- 1Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 2Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 3Department of Plant Sciences, University of Tennessee, Knoxville, Knoxville, TN, United States
Plant growth-defense tradeoffs are fundamental for optimizing plant performance and fitness in a changing biotic/abiotic environment. This process is thought to involve readjusting resource allocation to different pathways. It has been frequently observed that among secondary cell wall components, alteration in lignin biosynthesis results in changes in both growth and defense. How this process is regulated, leading to growth or defense, remains largely elusive. In this article, we review the canonical lignin biosynthesis pathway, the recently discovered tyrosine shortcut pathway, and the biosynthesis of unconventional C-lignin. We summarize the current model of the hierarchical transcriptional regulation of lignin biosynthesis. Moreover, the interface between recently identified transcription factors and the hierarchical model are also discussed. We propose the existence of a transcriptional co-regulation mechanism coordinating energy allowance among growth, defense and lignin biosynthesis.
Introduction
Lignin is a heterogeneous polymer of monolignols and is polymerized at the surface of the cell walls. The three essential monolignols of plant lignin are p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units (Boerjan et al., 2003). Lignin is important for terrestrial plants by providing structural support for the upward growth of plants and enabling the long-distance water transportation, which are essential for the evolutionary adaptation of plants from the aquatic to terrestrial environment. Lignin also provides physical and chemical protection for plants against pathogen invasion.
The biosynthesis process of lignin has attracted much attention as lignin is the major contributor to the recalcitrance of biomass feedstocks (Studer et al., 2011), dramatically increasing the cost of biomass deconstruction and biofuels production. To increase the digestibility of biomass, genetic engineering of lignin has been an active research area in the past decade (Vanholme et al., 2008) and efforts have been guided toward reducing the cost of biofuel production. On the other hand, the lignin residues after biomass saccharification can be used to produce biodegradable plastic and lignin-derived value-added solvents and chemicals (Doherty et al., 2011; Ragauskas, 2016).
Given the extensive biological and industrial importance of lignin, the understanding of lignin biosynthesis in plants is beneficial for both agricultural and industrial purposes. In this review, we summarize the current understanding of the lignin biosynthesis process and its transcriptional regulation. We then offer a working hypothesis on the transcriptional coordination of energy flux among plant growth, defense, and lignin biosynthesis.
Canonical Lignin Biosynthesis Pathway
In plants, there are two major steps to produce lignin: monolignol biosynthesis and monolignol polymerization via free radical coupling. Enzymes catalyzing monolignol biosynthesis have been well defined in the model plant Arabidopsis. Genetic modulation of these enzymes has been shown to dramatically alter the accumulation and/or composition of lignin.
In Arabidopsis, monolignols are synthesized from phenylalanine via the phenylpropanoid pathway. Therefore, most key phenylpropanoid biosynthetic enzymes are also critical for lignin biosynthesis. A recent study of 221 independent transgenic Populus lines has demonstrated the importance of phenylpropanoid biosynthetic enzymes for lignin biosynthesis in Populus (Wang et al., 2018). The phenylpropanoid pathway is essential in plants, providing precursors for numerous secondary metabolites, including monolignols, flavonoids, and coumarins (Fraser and Chapple, 2011). In the phenylpropanoid pathway, three enzymes: phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate: CoA ligase (4CL), catalyze the first three steps in sequence to provide precursors for all of the downstream metabolites (Fraser and Chapple, 2011). In Arabidopsis and Populus, genetic inhibition of PAL, C4H, and 4CL genes has been shown to significantly decrease lignin content (Rohde et al., 2004; Chen et al., 2006; Vanholme et al., 2008; Wang et al., 2018). In addition to these three enzymes, other phenylpropanoid biosynthetic enzymes, such as quinate/shikimate p-hydroxycinnamoyltransferase (HCT), p-coumaroylshikimate 3′-hydroxylase (C3′H), caffeoyl shikimate esterase (CSE), caffeic acid O-methyltransferase (COMT), and caffeoyl-CoA O-methyltransferase (CCoAOMT), which work downstream of 4CL, are also indispensable for normal lignin biosynthesis (Figure 1). In Arabidopsis, the down-regulation of HCT and C3′H has been shown to induce the enrichment of H units in lignin polymers (Shadle et al., 2007; Pu et al., 2009). Similarly, the down-regulation of HCT gene families dramatically increased the accumulation of H units in Populus (Wang et al., 2018). In Arabidopsis, Populus, and Medicago truncatula, CSE mutants were shown to deposit less lignin (Vanholme et al., 2013; Ha et al., 2016; Saleme et al., 2017). COMT has been found to be critical for the synthesis of S units (Goujon et al., 2003). The Arabidopsis null allele of CCoAOMT (ccomt1) also exhibits reduced lignin content, as well as reduced G units (Do et al., 2007). It is notable that these early steps of monolignol biosynthesis may have variations in monocots. A systematic study of lignin biosynthesis in switchgrass has demonstrated that the conversion of p-coumaroyl CoA to caffeoyl CoA in switchgrass may have an alternative route, which involves the formation of quinate esters catalyzed by HCT-like enzymes (Shen et al., 2013). In addition, the direct involvement of CCoAOMT in lignin biosynthesis was questioned because the knockdown of CCoAOMT genes (CCoAOMT1 and CCoAOMT2) did not change lignin content in switchgrass (Shen et al., 2013).
