
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 14 March 2019
Sec. Plant Biotechnology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00313
RNA interference (RNAi) is a promising method for controlling pest insects by silencing the expression of vital insect genes to interfere with development and physiology; however, certain insect Orders are resistant to this process. In this study, we set out to test the ability of in planta-expressed dsRNA synthesized within the plastids to silence gene expression in an insect recalcitrant to RNAi, the lepidopteran species, Manduca sexta (tobacco hornworm). Using the Manduca vacuolar-type H+ ATPase subunit A (v-ATPaseA) gene as the target, we first evaluated RNAi efficiency of two dsRNA products of different lengths by directly feeding the in vitro-synthesized dsRNAs to M. sexta larvae. We found that a long dsRNA of 2222 bp was the most effective in inducing lethality and silencing the v-ATPaseA gene, when delivered orally in a water droplet. We further transformed the plastid genome of the M. sexta host plant, Nicotiana tabacum, to produce this long dsRNA in its plastids and performed bioassays with M. sexta larvae on the transplastomic plants. In the tested insects, the plastid-derived dsRNA had no effect on larval survival and no statistically significant effect on expression of the v-ATPaseA gene was observed. Comparison of the absolute quantities of the dsRNA present in transplastomic leaf tissue for v-ATPaseA and a control gene, GFP, of a shorter size, revealed a lower concentration for the long dsRNA product compared to the short control product. We suggest that stability and length of the dsRNA may have influenced the quantities produced in the plastids, resulting in inefficient RNAi in the tested insects. Our results imply that many factors dictate the effectiveness of in planta RNAi, including a likely trade-off effect as increasing the dsRNA product length may be countered by a reduction in the amount of dsRNA produced and accumulated in the plastids.
Utilization of the RNA interference (RNAi) pathway, first discovered in Caenorhabditis elegans (Fire et al., 1998) and later observed in a wide variety of species (Bellés, 2010), to knockdown gene expression is a popular tool for biological research involving insects (Dietzl et al., 2007). Initially used to observe phenotypic effects of gene knockdowns, using RNAi knockdown of vital genes to induce a lethal response has been suggested as a means for developing species-specific pest control methods that are alternatives to potentially harmful chemical methods (Whyard et al., 2009). In insects, the RNAi pathway can be activated through environmental RNAi, whereby exogenous double-stranded RNA (dsRNA) molecules taken up from the environment, trigger the post-transcriptional silencing of endogenous mRNA molecules (Whangbo and Hunter, 2008). This process is accomplished by uptake of ingested dsRNA molecules from the midgut lumen into the insect cells through SID-1 channels and receptor-mediated endocytosis (Shih and Hunter, 2011; Cappelle et al., 2016). Later, the dsRNA molecules are processed into small interfering RNA (siRNA) molecules that induce gene knockdown (Meister and Tuschl, 2004). Thus, oral consumption is considered an effective tool for introducing dsRNA into insect cells for developing RNAi-based pest control strategies.
The dsRNA to be delivered to insects is commonly synthesized in vitro or expressed in bacteria (Timmons et al., 2001; Flores-Escobar et al., 2013). However, a more useful strategy for application in a greenhouse or field setting is in planta expression by the crop itself. Nuclear dsRNA expression was initially tested and has been successful for pest insects such as western corn rootworm (Baum et al., 2007). Yet, success for insects within the crop-devastating clade Lepidoptera is generally less pronounced, and complete plant resistance to feeding through nuclear dsRNA expression has not been achieved (Terenius et al., 2011). Plants, like other eukaryotes, possess an innate RNAi pathway (Melnyk et al., 2011), and thus process dsRNAs into siRNA prior to insect ingestion. In fact, gene knockout of RNAi processing pathway components within nuclear dsRNA-expressing plants increases the ability of these plants to resist feeding by cotton bollworm (Mao et al., 2007), a result which suggests that the plant’s intrinsic ability to process dsRNA into siRNA is responsible for the reduced RNAi efficiency in insects, because siRNA uptake in the insect midgut lumen is less efficient than long dsRNA uptake (Bolognesi et al., 2012).
Expression of dsRNA within the plastid presents a possible solution for overcoming this problem. Plastids are prokaryotic in origin and do not maintain an RNAi pathway, thus dsRNA should remain intact prior to insect consumption. This strategy was successful in knocking down gene expression in Colorado potato beetle (Zhang et al., 2015) and cotton bollworm (Jin et al., 2015; Bally et al., 2016). These results indicate that this strategy could potentially prove effective in imparting in planta protection from insect herbivory, but the full range of properties that influence success are yet to be elucidated. For instance, dsRNA molecule size and sequence play a role in effectiveness of the approach and must be examined individually for any specific research goal. With respect to molecule size, longer dsRNA products are generally associated with greater uptake and utilization by the insect RNAi pathway (Miller et al., 2012). Previously, a study comparing the effectiveness of in vitro synthesized long dsRNA and an artificial microRNA (miRNA) targeting chitin synthase gene A in the brown planthopper revealed that long dsRNA was more effective in gene silencing, resulting in more significant effects on fecundity and ovary development compared to miRNA (Li et al., 2017). Also, although several other studies using in planta expressed miRNA-based approaches showed positive results (Agrawal et al., 2015; Jiang et al., 2017; He et al., 2019), complete plant resistance to insect herbivory was not accomplished in most cases. With regard to molecule sequence, the range of insect species affected by a single dsRNA product depends on the gene target. For instance, dsRNAs targeting western corn rootworm vacuolar-type H+ ATPase subunits A and E (v-ATPaseA and v-ATPaseE) genes cause significant mortality in Colorado potato beetle (Baum et al., 2007). Similarly, dsRNAs that target two midgut genes in tobacco hornworm, Manduca sexta, silence homologous genes in tomato hornworm (Poreddy et al., 2017). These results imply that dsRNA molecules complementary to conserved genes offer a greater range of possible insect targets, which could be beneficial for field application.
In this study, we set out to evaluate the effect of dsRNA length on RNAi efficiency by first comparing the effectiveness of two in vitro-synthesized dsRNA molecules of different sizes (long, 2222 bp and short, 259 bp) at inducing gene silencing and mortality in larvae of the tobacco hornworm, M. sexta, a widely used insect model in molecular biology. The dsRNAs target the v-ATPaseA gene, which has been demonstrated to be an effective target for achieving lethality in numerous insect species (Baum et al., 2007; Ulrich et al., 2015; Yu et al., 2016). Then, we transformed the plastid genome of tobacco, Nicotiana tabacum, with an expression cassette producing the long v-ATPAseA dsRNA, as this fragment was the most effective at inducing RNAi when fed as in vitro-synthesized products. This product is larger than dsRNAs produced in chloroplasts in previous studies (Jin et al., 2015; Zhang et al., 2015; Bally et al., 2016). Bioassays with M. sexta larvae were then performed on the transplastomic plants to determine whether in planta-expressed dsRNA could induce lethality in feeding insects. Finally, we quantified expression of the long dsRNA product and compared with expression of a shorter 223 bp dsRNA product to determine whether the size of the dsRNA had any effect on the amount of dsRNA produced in the plastids.
The tobacco plants used in this study were of the variety “81V9,” a low-alkaloid cultivar (Menassa et al., 2001). Tobacco seeds were sterilized by a 5-min wash with 70% ethanol and grown in Magenta vessels containing Murashige and Skoog (MS) medium (4.4 g/L Murashige and Skoog Basal Medium with Vitamins, 3% sucrose and 0.7% agar). Plants for bioassays were transferred to soil and grown under 16L:8D light cycles in greenhouses under ambient conditions. All tissue culture stages were performed at room temperature under a 16L:8D photoperiod.
