
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 01 November 2018
Sec. Plant Membrane Traffic and Transport
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01610
Phenylpropanoids fulfill numerous physiological functions, essential for plant growth and development, as well as plant–environment interactions. Over the last few decades, many studies have shown that exquisite regulatory mechanisms at multiple levels control the phenylpropanoid metabolic pathway. Deciphering this pathway not only provides a greater, basic understanding of plant specialized metabolism, but also enhances our ability to rationally design plant metabolic pathways for future applications. Despite the identification of the participating enzymes of this complex, biosynthetic machinery, we still lack a complete picture of other genes, enzymes, and metabolites essential for regulation and compartmentation/distribution of phenylpropanoids. Compartmentation, as well as distribution, are critical for the fate/functioning of those molecules, and their effective biosynthesis. At the cellular level, we have narrowed down our understanding of these processes to organelles. Furthermore, various, overlapping, but not exclusive scenarios of phenylpropanoid distribution within the cell have also been described. The cross-membrane dynamics, but also intercellular communication of different branches from phenylpropanoid biosynthesis have become an exciting research frontier in plant science. The intra- and intercellular channeling of intermediates by various transport mechanisms and notably membrane transporters could be a meaningful tool that ensures, inter alia, efficient metabolite production.
The phenylpropanoid pathway is one of the most frequently investigated metabolic routes, among secondary metabolites. The existence of enzymatic reactions, where phenylalanine is converted to hydroxycinnamic acid, dates back to 450 million years ago (Fellenberg and Vogt, 2015), coinciding with colonization of the terrestrial environment by plants (Lanfranco et al., 2016). Products of the phenylpropanoid pathway are involved in many aspects of plant growth, structural support, and response to the stimuli inextricably associated with the life on land. Not only do they play a crucial role in stress response upon variation of light (Yang et al., 2018) and mineral shortage (Clemens and Weber, 2016), but they are also key mediators of the plant interactions with other organisms (Naoumkina et al., 2010; Shalaby and Horwitz, 2015; Liu and Murray, 2016). The utility of phenylpropanoids is a matter of being in the right place at the right time. This is tightly controlled, not only at the biosynthesis level, but also by various distribution systems. The latter comprise, inter alia, membrane transporters which participate in the circulation of both theintermediates and final products. This mini review is intended to position transport as a notable part of the regulatory network, tuning the phenylpropanoid biosynthesis according to the plant’s needs.
Phenylpropanoid metabolism generates an enormous array of secondary metabolites, based on the few intermediates of the shikimate pathway (Vogt, 2010). The shikimate pathway is a source of phenylalanine and the entry point leading to the biosynthesis of phenylpropanoids. The so-called central phenylpropanoid pathway is defined by three enzymatic activities: (i) the phenylalanine deamination by phenylalanine ammonia-lyase (PAL) to the trans-cinnamic acid, (ii) the trans-cinnamic acid hydroxylation to the 4-coumarate, as a resulting from cinnamic acid 4-hydroxylase (C4H) activity, and finally (iii) the 4-coumarate conversion to the 4-coumaroyl-CoA by 4-coumarate-CoA ligase (4CL). In many cases, genes from the central phenylpropanoid pathway are present in multiple copies. For instance, the PAL genes include six isoforms in Medicago (Medicago truncatula), five in poplar (Populus trichocarpa), nine in rice (Oryza sativa), and four in Arabidopsis thaliana. Various isoforms differ in terms of localization and activity. For instance, the Arabidopsis PAL1, PAL2, and PAL4 are expressed at relatively high levels in stems during the later stages of development, with PAL1 expression localized in the vascular tissue, and PAL2 and PAL4 both expressed in seeds. PAL1 and PAL2 were shown to be involved in flavonoid biosynthesis (Olsen et al., 2008), while the two others were suggested to participate in lignin formation (Huang et al., 2010). Similarly to PAL in Arabidopsis, 4CL possesses four isoforms. The 4CL3 is expressed in a broad range of cell types, and is predominantly associated with flavonoid biosynthesis. 4CL1, 4CL2, and 4CL4 revealed co-expression with lignin biosynthetic genes (Ehlting et al., 1999; Li et al., 2015). The product of 4CL activity, the p-Coumaroyl-CoA is a crucial intermediate in the phenylpropanoid pathway. It is a precursor for: (i) monolignol, (ii) coumarin, (iii) stilbene, as well as (iv) (iso)flavonoid biosynthesis (Kutchan et al., 2015).
