
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 September 2018
Sec. Plant Metabolism and Chemodiversity
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01348
This article is part of the Research Topic Single Plant Cell Metabolomics View all 9 articles
 Patrick A. McVey1,2†
Patrick A. McVey1,2† Liza E. Alexander2,3,4†
Liza E. Alexander2,3,4† Xinyu Fu2,3,4
Xinyu Fu2,3,4 Bo Xie2
Bo Xie2 Katherine-Jo Galayda1,2‡
Katherine-Jo Galayda1,2‡ Basil J. Nikolau2,3,4
Basil J. Nikolau2,3,4 Robert S. Houk1,2*
Robert S. Houk1,2*The visualization of foliage color in plants provides immediate insight into some of the compounds that exist in the leaf. However, many non-colored compounds are also present; their cellular distributions are not readily identifiable optically. In this study we evaluate the applicability of mass spectrometry imaging (MSI) via electrospray laser desorption ionization (ELDI) to reveal the spatial distribution of metabolites. ELDI-MSI is a matrix free, atmospheric pressure ionization method that utilizes a UV laser coupled with supplemental ionization by electrospray. We specifically applied ELDI-MSI to determine the spatial distribution of metabolites in Coleus Henna half leaves that were grown with half-sections either fully illuminated or shaded. We monitored dynamic changes in the spatial distribution of metabolites in response to the change of illumination every 7 days for a 28 day period. A novel source-sink relationship was observed between the 2 halves of the experimental leaf. Furthermore, Coleus Henna leaves present visually recognizable sectors associated with the differential accumulation of flavonoids. Thus, we correlated the effect of differential illumination and presence or absence of flavonoids with metabolic changes revealed by the accumulation of carbohydrates, amino acids, and organic acids. The results show the potential of ELDI-MSI to provide spatial information for a variety of plant metabolites with little sample preparation.
Traditional mass spectrometry (MS) based methods of metabolite analysis require extraction of metabolites from biological samples, followed by chromatography. Spatial distribution data on metabolites are lost in these procedures. Recently, mass spectrometry imaging (MSI) has made a significant impact in filling this gap and has contributed to more refined understanding of many areas of biology (Amstalden van Hove et al., 2011; Lee et al., 2012; Angel and Caprioli, 2013; Nilsson et al., 2015; Shroff et al., 2015). Interrogating the location of metabolites in multicellular organisms provides insight into the “sharing” of metabolic processes among different cell types.
Several ionization sources for MSI have been introduced. Each has its own unique advantages and limitations. Matrix assisted laser desorption ionization (MALDI) is the most widely used MSI technique in biology and can obtain spatial resolution as high as ~1 μm (Zavalin et al., 2015; Kompauer et al., 2017). MALDI performed in vacuum, coupled with different applied matrices, allows for the detection of a variety of metabolites (Klein et al., 2015; Shroff et al., 2015). However, the application of the matrix can complicate the use of MALDI especially for metabolites with low m/z values (<1000 Da) (Boughton et al., 2016), such as organic acids, sugars and amino acids. Laser ablation electrospray ionization (LAESI) generally uses an infrared laser to ablate particles from a biological surface, with water in the sample being the main IR absorber. The ablated plant particles undergo post-ionization via interaction with the ESI droplets before entering the mass spectrometer. As generally practiced, LAESI allows ~200 μm lateral resolution, while requiring little sample preparation because there is no applied matrix (Nemes and Vertes, 2007). Similar to the aforementioned methods, matrix assisted laser desorption electrospray ionization (MALDESI) utilizes an applied matrix to absorb radiation from an IR laser coupled with post-ionization via ESI; water ice is an effective matrix for MALDESI (Robichaud et al., 2014).
In the present study, electrospray laser desorption ionization (ELDI) with an ultraviolet laser (wavelength 355 nm) coupled with electrospray (Shiea et al., 2005), was used to analyze the spatial distribution of metabolites in plant samples. In this study, we applied ELDI to Coleus (Solenostemon scuttellarioides) leaves that were grown with only half of the leaf surface illuminated. Coleus is a member of the Labiatae (mint) family, and different varieties are generally grown as ornamentals. The Coleus variety Henna is known for its serrated foliage. Its upper side is a unique uniform chartreuse to copper color. The underside is dark burgundy. These colors are primarily due to anthocyanin pigments that accumulate in these plants. The color intensity can be affected by illumination, temperature, and other environmental conditions (Lebowitz, 2011).
The present work evaluates ELDI-MSI to study the distribution of anthocyanins, flavonoids, and small metabolites (<1000 Da), including carbohydrates, organic acids and amino acids in Coleus leaves. The effects of different states of illumination during growth are assessed.
Solenostemon scuttellariodes (Coleus Henna spp.) adult plants were obtained from Stam Greenhouse (Oskaloosa, IA). All plants were transferred to LC1 Sunshine Mix soil (Sun Gro Horticulture, Bellevue, WA), watered weekly, and grown in a growth room at 22°C under continuous illumination (2,568 Lux or photosynthetic photon flux density 100 μmol of photons m–2 sec–1).
For the light regulated experiments, half (through the leaf vein) of each leaf was covered length-wise with aluminum foil (shiny side up). The edges were sealed with scotch tape to ensure the covered region had limited light exposure. The leaves were harvested from the same plant at 1-week intervals for 4 weeks. In all imaging and profiling experiments, three biological replicates (each replicate being images of three plants) were used for MS imaging, optical imaging and metabolic profiling of all metabolites.
Leaves for anthocyanin and non-targeted metabolite profile analysis were harvested each week, cut through the middle vein to separate the light-exposed surface from the shaded/dark-surface, and immediately flash-frozen using liquid nitrogen. The samples were then dried using a vacuum lyophilizer and pulverized using a Mixer Mill 301 (Retsch GmbH, Germany) in 2 mL Eppendorf tubes prior to extraction.
For MS imaging, samples were gently cut from both the foil-covered and uncovered edge of the leaf. Sections measuring ~12 mm by 8 mm were immediately analyzed using ELDI. Leaf edges were also visualized under a light microscope.
For microscopy only, leaf edges were hand-sectioned using a vibratome (TPI-3000; www.tedpella.com) at 70–100 μm. The sections were mounted in water and visualized under bright-field using a BH40 compound microscope (Olympus, www.olympus-global.com) equipped with Axio Vision software (Carl Zeiss Inc., Thorwood, NY). Additionally, a fresh leaf was ablated and the depth of the ablation trenches were measured using a Keyence VHX-5000 Digital Microscope.
Compounds were extracted from 5 mg of lyophilized Henna leaf tissue using 300 μL of methanol/water/acetic acid (85:15:0.5; v/v/v) and sonicated for 1 h. Samples were then incubated in dark at 4°C for 2 h (Wu et al., 2004). The samples were centrifuged for 5 min at 13,000 rpm and the liquid phase was filtered twice using 13 mm × 0.45 μm Teflon Syringe filters (Supelco, PA) for HPLC MS analysis.
The chromatographic separations were performed with an Agilent Technologies 1100 series HPLC. This was coupled with an Agilent Technologies Mass Selective Trap SL detector, equipped with an electrospray ionization (ESI) source with an autosampler/injector and diode array detector (DAD) for LC-MS analysis. A silica-based reverse-phase C18 Atlantis T3 column (2.1 × 150 mm, 3 μm, Waters, Milford, MA) was used for separation. Elution was performed using mobile phases containing 5% formic acid in LC-MS grade water or methanol (MeOH). The flow rate was kept at 0.2 mL/min and the DAD detection was at UV Vis wavelength 520 nm. After 5 μL of sample injection, a gradient was used as follows: 0–6 min, 0–20% MeOH; 6–40 min, 20–50% MeOH; 40–44 min, 50–50% MeOH; 44–48 min, 50–00% MeOH 48–52 min, 100% MeOH; 52–56 min, 100–0% MeOH. The column effluent was then introduced via ESI into an Agilent 6210 MSD time-of- flight mass spectrometer operating in positive mode. The ESI capillary voltage was +3.0 kV, nitrogen gas temperature was set to 350°C, drying gas flow rate was 11 L/min, nebulizer gas pressure was 35 psi, skimmer was 65 V, and OCT RF was 250 V. Mass spectra from m/z 100 to 2000 were collected and analyzed using Agilent ChemStation Data Analysis. Individual anthocyanin peak areas were generated by Quant Analysis and used to compare different levels of anthocyanins quantitatively. The structure for each anthocyanin was identified from detailed MS/MS analysis performed at various collision energies.
Metabolite extracts were prepared as described previously (Schmidt et al., 2011). Extracts were prepared from 2 mg of lyophilized leaf tissue. The extracts were spiked with two internal standards: 10 μg of ribitol and 5 μg nonadecanoic acid for polar and non-polar fractions, respectively. To 5 mg of lyophilized tissue, 0.35 ml of hot methanol (60°C) was added. The sample was incubated at the same temperature for 10 min, followed by sonication for 10 min at full power. To this slurry, 0.35 ml of chloroform and 0.3 ml of water were added and the mixture was vortexed for 1–3 min. After centrifugation for 5 min at 13,000 g, 200 μl of the upper phase (polar fraction) and 200 μl of the lower phase (non-polar fraction) were separately removed into 2 ml GC-MS vials, and dried in a Speed-Vac concentrator (model SVC 100H, Savant, NY).
The samples were then methoximylated and silylated. For methoximation, 50 μl of 20 mg/mL methoxyamine hydrochloride dissolved in dry pyridine was added. The reaction mixture was shaken at 30°C for 1.5 h. Silylation was performed by adding 70 μL of N, O-Bis (trimethylsilyl) trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) and incubating at 65°C for 30 min.
