
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 23 November 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01732
This article is part of the Research Topic Forest Genomics and Biotechnology View all 20 articles
The CRISPR technology continues to diversify with a broadening array of applications that touch all kingdoms of life. The simplicity, versatility and species-independent nature of the CRISPR system offers researchers a previously unattainable level of precision and control over genomic modifications. Successful applications in forest, fruit and nut trees have demonstrated the efficacy of CRISPR technology at generating null mutations in the first generation. This eliminates the lengthy process of multigenerational crosses to obtain homozygous knockouts (KO). The high degree of genome heterozygosity in outcrossing trees is both a challenge and an opportunity for genome editing: a challenge because sequence polymorphisms at the target site can render CRISPR editing ineffective; yet an opportunity because the power and specificity of CRISPR can be harnessed for allele-specific editing. Examination of CRISPR/Cas9-induced mutational profiles from published tree studies reveals the potential involvement of multiple DNA repair pathways, suggesting that the influence of sequence context at or near the target sites can define mutagenesis outcomes. For commercial production of elite trees that rely on vegetative propagation, available data suggest an excellent outlook for stable CRISPR-induced mutations and associated phenotypes over multiple clonal generations.
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-based genome editing is rapidly becoming the system of choice for targeted mutagenesis in a growing variety of woody species, including forest trees. Forest trees are an invaluable commodity, providing fiber, energy, materials and climate buffering to the global community, and CRISPR has the potential to further enhance these important traits. Previous-generation methods for gene silencing in plants rely on expression of antisense RNAs, small interfering RNAs or microRNAs to base-pair with target mRNAs for degradation, often with unpredictable and unstable outcomes (Alessandra and Shihshieh, 2010). The specificity and efficiency of CRISPR for targeted DNA mutations, and the ease of adoption in virtually any species are behind the current revolution in genomic editing (Jiang and Doudna, 2017). Meanwhile, CRISPR’s popularity is driving the discovery and characterization of new CRISPR-associated (Cas) endonucleases with novel properties that make the system even more versatile (Burstein et al., 2016; Murovec et al., 2017). This review will focus on recent applications of CRISPR in woody species, with a special focus on forest trees, the mutation patterns observed at target sites, and the long-term stability of CRISPR/Cas9-edited outcomes.
Phytoene desaturase (PDS) has been a popular marker for evaluating CRISPR in new study systems (Table 1). Its mutation disrupts chlorophyll biosynthesis, allowing for visual assessment of knockout (KO) efficiency. CRISPR/Cas9-induced albino mutants have been reported in poplar (Fan et al., 2015), citrus (Jia and Wang, 2014; Zhang et al., 2017), apple (Nishitani et al., 2016), grape (Nakajima et al., 2017), cassava (Odipio et al., 2017), coffee (Breitler et al., 2018), and kiwifruit (Wang Z. et al., 2018). Successful implementation of CRISPR has also been demonstrated by targeting potential developmental and biosynthesis pathway genes in grape (Ren et al., 2016) and the tropical tree Parasponia andersonii (van Zeijl et al., 2018; Table 1). New CRISPR reagents have been developed to expand genome editing capabilities. One such reagent, SaCas9 from Staphylococcus aureus, was shown to effectively generate mutations in Duncan grapefruit (Jia et al., 2017a). Compared to the most commonly used SpCas9 from Streptococcus pyogenes, SaCas9 is considerably smaller and recognizes a distinct 5′-NNGRRT protospacer adjacent motif (PAM) sequence (versus 5′-NGG of SpCas9). Using alternative CRISPR/Cas systems such as SaCas9 can increase the number of potential guide-RNA (gRNA) target sites, especially in AT-rich regions which may facilitate promoter editing.
Besides proof-of-concept studies, the CRISPR/Cas9 system has been used to develop disease resistant fruit trees with promising results (Table 1). The devastating citrus canker disease is caused by Xanthomonas citri subsp. citri (Xcc) through effector-activation of a canker susceptibility gene LOB1 of the Lateral Organ Boundaries transcription factor family (Hu et al., 2014). When the LOB1 promoter was targeted by CRISPR/Cas9 to disrupt the effector-binding element, canker symptoms after Xcc infection were reduced in Duncan grapefruit (Jia et al., 2016) and Wanjincheng orange (Peng et al., 2017). CRISPR-KO of LOB1 also increases Xcc resistance in Duncan grapefruit (Jia et al., 2017b). KO-mutations in other susceptibility genes for powdery mildew and fire blight disease have also been achieved in grape and apple protoplasts, respectively (Malnoy et al., 2016), potentially allowing for the regeneration of disease-resistant plants. Several WRKY transcription factors involved in defense regulation have also been targeted for mutagenesis. CRISPR-KO of two positive regulators PtrWRKY18 and PtrWRKY35 compromised resistance to Melampsora rust in Populus (Jiang et al., 2017), whereas KO of grape VvWRKY52 increased resistance to necrotrophic Botrytis cinerea (Wang X. et al., 2018).
