- 1Division of Life Sciences, Korea University, Seoul, South Korea
- 2Division of Life Science, Chonbuk National University, Jeonju, South Korea
During pre-mRNA splicing, U2 small nuclear ribonucleoprotein auxiliary factor 65 (U2AF65) interacts with U2AF35 and splicing factor 1 (SF1), allowing for the recognition of the 3′-splice site by the ternary complex. The functional characterization of U2AF65 homologs has not been performed in Arabidopsis thaliana yet. Here, we show that normal plant development, including floral transition, and male gametophyte development, requires two Arabidopsis U2AF65 isoforms (AtU2AF65a and AtU2AF65b). Loss-of-function mutants of these two isoforms displayed opposite flowering phenotypes: atu2af65a mutants showed late flowering, whereas atu2af65b mutants were characterized by slightly early flowering, as compared to that in the wild-type (Col-0) plants. These abnormal flowering phenotypes were well-correlated with the expression patterns of the flowering time genes such as FLOWERING LOCUS C (FLC) and FLOWERING LOCUS T (FT). However, the two atu2af65 mutants did not display any morphological abnormalities or alterations in abiotic stress tests. Double mutation of the AtU2AF65a and AtU2AF65b genes resulted in non-viable seeds due to defective male gametophyte. In vitro pollen germination test revealed that mutations in both AtU2AF65a and AtU2AF65b genes significantly impaired pollen tube growth. Collectively, our findings suggest that two protein isoforms of AtU2AF65 are differentially involved in regulating flowering time and display a redundant role in pollen tube growth.
Introduction
In eukaryotic cells, pre-messenger RNA (pre-mRNA) splicing is important for processes such as expression of intron-containing genes, remodeling of protein-protein interaction networks, and regulation of transcript abundance (Nilsen and Graveley, 2010). Spliceosomes, consisting of five small nuclear ribonucleoproteins (snRNPs) and numerous non-snRNPs, mediate these key cellular functions (Wahl et al., 2009). The U2 snRNP auxiliary factor (U2AF) is one such non-snRNP component. In mammalian cells, U2AF is composed of two subunits, U2AF65 and U2AF35, representing the large and the small subunits, respectively. During assembly of the spliceosome, U2AF65 directly binds to the polypyrimidine (Py) tracts upon interaction with SF1 through its third RNA recognition motif (RRM) (Rain et al., 1998; Thickman et al., 2006). Meanwhile, U2AF35 binds to the 3′-splice sites and may also promote the binding of U2AF65 to the Py tract through its interaction with the U2AF65 and serine-arginine (SR)-rich proteins (Zuo and Maniatis, 1996). These molecular interactions are essential for the initial recognition of the intronic 5′- and 3′-splice sites by the spliceosome. Although plant introns contain neither conserved branch-point sequences nor Py tracts (Luehrsen et al., 1994; Simpson and Filipowicz, 1996), similar interactions found in mammals may also be present in plants. Two U2AF65 homologs purified from wild tobacco (Nicotiana plumbaginifolia) can rescue the in vitro splicing of adenovirus pre-mRNA in the extracts of U2AF factor-deficient HeLa cells (Domon et al., 1998). In addition, the Arabidopsis genome has U2AF35, U2AF65, and SF1 homologs (Wang and Brendel, 2006; Jang et al., 2014), and Arabidopsis U2AF65 (AtU2AF65) and SF1 (AtSF1) proteins interact with their rice homologs (Jang et al., 2014; Park et al., 2017). These results suggest that the interactions among splicing factors during intron 3′-splice site recognition is conserved in plants; however, plant introns do not contain conserved regions such as the branch point sequences and Py tracts at the 3′-splice proximal sites found in mammalians.
Although the detailed splicing mechanism is poorly understood in plants, some components of the plant spliceosome play essential roles in plant growth and development, such as in the flowering time. Mutations in AtU2AF35a or AtU2AF35b result in pleiotropic phenotypes including altered flowering time and abnormal morphological defects in leaves, flowers, and siliques (Wang and Brendel, 2006). A loss of AtSF1 activity also causes pleiotropic developmental defects such as early flowering and hypersensitivity to abscisic acid (ABA) (Jang et al., 2014). Furthermore, these mutations in spliceosomal components result in altered splicing patterns of specific genes in plants. For instance, altered AtU2AF35 expression generates novel splicing isoforms of the FCA pre-mRNA due to the splicing of non-canonical introns (Wang and Brendel, 2006). The mutation of AtSF1 affects the alternative splicing pattern of the heat shock transcription factor HsfA2 (Jang et al., 2014; Lee et al., 2017). These results suggest that lesions in spliceosomal components affect alternative splicing of a specific set of genes. Recent reports have also shown that ambient temperature shifts lead to the alternative splicing of splicing-related genes (Verhage et al., 2017) and the cyclin-dependent kinase G1 (CDKG1) affects the alternative splicing of AtU2AF65a in response to the changes in ambient temperature (Cavallari et al., 2018). These results suggest that the components of the spliceosome may be the target of ambient temperature-induced alternative splicing, thereby coordinating plant growth and development under continuously fluctuating temperature conditions.
The mutations in spliceosomal components also result in defects in female- or male-gametophytes. Loss-of-function mutations in genes encoding components of snRNPs such as the U2 snRNP-specific proteins ATROPOS (ATO)/Prp9 (Moll et al., 2008) and SF3b130 (Aki et al., 2011), the U5 snRNP-specific protein GAMETOPHYTIC FACTOR 1 (GFA1)/CLOTHO (CLO)/VAJRA (VAJ)/Snu114 (Moll et al., 2008; Yagi et al., 2009), and the U4/U6 snRNP-specific protein LACHESIS (LIS)/Prp4 (Gross-Hardt et al., 2007; Volz et al., 2012) lead to abolished or impaired genetic transmission through the female gametophyte. Mutations in the genes for the DEAH-box RNA-dependent ATPases Prp22/ROOT INITIATION DEFECTIVE1 (RID1) and Prp16/CLUMSY VEIN (CUV) cause defects in the female and male gametophyte, respectively (Ohtani et al., 2013; Tsugeki et al., 2015). Notably, mutants of ATO, SF3b130, GFA1/CLO/VAJ, and RID1 show less severe defects in male gametophyte development. However, a lesion in the SHOOT REDIFFERENTIATION DEFECTIVE 2 (SRD2) gene, causing defective expression of all the snRNAs, does not result in abnormal transmission through the male gametophyte (Onouchi et al., 2000; Ohtani and Sugiyama, 2005; Onodera et al., 2008). These results suggest that loss-of-activity mutations of spliceosomal components differentially affect male and female gametophyte development.
Although U2AF65 genes have been identified in plants such as Arabidopsis, rice, and tobacco (Gniadkowski et al., 1996; Domon et al., 1998), the function of U2AF65 in plants has not yet been reported. Four U2AF65 homologs are predicted in the Arabidopsis genome and only two of these (hereafter defined as AtU2AF65a and AtU2AF65b) have a U2AF homology motif (UHM), i.e., the third RNA recognition motif (RRM3) (Wang and Brendel, 2006; Jang et al., 2014). Here we show that defects in AtU2AF65a and AtU2AF65b may cause abnormal flowering time and pollen tube growth during the male gametophyte development.
Materials and Methods
Plant Materials and Growth Conditions
All mutants used in this study were Arabidopsis plants in the Columbia (Col-0) background, except for fy-1 (Ler) (Simpson et al., 2003; Henderson et al., 2005). Various T-DNA alleles of AtU2AF65a (SALK_075828, SALK_128420C, SALK_142561, and SALK_144790) and AtU2AF65b (SALK_055049C and SALK_104128) were obtained from the Arabidopsis Biological Resource Center (ABRC) (Alonso et al., 2003). The T-DNA homozygous lines and the double mutants used in this study were verified by DNA-based PCR genotyping using gene-specific and T-DNA specific primers. The oligonucleotide sequences used for genotyping are provided in Supplementary Table S1. The plants were grown in Sunshine Mix 5 (Sun Gro Horticulture, United States) or Murashige and Skoog (MS) medium at 23 or 16°C under long-day (LD) [16/8 h (light/dark)] or short-day (SD) [8/16 h (light/dark)] conditions with light supplied at an intensity of 120 μmol m-2 s-1.
Plasmid Construction
To check the β-glucuronidase (GUS) expression patterns driven under AtU2AF35 and AtU2AF65 promoter, we generated the pAtU2AF35a0.9 kb::GUS, pAtU2AF35b0.7 kb::GUS, pAtU2AF65a1.3 kb::GUS, and pAtU2AF65b1.5 kb::GUS constructs. For AtU2AF35a, the genomic region from 876 nucleotides (nt) before the ATG start codon was cloned, as previously described (Wang and Brendel, 2006). For AtU2AF35b, the genomic region from 700 nt before the start codon was newly cloned, because the previous report has shown that the promoter region of different lengths (555 and 982 nt) have similar GUS staining patterns (Wang and Brendel, 2006). For AtU2AF65a, the cloned region is from 1,300 nt before the start codon and includes the 5′-untranslated region (UTR) region of AtU2AF65a, and the 3′-UTR region and part of the exon of At4G36700. For AtU2AF65b, we had tried to clone constructs with the promoter region of different lengths, which begin 1,500 nt before the start codon, because this gene has an intergenic region (approximately 6,800 nt). However, we were not able to detect GUS staining-positive clones from any of the promoter-GUS constructs. We are going to characterize this unusual trait of the AtU2AF65b promoter in the near future. We amplified these promoter regions of each gene using specific primers and cloned them into the pGWB433 vector (gift from Tsuyoshi Nakagawa at the Research Institute of Molecular Genetics, Shimane University, Japan). To make the pAtU2AF65a890 bp::AtU2AF65a construct, the full-length coding sequence (CDS) of AtU2AF65a and the genomic region from 890 nt before the start codon including the 5′-UTR region of AtU2AF65a and the 3′-UTR region of At4G36700 were amplified and cloned into a pCHF3 vector. The reason we used the shorter AtU2AF65a promoter for the complementation test is that we wanted to find out the shorter promoter required for AtU2AF65a’s function. The construct with 890 nt promoter construct was our first and single one in order to check its usefulness for the complementation test. To produce the p35S::AtU2AF65b construct, the full-length CDS of AtU2AF65b was amplified and cloned into a pCHF3 vector. The resulting recombinant plasmid was sequenced to verify the absence of PCR errors during amplification. The oligonucleotide sequences used in this study are listed in Supplementary Table S1.
