
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 13 September 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01368
This article is part of the Research Topic Flowering Phenology in Trees: from Molecular Control to Mathematical Models View all 5 articles
During autumn perennial trees cease growth and form structures called buds in order to protect meristems from the unfavorable environmental conditions, including low temperature and desiccation. In addition to increased tolerance to these abiotic stresses, reproductive buds modulate developmental programs leading to dormancy induction to avoid premature growth resumption, and flowering pathways. Stress tolerance, dormancy, and flowering processes are thus physically and temporarily restricted to a bud, and consequently forced to interact at the regulatory level. We review recent genomic, genetic, and molecular contributions to the knowledge of these three processes in trees, highlighting the role of epigenetic modifications, phytohormones, and common regulatory factors. Finally, we emphasize the utility of transcriptomic approaches for the identification of key structural and regulatory genes involved in bud processes, illustrated with our own experience using peach as a model.
Boreal and temperate tree species cope with harsh environmental conditions during autumn and winter, including low and freezing temperatures, in a quiescent state named dormancy. Dormancy has been originally defined as the absence of visible growth in a meristematic structure. Traditionally, three types of dormancy have been distinguished according to physiological cues leading to growth inhibition: endodormancy, when signals are intrinsic to the meristem; paradormancy, imposed by another part of the plant; and ecodormancy, due to environmental factors (Lang, 1987). More recently, dormancy has been reformulated as “the inability to initiate growth from meristems under favorable conditions" (Singh et al., 2017). When applied to buds, this general definition covers endodormancy, and axillary bud paradormancy imposed by apical dominance, but not bud growth inhibition by environmental factors after fulfillment of chilling requirements (ecodormancy). In this review, we use the term dormancy referring to this second meaning.
Previously, prominent reviews have also addressed the known mechanisms of bud dormancy control in perennial plants from a molecular perspective (Rohde and Bhalerao, 2007; Allona et al., 2008; Anderson et al., 2010; Yamane, 2014; Maurya and Bhalerao, 2017; Singh et al., 2017). Several of these studies have focused on the molecular control of growth arrest in apical vegetative meristems. Growth cessation and dormancy induction in those meristems are regulated by endogenous and environmental signals, being photoperiod shortening and temperature lowering major determinants of dormancy setup in forest species and Rosaceae fruit trees, respectively (Heide and Prestrud, 2005; Cooke et al., 2012). By contrast, the development of flower lateral meristems is usually determined by apical dominance and other factors to overwinter in a differentiated immature stage, preceding dormancy release, and growth resumption on next season.
Even though diverse anatomical and physiological particularities are found, differentiated vegetative and reproductive meristems cease growth in a well defined stage and form a bud surrounded by protective scales in a similar fashion in different tree species. However, in spite of growth cessation and dormancy, overwintering buds do not remain in a completely inactive state, as verified by extensive transcriptomic and hormonal changes observed during bud development (Howe et al., 2015; Chao et al., 2017). In fact, three major processes including dormancy, cold acclimation, and flowering converge spatially and temporally in a reproductive bud, where they play an active and relevant role in bud dynamics and determine plant survival and growth resumption under favorable conditions. These processes determine bud phenology and development through their reciprocal interaction, integrating temperature, and photoperiod effects (Kurokura et al., 2013; Vitasse et al., 2014; Singh et al., 2017). Since flowering development requires cell division and expansion, and dormancy and cold acclimation are essentially non-growing processes, the control of cell cycle and growth is expected to influence the succession of the mutually incompatible periods of flowering and dormancy (Horvath et al., 2003).
The transition to reproductive growth starts around early spring in Populus (Boes and Strauss, 1994) and summer in many Rosaceae (Kurokura et al., 2013) when flower bud induction occurs in axillary meristems. Then flower organ differentiation starts and is substantially accomplished before dormancy initiates. Growth arrest and seasonal dormancy are induced specifically by either photoperiod or temperature in different species (Cooke et al., 2012). In parallel, cold, freezing, and desiccation tolerances are increased by an acclimation mechanism (Welling and Palva, 2006). Subsequent production of reproductive gametes and resumption of flower organ growth requires dormancy release triggered by the quantitative perception of chilling accumulated during the dormancy period (Coville, 1920; Couvillon and Erez, 1985). After dormancy release, buds remain cold-acclimated until a period of warm temperatures results in deacclimation and bud break (Welling and Palva, 2006). The whole succession of events from flower bud induction to blooming can be interpreted as a trade-off between defense factors leading to cold acclimation and dormancy and growth factors leading to dormancy release and flowering. In this context, photoperiod and temperature are important environmental inputs acting occasionally on opposing ways.
A more detailed review of recent progresses in the regulation of dormancy, cold acclimation and flowering processes in buds of temperate trees is shown below, with dormancy and growth promoting pathways addressed in separated sections. In addition, the utility of genomic approaches for the identification of genes related to these processes is illustrated with our own transcriptomic studies performed in flower buds of peach (Prunus persica) across dormancy release.
Reduction in daylength below a critical value induces growth cessation in several trees through the participation of orthologs of well known elements of the photoperiodic control of flowering in Arabidopsis, particularly photoreceptors and the circadian clock. One of them is phytochrome A (PHYA) gene that codes for a red/far-red light receptor and causes, respectively, impaired and faster growth cessation responses to short-days in hybrid aspen (Populus tremula × Populus tremuloides) plants overexpressing and down-regulating the gene (Olsen et al., 1997; Kozarewa et al., 2010). In addition, antisense inhibition of PHYA alters the expression of clock components, suggesting a link with circadian rhythms. This has been confirmed by down-regulating the circadian clock genes LATE ELONGATED HYPOCOTYL1 (PttLHY1), PttLHY2, and TIMING OF CAB EXPRESSION 1 (PttTOC1) in hybrid aspen, leading to a reduced critical daylength required for growth cessation and additional effects on winter hardiness and bud burst (Ibáñez et al., 2010). The regulatory module CONSTANS (CO)/FLOWERING LOCUS T (FT), which mediates the effect of photoreceptors and circadian clock on flowering initiation in Arabidopsis (Valverde et al., 2004), has been also proposed to control growth cessation and bud set in Populus trees (Böhlenius et al., 2006). In fact, two paralogs of FT act coordinately to determine vegetative and reproductive growth (Hsu et al., 2011); whereas FT1 induces the reproductive onset, FT2 promotes vegetative growth and inhibits bud formation under warm temperature and long photoperiod conditions. Also the FT/TERMINAL FLOWER1-Like2 (PaFTL2) gene induces bud set under transgenic inducible expression in the conifer Norway spruce (Picea abies), resembling the FT-antagonistic role of TERMINAL FLOWER1 (TFL1) in Arabidopsis (Karlgren et al., 2013). Finally, further evidences of the functional diversification of CO/FT module in flowering and dormancy processes arise from the study of other components of the pathway: the FT protein interactor gene FLOWERING LOCUS D is involved in growth cessation and bud formation (Tylewicz et al., 2015; Parmentier-Line and Coleman, 2016); and both overexpression and RNAi studies show that a tree ortholog of the flowering pathway integrator APETALA1 (AP1) mediates photoperiod-dependent growth cessation in hybrid aspen (Azeez et al., 2014).
Besides photoperiod, temperature also affects seasonal growth arrest in many species and ecotypes, although signaling pathways mediating temperature-dependent effects are far less known (Heide and Prestrud, 2005; Heide, 2011; Rohde et al., 2011; Cooke et al., 2012). According to Tanino et al. (2010), the existence of independent short photoperiod and low temperature pathways for growth cessation and dormancy induction ensures a higher plasticity for adaptation to changing conditions. DORMANCY-ASSOCIATED MADS-BOX (DAM) genes are known regulators of growth cessation and dormancy induction in many perennial species (Bielenberg et al., 2008; Horvath et al., 2010; Sasaki et al., 2011; Niu et al., 2016; Wu et al., 2017a). Low temperature has been proposed to activate DAM promoters for dormancy induction by direct binding of the cold-dependent C-Repeat Binding Factor (CBF), as confirmed by yeast one-hybrid and transient expression experiments in pear and Japanese apricot (Saito et al., 2015; Niu et al., 2016; Zhao et al., 2018). DAM genes, in turn, have been proposed to directly repress FT in pear (Pyrus pyrifolia) and leafy spurge (Hao et al., 2015; Niu et al., 2016) and activate a 9-cis-epoxycarotenoid dioxygenase gene (PpNCED3) in pear (Tuan et al., 2017), providing specific mechanisms for growth inhibition and abscisic acid (ABA) accumulation in dormant buds. Low temperature mediates a transient increase in ABA content in a photoperiod-independent manner (Welling et al., 2002), however, ABA has been shown to affect dormancy induction instead of growth arrest in hybrid aspen (Tylewicz et al., 2018). Interestingly, winter temperatures and even short treatments at 4°C disrupt the circadian oscillations of CsTOC1 and CsLHY expression in chestnut (Castanea sativa) (Ramos et al., 2005), and chilling treatments alter the expression pattern of GIGANTEA (GI) in almond, a known mediator of circadian effects on flowering in Arabidopsis (Barros et al., 2017). Recently, GI has been postulated to regulate photoperiod-dependent growth cessation in Populus through the activation of FT2, in a protein complex with Flavin-binding, Kelch repeat, F-BOX 1 (FKF1), and CYCLING DOF FACTOR (CDF) gene products (Ding et al., 2018). These and other studies suggest that temperature signals converge with photoperiod on circadian clock elements to modulate seasonal growth cessation (Cooke et al., 2012), although additional effects of temperature on carbon metabolism and hormone signaling have been also proposed to contribute to growth arrest (Wingler, 2015).
