promotes DAMs expression through activating CBF, which is also induced by low temperatures and SD, and as dormancy progresses, ABA inhibits DAMs activity through AREB-mediated negative feedback mechanism, and the reduced expression of DAMs at this stage should in turn repress ABA synthesis by downregulating NCED (Figure 1). In addition to ABA, SVL was also found to negatively affect GA as the GA biosynthesis gene GA20ox was reduced by the overexpression of SVL, but enhanced by its gene silencing after low temperature treatment (Singh et al., 2018). The finding of the versatility of SVL in targeting multiple dormancy-related genes raised the hypnosis that SVL may function as a hub gene that dictates both dormancy establishment and release by connecting ABA and GA, and cold perception pathways (Busov, 2019).
In addition to the environmental and hormonal regulation, epigenetic modification has also been found to play an important role in regulating DAM expression during the dormancy phase transition. In the chromatin modification, histone H3 trimethylation at K4 (H3K4me3) and H3 acetylation (H3ac) are associated with activation of the nearby genes, whereas histone trimethylation at K27 (H3K27me3) represses transcription (Shilatifard, 2006). Concomitant with cold accumulation, several chromatin regions including the promoter of DAM6 genes in peach are marked by the enrichment of H3K27me3 and removal of H3K4me3 and H3ac (Leida et al., 2012). In Japanese pear, Saito et al. (2015) showed that the reduction of expression of DAM homolog (PpMADS13-1) towards the dormancy release can be attributed to the decrease of H3K4me3, but not H3K27me3, which remained largely unaltered. Similarly, Singh et al. (2018) observed no significant increase of H3K27me3 at the SVL locus in low-temperature treated aspen. It was reported that H3K27me3 was specific to peach cultivars with high chilling requirement (Leida et al., 2012), and thus it appears that downregulation of SVL/DAM genes may be achieved predominantly via the reduction of H3K4me3, whereas H3K27me3 modification is reserved for species or cultivars that require high chill accumulation. This histone modification in DAM genes is highly reminiscent of that in FLC locus in Arabidopsis during winter vernalization (Bastow et al., 2004; He et al., 2004). In sweet cherry, chill accumulation leads to DNA methylation in the promoter of DAM genes and increase of the small interfering RNAs (siRNA) that match this region, concomitant with the upregulation of FT (Rothkegel et al., 2017). Similar DAM genes degradation mediated by siRNA (miR6390) and the subsequent release of FT2 was also found in pear (Niu et al., 2016). These findings indicate the epigenetic modification is a main mechanism that regulates DAM genes and highlight the involvement and importance of DAM genes during dormancy.
Although many plant hormones are implicated in dormancy regulation, some aspects are still open to questions. ABA is the central regulator of dormancy, and its repression of cell cycle and intercellular communication via plasmodesmata appears to be important mechanisms leading to dormancy induction. The role of GA is largely within its conventionally defined function: promotion of cell division and elongation, which is an essential step in dormancy release. However, whether GA directly unlock dormancy still needs further investigations. ET acts antagonistically with GA during dormancy induction, but the byproduct of its metabolism seems able to promote dormancy release. The direct interaction between DAM/SVL genes and ABA opens the door to hypothesize that DAM/SVL may regulate dormancy through modifying the metabolism and/or signaling of other hormones, such as GA and CK biosynthesis. The interaction between DAM/SVL genes and other plant hormones is expected to be further revealed in the coming years. The potential roles of DAM and SVL in the mediation of dormancy, though seemingly distinct, are expected to be integrated in a broader context of an overarching theme of growth inhibition, rather than dormancy per se. Thanks to the fast-growing research on bud dormancy in recent years, much have been learned about the hormone induction and repression kinetics during dormancy, the role of hormone biosynthesis and signaling-related genes, effects of hormones and their cross talks during the initiation, progression and release of bud dormancy. However, the fundamental molecular and cellular mechanisms that initiate the transitions between meristematic growth, bud arrest, dormancy and resumption of metabolic activities still require further elucidation.
JL and SS have contributed equally to the writing, editing and preparation of this review article.
The authors are grateful for the funding to SS from The Virginia Agricultural Council (#449846) and The Virginia Apple Research Program (#459952).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful for the funding to SS from The Virginia Agricultural Council (#449846) and The Virginia Apple Research Program (#459952).
ABA, abscisic acid; CALS1, callose synthase; CDK, cyclin-dependent kinases; CK, cytokinin; DAM, dormancy-associated MADS genes; ET, ethylene; FT, flowering locus T; GA, gibberellins; GA2ox, GA 2-oxidase; GA3ox, 3-oxidase; GA20ox, dioxygenases 20-oxidase; HC, hydrogen cyanamide; IAA, indole-3-acetic acid; JA, jasmonate; NCED, 9-cis-epoxycarotenoid dioxygenase; PP2Cs, protein phosphate 2Cs; ROS, reactive oxygen species; SD, short day; SVP, short vegetative phase.
