
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 November 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01711
This article is part of the Research Topic Interactions of Plants with Bacteria and Fungi: Molecular and Epigenetic Plasticity of the Host View all 14 articles
The bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo) is the most devastating bacterial disease of rice worldwide. A number of dominant major disease resistance (MR) genes and recessive MR genes against Xoo have been cloned and molecularly characterized in the last two decades. However, how these MR genes mediated-resistances occur at the cytological level is largely unknown. Here, by ultrastructural examination of xylem parenchyma cells, we show that resistances to Xoo conferred by dominant MR genes and recessive MR genes resulted in different types of programmed cell death (PCD). Three dominant MR genes Xa1, Xa4, and Xa21 and two recessive MR genes xa5 and xa13 that encode very different proteins were used in this study. We observed that Xa1-, Xa4-, and Xa21-mediated resistances to Xoo were associated mainly with autophagy-like cell death featured by the formation of autophagosome-like bodies in the xylem parenchyma cells. In contrast, the xa5- and xa13-mediated resistances to Xoo were associated mainly with vacuolar-mediated cell death characterized by tonoplast disruption of the xylem parenchyma cells. Application of autophagy inhibitor 3-methyladenine partially compromised Xa1-, Xa4-, and Xa21-mediated resistances, as did Na2HPO4 alkaline solution to xa5- and xa13-mediated resistances. These results suggest that autophagy-like cell death is a feature of the dominant MR gene-mediated resistance to Xoo and vacuolar-mediated cell death is a characteristic of the recessive MR gene-mediated resistance.
Plant resistance against pathogens can be genetically classified into two classes based on the strength of resistance: qualitative or complete resistance conferred by major disease resistance (MR) genes and quantitative or partial resistance mediated by multiple genes or quantitative trait loci (Kou and Wang, 2010; Zhang and Wang, 2013). The molecular mechanisms of plant disease resistance are explained, in general, with a two-tiered innate immune system: pathogen-associated molecular pattern-triggered immunity (PTI) or plant-derived damage-associated molecular PTI or basal resistance, and effector-triggered immunity (ETI) or gene-for-gene resistance (Jones and Dangl, 2006; Thomma et al., 2011; Monaghan and Zipfel, 2012). PTI is initiated by plasma membrane-localized plant pattern recognition receptors (PRRs), which are receptor-kinase proteins or receptor-like proteins, and ETI is initiated by cytoplasmic nucleotide-binding (NB)–leucine-rich repeat (LRR)-type resistance proteins (Jones and Dangl, 2006; Macho and Zipfel, 2014). Thus, in general, PTI is quantitative resistance and ETI is qualitative resistance in many plant–pathogen pathosystems (Zhang and Wang, 2013). However, rice and biotrophic Xanthomonas oryzae pv. oryzae (Xoo), which causes the most devastating bacterial disease of rice worldwide, are a unique pathosystem for rice qualitative resistance against Xoo (Zhang and Wang, 2013). The rice MR genes resistant to Xoo can be an ETI or a PTI or other mechanisms that cannot be explained by ETI and PTI (Zhang and Wang, 2013; Ke et al., 2017).
Except for the rice-Xoo pathosystem, earlier studies have shown that plant qualitative resistance to biotrophic pathogens frequently featured a rapid hypersensitive response (HR), which is characterized by rapid and localized cell death to restrict pathogen replication during the early stage of the plant–pathogen interaction (Pontier et al., 1998; Mur et al., 2008). Further studies have revealed that HR is often, but not always, a part of ETI initiated by NB-LRR proteins (Coll et al., 2011). HR-associated cell death is a kind of programmed cell death (PCD). Evolutionarily conserved autophagy, which is intracellular self-digestion of cytoplasmic components characterized by the formation of membrane-bound autophagosomes carrying a portion of the cytoplasm to be degraded or organelle permeabilization, has been observed to be involved in plant PCD (Dickman and Fluhr, 2013; Kabbage et al., 2017). The autophagosomes have different ultrastructures: (1) the vacuolar membrane (tonoplast)-bound body (microautophagy) formed by a portion of the cytoplasm, cytoplasmic vesicles or organelles in the vacuole; (2) the double-membrane body (macroautophagy) formed by a portion of cytoplasm bound by an endoplasmic reticulum-like tubule derived double-membrane in the cytoplasm; (3) the multilamellar body formed by many membranes bound by a single membrane in the cytoplasm (van Doorn and Papini, 2013). In addition, vacuolar-mediated PCD, in which the tonoplast integrity is compromised or the tonoplast is fused with the plasma membrane resulting in the release of vacuolar components into the cytoplasm or the extracellular space leading to cell death, also occurs in HR (Hara-Nishimura and Hatsugai, 2011; Dickman and Fluhr, 2013). In contrast, plant necrosis is characterized by shrinkage of the protoplast and rupture of the plasma membrane (van Doorn et al., 2011).
One of the features that makes the qualitative resistance of rice to Xoo unique from other pathosystems is that one third of the 41 MR genes identified thus far are genetically recessive (Zhang and Wang, 2013; Ke et al., 2017). Eleven (Xa1, Xa3/Xa26, Xa4, xa5, Xa10, xa13, Xa21, Xa23, xa25, Xa27, and xa41) of these 41 genes, which have been cloned and molecularly characterized at present, are shown to encode diverse types of proteins. Among the dominant genes, Xa1 encodes a classic NB-LRR-type protein (Yoshimura et al., 1998), Xa3/Xa26 and Xa21 encode plasma membrane-localized LRR receptor kinase-type proteins (Song et al., 1995; Sun et al., 2004), Xa4 encodes a cell wall-associated protein kinase (Hu et al., 2017). The recessive gene xa5 encodes a mutated basal transcriptional factor IIA gamma (TFIIAγ) subunit 5 (TFIIAγ5V39E) (Iyer and McCouch, 2004; Yuan et al., 2016), and xa13, xa25, and xa41 encode MtN3/saliva/SWEET-type membrane proteins with XA13 and XA25 localized in the plasma membrane (Chu et al., 2006; Liu et al., 2011; Hutin et al., 2015; Cheng et al., 2017). The dominant MR genes Xa1, Xa4, Xa21 and the recessive MR genes xa5, xa13 are race-specifically resistant to Xoo, while the recessive genes xa1, xa4, xa21 and the dominant Xa5, Xa13 are susceptible to Xoo (Zhang and Wang, 2013).
Xa27, xa13, and xa5 can trigger a HR (although the HR of xa5 is weak) leading to the brown symptoms on infected rice leaves after infiltrating inoculation with avirulent Xoo strains (Gu et al., 2005; Yang et al., 2006; Iyer-Pascuzzi et al., 2008). The ectopically expressed Xa10 and Xa23 can only induce HR in Nicotiana benthamiana (Tian et al., 2014; Wang et al., 2015). Nonetheless, the formation of HR in rice xylem vessel tissue against Xoo bacteria, which multiply in xylem vessels of rice leaves under natural infection conditions (Kou and Wang, 2010), needs to be investigated.
To address the HR of rice MR gene-mediated resistance to Xoo, we compared tissue phenotypes and ultrastructural morphologies of rice leaves from lines containing the dominant Xa1, Xa4, and Xa21 MR genes and the recessive xa5 and xa13 MR genes. We found that autophagy-like cell death is the major characteristic in dominant MR gene-mediated resistance and that vacuolar-mediated cell death is the main feature in recessive MR gene-mediated resistance. These findings suggest that the different types of HR-PCD contribute to the resistance of rice against Xoo by different MR genes.
IRBB1, IRBB4, IRBB21, IRBB5, and IRBB13 are near-isogenic rice lines (NILs) carrying dominant MR genes Xa1, Xa4, and Xa21 and recessive MR genes xa5 and xa13, respectively, in the genetic background of the rice variety IR24. Each of these five lines confers race-specific resistance to Xoo bacteria with different resistance spectra (Zhang and Wang, 2013; Hu et al., 2017).
Rice plants were inoculated with 109 cells ml-1 of Xoo strains T7174 (Japanese race (1), PXO61, PXO86, PXO112, PXO99, or PXO341 (Philippine race 1, 2, 5, 6 or 10) suspension at either the 4-leaf stage (rice lines IRBB4, IRBB5, IRBB13, and IR24) or 7-leaf stage (rice lines IRBB1, IRBB21, and IR24) by the leaf-clipping method (Chen et al., 2002). For mock treatment, water without Xoo was used by clipping rice leaves. Disease was scored by measuring the lesion length at 2 weeks after inoculation. To study the cell responses to Xoo and the effects of 3-methyladenine (3-MA) and Na2HPO4 on resistance, rice plants were inoculated by infiltrating leaves with a bacterial suspension of Xoo using a needleless syringe method (Schaad et al., 1996). The bacterial suspension with 109 cells ml-1 contained 5 mM 3-MA (Sigma, SIGMA-ALDRICH, St. Louis, MO, United States) or 2 mM Na2HPO4 (Sinopharm, Signopharm Chemical Reagent Co., Ltd., Shanghai, China). For mock treatment, leaves were infiltrated only by 5 mM 3-MA or 2 mM Na2HPO4 solution. Disease was scored by counting the number of infiltrating sites with water-soaked symptom at 3 days after inoculation. The inoculated leaves were photographed using scientific scanner (Image Scanner III, GE, Sweden). All the inoculation of plants with Xoo was biologically repeated at least twice with similar results, and one replicate was shown.
