
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 18 January 2019
Sec. Plant Nutrition
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00006
This article is part of the Research TopicIron Nutrition and Interactions in PlantsView all 28 articles
Iron is one of the most important micronutrients in plants as it is involved in many cellular functions (e.g., photosynthesis and respiration). Any defect in iron availability will affect plant growth and development as well as crop yield and plant product quality. Thus, iron homeostasis must be tightly controlled in order to ensure optimal absorption of this mineral element. Understanding mechanisms governing iron homeostasis in plants has been the focus of several studies during the past 10 years. These studies have greatly improved our understanding of the mechanisms involved, revealing a sophisticated iron-dependent transcriptional regulatory network. Strikingly, these studies have also highlighted that this regulatory web relies on the activity of numerous transcriptional regulators that belong to the same group of transcription factors (TF), the bHLH (basic helix-loop-helix) family. This is best exemplified in Arabidopsis where, to date, 16 bHLH TF have been characterized as involved in this process and acting in a complex regulatory cascade. Interestingly, among these bHLH TF some form specific clades, indicating that peculiar function dedicated to the maintenance of iron homeostasis, have emerged during the course of the evolution of the green lineage. Within this mini review, we present new insights on the control of iron homeostasis and the involvement of bHLH TF in this metabolic process.
Iron (Fe) is one of the most important micronutrient elements in plants as it is involved in many cellular functions (e.g., photosynthesis and respiration). Any defect in Fe availability will impact plant growth and development as well as crop yield and plant product quality (Briat et al., 2015).
In order to cope with Fe shortage and recover Fe from soil, where it is in poorly available forms, plants have evolved two strategies. The first one, strategy I, is used by all dicots and non-graminaceous monocots. This strategy consists in rhizosphere acidification via proton extrusion in order to promote Fe solubility and involves proton-ATPase such as AHA2. The secretion by the root of Fe-mobilizing phenolic compounds facilitates this process (Fourcroy et al., 2014, 2016). Fe3+ is thus reduced into Fe2+ by ferric chelate reductases, such as FRO2 (FERRIC REDUCTION OXIDASE 2), prior to being transported across the rhizodermis cell membranes by IRT1 (IRON-REGULATED TRANSPORTER 1) (Brumbarova et al., 2015). The second strategy, strategy II, is used by graminaceous species. This strategy consists in releasing phytosiderophores into the rhizosphere to chelate Fe3+ (Nozoye et al., 2011). Fe3+-phytosiderophores chelates are then transported into the roots by the YELLOW STRIPE 1 transporter (Curie et al., 2001). If the machinery allowing plant Fe uptake from the soil is central for the maintenance of Fe homeostasis, this is indeed not the sole mechanism involved in this process. It also necessitates several genes encoding proteins involved in Fe transport, compartmentation and storage, at the cellular and subcellular levels, throughout the entire plant body. Such complex mechanism must thus be tightly regulated in order to avoid any physiological situation that would be deleterious to the plant.
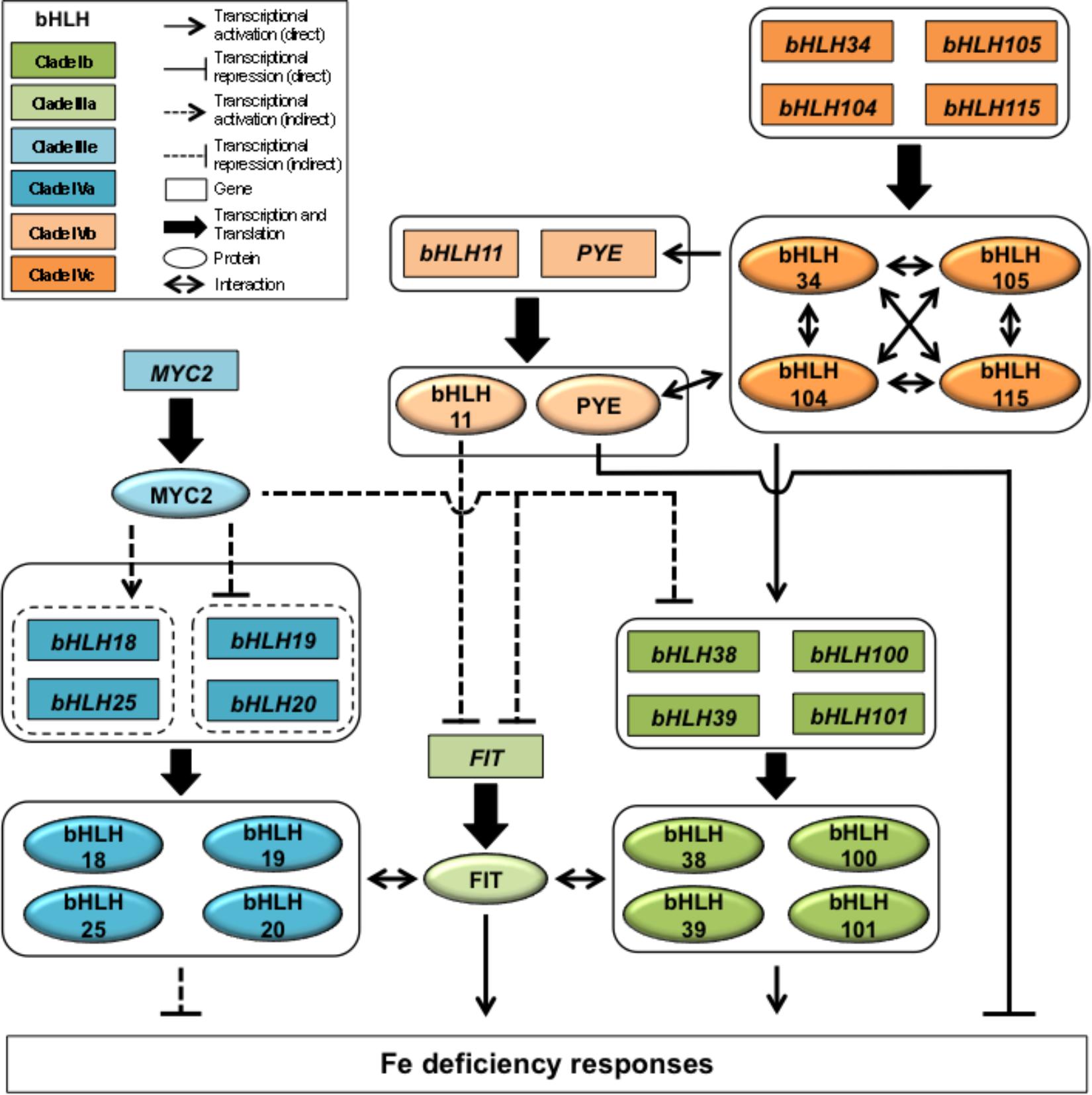
Figure 1. bHLH-dependent transcriptional regulatory network controlling the iron deficiency response in Arabidopsis thaliana.