The cooperating enzymes from the phenylpropanoid pathway were proposed to be organized into complexes called metabolons, and a number of reviews may serve as evidence in this respect (Winkel-Shirley, 1999; Jørgensen et al., 2005; Sweetlove and Fernie, 2013; Laursen et al., 2015; Bassard and Halkier, 2018). The term “metabolon” encompasses multienzymatic complexes bound to the cellular structural elements – membranes. Most metabolon models are based on a dynamic, non-covalent aggregation of components on the endoplasmic reticulum (ER) surface. Enzymes like chalcone synthase (CHS), chalcone reductase (CHR) are cytoplasmic enzymes however, other ones, like C4H or isoflavones synthase (IFS), are lodged in the ER, anchoring the biosynthetic enzyme complex (Dastmalchi et al., 2016). Organization of enzymes in metabolons is, at the cellular level, a way to optimize biosynthesis. It provides: (i) direct transport of intermediates between successive enzymes, hence increasing local concentration of the substrate around the enzyme active center, (ii) minimization of highly biologically active and potentially toxic intermediates within the cell, as well as (iii) coordination of reactions leading to different branches of pathways with shared enzymes or intermediates (Jørgensen et al., 2005; Bassard and Halkier, 2018). In the phenylpropanoid pathway, intracellular interactions between biosynthetic enzymes were shown for the central phenylpropanoid pathway – where PAL and C4H colocalize in the ER (Achnine et al., 2004), as well as for particular branches leading to the formation of (iso)flavonoids, monolignols, and anthocyanins (Figure 1). Key flavonoid enzymes exhibit multidirectional interactions: CHS-chalcone isomerase (CHI) (Saslowsky and Winkel-Shirley, 2001), CHS-CHR, IFS-upstreaming enzymes (Dastmalchi et al., 2016) and IFS- isoflavone O-methyltransferase (IOMT) (Liu and Dixon, 2001). In the anthocyanin route, flavonol synthase (FLS), and dihydroflavonol 4-reductase (DFR) were shown to interact with CHS in a competitive manner (Crosby et al., 2011), while flavanone 3-hydroxylase (F3′H) was indicated to interact with CHI (Burbulis and Winkel-Shirley, 1999). Additionally, flavone synthase II (FNS II) was shown to interact with DFR, as well as with the upstreaming enzymes like CHS and CHI, while the latter also interacts with CHS and DFR (Fujino et al., 2018). In the branch leading to lignin precursors, C4H and p-coumaroylshikimate 3′-hydroxylase (C3′H) were found to colocalize in the ER and being connected to shikimate hydroxycinnamoyl transferase (HCT) (Chen et al., 2011; Bassard et al., 2012). The existence of metabolons was also demonstrated in primary metabolism, for instance in fatty acid biosynthesis (Kwiatkowska et al., 2015), purine synthesis (Kyoung et al., 2015), or Krebs cycle (Wu et al., 2015). These systems involve stable enzyme associations that are suitable for experimental analysis. Potential metabolons formed in metabolic channeling of secondary metabolites in plants seem to be more dynamic and transient, according to the required plasticity in response to wide spectrum of stimuli, thus their existence is much more difficult to investigate and demonstrate.
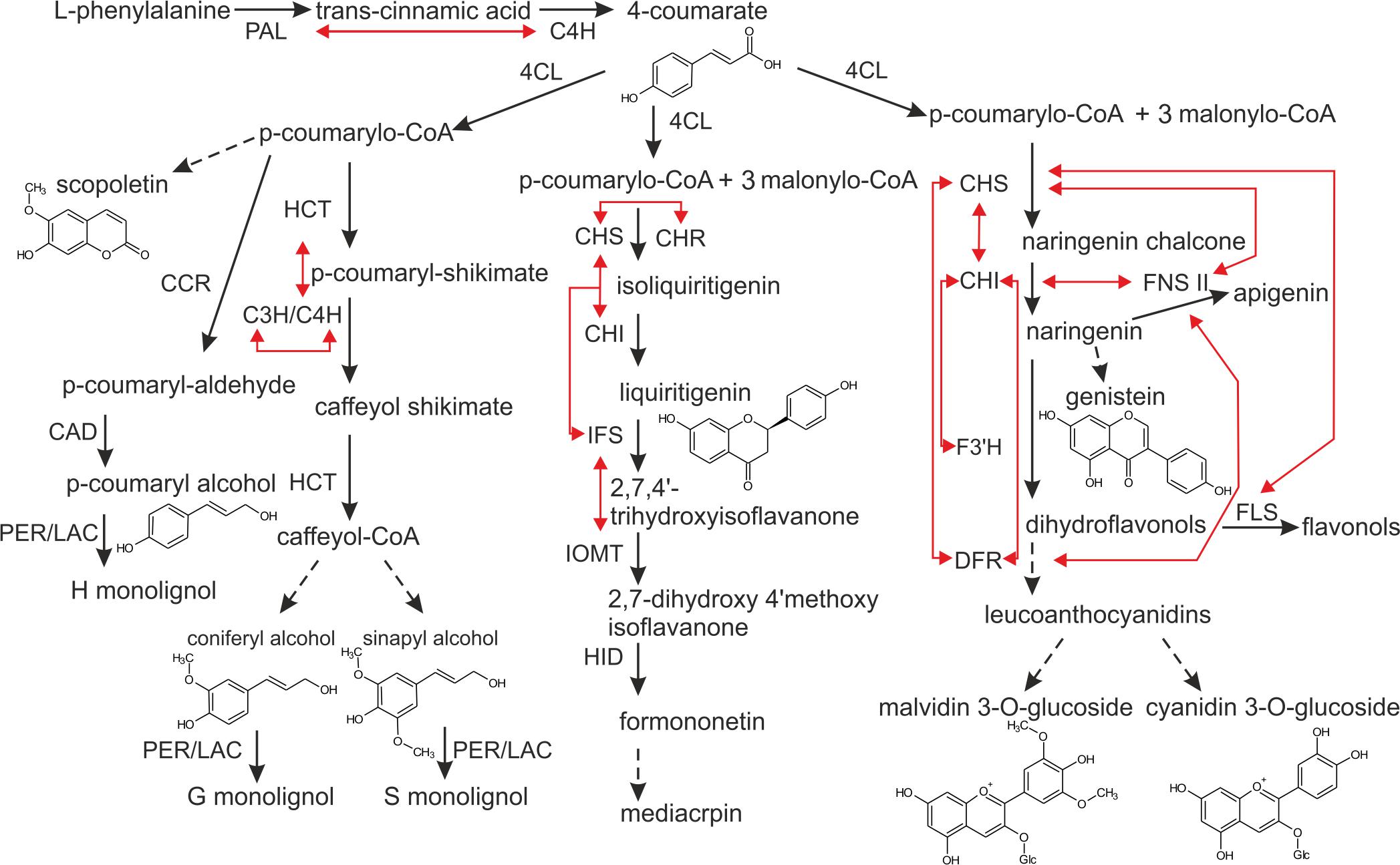
FIGURE 1. Scheme of the phenylpropanoid pathway. Direct interactions between particular biosynthetic enzymes, demonstrated by means of co-immunoprecipitation, BiFC or FRET, are illustrated by red arrows. Discontinuous lines indicate that certain steps along the pathway are not included in the figure. The enzymes are: PAL, phenylalanine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CCR, cinnamoyl CoA reductase; CAD, cinnamoyl alcohol dehydrogenase; PER/LAC, peroxidase/laccase; HCT, shikimate hydroxycinnamoyl transferase; C3H/C4H, 4-coumarylshikimate 3-hydroxylase/cinnamate 4-hydroxylase; CHS, chalcone synthase; CHR, chalcone reductase; CHI, chalcone isomerase; IFS, isoflavone synthase; IOMT, isoflavone O-methyltransferase; HID, hydroxyisoflavanone dehydratase; F3H, flavanone 3 hydroxylase; DFR, dihydroflavonol 4-reductase; FNSII, flavanone synthase II; FLS, flavonol synthase.
