
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 19 June 2019
Sec. Virology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00803
Plant molecular pharming has emerged as a reliable platform for recombinant protein expression providing a safe and low-cost alternative to bacterial and mammalian cells-based systems. Simultaneously, plant viruses have evolved from pathogens to molecular tools for recombinant protein expression, chimaeric viral vaccine production, and lately, as nanoagents for drug delivery. This review summarizes the genesis of viral vectors and agroinfection, the development of non-enveloped viruses for various biotechnological applications, and the on-going research on enveloped plant viruses.
Virology began in 1892 with D. Ivanovsky’s paper describing the retention of virulence in leaf sap extracted from Crimean tobacco with mosaic leaf disease (Lustig and Levine, 1992). Even though the extracted sap had been passed through bacteria-retaining filters, the filtrate retrained the ability to replicate within living plants (Beijerinck, 1898). Later, Vinson (1927) succeeded in precipitating the pathogen of tobacco mosaic disease, and experiments in 1936 by Bawden and Pirie revealed that this pathogen contained RNA and protein components (Bawden et al., 1936; Bawden and Pirie, 1937). Further experiments in 1956 demonstrated that genetic information was stored in RNA molecules (Fraenkel-Conrat et al., 1957) and later the concept of self-assembly of RNA and coat protein (CP) into particles was established (Butler and Klug, 1971). This pathogen is known as tobacco mosaic virus (TMV) and is one of the prominent viruses in plant molecular pharming.
Tobacco mosaic virus belongs to the Tobamovirus genus with a positive sense, single-stranded genomic RNA (gRNA) with a 7-methylguanosine-5-triphosphate cap at the 5′ terminus (Dunigan and Zaitlin, 1990) and a 3′ untranslated region (UTR) harboring a transfer RNA (tRNA)-like structure (Takamatsu et al., 1990) as shown in Figure 1A. It encodes a total of four proteins two of which are involved in RNA replication plus a movement protein (MP) and a CP (Goelet et al., 1982). Inside the plant cell, the gRNA acts as a messenger RNA (mRNA) template for expressing a 126 kDa protein containing methyltransferase and helicase domains plus a 183 kDa (readthrough) protein containing a polymerase domain (Osman and Buck, 1996; Lewandowski and Dawson, 2000). The 126 kDa and the 183 kDa replication proteins bind to the terminal tRNA-like structure initiating transcription of complementary (negative-sense) template (Lewandowski and Dawson, 2000; Osman and Buck, 2003). This negative-sense RNA acts as a template for the synthesis of full-length positive strands and subgenomic RNAs containing MP and CP open reading frames (ORFs) (Ishikawa et al., 1991). The MP is an RNA binding protein involved in cell-to-cell spreading of the virus (Citovsky et al., 1990; Chen et al., 2000) while the CP enhances the formation of replication complexes (Asurmendi et al., 2004), long-distance movement (Saito et al., 1990; Hilf and Dawson, 1993), and viral particle assembly (Bloomer et al., 1978; Butler, 1999).
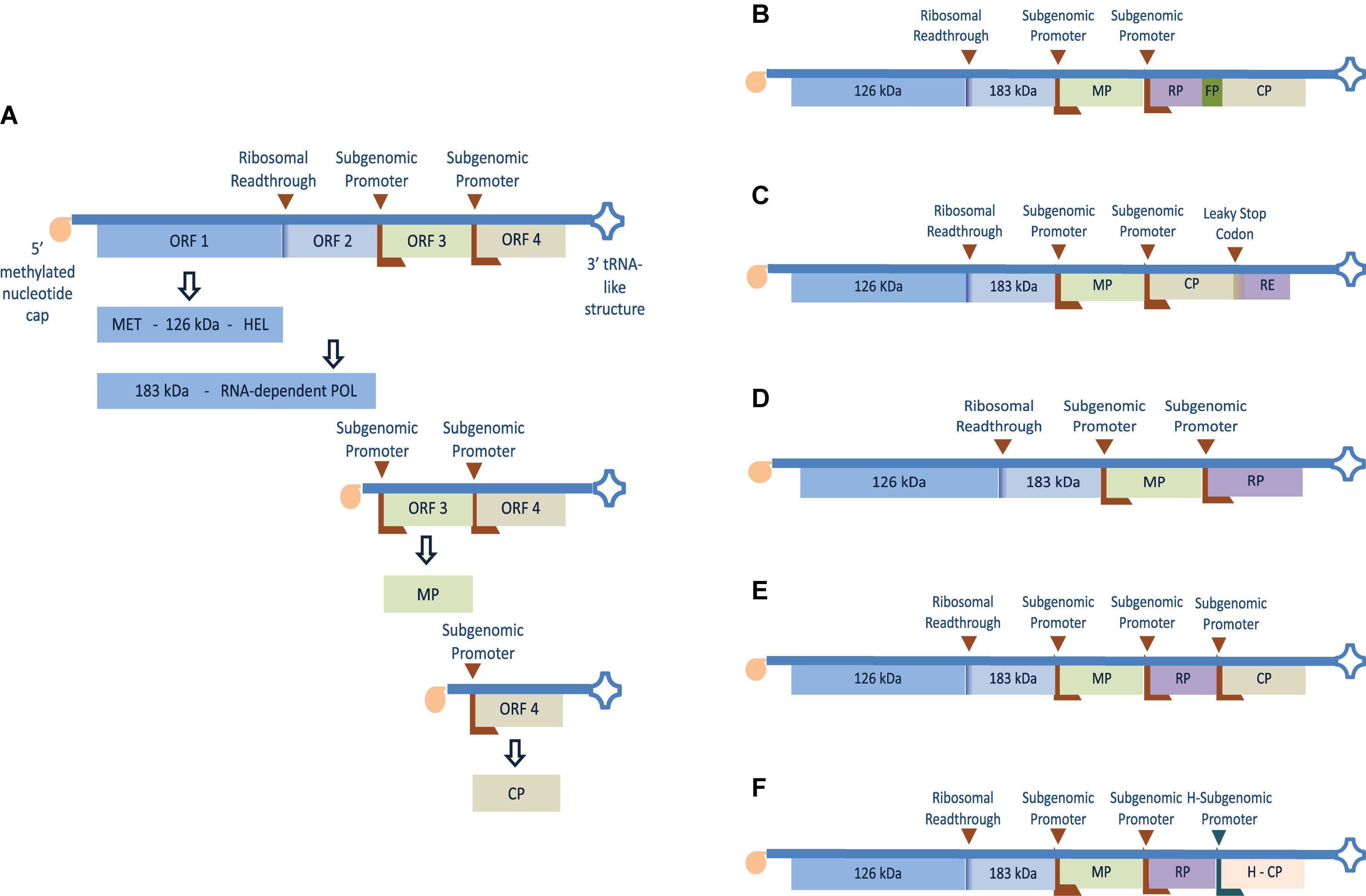
Figure 1. Genomic organization and expression strategy of TMV and different strategies adapted to express recombinant protein or heterologous epitope using the full virus genome. (A) The positive single-stranded RNA genome has four separate ORF(s) with a 5′ terminus methylated nucleotide cap (m7G5’pppG) and 3′-terminus tRNA-like structure. The first two 5′ proximal ORF(s) encode 126 and 183 kDa readthrough proteins containing methyltransferase (MET), helicase (HEL), and polymerase (POL) domains which are involved in replication and transcription of the genome. ORF 3 and 4 are translated from separate subgenomic promoters and encode the 30 kDa movement protein (MP) and 17 kDa coat protein (CP) respectively. (B) Recombinant protein (RP) fused to the CP N-terminus using fusion peptide (FP) (Roder et al., 2017). (C) Recombinant epitope (RE) fused to the CP C-terminus using leaky UAG stop codon (Sugiyama et al., 1995). (D) Coding region of RP cloned in place of the virus CP gene (Takamatsu et al., 1987). (E) Coding region of RP downstream a subgenomic RNA promoter and placed between the MP and the CP genes (Dawson et al., 1989). (F) Coding region of RP cloned downstream TMV subgenomic RNA promoter and placed between TMV MP gene and a heterogeneous CP (H-CP) gene. The latter was cloned into the TMV vector together with its heterogeneous subgenomic promoter from odontoglossum ringspot virus (ORSV) (Donson et al., 1991).
During the 1980s, the field of plant molecular pharming was born (Franken et al., 1997) and various pharmaceuticals such as human hormones (Barta et al., 1986), antibodies (Hiatt et al., 1989), and vaccines (Thanavala et al., 1995), were produced using transgenic plants. Currently, several proteins manufactured in plants are commercialized such as bovine trypsin TrypZean expressed in maize and commercialized by Sigma-Aldrich (#T3568, Sigma-Aldrich Corporation, United States), the 2006 USDA approved Newcastle disease virus for poultry produced in tobacco cell-suspension by Dow AgroSciences (Vermij and Waltz, 2006), and the 2012 FDA approved taliglucerase alfa (Elelyso®) for the management of type 1 Gaucher’s disease produced in carrot cells by Protalix Biotherapeutics Inc. (Maxmen, 2012).
However, the generation and selection of stably transformed plants for heterologous protein expression is quite elaborate and time-consuming which lead scientists to research exploiting viruses for this purpose. Plant viruses such as TMV (Takamatsu et al., 1987) and cowpea mosaic virus (CPMV) (Gopinath et al., 2000) were first adapted as full-virus vector then as a deconstructed-virus vector for recombinant protein expression. Later on, as we understood more of viral structure and capsid assembly, non-enveloped plant viruses were also manipulated for the generation of chimeric viral particles, nanoagents for carriage of various compounds and nanostructures building blocks. Furthermore, recently enveloped plant viruses, such as rhabdoviruses, have been recovered from agroinfiltrated cDNA, adding a viral particle with an amendable lipid envelope to the arsenal of available systems.
Adopting plant viruses as vectors for transient expression offered many advantages over the transgenic system such as ability for application in various plant species (Hamamoto et al., 1993), reduced gene-to-product time (Hendy et al., 1999), and increased yields (Maclean et al., 2007). At first, the host cells were infected with a full copy of plant virus (either DNA or in vitro transcribed RNA) into which the heterogenous sequence or gene of interest (GOI) was cloned. The first published virus-based vector was a gene-replacement model in which the GOI replaced the CP of brome mosaic virus (BMV) (French et al., 1986). However, lacking the CP, the recombinant virus was unable to spread, and infection was limited to inoculated cells. Further attempts followed using TMV-based vectors in which the GOI was inserted upstream the inherent CP gene and controlled by an additional subgenomic RNA promoter (the inherent CP subgenomic promoter was duplicated) (Dawson et al., 1988). Still this design was unstable and homologous recombination resulted in the loss of inserted sequence and reversion to the wild-type virus (Dawson et al., 1989). A third approach was to create a hybrid virus-vector containing different subgenomic promoter sequences from two tobamoviruses [TMV and odontoglossum ringspot virus (ORSV)]. This hybrid design resulted in a more stable vector that succeeded in the systemic expression of the recombinant protein (Donson et al., 1991). Other designs included fusing the recombinant protein to the TMV CP either at the C-terminal (Roder et al., 2017) or at the N-terminal downstream a leaky stop codon (Sugiyama et al., 1995). Figures 1B–F, show the above described modification approaches applied to TMV. Table 1 lists a number of plant-made pharmaceutical proteins expressed using full-viral vectors.
Although viral vectors based on a full-genome demonstrated success in producing recombinant proteins, constructs with large inserts showed instability and low systemic spread (Shivprasad et al., 1999; Avesani et al., 2007). Together with biosafety concerns (Scholthof et al., 1996), these limitations lead to the development of second-generation vectors in which the virus genome was deconstructed into a replicon containing the essential viral genomic components for replication and gene expression while plant-infection was initiated exogenously. In this system, viral MP and/or CP genes were replaced by the recombinant GOI and the vector was introduced into plants as part of Agrobacterium delivered T-DNA (Gleba et al., 2004), biolistic bombardment (Komarova et al., 2006) or as chromosome-inserted replicon (Cañizares et al., 2006). Lacking the CP, the deconstructed-virus based vectors lacked the ability to encapsidate into viral particles, however, they retained the ability to replicate, transcribe, and translate as shown in Figure 2.
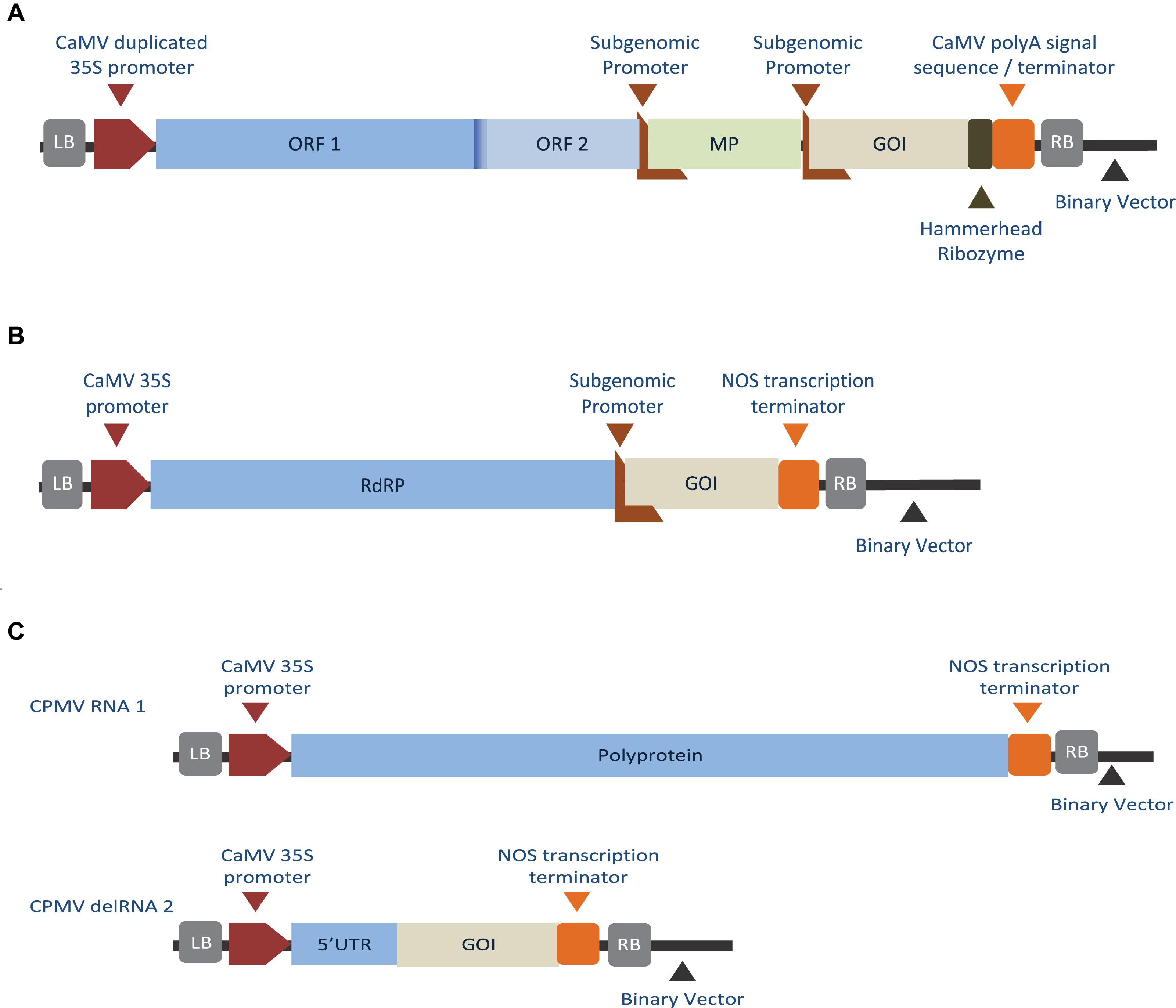
Figure 2. Diagrams showing different replicon-based expression systems. (A) Deconstructed monopartite RNA virus, TMV-based system, where the viral CP was substituted with the GOI under the control of a subgenomic promoter (Lindbo, 2007). (B) Deconstructed monopartite RNA virus, PVX-based system, where the viral CP and MP were substituted with the GOI under the control of a subgenomic promoter (Komarova et al., 2006). (C) Deconstructed bipartite RNA plant virus, CPMV-based system, where the RNA-1 component supplies the viral RdRp, virus genome-linked protein, helicase, and protease, while the modified RNA-2 contains the GOI, replacing those of the viral MP and CP (Cañizares et al., 2006). (A) Described replicon design is capable of replication, cell-to-cell movement, transcription, and replication, while designs (B,C) are capable of all, but deficient in cell-to-cell movement. LB, left border; RB, right border of the T-DNA region.