Generation of Transgenic Plants and Measurement of Flowering Time
Transgenic plants were generated using the floral dip method with minor modifications (Weigel and Glazebrook, 2002). The Agrobacterium tumefaciens strain GV3101, harboring the gene constructs, was infiltrated into the AtU2AF65 mutant backgrounds. Transgenic seedlings were first selected for pCHF3 vector content using kanamycin and then verified by PCR-based genotyping. A number of T1 seedlings between 20 and 30 were analyzed for each construct.
To assess the flowering time, the total numbers of rosette and cauline leaves of at least five or six independent transgenic lines (at least 16 individual plants per transgenic line) were counted in the T2 or T3 generation (when we checked flowering time in T3 generation, all experiments were repeated three times). To determine whether the flowering time of the transgenic plants significantly differed from that of the wild-type (Col-0) plants, the data were analyzed using the SPSS software version 12.0 (Michaels, 2009). The error bars indicate standard error of the mean (SEM) of three biological replicates consisting of independently harvested samples. The leaf number ratio (16°C/23°C, LNR) under LD conditions was used as an indicator of temperature-responsive flowering (Lee et al., 2007). A hypothetical temperature-sensitive plant produces a different total number of leaves at 23 and 16°C; thus, its LNR is near 2.0.
Germination Assays Under Hormone Treatment or Abiotic Stress Conditions
The sensitivity of the plants to hormones and abiotic treatments was determined as previously described (Hugouvieux et al., 2001). Seeds were sown on half-strength MS medium containing 1% sucrose, and the MS medium was maintained at 4°C for 2 days in the dark, after which it was transferred to a normal growth chamber kept at 23°C under LD conditions at a light intensity of 120 μmol m-2 s-1. Seed germination was analyzed in MS medium supplemented with various concentrations of ABA, NaCl, and mannitol. The germination rate of the seeds was measured at the indicated days. All experiments were repeated three times.
RNA Expression Analyses
Thirty micrograms of total RNA was used for northern blot analysis as described previously (Jang et al., 2014), and 32P-labeled AtU2AF65a and AtU2AF65b cDNA, which are approximately 500 nt fragment of CDSs starting from the ATG start codon in both AtU2AF65a and AtU2AF65b, were used as probes due to the unique nucleotide sequences for each gene. For reverse transcription-polymerase chain reaction (RT-PCR) and real-time quantitative PCR (RT-qPCR) analyses, total RNA was extracted from a variety of tissues or from 8-day-old whole seedlings of Arabidopsis plants using QIAzol Lysis reagent (Qiagen, Germany). The RNA quality was determined using a Nanodrop ND-2000 spectrophotometer (Nanodrop Technologies, Waltham, MA, United States), and only the high-quality RNA samples (A260/A230 > 2.0 and A260/A280 > 1.8) were used for subsequent experiments. The first-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA in accordance with the manufacturer’s instructions (Invitrogen, United States). The RT-qPCR analysis was carried out in 384-well plates with a LightCycler 480 (Roche Applied Science, United States) using SYBR green. RT-qPCR experiments were carried out using Roche SYBR Green Master mixture (Roche Applied Science, United States). In accordance with the best established RT-qPCR practices (Hong et al., 2010), PP2AA3 was included as a reference gene due to its stable expression in Arabidopsis. All RT-PCR and RT-qPCR experiments were performed in three biological replicates consisting of independently harvested samples with three technical replicates each. The error bars indicate the SEM of three biological replicates consisting of independently harvested samples. The oligonucleotide sequences used for expression analysis are provided in Supplementary Table S1. Changes in gene expression were calculated via the ΔΔCT method (Ramakers et al., 2003). For GUS expression analysis, GUS staining was carried out as described by Chen et al. (1998).
Characterization of Pollen Grains and Pollen Tubes
Pollen viability was assayed as described by Alexander (1969). For in vitro pollen tube growth assay, the pollen grains from mutant and wild-type flowers were plated onto the surface of agar plates and cultured at 23°C with a relative humidity of 100% for 3 h and then examined using a light microscope (OLYMPUS model BX-51). The detailed procedure has been previously reported (Jiang et al., 2005). All experiments were repeated three times. To determine whether the proportion data such as pollen morphology and pollen tube length significantly differ between mutants and wild-type (Col-0) plants, the data were analyzed by χ2 test.
Results
Isolation and Characterization of the Single Mutants of AtU2AF65a and AtU2AF65b
We have recently revealed that AtSF1 and AtU2AF65 interact with each other and that the alteration of AtSF1 activity affects the flowering time (Jang et al., 2014; Lee et al., 2017). Therefore, we analyzed the phenotypes of the T-DNA alleles of AtU2AF65a and AtU2AF65b to study their biological function. We obtained four T-DNA alleles (SALK_128420C, SALK_142561, SALK_075828, and SALK_144790) for AtU2AF65a and two T-DNA alleles (SALK_055049C and SALK_104128) for AtU2AF65b to isolate homozygous T-DNA insertional mutants for each gene. PCR genotyping and sequencing analyses confirmed the T-DNA insertion sites of each T-DNA allele (Figures 1A,B). They were renamed atu2af65a-1 (SALK_128420C), atu2af65a-2 (SALK_142561), atu2af65a-3 (SALK_075828), atu2af65a-4 (SALK_144790), atu2af65b-1 (SALK_055049C), and atu2af65b-2 (SALK_104128), respectively. Northern blot analysis revealed that AtU2AF65a and AtU2AF65b expression did not occur in atu2af65a-4 and atu2af65b-1 mutants, respectively (Figures 1A,B), whereas the other AtU2AF65a and AtU2AF65b T-DNA alleles displayed significantly lower or similar expression than the wild-type (Col-0) plants, suggesting that they were knock-down mutants. We also performed RT-PCR analyses using primer sets that spanned the T-DNA insertion sites in three atu2af65a and two atu2af65b mutant alleles. PCR fragments of the expected size were detected in wild-type (Col-0) plants, but no amplification was observed for the regions spanning the T-DNA insertion sites in these mutant alleles (Figure 1C). Based on these observations, we used the atu2af65a-4 and atu2af65b-1 knock-out mutants for further studies. As shown in Figure 2, the single atu2af65a and atu2af65b mutants did not show major defects in growth and development, except for flowering. The flowering time of atu2af65a-4 mutants was delayed at 23°C under LD conditions as compared to that of wild-type (Col-0) plants, whereas that of atu2af65b-1 mutants was slightly accelerated [atu2af65a-4, atu2af65b-1, and wild-type (Col-0) plants = 23.7, 14.0, and 17.2 leaves, respectively)]. These flowering phenotypes of atu2af65a and atu2af65b mutants were consistent with the findings (Verhage et al., 2017; Xiong et al., 2019). Notably, the expression of the AtU2AF65a gene (pAtU2AF65a890 bp::AtU2AF65a) in the atu2af65a-4 mutant background rescued the mutant phenotype, whereas AtU2AF65b overexpression (p35S::AtU2AF65b) in the atu2af65b-1 mutant background was not able to restore the wild-type (Col-0) phenotype (Supplementary Figures S1, S2). In order to establish whether different light or temperature conditions affect the flowering time, we further examined the impact on this parameter of plant incubation either at 23°C under SD or at 16°C under LD conditions. The flowering time phenotypes of these mutants were not significantly affected by the change in light/temperature conditions and the relative ratios remained substantially unchanged. For instance, atu2af65a-4 plants produced more leaves at 16°C (37.9 leaves) than at 23°C (23.2 leaves) (LNR = 1.6). Under same conditions, wild-type (Col-0) plants exhibited a delayed flowering time [16°C (28.7 leaves) vs. 23°C (16.1 leaves); LNR = 1.8]. This indicated that the flowering of atu2af65a-4 plants was sensitive to differences in temperature. More specifically, at 23°C under SD conditions, the leaf counts were 67.3, 45.6, and 54.9 for atu2af65a-4, atu2af65b-1, and wild-type (Col-0) plants, respectively. Furthermore, vernalization treatment showed that both the atu2af65 mutants and wild-type (Col-0) plants responded similarly to this condition (Supplementary Figure S3). However, the atu2af65a-4 and atu2af65b-1 single mutants did not respond to several abiotic stresses including exposure to various concentrations of ABA, NaCl, and mannitol (Supplementary Figure S4). These flowering time data indicated that AtU2AF65a and AtU2AF65b differentially affected the flowering time control in Arabidopsis.
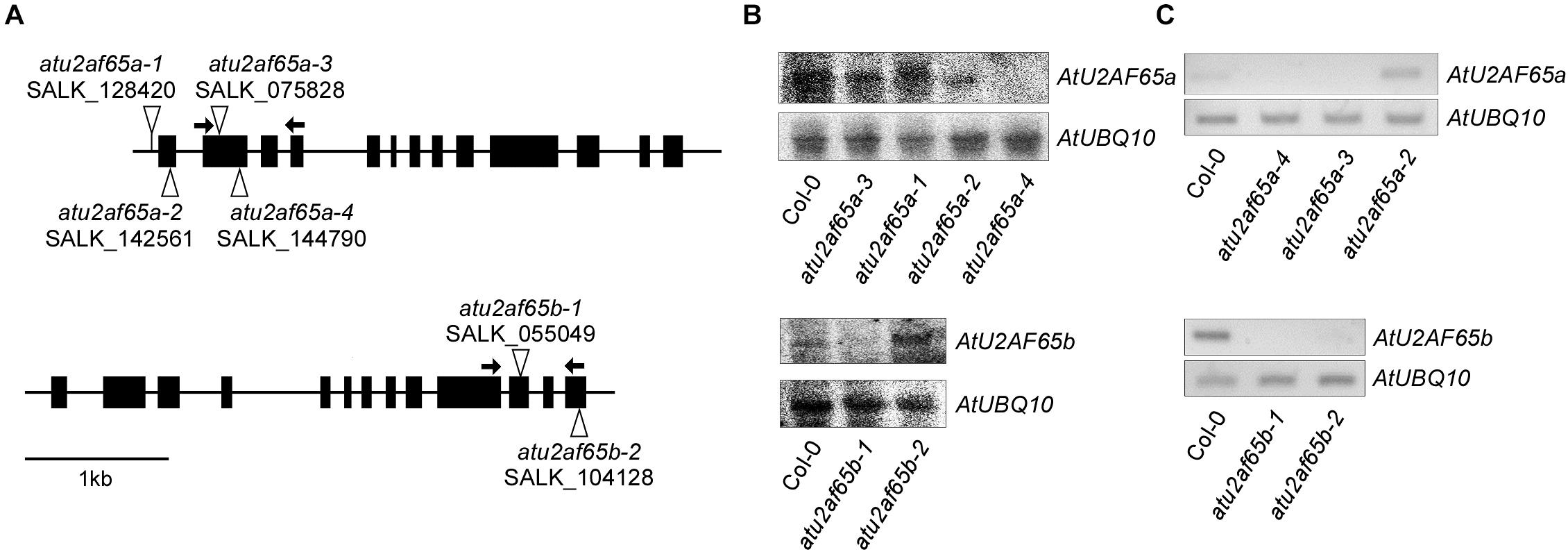
Figure 1. Characterization of T-DNA insertion alleles of AtU2AF65a and AtU2AF65b. (A) Schematic diagrams of T-DNA insertions in atu2af65a and atu2af65b mutants. The triangles and the black boxes indicate the T-DNA insertions and the exons of AtU2AF65a and AtU2AF65b genes, respectively. The arrows indicate the primers used for the RT-PCR analysis to detect the transcripts from each T-DNA mutant allele. (B) Northern blot analysis of several T-DNA alleles in atu2af65a and atu2af65b mutants. The expression levels of the AtU2AF65a and AtU2AF65b genes were checked in 10-day-old SALK T-DNA allele lines and wild-type (Col-0) plants grown at 23°C under LD conditions. The location of probes used in northern blot analysis was as follows: approximately 500 nucleotides from the start codon of the coding sequence (CDS) of both AtU2AF65a and AtU2AF65b. AtUBQ10 expression served as a loading control. (C) RT-PCR analysis of several T-DNA alleles in atu2af65a and atu2af65b mutants.