One microliter of the derivatized samples was injected into the GC-MS in splitless mode. GC-MS analysis was performed using an Agilent 6890 GC interfaced to an Agilent 5973 quadrupole MS with a HP-5ms (5%-Phenyl)-methylpolysiloxane column (30 m × 0.25 mm × 0.25 μm, Agilent). The temperature was programmed from 70 to 320°C at 5°C/min with helium flow rate at 1.0 mL/min and inlet temperature at 280°C. EI-MS ionization energy was set to 70 eV and the interface temperature was 280°C.
The GC-MS data files were deconvoluted and searched against an in-house MS-library, the NIST 14 Mass Spectral Library using NIST AMDIS software (Stein, 1999) and the Golm Metabolome Database (Hummel et al., 2010).
Non-targeted metabolite profiling data obtained by GC-MS analysis, and anthocyanin data obtained from LC-fluorescence are publicly available in the PMR database (http://metnetdb.org/PMR/; Hur et al., 2013). They are available at: metnetweb.gdcb.iastate.edu/PMR/experiments/?expid=279.
A Waters Synapt G2-S quadrupole time-of-flight mass spectrometer was used for data collection for all MSI. The Waters ESI source was removed and a homemade open-air ESI source (Galayda, 2017) was used with samples at ambient pressure. Data were acquired in the mass range from m/z 50 to 1200, spectra were summed for 0.3 s. The time-of-flight reflectron operated in single-pass mode with a resolution of ~10,000 FWHM for MS images. The TOF was then operated in double-pass mode with a resolution of ~40,000 FWHM, or “high resolution mode,” to confirm compound identifications by accurate m/z measurements. These confirmatory measurements were done on a different segment taken from the same leaf. Tandem MS quadrupole resolution varied from Δm = 5 to 12 depending on the analyte with a nominal collision energy of 20 eV. The Synapt was operated using Waters MassLynx V4.1 (SCN851) software.
Spectra were generated from total ion chromatograms (TIC) combined by the MassLynx software. The “.raw” Waters data files were converted to mzML files by Proteowizard Mass Converter Tool. The mzML files were then combined into an imzML file using imzML Converter. This combined image file was then viewed and images were generated from MSiReader V0.06 via the W.M. Keck FTMS Laboratory. All images made within MSiReader had Linear2 interpolation for image clarity, and used the “Jet” colormap/false color appearance. Co-localization and 3D images were created with MSiReader V1.00. For a few samples in the early stages of this study, images were generated after normalizing individual ion signals to the total ion signal. This normalization did not affect the images noticeably, so normalization was not used for any of the images shown below.
Compounds were identified from the ELDI spectra in a multiple step process to ensure confident assignments. Initial ELDI measurements were done in so-called “sensitivity” mode, m/Δm ~10,000 FWHM. Based on these results, high resolution spectra at ~40,000 were acquired on a different segment of the same leaf for accurate mass measurements. Next, if the compound was of high enough abundance and without background interferences, tandem mass spectra were taken to provide a fragmentation pattern. These high resolution m/z values were then put into online metabolite databases (i.e., METLIN) to generate possible compound matches based on accurate mass and tandem MS (if available from both ELDI data and the online database). ELDI data were also compared to the corresponding GC-MS and LC-MS data acquired from Coleus Henna leaves for identity overlap. Every possible ID was given a Δppm value based on the experimental m/z value compared with the true m/z value of the compound. These Δppm values were generally below 10 ppm. Some compounds were given identifications with greater than 10 Δppm values based on matches with the GC-MS data and/or tandem MS matches with the online database. Some flavonoid peaks were assigned as cyanidin or apigenin derivatives despite no database matches due to highly abundant tandem MS product peaks at m/z 287.0585 or 271.0627, respectively. Many METLIN database entries of flavonoids did not have supporting MS/MS data for comparison, with some having only predicted MS/MS spectra.
Acquiring tandem MS data with ELDI was successful for most peaks of interest, but was not always possible. It was difficult to acquire tandem MS spectra for highly spatially localized compounds, labeled “localized signal” in subsequent data tables. Compounds with a relatively low abundance did not give a satisfactory fragmentation pattern for identification purposes (labeled “too low abun.” in subsequent data tables). Analyte peaks near major background peaks had interferences with their tandem MS spectra. The quadrupole resolution could sometimes be increased to eliminate these interferences, but a subsequent drop in peak intensity sometimes resulted in poor tandem MS results.
The apparatus has been described (Galayda, 2017). Samples were ablated with a Nd:YAG laser (ULTRA, Big Sky Laser Tech, Inc. Bozeman, MT). The third harmonic was used at 355 nm. The laser was operated at a pulse repetition rate of 10 Hz, with a 5 ns pulse width, and an energy of ~250 μJ/pulse (before focusing). This pulse energy was just above the ablation threshold for these samples. The beam was focused onto the sample by a single plano-convex focusing lens (fused-silica, focal length 75 mm), with a nominal spot size of ~125 μm.
Samples were cut with a scalpel. Immediately after cutting, the sample segments were mounted on a glass slide using double sided tape. The laser did not penetrate completely through the sample; the underlying tape is not ablated. No matrix was applied, and leaves were pressed lightly to create an even surface. The optical images shown below were photographed before this pressing step. Thus, the dimensions of the optical images are slightly smaller than those of the MS images in some of the results shown below.
Plant samples were then placed 8 mm below the ESI-sample inlet axis on a computer-controlled translation stage (Z825B, Thorlabs, Inc. Newton, NJ). Samples were translated at 0.4 mm/s beneath the 10 Hz laser beam down the surface of the leaf to insure fresh tissue was constantly being ablated. The distance between the centers of adjacent ablation tracks was 125 μm, providing a lateral resolution of 125 μm. The ablation trench was ~ 30 μm deep. The underlying tape was not ablated. Mass spectra were averaged over 0.3 s intervals. Thus, the ablated volume was 125 × 120 × 30 μm deep. The leaves were irradiated normal to the sample surface with the laser beam axis ~2 mm downstream from the ESI capillary. The ESI tip was ~10 mm from the sample inlet (Galayda, 2017).
A solution of 50% methanol with 0.1% formic acid (99.5% purity, Fisher Scientific) was pumped through a 53 μm ID polyimide coated capillary as the ESI solution (pump: model Z2, Harvard Apparatus, South Natick, MA). Leucine enkephalin was added at 0.1 ppm to the ESI solution for use as a mass calibrant. All data were acquired in positive ESI mode. The ESI voltage for ELDI was +2.5 kV applied to a stainless steel union in the liquid flow line, with the sample cone completing the ESI circuit. The sample inlet was kept at 100°C with a N2 curtain gas flow of 1 L/h.
The mid-rib (Figure 1) was used as the boundary between the two halves of each Coleus Henna leaf. One half of a leaf was covered with aluminum foil to generate leaf tissue that was grown under lower illumination conditions, and the uncovered half of the leaf was used as a control for normal levels of illumination. Each leaf was dissected transversely. Segments ~12 mm × 8 mm were collected from both the illuminated and shaded sides of the leaf. These segments were placed on a glass slide and used in ELDI imaging experiments.
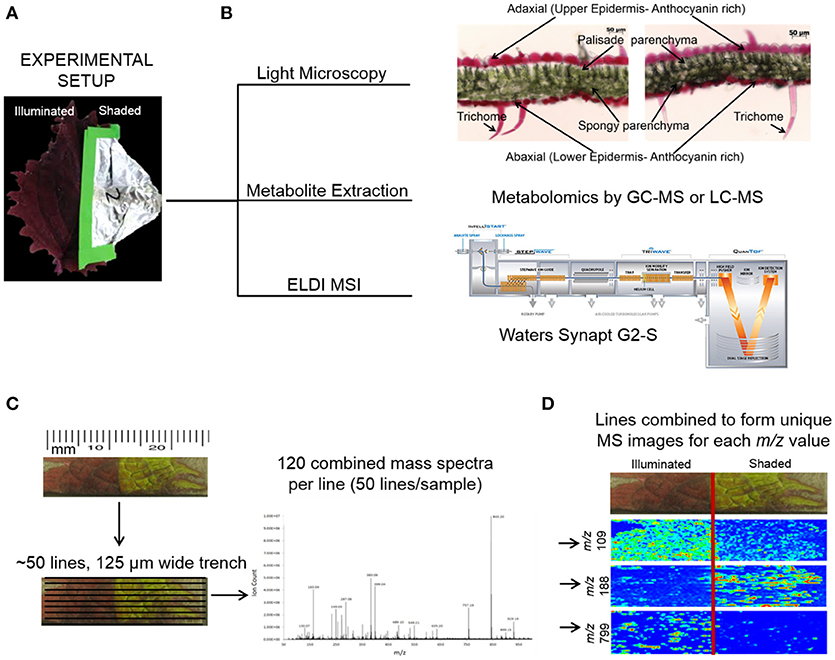
Figure 1. ELDI-MSI experimental workflow. (A) Individual Coleus Henna leaves were treated for a period of up to 28 days by shading half the leaf and leaving the other half fully illuminated. (B) Leaf samples were selected for light microscopy, metabolite extraction and metabolomics analysis, and ELDI-MSI analyses with a Waters Synapt G2-S mass spectrometer. (C) ELDI-MSI data were generated and validated by metabolite profiling of extracts by GC-MS or LC-MS analysis or METLIN database. (D) Mass spectrometric images were generated for individual ions and aligned with visual images of leaves.
