- 1National Key Laboratory of Crop Genetic Improvement, National Center of Oil Crop Improvement (Wuhan), College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Crop Research Institute, Sichuan Academy of Agricultural Sciences, Chengdu, China
Novel Brassica napus cytoplasmic male sterility (CMS) with carpelloid stamens (inap CMS) was produced by intertribal somatic hybridization with Isatis indigotica (Chinese woad), but its RF (restorer of fertility) gene(s) existed in one particular woad chromosome that was carried by one fertile monosomic alien addition line (MAAL) of rapeseed. Herein, the selfed progenies of this MAAL were extensively selected and analyzed to screen the rapeseed-type plants (2n = 38) with good male fertility and to produce their doubled haploid (DH) lines by microspore culture. From the investigation of fertility restoration in the F1 hybrids with inap CMS, one DH line (RF 39) was identified to adequately restore male fertility and likely carried one dominant RF gene. Specifically, this restorer produced brown pollen grains, similar to the woad and the MAAL, suggesting that this trait is closely linked with the RF gene(s) and serves as one phenotypic marker for the restorer. This restorer contained 38 chromosomes of rapeseed and no intact chromosomes of woad, but some DNA fragments of woad origin were detected at low frequency. This restorer was much improved for pollen and seed fertility and for low glucosinolate content. The successful breeding of the restorer for inap CMS rendered this new pollination control system feasible for rapeseed hybrid production.
Introduction
Male sterility in plants can be caused either by mitochondrial genes coupled with nuclear genes or by nuclear genes alone, known as cytoplasmic male sterility (CMS) and genic male sterility (GMS), respectively (Chen and Liu, 2014). As the CMS trait is maternally inherited and easy to maintain (Hanson and Bentolila, 2004), a CMS fertility restoration system has been widely used as an excellent pollination control system to facilitate hybrid seed production for many crops, particularly for predominantly self-fertilized crops, including oilseed rape (Brassica napus L.), thus allowing breeders to realize yield gains resulting from the expression of heterosis.
Depending on the origin of the sterility inducing the cytoplasm, two types of CMS lines are usually distinguished: autoplasmy CMS, which arises spontaneously due to mutation in the genome of one species, and alloplasmy CMS, which occurs following cytoplasmic substitutions between two species due to nuclear-mitochondrial incompatibility (Prakash et al., 2009). The most popular autoplasmy CMS of rapeseed (B. napus L.) is polima CMS, which has been widely used worldwide during past decades (Fu et al., 1990) and is still dominant in hybrid seed production in China. As Brassica coenospecies provide rich and diverse mitochondrial genomes (Pradhan et al., 1992), alloplasmy CMS lines have been developed following backcrossing of either sexually synthesized allopolyploids or somatic hybrids between wild and crop species to diversify the CMS sources for breeding (For review see Prakash et al., 2009). Expectedly, CMS lines from sexual hybridizations maintain unaltered organellar genomes from cytoplasmic donor species because of exclusive maternal inheritance, but those from somatic hybrids likely contain a different cytoplasmic constitution because various possible combinations of mitochondrial and chloroplast genomes have been revealed in different CMS lines due to frequent organelle assortment and intergenomic mitochondrial recombinants in Brassicaceae (Prakash et al., 2009). The Ogura CMS found in a wild population of radish (Raphanus sativus) (Ogura, 1968) was introduced into Brassica species through interspecific crosses (Heyn, 1976; Bannerot et al., 1977) and subsequent protoplast fusion (Pelletier et al., 1983). Since then, a large number of alloplasmics have been produced, mostly for B. napus and B. juncea (Prakash et al., 2009). However, only Ogura-INRA B. napus CMS (Friedt et al., 2018) and Mori B. juncea CMS, produced from somatic fusion with Moricandia arvensis (Singh, 2008), were successfully exploited for commercial seed production on a large scale after systematic improvement of developmental abnormalities, including leaf chlorosis (Pelletier et al., 1983; Kirti et al., 1998).
The development of a fertility restorer for alloplasmic CMS is a challenging task because its fertility restoration gene(s) has been shown to exist in one particular chromosome of the nuclear genome of the cytoplasm donor species (Prakash et al., 1998; Bang et al., 2002; Leino et al., 2004; Budahn et al., 2008; Wei et al., 2010; Kang et al., 2014), owing to the co-evolutionary relationship between the nucleus and cytoplasm (Fujii et al., 2011; Chen and Liu, 2014). The breeding process for restorer lines involves the introgression of the alien chromosomal segments carrying the restorer of fertility (RF, or restorer) gene(s) and selection of cytologically stable lines with the same chromosome number as the recipient species and with good agronomic value. Theoretically, these Rf genes could be introgressed into the recipient species by taking advantage of non-homologous allosyndetic recombination in backcrossing or selfing generations of sexual or somatic hybrids. The genetic improvement of the introgression lines, which meet the breeding requirements, is demonstrated to be very time and effort consuming, especially when a large amount of alien element is introduced and associated with linkage drag, leading to deleterious characters, such as poor agronomic performance (Delourme et al., 1998; Giancola et al., 2003).
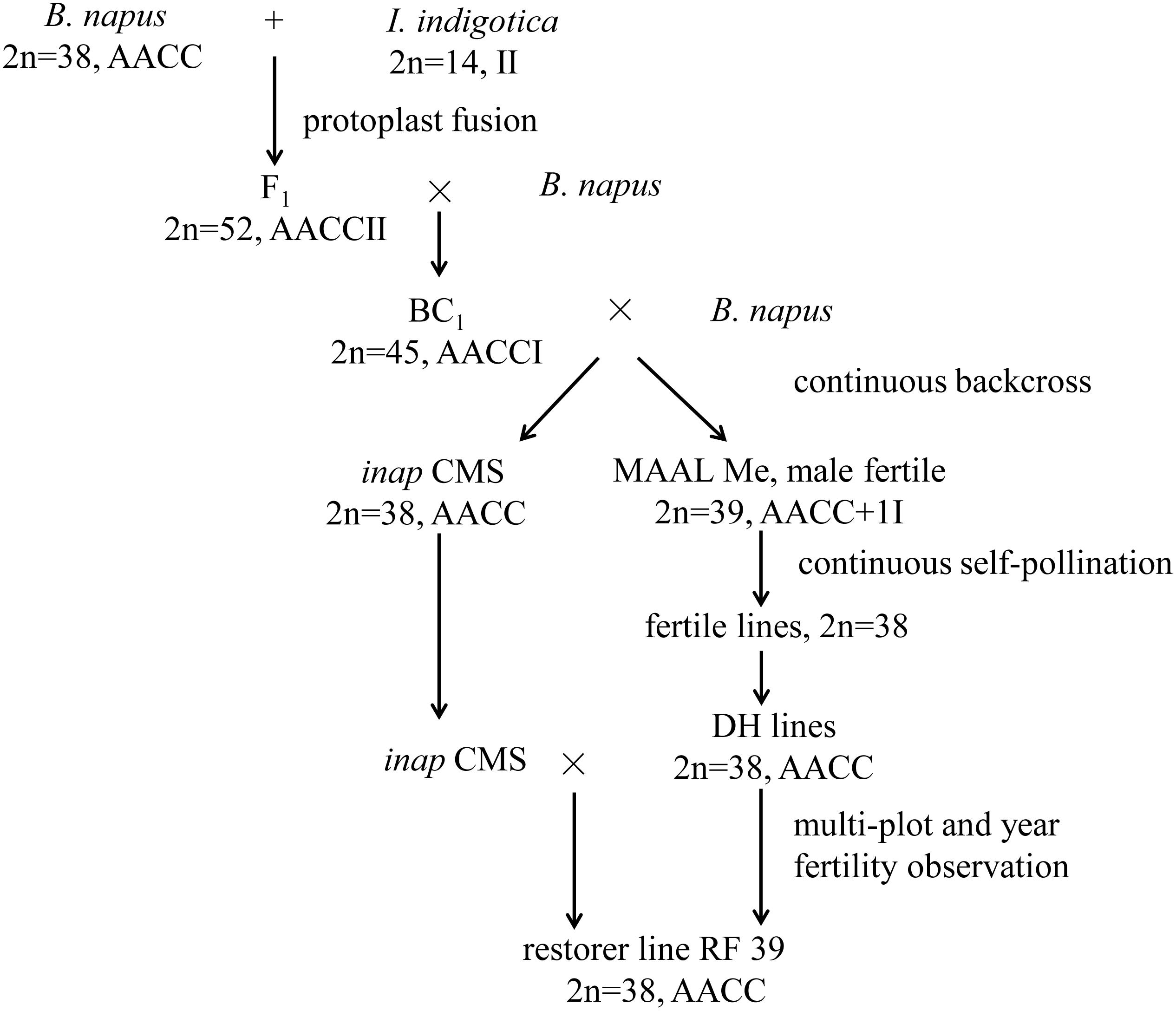
