- National Agri-Food Biotechnology Institute, Mohali, India
Wheat is one of the most important staple crops in the world and good source of calories and nutrition. Its flour and dough have unique physical properties and can be processed to make unique products like bread, cakes, biscuits, pasta, noodles etc., which is not possible from other staple crops. Due to domestication, the genetic variability of the genes coding for different economically important traits in wheat is narrow. This genetic variability can be increased by utilizing its wild relatives. Its closest relative, genus Aegilops can be an important source of new alleles. Aegilops has played a very important role in evolution of tetraploid and hexaploid wheat. It consists of 22 species with C, D, M, N, S, T and U genomes with high allelic diversity relative to wheat. Its utilization for wheat improvement for various abiotic and biotic stresses has been reported by various scientific publications. Here in, for the first time, we review the potential of Aegilops for improvement of processing and nutritional traits in wheat. Among processing quality related gluten proteins; high molecular weight glutenins (HMW GS), being easiest to study have been explored in highest number of accessions or lines i.e., 681 belonging to 13 species and selected ones like Ae. searsii, Ae. geniculata and Ae. longissima have been linked with improved bread making quality of wheat. Gliadins and low molecular weight glutenins (LMW GS) have also been extensively explored for wheat improvement and Ae. umbellulata specific LMW GS have been linked with wheat bread making quality improvement. Aegilops has been explored for seed texture diversity and proteins like puroindolins (Pin) and grain softness proteins (GSP). For nutrition quality improvement, it has been screened for essential micronutrients like Fe, Zn, phytochemicals like carotenoids and dietary fibers like arabinoxylan and β-glucan. Ae. kotschyi and Ae. biuncialis transfer in wheat have been associated with higher Fe, Zn content. In this article we have tried to compile information available on exploration of nutritional and processing quality related traits in Aegilops section and their utilization for wheat improvement by different approaches.
Introduction
Some of the most important cereal crops in the world are the members of the grass (Poaceae) family and belong to three major subfamilies – Pooideae, Oryzoideae and Panicoideae. These subfamilies diverged from a common ancestor around 50–70 million years ago (Bolot et al., 2009) (Figure 1A). Genus Aegilops is the closest relative of wheat followed by rye, barley, oats and brome in the Pooideae subfamily, rice in Oryzoideae, millets, sorghum and maize in Panicoideae (Figure 1A). Among the Pooideae, wheat (Triticum L.) is one of the major staple foods in the world. Due to its unique flour composition and viscoelastic properties, wheat is more suitable for industrialized food production than any other crop. Recently, demand for wheat based convenience foods (fast, ready to eat, frozen etc.) have increased due to the rise in urban population and changing lifestyles. Therefore, the end product quality of wheat has become important. With an increasing concern for texture and taste, there have been a lot of challenges for breeders to develop cultivars that satisfy specific end product requirements. Nutrition is another important aspect of wheat research. There are approximately two billion people in the world that suffer from nutrient deficiency also known as hidden hunger (World Health Organization, 2006). Since wheat provides around one fifth of calorific input to people across the world (Food and Agriculture Organization of United Nations [FAO], 2014), enhancing its nutritive value becomes of great importance.
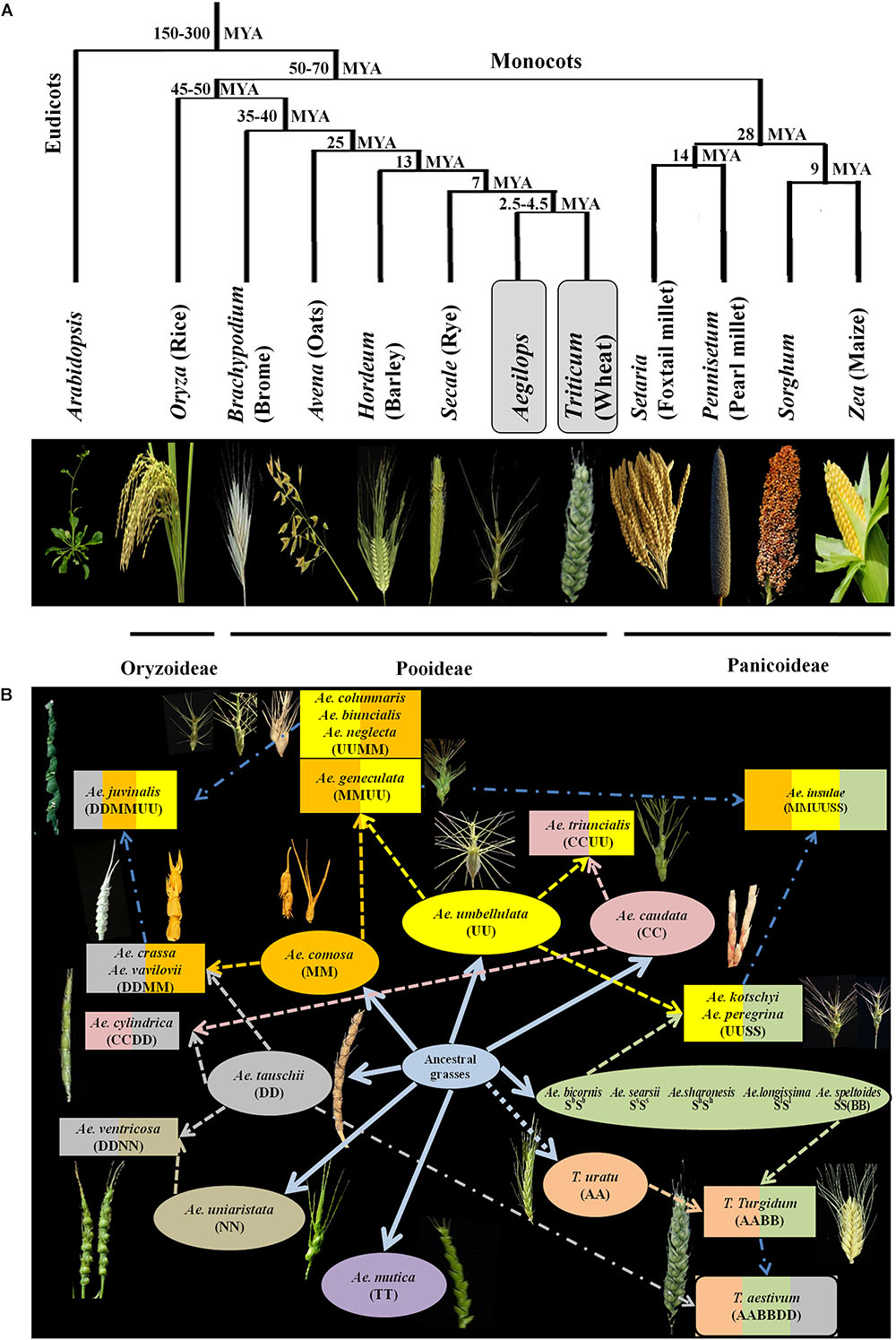
Figure 1. (A) Evolutionary relationship among different cereals. Aegilops is the closest relative of wheat. Divergence times from a common ancestor are indicated on the branches of the phylogenetic tree in million years (MYA). Modified from Bolot et al. (2009). (B) Hypothesized evolution of wheat and species of Aegilops. Seven different genomes of Aegilops evolved from common ancestor (color coded). Colored dash arrows indicate the involvement of species for formation of other species of Aegilops. Blue colored dash dot arrows indicate hypothetical involvement of species. Hypothetical wheat evolution is also explained, cross between Triticum urartu and Ae. speltoides led to formation of Triticum turgidum which further hybridized with Ae. tauschii to form cultivated Triticum aestivum. Modified from Meimberg et al. (2009).
A lot of breeding programs have been initiated to select or develop varieties with improved nutrient content and specific end product quality. The existing germplasm of wheat have been extensively explored for traits related to end product quality and nutrition. The Green revolution has resulted in the development of high yielding and disease resistant varieties and most of the varieties grown today consist of an assembly of genes pyramided by breeders (Lopes et al., 2015). The breeding programs thus have relied on limited number of parent lines for development of wheat germplasm. A report has suggested that due to this genetic bottleneck the population size of wheat has been reduced by 6% (Cavanagh et al., 2013). This narrow genetic diversity often limits the improvement of many traits in wheat. Therefore, the need to explore secondary and tertiary gene pools of wheat has grown. Secondary and tertiary gene pools of wheat mainly consist of wild varieties that are outstanding sources of genetic variability. The secondary gene pool of wheat mainly consists of polyploid Triticum and some of Aegilops species that share at least one of the A, B and D genomes of hexaploid wheat. The tertiary gene pool consists of wild species with genomes other than A, B and D of wheat. The relationship within and between Aegilops and Triticum has been a matter of debate and many classification systems exist (Kilian et al., 2011). The latest monograph of Van Slageren (1994) which is based on morphological studies is mostly followed for classification and nomenclature of Aegilops and same has been followed in this review article. For wheat the classification system by Dorofeev et al. (1979) is mainly followed. The Aegilops genus consists of 11 diploid, 10 tetraploid and 2 hexaploid species (Figure 1B). Species of Aegilops occur in Eurasia and North America, but most species are found near the center of origin, the Fertile Crescent in the Middle East, and around the Mediterranean Sea (Figure 2). These species consist of C, D, M, N, S, T and U genomes which have evolved from a common ancestor (Figure 1B) and can be used to incorporate genetic material from the wider gene pool into newly developed cultivars of wheat, thus increasing its genetic diversity.
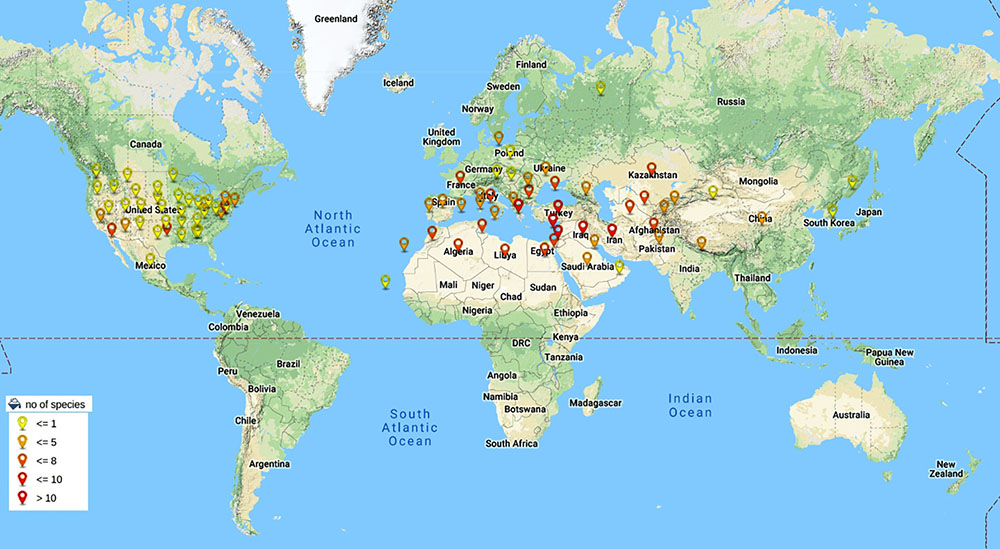
Figure 2. World wide distribution of Aegilops species. Species of Aegilops are mainly distributed in Eurasia and North America with highest density of occurrence in Fertile Crescent near Middle East (Colors of pins indicate number of species of Aegilops found in that area). Data taken from Kew (RBG Grassbase) database (Clayton et al., 2006).
There have been many reports of species of Aegilops being utilized for the improvement of agronomic traits such as rust resistance, powdery mildew resistance and tolerance against other abiotic stresses. More than 41 resistance genes for various biotic and abiotic stresses have been transferred from Aegilops to wheat via chromosome translocations or homoeologous recombination (Zhang et al., 2015) and many of these genes have been fairly successful in many breeding programs (Jahier et al., 1989; Ambrozkova et al., 2002; Zhang et al., 2015). This review summarizes the potential of Aegilops species for utilization in improvement of end product and nutritional quality of wheat.
Utilization of Aegilops for Improvement of End Product Quality of Wheat
The end product quality of wheat is affected by a number of factors such as: total protein content, grain texture and seed storage proteins composition. Seed storage proteins are the major determinants of end product quality and mainly consist of glutenins and gliadins. A large number of alleles of glutenins and gliadins have been explored in Aegilops species with their implications on end product quality. Grain texture related puroindolins, grain softness protein (GSP) and many other grain quality related genes have also been reported from Aegilops.
High Molecular Weight Glutenins (HMW GS)
High molecular weight glutenins are the major determinants of bread making quality of wheat. Their importance can be attributed to the fact that though they constitute only about 12% of total seed storage proteins, up to 60% of alterations in baking parameters are affected by them (Payne et al., 1987). HMW GS are coded by Glu1 loci present on the long arms of homoeologous group 1 chromosomes (1A, 1B and 1D) named as Glu A1, Glu B1 and Glu D1, respectively. Each locus produces two subunits of different size; called x-type (larger) and y-type (smaller) subunits i.e., 1Ax, 1Ay; 1Bx, 1By and 1Dx, 1Dy. Subunits 1Bx, 1Dx and 1Dy are expressed in most of the bread wheat cultivars while 1By and 1Ax are expressed in some wheat cultivars. The gene coding 1Ay generally remains silent in most of bread wheat cultivars (Halford et al., 1989). Only 21 alleles have been reported for Glu A1 locus, while for Glu B1 more than 69 alleles and for Glu D1 only 29 alleles have been documented in bread wheat germplasm (McIntosh et al., 2013).
