
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 18 December 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01860
This article is part of the Research Topic Metabolic Adjustments and Gene Expression Reprogramming for Symbiotic Nitrogen Fixation in Legume Nodules View all 12 articles
Members of the plant family Leguminosae (Fabaceae) are unique in that they have evolved a symbiotic relationship with rhizobia (a group of soil bacteria that can fix atmospheric nitrogen). Rhizobia infect and form root nodules on their specific host plants before differentiating into bacteroids, the symbiotic form of rhizobia. This complex relationship involves the supply of C4-dicarboxylate and phosphate by the host plants to the microsymbionts that utilize them in the energy-intensive process of fixing atmospheric nitrogen into ammonium, which is in turn made available to the host plants as a source of nitrogen, a macronutrient for growth. Although nitrogen-fixing bacteroids are no longer growing, they are metabolically active. The symbiotic process is complex and tightly regulated by both the host plants and the bacteroids. The metabolic pathways of carbon, nitrogen, and phosphate are heavily regulated in the host plants, as they need to strike a fine balance between satisfying their own needs as well as those of the microsymbionts. A network of transporters for the various metabolites are responsible for the trafficking of these essential molecules between the two partners through the symbiosome membrane (plant-derived membrane surrounding the bacteroid), and these are in turn regulated by various transcription factors that control their expressions under different environmental conditions. Understanding this complex process of symbiotic nitrogen fixation is vital in promoting sustainable agriculture and enhancing soil fertility.
Leguminosae (Fabaceae) is the third largest family of angiosperms with 750 genera and around 19,500 species (The Legume Phylogeny Working Group, 2013). Most legumes can establish a mutualistic association with alpha- and beta-proteobacteria to obtain biological nitrogen (reviewed by Andrews and Andrews, 2017; Sprent et al., 2017). Rhizobia are soil bacteria known for being able to establish symbiosis with legume plants. Symbiotic nitrogen fixation (SNF) can be carried out once rhizobia are established inside the cells of root nodules formed from newly differentiated tissue in the roots of host plants. The host plant provides the microsymbiont with dicarboxylates together with other nutrients, in exchange for fixed nitrogen in the form of ammonium and amino acids (Udvardi and Day, 1997). Nitrogen-fixing legumes contribute to nitrogen enrichment of the soil and therefore are valuable in improving soil fertility. The legume–rhizobium association has an important impact on sustainable agriculture since it provides more than 65% of the biologically fixed nitrogen in agricultural systems (Herridge et al., 2008).
Studies on legume–rhizobium symbiosis have covered a large number of legume species, such as soybean (Glycine max), bird’s foot trefoil (Lotus japonicus), alfalfa (Medicago sativa), barrelclover (Medicago truncatula), common bean (Phaseolus vulgaris), garden pea (Pisum sativum), common vetch (Vicia sativa), and narrowleaf lupin (Lupinus angustifolius) (Dupont et al., 2012). Although M. sativa–Sinorhizobium meliloti and P. sativum–Rhizobium leguminosarum associations are also well-studied symbiotic system (Kneen and LaRue, 1984; Jones et al., 2007), the genetic models of legume–rhizobium symbiosis are mainly focused on L. japonicus–Mesorhizobium loti and M. truncatula–S. meliloti associations, due to the small diploid genomes, high levels of genetic diversity, and well-established transformation systems of L. japonicus and M. truncatula (Oldroyd and Geurts, 2001; Oldroyd, 2005). L. japonicus possesses determinate root nodules, which are spherical with a well-defined, homogeneous central fixation zone composed of infected rhizobia-filled cells surrounded by uninfected cells (Schultze and Kondorosi, 1998). In contrast, M. truncatula forms indeterminate nodules which are cylindrical and consist of a gradient of developmental zones (Timmers et al., 1999).
Owing to their complex makeup, legume nodules have been extensively studied with respect to their metabolism and regulation. After the SNF process has been established in mature nodules, several biological processes occur simultaneously inside the nodules, including biological nitrogen fixation carried out by bacteroids, carbon–nitrogen metabolism, and exchange between host plants and bacteroids, and metabolite transport across cell membranes (Resendis-Antonio et al., 2011; Udvardi and Poole, 2013; Clarke et al., 2014). Root nodules mediate the influx of carbon sources and efflux of nitrogen compounds. The host plant supplies sucrose which will be converted to sources of energy and organic acids for fixation of atmospheric nitrogen, while the endosymbiont returns organic fixation products to the host (Day et al., 2001). Moreover, SNF in legume nodules exerts a high demand for phosphates (Gunawardena et al., 1992; Saad and Lam-Son, 2017). The phosphate concentration in nodules is threefold higher than in other organs or tissues (Sa and Israel, 1991). Under phosphate starvation, the phosphate acquisition rate increases in nodules to make them less vulnerable compared to other organs (Thuynsma et al., 2014; Saad and Lam-Son, 2017). Optimal plant phosphate requirement is about 0.05–0.30% of total dry weight, and inorganic phosphate is the main form absorbed by plants (Temple et al., 1998; Saad and Lam-Son, 2017).
Better understanding of the metabolic components participating in N2 fixation and the key regulators that control the processes in legume nodules are a crucial step in seeking possible ways to enhance SNF and further increase legume productivity. In this review, we focus on the latest knowledge on root nodules including the nodule types (the section “Legume Nodule Types and Their Compatible Rhizobia”), the metabolic changes, transportation and regulation mechanisms in host plant (the section “Overview of Metabolism and Regulation in the Host Plant”) and bacteroids (the section “Metabolism and Transport in Bacteroids”).
About 90% of the species within the family Leguminosae (Fabaceae) can fix atmospheric nitrogen through a symbiotic association with soil bacteria known as rhizobia (Rascio and La Rocca, 2013; Andrews and Andrews, 2017). Rhizobia are diazotrophic Gram-negative bacteria able to form nitrogen-fixing nodules on the roots of legumes, where they differentiate into bacteroids (Rosenberg et al., 2014). Rhizobia present different specificities toward different host plant species based on the recognition of specific signal molecules (Nod factors) (Spaink, 2000; Poole et al., 2018). However, promiscuity has been observed in some cases. For example, Sinorhizobium fredii strain NGR234 is able to establish symbiosis with over 100 plant legumes such as cultivated soybean (G. max), wild soybean (Glycine soja), pigeon pea (Cajanus cajan), and cowpea (Vigna unguiculata) (reviewed in Brenner et al., 2009). Recently, analysis on legume–rhizobia symbioses was published where specificity (reviewed in Andrews and Andrews, 2017) and biogeography distribution (reviewed in Sprent et al., 2017) of nodulating legumes and their symbionts were discussed in detail.
Nodules are divided into two types: determinate and indeterminate. Determinate nodules are characteristic of soybean and common bean. Legumes such as alfalfa and garden pea present indeterminate nodules. Indeterminate nodules display five developmental zones: meristem (Zone I), infection and differentiation (Zone II), transition region between Zone II and Zone III in which bacteria are engulfed by plant cells (interzone II–III), nitrogen fixation (Zone III), and senescence (Zone IV) (Vasse et al., 1990). The meristem is active in indeterminate nodules and continues to grow, unlike in determinate nodules where meristem cells die once the nodules are mature. The meristem is the distal zone to the root and Zone IV is the proximal to the root attachment site. Zone-specific transcriptional and metabolic changes have been reported in the nodules formed between M. truncatula with S. meliloti and S. medicae, respectively (Roux et al., 2014; Ogden et al., 2017). A fifth zone (Zone V) has been described in alfalfa nodules (Timmers et al., 2000). This zone is proximal to the senescence zone and contains saprophytic intracellular rhizobia that do not undergo bacteroid differentiation. Table 1 summarizes examples of legume–rhizobium associations and nodule types.
Sucrose is the primary carbon resource for energy supply and carbon skeletons for SNF and is supplied to legume nodules by transportation from the shoots (Gordon et al., 1998). However, sucrose is not used directly as a substrate by isolated nitrogen-fixing bacteroids to support nitrogenase activity, as demonstrated in the cases of soybean (Stovall and Cole, 1978), garden pea (Glenn and Dilworth, 1981), lupin (Tomaszewska et al., 1991), and alfalfa (Miller et al., 1988). In nodules, sucrose can be processed by one of two enzymes, sucrose synthase (SS; EC 2.4.1.13) and alkaline invertase (AI; EC 3.2.1.26), as shown in Figure 1. SS performs a reversible UDP-dependent cleavage of sucrose into UDP-glucose (UDP-Glc) and fructose (Akazawa and Okamoto, 1980). SS activity is reduced in the SNF-defective mutant, rug4, of P. sativum, indicating that SS plays an important role in nodule functions (Gordon et al., 1998, 1999). Moreover, overexpressing the antisense of MtSucS1 (encoding enzyme SS) in M. truncatula impaired the plant growth and nodulation in the MtSucS1-reduced nodules, and the contents of amino acids and their derivatives were also found to be reduced, which suggest SS is a crucial player in the establishment and maintenance of an efficient SNF process (Baier et al., 2007). Six SS isoforms have been identified, and the mutants of SS isoform sus1-1 and sus3-1 showed 38% and 67% reduction of SS activity, respectively, in nodules formed by inoculating L. japonicus with M. loti strain Tono, while the double mutant plants of sus1-1/sus3-1 had significantly impaired growth and their nodule SS activities were reduced by 94% compared to the wild type under the same growth conditions. This further shows that SS is essential in nodulation maintenance (Horst et al., 2007). In addition, when exposed to drought stress, nitrogen fixation in nodules declined, and the activity of SS was severely inhibited in various species (M. sativa, P. sativum, and P. vulgaris), and led to the further limitation of carbon availability in bacteroids (Ramos et al., 1999; Gálvez et al., 2005; Naya et al., 2007).
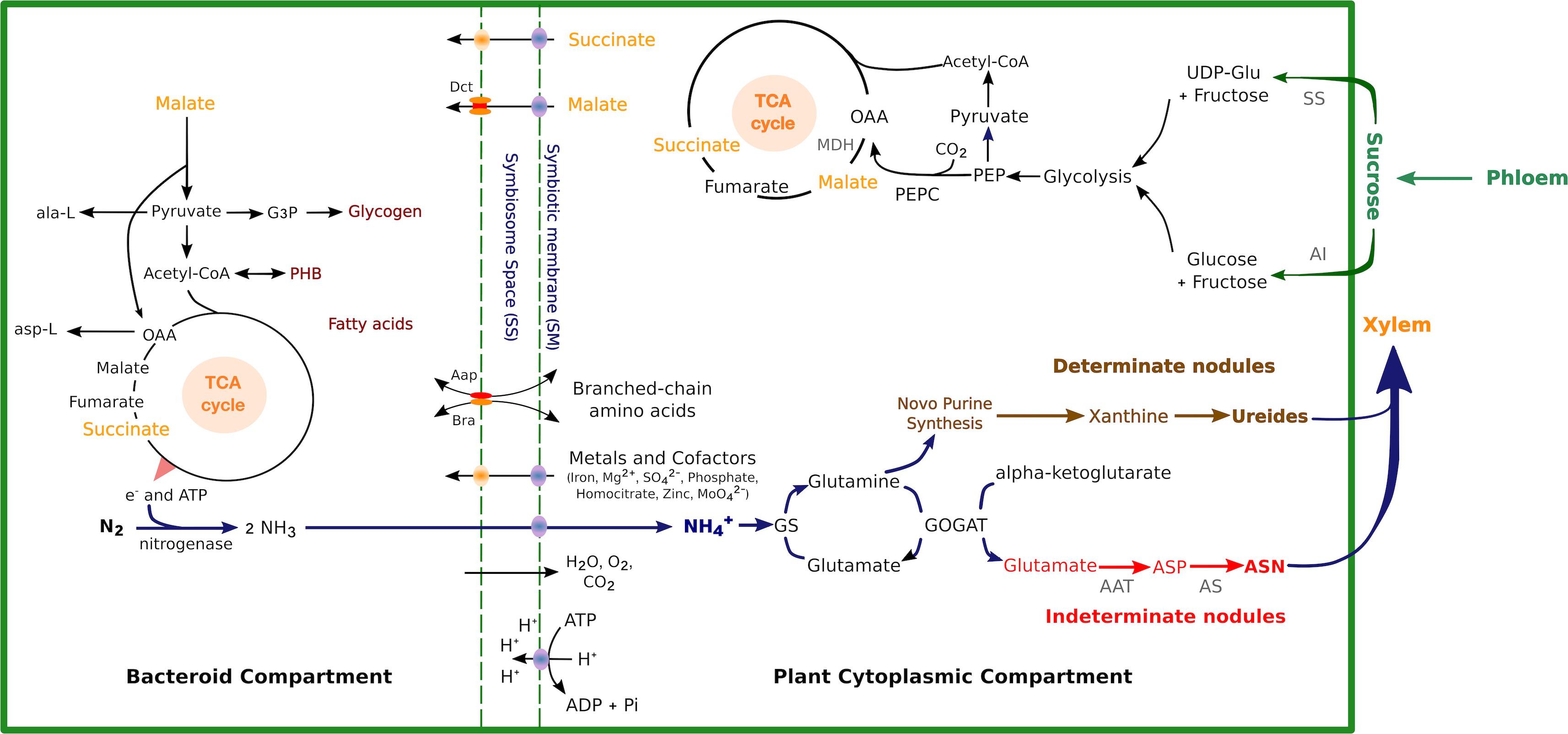
Figure 1. Schematics of carbon and nitrogen metabolic pathways with key enzymes, metabolites, and transporters in determinate nodules and indeterminate nodules. Sucrose in the plant cytosol is split into glucose and fructose by AI or UDP-Glc and fructose via SS, which is then catabolized via glycolysis to PEP. Carbon from PEP and carbonic acid is diverted to OAA and then malate by PEPC and the neMDH, respectively. OAA may be further converted to succinate or fumarate. Carbon sources are transported across the peribacteroid and bacteroid membranes and enter the TCA cycle in the bacteroid to be metabolized. Transport of inorganic ions and cofactors required for SNF across the SM is indicated. The ammonia produced by the SNF is transported back to the plant and assimilated by GS and GOGAT into Gln and Glu (blue arrows). In indeterminate nodules, Glu and Gln are further converted to Asp and Asn by AAT and AS, respectively (red arrows). In determinate nodules, Gln enters purine synthesis pathway and is converted to ureides (brown arrows). AI, alkaline invertase; UDP-Glc, UDP-glucose; SS, sucrose synthase; PEP, phosphoenolpyruvate; OAA, oxaloacetate; PEPC, PEP-carboxylase; MDH, malate dehydrogenase; PHB, polyhydroxybutyrate; AAT, aspartate aminotransferase; AS, asparagine synthetase; ASP, aspartate; ASN, asparagine.
