- 1Laboratory of Plant Nutrition and Biology, Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Plant Nutrition, College of Resources and Environmental Sciences, China Agricultural University, Beijing, China
- 3Jiujiang Academy of Agricultural Sciences, Jiujiang, China
Alkaline stress (AS) is one of the abiotic stressful factors limiting plant’s growth and development. Inorganic pyrophosphatase is usually involved in a variety of biological processes in plant in response to the abiotic stresses. Here, to clarify the responsive regulation of inorganic pyrophosphatase in rice under AS, the mutagenesis of the OsPPa6 gene encoding an inorganic pyrophosphatase in rice cv. Kitaake (Oryza sativa L. ssp. japonica) was performed by the CRISPR/Cas9 system. Two homozygous independent mutants with cas9-free were obtained by continuously screening. qPCR reveals that the OsPPa6 gene was significantly induced by AS, and the mutagenesis of the OsPPa6 gene apparently delayed rice’s growth and development, especially under AS. Measurements demonstrate that the contents of pyrophosphate in the mutants were higher than those in the wild type under AS, however, the accumulation of inorganic phosphate, ATP, chlorophyll, sucrose, and starch in the mutants were decreased significantly, and the mutagenesis of the OsPPa6 gene remarkably lowered the net photosynthetic rate of rice mutants, thus reducing the contents of soluble sugar and proline, but remarkably increasing MDA, osmotic potentials and Na+/K+ ratio in the mutants under AS. Metabonomics measurement shows that the mutants obviously down-regulated the accumulation of phosphorylcholine, choline, anthranilic acid, apigenin, coniferol and dodecanoic acid, but up-regulated the accumulation of L-valine, alpha-ketoglutarate, phenylpyruvate and L-phenylalanine under AS. This study suggests that the OsPPa6 gene is an important osmotic regulatory factor in rice, and the gene-editing of CRISPR/Cas9-guided is an effective method evaluating the responsive regulation of the stress-induced gene, and simultaneously provides a scientific support for the application of the gene encoding a soluble inorganic pyrophosphatase in molecular breeding.
Introduction
Soils with highly salinity-alkalinity are widely distributed in the world, and account for 10% of the arable land. Statistics shows that saline-alkaline soils of nearly 8.3 million ha are universally used for rice planting (Haefele et al., 2014). Alkaline stress (AS) usually hinders plant’s growth and development, leading to a decrease in biomass by triggering a series of physiological damage to the plants, and these damages are mainly demonstrated by affecting photosynthesis, respiration, enzymatic activity and accumulation profile of ions in plants (Yang et al., 2008; Bu et al., 2012). Particularly, AS not only alters the distribution of Cl-, NO3-, and H2PO4- ions, but also exerts an influence on the balance of Na+ and K+ ions in plant cells (Chen et al., 2011). Study shows that most of the translocation and distribution of ions are closely associated with the H+-translocating inorganic pyrophosphatase (H+-PPase) widely presenting in plants (Davies et al., 1992; Tsai et al., 2014). Therefore, to better understand whether the soluble inorganic pyrophosphatase (sPPase) has a similar regulatory role in the transport and translocation of ions to the H+-PPase, evaluating the genes encoding sPPase is imperative to better exploit these genes in cultivating alkaline tolerant crop.
In plant kingdom, two typical inorganic pyrophosphatase (PPases), the H+-PPase and the sPPase have been identified to play important regulatory roles in response to abiotic stresses (Ferjani et al., 2011; Gutiérrez-Luna et al., 2018). Usually, the H+-PPase catalyzes a coupled reaction of PPi hydrolysis and active proton transport across membranes (Segami et al., 2014). Previous studies have found that the genes encoding the H+-PPase have positive responses to the abiotic stresses. For example, the transcripts of the ZmVP1 gene in Zoysia matrella is significantly induced by salinity, drought and cold, and the overexpression of the ZmVP1 gene in Arabidopsis thaliana reveals strong salt tolerance (Chen et al., 2015). Similarly, the KfVP1 gene in Kalidium foliatum is induced by salt and drought, and the transgenic A. thaliana overexpressing the KfVP1 gene enhances the salt and drought tolerance (Yao et al., 2012). Wang et al. (2016) reported that the ScHP1 gene in rye is induced by cold, salt, drought and phosphate (P) starvation. Like the genes encoding the H+-PPase, the genes encoding the sPPase were also functionally explored in diverse plants. Study showed that both tobacco and potato overexpressing the gene encoding a sPPase of E. coli delays plant growth and demonstrated thicker leaves with discolored and small size compared to the wild type (Sonnewald, 1992). George et al. (2010) found that silencing the psPPase gene encoding a plastidial soluble inorganic pyrophosphatase in tobacco inhibits the tobacco growth, and alters the accumulation profiles of starch and chlorophyll, and affects carotenoid biosynthesis in tobacco leaves, thus limiting photosynthesis of tobacco and leading to a moderate wilting phenotype under drought stress. Hernández-Domíguez et al. (2012) confirmed that the transcripts of the PvPPa1 gene encoding a sPPase are increased in the common bean leaves, and the transcripts of the PvPPa4 are highly accumulated in both the oldest and the youngest leaves under P starvation. Moreover, May et al. (2011) found that the AtPPsPase1 gene encoding a pyrophosphatase in the HAD superfamily reveals an optimal catalytic activity under alkalinity with higher pH, and that the AtPPsPase1 gene is strongly induced and up-regulated by P starvation (Müller et al., 2007), indicating a direct participation of the PPases in phosphate metabolism in plant. While glutamine synthetase (GS2) in plant participates in nitrogen metabolism by in charge of ammonium (NH4+) fixation through a pathway of ATP-dependent (Kaminski et al., 2015). Hoshida et al. (2000) found that overexpressing the GS2 gene demonstrates an enhanced increase in photorespiration capacity in the transgenic rice under salt stress. These reports show that alteration of photosynthesis system seems to have a closely association with the metabolism of nitrogen in plant (Fuentes et al., 2001; Oliveira et al., 2002). Therefore, investigating the transcript profile of the OsGS2 gene under AS is beneficial to better understanding whether the physiological changes of the sPPase-mediated affect the GS2 activity.
Currently, a total of seven of putative isoforms of sPPase in rice are determined to exhibit similar protein structures to the sPPases in Arabidopsis, and six isoforms of sPPases share similarity in the predicted protein sequences, only the sPPase (Os02g0768600) protein demonstrates a lower similarity in sequences of amino acids, but has higher similarity with the AtPPa6 in Arabidopsis. Referring to the nomenclature of the gene encoding sPPase in Arabidopsis (Schulze et al., 2004), the sPPase (Os02g0768600) in rice was named as the OsPPa6. Multiple alignment shows that the OsPPa6 gene (accession: AK059725) in rice shares highly homology with the ThPP1 gene (accession: KC250018) in Thellungiella halophila, and a previous report showed that the ThPP1 gene, encodes a sPPase in T. halophila, is induced by AS, and obviously confers an enhanced alkaline tolerance in the transgenic rice (He et al., 2017).
At present, the genes encoding the sPPase were mainly characterized by overexpressing or silencing the target gene of RNAi-mediated in the plant. However, the regulatory role of the gene encoding sPPase in rice is not complete clear under AS. Particularly, the mutagenesis of the gene encoding the sPPase has not been reported yet. In this study, we used the CRISPR/Cas9 gene-editing system performed the mutagenesis of the OsPPa6 gene in rice Oryza sativa japonica L. cv. Kitaake, and obtained two independent mutants of T3 generation with base mutation or insertion. To identify the regulatory role of the OsPPa6 gene in response to AS, both the rice mutants and the wild type were subjected to AS, and investigated by profiling the physiological changes and the metabonomics-based metabolites differences. We are expected that manipulating the CRISPR/Cas9 gene-editing system not only is an effective pathway in identifying the functional genes, but also provides a scientific evidence for the application of the alkali-induced OsPPa6 gene.
Materials and Methods
Vector Construction and Transformants Generation
A specific target fragment of 20-bp at the upstream of the PAM (NGG) motif in the cDNA of the OsPPa6 gene (accession: AK059725) was identified by the Cas-OFFinder1 to avoid occurrence of the off-target effect. A pair of specific primers for the OsPPa6 gene-editing were designed by the online guide design tool2, and connected by the two adapters, 5′-GGCA-3′ at the 5′ terminus and 5′-AAAC-3′ at the 3′ terminus, respectively (sgRNA-F/R, Supplementary Table S1). The annealing reaction was performed in a 20-μL of reaction solution containing 1 μL of annealing buffer, 4.5 μL of 100 μM of Editing-F/R, respectively, and 10 μL of ddH2O to form sgRNA fragments with complementary base pairing. The reaction procedure was performed by culturing at 95°C for 5 min, then following a gradient culture from 95°C to 70°C, which is decreased by 1°C in 1 min, and stored at 10°C. The pCXUN-U3 vector used for gene-editing was digested with AarI at 37°C for 3–6 h, and the generated vector fragments were recovered and sequenced. The sgRNA fragments were ligated with the vector fragments in a reaction solution containing 1 μL of T4 ligase buffer, 2 μL of pCXUN-U3 vector, 3 μL of 10 μM of sgRNA, 0.5 μL of T4 ligase (5U μL-1) and 3.5 μL of ddH2O, then thoroughly mixed and incubated at 25°C over 10 min, and the fusion vector with sgRNA was transformed into the E. coli DH5α, and the generating transformants were cultured in a culture solution of 500 μL LB medium lacking antibiotics with 200 rpm at 37°C for 1 h. Then, total 100 μL of the culture medium were evenly spread on the LB plate containing Kan and cultured at 37°C under darkness overnight. One independent clone was chosen to further culture and detected by PCR using the universal primers U3-F/R (Supplementary Table S1). The plasmid DNA of transformants was extracted, and subsequently transferred into the A. tumefaciens competent cells of EHA105, and the generated transformants were cultured in a culture solution of 500 μL liquid medium lacking antibiotics with 230 rpm at 28°C overnight. A total of 100 μL of the culture solution was evenly spread on the LB plate containing Kan and rifampicin and cultured at 28°C under darkness overnight. Finally, a single plaque was picked up and cultured by the same condition, the generating bacterial culture solution was used for PCR detection using the specific primers OsPPa6-F/R (Supplementary Table S1), and the PCR products were sequenced to determine the positive strains.