Among the monopartite RNA viruses, potato virus X (PVX) (Komarova et al., 2006) and TMV (Lindbo, 2007) were first to be adopted as a deconstructed-virus vectors. Deconstructed-TMV based vectors were further commercially pursued by Icon Genetics as magnifection technology (trademarked as magnICON®) (Gleba et al., 2005). The modifications included using a hybrid RdRP [from turnip vein-clearing virus (TVCV)] and Arabidopsis actin 2 (ACT2) as a promoter together with removal of cryptic thymine-rich intron sites plus selective introduction of introns (Marillonnet et al., 2004, 2005). Magnifection of hybrid TMV-based vectors yielded 4 g/kg of fresh weight tissue (FWT) of recombinant protein and 4.8 g/kg FWT of full immunoglobulin G (IgG) in less than 2 weeks (Bendandi et al., 2010).
Similarly, a deconstructed virus strategy was developed for the bipartite RNA1/RNA2 CPMV. Early research was based on maintaining an unmodified RNA-1 while introducing the recombinant gene to the RNA-2 construct. Recombinant expression was achieved using this methodology with both full-length and defective versions of RNA-2 coding plasmids (Liu et al., 2005). Surprisingly, vector systems based on deleted regions of RNA-2 (delRNA-2) achieved higher expression yields than those obtained with full-length RNA-2 vectors (Sainsbury et al., 2008). Further experiments revealed that replication of delRNA-2 based molecules was not essential and high yields of recombinant protein expression was still achieved in the absence of RNA-1 (Sainsbury et al., 2008). The modified delRNA-2-based mRNA was “hyper-translated” providing approximately 0.3 g/kg FWT of fully assembled monoclonal antibody within 6 days after agroinfection (Sainsbury and Lomonossoff, 2008). Table 2 lists a number of plant-made pharmaceutical proteins expressed using deconstructed-viral vectors.
The CPMV RNA2-based expression system allowed an elegant extension in which the genome-integrated cDNA of RNA-2 was amplified using agroinfiltrated RNA-1 constructs or by crossing with RNA-1 transgenic plants (Cañizares et al., 2006). This method of an inducible-replicon system, or dormant viral cassette, was also applied with a tomato mosaic virus (ToMV) system in suspension-cultured plant cells (Dohi et al., 2006). Other applications included development of dormant viral cassettes which can be activated at a custom-chosen stage using chemical inducers, such as ethanol (Zhang and Mason, 2006), or via a Cre-LoxP recombination system (Tremblay et al., 2007).
The plant viruses first exploited in biotechnology were non-enveloped and consist of CP subunits that have the ability to self-assemble into filamentous structures such as TMV (Alonso et al., 2013) or hollow symmetric icosahedral structures such as CCMV (Zandi et al., 2004) as shown in Figures 3A,B.
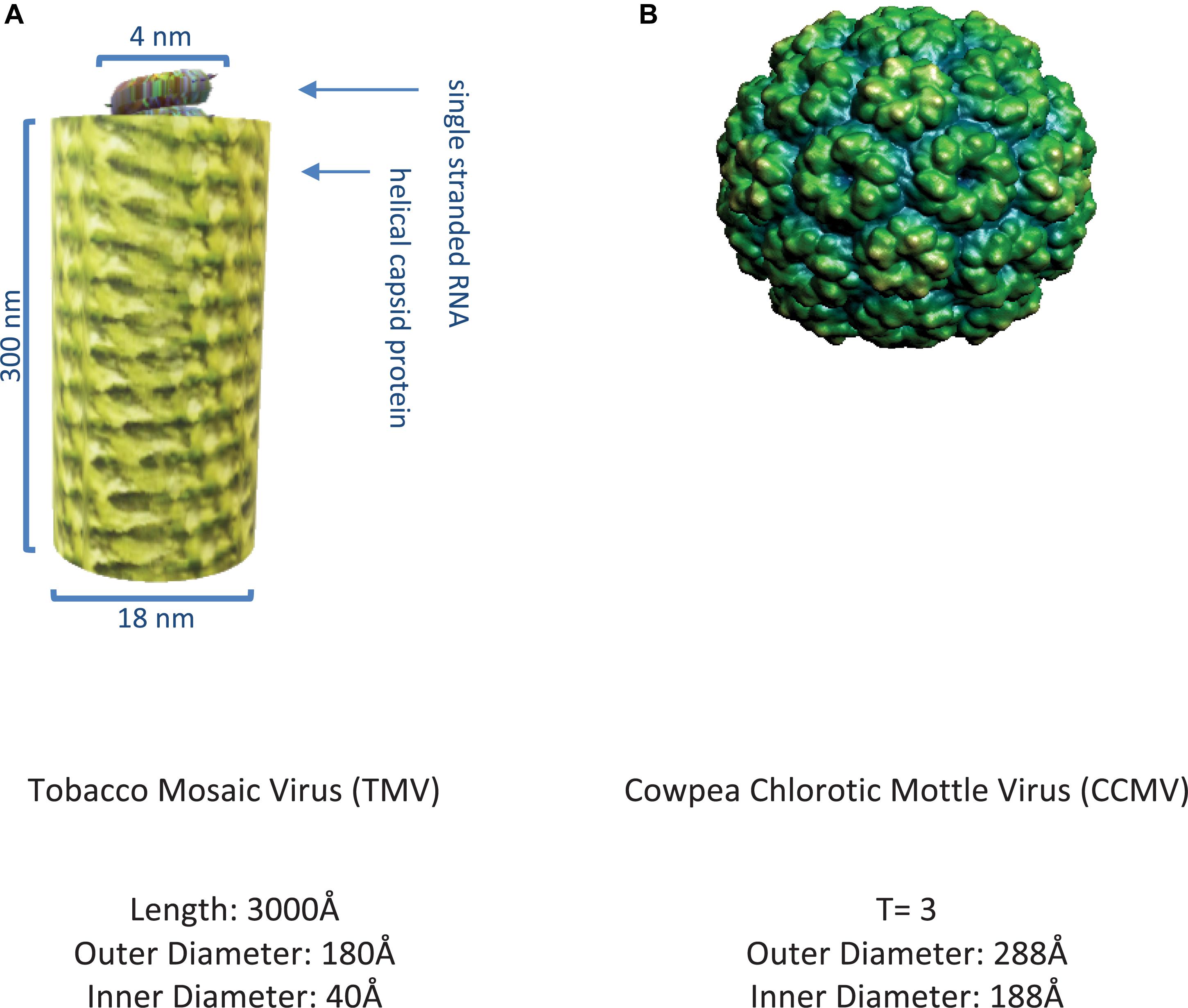
Figure 3. Representative plant viruses. (A) Image of tobacco mosaic virus (Namba and Stubbs, 1986) rendered on MS PowerPoint. (B) Image of cowpea chlorotic mottle virus (Speir et al., 1995) as obtained from VIPER (Carrillo-Tripp et al., 2009).
With better understanding of particle architecture, biophysical properties, and ability to manipulate their genomic material, these viruses also became exploited as virus-like particles (VLP), self-assembled structures devoid of any genomic material (Huang et al., 2009) and further developed as virus-based nanoparticles (VNP) (Steinmetz et al., 2009). For instance, the detailed molecular structure knowledge of filamentous viruses such as TMV (Namba et al., 1989), PVX (Parker et al., 2002), TVCV (Lartey et al., 1994), and PapMV (Yang et al., 2012) permitted their use for various applications such as vaccines (Smolenska et al., 1998; Petukhova et al., 2013; Therien et al., 2017), fluorescent markers (Yi et al., 2005), biocatalysts (Carette et al., 2007), nanoparticles for biologics purification (Werner et al., 2006), nanoparticles for in vivo imaging (Niehl et al., 2015), and assembly units for memory devices (Tseng et al., 2006), as shown in Figure 4.
Non-enveloped plant viruses have been engineered in three different ways: modification of the outer capsid (whether through genetic manipulation, chemical modification of capsid’s amino acids moieties, or through a combination of both), incorporation of heterologous cargo in the inner cavity, or particle integration into multivalent structures. Table 3 provides examples of some current VNP that are being developed as agents used in gene delivery (Azizgolshani et al., 2013), chemotherapy (Sánchez-Sánchez et al., 2014), immunotherapy (Venuti et al., 2015), vaccines (Phelps et al., 2007), and plant virus-assisted sensors (reviewed in Eiben et al., 2018).
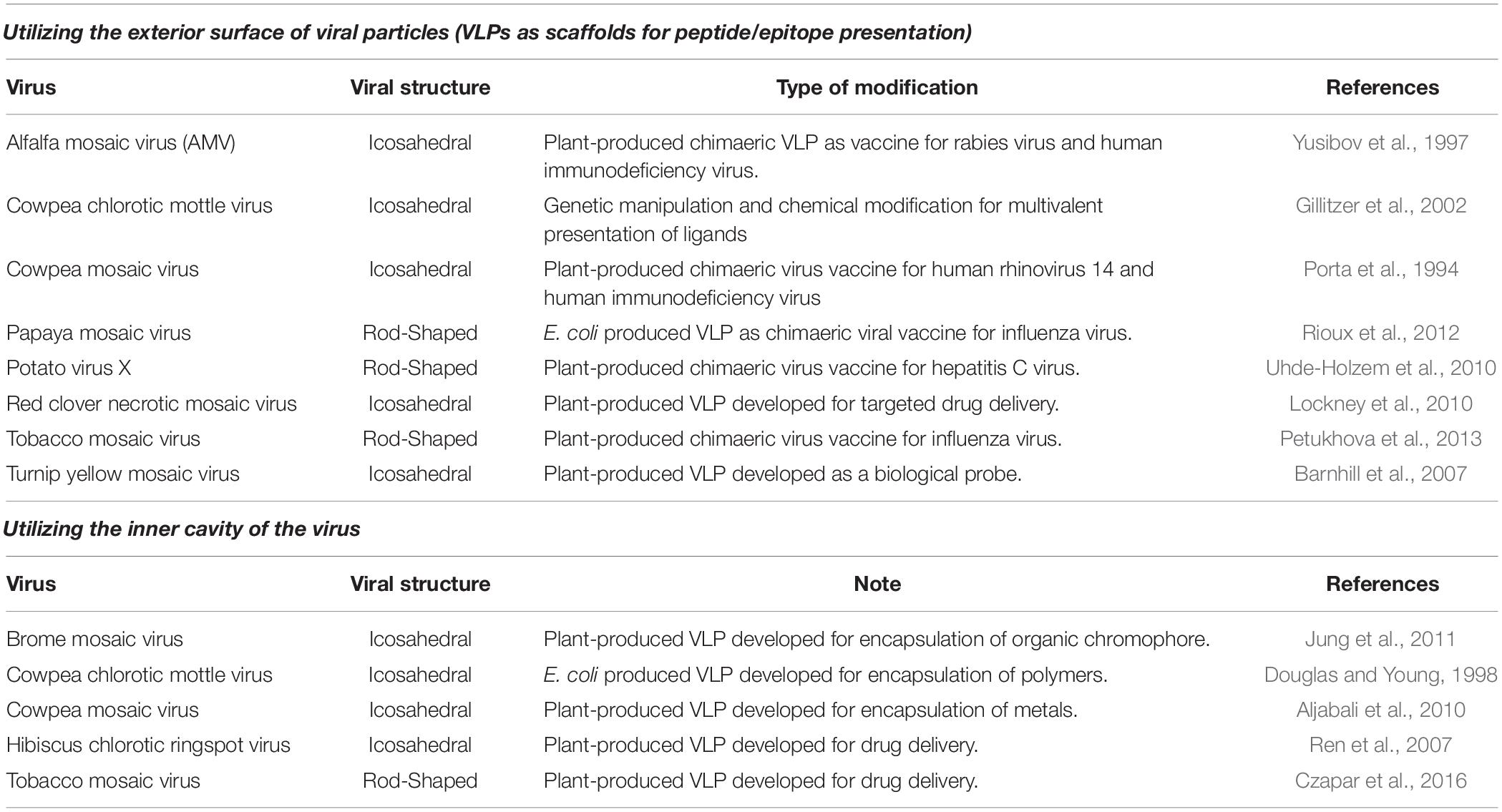
Table 3. List of some of the non-enveloped plant viruses that have been produced for VLP or VNP applications.