In our previous study, the intertribal somatic hybrids (2n = 52, AACCII) between B. napus (2n = 38, AACC) and Isatis indigotica Fort. (Chinese woad; 2n = 14, II) of the Isatideae tribe within the Brassicaceae family were obtained (Du et al., 2009) and backcrossed continuously to B. napus, resulting in the development of one novel B. napus CMS line with carpelloid stamens (named inap CMS) (Kang et al., 2017; Figure 1). The inap CMS contained the majority of mitochondrial genes from woad and one recombinant of two species. No fertility restorer for inap CMS was found among the restorers of other autoplasmic and alloplasmic CMS systems and among a large number of B. napus genotypes with extensive resources. Alternatively, a complete set of monosomic alien addition lines (MAALs) of B. napus, with one of seven woad chromosomes and with the same cytoplasm as inap CMS (artificially designated as Ma-Mg), was also established and characterized among the backcrossing progenies of the somatic hybrids (Figure 1; Kang et al., 2014). Only MAAL Me produced normal anthers with a brown color, a trait of woad origin, which contained viable pollen grains, but six other MAALs had carpelloid stamens, as did inap CMS. This suggested that this woad chromosome harbored the gene(s) that should rectify the feminized development of the stamens and restore the male fertility of inap CMS. Then the restorer line with the B. napus-like chromosome complement could probably be selected among the selfed progenies of Me when the chromosomal segment, related to floral development, was introgressed via certain pathways. In the present study, extensive selections and investigations of phenotype, male fertility, cytology, and genomic composition were conducted for the progenies of Me with the aims to breed a restorer line for inap CMS and to improve its fertility and agronomic performance.
Materials and Methods
Plant Materials
As only one B. napus-I. indigotica MAAL (Me) among all seven MAALs (in the presence of the sterility-inducing cytoplasm of inap CMS) had normal flowers with brown anthers, as did I. indigotica (Kang et al., 2014), its progenies by self-pollination from three generations were screened for plants with high pollen fertility and with the same 2n = 38 as B. napus during 2012–2014 in Wuhan, Hubei Province and Xining, Qinghai Province. Individuals with normal stamens were selected for microspore culture in the spring of 2014. Subsequently, doubled haploid (DH) plants with normal stamens and high pollen fertility were chosen for testcrossing with inap CMS in the spring of 2015. Then, DH lines and their F1 hybrids were planted together with the maintainer line B. napus cv. Huashuang 3 (H3) with double-low quality and the inap CMS line in Wuhan, Chengdu, Changsha (winter-type growing regions) and Xining (spring-type region) for fertility evaluation from 2015 to 2018. One restorer selected among DH lines was crossed with thirty rapeseed cultivars of wide origins to improve its fertility and agronomic performance.
Microspore Culture
To obtain pure lines of the selfed progenies from Me, a microspore culture regenerative technique was performed in March 2014 according to Lichter (1982) with minor modifications. Flower buds, all with a length of 2.5–4 mm, were picked out from the main inflorescence of fertile individuals between 8:00 and 10:00 AM to extract microspores. The isolated microspores were resuspended in 10 ml of NLN-16 medium with the addition of 60 mg l-1 colchicine at 32°C in darkness for 53 h and then incubated at 25°C in darkness until cotyledonous embryoids appeared. The plants generated from microspore culture were transplanted in the field in October 2014.
Plant Phenotype and Seed Quality
The maintainer, restorer, and F1 hybrid were compared for the following parameters: flower organs, including pistil length, flower diameter, sepal length, long stamen length, short stamen length, petal length, and petal width, by measuring 10 fully opened flowers on the same sunny day for each line. Their flowering and maturing times were counted from planting in 2016–2017 and 2017–2018 in Wuhan. At maturity, 10 representative plants of these lines were observed for agronomic traits. For the fatty acid compositions, mature seeds were measured by using near infra-red spectroscopy (NIRS) (Foss NIRS Systems 5000, KWS UK). The glucosinolate (GSL) profiles of the maintainer, restorer, and woad with two replicates were analyzed by the Institute of Oil Crops Research, Chinese Academy of Agricultural Sciences, Wuhan, with high-performance liquid chromatography (HPLC, YLSB077, 1200).
Cytology and Pollen Viability Analysis
To observe the chromosome numbers and meiosis of fertility restored plants and testcross progenies, young flower buds were collected in the morning. For mitosis, the ovaries were treated with 2 mM 8-hydroxyquinoline solution for 3 h at room temperature before being fixed in Carnoy’s solution I (3:1 ethanol:glacial acetic acid, v/v) and stored at -20°C for further chromosome counting of somatic cells. For meiosis, the young flower buds were fixed directly in Carnoy’s solution I and stored at -20°C. Cytogenetic observations were carried out as described by Li et al. (1995). Pollen grains of each plant were stained with acetocarmine (1%) and more than 300 were counted; the percentage of stainable pollen grains was calculated to evaluate pollen fertility.
Probe Labeling and GISH (Genomic in situ Hybridization) Analysis
Total genomic DNA was extracted from young leaves as described by Dellaporta et al. (1983). Genomic DNA of I. indigotica was labeled with Bio-11-dUTP (Fermentas) by random priming using the BioPrime DNA Labeling System kit (Invitrogen, Life Technologies). Total genomic DNA of B. napus was boiled twice, 15 min each time, to obtain DNA fragments of 100–500 bp that were used as blocks.
Chromosomes for GISH were prepared as described by Ge and Li (2007), and in situ hybridization was carried out according to Zhu et al. (2016). Pictures were taken using a fluorescence microscope (Axio Scope A1, Zeiss, Germany) with a CCD camera. Images were processed by Adobe Photoshop CS5 to adjust contrast and brightness.
