
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 August 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00996
 Le Wang1†
Le Wang1† Huiyu Wang1†
Huiyu Wang1† Shengfu He1†
Shengfu He1† Fanshan Meng1†
Fanshan Meng1† Chuanzhong Zhang1†
Chuanzhong Zhang1† Sujie Fan1,2
Sujie Fan1,2 Junjiang Wu3
Junjiang Wu3 Shuzhen Zhang1*
Shuzhen Zhang1* Pengfei Xu1*
Pengfei Xu1*Phytophthora root and stem rot, a destructive disease of soybean [Glycine max (L.) Merr.], is caused by the oomycete Phytophthora sojae. However, how the disease resistance mechanisms of soybean respond to P. sojae infection remains unclear. Previously, we showed that GmWRKY31, which interacts with a sucrose non-fermenting-1(SNF1)-related protein kinase (SnRK), enhances resistance to P. sojae in soybean. Here, we report that the membrane-localized SnRK GmSnRK1.1 is involved in the soybean host response to P. sojae. The overexpression of GmSnRK1.1 (GmSnRK1.1-OE) increased soybean resistance to P. sojae, and the RNA interference (RNAi)-mediated silencing of GmSnRK1.1 (GmSnRK1.1-R) reduced resistance to P. sojae. Moreover, the activities and transcript levels of the antioxidant enzymes superoxide dismutase and peroxidase were markedly higher in the GmSnRK1.1-OE transgenic soybean plants than in the wild type (WT), but were reduced in the GmSnRK1.1-R plants. Several isoflavonoid phytoalexins related genes GmPAL, GmIFR, Gm4CL and GmCHS were significantly higher in “Suinong 10” and GmSnRK1.1-OE lines than these in “Dongnong 50,” and were significantly lower in GmSnRK1.1-R lines. In addition, the accumulation of salicylic acid (SA) and the expression level of the SA biosynthesis-related gene were significantly higher in the GmSnRK1.1-OE plants than in the WT and GmSnRK1.1-R plants, moreover, SA biosynthesis inhibitor treated GmSnRK1.1-R lines plants displayed clearly increased pathogen biomass compared with H2O-treated plants after 24 h post-inoculation. These results showed that GmSnRK1.1 positively regulates soybean resistance to P. sojae, potentially functioning via effects on the expression of SA-related genes and increased accumulation of SA.
The sucrose non-fermenting-1(SNF1)-related protein kinases (SnRKs) are key factors in the regulation of energy metabolism, sugar signaling, seed germination, and seedling growth in plants, in addition to stress signaling in a diverse array of eukaryotes (Halford and Hey, 2009; Hey et al., 2010; Coello et al., 2011; Tsai and Gazzarrini, 2014). The SnRK1 subfamily comprises SnRK1.1, SnRK1.2, and SnRK1.3 (also named KIN10/AKIN10, KIN11/AKIN11, and KIN12/AKIN12, respectively), of which only SnRK1.1 and SnRK1.2 appear to be expressed in plants (Baena-González et al., 2007). SnRK1 is a heterotrimeric complex composed of an α-catalytic subunit, a γ subunit, and a β subunit that bridges the α and γ subunits (Polge and Thomas, 2007; Hedbacker and Carlson, 2008; Smeekens et al., 2010; Carling et al., 2012).
SnRK1 regulates carbon metabolism (Halford and Hardie, 1998; Fragoso et al., 2009; Nunes et al., 2013; Zhai et al., 2017) and responds to hormonal signals, particularly abscisic acid (ABA), providing a possible link between the hormone and sugar signaling pathways (Radchuk et al., 2006, 2010; Jossier et al., 2009; Coello et al., 2012; Tsai and Gazzarrini, 2012; Rodrigues et al., 2013). ABA negatively regulates resistance to P. sojae and active levels are depleted as part of the response to incompatible soybean genotypes (McDonald and Cahill, 1999; Mohr and Cahill, 2001; Asselbergh et al., 2007). Moreover, in wheat, SnRK1 is negatively regulated by ABA (Patricia et al., 2012). Moreover, SnRK1 regulates plant metabolism in response to stresses such as darkness and flooding, as well as developmental changes such as flowering, seed germination, and seedling growth (Baena-González et al., 2007; Jossier et al., 2009; Lee et al., 2009; Coello et al., 2011; Cho et al., 2012; Wu et al., 2017). In Arabidopsis thaliana, SnRK1 is involved in the responses to sugar and darkness by regulating the expression of stress-responsive genes and ABA signaling (Baena-González et al., 2007; Jossier et al., 2009). SnRK1 activities in rice (Oryza sativa) and Arabidopsis have a decisive influence on the expression of stress-inducible genes and the induction of stress-tolerance processes (Cho et al., 2012); for example, the rice protein kinase CIPK15 regulates carbohydrate catabolism and fermentation via the SnRK1A-MYBS1-mediated sugar signaling pathway, enabling rice to grow under floodwater (Lee et al., 2009). In Arabidopsis, FUS3 interacts with SnRK1.1 to regulate lateral organ development (Tsai and Gazzarrini, 2012), but also promotes dormancy and inhibits germination through cross-regulation of the ABA and gibberellin pathways (Gazzarrini and Tsai, 2015). Under low-sugar conditions, Arabidopsis SnRK1 was triggered to phosphorylate and inactivate the INDETERMINATE DOMAIN (IDD)-containing transcription factor IDD8, thereby leading to delayed flowering (Jeong et al., 2015). These discoveries show that SnRK1 coordinates the responses to a wide array of abiotic stresses (Baena-González et al., 2007; Lee et al., 2009; Cho et al., 2012; Jeong et al., 2015). Relatively little is known about the mechanisms by which SnRK1 functions in the responses to biotic stress. The overexpression of SnRK1 in tobacco (Nicotiana sp.) made the transgenic plants more resistant to geminivirus infection (Hao et al., 2003). SnRK1 interacts with the effector AvrBsT, which is involved in suppression of the AvrBs1-specific hypersensitive response in pepper (Capsicum annuum) plants (Szczesny et al., 2010). The rice SnRK1b gene OSK35 was enhanced the plant resistance to fungal and bacterial pathogens (Kim et al., 2015). Despite these insights, no systematic research on the disease-related roles of SnRK1 in another major crop species, soybean (Glycine max), has been reported.
In a previous study, we showed that a novel WRKY transcription factor, GmWRKY31, enhances soybean resistance to P. sojae, and identified 19 putative GmWRKY31-interacting proteins (Fan et al., 2017), of which a Sucrose non-Fermenting-1-Related Protein Kinase (SnRK1) was selected for further study. In the present study, we isolated a GmWRKY31-interacting GmSnRK1.1 (GenBank accession no. XM_006585690), and generated transgenic soybean plants either overexpressing GmSnRK1.1 (GmSnRK1.1-OE) or with an RNA-interference (RNAi)-mediated reduced expression of this gene (GmSnRK1.1-R). Overexpression and RNA interference analysis demonstrates that GmSnRK1.1 positively regulates of soybean resistance to this pathogen, likely via a SA-signaling pathway.
The soybean cultivar used for the various treatments and gene isolation was “Suinong 10,” which is highly resistant to P. sojae race 1 (PSR01) isolated in Heilongjiang, China (Zhang et al., 2010). The susceptible soybean cultivar “Dongnong 50” was used for the gene transformation experiments. These lines were obtained from the Key Laboratory of Soybean Biology in the Chinese Ministry of Education, Harbin. PSR01 was previously isolated from infected soybean plants in Heilongjiang, China (Zhang et al., 2010). This isolate was propagated at 25°C for 7 days on V8 juice agar in a glass dish. The seeds of “Suinong 10” and “Dongnong 50” were grown at 25°C and 60% relative ambient humidity in a growth cabinet, with a 16-h light/8-h dark photoperiod. For the P. sojae infection, the hypocotyls of soybean cultivars “Suinong 10” and “Dongnong 50” were inoculated at the first-node stage (V1) (Fehr et al., 1971) using either zoospores of P. sojae or a mock inoculation with sterile water following the procedure described by Kaufmann and Gerdemann (1958), with minor modifications. The P. sojae zoospores were induced as described by Ward et al. (1979), and the concentration of zoospores was estimated to be about 1 × 105 spores mL–1 using a hemacytometer. The leaves of the inoculated plants were harvested and immediately frozen in liquid nitrogen at 0, 1, 3, 6, 9, 12, 24, and 48 h after the treatment, and stored at −80°C until required for RNA extraction.
The full-length cDNA of GmSnRK1.1 (GenBank accession no. XM_006585690) was isolated from soybean “Suinong 10” using RT-PCR with the primers GmSnRK1.1-F/R (see Supplementary Table S1). The extraction of total RNA and reverse transcription were performed using TRIzol reagent (Invitrogen, China) and ReverTra Ace Kit (Toyobo, Japan). The products of the RT-PCR amplification were cloned into a pMD-18T vector (Takara Bio, Japan), transformed into Escherichia coli DH5α cells (TransGen Biotech, China), and sequenced by GENEWIZ (China). DNAMAN software1 was used for the sequence alignments, and a phylogenetic analysis of GmSnRK1.1 was carried out using MEGA5 software. The GmSnRK1.1 protein structure was analyzed using the online program Phyre22.
A qRT-PCR analysis was performed to confirm the transcript levels of GmSnRK1.1 using a LightCycler96 instrument (Roche, Switzerland) with a real-time PCR kit (TOYOBO, Japan). GmEF1β (GenBank accession no. NM_001248778) was used as the internal control (see Supplementary Table S1 for primers). The relative transcript abundance of the target gene was calculated using the 2–ΔΔCT method. Three biological replications were performed for each line in each analysis.
The coding sequence of GmSnRK1.1 was amplified and inserted into pGADT7 (Takara Bio), after which the plasmids pGADT7-GmSnRK1.1 and pGBKT7-GmWRKY31 were Co-transferred into the yeast strain Y2HGold (Takara Bio). The protein-protein interactions were determined by growth on three types of medium: SD (–Trp/–Leu) medium, SD (–Trp/–Leu/–His/–Ade) medium, and SD (–Trp/–Leu/–His/–Ade/X-α-gal) medium. Yeast cells carrying the pGBKT7-53 and pGADT7-SV40 plasmids were used as the positive control, and pGADT7-GmSnRK1.1: pGBKT7 and pGADT7:pGBKT7-GmWRKY31 were used as the negative control.