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. U.S.A. 104, 6484–6489. doi: 10.1073/pnas.0610717104
Addicott, F. T., Lyon, J. L. (1969). Physiology of abseisic acid and related substances. Ann. Rev. Plant Physiol. 20, 139–164. doi: 10.1146/annurev.pp.20.060169.001035
Aloni, R., Peterson, C. A. (1997). Auxin promotes dormancy callose removal from the phloem of Magnolia kobus and callose accumulation and earlywood vessel differentiation in Quercus robur. J. Plant Res. 110, 37. doi: 10.1007/BF02506841
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Ariizumi, T., Hauvermale, A. L., Nelson, S. K., Hanada, A., Yamaguchi, S., Steber, C. M. (2013). Lifting della repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol. 162, 2125–2139. doi: 10.1104/pp.113.219451
Arora, R., Rowland, L. J., Tanino, K. (2003). Induction and release of bud dormancy in woody perennials: a science comes of age. HortScience 38, 911–921. doi: 10.21273/HORTSCI.38.5.911
Baba, K., Karlberg, A., Schmidt, J., Schrader, J., Hvidsten, T. R., Bako, L., et al. (2011). Activity–dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc. Natl. Acad. Sci. U.S.A. 108, 3418–3423. doi: 10.1073/pnas.1011506108
Bai, S., Saito, T., Sakamoto, D., Ito, A., Fujii, H., Moriguchi, T. (2013). Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 54, 1132–1151. doi: 10.1093/pcp/pct067
Barrero, J. M., Piqueras, P., Gonzalez-Guzman, M., Serrano, R., Rodriguez, P. L., Ponce, M. R., et al. (2005). A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J. Exp. Bot. 56, 2071–2083. doi: 10.1093/jxb/eri206
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Beauvieux, R., Wenden, B., Dirlewanger, E. (2018). Bud dormancy in perennial fruit tree species: a pivotal role for oxidative cues. Front Plant Sci. 9, 657. doi: 10.3389/fpls.2018.00657
Bielenberg, D. G., Wang, Y., Fan, S., Reighard, G. L., Scorza, R., Abbott, A. G. (2004). A deletion affecting several gene candidates is present in the Evergrowing peach mutant. J. Hered. 95, 436–444. doi: 10.1093/jhered/esh057
Bielenberg, D. G., Wang, Y., Li, Z., Zhebentyayeva, T., Fan, S., Reighard, G., et al. (2008). Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507. doi: 10.1007/s11295-007-0126-9
Bittner, F., Oreb, M., Mendel, R. R. (2001). ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J. Biol. Chem. 276, 40381–40384. doi: 10.1074/jbc.C100472200
Bleecker, A. B., Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Bohlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Boneh, U., Biton, I., Zheng, C., Schwartz, A., Ben-Ari, G. (2012). Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 31, 311–321. doi: 10.1007/s00299-011-1166-z
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205. doi: 10.1146/annurev.arplant.043008.092007
Busov, V. B. (2019). Plant development: dual roles of poplar SVL in vegetative bud dormancy. Curr. Biol. 29, R68–R70. doi: 10.1016/j.cub.2018.11.061
Cheng, W. H., Endo, A., Zhou, L., Penney, J., Chen, H. C., Arroyo, A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. doi: 10.1105/tpc.006494
Chmielewski, F. M., Baldermann, S., Götz, K. P., Homann, T., Gödeke, K., Schumacher, F., et al. (2018). Abscisic acid related metabolites in sweet cherry buds (Prunus avium L.). J. Hortic. 5, 1–9.
Choubane, D., Rabot, A., Mortreau, E., Legourrierec, J., Peron, T., Foucher, F., et al. (2012). Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J. Plant Physiol. 169, 1271–1280. doi: 10.1016/j.jplph.2012.04.014
Chow, B., Mccourt, P. (2006). Plant hormone receptors: perception is everything. Genes Dev. 20, 1998–2008. doi: 10.1101/gad.1432806
Considine, M. J., Considine, J. A. (2016). On the language and physiology of dormancy and quiescence in plants. J. Exp. Bot. 67, 3189–3203. doi: 10.1093/jxb/erw138
Cooke, J. E., Eriksson, M. E., Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Cutting, J., Strydom, D., Jacobs, G., Bellstedt, D., Van Der Merwe, K., Weiler, E. (1991). Changes in xylem constituents in response to rest-breaking agents applied to apple before budbreak. J. Am. Soc. Hortic. Sci. 116, 680–683. doi: 10.21273/JASHS.116.4.680
Davière, J.-M., Achard, P. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Plant Hormones 9, 10–20. doi: 10.1016/j.molp.2015.09.011
Davies, P. J. (2010). “The plant hormones: their nature, occurrence, and functions,” in Plant hormones. (Dordrecht, Netherlands: Springer), 1–15. doi: 10.1007/978-1-4020-2686-7_1
De La Fuente, L., Conesa, A., Lloret, A., Badenes, M. L., Ríos, G. (2015). Genome-wide changes in histone H3 lysine 27 trimethylation associated with bud dormancy release in peach. Tree Genet. Genomes 11, 45. doi: 10.1007/s11295-015-0869-7
Delker, C., Stenzel, I., Hause, B., Miersch, O., Feussner, I., Wasternack, C. (2006). Jasmonate biosynthesis in Arabidopsis thaliana-enzymes, products, regulation. Plant Biol. 8, 297–306. doi: 10.1055/s-2006-923935
Destefano-Beltran, L., Knauber, D., Huckle, L., Suttle, J. C. (2006). Effects of postharvest storage and dormancy status on ABA content, metabolism, and expression of genes involved in ABA biosynthesis and metabolism in potato tuber tissues. Plant Mol. Biol. 61, 687–697. doi: 10.1007/s11103-006-0042-7
Devitt, M. L., Stafstrom, J. P. (1995). Cell cycle regulation during growth–dormancy cycles in pea axillary buds. Plant Mol. Biol. 29, 255–265. doi: 10.1007/BF00043650
Di, D.-W., Zhang, C., Luo, P., An, C.-W., Guo, G.-Q. (2016). The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regul. 78, 275–285. doi: 10.1007/s10725-015-0103-5
Dobisova, T., Hrdinova, V., Cuesta, C., Michlickova, S., Urbankova, I., Hejatkova, R., et al. (2017). Light controls cytokinin signaling via transcriptional regulation of constitutively active sensor histidine kinase CKI1. Plant Physiol. 174, 387–404. doi: 10.1104/pp.16.01964
Duan, C., Li, X., Gao, D., Liu, H., Li, M. (2004). Studies on regulations of endogenous ABA and GA3 in sweet cherry flower buds on dormancy. Acta Hortic. Sinica 31, 149–154.
