
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 February 2019
Sec. Plant Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00171
This article is part of the Research Topic New Insights into Mechanisms of Epigenetic Modifiers in Plant Growth and Development View all 15 articles
The circadian clock synchronizes endogenous rhythmic processes with environmental cycles and maximizes plant fitness. Multiple regulatory layers shape circadian oscillation, and chromatin modification is emerging as an important scheme for precise circadian waveforms. Here, we report the role of an evolutionarily conserved Sin3-histone deacetylase complex (HDAC) in circadian oscillation in Arabidopsis. SAP30 FUNCTION-RELATED 1 (AFR1) and AFR2, which are key components of Sin3-HDAC complex, are circadianly-regulated and possibly facilitate the temporal formation of the Arabidopsis Sin3-HDAC complex at dusk. The evening-expressed AFR proteins bind directly to the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and PSEUDO-RESPONSE REGULATOR 9 (PRR9) promoters and catalyze histone 3 (H3) deacetylation at the cognate regions to repress expression, allowing the declining phase of their expression at dusk. In support, the CCA1 and PRR9 genes were de-repressed around dusk in the afr1-1afr2-1 double mutant. These findings indicate that periodic histone deacetylation at the morning genes by the Sin3-HDAC complex contributes to robust circadian maintenance in higher plants.
The circadian clock is an internal time-keeper mechanism that ensures endogenous biological rhythms with a period of approximately 24 h, coinciding with daily environmental cycles. A large fraction of the plant transcriptome is clock-controlled, and thus the clock is globally linked to diverse signaling and metabolic pathways to ensure optimal biological functions at a specific time of day (Covington et al., 2008; Mizuno and Yamashino, 2008; Hsu and Harmer, 2012). Synchronization of the clock with the environment is closely associated with plant growth and fitness (Dodd et al., 2005; Fujiwara et al., 2008; Nusinow et al., 2011; Yoo et al., 2011; Lu et al., 2012; Nagel and Kay, 2012; Haydon et al., 2013; Zhang et al., 2013).
The circadian clock is a highly conserved system in higher eukaryotes. In Arabidopsis, the central oscillator is known to consist of an array of transcriptional loops. Two single-MYB transcription factors, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), establish the central loop by repressing transcription of TIMING OF CAB EXPRESSION 1 (TOC1) that in turn, represses CCA1 and LHY expression (Alabadi et al., 2001; Huang et al., 2012; Pokhilko et al., 2013). The central loop is further regulated by PSEUDO-RESPONSE REGULATORs (PRR5, PRR7, and PRR9) (Nakamichi et al., 2005, 2010; Salome et al., 2010) and the evening complex (EC) consisting of EARLY FLOWERING 3 (ELF3), ELF4, and LUX ARRYTHMO/PHYTOCLOCK 1 (LUX/PCL1) (Nusinow et al., 2011; Chow et al., 2012; Herrero et al., 2012). Moreover, the TOC1 protein also plays widespread roles in transcriptionally repressing multiple core clock components, underscoring the biological importance of transcriptional regulation in circadian homeostasis (Gendron et al., 2012; Huang et al., 2012).
Accumulating evidence suggests that circadian oscillation is further shaped by additional regulatory mechanisms (Seo and Mas, 2014). In particular, chromatin modification is an important regulatory scheme underlying precise circadian waveforms (Mas, 2008; Stratmann and Mas, 2008; Kusakina and Dodd, 2012; Nagel and Kay, 2012). Transcript accumulation of core clock components correlates with rhythmic changes in accumulation of histone H3 acetylation (H3ac) in Arabidopsis (Hemmes et al., 2012; Malapeira et al., 2012; Song and Noh, 2012). Consistent with the fact that histone acetylation status is dynamically regulated by the antagonistic action of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (Kuo and Allis, 1998; Yang and Seto, 2007), temporal association of specific sets of HATs and HDACs occurs at the loci of core clock components to shape rhythmic expression (Hemmes et al., 2012; Malapeira et al., 2012; Song and Noh, 2012). For instance, the midday-expressed HISTONE ACETYLTRANSFERASE OF THE TAFII250 FAMILY 2 (HAF2) protein catalyzes H3ac at the PRR5 and LUX loci to activate expression and is responsible for the rising phase of PRR5 and LUX circadian expression (Lee and Seo, 2018). In addition, the HDA6 and HDA19 proteins form protein complexes together with the TOPLESS (TPL) and PRR proteins, and repress expression of CCA1 and LHY during the daytime (Wang et al., 2013). Despite the importance of diurnal histone acetylation states of core clock genes in stable circadian oscillation, the responsible epigenetic modifiers are yet to be fully characterized.
Histone deacetylase complex often form diverse types of multiprotein co-repressor complexes and play a variety of roles during plant growth and development (Buszewicz et al., 2016; Kim et al., 2016; Hung et al., 2018; Park et al., 2018; Tasset et al., 2018). One well-characterized HDAC complex in eukaryotes is the Sin3-HDAC complex (Alland et al., 2002; Kuzmichev et al., 2002; Silverstein and Ekwall, 2005; Clark et al., 2015). In Arabidopsis, the Sin3-HDAC complex participates in photoperiodic flowering through the periodic acetylation of the FLOWERING LOCUS T (FT) locus (Gu et al., 2013). The Sin3-HDAC complex is activated at the end of the day and is recruited to the FT locus by AGAMOUS LIKE 18 (AGL18) in a CONSTANS (CO)-dependent manner under long-day conditions (Gu et al., 2013). In this study, we report that the Arabidopsis Sin3-HDAC complex also temporally regulates CCA1 and PRR9 expression through catalyzing H3 deacetylation and facilitates the declining phase of their circadian expression during the evening time. These results reveal that temporal association of chromatin modifiers underlies robust rhythmic expression of clock genes and thereby stable circadian oscillation.
Histone deacetylase complex often form multiprotein co-repressor complexes, as exemplified by the Sin3-HDAC complex that consists of the master scaffold protein Sin3, the Reduced Potassium Dependency 3 (RPD3)-type HDAC, and Sin3-associated structural components, such as SIN3-ASSOCIATED POLYPEPTIDE 18 (SAP18) and SAP30 (Zhang et al., 1997; Laherty et al., 1998; Wu et al., 2000; Scott and Plon, 2003; Song and Galbraith, 2006). The Arabidopsis genome contains six Sin3 homologs, SIN3-LIKE 1-6 (SNL1-6), four RPD3 homologs (HDA19, HDA9, HDA7, and HDA6), one SAP18 homolog, and two SAP30 homologs (SAP30 FUNCTION-RELATED 1 (AFR1) and AFR2) (Wu et al., 2000; Murfett et al., 2001; Pandey et al., 2002; Gu et al., 2013).
Notably, AFR1 and AFR2 have been identified as regulators of photoperiodic flowering, which facilitate periodic histone deacetylation at the FT locus (Gu et al., 2013). Considering their roles in temporal histone deacetylation, we hypothesized that the Arabidopsis Sin3-HDAC complex may also be implicated in circadian control. To examine the possible involvement of the HDAC complex in circadian oscillation, we first checked transcript accumulation of key components of the Sin3-HDAC complex in seedlings entrained under neutral day (ND) conditions. Quantitative real-time RT-PCR (RT-qPCR) analysis revealed that only the AFR1 and AFR2 genes are circadianly-regulated (Figure 1A), while the other components are not under the control of the circadian clock (Figure 1B). The AFR genes peaked at dusk (Figure 1A), as reported previously (Gu et al., 2013), suggesting that clock-controlled AFRs presumably lead to diurnal formation of the HDAC complex.