PCR primers (Supplementary Table 1) were designed to amplify product sizes of 2222 bp (v-ATPAseA long) and 259 bp (v-ATPaseA short) from M. sexta cDNA. The resulting dsRNA sequences are listed in Supplementary Table 2. The primers were designed such that v-ATPAseA short was contained within v-ATPAseA long (Supplementary Figure 1) and sequence identity with homologs in other organisms is shown in Supplementary Figure 2. A 223 bp product of green fluorescent protein (GFP) from Aequorea victoria was used as a dsRNA control. All primers contained T7 RNA polymerase promoter sequences at their 5′ end for in vitro dsRNA synthesis. Templates for in vitro dsRNA synthesis were prepared by PCR amplification, followed by gel purification using the QIAquick Gel Extraction Kit (Qiagen). Double-stranded RNA targeting v-ATPaseA was synthesized using the MEGAscript T7 Transcription Kit (Ambion) while dsRNA targeting GFP was synthesized using the RiboMAX Express RNAi System (Promega) following the manufacturer’s instructions.
With a long-term goal of creating pest-resistant tomato plants via plastome engineering for dsRNA production, we designed and constructed a tomato plastome transformation vector pTomCT. Given the prolonged reported timeframe for generating transplastomic tomato plants (Ruf et al., 2001) and the existence of much shorter and relatively routine procedures for tobacco plastid transformation, we initially tested the utility of the pTomCT vector by transforming tobacco chloroplasts. We hypothesized that the high level of homology between tomato and tobacco plastomes (99% homology in the targeted region) would facilitate efficient transformation and regeneration of transplastomic clones in tobacco.
The tomato plastome transformation construct pTomCT was designed to insert/integrate the expression cassette into a transcriptionally silent intergenic spacer between the rps12 and trnV genes of the tomato plastome NC_007898 (Figure 1), thus eliminating potential interference from endogenous transcriptional activity. For this purpose, DNA sequences between nucleotides 99,339 – 100,883 and 100,884 – 102,396 of the tomato plastome were used as the left and right flanking sequences of the expression cassette, respectively. The synthetic DNA comprising the flanking regions and the expression cassette was directionally cloned into Kas I and Hind III restriction sites on the pUC57 (AMPR) plasmid, which served as the backbone vector.
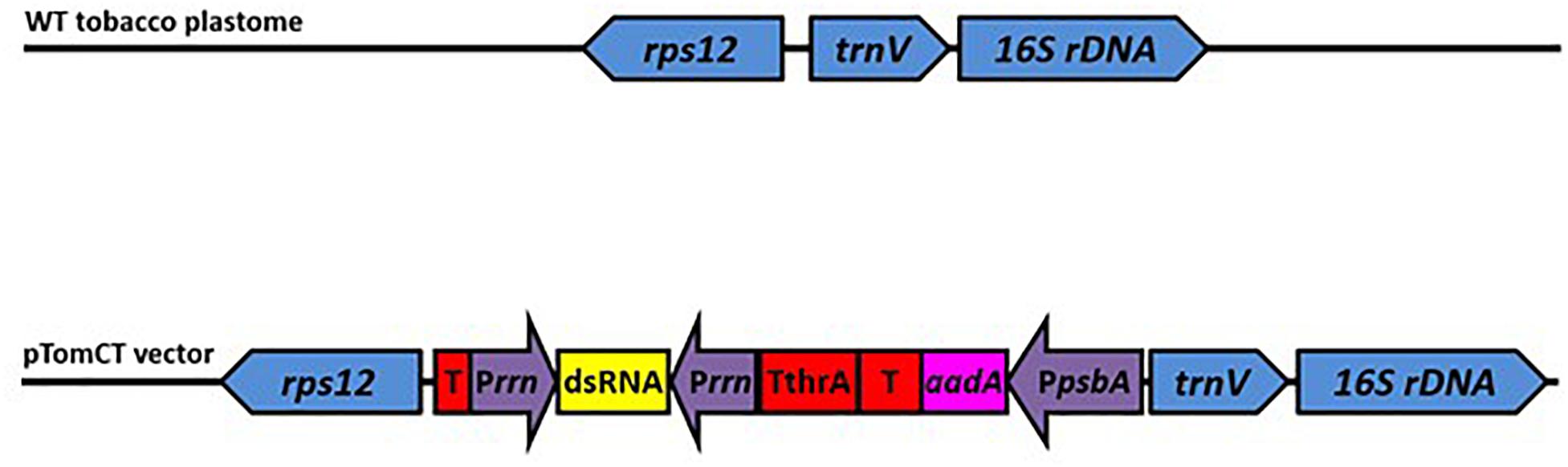
Figure 1. Schematic representation of the tobacco plastome integration site (top) and the transformation construct pTomCT (bottom). Blue: rps12-trnV-16s rDNA integration site; Red: Transcription terminators (T), including the threonine attenuator (TthrA); Yellow: Region where sequences from Manduca sexta v-ATPaseA and GFP genes were cloned for dsRNA production; Purple: promoters including PpsbA – psbA gene promoter for aadA gene expression, Prrn – chloroplast rrn operon promoter; Pink: aadA – gene encoding aminoglycoside 3′ adenylyltransferase for spectinomycin resistance.
The expression cassette (Figure 1) consists of two cis-linked expression modules, separated by an efficient transcriptional terminator of the Escherichia coli threonine attenuator (TThrA) (Gardner, 1982; Chen and Orozco, 1988), designed to disjoin the transcriptional modules in the expression cassette. The first transcriptional module in the cassette was designed for expression of a selective marker and contains the core promoter and the 5′-untranslated region of the plastid psbA gene (PpsbA), linked to the aadA gene open reading frame with a stop codon, followed by a heterologous untranslated sequence found on the 3′ end of the psbC gene of the Populus alba plastome (Kolotilin et al., 2013). The second transcriptional module contains two inward facing rrn operon promoters (Prrn) with a multiple cloning site between them (Zhang et al., 2015). Both promoters were engineered with two triplet nucleotide mutations (“-58 - -56” ATG to TAC; and “-22 – -20” AGG to TCC) (Suzuki et al., 2003) in order to reduce the homology to the endogenous Prrn, thus reducing the probability of deleterious homologous recombination. Upstream of the endogenous rps12 gene a heterologous untranslated DNA sequence (T2) found at the 3′-end of the rbcL gene of the P. alba plastome (Kolotilin et al., 2013) was introduced in order to eliminate possible transcriptional interference from the expression cassette.
To clone dsRNA encoding sequences, primers containing Not I and Sal I restriction sites at their 5′-ends were used to generate PCR products, which were then ligated into the multi-cloning site between the two convergent Prrn promoters on the expression cassette. Two dsRNA fragments [v-ATPAseA (2222 bp) and GFP (223 bp)] were cloned into pTomCT for plastid transformation. Primers used for cloning of the fragments are listed in Supplementary Table 1.
Plastid transformation was accomplished via the biolistic method (Daniell et al., 2005; Verma et al., 2008). After bombardment, leaves were cut into approximately 1 cm2 pieces and distributed onto petri dishes containing regeneration medium (MS medium containing 500 mg/L spectinomycin, 1 mg/L 6-benzylaminopurine, 0.1 mg/L 1-naphthaleneacetic acid, 1 mg/L thiamine hydrochloride, and 0.1 g/L myo-inositol). Potential transformants were detected by the growth of small, green shoots from the explants. Shoots were cut and returned to regeneration medium three times to ensure homoplastomy of the transgenic lines. Following the third round of selection, tissue was placed on MS medium with 500 mg/L spectinomycin for induction of root growth. Following the appearance of roots, plants were moved to soil for further growth.
Tissue samples from tobacco leaves were frozen in liquid nitrogen and disrupted using a TissueLyser II (QIAGEN). Total DNA was extracted using the CTAB DNA extraction protocol (Porebski et al., 1997), while total RNA was extracted using an acid phenol:chloroform extraction method (Deepa et al., 2014). To extract RNA from insects, RNeasy Mini Kit (QIAGEN) was used following the manufacturer’s protocol. All RNA samples were treated with Turbo RNase-Free DNase (Ambion) to prevent genomic DNA contamination in subsequent steps.