Overwintering buds must deal with low and freezing temperatures leading to different forms of physiological and cellular injury. In addition to physical damage caused by ice nucleation and propagation, a dehydration stress is induced by changes in water potential due to the formation of extracellular ice, and the water loss inherent to bud dormancy progress. Plants may actively enhance their tolerance to low temperatures and desiccation via gene expression modification by a cold acclimation process (Wisniewski et al., 2003). Several reviews describe in detail the molecular and genetic control of cold acclimation in trees (Welling and Palva, 2006; Preston and Sandve, 2013; Fennell, 2014; Wisniewski et al., 2014), which is broadly similar to cold acclimation mechanisms reported in herbaceous plants (Thomashow, 1999; Thomashow, 2010; Knight and Knight, 2012).
Seasonal cold acclimation and bud dormancy are related processes since both are induced by similar low temperature and photoperiod conditions (Welling et al., 2002), and both are incompatible with active plant growth, which suggests the presence of common regulatory mechanisms. In fact, impairment of the photoperiodic response by overexpression of PHYA and down-regulation of clock LHY genes reduces the critical daylength for growth cessation and also prevents cold acclimation in hybrid aspen (Olsen et al., 1997; Ibáñez et al., 2010). In addition, the effect of temperature on seasonal growth cessation and cold acclimation invoke the same cold responsive (COR) pathway (Wingler, 2015). However, cold deacclimation and bud dormancy release are not concurrent events; winter buds remain cold-acclimated after dormancy release under appropriate low temperature conditions as long as meristem growth is not resumed, after which deacclimation is not any longer reversible (Kalberer et al., 2006).
In Arabidopsis, MYC transcription factors encoded by INDUCER OF CBF EXPRESSION 1-2 (ICE1-2) are activated by specific cold-dependent post-translational modifications, causing up-regulation of CBF1-3 genes. Subsequently, CBFs regulate most of cold responsive targets by binding to the C-repeat/drought-responsive element (CRT/DRE) (Knight and Knight, 2012). Although COR pathway has been essentially described in Arabidopsis, COR components and functions are conserved in perennials (Fennell, 2014; Wingler, 2015). The ectopic expression of Arabidopsis CBF1 increases freezing tolerance in poplar and induces transcriptomic changes overlapping with Arabidopsis COR regulon (Benedict et al., 2006). On the other side, constitutive expression of birch BpCBF1 increases freezing tolerance and induces known targets of CBF genes in Arabidopsis (Welling and Palva, 2008). Moreover, the ectopic expression of a peach CBF gene in apple induces short-day dependent dormancy, improves freezing tolerance, and delays bud break in field studies (Wisniewski et al., 2011; Artlip et al., 2014). Interestingly, this apple line overexpressing peach CBF causes an altered expression of DAM-like and EBB-like genes in buds, providing an explanation for its prolonged dormancy period through the regulation of key transcription factors involved in dormancy regulation (Wisniewski et al., 2015).
CBF-dependent cold acclimation response includes synthesis of chaperones, dehydrins, and other protective proteins, change in lipid composition of membranes, alteration of sugars metabolism, and production of storage and antioxidant compounds, among other responses aiming at alleviate cold, drought, and oxidative stresses (Welling and Palva, 2006). Dehydrins are abundant cold-responsive proteins belonging to the late embryogenesis abundant (LEA) family that have been proposed to protect cell structures and enzymes against freezing and dehydration (Graether and Boddington, 2014). Seasonal up-regulation of a dehydrin gene in bark tissue is lower and restricted to a shorter period in the evergrowing (evg) mutant of peach having a deletion in DAM genes, in concordance with its lower cold tolerance (Arora et al., 1992; Arora and Wisniewski, 1994; Artlip et al., 1997). Diverse chitinases have been also suggested to act as antifreeze, storage, and defense proteins induced during the transition to dormancy in spruce (González et al., 2015).
Soluble sugars and other compounds potentially able to act as compatible solutes accumulate in dormant tissues in order to confer tolerance to cold and desiccation stresses. Low temperature up-regulates DUAL SPECIFICITY PROTEIN PHOSPHATASE 4 (DSP4), most likely involved in starch dephosphorylation and degradation, to increase the synthesis of oligosaccharides during winter dormancy in chestnut (Berrocal-Lobo et al., 2011). Raffinose family oligosaccharides (RFOs) including raffinose and stachyose are compatible solutes synthesized in seeds and plant tissues undergoing abiotic stresses (Sengupta et al., 2015). Genes coding for the enzyme galactinol synthase (GolS) catalyzing the first step in the synthesis of RFOs are up-regulated in dormant buds and other tissues of woody perennials (Ko et al., 2011; Ibáñez et al., 2013), and apple MdGolS2 gene confers tolerance to water deficit when expressed in Arabidopsis (Falavigna et al., 2018).
Epigenetic mechanisms have been postulated to participate in the control of both, bud phenology and cold acclimation traits. In Norway spruce, the environmental temperature during embryogenesis and seed maturation affects the duration and intensity of bud dormancy and cold acclimation in the progeny, by an “epigenetic memory” process (Johnsen et al., 2005). This epigenetic mechanism has been proposed to modify the expression of certain microRNAs and genes related to bud break, such as EBB1, leading to different epitypes with the same genotype (Yakovlev et al., 2010, 2011; Carneros et al., 2017).
The popular view of bud dormancy as a dynamic state of meristems moving between phases with varying depth during the low temperature period (Cooke et al., 2012) suggests that environmental and intrinsic signals are constantly interacting to determine such dormancy state, like a trade-off between cell growth and quiescence factors. For that reason, cell cycle and expansion pathways are hypothetical targets of those factors modulating dormancy induction and maintenance. In fact, Horvath et al. (2003) have brilliantly reviewed bud dormancy regulation under the perspective of cell cycle regulation and phytohormones action, substantiated on transcriptomic studies and the Arabidopsis model. Few more recent functional studies have provided new insights into that picture. AINTEGUMENTALIKE1-4 (AIL1-AIL4) genes code for transcription factors of the AP2 family in hybrid aspen that mediate short-day dependent growth cessation (Karlberg et al., 2011). This effect on growth has been explained by direct regulation of cell cycle since AIL1 protein interacts with the promoter of CYCD3.2, and AIL1 overexpression prevents down-regulation of D-type cyclins under short-day treatment. In addition, AIL genes are proposed to act downstream of FT and AP1 genes in the photoperiodic pathway (Karlberg et al., 2011; Azeez et al., 2014), providing an interesting link of photoperiod perception with cell cycle control. More evidences of this link have been obtained from the study of cell cycle genes promoters. Populus plants transformed with the promoters of Arabidopsis cell cycle genes CYC1 and CDC2a fused to a reporter gene have shown that both promoters respond to release of apical dominance by shoot decapitation, and CYC1 promoter activity associates with daylength (Rohde et al., 1997).
A recent reappraisal of public genomic data from buds of Arabidopsis, grapevine, and Populus undergoing the growth to dormancy transition has found a common regulatory network that resembles the low energy syndrome (LES), a response triggered under carbon starvation and energy limiting conditions (Tarancón et al., 2017). LES is mediated by Sucrose Non-Fermenting-1-Related Protein Kinase (SnRK1), which ultimately results in cell division arrest and metabolic reprogramming (Martín-Fontecha et al., 2018). This idea is in remarkable agreement with the indirect theory of apical dominance postulating that stem growth inhibits axillary bud outgrowth by diverting sugars away from buds (Mason et al., 2014; Kebrom, 2017).
Also plant hormones play an important role in LES and the growth-dormancy trade-off, with gibberellins (GAs) and auxins acting as promoters of cell growth, whereas ABA associates with dormancy maintenance. GA content in Prunus mume changes across bud dormancy phases, in concordance with the expression of biosynthetic GA20ox genes (Wen et al., 2016). Moreover, application of exogenous active GA increases bud break in Prunus mume (Zhuang et al., 2013), and induces shoot elongation under short-days in Salix pentandra (Junttila and Jensen, 1988). A set of transgenic Populus plants with altered GA metabolism and signaling show faster growth cessation in response to short photoperiod, early bud set and delayed bud break as compared with the wild type (Zawaski et al., 2011; Zawaski and Busov, 2014). On the contrary, hybrid aspen plants with increased GA concentration by overexpression of AtGA20ox1 continue to grow under short-day conditions (Eriksson et al., 2015). In addition to its role in apical dominance/paradormancy induction, gene expression studies associate auxin signaling with bud dormancy release and growth resumption (Anderson et al., 2005; El Kayal et al., 2011; Noriega and Pérez, 2017). On the other hand, modification of ABA signaling by overexpression and down-regulation of a poplar ortholog of ABA INSENSITIVE 3 (ABI3) alters bud formation in response to short-days (Rohde et al., 2002; Ruttink et al., 2007). Interestingly, ABI3 protein interacts with FLOWERING LOCUS D 1 (FDL1), pointing to an orchestrated control of bud development by photoperiodic and ABA pathways (Tylewicz et al., 2015; Singh et al., 2017). In addition, hybrid aspen plants with a reduced ABA response by expressing the dominant allele abi1-1 of the ABA signaling gene ABI1 show growth cessation and form buds under short photoperiod, but remain in a non-dormant state, arguing for a specific effect of ABA on dormancy induction (Tylewicz et al., 2018). In grapevine, ABA has been postulated to affect bud dormancy development through the modulation of the expression of cell cycle genes (Vergara et al., 2017). Similarly to ABI3 overexpressing lines, birch (Betula pendula) plants made insensitive to ethylene by expressing the dominant mutation etr1-1 of the ethylene receptor ETR1 show alterations in bud formation (Ruonala et al., 2006). In the same study, etr1-1 plants fail to accumulate ABA in response to short-days, which suggests an interplay of both hormones in bud development mechanisms. The role of these hormones in bud dormancy pathways have been largely supported by transcriptomic studies in different species (Bai et al., 2013; Doğramaci et al., 2013; Howe et al., 2015) and metabolic profiling (Chao et al., 2016).