Metabolite distribution maps were created for 77 chemically identified ions detected by ELDI-MS. Thirteen of these identified ions were observed to decrease in abundance in the shaded side of the leaf, while the abundances of 11 compounds increased in the shaded side. The relative abundances of the remaining 53 ions were unaffected by shading (Tables 1–3). In-parallel, targeted anthocyanin analysis via LC-MS, and non-targeted global metabolomic analysis via GC-MS were performed on separate leaf samples, which were sampled in triplicate.
Under normal illumination conditions both the adaxial (upper) and abaxial (lower) surfaces of the Coleus Henna leaf are a burgundy red-purple color. In the shaded condition, the foliage color changes from burgundy red-purple to chartreuse green (Figure 2). Microscopic cross-sections of leaves reveal that the abaxial and adaxial epidermal cells are heavily red-pigmented, and the mesophyll and palisade layers are green pigmented with chlorophylls. Epidermal cells range from 20 to 50 μm deep with cross-sections in the range of 40 to 55 μm long. These cell dimensions are unaffected by shading. The red pigmentation was decreased in the adaxial epidermis, and unaltered in the abaxial epidermis. This loss of the red pigment from the adaxial epidermis revealed the underlying chlorophyll pigments, imparting the green color to the shaded half of the adaxial surface of the leaf.
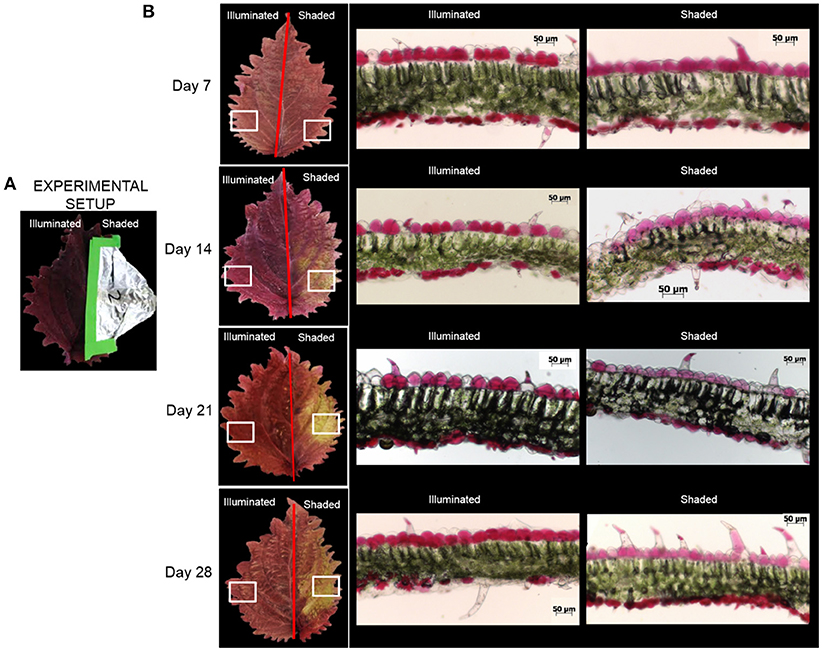
Figure 2. Optical images of Coleus Henna leaves over a 28 day period. (A) The right-side of each leaf was shaded by wrapping with aluminum foil, whereas the left side was fully illuminated. White boxes denote areas of the leaves that were sectioned for optical microscopic examination in cross section (B). Scale bar = 50 μm.
The GC-MS analyses of metabolite extracts from leaf tissue detected 156 polar and non-polar analytes; 67 of these were identified chemically. These metabolites include alcohols, polyols, sugars, lipids, fatty acids, esters, sterols, hydrocarbons, organic acids, and nitrogenous metabolites. The abundances of most of the chemically identified metabolites remained unaltered irrespective of the illumination conditions (Figure 3). The abundances of a few metabolites, such as gluconic acid, fructose, arachidonic acid, 1-monopalmitin and a hexose sugar, increased in the shaded side of the leaf as time progressed. The abundance of fructose increased in the shaded side of the leaf and became higher than in the illuminated side by the 28th day of the experiment. Benzoic acid showed a unique profile, with abundance decreasing through the time-line of the experiment; its abundance was always higher in the shaded side of the leaf.
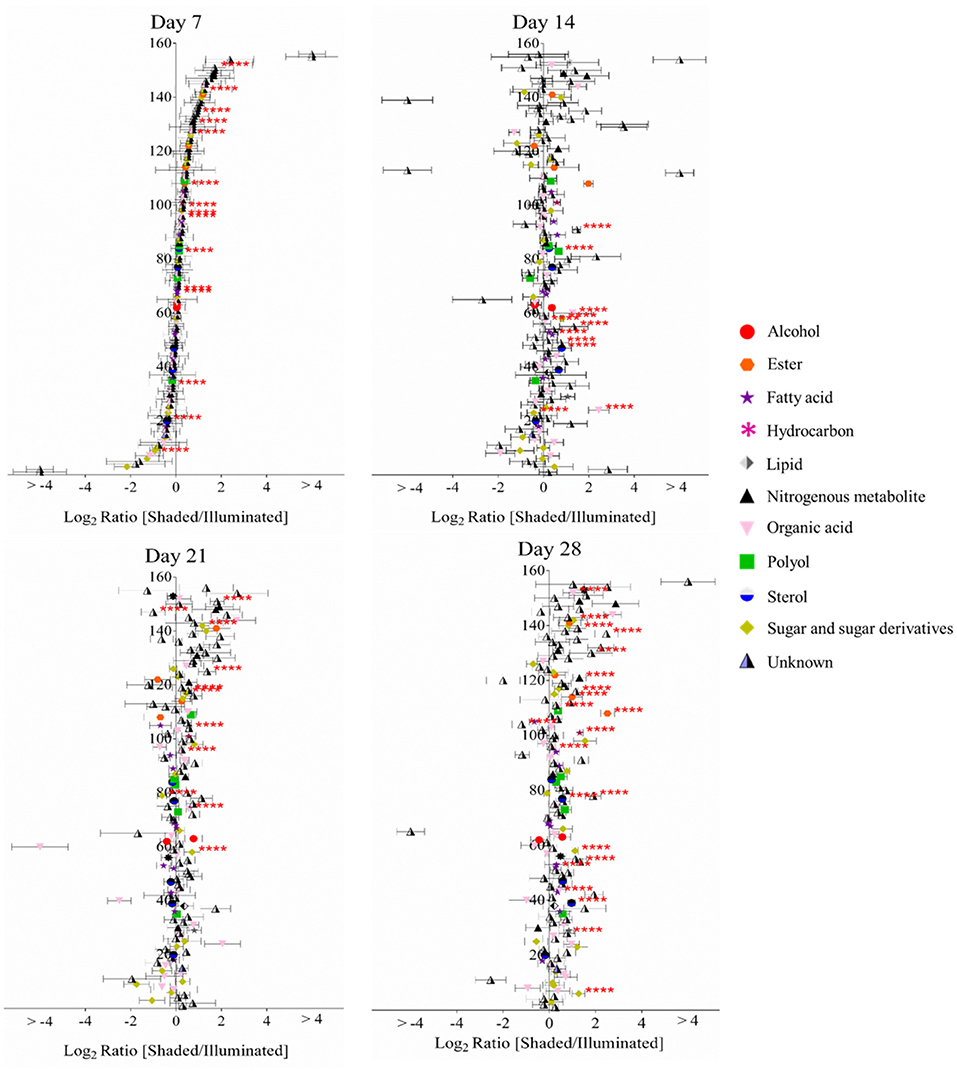
Figure 3. Log-ratio comparison of the differential metabolomes between illuminated and shaded sides of the leaf. The x-axis plots the log-transformed relative abundance ratio of each metabolite in illuminated versus shaded sides. The order of the metabolites (156 analytes, 67 chemically identified) on the y-axis is identical in all four plots, and they are ordered from the lowest to highest value on the x-axis as determined for the 7-day time-point. **** denote p-value < 0.05.
Targeted LC-MS analyses measured changes in anthocyanin concentrations during these experiments. Nine cyanidin-based anthocyanins (Figure 4) were identified. Two of these showed differential accumulation between the illuminated and shaded sides of the leaf: cyanidin-coumaroylglucoside-malonylglucoside at m/z 843.1970 and cyanidin-coumaroylglucoside-dimalonylglucoside at m/z 929.1966. The abundance of both these anthocyanins decreased significantly in the shaded side of the leaf after 21 days of the experiment. Day 21 appears to be anomalous compared to the other time points. We suggest that day 21 may represent a metabolic switch, as the leaf is depleted of carbon and subsequently is re-accessed from sink tissues. The accumulation of the other 7 anthocyanins was unaffected by the difference in illumination.
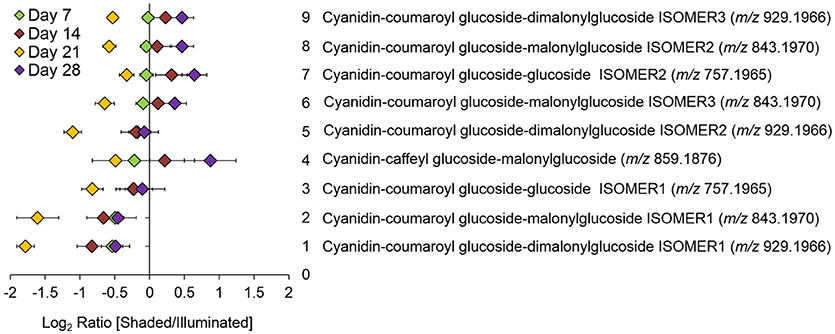
Figure 4. Log-ratio comparison of the differential accumulation of anthocyanins in Coleus Henna half-leaves maintained under full-illumination or shaded for the indicated time-periods. The table names each of the 9 anthocyanins that were identified by LC-MS analysis of extracts prepared from the two leaf halves. The order of the metabolites on the y-axis of the plot is from the lowest to the highest log-ratio values as determined for the 7-day time point.
