- 1Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences/Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, Nanjing, China
- 2Department of Plant Pathology, Nanjing Agricultural University, Nanjing, China
- 3Department of Crop and Soil Sciences, Washington State University, Pullman, WA, United States
- 4National Center for Soybean Improvement, Nanjing Agricultural University, Nanjing, China
Initially identified as a mammalian apoptosis suppressor, defender against apoptotic death 1 (DAD1) protein has conserved plant orthologs acting as negative regulators of cell death. The potential roles and action mechanisms of plant DADs in resistance against Phytophthora pathogens are still unknown. Here, we cloned GmDAD1 from soybean and performed functional dissection. GmDAD1 expression can be induced by Phytophthora sojae infection in both compatible and incompatible soybean varieties. By manipulating GmDAD1 expression in soybean hairy roots, we showed that GmDAD1 transcript accumulations are positively correlated with plant resistance levels against P. sojae. Heterologous expression of GmDAD1 in Nicotiana benthamiana enhanced its resistance to Phytophthora parasitica. NbDAD1 from N. benthamiana was shown to have similar role in conferring Phytophthora resistance. As an endoplasmic reticulum (ER)-localized protein, GmDAD1 was demonstrated to be involved in ER stress signaling and to affect the expression of multiple defense-related genes. Taken together, our findings reveal that GmDAD1 plays a critical role in defense against Phytophthora pathogens and might participate in the ER stress signaling pathway. The defense-associated characteristic of GmDAD1 makes it a valuable working target for breeding Phytophthora resistant soybean varieties.
Introduction
As sessile organisms, plants are continually exposed to various biotic and abiotic stresses. Therefore, complex stress perception, signal transduction and adaptation strategies have evolved in plants to cope with adverse environmental conditions. In particular, the programmed cell death (PCD) pathway has been demonstrated to play key roles in plant responses to both abiotic and biotic stresses (Dickman et al., 2001; Lam et al., 2001; Williams et al., 2010). In plant defense against pathogens, PCD restricts microbe growth and spreading in host tissue by eliminating excessive damaged cells (Kimchi, 2007).
Several PCD repressors have been identified in plants, including Bax inhibitor 1 (BI-1), B-cell lymphoma2 (Bcl-2)-associated athanogene (BAG), ER-luminal binding immunoglobulin protein (BiP), and defender against apoptotic death 1 (DAD1) (Gallois et al., 1997; Matsumura et al., 2003; Doukhanina et al., 2006; Williams et al., 2010; Jing et al., 2016; Li et al., 2016a,b). These repressors may increase or decrease plant resistance to different pathogens (Kawai-Yamada et al., 2004, 2009; Babaeizad et al., 2009; Watanabe and Lam, 2009; Eichmann et al., 2010; Ishikawa et al., 2011).
Among these PCD repressors, DAD1 is unique as it is conserved from yeast to mammals (Nakashima et al., 1993). Initially identified in a temperature-sensitive mutant hamster tsBN7 cell line, DAD1 is a subunit in the oligosaccharyltransferase (OST) complex, which is a core component for catalyzing N-glycosylation in ER (Yan et al., 2005; Peristera and Stephen, 2012). N-glycosylation is the attachment of oligosaccharides to certain asparagine residues of specific nascent proteins, which ensures their successful folding and export from ER. In Drosophila melanogaster, DmDAD1 is essential for efficient N-glycosylation in developing tissues (Zhang et al., 2016). Disruption of DmDAD1 increases accumulation of unfolded or misfolded proteins, which triggers stress signaling in ER and initiates PCD. In contrast, its overexpression stabilizes or increases N-glycosylation (Zhang et al., 2016).
Different hypotheses have been proposed for the roles of DAD1 in maintaining cell viability. DAD1 may facilitate the targeting of OST complex to proteins directly responsible for cell viability. On the other hand, since DAD1 interacts with Mcl1, a Bcl2-family protein acting as an apoptosis inhibitor (Makishima et al., 2000), DAD1 may also affect cell viability in an OST-independent manner.
Plant DAD1 orthologs from Arabidopsis thaliana and rice can rescue hamster tsBN7 cells from apoptosis (Gallois et al., 1997; Tanaka et al., 1997), which indicates they may also function as cell death repressors. Subsequent studies demonstrate that AtDAD1 protects Arabidopsis protoplast cells against ultraviolet-C-induced PCD (Danon et al., 2004) and DAD1 expression in Gladiolus decreases drastically during petal senescence (Yamada et al., 2004). Regarding the roles of DAD1 proteins in plant defense, Wang X. J. et al. (2011) reported that TaDAD2-silenced wheat leaves have attenuated resistance to Puccinia striiformis with down-regulated expression of several defense-related genes. However, how this protein modulates plant-pathogen interactions has not been well characterized overall.
In this study, a DAD1 orthologous gene was identified from soybean (Glycine max). Spatial and temporal expression of GmDAD1 upon P. sojae infection, as well as its protein subcellular localization, were investigated. The function of GmDAD1 in conferring Phytophthora resistance was dissected in soybean hairy roots with GmDAD1 specifically silenced by RNAi, and Nicotiana benthamiana transgenic lines overexpressing GmDAD1 or suppressing native NbDAD1. Our findings demonstrate that GmDAD1 plays a critical role in Phytophthora resistance probably via regulating ER stress signaling.
Materials and Methods
Plant Materials and Growth Conditions
Two soybean varieties were used in this research: Williams 82 carrying the gene Rps1k, which confers resistance to P. sojae race 2 (Bernard and Cremeens, 1988) and Williams which does not carry any known Rps resistance gene (Bernard and Lindahl, 1972). Seeds of Williams 82 and Williams were sown in small plastic pots containing disinfected soil and maintained in greenhouse at 25°C and 16h:8h light/dark photoperiod. N. benthamiana plants were grown under identical conditions as described above.
Culture of Phytophthora Pathogens
Phytophthora sojae isolates P6497 and P6497-RFP, which is a P. sojae strain constitutively expressing red fluorescence protein (RFP) (Xiong et al., 2014) were routinely cultured on 10% V8 juice agar plates at 25°C in the dark. Phytophthora parasitica was grown under the same conditions.
P. sojae Inoculation and Soybean Samples Collection
Root, stem and leaf samples of the soybean varieties Williams 82 and Williams were collected at seedling and pod-filling stages. Hypocotyl inoculation of P. sojae was performed on Williams 82 and Williams plants as described previously (Sun et al., 2014). Agar disks containing hyphae were cut from fresh cultures and inoculated onto hypocotyl incision. After inoculation, the seedlings were placed in growth chamber to keep moisture. Inoculated stems were collected at 0, 6, 12, 24, and 48 h post inoculation (hpi). All samples were frozen immediately in liquid nitrogen and stored at -70°C. Three biological replicates were performed for each time point.
DNA and RNA Extraction and RT-qPCR
Following supplier instructions, all DNA and RNA samples were extracted using the Hi-DNAsecure plant kit and the RNA simple Total RNA kit (Tiangen, China), respectively. For RNA samples, elimination of genomic DNA contamination and reverse transcription were performed using the HiScript II Q RT SuperMix reagent Kit (Vazyme, China).
qPCR reactions were performed on an ABI PRISM 7500 real-time PCR system (Applied Biosystems, United States) using the ChamQTM SYBR qPCR Master Mix reagent (Vazyme, China). Relative gene expression levels were calculated using the comparative 2-ΔΔCT method (Livak and Schmittgen, 2001). Statistical analysis was conducted using the Student’s t-test with Excel 2010 software and the data were considered statistically significant for P < 0.05. qPCR primers for GmDAD1 were designed from its conserved region. PsTEF (GenBank ID EU079791) was selected for determining P. sojae biomass (Yan et al., 2014). GmCons4 (GenBank ID BU578186.1) was selected as endogenous reference in soybean (Libault et al., 2008). NbEF1a (GenBank ID AY206004) was used as N. benthamiana reference in the VIGS (virus-induced gene silencing) assay.
Defense-related genes analyzed in this research include five pathogenesis-related (PR) genes: PR1a, PR2, PR3, PR4 and PR5 (Bertini et al., 2003; Chen et al., 2007; Mazarei et al., 2007; Maldonado et al., 2014); the JA-regulated defense gene plant defensin 1.2 (PDF1.2) (Lorenzo and Solano, 2005); the ethylene (ET) signaling marker gene ethylene response factor 1 (ERF1) (Lorenzo et al., 2003); the reactive oxygen species (ROS) biosynthetic gene NADP oxidase (NADPHOX) and two ROS scavenging genes: catalase (CAT) and ascorbate peroxidase (APX) (Perez and Brown, 2014). We employed the sequences of G. max if the genes have been reported already, or obtained them by searching in the soybean EST and genome databases1 using orthologous sequences from A. thaliana as queries. All primers were designed using the Primer Premier 5 software. Primer specificity was evaluated by sequence similarity comparison and melting curve results of RT-qPCR. The primers of ER related genes were designed used the same strategy. The analyzed ER-stress related genes were the binding immunoglobulin protein (Bip), the protein disulphide isomerase (PDI), the calnexin1 (CNX1), the ER lumen-localized Dnaj protein3a (ERdj3A), the luminal binding domain/glucose-regulated protein 94 (GRP94), the basic region/leucine zipper motif 17 (bZIP17) and the downstream gene vacuolar processing enzyme (VPE) (Rojo et al., 2004; Cai et al., 2014; Tiziana and Roberto, 2014). All primers used in this study and detailed information were listed in Supplementary Table S1.
Subcellular Localization of the GmDAD1 Protein
For subcellular localization, the full-length coding sequence (CDS) of GmDAD1 was amplified from cDNAs of the Williams variety using primer pair pBIN-G-DAD-F/R (Supplementary Table S1). The 351-bp GmDAD1 CDS was then translationally fused with GFP after cloning into pBIN-GFP (Zhang et al., 2014) using KpnI and XbaI sites. After sequencing validation, GmDAD1-GFP and mCherry-HDEL constructs were introduced into Agrobacterium tumefaciens stain GV3101. The two Agrobacterium liquid cultures were mixed and co-infiltrated into N. benthamiana leaves using a blunt syringe. After maintained for 48 h in greenhouse, agroinfiltrated leaves were detached and visualized with a laser scanning confocal microscope (Zeiss, GERMANY) at 488 and 591 nm for GFP and mCherry detection, respectively.