Transformation and Screening of Mutant Plants
The transformants of A. tumefaciens EHA105 strains were transferred into immature embryo calli of rice cv. Kitaake (O. sativa, L. japonica). After two rounds of screening on the N6 solid medium (Chu et al., 1975) containing 2,4-D (2,4-Dichlorophenoxyacetic acid) and hygromycin (Nishimura et al., 2006), the infected calli were cultured on the N6 solid medium lacking 2,4-D under darkness of 8 h and photolight of 16 h with a 70% humidity at 26°C for 1 month for generation of the adventitious buds, then the calli with adventitious buds were further cultured on the ½ MS solid medium for 1 month to induce roots regeneration. Regenerated seedlings were further cultured in peat soils to harvest the seeds of T0 generation under natural condition. The mutants of T1 generation were generated by self-bred of T0 generation seeds. The leaf genomic DNA of the mutants of T1 generation was extracted and used for PCR detection by the specific primers OsPPa6-F/R and Cas9 primers Cas9-F/R (Supplementary Table S1). Both the line 9 and line 13 were confirmed to be two positive homozygous mutants with cas9-free, and the mutant seeds of T3 generation were obtained by continuously screening under the same culture condition.
Plant Culture and Stress Treatment
Rice seeds were germinated on moist filter paper at 30°C under darkness. A total of 50 of grains from each line (WT and mutants) were cultured to investigate the germination rates, and the generating seedlings were transplanted into the mixture of peat soil and vermiculite (2:1) for further culture in a greenhouse with a humidity of 60%∼70% at 28°C under a 16 h light intensity of 350∼400 μmol m-2 s-1 and at 22°C for 8 h in the dark (Thimijan and Royal, 1983). The seedlings with 3∼4 leaves were subjected to AS (AS, 50 mM, molar ratio of NaHCO3/Na2CO3 9:1; pH 9.17) (Montesinos-Pereira et al., 2018) for 2 weeks, then the seedlings showing phenotype differences were sampled for physiological measurements and molecular detection, and the remained seedlings were continuously cultured to investigate rice grain yield of per plant at mature stage. Plant samples were collected at required time points and used for further analyses or total RNA extraction.
Isolation of Nucleotides and Quantitative RT-PCR
Genomic DNA of rice leaves was prepared by the method of CTAB (Allen et al., 2006). Total RNA of leaves was extracted by Easy Pure Plant RNA Kit (Trans Gene Biotech, Beijing, China). Quantitative RT-PCR procedures were carried out according to the manufacturer’s instructions by the Light Cycler System (Bio-Rad, Richmond, CA, United States) with the SYBR® qPCR Master Mix Kit (Trans Gene Biotech, Beijing, China) by a pair of primers (Supplementary Table S1). The relative expression levels were calculated by the 2-ΔΔCT formula (Livak and Schmittgen, 2001) using the reference gene (accession: XM_015774830) as an internal standard.
Physiological Measurements
The contents of phosphate, Na+ and K+ were determined as described in the following procedures, respectively. Approximately, 250 mg of dried matter were digested by 5 mL of the concentrated sulfuric acid and 2 mL of 30% H2O2, and diluted to a volume of 100 mL in a volumetric flask by ultrapure water. The phosphate was determined by the vanadate–molybdate-yellow colorimetric method using a Continuous Flowing Analyzer (SEAL Analytical AA3, Germany), and the contents of Na+ and K+ were determined by atomic absorption (ZEEnit®700P, Germany). The ATP contents in plant tissues were extracted by ddH2O and measured by the method described previously with HPLC system (Agilent 1290 Infinity II, United States) (Surova et al., 2016). Pyrophosphate was extracted by a previous method (Dancer and Rees, 1989) and determined by the oxidation of NADH at 340 nm using a spectrophotometer (Thermo Scientific GENESYS10S, United States) (Smyth and Black, 1984). The photosynthetic parameters of the first flag leaf of rice were measured by a portable photosynthesis measurer Li-6400XT (Li-COR, United States). The chlorophyll completely was extracted by 80% acetone (Lichtenthaler, 1987) and determined at 645 nm and 663 nm by spectrophotometer, respectively, and calculated by a formula of C (mg g-1 FW) = (20.29 × OD645 + 8.05 × OD663) × V × 10-3/FW, where FW means fresh weight and V means extract volume (Croft et al., 2017). The D (+)-sucrose was extracted by a solution of 4 mL of 80% ethanol and determined at 480 nm using a spectrophotometer by m-dihydroxybenzene method (Hendrix, 1993). Both the starch and soluble sugar in rice were quantitatively measured at 620 nm by the anthrone-sulfuric assay using a spectrophotometer (Yemm and Willis, 1954). The content of free proline was determined as the described method by Bates et al., 1973 in. The malondialdehyde (MDA) was extracted by a previous method (Hodges et al., 1999), and separately measured at 450, 532, and 600 nm and calculated by a formula of C (μmol g-1 FW) = [6.45 × (OD532 - OD660) - 0.56 × OD450]/(FW × 1000) (Khan et al., 2017). The osmotic potentials of leaves were measured using an osmotic pressure dew point meter (Wescor 5520, Logan, UT, United States) by a formula of Ψs (MPa) = -Ci × 0.008314 × (273+T) × 10-3, where Ci means instrument reading and T represents environment temperature (Ball and Oosterhuis, 2005).
Metabolites Extraction and LC-MS/MS Analysis
The metabolites of rice were extracted and analyzed by the method described previously (Zhang Y. et al., 2017). Briefly, a total of 50 mg of fresh leaves was ground in liquid nitrogen, and transferred into the new EP tube containing 1 mL of an internal target substance (Vmethanol:Vacetonitrile:Vwater = 2:2:1, which was kept at -20°C in advance), and homogenized by ball mill at 45 Hz for 4 min, then treated on ice bath by continuous three times of ultrasound for 5 min, then incubated at -20°C for 1 h to precipitate proteins, then centrifuged by 14500 × g at 4°C for 15 min. A total of 500 μL of supernatants were transferred into the new EP tubes, and dried in a vacuum concentrator without heating, and was supplemented by a reconstitution with 100 μL of extraction solution (Vacetonitrile:Vwater = 1:1), and mixed for 30 sec, and sonicated in a water bath at 4°C for 10 min, and centrifuged by 14500 × g at 4°C for 15 min. The supernatants were filtered through the PTFE membrane of 0.22 mm (Sigma, United States), and subjected to analyses on the UHPLC-QTOF-MS (1290, Agilent Technologies) equipped with a UPLC BEH Amide column (1.7 μm, 2.1 mm × 100 mm, Waters Corporation, United States) using optimized mobile phases. The extraction and analysis of metabolites were assisted by the Allwegene Technology Co., Ltd. (Beijing, China).
Statistics Analysis
MS data preprocessing such as alignment, peak intensity, retention time (RT) correction and mass-to-charge ratio (m/z) values was performed using Markerlynx XSTM software (Waters Corporation, United States). Orthogonal projection to latent structures-discriminant analysis (OPLS-DA) was applied to the supervised multivariate analysis using SIMCA-P software (version 12.0). The supervised OPLS-DA was used to reconstruct the metabonomics data and scientifically evaluate the differential variables expressions of separation by reasonably amplifying differences between the experimental and control groups (Zhang Y. et al., 2017). Moreover, a completely randomized design involving an arrangement of 3 (WT and two mutants) × 2 (control and AS) factor was established in this experiment. Univariate analyses were statistically performed by using one-way ANOVA in the statistical software SPSS 17.0 (SPSS, Chicago, IL, United States). Based on the results of ANOVA, a DUNCAN-test was analyzed. All data were represented by an average with a standard error of three replicates. The significance level was expressed by P ≤ 0.05 or 0.01.
Results
OsPPa6 Is a Member of sPPase Encoding a Soluble Inorganic Pyrophosphatase
The OsPPa6 gene encodes a protein of 286 amino acid and shares more than 90% of homology with the genes encoding the sPPase in Dichanthelium oligosanthes, Zea mays, and Aegilops tauschii. The OsPPa6 protein also shares homology of 71.2% with the ThPP1 in T. halophila and 68.0% with the AtPPa6 in A. thaliana (Figure 1A). Bioinformatics analysis shows that the OsPPa6 has a closely familiarity with the DoPPase and ZmPPase in diverse plants (Figure 1B). Moreover, a total of six of putative protein encoding the sPPase in rice were retrieved by the Rice Data database3 and are represented by Os01g0974800, Os01g0866500, Os02g0704900, Os04g0687100, Os05g0114000, and Os05g0438500, respectively. These six proteins share more than 75% of homogeneity, however, the OsPPa6 (Os02g0768600) only shares less than 30% of homology with these six putative proteins (Supplementary Figure S1), indicating that the OsPPa6 gene exhibits structural specificity in the sPPase family.
