- 1Department of Plant, Soil and Microbial Sciences, Michigan State University, East Lansing, MI, United States
- 2Department of Plant Biology, Michigan State University, East Lansing, MI, United States
- 3Plant Resilience Institute, Michigan State University, East Lansing, MI, United States
- 4AgBioResearch, Michigan State University, East Lansing, MI, United States
Potato breeding can be redirected to a diploid inbred/F1 hybrid variety breeding strategy if self-compatibility can be introduced into diploid germplasm. However, the majority of diploid potato clones (Solanum spp.) possess gametophytic self-incompatibility that is primarily controlled by a single multiallelic locus called the S-locus which is composed of tightly linked genes, S-RNase (S-locus RNase) and multiple SLFs (S-locus F-box proteins), which are expressed in the style and pollen, respectively. Using S-RNase genes known to function in the Solanaceae gametophytic SI mechanism, we identified S-RNase alleles with flower-specific expression in two diploid self-incompatible potato lines using genome resequencing data. Consistent with the location of the S-locus in potato, we genetically mapped the S-RNase gene using a segregating population to a region of low recombination within the pericentromere of chromosome 1. To generate self-compatible diploid potato lines, a dual single-guide RNA (sgRNA) strategy was used to target conserved exonic regions of the S-RNase gene and generate targeted knockouts (KOs) using a Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 (Cas9) approach. Self-compatibility was achieved in nine S-RNase KO T0 lines which contained bi-allelic and homozygous deletions/insertions in both genotypes, transmitting self compatibility to T1 progeny. This study demonstrates an efficient approach to achieve stable, consistent self-compatibility through S-RNase KO for use in diploid potato breeding approaches.
Introduction
Cultivated potato (Solanum tuberosum L.) is the third most important food crop after rice and wheat (Devaux et al., 2014) and plays an essential role in human nutrition as a primary source of carbohydrates. Although global production of potato totaled 376 million tons in 2016 (FAOSTAT, 2019), potatoes face high production losses due to biotic and abiotic stresses that will increase with global warming (Raymundo et al., 2017). While improvement of cultivated potatoes (2n = 4x = 48) relies on the discovery and introgression of genes from wild species for traits such as disease resistance, the polyploid nature of cultivated tetraploid potato hampers the fixation of desirable alleles in new cultivars. For example, the introgression of critical dominant alleles such as the potato virus Y (PVY) disease-resistance gene in a triplex or quadruplex allelic configuration can take up to 15 years (Mendoza et al., 1996). Re-inventing potato as a diploid inbred/F1 hybrid variety (2n = 2x = 24) would allow the application of efficient breeding methods (Jansky et al., 2016) as inbred potatoes would accelerate the generation of new varieties with favorable allelic combinations targeting yield, tuber quality, and resistance traits. A significant barrier to this approach is the occurrence of gametophytic self-incompatibility (SI) in a majority of the diploid potato germplasm, thereby preventing the ability to generate diploid homozygous lines.
In diploid potato, the gametophytic SI system is controlled by a single multiallelic locus called the S-locus (Porcher and Lande, 2005). This locus is composed of tightly linked genes, S-RNase (S-locus RNase) and SLFs (S-locus F-box) genes known also as S-haplotype-specific F-box brothers (SFBB), expressed in the style and pollen, respectively (Takayama and Isogai, 2005; Sassa et al., 2007; Kubo et al., 2010; Bush and Moore, 2012). The S-RNase protein produces cytotoxic effects that inhibit the elongation of self-pollen tubes via degradation of RNA from the pollen whereas SLF functions as a component of a detoxification complex that mediates ubiquitination of non-self S-RNase proteins leading to degradation via the proteasome pathway (Sijacic et al., 2004; Kubo et al., 2015). Hence, when self-pollination occurs in self-incompatible individuals, the SLFs genes do not recognize their native S-RNase and consequently, pollen tube growth in the style is inhibited due to the ribonuclease activity of the S-RNase (Hua et al., 2008).
In an effort to develop diploid self-compatible (SC) potato lines, the inbred line M6 was generated from the wild tuber-bearing species, Solanum chacoense (Jansky et al., 2014). In M6, a dominant allele of the S-locus inhibitor (Sli) inactivates the gametophytic SI system (Hosaka and Hanneman, 1998) leading to self-compatibility. However, introgression of Sli into other germplasm is time-consuming and could lead to linkage drag and fixation of undesirable traits such as high tuber glycoalkaloid content from the donor S. chacoense. An alternative strategy to Sli introgression is the use of genome editing to accelerate the generation of SC diploid lines by targeting genes involved in SI.
Genome editing by Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated protein 9 (Cas9) system has been widely used to generate gene knockouts (KOs) of candidate genes related to agronomic traits in important crops (Jaganathan et al., 2018). Cas9 induces double-strand breaks (DSBs) in the DNA at the target site, triggering the response of endogenous cell repair mechanisms. One of the cellular mechanisms to repair DSBs is non-homologous end joining (NHEJ), which can generate insertions and deletions in the coding region resulting in a KO of gene function (Bortesi and Fischer, 2015; Pellagatti et al., 2015). The target-DNA recognition is mediated by a single guide RNA (sgRNA) bearing a 20 bp target-site complementary to the region adjacent to a protospacer-adjacent motif (PAM), 5′-NGG-3′, resulting in the generation of a DSB (Doudna and Charpentier, 2014; Sternberg et al., 2014; Pellagatti et al., 2015) which can be leveraged to generate DSB of target genes.
Previous studies in tomato wild relatives demonstrated that missense mutations and gene loss prevent S-RNase ribonuclease activity in S. peruvianum and S. pennellii, leading to self-compatibility (Kowyama et al., 1994; Royo et al., 1994;Covey et al., 2010; Li and Chetelat, 2015). Considering that S-RNase is the gametophytic SI component directly implicated in degradation of RNA in self-pollen tubes, inhibiting the S-RNase function is a straightforward strategy to confer self compatibility in potato. In an effort to contribute to the development of diploid inbred potato lines, we generated SC diploid lines by targeted mutagenesis of S-RNase using CRISPR-Cas9, obtaining stable self-compatibility in T0 and T1 generations. Contemporaneous with the writing of this manuscript, Ye et al. (2018) published their findings using a similar approach. However, this study provides further insight into SI in diploid potatoes, reporting three new S-RNase alleles, their localization within a low recombination pericentromeric region consistent with the location of the S-locus, generation of stable SC KO lines, and documentation of plasticity in the phenotype of SI in two diploid lines.
Materials and Methods
Plant Material
After an initial test of self-compatibility with more than 50 self-pollinations, the SI diploid potato lines (2n = 2x = 24) DRH-195 and DRH-310 F1 lines were generated from a cross between S. tuberosum Gp. Phureja DM 1-3 516 R44 (DM) and S. tuberosum Gp Tuberosum RH 89-039-16 (RH) at Virginia Tech and used in this study. Plants were maintained in vitro, propagated on Murashige and Skoog (MS) medium (MS basal salts plus vitamins, 3% sucrose, 0.7% plant agar, pH 5.8) (Murashige and Skoog, 1962) and cultured in growth chambers with 16-h-light/8-h-dark photoperiod at 22°C and average light intensity of 200 μmoles m-2 s-1.
Allelic Identification, Annotation, and Phylogenetic Analysis of S-RNase
TBLASTN [basic local alignment search tool (BLAST)] searches were performed using reported S-RNase protein sequences (Table 1) from the Solanaceae family against the DM v4.04 assembly (Hardigan et al., 2016) using BLAST v2.2.31 (Altschul et al., 1990) with default parameters. A candidate S-RNase gene was selected using the top blast hits. Expression abundances across a range of developmental stages, tissues, and organs were determined using available gene expression atlases for DM and RH (The Potato Genome Sequencing Consortium, 2011). To identify S-RNase allelic variants in the diploid potato clones, genomic and complementary DNA sequence data from DRH-195 and DRH-310 leaf and tuber tissues were retrieved from the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (Supplementary Table S1) and aligned to the DM v4.04 assembly using BWA-MEM (Li, 2013). Duplicate reads were removed using Picard Tools v1.1131 and consensus sequences were obtained using the mpileup utility from Samtools v1.2 (Li et al., 2009) with the consensus option from bcftools v1.2 (Li, 2011).
A primer set was designed to amplify the predicted ORF of the S-RNase gene in DRH-195 and DRH-310 using the detected S-RNase variants (Supplementary Table S2). S-RNase amino acid sequences reported in Table 1, including the alleles reported by Ye et al. (2018), along with the deduced amino acid sequences from the S-RNase variants identified in this study were aligned using Clustal Omega (Sievers et al., 2011). A phylogenetic tree was constructed using the Neighbor joining method with 1000 bootstrap replicates in MEGA version 7.0 (Kumar et al., 2016). Amino acid similarities percentages were calculated using BioEdit (Hall, 1999).
