- Department of Entomology, Animal Pests and Biotechnology, Institute of Plant Protection – National Research Institute, Poznań, Poland
Positive-sense single-stranded plant RNA viruses are obligate intracellular parasites that infect many agriculturally important crops. Most known plant RNA viruses are characterized by small genomes encoding a limited number of multifunctional viral proteins. Viral pathogens are considered to be absolutely dependent on their hosts, and viruses must recruit numerous host proteins and other factors for genomic RNA replication. Overall, the replication process depends on virus–plant protein-protein, RNA–protein and protein–lipid interactions. Recent publications provide strong evidence for the important role of chloroplasts in viral RNA synthesis. The chloroplast is considered to be a multifunctional organelle responsible for photosynthesis and for the generation of plant defense signaling molecules. High-throughput technologies (genomics and proteomics), and electron microscopy, including three-dimensional tomography, have revealed that several groups of plant RNA viruses utilize chloroplast membranes to assemble viral replication complexes (VRCs). Moreover, some chloroplast-related proteins reportedly interact with both viral proteins and their genomic RNAs and participate in trafficking these molecules to the chloroplast, where replication occurs. Here, we present the current knowledge on the important role of chloroplasts in the viral replication process.
Introduction
Positive-sense, single-stranded [(+)ssRNA] viruses are the main agents of many serious diseases of agriculturally important crops. Despite their relatively small genomes, these viruses that encode only a few proteins can efficiently replicate in their host cells (Hyodo and Okuno, 2016). Viruses have evolved the ability to harness and reprogramme host cell metabolism, facilitating their propagation and the infection process and suppressing plant defense mechanisms (Nagy and Pogany, 2012; Wang and Li, 2012; de Castro et al., 2013). Indeed, viruses are able to hijack host factors, such as proteins, intracellular membranes and their modifiers, lipids, and metabolites, leading to the establishment of viral replication complexes (VRCs), sites where viruses assemble and mature (Nagy and Pogany, 2012). These viral factories may utilize organelles, such as the endoplasmic reticulum (ER), mitochondria, vacuoles, Golgi apparatus, peroxisomes, tonoplasts, and chloroplasts, as well as the plasma membrane (Sanfaçon, 2005; Nagy and Pogany, 2012; Xu and Nagy, 2014; Hyodo and Okuno, 2016). Xu and Nagy (2014) reviewed that members of virus genera, including tobamoviruses (tomato mosaic virus), potyviruses (tobacco etch virus, TEV; turnip mosaic virus, TuMV), and tombusviruses (tomato bushy stunt virus, TBSV), exhibit flexibility in the selection of subcellular compartments for viral factory biogenesis, suggesting that these viruses can utilize multiple organelles for replication, depending on the conditions or host species. Notably, results obtained mainly using plant viruses that are able to replicate in yeast, as exemplified by TBSV, showed that in the absence of peroxisomes, which are considered the primary site of replication, the ER provides an alternative subcellular membrane and that viruses are able to replicate on the ER surface. These findings highlight that membrane availability is a critical factor for virus replication (Jonczyk et al., 2007).
Despite their various locations, VRCs share common features such as comprising invaginated spherules/vesicles that are connected to the cytoplasm via a neck-like opening and single- and/or double-membrane systems (Grangeon et al., 2012; Jin et al., 2018b). Due to the development of advanced technologies in cellular, molecular, and structural biology, the identification of co-opted host factors and the biogenesis of viral factories are being widely studied, and chloroplasts have been found to be among the most important organelles for viral RNA synthesis. Accordingly, there is increasing interest in understanding the role of chloroplasts in virus replication.
The chloroplast is considered the most dynamic organelle in a plant cells. It is responsible for photosynthesis and plays an active role in antiviral defense involving the synthesis of phytohormones, including salicylic acid, and jasmonic acid, as well as the production of reactive oxygen species (ROS); chloroplasts are also crucial for interorganellar signaling (Bhattacharyya and Chakraborty, 2018). Zhao et al. (2016) reviewed a number of studies showing that chloroplasts are susceptible to damage during virus infection, leading to disease symptoms such as mosaics and chlorosis resulting from cytopathogenic effects in chloroplast structure or function. Additionally, chloroplast factors are known to participate in the movement of viral particles.
In this review, we describe chloroplast–virus interactions on the basis of recent evidence highlighting the direct function of this organelle in virus RNA synthesis. We also list the most useful molecular approaches for studying viral RNA replication in this context.
