
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 19 September 2018
Sec. Plant Metabolism and Chemodiversity
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01369
This article is part of the Research TopicHydrogen Sulfide Metabolism and Signalling in PlantsView all 11 articles
Hydrogen sulfide (H2S) has been largely referred as a toxic gas and environmental hazard, but recent years, it has emerged as an important gas-signaling molecule with effects on multiple physiological processes in both animal and plant systems. The regulatory functions of H2S in plants are involved in important processes such as the modulation of defense responses, plant growth and development, and the regulation of senescence and maturation. The main signaling pathway involving sulfide has been proven to be through protein persulfidation (alternatively called S-sulfhydration), in which the thiol group of cysteine (-SH) in proteins is modified into a persulfide group (-SSH). This modification may cause functional changes in protein activities, structures, and subcellular localizations of the target proteins. New shotgun proteomic approaches and bioinformatic analyses have revealed that persulfidated cysteines regulate important biological processes, highlighting their importance in cell signaling, since about one in 20 proteins in Arabidopsis is persulfidated. During oxidative stress, an increased persulfidation has been reported and speculated that persulfidation is the protective mechanism for protein oxidative damage. Nevertheless, cysteine residues are also oxidized to different post-translational modifications such S-nitrosylation or S-sulfenylation, which seems to be interconvertible. Thus, it must imply a tight cysteine redox regulation essential for cell survival. This review is aimed to focus on the current knowledge of protein persulfidation and addresses the regulation mechanisms that are disclosed based on the knowledge from other cysteine modifications.
Hydrogen sulfide (H2S) is an inorganic, flammable, water-soluble gas with a noticeable odor of rotten eggs. H2S has been historically considered as a pollutant and a toxic gas for life. Nevertheless, in the past decade, it has emerged as a new gaseous signaling molecule (gasotransmitter) in animal and plant cells and is as important as nitric oxide (NO), carbon monoxide (CO), and hydrogen peroxide (H2O2) (Garcia-Mata and Lamattina, 2010; Vandiver and Snyder, 2012; Kimura, 2014). Since 1996, when H2S was first described as an endogenous neuromodulator in animals (Abe and Kimura, 1996), an increasing number of articles have described its physiological effects on plants and animals. H2S is involved in many physiological and pathological processes in animals, such as apoptosis, inflammatory processes, the protective effects against hypoxia, neuromodulation, cell proliferation and cardioprotection, among others, as described in several recent reports and reviews (Wang, 2014; Olas, 2015; Paul and Snyder, 2015a).
In plants, the first descriptions of H2S effects are dated to the 1960s, when H2S was reported to influence the growth of vegetative plants and to affect disease resistance (Rodriguez-Kabana et al., 1965; Thompson and Kats, 1978). Mainly based on pharmacological approaches, H2S has been documented for its protective effects against different stresses, such as oxidative and metal stresses (Zhang et al., 2008; Wang et al., 2010; Zhang et al., 2010b; Li L. et al., 2012; Shen et al., 2013; Sun et al., 2013; Fang et al., 2016), drought and heat tolerance (Li Z.G. et al., 2012; Shen et al., 2013), and osmotic and saline stresses (Shi et al., 2013). H2S was also found to regulate important physiological processes in plants, such as stomatal closure/aperture (Garcia-Mata and Lamattina, 2010; Lisjak et al., 2010; Jin et al., 2013; Scuffi et al., 2014; Papanatsiou et al., 2015), the modulation of photosynthesis (Chen et al., 2011) and autophagy regulation (Álvarez et al., 2012a; Gotor et al., 2013; Romero et al., 2014; Laureano-Marin et al., 2016). However, an endogenous production of H2S was also observed in plants, and a possible cross-talk with NO was proposed due to the similarity of their physiological effects (Kolluru et al., 2015). All these observations have prompted interest in the scientific community to address the outstanding questions related to H2S signaling and its interaction with other gasotransmitters and hormones in plants.
This review is aimed to focus on the current knowledge of H2S as a signaling molecule, its mechanism of action and its role in plant physiology.
In plant systems, H2S production occurs mainly via the photosynthetic sulfate-assimilation pathway in chloroplasts in the reaction catalyzed by the sulfite reductase (SiR) (Takahashi et al., 2011; Garcia et al., 2015). During the synthesis of β-cyanoalanine, the enzyme cyanoalanine synthase c1 (CAS-C1) also generates sulfide in the mitochondria (Yamaguchi et al., 2000; Álvarez et al., 2012b). The enzyme L-cysteine desulfhydrase 1 (DES1) is the responsible of the major production of endogenous cytosolic H2S (Álvarez et al., 2010, 2012a; Gotor et al., 2010), nevertheless other enzymes have also been reported to produce sulfide, such as D-cysteine desulfhydrase (Riemenschneider et al., 2005) and Nifs-like proteins (ABA3) (Heidenreich et al., 2005). The sulfide concentration in chloroplasts is higher than that in the cytosol (125 and 55 μM, respectively) (Krueger et al., 2009), however, this sulfide is dissociated into its ionized forms due the basic pH inside organelles and therefore is unable to pass through the membranes to the cytosol (Kabil and Banerjee, 2010).
Hydrogen sulfide (H2S) is a particularly reactive molecule, and there is plenty of evidences that H2S interacts with other signaling molecules to modifying their signal. There is increasing interest in the interaction of sulfide with plant hormones such as abscisic acid (ABA) (Jin et al., 2013; Scuffi et al., 2014), giberellic acid (GA) (Xie et al., 2014), and ethylene (Liu et al., 2011; Liu et al., 2012). Further recent evidence suggests that H2S also plays a role in the hydrogen peroxide (H2O2) (Zhang et al., 2010a) and nitric oxide (NO) signaling pathways (Lisjak et al., 2011). Although numerous reports highlight the importance of H2S as a signaling molecule, its primary mechanism of action has been recently deciphered (Mustafa et al., 2009; Aroca et al., 2015). It has been explained through a new post-translational modification (PTM) of proteins, named persulfidation, where reactive cysteine residues on target proteins are modified via conversion of the thiol group (-SH) into a persulfide group (-SSH). This modification was inaccurately referred to as “S-sulfhydration”; however, since it does not imply any “hydration,” it was renamed as persulfidation (Filipovic, 2015). Persulfide adducts show an increased nucleophilicity compared to the thiol group and therefore, modified cysteines demonstrate a challenging greater reactivity (Paul and Snyder, 2012). That could be the reason persulfidation is widespread in nature and affects a higher percentage of proteins than reactive oxygen and nitrogen species (Mustafa et al., 2009; Ida et al., 2014).