Bud dormancy release integrates cumulative chilling perception into a molecular calendar mechanism that triggers growth resumption after fulfillment of the specific chilling requirements of a given species or genotype. Our current knowledge about both seasonal temperature sensing and calendar mechanisms is still scarce and fragmentary, although some common and specific elements of these regulatory circuits have been already described in different species. FT gene and the growth-promoting hormones GAs have been postulated as main factors leading to bud dormancy release in trees (Brunner et al., 2014; Maurya and Bhalerao, 2017; Singh et al., 2017). Exposure to seasonal low temperature leads to up-regulation of GA biosynthetic genes and down-regulation of GA catabolic genes during the dormancy-activity transition in hybrid aspen (Karlberg et al., 2010), and up-regulation of FT1 gene in poplar (Hsu et al., 2011; Rinne et al., 2011). Furthermore, seasonal chilling induces the GA-dependent expression of 1,3-β-D-glucanase genes, involved in removal of callose sphincters on plasmodesmata and the subsequent reopening of cell-to-cell communication in meristematic cells (Rinne et al., 2001; Rinne et al., 2011). The key role of cell-to-cell communication closure in dormancy induction has been recently confirmed by studying hybrid aspen plants with altered sensitivity to ABA (Tylewicz et al., 2018). Mobile peptides such as FT and CENTRORADIALIS (CEN) are possible candidates moving through those open plasmodesmata to control cell proliferation (Rinne et al., 2011; Tylewicz et al., 2018). Poplar transgenic plants overexpressing CEN1 gene require an extended chilling time for bud break (Mohamed et al., 2010), suggesting that CEN1 counteracts the flowering promoting effect of FT1 gene, and that relative levels of FT1 and CEN1 could determine dormancy release (Brunner et al., 2014). Similar results have been obtained in kiwifruit (Varkonyi-Gasic et al., 2013).
Besides, the EARLY BUD-BREAK 1 (EBB1) gene codes for an AP2 type transcription factor that has been associated with bud break events in different species. EBB1 has been identified as the tagged gene in a dominant mutant of poplar showing early bud break, whereas down-regulation of EBB1 delays bud break (Yordanov et al., 2014). EBB1 sequence and expression profile is conserved in other perennials, which suggests its positive participation in bud break across a wide range of tree species (Busov et al., 2016). In fact, Japanese pear PpEBB gene is up-regulated during the rapid enlargement stage in ecodormant buds prior to bud break events and induces the expression of several cyclin PpCYCD3 genes in transient expression assays, providing a link with cell division mechanisms required for bud break and blooming (Tuan et al., 2016).
In Rosaceae species and leafy spurge (Euphorbia esula), DAM genes are also considered major chilling-dependent regulators of bud dormancy, and thus are also putative components of their respective molecular calendars. The evg mutant of peach, showing a non-dormant phenotype, contains a partial deletion of a tandemly repeated family of DAM genes (Bielenberg et al., 2008). DAM genes are specifically expressed in dormant vegetative and reproductive buds, and down-regulated concomitantly with dormancy release events, although several DAM family members show gene expression particularities (Li et al., 2009; Jiménez et al., 2010b; Yamane et al., 2011; Kitamura et al., 2016). Other MADS-box domain genes (i.e., FLOWERING LOCUS C-like and SHORT VEGETATIVE PHASE-like) have been related to chilling requirements and dormancy release in apple (Malus × domestica) and kiwifruit (Actinidia deliciosa) among other species (Porto et al., 2015; Wu et al., 2017a,b).
Resembling the vernalization-dependent flowering in Arabidopsis, epigenetic modifications including chromatin histone methylation and acetylation, DNA methylation and small RNA regulation have been postulated to mediate chilling dependent release of dormancy (Horvath et al., 2003; Ríos et al., 2014). Concomitantly with cold accumulation and gene down-regulation, the chromatin in regulatory regions of DAM genes in leafy spurge and peach show a decrease in trimethylation of histone H3 at lysine 4 (H3K4me3) and an increase of trimethylated H3 at lysine 27 (H3K27me3), which are modifications associated with gene repression and silencing (Horvath et al., 2010; Leida et al., 2012a). However, additional functional approaches are required in order to state a role of these chromatin marks in DAM-dependent regulation of dormancy release by chilling. Down-regulation of the chromodomain/helicase/DNA-binding domain (CHD3) PICKLE, a known antagonist of H3K27me3 modification in Arabidopsis (Aichinger et al., 2009), restores plasmodesmata closure and photoperiod-dependent bud dormancy in ABA response defective plants, suggesting that ABA promotes bud dormancy by repressing PICKLE (Tylewicz et al., 2018). Also, methylation of DNA affects chromatin structure and gene-specific expression, and thus it may potentially account for large transcriptomic rearrangements observed in developmental transitions. In effect, global and specific levels of genomic DNA cytosine methylation change during bud development in chestnut (Santamaría et al., 2009) and apple (Kumar et al., 2016), and recent functional studies reveal the important role of DNA methylation enzymes in seasonal dormancy regulation: overexpression of a chestnut DEMETER-like (CsDML) DNA demethylase accelerates photoperiodic-dependent bud formation (Conde et al., 2017b), whereas down-regulation of poplar DEMETER-like (PtaDML10) delays bud break (Conde et al., 2017a) in poplar. In sweet cherry (Prunus avium), specific DNA methylations and siRNAs are associated with silencing of the DAM-like gene PavMADS1 during dormancy release (Rothkegel et al., 2017). Modification of transcript stability by microRNA action has been also hypothesized to participate in bud dormancy regulation. The aspen microRNA ptr-MIR169 represses the expression of Heme Activator Protein 2 (ptrHAP2) in dormant buds (Potkar et al., 2013), a component of nuclear factor Y (NF-Y) complexes involved in regulation of flowering in Arabidopsis by modulating the epigenetic state of target genes (Hou et al., 2014), which provides a potential way for regulation of FT. In pear, DAM transcripts are targeted and degraded by miR6390 microRNA, thus contributing to DAM down-regulation in the bud dormancy release transition (Niu et al., 2016).
Flowering pathways and genes are broadly conserved between herbaceous and perennial plants, in spite of their evident phenological particularities. In perennials, a period of seasonal dormancy usually interposes between flower induction and blooming (Boes and Strauss, 1994; Kurokura et al., 2013), which forces the mutual coordination of flowering, dormancy and cold acclimation processes. Under these circumstances, pre-existing components of flowering pathways have apparently evolved to acquire new functionalities adapted to the growth of perennials in temperate climates. The proposed functions of FT1 in flower induction and dormancy release and FT2 in the regulation of photoperiodic growth cessation in poplar constitute a paradigmatic case of neo-functionalization after a gene duplication event in trees, in contrast to the main role of FT in the transition to flowering in Arabidopsis (Pin and Nilsson, 2012). A role for FT and the similar but functionally antagonist TFL genes in flower induction has been also postulated in other perennial species different from poplar, based on expression and transgenic studies (Kotoda and Wada, 2005; Jones et al., 2011; Ziv et al., 2014; Bai et al., 2017; Reig et al., 2017). Similarly, orthologs of Arabidopsis flowering genes LEAFY (LFY) and AP1 perform a function related to flowering transition in perennial species. LFY-like genes from trees are preferentially expressed during flower induction and accelerate flowering when ectopically expressed in Arabidopsis, however no evidences of their flowering promoting effect have been observed when overexpressed in poplar (Rottmann et al., 2000). On the contrary, RNAi of PtLFY induces sterility and delays bud break in poplar (Klocko et al., 2016). On the other hand, a dominant negative mutation of AP1 from Arabidopsis modifies the regulation of flowering related genes in poplar (Chen et al., 2015), and overexpression of AP1-like gene from Salix integra induces early flowering in haploid poplar (Yang et al., 2018). In addition to homologs of known flowering genes, miRNAs and hormone signaling pathways have been proposed to integrate developmental and environmental cues affecting flower induction (Xing et al., 2015; Guo et al., 2017).
The reproductive development in perennials is closely associated with phenology. Following flower induction, reproductive organs differentiate and continue growing until a given developmental stage is reached before the dormancy period. In peach and apricot, dormant anthers are arrested in the form of sporogenous tissue (Julian et al., 2011; Ríos et al., 2013). Then after dormancy release, pollen mother cells undergo meiosis followed by pollen development and maturation, and ovaries start to form ovules (Luna et al., 1990; Julian et al., 2011). The harmful effect of cold and other environmental stresses on microsporogenesis, leading to ploidy alterations in male gametes and sterility (De Storme and Geelen, 2014), suggests that dormancy arrest in a pre-meiosis stage may serve to ensure a proper production of male gametes under more favorable environmental conditions.
