
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 28 August 2019
Sec. Plant Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01021
This article is part of the Research Topic Organelle Autophagy in Plant Development View all 9 articles
Peroxisomes are dynamic organelles involved in multiple functions, including oxygen and nitrogen reactive species metabolism. In plants, these organelles have a close relationship with chloroplasts and mitochondria, characterized by intense metabolic activity and signal transduction. Peroxisomes undergo rapid changes in size, morphology, and abundance depending on the plant development stage and environmental conditions. As peroxisomes are essential not only for redox homeostasis but also for sensing stress, signaling transduction, and cell survival, their formation and degradation need to be rigorously regulated. In this review, new insights into the regulation of plant peroxisomes are briefly described, with a particular emphasis on pexophagy components and their regulation.
Peroxisomes are small round organelles surrounded by a single lipid bilayer present in most eukaryotes (Sandalio and Romero-Puertas, 2015; Kao et al., 2018). Despite their morphological similarity and the conservation of their functions in all eukaryotes, major differences have been found between peroxisomes in plants and peroxisomes in fungi and animals (Kamada et al., 2003). Initially, peroxisomes were regarded as H2O2 homeostasis-regulating cellular compartments containing peroxide-producing as well as peroxide-decomposing enzymes such as catalase (CAT). An increasing number of functions have been identified in plant peroxisomes, which contain more than 50 different enzymes (Hu et al., 2012; Reumann and Bartel, 2016; Kao et al., 2018).
Fatty acid β-oxidation is an important source of metabolic energy and carbohydrates. In higher plants, this peroxisomal pathway is linked to the glyoxylate cycle, which plays a particularly significant role in germinating seeds (Hu et al., 2012). Other essential peroxisomal plant functions include glycolate metabolism involved in photorespiration, hormone biosynthesis (auxins, jasmonic acid, and salicylic acid), and purine and polyamine catabolism (Sandalio and Romero-Puertas, 2015). Most of these processes give rise reactive oxygen species (ROS) production, whereas peroxisomes also produce nitric oxide (NO) and its derivative nitrogen reactive species (RNS) (Sandalio and Romero-Puertas, 2015). The great versatility of peroxisomes enables them to develop and share many metabolic and signaling pathways with other organelles, including chloroplasts and mitochondria, which frequently exchange metabolites and signals to coordinate their functions (Linka and Theodoulou, 2013; Shai et al., 2016).
Higher plants greatly depend on the regulation of biogenesis and recycling of available nutrients and energy sources to withstand and adapt to constant environmental changes. Despite their crucial role in rapid responses to these changes by synthesizing ROS and RNS, which are signaling molecules that regulate developmental processes and stress responses, many peroxisomes cause severe oxidative and nitrosative damage (Marinho et al., 2014; Mittler, 2017; Castillo et al., 2018). Peroxisomes experience highly dynamic, mostly ROS-regulated, changes in metabolism, size, morphology, velocity, and abundance depending on cell type, developmental stage, and environmental stimuli (Rodriguez-Serrano et al., 2016). Peroxisomal homeostasis, regulated by peroxisomal biogenesis and degradation, is essential for cell viability. Excess, damaged, and obsolete peroxisomes can be degraded by protease-dependent pathways or selective autophagy termed as pexophagy. Although much research has been devoted to deciphering pexophagy in yeast and mammal cells, less is known about plant pexophagy. In this review, we focus on the regulation of plant peroxisomal populations, with particular emphasis on the function and regulation of pexophagy under developmental and stress conditions.
The regulation of peroxisome abundance is governed by the biogenesis, proliferation, and degradation of these tightly controlled organelles. Proteins involved in peroxisome biogenesis and its maintenance are called peroxins (PEXs) (Kao et al., 2018). Although these proteins have been well characterized in yeast and mammals, many questions regarding their role in plants remain unanswered.
In yeast, peroxisome biogenesis occurs according to two models: 1) de novo biogenesis from the endoplasmic reticulum (ER) and 2) directly through preexisting peroxisomes by growth and division (Aksit and van der Klei, 2018). Although not fully understood in plants, this process resembles that observed in mammals. Thus, as in mammals, PEX16 is delivered to peroxisomes from the ER and then recruits other peroxisomal membrane proteins (PMPs). The earlier acting PEX3, PEX16, and PEX19 directly insert PMPs into the peroxisomal membrane or a peroxisome-destined region of the ER (reviewed by Hu et al., 2012; Kao et al., 2018).
Although peroxisomes can divide by fission during cell division, they can also proliferate under various stress conditions. Excessive light (Desai and Hu, 2008), salinity (Mitsuya et al., 2011), ozone (Oksanen et al., 2003), H2O2 application (Lopez-Huertas et al., 2000), and xenobiotics, such as clofibrate (Castillo et al., 2008) and the heavy metal Cd (Rodriguez-Serrano et al., 2016), can rapidly increase plant peroxisome abundance. Peroxisome proliferation involves PEX11-regulated elongation and dynamin-related proteins and fission protein (FIS1)-regulated fission. Some evidence show that peroxisomal proliferation is regulated by ROS (Lopez-Huertas et al., 2000; Rodriguez-Serrano et al., 2016). Although peroxisome proliferation could be a protective response to stress conditions, its function and regulation are not fully understood.