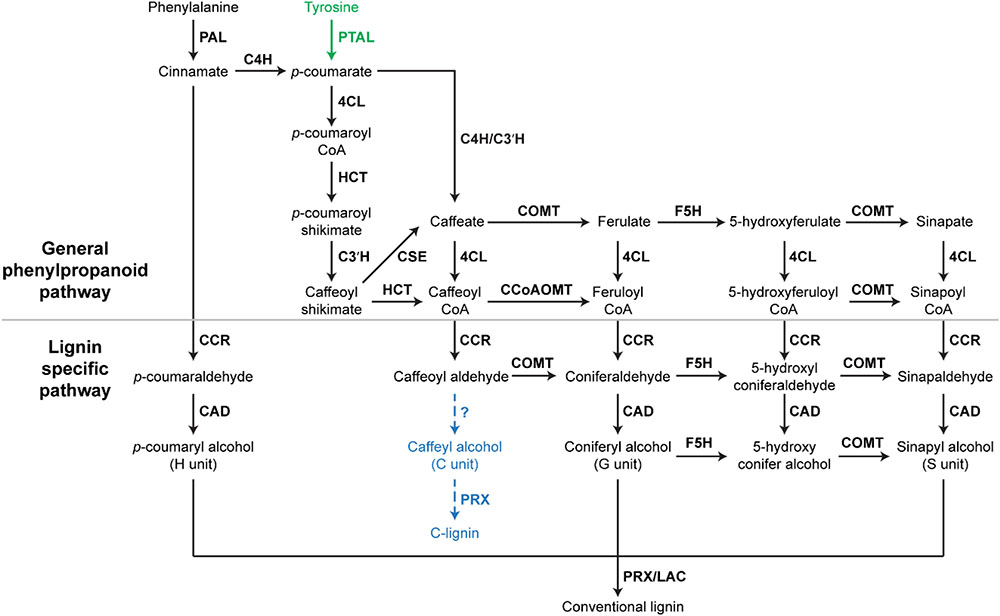
FIGURE 1. A scheme of lignin biosynthesis in plants. Black arrow indicates the canonical lignin biosynthesis in plants. Green arrow indicates the tyrosine shortcut pathway found in Brachypodium distachyon. Blue dashed arrow indicates the putative C-lignin biosynthesis pathway found in seed coats of vanilla orchid and most Cactoidae genera. Bold font indicates enzymes. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; HCT, quinateshikimate p-hydroxycinnamoyltransferase; C3′H, p-coumaroylshikimate 3′-hydroxylase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoAreductase; F5H, ferulate 5-hydroxylase; CAD, cinnamyl alcohol dehydrogenase; COMT, caffeic acid O-methyltransferase; CSE, caffeoyl shikimate esterase; PRX, peroxidase; LAC, laccase.
In addition to enzymes involved in the general phenylpropanoid pathway, enzymes specific for lignin biosynthesis have been identified and characterized, including cinnamoyl-CoA reductase (CCR), ferulate 5-hydroxylase (F5H), cinnamyl alcohol dehydrogenase (CAD) (Barros et al., 2015). These lignin biosynthesis-specific enzymes act downstream of phenylpropanoid biosynthetic enzymes and catalyze the biosynthesis of specific monolignols (Figure 1). The down-regulation of F5H can result in significant increase of G units in lignin (Chen et al., 2006). Arabidopsis mutants of CAD exhibit reduced lignin content, but increased accumulation of G and S units (Sibout et al., 2005). Besides CCR, F5H, and CAD, several lignin biosynthesis-specific steps are catalyzed by COMT, including the conversions of caffeoyl aldehyde to coniferaldehyde, 5-hydroxyl coniferaldehyde to sinapaldehyde, and 5-hydroxy conifer alcohol to sinapyl alcohol (Figure 1).
After biosynthesis, monolignols are polymerized to form lignin. It has been suggested that peroxidases and laccases are key enzymes catalyzing monolignol polymerization though experimental evidence is incomplete. Genetic studies of genes encoding peroxidase and laccase in Arabidopsis have illustrated the close relationship between these enzymes and lignin accumulation in secondary cell walls. Shigeto et al. (2015) found that double mutants of Arabidopsis peroxidases (atprx2/atprx25, atprx2/atprx71, and atprx25/atprx71) have 11–25% reduction in lignin content (Shigeto et al., 2015). Additionally, another peroxidase PRX17 has been found to be critical for lignin accumulation in a recent genetic study in Arabidopsis (Cosio et al., 2017). Similarly, laccases have been shown to affect lignin accumulation. A 20–40% reduction in lignin content has been observed in knockout mutants of two laccase genes (LAC4 and LAC17) in Arabidopsis (Berthet et al., 2011). Moreover, the triple mutant of LAC4, LAC11, and LAC17 completely lost lignin deposition in roots (Zhao et al., 2013).
Tyrosine Shortcut Pathway
A recent study of the model grass Brachypodium distachyon defined a monolignol biosynthesis process with fewer steps (Barros et al., 2016). In this process, monolignols are produced from tyrosine, which is directly converted into p-coumarate by a grass bifunctional phenylalanine and tyrosine ammonia-lyase (PTAL) (Barros et al., 2016). Consequently, this tyrosine shortcut of monolignol biosynthesis in grasses does not contain the steps catalyzed by C4H (Figure 1). With fewer steps, the tyrosine shortcut pathway is energetically more efficient than the canonical lignin biosynthesis pathway. The tyrosine shortcut pathway is capable of guiding carbon and electrons into the biosynthesis of lignin via skipping the production of cinnamate, which is the essential precursor of benzenoid volatiles and salicylic acid. Therefore, the discovery of the tyrosine shortcut pathway provides an alternative approach to optimize the energy investment of lignin production. Although PTAL activity has been detected in some non-grass species, such as tobacco callus and castor bean endosperm (Gregor, 1976; Beaudoin-Eagan and Thorpe, 1985), the involvement of tyrosine shortcut pathway in lignin biosynthesis of non-grass species remains unstudied.