Requirement of the Chloroplast Membrane for Plant ss(+)Rna Virus Replication
Although ss(+)RNA viruses display various genetic organization and expression strategies, they all replicate in a similar manner. All known plant ss(+)RNA viruses encode RNA-dependent RNA polymerase (RdRp) and ancillary replication factors. These viral proteins, which are produced in the early stage of infection, are involved in the assembly of VRCs and help to stabilize and facilitate optimal conditions for RNA synthesis. Viral and host proteins are selectively bound to the ss(+)RNA template and target subcellular membranes, which are later reorganized around the favorable microenvironment to form VRCs (Jin et al., 2018a). Within VRCs, negative-strand RNA intermediates are synthetized and then used as templates for the synthesis of new (+)RNA strands (Figure 1). The progeny RNAs are then released into the cytosol, where they might again be translated and/or replicated or encapsidated and transferred to adjacent cells. Indeed, the organellar compartmentalization of VRCs protects viral RNAs from being destroyed by host defense RNA degradation while simultaneously maximizing the concentration of viral and host factors required for replication (Nagy and Pogany, 2012). Previously published data reveal that chloroplasts serve as replication sites for several viruses, including hordeiviruses (barley stripe mosaic virus, BSMV) (Carroll, 1970; Torrance et al., 2006), potyviruses [maize draft mosaic virus (MDMV) (Mayhew and Ford, 1974), plum pox virus (Martin et al., 1995), TEV (Gadh and Hari, 1986)], TuMV (Kitajima and Costa, 1973), and tymoviruses (turnip yellow mosaic virus, TYMV) (Lafleche et al., 1972; Dreher, 2004). De Graaff et al. (1993) reported that VRCs of alfalfa mosaic virus (AMV), belonging to the Alfamovirus genus, are also associated with the chloroplast envelope. However, further studies performed by van der Heijden et al. (2001) and Ibrahim et al. (2012) using live cell imaging of Arabidopsis thaliana protoplasts showed that AMV replicates on tonoplast membranes.
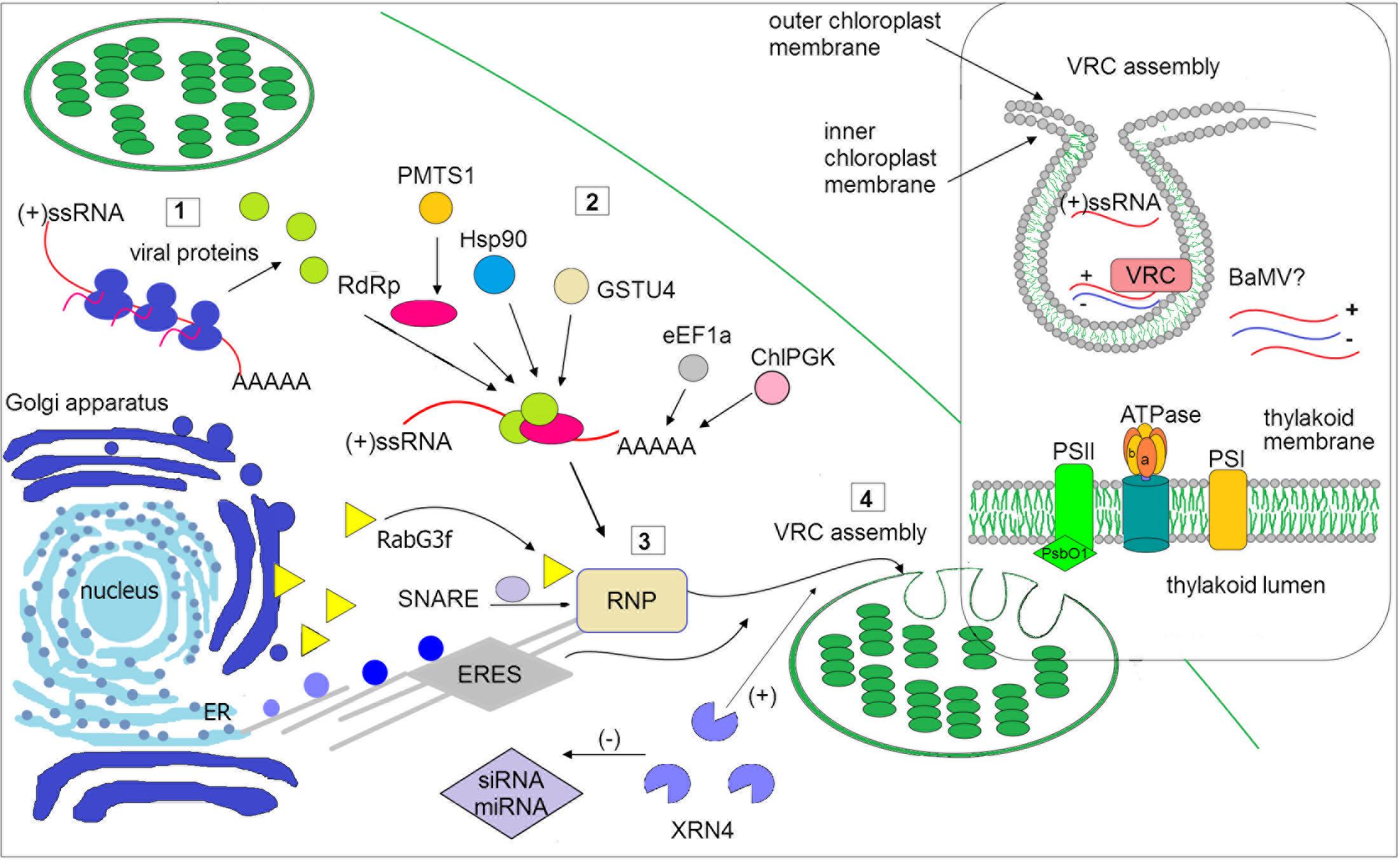
FIGURE 1. Schematic illustration of selected host factors affecting viral RNA replication in chloroplasts. Synthesis of viral genomic RNA [(+)ssRNA] requires viral proteins: RdRp (pink circle) and auxiliary factors (green circles) that are translated at the early stage of infection (1). Then, virus recruits host factors (2) such as chaperone (Hsp90), methyltransferase (e.g., PMTS1), chloroplast phosphoglycerate kinase (chlPGK), which participate in viral replication complexes (VRC) targeting (in potexvirus). The 3′UTR of viral RNAs (in potexviruses, e.g., BaMV) interacts with several cellular proteins, including elongation factor 1a (eEF1a), glutathione S-transferase (GSTU4), exonuclease (XRN4). Exonuclease negatively regulates the activity of silencing pathway (miRNA/siRNA). Subsequently, the hijacked host factors, viral genomic RNA and viral proteins are trafficked to chloroplast membranes (3). The ribonucleoprotein complex (RNP) is transported from endoplasmic reticulum (ER) exit sites ERES (in potyvirus) or via targeting proteins, such as Rabs or SNARE (in potexvirus) and actomyosin motility system. Chloroplast membranes are later reorganized to form VRC (4). The panel on the right depicts spherule where the viral RNA is synthesized. The host photosynthetic protein PsbO1 from photosystem II is recruited by potyvirus proteins playing a role in its replication (TVBMV, for details refer to the text). In thylakoid membranes, the β subunit of ATPase has been also involved in VRC assembly (potexvirus and tymovirus). Among viruses, viral RNA of BaMV was observed inside the chloroplast. The presented host factors participate in trafficking to the site of replication or affect (directly or indirectly) the synthesis of viral RNA, that is associated with chloroplast.