How, at the molecular level, plants control Fe homeostasis has thus been a critical question for several years. This question has been mostly addressed by studying plant response to Fe deficiency, in particular in the model plant Arabidopsis thaliana. These studies have highlighted that such response involves an intricate network of basic helix-loop-helix (bHLH) transcription factors (TF) (Figure 1). bHLH proteins form one of the largest family of TFs found in plants that act as homo- or heterodimers to regulate the expression of their target genes. In Arabidopsis, 133 members have been identified and divided into 12 clades (Heim et al., 2003). From what is known on the role played by several members of this family of TFs in plants, it appears that their participation in the control of Fe homeostasis is unique by the number of individual TFs and different clades that are involved as well as by the intricacy of the network they form.
Upstream from the regulatory network involved in Arabidopsis Fe deficiency response are four bHLH TFs belonging to the clade IVc, namely bHLH34, bHLH104, bHLH105/ILR3 (IAA-LEUCINE RESISTANT3), and bHLH115 (Figure 1). These four TFs, shown to interact in vivo in the form of homo- or heterodimers, act as transcriptional activators of the plant response to Fe deficiency and display partial redundant activities (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). Clade IVc bHLH targets consist of bHLH47/PYE (POPEYE; clade IVb) and four clade Ib bHLH genes (bHLH38, bHLH39, bHLH100, and bHLH101) (Colangelo and Guerinot, 2004; Wang et al., 2007; Yuan et al., 2008; Wang N. et al., 2013; Zhang et al., 2015). PYE acts as a transcriptional repressor. For example, PYE was shown to inhibit the expression of NAS4 (NICOTIANAMINE SYNTHASE 4), a key gene involved in phloem-based transport of Fe to sink organs, and FRO3, a Fe reductase located in root vasculature mitochondria (Jeong and Connolly, 2009; Klatte et al., 2009; Long et al., 2010). Interestingly, PYE can interact in vivo with bHLH104, ILR3, and bHLH115 (Long et al., 2010; Zhang et al., 2015). Whether or not these interactions play a role in the plant response to Fe deficiency or in the control of Fe homeostasis is an important question that remains to be elucidated. In contrast, bHLH38, bHLH39, bHLH100, and bHLH101 are partially redundant proteins that function at the root epidermis as positive regulators of FRO2 and IRT1. This activity relies on their interaction with FIT (Fe-deficiency induced transcription factor), a clade IIIa bHLH (Colangelo and Guerinot, 2004; Yuan et al., 2008; Wang N. et al., 2013; Maurer et al., 2014). FIT expression is likely controlled, at least in part, by a feed forward regulatory loop involving bHLH39 (Naranjo-Arcos et al., 2017). bHLH6/MYC2 (clade IIIe) is a master regulator of the jasmonic acid (JA) signaling pathway whose activity differentially affects the expression clade IVa bHLH (bHLH18, bHLH19, bHLH20, and bHLH25) genes to modulate FIT protein accumulation (Cui et al., 2018). This mechanism relies on the direct interaction of clade IVa bHLHs with FIT in order to promote its degradation via the 26S proteasome pathway (Cui et al., 2018). In addition, MYC2 is a JA-dependent repressor of FIT and clade Ib bHLH genes expression, hence inhibiting FIT-dependent Fe uptake machinery at the transcriptional and posttranslational levels (Cui et al., 2018). Interestingly, bHLH11, another clade IVb bHLH, was recently proposed to be a negative regulator of FIT-dependent Fe uptake mechanism effecting Fe levels in Arabidopsis plants (Tanabe et al., 2018).