The ultrastructure of rice leaf cells was studied by transmission electron microscopy. The leaves were sampled at 0 day after inoculation (sampling at 1 h after inoculation) (0 DAI), 3 DAI, 5 DAI, and 14 DAI. The leaf tissues at the inoculation sites were cut into 1 × 3 mm pieces and fixed in 2.5% (w/v) glutaraldehyde in 0.1 M phosphate buffer solution (PBS) (pH 7.2) at 4°C overnight. The fixed tissues were washed in PBS three times for 30 min each at room temperature (20–25°C), postfixed for 2 h in 1% osmium tetroxide, dehydrated in a graded series of acetone, infiltrated with Spurr resin (SPI, SPI Chem, West Chester, PA, United States), and polymerized at 65°C for 48 h. The samples were cut into ultrathin sections (60–70 nm thick), stained with 2% uranyl acetate, and examined with a Hitachi transmission electron microscope (H-7650; Hitachi, Japan) at 80 kv. Each sample had 3 biological replicates with each replicate having at least 3 ultrathin sections observed under the electron microscope. To quantify the cells containing autophagosome-like body, tonoplast disruption and protoplast shrinkage, 208–520 xylem parenchyma cells were observed from at least six or nine leaf xylem veins of six or nine plants in two or three independent inoculations. In each xylem vein, the total xylem parenchyma cells of one vein were observed, then the frequencies (%) of the cells with the above three structures in the total xylem parenchyma cells were calculated. 57–80 mesophyll cells (approximately 10 cells from each plant) were observed and calculated for the frequencies (%) of cells with the above three structures from six leaves of six plants in two independent inoculations.
The 3-cm leaf fragments next to the inoculation sites were used for RNA isolation. Quantitative reverse transcription-PCR (qRT-PCR) was conducted as described previously using gene-specific primers (Supplementary Table S1; Qiu et al., 2007). The expression level of actin gene was used to standardize the RNA sample for each qRT-PCR, and then the expression level relative to the control was calculated. The assays were biologically repeated twice with similar results, and only one replicate was presented.
The significant differences of lesion length, gene expression level and the number of cells with autophagosome-like body, tonoplast disruption or protoplast shrinkage ultrastructures between resistant and susceptible plants, were assessed using pairwise Student’s t-test in Excel (Microsoft1).
Leaf tissue is a major infection site of Xoo (Niño-Liu et al., 2006). To study whether different types of rice MR gene-mediated resistance against Xoo have different types of cell death, we first analyzed the leaf morphology of rice–Xoo interaction sites in resistant IRBB1, IRBB4, IRBB5, IRBB13 and IRBB21 lines each carrying a different MR gene and susceptible line IR24 against Xoo strains T7174, PXO61, or PXO99 (Figure 1). At 5 days after infection (DAI), the brown HR-like lesions appeared on all the inoculation sites of rice leaves of the NILs carrying MR genes, while the inoculation sites of susceptible IR24 leaves formed approximately 0.5-cm-long chlorotic water-soaked symptoms (Figure 1A). At 14 DAI, all the infection sites of leaf tissue turned into yellow lesions in the resistant rice lines and the susceptible rice line (Figure 1B). The average lesion length of IR24 was about 6-fold longer than that of IRBB1, IRBB4, IRBB5, IRBB13, and IRBB21 at 14 DAI (Figure 1C).
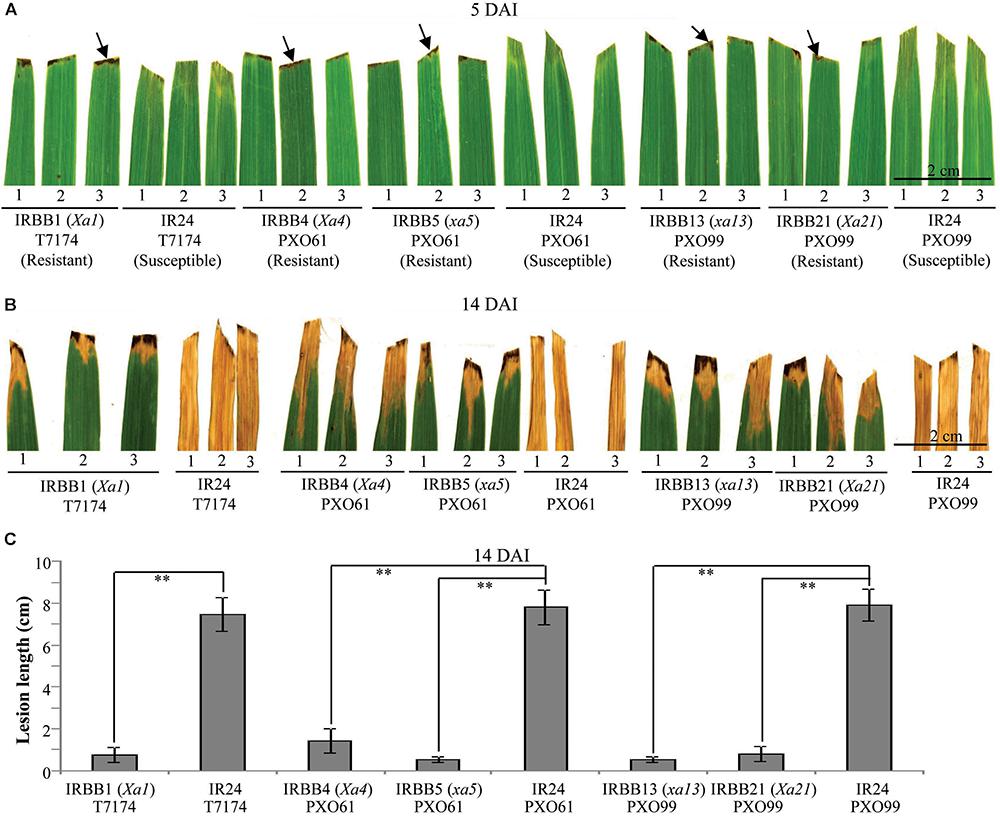
FIGURE 1. Phenotypes of different rice line–Xoo strain interactions. DAI, day after infection; arrow, hypersensitive response-like lesion. (A,B) Leaf responses of resistant rice lines IRBB1, IRBB4, IRBB5, IRBB13, and IRBB21 and susceptible rice line IR24 inoculated with Xoo strain T7174, PXO61, or PXO99 at 5 and 14 DAI. (C) Lesion length of different rice lines infected with Xoo at 14 DAI. Bars represent mean (12 to 16 leaves from four plants) ± standard deviation (SD). The double asterisk (∗∗) stands for the significant difference between resistant and susceptible plants at P < 0.01.
Xoo multiply and spread in the xylem vessels of rice leaves (Kou and Wang, 2010). The leaf xylem parenchyma cells surrounding xylem vessels are in direct contact with Xoo (Niño-Liu et al., 2006). Thus, we examined the ultrastructure of xylem parenchyma cells in the process of cell death after Xoo infection in NILs by transmission electron microscopy. We observed three types of abnormal ultrastructures in xylem parenchyma cells of infection sites at 3 DAI (Figures 2A–C). The first type of abnormal ultrastructure was observed mostly in resistant IRBB4 and IRBB21 lines, which showed autophagosome-like bodies formed by autophagy processes including double-membrane-like bodies in the cytoplasm, single-membrane-bound bodies containing multiple small vesicles in the cytoplasm, and tonoplast-bound bodies in vacuoles (Figure 2A). The second type of abnormal ultrastructure was tonoplast disruption that was commonly observed in the resistant IRBB13 line (Figure 2B). The third type of abnormal structure was protoplast shrinkage and rupture of the plasma membrane observed in the susceptible IR24 line (Figure 2C).