To date, the greatest progress in woody species has been made with poplar, the first stably transformed tree to be genome-edited by CRISPR with high efficiency (Zhou et al., 2015). Allele-sensitive bioinformatics resources to facilitate genome editing in heterozygous species quickly followed, again based on the poplar system (Xue and Tsai, 2015; Xue et al., 2015). The majority of CRISPR studies in poplar have targeted phenylpropanoid metabolism or cell wall traits (Table 1). Mutations of individual 4-coumarate:CoA ligase (4CL) genes decreased the levels of structural (lignin) or non-structural (proanthocyanidin) phenylpropanoid polymers. CRISPR-KO of MYB transcription factors either increased (PtoMYB156 and PtrMYB57) or decreased (PtoMYB115 and PtoMYB170) phenylpropanoid flux, affecting in turn lignin deposition (PtoMYB156 and PtoMYB170) or flavonoid accrual (PtrMYB57 and PtoMYB115), respectively (Wan et al., 2017; Wang et al., 2017; Xu et al., 2017; Yang et al., 2017). Secondary cell wall synthesis was also compromised by CRISPR-KO of a brassinosteroid biosynthetic gene, supporting a role for brassinosteroids in wood formation (Shen et al., 2018). CRISPR-KO of BRANCHED1-1 (BRC1-1) and BRC1-2 belonging to the TCP family of transcription factors resulted in altered shoot architecture, and revealed an additional role of BRC2 in leaf development not previously reported for its Arabidopsis ortholog (Muhr et al., 2018). A recent study reported successful mutation of essential flowering genes in both male and female poplar genotypes (Elorriaga et al., 2018). The study also collated a large mutation dataset from over 500 transgenic events (Elorriaga et al., 2018) which should prove of value to understanding CRISPR/Cas editing patterns (see below). Although phenotypic evaluation of the flowering traits will require follow-on and multiyear studies in the field, the work underscores a powerful social application of CRISPR in containment of transgenic trees.
Small frameshift indels are the most common repair outcomes of single gRNA-directed Cas9 cleavage in trees, with 1 bp insertions (+1), especially +T and +A, predominant in many cases, similar to findings from other plants and animals (Bortesi et al., 2016). However, considerable variations and case-dependent repair outcomes are also noted, suggesting potential influences of target site sequences and/or their genomic contexts (Jacobs et al., 2015; Xu et al., 2015). Meta-analysis of mutation patterns across published tree studies is necessary to gain further insight, but that is made difficult by different reporting formats (not all studies report multi-allele data), and by the use of detection methods that differ in their sensitivity, accuracy, and allele discrimination (Sentmanat et al., 2018). We combined amplicon sequencing data from CRISPR-edited P. tremula x alba IRNA 717-1B4 (717) generated in our lab (Zhou et al., 2015) with the large 717 dataset from Elorriaga et al. (2018), along with manual inspection of other published tree studies for mutation profile analysis (Figure 1). In aggregate, +1 insertions constituted the greatest fraction of mutation types, followed by -1, and then -2, although stereotyped repair patterns are evident (Figure 1A). Interestingly, insertions were limited to +1 and +2 across all sites, whereas deletions spanned a much broader size range, though with decreasing frequencies for larger deletions.
