
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 19 March 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00206
This article is part of the Research TopicRoot Branching: from Lateral Root Primordium Initiation and Morphogenesis to FunctionView all 10 articles
Morphogenetic processes are the basis of new organ formation. Lateral roots (LRs) are the building blocks of the root system. After LR initiation and before LR emergence, a new lateral root primordium (LRP) forms. During this period, the organization and functionality of the prospective LR is defined. Thus, proper LRP morphogenesis is a decisive process during root system formation. Most current studies on LRP morphogenesis have been performed in the model species Arabidopsis thaliana; little is known about this process in other angiosperms. To understand LRP morphogenesis from a wider perspective, we review both contemporary and earlier studies. The latter are largely forgotten, and we attempted to integrate them into present-day research. In particular, we consider in detail the participation of parent root tissue in LRP formation, cell proliferation and timing during LRP morphogenesis, and the hormonal and genetic regulation of LRP morphogenesis. Cell type identity acquisition and new stem cell establishement during LRP morphogenesis are also considered. Within each of these facets, unanswered or poorly understood questions are identified to help define future research in the field. Finally, we discuss emerging research avenues and new technologies that could be used to answer the remaining questions in studies of LRP morphogenesis.
A key function of roots—water and mineral uptake and transport—is strongly related to the root system surface area. Root branching promotes and underlies the increase in root surface area, and therefore a single lateral root (LR) constitutes a building block of the root system. Thus, root branching is a ubiquitous and widely distributed process in vascular plants. A classic example of the abundance of roots is the extended root system of a single rye plant (Secale cereale). During only one growth season of approximately 4 months, a single plant formed 13,815,672 roots (Dittmer, 1937), most of which were LRs. LRs are initiated in the pericycle (Laskowski et al., 1995; Dubrovsky et al., 2000, 2008; Beeckman et al., 2001; Dubrovsky and Rost, 2012; Beeckman and De Smet, 2014), and comprehensive analysis of LR development has been performed on a model species, Arabidopsis thaliana (hereafter Arabidopsis). In this species, it has been recognized that LR formation is a process that includes multiple steps: (a) pericycle priming; (b) founder cell specification; (c) the first divisions in pericycle founder cells leading to LR formation, processes defined as LR initiation; (d) lateral root primordium (LRP) formation, comprising developmental processes from the first derivatives of the founder cells to formation of the dome-shaped LRP; (e) LR emergence, i.e., protrusion of the LRP through the external root tissues, including ground tissues and epidermis; (f) activation of the apical meristem in the nascent LR; and (g) LR growth (Malamy and Benfey, 1997; De Smet et al., 2003; Péret et al., 2009; Malamy, 2010; Stoeckle et al., 2018). The details of these processes are studied at different levels, from developmental anatomy to hormonal and genetic control (Casimiro et al., 2003; De Smet et al., 2006a; Fukaki and Tasaka, 2009; De Smet, 2012; Atkinson et al., 2014; Du and Scheres, 2017a; Dubrovsky and Laskowski, 2017; Ötvös and Benková, 2017). However, not all these steps are equally understood. One of the less understood steps comprises morphogenetic processes from LR initiation to LR emergence (Figure 1). Indeed, the mechanisms underlying the remarkably stable and reproducible process of formation of the three-dimensional (3D) LRP structure from a 2D plate of founder cell derivatives are a mystery. In this review, we summarize what is known about the essential elements underlying LRP morphogenesis in angiosperms and attempt to identify the basic questions related to LRP morphogenesis that remain to be answered or better understood.
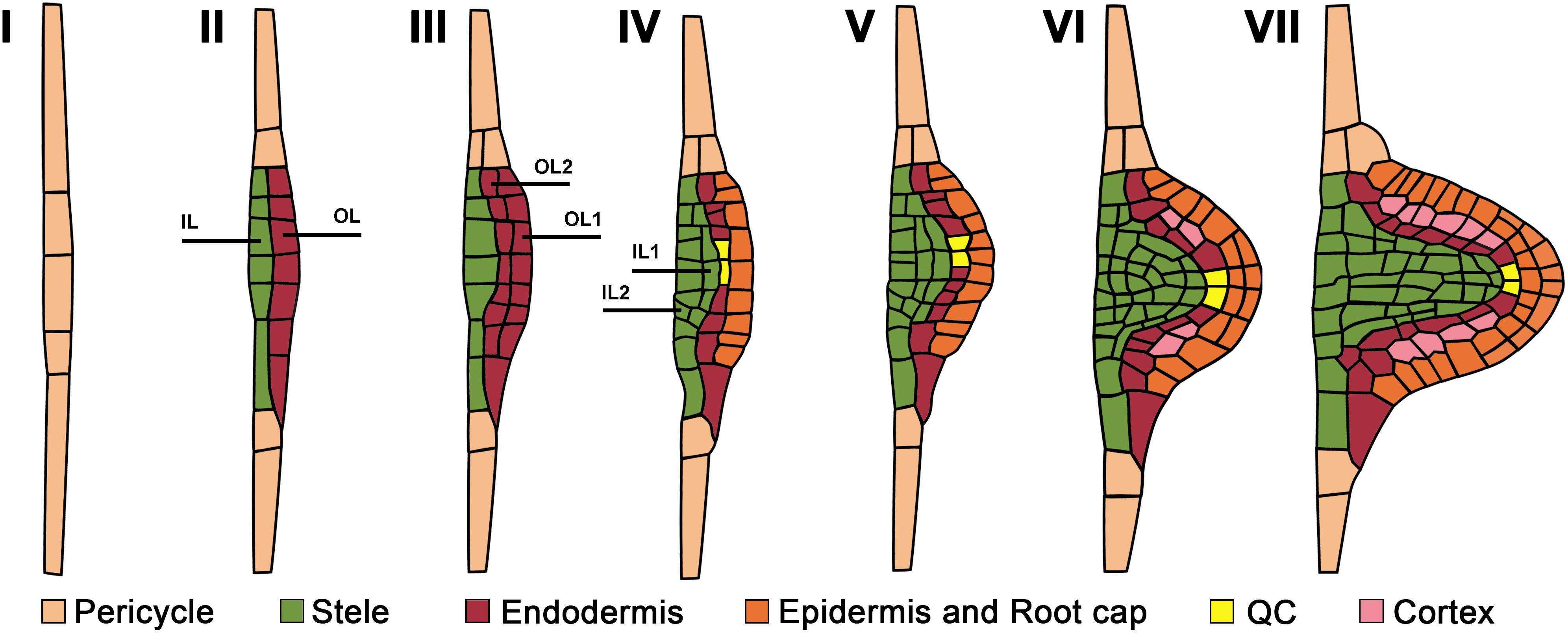
Figure 1. Developmental stages and cell type identity acquisition during lateral root primordium morphogenesis from initiation to lateral root emergence. Numbers correspond to the developmental stages as defined by Malamy and Benfey (1997). Emerging cell identity recognized based on cell type reporters is color-coded. Pericycle cell identity corresponds to that of the parent root. OL and IL are outer and internal layers. See text for details.
Although the pericycle is a principal tissue giving rise to LRs in angiosperms, other parent root tissues, including the endodermis, cortex and vascular parenchyma, participate in LRP morphogenesis.
The specification of pericycle founder cells and other pre-initiation events take place before LRP initiation (De Smet et al., 2007; Moreno-Risueno et al., 2010; Van Norman et al., 2013) and are therefore beyond the scope of this review. In Arabidopsis, two types of LRP initiation have been recognized: longitudinal unicellular and longitudinal bi-cellular, in which a single or two adjacent pericycle founder cells in the longitudinal plane, respectively, participate in LRP initiation (Dubrovsky et al., 2001, 2008). The most common type of initiation is considered to be longitudinal bi-cellular (Casero et al., 1995; Lloret and Casero, 2002; De Rybel et al., 2010; von Wangenheim et al., 2016). However, it cannot be excluded that the longitudinal bi-cellular type is a result of the cell division of the founder cell following the longitudinal unicellular type of initiation. Even for a model species such as Arabidopsis, it is not known how common each initiation type is.
When viewed in a transversal plane, the number of pericycle cell files that participate in the specification of the LRP founder cells varies among species; for instance, 4 to 6 phloem-adjacent files in wheat (Triticum aestivum; Demchenko and Demchenko, 2001) and 6–8 xylem-adjacent cell files in Arabidopsis (von Wangenheim et al., 2016) are involved in LRP formation. In most species, the pericycle is a unicellular tissue layer. Nevertheless, in Cucurbitaceae, two pericycle layers, internal and external, are formed in the xylem pole, and both participate in LRP formation (Dubrovsky, 1986a; Ilina et al., 2018). The most detailed analysis of pericycle participation in LRP morphogenesis has been performed in Arabidopsis. In this species, the first few divisions in the pericycle leading to LRP formation are anticlinal formative (asymmetrical) divisions (De Smet and Beeckman, 2011). Anticlinal divisions are perpendicular to the nearest root surface. As these divisions take place in few tangentially (i.e., in the direction perpendicular to the radius of the parent root) adjacent founder cells (Dubrovsky et al., 2001; Casimiro et al., 2003; von Wangenheim et al., 2016), a plate of on average 26 pericycle-derived cells is formed (von Wangenheim et al., 2016), corresponding to Stage (St) I, as defined by Malamy and Benfey (1997) (Figure 1). This cell plate has 2D organization and, at this point, the transition to the formation of the new growth axis that permits the 3D LRP organization is defined. The first event leading to this transition is the radial growth of StI LRP cells, resulting in the formation of the apical–basal axis of the new LR (Figure 2). This new growth direction is controlled by the adjacent endodermis through auxin signaling mediated by SHORT HYPOCOTYL2, SHY2/IAA3 (Vermeer et al., 2014). Radially expanded LRP cells eventually divide periclinally (Malamy and Benfey, 1997), i.e., parallel to the nearest root surface (Figure 1), starting in the central xylem-adjacent cell files of the plate. This division follows the established Errera’s rule, which states that cells divide preferentially along the shortest distance between cell walls (Besson and Dumais, 2011). Concurrently, the tangentially flanking cells of the plate divide in an oblique orientation, impacting the formation of the oval-shaped basal portion of the prospective LRP (Lucas et al., 2013). Starting from the two-layered LRP, 3D morphogenesis continues along the axis of the future LR. The number of cells at a given developmental stage and the division patterns vary, even though the overall LRP shape changes are conserved (Lucas et al., 2013; von Wangenheim et al., 2016). The developmental stages recognized for Arabidopsis (Malamy and Benfey, 1997; Napsucialy-Mendivil and Dubrovsky, 2018), depicted in Figure 1, are frequently applied to other species (Yu et al., 2016). In most angiosperms examined, pericycle participation in LRP formation is similar, at least during the early stages(Lloret and Casero, 2002).
As early as the 1870s, it was documented that in addition to the pericycle, other tissues participate in LRP morphogenesis (Janczewski, 1874; Van Tieghem and Douliot, 1888; Von Guttenberg, 1968). In most dicots and monocots, the endodermis is also involved in LRP formation (Van Tieghem and Douliot, 1888; Kawata and Shibayama, 1965; Bell and McCully, 1970; Seago, 1973). In some orders—for example, Poales—endodermis participation in LRP formation requires cell dedifferentiation (Danilova and Serdyuk, 1982). The first few divisions in the endodermal layer, like in the pericycle, are anticlinal (Seago, 1973; Demchenko and Demchenko, 2001). Next, in maize (Zea mays; Bell and McCully, 1970), T. aestivum (Demchenko and Demchenko, 2001), tomato (Solanum lycopersicum; Ivanchenko et al., 2006) and other species, the endodermal derivatives undergo periclinal divisions and form a two-layered structure. In many angiosperm taxa, even more than two layers of endodermal origin can be formed (Figure 3). The LRP tissues of endodermal origin form a temporary structure called Tasche in the German literature and Poche in the French (Clowes, 1978a). No specific term for this structure is used in the English literature. This temporary structure has some features of the root cap and sloughs off after LR emergence. Here we call this temporary structure the Cap-Like Structure (CLS). The CLS results from both anticlinal and periclinal divisions of the endodermis and sometimes cortex (see below). We should note here that in some cases the CLS is not temporary but a permanent structure (see below). Anatomical studies of Z. mays LRPs showed that the endodermis contributes to the formation of the LR’s permanent tissues, the epidermis and the root cap (Bell and McCully, 1970; Karas and McCully, 1973; McCully, 1975). This interpretation results from the fact that the epidermis of a recently emerged LR can be traced back to the endodermis of the parent root and that endodermal derivative cells in the central apical domain of the LRP start to divide periclinally and form a root cap (Bell and McCully, 1970; Karas and McCully, 1973). Particularly, when a Z. mays LRP protrudes about half the width of the parent root cortex, endodermal derivatives of the LRP contain abundant starch grains (Bell and McCully, 1970). By analyzing colchicine-treated chimeric LRPs that contain cells of different ploidy, Clowes (1978a) showed that the whole LR in Z. mays plants is of pericyclic origin and that the endodermis forms the CLS, which is maintained only for a short period after LR emergence. Similar conclusions were reached for more complex scenarios in which not only the endodermis but also the cortex participates in LRP formation (Dubrovsky, 1986a; Demchenko et al., 2001; Ilina et al., 2018), confirming the earlier view that the permanent body of the LR is entirely of pericyclic origin (Van Tieghem and Douliot, 1888).
