- 1International Maize and Wheat Improvement Center (CIMMYT), Nairobi, Kenya
- 2International Maize and Wheat Improvement Center (CIMMYT), Mexico City, Mexico
- 3Campo Experimental Valle de México INIFAP, Mexico City, Mexico
- 4Cereal Disease Laboratory, United States Department of Agriculture-Agricultural Research Service, St. Paul, MN, United States
- 5Colegio de Postgraduados, Texcoco, Mexico
Races belonging to Ug99 lineage of stem rust fungus Puccinia graminis f. sp. tritici (Pgt) continue to pose a threat to wheat (Triticum aestivum L.) production in various African countries. Growing resistant varieties is the most economical and environmentally friendly control measure. Recombinant inbred line (RIL) populations from the crosses of susceptible parent ‘Cacuke’ with the resistant parents ‘Huhwa’ and ‘Yaye’ were phenotyped for resistance at the seedling stage to Pgt race TTKSK (Ug99) and in adult plants in field trials at Njoro, Kenya for two seasons in 2016. Using the Affymetrix Axiom breeders SNP array, two stem rust resistance genes, temporarily designated as SrH and SrY, were identified and mapped on chromosome arm 2BL through selective genotyping and bulked segregant analysis (BSA), respectively. Kompetitive allele specific polymorphism (KASP) markers and simple sequence repeat (SSR) markers were used to saturate chromosome arm 2BL in both RIL populations. SrH mapped between markers cim109 and cim114 at a distance of 0.9 cM proximal, and cim117 at 2.9 cM distal. SrY was flanked by markers cim109 and cim116 at 0.8 cM proximal, and IWB45932 at 1.9 cM distal. Two Ug99-effective stem rust resistance genes derived from bread wheat, Sr9h and Sr28, have been reported on chromosome arm 2BL. Infection types and map position in Huhwa and Yaye indicated that Sr28 was absent in both the parents. However, susceptible reactions produced by resistant lines from both populations against Sr9h-virulent race TTKSF+ confirmed the presence of a common resistance locus Sr9h in both lines. Test of allelism is required to establish genetic relationships between genes identified in present study and Sr9h. Marker cim117 linked to SrH was genotyped on set of wheat lines with Huhwa in the pedigree and is advised to be used for marker assisted selection for this gene, however, a combination of phenotypic and genotypic assays is desirable for both genes especially for selection of Sr9h in breeding programs.
Introduction
Stem rust, caused by Puccinia graminis f. sp. tritici (Pgt), is one of the most damaging fungal diseases of bread and durum wheat (Triticum aestivum L. and Triticum turgidum ssp. durum, (Desf.) Husnot., respectively) (Roelfs et al., 1992). It can cause complete yield losses under severe epidemics if susceptible cultivars are grown in rust hot-spot areas (Saari and Prescott, 1985; Dean et al., 2012). Global wheat production was threatened by stem rust when a highly virulent Pgt race known as Ug99 or TTKSK that combined virulence to Sr31 and various other commonly deployed resistance genes was detected in 1998 in Uganda (Pretorius et al., 2000; Jin et al., 2007, 2008). After its appearance, evaluations of international wheat germplasm and varieties in both field and greenhouse screenings revealed the predominance of wheat susceptibility to race TTKSK (Jin and Singh, 2006; Singh et al., 2006, 2011). In subsequent years, new variants of Ug99 emerged that carry additional virulence to Sr24 (Jin et al., 2008; Pretorius et al., 2010; Visser et al., 2011), Sr36 (Jin et al., 2009), and SrTmp (Newcomb et al., 2016) placing an even greater number of wheat varieties at risk. Races of the Ug99 race group have already spread over a wide geographical area including 13 countries in the East African highlands, Southern Africa, Yemen, Egypt and Iran, and there is a high chance of further spread into large wheat growing belts of Asia and beyond (Singh et al., 2015). Consequently, the new variants belonging to Ug99 race group and their geographical spread have further reduced the number of effective genes that can be used by breeding programs.
Sixty wheat stem rust resistance genes have a designated gene symbol and a few more carry temporary designations (McIntosh et al., 2016). Five genes, namely Sr2, Sr55 (Lr67/Yr46/Pm46), Sr56, Sr57 (Lr34/Yr18/Pm38), and Sr58 (Lr46/Yr29/Pm39), confer adult plant resistance (APR) (Singh et al., 2015), and the 34 genes that impart all stage resistance (ASR) to Pgt race TTKSK are Sr9h, Sr13a, Sr13b, Sr15, Sr21, Sr22, Sr24, Sr25, Sr26, Sr27, Sr28, Sr32, Sr33, Sr35, Sr36, Sr37, Sr39, Sr40, Sr42, Sr44, Sr45, Sr46, Sr47, Sr50, Sr51, Sr52, Sr53, Sr59, SrCad, SrTA10171, SrTA10187, SrTA1662, SrTmp, and Sr1RSAmigo (Jin and Singh, 2006; Jin et al., 2007; Faris et al., 2008; Hiebert et al., 2010; Kolmer et al., 2011; Liu et al., 2011a,b; Qi et al., 2011; Rouse et al., 2011a, 2014; Rouse and Jin, 2011; Ghazvini et al., 2012; Olson et al., 2013a,b; Bansal et al., 2014; Moore et al., 2015; Singh et al., 2015; Rahmatov et al., 2016; Zhang et al., 2017). Interestingly, Sr8155B1 does not confer resistance to race TTKSK, but does confer resistance to the more recent variants in the Ug99 race group (Nirmala et al., 2017). Only a few resistance genes (Sr2, Sr13a, Sr13b, Sr25, Sr26, Sr57, Sr58, SrCad, SrTmp, SrND643, Sr1RSAmigo, and Sr8155B1) were found to confer resistance in adapted germplasm (Jin and Singh, 2006; Singh et al., 2008, 2015; Njau et al., 2010; Hiebert et al., 2011; Basnet et al., 2015; Nirmala et al., 2017; Zhang et al., 2017). The continuous search for new resistance sources and following through with their genetic characterization and strategic deployment is necessary for long-term control of the fast-evolving races of the stem rust fungus.
Bringing widely effective gene combinations into new cultivars is important to avoid the evolution of Ug99 races and minimize potential yield losses. Multiple gene combinations can be achieved more efficiently if DNA markers, tightly linked to target genes, are available. Markers linked to some stem rust resistance genes have been identified which can be useful in marker assisted selection (MAS). Cloning of genes Sr13a, Sr21, Sr22, Sr33, Sr35, Sr45, Sr50, Sr55, and Sr57 has not only enhanced the knowledge of resistance mechanisms but also resulted in the development of diagnostic gene-based markers (Krattinger et al., 2009; Periyannan et al., 2013, 2014; Saintenac et al., 2013; Mago et al., 2015; Moore et al., 2015; Zhang et al., 2017; Chen et al., 2018). Several genes effective against Ug99 were tagged using DNA markers, to name some; Sr2 (Hayden et al., 2004; Mago et al., 2011), Sr9h (Rouse et al., 2014), SrND643 (Basnet et al., 2015), Sr22 (Periyannan et al., 2011), Sr26 (Mago et al., 2005), Sr25 and Sr26 (Liu et al., 2010), Sr28 (Rouse et al., 2012), Sr35 (Saintenac et al., 2013), Sr45 (Periyannan et al., 2014), Sr48 (U. Bansal, University of Sydney, personal communication), Sr56 (Bansal et al., 2014), Sr55 (Moore et al., 2015), and Sr57 (Lagudah et al., 2006). In the last decade, new fast and cost-effective marker technologies emerged as powerful tools, which are very helpful in identifying rust resistance genes and their strategic deployment in breeding.
Observations on ‘Huhwa’ (H), ‘Yaye’ (Y) and their progenies in International Maize and Wheat Improvement Center (CIMMYT) germplasm indicated the presence of ASR to Pgt races belonging to the Ug99 group. Furthermore, several high yielding wheat lines derived from Huhwa and Yaye have been distributed in CIMMYT international nurseries. Despite the fact that Huhwa and Yaye carry stem rust resistance genes effective to Ug99 race group, their genetic and molecular characterization remains unclear. Therefore, the present study was undertaken to characterize stem rust resistance in these cultivars using recombinant inbred lines (RIL) populations, and to develop and validate SNP markers closely linked to these genes.