The pigmentation on the abaxial side of the leaf was unaffected by shading, whereas the pigmentation pattern was altered on the adaxial side of the leaf. Therefore, we focused on imaging the distribution of the metabolites on the adaxial surface using ELDI-MSI. Many of the ions detected in these ELDI spectra were attributed to flavonoids. Their chemical identity was confirmed by MS/MS experiments that generated fragment ions characteristic of the flavonoid backbones. The identities of these fragment ions were confirmed by matching accurate mass measurements with entries in the Metlin database (Smith et al., 2005). This strategy identified two main classes of flavonoids, based on the aglycone cores: (a) apigenin, a flavone (Figure 5A), and (b) cyanidin, an anthocyanin (Figure 5B).
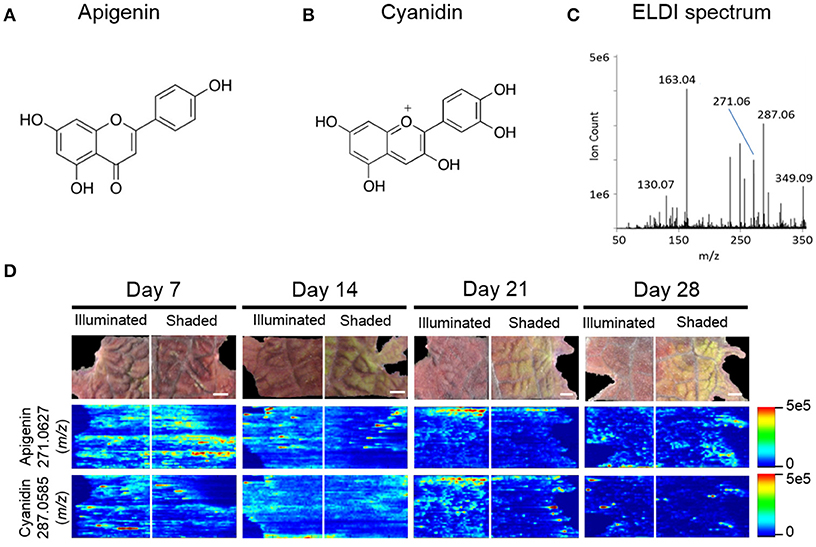
Figure 5. ELDI-MSI analysis of flavonoid backbones. Molecular structure of two aglycone flavonoids. (A) apigenin; and (B) cyanidin. (C) ELDI mass spectrum showing the relative abundance of apigenin ([M+H]+ at m/z 271.0627) and cyanidin ([M]+ at m/z 287.0585) ions. (D) Spatial distribution of apigenin and cyanidin in shaded or fully illuminated leaf-halves, at different time-points after initiation of the shading treatment. Scale bar = 2 mm. The MSI intensity scale bar is color coded: red is maximum signal and blue is minimum signal, in this and subsequent figures.
Apigenin was identified as its protonated ion [M+H]+ at m/z 271.0627. All analytes that generated this fragment ion by tandem MS were inferred to be apigenin-based flavones. Cyanidin has a permanent +1 charge and was identified as an [M]+ ion at m/z 287.0585. Cyanidin was distinguished from isobaric compounds (e.g., luteolin or kaempferol) based on MS/MS fragments derived from this [M]+ ion (Table 1). Cyanidin-based anthocyanins were identified by cyanidin [M]+ product ions at m/z 287.0585. The high intensities of the apigenin-aglycone (m/z 271.0627) and cyanidin-aglycone (m/z 287.0585) ions in the overall ELDI spectrum (Figure 5C) show that the aglycones have a high relative abundance compared to other low mass molecules observed.
The cyanidin-based anthocyanins are primarily glycosylated and further biochemically modified by malonylation or coumarylation (Table 1). These anthocyanins generated common fragment ions at m/z 163.0620 and 147.0485, which were identified as the protonated water-loss ion [M-H2O+H]+ of a hexoglycoside, and the protonated water-loss ion [M-H2O+H]+ of coumaric acid, respectively. The glycone moiety was not identified directly. However, based on the METLIN and the KEGG pathway databases (Smith et al., 2005; Guijas et al., 2018), and prior characterization of these metabolites in other Coleus lines (Boldt, 2013), these glycosides are subsequently referred to as glucosides. Collectively therefore, we identified 15 cyanidin-based anthocyanins (Table 1).
Similar characterizations identified four apigenin-based flavones, including the apigenin aglycone. Tandem MS generated the apigenin backbone product ion, but the specific chemical structures of these apigenin-based flavones were not determined. The ELDI experiments identified five apigenin-based flavones at m/z 447.0918, m/z 489.1057, m/z 619.2228, m/z 635.2041, and m/z 743.1454 (Table 1).
The 15 chemically identified cyanidin-derived anthocyanins can be categorized into three classes. Eight are less abundant in the shaded side of the leaves, one is more abundant in the shaded side, and six are not affected by the difference in illumination (Table 1). This last category includes the cyanidin aglycone, whose abundance are unaffected by shading during the entire 28-day period of the study, same as the apigenin aglycone (Figure 5D). Additional example images are presented in Figure 6, which shows the spatial distribution of five chemically identified cyanidin-based anthocyanins and three apigenin-based flavonoids. These include cyanidin-malonylglucoside (m/z 535.1069 [M]+), cyanidin-coumaroyl glucoside (m/z 595.1447 [M]+), cyanidin-coumaroylglucoside-glucoside (m/z 757.1965 [M]+), cyanidin-coumaroylglucoside-malonylglucoside (m/z 843.1970 [M]+), and cyanidin-coumaroylglucoside-dimalonylglucoside (m/z 929.1966 [M]+); and the apigenin flavonoids at m/z 447.0918 [M+H]+, m/z 619.2228 [M+H]+, and m/z 743.1454 [M+H]+.
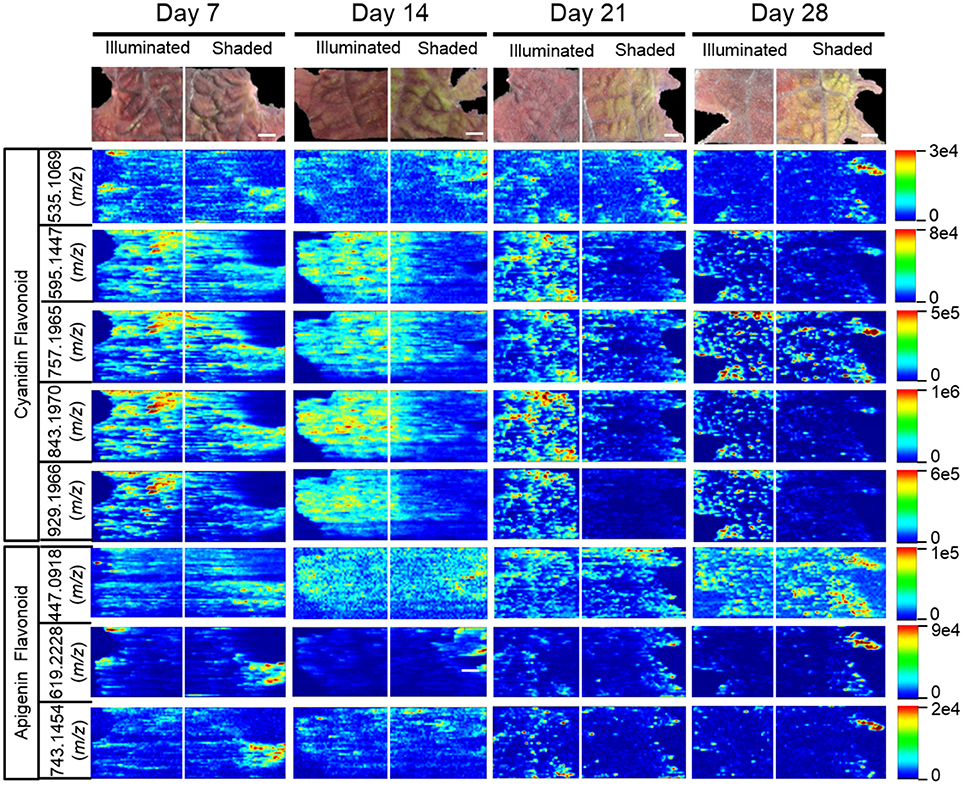
Figure 6. Spatial distribution between shaded and illuminated halves of leaves of cyanidin and apigenin-based flavonoids as affected by up to 28 days of shading. The cyanidin-based flavonoids were detected as [M]+ ions, and apigenin-based flavonoids were detected as [M+H]+ ions. The assigned IDs of the cyanidin-based flavonoids are as follows: cyanidin malonylglucoside (m/z 535.1069), cyanidin coumaroylglucoside (m/z 595.1447), cyanidin-coumaroylglucoside-glucoside (m/z 757.1965), cyanidin-coumaroyl glucoside- malonylglucoside (m/z 843.1970), cyanidin-coumaroyl glucoside-dimalonylglucoside (m/z 929.1966). The apigenin-based flavonoids are tentatively identified as apigenin glucuronide (m/z 447.0918), apigenin flavonoid (m/z 619.2228), and apigenin flavonoid (m/z 743.1454). Scale bar = 2 mm.