As peroxisomes house several oxidative metabolic pathways, their proteins are prone to oxidative damage and thus require regulated turnover. Peroxisomes containing obsolete or dysfunctional proteins need to be eliminated to ensure that cellular redox homeostasis is maintained to avoid excessive ROS accumulation. These situations, which are particularly frequent under biotic and abiotic stress conditions, can also occur during cell differentiation or development and even under normal cellular conditions (Luo and Zhuang, 2018). Like other eukaryotes, through a process called autophagy, plants remove, degrade, and recycle damaged or unnecessary cell components and organelles (Avin-Wittenberg et al., 2018). There are several types of autophagy: microautophagy, macroautophagy, chaperone-mediated autophagy, and cytoplasm-to-vacuole transport, but so far only microautophagy and macroautophagy have been identified in plants (Zientara-Rytter and Sirko, 2014; Galluzzi et al., 2017; Avin-Wittenberg et al., 2018). Microautophagy involves a direct invagination of the tonoplast engulfing the cellular components to be degraded in the vacuole, whereas, in macroautophagy, the de novo formation of a double-membrane organelle, called autophagosome, engulfing these components is required. In plants, both microautophagy and macroautophagy (hereafter referred to as autophagy) can be selective or non-selective. Depending on the cargo, different types of selective autophagy have been reported: pexophagy (peroxisomes), mitophagy (mitochondria), chlorophagy (chloroplasts), reticulophagy (ER), and xenophagy (plant pathogens) (Till et al., 2012; Yoshimoto, 2012; Liu and Bassham, 2013; Zientara-Rytter and Sirko, 2014; Michaeli et al., 2016; Barros et al., 2017; Hafren et al., 2017; Izumi et al., 2017; Avin-Wittenberg et al., 2018).
A conserved set of proteins encoded by autophagy-related genes (ATGs) are involved in regulating autophagy in all eukaryotic cells and have also been reported in crop species (Yoshimoto, 2012; Young et al., 2019).
Unlike in other eukaryotes, more than 30 ATG proteins have been identified in plants, suggesting a higher diversity of gene families, probably due to their sessile properties. ATGs are organized in four main cores for autophagosome initiation and formation: 1) the ATG1 complex containing ATG1, ATG13, ATG11/17, and ATG101 for autophagosome initiation and formation; 2) the VPS34 complex containing ATG6, VPS34, VPS15, and ATG14; 3) the ATG9 complex containing ATG9, ATG18, and ATG2 for phagophore expansion; and 4) the ATG8 conjugation system containing ATG4, ATG8, ATG7, ATG10, ATG12, ATG16, and ATG5 (Avin-Wittenberg et al., 2018).
The ubiquitin-like protein ATG8, which binds the growing autophagosome membrane by covalently bonding to phosphatidylethanolamine, is essential for selective cargo recruitment and autophagosome-tonoplast fusion (Kellner et al., 2017; Avin-Wittenberg et al., 2018). Whereas yeast and fungi contain a single ATG8, higher eukaryotes have several ATG8 isoforms (nine in Arabidopsis), and it is unclear whether each isoform has a different function (Kellner et al., 2017). The residues forming W and L pockets of ATG8 are considerably conserved, whereas those neighboring the core pockets may determine ATG8 binding specificity (Kellner et al., 2017). Autophagy receptors bind ATG8 via an ATG8-interacting motif (AIM), currently referred to as the xLIR motif, with the consensus core sequence (ADEFGLPRSK)(DEGMSTV)(WFY)(DEILQTV)(ADEFHIKLMPSTV) (ILV), where the residues in positions 3 and 6 are the most crucial for interaction with ATG8 family proteins (Jacomin et al., 2016). Given this strong binding and evidence of ATG8 cross-kingdom conservation, this protein has been widely used to monitor autophagy (reviewed by Avin-Wittenberg et al., 2018; Young et al., 2019).
Arabidopsis ATG mutants have played a critical role in the study of pexophagy in plants, which was first reported in 2013 (Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013; Lee et al., 2014; Yoshimoto et al., 2014). By analyzing Arabidopsis mutants with peroxisomes abnormally clustered (peups) in leaves, Shibata et al. (2013) found that ATG8 proteins were localized on the surface of peroxisomes, with ATG2, ATG18a, and ATG7 proteins involved in specific peroxisome degradation processes. Arabidopsis mutants in these genes contain high levels of insoluble inactive CAT (Shibata et al., 2013). Recently, peroxisome accumulation was observed in atg5 and atg7 mutants in leaves from Arabidopsis plants exposed to Cd, whereas phagophores surrounding peroxisomes were observed in wild-type (WT) Arabidopsis plants expressing peroxisomal markers ATG8a-GFP and SKL-CFP (Calero-Muñoz et al., 2019). The application of exogenous H2O2 produces peroxisome clustering, suggesting that H2O2-induced oxidative damage in peroxisomal proteins could signal pexophagy induction (Shibata et al., 2013). However, although the role of CAT in autophagy induction has been under debate, the analysis of CAT-defective mutants has shown that CAT is dispensable for peroxisome degradation (Shibata et al., 2013). Some evidence suggest that CAT activity is involved in ROS-regulated autophagy-dependent programmed cell death induced by hydroxyurea and avrRpm1 (Hackenberg et al., 2013) and starvation-induced pexophagy (Tyutereva et al., 2017). Calero-Muñoz et al. (2019) showed that ATG8a co-localizes with CAT in the electron-dense peroxisomal core where inactive CAT is accumulated. Further studies of Arabidopsis ATG mutants have revealed that clustered peroxisomes mainly accumulate in the aerial parts of plants, where oxidative pathways, mainly β-oxidation and photorespiration, take place rather than in roots (Yoshimoto et al., 2014). However, pexophagy has recently been reported to be also involved in peroxisomal quality control in Arabidopsis root meristems (Huang et al., 2019).