Production of plant virus based VLPs as vaccines received the attention since the early 1990 (Usha et al., 1993). Plant RNA viruses such as TMV (Koo et al., 1999), CPMV (Mclain et al., 1995), CMV (Nuzzaci et al., 2007), plum pox potyvirus (PPV) (Fernández-Fernández et al., 1998), potato virus X (PVX) (Lico et al., 2009), and tomato bushy stunt virus (TBSV) (Joelson et al., 1997) have been adapted for the production of vaccines and tested in various animal models. Studies conducted on the biodistribution and clearance of TMV (Bruckman et al., 2014), PVX and tomato bushy stunt virus (TBSV) (Blandino et al., 2015) in animal models showed no VLP induced pathological damage. Plant viruses are not pathogenic to mammals but are recognized by the pathogen associated molecular pattern (PAMP) receptors of the innate immune system (Acosta-Ramírez et al., 2008). They were shown to elicit a humoral response when administered by parenteral (Brennan et al., 1999b) or mucosal routes (Brennan et al., 1999a) and a cell mediated response (Yusibov et al., 2005; Kemnade et al., 2014). For instance, PapMV-CP-M2e VLP can induced murine anti-M2e antibodies that recognized influenza-infected cells and provided 100% protection (Denis et al., 2008) without any PapMV VLP induced cellular toxicity (Rioux et al., 2014).
The production of plant virus based VLP(s) were not limited to plants, and a variety of heterologous systems such as insect cells (Lamb et al., 1996), yeast (Brumfield et al., 2004) or Escherichia coli (Denis et al., 2008) have been employed to produce recombinant potato leafroll virus (PLRV), CCMV, and PapMV CP that assemble into viral particles indistinguishable from their plant produced counterparts. Nevertheless, plant-produced VLPs remain the most economical choice, as plant production is scalable and cost-effective with production costs lower than those in E. coli or eukaryotic-based systems (Kusnadi et al., 1997; Tusé et al., 2014), and has substantial safety advantage compared to mammalian-cells produced alternatives (Ma et al., 2003).
In the last decade, VLPs have been extensively developed as recombinant vaccines in different systems whether in plants or mammalian (Soulie et al., 1991), insects (Le Tallec et al., 2009), yeast (Agnandji et al., 2011), and E. coli (Fiers et al., 2009). They have proven to be immunogenic and few are currently available on the market such as Engerix® [for hepatitis B virus (HBV)] (Keating and Noble, 2003), and Cervarix® [for human papillomavirus (HPV)] (Szarewski, 2010), produced by GlaxoSmithKline, or their respective equivalents Recombivax HB® (Venters et al., 2004), and Gardasil® (Tomljenovic and Shaw, 2012) produced by Merck.
Nevertheless, producing VLPs whether through genetic fusion or chemical conjugation remains challenging, especially for complex multimeric antigens (Jagu et al., 2013; Benen et al., 2014), full length glycoproteins that adopt different conformations (Kwong et al., 2002; Cullen et al., 2017), large epitopes (Werner et al., 2006), or have termini that are involved in protein folding (Pejawar-Gaddy et al., 2010) and might interrupt the capsid synchronized assembly (Zlotnick, 1994; Roldão et al., 2012), while post-expression chemical conjugation has the drawback of further increasing downstream processing costs (Buonaguro and Butler-Ransohoff, 2010; Wilken and Nikolov, 2012; Sabalza et al., 2014; Buyel et al., 2015). Furthermore, chimeric VLPs expressing recombinant peptides were found to induce reduced neutralizing antibodies titre when compared to the full domain vaccines (Gedvilaite et al., 2015). Even among plant-virus based VLP(s), for instance, alfalfa mosaic virus-based Plasmodium falciparum Pfs25 VLP, although it was shown to induce P. falciparum blocking antibodies in mice (Jones et al., 2013), in Phase I clinical trial it was shown to provide a low protection level (Chichester et al., 2018).
In line with those difficulties and limitations, and considering that some of these are not encountered with enveloped VLPs (eVLPs), there has been a growing trend toward the use of eVLPs as production platform (Gheysen et al., 1989). In this system, structural viral proteins are expressed and incorporated into host membranes released as particles. The envelope provides the flexibility for integration of complete envelope-proteins and glycoproteins resembling the native virus (Yamshchikov et al., 1995; Baumert et al., 1998; Latham and Galarza, 2001). Along this idea attempts have been made to produce various eVLPs for vaccine purposes, e.g., against influenza virus (production of eVLPs in insect cells expressing influenza virus structural proteins) (Khurana et al., 2011), HIV (based on virosome, in vitro associated/spiked with an HIV1 gp41-derived peptide) (Leroux-Roels et al., 2013) and breast cancer (based on in vitro association of antigenic peptides to synthesized phospholipid membranes) (Wiedermann et al., 2010).
Influenza eVLPs have been produced from the co-expression of the two major antigenic envelope proteins hemagglutinin (HA) and neuraminidase (NA) plus matrix 1 (M1) (Pushko et al., 2005) or from the co-expression of HA and M1 to enable budding (Galarza et al., 2005). Most of these attempts have been performed in animal cell systems, but recently plants have also been used as platform for producing eVLPs against avian H5N1 Influenza (Landry et al., 2010). HA-based influenza VLPs were found to be budding from the plasma membrane of plant cells expressing HA only, without the need of further viral proteins (D’Aoust et al., 2008), due to the absence of glycoprotein sialylation in plants (Séveno et al., 2004). Moreover, and importantly, these plant-produced HA based eVLP vaccines were found to elicit durable and cross-reactive T cell responses and are currently undergoing clinical trials by Medicago (Landry et al., 2014). These findings support and strengthen the development and exploitation of enveloped plant virus-based vector systems, amenable for genetic manipulation, in plant molecular pharming. Few plant viruses exist that have a naturally occurring envelope. They classify within the orders Bunyavirales and Mononegavirales, of which the family Rhabdoviridae, and have a negative strand RNA genome (King et al., 2018). Considering that monopartite rhabdoviruses are easier to handle and a reverse genetics system has recently been established for a plant infecting rhabdovirus, they present the most attractive platform for development of pseudotyped viral-vaccines upon which their inherent envelope will provide the needed flexibility to incorporate large and complex antigens.
The Rhabdoviridae family contains an ecologically diverse group of viruses infecting hosts from a wide plethora of aquatic and terrestrial vertebrates and plants (Kuzmin et al., 2009); rhabdoviruses from different kingdoms are listed Table 4. Plant rhabdoviruses, like all other viruses of the Rhabdoviridae family are enveloped, negative-sense RNA viruses (Jackson et al., 2005). They have a bacilliform shape defined by two major structural components: an outer envelope made of the host lipid bilayer embedded with surface projections of the virus glycoprotein and a tightly coiled internal nucleocapsid composed of genomic RNA (gRNA) together nucleoprotein, phosphoprotein, and the polymerase protein (Wolanski et al., 1967) (Figure 5, traditionally, plant rhabdoviruses were divided into Cytorhabdovirus or Nucleorhabdovirus genera, depending on the virus propagation sites within the cell while recently two new genera Dichorhavirus and Varicosavirus, with bipartite genomes, were added (Dietzgen et al., 2017; Whitfield et al., 2018).
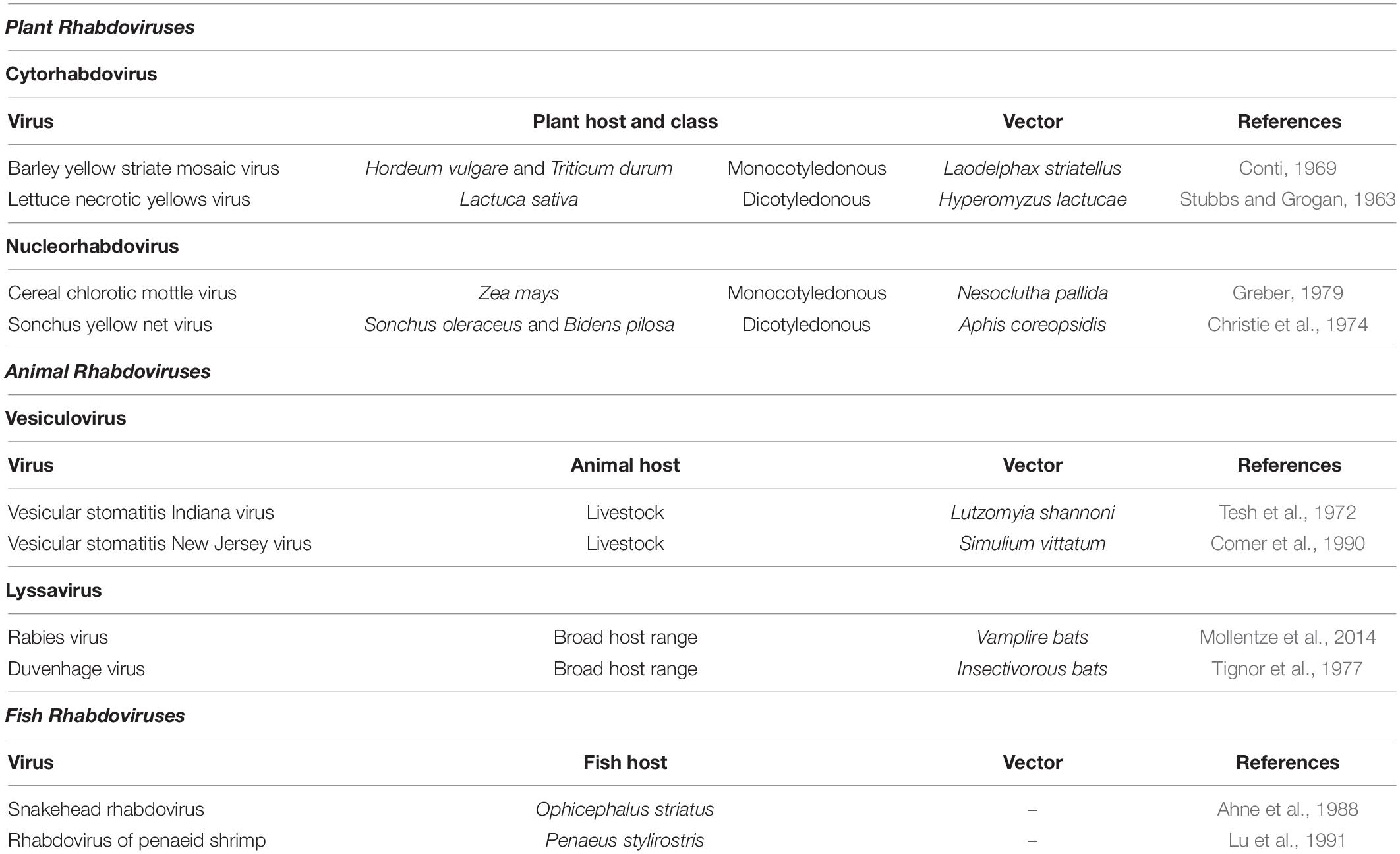
Table 4. List of some rhabdoviruses including plant Cytorhabdoviruses and Nucleorhabdoviruses together with their host, host class, and vectors.
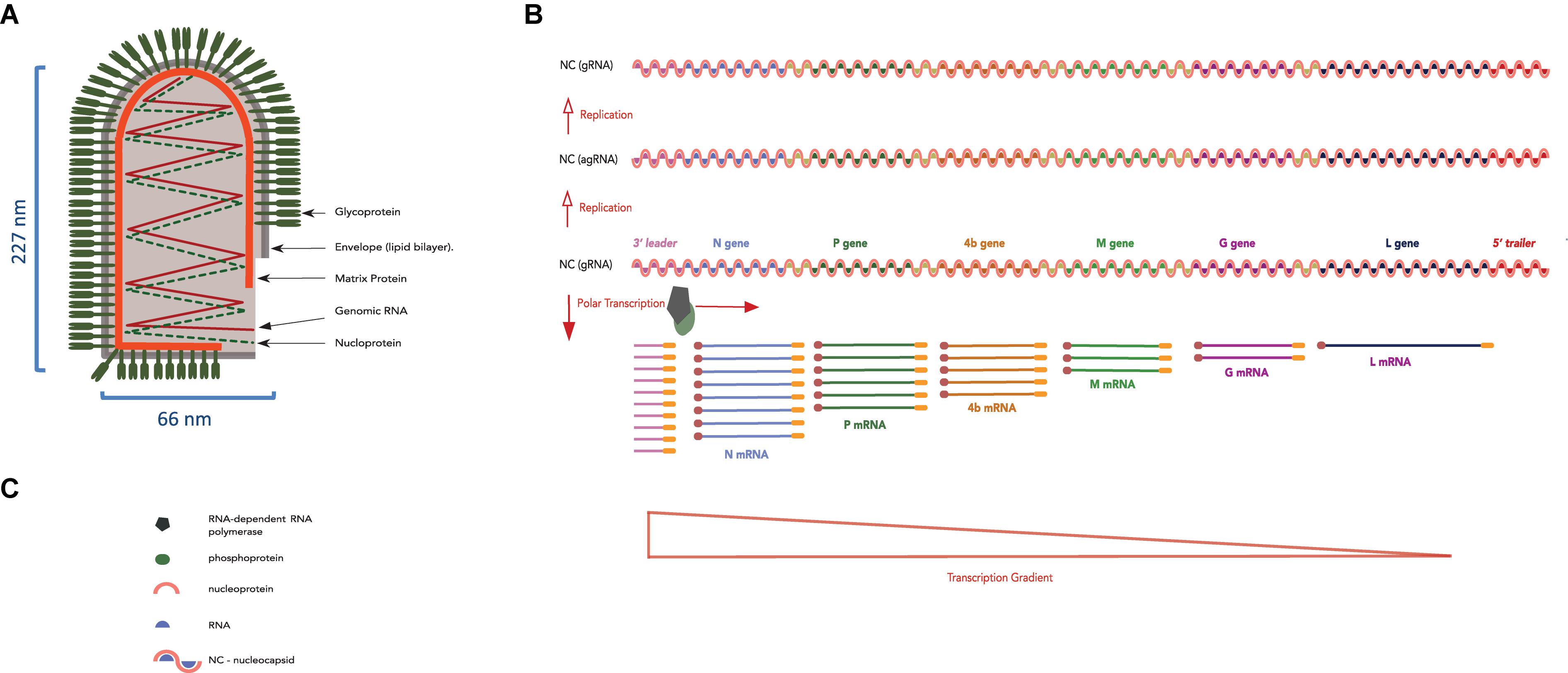
Figure 5. LNYV bullet-shape structure and its genomic RNA organization, replication and transcription strategy. (A) Diagram representing an LNYV particle showing the transmembrane glycoprotein, lipid bilayer, matrix protein, and genomic RNA together with the nucleoprotein forming the ribonucleoprotein coil. (B) LNYV gRNA (negative-sense) showing the 3′ leader, the N, P, 4b, M, G, and L genes, the 5′ trailer (sequential genes separated by a gene junction) and the transcription gradient. Genomic RNA is first replicated into agRNA (positive-sense), which is used as template for the production of (accumulating amounts of) progeny gRNA (negative-sense). (C) Legend of panel (B).