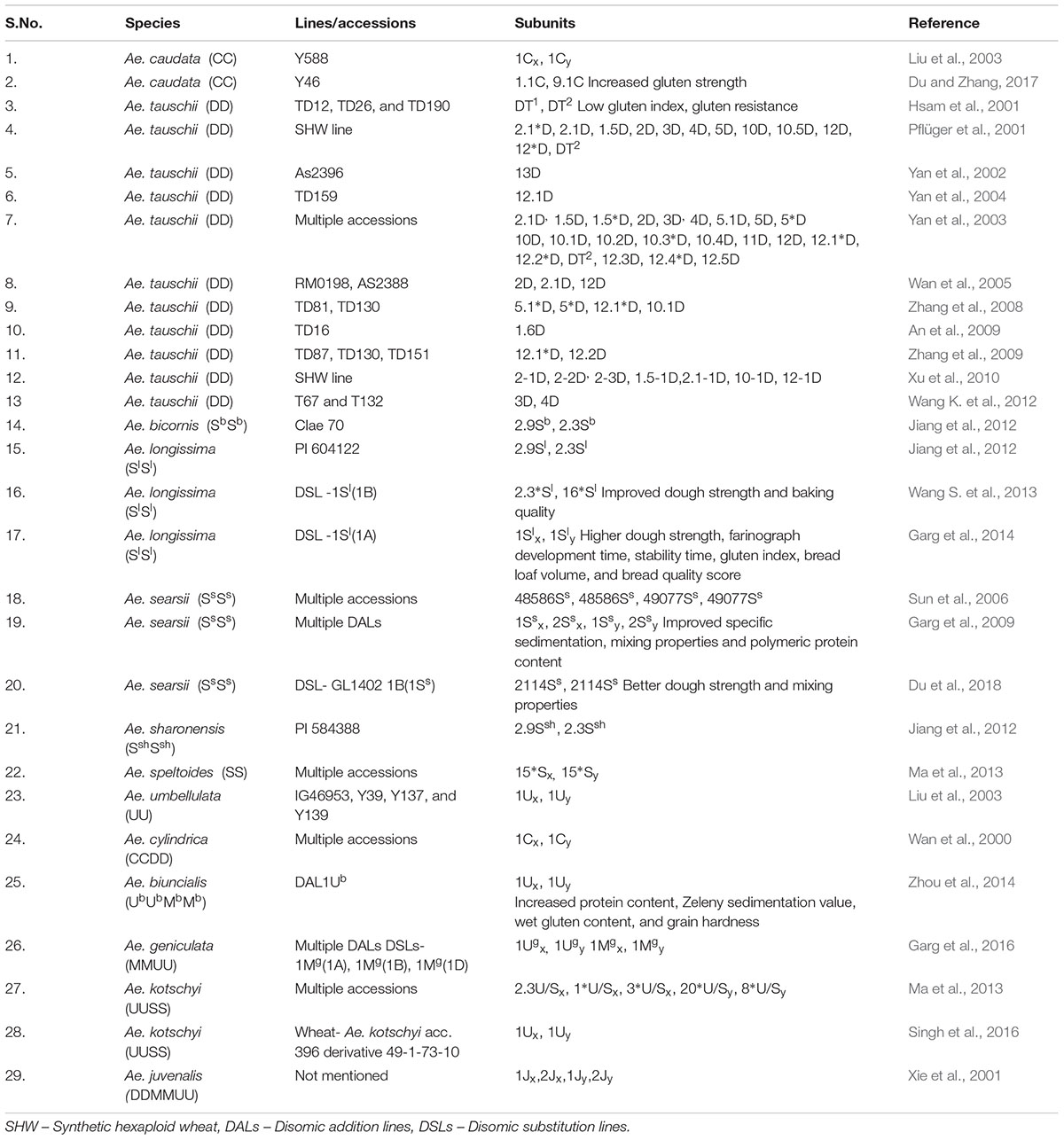
Due to this limited genetic diversity, high levels of allelic variations at Glu 1 loci are required in the quality wheat breeding practice. These are easiest to study as they can be conveniently resolved and identified by electrophoresis. Among the traits explored here, more than 600 lines and accessions of Aegilops have been studied across the world for their rich genetic diversity for HMW GS (Figure 3). Fairly large numbers of countries are involved in the exploration of HMW GS and their distribution across countries is also uniform (Figure 4). Primary structures of most of the Aegilops specific HMW GS are similar to wheat subunits. They contain conserved N-, C-terminals and a central variable repetitive region (Mackie et al., 1996; Wan et al., 2000; Xie et al., 2001). More than 30 subunits of HMW GS from Ae. bicornis, Ae. longissima, Ae. sharonensis, Ae. searsii, Ae. cylindrica, Ae. umbellulata, Ae. caudata, Ae. juvenalis, Ae. kotschyi, Ae. comosa, Ae. uniaristata, Ae. crassa, Ae. ventricosa and Ae. speltoides have been reported and studied (Table 1) (Wan et al., 2000, 2002; Xie et al., 2001; Liu et al., 2003; Sun et al., 2006; Farkhari et al., 2007; Jiang et al., 2012; Ma et al., 2013). Many of these HMW GS have been cloned and their sequence information is available.
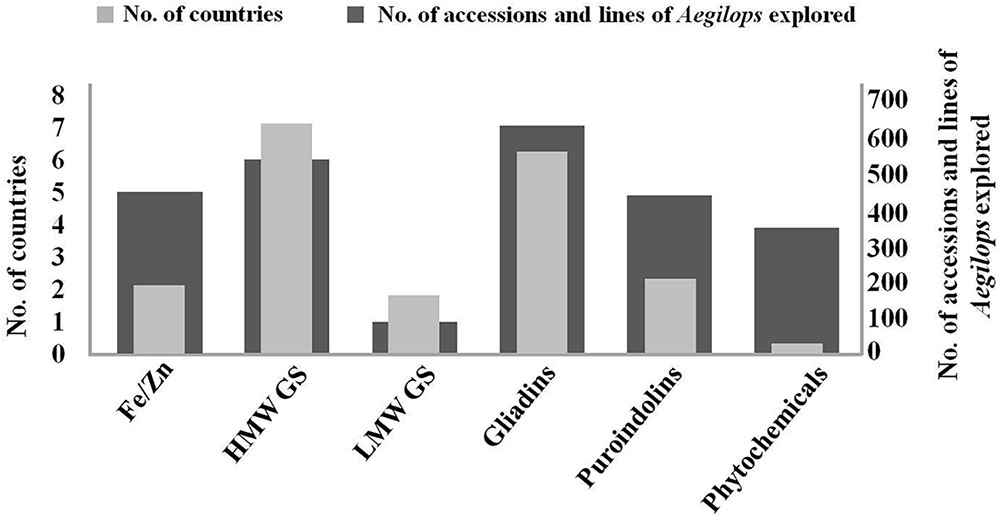
Figure 3. Number of accessions/lines of Aegilops explored along with number of countries involved in their exploration for improvement of quality and nutritional traits in wheat. HMW GS are most explored, while phytochemicals are least explored among different groups across the world. Gliadin exploration is being carried out by highest number of countries with LMW and phytochemicals being in the lowest category.
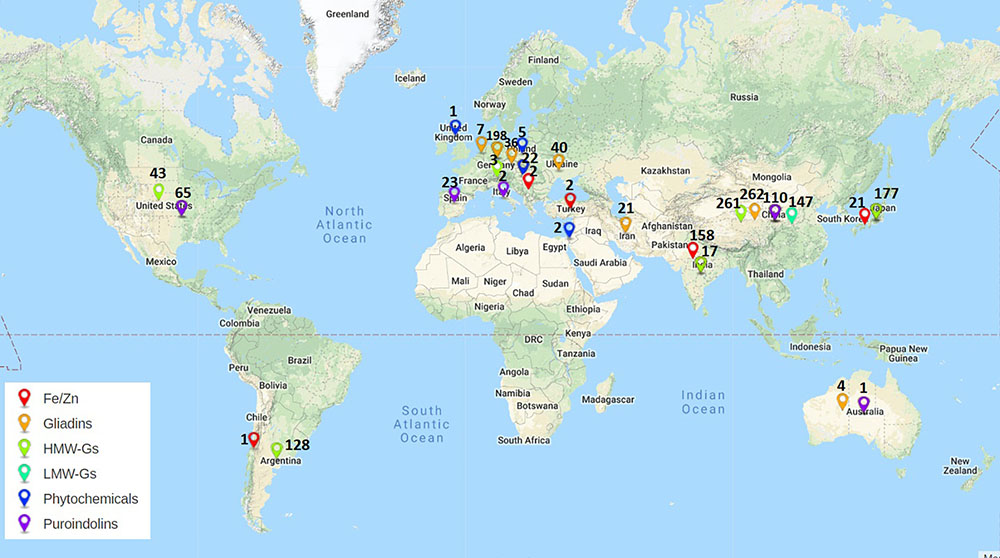
Figure 4. Work done across the world on Aegilops for improvement of quality and nutrition. Colors of pins indicate different traits. Numbers along pins indicate total number of accessions/lines explored.
Aegilops tauschii is regarded as D genome donor of wheat and its many accessions for HMW GS have been explored. For HMW GS, extensive studies have been done on the Glu D1 loci from Ae. tauschii as variation in this locus is very important in determining dough strength and other end product qualities. More than 40 HMW GS allelic variants have been reported from multiple accessions of Ae. tauschii (Yan et al., 2002, 2003, 2004; Wan et al., 2005; Zhang et al., 2008; An et al., 2009; Wang K. et al., 2012). Many D genome synthetic hexaploids have been generated by crossing tetraploid durum wheat with Ae. tauschii and thus HMW GS alleles 2.1∗D, 2.1D, 1.5D, 2D, 3D, 4D, 5D, 10D, 10.5D, 12D, 12∗D, T2 (Pflüger et al., 2001), 2-1D, 2-2D, 2-3D, 1.5-1D, 2.1-1D, 10-1D, and 12-1D (Xu et al., 2010) have been transferred to wheat. D genome specific subunits of 5Dx+10Dy have been reported to be most important for bread making quality of wheat (Branlard and Dardevet, 1985b). Attempts have been made to replace null Glu A1 allele of wheat with Glu D1 allele carrying 5Dx+10Dy subunits (Ceoloni et al., 1996; Ammar et al., 1997). Substitution of chromosome 1A with 1D has shown improvement in dough strength (Liu and Shepherd, 1995; Garg et al., 2007). A chromosomal translocation line 1AS.1AL-1DL carrying Glu D1d alleles (5Dx+10Dy) was generated in durum wheat background and was reported to possess improved mixing properties (Klindworth et al., 2005). Transfer of Glu D1 locus to chromosome 1R and 1A of Triticale has also been shown to improve bread making properties (Lukaszewski, 2006).
Implications of many HMW GS from Aegilops species on product quality have been studied. Subunits 1.1C and 9C from Ae. caudata led to increased gluten strength (Du and Zhang, 2017) while 2D+T1+T2 subunits from Ae. tauschii are associated with low gluten index and gluten resistance (Hsam et al., 2001). Disomic addition lines (DALs) from Ae. searsii have been used to transfer HMW GS subunits 1Ssx1, 1Ssx2, 1Ssy1and 1Ssy2 into wheat (Garg et al., 2009). These addition lines showed improved specific sedimentation, mixing properties and polymeric protein content. Similarly, DAL-1Ub of Ae. biuncialis (Zhou et al., 2014) were generated to transfer 1Ubx and 1Uby subunits to wheat and these lines showed increased protein content, Zeleny sedimentation value, wet gluten content, and grain hardness. Addition lines of Ae. umbellulata showed negative impact of its HMW GS on dough strength (Garg et al., 2009). Addition of 1Ug chromosome to transfer 1Ugx and 1Ugy subunits from Ae. geniculata led to reduced dough strength (Garg et al., 2016). Addition of 1Mg chromosome from Ae. geniculata to Chinese Spring background of wheat improved dough strength significantly (Garg et al., 2016). Many disomic substitution lines (DSLs) have also been generated from DALs. Addition line of 1Mg chromosome from Ae. geniculata was used to generate chromosome specific DSLs- 1Mg(1A), 1Mg(1B) and 1Mg(1D). DSLs- 1Mg(1A) and 1Mg(1B) showed improved dough strength and mixing properties but 1Mg(1D) showed reduced dough strength (Garg et al., 2016). Substitution of chromosome 1Sl from Ae. longissima with chromosomes 1A (Garg et al., 2014) and 1B (Wang S. et al., 2013) significantly improved bread making qualities of wheat. Similarly substituting chromosome 1Ss from Ae. searsii with 1B led to better dough strength and mixing properties (Du et al., 2018). All these addition and substitution lines that improved dough strength can be utilized to transfer HMW GS alleles into wheat in form of fine translocations with least linkage drag.