Altogether, this is 16 bHLH TFs out of the 133 present in Arabidopsis that have been identified as involved in the control of the Fe deficiency response, which represent more than 12% of the members of this large family of TFs (Heim et al., 2003). Indeed, orthologous members from the above-described clades were identified in other strategy I plant species such as tomato, apple, or soybean (Ling et al., 2002; Du et al., 2015; Zhao et al., 2016b; Li et al., 2018). In soybean, it is likely that the orthologs of FIT (GmbHLH57) and clade Ib bHLH (GmbHLH300) genes may also play a role in nodules, a tissue where several enzymes involved in symbiotic nitrogen fixation require Fe for their activity (Tang et al., 1990; O’Hara, 2001; Li et al., 2018). With the exception of FIT, which is specific to strategy I plants, orthologs of several bHLH are present in strategy II plants, indicating that the regulatory cascade controlling plant response to Fe deficiency is mostly conserved within the plant kingdom. For instance, orthologs of clade Ib (OsIRO2), IVc (OsPRI1), PYE (OsIRO3) and MYC2 (OsMYC2) genes have been characterized in rice (Ogo et al., 2007; Zheng et al., 2010; Ogawa et al., 2017; Zhang et al., 2017). However, no orthologous bHLH TF in strategy I plants has been described so far for OsbHLH133, another regulator of the Fe deficiency response in rice (Wang L. et al., 2013). Is OsbHLH133 function specific to strategy II plants as it is the case for FIT in strategy I plants? Protein sequence analysis tends to indicate that OsbHLH133 is closely related to the bHLH clade VIIIc and thus it might be that this clade plays a role in the control of Fe homeostasis in both strategy I and II plants. If this hypothesis is verified, it will certainly render more complicated our understanding of this transcriptional regulatory network. Indeed, it is not the complexity of this network that is intriguing considering that any defect in the control of Fe homeostasis might be deleterious to the plants. The main question is why so many bHLHs are involved in this process? If it is difficult to answer this question, the involvement of a large number of TFs from one family in a specific process has already been described. This is the case with the R2R3-MYB family where at least 19 members out of 122 (about 16%) are involved in the control of the phenylpropanoid pathway (Dubos et al., 2010; Zamioudis et al., 2014; Xu et al., 2015). From these observations it would be tempting to speculate that during the course of the evolution of the green lineage, TF families have evolved specialized roles, in which plant Fe homeostasis would be mostly regulated by TFs belonging to the bHLH gene family.
How these bHLH TFs interact with the cis-regulatory sequences usually present in the promoter of their target genes is an important question considering that (i) each bHLH must specifically recognized its own target and (ii) the number of bHLH involved in this complex network. Indeed, it is well known that bHLH TF bind to specific DNA motifs (CANNTG) named E-box, and in particular to the canonical CACGTG sequence, named G-box (De Masi et al., 2011). For instance, chromatin immunoprecipitation assays showed that PYE, bHLH104, bHLH115, ILR3, and FIT preferentially bound to the promoter of their target genes in region that contain E-box or G-box (Long et al., 2010; Zhang et al., 2014, 2015; Liang et al., 2017). Similar observations were made, using biochemical approaches, for MdbHLH104 and OsPRI1 (Zhao et al., 2016a; Zhang et al., 2017). Interestingly, it was demonstrated that the genomic regions flanking E-box binding sites influence the DNA binding specificity of TFs (Gordan et al., 2013; Ezer et al., 2017). This is for example the case for OsIRO2 that binds preferentially to CACGTGG motif (Ogo et al., 2007). Hence, despite the fact that several direct target genes have been identified for most of the bHLH involved in the transcriptional control of Fe homeostasis and the fact that it is possible to infer the E-box sequence recognized by a given bHLH dimer based on bHLH domain compositions (De Masi et al., 2011), very little is known on the actual bHLH/DNA interactions.
Additional TFs, from several gene families, involved in the control of Fe homeostasis in both strategy I and strategy II plant species, have also been characterized.
MYB10 and MYB72, two R2R3-MYB TFs whose expression is partially dependent on FIT, are involved in Fe acquisition and distribution by notably regulating the expression of BGLU42 and NAS4 (Palmer et al., 2013; Stringlis et al., 2018). A closely related apple R2R3-MYB, MdMYB58, was recently reported as potentially involved in the control of Fe transport and tissue partitioning. It is proposed that MdMYB58 activity is repressed by its heterodimerization with MdSAT1, a clade IVa bHLH (Wang et al., 2018). WRKY46 plays a critical role in Fe translocation from root to shoot by directly repressing the expression of VITL1/VTL1 (vacuolar iron transporter like 1) (Yan et al., 2016). ERF4 and ERF72 (AP2/ERF TFs) play negative roles in plant response to Fe deficiency by inhibiting the expression of genes involved in Fe uptake such as IRT1 or AHA2 (Liu et al., 2017a,b). Two TFs (EIL family) involved in ethylene signaling (EIN3, ETHYLENE INSENSITIVE 3 and EIL1, ETHYLENE INSENSITIVE 3 LIKE 1) and ZAT12 (a C2H2-type plant-specific zinc finger TF) are also involved by modulating FIT stability (Lingam et al., 2011; Le et al., 2016). Two MYB-CC TFs (PHR1, PHOSPHATE STARVATION RESPONSE 1 and PHL1, PHR1-LIKE 1), that play a central role in the phosphate deficiency response, regulate the expression of the main Fe storage ferritin gene in Arabidopsis (AtFER1), indicating that they act as integrators of both the phosphate and Fe signaling pathways (Bournier et al., 2013).