The replication complex of the aforementioned virus families is associated with the outer chloroplast membrane, particularly with peripheral vesicles and cytoplasmic invaginations (CIs) on the surface. Among recently identified host factors participating in the viral replication process are several chloroplast and chloroplast-related proteins (Table 1). Virus-induced membrane proliferation and remodeling are very complex processes, and all eukaryotic (+)RNA viruses induce an interaction network between host and viral factors to form VRCs in subcellular membranes (Nagy and Pogany, 2012; Hyodo and Okuno, 2014). In addition to viral RNA and ancillary replication factors, viruses recruit multiple host RNA-modifying enzymes, translation factors, RNA helicases, including DEAD-Box RNA helicases (in plants and yeast), and methyltransferase. The VRCs of different plant virus families reportedly might contain the following components: chaperones (facilitating proper folding of RNA), viral RNA, and proteins, host endosomal sorting complex required for transport (ESCRT) that participates also in membrane deformation (Nagy and Pogany, 2012; Hyodo and Okuno, 2014; Geng et al., 2017), as well as other cellular proteins involved in lipid metabolism and transport (Barajas et al., 2014).
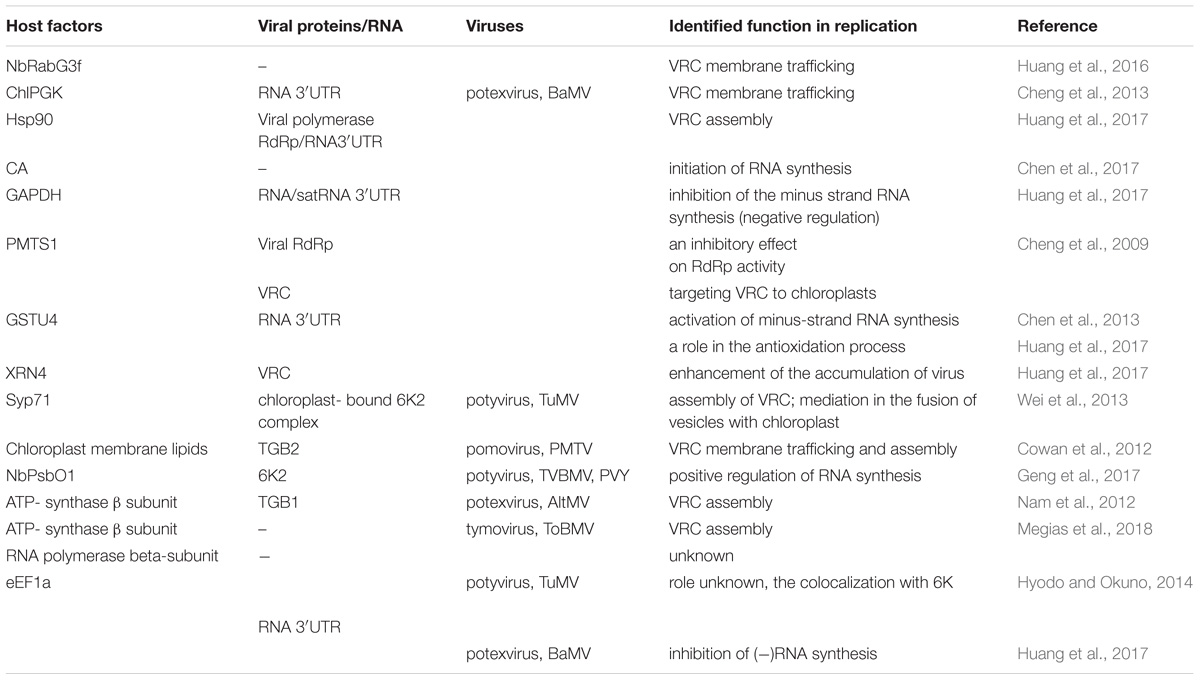
TABLE 1. List of host chloroplast and chloroplast-related factors and viral protein interactions involved in Synthesis of viral genomic RNA [(+)ssRNA] plant virus replication.