Genomic Analysis
To identify the I. indigotica DNA segments introgressed into the restorer lines, 21 SSR markers specific for MAAL Me identified by Kang et al. (2014) and one centromeric marker were used. PCR amplifications were performed in a volume of 10 μl containing 20 ng genomic DNA, 1×Taq buffer, 2 mM MgCl2, 0.2 U Taq DNA polymerase (Fermentas), 2 mM dNTPs (Fermentas), 5 μM forward and reverse primers. The PCR program was as follows: 94°C for 5 min; 10 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 45 s, with a 0.5°C decrease in annealing temperature in each cycle; 30 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension at 72°C for 10 min. PCR products were separated on 1% agarose gels.
AFLP analysis was performed on the maintainer (Huashuang 3), restorer, inap CMS, woad, additional line Me, and F1 plants following the procedures of Vos et al. (1995), with slight modifications. Total purified genomic DNA (50 ng) was digested by the restriction endonucleases EcoRI and MseI 6 h at 37°C. Inactivation was carried out at 65°C for 1 h, followed by ligation to EcoRI and MseI adapters at 4°C overnight. The resulting ligation products were then amplified by pre-selective PCR with primers matching the adapters. The pre-selective PCR products were used as templates for selective PCR as follows: 94°C for 3 min; 13 cycles at 94°C for 30 s, 65°C for 45 s, 72°C for 1 min, with a 0.7°C decrease in annealing temperature in each cycle; 26 cycles at 94°C for 30 s, 56°C for 45 s, 72°C for 1 min, and a final extension at 72°C for 5 min. Amplification products were separated on 6% denaturing polyacrylamide gels, and photographs were taken.
Statistical Data Analysis
The statistical significance of the data was calculated, including a t-test, an LSD test, and a χ2 test, using R statistical software.
Results
Development of a Restorer for inap CMS
Theoretically, MAAL Me, with the male sterility-inducing cytoplasm, should produce three types of progenies after selfing pollination: monosomic and disomic additions, both with small anthers containing viable brown pollen grains, and B. napus plants with carpelloid stamens, if the additional chromosome remained intact. These three types of progeny plants were easy to distinguish from their specific floral phenotypes but were not targeted to select the restorer. As the flowers of MAAL Me plants had anthers with a much smaller size than the B. napus recipient cultivar H3 (Kang et al., 2014), artificial supplementary pollination was performed to obtain more seeds. Thousands of seeds were harvested but some were found to have germinated in pods prior to maturity, probably resulting from development disturbance by alien chromosome or cytoplasm. Subsequently, these seeds were sown during three growing seasons in Wuhan and Xining to screen the individuals with male fertility and 2n = 38. In total, 60 plants were primarily selected by their larger flowers and larger anthers, compared with MAAL Me, but showed variable seed-sets from low to high, likely because they had shorter and thinner stamens with low pollen fertility compared to the maintainer line (H3). Certain parts of the seeds from these plants germinated in pods.
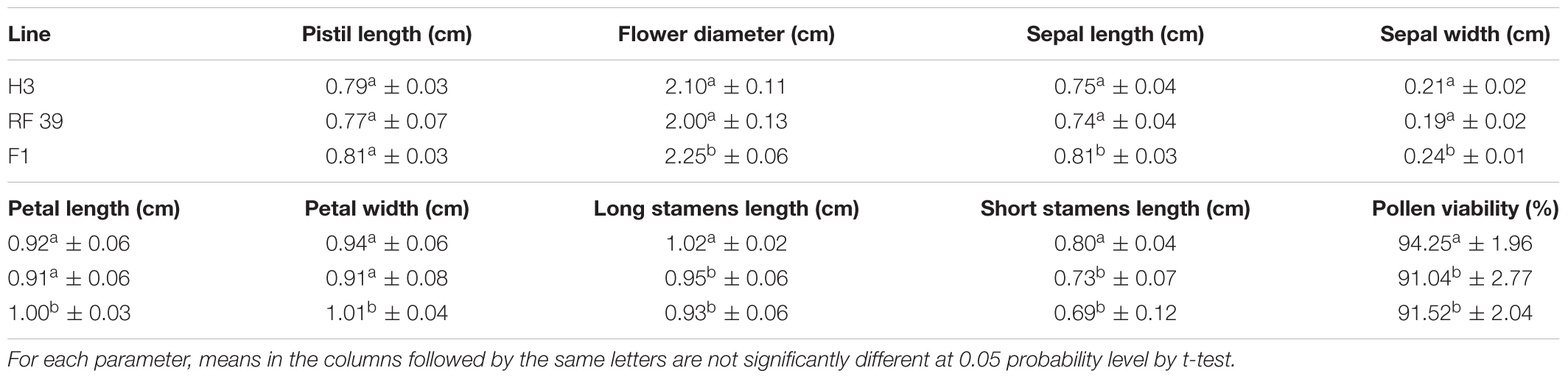
To stabilize these fertile progenies of the potential restorer, microspore culture was performed for 60 screened strains and 625 plants were obtained; in total, 192 DH plants, originating from all of the parental lines, were established and investigated for male development. While most of these DH plants (140/192) exhibited carpelloid stamens, 52 developed flowers with different numbers of stamens, mostly with large tetradynamous stamens but 2 tiny stamens, and only a few with 6 well-developed stamens. Although the stamens of these DH plants produced some stainable pollen grains, they contained obviously fewer pollen grains than B. napus and also showed poorer dehiscence. Therefore, the seed sets varied greatly for these plants. F1 hybrids between 52 DH plants and the inap CMS line were produced in spring 2015 and were observed for fertility restoration in four locations (Wuhan, Chengdu, Changsha and Xining) from 2015 to 2018. These 52 hybrids showed different degrees of male fertility by producing flowers with variable numbers of anthers (1–6) or a mixture of normal and carpelloid stamens. Only hybrids with DH plant No. 39 developed six normal stamens and consistently had good seed sets at four locations over the years. Plant No. 39 and its hybrids had high pollen stainability (91.04 and 91.52%, respectively) (Table 1), and produced long siliques with good seed sets after being bagged for self-pollination (Figures 2H,I). However, the seed number per pod was 11.8 for Plant No. 39 and 16.4 for the hybrid, lower than 19.5 for the maintainer H3. The phenomenon of the seed germination in pods prior to maturity was not observed for Plant No. 39 and its hybrid. Therefore, DH Line 39 had the ability to rectify the feminized development of the stamens in inap CMS and to restore male fertility in their F1 hybrids, which served as a restorer line and was designated as RF 39.

Figure 2. Flowers and inflorescences of inap CMS and restorer (RF 39). (A) One flower of the B. napus maintainer with yellow anthers and pollen grains. (B) One flower of I. indigotica with small anthers containing brown pollen grains. (C) One flower of inap CMS with carpelloid anthers (left) and the same flower with only carpelloid anthers and pistil (right). With the increase of backcrossing generations to the maintainer, the size of the carpelloid anthers reduces and their feminized structure degenerates. (D,E) One flower of RF 39 and F1 hybrid with some sepals and petals removed, respectively, to show the six well developed anthers. Notice the brownish yellow color of the pollen grains. (F) The comparison of the stamens from I. indigotica, B. napus, RF 39 and F1 (left to right) with the focus on the different colors of the pollen grains. Bars: (A–F) 1 cm. (G) Sterile and fertile inflorescences of the hybrid plants after inap CMS and RF 39 are pollinated by the maintainer as male and female, respectively. (H,I) Mature bagged inflorescences of RF 39 and F1 hybrid with good seed-sets, respectively. Bars: (G–I) 20 cm.