C-Lignin Pathway
In addition to the H, G, and S units of lignin, a natural lignin (called C-lignin) solely containing an unusual Catechyl (C) unit was found in seed coats of vanilla orchid and most Cactoidae genera (Chen et al., 2012, 2013). In C-lignin, caffeyl alcohols are linearly connected by benzodioxane bonds via radical coupling reactions putatively catalyzed by a peroxidase (Figure 1; Chen et al., 2013). Although the detailed caffeyl alcohol biosynthesis and polymerization processes remain unclear, the study in dicot plants demonstrated that the biosynthesis of C-lignin and conventional lignin may be controlled differently (Tobimatsu et al., 2013). C-lignin and conventional lignin were found to be synthesized in a spatially and/or temporally separated manner in seed coats of several dicot plants (Tobimatsu et al., 2013). Without side chains, the linear lignin has less cross-linking with cellulose and hemicellulose, and is capable of enhancing the digestibility of biomass. Further understanding of the mechanism of C-lignin biosynthesis will provide an alternative bioengineering approach to generate better biomass for biofuel production. In addition, the linear C-lignin may be an ideal natural material to replace fossil-fuel based materials for the production of carbon fibers. Compared with fossil-fuel-based feedstocks, the renewable and degradable features of C-lignin make it more environmentally attractive.
Transcriptional Regulation of Lignin Biosynthesis
A key finding of lignin biosynthesis studies is that AC elements widely exist in the promoters of major phenylpropanoid and lignin biosynthetic genes (Rogers and Campbell, 2004). AC elements are DNA motifs rich in adenosine and cytosine. Such significant enrichment of AC elements suggests that the phenylpropanoid and lignin biosynthesis may be under the control of specific types of transcription factors. In the past two decades, key transcription factors regulating the carbon flux into phenylpropanoid and lignin biosynthesis pathways have been identified and a hierarchical transcriptional network connecting these transcription factors has been established (Nakano et al., 2015; Figure 2).
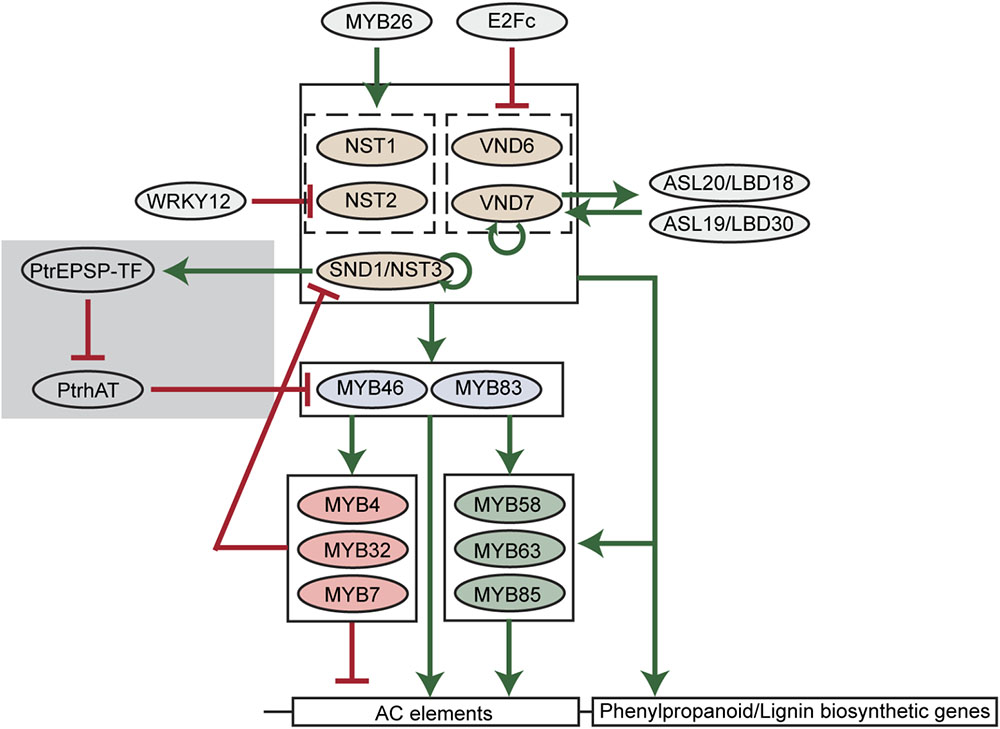
FIGURE 2. A scheme of transcriptional regulation of lignin biosynthetic genes in plants. Green arrows indicate transcriptional activation. Red blunt arrows indicate transcriptional repression. The gray shade indicates the woody plant-specific regulatory loop.
Among transcription factors regulating phenylpropanoid and lignin biosynthesis, MYB46 and its close homolog MYB83 are well-studied in various plant species. In Arabidopsis, MYB46 and MYB83 have been found to activate the expression of PAL1, C4H, 4CL1, C3′H1, HCT, CCoAOMT, CCR1, F5H1, CAD6 genes via binding to AC elements in the promoters of these genes (Zhong and Ye, 2012; Kim et al., 2014). The pine and Eucalyptus MYB46 homologs (PtMYB4 and EgMYB2) were also found to be functional in the regulation of lignin biosynthesis (Patzlaff et al., 2003; Goicoechea et al., 2005). Moreover, four MYB46 homologs in Populus (PtrMYB002, PtrMYB003, PtrMYB020, and PtrMYB021) were capable of triggering ectopic lignin deposition in Arabidopsis and Populus plants (Wilkins et al., 2009; McCarthy et al., 2010; Zhong et al., 2013). In addition to lignin, the biosynthesis of other secondary cell wall components, cellulose and xylan, can also be activated by MYB46 (Zhong and Ye, 2012). Downstream of MYB46/MYB83, multiple MYB transcription factors have been identified as specific regulators of lignin biosynthesis in Arabidopsis, including MYB58, MYB63, MYB85, MYB4, MYB32, and MYB7. Among them, MYB58 and MYB63 are close homologs and specifically activate lignin biosynthesis via targeting AC elements (Zhou et al., 2009). By inducing 4CL expression, MYB85 is also capable of activating monolignol biosynthesis (Zhong et al., 2008). In contrast, MYB4, MYB32, and MYB7 were found to negatively regulate the expression of lignin biosynthetic genes (Jin et al., 2000; Preston et al., 2004; Wang and Dixon, 2012).