Materials and Methods
Plant Material and Development of Mapping Populations
The two RIL populations were developed by crossing stem rust resistant bread wheat lines, Huhwa [CIMMYT germplasm identification number (GID): 5552006] and Yaye (GID: 5343322) each with the susceptible parent Cacuke (GID: 5347441) as the female parent during wheat seasons 2007-08 and 2008-09 at Obregon, Mexico, respectively. The Pedigrees of Huhwa and Yaye are ‘HUW234+Lr34/Prinia//Kronstad F2004’ and ‘Yanac/3/Parula/Sara//TSI/Veery#5/4/Croc_1/Ae. squarrosa (224)//Opata’, respectively. The pedigrees of HUW234 and Yanac are ‘HUW12/SPRW//HUW12’ and ‘Jabiru/M-5392-1//M-5392/3/Cook,’ respectively, whereas Cacuke is ‘Canadian/Cunningham//Kennedy.’ Hereafter, Cacuke × Huhwa and Cacuke × Yaye populations are abbreviated as C × H and C × Y, respectively. The F5 RIL populations C × H (148 RILs) and C × Y (198 RILs) were developed using single spike descent method modified from the single seed descent method. To develop the population, a single spike from each F2 plant, generated from three different F1 plants, was harvested under fungicide-protected conditions and advanced to the F4 generation by harvesting a single spike in subsequent generations. The F5 plots, derived from single F4 spikes were then harvested in bulk to obtain sufficient seed of the F5 RILs. C × H (3 RILs) and C × Y (27 RILs) did not germinate when populations were planted for seed multiplication at El Batan in 2015. Finally seeds of C × H (145 RILs) and C × Y (171 RILs) were used for all genotypic and phenotypic analyses.
Greenhouse Evaluations
The parents and RILs of the two populations, C × H and C × Y, were tested against Pgt race TTKSK (isolate 04KEN156/04) at the seedling stage in a greenhouse at the United States Department of Agriculture- Agricultural Research Service (USDA-ARS) Cereal Disease Laboratory (CDL), St. Paul, MN, United States following the procedure described in Rouse et al. (2011b). The avirulence/virulence formula of race TTKSK is Sr24,36,Tmp/Sr5,6,7b,8a,9a,9b,9d,9e,9g,10,11,17,30,31,38,McN (Jin et al., 2007). Between 20 and 30 seeds of each RIL and the parents were grown for seedling tests. Eight-day old seedlings were inoculated with urediniospores of Pgt race TTKSK. Infection types (ITs) on seedlings were recorded 14 days post-inoculation using 0 to 4 scale as described by Stakman et al. (1962). Infection types ‘0,’ ‘1,’ and ‘2’ were considered resistant, whereas ITs ‘3’ to ‘4’ were considered susceptible. RILs were classified in three categories homozygous resistant (HR), homozygous susceptible (HS) and segregating. Two HR and HS RILs from both populations along with respective parents were also tested against Sr9h-virulent race TTKSF+ (isolate 09ZIM01-2; Pretorius et al., 2012; Rouse et al., 2014). Tests were repeated twice to avoid any chances of misclassification of RILs in each population.
Field Evaluations
The two RIL populations and the parents were phenotyped for response to stem rust in field trials at the Njoro research station of Kenya Agricultural and Livestock Research Organization (KALRO) during two wheat growing seasons (2016- off and main seasons). About 5 g seed of each entry was planted in 0.7 m long paired-row flat beds, with 0.3 m spacing between them. Spreaders comprised of a mixture of stem rust susceptible cultivars ‘Cacuke’ and ‘Robin,’ and six-Sr24 carrying lines (GIDs: 5391050, 5391052, 5391056, 5391057, 5391059, and 5391061) were planted as hill plots on one side of each test plot in the middle of the 0.3 m-wide pathways. Spreaders were also planted along the borders of the experimental field in 1 m plots. Spreaders were inoculated with a field collection of races TTKST and TTKTT belonging to the Ug99 race group (Jin et al., 2008; Singh et al., 2015; Newcomb et al., 2016) by spraying a mixture of urediniospores suspended in water plus Tween 20 suspensions, and needle inoculations with the same suspension as described in Njau et al. (2013). Stem rust responses of each RIL and parents were recorded at post-flowering stage when the susceptible parent displayed moderately susceptible to susceptible (MS to S) responses (Roelfs et al., 1992) with 70% disease severity (DS) following the Modified Cobb’s Scale (Peterson et al., 1948). Responses were recorded a second time about 8 days later.
Molecular Marker Analysis
Leaf tissues were harvested from young leaves and genomic DNA was extracted using CIMMYT’s laboratory protocols (Dreisigacker et al., 2015). The quality and quantity of DNA was assessed on 1% agarose gels and using a NanoDrop 8000 spectrophotometer (Thermo scientific). Selective genotyping was carried out for parents, 47 resistant and 47 susceptible RILs of C × H population, whereas bulk-segregant analysis (BSA; Michelmore et al., 1991) was performed on pooled DNA of 10 resistant and 10 susceptible RILs along with parents of the C × Y population. Genotyping was conducted using the 35K Affymetrix Axiom Breeders array (Allen et al., 2017) by outsourcing to TraitGenetics GmbH (Germany). The SNPs closely linked to stem rust resistance locus in each population were converted into Kompetitive allele-specific polymorphic (KASP) markers using the automated bioinformatics pipeline, PolyMarker (Ramirez-Gonzalez et al., 2015). KASP markers were then used to verify the targeted SNP polymorphism in the parents and polymorphic markers were applied on the respective mapping population. In addition, 3 KASP markers derived from the SNP markers IWB45296, IWB3891 and IWB45932 (Wang et al., 2014), linked to Sr9h were tested on the respective parents of both populations. Oligos were synthesized in 0.025 μM concentration from Sigma-Aldrich, United States. SNP markers were carried out in a final volume of 8 μl consisting of 3 μl of genomic DNA (30 ng/μl), 4 μl of 2x KASP mix [a mixture of FAM and HEX Fluorescence Resonance Energy Transfer (FRET) cassettes, ROXTM passive dye, an optimized buffer containing MgCl2, Taq polymerase and dNTPs], 0.11 μl primer mix containing two allele specific forward primers each with 12 and 30 μM of common primer and 0.89 μl of sterile water. PCR amplification was performed in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems) using the cycling conditions: 95°C for 15 min for hot-start Taq DNA polymerase activation, followed by a touchdown profile of 9 cycles at 94°C for 20 s and 61°C for 1 min with a 0.6°C reduction per cycle, followed by 38 cycles at 94°C for 20 s and 55°C for 1 min. The reactions were held at 12°C for 3 min. End-point fluorescent images were visualized using the PHERAstarPlus (BMG LABTECH, Germany), and the data were analyzed using KlusterCallerTM software (LGC genomics).
Similarly, 22 simple sequence repeat (SSR) markers namely; barc101 and barc159 from Song et al. (2005); cfd70 and cfd73 (Sourdille et al., 2001); gwm16, gwm47, gwm120, gwm382, gwm388, gwm501 and gwm526 (Roder et al., 1995, 1998); wmc51, wmc149, wmc175, wmc317, wmc332, wmc356, wmc361, wmc441, wmc500, and wmc627 (Somers et al., 2004), located on chromosome arm 2BL were also tested on parents, Cacuke, Huhwa and Yaye, to survey polymorphisms between parents and the respective RIL population was genotyped with polymorphic SSRs using respective PCR protocols. In addition, Sr26-linked marker Sr26#43 (Mago et al., 2005) was tested on ten RILs each of populations C × H and C × Y along with respective parents.
Data Analyses and Genetic Mapping
Chi-squared analyses were performed to determine the goodness-of-fit of observed segregation with the expected genetic ratios for 1 gene segregation in F4:5 RIL population using only the homozygous lines (HR: HS = 1:1). Genetic linkage maps for the stem rust resistance loci were constructed using genotypic data of molecular markers and seedling stem rust data using Inclusive Composite Interval Mapping (ICIM) software (Li et al., 2008). The Kosambi mapping function (Kosambi, 1943) was applied to convert recombination fractions into centimorgans (cM). Markers and respective resistance loci were grouped using a logarithm of odds (LOD) threshold of 10, the linkage maps were ordered and rippled using nearest-neighbor two-opt (nnTwoOpt) and sum of adjacent recombination (SAR) fraction algorithms, respectively.
Associations between markers on chromosome arm 2BL and quantitative resistance were revealed using the DS data recorded during 2016 off and main seasons using the ICIM software. A quantitative trait locus (QTL) was designated to be significant with a LOD value above a threshold calculated through 1000 permutation tests. Linkage map diagrams were prepared and aligned for a visual inspection of map order using MapChart 2.2 software (Voorrips, 2002).
Validation of Flanking Markers for Marker-Assisted Selection
Closely linked markers to stem rust resistance were used in validation studies by testing on a set of wheat genotypes carrying Huhwa and Yaye in their pedigree, respectively, and the efficacy of the reported markers for marker assisted selection (MAS). Sixty-seven and 44 lines with Huhwa and Yaye in their pedigree respectively, were chosen for validation of flanking markers. These lines were selected from the CIMMYT 9th and 10th Stem Rust Resistance Screening Nursery (SRRSN); the 26th High Rainfall Wheat Screening Nursery (HRWSN); 32nd, 33rd, and 34th Semi-Arid Wheat Screening Nursery (SAWSN); 47th, 48th, and 49th International Bread Wheat Screening Nursery (IBWSN) (Supplementary Tables 1, 2). Seedling IT’s in response to races TTKSK, TTKSF+ and TTKTT (isolate 14KEN58-1, avirulent to Sr9h) were assessed at the USDA-ARS Cereal Disease Laboratory (CDL), St. Paul, MN following procedures described in Rouse et al. (2011b). Infection type data were recorded on the 0 to 4 scale of Stakman et al. (1962), where ITs 0, 1, and 2 (or combinations thereof) were considered resistant and ITs 3 and 4 were susceptible. For all wheat lines, stem rust infection response and severity data were also obtained from the international stem rust nurseries evaluated in 2013, 2014, and the 2015 off season in Kenya. The marker haplotype and stem rust phenotype in each line were compared to evaluate the efficiency of the markers in selecting resistance gene.