To further evaluate the interaction between GmSnRK1.1 and GmWRKY31, a BiFC assay based on yellow fluorescent protein (YFP) was performed. To construct the vectors, the coding region of GmSnRK1.1 was cloned using the primers GmSnRK1.1-bF/R and cloned into the pSAT6-cEYFP-N1 vector. The coding region of GmWRKY31 was amplified and cloned into the pSAT6-nEYFP-N1 vector (Fan et al., 2017). The plasmids were transformed into Arabidopsis protoplasts using polyethylene glycol (PEG)-mediated transfection (Yoo et al., 2007). The GmSnRK1.1-cEYFP-N1/pSAT6-nEYFP-N1 and GmWRKY31-nEYFP-N1/pSAT6-cEYFP-N1 vector combinations were used as negative controls, GmWRKY31-YEPN/GmHDL56-YEPC were used as positive controls (Fan et al., 2017). The transfected cells were imaged using a TCS SP2 confocal spectral microscope imaging system (Leica Microsystems, Germany). The 514 nm Ar/ArKr laser was used for YFP and Chlorophyll. YFP and Chlorophyll were excitated at 514 nm and 488 nm, respectively. The wavelength range of captured light was 530–560 nm for YFP, and 650–750 nm for Chlorophyll.
GmSnRK1.1 was cloned into the pET29b (+) expression vector (Merck Millipore, United States), while GmWRKY31 was cloned into the pGEX-4T-1 expression vector (GE Healthcare, United States). The His-GmSnRK1.1 and glutathione S-transferase-GmWRKY31 proteins were separately produced in E. coli BL21 (DE3) cells, then harvested and purified using a GST-Sefinose kit (Sangon, China) or a His-bind Purification Kit (Merck Millipore). The pull-down assay was performed as described by Yang et al. (2008), with minor modifications. In a total volume of 1 mL GST binding buffer (Sangon), the GST or GmWRKY31-GST recombinant proteins were incubated for 1 h at 4°C with 400 μL GST resin (Sangon), after which equal volumes of the GmSnRK1.1-His recombinant protein were added and incubated for 6 h at 4°C. The binding reaction was washed five times with binding buffer, each for 10 min at 4°C, then the pulled-down proteins were eluted by boiling, separated on a 12% SDS-PAGE gel, and immunoblotted with anti-His antibody and anti-GST antibody (Abmart, United States).
The full-length GmSnRK1.1 sequence was cloned using RT-PCR with the primers GmSnRK1.1GF and GmSnRK1.1GR (listed in Supplementary Table S1). The coding sequence under the control of the constitutive CaMV 35S promoter was fused to the N-terminus of the green fluorescent protein (GFP). The resulting 35S:GmSnRK1.1-GFP expression plasmid (or the 35S:GFP control) was transformed into Arabidopsis protoplast cells using a PEG-mediated transfection, as described by Yoo et al. (2007). The fluorescence signal was mapped using a TCS SP2 spectral confocal microscopic imaging system (Leica Microsystems). The 514 nm Ar/ArKr laser was used for GFP and Chlorophyll. GFP and Chlorophyll were excitated at 488 nm. The wavelength range of captured light was 500–530 nm for GFP, and 650–750 nm for Chlorophyll.
For the generation of the overexpression lines, a 4 × myc sequence was synthesized (GENEWIZ) and inserted into a pCAMBIA3301 vector to generate a pCAMBIA3301-4 × Myc plasmid, the GmSnRK1.1 coding sequence was inserted into the BglII/SpeI site of the plasmid, and the 4 × myc and bar sequences were later used as markers. The GmSnRK1.1 cDNA fragment was amplified using the primers GmSnRK1.1-R-F/R and inserted into the vector PFGC5941 (Kerschen et al., 2004). The constructs were transformed into Agrobacterium tumefaciens (strain LBA4404) using the freeze-thaw method (Holsters et al., 1978). “Dongnong 50” was previously used for Agrobacterium-mediated genetic transformation (Paz et al., 2004). Transgenic soybean plants were preliminarily verified using a PCR amplification and qPCR analysis, after which a Western blot with an anti-myc antibody (Abmart) was used to identify the plants overexpressing GmSnRK1.1.
For pathogen infection, the living cotyledons of the WT and transgenic soybean plants at the V1 stage of development were infected with P. sojae zoospores (approximately 1 × 105 spores mL–1) using the methods described by Morrison and Thorne (1978), and the roots inoculation was performed using the procedure described by Zhang et al. (2017). The disease symptoms on the infected cotyledons and roots were observed and photographed with a Nikon D7000 camera. ImageJ3 was used to measure the lesions of the infected cotyledons. The P. sojae biomass was quantified based on the accumulation of P. sojae TEF1 (GenBank accession no. EU079791) in the soybean plants, relative to the levels of GmEF1β, as previously described by Chacón et al. (2010). The pathogen response assays were performed on three biological replicates, each with three technical replicates.
For the enzyme assays, the total proteins were extracted from approximately 0.1 g of leaves using 1 mL ice-cold 25 mM HEPES buffer (pH 7.8) containing 0.2 mM EDTA, 2 mM ascorbate, and 2% polyvinylpyrrolidone. The homogenates were centrifuged at 4°C for 15 min at 12,000 × g, after which the supernatants were carefully removed and used for the enzymatic activity measurements. The superoxide dismutase (SOD) and peroxidase (POD) activities were assayed as described by Wang et al. (2011).
Salicylic acid (SA) was extracted from the T3 GmSnRK1.1 transgenic soybean leaves and quantified using HPLC-mass spectrometry, as previously described (Aboul-Soud et al., 2004, Pan et al., 2010).
Using yeast two-hybrid assays, GmWRKY31 was found to interact with GmSnRK1.1 (Figure 1A), which was further confirmed using a BiFC assay demonstrating that GmSnRK1.1 can interact with GmWRKY31 in the nuclei of Arabidopsis protoplast cells (Figure 1C). In accordance with the results of the BiFC assay, a glutathione S-transferase pull-down assay showed that the His-tagged GmSnRK1.1 recombinant protein was pulled down by GST-GmWRKY31, but not by GST alone (Figure 1B), further indicating that GmWRKY31 interacts with GmSnRK1.1 in vitro. These results suggest that GmWRKY31 directly interacts with GmSnRK1.1.
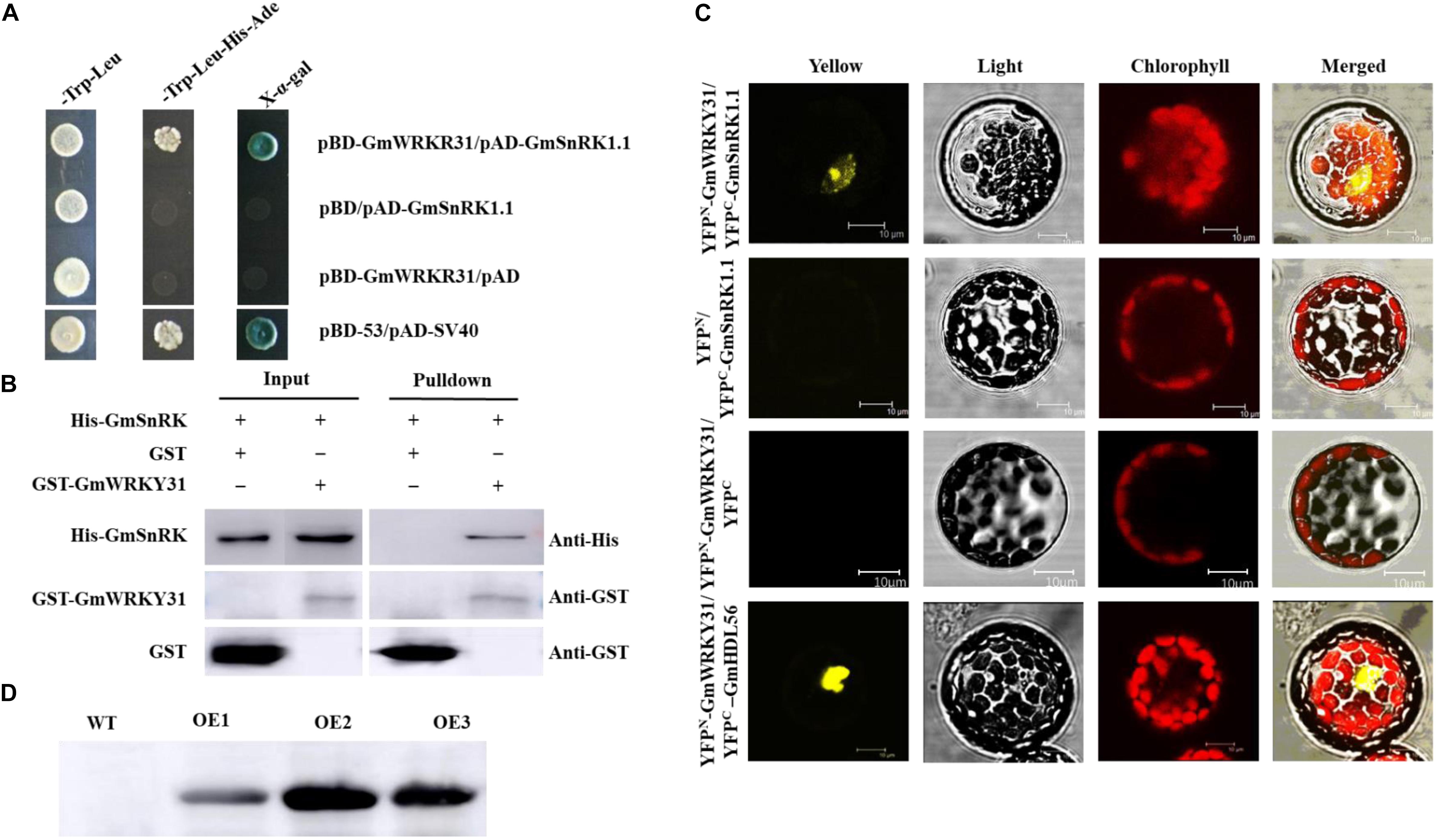
Figure 1. Interaction of GmSnRK1.1 with GmWRKY31 in vitro and in vivo and western blot analysis of the expression of GmSnRK1.1. (A) Analysis of interactions between GmSnRK1.1 and GmWRKY31 protein in yeast cells. The yeast cells of strain Y2H harboring pAD-GmSnRK1.1 and pBD-GmWRKY31 plasmid combinations were grown on either SD/-Trp/-Leu media or SD/-Trp/-Leu/-His/-Ade media, followed by b-galactosidase assay (SD/-Trp/-Leu/-His/-Ade/X-a-gal media). (B) Bimolecular fluorescence complementation (BiFC) analysis of interaction between GmSnRK1.1 and GmWRKY31 in Arabidopsis protoplast cells. The plasmid combinations are indicated on top. The fluorescence of YFP was observed by confocal laser microscopy 16 h after transfection. Bars, 10 μm. (C) Pull-down assay of GmSnRK1.1 interaction with GmWRKY31. His-GmSnRK1.1 protein was incubated with immobilized GST or GST-GmWRKY31 proteins, and immunoprecipitated fractions were detected by anti-His antibody. (D) western blot analysis of the expression of GmSnRK1.1 in three positive overexpressing transgenic soybean lines (OE1, OE2 and OE3).
