
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 July 2019
Sec. Plant Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00935
This article is part of the Research Topic Organelle Autophagy in Plant Development View all 9 articles
Autophagy is a catabolic process for bulk and selective degradation of cytoplasmic components in the vacuole/lysosome. In Saccharomyces cerevisiae, ATG genes were identified as essential genes for autophagy, and most ATG genes are highly conserved among eukaryotes, including plants. Although reverse genetic analyses have revealed that autophagy is involved in responses to abiotic and biotic stresses in land plants, our knowledge of its molecular mechanism remains limited. This limitation is partly because of the multiplication of some ATG genes, including ATG8, in widely used model plants such as Arabidopsis thaliana, which adds complexity to functional studies. Furthermore, due to limited information on the composition and functions of the ATG genes in basal land plants and charophytes, it remains unclear whether multiplication of ATG genes is associated with neofunctionalization of these genes. To gain insight into the diversification of ATG genes during plant evolution, we compared the composition of ATG genes in plants with a special focus on a liverwort and two charophytes, which have not previously been analyzed. Our results showed that the liverwort Marchantia polymorpha and the charophytes Klebsormidium nitens and Chara braunii harbor fundamental sets of ATG genes with low redundancy compared with those of A. thaliana and the moss Physcomitrella patens, suggesting that multiplication of ATG genes occurred during land plant evolution. We also attempted to establish an experimental system for analyzing autophagy in M. polymorpha. We generated transgenic plants expressing fluorescently tagged MpATG8 to observe its dynamics in M. polymorpha and produced autophagy-defective mutants by genome editing using the CRISPR/Cas9 system. These tools allowed us to demonstrate that MpATG8 is transported into the vacuole in an MpATG2-, MpATG5-, and MpATG7-dependent manner, suggesting that fluorescently tagged MpATG8 can be used as an autophagosome marker in M. polymorpha. M. polymorpha can provide a powerful system for studying the mechanisms and evolution of autophagy in plants.
Autophagy is a highly conserved catabolic process in eukaryotes for degrading and recycling cytoplasmic components. Among several types of autophagy reported thus far, the molecular mechanisms and physiological significance of macroautophagy have been the most intensively studied (Mizushima et al., 2011). During macroautophagy (hereafter simply referred to as “autophagy”), a cup-shaped membrane sac called the isolation membrane or phagophore elongates and sequesters cytoplasmic components, and its edge is closed to form a double-membrane-bounded structure called the autophagosome (Baba et al., 1994). The outer membrane of the autophagosome fuses with the vacuolar/lysosomal membrane, and a single-membraned structure called the autophagic body is released into the luminal space of the vacuole/lysosome to be degraded by lytic enzymes (Takeshige et al., 1992; Baba et al., 1994, 1995). In addition to bulk degradation, certain substrates, including organelles, are selectively recognized and degraded by autophagy (Johansen and Lamark, 2011). In Saccharomyces cerevisiae, autophagy-related processes are also involved in biosynthetic delivery; the newly synthesized precursor form of aminopeptidase I is transported into the vacuole by small double-membrane-bounded vesicles (Baba et al., 1997), which are formed through a mechanism similar to autophagy (Harding et al., 1996; Scott et al., 1996).
In the 1990s, several groups identified APG/AUT/CVT genes (later unified under the name ATG) as essential genes for autophagy or autophagy-related processes in S. cerevisiae (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding, 1995; Klionsky et al., 2003). Atg proteins are initially recruited to and function at the preautophagosomal structures or phagophore assembly site (PAS) in a hierarchical manner (Suzuki et al., 2001, 2007). Based on their functions, the essential genes for autophagosome formation in S. cerevisiae are classified into four groups: (1) the Atg1 complex, (2) phosphatidylinositol 3-kinase (PI3K) complex, (3) Atg9 cycling system, and (4) ubiquitin-like conjugation systems. The Atg1 complex, consisting of Atg1, Atg13, Atg17, Atg29, and Atg31, is one of the most upstream Atg factors and is recruited to the PAS under the regulation of various cellular signals (Noda and Ohsumi, 1998; Suzuki et al., 2007; Kamada et al., 2010). The Atg1 complex phosphorylates various substrates, including Atg9, and acts as a scaffold for downstream Atg proteins (Suzuki et al., 2007, 2015; Papinski et al., 2014; Harada et al., 2019). The PI3K complex comprises Atg6, Atg14, Vps15, and Vps34. The PI3K complex generates phosphatidylinositol 3-phosphate (PI3P) at the PAS, which leads to recruitment of PI3P-binding proteins such as the Atg2–Atg18 complex (Kihara et al., 2001; Obara et al., 2008). The Atg9 cycling system involves Atg2, Atg9, and Atg18. Atg9 is a multimembrane-spanning protein that shuttles between the PAS and mobile structures derived from the Golgi and provides a membrane source during the early step of autophagosome formation (Mari et al., 2010; Yamamoto et al., 2012). The Atg2–Atg18 complex is recruited to the edge of the isolation membrane with Atg9 and probably regulates recycling of Atg9 from the PAS (Reggiori et al., 2004; Graef et al., 2013; Suzuki et al., 2013). Atg2 was also recently shown to have membrane tethering and lipid transfer activities, which are crucial for expansion of the isolation membrane (Chowdhury et al., 2018; Gomez-Sanchez et al., 2018; Kotani et al., 2018; Osawa et al., 2019). Furthermore, mammalian WIPI2b, which is homologous to Atg18, interacts with and recruits ATG16L1a (homologous to Atg16) to regulate the site of lipidation of LC3 (homologous to Atg8) (Fujita et al., 2008b; Dooley et al., 2014). Two ubiquitin-like conjugation systems involve Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12, and Atg16. Atg12 is conjugated to Atg5 by E1-like Atg7 and E2-like Atg10, and Atg12–Atg5 forms a complex with Atg16 (Mizushima et al., 1998, 1999; Shintani et al., 1999; Tanida et al., 1999; Kuma et al., 2002). Atg8 is cleaved by Atg4, and glycine is exposed at its C-terminus (Kirisako et al., 2000). This glycine is conjugated with phosphatidylethanolamine through the actions of E1-like Atg7, E2-like Atg3, and the E3-like Atg12–Atg5–Atg16 complex (Ichimura et al., 2000; Hanada et al., 2007). Lipidated Atg8 is recruited to the autophagosomal membrane, which functions in expansion and closure of the isolation membrane (Abeliovich et al., 2000; Nakatogawa et al., 2007; Fujita et al., 2008a; Xie et al., 2008; Tsuboyama et al., 2016). Atg8 also interacts with cargo receptors/adaptors via Atg8-interacting motifs (AIMs) or LC3-interacting regions (LIRs), which mediate the effective degradation of cargos (Noda et al., 2010; Nguyen et al., 2016; Padman et al., 2019). A recent study also demonstrated that ubiquitin-interacting motif (UIM)-like sequences are also recognized by Atg8 (Marshall et al., 2019).
Most ATG genes are also conserved in plants, and deletion of many ATG genes in Arabidopsis thaliana results in defective autophagy, suggesting that ATG proteins in A. thaliana have the same functions as those in S. cerevisiae and mammals (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004; Thompson et al., 2005; Xiong et al., 2005; Inoue et al., 2006; Phillips et al., 2008; Chung et al., 2010; Suttangkakul et al., 2011; Li et al., 2014; Young et al., 2019). Genetic analyses have revealed that autophagy is involved in responses to abiotic and biotic stresses (Doelling et al., 2002; Liu et al., 2005, 2009; Xiong et al., 2007; Zhou et al., 2013; Chen et al., 2015). Whereas almost all atg mutants of A. thaliana are fertile under favorable growth conditions, autophagy is required for male gamete differentiation in Oryza sativa and Physcomitrella patens (Kurusu et al., 2014; Sanchez-Vera et al., 2017). Furthermore, selective degradation of various organelles, including peroxisomes, by autophagy has been reported (Kim et al., 2013; Shibata et al., 2013; Yoshimoto et al., 2014); the molecular mechanisms of this phenomenon, however, remain largely unknown (Yoshimoto and Ohsumi, 2018). The difficulty in studying autophagy in plants is partly due to the genetic redundancy of key ATG genes in model plants, including A. thaliana. For example, A. thaliana, O. sativa, and P. patens harbor nine, four, and six ATG8 homologs, respectively, which makes it difficult to unravel the functions of ATG8 proteins in these plants (Kellner et al., 2017). Although functional differentiation among mammalian ATG8 homologs has been reported (Weidberg et al., 2010), it remains completely unknown whether each ATG8 homolog in plants acts at a different step in autophagy. The significance of the duplication of ATG genes during plant evolution also remains obscure, since information from basal land plants and algal species remains sparse. To obtain more insights into the diversification and evolution of autophagy in the plant lineage, information from charophytes, the closest living relatives of land plants, and from additional bryophytes would be needed.
In this study, we identified homologs of ATG genes in the basal land plants Marchantia polymorpha and P. patens and the charophytes Klebsormidium nitens and Chara braunii. A comparison among these species, as well as Chlamydomonas reinhardtii and A. thaliana, indicated that M. polymorpha shares a common set of ATG genes with low redundancy compared with those of other land plants; this finding suggested that M. polymorpha would be a good system to investigate the molecular mechanisms and physiological significance of autophagy in land plants. Many molecular genetic techniques and cell biological tools have been established for M. polymorpha (Era et al., 2009; Ishizaki et al., 2016; Kanazawa et al., 2016; Minamino et al., 2018), which would also be a good reason to use this plant for autophagy studies. In this study, as the first step toward the study of autophagy in M. polymorpha, we generated transgenic plants expressing fluorescently tagged MpATG8 proteins to monitor autophagosomes and produced the autophagy-defective mutants Mpatg2, Mpatg5, and Mpatg7 to analyze the effect of defective autophagy on thallus development and their responses to nutrient starvation. Our results indicated that the number of ATG genes gradually increased during plant evolution. We also succeeded in monitoring the dynamics of MpATG8, which was transported into the vacuole in an MpATG2-, MpATG5-, and MpATG7-dependent manner. These tools would be useful for future studies to understand the basic mechanisms and physiological significance of autophagy in plants.