The selective autophagy pathways in eukaryotes require specific cargo receptor(s) and/or adaptors. Two kinds of autophagy cargo receptor have been described in yeast and mammals, which differ in their capacity to bind ubiquitylated cargos (Zientara-Rytter and Sirko, 2016). Four pexophagy receptors/adaptors have been reported in eukaryotes: the yeast Atg36 (Saccharomyces cerevisiae) and Atg30 (Pichia pastoris), unable to bind ubiquitin, and the mammalian NBR1 (neighbor of Brca1 gene) and p62, which recognize ubiquitinated proteins (Zientara-Rytter and Sirko, 2016). ScAtg36 interacts with Pex3 PMPs as well as with Atg8 and Atg11 via AIM and Atg11-binding site, respectively (Farre et al., 2013; Honsho et al., 2016). PpAtg30 requires both Atg37 and Pex3 to recruit Atg11 and Atg8 to the pexophagic receptor-protein complex (Zientara-Rytter et al., 2018). PpAtg30 also interacts with Pex14 (Farre et al., 2008), although only phosphorylated PEX14 is involved in pexophagy in Hansenula polymorpha (van Zutphen et al., 2008). No homologs of Atg30 or Atg36 are known in plants. However, plants have NBR1-like protein (NBR1 from Arabidopsis and Joka2 from tobacco), which is a functional hybrid of the two mammalian ubiquitin-binding autophagy receptors, NBR1 and p62, required for targeting ubiquitinated peroxisomes to autophagosomes for degradation (Deosaran et al., 2013; Walter et al., 2014; Yamashita et al., 2014). However, its role in pexophagy needs further investigation (Zhou et al., 2013; Hafren et al., 2017; Young et al., 2019). The co-localization of NBR1 and ATG8 in electron-dense peroxisomal cores in Arabidopsis plants exposed to Cd has recently been reported (Calero-Muñoz et al., 2019). However, using forward genetic screening, which consistently recovers Arabidopsis atg mutations by exploiting LON2 (peroxisomal matrix protease)-defective mutants (lon2), Young et al. (2019) showed that lon2 peroxisome autophagy does not require NBR1, which, however, may play an important role in LON2-independent pexophagy (Young et al., 2019). As reported in yeast and mammals, the ubiquitination of PEXs, such as PEX3, PEX5, and PEX14, may be also involved in plant pexophagy (Michaeli et al., 2016; Young and Bartel, 2016). Interestingly, CAT is ubiquitinated and preferentially accumulated in Arabidopsis nbr1 mutants, which support possible NBR1/CAT interactions (Zhou et al., 2013). Dominant suppressor of KAR2 (DSK2) is another ubiquitin receptor candidate in plants, which binds the transcription factor BES1 (BRI1-EMS-suppressor 1) for its autophagic degradation (Nolan et al., 2017). In Arabidopsis, DSK2 interacts with the ring PEX2 and PEX12, which act as E3 ligases (Kaur et al., 2013). However, PEXs could also interact directly with ATG8 without involving a bridging receptor. Using bioinformatics and bimolecular fluorescence complementation techniques, Xie et al. (2016) found that PEX6 and PEX10 may interact with ATG8. However, Gonzalez et al. (2018) have reported that the physiological defects of pex6-1 in Arabidopsis mutants are partially restored by preventing autophagy (disabling ATG7), which would rule out a role for PEX6 in promoting autophagy.
Although the signals determining plant peroxisome degradation have not been fully identified, ROS and oxidative stress factors, such as CAT and peroxisomal oxidation, are commonly cited (Shibata et al., 2013; Yoshimoto et al., 2014; Calero-Muñoz et al., 2019). Moreover, Arabidopsis atg mutants are more hypersensitive to oxidative stress, suggesting that pexophagy could be an important component in oxidative stress responses (Zhou et al., 2013). In mammalian cells, ROS induce pexophagy by promoting PEX5 phosphorylation and ubiquitination (Zhang et al., 2015). Although several PEXs have been reported to be phosphorylated in Arabidopsis (Kataya et al., 2019), a direct relationship between ROS, PEX phosphorylation and pexophagy has not been established.
ROS induce chloroplast extensions (stromules), which are associated with chlorophagy involving Rubisco-containing bodies (Hanson and Hines, 2018). Similar ROS-dependent extensions (peroxules) have been observed in peroxisomes under stress conditions (Sinclair et al., 2009; Jaipargas et al., 2016; Rodriguez-Serrano et al., 2016), although no evidence of their role in vesicle formation to recycle peroxisomal components has been reported.