Low Molecular Weight Glutenins (LMW GS)
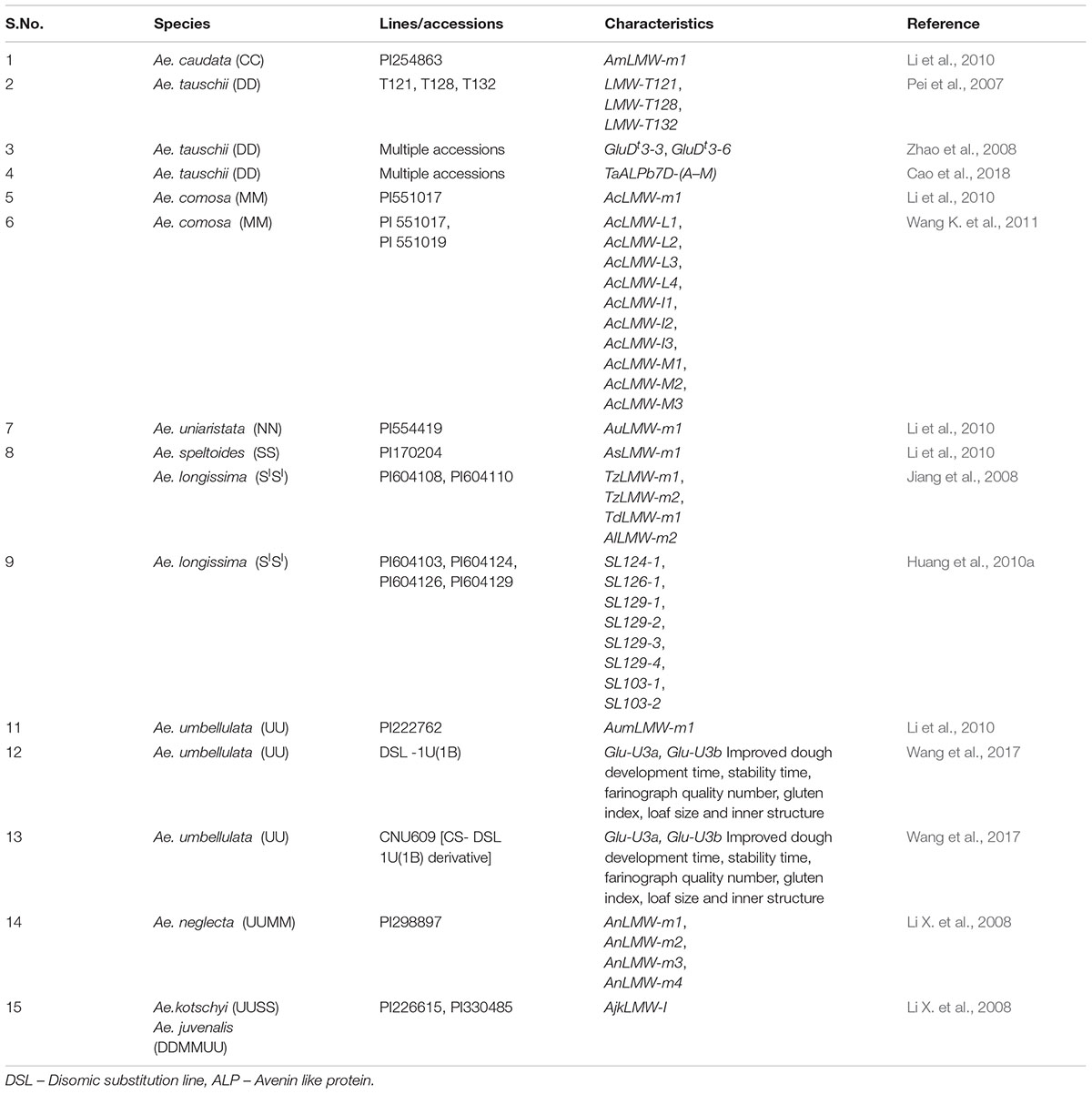
Low molecular weight glutenins account for 60% of total glutenins and one third of seed storage proteins. Genes that code for LMW GS (Glu A3, Glu B3 and Glu D3) are present on the short arms of group 1 homoeologous chromosomes (Singh and Shepherd, 1988; Sreeramulu and Singh, 1997). Only six alleles at Glu A3, nine at Glu B3 and five at Glu D3 have been reported in wheat germplasm (McIntosh et al., 2013). There are additional three loci (Glu 2, Glu 4 and Glu 5) present on chromosomes 1B, 1D and 7D (Jackson et al., 1985; Liu and Shepherd, 1995; Sreeramulu and Singh, 1997). On the basis of SDS PAGE mobility LMW GS can be classified into B, C and D types (Jackson et al., 1983). B type LMW GS are further classified into m, s and i type on the basis of first amino acid methionine, serine and isoleucine, respectively (Muccilli et al., 2010). Besides these three types, a novel LMW GS, l type was identified specifically in Aegilops with first amino acid being leucine (Wang K. et al., 2011).
Low molecular weight glutenins provide viscoelastic properties to the dough and some of their alleles have been reported to be associated with good bread making quality. Aegilops species serve as rich source of genetic diversity of LMW GS. More than 13 alleles of LMW GS from Ae. tauschii (Pei et al., 2007; Zhao et al., 2008; Cao et al., 2018), 12 alleles from Ae. longissima (Jiang et al., 2008; Huang et al., 2010a), 11 alleles from Ae. comosa (Wang K. et al., 2011), 4 alleles from Ae. neglecta (Li X. et al., 2008), 3 alleles from Ae. umbellulata and one from Ae. kotschyi (Li X. et al., 2008), Ae. uniaristata, Ae. caudata and Ae. speltoides each (Table 2) (Li et al., 2010) have been identified and characterized (Table 2). Most of these LMW GS genes have been cloned and their sequence information is available in NCBI. There is large amount of variability present in Aegilops specific LMW GS. Ae. tauschii exhibits even greater variation in LMW GS sequences than wheat (Rehman et al., 2008). There have been reports of novel LMW GS genes Glu U3a and Glu U3b from wheat-Ae. umbellulata 1U(1B) substitution line showing improved bread making and mixing properties. This substitution line was used to transfer the LMW GS genes to wheat. The line thus developed showed improvement in dough development time, stability time, farinograph quality number, gluten index, loaf size and inner structure (Wang et al., 2017). The variability in LMW GS genes found in Aegilops species indicates a large potential for their utilization in improvement of end product qualities of wheat. In comparison to HMW GS, works on transfer of LMW GS alleles from Aegilops species to wheat cultivars have been limited. As per literature only 147 accessions/lines have been explored for LMW GS, which too mainly in China (Figures 3, 4) and further exploration is needed.
Gliadins
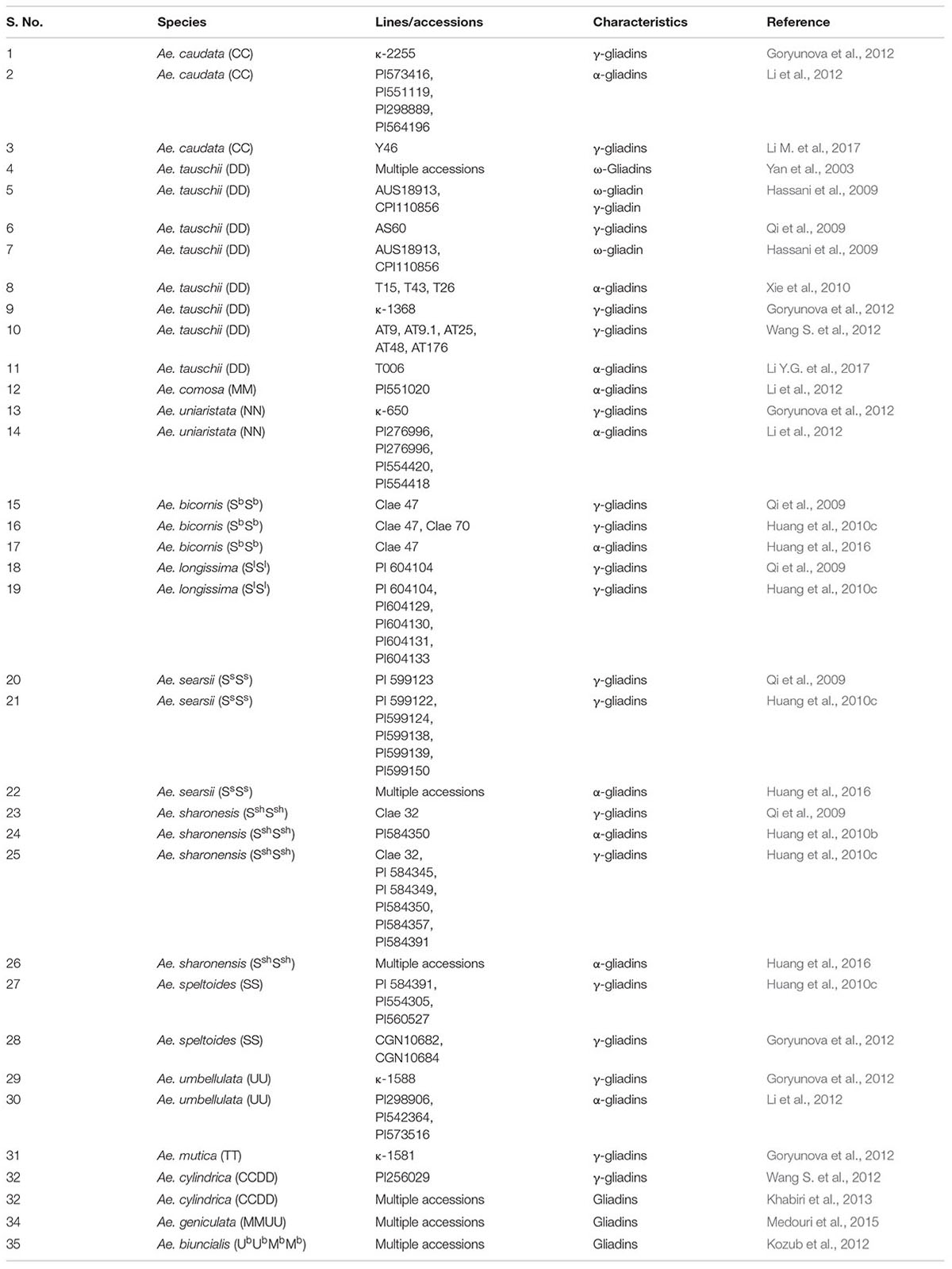
Gliadins account for 40–50% of total seed storage proteins. They have impacts on both processing and nutritional quality. Gliadins can be separated into α-/β-, γ-, and ω-gliadins based on differences in their mobility on SDS PAGE gel. Gli 1 loci present on short arms of homoeologous group 1 chromosomes code for all ω- and most of γ-gliadins, while, Gli 2 loci on the short arms of homoeologous group 6 chromosomes code for all α-, most of the β-, and some of the γ-gliadins (Payne, 1987; Metakovsky et al., 1990; Metakovsky, 1991). The effect of gliadins on rheological properties of dough has been studied (Branlard and Dardevet, 1985a). Due to lack of free cysteine residues in most of the gliadins, they are unable to form intermolecular S-S linkages. Hence, their overall impact on processing quality is small as compared to glutenins (Qi et al., 2009). Gliadins may act as chain terminators for gluten polymer. They therefore might limit the size of gluten complex and hence affect end product quality (Muccilli et al., 2005). However, many gliadins with odd number of cysteins also exist (Anderson et al., 2001; Goryunova et al., 2012). So some gliadins might also participate in gluten polymerization. It has been hypothesized that gliadins proteins contribute mostly toward dough cohesiveness (Uthayakumaran et al., 2000) and viscosity (Pistón et al., 2011) rather than resistance and extension. Studies on effect of Aegilops specific gliadins on product quality are limited. Multiple accessions of Ae. biuncialis and Ae. umbellulata have been reported to possess high gluten quality indices due to gliadins (Ahmadpoor et al., 2014) (Table 3). Gliadins from Ae. cylindrica (Khabiri et al., 2013), Ae. biuncialis (Kozub et al., 2012) and Ae. geniculata (Medouri et al., 2015) have been characterized on the basis of mobility on SDS PAGE (Table 3). Many ω-gliadins have been sequenced and characterized from Ae. tauschii (Yan et al., 2003; Hassani et al., 2009). γ-gliadins have been characterized from Ae. caudata, Ae. uniaristata, Ae. mutica, Ae. umbellulata (Goryunova et al., 2012), Ae. bicornis, Ae. searsii, Ae. sharonensis (Qi et al., 2009; Huang et al., 2010b), Ae. longissima (Qi et al., 2009), Ae. tauschii (Qi et al., 2009; Goryunova et al., 2012; Wang S. et al., 2012), Ae. speltoides (Huang et al., 2010b; Goryunova et al., 2012), Ae. markgrafii (Li M. et al., 2017) and Ae. cylindrica (Wang S. et al., 2012) (Table 3).
Although fairly large number of lines and accessions (more than 400) of Aegilops have been explored for gliadins (Figure 3) and their exploration is quite distributed across several countries of the world (Figure 4), most of the research conducted on gliadins of Aegilops is related to identification and characterization of allergic epitopes of celiac disease (Juhász et al., 2018). α-Gliadins are considered to be most allergic and are mostly responsible for inflammatory responses to celiac disease. α-Gliadins from Ae. speltoides (Spaenij-Dekking et al., 2004) and Ae. tauschii (Xie et al., 2010; Li et al., 2012, 2013) have been reported to be less allergic than corresponding wheat alleles. Novel α-gliadins have been reported from Ae. bicornis, Ae. searsii, Ae. sharonensis (Huang et al., 2010c, 2016), Ae tauschii (Xie et al., 2010; Li Y.G. et al., 2017), Ae. comosa, Ae. umbellulata, Ae. markgrafii and Ae. uniaristata (Li et al., 2012) (Table 3). These gliadins could contain useful variation and can be replaced from more allergic gliadins in wheat.