Phenotype of Restorer RF 39
With respect to the floral organs, RF 39 with the same cytoplasm as inap CMS-produced flowers with a slightly smaller size than those of the maintainer (Table 1), but its flowers were distinguishable by some curled petals, instead of the very smooth petals observed in the maintainer (Figures 2A,D). The four long stamens and two short stamens were shorter than those of the maintainer, were less plump and released few pollen grains due to poorer dehiscence (Figure 2D). In particular, the pollen grains of RF 39 and F1 hybrids were brown, similarly to those of Chinese woad and MAAL Me, while those of the maintainer were yellow (Figures 2A,B,D–F). The brown pollen color trait, which was associated with anther development on the woad chromosome, was dominant over the yellow pollen of B. napus in the somatic hybrids (Du et al., 2009), MAAL Me (Kang et al., 2014) and the introgressant restorer (present study). This revealed that the gene (s) responsible for the brown pollen color was (were) closely linked with the RF gene(s) and provided one phenotypic marker for the restorer line.
The whole plant architecture of RF 39 with the sterility-inducing cytoplasm was similar to that of the inap CMS but it was smaller than that of the maintainer (data unshown), except for the different floral organs and male fertilities (Figures 2A,C,D). It showed an obviously poorer seed set under selfing or open pollination than the maintainer, as its pods had fewer seeds (11.8 vs. 19.5). This most likely resulted from the negative effect of the sterility-inducing cytoplasm, as its hybrid with the maintainer as the female showed normal seed fertility (Figure 2G). Notably, the plants of RF 39 flowered almost 2 weeks later than the maintainer Huashuang 3 when sown in the autumn in Wuhan, similarly to the inap CMS (Kang et al., 2017). The delayed development of inap CMS and RF 39 might result from the detrimental cytoplasmic effect related to aberrant mitochondrial functions.
Seed Quality of Restorer RF 39
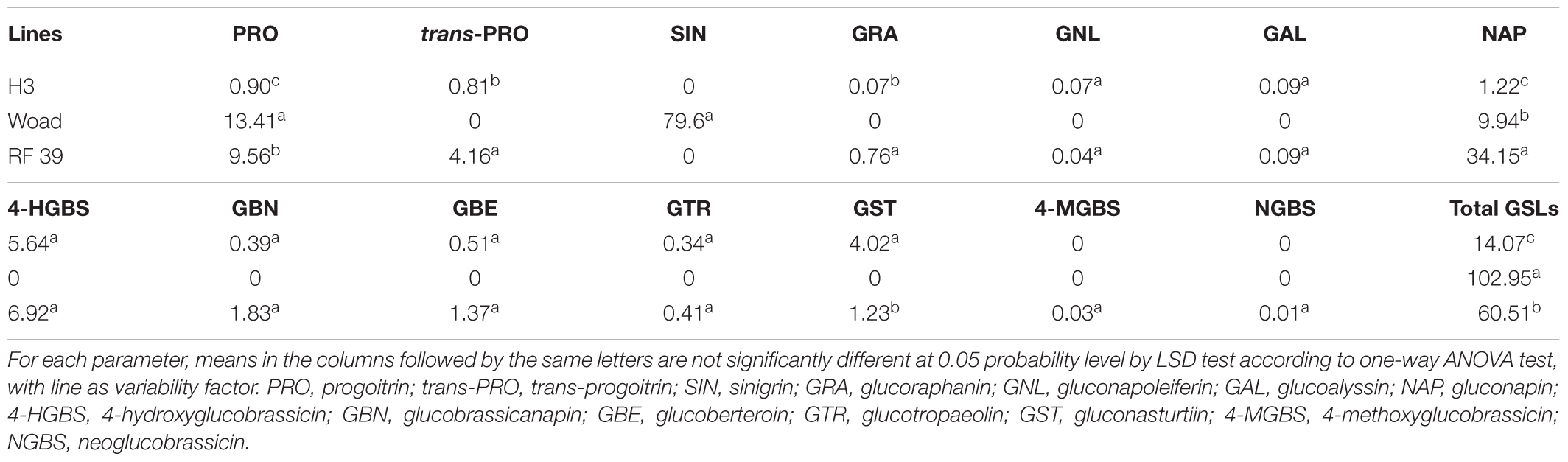
The seeds of I. indigotica had much higher glucosinolate (GSL) content (102.95 μmol/g) than those of the B. napus maintainer (14.07 μmol/g) and also quite different compositions of GSLs (Table 2), as they contained only three kinds of GSLs (sinigrin, SIN; progoitrin, PRO; gluconapin, NAP), and SIN was the predominant component (77%). In the seeds of the maintainer with a profile of 11 GSLs, 4-hydroxyglucobrassicin (4HGBS) (40%) and gluconasturtiin (GST) (29%) were dominant, but the others appeared at very low percentages. The major SIN in the woad was absent in the maintainer, and the main 4HGBS and GST in the maintainer were not detected in the woad. The seeds of RF 39 had a higher GSL content (60.51 μmol/g) than those of the maintainer and mainly contained NAP (56%), PRO (16%), and 4HGBS (11%), while their types of GSLs were very similar to each other. Intriguingly, NAP (34.15 μmol/g, 56%) was the overwhelming component among 13 GSLs of RF 39, while its content (1.22 μmol/g, 9%) was lower in the maintainer, but it was the third most common (9.94 μmol/g, 10%) in the woad. The rate of PRO in RF 39 (16%) was higher than that in the maintainer (6%) and the woad (13%), but the rate of 4HGBS (11%) was much lower than that in the maintainer (40%). RF 39 did not produce the predominant SIN in the woad, in contrast to the maintainer. Therefore, compared to the maintainer, the increase in the GSL content in RF 39 was largely attributable to the enhanced contents of NAP (34.15 vs. 1.22) and PRO (9.56 vs. 0.9).
However, the fatty acid compositions of RF 39 showed no obvious differences from those of the maintainer, and a low erucic acid content (∼1%) was maintained, but those of the woad included 45% erucic acid.
Chromosomal and Genomic Complements of Restorer RF 39
Chromosome number and the genomic complement of RF 39 were determined in mitotic and meiotic cells by conventional and molecular cytogenetic methods. In the somatic hybrids between B. napus, I. indigotica and MAALs, the chromosomes from I. indigotica were characterized by their much smaller sizes than those of B. napus (Du et al., 2009; Kang et al., 2014). In MAAL Me, one alien chromosome of woad origin was clearly identified to be of small size by GISH analysis with its labeled genomic DNA as a probe (Figure 3A). In some pollen mother cells at the meiotic zygotene/pachytene stage, this woad chromosome was associated with the other from B. napus (Figure 3B), likely indicating homoeologous pairing and exchange, and providing some explanation for the DNA introgressions detected. All of the observed somatic and meiotic cells of RF 39 included only 38 chromosomes, originating from B. napus, and no intact chromosome or chromosomal segment from I. indigotica, as no obvious signals of the woad probe were even detected on these chromosomes (Figures 3C–E). These chromosomes were paired as 19 bivalents at diakinesis and segregated equally as 19:19 at anaphase I (Figures 3D,E). No laggards occurred during meiosis I and II. Regular meiosis ensured the formation of pollen grains with high viability.