The full-length GmSnRK1.1 cDNA is 1,990 bp long and contains a 1,533 bp open reading frame, which encodes a polypeptide of 510 amino acids (Supplementary Figure S2). Phylogenetic tree and alignment analyses revealed that GmSnRK1.1 shares 67.91–93.02% identity in overall amino acid sequence with its other plant species homologs, including Lotus japonicus LjSnRK (BAD95888), Manihot esculenta MeSnRK (XP_021604368), Fragaria vesca FvSnRK (XP_004304271), Cucumis sativus CsSnRK (XP_004145003), Vitis vinifera VvSnRK (XM_002283963.1), Cucumis melo CmSnRK (XP_008460108), Pyrus bretschneideri PbSnRK (XP_009360590), Populus trichocarpa PtSnRK (XP_002306053), Morus notabilis MnSnRK (XP_024016886), Vicia faba VfSnRK (AJ971809.1), Pisum sativum PsSnRK (CAI96819.1), Nicotiana attenuate NaSnRK (AAS18877), Populus euphratica PeSnRK (XP_011010304), Arabidopsis thaliana AtSnRK (M93023.1), Daucus carota DcSnRK (XP_017242374), Sorghum bicolor SbSnRK (EF544393.1), Zea mays ZmSnRK (AY486125.1), Solanum tuberosum StSnRK (CAA65244.1), Solanum lycopersicum SlSnRK (NP_001234325.1), and GmSnRK1.1 has the highest similarity with LjSnRK (Supplementary Figures S1B,C). The structure of GmSnRK1.1 was analyzed using Phyre, predicting that it functions as a heterotrimer complex, in which the catalytic α subunit combines with a β regulatory subunit and an activating γ subunit (Supplementary Figure S1C).
To evaluate the responsiveness of GmSnRK1.1 to biotic stresses in the “Dongnong 50” and “Suinong 10” soybean cultivars, its temporal and spatial patterns were investigated using qRT-PCR. The examination of the tissue-specific transcript levels in these cultivars revealed that GmSnRK1.1 was highly expressed in the stems, followed by the roots and cotyledons (Figure 2A). In “Suinong 10” plants inoculated with P. sojae, the GmSnRK1.1 mRNA levels increased to a peak level at 9 h after inoculation, followed by a decline (Figure 2B). A similar pattern was observed in “Dongnong 50,” although the relative expression level of GmSnRK1.1 was significantly higher in “Suinong 10” than in “Dongnong 50” (Figure 2B).
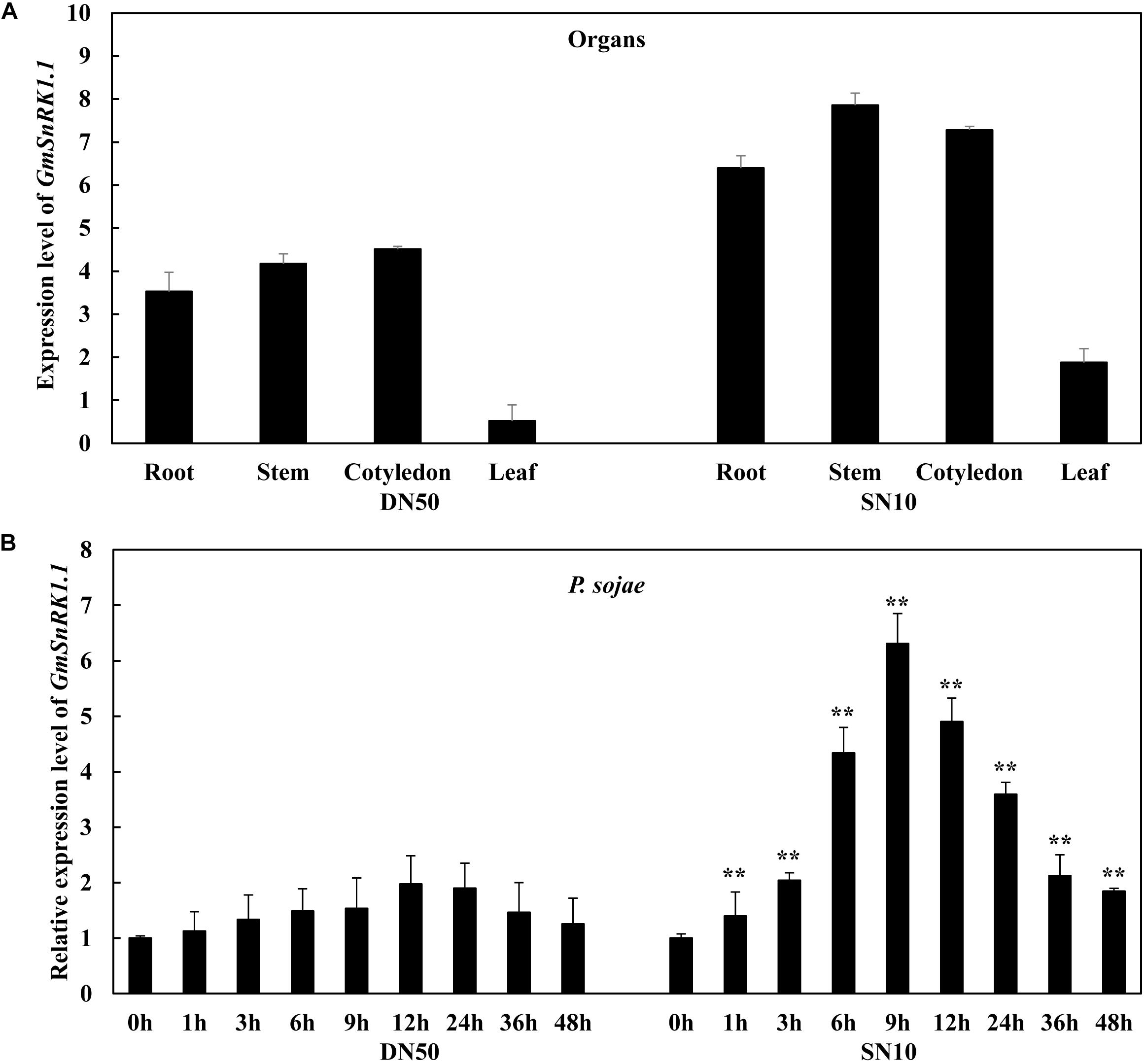
Figure 2. Expression patterns of GmSnRK1.1 in Phytophthora sojae-resistant and P. sojae-susceptible soybean cultivars. (A) The tissue-specific expression patterns of GmSnRK1.1 in resistant cultivar “Suinong 10” (SN10) and susceptible cultivar “Dongnong 50” (DN50) under normal conditions. (B) Relative expression of GmSnRK1.1 in soybean cultivars ‘Suinong 10” and “Dongnong 50” on P. sojae infection. The infected samples were collected at 0, 1, 3, 6, 9, 12, 24, 36 and 48 h after P. sojae infection. The relative expression levels of GmSnRK1.1 were compared with those of mock-treated plants (plants treated with sterile water) at the same time point. Fourteen-day-old soybean plants were used for analysis. The housekeeping gene of soybean GmEF1β (NM_001248778) was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean.
The subcellular localization of the GmSnRK1.1 protein was analyzed in Arabidopsis protoplasts producing a GmSnRK1.1-GFP fusion protein under the control of the 35S promoter. As shown in Figure 3, GFP fluorescence was distributed throughout the cells expressing the 35S:GFP control plasmid. In contrast, the GmSnRK1.1-GFP fusion protein was exclusively localized to the Arabidopsis cell membrane, resembling the pattern of the membrane-localized GmDIR22-GFP fusion protein (Li et al., 2017) used as a control. These results indicated that GmSnRK1.1 is a membrane-localized protein.
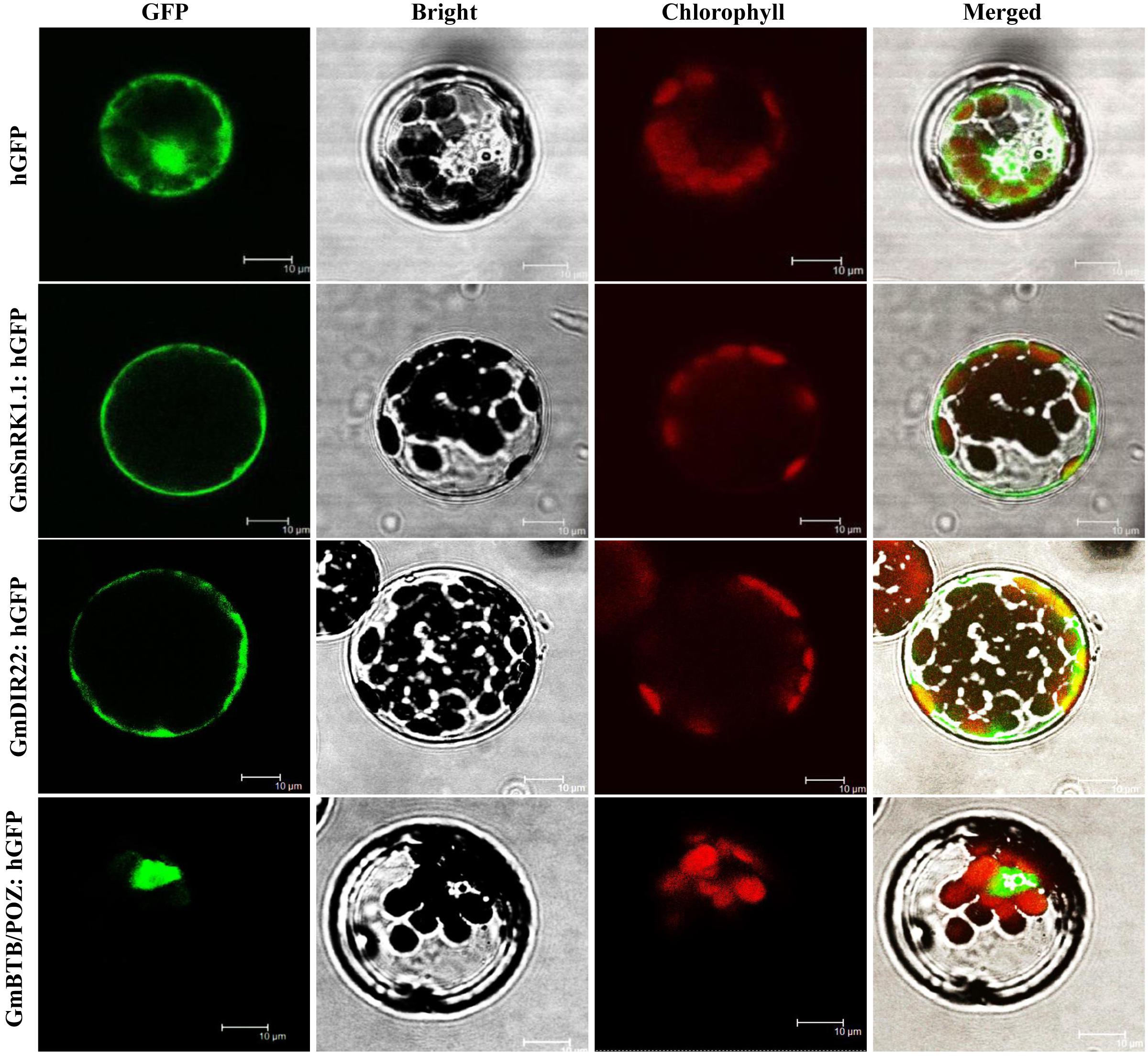
Figure 3. Analysis of the subcellular localization of GmSnRK1.1-GFP protein in Arabidopsis protoplasts. Subcellular localization was investigated in Arabidopsis mesophyll protoplasts under a confocal microscope. The fluorescent distribution of humanized hGFP, the fusion protein GmSnRK1.1-hGFP, GmDIR22-hGFP and GmBTB/POZ-hGFP were observed under white light, UV light, and red light, respectively. Bars, 10 μm.