S-RNase Linkage Mapping
The previously reported diploid DRH mapping population was used to genetically map the S-RNase gene (Manrique-Carpintero et al., 2015). DNA was isolated from DRH-195 and DRH-310 young leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany), and used for PCR with a Q5® High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, United States) with the following thermocycler conditions: one cycle of initial denaturation for 4 min at 94°C, followed by 34 cycles for 15 s at 94°C, 45 s at 56°C and 1 min at 72°C and a final extension of 5 min at 72°C. S-RNase amplicons were gel-purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and cloned into the Zero Blunt TOPO PCR Cloning vector (Thermo Fisher, Carlsbad, CA, United States). Ten colonies for each line were sequenced by the Sanger method and aligned using Clustal Omega (Sievers et al., 2011). DM and RH allelic sequences were confirmed and used to design S-RNase RH-allele specific primers (Supplementary Table S2). These primers were screened across 80 individuals of the DRH mapping population. The genotype from the presence/absence of an RH allele was coded as nnxnp and used for mapping in JoinMap4.1 with the same parameters as previously reported by Manrique-Carpintero et al. (2015).
sgRNA Identification, Assembly, and Validation
A double sgRNA construct targeting predicted conserved regions from the first (sgRNA 1) and second (sgRNA 2) S-RNase exons were designed using CRISPR RGEN tools (Supplementary Table S2, Park et al., 2015). A gene KO construct containing the sgRNA combination (sgRNA 1–2) was assembled in the pHSE40 vector containing the CRISPR-Cas9 cassette as described by Xing et al. (2014) and transferred into Agrobacterium tumefaciens strain GV3101 pMP90 (Koncz et al., 1994) by electroporation.
Agrobacterium-Mediated Transformation
Agrobacterium-mediated transformation was performed using leaf segments from 4-week-old tissue culture plants of DRH 195 and DRH 310 as described by Li et al. (1999). Briefly, explants were pre-cultured on a step I media (MS salts, 3% sucrose, 5 g/l phytoagar, 1 mg/l thiamine-HCl, 0. 8 mg/l zeatin-riboside and 2 mg/l 2,4-D) for 4 days and inoculated with Agrobacterium. After 3 days, explants were rinsed with sterile distilled water containing 250 mg/l cefotaxime and 200 mg/l carbenicillin and placed onto step II media (MS salts, 3% sucrose, 5 g/l phytoagar, 1 mg/l thiamine-HCl, 0.8 mg/l zeatin- riboside, 2 mg/l gibberellic acid, 20 mg/l hygromycin and 150 mg/l ticarcillin disodium and clavulanate potassium). Explants were transferred to fresh step II media every week. After approximately 30 days, transformation events (T0 lines) were selected from step II media and transferred to root induction media containing MS medium supplemented with antibiotics for selection as described above.
Molecular Characterization of KO Lines
DNA from T0 plants was isolated as described above. PCR was carried out using the GoTaq DNA polymerase (Promega, Fitchburg, WI, United States) with the following thermocycler conditions: one cycle of initial denaturation for 4 min at 94°C, followed by 34 cycles for 15 s at 94°C, 45 s at 56°C and 1 min at 72°C and a final extension of 5 min at 72°C. Amplicons were visualized on 1% (w/v) agarose gels. Allelic mutations of positive transformation events were identified by insertion/deletion presence. Selected transformation events were amplified with the Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, United States). Then, purified PCR products were cloned into the Zero Blunt TOPO PCR Cloning vector (Thermo Fisher, Carlsbad, CA, United States), and transformed into DH5α competent cells (Thermo Fisher, Carlsbad, CA, United States). Colonies carrying the alleles from each event were Sanger sequenced.
Assessment of Self-Compatibility
One month old in vitro plants were planted in one gallon plastic pots with a peat and perlite growth medium mixture (Bacto professional planting mix) and placed into a greenhouse with a light intensity of 250 μmoles m-2 s-1, 16/8-h light/dark photoperiod and a temperature of 25°C. Plants were fertilized with Peters Professional® 20: 20: 20 fertilizer (The Scotts, Co., Marysville, OH, United States) at a rate of 500 mg/l twice a week. Around 50 flowers per plants were hand self-pollinated to test for self compatibility. Pollen staining with acetocarmine-glycerol (Ordoñez, 2014b) and cross-pollination were also done to test male and female viability, respectively. T0 fruits were harvested 3–4 weeks after self-pollination and kept at room temperature for 2 weeks. Extracted T1 seeds were sterilized and subjected to overnight treatment with 1500 ppm of gibberellic acid then allowed to germinate. T1 seedlings were transferred to greenhouse and self-pollinated as described above. Additionally, chloroplast counting of guard cells was performed according to Ordoñez (2014a) to discard possible chromosome doubling in each selected S-RNase KO line.
S-RNase Expression Analysis
Twenty-five flowers from wild-type (WT) and DRH-195/310-derived T0 KO lines (DRH-195.158 and DRH-310.21) were self-pollinated at anthesis. Pollinated pistils were excised 24-hour post pollination (hpp) and preserved in -80°C until use. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and DNase treated using the TURBO DNA-free kit (Thermo Fisher, Carlsbad, CA, United States) following manufacturer’s instructions. RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Grand Island, NY, United States) and a reverse-transcription polymerase chain reaction (RT-PCR) was carried-out with 1 μg of total RNA using the Super-Script One-Step RT-PCR kit (Thermo Fisher Scientific, Carlsbad, CA, United States). Primers designed to amplify the S-RNase ORF and the elongation-factor one alpha (EF1α) housekeeping internal control were used for the RT-PCR reaction (Supplementary Table S2).
Results
Identification of the S-RNase Gene in Potato
For this study, we used available genomic and gene expression data from the sequenced doubled monoploid DM and the SC heterozygous breeding line RH (The Potato Genome Sequencing Consortium, 2011). To identify the S-RNase gene, the DM genome sequence was selected as the reference genome and utilized in sequence similarity searches using seven Solanaceae S-RNase genes (Table 1). Candidate genomic regions encoding the S-RNase gene were identified and located within 3,948,850–3,949,581 bp of the unanchored scaffold PGSC0003DMB000000091 and the annotated DM S-RNase allele, PGSC0003DMG400026738, which encodes a 738 bp open reading frame and a 216 amino acids (aa) predicted protein composed of five conserved and two hypervariable regions, characteristic of S-RNases (Figure 1A; Ioerger et al., 1991). The detected DM and RH S-RNase alleles resemble class III S-RNases (Figure 1B) and are comprised of two exons and one small intron, which is located at position five of the 11 recognized intron positions for this gene family (Igic and Kohn, 2001; Ramanauskas and Igić, 2017).
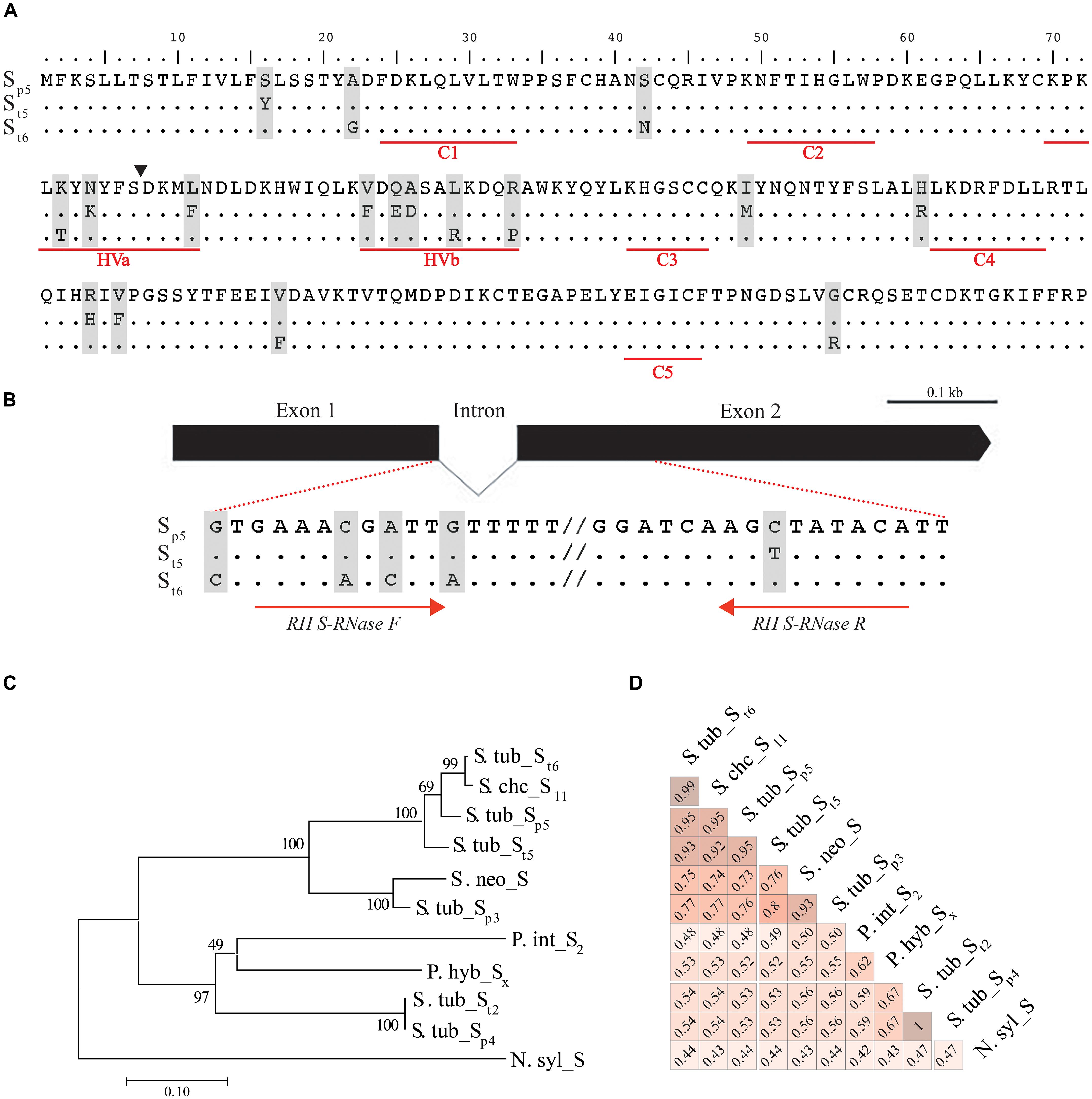
Figure 1. S-RNase gene structure and allelic variants in diploid potato and related species. (A) S-RNase predicted amino-acid sequence alignment of the DM (Sp5) and RH (St5 and St6) alleles. Underlined regions in red represent the typical five conserved regions (C1 to C5) and two hypervariable regions (HVa and HVb) of the S-RNase gene family. Exon/intron boundary is indicated with a filled triangle within the HVa region (B) S-RNase gene structure. The S-RNase open reading frame is composed of two exons separated by one small intron. Zoomed-in regions are shown within dotted lines indicating the intronic and exonic regions used for RH-specific primer design within the reported S-RNase alleles. (C) Phylogenetic tree constructed using the Neighbor Joining method based in the proportion of S-RNase amino acid differences. S-RNase from Nicotiana sylvestris was used as out-group. Numbers above each branch represent bootstrapping percentages from1000 replications. (D) Pairwise amino acid similarity of S-RNase in Solanum species and detected S-RNase alleles. S-RNase alleles are represented first by a species name abbreviation followed by S and the allele number for Tuberosum (t) or Phureja (p) group in Solanum tuberosum. For other species, a similar pattern is used, and allele numbers or letters (i.e., Sx for P. hybrida) are added if reported. S. tub: S. tuberosum, S. chc: S. chacoense, S. neo: S. neorickii, P. int: P. integrifolia, P. hyb: P. hybrida, N. syl: N. sylvestris.