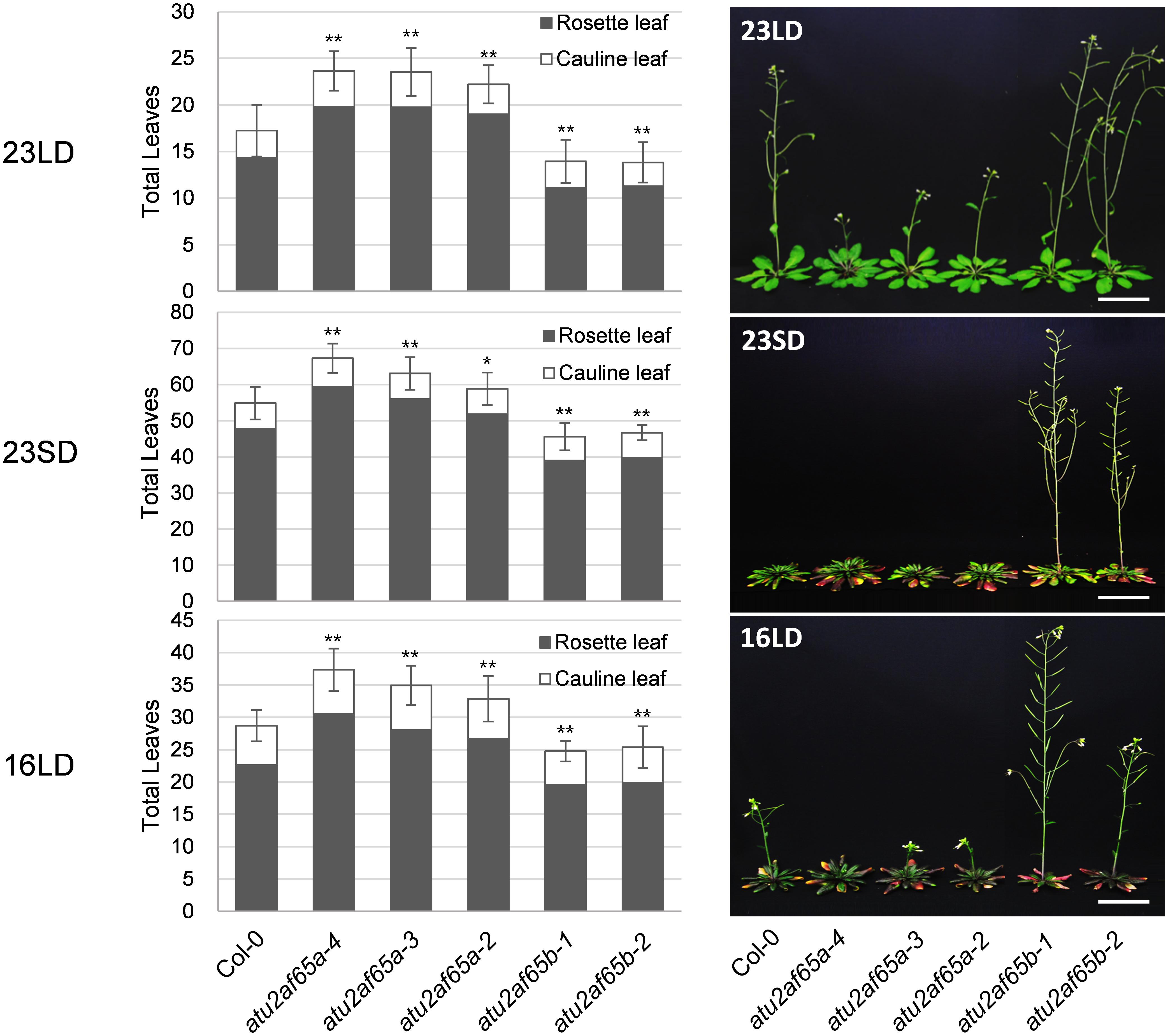
Figure 2. Flowering phenotypes of atu2af65a and atu2af65b mutants under different growth conditions. Plants were grown at 23°C under long-day (LD) condition (23LD), at 23°C under short-day (SD) condition (23SD) or at 16°C under long-day (LD) condition (16LD). The number of total leaves (rosette plus cauline leaves) shown in the bar graphs represents the flowering phenotype of each plant. As a control, the wild-type (Col-0) plants were used. The photos were taken upon atu2af65b mutant flowering. The error bars indicate the standard error of the mean (SEM) of three biological replicates consisting of independent samples. The asterisks denote a significant difference in flowering time between transgenic and wild-type (Col-0) plants (Student’s t-test, ∗P < 0.05, ∗∗P < 0.01). Scale bars 3 cm.
Expression Patterns of AtU2AF65a and AtU2AF65b
We analyzed the expression patterns of endogenous AtU2AF65a and AtU2AF65b by RT-PCR analysis. The AtU2AF65a and AtU2AF65b transcripts were abundantly expressed in a variety of tissues (Figures 3A,B) and their expression levels were very similar in most tissues, such as vegetative tissues and flowers. However, the expression level of AtU2AF65a was approximately twofold higher than that of AtU2AF65b in siliques. These ubiquitous expression patterns were similar to those of other splicing factor genes such as AtSF1 and AtU2AF35 in Arabidopsis (Wang and Brendel, 2006; Jang et al., 2014).
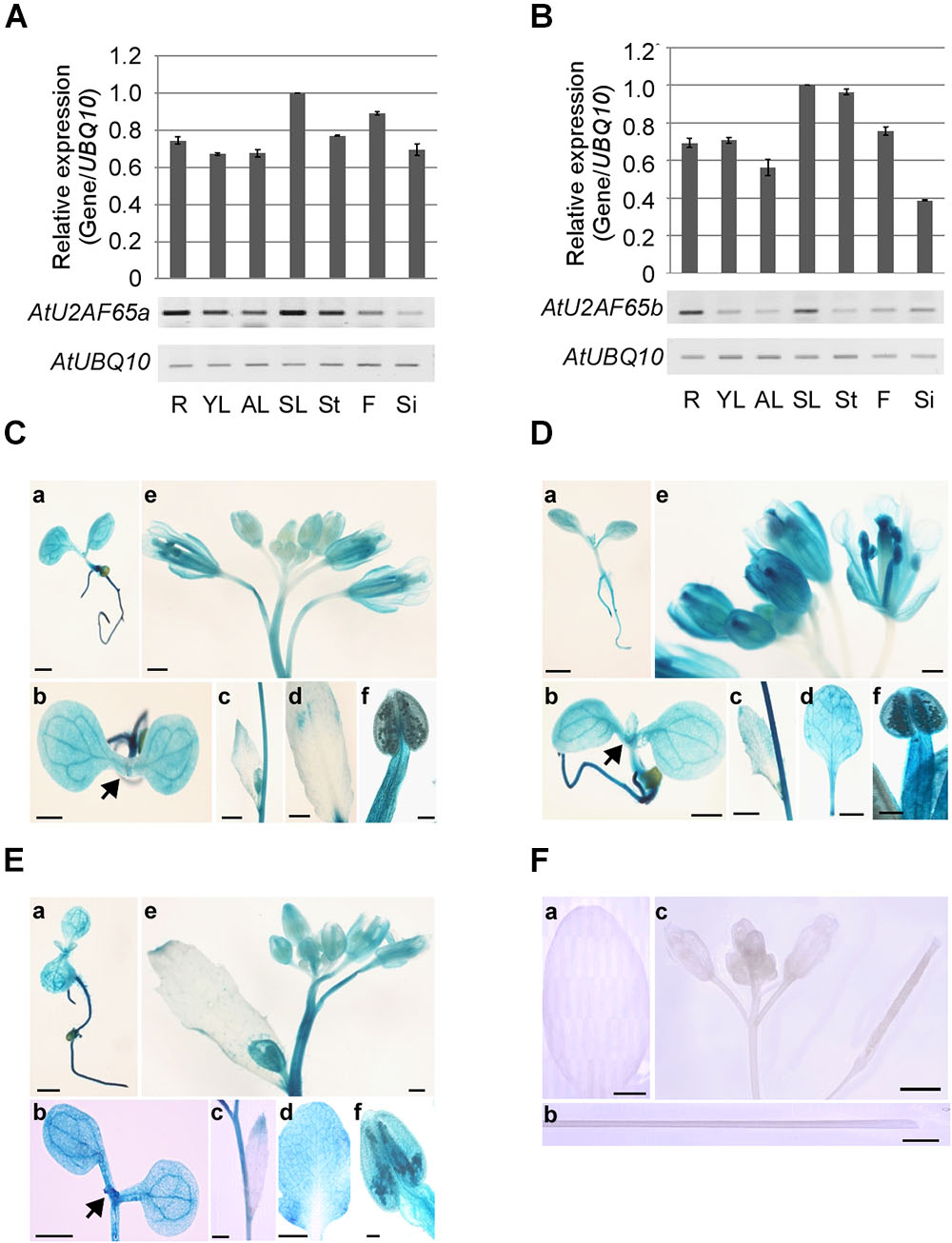
Figure 3. Expression patterns of AtU2AF65a and AtU2AF65b genes. (A,B) RT-PCR analysis of AtU2AF65a and AtU2AF65b expression in a variety of tissues [roots (R), young leaves (YL), adult leaves (AL), senescent leaves (SL), stems (St), flowers (F), and siliques (Si)] of wild-type (Col-0) plants grown at 23°C under LD conditions. The expression level of each AtU2AF65 gene in SL was set to 1. AtUBQ10 expression served as a loading control. The error bars indicate the SEM of three biological replicates consisting of independently harvested samples, with technical replicates for each sample. (C–F) Histochemical GUS staining in pAtU2AF35a0.9 kb::GUS (C), pAtU2AF35b0.7 kb::GUS (D), pAtU2AF65a1.3 kb::GUS (E), and GUS (F) plants under LD conditions. GUS staining was performed in a variety of tissues [seedlings (a,b), cauline leaves (c), adult leaves (d), inflorescences (e), and stigmas (f)]. Arrows indicate shoot apex regions. GUS expression was not detected in transgenic plants harboring GUS constructs without a promoter region [F, adult leaves (a), stems (b), and inflorescences (c)]. Scale bars 100 μm. Note that the GUS expression patterns of pAtU2AF35b0.7 kb::GUS were similar to those of pAtU2AF35b550 bp::GUS or pAtU2AF35b982 bp::GUS, as previously described (Wang and Brendel, 2006).