Plasmid Construction for Soybean Cotyledon Transformation
The pBIN-GFP-GmDAD1 construct which was used to determine GmDAD1 subcellular localization was also used to overexpress GmDAD1 in soybean hairy roots, and the pBIN-GFP empty vector was used as control which allows expression of the GFP only. To make the GmDAD1-RNAi construct, partial GmDAD1 gene was amplified (using primers p12-DAD-F and p12-DAD-R) and cloned into pDONR221 (Invitrogen, United States) and then entered in pHellsGate12:GFP via Gateway LR reaction. Modified from pHellsGate12 (Wesley et al., 2001), pHellsGate12:GFP harbors a 35S:GFP:nos expression cassette (Yan et al., 2014). After sequence validation, the pBIN-GFP-GmDAD1, GmDAD1-RNAi, the empty pBIN-GFP and pHellsGate12:GFP vectors were introduced into Agrobacterium rhizogenes strain K599 by electroporation.
Plasmid Construction for N. benthamiana Transformation
To overexpress GmDAD1 in N. benthamiana, the full length of GmDAD1 CDS was obtained from cDNAs of the Williams variety using primer pair pDONR-DAD-F/R (Supplementary Table S1) and then cloned into the entry vector pDONR221 via Gateway BP reaction. After sequencing validation, the fragment was then entered in pEarlyGate202 via LR recombination reaction between the entry clone and the destination vector (Invitrogen, United States) (Earley et al., 2006). To make Tobacco Rattle Virus (TRV)-based VIGS construct targeting NbDAD1, partial fragment of NbDAD1 was amplified using primer pair TRV:NbDAD-F/R and cloned into pTRV2 (Liu et al., 2002) using KpnI and EcoRI sites. All constructs were validated by sequencing and transformed into A. tumefaciens strain EHA105 for N. benthamiana transformation and GV3101 for VIGS experiment.
Soybean Cotyledon Transformation
Surface-sterilized soybean seeds were soaked in sterilized water overnight and then germinated on medium containing 0.5% sucrose and 1.2% agar in growth chamber with 16h:8h light/dark photoperiod. About 5 days after germination, unblemished cotyledons were harvested for A. rhizogenes-mediated transformation. Transformation was performed as described previously (Yan et al., 2014). After about 3 weeks of cultivation, transformed hairy roots became abundant at inoculated cotyledons. Positive transformants were selected by detecting GFP signal under fluorescence microscopy, cut off from cotyledons, and cultivated on White medium (Supplementary Table S2) for further verification and resistance level test.
N. benthamiana Transformation and Virus-Induced Gene Silencing (VIGS)
Nicotiana benthamiana plants overexpressing GmDAD1 were generated via A. tumefaciens mediated leaf disk transformation (Horsch et al., 1985). The T1 seeds harvested from self-pollinated T0 plants were surface-sterilized with 70% ethanol for 30 s, and 10% sodium hypochlorite solution for 5 min, then washed by sterilized water for five times. The sterilized seeds were germinated on MS medium with 100 mg/L glufosinate ammonium (Sigma, United States). T2 seeds were collected and sown in small plastic pots. After 2 weeks, the seedlings were sprayed with 100 mg/L glufosinate ammonium solution. Resistant were transplanted to new pots and confirmed by both genomic DNA and cDNA PCR using gene-specific primers (DAD-Test-F/R). The T2 plants were used for functional characterization.
For TRV-VIGS assay, Agrobacterium cultures harboring pTRV1 and pTRV2-VIGS (TRV2-NbDAD1, TRV2 empty vector or TRV2-NbPDS used as positive control of silencing) were mixed and infiltrated into N. benthamiana leaves using a blunt syringe (Fu et al., 2002). Inoculated plants were maintained at 20°C in greenhouse for effective virus infection and spread.
Resistance Assay of N. benthamiana Against Phytophthora parasitica
Leaves from 5 to 6-week-old N. benthamiana plants were detached and inoculated with 20 μl P. parasitica zoospores (104 ml-1) per leaf. Inoculated leaves were then kept in a moist chamber and lesion diameters were measured at 36 and 60 hpi. Representative infected leaves were photographed at 60 hpi under a UV lamp and then stained with trypan blue to visualize the infected area. The experiment was repeated three times with similar results and at least 20 leaves were inoculated for each biological replicate. Two weeks after infiltration, leaves from TRV and NbDAD1-VIGS plants were inoculated with P. parasitica using the same strategy. Lesion diameters were measured at 36 and 48 hpi due to the semi-dwarf phenotype of NbDAD1-VIGS plants. At least 10 lesions per construct were measured with three biological repeats. Student’s t-test was used to analyze the significance of differences. Difference were considered as significant when P < 0.05.
Root Infection and Observation
After verification by detection of GFP fluorescence and qPCR, transgenic hairy roots of similar length (approximately 3 cm) were excised and dipped in the zoospore suspension (104 zoospores per ml) of P. sojae race P6497-RFR for 5 min as described previously (Xiong et al., 2014). Inoculated roots were placed in Petri dishes containing 0.6% agar in the dark at room temperature. At 12, 24, and 36 hpi, the infection progression was monitored under an OLYMPUS MVX10 (OLYMPUS, Japan) fluorescence microscope via RFP fluorescence detection at 535 nm. The P. sojae-specific gene PsTEF was used for qPCR quantification of the relative biomass of P. sojae. For each sample, about 10 infected hairy roots were collected and pooled for DNA/RNA extraction which helps to reduce bias and increase statistical accuracy (Graham, 1991; Subramanian et al., 2005; Graham et al., 2007).
Western Blotting Assay
About 10 transgenic roots with GFP fluorescence were collected and ground in liquid nitrogen. Total proteins were extracted with the extraction buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 2 mM DTT, 1% triton, 2% polyvinylpolypyrrolidone and Roche complete protein inhibitor tablets). The samples were boiled for 10 min in 6× sodium dodecyl sulfate (SDS) loading buffer. SDS-PAGE and immunoblotting were performed in a mini-gel apparatus and submarine gel transfer systems (Bio-Rad, United States), respectively. Proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes and then membranes were blocked with 5% non-fat dry milk in 0.01 M PBST for 1 h and then incubated with anti-GFP (1:1,000) (Sigma, United States) for 2 h at room temperature. After washing by TBST three times, the membrane was incubated with IRDye®800CW Goat anti-rabbit IgG (LI-COR, United States) secondary antibody at room temperature for 1 h. Protein bands were detecting using the Odyssey® CLx quantitative fluorescence imaging system (LI-COR, United States).
Sequence Analysis and Alignment
The conserved and transmembrane domains of GmDAD1 were analyzed with InterProScan and TMPRED respectively (Hofmann and Stoel, 1993; Jones et al., 2014). Multiple sequence alignment was performed using the BioEdit software (Hall, 1999).
Results
ER-Located GmDAD1 Shares Conserved Regions With Other Plant DAD1 Orthologs
GmDAD1 (Gma.7542.2.S1_at) was identified from an Affymetrix Genechip microarray data analysis on soybean and P. sojae interaction (Zhou et al., 2009). GmDAD1 was up-regulated in soybean varieties with different degrees of resistance to P. sojae (Zhou et al., 2009). Sequence analysis of GmDAD1 (cloned from the Williams variety) revealed that its open reading frame (ORF) encodes a protein of 117 amino acid residues. GmDAD1 shares 91, 54, and 36% identities with DAD1 orthologs in Arabidopsis thaliana, Homo sapiens, and Saccharomyces cerevisiae, respectively. Similar to other plant DAD1 orthologs, GmDAD1 contains three transmembrane regions (residues 27–52, 61–81, and 95–115) and a subunit of OST (residues 13–116) (Figure 1A). To investigate the subcellular localization of GmDAD1, a GmDAD1-GFP fusion construct driven by the CaMV 35S promoter was expressed in N. benthamiana leaves. GmDAD1-GFP co-localized in the cytoplasm with mCherry-HDEL, an endoplasmic reticulum (ER) marker, demonstrating the ER localization of GmDAD1 (Figure 1B).
