
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 March 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00253
Part of this article's content has been mentioned in:
EjTFL1 Genes Promote Growth but Inhibit Flower Bud Differentiation in Loquat
The MADS-box transcription factor SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) integrates environmental and endogenous signals to promote flowering in Arabidopsis. However, the role of SOC1 homologs in regulating flowering time in fruit trees remains unclear. To better understand the molecular mechanism of flowering regulation in loquat (Eriobotrya japonica Lindl.), two SOC1 homologs (EjSOC1-1 and EjSOC1-2) were identified and characterized in this work. Sequence analysis showed that EjSOC1-1 and EjSOC1-2 have conserved MADS-box and K-box domains. EjSOC1-1 and EjSOC1-2 were clearly expressed in vegetative organs, and high expression was detected in flower buds. As observed in paraffin-embedded sections, expression of the downstream flowering genes EjAP1s and EjLFYs started to increase at the end of June, a time when flower bud differentiation occurs. Additionally, high expression of EjSOC1-1 and EjSOC1-2 began 10 days earlier than that of EjAP1s and EjLFYs in shoot apical meristem (SAM). EjSOC1-1 and EjSOC1-2 were inhibited by short-day (SD) conditions and exogenous GA3, and flower bud differentiation did not occur after these treatments. EjSOC1-1 and EjSOC1-2 were found to be localized to the nucleus. Moreover, ectopic overexpression of EjSOC1-1 and EjSOC1-2 in wild-type Arabidopsis promoted early flowering, and overexpression of both was able to rescue the late flowering phenotype of the soc1-2 mutant. In conclusion, the results suggest that cultivated loquat flower bud differentiation in southern China begins in late June to early July and that EjSOC1-1 and EjSOC1-2 participate in the induction of flower initiation. These findings provide new insight into the artificial regulation of flowering time in fruit trees.
Plant evolution has resulted in a variety of endogenous and exogenous factors that form a complex and sophisticated regulatory network to accurately respond to internal and external signals and integrate them to promote blooming at the most favorable time. The molecular genetic mechanisms at play in annual flowering plants, such as Arabidopsis thaliana, involve multiple regulatory pathways, including photoperiod, age, autonomic, vernalization, and gibberellin pathways (Moon et al., 2005; Amasino, 2010; Srikanth and Schmid, 2011; Teotia and Tang, 2015). These pathways precisely regulate flowering in Arabidopsis through major integrated genes such as FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1).
MADS-box genes are a key components of flower development networks. In addition to the MADS-box domain, MIKCC-type MADS-box genes contain three other domains, the I-domain, K-box and C-terminal domain; although the MADS-box is highly conserved, the degree of conservation of the I-domain and C-domain is relatively low (Theissen et al., 1996; Parenicova et al., 2003; Vandenbussche et al., 2003; Smaczniak et al., 2012; Chen et al., 2017). SOC1 is a member of the MIKCC-type gene family and encodes a type II MADS-box protein that contains the highly conserved MADS-box, K-box, and a C-terminal SOC1 motif (Vandenbussche et al., 2003). SOC1 plays a vital role in regulating plant development and flower organogenesis by integrating photoperiod, age, and gibberellin signals (Parcy, 2005; Lee and Lee, 2010; Teotia and Tang, 2015).
SOC1 is also found in other plants, such Oryza sativa (Tadege et al., 2003), Petunia hybrida (Ferrario et al., 2004), Citrus sinensis (Tan and Swain, 2007), Glycine max (Zhong et al., 2012), Fragaria vesca (Mouhu et al., 2013), Zea mays (Zhao et al., 2014), Brassica juncea (Sri et al., 2015), Actinidia spp. (Voogd et al., 2015), Kalanchoe daigremontiana (Liu et al., 2016), and Mangifera indica L. (Wei et al., 2016). SOC1 not only promotes flowering but also regulates other biological functions, such as floral organ identity deterioration in Gerbera hybrid (Ruokolainen et al., 2011), repression of flowering and promotion of vegetative growth in F. vesca (Mouhu et al., 2013), and dormancy duration in kiwifruit (Voogd et al., 2015). SOC1 function can vary among different plant species, though the function of EjSOC1 in loquat has not been studied.
Loquat (Eriobotrya japonica Lindl.) is an evergreen fruit tree belonging to the family Rosaceae that is cultivated mainly in tropical and subtropical regions. In Rosaceae, flower initiation and flowering typically occur in different years in species including apple, pear, plum, strawberry, and raspberry (Kurokura et al., 2013). However, flower bud initiation and flowering occur within the same year in loquat, with the former generally occurring from July to September in China (Lin, 2007) and the latter mainly from October to January; there is also slight variability depending on the cultivar and environment. To date, 26 Eriobotrya species have been identified, and each wild species has a different flowering time that includes the months of November to June of the next year for some (Lin, 2017). For example, cultivated loquat (E. japonica Lindl.) blooms in fall or early winter, whereas E. deflexa Nakai blooms from May to June (Gu and Spongberg, 2003).
Although the flowering of loquat has the above characteristics, there have been few reports on it. To date, several flower-related genes, such as EjAP1 (APETALA1), EjFT, EjLFY, and EjTFL1 (TERMINAL FLOWER1) (Esumi et al., 2005; Liu et al., 2013, 2017; Reig et al., 2017), have been cloned from cultivated loquat, with EdFT and EdFD (FLOWERING LOCUS D) cloned from wild loquat (E. deflexa Nakai forma koshunensis) (Zhang L. et al., 2016).
In this study, the flower initiation time of cultivated loquat (“Jiefangzhong”) in Southern China was confirmed. Two SOC1-like genes from cultivated loquat were identified and named EjSOC1-1 and EjSOC1-2. To elucidate their roles in regulating flowering time in loquat, their expression patterns and subcellular localizations were analyzed. In addition, we examined their function using transgenic Arabidopsis and explored the effects of short-day (SD) and GA3 treatments on bud differentiation.
Material was collected from 12-year-old “Jiefangzhong” loquat (E. japonica Lindl.) trees grown under natural conditions in the loquat germplasm resource preservation garden, South China Agricultural University, Guangzhou, China (N23°09′N,113°20′E). The trees used in the experiments were grafted, and they had grown to the flowering stage. Leaf and shoot apical meristem (SAM) tissues were randomly sampled from three sites on the trees, and tissues was collected at 16:00. Wild-type A. thaliana ecotype Col-0 and the soc1-2 mutant were used for genetic transformation. Nicotiana benthamiana was grown for transient expression. Arabidopsis and Nicotiana were grown under long-day conditions (16 h light/8 h dark) at 22°C.
Frozen mature loquat leaves or other tissues were ground to a powder in a mortar with liquid nitrogen. Total RNA was extracted using EasySpin Plus (Aidlab, China) and digested with recombinant RNase-free DNase I (Aidlab, China). First-strand cDNA was generated from loquat leaf RNA using the PrimeScriptTM RT (TAKARA, Japan) reagent kit and gDNA Eraser (TAKARA, Japan), the experiment was proceeded according to the manufacturer’s instructions.
The full-length coding sequences of EjSOC1-1 and EjSOC1-2 were obtained from the completed loquat de novo genome sequencing project, which has not yet been published. The two sequences were isolated from mature loquat leaf cDNA using Phusion DNA Polymerase (TAKARA, Japan). The gene-specific primers used for cloning were listed in Supplementary Table S1. Alignment of the deduced protein sequences was performed using ClustalX 2.0.12 and GeneDoc 2.7. Phylogenetic trees were constructed with MEGA 6.06 using the Neighbor-Joining (N-J) method with 1,000 bootstrap replicates.