When the concept of metabolons was conjectured the main feature of the model was the assumption that all stages of biosynthesis/intermediates sharing occur in a single cell. Recently, transient and dynamic metabolons formation/cooperation has been proposed as means of swift adaptation of the metabolite profile to environmental changes (Bassard and Halkier, 2018). Interestingly, in the opium poppy (Papaver somniferum), the spatial distribution of alkaloid biosynthesis and the cell-specific localization of key enzymes, which represent different pathway branches of the alkaloid, have been demonstrated. Such a separation of the intermediates in different cell types might help to avoid undesired secondary modifications (e.g., acetylation or methylation) of the alkaloids (Weid et al., 2004). Likewise, in Arabidopsis, the key steps of the glucosinolate biosynthesis are localized in distinct cells, in order to minimize the risk of self-toxication (Fuchs et al., 2016; Nintemann et al., 2018). In Medicago enzymes like PAL and IFS represent enzymatic crossroads in the central phenylpropanoid pathway and isoflavonoid biosynthesis, respectively. Upon biotic stress/elicitation, their activity is crucial for de novo production of medicarpin, a major phytoalexin of Medicago (Naoumkina et al., 2007). Among the six PAL and three IFS genes present in the Medicago genome, those expression of which is highly and peculiarly induced by fungal elicitor, exhibit differential tissue localization in root. The upregulated PAL isoforms (PAL4, PAL6) were mainly expressed in the vascular bundles, while the IFSs (IFS1, IFS3) were generally present in the root cortex (Biala et al., 2017). Spatial separation of enzymatic steps leading to desired product suggests that cooperation between various metabolons in different cells cannot be excluded and sharing of the intermediates in such scenario could also be considered.
In addition to the possibility that certain low-molecular-weight molecules, e.g., 4-coumarate, can disperse by membrane diffusion, three various overlapping, but not exclusive, scenarios of phenylpropanoid distribution exist in planta: (i) vesicle trafficking, (ii) gluthatione S-transferases (GSTs)–supported, and (iii) membrane transporter mediated, fulfilled mostly by members of two protein families, namely the MATE (Multidrug and Toxic Compound Extrusion) and ABC (ATP binding cassette) (Figure 2) (Zhao and Dixon, 2010).
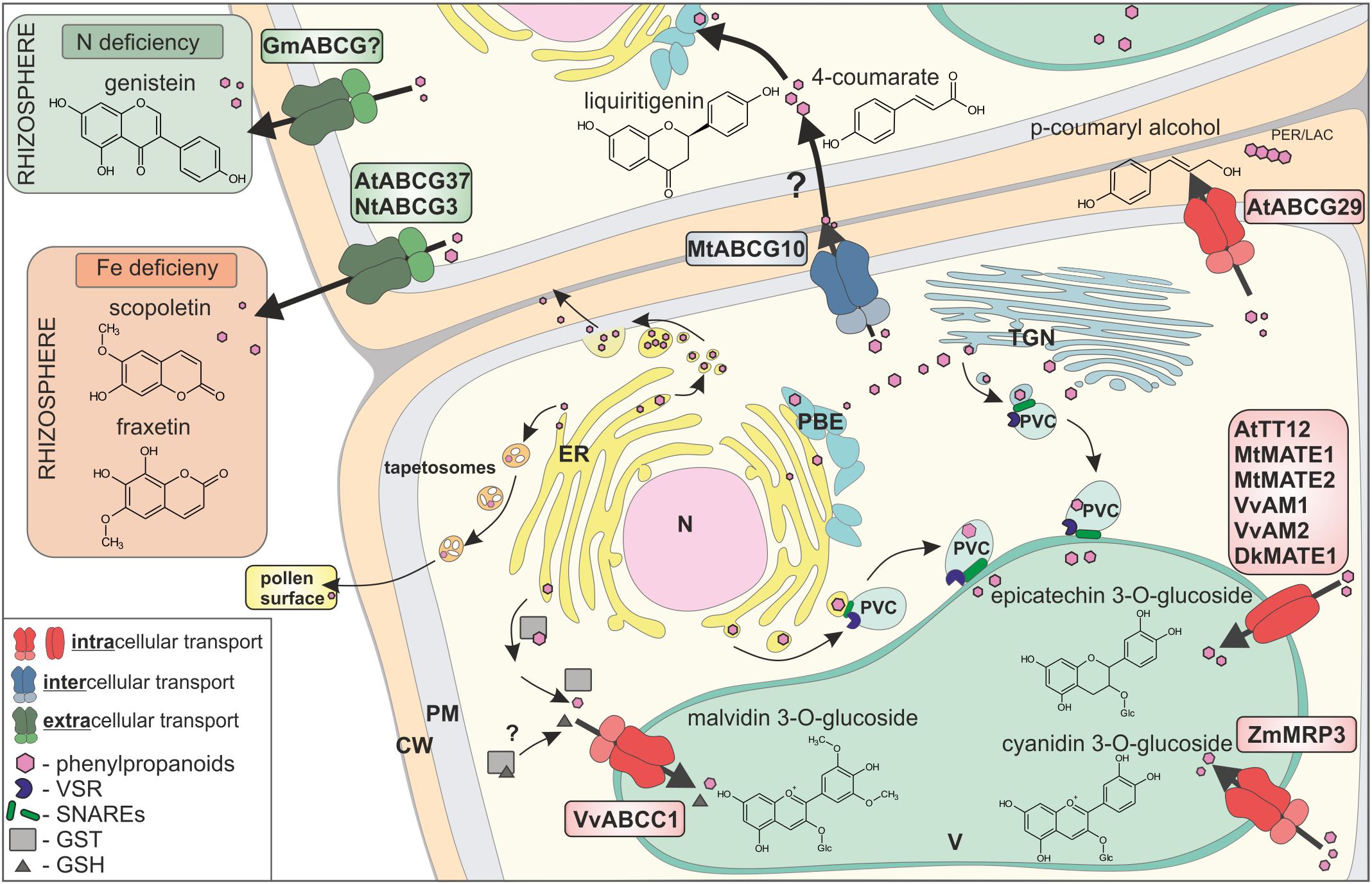