The other sucrose-catabolizing enzyme, AI, is a hydrolase, first identified in soybean nodules, that irreversibly cleaves sucrose into fructose and glucose (Copeland and Morell, 1985). The isolation and purification of AI was also carried out from chickpea nodules (Asthir and Singh, 1997). In L. japonicus, the expression level of gene LjInv1 encoding enzyme AI was twofold higher in mature nodules than in uninfected roots. Enzyme activity assays showed that LjInv1 contributed to the production of hexoses and other biosynthetic processes in developing nodules (Flemetakis et al., 2006). In G. max and M. truncatula root nodules, an alkaline/neutral Inv gene was found to have enhanced transcription levels in developing root nodules inferring an increased need for sucrose degradation (Flemetakis et al., 2006; Tesfaye et al., 2006). However, there is not much direct evidence to prove that AI is essential for the regulation of nodule metabolism.
The resulting UDP-Glc and free hexoses (glucose and fructose) are phosphorylated by hexokinase (EC 2.7.1.1) and enter glycolysis or oxidative pentose phosphate pathways. In nodules, the host plant provides carbon sources for bacteroid activities in the form of dicarboxylates, particularly malate and succinate (Day, 1991). Phosphoenolpyruvate carboxylase (PEPC; EC 4.1.1.31) and malate dehydrogenase (MDH; EC 1.1.1.82) convert the carbon flux from glycolysis to form malate (Figure 1). PEPC catalyzes the conversion of PEP into oxaloacetate (OAA). The active enzymes are less sensitive to malate, which act as a negative feedback control (Nimmo, 2000). The nodule PEPC was found to be activated with phosphorylation and its sensitivity was inhibited by L-malate in vitro and in vivo (Schuller and Werner, 1993; Zhang et al., 1995). In L. japonicus, abundant transcript of LjPEPC1 was found in the vascular bundles of nodules and bacteroid-infected cells, while LjPEPC2 was expressed in roots and shoots at low levels and postulated as the housekeeping isoform. Moreover, regulatory phosphorylation of PEPC is thought to be mainly controlled by the PEPC kinase (PEPC-PK) (Vidal and Chollet, 1997), and the expression of LjPEPC-PK shares a similar pattern with LjPEPC1 in mature nodules (Nakagawa et al., 2003). The OAA is then converted to malate by MDH in a reversible reaction, and this can then be uptaken by bacteroids directly (Figure 1). In alfalfa, five species of cDNAs were found to encode MDH, but only one of them had a nodule-enhanced expression pattern (Miller et al., 1998). One nodule-enhanced MDH (neMDH) was found in garden pea (Fedorova M. et al., 1999). Two MDH isoforms were found in the L. japonicus nodule by transcriptome profiling, one of which is a cytosolic MDH (cMDH) and the other a neMDH (Takanashi et al., 2012). The activity of cMDH in L. angustifolius nodules was shown to increase under P-deficient conditions, supporting the involvement of cMDH in nodules (Le Roux et al., 2014).
Besides the enzymes described above, some other carbon metabolic enzymes also play a role in legume nodules. MsSPSA, encoding sucrose phosphate synthase (SPS; EC 2.3.1.14) in M. sativa L., is involved in the first step of sucrose synthesis, and the expression of MsSPSA showed a nodule-enhanced pattern. SPS showed higher activities in the nodules formed by wild-type S. meliloti, compared to nodules inoculated with a N-deficient Fix- strain (a Tn5 insertional mutation in nifH; Hirsch et al., 1983), suggesting an important role in maintaining a stable carbohydrate/energy supply for the actively nitrogen-fixing symbiont (Aleman et al., 2010).
Multiple environmental stresses result in the inhibition of carbon metabolism in host legumes, resulting in the suppression of SNF efficiency in nodules. Under salt stress, the concentration of sucrose was decreased, and major carbon metabolic enzymes such as SS and AI showed lower activity in both M. truncatula nodules (indeterminate nodules) and L. japonicus nodules (determinate nodules) (López et al., 2008). When garden pea plants were cultured under drought stress, the SS activity was down-regulated, indicating the normal carbon metabolic pathway was inhibited. The NADP-dependent isocitrate dehydrogenase (ICDH; EC1.1.1.42) was reported to balance C/N metabolic fluxes in leaves (Gallardo et al., 1995; Gálvez and Gadal, 1995). The enhancement of ICDH activity in nodules was probably the result of compensating for the C and N imbalance or supplying additional NADPH to neutralize the effects of the increased production of reactive oxygen species (ROS) as a consequence of drought stress (Gálvez et al., 2005).
Ammonia exported from the bacteroids diffuses into the cytosol of the infected host cells to be rapidly assimilated. Amino acids and/or ureides are then synthesized and exported from the legume nodules to the shoots (Figure 1). The determinate-nodule legumes primarily transport allantoin and allantoate (ureides) as fixed-N compounds, while the indeterminate nodules assimilate amides in the form of asparagine (Asn) and glutamine (Gln) (Sprent, 2009). The first step of ammonium assimilation is apparently the same in amide- and ureide-exporting legume nodules (Todd et al., 2006). In both cases, nitrogen fixed in the bacteroids is exported into the host cell cytosol, where ammonia is converted to Gln and glutamate (Glu) by the enzymes Gln synthetase (GS; EC 6.3.1.2) and Glu synthase (NADH-GOGAT; EC 1.4.1.14). In indeterminate nodules, Glu and Gln are further converted to aspartate (Asp) and Asn by Asp aminotransferase (AAT EC 2.6.1.1) and Asn synthetase (AS; EC 6.3.5.4). While for determinate nodules, Gln further entry purine synthesis pathway and convert to ureides as final product exported in to host plant (Figure 1).
Glutamine synthetase, as the enzyme for the first step of ammonia assimilation in the infected root cells, is a key component of nitrogen metabolism in nodules. It catalyzes the ATP-dependent amination of Glu to Gln. GS possesses two major isoforms largely separated by their subcellular localization, plastidic GS2 and cytosolic GS1 (Temple et al., 1998; Lea and Miflin, 2003). In M. truncatula nodules, three GS genes have been identified: cytosolic MtGS1a and MtGS1b, and plastid MtGS2, where MtGS1a accounts for 90% of the GS activity in nodules (Carvalho et al., 1997, 2000). Impairment of GS activity using phosphinothricin resulted in the inhibition of nodule growth, and the promotion of nodule senescence. The reduced activity of GS also suppressed Asn biosynthesis, as demonstrated by the lower expression of AS transcripts and lower Asn content (Seabra et al., 2012). NADPH-GOGAT is another enzyme involved in the assimilation of , by transferring the amide group from Gln to α-ketoglutarate (Figure 1). In transgenic M. sativa expressing antisense NADPH-GOGAT, GOGAT activities were much reduced in the nodules. The transgenic M. sativa inoculated with S. meliloti exhibited moderate chlorosis, lower nodule weight, and reduction of N2 fixation efficiency compared to the control (Schoenbeck et al., 2000).
In addition to primary nitrogen assimilation by converting ammonia into amino acids, ureide biosynthesis is also a common pathway among tropical legumes such as soybean, common bean, and cowpea, which generally form determinate nodules (Tajima, 2004). The allantoin and allantoate are the final nitrogen forms exported from soybean nodules to the shoots (McClure and Israel, 1979). Before ureide synthesis, ammonia enters a purine synthesis pathway, which is catalyzed by xanthine oxidase and xanthine dehydrogenase into urate, whose activities have been detected in both infected cells and uninfected cells in cowpea nodules. While uricase, which catalyzes the irreversible conversion of urate to allantoin, was detected in uninfected cells but not infected cells (Atkins et al., 1997). Moreover, the genes GmALN3 and GmALN4, encoding the enzyme allantoinase (EC 3.5.2.5) that converts allantoin to allantoate, were expressed in soybean nodules (Duran and Todd, 2012). Gln may also be enzymatically transformed into xanthine through the de novo purine synthesis pathway (Werner and Witte, 2011; Figure 1).
Phosphate is the second most limiting macronutrient element required for crop growth after nitrogen, and there is a particularly high demand for phosphates in the nodules of N2-fixing legumes (Gunawardena et al., 1992; Vance et al., 2000). Phosphate, as an essential macronutrient, is incorporated into organic compounds such as nucleic acids (DNAs and RNAs), enzymes, ATP, sugar phosphates, phospholipids, and so on. Such organic phosphates are involved in many plant biochemical processes such as nutrient transport and photosynthesis. As a component of chromosomes, phosphates play an important role in cell division and organogenesis, and help to transfer the genetic information from one generation to the next (Ahemad and Oves, 2011). The soluble inorganic phosphate (Pi) is the most abundant metabolite in Bradyrhizobium japonicum cells. The Pi concentration in free-living B. japonicum cells is 8.1 μmol g-1 FW and 29.2 μmol g-1 FW in bacteroids. Massive amounts of Pi are transported into bacteroids through phosphate transporters and are highly involved in the metabolism of bacteroids. Indeed, SNF expends a great quantity of energy involving phosphorylated intermediates (Vauclare et al., 2013).
Deprivation of Pi can lead to an ATP shortage and a decrease in key metabolic enzymes (e.g., those involved in photosynthesis), and therefore is tightly correlated with plant growth (Czarnecki et al., 2013; Saad and Lam-Son, 2017). Although low Pi availability causes a decline in SNF, the responses to Pi deficiency vary among different legumes. There are two main strategies: one is to make changes to nitrogen and carbon metabolism, such as N acquisition and assimilation as well as carbon source adjustments, and the other is to enhance Pi uptake and recycling in nodules (Wang et al., 2010; Qin et al., 2012; Udvardi and Poole, 2013).
To investigate the metabolic changes in legume nodules under low-Pi stress, metabolic profiling has been done in common bean and chickpea (Hernandez et al., 2009; Zhang N. et al., 2012). Hernandez et al. (2009) performed a metabolic profiling using common bean plants inoculated with Rhizobium tropici CIA899 cultivated under Pi-deficient and Pi-sufficient conditions. The levels of 13 metabolites were significantly changed when comparing between the two conditions. Among these, amino acids and other nitrogen metabolites, such as urea, spermidine, and putrescine, were decreased in nodules under low-Pi conditions while most of the carbon metabolites including organic acids and polyhydroxy acids were increased (Hernandez et al., 2009). Similar changes were also found in chickpea inoculated with two different Mesorhizobium strains, M. ciceri CP-31 (McCP-31) and M. mediterranum SWRI9 (MmSWRI9) (Nasr Esfahani et al., 2016). These two Mesorhizobium strains inoculated on chickpea showed a differential symbiotic performance under both control and low-Pi condition. McCp-31-chickpea exhibited a higher symbiotic efficiency and higher Pi concentration than MmSWRI9–chickpea under normal and low-Pi conditions. The differences in SNF efficiency between the two types of chickpea–Mesorhizobium associations were correlated with various changes in carbon and nitrogen metabolites. For instance, the activities of most carbon and nitrogen metabolic enzymes, such as AAT, GS, and MDH, were increased in McCp-31–chickpea nodules, while the activities of these same enzymes were either the same or reduced in MmSWRI9–chickpea nodules. These results provide evidence that host plants adjust their carbon and nitrogen metabolic pathway to maintain the SNF efficiency under Pi deficiency condition (Nasr Esfahani et al., 2016).
According to previous studies, under low Pi availability, nodules would largely decrease the utilization of atmospheric nitrogen as the nitrogen source and utilize more soil nitrogen (such as and ) instead, as shown in Figure 2 (Valentine et al., 2017). The reason for such a shift is not only that itself is a strong inhibitor of nodulation and SNF, but also that the consumption of carbon as an energy source for nitrogen uptake from soil is less than that for SNF (Minchin and Witty, 2005). In Virgilia divaricata, the carbon cost related to SNF ranges from 3.3 to 6.6 gCg-1-N (gram of carbon was used when fixed each gram of N2), while the carbon cost of reduction is not more than 2.5 gCg-1-N. Hence, the assimilation of nitrogen from the soil would save more organic carbon for plant growth (Minchin and Witty, 2005). In SNF nodules, the host plant provides photosynthetically fixed carbon to symbiosomes as the major energy source. In turn, bacteria in the nodule fix N2 by the activities of nitrogenases and release to the host cells. The carbon transported to symbiosomes is mainly in the form of TCA cycle dicarboxylates (such as malate) and is also used as the carbon skeletons for the syntheses of amino acids from N2 (Rosendahl et al., 1990; Valentine et al., 2017; Figure 2). For example, in L. angustifolius cv. Wonga, there is an increased rate of malate synthesis via PEPC and MDH under long-term Pi deficiency. Although malate is the major energy source for symbiosomes, high malate accumulation could inhibit N2 fixation and nitrogen assimilation (Le Roux et al., 2008).