The way H2S modifies specific targets is still unclear, since direct reaction of H2S and a thiol is thermodynamically unfavorable (Filipovic, 2015). Sulfane sulfur is a sulfur atom that has the unique ability to bind reversibly to other sulfur atoms to form hydropersulfides (R-S-SH) and polysulfides (-S-Sn-S-). These polysulfides seems to be much more effective in persulfidation since they are more nucleophilic than H2S (Toohey, 1989, 2011). New low molecular weight (LMW) persulfides have recently emerged as potential mediators in sulfide signaling. In this regard, cysteine-persulfide (Cys-SSH), glutathione persulfide (GSSH) and its persulfurated species Cys-SSnH and GSSnH have been recognized as redox regulators (Kasamatsu et al., 2016; Kimura et al., 2017). Recently, the endogenous Cys-SSH production synthetized by prokaryotic and mammalian cysteinyl-tRNA synthetases (CARSs) using L-cysteine as substrate has been described. The cysteine polysulfides bound to tRNA are incorporated into polypeptides that are synthesized de novo in the ribosomes, suggesting that these enzymes are the principal cysteine persulfide synthases in vivo (Akaike et al., 2017).
Two extensive reviews have been recently published further explaining the chemical properties of persulfides and polysulfides (Cuevasanta et al., 2017; Filipovic et al., 2017). Due to the instability and high reactivity of persulfides, and its similarity to thiols, the efforts on developing detection methods have been a challenge.
The modified biotin switch assay (mBSM) was the first method used for a proteomic approach based on the analysis of persulfidated proteins in mammals (Mustafa et al., 2009). This method uses S-methyl-methanothiosulfonate (MMTS) to block free thiols. While the persulfide residues remain unreacted and are therefore available for subsequent reaction with the thiol-specific biotinylating agent biotin-HPDP. Lately, was used to detect for the first time the persulfidated proteins in plant systems (Aroca et al., 2015). A total of 106 persulfidated proteins were identified by liquid chromatography-mass spectrometry (LC-MS/MS) in Arabidopsis plants, which were mainly involved in photosynthesis, protein synthesis, and cell organization according to MapMan classification. Some of these identified proteins in plants were previously described to undergo persulfidation in mammals. Furthermore, the low concentration of sulfide produced an inactivation/activation effect on enzyme activities, which was reversible by reductants, demonstrating that sulfide had a biological role in plants through persulfidation, similar to mammalian systems.
Nevertheless, MMTS was questioned as a good blocking reagent since it could also react with persulfides and thus protein identification was understimated.
Several reactives have been recently reported and used in animal systems for the detection of persulfides. These are (i) the fluorescent Cy5-maleimide (Sen et al., 2012), (ii) maleimide-PEG2-biotin (Dóka et al., 2016), and (iii) iodoacetyl-PEG2-biotin (Gao et al., 2015); among others. However, most of the methods hitherto described have shown a weakness by lacking in specificity.
In plants, a new approach to detect persulfidated proteins was recently reported (Aroca et al., 2017a) based on the method previously described and named the tag-switch method (Zhang et al., 2014), which showed higher specificity than other methods described. This method employs methylsulfonylbenzothiazole (MSBT) to block both thiols and persulfide groups; then, a nucleophilic attack by the cyanoacetate-based reagent CN-biotin is performed labeling only the persulfide groups, which are purified with streptavidin conjugates and analyzed by Western blot, or directly by LC-MS/MS. This study revealed that 2,015 proteins (5% of the Arabidopsis proteome) were modified by persulfidation and that approximately 3,200 proteins were potentially targets for this modification in mature plants. This new method increased the number of persulfidated targets in plants from 106 to more than 2,000 (Aroca et al., 2015, 2017a). These proteins were involved in the regulation of important biochemical pathways for cell survival. However, these data were obtained from plants grown under physiological conditions. Therefore, the number of persulfidated proteins may be higher under stress conditions where sulfide plays a signaling role.
Although a high number of persulfidated proteins have been identified in both plant and animal systems, the functional impact of this modification in cells is starting to be clarified. Proteins modified by persulfidation show functional changes in enzymatic activities, structures and in subcellular localizations (Mustafa et al., 2009; Aroca et al., 2015, 2017b; Kimura, 2015; Paul and Snyder, 2015b). The biological importance of this protein modification in plant systems was initially demonstrated by the enzymatic activity of chloroplastic glutamine synthetase (GS2), cytosolic ascorbate peroxidase (APX1), and cytosolic glyceraldehyde 3-phosphate dehydrogenase (GapC1) (Aroca et al., 2015). The effect of persulfidation of these enzymes showed activity activation for APX1 and GapC1, while decreased activity was found for GS2; this effect was reversible by reducing agents.
The GO categorization of persulfidated proteins identified in Arabidopsis by a shot-gun proteomic assay revealed that persulfidation is involved in the regulation of important biological processes, such as carbon metabolism, plant responses to abiotic stresses, plant growth and development, and RNA translation (Aroca et al., 2017a). Moreover, the cytosolic or nuclear localization of GapC1 is regulated by persulfidation, and the persulfidated cysteine residue has been identified in nuclear fraction (Aroca et al., 2017b). In addition to the metabolic functions, GapC plays roles in mRNA regulation, transcriptional activation and apoptosis depending on its nuclear translocation outcome (Ortiz-Ortiz et al., 2010). Thus, persulfidation plays a key role not only in regulating the activity of modified proteins; it also may regulate cellular localization of proteins with significant consequences in plant systems.