Over the last few years, a pleiad of transcriptomic studies have provided abundant data about gene expression across bud development in white spruce (El Kayal et al., 2011), poplar (Ruttink et al., 2007), oak (Ueno et al., 2013), raspberry (Mazzitelli et al., 2007), apple (Falavigna et al., 2014), pear (Bai et al., 2013), Japanese apricot (Habu et al., 2014), peach (Jiménez et al., 2010a), Vitis riparia (Mathiason et al., 2009), and leafy spurge (Horvath et al., 2008) among other perennial species. In our laboratory, we have initiated a transcriptomic approach using flower buds of peach at different dormancy stages and cultivars with different chilling requirements (Leida et al., 2010). The systematic study of differentially expressed transcripts identified in this study has provided a dynamic snapshot of biological processes taking place in a flower bud across dormancy release, including regulation of dormancy release, tolerance to abiotic stresses and flower development (Figure 1).
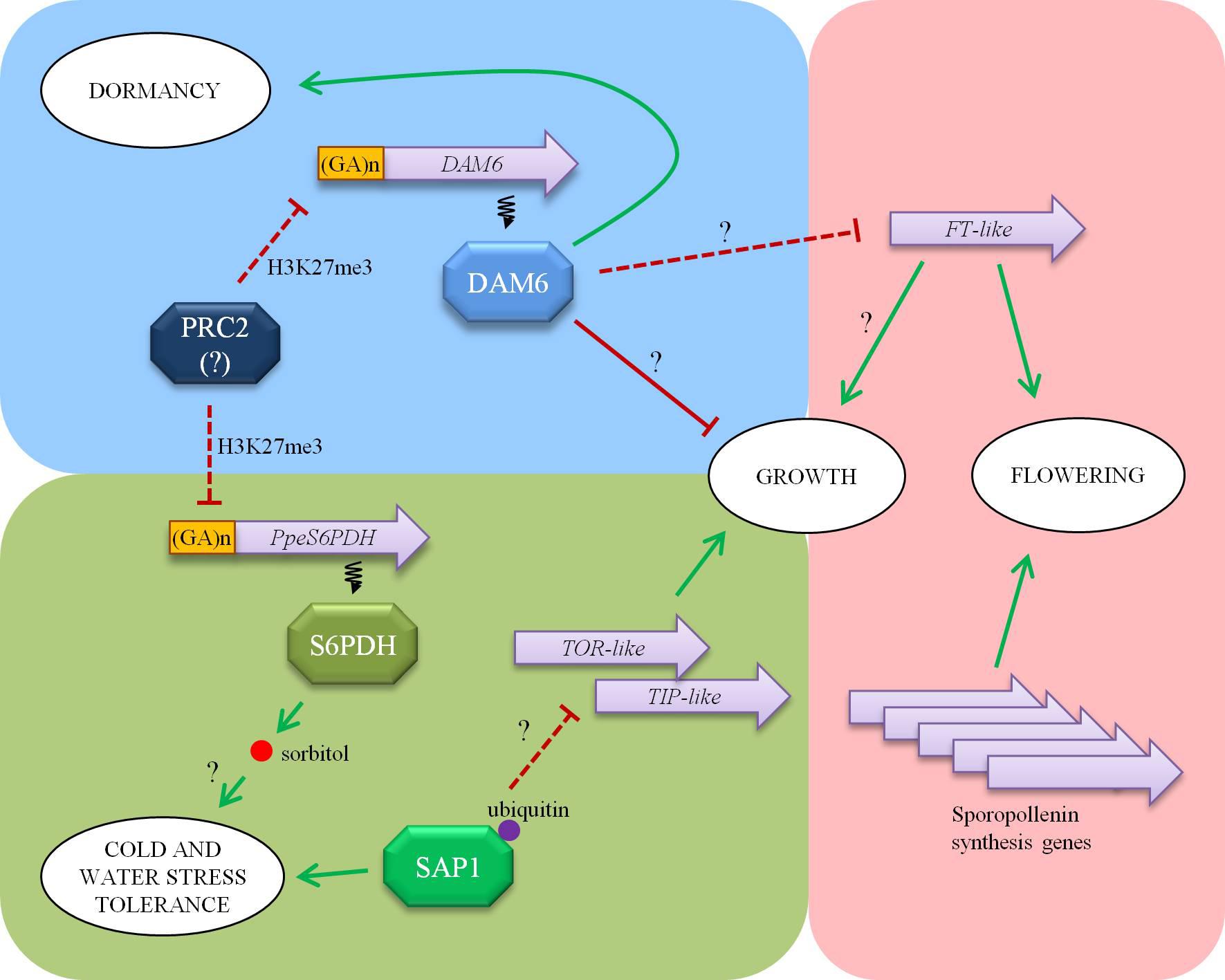
FIGURE 1. An overview of main processes converging in a flower bud of peach: dormancy (blue background), stress tolerance (green), and flowering (pink). Dormancy-Associated MADS-box 6 (DAM6) favors dormancy maintenance while repressing growth resumption and flowering. Sorbitol-6-phosphate dehydrogenase (S6PDH) enzyme synthesizes sorbitol in dormant buds, performing a putative role as cryoprotectant and compatible solute. Polycomb Repressive Complex 2 (PRC2) has been proposed to repress DAM6 and PpeS6PDH genes by H3K27me3 modification of a chromatin stretch enriched in GA repeats. Stress associated protein 1 (SAP1) is a regulatory protein with ubiquitin binding ability that modulates water loss and cell growth. This last function could be partially mediated by down-regulation of cell growth regulatory (TOR-like) and vacuolar aquaporin (TIP-like) genes. FLOWERING LOCUS T (FT)-like is a main regulator of growth and flowering processes, and hypothetically integrates environmental and physiological signals. Other flowering related genes, such as the ones involved in sporopollenin biosynthesis and pollen maturation are also linked to dormancy release and growth resumption events. Arrows indicate genetic or biochemical activation while lines ending in a bar indicate repression. Transcriptional interactions are depicted with discontinuous lines. Question marks tag the relations that have not been confirmed yet.
Dormancy-associated genes DAM1, 4, 5, and 6, belonging to the family of 6 tandemly arrayed DAM genes, have been found down-regulated in flower buds of peach following dormancy release, and differentially expressed in cultivars with different chilling requirements (Leida et al., 2010, 2012a). These genes share a common chromatin modification involving H3K27me3 enrichment after dormancy release (de la Fuente et al., 2015), suggesting thus a putative mechanism for gene silencing similar to the epigenetic regulation of the vernalization-responsive FLC gene in Arabidopsis (Ríos et al., 2014). H3K27me3 epigenetic mark in peach buds is associated with genomic (GA)n repeats, in concordance with the role of (GA)n binding proteins in recruiting the Polycomb repressive complex 2 (PRC2) involved in trimethylation at H3K27 in Arabidopsis (Xiao et al., 2017). FT-like gene is up-regulated in dormancy-released buds in peach, following an opposite pattern to DAM6 (Leida et al., 2012b), which resembles down-regulation of FT by DAM genes found in other species (Hao et al., 2015), and provides a mechanism by which DAM genes might mediate growth and dormancy responses.
In addition to DAM, many other genes are differentially enriched in the H3K27me3 mark in buds undergoing dormancy release. Among them, PpeS6PDH codifies a sorbitol-6-phosphate dehydrogenase involved in sorbitol synthesis that is expressed in dormant buds and down-regulated in dormancy released buds concomitantly with an increase in H3K27me3 modification (Lloret et al., 2017b). This correlates with sorbitol accumulation in dormant buds, and has prompted us to postulate a role of PpeS6PDH and sorbitol in protection against cold and hydric stresses (Figure 1). In that case, it would mean that bud dormancy and stress tolerance share common regulatory epigenetic mechanisms, which are apparently independent from the well known cold acclimation pathway since the H3K27me3 mark is concurrently established in DAM6 and PpeS6PDH, linked to the dormancy stage instead of the environmental temperature.
The study of gene expression in flower buds also has served to identify a stress associated protein (SAP)-like gene (PpSAP1) expressed in dormant buds and down-regulated concomitantly with dormancy release (Lloret et al., 2017a). SAP-like proteins containing Zn-finger domains A20 and AN1 have been found to regulate the abiotic stress response in different especies (Giri et al., 2013), most likely by an ubiquitin-related mechanism. The ectopic expression of PpSAP1 in plum alters water loss and leaf morphology, suggesting that has a dual role in stress tolerance and cell growth (Figure 1). This effect on cell growth could be mediated by down-regulation of target of rapamycin (TOR)-like, a key regulator of cell growth and metabolism in eukaryotic cells, and tonoplast intrinsic protein (TIP)-like, a tonoplast aquaporin affecting water permeability and cell turgor. This makes tempting to speculate that PpSAP1 might coordinate both growth inhibition and stress tolerance in dormant buds.
Finally, we have identified several genes transiently up-regulated after dormancy release that are specifically expressed in anthers (Ríos et al., 2013). Among these genes we have found some orthologs of Arabidopsis genes involved in synthesis of sporopollenin (a pollen cell wall component) and pollen maturation, which provides a molecular framework to characterize the mechanisms acting in growth resumption of reproductive organs and microsporogenesis initiated shortly after dormancy release. Altogether, the data obtained in these studies has contributed to outline a landscape of concerted cross-regulation of dormancy, stress response and flowering processes converging in a flower bud.