Puroindolins and Grain Softness Protein
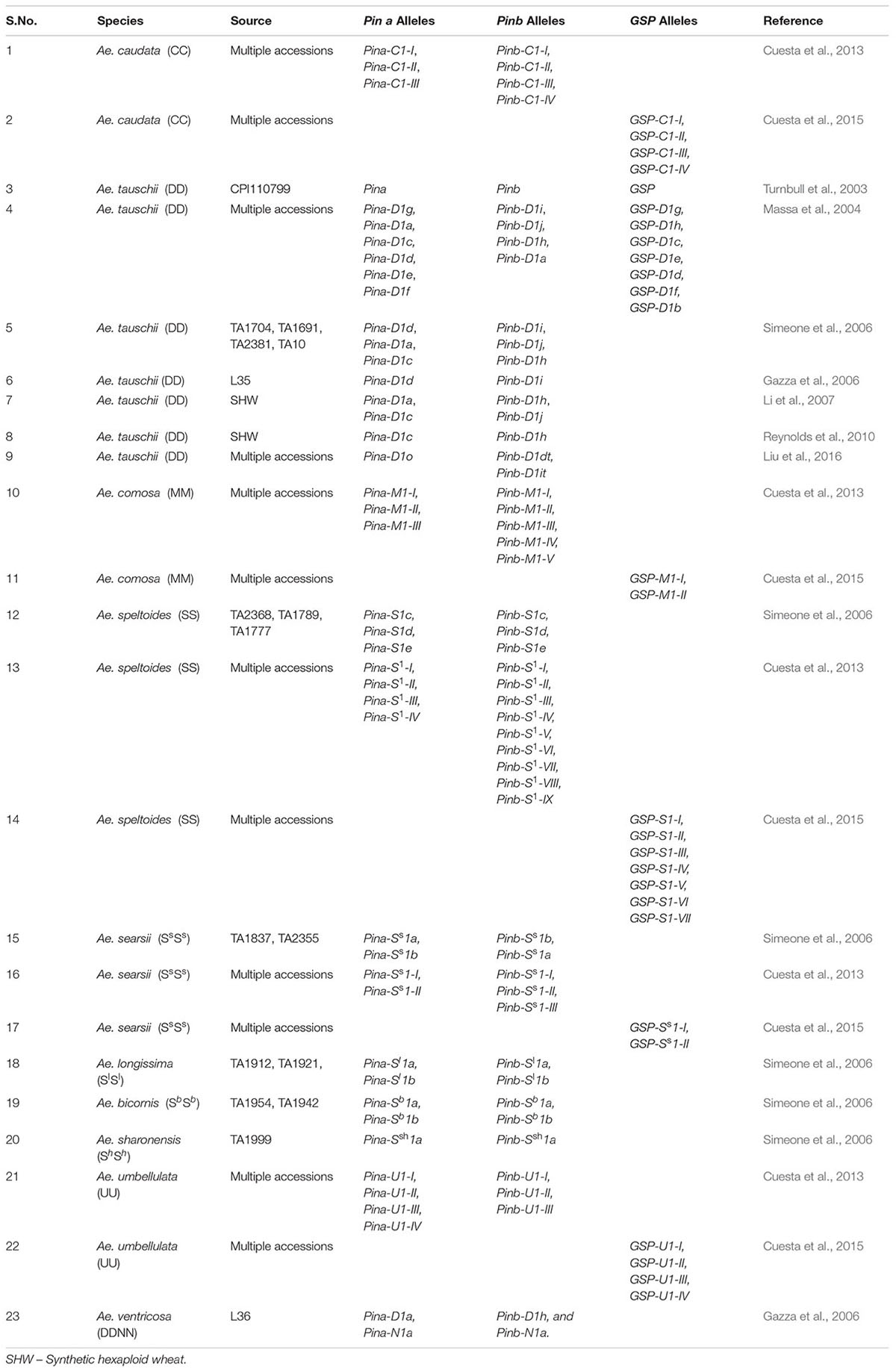
Grain texture plays important role in determining end product quality of wheat. Soft textured wheat is mostly used for pastries and biscuits, while hard textured wheat is used in making bread, pasta and noodles (Morris and Rose, 1996). Grain texture is determined by the hardness (Ha) locus present on the telomeric region of short arm of chromosome 5D of wheat which contains ten tightly linked genes (Chantret et al., 2005). Among them, three genes- puroindolin a (Pin a), puroindolin b (Pin b) and grain softness protein-1 (GSP) play major role in determining seed texture. These three genes code for the proteins which constitute a 15 kDa complex- friabilin, with Pin a, Pin b as major components and GSP-1 as minor component (Cuesta et al., 2015). This protein complex is found abundantly on the surface of starch granules of soft textured wheat and in very small amounts in hard textured wheat (Chen et al., 2005). Presence of this complex results in prevention of adhesion between starch granules and gluten matrix and hence soft texture (Greenwell and Schofield, 1986). Pin a and Pin b genes have also been associated with antimicrobial properties conferring protection to seed (Dubreil et al., 1998; Miao et al., 2012). Pin a especially has been hypothesized to have evolved in response to plant pathogens to enhance plant fitness (Massa and Morris, 2006). Soft seed texture is associated with wild type alleles of Pin a and Pin b (Pina-D1a and Pinb-D1a) and many mutations in those alleles have been linked with hard texture (Giroux and Morris, 1998). Pin a and Pin b genes are not present on A and B genome specific chromosomes (Li W. et al., 2008) and diploid species with A and B genomes as well as tetraploid durum wheat lack them, as a result of which durum has a very hard kernel texture (Chen et al., 2005). This also indicates Ae. tauschii as the donor of Pin genes in hexaploid wheat. Species of Aegilops have been explored for presence of different Pin alleles. Many novel Pin alleles have been reported from multiple accessions of Ae. tauschii (Table 4) (Massa et al., 2004; Gazza et al., 2006; Simeone et al., 2006; Liu et al., 2016). Many accessions of Ae. tauschii have been crossed with tetraploid durum wheat to produce synthetic wheat lines with different textures (Reynolds et al., 2010; Li et al., 2007). Many other Aegilops species have also been explored for variability in Pin a and Pin b gene alleles.19 alleles of puroindolins from Ae. speltoides, 9 alleles from Ae. searsii, 8 alleles from Ae. comosa, 7 from Ae. caudata and Ae. umbellulata each, 4 from Ae. longissima, Ae. ventricosa and Ae. bicornis each and 2 from Ae. sharonensis have been reported (Table 4) (Gazza et al., 2006; Simeone et al., 2006; Cuesta et al., 2013, 2015).
Unlike Pin a and Pin b, GSP genes are present on A and B genome specific chromosomes (5A, 5B). However, their deletion does not impact the grain texture (Chen et al., 2005). GSP genes have been characterized in many species of Aegilops. Many novel GSP alleles in Ae. tauschii, Ae. comosa, Ae. caudata, Ae. searsii, Ae. speltoides and Ae. umbellulata have been reported and characterized (Massa et al., 2004; Cuesta et al., 2015). Almost 100 alleles of Pin a, Pin b and GSP have been identified across 200 lines/accessions of Aegilops (Figure 3). Their exploration is quite uniform across different countries in the world (Figure 4). All these alleles can serve as useful source of variation and need to be evaluated and utilized in breeding programs for extending the textural characteristics of wheat.
Utilization of Aegilops for Improvement of Nutritional Quality of Wheat
Improvement of nutrition is a very important aspect of wheat research as there are over two billion people worldwide, suffering from deficiencies in proteins and micronutrients (World Health Organization, 2006). Nutritive value of wheat can be enhanced by increasing micronutrients like Fe and Zn, protein content, dietary fibers and many other phytochemicals such as carotenoids, vitamins etc. Aegilops genus can serve as important source for enhancing nutrition in wheat due to its high genetic variability.
Improvement of Grain Micronutrients Concentration
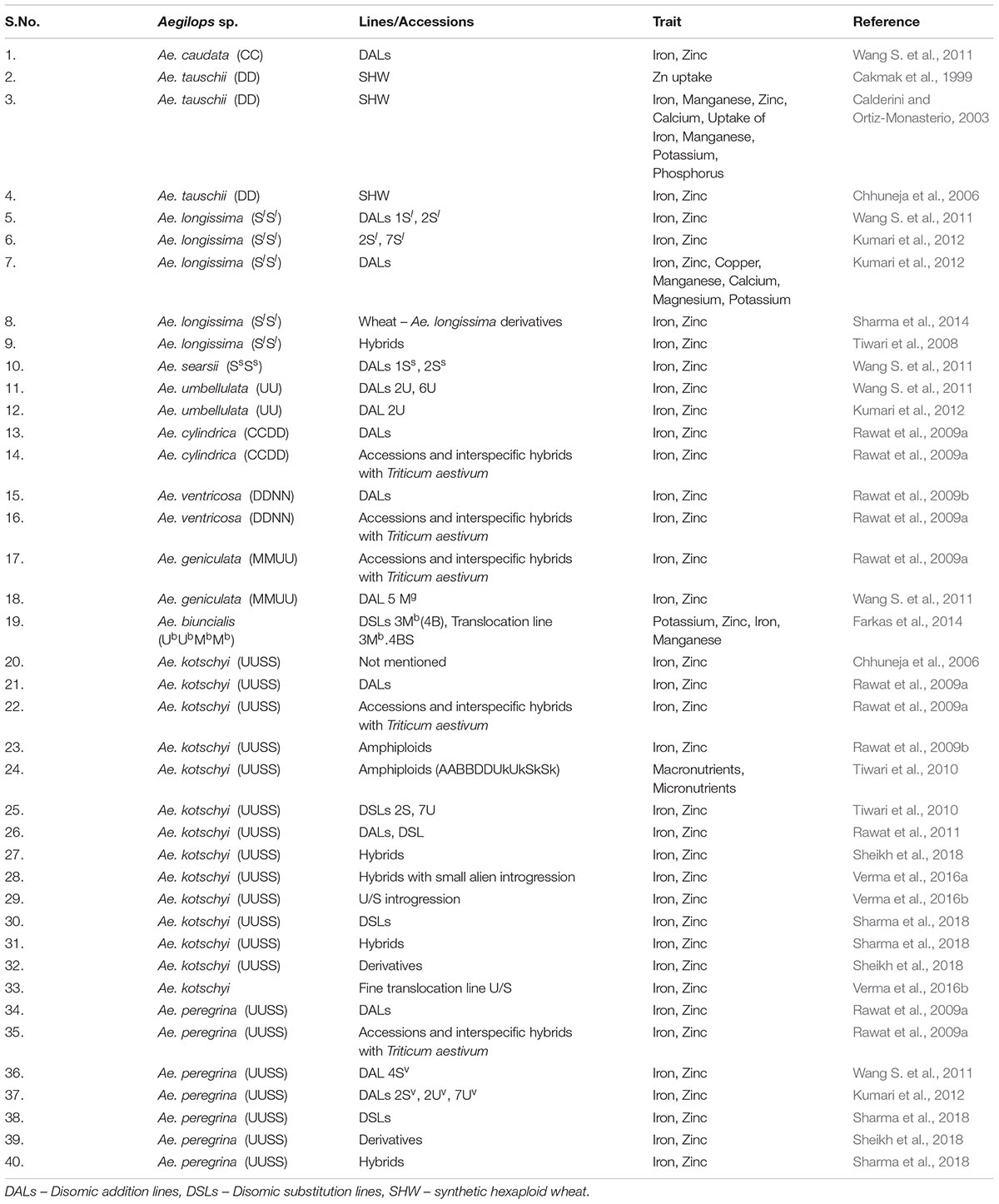
Micronutrients play very important role as health promoting factors. Since most of the world’s population especially developing nations depend on cereal based diet to fulfill their micronutrients requirements, it becomes very important to develop the varieties with improved micronutrients content. Iron and zinc are the most important components among micronutrients. Most varieties of wheat lack sufficient levels of iron and zinc due to low genetic variability. To overcome this limited genetic variability more than 180 lines/accessions of Aegilops have been explored (Figures 2, 3). Many accessions of Ae. kotschyi (Chhuneja et al., 2006; Rawat et al., 2009a,b, 2011), Ae. longissima (Kumari et al., 2013), Ae. tauschii, Ae. peregrina, Ae. cylindrica, Ae. ventricosa and Ae. geniculata (Rawat et al., 2009b) have been reported to have higher contents of iron and zinc in seeds (Table 5). These accessions can be exploited for increasing grain iron and zinc content. Amphiploids (Tiwari et al., 2010) and partial amphiploids (Rawat et al., 2009b) generated by crossing Ae. kotschyi accessions with wheat have been reported to have higher grain iron and zinc content. Many disomic and monosomic addition lines specific to various Aegilops species have been explored for higher micronutrient content. Fair exploration of grain micronutrient content has been carried out in many countries (Figure 3). Major exploration of Aegilops for Fe/Zn is from India (158 lines and accessions) as compared to other countries (Figure 4). Many disomic and monosomic addition lines of Ae. peregrina, Ae. longissima and Ae. umbellulata, in wheat have been explored for grain iron and zinc concentrations (Kumari et al., 2012). Addition of chromosome pairs 1Sl (Wang S. et al., 2011), 2Sl (Wang S. et al., 2011; Kumari et al., 2012) and 7Sl (Wang S. et al., 2011) of Ae. longissima into wheat showed increase in grain iron and zinc content. Similarly, DALs of chromosomes 2Sv, 2Uv, 7Uv (Kumari et al., 2012) and 4Sv (Wang S. et al., 2011) of Ae. peregrina, 2U (Kumari et al., 2012) and 6U (Wang S. et al., 2011; Kumari et al., 2012) of Ae. umbellulata, 1Ss and 2Ss of Ae. searsii (Wang S. et al., 2011), 5Mg of Ae. geniculata (Wang S. et al., 2011) and B chromosome additions from Ae. caudata (Wang S. et al., 2011) have been reported to increase the iron and zinc content in grains (Table 5). The addition lines can be used to produce DSLs which are better materials to study the compensation effect of alien chromosomes into wheat. Substitution of 4B chromosome of wheat with 3Mb chromosome of Ae. biuncialis (Farkas et al., 2014) also lead to increased iron and zinc content. Similarly, 2S(2A), 7U(7A) substitutions specific to Ae. kotschyi (Tiwari et al., 2010) have been reported with increased grain iron and zinc content.