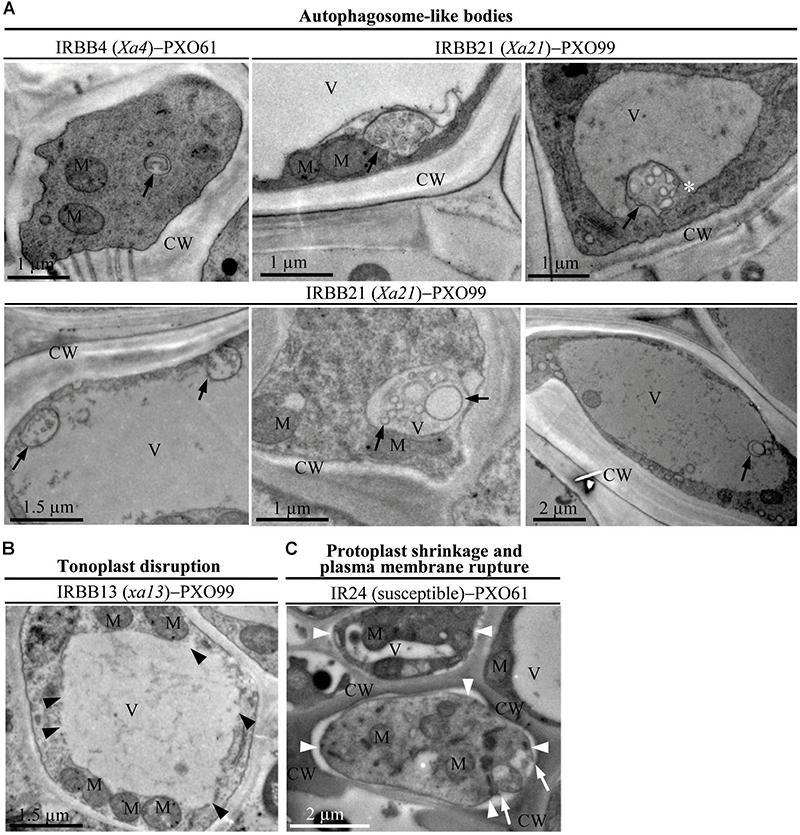
FIGURE 2. The abnormal ultrastructures in rice xylem parenchyma cells after Xoo infection at 3 DAI. XV, xylem vessel; V, vacuole; CW, cell wall; M, mitochondrion. (A) Autophagosome-like bodies (dark arrows showing double-membrane-like body or single-membrane-like body) in the cytoplasm and vacuoles during resistant reactions. (B) Tonoplast disruption (dark arrowheads) in the resistant reaction. (C) Protoplast shrinkage (white arrowheads) and plasma membrane rupture (white arrows) in the susceptible reaction.
To study whether the dominant or recessive MR gene-mediated resistance and the susceptible reaction are associated with different abnormal structures in rice-Xoo interactions, we counted the xylem parenchyma cells containing the three abnormal ultrastructures in all the rice lines at 0 and 5 DAI. There were integrated protoplasts and intact organelles in xylem parenchyma cells of all the rice lines and no significant difference in the numbers of xylem parenchyma cells with the three abnormal ultrastructures at 0 DAI among different lines (Figures 3A,C,E, 4A,C, 5A–C and Supplementary Figures S1, S2). However, at 5 DAI, autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage were observed in xylem parenchyma cells of all the rice lines, but the frequencies of cells with the three ultrastructures were very different among these rice–Xoo interactions (Figures 3–5 and Supplementary Figure S2). In IRBB1, IRBB4, and IRBB21 lines resistant to Xoo strains T7174, PXO61 or PXO112, PXO99, or PXO61, autophagosome-like bodies were the major feature in xylem parenchyma cells (Figures 3A,C,E and Supplementary Figures S2A–B). The number of rice cells containing autophagosome-like bodies was 4- and 3-fold higher than the number of cells containing tonoplast disruption and plasmolysis, respectively, in IRBB1; was 5- and 4-fold higher than the number of cells containing tonoplast disruption and plasmolysis, respectively, in IRBB4; and was 5- and 3-fold higher than the number of cells containing tonoplast disruption and plasmolysis, respectively, in IRBB21 (Figure 3G and Supplementary Figure S2D). However, protoplast shrinkage was the major features in xylem parenchyma cells in IRBB1, IRBB4 and IRBB21 lines susceptible to compatible Xoo strain PXO99, PXO341 (Figures 3B,D,F). The number of cells with protoplast shrinkage was 4- and 6-fold higher than the number of cells containing autophagosome-like bodies and tonoplast disruption (Figure 3G).
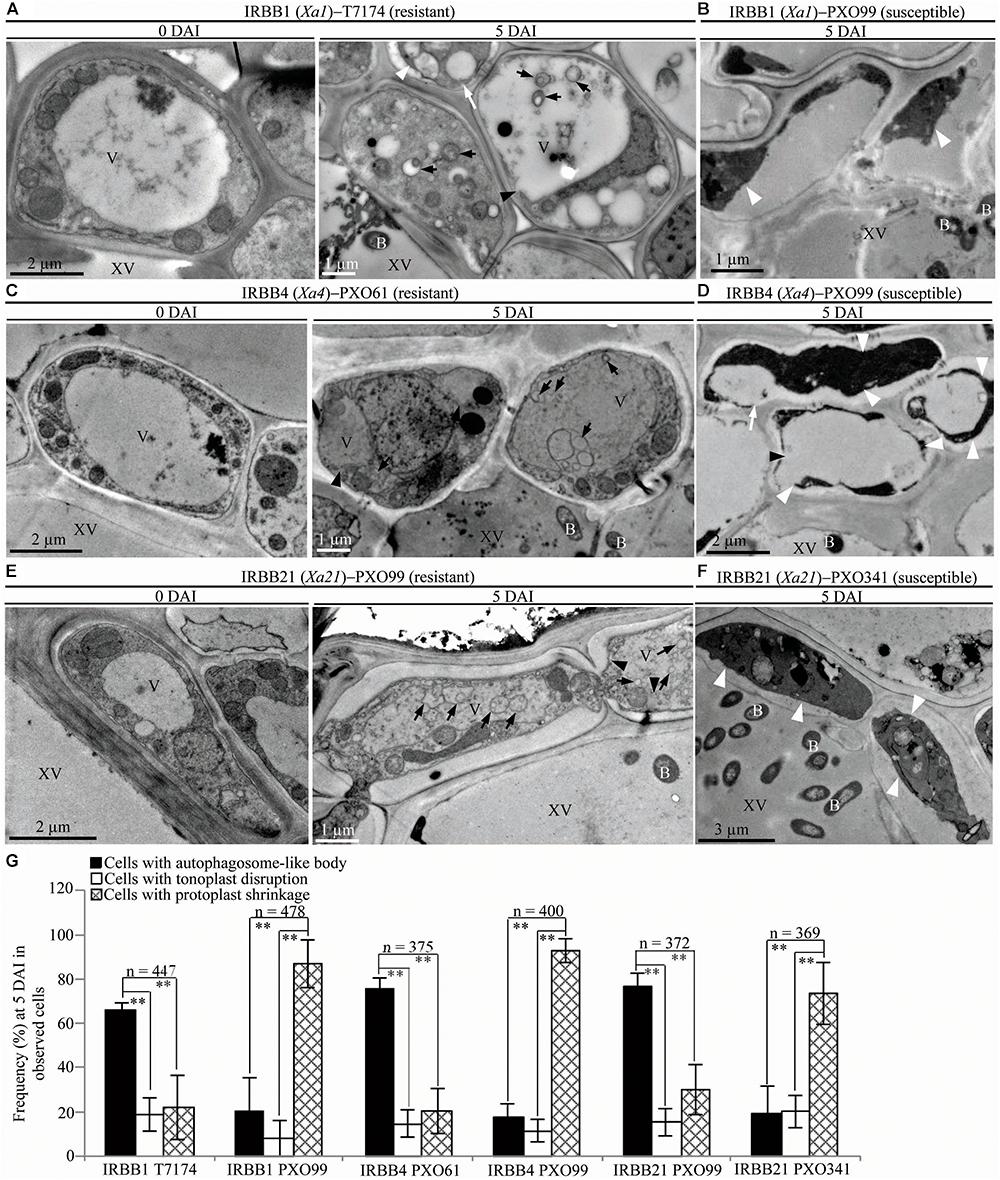
FIGURE 3. Autophagosome-like body ultrastructural feature of xylem parenchyma cells in dominant MR genes Xa1, Xa4, or Xa21 mediated-resistance. B, Xoo bacterium; V, vacuole; XV, xylem vessel; dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; white arrowhead, protoplast shrinkage; white arrow, rupture of plasma membrane. (A–F) Many autophagosome-like bodies in xylem parenchyma cells of IRBB1, IRBB4, and IRBB21 plants at 5 days after inoculation (DAI) with Xoo strains T7174, PXO61, or PXO99 comparison with plants at 0 DAI and plants susceptible reaction to compatible strains PXO99, PXO341. (G) The percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 5 DAI. Data represent mean (at least nine leaf xylem parenchyma cells were observed from nine different plants in two or three independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with autophagosome-like body and frequency of cells with tonoplast disruption or protoplast shrinkage at P < 0.01. n, the total number of observed cells.