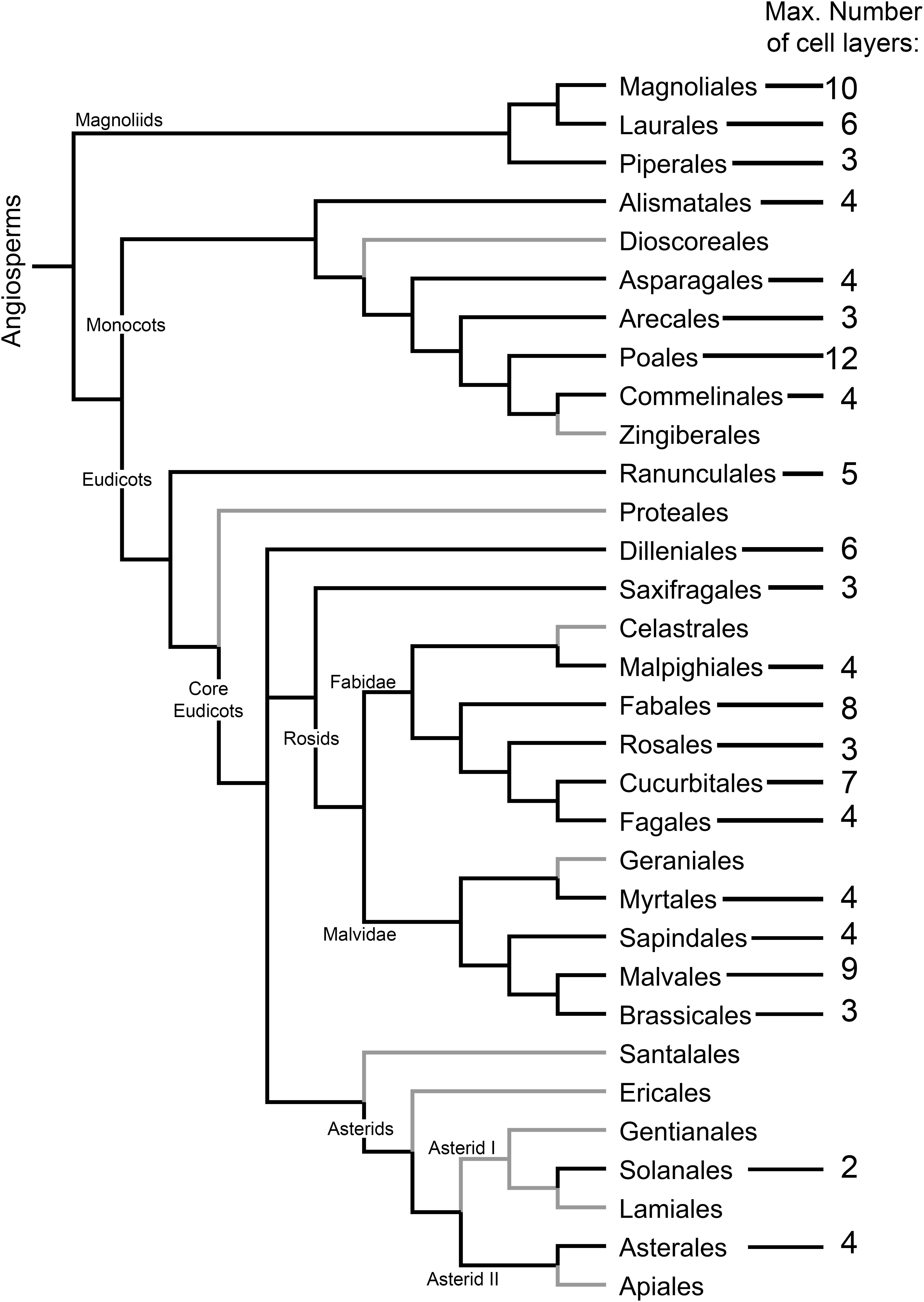
Figure 3. Distribution and extent of the development of the cap-like structure (CLS) of the lateral root primordium among angiosperm orders. The maximum number of CLS cell layers is indicated. Angiosperm orders for which no CLS has been found are depicted with gray branches. Cladogram topology was depicted using FigTree (http://tree.bio.ed.ac.uk/software/figtree/) following the phylogenetic relationship among angiosperms as proposed by the Angiosperm Phylogeny Group (Chase et al., 2016). The data were taken from Voronin’s (1957) analysis of Van Tieghem and Douliot (1888). Species included in the analysis were revised in accordance with contemporary taxonomic classification. Orders for which no data are available were not included in the cladogram. Additionally, data for Solanales were added (Seago, 1973; Ivanchenko et al., 2006).
As it is not always possible to deduce which LRP tissues are formed from the pericycle or endodermis based on anatomical observations alone, there is a need to develop cell type identity markers for this purpose. Nevertheless, anatomical studies are of great value. Based on the classical work of Philippe Van Tieghem and Douliot (1888), Voronin (1957, 1964) analyzed the types and distribution of the CLS among angiosperms. We incorporated Voronin’s data in the angiosperm phylogenetic tree proposed by the Angiosperm Phylogeny Group (Chase et al., 2016), in an effort to visualize the distribution and evolutionary trends of the appearance and types of the CLS in angiosperms (Figure 3). This analysis showed that the CLS is present in most angiosperm orders. The distribution of the CLS in angiosperms suggests an evolutionary trend toward CLS reduction until its complete disappearance, as inferred from the absence of a CLS in most orders of the recent Asterids clade (Figure 3). In Arabidopsis, a CLS is not found, even though it is present in other Brassicales. This ‘atypical’ pattern is perhaps a consequence of very simple root organization with a two-layered ground tissue composed of single layers of endodermis and cortex. It would be interesting to validate this hypothesis by analyzing LRP formation in Arabidopsis transgenic lines with supernumerary ground tissue layers that maintain endodermis identity (Nakajima et al., 2001).
In some species, the developing LRP is capable of inducing cell divisions in the adjacent cortex (Tschermak-Woess and Doležal, 1953; Demchenko et al., 2001) that are unrelated to LRP morphogenesis. The role of these divisions is unclear. In some taxa, cortex adjacent to the endodermis participates in CLS formation. This is commonly found in Fabaceae (Popham, 1955; Byrne et al., 1977) and Cucurbitaceae (Van Tieghem and Douliot, 1886; Dubrovsky, 1986a; Demchenko and Demchenko, 2001; Ilina et al., 2018), which form a massive CLS (Figure 4). Interestingly, the cortical and endodermal cells of the same files of the parent root that constitute the developing LRP participate in CLS formation and the cells outside the LRP of the same files do not divide (Dubrovsky, 1986a; Demchenko and Demchenko, 2001; Ilina et al., 2018). This endodermis continuity between the parent and lateral roots is not always maintained, and the CLS on the flanks can be destroyed before LR emergence, as in Z. mays (Clowes, 1978a). Few studies have examined how the ground tissue and epidermis of endodermal and cortical origin are replaced by the same cell types produced by the pericycle (Dubrovsky, 1986a).
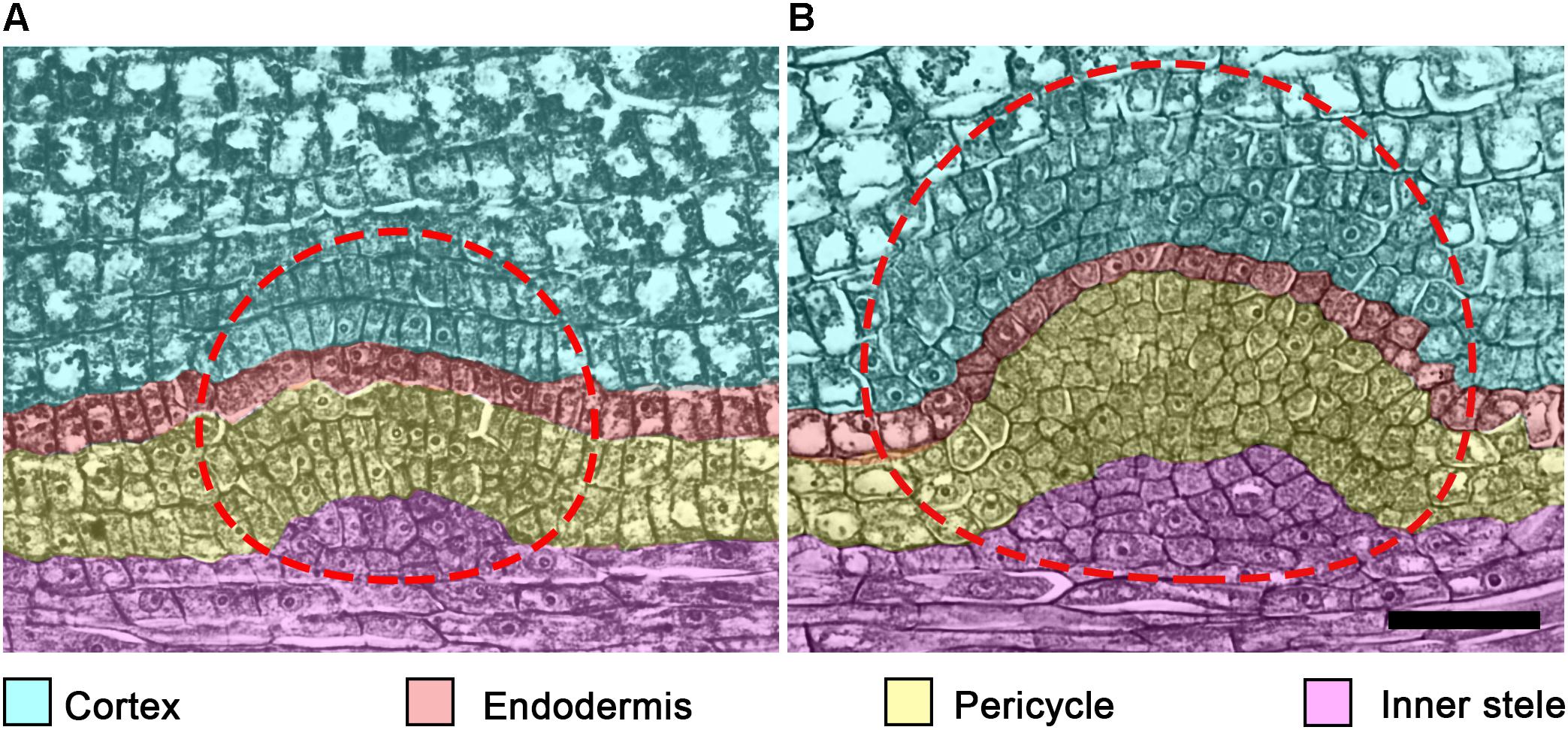
Figure 4. Tissues participating in lateral root primordium (LRP) formation in cucumber (Cucumis sativus) root. In C. sativus, several embryonic LRPs are formed during embryogenesis. Within the same parent root, the more apical primordium (A) is developed to a lesser extent than the most basal primordium (B). Note the different extent of each cell type participation in LRP morphogenesis in these primordia. The temporal cap-like structure (CLS) includes endodermis and cortex derivatives of the LRP. Cell types and their derivative cells produced within the LRP are color coded. The LRP is indicated by a dashed line. Seeds were imbibed for 18 h and fixed; histological sections of the radicle were prepared and stained as described (Dubrovsky, 1986a). Scale bar = 50 μm.
Surprisingly, studies of CLS function during and after LRP formation are scarce. It has been suggested that CLS cells secrete hydrolases that may facilitate LR emergence (Van Tieghem and Douliot, 1888; Bonnett, 1969; McCully, 1975). The early literature on CLSs suggests that this structure protects the pericycle derivatives from mechanical damage as the LRP protrudes through the parental root tissues before LR emergence. Despite the fact that in most angiosperms the CLS is displaced by the permanent cap of pericyclic origin post-emergence, in some hydrophytes (e.g., Hydrocharis, Lemna, Pistia, Eichornia, and Pontederia) the CLS is permanently maintained on the LRs (Voronin, 1957).
Another possible function of the CLS might be related to cell proliferation. Cell division in the endodermis and the pericycle start simultaneously during LRP initiation and may create a critical mass of cells required to sustain rapid cell divisions (see section “The Cell Cycle During Lateral Root Primordium Morphogenesis and Timing Aspects”). Also, cell proliferation of the CLS is important for quiescent center (QC) establishment (see Section “Cell Type Identity Acquisition”). Whether endodermal participation in LRP morphogenesis in angiosperms is evolutionarily linked to the ability of this tissue in ferns to form LRPs (Mallory et al., 1970; Hou et al., 2004) is an open question.
No participation of vascular parenchyma in LRP formation has been documented in Arabidopsis. Therefore, this aspect of LRP morphogenesis is seldom discussed in contemporary literature. Nonetheless, the vascular parenchyma participates in primordium formation in both monocots (Rywosch, 1909; Bunning, 1952; Bell and McCully, 1970; Demchenko and Demchenko, 1996b) and eudicots (Seago, 1973; Byrne et al., 1977) by contributing to the vascular connection of the nascent LR and the parent root. It has been documented that vascular parenchyma cells start to divide very early, in StI LRPs in monocots (e.g., T. aestivum; Demchenko and Demchenko, 1996a) and StII LRPs in eudicots (e.g., Glycine Max; Byrne et al., 1977). During LRP formation in G. max, vascular parenchyma derivatives divide periclinally and form files of 4–5 cells that contribute to the formation of vascular tissues connecting the parent and lateral root (Byrne et al., 1977). Similarly, during embryonic LRP morphogenesis in the cucumber (Cucumis sativus) radicle, vascular parenchyma cells of the parent root divide 2–3 times periclinally, forming several layers of derivative cells (Dubrovsky, 1986a) (Figure 4). The contribution of vascular tissues of the parent root to LRP formation was also documented by the analysis of ploidy chimeras in Z. mays roots (Clowes, 1978a). Whether the participation of vascular parenchyma during LRP development is related to transport of nutrients or hormones toward the developing LRP remains to be determined.
In most angiosperms, LR initiation takes place post-germination. However, there are well-documented cases in which LRs are initiated during embryogenesis, such as in Cucurbitaceae (Clowes, 1982; Dubrovsky, 1986a,b, 1987) and Polygonaceae (buckwheat, Fagopyrum sagittatum, O’Dell and Foard, 1969). The extent to which LRP morphogenesis proceeds during embryogenesis ranges from StII, as in F. sagittatum (O’Dell and Foard, 1969), to StVII, as in C. sativum (Dubrovsky, 1986a). A number of embryonic LRPs are formed within the embryo (O’Dell and Foard, 1969; Dubrovsky, 1987). Interestingly, in some Cucurbitaceae in which embryonic LRP morphogenesis is documented, post-germination LRP initiation takes place in the root apical meristem (Gulyaev, 1964; Dubrovsky, 1987; Demchenko and Demchenko, 2001; Ilina et al., 2018). Some other angiosperms, especially hydrophytes from Pontederiaceae, Araceae and Alismataceae, also begin LRP morphogenesis within the apical meristem of the parent root, as reviewed elsewhere (Dubrovsky and Rost, 2003; Ilina et al., 2018). Whether there is a correlation between the species capability to start LRP morphogenesis during embryogenesis and its capacity to initiate LRPs within the root apical meristem is an open question. In most angiosperm species, however, initiation starts post-germination within the differentiation zone, where LRP morphogenesis takes place. In this review, we consider mostly these cases.
The time from LR initiation to emergence ranges from 2.8 to 3.6 days in pea (Pisum sativum), faba bean (Vicia faba), Z. mays, and common bean (Phaseolus vulgaris) (MacLeod and Thompson, 1979), is about 2.5 days in radish (Raphanus sativus) (Blakely et al., 1982) and 1.6–2.2 days in Arabidopsis (Napsucialy-Mendivil et al., 2014; von Wangenheim et al., 2016). This suggests that the whole new organ can be formed during a relatively short period. Cell cycle studies in developing LRPs using labeled DNA precursors, e.g., tritiated thymidine (3H-thymidine), have restrictions because LRPs at advanced stages do not incorporate 3H-thymidine; for example, in V. faba LRPs of 1,500 or fewer cells incorporated 3H-thymidine, whereas LRPs that contained a greater number of cells did not (Davidson and MacLeod, 1968; MacLeod and Davidson, 1968). Similarly, in monocots (T. aestivum), LRPs at StIII and later did not incorporate 3H-thymidine (Demchenko and Demchenko, 1996a). Therefore, most earlier studies were based on estimating cell doubling time (Td), and contemporary studies use a time-lapse approach (von Wangenheim et al., 2016). Td estimations assume an exponential increase in cell number in the LRP (Thompson and MacLeod, 1981) to estimate the maximal duration of the cell cycle. As all LRP cells become polyploid when treated with colchicine (MacLeod and Davidson, 1968; Friedberg and Davidson, 1971), it is accepted that all the LRP cells proliferate, and the proliferation fraction is equal to one.