Disomic addition/substitution lines can be utilized to introgress useful variability of high grain Fe and Zn from Aegilops into wheat in form of short arm or fine chromosomal translocations through induced homoeologous pairing. Interspecific hybrids of Ae. longissima with T. turgidum (Tiwari et al., 2008) and Ae. kotschyi (Sheikh et al., 2018) produced after crossing addition /substitution lines with tetraploid and hexaploid wheat also showed elevated levels of grain iron and zinc content. Ae. biuncialis specific translocation line 3Mb.4BS (Farkas et al., 2014) and many U/S chromosome specific fine translocations of Ae. kotschyi in wheat (Verma et al., 2016a,b) have been produced with least linkage drag effect. These lines also showed significant increase in grain iron and zinc content.
Improvement in Phytochemicals Concentration
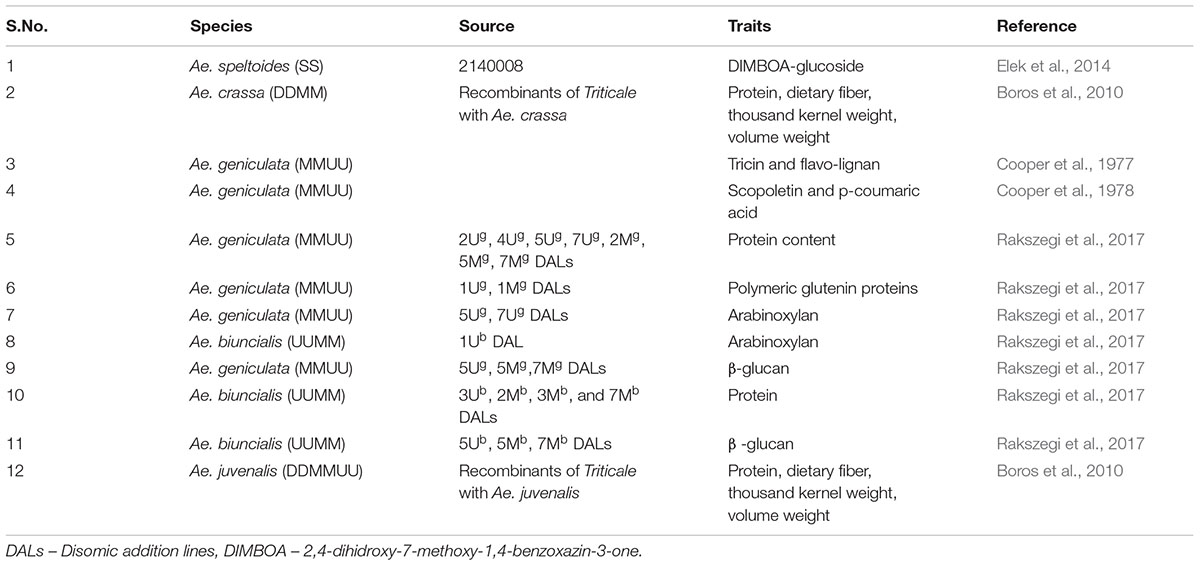
Studies on phytochemical contents of Aegilops species have been limited (Figure 3) with their work mainly being carried out in Europe (Figure 4). But given the rich genetic diversity of Aegilops, many phytochemicals such as phenolic acids, carotenoids, tocopherols, alkylresorcinols, benzoxazinoids, phytosterols and lignans can be explored in Aegilops species. Many phenolic diglycerides have been detected in Ae. geniculata (Cooper et al., 1978) (Table 6). p-hydroxybenzaldehyde, vanillin and mono-epoxylignanolide (MEL) have been detected in Ae. geniculata (Cooper et al., 1994). Alloplasmic lines derived from wheat and Ae. squarossa have been shown to increase the lutein content (Atienza et al., 2008). Synthetic hexaploid wheat (SHW) lines generated by crossing tetraploid durum wheat and Ae. tauschii also showed increased yellow pigment content and might be useful source for increasing carotenoids content in wheat (Li et al., 2015). DALs of Ae. geniculata and Ae. biuncalis showed increase in total protein content and polymeric proteins (Rakszegi et al., 2017) hence enhancing the nutritive value (Table 6).
Improvement in Dietary Fibers Concentration
Dietary fibers are important components of wheat which impact processing quality and have many health benefits. The major components of dietary fibers in wheat grain are cell wall polysaccharides, arabinoxylan (AX) and (1-3)(1-4)- β-D-glucan (β-glucan). Both of these occur in soluble and insoluble forms with different health benefits such as reduced risks of type II diabetes, coronary heart diseases and prevention of colon cancer. Soluble forms of dietary fibers also include FODMAPs (Fermentable oligosaccharides, disaccharides, monosaccharides and polyols) which are a group short chain carbohydrates. A diet rich in FODMAPs is often associated with diseases like Crohn disease and irritable bowel syndrome (IBS), which is a chronic gastrointestinal disease (Khan et al., 2015). Dietary fiber components have been reported to affect processing quality of wheat in terms of bread making and starch gluten separation. Arabinoxylan have effects on water absorption and development time of dough (Courtin and Delcour, 1998). β-glucan confers high viscosity, higher water absorption, lower loaf volume, height and stiffer dough (Symons and Brennan, 2004; Cleary et al., 2007; Skendi et al., 2009). From nutrition point of view higher levels of β-glucan are sought in food products as they lower serum cholesterol levels and regulate glucose levels in blood (McIntosh et al., 1991; Cavallero et al., 2002). Variability and composition of dietary fibers have been extensively studied in wheat and related cereal grains. Wheat primary gene pool has been explored in the European HEALTHGRAIN cereal diversity screening project1 for dietary fibers and other phytochemicals. However, such studies in wild species of wheat have been limited. There have been reports of recombinants of Triticale with Ae. crassa and Ae. juvenalis showing higher dietary fiber content along with increased values of total protein content, thousand kernel weight and volume weight (Boros et al., 2010) (Table 6). Both the species can be utilized for improving the nutrition value of wheat. Addition of 5Ug, 7Ug chromosome pairs of Ae. geniculata and 1Ub of Ae. biuncialis into wheat have resulted in increased arabinoxylan content (Rakszegi et al., 2017). Similarly, addition of 5Ug, 5Mg, and 7Mg chromosome pairs from Ae. geniculata and 5Ub, 5Mb, and 7Mb chromosomes from Ae. biuncialis have been reported to result in elevated levels of β-glucan content in wheat (Rakszegi et al., 2017). Since there is a large genetic diversity available in Aegilops species, they need to be explored for dietary fibers content and their potential use for enhancing nutritional value of wheat.
Conclusion
Quality and nutrition are two very important aspects of wheat research. Over the past few years, a lot of emphasis has been given by breeders worldwide to improve the end product quality of wheat and to develop varieties that meet specific end product and nutritional requirements. New sources of genetic variations in wheat are always sought after because of the narrow genetic diversity. Wild species of wheat can serve as excellent source of new variations that can be incorporated into wheat. Close relatedness to wheat makes Aegilops the most favorable genetic resource for wheat improvement through alien gene introgression. The basic approach for alien gene transfer is to cross the wild relative with wheat to generate interspecific hybrids followed by embryo rescue and colchicine treatment to double chromosomes. The amphiploids generated are then backcrossed multiple times with wheat to generate addition/substitution lines (Friebe et al., 1995, 1996, 1999). A large number of wheat-Aegilops amphiploids and chromosome addition/substitutions lines are available (Schneider et al., 2008). But these addition/substitution lines and amphiploids have no practical application in agriculture as the Aegilops chromosome segment carrying the gene of interest must be transferred to the wheat chromosome as translocation. The Ph1 locus, present at the long arm of chromosome 5B regulates chromosome pairing in wheat and ensures that only homologous chromosomes pair at metaphase. To generate translocations between wheat chromosome and alien chromosome, Ph1 mutants or Ph1 suppressors can be used to bypass the Ph1 control mechanism of homologous pairing. Translocations can also be generated via radiation induced chromosome breaks followed by random recombination. The recombinants generated then need to be screened using chromosome pairing, C banding pattern and in situ hybridization. Thus, the whole process of alien gene transfer is laborious and time consuming. However, with technological advancements and development of new high throughput marker technologies it is now possible to identify desirable recombinants from a large population with great precision and efficiency (Niu et al., 2011; Tiwari et al., 2014).
A large number of countries throughout the world are participating in the exploration of Aegilops. HMW GS are most explored, while phytochemicals are least explored among different research groups across the world. Gliadins have been explored by highest number of countries while, LMW GS and phytochemicals are least explored around the world (Figure 3). Based on this review we are aware that more than 95 subunits of HMW GS, 51 novel alleles of LMW GS, 34 alleles for Pin a, 40 alleles for Pin b and 26 alleles for GSP in Aegilops have been reported across multiple accessions, synthetic lines, addition/substitution lines and translocation lines (Figure 3). These can serve as excellent genetic sources of variation for wheat quality improvement. Large numbers of publications have arisen for Aegilops exploration for improvement of nutrition and processing quality. Highest exploration has been carried out in China and Europe followed by Japan and India (Figure 4). Major work on LMW GS has been carried out in China, Fe/Zn in India, others having good distribution across countries (Figure 4). More than 14 species of Aegilops have been proven to be excellent sources for the improvement of grain micronutrient content, protein content, dietary fiber content and phytochemical content. Many Aegilops species have already been incorporated in various breeding programs across the world. Still there is further need to explore Aegilops species to identify new variations. Though a large number of accessions are available in gene banks, many accessions of Aegilops species still remain unexploited. The real bottleneck for introgressing useful genes into wheat from Aegilops, however, is the generation of fine translocation lines containing the smallest possible segment of alien chromosome with the gene of interest. Although a lot of scientific exploration has been carried out, practically we still are nowhere in terms of introgressing and utilizing genes related to quality and nutrition from Aegilops species. There is still a long way to go. It is anticipated that the availability of the newly annotated wheat genome sequence (International Wheat Genome Sequencing Consortium, Appels et al., 2018) along with new genomic tools and genetic resources will aid the further exploration and exploitation of Aegilops species and the transfer of useful traits into wheat.
Author Contributions
AK and MG built the layout of article. AK, MG, and PK collected the literature. AK wrote the article. MG, VC, SS, and PK helped in manuscript editing. PK did the reference management. All authors prepared images and tables.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Department of Biotechnology, Government of India for the fellowship provided to AK. Contribution of Ms. Simran Bhatia, Ms. Neetu, and Ms. Anita Kumari in manuscript editing is truly acknowledged.
Footnote
References
Ahmadpoor, F., Asghari-Zakaria, R., Firoozi, B., and Shahbazi, H. (2014). Investigation of diversity in Aegilops biuncialis and Aegilops umbellulata by A-PAGE. Nat. Prod. Res. 28, 1626–1636. doi: 10.1080/14786419.2014.931392
Ambrozkova, M. O., Dedryver, F. R., Dumalasova, V. E., Hanzalova, A. L., and Bartos, P. (2002). Determination of the cluster of wheat rust resistance genes Yr17, Lr37, and Sr38 by a molecular marker. Plant Prot. Sci. 38, 41–45. doi: 10.17221/4849-PPS
Ammar, K., Lukaszewski, A. J., and Banowetz, G. M. (1997). Effect of Glu-1(5+10) on gluten strength and polymeric protein composition in durum wheat. Cereal Food World 42:610.
An, X. L., Li, X. H., Xiong, X. J., Yan, Y. M., Zhang, Y. Z., Gao, L. Y., et al. (2009). Identification and isolation of a new x-type HMW glutenin subunit 1Dx1.6t gene from Aegilops tauschii. Plant Breed. 128, 41–45. doi: 10.1111/j.1439-0523.2008.01543.x
Anderson, O. D., Hsia, C. C., and Torres, V. (2001). The wheat γ-gliadin genes: characterization of ten new sequences and further understanding of γ-gliadin gene family structure. Theor. Appl. Genet. 103, 323–330. doi: 10.1007/s00122-001-0551-3
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 361:eaar7191. doi: 10.1126/science.aar7191
Atienza, S. G., Martín, A., Pecchioni, N., Platani, C., and Cattivelli, L. (2008). The nuclear-cytoplasmic interaction controls carotenoid content in wheat. Euphytica 159, 325-331. doi: 10.1007/s10681-007-9511-6
Bolot, S., Abrouk, M., Masood-Quraishi, U., Stein, N., Messing, J., Feuillet, C., et al. (2009). The ‘inner circle’ of the cereal genomes. Curr. Opin. Plant Biol. 12, 119–125. doi: 10.1016/j.pbi.2008.10.011
Boros, D., Ploch, M., and Gruszecka, D. (2010). Possibility of utilization of two Aegilops sp. to enhance the nutritive value of triticale. J. Anim. Feed Sci. 19, 628–637. doi: 10.22358/jafs/66336/2010
Branlard, G., and Dardevet, M. (1985a). Diversity of grain protein and bread quality. I. Correlation between gliadin bands and technological characteristics. J. Cereal Sci. 3, 329–343. doi: 10.1016/S0733-5210(85)80006-0
Branlard, G., and Dardevet, M. (1985b). Diversity of grain protein and bread quality. II. Correlation between high molecular weight subunits of glutenin and flour quality characteristics. J. Cereal Sci. 3, 345–354. doi: 10.1016/S0733-5210(85)80007-2
Cakmak, I., Cakmak, O., Eker, S., Ozdemir, A., Watanabe, N., and Braun, H. J. (1999). Expression of high zinc efficiency of Aegilops tauschii and Triticum monococcum in synthetic hexaploid wheats. Plant Soil 215, 203–209.