To analyze the function of GmSnRK1.1 in response to infection by P. sojae, we generated GmSnRK1.1-OE and GmSnRK1.1-R transgenic soybean plants, which were developed into transgenic T3 lines. the expression of GmSnRK1.1 in three positive overexpressing transgenic soybean lines using Western blot (Figure 1D). The resistance of the T3 transgenic plants to P. sojae was tested in their cotyledons and roots. A notable difference was observed in the development of disease symptoms after a 96 h incubation with zoospores of P. sojae. In the GmSnRK1.1-R lines, the cotyledons exhibited clear water-soaked lesions and were softer than the WT, however, almost no disease symptoms were observed in the GmSnRK1.1-OE lines (Figure 4A). In addition, the P. sojae biomass (indicated by the relative abundance of TEF1 genomic sequence per area of infected living cotyledon) was significantly (P < 0.01) lower in the GmSnRK1.1-OE lines than in the WT plants, but higher in the GmSnRK1.1-R lines (Figure 4B). The lesion areas of the GmSnRK1.1-OE lines were significantly (P < 0.01) smaller than that of the WT (Figure 4C), but significantly larger in the GmSnRK1.1-R lines. Similar results were obtained after a 6-d incubation with P. sojae. The living roots of the WT soybean plants and GmSnRK1.1-R soybean lines exhibited watery lesions and even rotting, while those of the GmSnRK1.1-OE lines remained healthy (Figure 5). Similar to the results of infecting living cotyledon, the biomass of P. sojae after 6 days of roots infection was significantly reduced in the GmSnRK1.1-OE lines, but significantly increased in the GmSnRK1.1-R lines, relative to the WT. These results indicated that overexpression of GmSnRK1.1 in soybean plants enhances their resistance to P. sojae infection. In addition, we have constructed the overexpression vector of kinase-inactive GmSnRK1.1 by synthesizing the mutation sequence of phosphorylation site (Thr157Ala, Thr235Ala, Thr261Ala). Furthermore, the overexpression of kinase-inactive GmSnRK1.1, the overexpression of GmSnRK1.1, and vector control transgenic soybean hairy roots generated by Agrobacterium rhizogenes-mediated transformation will be used to investigate the effect of the GmSnRK1.1’s kinase activity on resistance to P. sojae in soybean. The results demonstrated that GmSnRK1.1’s kinase activity could increase the resistance to P. sojae in soybean (Supplementary Figure S5).
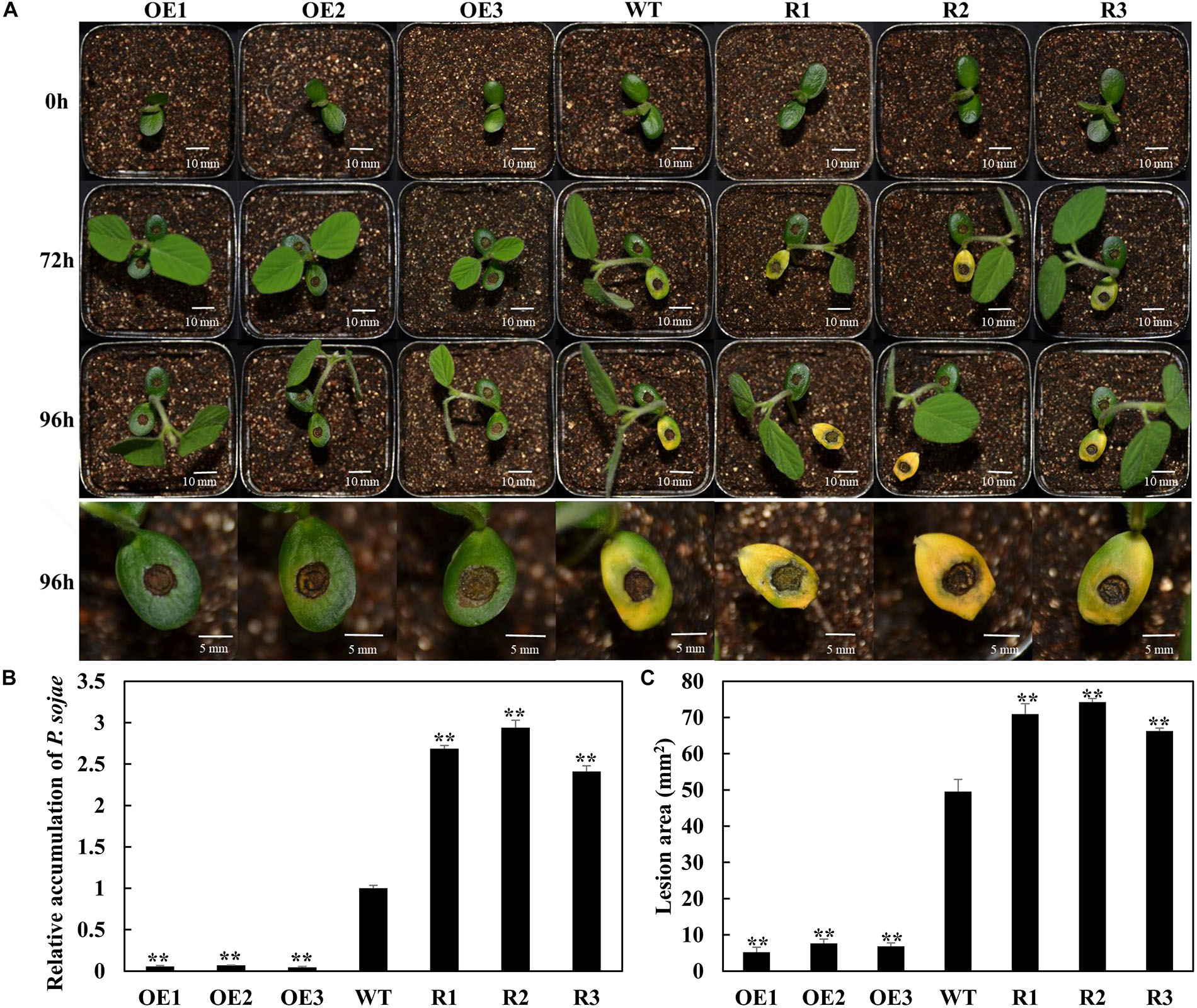
Figure 4. GmSnRK1.1 enhances resistance to Phytophthora sojae in transgenic soybean cotyledons. (A) Disease symptoms on living cotyledons of GmSnRK1.1-overexpressing (GmSnRK1.1-OE), GmSnRK1.1 RNA interference (RNAi)-mediated silencing (GmSnRK1.1-R) and wild-type (WT) plants at 96 h after inoculation with P. sojae. (B) Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of the relative biomass of P. sojae in GmSnRK1.1-OE, GmSnRK1.1-R transgenic lines and WT soybean based on P. sojae TEF1 transcript levels. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analyzed using Student’s t-test (∗P < 0.05,∗∗P < 0.01). Bars indicate the standard error of the mean. (C) Lesion size measured from photographed cotyledons of GmSnRK1.1-OE, GmSnRK1.1-R transgenic and WT plants at 96 h post-inoculation (hpi). The lesion size of each independent soybean line (n = 3) was calculated, and the lesion sizes are shown in the columns based on a comparison with the average lesion area on WT soybean.
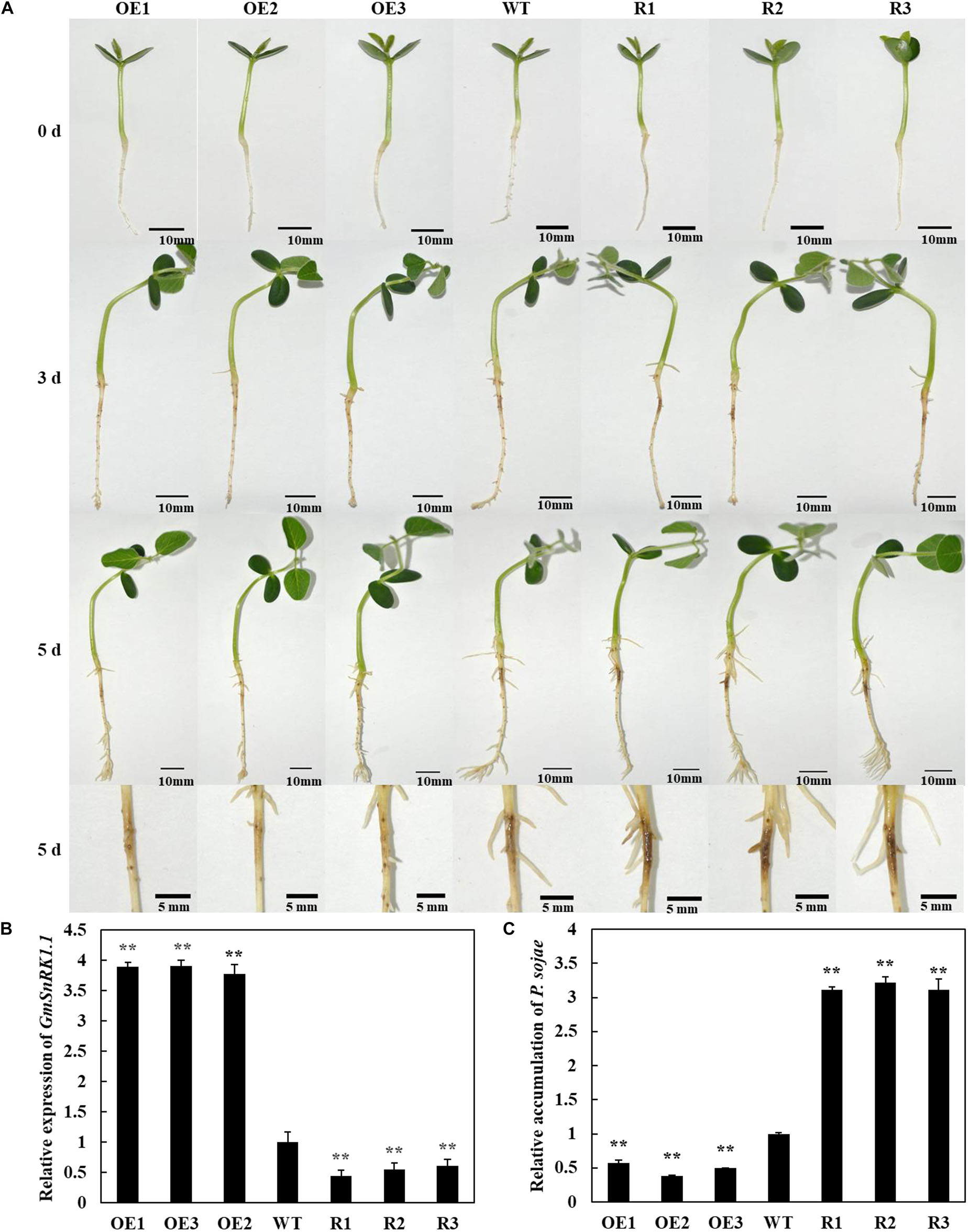
Figure 5. Resistance analysis of GmSnRK1.1 transgenic soybean plants. (A) Disease symptoms on the roots of GmSnRK1.1-overexpressing (GmSnRK1.1-OE), GmSnRK1.1 RNA interference (RNAi)-mediated silencing (GmSnRK1.1-R) and wild-type (WT) plants at 6 days after inoculation with Phytophthora sojae. (B) Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of the relative expression of GmSnRK1.1 in GmSnRK1.1-OE, GmSnRK1.1-R transgenic lines and WT soybean. (C) Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of the relative biomass of P. sojae in GmSnRK1.1-OE, GmSnRK1.1-R transgenic lines and WT soybean based on P. sojae TEF1 transcript levels. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analyzed using Student’s t-test (∗P < 0.05,∗∗P < 0.01). Bars indicate the standard error of the mean.