Upstream of MYB46/MYB83, the NAC transcription factor SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 1/NAC SECONDARY WALL THICKENING PROMOTING FACTOR 3 (SND1/NST3) and its close homologs NST1, NST2, VASCULAR-RELATED NAC DOMAIN6 (VND6), and VND7 regulate lignin biosynthesis in Arabidopsis (Kubo et al., 2005; Mitsuda et al., 2005; Zhong et al., 2006). Similar to MYB46/MYB83, these NAC transcription factors also regulate the biosynthesis of cellulose and xylan. In Arabidopsis, the regulation of secondary cell wall biosynthesis by these NAC transcription factors is cell-type specific. For example, VND6 and VND7 are specifically expressed in xylem vessel cells and regulate secondary cell wall thickening. Knockdown of VND6 or VND7 using a dominant chimeric repressor specifically inhibits the formation of metaxylem and protoxylem vessels (Kubo et al., 2005). In contrast, NST transcription factors were found to preferentially regulate the secondary cell wall biosynthesis of fiber cells [SND1/NST3 and NST1, (Zhong et al., 2006; Mitsuda et al., 2007)], silique cells [SND1/NST3 and NST1, (Mitsuda and Ohme-Takagi, 2008)], and anther endothecium [NST1 and NST2 (Mitsuda et al., 2005)]. However, the cell-type specificity seems to be absent in the woody plant Populus and monocots, suggesting distinct regulatory mechanisms may exist. In Populus, both VND and NST homologs were found to be expressed in vessels and fibers, though only homologs of VND transcription factors were found to be expressed in primary xylem vessels which have no secondary growth (Ohtani et al., 2011). In monocots, such as rice and maize, homologs of VND and NST transcription factors are named SECONDARY WALL-ASSOCIATED NAC (SWN) due to their regulatory roles in the secondary cell wall biosynthesis of vessel and fiber cells (Zhong et al., 2011).
Because of the essential regulatory roles during secondary cell wall formation, the hierarchical network comprised of NAC and MYB transcription factors is thought to be the major transcriptional regulatory mechanism of lignin and secondary cell wall biosynthesis (Nakano et al., 2015). In this network, feed-forward loops are prevalent. For example, NAC transcription factors directly activate MYB46/MYB83. Meanwhile, NAC transcription factors and MYB46/MYB83 directly activate the downstream MYB58, MYB63, MYB85, and lignin biosynthetic genes (Figure 2). Similarly, MYB46/MYB83 directly activates MYB58, MYB63, and MYB85, and they together directly activate lignin biosynthetic genes (Figure 2). These feed-forward loops are probably to ensure the robust regulation of lignin biosynthesis. Besides feed-forward loops, the NAC-MYB network also contains feed-back regulations to fine-tune lignin biosynthesis (Figure 2). For example, SND1 and VND7 were found to directly target and activate the expression of themselves in Arabidopsis, representing positive feed-back regulation (Wang et al., 2011; Endo et al., 2015). A negative feed-back regulatory loop, in which SND1 is targeted and down-regulated by downstream MYB transcription factors including MYB4, MYB7, and MYB32, was also defined in Arabidopsis (Wang et al., 2011).
Recent discoveries of new transcription factors in Arabidopsis, and other plant species, suggest that the regulatory network of lignin biosynthesis in plants extends beyond the established NAC-MYB network (Rao and Dixon, 2018). Among these new transcription factors, several regulate members of the NAC-MYB network. A transcriptional repressor E2Fc has been reported to directly target VND6 and VND7 in a large-scale yeast one hybrid screening (Taylor-Teeples et al., 2015). The knockdown of E2Fc using RNAi induced ectopic lignin deposition in Arabidopsis roots (Taylor-Teeples et al., 2015), further demonstrating that E2Fc is a negative regulator of lignin biosynthesis. Moreover, Arabidopsis WRKY12 was found to repress lignin biosynthesis via direct repression of the NST2 gene (Wang et al., 2010). Knockdown of WRKY12 in Medicago was found to enhance lignin deposition in cell walls (Wang et al., 2010). The positive regulators of SND1 and its close homologs have also been identified in Arabidopsis. ASYMMETRIC LEAVES2-LIKE20/LATERAL ORGAN BOUNDARIES DOMAIN18 (ASL20/LBD18) and ASL19/LBD30 genes are activated by VND6 and VND7 (Soyano et al., 2008). Overexpression of ASL20/LBD18 and ASL19/LBD30, in turn, enhances the expression of VND7, suggesting a positive feedback loop amplifying VND7 expression (Soyano et al., 2008). In addition, overexpression of MYB26 was found to increase lignin deposition and the expression of NST1 and NST2 (Yang et al., 2007). The discovery of these positive and negative regulators demonstrates that the NAC-MYB network is regulated to spatially and/or temporally switch on or off lignin biosynthesis.
Using genome-wide association studies (GWAS), a novel transcription factor was identified that regulates the expression of PtrMYB021 (homolog of MYB46) in Populus (Xie et al., 2018). This transcription factor (PtrEPSP-TF) is a Populus homolog of 5-enolpyruvylshikimate 3-phosphate (EPSP) synthase, an enzyme catalyzing the shikimate pathway. Aside from the well-established enzyme function of EPSP synthase, PtrEPSP-TF can act as a transcriptional repressor. Genetic and molecular studies further revealed that PtrEPSP-TF activates PtrMYB021 expression and lignin biosynthesis by inhibiting the expression of a transcriptional repressor of PtrMYB021, which is named PtrhAT (Xie et al., 2018). Different from the ancestral EPSP synthase, the PtrEPSP-TF protein contains an additional helix-turn-helix (HTH) motif at its N-terminus, which is indispensable for the nuclear accumulation and transcriptional function of PtrEPSP-TF (Xie et al., 2018). However, the HTH motif is almost entirely missing in EPSP synthases of non-vascular plants, algae, and monocots (Xie et al., 2018). As opposed to herbaceous plants (e.g., Arabidopsis), woody perennial plants (e.g., Populus) have extensive secondary cell wall thickening over multiple growing seasons. The discovery of the additional regulatory loop of MYB46 in Populus supports the existence of woody plant-specific regulatory mechanisms in lignin biosynthesis. Moreover, the discovery of an activator (PtrEPSP-TF) and repressor (PtrhAT) of MYB46 provides alternative approaches to fine tune lignin biosynthesis.