In order to investigate the spatial expression patterns of AtU2AF65a and AtU2AF65b in more detail, we analyzed β-glucuronidase (GUS) activity in transgenic pAtU2AF65a1.3 kb::GUS and pAtU2AF65b1.5 kb::GUS plants, harboring regions of about 1.3 and 1.5 kb upstream of the ATG of AtU2AF65, respectively. We also made pAtU2AF35a0.9 kb::GUS and pAtU2AF35b0.7 kb::GUS plants harboring about 0.9 or 0.7 kb regions upstream of the AtU2AF35 ATG, to compare the GUS staining patterns between AtU2AF65 and AtU2AF35. We were able to detect GUS activities in all the transgenic plants except for the pAtU2AF65b::GUS plants, because our attempt to clone the promoter of the AtU2AF65b gene was not successful. The spatial expression patterns of the GUS activities were very similar in most tissues. Strong GUS activities were detected in all parts of the young seedlings including shoot apex regions (Figures 3Ca,Cb,Da,Db,Ea,Eb). Older leaves also showed intense GUS staining in the pAtU2AF65a1.3 kb::GUS and pAtU2AF35b0.7 kb::GUS plants (Figures 3Dc,Dd,Ec,Ed). Particularly high GUS expression was observed in some parts of the flowers such as sepals, stamens, anthers, pollen, and stigma (Figures 3Ce,Cf,De,Df,Ee,Ef). Petals showed weak GUS expression. However, GUS activities were not observed in the tissues of the transgenic plants expressing only GUS constructs (Figure 3F). In general, the GUS staining patterns were consistent with the results of RT-PCR analysis (Figures 3A,B) and with the findings in pAtSF1::GUS and pAtU2AF35::GUS plants (Wang and Brendel, 2006; Jang et al., 2014).
Expression of Flowering Time Genes in atu2af65 Mutants
To examine whether the observed changes in the flowering phenotype of the atu2af65a-4 and atu2af65b-1 mutants were correlated with altered expression of flowering time genes, we performed RT-PCR and RT-qPCR analyses of the 8-day-old whole seedlings grown at 23°C under LD conditions. The expression of FLOWERING LOCUS C (FLC), SHORT VEGETATIVE PHASE (SVP), and FLOWERING LOCUS M-β (FLM-β) significantly increased in the atu2af65a-4 mutants, whereas the expression of only FLC was selectively decreased in the atu2af65b-1 mutants (Figure 4A). However, the expression of FLM-δ as another FLM spliced form was not altered in the atu2af65a-4 mutants. Moreover, the expression levels of FLOWERING LOCUS T (FT), which is a major downstream target of FLC, SVP, and FLM (Lee et al., 2013; Pose et al., 2013), were significantly altered (Figure 4B). We also observed changes in the spliced or unspliced isoforms of the FLC transcripts in the atu2af65a-4 and atu2af65b-1 mutants (Supplementary Figure S5), indicating that the altered alternative splicing of these FLC transcripts affected the changes of major FLC transcripts in these mutants. The reduced expression levels and altered splicing patterns of FLC observed in atu2af65b-1 mutants were consistent with the recent report (Xiong et al., 2019). However, we did not find significant changes in the alternative splicing of FCA or SVP transcripts (Wang and Brendel, 2006).
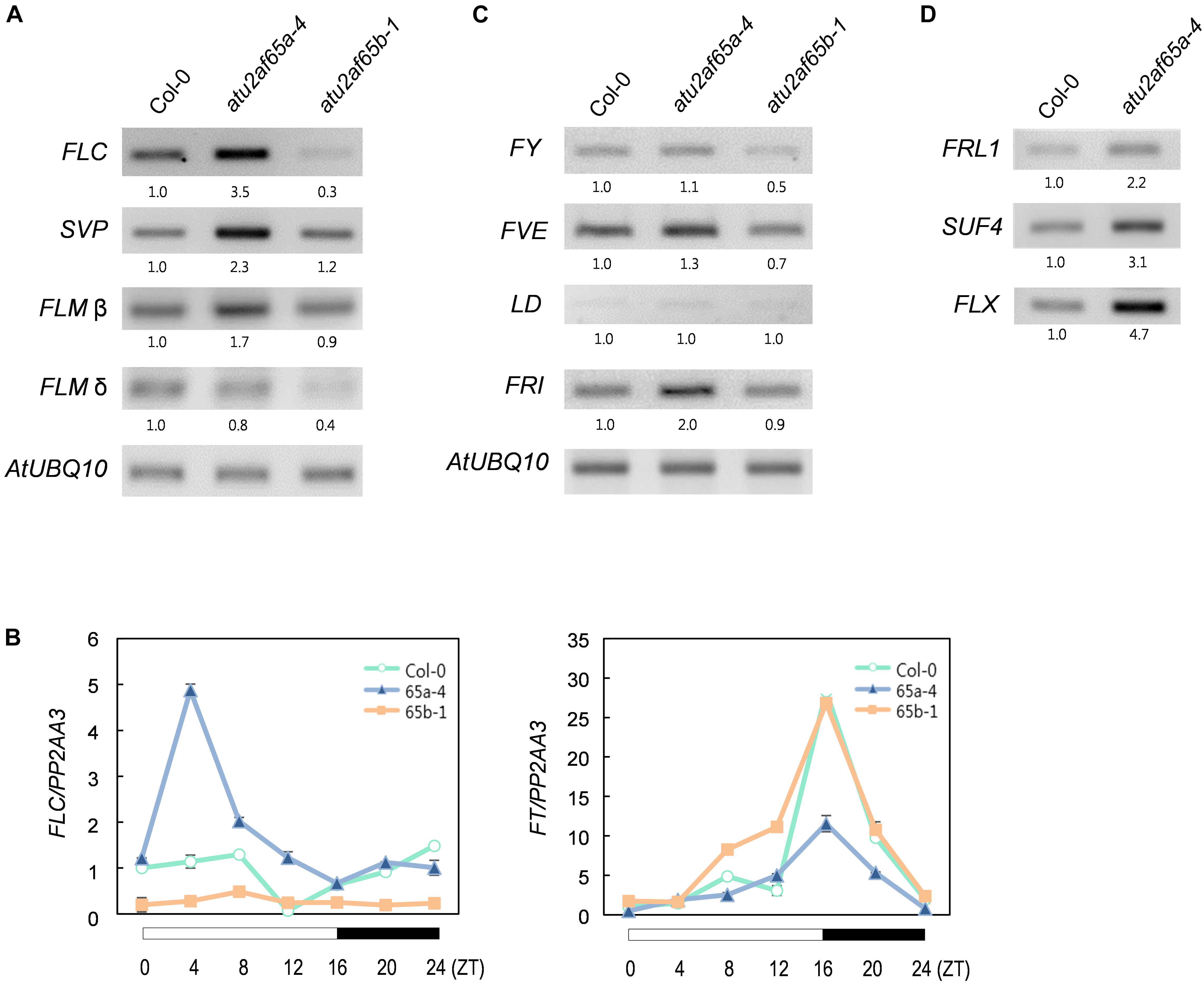
Figure 4. Expression of flowering time genes in the atu2af65a and atu2af65b mutants. (A) RT-PCR analysis of FLC, FLM, and SVP expression in the atu2af65a-4 and atu2af65b-1 mutants grown at 23°C under LD conditions. AtUBQ10 expression served as a loading control. The numbers below the gels denote the fold change relative to wild-type (Col-0) plants. (B) RT-qPCR analysis of FLC and FT expression in the atu2af65a-4 and atu2af65b-1 mutants grown at 23°C under LD conditions. PP2AA3 expression served as a loading control. The error bars indicate the SEM of three biological replicates consisting of independently harvested samples, with technical replicates for each sample. (C) RT-PCR analysis of FY, FVE, LD, and FRI expression in the atu2af65a-4 and atu2af65b-1 mutants grown at 23°C under LD conditions. (D) RT-PCR analysis of FRL1, SUF4, and FLX expression in the atu2af65a-4 and atu2af65b-1 mutants grown at 23°C under LD conditions. AtUBQ10 expression was shown in (C).
We next investigated the expression of negative and positive regulators of FLC in atu2af65 mutants. To this end, the transcriptional profiles of FLOWERING CONTROL LOCUS A (FCA), FY, FVE, LUMINIDEPENDENS (LD) (negative FLC regulators) as well as FRIGIDA (FRI) (a positive FLC regulator) were addressed (Ratcliffe et al., 2001). Although minor alterations in FY and FVE expression were observed in the atu2af65a-4 or atu2af65b-1 mutants (Figure 4C), these changes were too modest to explain the altered flowering phenotype shown by the atu2af65 mutants (Figure 2). Interestingly, FRI expression was significantly increased only in the atu2af65a-4 mutants. Because the genetic background of atu2af65a-4 mutants is Col-0, which includes a weak FRI allele (Gazzani et al., 2003), we further checked the expression levels of three other FLC-specific regulators (Choi et al., 2011), i.e., FRIGIDA LIKE1 (FRL1), SUPPRESSOR OF FRIGIDA4 (SUF4), and FLC EXPRESSOR (FLX). Their strong expression also observed in the atu2af65a-4 mutants (Figure 4D). Collectively, these results suggest that the late flowering phenotype of atu2af65a-4 mutants is likely due to increased FLC activity in an FRI complex-dependent manner, whereas the early flowering phenotype of atu2af65b-1 mutants is likely caused by decreased FLC activity in an FRI complex-independent manner.