Recently published data indicate the role of the 140K/98K protein domains of TYMV, which possess two amphipathic helices (αA and αB) with a 41-residue internal sequence described as the chloroplast envelope-targeting domain (CTD) (Moriceau et al., 2017). Based on the results of structural analyses involving nuclear magnetic resonance (NMR) spectroscopy and molecular dynamics simulations, it was reported that amphipathic helices within viral proteins might serve as membrane-anchoring domains able to promote changes in lipid organization, thereby modulating membrane bilayer properties. However, in TYMV, the detailed delivery mechanisms remain to be elucidated (Moriceau et al., 2017). Nonetheless, Jin et al. (2018b) published data for 3D electron tomography (ET) characterization of the architecture of BSMV replication factories and revealed that chloroplast invaginations are composed of inner chloroplast membrane-derived packets containing variable numbers of outer membrane-derived spherules. Furthermore, studies by Torrance et al. (2006) and immunolocalization experiments by Zhang et al. (2017) show that BSMV genomic RNAs, replicative dsRNA intermediates, triple gene block 2 (TGB2) and viral γb proteins accumulate at chloroplasts. Importantly, the γa protein possesses RdRp activity and interacts with λa to induce the replication process. λa, which contains highly conserved methyltransferase and helicase domains, also plays a significant role in chloroplast membrane rearrangement. It has been shown that λa binds directly to γb and that this interaction is absolutely necessary for chloroplast targeting. Moreover, the γb protein is reported to enhance the helicase activity of λa (Zhang et al., 2017; Jin et al., 2018b).
In potyviruses, for example TEV and TuMV, the transport of viral proteins and genomic RNA to sites of replication occurs via vesicles from ER-exit sites (ERESs) to the outer chloroplast membrane (Wei and Wang, 2008; Wei et al., 2010). In addition, a model of the biogenesis and trafficking of viral 6K2 vesicles has been proposed based on confocal microscopy imaging (Wei et al., 2010). Potyvirus 6K2, an integral membrane protein, induces the formation of ER-derived vesicles that traffic predominantly from the ER to the Golgi apparatus and then target chloroplasts depending on the activities of the coat protein complexes COPI and COPII as well as the actomyosin motility system (Wei et al., 2010, 2013; Hyodo and Okuno, 2014; Zhao et al., 2016; Bhattacharyya and Chakraborty, 2018; Jin et al., 2018a; Figure 1). Fusion of virus-induced vesicles with chloroplast membranes and also chloroplast amalgamation rely directly on SNARE proteins (soluble NSF (N-ethylmaleimide – sensitive factor) attachment protein receptor) that are essential host factors for successful virus infection. For example, the cellular SNARE protein Syp71 directly mediates the fusion of vesicles with chloroplasts (Wei et al., 2013; Figure 1).
Cowan et al. (2012), performed lipid interaction assays using confocal laser scanning microscopy and biochemical analysis to determine that chloroplast membrane lipids associate with viral TGB2 of potato mop-top virus (PMTV), facilitating localization of viral genomic RNA to chloroplast membranes for replication. These findings confirmed that PMTV induces large CIs in chloroplasts, though the detailed ultrastructure has not yet been determined (Cowan et al., 2012). Moreover, fluorescence in situ hybridization studies of alternanthera mosaic virus (AltMV, a member of the genus Potexvirus) showed that viral RNA and TGB3, which possesses a chloroplast-targeting signal, primarily accumulate near the surface of chloroplast membranes (Lim et al., 2010). Further analyses proved that the TGB1 protein interacts with host chloroplast β-ATPase in Nicotiana benthamiana and A. thaliana (Nam et al., 2012). These data suggest that AltMV replication occurs at the outer chloroplast membrane. Additionally, interaction with the ATP-synthase β subunit was indicated by chloroplast proteome analyses of N. benthamiana plants infected with a member of the Tymovirus genus, tomato blistering mosaic virus (ToBMV), with VRCs adjacent to the chloroplast membrane (Megias et al., 2018). Moreover, in plants infected with ToBMV, increased expression of the chloroplast RNA polymerase β subunit was also observed during ToBMV infection. Considering that viruses encode RNA polymerases, the role of the increased gene expression of chloroplast polymerase during ToBMV infection remains unknown (Megias et al., 2018). Importantly, nuclear-encoded chloroplast RNA polymerases have a direct function in the replication of viroids of the family Avsunviroidae (Navarro et al., 1999, 2000; Daròs and Flores, 2002). These viroids are noncoding, circular RNAs with hammerhead ribozyme activity that accumulate and replicate in chloroplasts (Flores et al., 2005). Overall, the mechanism of viroid transport is still being studied. Gómez and Pallás (2010, 2012) suggest that avsunviroids are at first translocated to the nucleus and then employ a nucleus-chloroplast transport mechanism to enter chloroplasts for replication. Members of the Avsunviroidae replicate through a symmetric rolling circle mechanism, in which RNA circularization is catalyzed by chloroplastic tRNA ligase (Nohales et al., 2012; Cordero et al., 2018). Viroids hijack host factors for efficient replication, as do viruses, and such host factors as an aminomethylotransferase, chaperones, a dynamin-like guanosine triphosphatases (GTP)-binding protein, ribosomal protein L5, and elongation factor eEF1a, have been identified as binding to peach latent mosaic viroid (PLMVd) in vitro (Dubé et al., 2009); however, their importance for pathogen replication remains unclear. The results obtained through analyses of the eEF1a–PLMVd interaction suggest its role in the initiation of viroid replication, as occurs for several viruses, e.g., TYMV (Joshi et al., 1986) and BMV (Bastin and Hall, 1976). In addition, Daròs and Flores (2002) showed that RNA-binding proteins PARBP33 and PARBP35 (from avocado) bind to the RNA of avocado sunblotch viroid, the type member of the Avsunviroidae, and act as chaperones that presumably assist and stimulate self-cleavage.