We found that the DM S-RNase (referred hereafter as S. tub_Sp5 from S. tuberosum S-RNase allele five of S. tuberosum Group Phureja) is highly expressed in mature flowers [245.5 Fragments Per Kilobase of exon model per Million mapped reads (FPKM)] compared with no expression in leaves or tubers in DM. High S-RNase expression levels were also detected in carpels (4342.7 FKPM) consistent with its role in preventing self compatibility (Kao and Tsukamoto, 2004). More limited gene expression data is available for RH (The Potato Genome Sequencing Consortium, 2011) and consistent with the expression of S-RNase in potato, it is expressed in flowers (167.04 FPKM) and not tubers or leaves. Together with the functional annotation, these results suggest we have identified the S-RNase gene involved in SI.
Identification of Allelic Variants of S-RNase in Diploid Potato Lines
The DRH-195 and DRH-310 F1 diploid self-incompatible lines derived from a cross between DM and RH were used to identify S-RNase allelic variants. Using whole genome resequencing data for these two lines, the DM and RH S-RNase alleles were identified in DRH-195 and DRH-310 and validated using Sanger sequencing. As previously observed for DM, the S. tub_Sp5 predicted 216 amino acid sequence was detected in both lines, beside one of the two RH S-RNase alleles in each line, hereafter referred as S. tub_St5 and S. tub_St6 (S. tuberosum S-RNase alleles five and six of S. tuberosum Gp Tuberosum, respectively, Figure 1A).
A phylogenetic tree was constructed using available S-RNase amino acid sequences from Solanum species and the allelic variants identified in this study (Figure 1C). Two main clades with high confidence bootstrap values were observed. The S. tub_Sp5, S. tub_St5, and S. tub_St6 alleles and the S. chacoense (S. chc_S11) allele exhibited highest percentage of similarity relative to the other species (Figure 1D and Supplementary Table S3), whereas the Petunia (P. int_S2 and P. hyb_Sx) and S. tuberosum (S. tub_St2) S-RNases clustered in two separate subgroups. Two S. tuberosum Gp Phureja alleles, S. tub_Sp3 and S. tub_Sp4, were located in separate clades (Figure 1C). Overall, the amino acid sequence identity between S-RNase alleles ranged from 42 to 100%, showing 92% similarity between the DM and RH S-RNase alleles and 93% between the two RH alleles (Figure 1D). Notably, the S. tub_Sp4 allele had 100% similarity with a previously reported S-RNase from S. tub_St2 (NCBI accession: Q01796), presumably representing the same allele. Moreover, S. tub_Sp5, S. tub_St5, and S. tub_St6 alleles had similar amino acid identity (53–54%) to the reported S. tub_Sp3 allele when compared with S. tub_Sp4 (Ye et al., 2018).
S-RNase Is Located Within a Pericentromeric Region of Chromosome 1 in Potato
Using the segregating DRH population, linkage mapping indicated that the S-RNase S. tub_St6 allele mapped to the pericentromeric region of chromosome 1 spanning a region between 13.7 and 17.8 cM (solcap_snp_c2_27882 and solcap_snp_c1_16425 markers, respectively), corresponding to 6.1 and 18.9 Mb of chromosome 1 in the physical map (Figure 2). These results are consistent with the region corresponding to the map location of the S-locus in potato (Gebhardt et al., 1991).
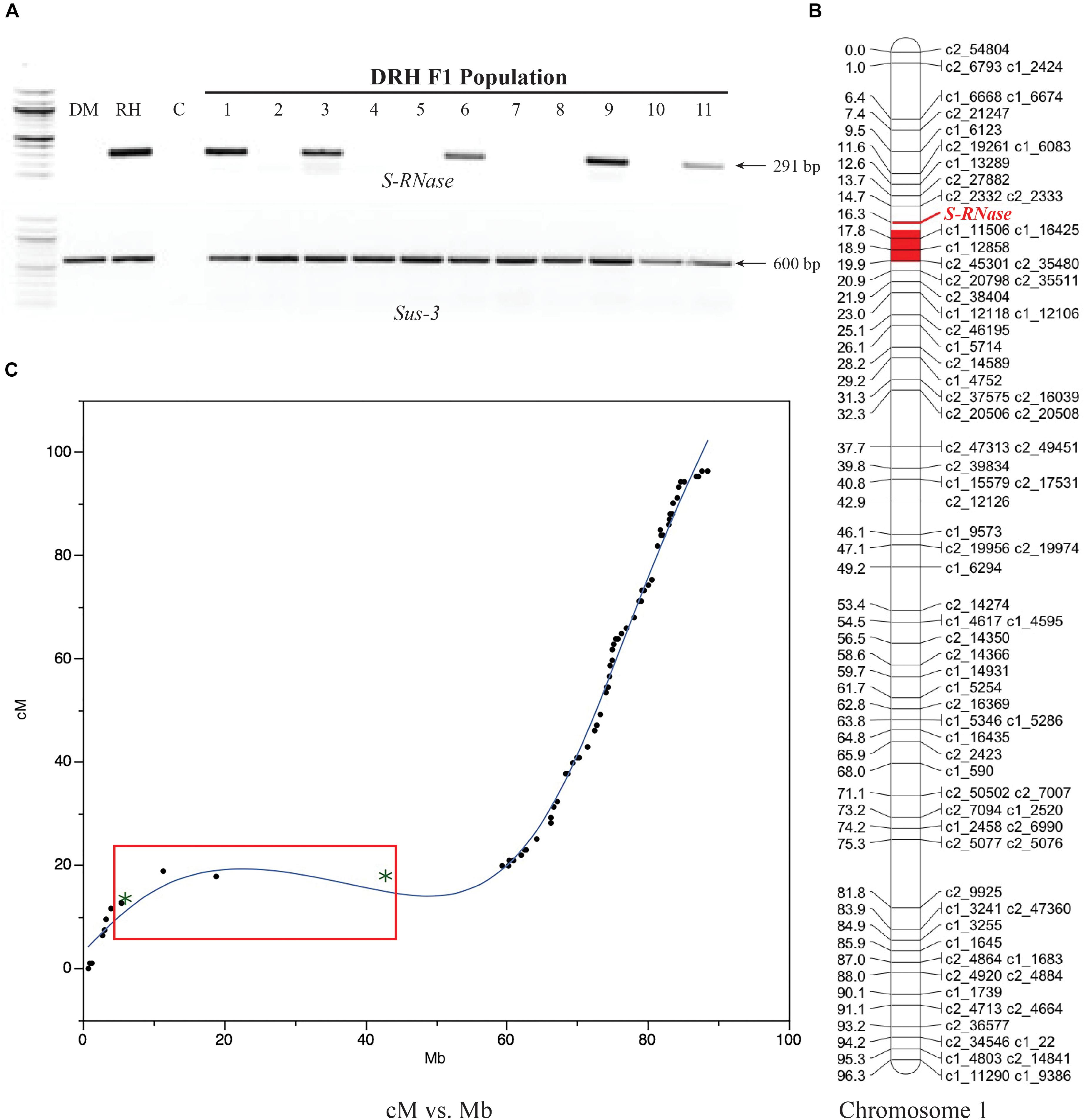
Figure 2. S-RNase gene mapping in diploid potato. (A) RH S-RNase (S. tub_St6) allelic screening on the DRH F1 population using the S-RNase and housekeeping Sucrose synthase 3 gene (Sus-3) primers. DM and RH parental lines are shown in the first two lanes followed by the negative control (C), and the RH-S-RNase segregation pattern of 11 F1-derived lines. (B) The S-RNase gene mapped to 16.3 cM on the short arm near the centromeric region of chromosome 1 (red). (C) Marey map of physical (Mb) versus genetic (cM) distances from chromosome I showing the S-RNase gene within a low-recombination region (red box). Asterisks within the red box represent SNPs spanning the region between to 6.1 and 18.9 Mb in the potato physical map (solcap_snp_c2_27882 and solcap_snp_c1_16425 markers, respectively).