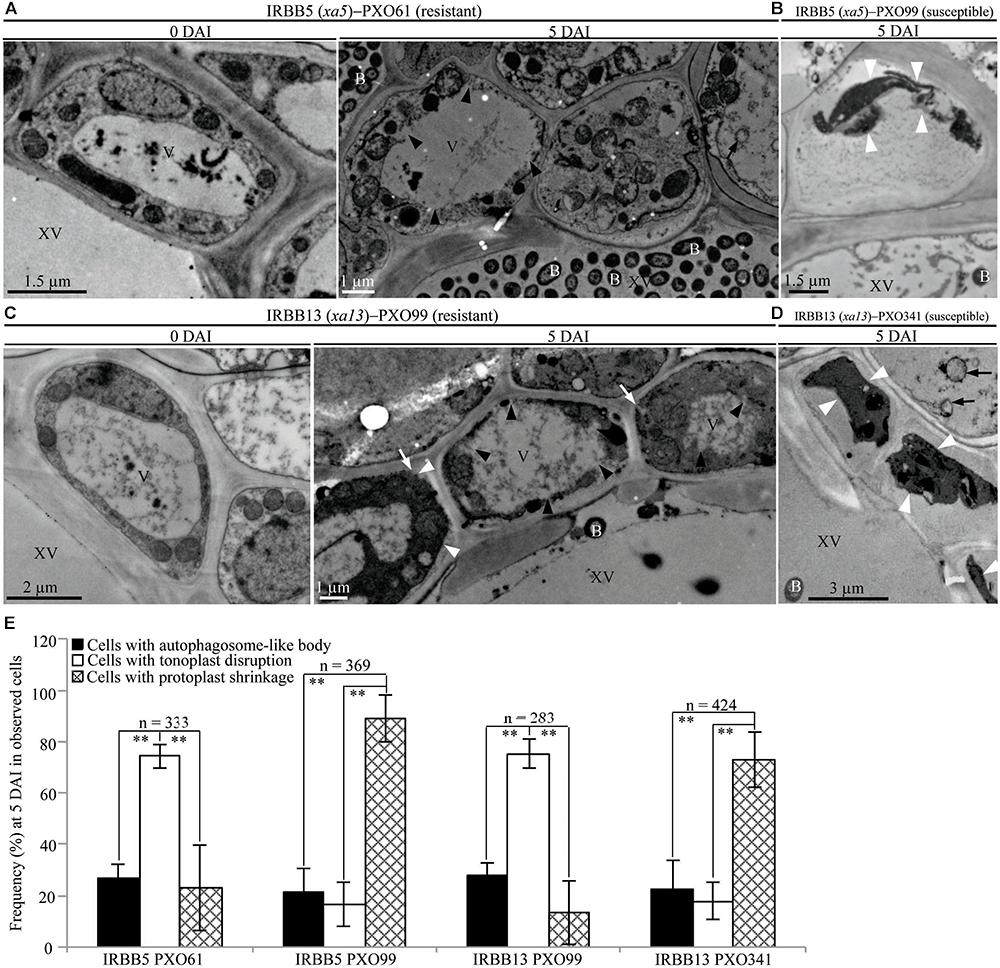
FIGURE 4. Tonoplast disruption ultrastructure of xylem parenchyma cells in recessive MR genes xa5 and xa13 mediated-resistance. V, vacuole; XV, xylem vessel; and B, Xoo bacterium. Dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; white arrowhead, protoplast shrinkage; and white arrow, rupture of plasma membrane. (A–D) Many xylem parenchyma cells with tonoplast disruption in IRBB5 and IRBB13 plants at 5 days after inoculation (DAI) with Xoo strains PXO61 and PXO99 comparison with plants at 0 DAI and plants susceptible reaction to compatible strains PXO99, PXO341. (E) The percentage of cells with autophagosome-like bodies, tonoplast disruption and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 5 DAI. Data represents mean (at least nine leaf xylem parenchyma cells were observed from nine different plants in two or three independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with tonoplast disruption and frequency of cells with autophagosome-like body or protoplast shrinkage at P < 0.01. n, the total number of observed cells.
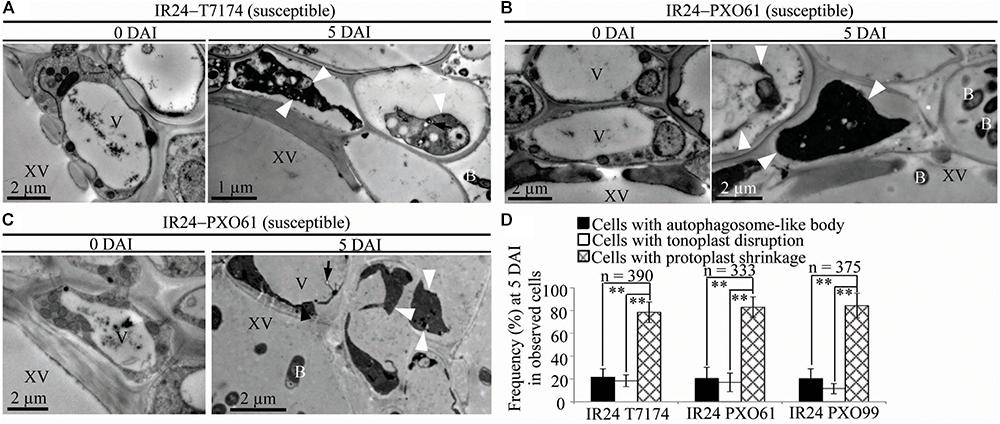
FIGURE 5. Necrosis ultrastructural features of xylem parenchyma cells in susceptible reactions after Xoo infection. V, vacuole; XV, xylem vessel; and B, Xoo bacterium. Dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; white arrowhead, protoplast shrinkage; and white arrow, rupture of plasma membrane. (A–C) Many xylem parenchyma cells with protoplast shrinkage in IR24 plants at 5 days after inoculation (DAI) susceptible to Xoo strains T7174, PXO61, PXO99 comparison with plants at 0 DAI. (D) The percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cells in rice leaves at 5 DAI. Data represent mean (at least nine leaf xylem parenchyma cells were observed from nine different plants in two or three independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with protoplast shrinkage and frequency of cells with tonoplast disruption or autophagosome-like body at P < 0.01. n, the total number of observed cells.
In contrast, tonoplast disruption was observed to be the major feature in xylem parenchyma cells during the IRBB5, IRBB13 lines resistant to Xoo strains PXO61 or PXO86, PXO99 (Figures 4A,C and Supplementary Figure S2C). The number of rice cells containing tonoplast disruption was 3-fold higher than the number of cells containing autophagosome-like bodies or cells showing protoplast shrinkage in IRBB5, and was 3-fold higher than the number of cells containing autophagosome-like bodies and 6-fold higher than cells showing protoplast shrinkage in IRBB13 (Figure 4E and Supplementary Figure S2D). However, protoplast shrinkage was also the major features in xylem parenchyma cells in IRBB5, IRBB13 lines susceptible to compatible Xoo strains PXO99, PXO341 (Figures 4B,D). The number of cells with protoplast shrinkage was 4- and 6-fold higher than the number of cells containing autophagosome-like bodies and tonoplast disruption (Figure 4E). Meanwhile, the average lesion length of rice lines containing MR genes inoculated with compatible strains all exceeded 6 cm at 14 DAI (Supplementary Figure S3).
Furthermore, protoplast shrinkage was observed to be the major ultrastructure feature in xylem parenchyma cells in rice susceptible reactions to Xoo (Figure 5). Many xylem parenchyma cells showed protoplast shrinkage in IR24 that was susceptible to Xoo strains T7174, PXO61, and PXO99 (Figures 5A–C). The number of cells with protoplast shrinkage was 4- and 7-fold higher than the number of cells containing autophagosome-like bodies and tonoplast disruption, respectively, in all IR24–Xoo interactions (Figure 5D).
Autophagy-related genes (ATG) control the formation of autophagosome-like body in plants (Liu et al., 2005; Xia et al., 2011). Vacuolar processing enzyme genes (VPE) regulate vacuolar mediated cell death in plant-pathogens interactions and H2O2-induced PCD (Hatsugai et al., 2004; Deng et al., 2011). We analyzed transcription level of autophagy-related genes OsATG5 and OsATG7, VPE gene OsVPE2 in IRBB1, IRBB4, IRBB21, and IRBB13 lines resistant to Xoo strains T7174, PXO61, and PXO99 (Supplementary Figure S4). On 8 and 24 h after inoculation, OsATG5 and OsATG7 were markedly induced to higher levels in IRBB1, IRBB4, and IRBB21 lines than that in susceptible IR24 control lines; OsVPE2 was not induced in IRBB1, IRBB21 lines but was markedly induced to higher levels in IRBB13 plants compared to IR24 (Supplementary Figure S4).