FIGURE 2. Transport of phenylpropanoids across biological membranes, by three different mechanisms: (i) vesicle trafficking, (ii) gluthatione S-transferases (GSTs)-supported, and (iii) membrane transporters. Membrane transporters are distinguished by respective colours, as involved in intracellular transport (red), intercellular transport (blue), and extracellular transport (green). CW, cell wall; PM, plasma membrane; ER, endoplasmic reticulum; N, nucleus; V, vacuole; TGN, trans-Golgi network; PVC, pre-vacuolar compartment; PBE, phenylpropanoid biosynthetic enzymes; PER/LAC, peroxidase/laccase; VSR, vacuolar sorting receptor; GST, glutathione S-transferase; GSH, glutathione.
All three scenarios are well-described in the intracellular distribution context and have been proposed to collaborate in the circulation of phenylpropanoids (Zhao, 2015). Vesicular transport may proceed in Golgi-dependent or independent manner, and it is controlled by adaptor proteins, like small GTPases and protein complexes such as SNARE (Žárský et al., 2009; Kulich and Žárský, 2014; Singh et al., 2014). It was shown that the distribution of, e.g., anthocyanins, into the vacuole occurs, inter alia, by vesicular transport (Winkel-Shirley, 2001; Hsieh and Huang, 2007; Poustka et al., 2007; Gomez et al., 2011; Ichino et al., 2014).
Gluthatione S-transferases from various plants, such as petunia (Petunia hybrida) Anthocyanin9 (AN9), maize (Zea mays) Bronze2 (BZ2), Arabidopsis Transparent Testa (TT) 19, and grapevine (Vitis vinifera) GST1/GST4 are also essential for anthocyanin and proanthocyanidin vacuolar accumulation (Conn et al., 2008). Two scenarios of GSTs involvement in anthocyanin accumulation were proposed: (i) GST activity and glutathione conjugation, followed by vacuolar sequestration of glutathione-conjugated anthocyanins, as well as (ii) just GST binding to anthocyanin and formation of GST-anthocyanin complexes protecting flavonoids from oxidation and/or guiding them to the central vacuole (Zhao and Dixon, 2010). Interestingly, it was also shown that free glutathione could be required for transport of the anthocyanin, malvidin 3-O-glucoside, into the vacuole (Francisco et al., 2013).
Membrane transporters belonging to MATE family from various species like V. vinifera (Gomez et al., 2009; Pérez-Díaz et al., 2014), persimmon (Diospyros kaki) (Yang et al., 2016), Arabidopsis (Marinova et al., 2007), as well as Medicago (Zhao and Dixon, 2009; Zhao et al., 2011) are engaged in the transport of phenylopropanoid glucosides into the vacuole. In Medicago, MATE1 transports epicatechin 3-O-glucoside associated with the synthesis of PAs, whereas MATE2 preferentially transports anthocyanin malonates (Zhao and Dixon, 2009; Zhao et al., 2011). Genetic studies have also shown that multidrug resistance-associated proteins (MRP)/C-type of ABC (ABCC) transporters, such as maize MRP3 and grapevine ABCC1, are involved in anthocyanin accumulation, with the assumption that they transport flavonoid conjugates through the tonoplast (Goodman et al., 2004; Francisco et al., 2013).