MS images in this paper are obtained by scanning the laser horizontally from left to right. Signals are not acquired between adjacent ablation tracks. This acquisition method generates images that appear to be elongated horizontally, largely due to the way in which the data are acquired. Additional horizontal stretching could occur because (a) there is no delay between pixels to allow recently ablated material to wash out of the ion source, and (b) ablation occurs into room air; there is no controlled gas flow in the ion source. Two examples are the two laterally-elongated features seen in Figure 5D for apigenin, day 14 shaded side, middle of image. The effect is intermittent; it is not always observed. When this elongation is observed, as in Figure 5D, usually multiple adjacent tracks are elongated, which indicate that stretched images are mainly due to the stretched analyte regions in the actual sample.
The anthocyanin with the highest relative abundance, cyanidin-coumaroylglucoside-malonylglucoside (m/z 843.1970 [M]+), is initially equally abundant in both illuminated and shaded sides of the leaf. However, starting at 14 days its abundance is reduced more rapidly in the shaded side of the leaf (Figure 6). A similar distribution pattern after 28 days of shading is observed for the structurally related cyanidin-coumaroylglucoside (m/z 595.1447 [M]+). In contrast, cyanidin-coumaroylglucoside-glucoside (m/z 757.1965 [M]+) is equally abundant in both the illuminated and shaded sides after 28 days (Figure 6). This anthocyanin had the second highest relative abundance and may be the immediate metabolic precursor to cyanidin-coumaroylglucoside-malonylglucoside (Guijas et al., 2018). Three-dimensional temporal distribution patterns of these structurally related anthocyanins (m/z 843.1970 [M]+, m/z 757.1965 [M]+, m/z 595.1447 [M]+, and m/z 287.0585 [M]+) after 28-days of shading are presented in Figure 7.
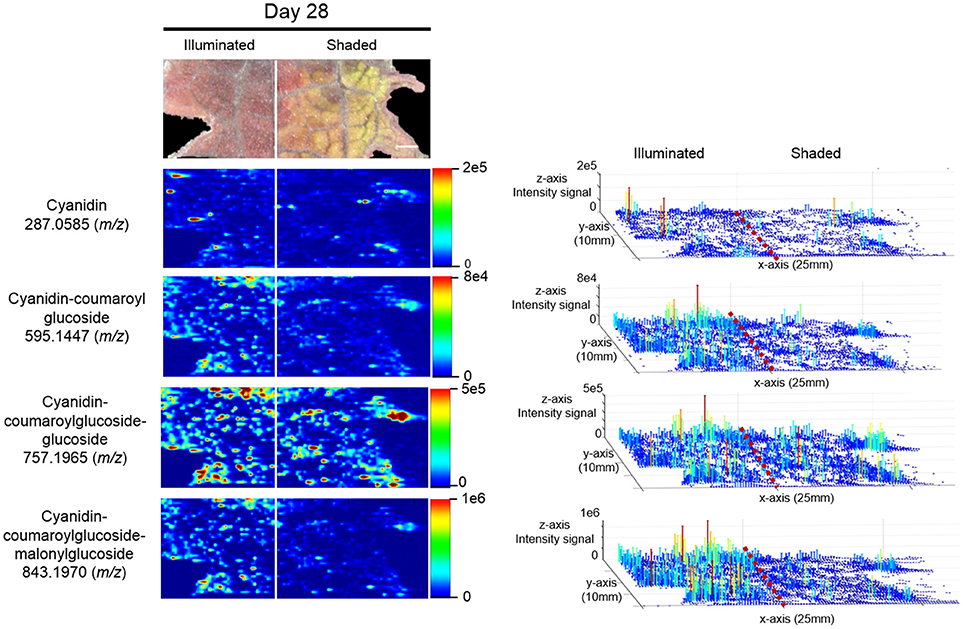
Figure 7. Spatial distributions of cyanidin-coumaroylglucoside-malonylglucoside and its metabolically related species determined by ELDI-MSI analysis of leaf-halves subjected to either full illumination or shaded for 28-days. Right panel represents the 3D representations of the abundance of these metabolically related ions. The x- and y-axes represent spatial coordinates (mm), and the z-axis maps ion intensity. Scale bar = 2 mm.
Similar to cyanidin-coumaroylglucoside-glucoside, the abundance of the dimalonylated derivative, cyanidin-coumaroylglucoside-dimalonylglucoside (m/z 929.1966 [M]+), is lower in the shaded side of the leaf (Figure 6). A precursor to cyanidin-coumaroylglucoside-dimalonylglucoside, the cyanidin-malonylglucoside (m/z 535.1069 [M]+) is equally abundant in both the illuminated and shaded sides until 28-days of shading, where its abundance is reduced in the illuminated side of the leaf (Figure 6). All these changes in relative abundance of cyanidin-based anthocyanins, revealed by ELDI-MSI, were confirmed by LC-MS analysis of extracts from these leaves (Figure 4).
The spatial redistribution of two apigenin-based metabolites was revealed by MSI. For example, the apigenin-based flavonoid at m/z 619.2228 occurs in both illuminated and shaded sides of the leaves and concentrates in the periphery of the shaded leaf-half (Figure 6). Another apigenin-based flavonoid at m/z 743.1454 is initially located in the periphery of the shaded leaf-half. It redistributes temporally to become more evenly dispersed among the two halves of the leaf (Figure 6).
Twelve sugars and sugar derivatives were chemically identified using three MS-based criteria: (a) accurate mass determination with ELDI; (b) ELDI-MS/MS fragmentation spectra; and (c) integrated retention index and electron impact fragmentation patterns from GC-MS analysis. Most of the 12 sugars were observed as 39K+ and/or Na+ adducts by ELDI (Table 2). Specifically, sucrose was identified by MS/MS fragmentation patterns upon ELDI, and also by GC-MS analysis of extracts. A hexose, possibly glucose, was identified as both Na+ (m/z 203.0560) and 39K+ (m/z 219.0310) adducts. The identity of a phosphorylated hexose (possibly glucose-phosphate) was inferred from accurate mass determination of the 39K+ adduct (m/z 312.9720). An organic acid glucoside (possibly coumaroyl glucoside) was identified by MS/MS experiments as both the Na+ (m/z 349.0923) and 39K+ (365.0647) adducts. A heptose was identified by accurate mass determination and by MS/MS fragmentation spectra, but the particular isomer could not be determined.
The abundances of eight of these sugar metabolites were unaffected by the illumination status of the leaf (Table 2). The spatial distributions of six typical metabolites are shown in Figure 8. Three of these sugar metabolites (m/z 203.0560, m/z 312.9720, m/z 349.0923) become more abundant over time in the shaded side of the leaf. Finally, a sugar-derivative, believed to be hexose-glycerol phosphate (m/z 357.0586), as determined by accurate mass, became less abundant across the entire leaf after 28 days of shading (Figure 8).
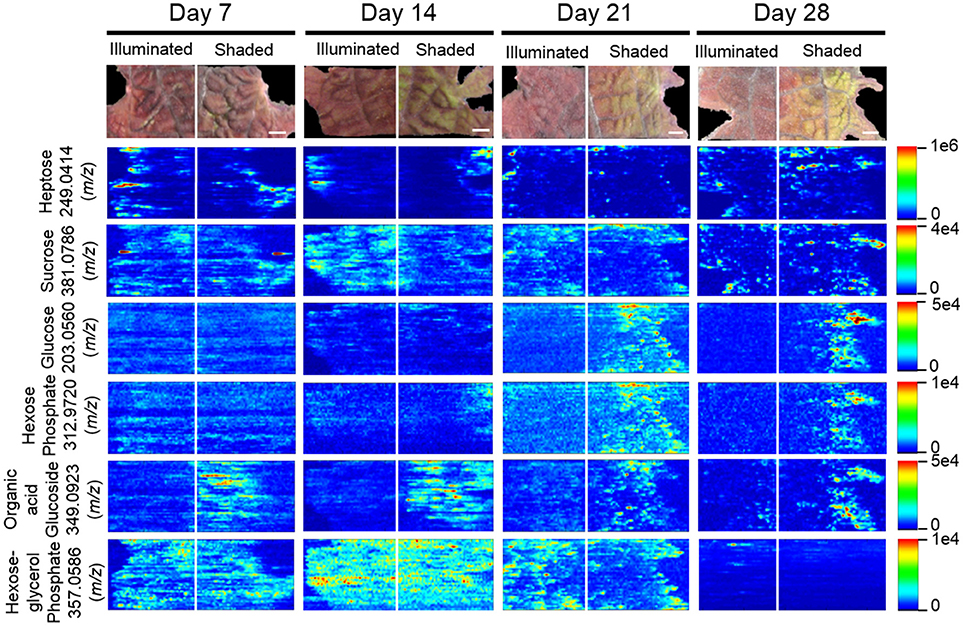
Figure 8. Spatial distributions of sugars determined by ELDI-MSI analysis of leaf-halves subjected to either full illumination or shading for up to 28-days. Each sugar was detected as [M+Na]+ or [M+K]+ adducts. Scale bar = 2 mm.
Table 3 lists the carboxylic acids and amino acids that were observed by ELDI-MSI. Seven of these metabolites were identified as H+ adducts, 15 as H+ adducts accompanied by water loss, two as Na+ adducts, and three as 39K+ adducts. Several of these compounds were observed as multiple adducts; individual molecules of a given compound had one of either H+, Na+ or 39K+ attached in the same spectrum.
Several analytical strategies were integrated to confirm the chemical identity of these organic acids, including accurate mass determination, MS/MS experiments, and GC-MS analysis of derivatized metabolite extracts. The latter strategy was also used to confirm the relative concentrations between the illuminated and shaded sides of the leaves. Collectively these analyses identified 23 organic acids. Eight were identified with ELDI-MS and the molecular images of these metabolites (Figure 9) indicate their abundances were unaffected by the shading of the leaf, except for glycolic acid seen only by GC-MS.