Many of the plant rhabdoviruses are transmitted by arthropods in a persistent propagative way in which the virus enters and replicates within the insect before transmission (Sylvester and Richardson, 1992; Creamer et al., 1997; Redinbaugh et al., 2002) while dichorhaviruses are transmitted by mites (Dietzgen et al., 2018) and varicosaviruses by soil-borne fungi (Latham et al., 2004). Their spread is correlated with that of the vector (Sylvester and Richardson, 1992), and no seed transmission has been reported. They infect both monocotyledonous (Conti and Appiano, 1973) and dicotyledonous (Salazar et al., 2000) plants, harming agricultural production (Chen et al., 1979; Fang et al., 1996). The typical symptoms of viral infection are necrosis, mosaic mottling of leaf tissue, and vein clearing (Jackson et al., 2009).
Plant rhabdovirus gRNA encodes the rhabdoviruses canonical genes: Nucleoprotein (N), Phosphoprotein (P), Matrix (M), Glycoprotein (G), and Large Polymerase (L) (Jackson et al., 2018), in addition to accessory genes (Walker et al., 2011) such as those coding for a cell-to-cell MP such as the 4b gene in lettuce necrotic yellow virus (LNYV) (Dietzgen et al., 2007), gene 3 of rice yellow stunt rhabdovirus (RYSV) (Huang et al., 2005). Furthermore, the gRNA is flanked by UTRs termed 3′ leader and 5′ trailer (Fu, 2005) (Figure 5B). The leader and trailer sequences are partially complementary and contain cis-acting signals involved in transcription and replication (Zuidema et al., 1986; Choi et al., 1994; Wang et al., 1999) while short intergenic sequences regulate mRNA synthesis and sequential transcription of the canonical genes (Wetzel et al., 1994; Bandyopadhyay et al., 2010).
Theoretical models that explain rhabdovirus transcription and replication were built upon experiments on mammalian rhabdoviruses, and one of the most well studied is Vesicular Stomatitis virus (VSV) (Emerson, 1987; Wertz et al., 1987). The ribonucleoprotein (RNP) unit, consisting of viral RNA associated with the N, L, and P proteins, acts as the template for viral transcription and replication (Yang et al., 1998, 1999; Kawai et al., 1999). Virion-associated L protein transcribes the NC into mRNA of distinct proteins (Moyer and Banerjee, 1975; Toriyama and Peters, 1980). The transcription direction and gradient follow the order of genes in the virus genome and are controlled by the untranslated 3′ – leader sequence (Emerson, 1982) and the non-translated inter-genomic regions respectively as shown in Figure 5B (Barr et al., 1997; Finke et al., 2000). Transcription of the viral genes results in the generation of monocistronic 5′-capped and polyadenylated mRNAs.
Later during the infection, the N-P complex inhibits transcription and switches the function of the L protein from that of transcription to replication (La Ferla and Peluso, 1989; Gupta and Banerjee, 1997). First, the gRNA is replicated into an antigenomic sense RNA (agRNA) template, which is full positive-sense RNA without a cap and poly (A) tail and is encapsidated by the N protein (Arnheiter et al., 1985; Zhang et al., 2008). Later, agRNA functions as a template for progeny negative-sense gRNA (Albertini et al., 2008; Ivanov et al., 2011). With the progeny gRNA assembled as RNP, the M protein converts the extended helical RNP into a condensed form (Kaptur et al., 1991), resulting in the bullet-shape morphology of rhabdoviruses (Mebatsion et al., 1999). Finally, the G protein tail is incorporated into the assembled M-RNP complex, aiding the budding of progeny virus particles (Whitt et al., 1989; Schnell et al., 1998).
Initial attempts to generate rhabdoviruses from cDNA were based on mammalian rhabdoviruses. Research on VSV revealed that genomic RNA must be encapsidated with the viral N protein to be a functional template for RNA-dependent RNA polymerase (Emerson and Wagner, 1972). This discovery was followed by a set of experiments that enabled the first rescue (reverse genetics system) of VSV defective interfering (DI) particles from cells co-transfected with the five canonical proteins (Pattnaik and Wertz, 1990, 1991). Research on rabies virus (RABV) demonstrated the expression of a reporter gene cloned between the viral 3′ and 5′ termini in cells co-transfected with plasmids encoding for the three helper proteins N, P, and L (Conzelmann and Schnell, 1994). This finding was followed by the recovery of infectious, enveloped rabies virus particles from cDNA (Schnell et al., 1994). Similarly, this strategy was subsequently used to rescue other vertebrate rhabdoviruses, such as VSV (Whelan et al., 1995), Snakehead rhabdovirus (SHRV) (Johnson et al., 2000), infectious haematopoietic necrosis virus (IIHNV) (Brémont, 2005), and haemorrhagic septicaemia virus (VHSV) (Kim et al., 2011).
Similar attempts to reconstitute a plant rhabdovirus started at Jackson’s laboratory, where N. benthamiana cellulose-digested protoplasts were found to be suitable for studying sonchus yellow net virus (SYNV) replication (Jones and Jackson, 1990). However, the real challenge was the co-delivery of all plasmids needed for the recovery of recombinant viruses into a single cell. This difficulty was circumvented by using Agrobacterium infiltration to co-deliver vectors expressing SYNV helper proteins into N. benthamiana leaves (Goodin et al., 2002). In 2013, the same group demonstrated the success of the first plant rhabdovirus SYNV minireplicon (MR) strategy (Ganesan et al., 2013). Plasmids harboring a positive-sense MR cassette containing two sequentially cloned reporter genes between the SYNV 3′ and 5′ termini together with SYNV helper proteins were co-infiltrated into N. benthamiana. The fluorescence signal was detected in 5–6 days post-infiltration and was restricted to single cells only. This promising success culminated in 2015 when infectious SYNV was recovered from the upper leaves of N. benthamiana plants agroinfiltrated with four plasmids harboring positive-sense SYNV cDNA together with helper protein plasmids (Wang et al., 2015).
As mentioned earlier, mammalian rhabdoviruses, and in particular vesicular stomatitis virus (VSV), remains the prototype for reverse engineering and molecular adaptation of rhabdoviruses. Like all other rhabdoviruses it has the canonical N, P, M, G, and L genes in a negative sense single stranded RNA genome (Figure 6A). VSV was successfully rescued from cDNA in 1995 (Whelan et al., 1995) and in 1996 it was first engineered to express a reporter gene placed between those of the viral glycoprotein and the polymerase (Schnell et al., 1996b). Later in the same year, recombinant VSV (rVSV) was successfully produced incorporating a foreign protein, in addition to the VSV inherent glycoprotein, in viral envelope (Schnell et al., 1996a). In these recombinant VSV particles, two designs were considered. The first was with the full foreign protein incorporated into the VSV envelope (Figure 6B) while the second was the exoplasmic domain of the foreign protein fused upstream of the transmembrane domain and the cytoplasmic tail of the VSV glycoprotein (Figure 6C). Furthermore, in 1997, recombinant VSV lacking its inherent glycoprotein and expressing CD4 receptor to infect and kill HIV-1 infected T-cells was successfully rescued (Schnell et al., 1997) (Figure 6D).
Figure 6. A schematic representation of wild type VSV and recombinant VSV designs. (A) 3′ to 5′ negative-strand genomic RNA organization of wild type VSV. (B) Recombinant VSV (rVSV) with foreign protein inserted between the VSV G and L genes. (C) rVSV with foreign protein fused to the inherent VSV glycoprotein. (D) Recombinant VSVΔG with foreign protein replacing the inherent VSV glycoprotein. (E) rVSV with foreign protein gene inserted upstream of the VSV N protein. (F) rVSV with the gene for foreign protein inserted upstream of the VSV N protein and with truncated VSV glycoprotein. le, leader; tr, trailer; insP, gene for inserted protein; Gtrc, truncated glycoprotein.
The ability to rescue VSV without its inherent G protein (VSVΔG) gave the opportunity to generate safer non-propagating VSV based vaccines (Roberts et al., 1999). Moreover, further attenuation strategies based on malleability of the rhabdovirus genome, strategies such as decreasing the expression level of viral genes by manipulating their position down the expression gradient (Flanagan et al., 2003) or by G gene truncations and N gene translocations (Clarke et al., 2007) were also considered. For instance, inserting the foreign gene upstream of the VSV N gene (Figure 6E), or inserting the foreign gene upstream the N gene while relocating the N gene downstream the M gene and truncating VSV G protein (Figure 6F) were also adapted to rescue a recombinant VSV based HIV-1 vaccine (Cooper et al., 2008).
The inherent genomic characteristics of rhabdoviruses, such as having well-defined transcription start/stop signals (Stillman and Whitt, 1997), and the ability to accommodate large inserts while retaining high level expression rates (Haglund et al., 2000), together with negligible seropositivity in the human population (Roberts et al., 1999) made VSV a lucrative candidate for vaccine development and as vector for a number of biomedical applications such as recombinant viral vaccines (Safronetz et al., 2015; Lauretti et al., 2016), gene delivery vectors (Beier et al., 2013), or oncolytic vectors (Bridle et al., 2009) as exemplified in Table 5. Currently, there are a number of on-going clinical trials based on VSV1, such as Phase I/II clinical trials for VSV based Ebola virus vaccines (rVSV-ZEBOV) (Huttner et al., 2018).
Table 5. List of some of the animal rhabdoviruses that have been reverse engineered for various medical applications.
Nowadays VSV is commonly considered a very successful and widely used platform for heterologous expression of more complex and interesting pharmaceuticals and biomolecules, and for the generation of (pseudo) typed eVLPs for vaccine purposes. It is expected that the availability and exploitation of enveloped plant viruses as biological particles, in analogy to VSV, will give a boost to the exploitation of plant-based production platforms for more complex (glyco)proteins.
Plant molecular pharming has materialized as a reliable and cost-effective platform for the production of pharmaceuticals, vaccines, and biobetter products with a number of plant-produced proteins starting to be commercialized (Ratner, 2010; Yusibov et al., 2011). Simultaneously, the development of viral vectors, together with agroinfection, culminated in rapid transient expression systems that successfully express high levels of large and complex pharmaceutical proteins and antimicrobial peptides (Fischer and Emans, 2000; Chen et al., 2013; Peyret and Lomonossoff, 2013; Loh et al., 2017; Leite et al., 2019) and enabled the production of plant-virus based VLPs against chronic and infectious diseases (Hefferon, 2018; Sahithi et al., 2019). However, as with their comparative systems counterparts, plant-virus based VLP(s) are inherently constrained in their ability to present complex antigens and glycoproteins. Moreover, in the case of rapidly evolving viruses, such as influenza, selective pressure will drive viral evasion of immune response induced by VLP vaccines based on few amino acids or antigens that cannot reproduce native conformational epitopes. Furthermore, attempts to produce plant-made eVLP, such as influenza HA VLP, has been so far proven successful only because of the inherent characteristics of the influenza virus HA and its ease of detachment as eVLP from plants cell surface (D’Aoust et al., 2010); a basis for success not necessary applicable in other cases. Therefore, developments in reverse engineering inherently enveloped plant-viruses such as rhabdoviruses is expected to give plant molecular pharming a platform for a wide range of biotechnological applications, most relevant of which is expressing enveloped VLPs exposing chimeric and complex antigens.
Considering VSV as prototype, the inherent rhabdovirus characteristics of being enveloped, with defined genomic transcription units (Schnell et al., 1996b), genome stability (Walker et al., 2015), and an ability to stably incorporate recombinant glycoprotein into their envelope (Schnell et al., 1996a) make them ideal for various biotechnological applications. Plant rhabdoviruses, share such inherent characteristics, together with dispensability of the G protein for systemic plant-infection as demonstrated with SNYV (Wang et al., 2015). The G protein is required for virus entry and subsequent propagation within the arthropods vector (Hogenhout et al., 2003), while only the carboxyl-terminal domain of the G protein, together with M protein are required for the morphogenesis and the budding of the enveloped particles (Sun et al., 2018). Hence these additional characteristics will enable large scale production of recombinant rhabdovirus based eVLP in which the (exposed) ectodomain of the G has been exchanged for another gene-of-interest (GOI) glycoprotein. Such chimeric viruses are contained within the plant and require less stringent containment controls. Similarly to VSV-recombinant design, plant rhabdovirus vector constructs can also be constructed containing a GOI at different gene positions for various expression purposes (Figure 6). All these advantages together with the establishment of plant molecular pharming as cost effective and reliable production platform, make plant rhabdoviruses promising candidates in biotechnology in general and in the field of plant-made recombinant viral vaccines in specific.
AI wrote the first draft of the manuscript. AI, VO, and RK contributed to manuscript revision, and also read and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acosta-Ramírez, E., Pérez-Flores, R., Majeau, N., Pastelin-Palacios, R., Gil-Cruz, C., Ramírez-Saldaña, M., et al. (2008). Translating innate response into long-lasting antibody response by the intrinsic antigen-adjuvant properties of papaya mosaic virus. Immunology 124, 186–197.
Agnandji, S. T., Lell, B., Soulanoudjingar, S. S., Fernandes, J. F., Abossolo, B. P., Conzelmann, C., et al. (2011). First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365, 1863–1875. doi: 10.1056/NEJMoa1102287
Ahne, W., Jørgensen, P. V., Olesen, N. J., and Wattanavijarn, W. (1988). Serological examination of a rhabdovirus isolated from snakehead (Ophicephalus striatus) in Thailand with ulcerative syndrome. J. Appl. Ichthyol. 4, 194–196.
Albertini, A., Schoehn, G., Weissenhorn, W., and Ruigrok, R. (2008). Structural aspects of rabies virus replication. Cell. Mol. Life Sci. 65, 282–294.
Aljabali, A. A., Sainsbury, F., Lomonossoff, G. P., and Evans, D. J. (2010). Cowpea mosaic virus unmodified empty viruslike particles loaded with metal and metal oxide. Small 6, 818–821.
Alonso, J. M., Gorzny, M. L., and Bittner, A. M. (2013). The physics of tobacco mosaic virus and virus-based devices in biotechnology. Trends Biotechnol. 31, 530–538. doi: 10.1016/j.tibtech.2013.05.013
Arnheiter, H., Davis, N. L., Wertz, G., Schubert, M., and Lazzarini, R. A. (1985). Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell 41, 259–267.
Asurmendi, S., Berg, R., Koo, J., and Beachy, R. (2004). Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc. Natl. Acad. Sci. U.S.A. 101, 1415–1420.
Avesani, L., Marconi, G., Morandini, F., Albertini, E., Bruschetta, M., Bortesi, L., et al. (2007). Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Res. 16, 587–597.
Azizgolshani, O., Garmann, R. F., Cadena-Nava, R., Knobler, C. M., and Gelbart, W. M. (2013). Reconstituted plant viral capsids can release genes to mammalian cells. Virology 441, 12–17. doi: 10.1016/j.virol.2013.03.001
Bandyopadhyay, A., Kopperud, K., Anderson, G., Martin, K., and Goodin, M. (2010). An integrated protein localization and interaction map for Potato yellow dwarf virus, type species of the genus Nucleorhabdovirus. Virology 402, 61–71. doi: 10.1016/j.virol.2010.03.013
Barnhill, H. N., Reuther, R., Ferguson, P. L., Dreher, T., and Wang, Q. (2007). Turnip yellow mosaic virus as a chemoaddressable bionanoparticle. Bioconjug. Chem. 18, 852–859.