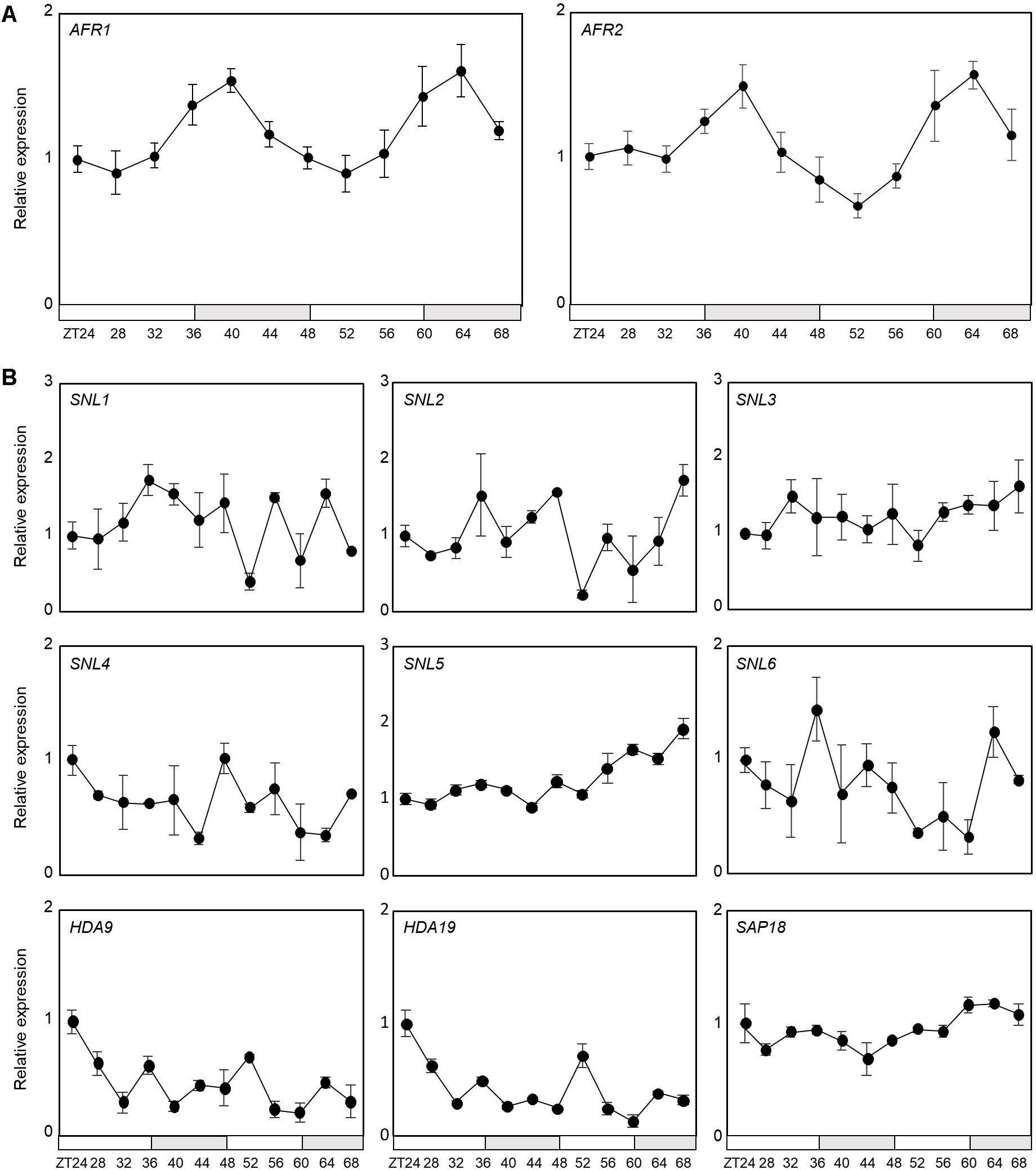
Figure 1. Circadian expression of AFR1 and AFR2. Seedlings grown under neutral day conditions (ND, 12 h light: 12 h dark) for 2 weeks were transferred to continuous light conditions (LL) at Zeitgeber Time 0 (ZT0). Whole seedlings were harvested from ZT24 to ZT68 to analyze transcript accumulation. Transcript levels were determined by quantitative real-time RT-PCR (RT-qPCR). Gene expression values were normalized to EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A) expression. Three independent biological replicates were averaged. Bars represent the standard error of the mean. The white and gray boxes indicate the subjective day and night, respectively. (A) Expression of AFR1 and AFR2. (B) Expression of other components of Sin3-HDAC.
To explore the circadian component responsible for regulation of the AFRs, we conducted analysis of the cis-elements present within the AFR promoters. AFRs have multiple CCA1-binding sites (CBSs, AAAATCT) and evening elements (EEs, AAATATCT) in the upstream promoters (Figure 2A), which are known to be bound by CCA1 and LHY (Wang et al., 1997; Harmer et al., 2000; Michael and McClung, 2003; Nagel et al., 2015). This observation raised the possibility that CCA1 may bind to the AFR promoters. To examine this possibility, a chromatin immunoprecipitation (ChIP) assay was performed using plants expressing epitope-tagged CCA1 under its own native promoter (pCCA1:CCA1-HA-YFP/cca1-1). Total protein extracts of samples collected at Zeitgeber Time 0 (ZT0) and ZT12 were immunoprecipitated with anti-HA antibody. ChIP-qPCR analysis showed that the proximal regions of transcriptional start sites (TSSs) on the AFR promoters containing CBS and/or EE elements were enriched following ChIP (Figure 2B). Binding of CCA1 to the AFR promoter was specifically observed at dawn, but not at dusk (Figure 2B), shaping circadian expression of the AFRs.
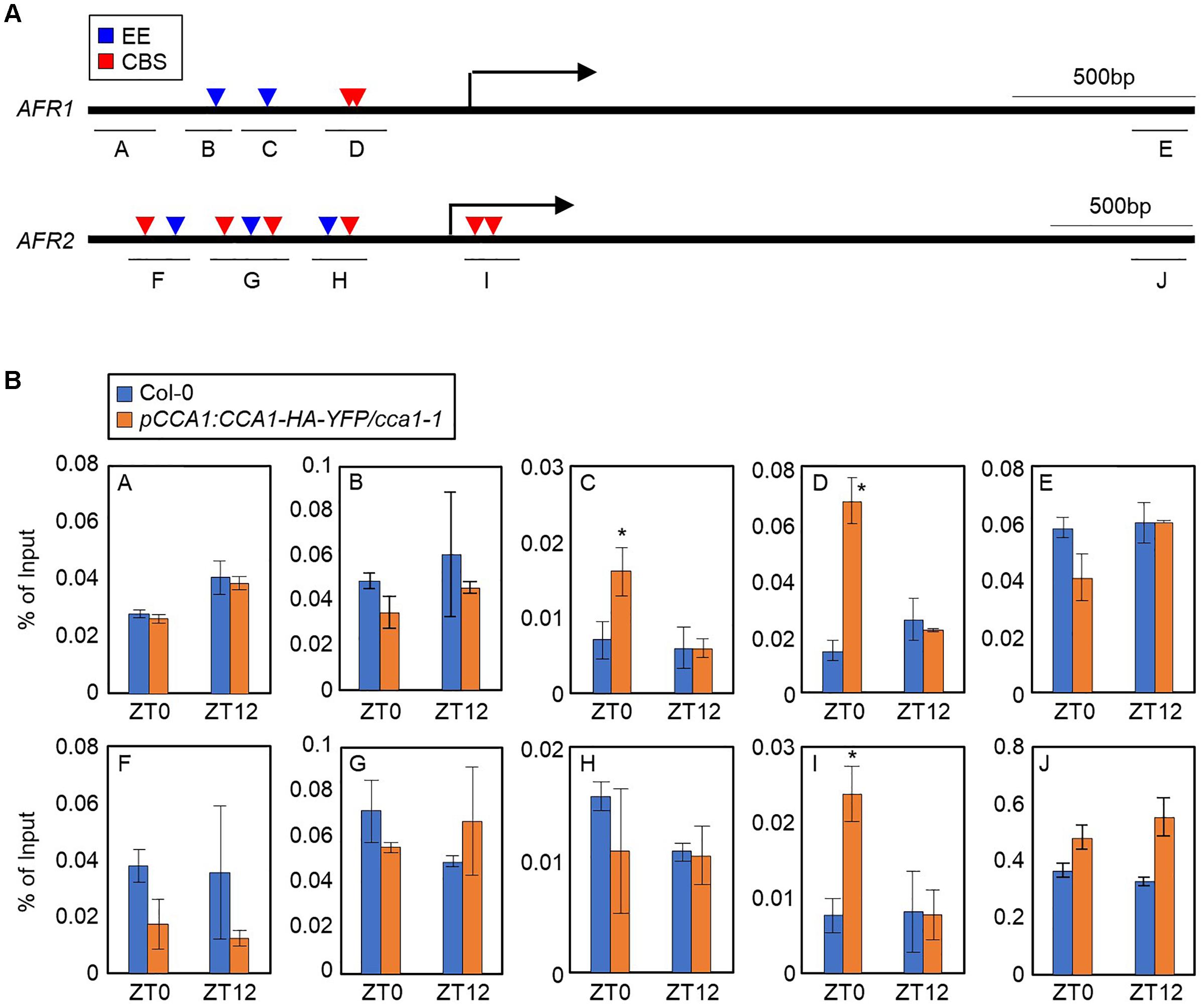
Figure 2. Binding of CCA1 to AFR promoters. (A) Promoter analysis of the AFR1 and AFR2 genes. Underbars indicate the regions amplified by PCR after chromatin immunoprecipitation (ChIP). CBS, CCA1-binding site; EE, evening element. (B) Binding of CCA1 to the AFR loci. Two-week-old plants entrained with ND cycles were subjected to LL. Plants were harvested at ZT0 and ZT12 for ChIP analysis with anti-HA antibody. Three independent biological replicates were averaged, and statistically significant differences (Student’s t-test, ∗P < 0.05) are indicated by asterisks. Bars indicate the standard error of the mean.
To support AFR regulation by the transcriptional regulator CCA1, we analyzed AFR expression in cca1-2 and cca1-1lhy-21 mutant seedlings grown under ND conditions. RT-qPCR analysis showed that the peak phase of AFR expression was delayed in cca1-2 and cca1-1lhy-21, and higher expression of AFRs around the end of night was observed in the cca1-2 and cca1-1lhy-21 mutants compared with wild-type (Figure 3A and Supplementary Figure S1). In contrast, AFR expression was dramatically reduced in CCA1-overexpressing lines (Figure 3B). To further support the repressive role of CCA1 in AFR expression, we performed transient expression assays using Arabidopsis mesophyll protoplasts. The GUS reporter plasmids and effector plasmids harboring 35S:CCA1-GFP fusion were co-transfected into protoplasts (Supplementary Figure S2). Co-transfection of a reporter construct with 35S:CCA1-GFP resulted in lower GUS activity than the control plasmid (Supplementary Figure S2). These results indicate that CCA1 shapes AFR expression and enables peak expression particularly during the evening time.
