
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 August 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01032
The unfolded protein response (UPR) is activated to sustain cell survival by reducing misfolded protein accumulation in the endoplasmic reticulum (ER). The UPR also promotes cell death when the ER stress is severe. However, the underlying molecular mechanisms of UPR activity regulation and cell death transition are less understood in plants. Arabidopsis GAAP1 and GAAP3 are involved in the regulation of UPR and cell death. Five GAAP gene members are found in Arabidopsis. Here, we analyzed the function of GAAP2 in addition to GAAP1 and GAAP3 in ER stress response using single, double, and triple mutants. Results showed that single or double or triple mutants reduced plant survival and enhanced cell death under ER stress. And the sensitivity increased with the number of mutation genes increase. Quantitative real-time polymerase chain reaction analysis showed that mutation in triple genes promoted UPR signaling when confronted with mild ER stress, advanced SA target genes upregulation when confronted with severe stress. Moreover, Quantitative detection by UPLC-ESI-MS/MS showed that ER stress upregulated salicylic acid (SA) content in plants. These data suggest that GAAP1 to GAAP3 played redundant roles in cell death resistance and fine tuning UPR activation. And the anti-cell death function of GAAPs might be achieved by impairing the up-regulation of the SA pathway under ER stress.
Plants as sessile beings have evolved sensitive detection systems and sophisticated signal transduction mechanisms that help them cope under numerous stress conditions. The endoplasmic reticulum (ER) is a central organelle for protein folding and maturation. Under various adverse conditions, protein folding process will be affected resulting in the accumulation unfolded or misfolded proteins in the ER, which named as ER stress. The unfolded protein response (UPR) is a cellular response that is highly conserved in eukaryotes to obviate ER stress (Liu et al., 2012). In plants, at least two UPR pathways have been identified. One UPR pathway involves the proteolytic cleavage and release of different bZIP transcription factors (TFs), such as bZIP28/17 from the ER membrane. The displaced TFs enter the nuclei and activate the transcription of specific genes involved in various ER quality-control processes under ER stress DDIN EN.CITE (Liu and Howell, 2010a; Walter and Ron, 2011). Another pathway is sensed by the molecule IRE1. Activated IRE1 catalyzes the splicing messenger RNA encoding transcription factor bZIP60 to up-regulate the UPR-related genes encoding factors that aid in protein folding and degradation. And IRE1 also degrades mRNAs encoding proteins in the secretory pathway referred to as regulated IRE1-dependent decay (RIDD) to reduce the amount of protein entering the ER in UPR. In addition, activated IRE1 can induce autophagy to mitigate ER stress TA (Deng et al., 2011; Nagashima et al., 2011; Humbert et al., 2012; Brewer, 2014). When cytoprotective outcomes are insufficient for ER homeostasis restoration under severe or chronic stress, the UPR triggers cell death program (Liu et al., 2007; Liu and Howell, 2010b; Yang et al., 2014). However, minimal information is known about the regulation of ER-stress sensor activities and about the determination of different outcomes between cell survival and death effects in plants. Under ER stress, IRE1 and bZIP28 can induce PCD by up-regulating NAC089, which promotes cell death by activating the expression of downstream genes, such as NAC094 and ATPase (Yang et al., 2014). In addition, the cell death domain (DCD)-containing asparagine-rich proteins (NRPs) have been identified in plant as transducers of a cell death signal derived from various stresses. In Arabidopsis, DCD/NRP-mediated signaling induces cell death by sequentially activating AtNRPs, ANAC036 and gVPE genes under osmotic or ER stress (Reis et al., 2016).
The UPR plays a fundamental role in plant immunity and abiotic stress responses (Moreno et al., 2012). Under biotic stress, plants trigger a robust disease resistance at the site of infection. Stimulation of defense responses occurs not only locally but also throughout the plant, known as systemic acquired resistance (SAR). Activation of the SAR pathway involves an increase in the cellular concentration of the immune signal salicylic acid (SA). SA induces pathogenesis-related (PR) genes and a large set of ER-resident genes to ensure proper folding and secretion of the PR proteins .CITE.DATA (Wang et al., 2005; Nishimura and Dangl, 2010). SA pathway also plays a key regulatory role in the resistance and promotion of PCD under stress (Yun and Chen, 2011; Gao et al., 2017). Exogenous SA has been reported to activate two plant UPR pathway genes (Nagashima et al., 2014). However, it is not clear whether cell death induced under ER stress is relevant to SA pathway. We have previously reported that Arabidopsis GAAP1 and GAAP3, a sub-family of BI-1 factors, are involved in UPR and PCD regulation. GAAP1/GAAP3 inhibits ER stress-induced cell death and promotes plant-growth recovery by attenuating the IRE1-mediated UPR process after the ER stress relief (Guo et al., 2018). Five GAAP gene members are found in Arabidopsis. Do the homologous members show similar function? How is the function of GAAPs in anti-cell death achieved? GAAP2, also named as BIL4, localizes to the trans-Golgi network/early endosome, late endosome, and vacuolar membranes. GAAP2/BIL4 regulates cell elongation and brassinosteroid (BR) signaling via the regulation of BRI1 localization. (Yamagami et al., 2017). We analyzed the function of GAAP2 in addition to GAAP1 and GAAP3 in ER stress. The analysis of double and triple mutants showed that GAAP1 to GAAP3 played redundant roles in inhibiting cell death and delaying the ER stress-induced UPR activation. Quantitative detection by UPLC-ESI-MS/MS showed that ER stress upregulated SA content in plants. The quantitative real-time reverse-transcription polymerase chain reaction assay showed that the anti-cell death function of GAAPs might be achieved by impairing the up-regulation of the SA pathway under ER stress.
A. thaliana of ecotype Columbia-0 (Col) plants and T-DNA insertion mutants in the Col-0 background were used. The mutants gaap2-1 (CS814747) and gaap2-2 (SALK_052507C) were isolated from the Salk T-DNA collection.
gaap1-1, gaap3, and gaap1gaap3 have been previously mentioned (Guo et al., 2018). The T-DNA insertion site was confirmed by plant genomic DNA polymerase chain reaction (PCR) amplification with T-DNA and gene-specific primers. The double and triple mutants gaap1gaap2, gaap2gaap3, and gaap1gaap2gaap3 were generated by hybridization. gaap1-1 and gaap2-1 were used in the double and triple mutants. Overexpression lines 35S::GAAP1, 35S::GAAP3, and 35S::GAAP3-FLAG have been previously reported (Guo et al., 2018).
Methods for plant growth and the ER stress seedling sensitivity have been previously described (Guo et al., 2018). Briefly, tunicamycin (TM), an inhibitor of N-linked glycosylation, and dithiothreitol (DTT), a redox reagent, were used to induce ER stress. Sterile seeds were cultured on medium containing low-concentration TM for various times. Alternatively, unless otherwise specified, 3-day-old seedlings grown on a 1/2 MS agar plate were transferred onto a plate containing different concentrations of TM or DTT for various times. To test growth recovery, 4-day-old seedlings that had been infiltrated with 1/2 MS liquid salt containing 0, 0.15, and 1.0 μg ml−1 TM for 6 h were transferred onto 1/2 MS solid medium. The fresh weight of seedlings was determined during recovery time.
For quantitative real-time reverse-transcription PCR (qPCR) assay, unless specifically noted, 7-day-old seedlings were incubated with 1/2 MS liquid medium containing different concentrations of TM for various times as indicated. To analyze the effect of various abiotic stresses on the expression of GAAP2 gene, the 7-day-old seedlings were incubated with liquid medium containing 0.5 μg ml−1 TM, 50 mmol L−1 ABA, or transferred to light incubator with 37°C (HT) or 15°C (LT) for 6 h, respectively. To analyze the induction of UPR marker genes under mild ER stress, seedlings were treated with 80 ng ml−1 TM for 6 h. Seedlings were treated with 2.0 or 5 μg ml−1 TM for 6 h, which were defined as acute ER stress condition.
All the data in the paper were from at least three independent experiments subjected to Student’s t-test or two-way ANOVA analysis. An α level of 0.05 was used for statistical significance.
To generate promoter-GUS (β-d-glucuronidase) constructs, we cloned genomic DNA sequence corresponding to 903 bp upstream of the ATG codon of the GAAP2 ORF into pBI101.1 vector. A DNA sequence of 1,925 bp in length, including the promoter and the GAAP2 gene, was cloned into pBI101.1 vector to produce the pGAAP2::GAAP2. The primer pairs are listed in Supplementary Table S1, and all generated constructs were confirmed by sequencing.
Agrobacterium tumefaciens strain GV3101 was used. Transformation was performed using the floral-dip method (Clough and Bent, 1998). The phenotypic effects of pGAAP2::GAAP2 in transgenic gaap2-1 plants were analyzed in more than four independent transgenic lines. Transgenic plants carrying GAAP2::GUS in Col were further selected for GUS activity detection as previously described (Li et al., 2009; Guo et al., 2018).
The 3-day-old seedlings that were vertically cultured were incubated with 1/2 MS liquid medium containing different concentrations of TM for various times as indicated. Propidium iodide (PI) staining as fluorescent indicator of cell-membrane permeability and ion leakage assay were performed as previously described (Watanabe and Lam, 2008; Duan et al., 2010). Briefly, the seedlings were immersed in the 1/2 MS liquid salt solution containing 100 μg ml−1 PI (Sigma) and incubated for 2 min on ice under dark. And then the seedlings were washed with 1/2 MS liquid salt several times. The pictures of staining roots were then taken under fluorescence microscope (Olympus BX53, excitation = 535 nm; emission = 615 nm) with exposure time 40 ms. The fluorescence intensity of the root was measured with Image J. Cell death detection by trypan blue staining was performed as previously described (Ning et al., 2002; Duan et al., 2010). Four to five biological replicates were conducted for each staining, and at least 20 samples were determined for each genotype every replicate.