Calderini, D. F., and Ortiz-Monasterio, I. (2003). Are synthetic hexaploids a means of increasing grain element concentrations in wheat? Euphytica 134, 169–178. doi: 10.1023/B:EUPH.0000003849.10595.ac
Cao, D., Wang, H., Zhang, B., Liu, B., Liu, D., Chen, W., et al. (2018). Genetic diversity of avenin-like b genes in Aegilops tauschii Coss. Genetica 146, 45–51. doi: 10.1007/s10709-017-9995-5
Cavallero, A., Empilli, S., Brighenti, F., and Stanca, A. M. (2002). High (1 → 3, 1 → 4)-β-glucan barley fractions in bread making and their effects on human glycemic response. J. Cereal Sci. 36, 59–66. doi: 10.1006/jcrs.2002.0454
Cavanagh, C. R., Chao, S., Wang, S., Huang, B. E., Stephen, S., Kiani, S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 8057–8062. doi: 10.1073/pnas.1217133110
Ceoloni, C., Biaggetti, M., Ciuffi, M., Forte, P., and Pasquini, M. (1996). Wheat chromosome engineering at the 4x level: the potential of different alien gene transfers into durum wheat. Euphytica 89, 87–97. doi: 10.1007/BF00015724
Chantret, N., Salse, J., Sabot, F., Rahman, S., Bellec, A., Laubin, B., et al. (2005). Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17, 1033–1045. doi: 10.1105/tpc.104.029181
Chen, M., Wilkinson, M., Tosi, P., He, G., and Shewry, P. (2005). Novel puroindoline and grain softness protein alleles in Aegilops species with the C, D, S, M and U genomes. Theor. Appl. Genet. 111, 1159–1166. doi: 10.1007/s00122-005-0047-7
Chhuneja, P., Dhaliwal, H. S., Bains, N. S., and Singh, K. (2006). Aegilops kotschyi and A. tauschii as a source for higher levels of grain iron and zinc. Plant Breed. 125, 531–539. doi: 10.1111/j.1439-0523.2006.01223.x
Clayton, W. D., Vorontsova, M. S., Harman, K. T., and Williamson, H. (2006). GrassBase- The Online World Grass flora. Available at: http://www.kew.org/data/grasses-db.html
Cleary, L. J., Andersson, R., and Brennan, C. S. (2007). The behavior and susceptibility to degradation of high and low molecular weight barley β-glucan in wheat bread during baking and in vitro digestion. Food Chem. 102, 889–897. doi: 10.1016/j.foodchem.2006.06.027
Cooper, R., Gottlieb, H. E., and Lavie, D. (1977). A new flavolignan of biogenetic interest from Aegilops ovata L.-Part I. Isr. J. Chem. 16, 12–15. doi: 10.1002/ijch.197700005
Cooper, R., Gottlieb, H. E., and Lavie, D. (1978). New phenolic diglycerides from Aegilops ovata. Phytochemistry 17, 1673–1675. doi: 10.1016/S0031-9422(00)94673-9
Cooper, R., Lavie, D., Gutterman, Y., and Evenari, M. (1994). The distribution of rare phenolic type compounds in wild and cultivated wheats. J. Arid. Environ. 27, 331–336. doi: 10.1006/jare.1994.1068
Courtin, C. M., and Delcour, J. A. (1998). Physicochemical and bread-making properties of low molecular weight wheat-derived arabinoxylans. J. Agric. Food Chem. 46, 4066–4073. doi: 10.1021/jf980339t
Cuesta, S., Alvarez, J. B., and Guzmán, C. (2015). Characterization and sequence diversity of the Gsp-1 gene in diploid species of the Aegilops genus. J. Cereal Sci. 63, 1–7. doi: 10.1016/j.jcs.2015.01.007
Cuesta, S., Guzmán, C., and Alvarez, J. B. (2013). Allelic diversity and molecular characterization of puroindoline genes in five diploid species of the Aegilops genus. J. Exp. Bot. 64, 5133–5143. doi: 10.1093/jxb/ert299
Dorofeev, V. F., Filatenko, A. A., Migushova, E. F., Udaczin, R. A., and Jakubziner, M. M. (1979). “Wheat,” in Flora of Cultivated Plants, Vol. 1, eds V. F. Dorofeev and O. N. Korovina (Leningrad: Springer-Verlag Wien).
Du, X., Ma, X., Min, J., Zhang, X., and Jia, Z. (2018). Development of a wheat-Aegilops searsii substitution line with positively affecting Chinese steamed bread quality. Breed. Sci. 68, 289–293. doi: 10.1270/jsbbs.17044
Du, X., and Zhang, X. (2017). Molecular cloning and functional characterization of two novel high molecular weight glutenin subunit genes in Aegilops markgrafii. J. Genet. 96, 563–570. doi: 10.1007/s12041-017-0799-2
Dubreil, L., Gaborit, T., Bouchet, B., Gallant, D. J., Broekaert, W. F., Quillien, L., et al. (1998). Spatial and temporal distribution of the major isoforms of puroindolines (puroindoline-a and puroindoline-b) and non specific lipid transfer protein (ns-LTP1e1) of Triticum aestivum seeds. Relationships with their in vitro antifungal properties. Plant Sci. 138, 121–135. doi: 10.1016/S0168-9452(98)00121-6
Elek, H., Smart, L., Ahmad, S., Anda, A., Werner, C., and Pickett, J. (2014). A comparison of the levels of hydroxamic acids in Aegilops speltoides and a hexaploid wheat and effects on Rhopalosiphum padi behavior and fecundity. Acta Biol. Hung. 65, 38–46. doi: 10.1556/ABiol.65.2014.1.4
Farkas, A., Molnár, I., Dulai, S., Rapi, S., Oldal, V., Cseh, A., et al. (2014). Increased micronutrient content (Zn, Mn) in the 3Mb(4B) wheat-Aegilops biuncialis substitution and 3Mb.4BS translocation identified by GISH and FISH. Genome 57, 61–67. doi: 10.1139/gen-2013-0204
Farkhari, M., Naghavi, M. R., and Pyghambari, S. A. (2007). Genetic variation of jointed goatgrass (Aegilops cylindrica Host.) from Iran using RAPD-PCR and SDS-PAGE of seed proteins. Pak. J. Biol. Sci. 10, 2868–2873. doi: 10.3923/pjbs.2007.2868.2873
Food and Agriculture Organization of United Nations [FAO] (2014). Food and Agriculture Organization of United Nations Available at: http://www.fao.org/resources/infographics/infographics-details/en/c/240943/ [accessed 28 January 2019; 16:00GMT]
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Friebe, B., Jiang, J., Tuleen, N., and Gill, B. S. (1995). Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor. Appl. Genet. 90, 150–156. doi: 10.1007/BF00221010
Friebe, B. R., Tuleen, N. A., and Gill, B. S. (1999). Development and identification of a complete set of Triticum aestivum-Aegilops geniculata chromosome addition lines. Genome 42, 374–380. doi: 10.1139/g99-011
Garg, M., Dhaliwal, H. S., Chhuneja, P., Kumar, D., Dou, Q. W., Tanaka, H., et al. (2007). Negative effect of chromosome 1A on dough strength shown by modification of 1D addition in durum wheat (Triticum durum). Theor. Appl. Genet. 114, 1141–1150. doi: 10.1007/s00122-007-0506-4
Garg, M., Kumar, R., Singh, R. P., and Tsujimoto, H. (2014). Development of an Aegilops longissima substitution line with improved bread-making quality. J. Cereal Sci. 60, 389–396. doi: 10.1016/j.jcs.2014.05.006
Garg, M., Tanaka, H., Ishikawa, N., Takata, K., Yanaka, M., and Tsujimoto, H. (2009). A novel pair of HMW glutenin subunits from Aegilops searsii improves quality of hexaploid wheat. Cereal Chem. 86, 26–32. doi: 10.1094/CCHEM-86-1-0026
Garg, M., Tsujimoto, H., Gupta, R. K., Kumar, A., Kaur, N., Kumar, R., et al. (2016). Chromosome specific substitution lines of Aegilops geniculata alter parameters of bread making quality of wheat. PLoS One 11:e0162350. doi: 10.1371/journal.pone.0162350
Gazza, L., Conti, S., Taddei, F., and Pogna, N. E. (2006). Molecular characterization of puroindolines and their encoding genes in Aegilops ventricosa. Mol. Breed. 17, 191–200. doi: 10.1007/s11032-005-4886-3
Giroux, M. J., and Morris, C. F. (1998). Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc. Natl. Acad. Sci. U.S.A. 95, 6262–6266. doi: 10.1073/pnas.95.11.6262
Goryunova, S. V., Salentijn, E. M., Chikida, N. N., Kochieva, E. Z., Vander, I. M., and Meer, L. J. et al. (2012). Expansion of the gamma-gliadin gene family in Aegilops and Triticum. BMC Evol. Biol. 12:215. doi: 10.1186/1471-2148-12-215
Greenwell, P., and Schofield, J. D. (1986). A starch granule protein associated with endosperm softness in wheat. Cereal Chem. 63, 379–380.
Halford, N. G., Forde, J., Shewry, P. R., and Kreis, M. (1989). Functional analysis of the upstream regions of a silent and an expressed member of a family of wheat seed protein genes in transgenic tobacco. Plant Sci. 62, 207–216. doi: 10.1016/0168-9452(89)90083-6
Hassani, M., Naghavi, M., Shariflou, M., and Sharp, P. (2009). Identification of novel ω-gliadin gene in Aegilops tauschii using RFLP. Cereal Res. Commun. 37, 75–82. doi: 10.1556/CRC.37.2009.1.9
Hsam, S. L. K., Kieffer, R., and Zeller, F. J. (2001). Significance of Aegilops tauschii glutenin genes on breadmaking properties of wheat. Cereal Chem. 78, 521–525. doi: 10.1094/CCHEM.2001.78.5.521
Huang, Z., Long, H., Jiang Q. T., Wei, Y. M., Yan Z. H., and Zheng, Y. L. (2010a). Molecular characterization of novel low molecular weight glutenin genes in Aegilops longissima. J. Appl. Genet. 51, 9–18. doi: 10.1007/BF03195705
Huang, Z., Long, H., Wei, Y., Qi, P., Yan, Z., and Zheng, Y. (2010b). Characterization and classification of γ-gliadin multigene sequences from Aegilops section Sitopsis. Cereal Res. Comm. 38, 1–14. doi: 10.1556/CRC.38.2010.1.1
Huang, Z., Long, H., Wei, Y., and Yan, Z. (2010c). Isolation and sequences analysis of the α-gliadin genes from Aegilops sharonensis. J. Plant Sci. 5, 256–262. doi: 10.3923/jps.2010.256.262
Huang, Z., Long, H., Wei, Y., Yan, Z., and Zheng, Y. (2016). Allelic variations of α-gliadin genes from species of Aegilops section Sitopsis and insights into evolution of α-gliadin multigene family among Triticum and Aegilops. Genetica 144, 213–222. doi: 10.1007/s10709-016-9891-4
Jackson, E. A., Holt, L. M., and Payne, P. I. (1983). Characterization of high molecular weight gliadin and low-molecular-weight glutenin subunits of wheat endosperm by two-dimensional electrophoresis and the chromosomal location of their controlling genes. Theor. Appl. Genet. 66: 29–37. doi: 10.1007/BF00281844
Jackson, E. A., Holt, L. M., and Payne, P. I. (1985). Glu B2, a storage protein locus controlling the D group of LMW glutenin subunits in bread wheat (Triticum aestivum). Genet. Res. 47, 11–17. doi: 10.1017/S0016672300022412
Jahier, J., Tanguy, A. M., and Doussinault, G. (1989). Analysis of the level of eyespot resistance due to genes transferred to wheat from Aegilops ventricosa. Euphytica 44, 55–59. doi: 10.1007/BF00022599
Jiang, C., Pei, Y., Zhang, Y., Li, X., Yao, D., Yan, Y., et al. (2008). Molecular cloning and characterization of four novel LMW glutenin subunit genes from Aegilops longissima, Triticum dicoccoides and T. zhukovskyi. Hereditas 145, 92–98. doi: 10.1111/j.0018-0661.2008.02035.x
Jiang, Q. T., Ma, J., Wei, Y. M., Liu, Y. X., Lan, X. J., Dai, S. F., et al. (2012). Novel variants of HMW glutenin subunits from Aegilops section Sitopsis species in relation to evolution and wheat breeding. BMC Plant Biol. 12:73. doi: 10.1186/1471-2229-12-73
Juhász, A., Belova, T., Florides, C. G., Maulis, C., Fischer, I., Gell, G., et al. (2018). Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 4:eaar8602. doi: 10.1126/sciadv.aar8602
Khabiri, T., Asghari-Zakaria R., Zare, N., and Sofalian, O. (2013). Assessing genetic diversity based on gliadin proteins in Aegilops cylindrica populations from northwest of Iran. Nat. Sci. Biol. 5, 109–113. doi: 10.15835/nsb518329
Khan, M. A., Nusrat, S., Khan, M. I., Nawras, A., and Bielefeldt, K. (2015). Low-FODMAP diet for irritable bowel syndrome: is it ready for prime time? Dig. Dis. Sci. 60, 1169–1177. doi: 10.1007/s10620-014-3436-4
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al. (2011). “Wild crop relatives: genomic and breeding resources, cereals,” in Aegilops, ed. Kole, C. (Berlin: Springer), 1–76. doi: 10.1007/978-3-642-14228-4_1
Klindworth, D. L., Hareland, G. A., Elias, E. M., and Xu, S. S. (2005). Agronomic and quality characteristics of 1AS.1AL-1DL translocation lines of durum wheat carrying Glu-D1d. Crop Sci. 45, 77–84. doi: 10.2135/cropsci2005.0077
Kozub, N. A., Sozinov, I. A., and Sozinov, A. A. (2012). Identification of alleles at the gliadin loci Gli-U1 and Gli-Mb1 in Aegilops biuncialis Vis. Russ. J. Genet. 48, 390–395. doi: 10.1134/S1022795412030052
Kumari, N., Rawat, N., Tiwari, V., Prasad, R., Tripathi, S., Randhawa, G., et al. (2012). Evaluation and identification of wheat-Aegilops addition lines controlling high grain iron and zinc concentration and mugineic acid production. Cereal Res. Commun. 40, 53–61. doi: 10.1556/CRC.40.2012.1.7
Kumari, N., Rawat, N., Tiwari, V. K., Gandhi, N., Arun, P. K., Kumar, S., et al. (2013). Development and molecular characterization of wheat- Aegilops longissima derivatives with high grain micronutrients. Aust. J. Crop Sci. 7, 508–514.
