
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 October 2018
Sec. Crop and Product Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01470
 Michela Chiumenti1†
Michela Chiumenti1† Claudia Rita Catacchio2†
Claudia Rita Catacchio2† Laura Miozzi3
Laura Miozzi3 Walter Pirovano4
Walter Pirovano4 Mario Ventura2
Mario Ventura2 Vitantonio Pantaleo1*
Vitantonio Pantaleo1*Plant viruses modify gene expression in infected tissues by altering the micro (mi)RNA-mediated regulation of genes. Among conserved miRNA targets there are transcripts coding for transcription factors, RNA silencing core, and disease-resistance proteins. Paralogs in these gene families are widely present in plant genomes and are known to respond differently to miRNA-mediated regulation during plant virus infections. Using genome-wide approaches applied to Solanum lycopersicum infected by a nuclear-replicating virus, we highlighted miRNA-mediated cleavage events that could not be revealed in virus-free systems. Among them we confirmed miR6024 targeting and cleavage of RX-coiled-coil (RX-CC), nucleotide binding site (NBS), leucine-rich (LRR) mRNA. Cleavage of paralogs was associated with short indels close to the target sites, indicating a general functional significance of indels in fine-tuning gene expression in plant–virus interaction. miR6024-mediated cleavage, uniquely in virus-infected tissues, triggers the production of several 21–22 nt secondary siRNAs. These secondary siRNAs, rather than being involved in the cascade regulation of other NBS–LRR paralogs, explained cleavages of several mRNAs annotated as defence-related proteins and components of the photosynthetic machinery. Outputs of these data explain part of the phenotype plasticity in plants, including the appearance of yellowing symptoms in the viral pathosystem.
RNA silencing refers to conserved pathways affecting gene expression through negative regulation mediated by non-coding RNAs, such as short interfering (si)RNAs. In plants, micro (mi)RNA comprises one of the most abundant classes of 21- to 24-nucleotide (nt)-long small RNAs that mediate post-transcriptional regulation of endogenous messenger (m)RNAs (Bartel, 2004). The biogenesis and activity of plant miRNAs are regulated by DICER-LIKE1 (DCL1) and Argonaute1 (AGO1), respectively. miRNAs function as a guide by base-pairing with their target RNAs, whereas AGO1 [mainly, but not exclusively (Adenot et al., 2006)] plays a role as effector, recruiting factors that induce mRNA translational repression and/or mRNA cleavage (Bartel, 2004; Voinnet, 2009; Iwakawa and Tomari, 2013). The 3′- and 5′-cleavage remnants of targeted mRNAs can be detected either by Northern blot analysis or rapid amplification of cDNA ends (RACE). The 5′-uncapped fragments detected by 5′RACE correspond to cleavage between the 10th and 11th nucleotides of miRNA/target site pairs. A genome-wide 5′-RACE analysis using next-generation sequencing, commonly known as parallel analysis of RNA ends (PARE), has been developed and used in plants for miRNA discovery and target validation (Addo-Quaye et al., 2008; German et al., 2008).
miRNAs may also initiate regulatory cascades that have a massive effect on the regulation of gene expression, reinforcing miRNA activity and ensuring gene expression homeostasis of large gene families. These cascades involve secondary siRNAs that associate with AGO proteins, similarly to miRNAs. Secondary siRNAs arise predominantly from RNAs that are targeted and cleaved by 22-nt- (Cuperus et al., 2010) or, if containing two target sites, 21-nt-miRNAs (Axtell et al., 2006). In the former case, the 22-nt miRNA duplex induces a conformational change in the AGO1 protein that allows the RNA-induced silencing complex (RISC) to recruit an RNA-dependent RNA polymerase (RDR6) and other components necessary for secondary siRNA production (Yoshikawa et al., 2005; Manavella et al., 2012). RDR6 converts the targeted RNA into long, double-stranded RNA which is then processed by Dicer-like protein 4 (DCL4) into secondary siRNAs mainly in a 21-nt-phased register starting from the miRNA cleavage site (Chen et al., 2007). The biogenesis of secondary siRNAs extends toward regions upstream or downstream of the initial target site, and the extension in the 5′- to 3′-direction is frequently observed (Aregger et al., 2012). Bioinformatics and molecular biology studies have shown that secondary siRNAs originate from non-coding, transacting siRNA loci such as those discovered at first in Arabidopsis (Peragine et al., 2004; Vazquez et al., 2004). In addition, secondary siRNAs also originate from coding genes of pentatricopeptide (Howell et al., 2007), auxin receptors (Si-Ammour et al., 2011; Windels and Vazquez, 2011), and NBS–LRR clades (Shivaprasad et al., 2012). These secondary siRNAs were shown to have roles in the expression of genes belonging to the same gene families since they can target paralogs in homologous regions.
Plant virus infections often result in onset of symptoms that are reconcilable with virus-induced alterations of RNA silencing-based endogenous pathways, due to: (i) the direct activity of viral silencing suppressors on endogenous sRNAs or on silencing related effectors (Csorba et al., 2015); (ii) the abundance of virus-deriving (v)-siRNAs in competition with endogenous sRNAs; (iii) the action of specific v-siRNAs entering into RNA silencing complexes and targeting specific host genes (Shimura and Pantaleo, 2011; Miozzi et al., 2013a); and (iv) potential functionalities of endogenous siRNAs triggered by viral infections (i.e., va-siRNAs, Cao et al., 2014). Tomato yellow leaf curl Sardinia virus (TYLCSV) is a Begomovirus of the family Geminiviridae. Geminiviruses are circular single-stranded (ss)DNA viruses forming mini-chromosomes associated with cellular histones and are transcribed and replicated in the nucleus through dsDNA intermediates (Rojas et al., 2005; Brown et al., 2012). Geminiviruses encode viral suppressors of RNA silencing that are known to interfere either with DNA methylation (Buchmann et al., 2009; Yang et al., 2011; Zhang et al., 2011) or with the biogenesis of RDR6-dependent secondary siRNAs (Glick et al., 2008). Geminiviruses belonging to the Begomovirus genus have been extensively studied for their capacity to alter the accumulation of specific conserved miRNAs in infected tissues, resulting in the modification of the translation landscape of genes involved in the development of plants (Naqvi et al., 2010; Amin et al., 2011). TYLCSV induces specific systemic yellowing in tomato through a mechanism that has yet to be fully characterized.
Plant miRNA targets fall within gene families whose members have largely redundant functions and are mostly present in plant genomes in several copies. Whole genome duplication is a significant aspect in the evolution of eukaryotes, and evidence shows that most, if not all, angiosperms have undergone at least one ancient genome-doubling event in their evolutionary history (Vanneste et al., 2014). As such, many paralogs show divergence in gene structure, expression pattern, and function. Furthermore, two paralogs may diverge in their miRNA binding sites and surroundings, which may impact their fine expression and function (Wang and Adams, 2015). Recently, it has been shown that Nicotiana benthamiana possesses two functional NbAGO1-like paralogs. One 18-nt-long insertion/deletion (indel), which is located in the immediate vicinity of the miR168 target site, considerably affects the miR168-guided, post-transcriptional regulation of NbAGO1 mRNAs. The indel effect is underpinned under conditions of viral infection. Indeed, NbAGO1 homeologs are redundantly involved in their susceptibility to viral infection but are divergently involved in the phenotype associated with the infection (Gursinsky et al., 2015).
In this report, we diagnosed miRNA-mediated cleaved transcripts in both healthy and virus-infected plants through PARE. We integrated the experimental data with publicly available annotations and gained insight into the parallel evolution of gene families and their miRNA-mediated transcriptional regulation. We focused our attention on those targets of conserved miRNAs (i.e., those commonly shared among flowering plants) that were exclusively revealed in virus-infected plants and identified their closest evolutionary paralogs. Concerning the capacity to reveal the miR168-mediated cleavage of a longer sly-AGO1 transcript (containing a 21-nt indel downstream the miRNA target site), we found that geminivirus infection in tomato parallels previous observations in tombusvirus-N. benthamiana (Gursinsky et al., 2015). Similarly, we revealed several cases of duplicated genes that are distinguished by 3-nt to 49-nt indels close to miRNA recognition sites and, in most cases, only paralogs lacking the indel were cleaved, being bona fide more vulnerable to miRNA-mediated cleavage. Within specific targets, our study revealed that one mRNA transcript coding for a CC–NBS–LRR protein holds both the miR482 and miR6024 target sites, which overlap to some extent. By mRNA-seq and PARE analysis we found that the transcript is significantly down-regulated and cleaved only in virus-infected plant tissues. Moreover, 21- to 22-nt-long secondary siRNAs were found associated with it either in a phased register or not. Unexpectedly, none of the secondary siRNAs from the miR6024-cleaved NBS–LRR were involved in cascade regulation of NBS–LRR paralogs; instead, some of them were involved in regulation of other disease proteins. Surprisingly, a group of these secondary siRNAs explained cleavages of several mRNAs coding for factors implicated in photosynthesis processes; this finding could explain the typical yellowing symptomatology induced by TYLCSV infection, thus suggesting a novel molecular mechanism involved in plant symptom development associated with biotic stresses.
Plants of S. lycopersicum L. cultivar “Moneymaker” were inoculated, as previously described, either by TYLCSV or by mock (Miozzi et al., 2013b). Plants were maintained in an insect-proof greenhouse at temperatures of 20–28°C/16–20°C (day/night), with a 16/8-h (light/dark) photoperiod and supplementary lighting. A minimum of six leaves per plant (from six plants, three biological replicates/thesis) were collected at the appearance of yellowing and curling symptoms typical of TYLCSV infection. Samples were ground in liquid nitrogen and used for total RNA extraction. RNAs were extracted using the Sigma RNA plant kit. DNA contaminants were removed using the Turbo DNA-free kit (Bio-Rad).
For Northern blot analysis, 10 μg of total RNA from either mock-inoculated or virus-infected leaf tissue were resolved on 12% PAGE gels. Once transferred to the positively charged nylon membrane Hybond N+ (Roche Diagnostics), miR171, and miR168 were detected as internal controls with oligoprobes, as previously described (Pantaleo et al., 2010b), whereas functional Solyc05g008070-derived siRNAs (Figure 7C) were detected using a mixture of P32-labeled DNA antisense oligoprobes. Radiolabeled signals were observed by Storm 860 Molecular Imager.
DNA either from TYLCSV-infected or from mock-inoculated tomato leaf tissues was extracted as previously described (Noris et al., 1996). Two micrograms of DNA were resolved on 1% Agarose, 0.5X Tris–borate–EDTA, and transferred to the positively charged nylon membrane Hybond N+. Viral replication intermediates were detected using a P32-labeled probe generated by random priming on a CP-amplicon, as previously reported (Accotto et al., 2000).
Two mRNA libraries for each of the two theses, virus-infected and mock-inoculated plant tissues, were obtained with Illumina strand-specific mRNA protocol sRNAs and sequenced. Libraries of sRNAs were produced using a TruSeq Small RNA Sample Kit (Illumina) and sequenced with standard sequencing oligos on the Illumina HiSeq 2500 platform. Differential expression of selected transcripts was confirmed by qRT-PCR (see later “Materials and Methods”).