Results
Greenhouse Evaluations
Huhwa and Yaye displayed resistant ITs 22+ and 2, respectively, in contrast to susceptible IT 3+ for the susceptible parent ‘Cacuke’ when tested at the seedling stage with Pgt race TTKSK (Figure 1A). Although both populations were classified into three categories (C × H: 52 HR, 60 HS and 33 segregating;C × Y: 81 HR, 65 HS and 25 segregating) based on seedling ITs, only two categories namely HR and HS, were used for analyses. Monogenic segregation of stem rust resistance was confirmed in C × H [χ2(1:1) = 0.58 non-significant and p = 0.45] and C × Y [χ2(1:1) = 1.76 non-significant and p = 0.19] (Table 1). Two HR and HS lines along with respective parents of both populations showed susceptible IT 3+ when tested against Sr9h-virulent race TTKSF+ at the seedling stage (Figure 1B). We refer to the single stem rust resistance genes segregating in the C × H and C × Y populations as “SrH” and “SrY,” respectively.
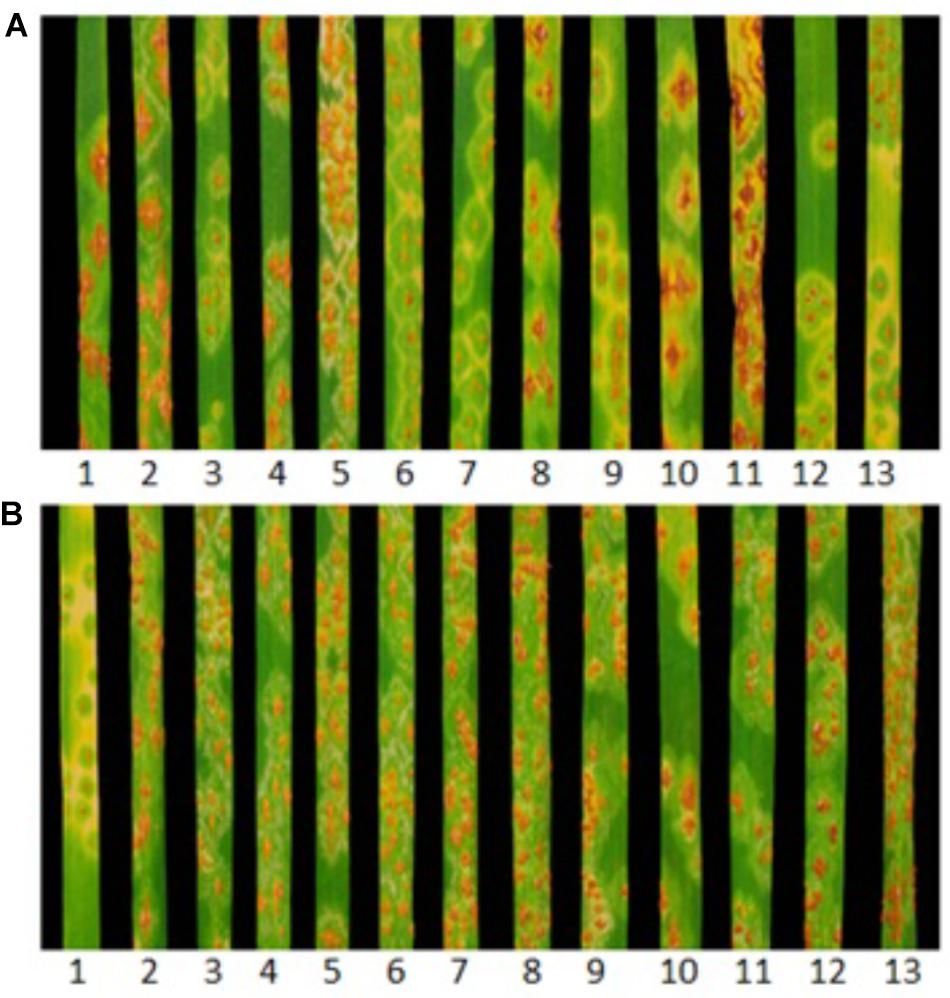
FIGURE 1. Infection type (IT) responses produced by (1) Sr31 differential (Sr31/6∗LMPG), (2) Cacuke, (3) Yaye, (4and 5) Cacuke × Yaye homozygous susceptible (HS) lines, (6 and 7) Cacuke × Yaye homozygous resistant (HR) lines, (8) Cacuke, (9) Huhwa, (10 and 11) Cacuke × Huhwa HS lines, (12 and 13) Cacuke × Huhwa HR lines when tested against Puccinia graminis f. sp. tritici races (A) TTKSK and (B) Sr9h-virulent TTKSF+.
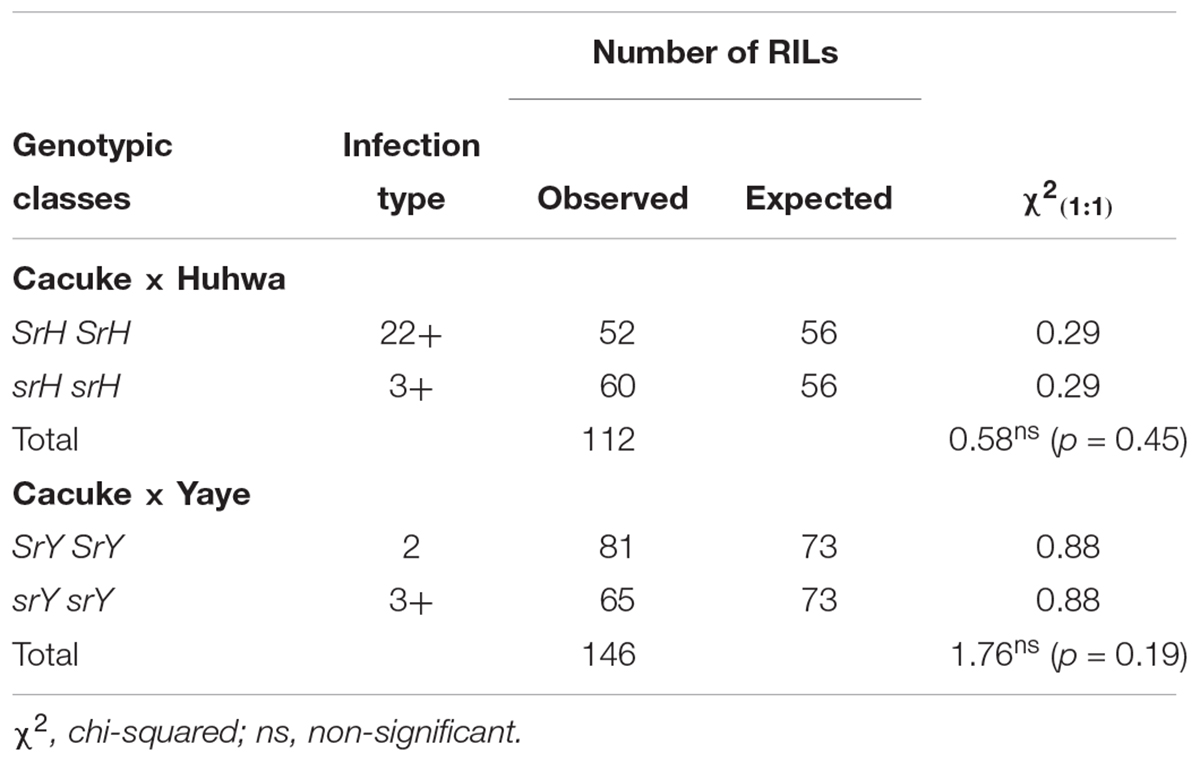
TABLE 1. Mendelian inheritance of stem rust resistance in Cacuke × Huhwa and Cacuke × Yaye homozygous F5 recombinant inbred lines (RILs) when tested with Puccinia graminis f. sp. tritici (Pgt) pathotype TTKSK at seedling stage.
Field Evaluations
Uniform disease development was observed in the field nurseries in both seasons in 2016. Terminal disease severity (TDS) and infection response for Cacuke was 70 S, whereas it ranged 10–20 MR for Huhwa and 5–15 RMR for Yaye over two seasons in the adult plant stage. A wide range of infection responses (MR to S) was observed among the RILs in both populations. TDS of RILs in C × H population ranged from 5 to 70% and 10 to 70% during 2016 off and main seasons, respectively (Figures 2A,B). In C × Y population, TDS varied from 10 to 100% and 10 to 80% during 2016 off and main seasons, respectively (Figures 2C,D). Only seedling stem rust data were used to classify lines carrying or not carrying the respective resistance locus in the two populations to avoid any chances of misclassification.