Barr, J. N., Whelan, S., and Wertz, G. W. (1997). cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 71, 8718–8725.
Barta, A., Sommergruber, K., Thompson, D., Hartmuth, K., Matzke, M. A., and Matzke, A. J. (1986). The expression of a nopaline synthase—human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol. Biol. 6, 347–357. doi: 10.1007/BF00034942
Baumert, T. F., Ito, S., Wong, D. T., and Liang, T. J. (1998). Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72, 3827–3836.
Bawden, F., and Pirie, N. (1937). The isolation and some properties of liquid crystalline substances from solanaceous plants infected with three strains of tobacco mosaic virus. Proc. R. Soc. Lond. Ser. B Biol. Sci. 123, 274–320.
Bawden, F., Pirie, N., Bernal, J., and Fankuchen, I. (1936). Liquid crystalline substances from virus-infected plants. Nature 138, 1051–1052.
Beier, K. T., Saunders, A. B., Oldenburg, I. A., Sabatini, B. L., and Cepko, C. L. (2013). Vesicular stomatitis virus with the rabies virus glycoprotein directs retrograde transsynaptic transport among neurons in vivo. Front. Neural Circuits 7:11. doi: 10.3389/fncir.2013.00011
Beijerinck, M. (1898). Concerning a contagium vivum fluidum as a cause of the spot-disease of tobacco leaves. Verhandelingen der koninklijke akademie wetenschappen te Amsterdam 1898; 65: 3–21. English edition: Beijerinck MW (trans: Johnson J). Phytopathol. Classics 65, 33–52.
Bendandi, M., Marillonnet, S., Kandzia, R., Thieme, F., Nickstadt, A., Herz, S., et al. (2010). Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin’s lymphoma. Ann. Oncol. 21, 2420–2427. doi: 10.1093/annonc/mdq256
Benen, T. D., Tonks, P., Kliche, A., Kapzan, R., Heeney, J. L., and Wagner, R. (2014). Development and immunological assessment of VLP-based immunogens exposing the membrane-proximal region of the HIV-1 gp41 protein. J. Biomed. Sci. 21:79. doi: 10.1186/s12929-014-0079-x
Blandino, A., Lico, C., Baschieri, S., Barberini, L., Cirotto, C., Blasi, P., et al. (2015). In vitro and in vivo toxicity evaluation of plant virus nanocarriers. Colloids Surf. B Biointerfaces 129, 130–136. doi: 10.1016/j.colsurfb.2015.03.039
Bloomer, A. C., Bricogne, G., Champness, J. N., Graham, J., Klug, A., and Staden, R. (1978). Structure of the protein disk of tobacco mosaic-virus - subunit interactions and implications for virus assembly. Acta Crystallogr. A 34, S396–S396.
Brémont, M. (2005). Reverse genetics on fish rhabdoviruses: tools to study the pathogenesis of fish rhabdoviruses. Curr. Top. Microbiol. Immunol. 292, 119–141.
Brennan, F. R., Bellaby, T., Helliwell, S. M., Jones, T. D., Kamstrup, S., Dalsgaard, K., et al. (1999a). Chimeric plant virus particles administered nasally or orally induce systemic and mucosal immune responses in mice. J. Virol. 73, 930–938.
Brennan, F. R., Jones, T., Longstaff, M., Chapman, S., Bellaby, T., Smith, H., et al. (1999b). Immunogenicity of peptides derived from a fibronectin-binding protein of S. aureus expressed on two different plant viruses. Vaccine 17, 1846–1857.
Bridle, B. W., Boudreau, J. E., Lichty, B. D., Brunellière, J., Stephenson, K., Koshy, S., et al. (2009). Vesicular stomatitis virus as a novel cancer vaccine vector to prime antitumor immunity amenable to rapid boosting with adenovirus. Mol. Ther. 17, 1814–1821. doi: 10.1038/mt.2009.154
Brisson, N., Paszkowski, J., Penswick, J., Gronenborn, B., Potrykus, I., and Hohn, T. (1984). Expression of a bacterial gene in plants by using a viral vector. Nature 310, 511–514.
Bruckman, M. A., Randolph, L. N., VanMeter, A., Hern, S., Shoffstall, A. J., Taurog, R. E., et al. (2014). Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano-rods and -spheres in mice. Virology 449, 163–173. doi: 10.1016/j.virol.2013.10.035
Brumfield, S., Willits, D., Tang, L., Johnson, J. E., Douglas, T., and Young, M. (2004). Heterologous expression of the modified coat protein of Cowpea chlorotic mottle bromovirus results in the assembly of protein cages with altered architectures and function. J. Gen. Virol. 85, 1049–1053.
Buonaguro, F. M., and Butler-Ransohoff, J.-E. (2010). PharmaPlant: the new frontier in vaccines. Expert Rev. Vaccines 9, 805–807.
Butler, P., and Klug, A. (1971). Assembly of the particle of tobacco mosaic virus from RNA and disks of protein. Nature 229, 47–50.
Butler, P. J. G. (1999). Self-assembly of tobacco mosaic virus: the role of an intermediate aggregate in generating both specificity and speed. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 354, 537–550. doi: 10.1098/rstb.1999.0405
Buyel, J., Twyman, R., and Fischer, R. (2015). Extraction and downstream processing of plant-derived recombinant proteins. Biotechnol. Adv. 33, 902–913. doi: 10.1016/j.biotechadv.2015.04.010
Cañizares, M. C., Liu, L., Perrin, Y., Tsakiris, E., and Lomonossoff, G. P. (2006). A bipartite system for the constitutive and inducible expression of high levels of foreign proteins in plants. Plant Biotechnol. J. 4, 183–193.
Carette, N., Engelkamp, H., Akpa, E., Pierre, S. J., Cameron, N. R., Christianen, P. C. M., et al. (2007). A virus-based biocatalyst. Nat. Nanotechnol. 2, 226–229. doi: 10.1038/nnano.2007.76
Carrillo-Tripp, M., Shepherd, C. M., Borelli, I. A., Venkataraman, S., Lander, G., Natarajan, P., et al. (2009). VIPERdb(2): an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 37, D436–D442. doi: 10.1093/nar/gkn840
Chen, M. H., Sheng, J. S., Hind, G., Handa, A. K., and Citovsky, V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19, 913–920. doi: 10.1093/emboj/19.5.913
Chen, Q., Lai, H., Hurtado, J., Stahnke, J., Leuzinger, K., and Dent, M. (2013). Agroinfiltration as an effective and scalable strategy of gene delivery for production of pharmaceutical proteins. Adv. Tech. Biol. Med. 1:103.
Chen, S., Yuan, Y., King, D., Chen, K., Lin, R., and Kao, T. (1979). Studies on the development and epiphytotics of the rice yellow stunt disease. Acta Phytopathol. Sin. 9, 41–71.
Chichester, J. A., Green, B. J., Jones, R. M., Shoji, Y., Miura, K., Long, C. A., et al. (2018). Safety and immunogenicity of a plant-produced Pfs25 virus-like particle as a transmission blocking vaccine against malaria: a Phase 1 dose-escalation study in healthy adults. Vaccine 36, 5865–5871. doi: 10.1016/j.vaccine.2018.08.033
Choi, T.-J., Wagner, J. D., and Jackson, A. (1994). Sequence analysis of the trailer region of sonchus yellow net virus genomic RNA. Virology 202, 33–40.
Christie, S., Christie, R., and Edwardson, J. (1974). Transmission of a bacilliform virus of sowthistle and Bidens pilosa. Phytopathology 64, 840–845.
Citovsky, V., Knorr, D., Schuster, G., and Zambryski, P. (1990). The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60, 637–647.
Clarke, D. K., Nasar, F., Lee, M., Johnson, J. E., Wright, K., Calderon, P., et al. (2007). Synergistic attenuation of vesicular stomatitis virus by combination of specific G gene truncations and N gene translocations. J. Virol. 81, 2056–2064.
Cobleigh, M. A., Buonocore, L., Uprichard, S. L., Rose, J. K., and Robek, M. D. (2010). A vesicular stomatitis virus-based hepatitis B virus vaccine vector provides protection against challenge in a single dose. J. Virol. 84, 7513–7522. doi: 10.1128/JVI.00200-10
Comer, J. A., Tesh, R. B., Modi, G. B., Corn, J. L., and Nettles, V. F. (1990). Vesicular stomatitis virus, New Jersey serotype: replication in and transmission by Lutzomyia shannoni (Diptera: Psychodidae). Am. J. Trop. Med. Hyg. 42, 483–490.
Conti, M. (1969). Investigations on a bullet-shaped virus of cereals isolated in Italy from planthoppers. J. Phytopathol. 66, 275–279.
Conti, M., and Appiano, A. (1973). Barley yellow striate mosaic virus and associated viroplasms in barley cells. J. Gen. Virol. 21, 315–322.
Conzelmann, K.-K., and Schnell, M. (1994). Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J. Virol. 68, 713–719.
Cooper, D., Wright, K. J., Calderon, P. C., Guo, M., Nasar, F., Johnson, J. E., et al. (2008). Attenuation of recombinant vesicular stomatitis virus-human immunodeficiency virus type 1 vaccine vectors by gene translocations and g gene truncation reduces neurovirulence and enhances immunogenicity in mice. J. Virol. 82, 207–219.
Creamer, R., He, X., and Styer, W. (1997). Transmission of sorghum stunt mosaic rhabdovirus by the leafhopper vector, Graminella sonora (Homoptera: Cicadellidae). Plant Dis. 81, 63–65. doi: 10.1094/PDIS.1997.81.1.63
Cullen, L. M., Schmidt, M. R., and Morrison, T. G. (2017). The importance of RSV F protein conformation in VLPs in stimulation of neutralizing antibody titers in mice previously infected with RSV. Hum. Vaccin. Immunother. 13, 2814–2823. doi: 10.1080/21645515.2017.1329069
Czapar, A. E., Zheng, Y.-R., Riddell, I. A., Shukla, S., Awuah, S. G., Lippard, S. J., et al. (2016). Tobacco mosaic virus delivery of phenanthriplatin for cancer therapy. ACS Nano 10, 4119–4126. doi: 10.1021/acsnano.5b07360
D’Aoust, M. A., Couture, M. M. J., Charland, N., Trepanier, S., Landry, N., Ors, F., et al. (2010). The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 8, 607–619. doi: 10.1111/j.1467-7652.2009.00496.x
D’Aoust, M. A., Lavoie, P. O., Couture, M. M. J., Trépanier, S., Guay, J. M., Dargis, M., et al. (2008). Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6, 930–940. doi: 10.1111/j.1467-7652.2008.00384.x
Dawson, W., Bubrick, P., and Grantham, G. (1988). Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology 78, 783–789.
Dawson, W., Lewandowski, D., Hilf, M., Bubrick, P., Raffo, A., Shaw, J., et al. (1989). A tobacco mosaic virus-hybrid expresses and loses an added gene. Virology 172, 285–292.
Denis, J., Acosta-Ramirez, E., Zhao, Y., Hamelin, M. E., Koukavica, I., Baz, M., et al. (2008). Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine 26, 3395–3403. doi: 10.1016/j.vaccine.2008.04.052
Dietzgen, R. G., Callaghan, B., and Campbell, P. R. (2007). Biology and genomics of lettuce necrotic yellows virus. Plant Viruses 1, 85–92.
Dietzgen, R. G., Freitas-Astúa, J., Chabi-Jesus, C., Ramos-González, P. L., Goodin, M. M., Kondo, H., et al. (2018). Dichorhaviruses in their host plants and mite vectors. Adv. Virus Res. 102, 119–148. doi: 10.1016/bs.aivir.2018.06.001
Dietzgen, R. G., Kondo, H., Goodin, M. M., Kurath, G., and Vasilakis, N. (2017). The family Rhabdoviridae: mono-and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 227, 158–170. doi: 10.1016/j.virusres.2016.10.010
Dohi, K., Nishikiori, M., Tamai, A., Ishikawa, M., Meshi, T., and Mori, M. (2006). Inducible virus-mediated expression of a foreign protein in suspension-cultured plant cells. Arch. Virol. 151, 1075–1084.
Donson, J., Kearney, C., Hilf, M., and Dawson, W. (1991). Systemic expression of a bacterial gene by a tobacco mosaic virus-based vector. Proc. Natl. Acad. Sci. U.S.A. 88, 7204–7208.
Douglas, T., and Young, M. (1998). Host–guest encapsulation of materials by assembled virus protein cages. Nature 393, 152–155.
Dunigan, D. D., and Zaitlin, M. (1990). Capping of tobacco mosaic virus RNA. Analysis of viral-coded guanylyltransferase-like activity. J. Biol. Chem. 265, 7779–7786.
Eiben, S., Koch, C., Altintoprak, K., Southan, A., Tovar, G., Laschat, S., et al. (2018). Plant virus-based materials for biomedical applications: trends and prospects. Adv. Drug Deliv. Rev. doi: 10.1016/j.addr.2018.08.011 [Epub ahead of print].
Emerson, S. U. (1982). Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell 31, 635–642.
Emerson, S. U. (1987). “Transcription of vesicular stomatitis virus,” in The Rhabdoviruses, ed. R. R. Wagner (Boston, MA: Springer), 245–269.
Emerson, S. U., and Wagner, R. R. (1972). Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J. Virol. 10, 297–309.
Fang, R.-X., Zhu, H.-T., Wang, Q., Mang, K.-Q., Gao, D.-M., Qin, W.-S., et al. (1996). Construction of transgenic rice plants resistant to rice yellow stunt virus, a plant rhabdovirus. Rice Genet. III, 201–205.
Fernández-Fernández, M. R., Martìnez-Torrecuadrada, J. L., Casal, J. I., and Garcìa, J. A. (1998). Development of an antigen presentation system based on plum pox potyvirus. FEBS Lett. 427, 229–235.
Fiers, W., De Filette, M., El Bakkouri, K., Schepens, B., Roose, K., Schotsaert, M., et al. (2009). M2e-based universal influenza A vaccine. Vaccine 27, 6280–6283. doi: 10.1016/j.vaccine.2009.07.007
Finke, S., Cox, J. H., and Conzelmann, K.-K. (2000). Differential transcription attenuation of rabies virus genes by intergenic regions: generation of recombinant viruses overexpressing the polymerase gene. J. Virol. 74, 7261–7269.
Fischer, R., and Emans, N. (2000). Molecular farming of pharmaceutical proteins. Transgenic Res. 9, 279–299.