Total RNA from different tissues was extracted from a frozen tissue using TRIZOL reagent (Invitrogen), and the first cDNA strand was generated in accordance with the instructions of Superscript RT (Toyobo, Japan). qPCR analysis was performed using three to six independent biological replicates, and data were analyzed as previously reported (Schmittgen and Livak, 2008; Li et al., 2013). The relative expression levels of UPR, cell death and salicylic acid (SA) target genes were the level of each gene in different plant genotypes normalized to the level in the wild-type control, both of which were normalized to ACTIN8 expression. The specific primers for each gene were described previously (Guo et al., 2018) or listed in Table S1. Two-way ANOVA was performed, and Tukey’s range (honest significant difference) test was used to determine significant differences among genotypes or different treatments.
Quantitative detection of SA content in samples was conducted by UPLC-ESI-MS/MS analysis. The mass spectrometry system uses the API4500 triple quadrupole tandem mass spectrometry detection system of AB Sciex Corporation of the United States. The determination of SA content was assisted by Shanghai Luming Biotechnology Co., Ltd. Total SA were extracted and measured from 3-week-old plants grown at 22°C or sprayed with 0.3 μg ml−1 TM for 48 h. To extract SA, the appropriate amount of lyophilized samples were put in 1.5 ml extraction liquid (methanol: water: formic acid = 7.9:2:0.1) with steel balls, and were crushed for 2 min followed by ultrasonication for 30 min at low temperature and then incubated overnight at 4°C. The supernatant was removed by centrifugation at 13,000 rpm for 1 min at 10°C, and the filter residue was added by 1.0 ml extraction liquid followed by ultrasonication for 30 min and further immersed for 60 min at 4°C. The supernatant by centrifugation was combined with the extraction at the first time and the methanol was evaporated. Then the extracted residue was purified by MAX solid phase extraction column. The eluate was concentrated and dried in vacuo. The last extracted residue was dissolved in 100 μl methanol and filtered with a 0.2 μm nylon membrane before detection. And the data was obtained from at least four biological replicates.
To illustrate the expression patterns of GAAP2, we generated three lines of GAAP2 promoter fusion GUS (pGAAP2::GUS) expression transformants and performed GUS staining analysis in addition to RT-PCR assay. GAAP2 was expressed in various tissues, including cotyledon, leaf, root and reproductive organs (Figures 1 and 2A). During vegetative development, strong expression signals were found in cotyledon tip, leaf primordia, root apical, and lateral root primordia, followed by the leaf vein (Figures 1A–G). With flower development, GUS activity was strong in the anther and ovules but not in the petals (Figures 1H–M).
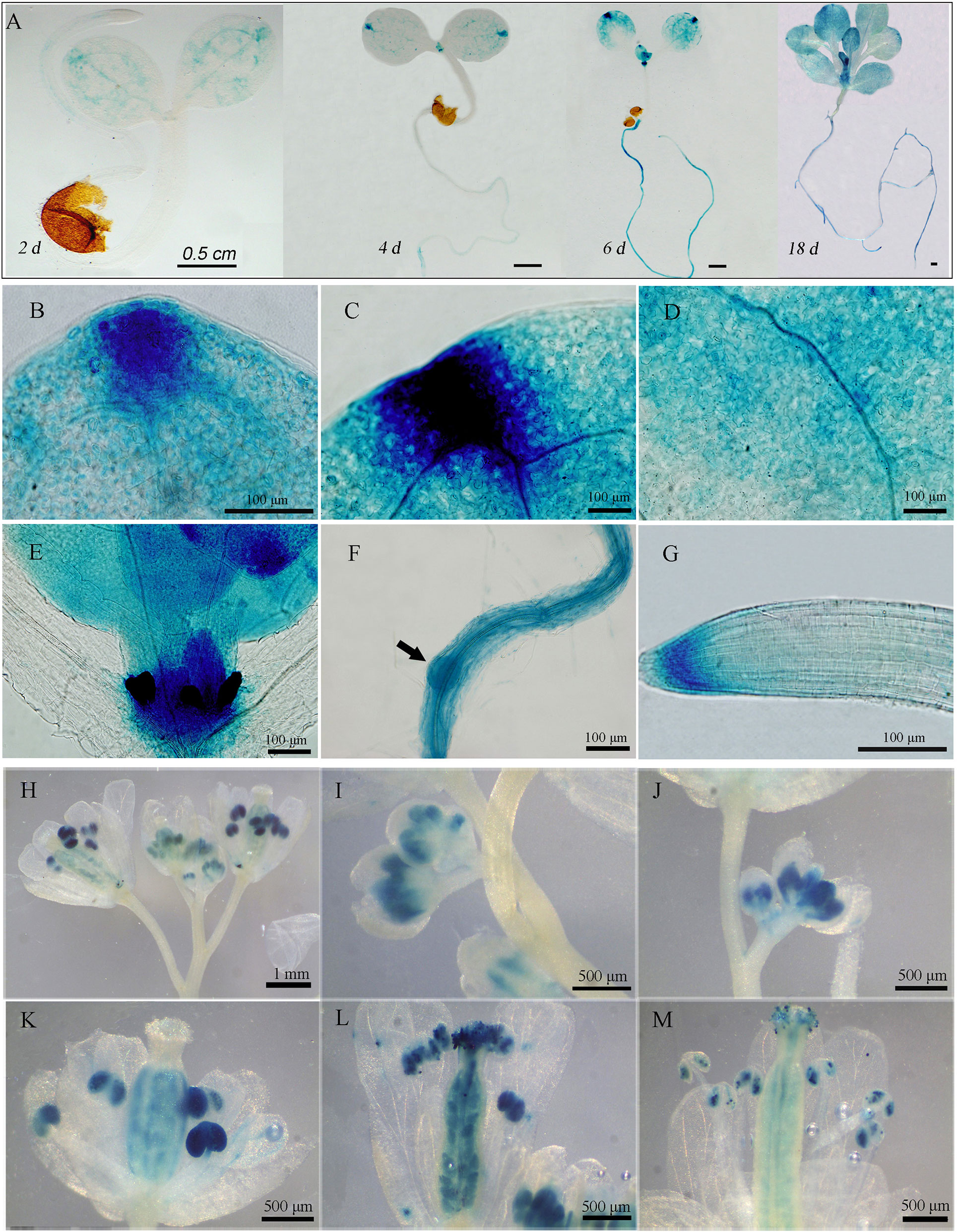
Figure 1 Expression pattern of GAAP2 assayed by GAAP2 promoter fusion GUS reporter assay. (A–G) The expression pattern of GAAP2 during the vegetative growth stage. The GUS staining of GAAP2 in the whole seedling at 2 days to 18 days (A), euphylla tip (B), cotyledon tip (C), cotyledon veins (D), leaf primordium and shoot apical (E), lateral root primordia, (F) and primary root tips (G) of 6 day-old pGAAP2::GUS transgenic plants. The expression pattern of GAAP2 during the reproductive growth stage (H–M). The GUS staining of GAAP2 in the whole inflorescence (H), flower buds at stages 9–11 (I and J), 12 (K), 14 (L), and 15 (M).
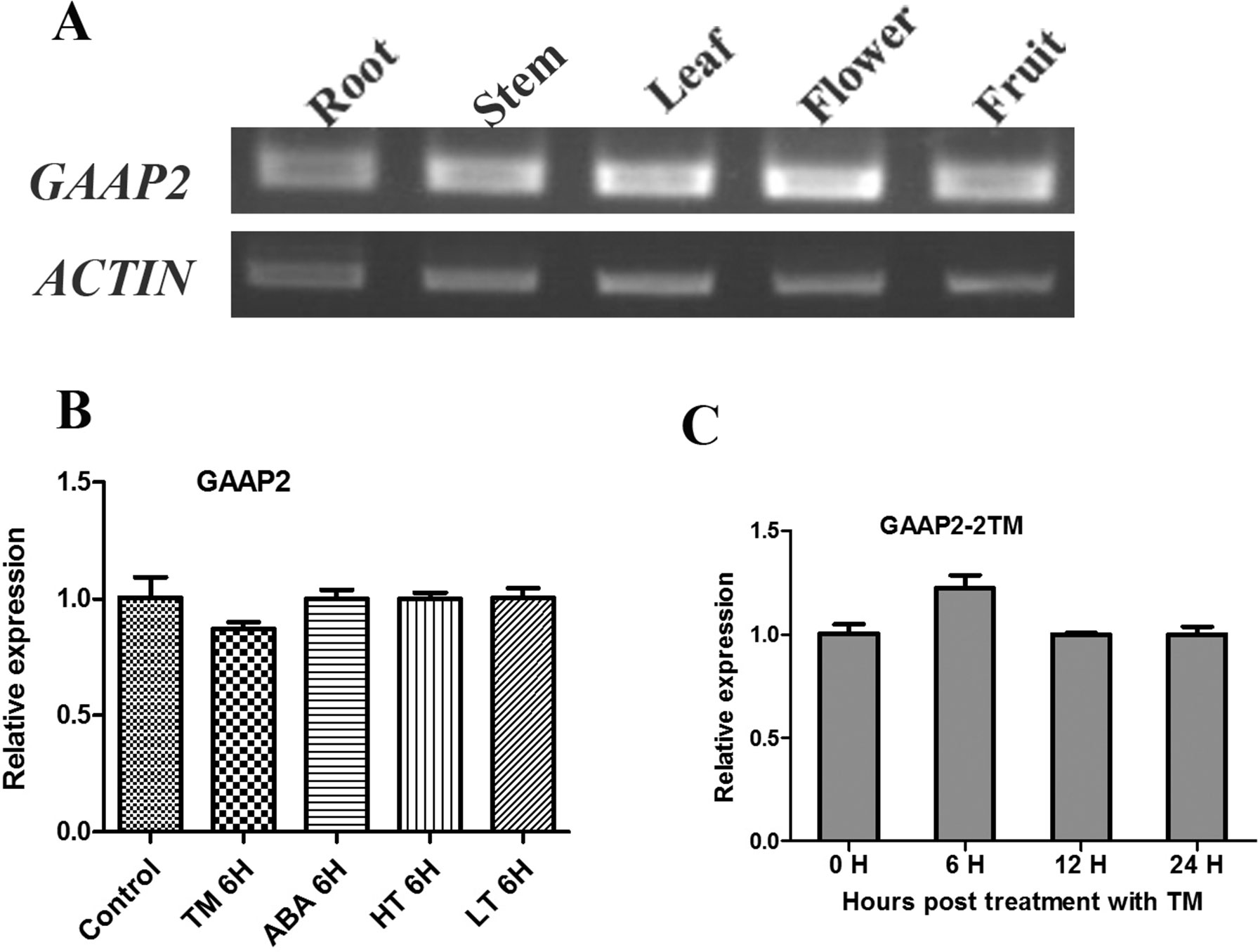
Figure 2 Transcript level of GAAP2 was not affected by various abiotic stresses. (A) The transcription level of GAAP2 in different tissues of 50-day-old plants assayed by RT-PCR. (B–C) The transcription level of GAAP2 in 7-day-old seedlings treated with different stresses assayed by qRT-PCR. The levels in seedlings in the absence or presence of 0.5 μg ml−1 TM, 50 mmol L−1 ABA, 37°C (HT) and 15°C (LT) for 6 h, respectively (B). The levels in seedlings in the presence of 2 μg ml−1 TM at different hours (C). The relative gene expression was normalized to the level of the control and was normalized to ACTIN8 expression. Data are from three biological replicates (± SD). Statistical significance was analyzed in accordance with Tukey’s range (honest significant difference) test and two-way ANOVA (p< 0.05).