Primers for qPCR were designed using Primer 5 software, and their specificity was confirmed by melting curve analysis and sequencing. qPCR was carried out in triplicate using a LightCycler® 480 system (Roche, United States) with iTaqTM universal SYBR Green Supermix (Bio-Rad, United States). The relative expression levels of target genes were evaluated using the ΔΔCt (cycle threshold) method. β-Actin was used as an internal reference gene for loquat (Shan et al., 2008). AtPP2AA3 (AT1G13320) was used as an internal control for Arabidopsis (Hong et al., 2010). Semi-quantitative reverse transcription PCR (RT-PCR) was used for detecting exogenous gene expression in transgenic Arabidopsis lines. The primers used for RT-PCR were identical to the cloning primers (removal of the stop codon). The primers used are listed in Supplementary Table S2.
For construction of 35S:EjSOC1-1/EjSOC1-2-6HA and 35S:EjSOC1-1/EjSOC1-2-GFP plasmids, coding regions without the stop codon were cloned into pGreen-35S-6HA (Hou et al., 2014) and pGreen-35S-GFP (Lee et al., 2012), respectively. All primers used for vector construction are listed in Supplementary Table S3. The constructed plasmids were verified by sequencing and introduced into Agrobacterium tumefaciens strain GV3101::psoup.
35S:EjSOC1-1-HA and EjSOC1-2-HA were introduced into Agrobacterium tumefaciens GV3101::psoup and then transformed into Arabidopsis Col-0 using the floral dip method (Zhang et al., 2006). Transgenic lines were screened on soil by Basta. For each construct, more than 10 independent transgenic lines were screened out, and two homozygous T3 generation lines of each genotype were used for checking ectopic gene expression.
An awning (Supplementary Figure S1) was set up to cover the tree to provide 8 h (10:00–18:00) of natural light and 16 h of darkness (18:00–10:00 [the next day]) each day. Control trees were grown under normal conditions. The experimental period lasted from May 18th to August 10th.
For GA3 treatment, trees were sprayed with 300 mg L-1 GA3 (Guangzhou DingGuo Biology Company, China) aqueous solution containing 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100 as a surfactant. Control trees were sprayed with a solution containing only 0.1% (v/v) phosphoric acid and 0.025% (v/v) Triton X-100. All leaves and top buds were sprayed every 2 weeks from May 18th to August 10th.
Agrobacterium-mediated transient transformation of N. benthamiana leaves (Sparkes et al., 2006) was used to observe the subcellular localization of EjSOC1-1 and EjSOC1-2. Green fluorescent protein (GFP) fluorescence signals were detected using a fluorescence microscope Observer D1 (Zeiss, Germany). A GFP-free construct was used as a negative control.
Significant differences between data were evaluated by Student’s t-test. Calculations were carried out using GraphPad Prism 6 software.
Continuous year-round observation of loquat SAM development was conducted. The panicle of “Jiefangzhong” loquat in Guangzhou was clearly visible from the end of August to early September (Figures 1A,B). In addition, analysis of paraffin-embedded sections of the SAM from June to September revealed no obvious inflorescence primordium formation in apical tissue before June 23rd, with multiple inflorescence primordia in the bottom of panicle appearing on July 7th (Figure 1A).
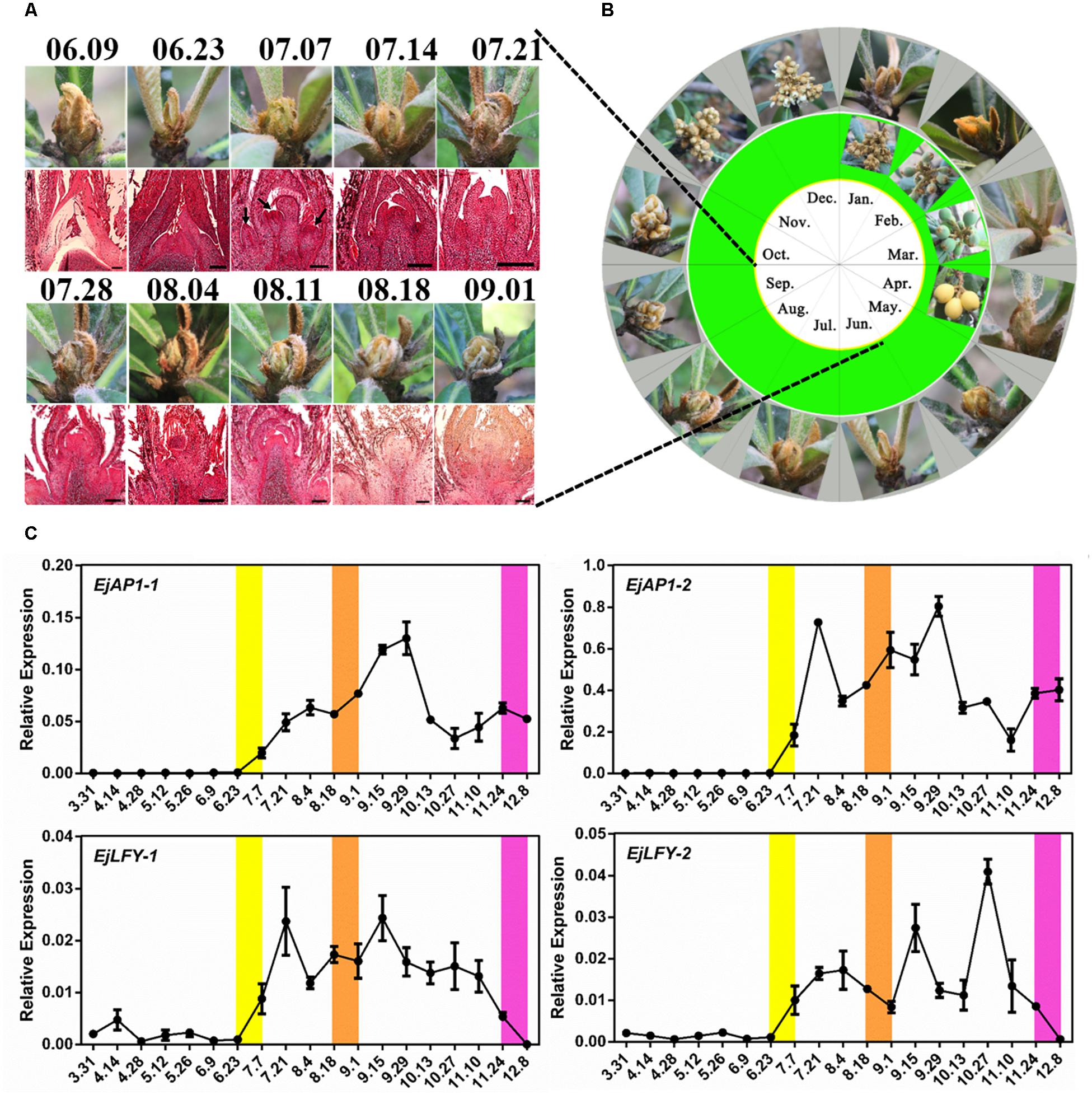
Figure 1. Determination of the flowering time of “Jiefangzhong” loquat. (A) The morphology of the shoot apex and its paraffin sections from June to September, with a scale of 200 μm. (B) The morphology of the shoot apex of loquat throughout the year. (C) Relative expression levels of the floral meristem identity genes EjAP1s and EjLFYs in the shoot apex (error bars indicating SD from three biological replicates). The yellow background represents the critical period of flower bud differentiation, and the orange background represents the period in which obvious inflorescence can be seen; the purple background represents the period of flower opening.