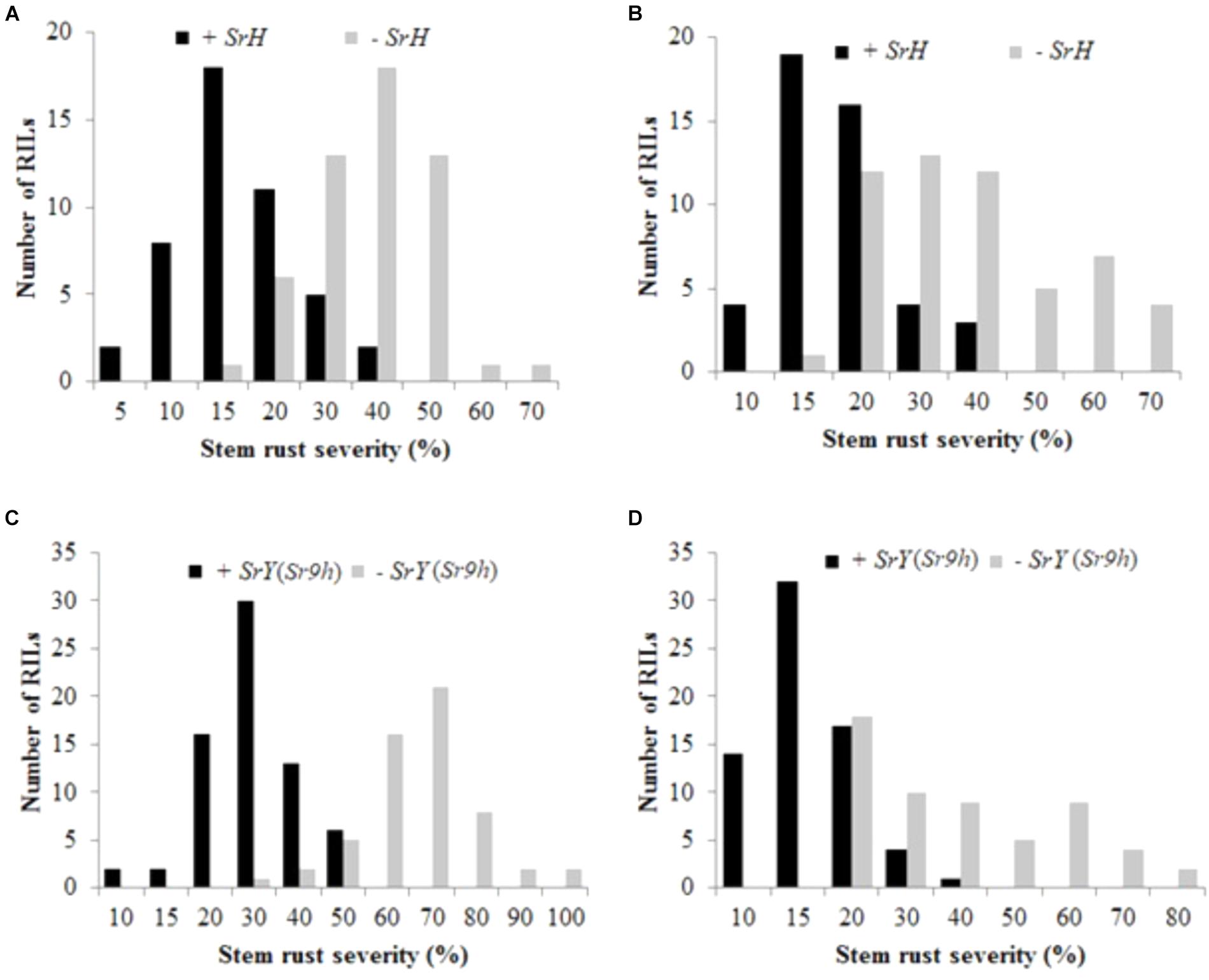
FIGURE 2. Distribution of stem rust disease severity among recombinant inbred lines of Cacuke × Huhwa with (+) or without (–) SrH (A,B); and Cacuke × Yaye with (+) or without (–) SrY (C,D). Data were collected during two seasons viz. 2016 off (A,C) and main (B,D) seasons at Njoro, Kenya.
Mapping of Stem Rust Resistance Locus
C × H Population
Selective genotyping using 47 each resistant and susceptible RILs along with the parents identified a total of 2,122 polymorphic SNPs distributed across 21 wheat chromosomes. Eighty-three SNPs located on chromosome arm 2BL showed close linkage with stem rust resistance in population C × H using a genetic map of 65.0 cM constructed using ICIM software. Nineteen SNPs linked to SrH within 10.0 cM distance were converted into KASP markers designated as cim (CIMMYT) namely; cim101, cim102, cim103, cim104, cim105, cim106, cim107, cim108, cim109, cim110, cim111, cim112, cim113, cim114, cim115, cim116, cim117, cim118, and cim119 (Table 2). In addition, 7 KASP markers (BS00004405, BS00003597, BS00009864, BS00003589, BS00003673, BS00010081, and BS00009460) previously mapped on chromosome arm 2BL (Wilkinson et al., 2012), 3 Sr9h-linked KASP markers, and 9 KASP markers developed for SrY (Table 2) were tested on parents, Cacuke and Huhwa, to verify their polymorphism. A total of 60 markers were screened (the 38 KASP above mentioned and 22 genomic SSRs). Out of them, 19 markers (cim102, cim103, cim104, cim106, cim107, cim109, cim114, cim116, cim117, cim128, BS00009864, barc159, cfd70, gwm47, gwm388, wmc149, wmc317, wmc332, and wmc627) showed clear differences between parents and were genotyped on the entire mapping population, C × H. Sr9h-linked markers were found to be monomorphic between Cacuke and Huhwa. Using the ICIM mapping software, a partial genetic map of chromosome arm 2BL spanning a distance of 49.8 cM was constructed in which SrH was flanked by markers cim109 and cim114 at 0.9 cM proximal, and a marker cim117 at 2.9 cM distal, respectively (Figure 3B). Independent assortment of seven markers (cim128, barc159, cfd70, gwm47, gwm388, wmc149, and wmc317) with respect to SrH was observed. The Sr9h map from previous study (Rouse et al., 2014) was included for comparison (Figure 3A).
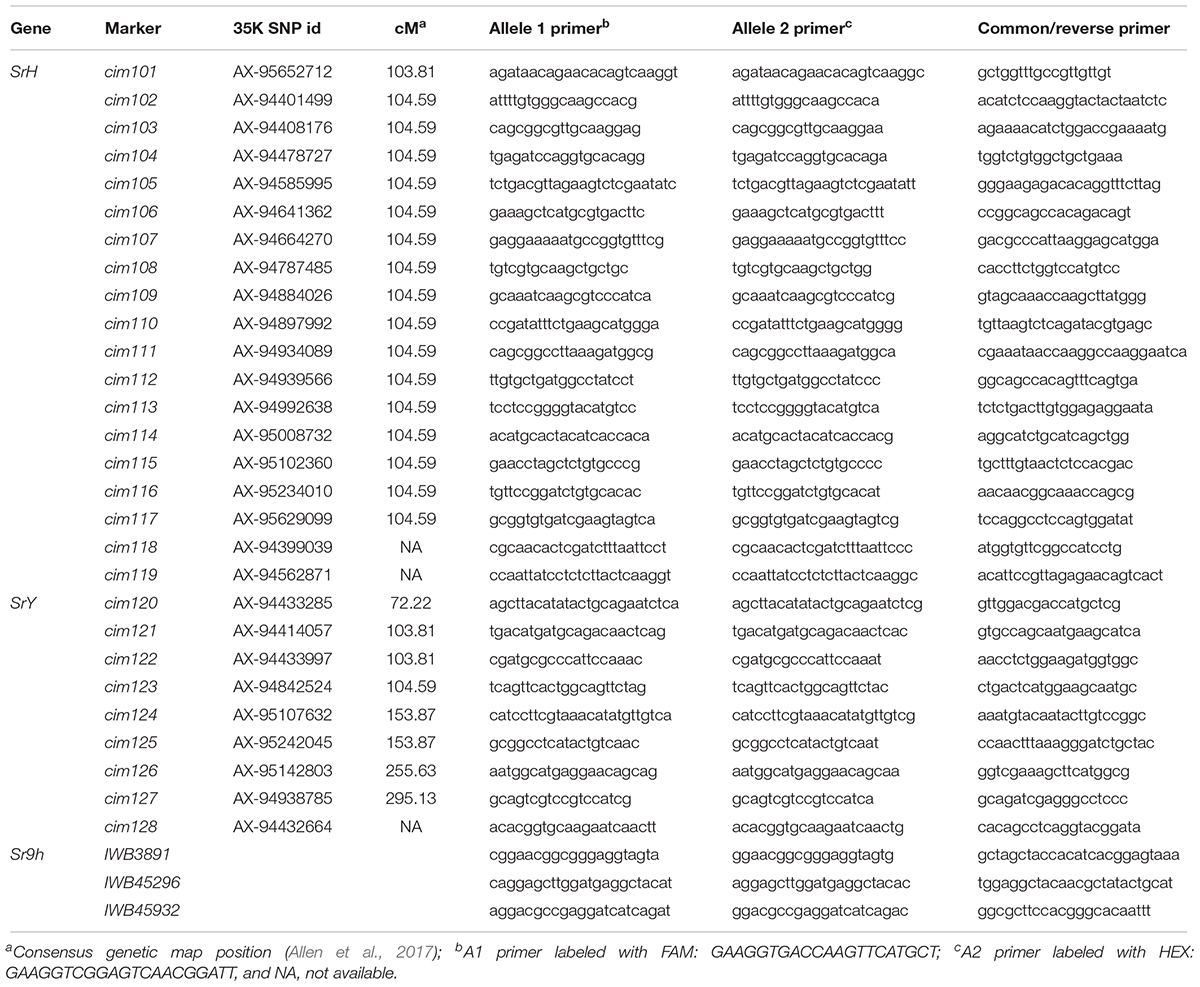
TABLE 2. Primer sequences of KASP markers used to saturate the SrH, SrY, and Sr9h regions on chromosome 2BL.