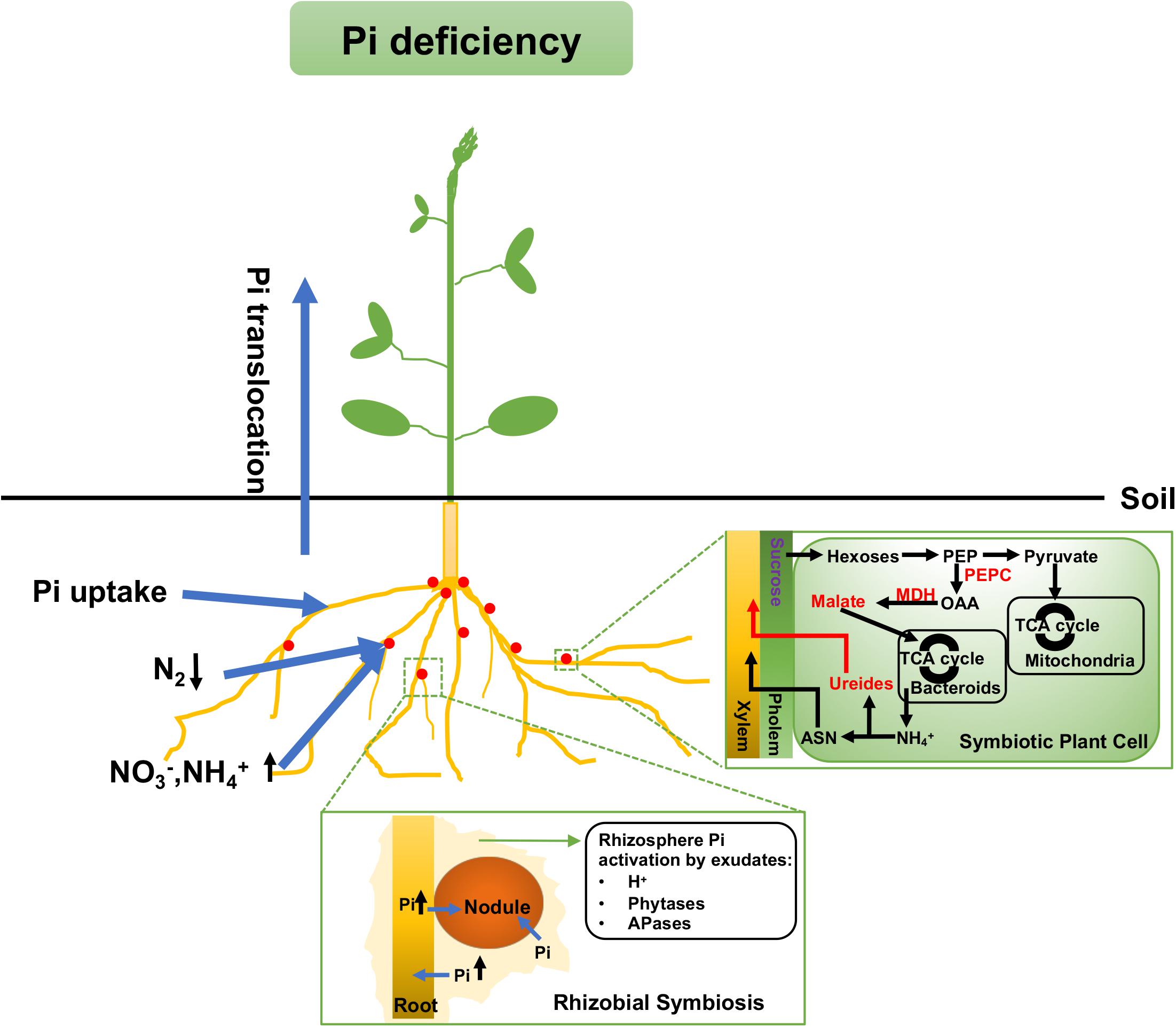
Figure 2. Mechanisms for maintaining Pi homeostasis under Pi deficiency in legumes. Legumes use more soil N (such as and ) as the main nitrogen source rather than through SNF. There is also a malate accumulation in both types of nodules and more ureides export in determine nodules under Pi deficiency. A red or purple label means the metabolite content or enzyme activity is up- or down-regulated, respectively. Arrows up (↑) and down (↓) mean the nutrient uptake is up- or down-regulated, respectively. SNF, symbiotic nitrogen fixation; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase; OAA, oxaloacetate; MDH, malate dehydrogenase; ASN, asparagine.
As aforementioned, in exchange for the nutrient supply, ammonia and/or alanine and Asp are translocated into the legume cytosol from the symbiosome and then assimilated into ureides or amides. Under Pi deprivation, legumes prefer exporting more ureides, due to the lower carbon cost (Figure 2). This was demonstrated through an experimental determination of C and N budgets in the tropical legume cowpea and the temperate legume lupin. The results were 1.4 gCg-1 fixed N in cowpea which forms ureide-exporting nodules, and a minimum of 3.9 gCg-1 fixed N in lupin which forms amide-exporting nodules. The result indicates that the export of ureides is more economical in the use of carbon (Atkins, 1991). The shift to higher amounts of ureide export under Pi stress was also observed in soybean, where there was higher accumulation of ureides relative to amino acids in nodules (Le Roux et al., 2009). The switch to ureide exports may reduce the carbon cost under Pi stress, which has a positive effect on legume growth.
Under Pi stress, legumes exhibit very flexible mechanisms to maintain Pi homeostasis in nodules. On the one hand, legumes increase Pi acquisition from the external environment. On the other hand, plants adopt flexible Pi recycling and internal Pi conservation mechanisms to improve Pi remobilization (Nasr Esfahani et al., 2016; Saad and Lam-Son, 2017).
Pi acquisition from the external environment by plants is affected by nitrogen metabolism. Pretreatment with and can increase Pi uptake from soil in Zea mays roots (Smith and Jackson, 1987). Acquisition of can lead to a release of proton (H+) that decreases the pH in the rhizosphere, which in turn stimulates Pi solubility and uptake (Zhao et al., 2009). Legumes tend to take up and from the soil through the roots in both Pi stress and Pi sufficient conditions (Dan and Brix, 2009). In turn, more acquisition of can improve the availability of Pi in the external environment.
Plant acid phosphatases (APases) can hydrolyze organic phosphates to improve soil Pi availability in legumes (Figure 2). Under Pi stress, APases become catalytically better using the same amount of nodule crude extract, which is an efficient way to utilize organic phosphates (Araújo et al., 2008). In addition, phytases can hydrolyze phytate into myoinositol and Pi to improve the Pi level in the rhizosphere for uptake through the roots (Araújo et al., 2008). The expression and activities of phytases are also induced by Pi deprivation in P. vulgaris nodules. Two recombinant inbred lines (RILs) of P. vulgaris, RILs 115 (P-efficient) and 147 (P-inefficient), were inoculated with R. tropici CIAT 899 strain and planted in Pi-deficient conditions. Under low Pi condition, the accumulation of phytase transcripts was observed in the nodules of both lines, and it is more enhanced in Line RIL115 than in Line RIL147. The increase in phytase and phosphatase enzyme activities and higher SNF efficiency were found in RIL115 under Pi-deficient treatment, indicating a possible role of phytase activity in nodules to maintain SNF under Pi deficiency condition (Lazali et al., 2013).
The uptake of Pi is also under the regulation of high-affinity Pi transporters. Plants have two identified Pi uptake mechanisms: a high-affinity system that may be up-regulated under low Pi condition and a low-affinity system which is constitutively expressed (Hernandez et al., 2009; Nasr Esfahani et al., 2016). In this review, we focus on the high-affinity Pi transporters. Most high-affinity Pi transporters are induced by Pi starvation and expressed in the root hairs and root epidermis which are directly exposed to soil Pi (Liu et al., 2008). Among Pi transporters, the Pht1 family is the most intensively studied in plants (Gu et al., 2016). Pht1, localized in the plasma membrane, is directly related to Pi uptake from soil and Pi translocation in the plant. Some Pht1s are directly under the control of the central regulators of Pi starvation signaling, the phosphate starvation response (PHR) transcription factors (Rubio et al., 2001; reviewed in Gu et al., 2016). In G. max, 15 Pht1 paralogs have been identified by bioinformatics and experimentation. The expressions of all 15 GmPht1s are up-regulated under low Pi condition compared to high Pi condition. By overexpressing each GmPHT1 paralog in the yeast Pi-transporter mutant PAM2 (Δpho84 Δpho89), GmPht1;1, GmPht1;2, GmPht1;5, GmPht1;7, and GmPht1;10 are found to be high-affinity Pi transporters and the others are lower affinity Pi transporters (Fan et al., 2013). A Pi starvation-induced high-affinity Pi transporter, GmPT5 (Glyma10g04230), was also identified, which was considered to play an important role in maintaining nodular Pi homeostasis. GmPT5 is expressed in the junction area between roots and young nodules, and mainly functions in transporting Pi from roots to nodules. Higher expression of GmPT5 leads to more Pi being transported from root to nodule, which is beneficial for maintaining SNF (Qin et al., 2012).
Besides the uptake of Pi from the external environment, legume nodules also develop phosphate recycling and phosphate conservation mechanisms under Pi deficiency (Figure 2) (Saad and Lam-Son, 2017). A report showed that chickpea could re-allocate Pi from roots to nodules when facing Pi stress. A sharp reduction in Pi (by around 78%) in roots was detected in chickpea, which the researchers assumed that the missing Pi was transported into nodules in order to prevent the complete depletion of nodular Pi (Nasr Esfahani et al., 2016). Common bean was also reported to mobilize Pi from nucleic acids and phospholipids (Hernandez et al., 2009). Under low Pi conditions, V. divaricata exhibited a lower Pi uptake rate but higher levels of phosphohydrolase exudation. This suggests that V. divaricata may prefer to recycle internal nodular Pi pools and use alternate bypass routes to conserve Pi rather than directly uptaking Pi from soil (Vardien et al., 2016).
It is an important challenge for legumes to maintain a balance between supplying the nodules with the amount of carbon required for nitrogen fixation while retaining sufficient carbon for growth, as well as keeping the nitrogen efflux from the nodules at an optimum level. The host plant regulates the symbiotic process by controlling nodule development and nodule numbers as well as by adjusting the nodule turnover and the level of nitrogen fixation (Ferguson et al., 2010; Sulieman and Schulze, 2010). The balance between plant nitrogen demand and nitrogen fixation rate is finely tuned to reach the nitrogen concentration that allows maximum plant growth. In common bean, nitrogen translocated from senescing lower leaves to nodules resulted in lower nitrogen fixation rate (Fischinger et al., 2006). Autoregulation of nodulation (AON) is the main systemic negative feedback mechanism of host plants to negatively regulate SNF in nodules (Ferguson et al., 2010). AON has been identified using several legume mutants with the super nodulation phenotype, such as the G. max mutant nts-1, M. truncatula mutant sunn, and L. japonicus mutant har1, were found to lack in a leucine-rich repeat receptor-like protein kinase for AON in shoots (Krusell et al., 2002; Nishimura et al., 2002; Schnabel et al., 2005). CLAVATA3/embryo-surrounding region (CLE) peptides are a group of small (12–13 amino acids) secreted peptides derived from the C-terminal region of preproproteins. In M. truncatula, it has been suggested that MtCLE12 or MtCLE13 plays a role for CLE signaling in controlling nodule numbers. These peptides are generated in roots and transported to leaves via the xylem to trigger the AON response (Mortier et al., 2010). LjCLE-RS1 and LjCLE-RS2 were also found to be involved in regulating nodule formation in L. japonicus (Magori and Kawaguch, 2010).
Legumes have evolved to adopt strategies to maintain the nodular Pi homeostasis (Sulieman and Tran, 2015). Multiple genes and proteins work as regulators (Rubio et al., 2001; Yao Z.F. et al., 2014; Sulieman and Tran, 2015). A MYB-CC-type transcription factor (PHR1), a small non-coding RNA (microRNA399), and proteins containing the SYG1/PHO81/XPR1 (SPX) domain all play important roles in Pi stability in nodules, and are considered to be vital regulators (Xue et al., 2017). DNA methylation may also be involved in the nodular Pi homeostasis regulation, but it still needs further research (Kim et al., 2015; Yong-Villalobos et al., 2015; Crampton et al., 2016).
In soybean, 35 GmPHR members have been identified. Among them, GmPHR25 is induced by Pi starvation and it in turn increased the transcripts of 11 out of 14 high-affinity Pi transporters as well as those of 5 other Pi starvation-responsive genes (Xue et al., 2017). This evidence indicated that GmPHR25 is a vital Pi homeostasis regulator in soybean (Xue et al., 2017). Besides small non-coding RNAs, long non-coding RNAs (lncRNAs) are also involved in Pi homeostasis. In the legume model plant, M. truncatula, three phosphate deficiency-induced LncRNAs (PDILs) have been characterized through their corresponding Tnt1 mutants. PDIL1 suppresses the degradation of MtPHO2, working as a positive Pi homeostasis regulator. However, PDIL2 and PDIL3 may directly inhibit Pi transporters, therefore working as negative regulators (Wang et al., 2017).
Proteins containing the SPX domain are considered to be vital regulators in the plant Pi signaling network (Yao Z.F. et al., 2014). In common bean, PvSPX1–PvSPX3 are all induced by Pi starvation, but PvSPX1 had higher sensitivity and faster response to Pi starvation than the other two PvSPXs. Ten Pi starvation-responsive genes are induced by overexpressing PvSPX1, even with an increase in Pi concentration at the same time. It was further shown that the overexpression of PvPHR1 resulted in a decrease in PvSPX1 transcripts. Therefore, PvSPX1 is a positive regulator in maintaining Pi homeostasis and is itself regulated by PvPHR1 (Yao Z.F. et al., 2014). But in soybean, GmSPX1 is a negative regulator in the Pi signaling network, and may be involved in phosphate starvation by inhibiting the expression of GmMYB48, a phosphate starvation-induced gene (Zhang et al., 2016). GmSPX3 is also induced by Pi starvation, and considered to be a positive regulator which induces seven Pi starvation-responsive genes in soybean hairy root (Yao Z. et al., 2014). Genes found to be associated with legume Pi homeostasis are listed in Table 2.
Symbiotic nitrogen fixation by rhizobia in legume root nodules is carried out through the nitrogenase enzyme complex. Nitrogenase catalyzes the following reaction:
This reaction describes the reduction of nitrogen to ammonia, and is associated with a high energetic cost (ATP). The enzymatic complex of nitrogenase consists of two enzymes: dinitrogenase reductase, a dimeric Fe-protein encoded by the nifH gene, and dinitrogenase, a tetrameric FeMo-protein encoded by the nifDK gene (Rascio and La Rocca, 2013; Rubio and Ludden, 2015). These enzymes are rapidly deactivated in the presence of atmospheric concentrations of oxygen (Dixon and Wheeler, 1986).
Nitrogenase activity is reduced at high oxygen concentrations. Nodules provide low oxygen environments for the N2-fixing bacteroids, as observed in soybean nodules where the oxygen concentration is 56 nM (Kuzma et al., 1993) and even lower concentrations have been registrated (Appleby, 1984). Therefore, oxygen tension is a key regulator of genes required for N2 fixation, nitrogenase synthesis (nif genes), and microoxic respiration (fix genes). The nitrogen fixation signaling pathway differs between rhizobium strains. In S. meliloti, the heme-containing FixL, a signal-transducing membrane protein, acts as the oxygen sensor for the system. In the absence of oxygen, FixL autophosphorylates and transfers the phosphate to FixJ (Gilles-Gonzalez et al., 1991; Lois et al., 1993). This transcriptional factor in turn regulates positively the expression of two regulatory genes, nifA and fixK. NifA is a transcriptional activator that controls the expressions of nitrogen fixation genes, fixABCX, nifN, nifB, and nifHDK (Gong et al., 2006). FixK upregulates the expression of fixNOQP and fixT (Batut et al., 1989; Foussard et al., 1997). FixT represses the expression of nifA (Foussard et al., 1997) and FixNOQP is a cytochrome terminal oxidase cbb3 with a high affinity for oxygen that allows ATP production required for the nitrogen fixation process (Preisig et al., 1993). However, in B. japonicum, the FixLJ system is not required to induce the expression of nifA (Fischer and Hennecke, 1987), but instead it is RegSR that mediates positive control to transcription of nifA (Lindemann et al., 2007).