The transcription of GAAP1 and GAAP3 genes are induced by ER stress and abiotic stress (Guo et al., 2018). The effects of tunicamycin (TM) or ABA, high or low temperature treatment on the transcription of GAAP2 gene during the seedling stage were tested. As shown in Figures 2B, C, GAAP2 expression was unchanged by the abiotic stresses.
To determine whether GAAP2 exhibits similar function as GAAP1 and GAAP3 in ER stress resistance, we obtained loss-of-function mutant gaap2-1 and gene knock-down mutant gaap2-2 in addition to transgenic line expressing GAAP2 driven by its own promoter in gaap2-1 (GAAP2 in gaap2-1) (Figures 3A–C). All mutants and transgenic plants did not exhibit evident aboveground growth defects under normal growth condition. When seedlings were transferred to a medium containing TM (0.5 μg ml−1) to grow for a couple of days, plant survival decreased with increasing time. The mortality rate was highest in gaap2-1, followed by gaap2-2 (Figures 3D, E), which were consistent with the GAAP2 gene level in the plant. The effect of GAAP2 on ER stress resistance at the cellular level was further analyzed. When seedlings of gaap2 mutants and complement lines was treated with 0.3 μg ml−1 TM for 48 h, cell death and membrane permeability were determined by trypan blue staining and electrical conductivity, respectively. The cells of gaap2-1 mutant showed the most damage, whereas gaap2-2 and GAAP2 in gaap2-1 showed similar cell damage as Col (Figure 4). These data suggested that the lost GAAP2 enhanced the cell death under ER stress induced by TM and GAAP2 conferred increased ER stress tolerance.
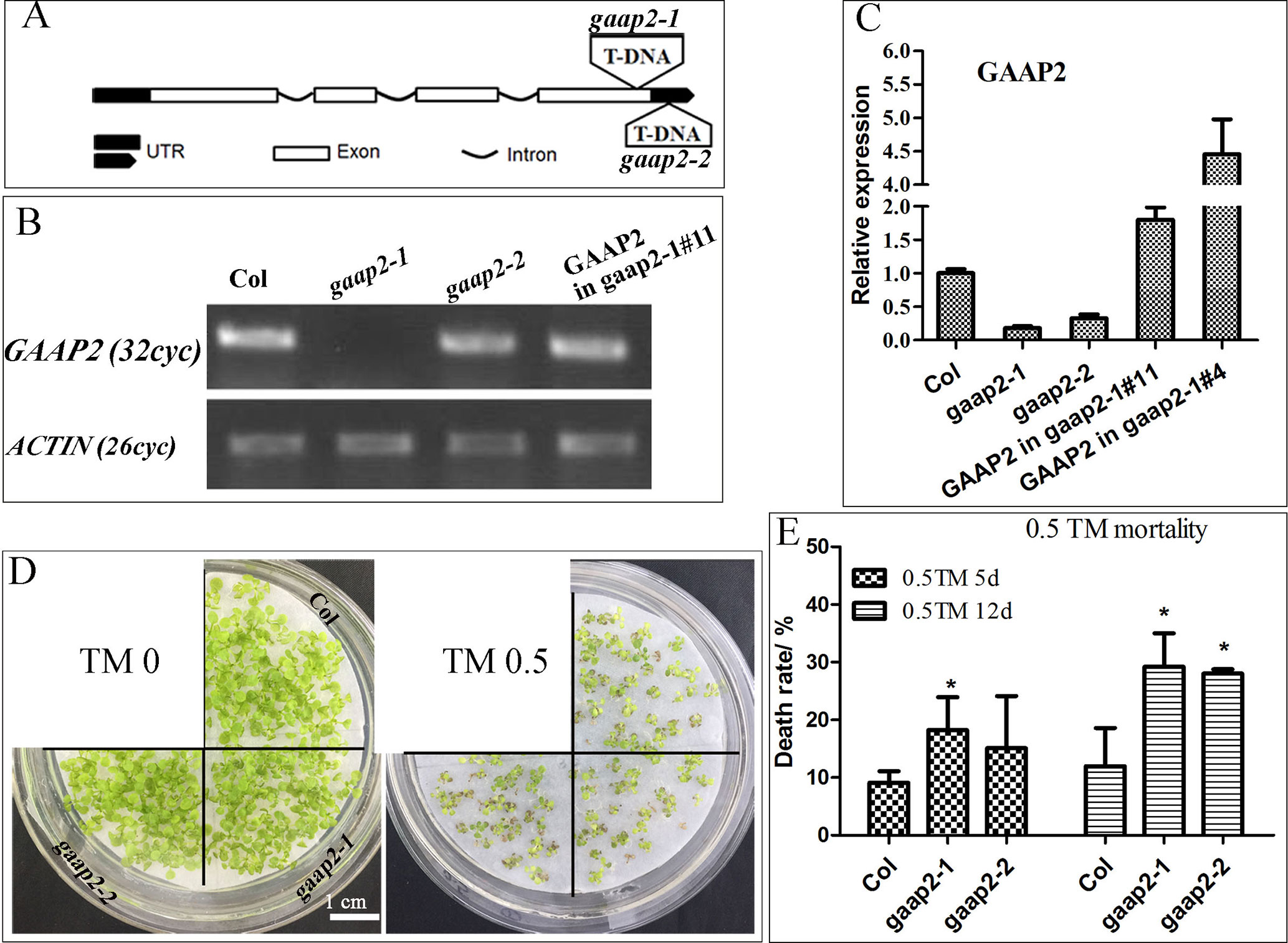
Figure 3 GAAP2 mutation increased the sensitivity of seedlings to TM treatment. (A) The schematic of T-DNA insertion sites in gaap2-1 and gaap2-2. (B–C) The expression of GAAP2 in 7-day-old seedlings of Col, gaap2-1, gaap2-2, and two transgenic lines with GAAP2 in gaap2-1was assayed by RT-PCR (B) and qRT-PCR (C). (D–E) GAAP2 mutation enhanced TM-induced plant death. Col, gaap2-1 and gaap2-2 seedlings were grown on 1/2 MS medium for 3 days and then transferred to 1/2 MS medium supplied with 0 or 0.50 μg ml−1 TM for another 12 days (D). The death rates of seedlings treated with 0.5 μg ml−1 TM for 5 and 12 days (E). Error bars depict a standard error of approximately three to four independent experiments. Significant differences compared with Col plants, as indicated by asterisks (χ2 test, *p < 0.05, n > 80).