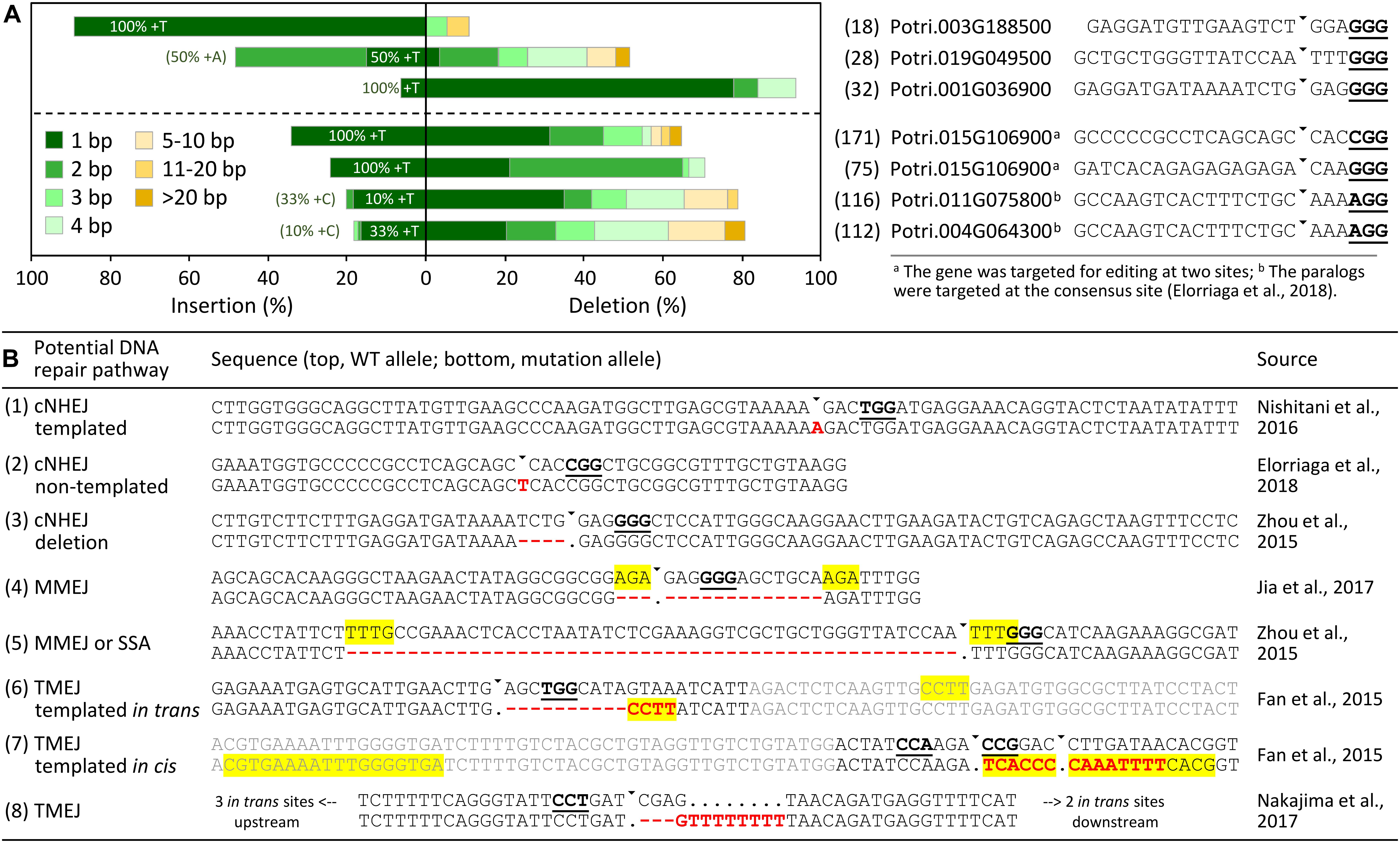
FIGURE 1. The mutation profiles and potential DNA repair pathways of CRISPR/Cas9-induced DSBs in Populus and other trees. (A) Distribution of mutation types at distinct genomic sites based on data from Zhou et al. (2015) (top panel) and Elorriaga et al. (2018) (bottom panel). The corresponding target genes and gRNA sequences are shown on the right, with allele number for each noted in parentheses. The percentages shown inside the +1 (1 bp insertion) bar indicate the fraction that were T insertions. The fraction of templated +1 insertions that deviate from T is shown in parentheses. (B) Representative examples of different mutation types and the potential DNA repair pathway involved in each case. PAM sequences are bold underlined, triangles denote predicted Cas9-cleavage sites, indels are shown in red, yellow-shaded regions denote microhomologies, and gray sequences in (6) and (7) were appended from P. tomentosa cDNA (GenBank accession KC954700) and P. tremula x alba 717 genomic sequences (Xue et al., 2015), respectively. Note, the region in (7) contains two overlapping target sites. For (8), there are five possible in trans template sites within introns of Phytozome (v12) gene model GSVIVG01016650001, the nearest one 640 bp upstream of the target site.