The metabolon formation together with intracellular distribution and, e.g., vacuolar storage, are important for the functioning of phenylpropanoids in particular cells. Less is known about the molecular determinants participating in the intercellular sharing/transport of those molecules. It is worth considering that export to the apoplast could be also a simple way to control, inter alia, the concentration of chemically/biologically active intermediates. The latter were shown to negatively modulate the biosynthesis of particular metabolites. For instance, the PAL activity is negatively regulated by trans-cinnamic acid and subsequent metabolites (Zhang and Liu, 2015). It was shown that in Arabidopsis, the double mutant deficient in UGT78D1 and UGT78D2 (UDP carbohydrate – dependent glycosylotransferases, responsible for flavonol-sugar conjugation) accumulating flavonols, repressed activity of flavonoid-related PAL isoforms as well as CHS, and inhibited flavonol synthesis (Yin et al., 2012). In this respect, the export to the apoplast could be recognized as a regulatory aspect of both the efficiency of cellular biosynthesis and intermediates dispersal.
Two transport mechanisms, (i) vesicular trafficking and (ii) membrane transporters, were shown to mediate phenylpropanoids transport out of the cell. In rape (Brassica napus) anthers, the ER derived vesicles – tapetosomes accumulate flavonoids, which are discharged to the pollen surface upon tapetum programmed cell death (Hsieh and Huang, 2007). Phenylpropanoids like, for instance, the hydroxycinnamic acid derivatives, were found in the cell walls, esterified to the wall polysaccharides. Hydroxycinnamic acid derivatives are synthesized at the ER, and from there, they are released as small membrane vesicles, which aggregate into bigger structures, fusing with the plasma membrane and releasing the content into the apoplast (Kulich and Žárský, 2014).
Membrane transporters of the G-type of ABC (ABCG) proteins have been shown to export phenylopropanoids out of the cell. Until recently, it was thought that most of the molecules translocated to the apoplast, by proteins like ABC transporters, are final products of individual biosynthetic pathway, fulfilling a particular biological role. For instance, the flavonoid genistein is suggested to be an exudate from soybean (Glycine max) roots, by the G-type of ABC (ABCG) transporters. Genistein, together with daidzein, were found in soybean root exudates, as signaling molecules mediating communication between plant and nitrogen fixing bacteria Bradyrhizobium japonicum (Sugiyama et al., 2007, 2008). Scopoletin, belonging to the coumarin subfamily, is synthesized in Arabidopsis roots and excreted to the rhizosphere by ABCG37, in order to facilitate Fe nutrition (Fourcroy et al., 2014, 2016; Ziegler et al., 2017). Similarly, in tobacco (Nicotiana tabacum), NtPDR3/ABCG3 is involved in secretion of O-methylated coumarins, such as fraxetin, to the rhizosphere (Lefèvre et al., 2018). For further reading, see Lefèvre and Boutry (2018).
Interestingly, it was recently shown that ABCG transporters might participate in transporting of the precursors from the phenylpropanoid pathway. The Medicago plasma membrane protein MtABCG10 is responsible for selective translocation of 4-coumarate, an early precursor of the core phenylpropanoid pathway, and liquiritigenin from the 5-deoxyflavonoid branch leading to medicarpin. Upon biotic stress, MtABCG10 action is strictly associated with the 5-deoxyflavonoid branch, even if early precursors like 4-coumarate are common for almost all of the phenylpropanoid products (Banasiak et al., 2013). Biotic stress driven expression of genes encoding enzymes crucial for de novo production of medicarpin like PAL and IFS goes along with MtABCG10. Moreover, PAL and MtABCG10 are concomitantly expressed in contrast to IFS present in different tissue. Thereby, it was proposed that MtABCG10 appears as a transporter facilitating allocation of common intermediates between various metabolons situated in diverse tissues, upon biotic stress (Biala et al., 2017). Another example is the AtABCG29, which in Arabidopsis has been described as a plasma membrane transporter of the p-coumaryl alcohol to the cell wall, where this molecule is further oxidized and finally polymerized to lignins (Miao and Liu, 2010; Alejandro et al., 2012). Moreover, it was also proposed that the translocation of the two other lignin precursors, namely sinapyl and coniferyl alcohol, is ABCG dependent (Takeuchi et al., 2018). The expression pattern of those putative transporters was similar to the transcription factor involved in lignin synthesis and lignin polymerization peroxidase (Takeuchi et al., 2018). Interestingly, the dedicated transcriptional network was also determined, underlying the production of benzenoid, as well as phenylpropanoid volatiles, in petunia flowers. This network involves: (i) the ODORANT1 transcription factor which controls biosynthesis in petunia flowers, (ii) a biosynthetic enzyme catalyzing an end-product, and (iii) ABC subfamily G member PhABCG1, a predicted plasma-membrane transporter that is expressed almost exclusively in petals of open flowers. However, the latter transporter is responsible for active transport of emitted volatiles across the plasma membrane, rather than the distribution of intermediates (Adebesin et al., 2017).