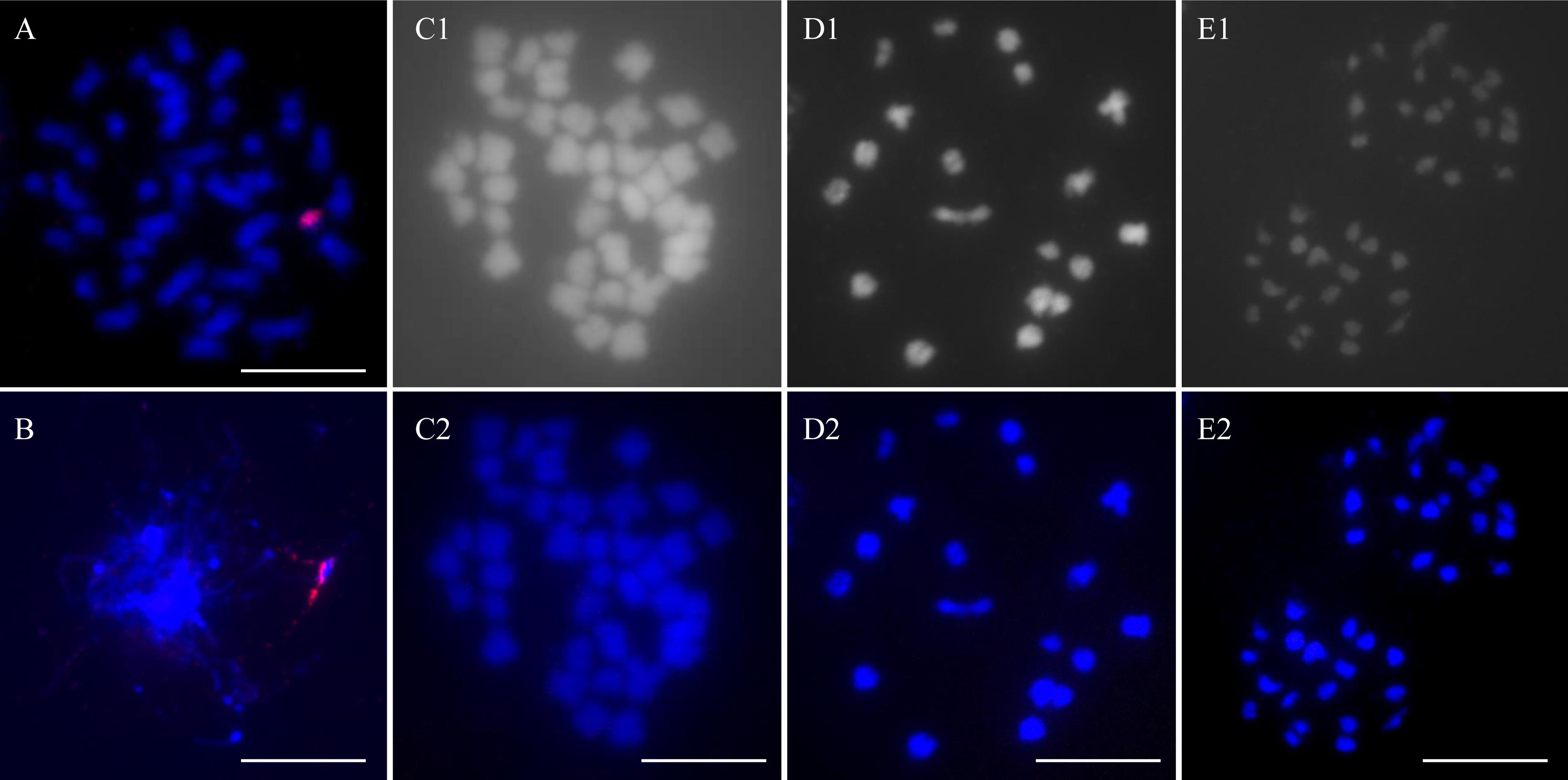
Figure 3. GISH analysis of the alien additional line and restorer. Blue color is from the DAPI staining of chromosomes. Red color is from the signal of the labeled genomic probe of I. indigotica. (A) One mitotic ovary cell of MAAL Me with one chromosome from I. indigotica (red). (B) One PMC of MAAL Me at meiotic pachytene with homoeologous association between the additional woad chromosome (red) and the other of rapeseed. (C1,C2) One mitotic ovary cell of RF 39 with only 38 unlabeled chromosomes from B. napus but no signals of I. indigotica on these chromosomes. (D1,D2) One diakinesis PMC of RF 39 with 19 bivalents unlabeled by the I. indigotica probe. (E1,E2) One PMC at anaphase of RF 39 with equal 19:19 segregation and with no labeled chromosomes in each polar group by the I. indigotica probe. Arrows indicate alien chromosomes. Bars: 10 μm.
The woad-specific SSR markers were allocated on each chromosome using our serial MAALs (Kang et al., 2014). Among those for MAAL Me, 21 SSR markers and 1 centromeric marker showed polymorphisms between the maintainer, woad, MAAL Me, RF 39, F1, and inap CMS. Their DNA bands were amplified only in woad and MAAL Me (Figures 4A,B), also revealing no existence of intact chromosome e in RF 39. From AFLP analysis with 256 primer combinations, only EA6/MC13, EA8/MC12, EA15/MC6, and EA15/MC12 amplified polymorphic bands in woad, MAAL Me, RF 39, and F1 but not in the maintainer and inap CMS (Figure 4C), showing the DNA introgression from I. indigotica into the genome of RF 39. Unfortunately, we failed to convert these four markers into SCAR markers. The majority of the consensus bands shared by the maintainer, RF 39, and inap CMS demonstrated the high similarities of their genomic constitutions.