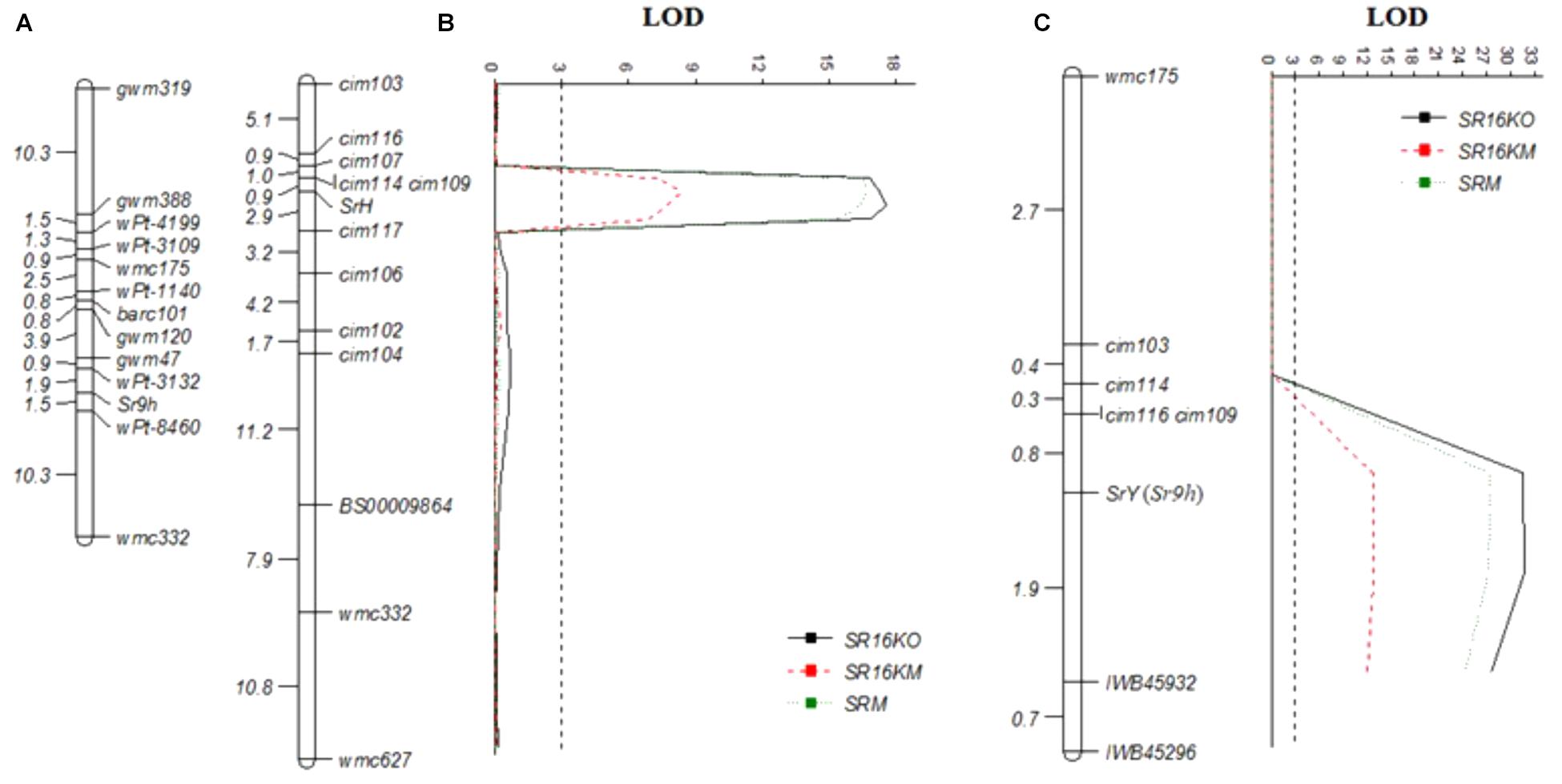
FIGURE 3. Genetic maps of the Sr9 region of chromosome 2BL from our data and previously published studies: (A) Rouse et al. (2014), (B) Cacuke × Huhwa and (C) Cacuke × Yaye. Genetic distances between the markers are given in centiMorgan (cM). LOD, Logarithm of odds.
C × Y Population
BSA results indicated association of 24 SNPs located on chromosome arm 2BL with stem rust resistance in population C × Y. Nine SNPs were converted into KASP markers cim120, cim121, cim122, cim123, cim124, cim125, cim126, cim127 and cim128 (Table 2). In total, 38 KASP markers (9 designed for SrY, 19 for SrH, 3 linked to Sr9h, 7 selected from Wilkinson et al. (2012) and 22 SSRs were tested on the parents, Cacuke and Yaye. Finally, 14 markers (cim103, cim109, cim114, cim116, cim128, BS00009864, IWB3891, IWB45296 and IWB45932, barc159, cfd70, gwm47, wmc149, and wmc175) showed clear polymorphism and were genotyped on the entire population Cauke × Yaye. A partial genetic map of chromosome arm 2BL spanned a genetic distance of 6.8 cM with SrY flanked by markers cim109 and cim116 at 0.8 cM proximal, and marker IWB45932 at 1.9 cM distal (Figure 3C). Seven markers (cim128, BS00009864, IWB3891, barc159, cfd70, gwm47, and wmc149) showed independent assortment with respect to SrY. The marker Sr26#43 linked to Sr26 did not amplify any product in Cacuke, Huhwa, and Yaye.
Effect of SrH and SrY on Stem Rust Disease Severity
To study the effect of SrH on disease severity, all RILs of the population C × H were reclassified into resistant (+SrH) and susceptible (-SrH) groups, based on the flanking markers cim109 and cim117. Among 112 RILs, two recombinants were observed between SrH and cim109 whereas 11 recombinants were identified between SrH and cim117. Based on the flanking markers, 100 non-recombinant RILs, (46 +SrH and 54 -SrH) were used for this analysis. In all cases, +SrH lines displayed MR or M infection responses in the field. Conversely, -SrH RILs displayed compatible infection responses (MS, MSS, or S). TDS of +SrH was 5 to 40% (mean = 17.2%) and 10 to 40% (mean = 19.3%) in off and main seasons in 2016, respectively, whereas it was 15–70% (mean = 38 and 38.4% during off and main seasons, respectively) in -SrH lines during both seasons (Figures 2A,B). The mean differences between the two groups were highly significant (t = 11.01, p < 0.0001 and t = 7.90, p < 0.0001 for 2016-off and 2016-main seasons, respectively) indicating that SrH confers significant effects on disease severity reduction in addition to conferring a resistant infection response.
Similarly, RILs of C × Y were reclassified into resistant (+SrY) and susceptible (-SrY) classes, based on the flanking markers cim109 and IWB45932. Among 146 RILs, 18 recombinants were observed between SrY and cim109 whereas 11 between SrY and IWB45932. Nine recombinants were common between SrY, and markers cim109 and IWB45932. Based on flanking markers, 126 non-recombinant RILs, (69 +SrY and 57 -SrY) were used for this analysis. In all cases, +SrY RILs displayed RMR or MR infection responses in the field. In contrast, -SrY RILs displayed compatible responses (MS, MSS, or S). The disease severity of +SrY was 10–50% (mean = 31%) and 10–40% (mean = 17.2%) in off and main seasons in 2016, respectively, whereas it was 30–100% (mean = 66.8%) and 20–80% (mean = 39.4%) in -SrY lines (Figures 2C,D). The mean differences between the two groups were highly significant (t = 17.1, P < 0.0001 and t = 9.13, P < 0.0001 for 2016-off and 2016-main seasons, respectively) showing that SrY also confers significant effects on disease severity reduction in addition to conferring a resistant infection response.
Validation of Flanking Markers
Five markers (cim103, cim109, cim114, cim116, cim117) closely linked to SrH were used to genotype 67 CIMMYT advanced breeding lines derived from Huhwa (Supplementary Table 1). Based on the seedling data, 35 and 32 lines were postulated to carry and not-carry SrH, respectively. Marker cim117 amplified SrH-linked positive allele A in 32 lines postulated to carry SrH whereas 3 lines showed negative allele G. Positive validation of marker cim117 in 32 lines (out of 35) lines indicated 91.4% efficiency of this marker to identify lines with SrH. The seedling ITs of the SrH postulated lines ranged from 2- to 2 IT response. Out of 32 lines not postulated to carry SrH, 31 lines amplified negative allele G except one line with positive allele A. The other 4 markers were not associated with SrH in the advanced breeding lines.