IDEF1 (ABI3/VP1 family) is an early regulator of Fe deficiency response in rice that directly binds to divalent metals suggesting that IDEF1 is a cellular sensor of metal ion balance caused by changes in Fe availability (Kobayashi et al., 2009, 2012). IDEF2 (NAC family) and OsARF16 (ARF family) play also critical roles in the control of Fe homeostasis in rice by modulating the expression of Fe-related genes and by integrating auxin signals, respectively (Ogo et al., 2008; Shen et al., 2015).
Iron deficiency results in a transcriptional response that leads to the activation of the Fe uptake machinery, which could lead to a Fe overload if it becomes suddenly available or to a toxic overaccumulation of other divalent metals (e.g., Zn, Mn, and Cd) due to the low specificity of IRT1. To cope with this, plants have developed posttranslational mechanisms such as the continuous recycling of IRT1 (Barberon et al., 2011, 2014). In addition, in Arabidopsis, IRT1 phosphorylation and ubiquitination leads to its internalization and subsequent degradation, a process that is triggered by direct binding of IRT1 to non-Fe metals (Ivanov et al., 2014; Dubeaux et al., 2018). However, maintaining Fe homeostasis requires also to tightly regulating, at the posttranslational level, the TFs involved in this process.
Fe-deficiency induced TF posttranslational regulation has been extensively investigated in Arabidopsis. For instance, it was shown that FIT heterodimerization with clade IVa bHLH promotes its degradation via the 26S proteasome pathway, whereas its interaction with clade Ib bHLH promotes its stability (Cui et al., 2018). FIT interaction with EIN3 or EIL1 also promotes its stability and contributes to Fe acquisition during the early stage of Fe deficiency (Lingam et al., 2011). In contrary, FIT interaction with ZAT12 has an inhibitory effect on its function when plants are grown under Fe sufficiency or prolonged Fe deficiency conditions (Le et al., 2016).
Several ubiquitin E3 ligases targeting bHLH TFs involved in the response to Fe deficiency have been characterized. BRUTUS (BTS), whose expression is induced upon Fe deficiency, is thought to be a Fe-sensing negative regulator in Arabidopsis (Selote et al., 2015). In vitro analyses suggest that BTS restricts the accumulation of ILR3 and bHLH115 through its RING E3 ligase activity and mediates their 26S proteasomal degradation (Selote et al., 2015). Whether the interaction of bHLH104 with BTS participate to its degradation remains to be demonstrated (Selote et al., 2015). The identification of two BTS ortologues in rice, namely OsHRZ1 [HEMERYTHRIN MOTIF-CONTAINING REALLY INTERESTING NEW GENE (RING) AND ZINC-FINGER PROTEIN 1] and OsHRZ2, suggests that BTS function is conserved in strategy II plants (Kobayashi et al., 2013). In addition to BTS, two closely related RING E3 ligases named BTSL1 and BTSL2 (BRUTUS LIKE 1 and 2) are proposed to negatively regulate the Fe deficiency responses by directly targeting FIT, leading to its degradation (Sivitz et al., 2011; Hindt et al., 2017; Rodriguez-Celma et al., 2018). Interestingly, it was shown in apple that a cullin-based E3 ligase mechanism, involving two BTB-TAZ proteins (MdBT1 and MdBT2) and MdCUL3, target MdbHLH104 for ubiquitin-dependent degradation via the 26S proteasome (Zhao et al., 2016a). Unlike BTS and the E3 ligase degrading FIT, MdBT expression and protein accumulation is induced when Fe availability is not limiting, leading to MdbHLH104 degradation (Zhao et al., 2016a). In addition, MdbHLH104 sumoylation, by the SUMO E3 ligase MdSIZ1, promotes MdbHLH104 stability, especially when Fe availability is scarce (Zhou et al., 2018). These findings suggest that the degradation of the bHLHs involved in the plant response to Fe availability may require two types of ubiquitin E3 ligases and that sumoylation may also have an important role in this process.