Cysteine-generated sulfide in the cytosol negatively regulates and inhibits autophagy in Arabidopsis plants by preventing ATG8 accumulation (Alvarez et al., 2012) probably through the persulfidation of enzymes involved in autophagosome formation (Alvarez et al., 2012). Although sulfide has not been shown to affect pexophagy, as some persulfidated peroxisomal proteins have been reported, most of which are associated with antioxidant defenses (Aroca et al., 2017), persulfidation could therefore be involved in pexophagy regulation.
During light-induced seed germination and seedling establishment, the transition from glyoxysome, which are present in seeds involved in the glyoxylate cycle and β-oxidation, to leaf-type peroxisomes containing photorespiration machinery is triggered. This transition degrades expendable glyoxylate cycle enzymes such as isocitrate lyase (ICL) and malate synthase (MLS), whereas photorespiration-related enzymes are imported into peroxisomes. Farmer et al. (2013) reported that peroxisomal protease LON2 plays an important role in the selective degradation of obsolete matrix proteins during this metabolic transition (Figure 1). These proteins are also exported to the cytoplasm and degraded by proteasome after ubiquitination (Figure 1). LON2 proteins are a conserved family of ATPases characterized by chaperone and protease activity (Goto-Yamada et al., 2014; Young and Bartel, 2016). Arabidopsis mutants expressing protease-deficient AAA-active LON2 in a lon2-null genetic background have lower pexophagy levels, demonstrating that LON2 chaperone activity inhibits pexophagy (Farmer et al., 2013; Young and Bartel, 2016). Stabilization studies of atg and lon2 mutants showed that both LON2 and autophagy affect ICL and MLS turnover, whereas pexophagy mainly affects the turnover of thiolase involved in β-oxidation (Young et al., 2019), suggesting that different pathways control peroxisomal components.
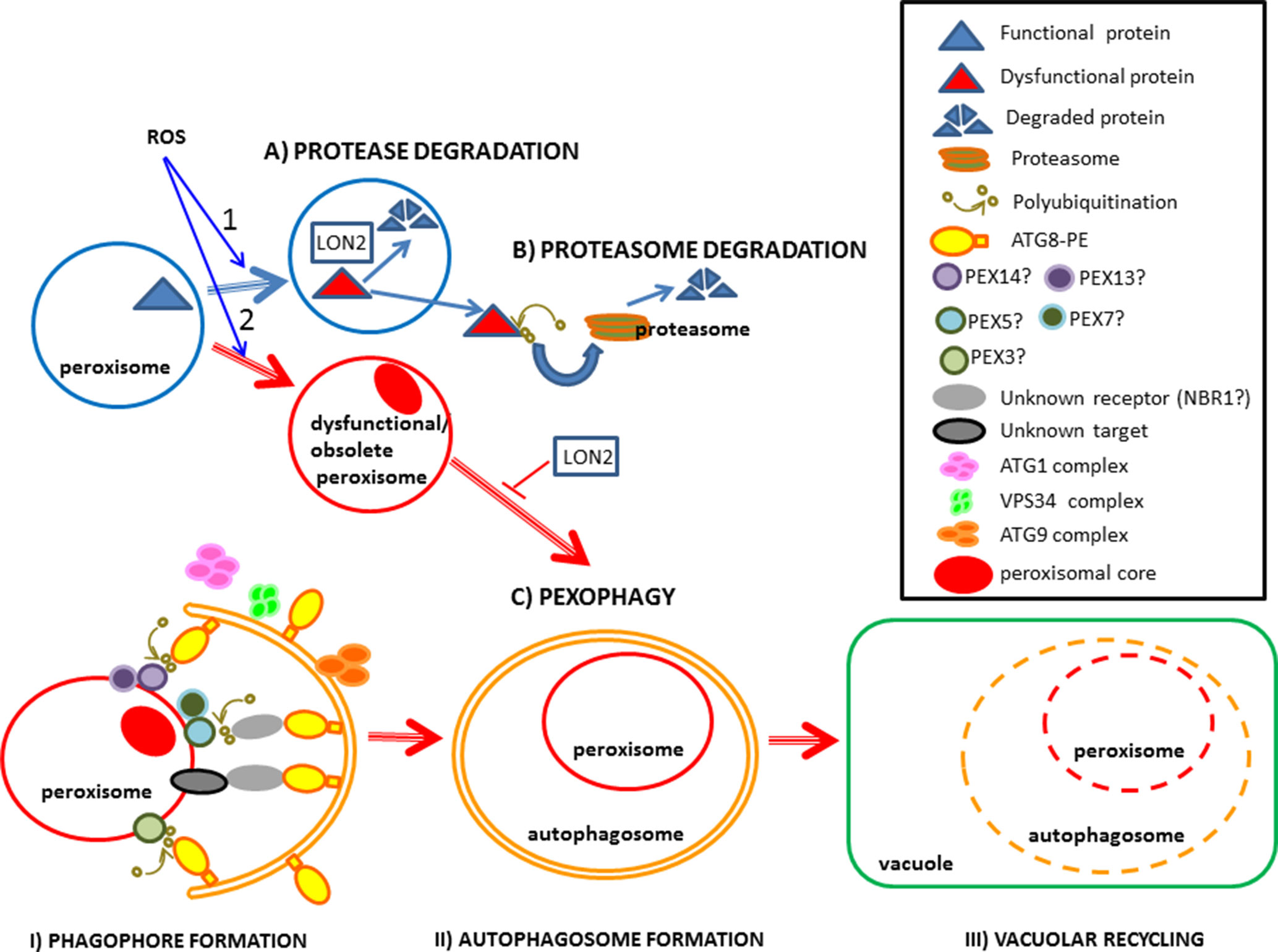
Figure 1 Diagram illustrating the elimination of peroxisomal proteins and obsolete/dysfunctional peroxisomes. 1) Peroxisomal proteins can be eliminated by: (A) endogenous peroxisomal proteases such as LON2 or (B) through proteasome degradation in the cytoplasm following ubiquitination. 2) Peroxisomes can be eliminated by: (C) pexophagy, involving three main steps: I) the formation of phagophores when they begin to surround the recyclable peroxisome; II) autophagosome formation when the peroxisome is completely engulfed in the autophagosome and III) vacuolar recycling when the peroxisome inside the vacuole starts to degrade. ROS produced during developmental processes, starvation and biotic or abiotic stresses induce the elimination of peroxisomal proteins, as well as obsolete and dysfuncional peroxisomes. Pexophagy components followed by a question mark in the diagram, whose precise role in plants remains unclear, require further study.