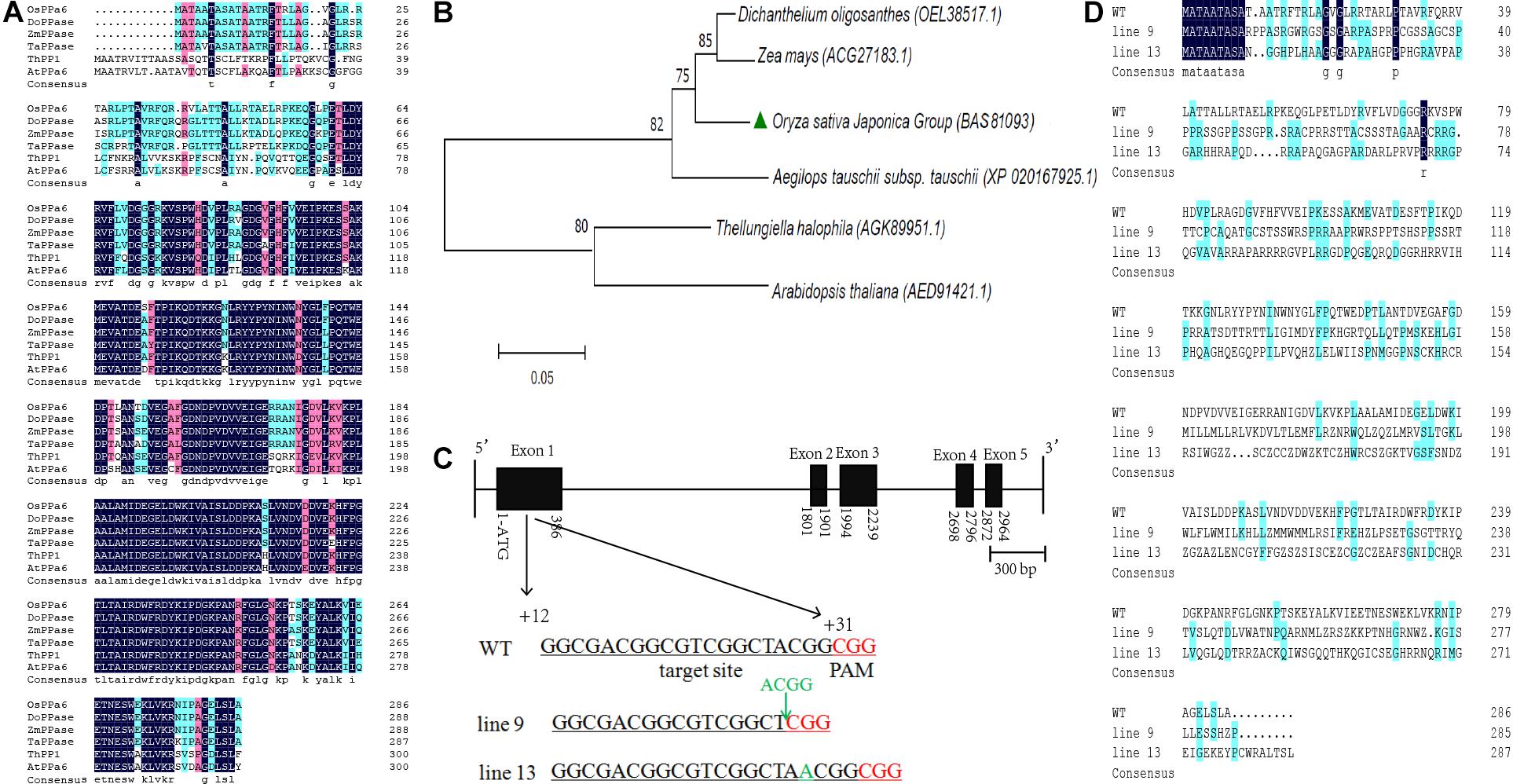
Figure 1. Bioinformatics analyses of the soluble inorganic pyrophosphatase and mutagenesis identification of the OsPPa6 gene. (A) Homology alignment of soluble inorganic pyrophosphatase. OsPPa6, O. sativa Japonica Group (accession: BAS81093); DoPPase, Dichanthelium oligosanthes (accession: OEL38517); ZmPPase, Zea mays (accession: ACG27183); TaPPase, Aegilops tauschii (accession: XP_020167925); ThPP1, Thellungiella halophila (accession: AGK89951); AtPPa6, Arabidopsis thaliana (accession: AED91421). Blue and pink color indicate the similarity; white color indicates the differences; (B) Phylogenetic tree analysis of the soluble inorganic pyrophosphatase. (C) Structural profiles of the OsPPa6 gene with the mutagenesis of the CRISPR/Cas9-mediated in the first exon. Black rectangle means the exon. Black line means the intron. The underlined nucleotides indicate the target sites. Nucleotides in red represent PAM sequences. PAM, protospacer adjacent motif. The target sequences are located at +12th to +31th of the gene; (D) Multiple alignments of the OsPPa6 proteins between the wild type and mutants of CRISPR/Cas9-edited.
The Mutagenesis of Nucleotides in the OsPPa6 Gene
Firstly, based on the specific recognition of the CRISPR/Cas9 gene editing system, the target sites in the clustered regularly interspaced short palindromic repeat sequences of the coding region of the OsPPa6 gene are determined, and located at the fragment between the +12th and +31th in the coding region (Figure 1C). PCR detection was continuously performed to confirm the mutagenesis of the target gene in the rice mutant. PCR products sequencing shows that four bases (A, C, G, G) at the target site in the line 9 were deleted and an A base was inserted into the +18th site of the target sequence in the line 13 compared with the WT (Figure 1C and Supplementary Figure S1), thus altering the sequences of amino acids in the OsPPa6 protein in the mutant lines (Figure 1D). Therefore, the encoded amino acids sequences in both line 9 and line 13 were obviously interfered by the deletion or insertion of a single base, thus triggering a loss of function of the OsPPa6 gene in rice. Homology analysis shows that the OsPPa6 shares lower similarity with the other six putative proteins of sPPase in rice, and exhibits obvious differences and specificity in nucleotide sequences of the CRISPR/Cas9 recognition target site compared to the other putative genes encoding the sPPase in rice (Supplementary Figure S2). PCR and sequencing also confirmed that no off-target effects were observed in the mutants, indicating that the other six putative genes encoding the sPPase were not edited by the CRISPR /Cas9 system.
The Mutagenesis of the OsPPa6 Gene Delayed Rice Growth
To observe the effect of the mutagenesis of the OsPPa6 gene on rice growth, we investigated rice germination rates in a growth chamber under favorable culture and performed pot experiment in a greenhouse under AS as described in the method. Data showed that both the line 9 and line 13 delayed germination time under favorable culture condition compared to the WT (Figure 2A). After 10-day of culture, both the line 9 and line 13 only showed 76 and 82% of germination rates, respectively, while the WT demonstrated 92% of germination rates (Figure 2B), suggesting that the mutagenesis of the OsPPa6 gene affected the seed germination. To investigate the growth change of rice in response to AS, both the mutants and the WT seedlings with 3∼4 leaves were cultured under AS for 2 weeks, and the line 9 and line 13 obviously showed a growth inhibition compared with the WT under both favorable culture and AS stress (Figure 2C-1). Investigation showed that the average plant heights of the line 9 and the line 13 were 35.64 cm and 37.16 cm, respectively, which significantly were lower than 43.45 cm of the WT at the seeding stage even at favorable culture. In the case of culture under AS, both the line 9 and line 13 revealed lower growth rates, their average plant heights were only 23.75 cm and 25.04 cm, respectively, and demonstrated obvious dwarf phenotype compared with the WT (Figure 2D). Similarly, the fresh weights of the line 9 and line 13 were decreased by 19.32 and 16.58% compared with the WT under favorable culture, respectively. In the case of the culture under AS, the line 9 and line 13 demonstrated a decrease of 21.29 and 19.20% in the fresh weights of per plant compared with the WT, respectively (Figure 2E). At the maturity stage, the two mutants still revealed a lower plumpness of grains compared to the WT even at favorable culture condition (Figures 2C-2,3), and that the seed-setting rates of 74.77% in the line 9 and 77.78% in the line 13 were significantly lower than the seed-setting rates of 93.22% in the WT. In accordance with favorable culture, the seed-setting rates of 63.82% in the line 9 and 66.07% in the line 13 were significantly lower than the seed-setting rates of 87.30% in the WT under AS (Figure 2F). Moreover, the grain yields of the line 9 and line 13 of per plant were decreased by 1.22 g and 0.97 g compared to the WT under favorable culture, respectively, and that the line 9 and line 13 led to a decrease of 40.26 and 33.99% in the grain yield of per plant compared with the WT under AS, respectively (Figure 2G).
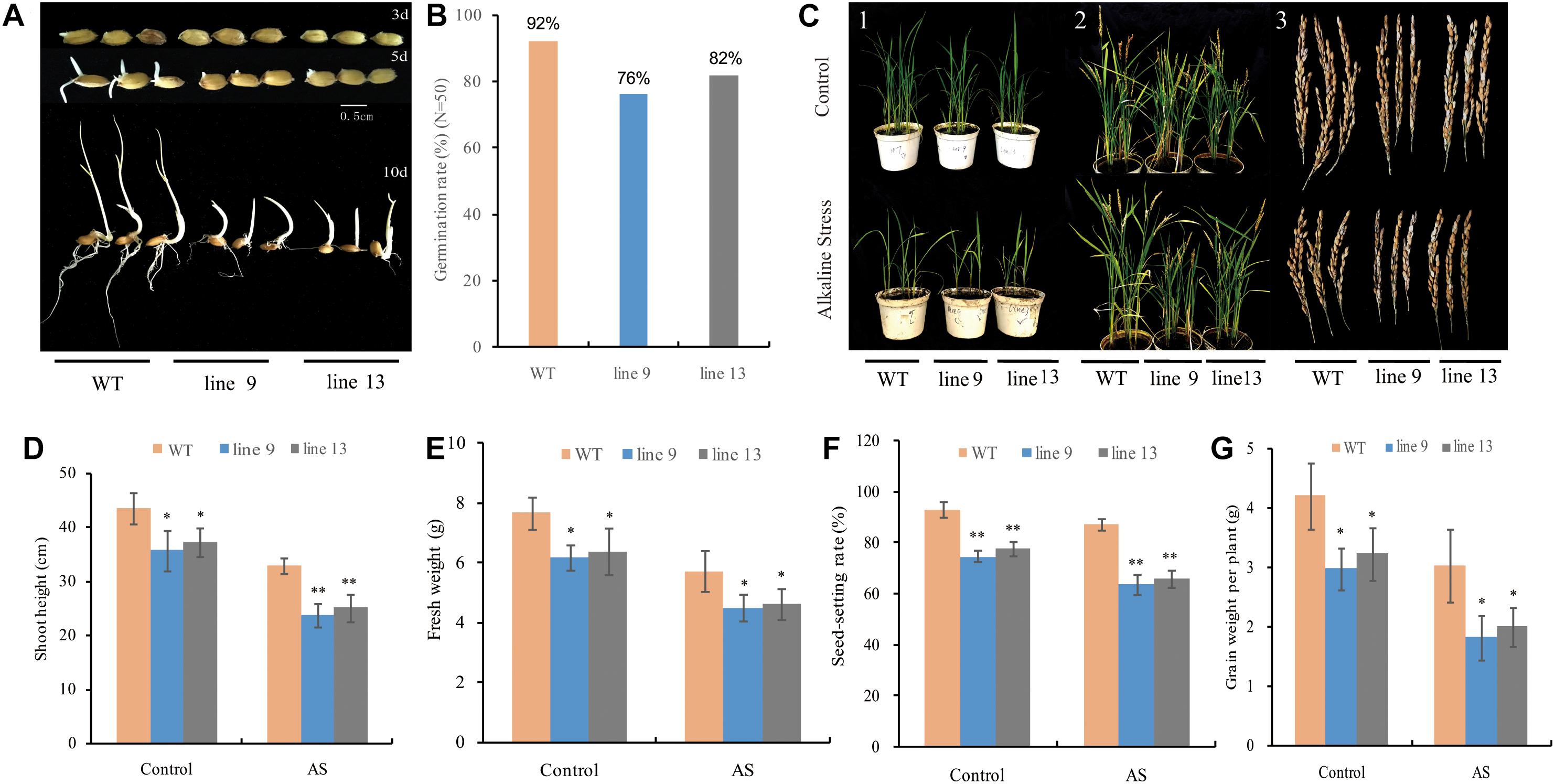
Figure 2. Changes in phenotype and biomass of rice under favorable condition or alkaline stress (AS). (A) Germination time; (B) Germination rate; (C) (1) Rice phenotype at seeding stage; (2) Rice phenotype at maturity stage; and (3) Seed differences in phenotype; (D) Shoot height of the seedlings; (E) Fresh weight of the seedlings; (F) Seed-setting rate; and (G) Grain weight of per plant. Values are mean ± SD of six replications and asterisks denote Student’s test significance at level of P ≤ 0.05 (∗) or P ≤ 0.01 (∗∗).