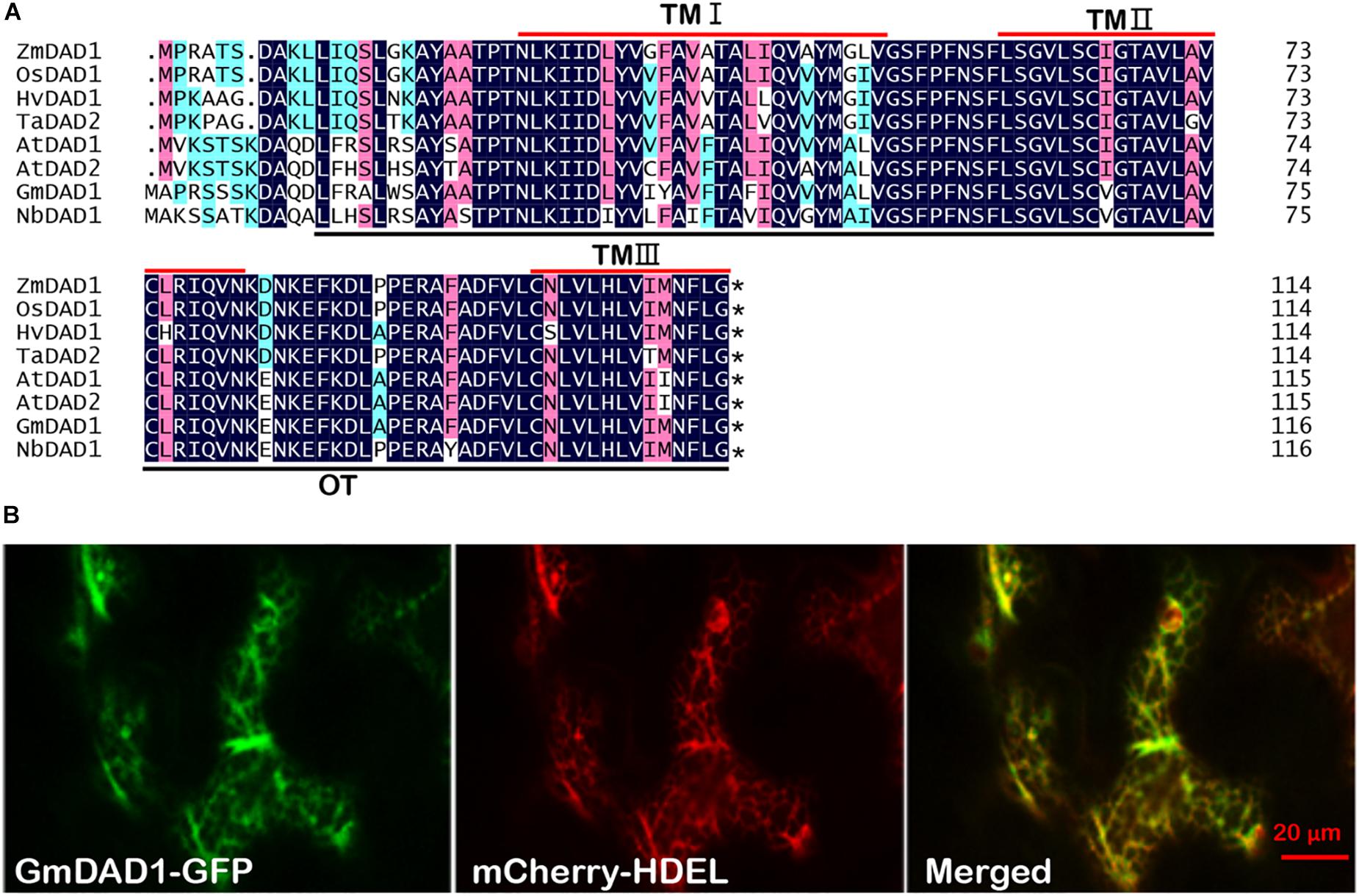
Figure 1. Molecular characterization and subcellular localization of GmDAD1 protein. (A) Sequence alignment of GmDAD1 and other defender against cell death (DAD) proteins. The darkblue (100%), pink (75%), and cyan (50%) boxes represent levels of amino acid identity or similarity. TM, transmembrane domain; OT, oligosaccharyltransferase domain. At, Arabidopsis thaliana; Hv, Hordeum vulgare; Zm, Zea mays; Os, Oryza sativa; Ta, Triticum aestivum. The asterisk indicates the stop codon. (B) Subcellular localization of GmDAD1 was performed via transient expression system in Nicotiana benthamiana. Green and red fluorescence represent the signal of GFP fusion protein and ER marker mCherry-HDEL, respectively. The reticulate fluorescence pattern of GmDAD1-GFP and its co-localization with mCherry-HDEL indicate accumulation in the ER.
GmDAD1 Expression Is Induced Upon P. sojae Infection
GmDAD1 transcript can be detected ubiquitously in roots, stems and leaves during plant development in cv Williams, with root being the organ exhibiting highest expression (Figure 2A). Interestingly, leaves showed much higher GmDAD1 transcript accumulation at pod filling stage than seedling stage (Figure 2A). Similar GmDAD1 expression pattern was detected in Williams 82 variety in the seedling stage (Supplementary Figure S1). On the contrary, the expression of GmDAD1 is higher in roots at the pod filling stage in Williams 82 than in Williams.
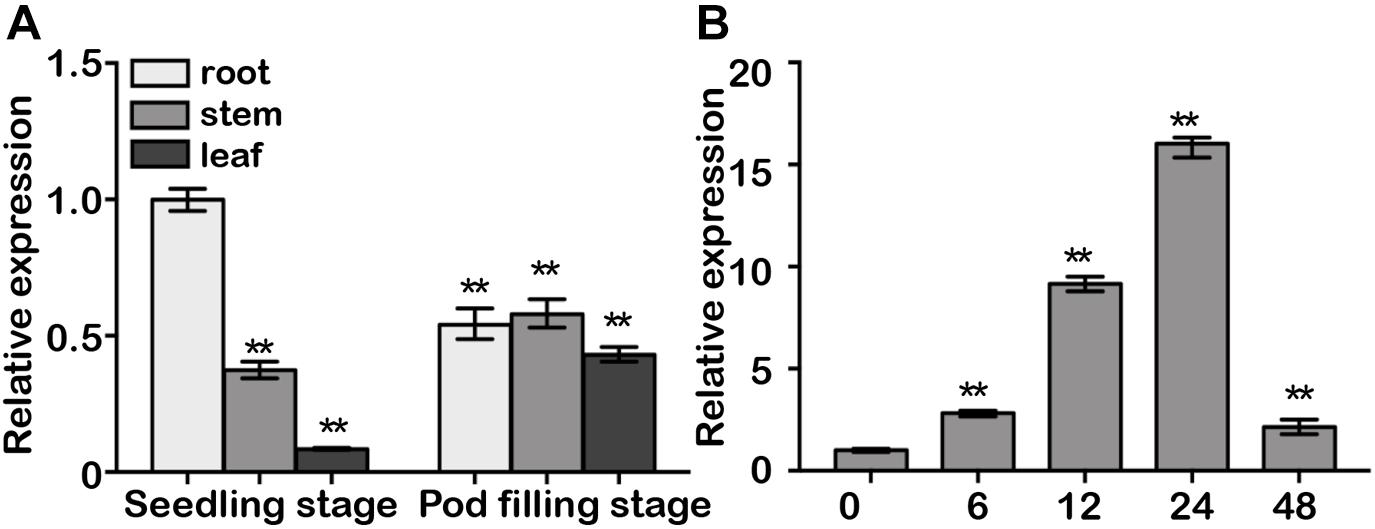
Figure 2. Gene expression analysis of GmDAD1. (A) GmDAD1 mRNA levels in various tissues of soybean cultivar Williams. Leaves, roots, and stems were harvested from plants at the seedling and pod filling stage. (B) Expression profiles of GmDAD1 in Williams (compatible interaction) at 0, 6, 12, 24, 48 h post inoculation (hpi) with P. sojae (P6497). The relative expression level was normalized to soybean GmCons4 (GenBank: BU578186.1). Means and standard deviations were calculated from three independent biological replicates. Data were analyzed by using Student’s t-tests (∗∗P < 0.01).
After inoculation with P6497, a P. sojae isolate of race 2, the compatible variety Williams showed elevated GmDAD1 expression which peaked at 24 hpi and subsequently decreased (Figure 2B). In the incompatible variety Williams 82, GmDAD1 was also significantly induced by P. sojae infection at 24 hpi (Supplementary Figure S1).
GmDAD1 Enhances Resistance to P. sojae in Soybean Hairy Roots
RT-qPCR analysis of ten mixed hairy roots displaying GFP fluorescence indicated that expression of GmDAD1 in GmDAD1-GFP overexpression (OE) plants was nearly 14-fold higher that in the control (GFP) (Figure 3A). Western blotting also showed the accumulation of the GmDAD1-GFP fusion protein (Figure 3B). When OE and GFP hairy roots were inoculated with P. sojae P6497-RFP (Xiong et al., 2014), the biomass of P. sojae was significantly and consistently less in OE hairy roots than in GFP samples at 12, 24, and 36 hpi (Figure 3C). In the GFP control, the invasion hyphae emerged at 12 hpi, rapidly extended at 24 hpi, and almost filled the entire tissue at 36 hpi (Figure 3D). In contrast, hyphal growth was limited and the invasion hyphae were much sparser in GmDAD1-GFP overexpression roots (Figure 3D), which is consistent with the lower accumulation of P. sojae biomass (Figure 3C).