Southern blotting was performed as described previously (Daniell et al., 2005; Southern, 2006). Briefly, tobacco DNA was digested using restriction enzymes Pfo I and Nde I, which cut regions within the flanking sequences used for homologous recombination. The hybridization probe was prepared using the PCR DIG Probe Synthesis Kit (Sigma-Aldrich) with primers (Supplementary Table 1) that generated a 1181 bp DIG-labeled probe from the homologous recombination regions. Digested DNA was separated on a 0.8% w/v agarose gel and transferred overnight to an Amersham Hybond membrane (GE Healthcare). The membrane was crosslinked at 1200V with a CL-1000 Crosslinker (UVP) and hybridized with the probe overnight at 50°C in DIG Easy Hyb buffer (Roche). After two washes in 2 × Saline Sodium Citrate (SSC, 20 × SSC is 3.0 M NaCl, 0.3 M sodium citrate, pH = 7.0) buffer + 0.1% SDS at room temperature and three washes in 0.5 × SSC + 0.1% SDS at 68°C, membrane was blocked with DIG blocking buffer (Roche). Hybridized probe was detected by soaking membrane in Anti-Digoxigenin-AP Fab antibody solution (Roche) followed by CSPD chemiluminescent substrate (Roche) detection. A MicroChemi 4.2 Bio Imaging System was used to visualize the bands after one hr exposure.
To detect dsRNA expression, cDNA was synthesized from 500 ng total RNA using a Superscript III First-Strand Supermix Kit (Invitrogen) with a minor modification to the manufacturer’s protocol. Following the addition of 50 ng random hexamers, RNA was incubated at 95°C for 5 min to denature dsRNA molecules, and then immediately transferred to ice. Subsequent steps followed the manufacturer’s protocol. No reverse transcriptase controls (NRT) were also performed to confirm absence of genomic DNA carryover. PCR was performed using primers specific for v-ATPAseA or GFP dsRNA (Supplementary Table 1) using Taq DNA polymerase followed by agarose gel electrophoresis to visualize the products.
To demonstrate that dsRNA molecules are formed after transcription from the convergent Prrn promoters, 2 μg of total RNA from wild type (WT), v-ATPAseA and GFP dsRNA-producing plants was digested with RNAseA (Promega) to remove all single-stranded RNA (ssRNA). The reaction mixtures were incubated at 37°C for 30 min, and dsRNA was precipitated using isopropanol following the protocol from the RiboMAX Express RNAi System (Promega). The dsRNA pellets were then dissolved in 15 μL nuclease-free water and 7 μL of the digested RNA was used for cDNA synthesis, followed by PCR amplification, as described above.
To quantify dsRNA expression, cDNA was synthesized from 500 ng RNA as described in the previous section and used as template. PCR was performed using Bio-Rad ddPCR Supermix for probes, forward and reverse primers at 900 nM, and probes at 250 nM in a 20 μL final reaction volume. NRT controls were also run to ensure that templates were free of DNA contamination. Droplets were generated using the Bio-Rad QX100 Droplet Generator, followed by cycling on a Bio-Rad T100 Thermocycler with the following temperature profile: initial denaturation at 95°C for 10 min, 40 cycles of 94°C for 30 s, 60°C for 1 min, and a final incubation at 98°C for 10 min. Droplets were read on a Bio-Rad QX100 Droplet reader and data was analyzed using Quantasoft 1.7.4 software. Absolute quantification of dsRNA was done using N. tabacum ribosomal protein L25 (RPL25) as a reference gene (Schmidt and Delaney, 2010). Quantities of dsRNA present in 500 ng total RNA were calculated using the guidelines from droplet digital PCR application guide (Bio-Rad, 2018). Statistical analysis of any differences between v-ATPaseA and GFP dsRNA levels was performed using a t-test.
Insect eggs and artificial diet were obtained from Reptile Feeders (Norwood, ON, Canada). The eggs were hatched on artificial diet for droplet feeding assays, and on WT tobacco leaves for in planta feeding assays. Insects were reared at 27°C and a 16L:8D light cycle. For droplet feeding assays, insects were first raised to second instar. Then, larvae were starved for 4–6 h and each presented with a 1 μL liquid droplet containing 2.5 μg of long or short v-ATPaseA dsRNA, GFP dsRNA, or water only. Any insect that did not consume the entire droplet was discarded from the bioassay. Insects were then moved to cups containing artificial diet and allowed to feed ad libitum. Survival was recorded daily and observed over a 7-day period. Additional insects were droplet-fed for RNA extraction after 3 days to confirm gene knockdown. For in planta feeding bioassays, first instar insects less than 24 h old were moved to cups containing 1% agarose and 3 cm diameter leaf disks from either WT, v-ATPAseA dsRNA-, or GFP dsRNA-expressing plants. Leaf disks were replaced daily and survival was recorded over a 7-day period. Survival of insects was compared using a Kaplan–Meier survival analysis and compared between treatments using log-rank tests.
To measure insect gene expression, cDNA was synthesized from 500 ng insect RNA using the SuperScript III First-Strand Synthesis SuperMix for RT-qPCR kit (Invitrogen) according to the manufacturer’s protocol. Reverse transcription quantitative PCR was performed using a SensiFAST SYBR No-ROX Mix Kit (Bioline) and a CFX96 Real-Time Detection System (Bio-Rad). Primers for RT-qPCR were designed to amplify an approximately 100 bp region of the M. sexta v-ATPaseA gene in a region outside of the dsRNA product to avoid spurious signal attributed to dsRNA carryover during extractions. Primers targeting the M. sexta elongation factor 1-alpha (EF1-α) gene were used as a reference (Supplementary Table 1). Gene expression knockdown was assessed between insect RNA samples using one-way ANOVA followed by Dunnett’s test.
To determine whether v-ATPaseA dsRNA can induce gene knockdown and lethality in second instar M. sexta larvae, we fed the larvae a 1 μL water droplet containing 2.5 μg of in vitro-synthesized dsRNA: v-ATPaseA dsRNA long, v-ATPaseA dsRNA short, GFP dsRNA, or no dsRNA. After 7 days, 56.8% of insects fed the long v-ATPaseA dsRNA died, compared to 6.8% (log rank χ2 = 17.1, df = 1, p = 0.000035, n = 44) and 27.2% (log rank χ2 = 5.46, df = 1, p = 0.019, n = 44) for insects fed water and GFP dsRNA, respectively (Figure 2A). In larvae fed v-ATPAseA dsRNA short, mortality was 47.8%, which was lower than that for the long dsRNA product, but not significantly different (p > 0.05). Mortality in v-ATPaseA short-fed larvae was also significantly higher in comparison to water-fed larvae (log rank χ2 = 11.12, df = 1, p = 0.00085, n = 44); however, it was not significantly different from the GFP dsRNA-fed larvae (p > 0.05) (Figure 2A). To ensure that knockdown of v-ATPaseA mRNA was taking place, additional insects were treated in the same way and RNA was extracted after 3 days. Reverse transcription-qPCR indicated that v-ATPaseA expression was reduced significantly, by 68.9%, in insects fed long v-ATPaseA dsRNA product compared to insects fed water (F = 3.70, p = 0.042) (Figure 2B). Relative v-ATPaseA gene expression in larvae fed v-ATPaseA dsRNA short or GFP dsRNA compared to insects fed water was reduced 62.5 and 29.1%, respectively, which were not significantly different from water (p > 0.05).