In trees from temperate climates, dormancy is a process required for survival during winter, but the molecular pathways that regulate it are poorly known. A better understanding of the molecular bases of bud dormancy will strongly facilitate plant breeding tasks aimed at assessing the potential for environmental adaptability of particular genotypes, and studies led to evaluate the impact of climate change on crop yields. In our opinion, as a result of our experience in the peach model, this will be better achieved by approaches involving the coordinate study of dormancy, flowering and stress pathways in buds, hence providing an added value to the molecular characterization of these processes separately. Moreover, integrating approaches will help to identify common regulatory mechanisms, thus contributing to decipher the time and spatial fitting of these processes.
According to the profuse literature mentioned in this review, numerous environmental inputs transmitting temperature and light data are found in different nodes of regulatory networks, ensuring a precise tuning of phenological transitions. On the other side, quantitative and delayed responses seem to be mediated by the epigenetic machinery, which employs common chromatin labels for dormancy release, stress acclimation and flowering induction in different species. In our opinion, epigenetic modifiers will become central to most molecular dormancy studies in the immediate future, with an increasingly important impact on development and environmental adaptation fields.
AL, MB, and GR wrote the manuscript. All the authors read and approved the final version of the manuscript.
This work was funded by the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA)-FEDER (RF2013-00043-C02-02) and the Ministry of Science and Innovation of Spain (AGL2010-20595).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aichinger, E., Villar, C. B., Farrona, S., Reyes, J. C., Hennig, L., and Köhler, C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:e1000605. doi: 10.1371/journal.pgen.1000605
Allona, I., Ramos, A., Ibáñez, C., Contreras, A., Casado, R., and Aragoncillo, C. (2008). Molecular control of winter dormancy establishment in trees. Span. J. Agric. Res. 6, 201–210. doi: 10.5424/sjar/200806S1-389
Anderson, J. V., Gesch, R. W., Jia, Y., Chao, W. S., and Horvath, D. P. (2005). Seasonal shifts in dormancy status, carbohydrate metabolism, and related gene expression in crown buds of leafy spurge. Plant Cell Environ. 28, 1567–1578. doi: 10.1111/j.1365-3040.2005.01393.x
Anderson, J. V., Horvath, D. P., Chao, W. S., and Foley, M. E. (2010). “Bud dormancy in perennial plants: A mechanism for survival,” in Dormancy and Resistance in Harsh Environments, eds E. Lubzens, J. Cerda, and M. Clark (Berlin: Springer), 69–90. doi: 10.1007/978-3-642-12422-8_5
Arora, R., and Wisniewski, M. E. (1994). Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica [L.] Batsch). II. A 60-kilodalton bark protein in cold-acclimated tissues of peach is heat stable and related to the dehydrin family of proteins. Plant Physiol. 105, 95–101. doi: 10.1104/pp.105.1.95
Arora, R., Wisniewski, M. E., and Scorza, R. (1992). Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica [L.] Batsch): I. seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiol. 99, 1562–1568. doi: 10.1104/pp.99.4.1562
Artlip, T. S., Callahan, A. M., Bassett, C. L., and Wisniewski, M. E. (1997). Seasonal expression of a dehydrin gene in sibling deciduous and evergreen genotypes of peach (Prunus persica [L.] Batsch). Plant Mol. Biol. 33, 61–70. doi: 10.1023/A:1005787909506
Artlip, T. S., Wisniewski, M. E., and Norelli, J. L. (2014). Field evaluation of apple overexpressing a peach CBF gene confirms its effect on cold hardiness, dormancy, and growth. Environ. Exp. Bot. 106, 79–86. doi: 10.1016/j.envexpbot.2013.12.008
Azeez, A., Miskolczi, P., Tylewicz, S., and Bhalerao, R. P. (2014). A tree ortholog of APETALA1 mediates photoperiodic control of seasonal growth. Curr. Biol. 24, 717–724. doi: 10.1016/j.cub.2014.02.037
Bai, S., Saito, T., Sakamoto, D., Ito, A., Fujii, H., and Moriguchi, T. (2013). Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 54, 1132–1151. doi: 10.1093/pcp/pct067
Bai, S., Tuan, P. A., Saito, T., Ito, A., Ubi, B. E., Ban, Y., et al. (2017). Repression of TERMINAL FLOWER1 primarily mediates floral induction in pear (Pyrus pyrifolia Nakai) concomitant with change in gene expression of plant hormone-related genes and transcription factors. J. Exp. Bot. 68, 4899–4914. doi: 10.1093/jxb/erx296
Barros, P. M., Cherian, S., Costa, M., Sapeta, H., Saibo, N. J. M., and Oliveira, M. M. (2017). The identification of almond GIGANTEA gene and its expression under cold stress, variable photoperiod, and seasonal dormancy. Biol. Plant. 61, 631–640. doi: 10.1007/s10535-017-0711-1
Benedict, C., Skinner, J. S., Meng, R., Chang, Y., Bhalerao, R., Huner, N. P., et al. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x
Berrocal-Lobo, M., Ibañez, C., Acebo, P., Ramos, A., Perez-Solis, E., Collada, C., et al. (2011). Identification of a homolog of Arabidopsis DSP4 (SEX4) in chestnut: its induction and accumulation in stem amyloplasts during winter or in response to the cold. Plant Cell Environ. 34, 1693–1704. doi: 10.1111/j.1365-3040.2011.02365.x
Bielenberg, D. G., Wang, Y., Li, Z., Zhebentyayeva, T., Fan, S., Reighard, G. L., et al. (2008). Sequencing and annotation of the evergrowing locus in peach (Prunus persica [L.] Batsch) reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet. Genomes 4, 495–507. doi: 10.1007/s11295-007-0126-9
Boes, T. K., and Strauss, S. H. (1994). Floral phenology and morphology of black cottonwood, Populus trichocarpa (Salicaceae). Am. J. Bot. 81, 562–567. doi: 10.2307/2445730
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Brunner, A. M., Evans, L. M., Hsu, C. Y., and Sheng, X. (2014). Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front. Plant Sci. 5:732. doi: 10.3389/fpls.2014.00732
Busov, V., Carneros, E., and Yakovlev, I. (2016). EARLY BUD-BREAK1 (EBB1) defines a conserved mechanism for control of bud-break in woody perennials. Plant Signal. Behav. 11:e1073873. doi: 10.1080/15592324.2015.1073873
Carneros, E., Yakovlev, I., Viejo, M., Olsen, J. E., and Fossdal, C. G. (2017). The epigenetic memory of temperature during embryogenesis modifies the expression of bud burst-related genes in Norway spruce epitypes. Planta 246, 553–566. doi: 10.1007/s00425-017-2713-9
Chao, W. S., Doğramaci, M., Horvath, D. P., Anderson, J. V., and Foley, M. E. (2016). Phytohormone balance and stress-related cellular responses are involved in the transition from bud to shoot growth in leafy spurge. BMC Plant Biol. 16:47. doi: 10.1186/s12870-016-0735-2
Chao, W. S., Doğramaci, M., Horvath, D. P., Anderson, J. V., and Foley, M. E. (2017). Comparison of phytohormone levels and transcript profiles during seasonal dormancy transitions in underground adventitious buds of leafy spurge. Plant Mol. Biol. 94, 281–302. doi: 10.1007/s11103-017-0607-7
Chen, Z., Ye, M., Su, X., Liao, W., Ma, H., Gao, K., et al. (2015). Overexpression of AtAP1M3 regulates flowering time and floral development in Arabidopsis and effects key flowering-related genes in poplar. Transgenic Res. 24, 705–715. doi: 10.1007/s11248-015-9870-z
Conde, D., Le Gac, A. L., Perales, M., Dervinis, C., Kirst, M., Maury, S., et al. (2017a). Chilling-responsive DEMETER-LIKE DNA demethylase mediates in poplar bud break. Plant Cell Environ. 40, 2236–2249. doi: 10.1111/pce.13019
Conde, D., Moreno-Cortés, A., Dervinis, C., Ramos-Sánchez, J. M., Kirst, M., Perales, M., et al. (2017b). Overexpression of DEMETER, a DNA demethylase, promotes early apical bud maturation in poplar. Plant Cell Environ. 40, 2806–2819. doi: 10.1111/pce.13056
Cooke, J. E. K., Eriksson, M. E., and Junttila, O. (2012). The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 35, 1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x
Couvillon, G. A., and Erez, A. (1985). Influence of prolonged exposure to chilling temperatures on bud break and heat requirement for bloom of several fruit species. J. Am. Soc. Hort. Sci. 110, 47–50.