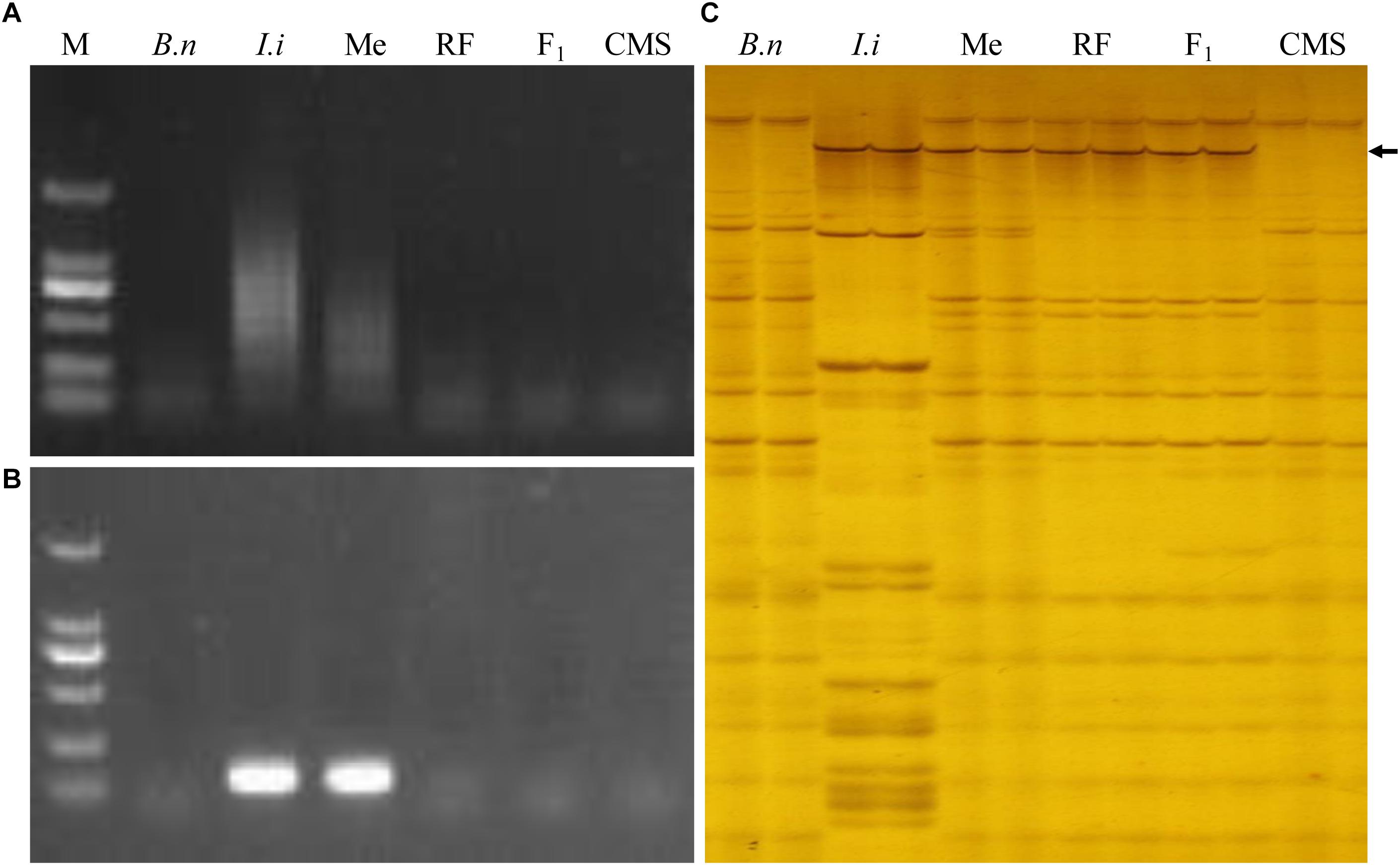
Figure 4. Molecular analysis for genomic complements of the restorer. (A) The PCR profile by the woad centromere specific marker with amplified bands only in woad and MAAL Me. (B) The woad chromosome specific marker SSR79 amplifies the bands only in woad and MAAL Me. (C) AFLP profile from marker EA15/MC6 with amplified bands in woad, MAAL Me, RF 39 and F1 (black arrow) but not in rapeseed. B. n represents for B. napus, I. i for I. indigotica, Me for MAAL Me, RF for RF 39, F1 for hybrid of RF 39 and inap CMS, CMS for inap CMS. M DL2000.
Genetic Control of Fertility Restoration
In one F2 population with 820 plants derived from the inap CMS × RF 39 F1 hybrid, which was male fertile, the 3:1 segregation ratio was presented by the male fertile plants (619) and male sterile plants (201) (χ2 = 0.08, p < 0.01), indicating that the Rf gene of RF 39 was a single dominant gene. Furthermore, all the plants in this F2 population, which were male fertile, had a brown pollen color, demonstrating that the gene(s) controlling brown pollen color was closely linked to the Rf gene of RF 39.
Genetic Improvement of Restorer RF 39
As the restorer RF 39 exhibited some limitations, such as the less plump anthers with fewer pollen grains, lower seed fertility than the maintainer and the increased GSL content, it was pollinated by 30 rapeseed cultivars of different origins, and their hybrids and selfed progenies were selected for pollen and seed fertility, and for double-low seed qualities and agronomic traits. Three lines were identified to produce plumper anthers with abundant pollen grains and to show the seed sets comparable to the maintainer, as well as the reduced GSL content (<30 μmol/g).
Discussion
As CMS can be associated with disease susceptibility, the diversification of cytoplasmic origin is beneficial for crop breeding and is more pressing for rapeseed in China, as no other efficient CMS systems other than the predominant polima CMS and shan 2A CMS, both with CMS gene orf224, have been developed in about 30 years. The polima CMS has been replaced by the “Ogu-INRA” CMS system and other systems in Europe and Canada (Friedt et al., 2018), while the M. arvensis-based CMS and fertility restoration system have also been widely adopted for B. juncea in India (Singh, 2008). From these two successful models of development of synthetic alloplasmic CMS and restorers, it is crucial to realize the introgression of Rf gene(s) via MAAL as the breeding bridge, where the additional chromosome carries the Rf gene(s), by selecting the target plants resulting from the homoeologous recombination or chromosomal translocation (Prakash et al., 1998; present study). In spite of the very distant relationship between rapeseed and Chinese woad from two tribes, the genetic introgression from the additional woad chromosome of MAAL Me into the rapeseed genome was obviously ascertained from the selection of the restorer line, which was also coupled with the woad trait of brown pollen grains. The presence of sterility-inducing cytoplasm in the fertile MAAL should help us to identify the target plants, as those without the introgressed RF segment were sterile (Prakash et al., 1998) and were easily identified by their carpelloid stamens in the present study. The frequency of introgression with the Rf gene seemed to be very low, as only a few plants with good fertility restoration were identified among a large progeny population.
Even after the successful development of primary restorers for alloplasmic CMS, the male or female fertilities of these restorer lines and other negative traits need further improvements to satisfy the production of commercial hybrids, due to the genetic linkage on the introgression region or the disturbance of genomic structure by the integration of an alien element. Although a single radish nuclear gene, Rfo, restored Ogura CMS in B. napus by altering the translational expression of orf138 (Bellaoui et al., 1999), the large size of the radish introgression of at least 50 cM carrying the Rfo locus led to a poor restorer with deleterious characters, including an increased GSL content (Delourme et al., 1991, 1995). Together with extensive backcrossing and pedigree breeding, molecular markers closely linked to the Rfo gene assisted with the selection of the restorer lines with shorter radish fragments and better agronomic performance (Delourme et al., 1991, 1995, 1998; Giancola et al., 2003; Primard-Brisset et al., 2005). In particular, a low GSL restorer line, R2000, which was developed through gamma ray irradiation, deleted radish introgression but recovered some B. oleracea sequences that were originally replaced by the introgression (Primard-Brisset et al., 2005; Hu et al., 2008). After the rectification of chlorosis in CMS (M. arvensis) B. juncea, genetic improvements were made for the parents in 1999–2005, before the release of the hybrids, including the elite NRCHB506 (Singh, 2008; Lei et al., 2018).