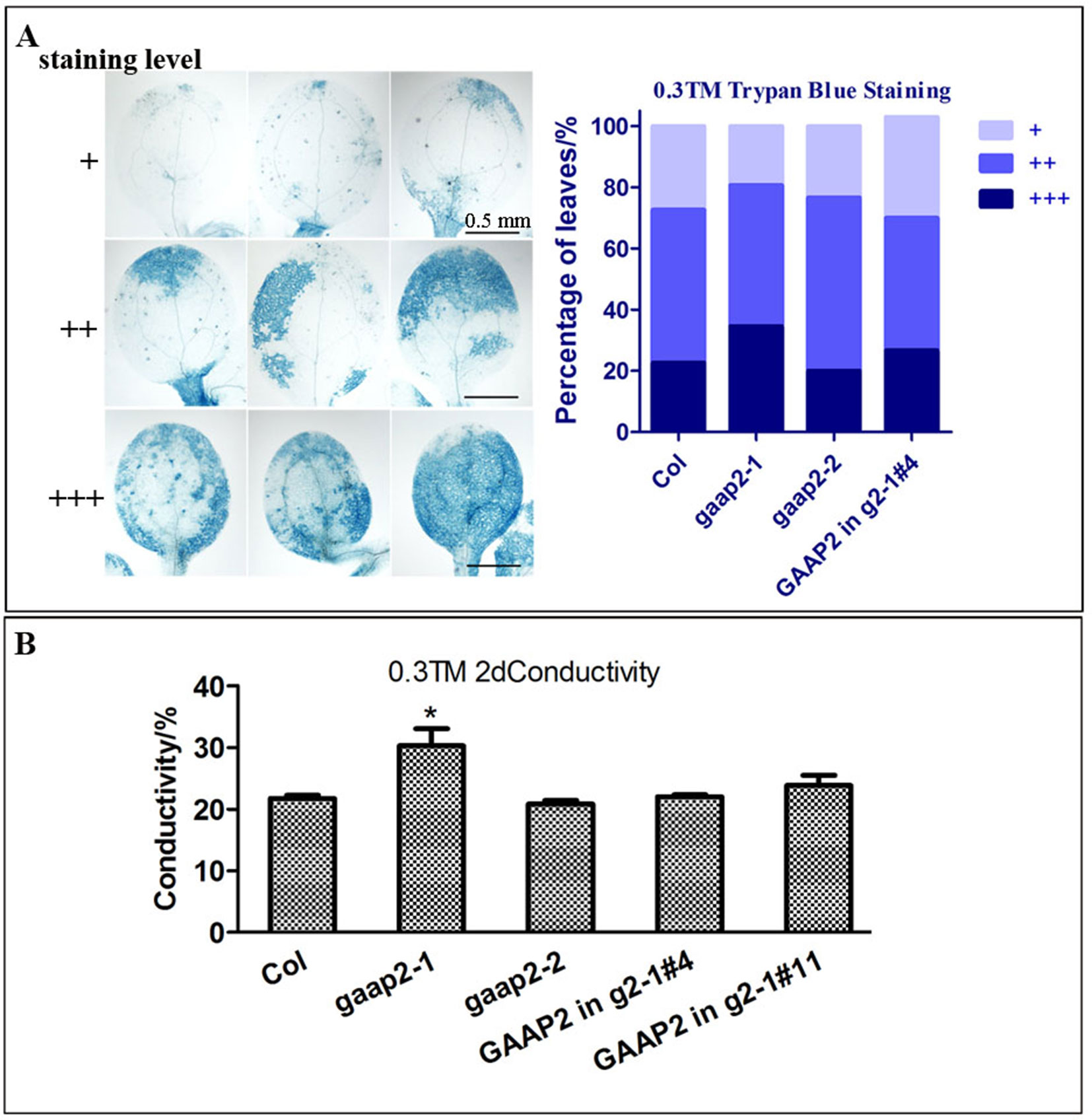
Figure 4 GAAP2 mutation enhanced cell death induced by ER stress. (A) The seedlings of 3-day-old Col, gaap2-1, gaap2-2 and GAAP2 in gaap2-1, which were soaked in 0.3 μg ml−1 TM for 48 h, were stained with trypan blue. The staining intensity of the cotyledons was classified in three levels, and cell death severity was analyzed by quantifying the staining degree. Data are shown as mean ± SD (n >25). (B) Shoots were collected from the samples obtained from the different sets of seedlings performed for (A) and then subjected to ion leakage measurements. Data are expressed as mean ± SD. Significant differences compared with Col plants are indicated by asterisks (*p < 0.05, n = 5).
When seedlings cultured in sterile medium or growing on soil were treated with DTT, the death rate and leaf etiolation rates were significantly higher in both gaap2 mutants than that in Col, GAAP2 in the mutant background partially restored the mortality rate and etiolation (Supplementary Figure S1).
GAAP1 and GAAP3 display redundant function in plants resistance to ER stress (Guo et al., 2018). To determine if GAAP1, GAAP2, and GAAP3 play functional redundant role in enhancing plant resistance to ER stress, we generated double mutants, gaap1gaap2, gaap2gaap3, and triple mutant gaap1gaap2gaap3 (Supplementary Figure S2). No obvious defects were found in double and triple mutants except for their slightly early bolting time. In terms of health rate, mortality and germination rate, both the seedling damage and growth inhibition by TM treatment were most severe in triple mutant, followed by double mutants (Figure 5 and Supplementary Figure S3), which conferred the function redundancy of the three GAAP homologous genes.
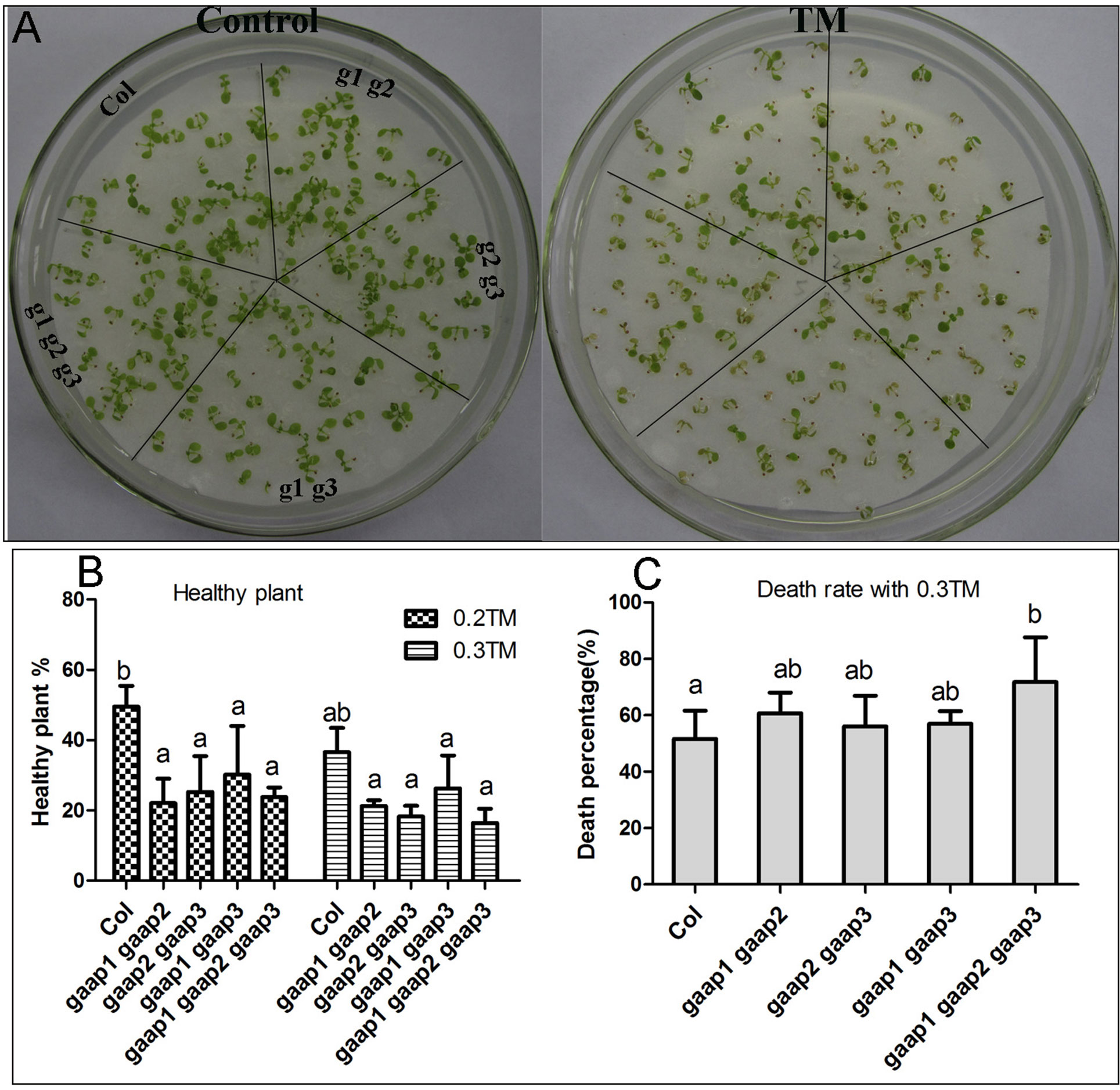
Figure 5 Plant sensitivity toward TM treatment increased with the increasing mutant gene number of GAAP1, GAAP2 and GAAP3. (A) The growth of Col, gaap1gaap2, gaap2gaap3, gaap1gaap3 and gaap1gaap2gaap3 seedlings on 1/2 MS medium for 3 days and then transferred on 1/2 MS medium supplied with 0–0.20 μg ml−1 TM for another 6 d. (B–C). Healthy plant (B) and death rates (C) of seedlings, which were treated with 0.2 and 0.3 μg ml−1 TM for 6 days. Error bars depict the standard error of four independent experiments, and different letters indicate significant differences between different plants subjected to χ test (*p < 0.05, n > 100).
In addition, when seedlings were treated with 0.3 and 0.5 μg ml−1 TM for 48 h, more dead cells were found in the cotyledon and root of gaap2gaap3 double mutants than in gaap2 single mutants, whereas the least was found in root cells of GAAP2 over-expression line (GAAP2 in gaap2-1 #4) (Supplementary Figures S4 and S5). The TM-induced cell viability of the triple mutant gaap1gaap2gaap3 and double mutants gaap2gaap3 were also evaluated with trypan blue and PI staining respectively. And the root cells of gaap1gaap2gaap3 triple mutant showed the strongest and largest area of signals, followed by double mutant (Figure 6), indicating that the higher order of the mutations, the more severe the cell damage under TM. Under dark condition, the degree of yellow leaves in vitro in the presence or absence of TM was enhanced in the triple mutant (Supplementary Figure S6). These data indicated that mutations in GAAP1, GAAP2, and GAAP3 played redundant roles in cell death resistance under ER stress.