FixLJ cascade system varies in some bacteria. FnrN, belonging to the CRP-Fnr family of global transcriptional factors found in bacteria, acts as the oxygen sensor in R. leguminosarum bv. viciae VF39 and Rhizobium etli CNPAF512 (Patschkowski et al., 1996; Moris et al., 2004). Both organisms synthesize FnrN and FixK. In R. leguminosarum, FnrN is the main oxygen sensor responsible for the induction of fixNOQP and is in turn induced by the FixL protein (Boesten and Priefer, 2004). In contrast, the expression of fnrN is autoregulated in R. etli CNPAF512 (Moris et al., 2004). Other cases of rhizobia oxygen-response cascade have been previously described in detail (Terpolilli et al., 2012).
Most rhizobia are obligate aerobes. The C4-dicarboxylic acids supplied by the host plant must be metabolized through the tricarboxylic acid (TCA) cycle in the bacteroid. Malate, the primary carbon source of bacteroids, is converted to pyruvate and CO2 via the NAD+-dependent malic enzyme (DME). Pyruvate is subsequently decarboxylated by pyruvate dehydrogenase to form acetyl-coenzyme A (acetyl-CoA) and enters the TCA cycle. DME is required to provide pyruvate to the TCA cycle in S. meliloti and Azorhizobium caulinodans (Zhang Y. et al., 2012). S. meliloti possesses two distinct malic enzymes, an NAD(P)+-dependent enzyme (DME) (EC 1.1.1.39) and a strictly NADP+-dependent enzyme (TME) (EC 1.1.1.40) (Driscoll and Finan, 1993, 1996; Voegele et al., 1999). In addition, acetyl-CoA can be produced alternatively by phosphoenolpyruvate carboxykinase (PCK) which catalyzes the decarboxylation of OAA to PEP. Symbiotic phenotypes of pck mutants vary depending on the host plant. pckA mutants of R. leguminosarum MNF3085 fix nitrogen at rates comparable to wild type, while pckA mutants of S. meliloti show a reduced N2 fixation capacity (McKay et al., 1985; Finan et al., 1991). In R. leguminosarum and S. fredii strain NGR234, DME or a pathway involving PCK and pyruvate kinase (PYK) can synthesize the precursors required for SNF (Zhang Y. et al., 2012). Enzymes participating in the TCA cycle have been identified in B. japonicum strain USDA110 (Sarma and Emerich, 2006), R. leguminosarum (McKay et al., 1985), S. meliloti (Djordjevic, 2004), and R. tropici (Romanov et al., 1994). However, TCA cycle involvement probably varies among rhizobia. Evidence shows that S. meliloti, R. tropici, and R. leguminosarum utilize the full oxidative TCA cycle to provide ATP, precursors of amino acid synthesis, as well as reducing equivalents for N2 fixation. Enzymes of the TCA cycle appear to be essential for nitrogen fixation in S. meliloti since mutations in succinate dehydrogenase (sdh), malate dehydrogenase (mdh), isocitrase dehydrogenase (icd), 2-oxoglutarate dehydrogenase, aconitase (acnA), and citrate synthase (gltA) abolish N2 fixation, despite that nodules were formed (Duncan and Fraenkel, 1979; McDermott and Kahn, 1992; Mortimer et al., 1999; Dymov et al., 2004; Koziol et al., 2009). In contrast, B. japonicum shows a higher metabolic plasticity. Mutations in fumarase (fumC), ICDH (idhA), alpha-ketoglutarate dehydrogenase (agdA), and acotinase (acnA) in B. japonicum USDA110 still show phenotypes capable of fixing nitrogen (Acuña et al., 1991; Thöny-Meyer and Künzler, 1996; Green and Emerich, 1997; Shah and Emerich, 2006). Transcriptome analyses of R. etli bacteroids have suggested that the TCA cycle is inactive in these bacteroids (Vercruysse et al., 2011), but additional experiments are required to explain how the bacteroids obtain the ATP necessary for nitrogen fixation in these cases. A comparative metabolic profiling study between free-living R. leguminosarum and pea bacteroids showed that the TCA cycle is not the only path to oxidize dicarboxylic acids derived from the host plant in the bacteroids (Terpolilli et al., 2016). Metabolic profiling and flux analysis revealed that pea bacteroids divert acetyl-CoA into TCA and the production of lipid or polyhydroxybutyrate (PHB). These findings suggest new pathways for electron allocation in nitrogen-fixing bacteroids where lipogenesis may be a requirement in legume nodules.
The legume–rhizobium symbiosis determines the accumulation of specific metabolites inside the microsymbiont. Excess carbon and reducing power provided by the host plant can be stored as polymers, glycogen, or lipids in bacteroids (Figure 1). PHB is accumulated in large cytoplasmic granules in the bacteroids that form determinate nodules (such as in common bean and soybean), but not in the indeterminate nodules of alfalfa, garden pea, and chickpea (Table 1; Tombolini and Nuti, 1989; Kim et al., 1996).
The PHB biosynthesis pathway consists of three steps. β-Ketothiolase (PhaA) catalyzes the first step in this pathway with the formation of acetoacetyl-CoA from acetyl-CoA. Acetoacetyl-CoA is then reduced to D-β-hydroxybutyryl-CoA by an NADH-dependent acetoacetyl-CoA (PhaB), and then PHB synthase (PhaC) catalyzes the final formation of PHB (Lodwig and Poole, 2003). In S. meliloti and R. etli, phaA and phaB form an operon, phaAB.
Fix+ symbiotic phenotypes have been observed in S. meliloti when mutations were introduced to disable the synthesis of PHB or alter the ability to utilize PHB cycle intermediates to support growth (Povolo et al., 1994; Aneja and Charles, 1999; Cai et al., 2000). Effective nodules were also observed when R. leguminosarum mutant strains defective in the phaC gene were used to inoculate bean (determinate nodules) or pea (indeterminate nodules) (Lodwig et al., 2005). However, a reduced nitrogen fixation capability was observed when PHB synthesis was abolished in A. caulinodans. In A. caulinodans, impaired PHB synthetase activity results in the loss of nitrogen fixation capacity both ex planta and in symbiosis with the tropical legume Sesbania rostrata (Mandon et al., 1998). These evidences suggest that PHB may play certain roles in nitrogen fixation of some legume–rhizobium systems.
Polyhydroxybutyrate synthesis is regulated in R. etli and S. meliloti by the PHA (polyhydroxyalkanoate) regulator (PhaR), previously called AniA (Povolo and Casella, 2000; Encarnación et al., 2002). PhaR homologs in other bacterial species bind to their own promoters and to the promoter of phaP (Maehara et al., 2001). PhaP, a phasin, binds to the surface of PHB granules and can control the size of the PHB granules (Kuchta et al., 2007; Mezzina et al., 2014). In Bradyrhizobium diazoefficiens, the regulation of PHB synthesis is mediated through the interaction between PhaR and FixK2, regulators associated with the synthesis of PHB and microoxic metabolism (Quelas et al., 2016).
Glycogen is co-produced with PHB in free-living rhizobia such as R. leguminosarum and S. meliloti under nutrient-limiting conditions (Povolo et al., 1994; Lodwig et al., 2005). The role of glycogen in SNF has been studied in some rhizobia. A glycogen synthase mutant (glgA) of R. tropici showed an enhanced symbiotic performance as measured by an increase in plant dry weight and nodule number, but the mechanisms behind this phenotype are uncertain (Marroquí et al., 2001). In garden pea bacteroids, a glgA mutant of R. leguminosarum did not alter nitrogen fixation rates, as indicated by the acetylene reduction assay for nitrogenase activity. Plants inoculated with a phaC/glgA double mutant resulted in similar phenotypes to wild-type-inoculated plants (Lodwig et al., 2005). In contrast, the glgA mutant of S. meliloti resulted in lower levels of N2 fixation in both M. truncatula and M. sativa (Wang et al., 2007). Thus, the SNF capability of S. meliloti is affected by both PHB and glycogen availability.
In Bradyrhizobium sp. ORS278, studies have shown that mutations in genes involved in the Calvin–Benson–Bassham (CBB) cycle result in deficiency in nitrogen fixation. The CBB cycle consumes 3 mol of ATP and 2 mol of reducing equivalents per mol of carbon dioxide. The symbiotic phenotypes suggest that Bradyrhizobium sp. ORS278 uses the Calvin cycle as a reductant store to regulate carbon flux. Specifically, phosphoglycerate kinase (CbbK) and ribulose 1,5-biphosphate carboxylase (RuBisCO) large chain (CbbL1) have been identified as being involved in the symbiotic process (Bonaldi et al., 2010; Gourion et al., 2011). Thus, the relevance of different carbon storage compounds in the symbiotic process varies depending on the rhizobial strain.
Mature bacteroids drive nitrogen fixation inside root nodules, releasing ammonia to the plant cell cytosol to be incorporated into nitrogenous compounds. Ammonia assimilation is severely reduced in bacteroids due to a coordinated response where growth-associated pathways are switched off and biosynthesis of amino acids is downregulated but still present (Li et al., 2013). However, bacteroids require the supply of some plant-derived amino acids to support bacteroid development (Prell et al., 2009; Mulley et al., 2011; reviewed in Dunn, 2014). The supply of amino acids is plant-type specific (Randhawa and Hassani, 2002). For example, alfalfa is not able to provide sufficient histidine to the histidine auxotrophs of S. meliloti in nodules (Malek and Kowalski, 1977) unlike the cowpea host plant where similar mutants of rhizobial strain IRC256 are able to fix nitrogen (McLaughlin et al., 1987). The transport of nutrients is regulated by the symbiosome membrane (SM). SM is a plant-derived membrane that controls the metabolite exchanges in the legume–rhizobia symbiosis via specific transport systems and encloses a single bacteroid in indeterminate nodules or several bacteroids in determinate nodules (Clarke et al., 2014). Different transport mechanisms have been identified across the legume SM such as carbon, nitrogen, and cation transport systems, but most of them are not characterized (Day et al., 2001; Clarke et al., 2015). The main metabolite transport systems are schematically represented in Figure 1.
C4-Dicarboxylates are the major metabolites that are transported across SM to generate ATP in the bacteroids. A dicarboxylate transporter for malate and succinate has been identified in the soybean SM (Udvardi et al., 1988). This transporter possesses a higher affinity for malate than for succinate. However, the gene encoding the transporter has not been identified in any legume. A dicarboxylate transporter has been characterized in the SM of a non-legume, Alnus glutinosa AgDCAT1 was shown to transport dicarboxylates (malate, succinate, fumarate, and OAA) when expressed in Escherichia coli (Jeong, 2004). AgDCAT1 belongs to the peptide transporter family (PTR) and proteins from this family are candidates for dicarboxylate transporters in legumes since experimental evidence shows that PTR-encoding genes are induced in legume nodules (Colebatch et al., 2004; Libault et al., 2010). Members of this family have been identified in the proteomic studies of the soybean SM, but their functions have not been characterized (Clarke et al., 2015). In rhizobia, a C4-dicarboxylate transport system (Dct) has been identified and characterized. Dct consists of a permease encoded by dctA and a two-component sensor-regulator system (DctBD) encoded by dctB and dctD. DctBD responds to the presence of C4-dicarboxylates and regulates dctA expression (reviewed in Yurgel and Kahn, 2004). dctA mutants of Rhizobium trifolii are unable to transport dicarboxylates and form ineffective nodules (Fix-) in Trifolium repens and Trifolium pratense. The resulting infected plant cells accumulated large amounts of starch (Ronson et al., 1981).
The ammonia produced by nitrogen fixation in bacteroids is probably protonated to ammonium after diffusing into the peribacteroid space (PBS) or symbiosome space (SS, the region between the SM and the bacteroid) (Figure 1). An H+-ATPase pumps H+ into the PBS, generating an acidic environment (Udvardi et al., 1991) and forming a membrane potential (Udvardi and Day, 1989). ATPase activity has been detected on the SM of soybean (Blumwald et al., 1985; Fedorova E. et al., 1999), lupin (Robertson et al., 1978; Domigan et al., 1988), and garden pea root nodules (Szafran and Haaker, 1995). Re-uptake of by bacteroids is prevented by the repression of the bacteroid ammonium carrier, Amt, during the symbiotic state (Howitt et al., 1986). At present, two pathways have been proposed for the transport of fixed nitrogen into the host plant: a monovalent cation channel for (Whitehead et al., 2008) and an aquaglyceroporin, nodulin 26 (Nod26), for NH3 transport. Nod26 is a transmembrane protein able to transport H2O, NH3, and other solutes (Weaver et al., 1994; Rivers et al., 1997; Hwang et al., 2010). It was first identified in the soybean SM (Fortin et al., 1987) and constitutes the major protein component of the SM (Rivers et al., 1997).
Host plants supply amino acids to the microsymbionts during the differentiation of rhizobia to bacteroids and for nitrogen fixation (Mulley et al., 2011). Recent analyses of the soybean proteome have revealed a putative amino acid transporter (GmAPC1), a homolog to members of the amino acid-polyamine-organocation (APC) family of transport proteins (Clarke et al., 2015). Previous studies have shown that the soybean SM is permeable to alanine and Asp (Whitehead et al., 1998). An Asp transporter has been identified in SM vesicles from root nodules of garden pea (Rudbeck et al., 1999).
In addition, rhizobia also encode transport systems that regulate the exchange of amino acids between the bacteroids and the host cell (Udvardi and Poole, 2013). For instance, while the bacteroids formed by R. leguminosarum bv. viciae in the indeterminate pea nodules are auxotrophs for branched amino acids, branch-chain amino acid transporters, Aap (AapJQMP) and Bra (BraDEFGC) are developed in the bacteroids (Prell et al., 2009). This symbiotic phenotype is known as symbiotic auxotrophy (Dunn, 2014) and occurs in both determinate and indeterminate nodules (de las Nieves Peltzer et al., 2008; Prell et al., 2010; di Cenzo et al., 2015).