Lignin Biosynthesis has Complex Crosstalk with Growth and Defense
Crosstalk among biological processes is widespread. Genetic studies in Arabidopsis have demonstrated crosstalk among lignin biosynthesis, growth, and defense. Meanwhile, the complexity of the crosstalk is demonstrated by these studies. To date, the relationship of lignin content with growth and defense remains unpredictable due to the limited understanding of the underlying mechanisms.
Lignin is thought to be indispensable for plant growth. The disruption of lignin biosynthesis by knocking down lignin biosynthetic genes, such as C4H and CCR1, was found to co-occur with the suppression of growth rate (Vanholme et al., 2012). However, recent studies suggest that lignin-growth crosstalk is more complex. A study of one C3′H mutant (ref8-1) illustrated that the growth suppression of ref8-1 might be due to the flavonoid hyperaccumulation, rather than the impaired lignin biosynthesis (Besseau et al., 2007; Bonawitz et al., 2014). On the other hand, increased lignin accumulation is also harmful for plant growth. The ectopic lignin deposition induced by the overexpression of NAC and MYB transcription factors, such as SND1, MYB46/83, and MYB58/63, was observed together with inhibited growth in Arabidopsis (Zhong et al., 2006, 2007; Zhou et al., 2009).
The crosstalk between lignin biosynthesis and defense is even more complicated. Lignin is a well-known defense polymer, which has antimicrobial activity, forms a physical barrier to block pathogen invasion, and prevents the ingress or diffusion of toxins from pathogens (Sattler and Funnell-Harris, 2013). It has been widely observed that lignin deposition and lignin biosynthetic genes are induced during the attack of various pathogens. However, the genetic suppression of multiple lignin biosynthesis-related genes was shown to enhance pathogen resistance. For example, Arabidopsis MYB46 mutants (with impaired lignin biosynthesis) displayed enhanced disease resistance, which was thought to be caused by cell wall integrity damage-triggered high immunity level (Ramírez et al., 2011). The repression of HCT in Arabidopsis and Medicago was found to trigger the accumulation of defense hormone salicylic acid (SA) and the expression of PATHOGENESIS-RELATED (PR) genes (Gallego-Giraldo et al., 2011a,b).
Studies on the Arabidopsis HCT mutant further illustrated the crosstalk among lignin biosynthesis, growth, and defense. HCT mutant has reduced lignin content, stunt growth, and enhanced defense (Gallego-Giraldo et al., 2011a). The reduced lignin content was found to trigger the accumulation of SA and the expression of PR genes, which subsequently enhance the pathogen resistance (Gallego-Giraldo et al., 2011a). Impaired lignin biosynthesis is generally thought to cause growth deficiency. However, in HCT mutant, the stunt growth was found to be unrelated with the reduced lignin content, but caused by the increased SA level (Gallego-Giraldo et al., 2011a).
The Transcriptional Co-Regulation of Lignin Biosynthesis, Growth, and Defense
Transcriptional co-regulation has been identified as a mechanism balancing growth and defense. In a recent study, Campos et al. (2016) found that transcriptional networks downstream of jasmonic acid (JA) and phytochrome B signaling form a hub to coordinate growth and defense. The down-regulation of both JA and phytochrome signaling (jazQ phyB double mutant) has been shown to uncouple growth and defense, resulting in plants displaying more growth and more defense (Campos et al., 2016).
Besides the co-regulation of growth and defense, evidence has accumulated that indicates the possible existence of transcriptional co-regulatory mechanisms coordinating growth and lignin biosynthesis, as well as defense and lignin biosynthesis. For example, the semi-dominant mutant (ref4-3) of one subunit of the transcriptional co-regulatory complex Mediator (MED5b) was found to exhibit the dwarfism phenotype, as well as repressed phenylpropanoid and lignin production (Bonawitz et al., 2012). Disruption of MED5b is capable of rescuing both the growth and lignin biosynthesis deficiencies of the C3′H mutant (Bonawitz et al., 2014), further demonstrating the involvement of transcriptional mechanisms in the co-regulation of growth and lignin biosynthesis. On the other hand, several transcription factors have been found to affect lignin biosynthesis, as well as defense responses. Overexpression of one Medicago WRKY transcription factor gene WRKY W109669 in tobacco was shown to enhance the accumulation of lignin and the transcription of PATHOGENESIS-RELATED 2 (PR2) gene (Naoumkina et al., 2008), which is a defense gene induced in response to virus infection (Ward et al., 1991). A recent genetic study of a Gossypium barbadense ethylene response-related factor gene (GbERF1-like) also demonstrated the transcriptional co-regulation of defense and lignin biosynthesis. Overexpression of GbERF1-like in Arabidopsis significantly enhanced the transcription of phenylpropanoid/lignin biosynthetic genes (e.g., PAL3, C4H, C3′H, HCT, CCoAOMT1, CCR1), as well as pathogenesis-related genes including PR3, PR4, and PLANT DEFENSIN1.2 (PDF1.2) (Guo et al., 2016). Moreover, the Arabidopsis myb46 mutants were found to be resistant to the necrotrophic pathogen Botrytis cinerea and to increase PR3 and PDF1.2 expression after Botrytis cinerea infection (Ramírez et al., 2011).