The atu2af65 Mutations Are Defective in Male Gametophyte Function
Because the atu2af65a and atu2af65b single mutants did not display noticeable defective phenotypes, except for flowering time (Figure 2), we decided to investigate the phenotype of atu2af65a atu2af65b double mutants. To generate these double mutants, the atu2af65a-4 homozygous mutants, either as male or female parents, were crossed with the atu2af65b-1 homozygous mutants. Genetic crossing was also performed using the atu2af65a-3 homozygous mutants, as a source of knock-down allele. The resulting F1 progeny [+/atu2af65a-4;+/atu2af65b-1 (65A/65a;65B/65b) or +/atu2af65a-3;+/atu2af65b-1] was subsequently allowed to self-pollinate and the genotypes of the F2 progeny were determined by PCR-based genotyping. No progeny of homozygous double mutants [atu2af65a-4/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65a/65a;65b/65b) or atu2af65a-3/atu2af65a-3;atu2af65b-1/atu2af65b-1] was identified, suggesting lethality of the homozygous double mutation as well as redundancy of AtU2AF65a and AtU2AF65b for plant growth and development. Only the heterozygous plants for one mutant allele and homozygous for the other [+/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) and atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b), or +/atu2af65a-3;atu2af65b-1/atu2af65b-1 and atu2af65a-3/atu2af65a-3;+/atu2af65b-1] were found in the F2 progeny. When these heterozygous double mutants were self-fertilized, no plants with the homozygous double mutants were identified in the progeny. Figure 5A shows the seed phenotype observed in the siliques of various combinations of mutants in the F3 progeny. Dissection of siliques from heterozygous double mutants [+/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) and atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b)] revealed a seed abortion rate (the frequencies of aborted seeds and unfertilized ovules in the analyzed seeds) of about 12.5%, whereas control plants [F3 progenies produced by crossing atu2af65a-4 or atu2af65b-1 homozygous mutants with wild-type (Col-0) plants] showed an abortion rate of about 2.2%, similar to that of wild-type (Col-0) plants (about 1.9%) (Figure 5B). However, the heterozygous double mutants [+/atu2af65a-3;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) and atu2af65a-3/atu2af65a-3;+/atu2af65b-1 (65a/65a;65B/65b)] using the atu2af65a-3 homozygous mutants as a knock-down allele showed a seed abortion rate of about 7.3%, suggesting that the residual activity of AtU2AF65a in the atu2af65a-3 mutants could affect seed development. Interestingly, atsf1-2 mutants showed a seed abortion rate of about 11.3%, which was similar with that observed in heterozygous double mutants [+/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) and atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b)] (Figure 5B). Previously characterized fy-1 mutants with defects in seed development (Henderson et al., 2005) also showed seed abortion rates of about 16.4%. However, silique morphology of heterozygous double mutants was not significantly affected and no embryo or seedling lethals were observed. These data indicated that the combinations of atu2af65a and atu2af65b null alleles lead to defects in genetic transmission through the male or female gametes.
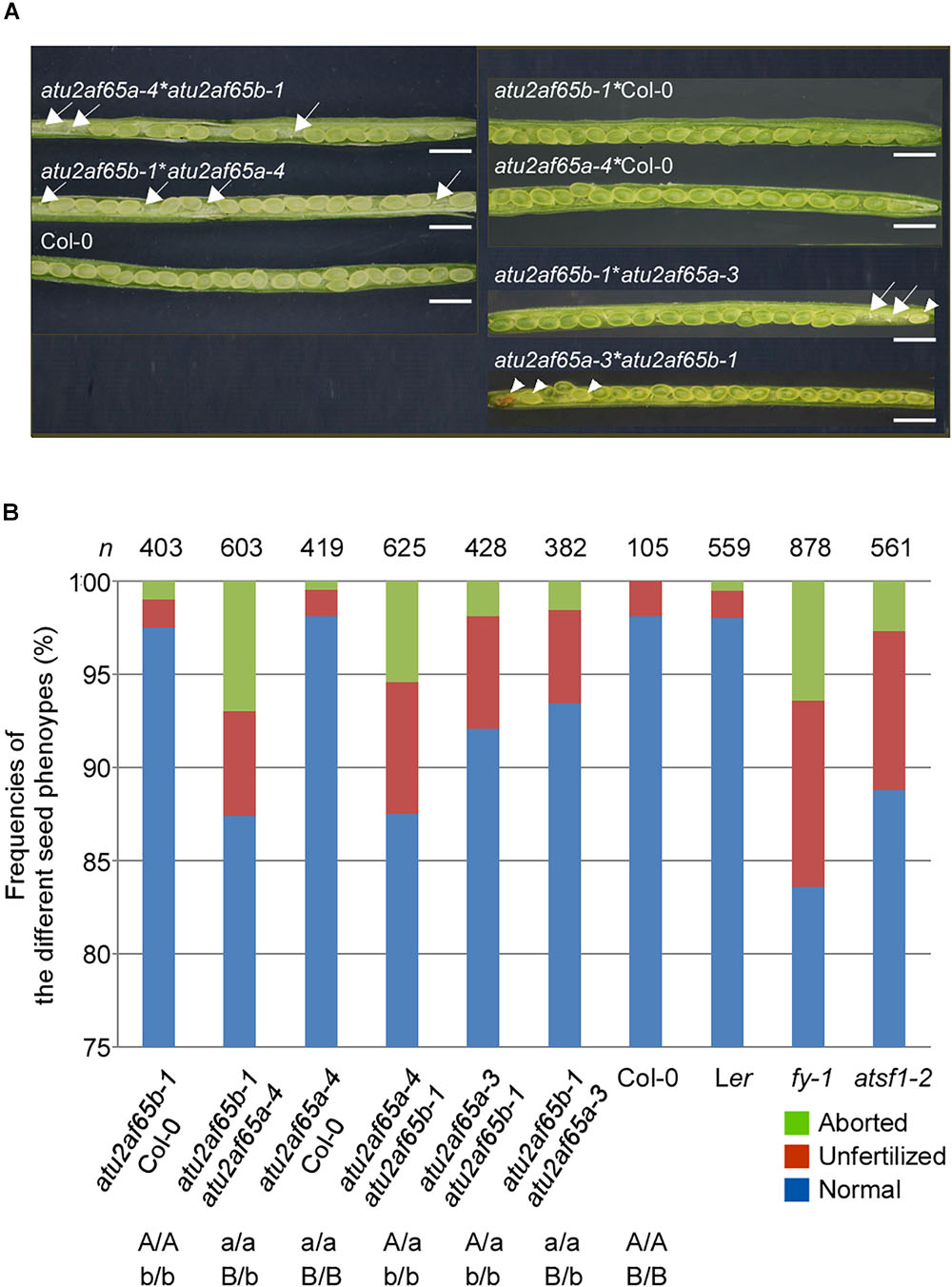
Figure 5. Seed developmental defects in siliques of various combinations of atu2af65a and atu2af65b mutants. (A) Seed phenotype of dissected siliques from various atu2af65a atu2af65b heterozygous double mutants in the F2 progeny. The plants obtained from the crosses between atu2af65a-4 or atu2af65a-3, and atu2af65b-1 single mutants as male parents and wild-type plants as female parents were used as controls. The arrows and arrow heads indicate the unfertilized ovules and aborted seeds, respectively. The classification of seed phenotypes in siliques has been described previously (Zhou et al., 2013). Scale bars 1.5 mm. (B) A histogram showing the fractions of normal and aborted seeds, and unfertilized ovules from various atu2af65a atu2af65b heterozygous double mutants in the F3 progeny. The genotypes of F3 progenies are indicated below the histogram. Other splicing factor mutants such as fy-1 and atsf1-2 were used to compare the seed developmental defects of atu2af65a atu2af65b heterozygous double mutants. Ler and Col-0 plants were used as controls for fy-1 mutants and other plants. n denotes the numbers of siliques used for the analysis of seed phenotype.
To investigate the impact of atu2af65a and atu2af65b mutations on male and female gametophytic functions, we performed reciprocal crosses between atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) and atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B), and tested transmission of the atu2af65b-1 allele in an atu2af65a-4 background, through both the male and the female haploid stage. Each genotype of progeny was confirmed by DNA-based PCR genotyping. atu2af65b-1 allele transmission through the male gametophyte was completely blocked [ratio of atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B): atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) = 0], whereas atu2af65b-1 allele transmission through the female gametophyte was normal [ratio of atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B): atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) = 1:1.2] (Table 1). Also, transmission of the atu2af65a-4 allele through the male gametophyte in reciprocal crosses between +/atu2af65a;atu2af65b/atu2af65b (65A/65a;65b/65b) was completely interrupted (Supplementary Table S2). This data indicate that mutations in AtU2AF65a and AtU2AF65b affect the genetic transmission through the male gametes.
The atu2af65 Mutations Redundantly Impair Pollen Tube Growth
To investigate how the mutations in AtU2AF65a and AtU2AF65b affect male gametophytic function, we phenotypically analyzed the AtU2AF65 mutants used as male parts in gametophytic function test. Alexander staining results showed that the pollen grains from atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B), +/+;atu2af65b-1/atu2af65b-1 (65A/65A;65b/65b), atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b), and +/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) plants were morphologically indistinguishable from wild-type (Col-0) pollen grains (Figure 6A), indicating that the atu2af65 mutations did not affect pollen grain formation. However, the fractions of approximately 31.6 and 32.1% of the pollen grains were short or burst in the single mutants [atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B) and +/+; atu2af65b-1/atu2af65b-1 (65A/65A;65b/65b)], respectively, whereas in the wild-type (Col-0) plants, this fraction was approximately 13.3% (Figure 6B). Furthermore, approximately 42.5% of the pollen grains was short or burst in atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) plants. Notably, the atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) pollen grains displayed a more profound impairment than the atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B) pollen grains. This data indicated that the mutant pollens are highly unstable under the employed in vitro conditions.