Cell proliferation dynamics impact the rate of primordium formation and LRP morphogenesis. It has been proposed that the rapid establishment of an LRP after initiation might have a role in lateral inhibition—i.e., preventing the initiation of new LRPs in the vicinity of ones already initiated (Dubrovsky et al., 2001). Therefore, the cell cycle in young LRPs is expected to be shorter than that in LRPs at a later developmental stage. Indeed, a few studies show that the shortest cell cycle is found at the earliest stages of LRP morphogenesis and increases at later stages. In V. faba, P. sativum, Z. mays, P. vulgaris (MacLeod and Thompson, 1979), and Arabidopsis (Dubrovsky et al., 2001), the Td from early to later stages of LRP development increases from 8.2, 2.9, 4.5, 6.9, and 2.7 h to 14.16, 9.96, 17.65, 11.4, and 4.9 h, respectively. For Arabidopsis, the average Td in LRP cells is 7.1 h (von Wangenheim et al., 2016), about half the average cell cycle duration observed in the primary-root apical meristem (reviewed in Zhukovskaya et al., 2018). Therefore, an overall short cell cycle and a gradual increase in cell cycle duration over time seems to be a general tendency. In species with LRPs already initiated during embryogenesis, the opposite trend is found post-germination. For instance, in C. sativus, Td is the longest (8.7 h) when pre-initiated LRP cells first enter the cell cycle soon after seed imbibition and decreases to 2.7 h in the LRP just before LR emergence (Dubrovsky, 1986b), explaining why LRs emerge rapidly after germination in this species.
When V. faba LRPs are about to emerge, their cells are less proliferatively active than during previous stages; after emergence, a sharp increase in proliferation is observed (Friedberg and Davidson, 1971; MacLeod, 1972, 1973). The rate of formation of individual LRPs within a parent root is variable, as documented for V. faba, P. sativum, Z. mays, and P. vulgaris (MacLeod and Thompson, 1979) and Arabidopsis (Dubrovsky et al., 2006; von Wangenheim et al., 2016). This is in agreement with the fact that, contrary to LR initiation, LRP formation along the parent root does not follow an acropetal pattern and is asynchronous. The heterogeneity in the rate of LRP formation explains why younger LRPs are found among the older ones or among emerged LRs, even in the zone where the vascular cambium and secondary tissues are formed (Napsucialy-Mendivil and Dubrovsky, 2018). Whether slow developing or delayed LRPs are capable of later resuming development is not well documented and remains an open question.
The processes of LR initiation and emergence are coordinated (Lucas et al., 2008b). The distance from the apex of the parent root to the site where the LR emerges depends on the site of LR initiation and on the rate of primordium formation. As mentioned above, in Arabidopsis, the time between LRP initiation and LR emergence is one of the shortest reported in Angiosperms. Nonetheless, the LR emerges a few centimeters from the apex. When LRs are initiated in the root apical meristem of the parent roots, as in Cucurbitaceae, LRs emerge at a shorter distance, e.g., 12–15 mm from the primary root apex in squash, Cucurbita pepo (Demchenko and Demchenko, 2001).
The role of cell cycle duration in LRP morphogenesis has not been extensively studied. The central domains of the LRP seem to develop faster than the flanking domains. Figure 2 shows the terminology used to describe the LRP domains. The progeny of central founder cells is characterized by an average interphase duration of 6.0 h, whereas the corresponding period in the progeny of peripheral founder cells is 7.2 h (von Wangenheim et al., 2016). This suggests that the difference between cell cycle duration in each cell lineage has a profound impact on morphogenesis. It is not understood how heterogeneity in cell cycle time is related to LRP morphogenesis, whether the differences in cell cycle duration in certain domains have a critical role in defining the dome shape of the developing LRP, or whether cell cycle time heterogeneity dictates the shape of the LRP or if the shape defines the cell cycle duration in different domains. Future studies should address these questions.
Since early studies, the importance of hormonal regulation in all aspects of LR development was recognized (Wightman et al., 1980). The role of auxin in LR development is well documented (Fukaki and Tasaka, 2009; Lavenus et al., 2013; Du and Scheres, 2017a) and LRP morphogenesis is known to depend on endogenous auxins up to StIV (Laskowski et al., 1995). In P. sativum plants treated with auxin transport inhibitors, normal LRP dome organization is lost and the primordium structure is transformed into a globular mass of cells (Hinchee and Rost, 1992), highlighting the significance of auxin in LRP morphogenesis. Accordingly, synthetic auxin-response promoter DIRECT REPEAT5 (DR5) (Ulmasov et al., 1997) activity is high during both LR initiation (Dubrovsky et al., 2008) and throughout LRP development (Benková et al., 2003; Dubrovsky et al., 2008). The auxin response maximum is present starting from StI in LRPs. From StIII, it is restricted to the central apical domain (Figure 2) of the LRP, corresponding to the prospective location of the QC (Dubrovsky et al., 2008). The auxin maximum apparently has a role in stem cell niche establishment and thus is important for normal LRP morphogenesis (see also the next section).
The localized auxin maximum response is present in different angiosperm orders, from monocots (Z. mays, Jansen et al., 2012) to eudicots (S. lycopersicum and Arabidopsis, Dubrovsky et al., 2008). It has been shown that while LR initiation does not depend on shoot-derived auxin, this source of auxin is essential for post-initiation LRP morphogenesis and LR emergence (Bhalerao et al., 2002; Ditengou et al., 2008; Swarup et al., 2008; Richter et al., 2009). A recent study strongly suggests that formation of an auxin maximum in the LRP also depends on local auxin synthesis in proliferating cells (Brumos et al., 2018). In most angiosperms, LR initiation starts in the young differentiation zone. Interestingly, when the LRP forms within the root apical meristem of the parent root, as in C. pepo, the auxin maximum is also established from the very early stages of LRP formation and is subsequently maintained throughout LRP development (Ilina et al., 2018). This suggests that LRP morphogenesis depends on auxin regardless of the differentiation state of the parent root cells giving rise to the LRP.
The auxin gradient with a maximum in the central apical domain (Figure 2) of the developing LRP is created by the auxin efflux carriers PIN-FORMED 1 (PIN1), PIN3, PIN4, PIN6, and PIN7 (Benková et al., 2003; Marhavý et al., 2014) and the auxin influx carriers AUX1 and LIKE AUX 3 (LAX3) (Marchant et al., 2002; Péret et al., 2012b). This gradient is formed by auxin flux from the flanking to the central domain and from the basal to the apical domain (Figure 2). Mutations that affect auxin transport lead to abnormal LRP formation. In the pin1 mutant, LRP formation is slow and DR5 promoter activity is extended to more cells (Benková et al., 2003). Similarly, LRP formation takes more time in a lax3 mutant (Swarup et al., 2008). Additionally, similar phenotypes are found in single and multiple mutants in LATERAL ORGAN BOUNDARIES-DOMAIN/ASYMMETRIC LEAVES2-LIKE (LBD) genes encoding transcription factors, such as lbd29 (Porco et al., 2016), lbd16 lbd18 double and lbd16 labd18 lbd29 triple mutants (Feng et al., 2012; Lee et al., 2015), which are targets of auxin signaling through AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 transcription factors (Okushima et al., 2007). These phenotypes are explained by the finding that LBDs directly or indirectly promote expression of auxin influx carriers (Feng et al., 2012; Lee et al., 2015; Porco et al., 2016).
In the pin1 pin3 pin4 triple mutant, treatment with exogenous auxin results in a massive division of pericycle cells and formation of a multilayered pericycle without a defined LRP structure, a phenotype similar to that of wild-type (Wt) seedlings treated with auxin transport inhibitor (Benková et al., 2003). Similarly, the role of auxin in correct cell division orientation during LRP morphogenesis is shown in pin2 pin3 pin7 triple mutants that form fused LRs (Laskowski et al., 2008). S. lycopersicum DIAGEOTROPICA (DGT) encodes Cyclophilin A, which negatively regulates PIN protein localization and thereby affects both LRP initiation and morphogenesis (Ivanchenko et al., 2015). When pericycle cell proliferation is induced by auxin treatment in a dgt mutant, massive cell proliferation without formation of a recognizable primordium is observed (Ivanchenko et al., 2006). Auxin response restriction to the central apical domain of the LRP depends on the APETALA2-class transcription factor PLETHORA (PLT) genes (Du and Scheres, 2017b), whose expression in turn depends on AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19 (Hofhuis et al., 2013). In the plt3 plt5 plt7 triple mutant, DR5 activity is more diffuse and PIN1 and PIN3 expression is low or absent. In addition, due to abolishment of the auxin gradient in the mutant, periclinal cell divisions in LRPs are delayed or absent from StII onwards (Du and Scheres, 2017b). This work underlines the importance of auxin in LRP morphogenesis and cell division orientation. In line with this, it was shown that the correct orientation of pericycle cell divisions leading to LRP formation is abolished when the adjacent endodermal cell is ablated, but it is restored when exogenous auxin is added (Marhavý et al., 2016).
The processes of LRP initiation and LRP morphogenesis are linked, as the first cell division of founder cells triggers a new developmental program that permits de novo organ formation (Dubrovsky et al., 2008). This triggering is dependent on ARF7, ARF19, and INDOLE-3 ACETIC ACID 14 (IAA14)/SOLITARY-ROOT, the latter of which represses ARFs (Fukaki et al., 2005). Importantly, in the lateral rootless mutant solitary root (slr)/iaa14 (Fukaki et al., 2002) and double mutant arf7 arf19 (Okushima et al., 2005; Wilmoth et al., 2005), LRP initiation is almost completely abolished, although some StI but no StII LRPs still form. This phenotype cannot be rescued by the application of exogenous auxin (Fukaki et al., 2002; Okushima et al., 2005; Wilmoth et al., 2005).
Overexpression of CYCLIND3;1 in the slr mutant background promotes cell proliferation in pericycle cells but no organized LRPs are formed beyond StI (Vanneste et al., 2005; De Smet et al., 2010). This LRP arrest is bypassed when the slr CYCD3;1OE line is treated with exogenous auxin. The treatment re-establishes normal LRP morphogenesis, which is accompanied by restoration of ARABIDOPSIS CRINKLY4 (ACR4) and PLT3 expression, which are downregulated in slr- CYCD3;1OE (De Smet et al., 2010). Therefore, activation of cell proliferation is necessary but not sufficient to trigger LRP morphogenesis, and both correct auxin signaling and cell proliferation are required for normal LRP morphogenesis (Figure 5). It seems that auxin is linked to cell proliferation in this context through an F-box protein S-Phase Kinase-Associated Protein 2A (SKP2A) (Jurado et al., 2010).
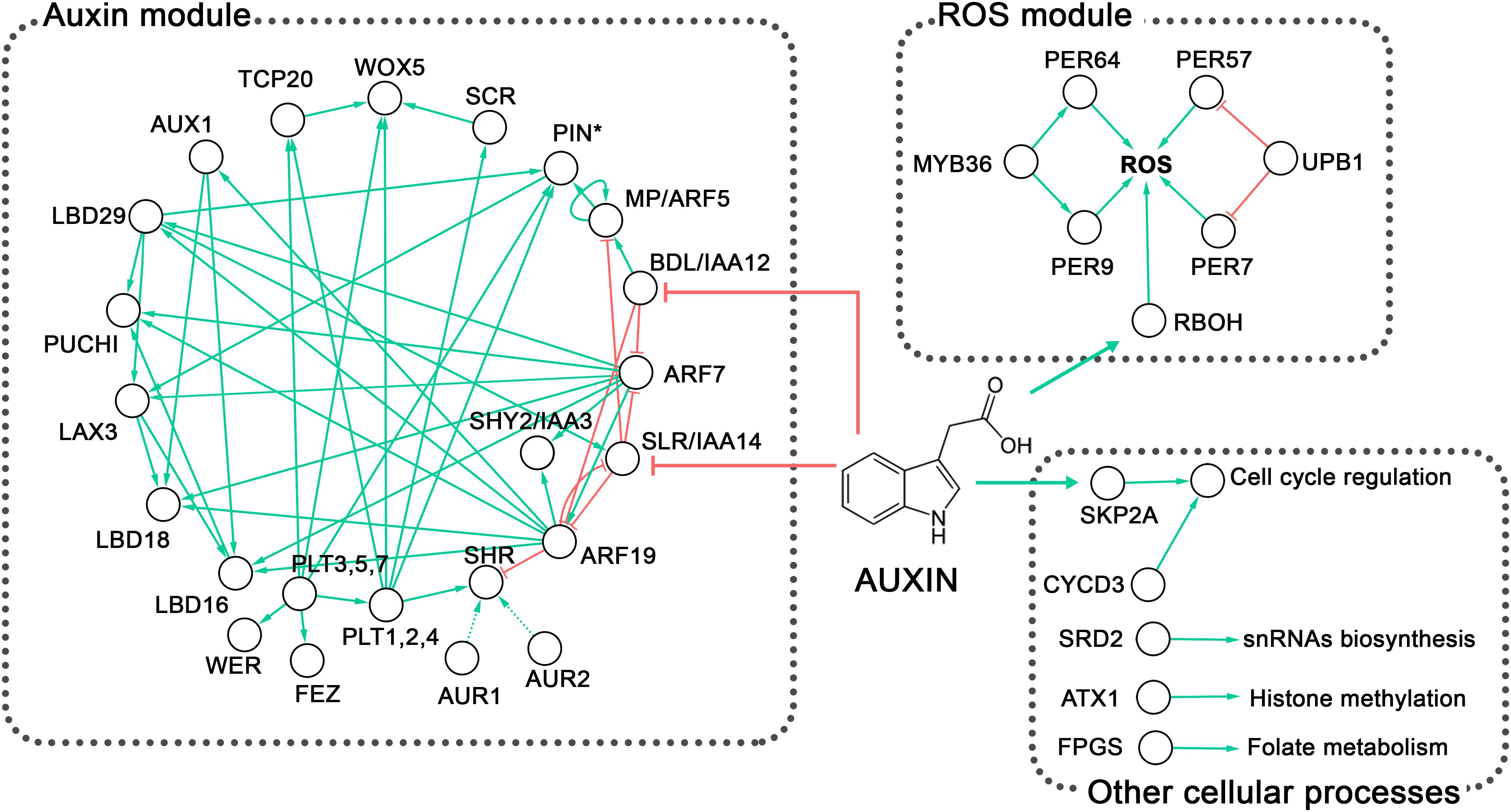
Figure 5. Overview of the genetic control of lateral root primordium (LRP) morphogenesis in Arabidopsis. Two well-defined regulatory modules are observed, the auxin regulatory and the reactive oxygen species (ROS) homeostasis modules (discussed in the text). The first module involving ARFs, IAAs, AUX/LAX and PIN proteins (PIN∗) also includes the PLT, PUCHI, FEZ, and WER transcription factors; the GRAS transcription cofactors SCR and SHR; and LBD proteins. The Ser/Thr kinases AUR1 and AUR2 are highly redundant and AlphaScreen assays showed that AUR1 interacts with SHR (Takagi et al., 2016); however, this interaction has not been proven in planta and hence is depicted with a discontinuous line. The second module includes the transcription factors MYB36 and UPB1, which control the expression of PER genes. Peroxidases together with RBOH control ROS homeostasis. Other cellular processes mentioned in the text and involved in LRP formation, such as the cell cycle, snRNA biosynthesis, histone methylation, and folate metabolism, are also depicted. Although particular genes involved in the regulation of these processes have been shown to affect LRP morphogenesis, their interactions with larger gene regulatory modules await to be discovered. Green arrows indicate activation or positive interaction; red lines with blunt ends indicate downregulation. The network was built from literature mining and visualized in Cytoscape v3.2.1 (Shannon et al., 2003). Complete names of the proteins are provided in the text.