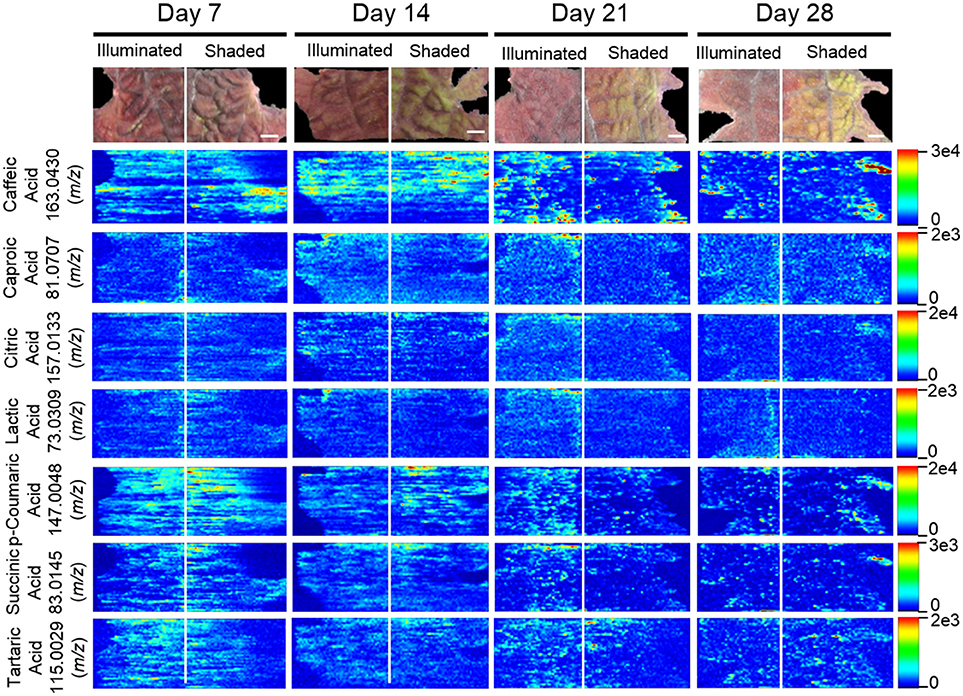
Figure 9. Spatial distributions of organic acids determined by ELDI-MSI analysis of leaf-halves subjected to either full illumination or shading for up to 28-days. Scale bars = 2 mm.
The locations of 12 proteogenic amino acids (alanine, arginine, asparagine, aspartic acid, glutamine, glycine, histidine, leucine, proline, serine, threonine, and valine) were determined by ELDI-MSI. The non-proteogenic amino acids were pyroglutamic acid and GABA (Figure 10). Arginine, aspartic acid, and histidine were considerably more abundant in the shaded side of the leaf, although in the case of histidine this increase occurred after the half-leaf was shaded for 21 days. The spatial distribution of seven amino acids were unaffected by shading, but the relative abundance of four amino acids changed as time progressed. Specifically, leucine abundance increased at 28 days, whereas the abundance of alanine (decreased from day 14 to 28), glycine (peaked day 14 decreased from day 21 onwards), and proline (depleted by day 21) was reduced during this time period.
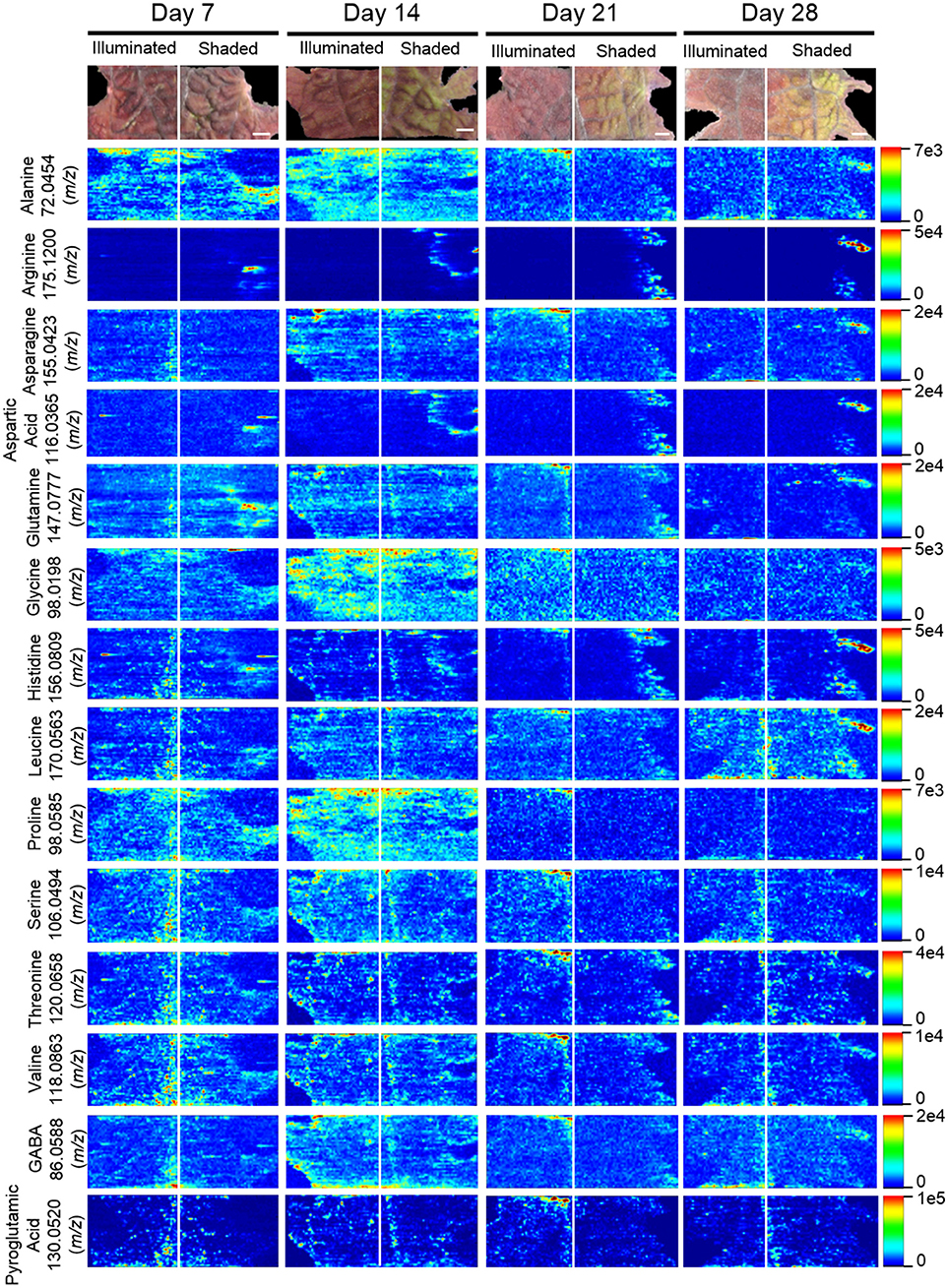
Figure 10. Spatial distribution of amino acids determined by ELDI-MSI analysis of leaf-halves subjected to either full illumination or shading for up to 28-days. Scale bar = 2 mm.
Several MSI methods have been utilized to obtain spatial distribution data of metabolites in plant samples. Each offers unique advantages and drawbacks (Lee et al., 2012; Boughton et al., 2016). The present work demonstrates the potential of applying ELDI for MSI. ELDI is analogous to LAESI, with the exception that ELDI uses ultraviolet radiation for ablation (e.g., 355 nm laser), whereas LAESI generally uses infrared radiation, for example a 2940 nm mid-IR laser (e.g., Nemes and Vertes, 2007). Although ELDI has previously been reported with fungi (i.e., Ganoderma lucidum and Antrodia camphorate) (Huang et al., 2012), there is only one other moderately extensive plant application study using ELDI, LAESI, or MALDESI (Etalo et al., 2015). The spatial resolution was limited to 500 μm in that work (Etalo et al., 2015). Many proof-of-concept experiments using LAESI-MSI on plants have been reported (Bartels and Svatoš, 2015).
Because plants accumulate large quantities of UV-absorbing molecules (e.g., flavonoids, chlorophylls, terpenes, phenolics) the plant leaf itself serves as a pseudo-matrix. Ablation of the plant pseudo-matrix enables ELDI-based observation of additional molecules that are not UV absorbers (e.g., sugars, amino acids, and organic acids). In both ELDI and LAESI methods, particles undergo post-ionization via interaction with ESI droplets, and the resulting ions are then extracted into the mass spectrometer. Although LDI without post-ionization via ESI can achieve much higher spatial resolution, issues associated with ionization transfer still need to be addressed (Hölscher et al., 2009).
One concern about our ELDI method is possible spatial heterogeneity of the UV-absorbing pseudo-matrix. Coupling electrospray and ablation with a UV laser appears to minimize these issues. Both UV and non-UV absorbing compounds can be measured at atmospheric pressure. For example, Figure 11 illustrates the localization of the UV-absorbing flavonoid cyanidin-coumaroylglucoside-glucoside (m/z 757.1965 [M+]) and the localization of the non-UV absorbing metabolites, histidine, sucrose, and glucose. Non-absorbing analytes can be observed readily in regions where the abundances of the pseudo-matrix flavonoids are relatively low (Figure 11). These comparisons indicate that ELDI-based MSI can be used to localize the distribution of non-UV absorbing metabolites despite large spatial changes in concentration of flavonoids or other possible pseudo-matrix compounds.