Furthermore, the expression levels of the floral meristem identity genes EjAP1-1, EjAP1-2, EjLFY-1, and EjLFY-2 at different developmental stages of apical tissues were analyzed by qPCR. The results showed a high level of expression for both EjAP1s and EjLFYs began on June 23rd that was maintained from July to September (Figure 1C), except for EjLFY-2, which maintained a peak until November. AP1 and LFY determine flower meristem characteristics and are key genes for flower induction and morphology (Lohmann et al., 2001). These results indicate that “Jiefangzhong” loquat flower bud differentiation in Guangzhou begins in late June to early July.
We cloned two genes homologous to SOC1, EjSOC1-1 and EjSOC1-2, using unpublished loquat genome sequence data. EjSOC1-1 and EjSOC1-2 CDSs are 642 and 648 bp and encode 213 and 215 amino acids, respectively (Supplementary Figure S2). Their sequences are highly similar, with nucleotide sequence identity of 93.36%. The predicted protein amino acid sequences of EjSOC1-1 and EjSOC1-2 are similar to those of other SOC1 orthologs from apple, soybean, rapeseed and Arabidopsis (Figure 2A). Sequence analysis showed that EjSOC1-1 and EjSOC1-2 harbor highly conserved MADS-box, K-box and SOC1-motif domains (Figure 2A).
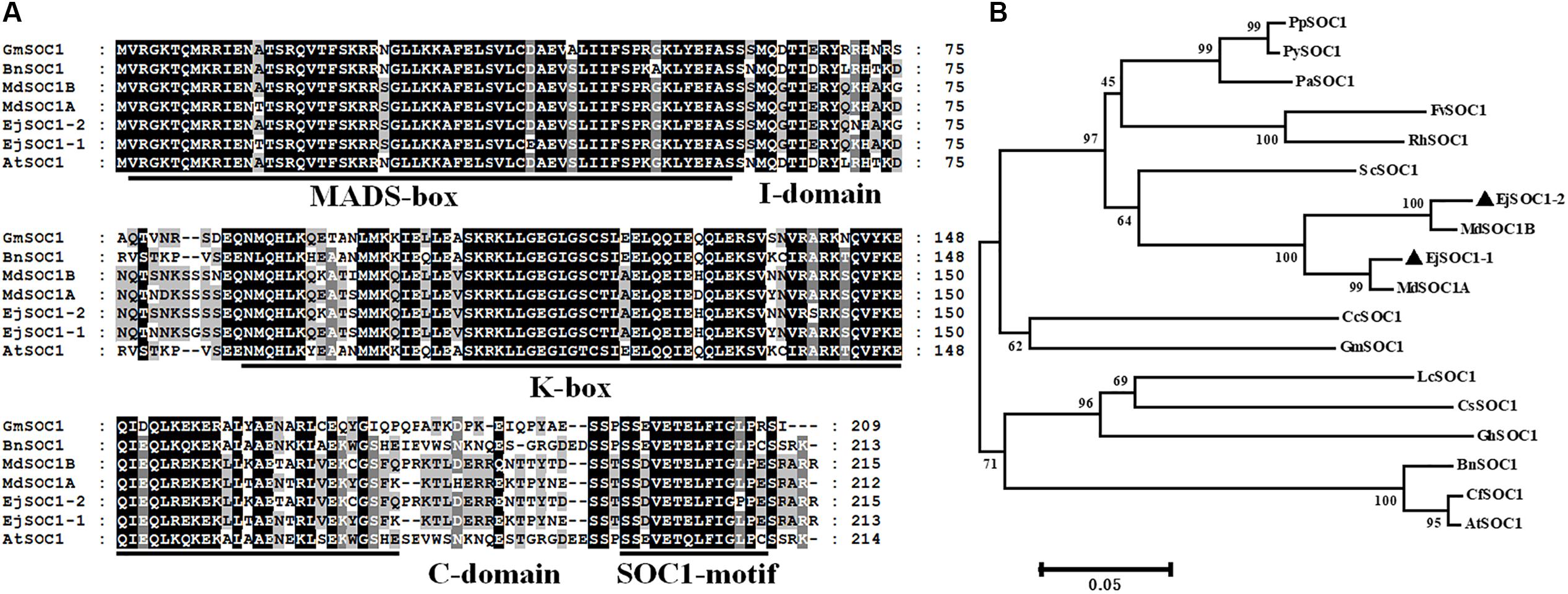
Figure 2. Sequence and phylogenetic analyses of EjSOC1-1 and EjSOC1-2. (A) Amino acid sequence alignment of several plant SOC1 family proteins. The highly conserved MADS-box, K-box, and SOC1-motif domains are marked by a solid line. The I-domain is located behind the MADS-box, and the C-domain is located behind the K-box. (B) Phylogenetic analysis of SOC1 family proteins from different species. The protein sequences of SOC1 genes aligned in this study were retrieved from NCBI. Accession IDs: AtSOC1 (NP_182090.1), BnSOC1 (NP_001303107.1), CcSOC1 (AHI85950.1), CsSOC1 (NP_001275772.1), CfSOC1 (AGN29205.1), FvSOC1 (AEO20231.1), GhSOC1 (AEA29618.1), GmSOC1 (NP_001236377.1), LcSOC1 (AGS32267.1), MdSOC1A (BAI49494.1), MdSOC1B (BAI49495.1), PaSOC1 (ACO40488.1), PpSOC1 (AJW29024.1), PySOC1 (AEO20233.1), and RhSOC1 (AEO20230.1).
Based on phylogenetic analysis of EjSOC1s and other plant SOC1 sequences, EjSOC1s and the other SOC1s from Rosaceae grouped into a large clade, with apple sequences forming a small clade with a high genetic relationship to the large clade (Figure 2B). EjSOC1s show the highest sequence similarity to MdSOC1 homologs (97.18% identity for EjSOC1-1 and MdSOC1A and 97.21% identity for EjSOC1-2 and MdSOC1B) (Figure 2A). These results confirm that EjSOC1-1 and EjSOC1-2 are MADS-box genes and SOC1 orthologs in loquat.
To understand the potential function of EjSOC1-1 and EjSOC1-2 in loquat, we employed qPCR to examine the expression patterns of EjSOC1-1 and EjSOC1-2 in various tissues of “Jiefangzhong” loquat, including roots (from rootstock), leaves, shoots, leaf buds, flower buds, flowers, and fruits (Figure 3A). EjSOC1-1 and EjSOC1-2 were mainly expressed in vegetative organs, and for both, the highest expression was observed in flower buds. In particular, the expression level of EjSOC1s in early flower buds was significantly higher than that in blooming flowers, with scant expression in fruits (Figure 3B). These results suggest that EjSOC1s participate in the development of vegetative organs and flower initiation.