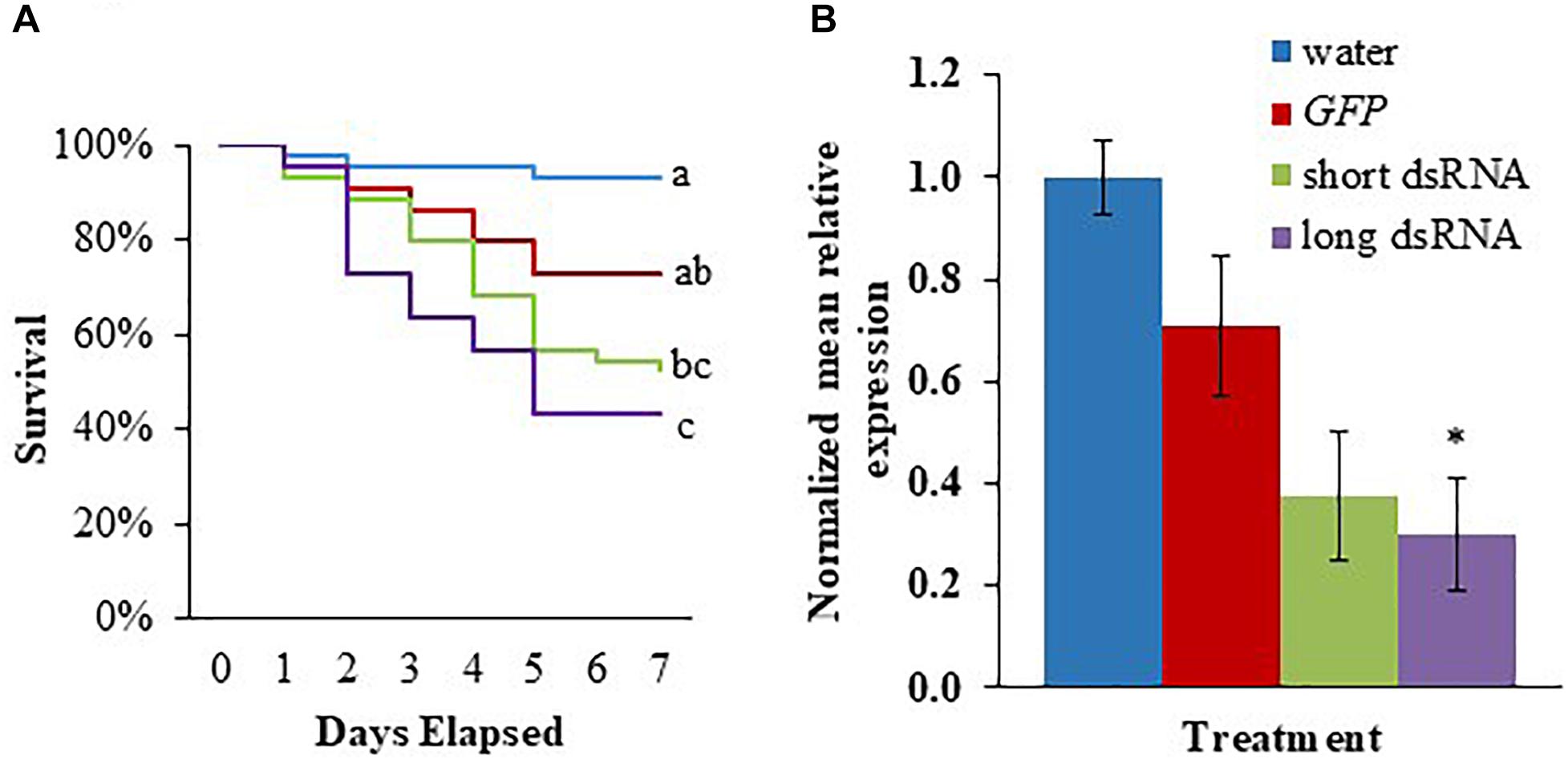
Figure 2. Survival of M. sexta second instar larvae and knockdown of v-ATPaseA gene after feeding with in vitro synthesized dsRNA. (A) Kaplan–Meier survival analysis over a 7-day period. Different letters represent significant difference according to log-rank tests (p < 0.05, n = 44), which were performed between each group. (B) Knockdown of v-ATPaseA gene in insects 3 days after feeding on dsRNA. Normalized mRNA quantity was set to one in water-fed larvae and relative differences in mRNA levels of treatment groups were calculated. Data are expressed as mean relative quantity ± SEM. Asterisks indicate significant difference from water control according to Dunnett’s test (∗p < 0.05, n = 4).
Based on the results of the droplet feeding assays, the long v-ATPaseA fragment and the GFP control fragment were selected for expression in transplastomic plants. Following biolistic delivery of gold particles carrying the appropriate recombinant pTomCT vectors, leaf explants were placed on regeneration medium containing spectinomycin for the selection of transformants. Three rounds of selective regeneration were performed to ensure homoplastomy of the transgenic plants, which was confirmed using a Southern blot (Figure 3A). In WT tobacco, the expected restriction fragment size was 2102 bp while in transplastomic plants, they were 6183 and 4184 bp for v-ATPaseA and GFP dsRNA expression cassettes, respectively. Both WT and transplastomic tobacco plants displayed single bands of the expected sizes, and absence of the WT band in transplastomic plant lines confirmed homoplastomy. The resulting transplastomic plants showed no significant observable differences in their phenotype compared to WT plants (Supplementary Figure 3).
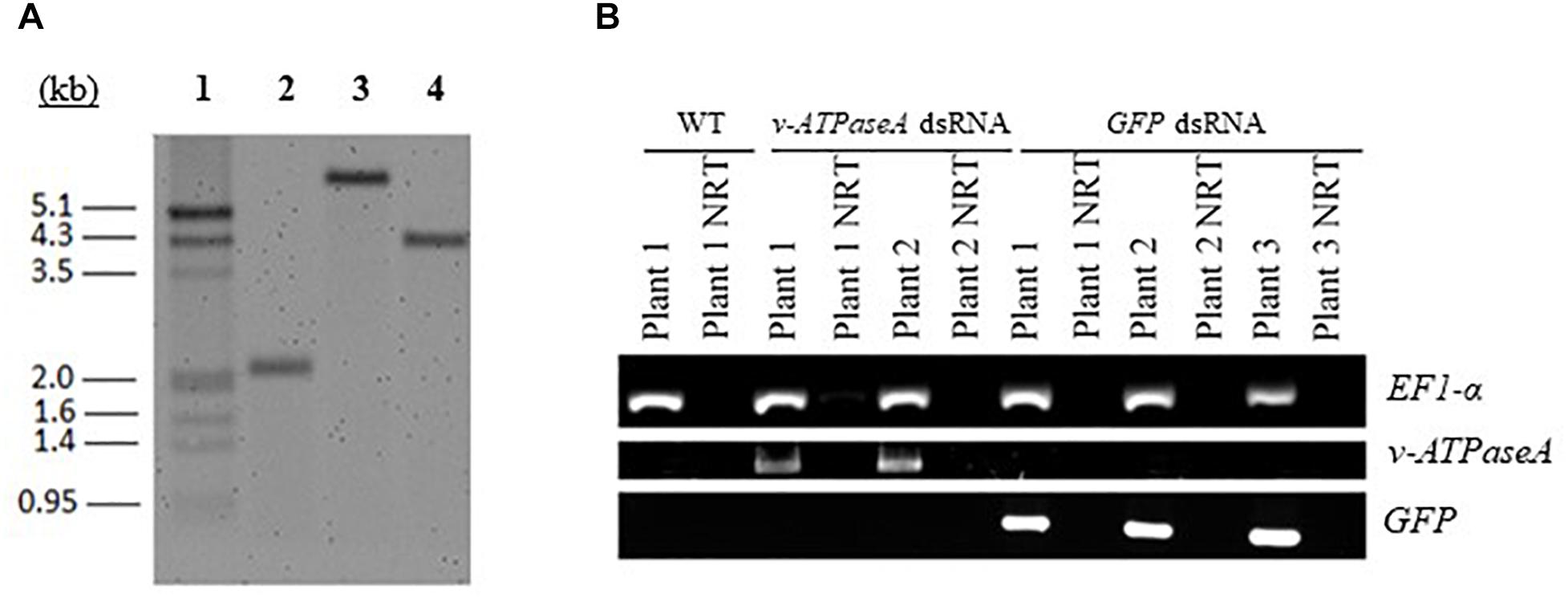
Figure 3. Characterization of transplastomic tobacco plants. (A) Southern blot confirming homoplastomy of the transformed tobacco lines. Tobacco DNA was cut with restriction enzymes and separated on an agarose gel for blotting. A probe hybridizing to homologous recombination regions in tobacco plastids was used to confirm insertion/integration of dsRNA expression cassettes into plastid DNA. A single band of the expected size was observed in each DNA sample; 1, DIG-labeled marker; 2 DNA from WT tobacco; 3, DNA from tobacco with v-ATPaseA dsRNA expression cassette; 4, DNA from tobacco with GFP dsRNA expression cassette. All DNA samples display single bands which confirm homoplastomy of transformed plants. (B) RT-PCR confirming expression of v-ATPaseA dsRNA and GFP dsRNA in transplastomic plants. Primers annealing to dsRNA sequences were used to confirm production of dsRNA in transplastomic tobacco plants. Tobacco EF1-α gene was used as a positive control and NRT reactions confirmed absence of DNA contamination in cDNA preparations.