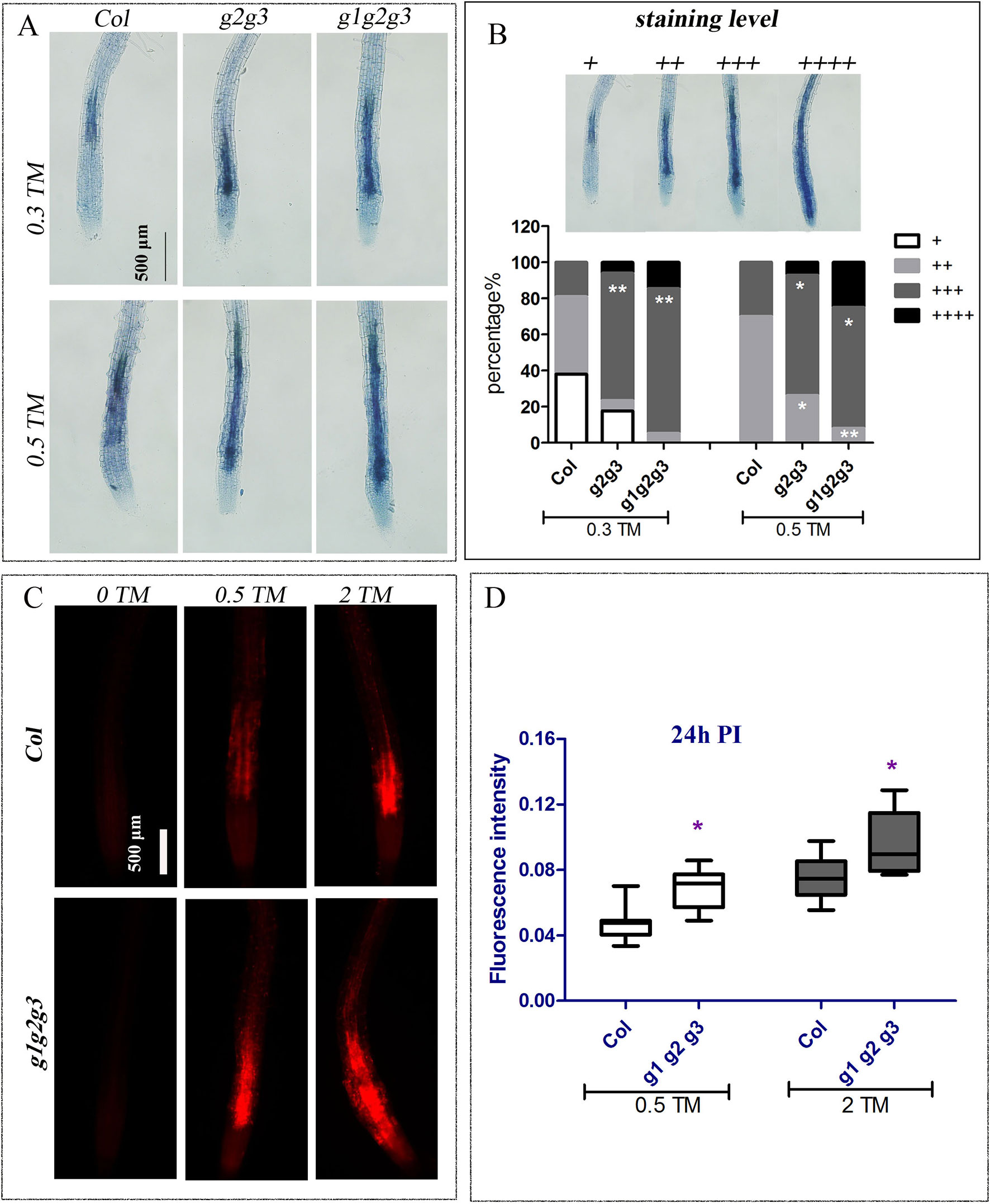
Figure 6 Mutation of GAAP1, GAAP2 and GAAP3 promoted ER stress-induced cell death on root. The seedlings of Col, gaap2gaap3 and gaap1gaap2gaap3 that were vertically cultured were transferred to a liquid medium containing different concentrations of TM (0, 0.30, 0.50, and 2.0 μg ml−1). (A–B) The root cells were assayed by trypan blue staining 48 h post treatment and representative images were shown (A). The classification of staining level and the cell death severity were analyzed by quantifying the staining degree (B). Data are from three biological replicates (± SE) and at least 20 samples for each plant line were used for each treatment (χ test, *p < 0.05, **p < 0.01). (C–D) The cell membrane permeability of root cells was assayed by PI staining 24 h post TM treatment (C), and the fluorescence intensity by Image J program (D). Significant differences compared with Col plants are indicated by asterisks (Student test, p < 0.05, n ≥ 30).
GAAP1 and GAAP3 are conductive for plant growth during mild ER stress recovery (Guo et al., 2018). We further examined the growth of the triple mutant after a brief ER stress. The 4-day-old seedlings were infiltrated with 0, 0.15, and 1.0 μg ml−1 TM for 6 h and then transferred to the normal solid medium. Consistent with previous result, the decreased fresh seedling weight of the GAAP3-overexpression line was remarkably lower than that of the wild type, whereas the inhibition rate of gaap1gaap2gaap3 seedling was higher (Guo et al., 2018) (Figure 7). These data showed that GAAP1 to GAAP3 played redundant roles in ER stress response.
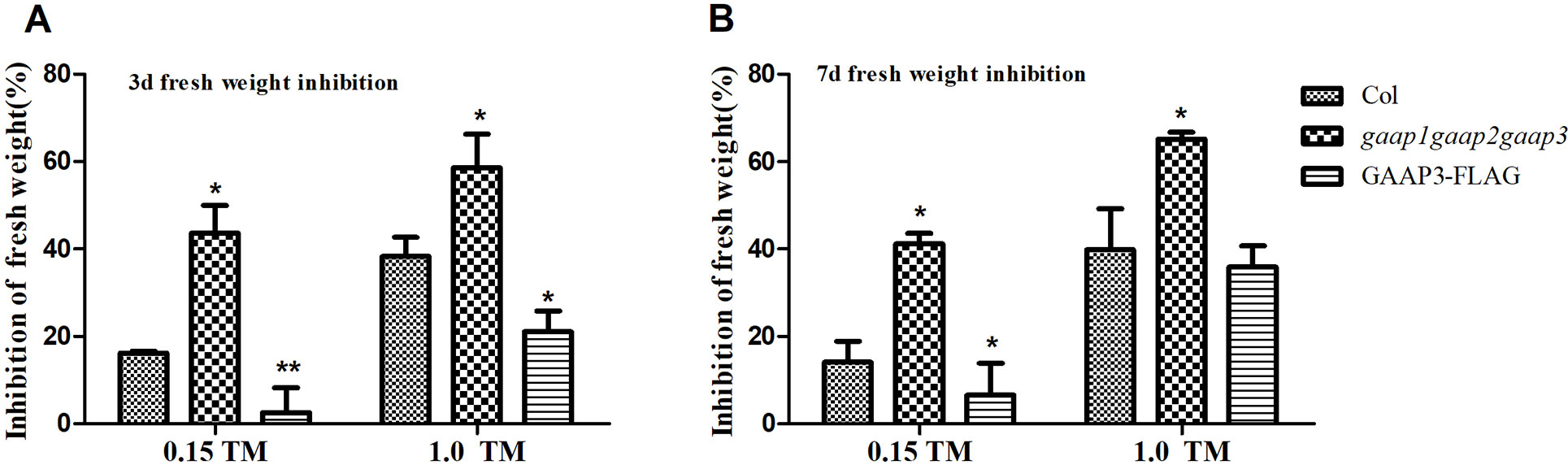
Figure 7 Mutation of GAAP1, GAAP2 and GAAP3 inhibited growth recovery after brief ER stress. (A–B) The 4-day-old seedlings that were infiltrated with 1/2 MS liquid salt containing 0, 0.15, and 1.0 μg ml−1 TM for 6 h were transferred to the 1/2 MS solid medium. The inhibition rates of the fresh weight of seedlings were determined after recovery for 3 and 7 days. Data are from three biological replicates (± SE), and at least 30 samples for each plant line were used for each treatment. Asterisks refer to significant differences from Col at the same time points and same conditions according to Tukey’s range (honest significant difference) test and two-way ANOVA (*p < 0.05, **p < 0.01).
Seedlings with mutations in GAAP1, GAAP2 and GAAP3 genes were more sensitive to ER stress than the wild type and double mutants. Under ER stress, UPR signaling pathways were initiated to alleviate stress (Walter and Ron, 2011; Deng et al., 2013). GAAP1 and GAAP3 slightly affect UPR under acute ER stress but play antagonistic roles in UPR gene induction at the beginning of mild ER stress (Guo et al., 2018). We further detected the transcription levels of the common target gene downstream of the UPR pathway in triple mutants treated with 80 ng ml−1 TM for 6 h, that is, short-term mild ER stress. The up-regulation of BIP3, bZIP60s, CNXI, SHP70, and SHD were all considerably higher in the gaap1gaap2gaap3 than in the wild type (Figure 8). These data suggested that GAAP1, GAAP2 and GAAP3 played redundant negative roles in the mild stress-activated UPR protective response. Upon acute ER stress caused by 2.0 or 5.0 μg ml−1 TM for 6 h, the induction values of the markers in Col, gaap2-1, gaap2gaap3, and gaap1gaap2gaap3 plants were similar (Supplementary Figures S7 and S8), which suggested that GAAP1 to GAAP3 mutations had little effect on the induction of UPR genes upon acute ER stress.