Constraint-based modeling enables the determination of the metabolic capabilities of an organism (Feist et al., 2009; Oberhardt et al., 2009). These metabolic models have been used successfully to bridge the gap between current knowledge and metabolic phenotypes (Schellenberger et al., 2011). Metabolic reconstructions are based on physiological and biochemical information on primary and specialized metabolic pathways, genome information, and available -omics data. These models have allowed for the prediction of the metabolic behavior of microorganisms under different nutrient availabilities and simulated gene deletions or over-expressions (Contador et al., 2015; Razmilic et al., 2018). In an attempt to get new insights into the genetic interactions that orchestrate the complex metabolic interactions in nitrogen-fixing bacteria, metabolic models have been used to study the rhizobial metabolism. Genome-scale models have been combined with experimental data to identify essential genes and to simulate the metabolic behaviors of rhizobia under nitrogen fixation conditions in legume nodules. High-throughput experimental data aimed to identify key genes, proteins and metabolites have been used in these studies to validate the model predictions.
At present, few manually curated rhizobial reconstructions are available despite their important roles in sustainable agriculture. These reconstructions cover different metabolic tasks from only the symbiotic process that takes place in the rhizobium to the complete set of metabolic reactions of the bacteria cell. iOR363, a reconstruction for R. etli CFN42, was the first model available for any rhizobium (Resendis-Antonio et al., 2007). This model describes the SNF process inside the determinate nodules of P. vulgaris and includes the main reactions associated with the nitrogen fixation process and the metabolic pathways, such as PHB synthesis, that allow simulating the accumulation of this polymer during the symbiotic process (Resendis-Antonio et al., 2007). Also, a symbiotic reaction was defined to represent this specific legume–rhizobia symbiosis. The symbiotic reaction describes the exchange of nutrients between the host plant and the bacteroid to establish symbiotic relationships and to obtain or produce chemical compounds required by the process of nitrogen fixation. A symbiotic auxotrophy hypothesis and nitrogenase requirements were included in its definition. This model has been updated to include new data sets available of this system (Resendis-Antonio et al., 2011, 2012).
On the other hand, the metabolic reconstructions for S. meliloti have different scopes. iHZ565 was designed to represent the SNF of S. meliloti 1021 with host plants such as alfalfa (Zhao et al., 2012). The reconstruction process gave insight about the missing context-specific information and genome annotation errors. Additionally, a new symbiotic reaction was proposed to capture specific SNF mechanisms of S. meliloti in indeterminate nodules. The first representation of the whole metabolism of a rhizobium cell was built for S. meliloti (iGD1575) to enable the characterization of the metabolic capabilities of rhizobia in bulk soil, rhizosphere, and nodule (di Cenzo et al., 2016). iGD1575 encompasses 94% of the genes present in iHZ565. Recently, iGD1575 and gene essentiality experiments were used to build a model of the core metabolism of S. meliloti (di Cenzo et al., 2018). Pfau et al. (2018) have described the symbiotic relationship of M. truncatula with S. meliloti through constraint-based modeling (Pfau et al., 2018). Metabolic models were used to study the metabolic exchange between the host plant and the rhizobial symbiont. An in silico representation of the metabolic network of B. diazoefficiens USDA110 (iYY1101) has also been constructed (Yang et al., 2017). This reconstruction was used to build context-specific models to describe the metabolic differences observed between the free-living state of B. diazoefficiens USDA110 and symbiotic bacteroid. These models will provide a starting point for building metabolic reconstructions of closely related organisms such as other rhizobial strains and modeling frameworks to study these symbiotic interactions (Baumler et al., 2011; Monk et al., 2013; Ong et al., 2014). Table 3 summarizes the available metabolic models of rhizobia.
The regulatory mechanisms regarding the formation and maintenance of root nodules for SNF are multi-faceted and association-specific, but they all involve the tight regulation of the flow of metabolites between the host plants and the bacteroids, as well as the different levels of controls over each step in the metabolic pathways through the metabolic enzymes involved and the transcription factors that regulate their expressions. Each host plant–rhizobium association has its own unique requirements and control mechanisms despite the overall common features. It is therefore critical that we have as much understanding as possible of the detailed mechanisms that make up this symbiotic relationship.
H-ML designed the conceptual framework of this paper and coordinated the writing. AL and KF wrote the part on metabolism and regulation of host plant. CC wrote the part on metabolism and regulation of bacteroids and metabolic models. AL, CC, and H-ML revised and polished the article.
This work was supported by the Hong Kong Research Grants Council General Research Fund (14108014) and Area of Excellence Scheme (AoE/M-403/16), CUHK VC Discretionary Fund VCF2014004, and the Lo Kwee-Seong Biomedical Research Fund to H-ML.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Jee-Yan Chu copy-edited this manuscript.
Acuña, G., Ebeling, S., and Hennecke, H. (1991). Cloning, sequencing, and mutational analysis of the Bradyrhizobium japonicum fumC-like gene: evidence for the existence of two different fumarases. J. Gen. Microbiol. 137, 991–1000. doi: 10.1099/00221287-137-4-991
Ahemad, M., and Oves, M. (2011). “Biological importance of phosphorus and phosphate solubilizing microbes,” in Characterization of Chromium (VI) Reducing, Plant Growth Promoting Rhizobacteria as a Potential Bio-Fertilizer in Cr (VI) Contaminated Soil View Project, eds M. S. Khan, and A. Zaidi (Hauppauge, NY: Nova Science Publishers)
Akazawa, T., and Okamoto, K. (1980). “Biosynthesis and metabolism of sucrose,” in The Biochemistry of Plants: A Comprehensiue Treatise: Carbohydrates: Structure and Function, ed. J. Preis (Amsterdam: Elsevier), 199–220.
Aleman, L., Ortega, J. L., Martinez-Grimes, M., Seger, M., Holguin, F. O., Uribe, D. J., et al. (2010). Nodule-enhanced expression of a sucrose phosphate synthase gene member (MsSPSA) has a role in carbon and nitrogen metabolism in the nodules of alfalfa (Medicago sativa L.). Planta 231, 233–244. doi: 10.1007/s00425-009-1043-y
Andrews, M., and Andrews, M. E. (2017). Specificity in legume-rhizobia symbioses. Int. J. Mol. Sci. 18:E705. doi: 10.3390/ijms18040705
Aneja, P., and Charles, T. C. (1999). Poly-3-hydroxybutyrate degradation in Rhizobium (Sinorhizobium) meliloti: isolation and characterization of a gene encoding 3-hydroxybutyrate dehydrogenase. J. Bacteriol. 181, 849–857.
Aparicio-Fabre, R., Guillén, G., Loredo, M., Arellano, J., Valdés-López, O., Ramírez, M., et al. (2013). Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol. 13:26. doi: 10.1186/1471-2229-13-26
Appleby, C. A. (1984). Leghemoglobin and rhizobium respiration. Annu. Rev. Plant Physiol. 35, 443–478. doi: 10.1146/annurev.pp.35.060184.002303
Araújo, A. P., Plassard, C., and Drevon, J. J. (2008). Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant Soil 312, 129–138. doi: 10.1007/s11104-008-9595-3
Asthir, B., and Singh, R. (1997). Purification and characterization of neutral invertase from chickpea nodules. Indian J. Biochem. Biophys. 34, 529–534.
Atkins, C. A. (1991). “Ammonia assimilation and export of nitrogen from the legume nodule,” in Biochemistry of Nitrogen Fixation, eds M. J. Dilworth, and A. R. Glenn (Amsterdam: Elsevier Science), 293–319.
Atkins, C. A., Smith, P., and Storer, P. J. (1997). Reexamination of the intracellular localization of de novo purine synthesis in cowpea nodules. Plant Physiol. 113, 127–135. doi: 10.1104/pp.113.1.127
Baier, M. C., Barsch, A., Kuster, H., and Hohnjec, N. (2007). Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol. 145, 1600–1618. doi: 10.1104/pp.107.106955
Batut, J., Daveran-Mingot, M. L., David, M., Jacobs, J., Garnerone, A. M., and Kahn, D. (1989). fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 8, 1279–1286. doi: 10.1002/J.1460-2075.1989.TB03502.X
Baumler, D. J., Peplinski, R. G., Reed, J. L., Glasner, J. D., and Perna, N. T. (2011). The evolution of metabolic networks of E. coli. BMC Syst. Biol. 5:182. doi: 10.1186/1752-0509-5-182
Blumwald, E., Fortin, M. G., Rea, P. A., Verma, D. P., and Poole, R. J. (1985). Presence of host-plasma membrane type H-ATPase in the membrane envelope enclosing the bacteroids in soybean root nodules. Plant Physiol. 78, 665–672. doi: 10.1104/pp.78.4.665
Boesten, B., and Priefer, U. B. (2004). The C-terminal receiver domain of the Rhizobium leguminosarum bv. viciae FixL protein is required for free-living microaerobic induction of the fnrN promoter. Microbiology 150, 3703–3713. doi: 10.1099/mic.0.27323-0
Bonaldi, K., Gourion, B., Fardoux, J., Hannibal, L., Cartieaux, F., Boursot, M., et al. (2010). Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium Sp. Strain ORS278 reveals new genetic loci putatively important for Nod-independent symbiosis with Aeschynomene indica. Mol. Plant Microbe Interact. 23, 760–770. doi: 10.1094/MPMI-23-6-0760
Brenner, D. J., Krieg, N. R., and Staley, J. T. (eds) (2009). Bergey’s Manual of Systematic Bacteriology: The Proteobacteria, 2nd Edn, Vol. 2. Berlin: Springer. doi: 10.1007/978-0-387-68489-5
Cai, G. Q., Driscoll, B. T., and Charles, T. C. (2000). Requirement for the enzymes acetoacetyl coenzyme A synthetase and poly-3-hydroxybutyrate (PHB) synthase for growth of Sinorhizobium meliloti on PHB cycle intermediates. J. Bacteriol. 182, 2113–2118. doi: 10.1128/JB.182.8.2113-2118.2000
Carvalho, H., Sunkel, C., Salema, R., and Cullimore, J. V. (1997). Heteromeric assembly of the cytosolic glutamine synthetase polypeptides of Medicago truncatula: complementation of a glnA Escherichia coli mutant with a plant domain-swapped enzyme. Plant Mol. Biol. 35, 623–632. doi: 10.1023/A:1005884304303
Carvalho, H. G., Lescure, N., de Billy, F., Chabaud, M., Lima, L. M., Salema, R., et al. (2000). Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol. Biol. 42, 741–756. doi: 10.1023/A:1006304003770
Clarke, V. C., Loughlin, P. C., Day, D. A., and Smith, P. M. C. (2014). Transport processes of the legume symbiosome membrane. Front. Plant Sci. 5:699. doi: 10.3389/fpls.2014.00699
Clarke, V. C., Loughlin, P. C., Gavrin, A., Chen, C., Brear, E. M., Day, D. A., et al. (2015). Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteomics 14, 1301–1322. doi: 10.1074/mcp.M114.043166
Colebatch, G., Desbrosses, G., Ott, T., Krusell, L., Montanari, O., Kloska, S., et al. (2004). Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 39, 487–512. doi: 10.1111/j.1365-313X.2004.02150.x
Contador, C. A., Rodríguez, V., Andrews, B. A., and Asenjo, J. A. (2015). Genome-scale reconstruction of Salinispora tropica CNB-440 metabolism to study strain-specific adaptation. Antonie Van Leeuwenhoek 108, 1075–1090. doi: 10.1007/s10482-015-0561-9
Copeland, L., and Morell, M. (1985). Hexose kinases from the plant cytosolic fraction of soybean nodules. Plant Physiol. 79, 114–117. doi: 10.1104/pp.79.1.114
Crampton, M., Sripathi, V. R., Hossain, K., and Kalavbacharla, V. (2016). Analyses of methylomes derived from Meso-American common ean (Phaseolus vulgaris L.) using MeDIP-Seq and whole genome sodium bisulfite-sequencing. Front. Plant Sci. 7:447. doi: 10.3389/fpls.2016.00447
Czarnecki, O., Yang, J., Weston, D. J., Tuskan, G. A., and Chen, J. G. (2013). A dual role of strigolactones in phosphate acquisition and utilization in plants. Int. J. Mol. Sci. 14, 7681–7701. doi: 10.3390/ijms14047681
Dan, T. H., and Brix, H. (2009). Growth responses of the perennial legume Sesbania sesban to NH4 and NO3 nutrition and effects on root nodulation. Aquat. Bot. 91, 238–244. doi: 10.1016/j.aquabot.2009.07.004
Day, D. A. (1991). Carbon metabolism and compartmentation in nitrogen fixing legume nodules. Plant Physiol. Biochem. 29, 185–201.
Day, D. A., Kaiser, B. N., Thomson, R., Udvardi, M. K., Moreau, S., and Puppo, A. (2001). Nutrient transport across symbiotic membranes from legume nodules. Aust. J. Plant Physiol. 28, 667–674. doi: 10.1071/PP01028
de las Nieves Peltzer, M., Roques, N., Poinsot, V., Aguilar, O. M., Batut, J., and Capela, D. (2008). Auxotrophy accounts for nodulation defect of most Sinorhizobium meliloti mutants in the branched-chain amino acid biosynthesis pathway. Mol. Plant Microbe Interact. 21, 1232–1241. doi: 10.1094/MPMI-21-9-1232
di Cenzo, G. C., Checcucci, A., Bazzicalupo, M., Mengoni, A., Viti, C., Dziewit, L., et al. (2016). Metabolic modelling reveals the specialization of secondary replicons for niche adaptation in Sinorhizobium meliloti. Nat. Commun. 7:12219. doi: 10.1038/ncomms12219
di Cenzo, G. C., Zamani, M., Cowie, A., and Finan, T. M. (2015). Proline auxotrophy in Sinorhizobium meliloti results in a plant-specific symbiotic phenotype. Microbiology 161, 2341–2351. doi: 10.1099/mic.0.000182
diCenzo, G. C., Benedict, A. B., Fondi, M., Walker, G. C., Finan, T. M., Mengoni, A., et al. (2018). Robustness encoded across essential and accessory replicons of the ecologically versatile bacterium Sinorhizobium meliloti. PLoS Genet. 14:e1007357. doi: 10.1371/journal.pgen.1007357
Djordjevic, M. A. (2004). Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4, 1859–1872. doi: 10.1002/pmic.200300802
Domigan, N. M., Farnden, K. J. F., Robertson, J. G., and Monk, B. C. (1988). Characterization of the peribacteroid membrane ATPase of lupin root nodules. Arch. Biochem. Biophys. 264, 564–573. doi: 10.1016/0003-9861(88)90322-0
Driscoll, B. T., and Finan, T. M. (1993). NAD(+)-dependent malic enzyme of Rhizobium meliloti is required for symbiotic nitrogen fixation. Mol. Microbiol. 7, 865–873. doi: 10.1111/j.1365-2958.1993.tb01177.x
Driscoll, B. T., and Finan, T. M. (1996). NADP+-dependent malic enzyme of Rhizobium meliloti. J. Bacteriol. 178, 2224–2231. doi: 10.1128/jb.178.8.2224-2231.1996
Duncan, M. J., and Fraenkel, D. G. (1979). Alpha-Ketoglutarate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 137, 415–419.