To study the ultrastructure of xylem parenchyma cells at the late stage of the rice–Xoo interaction and the ultrastucture of xylem parenchyma cells of rice lines at mock treatment (clipping leaf only with water), we examined the three abnormal structures, autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage, in the infection sites of the rice lines at 14 DAI and the mock inoculation sites at 3, 5, 14 DAI (Figure 6 and Supplementary Figure S5). Protoplast shrinkage was observed to be the major ultrastructural feature in xylem parenchyma cells at 14 DAI of all the rice–Xoo interactions and all the mock treatments (Figures 6A–H and Supplementary Figures S5A–F). The number of rice cells containing protoplast shrinkage was 3- to 6-fold higher than the number of cells with autophagosome-like bodies or tonoplast disruption, respectively, in all rice lines (Figure 6I and Supplementary Figures S5G,H). However, the number of rice cells containing autophagosome-like bodies was still 2- to 3-fold higher than the number of cells containing tonoplast disruption in dominant genes Xa1-, Xa4-, and Xa21-mediated resistance (Figure 6I). The number of rice cells containing tonoplast disruption was still 2- and 3-fold higher than the number of cells containing autophagosome-like bodies in recessive genes xa5- and xa13-mediated resistance (Figure 6I). Furthermore, at 3 and 5 DAI, there were integrated protoplasts and intact organelles in xylem parenchyma cells and no significant difference among the numbers of xylem parenchyma cell with the three abnormal ultrastructures at mock treatment (Supplementary Figure S5).
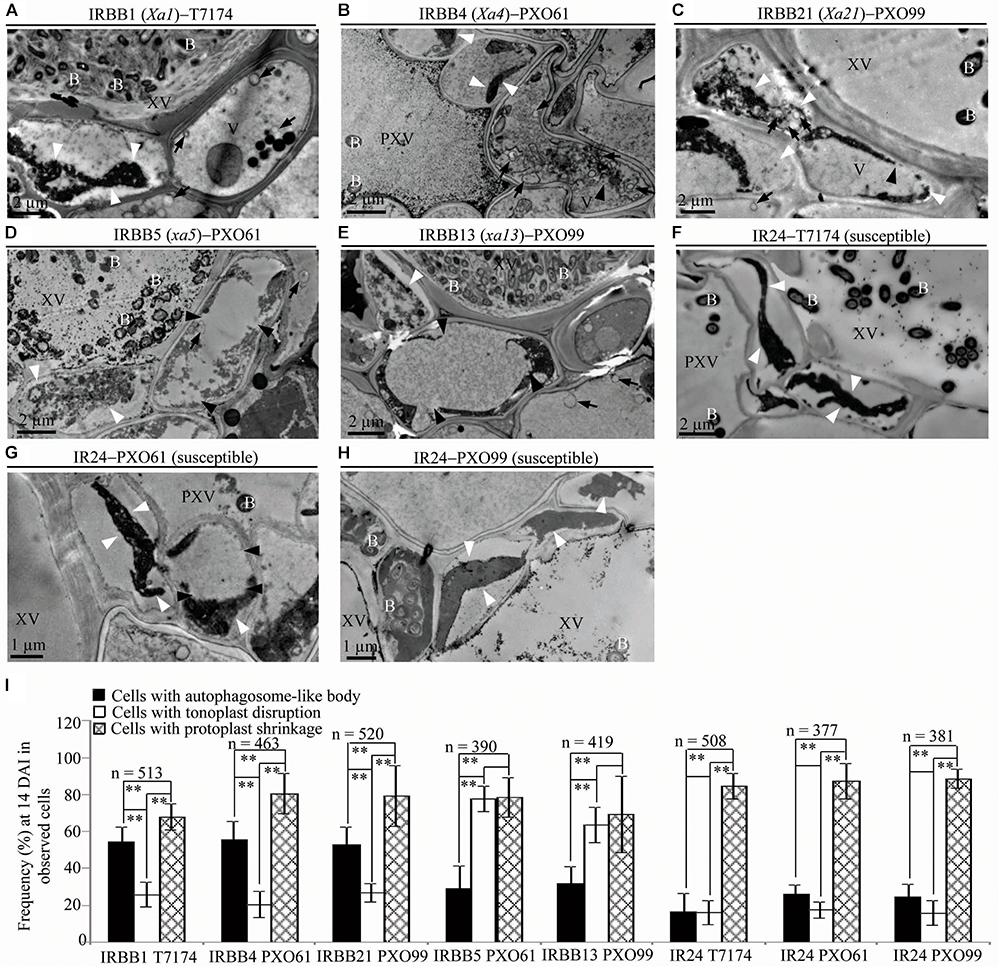
FIGURE 6. Ultrastructural features of xylem parenchyma cells in dominant MR genes Xa1, Xa4, and Xa21 and recessive MR genes xa5 and xa13 mediated-resistance compared with the susceptible control at 14 DAI with Xoo. B, Xoo bacterium; PXV, protoxylem vessel; V, vacuole; XV, xylem vessel; dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; and white arrowhead, protoplast shrinkage. (A–C) Xylem parenchyma cells with most autophagosome-like bodies and with most protoplast shrinkage in IRBB1, IRBB4, IRBB21 plants. (D,E) Xylem parenchyma cells with most tonoplast disruption and with most protoplast shrinkage in IRBB5 and IRBB13 plants. (F–H) Xylem parenchyma cells with most protoplast shrinkage in IR24 plants. (I) Percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 14 DAI with Xoo. Data represent mean (at least nine leaf xylem parenchyma cells were observed from nine different plants in two or three independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with protoplast shrinkage and frequency of cells with autophagosome-like body or tonoplast disruption, between frequency of cells with autophagosome-like body and frequency of cells with tonoplast disruption in resistant plants, at P < 0.01. n, the total number of observed cells.
The infiltrated inoculation sites on rice leaves with deep ink-colored water-soaked symptoms defined the rice susceptibility to Xoo (Sugio et al., 2005; Yang et al., 2005). Na2HPO4 alkaline solution could neutralize the low pH (5.2–6.0) liquid released from disrupted vacuole (Martiniere et al., 2013). To determine if the autophagy inhibitor 3-methyladenine (3-MA) and Na2HPO4 alkaline solution could influence the resistance of rice lines with different MR genes, we observed whether there were water-soaked symptoms on the infiltrated inoculation sites (susceptible reaction) when the rice lines with MR genes were inoculated with Xoo bacteria in 3-MA solution or Na2HPO4 alkaline solution at 3 DAI (Figures 7A–E). The infiltration sites appeared water-soaked (deep inky color) in IRBB1, IRBB4, and IRBB21 inoculated with Xoo in 3-MA solution and IRBB5 and IRBB13 inoculated with Xoo in Na2HPO4 alkaline solution (Figures 7A–E). However, all the corresponding IRBB lines with MR genes inoculated with only the incompatible Xoo strain bacteria (resistant reaction) did not have water-soaked symptoms (Figures 7A–E). Furthermore, all the numbers of infiltration site with water-soaked symptom (susceptible reaction) in the inoculation of IRBB1, IRBB4 and IRBB21 with Xoo in 3-MA solution and the inoculation of IRBB5 and IRBB13 with Xoo in Na2HPO4 alkaline solution were significantly (P < 0.01) more than that in each rice lines inoculated only with Xoo (Supplementary Table S2). In contrast, the susceptible line IR24, when inoculated with Xoo, Xoo in 3-MA solution and Xoo in Na2HPO4 alkaline solution, had water-soaked symptoms on all infiltration sites (Figures 7F–H). Whereas there was no water-soaked symptom on infiltration sites in the rice lines only injected with 3-MA solution or Na2HPO4 alkaline solution (Figure 7).
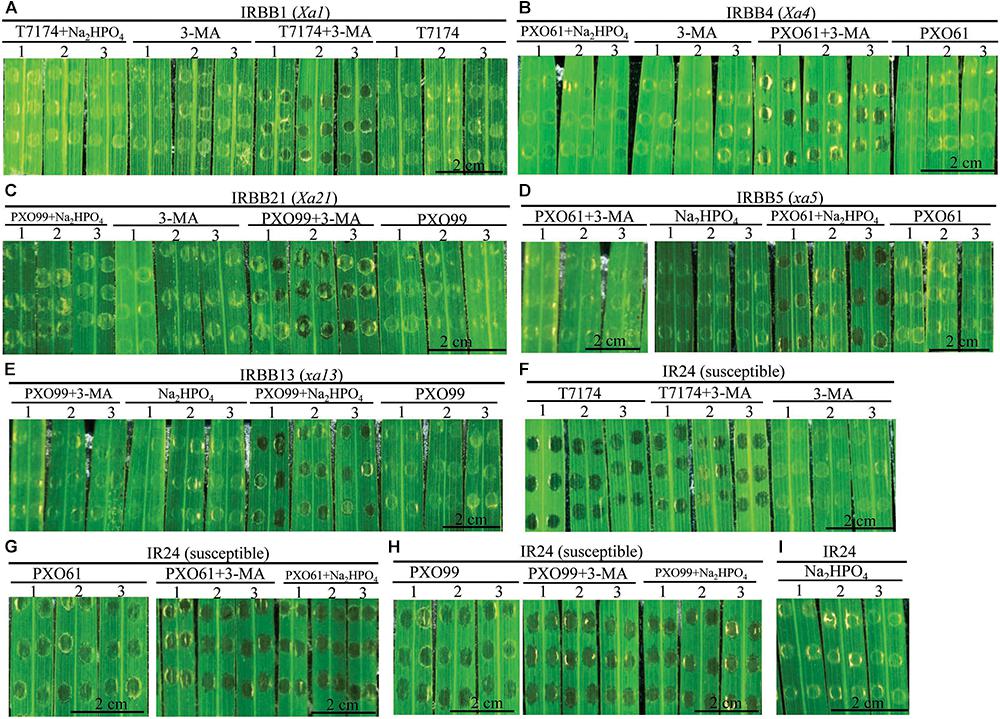
FIGURE 7. Effects of 3-methyladenine and Na2HPO4 on water-soaked symptoms in rice lines infiltrated with Xoo strains in different solutions at the 3rd day. (A–I) The symptom of IRBB1, IRBB4, IRBB21, IRBB5, IRBB13, and IR24 leaf infiltration sites with Xoo strains T7174, PXO61, and PXO99 (nephelometry) in H2O solution (T7174/PXO61/PXO99), in 5 mM 3-methyladenine (3-MA) solution (T7174/PXO61/PXO99 + 3-MA), in 2 mM Na2HPO4 solution (T7174/PXO61/PXO99 + Na2HPO4) and with only 5 mM 3-MA or 2 mM Na2HPO4 solution.