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). Ethylene response factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Dubois, M., Van Den Broeck, L., Claeys, H., Van Vlierberghe, K., Matsui, M., Inze, D. (2015). The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 169, 166–179. doi: 10.1104/pp.15.00335
Dutcher, R. D., Powell, L. E. (1972). Culture of apple shoots from buds in vitro. J. Am. Soc. Hortic. Sci. 97, 511–514.
Falavigna, V. D. S., Guitton, B., Costes, E., Andres, F. (2018). I want to (Bud) break free: the potential role of DAM and SVP-like genes in regulating dormancy cycle in temperate fruit trees. Front Plant Sci. 9, 1990. doi: 10.3389/fpls.2018.01990
Faust, M. (1997). Bud dormancy in perennial fruit trees: physiological basis for dormancy induction, maintenance, and release. HortScience 32, 623–629. doi: 10.21273/HORTSCI.32.4.623
Fendrych, M., Leung, J., Friml, J. (2016). TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. Elife 5, e19048. doi: 10.7554/eLife.19048
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11, e0166. doi: 10.1199/tab.0166
Finkelstein, R., Reeves, W., Ariizumi, T., Steber, C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. doi: 10.1146/annurev.arplant.59.032607.092740
Fleet, C. M., Sun, T.-P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85. doi: 10.1016/j.pbi.2004.11.015
Frigerio, M., Alabadi, D., Perez-Gomez, J., Garcia-Carcel, L., Phillips, A. L., Hedden, P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142, 553–563. doi: 10.1104/pp.106.084871
Friml, J., Palme, K. (2002). Polar auxin transport—old questions and new concepts? Plant Mol. Biol. 49, 273–284. doi: 10.1007/978-94-010-0377-3_2
Gao, Z., Chen, Y. F., Randlett, M. D., Zhao, X. C., Findell, J. L., Kieber, J. J., et al. (2003). Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 278, 34725–34732. doi: 10.1074/jbc.M305548200
Goossens, J., Fernandez-Calvo, P., Schweizer, F., Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. doi: 10.1007/s11103-016-0480-9
Guak, S., Fuchigami, L. H. (2015). Effects of applied ABA on growth cessation, bud dormancy, cold acclimation, leaf senescence and N mobilization in apple nursery plants. J. Hortic. Sci. Biotechnol. 76, 459–464. doi: 10.1080/14620316.2001.11511394
Gutierrez, C., Ramirez-Parra, E., Castellano, M. M., Del Pozo, J. C. (2002). G1 to S transition: more than a cell cycle engine switch. Curr. Opin. Plant Biol. 5, 480–486. doi: 10.1016/S1369-5266(02)00301-1
Hao, X., Chao, W., Yang, Y., Horvath, D. (2015). Coordinated Expression of FLOWERING LOCUS T and DORMANCY ASSOCIATED MADS-BOX-like genes in leafy Spurge. PLoS One 10, e0126030. doi: 10.1371/journal.pone.0126030
Hao, X., Yang, Y., Yue, C., Wang, L., Horvath, D. P., Wang, X. (2017). Comprehensive transcriptome analyses reveal differential gene expression profiles of Camellia sinensis axillary buds at para-, endo-, ecodormancy, and bud flush stages. Front Plant Sci. 8, 553. doi: 10.3389/fpls.2017.00553
Hartmann, U., Höhmann, S., Nettesheim, K., Wisman, E., Saedler, H., Huijser, P. (2000). Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 21, 351–360. doi: 10.1046/j.1365-313x.2000.00682.x
Hartmann, A., Senning, M., Hedden, P., Sonnewald, U., Sonnewald, S. (2011). Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiol. 155, 776–796. doi: 10.1104/pp.110.168252
Hauvermale, A. L., Ariizumi, T., Steber, C. M. (2014). The roles of the GA receptors GID1a, GID1b, and GID1c in sly1-independent GA signaling. Plant Signal. Behav 9, e28030. doi: 10.4161/psb.28030
He, Y., Doyle, M. R., Amasino, R. M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18, 2774–2784. doi: 10.1101/gad.1244504
Hedden, P., Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Hellman, E., Shelby, S., Lowery, C. (2006). Exogenously applied abscisic acid did not consistently delay budburst of deacclimating grapevines. J. Am. Pomol. Soc. 60, 178–186.