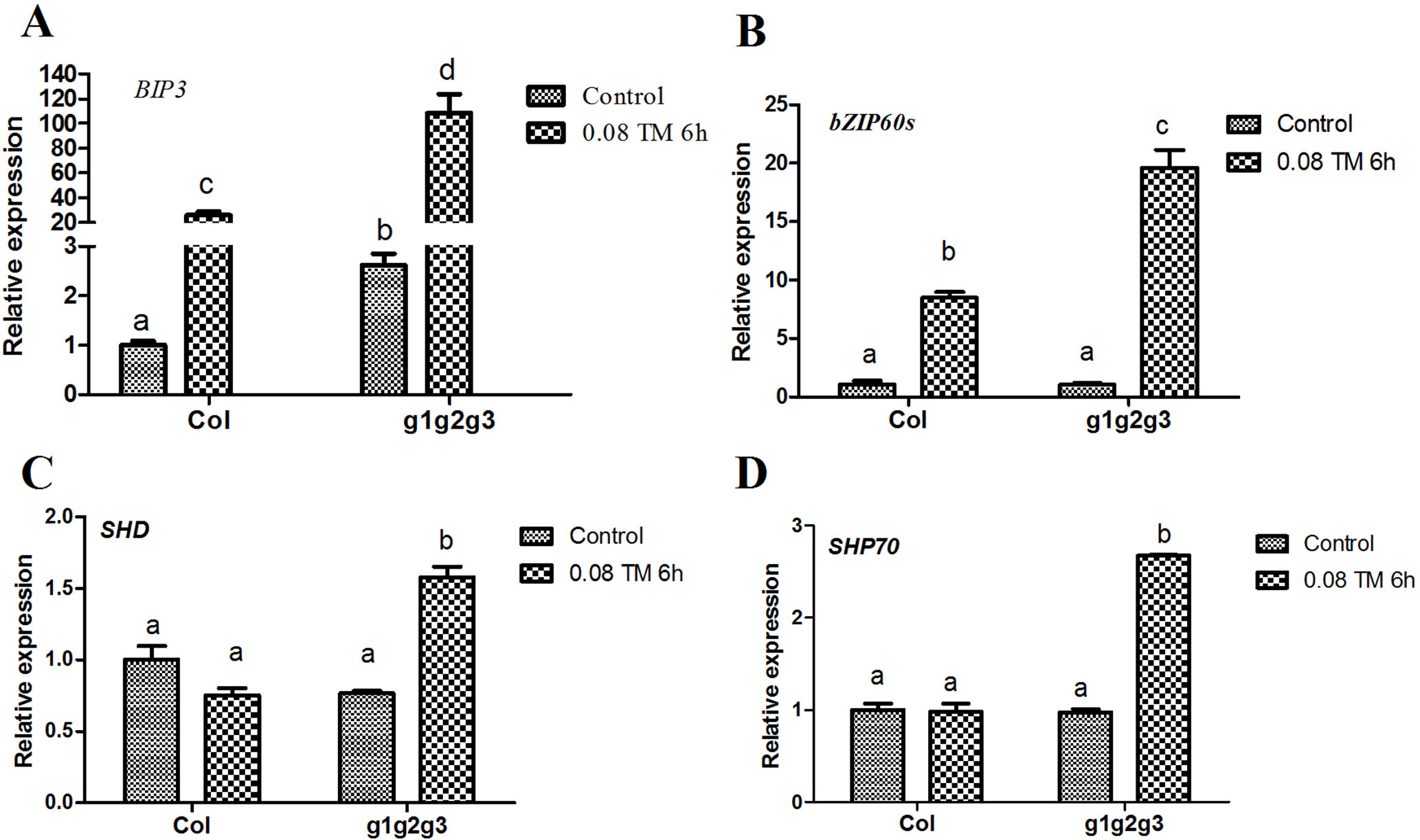
Figure 8 Mutations of GAAP1, GAAP2 and GAAP3 enhanced the up-regulation of mild ER stress-induced UPR gene. (A–D) The transcription levels of selected UPR marker genes in Col and gaap1gaap2gaap3 seedlings treated with 80 ng ml−1 TM for 6 h were quantified by qRT–PCR. The value of each control of Col was set at 1. Error bars represent the standard deviation. Different letters indicate significant differences between different plants according to Tukey’s range (honest significant difference) test and two-way ANOVA (p< 0.05, n = 3).
Under ER stress, IRE1 and bZIP28 can induce PCD by up-regulating NAC089, which promotes cell death by activating the expression of downstream PCD-related genes, such as NAC094 and ATPase (Yang et al., 2014). We have shown that cell death began to appear when 0.3 or 0.5 μg ml−1 TM for 48 h. To clarify the effect of GAAPs on NAC089 pathway-mediated cell death, the gene expression levels of NAC089 and its targets were analyzed at 24 and 48 h post TM treatment. The up-regulation of NAC089, NAC094 and ATPase genes in gaap1gaap2gaap3 was not remarkably different from that in wild type in response to TM treatment (Figures 9A–C). DCD/NRP also mediate cell death under ER stress (Reis et al., 2016). During ER stress, AtNRP1 activates downstream AtNRP2 and ANAC036, thereby ultimately activating the vacuolar processing enzyme VPEg to induce PCD (Reis et al., 2016). The expressions of AtNRP1 and ANAC036 genes were up-regulated in both triple mutant and wild type under TM treatment, but no significant difference of upregulation was observed between them (Figures 9D–F). These data suggest that GAAPs inhibiting PCD might not be mediated by NAC089 and DCD/NRP pathways.
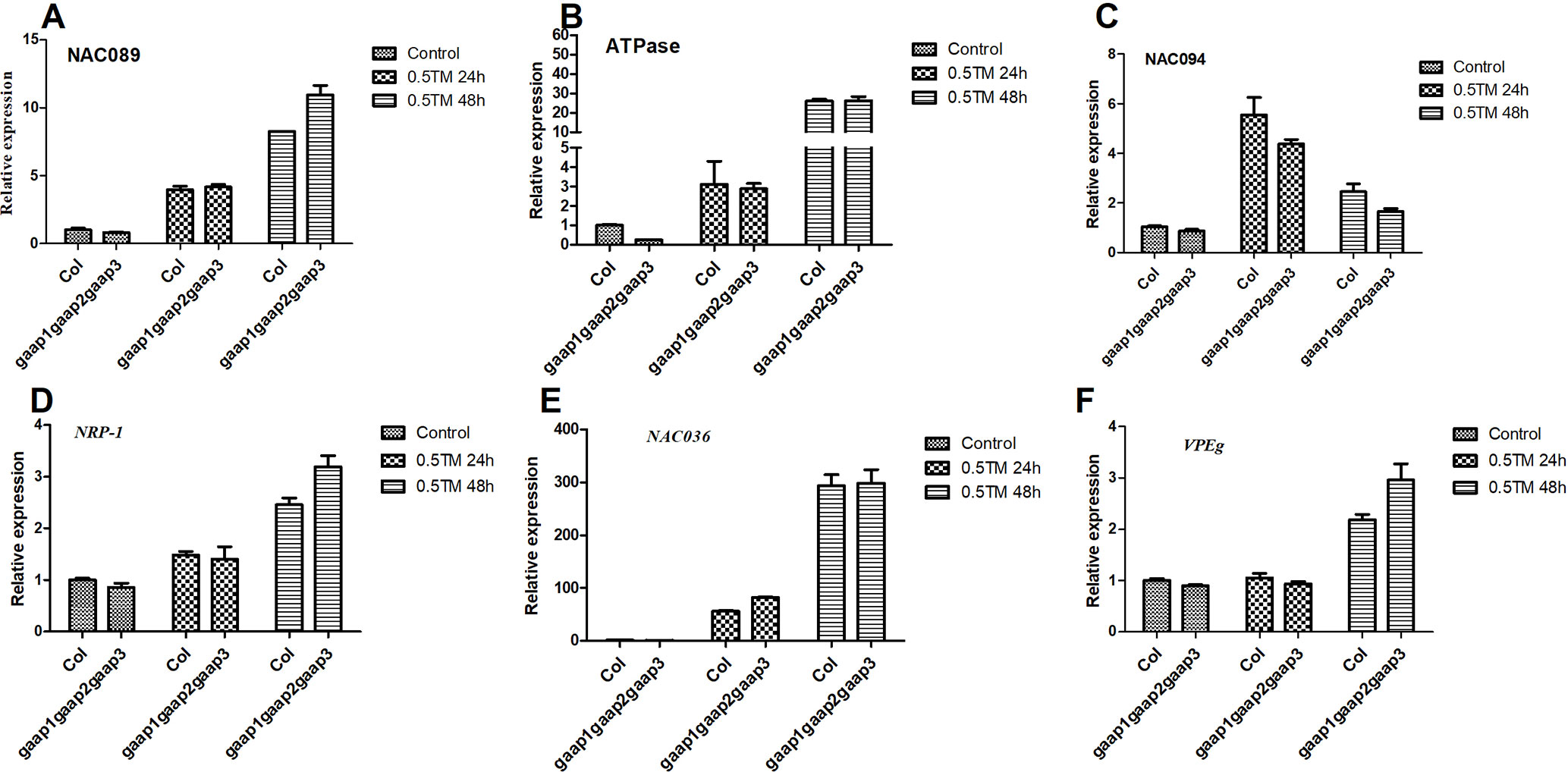
Figure 9 Upregulation of PCD genes of NAC089 pathway and DCD/NRP pathway by ER stress was not affected by mutations in GAAP1, GAAP2 and GAAP3. (A–F) The seedlings of Col and gaap1gaap2gaap3 treated with 0.5 μg ml−1 TM for 24 and 48 h. The expression levels of PCD genes in NAC089 pathway (A–C) and in DCD/NRP pathway (D–F) assayed by qRT-PCR. The value of each gene in Col under controlled condition was set at 1. Data were from three biological replicates (± SD). No significant differences compared with Col plants under the same condition were found (Student test, p < 0.05).