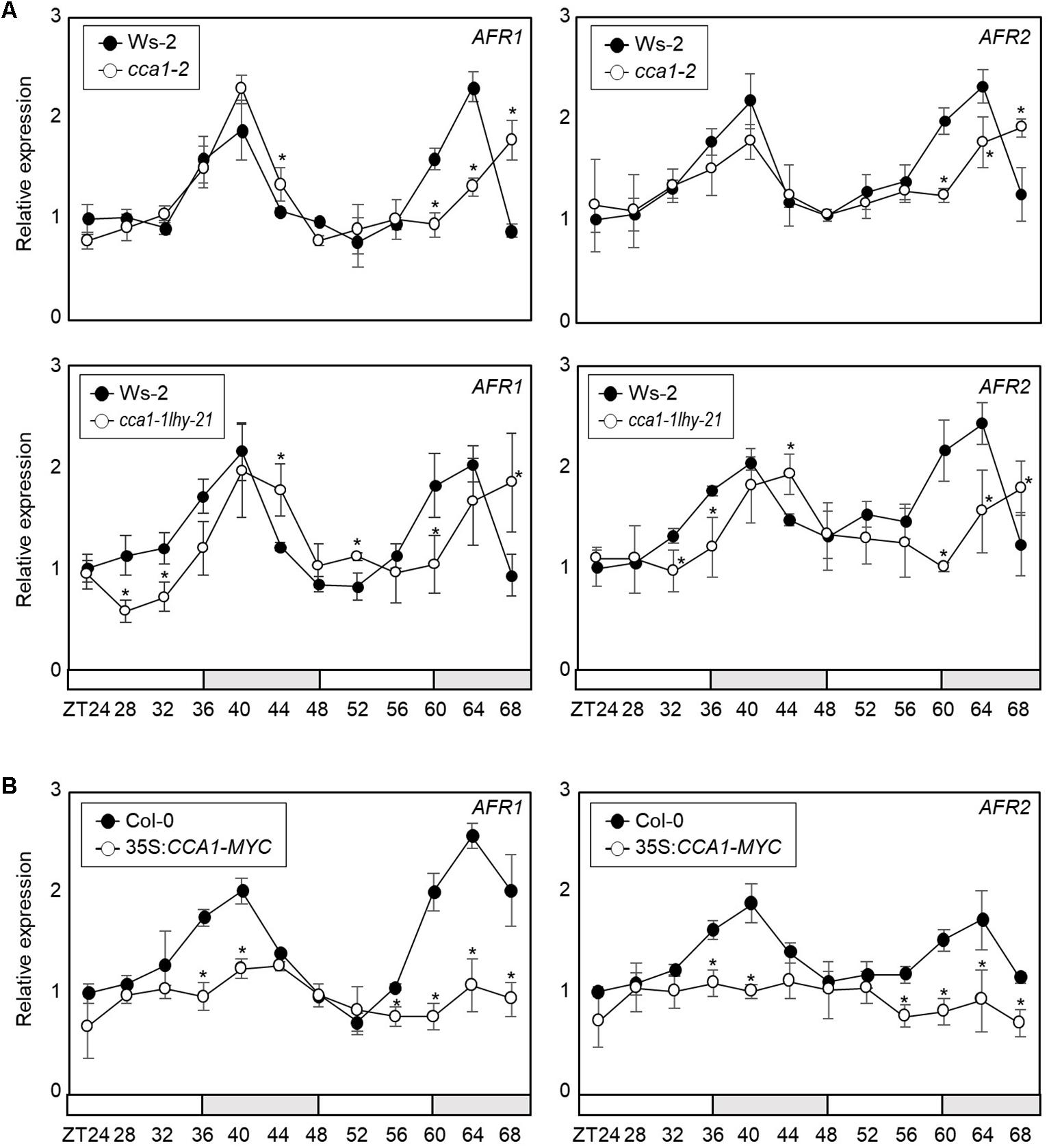
Figure 3. Circadian expression of AFRs in CCA1-misexpressing plants. In (A,B), seedlings grown under ND conditions for 2 weeks were transferred to LL conditions at ZT0. Whole seedlings were harvested from ZT24 to ZT68 to analyze transcript accumulation. Transcript levels were determined by RT-qPCR. Gene expression values were normalized to eIF4A expression. Three independent biological replicates were averaged, and statistically significant differences (Student’s t-test, ∗P < 0.05) are indicated by asterisks. Bars represent the standard error of the mean. The white and gray boxes indicate the subjective day and night, respectively. (A) Expression of AFRs in the cca1-2 and cca1-1lhy-21 mutant. (B) Expression of AFRs in 35S:CCA1-MYC transgenic plants.
Since the AFR proteins are core Sin3-HDAC components regulated by the circadian clock, we further investigated the role of AFRs in circadian oscillation. We employed the afr1-1afr2-1 double mutant and examined endogenous circadian behavior. RT-qPCR analysis showed that circadian output genes, COLD CIRCADIAN RHYTHM RNA BINDING 2 (CCR2) and CHLOROPHYLL A/B-BINDING PROTEIN 2 (CAB2), were altered in afr1-1afr2-1 mutant seedlings compared with wild-type (Figure 4A). We also checked several core circadian oscillator genes, including CCA1 and TOC1. Again, two genes were also differentially expressed in the afr1-1afr2-1 mutant compared with wild-type (Figure 4B). In particular, the morning gene expression was delayed in afr1-1afr2-1. The alteration patterns of the circadian genes were dissimilar in afr1-1afr2-1 mutant. This might be due to extensive circadian feedback network that balances 24 h clock oscillation, as observed in several previous studies (Somers et al., 2004; Ding et al., 2007; Hanano et al., 2008; Li et al., 2011).
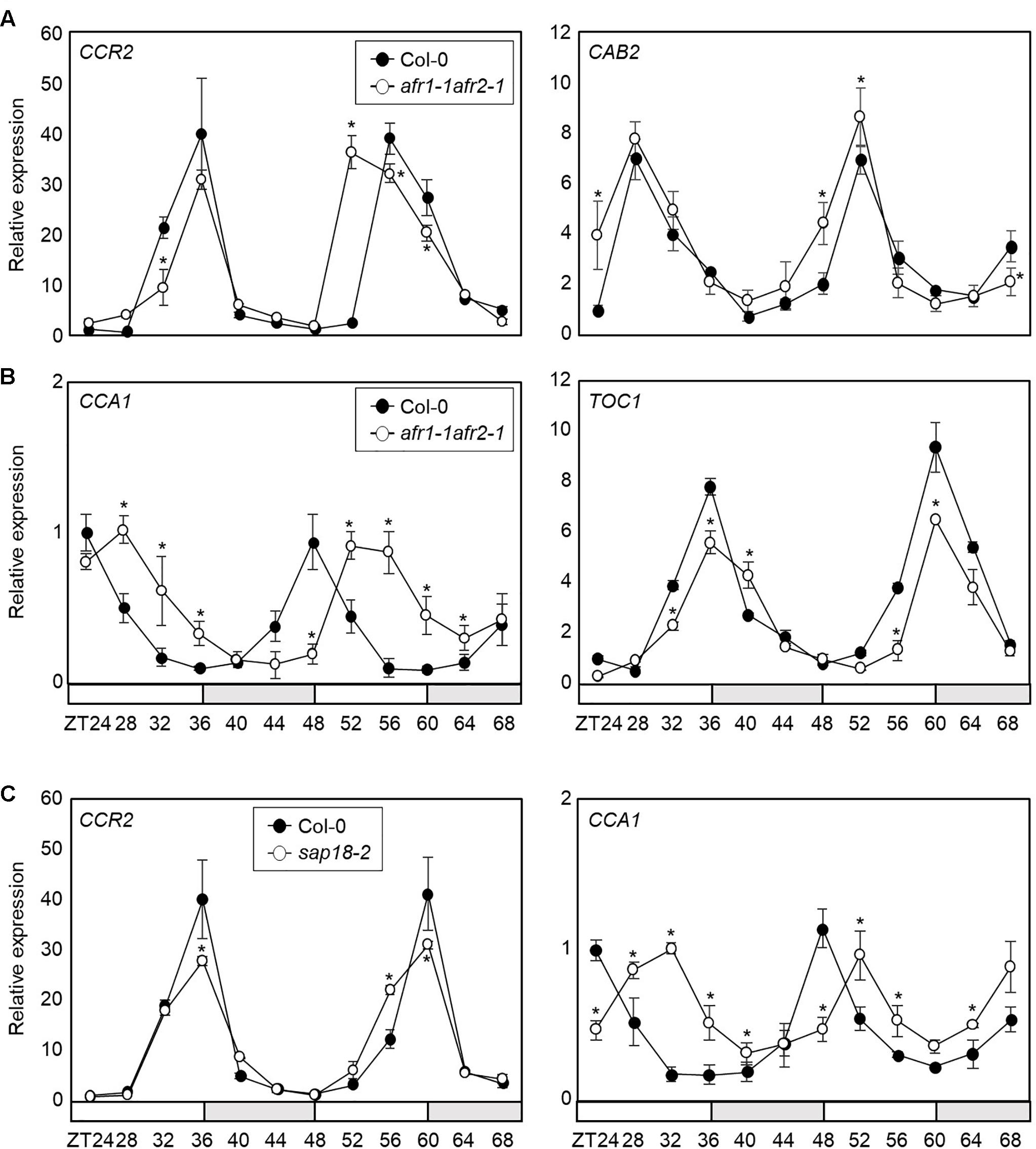
Figure 4. Altered circadian rhythm in the afr1-1afr2-1 mutant. In (A–C), seedlings grown under ND were transferred to LL at ZT0. Whole seedlings were harvested from ZT24 to ZT68 to analyze transcript accumulation. Gene expression values were normalized to eIF4A expression and represented as n-fold compared to the value of the wild-type sample at ZT24. Three independent biological replicates were averaged, and statistically significant differences (Student’s t-test, ∗P < 0.05) are indicated by asterisks. Bars indicate the standard error of the mean. The white and pale gray boxes indicate the subjective day and night, respectively. (A) Expression of CCR2 and CAB2 in afr1-1afr2-1. (B) Expression of CCA1 and TOC1 in afr1-1afr2-1. (C) Expression of CCA1 and CCR2 in sap18-2.