Another important auxin-signaling module involved in LRP morphogenesis was revealed based on phenotypic analysis of the bodenlos (bdl)/iaa12 and monopteros/arf5 mutants (De Smet et al., 2010). In these mutants, a multilayered pericycle and fused LRPs show clear abnormalities in morphogenesis (Figure 6), demonstrating again that auxin signaling is of paramount importance for organized LRP development. Similarly, in the triple mutant affected in proteins involved in auxin perception, TRANSPORT INHIBITOR RESPONSE1 (TIR1), AUXIN SIGNALING F-BOX2 (AFB2), and AFB3 (Dharmasiri et al., 2005; Kepinski and Leyser, 2005), LRP morphogenesis turns out to be abnormal, resulting either in a multilayered pericycle or unusually wide LRPs (Figure 6) (Dubrovsky et al., 2011).

Figure 6. Categories of lateral root primordium phenotype abnormalities in different Arabidopsis mutants. See text for details.
Cytokinins (CKs) are likewise important for LRP morphogenesis. CK biosynthesis takes place in the developing LRPs as shown using the PHOSPHATES-ISOPENTENYL TRANSFERASE pIPT5::GFP reporter (Takei et al., 2004). CKs are negative regulators of LR development (Li et al., 2006; Laplaze et al., 2007; Marhavý et al., 2011; Bielach et al., 2012). Specifically, CKs inhibit LRP initiation or some of the anticlinal divisions in StI LRPs (Li et al., 2006; Laplaze et al., 2007; Bielach et al., 2012). Later stages of LRP development are also inhibited, but they are less sensitive to CKs and, for this reason, time from initiation to emergence is increased (Li et al., 2006; Laplaze et al., 2007; Bielach et al., 2012). CK-treated plants show abnormal patterns of cell division throughout LRP morphogenesis. During the early stages, a series of periclinal divisions in external and internal layers occurs out of sequence (Laplaze et al., 2007). These irregularities at the cellular level result in a flattened LRP (Laplaze et al., 2007).
Cytokinins affect the auxin response maximum in developing LRPs, diffusing or abolishing it (Laplaze et al., 2007; Marhavý et al., 2011) through downregulation of PIN expression (Laplaze et al., 2007). From StIII onwards, low concentrations of exogenous N6-benzyladenine (a synthetic cytokinin) promote depletion of PIN1 in the plasma membrane to a greater extent at anticlinal cell walls than at periclinal ones (Marhavý et al., 2014). In this manner, cytokinin modulates the polarity of PIN1, allowing auxin to flow toward the LRP central apical domain (Figure 2) (Marhavý et al., 2014). In support of this notion, in Wt, PIN1 is predominantly found at periclinal cell walls, while in CK receptor mutants, PIN1 becomes localized at anticlinal walls (Marhavý et al., 2014). This CK-dependent redistribution of PIN1 in the mutants does not permit maintenance of the same number of cells in the external layer of the StIV LRP (Marhavý et al., 2014). This analysis clearly demonstrates that both auxins and CKs are involved in maintaining LRP morphogenesis and that crosstalk between these hormones is essential at all LRP developmental stages.
The role of other hormones in LRP morphogenesis is unclear. Ethylene promotes LR emergence through its effect on cell proliferation, at least in the outer LRP layers (Ivanchenko et al., 2008), but the mechanism has not been addressed. Abscisic acid (ABA) inhibits the emergence of LRPs and promotes its dormancy (De Smet et al., 2006b; Fukaki and Tasaka, 2009). In the parent root, a fraction of arrested or slowly developing LRPs is frequently found (Dubrovsky et al., 2006; Napsucialy-Mendivil and Dubrovsky, 2018). The most plausible scenario is that ABA inhibits cell proliferation in the developing LRP, keeping it ‘dormant,’ but this is yet to be shown.
Knowledge of the role of brassinosteroids and gibberellins (GA) in LRP morphogenesis is fragmentary (Fukaki and Tasaka, 2009). In poplar (Populus sp.), GA negatively regulates LRP initiation (Gou et al., 2010), but its role in LRP formation is unknown. The Gibberellic Acid Stimulated-Like (GAST-like) gene family, regulated by GA, is suggested to be involved in LRP development in rice (Oryza sativa) and Z. mays, but its exact role has yet to be established (Zimmermann et al., 2010). Nitric oxide was recently considered to be a phytohormone (Santner and Estelle, 2009). It inhibits LRP initiation but does not affect LR emergence (Lira-Ruan et al., 2013); however, its role in LRP morphogenesis is unknown. Many—if not all—of the hormone signaling pathways converge at one point or another. These interactions can potentially influence various aspects of LRP morphogenesis and further investigation is needed to address this complex cross-talk.
Even from early studies, it was known that developing LRPs experience mechanical stress imposed by the overlaying tissues (Pond, 1908). Additionally, the LRP is influenced by external factors such as substrate particles, soil compaction, and parent root curvatures. Roots in soil frequently meet mechanical barriers. The internal and external mechanical forces affect LRP morphogenesis, and we will briefly review what is known in this respect.
When roots were grown in beds of glass spheres, roots curved and LR initiation occurred on the external (convex) root side (Goss, 1977; Goss and Russell, 1980). Moreover, when a root is bent during the gravitropic response, or after permanent or transient manual bending that can be as short as 20 s, the LR is also formed on the convex root side (Ditengou et al., 2008; Laskowski et al., 2008; Lucas et al., 2008a,b; Richter et al., 2009; Kircher and Schopfer, 2016). At the bending site, auxin induces AUX1 expression within the root stele of the young differentiation zone, which together with PIN protein reorientation promotes increased auxin transport toward the convex side of the root, creating a positive feedback loop that results in greater auxin levels and eventually in LR initiation (Ditengou et al., 2008; Laskowski et al., 2008).
The possibility of inducing LR initiation at the desired time and place provides a useful experimental system for studying LRP morphogenesis. With this approach, it has been shown that shootward (basipetal) transport of solutes is important for LR initiation and LRP morphogenesis. After root apical meristem removal and experimental root bending (Ditengou et al., 2008) or only after bending (Lucas et al., 2008b), the LRP develops faster on the convex side compared to intact roots. Interestingly, manual root bending promotes LRP initiation in slr/iaa14 and arf7 arf19 mutants and they become capable of proceeding with LRP morphogenesis, though in the latter mutant only LRPs but not LRs are formed (Ditengou et al., 2008). Similarly, the ability of other auxin-related mutants to form LRPs and proceed with morphogenesis increases significantly after root bending (Richter et al., 2009). Specifically, these mutants are those affected in AUX1, AUXIN RESISTANT4 (AXR4) encoding an accessory protein involved in correct localization of AUX1 (Dharmasiri et al., 2006), and TRANSPORT INHIBITOR RESPONSE1 (TIR1) encoding the F-box protein auxin receptor (Tan et al., 2007). Overall, these experiments show a clear link between root bending, LR initiation, auxin accumulation at the convex root side, and the rate of LRP formation. The reasons for accelerated LRP formation in bent roots have not yet been addressed. A potential increase in auxin content during LR initiation could impact posterior development of the LRP. An increase of cytosolic Ca2+ in the pericycle on the convex side of bent root (Richter et al., 2009) could also impact LRP morphogenesis. Finally, mechanical forces affect cell wall properties in the root portion from which the LRP is emerging.
Cell wall remodeling and cell separation in tissues overlying the LRP are well documented and required for LRP protrusion. These processes depend on auxin signaling. Cell wall remodeling enzymes encoded by PECTATE LYASE1 (PLA1) and PLA2 are active during LRP protrusion (Laskowski et al., 2006). During this process, expression of EXPANSIN14 (EXPA14) (Lee et al., 2013) and EXPA17 (Lee and Kim, 2013), which encode cell wall remodeling proteins, is activated by LBD18 transcription factor in response to auxin. A loss-of-function mutant lbd18 is significantly affected in the progression of LRP morphogenesis, resulting in delayed LR emergence (Lee and Kim, 2013; Lee et al., 2013). This demonstrates an auxin signaling-dependent crosstalk in the tissues overlying the developing LRP. A similar delayed LR emergence phenotype is found in the loss-of-function auxin influx carrier mutant lax3 (Swarup et al., 2008). LAX3 is expressed in the cortex and epidermis overlying the LRP and is involved in regulating the expression of AUXIN INDUCED IN ROOT3 (AIR3), encoding a subtilisin-like protease (Neuteboom et al., 1999). Furthermore, POLYGALACTURONASES (PG) and a XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 6 (XTR6) were reported to be regulated by a signaling pathway mediated by ARF7, ARF19-IAA14 and LAX3 and by the peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) through the leucine-rich repeat receptor-like kinases HAESA (HAE) and HAESA-LIKE2 (HSL2) (Swarup et al., 2008; Kumpf et al., 2013). Consequently, mutations in IDA, HAE, and HSL2 result in both delayed LR emergence and flattened LRPs (Kumpf et al., 2013).
A more severe phenotype of no LR emergence and flattened LRPs is found in auxin-treated plants carrying the stabilized variant of SHY2/IAA3 expressed from the endodermis-specific promoter of CASPARIAN STRIP DOMAIN PROTEIN (CASP1) (Vermeer et al., 2014). In these transgenic lines, endodermal cells overlying the LRP were unable to decrease their turgor pressure and consequently the cell volume to permit LRP protrusion. The authors also showed that SHY2 activity in the endodermis and not in the cortex and epidermis is important for LR emergence (Vermeer et al., 2014). Furthermore, the aquaporin water channel PLASMA MEMBRANE INTRINSIC PROTEINS (PIPs) have essential roles in the turgor pressure control of both the LRP and the overlying cells (Péret et al., 2012a). As expected, in pip2 mutants, LRPs are flattened and LRs emerge at a slower rate (Péret et al., 2012a).
These data collectively demonstrate the importance of auxin signaling in the control of cell wall remodeling and turgor pressure in the overlying tissues that impact LRP morphogenesis and LR emergence. However, the mechanism by which the mechanical forces are perceived, the respective signal transduction pathways, the modes of communication between overlying tissues and LRP cells, and the morphogenesis mechanisms dependent on these factors, are still to be discovered.
In vascular plants, three main tissue systems form: dermal, ground, and vascular tissues, originating from the protoderm, periblem, and plerome histogens, respectively (Hanstein, 1870; Esau, 1977; Evert, 2006). The idea that cell types acquire their identity in the very early stages of LRP formation was first deduced from an anatomical retrospective analysis. Going back from advanced LRP stages, when cell types can be recognized, to earlier stages, Von Guttenberg (1960) proposed that the first periclinal division in StI primordium cells forms two layers, the internal and external layer, with different developmental fates. These layers are marked on Figure 1 as IL and OL following Malamy and Benfey (1997). The internal layer is already specified at this early stage as plerome, giving rise to the future vascular cylinder or stele. Next, the external layer of StII LRPs (OL) undergoes a second periclinal division that gives rise to two outer layers (the external OL1 and the internal outer OL2), again with different cell fates. The protoderm (prospective epidermis) and the root cap are specified from OL1 layer, and OL2 gives rise to the periblem (prospective ground tissue; Von Guttenberg, 1960). Thus, the main cell types can be recognized already in StIII LRPs. It seems that this holds true in different taxa (Von Guttenberg, 1960; Voronkina, 1978).
Remarkably, although this conclusion was based on anatomical studies, it was later confirmed using cell type-specific marker lines of Arabidopsis (Malamy and Benfey, 1997). Figure 1 illustrates the sequence of the cell type identity acquisitions in the developing LRP based on cell type reporters. It was reported that the enhancer trap markers for different cell types are expressed similarly in the primary and lateral roots. The stele-specific marker SHORT-ROOT (SHR) is transcribed in the stele of the primary root (Helariutta, 2000), and also in the internal cell layer of the StII LRP (IL), confirming its vascular (stele) identity (Tian et al., 2014; Goh et al., 2016; Du and Scheres, 2017b). The endodermis-specific SCARECROW (SCR; Di Laurenzio et al., 1996) is expressed in the external layer of the StII LRP (OL), confirming its endodermal identity (Tian et al., 2014; Goh et al., 2016; Du and Scheres, 2017b). After the second periclinal division in OL of StII LRP, both resulting outer layers (OL1 and OL2) maintain endodermal identity, as confirmed by pSCR::GFP:SCR expression (Goh et al., 2016). When the third periclinal division occurrs in the StIII LRP, it takes place in the innermost layer (IL). As evidenced by pSHR::SHR:GFP expression, both new layers maintain the stele identity (Goh et al., 2016). At this stage (StIV), the endormal cell indentity becomes restricted to the second outer layer (OL2).
When the internal layer of the StIII LRP (IL) divides periclinally, it forms the most internal (IL1) layer, presumably giving rise to pericycle, and the second layer (IL2), giving rise to other provascular tissues of the LRP (Malamy and Benfey, 1997). However, no pericycle-specific markers have been shown to be expressed within the central domains of an LRP. At stages V and VI, the external cells of the central domain of the LRP acquire root cap and epidermis identities, as evidenced by the promoter activity of the NAC domain transcription factor FEZ (Willemsen et al., 2008; Du and Scheres, 2017b) and of the MYB-related transcription factor WEREWOLF (Lee and Schiefelbein, 1999; Du and Scheres, 2017b), respectively. Therefore, the use of cell type-specific marker lines substantially enhanced our understanding of LRP morphogenesis and revealed that practically all meristematic cell type identitites are acquired before LR emergence (Malamy and Benfey, 1997; Du and Scheres, 2017b). These studies clearly demonstrate that differential gene expression involved in cell type identity acquisition starts very early in LRP morphogenesis.