The role of sulfide in another important biological process such as autophagy has also been described (Álvarez et al., 2012a; Laureano-Marin et al., 2016). Sulfide is able to inhibit the autophagy induced in Arabidopsis roots under nutrient deprivation; this repression is a mechanism independent of redox conditions. As described, H2S inhibits autophagy by preventing the ATG8 (autophagy-related ubiquitin-like protein) accumulation (Álvarez et al., 2012a). The mechanism of this inhibition by sulfide is still unclear, but Gotor and colleagues have speculated that this regulation might be through persulfidation of the enzymes involved in the autophagosome formation (Gotor et al., 2013). Furthermore, the ubiquitin-like systems ATG7-ATG10 and ATG7-ATG3, and the cysteine protease ATG4, which are essential proteins involved in autophagy, sense cellular redox alterations by their reactive Cys residues (Filomeni et al., 2010). Recently, a high throughput proteomic approach in Arabidopsis leaves revealed the susceptibility to be persulfidated of some autophagy (ATG)-related proteins, ATG18a, ATG3, ATG5, and ATG7 (Aroca et al., 2017a). In yeast and algae, ATG4 is regulated by thioredoxin, suggesting the involvement of a redox PTM in the regulation of ATG4 activity (Perez-Perez et al., 2014). In mammals, several PTMs play important roles in the regulation of autophagy. ATG4b and ATG1 are regulated by phosphorylation and S-nitrosylation (Li et al., 2017; Pengo et al., 2017; Sanchez-Wandelmer et al., 2017), and Caspase-3, which is also essential for autophagic activity, is persulfidated at Cys163 associating this modification to the cytoprotective effect of sulfide (Marutani et al., 2015).
The results in Arabidopsis suggest that persulfidation may be the molecular mechanism through which sulfide regulates autophagy in plant cells. However, the identification of other targets involved in autophagy requires further specific studies, and the functional role of persulfidation in autophagy has not been sufficiently studied.
Many other biological functions must be precisely regulated through sulfide and the crosstalk with other signaling molecules, including H2O2 and NO, and other antioxidant molecules such as glutathione, which has been integrated in redox signaling. In this sense, protein thiols are crucial players since they can be modified by different PTMs, showing an opposite regulation in some cases. H2O2 directly attacks the catalytic cysteine of GAPC, causing a strong inhibition of the enzyme activity (Hancock et al., 2005). S-nitrosylation of GAPC also abolishes its catalytic activity, whereas persulfidation increases its activity (Lindermayr et al., 2005; Aroca et al., 2015). Besides, this enzyme may also undergo S-glutathionylation, which also inhibits its activity (Zaffagnini et al., 2007; Bedhomme et al., 2012). Likewise, oxidation of glutamine synthetase by H2O2 inhibits its activity (Ortega et al., 1999) in a similar manner as S-nitrosylation and persulfidation (Melo et al., 2011; Aroca et al., 2015). Moreover, S-glutathionylation of glutamine synthetase can also regulate its activity (Dixon et al., 2005). Aroca et al. (2017a) showed that 639 proteins were targets for both S-nitrosylation and persulfidation. Although just a few proteins (91) have been identified in Arabidopsis as S-glutathionylated proteins in the literature (Lee et al., 2004; Dixon et al., 2005; Leferink et al., 2009; Lindermayr et al., 2010; Palmieri et al., 2010; Bedhomme et al., 2012), 85% of them may also undergo S-nitrosylation or persulfidation (Figure 1A). A large amount of the proteins modified by these three PTMs are common (a total of 690), meaning they can be modified by at least two of these PTMs. Functional classification of the GO terms categorized by biological processes shows that these common proteins are mainly involved in protein metabolism (20%). However, they are even more represented in amino acid metabolism (8.5%), glycolysis (3.1%), tricarboxylic acid cycle (3.6%), redox regulation (3.4%), and secondary metabolism (5.2%) in comparison with those proteins that are modified by just one of these PTMs (Figure 1B). Thus, these pathways must be finely regulated by different PTMs in a cysteine residue, suggesting a crosstalk among these signaling molecules under different stress conditions.
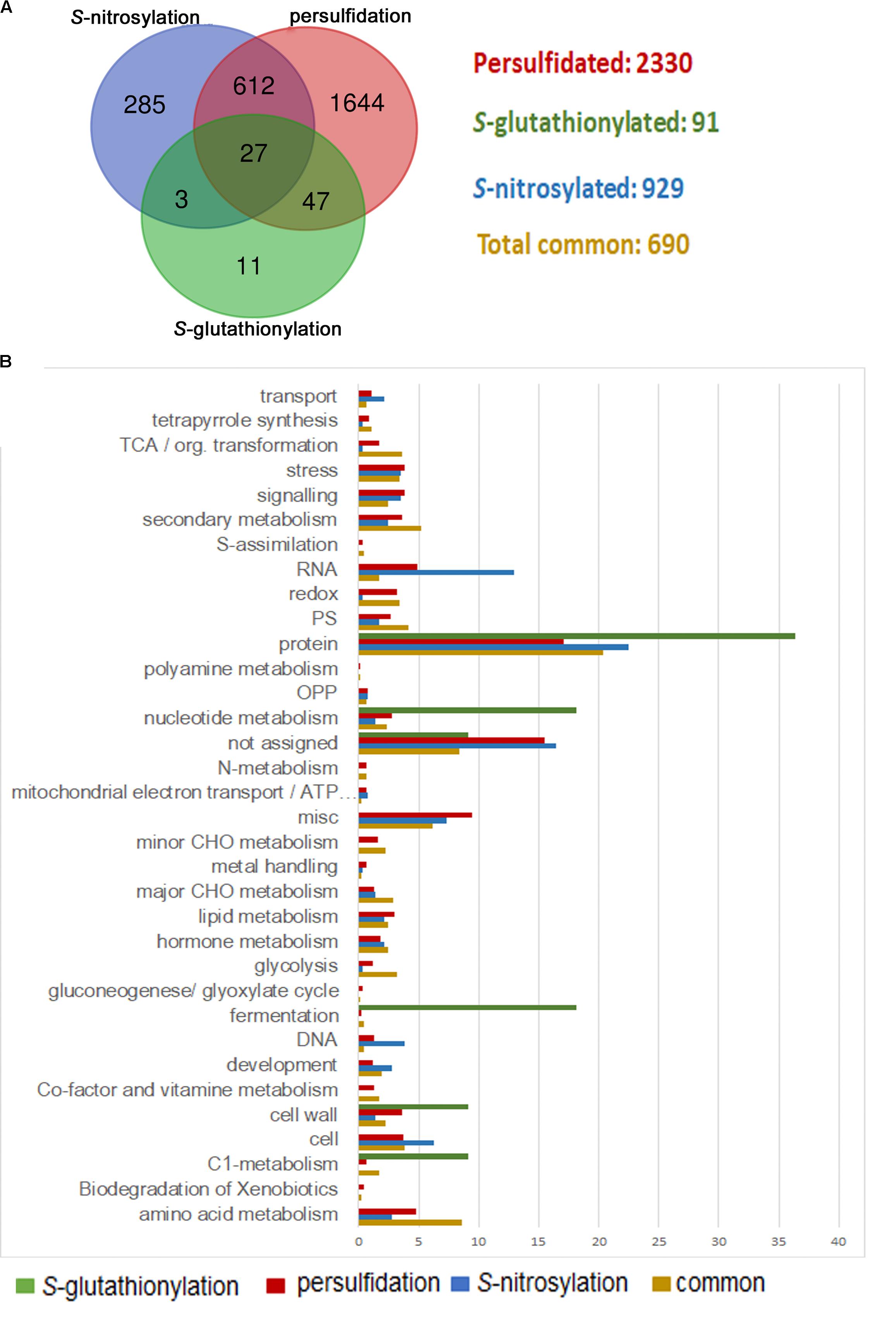