The poly-A fraction of total RNA extracted from leaf tissue was analyzed for the identification of target transcripts of conserved miRNAs. To generate miRNA-cleaved target libraries (RNA-end-dataset) from leaf tissue of either mock-inoculated or virus-infected, symptomatic tomato plants we applied the previously described high-throughput experimental approach that identifies mRNAs targeted by miRNAs, also known as “PARE” (Addo-Quaye et al., 2008; German et al., 2009). PARE libraries were sequenced using 5′-GAGATCTACACGTTCAGAGTTCTACAGTCCGA-3′ as oligo on the Illumina HiSeq 2500 platform.
A quality check of the transcriptome was performed with FASTX toolkit1. mRNAs were aligned to the selected paralogous genes using PatMaN aligner with 0 mismatch (Prufer et al., 2008). A raw count of mapped reads, uniquely aligned on each one of the two paralogous genes considered, was used as input for the differential expression (DESeq) analysis (Anders, 2010). sRNA-seq fastq datasets were converted to fasta format, adapters were removed and selected for molecules 16- to 26-nt in length using the FASTX toolkit. Genomic and viral-derived siRNAs were identified by aligning filtered sRNAs to the Tomato genome [retrieved at solgenomics.net, (Fernandez-Pozo et al., 2015)] and viral Ref_Seq database available at the NCBI. Bowtie software was used for alignment allowing 0 mismatches (Langmead et al., 2009). miRNAs were identified using miRProf (Stocks et al., 2012) with perfect matches to sequences from the miRNA repository [miRBASE release 21, (Kozomara and Griffiths-Jones, 2014)]. Alignment of viral siRNAs to the TYLCSV genome (Accession number NC_003828.1) was performed as above and SAM output files were used as input in Geneious® software 8.1.7 (Biomatters Ltd.) in order to obtain a graphical representation of viral siRNAs along the viral genome.
RNA-end libraries underwent the same bioinformatics flowchart indicated above for sRNAs, with the only difference being the 20- to 21-nt filter size that was used before performing the alignment.
5′-RACE was performed as previously described (Gursinsky et al., 2015), except for the following: the 5′ adapter oligo used was described by German et al. (German et al., 2009), whereas the reverse specific oligo used is 5′-WTGGGWTGCASCTGCTTCTGG-3′. Cleaved sly-AGO1a and sly-AGO1b were discriminated based on the indels and SNPs revealed by Sanger sequence downstream of the cleavage target site. PARE datasets were aligned with the NCBI seq IDs of sly-AGO1a and sly-AGO1b using Bowtie with 0 mismatches. The SAM file output was used in Geneious® software 8.1.7 (Biomatters Ltd.) to obtain the graphical representation.
cDNA targeted by sRNA were identified using PAREsnp software (Folkes et al., 2012). miRNAs selected from sRNA datasets by miRProf (see above) and PARE datasets from Tomato mock-inoculated or TYLCSV-inoculated plant tissues were used as input in both cases. PAREsnp was performed using cDNA library ITAG2.42.
The selected targeted cDNAs from PAREsnp analysis were searched in the Solgenomics platform (Fernandez-Pozo et al., 2015) and the SGNs associated with each gene model were then filtered out considering those from PAREsnp analysis. Orphan SGNs, i.e., those targeted by sRNAs but not associated with any gene model in ITAG2.4, were subsequently searched for in the Solgenomics datasets and queried with BLAST (Altschul et al., 1997) against the tomato genome cDNA (ITAG release 2.40) for a better annotation.
The gene orthology and paralogy predictions were downloaded from Ensembl (Ensembl Plants release 34—December 2016) [in EnsemblCompara GeneTrees: Analysis of complete, duplication-aware phylogenetic trees in vertebrates (Vilella et al., 2009)]. Both Plant Compara and Pan-Taxonomic Compara phylogenies were used to retrieve orthologous genes.
Protein domain predictions were obtained through the NCBI CD-Search (Marchler-Bauer and Bryant, 2004) searching against the CDD v3.14 database, and schematic representations were drawn by the CLC biogenomics workbench (QIAGEN Bioinformatics).
psRNATarget (Dai and Zhao, 2011) was used under default parameters. The preloaded small RNA “user-submitted transcripts” was chosen. Both cleavage and translation types of regulation were studied.
Centroid plain RNA secondary structure drawings and positional entropy were obtained using RNAfold with default parameters on the Vienna RNA website (Mathews et al., 2004; Gruber et al., 2008).
Identification of siRNAs with a phased register with the miRNA cleavage site on selected genes was obtained using the ta-si prediction tool from UEA small RNA workbench (Stocks et al., 2012) using as threshold a p-value of 0.01. A subtraction step of common sRNAs of mock-inoculated from TYLCSV-inoculated plant tissue dataset was run on Galaxy website3. The TYLCSV-infected plant tissue dataset filtered from the previous step was used in a PaTmaN alignment with 0 mismatches using sequences selected from mRNAs as reference (Prufer et al., 2008). TYLCSV-only mapping 21- and 22-nt-long siRNAs, either in a phased register or not, were used for a PAREsnp analysis as described above for miRNA target identification.
After quantification with Picodrop, the quality of RNA was checked using the Experion automated electrophoresis station (Bio-Rad). Total RNA (500 ng) was retro-transcribed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). All quantitative reverse-transcription PCR (qRT-PCR) assays were carried out using iTaq Universal SYBR Green Supermix (Bio-Rad, CA, United States) in an apparatus CFX Connect Real-Time PCR Detection System (Bio-Rad, CA, United States). The comparative threshold cycle method was employed to calculate relative expression levels using tomato UBC (ubiquitin-conjugating enzyme, SGN-U582847) as the reference gene (Miozzi et al., 2014). The list of primers used is shown in Supplementary Table S6. All qRT-PCR reactions were performed using three biological replicates and two technical replicates.
Small (s)RNA datasets were obtained from the leaf tissue of mock-inoculated and TYLCSV-infected plants. Primary quality checks yielded a total of 17.8 million reads in the range of 18–34-nt (Figure 1A and Supplementary Table S1). sRNAs not matching the tomato genome were used in order to retrieve v-siRNAs, i.e., 0.7 million redundant reads in the case of TYLCSV-infected plants. Traces of v-siRNAs found in mock-inoculated plants, i.e., 71 in total, were not considered reliable marks of viral infection: they are rather siRNAs in datasets that likely align randomly with 0 mismatches with the all viral reference sequence (Ref_Seq) dataset. In this case, we almost found a 0 level of redundancy (69 unique out of 71 redundant, Supplementary Table S1), whereas v-siRNAs from full replicative viruses in infected tissues were characterized by a high level of redundancy due at least to the replication process and the production of secondary v-siRNAs (Pantaleo et al., 2010a; Miozzi et al., 2013b; Pirovano et al., 2014; Ghasemzadeh et al., 2018; Figure 1B). Southern blot hybridization on the total DNA extracted from plant tissues confirmed the infection and the active replication of the virus. Indeed, all TYLCSV replicative forms were detected only in virus-infected plant tissues (i.e., ssDNA, dsDNA, and open circle, Figure 1C, lane 1 vs. lane 2), indicating an active TYLCSV infection in the tissues analyzed.
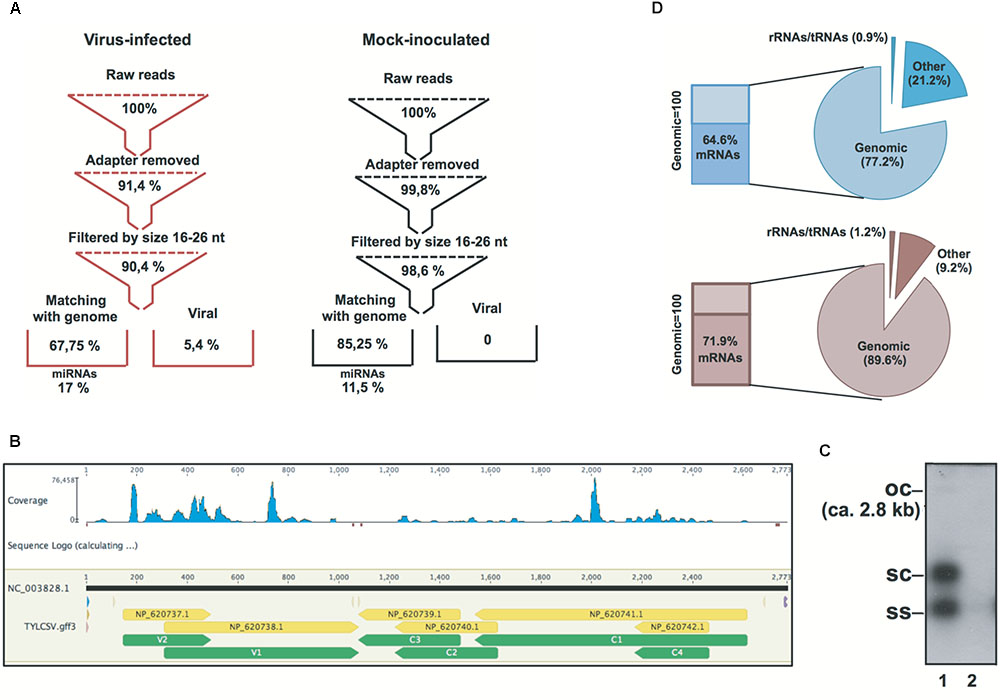
FIGURE 1. Statistics of Solanum lycopersicum genomic and non-genomic siRNAs in TYLCSV-infected and mock-inoculated plants. (A) Flowchart of sRNA datasets from raw data to genomic miRNAs and non-genomic viral sRNAs in virus-infected and mock-inoculated plants, respectively. (B) Distribution of TYLCSV-derived siRNAs in virus-infected plants.(C) Southern blot analysis of DNA from TYLCSV-infected tomato leaf tissue (lane 1) or from mock-inoculated tomato leaf tissue (lane 2). oc = open circle; sc = supercoiled; ss = single stranded. (D) Composition of PARE datasets in mock-inoculated and virus-infected plant tissues in terms of alignment or reads with rRNA, tRNAs, tomato genomic sequence, and tomato transcriptome (indicated in the figure as “mRNAs”).
PARE datasets were obtained from the same RNA preparations used for the sRNA libraries in line with previous reports (German et al., 2008; Arikit et al., 2014). RNA remnants of this size represent the 5′ ends of uncapped, poly-adenylated RNAs. Indeed, after initial processing, a small percentage of RNA remnants aligned to rRNAs/tRNAs (i.e., 0.9 and 1.2% for mock-inoculated and virus-infected plants, respectively) and the largest amount aligned to the S. lycopersicum genome (i.e., 77.2 and 89.6% for mock-inoculated and virus-infected plants, respectively). In addition, 64.6 and 71.9% of the reads aligning to the S. lycopersicum genomic sequence (for mock-inoculated and virus-infected plants, respectively) also aligned to the S. lycopersicum mRNA dataset (Figure 1D).
A total of 2.7 million redundant miRNAs were identified corresponding to only 250 and 251 unique miRNAs for TYLCSV-infected or mock-inoculated plants, respectively (Figure 1A). Some of the miRNAs analyzed were up-regulated in the presence of viral infection, while others were down-regulated in line with previous observations in Geminivirus-infected plants (Supplementary Tables S1–S3) (Naqvi et al., 2010; Amin et al., 2011).