Transcripts of OsPPa6 Gene and OsGS2 Gene
To profile the expression of the OsPPa6 gene in the WT, we performed analyses of qRT-PCR using the total RNA in leaves. The data showed that the OsPPa6 gene in the WT was significantly induced and its transcripts were increased by 1.81-fold after 12-h of AS relative to the transcripts in the treatment without AS (Supplementary Figure S3). Moreover, the expression levels of the OsPPa6 gene in the mutants were significantly lower than that in the WT, especially under AS (Figure 3A), suggesting that the mutagenesis of the OsPPa6 gene reduced the transcripts of the OsPPa6 gene. Unlike the OsPPa6 gene, theOsGS2 gene encoding a chloroplastic GS2 in the WT was significantly inhibited after 48 h of AS, and its relative expression levels lowered 35% compared to the measured value under the favorable culture (Supplementary Figure S3), suggesting that the OsGS2 was not induced by AS. Accordingly, as shown in Figure 3B, both the mutants and WT had no significant differences in the transcripts of the OsGS2 gene under favorable culture, but the transcript levels of the OsGS2 gene in the mutants were significantly lower than that in the WT with the prolongation of AS, indicating that the OsPPa6 gene and the OsGS2 gene demonstrated differential expression profiles in rice under AS.
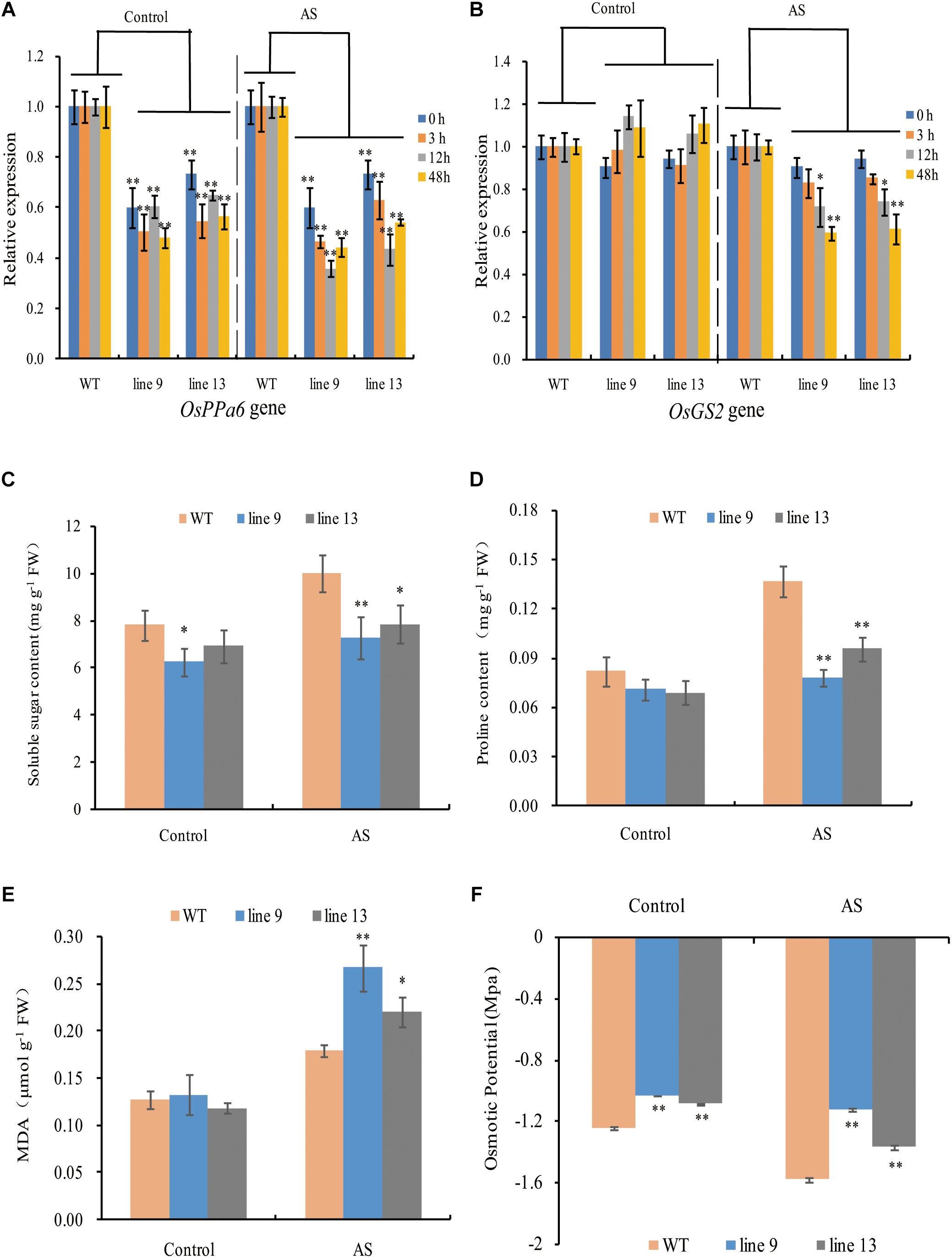
Figure 3. Gene relative expression levels and physiological changes under favorable culture or AS. (A) Relative expression levels of the OsPPa6 gene in rice shoots; (B) Relative expression levels of the OsGS2 gene in rice shoots; (C) Soluble sugar content; (D) Proline content; (E) MDA content; (F) Osmotic potential. Values are mean ± SD of three replications and asterisks denote Student’s test significance at level of P ≤ 0.05 (∗) or P ≤ 0.01 (∗∗).
A Loss of Function of the OsPPa6 Gene Lowered the Alkaline Tolerance in Rice
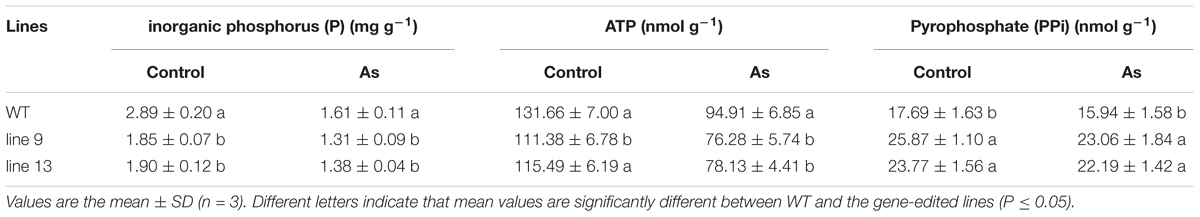
To characterize the physiological responses of the mutants and the WT to AS, we investigated the accumulation profiles of physiological compatible metabolites and performed photosynthetic measurements. Compatible metabolites are not only involved in the regulation of osmotic potentials, but also provide energy resource for the metabolism in plant, and thereby affecting the process of photosynthesis, conversely altering the accumulation of compatible metabolites such as soluble sugars and proline. Therefore, we measured inorganic phosphate (P), ATP, PPi, soluble sugar, proline, MDA, osmotic potential, photosynthesis, chlorophyll, sucrose, starch, Na+ and K+ in rice to evaluate the effects of the mutagenesis the OsPPa6 gene on accumulation of metabolites and photosynthetic process in response to AS. Measurements showed that when the rice were subjected to AS, the line 9 and line 13 led to a decrease of 0.30 mg g-1 and 0.23 mg g-1 in P content, and decrease of 19.63 and 17.68% in the ATP content compared to the WT, respectively, however, the line 9 and line 13 demonstrated an increase of 44.67 and 39.21% in the PPi content compared with that in the WT, respectively (Table 1). Data also showed that both the line 9 and line 13 lowered the contents of soluble sugar and proline than the WT under AS, and showed a decrease of 27.13 and 21.32% in the contents of soluble sugars, respectively (Figure 3C), and a decrease of 42.86 and 28.57% in the contents of proline compared with the WT, respectively (Figure 3D). However, in the case of AS, both the line 9 and line 13 obviously increased the accumulation of the MDA and the osmotic potentials in leaves, and showed an increase of 0.50- and 0.22-fold in the MDA contents compared to the WT, and an increase of 0.45 MPa and 0.21 MPa in the osmotic potentials of leaves compared with the WT, respectively (Figures 3E,F).
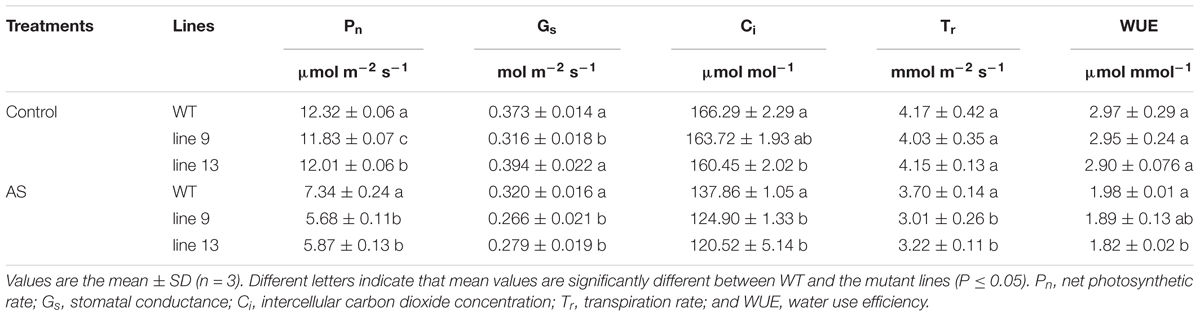
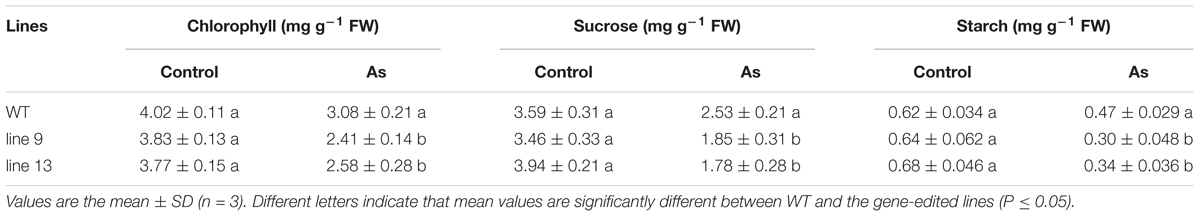
Photosynthetic measurements showed that the mutagenesis of the OsPPa6 gene significantly lowered the net photosynthetic rate (Pn) in the mutant lines under favorable treatment. Especially, the line 9 not only showed a decrease of 0.49 μmol m-2 s-1 in Pn, but also demonstrated a decrease of 0.057 mol m-2 s-1 in stomatal conductance (Gs), and the intercellular carbon dioxide concentration (Ci) in the line 13 was also significantly lower than that in the WT under favorable condition (Table 2). In the case of AS, the line 9 and line 13 lowered 22.62 and 20.03% of the Pn, and 16.88, and 12.81% of the Gs compared to the WT, respectively. Meanwhile, the Ci of the line 9 and line 13 were decreased by 12.96 μmol mol-1 and 17.34 μmol mol-1, and the transpiration rates (Tr) of the line 9 and line 13 were decreased by 18.65 and 12.97% compared to the WT under AS, respectively (Table 2). These data indicate that the mutagenesis of the OsPPa6 gene resulted in a remarkable decrease in photosynthetic capacity in the mutants compared with the WT under AS. In photosynthesis, the chlorophyll functions in the absorption and transformation of light energy. Therefore, we investigated the accumulation responses of the chlorophyll in the mutants because of the mutagenesis of the OsPPa6 gene under AS. Data show that no significant differences in the chlorophyll accumulation were observed between the mutants and the WT under favorable culture, but, in the case of culture under AS, the line 9 and line 13 significantly triggered a decrease of 0.67 mg g-1 and 0.5 mg g-1 of the chlorophyll compared to the WT, respectively (Table 3). Measurements also exhibit that chlorophyll accumulation and photosynthetic capacity affected the accumulation of sucrose and starch metabolites from plant photosynthesis because the accumulation of sucrose and starch in the mutants were significantly lowered compared with the WT under AS. The sucrose of 1.85 mg g-1 in the line 9 and 1.78 mg g-1 in the line 13 are significantly lower than the sucrose of 2.53 mg g-1 in the WT under AS, and the starch contents in the line 9 and line 13 were also decreased by 36.17 and 27.66% compared to the WT, respectively (Table 3). These results suggest that mutants obviously lowered the photosynthetic capacity and chlorophyll contents, consequently reducing the accumulation of sucrose and starch under AS.