AFRs are components of the Arabidopsis Sin3-HDAC complex (Gu et al., 2013). To provide further support that AFR function in circadian oscillation depends on formation of the Sin3-HDAC complex, we obtained a genetic mutant of SAP18 and analyzed circadian oscillation. Since SAP18 is the only member of the Sin3-HDAC components that exists as a single copy in the Arabidopsis genome (Zhang et al., 1997; Ahringer, 2000), we suspected that the sap18-2 mutant could be used to reflect the roles of the Arabidopsis Sin3-HDAC complex. Remarkably, the sap18-2 mutant exhibited altered circadian expression of CCA1 and CCR2 (Figure 4C and Supplementary Figure S3), similar to afr1-1afr2-1, indicating that the Arabidopsis Sin3-HDAC complex controls circadian oscillation.
AFRs most likely regulate the pace of the circadian clock possibly in association with the central oscillator(s). To identify which circadian components are regulated by the AFRs, we conducted ChIP assays using 35S:AFR1-MYC and 35S:AFR2-MYC transgenic plants. Plants were grown under ND conditions and harvested at ZT12, when AFR proteins highly accumulate (Gu et al., 2013). ChIP-qPCR analysis showed that the AFR proteins bind directly to the CCA1 and PRR9 loci (Figures 5A,B), while the other clock members examined were not targeted by the AFRs (Supplementary Figure S4). AFRs were primarily targeted around the TSSs of the CCA1 and PRR9 loci, rather than the 3′-regions of gene body (Figure 5B), which is consistent with previous observations that chromatin modification of core clock genes primarily occurs around TSSs (Hemmes et al., 2012; Malapeira et al., 2012). In addition, binding of AFRs to the CCA1 and PRR loci was prominent at ZT12 (Figure 5B), when peak expression of AFRs was observed (Figure 1A).
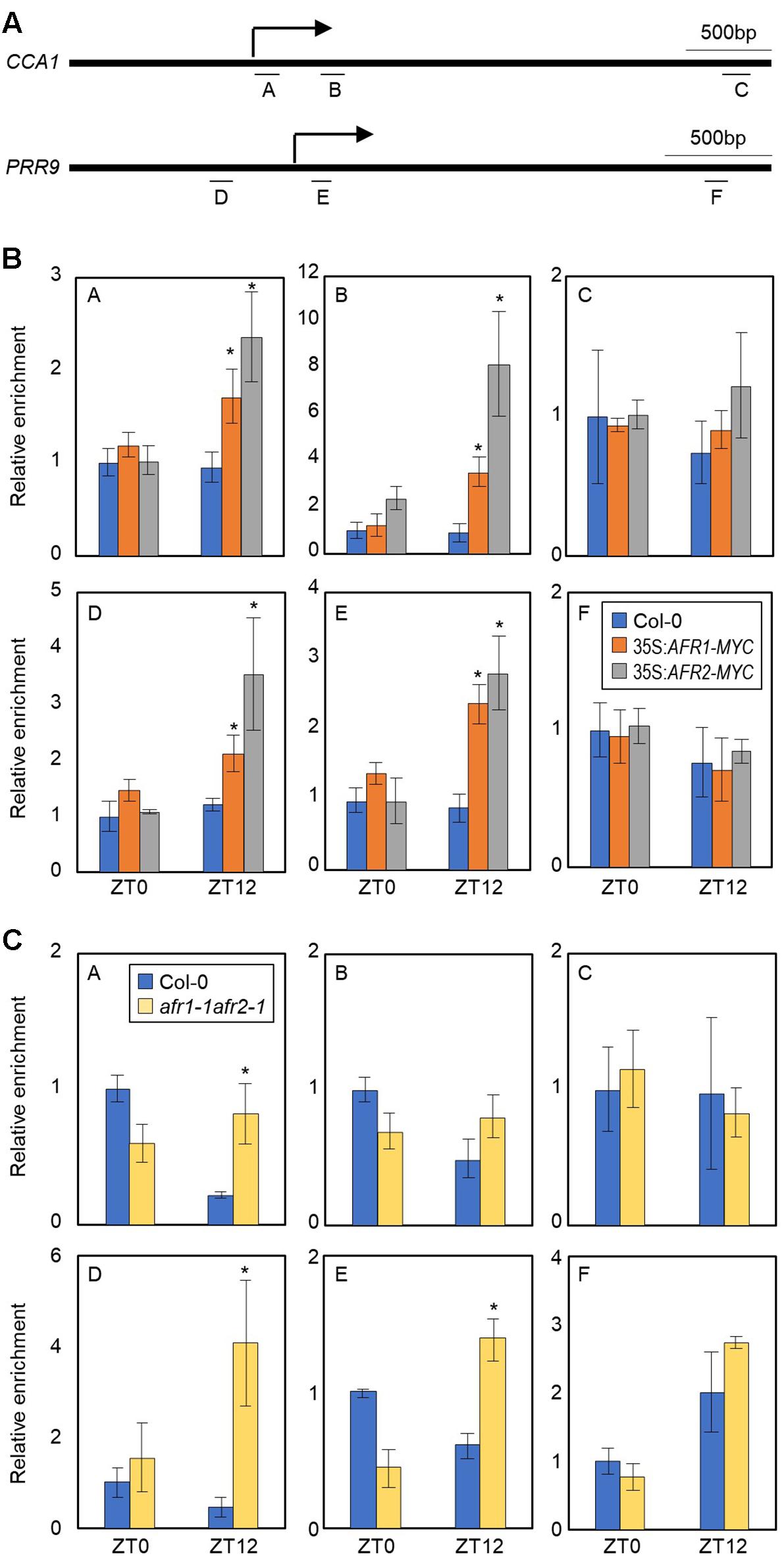
Figure 5. H3 deacetylation at the CCA1 and PRR9 loci during evening time by AFRs. In (B,C), 2-week-old seedlings grown under ND were transferred to LL and harvested at ZT0 and ZT12. Enrichment of putative binding regions of AFRs in promoters of the CCA1 and PRR9 genes was analyzed by ChIP-PCR. Three independent biological replicates were averaged, and statistical significance of the measurements was determined by a Student’s t-test (∗P < 0.05). Bars indicate the standard error of the mean. (A) Genomic regions for ChIP analysis. Underbars represent the amplified genomic regions. (B) Binding of AFRs to the CCA1 and PRR9 loci. (C) Accumulation of H3ac at the CCA1 and PRR9 loci in the afr1-1afr2-1 mutant. Anti-H3ac antibody was used for ChIP to assess H3ac accumulation at the loci.
The temporal recruitment of AFRs to the morning gene loci may cause periodic histone deacetylation. We examined H3 acetylation (H3ac) levels, which correlate to transcript accumulation of core clock genes (Hemmes et al., 2012; Malapeira et al., 2012), at the CCA1 and PRR9 promoters in wild-type and afr1-1afr2-1 seedlings. ChIP with anti-H3ac antibody revealed that H3ac levels of the CCA1 and PRR9 genes were elevated at ZT0 but reduced at ZT12 in wild-type (Figure 5C), as reported previously (Hemmes et al., 2012; Malapeira et al., 2012). However, the decline of H3ac accumulation at ZT12 was impaired in the afr1-1afr2-1 mutant (Figure 5C). Increased H3ac levels at the CCA1 and PRR9 loci were observed in the afr1-1afr2-1 mutant, particularly at ZT12 (Figure 5C). These results indicate that AFRs mediate histone deacetylation at the morning gene loci to stably downregulate expression during evening time.
Since the Sin3-HDAC complex catalyzes H3 deacetylation at the CCA1 and PRR9 loci, we speculated that circadian expression of the CCA1 and PRR9 genes may be shaped by diurnal H3ac accumulation. To test this possibility, we measured CCA1 and PRR9 expression in the afr1-1afr2-1 mutant. In wild-type seedlings, the CCA1 and PRR9 genes were highly expressed in the morning, but repressed during the afternoon (Figures 4B, 6A). In contrast, decrease of CCA1 and PRR9 expression during afternoon was compromised in the afr1-1afr2-1 mutant (Figures 4B, 6A). Circadian patterns of CCA1 and PRR9 expression were altered in the afr1-1afr2-1 mutant background, and the increased expression of CCA1 and PRR9 was clearly observed at afternoon (Figures 4B, 6A).