Extensive studies using S. cerevisiae have revealed that ATG1–10, 12–14, 16–18, 29, and 31, and VPS15 and VPS34 are required for autophagosome formation (Mizushima et al., 2011). Although some ATG genes are highly duplicated in angiosperms, the precise significance of the expansion of ATG genes remains obscure. To gain insight into this phenomenon, we searched the genome sequences of charophytes (K. nitens and C. braunii) and bryophytes (M. polymorpha and P. patens) for homologs of ATG genes, a number of which were then compared with those of A. thaliana and C. reinhardtii. As a query, we used the sequences of the ATG genes of A. thaliana (Shemi et al., 2015; Liu et al., 2018). We also examined ATG11 and ATG101. ATG11 is not required for starvation-induced bulk autophagy but is essential for selective autophagy in S. cerevisiae (Kim et al., 2001); Atg11 interacts with Atg1 and cargo receptors, which is crucial for selective autophagy (Yorimitsu and Klionsky, 2005; Farre et al., 2008; Okamoto et al., 2009). Atg101 forms a complex with ULK1 (homologous to Atg1) in mammalian cells, although S. cerevisiae does not harbor a homolog of ATG101 (Hosokawa et al., 2014; Mercer et al., 2014). In A. thaliana, ATG11 and ATG101 are thought to form a complex with ATG1, and the atg11 mutant exhibits similar phenotypes to those of other atg mutants (Li et al., 2014), suggesting that ATG11 and ATG101 are involved in general autophagy in plants. Therefore, ATG11 and ATG101 were also included among the “core autophagy machinery genes” in this study.
As shown in Table 1, almost all core autophagy machinery genes were highly conserved among the plants we investigated, although ATG16 homologs in C. reinhardtii (Shemi et al., 2015) and ATG2 and ATG10 homologs in C. braunii were not detected. Homologs of ATG17, ATG29, and ATG31 were not detected in this study, consistent with previous studies (Kawamata et al., 2005; Li et al., 2014). While a considerable number of ATG genes are present in A. thaliana and P. patens, C. reinhardtii, charophytes, and M. polymorpha possess only one gene for each core autophagy machinery component, with a few exceptions; multiple genes for ATG8 and ATG18 exist in the genomes of these plants with lower redundancy than A. thaliana and P. patens (Table 1). Thus, core autophagy machinery genes have seemingly expanded gradually during plant evolution.
Recently, Pang et al. (2019) reported that ATG10 has been lost in quite a few lineages of eukaryotes, including Pichiaceae and the SAR supergroup, which comprises stramenopiles, alveolates, and Rhizaria. As summarized in Table 1, we detected an ATG10 homolog in K. nitens but not in C. braunii. We also looked for ATG10 homologs in other charophytes whose genome and/or transcriptome information was available (Spirogyra pratensis, Nitella mirabilis, Coleochaete orbicularis, and Mesostigma viride) and found that these algae, except for N. mirabilis, possess ATG10 homologs. This distribution of ATG10 suggested that secondary loss of ATG10 occurred in Charophyceae (Table 2). The ATG10 protein is an E2-like enzyme required for covalent linkage between the C-terminal glycine residues of ATG12 and ATG5 (Mizushima et al., 1998; Shintani et al., 1999; Suzuki et al., 2005; Phillips et al., 2008; Chung et al., 2010). In Toxoplasma gondii and Komagataella phaffii, which do not possess ATG10 homologs, ATG12 forms a noncovalent complex with ATG5, which does not require the C-terminal glycine of ATG12 (Pang et al., 2019). Intriguingly, the ATG12 of C. braunii does not harbor glycine at the C-terminus, although the glycine residue is conserved at the C-terminus of N. mirabilis ATG12 (Table 2). These features could reflect a similar mechanism of complex formation between ATG12 and ATG5 in C. braunii; ATG12 might form a noncovalent complex with ATG5.
A. thaliana and Nicotiana tabacum are reported to possess two types of ATG18: conventional ATG18, which is similar to yeast and mammalian ATG18, and plant-unique ATG18, which harbors the BCAS3 domain at the C-terminus (Xiong et al., 2005; Zhou et al., 2015). All of the land plant species we analyzed in this study harbor both types of ATG18, whereas C. reinhardtii does not possess plant-unique ATG18 (Table 3). We then investigated whether plant-unique ATG18 is found in other chlorophytes. Although we did not detect ATG18 with the BCAS3 domain in Dunaliella salina, Volvox carteri, Micromonas pusilla, Ostreococcus lucimarinus, and Ostreococcus tauri, Coccomyxa subellipsoidea harbored this type of ATG18. Canonical ATG18 homologs were found in all of these species (Table 3). Therefore, ATG18 containing the BCAS3 domain seems to have been acquired before the emergence of Streptophyta, which was also supported by the result of phylogenetic analysis (Figure 1).
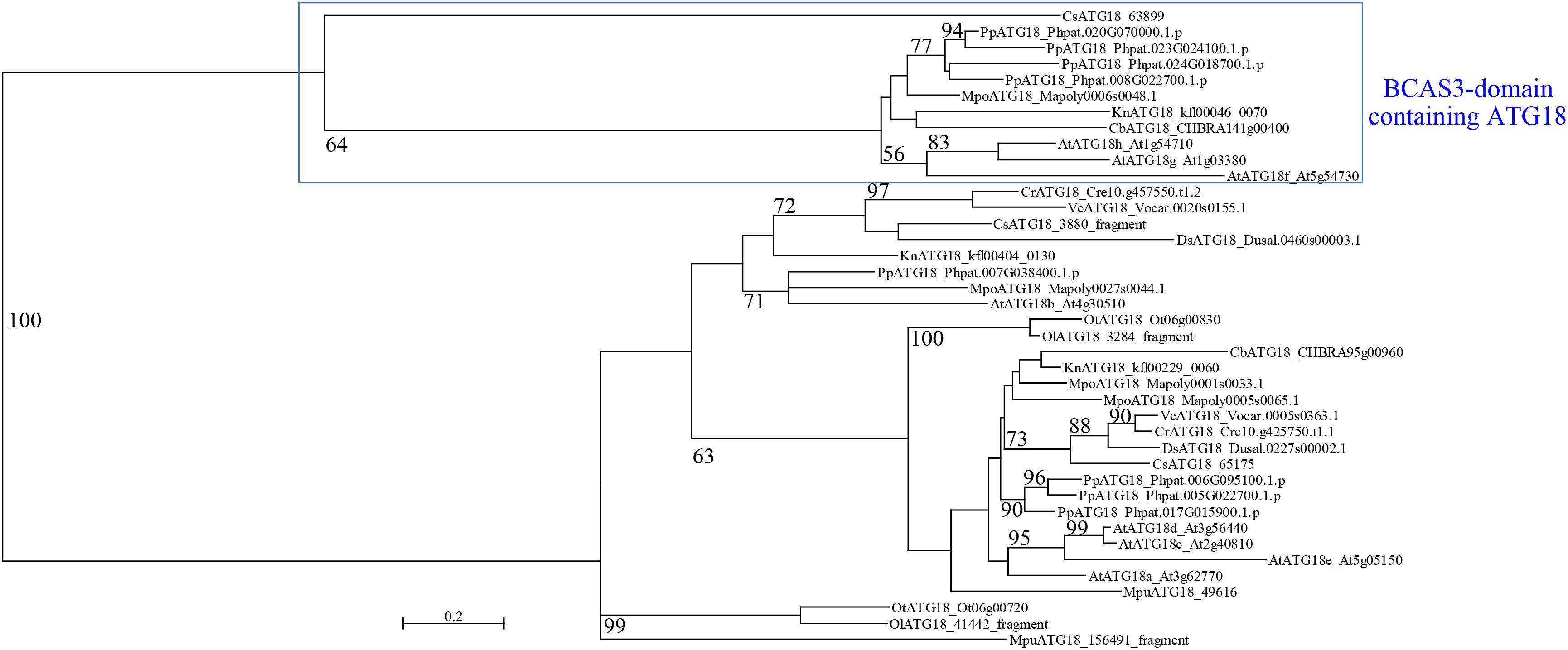
Figure 1. Phylogenetic analysis of ATG18 in Viridiplantae. The maximum-likelihood phylogenetic analysis was performed using amino acid sequences of ATG18 in Viridiplantae. Bootstrap probability with at least 50% support is indicated as percentage on each branch. At, A. thaliana; Pp, P. patens; Mpo, M. polymorpha; Cb, C. braunii; Kn, K. nitens; Cr, C. reinhardtii; Ds, D. salina; Vc, V. carteri; Cs, C. subellipsoidea; Mpu, M. pusilla; Ol, O. lucimarinus; Ot, O. tauri.
Because of the low genetic redundancy of ATG components (Table 1) and molecular genetic tools available (Ishizaki et al., 2016), M. polymorpha is expected to be a good model for understanding the fundamental molecular mechanisms of autophagy in land plants. As the first step of the autophagy study using M. polymorpha, we generated transgenic plants expressing fluorescently tagged MpATG8 proteins under the regulation of their own promoters. ATG8 localizes to the isolation membrane during autophagosome formation and stays on the inner membranes of autophagosomes until their degradation in the vacuole/lysosome in various organisms, including A. thaliana (Kirisako et al., 1999; Kabeya et al., 2000; Yoshimoto et al., 2004; Izumi et al., 2015). Therefore, fluorescently tagged MpATG8 is expected to be a good tool to visualize autophagosome dynamics in M. polymorpha. As shown in Figure 2A, monomeric Citrine (mCitrine)-tagged MpATG8a and MpATG8b were localized to punctate structures distributed throughout the cytosol. Fluorescence was also detected in the nucleus (asterisks in Figure 2A), which might reflect a function of the nucleus as a reservoir of MpATG8 as reported in mammalian cells (Huang et al., 2015). As autophagic bodies bearing ATG8 transported to the vacuolar lumen are generally degraded immediately by vacuolar enzymes (Kirisako et al., 1999; Huang et al., 2000), we treated transgenic M. polymorpha with concanamycin A (concA), which inhibits acidification of the vacuole and inactivates vacuolar lytic enzymes, to visualize autophagic bodies in the vacuole (Yoshimoto et al., 2004). After treatment with concA, both mCitrine-MpATG8a and mCitrine-MpATG8b were visible as punctate structures in the vacuole (Figure 2C). Vacuolar localization was not observed in mock-treated cells, suggesting that these punctate structures are autophagic bodies. We then investigated whether MpATG8a and MpATG8b are localized to the same structure. We expressed monomeric RFP (mRFP)-tagged MpATG8a and mCitrine-MpATG8b in the same plant and observed strong colocalization at the same punctate structures in the cytosol; colocalization was also observed in the vacuole in concA-treated cells, suggesting that MpATG8a and MpATG8b are localized to the same autophagosomes/autophagic bodies (Figures 2B,D). These observations indicated that MpATG8a and MpATG8b behave in a similar manner to ATG8 in other organisms, and these molecules with fluorescent tags would be useful as autophagosome markers.