Despite our knowledge on phenylpropanoid biosynthetic enzymes, we are still about to discover how they spatially interact and what are the molecular determinants enabling such interaction. One currently existing gap in the targeted intercellular distribution scenario is the presence/identification of dedicated importers. Indeed, importers are key regulators of intracellular channeling of simple phenolics and/or biosynthesis of different end products. For instance, the full size peroxisomal ABC transporter of Arabidopsis ABCD1/PXA1 is indirectly linked to the synthesis of various secondary metabolites such as: (i) benzoic acids, likely by transport of cinnamic acid/cinnamoyl-CoA into the peroxisome (Bussell et al., 2014), (ii) ubiquinone, probably by import of 4-coumarate/coumaroyl-CoA (Block et al., 2014), and (iii) flavonoids by mediation in fatty acids breakdown, which induces flavonoid biosynthetic enzymes (Carrera et al., 2007). From biosynthesis of other secondary metabolites, e.g., root-synthesized glucosinolates in Arabidopsis, we have learnt that modulating specific import activities at the plasma membrane level could be essential for shaping the distribution pattern/biosynthesis at the organismal level (Xu et al., 2017). However, our knowledge about import as well as plasma membrane localized importers of phenylpropanoids is still limited.
Revealing the transport mechanisms/transporters involved in targeted intermediates distribution will bridge the knowledge gaps regarding spatiotemporal phenylpropanoid production under various conditions. It will also facilitate more precise metabolic engineering of those compounds in plants, in order to improve agronomic traits or nutritional value. In the future, metabolons could incorporate switching mechanisms in which metabolic status is sensed, causing association or disassociation of the enzymatic complexes. Such a switching mechanism could rely, inter alia, on tightly controlled transport/dedicated transporters, being present in both inter and intracellular scenarios.
WB and MJ designed and wrote the manuscript.
This work was supported by National Science Centre (Grant No. 2011/03/B/NZ1/02840).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to apologize to the authors whose research results could not be included in this review due to space constraints.
Achnine, L., Blancaflor, E. B., Rasmussen, S., and Dixon, R. A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 6, 3098–3109. doi: 10.1105/tpc.104.024406
Adebesin, F., Widhalm, J. R., Boachon, B., Lefèvre, F., Pierman, B., Lynch, J. H., et al. (2017). Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 30, 1386–1388. doi: 10.1126/science.aan0826
Alejandro, S., Lee, Y., Tohge, T., Sudre, D., Osorio, S., Park, J., et al. (2012). AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 10, 1207–1212. doi: 10.1016/j.cub.2012.04.064
Banasiak, J., Biala, W., Staszkow, A., Swarcewicz, B., Kepczynska, E., Figlerowicz, M., et al. (2013). A Medicago truncatula ABC transporter belonging to subfamily G modulates the level of isoflavonoids. J. Exp. Bot. 64, 1005–1015. doi: 10.1093/jxb/ers380
Bassard, J. E., and Halkier, B. A. (2018). How to prove the existence of metabolons? Phytochem. Rev. 17, 211–227. doi: 10.1007/s11101-017-9509-1
Bassard, J. E., Richert, L., Geerinck, J., Renault, H., Duval, F., Ullmann, P., et al. (2012). Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24, 4465–4482. doi: 10.1105/tpc.112.102566
Biala, W., Banasiak, J., Jarzyniak, K., Pawela, A., and Jasinski, M. (2017). Medicago truncatula ABCG10 is a transporter of 4-coumarate and liquiritigenin in the medicarpin biosynthetic pathway. J. Exp. Bot. 68, 3231–3241. doi: 10.1093/jxb/erx059
Block, A., Widhalm, J. R., Fatihi, A., Cahoon, R. E., Wamboldt, Y., Elowsky, C., et al. (2014). The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in Arabidopsis. Plant Cell 26, 1938–1948. doi: 10.1105/tpc.114.125807
Burbulis, I. E., and Winkel-Shirley, B. (1999). Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. U.S.A. 26, 12929–12934. doi: 10.1073/pnas.96.22.12929
Bussell, J. D., Reichelt, M., Wiszniewski, A. A., Gershenzon, J., and Smith, S. M. (2014). Peroxisomal ATP-binding cassette transporter COMATOSE and the multifunctional protein abnormal INFLORESCENCE MERISTEM are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol. 164, 48–54. doi: 10.1104/pp.113.229807
Carrera, E., Holman, T., Medhurst, A., Peer, W., Schmuths, H., Footitt, S., et al. (2007). Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol. 143, 1669–1679. doi: 10.1104/pp.107.096057
Chen, H. C., Li, Q., Shuford, C. M., Liu, J., Muddiman, D. C., Sederoff, R. R., et al. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 27, 21253–21258. doi: 10.1073/pnas.1116416109
Clemens, S., and Weber, M. (2016). The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav. 11:e1114197. doi: 10.1080/15592324.2015.1114197
Conn, S., Curtin, C., Bézier, A., Franco, C., and Zhang, W. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59, 3621–3634. doi: 10.1093/jxb/ern217
Crosby, K. C., Pietraszewska-Bogiel, A., Gadella, T. W. Jr., and Winkel, B. S. (2011). Förster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 585, 2193–2198. doi: 10.1016/j.febslet.2011.05.066
Dastmalchi, M., Bernards, M. A., and Dhaubhadel, S. (2016). Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 85, 689–706. doi: 10.1111/tpj.13137