Dunn, M. F. (2014). Key roles of microsymbiont amino acid metabolism in rhizobia-legume interactions. Crit. Rev. Microbiol. 41, 411–451. doi: 10.3109/1040841X.2013.856854
Dupont, L., Alloing, G., Pierre, O., El, S., Hopkins, J., Hrouart, D., et al. (2012). The Legume Root Nodule: From Symbiotic Nitrogen Fixation to Senescence. London: INTECH Open Access Publisher. doi: 10.5772/34438
Duran, V. A., and Todd, C. D. (2012). Four allantoinase genes are expressed in nitrogen-fixing soybean. Plant Physiol. Biochem. 54, 149–155. doi: 10.1016/j.plaphy.2012.03.002
Dymov, S. I., Meek, D. J. J., Steven, B., and Driscoll, B. T. (2004). Insertion of transposon Tn5tac1 in the Sinorhizobium meliloti malate dehydrogenase (mdh) gene results in conditional polar effects on downstream TCA cycle genes. Mol. Plant. Microbe. Interact. 17, 1318–1327. doi: 10.1094/MPMI.2004.17.12.1318
Encarnación, S., Del Carmen Vargas, M., Dunn, M. F., Dávalos, A., Mendoza, G., Mora, Y., et al. (2002). Ania regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J. Bacteriol. 184, 2287–2295. doi: 10.1128/JB.184.8.2287-2295.2002
Fan, C., Wang, X., Hu, R., Wang, Y., Xiao, C., Jiang, Y., et al. (2013). The pattern of phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol. 13:48. doi: 10.1186/1471-2229-13-48
Fedorova, E., Thomson, R., Whitehead, L. F., Maudoux, O., Udvardi, M. K., and Day, D. A. (1999). Localization of H+-ATPases in soybean root nodules. Planta 209, 25–32. doi: 10.1007/s004250050603
Fedorova, M., Tikhonovich, I. A., and Vance, C. P. (1999). Expression of C-assimilating enzymes in pea (Pisum sativum L.) root nodules. In situ localization in effective nodules. Plant Cell Environ. 22, 1249–1262. doi: 10.1046/j.1365-3040.1999.00490.x
Feist, A. M., Herrgård, M. J., Thiele, I., Reed, J. L., and Palsson, B. Ø (2009). Reconstruction of biochemical networks in microorganisms. Nat. Rev. Microbiol. 7, 129–143. doi: 10.1038/nrmicro1949
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M. H., Lin, Y. H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Finan, T. M., McWhinnie, E., Driscoll, B., and Watson, R. (1991). Complex symbiotic phenotypes result from gluconeogenic mutations in Rhizobium meliloti. Mol. Plant Microbe Interact. 4, 386–392. doi: 10.1094/MPMI-4-386
Fischer, H.-M., and Hennecke, H. (1987). Direct response of Bradyrrhizobium japonicum nifA-mediated nif gene regulation to cellular oxygen status. Mol. Gen. Genet. 209, 621–626. doi: 10.1007/BF00331174
Fischinger, S. A., Drevon, J. J., Claassen, N., and Schulze, J. (2006). Nitrogen from senescing lower leaves of common bean is re-translocated to nodules and might be involved in a N-feedback regulation of nitrogen fixation. J. Plant Physiol. 163, 987–995. doi: 10.1016/j.jplph.2006.03.017
Flemetakis, E., Efrose, R. C., Ott, T., Stedel, C., Aivalakis, G., Udvardi, M. K., et al. (2006). Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Mol. Biol. 62, 53–69. doi: 10.1007/s11103-006-9003-4
Fortin, M. G., Morrison, N. A., and Verma, D. P. S. (1987). Nodulin-26, a peribacteroid membrane nodulin is expressed independently of the development of the peribacteroid compartment. Nucleic Acids Res. 15, 813–824. doi: 10.1093/nar/15.2.813
Foussard, M., Garnerone, A.-M., Ni, F., Soupène, E., Boistard, P., and Batut, J. (1997). Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol. Microbiol. 25, 27–37. doi: 10.1046/j.1365-2958.1997.4501814.x
Gallardo, F., Gálvez, S., Gadal, P., and Cánovas, F. M. (1995). Changes in NADP+-linked isocitrate dehydrogenase during tomato fruit ripening. Planta 196, 148–154. doi: 10.1007/BF00193228
Gálvez, L., González, E. M., and Arrese-Igor, C. (2005). Evidence for carbon flux shortage and strong carbon/nitrogen interactions in pea nodules at early stages of water stress. J. Exp. Bot. 56, 2551–2561. doi: 10.1093/jxb/eri249
Gálvez, S., and Gadal, P. (1995). On the function of the NADP-dependent isocitrate dehydrogenase isoenzymes in living organisms. Plant Sci. 105, 1–14. doi: 10.1016/0168-9452(94)04041-E
Gilles-Gonzalez, M. A., Ditta, G. S., and Helinski, D. R. (1991). A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350, 170–172. doi: 10.1038/350170a0
Glenn, A., and Dilworth, M. (1981). Oxidation of substrates by isolated bacteroids and free-living cells of Rhizobium leguminosarum 384 1. J. Gen. Microbiol. 126, 243–247.
Gong, Z. Y., He, Z. S., Zhu, J. B., Yu, G. Q., and Zou, H. S. (2006). Sinorhizobium meliloti nifA mutant induces different gene expression profile from wild type in alfalfa nodules. Cell Res. 16, 818–829. doi: 10.1038/sj.cr.7310096
Gordon, A. J., Minchin, F. R., James, C. L., and Komina, O. (1999). Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol. 120, 867–878. doi: 10.1104/pp.120.3.867
Gordon, A. J., Skot, L., Webb, K. J., Minchin, F. R., James, C. L., Wang, T. L., et al. (1998). “Down regulation of nodule sucrose synthase by mutation and antisense,” in Proceedings of the 11th International Congress on Biological Nitrogen Fixation for the 21st Century (Paris: Institute Pasteur), 473–473. doi: 10.1007/978-94-011-5159-7_288
Gourion, B., Delmotte, N., Bonaldi, K., Nouwen, N., Vorholt, J. A., and Giraud, E. (2011). Bacterial RuBisCO is required for efficient Bradyrhizobium/Aeschynomene symbiosis. PLoS One 6:e21900. doi: 10.1371/journal.pone.0021900
Green, L. S., and Emerich, D. W. (1997). The formation of nitrogen fixing bacteroids is delayed but not abolished in soybean infected by an a-ketoglutarate dehydrogenase deficient mutant of Bradyrhizobium japonicum. Plant Physiol. 114, 1359–1368. doi: 10.1104/pp.114.4.1359
Gu, M., Chen, A., Sun, S., and Xu, G. (2016). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Gunawardena, S. F. B. N., Danso, S. K. A., and Zapata, F. (1992). Phosphorus requirements and nitrogen accumulation by three mungbean (Vigna radiata (L) Welzek) cultivars. Plant Soil 147, 267–274. doi: 10.1007/BF00029078
Hernandez, G., Valdes-Lopez, O., Ramirez, M., Goffard, N., Weiller, G., Aparicio-Fabre, R., et al. (2009). Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151, 1221–1238. doi: 10.1104/pp.109.143842
Herridge, D. F., Peoples, M. B., and Boddey, R. M. (2008). Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311, 1–18. doi: 10.1007/s11104-008-9668-3
Hirsch, A. M., Bang, M., and Ausubel, F. M. (1983). Ultrastructural analysis of ineffective alfalfa nodules formed by nif:: Tn5 mutants of Rhizobium meliloti. J. Bacteriol. 155, 367–380.
Horst, I., Welham, T., Kelly, S., Kaneko, T., Sato, S., Tabata, S., et al. (2007). TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 144, 806–820. doi: 10.1104/pp.107.097063
Howitt, S. M., Udvardi, M. K., Day, D. A., and Gresshoff, P. M. (1986). Ammonia transport in free-living and symbiotic Rhizobium sp. ANU289. J. Gen. Microbiol. 132, 257–261. doi: 10.1099/00221287-132-2-257
Hwang, J. H., Ellingson, S. R., and Roberts, D. M. (2010). Ammonia permeability of the soybean nodulin 26 channel. FEBS Lett. 584, 4339–4343. doi: 10.1016/j.febslet.2010.09.033
Jeong, J. (2004). A nodule-specific dicarboxylate transporter from Alder is a member of the peptide transporter family. Plant Physiol. 134, 969–978. doi: 10.1104/pp.103.032102
Jones, K. M., Kobayashi, H., Davies, B. W., Taga, M. E., and Walker, G. C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633. doi: 10.1038/nrmicro1705
Kim, K., Do El Baidouri, M., Abernathy, B., Iwata-Otsubo, A., Chavarro, C., Gonzales, M., et al. (2015). A comparative epigenomic analysis of polyploidy-derived genes in soybean and common bean. Plant Physiol. 168, 1433–1447. doi: 10.1104/pp.15.00408
Kim, S. A., Copeland, L., and Copeland, L. E. S. (1996). Enzymes of poly-beta-hydroxybutyrate metabolism in soybean and chickpea bacteroids. Appl. Environ. Microbiol. 62, 4186–4190.
Kneen, B. E., and LaRue, T. A. (1984). Peas (Pisum sativum L.) with strain specificity for Rhizobium leguminosarum. Heredity 52, 383–389. doi: 10.1038/hdy.1984.46
Koziol, U., Hannibal, L., Rodríguez, M. C., Fabiano, E., Kahn, M. L., and Noya, F. (2009). Deletion of citrate synthase restores growth of Sinorhizobium meliloti 1021 aconitase mutants. J. Bacteriol. 191, 7581–7586. doi: 10.1128/JB.00777-09
Krusell, L., Madsen, L. H., Sato, S., Aubert, G., Genua, A., Szczyglowski, K., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. doi: 10.1038/nature01207
Kuchta, K., Chi, L., Fuchs, H., Pötter, M., and Steinbüchel, A. (2007). Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Raistonia eutropha H16. Biomacromolecules 8, 657–662. doi: 10.1021/bm060912e
Kuzma, M. M., Hunt, S., and Layzell, D. B. (1993). Role of oxygen in the limitation and inhibition of nitrogenase activity and respiration rate in individual soybean nodules. Plant Physiol. 101, 161–169. doi: 10.1104/pp.101.1.161
Lazali, M., Zaman-Allah, M., Amenc, L., Ounane, G., Abadie, J., and Drevon, J.-J. (2013). A phytase gene is overexpressed in root nodules cortex of Phaseolus vulgaris-rhizobia symbiosis under phosphorus deficiency. Planta 238, 317–324. doi: 10.1007/s00425-013-1893-1
Le Roux, M., Phiri, E., Khan, W., Şakiroǧlu, M., Valentine, A., and Khan, S. (2014). Expression of novel cytosolic malate dehydrogenases (cMDH) in Lupinus angustifolius nodules during phosphorus starvation. J. Plant Physiol. 171, 1609–1618. doi: 10.1016/j.jplph.2014.07.020
Le Roux, M. R., Khan, S., and Valentine, A. J. (2008). Organic acid accumulation may inhibit N2 fixation in phosphorus-stressed lupin nodules. New Phytol. 177, 956–964. doi: 10.1111/j.1469-8137.2007.02305.x
Le Roux, M. R., Khan, S., and Valentine, A. J. (2009). Nitrogen and carbon costs of soybean and lupin root systems during phosphate starvation. Symbiosis 48, 102–109. doi: 10.1007/BF03179989
Lea, P. J., and Miflin, B. J. (2003). Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 41, 555–564. doi: 10.1016/S0981-9428(03)00060-3
Li, Y., Tian, C. F., Chen, W. F., Wang, L., Sui, X. H., and Chen, W. X. (2013). High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS One 8:e70531. doi: 10.1371/journal.pone.0070531
Li, Z., Xu, H., Li, Y., Wan, X., Ma, Z., Cao, J., et al. (2018). Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.). Plant Mol. Biol. 96, 473–492. doi: 10.1007/s11103-018-0711-3
Libault, M., Farmer, A., Brechenmacher, L., Drnevich, J., Langley, R. J., Bilgin, D. D., et al. (2010). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552. doi: 10.1104/pp.109.148379
Lindemann, A., Moser, A., Pessi, G., Hauser, F., Friberg, M., Hennecke, H., et al. (2007). New target genes controlled by the Bradyrhizobium japonicum two-component regulatory system RegSR. J. Bacteriol. 189, 8928–8943. doi: 10.1128/JB.01088-07
Liu, J., Versaw, W. K., Pumplin, N., Gomez, S. K., Blaylock, L. A., and Harrison, M. J. (2008). Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 283, 24673–24681. doi: 10.1074/jbc.M802695200
Lodwig, E., and Poole, P. (2003). Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 22, 37–78. doi: 10.1080/713610850
Lodwig, E. M., Leonard, M., Marroqui, S., Wheeler, T. R., Findlay, K., Downie, J. A., et al. (2005). Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol. Plant Microbe Interact. 18, 67–74. doi: 10.1094/MPMI-18-0067
Lois, A. F., Weinstein, M., Ditta, G. S., and Helinski, D. R. (1993). Autophosphorylation and phosphatase activities of the oxygen-sensing protein FixL of Rhizobium meliloti are coordinately regulated by oxygen. J. Biol. Chem. 268, 4370–4375.