Small mutagenic indels have often been ascribed to the classical non-homologous end-joining (cNHEJ) DNA repair pathway, but recent studies have demonstrated involvement of the alternative end-joining (alt-EJ) pathway as well (Rodgers and McVey, 2016). It now appears that cNHEJ contributes to the most common +1 insertions and other small indels, whereas larger indels are due to alt-EJ. This is based on studies where impaired cNHEJ drastically changed the repair outcomes of CRISPR/Cas9 in yeast, human cells and Arabidopsis, such that the typically predominant +1 insertions as well as other small (<3 bp) indels were greatly reduced, while rates of large indels increased, apparently independently of cNHEJ (van Overbeek et al., 2016; Shen et al., 2017; Lemos et al., 2018). In yeast, the vast majority of the +1 insertions from cNHEJ were templated from 1 bp 5′overhangs at the Cas9 cleavage site (fourth base from the PAM), and dependent on POL4, a low-fidelity X-family DNA polymerase with terminal transferase activity (Lemos et al., 2018). POL4-deficient yeast also lost +2 and +3 insertions, many of which are homonucleotides and apparently templated from the Cas9 cleavage site as well (Lemos et al., 2018). Templated insertions could also explain the majority of +1 events in a large Cas9-induced indel dataset from human cells (van Overbeek et al., 2016), suggesting a conserved mechanism underlying +1 insertions in CRISPR/Cas9-edited organisms (Lemos et al., 2018). In the combined 717 dataset, the majority of +1 insertions were +T as reported in many CRISPR studies. However, evidence in support of templated +1 insertions was weak, and appeared to be target site-dependent (Figure 1A). Clearly, much more data with greater target site diversity and coverage are necessary before a conclusion can been drawn, but such data from trees will require significant and perhaps community-wide efforts. Regardless, the small target site collection used in our analysis supports involvement of more than one mechanism for the commonly observed +T insertions, at least in Populus.
cNHEJ-independent repair likely involves different alt-EJ pathways, including microhomology-mediated end-joining (MMEJ), single-strand annealing (SSA), or polymerase theta (POLQ)-mediated end-joining (TMEJ) (Rodgers and McVey, 2016). Both MMEJ and SSA require end resection or unwinding to expose short homologous sequences for annealing (up to ∼20–30 bp for MMEJ and longer for SSA) and subsequent repair, and always result in deletions (Sfeir and Symington, 2015). The presence of microhomologous sequences at the deletion junctions can therefore serve as evidence of MMEJ/SSA repair. Indeed, microhomologies of 1–5 bp are readily identifiable in most of the deletion (≥5 bp) alleles we examined, but are rarely found for small deletions (3–4 bp) that might have arisen from cNHEJ (Figure 1B). MMEJ has also been associated with abnormal chromosomal translocations and inversions (Sfeir and Symington, 2015). Such modifications have been reported in several studies – including two from Populus – where two or more gRNAs were designed to target the same gene to produce large deletions (Fan et al., 2015; Elorriaga et al., 2018). Moreover, large deletions are sometimes accompanied by small insertions (Fan et al., 2015; Nakajima et al., 2017), a pattern that is characteristic of the recently discovered TMEJ pathway (Koole et al., 2014). TMEJ depends on POLQ, an error-prone A-family DNA polymerase that can extend microhomologies in a template-dependent (either in cis or in trans) or independent manner (Kent et al., 2016). TMEJ is the essential repair pathway in animal germ cells, as embryos of zebrafish polq mutants are hypersensitive to DSB-inducing treatments, with low levels of repair producing only +1 insertions (Thyme and Schier, 2016). In Arabidopsis, TMEJ is required for T-DNA integration following Agrobacterium transformation of either flowers or roots (van Kregten et al., 2016). We found evidence of in cis or in trans templated insertions in the complex indels reported for poplar and grape (Figure 1B; Fan et al., 2015; Nakajima et al., 2017), supporting an active TMEJ pathway in somatic cells of plants.
Examination of published mutation profiles of Populus and other tree species suggests differential involvement of multiple repair pathways, probably with cNHEJ contributing to +1, +2 and small (1–4 bp) deletions, MMEJ (and SSA) to larger deletions, and TMEJ to complex indels (Figure 1B). The varying dependency of these pathways on sequence contexts (microhomologies) likely underpins the non-random nature of CRISPR/Cas9 repair outcomes reported in many studies, including trees (Jacobs et al., 2015; van Overbeek et al., 2016; Vu et al., 2017; Elorriaga et al., 2018). Incorporation of microhomology modeling into the gRNA design workflow (Bae et al., 2014; Segar et al., 2015) should enable prediction of potential DNA repair outcomes for informed selection of target sites.