Ehlting, J., Büttner, D., Wang, Q., Douglas, C. J., Somssich, I. E., and Kombrink, E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19, 9–20. doi: 10.1046/j.1365-313X.1999.00491.x
Fellenberg, C., and Vogt, T. (2015). Evolutionarily conserved phenylpropanoid pattern on angiosperm pollen. Trends Plant Sci. 20, 212–218. doi: 10.1016/j.tplants.2015.01.011
Fourcroy, P., Sisó-Terraza, P., Sudre, D., Savirón, M., Reyt, G., Gaymard, F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167. doi: 10.1111/nph.12471
Fourcroy, P., Tissot, N., Gaymard, F., Briat, J. F., and Dubos, C. (2016). Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity Root Fe(2+) transport system. Mol. Plant 9, 485–488. doi: 10.1016/j.molp.2015.09.010
Francisco, R. M., Regalado, A., and Ageorges, A. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 25, 1840–1854. doi: 10.1105/tpc.112.102152
Fuchs, R., Kopischke, M., Klapprodt, C., Hause, G., Meyer, A. J., Schwarzländer, M., et al. (2016). Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28, 130–145. doi: 10.1105/tpc.15.00887
Fujino, N., Tenma, N., Waki, T., Ito, K., Komatsuzaki, Y., Sugiyama, K., et al. (2018). Physical interactions among flavonoid enzymes in snapdragon and torenia reveal the diversity in the flavonoid metabolon organization of different plant species. Plant J. 94, 372–392. doi: 10.1111/tpj.13864
Gomez, C., Conejero, G., Torregrosa, L., Cheynier, V., Terrier, N., and Ageorges, A. (2011). In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant J. 67, 960–970. doi: 10.1111/j.1365-313X.2011.04648.x
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-Level, A., Verriès, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H + -dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
Goodman, C. D., Casati, P., and Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16, 1812–1826. doi: 10.1105/tpc.022574
Hsieh, K., and Huang, A. H. (2007). Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell 19, 582–596. doi: 10.1105/tpc.106.049049
Huang, J., Gu, M., Lai, Z., Fan, B., Shi, K., Zhou, Y. H., et al. (2010). Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 153, 1526–1538. doi: 10.1104/pp.110.157370
Ichino, T., Fuji, K., and Ueda, H. (2014). GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 80, 410–423. doi: 10.1111/tpj.12637
Jørgensen, K., Rasmussen, A. V., Morant, M., Nielsen, A. H., Bjarnholt, N., Zagrobelny, M., et al. (2005). Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 8, 280–291. doi: 10.1016/j.pbi.2005.03.014
Kulich, I., and Žárský, V. (2014). Autophagy-related direct membrane import from ER/cytoplasm into the vacuole or apoplast: a hidden gateway also for secondary metabolites and phytohormones? Int. J. Mol. Sci. 15, 7462–7474. doi: 10.3390/ijms15057462
Kutchan, T. M., Gershenzon, J., Lindberg Moller, B., and Gang, D. R. (2015). “Natural products,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchnan, W. Gruissem, and R. L. Jones (Blackwell: Wiley), 1132–1206.
Kwiatkowska, M., Polit, J. T., Stȩpiński, D., Popłońska, K., Wojtczak, A., Domínguez, E., et al. (2015). Lipotubuloids in ovary epidermis of Ornithogalum umbellatum act as metabolons: suggestion of the name ’lipotubuloid metabolon’. J. Exp. Bot. 66, 1157–1163. doi: 10.1093/jxb/eru469
Kyoung, M., Russell, S. J., Kohnhorst, C. L., Esemoto, N. N., and An, S. (2015). Dynamic architecture of the purinosome involved in human de novo purine biosynthesis. Biochemistry 54, 870–880. doi: 10.1021/bi501480d
Lanfranco, L., Bonfante, P., and Genre, A. (2016). The mutualistic interaction between plants and arbuscular Mycorrhizal fungi. Microbiol. Spectr. 4, 1–20. doi: 10.1128/microbiolspec
Laursen, T., Møller, B. L., and Bassard, J. E. (2015). Plasticity of specialized metabolism as mediated by dynamic metabolons. Trends Plant Sci. 20, 20–32. doi: 10.1016/j.tplants.2014.11.002
Lefèvre, F., and Boutry, M. (2018). Towards identification of the substrates of ATP-binding cassette transporters. Plant Physiol. 178, 18–39. doi: 10.1104/pp.18.00325
Lefèvre, F., Fourmeau, J., Pottier, M., Baijot, A., Cornet, T., Abadía, J., et al. (2018). The Nicotiana tabacum ABC transporter NtPDR3 secretes O-methylated coumarins in response to iron deficiency. J. Exp. Bot. J. 69, 4419–4431. doi: 10.1093/jxb/ery221
Li, Y., Kim, J. I., Pysh, L., and Chapple, C. (2015). Four isoforms of Arabidopsis 4-Coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Liu, C. J., and Dixon, R. A. (2001). Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents the formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13, 2643–2658. doi: 10.1105/tpc.13.12.2643
Liu, C. W., and Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants 5:E33. doi: 10.3390/plants5030033
Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J. M., et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H + -antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19, 2023–2038. doi: 10.1105/tpc.106.046029
Miao, Y. C., and Liu, C. J. (2010). ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S.A. 107, 22728–22733. doi: 10.1073/pnas.1007747108
Naoumkina, M., Farag, M. A., Sumner, L. W., Tang, Y., Liu, C. J., and Dixon, R. A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 104, 17909–17915. doi: 10.1073/pnas.0708697104