The antioxidant defense system is well-developed in plants, involving the scavenging of reactive oxygen species (ROS) by SOD and POD (Du et al., 2001). We analyzed the SOD and POD activities in the transgenic and WT soybean plants inoculated with P. sojae, as well as the expression of the associated genes GmSOD1 (NM_001248369) and GmPOD (XM_006575142). Under both the mock treatment and at 24 h after inoculation with P. sojae, the activity levels of SOD and POD, as well as the transcript abundance of the associated genes, were significantly higher in the GmSnRK1.1-OE lines than in the WT, but were significantly reduced in the GmSnRK1.1-R lines (Figure 6). These results suggested that GmSnRK1.1 increases the activities of the antioxidant enzymes in soybean plants in response to P. sojae infection.
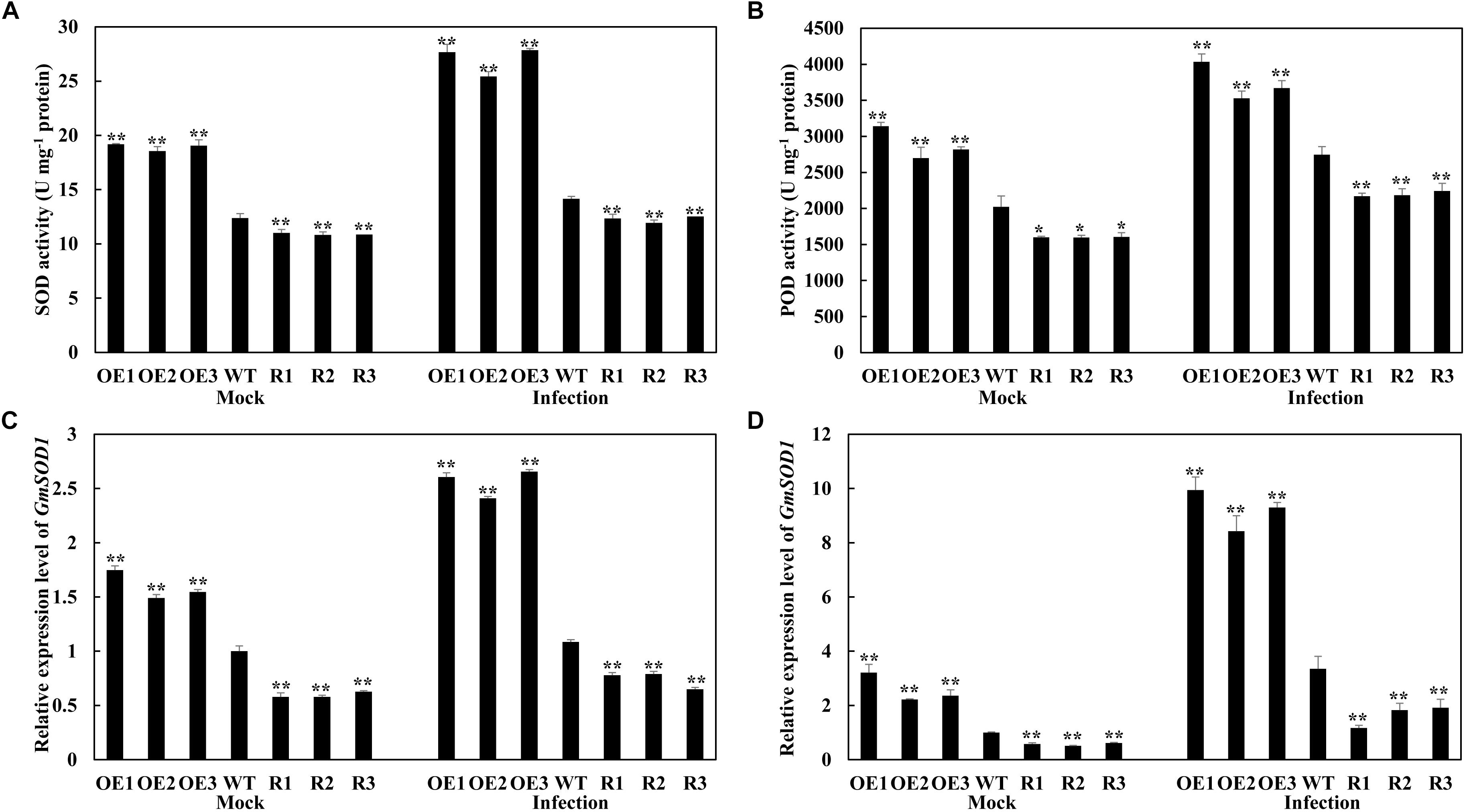
Figure 6. Analysis of antioxidant enzyme activity (A,B) and the relative expression of genes (C,D) under mock treatment and infected by Phytophthora sojae at 24 h post-inoculation (hpi). The activity of the control sample [mock-treated wild-type (WT) plants] was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean. POD, peroxidase; SOD, superoxide dismutase.
Race-specific resistance to P. sojae has been shown to be mediated by isoflavonoid phytoalexins in other soybean varieties (Subramanian et al., 2005; Graham et al., 2007; Cheng et al., 2015; Zhang et al., 2017). We measured the expressions of several isoflavonoid phytoalexins genes, including GmPAL (GenBank accession no. NM_001250027), GmIFR (GenBank accession no. NM_001254100), Gm4CL (GenBank accession no. NM_001256363.1) and GmCHS (GenBank accession no. XM_003518780). These results indicated that isoflavonoid phytoalexins related genes were significantly higher in “Suinong 10” and GmSnRK1.1-OE lines than these in “Dongnong 50,” and were significantly lower in GmSnRK1.1-R lines (Figure 7). Moreover, we next monitored the expression levels of GmSnRK1.1 during P. sojae infection using qRT-PCR. The transcript levels of GmSnRK1.1 were significantly higher in the GmSnRK1.1-OE plants than in the WT under both the mock treatment and a 24 h infection with P. sojae, but were significantly lower in the GmSnRK1.1-R lines (Supplementary Figure S4). Pathogen-related proteins are key members in the plant response to pathogen infection (Van Loon and Van Strien, 1999; Loon et al., 2006; Xu et al., 2014). To explore the possible mechanisms of the GmSnRK1.1-regulated resistance to P. sojae, we detected the transcriptional levels of various defense-response genes, including GmWRKY31, GmNPR1 (GenBank accession no. NM_001251745.1), GmPR1 (GenBank accession no. AF136636), and GmPR5 (GenBank accession no. M21297). As shown in Supplementary Figure S4, after 12 h incubation with P. sojae, the expression levels of these resistance-related genes were significantly more highly expressed in the GmSnRK1.1-OE plants than in the WT, but were significantly lower in the GmSnRK1.1-R plants. In contrast, no significant differences in the expression of GmPR10 (GenBank accession no. FJ960440) were detected between these lines. These results indicate that one mechanism by which GmSnRK1.1 enhances soybean defense against P. sojae is by regulating the expression of the defense-related genes.