The PARE datasets allowed us to assess the miRNA-mediated regulation of genes belonging to gene families coding for transcriptional factors and disease-resistance proteins, which are known to be altered under biotic stresses (Bartel, 2009). Therefore, we focused on transcriptional factors, such as Squamosa promoter binding proteins (SPL), Auxin response factors (ARF), Homeobox-leucine zipper proteins (HD-ZIPIII), nuclear transcription factors of the class of heme activator proteins (HAP), APETALA (AP) and related AP (RAP), and NBS–LRR-resistance factors. These gene families are regulated by miR156, miR160, miR166, miR169, miR172, and miR6024, respectively (Rhoades et al., 2002). Twenty-seven different genes from six different gene families were regulated by the above-mentionedsix miRNAs. Seventeen of them were cleaved in both mock-inoculated and virus-infected plants, while nine were targeted only in the virus-infected plants and only one was exclusively cleaved in the mock-inoculated plants (Table 1). We also interrogated psRNATarget (Dai and Zhao, 2011; Shao et al., 2014) and annotated the potential regulation of every single paralog belonging to each analyzed gene family. Twenty-five out of 27 genes from PARE analysis were common with psRNATarget (Supplementary Tables S5, S6).
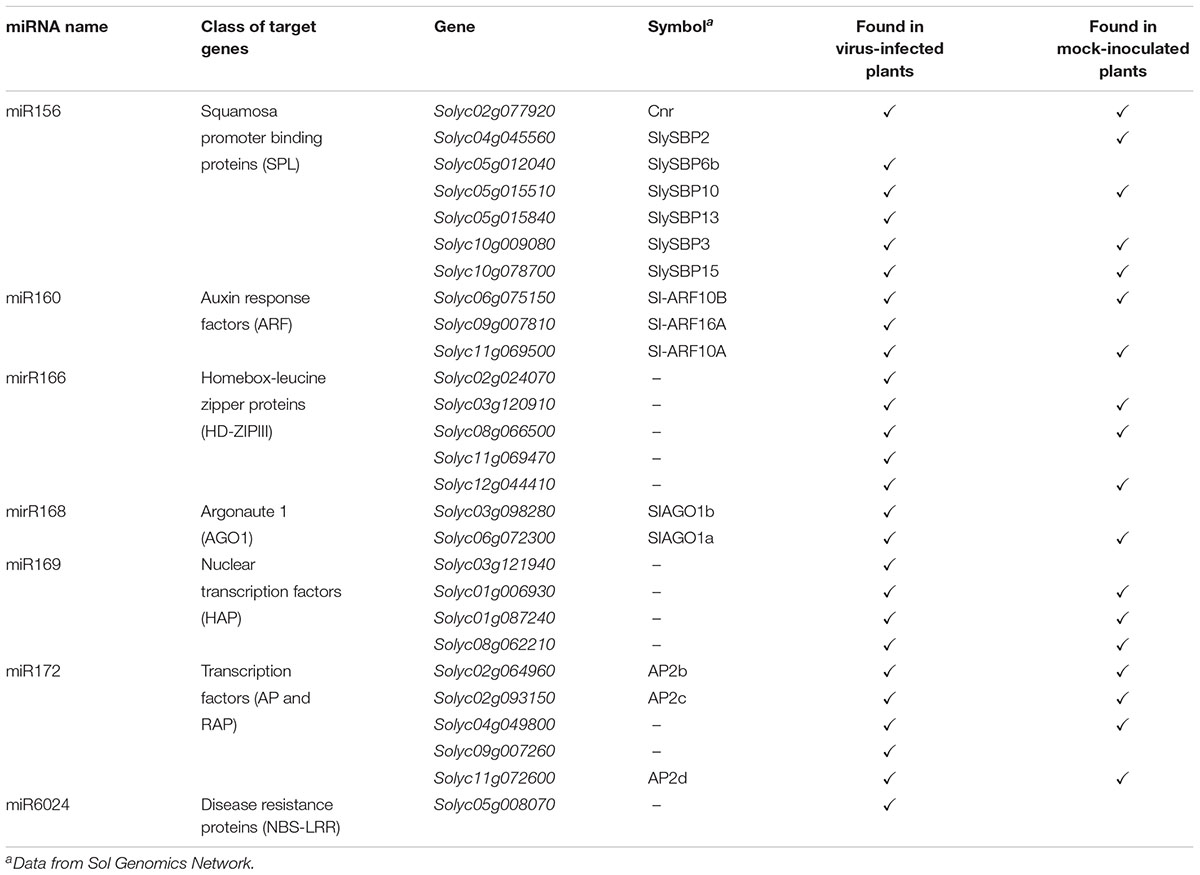
TABLE 1. Target genes found to be cleaved by miRNAs in the PARE analysis either in mock-inoculated or virus-infected tomato plants.
Regarding NBS-LRRs targeted by miR6024, one gene belonging to this large family was specifically cleaved only in the virus-infected tissues, i.e., Solyc05g008070 (Table 1). Worthy of note, Solyc05g008070 has been previously described as targeted by miR482 (Shivaprasad et al., 2012) and, indeed, the psRNATarget reports four genes targeted by either miR6024 or miR482, (Supplementary Table S3), although PARE analysis never found 5′ remnants explained by miR482 for this gene family.
As a proof of concept, we extended the analysis also to mRNAs coding for AGO1. We performed a 5′-RACE analysis of miR168-mediated cleavage of transcripts from sly-AGO1a and sly-AGO1b and found that both transcripts can be cleaved at the same position (Figure 2A). Cleaved sly-AGO1b could only be found in the case of TYLCSV-infected plants (Figure 2A, lower panel and Table 1) and not in mock-inoculated plants. It is noteworthy that, the “A” at the 3′-end of sly-miR168a-5p v2 ensures pairing with the target site in both sly-AGO1a and sly-AGO1b transcripts (in red, Figure 2A and Supplementary Table S2).
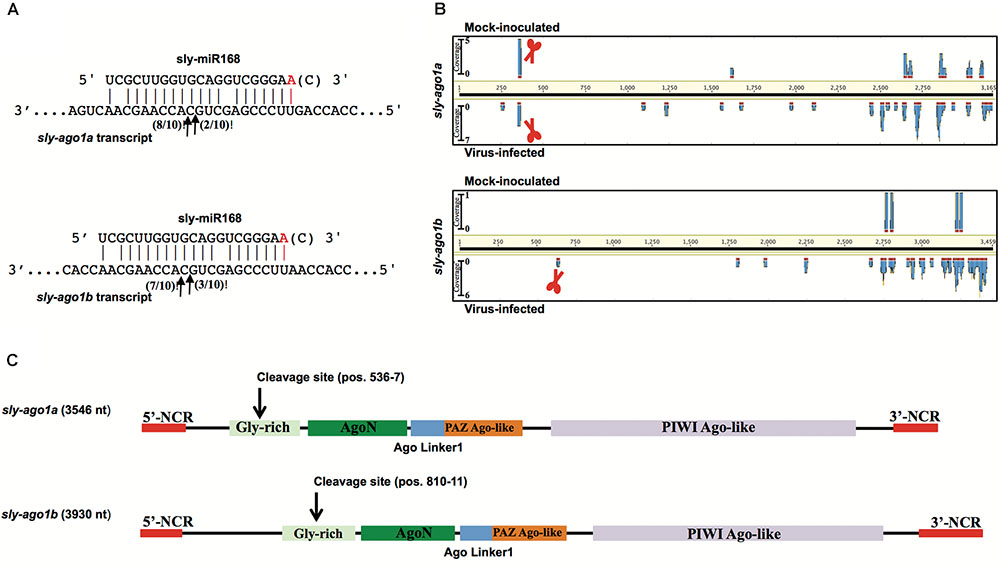
FIGURE 2. Degradation patterns of sly-AGO1a and sly-AGO1b. (A) 5′RACE of miR168-mediated cleavage of sly-AGO1a (NCBI seq ID NM_001279128.1) and sly-AGO1b (NCBI seqID NM_001279332.2) in TYLCSV-infected plants (upper panel) and sly-AGO1a in mock-inoculated plants. Arrows indicate 5′-remnants found in the analysis and with numbers are the clones sequenced. In red or in brackets sly-miR168 found in the datasets (Supplementary Table S1). (B) Degradation patterns of sly-AGO1a and sly-AGO1b (upper and bottom panels, respectively) in mock-inoculated and TYLCSV-infected tomato plants (upper and bottom profile in each panel, respectively). Schematic scissors indicate the miR168 cleavage sites. (C) Graph of sly-AGO1a and sly-AGO1b cDNAs showing the conserved protein domain and miR168 cleavage sites.
The high-throughput analysis of sly-AGO1a and sly-AGO1b by PARE analysis confirmed what was found with 5′-RACE. Figure 2B shows the distributions of 5′-remnants found in mock-inoculated and in virus-infected plants along transcripts of either sly-AGO1a or sly-AGO1b. The degradation patterns of the two transcripts revealed a peak at the cleavage position, which was absent in the case of sly-AGO1b in mock-inoculated plants (Figure 2B, bottom vs. upper frame). The two sly-AGO1 mRNAs differ in size: sly-AGO1a is 3,546 bp vs. 3,930 bp of sly-AGO1b; while the main difference is located at the 5′-terminal part of the coding sequence (Figure 2C).
miRNA/target RNA recognition is a critical feature for miRNA functionality, target cleavage occurrence, and regulation of gene expression. Sly-AGO1a and sly-AGO1b, which differ by one 21-nt-long indel immediately downstream of the miR168 target site, have been previously highlighted in S. lycopersicum (Gursinsky et al., 2015) and here we confirmed that miRNA-mediated control is associated with the indel. To describe the diversity of miRNA targets among tomato genes that could explain the occurrence of cleavage events, we reconstructed each gene family based on its own evolutionary history, including orthologs, when available. Each gene tree is identified by an Ensembl Plant GeneTree ID; AGOs and NBS-LRRs are reported in Figure 3, whereas SPLs, ARFs, HD-ZIPIIIs, and AP/RAP are in Supplementary Figure S1. Upon examination of the phylogenetic trees, we retrieved the closest evolutionary paralogs to those targeted in PARE analysis only in virus-infected plant tissue (i.e., located after the duplication node, red squares in Figure 3 and Supplementary Figure S1), and aligned their transcript sequences in pairs. This was carried out for all the gene families in Table 1, except for the HAP genes since the closest tomato paralog to HAP Solyc03g121940 (cleaved in PARE analysis) is not annotated in the region of the miR169 recognition site (i.e., 3′UTR of Solyc12g009050, Supplementary Figure S1A and Supplementary Table S3).