We tested six markers cim103, cim109, cim114, cim116, IWB45932 and IWB45296 closely linked to SrY on a set of 44 wheat lines derived from Yaye (Supplementary Table 2). Marker IWB45932 amplified SrY-linked positive allele T in 14 lines postulated to carry SrY whereas one line showed negative allele C. Positive validation of marker IWB45932 in 14 of the 15 lines showed 93.3% efficiency of this marker to identify lines carrying SrY. When tested on 29 lines not postulated to carry SrY, marker IWB45932 amplified SrY-linked negative allele C in 13 lines whereas 14 lines carried SrY-linked positive allele T. Therefore, IWB45932 alone cannot be used to accurately predict the presence of SrY.
Discussion
In this study, stem rust resistance genes SrH and SrY were identified in wheat lines Huhwa and Yaye, respectively, and mapped on chromosome arm 2BL using selective genotyping (SG) and bulked segregant analysis (BSA) methods, respectively. Both Huhwa and Yaye have been utilized as common parents in CIMMYT bread wheat improvement program. Therefore, it is expected that SrH and SrY are already widely present in recently developed wheat lines resistant to Pgt races in the Ug99 group.
To this date, five stem rust resistance genes Sr9, Sr16, Sr28, Sr47, and SrWeb have been mapped on chromosome arm 2BL. There are seven alleles characterized at the Sr9 locus namely; Sr9a, Sr9b, Sr9d, Sr9e, Sr9f, Sr9g, and Sr9h, and each allele demonstrates unique race specificities to Pgt races (Green et al., 1960; Knott, 1966; McIntosh and Luig, 1973; Loegering, 1975; Rouse et al., 2014). The gene designation Sr9c was originally reserved for a gene that was subsequently designated as Sr36 on chromosome arm 2BS (McIntosh et al., 1995). SrWeb was mapped to chromosome arm 2BL near the Sr9 locus (Tsilo et al., 2007; Hiebert et al., 2010) and named as Sr9h (Rouse et al., 2014).
Six characterized alleles at the Sr9 locus (Sr9a, Sr9b, Sr9d, Sr9e, Sr9f, and Sr9g) and Sr16 are not effective against Ug99 race, whereas Sr9h, Sr28, and Sr47 confer resistance (Jin et al., 2007; Rouse et al., 2014). Three Sr9h-linked KASP markers (IWB45296, IWB3891, and IWB45932) showed no polymorphism between Cacuke and Huhwa. Although SSR marker gwm47 closely linked to Sr9h was polymorphic between Cacuke and Huhwa, the marker could not be integrated into a genetic linkage map with SrH due to lack of linkage. No variation shown by Sr9h-linked markers between parents and independent assortment of marker gwm47 with respect to SrH suggested that the stem rust resistance locus in Huhwa was unlikely to be Sr9h. The Sr28-linked marker wmc332 (Rouse et al., 2012) was mapped more than 30 cM distal to SrH. Moreover, IT differences between Sr28 and SrH, indicate potential unlikeness of SrH to Sr28. SrH displayed IT 22+ whereas Sr28 displayed IT;13- (as reported in Rouse et al., 2014).
In contrast, Sr9h-linked markers (IWB45296 and IWB45932) and two SSR markers (wmc175 and gwm47) were found polymorphic between Cacuke and Yaye. Upon genotyping the entire RIL population C × Y, markers IWB45932 and IWB45296 were mapped at 1.9 cM proximal and 2.6 cM distal to SrY, respectively. Association of Sr9h-linked markers with SrY indicated possible similarity of SrY to Sr9h. Sr28-linked marker wmc332 was monomorphic in the parents Cacuke and Yaye, therefore indicating that segregation at the stem rust resistance locus was possibly not associated with Sr28. An Aegilops speltoides introgression carried Sr47 which ruled out potential similarity of either SrY or SrH with Sr47 (Klindworth et al., 2012) since neither parent carry Ae. speltoides in their pedigree. Both SrH and SrY were mapped on chromosome arm 2BL. There is no common marker between the SrY and SrH maps that suggests that the position of the genes is different. Both genes are slightly distal to cim114. Advanced wheat lines carrying HUW234 and Yanac in their pedigree showed stem rust resistance indicating the most likely source of SrH is HUW234 and SrY is Yanac. Based upon pedigree information, Yanac could carry stem rust resistance gene Sr26, however, marker Sr26#43 did not amplify any product in resistant parent Yaye, indicating that the gene in Yaye is different from Sr26.
Susceptible reactions displayed by seedlings of resistant lines from both populations when tested with Sr9h virulent pathotype TTKSF+ confirmed a common resistance locus in both cultivars Huhwa and Yaye which is most likely Sr9h identified in wheat cultivar Gabo 56 (Rouse et al., 2014). Gene Sr9h is effective against Pgt race TTKSK whereas it is ineffective against race TTKSF+. Race TTKSF+, detected in South Africa and Zimbabwe (Pretorius et al., 2012), is virulent on Sr9h (Rouse et al., 2014). Similar response shown by Huhwa and Yaye indicated presence of Sr9h in both lines.
Association of KASP marker cim117 on a set of advanced wheat lines postulated to carry SrH indicates that this marker could be useful for marker-assisted selection of this gene in breeding. During marker validation process, we concluded that previously known Sr9h-linked markers can be used for identification of Sr9h or SrY together with phenotypic markers as the behavior of Sr9h linked markers vary with genetic backgrounds.
RILs carrying SrH showed disease severities of 5–40% and 10–40% during 2016 off and main seasons, respectively. However, these lines still displayed incompatible reactions to Pgt with infection responses of MR or M. In contrast, in the absence of SrH, disease severities of the RILs were 15–70% with corresponding infection responses of MS or S during 2016 off and main seasons. Similar, results were also found for SrY-carrying and SrY -lacking RILs. As reported by Basnet et al. (2015), disease severity per se is not useful for determining the presence or absence of race-specific stem rust resistance genes. Basnet et al. (2015) reported SrND643-carrying lines expressing 5–40% disease severities accompanied by R, MR or M infection responses. Similarly, Singh et al. (2011) observed that CIMMYT advanced lines with race-specific genes Sha7 and SrTmp displayed 1–30 and 5–60% disease severities accompanied by R or MR infection responses. Stem rust variation observed in the present study may be due to the presence of quantitative adult plant resistance genes that interact with SrH or SrY.
Conclusion
This study identified a common stem rust resistance locus located on chromosome arm 2BL in wheat cultivars Huhwa and Yaye. A robust SNP marker cim117 closely linked to SrH was identified and validated on advanced wheat lines carrying Huhwa in their pedigree. This marker can serve as a breeder friendly tool for marker assisted selection of SrH in breeding programs to generate new gene combinations in breeding Ug99 resistant cultivars.
Kompetitive Allele-Specific Polymorphic (KASP) Markers Data Submission
We attempted to submit KASP markers generated in this study to European Variation Archive (EVA). EVA needs specific requirements including variant quality, filter, information, format etc. for each assay/variant that will only be available if we could have conducted axiom assay in our laboratory. As we outsourced genotyping using 35K axiom breeders’ arrays, this specific information is not available to be included in the submission file to EVA. Markers were generated from publicly available contig sequences developed by International wheat Genome Sequencing Consortium (IWGSC) and available on http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/axiom_download.php. Markers developed in this study are PCR based markers representing SNPs of contig sequences from 35K axiom breeder’s arrays. Therefore, we decided not to submit these markers to avoid any conflict of interest among wheat scientists.
Author Contributions
MSR phenotyped the population, did analysis, and wrote the manuscript. MSR, SD, JN, and MS-S genotyped the population with SSR and SNP markers. MSR, RS, SB, and JH-E phenotyped the population at adult plant stage. MNR phenotyped the population at seedling stage.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the financial support provided by the Bill & Melinda Gates foundation (BMFG) and Department of International Development, United Kingdom (DFID) through the Durable Rust Resistance in Wheat (DRRW) and the Delivering Genetic Gains in Wheat (DGGW) projects (Grant No. OPPGD1389), Grain Research and Development Corporation of Australia (GRDC), United States Department of Agriculture-Agricultural Research Service (USDA-ARS), and Wheat Consultative Group of International Agricultural Research (CGIAR) program (CRP) window 2 donors.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01788/full#supplementary-material
TABLE S1 | List of advanced wheat lines with Huhwa in their pedigree, their adult plant stem rust response, seedling infection types against Puccinia graminis f. sp. tritici (Pgt) races and haplotypes of SrH-linked markers.
TABLE S2 | List of advanced wheat lines with Yaye in their pedigree, their adult plant stem rust response, seedling infection types against Puccinia graminis f. sp. tritici (Pgt) races and haplotypes of SrY (Sr9h)-linked markers.