Li, G., Xia, X., He, Z., and Sun, Q. (2007). Allelic variations of puroindoline a and puroindoline b genes in new type of synthetic hexaploid wheats from CIMMYT. Acta Agron. Sin. 33, 242–249.
Li, J., Wang, S., Li, S., Ge, P., Li, X., Ma, W., et al. (2012). Variations and classification of toxic epitopes related to celiac disease among α-gliadin genes from four Aegilops genomes. Genome 55, 513–521. doi: 10.1139/g2012-038
Li, J., Wang, S. L., Cao, M., Lv, D. W., Subburaj, S., Li, X. L., et al. (2013). Cloning, expression and evolutionary analysis of α-gliadin genes from Triticum and Aegilops genomes. J. Appl. Genet. 54, 157–167. doi: 10.1007/s13353-013-0139-z
Li, M., Du, X., Ma, X., and Kong, L. (2017). Cloning and characterization of novel γ-gliadin genes from Aegilops markgrafii in relation to evolution and wheat breeding. Crop J. 5, 290–295. doi: 10.1016/j.cj.2017.03.006
Li, W., Huang, L., and Gill, B. S. (2008). Recurrent deletions of puroindoline genes at the grain hardness locus in four independent lineages of polyploid wheat. Plant Physiol. 146, 200–212. doi: 10.1104/pp.107.108852
Li, X., Ma, W., Gao, L., Zhang, Y., Wang, A., Ji, K., et al. (2008). A novel chimeric low-molecular-weight glutenin subunit gene from the wild relatives of wheat Aegilops kotschyi and Ae. juvenalis: evolution at the Glu-3 loci. Genetics 180, 93–101. doi: 10.1534/genetics.108.092403
Li, X. H., Wang, K., Wang, S. L., Gao, L. Y., Xie, X. X., Hsam, S. L. K., et al. (2010). Molecular characterization and comparative transcriptional analysis of LMW-m-type genes from wheat (Triticum aestivum L.) and Aegilops species. Theor. Appl. Genet. 121, 845–856. doi: 10.1007/s00122-010-1354-1
Li, Y. F., Wu, Y., Wang, T., Li, L. R., Lu, L., and Zhang, C. Y. et al. (2015). Polyphenol oxidase activity and yellow pigment content in Aegilops tauschii, Triticum turgidum, Triticum aestivum, synthetic hexaploid wheat and its parents. J. Cereal Sci. 65, 192–201 doi: 10.1016/j.jcs.2015.07.011
Li, Y. G., Liang, H. H., Bai, S. L., Zhou, Y., Sun, G., Su, Y. R., et al. (2017). Molecular characterization and variation of the celiac disease epitope domains among α-gliadin genes in Aegilops tauschii. J. Agric. Food Chem. 65, 3422–3429. doi: 10.1021/acs.jafc.7b00338
Liu, C. Y., and Shepherd, K. W. (1995). Inheritance of B subunits of glutenin and ω and γ-gliadins in tetraploid wheats. Theor. Appl. Genet. 90, 1149–1157. doi: 10.1007/BF00222936
Liu, D., Chen, W., Zhang, B., Liu, D., Liu, B., and Zhang, H. (2016). Diversity and distribution of puroindoline-D1 genes in Aegilops tauschii. Genet. Resour. Crop Evol. 63, 615-625. doi: 10.1007/s10722-015-0271-1
Liu, Z., Yan, Z., Wan, Y., Liu, K., Zheng, Y., and Wang, D. (2003). Analysis of HMW glutenin subunits and their coding sequences in two diploid Aegilops species. Theor. Appl. Genet. 106, 1368–1378. doi: 10.1007/s00122-002-1175-y
Lopes, M. S., Basyoni, I. E., Baenziger, P. S., Singh, S., Royo, C., Ozbek, K., et al. (2015). Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 66, 3477–3486. doi: 10.1093/jxb/erv122
Lukaszewski, A. J. (2006). Cytogenetically engineered rye chromosomes 1R to improve bread-making quality of hexaploid Triticale. Crop Sci. 46, 2183–2194. doi: 10.2135/cropsci2006.03.0135
Ma, C., Yang, Y., Li, X., Ge, P., Guo, G., Subburaj, S., et al. (2013). Molecular cloning and characterization of six novel HMW GS genes from Aegilops speltoides and Aegilops kotschyi. Plant Breed. 132, 284–289. doi: 10.1111/pbr.12046
Mackie, A. M., Sharp, P. J., and Lagudah, E. S. (1996). The nucleotide and derived amino acid sequence of a HMW glutenin gene from Triticum tauschii and comparison with those from the D genome of bread wheat. J. Cereal Sci. 24, 73–78. doi: 10.1006/jcrs.1996.0039
Massa, A. N., and Morris, C. F. (2006). Molecular evolution of the puroindoline-a, puroindoline-b, and grain softness protein-1 genes in the tribe Triticeae. J. Mol. Evol. 63, 526–536. doi: 10.1007/s00239-005-0292-z
Massa, A. N., Morris, C. F., and Gill, B. S. (2004). Sequence diversity of puroindoline-a, puroindoline-b, and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci. 44, 1808–1816. doi: 10.2135/cropsci2004.1808
McIntosh, G. H., Whyte, J., McAurthur, R., and Nestel, P. J. (1991). Barley and wheat foods: influence on plasma cholesterol concentrations in hypercholesterolemic men. Am. J. Clin. Nutr. 53, 1205–1209. doi: 10.1093/ajcn/53.5.1205
McIntosh, R., Yamazaki, Y., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X.C., (2013). “Catalogue of gene symbols for wheat,” in Proceedings of the 12th International Wheat Genetics Symposium (Minato: Yokohama).
Medouri, A., Bellil, I., and Khelifi, D. (2015). The genetic diversity of gliadins in Aegilops geniculata from Algeria. Czech. J. Genet. Plant Breed. 51, 9–15. doi: 10.17221/158/2014-CJGPB
Meimberg, H., Rice, K. J., Milan, N. F., Njoku, C. C., and McKay, J. K. (2009). Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). Am. J. Bot. 96, 1262–1273. doi: 10.3732/ajb.0800345
Metakovsky, E. V. (1991). Gliadin allele identification in common wheat II. Catalogue of gliadin alleles in common wheat. J. Genet. Breed. 45, 325–344.
Metakovsky, E. V., Wrigley, C. W., Bekes, F., and Gupta, R. B. (1990). Gluten polypeptides as useful genetic markers of dough quality in Australian wheats. Aust. J. Agr. Res. 41, 289–306. doi: 10.1071/AR9900289
Miao, Y., Chen, L., Wang, C., Wang, Y., Zheng, Q., Gao, C., et al. (2012). Expression, purification and antimicrobial activity of puroindoline A protein and its mutants. Amino Acids 43, 1689–1696. doi: 10.1007/s00726-012-1250-x
Morris, C. F., and Rose, S. P. (1996). Chapter 1. Wheat. in Cereal Grain Quality, eds R.J. Henry and P.S. Kettlewell (New York, NY: Chapman and Hall), 3–54. doi: 10.1007/978-94-009-1513-8_1
Muccilli, V., Cunsolo, V., Saletti, R., Foti, S., Margiotta, B., Scossa, F., et al. (2010). Characterization of a specific class of typical low molecular weight glutenin subunits of durum wheat by a proteomic approach. J. Cereal Sci. 51, 134–139. doi: 10.1016/j.jcs.2009.11.003
Muccilli, V., Cunsolo, V., Saletti, R., Foti, S., Masci, S., and Lafiandra, D. (2005). Characterization of B- and C-type low molecular weight glutenin subunits by electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry. Proteomics 5,719–728. doi: 10.1002/pmic.200401029
Niu, Z., Klindworth, D. L., Friesen, T. L., Chao, S., Jin, Y., Cai, X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi: 10.1534/genetics.110.123588
Payne, P. I. (1987). Genetics of wheat storage proteins and effect of allelic variation on bread making quality. Annu. Rev. Plant Physiol. 38, 141–153. doi: 10.1146/annurev.pp.38.060187.001041
Payne, P. I., Holt, L. M., Krattiger, A. F., and Carrillo, J. M. (1987). Relationships between seed quality characteristics and HMW glutenin subunit composition determined using wheats grown in Spain. J. Cereal Sci. 7, 229–235. doi: 10.1016/S0733-5210(88)80004-3
Pei, Y. H., Wang, A. L., An, X. L., Li, X. H., Zhang, Y. Z., Huang, X. Q., et al. (2007). Characterization and comparative analysis of three low molecular weight glutenin C-subunit genes isolated from Aegilops tauschii. Can. J. Plant Sci. 87, 273–280. doi: 10.4141/P06-152
Pflüger, L. A., D’Ovidio, R., Margiotta, B., Pena, R., Mujeeb-Kazi, A., and Lafiandra, D. (2001). Characterisation of high-and low-molecular weight glutenin subunits associated to the D genome of Aegilops tauschii in a collection of synthetic hexaploid wheats. Theor. Appl. Genet. 103, 1293–1301. doi: 10.1007/s001220100704
Pistón, F., Gil-Humanes, J., Rodríguez-Quijano, M., and Barro, F. (2011). Down-regulating γ-gliadins in bread wheat leads to non-specific increases in other gluten proteins and has no major effect on dough gluten strength. PLoS One 6:e24754. doi: 10.1371/journal.pone.0024754
Qi, P. F., Wei, Y. M., Ouellet, T., Chen, Q., Tan, X., and Zheng, Y. L. (2009). The γ-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genomics 10:168. doi: 10.1186/1471-2164-10-168
Rakszegi, M., Molnár, I., Lovegrove, A., Darkó,É., Farkas, A., Láng, L., et al. (2017). Addition of Aegilops U and M chromosomes affects protein and dietary fiber content of whole meal wheat flour. Front. Plant Sci. 8:1529. doi: 10.3389/fpls.2017.01529
Rawat, N., Neelam, K., Tiwari, V. K., Randhawa, G. S., Friebe, B., Gill, B. S., et al. (2011). Development and molecular characterization of wheat-Aegilops kotschyi addition and substitution lines with high grain protein, iron, and zinc. Genome 54, 943–953. doi: 10.1139/g11-059
Rawat, N., Tiwari, V. K., Neelam, K., Randhawa, G. S., Chhuneja, P., Singh, K., et al. (2009a). Development and characterization of Triticum aestivum-Aegilops kotschyi amphiploids with high grain iron and zinc contents. Plant Genet. Resour. 7, 271–280. doi: 10.1017/S1479262109356592
Rawat, N., Tiwari, V. K., Singh, N., Randhawa, G. S., Singh, K., Chhuneja, P., et al. (2009b). Evaluation and utilization of Aegilops and wild Triticum species for enhancing iron and zinc content in wheat. Genet. Resour. Crop Evol. 56, 53–64. doi: 10.1007/s10722-008-9344-8
Rehman, A., Evans, N., Gianibelli, M. C., and Rose, R. J. (2008). Allelic variations in high and low molecular weight glutenins at the Glu-Dt locus of Aegilops tauschii as a potential source for improving bread wheat quality. Aust. J. Agric. Res. 59, 399–405. doi: 10.1071/AR07229
Reynolds, N. P., Martin, J. M., and Giroux, M. J. (2010). Increased wheat grain hardness conferred by novel puroindoline haplotypes from Aegilops tauschii. Crop Sci. 50, 1718–1727. doi: 10.2135/cropsci2009.12.0700
Schneider, A., Molnar, I., and Molnar-Lang, M. (2008). Utilization of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Sharma, P., Imran, Sharma, P., Chugh, V., Dhaliwal, H. S., and Singh, D. (2014). Morphological, cytological and biochemical characterization of wheat Aegilops longissima derivatives BC1F6 and BC2F4 with high grain micronutrient. IJAEB 7, 191–204. doi: 10.5958/2230-732X.2014.00234.4
Sharma, P., Sheikh, I., Kumar, S., Verma, S. K., Kumar, R., Vyas, P., et al. (2018). Precise transfers for high grain iron and zinc from wheat-Aegilops substitution lines into wheat through pollen irradiation. Mol. Breed. 38:81. doi: 10.1007/s11032-018-0836-8