Targeted Mutagenesis of S-RNase in Diploid Potatoes Using CRISPR/Cas9 Results in Self-Compatibility
A dual gRNA strategy (sgRNA 1 and sgRNA 2) was used to efficiently generate S-RNase KOs and disrupt the S-RNase function in DRH-195 and DRH-310 (Figure 3A). Multiple T0 plants were recovered for each line due to a 98% regeneration and transformation efficiency for DRH-195 and 93% for DRH-310 (Table 2). Based upon PCR analysis using primers to the S-RNase and gel detection of insertion/deletion polymorphisms, biallelic S-RNase mutations were recovered in both the DRH-195 and DRH-310-derived T0 lines (Figures 3B,C). Specifically, seven S-RNase KOs exhibiting polymorphic deletions with up to 580 bp were detected for DRH-195-derived T0 lines. In contrast, for DRH-310, only three S-RNase KOs were detected with up to 524 bp monomorphic deletions. To further characterize the CRISPR-targeted regions in selected T0 lines, both T0 KOs and WT-like S-RNase amplicons were sequenced (Figures 3D,E). A distinct nucleotide deletion was detected in each KO line, ranging from small bi-allelic deletions (1 bp) to large homozygous deletions (527 bp) in both S-RNase alleles of each DRH-derived T0 lines. Insertions (1 to 18 bp) and inversions (486 bp) were also observed in a bi-allelic configuration. Similarly, besides the described mutation types, chimeric mutations were detected in T0 lines, which has been reported in other species subjected to CRISPR-mediated mutagenesis, potentially due to late embryogenesis editing (Gomez et al., 2018).
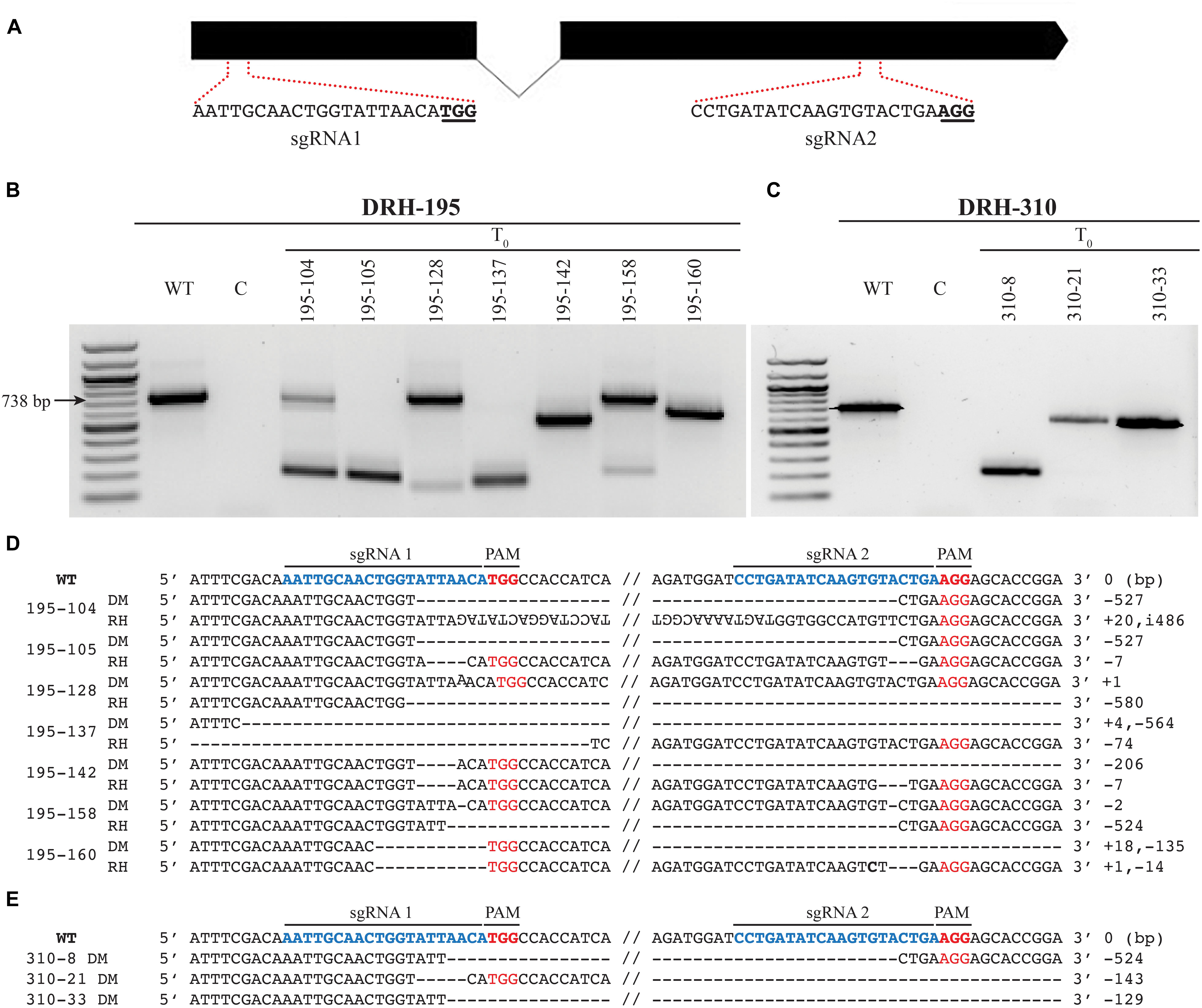
Figure 3. CRISPR/Cas9 mediated mutagenesis in two self incompatible diploid potato lines. (A) Single-guide RNAs (sgRNAs) designed to target the S-RNase exon1 (sgRNA 1) and exon 2 (sgRNA 2). Zoomed-in regions within the dotted lines show sgRNA and PAM sequences. (B) Seven and (C) three bi-allelic S-RNase knockouts (KOs) DRH-195 and DRH-310 T0 lines, respectively, detected by insertion/deletion polymorphisms compared to wild-type (WT) and negative controls (C). All DRH-195 KOs exhibit polymorphic deletions and DRH-310 monomorphic deletions (lines 195–105, 195–137, 195–142, and 195–160 presented a faint mutated or WT-like bands that is not observed in the figure). (D) Different mutation types detected by amplicon sequencing in selected KOs for DRH-195, and (E) DRH-310 T0 lines respectively. Wild-type S-RNase exhibiting selected sgRNAs (sgRNA1 and sgRNA2) and PAM sequences are shown at the top of each alignment. Different types of mutations including deletions (–), insertions (+), and inversions (i) detected in DM (DM) and RH (RH) S-RNase alleles of each T0 KO DRH-derived lines shown as ‘195-’ or ‘310-.’
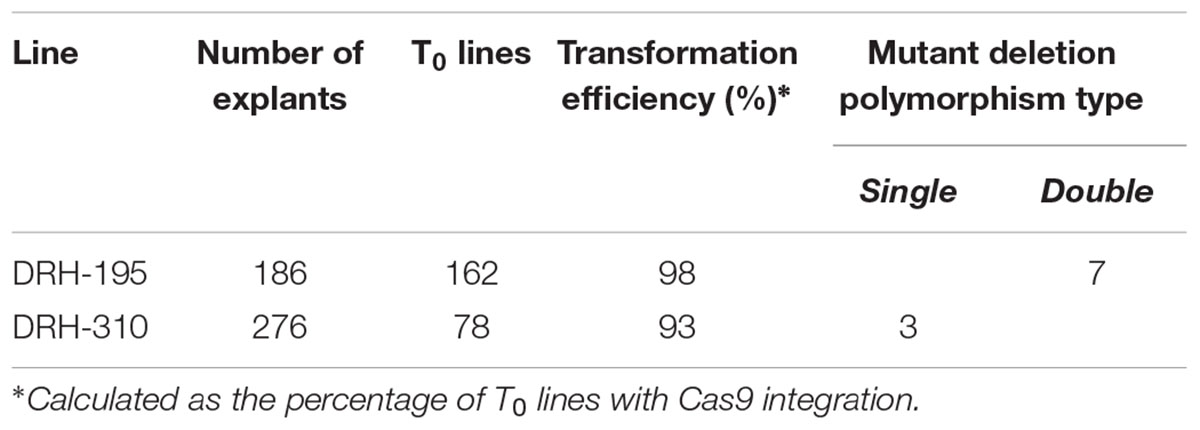
Table 2. DRH-195 and DRH-310-derived T0 and indel-based selected S-RNase KO lines with bi-allelic mutations.