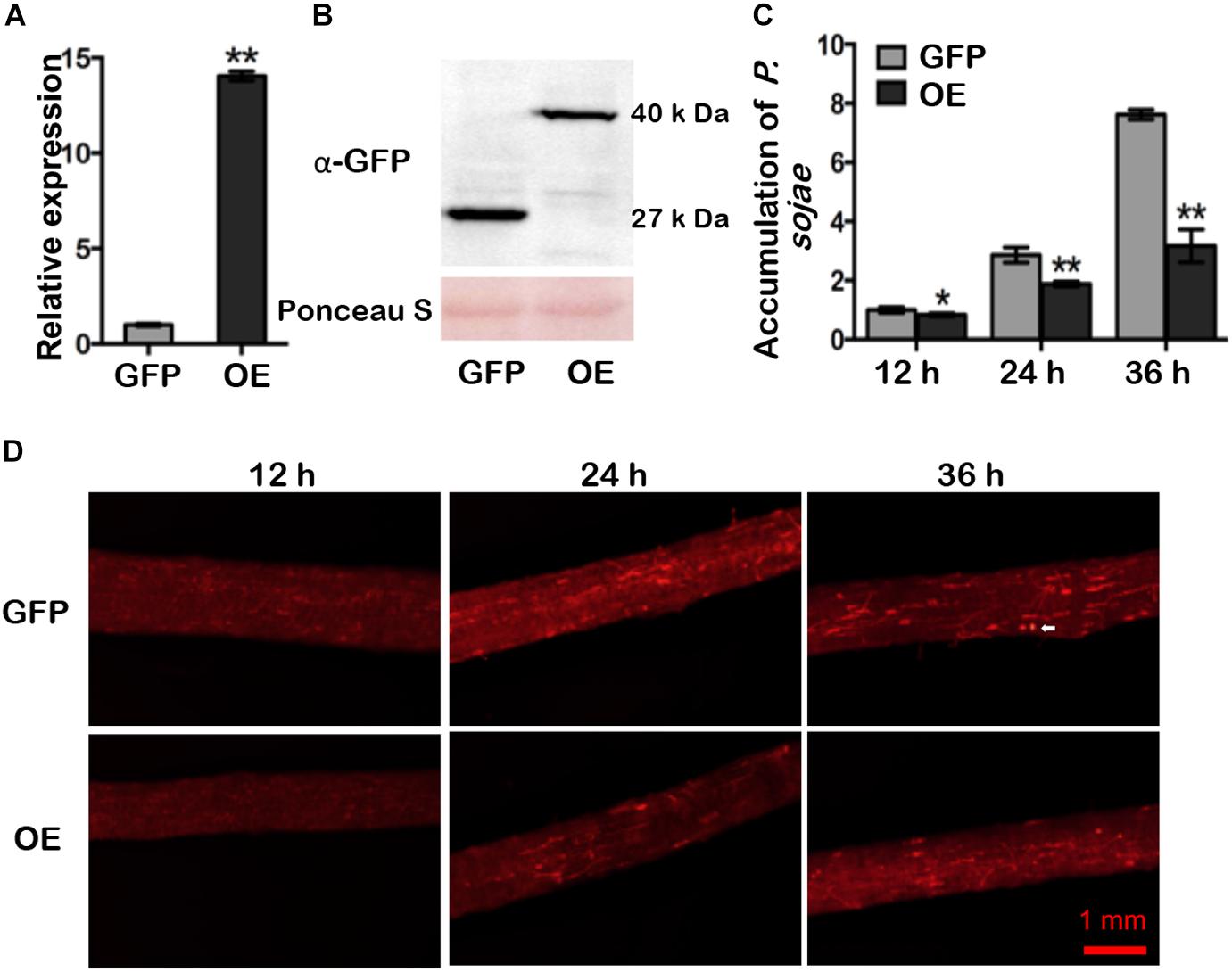
Figure 3. GmDAD1 overexpression enhances resistance to P. sojae in soybean hairy roots. (A) Expression of GmDAD1 in the GmDAD1-GFP overexpressing (OE) and control (GFP) hairy roots without P. sojae infection. Samples derived from different pooled root materials. Control has been transformed with the empty vector which allows expression of the GFP only. (B) Western blotting of proteins from hairy roots expressing GFP (control) and GmDAD1 fused with GFP tag. (C) Relative biomass of P. sojae determined by qPCR in inoculated OE and GFP hairy roots at 12, 24, and 36 hpi. Values represent the means of three replicates and 10 hairy roots were used for each biological replicate. Data were analyzed by using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01 compared with the control). (D) Microscopic analysis of P. sojae colonization in infected soybean hairy roots. The OE and control GFP hairy roots were inoculated with zoospore suspension (104 zoospore/ml) of the P. sojae P6497-RFP. Photos were taken at 12, 24, 36 hpi. The white arrow indicates a germinating oospore.
Silencing of GmDAD1 Reduces Resistance to P. sojae in Soybean Hairy Roots
RNAi-directed silencing of GmDAD1 in soybean hairy roots (Figure 4A) was performed as described previously (Yan et al., 2014). Both GmDAD1-RNAi (RNAi) and EV control (EV) roots were inoculated with P. sojae P6497-RFP. Compared with control, GmDAD1-RNAi roots showed gradually increased P. sojae biomass accumulation over time (Figure 4B). Furthermore, a greater hyphal growth and higher oospore germination can be observed in GmDAD1-RNAi roots (Figure 4C). Our results indicated that GmDAD1 is important for soybean resistance against P. sojae.
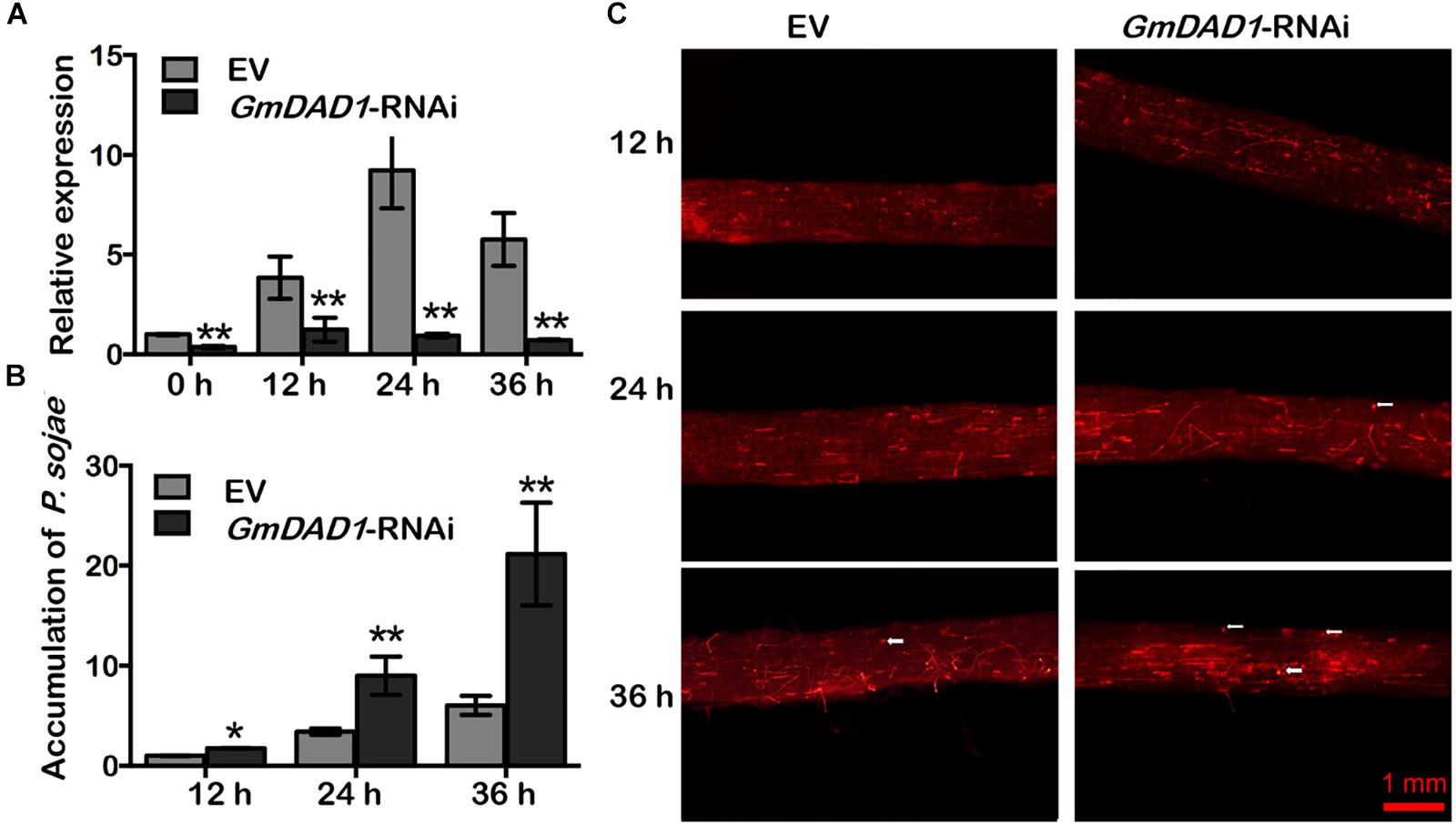
Figure 4. Silencing of GmDAD1 reduces resistance to P. sojae in soybean hairy roots. (A) Relative expression of GmDAD1 was determined by RT-qPCR in inoculated hairy roots in which GmDAD1 was silenced via RNAi (GmDAD1-RNAi) or empty vector (EV) at 0, 12, 24, and 36 hpi. (B) Relative biomass of P. sojae was determined in inoculated hairy roots GmDAD1-RNAi or EV at 12, 24, and 36 hpi. Values represent the means of three replicates ± SD. Data were analyzed by using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01 compared with the control). (C) Microscopic analysis of P. sojae colonization in soybean hairy roots. The control EV and GmDAD1-RNAi hairy roots were inoculated with zoospore suspension (104 zoospore/ml) of the P. sojae P6497-RFP. Photos were taken at 12, 24, 36 hpi. The white arrows indicate germinating oospores.
GmDAD1 Affects the Expression of Multiple Defense-Related Genes
To further determine whether the expression of defense-related genes was affected by GmDAD1 silencing, we assessed the expression of several genes in hairy roots inoculated with P. sojae, including the marker genes of SA, and JA/ET signaling pathways, ROS generation and scavenging. The expression of PR1a, PR2, PR3, PR5 and ERF1 were decreased in GmDAD1-RNAi roots after P. sojae inoculation. It is to note that the expression of PR1a was also dramatically suppressed without inoculation (Figure 5). In contrast, the expression of PDF1.2, PR4, and two ROS scavenging genes, CAT and APX, were induced in the GmDAD1 silencing roots infected with P. sojae (Figure 5). No significant change of NADPHOX expression was observed when GmDAD1 was silenced (Figure 5).