Higuchi, M., Pischke, M. S., Mahonen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. U.S.A. 101, 8821–8826. doi: 10.1073/pnas.0402887101
Horvath, D. P., Chao, W. S., Suttle, J. C., Thimmapuram, J., Anderson, J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9, 536. doi: 10.1186/1471-2164-9-536
Horvath, D. P., Sung, S., Kim, D., Chao, W., Anderson, J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73, 169–179. doi: 10.1007/s11103-009-9596-5
Howe, G. T., Horvath, D. P., Dharmawardhana, P., Priest, H. D., Mockler, T. C., Strauss, S. H. (2015). Extensive transcriptome changes during natural onset and release of vegetative bud dormancy in Populus. Front Plant Sci. 6, 989. doi: 10.3389/fpls.2015.00989
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Ionescu, I. A., Lopez-Ortega, G., Burow, M., Bayo-Canha, A., Junge, A., Gericke, O., et al. (2017). Transcriptome and metabolite changes during hydrogen cyanamide-induced floral bud break in sweet cherry. Front Plant Sci. 8, 1233. doi: 10.3389/fpls.2017.01233
Iqbal, N., Trivellini, A., Masood, A., Ferrante, A., Khan, N. A. (2013). Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol. Biochem. 73, 128–138. doi: 10.1016/j.plaphy.2013.09.011
Itoh, H., Matsuoka, M., Steber, C. M. (2003). A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 8, 492–497. doi: 10.1016/j.tplants.2003.08.002
Jimenez, S., Lawton-Rauh, A. L., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 9, 81. doi: 10.1186/1471-2229-9-81
Jimenez, S., Reighard, G. L., Bielenberg, D. G. (2010). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73, 157–167. doi: 10.1007/s11103-010-9608-5
Jin, Y., Zhai, S., Wang, W., Ding, X., Guo, Z., Bai, L., et al. (2018). Identification of genes from the ICE-CBF-COR pathway under cold stress in Aegilops–Triticum composite group and the evolution analysis with those from Triticeae. Physiol. Mol. Biol. Plants 24, 211–229. doi: 10.1007/s12298-017-0495-y
Johnson, P. R., Ecker, J. R. (1998). The ethylene gas signal transduction pathway: a molecular perspective. Annu. Rev. Genet. 32, 227–254. doi: 10.1146/annurev.genet.32.1.227
Juvany, M., Müller, M., Munné-Bosch, S. (2015). Bud vigor, budburst lipid peroxidation, and hormonal changes during bud development in healthy and moribund beech (Fagus sylvatica L.) trees. Trees 29, 1781–1790. doi: 10.1007/s00468-015-1259-3
Kaneko, M., Itoh, H., Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35, 104–115. doi: 10.1046/j.1365-313X.2003.01780.x
Kendall, S. L., Hellwege, A., Marriot, P., Whalley, C., Graham, I. A., Penfield, S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23, 2568–2580. doi: 10.1105/tpc.111.087643
Kieber, J. J., Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12, e0168. doi: 10.1199/tab.0168
Knight, H., Zarka, D. G., Okamoto, H., Thomashow, M. F., Knight, M. R. (2004). Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol. 135, 1710–1717. doi: 10.1104/pp.104.043562
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Le Bris, M., Michaux-Ferrière, N., Jacob, Y., Poupet, A., Barthe, P., Guigonis, J.-M., et al. (1999). Regulation of bud dormancy by manipulation of ABA in isolated buds of Rosa hybrida cultured in vitro. Funct. Plant Biol. 26, 273–281. doi: 10.1071/PP98133
Lee, J., Oh, M., Park, H., Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 55, 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Lee, K. H., Piao, H. L., Kim, H. Y., Choi, S. M., Jiang, F., Hartung, W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120. doi: 10.1016/j.cell.2006.07.034
Lee, S. H., Sakuraba, Y., Lee, T., Kim, K. W., An, G., Lee, H. Y., et al. (2015). Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 57, 562–576. doi: 10.1111/jipb.12276
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Leida, C., Conesa, A., Llácer, G., Badenes, M. L., Ríos, G. (2012). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193, 67–80. doi: 10.1111/j.1469-8137.2011.03863.x
Li, S., Dami, I. E. (2016). Responses of Vitis vinifera ‘Pinot gris’ grapevines to exogenous Abscisic Acid (ABA): I. Yield, fruit quality, dormancy, and freezing tolerance. J. Plant Growth Regul. 35, 245–255. doi: 10.1007/s00344-015-9529-2
Li, Z., Reighard, G. L., Abbott, A. G., Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530. doi: 10.1093/jxb/erp195
Li, J., Xu, Y., Niu, Q., He, L., Teng, Y., Bai, S. (2018). Abscisic Acid (ABA) promotes the induction and maintenance of pear (Pyrus pyrifolia White Pear Group) flower bud endodormancy. Int. J. Mol. Sci. 19(1): 310. doi: 10.3390/ijms19010310
Li, J., Yan, X., Yang, Q., Ma, Y., Yang, B., Tian, J., et al. (2019). PpCBFs selectively regulate PpDAMs and contribute to the pear bud endodormancy process. Plant Mol. Biol. 99, 575–586. doi: 10.1007/s11103-019-00837-7
Liao, X., Li, M., Liu, B., Yan, M., Yu, X., Zi, H., et al. (2018). Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. U.S.A. 115, E11542–E11550. doi: 10.1073/pnas.1812575115
Lim, G. H., Kachroo, A., Kachroo, P. (2016). Role of plasmodesmata and plasmodesmata localizing proteins in systemic immunity. Plant Signal. Behav 11, e1219829. doi: 10.1080/15592324.2016.1219829
Linkies, A., Leubner-Metzger, G. (2012). Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Rep. 31, 253–270. doi: 10.1007/s00299-011-1180-1
Lionakis, S. M., Schwabe, W. W. (1984). Bud dormancy in the kiwi fruit, Actinidia chinensis Planch. Ann. Bot. 54, 467–484. doi: 10.1093/oxfordjournals.aob.a086818
Liotenberg, S., North, H., Marion-Poll, A. (1999). Molecular biology of abscisic acid biosynthesis in plants. Plant Physiol. Biochem. 37, 341–350. doi: 10.1016/S0981-9428(99)80040-0
Lipavska, H., Maskova, P., Vojvodova, P. (2011). Regulatory dephosphorylation of CDK at G(2)/M in plants: yeast mitotic phosphatase cdc25 induces cytokinin-like effects in transgenic tobacco morphogenesis. Ann. Bot. 107, 1071–1086. doi: 10.1093/aob/mcr016
Liu, B., Zhao, S., Tan, F., Zhao, H., Wang, D., Si, H., et al. (2017). Changes in ROS production and antioxidant capacity during tuber sprouting in potato. Food Chem. 237, 205–213. doi: 10.1016/j.foodchem.2017.05.107
Mahonen, A. P., Higuchi, M., Tormakangas, K., Miyawaki, K., Pischke, M. S., Sussman, M. R., et al. (2006). Cytokinins regulate a bidirectional phosphorelay network in Arabidopsis. Curr. Biol. 16, 1116–1122. doi: 10.1016/j.cub.2006.04.030
Marin, E., Nussaume, L., Quesada, A. M., Sotta, B., Hugueney, P., Frey, A. (1996). Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 15, 2331–2342. doi: 10.1002/j.1460-2075.1996.tb00589.x
Mazzitelli, L., Hancock, R. D., Haupt, S., Walker, P. G., Pont, S. D., Mcnicol, J., et al. (2007). Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J. Exp. Bot. 58, 1035–1045. doi: 10.1093/jxb/erl266
Melcher, K., Zhou, X. E., Xu, H. E. (2010). Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr. Opin. Struct. Biol. 20, 722–729. doi: 10.1016/j.sbi.2010.09.007
Michaels, S. D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., Amasino, R. M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33, 867–874. doi: 10.1046/j.1365-313X.2003.01671.x
Mielke, E. A., Dennis, F. G. (1978). Hormonal control of flower bud dormancy in sour cherry (Prunus cerasus L.). III. Effects of leaves, defoliation and temperature on levels of abscisic acid in flower primordia. J. Am. Soc. Hortic. Sci. 103, 446–449.