Pexophagy mediates peroxisome turnover even under non-stress conditions, as demonstrated by peroxisome clustering in Arabidopsis atg mutants, which may regulate basal peroxisome levels, mainly in green tissues, where H2O2-producing photorespiration is highly active (Farmer et al., 2013; Kim et al., 2013; Shibata et al., 2013; Lee et al., 2014; Yoshimoto et al., 2014). Although Arabidopsis atg mutants accumulate more peroxisomes than their WT counterparts, this is not the case for other organelles such as the Golgi apparatus, mitochondria, ER, and chloroplasts. Maize atg mutants also accumulate PEX14, a peroxisomal marker (Li et al., 2015). In Arabidopsis atg5 mutants, analyses of peroxisomal redox status have shown a larger number of oxidized organelles than in WT plants (Yoshimoto et al., 2014). All these findings are consistent with the possibility that pexophagy clears damaged peroxisomes at a basal rate in the absence of stresses typically associated with autophagy induction (Yoshimoto et al., 2014).
Root growth and architecture are controlled by meristems, where stem cell activity is regulated by auxins, nutrient availability, and ROS homeostasis (Rellan-Alvarez et al., 2016). Pexophagy plays a key role in the glucose-mediated regulation of root meristem activity by maintaining ROS and auxin cellular homeostasis in Arabidopsis plants (Huang et al., 2019). Using yeast two-hybrid and co-immunoprecipitation assays, Huang et al. (2019) observed interactions between ATG8e and the peroxisomal ATP-binding cassette D1 (ABCD1) Walker B motif and that the glucose-tolerant root elongation phenotype of atg5 and atg7 mutants is partially rescued by ABCD1 overexpression.
Biochemical, cytological, and pharmacological analyses have provided evidence of peroxisome degradation by autophagy during carbohydrate starvation in Nicotiana tabacum cells, which is slowed down by the application of autophagy inhibitor 3-methyladenine (Voitsekhovskaja et al., 2014). Exposure to Cd induces peroxisome oxidation and pexophagy, whereas the reduction of ROS production in Arabidopsis mutants gox2 (glycolate oxidase 2) and rbohC (NADPH oxidase C) considerably reduces this process. Pexophagy is an important component in rapid plant responses to Cd, which prevents disturbances in peroxisomal populations and the cell redox balance (Calero-Muñoz et al., 2019).
Plants develop complex signaling mechanisms to adapt to and withstand constant environmental changes. Many cell/organelle cross-talk mechanisms are fine tuned by ROS. The regulation of peroxisome populations, which, in turn, affects peroxisomal metabolism and redox homeostasis, is essential for plant life. Although plant pexophagy is currently the subject of intense study, important aspects of this complex process remain unexplained. Questions regarding how, why, and when plants regulate peroxisome populations, the possible adaptive role of peroxisome proliferation in plants, and how exactly pexophagy is initiated and terminated remain unanswered. Little is known about the effect of other molecules, such as H2S, and NO, on pexophagy. Further studies are required to pinpoint the receptors that connect the peroxisome to be degraded to pexophagic machinery and to identify possible targets in the peroxisome to be recognized by putative receptor(s). Finally, the mechanisms enabling plants to induce and regulate pexophagy at a basal level also need to be deciphered.
The manuscript was prepared by AO and reviewed and rewritten by LMS.
This study was co-funded by ERDF grant BIO2015-67657-P and PGC2018-098372-B-I00 from MICINN, I-LINK1247 from CSIC, and TRANSAUTOPHAGY EU Framework Program Horizon 2020 COST grant OC-2015-1-19840. The authors wish to apologize to those colleagues whose work has not been cited due to space limitations. The authors wish to thank Michael O’Shea for proofreading the manuscript.