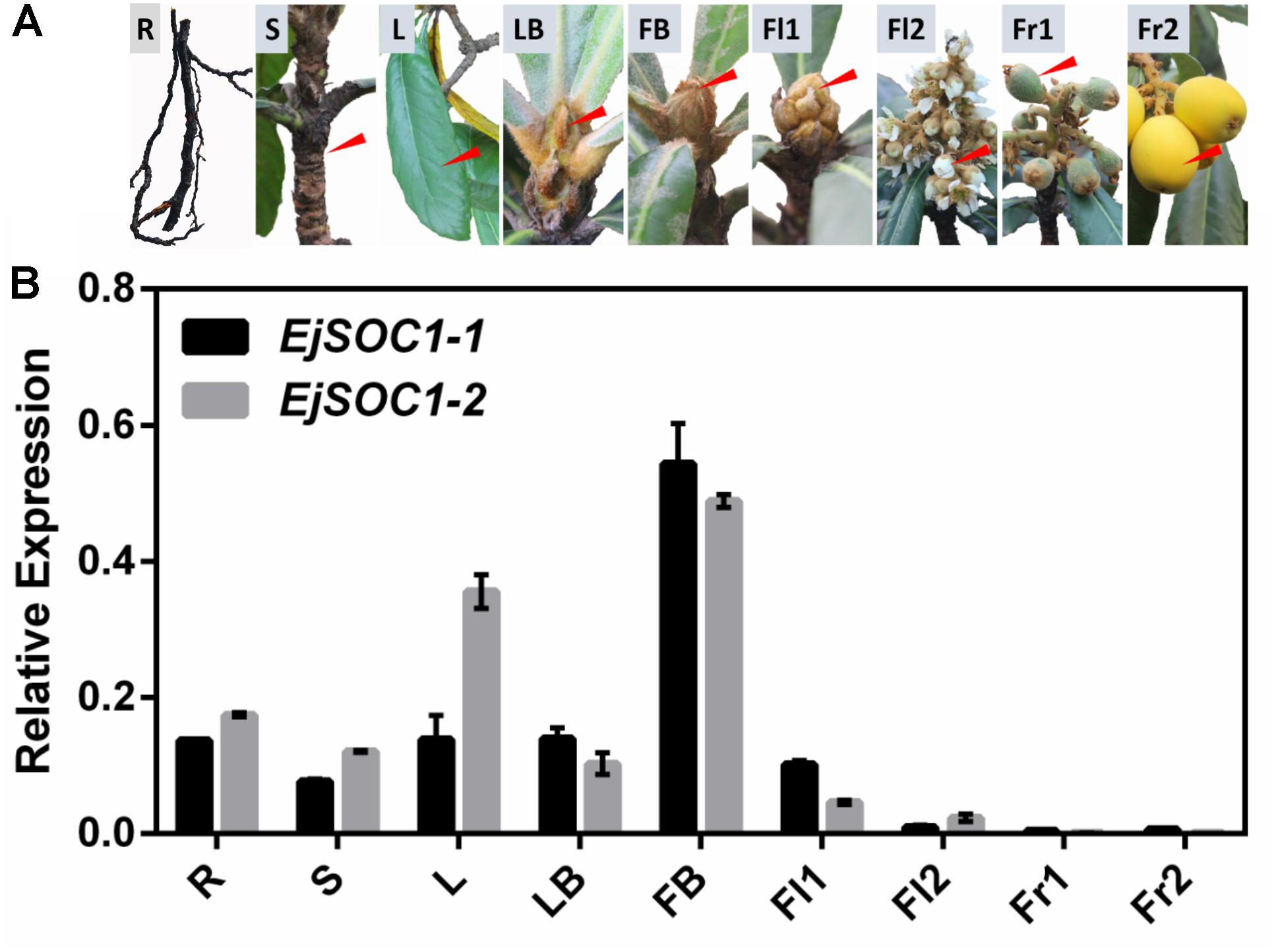
Figure 3. Tissue-specific expression of EjSOC1-1 and EjSOC1-2. (A) Different tissues from loquat (Eriobotrya japonica Lindl.) were analyzed. (B) Relative expression of EjSOC1s in different tissues shown in panel (A) (error bars indicating SD from three biological replicates). The β-actin gene served as an internal control. R, root (from rootstock); L, leaf; S, shoot; LB, leaf buds (April 28th); FB, flower bud (July 21st); Fl1, flower 1 (September 29th); Fl2, flower 2 (December 8th); Fr1, fruit 1 (February 2nd) and Fr2, fruit 2 (April 13th).
To further investigate the functions of EjSOC1-1 and EjSOC1-2 during vegetative and reproductive developmental processes, we examined their expression at different developmental stages of leaves, buds and flowers, as well as leaves of different maturities in the same period (Supplementary Figure S3), using qPCR.
In leaves, the tendency of EjSOC1-1 expression was similar to that of EjSOC1-2: their expression began to increase on June 23rd and reached the highest level by July 14th (Figure 4A). In addition, there was no significant difference in expression between the genes in mesophyll tissue or in veins (Figure 4C).

Figure 4. Relative expression patterns of EjSOC1-1 and EjSOC1-2. (A) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in different developmental stages of leaves. (B) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in leaves at different levels of maturity in the same period. (C) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in mesophyll tissue and veins. (D) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in the SAM in different developmental periods. (E) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in inflorescences at different developmental stages. (F) Relative expression patterns of EjSOC1-1 and EjSOC1-2 in different flower parts. Error bars indicating SD from three biological replicates.
With regard to the SAM in different periods, the expression levels of EjSOC1-1 and EjSOC1-2 began to increase sharply on June 23rd and reached the highest level around July 7th (Figure 4D). Moreover, expression of these genes gradually decreased as flower bud development progressed (Figure 4E).
For different flower parts (Supplementary Figure S4), EjSOC1-1 and EjSOC1-2 showed relatively high expression levels in receptacles, but only EjSOC1-2 was highly expressed in pistils. Little expression of either was found in petals and stamens (Figure 4F).
In summary, EjSOC1-1 and EjSOC1-2 may function to induce flowering and are involved in the growth and development of early flower organs in loquat.
Interestingly, we found that EjSOC1-1 and EjSOC1-2 exhibited different expression trends in leaves at different levels of maturity in the same period. EjSOC1-1 showed high expression in L3, L4 and L5 but relatively low expression in L1, L6, and L7 (Figure 4B), though EjSOC1-2 did not display this trend. The results indicate that EjSOC1-1 might also be involved in leaf development.
After we analyzed the possible roles of EjSOC1-1 and EjSOC1-2 in loquat flowering, further exploration of the function of EjSOC1s under SD and exogenous GA3 was proceeded. SOC1 can integrate the photoperiod and gibberellin pathways to regulate flower bud differentiation in Arabidopsis, and the loquat flower bud differentiation time coincides with the longest day of the year (summer solstice). Thus, we designed two experiments to alter growth conditions to explore whether EjSOC1s are affected by photoperiod and gibberellin. Interestingly, EjSOC1-1 and EjSOC1-2 were affected by SD and GA3 treatments, with lower levels of expression during the critical period of flower bud differentiation (late June and early July). EjSOC1-1 and EjSOC1-2 were abundantly expressed in the normal growth group and the control group at the end of June and early July (Figure 5A). More importantly, the SD-treated and GA3-treated loquat trees did not produce visible inflorescences in September compared to the trees in the normal growth group and the control group (Figure 5B). Furthermore, according to qPCR, the floral meristem identity genes EjAP1-1 and EjAP1-2 were hardly expressed (Figure 5A), as well as EjLFY-1 and EjLFY-2 were consistently expressed at a low level (Figure 5A). Based on the above results, we conclude that EjSOC1-1 and EjSOC1-2 are regulated by photoperiod and GA3 and that flower bud differentiation does not occur under SD conditions or after GA3 exposure. The results suggest that EjSOC1s can initiate flower bud differentiation by integrating photoperiod and gibberellin signaling.