Flanagan, E. B., Schoeb, T. R., and Wertz, G. W. (2003). Vesicular stomatitis viruses with rearranged genomes have altered invasiveness and neuropathogenesis in mice. J. Virol. 77, 5740–5748.
Fraenkel-Conrat, H., Singer, B., and Williams, R. (1957). Infectivity of viral nucleic acid. Biochim. Biophys. Acta 25, 87–96.
Franken, E., Teuschel, U., and Hain, R. (1997). Recombinant proteins from transgenic plants. Curr. Opin. Biotechnol. 8, 411–416.
French, R., Janda, M., and Ahlquist, P. (1986). Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science 231, 1294–1298.
Fu, Z. F. (2005). “Genetic comparison of the rhabdoviruses from animals and plants,” in The World of Rhabdoviruses, ed. Z. F. Fu (Berlin: Springer), 1–24.
Fujiki, M., Kaczmarczyk, J. F., Yusibov, V., and Rabindran, S. (2008). Development of a new cucumber mosaic virus-based plant expression vector with truncated 3a movement protein. Virology 381, 136–142. doi: 10.1016/j.virol.2008.08.022
Galarza, J. M., Latham, T., and Cupo, A. (2005). Virus-like particle (VLP) vaccine conferred complete protection against a lethal influenza virus challenge. Viral Immunol. 18, 244–251.
Ganesan, U., Bragg, J. N., Deng, M., Marr, S., Lee, M. Y., Qian, S., et al. (2013). Construction of a sonchus yellow net virus minireplicon: a step toward reverse genetic analysis of plant negative-strand RNA viruses. J. Virol. 87, 10598–10611. doi: 10.1128/JVI.01397-13
Gedvilaite, A., Kucinskaite-Kodze, I., Lasickiene, R., Timinskas, A., Vaitiekaite, A., Ziogiene, D., et al. (2015). Evaluation of trichodysplasia spinulosa-associated polyomavirus capsid protein as a new carrier for construction of chimeric virus-like particles harboring foreign epitopes. Viruses 7, 4204–4229. doi: 10.3390/v7082818
Gheysen, D., Jacobs, E., de Foresta, F., Thiriart, C., Francotte, M., Thines, D., et al. (1989). Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59, 103–112.
Gillitzer, E., Willits, D., Young, M., and Douglas, T. (2002). Chemical modification of a viral cage for multivalent presentation. Chem. Commun. 2002, 2390–2391.
Gils, M., Kandzia, R., Marillonnet, S., Klimyuk, V., and Gleba, Y. (2005). High-yield production of authentic human growth hormone using a plant virus-based expression system. Plant Biotechnol. J. 3, 613–620.
Giritch, A., Marillonnet, S., Engler, C., van Eldik, G., Botterman, J., Klimyuk, V., et al. (2006). Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl. Acad. Sci. U.S.A. 103, 14701–14706.
Gleba, Y., Klimyuk, V., and Marillonnet, S. (2005). Magnifection—a new platform for expressing recombinant vaccines in plants. Vaccine 23, 2042–2048.
Gleba, Y., Marillonnet, S., and Klimyuk, V. (2004). Engineering viral expression vectors for plants: the ‘full virus’ and the ‘deconstructed virus’ strategies. Curr. Opin. Plant Biol. 7, 182–188.
Goelet, P., Lomonossoff, G. P., Butler, P. J. G., Akam, M. E., Gait, M. J., and Karn, J. (1982). Nucleotide-sequence of tobacco mosaic-virus RNA. Proc. Natl. Acad. Sci. U.S.A. 79, 5818–5822. doi: 10.1073/pnas.79.19.5818
Goodin, M. M., Dietzgen, R. G., Schichnes, D., Ruzin, S., and Jackson, A. O. (2002). pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31, 375–383.
Gopinath, K., Wellink, J., Porta, C., Taylor, K. M., Lomonossoff, G. P., and van Kammen, A. (2000). Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267, 159–173.
Greber, R. (1979). Cereal chlorotic mottle virus—a rhabdovirus of Gramineae in Australia transmitted by Nesoclutha pallida (Evans). Aust. J. Agric. Res. 30, 433–443.
Gupta, A. K., and Banerjee, A. K. (1997). Expression and purification of vesicular stomatitis virus NP complex from Escherichia coli: role in genome RNA transcription and replication in vitro. J. Virol. 71, 4264–4271.
Haglund, K., Forman, J., Kräusslich, H.-G., and Rose, J. K. J. V. (2000). Expression of human immunodeficiency virus type 1 Gag protein precursor and envelope proteins from a vesicular stomatitis virus recombinant: high-level production of virus-like particles containing HIV envelope. Virology 268, 112–121.
Hamamoto, H., Sugiyama, Y., Nakagawa, N., Hashida, E., Matsunaga, Y., Takemoto, S., et al. (1993). A new tobacco mosaic-virus vector and its use for the systemic production of angiotensin-I-converting enzyme-inhibitor in transgenic tobacco and tomato. Biotechnology 11, 930–932. doi: 10.1038/nbt0893-930
Hefferon, K. (2018). Repurposing plant virus nanoparticles. Vaccines 6:E11. doi: 10.3390/vaccines6010011
Hendy, S., Chen, Z. C., Barker, H., Cruz, S. S., Chapman, S., Torrance, L., et al. (1999). Rapid production of single-chain Fv fragments in plants using a potato virus X episomal vector. J. Immunol. Methods 231, 137–146. doi: 10.1016/S0022-1759(99)00150-7
Hiatt, A., Caffferkey, R., and Bowdish, K. (1989). Production of antibodies in transgenic plants. Nature 342, 76–78.
Hilf, M. E., and Dawson, W. O. (1993). The tobamovirus capsid protein functions as a host-specific determinant of long-distance movement. Virology 193, 106–114. doi: 10.1006/viro.1993.1107
Hogenhout, S. A., Redinbaugh, M. G., and Ammar, E.-D. (2003). Plant and animal rhabdovirus host range: a bug’s view. Trends Microbiol. 11, 264–271.
Huang, Y.-W., Geng, Y.-F., Ying, X.-B., Chen, X.-Y., and Fang, R.-X. (2005). Identification of a movement protein of rice yellow stunt rhabdovirus. J. Virol. 79, 2108–2114.
Huang, Z., Chen, Q., Hjelm, B., Arntzen, C., and Mason, H. (2009). A DNA replicon system for rapid high-level production of virus-like particles in plants. Biotechnol. Bioeng. 103, 706–714. doi: 10.1002/bit.22299
Huttner, A., Agnandji, S. T., Combescure, C., Fernandes, J. F., Bache, E. B., Kabwende, L., et al. (2018). Determinants of antibody persistence across doses and continents after single-dose rVSV-ZEBOV vaccination for Ebola virus disease: an observational cohort study. Lancet Infect. Dis. 18, 738–748. doi: 10.1016/S1473-3099(18)30165-8
Ishikawa, M., Meshi, T., Ohno, T., and Okada, Y. (1991). Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic-virus infection. J. Virol. 65, 861–868.
Ivanov, I., Yabukarski, F., Ruigrok, R. W., and Jamin, M. (2011). Structural insights into the rhabdovirus transcription/replication complex. Virus Res. 162, 126–137. doi: 10.1016/j.virusres.2011.09.025
Jackson, A., Dietzgen, R., Fang, A. R., Hogenhout, S., Deng, M., and Bragg, J. (2009). “Plant rhabdoviruses,” in Desk Encyclopedia of Plant and Fungal Virology, eds M. V. Regenmortel and B. Mahy (Cambridge, MA: Academic Press), 270.
Jackson, A. O., Dietzgen, R. G., Goodin, M. M., Bragg, J. N., and Deng, M. (2005). Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43, 623–660.
Jackson, A. O., Dietzgen, R. G., Goodin, M. M., and Li, Z. (2018). Development of model systems for plant rhabdovirus research. Adv. Virus Res. 102, 23–57. doi: 10.1016/bs.aivir.2018.06.008
Jagu, S., Kwak, K., Karanam, B., Huh, W. K., Damotharan, V., Chivukula, S. V., et al. (2013). Optimization of multimeric human papillomavirus L2 vaccines. PLoS One 8:e55538. doi: 10.1371/journal.pone.0055538
Joelson, T., Akerblom, L., Oxelfelt, P., Strandberg, B., Tomenius, K., and Morris, T. J. (1997). Presentation of a foreign peptide on the surface of tomato bushy stunt virus. J. Gen. Virol. 78, 1213–1217.
Johnson, M. C., Simon, B. E., Kim, C. H., and Leong, J.-A. C. (2000). Production of recombinant snakehead rhabdovirus: the NV protein is not required for viral replication. J. Virol. 74, 2343–2350.
Jones, R. M., Chichester, J. A., Mett, V., Jaje, J., Tottey, S., Manceva, S., et al. (2013). A plant-produced Pfs25 VLP malaria vaccine candidate induces persistent transmission blocking antibodies against Plasmodium falciparum in immunized mice. PLoS One 8:e79538. doi: 10.1371/journal.pone.0079538
Jones, R. W., and Jackson, A. (1990). Replication of sonchus yellow net virus in infected protoplasts. Virology 179, 815–820.
Jung, B., Rao, A. L., and Anvari, B. (2011). Optical nano-constructs composed of genome-depleted brome mosaic virus doped with a near infrared chromophore for potential biomedical applications. Acs Nano 5, 1243–1252. doi: 10.1021/nn1028696
Kapadia, S. U., Simon, I. D., and Rose, J. K. J. V. (2008). SARS vaccine based on a replication-defective recombinant vesicular stomatitis virus is more potent than one based on a replication-competent vector. Virology 376, 165–172. doi: 10.1016/j.virol.2008.03.002
Kaptur, P. E., Rhodes, R. B., and Lyles, D. S. (1991). Sequences of the vesicular stomatitis virus matrix protein involved in binding to nucleocapsids. J. Virol. 65, 1057–1065.
Kawai, A., Toriumi, H., Tochikura, T. S., Takahashi, T., Honda, Y., and Morimoto, K. (1999). Nucleocapsid formation and/or subsequent conformational change of rabies virus nucleoprotein (N) is a prerequisite step for acquiring the phosphatase-sensitive epitope of monoclonal antibody 5-2-26. Virology 263, 395–407.
Keating, G. M., and Noble, S. (2003). Recombinant hepatitis B vaccine (Engerix-B®). Drugs 63, 1021–1051.
Kemnade, J. O., Seethammagari, M., Collinson-Pautz, M., Kaur, H., Spencer, D. M., and McCormick, A. A. (2014). Tobacco mosaic virus efficiently targets DC uptake, activation and antigen-specific T cell responses in vivo. Vaccine 32, 4228–4233. doi: 10.1016/j.vaccine.2014.04.051
Khurana, S., Wu, J., Verma, N., Verma, S., Raghunandan, R., Manischewitz, J., et al. (2011). H5N1 virus-like particle vaccine elicits cross-reactive neutralizing antibodies that preferentially bind to the oligomeric form of influenza virus hemagglutinin in humans. J. Virol. 85, 10945–10954. doi: 10.1128/jvi.05406-11
Kim, M. S., Kim, D. S., and Kim, K. H. (2011). Generation and characterization of NV gene-knockout recombinant viral hemorrhagic septicemia virus (VHSV) genotype IVa. Dis. Aquat. Organ. 97, 25–35. doi: 10.3354/dao02394
Kimpel, J., Urbiola, C., Koske, I., Tober, R., Banki, Z., Wollmann, G., et al. (2018). The oncolytic virus VSV-GP is effective against malignant melanoma. Viruses 10:E108. doi: 10.3390/v10030108
King, A. M. Q., Lefkowitz, E. J., Mushegian, A. R., Adams, M. J., Dutilh, B. E., Gorbalenya, A. E., et al. (2018). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2018). Arch. Virol. 163, 2601–2631. doi: 10.1007/s00705-018-3847-1
Komarova, T., Skulachev, M., Zvereva, A., Schwartz, A., Dorokhov, Y. L., and Atabekov, J. (2006). New viral vector for efficient production of target proteins in plants. Biochemistry 71, 846–850.
Koo, M., Bendahmane, M., Lettieri, G. A., Paoletti, A. D., Lane, T. E., Fitchen, J. H., et al. (1999). Protective immunity against murine hepatitis virus (MHV) induced by intranasal or subcutaneous administration of hybrids of tobacco mosaic virus that carries an MHV epitope. Proc. Natl. Acad. Sci. 96, 7774–7779.
Kumagai, M. H., Donson, J., della-Cioppa, G., and Grill, L. K. (2000). Rapid, high-level expression of glycosylated rice α-amylase in transfected plants by an RNA viral vector. Gene 245, 169–174.
Kusnadi, A. R., Nikolov, Z. L., and Howard, J. A. (1997). Production of recombinant proteins in transgenic plants: practical considerations. Biotechnol. Bioeng. 56, 473–484. doi: 10.1002/(SICI)1097-0290(19971205)56:5
Kuzmin, I. V., Novella, I. S., Dietzgen, R. G., Padhi, A., and Rupprecht, C. E. (2009). The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect. Genet. Evol. 9, 541–553.
Kwong, P. D., Doyle, M. L., Casper, D. J., Cicala, C., Leavitt, S. A., Majeed, S., et al. (2002). HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682.
La Ferla, F. M., and Peluso, R. W. (1989). The 1: 1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J. Virol. 63, 3852–3857.
Lamb, J., Duncan, G., Reavy, B., Gildow, F., Mayo, M., and Hay, R. (1996). Assembly of virus-like particles in insect cells infected with a baculovirus containing a modified coat protein gene of potato leafroll luteovirus. J. Gen. Virol. 77, 1349–1358.
Landry, N., Pillet, S., Favre, D., Poulin, J. F., Trepanier, S., Yassine-Diab, B., et al. (2014). Influenza virus-like particle vaccines made in Nicotiana benthamiana elicit durable, poly-functional and cross-reactive T cell responses to influenza HA antigens. Clin. Immunol. 154, 164–177. doi: 10.1016/j.clim.2014.08.003
Landry, N., Ward, B. J., Trepanier, S., Montomoli, E., Dargis, M., Lapini, G., et al. (2010). Preclinical and clinical development of plant-made virus-like particle vaccine against avian H5N1 influenza. PLoS One 5:e15559. doi: 10.1371/journal.pone.0015559
Lartey, R. T., Lane, L. C., and Melcher, U. (1994). Electron-microscopic and molecular characterization of turnip vein-clearing virus. Arch. Virol. 138, 287–298. doi: 10.1007/Bf01379132
Latham, L., Jones, R., and McKirdy, S. (2004). Lettuce big-vein disease: sources, patterns of spread, and losses. Aust. J. Agric. Res. 55, 125–130.