During the regeneration process the high rate of cell division can induce chromosome doubling. To test whether the selected T0 KOs lines underwent spontaneous chromosome doubling, chloroplasts were counted in stomatal guard cells. One out of 10 S-RNase KO lines (DRH-195.104) revealed chromosome doubling which has also been observed in a related S-RNase KO approach (Ye et al., 2018). This phenomenon, known as endopolyploidization, is frequently observed in potatoes subjected to regeneration processes, in which structural cell and chromosome rearrangements at mitosis results in increased chromosome numbers (Karp et al., 1984; Owen et al., 1988). The tetraploid KO line was not considered for further analysis.
To confirm the S-RNase mutant phenotype, T0 KO lines were self-pollinated in two separate replications under greenhouse conditions. In both replications, all T0 KO lines set fruit (Figure 4A and Supplementary Tables S4, S5). In one of the replications, both WT non-transformed lines (DRH-195 and DRH-310) also exhibited a limited number of specific self-pollination events with fruit set that either had complete development (DRH-195, Supplementary Table S4) or arrest of fruit set 2 weeks after self-pollination (DRH-310, Figure 5A). However, after 1 week, new self-pollinations of WT lines did not set fruit (Figure 4B) suggesting plasticity of self-compatibility, a phenomenon observed previously in Solanum (Saba-El-Leil et al., 1994; Mena-Ali and Stephenson, 2007). This plasticity however, represents an unreliable source of self-compatibility as was evident in the ratio of fruit set per pollination observed, with self-pollination success in S-RNase KO lines being an order of magnitude higher than the WT in both DRH-195 and DRH-310 KO lines (Supplementary Tables S4, S5). To further investigate if this result was associated with the suppression of S-RNase expression, a semi-quantitative RT-PCR was performed. As shown in Figure 4C, S-RNase transcripts were detected in both WT lines 24 hpp yet no expression was detected in T0 KOs, confirming S-RNase expression in WT but not mutant lines.
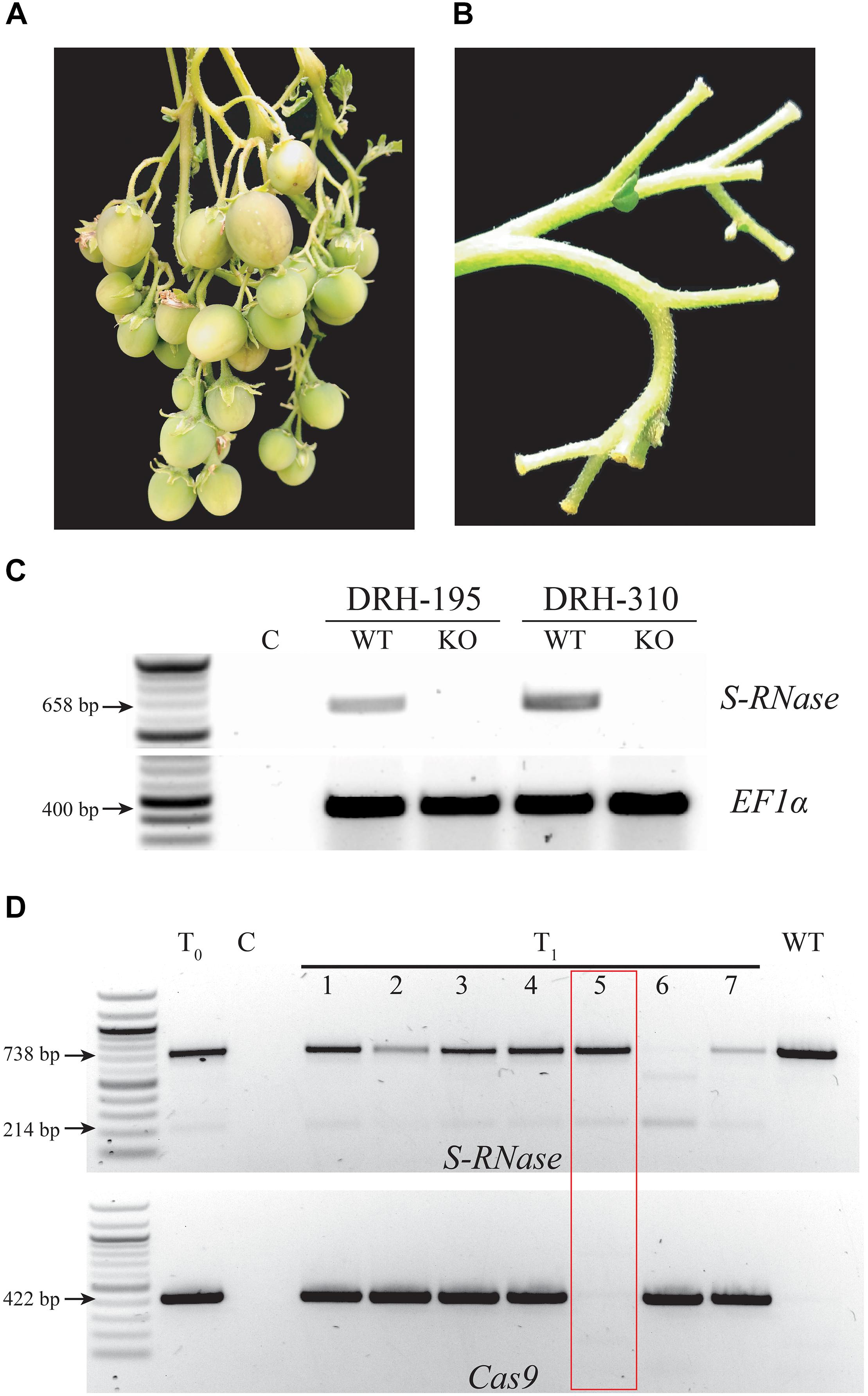
Figure 4. S-RNase expression and KO phenotype in self incompatible diploid potato lines. (A) Fruits obtained after 5 weeks of self-pollination in an S-RNase DRH-195-derived T0 mutant line. (B) Dropped flowers after self-pollination in the WT DRH-195. (C) Semi-quantitative reverse transcription PCR (RT-PCR) in DRH-195 and DRH-310 self-pollinated WT and KO T0 lines (DRH-195.158 and DRH-310-21, respectively). RNA was isolated from pistils 24 h after self-pollination revealing S-RNase expression in WT but not in KO lines as compared with the housekeeping gene control (EF1α). (D) T1 plants derived from the DRH-195.158 T0 line were screened with the S-RNase primers. WT-like bands with 1 bp deletion on each target site (causing frameshift leading to a premature stop codon, Supplementary Figure S1) are observed in T0 and T1 lines. Cas9 gene did not transmit to T1 line 5 (lane 5). A previously undetected band observed in lane 6 is potentially the result of transgenerational CRISPR/Cas9 activity. T0: DRH-195.158, C: Negative control. The red box is showing a T1 line segregating out Cas9 while maintaining the S-RNase KO.