Milborrow, B. V. (2001). The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52, 1145–1164. doi: 10.1093/jexbot/52.359.1145
Mimida, N., Saito, T., Moriguchi, T., Suzuki, A., Komori, S., Wada, M. (2015). Expression of DORMANCY-ASSOCIATED MADS-BOX (DAM)-like genes in apple. Biol. Plant. 59, 237–244. doi: 10.1007/s10535-015-0503-4
Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Mølmann, J. A., Asante, D. K. A., Jensen, J. B., Krane, M. N., Ernstsen, A., Junttila, O., et al. (2005). Low night temperature and inhibition of gibberellin biosynthesis override phytochrome action and induce bud set and cold acclimation, but not dormancy in PHYA overexpressors and wild-type of hybrid aspen. Plant Cell Environ. 28, 1579–1588. doi: 10.1111/j.1365-3040.2005.01395.x
Muller, D., Leyser, O. (2011). Auxin, cytokinin and the control of shoot branching. Ann. Bot. 107, 1203–1212. doi: 10.1093/aob/mcr069
Nagar, P. K., Sood, S. (2006). Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiol. Plant. 28, 165–169. doi: 10.1007/s11738-006-0043-9
Nakamura, S., Lynch, T. J., Finkelstein, R. R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. doi: 10.1046/j.1365-313x.2001.01069.x
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nambara, E., Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185. doi: 10.1146/annurev.arplant.56.032604.144046
Nambara, E., Nambara, E., Mccourt, P., Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636.
Niu, Q., Li, J., Cai, D., Qian, M., Jia, H., Bai, S., et al. (2016). Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 67, 239–257. doi: 10.1093/jxb/erv454
Noriega, X., Pérez, F. J. (2017). ABA biosynthesis genes are down-regulated while auxin and cytokinin biosynthesis genes are up-regulated during the release of grapevine buds from endodormancy. J. Plant Growth Regul. 36, 814–823. doi: 10.1007/s00344-017-9685-7
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Ophir, R., Pang, X., Halaly, T., Venkateswari, J., Lavee, S., Galbraith, D., et al. (2009). Gene-expression profiling of grape bud response to two alternative dormancy-release stimuli expose possible links between impaired mitochondrial activity, hypoxia, ethylene–ABA interplay and cell enlargement. Plant Mol. Biol. 71, 403–423. doi: 10.1007/s11103-009-9531-9
Or, E., Belausov, E., Popilevsky, I., Bental, Y. (2000). Changes in endogenous ABA level in relation to the dormancy cycle in grapevines grown in a hot climate. J. Hortic. Sci. Biotechnol. 75, 190–194. doi: 10.1080/14620316.2000.11511221
Parker, M. L., Clark, M. B., Campbell, C. (2012). ABSCISIC ACID APPLICATIONS IN PEACH. Acta Hortic. 962, 403–409. doi: 10.17660/ActaHortic.2012.962.55
Pierik, R., Cuppens, M. L., Voesenek, L. A., Visser, E. J. (2004). Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol. 136, 2928–2936. doi: 10.1104/pp.104.045120
Piotrowska, A., Bajguz, A. (2011). Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72, 2097–2112. doi: 10.1016/j.phytochem.2011.08.012
Qi, T., Wang, J., Huang, H., Liu, B., Gao, H., Liu, Y., et al. (2015). Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27, 1634–1649. doi: 10.1105/tpc.15.00110
Qiu, L., Xie, F., Yu, J., Wen, C. K. (2012). Arabidopsis RTE1 is essential to ethylene receptor ETR1 amino-terminal signaling independent of CTR1. Plant Physiol. 159, 1263–1276. doi: 10.1104/pp.112.193979
Qiu, Z., Wan, L., Chen, T., Wan, Y., He, X., Lu, S., et al. (2013). The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol. 199, 708–719. doi: 10.1111/nph.12301
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Riefler, M., Novak, O., Strnad, M., Schmulling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Rinne, P. L., Schoot, C. V. D. (2003). Plasmodesmata at the crossroads between development, dormancy, and defense. Can. J. Bot. 81 (12), 1182–1197. doi: 10.1139/b03-123
Rinne, P. L., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Rodriguez, J., Sherman, W. B., Scorza, R., Wisniewski, M., Okie, W. R. (1994). Evergreen peach, its inheritance and dormant behavior. J. Am. Soc. Hortic. Sci. 119 (4): 789–792. doi: 10.21273/JASHS.119.4.789
Rohde, A., Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Rohde, A., De Rycke, R., Beeckman, T., Engler, G., Van Montagu, M., Boerjan, W. (2000). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12, 35–52. doi: 10.1105/tpc.12.1.35
Rohde, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M., Boerjan, W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14, 1885–1901. doi: 10.1105/tpc.003186
Rohde, A., Van Montagu, M., Inzé, D., Boerjan, W. (1997). Factors regulating the expression of cell cycle genes in individual buds of Populus. Planta 201, 43–52. doi: 10.1007/BF01258679
Roman, H., Girault, T., Barbier, F., Péron, T., Brouard, N., Pěnčík, A., et al. (2016). Cytokinins are initial targets of light in the control of bud outgrowth. Plant Physiol. 172, 489–509. doi: 10.1104/pp.16.00530
Rothkegel, K., Sanchez, E., Montes, C., Greve, M., Tapia, S., Bravo, S., et al. (2017). DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.). Tree Physiol. 37, 1739–1751. doi: 10.1093/treephys/tpx055