The Role of Host Proteins in Regulating the Viral Replication Process in Chloroplasts
Cheng et al. (2013) have reported interesting results concerning virus replication sites, showing that the minus-strand RNA of bamboo mosaic virus (BaMV, a potexvirus) is found not only in the organellar membrane but also throughout the chloroplast. Chloroplast protein phosphoglycerate kinase (ChlPGK) may also participate in targeting of VRCs to chloroplasts. Indeed, PGK interacts with the 3′ untranslated region (UTR) of the BaMV genomic RNA, and, together with the host heat shock proteins (Hsp90), translocates the viral ribonucleoprotein complex (RNP) across chloroplast membranes to the stroma and then stimulates VRC assembly in association with the thylakoid membranes (Cheng et al., 2013). Screening analyses of differentially expressed genes in plants during BaMV infection have indicated that NbRabG3f is up-regulated. Rabs comprise a group of small GTPases that regulate membrane trafficking, including vesicle budding, delivery, tethering, and fusion with target compartment membranes (Huang et al., 2016). BaMV may employ NbRabG3f to form vesicles derived from the Golgi membrane for intracellular trafficking to deliver factors to its replication site (Figure 1). GTPase activity and membrane-targeting ability are both crucial for virus replication (Huang et al., 2017).
The available evidence indicates that some host factors involved in viral replication in chloroplasts may enhance or inhibit this process. Analyses of the host factors assisting in BaMV replication have revealed at least three proteins, together with the abovementioned ChlPGK and Hsp90 may interact with the 3’ end of the viral genomic RNA. Among them are the eEF1a, glutathione S-transferase NbGSTU4 (from N. benthamiana), and exonuclease XRN4, which all play a crucial role in the early stage of viral replication (Cheng et al., 2013; Chen et al., 2017; Huang et al., 2017; Figure 1). For instance, XRN4 functions in reducing the activities of siRNA- and miRNA-mediated RNA silencing pathways in Arabidopsis (Souret et al., 2004). Importantly, NbXRN4 acts as an antiviral agent that attenuates accumulation of tobacco mosaic virus and tomato bushy mosaic virus. In contrast, Lee et al. (2016) indicated an enhancer role for NbXRN4 in BaMV replication, which is also exhibited in the presence of the universal silencing suppressor P19. Indeed, Huang et al. (2017) suggested that during viral RNP trafficking to chloroplasts, NbXRN4 may reduce the activities of siRNA-mediated silencing and, consequently, positively regulate BaMV RNA accumulation; this view, however, should be confirmed experimentally. It is worth noting that VRCs require antioxidant enzymes once the viral BaMV RNP complex is transported to the chloroplast, which is the major producer of (ROS) in plant cells. Oxidative stress may be attenuated via enzymatic activity by glutathione transferases (GSTs), and the 3′UTR-associated protein NbGSTU4 that in the presence of glutathione is able to neutralize oxidative stress inside chloroplasts, enabling efficient minus-strand RNA synthesis (Huang et al., 2017). It has also been documented that the initiation and elongation of viral BaMV RNA synthesis is positively regulated by nuclear-encoded plastidic carbonic anhydrase (CA), with the knockdown of this gene reducing accumulation of the BaMV coat protein (Chen et al., 2017). Furthermore, an increased level of CA was also observed by Obrepalska-Steplowska et al. (2015) during peanut stunt virus infection in N. benthamiana, which suggests that the observed differences in the expression levels of chloroplast-related genes might be dependent on the virus infection process.
Involvement of the oxygen-evolving complex in the regulation of virus replication has also been indicated. Geng et al. (2017) experimentally proved that down-regulation of NbPsbO1 expression suppressed the replication of tobacco vein banding mosaic virus (TVBMV) as well as PVY. Additionally, the 6K2 protein was recently reported to recruit 6K1 and hijack the chloroplast protein NbPsbO1, a component of the photosystem II oxygen-evolving complex that regulates potyvirus replication; consistently virus accumulation is significantly decreased in NbPsbO1-silenced plants.
The abovementioned examples show that viral pathogens hijack host factors for genome replication and production of new virions to ensure successful infection.
Molecular Approaches to Identify and Characterize Host Factors Required for Plant ss(+)Rna Virus Replication
Identification of host factors involved in virus replication may be achieved via genome-wide screening approaches in combination with global proteomics analyses. A number of studies on viral–host protein interactions are based on a yeast two-hybrid approach, whereas host RNA-binding proteins may be identified using a viral RNA probe incubated with cellular extracts (with or without ultraviolet crosslinking), followed by mass spectrometry (Wang and Li, 2012). For identifying lipid factors, lipidomics assays demonstrating differences in lipid composition are very useful, and electron microscopy (EM) methods are extremely helpful for direct visualization of the particular steps of the life cycle of a given virus. Moreover, the development of three-dimensional EM (3D-EM) in combination with ET approaches provides unprecedented insights into how viruses remodel the intracellular architecture of the host cell. Indeed, 3D imaging techniques applied for viral replication factory reconstruction have been described for BSMV, TBSV, beet black scorch virus, melon necrotic spot virus (the Tombusviridae family), potyviruses TuMV and TEV, and Bromoviridae, as exemplified by brome mosaic virus (Jin et al., 2018a,b). All these innovative approaches together with molecular in situ mapping are powerful tools that provide essential architectural information regarding VRC biogenesis and the function of these structures (de Castro et al., 2016; Jin et al., 2018a).