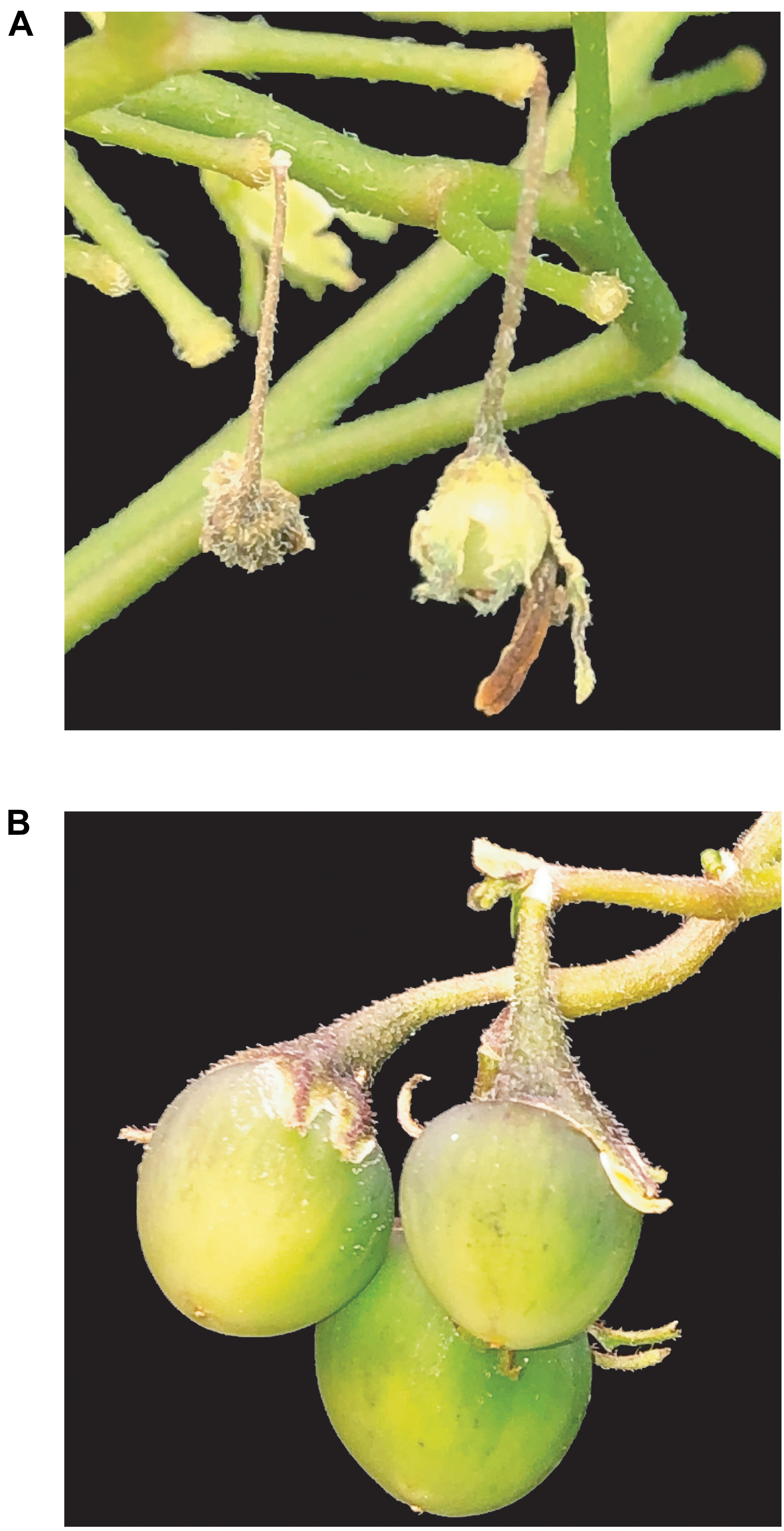
Figure 5. Fruit formation in WT and T1 S-RNase KO diploid potatoes lines. (A). Fruit setting arrest 2 weeks after self-pollination in the WT DRH-310. (B) Fruits obtained after 4 weeks of self-pollination in an S-RNase DRH-195-derived T1 mutant line.
Viable T1 seeds were obtained for each S-RNase T0 KO line. Self-compatibility was confirmed in T1 lines after self-pollination demostrating the inheritance and stablity of the S-RNase KO phenotype (Figure 5B). Cas9 inheritance in the T1 lines exhibited a segration ratio associated with a hemizygous multi-copy integration of Cas9 (4 out of 135), in addition to the segregation of the mutated S-RNase alleles (Figure 4D). Likewise, because of the activity of integrated Cas9, a potential transgenerational deletion was observed in a T1 line (DRH-195.158.6). These results demonstrate the advantage of using CRISPR/Cas9 to generate Cas9-free edited plants and the potential to transmit stable gene mutations through different generations.
Discussion
Self-incompatibility has been a limiting factor for inbred/F1 hybrid cultivar development in diploid potatoes because efforts involving crossing with wild SC relatives result in many undesirable traits segregating in the progeny. To redirect potato breeding toward an efficient inbred/F1 hybrid generation strategy, we exploited the S-RNase-based SI system in diploid potatoes and generated KO lines using CRISPR-based genome editing to achieve self-compatibility.
Amino acid sequence variation within S-RNase was observed among S. tub_Sp5, S. tub_St5, and S. tub_St6 alleles (Figure 1A). Nearly half of these variants were within the hypervariable domains (HVa and HVb) and not in conserved domains (C1-5) consistent with data that show the S-RNase variable regions are the determinants for allele specificity in different Solanum species (Matton et al., 1997, 1999; Brisolara-Corrêa et al., 2015). Specifically, four amino acids within these variable regions (T74, N76, Y77, and R101) have been reported as the sole factors for allele conversion of the pollen rejection phenotype in S. chacoense (Matton et al., 1997). Three of these amino acid changes were present within the S. tub_Sp5, S. tub_St5, and S. tub_St6 alleles indicating that these variations could be sufficient to confer allele specificity while preserving their catalytic activity which is associated with two of the five conserved domains (Kao and Tsukamoto, 2004).
Ioerger et al. (1990) observed that S-RNase inter-specific similarities were higher than intra-specific similarities in Solanaceae, concluding that S-RNase divergence pre-dates speciation in this clade. The results observed in our study further confirm this previous observation. A high degree of inter-specific S-RNase amino acid sequence similarity was observed in Solanum S-RNases (S. tuberosum and S. chacoense). Conversely, a clear intra-specific separation within the S. tuberosum S-RNase alleles (Figure 1C) was also observed, consistent with the hypothesis of a single ancestral origin of S-RNase and conservation of specific polymorphisms throughout evolution governing allelic diversity (Ioerger et al., 1991; Dzidzienyo et al., 2016).
The S-RNase gene mapped to chromosome 1 within a region of low recombination consistent with the hypothesis to promote outbreeding due to a reduction in recombination events between the S-RNase and SLF genes (Kubo et al., 2015; Fujii et al., 2016). This chromosome position has also been reported in other Solanaceae members. For instance, S-RNase is located on chromosome 1 in S. lycopersicum and S. peruvianum within highly complex and repetitive genomic regions (Kubo et al., 2015; Fujii et al., 2016). Furthermore, Kubo et al. (2015) mapped an SLF, the other component of the S-locus, also to chromosome 1 in potato genome within a repeat-rich sub-centromeric region, suggesting that the S-RNase location was at the same position since these genes are reported to be closely linked (Sijacic et al., 2004).
All edited T0 lines had a frameshift in the coding region close to sgRNA 1, leading to a premature stop codon. The resulting truncated sequence prevented the amplification of the S-RNase gene by removing the primer annealing site at the 3′ end (Supplementary Figure S1). Similarly, the consistent mutations generated by the two sgRNAs allowed detection of S-RNase size polymorphisms. It should be noted that this strategy was selected for the potential to use PCR for a quick and facile screen for large deletions in T0 lines. However, undetected insertions/deletions or inversions could be present in T0 lines. For instance, sequencing data revealed a single bp insertion and deletion in the DM S-RNase allele of DRH-195.128 and DRH-195.158, respectively, showing a similar amplicon size in agarose gels as WT (Figures 3B,D). These observations indicate that a large number of allelic KOs can be generated given the high transformation efficiency observed in both diploid lines.
The DRH-195.158 T0 KO line, which exhibited a single bp deletion at each sgRNA targeting site in the RH S. tub_Sp5 allele (Figure 3D), showed a new S-RNase deletion in a T1-derived line (Figure 4D). Given the Cas9 mismatching tolerance, this allele possibly underwent a new mutagenesis event, displaying a different mutation pattern in the T1 generation. In different plant species, it has been found that editing occurs at a higher frequency across generations, therefore new mutations segregate from WT alleles in heterozygous T0 as a result of constitutive expression of Cas9 (Feng et al., 2014; Xu et al., 2015; Wang et al., 2018).
Two independent self-pollination assays were conducted in DRH-195 and DRH-310 WT lines in 2015 and 2018 with a minimal of 50 flowers, demonstrating their SI. However, a third biological replicate in 2018 resulted in fruit formation suggesting plasticity in the strength of SI. Environmental effects along with plant phenology have been associated with unstable SI in angiosperms. For instance, temperature fluctuations, photoperiod, glucose starvation, and humidity significantly reduced SI in S. peruvianum after selfing (Webb and Williams, 1988). This process, known as pseudo-self-incompatibility, has also been reported in grasses in which artificial self-pollination techniques can contribute to SI breakdown (Do Canto et al., 2016). Similarly, sporadic fruit set has been observed across Solanaceae species such as Witheringia solanacea, S. carolinense, S. peruvianum, and N. alata in which floral age, flowering stage, and delayed floral abscission has been associated with fruit set in SI populations (Stone et al., 2006; Mena-Ali and Stephenson, 2007; Miller and Kostyun, 2011; Liao et al., 2016). This phenomenon has also been observed in species under sporophytic SI, in which floral age reduces the expression of the S-locus associated genes in Brassica oleracea resulting in SI breakdown (Hadj-Arab et al., 2010). In natural populations of Campanula rapunculoides, strong SI has been observed in young flowers. However, self-fruit formation is also evident in old flowers as a consequence of pollen scarcity and low fruit production from prior inflorescences (Stephenson et al., 2000). Therefore, environmental conditions favoring SI breakdown (plant age, plant health, and greenhouse conditions) could lead to fruit set in one of the WT biological replicates in this study.