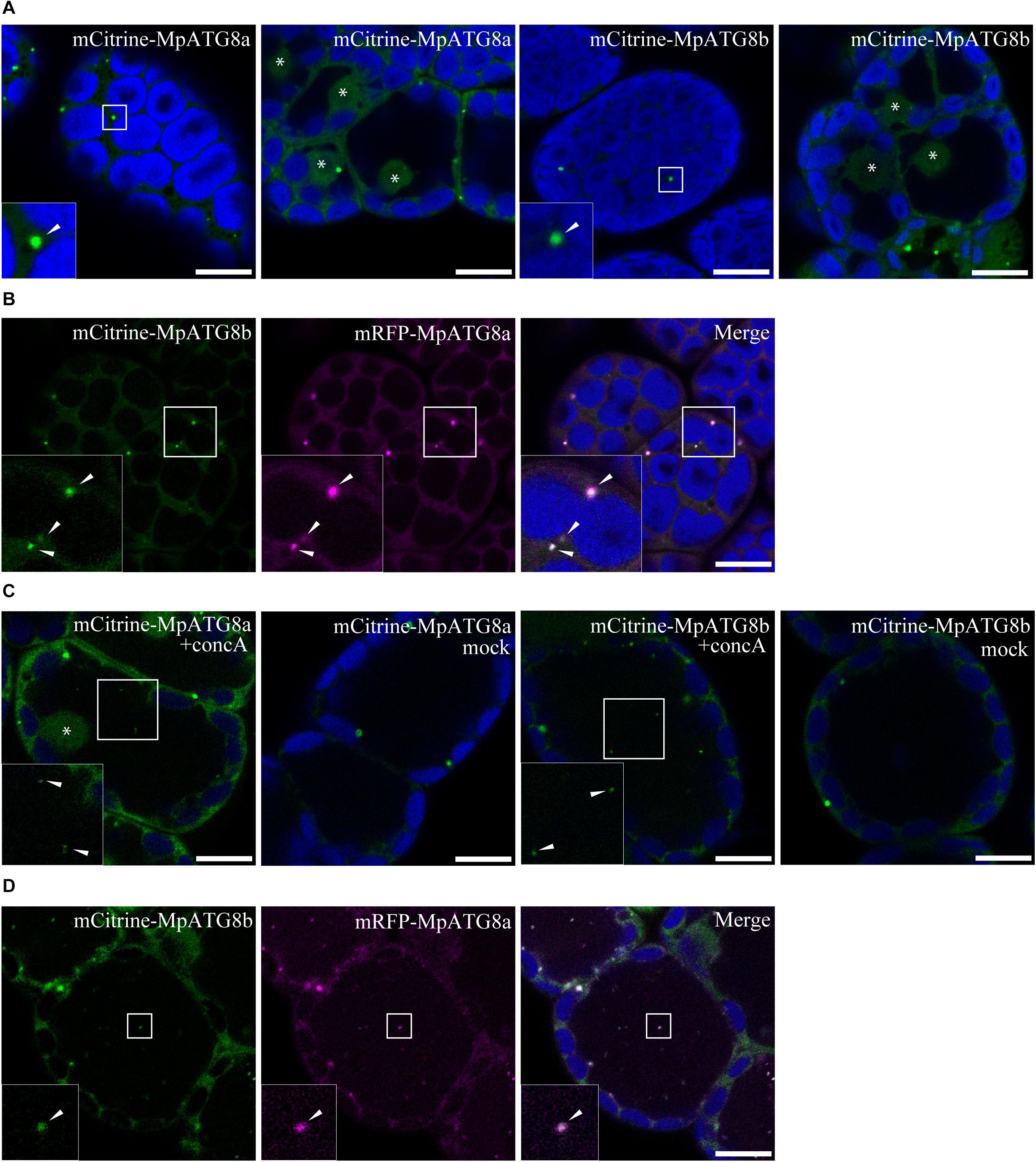
Figure 2. Subcellular localization of MpATG8 members. (A) MpATG8a and MpATG8b were localized to punctate structures in the cytosol, indicated by arrowheads. Fluorescence was also detected in the nucleus, indicated by asterisks. (B) mRFP-MpATG8a and mCitrine-MpATG8b colocalized to the punctate structures in the cytosol. Arrowheads indicate punctate compartments with mRFP-MpATG8a and mCitrine-MpATG8b. (C) MpATG8-positive puncta observed in the vacuoles upon concA treatment. The asterisk and arrowheads indicate the nucleus and puncta in the vacuole, respectively. (D) Colocalization of mRFP-MpATG8a and mCitrine-MpATG8b at punctate structures in the vacuole upon concA treatment. Arrowheads indicate punctate compartments with mRFP-MpATG8a and mCitrine-MpATG8b in the vacuole. The insets are magnified images of the boxed regions. Green, magenta, and blue show fluorescence from mCitrine, mRFP, and autofluorescence of chlorophyll, respectively. Scale bars = 10 μm.
To investigate the physiological significance of autophagy in M. polymorpha, we generated Mpatg5, Mpatg7, and Mpatg2 mutants (hereafter referred to as Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge, respectively) by genome editing using the clustered regularly interspaced short palindromic repeats (CRISPRs)-associated endonuclease Cas9 (CRISPR/Cas9) system (Sugano et al., 2014, 2018). ATG5, ATG7, and ATG2 are required for autophagosome formation, and deletion of these genes causes defects in autophagy in various organisms, including S. cerevisiae, mammals, and A. thaliana (Tsukada and Ohsumi, 1993; Mizushima et al., 2001; Doelling et al., 2002; Kuma et al., 2004; Komatsu et al., 2005; Thompson et al., 2005; Inoue et al., 2006; Velikkakath et al., 2012). The mutations detected in Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge result in frame shifts, and functional full-length proteins cannot be produced in these mutants (Figure 3). To investigate whether autophagy occurs in these mutants, we observed the dynamics of mCitrine-MpATG8a in these mutants. In wild-type (WT) plants, mCitrine-MpATG8a was localized to the punctate structures in the cytoplasm and vacuolar lumen (Figures 2A,C, 4A). In contrast, vacuolar localization of mCitrine-MpATG8a was not observed in any of the Mpatg mutants, while mCitrine-MpATG8a was observed as puncta in the cytoplasm (Figures 4A,B). We then performed a cleavage assay of mCitrine-MpATG8a in the Mpatg mutants. As ATG8 is rapidly degraded in the vacuole, whereas mCitrine/YFP is more resistant to lytic enzymes, translocation of mCitrine-MpATG8 to the vacuole can be monitored by examining the accumulation of free mCitrine by immunoblotting (Shintani and Klionsky, 2004; Chung et al., 2010). In WT, two bands were observed via immunoblotting using an anti-green fluorescent protein (GFP) antibody, at approximately 43 and 27 kDa (Figure 4C). The 43 kDa band represented full-length mCitrine-MpATG8a, and the 27 kDa product represented free mCitrine. In contrast, only the 43 kDa product was detected in Mpatg mutants, confirming that mCitrine-MpATG8a was not transported into the vacuole in these mutants (Figure 4C). This result indicated that Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge are defective in autophagy and that MpATG8 is transported into the vacuole in an MpATG5-, MpATG7-, and MpATG2-dependent manner in M. polymorpha.