Conclusion
In summary, the abovementioned research studies broaden our knowledge of chloroplasts and their important direct and indirect roles in the replication of several plant virus species. In recent years, increasing numbers of chloroplast and chloroplast-related proteins have been shown to be involved in the formation of membranous replication factories and RNA synthesis. However, it is worth noting that in many cases the mechanism of interactions between viral and the identified plant factors as well as their influence on the regulation of replication remains unknown. Importantly, it should be noted that despite being replication sites, chloroplasts are primarily implicated in plant defense against viruses. Therefore, molecular insights into pathways exploited by viral pathogens and the implementation of innovative powerful imaging techniques to study plant–virus interactions is of great importance. The results obtained might contribute to the development of new and effective antiviral strategies against existing and emerging plant virus diseases.
Author Contributions
MB and AO-S conceived the review. MB wrote a large part of the manuscript. AO-S critically reviewed and complemented the review. Both authors have made a substantial, direct and intellectual contributions to the work, and approved it for publication.
Funding
This work was supported by grant NN 2016/21D/NZ9/02468 from Polish National Science Centre.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Barajas, D., Xu, K., de Castro Martín, I. F., Sasvari, Z., Brandizzi, F., Risco, C., et al. (2014). Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog. 10:e1004388. doi: 10.1371/journal.ppat.1004388
Bastin, M., and Hall, T. C. (1976). Interaction of elongation factor 1 with aminoacylated brome mosaic virus and tRNA’s. J. Virol. 20, 117–122.
Bhattacharyya, D., and Chakraborty, S. (2018). Chloroplast: the Trojan horse in plant–virus interaction. Mol. Plant Pathol. 19, 504–518. doi: 10.1111/mpp.12533
Carroll, T. W. (1970). Relation of Barley stripe mosaic virus to plastids. Virology 42, 1015–1022. doi: 10.1016/0042-6822(70)90350-8
Chen, I., Tsai, A. Y., Huang, Y. P., Wu, I. F., Cheng, S. F., Hsu, Y. H., et al. (2017). Nuclear-encoded plastidal carbonic anhydrase is involved in replication of Bamboo mosaic virus RNA in Nicotiana benthamiana. Front. Microbiol. 8:2046. doi: 10.3389/fmicb.2017.02046
Chen, I. H., Chiu, M. H., Cheng, S. F., Hsu, Y. H., and Tsai, C. H. (2013). The glutathione transferase of Nicotiana benthamiana Nb GSTU 4 plays a role in regulating the early replication of Bamboo mosaic virus. New Phytol. 199, 749–757. doi: 10.1111/nph.12304
Cheng, C. W., Hsiao, Y. Y., Wu, H. C., Chuang, C. M., Chen, J. S., Tsai, C. H., et al. (2009). Suppression of bamboo mosaic virus accumulation by a putative methyltransferase in Nicotiana benthamiana. J. Virol. 83, 5796–5805. doi: 10.1128/JVI.02471-08
Cheng, S. F., Huang, Y. P., Chen, L. H., Hsu, Y. H., and Tsai, C. H. (2013). Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 163, 1598–1608. doi: 10.1104/pp.113.229666
Cordero, T., Ortolá, B., and Daròs, J. A. (2018). Mutational analysis of eggplant latent viroid RNA circularization by the eggplant tRNA ligase in Escherichia coli. Front. Microbiol. 9:635. doi: 10.3389/fmicb.2018.00635
Cowan, G. H., Roberts, A. G., Chapman, S. N., Ziegler, A., Savenkov, E. I., and Torrance, L. (2012). The potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Front. Plant Sci. 3:290. doi: 10.3389/fpls.2012.00290
Daròs, J. A., and Flores, R. (2002). A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 21, 749–759. doi: 10.1093/emboj/21.4.749
de Castro, I. F., Tenorio, R., and Risco, C. (2016). Virus assembly factories in a lipid world. Cur. Opin. Virol. 18, 20–26. doi: 10.1016/j.coviro.2016.02.009
de Castro, I. F., Volonte, L., and Risco, C. (2013). Virus factories: biogenesis and structural design. Cell Microbiol. 15, 24–34. doi: 10.1111/cmi.12029
De Graaff, M., Coscoy, L., and Jaspars, E. M. J. (1993). Localization and biochemical characterization of alfalfa mosaic virus replication complexes. Virology 194, 878–881. doi: 10.1006/viro.1993.1335
Dreher, T. W. (2004). Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol. Plant Pathol. 5, 367–375. doi: 10.1111/j.1364-3703.2004.00236.x
Dubé, A., Bisaillon, M., and Perreault, J. P. (2009). Identification of proteins from Prunus persica that interact with peach latent mosaic viroid. J. Virol. 83, 12057–12067. doi: 10.1128/JVI.01151-09
Flores, R., Hernández, C., Martinez de Alba, A. E., Daròs, J. A., and Di Serio, F. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43, 117–139. doi: 10.1146/annurev.phyto.43.040204.140243
Gadh, I. P. S., and Hari, V. (1986). Association of tobacco etch virus related RNA with chloroplasts in extracts of infected plants. Virology 150, 304–307. doi: 10.1016/0042-6822(86)90292-8