Total RNA was extracted from the transplastomic plants and RT-PCR was performed using primers specific for the inserted sequences to check for expression of the dsRNA. Bands were present when transplastomic cDNA was used as the template, but not when WT cDNA was used, confirming that the plants express the desired products (Figure 3B). In addition, to confirm that the products transcribed from both strands of the DNA are annealed and present as dsRNA in the transplastomic plants, RT-PCR was performed using cDNA made from total RNA that was first digested with RNaseA to remove all ssRNA. PCR products of the expected sizes resulted when using v-ATPAseA and GFP primers with cDNA from v-ATPAseA and GFP dsRNA producing plants, respectively, confirming the presence of dsRNA products for these sequences (Figure 4). On the other hand, PCR performed using primers for a housekeeping gene in the plant, tobacco EF1-α, resulted in no products in any of the RNaseA-digested samples, confirming the efficient elimination of ssRNA from all the digested samples (Figure 4).
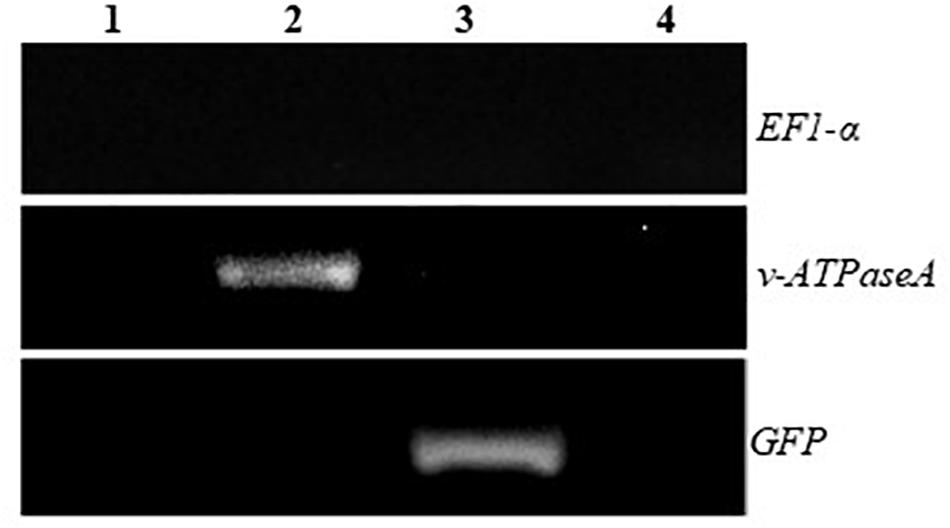
Figure 4. RT-PCR after removal of ssRNA using RNAseA to demonstrate the presence of annealed v-ATPaseA and GFP dsRNA in transplastomic plants. 1, WT plant; 2, v-ATPAseA dsRNA producing plant; 3, GFP dsRNA producing plant; and 4, No template control. Tobacco EF1-α gene specific primers were used to ensure that all ssRNA was removed by the RNAseA treatment.
First instar larvae were placed on leaf disks from either WT, v-ATPaseA dsRNA-, or GFP dsRNA-producing tobacco plants and survival was observed over 7 days. After 7 days, mortality of insects feeding on v-ATPaseA dsRNA plants was 16.0%, compared to 20.0% for GFP dsRNA and 13.0% for WT tobacco (p ≥ 0.05) (Figure 5A). Furthermore, expression of v-ATPaseA mRNA was not significantly reduced (p ≥ 0.05) in insects that were fed transplastomic plants compared to insects fed WT plants (Figure 5B).
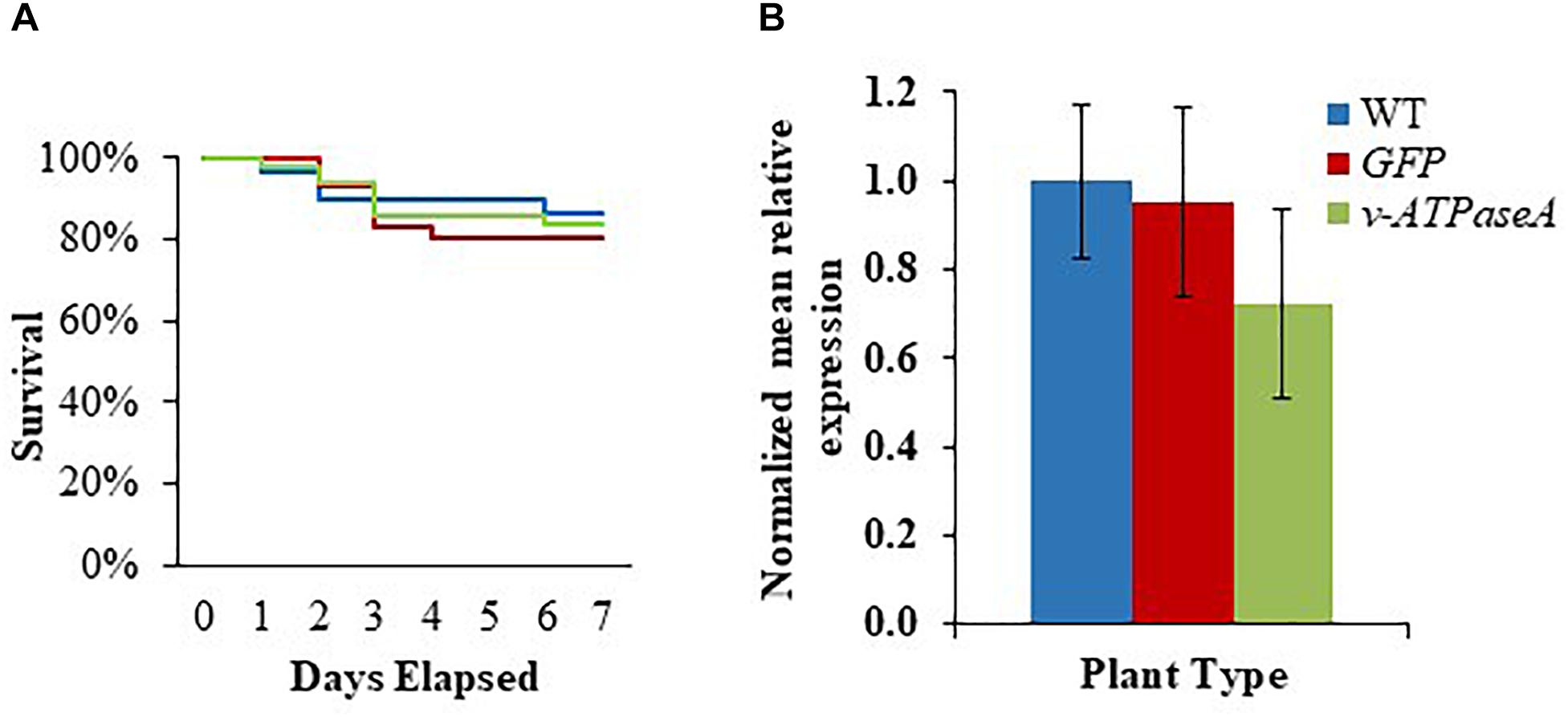
Figure 5. Survival of M. sexta first instar larvae and knockdown of v-ATPaseA gene after insects were fed either WT or transplastomic tobacco leaves expressing v-ATPaseA or GFP dsRNA over a 7-day period. (A) Kaplan–Meier survival analysis of insects over a 7-day period (p > 0.05, n = 44). (B) RT-qPCR showing relative expression of v-ATPaseA gene in insects after feeding on transplastomic and WT plants. Normalized mRNA quantity was set to one in WT tobacco-fed larvae and relative differences in mRNA levels in transplastomic tobacco-fed larvae were calculated. Data are expressed as mean relative quantity ± SEM (p > 0.05, n = 7 for GFP, n = 8 for v-ATPaseA and WT).