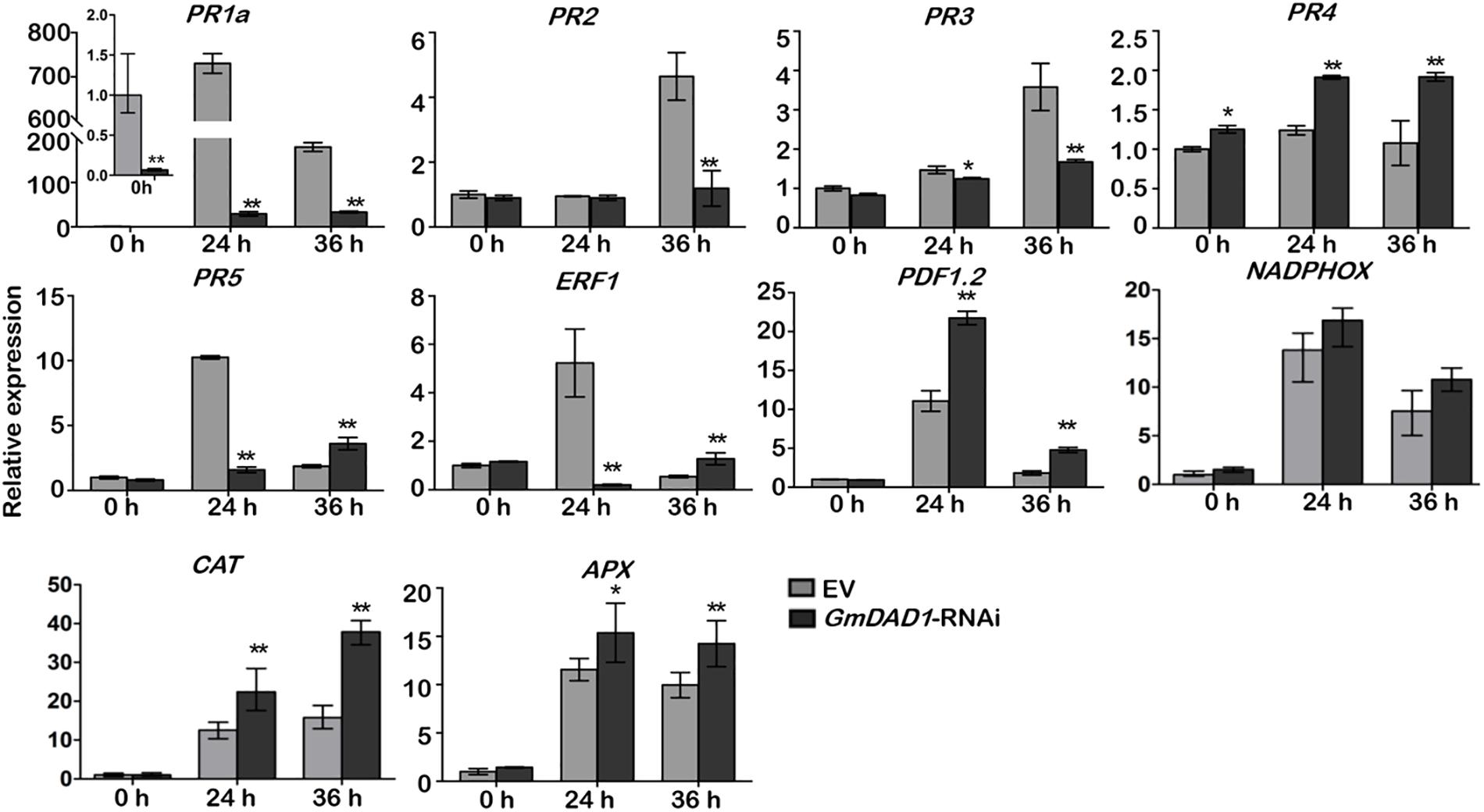
Figure 5. GmDAD1 affects the expression of multiple defense-related genes. RT-qPCR analysis of the expression patterns of defense-related genes in the EV and GmDAD1-RNAi transgenic hairy roots after inoculation with P. sojae. Values represent the means of three replicates ± SD. Data were analyzed by using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01 compared with the control).
GmDAD1 Is Involved in P. sojae-Activated ER Stress Signaling
Since DAD1 catalyzes the first step of protein N-linked glycosylation, disruption of GmDAD1 is expected to trigger unfolded protein response (UPR), which facilitates proper protein folding in ER via inducing the expression of a series of relevant genes (Li et al., 2011). After P. sojae inoculation, the transcript accumulations of six UPR marker genes were examined in soybean hairy roots, including Bip, PDI, CNX1, ERdj3A, GRP94, and bZIP17. All these genes are induced at the onset of ER stress and mark the activation of adaptive UPR. Expression changes of VPE were also monitored since its protein product possesses caspase-1-like activity and acts downstream of UPR and is part of the ER-PCD pathway. Compared to EV control, GmDAD1-RNAi roots showed significantly higher transcript accumulations of all seven UPR/ER stress marker genes at both 24 and 36 hpi (Figure 6). VPE was upregulated at 12 hpi and its expression decreased at 24 and 36 hpi in EV hairy roots, On the contrary, different trend was observed in GmDAD1 silencing hairy roots. The expression increased continuously through the selected time course, and was significantly higher at 24 and 36 hpi (Figure 6).
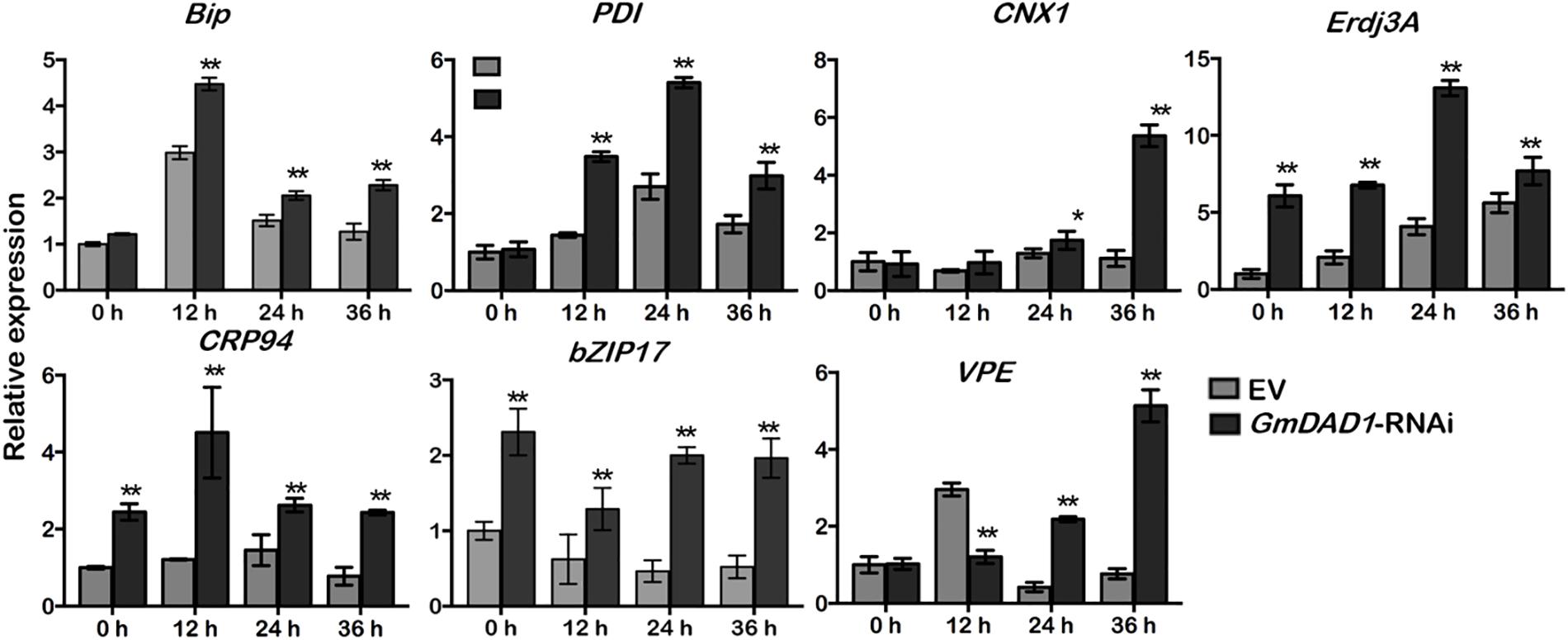
Figure 6. GmDAD1 is involved in P. sojae-activated ER stress signaling. Expression patterns of ER stress-related genes in the EV and GmDAD1-RNAi transgenic hairy roots after inoculation with P. sojae at 0, 12, 24, 36 hpi. Values represent the means of three replicates ± SD. Data were analyzed by using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01 compared with the control).
GmDAD1 Enhances Resistance to P. parasitica in N. benthamiana
To test whether GmDAD1 confers resistance against other Phytophthora pathogens, transgenic N. benthamiana plants overexpressing GmDAD1 were generated and verified (Supplementary Figure S2). Compared to wild-type (WT) and empty vector controls (EV) both GmDAD1 overexpression lines tested (4-1 and 8-4) showed reduced disease symptoms (Figures 7A,B) and significantly smaller lesion diameters on leaves (Figure 7C) when infected with P. parasitica zoospores. The results suggest that GmDAD1 overexpression enhances N. benthamiana resistance against P. parasitica.
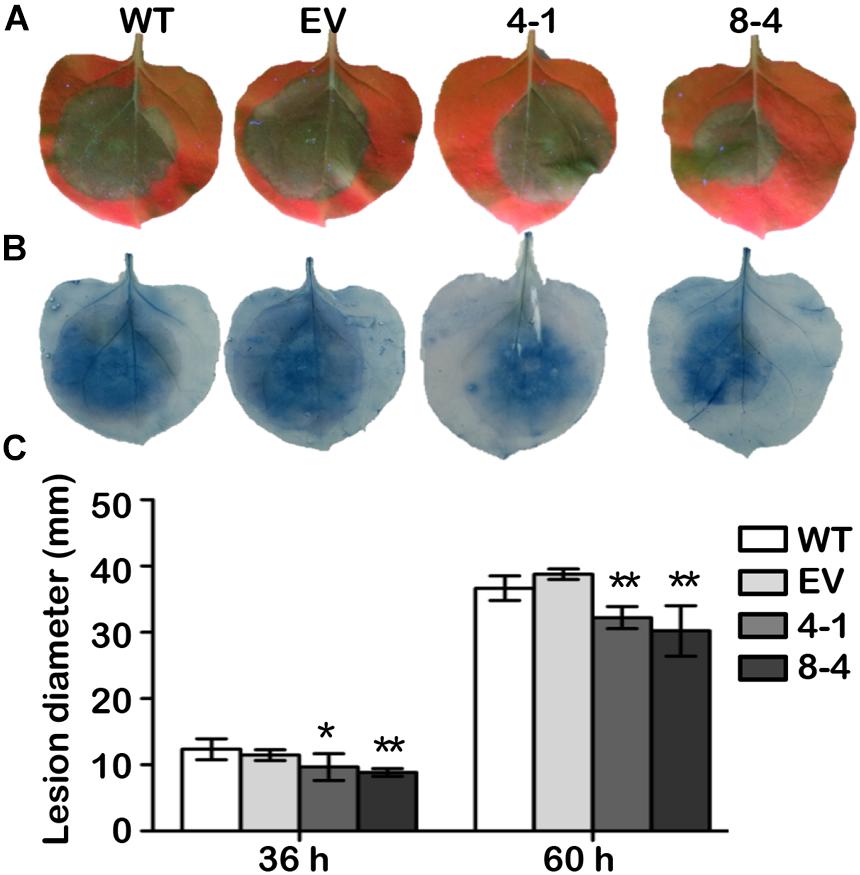
Figure 7. GmDAD1 enhances resistance to P. parasitica in N. benthamiana. (A) Detached leaves from wild type (WT), empty vector control (EV) and GmDAD1 overexpression plants (4-1 and 8-4) were inoculated with P. parasitica zoospores. Photographs were taken at 60 hpi under a UV lamp. (B) Trypan blue staining of the P. parasitica inoculated N. benthamiana leaves. (C) Lesion diameter of inoculated leaves measured at 36 and 60 hpi. The lesion size was calculated from 20 leaves ± SD with three biological repeats. Data were analyzed by using Student’s t-tests (∗P < 0.05, ∗∗P < 0.01 compared with the control).