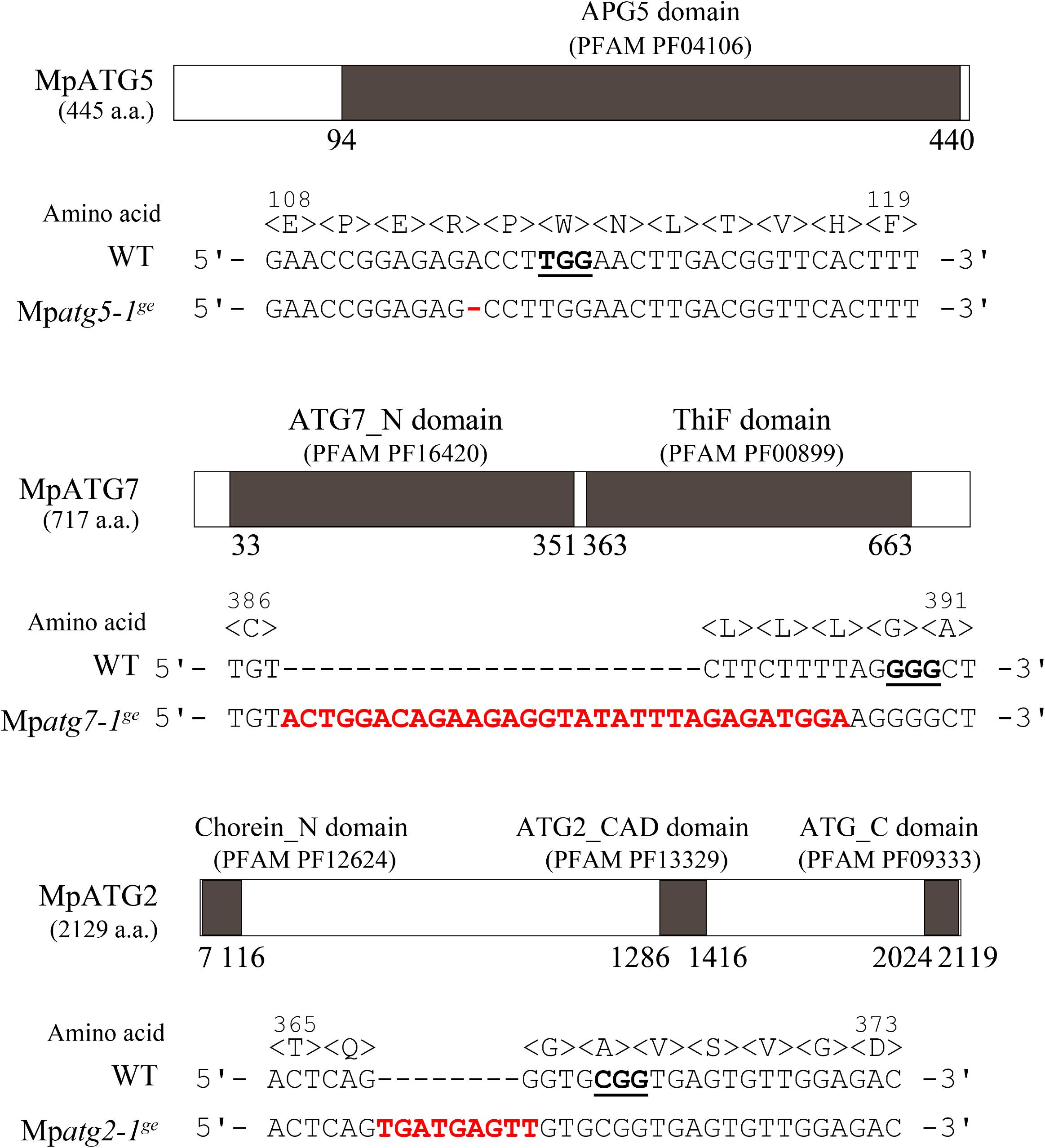
Figure 3. Mutations in Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge. Domain structures of the MpATG5, MpATG7, and MpATG2 proteins and sequences of the mutations in MpATG5, MpATG7, and MpATG2 introduced by genome editing. All three mutations result in frame shifts. Underlining indicates protospacer adjacent motif (PAM) sequences. Inserted, deleted, and substituted bases are shown in red.
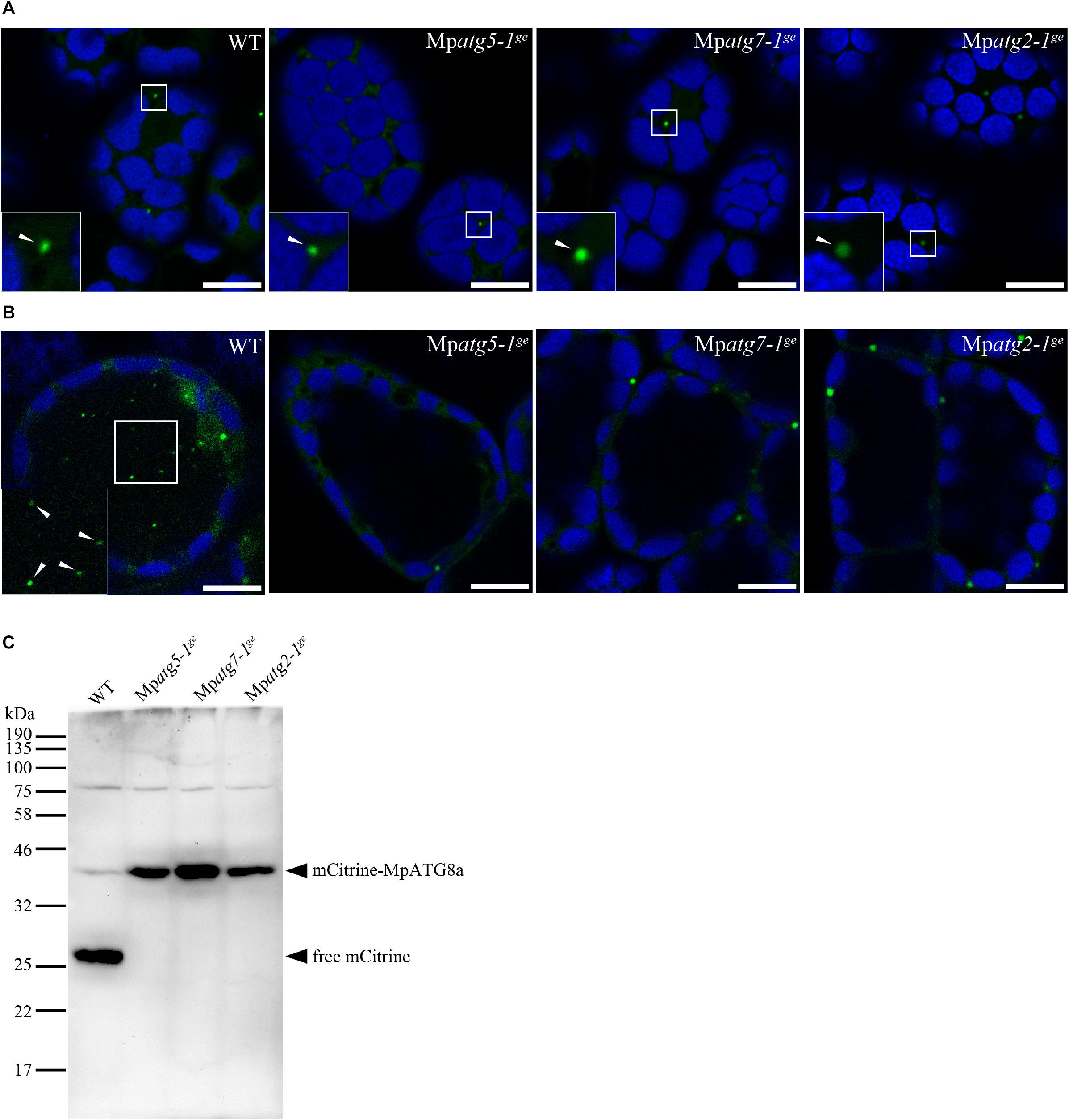
Figure 4. Generation of atg mutants of M. polymorpha. (A) MpATG8a was localized to punctate structures (arrowheads) throughout the cytosol in WT, Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge. Bars = 10 μm. (B) mCitrine-MpATG8a was not detected in the vacuole in Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge after concA treatment. Arrowheads indicate punctate structures in the vacuole. The insets are magnified images of the boxed regions. The green and blue pseudocolors indicate fluorescence from mCitrine and autofluorescence of chlorophyll, respectively. Scale bars = 10 μm. (C) Five-day-old thalli of WT, Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge expressing mCitrine-MpATG8a were subjected to immunoblotting using the anti-GFP antibody.
We then observed the macroscopic phenotypes of Mpatg mutants. Under normal laboratory conditions, the proximal regions of the thalli of all Mpatg mutants exhibited a yellowish chlorotic phenotype, which is similar to the early senescence phenotype of atg mutants of A. thaliana (Doelling et al., 2002; Hanaoka et al., 2002; Figure 5A). Thus, autophagy appears to also be required for preventing early senescence in M. polymorpha. We then investigated whether Mpatg mutants are hypersensitive to nutrient starvation, as reported for atg mutants of A. thaliana (Doelling et al., 2002; Hanaoka et al., 2002). We cultured thalli of M. polymorpha in liquid 1/2× Gamborg’s B5 medium under continuous light (control) or in 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) under the dark condition, which induces both bulk autophagy and piecemeal autophagy of chloroplasts in A. thaliana (Izumi et al., 2010), and measured chlorophyll contents. Consistent with the early senescence phenotype shown in Figure 5A, Mpatg mutants exhibited lower chlorophyll contents after incubation in 1/2× Gamborg’s B5 medium for 3 days under continuous light compared with WT, confirming that autophagy plays critical roles in preventing early senescence in M. polymorpha (Figures 5B,C). We also found that Mpatg mutants cultured in 10 mM MES for 3 days under dark condition exhibited significantly lower chlorophyll contents than that in WT cultured in 10 mM MES (Figure 5D). These data indicated that autophagy is required for normal response to nutrient starvation in M. polymorpha.