A GFP reporter of the enhancer trap line J0121 is expressed specifically in the protoxylem-adjacent pericycle of the parent root. J0121 GFP expression was detected in all layers of the developing LRP from StI to StIII (Laplaze et al., 2005; Dubrovsky et al., 2006). This suggests that the early LRP cells posess a mixed identity. In the J0121 line, GFP is not expressed in the root apical meristem, but it is expressed throughout the elongation and differentiation zones of the parent root. Importantly, starting from StIV, GFP expression in the J0121 line is excluded from the central domains of the LRP and is maintained in the flanking domains until LR emergence, suggesting that the central and flanking domains have different developmental fates. Furthermore, this pattern suggests that, starting from StIV, the central LRP domain acquires features of a root apical meristem, as it no longer expresses J0121 GFP. This observation is in line with the fact that, from StIV onwards, the LRP becomes less dependent on the parent root (Laskowski et al., 1995), which could be related to the beginning of autonomous auxin synthesis in the LRP. Additionally, a gene regulatory network analysis showed that the formation of the central and flanking domains is controlled by two distinct gene clusters (Lavenus et al., 2015).
As discussed, the first periclinal divisions are developmentally asymmetric (Scheres and Benfey, 1999), i.e., different developmental fates are aquired by the daughter cells. Indeed, each cell type origin is dependent on these asymmetric divisions and we still do not understand how these are regulated. We do not yet know how early pericycle, xylem, phloem, and vascular parenchyma cell type identities are acquired during LRP morphogenesis. Cell type identity acquisition studies with the aid of cell type reporters are limited to Arabidopsis. Extension to other angiosperms is needed.
The QC in roots is a population of slowly cycling cells that gives rise to all the cells of the apical meristem and serves as an organizing center with stem cell properties (Clowes, 1954, 1975; Barlow, 1997; Bennett and Scheres, 2010; Dubrovsky and Barlow, 2015). In some species, such as Malva sylvestris (Byrne, 1973) and V. faba (MacLeod and McLachlan, 1974; MacLeod, 1977), the QC is established after LR emergence. In other species, such as Eichornia, Pistia (Clowes, 1958), and Arabidopsis (Goh et al., 2016), the QC is established during LRP morphogenesis. In yet other species, e.g., Z. mays (Clowes, 1978b), the QC can be established either before or after LR emergence. The establishment of a functional QC requires a critical mass of proliferating cells within a developing LRP. In Z. mays, cell proliferation in the CLS of endodermal origin is essential for QC establishment (Clowes, 1978b). This early-established QC vanishes when the CLS cells are sloughed off. Soon after, the new root cap initial cells of pericyclic origin are produced and simultaneously a new cap and QC are established (Clowes, 1978b).
In Arabidopsis, the WUSCHEL-RELATED HOMEOBOX 5 (WOX5) gene is specifically expressed in the QC (Sarkar et al., 2007). The pWOX5::GFP reporter is expressed already at StI (Ditengou et al., 2008), when LRP initiation is triggered by a primary root bending, or at StII in intact roots (Tian et al., 2014; Du and Scheres, 2017b; Shimotohno et al., 2018). This expression pattern, which resembles the expression of WOX5 soon after the onset of embryogenesis (Sarkar et al., 2007), suggests that the cell lineage that will lead to QC establishment is specified early in root development. Moreover, the possibility exists that correct WOX5 expression is required and sufficient to define the central domain of the LRP and could provide a hallmark for the neigboring domains (Figure 2). Another QC marker, QC25 (Sabatini et al., 2003; ten Hove et al., 2010), starts to be expressed in the OL2 of StIV LRPs, when the cycle time increases by 70% in that layer compared to that in the most external layer, OL1 (Goh et al., 2016). QC establishment depends on SCR, as in the scr loss-of-function mutant, the specification of QC identity—as monitored by the pWOX5::n3GFP reporter—does not take place in the OL2 LRP layer but in the more internal layers that have stele identity (Goh et al., 2016). QC identity is completely lost in the plt3 plt5 plt7 mutant, underlying the importance of these transcription factors in QC establishment (Du and Scheres, 2017b). PLT1 or PLT3 and SCR form a protein complex mediated by TCP20/21 (a plant-specific Teosinte-branched-Cycloidea PCNA [Proliferating cell nuclear antigen]) transcription factor that binds to the WOX5 promoter, and this complex is important for QC specification and LRP morphogenesis (Shimotohno et al., 2018). Triple mutants in genes encoding members of this complex show LRP morphogenesis abnormalities in both the central and flanking domains (Shimotohno et al., 2018). This phenotype reveals a tight link between cell type identity acquisition and morphogenetic processes. The observation that the QC is established before LR emergence suggests that LRP morphogenesis culminates with a new root apical meristem that becomes functional post-emergence. An open question is how the stem cell identiy is acquired during LRP morphogenesis and what other factors are involved in this process. Again, our knowledge of this process for species other than Arabidopsis is limited.
A morphogenetic process must be considered from a 3D perspective. How gene regulatory networks define an organized structure is a central question for understanding morphogenesis. The 3D structure during embryogenesis was first acquired and fixed in evolution starting from gametophore development in bryophytes. In this basal land plant, rotation in the orientation of the cell division plane in stem cells permitted 3D morphogenesis (Harrison, 2017), an evolutionary novelty involving the CLAVATA (CLV) signaling pathway (Whitewoods et al., 2018). Time-lapse (4D) analyses of the genetic control of LRP morphogenesis are only beginning to be possible (Lucas et al., 2013; Vermeer et al., 2014; Goh et al., 2016; von Wangenheim et al., 2016) and few detailed studies of mutants affected in LRP morphogenesis have been performed. To examine the genetic control of LRP morphogenesis (Figure 5), we consider mutants that exhibit abnormal LRP formation and attempt to discern which morphogenetic processes are affected.
Some of the first mutants reported to be affected in LRP morphogenesis were shoot redifferentiation defective (srd2), root initiation defective (rid4), and root redifferentiation (rrd1 and rrd2) (Yasutani et al., 1994; Konishi and Sugiyama, 2003; Sugiyama, 2003). These mutants were isolated in a temperature-dependent mutant screen aimed at identifying genes involved in root development that could be essential, and thus conditionally lethal if absent, during early developmental stages. In the srd2 mutant, time from LRP initiation to LR emergence is significantly increased (Ohtani and Sugiyama, 2005; Ohtani et al., 2010). Furthermore, from StV onwards, LRP morphogenesis becomes abnormal. The altered cell division pattern results in the formation of an LRP with a wider dome than in the Wt (Ohtani et al., 2010) due to increased length and thickness of the flanking domains and a decrease in the apical–basal axis length (see the respective domains in Figure 2). This srd2 phenotype is related to the lack of auxin maximum establishment in the apical domain of the LRP in the mutant, which results from downregulation of the PIN proteins (Ohtani et al., 2010). SRD2 encodes a nuclear protein that shares sequence similarity with human SOLUBLE NSF ATTACHMENT PROTEIN 50 (SNAP50), a subunit of the SNAPc multiprotein complex required for small nuclear RNA transcription (Ohtani and Sugiyama, 2005; Ohtani et al., 2010). The precise mechanism of SRD2 action in LRP morphogenesis is unknown.
The phenotype of the rrd1, rrd2, and rid4 mutants during the LRP pre-emergence stages is similar to that described for srd2. Surprisingly, the post-emergence LR phenotype in these mutants is different from that of srd2. While in srd2, the LRs are globular without pronounced growth, those of the rrd1, rrd2, and rid4 mutants do grow, but produce fasciated (fused) roots (Konishi and Sugiyama, 2003; Sugiyama, 2003). During abnormal LRP development, the internal LRP cells form the prospective stele, which appears as a fusion of two adjacent LRPs. This was shown using pSHR:GFP and pPIN1:PIN1-GFP reporters. At the same time, pSCR:GFP expression revealed that the single external layer corresponding to the developing endodermis encloses the fused steles of the developing LRP (Otsuka and Sugiyama, 2012). The molecular function of genes affected in these mutants remains to be identified. The abnormal cell division pattern, which is more evident when seedlings are transferred from permissive to restrictive temperatures during the earlier stages of LRP morphogenesis, is a common feature of the mutants (Otsuka and Sugiyama, 2012). Furthermore, in the non-temperature-dependent atx1 mutant affected in a H3K4-methyltransferase, similar LRP phenotypes (wider LRP and fasciated primordia) were found (Napsucialy-Mendivil et al., 2014). Abnormalities in cell division pattern and proliferation in the LRP at different developmental stages are also reported in the shr (Lucas et al., 2011) and folylpolyglutamate synthetase1 (fpgs1) mutants (Reyes-Hernández et al., 2014). LRP morphogenesis in the srd2, rrd1, rrd2, rid4, atx1, shr, and fpgs1 mutants shows that correct cell division patterns and proliferation are critical factors in sustaining normal LRP development. In most of these cases, the abnormalities are equally distributed throughout all the LRP domains.
Lateral root primordium morphogenesis could also be affected either at specific stages or in specific LRP domains. Thus, in the puchi mutant, anticlinal and periclinal divisions are supernumerary in both the central and flanking domains, resulting in an increased thickness of the flanking domain and an overall flatter LRP at later stages of development (Hirota et al., 2007). PUCHI encodes an AP2/EREBP transcription factor that acts downstream of auxin signaling and is most strongly expressed in the flanking and basal domains (Hirota et al., 2007). Thus, PUCHI is apparently required to limit the extent of cell proliferation in the flanking domain and to restrict it to the central domain during LRP morphogenesis (Hirota et al., 2007). Similarly, myb36 mutant LRPs are flatter than those of the Wt and, from StIV onwards, a defect in the transition from the flat to dome-shaped LRP is observed (Fernández-Marcos et al., 2017). More cells along the central basal and flanking domains of the myb36 LRP are produced, resulting in wider LRPs than in the Wt (Fernández-Marcos et al., 2017). MYB36 is a transcription factor expressed in the LRP from StV onwards and is restricted to the central basal and flanking domains, where it controls the expression of peroxidases PER9 and PER64 (Fernández-Marcos et al., 2017). Thus, MYB36 apparently regulates LRP width through limiting cell proliferation mediated by changes in reactive oxygen species (ROS) balance.
In addition to the genetic control of morphogenesis at specific times and places, another aspect of morphogenesis is the control of cell division patterns and the orientation of the cell division plate. In the aurora (aur)1 aur2 double mutant, oblique or irregularly shaped divisions take place during StI, after the first 2–3 anticlinal divisions. Therefore, the typical layered structure of the LRP is not formed (Van Damme et al., 2011). AUR1 and AUR2 encode Ser/Thr kinases that phosphorylate Ser 10 of Histone H3 during mitosis (Demidov et al., 2005, 2009; Kawabe et al., 2005). A recent in vitro study showed that AUR1 interacts and phosphorylates the SHR transcription factor; however, this interaction has not been confirmed in planta (Takagi et al., 2016). Interestingly, despite altered orientation of cell division throughout LRP development, the overall shape of the primordium is not significantly affected (Lucas et al., 2013). Nevertheless, LR emergence is substantially delayed in the aur1 aur2 mutant (Van Damme et al., 2011; Lucas et al., 2013).
Other genes controlling the cell division orientation are those encoding PLT transcription factors. The triple mutant plt3 plt5 plt7 is characterized by multiple defects in LRP formation, including irregular cell shapes, aberrant LRP morphology, and a lack of layered LRP structure (Hofhuis et al., 2013; Du and Scheres, 2017b). PLT1, PLT2, and PLT4 are expressed at later stages during LRP development and their expression depends on the expression of PLT3, PLT5, and PLT7, which are expressed from StI onward. Therefore, the plt3 plt5 plt7 mutant is affected in these six PLT genes, highlighting their importance in LRP morphogenesis. An overview of the genetic control of LRP morphogenesis is presented in Figure 5, while categories of the outlined abnormal LRP phenotypes are shown in Figure 6.
Together with genetic analysis, transcriptomic approaches (Brady et al., 2007; Moreno-Risueno et al., 2010) have also been used recently to analyze the genetic control of LRP morphogenesis. Together, these two approaches permit construction of gene regulatory networks and thus contribute to the identification of genes involved in LR development (Lavenus et al., 2015; Voß et al., 2015). Recent papers report how the architecture of gene regulatory networks changes during LRP formation. In one such study, the gravistimulation-induced LRP system (Lucas et al., 2008a) was implemented and time-series expression data sets collected starting from LRP initiation. In this way, two mutually exclusive gene clusters regulated by auxin were identified that act in non-overlapping central and flanking domains of the LRP (Lavenus et al., 2015). One cluster involves regulation by ARF7 and ARF19 and first acts in both domains, but soon becomes restricted to the flanking domain, where it is maintained until LR emergence. The second cluster is regulated by MP, ARF6, and ARF8 and acts in the central LRP domain (Lavenus et al., 2015). This analysis confirmed the importance of previously known genes involved in LRP morphogenesis and allowed the identification of new gene regulatory network nodes that potentially participate in LRP morphogenesis (Lavenus et al., 2015). Experimental validation of the identified genes will settle their particular roles in this process.
A transcriptomic approach was also used to identify new genes involved in LRP formation. For this, cell sorting of roots expressing the pSKP2B:GFP reporter, which is active at all LRP stages (Manzano et al., 2012), was performed and used to identify SKP2B-coexpressed genes (Manzano et al., 2014). This analysis revealed genes involved in ROS signaling, among others. One of these genes, UPBEAT1 (UPB1), encodes a bHLH transcription factor and is expressed in the flanking LRP domain (Manzano et al., 2014). UPB1 regulates the expression of a subset of PEROXIDASE (PER) genes involved in maintaining the ROS balance (Tsukagoshi et al., 2010; Manzano et al., 2014). LR emergence is significantly delayed in per7 and per57 loss-of-function mutants and is promoted in the PER7 overexpression line, suggesting the importance of ROS in LRP morphogenesis through UPB1-mediated signaling (Manzano et al., 2014). In line with these studies, RESPIRATORY BURST OXIDASE HOMOLOGS (RBOH) NADPH oxidases that produce extracellular ROS are also involved in LRP development (Manzano et al., 2014; Orman-Ligeza et al., 2016). Interestingly, nitroblue tetrazolium (NBT) staining, employed for the localization of superoxide, was detected in the central domain, but staining was absent or much lower in the flanking LRP domains (Manzano et al., 2014). In double and triple rboh mutants, LR emergence was also delayed, while it was accelerated in RBOHD-overexpressor lines (Orman-Ligeza et al., 2016). Collectively, these studies demonstrate that ROS promote progression of LRP formation and that redox state is important for LRP morphogenesis, even though it is not known which specific morphogenetic processes are involved. Overall, transcriptomic approaches permit efficient identification of many new players involved in LRP formation and further studies should clarify their roles.