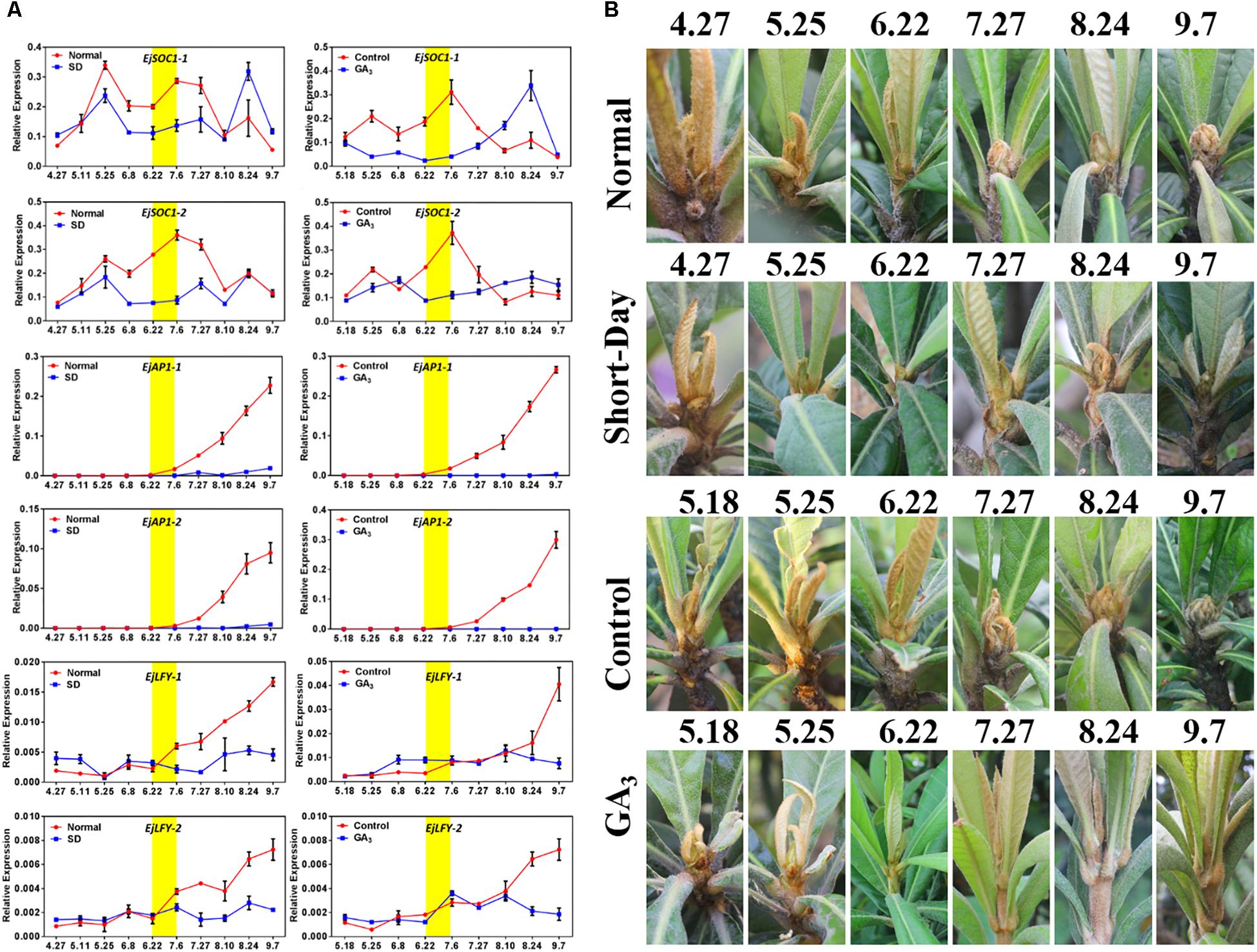
Figure 5. Effects of short-day and exogenous GA3 treatments on flowering. (A) Relative expression levels of EjSOC1s, EjAP1s, and EjLFYs in the shoot apex after different treatments (error bars indicating SD from three biological replicates). (B) The morphology of the shoot apex of loquat after different treatments.
To examine the subcellular localization of EjSOC1-1 and EjSOC1-2, 35S-EjSOC1-1-GFP, and 35S-EjSOC1-2-GFP fusion proteins were generated and transiently expressed in leaf epidermal cells of N. benthamiana. Fluorescence from the 35S-GFP control was detected in both the cytoplasm and nucleus, whereas fluorescence from the 35S-EjSOC1-1-GFP and 35S-EjSOC1-2-GFP fusions was detected only in the nucleus (Figure 6). These results indicate that EjSOC1-1 and EjSOC1-2 were nuclear-localized proteins. These subcellular localization patterns were similar to AtSOC1 in Arabidopsis (Lee et al., 2008).
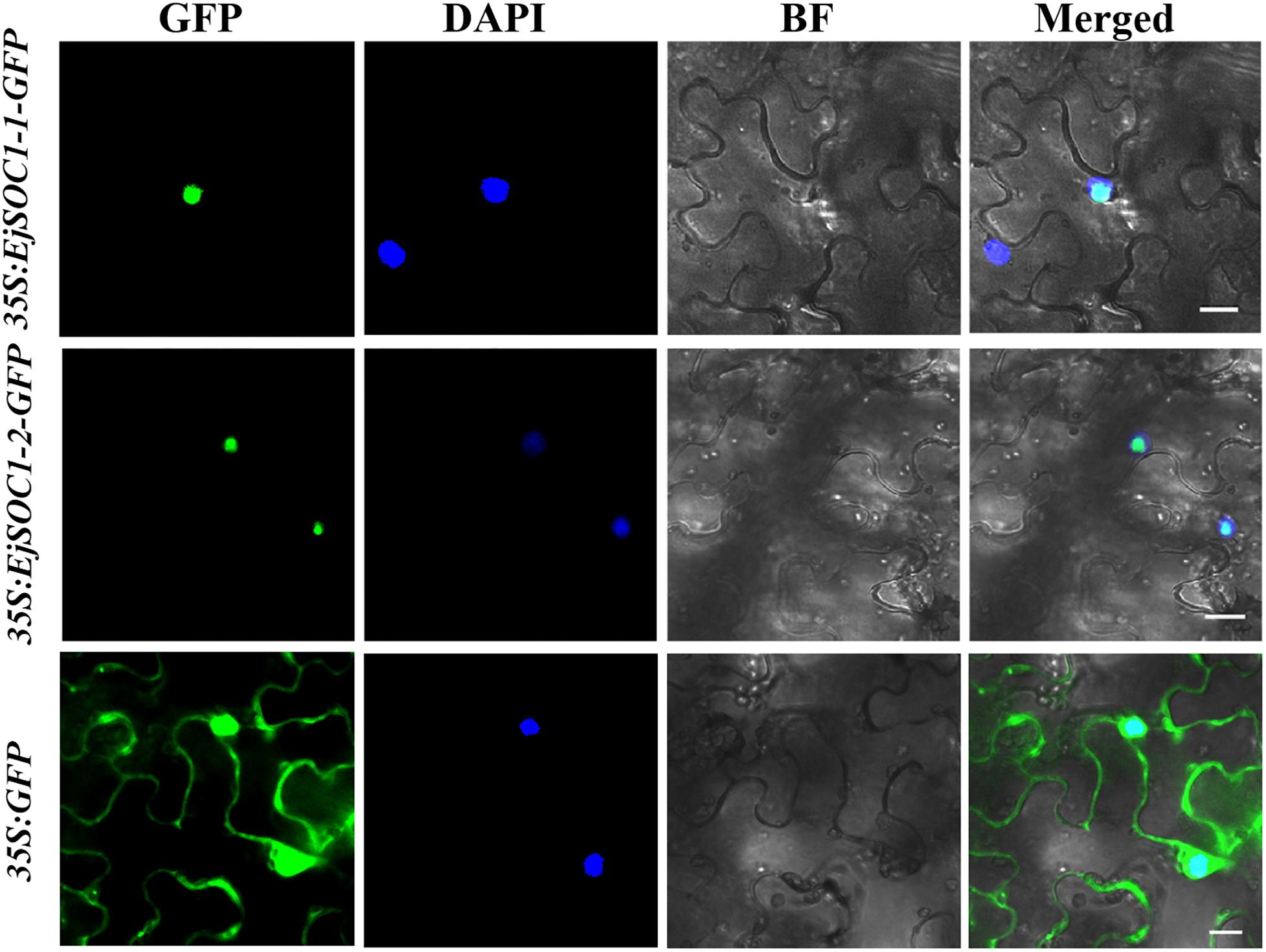
Figure 6. Subcellular localization of EjSOC1s. GFP, GFP fluorescence; 4,6-diamidino-2-phenylindole (DAPI) staining indicates nuclear localization; BF, bright-field; Merged, merged image of GFP and DAPI. Scale bars = 20 μm.