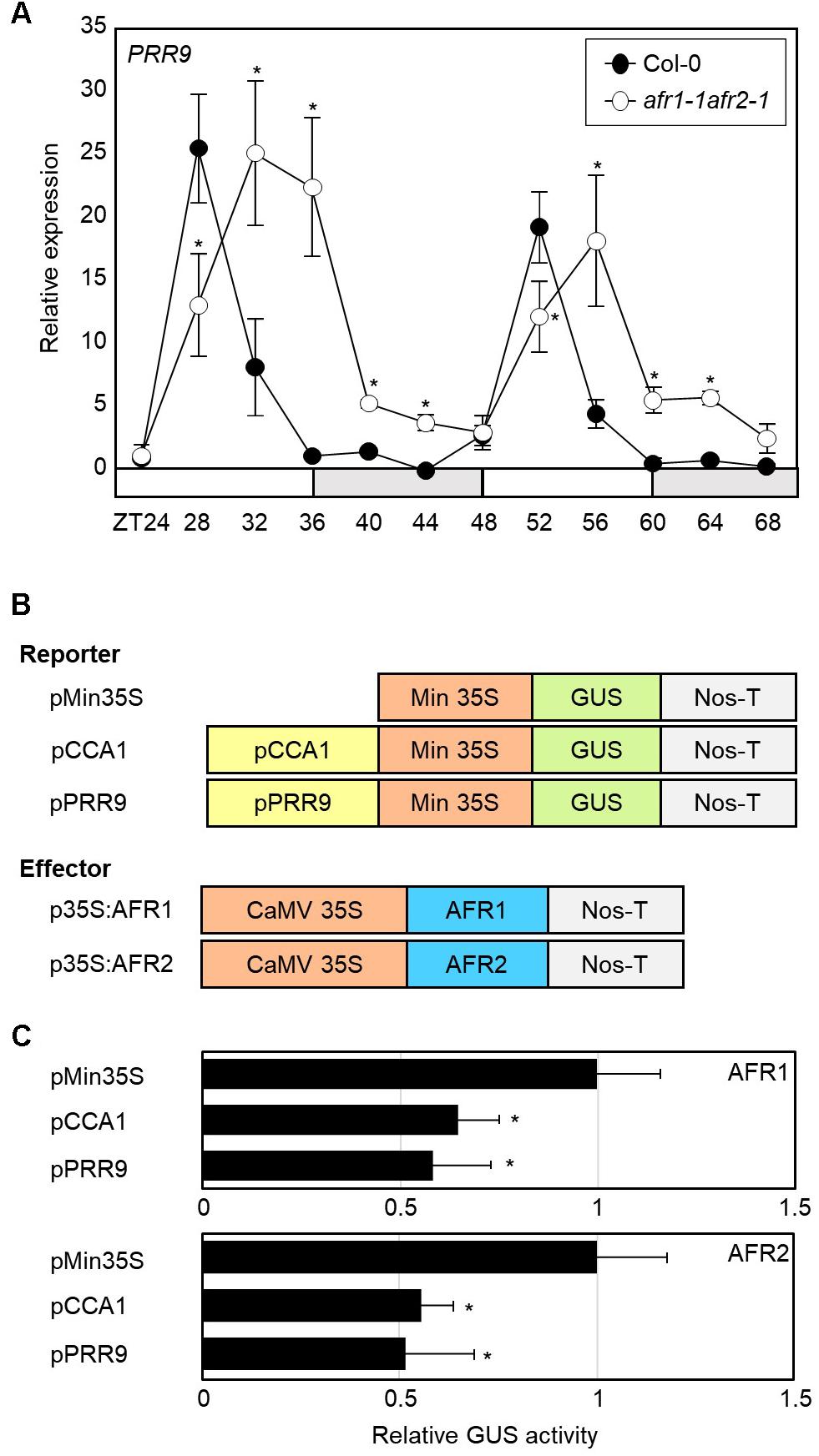
Figure 6. Increased expression of PRR9 at dusk in afr1-1afr2-1. (A) Transcript accumulation of PRR9. Seedlings grown under ND were transferred to LL at ZT0. Whole seedlings were harvested from ZT24 to ZT68 to analyze transcript accumulation. Gene expression values were normalized to eIF4A expression and represented as n-fold compared to the value of the wild-type sample at ZT24. Three independent biological replicates were averaged, and statistical significance of the measurements was determined by a Student’s t-test (∗P < 0.05). Bars indicate the standard error of the mean. The white and pale gray boxes indicate the subjective day and night, respectively. (B) Recombinant constructs used for transient expression assays. (C) Transient expression analysis using Arabidopsis protoplasts. The core elements of CCA1 and PRR9 genes were inserted into the reporter plasmid. A recombinant reporter was transiently coexpressed with an effector construct containing the 35S:AFR-MYC construct in Arabidopsis protoplasts, and GUS activity was fluorimetrically determined. Luciferase gene expression was used to normalize GUS activity. Three independent measurements were averaged. Statistical significance was determined by a Student’s t-test (∗P < 0.05). Bars indicate the standard error of the mean.
To further support the repressive role of AFRs in CCA1 and PRR9 expression, we examined the extent of AFR regulation of CCA1 and PRR9 transcription activity in Arabidopsis mesophyll protoplasts. The GUS reporter plasmids and effector plasmids harboring 35S:AFR-MYC fusion constructs were co-transfected into mesophyll protoplasts (Figure 6B). Co-transfection of a reporter construct with 35S:AFR1-MYC or 35S:AFR2-MYC led to lower GUS activity than the control plasmid (Figure 6C). These results indicate that AFR activity limits expression of morning genes, CCA1 and PRR9.
In yeast, SAP30 is a key player in recruitment of the SAP30-Sin3-HDAC co-repressor complex to target loci (Ahringer, 2000). It is possible that the yeast SAP30 protein interacts extensively with DNA-binding transcription factors. Consistently, the Arabidopsis AFR1 and AFR2 proteins also frequently associate with transcription factors and guide the Sin3-HDAC complex to cognate target chromatin regions (Gu et al., 2013). To identify the molecular components that recruit the Sin3-HDAC complex to the CCA1 and PRR9 loci, we performed yeast-two-hybrid (Y2H) assays. Clock genes were fused in-frame to the 3′-end of the activation domain (AD) of GAL4, and each construct was coexpressed in yeast cells with a recombinant plasmid containing the GAL4 DNA binding domain (BD)-AFR fusion construct. Cell growth on selective medium showed that the transcriptional corepressors NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED 1 (LNK1) and LNK2 specifically bind to AFR1 and AFR2 (Figure 7A and Supplementary Figure S5). The in vivo interactions of LNK and AFR proteins were verified by BiFC assays. Coexpression of AFR-nYFP and LNK-cYFP constructs allowed nuclear emission of YFP fluorescence, indicating physical interactions (Figure 7B). Given that the LNK corepressors act along with several DNA-binding proteins such as REVEILLE 4 (RVE4) and RVE8 (Xie et al., 2014; Perez-Garcia et al., 2015), AFRs may be recruited to the CCA1 and PRR9 loci at least by the DNA-binding RVE-LNK complex.
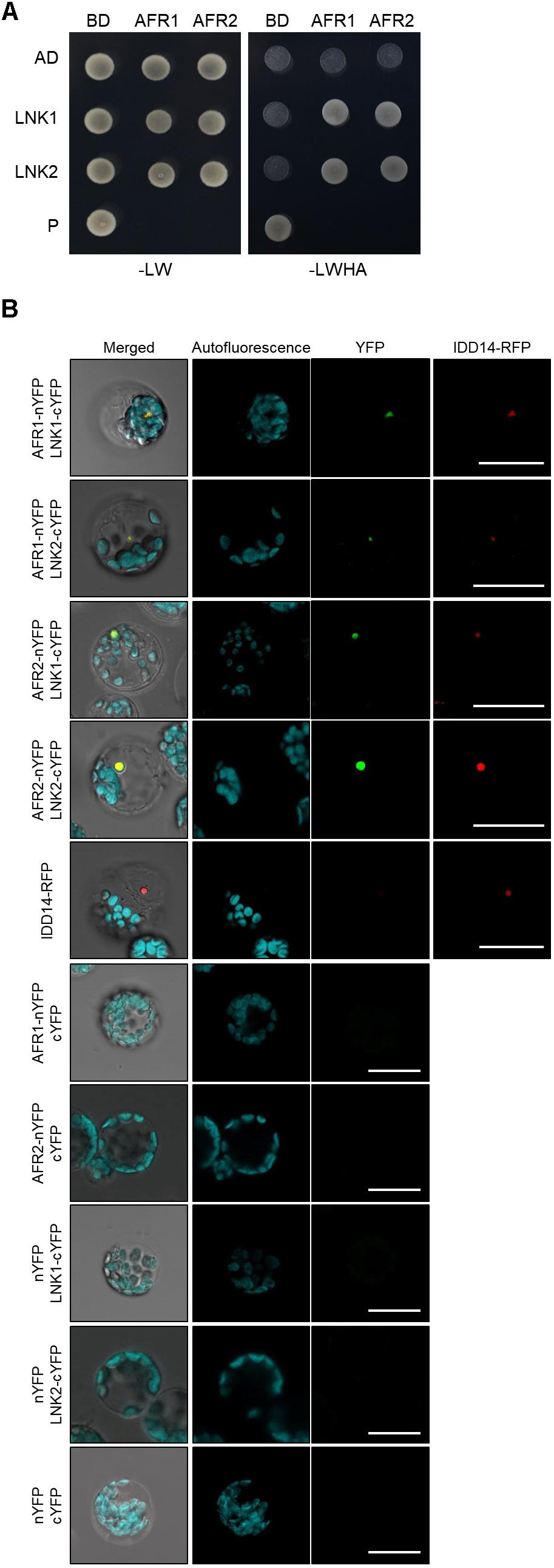
Figure 7. Interactions of AFRs with LNKs. (A) Y2H assays. Y2H assays were performed with AFR proteins fused to the DNA-binding domain (BD) of GAL4 and LNKs fused with the transcriptional activation domain (AD) of GAL4 for analysis of interactions. Interactions were examined by cell growth on selective media. -LWHA indicates Leu, Trp, His, and Ade drop-out plates. -LW indicates Leu and Trp drop-out plates. GAL4 was used as a positive control (P). (B) BiFC assays. Partial fragments of YFP protein were fused with AFRs and LNKs, and co-expressed in Arabidopsis protoplasts. The IDD14-RFP construct was used as a nuclear marker. Reconstituted fluorescence was examined by confocal microscopy. Scale bars: 20 μm.