A better understanding of LRP morphogenesis is important for both basic and applied science. Correct root primordium morphogenesis is the foundation of a healthy root system and appropriate root architecture. Abnormal primordium morphogenesis is a characteristic of rhizomania disease of sugar beet (Beta vulgaris) (D’ambra et al., 1972; Pollini and Giunchedi, 1989). This disease is induced by the beet necrotic yellow vein virus (BNYVV) and causes supernumerary LR formation on the taproot, leading to a dramatic decrease of root mass and yield. The viral P25 virulence factor mimics auxin action by deregulating BvAUX28. As a result, some root-specific LBD transcription factors and EXPANSINS are upregulated, which in turn promote uncontrolled LR formation (Gil et al., 2018) and probably cause abnormal morphogenesis. Early processes of LR development affected by this and other root diseases are underexplored, and studies of these diseases may suggest strategies to control and/or prevent abnormalities in LRP morphogenesis.
Here we outlined the main components of LRP morphogenesis in angiosperms. Each facet of LRP morphogenesis reflected in the respective sections of this review outlines specific open questions. Most data on the genetic control of LRP morphogenesis are available for Arabidopsis and can be used for comparative studies in angiosperms. Further understanding of LRP morphogenesis in crop species is needed to modulate or adjust root system architecture to specific growth conditions.
The main tendencies, important for further research in this field, are related to the development of new technologies that could be used to address the open questions. These tendencies are as follows:
(1) Addressing the genetic control of morphogenesis in mutants and Wt plants by 3D analysis in time (4D) can significantly advance our understanding of root system formation. New imaging technologies and new microscopy approaches (Ovečka et al., 2018) that could be used for this purpose are already accessible. Deciphering cell division patterns and developmental rules involved in morphogenesis (Yoshida et al., 2014; von Wangenheim et al., 2016) is an important goal.
(2) Gene regulatory networks uncover complex relationships between pathways involved in regulating different processes during LRP morphogenesis (Lavenus et al., 2015; Voß et al., 2015). Further studies of gene regulatory networks at the single cell level and implementation of plant systems biology approaches (Libault et al., 2017) will undoubtedly contribute to answering the questions of how different LRP domains and cell types are specified and maintained, how the overall shape of the developing LRP emerges, and how timing control is operated.
(3) LRPs develop under mechanical constraints imposed by the external parent root tissues. The role of mechanical forces during LRP morphogenesis was recognized in early studies, but only recently did their roles in both LR initiation (Vermeer et al., 2014) and morphogenesis (Stoeckle et al., 2018) begin to be understood. Models of auxin transport coupled to mechanical forces provide explanations for the robust morphogenesis observed in the Arabidopsis root (Romero-Arias et al., 2017) and application of these models to LRP morphogenesis should be promising. Developing new biophysical methods to monitor the mechanical properties of live cells (e.g., Elsayad et al., 2016) is challenging, but required to discern the role of the mechanical forces in LRP morphogenesis. LRP formation is closely linked to external and internal mechanical forces and the cytoskeleton (Eng and Sampathkumar, 2018), but it is unclear how the mechanical forces contribute to LRP morphogenesis.
(4) We outlined a possible role of the temporal CLS formed during LRP morphogenesis and attempted to visualize related evolutionary trends of this particular feature of LRP morphogenesis. Understanding the relationships between different facets of LRP morphogenesis (e.g., orientation of cell division, developmental rules, participation of different cell types) and how they change during evolution is challenging but feasible in the post-genomic era. Integration of different approaches from genomics and molecular to cell biology and anatomy could help reveal evo–devo relationships in LRP morphogenesis of angiosperms.
Here we reviewed the main aspects of LRP morphogenesis that have been under investigation for more than a century. Not all available information was discussed; for instance, we did not include the role of environmental factors and mineral nutrition. We hope that the historical perspective combined here with our overview of contemporary studies of LRP morphogenesis highlights key questions that will guide future research aimed at elucidating the morphogenetic processes that take place during LRP development. Such research would yield important insights into root biology and evolution, providing a framework to modulate root system architecture, root production and root adaptation to the environment in crop species.
HT-M and JD conceived the idea and designed the outlines of this review article. SS and JD conceptualized the content. HT-M and JD wrote the article. GR-A performed phylogenetic and gene regulatory network analyses. GR-A, HT-M, and JD prepared the illustrations. All authors participated in the editorial improvement of the text and approved the final manuscript.
This work was partially supported by Consejo Nacional de Ciencia y Tecnología, Mexico (CONACyT Grants 237430 to JD and 240055 to SS) and DGAPA-PAPIIT-UNAM (Grants IN200818 to JD and IN201318 to SS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A Ph.D. fellowship to HT-M from CONACyT and DGAPA-PAPIIT-UNAM and postdoctoral fellowship from CONACyT to GR-A are thankfully acknowledged. We thank the members of the Unidad de Cómputo. Instituto de Biotecnologia and S. Ainsworth for logistic help. We also thank K. Farquharson for editing the English text. We apologize to all those authors whose work was not cited due to time and space constraints.
Atkinson, J. A., Rasmussen, A., Traini, R., Voß, U., Sturrock, C. J., Mooney, S. J., et al. (2014). Branching out in roots: uncovering form, function and regulation. Plant Physiol. 166, 538–550. doi: 10.1104/pp.114.245423
Barlow, P. W. (1997). “Stem cells and founder zones in plants, particularly their roots,” in Stem Cells, ed. C. S. Potten (London: Academic Press), 29–57. doi: 10.1016/B978-012563455-7/50003-9
Beeckman, T., Burssens, S., and Inzé, D. (2001). The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52, 403–411.
Beeckman, T., and De Smet, I. (2014). Pericycle. Curr. Biol. 24, R378–R379. doi: 10.1016/j.cub.2014.03.031
Bell, J. K., and McCully, M. E. (1970). A histological study of lateral root initiation and development in Zea mays. Protoplasma 70, 179–205. doi: 10.1007/BF01276979
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Bennett, T., and Scheres, B. (2010). “Root development–Two meristems for the price of one?,” in Current Topics in Developmental Biology Plant Development, ed. C. P. T. Marja (Cambridge, MA: Academic Press), 67–102.
Besson, S., and Dumais, J. (2011). Universal rule for the symmetric division of plant cells. Proc. Natl. Acad. Sci. U.S.A. 108, 6294–6299. doi: 10.1073/pnas.1011866108
Bhalerao, R. P., Eklöf, J., Ljung, K., Marchant, A., Bennett, M., and Sandberg, G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29, 325–332. doi: 10.1046/j.0960-7412.2001.01217.x
Bielach, A., Podlešáková, K., Marhavý, P., Duclercq, J., Cuesta, C., Müller, B., et al. (2012). Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by Cytokinin. Plant Cell Online 24, 3967–3981. doi: 10.1105/tpc.112.103044
Blakely, L. M., Durham, M., Evans, T. A., and Blakely, R. M. (1982). Experimental studies on lateral root formation in radish seedling roots. I. General methods, developmental stages, and spontaneous formation of laterals. Bot. Gaz. 143, 341–352. doi: 10.1086/337308
Bonnett, H. T. (1969). Cortical cell death during lateral root formation. J. Cell Biol. 40, 144–159. doi: 10.1083/jcb.40.1.144
Brady, S. M., Orlando, D. A., Lee, J.-Y., Wang, J. Y., Koch, J., Dinneny, J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. doi: 10.1126/science.1146265
Brumos, J., Robles, L. M., Yun, J., Vu, T. C., Jackson, S., Alonso, J. M., et al. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318.e5. doi: 10.1016/j.devcel.2018.09.022
Bunning, E. (1952). Weitere Untersuchungen; uber die Differenzierungsvorgange in Wurzeln. Z. Bot. 40, 385–406.
Byrne, J. M. (1973). The root apex of malva sylvestris. III. lateral root development and the quiescent center. Am. J. Bot. 60, 657–662. doi: 10.1002/j.1537-2197.1973.tb05970.x
Byrne, J. M., Pesacreta, T. C., and Fox, J. A. (1977). Development and structure of the vascular connection between the primary and secondary root of glycine max (L.) Merr. Am. J. Bot. 64, 946–959. doi: 10.1002/j.1537-2197.1977.tb11939.x
Casero, P. J., Casimiro, I., and Lloret, P. G. (1995). Lateral root initiation by asymmetrical transverse divisions of pericycle cells in four plant species: Raphanus sativus, Helianthus annuus, Zea mays, and Daucus carota. Protoplasma 188, 49–58. doi: 10.1007/BF01276795
Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., et al. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8, 165–171. doi: 10.1016/S1360-1385(03)00051-7
Chase, M. W., Christenhusz, M., Fay, M., Byng, J., Judd, W., Soltis, D., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. doi: 10.1016/j.jep.2015.05.035
Clowes, F. A. L. (1954). The promeristem and the minimal constructional centre in grass root apices. New Phytol. 53, 108–116. doi: 10.1111/j.1469-8137.1954.tb05227.x
Clowes, F. A. L. (1958). Development of quiscent centres in root meristems. New Phytol. 57, 85–88. doi: 10.1111/j.1469-8137.1958.tb05918.x
Clowes, F. A. L. (1975). “The quiescent centre,” in Development and Function of Roots, eds J. G. Torrey and D. Clarkson (London: Academic Press), 3–19.
Clowes, F. A. L. (1978a). Chimeras and the origin of lateral root primordia in Zea mays. Ann. Bot. 42, 801–807. doi: 10.1093/oxfordjournals.aob.a085519
Clowes, F. A. L. (1978b). Origin of the quiescent centre in Zea mays. New Phytol. 80, 409–419. doi: 10.1111/j.1469-8137.1978.tb01575.x
Clowes, F. A. L. (1982). Changes in cell population kinetics in an open meristem during root growth. New Phytol. 91, 741–748. doi: 10.1111/j.1469-8137.1982.tb03353.x
D’ambra, V., Giulini, P., and Orsenigo, M. (1972). Ricerche anatomiche e istologiche sul fittone di bietole rizomani. Riv. Patol. Veg. 8, 359–372.
Danilova, M. F., and Serdyuk, E. M. (1982). Formation of the lateral roots in Hordeum vilgare (Poaceae) (The data of electron microscopy). Bot. Z. 67, 140–145.
Davidson, D., and MacLeod, R. D. (1968). Heterogeneity in cell behaviour in primordia of Vicia faba. Chromosoma 25, 470–474. doi: 10.1007/BF02327723
De Rybel, B., Vassileva, V., Parizot, B., Demeulenaere, M., Grunewald, W., Audenaert, D., et al. (2010). A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 20, 1697–1706. doi: 10.1016/j.cub.2010.09.007
De Smet, I. (2012). Lateral root initiation: one step at a time. New Phytol. 193, 867–873. doi: 10.1111/j.1469-8137.2011.03996.x
De Smet, I., and Beeckman, T. (2011). Asymmetric cell division in land plants and algae: the driving force for differentiation. Nat. Rev. Mol. Cell Biol. 12, 177–188. doi: 10.1038/nrm3064
De Smet, I., Lau, S., Voá, U., Vanneste, S., Benjamins, R., Rademacher, E. H., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 2705–2710. doi: 10.1073/pnas.0915001107
De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C. H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555. doi: 10.1046/j.1365-313X.2003.01652.x
De Smet, I., Tetsumura, T., De Rybel, B., Frei dit Frey, N., Laplaze, L., Casimiro, I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690. doi: 10.1242/dev.02753
De Smet, I., Vanneste, S., Inz, D., and Beeckman, T. (2006a). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60, 871–887.
De Smet, I., Zhang, H., Inze, D., and Beeckman, T. (2006b). A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11, 434–439.
Demchenko, K. N., and Demchenko, N. P. (1996a). Early stages of lateral root development in Triticum aestivum L. Acta Phytogeogr. Suec. 81, 71–75.
Demchenko, K. N., and Demchenko, N. P. (1996b). Lateral root initiation in seedlings of Triticum aestivum (Poaceae). Bot. Z. 81, 47–50.
Demchenko, K. N., and Demchenko, N. P. (2001). “Changes of root structure in connection with the development of lateral root primordia in wheat and pumpkins,” in Recent Advances of Plant Root Structure and Function, eds M. Čiamporová, I. Mistrík, and F. Baluška (Netherlands: Springer), 39–47.
Demchenko, K. N., Demchenko, N. P., and Danilova, M. F. (2001). Initiation and development of lateral root primordia in Trticum aestivum (Poaceae) Cucurbita pepo (Cucurbitacea) seedlings. Bot. Z. 86, 14–30.
Demidov, D., Hesse, S., Tewes, A., Rutten, T., Fuchs, J., Karimi Ashtiyani, R., et al. (2009). Aurora1 phosphorylation activity on histone H3 and its cross-talk with other post-translational histone modifications in Arabidopsis. Plant J. 59, 221–230. doi: 10.1111/j.1365-313X.2009.03861.x
Demidov, D., Van Damme, D., Geelen, D., Blattner, F. R., and Houben, A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17, 836–848. doi: 10.1105/tpc.104.029710
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Dharmasiri, S., Swarup, R., Mockaitis, K., Dharmasiri, N., Singh, S. K., Kowalchyk, M., et al. (2006). AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312, 1218–1220. doi: 10.1126/science.1122847
Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., et al. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. doi: 10.1016/S0092-8674(00)80115-4
Ditengou, F. A., Teale, W. D., Kochersperger, P., Flittner, K. A., Kneuper, I., van der Graaff, E., et al. (2008). Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 18818–18823. doi: 10.1073/pnas.0807814105
Dittmer, H. G. (1937). A quantitative study of the roots and root hairs of a winter rye plant ( Secale cereale ). Am. J. Bot. 24, 417–420. doi: 10.1002/j.1537-2197.1937.tb09121.x
Du, Y., and Scheres, B. (2017a). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. doi: 10.1093/jxb/erx223
Du, Y., and Scheres, B. (2017b). PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proc. Natl. Acad. Sci. U.S.A. 114, 11709–11714. doi: 10.1073/pnas.1714410114
Dubrovsky, J. G. (1986a). Origin of tissues of embryonic lateral root in the cucumber, tissue interactions, and positional control in development. Ontogenez 17, 176–189 (English translation from Russian appeared in Soviet Journal of Developmental Biology, New York, NY: Consultant Bureau 117, 119–128).
Dubrovsky, J. G. (1986b). Dynamics of cell reproduction and cell complexes (cell packets) in the embryonic lateral root primordium of the cucumber. Ontogenez 17, 525–534 (English translation from Russian appeared in Soviet Journal of Developmental Biology, New York, NY: Consultant Bureau, 517, 337–344).