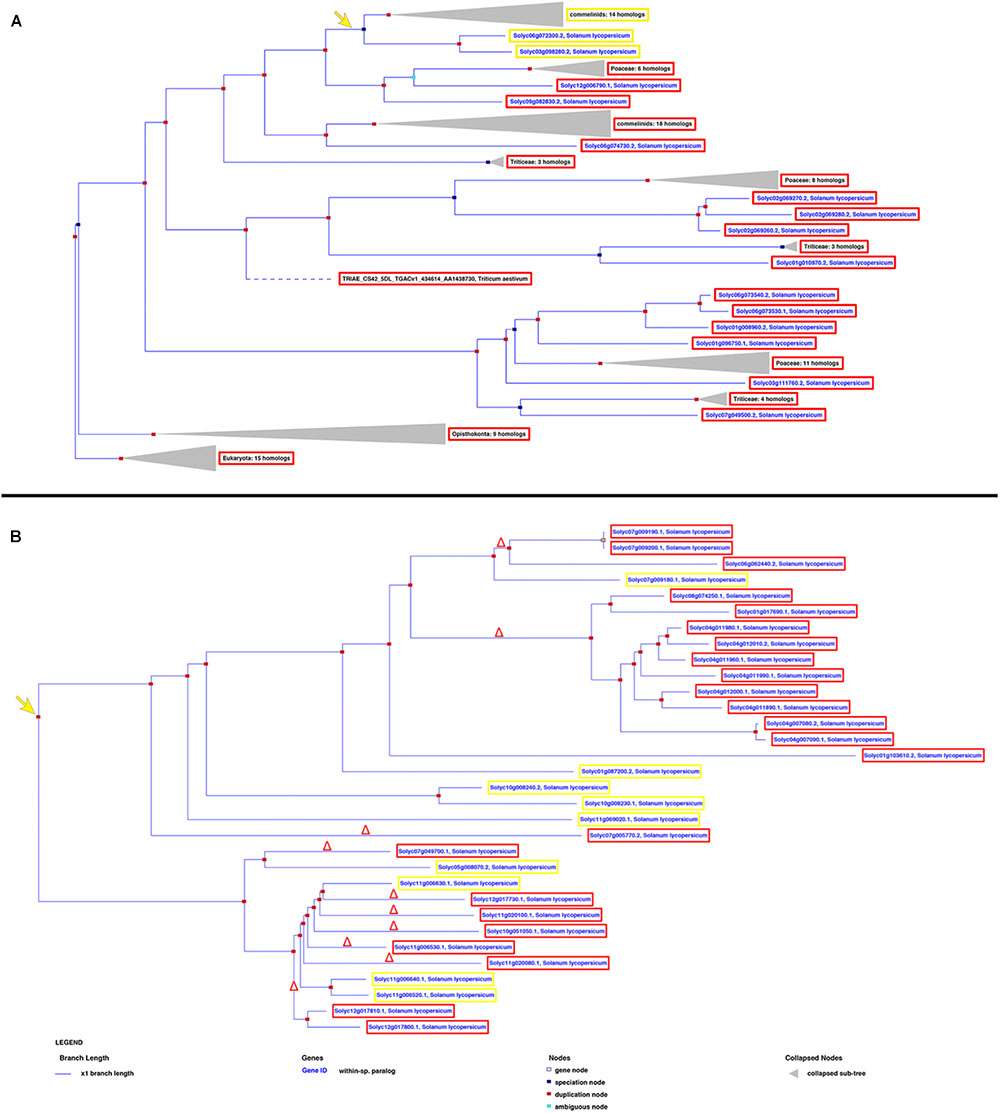
FIGURE 3. Phylogenetic trees showing the evolutionary history of AGOs (A) and NBS-LRRs (B) in Solanum lycopersicum. The yellow and red frames describe the genes harboring or not the miRNA recognition site, respectively, revealed by both PARE and in silico analyses. The yellow arrow indicates the evolutionary creation of the miRNA target site; the red delta symbol represents the deletion of the miRNA target site.
Sly-AGO1a and sly-AGO1b confirmed the presence of a 21-nt-long indel, 9-nt downstream of the miR168 target site (Figure 4A; Gursinsky et al., 2015). Surprisingly, also the pairwise alignment of all gene classes revealed the presence of indels close to the miRNA-target site. Indeed, we found (i) two consecutive indels of 11 and 49 nt in length, respectively, located 26 nt upstream the miR156 recognition site in the case of SPLs (Figure 4B); (ii) a 39-nt-long indel 48 nt upstream the miR160 cleavage site for ARFs (Figure 4C); (iii) a 3-nt-long indel, 113 nt upstream the miR166 recognition site in the case of HD-ZIPIIIs (Figure 4D); and (iv) a 12-nt-long indel, 24 nt downstream the miR172 recognition site in AP/RAP (Figure 4E).

FIGURE 4. Alignments of miRNA target sites and indels. Alignments of close paralogs in the miRNA target site vicinity showing the presence of short indels. Black arrows indicate the miRNA cleavage site found in the PARE analysis. (A) sly-AGO1a/sly-AGO1b alignment with respect to miR168 target site; (B) Solyc05g012040/Solyc12g038520 alignment with respect to miR156 target site; (C) Solyc09g007810/Solyc10g086130 alignment with respect to miR160 target site; (D) Solyc02g024070/Solyc02g069830 alignment with respect to miR166 target site; (E) Solyc09g007260/Solyc10g084340 alignment with respect to miR172 target site; and (F) Solyc05g008070/Solyc07g049700 alignment with respect to miR6024 target site.
Finally, alignment of Solyc05g008070 (cleaved in PARE analysis in virus-infected plants, Table 1) to Solyc07g049700 (its closest paralog in the NBS–LRR family, not cleaved in PARE analysis, Supplementary Table S3 and Supplementary Figure S1E) disclosed a 15-nt-long indel, 123 nucleotides upstream of the miR6024 recognition site (Figure 4F).
PARE analysis can verify target cleavage events, although it is not sufficient to exclude miRNA-mediated regulation of paralogs that were not found cleaved (Figure 4). We, therefore, set up transcriptome quantification via mRNA-seq, of selected target mRNAs, to confirm the presence/absence of miRNA targeting in the mock and infected plant tissues. All paralogs with no indels were significantly more expressed than those having indels in either mock or virus-infected tissues (Figure 5, blue and gray bars of histogram, respectively). When comparing the expression of each couple of paralogs in virus-infected vs. mock-inoculated plant tissues we found significant variations only in the case of sly-AGO1b and Solyc05g008070; the latter was down-regulated in virus-infected plant tissues (Figure 5, dark green with asterisk in the heatmap). Thus, we highlighted a possible correlation between the cleavage event (Table 1), the indel (Figure 4), and the down-regulation (Figure 5) in the case of the NBS–LRR transcript Solyc05g008070.
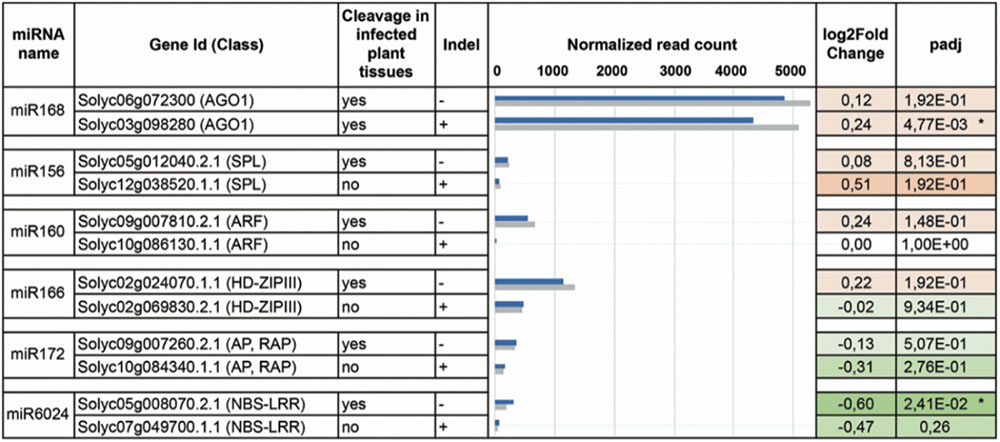
FIGURE 5. DEseq analysis. Paralogs for each miRNA-targeted (column 1) gene family (column 2) have been analyzed by DESeq. Corresponding transcripts are either cleaved in the virus-infested tissues or not (column 3). For each family, only one of the two paralogs contains the indel (column 4). In column 5 the normalized read counts for mRNAs purified from both virus-infected (gray bars) and mock-inoculated (blue bars) tissues. In column 6 and 7, log2FoldChange and adjusted p-values are displayed in the heatmap: red and green for up- and down-regulated transcripts. Asterisks indicate statistically significant variations that were also confirmed by qRT-PCR (three biological replicates and two technical replicates).
When predicting the secondary structure of the stretches of sequence in Figure 4F, which encompass both the 15nt indel and the miR6024 target site, we observed a sharp impact of the indel on the secondary structure and on the entropy at the target sites. The global-free energy of the indel-containing region was lower than in the case of the indel-lacking region (Supplementary Figure S2, panel B vs. A), and the artificial insertion of the indel in the latter strongly affected the structure (Supplementary Figure S2C). We also annotated the location of miRNA target sites in the gene structures with respect to the functional protein domains (Supplementary Figure S2, lower panels). Therefore, the functional indels described could impact secondary structures of the transcript rather than protein domains (see later in “Discussion”).
pecific plant endogenous cascade networks can be triggered by miRNAs associated exclusively with the virus infection. We, therefore, identified all the 21–22-nt secondary siRNAs deriving from mRNAs targeted by miRNAs and solely present in virus-infected plant tissues. We first searched for those conserved miRNAs that were present in the 22-nt-long form in our siRNA libraries (Supplementary Table S2) that explained cleavage events in our PARE analysis (Table 1). We thus restricted the analysis to HD-ZIPIII, HAP, AP/RAP, and NBS–LRR transcripts cleaved by miR166, miR169, miR172, and miR6024, respectively. Given that AGO1 mRNA is also known in Arabidopsis and tomato to have secondary siRNAs initiated by miR168 (Mallory and Vaucheret, 2009; Shivaprasad et al., 2012), we focused on sly-AGO1a and sly-AGO1b. We included the tomato transcript Solyc02g036270 coding for an NBS–LRR transcript targeted by miR482 as control for the effectiveness and reliability of the analysis. Indeed, Solyc02g036270 was previously described as a producer of massive functional phased secondary siRNAs in tomato plants (Shivaprasad et al., 2012).
We aligned the siRNA datasets uniquely from virus-infected plant tissues against selected mRNAs; the output is reported in Table 2. Massive amounts of secondary siRNAs derived from NBS–LRR genes and, as previously reported, a large number of siRNAs mapped to miR482-cleaved Solyc02g036270 (Shivaprasad et al., 2012), i.e., 1,049 (corresponding to 724 unique reads). Similarly, Solyc05g008070 targeted by miR6024 only in virus-infected tissues was covered by 936 siRNAs (corresponding to 665 unique reads). None or trifling reads mapped to Solyc02g024070 and Solyc11g069470 transcripts of the HD-ZIPIII clade, respectively. A modest number of siRNAs mapped to either sly-AGO1a or sly-AGO1b transcripts in agreement with a previous report (Mallory and Vaucheret, 2009). A similar situation was found for miR160-cleaved Solyc09g007810. In summary, we revealed secondary siRNAs associated mainly with those transcripts that were significantly affected in cleavage and expression in virus-infected plant tissues, i.e., sly-AGO1b and Solyc05g008070 (see previous paragraph and Figure 5). Within the selected mRNAs in Table 2, we identified only two cases of phased register with respect to the miRNA target site: miR482/Solyc02g036270 (Figure 6A) and miR6024/Solyc05g008070 (Figure 6B). Surprisingly, phased siRNAs from Solyc02g036270 were only present in mock-inoculated plant tissue datasets and not in the case of virus-infected tissues (histogram in Figure 6A). Phased siRNAs from Solyc05g008070 were detected only in TYLCSV-infected plant tissues where miR6024 was active (histogram in Figure 6B and Figure 6C), despite the fact that the majority of secondary siRNAs from the two NBS–LRR transcripts were not in phase (Figures 6A,B, blue lines vs. red lines). Interestingly, we found that the miR6024 target site overlapped to some extent with that of miR482 (Figure 6B), which is not functional according to PARE analysis.