Taken together, the Arabidopsis Sin3-HDAC complex facilitates temporal H3 deacetylation at the CCA1 and PRR9 loci to stably regulate circadian oscillation. The AFR proteins diurnally accumulate and possibly lead to temporal association of the Sin3-HDAC complex at evening time. The AFR proteins bind specifically to the morning gene loci and facilitate H3 deacetylation at the cognate regions at dusk. Binding of the Sin3-HDAC complex to the target promoter regions is likely specified by the RVE-LNK complex (Figure 8).
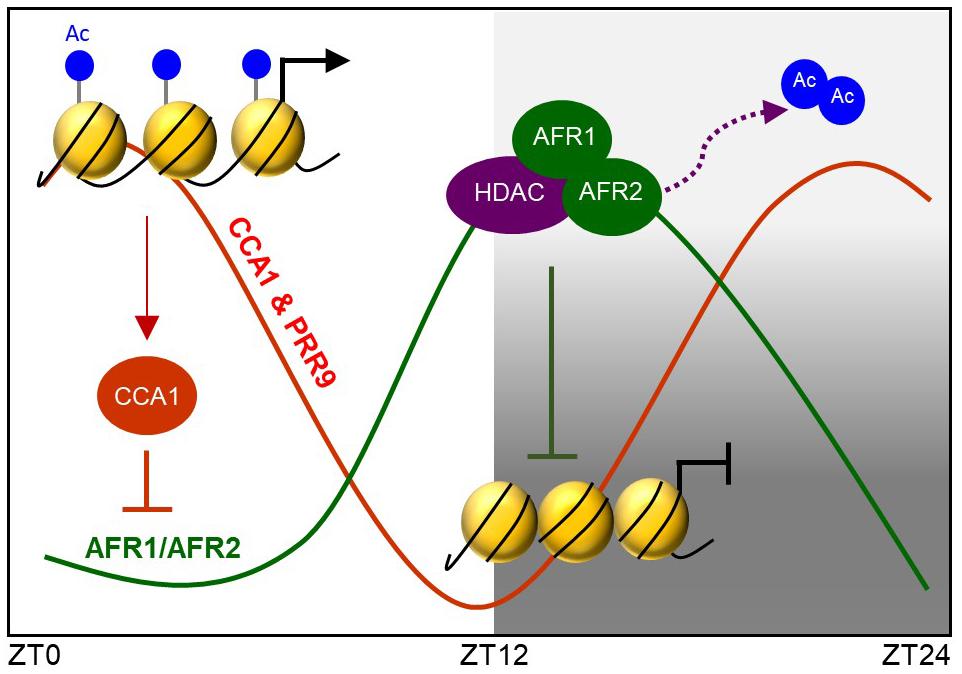
Figure 8. AFRs temporally regulate CCA1 and PRR9 genes during evening time. Arabidopsis Sin3-HDAC participates in regulating rhythmic expression of the CCA1 and PRR9 genes. The evening-expressed AFR proteins may temporally form the Sin3-HDAC corepressor complex and bind directly to the CCA1 and PRR9 promoters to catalyze H3 deacetylation at the cognate regions, allowing the declining phase of CCA1 and PRR9 expression during evening time. Binding regions of the Sin3-HDAC complex are likely specified by LNK-associated DNA-binding factors.
Rhythmic expression of core clock genes is intimately associated with the levels of histone modification, including H3ac and H3K4me3, at gene promoters in Arabidopsis (Hemmes et al., 2012; Malapeira et al., 2012). Dynamic cycles of histone modifications at the clock genes may result from transient binding of chromatin modifiers to the gene promoters. To date, several chromatin modifiers responsible for circadian control have been identified.
The SET DOMAIN GROUP 2 (SDG2)/ARABIDOPSIS TRITHORAX-RELATED 3 (ATXR3) protein is responsible for H3K4me3 deposition to activate multiple core clock genes. The H3K4me3 histone mark interferes with clock repressor binding at the core clock promoters, conferring correct timing of transcriptional repression to target clock genes (Hemmes et al., 2012; Malapeira et al., 2012). Accordingly, the SDG2/ATXR3-deficient mutants exhibit a global decrease in H3K4me3 levels and also a reduced amplitude of core clock gene expression (Berr et al., 2010; Malapeira et al., 2012; Yao et al., 2013; Pinon et al., 2017).
Circadian expression of the CCA1 and LHY genes is regulated by a couple of chromatin modifiers. The JMJ30/JMJD5 gene is clock-controlled and peaks at dusk (Lu et al., 2011). This pattern of JMJ30 expression is shaped by the central oscillators CCA1 and LHY, which directly bind to the JMJ30 promoter (Lu et al., 2011). In turn, JMJ30 promotes expression of CCA1 and LHY, presumably through its histone demethylase activity (Lu et al., 2011). In addition, HDA6 and HDA19 are also implicated in the Arabidopsis circadian system. The HDAC proteins form a protein complex with PRRs and TPL/TPRs (Wang et al., 2013), and repress expression of CCA1 and LHY by directly binding to the CCA1 and LHY promoters (Wang et al., 2013). Consistently, suppression of HDAC activity leads to circadian period lengthening and compromises the transcriptional repression activities of PRR5, PRR7, and PRR9 (Wang et al., 2013).
The Arabidopsis Sin3-HDAC complex is a different type of HDAC complex involved in circadian oscillation. Key members of the complex, AFR1 and AFR2, are under the control of the circadian clock and form a Sin3-HDAC complex possibly in a diurnal manner to mediate periodic histone deacetylation at the CCA1 and PRR9 loci. AFR-dependent H3 deacetylation at the CCA1 and PRR9 is relevant during the evening time and thereby dampens expression specifically at dusk. Notably, even though they share the same HDAC components, the AFR-containing Sin3-HDAC complex and HDA6/HDA19-PRR-TPL complex have different binding targets in the control of circadian oscillation. Different compositions of the protein complexes may lead to different abilities in interactive protein recognition, construction of protein interaction networks and thus target chromatin binding. For instance, the AFR proteins may specifically recruit transcriptional co-regulators, such as LNKs, and facilitate new repertoires of target gene regulation in circadian control.
A significant number of HATs and HDACs participate in circadian oscillation. Specific sets of HAT and HDAC shape circadian expression of core clock genes. For instance, HAF2 adds acetyl groups specifically to the PRR5 and LUX loci to facilitate the rising phase of expression (Lee and Seo, 2018), and the Sin3-HDAC complex removes the acetyl groups at the CCA1 and PRR9 loci to reset the acetylation state. This is likely not an exceptional case, and many biological responses are probably diurnally shaped by means of chromatin modifications (Kouzarides, 2007; Jang et al., 2011; Seo and Mas, 2014). The opposing activities of HAT and HDAC at specific genes conceivably modulate the acetylation dynamics of target chromatin regions during a day and set gene expression at the adequate level at the right time.
Histone acetyltransferases and HDACs are targeted to actively transcribed loci to control acetylation state and thereby gene expression at the genome level (Kuo and Allis, 1998; Wang et al., 2009; Peserico and Simone, 2011; Hemmes et al., 2012; Malapeira et al., 2012). However, since they have no selectivity to DNA elements, they are usually recruited to specific target loci by DNA-binding transcription factors (Todeschini et al., 2014; Bauer and Martin, 2017; Inukai et al., 2017). Interactions of chromatin modifiers with transcription factors allow elegant spatial and temporal modification of chromatin contexts (Munshi et al., 1998, 2001; Agalioti et al., 2000; Lomvardas and Thanos, 2002; Bauer and Martin, 2017).
Interactions of HDAC proteins with core clock components are crucial for refining circadian behavior in eukaryotes (Perales and Mas, 2007; Nakahata et al., 2008; Grimaldi et al., 2009). For example, in mammals, SIRT1 associates with a core transcription factor CLOCK, a positive regulator of the circadian machinery, and is recruited to the circadian gene promoters (Nakahata et al., 2008). Similarly, HDACs are associated with core clock components with DNA-binding activities in the control of circadian signaling in Arabidopsis (Perales and Mas, 2007). In the circadian expression of TOC1, the histone acetylation state seems to be regulated, at least in part, by the clock factors CCA1 and RVE8, as plants mis-expressing the MYB transcription factors exhibit an altered pattern of histone acetylation at the TOC1 locus (Perales and Mas, 2007). CCA1 may specify repressive chromatin structures at the TOC1 locus to regulate its expression at dawn, whereas RVE8, which has a high degree of sequence homology to CCA1, favors H3 acetylation in contrast to CCA1, most likely by antagonizing CCA1 function during the TOC1 raising phase (Farinas and Mas, 2011). Although chromatin modifiers responsible for accumulation of H3ac at the TOC1 locus are elusive so far, the oscillating H3ac levels are dependent on core clock transcription factors that will recruit HATs and/or HDACs to shape the waveform of TOC1.