Dubrovsky, J. G., and Barlow, P. W. (2015). The origins of the quiescent centre concept. New Phytol. 206, 493–496. doi: 10.1111/nph.13307
Dubrovsky, J. G., Doerner, P. W., Colón-Carmona, A., and Rost, T. L. (2000). Pericycle cell proliferation and lateral root initiation in Arabidopsis. Plant Physiol. 124, 1648–1657. doi: 10.1104/pp.124.4.1648
Dubrovsky, J. G., Gambetta, G. A., Hernández-Barrera, A., Shishkova, S., and González, I. (2006). Lateral root initiation in Arabidopsis: developmental window, spatial patterning, density and predictability. Ann. Bot. 97, 903–915. doi: 10.1093/aob/mcj604
Dubrovsky, J. G., and Laskowski, M. (2017). “Lateral Root Initiation,” in Encyclopedia of Applied Plant Sciences, 2nd Edn, eds B. G. Murray and D. J. Murphy (Oxford: Academic Press), 256–264. doi: 10.1016/B978-0-12-394807-6.00126-X
Dubrovsky, J. G., Napsucialy-Mendivil, S., Duclercq, J., Cheng, Y., Shishkova, S., Ivanchenko, M. G., et al. (2011). Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 191, 970–983. doi: 10.1111/j.1469-8137.2011.03757.x
Dubrovsky, J. G., and Rost, T. L. (2003). “Lateral root initiation,” in Encyclopedia of Applied Plant Sciences, eds B. Thomas, D. J. Murphy, and B. J. Murray (Oxford: Elsevier Academic Press), 1101–1107.
Dubrovsky, J. G., and Rost, T. L. (2012). Pericycle. Hoboken, NJ: John Wiley & Sons, Ltd. doi: 10.1002/9780470015902.a0002085.pub2
Dubrovsky, J. G., Rost, T. L., Colón-Carmona, A., and Doerner, P. W. (2001). Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214, 30–36. doi: 10.1007/s004250100598
Dubrovsky, J. G., Sauer, M., Napsucialy-Mendivil, S., Ivanchenko, M. G., Friml, J., Shishkova, S., et al. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. U.S.A. 105, 8790–8794. doi: 10.1073/pnas.0712307105
Elsayad, K., Werner, S., Gallemí, M., Kong, J., Guajardo, E. R. S., Zhang, L., et al. (2016). Mapping the subcellular mechanical properties of live cells in tissues with fluorescence emission–Brillouin imaging. Sci. Signal. 9:rs5. doi: 10.1126/scisignal.aaf6326
Eng, R. C., and Sampathkumar, A. (2018). Getting into shape: the mechanics behind plant morphogenesis. Curr. Opin. Plant Biol. 46, 25–31. doi: 10.1016/j.pbi.2018.07.002
Evert, R. F. (2006). Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: their Structure, Function, and Development. Hoboken, NJ: John Wiley & Sons. doi: 10.1002/0470047380
Feng, Z., Zhu, J., Du, X., and Cui, X. (2012). Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 236, 1227–1237. doi: 10.1007/s00425-012-1673-3
Fernández-Marcos, M., Desvoyes, B., Manzano, C., Liberman, L. M., Benfey, P. N., Pozo, J. C., et al. (2017). Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytol. 213, 105–112. doi: 10.1111/nph.14304
Friedberg, S., and Davidson, D. (1971). Cell population studies in developing root primordia. Ann. Bot. 35, 523–533. doi: 10.1093/oxfordjournals.aob.a084496
Fukaki, H., Nakao, Y., Okushima, Y., Theologis, A., and Tasaka, M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44, 382–395. doi: 10.1111/j.1365-313X.2005.02537.x
Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Fukaki, H., and Tasaka, M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69, 437–449. doi: 10.1007/s11103-008-9417-2
Gil, J. F., Liebe, S., Thiel, H., Lennfors, B. L., Kraft, T., Gilmer, D., et al. (2018). Massive up-regulation of LBD transcription factors and EXPANSIN s highlights the regulatory programs of rhizomania disease. Mol. Plant Pathol. 19, 2333–2348. doi: 10.1111/mpp.12702
Goh, T., Toyokura, K., Wells, D. M., Swarup, K., Yamamoto, M., Mimura, T., et al. (2016). Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143, 3363–3371. doi: 10.1242/dev.135319
Goss, M. (1977). Effects of mechanical impedance on root growth in barley (Hordeum vulgare L.) I. Effects on the elongation and branching of seminal root axes. J. Exp. Bot. 28, 96–111. doi: 10.1093/jxb/28.1.96
Goss, M., and Russell, R. S. (1980). Effects of mechanical impedance on root growth in barley (Hordeum vulgare L.) III. Observations on the mechanism of response. J. Exp. Bot. 31, 577–588. doi: 10.1093/jxb/31.2.577
Gou, J., Strauss, S. H., Tsai, C. J., Fang, K., Chen, Y., Jiang, X., et al. (2010). Gibberellins regulate lateral root formation in populus through interactions with auxin and other hormones. Plant Cell 22, 623–639. doi: 10.1105/tpc.109.073239
Gulyaev, V. A. (1964). Initiation and formation of lateral roots in some species of Cucurbitaceae family. Bot. Z. 49, 1482–1485.
Hanstein, J. (1870). Die Entwicklung des Keimes der Monocotylen und der Dikotylen. Bot. Abhandl. 1:1.
Harrison, C. J. (2017). Development and genetics in the evolution of land plant body plans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20150490. doi: 10.1098/rstb.2015.0490
Helariutta, Y. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. doi: 10.1016/S0092-8674(00)80865-X
Hinchee, M. A. W., and Rost, T. L. (1992). The control of lateral root development in cultured pea seedlings. II. Root fasciation induced by auxin inhibitors. Bot. Acta 105, 121–126. doi: 10.1111/j.1438-8677.1992.tb00276.x
Hirota, A., Kato, T., Fukaki, H., Aida, M., and Tasaka, M. (2007). The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19, 2156–2168. doi: 10.1105/tpc.107.050674
Hofhuis, H., Laskowski, M., Du, Y., Prasad, K., Grigg, S., Pinon, V., et al. (2013). Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 23, 956–962. doi: 10.1016/j.cub.2013.04.048
Hou, G., Hill, J., and Blancaflor, E. B. (2004). Developmental anatomy and auxin response of lateral root formation in Ceratopteris richardii. J. Exp. Bot. 55, 685–693. doi: 10.1093/jxb/erh068
Ilina, E. L., Kiryushkin, A. S., Semenova, V. A., Demchenko, N. P., Pawlowski, K., and Demchenko, K. N. (2018). Lateral root initiation and formation within the parental root meristem of Cucurbita pepo: is auxin a key player? Ann. Bot. 122, 873–888. doi: 10.1093/aob/mcy052
Ivanchenko, M. G., Coffeen, W. C., Lomax, T. L., and Dubrovsky, J. G. (2006). Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J. 46, 436–447. doi: 10.1111/j.1365-313X.2006.02702.x
Ivanchenko, M. G., Muday, G. K., and Dubrovsky, J. G. (2008). Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. 55, 335–347. doi: 10.1111/j.1365-313X.2008.03528.x
Ivanchenko, M. G., Zhu, J., Wang, B., Medvecká, E., Du, Y., Azzarello, E., et al. (2015). The cyclophilin A DIAGEOTROPICA gene affects auxin transport in both root and shoot to control lateral root formation. Development 142, 712–721. doi: 10.1242/dev.113225
Janczewski, E. (1874). Recherches sur le Developpement des Radicelles dans les Phanerogames. Ann. Sci. Nat. Bot. 20, 208–233.
Jansen, L., Roberts, I., De Rycke, R., and Beeckman, T. (2012). Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1525–1533. doi: 10.1098/rstb.2011.0239
Jurado, S., Abraham, Z., Manzano, C. Ý, López-Torrejón, G., Pacios, L. F., and del Pozo, J. C. (2010). The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22, 3891–3904. doi: 10.1105/tpc.110.078972
Karas, I., and McCully, M. E. (1973). Further studies of the histology of lateral root development in Zea mays. Protoplasma 77, 243–269. doi: 10.1007/BF01276762
Kawabe, A., Matsunaga, S., Nakagawa, K., Kurihara, D., Yoneda, A., Hasezawa, S., et al. (2005). Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58, 1–13. doi: 10.1007/s11103-005-3454-x
Kawata, I., and Shibayama, H. (1965). On the lateral root primordia formation in the crown roots of rice plants. Jpn. J. Crop Sci. 33, 423–431. doi: 10.1626/jcs.33.423
Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/nature03542
Kircher, S., and Schopfer, P. (2016). Priming and positioning of lateral roots in Arabidopsis. An approach for an integrating concept. J. Exp. Bot. 67, 1411–1420. doi: 10.1093/jxb/erv541
Konishi, M., and Sugiyama, M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130, 5637–5647. doi: 10.1242/dev.00794
Kumpf, R. P., Shi, C.-L., Larrieu, A., Stø, I. M., Butenko, M. A., Péret, B., et al. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. U.S.A. 110, 5235–5240. doi: 10.1073/pnas.1210835110
Laplaze, L., Benková, E., Casimiro, I., Maes, L., Vanneste, S., Swarup, R., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19, 3889–3900. doi: 10.1105/tpc.107.055863
Laplaze, L., Parizot, B., Baker, A., Ricaud, L., Martiniere, A., Auguy, F., et al. (2005). GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J. Exp. Bot. 56, 2433–2442. doi: 10.1093/jxb/eri236
Laskowski, M., Biller, S., Stanley, K., Kajstura, T., and Prusty, R. (2006). Expression profiling of auxin-treated Arabidopsis roots: toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 47, 788–792. doi: 10.1093/pcp/pcj043
Laskowski, M., Grieneisen, V. A., Hofhuis, H., Ten Hove, C. A., and Hogeweg, P. (2008). Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6:e307. doi: 10.1371/journal.pbio.0060307
Laskowski, M. J., Williams, M. E., Nusbaum, H. C., and Sussex, I. M. (1995). Formation of lateral root meristems is a two-stage process. Development 121, 3303–3310.
Lavenus, J., Goh, T., Guyomarc’h, S., Hill, K., Lucas, M., Voß, U., et al. (2015). Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. Plant Cell 27, 1368–1388. doi: 10.1105/tpc.114.132993
Lavenus, J., Goh, T., Roberts, I., Guyomarc’h, S., Lucas, M., De Smet, I., et al. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. doi: 10.1016/j.tplants.2013.04.006
Lee, H. W., Cho, C., and Kim, J. (2015). Lateral organ boundaries domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 168, 1792–1806. doi: 10.1104/pp.15.00578
Lee, H. W., and Kim, J. (2013). EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 54, 1600–1611. doi: 10.1093/pcp/pct105
Lee, H. W., Kim, M. J., Kim, N. Y., Lee, S. H., and Kim, J. (2013). LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 73, 212–224. doi: 10.1111/tpj.12013
Lee, M. M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. doi: 10.1016/S0092-8674(00)81536-6
Li, X., Mo, X., Shou, H., and Wu, P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47, 1112–1123. doi: 10.1093/pcp/pcj082
Libault, M., Pingault, L., Zogli, P., and Schiefelbein, J. (2017). Plant systems biology at the single-cell level. Trends Plant Sci. 22, 949–960. doi: 10.1016/j.tplants.2017.08.006
Lira-Ruan, V., Mendivil, S. N., and Dubrovsky, J. G. (2013). Heuristic aspect of the lateral root initiation index: a case study of the role of nitric oxide in root branching. Appl. Plant Sci. 1:1300029. doi: 10.3732/apps.1300029
Lloret, P. G., and Casero, P. J. (2002). “Lateral root initiation,” in Plant Roots: the Hidden Half, 3rd Edn, Vol. 2002, eds U. Kafkafi, Y. Waisel, and A. Eshel (Boca Raton, FL: CRC Press), 127–155. doi: 10.1201/9780203909423.ch8
Lucas, M., Godin, C., Jay-Allemand, C., and Laplaze, L. (2008a). Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J. Exp. Bot. 59, 55–66.
Lucas, M., Guédon, Y., Jay-Allemand, C., Godin, C., and Laplaze, L. (2008b). An auxin transport-based model of root branching in Arabidopsis thaliana. PLoS One 3:e3673. doi: 10.1371/journal.pone.0003673
Lucas, M., Kenobi, K., von Wangenheim, D., Voβ, U., Swarup, K., De Smet, I., et al. (2013). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc. Natl. Acad. Sci. U.S.A. 110, 5229–5234. doi: 10.1073/pnas.1210807110
Lucas, M., Swarup, R., Paponov, I. A., Swarup, K., Casimiro, I., Lake, D., et al. (2011). SHORT-ROOT regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol. 155, 384–398. doi: 10.1104/pp.110.165126
MacLeod, R. (1972). Lateral root formation in Vicia faba L. I. The development of large primordia. Chromosoma 39, 341–350. doi: 10.1007/BF00290792
MacLeod, R. (1977). Proliferating and quiescent cells in the apical meristem of elongating lateral roots of Vicia faba L. Ann. Bot. 41, 321–329. doi: 10.1093/oxfordjournals.aob.a085294
MacLeod, R., and Davidson, D. (1968). Delayed incorporation of H 3-thymidine by primordial cells. Chromosoma 24, 1–9. doi: 10.1007/BF00329602
MacLeod, R., and McLachlan, S. (1974). The development of a quiescent centre in lateral roots of Vicia faba L. Ann. Bot. 38, 535–544. doi: 10.1093/oxfordjournals.aob.a084839
MacLeod, R. D. (1973). The response of root meristems to colchicine and Indol-3yl-acetic acid in Vicia faba L. Ann. Bot. 37, 687–697. doi: 10.1093/oxfordjournals.aob.a084736
MacLeod, R. D., and Thompson, A. (1979). Development of lateral root primordia in Vicia faba, Pisum sativum, Zea mays and Phaseolus vulgaris: Rates of primordium formation and cell doubling times. Ann. Bot. 44, 435–449. doi: 10.1007/BF00384887
Malamy, J. E. (2010). “Lateral root formation,” in Root Development. Annual Plant Reviews, Vol. 37, ed. T. Beeckman (Hoboken, NJ: Blackwell Publishing), 83–126.
Malamy, J. E., and Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44.