To determine whether the amount of dsRNA produced by transplastomic tobacco is affected by the dsRNA size, we quantified dsRNA levels using ddPCR. GFP dsRNA (223 bp) levels were significantly higher (t = 5.61; p = 0.00497, n = 3) than v-ATPaseA dsRNA (2222 bp) (Figure 6). In 500 ng total plant RNA, we estimated that there were approximately 2.39 × 107 and 5.34 × 107 dsRNA molecules in long v-ATPaseA and GFP dsRNA producing tobacco, respectively, assuming 100% conversion to cDNA. Overall, there was an increase of >two-fold in dsRNA molecules when the plants were transformed with the cassette producing the shorter GFP dsRNA compared to the cassette producing the longer v-ATPase A dsRNA.
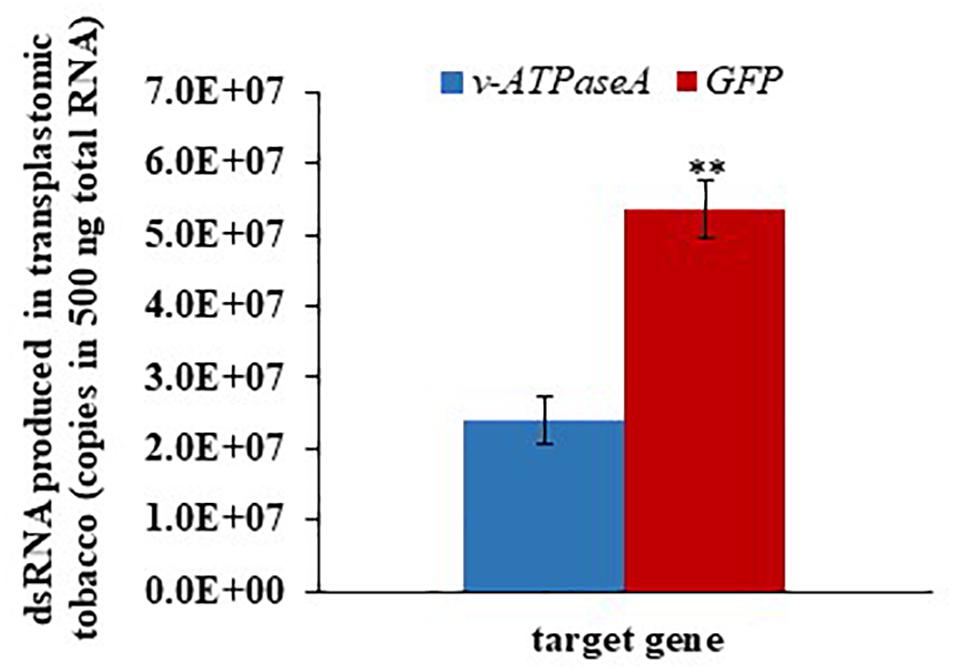
Figure 6. Absolute quantification of v-ATPaseA and GFP dsRNA in 500 ng transplastomic tobacco RNA. Data are expressed as mean quantity ± SEM. Asterisks represent significant changes in t-test (∗∗p ≤ 0.01), n = 3.
Development of plants expressing high quantities of insecticidal dsRNA within their plastids is a promising strategy for future application in pest control. In this study, we showed that targeting of the M. sexta v-ATPaseA gene using an in vitro synthesized long dsRNA (2222 bp) induces higher mortality and gene knockdown compared to a short dsRNA (259 bp). Then, we transformed the plastid genome of tobacco with an expression cassette to produce the long dsRNA product and confirmed its production in the transplastomic plants. To our knowledge, this is the first reported instance of a dsRNA product this large being produced in a transplastomic plant. In addition, for the first time, our study compared the absolute quantities of dsRNA produced in plastids to determine whether length of the dsRNA has any effects on the amounts produced, which could be important for future applications using this approach to induce RNAi in insects.
Messenger RNAs encoding subunits of the v-ATPase protein complex are common targets for studies which aim to reduce survival of feeding pests by gene expression knockdown, including in M. sexta (Whyard et al., 2009). This protein complex has an important function in maintaining pH homeostasis and/or in membrane energization (Wieczorek et al., 2009); hence, interfering with expression of any subunit in the complex results in severe developmental and physiological defects and death (Baum et al., 2007; Badillo-Vargas et al., 2015; Jin et al., 2015; Li et al., 2015). Our observation, that a long v-ATPaseA dsRNA product generated through in vitro synthesis induces higher lethality and gene knockdown compared to a short v-ATPaseA dsRNA, is consistent with previously published studies (Bolognesi et al., 2012; Miller et al., 2012; Li et al., 2015). Although we did observe significantly greater mortality in short v-ATPaseA dsRNA-fed insects compared to water-fed insects, the results were not significantly different from GFP dsRNA-fed insects. Longer dsRNAs should yield larger populations of overlapping siRNA molecules ranging in size and sequence (Nandety et al., 2015), and this would be expected to improve RNAi efficiency in comparison to shorter dsRNA products. Also, variation in RNAi efficiency due to different dsRNA lengths has been attributed to less efficient recognition of shorter dsRNA by cellular uptake mechanisms (Saleh et al., 2006). Both of these factors might contribute to a bias toward greater RNAi efficiency of longer dsRNAs.
Because we observed higher mortality with the long dsRNA, we transformed tobacco plants to produce the long dsRNA product within their plastids. We confirmed that annealed double-stranded products are present in the transplastomic plants using RT-PCR. Removal of ssRNA before cDNA synthesis provided evidence that dsRNA molecules are formed in the transplastomic plants after transcription. Previously, it was also shown that dsRNA produced in plastids accumulated stably, and was not processed into siRNA molecules, as there is no dsRNA-processing machinery present in these subcellular compartments (Zhang et al., 2015). However, despite the effectiveness of the droplet feeding approach, insects feeding upon transplastomic plants producing the long dsRNA neither showed a significant increase in mortality nor a drop in v-ATPaseA transcript level from controls, although the levels observed were suggestive of a trend to lower expression. There are several possible explanations for why the in planta approach failed to produce an effective dose of dsRNA. The most likely is that the in planta-produced dsRNA is simply present in lower quantities than those attained by feeding of in vitro-synthesized dsRNA. RNAi response can be dependent on dsRNA concentrations (Kumar et al., 2009; Tian et al., 2009), and in particular, high doses of dsRNA might be required when dsRNA is delivered orally (Luo et al., 2013; Scott et al., 2013).
When we compared the absolute quantities of dsRNA produced in plants transformed with the long v-ATPaseA gene (2222 bp) with those of the control plants producing the shorter GFP gene fragment (223 bp), we found that accumulation of the long dsRNA was lower by more than two-fold than that of the shorter dsRNA. This implies that expression of a very long product in planta may be a limiting factor in achieving high concentrations of dsRNA. One potential influence on the yield of dsRNA in our transplastomic plants stems from the efficiency with which such products would be synthesized. The strong chloroplast promoter, Prrn, used in our study to drive dsRNA expression was previously shown to result in high mRNA accumulation levels which led to exhaustion of the plastid translational machinery (Oey et al., 2009). However, the stability of the RNA transcripts produced will affect the final levels achieved. In fact, it was previously suggested that stability of RNA molecules plays an important role in RNA quantities in plastids (Mullet, 1993), and the sequence of the dsRNA may influence its stability (Zhang et al., 2015). The stability of RNA molecules can also be influenced by their length, as longer transcripts have been shown to be more prone to degradation by nucleases as well as mechanical damage compared to their shorter counterparts (Feng and Niu, 2007). Furthermore, it is also possible that long nascent RNA sequences could form secondary structures under cellular conditions (Sükösd et al., 2013), which could prevent stable dsRNA annealing, and cause more rapid polymerase dissociation from the template. It has been suggested that polymerase enzymes transcribing longer dsRNA products can dissociate from the template more often, which would result in truncated products prone to rapid degradation (Cagliero and Jin, 2013).