FIGURE 1. Comparison of persulfidated, S-nitrosylated, and S-glutathionylated proteins identified in Arabidopsis plants. (A) Venn diagram of total persulfidated (Aroca et al., 2017a), S-nitrosylated (Hu et al., 2015), and S-glutathionylated (Lee et al., 2004; Dixon et al., 2005; Lindermayr et al., 2010; Palmieri et al., 2010; Bedhomme et al., 2012) identified proteins in Arabidopsis plants. (B) Functional classification of gene ontology (GO) terms categorized by biological processes.
Arabidopsis plants exogenously treated with H2S also showed changes in the transcriptome (Huang et al., 2002; Álvarez et al., 2012a). Based on proteomic approaches, several transcription factors (TFs) and chromatin modifiers (CMs) such as histones, acetyltransferases, and methyltransferases, have been identified as targets for persulfidation (Sen et al., 2012; Aroca et al., 2017a). This modification affects their specificity to DNA and their binding affinity resulting in distinct transcriptional responses. In addition, this PTM of nuclear proteins can modulate transcription by affecting their subcellular localization or by regulating the association with their binding partners (Filipovic, 2015; Aroca et al., 2017b). Thus, sulfide is also involved in epigenetic regulation of chromatin by histone modification and chromatin structure alteration (Kamat et al., 2015; Yang, 2015), highlighting an interesting signaling role of sulfide through persulfidation that could also be functioning in plants.
Plants are exposed to several stresses, which cause oxidative stress due to the accumulation of reactive oxygen species (ROS) and nitrogen species (RNS) (Huang et al., 2012; Schieber and Chandel, 2014). Under these stress conditions, cysteine thiols may undergo different PTMs such as S-nitrosylation (SNO), S-glutathionylation (SSG), and S-sulfenylation (SOH). However, these oxidized compounds can be reduced in the cells by reducing agents, such as glutathione (GSH), thioredoxin (Trx), and glutaredoxin (Grx) (Sevilla et al., 2015). Nevertheless, when the stress remains, irreversible modifications of thiols occurs, such as sulfinic (RSO2H), and sulfonic acids (RSO3H). In that sense, persulfidation (SSH) is believed to account for the protective effect against ROS/RNS, since persulfidated proteins will react with ROS/RNS and form an adduct (RSSO3H) that may be restored by thioredoxin to free thiol (Wedmann et al., 2016; Filipovic and Jovanović, 2017). Most antioxidant enzymes are finely regulated by different PTMs under specific environmental stress conditions. In that sense, APX1 is inactivated by the oxidation of Cys32, while glutathionylation protects the enzyme from irreversible oxidation (Kitajima et al., 2008). The same site Cys32 can also be S-nitrosylated by NO and persulfidated by hydrogen sulfide, which increases the activity of the enzyme (Begara-Morales et al., 2014; Aroca et al., 2015). The fact that APX1 is modified by different PTMs means that this enzyme must be finely regulated under specific stress conditions.
Nitric Oxide increases the activities of antioxidant enzymes such as catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione reductase (GPX), and peroxidase (POD); therefore, NO may stimulate the antioxidant system to decrease oxidative stress (Lamattina et al., 2003; Laspina et al., 2005). NO levels increase in plants under drought stress, which helps plants to mitigate the negative effects of water deficit (Shi et al., 2014), and NO is an important player in ABA-induced stomatal closure, minimizing plant transpiration (Seabra and Oliveira, 2016). Exogenous H2S is found to induce stomatal closure through the regulation of ATP-binding cassette (ABC) transporters, while scavenging H2S can partially block ABA-dependent stomatal closure, indicating the protective role of H2S in plants against drought stress (Garcia-Mata and Lamattina, 2010). Sulfide-induced stomatal closure can be reverted by cPTIO (a NO-specific scavenger), also confirming that the function of sulfide in stomatal closure is mediated by NO. Furthermore, several proteins that play essential roles in the ABA-dependent regulation of stomatal movement, are modified by persulfidation and S-nitrosylation (Wang et al., 2015; Aroca et al., 2017a). These findings suggest a crosstalk between NO and H2S in drought stress mediated by S-nitrosylation and persulfidation, respectively.