Ruan, Y. L., Jin, Y., Yang, Y. J., Li, G. J., Boyer, J. S. (2010). Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol. Plant 3, 942–955. doi: 10.1093/mp/ssq044
Ruonala, R., Rinne, P. L., Baghour, M., Moritz, T., Tuominen, H., Kangasjarvi, J. (2006). Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 46, 628–640. doi: 10.1111/j.1365-313X.2006.02722.x
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. doi: 10.1105/tpc.107.052811
Saito, T., Bai, S., Imai, T., Ito, A., Nakajima, I., Moriguchi, T. (2015). Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38, 1157–1166. doi: 10.1111/pce.12469
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Santner, A., Estelle, M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. doi: 10.1038/nature08122
Santner, A., Calderon-Villalobos, L. I., Estelle, M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. doi: 10.1038/nchembio.165
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. doi: 10.1104/pp.111.181982
Sauter, A., Dietz, K. J., Hartung, W. (2002). A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ. 25, 223–228. doi: 10.1046/j.1365-3040.2002.00747.x
Schommer, C., Palatnik, J. F., Aggarwal, P., Chetelat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230
Seo, M., Hanada, A., Kuwahara, A., Endo, A., Okamoto, M., Yamauchi, Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48, 354–366. doi: 10.1111/j.1365-313X.2006.02881.x
Shan, X., Wang, J., Chua, L., Jiang, D., Peng, W., Xie, D. (2011). The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 155, 751–764. doi: 10.1104/pp.110.166595
Sherif, S., El-Sharkawy, I., Mathur, J., Ravindran, P., Kumar, P., Paliyath, G., et al. (2015). A stable JAZ protein from peach mediates the transition from outcrossing to self-pollination. BMC Biol. 13, 11. doi: 10.1186/s12915-015-0124-6
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269. doi: 10.1146/annurev.biochem.75.103004.142422
Simon, S., Petrášek, J. (2011). Why plants need more than one type of auxin. Plant Sci. 180, 454–460. doi: 10.1016/j.plantsci.2010.12.007
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkovic, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9, 4173. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Miskolczi, P., Maurya, J. P., Bhalerao, R. P. (2019). A tree ortholog of SHORT VEGETATIVE PHASE floral repressor mediates photoperiodic control of bud dormancy. Curr. Biol. 29, 128–133, e122. doi: 10.1016/j.cub.2018.11.006
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. doi: 10.1126/science.1215106
Sudawan, B., Chang, C. S., Chao, H. F., Ku, M. S., Yen, Y. F. (2016). Hydrogen cyanamide breaks grapevine bud dormancy in the summer through transient activation of gene expression and accumulation of reactive oxygen and nitrogen species. BMC Plant Biol. 16, 202. doi: 10.1186/s12870-016-0889-y
Sumitomo, K., Narumi, T., Satoh, S., Hisamatsu, T. (2008). Involvement of the ethylene response pathway in dormancy induction in chrysanthemum. J. Exp. Bot. 59, 4075–4082. doi: 10.1093/jxb/ern247
Sun, T. P. (2008). Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6, e0103. doi: 10.1199/tab.0103
Sun, T. P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21, R338–R345. doi: 10.1016/j.cub.2011.02.036
Suttle, J. C. (2004). Physiological regulation of potato tuber dormancy. Am. J. Potato Res. 81, 253–262. doi: 10.1007/BF02871767
Suttle, J. C., Huckle, L. L., Lulai, E. C. (2011). The effects of dormancy status on the endogenous contents and biological activities of jasmonic acid, n-(jasmonoyl)-isoleucine, and tuberonic acid in potato tubers. Am. J. Potato Res. 88, 283–293. doi: 10.1007/s12230-011-9192-5
Swiatek, A., Lenjou, M., Van Bockstaele, D., Inze, D., Van Onckelen, H. (2002). Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiol. 128, 201–211. doi: 10.1104/pp.010592
Takatsuka, H., Umeda, M. (2014). Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 65, 2633–2643. doi: 10.1093/jxb/ert485
Tamura, F., Tanabe, K., Itai, A. (2002). Regulation of endodormancy in Japanese pear. Acta Hortic. 587, 325–336. doi: 10.17660/ActaHortic.2002.587.44
Tanaka, M., Takei, K., Kojima, M., Sakakibara, H., Mori, H. (2006). Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45, 1028–1036. doi: 10.1111/j.1365-313X.2006.02656.x
Taylor, I. B., Burbidge, A., Thompson, A. J. (2000). Control of abscisic acid synthesis. J. Exp. Bot. 51, 1563–1574. doi: 10.1093/jexbot/51.350.1563
Torres Acosta, J. A., Fowke, L. C., Wang, H. (2011). Analyses of phylogeny, evolution, conserved sequences and genome-wide expression of the ICK/KRP family of plant CDK inhibitors. Ann. Bot. 107, 1141–1157. doi: 10.1093/aob/mcr034
Tran, L. S., Shinozaki, K., Yamaguchi-Shinozaki, K. (2010). Role of cytokinin responsive two-component system in ABA and osmotic stress signalings. Plant Signal. Behav 5, 148–150. doi: 10.4161/psb.5.2.10411
Tuan, P. A., Bai, S., Saito, T., Ito, A., Moriguchi, T. (2017). Dormancy-associated MADS-box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 58, 1378–1390. doi: 10.1093/pcp/pcx074
Tylewicz, S., Petterle, A., Marttila, S., Miskolczi, P., Azeez, A., Singh, R. K., et al. (2018). Photoperiodic control of seasonal growth is mediated by ABA acting on cell–cell communication. Science 360, 212–215. doi: 10.1126/science.aan8576