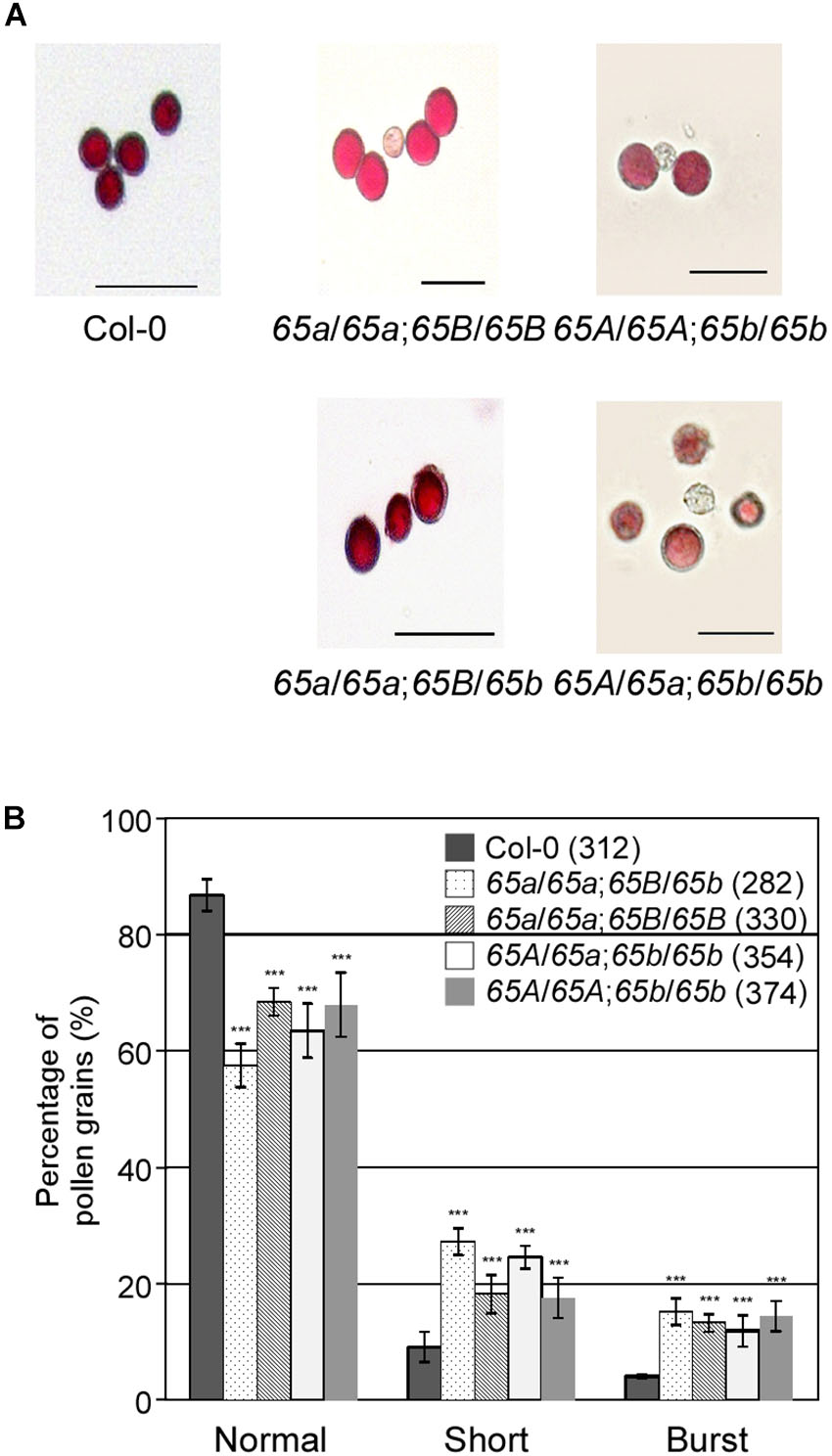
Figure 6. Pollen morphology in the F2 progeny containing the atu2af65a or atu2af65b mutant alleles. (A) Pollen morphology from wild-type (Col-0, 65A/65A;65B/65B), atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B), +/+; atu2af65b-1/atu2af65b-1 (65A/65A;65b/65b), atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b), +/atu2af65a-4/atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) plants. The atu2af65a mutant plants with the (65a/65a;65B/65b) genotype or without atu2af65b mutant alleles (65a/65a;65B/65B) and the AtU2AF65b mutant plants with (65A/65a;65b/65b) genotype or without AtU2AF65a mutant alleles (65A/65A;65b/65b) were selected from the F2 progeny population. Scale bars 50 μm. (B) Shape of pollen grains shown in panel (A). The shape of pollen was categorized into three groups (normal, short, and burst). The average of three biological replicates is shown in the histogram. Numbers in parenthesis denote the total number of pollen grains in three biological replicates of each genotype used for this analysis (approximately 110 pollen grains was used for each biological replicate). The error bars indicate the SEM of three biological replicates consisting of independently harvested samples. The asterisks denote a significant difference in pollen morphology between mutants and wild-type (Col-0) plants (χ2 test, ∗∗∗P < 0.001).
Next, we investigated the in vitro germination of pollen grains from the F2 progeny of various combinations of the atu2af65a and atu2af65b mutants. The pollen grains from both mutants germinated efficiently under the described in vitro conditions (data not shown), but the resulting pollen tubes were severely abnormal (Figure 7A). We also examined the length of pollen tubes after germination for 24 h. The mean length of the mutant pollen tubes was significantly reduced in comparison with that of the wild-type (Col-0) pollen tubes (Figure 7B): only 68.2 and 54.3% of the atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B) and +/+; atu2af65b-1/atu2af65b-1 (65A/65A;65b/65b) pollen tubes, respectively, were > 150 μm in length, whereas 75.0% of the wild-type (Col-0) pollen tubes were above 150 μm in length. Furthermore, the fraction of pollen tubes above 150 μm in length was smaller in the heterozygous double mutants [46.1 and 47.2% for the atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b) and +/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) mutants, respectively] than in the single mutants. Collectively, our results suggested that the mutations in AtU2AF65a and AtU2AF65b significantly impair the growth of pollen tubes, which may affect pollen tube growth during fertilization.
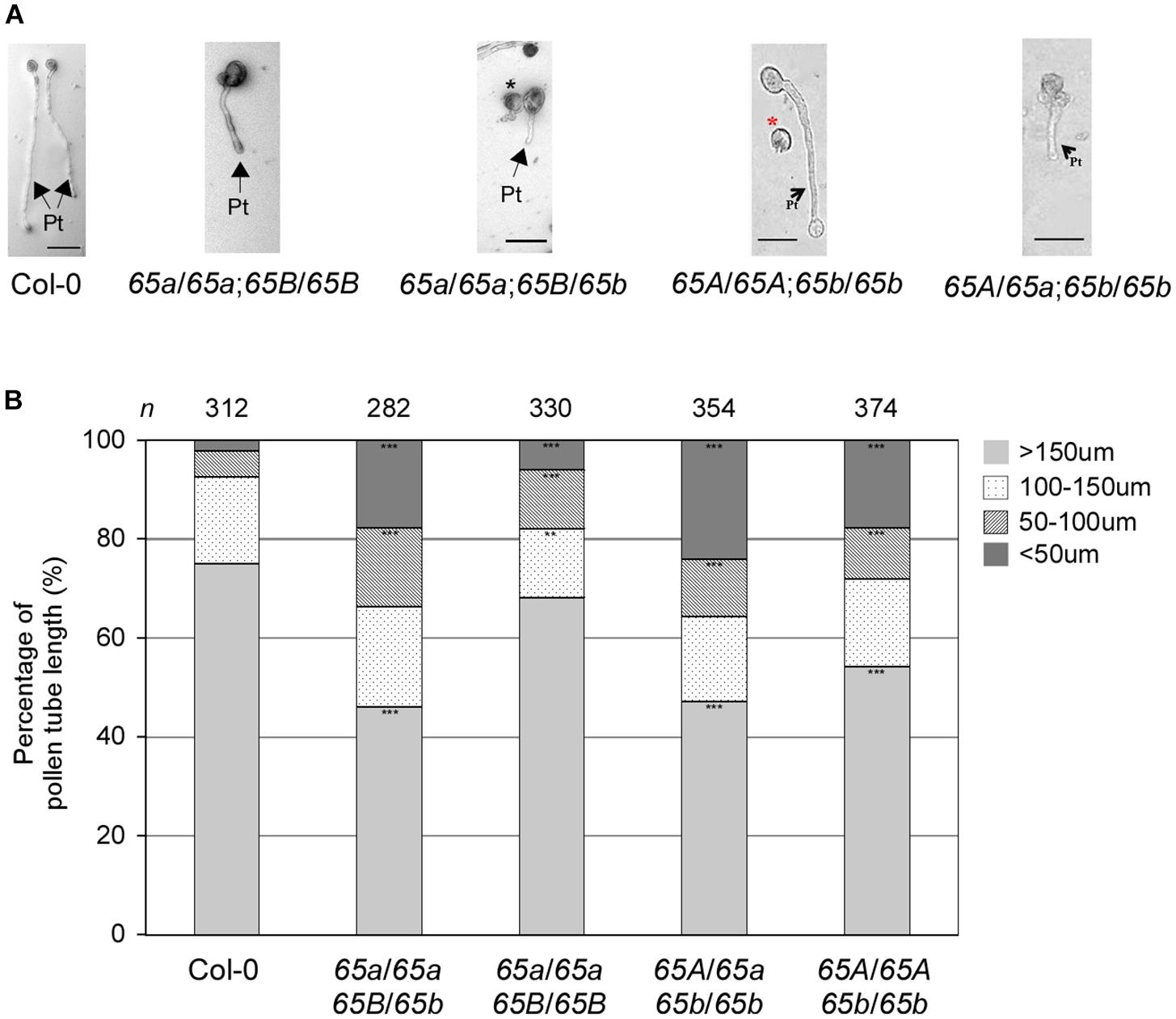
Figure 7. In vitro pollen tube growth defects in the F2 progeny containing the AtU2AF65a or AtU2AF65b mutant alleles. (A) Morphology of pollen tube growth from wild-type (Col-0, 65A/65A;65B/65B), atu2af65a-4/atu2af65a-4;+/+ (65a/65a;65B/65B), +/+; atu2af65b-1/atu2af65b-1 (65A/65A;65b/65b), atu2af65a-4/atu2af65a-4;+/atu2af65b-1 (65a/65a;65B/65b), +/atu2af65a-4;atu2af65b-1/atu2af65b-1 (65A/65a;65b/65b) plants. Pollen grains were germinated in vitro as in Figure 6. Arrows with Pt indicate pollen tubes germinated from pollens and asterisks (black and red) denote abnormal pollen grains. Scale bars 50 μm. (B) The length of pollen tube. Pollen tube length was measured after a 24-h incubation. The pollen tube length was categorized into four groups (shorter than 50 μm: <50 μm, between 50 and 100 μm: 50–100 μm, between 100 and 150 μm: 100–150 μm, longer than 150 μm: >150 μm). The average of three biological replicates is shown in the histogram. n indicates the total number of pollen grains in three biological replicates of each genotype used for this analysis (approximately 110 pollen grains was used for each biological replicate). The asterisks denote a significant difference in pollen morphology between mutants and wild-type (Col-0) plants (χ2 test, ∗∗P < 0.01, ∗∗∗P < 0.001).