Silencing of NbDAD1 in N. benthamiana Reduces Resistance to P. parasitica
Since plant DADs are highly conserved, the native NbDAD1 in N. benthamiana was silenced via TRV-based VIGS system for functional analysis. Compared to TRV-infected controls, plants infiltrated with TRV-NbDAD1 displayed a semi-dwarf phenotype with increased branching (Figures 8A,B), which implies a possible role of NbDAD1 in modulating growth and development. Three verified NbDAD1 knock-down lines and TRV-infected controls were challenged with P. parasitica zoospores on detached leaves (Figure 8C). Silencing of NbDAD1 led to significantly larger lesion diameters at both 36 and 48 hpi (Figures 8D–F), which indicates that NbDAD1 is similar as GmDAD1 in the function of conferring resistance against P. parasitica.
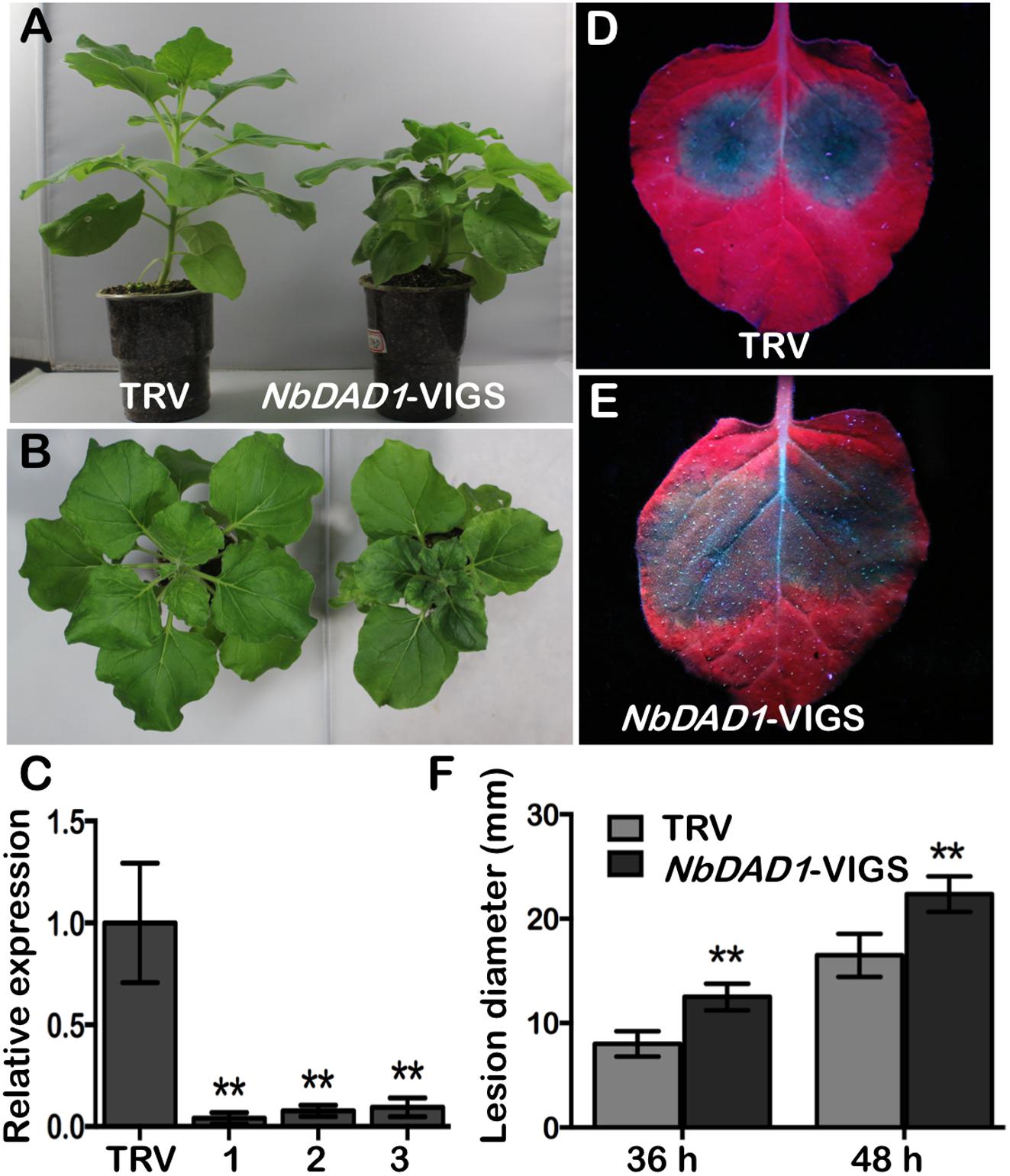
Figure 8. Silencing of NbDAD1 in N. benthamiana reduces resistance to P. parasitica. (A,B) Side- and top- view of the TRV-infected control and NbDAD1 silencing plants. (C) Relative expression of NbDAD1 in N. benthamiana plants inoculated with TRV and three NbDAD1 knock-down plants. The samples were collected 2 weeks after infection. N. benthamiana NbEF1α gene was used as reference for normalization. Data are the means ± SD calculated from three replicates. ∗∗P < 0.01. (D,E) Leaf phenotypes of TRV control and NbDAD1-VIGS after P. parasitica zoospores inoculation. Pictures were taken under UV illumination at 48 hpi. The experiments were repeated three times with similar results and representative images are shown. (F) Average lesion size of NbDAD1 knock-down leaves and TRV control after P. parasitica zoospores inoculation. Averages were calculated from at least 10 lesions per construct. Data are the means ± SD. Data were analyzed by using Student’s t-tests (∗∗P < 0.01).
Discussion
Being one of the most important crops worldwide, soybean can be infected by several major diseases, including the Phytophthora stem and root rot caused by P. sojae (Tyler, 2007). Continual efforts have been made to characterize novel defense genes against Phytophthora pathogens (Sugimoto et al., 2012). Here we identified GmDAD1, an ER-membrane protein from soybean, and dissected its function in plant–Phytophthora interactions.
Being evolutionary conserved across plant and animal species, DAD1 is a subunit of the OST complex, which catalyzes the first step of protein N-linked glycosylation in ER (Kelleher and Gilmore, 1997; Sanjay et al., 1998). In both animals and plants, the expression of DAD1 orthologs responds to a wide range of adverse environmental stimuli, including injury (Zhu et al., 2008), temperature (Lee et al., 2003), and pathogen infection (Wang X. J. et al., 2011). DAD1 inhibits undesired cell death triggered by host defense.
N-glycosylation has been reported to play a critical role in plant–pathogen interactions. For example, site-mutation on the N-glycosylation motif of A. thaliana receptor kinase EFR bleaches its ligand binding and results in oxidative burst elicitation capacity resulting in higher susceptibility of the plant to bacterial pathogens (Haweker et al., 2010). Several reports on the role of DAD proteins in plant defense have been published so far. The Arabidopsis dad1 mutant shows reduced secretion of PR proteins and resistance against pathogens (Wang et al., 2005). In wheat, knock-down of TaDAD2 suppresses the expression of PR1, PR2, and PR5 in response to the infection of Puccinia striiformis f. sp. tritici (Wang X. J. et al., 2011). We hence propose that GmDAD1 may also play a role in soybean disease resistance.
In soybean, GmDAD1 expression can be induced by P. sojae infection in both compatible and incompatible varieties, which indicates that GmDAD1 serves as a non-specific defense gene to some extent. However, GmDAD1 has consistently higher expression after P. sojae inoculation in the incompatible variety Williams 82, and its expression does not drop dramatically afterward at 48 hpi, as it happens in the compatible variety Williams. Therefore, GmDAD1 may be subjected to distinct transcriptional regulations in P. sojae compatible and incompatible soybean varieties.
Since GmDAD1 has highest transcript accumulation in roots, we adopted the soybean hairy root infection system for P. sojae resistance test. GmDAD1 gain- and loss-of-function mutants exhibit opposite P. sojae resistance phenotypes, which indicates that GmDAD1 contributes to the resistance of soybean against P. sojae. Similarly, knock-down of NbDAD1, the native DAD1 ortholog in N. benthamiana, reduces plant resistance to another Phytophthora pathogen, P. parasitica. Heterologous expression of GmDAD1 in N. benthamiana enhances resistance to P. parasitica. Our results reveal that DAD1 is a potential valuable defense gene against Phytophthora pathogens and this disease resistance function is conserved across plant species.