López, M., Herrera-Cervera, J. A., Iribarne, C., Tejera, N. A., and Lluch, C. (2008). Growth and nitrogen fixation in Lotus japonicus and Medicago truncatula under NaCl stress: nodule carbon metabolism. J. Plant Physiol. 165, 641–650. doi: 10.1016/j.jplph.2007.05.009
Maehara, A., Doi, Y., Nishiyama, T., Takagi, Y., Ueda, S., Nakano, H., et al. (2001). PhaR, a protein of unknown function conserved among short-chain-length polyhydroxyalkanoic acids producing bacteria, is a DNA-binding protein and represses Paracoccus denitrificans phaP expression in vitro. FEMS Microbiol. Lett. 200, 9–15. doi: 10.1016/S0378-1097(01)00182-3
Magori, S., and Kawaguch, M. (2010). Analysis of two potential long-distance signaling molecules, LjCLE-RS1/2 and jasmonic acid, in a hypernodulating mutant too much love. Plant Signal. Behav. 5, 403–405. doi: 10.4161/psb.5.4.10801
Malek, W., and Kowalski, M. (1977). Auxotrophic mutations related to symbiotic properties if Rhizobium meliloti strain L5-30. Acta Microbiol. Pol. 26, 345–350.
Mandon, K., Michel-reydellet, N., Encarnacio, S., Kaminski, P. A., Leija, A., Cevallos, M. A., et al. (1998). Poly-β-hydroxybutyrate turnover in Azorhizobium caulinodans is required for growth and affects nifA expression. J. Bacteriol. 180, 5070–5076.
Marroquí, S., Zorreguieta, A., Santamaría, C., Temprano, F., Soberón, M., Megías, M., et al. (2001). Enhanced symbiotic performance by Rhizobium tropici glycogen synthase mutants. J. Bacteriol. 183, 854–864. doi: 10.1128/JB.183.3.854-864.2001
McClure, P. R., and Israel, D. W. (1979). Transport of nitrogen in the xylem of soybean plants. Plant Physiol. 64, 411–416. doi: 10.1104/pp.64.3.411
McDermott, T. R., and Kahn, M. L. (1992). Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174, 4790–4797. doi: 10.1128/jb.174.14.4790-4797.1992
McKay, I., Glenn, A., and Dilworth, M. (1985). Gluconeogenesis in Rhizobium leguminosarum MNF3841. J. Gen. Microbiol. 131, 2067–2073. doi: 10.1099/00221287-131-8-2067
McLaughlin, W., Singh, I., and Ahmad, M. (1987). Characterization of Tn5-induced symbiotically defective mutants of cowpea rhizobia. Lett. FEMS Microbiol. 41, 331–336. doi: 10.1111/j.1574-6968.1987.tb02222.x
Mezzina, M. P., Wetzler, D. E., Catone, M. V., Bucci, H., Di Paola, M., and Pettinari, M. J. (2014). A phasin with many faces: structural insights on PhaP from Azotobacter sp. FA8. PLoS One 9:e103012. doi: 10.1371/journal.pone.0103012
Miller, R. W., Mcrae, D. G., Ai-Jobore, A., and Berndt, W. B. (1988). Respiration supported nitrogenase activity of isolated Rhizobium meliloti bacteroids. J. Cell. Biochem. 38, 35–49. doi: 10.1002/jcb.240380105
Miller, S. S., Driscoll, B. T., Gregerson, R. G., Gantt, J. S., and Vance, C. P. (1998). Alfalfa malate dehydrogenase (MDH): molecular cloning and characterization of five different forms reveals a unique nodule-enhanced MDH. Plant J. 15, 173–184. doi: 10.1046/j.1365-313X.1998.00192.x
Minchin, F. R., and Witty, J. F. (2005). “Respiratory/carbon costs of symbiotic nitrogen fixation in legumes,” in Plant Respiration, eds H. Lambers, and M. Ribas-Carbo (Dordrecht: Springer-Verlag), 195–205. doi: 10.1007/1-4020-3589-6_11
Monk, J. M., Charusanti, P., Aziz, R. K., Lerman, J. A., Premyodhin, N., Orth, J. D., et al. (2013). Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc. Natl. Acad. Sci. U.S.A. 110, 20338–20343. doi: 10.1073/pnas.1307797110
Moris, M., Dombrecht, B., Xi, C., Vanderleyden, J., and Michiels, J. (2004). Regulatory role of Rhizobium etli CNPAF512 fnrN during symbiosis. Appl. Environ. Microbiol. 70, 1287–1296. doi: 10.1128/AEM.70.3.1287-1296.2004
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D’haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237. doi: 10.1104/pp.110.153718
Mortimer, M. W., McDermott, T. R., York, G. M., Walker, G. C., and Kahn, M. L. (1999). Citrate synthase mutants of Sinorhizobium meliloti are ineffective and have altered cell surface polysaccharides. J. Bacteriol. 181, 7608–7613.
Mulley, G., White, J. P., Karunakaran, R., Prell, J., Bourdes, A., Bunnewell, S., et al. (2011). Mutation of GOGAT prevents pea bacteroid formation and N2 fixation by globally downregulating transport of organic nitrogen sources. Mol. Microbiol. 80, 149–167. doi: 10.1111/j.1365-2958.2011.07565.x
Nakagawa, T., Izumi, T., Banba, M., Umehara, Y., Kouchi, H., Izui, K., et al. (2003). Characterization and expression analysis of genes encoding phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase of Lotus japonicus, a model legume. Mol. Plant. Microbe. Interact. 16, 281–288. doi: 10.1094/MPMI.2003.16.4.281
Nasr Esfahani, M., Kusano, M., Nguyen, K. H., Watanabe, Y., Ha, C., Van Saito, K., et al. (2016). Adaptation of the symbiotic Mesorhizobium-chickpea relationship to phosphate deficiency relies on reprogramming of whole-plant metabolism. Proc. Natl. Acad. Sci. U.S.A. 113, E4610–E4619. doi: 10.1073/pnas.1609440113
Naya, L., Ladrera, R., Ramos, J., Gonzalez, E. M., Arrese-Igor, C., Minchin, F. R., et al. (2007). The response of carbon metabolism and antioxidant defenses of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiol. 144, 1104–1114. doi: 10.1104/pp.107.099648
Nimmo, H. G. (2000). The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 5, 75–80. doi: 10.1016/S1360-1385(99)01543-5
Nishimura, R., Hayashit, M., Wu, G. J., Kouchi, H., Imaizumi-Anrakull, H., Murakami, Y., et al. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420, 426–429. doi: 10.1038/nature01231
Oberhardt, M. A., Palsson, B. Ø, and Papin, J. A. (2009). Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol 5:320. doi: 10.1038/msb.2009.77
Ogden, A. J., Gargouri, M., Park, J. J., Gang, D. R., and Kahn, M. L. (2017). Integrated analysis of zone-specific protein and metabolite profiles within nitrogen-fixing Medicago truncatula-Sinorhizobium medicae nodules. PLoS One 12:e0180894. doi: 10.1371/journal.pone.0180894
Oldroyd, G. E. D. (2005). Peace talks and trade deals. Keys to long-term harmony in legume-microbe symbioses. Plant Physiol. 137, 1205–1210. doi: 10.1104/pp.104.057661
Oldroyd, G. E. D., and Geurts, R. (2001). Medicago truncatula, going where no plant has gone before. Trends Plant Sci. 6, 552–554. doi: 10.1016/S1360-1385(01)02153-7
Ong, W., Vu, T. T., Lovendahl, K. N., Llull, J. M., Serres, M. H., Romine, M. F., et al. (2014). Comparisons of Shewanella strains based on genome annotations, modeling, and experiments. BMC Syst. Biol. 8:31. doi: 10.1186/1752-0509-8-31
Patschkowski, T., Schlüter, A., and Priefer, U. B. (1996). Rhizobium leguminosarum bv. viciae contains a second fnr/fixK-like gene and an unusual fixL homologue. Microbiology 21, 267–280.
Pfau, T., Christian, N., Masakapalli, S. K., Sweetlove, L. J., Poolman, M. G., and Ebenhöh, O. (2018). The intertwined metabolism during symbiotic nitrogen fixation elucidated by metabolic modelling. Sci. Rep. 8:12504. doi: 10.1038/s41598-018-30884-x
Poole, P., Ramachandran, V., and Terpolilli, J. (2018). Rhizobia: from saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303. doi: 10.1038/nrmicro.2017.171
Povolo, S., and Casella, S. (2000). A critical role for aniA in energy-carbon flux and symbiotic nitrogen fixation in Sinorhizobium meliloti. Arch. Microbiol. 174, 42–49. doi: 10.1007/s002030000171
Povolo, S., Tombolini, R., Morea, A., Anderson, A. J., Casella, S., and Nuti, M. (1994). Isolation and characterization of mutants of Rhizobium meliloti unable to synthesize poly-beta-hydroxybutyrate. Can. J. Microbiol. 40, 823–829. doi: 10.1139/m94-131
Preisig, O., Anthamatten, D., and Hennecke, H. (1993). Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 90, 3309–3313. doi: 10.1073/pnas.90.8.3309
Prell, J., Bourdès, A., Kumar, S., Lodwig, E., Hosie, A., Kinghorn, S., et al. (2010). Role of symbiotic auxotrophy in the Rhizobium-legume symbioses. PLoS One 5:e13933. doi: 10.1371/journal.pone.0013933
Prell, J., White, J. P., Bourdes, A., Bunnewell, S., Bongaerts, R. J., and Poole, P. S. (2009). Legumes regulate bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. U.S.A. 106, 12477–12482. doi: 10.1073/pnas.0903653106
Qin, L., Zhao, J., Tian, J., Chen, L., Sun, Z., Guo, Y., et al. (2012). The High-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 159, 1634–1643. doi: 10.1104/pp.112.199786
Qin, S., Tang, Y., Chen, Y., Wu, P., Li, M., Wu, G., et al. (2016). Overexpression of the starch phosphorylase-like gene (PHO3) in Lotus japonicus has a profound effect on the growth of plants and reduction of transitory starch accumulation. Front. Plant Sci. 7:1315. doi: 10.3389/fpls.2016.01315
Quelas, J. I., Mesa, S., Mongiardini, E. J., Jendrossek, D., and Lodeiro, A. R. (2016). Regulation of polyhydroxybutyrate synthesis in the soil bacterium Bradyrhizobium diazoefficiens. Appl. Environ. Microbiol. 82, 4299–4308. doi: 10.1128/AEM.00757-16
Ramos, M. L. G., Gordon, A. J., Minchin, F. R., Sprent, J. I., and Parsons, R. (1999). Effect of water stress on nodule physiology and biochemistry of a drought tolerant cultivar of common bean (Phaseolus vulgaris L.). Ann. Bot. 83, 57–63. doi: 10.1006/anbo.1998.0792
Randhawa, G. S., and Hassani, R. (2002). Role of rhizobial biosynthetic pathways of amino acids, nucleotide bases and vitamins in symbiosis. Indian J. Exp. Biol. 40, 755–764.
Rascio, N., and La Rocca, N. (2013). Biological Nitrogen Fixation: Reference Module in Earth Systems and Environmental Sciences. Amsterdam: Elsevier Inc. doi: 10.1016/B978-0-12-409548-9.00685-0
Razmilic, V., Castro, J. F., Andrews, B., and Asenjo, J. A. (2018). Analysis of metabolic networks of Streptomyces leeuwenhoekii C34 by means of a genome scale model: prediction of modifications that enhance the production of specialized metabolites. Biotechnol. Bioeng. 115, 1815–1828. doi: 10.1002/bit.26598
Resendis-Antonio, O., Hernández, M., Mora, Y., and Encarnación, S. (2012). Functional modules, structural topology, and optimal activity in metabolic networks. PLoS Comput. Biol. 8:e1002720. doi: 10.1371/journal.pcbi.1002720
Resendis-Antonio, O., Hernández, M., Salazar, E., Contreras, S., Batallar, G. M., Mora, Y., et al. (2011). Systems biology of bacterial nitrogen fixation: high-throughput technology and its integrative description with constraint-based modeling. BMC Syst. Biol. 5:120. doi: 10.1186/1752-0509-5-120
Resendis-Antonio, O., Reed, J. L., Encarnación, S., Collado-Vides, J., and Palsson, B. (2007). Metabolic reconstruction and modeling of nitrogen fixation in Rhizobium etli. PLoS Comput. Biol. 3:e192. doi: 10.1371/journal.pcbi.0030192
Rivers, R. L., Dean, R. M., Chandy, G., Hall, J. E., Roberts, D. M., and Zeidel, M. L. (1997). Functional-analysis of nodulin-26, an aquaporin in soybean root-nodule symbiosomes. J. Biol. Chem. 272, 16256–16261. doi: 10.1074/jbc.272.26.16256
Robertson, J. G., Warburton, M. P., Lyttleton, P., Fordyce, A. M., and Bullivant, S. (1978). Membranes in lupin root nodules II. Preparation and properties of peribacteroid membranes and bacteroid envelope inner membranes from developing lupin nodules. J. Cell. Sci. 30, 151–174.
Romanov, V. I., Hernandezlucas, I., Martinezromero, E., Hernandez-Lucas, I., and Martinez-Romero, E. (1994). Carbon metabolism enzymes of Rhizobium tropici cultures and bacteroids. Appl. Environ. Microbiol. 60, 2339–2342.