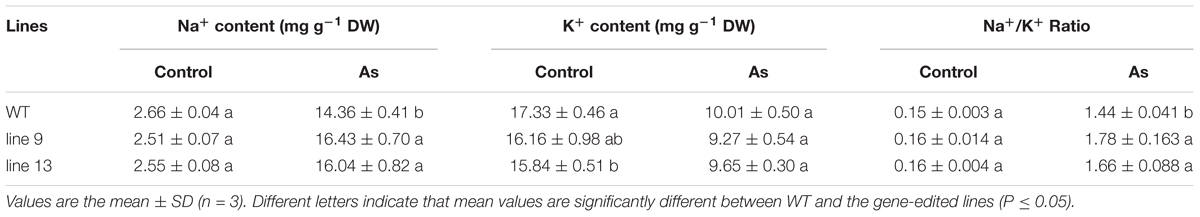
Except for the metabolites accumulation and photosynthesis, we also measured the contents of Na+ and K+ in rice leaves to investigate whether the mutagenesis of the OsPPa6 gene influence the distribution of Na+ and K+ under AS. Our data show that the mutagenesis of the OsPPa6 gene significantly affected the accumulation of Na+ and K+ in the mutants. Especially, when rice was exposed to AS, the mutants demonstrated an increase of 14.42% in line 9 and 11.70% in line 13 in the Na+ contents compared with the WT, but lowered 7.39% in line 9 and 3.60% in line 13 in the K+ contents compared with the WT, respectively, thus leading to an increase of 23.61% in Na+/K+ ratio in the line 9 and 15.28% in the line 13 comparing to the WT, respectively (Table 4). These data indicate that the mutagenesis of the OsPPa6 gene increased accumulation of Na+ and reduced the accumulation of K+ in leaves of the mutants, and lowered the alkaline tolerance because of the damage from excess uptake of Na+ in plant cells.
The Mutagenesis of the OsPPa6 Gene Revealed Metabolic Differences in Rice
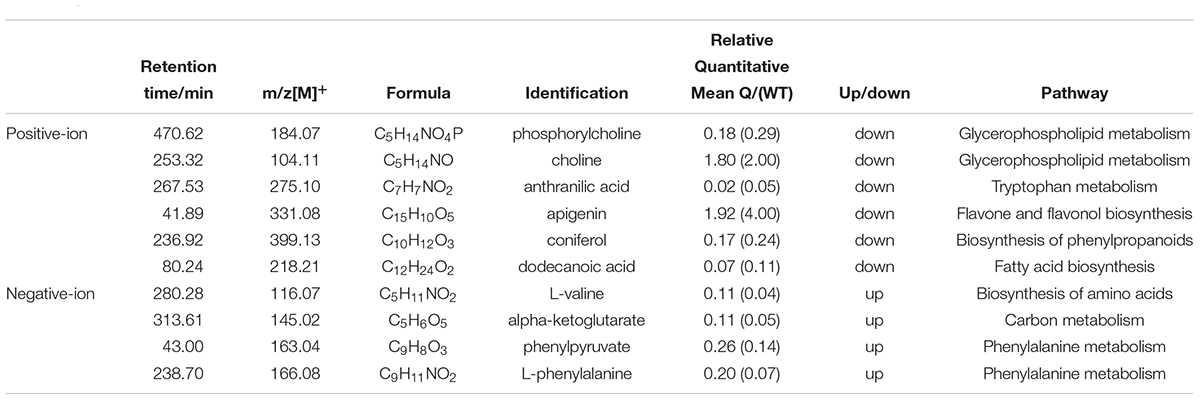
To better explore the differential metabolites between the mutants and the WT under AS, the mutant line 9 (experimental group) and the WT (control group) were analyzed by UPLC-MS to obtain a higher level of group separation and understand the metabolic differences triggered by the mutagenesis of the OsPPa6 gene. Our data show that the parameters of the OPLS-DA for the classification were expressed by the R2Y (cum) of 0.975 and Q2 (cum) of 0.782 representing the positive ion mode, and by the R2Y(cum) of 0.978 and Q2(cum) of 0.878 representing the negative ion mode, respectively. These parameters representing the two ion models exhibited a better stability and predictability, and effectively reflected the metabolic differences between the WT and the mutant in response to AS. The separation trends in both the experimental group and the control group were remarkable under positive or negative ion model (Figures 4A,B). The detection demonstrates that the mutagenesis of the OsPPa6 gene obviously altered the distribution of the metabolites in the mutant under AS. As shown in Figures 4C,D, the volcano plots expressing the differential metabolites revealed significant differences between the experimental and the control groups. Using VIP screening (variable importance in the projection) >1 at a level of P < 0.05, a total of 324 of the differential metabolites were isolated in the positive ion mode. In detail, the experimental group revealed a total of 117 of the up-regulated metabolites (red dot, Figure 4C) and a total of 207 of the down-regulated metabolites (blue dot, Figure 4C) relative to the control group. In the same way, a total of 482 of the differential metabolites were screened in the negative ion mode, in which a total of 215 of the differential metabolites were up-regulated (red dots, Figure 4D) and a total of 267 of the differential metabolites were down-regulated (blue dots, Figure 4D) in the experimental group compared with the control group, respectively. Furthermore, using screening conditions with more than 1 of VIP and fold change at a level of P less than 0.05, we isolated out a total of 10 of the differential metabolites that are mainly involved in different metabolic pathways. Analyses show that the mutagenesis of the OsPPa6 gene not only obviously down-regulated accumulation of phosphorylcholine, choline, anthranilic acid, apigenin, coniferol, and dodecanoic acid, but also up-regulated the accumulation of L-valine, alpha-ketoglutarate, phenylpyruvate, and L-phenylalanine in the mutants under AS (Table 5).
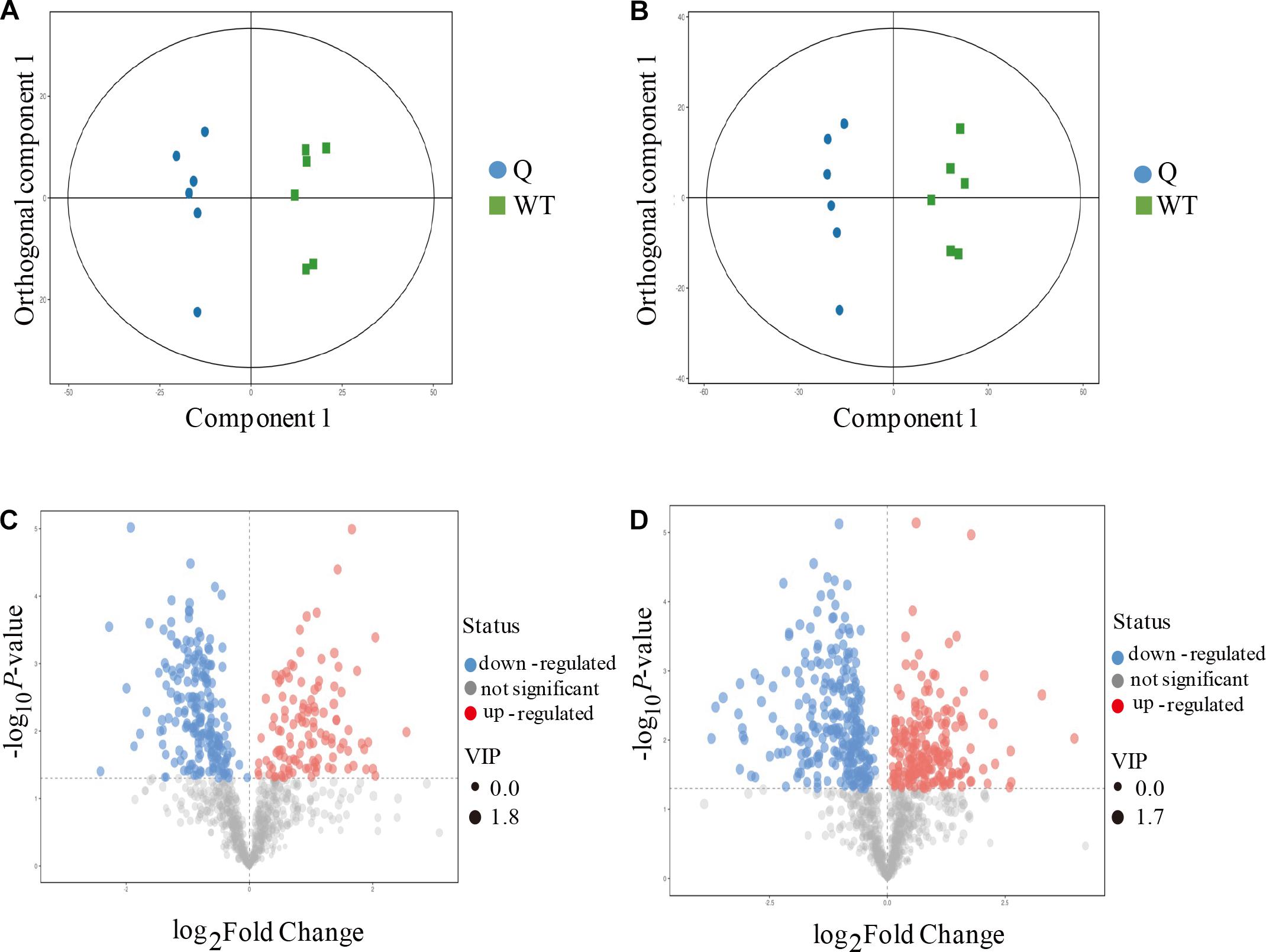
Figure 4. Metabolomics analyses of the mutants and the wild type. (A) Score scatter plot of OPLS-DA model for group Q vs WT (positive ion mode); (B) Score scatter plot of OPLS-DA model for group Q vs WT (negative ion mode); (C) Volcano plot for group Q vs WT (positive ion mode); (D) Volcano plot for group Q vs WT (negative ion mode).