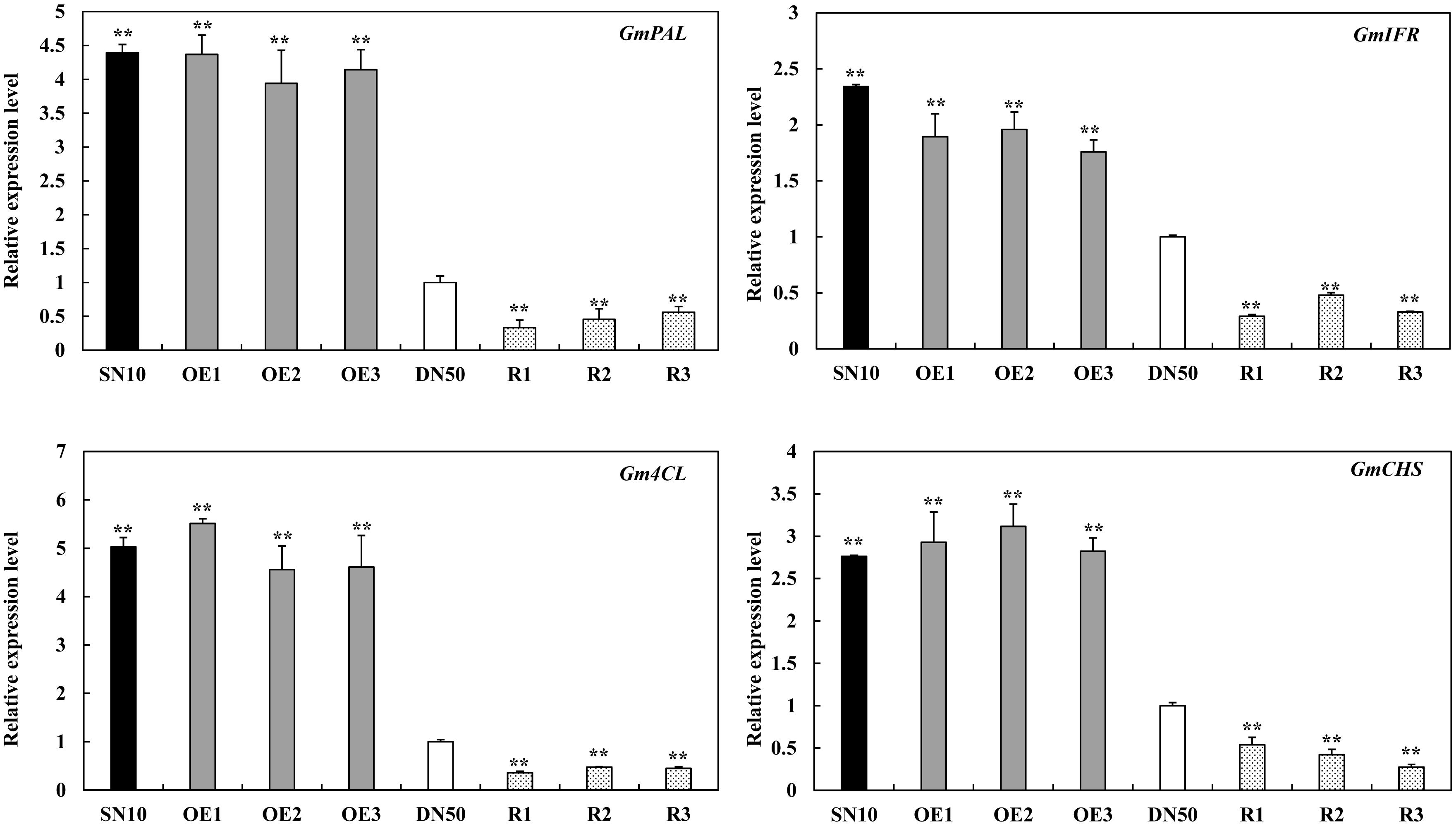
Figure 7. The relative transcript levels of isoflavonoid phytoalexins related genes in “Suinong 10” (SN10), “Dongnong 50” (DN50) and GmSnRK1.1 transgenic soybean plants. The housekeeping gene of soybean GmEF1β was used as an internal control to normalize the data. The experiment was performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean.
To test whether the GmSnRK1.1 could regulate the accumulation of SA, the SA contents of the T3 GmSnRK1.1 transgenic lines were evaluated. As shown in Figure 8A, the GmSnRK1.1-OE transgenic soybean leaves contained significantly more SA than the WT leaves, while the GmSnRK1.1-R plants accumulated significantly less SA. we also analyzed the transcript levels of GmICS1 (XM_003522145), which plays a key role in SA biosynthesis. GmICS1 was significantly more highly expressed in the GmSnRK1.1-OE transgenic lines than in the WT, while the GmSnRK1.1-R transgenic lines has a significantly lower level of GmICS1 expression (Figure 8B). Furthermore, whether or not the defense mechanism is dependent on SA, we measured the expression level of GmSnRK1.1 and the relative biomass of P. sojae in “Suinong 10,” “Dongnong 50” and GmSnRK1.1 transgenic soybean lines treat with sterile water and SA biosynthesis inhibitor (100 μM 1-aminobenzotriazole). As expected, SA biosynthesis inhibitor treated plants displayed clearly increased pathogen biomass compared with H2O-treated plants after 24 h post-inoculation (Figure 8D). These results suggested that GmSnRK1.1 plays a positive important role in the response to P. sojae infection, increasing the disease-resistance of soybean via a possible mechanism involving the SA signaling pathway.
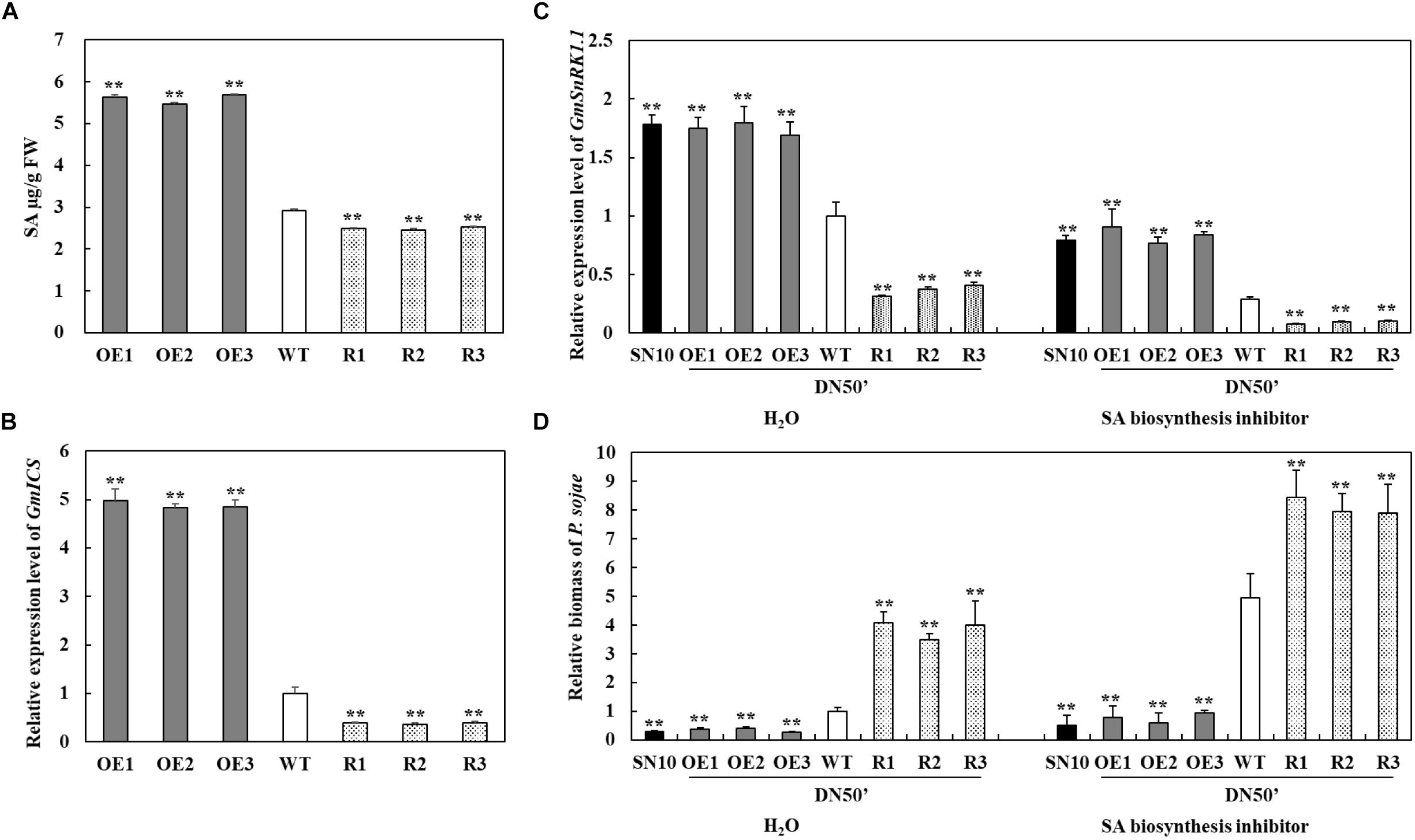
Figure 8. Investigation of the relationship between GmSnRK1.1 and the salicylic acid (SA) pathway in soybean. (A) SA contents in leaves of transgenic and wild type (WT) soybean. FW, fresh weight. (B) Relative transcript level of GmICS in GmSnRK1.1-overexpressing (GmSnRK1.1-OE), GmSnRK1.1 RNA interference (RNAi)-mediated silencing (GmSnRK1.1-R) transgenic and WT soybean. The level of the control sample (WT plants) was set to unity. (C) Relative transcript level of GmSnRK1.1 in “Suinong 10” (SN10), “Dongnong 50” (DN50) and GmSnRK1.1 transgenic soybean lines treated with H2O (mock) and SA biosynthesis inhibitor (100 μM ABT) under P. sojae infection. (D) The relative biomass of P. sojae in “Suinong 10” (SN10), “Dongnong 50” (DN50) and GmSnRK1.1 transgenic soybean lines treated with H2O (mock) and SA biosynthesis inhibitor (100 μM ABT) under P. sojae infection. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean.
Identification and characterization of genes involved in response to P. sojae infection in soybean has contributed to our understanding of the genetic mechanisms of resistance (Xu et al., 2014; Cheng et al., 2015; Kong et al., 2015; Zhang et al., 2017). In our previous study, GmWRKY31 was found to play a key role in increasing the disease resistance of soybean plants to P. sojae infection (Fan et al., 2017). In the present study, we identified and functionally characterized GmSnRK1.1 as an interacting partner of GmWRKY31, demonstrating that it plays a positive role in the response to P. sojae infection. The SnRK1 protein kinases are considered central regulators of the energy metabolism and stress signaling in plants (Baena-González and Sheen, 2008; Radchuk et al., 2010; Tsai and Gazzarrini, 2012), however, the biological functions of the SnRK1s in soybean are poorly understood. Here, we first discovered that the overexpression of GmSnRK1.1 significantly increased the plant responses to P. sojae infection, while RNA-interference mediated reduced expression of this gene significantly increased the susceptibility of the transgenic plants to this pathogen (Figures 4, 5).
Previous studies have confirmed that the SnRK1s play a role in the plant defense against viruses, fungi, and bacteria (Hao et al., 2003; Szczesny et al., 2010; Kim et al., 2015); for example, the overexpression of SnRK1 was shown to significantly increase the resistance of tobacco plants to geminivirus infection (Hao et al., 2003). In rice, OSK35 (a rice SnRK1b gene) positively regulates the disease resistance of plants subjected to the fungal pathogen Magnaporthe oryzae and the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo) (Kim et al., 2015). The wheat (Triticum aestivum) alpha subunit of TaSnRK1a interacts with TaFROG (Fusarium Resistance Orphan Gene) to increase the disease resistance response to the mycotoxigenic fungus Fusarium graminearum (Perochon et al., 2015). The hypersensitive response was induced by the Xanthomonas campestris pv. vesicatoria effector AvrBs1 in snrk1 mutant pepper plants (Szczesny et al., 2010). In this work, the temporal and spatial patterns of GmSnRK1.1 expression were analyzed in the P. sojae-resistant soybean cultivar “Suinong 10” and the susceptible cultivar “Dongnong 50,” revealing that GmSnRK1.1 was markedly expressed in the stems of these lines, with lower expression levels in the roots and cotyledons (Figure 2A). We detected that the expression levels of GmSnRK1.1 following P. sojae infection were much higher in “Suinong 10” than in the susceptible cultivar “Dongnong 50,” and that P. sojae infection markedly increased the expression levels of GmSnRK1.1 in both cultivars (Figure 2B). In addition, GmSnRK1.1 is negatively regulated by ABA (Supplementary Figure S3). We further demonstrated that the GmSnRK1.1-OE transgenic plants had an increased disease resistance to P. sojae infection, while the GmSnRK1.1-R transgenic plants exhibited increased susceptibility (Figures 4, 5).