The handling editor declared a past co-authorship with one of the authors LS.
Aksit, A., van der Klei, I. J. (2018). Yeast peroxisomes: how are they formed and how do they grow? Int. J. Biochem. Cell Biol. 105, 24–34. doi: 10.1016/j.biocel.2018.09.019
Alvarez, C., Garcia, I., Moreno, I., Perez-Perez, M. E., Crespo, J. L., Romero, L. C., et al. (2012). Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24 (11), 4621–4634. doi: 10.1105/tpc.112.105403
Aroca, A., Benito, J. M., Gotor, C., Romero, L. C. (2017). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68 (17), 4915–4927. doi: 10.1093/jxb/erx294
Avin-Wittenberg, T., Baluska, F., Bozhkov, P. V., Elander, P. H., Fernie, A. R., Galili, G., et al. (2018). Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69 (6), 1335–1353. doi: 10.1093/jxb/ery069
Barros, J. A. S., Cavalcanti, J. H. F., Medeiros, D. B., Nunes-Nesi, A., Avin-Wittenberg, T., Fernie, A. R., et al. (2017). Commonalities and differences in plants deficient in autophagy and alternative pathways of respiration on response to extended darkness. Plant Signal. Behav. 12 (11), e1377877. doi: 10.1080/15592324.2017.1377877
Calero-Muñoz, N., Exposito-Rodriguez, M., Collado-Arenal, A. M., Rodríguez-Serrano, M., Laureano-Marín, A. M., Santamaría, M. E., et al. (2019) Cadmium induces reactive oxygen species dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. doi: 10.1111/pce.13597
Castillo, M. C., Coego, A., Costa-Broseta, A., Leon, J. (2018). Nitric oxide responses in Arabidopsis hypocotyls are mediated by diverse phytohormone pathways. J. Exp. Bot. 69 (21), 5265–5278. doi: 10.1093/jxb/ery286
Castillo, M. C., Sandalio, L. M., Del Rio, L. A., Leon, J. (2008). Peroxisome proliferation, wound-activated responses and expression of peroxisome-associated genes are cross-regulated but uncoupled in Arabidopsis thaliana. Plant Cell Environ. 31 (4), 492–505. doi: 10.1111/j.1365-3040.2008.01780.x
Deosaran, E., Larsen, K. B., Hua, R., Sargent, G., Wang, Y., Kim, S. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126 (Pt 4), 939–952. doi: 10.1242/jcs.114819
Desai, M., Hu, J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146 (3), 1117–1127. doi: 10.1104/pp.107.113555
Farmer, L. M., Rinaldi, M. A., Young, P. G., Danan, C. H., Burkhart, S. E., Bartel, B. (2013). Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell 25 (10), 4085–4100. doi: 10.1105/tpc.113.113407
Farre, J. C., Burkenroad, A., Burnett, S. F., Subramani, S. (2013). Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 14 (5), 441–449. doi: 10.1038/embor.2013.40
Farre, J. C., Manjithaya, R., Mathewson, R. D., Subramani, S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14 (3), 365–376. doi: 10.1016/j.devcel.2007.12.011
Galluzzi, L., Baehrecke, E. H., Ballabio, A., Boya, P., Bravo-San Pedro, J. M., Cecconi, F., et al. (2017). Molecular definitions of autophagy and related processes. EMBO J. 36 (13), 1811–1836. doi: 10.15252/embj.201796697
Gonzalez, K. L., Ratzel, S. E., Burks, K. H., Danan, C. H., Wages, J. M., Zolman, B. K., et al. (2018). A pex1 missense mutation improves peroxisome function in a subset of Arabidopsis pex6 mutants without restoring PEX5 recycling. Proc. Natl. Acad. Sci. U.S.A. 115 (14), E3163–E3172. doi: 10.1073/pnas.1721279115
Goto-Yamada, S., Mano, S., Nakamori, C., Kondo, M., Yamawaki, R., Kato, A., et al. (2014). Chaperone and protease functions of LON protease 2 modulate the peroxisomal transition and degradation with autophagy. Plant Cell Physiol. 55 (3), 482–496. doi: 10.1093/pcp/pcu017
Hackenberg, T., Juul, T., Auzina, A., Gwizdz, S., Malolepszy, A., Van Der Kelen, K., et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25 (11), 4616–4626. doi: 10.1105/tpc.113.117192
Hafren, A., Macia, J. L., Love, A. J., Milner, J. J., Drucker, M., Hofius, D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. U.S.A. 114 (10), E2026–E2035. doi: 10.1073/pnas.1610687114
Hanson, M. R., Hines, K. M. (2018). Stromules: probing formation and function. Plant Physiol. 176 (1), 128–137. doi: 10.1104/pp.17.01287
Honsho, M., Yamashita, S., Fujiki, Y. (2016). Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Biochim. Biophys. Acta 1863 (5), 984–991. doi: 10.1016/j.bbamcr.2015.09.032
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24 (6), 2279–2303. doi: 10.1105/tpc.112.096586