Unlike pseudo-self-incompatibility, the S-RNase KO proved to be both stable and consistent across different replications and generations, presenting a higher ratio of fruit set per pollination when compared with self-fruit WT lines (Supplementary Tables S4, S5). Although the SC phenotype appears to be line dependent, distinctive S-RNase KO lines exhibited either high fruit set or seed formation. These results also indicated that genes other than S-RNase could be contributing to the strength of the SI response in both WT and S-RNase KOs. In fact, besides the S-RNase gene, other SI modifier loci can modulate the pollen rejection response in several Solanum species (McClure et al., 1999; O’Brien et al., 2002; Goldraij et al., 2006). For instance, S-RNase-independent stylar factors such as eEF1A or High Top-Band (HT-B) proteins, can directly or indirectly interact with S-RNase contributing to the SI response (Goldraij et al., 2006; Soulard et al., 2014). Similarly, unintended somaclonal variation and chromosomal rearrangements associated with the potato regeneration processes and Cas9 activity, respectively, could also contribute to variations in the observed ratio of fruit set per pollination within the S-RNase KOs.
This hypothesis is further supported by Peterson et al. (2016) which identified several genomic regions associated with self-fertility in a DRH F1 population, located on chromosomes IV, IX, XI, and XII. They also found that a specific Single Nucleotide Polymorphism (SNP) associated with the RH allele, fixed in selfed populations, is likely the primary factor for self-fertility in the DRH F1 progeny. Overall, this study demonstrates that S-RNase is the primary component for self-pollen rejection in DRH-195 and DRH-310. However, external evidence suggests that besides RH self-fertility mechanisms, S-RNase-independent stylar factors and environmental conditions could play a role in spontaneous self-compatibility observed in the WT lines in this study.
In this study, we generated self-compatible potato diploid lines by targeting the S-RNase gene using the CRISPR-Cas9 system. We first computationally identified three new S-RNase alleles in self incompatible diploid lines (a DM and two RH alleles, each inherited to DRH-195 and DRH-310, respectively) and mapped this gene to chromosome 1 within the peri-centromeric region consistent with the localization of the S-locus to a low recombination region on chromosome 1. S-RNase KO lines were obtained using a dual sgRNA strategy in which premature stop codons were generated. After self-pollination, fruits were set in selected KO lines in T0 and T1 lines. Cas9-free KO lines were also identified in T1 lines. Our results demonstrated the inheritance and stablity of the S-RNase KO phenotype, which can contribute to utilization of SC as a first step for the generation of commercial diploid cultivars.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found in Supplementary Table S1.
Author Contributions
FE-R and DD conceived the research idea. FE-R, NM-C, SN, CB, DZ, and DD designed the research experiments, analyzed the data, and wrote the manuscript. FE-R performed laboratory experiments. FE-R and NM-C performed self-pollination assays.
Funding
FE-R received support from a Colciencias-Fulbright Fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors want to thank Kate Shaw and Chen Zhang for chloroplast counting.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00376/full#supplementary-material
FIGURE S1 | S-RNase open reading-frames in T0 knockout (KO) lines derived in two SI diploid potato lines. Sequences shown within the dotted lines contain 5′ and 3′ mRNA borders of this gene. Primer sequences designed to amplify the S-RNase ORF are shown as well as the sgRNA 1 located in exon 1. Individual dots represent nucleotides within the S-RNase gene. All T0 KO lines had a frameshift near the sgRNA 1 target region (blue dotted line) creating a premature stop codon (red dotted line). Black dotted line represents in frame S-RNase regions and nucleotides. Black dotted dash lines represent missing transcript sequence. S-RNase alleles from DM and RH are shown for each KO line.
TABLE S1 | Retrieved Illumina reads for genomic assembly of the DRH potato lines and assembly statistics.
TABLE S2 | Primers and sgRNAs used in this study.
TABLE S3 | Top TBLASTN hits of reported S-RNase proteins in the DM genome assembly.
TABLE S4 | Fruit set and seed count upon self-pollination in DRH-195 wild-type and S-RNase-derived KO lines.
TABLE S5 | Fruit set and seed count upon self-pollination in DRH-310 wild-type and S-RNase-derived KO lines.
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Brisolara-Corrêa, L., Thompson, C. E., Fernandes, C. L., and de Freitas, L. B. (2015). Diversification and distinctive structural features of S-RNase alleles in the genus Solanum. Mol.Genet. Genomics. 290, 987–1002. doi: 10.1007/s00438-014-0969-3
Bush, W. S., and Moore, J. H. (2012). Chapter 11: genome-wide association studies. PLoS Comput. Biol. 8:e1002822. doi: 10.1371/journal.pcbi.1002822
Covey, P. A., Kondo, K., Welch, L., Frank, E., Sianta, S., Kumar, A., et al. (2010). Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade. Plant J. 64, 367–378. doi: 10.1111/j.1365-313X.2010.04340.x
Devaux, A., Kromann, P., and Ortiz, O. (2014). Potatoes for sustainable global food security. Potato Res. 57, 185–199. doi: 10.1007/s11540-014-9265-1
Do Canto, J., Studer, B., and Lubberstedt, T. (2016). Overcoming self-incompatibility in grasses: a pathway to hybrid breeding. Theor. Appl. Genet. 129, 1815–1829. doi: 10.1007/s00122-016-2775-2
Doudna, J. A., and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Dzidzienyo, D. K., Bryan, G. J., Wilde, G., and Robbins, T. P. (2016). Allelic diversity of S-RNase alleles in diploid potato species. Theor. Appl. Genet. 129, 1985–2001. doi: 10.1007/s00122-016-2754-7
FAOSTAT (2019). FAOSTAT Database Collections. Rome: Food and Agriculture Organization of the United Nations.
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.-L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Fujii, S., Kubo, K. I., and Takayama, S. (2016). Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants 2:16130. doi: 10.1038/nplants.2016.130
Gebhardt, C., Ritter, E., Barone, A., Debener, T., Walkemeier, B., Schachtschabel, U., et al. (1991). RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor. Appl. Genet. 83, 49–57. doi: 10.1007/BF00229225
Goldraij, A., Kondo, K., Lee, C. B., Hancock, C. N., Sivaguru, M., Vazquez-Santana, S., et al. (2006). Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439:805. doi: 10.1038/nature04491
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2018). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant. Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Hadj-Arab, H., Chèvre, A.-M., Gaude, T., and Chable, V. (2010). Variability of the self-incompatibility reaction in Brassica oleracea L. with S15 haplotype. Sex. Plant. Reprod. 23, 141–151. doi: 10.1007/s00497-009-0119-y
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hardigan, M. A., Crisovan, E., Hamiltion, J. P., Kim, J., Laimbeer, P., Leisner, C. P., et al. (2016). Genome reduction uncovers a large dispensable genome and adaptive role for copy number variation in asexually propagated Solanum tuberosum. Plant Cell 28, 388–405. doi: 10.1105/tpc.15.00538
Hosaka, K., and Hanneman, R. E. (1998). Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 1. Detection of an S locus inhibitor (Sli) gene. Euphytica 99, 191–197. doi: 10.1023/A:1018353613431
Hua, Z.-H., Fields, A., and Kao, T. (2008). Biochemical models for S-RNase-based self-incompatibility. Mol. Plant 1, 575–585. doi: 10.1093/mp/ssn032
Igic, B., and Kohn, J. R. (2001). Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. U.S.A. 98, 13167–13171. doi: 10.1073/pnas.231386798
Ioerger, T. R., Clark, A. G., and Kao, T. H. (1990). Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc. Natl. Acad. Sci. U.S.A. 87, 9732–9735. doi: 10.1073/pnas.87.24.9732
Ioerger, T. R., Gohlke, J. R., Xu, B., and Kao, T. I. H. (1991). Sexual plant lleproduction primary structural features of the self-incompatibility protein in Solanaceae. Sex. Plant. Reprod. 4, 81–87. doi: 10.1007/BF00196492
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant. Sci. 9:985. doi: 10.3389/fpls.2018.00985
Jansky, S. H., Charkowski, A. O., Douches, D. S., Gusmini, G., Richael, C., Bethke, P. C., et al. (2016). Reinventing potato as a diploid inbred line–based crop. Crop Sci. 56, 1412–1422. doi: 10.2135/cropsci2015.12.0740
Jansky, S. H., Chung, Y. S., and Kittipadukal, P. (2014). M6: a diploid potato inbred line for use in breeding and genetics research. J. Plant. Regist. 8, 195–199. doi: 10.3198/jpr2013.05.0024crg
Kao, T., and Tsukamoto, T. (2004). The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16, S72–S83. doi: 10.1105/tpc.016154