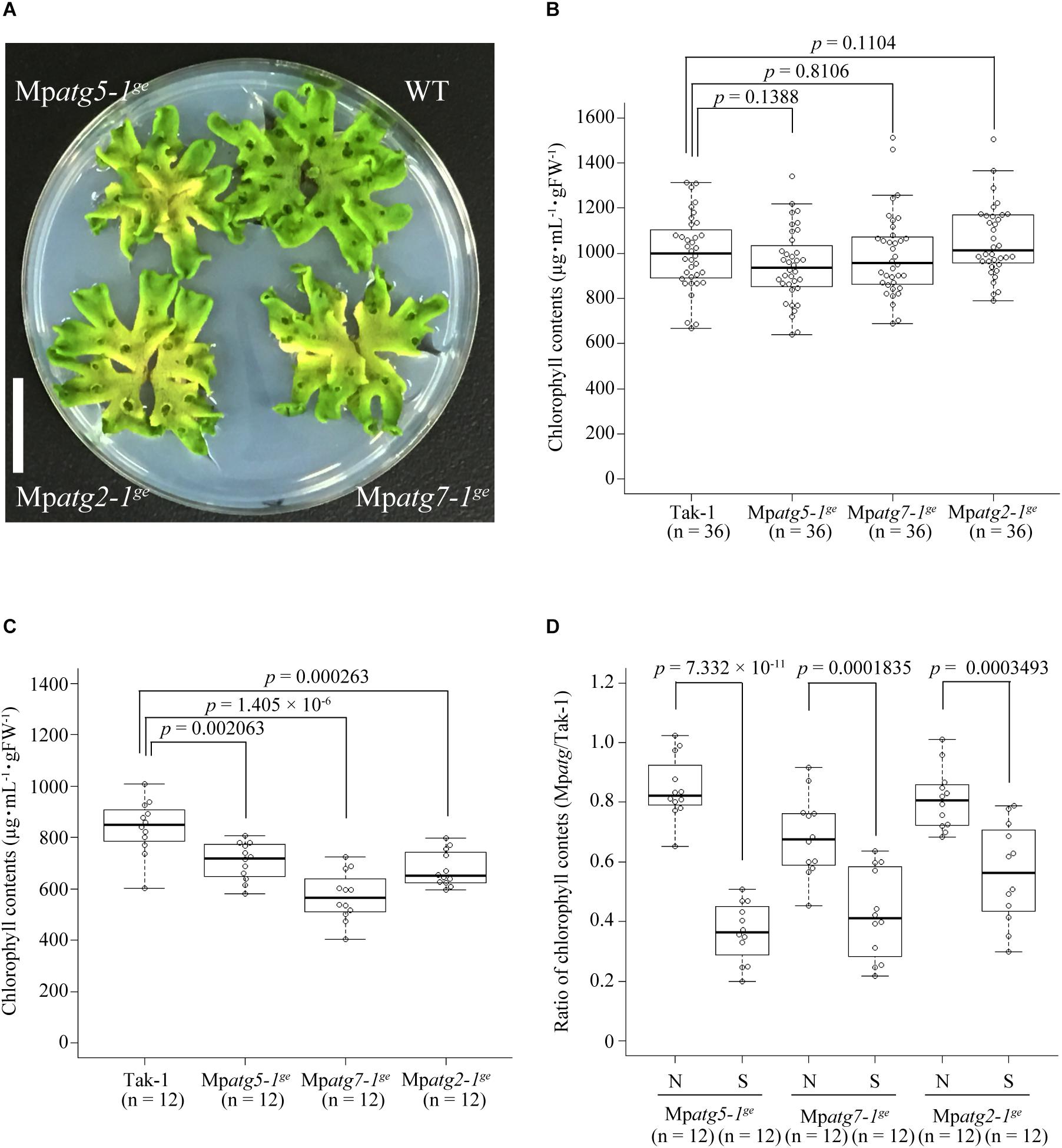
Figure 5. Phenotype of atg mutants of M. polymorpha. (A) 26-day-old thalli grown on 1/2× Gamborg’s B5 medium under continuous light. Scale bar = 2 cm. (B) Chlorophyll contents of 5-day-old WT or Mpatg mutant thalli before incubation in liquid 1/2× Gamborg’s B5 medium or 10 mM MES. (C) Chlorophyll contents of WT and Mpatg mutants after incubation in liquid 1/2× Gamborg’s B5 medium for 3 days under continuous light. (D) Ratios of chlorophyll contents in Mpatg mutants to those in WT, which were incubated in liquid 1/2× Gamborg’s B5 medium under continuous light for 3 days (N, non-starvation) or in 10 mM MES (pH 5.5) for 3 days under dark condition (S, starvation). The boxes and solid lines in the boxes show the first quartile and third quartile, and median values, respectively. The whiskers indicate 1.5× interquartile ranges. p-Values were calculated by Welch’s t-test. n, the number of samples (three thalli were treated as one sample for correct measurement of the fresh weight).
ATG genes were first identified in S. cerevisiae, and most ATG genes have now been shown to be conserved in various lineages of eukaryotes. In this study, we showed that charophyte species and M. polymorpha also possess a set of genes for the core autophagy machinery with lower redundancy than those of P. patens and A. thaliana. This result suggests that the core autophagy machinery has been expanded during land plant evolution. Some of the ATG genes, such as ATG8 and ATG18, are reported to have multiplied in various organisms, including land plants. Intriguingly, M. polymorpha also harbors multiple ATG8 and ATG18 genes, and the plant-unique ATG18 group comprising the BCAS3 domain is conserved in M. polymorpha, some charophytes, and the chlorophyte C. subellipsoidea. The distribution of the plant-unique ATG18 suggests that this type of ATG18 was acquired before the emergence of Streptophyta, and secondary losses of this gene occurred independently during plant evolution, although its molecular function remains unknown. Moreover, it remains unclear whether the members of ATG8 and ATG18 are functionally differentiated in plants. In A. thaliana, knockdown of ATG18a, one of eight ATG18 homologs, results in similar but weaker phenotypes to those of mutants of other ATG genes, suggesting that ATG18a plays a major role in autophagy, although the other seven ATG18 homologs could have a partly redundant function (Xiong et al., 2005; Kang et al., 2018). In addition to its localization to the PAS, Atg18 of S. cerevisiae is also localized to the vacuolar membrane via phosphatidylinositol 3,5-bisphosphate binding and acts in retrograde transport from the vacuole, which is independent of autophagy (Dove et al., 2004; Efe et al., 2007). ATG18 homologs in A. thaliana might also have autophagy-independent functions. In mammals, two groups of ATG8-related proteins, the LC3 and GABARAP/GATE-16 subfamilies, are both localized to the autophagosomes but play distinct roles in autophagosome formation (Kabeya et al., 2004; Weidberg et al., 2010). The LC3 group is involved in the elongation of the phagophore membrane, whereas GABARAP/GATE-16 plays an essential role in a later stage of autophagosome maturation (Weidberg et al., 2010). Caenorhabditis elegans possesses two ATG8 homologs: LGG-1 and LGG-2. LGG-1 is involved in the formation of autophagosomes. In contrast, LGG-2 mediates the maturation of autophagosomes and facilitates their tethering with lysosomes through interaction with VPS39 (Manil-Segalen et al., 2014). In plants, distinct binding affinities of potato ATG8 members with PexRD54, an effector protein in the pathogenic fungus Phytophthora infestans, have been reported (Dagdas et al., 2016). Thus, the plant ATG8 and ATG18 groups could comprise functionally differentiated members; this possibility should be verified in future studies.
Although most of the core machinery of autophagy is conserved in plants, ATG10 homologs were not detected in two charophyte species. Loss of ATG10 has been observed in various lineages of eukaryotes, some of which utilize a noncovalent complex of ATG5 and ATG12 without the C-terminal glycine residue (Pang et al., 2019). Given that ATG10 mediates covalent linkage between ATG12 and ATG5 at the C-terminal glycine residue of ATG12, secondary loss of ATG10 could be associated with conversion from covalent to noncovalent ATG12–ATG5 complexes during evolution, which could be followed by deletion of the glycine residue of the C-terminus of ATG12. Consistent with this notion, we did not detect ATG10 homologs in two charophyte species, one of which (C. braunii) possesses ATG12 without C-terminal glycine (Tables 1, 2). Further characterization of ATG12 and ATG5 in C. braunii would be needed to verify the possible convergence of the ubiquitin-like conjugation system involving ATG12 and ATG5 across a wide range of eukaryotic lineages.
M. polymorpha has been recognized as a new model for analyzing the developmental processes of land plants (Bowman, 2016; Bowman et al., 2017; Ishizaki, 2017). This plant would also be useful for analyses of molecular mechanisms of autophagy due to its low genetic redundancy and widely available molecular genetic tools (Ishizaki et al., 2016; Bowman et al., 2017). In the interest of understanding the molecular mechanisms of autophagy in plants, it would be beneficial to reveal the molecular functions of ATG8 members, given that nonplant ATG8 acts at various steps of autophagy, such as expansion and closure of the isolation membrane, cargo recognition, and transport of autophagosomes to the vacuole/lysosome (Abeliovich et al., 2000; Nakatogawa et al., 2007; Fujita et al., 2008a; Kimura et al., 2008; Xie et al., 2008; Noda et al., 2010; Manil-Segalen et al., 2014; Nguyen et al., 2016; Tsuboyama et al., 2016). Although several ATG8-interacting proteins, such as ATI1/2, ATI3, DSK2, NBR1, ORM1/2, PUX, RPN10, and TSPO, which are involved in autophagy induced by certain stresses, have been identified (Svenning et al., 2011; Vanhee et al., 2011; Honig et al., 2012; Zhou et al., 2013, 2014, 2018; Hachez et al., 2014; Michaeli et al., 2014; Marshall et al., 2015, 2019; Hafren et al., 2017; Nolan et al., 2017; Yang et al., 2019), it remains unclear how various substrates are selectively targeted by autophagy in plants (Yoshimoto and Ohsumi, 2018). Given that M. polymorpha possesses fewer copies of ATG8 than other model plants, this plant would be useful for revealing the molecular functions of ATG8 members and functional diversification of ATG8 in land plants.
In this study, we succeeded in visualizing autophagosomes using fluorescently tagged ATG8 proteins in M. polymorpha. Both MpATG8a and MpATG8b were localized to punctate structures in the cytosol and vacuole. Deletion of MpATG2, MpATG5, or MpATG7 resulted in defective transport of MpATG8a into the vacuole, indicating that vacuolar transport of ATG8 is autophagy-dependent in M. polymorpha, as reported in other organisms. Punctate localization of MpATG8a was also detected even in the Mpatg mutants, and similar localization is also observed in Arabidopsis atg mutants (Yoshimoto et al., 2004; Kang et al., 2018). ATG8/LC3 is reported to be incorporated into protein aggregates in an autophagy-independent manner in mammalian cells (Kuma et al., 2007; Tanida et al., 2008). Therefore, it is highly likely that MpATG8a also aggregates in the cytosol independently of autophagic activities in the Mpatg mutants. It would be also possible that some population of fluorescently tagged MpATG8a/b-positive puncta observed in WT plants represents unfunctional protein aggregates, which should be verified in future studies.
The autophagy-defective mutants of M. polymorpha exhibited an early senescence-like phenotype and hypersensitivity to nutrient starvation (Figure 5), which resembles the phenotypes observed in atg mutants of other plants (Doelling et al., 2002; Hanaoka et al., 2002; Mukae et al., 2015; Wada et al., 2015). In A. thaliana, salicylic acid (SA) signaling is involved in early senescence in atg mutants (Yoshimoto et al., 2009). Although the relevance of SA signaling to the senescence of Mpatg mutants remains unknown, autophagy might play a common role in preventing senescence among land plants.