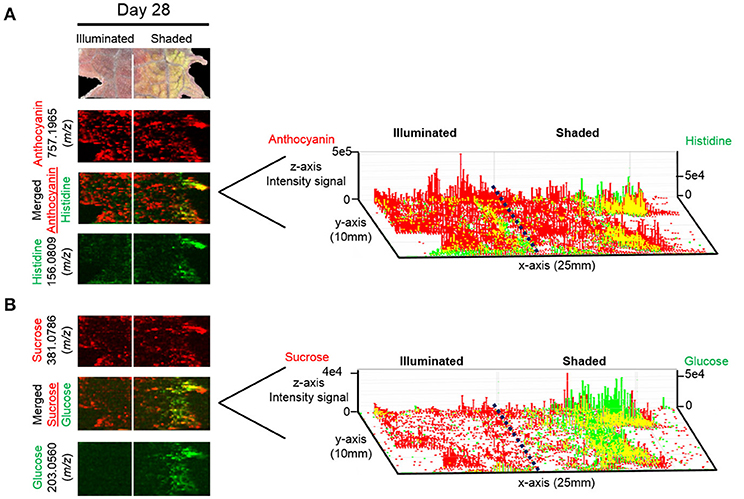
Figure 11. 3D representation of the localization of UV-absorbing and non-UV absorbing metabolites. Distributions of (A) anthocyanin 757.1965 (m/z) (red) along with histidine 156.0809 (m/z) (green) and (B) sucrose 381.0786 (m/z) (red) along with glucose 203.0560 (m/z) (green) are shown. Scale bar = 2 mm. Right panel represents the 3D representations of the abundances of these metabolically related ions. The x- and y-axes represent spatial coordinates (mm), and the z-axis maps ion intensity.
Coleus is convenient because its leaves are sectored visually. This reflects the spatial arrangement of the underlying flavonoids (Nguyen et al., 2008; Nguyen and Cin, 2009; Boldt, 2013). Moreover, in Coleus the sectoring of the adaxial surface is different from that of the abaxial surface. The adaxial surface is further modifiable by changes in the exposure to illumination (Nguyen and Cin, 2009; Logan et al., 2015). In addition, the depth of the leaf adaxial cells matches the penetration depth of the laser (~30 μm). Thus, the images presented herein reflect the abundance of metabolites in the epidermal cell layer of the leaf.
Correlated with the visible changes in the pigments of the leaf in response to shading, major metabolic changes in anthocyanins were visualized by ELDI-MS and quantitatively confirmed by LC-MS analysis of metabolite extracts. The cyanidin glycosides are the most abundant anthocyanins in the illuminated side of the leaves, which correlates with their photo-inhibitory potential (Steyn et al., 2002). Consistent with this photo-inhibitory attribute, the abundances of 17 of the 39 detected anthocyanins decreased upon shading of the leaf. Moreover, in response to the illumination status of the leaf, there is a coordinated change in the differential abundance of the structurally related anthocyanins, cyanidin-coumaroylglucoside-glucoside, cyanidin-malonyl glucoside, and the aglycone cyanidin. The ability to visualize this coordination at a spatial level provides additional insights to their potential metabolic interconnections. Namely, the enhanced accumulation of cyanidin-coumaroylglucoside-malonylglucoside, in response to illumination, may be associated with increased biosynthesis. The correlated changes in the levels of the potential precursors (i.e., cyanidin-malonyl glucoside) (Figure 7) support this hypothesis.
Multi-cellular photosynthetic plants are characterized by a series of source-sink tissues, that share the metabolic tasks of converting inorganic precursors, such as CO2, ammonia, phosphate etc., to organic constituents that are normally stored in sink tissues, such as seeds, tubers etc. (Turgeon, 1989; Basu et al., 1999; Roitsch, 1999; Paul and Foyer, 2001; Wardlaw, 2006; Lemoine et al., 2013; Osorio et al., 2014). Source-sink relationships can be genetically programmed and are further modified by environmental abiotic or biotic stimuli that are mediated by small molecules (e.g., sugars and amino acids) (Krapp and Stitt, 1995; Paul and Driscoll, 1997; Roitsch, 1999; McCormick et al., 2008; Lemoine et al., 2013). ELDI-MSI can image metabolic changes associated with the induction of an artificial new source-sink relationship between two halves of a leaf. The fully illuminated half of the leaf serves as the source tissue that fixes carbon, primarily in the form of sugars. These compounds are exported to the shaded half of the leaf that serves as the new sink tissue, whose strength increases with increasing time of shading (Figure 8). (Bagnall et al., 1988; Paul et al., 1992; Steyn et al., 2002; Islam et al., 2005).
ELDI revealed that the distributions of sugars (tetroses, pentoses, heptoses, and sucrose) were not significantly affected by the difference in the illumination between the two halves of the leaves. Only the distributions of glucose and a hexose phosphate (presumably glucose-6-phosphate) were affected; they increased on the shaded side (Figure 8). Therefore, the illuminated source-side of the leaf compensated for the reduced photosynthesis that was imposed by shading the other half of the leaf. This sugar-based interrelationship between source-sink tissues often manifests coordinated changes in amino acid metabolism, associated with the affiliation between carbon and nitrogen metabolism (Paul and Driscoll, 1997; McCormick et al., 2006). These changes in amino acid metabolism are usually associated with photosynthetic source tissues, where changes in RUBISCO levels (the major sink for amino acids) can drastically affect free amino acid pools (Paul and Driscoll, 1997; Nielsen et al., 2002; McCormick et al., 2008). Thus, in our studies we visualized increasing levels of Arg, Asp, His in the shaded side of the leaf (Figure 10), probably reflecting the turnover of RUBISCO as photosynthetic capacity was reduced in the dark, sink-side of the leaf.
This study demonstrates the capabilities of ELDI-MSI for identification and spatial characterization of a wide variety of compounds in plant tissues with minimal sample preparation requirement. The ability to generate spatial distribution data that are consistent with biological explanations provides confidence in the validity of the observations. Future experiments include a) separation of isobaric ions by ion mobility, and b) implementation of procedures for quantification with spatial resolution by ELDI.
Compared to MALDI or other methods that require an added matrix, ELDI has the advantage of less sample preparation; however its sensitivity and spatial resolution are poorer. Much of the plant sample remains intact after ELDI analysis. Compared to LAESI with the commercial source, their spot size and spatial resolution are ~200 μm, while ELDI as shown here is ~125 μm. All plants that we have analyzed absorb at 355 nm well enough for laser ablation analysis. While the classes of compounds we report here were readily observed, other classes, such as lipids and peptides, were difficult to observe in our ELDI experiments.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by funds from the U.S. Department of Energy, Office of Biological and Environmental Research through award DESC0014038 to Iowa State University. BJN and LEA were partially supported by the Center for Metabolic Biology, Iowa State University (metabolicbiology.iastate.edu). The ELDI-MSI instrumentation was funded by the Office of Basic Energy Sciences, Separations and Analysis Program, U. S. Department of Energy. Metabolomics analyses were conducted at the Iowa State University's W. M. Keck Metabolomics Research Laboratory (metabolomics.biotech.iastate.edu). We thank Dr. Lucas J. Showman and Dr. Ann M. Perera for their expert advice. Microscopic data were gathered at Iowa State University's Microscopy and NanoImaging Facility, and we thank Ms. Tracey Stewart and Dr. Harry T. Horner for their expert advice and assistance.
Amstalden van Hove, E. R., Smith, D. F., Fornai, L., Glunde, K., and Heeren, R. M. A. (2011). An alternative paper based tissue washing method for mass spectrometry imaging: localized washing and fragile tissue analysis. J. Am. Soc. Mass Spectromet. 22, 1885–1890. doi: 10.1007/s13361-011-0203-z
Angel, P. M., and Caprioli, R. M. (2013). Matrix-assisted laser desorption ionization imaging mass spectrometry: in situ molecular mapping. Biochemistry 52, 3818–3828. doi: 10.1021/bi301519p
Bagnall, D. J., King, R. W., and Farquhar, G. D. (1988). Temperature-dependent feedback inhibition of photosynthesis in peanut. Planta 175:348. doi: 10.1007/BF00396340
Bartels, B., and Svatoš, A. (2015). Spatially resolved in vivo plant metabolomics by laser ablation-based mass spectrometry imaging (MSI) techniques: LDI-MSI and LAESI. Front Plant Sci. 6:471. doi: 10.3389/fpls.2015.00471
Basu, P. S., Sharma, A., Garg, I. D., and Sukumaran, N. P. (1999). Tuber sink modifies photosynthetic response in potato under water stress1Publication No. 1408, CPRI, Shimla.1. Environ. Exp. Botany. 42, 25–39. doi: 10.1016/S0098-8472(99)00017-9
Boldt, J. K. (2013). Foliar Anthocyanins in Coleus and Ornamental Grasses: Accumulation, Localization, and Function. University of Minnesota Digital Conservancy. Available online at: http://hdl.handle.net/11299/150590
Boughton, B. A., Thinagaran, D., Sarabia, D., Bacic, A., and Roessner, U. (2016). Mass spectrometry imaging for plant biology: a review. Phytochem Rev. 15, 445–488. doi: 10.1007/s11101-015-9440-2
Etalo, D. W., De Vos, R. C. H., Joosten, M. H., and Hall, R. D. (2015). Spatially resolved plant metabolomics: some potentials and limitations of laser-ablation electrospray ionization mass spectrometry metabolite imaging. Plant Physiol. 169, 1424–1435. doi: 10.1104/pp.15.01176
Galayda, K. (2017). Use of Ultraviolet Laser Ablation Electrospray Ionization Mass Spectrometry for Plant Metabolite Imaging. Graduate Theses and Dissertations, 15517. Available online at: https://lib.dr.iastate.edu/etd/15517
Guijas, C., Montenegro-Burke, J. R., Domingo-Almenara, X., Palermo, A., Warth, B., Hermann, G., et al. (2018). METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 90, 3156–3164. doi: 10.1021/acs.analchem.7b04424
Hölscher, D., Shroff, R., Knop, K., Gottschaldt, M., Crecelius, A., Schneider, B., et al. (2009). Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 60, 907–918. doi: 10.1111/j.1365-313X.2009.04012.x
Huang, M.-Z., Cheng, S.-C., Jhang, S.-S., Chou, C.-C., Cheng, C.-N., Shiea, J., et al. (2012). Ambient molecular imaging of dry fungus surface by electrospray laser desorption ionization mass spectrometry. Int. J. Mass Spectromet. 325–327, 172–182. doi: 10.1016/j.ijms.2012.06.015
Hummel, J., Strehmel, N., Selbig, J., Walther, D., and Kopka, J. (2010). Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 6, 322–333. doi: 10.1007/s11306-010-0198-7
Hur, M., Campbell, A. A., Almeida-de-Macedo, M., Li, L., Ransom, N., Jose, A., et al. (2013). A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Natural Product Rep. 30, 565–583. doi: 10.1039/c3np20111b
Islam, M. S., Jalaluddin, M., Garner, J. O., Yoshimoto, M., and Yamakawa, O. (2005). Artificial shading and temperature influence on anthocyanin compositions in sweetpotato leaves. HortScience 40, 176–180.