Geng, C., Yan, Z. Y., Cheng, D. J., Liu, J., Tian, Y. P., Zhu, C. X., et al. (2017). Tobacco vein banding mosaic virus 6K2 protein hijacks NbPsbO1 for virus replication. Sci. Rep. 7:43455. doi: 10.1038/srep43455
Gómez, G., and Pallás, V. (2010). Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One 5:e12269. doi: 10.1371/journal.pone.0012269
Gómez, G., and Pallás, V. (2012). Studies on subcellular compartmentalization of plant pathogenic non coding RNAs give new insights into the intracellular RNA-traffic mechanisms. Plant Physiol. 159, 558–564. doi: 10.1104/pp.112.195214
Grangeon, R., Jiang, J., and Laliberté, J. F. (2012). Host endomembrane recruitment for plant RNA virus replication. Curr. Opin. Virol. 2, 683–690. doi: 10.1016/j.coviro.2012.10.003
Huang, Y. P., Chen, I., and Tsai, C. H. (2017). Host factors in the infection cycle of Bamboo mosaic virus. Front. Microbiol. 8:437. doi: 10.3389/fmicb.2017.00437
Huang, Y. P., Jhuo, J. H., Tsai, M. S., Tsai, C. H., Chen, H. C., Lin, N. S., et al. (2016). NbRABG 3f, a member of Rab GTPase, is involved in Bamboo mosaic virus infection in Nicotiana benthamiana. Mol. Plant Pathol. 17, 714–726. doi: 10.1111/mpp.12325
Hyodo, K., and Okuno, T. (2014). Host factors used by positive-strand RNA plant viruses for genome replication. J. Gen. Plant Pathol. 80, 123–135. doi: 10.1007/s10327-014-0505-7
Hyodo, K., and Okuno, T. (2016). Pathogenesis mediated by proviral host factors involved in translation and replication of plant positive-strand RNA viruses. Curr. Opin. Virol. 17, 11–18. doi: 10.1016/j.coviro.2015.11.004
Ibrahim, A., Hutchens, H. M., Berg, R. H., and Loesch-Fries, L. S. (2012). Alfalfa mosaic virus replicase proteins, P1 and P2, localize to the tonoplast in the presence of virus RNA. Virology 433, 449–461. doi: 10.1016/j.virol.2012.08.018
Jin, X., Cao, X., Wang, X., Jiang, J., Wan, J., Laliberté, J. F., et al. (2018a). Three-Dimensional architecture and biogenesis of membrane structures associated with plant virus replication. Front. Plant Sci. 9:57. doi: 10.3389/fpls.2018.00057
Jin, X., Jiang, Z., Zhang, K., Wang, P., Cao, X., Yue, N., et al. (2018b). Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiol. 176, 282–294. doi: 10.1104/pp.17.00871
Jonczyk, M., Pathak, K. B., Sharma, M., and Nagy, P. D. (2007). Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362, 320–330. doi: 10.1016/j.virol.2007.01.004
Joshi, R. L., Ravel, J. M., and Haenni, A. L. (1986). Interaction of turnip yellow mosaic virus Val-RNA with eukaryotic elongation factor EF-1 [alpha]. Search for a function. EMBO J. 5, 1143–1148. doi: 10.1002/j.1460-2075.1986.tb04339.x
Kitajima, E. W., and Costa, A. S. (1973). Aggregates of chloroplasts in local lesions induced in Chenopodium quinoa Wild. by Turnip mosaic virus. J. Gen. Virol. 20, 413–416. doi: 10.1099/0022-1317-20-3-413
Lafleche, D., Bové, C., Dupont, G., Mouches, C., Astier, T., Garnier, M., et al. (1972). Site of viral RNA replication in the cells of higher plants: TYMV-RNA synthesis on the chloroplast outer membrane system. Proc. Fed. Eur. Biochem. Soc. 27, 43–71.
Lee, C. C., Lin, T. L., Lin, J. W., Han, Y. T., Huang, Y. T., Hsu, Y. H., et al. (2016). Promotion of Bamboo Mosaic Virus accumulation in Nicotiana benthamiana by 5’→3’ exonuclease NbXRN4. Front. Microbiol. 6:1508. doi: 10.3389/fmicb.2015.01508
Lim, H. S., Vaira, A. M., Bae, H., Gim, Y. G., Kim, H. G., Cho, I. S., et al. (2010). Mutation of a chloroplast-targeting signal in Alternanthera mosaic virus TGB3 impairs cell-to-cell movement and eliminates long-distance virus movement. J. Gen. Virol. 91, 2102–2115. doi: 10.1099/vir.0.019448-0
Martin, M. T., Cervera, M. T., and Garcia, J. A. (1995). Properties of the active plum pox potyvirus RNA polymerase complex in defined glycerol gradient fractions. Virus Res. 37, 127–137. doi: 10.1016/0168-1702(95)00028-O
Mayhew, D. E., and Ford, R. E. (1974). Detection of ribonuclease-resistant RNA in chloroplasts of corn leaf tissue infected with maize dwarf mosaic virus. Virology 57, 503–509. doi: 10.1016/0042-6822(74)90189-5
Megias, E., do Carmo, L. S. T., Nicolini, C., Silva, L. P., Blawid, R., Nagata, T., et al. (2018). Chloroplast proteome of Nicotiana benthamiana infected by tomato blistering mosaic virus. Protein J. 37, 290–299. doi: 10.1007/s10930-018-9775-9
Moriceau, L., Jomat, L., Bressanelli, S., Alcaide-Loridan, C., and Jupin, I. (2017). Identification and molecular characterization of the chloroplast targeting domain of Turnip yellow mosaic virus replication proteins. Front. Plant Sci. 8:2138. doi: 10.3389/fpls.2017.02138
Nagy, P. D., and Pogany, J. (2012). The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 10, 137–149. doi: 10.1038/nrmicro2692
Nam, J., Jang, C., Li, M., Kim, H., Cho, S., Kim, H., et al. (2012). Alternanthera mosaic virus TGB1 interaction with chloroplast β ATPase is necessary for viral replication. Phytopathology 102(Suppl. 4):S4.85.