Discussion
The mutagenesis of the target gene by the CRISPR/Cas9 system-guided usually results in the deletion of the target bases or insertion of a single base in the target fragment, while the insertion mutation is mainly caused by inserting an A or a T base (Zhang H. et al., 2017). In our study, with a help of the CRISPR/Cas9 technology, the bases deletion in the line 9 and an insertion of an A base in the line 13 effectively generated the mutants with the mutagenesis of the OsPPa6 gene. Highly homology between the OsPPa6 protein and the sPPases in Z. mays, T. halophila, and A. thaliana seems to indicate a possible regulatory role of the OsPPa6 gene in response to abiotic stresses. As previously reported, the gene encoding the sPPase is induced by salt stress and involved in energy metabolism in young leaf tissues of the Z. mays, and maize growing cells has the cell-age specificity in response to salinity stress (Kravchik and Bernstein, 2013). He et al. (2017) found that the ThPP1 gene in T. halophila is induced by AS and confers an enhanced the alkaline tolerance in rice. In Arabidopsis, the AtPPa6 is an exclusively chloroplastic enzyme, and truly essential for plant survival under abiotic stress (Gutiérrez-Luna et al., 2018). Similarly, the up-regulation of the OsPPa6 gene transcripts might play an important regulatory role in enhancing alkaline tolerance in rice.
Not only responding to abiotic stresses, the sPPases also exert an influence on the plant phenotype. de Graaf et al. (2006) believed that a lower sPPase activity severely hinders the growth of pollen tube in the Papaver, and silencing the gene encoding sPPase displays a stunting and bushiness phenotype of potato (Eamens et al., 2014). Our finding basically suggests that the delayed growth of rice mutant obviously is associated with the loss of the activity of the OsPPa6 protein, and led to decreases in both biomass and grain yield, indicating the loss of function of the OsPPa6 gene, and thereby increased the accumulation of excess PPi in plant cells. However, Jelitto et al. (1992) tentatively transformed the E. coli pyrophosphatase gene into potato and tobacco, and observed an apparent growth inhibition of the transgenic plants accompanying an occurrence of yellow leaves. Accordingly, the inhibition or promotion in the sPPase activity would change the PPi/Pi ratio in plant cells and affect the plant growth. Therefore, maintaining PPi/Pi equilibrium is essential for plant’s growth and development.
Maintenance of PPi/Pi equilibrium in plant cells is closely with PPi hydrolysis which is essential for maintaining plant growth, and PPi is usually catalyzed to produce Pi by the sPPase, while Pi provides an important substrate for ATP synthesis by promoting ADP phosphorylation in plant metabolism (Pietropaolo et al., 2016). In our study, more accumulation of PPi in rice mutants is associated with the mutagenesis of the OsPPa6 gene, because the weak activity of the OsPPa6 protein reduced the accumulation of Pi and ATP. Particularly, a lower accumulation of ATP not only interferes in gluconeogenesis, amino acid activation and DNA replication, but also influences the synthesis and conversion of carbohydrate by energy supply in plants under AS (Vanegas et al., 2015). Generally, alteration of PPi metabolism affects the metabolism of carbohydrates, which conversely change the distribution of carbon in both source and sink organs. Report showed that the specific overexpression of a bacterial pyrophosphatase increases the contents of sucrose and glucose in tomato fruit (Osorio et al., 2013). Geigenberger et al. (1996) found that the transgenic tobacco expressing the gene encoding a E. coli PPase increases starch accumulation. Study also proved that the changes of the PPi contents in the transgenic potato overexpressing E. coli pyrophosphatase gene affects the metabolism of starch and sucrose (Farré et al., 2000). In this study, lower accumulation of sucrose and starch in the mutants might result from the alteration of PPi enrichment and the reduction of ATP energy supply, lowering the conversion efficiency of carbohydrate in rice. Generally, the carbohydrates conversion is controlled by the photosynthetic process, while both the efficiency of photosynthetic electron transport and PSII light capture in photosynthesis are limited by a higher pH, thus directly affecting Pn (Wu et al., 2014). The ThPP1 protein could specifically interact with a PSII light-harvesting-Chl-binding protein, and the transgenic rice overexpressing the ThPP1 gene encoding the sPPase increased the Pn and the accumulation of chlorophyll under AS (He et al., 2017). This study gave a fact that the decreases in the photosynthesis and the chlorophyll contents in the mutants has a closely association with the mutagenesis of the OsPPa6 gene because of shortage of energy supply of the ATP-mediated, thus lowering the adaptability of rice mutant to high alkalinity. In photosynthesis, photorespiration is an important regulatory factor participating in the re-assimilation of ammonia, and directly affects nitrogen assimilation that is usually controlled by GS2 in plants, and the transgenic rice overexpressing the OsGS2 gene enhanced the photosynthetic intensity and reduced the photorespiration, and conferred an enhanced salt tolerance (Hoshida et al., 2000), indicating that an enhanced increases in the GS activity is beneficial to the reuse of nitrogen in plants, thus improving plant salt tolerance. In this study, the lower transcripts of the OsGS2 gene in the mutants played a negative regulatory role in improving rice alkali tolerance because excess Na+ accumulation from AS severely affected the photosynthesis in rice.
Both soluble sugar and proline play important regulatory roles in the osmotic regulation of cells and alleviating the alkaline damage to the plants (Székely et al., 2008). Study also confirmed that the mutants Lotus japonicus lacking GS2 activity lower proline accumulation, and exhibit more sensitive to drought stress (Díaz et al., 2010). Our data also demonstrate a closely association between the OsGS2 activity and proline accumulation in rice in response to AS, and both soluble sugar and MDA exhibit the differentially accumulated responses to the abiotic stresses, and leading to the changes of osmotic potentials in plant cells (Mittler, 2002; Lv et al., 2013). Both the transgenic Eleusine coracana and Brahmi overexpressing a SbVPPase gene, encodes a vacuolar proton pyrophosphatase in Sorghum bicolor, increases the accumulation of proline and soluble sugar, but reduces the MDA contents under salt stress (Anjaneyulu et al., 2014; Ahire et al., 2018). In our study, the mutagenesis of the OsPPa6 gene might be an important modulator lowering the contents of soluble sugar and proline, and increasing osmotic potentials and MDA accumulation in the mutants because of the activity changes of the sPPase under AS, indicating that the loss of function of the OsPPa6 gene affected the accumulation of compatible solutes in rice and exerted negative effects on the maintenance of the osmotic equilibrium in plant cells, and aggravated the alkali damage to the plant cell membranes. The osmotic equilibrium in leaves is closely associated with the accumulation of the Na+ and K+ under abiotic stresses, and directly affects the plant tolerance to the AS (Yang et al., 2008; Chen et al., 2011). The changes of osmotic potentials in leaves are generally related to the intracellular ion homeostasis which is usually reflected by the acquisition and distribution of K+ and Na+, since the accumulation of K+ has a balance role in lowering the toxicity damage of excess Na+ (Deinlein et al., 2014). The homoeostasis of Na+ and K+ in plant cells is usually controlled by the H+-PPase. Lv et al. (2015) reported that the Arabidopsis overexpressing the SeVP1 and SeVP2 gene increased K+ accumulation, but decreased Na+ contents, maintaining higher K+/Na+ ratio in leaves under salt and low nitrogen stresses. Our measurements indicate that like the H+-PPase, the OsPPa6 gene encoding a sPPase might have similar regulatory role in the acquisition and transport of K+ and Na+, because the mutagenesis of the OsPPa6 gene significantly exhibited a negative regulatory role in maintaining homoeostasis of Na+ and K+ in the mutants under AS, thereby lowering the transport ability and compartmentation of Na+, increasing the accumulation of excess Na+ in plant cells.
The intracellular imbalance of Na+ and K+ usually changes the distribution of metabolites and disturbs the metabolism in plants (Wu, 2018). In this work, the differential metabolites between the mutant and the WT show that the mutagenesis of the OsPPa6 gene altered accumulation profiles of these metabolites in rice (Table 5). Reportedly, both phosphorylcholine and choline are two components of cell phospholipids, and mainly involved in the glycerophospholipid metabolism pathway by maintaining the cell’s structure and physiological function in plants, and confer a regulation of the osmotic pressure (Devaux, 1991). In our measurement, the obvious down-regulations of the phosphorylcholine and choline in the mutant imply that increases of osmotic potentials triggered Na+ ion damage to the cell membrane under AS. Except the phosphorylcholine and choline, as an important primary metabolite, anthranilic acid participates in the biosynthesis of tryptophan in plants. Jing et al. (2018) reported that the optimized anthranilic acid feeding to the engineered E. coli strain would improve L-tryptophan production, while tryptophan not only involves in the protein synthesis, but also is an important precursor substance for biosynthesis of auxin and alkaloid in diverse plants (Hibino and Choshi, 2001). In this study, a lower accumulation of anthranilic acid in the mutant might reduce the tryptophan contents, thus affecting rice growth and development. As an important bioactive plant flavonoid, apigenin is mainly involved in the flavone and flavonol biosynthesis pathway, and increases the resistance of the fungal and oomycetes pathogens in soybean (Jiang et al., 2012), and reduces the damage to the biomembrane by scavenging lipid peroxide, NO free radical, oxygen free radical, and superoxide anion radical (Husain et al., 1987). In this study, little accumulation of the apigenin in the mutant suggests that the mutagenesis of the OsPPa6 gene increased the harmful free radicals in the cell membrane, which is also suffered from the more damage of MDA. In higher plants, the biosynthesis process of lignin is elucidated by coniferol (Boerjan et al., 2003). Hänninen et al. (2011) found that the amounts of lignin in two similar wood species are dependent on the accumulation profiles of coniferyl alcohol. Also, the lignin plays an important regulatory role in enhancing the mechanical strength of cells and tissues under abiotic stresses (Taylor-Teeples et al., 2015). Our data indicates that the mutagenesis of the OsPPa6 gene weaken the lignin synthesis and cells mechanical strength, leading to the down-regulation of the coniferol contents in the mutant under AS. Meanwhile, the dodecanoic acid from secondary metabolism in plant also functions in improving the stress resistance through a pathway of synergistic reaction (Peñuelas et al., 1996; López-Villalobos et al., 2011). Our study reveals that lower alkaline tolerance in the mutant might have an association with the reduction of dodecanoic acid. While L-valine has a feedback inhibitory role on the acetolactate synthase (ALS) catalyzing the first step in the biosynthesis of valine, leucine, and isoleucine (Pang and Duggleby, 1999). Our data demonstrate that a lower ALS activity possibly inhibits the synthesis of these amino acids, and thereby affected the accumulation of related protein in the mutant. Ferrario-Méry et al. (2001) reported that both the α-ketoglutarate and glutamine are two metabolite signal molecules participating in ammonia assimilation and transcripts of the gene encoding a nitrate reductase in the transgenic tobacco, and the negative effect of glutamine on the abundance of the nitrate reductase was offset by highly enrichment of α-ketoglutarate mainly deriving from respiration, thus affecting the carbon-nitrogen metabolism in plant. In this study, the up-regulation of the α-ketoglutarate in the mutant is likely associated with the increases of respiration rate in rice at the beginning of AS. Study confirmed that the phenylpropanoid synthesis is an important metabolism pathway in plants, and all substances containing phenylpropane skeleton such as phenolic compounds, alkaloids and flavonoids, are almost produced directly or indirectly by a catalyzation reaction of phenylalanine ammonialyase (PAL) that links primary and secondary metabolism (Jones, 1984). In present study, more accumulation of phenylpyruvate and L-phenylalanine in the mutant might be caused by a lower PAL activity, thus influencing the synthesis of secondary metabolites in rice.