In addition, the SA pathway also plays a key regulatory role in the resistance and promotion of PCD under stress (Yun and Chen, 2011; Gao et al., 2017). PR-1 and PR-2 are key target genes in the SA pathway. Quantitative detection showed that the expression levels of PR-1 and PR-2 in the triple mutant gaap1gaap2gaap3 were considerably higher than those in the wild type and GAAP3 overexpressing lines at 24 h post TM treatment (Figures 10A, B). This finding suggests that the promotion of PCD by mutations in GAAP1 to GAAP3 under ER stress might be mediated by the SA pathway. To further determine whether SA content can be affected under ER stress, The total content of SA was measured from 5-week-old plants grown at 22°C or treated with 0.3 μg ml−1 TM for 48 h. And the result showed that SA level was upregulated by ER stress (Figure 10C).
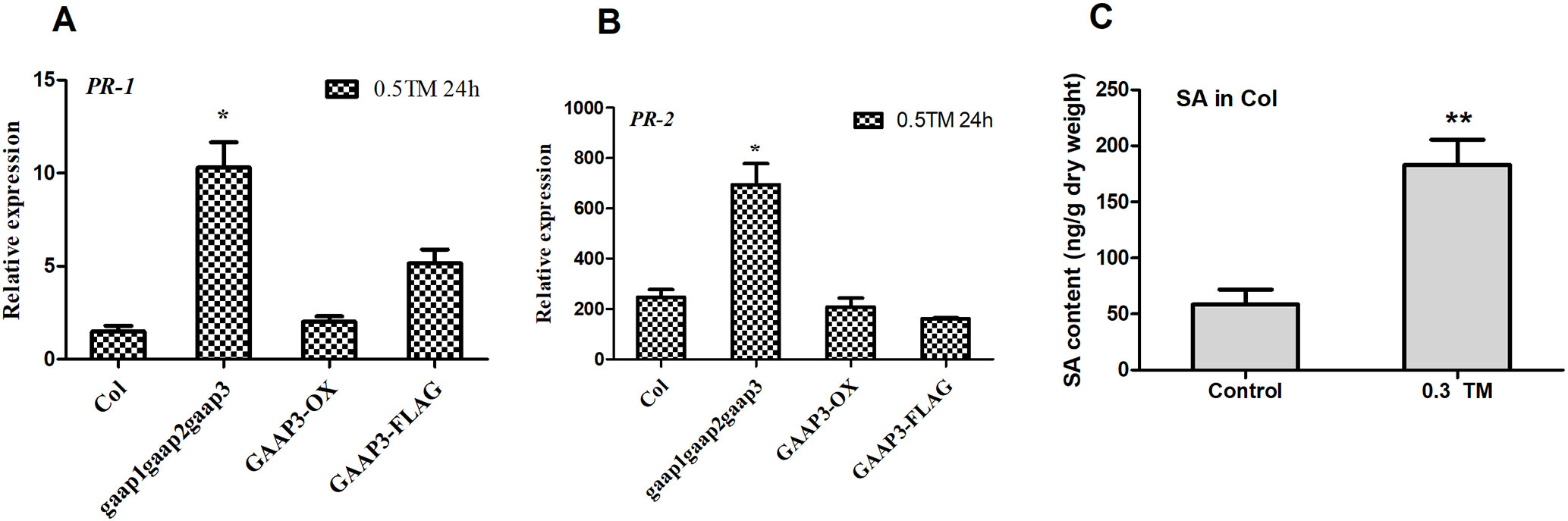
Figure 10 Mutations of GAAP1, GAAP2, and GAAP3 enhanced the upregulation of the target genes downstream SA pathway under ER stress. (A–B) The seedlings of Col, gaap1gaap2gaap3, GAAP3-OX, and GAAP3-FLAG were treated with 0.5 μg ml−1 TM for 24 h, and the expression levels of SA pathway genes were quantified by qRT–PCR. The value of each gene in Col at 0 h was set at 1. (C) SA accumulation under ER stress. Total SA were extracted and measured from 5-week-old plants grown at 22°C or treated with 0.3 μg ml−1 TM for 48 h. Data are from four biological replicates (± SD). Data were from four biological replicates (± SD). Significant differences compared with Col plants were indicated by stars (Student test, *p < 0.05, ** p < 0.01)
GAAPs are broadly conserved cytoprotective proteins that are localized in the ER and Golgi apparatus membranes (Henke et al., 2011; Guo et al., 2018). Arabidopsis GAAP1 and GAAP3 played roles in resistance to ER stress and also in the delayed activation of UPR signaling pathway by interacting with UPR receptor IRE1 (Guo et al., 2018). The sensitivity of gaap2 single, double or triple mutants with gaap1 or gaap3 to ER stress increased as the number of gene mutations increases assayed by plant or cell survival rate (Figures 3–6 and Supplementary Figure S4), indicating that GAAP1 to GAAP3 played redundant roles in maintaining plant growth and survival under ER stress, at least by partially attenuating cell death. GAAP1 and GAAP3 genes were mainly expressed in the young seedlings and preferentially expressed in the reproductive organs, whereas GAAP2 was strongly expressed in all parts throughout the entire development process (Guo et al., 2018)(Figures 1 and 2). The data suggested that different GAAPs might be involved in resistance to stress with tissue and developmental specificity. It’s worthy to note that GAAP2 in gaap2-1 showed only partial restoration under ER stress induced by DTT (Supplementary Figure S1). There are two possible reasons. One is that the gene driven by its own promoter without 3’-UTR region is not fully regulated. Another possible reason is that DTT, being a redox reagent, affects not only protein processing but also many other biological pathways dependent on the cellular redox state. And GAAP2 might also participate in the regulation of other biological process with accurate expression levels.
IRE1A/B and bZIP28 are the sensors mediated two main pathways of UPR in plants (Liu et al., 2007; Mishiba et al., 2013). Unmitigated ER stress induces PCD in animals and plants (Lisbona et al., 2009; Yang et al., 2014). However, there is limited knowledge regarding how the activities of UPR sensors are regulated to ensure plant growth under different stress intensities. Arabidopsis BI-1 attenuates the pro-survival function of bZIP28, but it does not temper the ribonuclease activity of IRE1 in recovery roles from temporary ER stress (Ruberti et al., 2018). GAAP1/GAAP3 function as the inhibitor of UPR dependent on IRE1 to reject the protective pathway at stress mitigation (Guo et al., 2018). Here, we showed that the upregulation of protective marker genes of both UPR pathways were considerably enhanced in triple mutant gaap1gaap2gaap3 under mild ER stress (Figure 8) and no significantly different induction levels in most UPR genes were observed when plants were treated with high doses of TM, namely, 2–5 μg ml−1, for a few hours (Supplementary Figures S7 and S8), indicating that GAAP1 to GAAP3 are fine modulator of UPR and might function as inhibitor in UPR protective response. The level of GAAPs might determine the sensitivity of cells to stress response to some extent. The mechanism needs further elucidation.
Under severe or persistent ER stress, cell death follows the induction of UPR (Watanabe and Lam, 2008; Ishikawa et al., 2011). The transcription factor NAC089 can be induced by IRE1 and bZIP28 to promote cell death by activating the expression of downstream PCD-related genes, such as NAC094 and ATPase (Yang et al., 2014). DCD/NRP also mediates cell death under ER stress (Reis et al., 2016). The changes in gene expression levels in both pathways did not differ between wild type and triple mutant in response to TM treatment (Figure 9). However, the expression levels of PR-1 and PR-2, which are key target genes in the SA pathway, were enhanced in the triple mutant after TM treatment (Figure 10). SA pathway also plays a key regulatory role in plant immunity and promotes PCD under stress (Yun and Chen, 2011; Pajerowska-Mukhtar et al., 2012; Gao et al., 2017). Being the production site of antimicrobial proteins and immune signaling components, ER functions as the central regulator in the execution of immune responses in plants (Pajerowska-Mukhtar et al., 2012). The UPR plays a fundamental role not only in abiotic stress responses but also in plant immunity (Moreno et al., 2012). Salicylic acid induces the expression of UPR genes, which in turn can induce the up-regulation of downstream genes of salicylic acid E (Watanabe and Lam, 2008; Moreno et al., 2012; Nagashima et al., 2014). Here we further showed that SA level was upregulated by ER stress (Figure 10C). These data suggested that the function of GAAP1 to GAAP3 against PCD might be achieved by reducing the upregulation of SA pathway under ER stress. AtBI-1 is involved in the inhibition of PCD in Arabidopsis by suppressing the ER-dependent reactive oxygen species (ROS) production or by regulating cell death-associated ER Ca2+ homeostasis (Watanabe and Lam, 2008; Watanabe and Lam, 2009; Ozgur et al., 2018). A close mutual induction relationship was observed between SA signaling pathway and ROS production. The detailed information regarding GAAP inhibiting PCD must be further studied.
In summary, GAAP1 to GAAP3 played redundant roles in maintaining plant growth and survival under ER stress, at least by partially resisting cell death. Upon mild ER stress, GAAP1 to GAAP3 negatively activated UPR pathway. ER stress also upregulated SA level in cells. GAAP1 to GAAP3 suppressed the upregulation of SA pathway against cell death under ER stress.