Coville, F. V. (1920). The influence of cold in stimulating the growth of plants. Proc. Natl. Acad. Sci. U.S.A. 6, 434–435. doi: 10.1073/pnas.6.7.434
de la Fuente, L., Conesa, A., Lloret, A., Badenes, M. L., and Ríos, G. (2015). Genome-wide changes in histone H3 lysine 27 trimethylation associated with bud dormancy release in peach. Tree Genet. Genomes 11:45. doi: 10.1007/s11295-015-0869-7
De Storme, N., and Geelen, D. (2014). The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant Cell Environ. 37, 1–18. doi: 10.1111/pce.12142
Ding, J., Böhlenius, H., Rühl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). GIGANTEA-like genes control seasonal growth cessation in Populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Doğramaci, M., Foley, M. E., Chao, W. S., Christoffers, M. J., and Anderson, J. V. (2013). Induction of endodormancy in crown buds of leafy spurge (Euphorbia esula L.) implicates a role for ethylene and cross-talk between photoperiod and temperature. Plant Mol. Biol. 81, 577–593. doi: 10.1007/s11103-013-0026-3
El Kayal, W., Allen, C. C., Ju, C. J., Adams, E., King-Jones, S., Zaharia, L. I., et al. (2011). Molecular events of apical bud formation in white spruce. Picea glauca. Plant Cell Environ. 34, 480–500. doi: 10.1111/j.1365-3040.2010.02257.x
Eriksson, M. E., Hoffman, D., Kaduk, M., Mauriat, M., and Moritz, T. (2015). Transgenic hybrid aspen trees with increased gibberellin (GA) concentrations suggest that GA acts in parallel with FLOWERING LOCUS T2 to control shoot elongation. New Phytol. 205, 1288–1295. doi: 10.1111/nph.13144
Falavigna, V. S., Porto, D. D., Buffon, V., Margis-Pinheiro, M., Pasquali, G., and Revers, L. F. (2014). Differential transcriptional profiles of dormancy-related genes in apple buds. Plant Mol. Biol. Rep. 32, 796–813. doi: 10.1007/s11105-013-0690-0
Falavigna, V. S., Porto, D. D., Miotto, Y. E., Santos, H. P. D., Oliveira, P. R. D., Margis-Pinheiro, M., et al. (2018). Evolutionary diversification of galactinol synthases in Rosaceae: adaptive roles of galactinol and raffinose during apple bud dormancy. J. Exp. Bot 69, 1247–1259. doi: 10.1093/jxb/erx451
Fennell, A. (2014). Genomics and functional genomics of winter low temperature tolerance in temperate fruit crops. Crit. Rev. Plant Sci. 33, 125–140. doi: 10.1080/07352689.2014.870410
Giri, J., Dansana, P. K., Kothari, K. S., Sharma, G., Vij, S., and Tyagi, A. K. (2013). SAPs as novel regulators of abiotic stress response in plants. Bioessays 35, 639–648. doi: 10.1002/bies.201200181
González, L. M. G., El Kayal, W., Morris, J. S., and Cooke, J. E. K. (2015). Diverse chitinases are invoked during the activity-dormancy transition in spruce. Tree Genet. Genomes 11:41. doi: 10.1007/s11295-015-0871-0
Graether, S. P., and Boddington, K. F. (2014). Disorder and function: a review of the dehydrin protein family. Front. Plant Sci. 5:576. doi: 10.3389/fpls.2014.00576
Guo, X., Ma, Z., Zhang, Z., Cheng, L., Zhang, X., and Li, T. (2017). Small RNA-sequencing links physiological changes and RdDM process to vegetative-to-floral transition in apple. Front. Plant Sci. 8:873. doi: 10.3389/fpls.2017.00873
Habu, T., Yamane, H., Sasaki, R., Yano, K., Fujii, H., Shimizu, T., et al. (2014). Custom microarray analysis for transcript profiling of dormant vegetative buds of Japanese apricot during prolonged chilling exposure. J. Jpn. Soc. Hortic. Sci. 83, 1–16. doi: 10.2503/jjshs1.CH-077
Hao, X., Chao, W., Yang, Y., and Horvath, D. (2015). Coordinated expression of FLOWERING LOCUS T and DORMANCY ASSOCIATED MADS-BOX-like genes in leafy spurge. PLoS One 10:e0126030. doi: 10.1371/journal.pone.0126030
Heide, O. M. (2011). Temperature rather than photoperiod controls growth cessation and dormancy in Sorbus species. J. Exp. Bot. 62, 5397–5404. doi: 10.1093/jxb/err213
Heide, O. M., and Prestrud, A. K. (2005). Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 25, 109–114. doi: 10.1093/treephys/25.1.109
Horvath, D. P., Anderson, J. V., Chao, W. S., and Foley, M. E. (2003). Knowing when to grow: signals regulating bud dormancy. Trends Plant Sci. 8, 534–540. doi: 10.1016/j.tplants.2003.09.013
Horvath, D. P., Chao, W. S., Suttle, J. C., Thimmapuram, J., and Anderson, J. V. (2008). Transcriptome analysis identifies novel responses and potential regulatory genes involved in seasonal dormancy transitions of leafy spurge (Euphorbia esula L.). BMC Genomics 9:536. doi: 10.1186/1471-2164-9-536
Horvath, D. P., Sung, S., Kim, D., Chao, W., and Anderson, J. (2010). Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Mol. Biol. 73, 169–179. doi: 10.1007/s11103-009-9596-5
Hou, X., Zhou, J., Liu, C., Liu, L., Shen, L., and Yu, H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5:4601. doi: 10.1038/ncomms5601
Howe, G. T., Horvath, D. P., Dharmawardhana, P., Priest, H. D., Mockler, T. C., and Strauss, S. H. (2015). Extensive transcriptome changes during natural onset and release of vegetative bud dormancy in Populus. Front. Plant Sci. 6:989. doi: 10.3389/fpls.2015.00989
Hsu, C. Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U.S.A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Ibáñez, C., Collada, C., Casado, R., González-Melendi, P., Aragoncillo, C., and Allona, I. (2013). Winter induction of the galactinol synthase gene is associated with endodormancy in chestnut trees. Trees 27, 1309–1316. doi: 10.1007/s00468-013-0879-8
Ibáñez, C., Kozarewa, I., Johansson, M., Ögren, E., Rohde, A., and Eriksson, M. E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153, 1823–1833. doi: 10.1104/pp.110.158220
Jiménez, S., Li, Z., Reighard, G. L., and Bielenberg, D. G. (2010a). Identification of genes associated with growth cessation and bud dormancy entrance using a dormancy-incapable tree mutant. BMC Plant Biol. 10:25. doi: 10.1186/1471-2229-10-25
Jiménez, S., Reighard, G. L., and Bielenberg, D. G. (2010b). Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol. Biol. 73, 157–167. doi: 10.1007/s11103-010-9608-5
Johnsen,Ø., Fossdal, C. G., Nagy, N., Mølmann, J., Dæhlen, O. G., and Skrøppa, T. (2005). Climatic adaptation in Picea abies progenies is affected by the temperature during zygotic embryogenesis and seed maturation. Plant Cell Environ. 28, 1090–1102. doi: 10.1111/j.1365-3040.2005.01356.x
Jones, R. C., Hecht, V. F. G., Potts, B. M., Vaillancourt, R. E., and Weller, J. L. (2011). Expression of a FLOWERING LOCUS T homologue is temporally associated with annual flower bud initiation in Eucalyptus globulus subsp. globulus (Myrtaceae). Aust. J. Bot. 59, 756–769. doi: 10.1071/BT11251
Julian, C., Rodrigo, J., and Herrero, M. (2011). Stamen development and winter dormancy in apricot (Prunus armeniaca). Ann. Bot. 108, 617–625. doi: 10.1093/aob/mcr056
Junttila, O., and Jensen, E. (1988). Gibberellins and photoperiodic control of shoot elongation in Salix. Physiol. Plant. 74, 371–376. doi: 10.1111/j.1399-3054.1988.tb00645.x
Kalberer, S. R., Wisniewski, M., and Arora, R. (2006). Deacclimation and reacclimation of cold-hardy plants: current understanding and emerging concepts. Plant Sci. 171, 3–16. doi: 10.1016/j.plantsci.2006.02.013
Karlberg, A., Bako, L., and Bhalerao, R. P. (2011). Short day-mediated cessation of growth requires the downregulation of AINTEGUMENTALIKE1 transcription factor in hybrid aspen. PLoS Genet. 7:e1002361. doi: 10.1371/journal.pgen.1002361
Karlberg, A., Englund, M., Petterle, A., Molnar, G., Sjödin, A., Bakó, L., et al. (2010). Analysis of global changes in gene expression during activity-dormancy cycle in hybrid aspen apex. Plant Biotechnol. 27, 1–16. doi: 10.5511/plantbiotechnology.27.1
Karlgren, A., Gyllenstrand, N., Clapham, D., and Lagercrantz, U. (2013). FLOWERING LOCUS T/TERMINAL FLOWER1-like genes affect growth rhythm and bud set in Norway spruce. Plant Physiol. 163, 792–803. doi: 10.1104/pp.113.224139
Kebrom, T. H. (2017). A growing stem inhibits bud outgrowth - the overlooked theory of apical dominance. Front. Plant Sci. 8:1874. doi: 10.3389/fpls.2017.01874
Kitamura, Y., Takeuchi, T., Yamane, H., and Tao, R. (2016). Simultaneous down-regulation of DORMANCY-ASSOCIATED MADS-box6 and SOC1 during dormancy release in Japanese apricot (Prunus mume) flower buds. J. Hortic. Sci. Biotechnol. 91, 476–482. doi: 10.1080/14620316.2016.1173524
Klocko, A. L., Brunner, A. M., Huang, J., Meilan, R., Lu, H., Ma, C., et al. (2016). Containment of transgenic trees by suppression of LEAFY. Nat. Biotechnol. 34, 918–922. doi: 10.1038/nbt.3636