Mallory, T. E., Chiang, S., Cutter, E. G., and Gifford, E. M. Jr. (1970). Sequence and pattern of lateral root formation in five selected species. Am. J. Bot. 57, 800–809. doi: 10.1093/jxb/erv346
Manzano, C., Pallero-Baena, M., Casimiro, I., De Rybel, B., Orman-Ligeza, B., Van Isterdael, G., et al. (2014). The emerging role of reactive oxygen species signaling during lateral root development. Plant Physiol. 165, 1105–1119. doi: 10.1104/pp.114.238873
Manzano, C., Ramirez-Parra, E., Casimiro, I., Otero, S., Desvoyes, B., De Rybel, B., et al. (2012). Auxin and epigenetic regulation of SKP2B, an F-Box that represses lateral root formation. Plant Physiol. 160, 749–762. doi: 10.1104/pp.112.198341
Marchant, A., Bhalerao, R., Casimiro, I., Eklöf, J., Casero, P. J., Bennett, M., et al. (2002). AUX1 promotes lateral root formation by facilitating Indole-3-Acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14, 589–597. doi: 10.1105/tpc.010354
Marhavý, P., Bielach, A., Abas, L., Abuzeineh, A., Duclercq, J., Tanaka, H., et al. (2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21, 796–804. doi: 10.1016/j.devcel.2011.08.014
Marhavý, P., Duclercq, J., Weller, B., Feraru, E., Bielach, A., Offringa, R., et al. (2014). Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 24, 1031–1037. doi: 10.1016/j.cub.2014.04.002
Marhavý, P., Montesinos, J. C., Abuzeineh, A., Van Damme, D., Vermeer, J. E., Duclercq, J., et al. (2016). Targeted cell elimination reveals an auxin-guided biphasic mode of lateral root initiation. Genes Dev. 30, 471–483. doi: 10.1101/gad.276964.115
McCully, M. E. (1975). “The development of lateral roots,” in The Development and Function of Roots, eds J. G. Torrey and D. T. Clarkson (Cambridge, MA: Academic Press).
Moreno-Risueno, M. A., Van Norman, J. M., Moreno, A., Zhang, J., Ahnert, S. E., and Benfey, P. N. (2010). Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311. doi: 10.1126/science.1191937
Nakajima, K., Sean, G., Nawy, T., and Benfey, P. N. (2001). Intercellular movement of the putative transcriprion factor SHR in root patterning. Nature 413, 307–311. doi: 10.1038/35095061
Napsucialy-Mendivil, S., Alvarez-Venegas, R., Shishkova, S., and Dubrovsky, J. G. (2014). ARABIDOPSIS HOMOLOG of TRITHORAX1 (ATX1) is required for cell production, patterning, and morphogenesis in root development. J. Exp. Bot. 65, 6373–6384. doi: 10.1093/jxb/eru355
Napsucialy-Mendivil, S., and Dubrovsky, J. G. (2018). Genetic and phenotypic analysis of lateral root development in Arabidopsis thaliana. Methods Mol. Biol. 1761, 47–75. doi: 10.1007/978-1-4939-7747-5_4
Neuteboom, L. W., Ng, J. M., Kuyper, M., Clijdesdale, O. R., Hooykaas, P. J., and Van Der Zaal, B. J. (1999). Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol. Biol. 39, 273–287. doi: 10.1023/A:1006104205959
O’Dell, D. H., and Foard, D. E. (1969). Presence of lateral root primordia in the radicle of buckwheat embryos. Bull. Torrey Bot. Club 96, 1–3. doi: 10.2307/2484002
Ohtani, M., Demura, T., and Sugiyama, M. (2010). Particular significance of SRD2-dependent snRNA accumulation in polarized pattern generation during lateral root development of Arabidopsis. Plant Cell Physiol. 51, 2002–2012. doi: 10.1093/pcp/pcq159
Ohtani, M., and Sugiyama, M. (2005). Involvement of SRD2-mediated activation of snRNA transcription in the control of cell proliferation competence in Arabidopsis. Plant J. 43, 479–490. doi: 10.1111/j.1365-313X.2005.02469.x
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL Genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Orman-Ligeza, B., Parizot, B., De Rycke, R., Fernandez, A., Himschoot, E., Van Breusegem, F., et al. (2016). RBOH-mediated ROS production facilitates lateral root emergence in Arabidopsis. Development 143, 3328–3339. doi: 10.1242/dev.136465
Otsuka, K., and Sugiyama, M. (2012). Tissue organization of fasciated lateral roots of Arabidopsis mutants suggestive of the robust nature of outer layer patterning. J. Plant Res. 125, 547–554. doi: 10.1007/s10265-011-0471-5
Ötvös, K., and Benková, E. (2017). Spatiotemporal mechanisms of root branching. Curr. Opin. Genet. Dev. 45, 82–89. doi: 10.1016/j.gde.2017.03.010
Ovečka, M., von Wangenheim, D., Tomančák, P., Šamajová, O., Komis, G., and Šamaj, J. (2018). Multiscale imaging of plant development by light-sheet fluorescence microscopy. Nat. Plants 4, 639–650. doi: 10.1038/s41477-018-0238-2
Péret, B., De Rybel, B., Casimiro, I., Benková, E., Swarup, R., Laplaze, L., et al. (2009). Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 14, 399–408. doi: 10.1016/j.tplants.2009.05.002
Péret, B., Li, G., Zhao, J., Band, L. R., Voß, U., Postaire, O., et al. (2012a). Auxin regulates aquaporin function to facilitate lateral root emergence. Nat. Cell Biol. 14, 991. doi: 10.1038/ncb2573
Péret, B., Swarup, K., Ferguson, A., Seth, M., Yang, Y., Dhondt, S., et al. (2012b). AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell Online 24, 2874–2885. doi: 10.1105/tpc.112.097766
Pollini, C. P., and Giunchedi, L. (1989). Comparative histopathology of sugar beets that are susceptible and partially resistant to rhizomania. Phytopathol. Mediterr. 28, 16–21.
Popham, R. A. (1955). Zonation of primary and lateral root apices af Pisum sativum. Am. J. Bot. 42, 267–273. doi: 10.1002/j.1537-2197.1955.tb11118.x
Porco, S., Larrieu, A., Du, Y., Gaudinier, A., Goh, T., Swarup, K., et al. (2016). Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulating auxin influx carrier LAX3. Development 143, 3340–3349. doi: 10.1242/dev.136283
Reyes-Hernández, B. J., Srivastava, A. C., Ugartechea-Chirino, Y., Shishkova, S., Ramos-Parra, P. A., Lira-Ruan, V., et al. (2014). The root indeterminacy-to-determinacy developmental switch is operated through a folate-dependent pathway in Arabidopsis thaliana. New Phytol. 202, 1223–1236. doi: 10.1111/nph.12757
Richter, G. L., Monshausen, G. B., Krol, A., and Gilroy, S. (2009). Mechanical stimuli modulate lateral root organogenesis. Plant Physiol. 151, 1855–1866. doi: 10.1104/pp.109.142448
Romero-Arias, J. R., Hernández-Hernández, V., Benítez, M., Alvarez-Buylla, E. R., and Barrio, R. A. (2017). Model of polar auxin transport coupled to mechanical forces retrieves robust morphogenesis along the Arabidopsis root. Phys. Rev. E 95:032410. doi: 10.1103/PhysRevE.95.032410
Rywosch, S. (1909). Untersuchungen uber die Entwicklungsgeschichte der Seitenwurzeln der Monocotylen. Z. Bot. 1, 253–283.
Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358. doi: 10.1101/gad.252503
Santner, A., and Estelle, M. (2009). Recent advances and emerging trends in plant hormone signalling. Nature 459, 1071–1078. doi: 10.1038/nature08122
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Scheres, B., and Benfey, P. N. (1999). Asymmetric cell division in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 505–537. doi: 10.1146/annurev.arplant.50.1.505
Seago, J. L. (1973). Developmental anatomy in roots of Ipomoea purpurea. II. initiation and development of secundary roots. Am. J. Bot. 60, 607–618. doi: 10.1002/j.1537-2197.1973.tb05965.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shimotohno, A., Heidstra, R., Blilou, I., and Scheres, B. (2018). Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 32, 1085–1100. doi: 10.1101/gad.314096.118
Stoeckle, D., Thellmann, M., and Vermeer, J. E. (2018). Breakout—lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 41, 67–72. doi: 10.1016/j.pbi.2017.09.005
Sugiyama, M. (2003). Isolation and initial characterization of temperature-sensitive mutants of Arabidopsis thaliana that are impaired in root redifferentiation. Plant Cell Physiol. 44, 588–596. doi: 10.1093/pcp/pcg077
Swarup, K., Benkova, E., Swarup, R., Casimiro, I., Peret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Takagi, M., Sakamoto, T., Suzuki, R., Nemoto, K., Obayashi, T., Hirakawa, T., et al. (2016). Plant Aurora kinases interact with and phosphorylate transcription factors. J. Plant Res. 129, 1165–1178. doi: 10.1007/s10265-016-0860-x
Takei, K., Ueda, N., Aoki, K., Kuromori, T., Hirayama, T., Shinozaki, K., et al. (2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant Cell Physiol. 45, 1053–1062. doi: 10.1093/pcp/pch119
Tan, X., Calderon-Villalobos, L. A., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
ten Hove, C. A., Willemsen, V., de Vries, W. J., van Dijken, A., Scheres, B., and Heidstra, R. (2010). SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20, 452–457. doi: 10.1016/j.cub.2010.01.018
Thompson, A., and MacLeod, R. D. (1981). Increase in size and cell number of lateral root primordia in the primary of intact plants and in excised roots of Pisum sativum and Vicia faba. Am. J. Bot. 68, 955–964. doi: 10.1002/j.1537-2197.1981.tb07812.x
Tian, H., Jia, Y., Niu, T., Yu, Q., and Ding, Z. (2014). The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep. 33, 745–753. doi: 10.1007/s00299-014-1575-x
Tschermak-Woess, E., and Doležal, R. (1953). Durch Seitenwurzelbildung induzierte und spontane Mitosen in den Dauergeweben der Wurzel. Österreichische Bot. Z. 100, 358–402. doi: 10.1007/BF01805779
Tsukagoshi, H., Busch, W., and Benfey, P. N. (2010). Transcriptional Regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. doi: 10.1016/j.cell.2010.10.020
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Van Damme, D., De Rybel, B., Gudesblat, G., Demidov, D., Grunewald, W., De Smet, I., et al. (2011). Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23, 4013–4024. doi: 10.1105/tpc.111.089565
Van Norman, J. M., Xuan, W., Beeckman, T., and Benfey, P. N. (2013). To branch or not to branch: the role of pre-patterning in lateral root formation. Development 140, 4301–4310. doi: 10.1242/dev.090548
Van Tieghem, P., and Douliot, H. (1886). Origine des Radicelles et des Racines Laterales Chez les Legumineuses et les Cucurbitacees. Bull. Soc. Bot. France 8, 494–501. doi: 10.1080/00378941.1886.10828482
Van Tieghem, P., and Douliot, H. (1888). Recherches comparatives sur l’origine des membres endogénes dans les plantes vasculaires. Ann. Sci. Natur. Bot. Ser. 8:660.
Vanneste, S., De Rybel, B., Beemster, G. T. S., Ljung, K., De Smet, I., Van Isterdael, G., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050. doi: 10.1105/tpc.105.035493
Vermeer, J. E., von Wangenheim, D., Barberon, M., Lee, Y., Stelzer, E. H., Maizel, A., et al. (2014). A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343, 178–183. doi: 10.1126/science.1245871
Von Guttenberg, H. (1960). Grundzüge der Histogenese höherer Pflanzen. I Die Angiospermen. Berlin: Gebruder Bomtraeger.
Von Guttenberg, H. (1968). Der Primare Bau der Angiospermenwurzel VIII/5. Berlin: Gebruder Bomtraeger.
von Wangenheim, D., Fangerau, J., Schmitz, A., Smith, R. S., Leitte, H., Stelzer, E. H., et al. (2016). Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr. Biol. 26, 439–449. doi: 10.1016/j.cub.2015.12.047
Voronin, N. S. (1957). On evolution of plant roots. 2, Evolution of the root’s origin. Bull. Moscow Soc. Nat. Biol. Ser. Russ. 62, 35–49.
Voronin, N. S. (1964). Evolution of the primary structures in plant roots. Uchenye Zapiski Kaluzhskogo Gosudarstvennogo Pedagogicheskogo Instituta 13, 3–179.
Voronkina, N. (1978). Early ontogenetic stages of lateral root development and their significance for understanding of the evolution of root hystogenesis. Bot. Z. 63, 205–214.
Voß, U., Wilson, M. H., Kenobi, K., Gould, P. D., Robertson, F. C., Peer, W. A., et al. (2015). The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 6:7641. doi: 10.1038/ncomms8641
Whitewoods, C. D., Cammarata, J., Venza, Z. N., Sang, S., Crook, A. D., Aoyama, T., et al. (2018). CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Curr. Biol. 28, 2365–2376.e5. doi: 10.1016/j.cub.2018.05.068
Wightman, F., Schneider, E. A., and Thimann, K. V. (1980). Hormonal factors controlling the initiation and development of lateral roots II. effects of exogenous growth factors on lateral root formation in pea roots. Physiol. Plant. 49, 304–314. doi: 10.1111/j.1399-3054.1980.tb02669.x
Willemsen, V., Bauch, M., Bennett, T., Campilho, A., Wolkenfelt, H., Xu, J., et al. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15, 913–922. doi: 10.1016/j.devcel.2008.09.019
Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130. doi: 10.1111/j.1365-313X.2005.02432.x
Yasutani, I., Ozawa, S., Nishida, T., Sugiyama, M., and Komamine, A. (1994). Isolation of temperature-sensitive mutant of Arabidopsis thaliana that are in the redifferentiation of shoots. Plant Physiol. 105, 815–822. doi: 10.1104/pp.105.3.815
Yoshida, S., de Reuille, P. B., Lane, B., Bassel, G. W., Prusinkiewicz, P., Smith, R. S., et al. (2014). Genetic control of plant development by overriding a geometric division rule. Dev. Cell 29, 75–87. doi: 10.1016/j.devcel.2014.02.002
Yu, P., Gutjahr, C., Li, C., and Hochholdinger, F. (2016). Genetic control of lateral root formation in cereals. Trends Plant Sci. 21, 951–961. doi: 10.1016/j.tplants.2016.07.011
Zhukovskaya, N. V., Bystrova, E. I., Dubrovsky, J. G., and Ivanov, V. B. (2018). Global analysis of an exponential model of cell proliferation for estimation of cell cycle duration in the root apical meristem of angiosperms. Ann. Bot. 122, 811–822. doi: 10.1093/aob/mcx216
Keywords: root development, lateral root primordium, morphogenesis, root architecture, crop species, Arabidopsis, cell proliferation, stem cells
Citation: Torres-Martínez HH, Rodríguez-Alonso G, Shishkova S and Dubrovsky JG (2019) Lateral Root Primordium Morphogenesis in Angiosperms. Front. Plant Sci. 10:206. doi: 10.3389/fpls.2019.00206
Received: 29 November 2018; Accepted: 07 February 2019;
Published: 19 March 2019.
Edited by:
José Manuel Pérez-Pérez, Universidad Miguel Hernández de Elche, SpainReviewed by:
Juan Carlos Del Pozo, Centro de Biotecnología y Genómica de Plantas (CBGP), SpainCopyright © 2019 Torres-Martínez, Rodríguez-Alonso, Shishkova and Dubrovsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph G. Dubrovsky, amR1YnJvdkBpYnQudW5hbS5teA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.