For many herbaceous species where CRISPR editing efficiencies are low, or where monoallelic/mosaic mutations predominate in the first-generation (T0) transformants, multi-generation progeny screening is necessary to obtain homozygous mutants (Xu et al., 2015). Although initial transmission rates vary depending on the study system and the nature of CRISPR-induced (somatic or germinal) mutations carried by the founder plant, stable mutation inheritance can be expected once homozygous lines are obtained, as reported for Arabidopsis, rice, tomato and potato (Brooks et al., 2014; Feng et al., 2014; Zhou et al., 2014; Butler et al., 2015). For woody perennials, however, the issues are rather different. Cross-generational screening is difficult to implement for transgenic trees owing to their long generation times and strict regulation of flowering transgenic trees (Strauss et al., 2015). The predominantly outcrossing nature of trees, many of which are also dioecious, adds further challenge to rapid-cycle breeding and introgression of CRISPR-derived mutations into elite germplasms. While advances of early-flowering induction in contained environments (Hoenicka et al., 2014; Klocko et al., 2016) promise to accelerate progress, commercial production of many forest, fruit and nut trees relies on clonal propagation of elite genotypes. For woody perennials, therefore, it is pertinent to address long-term stability of CRISPR editing, both on-target and off-target, in vegetatively-propagated T0 transformants.
In theory, CRISPR-induced DNA modifications should lead to permanent mutations in edited cells that can be inherited mitotically during clonal propagation, yet experimental data are rare. One study used tissue culture to clone CRISPR-derived mutations from T0 diploid and tetraploid potato, and reported stable maintenance of targeted mutations across clonal generations, and in three selected cases, through the germline as well (Butler et al., 2015). In that study, however, somatic mutations were prevalent in T0 plants, as fewer than half of the originally-detected mutation types were captured as single mutations in clonally-propagated plants (Butler et al., 2015). The high levels of somatic mutations likely reflect a high proportion of chimeras, a common problem in tissue culture when plants are regenerated from multiple cells, in this case, with heterogenomic modifications. Fortunately for Populus, the proven efficiency of CRISPR (Zhou et al., 2015; Elorriaga et al., 2018) means null mutations with biallelic KO can be readily obtained in T0 transformants and stably inherited through clonal propagation. Wang et al. (2017) reported faithful maintenance of PtoMYB115 mutations in tissue culture-propagated Populus tomentosa somaclones, though in one case low frequencies of new mutations not seen in the parent line were detected, indicative of chimeras. Similarly, the CRISPR editing outcomes of BRC1-1 and BRC2-1 were also stable over multiple cycles of vegetative propagation in tissue culture (Muhr et al., 2018). We have maintained a subset of the previously-characterized 717 mutants (Zhou et al., 2015) in the greenhouse for over 4 years by repeatedly cutting back the original transformants and/or propagation using rooted cuttings. The reddish-brown wood discoloration of lignin-reduced 4cl1 mutants has been stable in all re-sprouted shoots or clonally-propagated plants. Repeated amplicon-sequencing of randomly selected lines re-confirmed the targeted 4CL1 mutations 4 years later, with no off-target changes to the paralogous 4CL5 (Supplementary Table S1). Another group of transgenic plants harbors a non-functional gRNA for 4CL5 due to SNPs (one per allele) between the genome-sequenced P. trichocarpa and the transformation host 717 that prevented Cas9 cleavage as confirmed by amplicon sequencing (Zhou et al., 2015). It should be noted that one of the 717 SNPs alters the PAM site from NGG to NGA, the latter is a non-canonical PAM of SpCas9 thought to cause off-target cleavage in human cells (Zhang Y. et al., 2014b). Reanalyzing this group of plants will inform as to whether the imperfectly-matched 4CL5-gRNA exhibited any off-target activity over the long term. We found no evidence of CRISPR/Cas9 cleavage after 4 years of coppicing and regrowth (Supplementary Table S1). These findings echo other tree studies that showed no or very rare off-targeting (Jia et al., 2017a; Nakajima et al., 2017; Elorriaga et al., 2018; see also Table 1), as well as reports from Arabidopsis, rice and tomato based on whole-genome re-sequencing (Feng et al., 2014; Zhang et al., 2014a; Peterson et al., 2016; Rodríguez-Leal et al., 2017). The data provide support for long-term stability and specificity of CRISPR/Cas9-mediated mutagenesis, with extremely low off-target potential during vegetative propagation in poplar.
Nullizygous mutations harboring either identical (homozygous) or distinct (heterozygous) mutations in all alleles of the genome are the ideal repair outcomes for gene KO investigation. However, monoallelic, in-frame and/or mosaic mutations can expand the phenotypic spectrum to enhance the power of functional analysis. For instance, transgenic grapevine with monoallelic mutations of a defense-related WRKY gene exhibited intermediate levels of disease resistance between WT and biallelic mutants (Wang X. et al., 2018). Similarly, monoallelic or in-frame mutations of PDS led to partial albino phenotypes in both poplar and apple (Fan et al., 2015; Nishitani et al., 2016). Given the abundance of duplicate genes in plant genomes, and the proven successes of CRISPR in multi-allele as well as allele-specific editing (Jia et al., 2016; Elorriaga et al., 2018), there is exciting potential to exploit CRISPR for development of allelic series mutations to address functional redundancy of duplicate genes or tandem repeats, and to investigate the allele-dose response of agronomic traits. Thus, the ability to generate novel germplasms is invaluable not only for tree improvement but also for basic functional genomics research.