Discussion
Pre-mRNA splicing plays an essential role in various growth and developmental processes in plants, but as yet little is known about the function of many of the spliceosomal components. Here, we provided evidence that two isoforms of AtU2AF65 are important for flowering and for the growth of pollen tubes during male gametophyte development.
Since plant introns do not contain the conserved branch point sequences and Py tracts at the 3′-splice proximal sites found in mammalians (Wahl et al., 2009), the mechanism of 3′-splice site recognition in plants may differ. Our previous findings, however, showed that AtU2AF65 proteins physically interact with AtU2AF35 and AtSF1 proteins in vitro and in planta (Jang et al., 2014; Park et al., 2017), as already known for yeasts and metazoans. Therefore, the initial step consisting in 3′-splice site recognition may be functionally similar in mammalians and plants, despite the above mentioned structural differences in plant introns. This notion is supported by the finding that two tobacco U2AF65 homologs (NpU2AF65a and NpU2AF65b) can splice adenoviral pre-mRNA in HeLa cell extracts depleted of endogenous U2AF factors (Domon et al., 1998). However, some differences in the mechanism of intron recognition may occur in plants. Firstly, several copies of U2AF genes such as U2AF65 and U2AF35 exist in nearly all higher plant genomes (Wang and Brendel, 2006; Jang et al., 2014). Secondly, plant SF1 homologs have an RRM domain, which is not found in their fungal and metazoan counterparts (Worden et al., 2009). Thirdly, neither the branch point nor the 3′-splice site regions significantly contribute to the interaction between RNA and U2AF65 homologs, as is the case with tobacco (Domon et al., 1998). Finally, we observed differential binding affinities of AtU2AF65a and AtU2AF65b for AtU2AF35 proteins (Park et al., 2017). These differences can account for the diversity and flexibility of molecular interactions between various components of the spliceosome in the initial recognition of the 3′-splice site in plants.
Several lines of evidence suggest that mutations in both AtU2AF65a and AtU2AF65b may result in the changes in flowering time. Our and other groups have previously reported that the AtU2AF65 and AtU2AF35 proteins interact (Park et al., 2017) and both T-DNA mutation in AtU2AF35a as well as RNA interference against AtU2AF35b cause late flowering phenotypes (Wang and Brendel, 2006; Jang et al., 2014). Moreover, we found that the expression patterns of AtU2AF35 and AtU2AF65 were similar (Figure 3; Wang and Brendel, 2006). For instance, RT-PCR analyses showed that AtU2AF35 and AtU2AF65 expression was observed in all vegetative and reproductive tissues; however, the abundance of their transcripts was different in the tissues tested. The GUS assays also revealed that the staining patterns of AtU2AF65a promoter activity were similar to those of AtU2AF35a and AtU2AF35b in all the tissues analyzed; however, their GUS staining intensities were slightly different. However, T-DNA alleles of AtU2AF65a were associated with a late flowering phenotype, whereas T-DNA alleles of AtU2AF65b resulted in early flowering (Figure 2). Due to this partial inconsistency, the phenotype of atu2af65b mutants should be interpreted with caution, especially since the overexpression of AtU2AF65b in the atu2af65b mutant background was not able to rescue the mutant phenotype (Supplementary Figure S2). It is therefore difficult to infer the AtU2AF65b function from the results obtained with the T-DNA mutant alleles. One possible explanation is that AtU2AF65b does not affect the flowering time via a dominant negative effect when it is ectopically overexpressed, because overexpressed AtU2AF65b could interrupt the formation of functional complexes, which does not objectively reflect the flowering processes in wild-type plants. However, we cannot exclude the possibility that AtU2AF65b regulates flowering time under specific conditions such as abiotic or biotic stresses. A recent report showed that the expression of AtU2AF65b is increased by ABA and AtU2AF65b modulated ABA-mediated flowering (Xiong et al., 2019). Furthermore, another T-allele (WiscDsLox321F12) of AtU2AF65b has an early flowering phenotype under LD and SD conditions, which was confirmed our observations (Figure 2).
AtU2AF65a and AtU2AF65b appear to differentially control FLC expression, and this might result in the opposite flowering phenotypes observed. However, considering the expression patterns of flowering time-related genes, including positive and negative regulators of FLC, in the atu2af65a and atu2af65b mutants (Figure 4), and the effect of vernalization on the flowering time in these mutants (Supplementary Figure S3), it is likely that AtU2AF65a and AtU2AF65b control flowering time by modulating FLC expression in FRI complex–dependent and –independent manners, respectively. Similarly, the mutations in CAP-BINDING PROTEIN 80 (CBP80)/ABA HYPERSENSITIVE 1 (ABH1) affect flowering time via FRI-mediated FLC regulation (Bezerra et al., 2004). However, because the genetic background (Col-0) of the atu2af65a mutants includes a weak FRI allele (Gazzani et al., 2003), further investigations are needed to ascertain whether an altered AtU2AF65 function may have an impact on FLC expression in the FRI FLC background. Furthermore, genome-wide RNA-seq analysis in the fri FLC and FRI FLC backgrounds is required to identify the general or specific effect of atu2af65 mutation on splicing regulation in flowering. These information would result in a better understanding of the control exerted by AtU2AF65 on flowering time.
Our study strongly suggests redundant regulation of pollen tube growth by different AtU2AF65 isoforms during male gametophyte development. Firstly, GUS expression patterns of AtU2AF65 were strongly observed in pollen (Figure 3). Secondly, atu2af65a atu2af65b heterozygous double mutants (65A/65a;65b/65b and 65a/65a;65B/65b) showed higher seed abortion rates (about 12.5%) than single atu2af65 mutants (about 2.2%) (Figure 5B). Thirdly, reciprocal cross test revealed that atu2af65b-1 allele transmission through the male gametophyte was completely interrupted (Table 1 and Supplementary Table S2). Finally, the instability of pollen shape and impaired pollen tube growth observed in atu2af65a atu2af65b heterozygous double mutants (65A/65a;65b/65b and 65a/65a;65B/65b) were more pronounced than those induced by the single atu2af65 mutants (Figures 6, 7).
The importance of splicing factors during male gametophyte development has been reported by several research groups (Tsukaya et al., 2013). For instance, the mutations in ATO, SF3b130, GFA1/CLO/VAJ, and RID1 cause defects in male gametophyte development (Moll et al., 2008; Yagi et al., 2009; Ohtani et al., 2013). Since these genes are involved in cell wall synthesis, cytoskeletal functions, metabolism, cellular transport, signaling, and secretion, all important elements for pollen tube growth, one possible scenario is that AtU2AF65 is involved in the pre-mRNA splicing of a specific set of genes for pollen tube development. This notion is supported by previous evidence showing that mutations in genes involved in a number of processes, including potassium transport, membrane trafficking, carbohydrate metabolism, signaling cascades, and cell wall functions, affect pollen germination or pollen tube growth (Mouline et al., 2002; Johnson et al., 2004; Lalanne et al., 2004; Lobstein et al., 2004; Jiang et al., 2005). Notably, as the U2AF65-associated protein (UAP56) plays an important role in the nuclear export of mRNAs in mammals (Fleckner et al., 1997; Gatfield et al., 2001), another possible scenario is that AtU2AF65 affects the export of nuclear mRNAs for pollen tube development. This hypothesis is supported by the observation that Arabidopsis UAP56 interacts with mRNA export factors such as the RNA and export factor-binding proteins (REF)/ALY2 and MODIFIER OF SNC1 (MOS11) (Kammel et al., 2013). Therefore, AtU2AF65 may affect the splicing or nuclear export of the pre-mRNAs of a specific set of genes, thereby ensuring normal pollen tube stability and growth during male gametophyte development. However, direct evidence in support of this hypothesis is lacking and further work is needed to definitely establish the role of the AtU2AF65 in male gametophyte development.
Author Contributions
J-KK conceived and designed the research. H-YP and HL conducted the experiments. JL and J-KK analyzed the data. JL and J-KK wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIT) (No. R1429203) (to J-KK) and a Korea University Grant (to J-KK) and by the BK 21 program (to H-YP).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00569/full#supplementary-material
References
Aki, S., Nakai, H., Aoyama, T., Oka, A., and Tsuge, T. (2011). AtSAP130/AtSF3b-3 function is required for reproduction in Arabidopsis thaliana. Plant Cell. Physiol. 52, 1330–1339. doi: 10.1093/pcp/pcr077
Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. doi: 10.3109/10520296909063335
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Bezerra, I. C., Michaels, S. D., Schomburg, F. M., and Amasino, R. M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40, 112–119. doi: 10.1111/j.1365-313X.2004.02194.x
Cavallari, N., Nibau, C., Fuchs, A., Dadarou, D., Barta, A., and Doonan, J. H. (2018). The cyclin-dependent kinase G group defines a thermosensitive alternative splicing circuit modulating the expression of Arabidopsis ATU2AF65A. Plant J. 94, 1010–1022. doi: 10.1111/tpj.13914
Chen, R., Silver, D. L., and de Bruijn, F. J. (1998). Nodule parenchyma-specific expression of the sesbania rostrata early nodulin gene SrEnod2 is mediated by its 3′ untranslated region. Plant Cell 10, 1585–1602. doi: 10.1105/tpc.10.10.1585
Choi, K., Kim, J., Hwang, H. J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
Domon, C., Lorkovic, Z. J., Valcarcel, J., and Filipowicz, W. (1998). Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem. 273, 34603–34610. doi: 10.1074/jbc.273.51.34603
Fleckner, J., Zhang, M., Valcarcel, J., and Green, M. R. (1997). U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11, 1864–1872. doi: 10.1101/gad.11.14.1864
Gatfield, D., Le Hir, H., Schmitt, C., Braun, I. C., Kocher, T., Wilm, M., et al. (2001). The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11, 1716–1721. doi: 10.1016/s0960-9822(01)00532-2
Gazzani, S., Gendall, A. R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132, 1107–1114. doi: 10.1104/pp.103.021212
Gniadkowski, M., Hemmings-Mieszczak, M., Klahre, U., Liu, H. X., and Filipowicz, W. (1996). Characterization of intronic uridine-rich sequence elements acting as possible targets for nuclear proteins during pre-mRNA splicing in Nicotiana plumbaginifolia. Nucleic Acids Res. 24, 619–627. doi: 10.1093/nar/24.4.619
Gross-Hardt, R., Kagi, C., Baumann, N., Moore, J. M., Baskar, R., Gagliano, W. B., et al. (2007). LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis. PLoS Biol. 5:e47. doi: 10.1371/journal.pbio.0050047
Henderson, I. R., Liu, F., Drea, S., Simpson, G. G., and Dean, C. (2005). An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132, 3597–3607. doi: 10.1242/dev.01924
Hong, S. M., Bahn, S. C., Lyu, A., Jung, H. S., and Ahn, J. H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51, 1694–1706. doi: 10.1093/pcp/pcq128
Howden, R., Park, S. K., Moore, J. M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149, 621–631.