Phytohormone signaling, which is mediated by SA during biotrophic and hemibiotrophic plant–pathogen interactions and JA and ET for necrotrophic plant pathogens, plays important roles in plant resistance (Glazebrook, 2005). Previously studies demonstrated that the resistance to P. sojae is mediated by the SA and ET signaling pathways (Moy et al., 2004; Sugano et al., 2014). Therefore, we assessed the expression of several key defense related genes by RT-qPCR. When GmDAD1 silencing hairy roots were inoculated with P. sojae, the transcription of PR1a, PR2, PR3, PR5, and ERF1 were significantly reduced. Since the PR genes are generally regarded as early markers of resistance response, the suppressed expression of these genes may be responsible for the compromised resistance at the begin of the infection process (from 0 to 24 hpi). Moreover, the two JA-dependent signal marker genes PDF1.2 and PR4 were up-regulated after P. sojae infection in the silenced hairy roots (later than 24 hpi). We inferred that this JA resistance signaling activation might be lately induced, and the up-regulation might be caused by the antagonistic effect of JA and SA pathways.
Reactive oxygen species are important messenger molecules in defense signal regulation. The expression of ROS-generating gene NADPHOX showed no difference between EV and GmDAD1-RNAi hairy roots, however, the ROS-scavenging genes CAT and APX were statistically significant up-regulated after P. sojae infection in the silencing roots, this means that the ROS signaling was not completely affected by GmDAD1 silencing.
AS a core subunit of OST complex, DAD1 plays an important role in protein N-glycosylation (Peristera and Stephen, 2012), the defeat of protein N-glycosylation cause accumulation of misfolded proteins in ER and subsequently ER stress (Li et al., 2011; Cai et al., 2014). In soybean hairy roots infected by P. sojae, we found that GmDAD1 acts as a repressor for multiple UPR marker genes. In detail, all tested genes become up-regulated at later stages of the infection when GmDAD1 is silenced, indicating severe ER stress. We believe that this situation is caused by a less efficient or delayed defense signaling transduction. However, whether the suppression of defense-related genes was directly caused by the ER stress due to GmDAD1 silencing need to be further investigated.
Under extreme condition such as pathogen infection, a prolonged ER stress is known to eventually activate the ER-PCD pathway. Phytophthora pathogens are hemibiotrophic. They initially establish a biotrophic relationship with their hosts, and switch to necrotrophic phase later than 15 hpi (Enkerli et al., 1997). In EV hairy roots, a sharp increase of VPE, a cystein proteinase mediating PCD via the maturation and activation of vacuolar proteins, was observed at 12 hpi most likely to limit and overcome the biotrophic phase of P. sojae infection. In GmDAD1-RNAi roots, VPE expression was relatively suppressed at the same infection stage, suggesting the failure of PCD induction. However, elevated expression of VPE was detected at 24 and 36 hpi indicating a later activation of ER-PCD pathway. This late apoptosis overlaps with the necrotrophic phase of P. sojae, which may be one of the reasons of the increased P. sojae accumulation in GmDAD1 silencing hairy roots.
Disruption of DAD1 causes growth defect or even embryonic lethality in animal systems (Brewster et al., 2000; Zhang et al., 2016). In this study, we have observed significantly reduced transformation rate when silencing GmDAD1 in soybean hairy roots (Supplementary Figure S3). Moreover, knock-down of NbDAD1 by VIGS caused a semi-dwarf phenotype in N. benthamiana. These results suggest that DAD1 may play a similar role of regulating growth in plants most likely by acting on the N-glycosylation pathway of key proteins involved in plant development.
Conclusion
We observed that GmDAD1, a conserved component of the OST complex, via participating in the ER-PCD and UPR pathways and affecting the expression of multiple defense-related genes, confers resistance to Phytophthora pathogens. Moreover, GmDAD1 regulates plant growth and development likely by the effect on the N-glycosylation pathway. Taken together, GmDAD1 can be considered as a promising target for the molecular breeding of Phytophthora-resistant soybean varieties.
Author Contributions
DD and QY designed the project. QY, JS, and XaC performed the experiments and analyzed the data. XnC, HX, and DD guided the experimental work. DD, QY, HP, and MJ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31625023, 31721004, and 31672008) and Special Fund for Agro-scientific Research in the Public Interest (201503112).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00107/full#supplementary-material
Footnotes
References
Babaeizad, V., Imani, J., Kogel, K. H., Eichmann, R., and Huckelhoven, R. (2009). Over-expression of the cell death regulator BAX inhibitor-1 in barley confers reduced or enhanced susceptibility to distinct fungal pathogens. Theor. Appl. Genet. 118, 455–463. doi: 10.1007/s00122-008-0912-2
Bernard, R. L., and Cremeens, C. R. (1988). Registration of ‘Williams 82’ soybean. Crop Sci. 28, 1027–1028. doi: 10.2135/cropsci1988.0011183X002800060049x
Bernard, R. L., and Lindahl, D. A. (1972). Registration of williams soybean1 (reg. no. 94). Crop Sci. 12:716. doi: 10.2135/cropsci1972.0011183X001200050067x
Bertini, L., Leonardi, L., Caporale, C., Tucci, M., Cascone, N., Di Berardino, I., et al. (2003). Pathogen-responsive wheat PR4 genes are induced by activators of systemic acquired resistance and wounding. Plant Sci. 164, 1067–1078. doi: 10.1016/s0168-9452(03)00112-2
Brewster, J. L., Martin, S. L., Toms, J., Goss, D., Wang, K., Zachrone, K., et al. (2000). Deletion of Dad1 in mice induces an apoptosis-associated embryonic death. Genesis 26, 271–278. doi: 10.1002/(SICI)1526-968X(200004)26:4<271::AID-GENE90>3.0.CO;2-E
Cai, Y. M., Jia, Y., and Patrick, G. (2014). Endoplasmic reticulum stress-induced PCD and caspase-like activities involved. Front. Plant Sci. 5:41. doi: 10.3389/fpls.2014.00041
Chen, Y. P., Xing, L. P., Wu, G. J., Wang, H. Z., Wang, X. E., Cao, A. Z., et al. (2007). Plastidial glutathione reductase from Haynaldia villosa is an enhancer of powdery mildew resistance in wheat (Triticum aestivum). Plant Cell Physiol. 48, 1702–1712. doi: 10.1093/pcp/pcm142
Danon, A., Rotari, V. I., Gordon, A., Mailhac, N., and Gallois, P. (2004). Ultraviolet-C overexposure induces programmed cell death in Arabidopsis, which is mediated by caspase-like activities and which can be suppressed by caspase inhibitors, p35 and defender against apoptotic death. J. Biol. Chem. 279, 779–787. doi: 10.1074/jbc.M304468200
Dickman, M. B., Park, Y. K., Oltersdorf, T., Li, W., Clemente, T., and French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. U.S.A. 98, 6957–6962. doi: 10.1073/Pnas.091108998
Doukhanina, E. V., Chen, S., Van Der Zalm, E., Godzik, A., Reed, J., and Dickman, M. B. (2006). Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281, 18793–18801. doi: 10.1074/jbc.M511794200
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Eichmann, R., Bischof, M., Weis, C., Shaw, J., Lacomme, C., Schweizer, P., et al. (2010). BAX INHIBITOR-1 is required for full susceptibility of barley to powdery mildew. Mol. Plant Microbe Interact. 23, 1217–1227. doi: 10.1094/Mpmi-23-9-1217
Enkerli, K., Mims, C. W., and Hahn, M. G. (1997). Ultrastructure of compatible and incompatible interactions of soybean roots infected with the plant pathogenic oomycete Phytophthora sojae. Can. J. Bot. 75, 1493–1508. doi: 10.1139/b97-864
Fu, D. Q., Zhu, B. Z., Zhu, H. L., Jiang, W. B., and Luo, Y. B. (2002). Virus-induced gene silencing in tomato. Plant J. 43, 299–308. doi: 10.1111/j.1365-313X.2005.02441.x
Gallois, P., Makishima, T., Hecht, V., Despres, B., Laudie, M., Nishimoto, T., et al. (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J. 11, 1325–1331. doi: 10.1046/j.1365-313X.1997.11061325.x
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Graham, T. L. (1991). A rapid, high resolution high performance liquid chromatography profiling procedure for plant and microbial aromatic secondary metabolites. Plant Physiol. 95, 584–593. doi: 10.1104/pp.95.2.584
Graham, T. L., Graham, M. Y., Subramanian, S., and Yu, O. (2007). RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues. Plant Physiol. 144, 728–740. doi: 10.1104/pp.107.097865
Hall, T. A. (1999). Bioedit: a user-friendly biological sequence algnment editor and analysis program for windows 95/98/nt. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Haweker, H., Rips, S., Koiwa, H., Salomon, S., Saijo, Y., Chinchilla, D., et al. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636. doi: 10.1074/jbc.M109.063073
Hofmann, K., and Stoel, W. (1993). Tmbase-A database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler 374:166. doi: 10.1056/NEJM199001043220121
Horsch, R. B., Rogers, S. G., and Fraley, R. T. (1985). Transgenic Plants. Cold Spring Harb. Symp. Quant. Biol. 50, 433–437. doi: 10.1101/SQB.1985.050.01.054
Ishikawa, T., Watanabe, N., Nagano, M., Kawai-Yamada, M., and Lam, E. (2011). Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 18, 1271–1278. doi: 10.1038/cdd.2011.59
Jing, M., Guo, B., Li, H., Bo, Y., Wang, H., Kong, G., et al. (2016). A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nat. Commun. 7:11685. doi: 10.1038/ncomms11685