In contrast to CRISPR-mediated KO, site-specific gene targeting or replacement remains a major challenge in plants, due to the inefficient homology-directed repair pathway. Geminivirus replicons have been shown to increase site-specific gene knockin (KI) efficiencies by orders of magnitude in tobacco, tomato and hexaploid wheat (Baltes et al., 2014; Čermák et al., 2015; Gil-Humanes et al., 2017). In animal systems, the MMEJ and SSA pathways, along with a new CRISPR/Cpf1 system have been harnessed for targeted KI with success (Sakuma et al., 2016; Tóth et al., 2016). These and other emerging approaches represent promising options for developing efficient KI systems in trees. Finally, many economically important tree species or genotypes remain recalcitrant to transformation and/or tissue culture regeneration, hindering applications of CRISPR. Recent breakthroughs in morphogenic regulator-mediated regeneration (Lowe et al., 2016, 2018) have already stimulated similar research in trees. Direct delivery of pre-assembled Cas9-gRNA ribonucleoproteins into protoplasts for genome editing as already deployed in apple and grape offers a transgene-free alternative to Agrobacterium transformation (Malnoy et al., 2016). At the present time, however, protoplast regeneration for other tree species remains a challenge. There is strong incentive to overcome this challenge since avoiding the footprint of foreign DNA and the associated negative perceptions will improve the outlook for integration of CRISPR technology with commercial deployment of designer trees.
C-JT conceived the idea. WPB, DC, and C-JT collected background information and analyzed data. WPB and C-JT wrote the manuscript with contributions from DC. All authors approved the manuscript.
The CRISPR research and associated genomic resource development in the Tsai Lab were supported by the National Institute of Food and Agriculture in the Department of Agriculture (2015-67013-22812), the National Science Foundation (IOS-1546867), and The Center for Bioenergy Innovation, a Department of Energy’s Research Center funded by the Office of Biological and Environmental Research in the DOE Office of Science. DC was supported by the Fundamental Research Funds for the Central Universities (BLYJ201603) from China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank current and former lab members and collaborators who contributed to the CRISPR research, Gilles Pilate of INRA, France for providing poplar clone 717, Estefania Elorriaga and Steve Strauss at Oregon State University for sharing their data, and Scott Harding for critical review of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01732/full#supplementary-material
Alessandra, F., and Shihshieh, H. (2010). Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol. J. 8, 655–677. doi: 10.1111/j.1467-7652.2010.00505.x
Bae, S., Kweon, J., Kim, H. S., and Kim, J.-S. (2014). Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11:705. doi: 10.1038/nmeth.3015
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A., and Voytas, D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26, 151–163. doi: 10.1105/tpc.113.119792
Bortesi, L., Zhu, C., Zischewski, J., Perez, L., Bassié, L., Nadi, R., et al. (2016). Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14, 2203–2216. doi: 10.1111/pbi.12634
Breitler, J.-C., Dechamp, E., Campa, C., Zebral Rodrigues, L. A., Guyot, R., Marraccini, P., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. Plant Cell Tissue Organ Cult. 134, 383–394. doi: 10.1007/s11240-018-1429-2
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Burstein, D., Harrington, L. B., Strutt, S. C., Probst, A. J., Anantharaman, K., Thomas, B. C., et al. (2016). New CRISPR–Cas systems from uncultivated microbes. Nature 542, 237–241. doi: 10.1038/nature21059
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591. doi: 10.1371/journal.pone.0144591
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16:232. doi: 10.1186/s13059-015-0796-9
Elorriaga, E., Klocko, A. L., Ma, C., and Strauss, S. H. (2018). Variation in mutation spectra among CRISPR/Cas9 mutagenized poplars. Front. Plant Sci. 9:594. doi: 10.3389/fpls.2018.00594
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. doi: 10.1038/srep12217
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D.-L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Fister, A. S., Landherr, L., Maximova, S. N., and Guiltinan, M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 9:268. doi: 10.3389/fpls.2018.00268