Ubi, B. E., Sakamoto, D., Ban, Y., Shimada, T., Ito, A., Nakajima, I., et al. (2010). Molecular cloning of dormancy-associated MADS-box gene homologs and their characterization during seasonal endodormancy transitional phases of Japanese pear. J. Am. Soc. Hortic. Sci. 132, 174–182. doi: 10.21273/JASHS.135.2.174
Ueda, J., Kato, J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66, 246–249. doi: 10.1104/pp.66.2.246
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Van De Poel, B., Van Der Straeten, D. (2014). 1-Aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front Plant Sci. 5, 640. doi: 10.3389/fpls.2014.00640
Varbanova, M., Yamaguchi, S., Yang, Y., Mckelvey, K., Hanada, A., Borochov, R., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45. doi: 10.1105/tpc.106.044602
Vergara, R., Noriega, X., Aravena, K., Prieto, H., Perez, F. J. (2017). ABA represses the expression of cell cycle genes and may modulate the development of endodormancy in grapevine buds. Front Plant Sci. 8, 812. doi: 10.3389/fpls.2017.00812
Verkest, A., Manes, C. L., Vercruysse, S., Maes, S., Van Der Schueren, E., Beeckman, T., et al. (2005). The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. Plant Cell 17, 1723–1736. doi: 10.1105/tpc.105.032383
Wang, D., Gao, Z., Du, P., Xiao, W., Tan, Q., Chen, X., et al. (2015). Expression of ABA metabolism-related genes suggests similarities and differences between seed dormancy and bud dormancy of peach (Prunus persica). Front Plant Sci. 6, 1248. doi: 10.3389/fpls.2015.01248
Wang, Y., Georgi, L. L., Reighard, G. L., Scorza, R., Abbott, A. G. (2002b). Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch]. J. Hered. 93, 352–358. doi: 10.1093/jhered/93.5.352
Wang, D. Z., Jin, Y. N., Ding, X. H., Wang, W. J., Zhai, S. S., Bai, L. P., et al. (2017). Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry (Mosc.) 82, 1103–1117. doi: 10.1134/S0006297917100030
Wang, K. L., Li, H., Ecker, J. R. (2002a). Ethylene biosynthesis and signaling networks. Plant Cell 14 Suppl, S131–S151. doi: 10.1105/tpc.001768
Welling, A., Kaikuranta, P., Rinne, P. (1997). Photoperiodic induction of dormancy and freezing tolerance in Betula pubescens. Involvement of ABA and dehydrins. Physiol. Plant. 100, 119–125. doi: 10.1111/j.1399-3054.1997.tb03461.x
Wen, L. H., Zhong, W. J., Huo, X. M., Zhuang, W. B., Ni, Z. J., Gao, Z. H. (2016). Expression analysis of ABA- and GA-related genes during four stages of bud dormancy in Japanese apricot (Prunus mume Sieb. et Zucc). J. Hortic. Sci. Biotechnol. 91, 362–369. doi: 10.1080/14620316.2016.1160546
Wigge, P. A. (2011). FT, a mobile developmental signal in plants. Curr. Biol. 21, R374–R378. doi: 10.1016/j.cub.2011.03.038
Wisniewski, M., Norelli, J., Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci. 6, 85. doi: 10.3389/fpls.2015.00085
Wolbang, C. M., Ross, J. J. (2001). Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta 214, 153–157. doi: 10.1007/s004250100663
Wolbang, C. M., Chandler, P. M., Smith, J. J., Ross, J. J. (2004). Auxin from the developing inflorescence is required for the biosynthesis of active gibberellins in barley stems. Plant Physiol. 134, 769–776. doi: 10.1104/pp.103.030460
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017a). SVP-like MADS box genes control dormancy and budbreak in apple. Front Plant Sci. 8, 477. doi: 10.3389/fpls.2017.00477
Wu, R., Wang, T., Warren, B. A. W., Allan, A. C., Macknight, R. C., Varkonyi-Gasic, E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68, 1071–1082. doi: 10.1093/jxb/erx014
Wu, R., Wang, T., Warren, B. A. W., Thomson, S. J., Allan, A. C., Macknight, R. C., et al. (2018a). Kiwifruit SVP2 controls developmental and drought-stress pathways. Plant Mol. Biol. 96, 233–244. doi: 10.1007/s11103-017-0688-3
Wu, R. M., Walton, E. F., Richardson, A. C., Wood, M., Hellens, R. P., Varkonyi-Gasic, E. (2012). Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. J. Exp. Bot. 63, 797–807. doi: 10.1093/jxb/err304
Wu, S. W., Kumar, R., Iswanto, A. B. B., Kim, J. Y. (2018b). Callose balancing at plasmodesmata. J. Exp. Bot. 69, 5325–5339. doi: 10.1093/jxb/ery317
Xiong, L., Zhu, J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36. doi: 10.1104/pp.103.025395
Xiong, L., Lee, H., Ishitani, M., Zhu, J. K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596. doi: 10.1074/jbc.M109275200
Xu, Z. Y., Lee, K. H., Dong, T., Jeong, J. C., Jin, J. B., Kanno, Y., et al. (2012). A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24, 2184–2199. doi: 10.1105/tpc.112.095935
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamaguchi, M., Fabian, T., Sauter, M., Bhalerao, R. P., Schrader, J., Sandberg, G., et al. (2000). Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J. 24, 11–20. doi: 10.1046/j.1365-313x.2000.00846.x
Yamane, H., Ooka, T., Jotatsu, H., Hosaka, Y., Sasaki, R., Tao, R. (2011). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62, 3481–3488. doi: 10.1093/jxb/err028
Yamane, H., Wada, M., Honda, C., Matsuura, T., Ikeda, Y., Hirayama, T., et al. (2019). Overexpression of Prunus DAM6 inhibits growth, represses bud break competency of dormant buds and delays bud outgrowth in apple plants. PLoS One 14, e0214788. doi: 10.1371/journal.pone.0214788