A forward genetic approach using M. polymorpha would also be effective to reveal the molecular mechanisms of autophagy in land plants, as in other systems (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding, 1995; Tian et al., 2010; Morita et al., 2018). Its haploid-dominant life cycle and low genetic redundancy could make this plant even more amenable to forward genetic analyses than other model plants, including A. thaliana (Ishizaki et al., 2016). Screening of mutants defective in autophagy and functional analyses of obtained factors would facilitate understanding of the molecular mechanisms of autophagy in land plants. In conclusion, M. polymorpha is a suitable system for analyzing autophagy in land plants. Further studies in this plant will contribute to revealing the molecular mechanisms of autophagy in plants, which would also be useful to gain insights into how the autophagy machinery has been functionally diversified and how autophagy has been recruited to support plant physiology.
Amino acid sequences of core autophagy machinery in K. nitens, P. patens, and M. polymorpha were obtained in MarpolBase1 using ATG genes of A. thaliana (Li et al., 2014; Shemi et al., 2015; Liu et al., 2018) as queries. ATG genes in C. braunii were searched in the C. braunii portal site2. For core autophagy machinery orthologs in C. reinhardtii except for ATG11, we referred to the previous study (Jiang et al., 2012; Shemi et al., 2015). The ATG11 homolog in C. reinhardtii was searched in MarpolBase. For ATG17, ATG29, and ATG31, whose homologs have not been identified in A. thaliana thus far, ATG genes in S. cerevisiae were used as queries. ATG10 and ATG12 homologs in S. pratensis, N. mirabilis, C. orbicularis, and M. viride were searched in the transcriptome database in MarpolBase. ATG18 homologs in D. salina, V. carteri, C. subellipsoidea C-169, M. pusilla CCMP1545, and O. lucimarinus were searched in Phytozome v12.1.63. ATG18 homologs in O. tauri were searched in MarpolBase. A domain search was performed using SMART4 (Letunic et al., 2014; Letunic and Bork, 2017). The accession numbers and amino acid sequences analyzed in this study are included in the Supplementary Material. We followed the nomenclature proposed in Bowman et al. (2016) for nomenclature of genes, proteins, and mutants of M. polymorpha.
Amino acid sequences of ATG18 in various plant species were aligned with ClustalX 2.1 (Larkin et al., 2007), and alignment gaps were removed using Gblocks5. Phylogenetic analysis was performed using PhyML 3.06 (Guindon et al., 2010) under the LG+G+I+F model, which was selected by Smart Model Selection in PhyML (Lefort et al., 2017). Bootstrap analysis was performed by resampling 1,000 sets. The sequences used in the phylogenetic analysis and the alignment from which gaps were removed were included in the Supplementary Material.
Genomic sequences of MpATG8a (Mapoly0001s0494.1) and MpATG8b (Mapoly0027s0034.1) were amplified by PCR from genomic DNA prepared from gemmae of M. polymorpha accession Takaragaike-1 (Tak-1, male) (Ishizaki et al., 2008), and the amplified products were subcloned into pENTR/D-TOPO (Invitrogen) according to the manufacturer’s instructions. To construct mCitrine- and mRFP-MpATG8, genomic sequences comprising the protein-coding regions and 3′-flanking sequences (2 kb) were amplified with the SmaI site followed by a flexible linker sequence (Gly–Gly–Ser–Gly) attached at the 5′-end and subcloned into the pENTR vector. Then, cDNA for mRFP or mCitrine containing the SmaI site at the 5′-end was inserted into the SmaI site of pENTR vectors containing the MpATG8 genes using the In-Fusion HD Cloning System (Clontech) according to the manufacturer’s instructions. The 5 kb 5′-sequence [promoter + 5′-untranslated region (UTR)] of each MpATG8 was then amplified and inserted into the SmaI site of the mRFP/mCitrine-MpATG8 vectors. The resultant chimeric genes were then introduced into pMpGWB301 (mCitrine-tagged MpATG8a and MpATG8b) or pMpGWB101 (mRFP-MpATG8a) (Ishizaki et al., 2015) using the Gateway LR ClonaseTM II Enzyme Mix (Invitrogen) according to the manufacturer’s instructions. To construct CRISPR/Cas9 vectors, two complementary oligonucleotides in the sequences of MpATG2, MpATG5, and MpATG7 were synthesized and annealed, and the resulting double-stranded fragments were subcloned at the BsaI site of the pMpGE_En03 vector (Sugano et al., 2018). The resultant gRNA cassette flanked by the attL1 and attL2 sequences in pMpGE_En03 were then introduced into the pMpGE010 vector (Sugano et al., 2018) using the Gateway LR Clonase II Enzyme Mix. The list of primer sequences used in this study is included in the Supplementary Material.
The M. polymorpha accession Tak-1 was grown asexually and maintained on 1/2× Gamborg’s B5 medium containing 1.4% agar at 22°C under continuous white light. Transformation was performed as previously described (Kubota et al., 2013). Transformants were selected on plates containing 10 mg/l hygromycin B and 250 mg/l cefotaxime for the pMpGWB101 and pMpGE010 vectors and 0.5 μM chlorsulfuron and 250 mg/l cefotaxime for the pMpGWB301 vector.
Five-day-old thalli grown on 1/2× Gamborg’s B5 medium containing 1.4% agar at 22°C under continuous white light were used for observation. Dorsal thallus tissues were observed using an LSM 780 confocal microscope (Carl Zeiss) as previously described (Kanazawa et al., 2016). For concA treatment, 4-day-old thalli were incubated in liquid 1/2× Gamborg’s B5 medium plus 1 μM concA (Santa Cruz Biotechnology, sc-202111) for 14 h at 22°C under continuous white light. concA was dissolved in dimethyl sulfoxide (DMSO) at 1 mM as a stock solution. For the mock treatment, samples were treated with DMSO at a concentration equal to that used for the inhibitor-treated samples.
For genotyping of mutants generated by CRISPR/Cas9, total RNA was extracted from 5-day-old thalli of Mpatg5-1ge, Mpatg7-1ge, and Mpatg2-1ge using the RNeasy Plant Mini Kit (Qiagen) and used as a template for reverse transcription using SuperScript III Reverse Transcriptase (Invitrogen) and the oligo (dT) (18-mer) primer according to the manufacturer’s instructions. Mutations in the obtained cDNA fragments were analyzed by direct sequencing.
Five-day-old thalli were used for the immunoblot analysis. One hundred milligrams of plants were homogenized in 200 μl of grinding buffer [50 mM HEPES–KOH, pH 7.5, 340 mM sorbitol, 5 mM MgCl2, and 1× CompleteTM Protease Inhibitor Cocktail (Roche)] for each genotype and centrifuged at 1,000 × g for 10 min. The supernatants were centrifuged at 3,000 × g for 10 min, and the resulting supernatants were used for immunoblotting. The polyclonal anti-GFP antibody (Kanazawa et al., 2016) was purified by affinity column chromatography using the GST-mCitrine protein bound to the HiTrapTM NHS-activated HP Column (GE Healthcare) and used at 500× dilution. The peroxidase-conjugated donkey anti-rabbit immunoglobulin antibody (GE Healthcare) was used as the secondary antibody. Signals were detected using Immobilon Western Chemiluminescent HRP Substrate (Merck).
Five-day-old thalli were incubated in 1 ml of liquid 1/2× Gamborg’s B5 medium for 3 days under continuous light or in 10 mM MES (pH 5.5) for 3 days under dark condition, and chlorophyll was extracted by soaking in 500 μl N,N-dimethylformamide overnight. Calculation of chlorophyll concentrations was done according to Porra et al. (1989). Three thalli were treated as one sample for correct measurement of fresh weights.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
TN and TU designed the research and wrote the manuscript. TN performed a major part of the experiment and analyzed the data. TK and NM prepared the anti-GFP antibody. TK assisted with vector construction. HT and TU supervised the study.
This study was financially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [Grant Nos. 24114003, 17K19412, and 18H02470 (to TU) and 17H07333 and 18K14738 (to TK)], a Grant-in-Aid for the Japan Society for the Promotion of Science (JSPS) fellows (to TN, Grant No. 19J13751), the BIO-NEXT project from the Okazaki Institute for Integrative Bioscience (to HT), the Mitsubishi Foundation, and the Yamada Science Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Takayuki Kohchi and Dr. Ryuichi Nishihama (Kyoto University), Dr. Katsuyuki T. Yamato (Kindai University), Dr. Kimitsune Ishizaki (Kobe University), and Dr. Shigeo S. Sugano (Ritsumeikan University) for sharing the research materials. This work was supported by the Model Plant Research Facility, NIBB BioResource Center.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00935/full#supplementary-material
DATA SET S1 | This data set contains all amino acid sequences analyzed in this study, the alignment generated for phylogenetic analysis, and lists of primers and accession numbers.