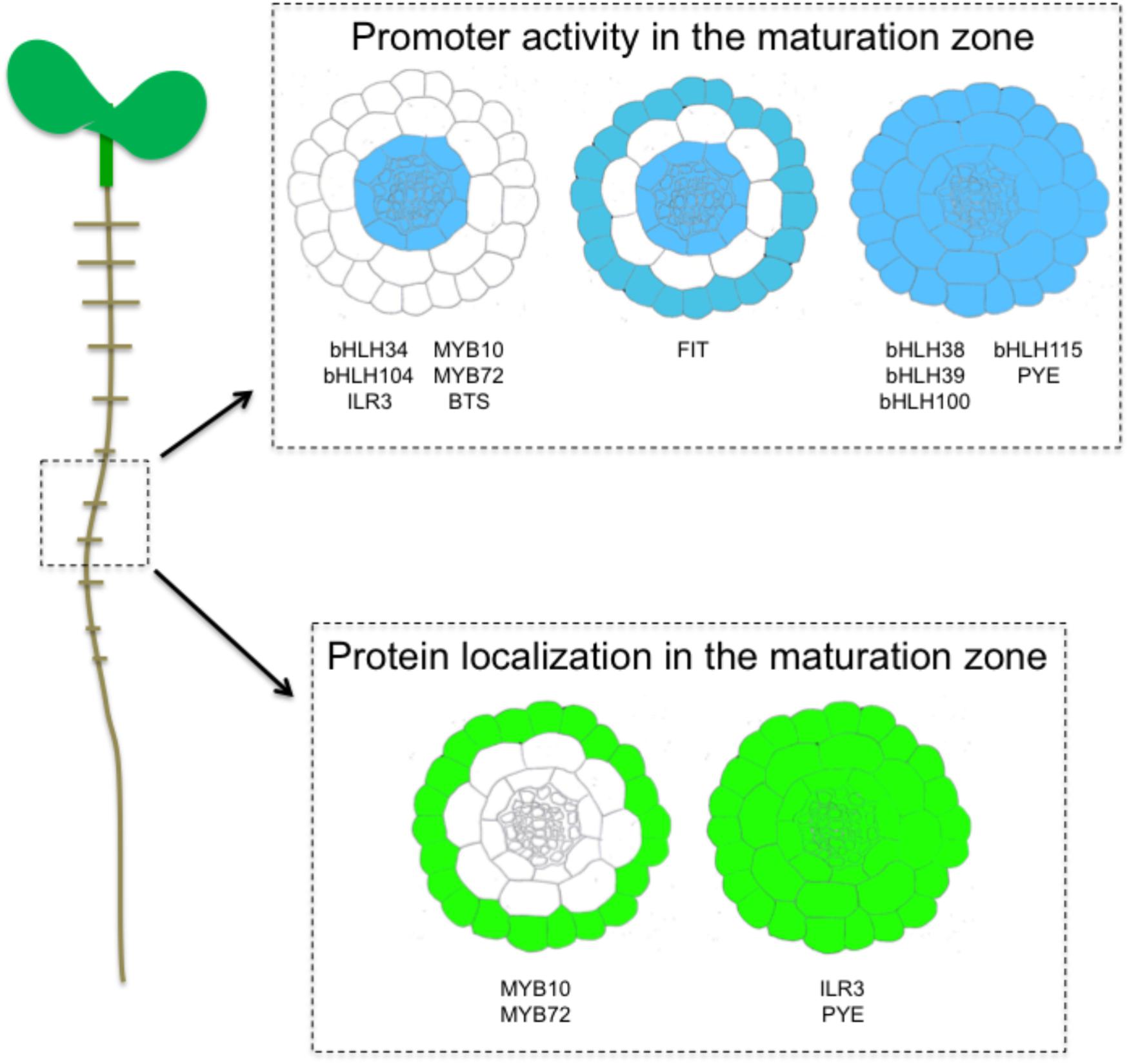
Figure 2. Schematic representation, based on previous studies, of expression (promoter activity) and localization (GFP fusion) pattern of transcription factors involved in the response to iron deficiency, in the maturation zone of the A. thaliana roots.
Iron uptake and translocation to the whole plant body requires coordinating the expression of several structural genes within and between the different cell types of roots that is achieved by the involvement of several TFs, in particular bHLHs (Figure 2).
Clade IVc bHLHs analysis of promoter activity revealed a specific expression in the stele (pericycle) of the root in the maturation zone (Li et al., 2016; Liang et al., 2017). Root tip expression was also detected except for bHLH115, whereas ILR3 was the sole bHLH from this clade to be expressed in the elongation zone of root tips and early lateral roots. These differences suggest that non-redundant biological functions may exist between the members of this clade (Liang et al., 2017). For instance, ILR3 was shown to participate in the plant response to biotic and abiotic stresses (Samira et al., 2018). Perivascular expression has also been observed in the aerial part of the plants (i.e., hypocotyl and leaves) (Rampey et al., 2006; Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). Promoter activity of PYE is similar in roots to that of bHLH115. In these cells PYE represses the expression of NAS4 and FRO3, modulating Fe translocation to the above ground part of the plant (Long et al., 2010). Clade Ib bHLHs (except bHLH101 whose promoter is not active in roots) were expressed throughout the root including the stele cells, which was in contrast with the expression pattern of their regulators, the clade IVc bHLH TFs (Wang et al., 2007). Clade Ib bHLHs promoter activity was essentially detected in the epidermis in the maturation zone, in the epidermis and inside the roots in the upper part zone (but not in the stele), and at the lateral root emergence site. However, a comparable pattern of expression with clade IVc bHLHs was observed in leaves. The apparent discrepancy observed in roots between the expression of clade Ib bHLHs and their transcriptional regulators suggests that clade IVc bHLH TFs may act in various cell types and thus have the ability to move from one cell type to another. In a recent study, it was shown that ILR3 protein is present in all root cell type when Fe availability is scarce (Samira et al., 2018). If this observation supports the hypothesis that ILR3 may move from the stele to other cell layers, it cannot be excluded that the promoter of ILR3 is not sufficient for proper ILR3 expression, and that sequences present in the coding region (e.g., introns) might be required. Interestingly, the pattern of PYE accumulation in root cells is similar to that of ILR3 (Long et al., 2010). The fact that ILR3 and PYE are present in the same cell layers and interact in vivo suggest that PYE function in the plant response to Fe deficiency might be tightly connected to ILR3 activity (Long et al., 2010; Selote et al., 2015). What would be the role of such heterodimers is still a matter of debate. Nevertheless, if the inter cellular trafficking of these two TFs was proven to be true, decrypting the underlying mechanisms would be the next challenge.