Ronson, C. W., Lyttleton, P., and Robertson, J. G. (1981). C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc. Natl. Acad. Sci. U.S.A. 78, 4284–4288. doi: 10.1073/pnas.78.7.4284
Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F. (2014). The Prokaryotes, 4th Edn. Berlin: Springer-Verlag. doi: 10.1007/978-3-642-30138-4
Rosendahl, L., Vance, C. P., and Pedersen, W. B. (1990). Products of dark CO2 fixation in pea root nodules support bacteroid metabolism. Plant Physiol. 93, 12–19. doi: 10.1104/pp.93.1.12
Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., et al. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. doi: 10.1111/tpj.12442
Rubio, L. M., and Ludden, P. W. (2015). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111. doi: 10.1002/9781119053095.ch7
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Rudbeck, A., Mouritzen, P., and Rosendahl, L. (1999). Characterization of aspartate transport across the symbiosome membrane in pea root nodules. J. Plant Physiol. 155, 576–583. doi: 10.1016/S0176-1617(99)80057-1
Sa, T. M., and Israel, D. W. (1991). Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 97, 928–935. doi: 10.1104/PP.97.3.928
Saad, S., and Lam-Son, P. T. (2017). Legume Nitrogen Fixation in Soils with Low Phosphorus Availability. Berlin: Springer International Publishing. doi: 10.1007/978-3-319-55729-8
Sarma, A. D., and Emerich, D. W. (2006). A comparative proteomic evaluation of culture grown vs nodule isolated Bradyrhizobium japonicum. Proteomics 6, 3008–3028. doi: 10.1002/pmic.200500783
Schellenberger, J., Que, R., Fleming, R. M. T., Thiele, I., Orth, J. D., Feist, A. M., et al. (2011). Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307. doi: 10.1038/nprot.2011.308
Schnabel, E., Journet, E. P., De Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58, 809–822. doi: 10.1007/s11103-005-8102-y
Schoenbeck, M. A., Temple, S. J., Trepp, G. B., Blumenthal, J. M., Samac, D. A., Gantt, J. S., et al. (2000). Decreased NADH glutamate synthase activity in nodules and flowers of alfalfa (Medicago sativa L.) transformed with an antisense glutamate synthase transgene. J. Exp. Bot. 51, 29–39. doi: 10.1093/jxb/51.342.29
Schuller, K. A., and Werner, D. (1993). Phosphorylation of soybean (Glycine max L.) nodule phosphoenolpyruvate carboxylase in vitro decreases sensitivity to inhibition by L-malate. Plant Physiol. 101, 1267–1273. doi: 10.1104/pp.101.4.1267
Schultze, M., and Kondorosi, A. (1998). Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32, 33–57. doi: 10.1146/annurev.genet.32.1.33
Seabra, A. R., Pereira, P. A., Becker, J. D., and Carvalho, H. G. (2012). Inhibition of glutamine synthetase by phosphinothricin leads to transcriptome reprograming in root nodules of Medicago truncatula. Mol. Plant Microbe Interact. 25, 976–992. doi: 10.1094/MPMI-12-11-0322
Shah, R., and Emerich, D. W. (2006). Isocitrate dehydrogenase of Bradyrhizobium japonicum is not required for symbiotic nitrogen fixation with soybean. J. Bacteriol. 188, 7600–7608. doi: 10.1128/JB.00671-06
Smith, F. W., and Jackson, W. A. (1987). Nitrogen enhancement of phosphate transport in roots of Zea mays L.: i. Effects of ammonium and nitrate pretreatment. Plant Physiol. 84, 1314–1318. doi: 10.1104/pp.84.4.1314
Spaink, H. P. (2000). Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 54, 257–288. doi: 10.1146/annurev.micro.54.1.257
Sprent, J. I. (2009). Legume Nodulation: A Global Perspective. Hoboken, NJ: John Wiley & Sons. doi: 10.1002/9781444316384
Sprent, J. I., Ardley, J., and James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56. doi: 10.1111/nph.14474
Stovall, I., and Cole, M. (1978). Organic acid metabolism by isolated Rhizobium japonicum bacteroids. Plant Physiol. 61, 787–790. doi: 10.1104/pp.61.5.787
Sulieman, S., and Schulze, J. (2010). The efficiency of nitrogen fixation of the model legume Medicago truncatula (Jemalong A17) is low compared to Medicago sativa. J. Plant Physiol. 167, 683–692. doi: 10.1016/j.jplph.2009.12.016
Sulieman, S., and Tran, L. S. P. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Szafran, M. M., and Haaker, H. (1995). Properties of the peribacteroid membrane ATPase of pea root modules and its effect on the nitrogenase activity. Plant Physiol. 108, 1227–1232. doi: 10.1104/PP.108.3.1227
Tajima, S. (2004). Ureide biosynthesis in legume nodules. Front. Biosci. 9, 1374–1381. doi: 10.2741/1345
Takanashi, K., Takahashi, H., Sakurai, N., Sugiyama, A., Suzuki, H., Shibata, D., et al. (2012). Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant. Microbe. Interact. 25, 869–876. doi: 10.1094/MPMI-01-12-0011-R
Temple, S. J., Vance, C. P., and Gantt, J. S. (1998). Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 3, 51–56. doi: 10.1016/S1360-1385(97)01159-X
Terpolilli, J. J., Hood, G. A., and Poole, P. S. (2012). What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 60, 325–389. doi: 10.1016/B978-0-12-398264-3.00005-X
Terpolilli, J. J., Masakapalli, S. K., Karunakaran, R., Webb, I. U. C., Green, R., Watmough, N. J., et al. (2016). Lipogenesis and redox balance in nitrogen-fixing pea bacteroids. J. Bacteriol. 198, 2864–2875. doi: 10.1128/JB.00451-16
Tesfaye, M., Samac, D. A., and Vance, C. P. (2006). Insights into symbiotic nitrogen fixation in Medicago truncatula. Mol. Plant microbe Interact. 19, 330–341. doi: 10.1094/MPMI-19-0330
The Legume Phylogeny Working Group (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. doi: 10.12705/622.8
Thöny-Meyer, L., and Künzler, P. (1996). The Bradyrhizobium japonicum aconitase gene (acnA) is important for free-living growth but not for an effective root nodule symbiosis. J. Bacteriol. 178, 6166–6172. doi: 10.1128/jb.178.21.6166-6172.1996
Thuynsma, R., Valentine, A., and Kleinert, A. (2014). Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J. Plant Physiol. 171, 285–291. doi: 10.1016/J.JPLPH.2013.09.001
Timmers, A. C. J., Auriac, M.-C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628.
Timmers, A. C. J., Soupene, E., Auriac, M.-C., de Billy, F., Vasse, J., Boistard, P., et al. (2000). Saprophytic intracellular rhizobia in alfalfa nodules. Mol. Plant Microbe Interact. 13, 1204–1213. doi: 10.1094/MPMI.2000.13.11.1204
Todd, C. D., Tipton, P. A., Blevins, D. G., Piedras, P., Pineda, M., and Polacco, J. C. (2006). Update on ureide degradation in legumes. J. Exp. Bot. 57, 5–12. doi: 10.1093/jxb/erj013
Tomaszewska, B., Jarmuszkiewicz, W., and Schrmm, R. W. (1991). The role of malate in the metabolism of mitochondria and symbiosomes from lupin root nodules. Plant Physiol. Biochem. 29, 489–496.
Tombolini, R., and Nuti, M. P. (1989). Poly (β-hydroyxalkanoate) biosynthesis and accumulation by different Rhizobium species. FEMS Microbiol. Lett. 60, 299–304.
Udvardi, M., Lister, D. L., and Day, D. A. (1991). ATPase activity and anion transport across the peribacteroid membrane of isolated soybean symbiosomes. Microbiology 156, 362–366.
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Udvardi, M. K., and Day, D. A. (1989). Electrogenic ATPase activity on the peribacteroid membrane of soybean (Glycine max L.) root nodules. Plant Physiol. 90, 982–987. doi: 10.1104/pp.90.3.982
Udvardi, M. K., and Day, D. A. (1997). Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 493–523. doi: 10.1146/annurev.arplant.48.1.493
Udvardi, M. K., Price, G. D., Gresshoff, P. M., and Day, D. A. (1988). A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett. 231, 36–40. doi: 10.1016/0014-5793(88)80697-5
Valdés-López, O., Arenas-huertero, C., Ramírez, M., Girard, L., Sánchez, F., Vance, C. P., et al. (2008). Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 31, 1834–1843. doi: 10.1111/j.1365-3040.2008.01883.x
Valdés-López, O., and Hernández, G. (2008). Transcriptional regulation and signaling in phosphorus starvation: what about legumes? J. Integr. Plant Biol. 50, 1213–1222. doi: 10.1111/j.1744-7909.2008.00758.x
Valentine, A. J., Kleinert, A., and Benedito, V. A. (2017). Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci. 256, 46–52. doi: 10.1016/j.plantsci.2016.12.010
Vance, C. P., Graham, P. H., and Allan, D. L. (2000). “Biological nitrogen fixation: phosphorus-a critical future need?,” in Nitrogen Fixation: From Molecules to Crop Productivity, eds F. O. Pedrosa, M. Hungria, G. Yates, and W. E. Newton (Dordrecht: Kluwer Academic Publishers), 509–514. doi: 10.1007/0-306-47615-0_291
Vardien, W., Steenkamp, E. T., and Valentine, A. J. (2016). Legume nodules from nutrient-poor soils exhibit high plasticity of cellular phosphorus recycling and conservation during variable phosphorus supply. J. Plant Physiol. 191, 73–81. doi: 10.1016/j.jplph.2015.12.002
Vasse, J., de Billy, F., Camut, S., and Truchet, G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172, 4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990
Vauclare, P., Bligny, R., Gout, E., and Widmer, F. (2013). An overview of the metabolic differences between Bradyrhizobium japonicum 110 bacteria and differentiated bacteroids from soybean (Glycine max) root nodules: an in vitro 13C- and 31P-nuclear magnetic resonanc. FEMS Microbiol. Lett. 343, 49–56. doi: 10.1111/1574-6968.12124
Vercruysse, M., Fauvart, M., Beullens, S., Braeken, K., Cloots, L., Engelen, K., et al. (2011). A comparative transcriptome analysis of Rhizobium etli bacteroids: specific gene expression during symbiotic nongrowth. Mol. Plant Microbe Interact. 24, 1553–1561. doi: 10.1094/MPMI-05-11-0140
Vidal, J., and Chollet, R. (1997). Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci. 2, 230–237. doi: 10.1016/S1360-1385(97)01046-7
Voegele, R. T., Mitsch, M. J., and Finan, T. M. (1999). Characterization of two members of a novel malic enzyme class. Biochim. Biophys. Acta 1432, 275–285. doi: 10.1016/S0167-4838(99)00112-0
Wang, C., Saldanha, M., Sheng, X., Shelswell, K. J., Walsh, K. T., Sobral, B. W. S., et al. (2007). Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 153, 388–398. doi: 10.1099/mic.0.29214-0
Wang, T., Zhao, M., Zhang, X., Liu, M., Yang, C., Chen, Y., et al. (2017). Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J. Exp. Bot. 68, 5937–5948. doi: 10.1093/jxb/erx384
Wang, X., Shen, J., and Liao, H. (2010). Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 179, 302–306. doi: 10.1016/j.plantsci.2010.06.007
Weaver, C. D., Shomer, N. H., Louis, C. F., and Roberts, D. M. (1994). Nodulin 26, a nodule-specific symbiosome membrane protein from soybean, is an ion channel. J. Biol. Chem. 269, 17858–17862.
Werner, A. K., and Witte, C. P. (2011). The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci. 16, 381–387. doi: 10.1016/j.tplants.2011.03.012
Whitehead, L. F., Day, D. A., and Tyerman, S. D. (2008). Divalent cation gating of an ammonium permeable channel in the symbiotic membrane from soybean nodules. Plant J. 16, 313–324. doi: 10.1046/j.1365-313X.1998.00298.x
Whitehead, L. F., Young, S., and Day, D. A. (1998). Aspartate and alanine movement across symbiotic membranes of soybean nodules. Soil Biol. Biochem. 30, 1583–1589. doi: 10.1016/S0038-0717(97)00229-0
Xue, Y., Bin Xiao, B. X., Zhu, S. N., Mo, X. H., Liang, C. Y., Tian, J., et al. (2017). GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 68, 4951–4967. doi: 10.1093/jxb/erx292
Yang, Y., Hu, X. P., and Ma, B. G. (2017). Construction and simulation of the Bradyrhizobium diazoefficiens USDA110 metabolic network: a comparison between free-living and symbiotic states. Mol. Biosyst. 13, 607–620. doi: 10.1039/C6MB00553E
Yao, Z., Tian, J., and Liao, H. (2014). Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann. Bot. 114, 477–488. doi: 10.1093/aob/mcu147
Yao, Z. F., Liang, C. Y., Zhang, Q., Chen, Z. J., Xiao, B. X., Tian, J., et al. (2014). SPX1 is an important component in the phosphorus signalling network of common bean regulating root growth and phosphorus homeostasis. J. Exp. Bot. 65, 3299–3310. doi: 10.1093/jxb/eru183
Yong-Villalobos, L., González-Morales, S. I., Wrobel, K., Gutiérrez-Alanis, D., Cervantes-Peréz, S. A., Hayano-Kanashiro, C., et al. (2015). Methylome analysis reveals an important role for epigenetic changes in the regulation of the Arabidopsis response to phosphate starvation. Proc. Natl. Acad. Sci. U.S.A. 112, E7293–E7302. doi: 10.1073/pnas.1522301112
Yurgel, S. N., and Kahn, M. L. (2004). Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28, 489–501. doi: 10.1016/j.femsre.2004.04.002
Zhang, J., Zhou, X., Xu, Y., Yao, M., Xie, F., Gai, J., et al. (2016). Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 248, 82–91. doi: 10.1016/j.plantsci.2016.04.010
Zhang, N., Venkateshwaran, M., Boersma, M., Harms, A., Howes-Podoll, M., Den Os, D., et al. (2012). Metabolomic profiling reveals suppression of oxylipin biosynthesis during the early stages of legume-rhizobia symbiosis. FEBS Lett. 586, 3150–3158. doi: 10.1016/j.febslet.2012.06.046
Zhang, X. Q., Li, B., and Chollet, R. (1995). In vivo regulatory phosphorylation of soybean nodule phosphoenolpyruvate carboxylase. Plant Physiol. 108, 1561–1568. doi: 10.1104/pp.108.4.1561
Zhang, Y., Aono, T., Poole, P., and Finan, T. M. (2012). NAD(P)+-malic enzyme mutants of Sinorhizobium sp. strain ngr234, but not Azorhizobium caulinodans ors571, maintain symbiotic N2 fixation capabilities. Appl. Environ. Microbiol. 78, 2803–2812. doi: 10.1128/AEM.06412-11
Zhao, H., Li, M., Fang, K., Chen, W., and Wang, J. (2012). In silico insights into the symbiotic nitrogen fixation in Sinorhizobium meliloti via metabolic reconstruction. PLoS One 7:e31287. doi: 10.1371/journal.pone.0031287
Keywords: rhizobia, bacteroids, nitrogen fixation, legumes, root nodule, phosphate homeostasis, metabolism, symbiosis
Citation: Liu A, Contador CA, Fan K and Lam H-M (2018) Interaction and Regulation of Carbon, Nitrogen, and Phosphorus Metabolisms in Root Nodules of Legumes. Front. Plant Sci. 9:1860. doi: 10.3389/fpls.2018.01860
Received: 21 September 2018; Accepted: 30 November 2018;
Published: 18 December 2018.
Edited by:
Benjamin Gourion, UMR2594 Plant Interactions Laboratory Microorganisms (LIPM), FranceReviewed by:
George Colin DiCenzo, Università degli Studi di Firenze, ItalyCopyright © 2018 Liu, Contador, Fan and Lam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hon-Ming Lam, aG9ubWluZ0BjdWhrLmVkdS5oaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.