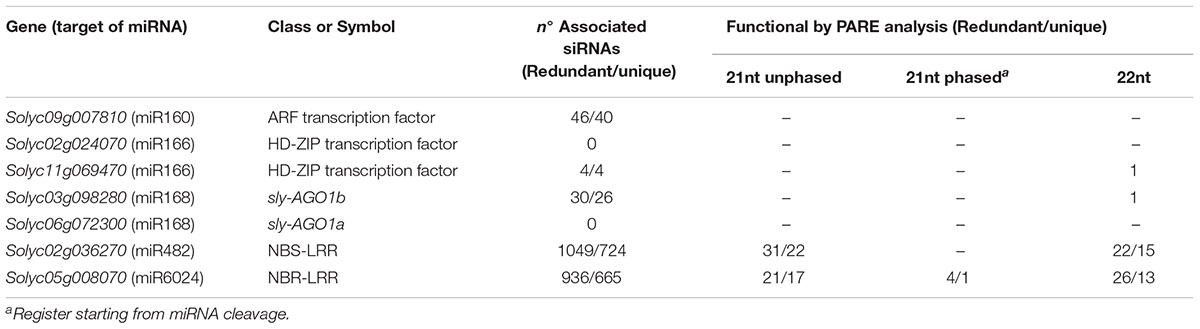
TABLE 2. Total and phased siRNAs associated with selected targets of miR160, miR168, miR169, miR482, and miR6024 and their functionality by PARE analysis in TYLCS-infected plants.
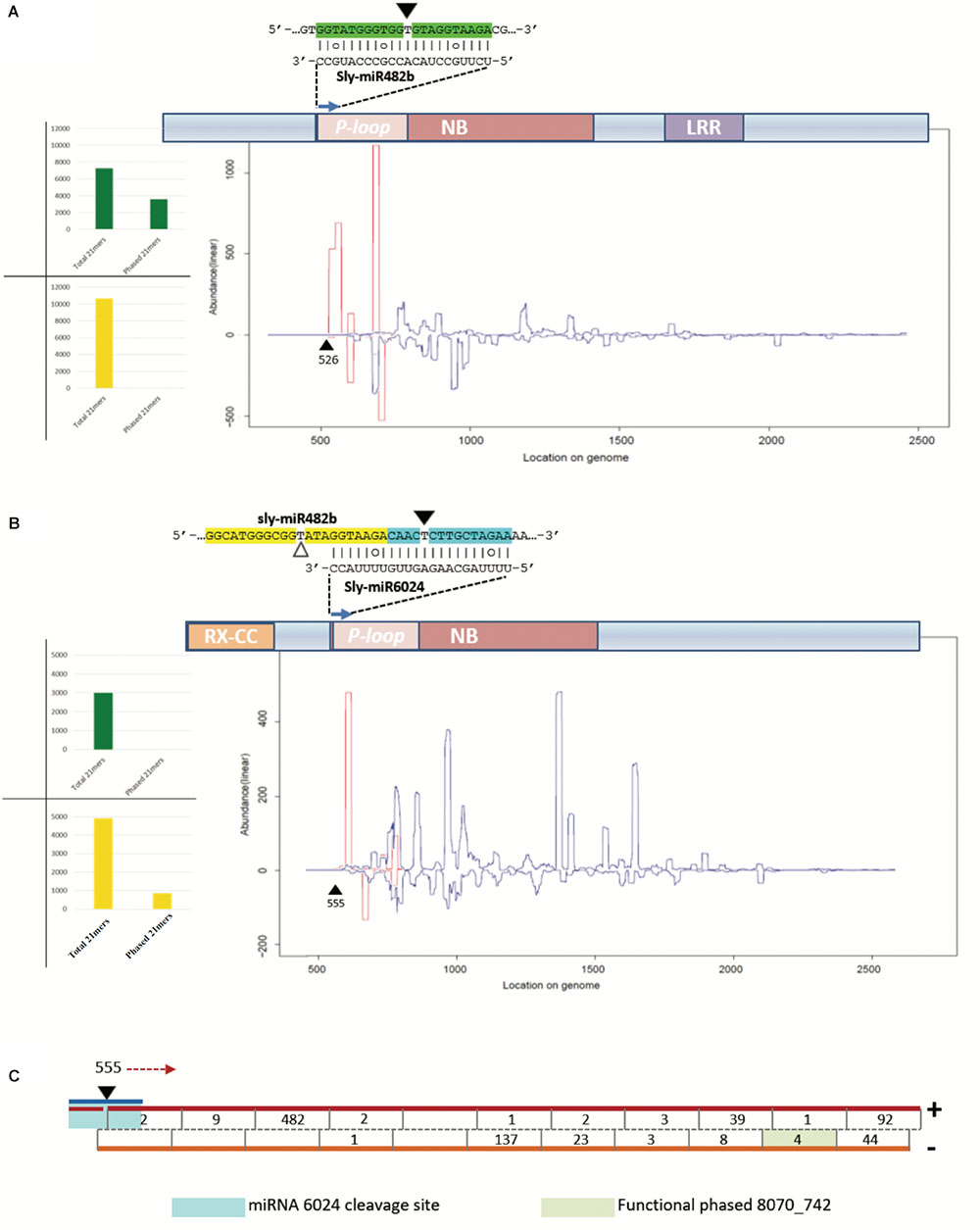
FIGURE 6. Secondary siRNAs initiated by miRNAs. (A) Secondary siRNAs initiated by miR482 on Solyc02g036270 mRNA in mock-inoculated plant tissues; (B) secondary siRNAs initiated by miR6024 on Solyc008070 mRNA in TYLCSV-infected plant tissues; (C) and portion of Solyc05g008070 that is recognized as a source of phased secondary siRNAs. In (A,B) graphics of NBS–LRR genes are shown, which include conserved domains P-loop, nucleotide binding site (NBS), leucine-rich repeat (LRR), and RX-coiled-coil (RX-CC). In the graphic below the transcripts are indicated siRNAs of 21 nt in length, starting from the miRNA cleavage site (black triangle). White triangles indicate uncleaved miRNA target sites. Red and blue lines represent the abundance of each nucleotide position of phased and unphased 21mers, respectively. Histograms in (A,B) show total numbers of phased and unphased 21mers mapping to each gene locus in mock-inoculated (green) and virus-infected (yellow) plant tissues. In (C), the blue line indicates miR6024, the black triangle the cleavage site in position 555, and red arrow indicates the direction of precursor processing into phased secondary siRNAs which are indicated by a black line in the double-stranded red/orange precursors. Numbers indicate abundance of each phased siRNA in the dataset.
Secondary siRNAs including those in a phased register are known to be functional upon incorporation into RISC complex (Vazquez and Hohn, 2013). Being PARE datasets a global analysis of the cleaved transcripts, we performed a dedicated analysis searching 5′-remnants of mRNAs explained by siRNAs in Table 2. Functional siRNAs of 21 and 22 nt are detailed in Supplementary Tables S4, S5, respectively, and are summarized in Table 2. Among functional secondary siRNAs in TYLCS-infected plant tissues deriving from Solyc02g036270, we could find 22 and 15 unique functional siRNAs of 21 and 22 nt in length, respectively. Instead, one phased, seventeen 21-nt-long un-phased and thirteen 22-nt-long non-redundant functional siRNAs were associated with Solyc05g008070 (Table 2 and Supplementary Tables S4,S5).
The 21-nt-long siRNA indicated as siRNA 8070_742 in Figure 6C (pale green box) derived from a highly statistically significant (P-value = 1,068e-8) phased locus starting at position 555 of Solyc05g008070, which corresponds to the miR6024 cleavage site (Figures 6B,C). siRNA 8070_742 explained the cleavage of Solyc03g083250 coding for a putative ATP-dependent CIp protease.
At least one secondary siRNA deriving from NBS–LRR genes has been shown to target other mRNAs of defense-related proteins (Shivaprasad et al., 2012); this was confirmed in our analysis, but we could not find any NBS–LRR transcript among the targets. Indeed, we identified targeted host factors previously described in literature as involved in plant pathogen-related molecular patterns, such as glucan-endo-1-3-beta-glucosidase (Bucher et al., 2001), polyphenol oxidase (Kikuchi and Yamaguchi, 1960), and aquaporin-like proteins (Sade et al., 2014; Supplementary Tables S4, S5).
Worthy of note, 11 out of 42 targeted mRNAs were key players of plant photosynthetic machinery. In Figure 7A, we report eight secondary siRNAs from Solyc05g008070 originated downstream of the miR6024 cleavage site uniquely present in virus-infected plant tissues that target and cleave core members of photosystem I and II, chlorophyll-binding proteins, and ribulose bisphosphate carboxylases (RuBiSCO). Conventional and specific Sanger sequenced 5′-RACE were also performed on the library besides the high-throughput sequencing. Validation by Northern blot analysis revealed the expression of the secondary functional siRNAs only in the case of RNAs from TYLCSV-inoculated plant tissues showing typical yellowing symptoms (Figures 7B,C, lane 4). No secondary functional siRNAs were detected in either mock-inoculated (Figure 7C, lanes 1 and 3) or TYLCSV-infected plant tissues not-showing yellowing symptoms (Figure 7C, lane 2). miR168 and miR171, used as a control of the Northern analysis, were detected in all the samples under investigation (Figure 7C). Finally, DESeq analysis resulted in down-regulation of all targeted transcripts in TYLCSV-infected plant tissues. For those transcripts that could be investigated by qRT-PCR, a significant 1.5- to 2-fold down-regulation in virus-infected vs. mock-inoculated plant tissues, thus confirming the functionality of these siRNA cleaving targets solely associated with virus infection (Figure 7D).