Huang, L., Yu, L. J., Zhang, X., Fan, B., Wang, F. Z., Dai, Y. S., et al. (2019). Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 15 (3), 407–422. doi: 10.1080/15548627.2018.1520547
Izumi, M., Ishida, H., Nakamura, S., Hidema, J. (2017). Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29 (2), 377–394. doi: 10.1105/tpc.16.00637
Jacomin, A. C., Samavedam, S., Promponas, V., Nezis, I. P. (2016). iLIR database: a web resource for LIR motif-containing proteins in eukaryotes. Autophagy 12 (10), 1945–1953. doi: 10.1080/15548627.2016.1207016
Jaipargas, E. A., Mathur, N., Bou Daher, F., Wasteneys, G. O., Mathur, J. (2016). High light intensity leads to increased peroxule-mitochondria interactions in plants. Front. Cell Dev. Biol. 4, 6. doi: 10.3389/fcell.2016.00006
Kamada, T., Nito, K., Hayashi, H., Mano, S., Hayashi, M., Nishimura, M. (2003). Functional differentiation of peroxisomes revealed by expression profiles of peroxisomal genes in Arabidopsis thaliana. Plant Cell Physiol. 44 (12), 1275–1289. doi: 10.1093/pcp/pcg173
Kao, Y. T., Gonzalez, K. L., Bartel, B. (2018). Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 176 (1), 162–177. doi: 10.1104/pp.17.01050
Kataya, A. R. A., Muench, D. G., Moorhead, G. B. (2019). A framework to investigate peroxisomal protein phosphorylation in Arabidopsis. Trends Plant Sci. 24 (4), 366–381. doi: 10.1016/j.tplants.2018.12.002
Kaur, N., Zhao, Q., Xie, Q., Hu, J. (2013). Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins(F). J. Integr. Plant Biol. 55 (1), 108–120. doi: 10.1111/jipb.12014
Kellner, R., De la Concepcion, J. C., Maqbool, A., Kamoun, S., Dagdas, Y. F. (2017). ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 22 (3), 204–214. doi: 10.1016/j.tplants.2016.11.015
Kim, J., Lee, H., Lee, H. N., Kim, S. H., Shin, K. D., Chung, T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25 (12), 4956–4966. doi: 10.1105/tpc.113.117960
Lee, H. N., Kim, J., Chung, T. (2014). Degradation of plant peroxisomes by autophagy. Front. Plant Sci. 5, 139. doi: 10.3389/fpls.2014.00139
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27 (5), 1389–1408. doi: 10.1105/tpc.15.00158
Linka, N., Theodoulou, F. L. (2013). Metabolite transporters of the plant peroxisomal membrane: known and unknown. Subcell. Biochem. 69, 169–194. doi: 10.1007/978-94-007-6889-5_10
Liu, Y., Bassham, D. C. (2013). Degradation of the endoplasmic reticulum by autophagy in plants. Autophagy 9 (4), 622–623. doi: 10.4161/auto.23559
Lopez-Huertas, E., Charlton, W. L., Johnson, B., Graham, I. A., Baker, A. (2000). Stress induces peroxisome biogenesis genes. EMBO J. 19 (24), 6770–6777. doi: 10.1093/emboj/19.24.6770
Luo, M., Zhuang, X. (2018). Review: selective degradation of peroxisome by autophagy in plants: mechanisms, functions, and perspectives. Plant Sci. 274, 485–491. doi: 10.1016/j.plantsci.2018.06.026
Marinho, H. S., Real, C., Cyrne, L., Soares, H., Antunes, F. (2014). Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2, 535–562. doi: 10.1016/j.redox.2014.02.006
Michaeli, S., Galili, G., Genschik, P., Fernie, A. R., Avin-Wittenberg, T. (2016). Autophagy in plants—What’s new on the menu? Trends Plant Sci. 21 (2), 134–144. doi: 10.1016/j.tplants.2015.10.008
Mitsuya, S., El Shami, M., Sparkes, I. A., Charlton, W. L., Marcos, L. C., et al. (2011). Salt stress causes peroxisome proliferation, but inducing peroxisome proliferation does not improve NaCl tolerance in Arabidopsis thaliana. PLoS One 5, e9408. doi: 10.1371/journal.pone.0009408
Mittler, R. (2017). ROS are good. Trends Plant Sci. 22 (1), 11–19. doi: 10.1016/j.tplants.2016.08.002
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41 (1), P33-46.e37. doi: 10.1016/j.devcel.2017.03.013
Oksanen, E., Häikiö, E., Sober, J., Karnosky, D. F. (2003). Ozone-induced H2O2 accumulation in field-grown aspen and birch is linked to foliar ultrastructure and peroxisomal activity. New Phytol. 161, 791–799. doi: 10.1111/j.1469-8137.2003.00981.x
Rellan-Alvarez, R., Lobet, G., Dinneny, J. R. (2016). Environmental control of root system biology. Annu. Rev. Plant Biol. 67, 619–642. doi: 10.1146/annurev-arplant-043015-111848
Reumann, S., Bartel, B. (2016). Plant peroxisomes: recent discoveries in functional complexity, organelle homeostasis, and morphological dynamics. Curr. Opin. Plant Biol. 34, 17–26. doi: 10.1016/j.pbi.2016.07.008