Abeliovich, H., Dunn, W. A., Kim, J., and Klionsky, D. J. (2000). Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034. doi: 10.1083/jcb.151.5.1025
Baba, M., Osumi, M., and Ohsumi, Y. (1995). Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 20, 465–471. doi: 10.1247/csf.20.465
Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687–1695. doi: 10.1083/jcb.139.7.1687
Baba, M., Takeshige, K., Baba, N., and Ohsumi, Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. doi: 10.1083/jcb.124.6.903
Bowman, J. L. (2016). A brief history of Marchantia from Greece to genomics. Plant Cell Physiol. 57, 210–229. doi: 10.1093/pcp/pcv044
Bowman, J. L., Araki, T., Arteaga-Vazquez, M. A., Berger, F., Dolan, L., Haseloff, J., et al. (2016). The naming of names: guidelines for gene nomenclature in Marchantia. Plant Cell Physiol. 57, 257–261. doi: 10.1093/pcp/pcv193
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha Genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W. J., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Chowdhury, S., Otomo, C., Leitner, A., Ohashi, K., Aebersold, R., Lander, G. C., et al. (2018). Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A–WIPI4 complex. Proc. Natl. Acad. Sci. U.S.A. 115, E9792–E9801. doi: 10.1073/pnas.1811874115
Chung, T., Phillips, A. R., and Vierstra, R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62, 483–493. doi: 10.1111/j.1365-313X.2010.04166.x
Dagdas, Y. F., Belhaj, K., Maqbool, A., Chaparro-Garcia, A., Pandey, P., Petre, B., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5:e10856. doi: 10.7554/eLife.10856
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Dooley, H. C., Razi, M., Polson, H. E., Girardin, S. E., Wilson, M. I., and Tooze, S. A. (2014). WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 55, 238–252. doi: 10.1016/j.molcel.2014.05.021
Dove, S. K., Piper, R. C., McEwen, R. K., Yu, J. W., King, M. C., Hughes, D. C., et al. (2004). Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23, 1922–1933. doi: 10.1038/sj.emboj.7600203
Efe, J. A., Botelho, R. J., and Emr, S. D. (2007). Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol. Biol. Cell 18, 4232–4244. doi: 10.1091/mbc.e07-04-0301
Era, A., Tominaga, M., Ebine, K., Awai, C., Saito, C., Ishizaki, K., et al. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50, 1041–1048. doi: 10.1093/pcp/pcp055
Farre, J. C., Manjithaya, R., Mathewson, R. D., and Subramani, S. (2008). PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376. doi: 10.1016/j.devcel.2007.12.011
Fujita, N., Hayashi-Nishino, M., Fukumoto, H., Omori, H., Yamamoto, A., Noda, T., et al. (2008a). An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651–4659. doi: 10.1091/mbc.E08-03-0312
Fujita, N., Itoh, T., Omori, H., Fukuda, M., Noda, T., and Yoshimori, T. (2008b). The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100. doi: 10.1091/mbc.E07-12-1257
Gomez-Sanchez, R., Rose, J., Guimaraes, R., Mari, M., Papinski, D., Rieter, E., et al. (2018). Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 217, 2743–2763. doi: 10.1083/jcb.201710116
Graef, M., Friedman, J. R., Graham, C., Babu, M., and Nunnari, J. (2013). ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 24, 2918–2931. doi: 10.1091/mbc.E13-07-0381
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hachez, C., Veljanovski, V., Reinhardt, H., Guillaumot, D., Vanhee, C., Chaumont, F., et al. (2014). The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 26, 4974–4990. doi: 10.1105/tpc.114.134080
Hafren, A., Macia, J. L., Love, A. J., Milner, J. J., Drucker, M., and Hofius, D. (2017). Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. U.S.A. 114, E2026–E2035. doi: 10.1073/pnas.1610687114
Hanada, T., Noda, N. N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao, T., et al. (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302. doi: 10.1074/jbc.C700195200
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Harada, K., Kotani, T., Kirisako, H., Sakoh-Nakatogawa, M., Oikawa, Y., Kimura, Y., et al. (2019). Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. eLife 8:e43088. doi: 10.7554/eLife.43088
Harding, T. M. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602. doi: 10.1083/jcb.131.3.591
Harding, T. M., Hefner-Gravink, A., Thumm, M., and Klionsky, D. J. (1996). Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 271, 17621–17624. doi: 10.1074/jbc.271.30.17621
Honig, A., Avin-Wittenberg, T., Ufaz, S., and Galili, G. (2012). A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24, 288–303. doi: 10.1105/tpc.111.093112
Hosokawa, N., Sasaki, T., Iemura, S.-I., Natsume, T., Hara, T., and Mizushima, N. (2014). Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5, 973–979. doi: 10.4161/auto.5.7.9296
Huang, R., Xu, Y., Wan, W., Shou, X., Qian, J., You, Z., et al. (2015). Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466. doi: 10.1016/j.molcel.2014.12.013
Huang, W. P., Scott, S. V., Kim, J., and Klionsky, D. J. (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845–5851. doi: 10.1074/jbc.275.8.5845
Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. doi: 10.1038/35044114
Inoue, Y., Suzuki, T., Hattori, M., Yoshimoto, K., Ohsumi, Y., and Moriyasu, Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47, 1641–1652. doi: 10.1093/pcp/pcl031
Ishizaki, K. (2017). Evolution of land plants: insights from molecular studies on basal lineages. Biosci. Biotechnol. Biochem. 81, 73–80. doi: 10.1080/09168451.2016.1224641
Ishizaki, K., Chiyoda, S., Yamato, K. T., and Kohchi, T. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. doi: 10.1093/pcp/pcn085
Ishizaki, K., Nishihama, R., Ueda, M., Inoue, K., Ishida, S., Nishimura, Y., et al. (2015). Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One 10:e0138876. doi: 10.1371/journal.pone.0138876
Ishizaki, K., Nishihama, R., Yamato, K. T., and Kohchi, T. (2016). Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–270. doi: 10.1093/pcp/pcv097
Izumi, M., Hidema, J., Wada, S., Kondo, E., Kurusu, T., Kuchitsu, K., et al. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167, 1307–1320. doi: 10.1104/pp.114.254078
Izumi, M., Wada, S., Makino, A., and Ishida, H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154, 1196–1209. doi: 10.1104/pp.110.158519
Jiang, Q., Zhao, L., Dai, J., and Wu, Q. (2012). Analysis of autophagy genes in microalgae: chlorella as a potential model to study mechanism of autophagy. PLoS One 7:e41826. doi: 10.1371/journal.pone.0041826
Johansen, T., and Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. doi: 10.1093/emboj/19.21.5720
Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y., and Yoshimori, T. (2004). LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117(Pt 13), 2805–2812. doi: 10.1242/jcs.01131
Kamada, Y., Yoshino, K., Kondo, C., Kawamata, T., Oshiro, N., Yonezawa, K., et al. (2010). Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 30, 1049–1058. doi: 10.1128/MCB.01344-09
Kanazawa, T., Era, A., Minamino, N., Shikano, Y., Fujimoto, M., Uemura, T., et al. (2016). SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57, 307–324. doi: 10.1093/pcp/pcv076
Kang, S., Shin, K. D., Kim, J. H., and Chung, T. (2018). Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 37, 653–664. doi: 10.1007/s00299-018-2258-9
Kawamata, T., Kamada, Y., Suzuki, K., Kuboshima, N., Akimatsu, H., Ota, S., et al. (2005). Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 338, 1884–1889. doi: 10.1016/j.bbrc.2005.10.163
Kellner, R., De la Concepcion, J. C., Maqbool, A., Kamoun, S., and Dagdas, Y. F. (2017). ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 22, 204–214. doi: 10.1016/j.tplants.2016.11.015
Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3–kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. doi: 10.1083/jcb.152.3.519
Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., et al. (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. doi: 10.1083/jcb.153.2.381
Kim, J., Lee, H., Lee, H. N., Kim, S. H., Shin, K. D., and Chung, T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25, 4956–4966. doi: 10.1105/tpc.113.117960
Kimura, S., Noda, T., and Yoshimori, T. (2008). Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct. Funct. 33, 109–122. doi: 10.1247/csf.08005
Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., et al. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. doi: 10.1083/jcb.147.2.435
Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., et al. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. doi: 10.1083/jcb.151.2.263
Klionsky, D. J., Cregg, J. M., Dunn, W. A. Jr., Emr, S. D., Sakai, Y., Sandoval, I. V., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. doi: 10.1016/s1534-5807(03)00296-x
Komatsu, M., Waguri, S., Ueno, T., Iwata, J., Murata, S., Tanida, I., et al. (2005). Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434. doi: 10.1083/jcb.200412022
Kotani, T., Kirisako, H., Koizumi, M., Ohsumi, Y., and Nakatogawa, H. (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U.S.A. 115, 10363–10368. doi: 10.1073/pnas.1806727115
Kubota, A., Ishizaki, K., Hosaka, M., and Kohchi, T. (2013). Efficient agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Biochem. 77, 167–172. doi: 10.1271/bbb.120700
Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., et al. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. doi: 10.1038/nature03029
Kuma, A., Matsui, M., and Mizushima, N. (2007). LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3, 323–328. doi: 10.4161/auto.4012
Kuma, A., Mizushima, N., Ishihara, N., and Ohsumi, Y. (2002). Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277, 18619–18625. doi: 10.1074/jbc.M111889200
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lefort, V., Longueville, J. E., and Gascuel, O. (2017). SMS: smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424. doi: 10.1093/molbev/msx149
Letunic, I., and Bork, P. (2017). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Doerks, T., and Bork, P. (2014). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, F., Chung, T., and Vierstra, R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807. doi: 10.1105/tpc.113.120014
Liu, F., Hu, W., and Vierstra, R. D. (2018). The vacuolar protein sorting-38 subunit of the arabidopsis phosphatidylinositol-3-kinase complex plays critical roles in autophagy, endosome sorting, and gravitropism. Front. Plant Sci. 9, 781. doi: 10.3389/fpls.2018.00781
Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Manil-Segalen, M., Lefebvre, C., Jenzer, C., Trichet, M., Boulogne, C., Satiat-Jeunemaitre, B., et al. (2014). The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell 28, 43–55. doi: 10.1016/j.devcel.2013.11.022
Mari, M., Griffith, J., Rieter, E., Krishnappa, L., Klionsky, D. J., and Reggiori, F. (2010). An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022. doi: 10.1083/jcb.200912089