AFR1 and AFR2 are recruited to the CCA1 and PRR9 chromatin for H3 deacetylation possibly by LNKs, although further experiments are required to prove the putative interactions. The morning-expressed LNK1 and LNK2 transcriptional coactivators lack DNA binding domains, but they interact with the bona fide DNA-binding proteins including CCA1, LHY, RVE4, and RVE8 to bind to core clock genes (Xie et al., 2014). Although it is unclear so far, the LNK1/2-interacting CCA1/RVEs and/or as-yet-unidentified DNA-binding proteins may transcriptionally activate CCA1 and PRR9 expression in the morning and also enable recruitment of the Sin3-HDAC complex to the morning gene loci to subsequently dampen expression after peak phase. The dynamic nature of histone acetylation and deacetylation depends on sophisticated interactions with transcription factors, and protein interaction networks further diversify the molecular mechanisms underlying rhythmic expression of core clock genes and thus circadian oscillation.
Arabidopsis thaliana (Columbia-0 ecotype) was used for all experiments described, unless specified otherwise. Plants were grown under neutral day conditions (NDs; 12-h light/12-h dark cycles) with cool white fluorescent light (120 μmol photons m-2 s-1) at 22-23°C. The afr1-1afr2-1 mutant was previously reported (Gu et al., 2013). sap18-2, cca1-1lhy-21, and cca1-2 mutants were obtained from Arabidopsis Biological Resource Center (ABRC). The lack of gene expression in mutants was verified by means of RT-PCR.
Total RNA was extracted using the TRI reagent (TAKARA Bio, Singa, Japan) according to the manufacturer’s recommendations. Reverse transcription (RT) was performed using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Dr. Protein, Seoul, South Korea) with oligo(dT18) to synthesize first-strand cDNA from 2 μg of total RNA. Total RNA samples were pretreated with an RNAse-free DNAse. cDNAs were diluted to 100 μL with TE buffer, and 1 μL of diluted cDNA was used for PCR amplification.
Quantitative RT-PCR reactions were performed in 96-well blocks using the Step-One Plus Real-Time PCR System (Applied Biosystems). The PCR primers used are listed in Supplementary Table S1. The values for each set of primers were normalized relative to the EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A) gene (At3g13920). All RT-qPCR reactions were performed in three independent biological replicates using total RNA samples extracted from three independent replicate samples. The comparative ΔΔCT method was employed to evaluate the relative quantities of each amplified product in the samples. The threshold cycle (CT) was automatically determined for each reaction by the system set with default parameters. Specificity of the RT-qPCR reactions was determined by melt curve analysis of the amplified products using the standard method installed in the system.
Yeast two-hybrid (Y2H) assays were performed using the BD Matchmaker system (Clontech, Mountain View, CA, United States). The pGADT7 vector was used for GAL4-AD fusion, and the pGBKT7 vector was used for GAL4-BD fusion. The yeast strain AH109 harboring the LacZ and His reporter genes was used. PCR products were subcloned into the pGBKT7 and pGADT7 vectors. The expression constructs were cotransformed into yeast AH109 cells and transformed cells were selected by growth on SD/-Leu/-Trp medium.
The LNK genes were fused in-frame to the 5′ end of a gene sequence encoding the C-terminal half of EYFP in the pSATN-cEYFP-C1 vector (E3082). The AFR cDNA sequences were fused in-frame to the 5′ end of a gene sequence encoding the N-terminal half of EYFP in the pSATN-nEYFP-C1 vector (E3081). The IDD14-RFP construct was used as a nuclear marker (Seo et al., 2011). The expression constructs were cotransformed into Arabidopsis protoplasts. Expression of the fusion constructs was monitored by fluorescence microscopy using a Zeiss LSM510 confocal microscope (Carl Zeiss, Jena, Germany).
pCCA1:CCA1-HA-YFP/cca1-1 and 35S:AFR-MYC transgenic plants were used for ChIP. Anti-MYC (06-599, Millipore), anti-HA (ab9110, Abcam), and anti-H3ac (05-724, Millipore) antibodies and salmon sperm DNA/protein A agarose beads (Millipore, Billerica, MA, United States) were used for chromatin immunoprecipitation. DNA was purified using phenol/chloroform/isoamyl alcohol and sodium acetate (pH 5.2). The level of eluted DNA fragments was quantified by quantitative real-time PCR using specific primer sets (Supplementary Table S2). The values were normalized to the input DNA level.
For transient expression assays using Arabidopsis protoplasts, reporter and effector plasmids were constructed. The core elements of the CCA1 and PRR9 promoters were inserted into the reporter plasmid, which contains a minimal 35S promoter sequence and the GUS gene. To construct the p35S:AFR effector plasmids, the AFR1 and AFR2 cDNAs were inserted into the effector vector containing the CaMV 35S promoter. Recombinant reporter and effector plasmids were cotransformed into Arabidopsis protoplasts by polyethylene glycol-mediated transformation. GUS activity was measured by a fluorometric method. A CaMV 35S promoter-luciferase construct was also cotransformed as an internal control. The luciferase assay was performed using the Luciferase Assay System kit (Promega,1).
PJS conceived and designed the experiments. PJS wrote the manuscript with the help of HGL. HGL and CH conducted the experiments and contributed to the study design.
This work was supported by the Basic Science Research (NRF-2016R1D1A1B03931139) and Basic Research Laboratory (NRF-2017R1A4A1015620) programs provided by the National Research Foundation of Korea and by the Next-Generation BioGreen 21 Program (PJ01319304) provided by the Rural Development Administration.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Yuehui He for kindly providing afr1-1afr1-2 seeds and Dr. Rachel Green for pCCA1:CCA1-HA-YFP/cca1-1 seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00171/full#supplementary-material
Agalioti, T., Lomvardas, S., Parekh, B., Yie, J., Maniatis, T., and Thanos, D. (2000). Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103, 667–678. doi: 10.1016/S0092-8674(00)00169-0
Ahringer, J. (2000). NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16, 351–356. doi: 10.1016/S0168-9525(00)02066-7
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Alland, L., David, G., Shen-Li, H., Potes, J., Muhle, R., Lee, H. C., et al. (2002). Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 22, 2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002
Bauer, A. J., and Martin, K. A. (2017). Coordinating regulation of gene expression in cardiovascular disease: interactions between chromatin modifiers and transcription factors. Front. Cardiovasc. Med. 4:19. doi: 10.3389/fcvm.2017.00019
Berr, A., McCallum, E. J., Ménard, R., Meyer, D., Fuchs, J., Dong, A., et al. (2010). Arabidopsis SET DOMAIN GROUP2 is required for H3K4 trimethylation and is crucial for both sporophyte and gametophyte development. Plant Cell 22, 3232–3248. doi: 10.1105/tpc.110.079962
Buszewicz, D., Archacki, R., Palusiñski, A., Kotliñski, M., Fogtman, A., Iwanicka-Nowicka, R., et al. (2016). HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 39, 2108–2122. doi: 10.1111/pce.12756
Chow, B. Y., Helfer, A., Nusinow, D. A., and Kay, S. A. (2012). ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7, 170–173. doi: 10.4161/psb.18766
Clark, M. D., Marcum, R., Graveline, R., Chan, C. W., Xie, T., Chen, Z., et al. (2015). Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc. Natl. Acad. Sci. U.S.A. 112, E3669–E3678. doi: 10.1073/pnas.1504021112
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-r130
Ding, Z., Millar, A. J., Davis, A. M., and Davis, S. J. (2007). TIME FOR COFFEE encodes a nuclear regulator in the Arabidopsis thaliana circadian clock. Plant Cell 19, 1522–1536. doi: 10.1105/tpc.106.047241
Dodd, A. N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.1115581
Farinas, B., and Mas, P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66, 318–329. doi: 10.1111/j.1365-313X.2011.04484.x
Fujiwara, S., Wang, L., Han, L., Suh, S. S., Salomé, P. A., McClung, C. R., et al. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23073–23083. doi: 10.1074/jbc.M803471200
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Grimaldi, B., Nakahata, Y., Kaluzova, M., Masubuchi, S., and Sassone-Corsi, P. (2009). Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int. J. Biochem. Cell Biol. 41, 81–86. doi: 10.1016/j.biocel.2008.08.035
Gu, X., Wang, Y., and He, Y. (2013). Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 11:e1001649. doi: 10.1371/journal.pbio.1001649
Hanano, S., Stracke, R., Jakoby, M., Merkle, T., Domagalska, M. A., Weisshaar, B., et al. (2008). A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics 9:182. doi: 10.1186/1471-2164-9-182
Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Haydon, M. J., Mielczarek, O., Robertson, F. C., Hubbard, K. E., and Webb, A. A. (2013). Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 502, 689–692. doi: 10.1038/nature12603
Hemmes, H., Henriques, R., Jang, I. C., Kim, S., and Chua, N. H. (2012). Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 53, 2016–2029. doi: 10.1093/pcp/pcs148