Rodriguez-Serrano, M., Romero-Puertas, M. C., Sanz-Fernandez, M., Hu, J., Sandalio, L. M. (2016). Peroxisomes extend peroxules in a fast response to stress via a reactive oxygen species-mediated induction of the peroxin PEX11a. Plant Physiol. 171 (3), 1665–1674. doi: 10.1104/pp.16.00648
Sandalio, L. M., Romero-Puertas, M. C. (2015). Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. 116 (4), 475–485. doi: 10.1093/aob/mcv074
Shai, N., Schuldiner, M., Zalckvar, E. (2016). No peroxisome is an island — Peroxisome contact sites. Biochim. Biophys. Acta 1863 (5), 1061–1069. doi: 10.1016/j.bbamcr.2015.09.016
Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25 (12), 4967–4983. doi: 10.1105/tpc.113.116947
Sinclair, A. M., Trobacher, C. P., Mathur, N., Greenwood, J. S., Mathur, J. (2009). Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 59 (2), 231–242. doi: 10.1111/j.1365-313X.2009.03863.x
Till, A., Lakhani, R., Burnett, S. F., Subramani, S. (2012). Pexophagy: the selective degradation of peroxisomes. Int. J. Cell Biol. 2012, 512721. doi: 10.1155/2012/512721
Tyutereva, E. V., Dobryakova, K. S., Schiermeyer, A., Shishova, M. F., Pawlowski, K., Demidchik, V. (2017). The levels of peroxisomal catalase protein and activity modulate the onset of cell death in tobacco BY-2 cells via reactive oxygen species levels and autophagy. Funct. Plant Biol. 45, 247–258. doi: 10.1071/FP16418
van Zutphen, T., van der Klei, I. J., Kiel, J. A. (2008). Pexophagy in Hansenula polymorpha. Methods Enzymol. 451, 197–215. doi: 10.1016/S0076-6879(08)03214-X
Voitsekhovskaja, O. V., Schiermeyer, A., Reumann, S. (2014). Plant peroxisomes are degraded by starvation-induced and constitutive autophagy in tobacco BY-2 suspension-cultured cells. Front. Plant Sci. 5, 629. doi: 10.3389/fpls.2014.00629
Walter, K. M., Schonenberger, M. J., Trotzmuller, M., Horn, M., Elsasser, H. P., Moser, A. B., et al. (2014). Hif-2alpha promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 20 (5), 882–897. doi: 10.1016/j.cmet.2014.09.017
Xie, Q., Tzfadia, O., Levy, M., Weithorn, E., Peled-Zehavi, H., Van Parys, T., et al. (2016). hfAIM: a reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 12 (5), 876–887. doi: 10.1080/15548627.2016.1147668
Yamashita, S., Abe, K., Tatemichi, Y., Fujiki, Y. (2014). The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10 (9), 1549–1564. doi: 10.4161/auto.29329
Yoshimoto, K. (2012). Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol. 53 (8), 1355–1365. doi: 10.1093/pcp/pcs099
Yoshimoto, K., Shibata, M., Kondo, M., Oikawa, K., Sato, M., Toyooka, K. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 127 (Pt 6), 1161–1168. doi: 10.1242/jcs.139709
Young, P. G., Bartel, B. (2016). Pexophagy and peroxisomal protein turnover in plants. Biochim. Biophys. Acta 1863 (5), 999–1005. doi: 10.1016/j.bbamcr.2015.09.005
Young, P. G., Passalacqua, M. J., Chappell, K., Llinas, R. J., Bartel, B. (2019). A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy, 30, 1–19. doi: 10.1080/15548627.2019.1569915
Zhang, J., Tripathi, D. N., Jing, J., Alexander, A., Kim, J., Powell, R. T., et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17 (10), 1259–1269. doi: 10.1038/ncb3230
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9 (1), e1003196. doi: 10.1371/journal.pgen.1003196
Zientara-Rytter, K., Sirko, A. (2014). Selective autophagy receptor Joka2 co-localizes with cytoskeleton in plant cells. Plant Signal. Behav. 9 (3), e28523. doi: 10.4161/psb.28523
Zientara-Rytter, K., Sirko, A. (2016). To deliver or to degrade — an interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 283 (19), 3534–3555. doi: 10.1111/febs.13712
Keywords: autophogy-related genes, peroxins, pexophagy, plants, reactive oxygen species
Citation: Olmedilla A and Sandalio LM (2019) Selective Autophagy of Peroxisomes in Plants: From Housekeeping to Development and Stress Responses. Front. Plant Sci. 10:1021. doi: 10.3389/fpls.2019.01021
Received: 17 April 2019; Accepted: 22 July 2019;
Published: 28 August 2019.
Edited by:
Henri Batoko, Catholic University of Louvain, BelgiumReviewed by:
Katarzyna Zientara-Rytter, University of California, United StatesCopyright © 2019 Olmedilla and Sandalio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Adela Olmedilla, YWxkZWxhLm9sbWVkaWxsYUBlZXouY3NpYy5lcw==; Luisa M. Sandalio, bHVpc2FtYXJpYS5zYW5kYWxpb0BlZXouY3NpYy5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.