References
Allen, A. M., Winfield, M. O., Burridge, A. J., Downie, R. C., Benbow, H. R., Barker, G. L. A., et al. (2017). Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. doi: 10.1111/pbi.12635
Bansal, U., Bariana, H., Wong, D., Randhawa, M., Wicker, T., Hayden, M., et al. (2014). Molecular mapping of an adult plant stem rust resistance gene Sr56 in winter wheat cultivar Arina. Theor. Appl. Genet. 127, 1441–1448. doi: 10.1007/s00122-014-2311-1
Basnet, B. R., Singh, S., Lopez-Vera, E. E., Huerta-Espino, J., Bhavani, S., Jin, Y., et al. (2015). Molecular mapping and validation of SrND643: a new wheat gene for resistance to the stem rust pathogen Ug99 race group. Phytopathology 105, 470–476. doi: 10.1094/Phyto-01-14-0016-R
Chen, S., Zhang, W., Bolus, S., Rouse, M. N., and Dubcovsky, J. (2018). Identification and characterization of wheat stem rust resistance gene Sr21 effective against the Ug99 race group at high temperature. PLoS Genet. 14:e1007287. doi: 10.1371/journal.pgen.1007287
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dreisigacker, S., Sehgal, D., Luna, B., Reyes, A. E., and Mollins, J. (2015). CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding. Mexico: CIMMYT.
Faris, J. D., Xu, S. S., Cai, X., Friesen, T. L., and Jin, Y. (2008). Molecular and cytogenetic characterization of a durum wheat-Aegilops speltoides chromosome translocation conferring resistance to stem rust. Chromosome Res. 16, 1097–1105. doi: 10.1007/s10577-008-1261-3
Ghazvini, H., Hiebert, C. W., Zegeye, T., Liu, S., Dilawari, M., Tsilo, T., et al. (2012). Inheritance of resistance to Ug99 stem rust in wheat cultivar Norin 40 and genetic mapping of Sr42. Theor. Appl. Genet. 125, 817–824. doi: 10.1007/s00122-012-1874-y
Green, G. J., Knott, D. R., Watson, I. A., and Pugsley, A. T. (1960). Seedling reactions to stem rust of lines of Marquis wheat with substituted genes for rust resistance. Can. J. Plant. Sci. 40, 524–538. doi: 10.4141/cjps60-069
Hayden, M. J., Kuchel, H., and Chalmers, K. J. (2004). Sequence tagged microsatellites for the Xgwm533 locus provide new diagnostic markers to select for the presence of stem rust resistance gene Sr2 in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1641–1647. doi: 10.1007/s00122-004-1787-5
Hiebert, C. W., Fetch, T. G., and Zegeye, T. (2010). Genetics and mapping of stem rust resistance to Ug99 in the wheat cultivar Webster. Theor. Appl. Genet. 121, 65–69. doi: 10.1007/s00122-010-1291-z
Hiebert, C. W., Fetch, T. G., Zegeye, T., Thomas, J. B., Somers, D. J., Humphreys, D. G., et al. (2011). Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ‘Peace’ and ‘AC Cadillac’. Theor. Appl. Genet. 122, 143–149. doi: 10.1007/s00122-010-1430-6
Jin, Y., and Singh, R. P. (2006). Resistance in U.S. wheat to recent Eastern African isolates of Puccinia graminis f. sp. tritici with virulence to resistance gene Sr31. Plant Dis. 90, 476–480. doi: 10.1094/PD-90-0476
Jin, Y., Singh, R. P., Ward, R. W., Wanyera, R., Kinyua, M., Njau, P., et al. (2007). Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 91, 1096–1099. doi: 10.1094/PDIS-91-9-1096
Jin, Y., Szabo, L. J., Pretorius, Z. A., Singh, R. P., Ward, R., and Fetch, T. (2008). Detection of virulence to resistance gene Sr24 with race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 92, 923–926. doi: 10.1094/PDIS-92-6-0923
Jin, Y., Szabo, L. J., Rouse, M. N., Fetch, T. Jr., Pretorus, Z. A., Wanyera, R., et al. (2009). Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinia graminis f. sp. tritici. Plant Dis. 93, 367–370. doi: 10.1094/PDIS-93-4-0367
Klindworth, D. L., Niu, Z., Chao, S., Friesen, T. L., Jin, Y., Faris, J. D., et al. (2012). Introgression and characterization of a goatgrass gene for a high level of resistance to Ug99 stem rust in tetraploid wheat. G3 2, 665–673. doi: 10.1534/g3.112.002386
Knott, D. R. (1966). “The inheritance of stem rust resistance in wheat,” in Proceedings of the Second International Wheat Genetics Symposium, ed. J. MacKey (Lund: Heriditas Supplement 2), 156–166.
Kolmer, J. A., Garvin, D. F., and Jin, Y. (2011). Expression of a Thatcher wheat adult plant stem rust resistance QTL on chromosome arm 2BL is enhanced by Lr34. Crop Sci. 51, 526–533. doi: 10.2135/cropsci2010.06.0381
Kosambi, D. D. (1943). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lagudah, E. S., McFadden, H., Singh, R. P., Huerta-Espino, J., Bariana, H. S., and Spielmeyer, W. (2006). Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor. Appl. Genet. 114, 21–30. doi: 10.1007/s00122-006-0406-z
Li, H., Ribaut, J.-M., Li, Z., and Wang, J. (2008). Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet. 116, 243–260. doi: 10.1007/s00122-007-0663-5
Liu, S., Yu, L.-X., Singh, R. P., Jin, Y., Sorrells, M. E., and Anderson, J. A. (2010). Diagnostic and co-dominant PCR markers for wheat stem rust resistance genes Sr25 and Sr26. Theor. Appl. Genet. 120, 691–697. doi: 10.1007/s00122-009-1186-z
Liu, W., Jin, Y., Rouse, M., Friebe, B., Gill, B., and Pumphrey, M. O. (2011a). Development and characterization of wheat-Ae. searsii Robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor. Appl. Genet. 122, 1537–1545. doi: 10.1007/s00122-011-1553-4
Liu, W., Rouse, M., Friebe, B., Jin, Y., Gill, B., and Pumphrey, M. O. (2011b). Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosome Res. 19, 669–682. doi: 10.1007/s10577-011-9226-3
Loegering, W. Q. (1975). An allele for low reaction to Puccinia graminis tritici in Chinese Spring wheat. Phytopathology 65:925. doi: 10.1094/Phyto-65-925
Mago, R., Bariana, H. S., Dundas, I. S., Spielmeyer, W., Lawrence, G. J., Pryor, A. J., et al. (2005). Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504. doi: 10.1007/s00122-005-2039-z
Mago, R., Brown-Guedira, G., Dreisigacker, S., Breen, J., Jin, Y., Singh, R., et al. (2011). An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theor. Appl. Genet. 122, 735–744. doi: 10.1007/s00122-010-1482-7
Mago, R., Zhang, P., Vautrin, S., Simkova, H., Bansal, U., Luo, M. C., et al. (2015). The wheat Sr50 reveals a rich diversity at a cereal disease resistance locus. Nat. Plants 1:15186. doi: 10.1038/nplants.2015.186
McIntosh, R. A., Dubcovsky, J., Rogers, J., Morris, C., Appels, R., and Xia, X. C. (2016). Catalogue of Gene Symbols for Wheat: 2015-16 Supplement. Available at: https://shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2015.pdf.
McIntosh, R. A., and Luig, N. H. (1973). “Recombination between genes for reaction to P. graminis at or near the Sr9 locus,” in Proceedings of The Fourth International Wheat Genetics Symposium. Agricultural Experiment Station, eds E. R. Sears and L. M. S. Sears (Columbia, MO: University of Missouri), 425–432.