More importantly, genetic studies in Arabidopsis have demonstrated the possibility of the transcriptional co-regulation of growth, defense, and lignin biosynthesis. In mutants of C4H and CCR1, which display significantly reduced lignin accumulation, many defense-responsive genes and many growth-related genes (i.e., auxin response genes) are down-regulated (Vanholme et al., 2012). A MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) was found to regulate the expression of miRNA156 (Serivichyaswat et al., 2015), which negatively regulates inflorescence development and positively regulates tolerance to recurring environmental stress (Wang et al., 2009; Stief et al., 2014). A recent study further identified the regulatory role of AGL15 in lignin biosynthesis. In a study by Cosio et al. (2017), AGL15 was found to directly repress PRX17 expression to regulate lignin formation.
Collectively, genetic studies have illustrated the existence of the transcriptional co-regulation mechanism coordinating growth, defense, and lignin biosynthesis. It is generally recognized that changes in lignin biosynthesis affect the structure and integration of cell walls, which in turn affects growth and defense. However, little is known about the genetic factors co-regulating growth, defense, and lignin biosynthesis or factors bridging the transcriptional co-regulations of growth-lignin biosynthesis and defense-lignin biosynthesis. This is a much-needed research area for future studies.
A Proposed Strategy to Mitigate Growth-Defense Tradeoffs by Manipulating Lignin Biosynthesis
Growing in a dynamic environment, plants have evolved sophisticated strategies to determine the energy allowance between growth (to compete for light) and defense (to fight against pathogens). The competition for energy suggests that growth and defense of plants may have a negative relationship, which means the activation of defense processes negatively affect the growth and reproduction of plants. Such tradeoffs between growth and defense have been observed in many studies (Agrawal et al., 2012; Züst et al., 2015). Although the tradeoff between growth and defense is fundamental for plant survival in a changing environment, it is detrimental for plant yield.
Lignin biosynthesis is traditionally thought as one indivisible part of plant growth and defense, to provide structural support, transport water, and act as physical barrier. However, discoveries of transcriptional mechanisms underlying the crosstalk of lignin biosynthesis, growth, and defense suggest the possibility to unlock lignin biosynthesis from growth and defense. On the other hand, lignin accounts for approximately 30% of the organic carbon in the biosphere (Cesarino et al., 2012). This large amount of energy invested in lignin and its precursors has the potential to compensate the costly expenditure of defense, which consequently would mitigate the tradeoff between growth and defense. Collectively, the genetic modification of the co-regulatory mechanism represents a potential strategy to overcome growth-defense tradeoffs.
Prospective
To date, the lignin biosynthesis process and its regulatory mechanism in the model plant Arabidopsis are well established. Although the lignin biosynthesis process has been well-studied and monolignol biosynthetic enzymes have been systematically analyzed in woody plants, such as Populus, the regulation of lignin biosynthesis remains poorly understood. The regulatory mechanism in a woody perennial plant is much more complex than that in Arabidopsis, due to the complex genome and long life-cycle. With the application of genome-wide approaches, such as GWAS, expression quantitative trait loci (eQTL), and expression quantitative trait nucleotide (eQTN) mapping, in plant studies, it is plausible to identify causal genes affecting complex traits including lignin biosynthesis. Together with efficient post-GWAS characterization and validation, species-specific mechanisms of lignin biosynthesis can be defined.
The concept of a transcriptional hub to coordinate carbon and energy flux into growth, defense, and lignin biosynthesis is just emerging based on studies in Arabidopsis. A comprehensive understanding of this transcriptional hub will have significant impacts on the field of bioengineering. Current studies of NAC-MYB network are restricted in secondary cell wall biosynthesis, although RNA-seq analyses have illustrated that NAC and MYB transcription factors can induce broader gene expression changes. By identifying genome-wide targets of secondary cell wall-related NAC and MYB transcription factors using ChIP-seq, molecular mechanisms linking lignin biosynthesis, growth, and defense are expected to be discovered. In the woody plant Populus, although data is relatively limited, our recent study demonstrated the transcriptional co-regulation of lignin biosynthesis and defense. A defense-responsive WRKY transcription factor was found to regulate the expression of a Populus HCT gene (PtHCT2) (Zhang et al., 2018). With the identification and characterization of transcription factors with multiple functions in the regulation of growth, defense, and lignin biosynthesis, a transcriptional network needs to be established to unveil how growth, defense, and lignin biosynthesis are co-regulated. With this knowledge, the resource flux in plants can be fine-tuned depending on human demand, which will greatly reduce the cost of agricultural and forestry biofuels production.
Author Contributions
MX drafted the manuscript. JZ, TT, GT, J-GC, and WM revised the manuscript.
Funding
This work was supported by the Center for Bioenergy Innovation and the Plant-Microbe Interfaces Scientific Focus Area by the Office of Biological and Environmental Research in the U.S. Department of Energy Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
References
Agrawal, A. A., Hastings, A. P., Johnson, M. T., Maron, J. L., and Salminen, J.-P. (2012). Insect herbivores drive real-time ecological and evolutionary change in plant populations. Science 338, 113–116. doi: 10.1126/science.1225977
Barros, J., Serk, H., Granlund, I., and Pesquet, E. (2015). The cell biology of lignification in higher plants. Ann. Bot. 115, 1053–1074. doi: 10.1093/aob/mcv046
Barros, J., Serrani-Yarce, J. C., Chen, F., Baxter, D., Venables, B. J., and Dixon, R. A. (2016). Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat. Plants 2:16050. doi: 10.1038/nplants.2016.50
Beaudoin-Eagan, L. D., and Thorpe, T. A. (1985). Tyrosine and phenylalanine ammonia lyase activities during shoot initiation in tobacco callus cultures. Plant Physiol. 78, 438–441.