Hugouvieux, V., Kwak, J. M., and Schroeder, J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. doi: 10.1016/s0092-8674(01)00460-3
Jang, Y. H., Park, H. Y., Lee, K. C., Thu, M. P., Kim, S. K., Suh, M. C., et al. (2014). A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 78, 591–603. doi: 10.1111/tpj.12491
Jiang, L., Yang, S. L., Xie, L. F., Puah, C. S., Zhang, X. Q., Yang, W. C., et al. (2005). Vanguard1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17, 584–596. doi: 10.1105/tpc.104.027631
Johnson, M. A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., et al. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971–982. doi: 10.1534/genetics.104.029447
Kammel, C., Thomaier, M., Sorensen, B. B., Schubert, T., Langst, G., Grasser, M., et al. (2013). Arabidopsis DEAD-box RNA helicase UAP56 interacts with both RNA and DNA as well as with mRNA export factors. PLoS One 8:e60644. doi: 10.1371/journal.pone.0060644
Lalanne, E., Michaelidis, C., Moore, J. M., Gagliano, W., Johnson, A., Patel, R., et al. (2004). Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics 167, 1975–1986. doi: 10.1534/genetics.104.030270
Lee, J. H., Ryu, H. S., Chung, K. S., Pose, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632. doi: 10.1126/science.1241097
Lee, J. H., Yoo, S. J., Park, S. H., Hwang, I., Lee, J. S., and Ahn, J. H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21, 397–402. doi: 10.1101/gad.1518407
Lee, K. C., Jang, Y. H., Kim, S. K., Park, H. Y., Thu, M. P., Lee, J. H., et al. (2017). RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 36, 1083–1095. doi: 10.1007/s00299-017-2140-1
Lobstein, E., Guyon, A., Ferault, M., Twell, D., Pelletier, G., and Bonhomme, S. (2004). The putative Arabidopsis homolog of yeast vps52p is required for pollen tube elongation, localizes to Golgi, and might be involved in vesicle trafficking. Plant Physiol. 135, 1480–1490. doi: 10.1104/pp.103.037747
Luehrsen, K. R., Taha, S., and Walbot, V. (1994). Nuclear pre-mRNA processing in higher plants. Prog. Nucleic Acid. Res. Mol. Biol. 47, 149–193. doi: 10.1016/s0079-6603(08)60252-4
Michaels, S. D. (2009). Flowering time regulation produces much fruit. Curr. Opin. Plant Biol. 12, 75–80. doi: 10.1016/j.pbi.2008.09.005
Moll, C., von Lyncker, L., Zimmermann, S., Kagi, C., Baumann, N., Twell, D., et al. (2008). CLO/GFA1 and ATO are novel regulators of gametic cell fate in plants. Plant J. 56, 913–921. doi: 10.1111/j.1365-313X.2008.03650.x
Mouline, K., Very, A. A., Gaymard, F., Boucherez, J., Pilot, G., Devic, M., et al. (2002). Pollen tube development and competitive ability are impaired by disruption of a Shaker K(+) channel in Arabidopsis. Genes Dev. 16, 339–350. doi: 10.1101/gad.213902
Nilsen, T. W., and Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463. doi: 10.1038/nature08909
Ohtani, M., Demura, T., and Sugiyama, M. (2013). Arabidopsis root initiation defective1, a DEAH-box RNA helicase involved in pre-mRNA splicing, is essential for plant development. Plant Cell 25, 2056–2069. doi: 10.1105/tpc.113.111922
Ohtani, M., and Sugiyama, M. (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 43, 479–490. doi: 10.1111/j.1365-313X.2005.02469.x
Onodera, Y., Nakagawa, K., Haag, J. R., Pikaard, D., Mikami, T., Ream, T., et al. (2008). Sex-biased lethality or transmission of defective transcription machinery in Arabidopsis. Genetics 180, 207–218. doi: 10.1534/genetics.108.090621
Onouchi, H., Igeno, M. I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12, 885–900. doi: 10.1105/tpc.12.6.885
Park, H. Y., Lee, K. C., Jang, Y. H., Kim, S. K., Thu, M. P., Lee, J. H., et al. (2017). The Arabidopsis splicing factors, AtU2AF65, AtU2AF35, and AtSF1 shuttle between nuclei and cytoplasms. Plant Cell Rep. 36, 1113–1123. doi: 10.1007/s00299-017-2142-z
Pose, D., Verhage, L., Ott, F., Yant, L., Mathieu, J., Angenent, G. C., et al. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503, 414–417. doi: 10.1038/nature12633
Rain, J. C., Rafi, Z., Rhani, Z., Legrain, P., and Kramer, A. (1998). Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4, 551–565. doi: 10.1017/s1355838298980335
Ramakers, C., Ruijter, J. M., Deprez, R. H., and Moorman, A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66. doi: 10.1016/s0304-3940(02)01423-4
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L., and Riechmann, J. L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. doi: 10.1104/pp.126.1.122
Simpson, G. G., Dijkwel, P. P., Quesada, V., Henderson, I., and Dean, C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787. doi: 10.1016/s0092-8674(03)00425-2
Simpson, G. G., and Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1–41. doi: 10.1007/978-94-009-0353-1_1
Thickman, K. R., Swenson, M. C., Kabogo, J. M., Gryczynski, Z., and Kielkopf, C. L. (2006). Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 356, 664–683. doi: 10.1016/j.jmb.2005.11.067
Tsugeki, R., Tanaka-Sato, N., Maruyama, N., Terada, S., Kojima, M., Sakakibara, H., et al. (2015). CLUMSY VEIN, the Arabidopsis DEAH-box Prp16 ortholog, is required for auxin-mediated development. Plant J. 81, 183–197. doi: 10.1111/tpj.12721
Tsukaya, H., Byrne, M. E., Horiguchi, G., Sugiyama, M., Van Lijsebettens, M., and Lenhard, M. (2013). How do ‘housekeeping’ genes control organogenesis?–Unexpected new findings on the role of housekeeping genes in cell and organ differentiation. J. Plant Res. 126, 3–15. doi: 10.1007/s10265-012-0518-2
Verhage, L., Severing, E. I., Bucher, J., Lammers, M., Busscher-Lange, J., Bonnema, G., et al. (2017). Splicing-related genes are alternatively spliced upon changes in ambient temperatures in plants. PLoS One 12:e0172950. doi: 10.1371/journal.pone.0172950
Volz, R., von Lyncker, L., Baumann, N., Dresselhaus, T., Sprunck, S., and Gross-Hardt, R. (2012). LACHESIS-dependent egg-cell signaling regulates the development of female gametophytic cells. Development 139, 498–502. doi: 10.1242/dev.075234
Wahl, M. C., Will, C. L., and Luhrmann, R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718. doi: 10.1016/j.cell.2009.02.009
Wang, B. B., and Brendel, V. (2006). Molecular characterization and phylogeny of U2AF35 homologs in plants. Plant Physiol. 140, 624–636. doi: 10.1104/pp.105.073858
Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Worden, A. Z., Lee, J. H., Mock, T., Rouze, P., Simmons, M. P., Aerts, A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes micromonas. Science 324, 268–272. doi: 10.1126/science.1167222
Xiong, F., Ren, J.-J., Yu, Q., Wang, Y.-Y., Lu, C.-C., Kong, L.-J., et al. (2019). AtU2AF65b functions in abscisic acid mediated flowering via regulating the precursor messenger RNA splicing of ABI5 and FLC in Arabidopsis. New Phytol. doi: 10.1111/nph.15756 [Epub ahead of print].
Yagi, N., Takeda, S., Matsumoto, N., and Okada, K. (2009). VAJ/GFA1/CLO is involved in the directional control of floral organ growth. Plant Cell Physiol. 50, 515–527. doi: 10.1093/pcp/pcp011
Zhou, J.-J., Liang, Y., Niu, Q.-K., Chen, L.-Q., Zhang, X.-Q., and Ye, D. (2013). The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J. Exp. Bot. 64, 2205–2218. doi: 10.1093/jxb/ert078
Keywords: AtU2AF65a, AtU2AF65b, flowering time, male gametophyte development, pollen tube growth, premRNA splicing
Citation: Park H-Y, Lee HT, Lee JH and Kim J-K (2019) Arabidopsis U2AF65 Regulates Flowering Time and the Growth of Pollen Tubes. Front. Plant Sci. 10:569. doi: 10.3389/fpls.2019.00569
Received: 27 September 2018; Accepted: 15 April 2019;
Published: 03 May 2019.
Edited by:
Zhong-Nan Yang, Shanghai Normal University, ChinaReviewed by:
Tomohiko Tsuge, Kyoto University, JapanXin-Qi Gao, Shandong Agricultural University, China
Copyright © 2019 Park, Lee, Lee and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong Hwan Lee, amh3YW5sZWU5MEBqYm51LmFjLmty Jeong-Kook Kim, amtraW1Aa29yZWEuYWMua3I=
 Hyo-Young Park1
Hyo-Young Park1 Hee Tae Lee
Hee Tae Lee Jeong Hwan Lee
Jeong Hwan Lee Jeong-Kook Kim
Jeong-Kook Kim