Fe-deficiency induced TF expression is mostly restricted to root tissues. At the rhizodermis, FIT expression overlaps those of its bHLH interacting partners from clade Ib (Colangelo and Guerinot, 2004; Jakoby et al., 2004). This is in agreement with the heterodimerization of FIT with clade Ib bHLHs to regulate the expression of Fe mobilization genes such as IRT1 and FRO2. Two other TFs whose expression is partly regulated by FIT in response to Fe deficiency, namely MYB10 and MYB72, are mainly localized at the rhizodermis where they control the expression of BGLU42 (Zamioudis et al., 2014). In the maturation zone, FIT is expressed in the epidermis and throughout the stele (Jakoby et al., 2004). Clade IVa bHLHs promoter activity is specific to the stele, a tissues where the repressive role of the encoded proteins on FIT stability is counteracted by FIT heterodimerization with clade Ib bHLHs (Cui et al., 2018). In the aerial part of the plant, clade IVa expression follows the vasculature as it is the case for ILR3 or clade Ib bHLHs.
To date several TFs involved in the control of Fe homeostasis have been characterized and several molecular connections have been identified. However we still do not know how the expression of the most upstream TFs of this network is regulated (i.e., FIT, clade IVc bHLH), indicating that we still do not have the full set of TFs involved in this network. Whether or not the Fe deficiency and Fe excess responses are controlled by an integrated pathway involving common players is still an open question that remains to be addressed. Last, deciphering the precise localization, at the protein level, of all the TFs involved in the control of Fe homeostasis is an important task that should be achieved if one aims at fully decrypting the functioning of this intricate molecular network.
FeG, KR, FrG, EI, and CD wrote the manuscript. FeG, KR, and CD made the figures.
KR and FeG were supported by a fellowship from the Agence Nationale pour la Recherche (ANR-17-CE20-0008-01) and from the China Scholarship Council (CSC), respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. N. Berger for help in preparing this manuscript.
Barberon, M., Dubeaux, G., Kolb, C., Isono, E., Zelazny, E., and Vert, G. (2014). Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, 8293–8298. doi: 10.1073/pnas.1402262111
Barberon, M., Zelazny, E., Robert, S., Conejero, G., Curie, C., Friml, J., et al. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, E450–E458. doi: 10.1073/pnas.1100659108
Bournier, M., Tissot, N., Mari, S., Boucherez, J., Lacombe, E., Briat, J. F., et al. (2013). Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288, 22670–22680. doi: 10.1074/jbc.M113.482281
Briat, J. F., Dubos, C., and Gaymard, F. (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40. doi: 10.1016/j.tplants.2014.07.005
Brumbarova, T., Bauer, P., and Ivanov, R. (2015). Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 20, 124–133. doi: 10.1016/j.tplants.2014.11.004
Colangelo, E. P., and Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412. doi: 10.1105/tpc.104.024315
Cui, Y., Chen, C. L., Cui, M., Zhou, W. J., Wu, H. L., and Ling, H. Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 11, 1166–1183. doi: 10.1016/j.molp.2018.06.005
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
De Masi, F., Grove, C. A., Vedenko, A., Alibes, A., Gisselbrecht, S. S., Serrano, L., et al. (2011). Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 39, 4553–4563. doi: 10.1093/nar/gkr070
Du, J., Huang, Z., Wang, B., Sun, H., Chen, C., Ling, H. Q., et al. (2015). SlbHLH068 interacts with FER to regulate the iron-deficiency response in tomato. Ann. Bot. 116, 23–34. doi: 10.1093/aob/mcv058
Dubeaux, G., Neveu, J., Zelazny, E., and Vert, G. (2018). Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69, 953.e5–964.e5. doi: 10.1016/j.molcel.2018.02.009
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Ezer, D., Shepherd, S. J. K., Brestovitsky, A., Dickinson, P., Cortijo, S., Charoensawan, V., et al. (2017). The G-Box transcriptional regulatory code in Arabidopsis. Plant Physiol. 175, 628–640. doi: 10.1104/pp.17.01086
Fourcroy, P., Siso-Terraza, P., Sudre, D., Saviron, M., Reyt, G., Gaymard, F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167. doi: 10.1111/nph.12471
Fourcroy, P., Tissot, N., Gaymard, F., Briat, J. F., and Dubos, C. (2016). Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+) transport system. Mol. Plant 9, 485–488. doi: 10.1016/j.molp.2015.09.010
Gordan, R., Shen, N., Dror, I., Zhou, T., Horton, J., Rohs, R., et al. (2013). Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 3, 1093–1104. doi: 10.1016/j.celrep.2013.03.014
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hindt, M. N., Akmakjian, G. Z., Pivarski, K. L., Punshon, T., Baxter, I., Salt, D. E., et al. (2017). BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890. doi: 10.1039/c7mt00152e
Ivanov, R., Brumbarova, T., Blum, A., Jantke, A. M., Fink-Straube, C., and Bauer, P. (2014). SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26, 1294–1307. doi: 10.1105/tpc.113.116244