Collectively, our study shows that the mutagenesis of the OsPPa6 gene encoding a sPPase obviously reveals a lower alkaline tolerance in rice although more detailed and in depth analyses are required. This study provides an inspiration that it would be interesting to investigate whether both the OsPPa6 itself and its homologous confers a regulatory role in improving rice tolerance in response to the other abiotic stresses such as high salinity and drought, and better utilize the stress-resistant genes for the cultivation of stress tolerant crops.
Conclusion
In summary, the OsPPa6 gene is an important regulatory factor conferring an adaptive acclimation of alkaline tolerance in rice. The mutants with the mutagenesis of the OsPPa6 gene not only showed dwarf phenotype, but also triggered a series of adverse physiological changes, thus resulting in a lower biomass, especially under AS. The mutagenesis of the OsPPa6 gene led to an imbalance of osmotic potentials and disorder of physiological metabolites in plant cells under AS, indicating that when the OsPPa6 gene normally functions, a beneficial alteration of physiological metabolites positively contribute to an enhanced increase of alkali tolerance in rice. This study provides an insight into the regulatory role of the OsPPa6 gene encoding a soluble inorganic pyrophosphatase in adaptive acclimation of plants to AS.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
BW performed entire experiment and the preliminary manuscript. GX, ZL, RH, JH, and SH participated in the physiological measurements. LL and XC designed the experiment. XC corrected the manuscript.
Funding
This work was supported by the National Key Project of 973 Fundamental Research (2015CB150800), the National Key Research and Development Projects (2016YFC0501203), and the Transgenic Technology Major Project (2016ZX08010005-9).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Lanqin Xia for providing the pCXUN-U3 vector and technical support. Special thanks to Prof. Wan Jianmin’s team for rice transformation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00759/full#supplementary-material
Footnotes
References
Ahire, M. L., Anil Kumar, S., Punita, D. L., Mundada, P. S., Kishor, P. B. K., and Nikam, T. D. (2018). The vacuolar proton pyrophosphatase gene (SbVPPase) from the Sorghum bicolor confers salt tolerance in transgenic Brahmi [Bacopa monnieri (L.) Pennell]. Physiol. Mol. Biol. Plants 24, 809–819. doi: 10.1007/s12298-018-0586-4
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethyl ammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Anjaneyulu, E., Reddy, P. S., Sunita, M. S., Kishor, P. B., and Meriga, B. (2014). Salt tolerance and activity of antioxidative enzymes of transgenic finger millet overexpressing a vacuolar H(+)-pyrophosphatase gene (SbVPPase) from Sorghum bicolor. J. Plant. Physiol. 171, 789–798. doi: 10.1016/j.jplph.2014.02.001
Ball, R. A., and Oosterhuis, D. M. (2005). Measurement of root and leaf osmotic potential using the vapor-pressure osmometer. Environ. Exp. Bot. 53, 77–84. doi: 10.1016/j.envexpbot.2004.03.003
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bu, N., Li, X. M., Li, Y. Y., Ma, C. Y., Ma, L. J., and Zhang, C. (2012). Effects of Na2CO3 stress on photosynthesis and antioxidative enzymes in endophyte infected and non-infected rice. Ecotox. Environ. Saf. 78, 35–40. doi: 10.1016/j.ecoenv.2011.11.007
Chen, W., Feng, C., Guo, W., Shi, D., and Yang, C. (2011). Comparative effects of osmotic-, salt- and alkaline stress on growth, photosynthesis, and osmotic adjustment of cotton plants. Photosynthetica 49, 417–425. doi: 10.1007/s11099-011-0050-y
Chen, Y., Li, L. L., Zong, J. Q., Chen, J. B., Guo, H. L., Guo, A. G., et al. (2015). Heterologous expression of the halophyte Zoysia matrella H+-pyrophosphatase gene improved salt tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 91, 49–55. doi: 10.1016/j.plaphy.2015.04.004
Chu, C. C., Wang, C. C., Sun, C. S., Hsu, C., Yin, K. C., Chu, C. Y., et al. (1975). Establishment of an efficient medium for anther culture of rice through comparative experiments of the nitrogen sources. Sci. Sin. 18, 659–668.
Croft, H., Chen, J. M., Luo, X. Z., Bartlett, P., Chen, B., and Staebler, R. M. (2017). Leaf chlorophyll content as a proxy for leaf photosynthetic capacity. Glob. Chang. Biol. 23, 3513–3524. doi: 10.1111/gcb.13599
Dancer, J. E., and Rees, T. A. (1989). Phosphoribosyl pyrophosphate and the measurement on inorganic pyrophosphate in plant tissues. Planta 177, 261–264. doi: 10.1007/BF00392814
Davies, J. M., Poole, R. J., Rea, P. A., and Sanders, D. (1992). Potassium transport into plant vacuoles energized directly by a proton-pumping inorganic pyrophosphatase. Proc. Natl. Acad. Sci. U.S.A. 89, 11701–11705. doi: 10.1073/pnas.89.24.11701
de Graaf, B. H., Rudd, J. J., Wheeler, M. J., Perry, R. M., Bell, E. M., Osman, K., et al. (2006). Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature 444, 490–493. doi: 10.1038/nature05311
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G. H., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Devaux, P. F. (1991). Static and dynamic lipid asymmetry in cell membranes. Biochemistry 30, 1163–1173. doi: 10.1021/bi00219a001
Díaz, P., Betti, M., Sánchez, D. H., Udvardi, M. K., Monza, J., and Márquez, A. J. (2010). Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 188, 1001–1013. doi: 10.1111/j.1469-8137.2010.03440.x
Eamens, A. L., Smith, N. A., Dennis, E. S., Wassenegger, M., and Wang, M. B. (2014). In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450-451, 266–277. doi: 10.1016/j.virol.2013.12.019
Farré, E. M., Geigenberger, P., Willmitzer, L., and Trethewey, R. N. (2000). A possible role for pyrophosphate in the coordination of cytosolic and plastidial carbon metabolism within the potato tuber. Plant Physiol. 23, 681–688. doi: 10.1104/pp.123.2.681
Ferjani, A., Segami, S., Horiguchi, G., Muto, Y., Maeshima, M., and Tsukaya, H. (2011). Keep an eye on PPi: the vacuolar-type H+-pyrophosphatase regulates post germinative development in Arabidopsis. Plant Cell 23, 2895–2908. doi: 10.1105/tpc.111.085415
Ferrario-Méry, S., Masclaux, C., Suzuki, A., Valadier, M. H., Hirel, B., and Foyer, C. H. (2001). Glutamine and alpha-ketoglutarate are metabolite signals involved in nitrate reductase gene transcription in untransformed and transformed tobacco plants deficient in ferredoxin-glutamine- alpha-ketoglutarate aminotransferase. Planta 213, 265–271. doi: 10.1007/s004250000504
Fuentes, S. I., Allen, D. J., Ortiz-Lopez, A., and Hernández, G. (2001). Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J. Exp. Bot. 52, 1071–1081. doi: 10.1093/jexbot/52.358.1071
Geigenberger, P., Lerchi, J., Stitt, M., and Sonnewald, U. (1996). Phloem-specific expression of pyrophosphatase inhibits long distance transport of carbohydrates and amino acids in tobacco plants. Plant Cell Environ. 19, 43–55. doi: 10.1111/j.1365-3040.1996.tb00225.x
George, G. M., Merwe, M. J., Nunes-Nesi, A., Bauer, R., Fernie, A. R., Kossmann, J., et al. (2010). Virus-induced gene silencing of plastidial soluble inorganic pyrophosphatase impairs essential leaf anabolic pathways and reduces drought stress tolerance in Nicotiana benthamiana. Plant Physiol. 154, 55–66. doi: 10.1104/pp.110.157776
Gutiérrez-Luna, F. M., Hernández-Domínguez, E. E., Valencia-Turcotte, L. G., and Rodríguez-Sotres, R. (2018). Review: “pyrophosphate and pyrophosphatases in plants, their involvement in stress responses and their possible relationship to secondary metabolism”. Plant Sci. 267, 11–19. doi: 10.1016/j.plantsci.2017.10.016
Haefele, S. M., Nelson, A., and Hijmans, R. J. (2014). Soil quality and constraints in global rice production. Geoderma 235-236, 250–259. doi: 10.1016/j.geoderma.2014.07.019
Hänninen, T., Kontturi, E., and Vuorinen, T. (2011). Distribution of lignin and its coniferyl alcohol and coniferyl aldehyde groups in Piceaabies and Pinus sylvestris as observed by Raman imaging. Phytochemistry 72, 1889–1895. doi: 10.1016/j.phytochem.2011.05.005
He, R., Yu, G. H., Han, X. R., Han, J., Li, W., Wang, B., et al. (2017). ThPP1 gene, encodes an inorganic pyrophosphatase in Thellungiella halophila, enhanced the tolerance of the transgenic rice to alkaline stress. Plant Cell Rep. 36, 1929–1942. doi: 10.1007/s00299-017-2208-y
Hendrix, D. L. (1993). Rapid extraction and analysis of non-structural carbohydrates in plant tissues. Crop Sci. 33, 1306–1311. doi: 10.2135/cropsci1993.0011183X003300060037x
Hernández-Domíguez, E. E., Valencia-Turcotte, L. G., and Rodríguez-Sotres, R. (2012). Changes in expression of soluble inorganic pyrophosphatases of Phaseolus vulgaris under phosphate starvation. Plant Sci. 187, 39–48. doi: 10.1016/j.plantsci.2012.01.009