To examine whether EjSOC1-1 and EjSOC1-2 encode functional homologs of AtSOC1, we generated 35S:EjSOC1-1-HA and 35S:EjSOC1-2-HA constructs and introduced them into Arabidopsis wild-type ecotype Col-0 and the late flowering mutant soc1-2. Although the wild-type plants flowered when 13 or 14 rosette leaves appeared, the 35S:EjSOC1-1-HA and 35S:EjSOC1-2-HA transgenic lines flowered with only 6–10 rosette leaves (Figures 7A–D). In addition, soc1-2 mutant, which showed an obviously delayed phenotype compared to wild-type, flowered with 17 or 18 rosette leaves. However, in 35S:EjSOC1-1-HA/+soc1-2- and 35S:EjSOC1-2-HA/+soc1-2-overexpressing lines, flowering occurred at a comparable or even lower number of rosette leaves compared to wild-type (Figures 7E–H). We also detected expression of EjSCO1s in the transgenic plants and found high levels in the respective lines (10 and 20 days) (Supplementary Figures S5A–C). Compared to Col-0, both AtAP1 and AtLFY were relatively highly expressed in the 35S:EjSOC1s-HA transgenic lines (Supplementary Figures S5D,E), and the expression levels of AtAP1 and AtLFY in the 35S:EjSOC1s-HA/+soc1-2 transgenic line and Col-0 were similar or even higher than the expression level in Col-0 (Supplementary Figures S5F,G). These results suggest that EjSOC1-1 and EjSOC1-2 both have a conserved role in accelerating flowering in Arabidopsis and that they may have a significant function in inducing flowering in loquat.
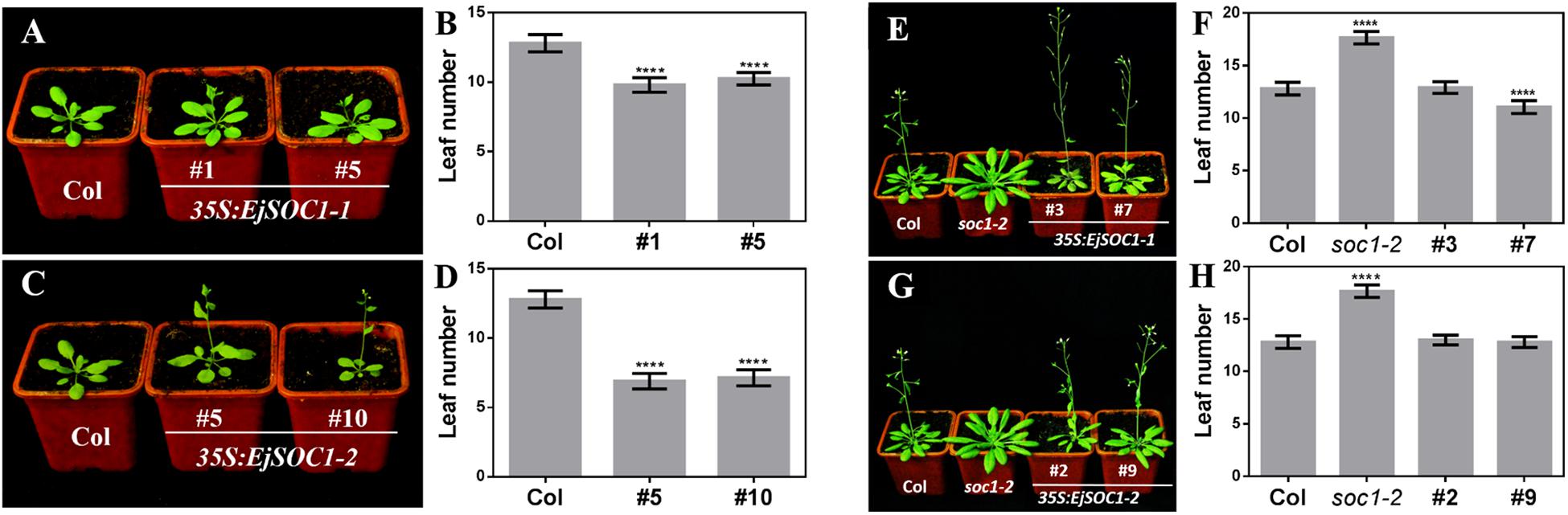
Figure 7. Overexpression of EjSOC1s in Arabidopsis accelerates flowering. (A) 35S:EjSOC1-1-HA transgenic plants exhibit earlier flowering than wild-type Col-0 plants. (B) Rosette leaf number of Col-0 and 35S:EjSOC1-1-HA transgenic plants. (C) 35S:EjSOC1-2-HA transgenic plants exhibit earlier flowering than wild-type Col-0 plants. (D) Rosette leaf number of Col-0 and 35S:EjSOC1-1-HA transgenic plants. (E) 35S:EjSOC1-1-HA/+soc1-2 transgenic plants exhibit earlier flowering than soc1-2 mutant plants. (F) Rosette leaf number of Col-0, soc1-2 mutant and 35S:EjSOC1-1-HA/+soc1-2 transgenic plants. (G) 35S:EjSOC1-2-HA/+soc1-2 transgenic plants exhibit earlier flowering than soc1-2 mutant plants. (H) Rosette leaf number of Col-0, soc1-2 mutant and 35S:EjSOC1-2-HA/+soc1-2 transgenic plants. Asterisks indicate significant differences between Col-0, soc1-2 mutant and transgenic plants (n ≥ 20; error bars indicating SD from three biological replicates; ∗∗∗∗are significantly different from WT at p < 0.0001, by Student’s t-test).
Interestingly, different from the Col-0 phenotypes (Figures 8A,E), a number of flower and silique phenotypes were observed in the 35S:EjSOC1-2-HA transgenic lines. For example, some of the petals were green or lilac in color; hypogenetic stamens were also observed, and the calyx showed anomalous growth (Figures 8B–D) and was not shed after maturity (Figure 8F). Additionally, the surface of some siliques was lilac (Figures 8D,F).

Figure 8. Phenotypes of flowers and siliques of Col-0 and 35S:EjSOC1-2-HA transgenic plants. (A) Normal flower development of wild-type Arabidopsis Col-0. (B–D) Abnormal phenotypes of flowers and siliques of transgenic plants. (E) Normal phenotypes of wild-type Arabidopsis Col-0 siliques. (D,F) Abnormal phenotypes of transgenic plant siliques. Scale bars = 5 mm.
The phenomenon of flowering in autumn and harvesting in spring is very unique in rosaceous plants. In spring, other fresh fruits are rarely sold, or the price of fruit is relatively high, and the range of choices for fruits is greatly reduced. Loquat is undoubtedly a relatively healthy and delicious choice in the fruit market at this time of year. In addition, loquat fruit is affected by storage and transportation. However, loquat fruits are easily injured and only remain fresh for 10 days at normal temperatures (Lin, 2007). Therefore, the price of loquat fruits is usually high, and their transport time to market is short. Overall, determining the flowering time and flowering mechanism of loquat will provide a means to successfully advance or delay it. If that were the case, loquat may be cultivated for a longer period of time and would be more stable in the fruit market for a longer duration. Notably, Eriobotrya species vary in flowering time, but each species of this genus can be hybridized, and fertile offspring can be obtained (Lin, 2017). This fact suggests that the flowering period of loquat is flexible. Research on the flowering mechanism of loquat can greatly benefit loquat growers and, more importantly, provide us with new insight into perennial fruit breeding.