Another potential factor influencing the effective dose of dsRNA to a feeding insect is the presence of nucleases that degrade dsRNA. The importance of dsRNA degrading nucleases in RNAi efficiency using orally delivered dsRNA was highlighted by a study that demonstrated improved RNAi response upon silencing of nucleases in the Colorado potato beetle midgut (Spit et al., 2017), an insect known to be sensitive to RNAi (Palli, 2014). The effect of nucleases on RNAi efficiency is dependent on the species involved (Joga et al., 2016; Garcia et al., 2017; Guan et al., 2018), and several studies have demonstrated that dsRNA molecules are degraded faster in the midgut and hemolymph of lepidopteran insects compared to other Orders (Garbutt et al., 2013; Wang et al., 2016; Singh et al., 2017). When we fed in vitro synthesized dsRNA to M. sexta larvae, we first starved the insects for 4–6 h; whereas, the plant fed larvae were not starved and were provided with leaves continuously to deliver the dsRNA. Previously, it has been shown that starvation can influence the midgut environment and reduce the nuclease activities in the insect, resulting in increased dsRNA stability (Rodríguez-Cabrera et al., 2010; Cooper et al., 2018). This difference presents another potential explanation for the lower RNAi effectiveness we observed using in planta delivered dsRNA. Additionally, a lepidopteran-specific nuclease has been shown to be upregulated in response to dsRNA exposure in another lepidopteran insect, Asian corn borer (Guan et al., 2018). Continuous feeding by the larvae on dsRNA-producing plants may result in detection of low quantities of dsRNA early by the midgut cells, resulting in upregulation of nucleases to degrade the dsRNA molecules. Therefore, slow consumption of the dsRNA in planta over a longer period of time may produce both a reduced dsRNA dose level compared to a one-time ingestion of high quantities of dsRNA by starved insects, as well as lead to greater presence of nucleases in the insect gut.
Taken together, our results suggest that lower amounts of long dsRNA produced in our transplastomic plants were insufficient, in comparison with in vitro produced material fed as a bolus, to induce RNAi in M. sexta larvae. It is possible that the level of dsRNA produced in our transplastomic plants would be sufficient to induce lethality in insects that are more susceptible to dsRNA, and representatives of other Orders are worth investigating in the future. However, in general, the results of this study suggest that a size-dependent limit on high level dsRNA production in plastids exists, and future research needs to be directed to determining the point at which the benefits of more efficient long dsRNA uptake are balanced with the decline in production efficiency for long dsRNA products in the plastid.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
WB, CD, and RM contributed conception and design of the study. WB and EK performed the experiments. CD, IK, and RM contributed reagents, materials, and analysis tools. WB wrote the first draft of the manuscript. EK, IK, and CD wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This study was funded by Agriculture and Agri-Food Canada.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. David Smith and Dr. Graham Thompson (The University of Western Ontario) for their advice and comments throughout the duration of this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00313/full#supplementary-material
Agrawal, A., Rajamani, V., Reddy, V. S., Mukherjee, S. K., and Bhatnagar, R. K. (2015). Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: an alternative to Bt-toxin technology. Transgenic Res. 24, 791–801. doi: 10.1007/s11248-015-9880-x
Badillo-Vargas, I. E., Rotenberg, D., Schneweis, B. A., and Whitfield, A. E. (2015). RNA interference tools for the western flower thrips, Frankliniella occidentalis. J. Insect Physiol. 76, 36–46. doi: 10.1016/j.jinsphys.2015.03.009
Bally, J., McIntyre, G. J., Doran, R. L., Lee, K., Perez, A., Jung, H., et al. (2016). In-plant protection against Helicoverpa armigera by production of long hpRNA in chloroplasts. Front. Plant Sci. 7:1453. doi: 10.3389/fpls.2016.01453
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Bellés, X. (2010). Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 55, 111–128. doi: 10.1146/annurev-ento-112408-085301
Bio-Rad (2018). Droplet Digital PCR Application Guide (Bulletin 6407 Ver B). Available at: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf
Bolognesi, R., Ramaseshadri, P., Anderson, J., Bachman, P., Clinton, W., Flannagan, R., et al. (2012). Characterizing the mechanism of action of double-stranded RNA activity against Western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7:e47534. doi: 10.1371/journal.pone.0047534
Cagliero, C., and Jin, D. J. (2013). Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res. 41, 315–326. doi: 10.1093/nar/gks988
Cappelle, K., de Oliveira, C. F. R., Van Eynde, B., Christiaens, O., and Smagghe, G. (2016). The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in double-stranded RNA uptake in the Colorado potato beetle midgut. Insect Mol. Biol. 25, 315–323. doi: 10.1111/imb.12222
Chen, L.-J., and Orozco, E. M. (1988). Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 16, 8411–8431. doi: 10.1093/nar/16.17.8411
Cooper, A. M. W., Silver, K., Zhang, J., Park, Y., and Zhu, K. Y. (2018). Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 75, 18–28. doi: 10.1002/ps.5126
Daniell, H., Ruiz, O. N., and Dhingra, A. (2005). “Chloroplast genetic engineering to improve agronomic traits,” in Transgenic Plants: Methods and Protocols, ed. L. Pena (Totowa, NJ: Humana Press Inc.), 111–138.
Deepa, K., Sheeja, T. E., Santhi, R., Sasikumar, B., Cyriac, A., Deepesh, P. V., et al. (2014). A simple and efficient protocol for isolation of high quality functional RNA from different tissues of turmeric (Curcuma longa L.). Physiol. Mol. Biol. Plants 20, 263–271. doi: 10.1007/s12298-013-0218-y
Dietzl, G., Chen, D., Schnorrer, F., Su, K. C., Barinova, Y., Fellner, M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. doi: 10.1038/nature05954
Feng, L., and Niu, D. K. (2007). Relationship between mRNA stability and length: an old question with a new twist. Biochem. Genet. 45, 131–137. doi: 10.1007/s10528-006-9059-5
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Flores-Escobar, B., Rodríguez-Magadan, H., Bravo, A., Soberón, M., and Gómez, I. (2013). Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl. Environ. Microbiol. 79, 4543–4550. doi: 10.1128/AEM.01062-13
Garbutt, J. S., Bellés, X., Richards, E. H., and Reynolds, S. E. (2013). Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 59, 171–178. doi: 10.1016/j.jinsphys.2012.05.013
Garcia, R. A., Pepino Macedo, L. L., Do Nascimento, D. C., Gillet, F. X., Moreira-Pinto, C. E., Faheem, M., et al. (2017). Nucleases as a barrier to gene silencing in the cotton boll weevil, Anthonomus grandis. PLoS One 12:e0189600. doi: 10.1371/journal.pone.0189600
Gardner, J. F. (1982). Initiation, pausing, and termination of transcription in the threonine operon regulatory region of Escherichia coli. J. Biol. Chem. 257, 3896–3904.