Of interest, the increased GSL content also appeared in our restorer for inap CMS, compared with the maintainer (Table 2), as observed in the restorer of Ogura CMS B. napus (Delourme et al., 1995; Primard-Brisset et al., 2005). Although the major GSL component SIN in woad was absent in the restorer RF 39, the extensive increase in NAP from 1.22 μmol/g in the maintainer to 34.15 μmol/g in RF 39 was most likely caused by the genetic introgression or the following interaction, as the woad showed a higher NAP content (9.94 μmol/g). The increased content of PRO from 0.9 μmol/g in the maintainer to 9.56 μmol/g in RF 39 likely resulted from the alien genetic element, as the woad presented the highest PRO content (13.41 μmol/g). The changed GSL profile of RF 39 was probably due to the disturbance of complex GSL biosynthesis, transport, and degradation pathways by alien introgression (Lu et al., 2018). The close association between fertility restoration and the GSL content in the radish introgression was quite difficult to break, while the woad introgression resulted in a looser association that could be more easily modified for the reduced GSL content. This difference was likely not only related to the fact that the B. napus donor and the alien introgression restorer did not produce the major GSL type of SIN in the woad but also suggested that the RF gene(s) in these two cases were linked with different genes controlling GSL synthesis. This further reflected the structural difference in the genomes of radish and woad, as woad has a much simpler and smaller genome (Lysak and Koch, 2011) than radish (Kitashiba et al., 2014; Jeong et al., 2016), and its genome is about two times greater than that of Arabidopsis thaliana (Lysak et al., 2009) or nearly one half of that of Brassica diploids. The genomic sequences of the radish and B. rapa showed that no significant difference existed in the gene contents related to GSL synthesis (Jeong et al., 2016), except that the radish had fewer myrosinase and thioglucoside glucohydrolase1 genes in the GSL breakdown pathway (Jeong et al., 2016). Specifically, the radish genome had intermediate characteristics between the Brassica A/C and B genomes in the triplicated segments, and its nine chromosomes were rearranged from the chromosomes of the hexaploid progenitor, suggesting an internal origin from the genus Brassica (Jeong et al., 2016). However, the Isatideae tribe had a rearranged proto-Calepine karyotype (PCK, n = 7) with a single copy of all 24 blocks derived from an ancestral cruciferous karyotype (ACK, n = 8) (Lysak and Koch, 2011). Since the genetic mapping of the RF gene for our restorer line and the development of the linked molecular markers lagged behind, it is now impossible to figure out the genomic region of the RF locus and the size of the woad fragment introduced. The ongoing genome sequencing of the Chinese woad genotype used in the present study will provide new opportunities to unravel the genetic structure of the restorer harboring the certain alien fragment and to elucidate the reasons for the linkage with the glucosinolate content, as well as the poor anther dehiscence and brown pollen color.
Conclusion
In conclusion, the restorer for inap CMS of rapeseed was capable of restoring its male fertility by converting the carpelloid stamens into normal stamens with viable pollen grains. The alien introgression of RF gene was visibly accompanied by the expression of the brown pollen trait specific to one woad chromosome. The restorer was improved for seed fertility, low glucosinolate content. Thus, a new alloplasmic CMS/fertility restoration system was established for hybrid seed production in B. napus, after Ogura CMS system in B. napus and Mor CMS system in B. juncea.
Ethics Statement
The authors declare that the experiments complied with current laws of the country in which they were performed.
Author Contributions
ZL and LK conceived the experiments. PL performed the research. AW, SG, and XG contributed to phenotypic measurements. CC, LJ, and ZL contributed to restorer improvement. PL and ZL wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Funding
This work was supported by Grants from NSFC (No. 31571701) and National Key Research and Development Program of China (Grant No. 2016YFD0100202).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bang, S. W., Kaneko, Y., Matsuzawa, Y., and Bang, K. S. (2002). Breeding of Moricandia arvensis monosomic chromosome addition lines (2n = 19) of alloplasmic (M. arvensis) Raphanus sativus. Breed. Sci. 52, 193–199. doi: 10.1270/jsbbs.52.193
Bannerot, H., Boulidard, L., and Chupeau, Y. (1977). Unexpected difficulties met with the radish cytoplasm. Eucarpia Cruciferae Newslett. 2:16.
Bellaoui, M., Grelon, M., Pelletier, G., and Budar, F. (1999). The restorer Rfo gene acts post-translationally on the stability of the ORF138 Ogura CMS-associated protein in reproductive tissues of rapeseed cybrids. Plant Mol. Biol. 40, 893–902. doi: 10.1023/A:1006223908044
Budahn, H., Schrader, O., and Peterka, H. (2008). Development of a complete set of disomic rape-radish chromosome-addition lines. Euphytica 162, 117–128. doi: 10.1007/s10681-007-9609-x
Chen, L. T., and Liu, Y. G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Dellaporta, S. L., Wood, J., and Hicks, J. B. (1983). A plant DNA mini preparation: version II. Plant Mol. Biol. Rep. 1, 19–21. doi: 10.1007/BF02712670
Delourme, R., Eber, F., and Renard, M. (1991). “Radish cytoplasmic male sterility in rapeseed: breeding restorer lines with a good female fertility,” in Proceedings of the 8th International Rapeseed Conference, (Saskatoon, SK), 1506–1510.
Delourme, R., Eber, F., and Renard, M. (1995). “Breeding double low restorer lines in radish cytoplasmic male sterility of rapeseed (Brassica napus L.),” in Proceedings of the 9th International Rapeseed Conference, (Cambridge), 6–8.
Delourme, R., Foisset, N., Horvais, R., Barret, P., Champagne, G., Cheung, W. Y., et al. (1998). Characterisation of the radish introgression carrying the Rfo restorer gene for the Ogu-INRA cytoplasmic male sterility in rapeseed (Brassica napus L.). Theor. Appl. Genet. 97, 129–134. doi: 10.1007/s00122-005-0876
Du, X. Z., Ge, X. H., Yao, X. C., Zhao, Z. G., and Li, Z. Y. (2009). Production and cytogenetic characterization of intertribal somatic hybrids between Brassica napus and Isatis indigotica and backcross progenies. Plant Cell Rep. 28, 1105–1113. doi: 10.1007/s00299-009-0712-4
Friedt, W., Tu, J. X., and Fu, T. D. (2018). “Academic and economic importance of Brassica napus rapeseed,” in The Brassica napus Genome, eds S. Y. Liu, S. Rod, and B. Chalhoub (Basel: Springer Nature Switzerland AG), 1–20.
Fu, T. D., Yang, G. S., and Yang, X. N. (1990). Studies on “three line” polima cytoplasmic male sterility developed in Brassica napus L. Plant Breed. 104, 115–120. doi: 10.1111/j.1439-0523.1990.tb00412.x
Fujii, S., Bond, C. S., and Small, I. D. (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 1723–1728. doi: 10.1073/pnas.1007667108
Ge, X. H., and Li, Z. Y. (2007). Intra- and intergenomic homology of B-genome chromosomes in trigenomic combinations of the cultivated Brassica species revealed by GISH analysis. Chromosome Res. 15, 849–861. doi: 10.1007/s10577-007-1168-4
Giancola, S., Marhadour, S., Desloire, S., Clouet, V., Falentin-Guyomarc’h, H., Laloui, W., et al. (2003). Characterization of a radish introgression carrying the ogura fertility restorer gene Rfo in rapeseed, using the Arabidopsis genome sequence and radish genetic mapping. Theor. Appl. Genet. 107, 1442–1451. doi: 10.1007/s00122-003-1381-2
Hanson, M., and Bentolila, S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, 154–170. doi: 10.1105/tpc.015966.Indeed
Heyn, F. W. (1976). Transfer of restorer genes from Raphanus to cytoplasmic male sterile Brassica napus. Cruciferae Newslett. 1, 15–16.