An increasing number of articles regarding persulfidation have been published in the last years on different organisms, and many proteins have been found to be modified by sulfide through persulfidation. Nevertheless, the functional role of this modification in specific targets must be studied to validate these data. Furthermore, the specificity on cysteine residues is still unclear since, in some targets, the modified cysteine is the active site, but not in others. Additionally, the extent of the interaction between several signaling molecules into the same cysteine residue deserves more investigation. Understanding the interplay among them will enlarge our knowledge on the biochemical cascade triggered in plant cells against different stresses.
A large number of laboratory experiments have revealed the protective effect of sulfide in plants to overcome different environmental stresses. A better understanding of persulfidation in these stress events would help to address biotechnological applications in order to improve the productivity of crops and protection against abiotic stress processes.
AA organized and prepared all this manuscript. CG and LR contributed for writing and reviewing the major part of the manuscript.
This work was supported in part by the European Regional Development Fund through the Agencia Estatal de Investigación grant BIO2016-76633-P. We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abe, K., and Kimura, H. (1996). The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 16, 1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996
Akaike, T., Ida, T., Wei, F.-Y., Nishida, M., Kumagai, Y., Alam, M. M., et al. (2017). Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 8:1177. doi: 10.1038/s41467-017-01311-y
Álvarez, C., Calo, L., Romero, L. C., García, I., and Gotor, C. (2010). An O-Acetylserine(thiol)lyase homolog with l-Cysteine Desulfhydrase activity regulates Cysteine Homeostasis in Arabidopsis. Plant Physiol. 152, 656–669. doi: 10.1104/pp.109.147975
Álvarez, C., Garcia, I., Moreno, I., Perez-Perez, M. E., Crespo, J. L., Romero, L. C., et al. (2012a). Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24, 4621–4634. doi: 10.1105/tpc.112.105403
Álvarez, C., Garcia, I., Romero, L. C., and Gotor, C. (2012b). Mitochondrial sulfide detoxification requires a functional isoform O-acetylserine(thiol)lyase C in Arabidopsis thaliana. Mol. Plant 5, 1217–1226. doi: 10.1093/mp/sss043
Aroca, A., Benito, J. M., Gotor, C., and Romero, L. C. (2017a). Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 68, 4915–4927. doi: 10.1093/jxb/erx294
Aroca, A., Schneider, M., Scheibe, R., Gotor, C., and Romero, L. C. (2017b). Hydrogen sulfide regulates the Cytosolic/Nuclear partitioning of Glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 58, 983–992. doi: 10.1093/pcp/pcx056
Aroca, A., Serna, A., Gotor, C., and Romero, L. C. (2015). S-sulfhydration: a cysteine posttranslational modification in plant systems. Plant Physiol. 168, 334–342. doi: 10.1104/pp.15.00009
Bedhomme, M., Adamo, M., Marchand, C. H., Couturier, J., Rouhier, N., Lemaire, S. D., et al. (2012). Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 445, 337–347. doi: 10.1042/BJ20120505
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Valderrama, R., Mata-Pérez, C., López-Jaramillo, J., et al. (2014). Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 65, 527–538. doi: 10.1093/jxb/ert396
Chen, J., Wu, F. H., Wang, W. H., Zheng, C. J., Lin, G. H., Dong, X. J., et al. (2011). Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. J. Exp. Bot. 62, 4481–4493. doi: 10.1093/jxb/err145
Cuevasanta, E., Möller, M. N., and Alvarez, B. (2017). Biological chemistry of hydrogen sulfide and persulfides. Arch. Biochem. Biophys. 617, 9–25. doi: 10.1016/j.abb.2016.09.018
Dixon, D. P., Skipsey, M., Grundy, N. M., and Edwards, R. (2005). Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138, 2233–2244. doi: 10.1104/pp.104.058917
Dóka,É, Pader, I., Bíró, A., Johansson, K., Cheng, Q., Ballagó, K., et al. (2016). A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci. Adv. 2:e1500968. doi: 10.1126/sciadv.1500968
Fang, H., Liu, Z., Jin, Z., Zhang, L., Liu, D., and Pei, Y. (2016). An emphasis of hydrogen sulfide-cysteine cycle on enhancing the tolerance to chromium stress in Arabidopsis. Environ. Pollut. 213, 870–877. doi: 10.1016/j.envpol.2016.03.035
Filipovic, M. R. (2015). Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 230, 29–59. doi: 10.1007/978-3-319-18144-8_2
Filipovic, M. R., and Jovanović, V. M. (2017). More than just an intermediate: hydrogen sulfide signalling in plants. J. Exp. Bot. 68, 4733–4736. doi: 10.1093/jxb/erx352
Filipovic, M. R., Zivanovic, J., Alvarez, B., and Banerjee, R. (2017). Chemical biology of H2S signaling through persulfidation. Chem. Rev. 118, 1253–1337. doi: 10.1021/acs.chemrev.7b00205
Filomeni, G., Desideri, E., Cardaci, S., Rotilio, G., and Ciriolo, M. R. (2010). Under the ROS.thiol network is the principal suspect for autophagy commitment. Autophagy 6, 999–1005. doi: 10.4161/auto.6.7.12754
Gao, X. H., Krokowski, D., Guan, B. J., Bederman, I., Majumder, M., Parisien, M., et al. (2015). Quantitative HS-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. eLife 4:e10067. doi: 10.7554/eLife.10067
Garcia, I., Gotor, C., and Romero, L. C. (2015). “Cysteine homeostasis,” in Amino Acids in Higher Plants, ed. J. P. F. D’mello (Wallingford: CABI Publishing), 219–233.