Herrero, E., Kolmos, E., Bujdoso, N., Yuan, Y., Wang, M., Berns, M. C., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443. doi: 10.1105/tpc.111.093807
Hsu, P. Y., and Harmer, S. L. (2012). Circadian phase has profound effects on differential expression analysis. PLoS One 7:e49853. doi: 10.1371/journal.pone.0049853
Huang, W., Pérez-García, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Hung, F. Y., Chen, F. F., Li, C., Chen, C., Lai, Y. C., Chen, J. H., et al. (2018). The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 46, 10669–10681. doi: 10.1093/nar/gky749
Inukai, S., Kock, K. H., and Bulyk, M. L. (2017). Transcription factor-DNA binding: beyond binding site motifs. Curr. Opin. Genet. Dev. 43, 110–119. doi: 10.1016/j.gde.2017.02.007
Jang, I. C., Chung, P. J., Hemmes, H., Jung, C., and Chua, N. H. (2011). Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23, 459–470. doi: 10.1105/tpc.110.080481
Kim, Y. J., Wang, R., Gao, L., Li, D., Xu, C., Mang, H., et al. (2016). POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863. doi: 10.1073/pnas.1618618114
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Kuo, M. H., and Allis, C. D. (1998). Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20, 615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H
Kusakina, J., and Dodd, A. N. (2012). Phosphorylation in the plant circadian system. Trends Plant Sci. 17, 575–583. doi: 10.1016/j.tplants.2012.06.008
Kuzmichev, A., Zhang, Y., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2002). Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell Biol. 22, 835–848. doi: 10.1128/MCB.22.3.835-848.2002
Laherty, C. D., Billin, A. N., Lavinsky, R. M., Yochum, G. S., Bush, A. C., Sun, J. M., et al. (1998). SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2, 33–42. doi: 10.1016/S1097-2765(00)80111-2
Lee, K., and Seo, P. J. (2018). The HAF2 protein shapes histone acetylation levels of PRR5 and LUX loci in Arabidopsis. Planta 248, 513–518. doi: 10.1007/s00425-018-2921-y
Li, G., Siddiqui, H., Teng, Y., Lin, R., Wan, X. Y., Li, J., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13, 616–622. doi: 10.1038/ncb2219
Lomvardas, S., and Thanos, D. (2002). Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110, 261–271. doi: 10.1016/S0092-8674(02)00822-X
Lu, S. X., Knowles, S. M., Webb, C. J., Celaya, R. B., Cha, C., Siu, J. P., et al. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155, 906–915. doi: 10.1104/pp.110.167015
Lu, S. X., Webb, C. J., Knowles, S. M., Kim, S. H. J., Wang, Z., and Tobin, E. M. (2012). CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158, 1079–1088. doi: 10.1104/pp.111.189670
Malapeira, J., Khaitova, L. C., and Mas, P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 21540–21545. doi: 10.1073/pnas.1217022110
Mas, P. (2008). Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 18, 273–281. doi: 10.1016/j.tcb.2008.03.005
Michael, T. P., and McClung, C. R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132, 629–639. doi: 10.1104/pp.021006
Mizuno, T., and Yamashino, T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49, 481–487. doi: 10.1093/pcp/pcn008
Munshi, N., Agalioti, T., Lomvardas, S., Merika, M., Chen, G., and Thanos, D. (2001). Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293, 1133–1136. doi: 10.1126/science.293.5532.1133
Munshi, N., Merika, M., Yie, J., Senger, K., Chen, G., and Thanos, D. (1998). Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2, 457–467. doi: 10.1016/S1097-2765(00)80145-8
Murfett, J., Wang, X. J., Hagen, G., and Guilfoyle, T. J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. doi: 10.1105/tpc.13.5.1047
Nagel, D. H., Doherty, C. J., Pruneda-Paz, J. L., Schmitz, R. J., Ecker, J. R., and Kay, S. A. (2015). Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, E4802–E4810. doi: 10.1073/pnas.1513609112
Nagel, D. H., and Kay, S. A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–R657. doi: 10.1016/j.cub.2012.07.025
Nakahata, Y., Kaluzova, M., Grimaldi, B., Sahar, S., Hirayama, J., Chen, D., et al. (2008). The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340. doi: 10.1016/j.cell.2008.07.002
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Nakamichi, N., Kita, M., Ito, S., Yamashino, T., and Mizuno, T. (2005). Pseudo-response regulators, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46, 686–698. doi: 10.1093/pcp/pci086
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Park, J., Lim, C. J., Shen, M., Park, H. J., Cha, J. Y., Iniesto, E., et al. (2018). Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. U.S.A. 115, E5400–E5409. doi: 10.1073/pnas.1721241115
Perales, M., and Mas, P. (2007). A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19, 2111–2123. doi: 10.1105/tpc.107.050807
Perez-Garcia, P., Ma, Y., Yanovsky, M. J., and Mas, P. (2015). Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 5249–5253. doi: 10.1073/pnas.1420792112
Peserico, A., and Simone, C. (2011). Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011:371832. doi: 10.1155/2011/371832
Pinon, V., Yao, X., Dong, A., and Shen, W. H. (2017). SDG2-mediated H3K4me3 is crucial for chromatin condensation and mitotic division during male gametogenesis in Arabidopsis. Plant Physiol. 174, 1205–1215. doi: 10.1104/pp.17.00306
Pokhilko, A., Mas, P., and Millar, A. J. (2013). Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 7:23. doi: 10.1186/1752-0509-7-23
Salome, P. A., Weigel, D., and McClung, C. R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22, 3650–3661. doi: 10.1105/tpc.110.079087
Scott, K. L., and Plon, S. E. (2003). Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 4522–4531. doi: 10.1128/MCB.23.13.4522-4531.2003
Seo, P. J., Kim, M. J., Ryu, J. Y., Jeong, E. Y., and Park, C. M. (2011). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2:303. doi: 10.1038/ncomms1303
Seo, P. J., and Mas, P. (2014). Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26, 79–87. doi: 10.1105/tpc.113.119842
Silverstein, R. A., and Ekwall, K. (2005). Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 47, 1–17. doi: 10.1007/s00294-004-0541-5
Somers, D. E., Kim, W. Y., and Geng, R. (2004). The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16, 769–782. doi: 10.1105/tpc.016808
Song, C. P., and Galbraith, D. W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257. doi: 10.1007/s11103-005-3880-9
Song, H. R., and Noh, Y. S. (2012). Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells 34, 279–287. doi: 10.1007/s10059-012-0103-5
Stratmann, T., and Mas, P. (2008). Chromatin, photoperiod and the Arabidopsis circadian clock: a question of time. Semin. Cell Dev. Biol. 19, 554–559. doi: 10.1016/j.semcdb.2008.07.012
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14:e1007280. doi: 10.1371/journal.pgen.1007280
Todeschini, A. L., Georges, A., and Veitia, R. A. (2014). Transcription factors: specific DNA binding and specific gene regulation. Trends Genet. 30, 211–219. doi: 10.1016/j.tig.2014.04.002
Wang, L., Kim, J., and Somers, D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766. doi: 10.1073/pnas.1215010110
Wang, Z., Zang, C., Cui, K., Schones, D. E., Barski, A., Peng, W., et al. (2009). Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031. doi: 10.1016/j.cell.2009.06.049
Wang, Z. Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M. S., and Tobin, E. M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. doi: 10.1105/tpc.9.4.491
Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000). Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. doi: 10.1023/A:1006498413543
Xie, Q., Wang, P., Liu, X., Yuan, L., Wang, L., Zhang, C., et al. (2014). LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26, 2843–2857. doi: 10.1105/tpc.114.126573
Yang, X. J., and Seto, E. (2007). HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318. doi: 10.1038/sj.onc.1210599
Yao, X., Feng, H., Yu, Y., Dong, A., and Shen, W. H. (2013). SDG2-mediated H3K4 methylation is required for proper Arabidopsis root growth and development. PLoS One 8:e56537. doi: 10.1371/journal.pone.0056537
Yoo, S. K., Hong, S. M., Lee, J. S., and Ahn, J. H. (2011). A genetic screen for leaf movement mutants identifies a potential role for AGAMOUS-LIKE 6 (AGL6) in circadian-clock control. Mol. Cells 31, 281–287. doi: 10.1007/s10059-011-0035-5
Zhang, C., Xie, Q., Anderson, R. G., Ng, G., Seitz, N. C., Peterson, T., et al. (2013). Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 9:e1003370. doi: 10.1371/journal.ppat.1003370
Keywords: circadian clock, chromatin modification, histone deacetylase (HDAC), Sin3 histone deacetylase and corepressor complex, CCA1, PRR9
Citation: Lee HG, Hong C and Seo PJ (2019) The Arabidopsis Sin3-HDAC Complex Facilitates Temporal Histone Deacetylation at the CCA1 and PRR9 Loci for Robust Circadian Oscillation. Front. Plant Sci. 10:171. doi: 10.3389/fpls.2019.00171
Received: 30 October 2018; Accepted: 01 February 2019;
Published: 18 February 2019.
Edited by:
Jean-Benoit Charron, McGill University, CanadaReviewed by:
Matt Jones, University of Essex, United KingdomCopyright © 2019 Lee, Hong and Seo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pil Joon Seo, cGpzZW8xQHNudS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.