Gil-Humanes, J., Wang, Y., Liang, Z., Shan, Q., Ozuna, C. V., Sánchez-León, S., et al. (2017). High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89, 1251–1262. doi: 10.1111/tpj.13446
Hoenicka, H., Lehnhardt, D., Nilsson, O., Hanelt, D., and Fladung, M. (2014). Successful crossings with early flowering transgenic poplar: interspecific crossings, but not transgenesis, promoted aberrant phenotypes in offspring. Plant Biotechnol. J. 12, 1066–1074. doi: 10.1111/pbi.12213
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Jacobs, T. B., Lafayette, P. R., Schmitz, R. J., and Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. doi: 10.1186/s12896-015-0131-2
Jia, H., Orbovic, V., Jones, J. B., and Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, H., and Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One 9:e93806. doi: 10.1371/journal.pone.0093806
Jia, H., Xu, J., Orbovic, V., Zhang, Y., and Wang, N. (2017a). Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 8:2135. doi: 10.3389/fpls.2017.02135
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017b). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jiang, F., and Doudna, J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Kent, T., Mateos-Gomez, P. A., Sfeir, A., and Pomerantz, R. T. (2016). Polymerase 𝜃 is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife 5:e13740. doi: 10.7554/eLife.13740
Klocko, A. L., Ma, C., Robertson, S., Esfandiari, E., Nilsson, O., and Strauss, S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14, 808–819. doi: 10.1111/pbi.12431
Koole, W., Van Schendel, R., Karambelas, A. E., Van Heteren, J. T., Okihara, K. L., and Tijsterman, M. (2014). A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 5:3216. doi: 10.1038/ncomms4216
Lemos, B. R., Kaplan, A. C., Bae, J. E., Ferrazzoli, A. E., Kuo, J., Anand, R. P., et al. (2018). CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. U.S.A. 115, E2040–E2047. doi: 10.1073/pnas.1716855115
Lowe, K., La Rota, M., Hoerster, G., Hastings, C., Wang, N., Chamberlin, M., et al. (2018). Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. 54, 240–252. doi: 10.1007/s11627-018-9905-2
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M.-J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Muhr, M., Paulat, M., Awwanah, M., Brinkkötter, M., and Teichmann, T. (2018). CRISPR/Cas9-mediated knockout of Populus BRANCHED1 and BRANCHED2 orthologs reveals a major function in bud outgrowth control. Tree Physiol. 38, 1588–1597. doi: 10.1093/treephys/tpy088
Murovec, J., Pirc,Ž, and Yang, B. (2017). New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol. J. 15, 917–926. doi: 10.1111/pbi.12736
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS One 12:e0177966. doi: 10.1371/journal.pone.0177966
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6:31481. doi: 10.1038/srep31481
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8:1780. doi: 10.3389/fpls.2017.01780
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Peterson, B. A., Haak, D. C., Nishimura, M. T., Teixeira, P. J., James, S. R., Dangl, J. L., et al. (2016). Genome-wide assessment of efficiency and specificity in CRISPR/Cas9 mediated multiple site targeting in Arabidopsis. PLoS One 11:e0162169. doi: 10.1371/journal.pone.0162169
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., et al. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 6:32289. doi: 10.1038/srep32289
Rodgers, K., and McVey, M. (2016). Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 231, 15–24. doi: 10.1002/jcp.25053
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470.e–480.e. doi: 10.1016/j.cell.2017.08.030
Sakuma, T., Nakade, S., Sakane, Y., Suzuki, K.-I. T., and Yamamoto, T. (2016). MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 11, 118–133. doi: 10.1038/nprot.2015.140
Segar, M. W., Sakofsky, C. J., Malkova, A., and Liu, Y. (2015). MMBIRFinder: a tool to detect microhomology-mediated break-induced replication. IEEE/ACM Trans. Comput. Biol. Bioinform. 12, 799–806. doi: 10.1109/TCBB.2014.2359450
Sentmanat, M. F., Peters, S. T., Florian, C. P., Connelly, J. P., and Pruett-Miller, S. M. (2018). A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8:888. doi: 10.1038/s41598-018-19441-8
Sfeir, A., and Symington, L. S. (2015). Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci. 40, 701–714. doi: 10.1016/j.tibs.2015.08.006
Shen, H., Strunks, G. D., Klemann, B. J. P. M., Hooykaas, P. J. J., and De Pater, S. (2017). CRISPR/Cas9-induced double-strand break repair in Arabidopsis nonhomologous end-joining mutants. G3 7, 193–202. doi: 10.1534/g3.116.035204