Latham, T., and Galarza, J. M. (2001). Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75, 6154–6165. doi: 10.1128/jvi.75.13.6154-6165.2001
Lauretti, F., Chattopadhyay, A., de Oliveira França, R. F., Castro-Jorge, L., Rose, J., and da Fonseca, B. A. (2016). Recombinant vesicular stomatitis virus-based dengue-2 vaccine candidate induces humoral response and protects mice against lethal infection. Hum. Vaccin. Immunother. 12, 2327–2333. doi: 10.1080/21645515.2016.1183857
Le Tallec, D., Doucet, D., Elouahabi, A., Harvengt, P., Deschuyteneer, M., and Deschamps, M. (2009). CervarixTM, the GSK HPV-16/HPV-18 AS04-adjuvanted cervical cancer vaccine, demonstrates stability upon long-term storage and under simulated cold chain break conditions. Hum. Vaccin. 5, 467–474.
Leite, M. L., Sampaio, K. B., Costa, F. F., Franco, O. L., Dias, S. C., and Cunha, N. B. (2019). Molecular farming of antimicrobial peptides: available platforms and strategies for improving protein biosynthesis using modified virus vectors. An. Acad. Bras. Cienc. 91 (Suppl. 1):e20180124. doi: 10.1590/0001-3765201820180124
Leroux-Roels, G., Maes, C., Clement, F., van Engelenburg, F., van den Dobbelsteen, M., Adler, M., et al. (2013). Randomized phase I: safety, immunogenicity and mucosal antiviral activity in young healthy women vaccinated with HIV-1 Gp41 P1 peptide on virosomes. PLoS One 8:e55438. doi: 10.1371/journal.pone.0055438
Lewandowski, D. J., and Dawson, W. O. (2000). Functions of the 126-and 183-kDa proteins of tobacco mosaic virus. Virology 271, 90–98. doi: 10.1006/viro.2000.0313
Lico, C., Mancini, C., Italiani, P., Betti, C., Boraschi, D., Benvenuto, E., et al. (2009). Plant-produced potato virus X chimeric particles displaying an influenza virus-derived peptide activate specific CD8+ T cells in mice. Vaccine 27, 5069–5076. doi: 10.1016/j.vaccine.2009.06.045
Lindbo, J. A. (2007). TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 145, 1232–1240.
Liu, L., Cañizares, M. C., Monger, W., Perrin, Y., Tsakiris, E., Porta, C., et al. (2005). Cowpea mosaic virus-based systems for the production of antigens and antibodies in plants. Vaccine 23, 1788–1792.
Lockney, D. M., Guenther, R. N., Loo, L., Overton, W., Antonelli, R., Clark, J., et al. (2010). The Red clover necrotic mosaic virus capsid as a multifunctional cell targeting plant viral nanoparticle. Bioconjug. Chem. 22, 67–73. doi: 10.1021/bc100361z
Loh, H.-S., Green, B. J., and Yusibov, V. (2017). Using transgenic plants and modified plant viruses for the development of treatments for human diseases. Curr. Opin. Virol. 26, 81–89. doi: 10.1016/j.coviro.2017.07.019
Lu, Y., Cesar, E., Nadala, B. Jr., Brock, J. A., and Loh, P. C. (1991). A new virus isolate from infectious hypodermal and hematopoietic necrosis virus (IHHNV)-infected penaeid shrimps. J. Virol. Methods 31, 189–195.
Ma, J. K., Drake, P. M., and Christou, P. (2003). The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 4, 794–805.
Maclean, J., Koekemoer, M., Olivier, A. J., Stewart, D., Hitzeroth, I. I., Rademacher, T., et al. (2007). Optimization of human papillomavirus type 16 (HPV-16) L1 expression in plants: comparison of the suitability of different HPV-16 L1 gene variants and different cell-compartment localization. J. Gen. Virol. 88(Pt 5), 1460–1469. doi: 10.1099/vir.0.82718-0
Majid, A. M., Ezelle, H., Shah, S., and Barber, G. N. (2006). Evaluating replication-defective vesicular stomatitis virus as a vaccine vehicle. J. Virol. 80, 6993–7008.
Marillonnet, S., Giritch, A., Gils, M., Kandzia, R., Klimyuk, V., and Gleba, Y. (2004). In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 101, 6852–6857.
Marillonnet, S., Thoeringer, C., Kandzia, R., Klimyuk, V., and Gleba, Y. (2005). Systemic Agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 23, 718–723.
McCormick, A. A., Reinl, S. J., Cameron, T. I., Vojdani, F., Fronefield, M., Levy, R., et al. (2003). Individualized human scFv vaccines produced in plants: humoral anti-idiotype responses in vaccinated mice confirm relevance to the tumor Ig. J. Immunol. Methods 278, 95–104.
Mclain, L., Porta, C., Lomonossoff, G. P., Durrani, Z., and Dimmock, N. J. (1995). Human-immunodeficiency-virus type-1-neutralizing antibodies raised to a glycoprotein-41 peptide expressed on the surface of a plant-virus. AIDS Res. Hum. Retroviruses 11, 327–334. doi: 10.1089/aid.1995.11.327
Mebatsion, T., Weiland, F., and Conzelmann, K.-K. (1999). Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73, 242–250.
Mollentze, N., Biek, R., and Streicker, D. G. (2014). The role of viral evolution in rabies host shifts and emergence. Curr. Opin. Virol. 8, 68–72. doi: 10.1016/j.coviro.2014.07.004
Moore, L., Hamorsky, K., and Matoba, N. (2016). Production of recombinant cholera toxin b subunit in Nicotiana benthamiana using GENEWARE® tobacco mosaic virus vector. Methods Protoc. 1385, 129–137. doi: 10.1007/978-1-4939-3289-4_9
Moyer, S. A., and Banerjee, A. K. (1975). Messenger RNA species synthesized in vitro by the virion-associated RNA polymerase of vesicular stomatitis virus. Cell 4, 37–43.
Namba, K., Pattanayek, R., and Stubbs, G. (1989). Visualization of protein-nucleic acid interactions in a virus - refined structure of intact tobacco mosaic-virus at 2.9-a resolution by X-ray fiber diffraction. J. Mol. Biol. 208, 307–325. doi: 10.1016/0022-2836(89)90391-4
Namba, K., and Stubbs, G. (1986). Structure of tobacco mosaic-virus at 3.6-a resolution - implications for assembly. Science 231, 1401–1406. doi: 10.1126/science.3952490
Niehl, A., Appaix, F., Bosca, S., van der Sanden, B., Nicoud, J. F., Bolze, F., et al. (2015). Fluorescent tobacco mosaic virus-derived bio-nanoparticles for intravital two-photon imaging. Front. Plant Sci. 6:1244. doi: 10.3389/fpls.2015.01244
Nuzzaci, M., Piazzolla, G., Vitti, A., Lapelosa, M., Tortorella, C., Stella, I., et al. (2007). Cucumber mosaic virus as a presentation system for a double hepatitis C virus-derived epitope. Arch. Virol. 152, 915–928.
Osman, T. A. A., and Buck, K. W. (2003). Identification of a region of the tobacco mosaic virus 126-and 183-kilodalton replication proteins which binds specifically to the viral 3 ’-Terminal tRNA-like structure. J. Virol. 77, 8669–8675. doi: 10.1128/Jvi.77.16.8669-8675.2003
Osman, T. A. M., and Buck, K. W. (1996). Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70, 6227–6234.
Parker, L., Kendall, A., and Stubbs, G. (2002). Surface features of potato virus X from fiber diffraction. Virology 300, 291–295. doi: 10.1006/viro.2002.1483
Pattnaik, A. K., and Wertz, G. W. (1990). Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J. Virol. 64, 2948–2957.
Pattnaik, A. K., and Wertz, G. W. (1991). Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc. Natl. Acad. Sci. U.S.A. 88, 1379–1383.
Pejawar-Gaddy, S., Rajawat, Y., Hilioti, Z., Xue, J., Gaddy, D. F., Finn, O. J., et al. (2010). Generation of a tumor vaccine candidate based on conjugation of a MUC1 peptide to polyionic papillomavirus virus-like particles. Cancer Immunol. Immunother. 59, 1685–1696. doi: 10.1007/s00262-010-0895-0
Petukhova, N. V., Gasanova, T. V., Stepanova, L. A., Rusova, O. A., Potapchuk, M. V., Korotkov, A. V., et al. (2013). Immunogenicity and protective efficacy of candidate universal influenza A nanovaccines produced in plants by tobacco mosaic virus-based vectors. Curr. Pharm. Des. 19, 5587–5600. doi: 10.2174/13816128113199990337
Peyret, H., and Lomonossoff, G. P. (2013). The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83, 51–58. doi: 10.1007/s11103-013-0036-1
Phelps, J. P., Dang, N., and Rasochova, L. (2007). Inactivation and purification of cowpea mosaic virus-like particles displaying peptide antigens from Bacillus anthracis. J. Virol. Methods 141, 146–153.
Porta, C., Spall, V. E., Loveland, J., Johnson, J. E., Barker, P. J., and Lomonossoff, G. P. (1994). Development of cowpea mosaic virus as a high-yielding system for the presentation of foreign peptides. Virology 202, 949–955.
Pushko, P., Tumpey, T. M., Bu, F., Knell, J., Robinson, R., and Smith, G. (2005). Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine 23, 5751–5759.
Ratner, M. (2010). Pfizer stakes a claim in plant cell-made biopharmaceuticals. Nat. Biotechnol. 28, 107–108.
Redinbaugh, M., Seifers, D., Meulia, T., Abt, J., Anderson, R., Styer, W., et al. (2002). Maize fine streak virus, a new leafhopper-transmitted rhabdovirus. Phytopathology 92, 1167–1174. doi: 10.1094/PHYTO.2002.92.11.1167
Regules, J. A., Beigel, J. H., Paolino, K. M., Voell, J., Castellano, A. R., Hu, Z., et al. (2017). A recombinant vesicular stomatitis virus Ebola vaccine. N. Engl. J. Med. 376, 330–341. doi: 10.1056/NEJMoa1414216
Ren, Y., Wong, S. M., and Lim, L.-Y. (2007). Folic acid-conjugated protein cages of a plant virus: a novel delivery platform for doxorubicin. Bioconjug. Chem. 18, 836–843.
Rioux, G., Babin, C., Majeau, N., and Leclerc, D. (2012). Engineering of papaya mosaic virus (PapMV) nanoparticles through fusion of the HA11 peptide to several putative surface-exposed sites. PLoS One 7:e31925. doi: 10.1371/journal.pone.0031925
Rioux, G., Mathieu, C., Russell, A., Bolduc, M., Laliberte-Gagne, M. E., Savard, P., et al. (2014). PapMV nanoparticles improve mucosal immune responses to the trivalent inactivated flu vaccine. J. Nanobiotechnology 12:19. doi: 10.1186/1477-3155-12-19
Roberts, A., Buonocore, L., Price, R., Forman, J., and Rose, J. K. (1999). Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 73, 3723–3732.
Roder, J., Fischer, R., and Commandeur, U. (2017). Adoption of the 2A ribosomal skip principle to tobacco mosaic virus for peptide display. Front. Plant Sci. 8:1125. doi: 10.3389/fpls.2017.01125
Roldão, A., Mellado, M. C. M., Lima, J., Carrondo, M. J., Alves, P. M., and Oliveira, R. (2012). On the effect of thermodynamic equilibrium on the assembly efficiency of complex multi-layered virus-like particles (VLP): the case of rotavirus VLP. PLoS Comput. Biol. 8:e1002367. doi: 10.1371/journal.pcbi.1002367
Rose, N. F., Marx, P. A., Luckay, A., Nixon, D. F., Moretto, W. J., Donahoe, S. M., et al. (2001). An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106, 539–549.
Ryder, A. B., Buonocore, L., Vogel, L., Nachbagauer, R., Krammer, F., and Rose, J. K. (2015). A viable recombinant rhabdovirus lacking its glycoprotein gene and expressing influenza virus hemagglutinin and neuraminidase is a potent influenza vaccine. J. Virol. 89, 2820–2830. doi: 10.1128/JVI.03246-14
Sabalza, M., Christou, P., and Capell, T. (2014). Recombinant plant-derived pharmaceutical proteins: current technical and economic bottlenecks. Biotechnol. Lett. 36, 2367–2379. doi: 10.1007/s10529-014-1621-3
Safronetz, D., Mire, C., Rosenke, K., Feldmann, F., Haddock, E., Geisbert, T., et al. (2015). A recombinant vesicular stomatitis virus-based Lassa fever vaccine protects guinea pigs and macaques against challenge with geographically and genetically distinct Lassa viruses. PLoS Negl. Trop. Dis. 9:e0003736. doi: 10.1371/journal.pntd.0003736
Sahithi, K. D., Nancy, P. A., Vardhan, G. P. V., Kumanan, K., Vijayarani, K., and Hema, M. (2019). Detection of infectious bursal disease virus (IBDV) antibodies using chimeric plant virus-like particles. Vet. Microbiol. 229, 20–27. doi: 10.1016/j.vetmic.2018.12.008
Sainsbury, F., Lavoie, P. O., D’Aoust, M. A., Vézina, L. P., and Lomonossoff, G. P. (2008). Expression of multiple proteins using full-length and deleted versions of cowpea mosaic virus RNA-2. Plant Biotechnol. J. 6, 82–92.
Sainsbury, F., and Lomonossoff, G. P. (2008). Extremely high-level and rapid transient protein production in plants without the use of viral replication. Plant Physiol. 148, 1212–1218. doi: 10.1104/pp.108.126284
Saito, T., Yamanaka, K., and Okada, Y. (1990). Long-distance movement and viral assembly of tobacco mosaic-virus mutants. Virology 176, 329–336. doi: 10.1016/0042-6822(90)90002-9
Salazar, L., Müller, G., Querci, M., Zapata, J., and Owens, R. (2000). Potato yellow vein virus: its host range, distribution in South America and identification as a crinivirus transmitted by Trialeurodes vaporariorum. Ann. Appl. Biol. 137, 7–19.
Sánchez-Sánchez, L., Cadena-Nava, R. D., Palomares, L. A., Ruiz-Garcia, J., Koay, M. S., Cornelissen, J. J., et al. (2014). Chemotherapy pro-drug activation by biocatalytic virus-like nanoparticles containing cytochrome P450. Enzyme Microb. Technol. 60, 24–31. doi: 10.1016/j.enzmictec.2014.04.003
Schnell, M. J., Buonocore, L., Boritz, E., Ghosh, H. P., Chernish, R., and Rose, J. K. (1998). Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17, 1289–1296.
Schnell, M. J., Buonocore, L., Kretzschmar, E., Johnson, E., and Rose, J. K. (1996a). Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc. Natl. Acad. Sci. U.S.A. 93, 11359–11365.
Schnell, M. J., Buonocore, L., Whitt, M. A., and Rose, J. K. (1996b). The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 70, 2318–2323.
Schnell, M. J., Johnson, J. E., Buonocore, L., and Rose, J. K. (1997). Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell 90, 849–857.