Guan, R. B., Li, H. C., Fan, Y. J., Hu, S. R., Christiaens, O., Smagghe, G., et al. (2018). A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 293, 6011–6021. doi: 10.1074/jbc.RA117.001553
He, K., Xiao, H., Sun, Y., Ding, S., Situ, G., and Li, F. (2019). Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 17, 461–471. doi: 10.1111/pbi.12990
Jiang, S., Wu, H., Liu, H., Zheng, J., Lin, Y., and Chen, H. (2017). The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (Chilo suppressalis). Pest Manag. Sci. 73, 1453–1461. doi: 10.1002/ps.4477
Jin, S., Singh, N. D., Li, L., Zhang, X., and Daniell, H. (2015). Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnol. J. 13, 435–446. doi: 10.1111/pbi.12355
Joga, M. R., Zotti, M. J., Smagghe, G., and Christiaens, O. (2016). RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: what we know so far. Front. Physiol. 7:553. doi: 10.3389/fphys.2016.00553
Kolotilin, I., Kaldis, A., Pereira, E. O., Laberge, S., and Menassa, R. (2013). Optimization of transplastomic production of hemicellulases in tobacco: effects of expression cassette configuration and tobacco cultivar used as production platform on recombinant protein yields. Biotechnol. Biofuels 6, 1–15. doi: 10.1186/1754-6834-6-65
Kumar, M., Gupta, G. P., and Rajam, M. V. (2009). Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J. Insect Physiol. 55, 273–278. doi: 10.1016/j.jinsphys.2008.12.005
Li, H., Khajuria, C., Rangasamy, M., Gandra, P., Fitter, M., Geng, C., et al. (2015). Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. J. Appl. Entomol. 139, 432–445. doi: 10.1111/jen.12224
Li, T., Chen, J., Fan, X., and Zhang, W. (2017). MicroRNA and dsRNA targeting chitin synthase a reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag. Sci. 73, 1529–1537. doi: 10.1002/ps.4492
Luo, Y., Wang, X., Wang, X., Yu, D., Chen, B., and Kang, L. (2013). Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. Insect Mol. Biol. 22, 574–583. doi: 10.1111/imb.12046
Mao, Y.-B. B., Cai, W.-J. J., Wang, J.-W. W., Hong, G.-J. J., Tao, X.-Y. Y., Wang, L.-J. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349. doi: 10.1038/nature02873
Melnyk, C. W., Molnar, A., and Baulcombe, D. C. (2011). Intercellular and systemic movement of RNA silencing signals. EMBO J. 30, 3553–3563. doi: 10.1038/emboj.2011.274
Menassa, R., Nguyen, V., Jevnikar, A., and Brandle, J. (2001). A self-contained system for the field production of plant recombinant interleukin-10. Mol. Breed. 8, 177–185. doi: 10.1023/A:1013376407362
Miller, S. C., Miyata, K., Brown, S. J., and Tomoyasu, Y. (2012). Dissecting Systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS One 7:e47431. doi: 10.1371/journal.pone.0047431
Mullet, J. E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103, 309–313. doi: 10.1104/pp.103.2.309
Nandety, R. S., Kuo, Y. W., Nouri, S., and Falk, B. W. (2015). Emerging strategies for RNA interference (RNAi) applications in insects. Bioengineered 6, 8–19. doi: 10.4161/21655979.2014.979701
Oey, M., Lohse, M., Kreikemeyer, B., and Bock, R. (2009). Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 57, 436–445. doi: 10.1111/j.1365-313X.2008.03702.x
Palli, S. R. (2014). RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr. Opin. Insect Sci. 6, 1–8. doi: 10.1016/j.cois.2014.09.011
Porebski, S., Bailey, L. G., and Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 15, 8–15. doi: 10.1007/BF02772108
Poreddy, S., Li, J., and Baldwin, I. T. (2017). Plant-mediated RNAi silences midgut-expressed genes in congeneric lepidopteran insects in nature. BMC Plant Biol. 17:199. doi: 10.1186/s12870-017-1149-5
Rodríguez-Cabrera, L., Trujillo-Bacallao, D., Borrás-Hidalgo, O., Wright, D. J., and Ayra-Pardo, C. (2010). RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ. Microbiol. 12, 2894–2903. doi: 10.1111/j.1462-2920.2010.02259.x
Ruf, S., Hermann, M., Berger, I. J., Carrer, H., and Bock, R. (2001). Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 19, 870–875. doi: 10.1038/nbt0901-870
Saleh, M.-C., van Rij, R. P., Hekele, A., Gillis, A., Foley, E., O’Farrell, P. H., et al. (2006). The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802. doi: 10.1038/ncb1439
Schmidt, G. W., and Delaney, S. K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 283, 233–241. doi: 10.1007/s00438-010-0511-1
Scott, J. G., Michel, K., Bartholomay, L. C., Siegfried, B. D., Hunter, W. B., Smagghe, G., et al. (2013). Towards the elements of successful insect RNAi. J. Insect Physiol. 59, 1212–1221. doi: 10.1016/j.jinsphys.2013.08.014
Shih, J. D., and Hunter, C. P. (2011). SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 1057–1065. doi: 10.1261/rna.2596511
Singh, I. K., Singh, S., Mogilicherla, K., Shukla, J. N., and Palli, S. R. (2017). Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-17134-2
Spit, J., Philips, A., Wynant, N., Santos, D., Plaetinck, G., and Vanden Broeck, J. (2017). Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 81, 103–116. doi: 10.1016/j.ibmb.2017.01.004
Sükösd, Z., Knudsen, B., Anderson, J. W., Novák,Á, Kjems, J., and Pedersen, C. N. (2013). Characterising RNA secondary structure space using information entropy. BMC Bioinformatics 14:S22. doi: 10.1186/1471-2105-14-S2-S22
Suzuki, J. Y., Sriraman, P., Svab, Z., and Maliga, P. (2003). Unique architecture of the plastid ribosomal RNA operon promoter recognized by the multisubunit RNA Polymerase in tobacco and other higher plants. Plant Cell 15, 195–205. doi: 10.1105/tpc.007914
Terenius, O., Papanicolaou, A., Garbutt, J. S., Eleftherianos, I., Huvenne, H., Kanginakudru, S., et al. (2011). RNA interference in lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 57, 231–245. doi: 10.1016/j.jinsphys.2010.11.006
Tian, H., Peng, H., Yao, Q., Chen, H., Xie, Q., Tang, B., et al. (2009). Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4:e6225. doi: 10.1371/journal.pone.0006225
Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. doi: 10.1016/S0378-1119(00)00579-5
Ulrich, J., Dao, V. A., Majumdar, U., Schmitt-Engel, C., Schwirz, J., Schultheis, D., et al. (2015). Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics 16:674. doi: 10.1186/s12864-015-1880-y
Verma, D., Samson, N. P., Koya, V., and Daniell, H. (2008). A protocol for expression of foreign genes in chloroplasts. Nat. Protoc. 3, 739–758. doi: 10.1038/nprot.2007.522
Wang, K., Peng, Y., Pu, J., Fu, W., Wang, J., and Han, Z. (2016). Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 77, 1–9. doi: 10.1016/j.ibmb.2016.07.007
Whangbo, J. S., and Hunter, C. P. (2008). Environmental RNA interference. Trends Genet. 24, 297–305. doi: 10.1016/j.tig.2008.03.007
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Wieczorek, H., Beyenbach, K. W., Huss, M., and Vitavska, O. (2009). Vacuolar-type proton pumps in insect epithelia. J. Exp. Biol. 212, 1611–1619. doi: 10.1242/jeb.030007
Yu, X.-D., Liu, Z.-C., Huang, S.-L., Chen, Z.-Q., Sun, Y.-W., Duan, P.-F., et al. (2016). RNAi-mediated plant protection against aphids. Pest Manag. Sci. 72, 1090–1098. doi: 10.1002/ps.4258
Keywords: RNA interference, Manduca sexta, tobacco hornworm, Nicotiana tabacum, plastid transformation, v-ATPaseA, long dsRNA
Citation: Burke WG, Kaplanoglu E, Kolotilin I, Menassa R and Donly C (2019) RNA Interference in the Tobacco Hornworm, Manduca sexta, Using Plastid-Encoded Long Double-Stranded RNA. Front. Plant Sci. 10:313. doi: 10.3389/fpls.2019.00313
Received: 10 December 2018; Accepted: 26 February 2019;
Published: 14 March 2019.
Edited by:
Goetz Hensel, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Huipeng Pan, South China Agricultural University, ChinaCopyright © 2019 Burke, Kaplanoglu, Kolotilin, Menassa and Donly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cam Donly, Y2FtLmRvbmx5QGNhbmFkYS5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.