To investigate whether the 3-MA and Na2HPO4 alkaline solution affected the ultrastructure of mesophyll cell in rice–Xoo interaction, we analyzed the mesophyll cell with the three abnormal structures in rice lines with MR genes inoculated with Xoo bacteria in 3-MA solution or Na2HPO4 alkaline solution at 3 DAI (Supplementary Figure S6). In rice lines with MR genes infiltrated by only 3-MA or Na2HPO4 alkaline solution, the mesophyll cells represented intact protoplast and the numbers of mesophyll cell with three abnormal ultrastructures did not have difference (Supplementary Figures S6A–E,I–J). However, in IRBB1, IRBB4, IRBB21 lines inoculated with Xoo strains, the auphagosome-like bodies were the major ultrastructural features in mesophyll cells and the numbers of mesophyll cells with autophagosome-like bodies were 3- and 4-fold higher than the number of cells containing tonoplast disruption or protoplast shrinkage (Supplementary Figures S6A–C,I). In the IRBB1, IRBB4, and IRBB21 lines inoculated with Xoo strains in 3-MA solution, the IRBB5, IRBB13 lines inoculated with Xoo strains in Na2HPO4 alkaline solution and the IR24 lines only inoculated with Xoo strains, there were significantly more mesophyll cells with protoplast shrinkage than the mesophyll cells with autophagosome-like body or tonoplast disruption (Supplementary Figure S6). More mesophyll cells with tonoplast disruption were observed in IRBB5 and IRBB13 lines inoculated with only Xoo strains, even in IRBB5 and IRBB13 lines inoculated with Xoo strains in Na2HPO4 alkaline solution (Supplementary Figures S6D–E,J).
These results indicated that the mixing Xoo bacteria in 3-MA and Na2HPO4 alkaline solution significantly reduced the dominant genes Xa1-, Xa4-, and Xa21- mediated resistance and the recessive genes xa5- and xa13- mediated resistance, respectively.
A wide variety of pathogens can lead to lesion formation on infected plant tissue and trigger hypersensitive response-programmed cell death (HR-PCD) during plant resistance against pathogens (Mur et al., 2008; Kabbage et al., 2017). Most research on HR-PCD focused on the pathogens which grow and spread in the intercellular spaces of plant cells (Kabbage et al., 2017). However, the Xoo bacteria multiply and spread in the xylem vessels, a vascular structure surrounded by xylem parenchyma cells in rice leaves (Niño-Liu et al., 2006; Kou and Wang, 2010). In comparison with the chlorotic water-soaked symptoms of inoculated sites of susceptible rice line, the brown HR-like lesions on the Xoo-infected leaves of resistant rice lines (Figure 1) indicate that Xoo triggered the HR-PCD of resistant rice leaf cells. The xylem parenchyma cells, which directly interact with the Xoo bacterium (Niño-Liu et al., 2006), and the mesophyll cells appeared to have many classic autophagosome-like bodies and tonoplast rupture structures in resistant rice lines–Xoo interactions (Figures 2–6 and Supplementary Figures S2, S6). OsATG5 and OsATG7 were only markedly induced expression in Xa1-, Xa4-, and Xa21-mediated resistance, as did OsVPE2 only in xa13-mediated resistances (Supplementary Figure S4). Autophagy inhibitor 3-MA partially impaired the Xa1-, Xa4-, and Xa21-mediated resistances through reducing the number of mesophyll cells with autophagosome-like bodies (Figure 7 and Supplementary Figure S6). Meanwhile, in all the susceptible reactions, the xylem parenchyma cells showed protoplast shrinkage and plasma membrane disruption (Figures 3–6). In all the resistant rice lines, there were little cells with autophagosome-like bodies and tonoplast rupture structures in control treatment (0 DAI) and mock treatment (3, 5, 14 DAI) (Figures 3–5 and Supplementary Figures S1, S5), Therefore, the HR-PCD of xylem parenchyma cells belongs to autophagy-like cell death in Xa1-, Xa4-, and Xa21-mediated resistance and vacuolar-mediated cell death in xa5- and xa13-mediated resistance. However, there were approximately 70% of xylem parenchyma cells that had autophagy-like cell death and vacuolar-mediated cell death at 5 DAI (Figures 3, 4 and Supplementary Figure S2). Thus, these findings suggest that autophagy-like cell death and vacuolar-mediated cell death are the major types partly mixed with other types in resistances against Xoo.
The dominant Xa1, Xa4, and Xa21 encode a NB-LRR-type protein, a cell wall-associated kinase and a plasma membrane-localized LRR receptor kinase, respectively (Yoshimura et al., 1998; Chen et al., 2010; Hu et al., 2017). The Arabidopsis thaliana MR genes Cf9, Pto, PRS2, and RPS4 encode a membrane-anchored glycoprotein, a cytoplasmic serine-threonine protein kinase, a CC-NB-LRR-type protein and a TIR-NB-LRR-type protein, respectively (Pedley and Martin, 2003; Hofius et al., 2009; Chakrabarti et al., 2016). The Cf9, Pto, PRS2 and RPS4 proteins as receptors can trigger autophagy cell death to mediate A. thaliana resistance to Pseudomonas syringae pathovar (pv) tomato, Pst (Liu et al., 2005; Hofius et al., 2009; Gururani et al., 2012). Although XA1, XA4, and XA21 belong to different types of receptor proteins, they can accept and transfer resistance signals into rice cells during the resistance response (Yoshimura et al., 1998; Wang et al., 2006; Hu et al., 2017). As receptors, XA1, XA4, and XA21 proteins presumably have triggered autophagy-like cell death to partially mediate rice resistance against Xoo. The xylem parenchyma cells with autophagy-like cell death still had intact morphologies in Xa1-, Xa4-, and Xa21-mediated resistance (Figure 3 and Supplementary Figure S2). The intact xylem parenchyma cells possibly provide a vital cell environment to facilitate rice resistance against Xoo.
The low expression level of susceptible genes such as SWEET11/Xa13, SWEET13, and SWEET14/Xa41 limits the growth of Xoo bacteria in xa5-, xa13-, and xa41- mediated resistances (Chen et al., 2010; Hutin et al., 2015; Huang et al., 2016; Yuan et al., 2016). The above SWEET genes encode glucose transporters which localize on plasma membrane and take part in pumping glucose to extracellular space (Chen et al., 2010; Hutin et al., 2015; Huang et al., 2016). A lot of other glucose transporters on tonoplast play important roles in uptake glucose into vacuole in A. thaliana and rice plant cells (Cho et al., 2010; Hedrich et al., 2015). The SWEET genes such as wheat leaf rust R genes Lr67 and Lr34 can lead to the intracellular glucose accumulation and the leaf senescence in resistance reactions (Krattinger et al., 2009; Chen et al., 2010; Moore et al., 2015). Therefore, in xa5- and xa13-mediated resistances, glucose may accumulate in xylem parenchyma cell where it is pumped into vacuole. Consequently, the tonoplast is disrupted by the high concentration of glucose in vacuole. The vacuole of plants has a low pH (5.2–6.0) to maintain the activity of acid hydrolytic enzymes and defense proteins (Neuhaus et al., 1991; Martiniere et al., 2013). These hydrolytic enzymes can inhibit Pst bacteria growth after being released into the extracellular matrix by tonoplast fusion with the plasma membrane in A. thaliana (Hatsugai et al., 2009). Meanwhile, the destructive vacuolar-mediated cell death mediated by VPE, which is characterized by vacuole collapse by tonoplast disruption and vacuole collapse leading to cytoplasmic content degradation and rapid cell death, is involved in N. benthamiana resistance to tobacco mosaic virus and A. thaliana resistance to Pst or Botrytis cinerea (Hatsugai et al., 2004; Rojo et al., 2004). We also found the tonoplast disintegration of most xylem parenchyma cells in xa5- and xa13-mediated resistances and the higher expression level of OsVPE2 in xa13-mediated resistance (Figures 4, 6 and Supplementary Figure S4). Based on our results, vacuolar-mediated cell death is a form of destructive cell death. The Na2HPO4 alkaline solution (pH 9.0) dramatically increased the susceptibility but did not change the number of mesophyll cells with tonoplast disruption in xa5- and xa13-mediated resistances (Figure 7, Supplementary Figure S6 and Supplementary Table S2), which suggests that the alkaline solution partially impairs xa5- and xa13-mediated resistance against Xoo. Destructive vacuolar-mediated cell death may depend on the low pH created by vacuole collapse to inhibit Xoo growth. Alternatively, hydrolytic enzymes and defense proteins released into the cytoplasm possibly participate in xa5- and xa13-mediated resistance.