Karp, A., Risiott, R., Jones, M. G. K., and Bright, S. W. J. (1984). Chromosome doubling in monohaploid and dihaploid potatoes by regeneration from cultured leaf explants. Plant Cell Tissue Organ. Cult. 3, 363–373. doi: 10.1007/BF00043089
Koncz, C., Martini, N., Szabados, L., Hrouda, M., Bachmair, A., and Schell, J. (1994). “Specialized vectors for gene tagging and expression studies,” in Plant Molecular Biology Manual, eds S. B. Gelvin and R. A. Schilperoort (Dordrecht: Springer), 53–74. doi: 10.1007/978-94-011-0511-8_4
Kowyama, Y., Kunz, C., Lewis, I., Newbigin, E., Clarke, A. E., and Anderson, M. A. (1994). Self-compatibility in aLycopersicon peruvianum variant (LA2157) is associated with a lack of style S-RNase activity. Theor. Appl. Genet. 88, 859–864. doi: 10.1007/BF01253997
Kubo, K., Entani, T., Takara, A., Wang, N., Fields, A. M., Hua, Z., et al. (2010). Collaborative non-self recognition system in S-RNase based self-incompatibility. Science 330, 796–799. doi: 10.1126/science.1195243
Kubo, K., Paape, T., Hatakeyama, M., Entani, T., Takara, A., Kajihara, K., et al. (2015). Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat. Plants 1:14005. doi: 10.1038/nplants.2014.5
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv [Preprint]. arXiv:1303.3997
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, W., and Chetelat, R. T. (2015). Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl. Acad. Sci. U.S.A. 112, 4417–4422. doi: 10.1073/pnas.1423301112
Li, W., Zarka, K. A., Douches, D. S., Coombs, J. J., Pett, W. L., and Grafius, E. J. (1999). Coexpression of potato PVYo coat protein and cryV-Bt genes in potato. J. Am. Soc. Hort. Sci. 124, 218–223. doi: 10.21273/JASHS.124.3.218
Liao, J., Dai, J., Kang, H., Liao, K., Ma, W., Wang, J., et al. (2016). Plasticity in the self-incompatibility system of cultivated Nicotiana alata. Euphytica 208, 129–141. doi: 10.1007/s10681-015-1606-x
Manrique-Carpintero, N. C., Coombs, J. J., Cui, Y., Veilleux, R. E., Buell, C. R., and Douches, D. (2015). Genetic map and QTL analysis of agronomic traits in a diploid potato population using single nucleotide polymorphism markers. Crop Sci. 55, 2566–2579. doi: 10.2135/cropsci2014.10.0745
Matton, D. P., Luu, D. T., Xike, Q., Laublin, G., O’Brien, M., Maes, O., et al. (1999). Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11, 2087–2097. doi: 10.1105/tpc.11.11.2087
Matton, D. P., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D., et al. (1997). Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9, 1757–1766. doi: 10.1105/tpc.9.10.1757
McClure, B., Mou, B., Canevascini, S., and Bernatzky, R. (1999). A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. U.S.A. 96, 13548–13553. doi: 10.1073/pnas.96.23.13548
Mena-Ali, J. I., and Stephenson, A. G. (2007). Segregation analyses of partial self-incompatibility in self and cross progeny of Solanum carolinense reveal a leaky S-allele. Genetics 177, 501–510. doi: 10.1534/genetics.107.073775
Mendoza, H. A., Mihovilovich, E. J., and Saguma, F. (1996). Identification of triplex (YYYy) potato virus Y (PVY) immune progenitors derived from Solanum tuberosum ssp.andigena. Am. Potato J. 73, 13–19. doi: 10.1007/BF02849300
Miller, J. S., and Kostyun, J. L. (2011). Functional gametophytic self-incompatibility in a peripheral population of Solanum peruvianum (Solanaceae). Heredity 107, 30–39. doi: 10.1038/hdy.2010.151
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
O’Brien, M., Kapfer, C., Major, G., Laurin, M., Bertrand, C., Kondo, K., et al. (2002). Molecular analysis of the stylar-expressed Solanum chacoense small asparagine-rich protein family related to the HT modifier of gametophytic self-incompatibility in Nicotiana. Plant J 32, 985–996. doi: 10.1046/j.1365-313X.2002.01486.x
Ordoñez, B. (2014a). Assessment of Ploidy by Chloroplast Count in Stomatal Guard Cells. Lima: International Potato Center, 4.
Owen, H. R., Veilleux, R. E., Levy, D., and Ochs, D. L. (1988). Environmental, genotypic, and ploidy effects on endopolyploidization within a genotype of Solanum phureja and its derivatives. Genome 30, 506–510. doi: 10.1139/g88-085
Park, J., Bae, S., and Kim, J. (2015). Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. doi: 10.1101/005074.Bae
Pellagatti, A., Dolatshad, H., Valletta, S., and Boultwood, J. (2015). Application of CRISPR/Cas9 genome editing to the study and treatment of disease. Arch. Toxicol. 89, 1023–1034. doi: 10.1007/s00204-015-1504-y
Peterson, B. A., Holt, S. H., Laimbeer, F. P. E., Doulis, A. G., Coombs, J., Douches, D. S., et al. (2016). Self-fertility in a cultivated diploid potato population examined with the infinium 8303 potato single-nucleotide polymorphism array. Plant Genome 9, 1–13. doi: 10.3835/plantgenome2016.01.0003
Porcher, E., and Lande, R. (2005). Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution 59, 46–60. doi: 10.1111/j.0014-3820.2005.tb00893.x
Ramanauskas, K., and Igić, B. (2017). The evolutionary history of plant T2/S-type ribonucleases. PeerJ 5:e3790. doi: 10.7717/peerj.3790
Raymundo, R., Asseng, S., Robertson, R., Petsakos, A., Hoogenboom, G., Quiroz, R., et al. (2017). Climate change impact on global potato production. Eur. J. Agron. 100, 87–98. doi: 10.1016/j.eja.2017.11.008
Royo, J., Kunz, C., Kowyama, Y., Anderson, M., Clarke, A. E., and Newbigin, E. (1994). Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc. Natl. Acad. Sci U.S.A. 91, 6511–6514. doi: 10.1073/pnas.91.14.6511
Saba-El-Leil, M. K., Rivard, S., Morse, D., and Cappadocia, M. (1994). The S11 and S13 self incompatibility alleles in Solanum chacoense Bitt. are remarkably similar. Plant. Mol. Biol. 24, 571–583. doi: 10.1007/BF00023555
Sassa, H., Kakui, H., Miyamoto, M., Suzuki, Y., Hanada, T., Ushijima, K., et al. (2007). S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175, 1869–1881. doi: 10.1534/genetics.106.068858
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sijacic, P., Wang, X., Skirpan, A. L., Wang, Y., Dowd, P. E., McCubbin, A. G., et al. (2004). Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429, 302–305. doi: 10.1038/nature02523
Soulard, J., Boivin, N., Morse, D., and Cappadocia, M. (2014). eEF1A Is an S-RNase binding factor in Self-incompatible Solanum chacoense. PLoS One 9:e90206. doi: 10.1371/journal.pone.0090206
Stephenson, A. G., Good, S. V., and Vogler, D. W. (2000). Interrelationships among inbreeding depression, plasticity in the self-incompatibility system, and the breeding system of Campanula rapunculoides L. (Campanulaceae). Ann. Bot. 85, 211–219. doi: 10.1006/anbo.1999.1033
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi: 10.1038/nature13011
Stone, J. L., Sasuclark, M. A., and Blomberg, C. P. (2006). Variation in the self-incompatibility response within and among populations of the tropical shrub Witheringia solanacea (Solanaceae). Am. J. Bot. 93, 592–598. doi: 10.3732/ajb.93.4.592
Takayama, S., and Isogai, A. (2005). Self-Incompatibility in Plants. Annu. Rev. Plant. Biol. 56, 467–489. doi: 10.1146/annurev.arplant.56.032604.144249
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Wang, W., Pan, Q., He, F., Akhunova, A., Chao, S., Trick, H., et al. (2018). Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 1, 65–74. doi: 10.1089/crispr.2017.0010
Webb, M. C., and Williams, E. G. (1988). Effects of temperature, light, nutrients and carbon dioxide on the strength of the self-incompatibility response in detached flowers of Lycopersicon peruvianum. Ann. Bot. 61, 395–404. doi: 10.1093/oxfordjournals.aob.a087570
Xing, H.-L., Dong, L., Wang, Z.-P., Zhang, H.-Y., Han, C.-Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant. Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xu, R.-F., Li, H., Qin, R.-Y., Li, J., Qiu, C.-H., Yang, Y.-C., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5:11491. doi: 10.1038/srep11491
Keywords: self-incompatibility, diploid potato, S-RNase, CRISPR-Cas9, gene editing
Citation: Enciso-Rodriguez F, Manrique-Carpintero NC, Nadakuduti SS, Buell CR, Zarka D and Douches D (2019) Overcoming Self-Incompatibility in Diploid Potato Using CRISPR-Cas9. Front. Plant Sci. 10:376. doi: 10.3389/fpls.2019.00376
Received: 22 December 2018; Accepted: 12 March 2019;
Published: 02 April 2019.
Edited by:
Jose M. Seguí-Simarro, Universitat Politècnica de València, SpainReviewed by:
Roger T. Chetelat, University of California, Davis, United StatesCarlos Romero, Instituto de Biología Molecular y Celular de Plantas (IBMCP), Spain
Copyright © 2019 Enciso-Rodriguez, Manrique-Carpintero, Nadakuduti, Buell, Zarka and Douches. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Douches, ZG91Y2hlc2RAbXN1LmVkdQ==
 Felix Enciso-Rodriguez
Felix Enciso-Rodriguez Norma C. Manrique-Carpintero
Norma C. Manrique-Carpintero Satya Swathi Nadakuduti
Satya Swathi Nadakuduti C. Robin Buell
C. Robin Buell Daniel Zarka
Daniel Zarka David Douches
David Douches