The Arabidopsis Genome Initiative accession numbers for the proteins referred to in the paper are At4G14730 (GAAP1), AT3G63310 (GAAP2), At4G02690 (GAAP3), At3G10800 (bZIP28), At1g49240 (ACTIN8), At1G09080 (BIP3), At1G42990 (bZIP60), At5G24360 (IRE1B), At4G16660 (HSP70), At4g24190 (SHD), At5g61790 (CNX1), AT2G14610 (PR-1), AT3G57260 (PR-2), AT5G22290 (NAC089), AT5G39820 (NAC094), AT5G40010 (ATPase), AT2G03440 (AtNRP1), AT2G17040 (ANAC036), and AT4G32940 (GAMMA-VPE).
All datasets generated for this study are included in the manuscript/supplementary files.
No human studies are presented in this manuscript. No animal studies are presented in this manuscript. No potentially identifiable human images or data is presented in this study.
WW and MZ performed the assay about the plant sensitivity to ER stress. XinL performed the expression pattern and cell death pathway assay. WW, MZ and XT conducted the UPR assay. XT conducted the SA level detection. KG and ZW generated the mutants and transgenic plants. YZ performed gene expression assay. YS and WZ supervised the physiological experiments and participated in interpreting the morpho-physiological data. XL designed the experiment, supervised the study, analyzed the data and wrote the paper.
This work was supported by the National Natural Science Foundation of China (No. 31670271).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Hong-Li Lian (Shanghai Jiao-Tong University) for kindly supplying the pHB vector.
ER, endoplasmic reticulum; UPR, unfolded protein response, Bax, Bcl-2 associated X protein; PCD, programmed cell death; qRT-PCR, Quantitative real-time PCR; TM, tunicamycin; DTT, dithiothreitol; ROS, reactive oxygen species; BiFC, bimolecular fluorescence complementation; GUS, beta-Dglucuronidase; PI, Propidium iodide.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01032/full#supplementary-material
Brewer, J. W. (2014). Regulatory crosstalk within the mammalian unfolded protein response. Cell Mol. Life Sci. 71, 1067–1079. doi: 10.1007/s00018-013-1490-2
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Deng, Y., Humbert, S., Liu, J. X., Srivastava, R., Rothstein, S. J., Howell, S. H. (2011). Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7247–7252. doi: 10.1073/pnas.1102117108
Deng, Y., Srivastava, R., Howell, S. H. (2013). Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 19633–19638. doi: 10.1073/pnas.1314749110
Duan, Y., Zhang, W., Li, B., Wang, Y., Li, K., Sodmergen, et al. (2010). An endoplasmic reticulum response pathway mediates programmed cell death of root tip induced by water stress in Arabidopsis. New Phytol. 186, 681–695. doi: 10.1111/j.1469-8137.2010.03207.x
Gao, Y., Wu, Y., Du, J., Zhan, Y., Sun, D., Zhao, J., et al. (2017). Both light-induced SA accumulation and ETI mediators contribute to the cell death regulated by BAK1 and BKK1. Front Plant Sci. 8, 622. doi: 10.3389/fpls.2017.00622
Guo, K., Wang, W., Fan, W., Wang, Z., Zhu, M., Tang, X., et al. (2018). Arabidopsis GAAP1 and GAAP3 modulate the unfolded protein response and the onset of cell death in response to ER Stress. Front. Plant Sci. 9, 348 doi: 10.3389/fpls.2018.00348
Henke, N., Lisak, D. A., Schneider, L., Habicht, J., Pergande, M., Methner, A. (2011). The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 50, 251–260. doi: 10.1016/j.ceca.2011.05.005
Humbert, S., Zhong, S., Deng, Y., Howell, S. H., Rothstein, S. J. (2012). Alteration of the bZIP60/IRE1 pathway affects plant response to ER stress in Arabidopsis thaliana. PLoS One 7, e39023. doi: 10.1371/journal.pone.0039023
Ishikawa, T., Watanabe, N., Nagano, M., Kawai-Yamada, M., Lam, E. (2011). Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 18, 1271–1278. doi: 10.1038/cdd.2011.59
Li, X. F., Jia, L. Y., Xu, J., Deng, X. J., Wang, Y., Zhang, W., et al. (2013). FT-like NFT1 gene may play a role in flower transition induced by heat accumulation in Narcissus tazetta var. chinensis. Plant Cell Physiol. 54, 270–281. doi: 10.1093/pcp/pcs181
Li, X. F., Li, Y. J., An, Y. H., Xiong, L. J., Shao, X. H., Wang, Y., et al. (2009). AKINbeta1 is involved in the regulation of nitrogen metabolism and sugar signaling in Arabidopsis. J. Integr. Plant Biol 51, 513–520. doi: 10.1111/j.1744-7909.2009.00811.x
Lisbona, F., Rojas-Rivera, D., Thielen, P., Zamorano, S., Todd, D., Martinon, F., et al. (2009). BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell 33, 679–691. doi: 10.1016/j.molcel.2009.02.017
Liu, J. X., Howell, S. H. (2010a). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796. doi: 10.1105/tpc.109.072173
Liu, J. X., Howell, S. H. (2010b). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22, 2930–2942. doi: 10.1105/tpc.110.078154
Liu, J. X., Srivastava, R., Che, P., Howell, S. H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4119. doi: 10.1105/tpc.106.050021
Liu, Y., Burgos, J. S., Deng, Y., Srivastava, R., Howell, S. H., Bassham, D. C. (2012). Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24, 4635–4651. doi: 10.1105/tpc.112.101535
Mishiba, K., Nagashima, Y., Suzuki, E., Hayashi, N., Ogata, Y., Shimada, Y., et al. (2013). Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl. Acad. Sci. U.S.A 110, 5713–5718. doi: 10.1073/pnas.1219047110
Moreno, A. A., Mukhtar, M. S., Blanco, F., Boatwright, J. L., Moreno, I., Jordan, M. R., et al. (2012). IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One 7, e31944. doi: 10.1371/journal.pone.0031944
Nagashima, Y., Iwata, Y., Ashida, M., Mishiba, K., Koizumi, N. (2014). Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55, 1772–1778. doi: 10.1093/pcp/pcu108
Nagashima, Y., Mishiba, K., Suzuki, E., Shimada, Y., Iwata, Y., Koizumi, N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1, 29. doi: 10.1038/srep00029
Ning, S. B., Wang, L., Song, Y. C. (2002). Identification of programmed cell death in situ in individual plant cells in vivo using a chromosome preparation technique. J. Exp. Bot. 53, 651–658. doi: 10.1093/jexbot/53.369.651
Nishimura, M. T., Dangl, J. L. (2010). Arabidopsis and the plant immune system. Plant J. 61, 1053–1066. doi: 10.1111/j.1365-313X.2010.04131.x
Ozgur, R., Uzilday, B., Iwata, Y., Koizumi, N., Turkan, I. (2018). Interplay between the unfolded protein response and reactive oxygen species: a dynamic duo. J. Exp. Bot. 69, 3333–3345. doi: 10.1093/jxb/ery040
Pajerowska-Mukhtar, K. M., Wang, W., Tada, Y., Oka, N., Tucker, C. L., Fonseca, J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22, 103–112. doi: 10.1016/j.cub.2011.12.015
Reis, P. A., Carpinetti, P. A., Freitas, P. P., Santos, E. G., Camargos, L. F., Oliveira, I. H., et al. (2016). Functional and regulatory conservation of the soybean ER stress-induced DCD/NRP-mediated cell death signaling in plants. BMC Plant Biol. 16, 156. doi: 10.1186/s12870-016-0843-z
Ruberti, C., Lai, Y., Brandizzi, F. (2018). Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. 93, 155–165. doi: 10.1111/tpj.13768
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Walter, P., Ron, D. (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. doi: 10.1126/science.1209038
Wang, D., Weaver, N. D., Kesarwani, M., Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. doi: 10.1126/science.1108791
Watanabe, N., Lam, E. (2008). BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 283, 3200–3210. doi: 10.1074/jbc.M706659200
Watanabe, N., Lam, E. (2009). Bax inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int. J. Mol. Sci. 10, 3149–3167. doi: 10.3390/ijms10073149
Yamagami, A., Saito, C., Nakazawa, M., Fujioka, S., Uemura, T., Matsui, M., et al. (2017). Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation. Sci. Rep. 7, 5739. doi: 10.1038/s41598-017-06016-2
Yang, Z. T., Lu, S. J., Wang, M. J., Bi, D. L., Sun, L., Zhou, S. F., et al. (2014). A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 79, 1033–1043. doi: 10.1111/tpj.12604
Keywords: Arabidopsis thaliana, GAAP, endoplasmic reticulum stress, cell death, unfolded-protein response, salicylic acid
Citation: Wang W, Li X, Zhu M, Tang X, Wang Z, Guo K, Zhou Y, Sun Y, Zhang W and Li X (2019) Arabidopsis GAAP1 to GAAP3 Play Redundant Role in Cell Death Inhibition by Suppressing the Upregulation of Salicylic Acid Pathway Under Endoplasmic Reticulum Stress. Front. Plant Sci. 10:1032. doi: 10.3389/fpls.2019.01032
Received: 22 May 2019; Accepted: 24 July 2019;
Published: 27 August 2019.
Edited by:
Eric Ruelland, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Tibor Janda, Centre for Agricultural Research (MTA), HungaryCopyright © 2019 Wang, Li, Zhu, Tang, Wang, Guo, Zhou, Sun, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofang Li, eGZsaUBiaW8uZWNudS5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.