Shen, Y., Li, Y., Xu, D., Yang, C., Li, C., and Luo, K. (2018). Molecular cloning and characterization of a brassinosteriod biosynthesis-related gene PtoDWF4 from Populus tomentosa. Tree Physiol. 38, 1424–1436. doi: 10.1093/treephys/tpy027
Strauss, S. H., Costanza, A., and Séguin, A. (2015). Genetically engineered trees: paralysis from good intentions. Science 349, 794–795. doi: 10.1126/science.aab0493
Thyme, S. B., and Schier, A. F. (2016). Polq-mediated end joining is essential for surviving DNA double-strand breaks during early zebrafish development. Cell Rep. 15, 707–714. doi: 10.1016/j.celrep.2016.03.072
Tóth, E., Weinhardt, N., Bencsura, P., Huszár, K., Kulcsár, P. I., Tálas, A., et al. (2016). Cpf1 nucleases demonstrate robust activity to induce DNA modification by exploiting homology directed repair pathways in mammalian cells. Biol. Direct 11:46. doi: 10.1186/s13062-016-0147-0
van Kregten, M., De Pater, S., Romeijn, R., Van Schendel, R., Hooykaas, P. J. J., and Tijsterman, M. (2016). T-DNA integration in plants results from polymerase-𝜃-mediated DNA repair. Nat. Plants 2:16164. doi: 10.1038/nplants.2016.164
van Overbeek, M., Capurso, D., Carter, M. M., Thompson, M. S., Frias, E., Russ, C., et al. (2016). DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell. 63, 633–646. doi: 10.1016/j.molcel.2016.06.037
van Zeijl, A., Wardhani, T. A. K., Seifi Kalhor, M., Rutten, L., Bu, F., Hartog, M., et al. (2018). CRISPR/Cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Front. Plant Sci. 9:284. doi: 10.3389/fpls.2018.00284
Vu, G. T. H., Cao, H. X., Fauser, F., Reiss, B., Puchta, H., and Schubert, I. (2017). Endogenous sequence patterns predispose the repair modes of CRISPR/Cas9-induced DNA double-stranded breaks in Arabidopsis thaliana. Plant J. 92, 57–67. doi: 10.1111/tpj.13634
Wan, S., Li, C., Ma, X., and Luo, K. (2017). PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 36, 1263–1276. doi: 10.1007/s00299-017-2151-y
Wang, L., Ran, L., Hou, Y., Tian, Q., Li, C., Liu, R., et al. (2017). The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol. 215, 351–367. doi: 10.1111/nph.14569
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wang, Z., Wang, S., Li, D., Zhang, Q., Li, L., Zhong, C., et al. (2018). Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 16, 1424–1433. doi: 10.1111/pbi.12884
Xu, C., Fu, X., Liu, R., Guo, L., Ran, L., Li, C., et al. (2017). PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 37, 1713–1726. doi: 10.1093/treephys/tpx093
Xu, R.-F., Li, H., Qin, R.-Y., Li, J., Qiu, C.-H., Yang, Y.-C., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5:11491. doi: 10.1038/srep11491
Xue, L.-J., Alabady, M. S., Mohebbi, M., and Tsai, C.-J. (2015). Exploiting genome variation to improve next-generation sequencing data analysis and genome editing efficiency in Populus tremula x alba 717-1B4. Tree Genet. Genomics 11:82. doi: 10.1007/s11295-015-0907-5
Xue, L.-J., and Tsai, C.-J. (2015). AGEseq: analysis of genome editing by sequencing. Mol. Plant 8, 1428–1430. doi: 10.1016/j.molp.2015.1006.1001
Yang, L., Zhao, X., Ran, L., Li, C., Fan, D., and Luo, K. (2017). PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci. Rep. 7:41209. doi: 10.1038/srep41209
Zhang, F., Leblanc, C., Irish, V. F., and Jacob, Y. (2017). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36, 1883–1887. doi: 10.1007/s00299-017-2202-4
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014a). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhang, Y., Ge, X., Yang, F., Zhang, L., Zheng, J., Tan, X., et al. (2014b). Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 4:5405. doi: 10.1038/srep05405
Zhou, H., Liu, B., Weeks, D. P., Spalding, M. H., and Yang, B. (2014). Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 42, 10903–10914. doi: 10.1093/nar/gku806
Keywords: mutagenesis, genome engineering, allele dose effect, Populus, biallelic, monoallelic, knockout
Citation: Bewg WP, Ci D and Tsai C-J (2018) Genome Editing in Trees: From Multiple Repair Pathways to Long-Term Stability. Front. Plant Sci. 9:1732. doi: 10.3389/fpls.2018.01732
Received: 06 July 2018; Accepted: 07 November 2018;
Published: 23 November 2018.
Edited by:
Ronald Ross Sederoff, North Carolina State University, United StatesReviewed by:
Eva Stoger, University of Natural Resources and Life Sciences Vienna, AustriaCopyright © 2018 Bewg, Ci and Tsai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chung-Jui Tsai, Y2p0c2FpQHVnYS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.