Sheikh, I., Sharma, P., Verma, S. K., Kumar, S., Kumar, N., Kumar, S., et al. (2018). Development of intron targeted amplified polymorphic markers of metal homeostasis genes for monitoring their introgression from Aegilops species to wheat. Mol. Breed. 38:47. doi: 10.1007/s11032-018-0809-y
Simeone, M. C., Gedye, K. R., Mason-Gamer, R., Gill, B. S., and Morris, C. F. (2006). Conserved regulatory elements identified from a comparative puroindoline gene sequence survey of Triticum and Aegilops diploid taxa. J. Cereal Sci. 44, 21–33. doi: 10.1016/j.jcs.2006.02.002
Singh, J., Sheikh, I., Sharma, P., Kumar, S., Verma, S. K., Kumar, R. et al. (2016). Transfer of HMW glutenin subunits from Aegilops kotschyi to wheat through radiation hybridization. J. Food Sci. Technol. 53, 3543–3549. doi: 10.1007/s13197-016-2333-6
Singh, N. K., and Shepherd, K. W. (1988). Linkage mapping of the genes controlling endosperm proteins in wheat. 1. Genes on the short arms of group 1 chromosomes. Theor. Appl. Genet. 66, 628–641. doi: 10.1007/BF00289132
Skendi, A., Papageorgiou, M., and Biliaderis, C. G. (2009). Effect of barley β-glucan molecular size and level on wheat dough rheological properties. J. Food Eng. 91, 594–601. doi: 10.1016/j.jfoodeng.2008.10.009
Spaenij-Dekking, E. H. A., Kooy-Winkelaar, E. M. C., Nieuwenhuizen, W. F., Drijfhout, J. W., and Koning, F. (2004). A novel and sensitive method for the detection of T cell stimulatory epitopes of α/β-and γ-gliadin. Gut 53, 1267–1273. doi: 10.1136/gut.2003.037952
Sreeramulu, G., and Singh, N. K. (1997). Genetic and biochemical characterization of novel low molecular weight glutenin subunits in wheat (Triticum aestivum L.). Genome 40, 41–48. doi: 10.1139/g97-006
Sun, X., Hu, S., Liu, X., Qian, W., Hao, S., Zhang, A. et al. (2006). Characterization of the HMW glutenin subunits from Aegilops searsii and identification of a novel variant HMW glutenin subunit. Theor. Appl. Genet. 113, 631–641. doi: 10.1007/s00122-006-0327-x
Symons, L. J., and Brennan, C. S. (2004). The influence of (1 → 3) (1 → 4)-β-D-glucan-rich fractions from barley on the physicochemical properties and in vitro reducing sugar release of white wheat breads. J. Food Sci. 69, C463–C467. doi: 10.1111/j.1365-2621.2004.tb10989.x
Tiwari, V. K., Rawat, N., Neelam, K., Kumar, S., Randhawa, G. S., and Dhaliwal, H. S. (2010). Random chromosome elimination in synthetic Triticum-Aegilops amphiploids leads to development of a stable partial amphiploid with high grain micro and macronutrient content and powdery mildew resistance. Genome 53, 1053–1065. doi: 10.1139/G10-083
Tiwari, V. K., Rawat, N., Neelam, K., Randhawa, G. S., Singh, K., Chhuneja, P., et al. (2008). Development of Triticum turgidum subsp. durum-Aegilops longissima amphiploids with high iron and zinc content through unreduced gamete formation in F1 hybrids. Genome 51, 757–766. doi: 10.1139/G08-057
Tiwari, V. K., Wang, S., Sehgal, S., Vrána, J., Friebe, B., Kubaláková, M., et al. (2014). SNP discovery for mapping alien introgressions in wheat. BMC Genomics 15:273. doi: 10.1186/1471-2164-15-273
Turnbull, K. M., Turner, M., Mukai, Y., Yamamoto, M., Morell, M. K., Appels, R., et al. (2003). The organization of genes tightly linked to Ha locus in Aegilops tauschii, the D genome donor to wheat. Genome 46, 330–338. doi: 10.1139/g02-124
Uthayakumaran, S., Newberry, M., Keentok, M., Stoddard, F. L., and Békés, F. (2000). Basic rheology of bread dough with modified protein content and glutenin to gliadin ratio. Cereal Chem. 77, 744–749. doi: 10.1094/CCHEM.2000.77.6.744
Van Slageren, M. W. (1994). Wild wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Paper Presented Agricultural University, 94-7, Wageningen.
Verma, S. K., Kumar, S., Sheikh, I., Malik, S., Mathpal, P., Chugh, V., et al. (2016a). Transfer of useful variability of high grain iron and zinc from Aegilops kotschyi into wheat through seed irradiation approach. Int. J. Radiat. Biol. 92, 132–139. doi: 10.3109/09553002.2016.1135263
Verma, S. K., Kumar, S., Sheikh, I., Sharma, P., Mathpal, P., Malik, S., et al. (2016b). Induced homoeologous pairing for transfer of useful variability for high grain Fe and Zn from Aegilops kotschyi into wheat. Plant Mol. Biol. Rep. 34, 1083–1094. doi: 10.1007/s11105-016-0989-8
Wan, Y., Liu, K., Wang, D., and Shewry, P. R. (2000). High-molecular-weight glutenin subunits in the Cylindropyrum and Vertebrata section of the Aegilops genus and identification of subunits related to those encoded by the Dx alleles of common wheat. Theor. Appl. Genet. 101, 879–884. doi: 10.1007/s001220051556
Wan, Y., Wang, D., Shewry, P., and Halford, N. (2002). Isolation and characterization of five novel high molecular weight subunits of glutenin genes from Triticum timopheevi and Aegilops cylindrica. Theor. Appl. Genet. 104, 828–839. doi: 10.1007/s00122-001-0793-0
Wan, Y., Yan, Z., Liu, K., Zheng, Y., D’Ovidio, R., Shewry, P. R., et al. (2005). Comparative analysis of the D genome-encoded high-molecular weight subunits of glutenin. Theor. Appl. Genet. 111, 1183–1190. doi: 10.1007/s00122-005-0051-y
Wang, D. W., Li, D., Wang, J., Zhao, Y., Wang, Z., Yue, G., et al. (2017). Genome-wide analysis of complex wheat gliadins, the dominant carriers of celiac disease epitopes. Sci. Rep. 7:44609. doi: 10.1038/srep44609
Wang, K., An, X. L., Pan, L. P., Dong, K., Gao, L. Y., Wang, S. L. et al. (2012). Molecular characterization of HMW GS 1Dx3t and 1Dx4t genes from Aegilops tauschii and their potential value for wheat quality improvement. Hereditas 149, 41–49. doi: 10.1111/j.1601-5223.2011.02215.x
Wang, S., Shen, X., Ge, P., Li, J., Subburaj, S., Li, X., et al. (2012). Molecular characterization and dynamic expression patterns of two types of γ-gliadin genes from Aegilops and Triticum species. Theor. Appl. Genet. 125, 1371–1384. doi: 10.1007/s00122-012-1917-4
Wang, K., Gao, L., Wang, S., Zhang, Y., Li, X., Zhang, M., et al. (2011). Phylogenetic relationship of a new class of LMW-GS genes in the M genome of Aegilops comosa. Theor. Appl. Genet. 122, 1411–1425. doi: 10.1007/s00122-011-1541-8
Wang, S., Yin, L., Tanaka, H., Tanaka, K., and Tsujimoto, H. (2011). Wheat-Aegilops chromosome addition lines showing high iron and zinc contents in grains. Breed. Sci. 61, 189–195. doi: 10.1270/jsbbs.61.189
Wang, S., Yu, Z., Cao, M., Shen, X., Li, N., Li, X., et al. (2013). Molecular mechanisms of HMW glutenin subunits from 1Sl genome of Aegilops longissima positively affecting wheat breadmaking quality. PLoS One 8:e58947. doi: 10.1371/journal.pone.0058947
World Health Organization (2006). “Micronutrient malnutrition: a public health problem,” in Guidelines on Food Fortification with Micronutrients. FAO-WHO, eds A. Lindsay, B. de Benoist, D. Omar, and Rı. Hurrell (Geneva: WHO).
Xie, R., Wan, Y., Zhang, Y., and Wang, D. (2001). HMW glutenin subunits in multiploid Aegilops species: composition analysis and molecular cloning of coding sequences. Chin. Sci. Bull. 46, 309–313. doi: 10.1007/BF03187192
Xie, Z. Z., Wang, C. Y., Wang, K., Wang, S. L., Li, X. H., and Zhang, Z. (2010). Molecular characterization of the celiac disease epitope domains in α-gliadin genes in Aegilops tauschii and hexaploid wheats (Triticum aestivum L.). Theor. Appl. Genet. 121, 1239–1251. doi: 10.1007/s00122-010-1384-8
Xu, S. S., Khan, K., Klindworth, D. L., and Nygard, G. (2010). Evaluation and characterization of high molecular weight 1D glutenin subunits from Aegilops tauschii in synthetic hexaploid wheats. J. Cereal. Sci. 52, 333–336 doi: 10.1016/j.jcs.2010.05.004
Yan, Y., Hsam, S. L. K., Yu, J., Jiang, Y., and Zeller, F. J. (2003). Allelic variation of the HMW glutenin subunits in Aegilops tauschii accessions detected by sodium dodecyl sulphate (SDS-PAGE), acid polyacrylamide gel (A-PAGE) and capillary electrophoresis. Euphytica 130, 377–385. doi: 10.1023/A:1023062316439
Yan, Y., Zheng, J., Xiao, Y., Yu, J., Hu, Y., Cai, M., et al. (2004). Identification and molecular characterization of a novel y-type Glu-Dt1 glutenin gene of Aegilops tauschii. Theor. Appl. Genet. 108, 1349–1358. doi: 10.1007/s00122-003-1547-y
Yan, Z., Wan, Y., Liu, K., Zheng, Y., and Wang, D. (2002). Identification of a novel HMW glutenin subunit and comparison of its amino acid sequence with those of homologous subunits. Chin. Sci. Bull. 47, 222–226. doi: 10.1360/02tb9053
Zhang, P., Dundas, I. S., McIntosh, R. A., Xu, S. S., Park, R. F., Gill, B. S., et al. (2015). “Wheat-Aegilops introgressions,” in Alien Introgression in Wheat: Cytogenetics, Molecular Biology and Genomics, eds M. Molnár-Láng,, C. Ceoloni, and J. Doležel (Springer Science-Business Media: Berlin),221–243.
Zhang, Y., An, X., Li, X., Chen, S., Gao, L., Wang, K., et al. (2009). Isolation and expression of a new high molecular weight glutenin subunit gene at the Glu-D-1-2 locus from Aegilops tauschii. Cereal Res. Commun. 37, 449–457. doi: 10.1556/CRC.37.2009.3.14
Zhang, Y., Li, X., Wang, A., An, X., Zhang, Q., Pei, Y., et al. (2008). Novel x-type high-molecular-weight glutenin genes from Aegilops tauschii and their implications on the wheat origin and evolution mechanism of Glu-D1-1 proteins. Genetics 178, 23–33. doi: 10.1534/genetics.107.077412
Zhao, X., Yang, Y., He, Z., Lei, Z., Ma, W., Sun, Q., et al. (2008). Characterization of novel LMW GS genes at Glu-D3 locus on chromosome 1D in Aegilops tauschii. Hereditas 145, 238–250. doi: 10.1111/j.1601-5223.2008.02046.x
Keywords: Aegilops, grain micronutrients, puroindolins, gliadins, dietary fiber, glutenins, phytochemicals
Citation: Kumar A, Kapoor P, Chunduri V, Sharma S and Garg M (2019) Potential of Aegilops sp. for Improvement of Grain Processing and Nutritional Quality in Wheat (Triticum aestivum). Front. Plant Sci. 10:308. doi: 10.3389/fpls.2019.00308
Received: 30 August 2018; Accepted: 26 February 2019;
Published: 18 March 2019.
Edited by:
Peter Shewry, Rothamsted Research (BBSRC), United KingdomReviewed by:
Heinrich Grausgruber, University of Natural Resources and Life Sciences, Vienna, AustriaMarinus J. M. Smulders, Wageningen University & Research, Netherlands
Copyright © 2019 Kumar, Kapoor, Chunduri, Sharma and Garg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monika Garg, bWthamdhcmdAZ21haWwuY29t; bW9uaWthZ2FyZ0BuYWJpLnJlcy5pbg==; bWdhcmcxMDBAeWFob28uY29t
 Aman Kumar
Aman Kumar Payal Kapoor
Payal Kapoor Venkatesh Chunduri
Venkatesh Chunduri Saloni Sharma
Saloni Sharma Monika Garg
Monika Garg