SOC1, which can integrate the gibberellin pathway, photoperiod pathway and age pathway, is a key integration factor in flowering. In the age pathway, miRNA156 is highly expressed in juveniles and inhibits transcription of the SBP family transcription factor SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL). As plants mature, expression of miRNA156 decreases, the transcriptional level of SPLs increases, and SPL15 binds to SOC1 in a GA-dependent manner, recruiting MED18 and RNAPII to induce expression of the downstream MADS-box flowering gene FRUITFULL (FUL), which promotes flowering (Hyun et al., 2016). In the photoperiod pathway, FT activates expression of the flowering integration gene SOC1 and the floral meristem gene APETALA (AP1) to initiate flower bud differentiation and flower development (Abe et al., 2005; Wigge et al., 2005). In the gibberellic acid (GA) pathway, the MADS-box transcription factor AGAMOUS-LIKE24 (AGL24) interacts with SOC1, resulting in direct transcriptional upregulation of both (Liu et al., 2008). Under SD conditions, FT expression is low, and AGL24 interacts with SOC1 to promote Arabidopsis flowering (Liu et al., 2008; Tao et al., 2012). In addition, nuclear factor Y (NF-Y) interacts with the photoperiod transcription factor CO and the GA pathway transcription factor DELLA, directly binding to a unique cis-element within the SOC1 gene, regulates H3K27 methylation levels of SOC1, and affects flowering time (Hou et al., 2014). Unlike in Arabidopsis, studies on woody fruit trees have shown that GA3 inhibits floral bud induction (García-Pallas et al., 2001; Lenahan et al., 2006; Nakagawa et al., 2012; Goldberg-Moeller et al., 2013; Zhang S. et al., 2016).
In this study, the possibility that EjSOC1s are involved in loquat flower formation was identified, and we found that EjSOC1 expression is regulated by photoperiod and GA3. More importantly, our results show that SD and GA3 treatment can inhibit flower differentiation in loquat. A detailed analysis of the gibberellin pathway and photoperiod pathway will help us to better understand the biological mechanism of flower bud differentiation in loquat and provide new insight into artificially delaying flowering in woody fruit trees.
There is increasing evidence that the initiation of flower buds is mainly regulated by AP1 and LFY, of which AP1 is mainly regulated by FT and LFY mainly by SOC1 (Abe et al., 2005; Lee et al., 2008). Furthermore, chromatin immunoprecipitation analysis indicated that the SOC1 protein can directly bind to the CArG domain in the LFY promoter (Lee et al., 2008; Liu et al., 2008). In this study, expression of EjAP1-1, EjAP1-2, EjLFY-1, and EjLFY-2 began to increase in late June and early July, and observation of paraffin-embedded sections showed that the leaf buds began to differentiate into flower buds from late June to early July. These results are consistent and show that EjAP1-1, EjAP1-2, EjLFY-1, or EjLFY-2 may be used as markers for identifying flower bud differentiation in loquat. In addition, the expression trends of EjSOC1s (Figures 4D,E) and EjLFYs (Figure 1C) in the SAM were similar, with EjSOC1-1 and EjSOC1-2 beginning to be highly expressed only 10 days earlier than EjLFYs. In addition, heterologous overexpression of EjSOC1s in Arabidopsis significantly upregulated expression of AtLFY.
Furthermore, it is worth noting that, EjSOC1-1 and EjSOC1-2 were differentially expressed at different developmental stages in leaves, EjSOC1-1 transcription level increased obviously as the young leaves getting mature, and decreased in the late stage of leaf development, this implied EjSOC1-1 might attend the regulation of leaf development, however, the expression of EjSOC1-2 did not show distinct variation. It was reported that aging transcription factor AtSPLs can up-regulate AtSOC1, and therefore promote flowering in Arabidopsis (Wang et al., 2009). In loquat flower, EjSOC1-1 and EjSOC1-2 were mainly expressed in receptacle and pistil, respectively. We speculated that EjSOC1s have a positive effect on the development of floral organs. In the overexpressed transgenic Arabidopsis, phenotypes including color changed petal and silique suggested that EjSOC1-2 might interrupt normal flower development through the abnormal regulation, in addition, the changed color of petals showed possible function of EjSOC1-2 on secondary metabolism, which is worth to investigate in the future. These results provide a theoretical basis for further exploration of the function and mechanism of EjSOC1s in loquat growth and development.
In recent research, EjFT1 has been shown to possibly have to do with bud sprouting and leaf development, whereas EjFT2 has been shown to possibly be involved in flower bud induction (Reig et al., 2017). In this study, EjSOC1-1 and EjSOC1-2 showed different expression trends in leaves with different levels of maturity in the same period (Figure 4B). Therefore, it is speculated that EjSOC1-1 is involved in the processes of flower development and leaf growth. In addition, the expression levels of EjSOC1-1 and EjSOC1-2 differed in various flower tissues (Figure 4F). Clearly, EjSOC1-1 and EjSOC1-2 have some different functions. In the model plant Arabidopsis, SOC1 integrates the photoperiod pathway through FT, which is transported to the SAM and interacts with FD to upregulate SOC1 (Lee and Lee, 2010). Similarly, in loquat (Eriobotrya deflexa Nakai f. koshunensis), EdFT can interact with both EdFD1 and EdFD2 (Zhang L. et al., 2016). These interesting and meaningful findings provide a basis for further studies on the growth and development of loquat and a reference for such studies in other species.
YyJ, SL, and YG designed the research. YyJ mainly performed the research. JP, YZ, WS, LZ, and YiJ finished some parts of the experiments. YyJ wrote the manuscript. SL and YG revised and approved the manuscript.
This study was financially supported by Natural Science Foundation of Guangdong Province (2017A030313129), The Key Areas of Science and Technology Planning Project of Guangdong Province (2018B020202011), National Natural Science Foundation of China (No. 31560091), The Earmarked Fund for Guangdong Modern Agro-industry Technology Research System (2018LM1128), and Ministry of Agriculture Industry Technology Special Project (201003073).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00253/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Chen, F., Zhang, X., Liu, X., and Zhang, L. (2017). Evolutionary analysis of MIKC(c)-Type MADS-box genes in gymnosperms and angiosperms. Front. Plant Sci. 8:895. doi: 10.3389/fpls.2017.00895
Esumi, T., Tao, R., and Yonemori, K. (2005). Isolation of leafy and terminal flower 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sex. Plant Reprod. 17, 277–287. doi: 10.1007/s00497-004-0239-3
Ferrario, S., Busscher, J., Franken, J., Gerats, T., Vandenbussche, M., Angenent, G. C., et al. (2004). Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner. Plant Cell 16, 1490–1505. doi: 10.1105/tpc.019679
García-Pallas, I., Val, J., and Blanco, A. (2001). The inhibition of flower bud differentiation in ‘Crimson Gold’ nectarine with GA3 as an alternative to hand thinning. Sci. Horticult. 90, 265–278. doi: 10.1016/S0304-4238(01)00229-1
Goldberg-Moeller, R., Shalom, L., Shlizerman, L., Samuels, S., Zur, N., Ophir, R., et al. (2013). Effects of gibberellin treatment during flowering induction period on global gene expression and the transcription of flowering-control genes in Citrus buds. Plant Sci. 198, 46–57. doi: 10.1016/j.plantsci.2012.09.012
Gu, C., and Spongberg, S. A. (2003). “ERIOBOTRYA lindley,” in Flora of China, eds W. Zhengyi, P. H. Raven, and D. Y. Hong (Jefferson, MO: Missouri Botanical Garden Press).