Jakoby, M., Wang, H. Y., Reidt, W., Weisshaar, B., and Bauer, P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534. doi: 10.1016/j.febslet.2004.10.062
Jeong, J., and Connolly, E. L. (2009). Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci. 176, 709–714. doi: 10.1016/j.plantsci.2009.02.011
Klatte, M., Schuler, M., Wirtz, M., Fink-Straube, C., Hell, R., and Bauer, P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150, 257–271. doi: 10.1104/pp.109.136374
Kobayashi, T., Itai, R. N., Aung, M. S., Senoura, T., Nakanishi, H., and Nishizawa, N. K. (2012). The rice transcription factor IDEF1 directly binds to iron and other divalent metals for sensing cellular iron status. Plant J. 69, 81–91. doi: 10.1111/j.1365-313X.2011.04772.x
Kobayashi, T., Itai, R. N., Ogo, Y., Kakei, Y., Nakanishi, H., Takahashi, M., et al. (2009). The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J. 60, 948–961. doi: 10.1111/j.1365-313X.2009.04015.x
Kobayashi, T., Nagasaka, S., Senoura, T., Itai, R. N., Nakanishi, H., and Nishizawa, N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4:2792. doi: 10.1038/ncomms3792
Le, C. T., Brumbarova, T., Ivanov, R., Stoof, C., Weber, E., Mohrbacher, J., et al. (2016). Zinc finger of Arabidopsis thaliana 12 (ZAT12) interacts with FER-LIKE Iron Deficiency-Induced Transcription Factor (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 170, 540–557. doi: 10.1104/pp.15.01589
Li, L., Gao, W., Peng, Q., Zhou, B., Kong, Q., Ying, Y., et al. (2018). Two soybean bHLH factors regulate response to iron deficiency. J. Integr. Plant Biol. 60, 608–622. doi: 10.1111/jipb.12651
Li, X., Zhang, H., Ai, Q., Liang, G., and Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Liang, G., Zhang, H., Li, X., Ai, Q., and Yu, D. (2017). bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68, 1743–1755. doi: 10.1093/jxb/erx043
Ling, H. Q., Bauer, P., Bereczky, Z., Keller, B., and Ganal, M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. U.S.A. 99, 13938–13943. doi: 10.1073/pnas.212448699
Lingam, S., Mohrbacher, J., Brumbarova, T., Potuschak, T., Fink-Straube, C., Blondet, E., et al. (2011). Interaction between the bHLH transcription factor FIT and Ethylene Insensitive3/Ethylene Insensitive 3-Like1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23, 1815–1829. doi: 10.1105/tpc.111.084715
Liu, W., Karemera, N. J. U., Wu, T., Yang, Y., Zhang, X., Xu, X., et al. (2017a). The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS One 12:e0186580. doi: 10.1371/journal.pone.0186580
Liu, W., Li, Q., Wang, Y., Wu, T., Yang, Y., Zhang, X., et al. (2017b). Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency. Biochem. Biophys. Res. Commun. 491, 862–868. doi: 10.1016/j.bbrc.2017.04.014
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., and Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Maurer, F., Naranjo Arcos, M. A., and Bauer, P. (2014). Responses of a triple mutant defective in three iron deficiency-induced Basic Helix-Loop-Helix genes of the subgroup Ib(2) to iron deficiency and salicylic acid. PLoS One 9:e99234. doi: 10.1371/journal.pone.0099234
Naranjo-Arcos, M. A., Maurer, F., Meiser, J., Pateyron, S., Fink-Straube, C., and Bauer, P. (2017). Dissection of iron signaling and iron accumulation by over expression of subgroup Ib bHLH039 protein. Sci. Rep. 7:10911. doi: 10.1038/s41598-017-11171-7
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Uozumi, N., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Ogawa, S., Kawahara-Miki, R., Miyamoto, K., Yamane, H., Nojiri, H., Tsujii, Y., et al. (2017). OsMYC2 mediates numerous defence-related transcriptional changes via jasmonic acid signalling in rice. Biochem. Biophys. Res. Commun. 486, 796–803. doi: 10.1016/j.bbrc.2017.03.125
Ogo, Y., Itai, R. N., Nakanishi, H., Kobayashi, T., Takahashi, M., Mori, S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377. doi: 10.1111/j.1365-313X.2007.03149.x
Ogo, Y., Kobayashi, T., Nakanishi Itai, R., Nakanishi, H., Kakei, Y., Takahashi, M., et al. (2008). A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 283, 13407–13417. doi: 10.1074/jbc.M708732200
O’Hara, G. (2001). Nutritional constraints on root nodule bacteria affecting symbiotic nitrogen fixation: a review. Anim. Prod. Sci. 41, 417–433. doi: 10.1071/EA00087
Palmer, C. M., Hindt, M. N., Schmidt, H., Clemens, S., and Guerinot, M. L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9:e1003953. doi: 10.1371/journal.pgen.1003953
Rampey, R. A., Woodward, A. W., Hobbs, B. N., Tierney, M. P., Lahner, B., Salt, D. E., et al. (2006). An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174, 1841–1857. doi: 10.1534/genetics.106.061044
Rodriguez-Celma, J., Green, R. T., Connorton, J. M., Kruse, I., Cui, Y., Ling, H. Q., et al. (2018). BRUTUS-LIKE proteins moderate the transcriptional response to iron deficiency in roots. bioRxiv [Preprint]. doi: 10.1101/231365
Samira, R., Li, B., Kliebenstein, D., Li, C., Davis, E., Gillikin, J. W., et al. (2018). The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. doi: 10.1007/s11103-018-0735-8 [Epub ahead of print].