Hibino, S., and Choshi, T. (2001). Simple indole alkaloids and those with a non-rearranged monoterpenoid unit. Nat. Prod. Rep. 18, 66–87. doi: 10.1039/b004055j
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1017/S0031182002002585
Hoshida, H., Tanaka, Y., Hibino, T., Hayashi, Y., Tanaka, A., Takabe, T., et al. (2000). Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 43, 103–111. doi: 10.1023/A:1006408712416
Husain, S. R., Cillard, J., and Cillard, P. (1987). Hydroxyl radical scavenging activity of flavonoids. Phytochemistry 26, 2489–2491. doi: 10.1016/s0031-9422(00)83860-1
Jelitto, T., Sonnewald, U., Willmitzer, L., Hajirezeai, M., and Stitt, M. (1992). Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188, 238–244. doi: 10.1007/BF00216819
Jiang, Y. N., Haudenshield, J. S., and Hartman, G. L. (2012). Response of soybean fungal and oomycete pathogens to apigenin and genistein. Mycology 3, 153–157. doi: 10.1080/21501203.2012.684360
Jing, K., Tang, Y. W., Yao, C. Y., Chanona, E. A., Ling, X. P., and Zhang, D. D. (2018). Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: kinetic modeling and process scale-up. Biotechnol. Bioeng. 115, 371–381. doi: 10.1002/bit.26398
Jones, D. H. (1984). Phenylalanine ammonia-lyase: regulation of its induction, and its role in plant development. Phytochemisty 23, 1349–1359. doi: 10.1016/S0031-9422(00)80465-3
Kaminski, K. P., Kørup, K., Andersen, M. N., Sønderkær, M., Andersen, M. S., Kirk, H. G., et al. (2015). Cytosolic glutamine synthetase is important for photosynthetic efficiency and water use efficiency in potato as revealed by high-throughput sequencing QTL analysis. Theor. Appl. Genet. 128, 2143–2153. doi: 10.1007/s00122-015-2573-2
Khan, F., Upreti, P., Singh, R., Shukla, P. K., and Shirke, P. A. (2017). Physiological performance of two contrasting rice varieties under water stress. Physiol. Mol. Biol. Plants 23, 85–97. doi: 10.1007/s12298-016-0399-2
Kravchik, M., and Bernstein, N. (2013). Effects of salinity on the transcriptome of growing maize leaf cells point at cell-age specificity in the involvement of the antioxidative response in cell growth restriction. BMC Genom. 14:24. doi: 10.1186/1471-2164-14-24
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Villalobos, A., Dodds, P. F., and Hornung, R. (2011). Lauric acid improves the growth of zygotic coconut (Cocos nucifera L.) embryos in vitro. Plant Cell Tiss. Organ. Cult. 106, 317–327. doi: 10.1007/s11240-011-9924-8
Lv, B. S., Li, X. W., Ma, H. Y., Sun, Y., Wei, L. X., Jiang, C. J., et al. (2013). Differences in growth and physiology of rice in response to different saline-alkaline stress factors. Agron. J. 105, 1119–1128. doi: 10.2134/agronj2013.0017
Lv, S. L., Jiang, P., Nie, L. L., Chen, X. Y., Tai, F., Wang, D., et al. (2015). H(+)-pyrophosphatase from Salicornia europaea confers tolerance to simultaneously occurring salt stress and nitrogen deficiency in Arabidopsis and wheat. Plant Cell Environ. 38, 2433–2449. doi: 10.1111/pce.12557
May, A., Berger, S., Hertel, T., and Köck, M. (2011). The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim. Biophys. Acta 1810, 178–185. doi: 10.1016/j.bbagen.2010.11.006
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Montesinos-Pereira, D., Torre-González, A., Blasco, B., and Ruiz, J. M. (2018). Hydrogen sulphide increase the tolerance to alkalinity stress in cabbage plants (Brassica oleracea L. ’Bronco’). Sci. Hortic. 235, 349–356. doi: 10.1016/j.scienta.2018.03.021
Müller, R., Morant, M., Jarmer, H., Nilsson, L., and Nielsen, T. H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171. doi: 10.1104/pp.106.090167
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Oliveira, I. C., Brears, T., Knight, T. J., Clark, A., and Coruzzi, G. M. (2002). Overexpression of cytosolic glutamine synthetase. Relation to nitrogen, light, and photorespiration. Plant Physiol. 129, 1170–1180. doi: 10.1104/pp.020013
Osorio, S., Nunes-Nesi, A., Stratmann, M., and Fernie, A. R. (2013). Pyrophosphate levels strongly influence ascorbate and starch content in tomato fruit. Front. Plant Sci. 4:308. doi: 10.3389/fpls.2013.00308
Pang, S. S., and Duggleby, R. G. (1999). Expression, purification, characterisation, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry 38, 5222–5231. doi: 10.1021/bi983013m
Peñuelas, J., Ribas-Carbo, M., and Giles, L. (1996). Effects of allelochemicals on plant respiration and oxygen isotope fractionation by the alternative oxidase. J. Chem. Ecol. 22, 801–805. doi: 10.1007/BF02033587
Pietropaolo, A., Pierri, C. L., Palmieri, F., and Klingenberg, M. (2016). The switching mechanism of the mitochondrial ADP/ATP carrier explored by free-energy landscapes. Biochim. Biophys. Acta. 1857, 772–781. doi: 10.1016/j.bbabio.2016.02.006
Schulze, S., Mant, A., Kossmann, J., and Lloyd, J. R. (2004). Identification of an Arabidopsis inorganic pyrophosphatase capable of being imported into chloroplasts. FEBS Lett. 565, 101–105. doi: 10.1016/j.febslet.2004.03.080
Segami, S., Makino, S., Miyake, A., Asaoka, M., and Maeshima, M. (2014). Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell 26, 3416–3434. doi: 10.1105/tpc.114.127571
Smyth, D. A., and Black, C. C. (1984). Measurement of the pyrophosphate content of plant tissues. Plant Physiol. 75, 862–864.
Sonnewald, U. (1992). Expression of E.coli inorganic pyrophosphatase in transgenic plants alters photoassimilate partitioning. Plant J. 2, 571–581. doi: 10.1046/j.1365-313x.1992.t01-26-00999.x
Surova, L., Sherstneva, O., Vodeneev, V., Katicheva, L., Senina, M., and Sukhov, V. (2016). Variation potential-induced photosynthetic and respiratory changes increase ATP content in pea leaves. J. Plant Physiol. 202, 57–64. doi: 10.1016/j.jplph.2016.05.024
Székely, G., Abrahám, E., Cséplo, A., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Thimijan, R. W., and Royal, D. H. (1983). Photometric, radiometric, and quantum light units of measure: a review of procedures for interconversion. Hortscience 18, 818–822.
Tsai, J. Y., Kellosalo, J., Sun, Y. J., and Goldman, A. (2014). Proton/sodium pumping pyrophosphatases: the last of the primary ion pumps. Curr. Opin. Struct. Biol. 27, 38–47. doi: 10.1016/j.sbi.2014.03.007
Vanegas, D. C., Clark, G., Cannon, A. E., Roux, S., Chaturvedi, P., and Mclamore, E. S. (2015). A self-referencing biosensor for real-time monitoring of physiological ATP transport in plant systems. Biosens. Bioelectron. 74, 37–44. doi: 10.1016/j.bios.2015.05.027
Wang, C. S., Jiang, Q. T., Ma, J., Wang, X. Y., Wang, J. R., Chen, G. Y., et al. (2016). Characterization and expression analyses of the H+-pyrophosphatase gene in rye. J. Genet. 95, 565–572. doi: 10.1007/s12041-016-0664-8
Wu, H. H. (2018). Plant salt tolerance and Na+ sensing and transport. Crop J. 6, 215–225. doi: 10.1016/j.cj.2018.01.003
Wu, Z. H., Yang, C. W., and Yang, M. Y. (2014). Photosynthesis, photosystem II efficiency, amino acid metabolism and ion distribution in rice (Oryza sativa,L.) in response to alkaline stress. Photosynthetica 52, 157–160. doi: 10.1007/s11099-014-0002-4
Yang, C. W., Shi, D. C., and Wang, D. L. (2008). Comparative effects of salt and alkaline stresses on growth, osmotic adjustment and ionic balance of an alkaline-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 56, 179–190. doi: 10.1007/s10725-008-9299-y
Yao, M. H., Zeng, Y. L., Liu, L., Huang, Y. L., Zhao, E. F., and Zhang, F. C. (2012). Overexpression of the halophyte Kalidium foliatum H+-pyrophosphatase gene confers salt and drought tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 39, 7989–7996. doi: 10.1007/s11033-012-1645-5
Yemm, B. E. W., and Willis, A. J. (1954). The estimation of carbohydrate in plant extracts by anthrone. Biochem. J. 57, 508–514. doi: 10.1042/bj0570508
Zhang, H., Zhang, J. S., Lang, Z. B., Botella, J. R., and Zhu, J. K. (2017). Genome editing-principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36, 291–309. doi: 10.1080/07352689.2017.1402989
Keywords: alkaline stress, CRISPR/Cas9, inorganic pyrophosphatase, metabonomics, mutagenesis, OsPPa6 gene
Citation: Wang B, Xie G, Liu Z, He R, Han J, Huang S, Liu L and Cheng X (2019) Mutagenesis Reveals That the OsPPa6 Gene Is Required for Enhancing the Alkaline Tolerance in Rice. Front. Plant Sci. 10:759. doi: 10.3389/fpls.2019.00759
Received: 22 March 2019; Accepted: 24 May 2019;
Published: 11 June 2019.
Edited by:
Paulo Arruda, University of Campinas, BrazilReviewed by:
Suriyan Cha-um, National Science and Technology Development Agency (NSTDA), ThailandKazuo Nakashima, Japan International Research Center for Agricultural Sciences, Japan
Copyright © 2019 Wang, Xie, Liu, He, Han, Huang, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianguo Cheng, Y2hlbmd4aWFuZ3VvQGNhYXMuY24=
†Present address: Jiao Han, Guangling No. 5 Middle School, Shanxi, China
 Bing Wang
Bing Wang Guoqiang Xie3
Guoqiang Xie3 Laihua Liu
Laihua Liu Xianguo Cheng
Xianguo Cheng