Hong, S. M., Bahn, S. C., Lyu, A., Jung, H. S., and Ahn, J. H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 51, 1694–1706. doi: 10.1093/pcp/pcq128
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Hyun, Y., Richter, R., Vincent, C., Martinez-Gallegos, R., Porri, A., and Coupland, G. (2016). Multi-layered regulation of SPL15 and cooperation with SOC1 integrate endogenous flowering pathways at the Arabidopsis shoot meristem. Dev. Cell 37, 254–266. doi: 10.1016/j.devcel.2016.04.001
Kurokura, T., Mimida, N., Battey, N. H., and Hytonen, T. (2013). The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 64, 4131–4141. doi: 10.1093/jxb/ert233
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lee, J., Oh, M., Park, H., and Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55, 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Lee, L. Y., Hou, X., Fang, L., Fan, S., Kumar, P. P., and Yu, H. (2012). STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139, 1568–1576. doi: 10.1242/dev.079426
Lenahan, O. M., Whiting, M. D., and Elfving, D. C. (2006). Gibberellic acid inhibits floral bud induction and improves ‘Bing’ sweet cherry fruit quality. Hortscience 41, 654–659.
Lin, S. Q. (2007). World loquat production and research with special reference to China. Acta Hortic. 750, 37–44. doi: 10.17660/ActaHortic.2007.750.2
Lin, S. Q. (2017). A review on research of the wild species in genus eriobotrya germplasm and their innovative utilization. Acta Hortic. Sin. 44, 1704–1716. doi: 10.16420/j.issn.0513-353x
Liu, C., Chen, H., Er, H. L., Soo, H. M., Kumar, P. P., Han, J. H., et al. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135, 1481–1491. doi: 10.1242/dev.020255
Liu, C., Zhu, C., and Zeng, H. M. (2016). Key KdSOC1 gene expression profiles during plantlet morphogenesis under hormone, photoperiod, and drought treatments. Genet Mol. Res. 15, gmr7579. doi: 10.4238/gmr.15017579
Liu, Y., Song, H., Liu, Z., Hu, G., and Lin, S. (2013). Molecular characterization of loquat EjAP1 gene in relation to flowering. Plant Growth Regul. 70, 287–296. doi: 10.1007/s10725-013-9800-0
Liu, Y., Zhao, Q., Meng, N., Song, H., Li, C., Hu, G., et al. (2017). Over-expression of EjLFY-1 leads to an early flowering habit in strawberry (Fragaria x ananassa) and Its asexual progeny. Front. Plant Sci. 8:496. doi: 10.3389/fpls.2017.00496
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/s0092-8674(01)00384-1
Moon, J., Lee, H., Kim, M., and Lee, I. (2005). Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299. doi: 10.1093/pcp/pci024
Mouhu, K., Kurokura, T., Koskela, E. A., Albert, V. A., Elomaa, P., Hytonen, T., et al. (2013). The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth. Plant Cell 25, 3296–3310. doi: 10.1105/tpc.113.115055
Nakagawa, M., Honsho, C., Kanzaki, S., Shimizu, K., and Utsunomiya, N. (2012). Isolation and expression analysis of FLOWERING LOCUS T-like and gibberellin metabolism genes in biennial-bearing mango trees. Sci. Hortic. 139, 108–117. doi: 10.1016/j.scienta.2012.03.005
Parcy, F. (2005). Flowering: a time for integration. Int. J. Dev. Biol. 49, 585–593. doi: 10.1387/ijdb.041930fp
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Reig, C., Gil-Munoz, F., Vera-Sirera, F., Garcia-Lorca, A., Martinez-Fuentes, A., Mesejo, C., et al. (2017). Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta 246, 915–925. doi: 10.1007/s00425-017-2740-6
Ruokolainen, S., Ng, Y. P., Albert, V. A., Elomaa, P., and Teeri, T. H. (2011). Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 107, 1491–1499. doi: 10.1093/aob/mcr112
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., Sun, C. D., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. doi: 10.1007/s00425-008-0696-2
Smaczniak, C., Immink, R. G., Angenent, G. C., and Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sri, T., Mayee, P., and Singh, A. (2015). Sequence and expression variation in suppressor of overexpression of constans 1 (SOC1): homeolog evolution in Indian Brassicas. Dev. Genes Evol. 225, 287–303. doi: 10.1007/s00427-015-0513-4
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Tadege, M., Sheldon, C. C., Helliwell, C. A., Upadhyaya, N. M., Dennis, E. S., Peacock, W. J., et al. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1, 361–369. doi: 10.1046/j.1467-7652.2003.00034.x
Tan, F. C., and Swain, S. M. (2007). Functional characterization of AP3, SOC1 and WUS homologues from citrus (Citrus sinensis). Physiol. Plant 131, 481–495. doi: 10.1111/j.1399-3054.2007.00971.x
Tao, Z., Shen, L. S., Liu, C., Liu, L., Yan, Y. Y., Yu, H., et al. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70, 549–561. doi: 10.1111/j.1365-313X.2012.04919.x
Teotia, S., and Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Theissen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516.
Vandenbussche, M., Theissen, G., Van de Peer, Y., and Gerats, T. (2003). Structural diversification and neo-functionalization during floral MADS-box gene evolution by C-terminal frameshift mutations. Nucleic Acids Res. 31, 4401–4409. doi: 10.1093/nar/gkg642
Voogd, C., Wang, T., and Varkonyi-Gasic, E. (2015). Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 66, 4699–4710. doi: 10.1093/jxb/erv234
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wei, J., Liu, D., Liu, G., Tang, J., and Chen, Y. (2016). Molecular cloning, characterization, and expression of MiSOC1: a homolog of the flowering gene suppressor of overexpression of constans1 from mango (Mangifera indica L). Front. Plant Sci. 7:1758. doi: 10.3389/fpls.2016.01758
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Zhang, L., Yu, H., Lin, S., and Gao, Y. (2016). Molecular characterization of FT and FD homologs from eriobotrya deflexa nakai forma koshunensis. Front. Plant Sci. 7:8. doi: 10.3389/fpls.2016.00008
Zhang, S., Zhang, D., Fan, S., Du, L., Shen, Y., Xing, L., et al. (2016). Effect of exogenous GA3 and its inhibitor paclobutrazol on floral formation, endogenous hormones, and flowering-associated genes in ‘Fuji’ apple (Malus domestica Borkh.). Plant Physiol. Biochem. 107, 178–186. doi: 10.1016/j.plaphy.2016.06.005
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhao, S., Luo, Y., Zhang, Z., Xu, M., Wang, W., Zhao, Y., et al. (2014). ZmSOC1, a MADS-box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int. J. Mol. Sci. 15, 19987–20003. doi: 10.3390/ijms151119987
Keywords: loquat, flowering time, GA3, short-day, EjSOC1, EjAP1, EjLFY
Citation: Jiang Y, Peng J, Zhu Y, Su W, Zhang L, Jing Y, Lin S and Gao Y (2019) The Role of EjSOC1s in Flower Initiation in Eriobotrya japonica. Front. Plant Sci. 10:253. doi: 10.3389/fpls.2019.00253
Received: 22 November 2018; Accepted: 15 February 2019;
Published: 04 March 2019.
Edited by:
Zhongchi Liu, University of Maryland, United StatesReviewed by:
Lee Jeong Hwan, Chonbuk National University, South KoreaCopyright © 2019 Jiang, Peng, Zhu, Su, Zhang, Jing, Lin and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shunquan Lin, bG9xdWF0QHNjYXUuZWR1LmNu Yongshun Gao, eW9uZ3NodW5nYW9AMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.