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cezard, L., Le Bris, P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/tpc.110.082792
Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162. doi: 10.1105/tpc.106.044495
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Bonawitz, N. D., Kim, J. I., Tobimatsu, Y., Ciesielski, P. N., Anderson, N. A., Ximenes, E., et al. (2014). Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380. doi: 10.1038/nature13084
Bonawitz, N. D., Soltau, W. L., Blatchley, M. R., Powers, B. L., Hurlock, A. K., Seals, L. A., et al. (2012). REF4 and RFR1, subunits of the transcriptional coregulatory complex mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287, 5434–5445. doi: 10.1074/jbc.M111.312298
Campos, M. L., Yoshida, Y., Major, I. T., De Oliveira Ferreira, D., Weraduwage, S. M., Froehlich, J. E., et al. (2016). Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat. Commun. 7:12570. doi: 10.1038/ncomms12570
Cesarino, I., Araújo, P., Domingues Júnior, A. P., and Mazzafera, P. (2012). An overview of lignin metabolism and its effect on biomass recalcitrance. Braz. J. Bot. 35, 303–311. doi: 10.1016/j.biotechadv.2012.03.003
Chen, F., Srinivasa Reddy, M. S., Temple, S., Jackson, L., Shadle, G., and Dixon, R. A. (2006). Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. 48, 113–124. doi: 10.1111/j.1365-313X.2006.02857.x
Chen, F., Tobimatsu, Y., Havkin-Frenkel, D., Dixon, R. A., and Ralph, J. (2012). A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. U.S.A. 109, 1772–1777. doi: 10.1073/pnas.1120992109
Chen, F., Tobimatsu, Y., Jackson, L., Nakashima, J., Ralph, J., and Dixon, R. A. (2013). Novel seed coat lignins in the cactaceae: structure, distribution and implications for the evolution of lignin diversity. Plant J. 73, 201–211. doi: 10.1111/tpj.12012
Cosio, C., Ranocha, P., Francoz, E., Burlat, V., Zheng, Y., Perry, S. E., et al. (2017). The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 213, 250–263. doi: 10.1111/nph.14127
Do, C. T., Pollet, B., Thevenin, J., Sibout, R., Denoue, D., Barriere, Y., et al. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129. doi: 10.1007/s00425-007-0558-3
Doherty, W. O. S., Mousavioun, P., and Fellows, C. M. (2011). Value-adding to cellulosic ethanol: lignin polymers. Ind. Crops Prod. 33, 259–276. doi: 10.1016/j.indcrop.2010.10.022
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Fraser, C. M., and Chapple, C. (2011). The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 9:e0152. doi: 10.1199/tab.0152
Gallego-Giraldo, L., Escamilla-Trevino, L., Jackson, L. A., and Dixon, R. A. (2011a). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. U.S.A. 108, 20814–20819. doi: 10.1073/pnas.1117873108
Gallego-Giraldo, L., Jikumaru, Y., Kamiya, Y., Tang, Y., and Dixon, R. A. (2011b). Selective lignin downregulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 190, 627–639. doi: 10.1111/j.1469-8137.2010.03621.x
Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., et al. (2005). EgMYB2, a new transcriptional activator from eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43, 553–567. doi: 10.1111/j.1365-313X.2005.02480.x
Goujon, T., Sibout, R., Pollet, B., Maba, B., Nussaume, L., Bechtold, N., et al. (2003). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51, 973–989.
Gregor, H. D. (1976). Studies on tyrosine ammonia-lyase activity in castor bean endosperm. Zeitschrift Fur Pflanzenphysiologie 77, 372–375.
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91, 305–318. doi: 10.1007/s11103-016-0467-6
Ha, C. M., Escamilla-Trevino, L., Yarce, J. C., Kim, H., Ralph, J., Chen, F., et al. (2016). An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 86, 363–375. doi: 10.1111/tpj.13177
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Kim, W. C., Kim, J. Y., Ko, J. H., Kang, H., and Han, K. H. (2014). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 85, 589–599. doi: 10.1007/s11103-014-0205-x
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
McCarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. doi: 10.1093/pcp/pcq064
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitsuda, N., and Ohme-Takagi, M. (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56, 768–778. doi: 10.1111/j.1365-313X.2008.03633.x
Mitsuda, N., Seki, M., Shinozaki, K., and Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. doi: 10.1105/tpc.105.036004
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., and Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6:288. doi: 10.3389/fpls.2015.00288
Naoumkina, M. A., He, X., and Dixon, R. A. (2008). Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 8:132. doi: 10.1186/1471-2229-8-132
Ohtani, M., Nishikubo, N., Xu, B., Yamaguchi, M., Mitsuda, N., Goue, N., et al. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67, 499–512. doi: 10.1111/j.1365-313X.2011.04614.x
Patzlaff, A., Mcinnis, S., Courtenay, A., Surman, C., Newman, L. J., Smith, C., et al. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36, 743–754. doi: 10.1046/j.1365-313X.2003.01916.x
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Pu, Y. Q., Chen, F., Ziebell, A., Davison, B. H., and Ragauskas, A. J. (2009). NMR characterization of C3H and HCT down-regulated alfalfa lignin. Bioenergy Res. 2, 198–208. doi: 10.1007/s12155-009-9056-8
Ragauskas, A. J. (2016). Challenging/interesting lignin times. Biofuels Bioprod. Biorefining Biofpr 10, 489–491. doi: 10.1002/bbb.1714
Ramírez, V., García-Andrade, J., and Vera, P. (2011). Enhanced disease resistance to Botrytis cinerea in myb46 Arabidopsis plants is associated to an early down-regulation of CesA genes. Plant Signal. Behav. 6, 911–913.