The plant cells with shrinkage of protoplast and rupture of plasma membrane is regarded as necrosis cell death (van Doorn et al., 2011). The necrosis cell death typically happens under abiotic stress (van Doorn et al., 2011; Gao et al., 2015). However, the protoplast shrinkage also occurs in fungal toxin victorin-induced cell death (van Doorn et al., 2011). Meanwhile, the cell wall deformation, protoplast shrinkage and swelling of chloroplasts are observed in susceptible reaction of a black-rot-susceptible cultivar (Golden Acre) inoculated with Xanthomonas campestris pv.campestri (Bretschneider et al., 1989). At 3 and 5 DAI, the xylem parenchyma cells appeared protoplast shrinkage and plasma membrane rupture only in rice susceptible line IR24–Xoo interactions and the rice lines with MR genes but susceptible to compatible Xoo strains (Figures 3–5). Therefore, the cell death of xylem parenchyma cells in rice lines susceptible to Xoo can be categorized into necrosis at early stage of infection. At 14 DAI of resistant reaction, susceptible reaction or mock treatment, most parenchyma cells of rice line all represented protoplast shrinkage (Figure 6 and Supplementary Figure S5), which indicates that the necrosis may be the general features of cell death in rice during late stages of pathogen infection and wound stress. In rice susceptible to Xoo, there are more bacteria in infection site than that in resistant reaction (Song et al., 1995; Yoshimura et al., 1998; Iyer and McCouch, 2004; Chu et al., 2006; Hu et al., 2017). The high bacterial population in xylem vessel of susceptible rice line may damage the xylem parenchyma cells to form necrosis. The necrosis of xylem parenchyma cells with plasma membrane rupture can possibly release nutrients into xylem vessels where Xoo bacteria multiply.
Plant cell often represents protoplast shrinkage and cell corpse at late stage of PCD (van Doorn et al., 2011). At 14 DAI, the leaves of infection site becoming yellow lesion (Figure 1B) indicated that the cell death of xylem parenchyma cells reached to late stage in resistant rice plants. Meanwhile, most xylem parenchyma cells appeared protoplast shrinkage and cell corpse in resistant rice plants at 14 DAI (Figure 6). But, most xylem parenchyma cells also represented autophagosome-like bodies and tonoplast disruption ultrastructures in resistant reactions (Figures 6A–E,I). Based on our results, the cell death progression in resistant rice lines happens as the following steps. Firstly, the xylem parenchyma cells always keep autophagy-like cell death or vacuolar-mediated cell death from 3 DAI to 14 DAI. Secondly, the long-time autophagosome-like body formation or tonoplast disruption processes possibly lead to the rupture of plasma membrane. Lastly, the protoplast shrinkage leads to cell corpse at late stage. However, the cell death progression of xylem parenchyma cells in susceptible rice line is always keeping necrosis i.e., protoplast shrinkage from 3 DAI to 14 DAI (Figures 2, 5, 6F–H).
JC designed and performed most of the experiments, analyzed the data, and drafted the manuscript. MZ performed the qRT-PCR assays. JX and XL provided biochemical analysis support and field management. MY revised part of the manuscript. SW supervised the project, interpreted the data, and revised the manuscript. All authors read and approved the manuscript.
This work was supported by grants from the National Key Research and Development Program of China (2016YFD0100903) and the National Natural Science Foundations of China (31500977 and 31821005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01711/full#supplementary-material
FIGURE S1 | Percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cells in rice leaves at 0 DAI. Data represent mean (at least nine leaf xylem parenchyma cells were observed from nine different plants in two or three independent inoculations) ± SD. No difference between frequency of cells with protoplast shrinkage and frequency of cells with tonoplast disruption or autophagosome-like body at P < 0.05. n, the total number of observed cells.
FIGURE S2 | Autophagosome-like body and tonoplast disruption ultrastructural features of xylem parenchyma cells in rice lines with MR genes resistant to another incompatible Xoo strains. V, vacuole; XV, xylem vessel; and B, Xoo bacterium. Dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; white arrowhead, protoplast shrinkage; and white arrow, rupture of plasma membrane. (A–C) Many autophagosome-like bodies and tonoplast disruption in xylem parenchyma cells of IRBB4, IRBB21, and IRBB5 plants at 5 days after inoculation (DAI) with Xoo strains PXO112, PXO61, and PXO86 comparison with plants at 0 DAI. (D) The percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 0 and 5 DAI. Data represent mean (at least six leaf xylem parenchyma cells were observed from six different plants in two independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with autophagosome-like body and frequency of cells with tonoplast disruption or protoplast shrinkage in at P < 0.01. n, the total number of observed cells.
FIGURE S3 | Lesion length of rice lines with MR genes Xa1, Xa4, xa5, xa13, and Xa21 infected by compatible Xoo strains at 14 DAI. Bars represent mean (10 to 15 leaves from four plants) ± standard deviation (SD).
FIGURE S4 | Expression pattern of OsATG5, OsATG7, and OsVPE2 in resistant (IRBB1, IRBB21, IRBB4, and IRBB13) and susceptible (IR24) rice lines–Xoo interactions. The expression of autophagy-related genes (OsATG5, OsATG7) and a vacuolar processing enzyme (OsVPE2) were analyzed by qRT-PCR in IRBB1/IR24 rice plants inoculated with Xoo strain T7174, IRBB21/IR24, and IRBB13/IR24 rice plants inoculated with Xoo strain PXO99, IRBB4/IR24 rice plants inoculated with Xoo strain PXO61. The rice plants were sampled on ck, 8 and 24 h after inoculation. Data are means (three replicates) ± standard deviations. ck, before inoculation. The letters “a” indicates statistically significant differences between ck and inoculated plants of the same rice plant at P < 0.01. Double astericks (∗∗P < 0.01) indicate statistically significant differences resistant rice plant and susceptible rice plant inoculated at same time.
FIGURE S5 | Ultrastructural features of xylem parenchyma cell in rice lines with mock treatment. V, vacuole; XV, xylem vessel; and B, Xoo bacterium. White arrowhead, protoplast shrinkage; and white arrow, rupture of plasma membrane. (A–F) Many xylem parenchyma cells with protoplast shrinkage at 14 day after inoculation (DAI) comparison with the normal xylem parenchyma cells without three abnormal ultrastructures at 3 DAI and 5 DAI in IRBB1, IRBB4, IRBB21, IRBB5, IRBB13, and IR24 plants. (G,H) Percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 3, 5, and 14 DAI. Data represent mean (at least six leaf xylem parenchyma cells were observed from six different plants in two independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with protoplast shrinkage and frequency of cells with tonoplast disruption or autophagosome-like body at P < 0.01. n, the total number of observed cells.
FIGURE S6 | Effects of 3-methyladenine and Na2HPO4 on the ultrastructural features of mesophyll cell in rice lines infiltrated with Xoo strains in different solutions at the 3rd day. B, Xoo bacterium; V, vacuole; IS, intercellular space; Ch, chloroplast; N, nucleus; dark arrow, autophagosome-like body; dark arrowhead, tonoplast disruption; white arrowhead, protoplast shrinkage; and white arrow, rupture of plasma membrane. (A–H) the ultrastructural features of mesophyll cell in IRBB1, IRBB4, IRBB21, IRBB5, IRBB13, and IR24 leaf infiltration sites with Xoo strains T7174, PXO61, and PXO99 in H2O solution (T7174/PXO61/PXO99), in 5 mM 3-methyladenine (3-MA) solution (T7174/PXO61/PXO99 + 3-MA), in 2 mM Na2HPO4 solution (T7174/PXO61/PXO99 + Na2HPO4) and with only 5 mM 3-MA or 2 mM Na2HPO4 solution. (I,J) percentage of cells with autophagosome-like bodies, tonoplast disruption, and protoplast shrinkage in micrographs of xylem parenchyma cell in rice leaves at 14 DAI with Xoo. Data represent mean (at least six leaf mesohyll cells were observed from six different plants in two independent inoculations) ± SD. The double asterisk (∗∗) stands for a significant difference between frequency of cells with protoplast shrinkage and frequency of cells with autophagosome-like body or tonoplast disruption, between frequency of cells with autophagosome-like body and frequency of cells with tonoplast disruption in resistant plants, at P < 0.01. n, the total number of observed cells.