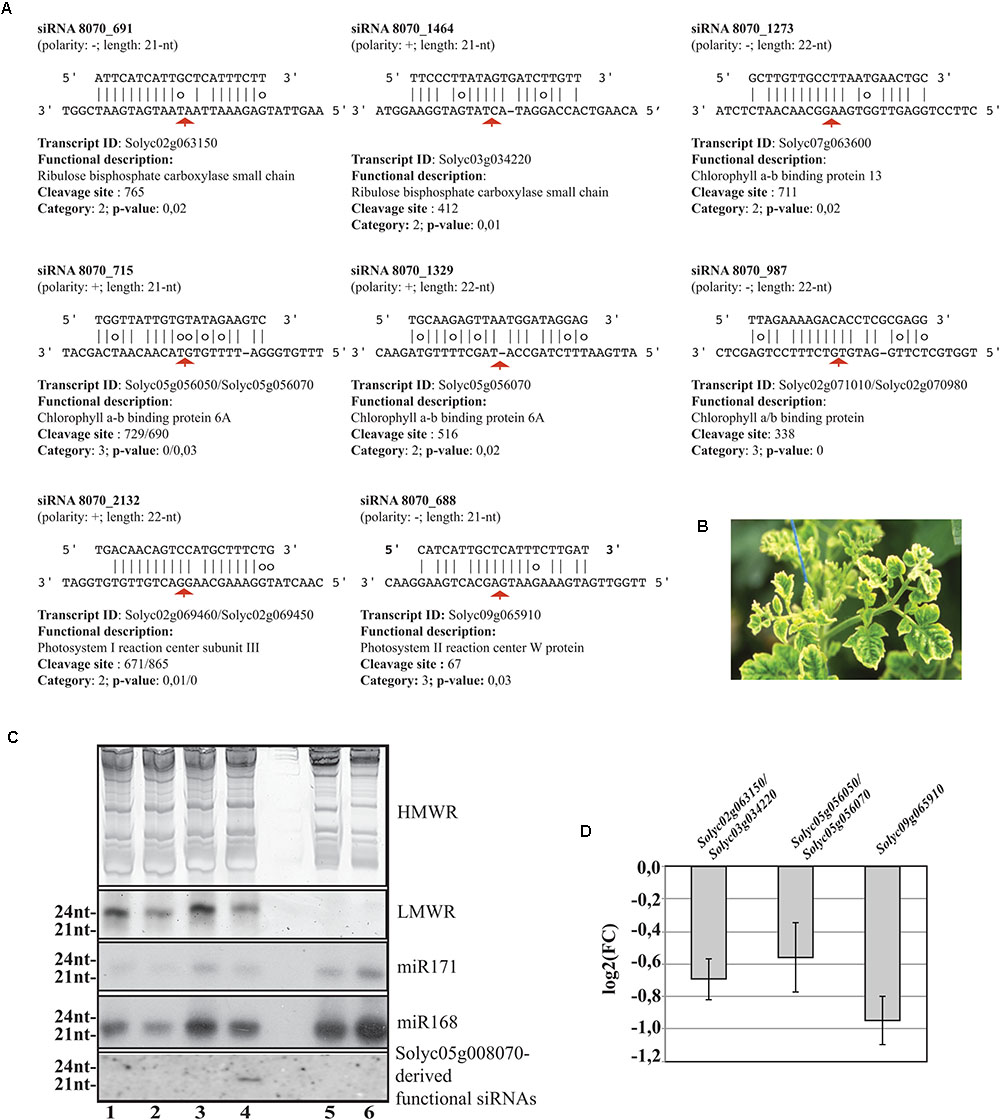
FIGURE 7. Functional secondary siRNAs and expression of genes involved in photosynthesis. (A) Functional 21nt and 22nt-long secondary siRNAs from the NBS–LRR transcript Solyc05g008070 and targeted transcripts. Transcripts sharing conserved miRNA target sites are indicated and separated by “/”. Red arrows indicate the cleavage sites. (B) Typical leaf yellowing induced by TYLCSV in virus-infected tissues. (C) Short RNA Northern blot analysis. High- or low-molecular weight RNAs patterns (HMWR and LMWR, respectively). Lanes 1 and 3: mock-inoculated Solanum lycoperscum, Lane 2: total RNA from TYLCSV-inoculated plant tissues not showing yellowing sympthoms, Lane 4: total RNA from TYLCSV-inoculated plant tissues showing typical yellowing symptoms. Lanes 5 and 6, Arabidopsis thaliana and Nicotiana benthamiana total RNA, respectively. (D) qRT-PCR (three biological replicates and two technical replicates) for the expression of Solyc02g063150/Solyc03g034220 (1), Solyc05g056050/Solyc05g056070 (2), and Solyc09g065910 (3) transcripts in TYLCSV-infected tissues from tomato plants. Expression values (bars ± SE) for each transcript or paralogs are given as log2 of fold change (FC) relative to control tissues from mock-inoculated plants.
Plant viruses are infectious entities that alter gene expression and are therefore recognized as “phenotype extenders.” For instance, it has been shown that virus infection can enhance plants’ attractiveness to pollinators (Groen et al., 2016) as well as the resilience of susceptible plants to drought (Xu et al., 2008; Westwood et al., 2013; Pantaleo et al., 2016) and that they defend plants from several species of herbivores (Gibbs, 1980; Pan et al., 2013). The fundamental reasons for this gene regulation are not yet fully understood, but many phenomena can be ascribed to a mechanistic role for miRNAs in regulating innate immunity in the case of plant–pathogen interaction (Katiyar-Agarwal and Jin, 2007; Deng et al., 2018). Moreover, plant endogenous siRNAs associated with viral infection denoted as va-siRNAs are surely implicated in the regulation of genes in plants under virus attack (Cao et al., 2014).
In plants, fungi, and animals, indel studies have led to understand how proteins function as well as to the discovery of useful experimental and drug targets. Moreover, indels are recognized as having significant value as phylogenetic markers. For selected miRNA targets, we show that short indels upstream or downstream in the vicinity of miRNA targets can discriminate between the miRNA cleavage events of close paralogs. Indeed, S. lycopersicum possesses two AGO1 homologs named sly-AGO1a and sly-AGO1b, differing for 21-nt-long indels just downstream of the miR168 target site. Sly-AGO1b (the transcript containing the insertion) is cleaved in TYLCSV-infected plants, whereas it is not in mock-inoculated plants. Consistent data came from mRNA-seq analysis since the sly-AGO1b appeared significantly up-regulated in virus-infected tissues, which is in agreement with our previous observations (Gursinsky et al., 2015). This provides further support to specific roles ascribed to the two AGO1 paralogs even in a different plant-virus system. Herein, we reveal that several nuclear transcription factors are also cleaved by specific miRNAs only in the presence of the viral infection. Among these transcriptional factors, there are two SPL genes cleaved by miR156 (i.e., sly-SPB6b and sly-SPB13), one ARF gene cleaved by miR160 (Sl-ARF16A), two HD-ZIPIII genes cleaved by miR166 (Solyc02g024070 and Solyc11g069470), one AP transcription factor cleaved by miR172 (Solyc09g007260), and one NBS–LRR disease-resistance protein-coding gene cleaved by miR6024 (Solyc05g008070). We show that 12-, 15-, 3-, and 39-nt-long single indels are in close vicinity to the target sites of miR172, miR6024, miR166, and miR160, respectively. Nevertheless, in all these cases, the indels discriminate between cleavages of close paralogs: the version containing the insertion was never cleaved, whereas the other was found cleaved in TYLCSV-infected plant tissues. All paralogs with no indels were significantly more expressed than those having indels in either virus-infected or mock plant tissue. A significant differential expression of paralogs in mock-inoculated tissues suggests fine and independent gene regulation that renders unclear the interpretation of the role of indels in miRNA-dependent regulation. However, Solyc05g008070 lacking one 15-nt-long indel, 123 nucleotides upstream of the miR6024 recognition site, is cleaved and is down-regulated solely in virus-infected plant tissues. All the analyzed genes are homologs, thus they derive from a whole genome duplication and belong to different subgenomes (Tomato Genome, 2012); moreover, they are likely to have a redundant role. Hence, we hypothesized the possibility to regulate them differently, mediated by a distinctive sensitivity to the miRNA-mediated cleavage, might be skilfully exploited by the plant to convert an “excess of function” in a stress-resistance tool. Short indels are unlikely to have a significant impact on protein size and we show that they do not impair the recognition of functional domains. Instead, they are confirmed to play a role in the secondary structure of RNA influencing the access of RISC to target sites in line with previous in vitro and in vivo observations either in animal or plant systems (Ameres et al., 2007; Long et al., 2007; Schuck et al., 2013; Gursinsky et al., 2015). In the case of the indel-lacking paralog, Solyc05g008070, this is further corroborated by the in silico prediction of RNA folding and entropy at target site level.
The CC–NBS–LRR class of genes confers resistance to several plant viruses in an effector-specific interaction (Kachroo et al., 2006). miR482 targets tomato mRNAs for CC–NBS–LRR (i.e., Solyc02g036270) and the mRNA decay causes the production of secondary siRNAs that in turn down-regulate other mRNAs of disease-resistance genes: this has been proposed as a novel pathogen-inducible layer of plant defense (Shivaprasad et al., 2012). Here, we analyzed the production of secondary siRNAs from Solyc02g036270 and we confirm that it produces phased register secondary siRNAs with respect to the miRNA target site, likely induced by the 22-nt-long miR482 found in our dataset. Importantly, phased siRNAs from Solyc02g036270 were only present in mock-inoculated and not in virus-infected plant tissue datasets. The production of secondary siRNAs from Solyc02g036270 requires RDR6 that is engaged in converting targeted RNA in dsRNA (Shivaprasad et al., 2012). In this pathway, suppressor of gene silencing 3 (SGS3) is also involved (Mourrain et al., 2000). Tomato yellow leaf curl virus (TYLCV), a close relative of TYLCSV, is known to produce the V2 protein that is recognized as a viral suppressor of RNA silencing (Zrachya et al., 2007); V2 directly binds SGS3 in plants and this interaction is required for V2 RNA silencing suppression activity (Glick et al., 2008). V2 is produced by the virus to counteract plant antiviral silencing but to date, no specific action has been described. Glick and colleagues (Glick et al., 2008) advanced the hypothesis that V2-mediated inactivation of SGS3 in infected tissues could explain “leaf curling” symptoms. Here, we reveal for the first time that the miRNA-mediated cleavage of the NBS–LRR transcript is not relieved in TYLCSV-infected plant tissues; instead, the production of secondary siRNAs in phased register is abolished, likely through the inactivation of SGS3/RDR6 by V2. The availability of genome-wide 5′-RACE analysis allowed us to show that in the absence of phased secondary siRNAs in TYLCSV-infected tissues no transcript of the NBS–LRR clade was found cleaved except by miRNAs. The loss of biogenesis of phased secondary siRNAs would reinforce the defensive effect and, noteworthily, our data suggest that such inactivation could happen downstream the miRNA-mediated cleavage hampering dsRNA synthesis by geminivirus viral suppression. Mechanisms of production of functional unphased siRNAs are at the moment unknown and deserve further investigations.
The data from the present study suggest for the first time that CC–NBS–LRR class of genes includes many resistance (R) genes could be directly implicated in symptom development through secondary cascade pathways. Indeed, one NBS–LRR transcript releases secondary siRNAs that target transcripts coding for RuBISCO, chlorophyll a/b binding protein 6A, and photosystem II reaction center W protein. These are significantly down-expressed in infected tissues at least 1.5-fold. In addition, Northern blot analysis unveiled the expression of these secondary sRNAs exclusively in plant virus-infected tissues showing yellowing. Thus, here we reveal a novel mechanism that could be added to earlier findings in virus–host interaction. Leaf chlorosis is often seen in plants infected with viruses and viroids and is known to be associated with a reduced content of chlorophyll in leaf cells (Dawson, 1992). As chlorophyll is responsible for the green color of leaves, chlorotic leaves range from pale to yellow, through yellow–white. Cucumber mosaic virus (CMV) yellowing (Y) satellite RNA (Y-sat) is a non-coding subviral RNA and modifies the typical symptom induced by CMV in specific hosts; direct evidence identified one 22-nt-long siRNA derived from Y-sat that modulates the yellowing symptom by RNA silencing-based regulation of tobacco magnesium protoporphyrin chelatase subunit I (ChlI), a key gene involved in chlorophyll synthesis (Shimura et al., 2011). In the case of grapevine infected by Grapevine fleck virus, v-siRNAs were able to target and cleave transcripts of the photosynthetic system Y (Miozzi et al., 2013a). However, the v-siRNAs-dependent pathogenesis mechanism is not general. Indeed, alteration of chloroplast development induced by Peach latent mosaic viroid (PLMVd) leads to albino phenotype (an extreme chlorosis symptom) (Rodio et al., 2007). In the case of PLMVd, the block of chloroplast development is likely due to the RNA silencing of one chloroplastic heat shock protein 90 (Navarro et al., 2012). Chlorosis is also due to the localization in chloroplasts of either viral proteins, such as in the case of Tobacco mosaic virus coat protein and Rice stripe virus disease-specific protein that hamper photosynthetic system II (Reinero and Beachy, 1989; Kong et al., 2014), or subviral RNAs such as in the case of Geminivirus betasatellites that severely alter chloroplast ultrastructure and induce vein clearing (Bhattacharyya et al., 2015).