Knight, M. R., and Knight, H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195, 737–751. doi: 10.1111/j.1469-8137.2012.04239.x
Ko, J. H., Prassinos, C., Keathley, D., and Han, K. H. (2011). Novel aspects of transcriptional regulation in the winter survival and maintenance mechanism of poplar. Tree Physiol. 31, 208–225. doi: 10.1093/treephys/tpq109
Kotoda, N., and Wada, M. (2005). MdTFL1, a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis. Plant Sci. 168, 95–104. doi: 10.1016/j.plantsci.2004.07.024
Kozarewa, I., Ibáñez, C., Johansson, M., Ögren, E., Mozley, D., Nylander, E., et al. (2010). Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Mol. Biol. 73, 143–156. doi: 10.1007/s11103-010-9619-2
Kumar, G., Rattan, U. K., and Singh, A. K. (2016). Chilling-mediated DNA methylation changes during dormancy and its release reveal the importance of epigenetic regulation during winter dormancy in apple (Malus x domestica Borkh.). PLoS One 11:e0149934. doi: 10.1371/journal.pone.0149934
Kurokura, T., Mimida, N., Battey, N. H., and Hytönen, T. (2013). The regulation of seasonal flowering in the Rosaceae. J. Exp. Bot. 64, 4131–4141. doi: 10.1093/jxb/ert233
Leida, C., Conesa, A., Llácer, G., Badenes, M. L., and Ríos, G. (2012a). Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol. 193, 67–80. doi: 10.1111/j.1469-8137.2011.03863.x
Leida, C., Terol, J., Llácer, G., Badenes, M. L., Ríos, G., Martí, G., et al. (2012b). Gene expression during bud dormancy release in Prunus persica. Acta Hortic. 929, 27–31. doi: 10.17660/ActaHortic.2012.929.2
Leida, C., Terol, J., Martí, G., Agustí, M., Llácer, G., Badenes, M. L., et al. (2010). Identification of genes associated with bud dormancy release in Prunus persica by suppression subtractive hybridization. Tree Physiol. 30, 655–666. doi: 10.1093/treephys/tpq008
Li, Z., Reighard, G. L., Abbott, A. G., and Bielenberg, D. G. (2009). Dormancy-associated MADS genes from the EVG locus of peach (Prunus persica [L.] Batsch) have distinct seasonal and photoperiodic expression patterns. J. Exp. Bot. 60, 3521–3530. doi: 10.1093/jxb/erp195
Lloret, A., Conejero, A., Leida, C., Petri, C., Gil-Muñoz, F., Burgos, L., et al. (2017a). Dual regulation of water retention and cell growth by a stress-associated protein (SAP) gene in Prunus. Sci. Rep. 7, 332. doi: 10.1038/s41598-017-00471-7
Lloret, A., Martínez-Fuentes, A., Agustí, M., Badenes, M. L., and Ríos, G. (2017b). Chromatin-associated regulation of sorbitol synthesis in flower buds of peach. Plant Mol. Biol. 95, 507–517. doi: 10.1007/s11103-017-0669-6
Luna, V., Lorenzo, E., Reinoso, H., Tordable, M. C., Abdala, G., Pharis, R. P., et al. (1990). Dormancy in peach (Prunus persica L.) flower buds: I. Floral morphogenesis and endogenous gibberellins at the end of the dormancy period. Plant Physiol. 93, 20–25. doi: 10.1104/pp.93.1.20
Martín-Fontecha, E. S., Tarancón, C., and Cubas, P. (2018). To grow or not to grow, a power-saving program induced in dormant buds. Curr. Opin. Plant Biol. 41, 102–109. doi: 10.1016/j.pbi.2017.10.001
Mason, M. G., Ross, J. J., Babst, B. A., Wienclaw, B. N., and Beveridge, C. A. (2014). Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. U.S.A. 111, 6092–6097. doi: 10.1073/pnas.1322045111
Mathiason, K., He, D., Grimplet, J., Venkateswari, J., Galbraith, D. W., Or, E., et al. (2009). Transcript profiling in Vitis riparia during chilling requirement fulfillment reveals coordination of gene expression patterns with optimized bud break. Funct. Integr. Genomics 9, 81–96. doi: 10.1007/s10142-008-0090-y
Maurya, J. P., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Mazzitelli, L., Hancock, R. D., Haupt, S., Walker, P. G., Pont, S. D., McNicol, J., et al. (2007). Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J. Exp. Bot. 58, 1035–1045. doi: 10.1093/jxb/erl266
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Niu, Q., Li, J., Cai, D., Qian, M., Jia, H., Bai, S., et al. (2016). Dormancy-associated MADS-box genes and microRNAs jointly control dormancy transition in pear (Pyrus pyrifolia white pear group) flower bud. J. Exp. Bot. 67, 239–257. doi: 10.1093/jxb/erv454
Noriega, X., and Pérez, F. J. (2017). ABA biosynthesis genes are down-regulated while auxin and cytokinin biosynthesis genes are up-regulated during the release of grapevine buds from endodormancy. J. Plant Growth Regul. 36, 814–823. doi: 10.1007/s00344-017-9685-7
Olsen, J. E., Junttila, O., Nilsen, J., Eriksson, M. E., Martinussen, I., Olsson, O., et al. (1997). Ectopic expression of oat phytochrome A in hybrid aspen changes critical day length for growth and prevents cold acclimation. Plant J. 12, 1339–1350. doi: 10.1046/j.1365-313x.1997.12061339.x
Parmentier-Line, C. M., and Coleman, G. D. (2016). Constitutive expression of the Poplar FD-like basic leucine zipper transcription factor alters growth and bud development. Plant Biotechnol. J. 14, 260–270. doi: 10.1111/pbi.12380
Pin, P. A., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Porto, D. D., Bruneau, M., Perini, P., Anzanello, R., Renou, J. P., dos Santos, H. P., et al. (2015). Transcription profiling of the chilling requirement for bud break in apples: a putative role for FLC-like genes. J. Exp. Bot. 66, 2659–2672. doi: 10.1093/jxb/erv061
Potkar, R., Recla, J., and Busov, V. (2013). ptr-MIR169 is a posttranscriptional repressor of PtrHAP2 during vegetative bud dormancy period of aspen (Populus tremuloides) trees. Biochem. Biophys. Res. Commun. 431, 512–518. doi: 10.1016/j.bbrc.2013.01.027
Preston, J. C., and Sandve, S. R. (2013). Adaptation to seasonality and the winter freeze. Front. Plant Sci. 4:167. doi: 10.3389/fpls.2013.00167
Ramos, A., Pérez-Solís, E., Ibáñez, C., Casado, R., Collada, C., Gómez, L., et al. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. U.S.A. 102, 7037–7042. doi: 10.1073/pnas.0408549102
Reig, C., Gil-Muñoz, F., Vera-Sirera, F., García-Lorca, A., Martínez-Fuentes, A., Mesejo, C., et al. (2017). Bud sprouting and floral induction and expression of FT in loquat [Eriobotrya japonica (Thunb.) Lindl.]. Planta 246, 915–925. doi: 10.1007/s00425-017-2740-6
Rinne, P. L., Kaikuranta, P. M., and van der Schoot, C. (2001). The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 26, 249–264. doi: 10.1046/j.1365-313X.2001.01022.x
Rinne, P. L., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjärvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Ríos, G., Leida, C., Conejero, A., and Badenes, M. L. (2014). Epigenetic regulation of bud dormancy events in perennial plants. Front. Plant Sci. 5:247. doi: 10.3389/fpls.2014.00247
Ríos, G., Tadeo, F. R., Leida, C., and Badenes, M. L. (2013). Prediction of components of the sporopollenin synthesis pathway in peach by genomic and expression analyses. BMC Genomics 14:40. doi: 10.1186/1471-2164-14-40
Rohde, A., Bastien, C., and Boerjan, W. (2011). Temperature signals contribute to the timing of photoperiodic growth cessation and bud set in poplar. Tree Physiol. 31, 472–482. doi: 10.1093/treephys/tpr038
Rohde, A., and Bhalerao, R. P. (2007). Plant dormancy in the perennial context. Trends Plant Sci. 12, 217–223. doi: 10.1016/j.tplants.2007.03.012
Rohde, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M., and Boerjan, W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14, 1885–1901. doi: 10.1105/tpc.003186
Rohde, A., Van Montagu, M., Inzé, D., and Boerjan, W. (1997). Factors regulating the expression of cell cycle genes in individual buds of Populus. Planta 201, 43–52. doi: 10.1007/BF01258679
Rothkegel, K., Sánchez, E., Montes, C., Greve, M., Tapia, S., Bravo, S., et al. (2017). DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.). Tree Physiol. 37, 1739–1751. doi: 10.1093/treephys/tpx055
Rottmann, W. H., Meilan, R., Sheppard, L. A., Brunner, A. M., Skinner, J. S., Ma, C., et al. (2000). Diverse effects of overexpression of LEAFY and PTLF, a poplar (Populus) homolog of LEAFY/FLORICAULA, in transgenic poplar and Arabidopsis. Plant J. 22, 235–245. doi: 10.1046/j.1365-313x.2000.00734.x
Ruonala, R., Rinne, P. L., Baghour, M., Moritz, T., Tuominen, H., and Kangasjärvi, J. (2006). Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 46, 628–640. doi: 10.1111/j.1365-313X.2006.02722.x
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. doi: 10.1105/tpc.107.052811
Saito, T., Bai, S., Imai, T., Ito, A., Nakajima, I., and Moriguchi, T. (2015). Histone modification and signalling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ. 38, 1157–1166. doi: 10.1111/pce.12469