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kawai-Yamada, M., Hori, Z., Ogawa, T., Ihara-Ohori, Y., Tamura, K., Nagano, M., et al. (2009). Loss of calmodulin binding to Bax inhibitor-1 affects Pseudomonas-mediated hypersensitive response-associated cell death in Arabidopsis thaliana. J. Biol. Chem. 284, 27998–28003. doi: 10.1074/jbc.M109.037234
Kawai-Yamada, M., Ohori, Y., and Uchimiya, H. (2004). Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16, 21–32. doi: 10.1105/tpc.014613
Kelleher, D. J., and Gilmore, R. (1997). DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc. Natl. Acad. Sci. U.S.A. 94, 4994–4999. doi: 10.1073/pnas.94.10.4994
Kimchi, A. (2007). Programmed cell death: from novel gene discovery to studies on network connectivity and emerging biomedical implications. Cytokine Growth Factor Rev. 18, 435–440. doi: 10.1016/j.cytogfr.2007.06.004
Lam, E., Kato, N., and Lawton, M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. doi: 10.1038/35081184
Lee, K. S., Han, J. H., Sohn, H. D., and Jin, B. R. (2003). cDNA cloning of a defender against apoptotic cell death 1 (DAD1) homologue, responsive to external temperature stimulus from the spider, Araneus ventricosus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 135, 117–123. doi: 10.1016/S1096-4959(03)00055-1
Li, K., Ouyang, H., Lü, Y., Liang, J., Wilson, I. B., and Jin, C. (2011). Repression of N-glycosylation triggers the unfolded protein response (UPR) and overexpression of cell wall protein and chitin in Aspergillus fumigatus. Microbiology 157:1968. doi: 10.1099/mic.0.047712-0
Li, Y., Kabbage, M., Liu, W., and Dickman, M. B. (2016a). Aspartyl protease mediated cleavage of AtBAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28, 233–247. doi: 10.1105/tpc.15.00626
Li, Y., Williams, B., and Dickman, M. (2016b). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214, 695–705. doi: 10.1111/nph.14388
Libault, M., Thibivilliers, S., Bilgin, D. D., Radwan, O., Benitez, M., Clough, S. J., et al. (2008). Identification of four soybean reference genes for gene expression normalization. Plant Genome 1, 44–54. doi: 10.3835/plantgenome2008.02.0091
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCTmethod. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenzo, O., Piqueras, R., Sánchezserrano, J. J., and Solano, R. (2003). Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lorenzo, O., and Solano, R. (2005). Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 8, 532–540. doi: 10.1016/j.pbi.2005.07.003
Makishima, T., Yoshimi, M., Komiyama, S., Hara, N., and Nishimoto, T. (2000). A Subunit of the mammalian oligosaccharyltransferase, DADI, interacts with Mcl-1, one of the bcl-2 protein family1. J. Biochem. 128, 399–405. doi: 10.1093/oxfordjournals.jbchem.a022767
Maldonado, A., Youssef, R., Mcdonald, M., Brewer, E., Beard, H., Matthews, B. J. P., et al. (2014). Overexpression of four Arabidopsis thaliana NHL genes in soybean (Glycine max) roots and their effect on resistance to the soybean cyst nematode (Heterodera glycines). Physiol. Mol. Plant P 86, 1–10. doi: 10.1016/j.pmpp.2014.02.001
Matsumura, H., Nirasawa, S., Kiba, A., Urasaki, N., Saitoh, H., Ito, M., et al. (2003). Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. Plant J. 33, 425–434. doi: 10.1046/J.1365-313x.2003.01639.X
Mazarei, M., Elling, A. A., Maier, T. R., Puthoff, D. P., and Baum, T. J. (2007). GmEREBP1 is a transcription factor activating defense genes in soybean and Arabidopsis. Mol. Plant Microbe Interact. 20, 107–119. doi: 10.1094/MPMI-20-2-0107
Moy, P., Qutob, D., Chapman, B. P., Atkinson, I., and Gijzen, M. (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant Microbe Interact. 17, 1051–1062. doi: 10.1094/MPMI.2004.17.10.1051
Nakashima, T., Sekiguchi, T., Kuraoka, A., Fukushima, K., Shibata, Y., Komiyama, S., et al. (1993). Molecular-cloning of a human cDNA-encoding a novel protein, Dad1, whose defect causes apoptotic cell-death in hamster BHK-21-cells. Mol. Cell Biol. 13, 6367–6374. doi: 10.1128/MCB.13.10.6367
Perez, I. B., and Brown, P. J. (2014). The role of ROS signaling in cross-tolerance: from model to crop. Front. Plant Sci. 5:754. doi: 10.3389/fpls.2014.00754
Peristera, R., and Stephen, H. (2012). The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J. Cell Sci. 125, 3474–3484. doi: 10.1242/jcs.103952
Rojo, E., Martin, R., Carter, C., Zouhar, J., Pan, S., Plotnikova, J., et al. (2004). VPE gamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14, 1897–1906. doi: 10.1016/j.cub.2004.09.056
Sanjay, A., Fu, J., and Kreibich, G. (1998). DAD1 is required for the function and the structural integrity of the oligosaccharyltransferase complex. J. Biol. Chem. 273, 26094–26099. doi: 10.1074/jbc.273.40.26094
Subramanian, S., Graham, M. Y., Yu, O., and Graham, T. L. (2005). RNA interference of soybean isoflavone synthase genes leads to silencing in tissues distal to the transformation site and to enhanced susceptibility to Phytophthora sojae. Plant Physiol. 137, 1345–1353. doi: 10.1104/pp.104.057257
Sugano, S., Sugimoto, T., Takatsuji, H., and Jiang, C. J. (2014). Induction of resistance to Phytophthora sojae in soyabean (Glycine Max) by salicylic acid and ethylene. Plant Pathol. 62, 1048–1056. doi: 10.1111/ppa.12011
Sugimoto, T., Kato, M., Yoshida, S., Matsumoto, I., Kobayashi, T., Kaga, A., et al. (2012). Pathogenic diversity of Phytophthora sojae and breeding strategies to develop phytophthora-resistant soybeans. Breed. Sci. 61, 511–522. doi: 10.1270/jsbbs.61.511
Sun, J., Li, L., Zhao, J., Huang, J., Yan, Q., Xing, H., et al. (2014). Genetic analysis and fine mapping of RpsJS, a novel resistance gene to Phytophthora sojae in soybean [Glycine max (L.) Merr]. Theor. Appl. Genet. 127, 913–919. doi: 10.1007/s00122-014-2266-2
Tanaka, Y., Makishima, T., Sasabe, M., Ichinose, Y., Shiraishi, T., Nishimoto, T., et al. (1997). dad-1, a putative programmed cell death suppressor gene in rice. Plant Cell Physiol. 38, 379–383. doi: 10.1093/oxfordjournals.pcp.a029179
Tiziana, A., and Roberto, S. (2014). Protein quality control in the early secretory pathway. Embo J. 27, 315–327. doi: 10.1038/sj.emboj.7601974
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Wang, D., Weaver, N. D., Kesarwani, M., and Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. doi: 10.1126/science.1108791
Wang, X. J., Tang, C. L., Zhang, H. C., Xu, J. R., Liu, B., Lv, J., et al. (2011). TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus. Mol. Plant Microbe Interact. 24, 79–90. doi: 10.1094/MPMI-06-10-0131
Watanabe, N., and Lam, E. (2009). Bax Inhibitor-1, a conserved cell death suppressor, is a key molecular switch downstream from a variety of biotic and abiotic stress signals in plants. Int. J. Mol. Sci. 10, 3149–3167. doi: 10.3390/ijms10073149
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M., Rouse, D. T., Liu, Q., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313X.2001.01105.x
Williams, B., Kabbage, M., Britt, R., and Dickman, M. B. (2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 107, 6088–6093. doi: 10.1073/pnas.0912670107
Xiong, Q., Ye, W., Choi, D., Wong, J., Qiao, Y., Tao, K., et al. (2014). Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol. Plant Microbe Interact. 27, 1379–1389. doi: 10.1094/MPMI-06-14-0190-R
Yamada, T., Takatsu, Y., Kasumi, M., Marubashi, W., and Ichimura, K. (2004). A homolog of the defender against apoptotic death gene (DAD1) in senescing gladiolus petals is down-regulated prior to the onset of programmed cell death. J. Plant Physiol. 161, 1281–1283. doi: 10.1016/j.jplph.2004.06.005
Yan, A., Wu, E., and Lennarz, W. J. (2005). Studies of yeast oligosaccharyl transferase subunits using the split-ubiquitin system: topological features and in vivo interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 7121–7126. doi: 10.1073/pnas.0502669102
Yan, Q., Cui, X., Su, L., Xu, N., Guo, N., Xing, H., et al. (2014). GmSGT1 is differently required for soybean Rps genes-mediated and basal resistance to Phytophthora sojae. Plant Cell Rep. 33, 1275–1288. doi: 10.1007/s00299-014-1615-6
Zhang, M., Rajput, N. A., Shen, D., Sun, P., Zeng, W., Liu, T., et al. (2014). A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana. Sci. Rep. 5:10837. doi: 10.1038/srep10837
Zhang, Y., Chang, C., and Lai, Z. C. (2016). The defender against apoptotic cell death 1 gene is required for tissue growth and efficient N-glycosylation in Drosophila melanogaster. Dev. Biol. 420:186. doi: 10.1016/j.ydbio.2016.09.021
Zhou, L., Mideros, S. X., Bao, L., Hanlon, R., Arredondo, F. D., Tripathy, S., et al. (2009). Infection and genotype remodel the entire soybean transcriptome. BMC Genomics 10:49. doi: 10.1186/1471-2164-10-49
Keywords: Glycine max, Phytophthora resistant, defender against apoptotic death 1 (DAD1), programmed cell death (PCD), ER stress
Citation: Yan Q, Si J, Cui X, Peng H, Jing M, Chen X, Xing H and Dou D (2019) GmDAD1, a Conserved Defender Against Cell Death 1 (DAD1) From Soybean, Positively Regulates Plant Resistance Against Phytophthora Pathogens. Front. Plant Sci. 10:107. doi: 10.3389/fpls.2019.00107
Received: 10 September 2018; Accepted: 23 January 2019;
Published: 08 February 2019.
Edited by:
Valentina Fiorilli, University of Turin, ItalyReviewed by:
Fabiano Sillo, University of Turin, ItalyMarino Moretti, Max-Planck-Institut für Terrestrische Mikrobiologie, Germany
Ivan Fernandez Lopez, Helmholtz-Zentrum für Umweltforschung (UFZ), Germany
Copyright © 2019 Yan, Si, Cui, Peng, Jing, Chen, Xing and Dou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daolong Dou, ZGRvdUBuamF1LmVkdS5jbg==
 Qiang Yan
Qiang Yan Jierui Si
Jierui Si Xiaoxia Cui1
Xiaoxia Cui1 Hao Peng
Hao Peng Maofeng Jing
Maofeng Jing Xin Chen
Xin Chen Daolong Dou
Daolong Dou