Hu, X. Y., Sullivan-Gilbert, M., Kubik, T., Danielson, J., Hnatiuk, N., Marchione, W., et al. (2008). Mapping of the ogura fertility restorer gene Rfo and development of Rfo allele-specific markers in canola (Brassica napus L.). Mol. Breed. 22, 663–674. doi: 10.1007/s11032-008-9207-1
Jeong, Y. M., Kim, N., Ahn, B. O., Oh, M., Chung, W. H., Chung, H., et al. (2016). Elucidating the triplicated ancestral genome structure of radish based on chromosome-level comparison with the Brassica genomes. Theor. Appl. Genet. 129, 1357–1372. doi: 10.1007/s00122-016-2708-0
Kang, L., Du, X. Z., Zhou, Y. Y., Zhu, B., Ge, X. H., and Li, Z. Y. (2014). Development of a complete set of monosomic alien addition lines between Brassica napus and Isatis indigotica (Chinese woad). Plant Cell Rep. 33, 1355–1364. doi: 10.1007/s00299-014-1621-8
Kang, L., Li, P. F., Wang, A. F., Ge, X. H., and Li, Z. Y. (2017). A novel cytoplasmic male sterility in Brassica napus (inap CMS) with carpelloid stamens via protoplast fusion with Chinese woad. Front. Plant Sci. 8:529. doi: 10.3389/fpls.2017.00529
Kirti, P. B., Prakash, S., Gaikwad, K., Bhat, S. R., Dineshkumar, V., and Chopra, V. L. (1998). Chloroplast substitution overcomes leaf chlorosis in Moricandia arvensis based cytoplasmic male sterile Brassica juncea. Theor. Appl. Genet. 97, 1179–1182. doi: 10.1007/s001220051007
Kitashiba, H., Li, F., Hirakawa, H., Kawanabe, T., Zou, Z., Hasegawa, Y., et al. (2014). Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 21, 481–490. doi: 10.1093/dnares/dsu014
Lei, S. L., Chen, L., Yang, W. Z., Guo, G. M., Qin, X. R., Zhang, X. X., et al. (2018). Moricandia arvensis cytoplasmic male sterile system in Brassica juncea. Chin. J. Oil Crop Sci. 40, 297–301.
Leino, M., Thyselius, S., Landgren, M., and Glimelius, K. (2004). Arabidopsis thaliana chromosome III restores fertility in a cytoplasmic male-sterile Brassica napus line with A. thaliana mitochondrial DNA. Theor. Appl. Genet. 109, 272–279. doi: 10.1007/s00122-004-1644-6
Li, Z. Y., Liu, H. L., and Luo, P. (1995). Production and cytogenetics of ntergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor. Appl. Genet. 91, 131–136. doi: 10.1007/BF00220869
Lichter, R. (1982). Induction of haploid plants fromisolated pollen of Brassica napus. Plant Breed. 105, 427–434. doi: 10.1016/S0044-328X(82)80040-8
Lu, K., Snowdon, R., and Li, J. N. (2018). “Case study for trait-related gene evolution: glucosinolates,” in The Brassica napus Genome, eds S. Y. Liu, S. Rod, and B. Chalhoub (Basel: Springer Nature Switzerland AG), 198–222.
Lysak, M. A., and Koch, M. A. (2011). “Phylogeny, genome, and karyotype evolution of crucifers (Brassicaceae),” in Genetics and Genomics of the Brassicaceae, eds R. Schmidt and I. Bancroft (New York, NY: Springer Science+Business Media), 1–32. doi: 10.1007/978-1-4419-7118-0_1
Lysak, M. A., Koch, M. A., Beaulieu, J. M., Meister, A., and Leitch, I. J. (2009). The dynamic ups and downs of genome size evolution in Brassicaceae. Mol. Biol. Evol. 26, 85–98. doi: 10.1093/molbev/msn223
Ogura, H. (1968). Studies on the new male-sterility in Japanese radish, with special reference to the utilization of this sterility towerds the practical raising of hybrid seeds. Mem. Fac. Agric. Kagoshima Univ. 6, 39–78.
Pelletier, G., Primard, C., Vedel, F., Che’trit, P., Re’my, R., Rousselle, P., et al. (1983). Intergeneric cytoplasmic hybridization in Cruciferae by protoplast fusion. Mol. Gen. Genet. 191, 244–250. doi: 10.1007/BF00334821
Pradhan, S. K., Prakash, S., Mukhopadhyay, A., and Pental, D. (1992). Phylogeny of Brassica and allied genera based on variation in chloroplast and mitochondrial DNA patterns: molecular and taxonomical classifications are incongruous. Theor. Appl. Genet. 85, 331–340. doi: 10.1007/BF00222878
Prakash, S., Bhat, S. R., Quiros, C. F., Kirti, P. B., and Chopra, V. L. (2009). Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31, 21–187. doi: 10.1002/9780470593783.ch2
Prakash, S., Kirti, P. B., Bhat, S. R., Gaikwad, K., Kumar, V. D., and Chopra, V. L. (1998). A Moricandia arvensis - based cytoplasmic male sterility and fertility restoration system in Brassica juncea. Theor. Appl. Genet. 97, 488–492. doi: 10.1007/s001220050921
Primard-Brisset, C., Poupard, J. P., Horvais, R., Eber, F., Pelletier, G., Remard, M., et al. (2005). A new recombined double low restorer line for the Ogu-INRA cms in rapeseed (Brassica napus L.). Theor. Appl. Genet. 111, 736–746. doi: 10.1007/s00122-005-2059-8
Singh, K. H. (2008). Hybrid development in mustard (Brassica juncea L.), a success story. Sarson News 12, 2–3.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Wei, W. H., Li, Y. C., Wang, L. J., Liu, S. Y., Yan, X. H., Mei, D. S., et al. (2010). Development of a novel sinapis arvensis disomic addition line in Brassica napus containing the restorer gene for Nsa CMS and improved resistance to Sclerotinia sclerotiorum and pod shattering. Theor. Appl. Genet. 120, 1089–1097. doi: 10.1007/s00122-009-1236-6
Keywords: Brassica napus, cytoplasmic male sterility, restorer of fertility, nuclear-mitochondrial incompatibility, monosomic alien addition line
Citation: Li P, Kang L, Wang A, Cui C, Jiang L, Guo S, Ge X and Li Z (2019) Development of a Fertility Restorer for inap CMS (Isatis indigotica) Brassica napus Through Genetic Introgression of One Alien Addition. Front. Plant Sci. 10:257. doi: 10.3389/fpls.2019.00257
Received: 11 December 2018; Accepted: 18 February 2019;
Published: 05 March 2019.
Edited by:
Guijun Yan, The University of Western Australia, AustraliaReviewed by:
Zhongyun Piao, Shenyang Agricultural University, ChinaXiaodong Wang, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2019 Li, Kang, Wang, Cui, Jiang, Guo, Ge and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Kang, kang5371@sina.com Zaiyun Li, lizaiyun@mail.hzau.edu.cn
†Present address: Lei Kang, College of Agronomy, Hunan Agricultural University, Changsha, China
 Pengfei Li
Pengfei Li Lei Kang
Lei Kang Aifan Wang
Aifan Wang Cheng Cui
Cheng Cui Liangcai Jiang
Liangcai Jiang Shizhen Guo
Shizhen Guo Xianhong Ge
Xianhong Ge Zaiyun Li
Zaiyun Li