Garcia-Mata, C., and Lamattina, L. (2010). Hydrogen sulphide, a novel gasotransmitter involved in guard cell signalling. New Phytol. 188, 977–984. doi: 10.1111/j.1469-8137.2010.03465.x
Gotor, C., Álvarez, C., Bermúdez, M. Á, Moreno, I., García, I., and Romero, L. C. (2010). Low abundance does not mean less importance in cysteine metabolism. Plant Signal. Behav. 5, 1028–1030. doi: 10.1105/tpc.109.071985
Gotor, C., Garcia, I., Crespo, J. L., and Romero, L. C. (2013). Sulfide as a signaling molecule in autophagy. Autophagy 9, 609–611. doi: 10.4161/auto.23460
Hancock, J. T., Henson, D., Nyirenda, M., Desikan, R., Harrison, J., Lewis, M., et al. (2005). Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Biochem. 43, 828–835. doi: 10.1016/j.plaphy.2005.07.012
Heidenreich, T., Wollers, S., Mendel, R. R., and Bittner, F. (2005). Characterization of the NifS-like domain of ABA3 from Arabidopsis thaliana provides insight into the mechanism of molybdenum cofactor sulfuration. J. Biol. Chem. 280, 4213–4218. doi: 10.1074/jbc.M411195200
Hu, J., Huang, X., Chen, L., Sun, X., Lu, C., Zhang, L., et al. (2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 167, 1731–1746. doi: 10.1104/pp.15.00026
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Huang, X., Von Rad, U., and Durner, J. (2002). Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215, 914–923. doi: 10.1007/s00425-002-0828-z
Ida, T., Sawa, T., Ihara, H., Tsuchiya, Y., Watanabe, Y., Kumagai, Y., et al. (2014). Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 7606–7611. doi: 10.1073/pnas.1321232111
Jin, Z., Xue, S., Luo, Y., Tian, B., Fang, H., Li, H., et al. (2013). Hydrogen sulfide interacting with abscisic acid in stomatal regulation responses to drought stress in Arabidopsis. Plant Physiol. Biochem. 62, 41–46. doi: 10.1016/j.plaphy.2012.10.017
Kabil, O., and Banerjee, R. (2010). Redox biochemistry of hydrogen sulfide. J. Biol. Chem. 285, 21903–21907. doi: 10.1074/jbc.R110.128363
Kamat, P. K., Kalani, A., Tyagi, S. C., and Tyagi, N. (2015). Hydrogen sulfide epigenetically attenuates homocysteine-induced mitochondrial toxicity mediated through NMDA receptor in mouse brain endothelial (bEnd3) cells(). J. Cell. Physiol. 230, 378–394. doi: 10.1002/jcp.24722
Kasamatsu, S., Nishimura, A., Morita, M., Matsunaga, T., Abdul Hamid, H., and Akaike, T. (2016). Redox signaling regulated by cysteine persulfide and protein polysulfidation. Molecules 21:E1721. doi: 10.3390/molecules21121721
Kimura, H. (2014). The physiological role of hydrogen sulfide and beyond. Nitric Oxide 41, 4–10. doi: 10.1016/j.niox.2014.01.002
Kimura, H. (2015). Signaling of hydrogen sulfide and polysulfides. Antioxid. Redox Signal. 22, 347–349. doi: 10.1089/ars.2014.6082
Kimura, Y., Koike, S., Shibuya, N., Lefer, D., Ogasawara, Y., and Kimura, H. (2017). 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci. Rep. 7:10459. doi: 10.1038/s41598-017-11004-7
Kitajima, S., Kurioka, M., Yoshimoto, T., Shindo, M., Kanaori, K., Tajima, K., et al. (2008). A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase. FEBS J. 275, 470–480. doi: 10.1111/j.1742-4658.2007.06214.x
Kolluru, G. K., Yuan, S., Shen, X., and Kevil, C. G. (2015). “Chapter fifteen - H2S regulation of nitric oxide metabolism,” in Methods in Enzymology, eds C. Enrique and P. Lester (Cambridge, MA: Academic Press), 271–297.
Krueger, S., Niehl, A., Martin, M. C., Steinhauser, D., Donath, A., Hildebrandt, T., et al. (2009). Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 32, 349–367. doi: 10.1111/j.1365-3040.2008.01928.x
Lamattina, L., Garcia-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Laspina, N. V., Groppa, M. D., Tomaro, M. L., and Benavides, M. P. (2005). Nitric oxide protects sunflower leaves against Cd-induced oxidative stress. Plant Sci. 169, 323–330. doi: 10.1016/j.plantsci.2005.02.007
Laureano-Marin, A. M., Moreno, I., Romero, L. C., and Gotor, C. (2016). Negative regulation of autophagy by sulfide is independent of reactive oxygen species. Plant Physiol. 171, 1378–1391. doi: 10.1104/pp.16.00110
Lee, K., Lee, J., Kim, Y., Bae, D., Kang, K. Y., Yoon, S. C., et al. (2004). Defining the plant disulfide proteome. Electrophoresis 25, 532–541. doi: 10.1002/elps.200305677
Leferink, N. G., Van Duijn, E., Barendregt, A., Heck, A. J., and Van Berkel, W. J. (2009). Galactonolactone dehydrogenase requires a redox-sensitive thiol for optimal production of vitamin C. Plant Physiol. 150, 596–605. doi: 10.1104/pp.109.136929
Li, L., Wang, Y., and Shen, W. (2012). Roles of hydrogen sulfide and nitric oxide in the alleviation of cadmium-induced oxidative damage in alfalfa seedling roots. Biometals 25, 617–631. doi: 10.1007/s10534-012-9551-9
Li, Z. G., Gong, M., Xie, H., Yang, L., and Li, J. (2012). Hydrogen sulfide donor sodium hydrosulfide-induced heat tolerance in tobacco (Nicotiana tabacum L) suspension cultured cells and involvement of Ca(2+) and calmodulin. Plant Sci. 185–186, 185–189. doi: 10.1016/j.plantsci.2011.10.006