Santamaría, M. E., Hasbún, R., Valera, M. J., Meijón, M., Valledor, L., Rodríguez, J. L., et al. (2009). Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J. Plant Physiol. 166, 1360–1369. doi: 10.1016/j.jplph.2009.02.014
Sasaki, R., Yamane, H., Ooka, T., Jotatsu, H., Kitamura, Y., Akagi, T., et al. (2011). Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 157, 485–497. doi: 10.1104/pp.111.181982
Sengupta, S., Mukherjee, S., Basak, P., and Majumder, A. L. (2015). Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6:656. doi: 10.3389/fpls.2015.00656
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Tanino, K. K., Kalcsits, L., Silim, S., Kendall, E., and Gray, G. R. (2010). Temperature-driven plasticity in growth cessation and dormancy development in deciduous woody plants: a working hypothesis suggesting how molecular and cellular function is affected by temperature during dormancy induction. Plant Mol. Biol. 73, 49–65. doi: 10.1007/s11103-010-9610-y
Tarancón, C., González-Grandío, E., Oliveros, J. C., Nicolas, M., and Cubas, P. (2017). A conserved carbon starvation response underlies bud dormancy in woody and herbaceous species. Front. Plant Sci. 8:788. doi: 10.3389/fpls.2017.00788
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Tuan, P. A., Bai, S., Saito, T., Imai, T., Ito, A., and Moriguchi, T. (2016). Involvement of EARLY BUD-BREAK, an AP2/ERF transcription factor gene, in bud break in Japanese pear (Pyrus pyrifolia Nakai) lateral flower buds: expression, histone modifications and possible target genes. Plant Cell Physiol. 57, 1038–1047. doi: 10.1093/pcp/pcw041
Tuan, P. A., Bai, S., Saito, T., Ito, A., and Moriguchi, T. (2017). Dormancy-associated MADS-box (DAM) and the abscisic acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 58, 1378–1390. doi: 10.1093/pcp/pcx074
Tylewicz, S., Petterle, A., Marttila, S., Miskolczi, P., Azeez, A., Singh, R. K., et al. (2018). Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science 360, 212–215. doi: 10.1126/science.aan8576
Tylewicz, S., Tsuji, H., Miskolczi, P., Petterle, A., Azeez, A., Jonsson, K., et al. (2015). Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. U.S.A. 112, 3140–3145. doi: 10.1073/pnas.1423440112
Ueno, S., Klopp, C., Leplé, J. C., Derory, J., Noirot, C., Léger, V., et al. (2013). Transcriptional profiling of bud dormancy induction and release in oak by next-generation sequencing. BMC Genomics 14:236. doi: 10.1186/1471-2164-14-236
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Varkonyi-Gasic, E., Moss, S. M., Voogd, C., Wang, T., Putterill, J., and Hellens, R. P. (2013). Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytol. 198, 732–746. doi: 10.1111/nph.12162
Vergara, R., Noriega, X., Aravena, K., Prieto, H., and Pérez, F. J. (2017). ABA represses the expression of cell cycle genes and may modulate the development of endodormancy in grapevine buds. Front. Plant Sci. 8:812. doi: 10.3389/fpls.2017.00812
Vitasse, Y., Lenz, A., and Körner, C. (2014). The interaction between freezing tolerance and phenology in temperate deciduous trees. Front. Plant Sci. 5:541. doi: 10.3389/fpls.2014.00541
Welling, A., Moritz, T., Palva, E. T., and Junttila, O. (2002). Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 129, 1633–1641. doi: 10.1104/pp.003814
Welling, A., and Palva, E. T. (2006). Molecular control of cold acclimation in trees. Physiol. Plant. 127, 167–181. doi: 10.1111/j.1399-3054.2006.00672.x
Welling, A., and Palva, E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147, 1199–1211. doi: 10.1104/pp.108.117812
Wen, L. H., Zhong, W. J., Huo, X. M., Zhuang, W. B., Ni, Z. J., and Gao, Z. H. (2016). Expression analysis of ABA- and GA-related genes during four stages of bud dormancy in Japanese apricot (Prunus mume Sieb. et Zucc). J. Hortic. Sci. Biotechnol. 91, 362–369. doi: 10.1080/14620316.2016.1160546
Wingler, A. (2015). Comparison of signaling interactions determining annual and perennial plant growth in response to low temperature. Front. Plant Sci. 5:794. doi: 10.3389/fpls.2014.00794
Wisniewski, M., Bassett, C., and Gusta, L. V. (2003). An overview of cold hardiness in woody plants: seeing the forest through the trees. HortScience 38, 952–959.
Wisniewski, M., Nassuth, A., Teulières, C., Marque, C., Rowland, J., Cao, P. B., et al. (2014). Genomics of cold hardiness in woody plants. Crit. Rev. Plant Sci. 33, 92–124. doi: 10.1080/07352689.2014.870408
Wisniewski, M., Norelli, J., and Artlip, T. (2015). Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front. Plant Sci. 6:85. doi: 10.3389/fpls.2015.00085
Wisniewski, M., Norelli, J., Bassett, C., Artlip, T., and Macarisin, D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233, 971–983. doi: 10.1007/s00425-011-1358-3
Wu, R., Tomes, S., Karunairetnam, S., Tustin, S. D., Hellens, R. P., Allan, A. C., et al. (2017a). SVP-like MADS box genes control dormancy and budbreak in apple. Front. Plant Sci. 8:477. doi: 10.3389/fpls.2017.00477
Wu, R., Wang, T., Warren, B. A. W., Allan, A. C., Macknight, R. C., and Varkonyi-Gasic, E. (2017b). Kiwifruit SVP2 gene prevents premature budbreak during dormancy. J. Exp. Bot. 68, 1071–1082. doi: 10.1093/jxb/erx014
Xiao, J., Jin, R., Yu, X., Shen, M., Wagner, J. D., Pai, A., et al. (2017). Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 49, 1546–1552. doi: 10.1038/ng.3937
Xing, L. B., Zhang, D., Li, Y. M., Shen, Y. W., Zhao, C. P., Ma, J. J., et al. (2015). Transcription profiles reveal sugar and hormone signaling pathways mediating flower induction in apple (Malus domestica Borkh.). Plant Cell Physiol. 56, 2052–2068. doi: 10.1093/pcp/pcv124
Yakovlev, I. A., Asante, D. K. A., Fossdal, C. G., Junttila, O., and Johnsen, Ø (2011). Differential gene expression related to an epigenetic memory affecting climatic adaptation in Norway spruce. Plant Sci 180, 132–139. doi: 10.1016/j.plantsci.2010.07.004
Yakovlev, I. A., Fossdal, C. G., and Johnsen, Ø (2010). MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol. 187, 1154–1169. doi: 10.1111/j.1469-8137.2010.03341.x
Yamane, H. (2014). Regulation of bud dormancy and bud break in Japanese apricot (Prunus mume Siebold & Zucc.) and peach [Prunus persica (L.) Batsch]: a summary of recent studies. J. Jpn. Soc. Hort. Sci. 83, 187–202. doi: 10.2503/jjshs1.CH-Rev4
Yamane, H., Ooka, T., Jotatsu, H., Hosaka, Y., Sasaki, R., and Tao, R. (2011). Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. J. Exp. Bot. 62, 3481–3488. doi: 10.1093/jxb/err028
Yang, J., Li, K., Li, C., Li, J., Zhao, B., Zheng, W., et al. (2018). In vitro anther culture and Agrobacterium-mediated transformation of the AP1 gene from Salix integra Linn. in haploid poplar (Populus simonii × P. nigra). J. For. Res. 29, 321–330. doi: 10.1007/s11676-017-0453-0
Yordanov, Y. S., Ma, C., Strauss, S. H., and Busov, V. B. (2014). EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc. Natl. Acad. Sci. U.S.A. 111, 10001–10006. doi: 10.1073/pnas.1405621111
Zawaski, C., and Busov, V. B. (2014). Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PLoS One 9:e86217. doi: 10.1371/journal.pone.0086217
Zawaski, C., Kadmiel, M., Pickens, J., Ma, C., Strauss, S., and Busov, V. (2011). Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 234, 1285–1298. doi: 10.1007/s00425-011-1485-x
Zhao, K., Zhou, Y., Ahmad, S., Yong, X., Xie, X., Han, Y., et al. (2018). PmCBFs synthetically affect PmDAM6 by alternative promoter binding and protein complexes towards the dormancy of bud for Prunus mume. Sci. Rep. 8:4527. doi: 10.1038/s41598-018-22537-w
Zhuang, W., Gao, Z., Wang, L., Zhong, W., Ni, Z., and Zhang, Z. (2013). Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 64, 4953–4966. doi: 10.1093/jxb/ert284
Keywords: bud dormancy, cold acclimation, epigenetic regulation, flowering, peach (Prunus persica)
Citation: Lloret A, Badenes ML and Ríos G (2018) Modulation of Dormancy and Growth Responses in Reproductive Buds of Temperate Trees. Front. Plant Sci. 9:1368. doi: 10.3389/fpls.2018.01368
Received: 24 April 2018; Accepted: 29 August 2018;
Published: 13 September 2018.
Edited by:
Henryk Flachowsky, Julius Kühn-Institut, GermanyReviewed by:
Michael Nicolas, Centro Nacional de Biotecnología (CNB), SpainCopyright © 2018 Lloret, Badenes and Ríos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabino Ríos, cmlvc19nYWJAZ3ZhLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.