TABLE S1 | PCR primers used for quantitative RT-PCR assays.
TABLE S2 | Effect of 3-methyladenine and Na2HPO4 on the percentage of infiltrating inoculation site with water-soaked symptoms.
Bretschneider, K. E., Gonella, M. P., and Robeson, D. J. (1989). A comparative light and electron microscopical study of compatible and incompatible interactions between Xanthomonas campestris pv. campestris and cabbage (Brassica oleracea). Mol. Plant Pathol. 34, 285–297. doi: 10.1016/0885-5765(89)90026-X
Chakrabarti, A., Velusamy, T., Tee, C. Y., and Jones, D. (2016). A mutational analysis of the cytosolic domain of the tomato Cf-9 disease-resistance protein shows that membrane-proximal residues are important for Avr9-dependent necrosis. Mol. Plant Pathol. 17, 565–576. doi: 10.1111/mpp.12315
Chen, H., Wang, S., and Zhang, Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology 92, 750–754. doi: 10.1094/PHYTO.2002.92.7.750
Chen, L. Q., Hou, B. H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X. Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Cheng, Q., Mao, W., Xie, W., Liu, Q., Cao, J., Yuan, M., et al. (2017). Characterization of a disease susceptibility locus for exploring an efficient way to improve rice resistance against bacterial blight. Sci. China Life Sci. 60, 298–306. doi: 10.1007/s11427-016-0299-x
Cho, J. I., Burla, B., Lee, D. W., Ryoo, N., Hong, S. K., Kim, H. B., et al. (2010). Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytol. 186, 657–668. doi: 10.1111/j.1469-8137.2010.03194.x
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Deng, M., Bian, H., Xie, Y., Kim, Y., Wang, W., Lin, E., et al. (2011). Bcl-2 suppresses hydrogen peroxide-induced programmed cell death via OsVPE2 and OsVPE3, but not via OsVPE1 and OsVPE4, in rice. FEBS J. 278, 4797–4810. doi: 10.1111/j.1742-4658.2011.08380.x
Dickman, M. B., and Fluhr, R. (2013). Centrality of host cell death in plant-microbe interactions. Annu. Rev. Phytopathol. 51, 543–570. doi: 10.1146/annurev-phyto-081211-173027
Gao, H. J., Yang, H. Y., Bai, J. P., Liang, X. Y., Lou, Y., Zhang, J. L., et al. (2015). Ultrastructural and physiological responses of potato (Solanum tuberosum L.) plantlets to gradient saline stress. Front. Plant Sci. 5:787. doi: 10.3389/fpls.2014.00787
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., and Park, S. W. (2012). Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78, 51–65. doi: 10.1016/j.pmpp.2012.01.002
Hara-Nishimura, I., and Hatsugai, N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18, 1298–1304. doi: 10.1038/cdd.2011.70
Hatsugai, N., Iwasaki, S., Tamura, K., Kondo, M., Fuji, K., Ogasawara, K., et al. (2009). A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 23, 2496–2506. doi: 10.1101/gad.1825209
Hatsugai, N., Kuroyanagi, M., Yamada, K., Meshi, T., Tsuda, S., Kondo, M., et al. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305, 855–858. doi: 10.1126/science.1099859
Hedrich, R., Sauer, N., and Neuhaus, H. E. (2015). Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr. Opin. Plant Biol 25, 63–70. doi: 10.1016/j.pbi.2015.04.008
Hofius, D., Schultz-Larsen, T., Joensen, J., Tsitsigiannis, D. I., Petersen, N. H., Mattsson, O., et al. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–783. doi: 10.1016/j.cell.2009.02.036
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009–17018. doi: 10.1038/nplants.2017.9
Huang, S., Antony, G., Li, T., Obasa, K., Yang, B., and White, F. F. (2016). The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 86, 186–194. doi: 10.1111/tpj.13164
Hutin, M., Perez-Quintero, A., Lopez, C., and Szurek, B. (2015). MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front. Plant Sci. 6:535. doi: 10.3389/fpls.2015.00535
Iyer, A. S., and McCouch, S. R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17, 1348–1354. doi: 10.1094/MPMI.2004.17.12.1348
Iyer-Pascuzzi, A. S., Jiang, H., Huang, L., and McCouch, S. R. (2008). Genetic and functional characterization of the rice bacterial blight disease resistance gene xa5. Phytopathology 98, 289–295. doi: 10.1094/PHYTO-98-3-0289
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kabbage, M., Kessens, R., Bartholomay, L. C., and Williams, B. (2017). The life and death of a plant cell. Annu. Rev. Plant Biol. 68, 375–404. doi: 10.1146/annurev-arplant-043015-111655
Ke, Y., Deng, H., and Wang, S. (2017). Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 90, 738–748. doi: 10.1111/tpj.13438
Kou, Y., and Wang, S. (2010). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185. doi: 10.1016/j.pbi.2009.12.010
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Liu, Q., Yuan, M., Zhou, Y., Li, X., Xiao, J., and Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x
Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell. 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Martiniere, A., Desbrosses, G., Sentenac, H., and Paris, N. (2013). Development and properties of genetically encoded pH sensors in plants. Front. Plant Sci. 4:523. doi: 10.3389/fpls.2013.00523
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Mur, L. A., Kenton, P., Lloyd, A. J., Ougham, H., and Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. doi: 10.1093/jxb/erm239
Neuhaus, J. M., Sticher, L., Meins, F. Jr., and Boller, T. (1991). A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc. Natl. Acad. Sci. U.S.A. 88, 10362–10366. doi: 10.1073/pnas.88.22.10362
Niño-Liu, D. O., Ronald, P. C., and Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Pedley, K. F., and Martin, G. B. (2003). Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41, 215–243. doi: 10.1146/annurev.phyto.41.121602.143032
Pontier, D., Balague, C., and Roby, D. (1998). The hypersensitive response. A programmed cell death associated with plant resistance. C. R. Acad. Sci. III 321, 721–734. doi: 10.1016/S0764-4469(98)80013-9
Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20, 492–499. doi: 10.1094/MPMI-20-5-0492
Rojo, E., Martin, R., Carter, C., Zouhar, J., Pan, S., Plotnikova, J., et al. (2004). VPEγ exhibits a caspase-like activity that contributes to defense against pathogens. Curr. Biol. 14, 1897–1906. doi: 10.1016/j.cub.2004.09.056
Schaad, N., Wang, Z., Di, M., McBeath, J., Peterson, G., and Bonde, M. (1996). An improved infiltration technique to test the pathogenicity of Xanthomonas oryzae pv. oryzae in rice seedlings. Seed Sci. Technol. 24, 449–456.
Song, W., Wang, G., Chen, L., Kim, H., Pi, L., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Sugio, A., Yang, B., and White, F. F. (2005). Characterization of the hrpF pathogenicity peninsula of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 18, 546–554. doi: 10.1094/MPMI-18-0546
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313X.2003.01976.x
Thomma, B. P., Nurnberger, T., and Joosten, M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15. doi: 10.1105/tpc.110.082602
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26, 497–515. doi: 10.1105/tpc.113.119255
van Doorn, W. G., Beers, E., Dangl, J., Franklin-Tong, V., Gallois, P., Hara-Nishimura, I., et al. (2011). Morphological classification of plant cell deaths. Cell Death Differ. 18, 1241–1246. doi: 10.1038/cdd.2011.36
van Doorn, W. G., and Papini, A. (2013). Ultrastructure of autophagy in plant cells. Autophagy 9, 1922–1936. doi: 10.4161/auto.26275
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wang, Y. S., Pi, L. Y., Chen, X. H., Chakrabarty, P. K., Jiang, J., De Leon, A. L., et al. (2006). Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. Plant Cell 18, 3635–3646. doi: 10.1105/tpc.106.046730
Xia, K., Liu, T., Quyang, J., Wang, R., Fan, T., and Zhang, M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Yang, B., Sugio, A., and White, F. F. (2005). Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 18, 142–149. doi: 10.1094/MPMI-18-0142
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S.A. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., et al. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5:e19605. doi: 10.7554/eLife.19605
Keywords: major disease resistance gene, bacteria blight, autophagy-like cell death, vacuolar-mediated cell death, ultrastructure
Citation: Cao J, Zhang M, Xiao J, Li X, Yuan M and Wang S (2018) Dominant and Recessive Major R Genes Lead to Different Types of Host Cell Death During Resistance to Xanthomonas oryzae in Rice. Front. Plant Sci. 9:1711. doi: 10.3389/fpls.2018.01711
Received: 09 August 2018; Accepted: 02 November 2018;
Published: 21 November 2018.
Edited by:
Marco Catoni, University of Birmingham, United KingdomReviewed by:
Philip Carella, University of Cambridge, United KingdomCopyright © 2018 Cao, Zhang, Xiao, Li, Yuan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiping Wang, c3dhbmdAbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.