Li, Y., Zhang, Y., Wang, L., Wang, P., Xue, Y., Li, X., et al. (2017). Autophagy impairment mediated by S-nitrosation of ATG4B leads to neurotoxicity in response to hyperglycemia. Autophagy 13, 1145–1160. doi: 10.1080/15548627.2017.1320467
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-Nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Lindermayr, C., Sell, S., Muller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Lisjak, M., Srivastava, N., Teklic, T., Civale, L., Lewandowski, K., Wilson, I., et al. (2010). A novel hydrogen sulfide donor causes stomatal opening and reduces nitric oxide accumulation. Plant Physiol. Biochem. 48, 931–935. doi: 10.1016/j.plaphy.2010.09.016
Lisjak, M., Teklić, T., Wilson, I. D., Wood, M., Whiteman, M., and Hancock, J. T. (2011). Hydrogen sulfide effects on stomatal apertures. Plant Signal. Behav. 6, 1444–1446. doi: 10.4161/psb.6.10.17104
Liu, J., Hou, L., Liu, G., Liu, X., and Wang, X. (2011). Hydrogen sulfide induced by nitric oxide mediates ethylene-induced stomatal closure of Arabidopsis thaliana. Chin. Sci. Bull. 56, 3547–3553. doi: 10.1007/s11434-011-4819-y
Liu, J., Hou, Z.-H., Liu, G.-H., Hou, L.-X., and Liu, X. (2012). Hydrogen sulfide may function downstream of nitric oxide in ethylene-induced stomatal closure in Vicia faba L. J. Integr. Agric. 11, 1644–1653. doi: 10.1016/S2095-3119(12)60167-1
Marutani, E., Yamada, M., Ida, T., Tokuda, K., Ikeda, K., Kai, S., et al. (2015). Thiosulfate mediates cytoprotective effects of hydrogen sulfide against neuronal ischemia. J. Am. Heart Assoc. 4:e002125. doi: 10.1161/JAHA.115.002125
Melo, P. M., Silva, L. S., Ribeiro, I., Seabra, A. R., and Carvalho, H. G. (2011). glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. doi: 10.1104/pp.111.186056
Mustafa, A. K., Gadalla, M. M., Sen, N., Kim, S., Mu, W., Gazi, S. K., et al. (2009). H2S signals through protein S-sulfhydration. Sci. Signal. 2:ra72. doi: 10.1126/scisignal.2000464
Olas, B. (2015). Hydrogen sulfide in signaling pathways. Clin. Chim. Acta 439, 212–218. doi: 10.1016/j.cca.2014.10.037
Ortega, J. L., Roche, D., and Sengupta-Gopalan, C. (1999). Oxidative turnover of soybean root glutamine synthetase. in vitro and in vivo studies. Plant Physiol. 119, 1483–1496. doi: 10.1104/pp.119.4.1483
Ortiz-Ortiz, M. A., Moran, J. M., Ruiz-Mesa, L. M., Bravo-San Pedro, J. M., and Fuentes, J. M. (2010). Paraquat exposure induces nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and the activation of the nitric oxide-GAPDH-Siah cell death cascade. Toxicol. Sci. 116, 614–622. doi: 10.1093/toxsci/kfq146
Palmieri, M. C., Lindermayr, C., Bauwe, H., Steinhauser, C., and Durner, J. (2010). Regulation of plant glycine decarboxylase by s-nitrosylation and glutathionylation. Plant Physiol. 152, 1514–1528. doi: 10.1104/pp.109.152579
Papanatsiou, M., Scuffi, D., Blatt, M. R., and García-Mata, C. (2015). Hydrogen sulfide regulates inward-rectifying K+ channels in conjunction with stomatal closure. Plant Physiol. 168, 29–35. doi: 10.1104/pp.114.256057
Paul, B. D., and Snyder, S. H. (2012). H2S signalling through protein sulfhydration and beyond. Nat. Rev. Mol. Cell Biol. 13, 499–507. doi: 10.1038/nrm3391
Paul, B. D., and Snyder, S. H. (2015a). Modes of physiologic H2S signaling in the brain and peripheral tissues. Antioxid. Redox Signal. 22, 411–423. doi: 10.1089/ars.2014.5917
Paul, B. D., and Snyder, S. H. (2015b). Protein sulfhydration. Methods Enzymol. 555, 79–90. doi: 10.1016/bs.mie.2014.11.021
Pengo, N., Agrotis, A., Prak, K., Jones, J., and Ketteler, R. (2017). A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat. Commun. 8:294. doi: 10.1038/s41467-017-00303-2
Perez-Perez, M. E., Zaffagnini, M., Marchand, C. H., Crespo, J. L., and Lemaire, S. D. (2014). The yeast autophagy protease Atg4 is regulated by thioredoxin. Autophagy 10, 1953–1964. doi: 10.4161/auto.34396
Riemenschneider, A., Wegele, R., Schmidt, A., and Papenbrock, J. (2005). Isolation and characterization of a D-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J. 272, 1291–1304. doi: 10.1111/j.1742-4658.2005.04567.x
Rodriguez-Kabana, R., Jordan, J. W., and Hollis, J. P. (1965). Nematodes: biological control in rice fields: role of hydrogen sulfide. Science 148, 524–526. doi: 10.1126/science.148.3669.524
Romero, L. C., Aroca, M. A., Laureano-Marin, A. M., Moreno, I., Garcia, I., and Gotor, C. (2014). Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant 7, 264–276. doi: 10.1093/mp/sst168
Sanchez-Wandelmer, J., Kriegenburg, F., Rohringer, S., Schuschnig, M., Gomez-Sanchez, R., Zens, B., et al. (2017). Atg4 proteolytic activity can be inhibited by Atg1 phosphorylation. Nat. Commun. 8:295. doi: 10.1038/s41467-017-00302-3
Schieber, M., and Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Scuffi, D., Nunez, A., Laspina, N., Gotor, C., Lamattina, L., and Garcia-Mata, C. (2014). Hydrogen sulfide generated by L-cysteine desulfhydrase acts upstream of nitric oxide to modulate ABA-dependent stomatal closure. Plant Physiol. 166, 2065–2076. doi: 10.1104/pp.114.245373