Klein, A. T., Yagnik, G. B., Hohenstein, J. D., Ji, Z., Zi, J., Reichert, M. D., et al. (2015). Investigation of the chemical interface in the soybean–aphid and rice–bacteria interactions using MALDI-mass spectrometry imaging. Anal. Chem. 87, 5294–5301. doi: 10.1021/acs.analchem.5b00459
Kompauer, M., Heiles, S., and Spengler, B. (2017). Atmospheric pressure MALDI mass spectrometry imaging of tissues and cells at 1.4-mum lateral resolution. Nat. Methods. 14, 90–96. doi: 10.1038/nmeth.4071
Krapp, A., and Stitt, M. (1995). An evaluation of direct and indirect mechanisms for the “sink-regulation” of photosynthesis in spinach: changes in gas exchange, carbohydrates, metabolites, enzyme activities and steady-state transcript levels after cold-girdling source leaves. Planta 195, 313–323.
Lebowitz, R. J. (2011). The genetics and breeding of coleus. Plant Breeding Rev. 3, 343–360 doi: 10.1002/9781118061008.ch9
Lee, Y. J., Perdian, D. C., Song, Z., Yeung, E. S., and Nikolau, B. J. (2012). Use of mass spectrometry for imaging metabolites in plants. Plant J. 70, 81–95. doi: 10.1111/j.1365-313X.2012.04899.x
Lemoine, R., Camera, S., Atanassova, R., Dédaldéchamp, F., Allario, T., and Durand, M. (2013). Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci. 4, 1–21. doi: 10.3389/fpls.2013.00272
Logan, B. A., Stafstrom, W. C., Walsh, M. J., Reblin, J. S., and Gould, K. S. (2015). Examining the photoprotection hypothesis for adaxial foliar anthocyanin accumulation by revisiting comparisons of green- and red-leafed varieties of coleus (Solenostemon scutellarioides). Photosynth. Res. 124, 267–274. doi: 10.1007/s11120-015-0130-0
McCormick, A. J., Cramer, M. D., and Watt, D. A. (2006). Sink strength regulates photosynthesis in sugarcane. New Phytol. 171, 759–770. doi: 10.1111/j.1469-8137.2006.01785.x
McCormick, A. J., Cramer, M. D., and Watt, D. A. (2008). Changes in photosynthetic rates and gene expression of leaves during a source-sink perturbation in sugarcane. Ann. Bot. 101, 89–102. doi: 10.1093/aob/mcm258
Nemes, P., and Vertes, A. (2007). Laser Ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal. Chem. 79, 8098–8106. doi: 10.1021/ac071181r
Nguyen, P., and Cin, V. D. (2009). The role of light on foliage colour development in coleus (Solenostemon scutellarioides (L.) Codd). Plant Physiol. Biochem. 47, 934–945. doi: 10.1016/j.plaphy.2009.06.006
Nguyen, P., Quesenberry, K., and Clark, D. (2008). Genetics of growth habit and development of new coleus (Solenostemon scutellarioides (L.) Codd) varieties with trailing habit and bright color. J. Heredity 99, 573–580. doi: 10.1093/jhered/esn054
Nielsen, T. H., Krapp, A., Röper-Schwarz, U., and Stitt, M. (2002). The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP glucose pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ. 21, 443–454. doi: 10.1046/j.1365-3040.1998.00295.x
Nilsson, A., Goodwin, R. J., Shariatgorji, M., Vallianatou, T., Webborn, P. J., and Andrén, P. E. (2015). Mass spectrometry imaging in drug development. Anal. Chem. 87, 1437–1455. doi: 10.1021/ac504734s
Osorio, S., Ruan, Y. L., and Fernie, A. R. (2014). An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 5, 1–11. doi: 10.3389/fpls.2014.00516
Paul, M. J., and Driscoll, S. P. (1997). Sugar repression of photosynthesis: the role of carbohydrates in signalling nitrogen deficiency through source:sink imbalance. Plant Cell Environ. 20, 110–116. doi: 10.1046/j.1365-3040.1997.d01-17.x
Paul, M. J., Driscoll, S. P., and Lawlor, D. W. (1992). Sink-regulation of photosynthesis in relation to temperature in sunflower and rape. J. Exp. Bot. 43, 147–153. doi: 10.1093/jxb/43.2.147
Paul, M. J., and Foyer, C. H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52, 1383–1400. doi: 10.1093/jexbot/52.360.1383
Robichaud, G., Barry, J. A., and Muddiman, D. C. (2014). IR-MALDESI mass spectrometry imaging of biological tissue sections using ice as a matrix. J. Am. Soc. Mass Spectro. 25, 319–328. doi: 10.1007/s13361-013-0787-6
Roitsch, T. (1999). Source-sink regulation by sugar and stress. Curr. Opin. Plant Biol. 2, 198–206. doi: 10.1016/S1369-5266(99)80036-3
Schmidt, M. A., Barbazuk, W. B., Sandford, M., May, G., Song, Z., Zhou, W., et al. (2011). Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiol. 156, 330–345. doi: 10.1104/pp.111.173807
Shiea, J., Huang, M. Z., Lee, C. Y., Yuan, C. H., Beech, I., and Sunner, J. (2005). Electrospray-assisted laser desorption/ionization mass spectrometry for direct ambient analysis of solids. Rapid Commun. Mass Spectrom. 19, 3701–3704. doi: 10.1002/rcm.2243
Shroff, R., Schramm, K., Jeschke, V., Nemes, P., Vertes, A., Gershenzon, J., et al. (2015). Quantification of plant surface metabolites by matrix-assisted laser desorption-ionization mass spectrometry imaging: glucosinolates on Arabidopsis thaliana leaves. Plant J. 81, 961–972. doi: 10.1111/tpj.12760
Smith, C. A., O'Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Ther Drug Monit. 27, 747–751.
Stein, S. E. (1999). An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectro. 10, 770–781. doi: 10.1016/S1044-0305(99)00047-1
Steyn, W. J., Wand, S. J. E., Holcroft, D. M., and Jacobs, G. (2002). In vegetative tissues : anthocyanins in unified function a proposed photoprotection. New Phytol. 155, 349–361. doi: 10.1046/j.1469-8137.2002.00482.x
Turgeon, R. (1989). The sink-source transition in leaves. Ann. Rev. Plant Physiol. Plant Mol. Biol. 40, 119–138. doi: 10.1146/annurev.pp.40.060189.001003
Wardlaw, I. F. (2006). Tansley review no. 27. The control of carbon partitioning in plants. New Phytol. 116, 341–381. doi: 10.1111/j.1469-8137.1990.tb00524.x
Wu, X., Gu, L., Prior, R. L., and McKay, S. (2004). Characterization of anthocyanins and proanthocyanidins in some cultivars of ribes, aronia, and sambucus and their antioxidant capacity. J. Agricult. Food Chem. 52, 7846–7856. doi: 10.1021/jf0486850
Keywords: matrix-free imaging, mass spectrometry imaging, electrospray laser desorption ionization, shading, Coleus (Solenostemon scuttellarioides), anthocyanin, flavonoid, source-to-sink
Citation: McVey PA, Alexander LE, Fu X, Xie B, Galayda K-J, Nikolau BJ and Houk RS (2018) Light-Dependent Changes in the Spatial Localization of Metabolites in Solenostemon scutellarioides (Coleus Henna) Visualized by Matrix-Free Atmospheric Pressure Electrospray Laser Desorption Ionization Mass Spectrometry Imaging. Front. Plant Sci. 9:1348. doi: 10.3389/fpls.2018.01348
Received: 16 May 2018; Accepted: 27 August 2018;
Published: 19 September 2018.
Edited by:
Aleš Svatoš, Max-Planck-Institut für Chemische Ökologie, GermanyReviewed by:
Berin A. Boughton, The University of Melbourne, AustraliaCopyright © 2018 McVey, Alexander, Fu, Xie, Galayda, Nikolau and Houk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert S. Houk, cnNob3VrQGlhc3RhdGUuZWR1
†These authors have contributed equally to this work
‡Present Address: Katherine-Jo, Galayda, Kemin Industries Inc., Des Moines, IA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.