Schnell, M. J., Mebatsion, T., and Conzelmann, K.-K. (1994). Infectious rabies viruses from cloned cDNA. EMBO J. 13, 4195–4203.
Scholthof, H. B., Scholthof, K.-B. G., and Jackson, A. O. (1996). Plant virus gene vectors for transient expression of foreign proteins in plants. Annu. Rev. Phytopathol. 34, 299–323.
Séveno, M., Bardor, M., Paccalet, T., Gomord, V., Lerouge, P., and Faye, L. (2004). Glycoprotein sialylation in plants? Nat. Biotechnol. 22, 1351–1352.
Shivprasad, S., Pogue, G. P., Lewandowski, D. J., Hidalgo, J., Donson, J., Grill, L. K., et al. (1999). Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255, 312–323.
Smolenska, L., Roberts, I. M., Learmonth, D., Porter, A. J., Harris, W. J., Wilson, T. M. A., et al. (1998). Production of a functional single chain antibody attached to the surface of a plant virus. FEBS Lett. 441, 379–382. doi: 10.1016/S0014-5793(98)01586-5
Soulie, J., Devillier, P., Santarelli, J., Goudeau, A., Vermeulen, P., Guellier, M., et al. (1991). Immunogenicity and safety in newborns of a new recombinant hepatitis B vaccine containing the S and pre-S2 antigens. Vaccine 9, 545–548.
Speir, J. A., Munshi, S., Wang, G., Baker, T. S., and Johnson, J. E. (1995). Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3, 63–78.
Steinmetz, N. F., Mertens, M. E., Taurog, R. E., Johnson, J. E., Commandeur, U., Fischer, R., et al. (2009). Potato virus X as a novel platform for potential biomedical applications. Nano Lett. 10, 305–312.
Stillman, E. A., and Whitt, M. A. (1997). Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J. Virol. 71, 2127–2137.
Stubbs, L., and Grogan, R. (1963). Necrotic yellows: a newly recognized virus disease of lettuce. Crop Pasture Sci. 14, 439–459.
Sugiyama, Y., Hamamoto, H., Takemoto, S., Watanabe, Y., and Okada, Y. (1995). Systemic production of foreign peptides on the particle surface of tobacco mosaic-virus. FEBS Lett. 359, 247–250. doi: 10.1016/0014-5793(95)00054-D
Sun, K., Zhou, X., Lin, W., Zhou, X., Jackson, A. O., and Li, Z. (2018). Matrix-glycoprotein interactions required for budding of a plant nucleorhabdovirus and induction of inner nuclear membrane invagination. Mol. Plant Pathol. 19, 2288–2301. doi: 10.1111/mpp.12699
Sylvester, E. S., and Richardson, J. (1992). Aphid-borne rhabdoviruses–relationships with their vectors. Adv. Dis. Vector Res. 9, 313–341.
Szarewski, A. (2010). HPV vaccine: cervarix. Expert Opin. Biol. Ther. 10, 477–487. doi: 10.1517/14712591003601944
Takamatsu, N., Ishikawa, M., Meshi, T., and Okada, Y. (1987). Expression of bacterial chloramphenicol acetyltransferase gene in tobacco plants mediated by TMV-RNA. EMBO J. 6, 307–311.
Takamatsu, N., Watanabe, Y., Meshi, T., and Okada, Y. (1990). Mutational analysis of the pseudoknot region in the 3’ noncoding region of tobacco mosaic-virus RNA. J. Virol. 64, 3686–3693.
Tesh, R. B., Chaniotis, B. N., and Johnson, K. M. J. S. (1972). Vesicular stomatitis virus (Indiana serotype): transovarial transmission by phlebotomine sandlies. Science 175, 1477–1479.
Tezuka, K., Okuma, K., Kuramitsu, M., Matsuoka, S., Tanaka, R., Tanaka, Y., et al. (2018). Control of human T-cell leukemia virus type 1 (HTLV-1) infection by eliminating envelope protein-positive cells with recombinant vesicular stomatitis viruses encoding HTLV-1 primary receptor. J. Virol. 92:e01885-17. doi: 10.1128/JVI.01885-17
Thanavala, Y., Yang, Y., Lyons, P., Mason, H., and Arntzen, C. (1995). Immunogenicity of transgenic plant-derived hepatitis B surface antigen. Proc. Natl. Acad. Sci. U.S.A. 92, 3358–3361.
Therien, A., Bedard, M., Carignan, D., Rioux, G., Gauthier-Landry, L., Laliberte-Gagne, M. E., et al. (2017). A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. J. Nanobiotechnology 15:54. doi: 10.1186/s12951-017-0289-y
Tignor, G., Murphy, F., Clark, H., Shope, R., Madore, P., Bauer, S., et al. (1977). Duvenhage virus: morphological, biochemical, histopathological and antigenic relationships to the rabies serogroup. J. Gen. Virol. 37, 595–611.
Tomljenovic, L., and Shaw, C. A. (2012). Too fast or not too fast: the FDA’s approval of Merck’s HPV vaccine Gardasil. J. Law Med. Ethics 40, 673–681. doi: 10.1111/j.1748-720X.2012.00698.x
Toriyama, S., and Peters, D. (1980). In vitro synthesis of RNA by dissociated lettuce necrotic yellows virus particles. J. Gen. Virol. 50, 125–134.
Tremblay, A., Beauchemin, C., Séguin, A., and Laliberté, J.-F. (2007). Reactivation of an integrated disabled viral vector using a Cre-loxP recombination system in Arabidopsis thaliana. Transgenic Res. 16, 213–222.
Tseng, R. J., Tsai, C. L., Ma, L. P., and Ouyang, J. Y. (2006). Digital memory device based on tobacco mosaic virus conjugated with nanoparticles. Nat. Nanotechnol. 1, 72–77. doi: 10.1038/nnano.2006.55
Tusé, D., Tu, T., and McDonald, K. A. (2014). Manufacturing economics of plant-made biologics: case studies in therapeutic and industrial enzymes. Biomed Res. Int. 2014:256135. doi: 10.1155/2014/256135
Uhde-Holzem, K., Schlösser, V., Viazov, S., Fischer, R., and Commandeur, U. (2010). Immunogenic properties of chimeric potato virus X particles displaying the hepatitis C virus hypervariable region I peptide R9. J. Virol. Methods 166, 12–20. doi: 10.1016/j.jviromet.2010.01.017
Urbiola, C., Santer, F. R., Petersson, M., van der Pluijm, G., Horninger, W., Erlmann, P., et al. (2018). Oncolytic activity of the rhabdovirus VSV-GP against prostate cancer. Int. J. Cancer doi: 10.1002/ijc.31556 [Epub ahead of print].
Usha, R., Rohll, J. B., Spall, V. E., Shanks, M., Maule, A. J., Johnson, J. E., et al. (1993). Expression of an animal virus antigenic site on the surface of a plant virus particle. Virology 197, 366–374.
Venters, C., Graham, W., and Cassidy, W. (2004). Recombivax-HB: perspectives past, present and future. Expert Rev. Vaccines 3, 119–129.
Venuti, A., Curzio, G., Mariani, L., and Paolini, F. (2015). Immunotherapy of HPV-associated cancer: DNA/plant-derived vaccines and new orthotopic mouse models. Cancer Immunol. Immunother. 64, 1329–1338. doi: 10.1007/s00262-015-1734-0
Vermij, P., and Waltz, E. (2006). USDA approves the first plant-based vaccine. Nat. Biotechnol. 24, 233–234.
Vitti, A., Piazzolla, G., Condelli, V., Nuzzaci, M., Lanorte, M., Boscia, D., et al. (2010). Cucumber mosaic virus as the expression system for a potential vaccine against Alzheimer’s disease. J. Virol. Methods 169, 332–340. doi: 10.1016/j.jviromet.2010.07.039
Walker, P. J., Dietzgen, R. G., Joubert, D. A., and Blasdell, K. R. (2011). Rhabdovirus accessory genes. Virus Res. 162, 110–125. doi: 10.1016/j.virusres.2011.09.004
Walker, P. J., Firth, C., Widen, S. G., Blasdell, K. R., Guzman, H., Wood, T. G., et al. (2015). Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog. 11:e1004664. doi: 10.1371/journal.ppat.1004664
Wang, Q., Chen, X., Luo, Z., and Fang, R. (1999). Sequence analysis of leader and trailer regions of rice yellow stunt rhabdovirus and characterization of their in vivo transcripts. Sci. China Ser. C Life Sci. 42, 50–56. doi: 10.1007/BF02881747
Wang, Q., Ma, X., Qian, S., Zhou, X., Sun, K., Chen, X., et al. (2015). Rescue of a plant negative-strand RNA virus from cloned cDNA: insights into enveloped plant virus movement and morphogenesis. PLoS Pathog. 11:e1005223. doi: 10.1371/journal.ppat.1005223
Werner, S., Marillonnet, S., Hause, G., Klimyuk, V., and Gleba, Y. (2006). Immunoabsorbent nanoparticles based on a tobamovirus displaying protein A. Proc. Nat. Acad. Sci. U.S.A. 103, 17678–17683. doi: 10.1073/pnas.0608869103
Wertz, G. W., Davis, N. L., and Patton, J. (1987). “The role of proteins in vesicular stomatitis virus RNA replication,” in The Rhabdoviruses, ed. R. R. Wagner (Boston, MA: Springer), 271–296.
Wetzel, T., Dietzgen, R., and Dale, J. (1994). Genomic organization of lettuce necrotic yellows rhabdovirus. Virology 200, 401–412.
Whelan, S., Ball, L. A., Barr, J. N., and Wertz, G. (1995). Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. U.S.A. 92, 8388–8392.
Whitfield, A. E., Huot, O. B., Martin, K. M., Kondo, H., and Dietzgen, R. G. (2018). Plant rhabdoviruses—their origins and vector interactions. Curr. Opin. Virol. 33, 198–207. doi: 10.1016/j.coviro.2018.11.002
Whitt, M., Chong, L., and Rose, J. (1989). Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J. Virol. 63, 3569–3578.
Wiedermann, U., Wiltschke, C., Jasinska, J., Kundi, M., Zurbriggen, R., Garner-Spitzer, E., et al. (2010). A virosomal formulated Her-2/neu multi-peptide vaccine induces Her-2/neu-specific immune responses in patients with metastatic breast cancer: a phase I study. Breast Cancer Res. Treat. 119, 673–683.
Wilken, L. R., and Nikolov, Z. L. (2012). Recovery and purification of plant-made recombinant proteins. Biotechnol. Adv. 30, 419–433. doi: 10.1016/j.biotechadv.2011.07.020
Wolanski, B., Francki, R., and Chambers, T. (1967). Structure of lettuce necrotic yellows virus: I. Electron microscopy of negatively stained preparations. Virology 33, 287–296.
Yamshchikov, G. V., Ritter, G. D., Vey, M., and Compans, R. W. (1995). Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology 214, 50–58. doi: 10.1006/viro.1995.9955
Yang, C.-D., Liao, J.-T., Lai, C.-Y., Jong, M.-H., Liang, C.-M., Lin, Y.-L., et al. (2007). Induction of protective immunity in swine by recombinant bamboo mosaic virus expressing foot-and-mouth disease virus epitopes. BMC Biotechnol. 7:62. doi: 10.1186/1472-6750-7-62
Yang, J., Hooper, D. C., Wunner, W. H., Koprowski, H., Dietzschold, B., and Fu, Z. F. (1998). The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology 242, 107–117.
Yang, J., Koprowski, H., Dietzschold, B., and Fu, Z. F. (1999). Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J. Virol. 73, 1661–1664.
Yang, S. Q., Wang, T., Bohon, J., Gagne, M. E. L., Bolduc, M., Leclerc, D., et al. (2012). Crystal structure of the coat protein of the flexible filamentous papaya mosaic virus. J. Mol. Biol. 422, 263–273. doi: 10.1016/j.jmb.2012.05.032
Yi, H. M., Nisar, S., Lee, S. Y., Powers, M. A., Bentley, W. E., Payne, G. F., et al. (2005). Patterned assembly of genetically modified viral nanotemplates via nucleic acid hybridization. Nano Lett. 5, 1931–1936. doi: 10.1021/nl051254r
Yusibov, V., Mett, V., Mett, V., Davidson, C., Musiychuk, K., Gilliam, S., et al. (2005). Peptide-based candidate vaccine against respiratory syncytial virus. Vaccine 23, 2261–2265.
Yusibov, V., Modelska, A., Steplewski, K., Agadjanyan, M., Weiner, D., Hooper, D. C., et al. (1997). Antigens produced in plants by infection with chimeric plant viruses immunize against rabies virus and HIV-1. Proc. Natl. Acad. Sci. U.S.A. 94, 5784–5788.
Yusibov, V., Streatfield, S. J., and Kushnir, N. (2011). Clinical development of plant-produced recombinant pharmaceuticals: vaccines, antibodies and beyond. Hum. Vaccin. 7, 313–321.
Zandi, R., Reguera, D., Bruinsma, R. F., Gelbart, W. M., and Rudnick, J. (2004). Origin of icosahedral symmetry in viruses. Proc. Nat. Acad. Sci. U.S.A. 101, 15556–15560. doi: 10.1073/pnas.0405844101
Zhang, X., Green, T. J., Tsao, J., Qiu, S., and Luo, M. (2008). Role of intermolecular interactions of vesicular stomatitis virus nucleoprotein in RNA encapsidation. J. Virol. 82, 674–682.
Zhang, X., Mao, G., and van den Pol, A. N. J. V. (2018). Chikungunya-vesicular stomatitis chimeric virus targets and eliminates brain tumors. Virology 522, 244–259. doi: 10.1016/j.virol.2018.06.018
Zhang, X., and Mason, H. (2006). Bean yellow dwarf virus replicons for high-level transgene expression in transgenic plants and cell cultures. Biotechnol. Bioeng. 93, 271–279.
Zlotnick, A. (1994). To build a virus capsid: an equilibrium model of the self assembly of polyhedral protein complexes. J. Mol. Biol. 241, 59–67.
Keywords: plant molecular pharming, non-enveloped viruses, plant rhabdoviruses, recombinant vaccines, virus like particles, biotechnology
Citation: Ibrahim A, Odon V and Kormelink R (2019) Plant Viruses in Plant Molecular Pharming: Toward the Use of Enveloped Viruses. Front. Plant Sci. 10:803. doi: 10.3389/fpls.2019.00803
Received: 28 February 2019; Accepted: 04 June 2019;
Published: 19 June 2019.
Edited by:
Helene Sanfacon, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
George Peter Lomonossoff, John Innes Centre (JIC), United KingdomCopyright © 2019 Ibrahim, Odon and Kormelink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard Kormelink, cmljaGFyZC5rb3JtZWxpbmtAd3VyLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.