Selote, D., Samira, R., Matthiadis, A., Gillikin, J. W., and Long, T. A. (2015). Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167, 273–286. doi: 10.1104/pp.114.250837
Shen, C., Yue, R., Sun, T., Zhang, L., Yang, Y., and Wang, H. (2015). OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 231, 148–158. doi: 10.1016/j.plantsci.2014.12.003
Sivitz, A., Grinvalds, C., Barberon, M., Curie, C., and Vert, G. (2011). Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J. 66, 1044–1052. doi: 10.1111/j.1365-313X.2011.04565.x
Stringlis, I. A., Yu, K., Feussner, K., de Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U.S.A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Tanabe, N., Noshi, M., Mori, D., Nozawa, K., Tamoi, M., and Shigeoka, S. (2018). The basic helix-loop-helix transcription factor, bHLH11 functions in the iron-uptake system in Arabidopsis thaliana. J. Plant Res. doi: 10.1007/s10265-018-1068-z [Epub ahead of print].
Tang, C., Robson, A., and Dilworth, M. J. (1990). The role of iron in nodulation and nitrogen fixation in Lupinus angustifolius L. New Phytol. 114, 173–182. doi: 10.1111/j.1469-8137.1990.tb00388.x
Wang, F. P., Wang, X. F., Zhang, J., Ma, F., and Hao, Y. J. (2018). MdMYB58 modulates Fe homeostasis by directly binding to the MdMATE43 promoter in plants. Plant Cell Physiol. 59, 2476–2489. doi: 10.1093/pcp/pcy168
Wang, H. Y., Klatte, M., Jakoby, M., Baumlein, H., Weisshaar, B., and Bauer, P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. doi: 10.1007/s00425-007-0535-x
Wang, L., Ying, Y., Narsai, R., Ye, L., Zheng, L., Tian, J., et al. (2013). Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ. 36, 224–236. doi: 10.1111/j.1365-3040.2012.02569.x
Wang, N., Cui, Y., Liu, Y., Fan, H., Du, J., Huang, Z., et al. (2013). Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 6, 503–513. doi: 10.1093/mp/sss089
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yan, J. Y., Li, C. X., Sun, L., Ren, J. Y., Li, G. X., Ding, Z. J., et al. (2016). A WRKY transcription factor regulates fe translocation under fe deficiency. Plant Physiol. 171, 2017–2027. doi: 10.1104/pp.16.00252
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Zamioudis, C., Hanson, J., and Pieterse, C. M. (2014). beta-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379. doi: 10.1111/nph.12980
Zhang, H., Li, Y., Yao, X., Liang, G., and Yu, D. (2017). Positive regulator of iron homeostasis1, OsPRI1, facilitates iron homeostasis. Plant Physiol. 175, 543–554. doi: 10.1104/pp.17.00794
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805. doi: 10.1105/tpc.114.132704
Zhang, Y., Wu, H., Wang, N., Fan, H., Chen, C., Cui, Y., et al. (2014). Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytol. 203, 770–783. doi: 10.1111/nph.12860
Zhao, Q., Ren, Y. R., Wang, Q. J., Wang, X. F., You, C. X., and Hao, Y. J. (2016a). Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 172, 1973–1988. doi: 10.1104/pp.16.01323
Zhao, Q., Ren, Y. R., Wang, Q. J., Yao, Y. X., You, C. X., and Hao, Y. J. (2016b). Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol. J. 14, 1633–1645. doi: 10.1111/pbi.12526
Zheng, L., Ying, Y., Wang, L., Wang, F., Whelan, J., and Shou, H. (2010). Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 10:166. doi: 10.1186/1471-2229-10-166
Keywords: basic helix loop helix, bHLH, iron, homeostasis, Arabidopsis thaliana
Citation: Gao F, Robe K, Gaymard F, Izquierdo E and Dubos C (2019) The Transcriptional Control of Iron Homeostasis in Plants: A Tale of bHLH Transcription Factors? Front. Plant Sci. 10:6. doi: 10.3389/fpls.2019.00006
Received: 16 November 2018; Accepted: 07 January 2019;
Published: 18 January 2019.
Edited by:
Thomas J. Buckhout, Humboldt-Universität zu Berlin, GermanyReviewed by:
Takanori Kobayashi, Ishikawa Prefectural University, JapanCopyright © 2019 Gao, Robe, Gaymard, Izquierdo and Dubos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Dubos, Q2hyaXN0aWFuLmR1Ym9zQGlucmEuZnI=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.