Naoumkina, M. A., Zhao, Q., Gallego-Giraldo, L., Dai, X., Zhao, P. X., and Dixon, R. A. (2010). Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11, 829–846. doi: 10.1111/j.1364-3703.2010.00648.x
Nintemann, S. J., Hunziker, P., Andersen, T. G., Schulz, A., Burow, M., and Halkier, B. A. (2018). Localization of the glucosinolate biosynthetic enzymes reveals distinct spatial patterns for the biosynthesis of indole and aliphatic glucosinolates. Physiol. Plant. 163, 138–154. doi: 10.1111/ppl.12672
Olsen, K. M., Lea, U. S., Slimestad, R., Verheul, M., and Lillo, C. (2008). Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J. Plant Physiol. 29, 1491–1499. doi: 10.1016/j.jplph.2007.11.005
Pérez-Díaz, R., Ryngajllo, M., Pérez-Díaz, J., Peña-Cortés, H., Casaretto, J. A., González-Villanueva, E., et al. (2014). VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep. 33, 1147–1159. doi: 10.1007/s00299-014-1604-9
Poustka, F., Irani, N. G., Feller, A., Lu, Y., Pourcel, L., Frame, K., et al. (2007). A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 145, 1323–1335. doi: 10.1104/pp.107.105064
Saslowsky, D., and Winkel-Shirley, B. (2001). Localization of flavonoid enzymes in Arabidopsis roots. Plant J. 27, 37–48. doi: 10.1046/j.1365-313x.2001.01073.x
Shalaby, S., and Horwitz, B. A. (2015). Plant phenolic compounds and oxidative stress: integrated signals in fungal-plant interactions. Curr. Genet. 61, 347–357. doi: 10.1007/s00294-014-0458-6
Singh, M. K., Krüger, F., Beckmann, H., Brumm, S., Vermeer, J. E., Munnik, T., et al. (2014). Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr. Biol. 16, 1383–1389. doi: 10.1016/j.cub.2014.05.005
Sugiyama, A., Shitan, N., and Yazaki, K. (2007). Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol. 144, 2000–2008. doi: 10.1104/pp.107.096727
Sugiyama, A., Shitan, N., and Yazaki, K. (2008). Signaling from soybean roots to rhizobium: an ATP-binding cassette-type transporter mediates genistein secretion. Plant Signal. Behav. 3, 38–40. doi: 10.4161/psb.3.1.4819
Sweetlove, L. J., and Fernie, A. R. (2013). The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 64, 723–746. doi: 10.1146/annurev-arplant-050312-120233
Takeuchi, M., Kegasa, T., Watanabe, A., Tamura, M., and Tsutsumi, Y. (2018). Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J. Plant Res. 131, 297–305. doi: 10.1007/s10265-017-0979-4
Weid, M., Ziegler, J., and Kutchan, T. M. (2004). The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proc. Natl. Acad. Sci. U.S.A. 101, 13957–13962. doi: 10.1073/pnas.0405704101
Winkel-Shirley, B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107, 142–149. doi: 10.1034/j.1399-3054.1999.100119.x
Winkel-Shirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Wu, F., Pelster, L. N., and Minteer, S. D. (2015). Krebs cycle metabolon formation: metabolite concentration gradient enhanced compartmentation of sequential enzymes. Chem. Commun. 51, 1244–1247. doi: 10.1039/c4cc08702j
Xu, D., Hanschen, F. S., Witzel, K., Nintemann, S. J., Nour-Eldin, H. H., Schreiner, M., et al. (2017). Rhizosecretion of stele-synthesized glucosinolates and their catabolites requires GTR-mediated import in Arabidopsis. J. Exp. Bot. 68, 3205–3214. doi: 10.1093/jxb/erw355
Yang, L., Wen, K. S., Ruan, X., Zhao, Y. X., Wei, F., and Wang, Q. (2018). Response of plant secondary metabolites to environmental factors. Molecules 23:E762. doi: 10.3390/molecules23040762
Yang, S., Jiang, Y., Xu, L., Shiratake, K., Luo, Z., and Zhang, Q. (2016). Molecular cloning and functional characterization of DkMATE1 involved in proanthocyanidin precursor transport in persimmon (Diospyros kaki Thunb.) fruit. Plant Physiol. Biochem. 108, 241–250. doi: 10.1016/j.plaphy.2016.07.016
Yin, R., Messner, B., Faus-Kessler, T., Hoffmann, T., Schwab, W., Hajirezaei, M. R., et al. (2012). Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J. Exp. Bot. 63, 2465–2478. doi: 10.1093/jxb/err416
Žárský, V., Cvrckova, F., Potocky, M., and Hala, M. (2009). Exocytosis and cell polarity in plants – exocyst and recycling domains. New Phytol. 183, 255–272. doi: 10.1111/j.1469-8137.2009.02880.x
Zhang, X., and Liu, C. J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant 8, 17–27. doi: 10.1016/j.molp.2014.11.001
Zhao, J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007
Zhao, J., and Dixon, R. A. (2009). MATE transporters facilitate vacuolar uptake of epicatechin 3’-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21, 2323–2340. doi: 10.1105/tpc.109.067819
Zhao, J., and Dixon, R. A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15, 72–80. doi: 10.1016/j.tplants.2009.11.006
Zhao, J., Huhman, D., Shadle, G., He, X. Z., Sumner, L. W., Tang, Y., et al. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23, 1536–1555. doi: 10.1105/tpc.110.080804
Keywords: phenylpropanoids, transport, intermediates, metabolons, membrane transporters
Citation: Biała W and Jasiński M (2018) The Phenylpropanoid Case – It Is Transport That Matters. Front. Plant Sci. 9:1610. doi: 10.3389/fpls.2018.01610
Received: 19 July 2018; Accepted: 17 October 2018;
Published: 01 November 2018.
Edited by:
Jan Petrášek, Charles University, CzechiaReviewed by:
Juergen Ehlting, University of Victoria, CanadaCopyright © 2018 Biała and Jasiński. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michał Jasiński, amFzaW5za2lAaWJjaC5wb3puYW4ucGw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.