Navarro, J. A., Daròs, J. A., and Flores, R. (1999). Complexes containing both polarity strands of Avocado sunblotch viroid: identification in chloroplasts and characterization. Virology 253, 77–85. doi: 10.1006/viro.1998.9497
Navarro, J. A., Vera, A., and Flores, R. (2000). A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of Avocado sunblotch viroid. Virology 268, 218–225. doi: 10.1006/viro.1999.0161
Nohales, M. Á., Molina-Serrano, D., Flores, R., and Daròs, J. A. (2012). Involvement of the chloroplastic isoform of tRNA ligase in the replication of the viroids belonging to the family Avsunviroidae. J. Virol. 86, 8269–8276. doi: 10.1128/JVI.00629-12
Obrepalska-Steplowska, A., Renaut, J., Planchon, S., Przybylska, A., Wieczorek, P., Barylski, J., et al. (2015). Effect of temperature on the pathogenesis, accumulation of viral and satellite RNAs and on plant proteome in peanut stunt virus and satellite RNA-infected plants. Front. Plant Sci. 6:903. doi: 10.3389/fpls.2015.00903
Sanfaçon, H. (2005). Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Botany 83, 1529–1549.
Souret, F. F., Kastenmayer, J. P., and Green, P. J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15, 173–183. doi: 10.1016/j.molcel.2004.06.006
Torrance, L., Cowan, G. H., Gillespie, T., Ziegler, A., and Lacomme, C. (2006). Barley stripe mosaic virus-encoded proteins triple-gene block 2 and γb localize to chloroplasts in virus-infected monocot and dicot plants, revealing hitherto-unknown roles in virus replication. J. Gen. Virol. 87, 2403–2411. doi: 10.1099/vir.0.81975-0
van der Heijden, M. W., Carette, J. E., Reinhoud, P. J., Haegi, A., and Bol, J. F. (2001). Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virology 75, 1879–1887. doi: 10.1128/JVI.75.4.1879-1887.2001
Wang, R., and Li, K. (2012). Host factors in the replication of positive-strand RNA viruses. Chang Gung Med. J. 35, 111–124. doi: 10.4103/2319-4170.106160
Wei, T., Huang, T. S., McNeil, J., Laliberté, J. F., Hong, J., Nelson, R. S., et al. (2010). Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. doi: 10.1128/JVI.01824-09
Wei, T., and Wang, A. (2008). Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI-and COPII-dependent manner. J. Virol. 82, 12252–12264. doi: 10.1128/JVI.01329-08
Wei, T., Zhang, C., Hou, X., Sanfaçon, H., and Wang, A. (2013). The SNARE protein Syp71 is essential for Turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 9:1003378. doi: 10.1371/journal.ppat.1003378
Xu, K., and Nagy, P. D. (2014). Expanding use of multi-origin subcellular membranes by positive-strand RNA viruses during replication. Curr. Opin. Virol. 9, 119–126. doi: 10.1016/j.coviro.2014.09.015
Zhang, K., Zhang, Y., Yang, M., Liu, S., Li, Z., Wang, X., et al. (2017). The Barley stripe mosaic virus γb protein promotes chloroplast-targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog. 13:e1006319. doi: 10.1371/journal.ppat.1006319
Keywords: replication, chloroplast, chloroplast membrane, viral replication complex (VRC), plant-virus interaction, viroids
Citation: Budziszewska M and Obrępalska-Stęplowska A (2018) The Role of the Chloroplast in the Replication of Positive-Sense Single-Stranded Plant RNA Viruses. Front. Plant Sci. 9:1776. doi: 10.3389/fpls.2018.01776
Received: 19 July 2018; Accepted: 15 November 2018;
Published: 27 November 2018.
Edited by:
Ricardo Flores, Instituto de Biología Molecular y Celular de Plantas (IBMCP), SpainReviewed by:
Ching-Hsiu Tsai, National Chung Hsing University, TaiwanJesús Sánchez-Navarro, Universitat Politècnica de València, Spain
Copyright © 2018 Budziszewska and Obrępalska-Stęplowska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleksandra Obrępalska-Stęplowska, b2xhb2JAbzIucGw=
 Marta Budziszewska
Marta Budziszewska Aleksandra Obrępalska-Stęplowska
Aleksandra Obrępalska-Stęplowska