Yan, Y., Christensen, S., Isakeit, T., Engelberth, J., Meeley, R., Hayward, A., et al. (2012). Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 24, 1420–1436. doi: 10.1105/tpc.111.094151
Yoshida, H., Ueguchi-Tanaka, M. (2014). DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds. Plant Signal. Behav. 9, e29726. doi: 10.4161/psb.29726
Yue, C., Cao, H., Hao, X., Zeng, J., Qian, W., Guo, Y., et al. (2018). Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity–dormancy transition of tea plants. Plant Cell Rep. 37, 425–441. doi: 10.1007/s00299-017-2238-5
Zawaski, C., Busov, V. B. (2014). Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PLoS One 9, e86217. doi: 10.1371/journal.pone.0086217
Zawaski, C., Kadmiel, M., Pickens, J., Ma, C., Strauss, S., Busov, V. (2011). Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 234, 1285–1298. doi: 10.1007/s00425-011-1485-x
Zentella, R., Zhang, Z. L., Park, M., Thomas, S. G., Endo, A., Murase, K., et al. (2007). Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. doi: 10.1105/tpc.107.054999
Zhang, Y., Turner, J. G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3, e3699. doi: 10.1371/journal.pone.0003699
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., Ogas, J. (2012a). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, K., Diederich, L., John, P. C. (2005). The cytokinin requirement for cell division in cultured Nicotiana plumbaginifolia cells can be satisfied by yeast Cdc25 protein tyrosine phosphatase. Implications for mechanisms of cytokinin response and plant development. Plant Physiol. 137, 308–316. doi: 10.1104/pp.104.051938
Zhang, Y., Mechlin, T., Dami, I. (2011). Foliar application of abscisic acid induces dormancy responses in greenhouse-grown grapes. HortScience 46, 1271–1277. doi: 10.21273/HORTSCI.46.9.1271
Zhang, L., Wang, Y., Zhang, X., Zhang, M., Han, D., Qiu, C., et al. (2012b). Dynamics of phytohormone and DNA methylation patterns changes during dormancy induction in strawberry (Fragaria × ananassa Duch.). Plant Cell Rep. 31, 155–165. doi: 10.1007/s00299-011-1149-0
Zhang, Z., Zhuo, X., Zhao, K., Zheng, T., Han, Y., Yuan, C., et al. (2018). Transcriptome profiles reveal the crucial roles of hormone and sugar in the bud dormancy of Prunus mume. Sci. Rep. 8, 5090. doi: 10.1038/s41598-018-23108-9
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zhao, M. L., Wang, J. N., Shan, W., Fan, J. G., Kuang, J. F., Wu, K. Q., et al. (2013). Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 36, 30–51. doi: 10.1111/j.1365-3040.2012.02551.x
Zhao, K., Zhou, Y., Ahmad, S., Yong, X., Xie, X., Han, Y., et al. (2018). PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume. Sci. Rep. 8, 4527. doi: 10.1038/s41598-018-22537-w
Zheng, C., Halaly, T., Acheampong, A. K., Takebayashi, Y., Jikumaru, Y., Kamiya, Y., et al. (2015). Abscisic acid (ABA) regulates grape bud dormancy, and dormancy release stimuli may act through modification of ABA metabolism. J. Exp. Bot. 66, 1527–1542. doi: 10.1093/jxb/eru519
Zheng, C., Kwame Acheampong, A., Shi, Z., Halaly, T., Kamiya, Y., Ophir, R., et al. (2018). Distinct gibberellin functions during and after grapevine bud dormancy release. J. Exp. Bot. 69, 1635–1648. doi: 10.1093/jxb/ery022
Zhong, W., Gao, Z., Zhuang, W., Shi, T., Zhang, Z., Ni, Z. (2013). Genome-wide expression profiles of seasonal bud dormancy at four critical stages in Japanese apricot. Plant Mol. Biol. 83, 247–264. doi: 10.1007/s11103-013-0086-4
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L., A, M., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544. doi: 10.1073/pnas.1103959108
Zhu, Y., Li, Y., Xin, D., Chen, W., Shao, X., Wang, Y., et al. (2015). RNA-Seq-based transcriptome analysis of dormant flower buds of Chinese cherry (Prunus pseudocerasus). Gene 555, 362–376. doi: 10.1016/j.gene.2014.11.032
Zhu, Y., Nomura, T., Xu, Y., Zhang, Y., Peng, Y., Mao, B., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18, 442–456. doi: 10.1105/tpc.105.038455
Zhuang, W., Gao, Z., Wang, L., Zhong, W., Ni, Z., Zhang, Z. (2013). Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 64, 4953–4966. doi: 10.1093/jxb/ert284
Zhuang, W., Gao, Z., Wen, L., Huo, X., Cai, B., Zhang, Z. (2015). Metabolic changes upon flower bud break in Japanese apricot are enhanced by exogenous GA4. Hortic. Res. 2, 15046. doi: 10.1038/hortres.2015.46
Keywords: phytohormones, woody species, bud dormancy, endodormancy, DAM genes, hormone signaling
Citation: Liu J and Sherif SM (2019) Hormonal Orchestration of Bud Dormancy Cycle in Deciduous Woody Perennials. Front. Plant Sci. 10:1136. doi: 10.3389/fpls.2019.01136
Received: 28 June 2019; Accepted: 19 August 2019;
Published: 18 September 2019.
Edited by:
Michael James Considine, University of Western Australia, AustraliaReviewed by:
Gabino Ríos, Instituto Valenciano de Investigaciones Agrarias, SpainCopyright © 2019 Liu and Sherif. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sherif M. Sherif, c3NoZXJpZkB2dC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.