Marshall, R. S., Hua, Z., Mali, S., McLoughlin, F., and Vierstra, R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781.e24. doi: 10.1016/j.cell.2019.02.009
Marshall, R. S., Li, F., Gemperline, D. C., Book, A. J., and Vierstra, R. D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066. doi: 10.1016/j.molcel.2015.04.023
Mercer, C. A., Kaliappan, A., and Dennis, P. B. (2014). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5, 649–662. doi: 10.4161/auto.5.5.8249
Michaeli, S., Honig, A., Levanony, H., Peled-Zehavi, H., and Galili, G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26, 4084–4101. doi: 10.1105/tpc.114.129999
Minamino, N., Kanazawa, T., Era, A., Ebine, K., Nakano, A., and Ueda, T. (2018). RAB GTPases in the basal land plant Marchantia polymorpha. Plant Cell Physiol. 59, 845–856. doi: 10.1093/pcp/pcy027
Mizushima, N., Noda, T., and Ohsumi, Y. (1999). Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18, 3888–3896. doi: 10.1093/emboj/18.14.3888
Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M. D., et al. (1998). A protein conjugation system essential for autophagy. Nature 395, 395–398. doi: 10.1038/26506
Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., et al. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668. doi: 10.1083/jcb.152.4.657
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Morita, K., Hama, Y., Izume, T., Tamura, N., Ueno, T., Yamashita, Y., et al. (2018). Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217, 3817–3828. doi: 10.1083/jcb.201804132
Mukae, K., Inoue, Y., and Moriyasu, Y. (2015). ATG5-knockout mutants of physcomitrella provide a platform for analyzing the involvement of autophagy in senescence processes in plant cells. Plant Signal. Behav. 10, e1086859. doi: 10.1080/15592324.2015.1086859
Nakatogawa, H., Ichimura, Y., and Ohsumi, Y. (2007). Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178. doi: 10.1016/j.cell.2007.05.021
Nguyen, T. N., Padman, B. S., Usher, J., Oorschot, V., Ramm, G., and Lazarou, M. (2016). Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 215, 857–874. doi: 10.1083/jcb.201607039
Noda, N. N., Ohsumi, Y., and Inagaki, F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385. doi: 10.1016/j.febslet.2010.01.018
Noda, T., and Ohsumi, Y. (1998). Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966. doi: 10.1074/jbc.273.7.3963
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46.e7. doi: 10.1016/j.devcel.2017.03.013
Obara, K., Sekito, T., Niimi, K., and Ohsumi, Y. (2008). The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283, 23972–23980. doi: 10.1074/jbc.M803180200
Okamoto, K., Kondo-Okamoto, N., and Ohsumi, Y. (2009). Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97. doi: 10.1016/j.devcel.2009.06.013
Osawa, T., Kotani, T., Kawaoka, T., Hirata, E., Suzuki, K., Nakatogawa, H., et al. (2019). Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288. doi: 10.1038/s41594-019-0203-4
Padman, B. S., Nguyen, T. N., Uoselis, L., Skulsuppaisarn, M., Nguyen, L. K., and Lazarou, M. (2019). LC3/GABARAPs drive ubiquitin-independent recruitment of Optineurin and NDP52 to amplify mitophagy. Nat. Commun. 10, 408. doi: 10.1038/s41467-019-08335-6
Pang, Y., Yamamoto, H., Sakamoto, H., Oku, M., Mutungi, J. K., Sahani, M. H., et al. (2019). Evolution from covalent conjugation to non-covalent interaction in the ubiquitin-like ATG12 system. Nat. Struct. Mol. Biol. 26, 289–296. doi: 10.1038/s41594-019-0204-3
Papinski, D., Schuschnig, M., Reiter, W., Wilhelm, L., Barnes, C. A., Maiolica, A., et al. (2014). Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell 53, 471–483. doi: 10.1016/j.molcel.2013.12.011
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Reggiori, F., Tucker, K. A., Stromhaug, P. E., and Klionsky, D. J. (2004). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90. doi: 10.1016/s1534-5807(03)00402-7
Sanchez-Vera, V., Kenchappa, C. S., Landberg, K., Bressendorff, S., Schwarzbach, S., Martin, T., et al. (2017). Autophagy is required for gamete differentiation in the moss Physcomitrella patens. Autophagy 13, 1939–1951. doi: 10.1080/15548627.2017.1366406
Scott, S. V., Hefner-Gravink, A., Morano, K. A., Noda, T., Ohsumi, Y., and Klionsky, D. J. (1996). Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. U.S.A. 93, 12304–12308. doi: 10.1073/pnas.93.22.12304
Shemi, A., Ben-Dor, S., and Vardi, A. (2015). Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy 11, 701–715. doi: 10.1080/15548627.2015.1034407
Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. doi: 10.1105/tpc.113.116947
Shintani, T., and Klionsky, D. J. (2004). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894. doi: 10.1074/jbc.M404399200
Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T., and Ohsumi, Y. (1999). Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18, 5234–5241. doi: 10.1093/emboj/18.19.5234
Sugano, S. S., Nishihama, R., Shirakawa, M., Takagi, J., Matsuda, Y., Ishida, S., et al. (2018). Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS One 13:e0205117. doi: 10.1371/journal.pone.0205117
Sugano, S. S., Shirakawa, M., Takagi, J., Matsuda, Y., Shimada, T., Hara-Nishimura, I., et al. (2014). CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55, 475–481. doi: 10.1093/pcp/pcu014
Suttangkakul, A., Li, F., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Suzuki, K., Akioka, M., Kondo-Kakuta, C., Yamamoto, H., and Ohsumi, Y. (2013). Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 126(Pt 11), 2534–2544. doi: 10.1242/jcs.122960
Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. doi: 10.1093/emboj/20.21.5971
Suzuki, K., Kubota, Y., Sekito, T., and Ohsumi, Y. (2007). Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 12, 209–218. doi: 10.1111/j.1365-2443.2007.01050.x
Suzuki, N. N., Yoshimoto, K., Fujioka, Y., Ohsumi, Y., and Inagaki, F. (2005). The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1, 119–126. doi: 10.4161/auto.1.2.1859
Suzuki, S. W., Yamamoto, H., Oikawa, Y., Kondo-Kakuta, C., Kimura, Y., Hirano, H., et al. (2015). Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl. Acad. Sci. U.S.A. 112, 3350–3355. doi: 10.1073/pnas.1421092112
Svenning, S., Lamark, T., Krause, K., and Johansen, T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311. doi: 10.1083/jcb.119.2.301
Tanida, I., Mizushima, N., Kiyooka, M., Ohsumi, M., Ueno, T., Ohsumi, Y., et al. (1999). Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379. doi: 10.1091/mbc.10.5.1367
Tanida, I., Yamaji, T., Ueno, T., Ishiura, S., Kominami, E., and Hanada, K. (2008). Consideration about negative controls for LC3 and expression vectors for four colored fluorescent protein-LC3 negative controls. Autophagy 4, 131–134. doi: 10.4161/auto.5233
Thompson, A. R., Doelling, J. H., Suttangkakul, A., and Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Thumm, M., Egner, R., Koch, B., Schlumpberger, M., Straub, M., Veenhuis, M., et al. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280. doi: 10.1016/0014-5793(94)00672-5
Tian, Y., Li, Z., Hu, W., Ren, H., Tian, E., Zhao, Y., et al. (2010). C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055. doi: 10.1016/j.cell.2010.04.034
Tsuboyama, K., Koyama-Honda, I., Sakamaki, Y., Koike, M., Morishita, H., and Mizushima, N. (2016). The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041. doi: 10.1126/science.aaf6136
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-e
Vanhee, C., Zapotoczny, G., Masquelier, D., Ghislain, M., and Batoko, H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805. doi: 10.1105/tpc.110.081570
Velikkakath, A. K., Nishimura, T., Oita, E., Ishihara, N., and Mizushima, N. (2012). Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909. doi: 10.1091/mbc.E11-09-0785
Wada, S., Hayashida, Y., Izumi, M., Kurusu, T., Hanamata, S., Kanno, K., et al. (2015). Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 168, 60–73. doi: 10.1104/pp.15.00242
Weidberg, H., Shvets, E., Shpilka, T., Shimron, F., Shinder, V., and Elazar, Z. (2010). LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802. doi: 10.1038/emboj.2010.74
Xie, Z., Nair, U., and Klionsky, D. J. (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298. doi: 10.1091/mbc.E07-12-1292
Xiong, Y., Contento, A. L., and Bassham, D. C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546. doi: 10.1111/j.1365-313X.2005.02397.x
Xiong, Y., Contento, A. L., Nguyen, P. Q., and Bassham, D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Yamamoto, H., Kakuta, S., Watanabe, T. M., Kitamura, A., Sekito, T., Kondo-Kakuta, C., et al. (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198, 219–233. doi: 10.1083/jcb.201202061
Yang, F., Kimberlin, A. N., Elowsky, C. G., Liu, Y., Gonzalez-Solis, A., Cahoon, E. B., et al. (2019). A plant immune receptor degraded by selective autophagy. Mol. Plant 12, 113–123. doi: 10.1016/j.molp.2018.11.011
Yorimitsu, T., and Klionsky, D. J. (2005). Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16, 1593–1605. doi: 10.1091/mbc.e04-11-1035
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.1105/tpc.109.068635
Yoshimoto, K., and Ohsumi, Y. (2018). Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59, 1337–1344. doi: 10.1093/pcp/pcy112
Yoshimoto, K., Shibata, M., Kondo, M., Oikawa, K., Sato, M., Toyooka, K., et al. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 127(Pt 6), 1161–1168. doi: 10.1242/jcs.139709
Young, P. G., Passalacqua, M. J., Chappell, K., Llinas, R. J., and Bartel, B. (2019). A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 15, 941–959. doi: 10.1080/15548627.2019.1569915
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 9, e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Wang, Z., Wang, X., Li, X., Zhang, Z., Fan, B., et al. (2018). Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 14, 487–504. doi: 10.1080/15548627.2017.1422856
Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J. Q., et al. (2014). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 10, e1004116. doi: 10.1371/journal.pgen.1004116
Zhou, X. M., Zhao, P., Wang, W., Zou, J., Cheng, T. H., Peng, X. B., et al. (2015). A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 22, 245–257. doi: 10.1093/dnares/dsv012
Keywords: autophagy, ATG, plant evolution, gene duplication, Marchantia polymorpha
Citation: Norizuki T, Kanazawa T, Minamino N, Tsukaya H and Ueda T (2019) Marchantia polymorpha, a New Model Plant for Autophagy Studies. Front. Plant Sci. 10:935. doi: 10.3389/fpls.2019.00935
Received: 25 April 2019; Accepted: 03 July 2019;
Published: 17 July 2019.
Edited by:
Masanori Izumi, RIKEN, JapanReviewed by:
Taijoon Chung, Pusan National University, South KoreaCopyright © 2019 Norizuki, Kanazawa, Minamino, Tsukaya and Ueda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Ueda, dHVlZGFAbmliYi5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.