The understanding of miRNA biology is still limited by our poor appreciation of their cell-type specific and spatio-temporal regulation. Many miRNAs exhibit discrete expression patterns and their regulation depends largely on the availability of particular AGO proteins and distinct target mRNAs within a given cell/tissue type. Next-generation sequence approaches to facilitate the simultaneous analysis of large gene sets but they typically provide a very limited spatio-temporal resolution of gene expression changes. Either in plants or animals (Kozomara et al., 2014), miRNA expression level is an unreliable indicator of miRNA repressive activity. In plants, laser microdissection coupled to RT-PCR showed that, on the contrary, the global pattern of mature conserved accumulation entails exquisite spatio-temporal variations in the transcription of nine distinct miRNA paralogs (Nogueira et al., 2007). The combinatorial analysis of small RNA sequencing and accurate expression data for target mRNA/proteins by the novel single-cell RNA-Seq analysis would solve either some discrepancies (such as miR6024 ∼2/3X less abundance in infected tissues than mock-inoculated tissues), or highlight the impact of secondary siRNAs controlling the expression of analyzed transcripts.
The sRNA and PARE datasets have been deposited in GEO Omnibus under the entry code GSE93648. The DESeq analysis of selected paralogs was carried out using alignments deposited in SRA under the entry code SRP133920.
(To review GEO accession GSE93648:
Go to https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93648
Enter token onsrwgugbxmpnwd into the box)
(To Access to metadata about SRA submission https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP133920).
VP designed the experiments and prepared PARE and sRNA datasets. LM prepared total RNA from tomato plants and performed quantification analysis. MC, CC, WP, and VP analyzed the data. MC, CC, and VP prepared the manuscript. All authors discussed the results and contributed to editing the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Director of the Istituto per la Protezione Sostenibile delle Piante (IPSP)–CNR, Dr. G.P Accotto for promoting the use of Ensembl for Evolutionary Genomics through dedicated training funding. We thank the data center ReCaS for hosting High Performance Computing analysis. We thank Dr. E. Noris for DNA samples from virus-infected tissues controls and Dr. G.P Accotto for pictures of yellowing associated with virus infections and for suggestions in virus detection by Southern blot analysis. We thank also Mrs. Antonia Antonacci for technical assistance. CC is Co-founded by Cohesion and Development Fund 2007-2013—APQ Research Puglia Region “Regional programme supporting smart specialization and social and environmental sustainability—FutureInResearch.” MC research was supported by SaveGraInPuglia project—“Progetti integrati per la biodiversità” PSR 2014-2020 Reg. (CE) 1698/2005. The IPSP-CNR was also supported by the MIUR “Progetto Premiale” Photosynthesis 2.0.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01470/full#supplementary-material
FIGURE S1 | Phylogenetic trees showing the evolutionary history of selected gene families targeted by conserved miRNAs in Solanum lycopersicum.
(A) EPlGT00140000000465 evolutionary history for HAP genes;
(B) EPlGT00850000106226 evolutionary history for ARF genes;
(C) EPlGT00140000000795 evolutionary history for HD ZIPIII genes;
(D) EPlGT00850000106232 evolutionary history for AP/RAP genes; and
(E) EPlGT00820000103254 evolutionary history for SPL genes.
FIGURE S2 | Impact of the functional indels on the secondary structures of the transcript. (A,B) Global-free energy of the indel-containing region (positions 360–622, Solyc05g008070) and of the indel-lacking region (positions 360–638, Solyc07g049700), respectively. (C) Impact of the artificial insertion of the indel in the indel-lacking region.
TABLE S1 | Statistics of Solanum lycopersicum genomic and non-genomic siRNAs in tomato yellow leaf curl sardinia virus-infected and mock-inoculated plant tissues.
TABLE S2 | Diversity of selected functional conserved miRNAs (expression profile in sRNA datasets from mock-inoculated and TYLCSV-infected tomato plants). Bold characters indicate nucleotides differing from the canonical miRNA sequence.
TABLE S3 |In silico analysis of miRNA targets among the selected gene families, validated by PARE analysis.
TABLE S4 | Output of PAREsnp listing 5′ RNA remnants of cDNA targeted by 21-nt-long secondary siRNAs in tomato TYLCSV-infected plant tissues.
TABLE S5 | Output of PAREsnp listing 5′ RNA remnants of cDNA targeted by 22-nt-long secondary siRNAs in tomato TYLCSV-infected plant tissues.
TABLE S6 | Oligonucleotides used for quantitative reverse transcription PCR experiments of selected transcripts in TYLCSV-infected vs. mock-inoculated tissues.
Accotto, G. P., Navas-Castillo, J., Noris, E., Moriones, E., and Louro, E. (2000). Typing of Tomato yellow leaf curl viruses in Europe. Eur. J. Plant Pathol. 106, 179–186. doi: 10.1023/A:1008736023293
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouche, N., Gasciolli, V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16, 927–932. doi: 10.1016/j.cub.2006.03.035
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Ameres, S. L., Martinez, J., and Schroeder, R. (2007). Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130, 101–112. doi: 10.1016/j.cell.2007.04.037
Amin, I., Patil, B. L., Briddon, R. W., Mansoor, S., and Fauquet, C. M. (2011). A common set of developmental miRNAs are upregulated in Nicotiana benthamiana by diverse begomoviruses. Virol. J. 8:143. doi: 10.1186/1743-422X-8-143
Aregger, M., Borah, B. K., Seguin, J., Rajeswaran, R., Gubaeva, E. G., Zvereva, A. S., et al. (2012). Primary and secondary siRNAs in geminivirus-induced gene silencing. PLoS Pathog. 8:e1002941. doi: 10.1371/journal.ppat.1002941
Arikit, S., Xia, R., Kakrana, A., Huang, K., Zhai, J., Yan, Z., et al. (2014). An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell 26, 4584–4601. doi: 10.1105/tpc.114.131847
Axtell, M. J., Jan, C., Rajagopalan, R., and Bartel, D. P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. doi: 10.1016/j.cell.2006.09.032
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bhattacharyya, D., Gnanasekaran, P., Kumar, R. K., Kushwaha, N. K., Sharma, V. K., Yusuf, M. A., et al. (2015). A geminivirus betasatellite damages the structural and functional integrity of chloroplasts leading to symptom formation and inhibition of photosynthesis. J. Exp. Bot. 66, 5881–5895. doi: 10.1093/jxb/erv299
Brown, J. K., Fauquet, C. M., Briddon, R. W., Zerbini, M., Moriones, E., and And Navas-Castillo, J. (2012). “Genminiviridae,” in Virus Taxonomy. Classification and Nomenclature of Viruses, ed. E. J. Lefkowitz (New York, NY: Elsevier).
Bucher, G. L., Tarina, C., Heinlein, M., Di, Serio F, Meins, F. Jr., and Iglesias, V. A. (2001). Local expression of enzymatically active class I beta-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 28, 361–369. doi: 10.1046/j.1365-313X.2001.01181.x
Buchmann, R. C., Asad, S., Wolf, J. N., Mohannath, G., and Bisaro, D. M. (2009). Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J. Virol. 83, 5005–5013. doi: 10.1128/JVI.01771-08
Cao, M., Du, P., Wang, X., Yu, Y. Q., Qiu, Y. H., Li, W., et al. (2014). Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14613–14618. doi: 10.1073/pnas.1407131111
Chen, H. M., Li, Y. H., and Wu, S. H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 3318–3323. doi: 10.1073/pnas.0611119104
Csorba, T., Kontra, L., and Burgyan, J. (2015). Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 47, 85–103. doi: 10.1016/j.virol.2015.02.028
Cuperus, J. T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R. T., Takeda, A., et al. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003. doi: 10.1038/nsmb.1866
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dawson, W. O. (1992). Tobamovirus-plant interactions. Virology 186, 359–367. doi: 10.1016/0042-6822(92)90001-6
Deng, Y., Wang, J., Tung, J., Liu, D., Zhou, Y., He, S., et al. (2018). A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 14:e1006756. doi: 10.1371/journal.ppat.1006756
Fernandez-Pozo, N., Menda, N., Edwards, J. D., Saha, S., Tecle, I. Y., Strickler, S. R., et al. (2015). The Sol Genomics Network (SGN)–from genotype to phenotype to breeding. Nucleic Acids Res. 43, D1036–D1041. doi: 10.1093/nar/gku1195
Folkes, L., Moxon, S., Woolfenden, H. C., Stocks, M. B., Szittya, G., Dalmay, T., et al. (2012). PAREsnip: a tool for rapid genome-wide discovery of small RNA/target interactions evidenced through degradome sequencing. Nucleic Acids Res. 40, e103. doi: 10.1093/nar/gks277
German, M. A., Luo, S., Schroth, G., Meyers, B. C., and Green, P. J. (2009). Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4, 356–362. doi: 10.1038/nprot.2009.8
German, M. A., Pillay, M., Jeong, D. H., Hetawal, A., Luo, S., Janardhanan, P., et al. (2008). Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946. doi: 10.1038/nbt1417
Ghasemzadeh, A., Małgorzata Ter Haar, M., Shams-Bakhsh, M., Pirovano, W., and Pantaleo, V. (2018). Shannon entropy to evaluate substitution rate variation among viral nucleotide positions in datasets of viral siRNAs. Methods Mol. Biol. 1746, 187–195. doi: 10.1007/978-1-4939-7683-6_15
Gibbs, A. (1980). A plant virus that partially protects its wild legume host against herbivores. Intervirology 13, 42–47. doi: 10.1159/000149105
Glick, E., Zrachya, A., Levy, Y., Mett, A., Gidoni, D., Belausov, E., et al. (2008). Interaction with host SGS3 is required for suppression of RNA silencing by Tomato yellow leaf curl virus V2 protein. Proc. Natl. Acad. Sci. U.S.A. 105, 157–161. doi: 10.1073/pnas.0709036105
Groen, S. C., Jiang, S., Murphy, A. M., Cunniffe, N. J., Westwood, J. H., Davey, M. P., et al. (2016). Virus infection of plants alters pollinator preference: a payback for susceptible hosts? PLoS Pathog. 12:e1005790. doi: 10.1371/journal.ppat.1005790
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neubock, R., and Hofacker, I. L. (2008). The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74. doi: 10.1093/nar/gkn188
Gursinsky, T., Pirovano, W., Gambino, G., Friedrich, S., Behrens, S. E., and Pantaleo, V. (2015). Homeologs of the Nicotiana benthamiana antiviral ARGONAUTE1 show different susceptibilities to microRNA168-mediated control. Plant Physiol. 168, 938–952. doi: 10.1104/pp.15.00070