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: an Atlas of Resistance Genes. East Melbourne: CSIRO Publications, 93–99. doi: 10.1007/978-94-011-0083-0
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Newcomb, M., Olivera, P. D., Rouse, M. N., Szabo, L. J., Johnson, J., Gale, S., et al. (2016). Kenyan isolates of Puccinia graminis f. sp. tritici from 2008 to 2014: virulence to SrTmp in the Ug99 race group and implications for breeding programs. Phytopathology 106, 729–736. doi: 10.1094/PHYTO-11-14-0302-FI
Nirmala, J., Saini, J., Newcomb, M., Olivera, P., Gale, S., Klindworth, D., et al. (2017). Discovery of a novel stem rust resistance allele in durum wheat that exhibits differential reactions to Ug99 isolates. G3 7, 3481–3490. doi: 10.1534/g3.117.300209
Njau, P. N., Bhavani, S., Huerta-Espino, J., Keller, B., and Singh, R. P. (2013). Identification of QTL associated with durable adult plant resistance to stem rust race Ug99 in wheat cultivar “Pavon 76”. Euphytica 190, 33–44. doi: 10.1007/s10681-012-0763-4
Njau, P. N., Jin, Y., Huerta-Espino, J., Keller, B., and Singh, R. P. (2010). Identification and evaluation of sources of resistance to stem rust race Ug99 in wheat. Plant Dis. 94, 413–419. doi: 10.1094/PDIS-94-4-0413
Olson, E. L., Rouse, M. N., Pumphrey, M. O., Bowden, R. L., Gill, B. S., and Poland, J. A. (2013a). Introgression of stem rust resistance genes SrTA10187 and SrTA10171 from Aegilops tauschii to wheat. Theor. Appl. Genet. 126, 2477–2484. doi: 10.1007/s00122-013-2148-z
Olson, E. L., Rouse, M. N., Pumphrey, M. O., Bowden, R. L., Gill, B. S., and Poland, J. A. (2013b). Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor. Appl. Genet. 126, 1179–1188. doi: 10.1007/s00122-013-2045-5
Periyannan, K. S., Bansal, U. K., Bariana, H. S., Pumphrey, M., and Lagudah, E. S. (2011). A robust molecular marker for the detection of shortened introgressed segment carrying the stem rust resistance gene Sr22 in common wheat. Theor. Appl. Genet. 122, 1–7. doi: 10.1007/s00122-010-1417-3
Periyannan, S., Bansal, U., Bariana, H., Deal, K., Luo, M. C., Dvorak, J., et al. (2014). Identification of a robust molecular marker for the detection of the stem rust resistance gene Sr45 in common wheat. Theor. Appl. Genet. 127, 947–955. doi: 10.1007/s00122-014-2270-6
Periyannan, S., Moore, J., Ayliffe, M., Bansal, U., Wang, X., Huang, L., et al. (2013). The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788. doi: 10.1126/science.1239028
Peterson, R. F., Campbell, A. B., and Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Pretorius, Z. A., Bender, C. M., Visser, B., and Terefe, T. (2010). First report of a Puccinia graminis f. sp. tritici race virulent to the Sr24 and Sr31 wheat stem rust resistance genes in South Africa. Plant Dis. 94:784. doi: 10.1094/PDIS-94-6-0784C
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., and Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/PDIS.2000.84.2.203B
Pretorius, Z. A., Szabo, L. J., Boshoff, W. H. P., Herselman, L., and Visser, B. (2012). First report of a new TTKSF race of wheat stem rust (Puccinia graminis f. sp. tritici) in South Africa and Zimbabwe. Plant Dis. 96:590. doi: 10.1094/PDIS-12-11-1027-PDN
Qi, L. L., Pumphrey, M. O., Friebe, B., Zhang, P., Qian, C., Bowden, R. L., et al. (2011). A novel Robertsonian translocation event leads to transfer of a stem rust resistance gene (Sr52) effective against race Ug99 from Dasypyrum villosum into bread wheat. Theor. Appl. Genet. 123, 159–167. doi: 10.1007/s00122-011-1574-z
Rahmatov, M., Rouse, M. N., Nirmala, J., Danilova, T., Friebe, B., Steffenson, B. J., et al. (2016). A new 2DS.2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. doi: 10.1007/s00122-016-2710-6
Ramirez-Gonzalez, R. H., Uauy, C., and Caccamo, M. (2015). PolyMarker: a fast polyploid primer design pipeline. Bioinformatics 31, 2038–2039. doi: 10.1093/bioinformatics/btv069
Roder, M. S., Korzun, V., Wendehake, K., Plaschke, J., Tixier, M. H., Leroy, P., et al. (1998). A microsatellite map of wheat. Genetics 149, 2007–2023.
Roder, M. S., Plaschke, J., Konig, S. U., Börner, A., Sorrells, M. E., Tanksley, S. D., et al. (1995). Abundance, variability and chromosomal location of microsatellites in wheat. Mol. Genet. Genomics 246, 327–333. doi: 10.1007/BF00288605
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Diseases of Wheat: Concepts and Methods of Disease Management. Mexico: CIMMYT.
Rouse, M. N., and Jin, Y. (2011). Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis. 95, 941–944. doi: 10.1094/PDIS-04-10-0260
Rouse, M. N., Nava, I. C., Chao, S., Anderson, J. A., and Jin, Y. (2012). Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor. Appl. Genet. 125, 877–885. doi: 10.1007/s00122-012-1879-6
Rouse, M. N., Nirmala, J., Jin, Y., Chao, S., Fetch, T. G. Jr., Pretorius, Z. A., et al. (2014). Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor. Appl. Genet. 127, 1681–1688. doi: 10.1007/s00122-014-2330-y
Rouse, M. N, Olson, E., Gill, B., Pumphrey, M., and Jin, Y. (2011a). Stem rust resistance in Aegilops tauschii germplasm. Crop Sci. 51, 2074–2078. doi: 10.2135/cropsci2010.12.0719
Rouse, M. N., Wanyera, R., Njau, P., and Jin, Y. (2011b). Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis. 95, 762–766. doi: 10.1094/PDIS-12-10-0940
Saari, E. E., and Prescott, J. M. (1985). “World distribution in relation to economic losses,” in The Cereal Rusts: Diseases, Distribution, Epidemiology and Control, Vol. 2, eds A. P. Roelfs and W. R. Bushnell (Orlando,FL: Academic Press), 259–298. doi: 10.1016/B978-0-12-148402-6.50017-1
Saintenac, C., Zhang, W., Salcedo, A., Rouse, M. N., Trick, H. N., Akhunov, E., et al. (2013). Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341, 783–786. doi: 10.1126/science.1239022
Singh, R. P., Hodson, D. P., Huerta-Espino, J., Jin, Y., Bhavani, S., Njau, P., et al. (2011). The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49, 465–481. doi: 10.1146/annurev-phyto-072910-095423
Singh, R. P., Hodson, D. P., Huerta-Espino, J., Jin, Y., Njau, P., Wanyera, R., et al. (2008). Will stem rust destroy the world’s wheat crop? Adv. Agron. 98, 271–309. doi: 10.1016/S0065-2113(08)00205-8
Singh, R. P., Hodson, D. P., Jin, Y., Huerta-Espino, J., Kinyua, M. G., Wanyera, R., et al. (2006). Current status, likely migration and strategies to mitigate the threat to wheat production from race Ug99 (TTKS) of stem rust pathogen. CAB Rev. Persp. Agric. Veter. Sci. Nutr. Nat. Resour. 1, 1–13. doi: 10.1079/PAVSNNR20061054
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Somers, D., Isaac, P., and Edwards, K. (2004). A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114. doi: 10.1007/s00122-004-1740-7
Song, Q. J., Shi, J. R., Singh, S., Fickus, E. W., Costa, J. M., Lewis, J., et al. (2005). Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110, 550–560. doi: 10.1007/s00122-004-1871-x
Sourdille, P., Guyomarc’h, H., Baron, C., Gandon, B., Chiquet, V., Artiguenave, F., et al. (2001). Improvement of the Genetic Maps of Wheat using New Microsatellite Markers. Plant & Animal Genome IX, Final Abstracts Guide. Foster City, CA: Applied Biosystems Press, 167.
Stakman, E. C., Stewart, D. M., and Loegering, W. Q. (1962). Identification of Physiologic Races of Puccinia graminis var. tritici. Maryland: United States Department of Agriculture, Agricultural Research Service.
Tsilo, T. J., Jin, Y., and Anderson, J. A. (2007). Microsatellite markers linked to stem rust resistance allele Sr9a in wheat. Crop Sci. 47, 2013–2020. doi: 10.2135/cropsci2007.02.0087
Visser, B., Herselman, L., Park, R. F., Karaoglu, H., Bender, C. M., and Pretorius, Z. A. (2011). Characterization of two new Puccinia graminis f. sp. tritici races within the Ug99 lineage in South Africa. Euphytica 179, 119–127. doi: 10.1007/s10681-010-0269-x
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, S. D., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wilkinson, P. A., Winfield, M. O., Barker, G. L. A., Allen, A. M., Burridge, A., Coghill, J. A., et al. (2012). CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinformatics 13:219. doi: 10.1186/1471-2105-13-219
Keywords: QTL mapping, SNP, Puccinia graminis f. sp. tritici, Triticum aestivum L., adult plant resistance
Citation: Randhawa MS, Singh RP, Dreisigacker S, Bhavani S, Huerta-Espino J, Rouse MN, Nirmala J and Sandoval-Sanchez M (2018) Identification and Validation of a Common Stem Rust Resistance Locus in Two Bi-parental Populations. Front. Plant Sci. 9:1788. doi: 10.3389/fpls.2018.01788
Received: 17 August 2018; Accepted: 16 November 2018;
Published: 30 November 2018.
Edited by:
Roberto Papa, Università Politecnica delle Marche, ItalyReviewed by:
Benoit Bertrand, Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), FranceDaniela Marone, Cereal Research Centre, CRA-CER, Italy
Copyright © 2018 Randhawa, Singh, Dreisigacker, Bhavani, Huerta-Espino, Rouse, Nirmala and Sandoval-Sanchez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mandeep S. Randhawa, bS5yYW5kaGF3YUBjZ2lhci5vcmc=
 Mandeep S. Randhawa
Mandeep S. Randhawa Ravi P. Singh
Ravi P. Singh Susanne Dreisigacker
Susanne Dreisigacker Sridhar Bhavani
Sridhar Bhavani Julio Huerta-Espino
Julio Huerta-Espino Matthew N. Rouse
Matthew N. Rouse Jayaveeramuthu Nirmala
Jayaveeramuthu Nirmala Maricarmen Sandoval-Sanchez
Maricarmen Sandoval-Sanchez