Rao, X., and Dixon, R. A. (2018). Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 9:399. doi: 10.3389/fpls.2018.00399
Rogers, L. A., and Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Rohde, A., Morreel, K., Ralph, J., Goeminne, G., Hostyn, V., De Rycke, R., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16, 2749–2771. doi: 10.1105/tpc.104.023705
Saleme, M. L. S., Cesarino, I., Vargas, L., Kim, H., Vanholme, R., Goeminne, G., et al. (2017). Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar. Plant Physiol. 175, 1040–1057. doi: 10.1104/pp.17.00920
Sattler, S. E., and Funnell-Harris, D. L. (2013). Modifying lignin to improve bioenergy feedstocks: strengthening the barrier against pathogens? Front. Plant Sci. 4:70. doi: 10.3389/fpls.2013.00070
Serivichyaswat, P., Ryu, H. S., Kim, W., Kim, S., Chung, K. S., Kim, J. J., et al. (2015). Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol. Cells 38, 259–266. doi: 10.14348/molcells.2015.2311
Shadle, G., Chen, F., Srinivasa Reddy, M. S., Jackson, L., Nakashima, J., and Dixon, R. A. (2007). Down-regulation of hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 68, 1521–1529. doi: 10.1016/j.phytochem.2007.03.022
Shen, H., Mazarei, M., Hisano, H., Escamilla-Trevino, L., Fu, C., Pu, Y., et al. (2013). A genomics approach to deciphering lignin biosynthesis in switchgrass. Plant Cell 25, 4342–4361. doi: 10.1105/tpc.113.118828
Shigeto, J., Itoh, Y., Hirao, S., Ohira, K., Fujita, K., and Tsutsumi, Y. (2015). Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 57, 349–356. doi: 10.1111/jipb.12334
Sibout, R., Eudes, A., Mouille, G., Pollet, B., Lapierre, C., Jouanin, L., et al. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17, 2059–2076. doi: 10.1105/tpc.105.030767
Soyano, T., Thitamadee, S., Machida, Y., and Chua, N. H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373. doi: 10.1105/tpc.108.061796
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Baurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Studer, M. H., Demartini, J. D., Davis, M. F., Sykes, R. W., Davison, B., Keller, M., et al. (2011). Lignin content in natural populus variants affects sugar release. Proc. Natl. Acad. Sci. U.S.A. 108, 6300–6305. doi: 10.1073/pnas.1009252108
Taylor-Teeples, M., Lin, L., De Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tobimatsu, Y., Chen, F., Nakashima, J., Escamilla-Trevino, L. L., Jackson, L., Dixon, R. A., et al. (2013). Coexistence but independent biosynthesis of catechyl and guaiacyl/syringyl lignin polymers in seed coats. Plant Cell 25, 2587–2600. doi: 10.1105/tpc.113.113142
Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y., Sundin, L., Goeminne, G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602
Vanholme, R., Morreel, K., Ralph, J., and Boerjan, W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11, 278–285. doi: 10.1016/j.pbi.2008.03.005
Vanholme, R., Storme, V., Vanholme, B., Sundin, L., Christensen, J. H., Goeminne, G., et al. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24, 3506–3529. doi: 10.1105/tpc.112.102574
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., and Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U.S.A. 107, 22338–22343. doi: 10.1073/pnas.1016436107
Wang, H. Z., and Dixon, R. A. (2012). On-off switches for secondary cell wall biosynthesis. Mol. Plant 5, 297–303. doi: 10.1093/mp/ssr098
Wang, H. Z., Zhao, Q., Chen, F., Wang, M. Y., and Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x
Wang, J. P., Matthews, M. L., Williams, C. M., Shi, R., Yang, C., Tunlaya-Anukit, S., et al. (2018). Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 9:1579. doi: 10.1038/s41467-018-03863-z
Wang, J. W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Ward, E. R., Payne, G. B., Moyer, M. B., Williams, S. C., Dincher, S. S., Sharkey, K. C., et al. (1991). Differential regulation of beta-1,3-glucanase messenger RNAs in response to pathogen infection. Plant Physiol. 96, 390–397.
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Xie, M., Muchero, W., Bryan, A. C., Yee, K., Guo, H. B., Zhang, J., et al. (2018). A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in populus. Plant Cell 30, 1645–1660. doi: 10.1105/tpc.18.00168
Yang, C., Xu, Z., Song, J., Conner, K., Vizcay Barrena, G., and Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548. doi: 10.1105/tpc.106.046391
Zhang, J., Yang, Y., Zheng, K., Xie, M., Feng, K., Jawdy, S. S., et al. (2018). Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT 2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in populus. New Phytol. doi: 10.1111/nph.15297 [Epub ahead of print].
Zhao, Q., Nakashima, J., Chen, F., Yin, Y., Fu, C., Yun, J., et al. (2013). Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987. doi: 10.1105/tpc.113.117770
Zhong, R., Demura, T., and Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., Mccarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/pcp/pcr123
Zhong, R., Lee, C., Zhou, J., Mccarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., Mccarthy, R. L., Haghighat, M., and Ye, Z. H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8:e69219. doi: 10.1371/journal.pone.0069219
Zhong, R., Richardson, E. A., and Ye, Z.-H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2792.
Zhong, R., and Ye, Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. doi: 10.1093/pcp/pcr185
Zhou, J., Lee, C., Zhong, R., and Ye, Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Keywords: phenylpropanoid, lignin, transcription factor, growth-defense tradeoffs, secondary cell wall, transcriptional co-regulation
Citation: Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen J-G and Muchero W (2018) Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 9:1427. doi: 10.3389/fpls.2018.01427
Received: 13 June 2018; Accepted: 07 September 2018;
Published: 28 September 2018.
Edited by:
Laura Elizabeth Bartley, The University of Oklahoma, United StatesReviewed by:
Huanzhong Wang, University of Connecticut, United StatesJavier Agusti, Instituto de Biología Molecular y Celular de Plantas (IBMCP), Spain
Copyright © 2018 Xie, Zhang, Tschaplinski, Tuskan, Chen and Muchero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Gui Chen, chenj@ornl.gov Wellington Muchero, mucherow@ornl.gov
 Meng Xie
Meng Xie Jin Zhang
Jin Zhang Timothy J. Tschaplinski1,2
Timothy J. Tschaplinski1,2 Gerald A. Tuskan
Gerald A. Tuskan Jin-Gui Chen
Jin-Gui Chen Wellington Muchero
Wellington Muchero