Howell, M. D., Fahlgren, N., Chapman, E. J., Cumbie, J. S., Sullivan, C. M., Givan, S. A., et al. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19, 926–942. doi: 10.1105/tpc.107.050062
Iwakawa, H. O., and Tomari, Y. (2013). Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52, 591–601. doi: 10.1016/j.molcel.2013.10.033
Kachroo, P., Chandra-Shekara, A. C., and Klessig, D. F. (2006). Plant signal transduction and defense against viral pathogens. Adv. Virus Res. 66, 161–191. doi: 10.1016/S0065-3527(06)66004-1
Katiyar-Agarwal, S., and Jin, H. (2007). Discovery of pathogen-regulated small RNAs in plants. Methods Enzymol. 427, 215–227. doi: 10.1016/S0076-6879(07)27012-0
Kikuchi, M., and Yamaguchi, A. (1960). Polyphenol oxidase activity of Nicotiana glutinosa leaves infected with tobacco mosaic virus. Nature 187, 1048–1049. doi: 10.1038/1871048a0
Kong, L., Wu, J., Lu, L., Xu, Y., and Zhou, X. (2014). Interaction between rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant 7, 691–708. doi: 10.1093/mp/sst158
Kozomara, A., and Griffiths-Jones, S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.1093/nar/gkt1181
Kozomara, A., Hunt, S., Ninova, M., Griffiths-Jones, S., and Ronshaugen, M. (2014). Target repression induced by endogenous microRNAs: large differences, small effects. PLoS One 9:e104286. doi: 10.1371/journal.pone.0104286
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Long, D., Lee, R., Williams, P., Chan, C. Y., Ambros, V., and Ding, Y. (2007). Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 14, 287–294. doi: 10.1038/nsmb1226
Mallory, A. C., and Vaucheret, H. (2009). ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10, 521–526. doi: 10.1038/embor.2009.32
Manavella, P. A., Koenig, D., and Weigel, D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U.S.A. 109, 2461–2466. doi: 10.1073/pnas.1200169109
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Mathews, D. H., Disney, M. D., Childs, J. L., Schroeder, S. J., Zuker, M., and Turner, D. H. (2004). Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U.S.A. 101, 7287–7292. doi: 10.1073/pnas.0401799101
Miozzi, L., Gambino, G., Burgyan, J., and Pantaleo, V. (2013a). Genome-wide identification of viral and host transcripts targeted by viral siRNAs in Vitis vinifera. Mol. Plant Pathol. 14, 30–43. doi: 10.1111/j.1364-3703.2012.00828.x
Miozzi, L., Pantaleo, V., Burgyan, J., Accotto, G. P., and Noris, E. (2013b). Analysis of small RNAs derived from tomato yellow leaf curl Sardinia virus reveals a cross reaction between the major viral hotspot and the plant host genome. Virus Res. 178, 287–296. doi: 10.1016/j.virusres.2013.09.029
Miozzi, L., Napoli, C., Sardo, L., and Accotto, G. P. (2014). Transcriptomics of the interaction between the monopartite phloem-limited geminivirus tomato yellow leaf curl Sardinia virus and Solanum lycopersicum highlights a role for plant hormones, autophagy and plant immune system fine tuning during infection. PLoS One 9:e89951. doi: 10.1371/journal.pone.0089951
Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. doi: 10.1016/S0092-8674(00)80863-6
Naqvi, A. R., Haq, Q. M., and Mukherjee, S. K. (2010). MicroRNA profiling of tomato leaf curl New Delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 7:281. doi: 10.1186/1743-422X-7-281
Navarro, B., Gisel, A., Rodio, M. E., Delgado, S., Flores, R., and Di Serio, F. (2012). Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 70, 991–1003. doi: 10.1111/j.1365-313X.2012.04940.x
Nogueira, F. T., Madi, S., Chitwood, D. H., Juarez, M. T., and Timmermans, M. C. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21, 750–755. doi: 10.1101/gad.1528607
Noris, E., Accotto, G. P., Tavazza, R., Brunetti, A., Crespi, S., and Tavazza, M. (1996). Resistance to tomato yellow leaf curl geminivirus in Nicotiana benthamiana plants transformed with a truncated viral C1 gene. Virology 224, 130–138. doi: 10.1006/viro.1996.0514
Pan, H., Chen, G., Li, F., Wu, Q., Wang, S., Xie, W., et al. (2013). Tomato spotted wilt virus infection reduces the fitness of a nonvector herbivore on pepper. J. Econ. Entomol. 106, 924–928. doi: 10.1603/EC12365
Pantaleo, V., Saldarelli, P., Miozzi, L., Giampetruzzi, A., Gisel, A., Moxon, S., et al. (2010a). Deep sequencing analysis of viral short RNAs from an infected Pinot Noir grapevine. Virology 408, 49–56. doi: 10.1016/j.virol.2010.09.001
Pantaleo, V., Szittya, G., Moxon, S., Miozzi, L., Moulton, V., Dalmay, T., et al. (2010b). Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 62, 960–976. doi: 10.1111/j.0960-7412.2010.04208.x
Pantaleo, V., Vitali, M., Boccacci, P., Miozzi, L., Cuozzo, D., Chitarra, W., et al. (2016). Novel functional microRNAs from virus-free and infected Vitis vinifera plants under water stress. Sci. Rep. 6:20167. doi: 10.1038/srep20167
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Pirovano, W., Miozzi, L., Boetzer, M., and Pantaleo, V. (2014). Bioinformatics approaches for viral metagenomics in plants using short RNAs: model case of study and application to a Cicer arietinum population. Front. Microbiol. 5:790. doi: 10.3389/fmicb.2014.00790
Prufer, K., Stenzel, U., Dannemann, M., Green, R. E., Lachmann, M., and Kelso, J. (2008). PatMaN: rapid alignment of short sequences to large databases. Bioinformatics 24, 1530–1531. doi: 10.1093/bioinformatics/btn223
Reinero, A., and Beachy, R. N. (1989). Reduced photosystem II activity and accumulation of viral coat protein in chloroplasts of leaves infected with Tobacco mosaic virus. Plant Physiol. 89, 111–116. doi: 10.1104/pp.89.1.111
Rhoades, M., Reinhart, B., Lim, L., Burge, C., Bartel, B., and Bartel, D. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Rodio, M. E., Delgado, S., De Stradis, A., Gomez, M. D., Flores, R., and Di Serio, F. (2007). A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 19, 3610–3626. doi: 10.1105/tpc.106.049775
Rojas, M. R., Hagen, C., Lucas, W. J., and Gilbertson, R. L. (2005). Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43, 361–394. doi: 10.1146/annurev.phyto.43.040204.135939
Sade, D., Sade, N., Shriki, O., Lerner, S., Gebremedhin, A., Karavani, A., et al. (2014). Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to Tomato yellow leaf curl virus. Plant Physiol. 165, 1684–1697. doi: 10.1104/pp.114.243402
Schuck, J., Gursinsky, T., Pantaleo, V., Burgyan, J., and Behrens, S. E. (2013). AGO/RISC-mediated antiviral RNA silencing in a plant in vitro system. Nucleic Acids Res. 41, 5090–5103. doi: 10.1093/nar/gkt193
Shao, Z. Q., Zhang, Y. M., Hang, Y. Y., Xue, J. Y., Zhou, G. C., Wu, P., et al. (2014). Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: understanding gained from and beyond the legume family. Plant Physiol. 166, 217–234. doi: 10.1104/pp.114.243626
Shimura, H., and Pantaleo, V. (2011). Viral induction and suppression of RNA silencing in plants. Biochim. Biophys. Acta 1809, 601–612. doi: 10.1016/j.bbagrm.2011.04.005
Shimura, H., Pantaleo, V., Ishihara, T., Myojo, N., Inaba, J., Sueda, K., et al. (2011). A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 7:e1002021. doi: 10.1371/journal.ppat.1002021
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Si-Ammour, A., Windels, D., Arn-Bouldoires, E., Kutter, C., Ailhas, J., Meins, F., et al. (2011). miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157, 683–691. doi: 10.1104/pp.111.180083
Stocks, M. B., Moxon, S., Mapleson, D., Woolfenden, H. C., Mohorianu, I., Folkes, L., et al. (2012). The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28, 2059–2061. doi: 10.1093/bioinformatics/bts311
Tomato Genome, C. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Vanneste, K., Baele, G., Maere, S., and Van De Peer, Y. (2014). Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res. 24, 1334–1347. doi: 10.1101/gr.168997.113
Vazquez, F., and Hohn, T. (2013). Biogenesis and biological activity of secondary siRNAs in plants. Scientifica 2013:783253. doi: 10.1155/2013/783253
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Vilella, A. J., Severin, J., Ureta-Vidal, A., Heng, L., Durbin, R., and Birney, E. (2009). EnsemblCompara genetrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335. doi: 10.1101/gr.073585.107
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, S., and Adams, K. L. (2015). Duplicate gene divergence by changes in microRNA binding sites in Arabidopsis and Brassica. Genome Biol. Evol. 7, 646–655. doi: 10.1093/gbe/evv023
Westwood, J. H., Mccann, L., Naish, M., Dixon, H., Murphy, A. M., Stancombe, M. A., et al. (2013). A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol. Plant Pathol. 14, 158–170. doi: 10.1111/j.1364-3703.2012.00840.x
Windels, D., and Vazquez, F. (2011). miR393: integrator of environmental cues in auxin signaling? Plant Signal. Behav. 6, 1672–1675. doi: 10.4161/psb.6.11.17900
Xu, P., Chen, F., Mannas, J. P., Feldman, T., Sumner, L. W., and Roossinck, M. J. (2008). Virus infection improves drought tolerance. New Phytol. 180, 911–921. doi: 10.1111/j.1469-8137.2008.02627.x
Yang, X., Xie, Y., Raja, P., Li, S., Wolf, J. N., Shen, Q., et al. (2011). Suppression of methylation-mediated transcriptional gene silencing by betaC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog. 7:e1002329. doi: 10.1371/journal.ppat.1002329
Yoshikawa, M., Peragine, A., Park, M. Y., and Poethig, R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. doi: 10.1101/gad.1352605
Zhang, Z., Chen, H., Huang, X., Xia, R., Zhao, Q., Lai, J., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23, 273–288. doi: 10.1105/tpc.110.081695
Keywords: functional indels, miRNA-mediated regulation, secondary siRNAs, viral symptoms, NBS-LRR clade
Citation: Chiumenti M, Catacchio CR, Miozzi L, Pirovano W, Ventura M and Pantaleo V (2018) A Short Indel-Lacking-Resistance Gene Triggers Silencing of the Photosynthetic Machinery Components Through TYLCSV-Associated Endogenous siRNAs in Tomato. Front. Plant Sci. 9:1470. doi: 10.3389/fpls.2018.01470
Received: 07 May 2018; Accepted: 19 September 2018;
Published: 11 October 2018.
Edited by:
Elodie Vandelle, Università degli Studi di Verona, ItalyReviewed by:
Jose Luis Reyes, Universidad Nacional Autónoma de México, MexicoCopyright © 2018 Chiumenti, Catacchio, Miozzi, Pirovano, Ventura and Pantaleo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vitantonio Pantaleo, dml0YW50b25pby5wYW50YWxlb0BjbnIuaXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.