Seabra, A. B., and Oliveira, H. C. (2016). How nitric oxide donors can protect plants in a changing environment: what we know so far and perspectives. AIMS Mol. Sci. 3, 692–718. doi: 10.3934/molsci.2016.4.692
Sen, N., Paul, B. D., Gadalla, M. M., Mustafa, A. K., Sen, T., Xu, R., et al. (2012). Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol. Cell 45, 13–24. doi: 10.1016/j.molcel.2011.10.021
Sevilla, F., Camejo, D., Ortiz-Espin, A., Calderon, A., Lazaro, J. J., and Jimenez, A. (2015). The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 66, 2945–2955. doi: 10.1093/jxb/erv146
Shen, J., Xing, T., Yuan, H., Liu, Z., Jin, Z., Zhang, L., et al. (2013). Hydrogen sulfide improves drought tolerance in Arabidopsis thaliana by microRNA expressions. PLoS One 8:e77047. doi: 10.1371/journal.pone.0077047
Shi, H., Ye, T., and Chan, Z. (2013). Exogenous application of hydrogen sulfide donor sodium hydrosulfide enhanced multiple abiotic stress tolerance in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiol. Biochem. 71, 226–234. doi: 10.1016/j.plaphy.2013.07.021
Shi, H., Ye, T., Zhu, J.-K., and Chan, Z. (2014). Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J. Exp. Bot. 65, 4119–4131. doi: 10.1093/jxb/eru184
Sun, J., Wang, R., Zhang, X., Yu, Y., Zhao, R., Li, Z., et al. (2013). Hydrogen sulfide alleviates cadmium toxicity through regulations of cadmium transport across the plasma and vacuolar membranes in Populus euphratica cells. Plant Physiol. Biochem. 65, 67–74. doi: 10.1016/j.plaphy.2013.01.003
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Thompson, C. R., and Kats, G. (1978). Effects of continuous hydrogen sulfide fumigation on crop and forest plants. Environ. Sci. Technol. 12, 550–553. doi: 10.1021/es60141a001
Toohey, J. I. (1989). Sulphane sulphur in biological systems: a possible regulatory role. Biochem. J. 264, 625–632. doi: 10.1042/bj2640625
Toohey, J. I. (2011). Sulfur signaling: is the agent sulfide or sulfane? Anal. Biochem. 413, 1–7. doi: 10.1016/j.ab.2011.01.044
Vandiver, M., and Snyder, S. (2012). Hydrogen sulfide: a gasotransmitter of clinical relevance. J. Mol. Med. 90, 255–263. doi: 10.1007/s00109-012-0873-4
Wang, B. L., Shi, L., Li, Y. X., and Zhang, W. H. (2010). Boron toxicity is alleviated by hydrogen sulfide in cucumber (Cucumis sativus L.) seedlings. Planta 231, 1301–1309. doi: 10.1007/s00425-010-1134-9
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wang, R. (2014). Gasotransmitters: growing pains and joys. Trends Biochem. Sci. 39, 227–232. doi: 10.1016/j.tibs.2014.03.003
Wedmann, R., Onderka, C., Wei, S., Szijártó, I. A., Miljkovic, J. L., Mitrovic, A., et al. (2016). Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem. Sci. 7, 3414–3426. doi: 10.1039/c5sc04818d
Xie, Y., Zhang, C., Lai, D., Sun, Y., Samma, M. K., Zhang, J., et al. (2014). Hydrogen sulfide delays GA-triggered programmed cell death in wheat aleurone layers by the modulation of glutathione homeostasis and heme oxygenase-1 expression. J. Plant Physiol. 171, 53–62. doi: 10.1016/j.jplph.2013.09.018
Yamaguchi, Y., Nakamura, T., Kusano, T., and Sano, H. (2000). Three Arabidopsis genes encoding proteins with differential activities for cysteine synthase and beta-cyanoalanine synthase. Plant Cell Physiol. 41, 465–476. doi: 10.1093/pcp/41.4.465
Yang, G. (2015). H2S epigenetic regulation of vascular cell functions. Cardiovasc. Regen. Med. 2:e967.
Zaffagnini, M., Michelet, L., Marchand, C., Sparla, F., Decottignies, P., Le Marechal, P., et al. (2007). The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 274, 212–226. doi: 10.1111/j.1742-4658.2006.05577.x
Zhang, D., Macinkovic, I., Devarie-Baez, N. O., Pan, J., Park, C. M., Carroll, K. S., et al. (2014). Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. Engl. 53, 575–581. doi: 10.1002/anie.201305876
Zhang, H., Hu, L. Y., Hu, K. D., He, Y. D., Wang, S. H., and Luo, J. P. (2008). Hydrogen sulfide promotes wheat seed germination and alleviates oxidative damage against copper stress. J. Integr. Plant Biol. 50, 1518–1529. doi: 10.1111/j.1744-7909.2008.00769.x
Zhang, H., Jiao, H., Jiang, C.-X., Wang, S.-H., Wei, Z.-J., Luo, J.-P., et al. (2010a). Hydrogen sulfide protects soybean seedlings against drought-induced oxidative stress. Acta Physiol. Plant. 32, 849–857. doi: 10.1007/s11738-010-0469-y
Keywords: Arabidopsis, cell signaling, cysteine, hydrogen sulfide, persulfidation, post-translational modification, proteomic
Citation: Aroca A, Gotor C and Romero LC (2018) Hydrogen Sulfide Signaling in Plants: Emerging Roles of Protein Persulfidation. Front. Plant Sci. 9:1369. doi: 10.3389/fpls.2018.01369
Received: 03 June 2018; Accepted: 29 August 2018;
Published: 19 September 2018.
Edited by:
Fumiya Kurosaki, University of Toyama, JapanReviewed by:
Masami Yokota Hirai, RIKEN Center for Sustainable Resource Science (CSRS), JapanCopyright © 2018 Aroca, Gotor and Romero. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angeles Aroca, YW5nZWxlcy5hcm9jYUBpYnZmLmNzaWMuZXM=; bWFuZ2VsZXNhcm9jYUB5YWhvby5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.