Further research showed that GmSnRK1.1 positively regulates the activities and transcription levels of antioxidant enzymes SOD and POD (Figure 6). SOD constitutes the first line of defense against ROS within a cell (Alscher et al., 2002), while POD plays a key role in scavenging ROS and preventing cellular damage (Tewari et al., 2006). Therefore, we further analysis the activities and transcription level of SOD and POD, In plants infected with P. sojae, GmSnRK1.1 positively enhanced the activities and transcription levels of SOD and POD, enabling them to scavenge the ROS and provide sufficient protection against oxidative damage.
The phytohormones SA, jasmonic acid, and ethylene play central roles in regulating the plant responses to pathogen attack (Reymond and Farmer, 1998; Kunkel and Brooks, 2002; Spoel et al., 2003; Robert-Seilaniantz et al., 2011; Alazem and Lin, 2015). SA mediates and activates the biotic stress response to pathogenic challenge (Pieterse et al., 2009; Sugano et al., 2013; Alazem and Lin, 2015), with the transcriptional cofactor NPR1 playing a key role in the SA-signaling pathway of several plant species (Vlot et al., 2009). In rice, OsSnRK1a positively regulates plant resistance by linking to the SA pathway (Filipe et al., 2018). In previous studies, the overexpression of GmWRKY31 was found to induce the expression of GmNPR1, increasing the disease resistance of soybean plants in response to P. sojae infection via the activation of the SA-signaling pathway (Fan et al., 2017). The results of this study supported these findings, as the expression levels of GmWRKY31 and GmNPR1 were markedly higher in the GmSnRK1.1-OE transgenic plants in comparison with the WT and lower in the GmSnRK1.1-R transgenic plants (Supplementary Figure S4). A downstream member of the SnRK1 signaling pathway, STOREKEEPER RELATED1/G-Element Binding Protein (STKR1), was previously found to display transcriptional changes which constitutively activated the SA-related defense in transformed Arabidopsis plants (Nietzsche et al., 2018). We also found that the overexpression of GmSnRK1.1 induced the expression of GmPR1 and GmPR5, which are effector genes for the systemic acquired resistance response, a process mediated by SA (Ward et al., 1991; He et al., 2007). The high expression levels of these genes indicated that SA signaling was activated in the GmSnRK1.1-OE plants, which was confirmed by our determination that SA accumulation and the expression of the SA biosynthesis gene GmICS1 were upregulated in these plants relative to the WT (Figure 8). In addition, we examined the expression level of GmSnRK1.1 and relative biomass of P. sojae in “Suinong 10,” “Dongnong 50” and GmSnRK1.1 transgenic soybean lines treated with H2O (mock) and SA biosynthesis inhibitor (100 μM ABT) under P. sojae infection, the results showed that SA is involved in GmSnRK1.1-mediating defense to P. sojae (Figure 8). These indicated that GmSnRK1.1 promotes the accumulation of SA and the expression of GmICS1, and suggests that GmSnRK1.1 acts as a positive regulator of the downstream defense pathways and SA-dependent defense signaling.
In Glycine soja, a ABA activated calcium independent SnRK-type kinase, GsAPK, was localized in the plasma membrane (Liang et al., 2012). In Arabidopsis, SnRK1 is localized to the plant nucleus and endoplasmic reticulum (Blanco et al., 2019). In this work, GmSnRK1.1 was localized in the plasma membrane (Figure 3), and GmWRKY31 was localized to the plant nucleus (Fan et al., 2017). The BiFC assay showed that GmSnRK1.1 interacted with GmWRKY31 in the nucleus (Figure 1), but the mechanism of nuclear interaction is not clear which require further study and discussion.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
PX and SZ designed the experiments. LW, HW, SH, and FM performed the experiments. FM, LW, SF, and JW analyzed the data. LW, PX, SZ, and CZ wrote the manuscript.
This work was supported by National Key Research and Development Program of China (2017YFD0101300), NSFC Projects (31671719), Outstanding Talents and Innovative Team of Agricultural Scientific Research, Key Research and Development Program of Heilongjiang (GJ2018GJ0028), Academic backbone of NEAU (17XG21), Natural Science Foundation of Heilongjiang Province (ZD2019C001), and Changjiang Scholar Candidates Program for Provincial Universities in Heilongjiang (2013CJHB003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00996/full#supplementary-material
FIGURE S1 | Sequence comparison of GmSnRK1.1 with orthologs from other plant species. (A) Phylogenetic analysis of GmSnRK1.1 with orthologs from other plant species. (B) Alignment of amino acid sequences of GmSnRK1.1 with orthologs from other plant species. (C) The tertiary structure of the GmSnRK1.1 protein.
FIGURE S2 | The open reading frame sequence and deduced polypeptide sequence of GmSnRK1.1.
FIGURE S3 | The relative transcript levels of GmSnRK1.1 at various time points post-treatment with ABA in “Suinong 10” and “Dongnong 50” soybean plants. Fourteen day-old plants were used for the treatments and analyses. The housekeeping gene of soybean GmEF1β was used as an internal control to normalize the data. The relative transcript levels of GmSnRK1.1 were quantified compared with mock plants at the same time points. The experiment was performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean.
FIGURE S4 | Relative expression levels of defense-associated genes in soybean plants under mock treatment and infected by Phytophthora sojae at 24 h post-inoculation (hpi). The housekeeping gene of soybean GmEF1β was used as an internal control to normalize the data. The expression level of the control sample [mock-treated wild-type (WT) plants] was set to unity. The experiment was performed on three biological replicates, each with three technical replicates, and was statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean.
FIGURE S5 | The effect of GmSnRK1.1 kinase assay on the resistance to P. sojae. (A) Mutation of GmSnRK1.1 activation sites. (B) Disease symptoms on the hairy roots of the overexpression of GmSnRK1.1 (GmSnRK1.1-OE), the overexpression of kinase-inactive GmSnRK1.1 (GmSnRK1.1 mutation-OE), and empty vector control (3301), GmSnRK1.1 RNA interference (RNAi)-mediated silencing (GmSnRK1.1-R) transgenic soybean at 4 days after inoculation with Phytophthora sojae. (C) Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of the relative biomass of P. sojae in GmSnRK1.1-OE, GmSnRK1.1 mutation-OE, GmSnRK1.1-R, and 3301 empty vector transgenic soybean hairy roots based on P. sojae TEF1 transcript levels. The experiment was performed on three biological replicates, each with three technical replicates, and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate the standard error of the mean. (D) Transgenic soybean hairy roots were tested using Liberty Link strips.
TABLE S1 | Primer sequences used in this study.
Aboul-Soud, M. A. M., Cook, K., and Loake, G. J. (2004). Measurement of salicylic acid by a high-performance liquid chromatography procedure based on ion-exchange. Chromatographia 59, 129–133. doi: 10.1365/s10337-003-0136-1
Alazem, M., and Lin, N. S. (2015). Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 16, 529–540. doi: 10.1111/mpp.12204
Alscher, R. G., Erturk, N., and Heath, L. S. (2002). Role of superoxide dismutases (sods) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jxb/53.372.1331
Asselbergh, B., Curvers, K., Franca, S. C., Audenaert, K., Vuylsteke, M., and Höfte, B. M. (2007). Resistance to botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144, 1863–1877. doi: 10.2307/40065584
Baena-González, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signaling. Nature 448, 938–942. doi: 10.1038/nature06069
Baena-González, E., and Sheen, J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13, 474–482. doi: 10.1016/j.tplants.2008.06.006
Blanco, N., Liebsch, D., Díaz, M., Strand, A., and Whelan, J. (2019). Dual and dynamic intracellular localization of Arabidopsis thaliana SnRK1.1. J. Exp. Bot. 70, 2325–2338. doi: 10.1093/jxb/erz023
Carling, D., Thornton, C., Woods, A., and Sanders, M. J. (2012). AMP-activated protein kinase: new regulation, new roles. Biochem. J. 445, 11–27. doi: 10.1042/BJ20120546
Chacón, O., González, M., López, Y., Portieles, R., Pujol, M., González, E., et al. (2010). Over-expression of a protein kinase gene enhances the defense of tobacco against Rhizoctonia solani. Gene 452, 54–62. doi: 10.1016/j.gene.2009.11.011
Cheng, Q., Li, N. H., Dong, L. D., Zhang, D. Y., Fan, S. J., Jiang, L. Y., et al. (2015). Overexpression of soybean isoflavone reductase (GmIFR) enhances resistance to Phytophthora sojae in soybean. Front. Plant Sci. 6:1024. doi: 10.3389/fpls.2015.01024
Cho, Y. H., Hong, J. W., Kim, E. C., and Yoo, S. D. (2012). Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158, 1955–1964. doi: 10.1104/pp.111.189829
Coello, P., Hey, S. J., and Halford, N. G. (2011). The sucrose non-fermenting-1-related(SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 62, 883–893. doi: 10.1093/jxb/erq331
Coello, P., Hirano, E., Hey, S. J., Muttucumaru, N., Martinez-Barajas, E., Parry, M. A., et al. (2012). Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 63, 913–924. doi: 10.1093/jxb/err320
Du, X. M., Yin, W. X., Zhao, Y. X., and Zhang, H. (2001). The production and scavenging of reactive oxygen species in plants. Sheng Wu Gong Cheng Xue Bao 17, 121–125.
Fan, S. J., Dong, L. D., Han, D., Zhang, F., Wu, J. J., and Jiang, L. Y. (2017). GmWRKY31 and GmHDL56 enhances resistance to Phytophthora sojae by regulating defense-related gene expression in soybean. Front. Plant Sci. 8:781. doi: 10.3389/fpls.2017.00781
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. (1971). Stage of development descriptions for soybeans, glycine max (L.). Merrill. Crop Sci. 11, 929–931. doi: 10.1007/BF02197963
Filipe, O., De Vleesschauwer, D., Haeck, A., Demeestere, K., and Höfte, M. (2018). The energy sensor OsSnRK1a confers broad-spectrum disease resistance in rice. Sci. Rep. 8:3864. doi: 10.1038/s41598-018-22101-6
Fragoso, S., Espíndola, L., Páez-Valencia, J., Gamboa, A., Camacho, Y., Martínez Barajas, E., et al. (2009). SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in arabidopsis plants under phosphate starvation. Plant Physiol. 149, 1906–1916. doi: 10.1104/pp.108.133298
Gazzarrini, S., and Tsai, Y. L. (2015). Hormone cross-talk during seed germination. Essays Biochem. 58, 151–164. doi: 10.1042/bse0580151
Graham, T. L., Graham, M. Y., Subramanian, S., and Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant physiol. 144, 728–740. doi: 10.1104/pp.107.097865
Halford, N. G., and Hardie, D. G. (1998). SNF1-related protein kinases: global regulators of carbon metabolism in plants. Plant Mol. Biol. 37, 735–748. doi: 10.1023/A:1006024231305
Halford, N. G., and Hey, S. J. (2009). Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signaling in plants. Biochem. J. 419, 247–259. doi: 10.1042/BJ20082408
Hao, L., Wang, H., Sunter, G., and Bisaro, D. M. (2003). Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15, 1034–1048. doi: 10.1105/tpc.009530
He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., et al. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115. doi: 10.1016/j.cub.2007.05.036
Hedbacker, K., and Carlson, M. (2008). SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408. doi: 10.2741/2854
Hey, S. J., Byrne, E., and Halford, N. G. (2010). The interface between metabolic and stress signaling. Ann. Bot. 105, 197–203. doi: 10.1093/aob/mcp285
Holsters, M., De, W. D., Depicker, A., Messens, E., Van, M. M., and Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163, 181–187. doi: 10.1007/bf00267408
Jeong, E. Y., Seo, P. J., Woo, J. C., and Park, C. M. (2015). AKIN10 delays flowering by inactivating IDD8 transcription factor through protein phosphorylation in Arabidopsis. BMC Plant Biol. 15:110. doi: 10.1186/s12870-015-0503-8
Jossier, M., Bouly, J. P., Meimoun, P., Arjmand, A., Lessard, P., Hawley, S., et al. (2009). Snrk1 (snf1-related kinase 1) has a central role in sugar and ABA signaling in Arabidopsis thaliana. Plant J. 59:316. doi: 10.1111/j.1365-313X.2009.03871.x
Kaufmann, M. J., and Gerdemann, J. W. (1958). Root and stem rot of soybean caused by Phytophthora sojae. Phytopathology 48, 201–208.
Kerschen, A., Napoli, C. A., Jorgensen, R. A., and Müller, A. E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566, 223–228. doi: 10.1016/j.febslet.2004.04
Kim, C. Y., Vo, K. T. X., An, G., and Jeon, J. S. (2015). A rice sucrose non-fermenting-1 related protein kinase 1, osk35, plays an important role in fungal and bacterial disease resistance. J. Korean Soc. Appl. Biol. Chem. 58, 669–675. doi: 10.1007/s13765-015-0089
Kong, G., Zhao, Y., Jing, M., Huang, J., Yang, J., Xia, Y., et al. (2015). The activation of phytophthora effector Avr3b by plant cyclophilin is required for the nudix hydrolase activity of Avr3b. PLoS Pathog. 11:e1005139. doi: 10.1371/journal.ppat.1005139
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/s1369-5266(02)00275-3
Lee, K. W., Chen, P. W., Lu, C. A., Chen, S., Ho, T. H. D., and Yu, S. M. (2009). Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2:ra61. doi: 10.1126/scisignal.2000333
Li, N., Zhao, M., Liu, T., Dong, L., Cheng, Q., Wu, J., et al. (2017). A novel soybean dirigent gene gmdir22 contributes to promotion of lignan biosynthesis and enhances resistance to phytophthora sojae. Front. Plant Sci. 8:1185. doi: 10.3389/fpls.2017.01185
Liang, Y., Wei, J., Peng, G., Yong, L., Hua, C., Xi, B., et al. (2012). GsAPK, an ABA-activated and calcium-independent SnRK2-type kinase from G. soja, mediates the regulation of plant tolerance to salinity and aba stress. PLoS One 7:e33838. doi: 10.1371/journal.pone.0033838
Loon, L. L. V., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
McDonald, K. L., and Cahill, D. M. (1999). Influence of abscisic acid and the abscisic acid biosynthesis inhibitor, norflurazon, on interactions between Phytophthora sojae and soybean (Glycine max). Eur. J. Plant Pathol. 105, 651–658. doi: 10.1023/a:1008705321113
Mohr, P. G., and Cahill, D. M. (2001). Relative roles of glyceollin, lignin and the hypersensitive response and the influence of ABA in compatible and incompatible interactions of soybeans with Phytophthora sojae. Physiol. Mol. Plant Pathol. 58, 31–41. doi: 10.1006/pmpp.2000.0306
Morrison, R. H., and Thorne, J. C. (1978). Inoculation of detached cotyledons for screening soybeans against two races of Phytophthora megasperma var. sojae. Crop Sci. 18, 1089–1091. doi: 10.2135/cropsci1978.0011183X001800060049x
Nietzsche, M., Guerra, T., Alseekh, S., Wiermer, M., Sonnewald, S., Fernie, A. R., et al. (2018). Storekeeper related1/g-element binding protein (STKR1) interacts with protein kinase SnRK1. Plant Physiol. 176, 1773–1792. doi: 10.1104/pp.17.01461
Nunes, C., O’Hara, L. E., Primavesi, L. F., Delatte, T. L., Schluepmann, H., Somsen, G. W., et al. (2013). The trehalose 6-phosphate/SnRK1 signaling pathway primes growth recovery following relief of sink limitation. Plant Physiol. 162, 1720–1732. doi: 10.1104/pp.113.220657
Pan, X., Welti, R., and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Patricia, C., Emi, H., Sandra, J., Muttucumaru, N., Martinez, B. E., Parry, M. A., et al. (2012). Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 63, 913–924. doi: 10.1093/jxb/err320
Paz, M. M., Shou, H., Guo, Z., Zhang, Z., Banerjee, A. K., and Wang, K. (2004). Assessment of conditions affecting agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136, 167–179. doi: 10.1023/B:EUPH.0000030670.36730.a4
Perochon, A., Jia, J., Kahla, A., Arunachalam, C., Scofield, S. R., Bowden, S., et al. (2015). TaFROG encodes a pooideae orphan protein that interacts with SnRK1 and enhances resistance to the mycotoxigenic fungus fusarium graminearum. Plant Physiol. 169, 2895–2906. doi: 10.1104/pp.15.01056
Pieterse, C. M., Leon-Reye, A., Van der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Polge, C., and Thomas, M. (2007). SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12, 20–28. doi: 10.1016/j.tplants.2006.11.005
Radchuk, R., Emery, R. J., Weier, D., Vigeolas, H., Geigenberger, P., Lunn, J. E., et al. (2010). Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 61, 324–338. doi: 10.1111/j.1365-313X.2009.04057.x
Radchuk, R., Radchuk, V., Weschke, W., Borisjuk, L., and Weber, H. (2006). Repressing the expression of the sucrose nonfermenting-1-related protein kinase gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140, 263–278. doi: 10.1104/pp.105.071167
Reymond, P., and Farmer, E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. doi: 10.1016/S1369-5266(98)80264-1
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Ann. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rodrigues, A., Adamo, M., Crozet, P., Margalha, L., Confraria, A., Martinho, C., et al. (2013). ABI1 and PP2C a phosphatases are negative regulators of snf1-related protein kinase1 signaling in arabidopsis. Plant Cell 25, 3871–3884. doi: 10.1105/tpc.113.114066
Smeekens, S., Ma, J., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 273–278. doi: 10.1016/j.pbi.2009.12.002
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Subramanian, S., Graham, M. Y., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353. doi: 10.1104/pp.104.057257
Sugano, S., Sugimoto, T., Takatsuji, H., and Jiang, C. (2013). Induction of resistance to Phytophthora sojae in soybean (Glycine max) by salicylic acid and ethylene. Plant Pathol. 62, 1048–1056. doi: 10.1111/ppa.12011
Szczesny, R., Büttner, D., Escolar, L., Schulze, S., Seiferth, A., and Bonas, U. (2010). Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 187, 1058–1074. doi: 10.1111/j.1469-8137.2010.03346.x
Tewari, R. K., Kumar, P., and Sharma, P. N. (2006). Magnesium deficiency induced oxidative stress and antioxidant responses in mulberry plants. Sci. Horticu. 108, 7–14. doi: 10.1016/j.scienta.2005.12.006
Tsai, A. Y., and Gazzarrini, S. (2012). AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 69, 809–821. doi: 10.1111/j.1365-313X.2011.04832.x
Tsai, A. Y., and Gazzarrini, S. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5:119. doi: 10.3389/fpls.2014.00119
Van Loon, L. C., and Van Strien, E. A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. doi: 10.1006/pmpp.1999.0213
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Ann. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, S. D., Zhu, F., Yuan, S., Yang, H., Xu, F., Shang, J., et al. (2011). The roles of ascorbic acid and glutathione in symptom alleviation to SA-deficient plants infected with RNA viruses. Planta 234, 171–181. doi: 10.1007/s00425-011-1391-2
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., et al. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. doi: 10.2307/3869297
Ward, E. W. B., Lazarovits, G., Unwin, C. H., and Buzzell, R. I. (1979). Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megaspuma var. sojae. Phytopathology 69, 951–955. doi: 10.1094/Phyto-69-951
Wu, P., Wang, W., Duan, W., Li, Y., and Hou, X. (2017). Comprehensive analysis of the CDPK-SnRK superfamily genes in chinese cabbage and its evolutionary implications in plants. Front. Plant Sci. 8:162. doi: 10.3389/fpls.2017.00162
Xu, P., Jiang, L., Wu, J., Li, W., Fan, S., and Zhang, S. (2014). Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 41, 4899–4909. doi: 10.1007/s11033-014-3356-6
Yang, L., Jiang, Y., Wu, S. F., Zhou, M. Y., Wu, Y. L., and Chen, G. Q. (2008). CCAAT/enhancer-binding protein alpha antagonizes transcriptional activity of hypoxia-inducible factor 1 alpha with direct protein-protein interaction. Carcinogenesis 29, 291–298. doi: 10.1093/carcin/bgm262
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhai, Z., Liu, H., and Shanklin, J. (2017). Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29, 871–889. doi: 10.1105/tpc.17.00019
Zhang, C., Wang, X., Zhang, F., Dong, L., Wu, J., Cheng, Q., et al. (2017). Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 7:7242. doi: 10.1038/s41598-017-07832-2
Keywords: Glycine max, GmSnRK1.1, enzymatic antioxidants, salicylic acid, Phytophthora sojae
Citation: Wang L, Wang H, He S, Meng F, Zhang C, Fan S, Wu J, Zhang S and Xu P (2019) GmSnRK1.1, a Sucrose Non-fermenting-1(SNF1)-Related Protein Kinase, Promotes Soybean Resistance to Phytophthora sojae. Front. Plant Sci. 10:996. doi: 10.3389/fpls.2019.00996
Received: 18 March 2019; Accepted: 16 July 2019;
Published: 02 August 2019.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Meixiang Zhang, Nanjing Agricultural University, ChinaCopyright © 2019 Wang, Wang, He, Meng, Zhang, Fan, Wu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuzhen Zhang, emhhbmdzaHV6aGVuQG5lYXUuZWR1LmNu; Pengfei Xu, eHVwZW5nZmVpQG5lYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.